Summary:
Cellular perturbations underlying Alzheimer’s disease are primarily studied in human postmortem samples and model organisms. Here we generated a single-nucleus atlas from a rare cohort of cortical biopsies from living individuals with varying degrees of Alzheimer’s disease pathology. We next performed a systematic cross-disease and cross-species integrative analysis to identify a set of cell states that are specific to early AD pathology. These changes–which we refer to as the Early Cortical Amyloid Response—were prominent in neurons, wherein we identified a transient state of hyperactivity preceding loss of excitatory neurons, which correlated with the selective loss of layer 1 inhibitory neurons. Microglia overexpressing neuroinflammatory-related processes also expanded as AD pathological burden increased. Lastly, both oligodendrocytes and pyramidal neurons upregulated genes associated with amyloid beta production and processing during this early hyperactive phase. Our integrative analysis provides an organizing framework for targeting circuit dysfunction, neuroinflammation, and amyloid production early in AD pathogenesis.
Introduction
The first pathological sign of AD in the human cortex is the gradual accumulation of amyloid beta plaques, followed by the appearance of gliosis, misfolded tau, and neurodegeneration. Of critical importance is understanding the coordinated activities of neurons and glia during the early phases of the disease that initiate this pathogenic cascade1,2. Several postmortem single-cell studies have begun identifying disease-associated cellular changes in AD, particularly at later histopathological disease stages3–8. Inference from postmortem samples can be complicated by peri-mortem transcriptional responses to agonal state, cessation of blood flow, hypoxia, and neuronal atrophy. Prior cytological9–11 and transcriptional12 analyses demonstrate a marked decline, particularly in neurons, of cell health within two to four hours postmortem. Consequently, several fundamental questions related to the early stages of AD remain unanswered, including which cell types are perturbed the most, what molecular mechanisms are dysregulated in neuronal types of different cortical layers, and how these early perturbations contribute to the production of misfolded proteins and progression of pathology in the human brain.
We reasoned that a deep analysis of samples from living individuals harboring various extents of amyloid deposits could provide an opportunity to comprehensively capture veridical states associated with early-stage AD pathology. We performed single-nucleus RNA-sequencing (snRNA-seq) on a rare set of surgical biopsy samples obtained from patients undergoing ventriculoperitoneal shunt placement for treatment of suspected normal pressure hydrocephalus (NPH). In a study of 335 individuals, 44% of these biopsies contained amyloid beta plaques13 and, most importantly, longitudinal follow up of multiple cohorts has indicated the presence of within NPH biopsies is strongly associated with a decline in cognitive performance and a future clinical diagnosis of AD13–15, demonstrating these biopsies capture early AD pathology. To ensure that our insights were not restricted to a single cohort and were specific to AD, we further developed an accurate integrative analysis framework to incorporate published postmortem and mouse model single-cell datasets to construct a compendium of 2.4 million uniformly annotated cell profiles across diseases and species. The resulting analyses revealed what we collectively term the Early Cortical Amyloid Response (ECAR): a suite of consistent tissue changes in specific cell types that co-occur with the initial onset of brain amyloidosis.
Results
A single-nucleus atlas of human brain biopsies to identify AD pathological perturbations
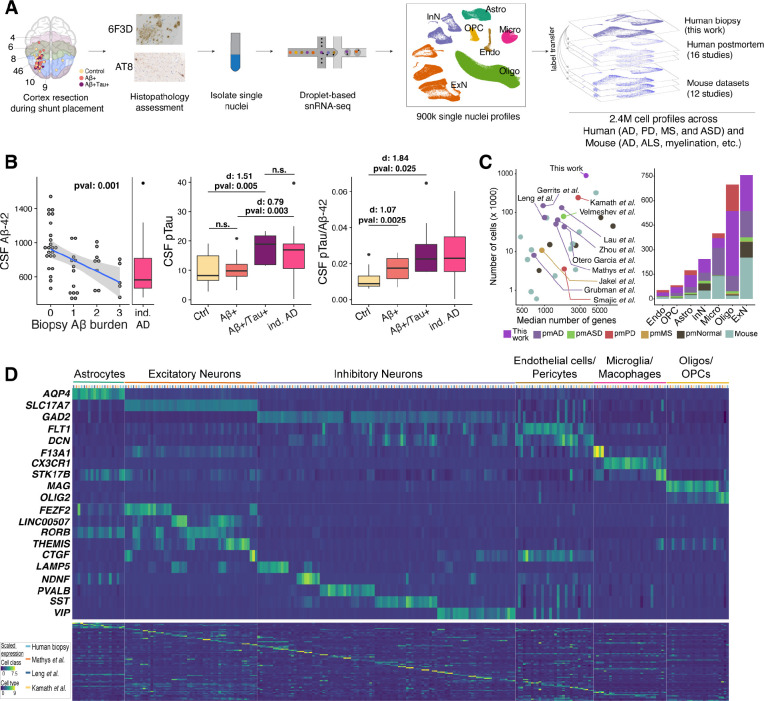
To capture cellular perturbations in cortex of living individuals in response to AD pathology, we collected biopsies–frozen within five minutes of surgical excision to ensure fidelity of in vivo transcriptional states–from the frontal cortices (Brodmann areas 8 and 9) of 52 patients with NPH (Figure 1A). Histopathological examination of the biopsies identified 19 with plaques , eight with both plaques and phosphorylated tau pathology , and 25 biopsies that had neither histopathology (Table S1). From the stereotactic position of the catheter insertion site recorded by post-surgical CT or MRI (Figure 1A), we determined that the anatomical location of sampling did not correlate with AD histopathological burden (Figure S1A). We further divided the biopsies into three groups by their level of plaque burden (Figure S1B). The extent of plaque and tau tangle burden within the biopsies correlated inversely with these patients’ CSF levels (p-value < 0.001; Figure 1B), and positively with CSF levels of phosphorylated tau (p-value < 0.005; Figure 1B), consistent with prior biomarker studies of AD progression16,17. Moreover, the CSF levels of phosphorylated tau in individuals were similar to individuals without histopathology (p-value >0.95; Student’s t-test) and significantly less than an independent cohort of 36 clinically diagnosed AD individuals (p-value < 0.004; Student’s t-test; Figure 1B). Collectively, these results suggest the severity of biopsy histopathology is representative of the overall burden in the brain.
Figure 1. A fresh-tissue atlas of cortical states associated with AD pathology.
A) Schematic of the frontal cortex brain biopsy sampling workflow. Samples were stained and quantitatively assessed for AD histopathology by the 6F3D and AT8 (phosphorylated tau) antibodies (Methods). Brodmann areas are color-coded in the first panel. B) CSF (left), phosphorylated tau (middle) and ratio of the two (right) in association with and tau burden scores (see Methods) in 49 subjects sampled. We have excluded three individuals for whom the CSF measurements were missing. The “ind. AD” refers to an independent cohort of 36 NPH patients who were clinically diagnosed with AD prior to, or within one year after, CSF collection. Cohen d (d) effect sizes are reported. C) A summary of datasets included in the integrative analysis. Case-control datasets of human brain diseases are labeled. pm, postmortem; ASD, autism spectrum disorder; PD, Parkinson’s disease; MS, multiple sclerosis. D) Expression of markers of cell classes (top), main neuronal classes (middle), and individual cell types (bottom) across four human studies of neurodegenerative disease from the integrative analysis. Each row indicates the normalized expression level of each gene across the select human postmortem datasets (color-coded on y-axis) and 82 cell types. A detailed analysis of cell types and associated markers can be found in Table S3.
To explore the cell-type-specific changes associated with and tau histopathology in the cortex, we obtained 892,828 high-quality nuclei profiles from this biopsy cohort, with a median of 17,082 nuclei per individual. By unsupervised clustering18 (Methods), we identified the seven major classes of cells in the cortex: excitatory neurons (ExN; 222,449 nuclei), inhibitory neurons (InN; 83,702 nuclei), microglia (Micro; 59,624 nuclei), astrocytes (Astro; 73,487 nuclei), endothelial cells/pericytes (Endo; 22,407 nuclei), oligodendrocytes (Oligo; 396,292 nuclei), and oligodendrocyte progenitor cells (OPC; 34,867 nuclei) (Figure 1A). To increase our resolution, we repeated our clustering analysis within each class to identify a total of 82 cell types with a median size of 3,586 nuclei per type.
An integrative analysis of biopsy and postmortem brain
Several studies have profiled brain cells under normal and disease conditions using human postmortem or mouse samples. However, a direct comparison of their results have been hampered by differences in sample qualities, dataset sizes, analysis pipelines, and cell type annotations. We reasoned that the size and quality of our biopsy dataset would be sufficiently analytically powered to conduct a comprehensive integrative analysis of these datasets with highly granular cell type specificity. For our integrative analysis, we considered 27 published single-cell/nuclei studies of the brain derived from both human disease studies and mouse disease models (Figure 1C). Human studies included postmortem samples from individuals with AD, Parkinson’s disease (PD), multiple sclerosis (MS), and autism spectrum disorder (ASD) (Table S2). Mouse datasets included models of AD and ALS, as well as de/re-myelination, aging, prenatal, and food deprivation conditions among others (Table S2). To accurately combine these datasets with our biopsy cohort, we developed an optimized single-cell integration framework that efficiently handled the substantial technical (e.g., sample preparation, sequencing platforms and depth) and biological (e.g., human vs mouse) variation that exists among these datasets (Methods). We employed three criteria to validate the quality of our integrative analysis results: 1) uniform mixing of the datasets across clusters; 2) cells expressing similar cortical cell type markers are aligned with each other across datasets and organisms; 3) reported cell type identities in each of the studies are preserved in the aligned space. A total of 2,406,980 cells were included in our integrative analysis after removing artifacts, low quality cells, and doublets. We next implemented a random walk method to transfer cell type annotations from our biopsy cohort to each of 27 other studies, thereby uniformly annotating all datasets to the 82 cell types (Figure 1D; Table S3). Comparison across human datasets demonstrated our biopsy cohort had among the highest number of cells sampled per cell type and minimal expression of artifactual genes often associated with sample quality and dissociation methods19,20 (Figures S2–S4). Congruently, our attempts failed to achieve similarly high integration resolution after exclusion of our biopsy dataset from the analysis (data not shown).
We then investigated how agonal states and the postmortem interval affected gene expression patterns in different cell types by comparing our biopsy dataset with postmortem data, and identifying recurrent correlates with postmortem interval across datasets. Our analysis demonstrated a small but statistically significant decrease in gene expression levels in both excitatory (p-value < 0.038; Meta analysis) and inhibitory neurons (p-value < 0.024; Meta analysis, Figure S5A), as well as a trend towards increased gene expression levels in microglia (Figure S5A). Consistently, the ratio of glial to neuronal gene expression was lowest in the biopsy dataset, and this ratio increased with longer postmortem intervals within postmortem datasets (Figures S5B–S5D). Together, our results, in combination with the expression patterns of artifact associated genes (Figures S3 and S4), indicate loss of transcriptional complexity in both inhibitory and excitatory neurons, as well as an increase in artifact-related genes in microglial cells in response to peri- and post-mortem events.
Meta-analysis reveals cortical cellular changes in early AD pathology
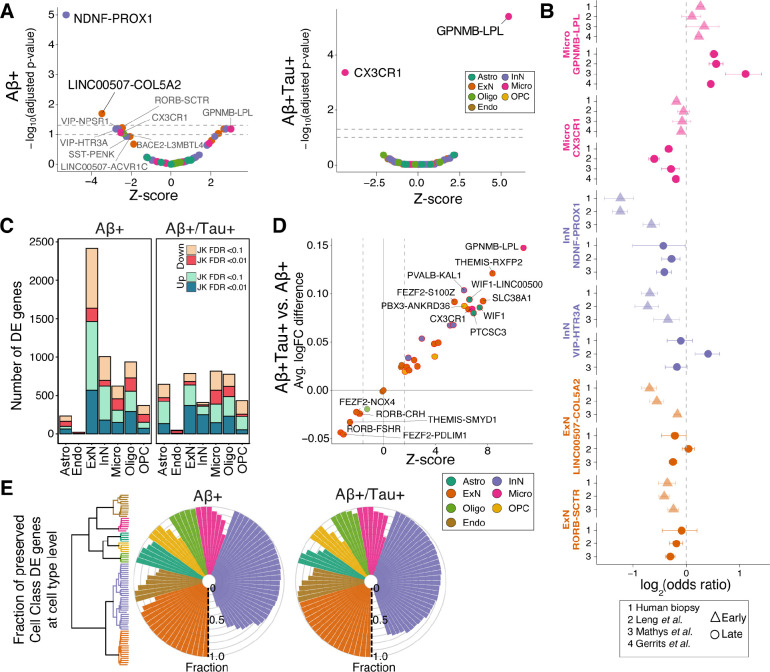
To identify cortical tissue changes across progression of AD pathology, we divided our biopsy samples into those with only pathology and those with both and tau pathology . In parallel, we also analyzed two AD postmortem studies3,4 that sampled both neuronal and glial cells from subjects with low Braak pathology staging and one dataset that only measured glia6 (Methods). We first tested for alterations in relative abundance of cell populations with increasing histopathological burden. A meta-analysis of cell proportions identified two neuron types–NDNF-PROX1 and LINC00507-COL5A2–that were significantly depleted (FDR-adjusted p-value < 0.05) (Figure 2A) in each of the cohorts with early amyloid pathology. The NDNF-PROX1 population expressed NDNF and RELN, markers of an interneuron type known to reside primarily in layer 1 (L1) of cortex21. The LINC0050s7-COL5A2 population expressed CUX2 and LINC00507, consistent with a layer 2/3 (L2/3) telencephalic identity21. Seven additional cell types showed a trend toward significant loss (0.05 < FDR-adjusted p-value < 0.12; Table S4): two upper layer excitatory types (RORB-SCTR, LINC00507-ACVR1C), three inhibitory types (VIP-HTR3A and SST-PENK that are upper-layer-enriched,and VIP-NPSR1 that spans cortical layers; Figure S6A), one microglia type (CX3CR1) and one oligodendrocyte type (BACE2-L3MBTL4), while one microglia type (GPNMB-LPL) showed a trend toward expansion (Figure 2A). We further confirmed that the observed changes in neuronal populations do not correlate with the severity of any iNPH symptoms within the biopsy cohort (Figure S6B). Most importantly, our refined integration strategy and meta-analysis revealed similar alterations in neuronal and microglial proportions within each of the published postmortem AD case-control cohorts (Figure 2B), underscoring the robustness of the observed cellular changes associated with early-stage AD pathology.
Figure 2. Identification of early- and late-stage cellular perturbations in AD.
A) Volcano plot of a meta-analysis of cell type proportional changes (Methods) in early- and late-stage AD-related samples. Cell types reaching significance are labeled. Colors indicate cell class assignment. Dashed lines represent FDR thresholds of 0.05 and 0.1. B) Individual log-odds ratios of six significant cell types in (triangles) and samples (circles) for our biopsy cohort and published postmortem AD case-control datasets. Whiskers indicate standard errors. C) Number of DE genes in each cell class, stratified by biopsy histopathology. JK: Jack-knife. D) Fold change pattern concordance of DE genes between and samples. The y-axis shows the average logFC difference between and . The Z-scores on x-axis are based on the transformation of p-values from a paired t-test analysis on the union of top 300 protein-coding genes (sorted by their jack-knifed p-value) from each condition. E) Fraction of DE genes in and biopsies that are similarly up- or down-regulated between the seven major cell classes and their associated subtypes in biopsy samples. The fraction was calculated by examining the top 300 protein-coding DE genes at the cell class. The dendrogram illustrates the subdivision of the seven major cell classes to a total of 82 subtypes.
In subjects with high histopathological burden, the proportional losses of the NDNF-PROX1 and LINC00507-COL5A2 neuronal populations were no longer significant (Figures 2A and 2B), likely due to additional loss of other cortical neurons, since the overall proportions of excitatory and inhibitory neurons were both lower in these subjects across cohorts (Figure S6C). Instead, we identified a significant (FDR-adjusted p-value < 0.05) expansion of the GPNMB-LPL microglia type and loss of the major homeostatic microglia population, marked by expression of CX3CR1 (Figures 2A and 2B). In addition, while not consistently altered in all three human studies (meta-analysis p-value < 0.11), one astrocyte population expressing CHI3L1 and GFAP increased in abundance in the late stages of disease in our biopsy cohort (p-value < 0.05; odds ratio (OR): 1.5), one postmortem cohort4 (p-value < 0.075; OR: 1.6), and in a mouse model of AD22 (p-value < 0.02; OR: 1.4) (Table S4). Together, these results indicate that gliosis becomes an increasingly prominent feature of cortical tissue as histopathology worsens.
Next, we examined how the transcriptional phenotype of each cell type changes across early and late histopathological stages of AD. We used a pseudocell-based strategy, coupled with mixed-effect modeling and jack-knifing (Methods), to robustly identify differentially expressed (DE) genes in both the and the biopsies (Figure 2C; Table S5). To better understand the association of gene perturbations with progression of AD pathology, we developed a metric to quantify the relative magnitude of transcriptional alteration across each cell type in early- versus late-stage samples (Methods). For most cell types and most notably in microglia populations, we found that the transcriptional alterations quantified in biopsies were consistent with, but stronger than, those changes measured in samples (Figure 2D). However, several excitatory neuron populations showed transcriptional perturbations in the samples that were absent in the biopsies (Figures 2C and 2D), indicating their passage through a distinct transcriptional state early in histopathological progression. To further assess the extent of overlap in dysregulated transcriptional programs among related cell populations, we calculated the fraction of DE genes in each of the seven major cell classes that show consistent DE within each of their constituent cell types. This comparison demonstrated that DE genes identified by the analysis of each of seven major cell classes exhibited highly preserved perturbation patterns (i.e., similar up- or down-regulation patterns) within their related cell types (Figures 2E and S6D). Collectively, our DE analyses demonstrated that: a) perturbation of the transcriptomes increases in magnitude as neuropathology worsens–with the exception of excitatory neurons, which show a distinct early phase response; and b) that individual cell type responses are largely similar within a major cell class.
Neuronal loss and hyperactivity in early AD pathology
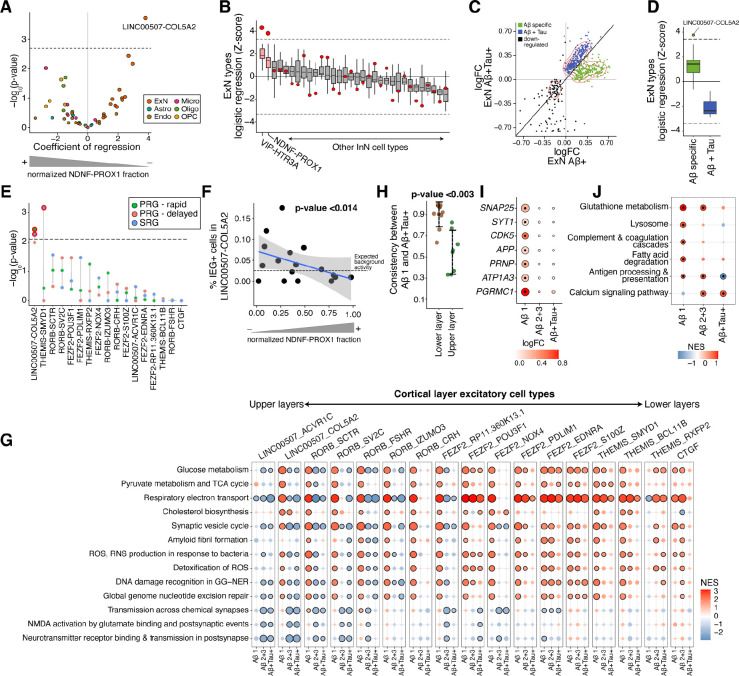
AD is increasingly recognized as a systems disease where interactions of different cell types define its pathological course. The strongest proportional change in our meta-analysis of the early AD pathological stage was the loss of NDNF-PROX1 inhibitory neurons (Figure 2A). We therefore wondered whether the loss of these inhibitory neurons could contribute to the onset and early progression of AD by induction of specific transcriptional states in other cortical cell types. To examine this, we correlated the fraction of NDNF-PROX1 inhibitory neurons with the extent of molecular perturbations in all other cell types (Methods). Intriguingly, applying this analysis to the biopsy samples identified a specific and significant (FDR-adjusted p-value < 0.01) correlation between NDNF-PROX1 depletion and upregulated ExN DE genes in the LINC00507-COL5A2 excitatory neurons (Figures 3A and S7A), which themselves are vulnerable to loss in early AD pathology (Figure 2A). Alternative analysis methods and robustness analyses confirmed the strength of association between the ExN DE signature in LINC00507-COL5A2 with the loss of NDNF+ expressing cells in individuals (Figures S7B–S7E). Moreover, this association was also significant (FDR- adjusted p-value < 0.05) within the control biopsy samples (Figures S7F and S7G), reinforcing that this pair of neuronal changes–loss of NDNF-PROX1 and induction of a specific transcriptional state in LINC00507-COL5A2–occurs early in disease. We next tested whether transcriptional response in LINC00507-COL5A2 neurons is specifically induced by loss of NDNF-PROX1 cells, or is also associated with the loss of other inhibitory neurons. Importantly, the relationship between the inhibitory neuron loss and ExN transcriptional state was specific to NDNF-PROX1 and VIP-HTR3A inhibitory neurons in biopsies (Figure 3B), the two most depleted inhibitory cell types in the early stage of AD. These results suggest a tight and specific coupling between layer 1 inhibitory neuron loss and transcriptional alterations in layer 2/3 excitatory neurons.
Figure 3. NDNF-PROX1 inhibitory neuron loss is associated with a hyperactivity signature in L2/3 excitatory neurons.
A) Logistic mixed-effect model regression of NDNF-PROX1 proportion versus cell type transcriptional signature in subjects. The dashed horizontal line represents the FDR threshold of 0.05. B) Associations (by logistic mixed-effect model) between the proportion of each inhibitory neuron type with each ExN type’s transcriptional signature in subjects. The red dots indicate the regression Z-score of the LINC00507-COL5A2 neurons with the corresponding inhibitory neuron cell type. The dashed line represents an FDR threshold of 0.05. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. See Figure S7H for more details. C) Scatter plot comparing the logFC in the ExNs of (x-axis) and samples (y-axis). Visualization is based on the union of top 300 protein-coding DE genes (sorted by jack-knifed p-value) in either group. D) Logistic mixed-effect model regression of NDNF-PROX1 proportion versus early-specific up-regulated DE genes (green dots in C) and up-regulated DE genes shared in both and samples (blue dots in C) for each ExN cell type. The dashed lines represent an FDR threshold of 0.05. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. E) Logistic mixed-effect model regression of NDNF-PROX1 cell fraction versus expression of neural activity signatures23 in each ExN type in samples (one-sided). The dashed line represents a one-sided FDR threshold of 0.05. PRG, primary response genes; SRG, secondary response genes. F) Scatter plot showing normalized NDNF-PROX1 fraction (x-axis) and the percent of LINC00507-COL5A2 ExNs with high expression of the core immediate early genes FOS, JUNB, ARC, NPAS4, ERG1, and ERG2 (y-axis, Methods) in subjects. A logistic mixed-effect model was used to calculate the p-value. G) GSEA of Reactome pathways on DE results from subjects with varying and tau burdens, across ExN types. Dots outlined in black denote significant terms (FDR-adjusted p-value < 0.05). individuals with burden scores of 2 and 3 are grouped together. H) Concordance of DE genes between different stages of AD pathology within excitatory neuron cell types. The LINC00507+ and RORB+ were selected as upper layer excitatory neurons and FEZF2+, CTGF+, and THEMIS+ populations as lower layer. I) ExN DE genes whose products are involved in synapse vesicle cycle and trafficking (SYT1, SNAP25, and CDK5), amyloid precursor protein (APP), and receptors of oligomeric (PRNP, ATP1A3, and PGRMC1) across different and tau burdens. The outlined dots represent DE genes with jack-knifed FDR-adjusted p-value < 0.01. J) GSEA of human KEGG gene sets using DE genes of WIF1+ homeostatic astrocytes across increasing and tau burden. Outlined dots represent significant terms (FDR-adjusted p-value < 0.1).
We next sought to better understand the association of the ExN DE signature with plaque pathology. Comparison of ExN DE genes between and biopsies revealed a bimodal pattern among upregulated DE genes (Figure 3C), in which one DE gene set was evident solely in the early stage of pathology while another was present in both early- and late-stage samples. Given their differences in expression trajectory, we asked whether these two sets of DE genes were differentially correlated with NDNF-PROX1 inhibitory loss. Only the DE genes specifically found in response to early AD pathology, particularly within the LINC00507-COL5A2 population, correlated with NDNF-PROX1 proportional loss (Figure 3D).
The activity of layer 1 NDNF-expressing inhibitory neurons has been shown to play crucial roles in the integration of long-range inputs into cortex, particularly through gain modulation of whole cortical columns24,25. We wondered if their loss may alter excitability of nearby L2/3 pyramidal cells. Indeed, we identified a significant association between NDNF-PROX1 loss and upregulation of neural activity response genes23 specifically within LINC00507-COL5A2 excitatory neurons in individuals (FDR-adjusted p-value < 0.01; Figure 3E). Furthermore, biopsy samples with a greater proportional loss of NDNF-PROX1 cells showed a higher percentage of LINC00507-COL5A2 cells expressing the core immediate early genes (FOS, JUNB, ARC, NPAS4, ERG1, and ERG2) that are induced after neuronal activity26 (p-value < 0.014; Figure 3F). Increased activity of excitatory neurons would be expected to affect their metabolism. Consistent with this, gene set enrichment analysis (GSEA) demonstrated increased expression of metabolism- and mitochondria-related gene sets (Methods) specifically in biopsies with the lowest level of plaque burden, further reinforcing the relevance of the hyperactivity phenotype to the early stages of AD pathology (Figure 3G). We also found an upregulation of gene sets indicating a cell-protective response to increased metabolism, including cholesterol biosynthesis, and responses to both reactive oxygen species (ROS) and DNA damage (Figure 3G). The enrichment of these terms was diminished in biopsies with higher burdens of and the presence of phosphorylated tau (Figure 3G), a pattern that was stronger in upper layer excitatory neurons. Consistently, comparing samples with lowest burden with demonstrated a significantly higher divergence of the DE patterns of upper layer neurons expressing LINC00507 and RORB compared to the lower layer excitatory neurons expressing FEZF2, THEMIS, and CTGF (p-value < 0.003; Student’s t-test; Figure 3I), demonstrating the specificity of this response to upper layer cortical neurons at the early stages of plaque formation. Collectively, our results demonstrate NDNF-PROX1 inhibitory neuron loss is correlated with hyperactivity and preferential loss of layer 2/3 excitatory neurons in the prefrontal cortex with low plaque burden.
Hyperactivity of neurons can trigger homeostatic pre- and postsynaptic mechanisms27. In subjects with a low burden, we identified upregulation of SNAP25, SYT1, and CDK5 in excitatory neurons, three genes whose products are involved in presynaptic vesicle release28–30 (Figures 3G and 3I). Increased activity of the presynaptic vesicle cycle can elevate production31. Congruently, we found upregulation of genes encoding for protein components involved in fibril formation, such as APP itself, only in the -low disease samples (Figures 3G and 3I). The oligomeric receptor genes PRNP, ATP1A3, and PGRMC1, whose protein products influence neuronal activity through the modulation of N-methyl-D-aspartate (NMDA) receptors32, were similarly upregulated in excitatory neurons at the early stages of AD pathology (Figure 3I). Homeostatic astrocytes also play critical roles in supporting synaptic function and coordinating antioxidant responses, especially in the context of neuronal hyperactivity33,34. In our integrative analysis of astrocytes, we identified one WIF1+ type with low expression of GFAP and high expression of EAAT1, EAAT2, and GSTP1 genes, which encode for critical components of glutamate/glutathione cycling (Figures S7I and S7J). The WIF1-expressing astrocytes showed enrichment of DE genes related to glutathione metabolism, lysosomal machinery, and fatty acid degradation specifically in subjects with low burden (Figure 3J), consistent with gene sets previously reported to be upregulated in the astrocytic response to hyperactive neurons34. Together, these results suggest that aberrant activity and metabolism of upper layer pyramidal cells perturb synapse homeostasis and astrocyte functioning in the brain.
Expanded microglia populations with AD-specific alterations
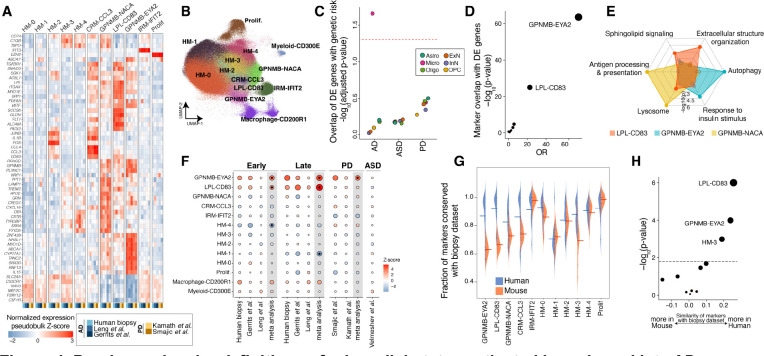
Our integrative analysis across four AD-related cohorts indicated a mild expansion of GPNMB-expressing microglia population at early stages of AD pathology that further expands and becomes the strongest signal in samples with high histopathological burden (Figure 2A; Table S4). Human genetics and transcriptome studies have strongly implicated microglia in the AD pathogenic process35–38. A reactive population expressing GPNMB was also identified as enriched in an AD animal model near amyloid plaques39, but its connection to human in vivo microglial states–in AD, normal aging, and other diseases–remains debated. To more deeply explore microglial states in AD pathology, we leveraged our well-powered integrative analysis of 400,743 microglia profiles across human and mouse studies from diverse brain regions and biological conditions, including 59,624 high-quality microglia nuclei (median number of genes per nucleus = 2,384) from our biopsy cohort (Table S2). We more deeply sub-clustered the microglia profiles into a total of 13 microglial states (Figures 4A and 4B), including five homeostatic (HM) states, a chemokine-enriched state (CRM-CCL3), three reactive states expressing GPNMB (GPNMB-NACA, LPL-CD83, and GPNMB-EYA2), an interferon gene-enriched state (IRM-IFIT2), and a proliferative (Prolif) state (Figures 4A and 4B; Table S3). Comparing the microglia populations, we observed the main microglial markers, including SLC2A5, CX3CR1, CSF1R, P2RY12 were downregulated in the three GPNMB-expressing populations relative to the homeostatic microglial cells, but were still expressed at higher levels than macrophages and myeloid cells (Figure 4A). One of the smaller homeostatic microglia populations, which we designated HM-2, also exhibited high expression of genes associated with technical dissociation artifacts, including FOS and JUNB19. All microglia states were well represented across datasets, biological conditions, and sequencing platforms. Moreover, we observed that markers of microglia states correlated strongly across different human brain regions, which is consistent with previous reports40,41 (Figures S8A–S8D).
Figure 4. Precise molecular definitions of microglial states activated in early and late AD.
A) Expression of select marker genes (Methods) across human neurodegeneration datasets in the microglia integrative analysis. The expression values represent pseudobulk expression of each marker in each cell state and dataset. B) Uniform manifold approximation and projection (UMAP) representation of microglia profiles from integrative analysis, colored by the 13 identified states. C) Dot plot of −log10-transformed p-values for MAGMA enrichment analysis42 (y-axis) of AD, PD, or ASD genetic risk in the up-regulated DE genes of each cell class. Dots are colored by cell class membership. Dashed line represents an FDR threshold of 0.05. D) Dot plot of −log10-transformed p-values for a Fisher’s exact test assessing the overlap between microglial DE genes with markers of each of the 13 microglial states (Methods). E) Radar plot representation of enriched gene sets in markers of the three GPNMB-LPL states. The marker analysis was conducted by comparing the three cell states against each other. See Table S6 for more details. F) Association of proportion of each microglial state with early and late AD pathology, as well as PD and ASD. In meta-analysis columns, black dots represent microglia states with significant changes in cell state proportions (FDR-adjusted p-value < 0.05). The scale of points is based on the absolute Z-score values. G) Distribution of the fraction of markers shared between the biopsy cohort and each other dataset (y-axis), in each microglial state (x-axis). Datasets are stratified by species. Mean values are denoted with a line. Only genes expressed in more than 1% of cells were considered in the analysis of each dataset. H) Statistical comparison of the differences in (G) by Student’s t-test. The dashed line represents an FDR threshold of 0.05.
Next, we examined how each of these 13 microglial states was affected by the presence of . Differential expression analysis across all microglia in our cohort identified a pattern that was highly similar in each of the 13 states (Figures S8E and S8F), suggesting that all microglia states respond to accumulation in a similar manner. This transcriptional pattern was also highly consistent across postmortem cohorts (Figure S8G). The DE signature showed upregulation of genes whose protein products are involved in microglia neuroinflammatory responses, including phagocytosis (COLEC12), antigen presentation (CD74 and HLA genes), lipoprotein metabolism and biosynthesis (APOE, OLR1, ATG7), fatty acid metabolism (ACSL1), autophagy (ATG7, ATG16L2), and lysosomal function (ASAH1, NPC2, SLC11A1, PSAP) (Table S5). Underscoring the pathological relevance of this common microglial DE signature, we found that it was significantly (FDR-adjusted p-value < 0.05) enriched for the expression of genes implicated in AD by common variant case-control studies35,36, including: APOE, MS4A6A/4A, TREM2, and INPP5D (Figure 4C, Methods). In addition, intersection of this DE signature with marker genes for each of the 13 states showed highly significant overlap with markers of GPNMB-EYA2 and LPL-CD83 microglia (FDR-adjusted p-value < 0.001; Fisher’s exact test; Figure 4D), indicating a transcriptional transition across microglia cells towards a state more resembling the GPNMB-EYA2 and LPL-CD83 populations. These results indicate that microglia cells collectively transition towards a transcriptional state with high expression of AD risk genes and neuroinflammatory-related processes as AD pathological burden increases in human brains.
We focused particularly on the three reactive GPNMB+ states, given their disease relevance. All three states highly expressed genes related to microglial reactivity, including APOE, ITGAX, MITF, and SGK1 (Figure 4A). However, comparative marker analysis between the three states revealed substantial differences. The GPNMB-NACA population preferentially expressed genes involved in antigen processing and presentation, as well as lysosomal and phagosomal function relative to the other two states (Figure 4E). By contrast, the GPNMB-EYA2 microglia preferentially expressed genes involved in autophagy (e.g., IGF1R, ATG7, and ATG16L2) and response to insulin (e.g., MYO5A, IGF1R, and PPARG) (Figure 4E). This cell state also expressed IL15, a key modulator of the nervous system inflammatory response43 (Figure 4A). The LPL-CD83 microglia expressed genes, including TGFBR1 and SMAD3, which encode for key proteins in TGF-β signaling (Figure 4A), and showed enrichment of genes involved in extracellular structure organization, response to cytokines, focal adhesion, and actin cytoskeleton (Figure 4E). Both IL-15 and TGF-β also mediate neuroinflammatory cross-talk between astrocytes and microglia44,45. Supporting this notion, we found a strong positive correlation between the expression of GFAP in astrocytes and the expansion of GPNMB-EYA2 and LPL-CD83 microglia states in our cohort (Figure S8H). Together, we find transcriptional heterogeneity among reactive microglia cells in human brains that points to specialized functional roles in responding to cues from their surrounding microenvironment.
We next conducted a meta-analysis to ask which of these microglial states is specifically enriched in AD, and how these states relate to those found in other neurodegenerative diseases and disease models. Across the three AD-related datasets with sufficient numbers of microglia to power proportional testing, we identified an expansion of the LPL-CD83 and GPNMB-EYA2 states in both early and late stages of AD histopathology (FDR-adjusted p-value < 0.05; Figure 4F). Interestingly, the GPMNB-EYA2 state was also enriched in a meta-analysis of two PD datasets (FDR-adjusted p-value < 0.002; Figure 4F), while LPL-CD83 was exclusively expanded across the AD datasets. Neither GPNMB-expressing microglia population was expanded in individuals with ASD, underscoring the specific role of these microglia in neurodegenerative diseases. In contrast to the human datasets, only the GPNMB-NACA state was consistently expanded in AD mouse models (Figure S8I). This state was also increased in several other mouse datasets, including a model of amyotrophic lateral sclerosis, in both juvenile and aged mice, and in response to demyelinating injury (Figure S8I). To better understand the underlying factors contributing to this apparent divergence in microglia response, we performed a systematic marker analysis of the 13 microglia states across the human and mouse datasets that are included in our integrative analysis. First, given the superior size, quality, and coverage of our human biopsy dataset, we used it as the base to reliably identify markers of each state. We next examined the concordance of marker genes in each of the remaining datasets. As expected, we found that microglia states are highly consistent across human datasets. Although human microglia states were less preserved in the mouse datasets in overall, preservation was notably lower for the mouse LPL-CD83 and GPNMB-EYA2 states (Figures 4G and 4H), suggesting these transcriptional states are less well recapitulated by laboratory mice. Collectively, our results demonstrate shared and AD-specific microglia responses to disease in the human brain, and selective divergence of the most disease-relevant states in mouse models.
Amyloidogenic cell populations in human frontal cortex
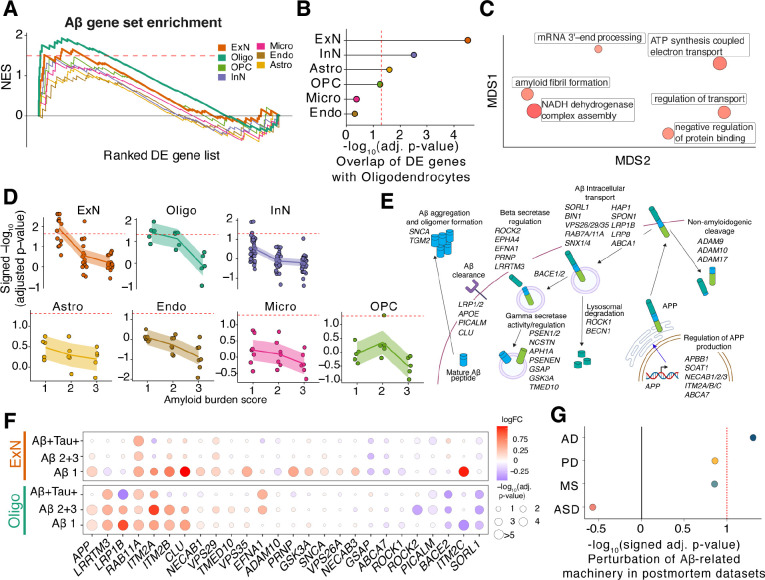
The production of amyloid in the brain has largely been assumed to be only in neurons but has been challenging to directly study in human tissue. We leveraged our high-quality surgical biopsy dataset to assess amyloidogenicity in each cell type using transcriptional signatures as a proxy. We took an unbiased approach, assessing the enrichment of a set of 49 genes known to regulate production and secretion in each cell type (Table S7). Interestingly, GSEA against an ordered list of DE genes for each of the seven cell classes identified not only excitatory neurons but also oligodendrocytes as having significant, positive enrichment for the amyloid gene set (FDR-adjusted p-value < 0.05, Figure 5A, Methods). The enrichment in these two cell classes was robust to the statistic used to order genes (Figure S9A). A leading-edge analysis identified specific genes that were highly up-regulated (including APP, LRRTM3, and ITM2B) and down-regulated (such as BACE2, SORL1, and PICALM) in biopsy samples with plaques in our cohort (Figure S9B).
Figure 5. Cell-type-specific dysregulation of amyloid formation in the human frontal cortex.
A) GSEA trace plot of amyloid-associated gene set ordered by their signed p-value from DE analysis across the seven cell classes. The x-axis shows the rank order of the DE genes in corresponding cell classes; the y-axis is the normalized enrichment scores (NES) from GSEA. Bold lines indicate GSEA traces for significant cell classes, oligodendrocytes and excitatory neurons. The dashed line indicates NES score corresponding to FDR threshold of 0.05. B) Dot plot of −log10-transformed FDR-adjusted p-values of GSEA results of the top 300 upregulated protein-coding genes (sorted by their jack-knifed p-values) from each cell class against an ordered list of DE genes in oligodendrocytes. Dotted red line indicates significance at FDR threshold of 0.05. C) Multidimensional scaling (MDS) low-dimension embedding of gene ontology terms significantly enriched in intersect of DE genes between oligodendrocytes and excitatory neurons from REVIGO46 (see Methods). Size of dots indicate significance values. D) GSEA of amyloid gene set against cell type level DE genes across increasing levels of and tau burden. Cell types are grouped based on their major cell class annotations. The dashed line represents a significance threshold of FDR-adjusted p-value < 0.05. E) Schematic of regulation of formation, intracellular transport, and degradation/clearance pathways, showing the substituent in each pathway genes that together comprise the amyloid gene set (Table S7). F) Excitatory neuron and oligodendrocyte DE results across increasing levels of and tau burden for genes found by the leading edge analysis in A. The size of each dot is scaled by p-values and the color of each dot denotes the logFC. G) Signed −log10-transformed p-values from GSEA results for the amyloid gene set on Oligo DE genes from postmortem AD, PD, MS, and ASD cohorts.
The unexpected enrichment of amyloid-related genes in oligodendrocytes prompted us to investigate whether they share a common DE gene signature with excitatory neurons.
Assessment of the degree of overlap between DE genes from oligodendrocytes and other cell classes revealed that the excitatory neuron DE genes had the most significant degree of overlap (FDR-adjusted p-value < 0.05; Figure 5B), which was consistent across a wide range of gene set sizes tested (Figure S9C). The high overlap of DE genes between oligodendrocytes and excitatory neurons is in contrast with the overall expression of genes in oligodendrocytes, which overlaps most with OPC populations (Figure S9D), indicating a selective dysregulation of shared processes in oligodendrocytes and excitatory neurons. A gene ontology analysis of the intersecting co-regulated genes identified enrichment for multiple terms, including those related to amyloid fibril formation (Figure 5C), further suggesting a similar, shared -related response. Remarkably, this signature was most prominent in the samples with lowest burden for both excitatory neurons and oligodendrocytes (FDR-adjusted p-value < 0.05, Figures 5D, S9E and S9F). The DE genes that made up the leading edge from GSEA were involved in multiple aspects of amyloid processing, including regulation of APP transcription, beta-secretase regulation, and degradation/clearance pathways for peptides (Figure 5E). These changes included the downregulation of genes known to decrease the production or else help clear peptides, such as SORL1, BACE2, and PICALM, as well as the upregulation of genes involved in amyloid formation like RAB11A, LRRTM3, and APP itself (Figure 5F). Crucially, the gene set was consistently enriched across oligodendrocytes in a meta-analysis of postmortem cohorts with low AD histopathology3,4 (Figure S9G), and not in DE genes from other disease states, including PD47,48, ASD49, and MS50 (Figure 5G), reinforcing its robust and specific association with AD across cohorts.
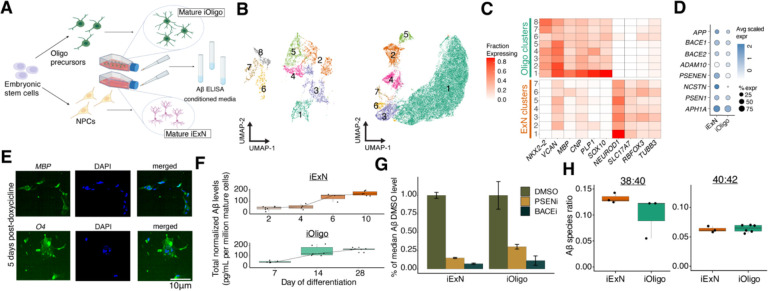
To experimentally assess the relative -forming potential of these two cell populations, we differentiated the H1 embryonic stem cell (ESC) line into mature oligodendrocytes (iOligos) and excitatory neurons (iExNs) (Figure 6A; Methods). In our iExN culture, we found nearly all cells expressed major excitatory neuron markers, as measured by single-cell RNA-sequencing (Figures S10A–S10C). Single-cell analysis (Figure 6B) of our iOligo cultures showed robust expression of numerous genes known to play roles in myelin function such as MBP, PLP1, and CNP, as well as transcription factors important for oligodendrocyte differentiation and maturation such as SOX10 and NKX2–2 (Figures 6C and S10D–S10F). These genes were not expressed at high levels in our excitatory neuron cultures, which instead were marked by canonical neuronal marker genes such as RBFOX3, SLC17A7, and TUBB3 (Figure 6C). Importantly, both cultures expressed appreciable levels of all the necessary machinery to produce amyloid beta protein (Figure 6D). Further, immunohistochemistry of key proteins defining the oligodendrocyte lineage, such as MBP and O4, showed a linear, significant increase upon induction of SOX10 (Figure 6E, Figure S10I, p-value < 0.05, linear mixed-effect model), while markers of other cell types were not significantly associated (Figures S10G and S10H).
Figure 6. Quantitation of production by human mature oligodendrocytes and excitatory neurons.
A) Schematic of differentiation of ESCs and ELISA-based quantification of from conditioned media. B) Two-dimension UMAP embeddings of single-cell expression profiling for ESC-derived iOligo (left) and iExN (right) cultures. C) Expression of key marker genes in ESC-derived iOligo and iExN cultures. D) Dot plot depicting scaled expression of essential machinery in ESC-derived cultures of iOligos and iExNs. E) Representative images of immunofluorescence stains of O4 and MBP in ESC-derived iOligo cultures five days after doxycycline addition. F) Normalized protein abundance for ESC-derived iExNs (top) and iOligos (bottom) across days of differentiation. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. G) Fractional abundance of protein levels relative to median protein levels in DMSO condition for PSEN inhibitor-treated and BACE inhibitor-treated conditioned media samples for ESC-derived iOligos and iExNs. Error bars indicate one standard deviation above and below the mean value. H) Ratio of to and to species from conditioned media obtained from ESC-derived cultures of iExNs (left) and iOligos (right). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers.
An ELISA-based quantification of amyloid-beta peptides from our iOligo cultures (Methods) showed a linear, three-fold increase in upon SOX10 induction (Figure 6F). We observed a similar fold increase after induction of differentiation in the iExN culture (Figure 6F). Indeed, the total abundance of was not significantly different (, Student’s t-test, Figure 6F) between iExN and iOligo at their respective differentiation endpoints, suggesting a similar intrinsic cell-autonomous capacity to produce amyloid beta. As predicted with our analysis of single-nucleus transcriptome data, we could not detect any in media taken from ESC-derived microglia cultures, with raw values similar to those found in an unconditioned media (Figure S10J). Further supporting the functionality of production and processing machinery in both populations, treatment of both iOligo and iExN cultures with a beta-secretase (BACE) inhibitor or gamma-secretase inhibitor caused a 10-fold reduction in total protein levels in both cell types (Figure 6G). Finally, we sought to determine whether the species composition of amyloid peptide production was significantly different between cell types given the well-established higher aggregation and amyloid formation potential of longer species. We found that species ratios were not significantly different (p-value = 0.64, Student’s t-test, Figure 6H) between iOligos and iExNs, suggesting that oligodendrocyte-derived peptides could contribute to AD-related amyloidosis in similar ways to excitatory neurons.
Discussion
Therapeutic trials of AD have made increasingly clear the importance of early intervention into the disease2, but identifying the cellular states occurring in human tissue at early disease stages has been challenging. Here, we leveraged a unique cohort of fresh human brain biopsy tissue to identify cellular perturbations–which we collectively refer to as the Early Cortical Amyloid Response (ECAR)--that are specifically present in tissue at the earliest stages of AD pathology. One prominent ECAR component was the identification of a hyperactive, hypermetabolic signature within excitatory neurons. This signature was associated with an astrocytic upregulation of glutathione metabolism and fatty acid degradation, suggesting dysregulation of synapse homeostasis in response to aberrant neuronal activity33,34. Furthermore, this upper layer hyperactivity phenotype was tightly coupled with the loss of a specific NDNF+ layer 1 interneuron population very early in disease progression. NDNF-expressing interneurons are most active in states of arousal25, and their activation is positively correlated with associative learning51, suggesting that their loss may directly affect memory formation. In addition, a recent study of NDNF-expressing neurons in the hippocampus found that their potentiation led to an inhibitory shift at excitatory synapses between entorhinal cortical projections and CA1 neuronal dendrites52. The loss of this cell type could thus help seed foci of aberrant excitation, further aggravating the acute effects of accumulation in the tissue. Hyperactivity has been observed in animal AD models that either overexpress APP or are exposed to -containing extracts from AD patients32,33,53,54. Our work in human tissue finds that hyperactivity is a prelude to subsequent neuronal loss, and postulates a mechanism–loss of a specific inhibitory neuron population–that contributes to its onset.
The second ECAR component is the expansion of two activated microglial states, one of which (GPNMB-EYA2) is shared between AD and PD, and the other (LPL-CD83) that is expanded only in AD. One means by which microglia protect against neurodegeneration is through the autophagy-mediated clearance of 55 and α-synuclein56, a convergence that could explain the expansion of the GPNMB-EYA2 population–enriched for autophagy-related genes–in both AD and PD. The LPL-CD83 population–whose expansion is AD-specific–shows high expression of TGF-β signaling components, including TGFBR1 and SMAD3, which both promote clearance by microglia57 and mediate tissue repair58. According to our integrative analysis, neither of these cell states was expanded in the examined AD mouse models, and the states themselves showed more molecular divergence between species than other microglial states, arguing for the importance of human samples when studying these highly disease-relevant cells. Interestingly, exposure of human ESC-derived microglia to diverse brain-related challenges was recently shown to induce in vitro cellular states that transcriptionally resemble our GPNMB+ states (e.g., high expression of GLDN, CD83, PPARG, and MYO1E)59. It will be important to more deeply characterize these states genomically, and to study their functional properties, such as capacity for phagocytosis, synaptic engulfment, and neuroinflammatory potential.
The last ECAR component we identified was a shared signature, in both oligodendrocytes and excitatory neurons, of differentially regulated genes associated with production. This signature peaked especially in the lowest-stage amyloid burden samples, suggesting a declining rate of amyloid production with the progression of the disease, consistent with rates of amyloid accumulation determined from pre-clinical non-invasive measurements made in patients with dominantly-inherited AD60. Because our signature derives from measurements made from human biopsy tissue, it provided us with a unique opportunity to uncover the molecular mechanisms underlying excess production and accumulation of in early stages of AD pathology in the human brain. Upregulated genes in this signature encoded for pro-amyloidogenic factors such as LRRTM3, and RAB11A, as well as APP itself, while ITM2B, SORL1, and BACE2 were downregulated. The dysregulation of these specific genes within human diseased tissue nominates them as especially promising targets for therapeutic intervention into early amyloid beta accumulation.
Our analyses establish oligodendrocytes as an amyloid-producing cell type in AD. Prior work in animal cells and models have suggested that other cell types, beyond excitatory neurons, could be sources of amyloid61,62. Our work–supported by both analyses of human tissue and human ESC-derived cultures–suggests that in humans, production is primarily in oligodendrocytes and excitatory neurons. Neuropathological studies have postulated an inverse relationship between myelination and AD pathology63, prompting hypotheses that myelin breakdown may play a causal role in the disease64. Additionally, white matter regions are some of the first to exhibit a high burden of oligomeric 65. These studies, coupled with our results, underscore the relevance of the interface between neuronal axons and oligodendrocytic bodies to AD pathogenesis.
Although our findings were enabled by the exceptional data quality obtained from a rare cohort of freshly-frozen brain biopsies, we utilized large-scale integrative analysis of many published datasets to corroborate many of our findings, and to assess their specificity for AD. Our integrative analysis illustrates that cell type identities are more resilient to peri- and post-mortem effects compared to expression patterns of individual genes, and can be accurately recovered by anchoring to high-quality datasets. From this work, we conclude that single-cell brain datasets are generally of sufficient consistency and quality that it is possible to conduct cumulative, highly informative meta-analyses. Such analyses will not only ensure the consistency of biological findings across multiple cohorts, but will also enable comparative analyses–as we performed here across many diseases and models with microglial expansion phenotypes–to assess the specificity of a state for a particular disease. To further facilitate this endeavor, we have established a web-based resource (available at https://braincelldata.org/resource) where individual scientists can explore our integrative analysis–which covers most published single-cell studies of the brain– to formulate and test mechanistic hypotheses. In addition, we have generalized the process of data integration to enable scientists to seamlessly integrate their own new datasets into our analysis, providing a common language for understanding cell-type-specific changes in different cortical diseases. We expect that the continued accrual of data from more donors, regions, species, and related conditions will provide additional crucial insights into the pathogenic process of AD and other diseases of the brain.
STAR Methods
Procurement of frontal cortex brain biopsies
Patients presenting to a clinic at the Kuopio University Hospital were evaluated for adult hydrocephalus with NPH symptoms: 47 with idiopathic normal pressure hydrocephalus, 3 with previously unrecognized congenital hydrocephalus and 2 with acquired hydrocephalus. Patients were consented for retrieval of brain biopsies during ventriculoperitoneal shunt placement for treatment of their symptomatic adult hydrocephalus. Biopsies were taken at the site where the shunt would penetrate the brain. Three cylindrical biopsies were taken approximately 2mm in diameter and 3–10mm in length using a disposable Temno Evolution TT146 (Merit Medical Systems) biopsy tool. The insertion point of the catheter was approximately 3 cm from the midline and anterior to the coronal suture66. Biopsies were immediately frozen with liquid nitrogen and stored at −80°C. One biopsy was sent for histopathological staining using the 6F3D and AT8 antibodies and evaluated by a neuropathologist for presence of plaques and tau tangles via light microscopy67. Biopsy plaques burden was further assessed semiquantitatively by a neuropathologist (T.R.) under light microscopy and assigned to mild (1), moderate (2), or severe amyloid burden (3) as described previously68. Our initial cohort included 58 individuals. We excluded one individual with tau-only pathology and another patient with a history of psychosis. We excluded four additional individuals (2 -free and 2 ) that, upon microscopic inspection of Nissl stained cryosections (see the biopsy tissue quality scoring methods), displayed decidedly poor tissue quality and a very high ratio of white matter to cortical matter. All of these excluded biopsies were more than 85% white matter tracts with the diminished cortical regions showing dysmorphic neuronal profiles. The biopsy procedure was approved by the Research Ethics Committee of the Northern Savo Hospital District (decision No. 276/13.02.00/2016).
Neuroanatomical localization of biopsy site
The stereotactic position (distances in millimeters) was measured from anatomically linked planes (transverse, sagittal, coronal) in a multiplanar reconstruction (MPR) produced from the postoperative CT/MRI DICOM image. After planar alignment (transverse and sagittal planes to the midline, and coronal plane in a 90-degree angle to the planum sphenoidale), the biopsy location was determined to be at proximal catheter’s cortex entry site at the catheter’s midline. Following distances were measured: In the transverse plane from the midline to the biopsy location (x). In the sagittal plane from the frontal bone’s internal cortex to the biopsy location’s coronal axis (at 90-degree angle) (y). In the sagittal plane, distance from the planum sphenoidale to the biopsy location’s transverse axis (at 90-degree angle) (z). EBRAINS Siibra-explorer was used to map and visualize each biopsy position69–71.
Biopsy tissue quality scoring
To ascertain tissue quality measurements (range from 1–10), we performed Nissl staining followed by semi-quantitative scoring of each biopsy slide image. For Nissl staining, briefly, fresh frozen tissue was thermally equilibrated to −20°C in a cryostat (Leica CM3050S) for 20 minutes. Tissue was mounted onto a cryostat chuck with Optimal Cutting Temperature compound (O.C.T. compound), aligned at a 5° cutting angle, and sectioned at 10 μm in thickness per tissue slice. Using a Superfrost plus slide that has been pre-cooled to −20°C, the tissue section was collected by carefully placing it and gently flattening it with the brush on top of the slide. Subsequently, slides were stained with 0.1% Cresyl Violet acetate in DiH2O, destained with ethanol, and 100% Xylene for 5 minutes. Slides were mounted by adding 2–3 drops of Permount around the tissue and coverslipped. Images were subsequently collected using a Keyence BZ-X810 series All-in-one Fluorescence microscope. With BZ-X800 viewer software, each stained slide was imaged on the Brightfield/Phase contrast channel using a 20X objective. The stained region of interest was selected by specifying the XY positions of the tissue outer edges and adjusting the Z-stack function to auto-focus prior to each image capture. The stitching of captured image series was made with BZ-X800 analyzer software. The images are exported as Big TIFF files and edited for cropping, contrast, and brightness with Photoshop software.
Measurement of iNPH grading scale
Severity of iNPH related symptoms was evaluated using the iNPH grading scale (iNPHGS)72, a clinician-rated scale that aims to assess the hallmark triad of the disease. Inversely-correlated with the severity of the disease, the 12-point iNPHGS has been shown to be clinically meaningful down to a single point72.
Generation of single-nuclei suspensions from frozen brain biopsies
Fresh-frozen brain biopsy tissue was cryosectioned at −15 to −20°C into 60-micron sections. Following microdissection, samples were placed on dry ice until nuclei isolation. To each cryosectioned sample, 1 mL of Extraction Buffer (ExB) was added into a 1.5-mL Eppendorf tube. Samples were briefly triturated before being placed in a six-well plate. Samples were then triturated 20 times with the ExB, every 2 minutes, until no large chunks of tissue were observed in each well. After the last trituration, samples were diluted with 45–50mL of wash buffer in a 50-mL Falcon tube, and then split into four 13–15 mL solutions in 50mL Falcon tubes. The diluted samples were then spun at 500g for 10 minutes at 4°C (pre-cooled) in a swing bucket benchtop centrifuge.
After centrifugation, a visible nuclei pellet was observed. Samples were then removed very gently from the centrifuge and placed in an ice bucket. The supernatant was aspirated until there was barely any liquid observed on top of the pellet (50–100μL of liquid left). To aspirate without disturbing the pellet, a serological pipette was first used till about 1mL was remaining, followed by serial aspiration with a P2000 and P200 pipette.
The pellets were then resuspended in 250μL of wash buffer (WB), mixed thoroughly by trituration and placed in an Eppendorf 1.5-mL tube.
Single-nucleus and single-cell RNA-sequencing and read pre-processing
For all single-nuclei experiments, the 10X Genomics (v3) kit was used according to the manufacturer’s protocol recommendations. Library preparation was performed according to the manufacturer’s recommendation. Libraries were pooled and sequenced on either a NovaSeq S2 or S4.
Sequencing reads from human brain biopsy experiments were demultiplexed and aligned to the hg19 reference using DropSeqTools (https://github.com/broadinstitute/Drop-seq) with the default settings. To reduce background noise from ambient RNA and potential UMI barcode swaps, we used Cellbender remove-background v273 with the default applied settings to all libraries. The Cellbender-corrected reads were used for downstream variable gene selection, dimensionality reduction, clustering, and differential expression. Cellbender was also used to distinguish cells from empty droplets.
Initial clustering of the biopsy cohort
Pre-processed Cellbender-corrected digital expression matrices were loaded into R per library as a digital gene expression matrix. All matrices were combined per individual and an initial variable gene selection was performed. A low-dimensional embedding was generated via rliger v1.0 at a and lambda = 5. Following integrative non-negative matrix factorization18, a shared nearest neighbors graph was generated and individual nuclei profiles were clustered according to the SLM (smart local moving) algorithm to identify broad cell classes. We used a recent large-scale survey of postmortem human brain21 to identify cell class markers and merged each cluster into one of eight cell classes (excitatory neurons, inhibitory neurons, astrocytes, microglia/macrophages, oligodendrocytes, oligodendrocyte precursor cells, endothelial cells/pericytes, and peripheral blood mononuclear cells (PBMCs)). PBMCs were excluded from downstream analysis.
For each cell class, individual nuclei were subsetted and the above clustering process was repeated to identify individual cell types. Marker genes were identified for neuronal populations based on a large-scale survey of neurons in the human neocortex21. Further, a recent survey of microglia/macrophage in the murine brain was used to identify cell type markers for microglia and macrophages74. For other non-neuronal types, we performed the Wilcoxon rank-sum test on SLM-defined cell clusters to find markers and thereby determine cell type annotations. We removed doublets identified as clusters that expressed markers of more than one cell class population. We also removed clusters whose markers contained high numbers of mitochondrial genes or heat shock related proteins.
Integrative analysis of the biopsy dataset with postmortem studies
We collected and uniformly processed all publicly available metadata on each dataset including the donor information (e.g., age, sex, diagnosis), sample information (e.g., brain region, sequencing protocol, batch structure), cell type identities, and quality metrics. All gene identifiers were mapped to Ensembl gene id. For mouse datasets, we further mapped Ensembl gene ids to their human orthologs75. However, we did retain non-orthologous mouse genes for normalization. We calculated the following quality metrics for every cell in each dataset: number of unique genes (nGene) and total unique molecular identifier (nUMI), percentage of mitochondrial genes (MT%), percentage of ribosomal genes (Ribo%), percentage of non-coding lncRNAs (lncRNA%), and percentage of dissociation-related artifact genes20. We used nGene and MT% quality metrics as our initial criteria to select cells for our integrative analyses and used the other quality metrics to identify and remove low quality cell clusters from the integrative analysis results. We retained cells with nGene >500 and MT% <5. For microglia cells, we used nGene >200 for two studies4,39 to compensate for the lower number of unique genes compared to other cell types. We further used the nGene >200 threshold for all cell classes in Mathys et al. dataset3. Finally, we removed donors with less than 50 cells within each cell class. Our integrative analyses across the seven cell classes included a total of 2,406,980 high quality cell profiles across 36 datasets from 28 studies on humans and mice. Distributions of the quality metrics are included in Figures S3 and S4. See data availability section for information about availability of the integrative analysis.
We performed our integrative analysis of each cell class individually to maximize the accuracy of cell state mapping across datasets. The seven major cell classes were: excitatory neurons (ExN), inhibitory neurons (InN), astrocytes (Astro), microglia/macrophages (Micro), oligodendrocytes (Oligo), Oligodendrocyte progenitor cells (OPC), and endothelial/pericyte cells (Endo). For ExNs and InNs, we limited our analysis to cortex brain region. However, glial cells were represented from across the brain regions. Table S2 summarizes the datasets that are included in integrative analysis of each of the seven cell classes. As outlined below, we developed a multi-step framework to efficiently handle substantial biological and technical variation that exists among the single cell and nuclei datasets.
Selecting highly variable genes.
We reasoned that the influence of batch effects on the cell embedding space would be minimized by selection of genes that recurrently show high variability across the human and mouse datasets. To achieve this, we implemented the following method: 1) Select the top 2000 variable genes within each donor of each dataset by the vst method in Seurat76. 2) Weight the selected genes in each donor so that the sum of the weights for each dataset add up to one. 3) Calculate an aggregate score for each gene by summing up their weighted scores. This procedure aims to minimize the participation of genes that show between dataset variability (hence likely influenced by batch effects) in the follow up analysis of cell embedding construction and clustering.
Principal component analysis.
To remove donor-specific batch effects (e.g., due to pre and post-mortem effects, sample preparation, and sequencing settings), we performed scaling (i.e., mean of zero and unit variance) of transcriptome data per gene and per donor and used this scaled data for principal component analysis. Comparison of different integrative solutions indicated the better quality after removal of donor specific effects. For all seven cell class analyses, we used the top 30 principal components, weighted by their variance explained.
Batch effect removal.
We used Harmony v1.077 to remove batch effects from the PCs with donor id and organism specified as the main source of batch effects. The default theta and lambda parameters were used for all analyses, except for the Endo cells with the theta parameter of four.
Assessing the quality integration solutions.
To assess the quality of the results, we developed multiple “cluster-free” quality metrics enabling us to perform a systematic and unbiased comparison of the solutions that is independent of the clustering method. These metrics can be grouped into three main categories. First, we required a uniform distribution of the datasets in the UMAP space. In addition to visual inspection, we developed a method that allowed us to do a quantitative evaluation of dataset distributions. Briefly, each of the two UMAP coordinates are split to 100 units, providing 10,000 bins. Within each bin a hypergeometric test is performed to assess whether or not cells from a specific dataset are over-represented. This analysis is performed for each dataset from each integrative solution (the related R functions and visualizations are provided at https://braincelldata.org/resource). Second, we examined whether cells expressing known cortical cell type markers are aligned with each other across datasets and organisms. To systematically test this, we repurposed the commonly used feature plot visualizations to represent donors instead of individual cells, thereby bypassing the effect of sample size variation between donors and datasets (Figure S4). Finally, we assessed if the initial clustering structure of each dataset is preserved in the aligned space. For this analysis, we used the reported clustering structure for each of the datasets individually. We also constructed confusion matrices to compare the cluster annotations between datasets (see the linked website for more details).
Cluster quality analysis.
On each cell class integrative analysis we performed Leiden clustering using the Seurat package76 with clustering resolutions of 0.6 or 0.8. We next used our calculated cell-based quality metrics (nGene, nUMI, MT%, Ribo%, lncRNA%, and %dissociation-related artifact genes) to identify and remove low quality clusters. We also performed marker analysis of each cluster per each dataset using FindAllMarkers() function in Seurat to identify and remove doublet clusters.
Uniform annotation of the datasets.
We modified a previously developed random walk algorithm78 to transfer cell type annotations from the biopsy dataset to each of 35 datasets (27 studies) in the aligned space, thereby uniformly annotating all datasets with cluster labels from the biopsy cohort. We next checked the consistency of cell type proportions among datasets and expression of marker genes across datasets and clusters (Table S3; See the linked website for more details).
Cell type marker analysis
Marker genes were identified in each dataset by running the ‘FindAllMarkers’ function in Seurat76. Significant genes (adjusted p-value < 0.1) with min.diff.pct of 0.1 were considered as markers. The heatmap in Figure 3A based on a select number of microglia markers based on the existing knowledge on microglia states and function. The full organized marker results are provided in Table S3.
To assess an overlap between cell state markers and DE genes in the microglia analysis (Figure 3D), we retained the top 100 upregulated markers (adjusted p-value < 0.01; sorted by p-value) that were markers with logFC >0 in less than a third of the cell states. This additional criterion was added to avoid spurious overlap of markers in non-homeostatic microglia that were driven by the large size of the homeostatic microglia.
Differential abundance
In our integrative analysis, a major analytic challenge was the wide variation in cell class compositions among the analyzed datasets. As an illustration, both of human PD datasets, one of human AD datasets, and all of mouse AD datasets included only glial cells and not neuronal cells from cortex. To address this, we conducted our meta-analysis of cell type proportional changes within each cell class separately. For our meta-analysis of early AD pathology, we included three out of 6 AD postmortem cohorts from the frontal lobe (more specifically prefrontal cortex or superior frontal gyrus)3,4,6. We excluded one cohort79 due to overlap of individuals with another included cohort3, and two that lacked sufficient numbers of early AD stage subjects5,7. We further included two PD postmortem datasets47,48, one ASD dataset49 and one MS dataset50 as contrast groups in our analyses.
We used a logistic mixed-effect model80 to identify differentially abundant cell populations in each dataset separately (Table S4). For all human datasets, we included sex as a fixed effect, and individual as a random effect in the model. We then tested the significance of association between the status with the clusters using a Wald test. For assessing cell type abundance associations with iNPH grading scales, we modified the method to allow for continuous independent variables, while preserving the Wald test for assigning significance.
We performed a meta-analysis of cell type abundance results across four cohorts with designations for earlier and later AD stages: (1) the biopsy cohort ( as early and as late; all cell classes), (2) Mathys et al. (Braak III-IV as early and Braak V-VI as late; all cell classes except endothelial cells), (3) Leng et al. (Braak III-IV as early and Braak V-VI as late; all cell classes) and (4) Gerrits et al. (CtrlPlus as early and AD as late). The p-values from individual analyses were combined together via the Stouffer’s method, with an additional consideration of the directionality of the change as determined via the odds ratio assessment from the Wald test. All Stouffer’s p-values were subsequently corrected for multiple hypothesis testing via the Benjamini-Hochberg correction.
To determine the relative cellular abundance changes at the cell class level we generated p-values by comparing proportions of the seven cell classes by a Wilcoxon rank-sum test. Meta-analysis p-values (and Z-scores) were generated using Stouffer’s method, taking into account the directionality of the abundance change.
For mouse datasets, we used Fisher’s exact test to examine expansion and loss of different microglia cell types and states (Table S4). The p-values were subsequently corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. For visualization purposes, z-scores were calculated by transformation of the p-values and signed by the directionality of the log odds ratio.
Differential gene expression analysis
We employed a pseudocell strategy coupled with mixed linear models and jack-knifing to robustly identify differentially expressed genes. To construct pseudocells, we aggregated the raw UMI count of, on average, every 30 cells per subject and cell type. We constructed one pseudocell for cell types that had between 15 to 45 cells in a donor and excluded cell types that had less than 15 cells. This reduces the impact of dropout and technical variability, while ameliorating low statistical power and high variation in sample size issues attributed to the pseudobulk approaches81. We used the Limma Trend82 approach with robust moderated t-statistic to identify DE genes within each cell class with sex, cell type, log2(pseudocell MT%) and log2(pseudocell nUMI) as covariates and subject id as a random effect. Cell type annotation was included as a covariate to account for the cell type-specific baseline expression of the genes and therefore to ameliorate the impact of cell type expansion on the DE patterns.
We further performed jack-knife resampling at two levels to identify robust DE genes that are shared among the majority of individuals. First, iterating on each of the 52 individuals in the biopsy cohort, we excluded one subject from the analysis of each cell class and then re-calculated the DE statistic for the remaining 51 individuals, retaining the maximum p-value (i.e., the least significant p-value) achieved for each gene. An adjusted jack-knife p-value was next calculated for genes with adjusted p-value < 0.1 in the main analysis using the Benjamini-Hochberg correction. Second, iterating 50 times, we randomly sampled 50% of cohort subjects (balanced by their pathological status) and re-calculated the logFC patterns. A consistency score was defined for each gene as the fraction of iterations in which the jack-knifed logFCs were consistent with the logFC pattern from the full cohort of 52 individuals (i.e., up- or down-regulated in both). Genes with jack-knifed adjusted p-value < 0.01 and jack-knifed consistency score ≥0.9 were deemed as significant. Comparison of the DE patterns with a pseudobulk approach using LimmaTrend indicated highly consistent (>99%) logFC patterns between the pseudobulk and our pseudocell strategy for the DE genes. We also found majority of identified DE gens by pseudocell approach remain significant (median >72% per cell class; pseudobulk FDR-adjusted p-value < 0.1) in the pseudobulk approach, while the identified DE genes by our pseudocell approach show much less sensitivity to variation in cohort size (data not shown).
To compare the DE genes between and individuals (Figure 1F), we performed a paired t-test based on values from below equation:
This equation will be positive only if the logFC from both and are in the same direction (i.e., gene is up or down regulated in both conditions) and are stronger in and negative otherwise. We excluded from this analysis cell types with less than 20 DE genes in either and conditions as they usually were small and their fold change patterns were not reliable. We used a paired t-test to determine if the outputs of this function are randomly distributed around zero or are biased towards positive (i.e., consistent but stronger logFC in ) or negative (i.e., discordant or stronger in ) values.
Gene set enrichment analysis
DE genes were filtered to protein-coding based on the gene biotype information from the ‘EnsDb.Hsapiens.v86’ package in R Bioconductor. Genes were next ordered based on their t-statistic from LimmaTrend mixed linear models. Curated GO Biological Process, KEGG and Reactome gene sets were retrieved from the EnrichR portal83. To identify enriched pathways, we ran the fGSEA package84 v1.16.0 with default setting while limiting the geneset sizes between 15 and 250 genes. For each cell class and cell type analysis, protein-coding genes expressed in more than one percent of cells in the corresponding group were used as background.
Correlating cell type proportional changes to transcriptome responses across cell types
To identify correlation between cell type abundances and transcriptional phenotypes, we used a logistic mixed-effect model80. Specifically, we constructed a meta-gene (referred to as DE signature in the main text) from the top 300 upregulated protein-coding genes (sorted by their jack-knifed p-value) in each individual cell by aggregating their corresponding UMI counts, as proposed before85. The meta-gene was next normalized over the total nUMI count of the cells and standardized to have a mean of zero and variance of one across cells from all subjects in our cohort. We then binarized cells as active or inactive for a meta gene based on a standardized score threshold of 2. In our analyses we required presence of at least two subjects with more than 3% transcriptionally active cells in each cell type and status category. For cell types that do not meet this criterion we set the association p-value to one to indicate transcriptional changes in the corresponding cell type are not associated with the interested cell type fractional variation. Finally, we fit a logistic mixed-effect model on the binarized scores to examine their association with the normalized cell type proportional changes with sex as a covariate and subject as a random effect. The cell type proportions were normalized by applying an empirical cumulative estimation using the ecdf() function in R. We used cell-class level DE genes to construct meta-genes since: 1) cell class level DE genes were highly conserved within cell types (Figure S6D); 2) DE genes were not driven by the variation in the cell counts of the cell types. As summarized below, we performed robustness analysis and alternative meta-gene construction schemes to further confirm the observed associations.
First, to assess the robustness of the results, we tested the sensitivity of the results to the presence of technical variation in cell gene counts by, iterating 30 times, adding Poisson noise to the transcriptome data of each individual cell before calculation of the meta-gene expressions (Figure S7B). In addition, iterating 30 times we randomly down sampled the cell types to examine the association of the cell type sizes on the results (Figure S7C).
Second, as an alternative analysis method to support our findings (Figure S7D), we constructed meta-genes through principal component analysis of the normalized and scaled expression of the top 300 upregulated protein-coding genes (sorted based on jack-knifed p-values). Similar to the WGCNA approach86, the first principal component was chosen as the meta-gene. The meta-gene scores were then binarized as above and the association with cell type proportional changes were examined using a logistic mixed-effects model similar to above.
Heritability enrichment of differentially expressed genes with MAGMA
We used MAGMA42 to determine the degree of enrichment of common variant risk in the list of differentially expressed genes across cell types. We first downloaded the summary statistics from a recent common variant meta-analysis of AD and related dementias35, PD87, and ASD88. Using the online FUMA tool89, we generated Z-scores for each gene, corresponding to the approximate degree of association between the gene and AD (SNP2GENE function). To determine gene sets for each cell class, we took the top 200 differentially expressed genes between biopsy samples with AD pathology versus those samples without, ordered by t-statistic at a significance value of p < 0.2 (to ensure enough genes were being captured per gene set). Significance values for the gene set of interest were calculated via MAGMA, wherein a regression is fit to determine whether those genes with membership for that set have a significant enrichment for heritable risk of the trait of interest.
Generation of a doxycycline-inducible SOX10 H1 stem cell line
We adapted a recently-published protocol to produce mature oligodendrocytes from the H1 embryonic stem cell precursors cell line90. First, we isolated and incorporated a SOX10 transcription factor (Addgene #115242) into the backbone of a doxycycline-inducible cassette (Addgene #105840) to generate pBR01.
H1 ESCs were plated in Matrigel (cat. #47743–716)-coated (30-minute incubation at 37°C prior to cell plating) plates in mTesR1 (cat. #85857) with supplements (ESC media, StemCell Technologies cat. #85857) and RevitaCell (cat. #A2644501). After plating, we performed daily media changes with ESC media without RevitaCell until plates were approximately 80% confluent with compact colonies. For routine passaging, ESCs were washed 1X with PBS (cat. #10010049) and incubated in Versene (cat. #BE17–711E) for 5 mins at room temperature, after which Versene was gently aspirated from the plate and replaced with ESC media. ESCs were gently dissociated into a cell suspension using a manual cell scraper and transferred as small colonies to a fresh Matrigel-coated plate at a 1:20 dilution. In order to generate a doxycycline-inducible SOX10 cell line, we performed TALEN-based integration as has been previously described91. Briefly, we electroporated (1050V, 50 ms pulse, two pulses total) 1 million ESCs with 4 μg pBR01, 2 μg TALEN-L (Addgene #59025), 2 μg TALEN-R (Addgene #59026), and 0.4 μg Bcl-XL (Addgene #8790) plasmids using a Neon Transfection System Pipette Station (Thermo Fisher). After 96 hours, cells were incubated with 2 μg/mL puromycin (cat. #A1113803) for 72 hours. After puromycin selection, polyclonal ESCs were expanded and stored in liquid nitrogen at 106 cells/mL in 10% DMSO + mTesR1.
Oligodendrocyte differentiation of ESCs
To differentiate the resulting cell line into mature oligodendrocyte lineage cells, we adapted the Garcia-Leon protocol with minor modifications90. Briefly, we used the H1 embryonic stem cell line with the integrated SOX10 cassette to generate neural progenitor cells which were subsequently differentiated into mature oligodendrocytes. Vials containing 1 million ESC precursors were thawed and plated into one well of a Matrigel-coated 6-well plate, supplementing the cells with RevitaCell to increase vitality. These cells were then allowed to grow to confluence with supplementation of 1mL E8 media (cat. #A2858501). Confluent cells were subsequently split with the following procedure. First, cells were washed with 1mL of Dulbecco’s PBS per well. Then, cells were subsequently treated with 1mL of ReleSR (cat. #05872) incubated at 37°C for five minutes. Cells were then spun down and split at a ratio of 1:14 into Matrigel-coated 6-well plates and grown in 1mL of E8 media (cat. #A2858501) supplemented with 1× RevitaCell solution. The cells were transitioned to mTeSR1 media by replacing with a 1:1 E8 to mTeSR1 solution on the first day, 75% mTeSR1 with 25% E8 on the second day, and a full 1mL of 100% mTeSR1 on the thirdday. Cells were then allowed to grow to confluence before being split again with 1mL ReleSR as above. The cells were replated onto 6-well matrigel-coated plates, supplemented with RevitaCell. An N2B27 media was made by mixing non-essential amino acid MEM (cat. #11140–050), 2-mercaptoethanol (cat. #21985023), N2 (cat. #17502048) and B27 (cat. #12587010) to 1× concentration plus insulin (cat. #I9278) at 25 μg/ml final concentration to Dulbecco’s modified essential media. The cells were then grown in 2 mL of the pre-made N2B27 medium supplemented with 0.1 μM retinoic acid (RA, cat. #R2625), 10 μM SB431542 (cat. #04–0010-10) and 1 μM LDN193189 (cat. #04–0074) for five days and an additional two days with 10 μM of smoothened agonist (SAG, cat. #566660).
After cells achieved confluence, they were passaged using a pre-warmed 1mL aliquot of Accutase (cat. #A1110501) for 1–2 minutes. The cells were seeded onto 6-well plates coated with poly-l-ornithine (cat. #P3655) and laminin (cat. #L2020–1MG)-coated plates at a density of 10,000 cells per square centimeter. The cells were fed a differentiation medium supplemented with 2 μg/mL of doxycycline (cat. #D9891–10G) to induce expression of SOX10 and allowed to grow for 10 days, at which time they are mature.
Neuronal differentiation of ESCs
Neuronal differentiation of ESCs into cortical glutamatergic neurons was carried out as previously described92. In brief, the differentiation was carried out by adding doxycycline hyclate (2 μg/mL) to N2 supplemented media (Thermo Fisher, 17502048) with patterning factors SB431542 (Tocris, 1614, 10 μM), XAV939 (Stemgent, 04–00046, 2 μM) and LDN-193189 (Stemgent, 04–0074, 100 nM), as described previously92–94. Puromycin selection was used (5μg/μL), from days 2 to 6 to remove non-transduced cells. At 4 days post induction, neuronal cells were resuspended into Neurobasal media (Gibco, 21103049) that was supplemented with B27 (Gibco, 17504044, 50X), doxycycline (2 μg/mL), brain-derived neurotrophic factor (BDNF), ciliary neurotrophic factor (CTNF), and glial cell-derived neurotrophic factor (GDNF) (R&D Systems 248-BD/CF, 257–33 NT/CF, and 212-GD/CF at 10 ng/mL each). From this point onwards the neurons were either co-cultured with murine glial cells that were derived from early postnatal (P1-P3) mouse brains as described previously95 or were left to grow as monocultures (mouse strain https://www.jax.org/strain/100012; animal ethical committee approval by Harvard University: Animal Experimentation Protocol (AEP) # 93–15).
Immunohistochemistry and imaging of ESCs
We performed immunohistochemistry on ESC-derived oligodendrocytes at 1, 5, and 10 days after doxycycline-based SOX10 induction. Briefly, cells were grown on a 6-well plate and fixed using 2% PFA and then permeabilized using Triton-X (cat.# T9284–1L), followed by multiple washes with 1× Dulbecco’s PBS at each step. We used the following primary antibodies for our immunohistochemistry experiments: anti-O4 (cat. #MAB1326), anti-MBP (cat. #AB9348), anti-NeuN (cat. #MAB377), and anti-PAX6 (cat. #AB78545). The primary antibodies were diluted in a solution of 10% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) supplemented with 1% Triton-X then added to the cells and allowed to incubate overnight at 4°C. Cells were then washed three times in PBS. Finally, the secondary antibodies were diluted in a solution of 10% BSA in PBS supplemented with 1% Triton-X then added to the cells and allowed to incubate for 1–2 hours at room temperature. After the secondary incubation, one to two drops of ProLong Glass AntiFade Mountant with NucBlue (cat.# P36981) was added into the wells and coverslips were added on top of each cell culture into wells for downstream imaging.
Imaging of immunohistochemical stains was performed on a Keyence BZ-800XE microscope with a Nikon Apo 20× objective. All images were acquired using the same light emission settings and all channels were set to the same LUTs before quantification. For quantification, we used CellProfiler’s IdentifyPrimaryObjects and MeasureObjectIntensity function to segment cells based on their DAPI signal. Subsequently, the average fluorescence value (mean intensity value) was normalized per cell to the average fluorescence intensity of the DAPI signal. To determine the significance of an intensity difference, a linear mixed-effect model was used to calculate the significance of a change in normalized intensity value across days of differentiation, treating each slice image as a random effect. Significance values were determined via a likelihood ratio test against the null model not containing the day of differentiation.
Generation of single-cell suspension from ESC-derived H1 iOligodendrocytes
To generate single-cell experiments, briefly we used oligodendrocytes at terminal differentiation (past day 8 post-doxycycline induction of SOX10). We isolated cells using the passaging protocol as mentioned above and measured cell concentrations in our isolate using a hemocytometer.
Read processing and clustering of iOligodendrocyte and iExcitatory Neuron scRNA-seq experiments
Sequencing reads from iOligo experiments were demultiplexed and aligned to the hg38 reference using CellRanger with default setting using the command CellRanger mkfastq, followed by count generation using the command CellRanger count.
Sequencing reads from iExN experiments were demultiplexed and aligned to the hg38 reference using DropSeqTools with default setting.
To analyze single-cell RNA-sequencing data from ESC-derived oligodendrocytes and neurons we first determined highly variable genes using LIGER. We further used non-negative matrix factorization (with k, number of factors, set to 20) to determine a low-dimension embedding followed by graph-based clustering using SLM. Marker genes were identified by a Wilcoxon rank-sum and cells were annotated based on known markers of mature cell types as identified from our biopsy dataset.
ELISA-based amyloid beta quantification
To quantitate amyloid beta peptide levels from cell culture, we used the MesoScale Discovery V-Plex Plus Peptide Panel 1 (6E10) ELISA kit (cat. #K15200G). Briefly, we extracted 1.5 mL of conditioned media per well replicate from isolates of ESC-derived oligodendrocytes, neurons, and microglia. Isolates were stored at −80°C till the ELISA assay was run at which time they were brought up to 4°C before being spun down at 10,000rpm for 15 minutes. The MSD ELISA was run according to the manufacturer’s guidelines. Absolute peptide abundances were quantified using the MSD Discovery Workbench Analysis Software.
Supplementary Material
Acknowledgments
We would like to thank Henrik Zetterberg, Nader Morshed, and Michael-John Dolan for helpful discussions. We are also grateful for the help of Marita Parviainen and Tiina Laaksonen with patient management and cognitive testing, and Andrea Jiang with performing the amyloid beta ELISA assays. This work was supported by the Alzheimer’s Association and the International Neuroimmune Consortium (to B.S. and E.Z.M.), the Pew Biomedical Scholars Program (to E.Z.M.), NIH NIGMS grants T32GM007753/T32GM144273 (to T.K.), NIH NIA grant F30AG069446 (to T.K.), and the Kuopio University Hospital VTR fund (grant 5252614 to V.L.); Academy of Finland (grants 339763, 334801 to TM; 338182, 328287 to MH); the Sigrid Juselius Foundation; the Strategic Neuroscience Funding of the University of Eastern Finland, the Finnish Cultural Foundation, and the North Savo Regional Fund and by the European Union (ERC, HUMANE, 101043584 to TM). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
Footnotes
Competing interests: The authors declare no competing interests.
Data availability
All generated snRNA-seq data and the results of our integrative analysis of 28 single cell/nuclei studies are publicly available at: https://braincelldata.org/resource. This includes sample annotations related to the dataset source (36 datasets across 28 studies), cell identifiers (e.g., cell barcodes), quality metrics, and cell type annotations from integrative analysis. We have also included functionality to perform several analyses in a fast and efficient way, including: examination of the integration solutions, performing marker analysis across all of the datasets, and exploring differentially expressed genes.
References
- 1.De Strooper B., and Karran E. (2016). The Cellular Phase of Alzheimer’s Disease. Cell 164, 603–615. [DOI] [PubMed] [Google Scholar]
- 2.Long J.M., and Holtzman D.M. (2019). Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 179, 312–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X., et al. (2019). Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leng K., Li E., Eser R., Piergies A., Sit R., Tan M., Neff N., Li S.H., Rodriguez R.D., Suemoto C.K., et al. (2021). Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci. 24, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau S.-F., Cao H., Fu A.K.Y., and Ip N.Y. (2020). Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 117, 25800–25809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrits E., Brouwer N., Kooistra S.M., Woodbury M.E., Vermeiren Y., Lambourne M., Mulder J., Kummer M., Möller T., Biber K., et al. (2021). Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol. 141, 681–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otero-Garcia M., Mahajani S.U., Wakhloo D., Tang W., Xue Y.-Q., Morabito S., Pan J., Oberhauser J., Madira A.E., Shakouri T., et al. (2022). Molecular signatures underlying neurofibrillary tangle susceptibility in Alzheimer’s disease. Neuron 110, 2929–2948.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinaro F., Haneklaus M., Zhang Z., Strano A., Evans L., Handfield L.-F., Ryan N.S., Fox N.C., Hemberg M., Ramanathan S., et al. (2020). Molecular and cellular pathology of monogenic Alzheimer’s disease at single cell resolution. bioRxiv, 2020.07.14.202317. 10.1101/2020.07.14.202317. [DOI] [Google Scholar]
- 9.Kramvis I., Mansvelder H.D., and Meredith R.M. (2018). Neuronal life after death: electrophysiologic recordings from neurons in adult human brain tissue obtained through surgical resection or postmortem. Handb. Clin. Neurol. 150, 319–333. [DOI] [PubMed] [Google Scholar]
- 10.Guo L., Rezvanian A., Kukreja L., Hoveydai R., Bigio E.H., Mesulam M.-M., El Khoury J., and Geula C. (2016). Postmortem Adult Human Microglia Proliferate in Culture to High Passage and Maintain Their Response to Amyloid-β. J. Alzheimers. Dis. 54, 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Groot C.J., Montagne L., Janssen I., Ravid R., Van Der Valk P., and Veerhuis R. (2000). Isolation and characterization of adult microglial cells and oligodendrocytes derived from postmortem human brain tissue. Brain Res. Brain Res. Protoc. 5, 85–94. [DOI] [PubMed] [Google Scholar]
- 12.Dachet F., Brown J.B., Valyi-Nagy T., Narayan K.D., Serafini A., Boley N., Gingeras T.R., Celniker S.E., Mohapatra G., and Loeb J.A. (2021). Selective time-dependent changes in activity and cell-specific gene expression in human postmortem brain. Sci. Rep. 11, 6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luikku A.J., Hall A., Nerg O., Koivisto A.M., Hiltunen M., Helisalmi S., Herukka S.-K., Junkkari A., Sutela A., Kojoukhova M., et al. (2019). Predicting Development of Alzheimer’s Disease in Patients with Shunted Idiopathic Normal Pressure Hydrocephalus. J. Alzheimers. Dis. 71, 1233–1243. [DOI] [PubMed] [Google Scholar]
- 14.Leinonen V., Koivisto A.M., Alafuzoff I., Pyykkö O.T., Rummukainen J., von Und Zu Fraunberg, M., Jääskeläinen J.E., Soininen H., Rinne J., and Savolainen S. (2012). Cortical brain biopsy in long-term prognostication of 468 patients with possible normal pressure hydrocephalus. Neurodegener. Dis. 10, 166–169. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton R., Patel S., Lee E.B., Jackson E.M., Lopinto J., Arnold S.E., Clark C.M., Basil A., Shaw L.M., Xie S.X., et al. (2010). Lack of shunt response in suspected idiopathic normal pressure hydrocephalus with Alzheimer disease pathology. Ann. Neurol. 68, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seeburger J.L., Holder D.J., Combrinck M., Joachim C., Laterza O., Tanen M., Dallob A., Chappell D., Snyder K., Flynn M., et al. (2015). Cerebrospinal fluid biomarkers distinguish postmortem-confirmed Alzheimer’s disease from other dementias and healthy controls in the OPTIMA cohort. J. Alzheimers. Dis. 44, 525–539. [DOI] [PubMed] [Google Scholar]
- 17.Strozyk D., Blennow K., White L.R., and Launer L.J. (2003). CSF Abeta 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology 60, 652–656. [DOI] [PubMed] [Google Scholar]
- 18.Welch J.D., Kozareva V., Ferreira A., Vanderburg C., Martin C., and Macosko E.Z. (2019). Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh S.E., Walker A.J., Kamath T., Dissing-Olesen L., Hammond T.R., de Soysa T.Y., Young A.M.H., Murphy S., Abdulraouf A., Nadaf N., et al. (2022). Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 25, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y.E., Pan L., Zuo Y., Li X., and Hong W. (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329.e6. [DOI] [PubMed] [Google Scholar]
- 21.Bakken T.E., Jorstad N.L., Hu Q., Lake B.B., Tian W., Kalmbach B.E., Crow M., Hodge R.D., Krienen F.M., Sorensen S.A., et al. (2021). Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habib N., McCabe C., Medina S., Varshavsky M., Kitsberg D., Dvir-Szternfeld R., Green G., Dionne D., Nguyen L., Marshall J.L., et al. (2020). Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyssowski K.M., DeStefino N.R., Cho J.-H., Dunn C.J., Poston R.G., Carty C.E., Jones R.D., Chang S.M., Romeo P., Wurzelmann M.K., et al. (2018). Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 98, 530–546.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim L.A., Huang S., Fernandez-Otero M., Sherer M., Qiu Y., Vemuri S., Xu Q., Machold R., Pouchelon G., Rudy B., et al. (2021). Bottom-up inputs are required for establishment of top-down connectivity onto cortical layer 1 neurogliaform cells. Neuron 109, 3473–3485.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malina K.C.-K., Tsivourakis E., Kushinsky D., Apelblat D., Shtiglitz S., Zohar E., Sokoletsky M., Tasaka G.-I., Mizrahi A., Lampl I., et al. (2021). NDNF interneurons in layer 1 gain-modulate whole cortical columns according to an animal’s behavioral state. Neuron. 10.1016/j.neuron.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 26.West A.E., and Greenberg M.E. (2011). Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 3. 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turrigiano G.G. (2008). The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135, 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verderio C., Pozzi D., Pravettoni E., Inverardi F., Schenk U., Coco S., Proux-Gillardeaux V., Galli T., Rossetto O., Frassoni C., et al. (2004). SNAP-25 modulation of calcium dynamics underlies differences in GABAergic and glutamatergic responsiveness to depolarization. Neuron 41, 599–610. [DOI] [PubMed] [Google Scholar]
- 29.Pozzi D., Condliffe S., Bozzi Y., Chikhladze M., Grumelli C., Proux-Gillardeaux V., Takahashi M., Franceschetti S., Verderio C., and Matteoli M. (2008). Activity-dependent phosphorylation of Ser187 is required for SNAP-25-negative modulation of neuronal voltage-gated calcium channels. Proc. Natl. Acad. Sci. U. S. A. 105, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S.H., and Ryan T.A. (2010). CDK5 serves as a major control point in neurotransmitter release. Neuron 67, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirrito J.R., Yamada K.A., Finn M.B., Sloviter R.S., Bales K.R., May P.C., Schoepp D.D., Paul S.M., Mennerick S., and Holtzman D.M. (2005). Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron 48, 913–922. [DOI] [PubMed] [Google Scholar]
- 32.Li S., and Selkoe D.J. (2020). A mechanistic hypothesis for the impairment of synaptic plasticity by soluble oligomers from Alzheimer’s brain. J. Neurochem. 154, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zott B., Simon M.M., Hong W., Unger F., Chen-Engerer H.-J., Frosch M.P., Sakmann B., Walsh D.M., and Konnerth A. (2019). A vicious cycle of β amyloid-dependent neuronal hyperactivation. Science 365, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ioannou M.S., Jackson J., Sheu S.-H., Chang C.-L., Weigel A.V., Liu H., Pasolli H.A., Xu C.S., Pang S., Matthies D., et al. (2019). Neuron-Astrocyte Metabolic Coupling Protects against Activity-Induced Fatty Acid Toxicity. Cell 177, 1522–1535.e14. [DOI] [PubMed] [Google Scholar]
- 35.Wightman D.P., Jansen I.E., Savage J.E., Shadrin A.A., Bahrami S., Holland D., Rongve A., Børte S., Winsvold B.S., Drange O.K., et al. (2021). A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellenguez C., Küçükali F., Jansen I.E., Kleineidam L., Moreno-Grau S., Amin N., Naj A.C., Campos-Martin R., Grenier-Boley B., Andrade V., et al. (2022). New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hägg S., Athanasiu L., et al. (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mich J.K., Graybuck L.T., Hess E.E., Mahoney J.T., Kojima Y., Ding Y., Somasundaram S., Miller J.A., Kalmbach B.E., Radaelli C., et al. (2021). Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 34, 108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 40.Li Q., Cheng Z., Zhou L., Darmanis S., Neff N.F., Okamoto J., Gulati G., Bennett M.L., Sun L.O., Clarke L.E., et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207–223.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopes K. de P., Snijders G.J.L., Humphrey J., Allan A., Sneeboer M.A.M., Navarro E., Schilder B.M., Vialle R.A., Parks M., Missall R., et al. (2022). Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet. 54, 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Leeuw C.A., Mooij J.M., Heskes T., and Posthuma D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Nicola D., Valle-Argos B., and Nieto-Sampedro M. (2010). Blockade of IL-15 activity inhibits microglial activation through the NFkappaB, p38, and ERK1/2 pathways, reducing cytokine and chemokine release. Glia 58, 264–276. [DOI] [PubMed] [Google Scholar]
- 44.Matejuk A., and Ransohoff R.M. (2020). Crosstalk Between Astrocytes and Microglia: An Overview. Frontiers in Immunology 11. 10.3389/fimmu.2020.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi S.X., Li Y.-J., Shi K., Wood K., Ducruet A.F., and Liu Q. (2020). IL (Interleukin)-15 Bridges Astrocyte-Microglia Crosstalk and Exacerbates Brain Injury Following Intracerebral Hemorrhage. Stroke 51, 967–974. [DOI] [PubMed] [Google Scholar]
- 46.Supek F., Bošnjak M., Škunca N., and Šmuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamath T., Abdulraouf A., Burris S.J., Langlieb J., Gazestani V., Nadaf N.M., Balderrama K., Vanderburg C., and Macosko E.Z. (2022). Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 25, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smajić S., Prada-Medina C.A., Landoulsi Z., Ghelfi J., Delcambre S., Dietrich C., Jarazo J., Henck J., Balachandran S., Pachchek S., et al. (2022). Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145, 964–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velmeshev D., Schirmer L., Jung D., Haeussler M., Perez Y., Mayer S., Bhaduri A., Goyal N., Rowitch D.H., and Kriegstein A.R. (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jäkel S., Agirre E., Mendanha Falcão A., van Bruggen D., Lee K.W., Knuesel I., Malhotra D., Ffrench-Constant C., Williams A., and Castelo-Branco G. (2019). Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abs E., Poorthuis R.B., Apelblat D., Muhammad K., Pardi M.B., Enke L., Kushinsky D., Pu D.-L., Eizinger M.F., Conzelmann K.-K., et al. (2018). Learning-Related Plasticity in Dendrite-Targeting Layer 1 Interneurons. Neuron 100, 684–699.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercier M.S., Magloire V., Cornford J.H., and Kullmann D.M. (2022). Long-term potentiation in neurogliaform interneurons modulates excitation-inhibition balance in the temporoammonic pathway. J. Physiol. 600, 4001–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah D., Gsell W., Wahis J., Luckett E.S., Jamoulle T., Vermaercke B., Preman P., Moechars D., Hendrickx V., Jaspers T., et al. (2022). Astrocyte calcium dysfunction causes early network hyperactivity in Alzheimer’s disease. Cell Rep. 40, 111280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busche M.A., Eichhoff G., Adelsberger H., Abramowski D., Wiederhold K.-H., Haass C., Staufenbiel M., Konnerth A., and Garaschuk O. (2008). Clusters of hyperactive neurons near amyloid plaques in a mouse model of Alzheimer’s disease. Science 321, 1686–1689. [DOI] [PubMed] [Google Scholar]
- 55.Tarasoff-Conway J.M., Carare R.O., Osorio R.S., Glodzik L., Butler T., Fieremans E., Axel L., Rusinek H., Nicholson C., Zlokovic B.V., et al. (2015). Clearance systems in the brain—implications for Alzheimer disease. Nature Reviews Neurology 11, 457–470. 10.1038/nrneurol.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi I., Zhang Y., Seegobin S.P., Pruvost M., Wang Q., Purtell K., Zhang B., and Yue Z. (2020). Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat. Commun. 11, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyss-Coray T., Lin C., Yan F., Yu G.Q., Rohde M., McConlogue L., Masliah E., and Mucke L. (2001). TGF-beta1 promotes microglial amyloid-beta clearance and reduces plaque burden in transgenic mice. Nat. Med. 7, 612–618. [DOI] [PubMed] [Google Scholar]
- 58.Brionne T.C., Tesseur I., Masliah E., and Wyss-Coray T. (2003). Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron 40, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 59.Dolan M.-J., Therrien M., Jereb S., Kamath T., Atkeson T., Marsh S.E., Goeva A., Lojek N.M., Murphy S., White C.M., et al. (2022). A resource for generating and manipulating human microglial states in vitro. bioRxiv, 2022.05.02.490100. 10.1101/2022.05.02.490100. [DOI] [Google Scholar]
- 60.Bateman R.J., Xiong C., Benzinger T.L.S., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., Blazey T.M., et al. (2012). Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 367, 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veeraraghavalu K., Zhang C., Zhang X., Tanzi R.E., and Sisodia S.S. (2014). Age-dependent, non-cell-autonomous deposition of amyloid from synthesis of β-amyloid by cells other than excitatory neurons. J. Neurosci. 34, 3668–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skaper S.D., Evans N.A., Evans N.A., Rosin C., Facci L., and Richardson J.C. (2009). Oligodendrocytes are a novel source of amyloid peptide generation. Neurochem. Res. 34, 2243–2250. [DOI] [PubMed] [Google Scholar]
- 63.Braak H., and Braak E. (1996). Development of Alzheimer-related neurofibrillary changes in the neocortex inversely recapitulates cortical myelogenesis. Acta Neuropathol. 92, 197–201. [DOI] [PubMed] [Google Scholar]
- 64.Bartzokis G., Lu P.H., and Mintz J. (2007). Human brain myelination and amyloid beta deposition in Alzheimer’s disease. Alzheimers. Dement. 3, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collins-Praino L.E., Francis Y.I., Griffith E.Y., Wiegman A.F., Urbach J., Lawton A., Honig L.S., Cortes E., Vonsattel J.P.G., Canoll P.D., et al. (2014). Soluble amyloid beta levels are elevated in the white matter of Alzheimer’s patients, independent of cortical plaque severity. Acta Neuropathol Commun 2, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Junkkari A., Luikku A.J., Danner N., Jyrkkänen H.K., Rauramaa T., Korhonen V.E., Koivisto A.M., Nerg O., Kojoukhova M., Huttunen T.J., et al. (2019). The Kuopio idiopathic normal pressure hydrocephalus protocol: initial outcome of 175 patients. Fluids Barriers CNS 16, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seppälä T.T., Nerg O., Koivisto A.M., Rummukainen J., Puli L., Zetterberg H., Pyykkö O.T., Helisalmi S., Alafuzoff I., Hiltunen M., et al. (2012). CSF biomarkers for Alzheimer disease correlate with cortical brain biopsy findings. Neurology 78, 1568–1575. [DOI] [PubMed] [Google Scholar]
- 68.Leinonen V., Koivisto A.M., Savolainen S., Rummukainen J., Sutela A., Vanninen R., Jääskeläinen J.E., Soininen H., and Alafuzoff I. (2012). Post-mortem findings in 10 patients with presumed normal-pressure hydrocephalus and review of the literature. Neuropathol. Appl. Neurobiol. 38, 72–86. [DOI] [PubMed] [Google Scholar]
- 69.Evans A.C., Janke A.L., Collins D.L., and Baillet S. (2012). Brain templates and atlases. Neuroimage 62, 911–922. [DOI] [PubMed] [Google Scholar]
- 70.Eickhoff S.B., Stephan K.E., Mohlberg H., Grefkes C., Fink G.R., Amunts K., and Zilles K. (2005). A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 25, 1325–1335. [DOI] [PubMed] [Google Scholar]
- 71.Amunts K., Mohlberg H., Bludau S., and Zilles K. (2020). Julich-Brain: A 3D probabilistic atlas of the human brain’s cytoarchitecture. Science 369, 988–992. 10.1126/science.abb4588. [DOI] [PubMed] [Google Scholar]
- 72.Kubo Y., Kazui H., Yoshida T., Kito Y., Kimura N., Tokunaga H., Ogino A., Miyake H., Ishikawa M., and Takeda M. (2008). Validation of grading scale for evaluating symptoms of idiopathic normal-pressure hydrocephalus. Dement. Geriatr. Cogn. Disord. 25, 37–45. [DOI] [PubMed] [Google Scholar]
- 73.Fleming S.J., Chaffin M.D., Arduini A., Akkad A.-D., Banks E., Marioni J.C., Philippakis A.A., Ellinor P.T., and Babadi M. (2019). Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. bioRxiv. 10.1101/791699. [DOI] [PubMed] [Google Scholar]
- 74.Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blake J.A., Baldarelli R., Kadin J.A., Richardson J.E., Smith C.L., Bult C.J., and Mouse Genome Database Group (2021). Mouse Genome Database (MGD): Knowledgebase for mouse-human comparative biology. Nucleic Acids Res. 49, D981–D987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baglaenko Y., Brenner M., Loh P., and Raychaudhuri S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D., et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Y., Song W.M., Andhey P.S., Swain A., Levy T., Miller K.R., Poliani P.L., Cominelli M., Grover S., Gilfillan S., et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fonseka C.Y., Rao D.A., Teslovich N.C., Korsunsky I., Hannes S.K., Slowikowski K., Gurish M.F., Donlin L.T., Lederer J.A., Weinblatt M.E., et al. (2018). Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci. Transl. Med. 10. 10.1126/scitranslmed.aaq0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimmerman K.D., Espeland M.A., and Langefeld C.D. (2021). A practical solution to pseudoreplication bias in single-cell studies. Nat. Commun. 12, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Law C.W., Chen Y., Shi W., and Smyth G.K. (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., and Sergushichev A. Fast gene set enrichment analysis. 10.1101/060012. [DOI] [Google Scholar]
- 85.Ding J., Adiconis X., Simmons S.K., Kowalczyk M.S., Hession C.C., Marjanovic N.D., Hughes T.K., Wadsworth M.H., Burks T., Nguyen L.T., et al. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langfelder P., and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. (2019). Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., et al. (2019). Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe K., Mirkov M.U., de Leeuw C.A., van den Heuvel M.P., and Posthuma D. (2019). Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.García-León J.A., García-Díaz B., Eggermont K., Cáceres-Palomo L., Neyrinck K., Madeiro da Costa R., Dávila J.C., Baron-Van Evercooren A., Gutiérrez A., and Verfaillie C.M. (2020). Generation of oligodendrocytes and establishment of an all-human myelinating platform from human pluripotent stem cells. Nat. Protoc. 15, 3716–3744. [DOI] [PubMed] [Google Scholar]
- 91.Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., and Ward M.E. (2018). Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr. Protoc. Cell Biol. 79, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S., et al. (2018). Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep. 23, 2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Busskamp V., Lewis N.E., Guye P., Ng A.H.M., Shipman S.L., Byrne S.M., Sanjana N.E., Murn J., Li Y., Li S., et al. (2014). Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di Giorgio F.P., Boulting G.L., Bobrowicz S., and Eggan K.C. (2008). Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3, 637–648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated snRNA-seq data and the results of our integrative analysis of 28 single cell/nuclei studies are publicly available at: https://braincelldata.org/resource. This includes sample annotations related to the dataset source (36 datasets across 28 studies), cell identifiers (e.g., cell barcodes), quality metrics, and cell type annotations from integrative analysis. We have also included functionality to perform several analyses in a fast and efficient way, including: examination of the integration solutions, performing marker analysis across all of the datasets, and exploring differentially expressed genes.