Abstract
Bone has a robust regenerative potential, but its capacity to repair critical-sized bone defects is limited. In recent years, stem cells have attracted significant interest for their potential in tissue engineering. Applying mesenchymal stem cells (MSCs) for enhancing bone regeneration is a promising therapeutic strategy. However, maintaining optimal cell efficacy or viability of MSCs is limited by several factors. Epigenetic modification can cause changes in gene expression levels without changing its sequence, mainly including nucleic acids methylation, histone modification, and non-coding RNAs. This modification is believed to be one of the determinants of MSCs fate and differentiation. Understanding the epigenetic modification of MSCs can improve the activity and function of stem cells. This review summarizes recent advances in the epigenetic mechanisms of MSCs differentiation into osteoblast lineages. We expound that epigenetic modification of MSCs can be harnessed to treat bone defects and promote bone regeneration, providing potential therapeutic targets for bone-related diseases.
Keywords: Mesenchymal stem cell, epigenetics, osteogenesis, tissue engineering
Introduction
Bone defects are usually caused by trauma, infection, congenital malformation and deterioration of related diseases. In recent years, the study of bone tissue regeneration has contributed to regenerative medicine development, but brought new challenges and directions to scientists and clinicians. Currently, the effective bone regeneration therapy includes autologous, allograft bone transplantation, and synthetic bone substitutes. 1 However, these techniques exhibit limitations, such as donor site defects, poor bone quality, and limited applications of graft materials. 2 Alternatively, these issues are a driving force for developing novel therapies tailored to individual patients or medical needs. Among these, somatic stem cell-based approaches are recognized as the most efficacious for maintaining physiological osteogenesis process and providing effective bone-inducing stimulation. 3 To date, five basic categories of stem cells have been identified: embryonic stem cells (ESCs), very small embryonic-like stem cells, nuclear transfer stem cells, reprogramed stem cells, and adult stem cells. 4
MSCs from adult tissues are superior to ESCs and can differentiate into multiple cell lineages with the appropriate differentiation factors. 5 The most important potential of MSCs is their capacity to differentiate into bone or cartilage cell lineages. 6 During intramembranous ossification simulation, MSCs are transformed into osteoblasts that create a bone-like matrix. On the other side, MSCs can differentiate into chondrocytes in vitro to form hypertrophic cartilage structures. After implantation, chondrocytes differentiated from MSCs can release many osteogenic factors to initiate the endochondral ossification process and achieve endochondral bone formation. 7 MSCs can be isolated from various tissues, most frequently from bone marrow and adipose tissue. Bone marrow mesenchymal stem cells (BMSCs) are usually isolated from bone marrow aspirates and have been widely investigated as a promising source of regenerative bone cells for research and clinical purposes. 8 Xu et al. reported that compared with adipose-derived stem cells (ADSCs), BMSCs possessed a more robust osteogenic differentiation capacity and lower adipogenic differentiation potential. 9 Nonetheless, BMSCs are rare in bone marrow, accounting for less than 0.01% of the total monocytes, 10 and bone marrow aspiration is a complicated invasive procedure. 11 ADSCs are nowadays regarded as another ideal cell source. Zuk et al. isolated and identified pluripotent stem cells from adipose tissue for the first time. 12 And the easy availability, high yield, and low donor morbidity account for their priority nowadays in tissue engineering. 13 However, aged MSCs gradually lose their “stemness” characteristic caused by increased DNA methylation status, accumulation of oxidative stress factors, and mitochondrial dysfunction. 14 MSCs exhibit low differentiation efficiency with prolonged incubation time, especially osteogenic damage. 15 Many researchers have focused on better maintaining the characteristics of MSCs and osteogenic differentiation. Biomaterials, chemical substances and physical stimuli (e.g. mechanical and photobiomodulation) have been identified as effective biological structures or methods to enhance the biological process.16–19 An increasing body of evidence from recent studies suggests that epigenetics is a new regulator of stem cell differentiation and has created a wider field of research on bone regeneration.
Ever since Waddington brought up the word “epigenetics” in 1942, 20 there has been a surge of interest in its scientific investigation in the last few decades, highlighting its role in regulating gene expression. Epigenetic modifications involve changes in the gene activity but not the sequence,21,22 which is susceptible to influence from various factors such as environmental conditions, nutritional intake, chemical exposure, and stress.23–26 It has been established that the core components of epigenetics include DNA methylation, histone modification, nucleosome positioning, non-coding RNAs, chromatin structural remodeling, and RNA N6-methyladenosine (m6A) modification.
In this review, we provide a summary of three mainstream epigenetic modifications, including nucleic acid methylation, histone modification and non-coding RNAs. We focus on the specific mechanisms of each epigenetic modification in MSCs osteogenesis and further explore the role of MSCs in bone tissue engineering through the interaction of different epigenetic modifications and their association with other factors.
Nucleic acids methylation
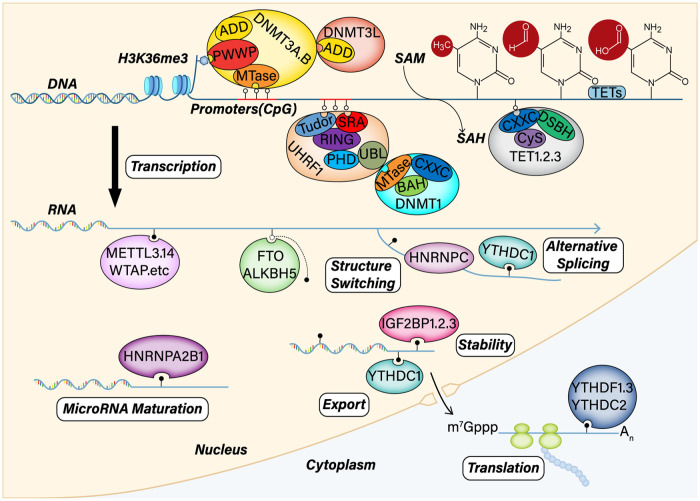
Until now, at least 17 and 160 types of chemical modifications have been identified in DNA and RNA. 27 The biological processes of nucleic acid methylation are mainly accomplished by related methyltransferases and demethylases. DNA methylation was first described in 1965. 28 This stable modification occurs on the cytosine nucleotide following the addition of methyl groups to form 5mC in mammals, primarily at CG and CH (CH=CA, CT, CC) sites. However, the 5mC alteration usually occurs at the CpG islands dense at the promoter, intergenic units, and transposable elements. 29 Compared with somatic cells, MSC genes are differentially methylated at 5mC in their promoter CpG sites. 30 The DNA methyltransferase family comprises a conserved set of DNA-modifying enzymes that play a central role in epigenetic gene regulation, including DNMT1, DNMT3A, DNMT3B, and DNMT3L. Generally, passive DNA demethylation occurs after consecutive cell divisions, while members of the Ten-Eleven Translocation family, including TET1, TET2, and TET3, mediate active demethylation.31–34 These enzymes can be served as regulators of 5mC or 5hmC to control gene expression (Figure 1). A study revealed that the epigenome-wide signature of human MSCs derived from osteoporotic fracture patients contained differentially DNA-methylated regions; these regions were associated with several genes involved in proliferation and differentiation. 35 Osteosarcoma is one of the most common primary bone tumors derived from mesenchymal cells and produces immature bone and osteoid. The pathogenesis of osteosarcoma has also been associated with the DNA methylation of multiple genes. 36
Figure 1.
DNA and RNA methylation machinery and mechanisms. DNA methylation modification includes three phases: de novo DNA methylation, maintenance methylation, and demethylation. DNMT3A and DNMT3B are responsible for de novo DNA methylation, which contain a highly conserved DNMT domain (the MTase domain) in the carboxy terminus and two chromatin reading domains, ADD and PWWP. DNMT3L also comprises ADD and interacts with and stimulates the activity of DNMT3A and DNMT3B. DNMT1 is critical for maintaining DNA methylation after replication through cell division. UHRF1 is also responsible for maintaining methylation. UHRF1 specifically binds CpG dinucleotides at replication forks through its SRA domain and H3K9me2 and H3K9me3 through its Tudor-PHD domain. UHRF1 recruits DNMT1 through its UBL domain, thereby relieving its auto-inhibition. The TET family carries out DNA demethylation via the oxidation of methylcytosine. The TET family includes TET1/2/3, which contain catalytic domains (Cysh-rich and DSBH). However, TET1 and TET3 also have CXXC regions. As a post transcriptional modification, m6A can influence RNA processing and metabolism via various methods, including alternative splicing, structure switching, stability, maturation, translocation, and translation.
Willyard commented that RNA modification was a new gene regulation mechanism in this golden age of epigenetics. 37 The first modified RNA nucleoside was discovered 60 years ago by analyzing salt-soluble RNAs from yeast. Since then, more than 100 different types of chemical biomarkers have been identified in RNA. 38 M6A, one of the most prevalent modifications, can be installed by m6A methyltransferases (“writers”), removed by m6A demethylase (“erasers”), and recognized by m6A binding protein (“readers”). This dynamical and reversible biological process occurs in sites of motif DRACH (D = G/A/U, R = G/A, H = A/U/C). This RNA marker can be written or erased in response to various stressors and biological variables. To date, m6A “writers” include METTL3, METTL14, METTL16, WTAP, KIAA299 and RMB15. m6A “erasers” include FTO, ALKBH5. And m6A “readers” include YTH domain family proteins, hnRNP family proteins, and IGF2BP domain family proteins (Figure 1).39–41
DNA methylation changes during osteogenesis
Recent research suggests that the osteogenic differentiation of MSCs is regulated by DNA methylation. MSCs derived from DNA methyltransferase knockout mice exhibit decreased colony forming capacity and osteogenic potential. At the same time, the identified differentially regulated genes have been associated with skeletal development, with few acting directly in MSCs to regulate osteogenic differentiation, namely, HIEVP3, GATA2, DLX3, etc. As cells proliferate, such epigenetic defects of genes may expand and eventually lead to impaired differentiation. 42 Moreover, the CpG sites are mainly located in the promotor and exon regions of these differentially expressed genes. The methylation of CpG is associated with most encoding genes in humans. A previous study showed that average genomic methylation levels and CpG methylation in transcriptional factor regions were increased along with induced osteogenic differentiation in BMSCs. It has also been found that the CpG methylation levels in the repeat elements, including DNA transposons, LINEs, SINEs, and LTRs, had highly methylated levels during osteogenic differentiation. Meanwhile, the promoters of several critical osteogenic genes, such as BMP1, PAX1, and RUNX3, showed significant methylation level changes. 43 This modification type also is also observed during the osteogenic differentiation of ADSCs. 5mC losses and 5hmC gains are observed in ADSCs that differentiated into osteogenic lineages. Compared with 5mC, 5hmC modification is more abundant, and accompanied by an increased expression of TET1. 44
Several regulators are essential for MSCs osteogenic differentiation. One of the most important osteogenic active factors, RUNX2, is activated to start transcription by two distinct promoters at different sites. The proximal promoter produces the type I isoform, whereas the distal one produces the type II isoform. The transcription start site of RUNX2-I, located on a CpG island, contains an abundance of cytosine bases that can be methylated to regulate osteogenesis. Similarly, Uehara et al. discovered a differentially methylated CpG site upstream of the RUNX2-I locus, where hypermethylation was associated with RUNX2 expression downregulation.45,46 Another osteogenic regulator, OCN, was upregulated, whereas its promoter region methylation level was attenuated over time during osteogenic differentiation. Meanwhile, PPARγ2 and SOX9, two negative regulators of osteoblastic differentiation, exhibited opposite expression trends. 47 In addition to the methylation of the osteogenic factor themselves, they were regulated by other methylated genes. For example, ATF4 and DLX3 were important regulators of bone development. The DNA methylation rate in the promoter region of ATF4 and DLX3 was decreased, leading to enhanced DLX3 and ATF4 transcriptions during osteoblastic differentiation of ADSCs. DLX3 controlled the co-expression of OCN and RUNX2 with a specific activity on their promoter regions. Moreover, the suppression of ATF4 could reduce bone formation capacity by inhibiting OCN expression. 48 The GADD45A belongs to the GADD45 family is a small (18 kDa) p53-regulated histone-fold protein. It is highly conserved and a key mediator of cellular stress responses to regulate cell proliferation, DNA repair, cell cycle, and apoptosis. 49 A later study found that GADD45A was involved in the DNA demethylation of osteogenic gene promoters such as DLX5, RUNX2, OCN, and OSX during the differentiation of ADSCs. Knockdown of GADD45A inhibited the demethylation of these gene promoters and suppressed osteogenesis. However, this study did not clarify the specific mechanism of demethylation affected by GADD45A, which needs to be further explored. 50 More details of the effect of DNA methylation modifiers on osteogenic differentiation and their cellular phenotypes based on the latest research are summarized in Table 1.
Table 1.
Effect of DNA methylation modifiers on osteogenesis and cellular phenotype.
| DNA modifier | Epigenetic modification | MSC lineage | Target genes | Effect on osteogenesis | Ref. |
|---|---|---|---|---|---|
| DNMT1 DNMT3A |
Methylation | ADSC | β-Catenin GSK-3β LEF1 |
Overexpression of DNMT1 and DNMT3A decreased the β-Catenin and LEF1 levels, but with an increase in GSK-3β, thus leading to inhibit osteogenesis. | Zhang et al. 51 |
| DNMT3A | Methylation | ADSC | β-Catenin LEF1 |
The highly expressed DNMT3A decreased the β-Catenin and LEF1 levels, resulting in the inhibition of osteogenic differentiation. | Zhang et al. 52 |
| DNMT3A | Methylation | BMSC | ALP RUNX2 |
Downregulation of DNMT3A induced the hypomethylation of ALP and RUNX2, resulting in the promotion of osteogenesis. | Li et al. 53 |
| DNMT3B | Methylation | BMSC | KLF5 | Hypermethylation of KLF5 mediated by DNMT3B resulted in impairing osteogenesis. | Li et al. 54 |
| DNMT1 | Methylation | ADSC | NOTCH1 NOTCH2 |
DNMT1 suppressed osteogenic differentiation by inducing the hypermethylation of NOTCH1 and NOTCH2. | Jia et al. 55 |
| TET1 | Demethylation | ADSC | POU5F1 | TET1 caused 5hmC changes on POU5F1 and then promoted osteogenesis. | Daniunaite et al. 44 |
| TET2 | Demethylation | BMSC | RUNX2 BMP2 |
TET2 increased 5hmC on the BMP2 and RUNX2 transcription start sites, resulting in the promotion of osteogenesis. | Cakouros et al. 56 |
DNA methylation and osteogenic differentiation can be affected by other factors, such as diseases and the environment. The reduced osteogenic differentiation of BMSCs has been found in pediatric aplastic anemia patients. A study explored this mechanism underlying the high expression of DNMT1 and low levels of lncRNA MEG3. MEG3 could increase the transcriptional activity of BMP4 and promote osteogenesis. Nevertheless, overexpression of DNMT1 could increase MEG3 promoter methylation and enhance binding between them to reduce MEG3 expression levels. 57 It has been found that type 2 diabetes mellitus (T2DM) can change the level of nucleic acid methylation. 58 For example, a study based on diabetic osteoporosis (DOP) showed that ADSCs from DOP mice had a lower osteogenesis potential than normal ADSCs, mainly due to the increased methylation of JKAMP, leading to its decreased expression levels. JKAMP, a seven-transmembrane protein, located primarily on the plasma membrane of cells, provided cells with apoptosis-related signals. The low expression of JKAMP inhibited the activation of the Wnt signaling pathway through JNK1, ultimately impairing the osteoblast ability of cells. 59 Besides, in the same disease model, lncRNA AK137033 expression and its DNA methylation level in the promoter region of sFrp2 were downregulated. Furthermore, lncRNA AK137033 silencing could inhibit Wnt signaling and reduce the osteogenic ability in ADSCs. Therefore, the expression of lncRNA AK137033 could be enhanced by increasing DNA methylation in sFrp2 to promote osteogenesis in vivo or in vitro in DOP-ADSCs. 60 By establishing a T2DM mice model and using IGV software, Wang et al. found that the methylation level of the CALCA promoter was increased, and its expression level decreased in ADSCs-T2DM. CALCA, an osteogenesis-related gene, could encode two peptides: calcitonin and α-CGRP. Moreover, CGRP could promote the formation of calcium nodules in the osteogenic differentiation of ADSCs-T2DM and upregulated osteogenic genes (e.g. ALP, RUNX2, COL1A1, OCN, and BMP) in a concentration-dependent manner. However, 5-AzaC could down regulate DNA methylation in the promoter region and decrease the expression of DNMT1, thus improving Calca expression and osteogenesis in T2DM. 61
A study investigating DNA methylation sites in the osteogenic gene promoter regions of MSCs first found that mechanical stimulation could effectively alter the cell epigenetic state and increase the expression of osteogenic abilities by reducing DNA methylation. 62 A follow-up study suggested that cyclic mechanical stretch could promote osteogenic differentiation earlier in human MSCs. Daily mechanical stretch could induce down regulation in the DNA methylation status of critical CpG sites of NESP and GNASXL isoforms. All CpG islands with notable methylation changes contained CTCF binding sites for transcriptional repressors. As a crucial component of epigenetic regulation, CTCF could inhibit the spread of CpG methylation by binding to a transcriptional activator, thus blocking communication between enhancers and upstream promoters. 63 At the same time, this stimulation activated SHH, a core gene in the Hedgehog signaling pathway, down regulating DNMT3B protein expression, which could reduce the hypermethylation of gene promoter caused by the binding of DNMT3B with SHH, thus stimulating SHH expression and promoting osteogenesis. 64
With an improved understanding of methylation, epigenetic modifiers have gradually attracted much attention and have been applied to osteogenic differentiation. These compounds, also known as epidrugs, target epigenetic marks which are responsible for epigenetic alterations or modifications. For example, methyltransferase inhibitors can potentially regulate the osteogenic differentiation of MSCs directly and effectively. The nucleoside analog 5-AzaC is the most widely used DNA methylation inhibitor. It has been established that pretreatment of MSCs with a suitable concentration of 5-AzaC reduces CpG island methylation of osteogenic genes promoters, including DLX5, RUNX2, COL1A1, OSX, and OCN, and increases their expressions.65,66 With aging, the proliferation of MSCs decreases, the osteogenic capacity is impaired, and the MSCs are more inclined to present adipogenic differentiation. 67 Research conducted by Yan et al. revealed that 5-AzaC significantly increased TET2 and TET3 gene expression and 5hmc levels, thus ameliorating the osteogenic ability in aged ADSCs. 68 The accumulation of oxidative stress in BMSCs results in impaired osteogenic differentiation caused by an imbalance between the generation and scavenging of reactive oxygen species. With the stimulation of oxidative stress, the elevated level of DNMT3B results in hypermethylation of the key osteogenic gene KLF5, inhibiting its expression. Given that KLF5 can promote the nuclear translocation of β-catenin, its hypermethylation impairs osteogenic differentiation by reducing the interaction with β-catenin. This process can be reversed by the methylation inhibitors 5-AzaC and nanomycin A, thus protecting BMSCs under oxidative stress. 54 However, another study suggested that moderate stimulation of oxidative stress promoted osteogenesis. Oxidative stress induced by bone repair material implantation could decrease DNMT3A expression, leading to hypomethylation of ALP and RUNX2 and increasing their expression. Meanwhile, the above process could be further enhanced by using 5-AzaC. 53 These studies suggested that combining MSCs with epigenetic drugs is a practical approach to bone regeneration and repair.
RNA m6A methylation changes during osteogenesis
As a post-transcriptional regulation, the m6A has been discovered on several transcripts in BMSCs, specifically those encoding regulators responsible for BMSCs differentiation into a specific lineage. A study revealed that the total m6A level significantly decreased during the osteogenic differentiation of ADSCs. FTO expression was significantly increased in the differentiation group on day 7. However, the expression of METTL3, METTL14, and ALKBH5 showed no significant differences. Further, 1145 differentially methylated peaks, 2261 differentially expressed genes, and 671 differentially methylated and expressed genes were identified. 69 Intriguingly, another study demonstrated that FTO was downregulated during osteogenic differentiation and upregulated during adipogenic differentiation in BMSCs. FTO could bind to the fat-related gene PPARγ and demethylate it, resulting in an increased level of PPARγ and promoting the shift of BMSCs fate to adipocytes. 70 However, research on MSCs osteogenesis revealed that FTO was markedly upregulated. Besides uncovering FTO’s potential to facilitate m6A demethylation in the 3′-UTR of PPARγ, this study also found that FTO lowered PPARγ mRNA stability in a YTHDF1 dependent manner. 71 Meanwhile, YTHDF1 overexpression improved the osteogenic capacity of mouse BMSCs, and its depletion decreased Zfp839 (a zinc finger protein) levels and bone mass. 72 The expression trend of FTO appears inconsistent among various types of MSCs. Whether this phenomenon is related to cell type and its specific mechanism remains to be investigated.
Huang and Wang believed that unlike ADSCs, m6A levels increased during the osteogenic differentiation of BMSCs, while METTL14 and METTL3 were significantly up-regulated, especially METTL14. METTL14 could promote the m6A methylation of SMAD1 through IGF2BP1, thereby regulating differentiation. 73 Yan et al. first unraveled that METTL3 could directly induce m6A methylation of RUNX2 mRNA to enhance its cellular stability and indirectly upregulate RUNX2 expression via m6A methylation of pre-miR-320. Moreover, they demonstrated that overexpression of METTL3 could increase m6A levels in total RNAs and significantly promote the osteogenic differentiation of BMSCs cells and bone formation. 74 Another experiment also substantiated that METTL3 knockdown could reduce the expression of RUNX2, OSX, and OCN and significantly decrease the phosphorylation of Akt, thus inhibiting the PI3K-Akt signaling pathway and osteogenic process. 75 This research further showed that the downregulated METTL3 inhibited the expression of VEGFA, especially its splicing variants VEGFA-164 and VEGFA-188, which could enhance bone regeneration due to its capacity to promote both osteogenic and angiogenic differentiation. 76 The other mechanisms of m6A modifiers on osteogenic differentiation and their cellular phenotypes are summarized in Table 2.
Table 2.
Effect of m6A modifiers on osteogenesis and cellular phenotype.
| m6A modifier | Epigenetic modification | MSC type | Target genes | Effect on osteogenesis | Ref. |
|---|---|---|---|---|---|
| METTL3 | Methylation | BMSC | β-Catenin GSK-3β LEF1 |
METTL3 promoted osteogenic differentiation by increasing β-Catenin, GSK-3β, and LEF1 expression. | Wu et al. 77 |
| METTL3 | Methylation | BMSC | RUNX2 | METTL3 increased the m6A level of RUNX2, which stabilized the its mRNA level and promoted osteogenesis. | Jiao et al. 78 |
| METTL3 | Methylation | BMSC | Lnc MIR99AHG | Lnc MIR99AHG methylation mediated by METLL3 could enhance the osteogenic differentiation via targeting miR-4660. | Li et al. 79 |
| METTL3 IGF2BP1 |
Methylation | BMSC | RUNX2 | METTL3 methylated RUNX2 at its 3′-UTR, and IGF2BP1 recognized the m6A site on RUNX2 to enhance its stability, thus promoting osteogenesis. | Zhou et al. 80 |
| METTL14 IGF2BP1 IGF2BP2 IGF2BP3 |
Methylation | BMSC | Beclin-1 | METTL14 enhanced osteogenic differentiation through regulating the expression of Beclin-1, and IGF2BP1/2/3 recognized the m6A-methylated Beclin-1 mRNA and promoted its translation. | He et al. 81 |
| METTL14 | Methylation | BMSC | PTPN6 | METTL14 increased PTPN6 expression by increasing PTPN6 mRNA stability in an m6A-dependent manner, leading to promoting osteogenesis. | Cheng et al. 82 |
| METTL14 | Methylation | BMSC | SIRT1 | METTL14 promoted osteogenic differentiation by m6A-dependent upregulation of SIRT1. | Wang et al. 83 |
| FTO | Demethylation | BMSC | RUNX2 | FTO inhibited osteogenic differentiation through demethylating RUNX2. | Wang et al. 84 |
| ALKBH5 | Demethylation | BMSC | MYD88 | ALKBH5 decreased the expression of MYD88 and promoted osteogenesis. | Yu et al. 85 |
As mentioned above, m6A modification directly modifies osteogenic factors and indirectly affects signaling pathways to regulate osteogenic differentiation. These signaling pathways can mediate the activation of specific genes that regulate MSCs osteogenic differentiation. PTH is essential for maintaining bone mass in the human body and promoting MSCs osteogenic differentiation. PTH can stimulate osteoclast formation and activity by increasing the receptor activator of NF-κB ligand/osteoprotegerin ratio through its receptor. PTH1R is a direct signaling mediator of PTH whose aberrant expression can lead to adipogenic differentiation. 86 Although the mRNA expression of PTH1R is hardly affected by METTL3 deletion, the translation efficiency of PTH1R is inhibited and then disrupts the PTH-induced osteogenic responses. 87 However, another study suggested that activation of NF-κB signaling could inhibit osteogenic differentiation. METTL3 knockdown blocked proteasome-mediated IκBα degradation and restrained the phosphorylation level of p65 at the S536 site, a crucial modification responsible for NF-κB activity. METTL3 simultaneously upregulated expression of MYD88, a regulator of NF-κB, through controlling its m6A methylation status, therefore leading to the activation of NF-κB signaling and suppressing the osteogenic differentiation of MSCs. And the above process could be reversed by ALKBH5. 85 A study indicated that demethylase ALKBH5 deletion in MSCs could increase osteogenesis. After knocking down ALKBH5 expression during the osteogenic differentiation of MSCs, 790 genes were differentially expressed; genes altered by ALKBH5 were clustered in ECM–receptor interactions, and the PI3K/Akt signaling pathway. Meanwhile, ALKBH5 could directly target its downstream gene PRMT6 and co-regulate the PI3K/Akt signaling pathway. 88
Histone modification
In eukaryotes, the fundamental component of chromatin is a nucleosomal core particle consisting of 145–147 bp of DNA looped around an octamer of the four core histones (H2A, H2B, H3, and H4). 89 Histones are globular proteins that contain two copies of H3 and H4, coupled to two H2A-H2B dimers. Each nucleosome is linked by histone H1 to form higher-order chromatin fiber. The H1 histone is a linker between inter-nucleosomal DNA and is essential for preserving chromatin’s structural and functional flexibility. 90 Histone modifications are necessary for most biological processes, especially for adjusting the activation and silencing of genes. 91 This modification influences molecular biology activities such as replication, splicing, repair, and chromatin condensation, eventually leading to individual aging, neurodevelopment, cancer, and chronic diseases. Histone modification, more specifically known as histone post-translation modification (PTM), can create a binding site for chromatin-associated proteins and directly modulate chromatin fiber dynamics. Histone PTM correlates with active, repressive, and poised gene expression states. PTMs, including phosphorylation, acetylation, glycosylation, methylation, ubiquitination, and citrullination, are mainly found on the N-terminal tail domains of histones. Nonetheless, recent research suggests that PTMs also occur in histones’ cores. The altered histone tails may contact with the adjacent nucleosome, disrupting interactions between nucleosomes and causing changes in chromatin structure. Among these, histone acetylation and methylation are the most common PTMs.92–94
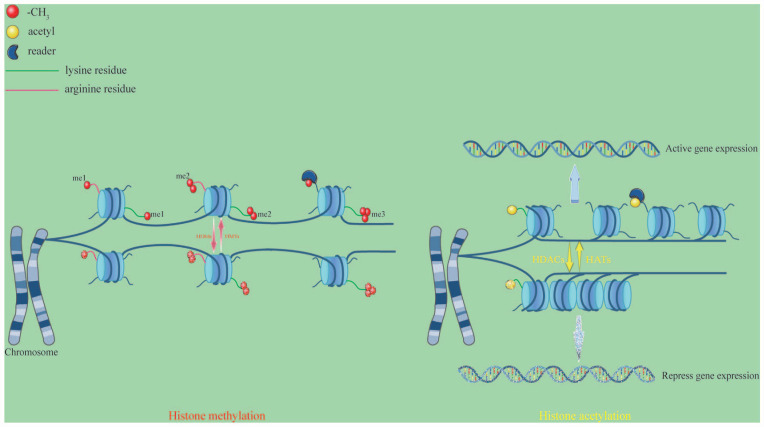
Histone methylation is a complicated epigenetic mark since it may signal for gene activation or repression depending on its degree of deposition. This progress has been confirmed to be dynamically regulated by histone methyltransferases (HMTs) or histone demethylases (HDMs or KDMs). HMTs transfer the methyl group from S-adenosine-L-methionine to the amino group of basic amino acids. Alternatively, HDMs can facilitate eliminating the methyl group from the proprer histone site. Two demethylase groups have been identified: jumonji C (JmjC) Fe-dependent dioxygenases and amine oxidases, such as lysine-specific demethylases (LSDs). 95 This modification only affects the nitrogen atoms of basic amino acids such as lysine and arginine. Histone H3 is the primary site of histone methylation, although the other core histones also display methylations. 96 Methylations of H3K4, H3K36, and H3K79 have been associated with gene activation, while H3K9, H3K27, and H4K20 have been associated with gene silencing. 97 HMTs and HDMs are modifiers that influence gene expression positively or negatively during differentiation. Except for these two types of enzymes, the “readers” are a group of proteins belonging to the royal superfamily, and they contain a chromodomain that regulates the binding of methylation marks. Given that none of the three methylation states changes the electronic charge of the amino-acid side chain, histone lysine methylation functions are mainly exerted by “readers” that specifically recognize the methylated site 98 (Figure 2).
Figure 2.
Histone methylation and acetylation. These two modification types constitute a hypothetical “histone code” for chromatin organization and gene regulation. HMTs and HDMs mainly activate the modification processes of histone methylation and demethylation. Histone methylation usually occurs on lysine or arginine residues at the N-terminus of H3 and H4 histones. Lysine residues can be modified by mono-methylated (me1), di-methylated (me2), and tri-methylated (me3), while arginine residues are modified by me1 and me2. HATs catalyze acetyl transfer to lysine residues in the histone tail, which neutralizes the positive charge of lysine, relaxes chromatin, and activates gene expression. Conversely, HDACs reverse the above process and pack chromatins, repressing gene expression. In some biological contexts, “readers”-mediated chromatin interaction aids in the recruitment and/or stabilization of the associated multi-protein complexes to specific loci to further modify chromatin structure and control DNA-dependent activities. Moreover, these readers play a crucial role in a variety of PTMs.
Histone acetylation occurs mainly on lysine residues on histones H3 and H4 by decreasing the positive charge of lysine residues upon adding anionic acetylation groups. It is associated with transcription activation, increasing chromatin accessibility and weakening the interactions between the DNA and histones, resulting in a higher-order configuration of open architectural chromatin. Lysine residues are acetylated by histone acetyltransferases (HATs), which loosen the chromatin structure and increase transcriptional activity. HATs can be classified into A-type HATs and B-type HATs. A-Type HATs mediate the transcriptional control mechanisms through the recognition and binding of acetyl-CoA. The B-Type HATs are located in the cytoplasm and acetylate newly formed histones, allowing them to move from the cytoplasm to the nucleus.99,100 Conversely, histone deacetylases (HDACs) remove histone acetylation and enhance histone-DNA binding to repress gene expression (Figure 2).
There is a close correlation between chromatin state dynamics and transcriptional regulation of the osteoblast differentiation from MSCs. It has been confirmed that genomic binding of transcription factors and histone modifications occur during osteogenic differentiation. Wu et al. outlined the epigenetic modification characteristic that occur during the differentiation of MSCs into specific cell lineages. They found that the absence of H3K4me3 modification at promoters defined a subset of osteoblast-specific upregulated genes. Meanwhile, the H3K27ac signal increased across the entire gene region and peaked on day 21 during differentiation, resulting in the activation of these genes. Conversely, downregulated genes displayed a decrease in the H3K27ac signal near the transcription start site during differentiation. H3K27me3 was removed from upregulated genes, but there was not a substantial gain of this signal to repress downregulated genes. 101 Current evidence indicates that histone modification regulates the expression of osteo-specific genes, and more detailed information is provided in Table 3. This review will mainly focus on these two modification types during the osteogenesis of MSCs.
Table 3.
Histone modification and mechanism during the osteogenic differentiation of MSCs.
| Histone modifier | Modification | MSC type | Target genes | Mechanism | Ref. |
|---|---|---|---|---|---|
| EHMT1 | Methylation | BMSC | RUNX2 | EHMT1 upregulated the H3K9me2 levels at the promoter of RUNX2, resulting in the repression of osteogenesis. | Huang et al. 102 |
| ASH1L | Methylation | BMSC | OSX RUNX2 |
ASH1L-mediated H3K4me3 occurred at the promoter regions of OSX and RUNX2, ultimately promoting osteogenic differentiation. | Yin et al. 103 |
| PRMT3 | Methylation | MSC | miR-3648 | PRMT3 activated the expression of miR-3648 by enhancing H4R3me2a levels at its promoter region, resulting in the promotion of osteogenic differentiation. | Min et al. 104 |
| CARM1 | Methylation | BMSC | OCT4 SOX2 ANOG |
CARM1 was enriched on OCT4, SOX2, and NANOG promoter sites by enhancing di-methylation of H3R17 of these genes, resulting in the promotion of osteogenesis. | Jo et al. 105 |
| LSD1 | Demethylation | BMSC | RUNX2 OCN |
LSD1 catalyzed the demethylation of di- and mono-methylation of H3K4 at RUNX2 and OCN promoter regions, resulting in the repression of osteogenic differentiation. | Lv et al. 106 |
| LSD1 | Demethylation | ADSC | BMP2 SMAD4 |
Silencing of LSD1 promoted the osteoblastic potential by stimulating BMP2/SMAD4 signaling pathway. | Ma et al. 107 |
| KDM1A | Demethylation | BMSC | circ_AFF4 FNDC5 |
KDM1A promoted circ_AFF4 and FNDC5 expression via reducing H3K9me3, thus inducing circ_AFF4 and FNDC5 expression and osteogenesis. | Liu et al. 108 |
| GCN5 | Acetylation | ADSC | NF-κB | GCN5 repressed NF-κB-dependent transcription and inhibited the NF-κB signaling pathway, thus promoting osteogenic differentiation | Zhang et al. 109 |
| HBO1 | Acetylation | ADSC | 14-3-3β | HBO1 suppressed the expression of 14-3-3β via increasing its acK49/51 acetylation, resulting in the promotion of osteogenic differentiation. | Frontini-López et al. 110 |
| SIRT6 | Deacetylation | MSC | BMP2 BMP4 BMPR1A BMPR1B |
SIRT6 promoted osteogenic differentiation through BMP signaling pathway. | Zhang et al. 111 |
| HDAC9 | Deacetylation | BMSC | pERK1/2 | Silencing of HDAC9 reduced the expression level of pERK1/2 and inhibited osteogenic differentiation. | Wang et al. 112 |
Histone methylation changes during osteogenesis
The genome-wide histone PTM landscape requires constantly changing to direct gene expression in pluripotent stem cells, giving rise to distinct pluripotent states. 113 Enzymes for methylation of histone H3 at lysine 4, 9, and 27 are essential for embryogenesis. H3K4me3 is one of the most recognized epigenetic marks of active transcription, and H3K27me3 may play a vital role in the early developmental stage of human ESCs cellular differentiation at CG-rich areas and decrease with aging.114–116 Notably, the combination of H3K4me3 and H3K27me3, named the bivalent modification, maintain the “stemness” of stem cells. 117
MSCs in the bone marrow lose their osteogenic capacity during aging and are more likely to develop into adipocytes, leading to osteoporosis. SET domain (SETD) proteins have been shown to methylate lysine residues in histones and other proteins. These enzymes can “read” the tails of histones and “write” a methyl group on specific lysine residues. 118 SETD2 is a methyltransferase responsible for H3K36me3. A study confirmed that SETD2 and H3K36me3 levels were decreased during adipocyte differentiation. On the contrary, SETD2 and H3K36me3 levels were increased during osteogenesis differentiation, while stable expression of H3K36me1 and H3K36me2 was observed. Furthermore, lipopolysaccharide-binding protein (LBP), a negative regulator of adipocyte differentiation, was identified as a downstream target of SETD2. Moreover, the loss of H3K36me3 occupancy might affect RNA polymerase II (Pol II) binding affinity around transcription start sites and gene body, impairing both mRNA transcriptional initiation and elongation of LBP. 119 Another methyltransferase, SETDB1, also known as ERG-associated protein with a SET domain, is key in MSCs osteogenic differentiation. It has been reported that SETDB1 could bind to RUNX2 and inhibit its activity. Intriguingly, the absence of SETDB1 also inhibited osteogenic differentiation. Since the deletion of SETD eliminated the inhibitory effect but also led to an oversupply of RUNX2 and an abnormal gene expression program that disrupted bone development. 120 Yin et al. revealed that SETD7 was involved in the osteogenesis process of BMSCs stimulated by boron mesoporous bioactive glass scaffolds. Boron, a trace element, was discovered in the human body playing an essential role in bone regeneration. SETD7 and H3K4me3 were highly expressed when BMSCs were cultured in liquids of boron scaffolds at a concentration of 100 μg/ml. At the same time, the Wnt/β-catenin signaling pathway was highly activated with boron scaffold stimulation. 121 Strikingly, this study did not clarify the specific molecular mechanism of interaction between boron and SETD7, emphasizing the need for more research.
Zeste homolog 2 (EZH2), one of the polycomb group (PcG) genes family, is a histone methyltransferase that acts as a critical epigenetic regulator that suppresses transcription. 122 EZH2 is responsible for catalyzing H3K27me3; it is decreased during osteogenic differentiation and increased during adipogenic differentiation in BMSCs. EZH2 is highly enriched in Wnt gene promoters (e.g. Wnt1, Wnt6, and Wnt10a) and increases H3K27me3 levels on their promoters by directly binding. Overexpression of EZH2 inhibits the Wnt signaling pathway and thus attenuates osteogenesis. However, this process can be reversed by H3K27me3 inhibitor DZNep, a potent inhibitor of S-adenosylhomocysteine hydrolase, thus improving osteoporosis and strengthening osteogenesis. 123 Compared with young MSCs, the expression level of EZH2 in aged MSCs was higher and could bind to the HDAC9c promoter to subsequently suppress its gene expression. This biological process activates PPARγ and FABP4 expression and attenuates RUNX2 levels, impairing osteogenesis. GSK343, an EZH2 inhibitor, can reverse this process, thus enhancing osteogenesis differentiation and preventing age-related osteoporosis. 124 Another EZH2 inhibitor, GSK126, applied with recombinant BMP2 during osteogenic differentiation, which could prevent the formation of new H3K27me3 marks and reduce the total H3K27me3 level in a concentration-dependent manner in human BMSCs. 125 Cytochalasin D has been identified as a potent osteogenic stimulant by redistributing the intracellular location of β-actin. It alters chromatin organization by suppressing the expression of EZH2 and decreasing H3K27me3, resulting in increased mineralization, bone extracellular matrix, and osteoblast-related gene expression (e.g. RUNX2). 126 Polycomb repressive complex 2 (PRC2) is one of the two PcG protein core complexes that mediate gene silencing primarily by chromatin structural modulation. EZH2 is an enzymatic catalytic subunit of PRC2 that can repress gene expression. A previous study revealed that EZH2 could be directly phosphorylated at Thr 487 by cyclin-dependent kinase 1. Interestingly, this biological process could disrupt EZH2 binding with other PRC2 components SUZ12 and EED, inhibiting EZH2 activity and decreasing H3K27me3 levels, thus promoting MSCs differentiation into osteoblasts. 127 However, this repressive mark can be removed by lysine demethylase 6A (KDM6A). A study performed by Hemming et al. showed that the knockdown of KDM6A expression in MSCs increased adipogenesis and decreased osteogenesis. 128 Then, they identified targets of EZH2 during the osteogenic differentiation of MSCs, including ZBTB16, MX1, and FHL1. These genes were directly targeted by EZH2 and activated in undifferentiated MSCs leading to suppression of gene expression. However, these genes were induced during osteogenic differentiation due to the loss of EZH2 and H3K27me3 but related to H3K4me3 modification. 129
A study conducted by Ge et al. first revealed that retinoblastoma binding protein 2 (RBP2), a histone demethylase, could inhibit osteogenic differentiation of human ADSCs in vitro and in vivo. RBP2 was recruited to the promoters of OSX and OCN to maintain the level of H3K4me3 during osteogenic induction. Conversely, RBP2 could functionally repress RUNX2 transcriptional activity through its H3K4 demethylase ability to inhibit osteogenic differentiation. 130 In a subsequent study, TiO2 nanotubes with a diameter of 70 nm were found to efficiently inhibit RBP2 expression and upregulate the level of H3K4me3 in the promoter regions of RUNX2 and OCN, leading to enhanced osteogenesis in ADSCs. 131 Moreover, the single-layer graphene promoted osteogenic differentiation by a similar mechanism. 132 Although these two studies showed that biomaterials could lead to better protein adsorption, cell adhesion, and proliferation, the specific interaction mechanism between biomaterials and methylation was not deeply elaborated. LSD1, another histone demethylase, is a key inhibitor in the osteogenic differentiation of human ADSCs. Similar to RBP2, LSD1 can also occupy OSX and OCN promoters to maintain the levels of H3K4me2/1. Moreover, Pargyline and CBB1007, inhibitors of LSD1, increase the levels of H3K4me2 at the promoter regions of osteogenesis-related genes to rescue the osteogenic differentiation ability of mouse BMSCs under osteoporotic conditions.106,133 LSD1, also known as KDM1A, belongs to the KDM1 family. Unlike the LSD family, all other lysine demethylases family members contain the JmjC domain. Among these, KDM4 family and KDM6 family have respectively specific catalytic activities on H3K9me3/me2, H3K36me3/me2 and H3K27me3/me2. Following the stimulation of BMP-4/7, the expression of KDM4B and KDM6B was increased and accompanied by the mineralization of MSCs, attributed to KDM6B and KDM4B activating HOX and DLX transcription by removing H3K27me3 and H3K9me3, respectively. Furthermore, HOX and DLX could promote RUNX2 and OSX transcription, thus promoting MSCs osteogenesis. 134 PHF8 has been identified as another histone demethylase linked to H3K4me3 nucleosomes via its PHD domain. It can demethylate H3K9, H3K27, and H4K20 at active promoter transcription start sites and then controls gene transcription. It has been shown that overexpression of PHF8 downregulates the binding of H3K9me1 at the transcription start sites region of SATB2. SATB2, a DNA-binding protein, enhances expressions of bone matrix proteins and osteogenic transcription factors in BMSCs. As a result, PHF8 epigenetically modulates SATB2 activity, triggering BMSC osteogenic differentiation. 135
Histone acetylation changes during osteogenesis
The regulation of histone acetylation by HAT and HDACs exhibits a balanced state under normal physiological conditions. However, during cell differentiation, the activity of these modification enzymes significantly changes, disrupting the original balance of gene expression and resulting in an imbalance of molecules that affect cell proliferation and regulate cell differentiation. Aged BMSCs are arrested at the G1 phase, and the proliferation capability is significantly suppressed. Accumulation of telomere DNA damage has been observed in the old BMSCs. Hu et al. found that old BMSCs experienced considerably impaired osteogenic differentiation capacity compared to young BMSCs due to elevated NAP1L2 gene expression. NAP1L2 could recruit SIRT1, a histone deacetylase involved in cellular senescence, to deacetylate H3K14ac on osteogenic gene promoters, finally suppressing the osteogenic differentiation of BMSCs. 136 Another study showed that with the aging of MSCs, the accumulation of reactive oxygen species (ROS) increased, leading to the reduction of osteogenic ability. The Nrf2/ARE signaling pathway is responsible for ROS scavenging. KAT6A, an acetyltransferase, could regulate the Nrf2/ARE signaling pathway and improve the acetylation level, thus inhibiting ROS accumulation and promoting osteogenic differentiation. 137 Acetylation of critical factors in specific signaling pathways is also required for osteogenesis. After SMAD signaling is activated, an H3K9 acetyltransferase, PCAF, acetylates histone H3K9 at promoters of BMP pathway genes (e.g. BMP2, BMP4, and BMPR1B) and activates BMP signaling in MSCs. 138 Other studies also substantiated that the significantly increased level of H3K9ac recruited onto the RUNX2 promoter could promote ADSCs osteogenic differentiation through BMP signaling.139,140 Wnt/β-catenin signaling is activated during osteogenesis of BMSCs but inhibited during osteoporosis. The expression of Wnt genes (e.g. Wnt1, Wnt6, Wnt10a, and Wnt10b) and their acetylation levels are decreased in osteoporosis BMSCs. The histone acetyltransferase GCN5 has been identified as one of the key factors. Downregulation of GCN5 decreases its recruitment onto Wnt promoter regions and impairs H3K9 acetylation, ultimately inhibiting β-catenin activation and osteogenesis in BMSCs. 141
Classified into four groups, the HDAC family encompasses a minimum of 18 genes, including class I (HDAC1, HDAC2, HDAC3, and HDAC8) and class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10). HDAC6 is significantly increased in BMSCs of aged mice. HDAC6 can directly target the transcription factor binding site (− 327 bp/− 1 bp) of RUNX2, thus causing hypoacetylation of H3K9/K14 and H4K12 to inhibit its expression. 142 Similarly, another study indicated that HDAC9 levels were also increased in aged BMSCs. Further results showed that HDAC9 enhanced autophagy in BMSCs by upregulating H3K9ac of autophagy-associated genes (ATG7, BECN1, LC3a, and LC3b), thereby improving bone mass loss. 143 It is well accepted that dexamethasone, a potent synthetic form of the steroid glucocorticoid, is usually included in the in-vitro osteogenic culture system. Low GC concentration promotes MSCs commitment and enhances differentiation, which may be closely related to acetylation modification. The main downstream effector of steroid glucocorticoid is the glucocorticoid receptor which positively regulates RUNX2 and OCN transcription through direct binding. However, a study by Rimando et al. suggested that long-term use of high concentrations of dexamethasone yielded different effects on the osteogenic induction of MSCs. The early osteogenic marker RUNX2 was highly responsive to dexamethasone. With time, RUNX2 became unresponsive to the treatment, and inhibition of OCN occurred in all concentrations of dexamethasone. A high concentration of dexamethasone caused a delay or blockade of cytoplasm-nucleus trafficking of the GR-ligand complex. Then HDAC6 could translocate to the nucleus and combine with GR. GR-HDAC6 complex negatively regulated OCN promoters to inhibit osteogenesis. 144 Meanwhile, another study first provided evidence that HDAC2 could bind with GR in the cytoplasm. The ChIP assay confirmed that GR was recruited to the nGRE element in the OCN promoter and inhibited its expression. 145 It has been established that HDAC1 expression is reduced during osteogenesis, and inhibiting HDAC1 activity results in transcription activation. In this respect, under the stimulation of cyclic mechanical stretch, the expression of HDAC1 protein is further downregulated, activating the Notch signaling pathway. Besides, this mechanical stimulation promotes the mRNA and protein expression of the ligand for the Notch receptor JAG1 and its histone H3 acetylation level, thus promoting osteogenesis. 146 HDAC inhibitors can improve the reprograming efficiency of induced pluripotent stem cells and enhance the conversion efficiency of MSCs in vitro, boosting the practicality and safety of stem cell treatment and tissue engineering. 147 HDAC8 can suppress the osteogenic differentiation of BMSCs by removing H3K9ac and inhibit the transcriptional activity of RUNX2 via direct binding with it. Conversely, valproic acid, an HDAC inhibitor, promotes the osteogenic differentiation of BMSCs via inhibiting HDAC8 expression and enhancing the H3K9ac level. 148 Trichostatin A(TSA) is another broadly used HDAC inhibitor. Man et al. reported that 5 nM TSA more effectively altered osteoblast epigenetic function and promoted its mineralizing capacity. When treated with TSA, extracellular vehicles (EVs) could promote the proliferation and migration of human BMSCs. TSA-EV-treated cells displayed a significantly enhanced collagen, extracellular matrix, and calcium deposition. Moreover, TSA-EV-treated human BMSCs increased H3K9ac levels in a time-dependent manner. 149
Non-coding RNA
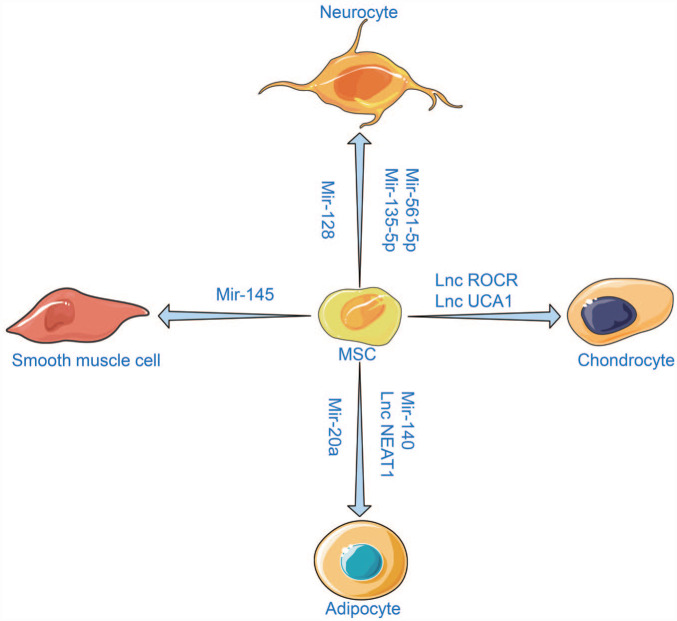
Upon the completion of the human genome project in June 2000, scientists concluded that only 1.5% of the human genome encodes for proteins, while the remaining regions initiate transcription to produce non-coding RNAs (ncRNAs). 150 NcRNAs can be classified and named according to the length, function, and position between the strand and coding gene. Generally, this category includes endogenous long non-coding RNAs (lncRNAs), long intergenic noncoding RNAs (lincRNAs), microRNAs (miRNAs), small interfering RNAs, PIWI-interacting RNAs, small nucleolar RNAs, tRNA-derived small RNAs, natural antisense transcripts, circular RNAs, enhancer noncoding RNAs, transcribed ultraconserved regions, or primate-specific pyknon transcripts. 151 These ncRNAs appear as “Jack of all trades,” inducing complicated transcriptional and posttranscriptional regulatory mechanisms and gene activation or depression. The emergence of newly discovered ncRNAs in recent years attributed to the next-generation sequencing methods has increased understanding of their biological roles in stem cells. For example, the ES cell-specific cell-cycle-regulating (ESCC) ncRNAs, activated by the core pluripotency proteins, are essential for maintaining the fate of ESCs. MiR-430 family members have emerged as critical regulators of ESCs pluripotency and self-renewal. Moreover, miR-let-7 has been implicated in repressing pluripotency in differentiating ESCs. Several lncRNAs are correlated with the expression of pluripotency markers. OCT4 and NANOG can bind more than 100 lincRNA promoters. Among them, HOTAIR, a lncRNA expressed from the HOXC cluster, controls gene expression in trans via acting as a scaffold for chromatin-modifying complexes. HOTAIR can interact with H3K27me3 PRC2 and the H3K4me2/3-demethylating LSD1–CoREST–REST complex to repress multiple target genes in the genome. 152 A study by Lin et al. suggested that lncRNA TUNA was essential for pluripotency and formed a complex with three RNA-binding proteins (RBPs). Furthermore, the TUNA-RBP complex was located in the promoters of NANOG, SOX2, and FGF4. The knockdown of TUNA or the individual RBPs inhibited the neural differentiation of ESCs. 153 In MSCs, ncRNAs are involved in the regulation of diverse differentiation lineages (Figure 3).
Figure 3.
MSCs have multi-differentiation potential and can be induced to differentiate into various tissue cells. Different ncRNAs are identified during the differentiation. At the same time, the differentiation of MSCs into specific cell lineages can be promoted or inhibited by regulating these ncRNAs.
Numerous studies have confirmed that ncRNAs play a crucial role in osteogenesis. Additionally, several in vivo and in vitro studies have been conducted to stimulate the regulatory environment of ncRNA on osteogenesis to propose new therapeutic targets for various associated diseases. In the following section, we will primarily summarize and discuss the role of miRNAs and lncRNAs related to MSCs osteogenesis.
Long non-coding RNA
Non-coding regions comprise a wide range of regulatory and functional units, mostly loci encoding lncRNAs. LncRNAs are defined as those having a length greater than 200 nucleotides, sometimes in the range of tens of kilobases, and are predominantly localized in the chromatin and nucleus. In contrast with protein-coding genes, lncRNAs tend to generate two-exon transcripts, allowing them to fold into potentially complex secondary and three-dimensional structures.154–156 Besides interacting with other cell components, lncRNAs direct the activity of the regulatory element and influence genome or gene activity during transcription. 157 LncRNAs execute their functions through diverse mechanisms, acting as scaffolds, decoys, signals, and guides. 158
Long non-coding RNAs in the osteogenic differentiation
A previous research revealed that after 14 days of in vitro osteogenic differentiation, 1206 lncRNAs were differentially expressed (at least twice folds) in human BMSCs compared to undifferentiated ones. 159 Another study using human whole transcriptome microarray on BMSCs showed that 1408 lncRNAs were differentially expressed at day 7 of osteogenic differentiation, including 785 upregulated and 623 downregulated transcripts. Interestingly, several pathways might be involved in this process, such as the MAPK signaling pathway, the Jak-STAT signaling pathway, the Toll-like receptor signaling pathway, and the TGF-beta signaling pathway. 160 In addition, Qiu et al. discovered that during the 21 days of osteogenic differentiation in human BMSCs, 433 and 232 lncRNAs were consistently elevated and downregulated, respectively. 161
Li et al. and his colleagues identified an aging-related lncRNA, BMNCR, which could maintain extracellular matrix protein fibromodulin and affect its local chromatin structure or transcription, thus leading to activation of the BMP2 pathway. BMNCR was considered as a scaffold to enhance interaction between TAZ and ABL resulted in facilitating the assembly of the TAZ and RUNX2/PPARG transcriptional complex, ultimately promoting osteogenesis while suppressing adipogenesis. 162 Compared to adipogenesis, osteogenesis requires more mitochondrial metabolites, oxygen consumption, and mitochondrial membrane potential. LncRNA NEAT1 has been reported as an essential marker gene during BMSCs differentiation and senescence since it could regulate the lineage fates of BMSCs by impairing mitochondrial function. 163 Moreover, increased cell proliferation and differentiation time gradually led to the accumulation of intracellular inflammatory factors. In the inflammatory setting, cytokines TNF-α and IL-17 inhibit osteoblast differentiation of ADSCs by activating NF-κB signaling. In this regard, the p65 subunit of NF-κB binds to the LncRNA MIR31HG promoter and significantly increases its activity, followed by high MIR31HG expression. MIR31HG directly interacts with subunit IκBα and participates in NF-κB activation. 164 With the stimulation of TNF-α, LncRNA MIAT expression is increased, resulting in inhibited osteogenic differentiation of human ADSCs both in vitro and in vivo. 165 High glucose levels can aggravate the inflammatory status and impair osteogenic differentiation. Although high glucose can decrease the expression of lncRNA AK028326 and CXCL13, as well as mineralization, ALP activity, and osteogenic gene expression, it can be reversed by overexpression of AK028326 or CXCL13. 166 LncRNAs contain miRNA recognition sites and can sequester them through sequence complementarity. Consequently, they can inhibit miRNA binding to target mRNA, which may increase the expression of coding transcripts targeted by those miRNAs. These lncRNAs are referred to as “ceRNAs.” 167 When treated with glucocorticoids, MSCs are more likely to experience lipid accumulation, which can be reversed by the overexpression of LncRNA TCONS_00041960. TCONS_00041960, acting as a ceRNA, can interact with miR-204-5p and miR-125a-3p to regulate RUNX2 and GILZ expression, respectively, and enhance osteogenic differentiation 168 (Figure 4).
Figure 4.
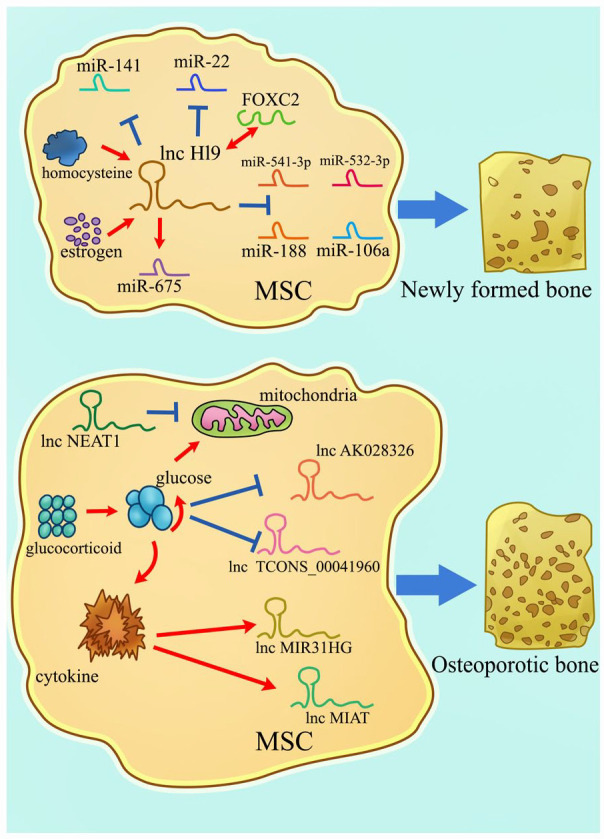
A schematic representation of lncRNAs involved in osteogenic differentiation. Different lncRNAs can promote or inhibit the osteogenesis of MSCs. LncRNAs containing the complementary sequence of miRNAs can act as ceRNAs to regulate gene expression, thus weakening the effect of miRNAs on mRNAs. At the same time, lncRNAs can be targeted by other factors in vivo. These factors amplify the regulatory role of lncRNAs, thus finally affecting the osteogenesis of MSCs.
Several studies have shown that lncRNA H19 is a pro-osteogenic gene (Figure 4). H19 enhances the expression of osteogenic genes (e.g. BMP2, OCN, RUNX2, and OSX) and inhibits adipogenic differentiation of BMSCs by directly suppressing miR-541-3p and miR-188.169,170 The exon1 of H19 is responsible for encoding miR-675. H19 works synergistically with miR-675 to inhibit the expression of TGF-β1 and HDAC4/5. The downregulation of TGF-β1 subsequently inhibits phosphorylation of SMAD3, thus increasing osteoblast marker gene expression. Conversely, miR-675-5p also directly targets H19, causing the process to proceed in the opposite direction. 171 Nonetheless, H19 acts as a natural molecular sponge of miR-141 and miR-22 and antagonizes their functions, leading to mitigation of their common target gene β-catenin, which eventually activates the Wnt/β-catenin pathway and hence potentiates osteogenesis. 172 Moreover, H19 can bind to FOXC2 and subsequently regulate the Wnt promoter, especially Wnt4 promoter transcription. H19/FOXC2 regulated the osteogenic differentiation of BMSCs synergistically. 173 One of the major contributing factors to osteoporosis brought on by various diseases is H19. Obesity and high-fat factors can inhibit the expression of osteogenic markers and decelerate fracture healing primarily because high-fat BMSCs generate limited exosomes that contain reduced H19 and HOXA10. H19 competitively binds to miR-467 by regulating the expression of HOXA10. Besides, the overexpression of miR-467 can reverse the pro-osteogenic differentiation process caused by H19. 174 It is widely thought that estrogen is a crucial factor in postmenopausal osteoporosis. A study revealed that the underlying mechanism might be connected with H19. Estrogen promoted osteogenic differentiation of BMSCs by upregulating H19. Meanwhile, the upregulation of H19 could inhibit miR-532-3p and upregulate SIRT1, finally alleviating osteoporosis. 175 Decreased bone mineral density in osteoporotic postmenopausal women is associated with high homocysteine levels. Furthermore, elevated homocysteine is related to the cystathionine β-synthase (CBS) enzyme mutation. Behera et al. found that exosomal H19 from CBS-heterozygous mice BMSCs promoted osteogenic differentiation by competitively binding to the miR-106a. Meanwhile, miR-106a directly regulated the angiogenic factor, Angpt1, that activated H19/Tie2-NO signaling and promoted cell osteogenesis. 176
Dysregulated osteogenic differentiation is the primary cause of bone loss in people with osteoporosis. Reduced bone mineral density and the degradation of the microarchitectural bone structure are its hallmarks, increasing the risk of bone fracture. Ovariectomized mice are generally applied as models to simulate postmenopausal osteoporosis patients. BMSCs extracted from ovariectomized mice typically have a lower level of BDNF, while its lncRNA antisense transcript (BDNF-AS) is highly expressed. The primary function of antisense transcripts is to regulate the expression of sense transcripts, and they may form double-stranded RNA structures with sense transcripts, which regulate the stability, splicing, or transport of sense transcripts. BDNF-AS upregulation decreases BDNF levels and inhibits osteogenic differentiation.177,178 Wang et al. identified that MEG3 was upregulated in BMSCs derived from ovariectomized mice, similar to those of patients with postmenopausal osteoporosis. The osteogenic induction-mediated downregulation of miR-133a-3p, accompanied by a significant decline in SLC39A1 expression, was reversed by MEG3 overexpression. Additionally, miR-133a-3p overexpression or silencing eliminated the effects of MEG3 on osteogenic differentiation through binding directly.167,179 Chen et al. suggested that the DEP domain containing mTOR interacting protein, an endogenous mTOR inhibitor, could bind to a specific region (− 1000 bp ~ 0) of the MEG3 promoter to inhibit its transcription, which subsequently deactivated BMP4 signaling. 180 Another study on osteoporosis suggested that the knockdown of lncRNA LNC_000052 could promote BMSCs osteogenesis via the PI3K/Akt signaling pathway. LNC_000052 and PIK3R1 shared a common miRNA target, miR-96-5p, whose downregulation could restrain the effects of LNC_000052 knockdown. 181 In another study on osteoporosis, PTEN could inhibit the activation of downstream proteins in Akt signaling. Moreover, lncRNA MIR22HG could downregulate PTEN, thus promoting osteogenic differentiation by activating Akt signaling. Therefore, MIR22HG played a crucial role in bone metabolism, making it a potential target for osteoporosis treatment. 182
Micro RNA
MiRNA is a short RNA fragment consisting of typically 19–25 nucleotides in length. The first miRNA (lin-4) was discovered in Caenorhabditis elegans in 1993, and 7 years later, let-7, the first human miRNA, was discovered. Currently, 2675 miRNAs have been identified in humans and regulate the expression of more than 60% of all protein-coding genes. A single miRNA can target numerous mRNAs and influence the expression of genes involved in a functional interacting pathway. Most miRNAs are transcribed by RNA polymerase II (Pol II) as large primary miRNAs. Then primary miRNAs are cleaved into a stem-loop structure called precursor miRNAs, which are transferred to the cytoplasm. They are cleaved into small double-stranded RNAs and then loaded into an argonaute protein, which promotes the ribonucleoprotein complex assembly. They can bind to the 3′-UTR of its target mRNAs to inhibit expression. Accordingly, the function of miRNAs is commonly perceived as a gene expression repressor. However, more roles have been discovered for miRNAs in recent years, such as gene promoter, mRNA degradation, cell communication, and differentiation.183–185 MiRNAs are crucial for the cell cycle regulation of stem cells. Their role in stem cell proliferation was first observed in knockout mice lacking miRNA Dicer and Dgcr8. Dicer knockout mice were embryonic lethal, and their ESCs exhibited defects in cell cycle progression. ESCs derived from Dgcr8-deficient mice showed cell cycle arrest due to the downregulation of genes involved in self-renewal.186,187 Recent studies suggest that miRNAs even have the potential to play critical roles in reprograming processes that lead to the generation of pluripotent stem cells, neurons, and cardiomyocytes. 188
Micro RNAs in the osteogenic differentiation
Growing evidence suggests miRNAs can act as a dynamic regulatory switch for adipogenic or osteogenic differentiation of MSCs. A previous study showed that miR-99a-5p was downregulated during osteogenic differentiation and raised adipogenic differentiation in the early stages of MSCs. 189 However, overexpression of miR-130a increased osteogenic differentiation and attenuated adipogenic differentiation in aging BMSCs. An aged mouse model further demonstrated that the decrease in miR-130a expression was the primary cause of bone decline, as miR-130a directly binds with the 3′-UTRs of SMURF2 and PPARγ to elevate their protein expression. 190 Pi et al. found that miR-34a might be implicated in the modulation of expansion-mediated MSCs replicative senescence and age-related MSC natural senescence. They identified NAMPT as a direct target gene of miR-34a. Overexpression miR-34a in young MSCs could accelerate their aging process by increasing NAMPT expression levels, NAD+ content, NAD+/NADH ratio, and SIRT1 activity, thus mitigating osteogenic differentiation. 191 MiRNAs are undoubtedly crucial for the osteogenic differentiation of MSCs. Therefore, based on the latest research, we summarized the effects of different miRNAs on the osteogenic differentiation of MSCs and their target mRNAs and related signaling pathways (Table 4).
Table 4.
Roles of different miRNAs in osteogenic differentiation of MSCs.
| MiRNA | Osteogenesis | Target mRNA | MRNA expression | MRNA binding site | Signaling pathway | Ref. |
|---|---|---|---|---|---|---|
| miR-143 | inhibit | k-Ras | ↓ | 3′-UTR | ERK1/2 | Zhang et al. 192 |
| miR-186 | inhibit | SIRT6 | ↓ | 3′-UTR | NF-κB | Xiao et al. 193 |
| miR-765 | inhibit | BMP6 | ↓ | 3′-UTR | SMAD | Wang et al. 194 |
| miR-483-5p | inhibit | RPL31 | ↓ | 3′-UTR | RAS/MEK/ERK | Peng et al. 195 |
| miR-137 | inhibit | LSD1 | ↑ | 3′-UTR | SMAD | Ma et al. 107 |
| miR-503-3p | inhibit | Wnt2/Wnt7 | ↓ | 3′-UTR | Wnt/β-catenin | Luo et al. 196 |
| miR-381 | inhibit | Wnt5/FZD3 | ↓ | 3′-UTR | Wnt/β-catenin | Long et al. 197 |
| miR-23 | inhibit | MEF2C | ↓ | 3′-UTR | MAPK | Jiang et al. 198 |
| miR-378 | inhibit | Wnt6 Wnt10a |
↓ | 3′-UTR | Wnt/β-catenin | Feng et al. 199 |
| miR-200c | promote | MYD88 | ↓ | 3′-UTR | AKT/β-Catenin | Xia et al. 200 |
| miR-200c | promote | SOX2/Klf4 | ↓ | 3′-UTR | Wnt | Akkouch et al. 201 |
| miR-224 | promote | Rac1 | ↓ | 3′-UTR | JAK/STAT3 Wnt/β-catenin |
Cai et al. 202 |
| miR-128 | promote | DKK2 | ↓ | 3′-UTR | Wnt/β-catenin | Wang et al. 203 |
| miR-483-3p | promote | DKK2 | ↓ | 3′-UTR | Wnt/β-catenin | Zhou et al. 204 |
| miR-21 | promote | P-Akt/HIF-1α | ↑ | N/A | PTEN/PI3K/Akt | Yang et al. 205 |
| miR-29b | promote | PTEN | ↓ | 3′-UTR | AKT/β -catenin | Xia et al. 206 |
| miR-211-5p | promote | DUSP6 | ↓ | 3′-UTR | ERK/SMAD/β-catenin | Wang et al. 207 |
MSCs have positive impacts on bone tissue healing via their immunomodulatory effects. MiR-146a has been identified as a negative regulator in some chronic inflammatory diseases. Besides, the inhibition of miR-146a enhanced the expression of immunomodulatory factors and promoted the chemotactic migration of dTHP-1 cells, a model for macrophages, in ADSCs. MiR-146a suppression could activate NF-κB signaling and stimulate the osteogenic differentiation of ADSCs. 208 Chemokines represent a class of small molecule secretory proteins (about 8–10 kDa) that play a pivotal role in immunomodulatory and immunotolerance. CXC chemokines are essential for recruiting MSCs during bone repair or osteogenic differentiation via interacting with miR-23a. Tian et al. found that CXCL13 attenuated the interaction of miR-23a with the RUNX2 3′-UTR by suppressing the expression of miR-23a, thus promoting RUNX2 expression and stimulating ALP activity. 209 MiR-23a, one of the osteogenic inhibitors, is downregulated during the osteogenic differentiation of BMSCs. Current evidence suggests that miR-23a directly represses luciferase activity through 3′-UTR of lipoprotein-receptor-related protein 5, thereby depressing the Wnt/β-catenin signaling pathway to regulate osteogenic differentiation of BMSCs.210,211 Another research indicated that miR-23b, an endogenous attenuator of RUNX2 in BMSCs, was significantly induced under the stimulation of TNF-α. And overexpression of miR-23b led to inhibiting osteogenic differentiation. 212 CXCL12 is an important angiogenic factor. It can promote endothelial progenitor cell migration and osteogenesis by producing SDF-1α. Meanwhile, miR-137-3p directly targets CXCL12 and RUNX2 to suppress their expression at the protein level. Overexpression of Runx2 and CXCL12 without the 3′-UTR partially rescued the effects of miR-137-3p on osteogenesis and angiogenesis, respectively, thus promoting osteogenesis of BMSCs and intraosseous blood supply. 213 The manipulation and alteration of the physical, chemical, and biological characteristics of 3D biological materials can amplify the attraction of MSCs, release of chemicals, and ultimately regulate inflammation to facilitate tissue regeneration. The titanium surface, for instance, had been proven to enhance the osseointegration. The modified titanium implants induce the recruitment of MSCs and macrophage to damaged sites and control the inflammatory response. 214 Furtherly, this specific mechanism was reported by Zhuang et al. When BMSCs cultured on nanostructured titanium surface, miR-23a silencing promoted their osteogenic differentiation ability while enhancing CXCL12 expression by directly targeting its 3′-UTR. Suppressing CXCL12, however, could inhibit osteogenic differentiation. 215
Besides their direct effect of intracellular miRNAs on related genes, EVs participate in intercellular signal transduction by carrying and transmitting higher concentrations of growth factors and regulatory miRNAs. At present, EVs have become a research hotspot (Figure 5). BMSCs-derived EVs loaded with miR-15b suppress WWP1 expression, attenuating KLF2 degradation by E3 ubiquitin ligase and inhibiting NF-κB activity. After the culture of ovariectomized mice with these EVs, their tibiae exhibited increased bone volume and trabecular number but reduced bone loss. 216 Liu et al. found that small EVs derived from BMSCs could act as nanoscale carriers. These carriers could wrap up miR-20a to promote cell migration, osteogenesis, and osteointegration in an osteoporotic rat model by targeting BAMBI. 217 Exosomes are classified as EVs and comprise miRNAs that can be transferred to nearby cells or throughout the body. 218 It has been established that miR-19b was enriched in BMSC-derived exosomes, and exosome treatment could accelerate fracture healing. MiR-19b facilitates the differentiation of human BMSCs into osteoblasts by inhibiting WWP1 and SMURF2, which degrade KLF5. The delay in fracture healing caused by KLF5 modulation through the Wnt/β-catenin signaling pathway is thus prevented. 219 Nevertheless, aged-exosomes display minimal effects on the in vitro osteogenic differentiation of MSCs or the in vivo fracture healing process. Xu et al. evaluated the level of miRNAs in exosomes from old and young MSCs by miRNA array analysis. It was found that miR-128-3p was markedly upregulated in aged-exosomes. Besides, miR-128-3p could directly target SMAD5 and act as an inhibitor in the process of fracture healing. 220 A similar study revealed that miR-101 in MSCs-derived exosomes was upregulated during osteogenic differentiation and specifically bound to the 3′-UTR of the FBXW7 gene to inhibit its expression. FBXW7 could ubiquitinate and degrade HIF1α, which could bind to the promoter region of FOXP3 and facilitate osteogenic differentiation. 221 Chen et al. designed exosomes overexpressing miR-375. The absorption of miR-375 by BMSCs resulted in the inhibition of IGFBP3 through binding to its 3′-UTR. As BMSCs were stimulated with these modified exosomes at 50μg/mL, the mRNA expression of osteogenesis-related genes was significantly upregulated. 222
Figure 5.
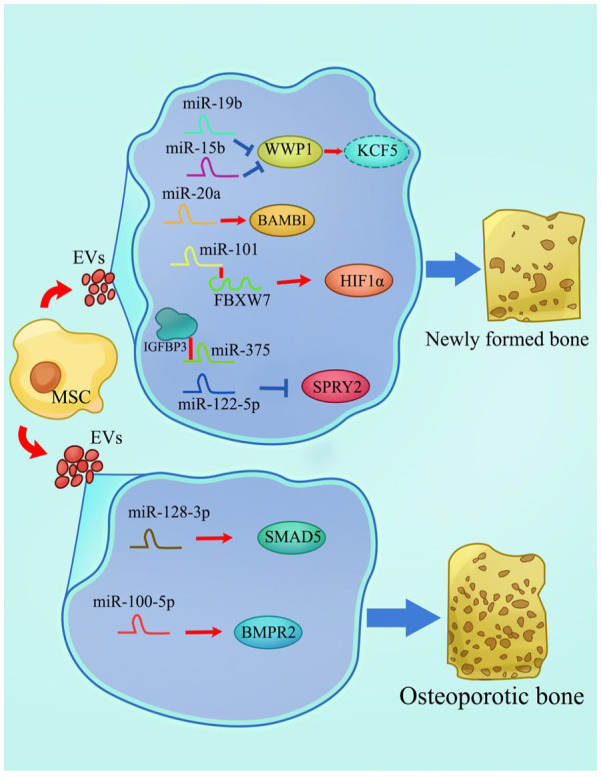
EVs as miRNA carriers in osteogenic differentiation of MSCs. Based on their size and origin, EVs can be classified into two main classes: exosomes and microvesicles. EV cargos regulate biological functions at autocrine, paracrine, and systemic levels and are transported to recipient cells in protected and directed manners. Multiple RNA types are found in EVs, where ncRNAs comprise the majority of EV-RNA transcripts. During the osteogenic differentiation of MSCs, EVs secreted from MSCs contain different miRNAs, which promote or inhibit osteogenesis by interacting with related mRNAs.
In addition to physiological osteogenic processes, miRNAs play an essential role in bone injury caused by related diseases. According to recent studies, abnormally expressed genes or molecules have been added to the in vitro osteogenesis of MSCs to simulate the ossification in vivo to provide a better understanding of different pathological factors and potential therapeutic targets or directions. The function of miRNAs in osteonecrosis of the femoral head (ONFH) is one of the critical focus areas. The pathogenesis of ONFH includes a decreased osteogenic capacity of MSCs. Liao et al. suggested that miR-122-5p might be a novel biomarker for ONFH treatment. They found that bone mineral density, trabecular bone volume, and mean trabecular plate thickness of the femoral head were increased after overexpressing miR-122-5p in MSCs exosomes. Moreover, miR-122-5p attenuated ONFH development by downregulating SPRY2 via the RTK/Ras/MAPK signaling pathway. 223 Yang et al. revealed the detailed pathogenesis of nontraumatic ONFH. They identified that miR-100-5p expression was upregulated in the nontraumatic ONFH exosomes and inhibited the osteogenesis of BMSCs by targeting BMPR2 and suppressing the BMPR2/SMAD1/5/9 signaling pathway. Inhibition of miR-100-5p could rescue the impaired osteogenesis of BMSCs caused by NONFH exosomes to a certain extent. 224 MiR-144-3p expression is also higher in ONFH samples. MiR-144-3p inhibits the proliferation and osteogenesis of MSCs via targeting FZD4 and finally decreases β-catenin nuclear translocation and the transcription of RUNX2 and COL1A1. 225 Recent studies have shown that the T2DM environment may impair bone fracture repair through miRNA regulation. Jiang et al. demonstrated that miR-222 displayed a dramatic increase in T2DM rats. Furthermore, miR-222 could directly bind with 3′-UTR regions of TIMP-3 to reduce both its mRNA and protein expression in MSCs, thereby suppressing osteogenic differentiation. 226 Another research suggested that BMSCs cultured in an osteogenic medium with high glucose could induce upregulation of miR-493-5p and decreased levels of its downstream target ZEB2, followed by inhibiting osteogenic differentiation. 227 Meanwhile, miR-221-3p and miR-222-3p were upregulated in osteogenic BMSCs cultured in a high-glucose environment. Both these two miRNAs could regulate ERK expression by targeting IGF-1 simultaneously and synergistically, eventually inhibiting osteogenic differentiation. 228
Multiple studies have previously explored the impact of a specific ncRNA on osteogenic differentiation. Recent research has indicated that various types of ncRNAs can work synergistically to co-regulate the osteogenic differentiation process of MSCs. Notably, the interaction between lncRNAs and miRNAs is significant due to the lengthy structure of lncRNA that can span thousands of nucleotides with introns. This characteristic facilitates the binding of numerous miRNAs. For instance, research has shown that the lncRNA DANCR is reduced in human BMSCs during osteogenic differentiation. The ncRNA modulates the growth and osteogenic differentiation of BMSCs by manipulating the p38 MAPK pathway. A later study reported that DANCR, miR-320a, and CTNNB1 were closely associated with osteoporosis. MiR-320a and DANCR could act independently, inhibit CTNNB1 expression, and deactivate the Wnt/β-catenin signaling pathway. The co-overexpression of miR-320a and DANCR resulted in an increased inhibitory effect and impaired the osteogenic capacity of BMSCs. In the meantime, silencing DANCR could promote miR-1301-3p expression via sponging, thus leading to downregulated PROX1 expression and enhanced osteogenesis.229–231 LncRNAs compete to occupy many miRNAs, which can act like a sponge to interfere with target mRNAs and their protein-encoding ability. Therefore, based on several studies on the osteogenesis of MSCs, we summarized the osteogenic roles of different lncRNAs and listed related miRNAs and their downstream mRNA genes in Table 5.
Table 5.
LncRNAs roles in osteogenic differentiation and interaction with miRNAs.
| LncRNA | Osteogenesis | Target miRNA | MiRNA binding sequence | MiRNA target gene | Ref. |
|---|---|---|---|---|---|
| XIST | promote | miR-9-5p | 3′-UUGGUUUC-5′ | ALP | Zheng et al. 232 |
| PCAT1 | promote | miR-145-5p | 3′-UUGACCU-5′ | TLR4 | Yu et al. 233 |
| SNHG16 | promote | miR-485-5p | 3′-GUCGGAG-5′ | BMP7 | Asila et al. 234 |
| NEAT1 | promote | miR-29b-3p | 3′-UUACCACGAU-5′ | BMP1 | Zhang et al. 235 |
| KCNQ1OT1 | promote | miR-205-5p | 3′-CUUACUUCC-5′ | RICTOR | Yang et al. 236 |
| LOC100126784 POM121L9P |
promote | miR-503-5p | 3′-CGACGA···GCGACGA-5′ | SORBS1 | Xu et al. 237 |
| GAS5 | promote | miR-135a-5p | 3′-TTTCGGTA-5′ | FOXO1 | Wang et al. 238 |
| H19 | promote | miR-149 | 3′-GGAGCCAGA-5′ | SDF-1 | Li et al. 239 |
| SNHG5 | promote | miR-212-3p | 3′-CUGACAAU-5′ | GDF5 | Han et al. 240 |
| HIF1A-AS2 | promote | miR-665 | 3′-GAGGACC-5′ | IL6 | Wu et al. 241 |
| MALAT1 LINC00657 |
promote | miR-214-3p | 3′-CAGAAC···GACGAC-5′ 3′-CAGA···GACGACA-5′ |
BMP2 | Li et al. 242 |
| SNHG1 | inhibit | miR-101 | 3′-CAUGACA-5′ | DKK1 | Xiang et al. 243 |
| ZFAS1 | inhibit | miR-499 | 3′-UCACAAUU-5′ | EPHA5 | Wu et al. 244 |
| GAS5(transcript variant 2) | inhibit | miR-382-3p | 3′-CUUACUA-5′ | TAF1 | Song et al. 245 |
| ORLNC1 | inhibit | miR-296 | - | PTEN | Yang et al. 246 |
Standing at the crossroad of epigenetic modifications
These three distinct types do not operate independently; instead, they interact with one another to impact gene regulation. Many studies have focused on single modifications and their corresponding effect on osteogenic differentiation. Nevertheless, this fact overlooks the possibility that a single factor may trigger a sequence of additional epigenetic alterations in genetic transcription or translation. As a result, scientists must stand at a “crossroad” to contemplate the mechanism and direction more comprehensively.
Nucleic acid methylation and histone acetylation regulate BMSC osteogenic differentiation. For example, the CCAAT/enhancer binding protein a (C/EBPa) has been shown to regulate the balance between osteogenesis and adipogenesis in BMSCs. C/EBPa expression is activated by PPARγ binding to the 1286 bp/−1065 bp region of the C/EBPa promoter. However, DNA hypermethylation in this region prevents PPARγ binding, and HDAC1 occupies the region to reduce histone acetylation to promote osteogenesis at the terminal stage. 247 Bone regeneration begins with macrophage inflammatory responses that interact with the skeletal system via regulating BMSC function. Macrophages are induced to differentiate into M1 pro-inflammatory macrophages by lipopolysaccharide treatment, and the expression of METTL3 is upregulated, promoting the differentiation of macrophages, as well as the migration and migration osteogenic ability of BMSCs. METTL3 can directly promote HDAC5 expression and synergistically regulate the osteogenic differentiation of BMSCs. 248
And to some extent, ncRNAs are believed to act as targets of epigenetic modifications and regulators of epigenetic modifiers to a certain degree. Despite lacking a direct method of epigenetic control, ncRNAs can impact the functioning of diverse epigenetic modifiers during osteoblast formation. MiR-29b can hypomethylate the Notch promoter region via the downregulation of DNMT1, which inhibits osteogenesis.249,250 MiR-149 was identified as an inhibitor of MSC osteogenic differentiation. The DNA methylation level of MiR-149 increased, leading to a reduction in its expression during the osteogenic differentiation of MSCs. Afterward, miR-149 could specifically bind to the 3′-UTR of SDF-1 and reduce SDF-1 expression at the posttranscriptional level, thus inhibiting osteogenic differentiation. This process can be reversed by DNMT inhibitor 5-aza-CdR. 251 A series of ncRNAs can modify the m6A modification enzyme. By binding to the 3′-UTR of FTO, overexpression of miR-149-3p can inhibit its expression and adipogenic differentiation, simultaneously stimulating the BMSCs into osteoblasts. 252 Zhang et al. discovered that 3′-UTR of FTO was also negatively regulated by miR-22-3p, which were encapsulated in EVs of BMSCs. FTO suppression might result in the accumulation of m6A on MYC transcripts, further impairing the stability of MYC mRNA and impeding the MYC pathway. Besides, miR-22-3p could also inhibit the PI3K/AKT signaling pathway to promote osteogenic differentiation. 253 On the other side, the above modification mechanism in the reverse direction is feasible. In this respect, many m6A modifications were detected on lncRNAs when MSCs underwent osteogenic differentiation. A study indicated that lncRNAs exhibiting differentially upregulated methylated peaks during ADSCs osteogenesis were mainly concentrated in the MAPK signaling pathway. Among these methylated lncRNAs, lncRNA RP1144 N12.5 was widely thought to be a critical gene that promoted osteogenesis. In addition, METTL3 could directly and positively regulate this lncRNA, while the latter regulated its downstream target gene STK3, thus affecting the MAPK signaling pathway and ultimately regulating osteogenic differentiation. 254 Research performed by Peng et al. suggested that METTL3 promoted osteogenic differentiation of BMSCs via m6A methylation of LINC00657 and further validated that biotin-labeled miR-144-3p increased the enrichment of LINC00657. By acting as a ceRNA and absorbing miR-144-3p, LINC00657 was able to increase BMPR1B expression, ultimately promoting osteogenic differentiation. 255 LncRNAs are transcribed from enhancers with various histone marks and are occupied by specific enhancer-binding proteins related to a particular lineage. For instance, lncRNA AK141205 is not only positively regulated by the osteogenic growth peptide, an osteogenic differentiation promoter, but also induces CXCL1378 expression by increasing H4 histone acetylation via suppressing HDAC1. 256 HnRNPK directly binds to the primer 4 region (−754 to −523) in the lncRNA-OG promoter and promotes lncRNA-OG transcription by increasing its promoter H3K27ac. LncRNA-OG combines with HnRNPK to form an RNA-protein complex in the nuclei to regulate the expression of the BMP family protein, resulting in an almost 12-fold surge in lncRNA-OG expression during the osteogenesis process of BMSCs. 257 Likewise, the combination of lncRNA PLXDC2-OT and the SIRT7/RBM6 protein complex diminishes its ability to bind and deacetylate in the OSX promoter, thus promoting the osteogenic potential of MSCs. 258
Conclusion
Overall, segmental bone defects in humans longer than 2 cm are considered critical-sized defects that will not heal spontaneously unless external interventions are applied. 259 In contrast to traditional methods, tissue engineering techniques have shifted their focus toward exploring the potential of utilizing MSCs in combination with tissue-engineered constructs specifically designed to facilitate bone regeneration. However, these breakthroughs about the fate determination of MSCs are still preliminary, and more in-depth studies are needed to fully understand the underlying mechanism. Harnessing the pluripotency differentiation of MSCs for treating bone defects and enhancing osteoporosis has enormous prospects. Moreover, the advancement of novel gene delivery methods augurs a more accurate regulation of the fate of MSCs. This review only covers a limited number of theories concerning the osteogenesis and epigenetics of MSCs, leaving many unexplored. It also remains unclear how biomaterials and chemicals affect the epigenetic modifications of MSCs, and further research is needed to fully understand the mechanisms of epigenetic drugs in vivo. Furthermore, more evidence is required to verify whether osteoblast epigenetic modifications play a role in other biological mechanisms. In conclusion, epigenetics opens a new gate for osteogenic differentiation of MSCs, but more epigenetic molecules, mechanisms and drugs need to be further discovered and applied.
Footnotes
Abbreviations: MSCs: mesenchymal stem cells; ESCs: embryonic stem cells; BMSCs: Bone marrow mesenchymal stem cells; ADSCs: adipose derived stem cells; m6A: N6-methyladenosine; 5mC:5-methyl cytosine; DNMT: DNA methyltransferase; TET: Ten-Eleven Translocation; OCT4: octamer-binding transcription factor 4; BMP: bone morphogenetic protein; RUNX2: runt-related transcription factor 2; SOX9: SRY-box transcription factor 9; ATF4: activating transcription factor 4; DLX3: distal-less homeobox 3; OCN: osteocalcin; OSX: osterix; VEGFA: vascular endothelial growth factor A; PTH: parathyroid hormone; T2DM: type 2 diabetes mellitus; DOP: osteoporosis; α-CGRP: α-calcitonin gene-related peptide; ALP: alkaline phosphatase; COL1A1: collagen type I alpha 1 chain; CTCF: CCCTC-binding factor; SHH: sonic hedgehog signaling molecule; 5-AzaC: 5-azacytidine; KLF5: KLF transcription factor 5; PTM: histone post-translation modification; HMTs: histone methyltransferase; KDMs: histone demethylases; JmjC: jumonji C; LSDs: lysine specific demethylases; HATs: histone acetyltransferases; HDACs: histone deacetylases; SETD: SET domain; LBP :lipopolysaccharide-binding protein; EZH2: Zeste homolog 2; PcG: polycomb group; FABP4: fatty acid binding protein 4; PRC2: Polycomb repressive complex 2; ZBTB16: zinc finger and BTB domain containing 16; MX1: MX dynamin like GTPase 1; FHL1: four and a half LIM domains 1; RBP2: retinoblastoma binding protein 2; SATB2: SATB homeobox 2; NAP1L2: nucleosome assembly protein 1 like 2; SIRT1: sirtuin; ROS: reactive oxygen species; JAG1: jagged canonical Notch ligand 1; TSA: Trichostatin A; EVs: extracellular vehicles; ncRNAs: no-coding RNAs; miRNAs: microRNAs; lncRNAs: long non-coding RNAs; RBPs: RNA-binding proteins; FGF4: fibroblast growth factor 4; TNF-α: tumor necrosis factor-α; IL-17: interleukin 17; CXCL13: C-X-C motif chemokine ligand 13; GILZ: glucocorticoid-induced leucine zipper; TGF-β1: transforming growth factor beta 1; CBS:cystathionine β-synthase; PTEN: phosphatase and tensin homolog; SMURF2: SMAD specific E3 ubiquitin protein ligase 2; NAMPT: nicotinamide phosphoribosyltransferase; SIRT1: sirtuin 1; WWP1: WW domain containing E3 ubiquitin protein ligase 1; BAMBI: BMP and activin membrane bound inhibitor; FBXW7: F-box and WD repeat domain containing 7; FOXP3: forkhead box P3; ONFH: osteonecrosis of the femoral head; SPRY2: sprouty RTK signaling antagonist 2; FZD4: frizzled class receptor 4; TIMP-3: TIMP metallopeptidase inhibitor 3; ZEB2: zinc finger E-box binding homeobox 2;IGF-1: insulin-like growth factor 1; CTNNB1: catenin beta 1; C/EBPa: CCAAT/enhancer binding protein a; SDF-1: stromal cell-derived factor-1; BMPR1B: bone morphogenetic protein receptor, type 1B; RBM6: RNA binding motif protein 6.
Authors’ contributions: Zhaohua Wang, Shude Yang and Shu Guo contributed to this review conception and direction. Zhaohua Wang, Si Wen, Ziming Yang, and Wei Xiong collect reasonably related literature materials. Zhaohua Wang, Si Wen, Kuo Zhang and Meiqi Zhong designed tables and figures. Zhaohua Wang drafted the manuscript. Shu Guo, Huizheng Li and Shude Yang revised the manuscript critically. All authors approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was sponsored by the National Natural Science Foundation of China (51872332).
Consent for publication: All authors approved the final manuscript and agreed to publish.
ORCID iD: Zhaohua Wang  https://orcid.org/0000-0001-8243-8046
https://orcid.org/0000-0001-8243-8046
References
- 1.Barba M, Di Taranto G, Lattanzi W. Adipose-derived stem cell therapies for bone regeneration. Expert Opin Biol Ther 2017; 17: 677–689. [DOI] [PubMed] [Google Scholar]
- 2.Zhu M, Liu Y, Qin H, et al. Osteogenically-induced exosomes stimulate osteogenesis of human adipose-derived stem cells. Cell Tissue Bank 2021; 22: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bueno EM, Glowacki J. Cell-free and cell-based approaches for bone regeneration. Nat Rev Rheumatol 2009; 5: 685–697. [DOI] [PubMed] [Google Scholar]
- 4.Liu G, David BT, Trawczynski M, et al. Advances in pluripotent stem cells: history, mechanisms, technologies, and Applications. Stem Cell Rev Rep 2020; 16: 3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macrin D, Joseph JP, Pillai AA, et al. Eminent sources of adult mesenchymal stem cells and their therapeutic imminence. Stem Cell Rev Rep 2017; 13: 741–756. [DOI] [PubMed] [Google Scholar]
- 6.Eslaminejad MB, Fani N, Shahhoseini M. Epigenetic regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in culture. Cell J 2013; 15: 1–10. [PMC free article] [PubMed] [Google Scholar]
- 7.Fu R, Liu C, Yan Y, et al. Bone defect reconstruction via endochondral ossification: A developmental engineering strategy. J Tissue Eng 2021; 12: 20417314211004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur A, Gronthos S. Clinical application of bone marrow mesenchymal stem/stromal cells to repair skeletal tissue. Int J Mol Sci 2020; 21: 9759. DOI: 10.3390/ijms21249759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Liu Y, Sun Y, et al. Tissue source determines the differentiation potentials of mesenchymal stem cells: a comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. Stem Cell Res Ther 2017; 8: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284: 143–147. [DOI] [PubMed] [Google Scholar]
- 11.Lindroos B, Suuronen R, Miettinen S. The potential of adipose stem cells in regenerative medicine. Stem Cell Rev Rep 2011; 7: 269–291. [DOI] [PubMed] [Google Scholar]
- 12.Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 2001; 7: 211–228. [DOI] [PubMed] [Google Scholar]
- 13.Naderi N, Combellack EJ, Griffin M, et al. The regenerative role of adipose-derived stem cells (ADSC) in plastic and reconstructive surgery. Int Wound J 2017; 14: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornicka K, Marycz K, Marędziak M, et al. The effects of the DNA methyltranfserases inhibitor 5-azacitidine on ageing, oxidative stress and DNA methylation of adipose derived stem cells. J Cell Mol Med 2017; 21: 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S, Greenberger JS, Epperly MW, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell 2008; 7: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayin E, Baran ET, Elsheikh A, et al. Evaluating oxygen tensions related to bone marrow and matrix for MSC differentiation in 2D and 3D biomimetic lamellar scaffolds. Int J Mol Sci 2021; 22: 4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitehead J, Griffin KH, Gionet-Gonzales M, et al. Hydrogel mechanics are a key driver of bone formation by mesenchymal stromal cell spheroids. Biomaterials 2021; 269: 120607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Díaz-Tocados JM, Herencia C, Martínez-Moreno JM, et al. Magnesium chloride promotes osteogenesis through notch signaling activation and expansion of mesenchymal stem cells. Sci Rep 2017; 7: 7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amaroli A, Pasquale C, Zekiy A, et al. Steering the multipotent mesenchymal cells towards an anti-inflammatory and osteogenic bias via photobiomodulation therapy: How to kill two birds with one stone. J Tissue Eng 2022; 13: 20417314221110192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature 2019; 571: 489–499. [DOI] [PubMed] [Google Scholar]
- 21.Topper MJ, Vaz M, Marrone KA, et al. The emerging role of epigenetic therapeutics in immuno-oncology. Nat Rev Clin Oncol 2020; 17: 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wątroba M, Dudek I, Skoda M, et al. Sirtuins, epigenetics and longevity. Ageing Res Rev 2017; 40: 11–19. [DOI] [PubMed] [Google Scholar]
- 23.Park C, Rosenblat JD, Brietzke E, et al. Stress, epigenetics and depression: a systematic review. Neurosci Biobehav Rev 2019; 102: 139–152. [DOI] [PubMed] [Google Scholar]
- 24.Muñoa-Hoyos I, Araolaza M, Urizar-Arenaza I, et al. Sex dependent alteration of epigenetic marks after chronic morphine treatment in mice organs. Food Chem Toxicol 2021; 152: 112200. [DOI] [PubMed] [Google Scholar]
- 25.D’Aquila P, De Rango F, Paparazzo E, et al. Impact of nutrition on age-related epigenetic RNA modifications in rats. Nutrients 2022; 14: 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu R, Li S, Guo S, et al. Environmental temperature and human epigenetic modifications: A systematic review. Environ Pollut 2020; 259: 113840. [DOI] [PubMed] [Google Scholar]
- 27.Zhao LY, Song J, Liu Y, et al. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 2020; 11: 792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peixoto P, Cartron PF, Serandour AA, et al. From 1957 to nowadays: a brief history of epigenetics. Int J Mol Sci 2020; 21: 7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansz N. DNA methylation dynamics at transposable elements in mammals. Essays Biochem 2019; 63: 677–689. [DOI] [PubMed] [Google Scholar]
- 30.Sui BD, Zheng CX, Li M, et al. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol 2020; 30: 97–116. [DOI] [PubMed] [Google Scholar]
- 31.Kim M, Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp Mol Med 2017; 49: e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Ngo V, Wang W. Deciphering the genetic code of DNA methylation. Brief Bioinform 2021; 22: bbaa424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18: 517–534. [DOI] [PubMed] [Google Scholar]
- 34.Calle-Fabregat C, Morante-Palacios O, Ballestar E. Understanding the relevance of DNA methylation changes in immune differentiation and disease. Genes 2020; 11: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Real A, Pérez-Campo FM, Fernández AF, et al. Differential analysis of genome-wide methylation and gene expression in mesenchymal stem cells of patients with fractures and osteoarthritis. Epigenetics 2017; 12: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L, Xia K, Cen X, et al. DNA methylation of noncoding RNAs: new insights into osteogenesis and common bone diseases. Stem Cell Res Ther 2020; 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willyard C. An epigenetics gold rush: new controls for gene expression. Nature 2017; 542: 406–408. [DOI] [PubMed] [Google Scholar]
- 38.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science 2016; 352: 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet 2014; 15: 293–306. [DOI] [PubMed] [Google Scholar]
- 40.Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 2021; 6: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018; 28: 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang CY, Lu RJ, Lee MK, et al. Transcriptome analysis of Dnmt3l knock-out mice derived multipotent mesenchymal stem/stromal cells during osteogenic differentiation. Front Cell Dev Biol 2021; 9: 615098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Y, Yang H, Jin L, et al. Genome-wide DNA methylation analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cells Int 2018; 2018: 8238496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniunaite K, Serenaite I, Misgirdaite R, et al. Epigenetic regulation of human adipose-derived stem cells differentiation. Mol Cell Biochem 2015; 410: 111–120. [DOI] [PubMed] [Google Scholar]
- 45.Wakitani S, Yokoi D, Hidaka Y, et al. The differentially DNA-methylated region responsible for expression of runt-related transcription factor 2. J Vet Med Sci 2017; 79: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uehara O, Abiko Y, Saitoh M, et al. Lipopolysaccharide extracted from Porphyromonas gingivalis induces DNA hypermethylation of runt-related transcription factor 2 in human periodontal fibroblasts. J Microbiol Immunol Infect 2014; 47: 176–181. [DOI] [PubMed] [Google Scholar]
- 47.Marofi F, Hassanzadeh A, Solali S, et al. Epigenetic mechanisms are behind the regulation of the key genes associated with the osteoblastic differentiation of the mesenchymal stem cells: the role of zoledronic acid on tuning the epigenetic changes. J Cell Physiol 2019; 234: 15108–15122. [DOI] [PubMed] [Google Scholar]
- 48.Rahimzadeh S, Rahbarghazi R, Aslani S, et al. Promoter methylation and expression pattern of DLX3, ATF4, and FRA1 genes during osteoblastic differentiation of adipose-derived mesenchymal stem cells. Bioimpacts 2020; 10: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan Q. Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat Res 2005; 569: 133–143. [DOI] [PubMed] [Google Scholar]
- 50.Zhang RP, Shao JZ, Xiang LX. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J Biol Chem 2011; 286: 41083–41094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Li Y, Rao P, et al. Blockade of receptors of advanced glycation end products ameliorates diabetic osteogenesis of adipose-derived stem cells through DNA methylation and Wnt signalling pathway. Cell Prolif 2018; 51: e12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Gao Y, Li Q, et al. Downregulation of DNA methyltransferase-3a ameliorates the osteogenic differentiation ability of adipose-derived stem cells in diabetic osteoporosis via Wnt/β-catenin signaling pathway. Stem Cell Res Ther 2022; 13: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, Ling Z, Dong W, et al. Dnmt3a-Mediated DNA methylation changes regulate osteogenic differentiation of hMSCs cultivated in the 3D scaffolds under oxidative stress. Oxid Med Cell Longev 2019; 2019: 4824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Wang H, Chen X, et al. Oxidative stress-induced hypermethylation of KLF5 promoter mediated by DNMT3B impairs osteogenesis by diminishing the interaction with β-Catenin. Antioxid Redox Signal 2021; 35: 1–20. [DOI] [PubMed] [Google Scholar]
- 55.Jia B, Chen J, Wang Q, et al. SIRT6 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells through antagonizing DNMT1. Front Cell Dev Biol 2021; 9: 648627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cakouros D, Hemming S, Gronthos K, et al. Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenet Chromatin 2019; 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Xu X, Wang D, et al. Hypermethylation-mediated downregulation of long non-coding RNA MEG3 inhibits osteogenic differentiation of bone marrow mesenchymal stem cells and promotes pediatric aplastic anemia. Int Immunopharmacol 2021; 93: 107292. [DOI] [PubMed] [Google Scholar]
- 58.Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther 2012; 92: 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng S, Shi S, Tao G, et al. JKAMP inhibits the osteogenic capacity of adipose-derived stem cells in diabetic osteoporosis by modulating the Wnt signaling pathway through intragenic DNA methylation. Stem Cell Res Ther 2021; 12: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng S, Gao Y, Shi S, et al. LncRNA-AK137033 inhibits the osteogenic potential of adipose-derived stem cells in diabetic osteoporosis by regulating Wnt signaling pathway via DNA methylation. Cell Prolif 2022; 55: e13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Ding F, Shi S, et al. Hypermethylation in Calca promoter inhibited ASC osteogenic differentiation in rats with type 2 diabetic mellitus. Stem Cells Int 2020; 2020: 5245294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnsdorf EJ, Tummala P, Castillo AB, et al. The epigenetic mechanism of mechanically induced osteogenic differentiation. J Biomech 2010; 43: 2881–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlaikou AM, Kouroupis D, Sgourou A, et al. Mechanical stress affects methylation pattern of GNAS isoforms and osteogenic differentiation of hAT-MSCs. Biochim Biophys Acta Mol Cell Res 2017; 1864: 1371–1381. [DOI] [PubMed] [Google Scholar]
- 64.Wang C, Shan S, Wang C, et al. Mechanical stimulation promote the osteogenic differentiation of bone marrow stromal cells through epigenetic regulation of Sonic Hedgehog. Exp Cell Res 2017; 352: 346–356. [DOI] [PubMed] [Google Scholar]
- 65.Zhou GS, Zhang XL, Wu JP, et al. 5-azacytidine facilitates osteogenic gene expression and differentiation of mesenchymal stem cells by alteration in DNA methylation. Cytotechnology 2009; 60: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han X, Li X, Zhong G, et al. Regulation of osteogenic differentiation by DNA methylation of the dishevelled gene in bone marrow mesenchymal stem cells. Am J Transl Res 2017; 9: 4848–4855. [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Wang L, Hou J, et al. Study on the dynamic biological characteristics of human bone marrow mesenchymal stem cell senescence. Stem Cells Int 2019; 2019: 92715951–92715959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan X, Ehnert S, Culmes M, et al. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS One 2014; 9: e90846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W, Song Y, Xia K, et al. Transcriptome-wide m(6)A methylome during osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res Ther 2021; 12: 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen GS, Zhou HB, Zhang H, et al. The GDF11-FTO-PPARγ axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim Biophys Acta Mol Basis Dis 2018; 1864: 3644–3654. [DOI] [PubMed] [Google Scholar]
- 71.Chen LS, Zhang M, Chen P, et al. The m(6)A demethylase FTO promotes the osteogenesis of mesenchymal stem cells by downregulating PPARG. Acta Pharmacol Sin 2022; 43: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu T, Zheng X, Wang C, et al. The m(6)A "reader" YTHDF1 promotes osteogenesis of bone marrow mesenchymal stem cells through translational control of ZNF839. Cell Death Dis 2021; 12: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang C, Wang Y. Downregulation of METTL14 improves postmenopausal osteoporosis via IGF2BP1 dependent posttranscriptional silencing of SMAD1. Cell Death Dis 2022; 13: 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan G, Yuan Y, He M, et al. m(6)A methylation of Precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids 2020; 19: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tian C, Huang Y, Li Q, et al. Mettl3 regulates osteogenic differentiation and alternative splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol Sci 2019; 20: 551. DOI: 10.3390/ijms20030551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carmeliet P, Ng YS, Nuyens D, et al. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nat Med 1999; 5: 495–502. [DOI] [PubMed] [Google Scholar]
- 77.Wu T, Tang H, Yang J, et al. METTL3-m(6) A methylase regulates the osteogenic potential of bone marrow mesenchymal stem cells in osteoporotic rats via the Wnt signalling pathway. Cell Prolif 2022; 55: e13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiao X, Sun X, Li W, et al. 3D-printed β-tricalcium phosphate scaffolds promote osteogenic differentiation of bone marrow-deprived mesenchymal stem cells in an N6-methyladenosine- dependent manner. Int J Bioprinting 2022; 8: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, Wang B, Zhou X, et al. METTL3-mediated long non-coding RNA MIR99AHG methylation targets miR-4660 to promote bone marrow mesenchymal stem cell osteogenic differentiation. Cell Cycle 2023; 22: 476–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou S, Zhang G, Wang K, et al. METTL3 potentiates osteogenic differentiation of bone marrow mesenchymal stem cells via IGF2BP1/m6A/RUNX2. Oral Dis. Epub ahead of print 27January2023. DOI: 10.1111/odi.14526 [DOI] [PubMed] [Google Scholar]
- 81.He M, Lei H, He X, et al. METTL14 regulates osteogenesis of bone marrow mesenchymal stem cells via inducing autophagy through m6A/IGF2BPs/Beclin-1 Signal Axis. Stem Cells Transl Med 2022; 11: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng C, Zhang H, Zheng J, et al. METTL14 benefits the mesenchymal stem cells in patients with steroid-associated osteonecrosis of the femoral head by regulating the m6A level of PTPN6. Aging 2021; 13: 25903–25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang C, Chen R, Zhu X, et al. METTL14 alleviates the development of osteoporosis in ovariectomized mice by upregulating m(6)A level of SIRT1 mRNA. Bone 2023; 168: 116652. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Fu Q, Yang J, et al. RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating runx2 mRNA and inhibiting osteogenic differentiation. Aging 2021; 13: 21134–21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu J, Shen L, Liu Y, et al. The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-κB signaling. Mol Cell Biochem 2020; 463: 203–210. [DOI] [PubMed] [Google Scholar]
- 86.Fan Y, Hanai JI, Le PT, et al. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab 2017; 25: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Y, Xie L, Wang M, et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun 2018; 9: 4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Z, Wang P, Li J, et al. The N(6)-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis 2021; 12: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389: 251–260. [DOI] [PubMed] [Google Scholar]
- 90.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 2015; 16: 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stillman B. Histone modifications: insights into their influence on gene expression. Cell 2018; 175: 6–9. [DOI] [PubMed] [Google Scholar]
- 92.Zhu D, Zhang Y, Wang S. Histone citrullination: a new target for tumors. Mol Cancer 2021; 20: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tolsma TO, Hansen JC. Post-translational modifications and chromatin dynamics. Essays Biochem 2019; 63: 89–96. [DOI] [PubMed] [Google Scholar]
- 94.Lin Y, Qiu T, Wei G, et al. Role of histone post-translational modifications in inflammatory diseases. Front Immunol 2022; 13: 852272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab 2010; 12: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 2019; 20: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng P, Chen QM, Hong C, et al. Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int J Oral Sci 2015; 7: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hyun K, Jeon J, Park K, et al. Writing, erasing and reading histone lysine methylations. Exp Mol Med 2017; 49: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem 2001; 70: 81–120. [DOI] [PubMed] [Google Scholar]
- 100.Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol 2022; 23: 329–349. [DOI] [PubMed] [Google Scholar]
- 101.Wu H, Gordon JA, Whitfield TW, et al. Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochim Biophys Acta Gene Regul Mech 2017; 1860: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang H, Dou L, Song J, et al. CBFA2T2 is required for BMP-2-induced osteogenic differentiation of mesenchymal stem cells. Biochem Biophys Res Commun 2018; 496: 1095–1101. [DOI] [PubMed] [Google Scholar]
- 103.Yin B, Yu F, Wang C, et al. Epigenetic control of mesenchymal stem cell fate decision via histone methyltransferase ash1l. Stem Cells 2019; 37: 115–127. [DOI] [PubMed] [Google Scholar]
- 104.Min Z, Xiaomeng L, Zheng L, et al. Asymmetrical methyltransferase PRMT3 regulates human mesenchymal stem cell osteogenesis via miR-3648. Cell Death Dis 2019; 10: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jo J, Song H, Park SG, et al. Regulation of differentiation potential of human mesenchymal stem cells by intracytoplasmic delivery of coactivator-associated arginine methyltransferase 1 protein using cell-penetrating peptide. Stem Cells 2012; 30: 1703–1713. [DOI] [PubMed] [Google Scholar]
- 106.Lv L, Ge W, Liu Y, et al. Lysine-specific demethylase 1 inhibitor rescues the osteogenic ability of mesenchymal stem cells under osteoporotic conditions by modulating H3K4 methylation. Bone Res 2016; 4: 16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma X, Fan C, Wang Y, et al. MiR-137 knockdown promotes the osteogenic differentiation of human adipose-derived stem cells via the LSD1/BMP2/SMAD4 signaling network. J Cell Physiol 2020; 235: 909–919. [DOI] [PubMed] [Google Scholar]
- 108.Liu A, Chen Y, Zhong D, et al. CircRNA AFF4 induced by KDM1A promotes osteogenic differentiation through FNDC5/Irisin pathway. Mol Med 2022; 28: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang P, Liu Y, Jin C, et al. Histone acetyltransferase GCN5 regulates osteogenic differentiation of mesenchymal stem cells by inhibiting NF-κB. J Bone Miner Res 2016; 31: 391–402. [DOI] [PubMed] [Google Scholar]
- 110.Frontini-López YR, Gojanovich AD, Del Veliz S, et al. 14-3-3β isoform is specifically acetylated at Lys51 during differentiation to the osteogenic lineage. J Cell Biochem 2021; 122: 1767–1780. [DOI] [PubMed] [Google Scholar]
- 111.Zhang P, Liu Y, Wang Y, et al. SIRT6 promotes osteogenic differentiation of mesenchymal stem cells through BMP signaling. Sci Rep 2017; 7: 10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang B, Gong S, Han L, et al. Knockdown of HDAC9 inhibits osteogenic differentiation of human bone marrow mesenchymal stem cells partially by suppressing the MAPK signaling pathway. Clin Interv Aging 2022; 17: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tee WW, Reinberg D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 2014; 141: 2376–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xie W, Schultz M, Lister R, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 2013; 153: 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park S, Kim GW, Kwon SH, et al. Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J 2020; 287: 2891–2902. [DOI] [PubMed] [Google Scholar]
- 116.Liu L, Cheung TH, Charville GW, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 2013; 4: 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hattori N, Niwa T, Kimura K, et al. Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res 2013; 41: 7231–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell 2002; 111: 5–7. [DOI] [PubMed] [Google Scholar]
- 119.Wang L, Niu N, Li L, et al. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol 2018; 16: e2006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lawson KA, Teteak CJ, Gao J, et al. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett 2013; 587: 3961–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin C, Jia X, Miron RJ, et al. Setd7 and its contribution to boron-induced bone regeneration in boron-mesoporous bioactive glass scaffolds. Acta Biomater 2018; 73: 522–530. [DOI] [PubMed] [Google Scholar]
- 122.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol 2020; 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jing H, Liao L, An Y, et al. Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol Ther 2016; 24: 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen YH, Chung CC, Liu YC, et al. Enhancer of zeste homolog 2 and histone deacetylase 9c regulate Age-Dependent mesenchymal stem cell differentiation into osteoblasts and adipocytes. Stem Cells 2016; 34: 2183–2193. [DOI] [PubMed] [Google Scholar]
- 125.Dudakovic A, Samsonraj RM, Paradise CR, et al. Inhibition of the epigenetic suppressor EZH2 primes osteogenic differentiation mediated by BMP2. J Biol Chem 2020; 295: 7877–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Samsonraj RM, Dudakovic A, Manzar B, et al. Osteogenic stimulation of human adipose-derived mesenchymal stem cells using a fungal metabolite that suppresses the polycomb group protein EZH2. Stem Cells Transl Med 2018; 7: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wei Y, Chen YH, Li LY, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011; 13: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hemming S, Cakouros D, Isenmann S, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014; 32: 802–815. [DOI] [PubMed] [Google Scholar]
- 129.Hemming S, Cakouros D, Vandyke K, et al. Identification of novel EZH2 targets regulating osteogenic differentiation in mesenchymal stem cells. Stem Cells Dev 2016; 25: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ge W, Shi L, Zhou Y, et al. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells 2011; 29: 1112–1125. [DOI] [PubMed] [Google Scholar]
- 131.Lv L, Liu Y, Zhang P, et al. The nanoscale geometry of TiO2 nanotubes influences the osteogenic differentiation of human adipose-derived stem cells by modulating H3K4 trimethylation. Biomaterials 2015; 39: 193–205. [DOI] [PubMed] [Google Scholar]
- 132.Liu Y, Chen T, Du F, et al. Single-layer graphene enhances the osteogenic differentiation of human mesenchymal stem cells in vitro and in vivo. J Biomed Nanotechnol 2016; 12: 1270–1284. [DOI] [PubMed] [Google Scholar]
- 133.Ge W, Liu Y, Chen T, et al. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials 2014; 35: 6015–6025. [DOI] [PubMed] [Google Scholar]
- 134.Ye L, Fan Z, Yu B, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012; 11: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Han Q, Yang P, Wu Y, et al. Epigenetically Modified Bone Marrow Stromal Cells in Silk Scaffolds Promote Craniofacial Bone Repair and Wound Healing. Tissue engineering Part A 2015; 21: 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu M, Xing L, Zhang L, et al. NAP1L2 drives mesenchymal stem cell senescence and suppresses osteogenic differentiation. Aging Cell 2022; 21: e13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fei D, Wang Y, Zhai Q, et al. KAT6A regulates stemness of aging bone marrow-derived mesenchymal stem cells through Nrf2/are signaling pathway. Stem Cell Res Ther 2021; 12: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang P, Liu Y, Jin C, et al. Histone H3K9 acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may Be involved in osteoporosis. Stem Cells 2016; 34: 2332–2341. [DOI] [PubMed] [Google Scholar]
- 139.Hu X, Zhang X, Dai L, et al. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev 2013; 22: 248–255. [DOI] [PubMed] [Google Scholar]
- 140.Zych J, Stimamiglio MA, Senegaglia AC, et al. The epigenetic modifiers 5-aza-2’-deoxycytidine and trichostatin A influence adipocyte differentiation in human mesenchymal stem cells. Braz J Med Biol Res 2013; 46: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jing H, Su X, Gao B, et al. Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis 2018; 9: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ma C, Gao J, Liang J, et al. HDAC6 inactivates runx2 promoter to block osteogenesis of bone marrow stromal cells in age-related bone loss of mice. Stem Cell Res Ther 2021; 12: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang L, Qi M, Chen J, et al. Impaired autophagy triggered by HDAC9 in mesenchymal stem cells accelerates bone mass loss. Stem Cell Res Ther 2020; 11: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rimando MG, Wu HH, Liu YA, et al. Glucocorticoid receptor and histone deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of mesenchymal stromal cells (MSCs). Sci Rep 2016; 6: 37371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.La Noce M, Mele L, Laino L, et al. Cytoplasmic interactions between the glucocorticoid receptor and HDAC2 regulate osteocalcin expression in VPA-treated MSCs. Cells 2019; 8: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang J, Wang CD, Zhang N, et al. Mechanical stimulation orchestrates the osteogenic differentiation of human bone marrow stromal cells by regulating HDAC1. Cell Death Dis 2016; 7: e2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao Q, Ji K, Wang T, et al. Effect of the histone deacetylases inhibitors on the differentiation of stem cells in bone damage repairing and regeneration. Curr Stem Cell Res Ther 2020; 15: 24–31. [DOI] [PubMed] [Google Scholar]
- 148.Fu Y, Zhang P, Ge J, et al. Histone deacetylase 8 suppresses osteogenic differentiation of bone marrow stromal cells by inhibiting histone H3K9 acetylation and RUNX2 activity. Int J Biochem Cell Biol 2014; 54: 68–77. [DOI] [PubMed] [Google Scholar]
- 149.Man K, Brunet MY, Fernandez-Rhodes M, et al. Epigenetic reprogramming enhances the therapeutic efficacy of osteoblast-derived extracellular vesicles to promote human bone marrow stem cell osteogenic differentiation. J Extracell Vesicles 2021; 10: e12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xue Y, Chen R, Qu L, et al. Noncoding RNA: from dark matter to bright star. Sci China Life Sci 2020; 63: 463–468. [DOI] [PubMed] [Google Scholar]
- 151.Fabbri M, Girnita L, Varani G, et al. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res 2019; 29: 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011; 12: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin N, Chang KY, Li Z, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014; 53: 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Isoda T, Morio T, Takagi M. Noncoding RNA transcription at enhancers and genome folding in cancer. Cancer Sci 2019; 110: 2328–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Long Y, Wang X, Youmans DT, et al. How do lncRNAs regulate transcription? Sci Adv 2017; 3: eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yin Y, Yan P, Lu J, et al. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 2015; 16: 504–516. [DOI] [PubMed] [Google Scholar]
- 158.Ju C, Liu R, Zhang YW, et al. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother 2019; 115: 108912. [DOI] [PubMed] [Google Scholar]
- 159.Wang L, Wang Y, Li Z, et al. Differential expression of long noncoding ribonucleic acids during osteogenic differentiation of human bone marrow mesenchymal stem cells. Int Orthop 2015; 39: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 160.Zhang W, Dong R, Diao S, et al. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther 2017; 8: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Qiu X, Jia B, Sun X, et al. The critical role of long noncoding RNA in osteogenic differentiation of human bone marrow mesenchymal stem cells. Biomed Res Int 2017; 2017: 5045827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li CJ, Xiao Y, Yang M, et al. Long noncoding RNA bmncr regulates mesenchymal stem cell fate during skeletal aging. J Clin Investig 2018; 128: 5251–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang H, Xu R, Li B, et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ 2022; 29: 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jin C, Jia L, Huang Y, et al. Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived Stem Cells. Stem Cells 2016; 34: 2707–2720. [DOI] [PubMed] [Google Scholar]
- 165.Jin C, Zheng Y, Huang Y, et al. Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells. Cell Biol Int 2017; 41: 33–41. [DOI] [PubMed] [Google Scholar]
- 166.Cao B, Liu N, Wang W. High glucose prevents osteogenic differentiation of mesenchymal stem cells via lncRNA AK028326/CXCL13 pathway. Biomed Pharmacother 2016; 84: 544–551. [DOI] [PubMed] [Google Scholar]
- 167.Silva AM, Moura SR, Teixeira JH, et al. Long noncoding RNAs: a missing link in osteoporosis. Bone Res 2019; 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shang G, Wang Y, Xu Y, et al. Long non-coding RNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis of rat bone marrow mesenchymal stem cell by targeting miR-204-5p and miR-125a-3p. J Cell Physiol 2018; 233: 6041–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Han H, Tian T, Huang G, et al. The lncRNA H19/miR-541-3p/Wnt/β-catenin axis plays a vital role in melatonin-mediated osteogenic differentiation of bone marrow mesenchymal stem cells. Aging 2021; 13: 18257–18273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y, Liu W, Liu Y, et al. Long noncoding RNA H19 mediates LCoR to impact the osteogenic and adipogenic differentiation of mBMSCs in mice through sponging miR-188. J Cell Physiol 2018; 233: 7435–7446. [DOI] [PubMed] [Google Scholar]
- 171.Huang Y, Zheng Y, Jia L, et al. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells 2015; 33: 3481–3492. [DOI] [PubMed] [Google Scholar]
- 172.Liang WC, Fu WM, Wang YB, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep 2016; 6: 20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou P, Li Y, Di R, et al. H19 and foxc2 synergistically promotes osteogenic differentiation of BMSCs via Wnt-β-catenin pathway. J Cell Physiol 2019; 234: 13799–13806. [DOI] [PubMed] [Google Scholar]
- 174.Wang Y, Chen W, Zhao L, et al. Obesity regulates miR-467/HoxA10 axis on osteogenic differentiation and fracture healing by BMSC-derived exosome LncRNA H19. J Cell Mol Med 2021; 25: 1712–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li T, Jiang H, Li Y, et al. Estrogen promotes lncRNA H19 expression to regulate osteogenic differentiation of BMSCs and reduce osteoporosis via miR-532-3p/SIRT1 axis. Mol Cell Endocrinol 2021; 527: 111171. [DOI] [PubMed] [Google Scholar]
- 176.Behera J, Kumar A, Voor MJ, et al. Exosomal lncRNA-H19 promotes osteogenesis and angiogenesis through mediating Angpt1/Tie2-NO signaling in CBS-heterozygous mice. Theranostics 2021; 11: 7715–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Feng X, Lin T, Liu X, et al. Long non-coding RNA BDNF-AS modulates osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Mol Cell Biochem 2018; 445: 59–65. [DOI] [PubMed] [Google Scholar]
- 178.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009; 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang Q, Li Y, Zhang Y, et al. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother 2017; 89: 1178–1186. [DOI] [PubMed] [Google Scholar]
- 180.Chen S, Jia L, Zhang S, et al. DEPTOR regulates osteogenic differentiation via inhibiting MEG3-mediated activation of BMP4 signaling and is involved in osteoporosis. Stem Cell Res Ther 2018; 9: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li M, Cong R, Yang L, et al. A novel lncRNA LNC_000052 leads to the dysfunction of osteoporotic BMSCs via the miR-96-5p–PIK3R1 axis. Cell Death Dis 2020; 11: 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jin C, Jia L, Tang Z, et al. Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via pten/ AKT pathway. Cell Death Dis 2020; 11: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019; 234: 5451–5465. [DOI] [PubMed] [Google Scholar]
- 184.Mens MMJ, Ghanbari M. Cell cycle regulation of stem cells by MicroRNAs. Stem Cell Rev Rep 2018; 14: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018; 141: 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet 2003; 35: 215–217. [DOI] [PubMed] [Google Scholar]
- 187.Wang Y, Medvid R, Melton C, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007; 39: 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pascale E, Caiazza C, Paladino M, et al. MicroRNA roles in cell reprogramming mechanisms. Cells 2022; 11: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Moura SR, Bras JP, Freitas J, et al. miR-99a in bone homeostasis: regulating osteogenic lineage commitment and osteoclast differentiation. Bone 2020; 134: 115303. [DOI] [PubMed] [Google Scholar]
- 190.Lin Z, He H, Wang M, et al. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif 2019; 52: e12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pi C, Ma C, Wang H, et al. MiR-34a suppression targets nampt to ameliorate bone marrow mesenchymal stem cell senescence by regulating NAD(+)-Sirt1 pathway. Stem Cell Res Ther 2021; 12: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang Y, Zhou K, Wu L, et al. Downregulation of microRNA-143 promotes osteogenic differentiation of human adipose-derived mesenchymal stem cells through the k-Ras/MEK/ERK signaling pathway. Int J Mol Med 2020; 46: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xiao J, Qin S, Li W, et al. Osteogenic differentiation of rat bone mesenchymal stem cells modulated by miR-186 via SIRT6. Life Sci 2020; 253: 117660. [DOI] [PubMed] [Google Scholar]
- 194.Wang T, Zhang C, Wu C, et al. MiR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/smad1/5/9 signaling pathway. Stem Cell Res Ther 2020; 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Peng H, Yu Y, Gu H, et al. MicroRNA-483-5p inhibits osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting the RPL31-mediated RAS/MEK/ERK signaling pathway. Cell Signal 2022; 93: 110298. [DOI] [PubMed] [Google Scholar]
- 196.Luo Y, Ding X, Ji H, et al. MicroRNA-503-3p affects osteogenic differentiation of human adipose-derived stem cells by regulation of wnt2 and wnt7b under cyclic strain. Stem Cell Res Ther 2020; 11: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Long H, Zhu Y, Lin Z, et al. MiR-381 modulates human bone mesenchymal stromal cells (BMSCs) osteogenesis via suppressing Wnt signaling pathway during atrophic nonunion development. Cell Death Dis 2019; 10: 470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 198.Jiang K, Teng GD, Chen YQ. microRNA-23 suppresses osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting the MEF2C-mediated MAPK signaling pathway. J Gene Med 2020; 22: e3216. [DOI] [PubMed] [Google Scholar]
- 199.Feng L, Zhang JF, Shi L, et al. MicroRNA-378 suppressed osteogenesis of MSCs and impaired bone formation via inactivating Wnt/β-catenin signaling. Mol Ther Nucleic Acids 2020; 21: 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xia P, Gu R, Zhang W, et al. MicroRNA-200c promotes osteogenic differentiation of human bone mesenchymal stem cells through activating the AKT/β-Catenin signaling pathway via downregulating Myd88. J Cell Physiol 2019; 234: 22675–22686. [DOI] [PubMed] [Google Scholar]
- 201.Akkouch A, Eliason S, Sweat ME, et al. Enhancement of MicroRNA-200c on osteogenic differentiation and bone regeneration by targeting Sox2-mediated Wnt signaling and klf4. Hum Gene Ther 2019; 30: 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cai Q, Zheng P, Ma F, et al. MicroRNA-224 enhances the osteoblastic differentiation of hMSCs via rac1. Cell Biochem Funct 2019; 37: 62–71. [DOI] [PubMed] [Google Scholar]
- 203.Wang C, Qiao X, Zhang Z, et al. MiR-128 promotes osteogenic differentiation of bone marrow mesenchymal stem cells in rat by targeting DKK2. Biosci Rep 2020; 40: BSR20182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou B, Peng K, Wang G, et al. miR-483-3p promotes the osteogenesis of human osteoblasts by targeting Dikkopf 2 (DKK2) and the Wnt signaling pathway. Int J Mol Med 2020; 46: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang C, Liu X, Zhao K, et al. miRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res Ther 2019; 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xia T, Dong S, Tian J. miR-29b promotes the osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue via the PTEN/AKT/β-catenin signaling pathway. Int J Mol Med 2020; 46: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang H, Shi X, Guo Z, et al. microRNA-211-5p predicts the progression of postmenopausal osteoporosis and attenuates osteogenesis by targeting dual specific phosphatase 6. Bioengineered 2022; 13: 5709–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wan S, Wu Q, Ji Y, et al. Promotion of the immunomodulatory properties and osteogenic differentiation of adipose-derived mesenchymal stem cells in vitro by lentivirus-mediated mir-146a sponge expression. J Tissue Eng Regen Med 2020; 14: 1581–1591. [DOI] [PubMed] [Google Scholar]
- 209.Tian F, Ji XL, Xiao WA, et al. CXCL13 promotes osteogenic differentiation of mesenchymal stem cells by inhibiting miR-23a Expression. Stem Cells Int 2015; 2015: 632305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li T, Li H, Wang Y, et al. MicroRNA-23a inhibits osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting LRP5. Int J Biochem Cell Biol 2016; 72: 55–62. [DOI] [PubMed] [Google Scholar]
- 211.Ren G, Sun J, Li MM, et al. microRNA-23a-5p regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting mitogen-activated protein kinase-13. Mol Med Rep 2018; 17: 4554–4560. [DOI] [PubMed] [Google Scholar]
- 212.Deng L, Hu G, Jin L, et al. Involvement of microRNA-23b in TNF-α-reduced BMSC osteogenic differentiation via targeting runx2. J Bone Miner Metab 2018; 36: 648–660. [DOI] [PubMed] [Google Scholar]
- 213.Kong L, Zuo R, Wang M, et al. Silencing MicroRNA-137-3p, which targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int J Biol Sci 2020; 16: 655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Batool F, Özçelik H, Stutz C, et al. Modulation of immune-inflammatory responses through surface modifications of biomaterials to promote bone healing and regeneration. J Tissue Eng 2021; 12: 20417314211041428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhuang XM, Zhou B, Yuan KF. Role of p53 mediated miR-23a/CXCL12 pathway in osteogenic differentiation of bone mesenchymal stem cells on nanostructured titanium surfaces. Biomed Pharmacother 2019; 112: 108649. [DOI] [PubMed] [Google Scholar]
- 216.Li Y, Wang J, Ma Y, et al. MicroRNA-15b shuttled by bone marrow mesenchymal stem cell-derived extracellular vesicles binds to WWP1 and promotes osteogenic differentiation. Arthritis Res Ther 2020; 22: 269. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 217.Liu W, Huang J, Chen F, et al. MSC-derived small extracellular vesicles overexpressing miR-20a promoted the osteointegration of porous titanium alloy by enhancing osteogenesis via targeting BAMBI. Stem Cell Res Ther 2021; 12: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Asgarpour K, Shojaei Z, Amiri F, et al. Exosomal microRNAs derived from mesenchymal stem cells: cell-to-cell messages. Cell Commun Signal 2020; 18: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Huang Y, Xu Y, Feng S, et al. miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/β-catenin signaling pathway. Exp Mol Med 2021; 53: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xu T, Luo Y, Wang J, et al. Exosomal miRNA-128-3p from mesenchymal stem cells of aged rats regulates osteogenesis and bone fracture healing by targeting Smad5. J Nanobiotechnol 2020; 18: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li Y, Wang J, Ma Y, et al. miR-101-loaded exosomes secreted by bone marrow mesenchymal stem cells requires the FBXW7/HIF1α/FOXP3 axis, facilitating osteogenic differentiation. J Cell Physiol 2021; 236: 4258–4272. [DOI] [PubMed] [Google Scholar]
- 222.Chen S, Tang Y, Liu Y, et al. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif 2019; 52: e12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liao W, Ning Y, Xu HJ, et al. BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin Sci 2019; 133: 1955–1975. [DOI] [PubMed] [Google Scholar]
- 224.Yang W, Zhu W, Yang Y, et al. Exosomal miR-100-5p inhibits osteogenesis of hBMSCs and angiogenesis of HUVECs by suppressing the BMPR2/Smad1/5/9 signalling pathway. Stem Cell Res Ther 2021; 12: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sun Z, Wu F, Yang Y, et al. MiR-144-3p inhibits BMSC proliferation and osteogenic differentiation via targeting FZD4 in steroid-associated osteonecrosis. Curr Pharm Des 2019; 25: 4806–4812. [DOI] [PubMed] [Google Scholar]
- 226.Jiang C, Xia W, Wu T, et al. Inhibition of microRNA-222 up-regulates TIMP3 to promotes osteogenic differentiation of MSCs from fracture rats with type 2 diabetes mellitus. J Cell Mol Med 2020; 24: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhai Z, Chen W, Hu Q, et al. High glucose inhibits osteogenic differentiation of bone marrow mesenchymal stem cells via regulating miR-493-5p/ZEB2 signalling. J Biochem 2020; 167: 613–621. [DOI] [PubMed] [Google Scholar]
- 228.Gan K, Dong GH, Wang N, et al. miR-221-3p and miR-222-3p downregulation promoted osteogenic differentiation of bone marrow mesenchyme stem cells through IGF-1/ERK pathway under high glucose condition. Diabetes Res Clin Pract 2020; 167: 108121. [DOI] [PubMed] [Google Scholar]
- 229.Wang CG, Hu YH, Su SL, et al. LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/β-catenin signaling pathway. Exp Mol Med 2020; 52: 1310–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhang J, Tao Z, Wang Y. Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int J Mol Med 2018; 41: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Weng W, Di S, Xing S, et al. Long non-coding RNA DANCR modulates osteogenic differentiation by regulating the miR-1301-3p/PROX1 axis. Mol Cell Biochem 2021; 476: 2503–2512. [DOI] [PubMed] [Google Scholar]
- 232.Zheng C, Bai C, Sun Q, et al. Long noncoding RNA XIST regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting miR-9-5p. Mech Dev 2020; 162: 103612. [DOI] [PubMed] [Google Scholar]
- 233.Yu L, Qu H, Yu Y, et al. LncRNA-PCAT1 targeting miR-145-5p promotes TLR4-associated osteogenic differentiation of adipose-derived stem cells. J Cell Mol Med 2018; 22: 6134–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Asila A, Yang X, Kaisaer Y, et al. SNHG16/miR-485-5p/BMP7 axis modulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. J Gene Med 2021; 23: e3296. [DOI] [PubMed] [Google Scholar]
- 235.Zhang Y, Chen B, Li D, et al. LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathol Res Pract 2019; 215: 525–531. [DOI] [PubMed] [Google Scholar]
- 236.Yang JJ, Peng WX, Zhang MB. LncRNA KCNQ1OT1 promotes osteogenic differentiation via miR-205-5p/RICTOR axis. Exp Cell Res 2022; 415: 113119. [DOI] [PubMed] [Google Scholar]
- 237.Xu Y, Xin R, Sun H, et al. Long non-coding RNAs LOC100126784 and POM121L9P derived from bone marrow mesenchymal stem cells enhance osteogenic differentiation via the miR-503-5p/SORBS1 Axis. Front Cell Dev Biol 2021; 9: 723759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang X, Zhao D, Zhu Y, et al. Long non-coding RNA GAS5 promotes osteogenic differentiation of bone marrow mesenchymal stem cells by regulating the miR-135a-5p/FOXO1 pathway. Mol Cell Endocrinol 2019; 496: 110534. [DOI] [PubMed] [Google Scholar]
- 239.Li G, Yun X, Ye K, et al. Long non-coding RNA-H19 stimulates osteogenic differentiation of bone marrow mesenchymal stem cells via the microRNA-149/SDF-1 axis. J Cell Mol Med 2020; 24: 4944–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Han Y, Yang Q, Huang Y, et al. Long non-coding RNA SNHG5 promotes the osteogenic differentiation of bone marrow mesenchymal stem cells via the miR-212-3p/GDF5/SMAD pathway. Stem Cell Res Ther 2022; 13: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wu R, Ruan J, Sun Y, et al. Long non-coding RNA HIF1A-AS2 facilitates adipose-derived stem cells (ASCs) osteogenic differentiation through miR-665/IL6 axis via PI3K/Akt signaling pathway. Stem Cell Res Ther 2018; 9: 348. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 242.Li J, Zhuang H, Wang Z, et al. lncRNAs MALAT1 and LINC00657 upstream to miR-214-3p/BMP2 regulate osteogenic differentiation of human mesenchymal stem cells. Mol Biol Rep 2022; 49: 6847–6857. [DOI] [PubMed] [Google Scholar]
- 243.Xiang J, Fu H, Xu Z, et al. lncRNA SNHG1 attenuates osteogenic differentiation via the miR-101/DKK1 axis in bone marrow mesenchymal stem cells. Mol Med Rep 2020; 22: 3715–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wu J, Lin T, Gao Y, et al. Long noncoding RNA ZFAS1 suppresses osteogenic differentiation of bone marrow-derived mesenchymal stem cells by upregulating miR-499-EPHA5 axis. Mol Cell Endocrinol 2022; 539: 111490. [DOI] [PubMed] [Google Scholar]
- 245.Song Y, Zhang H, Song Z, et al. Long noncoding RNA GAS5 inhibits osteogenic differentiation through MicroRNA 382-3p/TAF1 signaling. Mol Cell Biol 2022; 42: e0054120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yang L, Li Y, Gong R, et al. The long non-coding RNA-ORLNC1 regulates bone mass by directing mesenchymal stem cell fate. Mol Ther 2019; 27: 394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhao QH, Wang SG, Liu SX, et al. PPARγ forms a bridge between DNA methylation and histone acetylation at the C/EBPα gene promoter to regulate the balance between osteogenesis and adipogenesis of bone marrow stromal cells. FEBS J 2013; 280: 5801–5814. [DOI] [PubMed] [Google Scholar]
- 248.Lei H, He M, He X, et al. METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am J Transl Res 2021; 13: 4376–4388. [PMC free article] [PubMed] [Google Scholar]
- 249.Liu S, Liu D, Chen C, et al. MSC transplantation improves osteopenia via epigenetic regulation of Notch signaling in Lupus. Cell Metab 2015; 22: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shreya S, Malavika D, Priya VR, et al. Regulation of histone deacetylases by MicroRNAs in bone. Curr Protein Pept Sci 2019; 20: 356–367. [DOI] [PubMed] [Google Scholar]
- 251.Li G, An J, Han X, et al. Hypermethylation of microRNA-149 activates SDF-1/CXCR4 to promote osteogenic differentiation of mesenchymal stem cells. J Cell Physiol 2019; 234: 23485–23494. [DOI] [PubMed] [Google Scholar]
- 252.Li Y, Yang F, Gao M, et al. miR-149-3p regulates the switch between adipogenic and osteogenic differentiation of BMSCs by targeting FTO. Mol Ther Nucleic Acids 2019; 17: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhang X, Wang Y, Zhao H, et al. Extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cell promotes osteogenic differentiation via FTO inhibition. Stem Cell Res Ther 2020; 11: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Song Y, Pan Y, Wu M, et al. METTL3-Mediated lncRNA m(6)A modification in the osteogenic differentiation of human adipose-derived stem cells induced by NEL-Like 1 protein. Stem Cell Rev Rep 2021; 17: 2276–2290. [DOI] [PubMed] [Google Scholar]
- 255.Peng J, Zhan Y, Zong Y. METTL3-mediated LINC00657 promotes osteogenic differentiation of mesenchymal stem cells via miR-144-3p/BMPR1B axis. Cell Tissue Res 2022; 388: 301–312. [DOI] [PubMed] [Google Scholar]
- 256.Li H, Zhang Z, Chen Z, et al. Osteogenic growth peptide promotes osteogenic differentiation of mesenchymal stem cells mediated by LncRNA AK141205-induced upregulation of CXCL13. Biochem Biophys Res Commun 2015; 466: 82–88. [DOI] [PubMed] [Google Scholar]
- 257.Tang S, Xie Z, Wang P, et al. LncRNA-OG promotes the osteogenic differentiation of bone marrow-derived mesenchymal stem cells under the regulation of hnRNPK. Stem Cells 2019; 37: 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liu H, Hu L, Yu G, et al. LncRNA, PLXDC2-OT promoted the osteogenesis potentials of MSCs by inhibiting the deacetylation function of RBM6/SIRT7 complex and OSX specific isoform. Stem Cells 2021; 39: 1049–1066. [DOI] [PubMed] [Google Scholar]
- 259.Kim YS, Mikos AG. Emerging strategies in reprogramming and enhancing the fate of mesenchymal stem cells for bone and cartilage tissue engineering. J Control Release 2021; 330: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]