Abstract
The COVID-19 pandemic has necessitated the development of reliable diagnostic methods for accurately detecting the novel coronavirus and its variants. Deep learning (DL) techniques have shown promising potential as screening tools for COVID-19 detection. In this study, we explore the realistic development of DL-driven COVID-19 detection methods and focus on the fully automatic framework using available resources, which can effectively investigate various coronavirus variants through modalities. We conducted an exploration and comparison of several diagnostic techniques that are widely used and globally validated for the detection of COVID-19. Furthermore, we explore review-based studies that provide detailed information on synergistic medicine combinations for the treatment of COVID-19. We recommend DL methods that effectively reduce time, cost, and complexity, providing valuable guidance for utilizing available synergistic combinations in clinical and research settings. This study also highlights the implication of innovative diagnostic technical and instrumental strategies, exploring public datasets, and investigating synergistic medicines using optimised DL rules. By summarizing these findings, we aim to assist future researchers in their endeavours by providing a comprehensive overview of the implication of DL techniques in COVID-19 detection and treatment. Integrating DL methods with various diagnostic approaches holds great promise in improving the accuracy and efficiency of COVID-19 diagnostics, thus contributing to effective control and management of the ongoing pandemic.
Keywords: COVID-19, SARS-CoV2, Coronavirus variants, Diagnostic methods, Deep learning, Machine learning, Synergistic medicine
Graphical abstract
List of abbreviations
Abbreviation Definition
- DL
Deep Learning
- ML
Machine Learning
- AI
Artificial Intelligence
- COVNet
Convolutional
- CCN
Convolutional Neural Network
- DNN
Deep Neural Network
- LSTM
Long Short-Term Memory
- GAN
Generic Access Network
- COVID-19
Coronavirus Disease 2019
- ELISA
Enzyme-Linked Immunosorbent Assay
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus
- NAT
Network Address Translation
- CPU
Central Processing Unit
- PCR
Polymerase Chain Reaction
- ELM
Extreme Learning Machines
- RT-LAMP
Real-Time Loop-Mediated Isothermal Amplification
- GAN
Generative Adversarial Network
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- CXR
Chest X-Rays
- FDA
Food and Drug Administration
Abbreviation Definition
- SERS
Surface-enhanced Raman spectroscopy
- BT
Biosensor Tests
- NGS
Next-Generation Sequencing
- FAT
Fast Antigen Test
- WHO
World Health Organization
- MRI
Magnetic Resonance Imaging
- CT-Scans
Computed Tomography Scans
- PET
Positron Emission Tomography
- ANN
Artificial Neuron Network
- DTI
Deep Learning Identifies
- GCNs
Generic Code Numbers
- LFAs
Lateral Flow Assays
- NAATs
Nucleic Acid Amplification Tests
- LAMP
Loop-Mediated Isothermal Amplification
- RDTs
Rapid Diagnostic Tests
- RNN
Recurrent Neural Networks
- RT-PCR
Reverse Transcription Polymerase Chain Reaction
- CADD
Computer-Aided Drafting and Design
- EUA
Emergency Use Authorization
1. Introduction
The COVID-19 pandemic caused by the novel coronavirus has been a significant public health crisis since December 2019 [1]. The COVID-19 pandemic has significantly impacted the world, affecting millions of lives and causing widespread social and economic disruptions [2]. The World Health Organization (WHO) declared it a global health emergency in January 2020 [3]. COVID-19 patients typically experience respiratory symptoms like fever, cough, lung damage, and other symptoms such as myalgia, diarrhea, and fatigue [4,5]. In severe cases, pneumonia can lead to organ failure and death. Coronaviruses are a group of large, enveloped RNA viruses that cause significant human and animal diseases [6,7]. One of the key challenges in addressing this pandemic is the development of effective diagnostic methods for COVID-19 and its variants [8]. Deep learning (DL), a rapidly evolving field of artificial intelligence, has shown promising results in various applications, including medical diagnosis [9]. In recent years, DL methods have been employed to diagnose COVID-19 and its variants and identify synergistic medicine combinations for treating the disease [10,11].
The rapid spread of SARS-CoV-2 among humans has caused a surge in COVID-19 cases, posing a severe threat to the global economy and health [12]. Vaccines and antiviral drugs are urgently needed to combat this deadly disease, but their development can take months if not years [13]. Advanced technological methods can be used to control the outbreak [14], and many screening techniques have been developed to identify patients infected with COVID-19 and its mutants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.). However, the appropriate use of diagnostic tests still requires clarification, depending on patients' medical history or the examination's goal.
To effectively combat the COVID-19 outbreak, various tools, methodologies, and critical approaches are required [15]. Monitoring and testing methods are also necessary to detect the virus and its mutant variants. Traditional detection methods such as chest X-rays, PCR, and serologic assays have been refined to cater to COVID-19 and its variants [16]. Physical diagnostic tools based on biosensors have been developed, with electrochemical biosensors being the most popular and considered the first line of defence against COVID-19 [17]. In addition, artificial intelligence (AI) can play a critical role in combatting COVID-19 due to its potential advantages [18]. Machine learning (ML) and deep learning (DL) techniques are utilized to process vast datasets [19], while AI-centric technology can complement current conventional technologies to address global COVID-19 issues in healthcare systems. However, the effectiveness of AI technologies during the pandemic depends on human effort and collaboration, and the successful implementation of AI-based systems is subject to their codes and potential challenges.
The accurate diagnosis of COVID-19 is crucial in controlling the spread of the disease, and it requires various laboratory techniques [20]. However, these techniques can pose significant challenges that must be addressed to ensure consistent and reliable test results. Proper specimen collection, timely analysis, and adherence to safety measures in the laboratory are essential for achieving accurate diagnosis while ensuring the safety of laboratory personnel. Fig. 1 serves as a visual representation of the various laboratory techniques employed in diagnosing COVID-19, such as reverse transcription polymerase chain reaction (RT-PCR),enzyme-linked immunosorbent assay(ELISA), and other molecular diagnostic tools. The figure also highlights the utilization of DL strategies for COVID-19 diagnosis and explores the frameworks employed for their implementation.
Fig. 1.
AI-Based Deep Learning Models for Accurate Prediction and Monitoring of COVID-19 and its Variants.
1.1. Major research gaps
One of the leading research gaps in deep learning for diagnosing COVID-19, its variants, and synergistic medicine combinations is the lack of large-scale datasets. Deep learning models require vast amounts of data for practical training. Still, given that the pandemic is relatively new, there is a shortage of large-scale datasets for this purpose. Additionally, most studies on deep learning for COVID-19 diagnosis focus solely on the original strain of the virus, ignoring the different variants that have emerged. As such, there is a need for further studies that investigate the effectiveness of deep learning models in diagnosing COVID-19 variants and predicting the efficacy of different medicine combinations. Finally, the interpretability of deep learning models is limited, as they are often considered black boxes, making it difficult for doctors to understand how they make their diagnoses.
1.2. Motivations
The utilization of DL models has demonstrated their potential to enhance the accuracy and speed of diagnosing COVID-19 from medical images, such as chest X-rays and CT scans. As the pandemic continues to spread, timely and precise diagnosis is crucial for effectively managing the disease. DL models can potentially identify new variants and synergistic medicine combinations, which can be deployed to combat the disease. Given the emergence of new variants and the need for efficient and precise methods to identify these variants, along with predicting the efficacy of different medicine combinations, there is a growing demand for DL models to fill this gap. DL models can potentially reduce the burden on healthcare systems by facilitating early and accurate diagnosis and management of the disease. The COVID-19 pandemic has caused significant strain on healthcare systems worldwide, and any interventions that can alleviate this burden are highly sought after.
1.3. Research objectives
This review explores the impact of recent technological advancements in biosensors on the accuracy and speed of COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda Delta, Zeta and Theta etc.) diagnosis. We consider various diagnostic techniques, management strategies, and the efficiency of using deep learning (DL) to tackle the challenges and complexities of treating COVID-19 patients. We also evaluate several research studies that offer valuable insights into the potential of using synergistic drug combinations for COVID-19 and its variant treatment, where DL techniques can reduce time, cost, and complexity and provide reliable guidance for the appropriate use of synergistic drug combinations in clinical and research settings. Our study also highlights the implications of the latest diagnostic technical and instrumental technologies. It explores the use of open datasets and synergistic medicine research to aid future prospectives. The review aims to inform future research efforts and improve COVID-19 diagnosis and treatment by examining these implications. Our study offers a comprehensive and informative analysis of COVID-19 diagnosis methods, their challenges, and the application of deep learning strategies. The study sheds light on the latest advancements in COVID-19 diagnosis and treatment, which can aid healthcare professionals and researchers in their efforts to combat this global health crisis.
2. Diagnosis of COVID-19
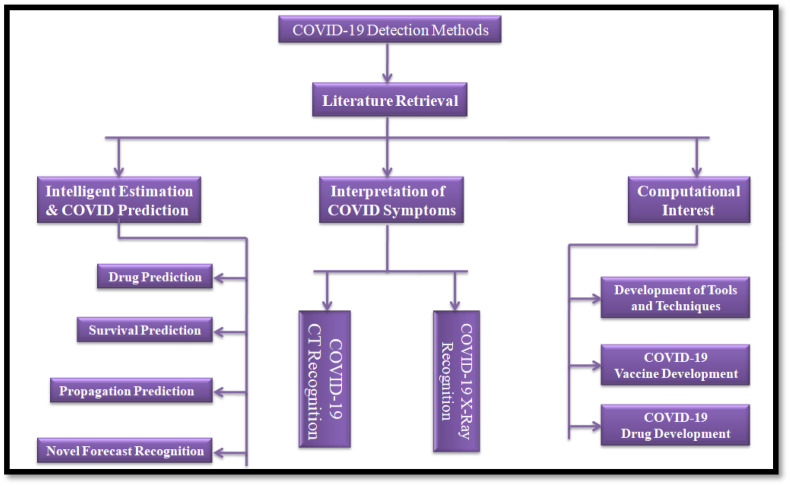
Toward point-of-care diagnostics of COVID-19 and its variants (Alpha, beta, gamma, omicron etc.), identification protocols are commonly founded on the pathogenic movement history to the influenced territories, just as an examination of their clinical manifestations and some further investigations [21]. Retrospective investigation reveals that the clinical symptoms of COVID-19 screening and containment techniques are remarkably unusual and fundamentally similar to legionnaires' diseases and other respiratory viral pneumonia, which are not comparable for immunocompetent people [22]. Fast and touchy determination is as yet not accessible, albeit some indicative techniques are accessible for infection location, each with fluctuating degrees of particularity and dependent on interesting objective atoms or, then again, numerous SARS-CoV-2. These strategies exploit neurotic changes in the patient's organs through imaging, for example, figured tomography or viral nucleic corrosive sort RT-PCR utilizing at least one quality, or entire genome sequencing of the up-and-coming age of immunological particles delivered by the patient or by the infection in the patient's body.
Tests dependent on the antigen-counter-acting agent reaction, for example, ELISA and utilizing all of these symptomatic techniques have their points of interest and weaknesses [23]. A few strategies have been set up and are considered Gold Standard techniques that can likewise be duplicated for this new infection. In contrast, others are a work in progress and assessment for the finding of this infection. Then again, there are different strategies; innovations/gadgets have likewise been grown; however, pending endorsement and proposed use in COVID-19 techniques are depicted here.
The detailed view of COVID-19 and its variants (Alpha, Beta, Gamma, Delta, Omicrons), innovative methods, identifications, utilizations of database search information and extractions procedures, and synergistic drug combinations in clinical and research settings are mentioned in the context of DL perspective (COVNet, CCN, DNN, LSTM, GAN, VGGc etc.) to examine and relate the outcomes are mentioned in Fig. 2 .
Fig. 2.
DL approaches for exploration of COVID-19 variant analysis and treatment optimization.
3. Databases and search strategy
We used electronic databases, including PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase (https://www.embase.com/), Cochrane Central Register of Controlled Trials (https://www.cochranelibrary.com/central/about-central), Chinese Biomedical Literature Database (http://allie.dbcls.jp/pair/CBM;Chinese+BioMedical+Disc.html), China National Knowledge Infrastructure (https://en.cnki.com.cn/), Chinese Science and Technology Periodical Database (http://www.nlc.cn/newen/periodicals/) and Wanfang database (http://www.wanfangdata.com/) are utilizing catchphrase mixes for potency and timing of antiviral therapy possible updating and identifications, retrieve terms from such databases as per query “new corona-virus [24], COVID-19 [25], 2019-nCoV [26], COVID-2019 pneumonia [26], SARS-CoV-2 variants [27], Alpha, beta, gamma, delta, omicron etc., conventional synergistic medication [28], and synergistic Medicine”.
Moreover, in the field of COVID-19 research, several practical and theoretical databases have been utilized to predict novel relationships and understand the intricacies of the disease and its variants. For instance, CovInter, as described by Ref. [29], is an interactive database specifically designed to explore the intricate connections between coronavirus RNAs and host proteins. The study conducted by Ref. [30], utilized multiple databases to investigate RNA-RNA interactions between SARS-CoV-2 and viral development, providing valuable insights into the exploration of COVID-19 infection. Additionally, the Therapeutic Target Database, as highlighted by Ref. [31], has played a pivotal role in facilitating target-oriented drug discovery efforts for COVID-19.
To facilitate the efficient exploration of electronic databases and retrieve comprehensive information related to COVID-19, its variants, and therapeutic approaches in a user-friendly manner, a comprehensive PubMed search methodology and comparative systems were also employed [32].By analyzing patient details, inquiries, and examinations, our aim to evaluate the connection between symptoms and persistent diseases as per [[29], [30], [31]]. To verify this relationship, individuals and examiners can visually examine encoded symbols obtained from a wide range of databases. These databases include gathering procedures, scholarly recognition initiatives, testing libraries, and national registration sites, collectively providing a diverse and extensive information collection.
4. Computational frameworks for COVID-19 diagnostics and therapeutics
Artificial intelligence (AI) and machine learning (ML) have emerged as crucial tools in combating the COVID-19 pandemic. These technologies have the potential to process vast amounts of data rapidly and accurately, offering valuable insights and complementing traditional methods in healthcare. However, it is to note that AI interventions' success relies heavily on human input and collaboration. The effectiveness of AI techniques is contingent upon the expertise and skills of the individuals who develop and program the AI-based systems. Therefore, the synergy between human knowledge and AI capabilities is crucial for achieving desired outcomes in the fight against COVID-19 [33].To achieve proficiency in cross-functional applications, including image analysis, data retrieval, and protein structure prediction, combining intensive training in medicine and wellness is essential. This approach promotes cutting-edge practices and improves supervised learning methods for accurate COVID-19 detection. DL has significantly impacted the COVID-19 epidemic, creating new research opportunities and applications for machine perception, semantic analysis, and medicine. These techniques can enhance accurate diagnosis, protein structure prediction, and drug repurposing [34,35]. However, the interpretation of computational models and conflicting statistical properties can pose challenges.
Supervised, unsupervised, and semi-supervised learning can all be applied in the context of COVID-19 and its variant detection, as mentioned in Fig. 3 . Supervised learning trains algorithms on labelled datasets to classify new data, and it can be used to identify patterns or images associated with the disease [36]. Unsupervised learning trains algorithms on unlabeled datasets to identify clusters or patterns in the data [37], which can be helpful in identifying groups of patients with similar symptoms or risk factors. Semi-supervised learning combines both, using labelled and unlabeled data to improve model accuracy for maintaining the topological structures to solve the multiple frameworks [38]. For example, semi-supervised learning can train models on a small set of labelled COVID-19 patient data and a more extensive set of unlabeled patient data to identify patterns and improve accuracy. By leveraging these learning techniques, researchers can improve the accuracy and speed of COVID-19 detection and ultimately improve patient outcomes.
Fig. 3.
Multiple computational techniques and applications for fighting COVID-19.
The application of systems medicine approaches in identifying differentially expressed biomarkers has been valuable in exploring potential biomarker signatures. However, crucial features may be overlooked. In earlier studies [39], researchers employed specific machine learning algorithms to reduce the dimensionality of the clinical feature space and identify clinical prognostic indicators for COVID-19. The models, combined with pertinent clinical studies, are essential for readiness against the emergence or resurgence of infectious diseases. Recent endeavours have concentrated on utilizing machine learning methods to explore biomarkers and clinical marker signatures for the optimal management of COVID-19 [39]. In numerous studies with identical numbers of patients in all clinical cohorts have demonstrated that the identified markers play a fundamental role in the pathogenesis and clinical manifestations of COVID-19 [39,40]. Furthermore, the clinical biomarker signature is distinct in terms of its unique feature combination and independent of patient demographics.
Convolutional neural networks have become well-known deep learning algorithms and efficient ways for recognizing inconsistencies, anomalies, and diagnoses in chest radiography [41]. Throughout the COVID-19 pandemic, scholars use a Convolutional Neural Network to analyze appropriate COVID-19 its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) diagnoses. Studies have shown deep learning algorithms may improve CT scan pictures' specificity, sensitivity, and diagnostic efficacy [41]. DL is a feasible, efficient, and dependable method for precise COVID-19 virus detection [42]. It highlights the possibility of enhancing image properties using artificial intelligence and identifying cost-effective and reliable imaging techniques for predicting deadly infections. Multiple researchers have recently utilized deep learning for the COVID-19 virus. Jamshidi et al. [34], Minaee et al. [43], Muhammad and Hossain [44], Campos-Ferreira et al. [45], Mostafa et al. [46], Velay et al. [47] and Zhang et al. [26]have made notable contributions to the literature. Although implementing deep learning approaches significantly influences diagnosing COVID infections using image data, scholars encounter difficulties implementing solutions effectively owing to intra-class correlation. Scale Variation, Occlusion, Illumination, Background Clutter and View-Point Variation [43].
Gathering, analyzing, and combining the data like that of patients’ physical and physiological measures comprises big data. Initially, training a machine requires preparing the big data to be mined, such as medical reports, registries, images, etc. Learning data involves understanding variables and recognizing significant features like the size of data and the number of attributes describing the data. Before processing and analysis, raw data are curated and remodelled, where data is reshaped, repaired, and integrated. Scientists intervene in ML by exploring and analyzing the data to extract the most delicate structures, patterns, and characteristics [48].
As shown in Fig. 4 , DL techniques work independently of man's interferences. DL, a subset of ML, comprises several algorithms' layers for inferences upon inputting data. Yet, DL differs from ML since the system has variable data representations. DL networks operate via artificial neural network (ANN) layers; meanwhile, ML algorithms often need structured data. Supervised learning is learning a function mapping an input to output. In contrast, unsupervised learning is unique to minor human supervision, and the machine learns by seeking unknown patterns in a dataset with no previous tags [49].
Fig. 4.
ANNs in Action for Unveiling the Versatile 5-Layered Applications to track Symptom based on speedy protocols.
Meanwhile, the database server is linked via the network and securely connected to the main central processing unit (CPU). The excessive microprocessors in the database software enables the database to transfer enormous amounts of data to the mainframe computer. The following selection layer is developed via a smart ANN and could take up among the leading imaging technologies in the system [50]. When a medical professional confirms the advice provided by this layer, the third layer's images are subjected to pathological applications, visual and automatic microscopic imaging technologies, computed tomography scans (CT-Scans), magnetic resonance imaging (MRI), and positron emission tomography (PET). The fourth layer is designed for enhancing and optimizing images. A DL approach was used to structure the network to achieve a sorting network for differentiating COVID-19 from influenza-A viral pneumonia, and the conventional ResNet was used to extract features [51]. The fifth layer is designed to finally diagnose using the stored data on the system where algorithms learn via an ANN technique. DL techniques, like a convolutional neural network (CNN), could reach such targets for their nonlinear modelling capability and be extensively applied to process medical images for diagnosis [41].
4.1. Computed tomography (CT) scan
Many studies advise using computed tomography as an additional diagnostic strategy for COVID-19 pneumonia since it has a high sensitivity for diagnosis and provides results even before symptoms and clinical symptoms are based on pre-trained convolutional neural networks [52,53]. Clinical evaluation and routine clinical practices for COVID-19 patients provide wide-ranging sensitive detection paths to understand the meta-analysis accuracy of diagnostic explanations [54]. According to a recent report from Wuhan, a CT scan is significantly more sensitive than PCR to the putative SARS-CoV-2 [55]. The results concluded tha more sensitive and accurate conclusion could be reached in patients with adverse RT-qPCR reports with a combination of CT-Scan and other standard techniques such as RT-qPCR or other sensitive diagnostic tests [56]. In addition, high-resolution chest computed tomography has also been shown to be an essential tool for the early detection of SARS-CoV-2 and rapid and necessary intervention [57]. Therefore, several studies have recently used CT images of breasts and lungs to diagnose COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) [58]. Previously, typical CT images of patients infected with SARS-CoV and MERS-CoV also exhibited symptoms similar to those of COVID-19 [59]. According to these results, CT scans have proven to be an excellent diagnostic tool for screening patients with COVID-19, especially in areas with a high prevalence of a pandemic. CT scans are indicative and confirmatory tools for the detection of pathogens in the diagnosis of COVID-19 its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc) [60], and some show lacks associated with specific deficiencies, such as the inability to separate cases from other pneumonia (viral or non-viral) which may misguide for exact treatments.
4.2. Radiographic images
Graphically designed diagnostic procedures in epidemics serve a fundamental function in identifying and identifying COVID-19 instances by filtering hitting origins that, on average, produce better performance than the main radioscopy strategies [61]. Chest X-rays (CXR) &radiographs are two examples of radiographic images that extensively use the DL methodology [62]. These radiographic scans include a plethora of information, including patterns & cluster-like configurations that can be used to identify epidemics similar to COVID-19 [63].
DL algorithms can be effectively used for medical image analysis. Many DL models have shown promising results for the detection of COVID-19 its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) [64]. Numerous methods have been developed to diagnose COVID-19 in X-ray images automatically. Most of the researchers used CNNs that were already in use and built to classify actual photographs for COVID-19 identification. Natural images frequently contain enormous, well-defined structures, in contrast to COVID-19 radiography patterns, typically characterized by oblique lung lines and areas of transparency and accumulation [65].
First, the lack of and low quality of COVID-19 radiography images appears to impact the categorization method significantly [66]. Most past studies relied on datasets that, at most, contained a few hundred COVID-19 CXR graphs that had been confirmed. Poor suggestions were generated as a result of over-fitting and higher generalized errors. The currently available radiographic image reporting is preferred to transferring deep features using fully convolutional models, such as those from ResNet utilizing the ImageNet dataset, which entirely differs from the aspects of medical images [67].
4.3. Chest X-rays
The chest X-ray (CXR) is one of the essential non-invasive therapeutic adjuvants crucial in the initial examination of various lung diseases. Having professional radiologists evaluate CXR pictures to check for contagious abnormalities linked with COVID-19 can serve as a substitute diagnostic tool for identifying COVID-19 or confirm the corresponding diagnostic [68].
Deep CNNs are often utilized in image processing applications because of their predictive solid modelling capabilities. Following the target medical image analysis, CNN uses a convolutional technique to benefit from the image's structural information and automatically create attribute hierarchies [69]. Using novel concepts in CNN design has increased the application of Deep learning in medical image processing, identification, and data classification tasks. A CXR is the most popular imaging method for determining SARS-CoV-2 infection. According to a few studies, CNN is effective at interpreting COVID-19 radiography images, according to a few studiAccording to a studies, CNN is effective at interpreting COVID-19 radiography images, and their usefulness justifies further research [70].
Analyzing these small distinguishing characteristics on CXR images is challenging and needs a domain expert. Additionally, the exponential increase in infected people makes it difficult for radiologists to establish an early diagnosis, leading to severe morbidity and mortality [71]; the process of visualizing c COVID-19 and its variants through x-ray image analysis with DL working details is mentioned in Fig. 5 .
Fig. 5.
Unveiling Insights through Visual Intelligence: Harnessing DL Methods to Visualize X-ray Images for Detection of COVID-19 and its Variants.
4.4. Computational frameworks and COVID-19 therapeutics: A key approaches to drug targets
Accessible research and data swapping can help data analysts to identify potential therapeutics, and computational scientists must seek realistic criteria for their “digital illusions” before publishing computational results. The coming contemporary research procedures may dramatically minimize the number of articles while increasing the number of computer-aided, evidence-based potential antiviral medications discoveries.
Non-experts now employ computer-aided drafting and design (CADD)approaches to datasets and biological targets crucial to SARS-CoV-2 drug development since computational resources and software are more readily available [72]. Dozens of drugs have been suggested as potential COVID-19 treatments, and many of these have entered pharmacological trials with little to no supporting evidence or explanation. Several have been approved by the Food and Drug Administration (FDA), for example, remdesivir and baricitinib, but none cure the disease. Per Serval virtual analysis, even computational skills cannot replace experimental approaches for developing excellent medical ideas. In contrast, using well-processed historical empirical findings and strict numerical methodologies can allow for the rapid practical identification of potentially active compounds.
Several Deep Docking (DD) and Quantitative Structure-Activity Relationships (QSAR)hits were picked and compared with experimental results by different research groups [73], resulting in the identification of several potential therapies and the repurposing of cenicriviroc and two more drugs, among others. In the case of combinations, the AI-derived hypothesis of baricitinib as a potential COVID-19 treatment resulted in the FDA awarding an Emergency Use Authorization (EUA) for its combination with Remdesivir.
Sixteen synergistic and eight antagonistic therapeutic interactions were found utilizing data mining algorithms and QSAR [74], including the most prominent being nitazoxanide - umifenovir for synergy and remdesivir - (hydroxy)chloroquine for antagonism, and then compared with experimental results. Amodiaquine, identified as a possible anti-COVID-19 repositioning option using knowledge-mining techniques, demonstrated preliminary antiviral efficacy in CPE and titer-lowering assays and animal investigations. Given its three-week half-life, amodiaquine might be an excellent answer, especially where Remdesivir, Favipiravir, and other antivirals are unavailable.
4.5. Key deep learning techniques and applications for COVID-19
4.5.1. Predicting the outbreak
ANN-centred techniques are an option to forecast the outbreak of COVID-19 its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc). The timely data on the epidemic [75] have been collected and arranged for forecasting transmission of the infection. Recurrent Neural Network (RNN) is a type of deep learning (DL) technique used to analyze and predict time series data and overview of details we, indicated in Fig. 6 . In the context of COVID-19, RNNs can be used to forecast the transmission of the virus based on spatial and medical big data. Clockwork RNN (CW-RNN), Gated Recurrent Unit RNN (GRU RNN), and Long Short-Term Memory (LSTM) networks are all variations of RNNs that are designed to handle complex, multi-step problems.
Fig. 6.
Navigating the Pandemic Landscape for Predicting COVID-19 Transmission with RNNs: An In-Depth Comparison of Clockwork RNN, GRU RNN, and LSTM Networks using Spatial and Medical Big Data.
These networks can be used to analyze large, complex data sets and make predictions about the spread of COVID-19. Comparing these three network types, the Clockwork RNN is designed to handle long-term dependencies and is especially useful for forecasting data with long-term patterns. GRU RNN and LSTM networks, on the other hand, are designed to handle short-term dependencies and are more effective at handling sequences with short-term patterns. Overall, the use of RNNs and its variants in analyzing and predicting the transmission of COVID-19 via spatial and medical big data provides a powerful tool for monitoring the spread of the virus and informing public health decision-making [75].
The RNN, also known as Feedback Network or Auto Associative Network (AAN), is a type of ANNs where a guided cycle is formed via linking units. RNNs are promising in providing outputs in many automatic and DL functions [76] especially analyzing qualitative inputs like locations or countries. It will likely upgrade the model with timely RNN data and learning capabilities. Such an ANN model could predict the viral epidemiological model in various locations with better speed and precision of recognition and classification [77].
4.5.2. Detecting the COVID-19 infectionits variants
Although there has not been enough experience either for technical experts or radiologists in responding to COVID-19, radiology scans, such as digital photography (DR) or computed tomography (CT), have been functioning well in COVID-19 its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) mentioned in Fig. 7 screening, diagnosing, and evaluating progress [78]. In the middle of this epidemic, a negative RT-PCR but a positive CT one might show COVID-19 infection showing the significance of fast detecting and addressing the disease clinically and socially [55]. According to the structure, the image sorting framework distinguishes various disorders. The framework employs relative distance-from-edge as an additional weight to learn the estimated position data of the patch on lung imaging [79]. However, radiologists adept at extracting diagnostic information from images in structured tags for training ML models bear the total burden of acquiring more medical images for ML applications. Expertly reading diagnostic imaging reports could mainly deal with scope, syntax, and specific terms required to translate the photos [80]. Ultimately, DL frameworks could be a valuable diagnostic tool through efficiently screening COVID-19 patients early [44].
Fig. 7.
Unraveling the mysteries of zoonotic origin delving into the intricacies of COVID-19 variants.
COVID-19 infection might destroy the respiratory epithelial cells. Lately, a study has displayed that visualizing and detecting COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.), which might be undetectable through conventional techniques, could be done via the preliminary disseminated airway discharges onto respiratory epithelial cells coupled with whole-genome sequencing and transmission electron microscopy of culture supernatant [81]. Fig. 8 shows the implemented process of recommended Generative Adversarial Network (GAN). GAN is a unique neural network framework where two networks are trained simultaneously, one produces images, and the other classifies them [82] via efficient modelling of the potential dissemination of the training data. GANs have been used to translate image-to-image [83], and fragmenting [84], among other applications [85].
Fig. 8.
Revolutionizing COVID-19 analysis: Harnessing the power of generative adversarial networks (GANs) to efficiently model transmission and dissemination.
4.5.3. Predicting complications
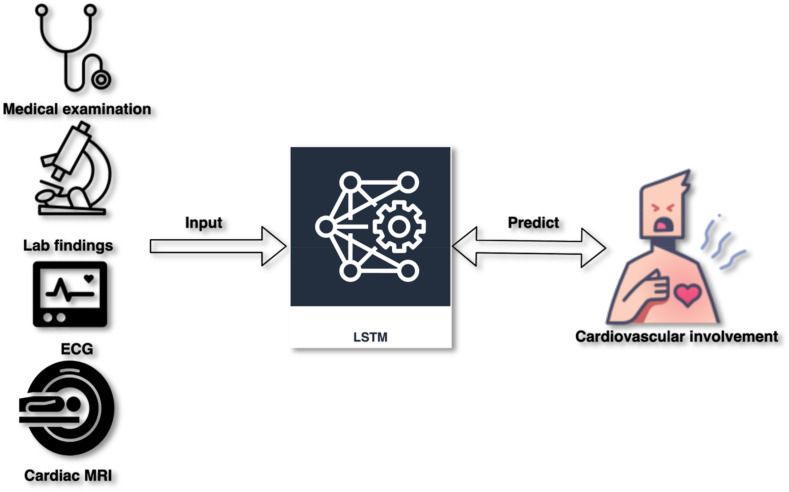
Lately, the adversarial training plan has gained significant interest for its capabilities in reversing domain shift and producing samples of new images, especially in text-to-image generation [86], super-resolution [87], and image-to-image translation [88]. COVID-19 might lead to acute myocarditis. Long/Short Term Memory (LSTM) network might estimate COVID-19-associated cardiovascular involvement, as shown in Fig. 9 .
Fig. 9.
Unveiling the future empowerment of Leveraging LSTM for prognosticating cardiovascular involvement in COVID-19.
In feed-forward neural networks, signals could move in one forward direction from the input to the output [89]. RNN enables signs to move in both directions, allowing loops in the network and intrinsic links between hidden units [90]. The RNN works out the sequential inputs via a recurrent hidden state where activation in every phase relies on the preceding one; thus, the network shows dynamic temporal behavior [91].
4.6. Molecular assays techniques for COVID-19 detection
4.6.1. RT-PCR
For example, RT-PCR depends on its capacity to enhance a limited quantity of hereditary viral material and is viewed as the best quality level for SARS-CoV-2 infection distinguishing proof [92]. Presently, RT-PCR is used to measure epidemiological inference ordinarily upper respiratory tract using segregating sites [93] and provides detailed information about the quantification of SARS-CoV-2 estimated rate of the global pattern of the virus [94]. Moreover, a few investigations have been performed on serum, faecal or visual emissions using the vitality of emerging technologies [95]. As of late, the Rutgers Clinical Genomics Laboratory [96] built up an RT-PCR test Combo Kit [97] for clinical diagnostics from RNA of patients’ blood products [98].Utilizing quick spit tests gathered and less agonizing than other examining techniques diminishes the hazard to parental figures and may consider more significant volume tests [97].
RT-PCR starts by in vitro transforming viral genomic RNA into DNA using RNA-subordinate SARS-CoV-2 to offer pharmacological elements in testing for epidemiology identifications [99]. This response depends on the little DNA arrangement of a molecular logic system intended to explicitly perceive correlative successions in the viral RNA genome and converse transcriptase to produce a short complementary DNA (cDNA) duplicate in viral RNA [100]. Constant RT-PCR and ongoing DNA enhancement are checked as the PCR response advances to detect SARS-CoV-2 VOC monitoring and its variants like alpha, beta, gamma and omicrons B1 and B2 [101]. According to the TaqMan measures, this is finished utilizing a fluorescent colour or DNA test explicit for a grouping marked with a fluorescent particle and a blurring atom [101]. The enhancement process is repeated using an automated system for around 40 cycles or until the viral cDNA can be identified, typically by fluorescent or electrical indicator.
RT-PCR is generally a couple-of-step technique for evaluating viral concentration from effective samples [102]. One-advance constant RT-PCR runs the entire RT-PCR reaction on a single cylinder that contains the essential preliminaries [102]. The two-advance continuous RT-PCR method uses multiple cylinders to conduct independent converse translation and intensification reactions. Yet, it is more adaptable and effective than the one-advance method [102], fewer raw ingredients are needed, and cDNA can be stored for later objective examination. The one-advance technique is frequently chosen for SARS-CoV-2 detection because it is swiftly balanced, comprises limited example handling, reduces track time, lowers the risk of error and contamination, and continuously combines RT and PCR stages. The majority of molecular diagnostic tests to date have targeted different SARS-CoV-2 genomic regions with real-time RT-PCR technologies, such as the ORF1b or ORF8 sections, the R-dependent nucleocapsid (N), S protein RNA polymerase (RdRP), or envelope (E) genes.
The first COVID-19 RT-PCR symptomatic tests began included COVID-19 RT-PCR diagnostic disease control and prevention (CDC) [103,104]. TaqPath COVID-19 Combo pack (ThermoFisher-Applied Biosystems), Allplex 2019-nCoV Assay and SARS-CoV - 2.RT-PCR tests are continually advancing with improved discovery strategies and progressively robotized techniques. For instance, the ePlex SARS-CoV-2 test created by GenMark Diagnostics [105] uses the ePlex instrument, “The real example answer for reacting”, to recognize SARS-CoV-2 in nasopharyngeal smears [106]. Each test cartridge contains reagents for attractive extraction of intense stage viral RNA, enhancement, and cDNA location consolidating electro-wetting and GenMarkeSensor innovation. Target DNA is blended in with iron-marked sign tests integral to explicit targets. The focus on DNA hybridizes to the sign and catches tests, the two of which are appended to gold-plated anodes. The proximity of an object is determined using voltammetry signals to detect COVID-19 its variants (Alpha, beta, gamma, omicron) [107].
Although RT-PCR is the most generally utilized technique for distinguishing SARS-CoV-2 diseases, it has the inconvenience of requiring significant level lab staff from costly lab instruments, which can take days to create results [108]. Subsequently, a few organizations and labs around the globe are attempting to improve the effectiveness and speed of RT-PCR advancements and to create different innovations.
4.6.2. Isothermal nucleic acid amplification
RT-PCR requires several temperature changes for each cycle, incorporating propelled warm gear [109,110]. Isothermal nucleic acid amplification is an optional method allowing consistent temperature enhancement while eliminating the need for a heated cyclize. As a result of this guideline, numerous strategies have been developed.
4.6.3. Reverse transcription loop-mediated isothermal amplification (RT-LAMP)
RT-LAMP was created as a quick and practical option in contrast to SARS-CoV-2. RT-LAMP requires four objective quality/area explicit preliminaries to improve affectability and consolidate LAMP with an opposite translation venture to empower the discovery of RNA. The response can be tested incrementally by measuring turbidity or fluorescence with varying colors. Because continuous RT-LAMP diagnostics require a warm-up and visual evaluation, its simplicity and applicability make it an attractive candidate for infection detection [111].
4.6.4. Transcription-mediated amplification (TMA)
TMA is a licensed isothermal intensification method that exploits a canister produced during retrovirus replication to significantly enhance specific RNA or DNA regions more effectively than RT-PCR. It uses retroviral reverse transcriptase and T7 RNA polymerase to differentiate nucleic acids from various microbes. Per this criterion, the Panther Fusion stage from Hologic may perform both RT-PCR and TMA [112]. The Panther combination stage stands apart for its high test effectiveness (up to 1,000 tests in 24 h) and its capacity to control other regular respiratory infections whose manifestations cover COVID-19 utilizing a similar example and a similar patient assortment bottle [113].
The fundamental advancement includes hybridizing the target viral RNA to a specific catch test and an additional oligonucleotide containing a T7, and it is captured in attractive micro-particles [114]. The connected RNA target hybridized to the T7 advertiser preliminary is then deciphered into a corresponding cDNA. RNase H-turn around transcriptase action at that point severs the objective RNA clone from half and half twofold RNA cDNA, leaving a solitary abandoned cDNA, which incorporates the T7 advertiser. An extra preliminary is utilized to produce two-fold abandoned DNA, which is then translated into RNA enhancers with T7 RNA polymerase [115]. These new RNA enhancers, at that point, go into the TMA procedure with the goal that this exponential intensification procedure can create billions of RPL in under 60 min. The discovery procedure includes the utilization of single-abandoned nucleic corrosive flares that explicitly hybridize to ongoing RNA amplicon. Every focal point is associated with a fluorescent light and a fire quencher. When the focal point is hybridisedwith a RNA amplicon, the fluorophore can transmit a sign upon excitation [116].
4.6.5. CRISPR-based tests or assay
The typically appropriated short palindromic rehashes (CRISPR) speak to a group of corrosive nucleic successions in prokaryotic creatures, such as microorganisms [117]. Many bacterial proteins, called compounds related to CRISPR, delineated by Cas9, Cas12, and Cas13, can be perceived and cut these arrangements. Certain chemicals of the Cas12 and Cas13 families can be modified to target and cut the viral RNA arrangements [118].
Two organizations, Mammoth Biosciences and Sherlock Biosciences, made by spearheading CRISPR scientists, are freely investigating the chance of utilizing the CRISPR quality-altering technique to distinguish SARS-CoV-2. The SHERLOCK technique created by Sherlock Biosciences uses Cas13, which can cut correspondent RNA arrangements because of the control of SARS-CoV-2 precise RNA actuation [119]. The DETECTOR trial of Mammoth Biosciences depends on the cleavage of correspondent RNA by Cas12a to explicitly identify the viral RNA successions of E and N qualities, trailed by isothermal enhancement of the objective, bringing about visual perusing with a fluorophore. These CRISPR-based techniques don't require complicated equipment and can be employed with paper strips to detect the presence of SARS-CoV-2 infection. These tests are both inexpensive and should be completed in under 60 min. These tests have enormous potential for determining the goal of care [117].
4.6.6. SHERLOCK
SHERLOCK is a different molecular detection approach, most notably the SHERLOCK test; SARS-CoV-2 might be identified using this [120]. SHERLOCK combines nucleic acid amplification with CRISPR/Cas enzymology to identify the target nucleic acid. Guide RNA integrates to Cas nuclease by CRISPR-Cas devices. CRISPR RNA, which correlates to the target RNA, and TRACR RNA operate as a scaffolding for the Cas nucleus in the guide RNA that recognizes the target sequence are the two primary components of the guide RNA. The programmable endonuclease function of CRISPR-Cas technology permits the identification of nucleic acids with exceptional accuracy and sensitivity [121].
In a nutshell, Cas13 or Cas12 nuclease is controlled by CRISPR RNA and its nonspecific endonuclease activity is activated using CRISPR RNA binding to a target gene, cleaving adjacent RNAs, including the reporter RNA, creating a signal and framework for sensitive and specific detection of RNA [122,123]. In the SHERLOCK detection technology, recombinase polymerase amplification duplicates the target RNA. Cas13a ribonuclease, a tiny fluorophore-quenching probe, and a guide RNA to connect the target gene are introduced in that order after amplification. When Cas13a ribonuclease returns to an active state after cleaving its target RNA, it attaches to and digests more RNA without specificity. The target RNA is broken down in the target gene presence, and then Cas13a′s non-specific activity breaks down for the fluorescent signal production by the fluorophore-quenching probe [124]. SHERLOCK can be run in a single or dual-step procedure, depending on the need for data delivery speed and accuracy. The dual-step reaction takes 30–60 min and has a sensitivity level of femtomolar. In contrast, the single-step reaction takes 15–30 min and has a sensitive range of femtomolar to attomole [121].
Cas13 is inactive if the target RNA has two or more errors; hence SHERLOCK can discriminate among SARS-CoV-2 and other viruses. Transporting freeze-dried items is inexpensive, which is another advantage. As there are no predesigned SHERLOCK tests, constructing the reaction mixture and nucleotides is complex, and nucleic acid amplification is another issue [120]. CRISPR technology and a graphene-based FET were coupled to create the CRISPR-Chip, enabling rapid and sensitive nucleic acid detection and fluorescent protein gene [121]. By functionalizing graphene with a catalytic CRISPR group, the electrical properties of the biosensor are altered, and an electrical signal is generated. The biosensor identified the target gene with a sensitivity of 1.7 FM in 15 min. SHERLOCK may identify SARS-CoV-2 as this 1-h treatment offers rapid COVID-19 diagnosis [120,125].
4.7. Immunoassays
Although RT-PCR-based viral RNA location has been generally utilized in the analysis of COVID-19its variants (Alpha, beta, gamma, omicron), it can't be used to screen the advancement of illness arranges and can't be applied to comprehensive distinguishing proof of past contamination and invulnerability. Immunoassays are biochemical tests that use antigens or antibodies to identify the presence and quantity of a particular biomarker. The detection process relies on a competitive affinity response between the target biomarker (antigen or antibody) and other molecules in the sample for restricted binding sites given by the immobilized capture reagent (antigen or antigen). Antigen tests evaluate the presence of the SARS-CoV-2 antigen, whereas serology tests measure the anti-SARS-coV-2 antibody created to combat SARS-coV-2 [126]. Numerous researchers and manufacturers of medical devices have designed and tested COVID-19 immunoassays to detect the presence of antigens or antibodies in COVID-19 patients [127].
4.7.1. Serological tests
Serological tests are characterized as an examination of serum or blood plasma. They have been precisely reached to incorporate testing of spit, sputum and other body liquids for the nearness of immunoglobulin M (IgM) antibodies [128]. Furthermore, the immunoglobulin G (IgG) in this study plays an essential role in the study of disease transmission and the advancement of immunizations. It assesses the pathways of the immunizer reaction on both short (days to weeks) and long (years or lasting), as well as the abundance and a good variety of antibodies. IgMs are first detected in serum a few days and hours after infection, followed by a progression to IgG [129]. Along these lines, IgM can be a pointer of beginning phase contamination, and IgG can be a marker of current or past disease. IgG can likewise be utilized to recommend the nearness of many insusceptible contaminations. As of late, the complexity and affectability of immunoassays have expanded not just for the location of the antibodies themselves but also for the utilization of antibodies (mostly monoclonal antibodies) for the identification of antigens obtained from microbes. These tests have tremendous potential for the study of disease transmission of COVID-19 [130]. Yet, the test outcomes can be influenced by, in any event, three circumstances: (1) a subset of individuals with a positive outcome from atomic hereditary testing for SARS-CoV-2 contaminations are harmful because of deferred counteracting agent creation after contamination, (2) people might be HIV constructive yet contrary for sub-atomic hereditary test outcomes reflecting prior and milder disease freedom, and (3) affectability and explicitness of the examines. The latter issue is critical since even a tiny percentage of false favourable outcomes due to inadequate specificity can lead to the deceptive predicted pervasiveness of antibodies in a particular community, negatively affecting financial decisions and genuine belief in the outcomes [131,132].
Assurance of presentation to SARS-CoV-2 is profoundly subject to the recognition of IgM or IgG antibodies explicit for different viral antigens, including, however not constrained to, the nail glycoprotein (S1 and S2 subunits, receptor restricting space) and the atomic protein [133]. These philosophies incorporate the standard catalyst-connected immune-sorbent test (ELISA), immune-chromatographic investigation of the parallel stream, balance bioassay, and explicit chemosensors. Each organization offers points of interest (speed, multiplexing, and computerization) and burdens (prepared workforce, devoted research centers). In serological examples, quick antigen tests utilizing antibodies to identify the nearness of viral antigen (s) supplement these counters-acting agent identification strategies. The improvement of high throughput serological tests is at the core of significant symptomatic organizations [134].
In addition to direct diagnosis, indirect SARS-CoV-2 detection can be done by analyzing an infected person's immunological response. A recent study has shown the serological identification of COVID-19 in several fluids, including saliva samples [135]. Serological diagnosis, instead of nasopharyngeal swabs, focuses mainly on blood samples or viral detection [136]. After seven days post-infection, 50% of infected patients have antibodies in their blood, and all infected individuals have antibodies after 14 days. This assessment of the fundamental immune response is essential for analyzing community transmission. IgM and IgG antibody analyses are the most crucial for detection. Immunoassay denotes the bio-analytical approach that relies on the interaction between antigen and antibody [137].
ELISA and LFA are the two widely prevalent serological diagnostic procedures based on immunoassays. For COVID-19, its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) diagnosis, particular antigens or antibodies are required. The infection stage is crucial for establishing the COVID-19 detection technique. Several studies indicate that less than five days is needed for significant viral load, and at least seven days is required for antibody formation. Nevertheless, following seven days of infection, the concentration of antibodies may decline. Consequently, numerous immunoassay techniques for rapidly detecting COVID-19 have been established [138,139].
4.7.2. Enzyme-linked immunosorbent assay (ELISA)
ELISA is a microwell, plate-based investigation procedure intended to recognize and evaluate substances, for example, peptides, proteins, antibodies and hormones [140]. The test can be subjective or quantitative, and the chance of results is generally 1–5 h. On account of SARS-CoV-2, the plate wells are typically covered with a viral protein. If present, antiviral antibodies stuck patient examples will imbroglio explicitly, and the bound immune response protein complex can be distinguished with an extra hint of counteracting agent to deliver a colourimetric or fluorescent perusing. ELISA is rapid, can test many samples, and may be changed to robotization for increased throughput. However, it may differ in affectability and is helpful for determining care purposes. An ELISA test distinguishes antibodies delivered in the patient's blood because of disease with SARS-CoV-2 [141]. The whole test can be acted in a cylinder or well and includes blending tolerant examples, antibodies, antigens and proteins with a shading evolving atom. The model underneath depicts a commonplace ELISA test for antibodies [142].
To date, IgM and IgG detection by ELISA has shown high specificity and sensitivity in detecting COVID-19. This approach is also known as an enzyme-linked immunosorbent assay (ELISA) [143] since particular antibodies-enzymes associations are generated during the ELISA procedure to identify different proteins and bacteria in the bloodstream. Identifying the COVID-19 antibodies or the COVID-19 viral antigen produced by the host's immune system is possible. The antibody is covered or fixed using 96-well microtiter plates before adding the sample containing the particular analyte (virus, protein, or antigen). The enzyme-tagged antibody recognizes the coupling of the fixed antibody with the analyte in the presence of a specific substrate that emits visible colour, luminescence or fluorescence [144]. Similar procedures are used to identify COVID-19 antibodies, IgG and IgM [145].
Several ELISAs have been developed recently to detect human IgA antibodies, which are the first to form in reaction to viral stimulation [146]. Their identification is crucial because it helps scientists understand how the body responds to the illness. In a short amount of time, multiple COVID-19 ELISAs were also created and are being utilized. And have received global approval. Usually, such tests are utilized to evaluate a patient's immunological health [143]. COVID-19 patients with a negative molecular nasopharyngeal swab are tested for IgM and IgG detection by ELISA [143,147].
Due to the quick formation of these molecules in clinical samples, ELISAs are used to identify the viral protein. It is feasible to use anti-SARS Cov-2 human IgA diagnosing or mass screening applications [148]. In reality, nasopharyngeal swabs must undergo confirmatory molecular testing during positive ELISA results [127]. The specificity and sensitivity of these tests have results between 75.6%, 100%, 85.7% and 100%, respectively, even though these values may vary significantly depending on the provider and the individual antibody or viral antigen examined [149]. In contrast, ELISA-based serological testing is more credible than fast antibody or antigen assays [150]. Searching for IgG is often more precise than searching for IgM or IgA [145]. Remember that the runtime of the test and the infection depend on sensitivity and specificity. The efficiency of SARS CoV- 2 ELISAs, the capacity to analyze several samples concurrently, and the accessibility of the automatic or semi-automated method can accurately quantify viral antigens or human antibodies. Therefore, this method became an important clinical way for extensive surveillance and monitoring initiatives mainly used for particular categories such as Covid19 diagnosis [143].
4.7.3. Lateral flow immunoassay (LFT)
This test is usually a subjective (positive or negative) chromatographic examination that is little, compact and utilized at the consideration site. The test is a kind of rapid diagnostic test (RDT) because the outcome can be obtained within 10–30 min. By and by, liquid examples are applied to a substrate material permitting the example to go through a piece of immobilized viral antigen, at the point when present, hostile to CoV antibodies gather in the band, where shading creates with the gathered following antibodies to show results. The test is modest and doesn't require qualified staff; however, it just gives subjective outcomes. When utilized related to symptomatology, a determination of contamination might be conceivable. Fast antigen test using anti-CoV antibodies rather than immobilized viral antigen, allowing for a more easy assessment of the current disease [151].
4.7.4. Neutralisation assay technique (NAT)
Balance tests decide a counteracting agent's capacity to restrain the viral contamination of cultured cells and the cytopathogenic impacts of viral replication. For this test, tolerant examples are weakened with whole blood, serum or plasma and added to diminishing focuses in cell societies. If killing antibodies are available, their levels can be estimated by deciding the edge by which they can forestall viral replication in societies of contaminated cells. The time for the balance test results is commonly 3–5 days; however, late advancement has diminished this to a couple of hours [152]. This test requires cell culture offices and, for SARS coronavirus, level 3 biosafety research facilities (BSL3). Despite these constraints, the assurance of killing antibodies in the present moment is significant for the remedial utilization of therapeutic plasma and, in the long haul, for improving immunizations [153].
4.7.5. Luminescent immunoassay (LI)
Glowing immunoassays incorporate techniques that bring down the furthest reaches of counteracting agent reagents. They, by and large, identify with chemiluminescence and fluorescence. Earlier researchers built up a peptide-based attractive chemiluminescence catalyst immunoassay for the analysis of COVID-19, and Diazyme Laboratories, Inc. (San Diego, California) declared the accessibility of two new completely mechanized serological tests for SARS-CoV-2 running on completely robotized Diazyme DZ-light 3000 Plus chemiluminescent [154].
4.8. Additional diagnostic methods
4.8.1. Biomarkers
Multiple biomarkers found in biofluids may potentially be utilized to identify SARS-CoV-2. Research has indicated that patients with COVID-19 have a high concentration of leukocytes, C-reactive protein, blood platelets, lymphocytes, and D-dimer [155]. A quantitative examination of molecular markers to distinguish severe COVID-19 individuals with severe symptoms were recognized due to elevated blood creatinine, urea, and cystatin C levels than patients with mild conditions [156]. All of these indicators may be associated with the function of glomerular filtration, which may be exploited for the prior identification and distinction between intense and mild instances. It is challenging to use these biomarkers to diagnose COVID-19 and correlate them with different disease severities. These biomarkers are not unique to COVID-19; an aberrant concentration of these biomarkers is also present in various disorders [157].
As a biomarker, sputum comprising reactive oxygen molecules (ROS) is utilized to develop a genuine electrochemical biosensor for COVID-19 detection [158]. As SARS-CoV-2 infects lung cells, mitochondrial ROS would be created in excess, corresponding to the considerable rise in cellular ROS in SARS-infected persons [159]. Consequently, a large concentration of ROS is utilized as a biomarker to diagnose COVID-19 and its variants (Alpha, beta, gamma, omicron). As COVID-19 and influenza are more prone to be misdiagnosed, ROS level may be a valuable biomarker for identifying COVID-19 patients and differentiating them from influenza.
4.8.2. Biosensor tests (BT)
It has been possible to develop a biosensor for the continual and real-time SARS-CoV-2 diagnosis that is clinically applicable [160,161]. The nucleic acid of SARS-CoV-2 is detected by the biosensor using photothermal effect and plasmon sensing. Surface conduction electron localized resonance oscillations close to the target biomarker is found using light by LSPR sensors. This binding and affinity alter the plasmonic material is refractive index [162]. Two-dimensional gold nanoislands (AuNIs) having a complementary sequence hybridize with the SARS-CoV-2 viral nucleic acid. The thermoplastic effect occurs when AuNIs, plasmonic nanoparticles with large optical cross-sections, transform incoming light into heat without emitting radiation [163]; this provides the procedure with an in-situ heat source. By having a detection limit of 0.22 pM, localized thermoplastic heating may raise the temperature of hybridization and enable accurate differentiation from related SARS-CoV-2 gene sequences [164].
Biosensor tests depend on changing the particular cooperation between biomolecules into a quantifiable estimation by optical, electrical, enzymatic and different strategies. Surface plasmon reverberation (SPR) is a method that estimates episode light impedance at a fixed limit because of nearby obstruction, for example, the adsorption of antibodies or antigens. An SPR-based biosensor was created to determine SARS with a coronaviral surface antigen tied to a gold base [165]. The SPR chip had a lower recognition breaking point of 200 ml against SCV antibodies in a short time. Also, as of late, PathSensors Inc. declared a canary biosensor to distinguish the new SARS coronavirus. This stage utilizes a smart immunosensor that joins the infection with the symptom enhancement to give an outcome within 3–5 min. The biosensor is relied upon to be accessible for research in May 2020.
Several electrochemical biosensing approaches have been utilized to quickly and precisely detect COVID-19 infection. These biosensor-based devices rely on electrochemical and impedance reactions when viral proteins or RNA bind to specific antibodies or probes. Many different types of biosensor technologies are now available for COVID-19 diagnosis. Localised surface plasmon resonance (LSPR) biosensors, crystal microbalance biosensors (QCB), fluorescence-based biosensors, colourimetric biosensors, electrochemical biosensors, quartz surface-enhanced Raman scattering (SERS) biosensors, and others are platforms used diagnosed COVID-19. SERS and Electrochemical biosensors are the most frequently deployed point-of-care platforms [45]because of their small size, simplicity of use, and cheap cost. Electrochemical biosensors, SARS CoV-2 viral RNA and proteins may be identified [164].
Additional nano biosensor research might exacerbate problems with residual detection. The application of polymer-coated biosensors for quick, precise detection has been shown [17]; this contrasts with the previously discussed nanoparticle-decorated biosensors. It is advised to use polymers containing acrylic groups for bulk biosensor manufacture. LFA biosensors are the efficient approach to detect SARS-CoV-2 on the commercial market, outperforming all other approaches. The public is interested in the powerful CRISPR-Cas technology combined with LFA. This biosensor is inexpensive, on-site monitoring equipment that allows non-specialists to conduct real-time testing and has excellent specificity and sensitivity. A fast antigen test with gold-standard RT-PCR is preferred since diagnoses cannot be made with absolute certainty at the service point [166].
4.8.3. Aptamer-based nano-biosensor
Aptamers are synthetic RNA or DNA molecules that can bind to specific targets, including proteins. In the context of SARS-CoV-2, aptamers can be designed to bind to the centre protein of the virus. This makes them a potential tool for quickly detecting the presence of the virus. The developers of an aptamer-based point-of-care (POC) test for SARS-CoV-2 are developing a test that can deliver results in just 30 s without the need for sample preparation steps. This could provide a fast and convenient method for detecting the virus in real-world settings [167].
The most recent advancements in aptameric nano biosensors for SARS-CoV-2 diagnosis are a crucial milestone. In the future, applying deep learning algorithms may impact the choice and accuracy of the nano-biosensor for COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) identification exploration is mentioned in Fig. 10 . Extensive medical validation tests and studying complex materials like blood, sweat, faeces, inhaled air, and a few others are needed to fully comprehend sensor robustness and commercial potential. The development of wearable diagnostic equipment may be helpful for continuous COVID-19 monitoring. Employing additive manufacturing, such as 3D or 4D printing, creates multiple items for various uses. In reality, 3D-edge-cutting technology can generate diverse nanocrystals for uniquely manufactured aptameric nano biosensors to diagnose COVID-19its variants (Alpha, beta, gamma, and omicron).
Fig. 10.
Detection of SARS-CoV-2 and its varients with Gold Nanoparticle Aptasensors: A Process Diagram.
4.8.4. Fast antigen test (FAT)
Notwithstanding sub-atomic genetic testing, quick antigen tests are utilized to distinguish viral antigens; these tests depend on explicit monoclonal antibodies to give an instrument to catch viral antigens from a logical example [168]. These investigations are not restricted to a particular configuration [169].
4.8.5. Paper-based detection
An elective paper-based strategy that utilizes wastewater as tests have been proposed by Kang [170]. Paper-put-together unit-based concerning the coordination of different applicable territories, for example, extraction, elution, sanitization, fortification and identification, all in a tiny, modest expendable paper and printed with wax on a superficial level as zones. It is very conceivable to finish the whole test process with no force source or force just by collapsing the paper in various modes, so it is more practical than the costly and messy multi-step procedures. These systematic gadgets give a top-notch, quick yet exceptionally exact microorganism recognition strategy, just as low assembling expenses and easy to use nature [171].
This strategy can fill in as an elective discovery instrument to rapidly distinguish the source or nearness of causative specialists, for example, COVID-19, in any pandemic territory. The dung and pee of transporters of Community sicknesses entering the sewage framework can contain numerous biomarkers for the infection. Another investigation affirmed the equivalent, demonstrating that these irresistible operators could stay dynamic. For a few days, considerably after they have been released from patients on the off chance that they locate a reasonable domain [171,172]. This paper machine can follow the transmission of COVID-19 in network wastewater by dissecting SARS-CoV-2 in defecation, pee and other human dung.
4.8.6. Digital PCR
The currently used RT-PCR techniques have been technologically improved by digital PCR [173]. Digital PCR is recently employed for several purposes, including detecting viral loads, mutational analysis, research, analysing liquid biopsy samples, single cell analysis, and identifying low-expression targets [174]. Various businesses have developed unique digital PCR systems. Droplet digital PCR (ddPCR) is the best method for determining individuals' viral infections. The ddPCR was designed for scientific work, particularly in the case of viruses, when assessing a patient's viral load is essential to assess pathogenicity [175]. Several disorders, including COVID-19 infection, can now be diagnosed using ddPCR, which was applied to other clinical domains [176]. It has become one of the most precise and sensitive methods available.
It should be noted that digital PCR uses DNA ultra-dilutions along with micro-sample divisions conducted on solid supports or with the reaction medium's emulsifier. The target nucleic acid, specific Taq polymerase, probes and primers, and the PCR amplification buffer are all included in the ddPCRnano-partitions of the PCR mixture into >20,000 oil-water droplets. Identical to RT-PCR, sample preparation is followed by the extraction of viral RNA using either custom or commercial techniques. Consequently, viral RNA may be captured automatically in ddPCR utilizing one-step techniques or analysed after an RT step to generate cDNA [177]. The creation of the reaction mixture is the first step in the ddPCRmethod. Next, emulsifier oil-water droplets are created by employing a droplet generator and specialized cartridges. The resulting droplets are replicated using a conventional RT-PCR amplification method, amplifying the target DNA of the droplet and producing a variety of amplification responses in a single experiment. The magnified droplets are then scanned using a droplet reader that uses capillary tubes that ensure that each droplet flows separately and is triggered by a laser. If the droplet reader detects a positive result, the fluorescence emission signal it produces is recorded by a CCD camera [178].
4.8.7. Next-generation sequencing
The role of next-generation sequencing (NGS) has been significant in identifying the SARS-CoV-2 sequence, ofidentifying the SARS-CoV-2 sequence, and improving the vast majority of current early diagnostic methods. NGS was used to characterize the whole genome of SARS-CoV-2, demonstrating its classification in the coronavirus family [179]. Using just a single, sequence-independent primer for amplification and nanopore technology, SARS CoV-2 was sequenced from scratch [180]. NGS is now used for identifying novel genetic variants and molecular techniques but never for diagnostic purposes. Its clinical use is reduced based on the high analysis cost, expensive equipment, and the requirement for highly educated bioinformaticians [181]. Although the substantial cost of the technique, several firms have produced commercial kits for SARS-CoV-2 sequencing utilizing NGS methodologies. In particular, most commercially available kits depend on NGS and mixed collection methods such as quick tests [182].
In addition, other sophisticated NGS approaches have been developed to discover mutations in the sequencing of the SARS-CoV- 2 genome, therefore; detecting fast-evolving strains that are significant for vaccine development and epidemiology. Among these methods, Amplicon-based metagenomic sequencing provides the most efficient method for rapidly detecting and analyzing SARS CoV-2 and specific other pathogens. For example, metagenomic sequencing detects a patient's microbial community, and SARS-CoV-2 virus RNA can be replicated and found using amplicon-based sequencing [46].
Consequently, metagenomic analysis and amplicon-based sequencing may correctly detect COVID-19 infection, thereby detecting secondary infection caused by other viruses that negatively impact patient health [183]. MinION and IDbyDNA are examples of these sequencing techniques [183,184]. With a 75-bp average length and complete coverage, the IDbyDNA technique offers >13 million readings, of which >8 million are unique. Shotgun sequencing promotes the discovery of SARS CoV- 2 sequence variants by enabling a high library score and Q score [185]. Furthermore, MinION technology gathers incredibly short and lengthy reads (4,000 bp), providing 30 Gb of output data. This portable approach allows low-cost, real-time clinical sample testing [186].
This method was used on the SARS CoV-2 genome, including primers targeting 16 conserved coronavirus binding sites [187], enabling reconstruction of the complete genome overlapped sections of 1,000 bp readings. Next-generation sequencing is the most powerful tool for the molecular investigation of SARS CoV-2, the discovery of novel mutations during genomic testing, and the creation of genome-based therapeutic alternatives [188].
Besides, in microbiology labs, NGS technology has grown to be generally regarded as a means for monitoring outbreaks and genetic epidemiology. Finding novel mutations in SARS-CoV-2 enables researchers to reconstruct previously unrecognized infection paths and provides a genetic foundation for SARS-CoV-2 medication discovery, vaccine design, and diagnostic development [189]. Monitoring the SARS-CoV-2 genome enables quick turnaround on patients with unknown origins of infection and a more efficient COVID-19 management approach [127,141].
There are several techniques used for the diagnosis of COVID-19, each with its advantages and disadvantages. CT scans are used for screening but require expensive equipment and technical expertise. RT-PCR is the gold standard for identifying active infections but requires a specialized laboratory setup and qualified personnel. Immune assays and ELISA tests are more sensitive and specific but require complex processes and culture mediums. Serological tests can identify previous infections and determine the stage of infection but have the problem of providing false-negative results in the early stages. CRISPR-based tests are easy to use and cost-effective but may lack sensitivity. Electrochemical biosensors quickly and precisely detect COVID-19 and its variants, but some lack sensitivity, selectivity, and electrode manufacturing. Comparative knowledge of different diagnostic techniques for COVID-19 and their advantages and disadvantages, are mentioned in Table 1 .
Table 1.
Comparative analysis of diverse diagnostic approaches for COVID-19: Unveiling the power of detection methods.
| Techniques | Procedure | Advantages | Disadvantages |
|---|---|---|---|
| CT scan |
|
A chest CT scan is more sensitive thanRT-PCR, particularly in the early stages. |
|
| RT-PCR |
|
|
|
| Immune essay |
|
|
|
| CRISPR-Based Tests |
|
|
|
| ELISA |
|
|
|
| Serological test |
|
|
|
| Electrochemical biosensors ‘ |
|
|
|
5. Synergistic medicine combinations for COVID-19 treatment
Synergistic combinations may increase therapeutic efficacy and potency by generating higher therapeutic benefits or lowering the needed dosage, minimizing adverse impacts. Identifying acceptable combinations of licensed chemicals to counter the COVID-19 outbreak and subsequent pandemics are considered more advantageous to discovering and developing a completely novel single-agent therapy: a faster time to clinical application. Numerous studies have debated the advantages of identifying novel synergistic medication combinations for treating COVID-19 and its variants (Alpha, beta, gamma, omicron) [72].
5.1. Databases retrieval and available resources for synergistic drug detection
In response to the outbreak, several identified research institutions have established particular COVID-19 datasets, publishing libraries, and various new tools have evolved. Among highlights a few, PubMed, the world's largest scientific and biosciences resource repository, has created a SARS-CoV-2 Data, which includes links to indexed publications in both Pubmed and Pubmed Central for indication of clinical trials. The ChEMBL database, one of the essential general databases of pharmacological outputs, has released a special edition with COVID-19-associated diagnostic data for about 20 000 molecules. The European Bioinformatics Institute (EBI), which maintains ChEMBL, has developed a comprehensive COVID-19 research gateway that includes data on viral and host protein-coding genes, viral-host protein associations, and several other sources. The NIH's National Center for Advancing Translational Studies (NCATS) has established a significant biochemical and functional genomics library, providing information on the therapeutic potential of pharmacological pools in SARS-CoV-2 target-specific and phenomenological studies.
In pharmacological research, the Diamond Synchrotron source has made accessible a library of 1500 resolved microstructures of lower mass segments bound to SARS-CoV-2 Mpro and their realistically estimated target proteins. As a result of these and similar endeavours, nearly 1100 protein structures spanning the bulk of SARS-CoV-2 RNA encodes were deposited into the Protein Data Bank (PDB). PostEra also used the Diamond incommensurate dataset as a starting point for crowdsourcing, community-sourced de novo ligand synthesis. As a result, over 1800 internal consistency dependability molecules have already been created, synthesised, and tested, and results have been released publicly.
Non-structured datasets are another significant source of information on the virus and the disease. As a result, most medical journals chose to make all COVID-19-related articles open to the public. Kaggle has made available the COVID-19 Open Research Dataset (CORD-19), which comprises over 200,00 research publications on new and related coronaviruses in over 100 000 full-text entries. Meanwhile, Elsevier developed the Coronavirus Information Center, a free resource with over 30 000 publications and chapters.
As in Fig. 11 , we show the process of identification of synergistic effects using DL database retrieval techniques. These approaches evaluated numerous plant compounds from available resources to collect many phytochemical compounds used for target-disease association and disease-target interaction. The combination may have an antiviral effect that can be evaluated using its molecular representation to develop novel drugs and vaccines to treat COVID-19 infection and its variants (Alpha, beta, gamma, omicron) [190].
Fig. 11.
Unveiling the power of deep Learning database retrieval techniques for unlocking the synergistic effects in COVID-19 treatment.
Due to the massive number of different chemical combinations, it is prohibitive to investigate the vast array of possibilities by high-throughput screening of even relatively large chemical libraries. Consequently, in silico screening employing various computational approaches is a desirable choice [191]. Chushak and Stone [191] employed knowledge-based methodologies to build potential medicine combinations and verified synergies for the SARS-COV2. Liu et al. [192] established a biological network proximity metric to evaluate the synergy between anti-cancer drugs and hypertension. Synergy has been predicted using machine learning techniques, particularly DL approaches [193,194]. In contrast, Goh et al. [195], constructed on a large screen deep neural network [196] and showed that deep learning is better than standard machine learning models such as SVMs and RFs [197].
A significant development in the DL approach is the construction of ComboNet [198]. ComboNetis a DL architecture that models biological targets and chemical structures to anticipate synergistic drug combinations. By precisely simulating interactions between medications and biological targets, the reliance on combination synergy data may be significantly reduced. Indeed, unlike prior methods [199], ComboNet learns to model drug-target interaction (DTI) based on molecular structures, which is advantageous compared to methods that utilize drug-target contact (DTI) as static characteristics [200].
The architecture of ComboNet comprises two major parts. The first part is a convolutional graph network (GCN) which generates a continuous molecule representation [201]. This depiction includes the structural characteristics of the molecule and expected targets (the molecule interacts with the biological target). The second component of ComboNet simulates the relationship between the target and the disease. It is a linear function that determines the relationship between biological targets, structural characteristics of compounds, synergy, and antiviral activity [202].
5.2. Implications of deep learning methods to target proteins and drug sites for COVID-19 therapeutics
Even though no quantum computing initiatives still have resulted in verified medicinal compounds, the mega combinatorial libraries have the potential to provide innovative suggestions for COVID-19 drug development. DL approaches were employed in a computational effort on the protein surface to resolve the discrepancy between the number of accessible database searches and traditional docking capabilities. The Deep Docking framework created Quantitative structure-activity relationships (QSAR) algorithms for molecular docking. Unlike traditional docking, which does an entire screening run and picks just a limited number of successfully docked molecules, this technique incorporates all docking data. Deep Docking predicted docking scores for 1.36 billion molecules from the ZINC15 database versus 12 primary protein targets in the initial stage. It demonstrates a 100-fold improvement in computational system performance and a 6000-fold boost in sensitivity for top-scoring compounds. When used for virtual screening against protein, Deep Docking may reduce >1.4B compounds (from the ZINC15 library) to 1000 potential hits in just one week. It used 640 CPU and 40 GPU units (GLIDE docking and DL computations, respectively). Surprisingly, conventional docking methods would need years of continuous work on this technology without DL improvement.
5.3. Knowledge-based AI tools for the discovery of synergistic drug combinations for COVID-19
The seriousness of coronaviral outbreaks encouraged scientists, institutions, publishers, organizations, and authorities to invest heavily in studies and data collection to better understand the illness and find an effective treatment as quickly as possible. Several structured and unstructured COVID-19 datasets have been made public, allowing for broader use of information extraction approaches and Intelligence technologies for COVID-19 drug development, with some notable instances mentioned in the following sections [203].
Medical Data Sets are intended to provide a high-level view of the links between diseases (symptomatology, taxonomies, etc.), biological entities (genomics, protein production, related proteins, polynucleotides), and natural chemicals (clinical and investigational drugs, tool compounds, etc.).These correlations can be inferred directly from hierarchical datasets, such as medical and pharmaceutical archives, or massive volumes of data, such as a corpus of academic research and inventions, using information retrieval techniques aided by machine learning algorithms. Natural language processing (NLP) approaches for entity recognition can generate a KG from unstructured data [204]. This establishes whether artefacts in the text correspond to the same underpinning enterprises; similarity excavation, which recognizes significant subject triplets in the text; and resemblance priority, which analyses the believability of the data collected programmatically or manually).
The synergistic effect of medications creates several significant therapeutic potentials. The significant efficacy of anti-HIV medication combinations and the synergism of multiple other medicines underscores the need to investigate COVID-19 treatment methods. Modern AI technologies can be employed as practical optimization algorithms to investigate medication combinations with synergistic activity against SARS-CoV2 [205].
The review-based findings [74,206] emphasize the need for biological models in antiretroviral medicine combinations and the use of data and text mining technologies in determining the mechanisms of action behind synergism/antagonism in COVID-19. These data also imply that a lack of biological models on pharmaceutical combinations before patient use may increase the probability of undesired side effects and poor outcomes. Furthermore, the well-established matrix diagnostic test provides a simple, data-driven way to identify synergistic COVID-19 treatment combinations and emphasize unfavourable medicine combinations [207].
6. Recommending medications to COVID patients using AI
COVID-19 could lead to cardiovascular complications, and intervening with COVID-19's vulnerable groups is a significant concern. ANN-based techniques could achieve better outcomes parallel to traditional techniques. Keeping patients' COVID-19 records showing clinical factors and cardiac complications would enable recognizing patterns to build cardiovascular complications' risk models forecasting therapeutic responses. Fig. 12 shows an Extreme Learning Machine (ELM) model suggesting proper medications for patients' cardiac complications [208]. ELM-ANN could apply prior model inputs to forecast required outputs where the training of the supervised model occurs via actual network data. Hence, regarding different viral infections for past patients, ELM would recommend potential medications for cardiovascular complications [34]. Compared to the traditional feed-forward network, ELM algorithms, such as back-propagation (BP), learn faster with improved generalization functionality [209]. Frequently, traditional tuning-based algorithms need fewer hidden neurons than ELM [210]. After the learning procedure, forecasting new data is done via the validation process. The proposed model applies data to forecast how COVID-19 affects the cardiovascular system and various therapeutic responses, thus lowering the risk of potential cardiac complications [211].
Fig. 12.
Illuminating the path: Harnessing LSTM-ANN networks to anticipate optimal therapy for COVID-19 through Long-term pattern sequence.
COVID-19 may cause cardiac arrest in the elderly, necessitating the interference of cardiologists; thus, experts are involved in developing a structured technique capable of providing early investigations or clinical trials worldwide that could deepen understanding of the disease's final stages [212]. Biomarkers should be used carefully, particularly in vulnerable elderly patients with comorbid structural heart diseases [213].
The level of COVID-19 infection, electrocardiography, and history of chronic diseases could be inputted to train the model. Adopting multiplicative gates would deal with the ongoing stream of flaws via the inward conditions of the unique entities of memory cells [214]. Long Short-Term Memory (LSTM)neural networks could address losing gradients in Recurrent Neural Networks (RNNs) [215]. Lately, LSTM-NN is standard and highly applied in controlling robotics, human handwriting, speech, speed, and other activities recognition and text classification [216,217].
7. Discussion
This review discusses the numerous COVID-19 variants (Alpha, beta, gamma, omicron) diagnosis approaches used in various clinical settings. Numerous kits and tools are currently available on the market; however, the absence of relevant qualities such as cost, run time and complexity of a particular method, thus; making it necessary to build robust point-of-care tools. Researchers have swiftly adapted available diagnostic tools to the COVID-19 virus and its variants (Alpha, beta, gamma, omicron) due to the availability of these technologies [218]. The lessons learned during the SARS epidemic 2002 have influenced the development of COVID-19 techniques. The SARS-CoV-2 genetic sequence was deduced in just three weeks after transmission electron microscopy virus detection, while SARS-CoV19 was identified after five months [219]. This is a result of the scientific community's swift reaction and advancements in diagnostic capability between 2002 and 2020, such as the availability of NGS (Next-Generation Sequencing)for speedy sequence Identification. However, the growing number of testing techniques needs constant optimization and regulation. In addition, various techniques require additional validation to assure precision, usability, accuracy, and widespread deployment. Additional investigation on these diagnostics for zoonotic monitoring might aid in preventing future outbreaks [220].
The gold standard RT-PCR is utilized to compare other detecting methods based on sensitivity, precision, measurement principle and cost. RT-PCR measures nucleic acids to diagnose an infection, while most detection procedures depend on serological testing. Serological assays detect both the antigen and the antibody. LFIA can be an effective point-of-care diagnostic tool to rapidly detect the blood antigen. Several rapid diagnostic tests (RDTs) were produced with less accuracy, and additional advancements in this sector can be employed to create point-of-care testing devices that are very sensitive and accurate [221].
Among the recently discovered diagnostic techniques for COVID-19 nucleic acid identification, LAMP (Loop-mediated isothermal amplification) yields more convincing findings than RT-PCR [222]. A lateral Flow-based test for detecting nucleic acid based on isothermal amplification is a new method for developing diagnostic instruments. In the COVID-19 diagnosis, lateral flow assay (LFA), which is paper-based, has gained people's assurance and acceptance [223]. Literature has demonstrated that less sensitivity is the critical concern linked with immune-related LFA. Several solutions exist, including utilizing Analyte pre-concentration, colloidal gold particles and Ion concentration polarisation techniques. Another novel method for enhancing the specificity, sensitivity, and speed of diagnostic testing is the proximity ligation assay, in which aptamers and antibodies are utilized to target the viral genome [224].
Serology tests have garnered significant interest since they are simpler to administer and give more insight into all infected persons than NAATs (Nucleic Acid Amplification Tests). The latest methods and techniques used for COVID-19 and its variants (Alpha, beta, gamma, omicron) detection are evaluated in terms of their operational principles, their worth for viral detection and potential limitations. Since diagnostic procedures are vital to managing outbreaks, it is essential to recognize the limitations of present methods, create more effective methods, and discover all infected persons, including asymptomatic COVID-19 carriers, swiftly and correctly in the early stages. With this objective in mind, Paper-based processes, LFAs (Lateral flow assays), and the application of AI approaches are a few of the suitable techniques that might be utilized to avoid and manage the future spread of COVID-19 and other possible pandemic breakouts [225].
During the pandemic, several additional biosensors were developed, most of which were dependent on probe-based technology, aptamer-based technology, nanotechnology, Barcoding, and CRISPR- Cas; all of these have relatively more straightforward usage and provide rapid screening for the SARS-CoV-2 virus. Biosensors are devices with organic and inorganic constituents that assess and deliver quantitative and qualitative information on a particular analyte in the samples. During pandemics, these biosensors are considered potential diagnostic and decision-making tools that might save millions of lives.COVID-19 may also be detected with DNA microarray-based methods [226]. Biosensors must simplify the basic concept to minimize costs and increase demand [227,228]. Biosensors have several options to increase sensitivity and effectiveness.
In addition, technologies in the research stage, including developing electrochemical biosensors and artificial intelligence to deliver data-driven insights, may be valuable for rapid and precise detection [229]. The development of nanotechnology and microfluidics is paving the way for the creation of biosensors having varied characteristics [230]. This benefits the miniaturization of biosensors and increases their availability during a pandemic. These biosensors are based on the microarray, CRISPR technology, and microfluidics, potential early-stage disease diagnosis techniques [231].
Similarly, using NGS technology in microbiology laboratories for epidemic monitoring has acquired general recognition. Detecting SARS-CoV-2 novel mutations has allowed researchers to rebuild undiscovered infection pathways and offers a genetic basis for developing SARS-CoV-2 vaccines, diagnostics, and therapeutics, and is an essential method in genomic epidemiology [232]. SARS-CoV-2 genome monitoring facilitates an efficient COVID-19 management approach and expedites the investigation of patients with unknown infection sources [233,234].
Additionally, during the COVID-19 pandemic, vaccine development or antiviral medicine took a long time. Therefore, drug combinations emerged to be significant in managing viral infections owing to their efficacy in significantly decreasing the danger of developing drug resistance. They have been very effective against several viral illnesses in the past [235]. According to the literature, antiviral drug combinations target various phases of the virus life cycle, have a variety of mechanisms of action, and are composed of different classes of antiviral drugs that have synergistic effects [236,237].
For drug designing, DL methods have shown efficacy [237]. It is standard practice for deep neural network training to c, toonduct virtual screenings of chemical databases in-silico, and suggest chemicals for laboratory testing. For these models to anticipate biological activities, a considerable number of training data is required. To generate correct recommendations, such information is often unavailable for developing drugs against pathogens like SARS-CoV-2. Therefore, the literature argues that complementing the limited task-specific data with a new biological understanding of these diseases is essential.
The fast improvement of GCNs in estimating molecular characteristics spurred the development of ComboNet [238]. Most of these approaches find molecular representations based chiefly on chemical structures, and it is argued that it does not imitate biological interaction directly [239]. While prior cheminformatics techniques have modelled DTI for properties prediction, most of these methods do not use molecular compounds such as GCNs. ComboNet suggests combining the advantages of the two approaches into a single deep-learning model. Therefore, deep learning approaches Are practical for predicting synergistic drug combinations.
8. Future direction and perspective
Even after distinct coronavirus mutations (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.), RT-PCR detecting numerous targets delivers accurate results (with decreased sensitivity) to novel variants. Nonetheless, Limitations of RT-PCR include the need for laboratory tools and the complexities of its setup and diagnostic processes. Further nucleic acid approaches, including microarray, may be employed to overcome these limitations and develop an affordable, quick, and efficient detection approach. Extensive research is being conducted to create correct procedures. However, more thorough research is required to translate laboratory approaches into clinical trials. Point-of-care diagnostics that are efficient and more accessible with fewer sample collection stages and faster detection rates may be developed via efforts with the proper attention and the provision of required research resources.
The techniques and progressions in nanotechnology and microfluidics can aid in employing a multifaceted approach to tackle many pandemic issues creating varied biosensors that will enable more quick diagnosis with high sensitivity and specificity at the point of treatment. It has been found that this virus mutates rapidly; hence, it is more likely to resurface from several mutant strains. In diagnostics, biosensors are in high demand and will be particularly useful for the early diagnosis of viruses. The biosensor-based instrument may fulfil the demand for earlier detection cheaply, even in distant places, allowing worldwide screening with effective results [17]. Even with RT-PCR, CT scans, and sequencing, user-friendly rapid diagnostic kits are needed for global viral screening and control. Given the benefits and shortcomings of existing COVID-19 screening methods, more methods, such as biosensors, are needed to manage and prevent the disease [240].
Epidemic control requires comprehensive monitoring and swift dissemination of epidemiological data [241]. Smartphones, whose use has expanded dramatically, may be used for this purpose since they have the connection, hardware and processing capacity, which may promote electronic reporting, epidemiology databasing, and Point-of-care diagnostics [242]. Integrating diagnostic technologies with a smartphone can improve care, prevent the spread of illness, and decrease mortality [221].
The safety of laboratory personnel doing COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) testing is also crucial [[243], [244], [245]]. When systems are overworked, it is more likely that shortcomings in inefficient microbiological processes and worse sanitation procedures may occur [246]. Optimizing laboratory worker protection procedures should proceed with the optimization of COVID-19 diagnosis. Also, more technological advancements are warranted because of future pandemic risks.
9. Advantages
Deep learning methods offer several advantages in diagnosing COVID-19, its variants and identifying synergistic medicine combinations. They enhance diagnostic accuracy by analyzing various modalities and detecting subtle abnormalities. Deep learning enables rapid screening and early detection, aiding in timely interventions and containment strategies. Automation and efficiency are improved as deep learning models quickly process large volumes of data. They contribute to identifying and characterising variant strains, facilitating targeted control measures. Deep learning algorithms predict effective medicine combinations, optimizing treatment strategies. These models can adapt and evolve, future-proofing their relevance. By complementing traditional diagnostic approaches, deep learning provides an additional layer of analysis, enhancing accuracy and comprehensive assessments.
9.1. Limitations
Despite the promise of deep learning methods in diagnosing COVID-19, its variants, and identifying synergistic medicine combinations, several limitations exist. Firstly, data availability and quality pose challenges, as obtaining well-annotated datasets for specific variants or medicine combinations can be difficult. Interpretability is another limitation, as deep learning models often operate as black boxes, making it hard to understand their decision-making processes. Generalizability is a concern, as models trained on specific datasets may struggle to perform well on diverse populations or settings. Ethical considerations, validation for clinical implementation, limited understanding of complex interactions, and resource requirements further add to the challenges. Overcoming these limitations will require collaborative efforts to ensure the responsible and effective use of deep learning in combating COVID-19.
9.2. Summary
The relentless impact of the COVID-19 pandemic necessitates novel approaches to tackle its challenges. This comprehensive review focuses on exploring and evaluating the potential of deep learning methods in diagnosing COVID-19, its variants and identifying synergistic medicine combinations. We begin by providing an overview of the challenges and limitations faced in diagnosing COVID-19 and its variants, emphasizing the urgent need for improved diagnostic accuracy. Deep learning, a subset of machine learning, holds great promise, offering innovative solutions to enhance disease diagnosis and treatment optimization.This review delves into the fundamental concepts and techniques of deep learning, tailored explicitly for COVID-19 diagnosis, variant detection, and medicine combination analysis. We shed light on the prevalent deep learning architectures employed in medical image analysis, sequence analysis, and data integration. By utilizing various modalities such as chest X-rays, CT scans, and molecular data, deep learning algorithms can significantly contribute to diagnosing COVID-19 with improved sensitivity, specificity, and interpretability. Moreover, we explore the potential of deep learning techniques in identifying and characterizing emerging variants of SARS-CoV-2. Leveraging genomic sequencing data, phylogenetic analysis, and relevant information, deep learning algorithms offer a promising avenue for accurate variant detection and classification.
Furthermore, we investigate how deep learning can revolutionize the prediction of effective drug combinations for treating COVID-19 and its variants. Through an extensive analysis of existing literature and studies, we highlight the role of deep learning algorithms in identifying synergistic drug combinations and optimizing treatment strategies. Addressing the current limitations of data availability, interpretability, and generalizability, we emphasize the need for further advancements in applying deep learning for COVID-19 diagnosis and medicine combination analysis. We discuss potential future developments, including integrating multimodal data, transfer learning, and explainable AI, which hold tremendous potential in enhancing the performance and robustness of deep learning models in COVID-19 research.
10. Conclusion
In conclusion, the COVID-19 pandemic has highlighted the importance of accurate diagnosis and effective treatment in managing the global health crisis. While RT-PCR-based tests remain the standard for COVID-19 and its variants (Alpha, beta, gamma, omicron, Kappa, Zeta, Lambda, Epsilon, Lambda, Delta, Zeta and Theta etc.) diagnosis, they have limitations in detecting infections with low viral loads. They are not easily accessible in low-income regions. Alternative methods such as CRISPR/CAS-based assays, isothermal amplification, ddPCR, biosensors, rapid antigen tests, and antibody tests have been developed to overcome these drawbacks and provide more sensitive and cost-effective diagnostic options. Additionally, next-generation and whole-genome sequencing are crucial in evaluating novel SARS-CoV-2 genetic mutations. Deep learning (DL) approaches have emerged as powerful tools for screening, prediction, contact tracing, drug development, and treatment management for COVID-19 and its variants. DL advancements have significantly improved various aspects of the pandemic response, reducing human intervention and enhancing clinical practices. Furthermore, DL has shown promise in identifying synergistic medicine combinations for COVID-19 treatment. DL and other innovative diagnostic and therapeutic approaches offer hope for effectively combating the COVID-19 pandemic and its variants.
Compliance with ethical standards
Not applicable.
Author's contribution
Conceptualization, Methodology, Investigation, data collection and Writing-original manuscript: Qandeel Rafique, Ali Rehman, Muhammad Sher Afghan, Hafiz Muhamad Ahmad; Editing and proofreading, Imran Zafar, Kompal Fayyaz, Quratul Ain, Rehab A. Rayan, Khadija Mohammed Al-Aidarous, Summya Rashid, Gohar Mushtaq; Supervision: Rohit Sharma, and Imran Zafar. All authors approved the submission of the final manuscript.
Declaration of competing interest
There is no conflict of interest in the present work.
Acknowledgement
This study is supported via funding from Prince sattam bin Abdulaziz University project number (PSAU/2023/R/1444).
References
- 1.Aggarwal A., Verma T., Rai D.C., Sharma R. Post-COVID-19 trends in indian dairy industry: current challenges, recoveries and future strategies. Indian Vet. J. 2022;99:7–15. [Google Scholar]
- 2.Asare P., Barfi R.J.E. The impact of Covid-19 pandemic on the Global economy: emphasis on poverty alleviation and economic growth. Economics. 2021;8(1):32–43. [Google Scholar]
- 3.Rahman M.M., Islam M.R., Akash S., Mim S.A., Rahaman M.S., Emran T.B., Akkol E.K., Sharma R., Alhumaydhi F.A., Sweilam S.H., Hossain M.E., Ray T.K., Sultana S., Ahmed M., Sobarzo-Sánchez E., Wilairatana P. In silicoinvestigation and potential therapeutic approaches of natural products for COVID-19: computer-aided drug design perspective. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.929430. PMID: 36072227; PMCID: PMC9441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marriam S., Afghan M.S., Nadeem M., Sajid M., Ahsan M., Basit A., Wajid M., Sabri S., Sajid M., Zafar I., Rashid S., Sehgal S.A., Alkhalifah D.H.M., Hozzein W.N., Chen K.-T., Sharma R. Elucidation of novel compounds and epitope-based peptide vaccine design against C30 endopeptidase regions of SARS-CoV-2 using immunoinformatics approaches. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P.A., Hall L.E., John J.N., Rapoport A.B. vol. 95. Elsevier; 2020. The early natural history of SARS-CoV-2 infection: clinical observations from an urban, ambulatory COVID-19 clinic; pp. 1124–1126. (Mayo Clinic Proceedings). 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauf A., Abu-Izneid T., Khalil A.A., Hafeez N., Olatunde A., Rahman M., Semwal P., Al-Awthan Y.S., Bahattab O.S., Khan I.N., Khan M.A., Sharma R. Nanoparticles in clinical trials of COVID-19: an update. Int. J. Surg. 2022 Aug;104 doi: 10.1016/j.ijsu.2022.106818. Epub 2022. PMID: 35953020; PMCID: PMC9359769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amarilla A.A., et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021;12(1):1–15. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma R., Kumar P., Rauf A., Chaudhary A., Prajapati P.K., Emran T.B., Gonçalves Lima C.M., Conte-Junior C.A. Mucormycosis in the COVID-19 environment: a multifaceted complication. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.937481. PMID: 35923801; PMCID: PMC9339637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aung Y.Y., Wong D.C., Ting D.S.J. The promise of artificial intelligence: a review of the opportunities and challenges of artificial intelligence in healthcare. Br. Med. Bull. 2021;139(1):4–15. doi: 10.1093/bmb/ldab016. [DOI] [PubMed] [Google Scholar]
- 10.Jin S., Liu G., Bai Q.J.M. vol. 11. 2023. p. 1279. (Deep Learning in COVID-19 Diagnosis, Prognosis and Treatment Selection). 6. [Google Scholar]
- 11.Jamshidi M., Moztarzadeh O., Jamshidi A., Abdelgawad A., El-Baz A.S., Hauer L.J.F.I. Future of drug discovery: the synergy of edge computing. Internet Med. Things, Deep Learn. 2023;15(4):142. [Google Scholar]
- 12.Santaniello A., et al. SARS-CoV-2 affects both humans and animals: what is the potential transmission risk? Lit. Rev. 2023;11(2):514. doi: 10.3390/microorganisms11020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohapatra R.K., et al. vol. 8. 2023. (The SARS‐CoV‐2 Omicron Variant and its Multiple Sub‐lineages: Transmissibility, Vaccine Development, Antiviral Drugs, Monoclonal Antibodies, and Strategies for Infection Control–A Review). 9. [Google Scholar]
- 14.Yu Z., Umar M., Rehman S.A.J. Adoption of technological innovation and recycling practices in automobile sector: under the Covid-19 pandemic. Operat. Manag. Res. 2022;15(1–2):298–306. [Google Scholar]
- 15.Choi T.-M.J. Fighting against COVID-19: what operations research can help and the sense-and-respond framework. Ann. Oper. Res. 2021:1–17. doi: 10.1007/s10479-021-03973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiroma H., Ezugwu A.E., Jauro F., Al-Garadi M.A., Abdullahi I.N., Shuib L.J.P.C.S. vol. 6. 2020. p. e313. (Early Survey with Bibliometric Analysis on Machine Learning Approaches in Controlling COVID-19 Outbreaks). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh B., Datta B., Ashish A., Dutta G. A comprehensive review on current COVID-19 detection methods: from lab care to point of care diagnosis. Sens. Int. 2021;2 doi: 10.1016/j.sintl.2021.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Sherif D.M., Abouzid M., Elzarif M.T., Ahmed A.A., Albakri A., Alshehri M.M. vol. 10. MDPI; 2022. Telehealth and Artificial Intelligence insights into healthcare during the COVID-19 pandemic; p. 385. (Healthcare). 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S., Mittal R., Goyal N.J.E.T. An Assessment of Machine Learning and Deep Learning Techniques with Applications. 2022;107(1):8979. [Google Scholar]
- 20.Fang B., Meng Q.H.J. The laboratory's role in combating COVID-19. Crit. Rev. Clin. Lab Sci. 2020;57(6):400–414. doi: 10.1080/10408363.2020.1776675. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty T., Ghosh I. Real-time forecasts and risk assessment of novel coronavirus (COVID-19) cases: a data-driven analysis. Chaos, Solit. Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields B.K., Demirjian N.L., Gholamrezanezhad A. Coronavirus Disease 2019 (COVID-19) diagnostic technologies: a country-based retrospective analysis of screening and containment procedures during the first wave of the pandemic. Clin. Imag. 2020;67:219–225. doi: 10.1016/j.clinimag.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S., Maurya S. Innovative Approaches in Diagnosis and Management of Crop Diseases. Apple Academic Press; 2021. Innovative diagnostic tools for plant pathogenic virus; pp. 101–165. [Google Scholar]
- 24.Raybould M.I., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37(5):734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q., Allot A., Lu Z. LitCovid: an open database of COVID-19 literature. Nucleic Acids Res. 2021;49(D1):D1534–D1540. doi: 10.1093/nar/gkaa952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., et al. Deep learning based drug screening for novel coronavirus 2019-nCov. Interdiscipl. Sci. Comput. Life Sci. 2020;12(3):368–376. doi: 10.1007/s12539-020-00376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang S., et al. GESS: a database of global evaluation of SARS-CoV-2/hCoV-19 sequences. Nucleic Acids Res. 2021;49(D1):D706–D714. doi: 10.1093/nar/gkaa808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6(1):1–6. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amahong K., et al. vol. 51. 2023. pp. D546–D556. (CovInter: Interaction Data between Coronavirus RNAs and Host Proteins). D1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S., et al. vol. 23. 2022. p. bbab397. (RNA–RNA Interactions between SARS-CoV-2 and Host Benefit Viral Development and Evolution during COVID-19 Infection). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu F., et al. vol. 40. 2012. pp. D1128–D1136. (Therapeutic Target Database Update 2012: a Resource for Facilitating Target-Oriented Drug Discovery). D1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge Y., Wu Q.J. Knowledge‐based planning for intensity‐modulated radiation therapy: a review of data‐driven approaches. Med. Phys. 2019;46(6):2760–2775. doi: 10.1002/mp.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alimadadi A., Aryal S., Manandhar I., Munroe P.B., Joe B., Cheng X. vol. 52. American Physiological Society; Bethesda, MD: 2020. pp. 200–202. (Artificial Intelligence and Machine Learning to Fight COVID-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamshidi M., et al. Artificial intelligence and COVID-19: deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmatizadeh S., Valizadeh-Haghi S., Dabbagh A. The role of artificial intelligence in management of critical COVID-19 patients. J. Cell. Molecul. Anesthesia. 2020;5(1):16–22. [Google Scholar]
- 36.Uddin S., Khan A., Hossain M.E., Moni M.A.J., making d. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inf. Decis. Making. 2019;19(1):1–16. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel A.A. O'Reilly Media; 2019. Hands-on Unsupervised Learning Using Python: How to Build Applied Machine Learning Solutions from Unlabeled Data. [Google Scholar]
- 38.Sheikhpour R., Berahmand K., Forouzandeh S.J.K.-B.S. vol. 269. 2023. (Hessian-based Semi-supervised Feature Selection Using Generalized Uncorrelated Constraint). [Google Scholar]
- 39.Saberi-Movahed F., et al. vol. 146. 2022. (Decoding Clinical Biomarker Space of Covid-19: Exploring Matrix Factorization-Based Feature Selection Methods). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apple F.S., et al. vol. 51. 2005. pp. 810–824. (Future Biomarkers for Detection of Ischemia and Risk Stratification in Acute Coronary Syndrome). 5. [DOI] [PubMed] [Google Scholar]
- 41.Kamnitsas K., et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Bolhasani H., Mohseni M., Rahmani A.M. Deep learning applications for IoT in health care: a systematic review. Inform. Med. Unlocked. 2021;23 [Google Scholar]
- 43.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muhammad G., Hossain M.S. A deep-learning-based edge-centric COVID-19-like pandemic screening and diagnosis system within a B5G framework using blockchain. IEEE Network. 2021;35(2):74–81. [Google Scholar]
- 45.Campos-Ferreira D., et al. COVID-19 challenges: from SARS-CoV-2 infection to effective point-of-care diagnosis by electrochemical biosensing platforms. Biochem. Eng. J. 2021;176 doi: 10.1016/j.bej.2021.108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mostafa H.H., et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio. 2020;11(6) doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velay A., et al. Evaluation of the performance of SARS-CoV-2 serological tools and their positioning in COVID-19 diagnostic strategies. Diagn. Microbiol. Infect. Dis. 2020;98(4) doi: 10.1016/j.diagmicrobio.2020.115181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsharif M., Alsharif Y., Yahya K., Alomari O., Albreem M., Jahid A. Deep learning applications to combat the dissemination of COVID-19 disease: a review. Eur. Rev. Med. Pharmacol. Sci. 2020;24(21):11455–11460. doi: 10.26355/eurrev_202011_23640. [DOI] [PubMed] [Google Scholar]
- 49.Bharati S., Podder P., Mondal M., Prasath V. Medical imaging with deep learning for COVID-19 diagnosis: a comprehensive review. arXiv preprint arXiv:2107.09602. 2021 [Google Scholar]
- 50.Nguyen K.A., Stewart R.A., Zhang H., Jones C. Intelligent autonomous system for residential water end use classification: autoflow. Appl. Soft Comput. 2015;31:118–131. [Google Scholar]
- 51.Pustokhin D.A., Pustokhina I.V., Dinh P.N., Phan S.V., Nguyen G.N., Joshi G.P. An effective deep residual network based class attention layer with bidirectional LSTM for diagnosis and classification of COVID-19. J. Appl. Stat. 2020:1–18. doi: 10.1080/02664763.2020.1849057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Axiaq A., Almohtadi A., Massias S.A., Ngemoh D., Harky A. The role of computed tomography scan in the diagnosis of COVID-19 pneumonia. Curr. Opin. Pulm. Med. 2021;27(3):163–168. doi: 10.1097/MCP.0000000000000765. [DOI] [PubMed] [Google Scholar]
- 53.Pham T.D. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-74164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vengesai A., et al. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst. Rev. 2021;10(1):1–23. doi: 10.1186/s13643-021-01689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ai T., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khatami F., et al. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-80061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishfaq A., et al. Role of High Resolution Computed Tomography chest in the diagnosis and evaluation of COVID-19 patients-A systematic review and meta-analysis. European J. Radiol. open. 2021;8 doi: 10.1016/j.ejro.2021.100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prokop M., et al. CO-RADS: a categorical CT assessment scheme for patients suspected of having COVID-19—definition and evaluation. Radiology. 2020;296(2):E97–E104. doi: 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabaan A.A., et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Inf. Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- 60.Liu J. A novel implementation of machine learning for the efficient, explainable diagnosis of COVID-19 from chest CT. arXiv preprint arXiv:2207.07117. 2022 [Google Scholar]
- 61.Verma A., Amin S.B., Naeem M., Saha M. Detecting COVID-19 from chest computed tomography scans using AI-driven android application. Comput. Biol. Med. 2022;143 doi: 10.1016/j.compbiomed.2022.105298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z., et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recogn. 2021;110 doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dey N., Zhang Y.-D., Rajinikanth V., Pugalenthi R., Raja N.S.M. Customized VGG19 architecture for pneumonia detection in chest X-rays. Pattern Recogn. Lett. 2021;143:67–74. [Google Scholar]
- 64.Bhattacharya S., et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: a survey. Sustain. Cities Soc. 2021;65 doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abumalloh R.A., et al. Medical image processing and COVID-19: a literature review and bibliometric analysis. J. Infect. Public Health. 2021 doi: 10.1016/j.jiph.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imag. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 68.Jaiswal A.K., Tiwari P., Kumar S., Gupta D., Khanna A., Rodrigues J.J. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement. 2019;145:511–518. [Google Scholar]
- 69.Joo H.S., Wong J., Naik V.N., Savoldelli G.L. The value of screening preoperative chest x-rays: a systematic review. Can. J. Anaesth. 2005;52(6):568–574. doi: 10.1007/BF03015764. [DOI] [PubMed] [Google Scholar]
- 70.Candemir S., Antani S. A review on lung boundary detection in chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019;14(4):563–576. doi: 10.1007/s11548-019-01917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khobahi S., Agarwal C., Soltanalian M. MedRxiv; 2020. Coronet: A Deep Network Architecture for Semi-supervised Task-Based Identification of Covid-19 from Chest X-Ray Images. [Google Scholar]
- 72.Muratov E.N., et al. Chemical Society Reviews; 2021. A Critical Overview of Computational Approaches Employed for COVID-19 Drug Discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Molicul. Info. 2020;39(8) doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bobrowski T., et al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol. Ther. 2021;29(2):873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan A., et al. HantavirusesDB: vaccinomics and RNA-based therapeutics database for the potentially emerging human respiratory pandemic agents. Microb. Pathog. 2021;160 doi: 10.1016/j.micpath.2021.105161. [DOI] [PubMed] [Google Scholar]
- 76.Unanue I.J., Borzeshi E.Z., Piccardi M. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. J. Biomed. Inf. 2017;76:102–109. doi: 10.1016/j.jbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 77.Indumathi N., Shanmuga Eswari M., Salau A.O., Ramalakshmi R., Revathy R. Intelligent Interactive Multimedia Systems for E-Healthcare Applications. Springer; 2022. Prediction of COVID-19 outbreak with current substantiation using machine learning algorithms; pp. 171–190. [Google Scholar]
- 78.Tello-Mijares S., Woo L. Computed tomography image processing analysis in COVID-19 patient follow-up assessment. J. Healthc. Eng. 2021;2021 doi: 10.1155/2021/8869372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi F., et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEE Rev. Biomed. Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 80.Steinkamp J.M., Chambers C., Lalevic D., Zafar H.M., Cook T.S. Toward complete structured information extraction from radiology reports using machine learning. J. Digit. Imag. 2019;32(4):554–564. doi: 10.1007/s10278-019-00234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.J. Cui, J. Cui, L. I. Bouwman, K. S. van Rooijen, J. P. van Putten, and M. R. de Zoete, "Campylobacter Releases High Levels of the Inflammatory Metabolite ADP-Heptose that Activate Pro-inflammatory Responses in Intestinal Epithelial Cells.".
- 82.Aggarwal A., Mittal M., Battineni G. Generative adversarial network: an overview of theory and applications. Int. J. Info. Manag. Data Insight. 2021;1(1) [Google Scholar]
- 83.Welander P., Karlsson S., Eklund A. Generative adversarial networks for image-to-image translation on multi-contrast mr images-a comparison of cyclegan and unit. arXiv preprint arXiv:1806.07777. 2018 [Google Scholar]
- 84.Armanious K., et al. MedGAN: medical image translation using GANs. Comput. Med. Imag. Graph. 2020;79 doi: 10.1016/j.compmedimag.2019.101684. [DOI] [PubMed] [Google Scholar]
- 85.Dash A., Ye J., Wang G. A review of generative adversarial networks (GANs) and its applications in a wide variety of disciplines--from medical to remote sensing. arXiv preprint arXiv:2110.01442. 2021 [Google Scholar]
- 86.Jabbar A., Li X., Omar B. A survey on generative adversarial networks: variants, applications, and training. ACM Comput. Surv. 2021;54(8):1–49. [Google Scholar]
- 87.Fekri M.N., Ghosh A.M., Grolinger K. Generating energy data for machine learning with recurrent generative adversarial networks. Energies. 2019;13(1):130. [Google Scholar]
- 88.Chen Y., Zhao Y., Jia W., Cao L., Liu X. Adversarial-learning-based image-to-image transformation: a survey. Neurocomputing. 2020;411:468–486. [Google Scholar]
- 89.Qin Z., Chen Q., Ding Y., Zhuang T., Qin Z., Choo K.-K.R. Segmentation mask and feature similarity loss guided GAN for object-oriented image-to-image translation. Inf. Process. Manag. 2022;59(3) [Google Scholar]
- 90.Lukoševičius M., Jaeger H. Reservoir computing approaches to recurrent neural network training. Cumput. Sci. Rev. 2009;3(3):127–149. [Google Scholar]
- 91.Hartmann S., Baumert M. Automatic a-phase detection of cyclic alternating patterns in sleep using dynamic temporal information. IEEE Trans. Neural Syst. Rehabil. Eng. 2019;27(9):1695–1703. doi: 10.1109/TNSRE.2019.2934828. [DOI] [PubMed] [Google Scholar]
- 92.Mattioli I.A., Hassan A., Oliveira O.N., Jr., Crespilho F.N. On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sens. 2020;5(12):3655–3677. doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- 93.Park Y., Martin M., Koelle K. Epidemiological inference for emerging viruses using segregating sites. bioRxiv. 2022;7:2021. doi: 10.1038/s41467-023-38809-7. 07.451508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patiño-Galindo J.Á., Filip I., Rabadan R. Global patterns of recombination across human viruses. Mol. Biol. Evol. 2021;38(6):2520–2531. doi: 10.1093/molbev/msab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asif M., Xu Y., Xiao F., Sun Y. Diagnosis of COVID-19, vitality of emerging technologies and preventive measures. Chem. Eng. J. 2021;423 doi: 10.1016/j.cej.2021.130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sharma Y., Sharma L., Agrawal P., Gupta M., Vardhan H. Covid-19 detection using qRT-PCR-A review. J. Res. Appl. Sci. Biotechnol. 2022;1(3):238–248. [Google Scholar]
- 97.T. Covid, "Combo Kit," Instructions for Use, Multiplex Real time RT-PCR Test Intended for the Qualitative Detection Fo Nucleic Acid from SARS-CoV-2. AppliedBiosystems. ThermoFisher Scientific. Catalog, no. A47814.
- 98.Andersson M.I., et al. SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res. 2020;5 doi: 10.12688/wellcomeopenres.16002.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhavaniramya S., Ramar V., Vishnupriya S., Palaniappan R., Sibiya A., Vaseeharan B. Comprehensive analysis of SARS-COV-2 drug targets and pharmacological aspects in treating the COVID-19. Curr. Mol. Pharmacol. 2022;15(2):393–417. doi: 10.2174/1874467214666210811120635. [DOI] [PubMed] [Google Scholar]
- 100.Lagerborg K.A. 2022. NGS-Based Engineering of Potent Muscle-Tropic Gene Therapy Vectors and Synthetic Controls for Viral Surveillance. [Google Scholar]
- 101.Wurtzer S., et al. From Alpha to Omicron BA. 2: new digital RT-PCR approach and challenges for SARS-CoV-2 VOC monitoring and normalization of variant dynamics in wastewater. Sci. Total Environ. 2022;848 doi: 10.1016/j.scitotenv.2022.157740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rusková M., et al. Useful molecular tools for facing next pandemic events: effective sample preparation and improved RT-PCR for highly sensitive detection of SARS-CoV-2 in wastewater environment. Int. J. Hyg Environ. Health. 2022;245 doi: 10.1016/j.ijheh.2022.114017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahmed W., et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Al-Rohaimi A.H., Al Otaibi F. Novel SARS-CoV-2 outbreak and COVID19 disease; a systemic review on the global pandemic. Genes Dis. 2020;7(4):491–501. doi: 10.1016/j.gendis.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eftekhari A., et al. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms. 2021;9(2):232. doi: 10.3390/microorganisms9020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng D., Shi X., Yu J., Zhang D. How does the Chinese economy react to uncertainty in international crude oil prices? Int. Rev. Econ. Finance. 2019;64:147–164. [Google Scholar]
- 107.Castañeda M.e.T., Alegret S., Merkoci A. Electrochemical sensing of DNA using gold nanoparticles. Electroanalysis: An International Journal Devoted to Fundamental and Practical Aspects of Electroanalysis. 2007;19(7‐8):743–753. [Google Scholar]
- 108.Verma M.K., et al. Rapid diagnostic methods for SARS-CoV-2 (COVID-19) detection: an evidence-based report. J. Med. Life. 2021;14(4):431. doi: 10.25122/jml-2021-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.R. H. Khalil and M. A. S. M. Cuse, "Validation of the Current RT-PCR Assay Used to Detect Leptospira DNA.".
- 110.O'Toole J., Sinclair M., Malawaraarachchi M., Hamilton A., Barker S.F., Leder K. Microbial quality assessment of household greywater. Water Res. 2012;46(13):4301–4313. doi: 10.1016/j.watres.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 111.Soliman H., El-Matbouli M. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of viral hemorrhagic septicaemia virus (VHS) Vet. Microbiol. 2006;114(3–4):205–213. doi: 10.1016/j.vetmic.2005.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stevens B.A., et al. Combined SARS-CoV-2 nucleic acid amplification testing and respiratory virus panel RT-PCR on the Hologic Panther Fusion system. J. Clin. Virol. 2021;138 doi: 10.1016/j.jcv.2021.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mertens P., et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front. Med. 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. 2017;14(5):637–650. doi: 10.1080/15476286.2016.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J., Khetarpal I., Sen S., Murray R.M. Synthetic circuit for exact adaptation and fold-change detection. Nucleic Acids Res. 2014;42(9):6078–6089. doi: 10.1093/nar/gku233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Su X., Xiao X., Zhang C., Zhao M. Nucleic acid fluorescent probes for biological sensing. Appl. Spectrosc. 2012;66(11):1249–1262. doi: 10.1366/12-06803. [DOI] [PubMed] [Google Scholar]
- 117.Javalkote V.S., et al. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods. 2020 doi: 10.1016/j.ymeth.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ganbaatar U., Liu C. CRISPR-based COVID-19 testing: toward next-generation point-of-care diagnostics. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.663949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peyravian N., Malekzadeh Kebria M., Kiani J., Brouki Milan P., Mozafari M. CRISPR-associated (CAS) effectors delivery via microfluidic cell-deformation Chip. Materials. 2021;14(12):3164. doi: 10.3390/ma14123164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Joung J., et al. MedRxiv; 2020. Point-of-care Testing for COVID-19 Using SHERLOCK Diagnostics. [Google Scholar]
- 121.de Puig H., et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021;7(32):eabh2944. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Swarts D.C., Jinek M. Mechanistic insights into the cis-and trans-acting DNase activities of Cas12a. Mol. Cell. 2019;73(3):589–600. doi: 10.1016/j.molcel.2018.11.021. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chenya Z., et al. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Targeted Ther. 2021;6(1) doi: 10.1038/s41392-021-00645-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu L., et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170(4):714–726. doi: 10.1016/j.cell.2017.06.050. e10. [DOI] [PubMed] [Google Scholar]
- 125.Kriegova E., Fillerova R., Kvapil P. Direct-RT-qPCR detection of SARS-CoV-2 without RNA extraction as part of a COVID-19 testing strategy: from sample to result in one hour. Diagnostics. 2020;10(8):605. doi: 10.3390/diagnostics10080605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Higgins V., Fabros A., Kulasingam V. Quantitative measurement of anti-SARS-CoV-2 antibodies: analytical and clinical evaluation. J. Clin. Microbiol. 2021;59(4) doi: 10.1128/JCM.03149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beavis K.G., et al. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bastos M.L., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Davern T.J., et al. Acute hepatitis E infection accounts for some cases of suspected drug-induced liver injury. Gastroenterology. 2011;141(5):1665–1672. doi: 10.1053/j.gastro.2011.07.051. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Isho B., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5(52):eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tantuoyir M.M., Rezaei N. Serological tests for COVID‐19: potential opportunities. Cell Biol. Int. 2021;45(4):740–748. doi: 10.1002/cbin.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Richardson M., Page I. Role of serological tests in the diagnosis of mold infections. Curr. Fungal Infect. Rep. 2018;12(3):127–136. doi: 10.1007/s12281-018-0321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Datta S. 2021. Aptamers for Detection and Diagnostics (ADD) Is a Proposed Mobile App Acquiring Optical Data from Conjugated Quantum Nanodots to Identify Molecules Indicating Presence of SARS-CoV-2 Virus: Why Public Health and Healthcare Need Smartphone Sensors as a Platform for Early Detection and Prevention. [Google Scholar]
- 134.Martinez W.C. How to get away with immunity: FDA's emergency use authorization scheme and PREP act liability protection in the context of COVID-19. Loyola Consum. Law Rev. 2021;33:1. [Google Scholar]
- 135.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fernandes L., et al. Saliva in the diagnosis of COVID-19: a review and new research directions. J. Dent. Res. 2020;99(13):1435–1443. doi: 10.1177/0022034520960070. [DOI] [PubMed] [Google Scholar]
- 137.Tatarewicz S.M., Mytych D.T., Manning M.S., Swanson S.J., Moxness M.S., Chirmule N. Strategic characterization of anti-drug antibody responses for the assessment of clinical relevance and impact. Bioanalysis. 2014;6(11):1509–1523. doi: 10.4155/bio.14.114. [DOI] [PubMed] [Google Scholar]
- 138.Giri B., Pandey S., Shrestha R., Pokharel K., Ligler F.S., Neupane B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2021;413(1):35–48. doi: 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arun Krishnan R., Elizabeth Thomas R., Sukumaran A., Paul J.K., Vasudevan D. COVID-19: current trends in invitro diagnostics. Indian J. Clin. Biochem. 2020;35(3):285–289. doi: 10.1007/s12291-020-00906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reen D.J. Enzyme-linked immunosorbent assay (ELISA) Basic Protein and Peptide Protocols. 1994:461–466. doi: 10.1385/0-89603-268-X:461. [DOI] [PubMed] [Google Scholar]
- 141.Alhamid G., Tombuloglu H., Rabaan A.A., Al-Suhaimi E. SARS-CoV-2 detection methods: a comprehensive review. Saudi J. Biol. Sci. 2022 doi: 10.1016/j.sjbs.2022.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Konstantinou G.N. Food Allergens. Springer; 2017. Enzyme-linked immunosorbent assay (ELISA) pp. 79–94. [DOI] [PubMed] [Google Scholar]
- 143.Nuccetelli M., Pieri M., Gisone F., Bernardini S. Combined anti-SARS-CoV-2 IgA, IgG, and IgM detection as a better strategy to prevent second infection spreading waves. Immunol. Invest. 2022;51(2):233–245. doi: 10.1080/08820139.2020.1823407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luvira V., et al. False positivity of anti-SARS-CoV-2 antibodies in patients with acute tropical diseases in Thailand. Tropical Med. Infect. Dis. 2022;7(7):132. doi: 10.3390/tropicalmed7070132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Melgoza‐González E.A., et al. Analysis of IgG, IgA and IgM antibodies against SARS‐CoV‐2 spike protein S1 in convalescent and vaccinated patients with the pfizer‐BioNTech and CanSinoBio vaccines. Transboundary Emerg. Dis. 2022;69(4):e734–e745. doi: 10.1111/tbed.14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li D., Calderone R., Nsouli T.M., Reznikov E., Bellanti J.A. vol. 43. OceanSide Publications, Inc; 2022. Salivary and serum IgA and IgG responses to SARS-CoV-2-spike protein following SARS-CoV-2 infection and after immunization with COVID-19 vaccines; pp. 419–430. (Allergy and Asthma Proceedings). 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zorgi N.E., Meireles L.R., Oliveira D.B.L., Araujo D.B., Durigon E.L., de Andrade Junior H.F. Isolated specific IgA against respiratory viruses, Influenza or SARS-CoV-2, present in the saliva of a fraction of healthy and asymptomatic volunteers. Clinics. 2022 doi: 10.1016/j.clinsp.2022.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.La Rosa G., et al. Key SARS-CoV-2 mutations of alpha, gamma, and eta variants detected in urban wastewaters in Italy by long-read amplicon sequencing based on nanopore technology. Water. 2021;13(18):2503. [Google Scholar]
- 149.Schrom J., et al. Comparison of SARS-CoV-2 reverse transcriptase polymerase chain reaction and BinaxNOW rapid antigen tests at a community site during an omicron surge: a cross-sectional study. Ann. Intern. Med. 2022 doi: 10.7326/L22-0257. [DOI] [PubMed] [Google Scholar]
- 150.Mostafa M., et al. Current trends in COVID-19 diagnosis and its new variants in physiological fluids: surface antigens, antibodies, nucleic acids, and RNA sequencing. TrAC, Trends Anal. Chem. 2022 doi: 10.1016/j.trac.2022.116750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Greenland-Bews C., et al. Evaluation of Eight Lateral Flow Tests for the detection of anti-SARS-CoV-2 antibodies in a vaccinated population, medRxiv. 2022 doi: 10.1186/s12879-023-08033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oh D.Y., et al. "Advancing precision vaccinology by molecular and genomic surveillance of severe acute respiratory syndrome coronavirus 2 in Germany, 2021,". Clin. Infect. Dis. 2022;75(Supplement_1):S110–S120. doi: 10.1093/cid/ciac399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elyanow R., et al. T cell receptor sequencing identifies prior SARS-CoV-2 infection and correlates with neutralizing antibodies and disease severity. JCI insight. 2022;7(10) doi: 10.1172/jci.insight.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu J., et al. Horseradish peroxidase-triggered direct in situ fluorescent immunoassay platform for sensing cardiac troponin I and SARS-CoV-2 nucleocapsid protein in serum. Biosens. Bioelectron. 2022;198 doi: 10.1016/j.bios.2021.113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ansari-Moghaddam B., et al. Screening role of complete blood cell count indices and C reactive protein in patients who are symptomatic for COVID-19. Dis. Emerg. Med. J. 2022 [Google Scholar]
- 156.Li Y., et al. Predictive value of serum cystatin C for risk of mortality in severe and critically ill patients with COVID-19. World J. Clin. Cases. 2020;8(20):4726. doi: 10.12998/wjcc.v8.i20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin L., et al. The predictive value of serum level of cystatin C for COVID-19 severity. Sci. Rep. 2021;11(1):1–8. doi: 10.1038/s41598-021-01570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shakked N.P., et al. Early prediction of COVID-19-associated acute kidney injury: are serum NGAL and serum Cystatin C levels better than serum creatinine? Clin. Biochem. 2022;102:1–8. doi: 10.1016/j.clinbiochem.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Law J.N. Development and application of network algorithms for prediction of gene function and response to viral infection and chemicals. Virginia Tech. 2020 [Google Scholar]
- 160.Iravani S. Nano-and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater. Adv. 2020;1(9):3092–3103. [Google Scholar]
- 161.Chaimayo C., et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2020;17(1):1–7. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Davis T.M., Wilson W.D. Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal. Biochem. 2000;284(2):348–353. doi: 10.1006/abio.2000.4726. [DOI] [PubMed] [Google Scholar]
- 163.Kim M., Lee J.H., Nam J.M. Plasmonic photothermal nanoparticles for biomedical applications. Adv. Sci. 2019;6(17) doi: 10.1002/advs.201900471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kumar N., Shetti N.P., Jagannath S., Aminabhavi T.M. Electrochemical sensors for the detection of SARS-CoV-2 virus. Chem. Eng. J. 2022;430 doi: 10.1016/j.cej.2021.132966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pandey P.S., Raghuwanshi S.K., Shadab A., Ansari M.T.I., Tiwari U.K., Kumar S. SPR based biosensing chip for COVID-19 diagnosis-A review. IEEE Sensor. J. 2022 doi: 10.1109/JSEN.2022.3181423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krüger L.J., et al. Evaluation of the accuracy and ease-of-use of Abbott PanBio-A WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2. medRxiv. 2020 doi: 10.1371/journal.pone.0247918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mukherjee S., et al. A graphene and aptamer based liquid gated FET-like electrochemical biosensor to detect adenosine triphosphate. IEEE Trans. NanoBioscience. 2015;14(8):967–972. doi: 10.1109/TNB.2015.2501364. [DOI] [PubMed] [Google Scholar]
- 168.Kunkel S.D., et al. Ursolic acid increases skeletal muscle and brown fat and decreases diet-induced obesity, glucose intolerance and fatty liver disease. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pelsers M.M., et al. A sensitive immunoassay for rat fatty acid translocase (CD36) using phage antibodies selected on cell transfectants: abundant presence of fatty acid translocase/CD36 in cardiac and red skeletal muscle and up-regulation in diabetes. Biochem. J. 1999;337(3):407–414. [PMC free article] [PubMed] [Google Scholar]
- 170.Ahmed S., Bui M.-P.N., Abbas A. Based chemical and biological sensors: engineering aspects. Biosens. Bioelectron. 2016;77:249–263. doi: 10.1016/j.bios.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 171.Umapathi R., et al. Colorimetric based on-site sensing strategies for the rapid detection of pesticides in agricultural foods: new horizons, perspectives, and challenges. Coord. Chem. Rev. 2021;446 [Google Scholar]
- 172.Al Mamun M., Wahab Y.A., Hossain M.M., Hashem A., Johan M.R. Electrochemical biosensors with aptamer recognition layer for the diagnosis of pathogenic bacteria: barriers to commercialization and remediation. TrAC, Trends Anal. Chem. 2021;145 [Google Scholar]
- 173.Mao X., Liu C., Tong H., Chen Y., Liu K. Principles of digital PCR and its applications in current obstetrical and gynecological diseases. Am. J. Tourism Res. 2019;11(12):7209. [PMC free article] [PubMed] [Google Scholar]
- 174.Tiwari A., et al. Science of The Total Environment; 2022. Application of Digital PCR for Public Health-Related Water Quality Monitoring. [DOI] [PubMed] [Google Scholar]
- 175.Matsuda K. PCR-based detection methods for single-nucleotide polymorphism or mutation: real-time PCR and its substantial contribution toward technological refinement. Adv. Clin. Chem. 2017;80:45–72. doi: 10.1016/bs.acc.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 176.Li C., Zhao C., Bao J., Tang B., Wang Y., Gu B. Laboratory diagnosis of coronavirus disease-2019 (COVID-19) Clin. Chim. Acta. 2020;510:35–46. doi: 10.1016/j.cca.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yao L., et al. Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chem. Soc. Rev. 2021;50(6):3656–3676. doi: 10.1039/d0cs00595a. [DOI] [PubMed] [Google Scholar]
- 178.Agronskaia A., Tertoolen L., Gerritsen H. High frame rate fluorescence lifetime imaging. J. Phys. D Appl. Phys. 2003;36(14):1655–1662. [Google Scholar]
- 179.Behjati S., Tarpey P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. 2013;98(6):236–238. doi: 10.1136/archdischild-2013-304340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Reis-Filho J.S. Next-generation sequencing. Breast Cancer Res. 2009;11(3):1–7. doi: 10.1186/bcr2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu L., et al. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012;2012 doi: 10.1155/2012/251364. 251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Levy S.E., Myers R.M. Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 2016;17(1):95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- 183.Chiara M., et al. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Briefings Bioinf. 2021;22(2):616–630. doi: 10.1093/bib/bbaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Aljabr W., et al. Amplicon and metagenomic analysis of middle east respiratory syndrome (MERS) coronavirus and the microbiome in patients with severe MERS. mSphere. 2021;6(4) doi: 10.1128/mSphere.00219-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hillier L.W., et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5(2):183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 186.Landgraff C., et al. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B. 1.1. 7 variant of concern from a Canadian municipality concurrent with an outbreak. medRxiv. 2021 [Google Scholar]
- 187.Clark S.C., Egan R., Frazier P.I., Wang Z. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics. 2013;29(4):435–443. doi: 10.1093/bioinformatics/bts723. [DOI] [PubMed] [Google Scholar]
- 188.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 2012;109(36):14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Arnaout R., et al. SARS-CoV2 testing: the limit of detection matters. bioRxiv. 2020 [Google Scholar]
- 190.Jin Z., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 191.Chushak Y., Stone M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009;37(12) doi: 10.1093/nar/gkp408. e87-e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu D.-Y., et al. Drug repurposing for COVID-19 treatment by integrating network pharmacology and transcriptomics. Pharmaceutics. 2021;13(4):545. doi: 10.3390/pharmaceutics13040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sadowski P., Fooshee D., Subrahmanya N., Baldi P. Synergies between quantum mechanics and machine learning in reaction prediction. J. Chem. Inf. Model. 2016;56(11):2125–2128. doi: 10.1021/acs.jcim.6b00351. [DOI] [PubMed] [Google Scholar]
- 194.Poggi M., Tosi F., Batsos K., Mordohai P., Mattoccia S. On the synergies between machine learning and binocular stereo for depth estimation from images: a survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021;44(9):5314–5334. doi: 10.1109/TPAMI.2021.3070917. [DOI] [PubMed] [Google Scholar]
- 195.Goh G.B., Hodas N.O., Vishnu A. Deep learning for computational chemistry. J. Comput. Chem. 2017;38(16):1291–1307. doi: 10.1002/jcc.24764. [DOI] [PubMed] [Google Scholar]
- 196.Titano J.J., et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med. 2018;24(9):1337–1341. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 197.Nikou M., Mansourfar G., Bagherzadeh J. Stock price prediction using DEEP learning algorithm and its comparison with machine learning algorithms. Intell. Syst. Account. Finance Manag. 2019;26(4):164–174. [Google Scholar]
- 198.Chiba Z., Abghour N., Moussaid K., Rida M. Intelligent approach to build a Deep Neural Network based IDS for cloud environment using combination of machine learning algorithms. Comput. Secur. 2019;86:291–317. [Google Scholar]
- 199.Miyamoto D.T. Harvard University; 2004. Probing the Functions of Kinesins in Mitosis. [Google Scholar]
- 200.Sharma A., Rani R. BE-DTI’: ensemble framework for drug target interaction prediction using dimensionality reduction and active learning. Comput. Methods Progr. Biomed. 2018;165:151–162. doi: 10.1016/j.cmpb.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 201.Zhou J., et al. Graph neural networks: a review of methods and applications. AI Open. 2020;1:57–81. [Google Scholar]
- 202.Gao K., et al. Methodology-centered review of molecular modeling, simulation, and prediction of SARS-CoV-2. Chem. Rev. 2022;122(13):11287–11368. doi: 10.1021/acs.chemrev.1c00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sangeetha S., Kumar M.S., Rajadurai H., Maheshwari V., Dalu G.T. An empirical analysis of an optimized pretrained deep learning model for COVID-19 diagnosis. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/9771212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cenikj G., Popovski G., Stojanov R., Seljak B.K., Eftimov T. 2020 IEEE International Conference on Big Data (Big Data) IEEE; 2020. BuTTER: BidirecTional LSTM for Food named-entity recognition; pp. 3550–3556. [Google Scholar]
- 205.Guo W.-F., Zhang S.-W., Feng Y.-H., Liang J., Zeng T., Chen L. Network controllability-based algorithm to target personalized driver genes for discovering combinatorial drugs of individual patients. Nucleic Acids Res. 2021;49(7) doi: 10.1093/nar/gkaa1272. e37-e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chou T.-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 207.Booth A.L., Abels E., McCaffrey P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021;34(3):522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yang H., et al. Application of extreme learning machine in the survival analysis of chronic heart failure patients with high percentage of censored survival time. Front. Cadiovac. Med. 2021;8 doi: 10.3389/fcvm.2021.726516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Khellal A., Ma H., Fei Q. 2018 37th Chinese Control Conference (CCC) IEEE; 2018. Convolutional neural network features comparison between back-propagation and extreme learning machine; pp. 9629–9634. [Google Scholar]
- 210.Li M.-B., Huang G.-B., Saratchandran P., Sundararajan N. Fully complex extreme learning machine. Neurocomputing. 2005;68:306–314. [Google Scholar]
- 211.Guzik T.J., et al. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020;116(10):1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mahmud E., et al. Management of acute myocardial infarction during the COVID-19 pandemic: a position statement from the society for cardiovascular angiography and interventions (SCAI), the American college of cardiology (ACC), and the American college of emergency physicians (ACEP) J. Am. Coll. Cardiol. 2020;76(11):1375–1384. doi: 10.1016/j.jacc.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mao G., et al. vol. 123. Parts A/B/C; 2021. Comprehensive comparison of artificial neural networks and long short-term memory networks for rainfall-runoff simulation. (Physics and Chemistry of the Earth). [Google Scholar]
- 214.Yu Y., Si X., Hu C., Zhang J. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019;31(7):1235–1270. doi: 10.1162/neco_a_01199. [DOI] [PubMed] [Google Scholar]
- 215.Li S., Li W., Cook C., Gao Y. Deep independently recurrent neural network (indrnn) arXiv preprint arXiv:1910.06251. 2019 [Google Scholar]
- 216.Liu Y., et al. Deep C-LSTM neural network for epileptic seizure and tumor detection using high-dimension EEG signals. IEEE Access. 2020;8:37495–37504. [Google Scholar]
- 217.Sharma S., Singh G., Sharma M. A comprehensive review and analysis of supervised-learning and soft computing techniques for stress diagnosis in humans. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104450. [DOI] [PubMed] [Google Scholar]
- 218.Moulahoum H., Ghorbanizamani F., Zihnioglu F., Turhan K., Timur S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta. 2021;222 doi: 10.1016/j.talanta.2020.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yeasmin M., et al. Routes of transmission of newly emerging SARS-CoV-2: a systematic review. Bangladesh J. Infect. Dis. 2020:S18–S31. [Google Scholar]
- 220.Morse S.S., et al. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380(9857):1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nayak S., Blumenfeld N.R., Laksanasopin T., Sia S.K. Point-of-care diagnostics: recent developments in a connected age. Anal. Chem. 2017;89(1):102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Subali A.D., Wiyono L. Reverse transcriptase loop mediated isothermal AmplifiCation (RT-LAMP) for COVID-19 diagnosis: a systematic review and meta-analysis. Pathog. Glob. Health. 2021;115(5):281–291. doi: 10.1080/20477724.2021.1933335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sadeghi P., et al. Lateral flow assays (LFA) as an alternative medical diagnosis method for detection of virus species: the intertwine of nanotechnology with sensing strategies. TrAC, Trends Anal. Chem. 2021;145 doi: 10.1016/j.trac.2021.116460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gustafsdottir S.M., et al. Proximity ligation assays for sensitive and specific protein analyses. Anal. Biochem. 2005;345(1):2–9. doi: 10.1016/j.ab.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 225.Alyafei K., Ahmed R., Abir F.F., Chowdhury M.E., Naji K.K. A comprehensive review of COVID-19 detection techniques: from laboratory systems to wearable devices. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Damin F., et al. CovidArray: a microarray-based assay with high sensitivity for the detection of Sars-Cov-2 in nasopharyngeal swabs. Sensors. 2021;21(7):2490. doi: 10.3390/s21072490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Newman J.D., Setford S.J. Enzymatic biosensors. Mol. Biotechnol. 2006;32(3):249–268. doi: 10.1385/MB:32:3:249. [DOI] [PubMed] [Google Scholar]
- 228.Srinivasan B., Tung S. Development and applications of portable biosensors. J. Lab. Autom. 2015;20(4):365–389. doi: 10.1177/2211068215581349. [DOI] [PubMed] [Google Scholar]
- 229.Mekonnen Y., Namuduri S., Burton L., Sarwat A., Bhansali S. Machine learning techniques in wireless sensor network based precision agriculture. J. Electrochem. Soc. 2019;167(3) [Google Scholar]
- 230.Bezinge L., Suea-Ngam A., deMello A.J., Shih C.-J. Nanomaterials for molecular signal amplification in electrochemical nucleic acid biosensing: recent advances and future prospects for point-of-care diagnostics. Molecul. Syst. Design Eng. 2020;5(1):49–66. [Google Scholar]
- 231.Bruch R., et al. CRISPR/Cas13a‐powered electrochemical microfluidic biosensor for nucleic acid amplification‐free miRNA diagnostics. Adv. Mater. 2019;31(51) doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 232.Quan C., Li C., Ma H., Li Y., Zhang H. Immunopathogenesis of coronavirus-induced acute respiratory distress syndrome (ARDS): potential infection-associated hemophagocytic lymphohistiocytosis. Clin. Microbiol. Rev. 2020;34(1) doi: 10.1128/CMR.00074-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kaushik A.K., et al. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl. Bio Mater. 2020;3(11):7306–7325. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 234.Zhang L., Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv. Biomarker Sci. Technol. 2020;2:1–23. doi: 10.1016/j.abst.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. Lancet Infect. Dis. 2003;3(11):722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xiao S., Tian Z., Wang Y., Si L., Zhang L., Zhou D. Recent progress in the antiviral activity and mechanism study of pentacyclic triterpenoids and their derivatives. Med. Res. Rev. 2018;38(3):951–976. doi: 10.1002/med.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jassim S.A., Naji M.A. Novel antiviral agents: a medicinal plant perspective. J. Appl. Microbiol. 2003;95(3):412–427. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 238.Tiwari J.N., Tiwari R.N., Singh G., Kim K.S. Recent progress in the development of anode and cathode catalysts for direct methanol fuel cells. Nano Energy. 2013;2(5):553–578. [Google Scholar]
- 239.Bergamo A., Sava G. Ruthenium anticancer compounds: myths and realities of the emerging metal-based drugs. Dalton Trans. 2011;40(31):7817–7823. doi: 10.1039/c0dt01816c. [DOI] [PubMed] [Google Scholar]
- 240.Antiochia R. Developments in biosensors for CoV detection and future trends. Biosens. Bioelectron. 2021;173 doi: 10.1016/j.bios.2020.112777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Oberholtzer K., Sivitz L., Mack A., Lemon S., Mahmoud A., Knobler S. Learning from SARS: Preparing for the Next Disease Outbreak: Workshop Summary. 2004. [PubMed] [Google Scholar]
- 242.Petryayeva E., Algar W.R. Toward point-of-care diagnostics with consumer electronic devices: the expanding role of nanoparticles. RSC Adv. 2015;5(28):22256–22282. [Google Scholar]
- 243.Varghese R., Kumar D., Sharma R. Global threat from novel SARS-CoV-2 variants, BF.7, XBB.1.5, BQ.1, and BQ.1.1: variants of concern? Hum. Cell. 2023;36(3):1218–1221. doi: 10.1007/s13577-023-00903-9. Epub 2023. PMID: 37000399; PMCID: PMC10063927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Varghese R., Pai S., Kumar D., Sharma R. SARS-CoV-2 XBB.1.16 variant: India in focus? J. Med. Virol. 2023;95(5) doi: 10.1002/jmv.28829. PMID: 37222492. [DOI] [PubMed] [Google Scholar]
- 245.Varghese R., Patel P., Kumar D., Sharma R. Climate change and glacier melting: risks for unusual outbreaks? J. Trav. Med. 2023 doi: 10.1093/jtm/taad015. taad015. Epub ahead of print. PMID: 36721991. [DOI] [PubMed] [Google Scholar]
- 246.Dalton K.R., Rock C., Carroll K.C., Davis M.F. One Health in hospitals: how understanding the dynamics of people, animals, and the hospital built-environment can be used to better inform interventions for antimicrobial-resistant gram-positive infections. Antimicrob. Resist. Infect. Control. 2020;9(1):1–17. doi: 10.1186/s13756-020-00737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]