Abstract
Inflammatory bowel diseases (IBD) are chronic disorders of the gastrointestinal tract with two subtypes: Crohn’s disease (CD) and ulcerative colitis (UC). To date, most IBD genetic associations were derived from individuals of European ancestries (EUR). Here we report the largest IBD study of individuals of East Asian ancestries (EAS), including 14,393 cases and 15,456 controls. We found 80 IBD loci in EAS alone and 320 when meta-analyzed with ~370,000 EUR individuals (~30,000 cases), among which 81 are novel. EAS enriched coding variants implicate many new IBD genes, including ADAP1 and GIT2. While IBD genetic effects are generally consistent across ancestries, genetics underlying CD appears more ancestry dependent than UC, driven by both allele frequency (NOD2) and effect (TNFSF15). We extended the IBD polygenic risk score (PRS) by incorporating both ancestries, greatly improving its accuracy and highlighting the importance of diversity for the equitable deployment of PRS.
Inflammatory bowel diseases (IBD) are a group of chronic, debilitating disorders of the gastrointestinal tract with the peak onset in adolescence and early adulthood1. As of 2017, there were 6.8 million people diagnosed with IBD globally2, with increasing incidence and prevalence worldwide, especially in recently industrialized countries, likely due to the modernization and westernization of the populations2,3. IBD have two etiologically related subtypes, Crohn’s disease (CD) and ulcerative colitis (UC). Genome-wide association studies (GWAS) have discovered over 240 genetic loci associated with IBD4.
However, to date, most IBD genetic associations have been derived using individuals of European ancestries (EUR)4,5, with only a few studies of much smaller sample sizes in non-European populations6–8. For example, the largest IBD GWAS in African9 and Asian7 populations included 2,345 and 3,195 cases, respectively, only about 10% of the number in the largest EUR IBD GWAS (29,336)4. Among the ImmunoChip samples, a cohort that was uniformly processed and drove several large-scale IBD genetics studies6,10–12, 87% of patients were of European ancestries, with the remaining 13% from Asian (7%), Indian (4%) and Iranian (2%) ancestries, respectively. This strong bias towards EUR severely limits our understanding of IBD biology and its application to most of the world’s population. First, because not all disease-causing variants are present in EUR, using EUR alone will miss important disease-causing variants that are absent or rare in EUR. For example, a schizophrenia GWAS in East Asian ancestries (EAS), with a sample size only 30% of its EUR counterpart, discovered a disease-associated variant implicating the calcium channel α2δ−2 subunit that was missed in EUR because this variant is 60× rarer in EUR13. Similarly, a PCSK9 missense variant (R93C) was found to strongly influence LDL-C levels in the Chinese population, which was missed in the GWAS in EUR with 10× sample size because R93C is 100× rarer in EUR14.
Further, genetic findings derived from EUR may not apply to non-Europeans, who collectively constitute 88% of the world population. For example, NOD2, the first reported and a well-established CD risk gene15,16, has a composite allele frequency (AF) of 13% across nine putative rare or low-frequency IBD causal variants in EUR12. The composite AF in EAS for these nine variants is only 0.06%, suggesting that substantially fewer individuals of EAS ancestries are affected by these well-established IBD causal alleles6. TNFSF15, in contrast, increases CD risk with a substantially greater magnitude in EAS than in EUR6 (OR 1.75 (95% CI: 1.57-1.91) vs. 1.15 (95% CI: 1.11-1.17)), an as yet unexplained difference perhaps driven by clinical heterogeneity or gene-environment/gene-gene interactions6. A comparative IBD genetic study across ancestries should therefore reveal interesting IBD biology and identify ancestry-specific and shared biological components and therapeutic targets to ensure that genetic discoveries may reduce, rather than expand, health disparities. In addition to specific IBD loci, over the genome, a polygenic risk score (PRS) trained using Korean individuals of a much smaller sample size outperformed those trained using EUR individuals in predicting CD risk in the Korean target population7, suggesting the importance of performing GWAS in global populations to accelerate the equitable deployment of PRS in clinical settings and maximize its healthcare potential17.
Here we present the largest IBD genetics study in East Asians with 14,393 cases and 15,456 controls, a 4× increase from the previous IBD genetic studies in EAS7. Through integrative and comparative analyses with resources in EUR, including 25,042 cases and 34,915 controls of non-Finnish European (NFE) ancestries from the International IBD Genetics Consortium (IIBDGC), and 5,671 cases and 303,191 controls from FinnGen18, a European population with a unique founding bottleneck, our study comprehensively investigates the comparative genetic architecture of IBD across East Asian and European ancestries with unprecedented statistical power from a total of 45,106 cases and 353,562 controls.
RESULTS
Study samples
We included individuals of East Asian and European ancestries in this study (Fig. 1). In the East Asian (EAS) ancestry analysis, we aggregated 14,393 IBD cases (7,372 CD, 6,862 UC, 159 IBD-Unclassified (IBD-U)) and 15,456 selected controls from sample collections from China (SHA1), Korea (KOR1), Japan (JPN1) and a multi-ancestry cohort genotyped on ImmunoChip (ICH1), a non-GWAS custom array (Fig. 1 and Methods). After rigorous quality control and association analyses (Supplementary Table 1 and Methods), quantile-quantile (QQ) plots of the test statistics, the genomic inflation factors (λ1000 = 0.99-1.05), and the LD score regression (LDSC) intercepts (1.02-1.06) all showed that population structure and other confounding factors were well controlled (Extended Data Fig. 1a–o). In the European (EUR) ancestry analysis, we aggregated 30,713 IBD cases (13,501 CD, 16,390 UC, 822 IBD-U) and 338,106 controls from Finnish (FIN, unpublished) and non-Finnish Europeans (NFE, published4) (Methods). For FIN individuals, QQ plots, genomic inflation factors (λ1000 = 1.01-1.02), and the LDSC intercepts (1.01-1.07) confirmed that population structure and other confounding factors were well controlled (Extended Data Fig. 1p–r).
Figure 1 |. Overview of the study design.
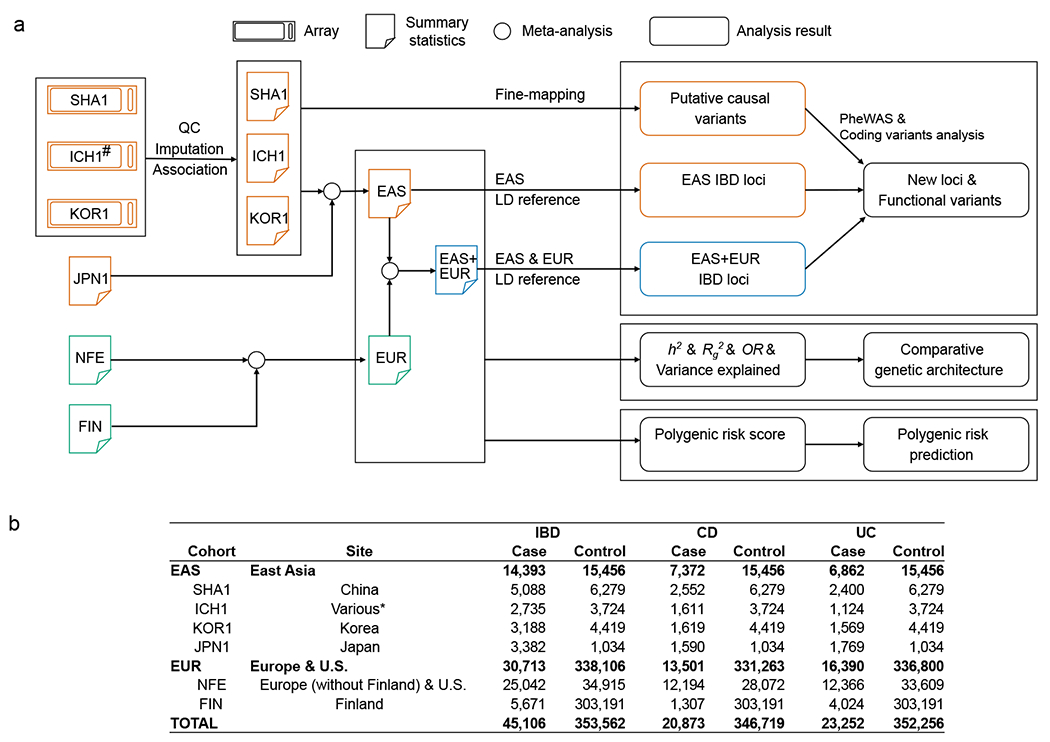
a, Data and analyses in this study. b, Post-QC sample size from each ancestry. #ICH1 was genotyped on ImmunoChip, a non-GWAS custom array (Methods). *ICH1 includes individuals recruited from Hong Kong SAR, China, Korea, and Japan.
Genetic loci associated with IBD
Using fixed-effect meta-analysis to combine EAS studies (Methods), we found 80 genetic loci significantly associated (P < 5 × 10−8) with CD, UC, or both (Fig. 2, Supplementary Tables 2 and 3, and Supplementary Data 1 and 2). For convenience, unless noted otherwise, we used “IBD-associated” or “associated with IBD” to broadly refer to loci associated with CD, UC or both in this study. Among the 80 IBD-associated loci, 54 were reported for the first time in EAS, while 38 of these 54 loci had been reported in NFE before, including seven in a recent CD exome sequencing study4,19, suggesting an overall convergence of IBD genetic architecture across ancestries. Altogether, we found 16 new IBD genetic associations in EAS that were not reported previously (Table 1). These new IBD genetic associations have elevated minor allele frequencies (MAF) in EAS compared with those in EUR (average MAF 0.25 vs. 0.17, two-sided paired t test P = 0.015; Extended Data Fig. 2), suggesting the importance of including global ancestries in genetic studies to ensure the relevance of the genetic findings. Twelve of these IBD genetic associations have pleiotropic associations with gene expressions and other complex traits and disorders, including immune cell counts in Biobank Japan, UK Biobank and FinnGen (Supplementary Table 4).
Figure 2 |. IBD genetic associations.
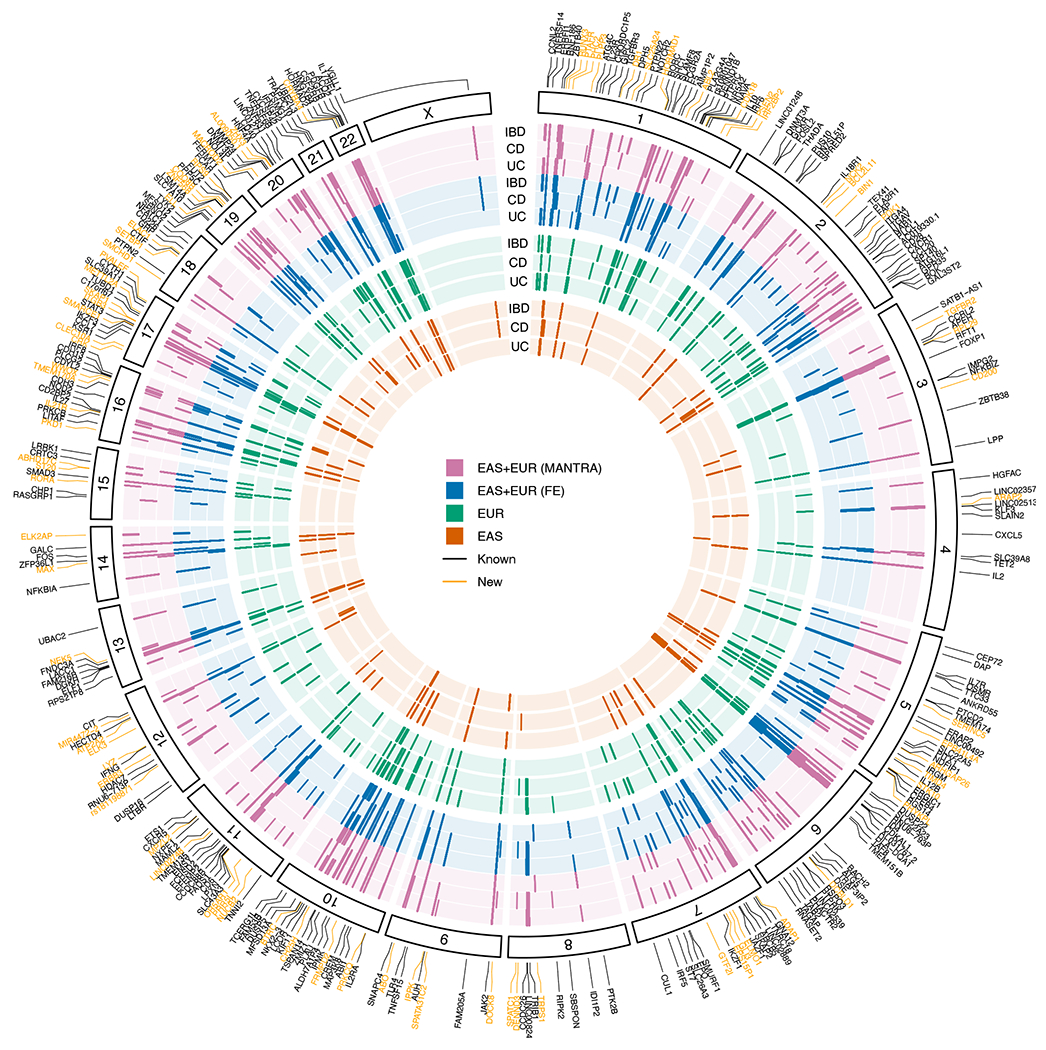
Each layer of the plot represents results from a GWAS analysis, with results from the same ancestry grouped by the color. Within each ancestry, UC, CD, and IBD are ordered from the inner to the outer layer. Genome-wide significant associations (P < 5 × 10−8) were plotted as short lines. Known indicates previously reported IBD loci4,7,8,34–43. New indicates new IBD loci from the meta-analysis (MANTRA and FE) and/or the EAS analysis.
Table 1 |.
New IBD-associated genetic loci in EAS
| Index varianta | rsID | EAb | EAFc | Subtype | ORd | P d | Genee |
|---|---|---|---|---|---|---|---|
| chr1:24977085:A:G | rs10903122 | G | 0.68 | CD | 0.86 | 2.84E-11 | RUNX3 |
| chr1:28131939:C:T | rs140466198 | T | 0.10 | CD | 1.30 | 1.05E-08 | PTAFR |
| chr3:112334718:G:A | rs1317244 | A | 0.36 | CD | 1.17 | 2.65E-08 | CD200 |
| chr6:22062256:C:A | rs4712651 | A | 0.65 | UC | 0.83 | 2.53E-11 | CASC15 |
| chr7:950350:G:A | rs77992257 | A | 0.20 | IBD | 0.83 | 1.04E-13 | ADAP1 |
| chr7:37417420:C:G | rs28581678 | G | 0.20 | CD | 1.22 | 3.02E-10 | ELMO1 |
| chr7:74711703:C:T | rs117026326 | T | 0.08 | CD | 1.37 | 8.91E-11 | GTF2I |
| chr8:141164479:C:T | rs438041 | T | 0.53 | CD | 1.17 | 4.10E-08 | DENND3 |
| chr11:118273990:A:G | rs141340254 | G | 0.03 | CD | 0.62 | 4.66E-08 | MPZL2 |
| chr12:96134457:C:G | rs11108429 | G | 0.60 | CD | 1.19 | 7.93E-10 | ELK3 |
| chr12:110259525:G:A | rs117121174 | A | 0.08 | CD | 0.74 | 1.35E-10 | ATP2A2 |
| chr12:116408331:C:T | rs113281820 | T | 0.27 | CD | 1.19 | 1.03E-08 | MIR4472-2 |
| chr16:27384341:C:CT | rs201121732 | CT | 0.02 | UC | 1.63 | 1.75E-08 | IL21R |
| chr17:47280984:G:C | rs11079770 | C | 0.44 | CD | 0.86 | 2.21E-08 | ITGB3 |
| chr18:44806588:T:C | rs16978179 | C | 0.15 | CD | 1.25 | 8.72E-09 | SETBP1 |
| chr19:54219677:C:T | rs255773 | T | 0.51 | CD | 1.24 | 5.28E-09 | LILRB3 |
Index variant chosen as the most significant variant in the locus and annotated as CHR:POS:A1:A2. CHR, chromosome; POS, genomic position in genome build 38; A1, reference allele; A2, effect allele.
EA, effect allele.
EAF, effect allele frequency.
OR and P-value are from the inverse-variance-weighted fixed-effect meta-analysis (two-tailed) including all EAS samples.
Nearest gene to the index variant.
We then performed fine-mapping of genome-wide significant loci in EAS (Supplementary Tables 5 and 6). High quality fine-mapping requires samples harmonized with consistent QC and imputation, and conducted with in-sample linkage disequilibrium (LD)20. We thus had to exclude JPN1 and only used SHA1, ICH1 and KOR1 in fine-mapping (Methods). As a result, only 50 out of the 80 total IBD-associated loci in EAS were fine-mapped because other loci dropped below the significance threshold after the removal of JPN1. Among them, five loci were mapped to variants with posterior inclusion probability (PIP) > 95%, and nine additional loci to variants with PIP > 50%, including those implicating new IBD genes and new putative causal variants in known IBD genes (Tiers 1 and 2; Table 2 and Box 1).
Table 2 |.
Putative causal variants and coding variants in LD (r2 > 0.5) with index variants in EAS loci
| Varianta | rsID | Subtype | ORb | P b | Gene | AA change | EAS_MAF | EUR_MAF | R2c | PIPd | Tiere |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1:67182913:G:A | rs76418789 | IBDf | 0.57 | 2.0E-34 | IL23R | G149R | 0.053 | 0.002 | 1.00 | 1.00 | 1 |
| chr1:154962487:G:A | rs3766920 | CD | 1.46 | 7.3E-13 | SHC1 h | 0.050 | 0.000 | 1.00 | 1.00 | 1 | |
| chr10:62710915:C:T | rs224136 | CD | 0.76 | 5.5E-27 | ADO | 0.303 | 0.182 | 1.00 | 1.00 | 1 | |
| chr19:3548233:A:G | rs2240751 | CD | 1.17 | 5.0E-09 | MFSD12 | Y182H | 0.269 | 0.010 | 1.00 | 0.99 | 1 |
| chr20:63744874:T:C | rs2427537 | CD | 0.71 | 1.2E-11 | ZBTB46 | 0.044 | 0.470 | 0.23 | 0.97 | 1 | |
| chr10:110426390:C:T | rs11195128 | CD | 1.33 | 6.0E-16 | DUSP5 | 0.148 | 0.320 | 1.00 | 0.95 | 2 | |
| chr2:190704810:C:T | rs142152795 | CD | 1.43 | 2.1E-09 | NAB1 | 0.021 | 0.000 | 1.00 | 0.93 | 2 | |
| chr7:74711703:C:T | rs117026326 | CD | 1.37 | 8.9E-11 | GTF2I | 0.073 | 0.016 | 1.00 | 0.91 | 2 | |
| chr16:85976134:T:C | rs16940186 | UC | 1.27 | 8.8E-18 | IRF8 | 0.258 | 0.175 | 1.00 | 0.89 | 2 | |
| chr4:38323415:T:C | rs6856616 | CD | 1.33 | 4.0E-29 | LINC02513 | 0.232 | 0.060 | 0.69 | 0.87 | 2 | |
| chr10:79286696:G:A | rs1250566 | CD | 0.82 | 1.9E-17 | ZMIZ1 | 0.340 | 0.282 | 1.00 | 0.55 | 2 | |
| chr17:39732988:C:CT | rs34372308 | UC | 0.86 | 3.8E-10 | MIEN1 | 0.440 | 0.305 | 1.00 | 0.52 | 2 | |
| chr22:36911669:C:T | rs12628495 | CD | 1.27 | 8.2E-20 | CSF2RB | 0.339 | 0.090 | 1.00 | 0.52 | 2 | |
| chr19:48709897:T:C | rs78966440 | CD | 1.17 | 1.5E-11 | FUT2 | 0.497 | 0.000 | 0.31 | 0.52 | 2 | |
| chr1:161509955:A:G | rs1801274 | UC | 0.84 | 8.5E-11 | FCGR2A | H167R | 0.278 | 0.489 | 0.93 | 0.29 | 2 |
| chr2:240630832:A:C | rs3749172 | IBDg | 0.81 | 6.4E-28 | GPR35 | S325R | 0.289 | 0.440 | 1.00 | 0.11 | 2 |
| chr1:21981045:C:T | rs7528405 | IBD | 1.14 | 2.1E-08 | CELA3B | R79W | 0.245 | 0.008 | 0.98 | - | 3 |
| chr1:24964519:A:T | rs6672420 | CD | 0.86 | 4.6E-10 | RUNX3 | I18N | 0.312 | 0.479 | 0.91 | - | 3 |
| chr1:28150351:G:T | rs5938 | CD | 1.27 | 3.2E-08 | PTAFR | A224D | 0.115 | 0.000 | 0.73 | - | 3 |
| chr1:154968787:G:A | rs8191981 | IBD | 1.32 | 1.6E-08 | SHC1 h | A205V | 0.050 | 0.000 | 1.00 | - | 3 |
| chr2:233274722:A:G | rs2241880 | CD | 1.15 | 3.0E-09 | ATG16L1 | T317A | 0.322 | 0.463 | 0.98 | - | 3 |
| chr7:955367:G:C | rs79805216 | IBD | 0.83 | 2.0E-13 | ADAP1 | P14R | 0.205 | 0.018 | 0.95 | - | 3 |
| chr7:1093055:C:T | rs1133041 | IBD | 0.86 | 1.2E-09 | GPER1 | L349F | 0.158 | 0.013 | 0.53 | - | 3 |
| chr7:1093188:TTC:T | rs3840681 | IBD | 0.86 | 1.2E-09 | GPER1 | FL393-394FX | 0.157 | 0.014 | 0.53 | - | 3 |
| chr12:109953174:T:C | rs925368 | CD | 0.73 | 3.8E-09 | GIT2 | N387S | 0.072 | 0.000 | 0.96 | - | 3 |
| chr13:43883789:A:G | rs3764147 | CD | 1.22 | 1.3E-17 | LACC1 | I254V | 0.347 | 0.227 | 1.00 | - | 3 |
| chr14:87941544:A:G | rs398607 | IBD | 1.15 | 4.6E-09 | GALC | I562T | 0.231 | 0.488 | 0.60 | - | 3 |
| chr14:88011538:A:C | rs3742704 | IBD | 1.19 | 2.8E-12 | GPR65 | I231L | 0.206 | 0.099 | 0.71 | - | 3 |
| chr16:28496323:C:G | rs180743 | CD | 1.27 | 3.9E-12 | APOBR | P428A | 0.104 | 0.348 | 0.78 | - | 3 |
| chr16:28502082:A:G | rs181206 | CD | 1.29 | 5.5E-13 | IL27 | L119P | 0.083 | 0.287 | 0.97 | - | 3 |
| chr16:28592334:T:G | rs1059491 | CD | 1.28 | 7.4E-13 | SULT1A2 | N235T | 0.074 | 0.317 | 0.81 | - | 3 |
| chr16:28595911:A:G | rs1136703 | CD | 1.29 | 3.6E-13 | SULT1A2 | I7T | 0.073 | 0.310 | 0.80 | - | 3 |
| chr17:39727784:C:G | rs1058808 | UC | 1.15 | 9.2E-09 | ERBB2 | P1170A | 0.402 | 0.327 | 0.93 | - | 3 |
| chr17:59886176:A:G | rs1292053 | IBD | 1.11 | 5.6E-09 | TUBD1 | M76T | 0.425 | 0.418 | 1.00 | - | 3 |
| chr19:48751247:G:A | rs2071699 | IBD | 1.13 | 3.2E-10 | FUT1 | A12V | 0.313 | 0.022 | 1.00 | - | 3 |
| chr19:54219486:A:G | rs255774 | CD | 1.23 | 2.4E-08 | LILRB3 | Splice donor | 0.486 | 0.482 | 1.00 | - | 3 |
| chr22:21628603:C:T | rs2298428 | IBD | 1.16 | 4.0E-14 | YDJC | A263T | 0.412 | 0.177 | 1.00 | - | 3 |
| chr22:36875840:T:C | rs2075939 | CD | 0.83 | 1.6E-14 | NCF4 | L272P | 0.318 | 0.164 | 0.50 | - | 3 |
Variant annotated as CHR:POS:A1:A2. CHR, chromosome; POS, genomic position in genome build 38; A1, reference allele; A2, effect allele.
OR and P-value are from the meta-analysis including all EAS samples. P-value is from the inverse-variance-weighted fixed-effect meta-analysis (two-tailed).
R2, LD with the index variant measured as r2 using the 1000 genomes EAS individuals.
PIP, posterior inclusion probability.
Tier, 1 (PIP > 95% in EAS); 2 (PIP > 50%, or PIP > 10% if missense or predicted loss-of-function); 3 (missense variants tagging the index variants with r2 > 0.5).
rs76418789 has PIP = 1 for both CD and UC and was therefore listed as PIP = 1 for IBD.
rs3749172 has PIP = 0.11 and 0.09 for CD and UC, respectively, and was therefore listed as PIP = 0.11 with IBD.
The two SHC1 variants are in incomplete LD in study samples and complete LD in 1000 genomes EAS (rs3766920 has higher significance). We boldfaced variants implicating new IBD loci for tiers 1 and 2, and variants that had not reached genome-wide significance nor been identified as putative causal in previous studies4,5,7,8,12,34–43 for tier 3.
Box 1 |. Notable genes implicated in this study.
Here we highlight a few genes discovered in this study with their further details in the Supplementary Note.
Tier 1 (PIP > 95% in EAS), 5 variants.
Tier 1 includes a coding variant in MFSD12 (Y128H, rs2240751, CD) that was reported before with smaller PIP (83%)7, a putatively causal coding variant in IL23R that was fine-mapped before in EUR but for the first time in EAS (G149R, rs76418789, IBD), two variants implicating the 3’UTR in SHC1 (rs3766920, CD) and ZBTB46 (rs2427537, CD), respectively, and an intergenic variant near ADO (rs224136, CD). While all genes in this tier have been reported previously, we mapped three of them to single variant resolution for the first time (SHC1, ZBTB46, ADO).
Tier 2 (PIP > 50% in EAS, or PIP > 10% in EAS if missense or predicted loss-of-function), 11 variants.
GTF2I is the only new IBD locus in this tier, which was implicated by an intronic variant, rs117026326, with PIP of 91% and MAF of 7.3% and 1.6% in EAS and EUR, respectively. For the remaining loci, this study mapped them to single variant resolution. For example, while the encompassing genetic locus was reported before, we implicated GPR35 through a coding variant with PIP of 11% for the first time. Interestingly, FUT2 was mapped to a protein truncating variant (W154X) in EUR with PIP of 17% in a previous study (MAF 48% in EUR and 0.2% in EAS), and to rs78966440, a variant 3.9 kb downstream with PIP of 52% in this study (MAF 0.1% in EUR and 50% in EAS), suggesting that including global populations leads to a deeper discovery of the allelic series underlying human complex disorders44.
Tier 3 (missense variants tagging the index variants with r2 > 0.5 in EAS), 22 variants.
We found a few new IBD-associated loci in this tier, including ADAP1 (rs79805216, P14R) and GIT2 (rs925368, N387S). Both coding variants are common in EAS (21% and 7%) and of low frequency in EUR (2% and < 0.01%). We also found new coding variants implicating known IBD genes, including CELA3B (R79W, MAF 24.5% in EAS and 2% in EUR) and SHC1 (A205V, MAF 5% in EAS and < 0.1% in EUR). Similar to the FUT2 variants in tier 2, rs5938 (PTAFR:A224D), a variant with MAF of 12% in EAS and monomorphic in EUR, complements another known coding variant identified in EUR, rs138629813 (PTAFR:N114S)19. Both variants implicate the same exon with ORs of 1.3 (A224D) and 1.7 (N114S).
Cross-ancestry meta-analysis, 81 new loci.
In addition to coding variants discovered in tiers 1-3, we found two missense variants and one frameshift variant that have r2 > 0.5 (highest across EAS and EUR) with the index variants in these new loci (Supplementary Table 9). These variants implicate ABO, HORMAD1 and CTSS. We note that the frameshift variant (rs8176719) implicating the ABO gene is the key variant in determining blood group type O status. A recent study showed that blood type O is a protective factor against CD in EAS45.
In EUR, many IBD genetic associations were mapped to coding variants implicating specific genes such as NOD2, IL23R and CARD9, leading to important insights into disease pathogenesis12,19,21–25. While the EAS sample size in this study is substantially larger than previous studies, it is still modest such that key coding variants may not yet have the power to be fine-mapped to high PIP. We thus searched for coding variants in LD (r2 > 0.5) with the index variants (variants with the most significant P-value in the corresponding IBD subtype GWAS) to capture coding variants that are suggestively causal. We found 24 coding variants in addition to the coding variants fine-mapped with PIP > 50%, among which 13 have not been reported previously in EAS nor EUR ancestries (Tiers 2 and 3; Table 2, Box 1, and Supplementary Table 7). Ten of the 13 coding variants have higher MAF in EAS than in EUR.
To discover IBD genetic associations across ancestries, we performed inverse-variance-weighted fixed-effect meta-analysis (FE) and MANTRA26, a Bayesian trans-ancestry meta-analysis that models the allelic heterogeneity across studies, to combine samples from EAS and EUR (Methods). Results from FE and MANTRA are largely consistent with each other (Extended Data Fig. 3). We found 255 genetic loci significantly associated with IBD through FE, and 12 additional loci through MANTRA (Fig. 2, Supplementary Data 1 and 3, and Supplementary Table 8). Four loci were significantly associated with IBD in EAS but dropped below the significance threshold in both FE and MANTRA, likely due to cross-ancestry heterogeneity that both methods failed to fully account for (Supplementary Table 8). Taken together, we identified 81 new genetic loci associated with IBD, increasing the number of IBD-associated genetic loci to 320 after including known IBD loci (Box 1, Methods, and Supplementary Tables 8 and 9). A network analysis found that new IBD genes are significantly more connected to known IBD genes (Methods): while, on average, a randomly picked gene has 0.95 ± 0.32 (mean ± standard deviation) connections to known IBD genes (from 1,000 random sampling), the average number of connections from new IBD genes to known IBD genes is 2.62 (empirical P-value = 0.001). We found that new IBD loci implicate known network clusters and also suggest new clusters (Extended Data Fig. 4). For example IL21R, the receptor of a known IBD-associated gene (IL21)27, implicates the IL23R signaling pathway (cluster 2). In contrast, RUNX3, with other new IBD genes, implicates a new cluster enriched in TGF-beta signaling (cluster 3). Defects in the TGF-beta pathway induce autoimmune disorders28. While the role of TGF-beta in IBD had been suggested previously29, we for the first time demonstrated its role through principled genetic analysis. We found many (52%, 42 of 81) of the new IBD loci are highly pleiotropic, e.g., HORMAD1 is associated with monocyte count, neutrophil percentage of white cells, and granulocyte percentage of myeloid white cells (Supplementary Table 10). We also note that a strikingly large proportion of loci associated with IgA nephropathy are associated with IBD (10 out of 25 not counting the major histocompatibility complex (MHC))30, suggesting convergence of pathogenesis pathways for these two disorders that appear unrelated.
To investigate the regulatory effect of the new IBD-associated loci, we searched for their index variants in GTEx-v8 eQTL variants that had PIP > 0.1 in fine-mapping using CaVEMaN and DAPG (Supplementary Table 11). Focusing on variants that both methods converged on, we found that rs72709461, a CD locus, is associated with expression of ABL2 in EBV-transformed lymphocytes cells and esophageal mucosa (PIP > 0.45 in both), and rs8176719, another CD locus, is associated with expression of ABO in colon sigmoid, esophagus, gastroesophageal junction and spleen (PIP > 0.5 in both).
Comparative genetic architecture across ancestries
We first compared the IBD genetic architecture for common variants across the three EAS samples: Chinese (SHA1), Korean (KOR1) and Japanese (JPN1). ICH1 does not have genome-wide coverage and was thus not included in this comparison. We found SNP-heritabilities on the liability scale are comparable in all three groups (CD heritability in SHA1: 0.168 ± 0.036, KOR1: 0.312 ± 0.060, and JPN1: 0.165 ± 0079; UC heritability in SHA1: 0.179 ± 0.037, KOR1: 0.176 ± 0.051, and JPN1: 0.183 ± 0.086; mean ± standard error (s.e.)), and their genetic correlations are not distinguishable from one for both CD and UC (CD genetic correlations in SHA1 vs. KOR1: 0.995 ± 0.136, SHA1 vs. JPN1: 1.242 ± 0.356, KOR1 vs. JPN1: 0.781 ± 0.246; UC genetic correlation in SHA1 vs. KOR1: 0.760 ± 0.185, SHA1 vs. JPN1: 0.769 ± 0.247, KOR1 vs. JPN1: 0.336 ± 0.274; mean ± s.e.; Extended Data Fig. 5 and Methods). At the locus level, we found that only MHC showed evidence of significant heterogeneity of the genetic effect across the three ancestries (P = 5 × 10−8, Cochrane’s Q test, two-sided). Removing the MHC appeared to leave no heterogeneity in the QQ plots (Extended Data Fig. 6). The MHC is a highly complex locus with long range LD that spans megabases. Therefore, the observed heterogeneity does not necessarily suggest different biology within EAS (Supplementary Note).
Across EAS and EUR ancestries, we found that IBD have comparable SNP-based heritability on the liability scale (CD heritability in EAS vs. EUR: 0.213 ± 0.027 vs. 0.196 ± 0.020, UC heritability in EAS vs. EUR: 0.137 ± 0.019 vs 0.134 ± 0.012, mean ± s.e.; Fig. 3a and Methods), suggesting that the amount of genetic contribution to IBD, relative to the total disease risk, is roughly the same across the two populations. Further, the genetic correlations (per allele) across ancestries, calculated using variants shared across EAS and EUR, are slightly smaller than one for CD (rg = 0.85 ± 0.056; mean ± s.e.) and not distinguishable from one for UC (rg = 1.03 ± 0.061; mean ± s.e.), indicating an overall consistency of genetic effect with a small amount of heterogeneity (Fig. 3b). The cross-ancestry genetic correlation appeared to be similar across functional annotations when partitioned (Extended Data Fig. 7 and Methods).
Figure 3 |. Comparative genetic architecture across EAS and EUR.
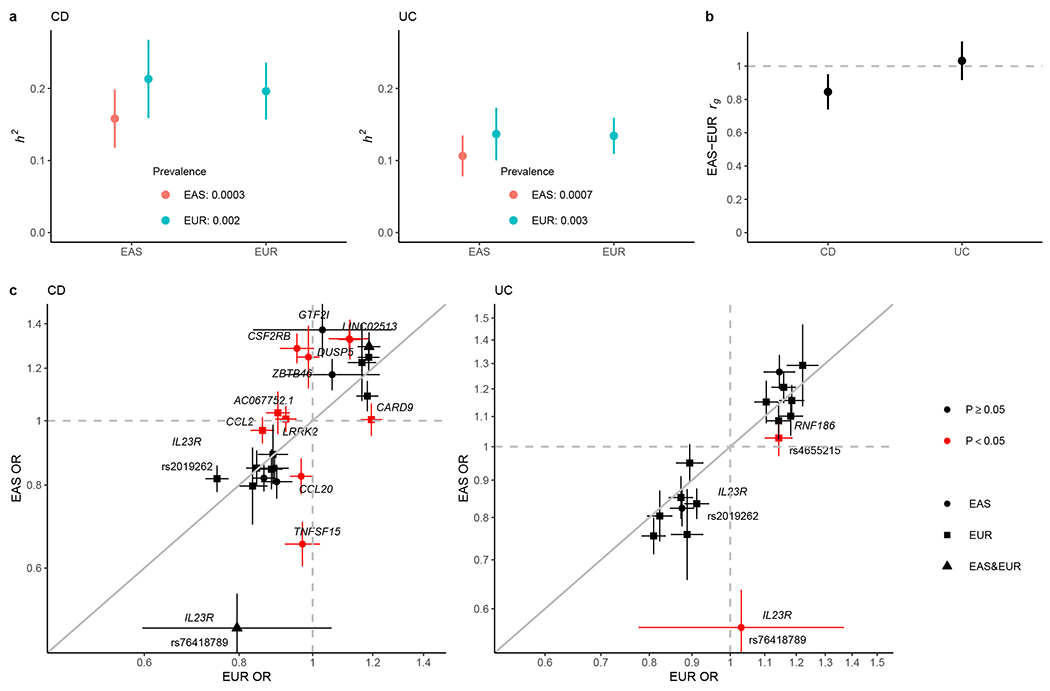
a, SNP-based heritability (h2) on the liability scale. As the population prevalence in EAS can be underestimated for under diagnosis in certain regions in Asia, we calculated the h2 also assuming the prevalence in EUR as the upper bound of the estimate. b, Genetic correlation (rg) between EAS and EUR for CD and UC, respectively. In a and b, the sample sizes used to derive EAS and EUR h2 and EAS-EUR rg were 17,493 and 40,266 for CD, and 17,470 and 45,975 for UC, respectively. Only NFE samples were used in the EUR analysis. c, Per allele genetic effect (OR) for IBD putative causal variants in EAS (from this study) and EUR (from ref. 4). OR is from conditional analysis if there are multiple genetic associations in the locus (Methods). OR was aligned such that the minor allele in EUR was the tested allele. The sample size used to derive OR in EAS and EUR were 22,828 and 40,266 for CD, and 22,318 and 45,975 for UC, respectively. Only NFE samples were used in the EUR analysis. Cochrane’s Q test (two-sided) was used for testing heterogeneity. Variants are colored according to their heterogeneity P-values, which are reported in Supplementary Table 12. P, Bonferroni corrected P-value threshold for coloring. We used the number of putative causal variants tested for the correction such that (P < 0.05 / 25 for CD and P < 0.05 / 16 for UC). Results are plotted as mean value ± 95% confidence interval (error bar).
At the locus level, we compared the conditional effect size (Methods) across EAS and EUR for IBD putative causal variants, including those published in EUR (PIP > 50%, Table 1 in ref. 12) and from this study in EAS (PIP > 40%, reduced threshold for power) (Fig. 3c). We found that genetic effects for many CD (60%, 15 of 25) and UC (88%, 14 of 16) loci included in this analysis are consistent across ancestries, with a few loci as clear exceptions (Fig. 3c, Supplementary Table 12, and Supplementary Data 4). For example, while the primary CD association in TNFSF15 was not included in this analysis as its PIP is below the inclusion threshold (but its EAS-EUR heterogeneity has been replicated in this study), we found a new CD association in TNFSF15, tagged by rs7043505, with conditional OR of 1.03 in EUR (95% CI: 0.98-1.09) and 1.53 (95% CI: 1.42-1.66) in EAS (heterogeneity P = 9 × 10−17, Cochrane’s Q test, two-sided). We also found new loci with allelic heterogeneity such as CSF2RB, which was only associated with CD in EAS with OR of 1.29 (95% CI: 1.22-1.35) compared with OR of 0.95 (95% CI: 0.90-1.00) in EUR (heterogeneity P = 9 × 10−16, Cochrane’s Q test, two-sided). Interestingly, in EUR, while most putative causal variants in IL23R are protective towards both CD and UC, G149R (rs76418789) was only found protective towards CD (OR = 0.77 and 1 in CD and UC, respectively)12. In this study, we found that G149R was also protective towards UC in EAS with OR of 0.57 (95% CI: 0.50-0.64), compared with OR of 1.03 (95% CI: 0.78-1.37) for UC in EUR (heterogeneity P = 1 × 10−4, Cochrane’s Q test, two-sided).
Genetic correlation and comparison of genetic effects can only be conducted for variants shared across EAS and EUR. Differences in genetic findings across ancestries can be driven by both the genetic effect and MAF. The former measures the contribution to an individual’s IBD risk from a single allele, and the latter measures the prevalence of the risk allele in the population. Variance explained, approximately calculated as 2f (1 − f) (logOR)2 / (π2 / 3), combines both and can be used as an approximate measure of the ‘importance’ of a causal variant in a population. We did a comparison of variance explained in EAS vs. EUR. Consistent with earlier discussions, we found that variance explained by IBD-associated loci differ across EAS and EUR, which was, to a greater extent, driven by MAF and less by the effect size (32% IBD associations have different MAF and 22% have different OR, P = 0.026, Fisher’s exact test; Extended Data Fig. 8). TNFSF15 and NOD2, both having a strong preference for CD, showed the largest difference in variance explained across EAS and EUR (5.5% and 1.9%, respectively, median difference: 0.3%).
Overall, UC-associated loci showed a better consistency in variance explained across EAS and EUR compared with CD-associated loci (mean variance explained difference = 0.002 and 0.001, and P = 0.009 and 0.16 for CD and UC, respectively, pairwise Wilcoxon rank order test), consistent with observations that genetic associations for UC tend to overlap more extensively among different ancestry groups than for CD, which shows well established ancestry-dependence. Locus-wise, NOD2 and ATG16L1, both in the autophagy pathway, are the top drivers of CD specificity in EUR. The intronic variant of IL23R (rs11581607), mapped to PIP of 49%12, has a preference for CD (OR = 0.44 and 0.60 for CD and UC, respectively). This variant is not present in EAS (MAF < 0.1%) and therefore is driving the CD specificity in EUR but not in EAS (Extended Data Fig. 9). In EAS, TNFSF15, despite being associated with both CD and UC, has a strong preference for CD (OR = 1.9 and 1.2 for CD and UC, respectively) and is a top driver for CD in EAS due to its greater effect size (Fig. 3 and Extended Data Fig. 9). In the MHC, while EAS and EUR have largely consistent genetic effects (−0.399 ± 0.015 and −0.434 ± 0.026, mean ± s.e.) and variance explained (2.9% vs. 2.4%) for the primary UC association (rs6927022), EAS hosts a CD association that explains ~6× greater amount of phenotypic variance than that in EUR (3.5% (rs9270965) vs. 0.6% (rs145568234); Supplementary Note).
Polygenic risk prediction
As the variance explained for IBD loci differs across ancestries, the ability to use genetic information to predict an individual’s disease risk can also differ. We empirically evaluated this using PRS, which measures an individual’s genetic risk for IBD aggregated over the genome (Methods). We found that, when trained using the NFE summary statistics, PRS calculated using PRS-CS31 explains about 3.3% of CD risk, 3% of UC risk and 3.5% of IBD risk on the liability scale when predicting into the Chinese population (20% of SHA1 samples as the target, assuming 0.02%, 0.02%, and 0.04% prevalence for CD, UC and IBD in the Chinese population32; Fig. 4a). In contrast, PRS explained about 6.4% of CD risk, 3.2% of UC risk and 4.7% of IBD risk on the liability scale when trained using the EAS summary statistics and predicting into the Chinese population in a leave-one-set-out manner, despite that the training sample size is much smaller compared to EUR (Methods and Fig. 4). Of note, PRS constructed using a novel method, PRS-CSx33, combining summary statistics across EAS and EUR as the training data, explained as much as 8.0% of CD, 5.5% of UC and 6.5% of IBD risk on the liability scale in the Chinese population (Fig. 4), leading to an average of 11.8-, 7.2-, and 5.4-fold increase in CD, UC, and IBD case proportions, respectively, when comparing the top 5% of the PRS distribution with the bottom 5%. We have released posterior variant effects and linear combination weights from this EAS+EUR combined ancestry PRS model to facilitate equitable deployment of genetic risk prediction (Data availability). As a validation, we performed similar analyses in a leave-one-country-out manner for all samples respectively (Methods). The UC prediction accuracy drops due to its low heritability (compared with CD) and the reduced sample size (due to the exclusion of a country in discovery). Our general findings hold with each cohort providing qualitatively similar results (Extended Data Fig. 10).
Figure 4 |. Polygenic risk prediction on the Chinese samples.
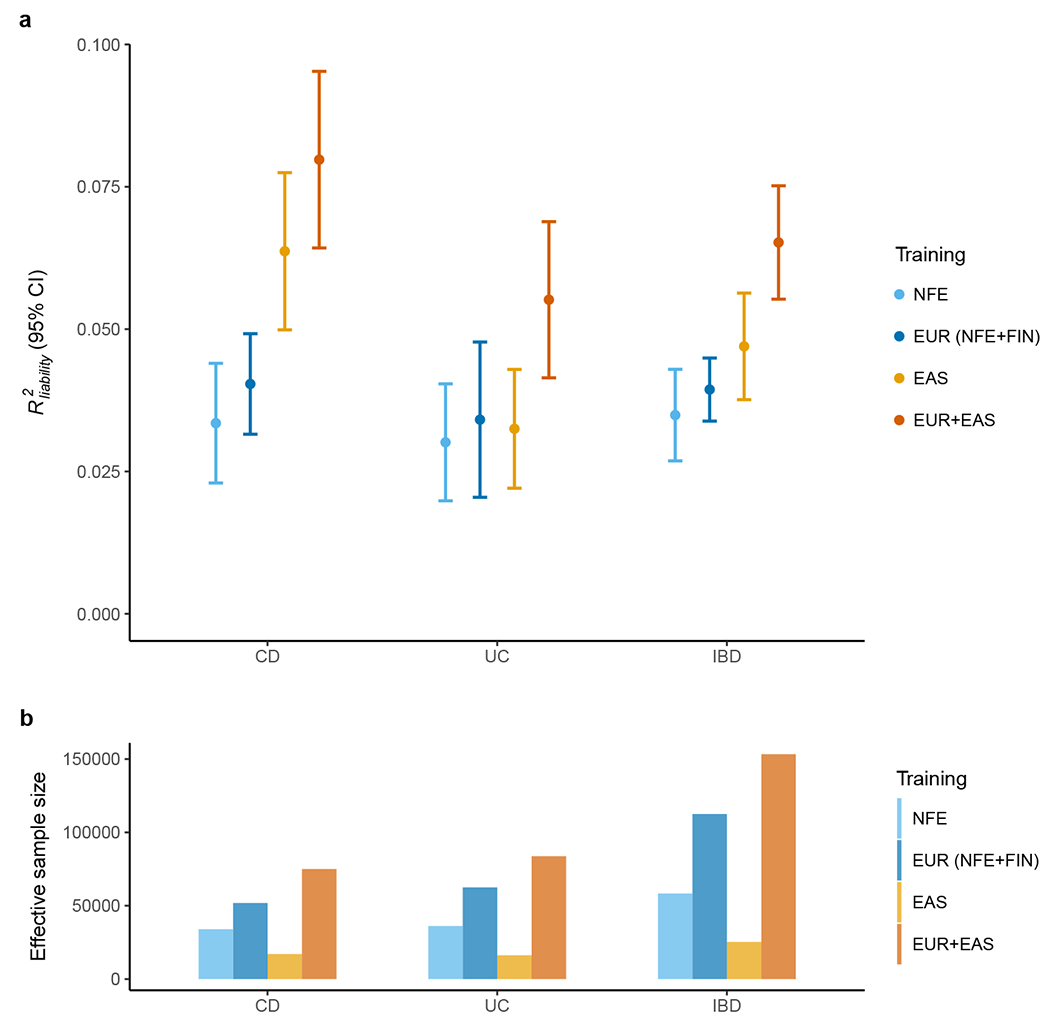
a, Prediction accuracy was measured as R2 on the liability scale using the population prevalence in China as an approximation for Asia. We randomly split SHA1 subjects into discovery, validation and testing 100 times (Methods). All other EAS samples were used as discovery. Results are plotted as mean value ± 95% confidence interval of R2 across the 100 replicates (error bar). b, Effective sample size of training datasets, calculated as 4 / (1 / ncase + 1 / ncontrol).
We note that, interestingly, while UC showed an overall higher consistency across EAS and EUR, the improvement in R2 was greater for CD (2.4× increase) than for UC (1.8× increase) when adding EAS to the EUR discovery samples. This could be driven by several factors: (1) EAS samples contribute more to CD than UC in EAS relatively because CD genetics is more population specific; (2) UC heritability is lower and thus the benefit from adding new samples is also lower; or (3) our ability to model the MHC, which is the most important UC locus, is limited. The MHC has the highest variance explained among all UC-associated loci, but due to its LD complexity, we only used the most significant variant in the MHC as the proxy in our PRS. Overall, both CD and UC had substantial improvements in prediction accuracy when data from both ancestries were used, suggesting the importance of appropriately modeling and integrating ancestrally diverse populations for equitable deployment of PRS in clinical and research settings.
DISCUSSION
We aggregated data from ~30,000 individuals to perform the largest IBD GWAS of East Asian ancestries to date, leading to the discovery of 80 genetic loci associated with IBD in EAS. Combined with over 30,000 IBD cases of European ancestries and controls, we found 81 new IBD-associated loci, increasing the total number of IBD-associated loci to 320. Many new IBD-associated loci discovered from this study were driven by variants with elevated MAF in EAS (e.g., ADAP1), demonstrating the value of including non-European individuals in genetics studies to identify new disease associations. In known IBD-associated loci, we directly implicated many genes for the first time through coding variants (e.g., GPR35, CELA3B, and SHC1). Analyses on this expanded list of IBD-associated loci suggested potentially new pathways, such as TGF-beta signaling.
Over the genome, with several exceptions (e.g., TNFSF15 and CSF2RB), we found that IBD genetic effects are comparable across EAS and EUR. MAF contributes, to a greater extent than genetic effects, to the heterogeneity in IBD genetic loci across EAS and EUR. Combining MAF and genetic effects, we found that CD in general has a greater ancestral dependency than UC, with NOD2 and ATG16L1 as top CD drivers in EUR (through MAF) and TNFSF15 in EAS (through genetic effect). The MHC also appears to make a greater contribution towards CD in EAS than in EUR, but a comprehensive investigation is needed to fully resolve this locus (Supplementary Note).
PRS trained in EUR has reduced accuracy in EAS as expected17. We showed that the accuracy could be improved by jointly modeling discovery samples of both ancestries, highlighting the importance of including global populations in GWAS for equitable deployment of PRS in clinical settings.
There are a couple of limitations in this study. While this study greatly improved the statistical power in EAS, our sample size is still modest compared to EUR studies. Fine-mapping resolution and the ability to compare genetic effects across ancestries, especially at loci hosting multiple independent associations, are therefore quite limited. Additionally, we were only able to compare genetic effects across ancestries for variants that are common in both. For example, NOD2 putative causal variants are ultra-rare in EAS and were therefore unable to be evaluated for their causal roles in EAS. Moving forward, sequencing technologies, with larger sample sizes, are needed to capture rare variants in non-European populations to enable comparative studies at a lower MAF.
Findings from this study can be affected by different ascertainment strategies or even clinical diagnosis practices across nations. We note that all three EAS collections (SHA1, KOR1 and JPN1) followed similar clinical diagnostic criteria so that differences are reasonably managed. We also attempted to reduce the impact from potential clinical heterogeneity by focusing on genetic discoveries shared across ancestries and from the broader diagnosis categories (CD, UC and IBD). We hope that, with larger sample sizes and detailed subphenotyping data, clinical heterogeneity in IBD genetics will be modeled in future studies.
Novel findings and resources from this study represent an advance in diversifying IBD genetics across global ancestries, and highlight the need for future efforts in increasing the sample size, diversity, genome coverage and clinical phenotyping in genetic studies of IBD and other human complex disorders.
METHODS
East Asian samples.
EAS samples included four collections. All sample sizes in this paragraph are post-QC. SHA1 included 2,552 CD patients, 2,400 UC patients, 136 IBD-U patients and 6,279 matched controls, all of Han Chinese descent. We recruited patients from inpatient and outpatient IBD centers in China. The diagnosis of IBD followed either the European Evidence-based Consensus46,47 or the ECCO-ESGAR Guideline for Diagnostic Assessment in IBD48,49. We used clinical characteristics, radiological and endoscopic examination, and histological features in the diagnosis. We excluded patients with infectious diseases, other autoimmune diseases, tumors, and indeterminate colitis. A small number of patients (136) were found to have inconsistent CD/UC diagnosis in our later analysis and were reassigned to and treated as IBD-U (discussed in Removing sample overlap). Controls were recruited from the outpatient service at each recruitment site, who typically visited for routine physical examinations during the period of study. DNA samples were purified from blood using RelaxGene Blood DNA System (Tiangen Biotech, Beijing Co., Ltd., Beijing, China), and genotyped at Beijing CapitalBio Technology Co., Ltd. (Beijing, China) using the Illumina Asian Screening Array (ASA). KOR1 included 1,619 CD patients, 1,569 UC patients and 4,419 selected control subjects recruited from Korea and genotyped using various Illumina arrays as described7. JPN1 included 1,590 CD patients, 1,769 UC patients, 23 IBD-U patients and 1,034 selected controls recruited at Tohoku University Hospital, Kyushu University and 16 affiliated hospitals, and genotyped in previous studies41,42,50,51. ICH1 included 1,611 CD patients, 1,124 UC patients and 3,724 selected controls recruited from Japan, Korea, and Hong Kong SAR China, and genotyped using the Illumina ImmunoChip. ImmunoChip is a custom genotyping array with 196,524 polymorphisms mostly in loci with known associations with major autoimmune and inflammatory diseases derived from individuals of European ancestry52. Beyond these loci, the genomic coverage is sparse, making the ImmunoChip ideal for replicating or fine-mapping known loci from EUR rather than discovering new loci from EAS. We also excluded ImmunoChip in some analyses (e.g., heritability) because of its lack of genome-wide coverage, as described later. Further details of this cohort are described in ref. 6.
European samples.
European samples included two collections: Finnish (FIN) and non-Finnish (NFE) Europeans. FIN study participants were from FinnGen, a public-private partnership project combining genotype data from Finnish biobanks and digital health record data from Finnish health registries. IBD cases were ascertained using the ICD codes. Quality control and analytic details are available from FinnGen (Code availability). The FIN summary statistics used in this study included 1,307 CD patients, 4,024 UC patients, and 303,191 controls (FinnGen R7, Data availability). NFE study participants were from ref. 4. Quality control and analytic details are available from ref. 4. The NFE summary statistics used in this study included 12,194 CD patients, 12,366 UC patients and 28,072-34,915 controls (Data availability).
Quality control.
QC was performed for SHA1, KOR1 and ICH1 using the RICOPILI pipeline (JPN1 is described separately). We first excluded individuals with a mismatch in their reported sex and sex imputed from chromosome X, and updated 98 individuals with no sex reported using the imputed sex. We then performed QC using the following steps. For autosomes, we excluded: (1) variants with a call rate below 95%; (2) individuals with a call rate below 98%; (3) monomorphic variants; (4) individuals with an inbred coefficient above 0.2 and below −0.2; (5) variants with missing rate differences > 2% between cases (IBD) and controls; (6) variants with a call rate < 98%; and (7) variants in violation of Hardy-Weinberg equilibrium (two-sided) with P < 10−6 in controls or P < 10−10 in cases (IBD). For chromosome X, we started with samples that passed the QC using autosomes. Variants in chromosome X from these samples were QC’ed by excluding: (1) monomorphic variants; (2) variants with a call rate below 98% in either male or female; (3) variants with missing rate differences > 2% between cases (IBD) and controls in either males or females; and (4) variants in violation of Hardy-Weinberg equilibrium with P < 10−6 in controls or P < 10−10 in cases (IBD) in females. The numbers of variants and individuals removed in each step are reported in Supplementary Table 1. Chromosome X data are only available for SHA1. KOR1 and ICH1 data had been QC’ed in previous studies7,12 and thus had fewer variants and samples removed in our QC, which was performed to align the studies.
Population structure and outliers.
To calculate the principal components (PC) for all study participants in SHA1, KOR1 and ICH1 (JPN1 is described separately), the following steps were performed on post-QC variants: (1) exclude variants with MAF < 5%; (2) exclude variants in violation of Hardy-Weinberg equilibrium with P < 10−3; (3) exclude variants with missing rate > 2%; (4) exclude strand ambiguous variants; (5) exclude variants in the MHC (chromosome 6, from 25 Mb to 35 Mb in hg19) and the chromosome 8 inversion region (chromosome 8, from 7 Mb to 13 Mb in hg19); (6) prune variants with an r2 threshold of 0.2, window of 200 variants, and step size of 100 variants; (7) repeat (6); and (8) perform EIGENSTRAT to calculate the PCs.
To identify population outliers, we conducted a visual inspection using the first two PCs. 44 individuals were identified as outliers and removed. Further population-level inspection was performed using PCs created from samples combining the study participants and the 1000 Genomes Project Phase 3 (1KG) East Asian panel. We found no outliers through the visual inspection.
Removing sample overlap.
To identify the within cohort sample overlap and relatedness, we computed the identity-by-descent matrix. We identified all sample pairs that had pi-hat > 0.9 as “duplicated” and pi-hat > 0.2 as “related”. We treated sample pairs as follows: (1) control-control pairs: randomly keep one individual; (2) case-control pairs: keep the case for “related” pairs and remove both individuals for “duplicated” pairs; (3) case-case pairs: randomly keep one individual and reassign the subtype to IBD-U for “duplicated” pairs that have different subtypes for the two individuals.
To identify cross-cohort sample overlap within EAS, we computed the identity-by-descent matrix across individuals from different cohorts (SHA1, KOR1 and ICH1). A small number of samples have been removed following the same approach as in the removal of within cohort overlaps (Supplementary Table 1).
Phasing and imputation.
The pre-imputation checks were performed on all samples in SHA1, KOR1 and ICH1 (JPN1 is described separately) before imputation using the following steps: (1) remove variants not mapped and not aligned to the GRCh37 genome build using bcftools; (2) liftover the variants to GRCh38 and perform strand alignment using bcftools; (3) remove strand ambiguous variants; (4) remove variants not matched to the TOPMed reference panel (R2 2020) using HRC-1000G-check-bim; and (5) create VCF file using VcfCooker. Additionally, to account for known issues in the ImmunoChip design, for the ICH1 sample we removed variants having MAF > 10× or < 0.1× compared with the MAF from 1KG EAS individuals12. These validated samples were phased using Eagle2 and imputed to the TOPMed reference panel (R2 2020) using Minimac4. The imputation panel has 97,256 individuals, among which 90,339 were assigned to a super population, including 24,267 African individuals, 17,085 admixed American individuals, 47,159 European individuals, 1,184 East Asian individuals and 644 South Asian individuals53. The post-imputation VCF files were converted to the PLINK2 dosage file format for the association and PRS analyses, and to the PLINK best-guess genotypes for LD calculation (dosage 0-0.1 for homozygous major, 0.9-1.1 for heterozygous, and 1.9-2.0 for homozygous minor genotypes, other dosage values will be converted to missing).
Association analysis.
Association analysis was performed by PLINK2 using the genotype dosage for SHA1, KOR1 and ICH1 (JPN1 is described separately). Only variants with imputation quality r2 > 0.6 and MAF > 0.1% were included in the analysis. We tested the genetic associations with CD, UC and IBD separately using logistic regression with the first ten PCs as covariates. For chromosome X, we did dosage compensation for males and included sex in addition to the first ten PCs as a covariate.
Processing of JPN1.
All JPN1 samples were genotyped on the Japonica Array V154. DNA samples were sent to an outsourced laboratory (Toshiba Inc.) and the raw data were received in CEL format. Genotypes were obtained by an in-house data analysis pipeline. Data analysis was performed following the manufacturer’s recommendations using apt software (ver. 2.10.2.2, Thermo Fisher Scientific). Of the 4,701 CEL files analyzed, 55 samples with a sample call rate < 97% were excluded from the analysis. The genotypes of 645,843 autosomal variants were obtained. Of these, variants that were not classified as Recommendations by SNPolisher (Affymetrix, Inc.) were removed, and the remaining 645,708 variants were used for later analysis. Before genotype imputation, GWAS was performed for quality control. Variants with P < 10−6 were visually checked for clustering results, and seven variants which showed poor cluster resolution were excluded. Then, we excluded 27,398 variants with P < 10−5 in Hardy-Weinberg equilibrium test (two-sided) and prepared VCF consisting only of autosomal polymorphic sites. The imputation was performed using BEAGLE 5.1. The imputation panel was an in-house constructed haplotype panel comprising the haplotypes of 5,765 individuals from diverse populations including 2,493 individuals from 1KG, 820 individuals from the Human Genome Diversity Project, 278 individuals from the Simons Genome Diversity Project, 90 samples from the Korean Personal Genome Diversity Project, and 1,634 Japanese individuals. The Japanese data includes genomic data from 608 individuals that we collected from volunteers and those of 1,026 participants of BioBank Japan which we received from NBDC human database (accession ID: JGAS000114). After removing variants that did not match alleles in the reference panel using the comfort-gt program distributed with BEAGLE, we ran the imputation in BEAGLE 5.1 with default parameters.
Before the association analysis, we performed final QC excluding: (1) variants with a call rate < 95%; (2) individuals with a call rate < 98%; (3) variants with MAF < 0.005%; (4) individuals with pi-hat > 0.25; (5) variants in violation of Hardy-Weinberg equilibrium with P < 10−6; (6) variants with imputation quality r2 < 0.5; and (7) variants in sex chromosomes. Finally, association analysis was performed with 8,944,430 variants by PLINK2 using logistic regression with the first three PCs as covariates.
Meta-analysis.
We used METAL to perform inverse-variance-weighted fixed-effect meta-analysis to combine samples within EAS (SHA1, KOR1, ICH1 and JPN1) and cross-ancestry (EAS and EUR). For the cross-ancestry meta-analysis, we additionally performed MANTRA26, a Bayesian trans-ancestry meta-analysis, with log10(Bayer Factor) > 6 as the “significance” cutoff.
Locus definition.
Genetic loci in this study were defined following the same manner as in a previous IBD genetic study4. For each trait (CD, UC or IBD) in EAS, genome-wide significant variants in EAS were clumped with an r2 threshold of 0.6 using the 1KG EAS reference panel. The LD window was then defined by the downstream-most and upstream-most variants that are in LD with the index variant with r2 > 0.6 and capped at 1 Mb from the index variant. Loci with overlapping LD windows and loci of which index variants were separated by < 500 kb were subsequently merged, and the variant with the most significant P-value was kept as the index variant for each merged locus. We then further merged loci from CD, UC and IBD using the same method.
In the meta-analysis combining all participants (EAS and EUR), we first defined the LD windows in EAS and EUR separately (using the respective 1KG reference panel) as described in the last paragraph. We then merged the LD windows across EAS and EUR if they overlapped or their index variants were separated by < 500 kb (the same criteria as in the ancestry-specific locus definition). A locus was defined as known, very conservatively, if it or the region of its index variants padded with 500 kb upstream and downstream, overlapped with one of the 241 reported loci in ref. 4 or included any variant previously reported as genome-wide significant in EAS samples7,8,34–43. A handful of new loci from this study had genome-wide significance in the NFE summary statistics but were manually censored in the publication4 because they had relatively low imputation quality and/or P-values close to the threshold. We reported them as new findings if and only if the meta-analysis with EAS and/or FIN samples reached genome-wide significance.
We assigned a locus as “IBD” associated if the index variant was significantly associated with both CD and UC, or was only significantly associated with IBD (but with neither CD nor UC individually). For the remaining loci, we assigned them “CD” or “UC” if their index variant was significantly associated with CD or UC, respectively. For the CD/UC/IBD assignment only, we used the Bonferroni corrected P-value threshold as the significance cutoff (0.05/n, where n is the total number of loci tested).
Fine-mapping and conditional genetic effects.
We performed fine-mapping on genome-wide significant loci in EAS (including CD, UC and IBD phenotypes). We used the summary statistics and the in-sample LD calculated using hard-called genotypes merged across post-imputation EAS subjects. We used Sum of Single Effects (SuSiE)55 for the fine-mapping analysis (Code availability) using the following parameters: minimum purity = 0.5, algorithmic convergence tolerance = 10−4, and the maximum number of iterations = 100. The initial analysis was performed using a maximum of 5 signals. Loci with 5 signals identified were re-run with a maximum of 10. Loci that failed to converge were rerun with the maximum number of signals reduced by one iteratively until convergence.
While SuSiE models multiple independent associations in a locus, it does not output the genetic effect estimate conditional on other associations in the locus. We do so by first filtering out credible sets with marginal P > 10−9, as they represent less reliable findings. We then computed the conditional effects and P-values for variants with the best PIP in each credible set using COJO56 and LD from the study samples. Lastly, we filtered out the credible sets with conditional P > 5 × 10−8. While we only discussed credible sets with conditional P < 5 × 10−8 and marginal P < 10−9 as they represent the most reliable findings, we retained all credible sets for reporting purposes. Credible sets filtered out are flagged with ‘rm_p_sig’ and ‘rm_p_cond’ for being removed due to marginal and conditional P-values, respectively (Supplementary Table 5).
Heritability analysis.
We calculated the heritability in the observed scale using LDSC57. Only autosomal variants with MAF > 5% in their respective populations were used in the analysis, with variants in MHC removed for its long-range LD. We used the genome-wide summary statistics from each ancestry, with the exception that ICH1 in EAS was not included because it does not have genome-wide coverage (ImmunoChip). We used the pre-computed LD scores on the 1KG reference panel. The heritability in the observed scale was converted to the liability scale with the prevalence in either the respective or the European population by using a published method32,58. The assumed prevalence is shown in Figure 3 and Extended Data Figure 5.
Genetic correlation.
We computed the per allele IBD genetic correlations across EAS samples (SHA1, KOR1 and JPN1) using LDSC57. We used LD scores pre-computed on the 1KG reference panel. We computed the genetic correlation across EUR and EAS using S-LDXR59, with the LD scores from EUR and EAS provided by S-LDXR. S-LDXR also computes the enrichment of squared genetic correlation stratified across genomic annotations. For all analyses, only autosomal variants with MAF > 5% and outside of the MHC were used.
Conditional effect size for putative causal variants.
To properly compare the effect size across EAS and EUR, for each locus, we calculated the effect size for putative causal variants conditional on variants with the best PIP in their respective credible sets from all ancestries. Credible sets were taken from ref. 12 for EUR. Putative causal variants were those with PIP > 50% in EUR or PIP > 40% in EAS. We used COJO and the 1KG reference panel respective to the ancestry which the effect size was calculated in.
Polygenic risk analysis.
Single-population PRS.
We constructed PRS using PRS-CS31 for EAS individuals with training summary statistics from NFE, EUR (NFE+FIN) and EAS. For leave-one-set-out PRS analysis (for retaining the largest sample size as discovery), we randomly split the SHA1 sample such that 60% of the dataset was used as discovery, 20% as validation and 20% as testing. We performed association analysis on the 60% discovery samples, and meta-analyzed them with KOR1, ICH1 and JPN1 samples to create the discovery summary statistics. In parallel, we performed a leave-one-country-out PRS analysis (for evaluating results across countries). For the country being tested, we split its samples 50% as validation and 50% as testing. We then meta-analyzed the remained EAS cohorts to create the discovery summary statistics. The random split was repeated 100 times. We filtered variants to HapMap3 variants with MAF > 1% in each respective population, and removed indels and strand-ambiguous variants. For the MHC, we included only the top significant variant due to its LD complexity. Only variants with imputation quality r2 > 0.6 were included. Multi-population PRS. We constructed PRS using PRS-CSx33 for EAS individuals with training summary statistics from EUR (FIN+NFE through meta-analysis) and EAS individuals. We followed all details in the “Single-population PRS” except that NFE, FIN and EAS summary statistics were all used in the PRS construction by PRS-CSx.
For evaluation, we computed R2 on the observed scale by comparing the full model with PRS and ten PCs with a null model excluding the PRS. We then converted the R2 from the observed scale to the liability scale assuming 0.02%, 0.02%, and 0.04% population prevalence, the respective prevalence in China for CD, UC, and IBD; 0.03%, 0.08%, and 0.11% in Korea; and 0.06%, 0.17%, and 0.23% in Japan32,60.
Network analysis.
We created the IBD gene network from tiers 1-3 genes (Table 2) and the nearest genes to index variants in all IBD loci (Supplementary Table 8). We defined edges as those have gene-gene interaction score > 0.4 in the STRING functional protein association networks61 (Data availability). We excluded edges that only have text mining, neighborhood, and gene fusion as evidences. We used Community Clustering Glay with default parameters in Cytoscape to perform the clustering (Code availability). Clusters including only one IBD gene were not shown. For clusters with more than two genes or with new IBD genes, we performed pathway enrichment analyses for the GO Biological Process, GO Cellular Component, GO Molecular Function, KEGG pathways, Reactome Pathways, Reactome Pathway and WikiPathways. Pathways whose enriched genes significantly overlap with a more significant pathway (Jaccard similarity > 0.5) were excluded.
Ethics.
Written Informed consent and permission to share the data were obtained from all study participants, in compliance with the guidelines specified by the recruiting center’s institutional review board. SHA1 was approved by the Institutional Review Board for Clinical Research of the Shanghai Tenth People’s Hospital of Tongji University (SHSY-IEC-4.0/18-33/01). Samples recruited in mainland China were processed and analyzed in a Chinese server by the Chinese co-authors to comply with the Administrative Regulations on Human Genetic Resources (a regulation from the Ministry of Science and Technology of the People’s Republic of China). Results from the analyses as aggregated information (e.g., summary statistics), which contain no individual-level nor identifiable data, were used in this study. KOR1 was approved by the Institutional Review Board of Asan Medical Center (2017-0456). JPN1 was approved by the Ethics Committee of Tohoku University School of Medicine (2020-1-608). MGH Institutional Review Board reviewed and approved this study (2013P002634), including the use of ICH1. Patients and controls in FinnGen provided informed consent for biobank research, based on the Finnish Biobank Act. Alternatively, separate research cohorts, collected prior the Finnish Biobank Act came into effect (in September 2013) and start of FinnGen (August 2017), were collected based on study-specific consents and later transferred to the Finnish biobanks after approval by Fimea (Finnish Medicines Agency), the National Supervisory Authority for Welfare and Health. Recruitment protocols followed the biobank protocols approved by Fimea. The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) statement number for the FinnGen study is Nr HUS/990/2017. The FinnGen study is approved by Finnish Institute for Health and Welfare (permit numbers: THL/2031/6.02.00/2017, THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019, THL/1524/5.05.00/2020, and THL/2364/14.02/2020), Digital and population data service agency (permit numbers: VRK43431/2017-3, VRK/6909/2018-3, VRK/4415/2019-3), the Social Insurance Institution (permit numbers: KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, KELA 138/522/2019, KELA 2/522/2020, KELA 16/522/2020, Findata THL/2364/14.02/2020 and Statistics Finland (permit numbers: TK-53-1041-17 and TK/143/07.03.00/2020 (earlier TK-53-90-20). The Biobank Access Decisions for FinnGen samples and data utilized in FinnGen Data Freeze 7 include: THL Biobank BB2017_55, BB2017_111, BB2018_19, BB_2018_34, BB_2018_67, BB2018_71, BB2019_7, BB2019_8, BB2019_26, BB2020_1, Finnish Red Cross Blood Service Biobank 7.12.2017, Helsinki Biobank HUS/359/2017, Auria Biobank AB17-5154 and amendment #1 (August 17 2020), Biobank Borealis of Northern Finland_2017_1013, Biobank of Eastern Finland 1186/2018 and amendment 22 § /2020, Finnish Clinical Biobank Tampere MH0004 and amendments (21.02.2020 & 06.10.2020), Central Finland Biobank 1-2017, and Terveystalo Biobank STB 2018001.
Extended Data
Extended Data Fig. 1. Quantile-Quantile plots for IBD genetic associations.
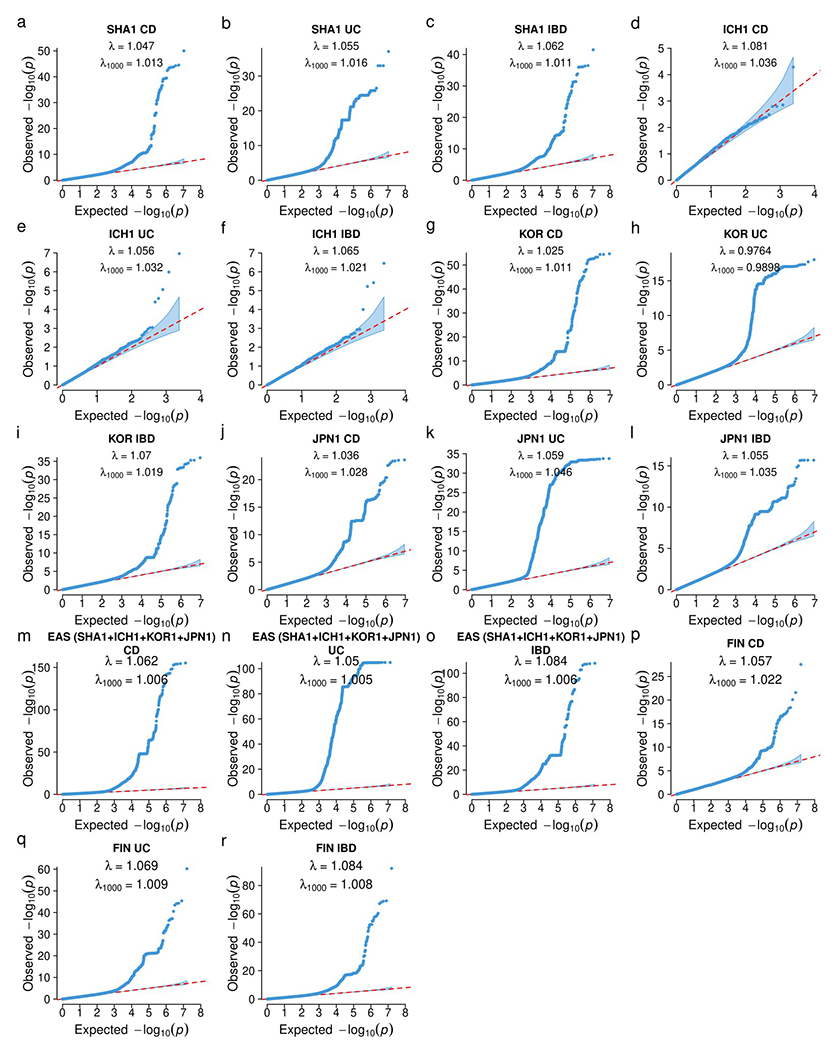
λ: genomic inflation factor; λ1000 : scaled inflation factor for an equivalent study of 1000 cases and 1000 controls. The dots indicate variants. Shaded area indicates the 95% confidence interval under the null distribution. a-c, SHA1. d-f, ICH1 (only the designated null variants in ImmunoChip were used). g-i, KOR1. j-l, JPN1. m-o, meta-analysis including all EAS samples (SHA1, ICH1, KOR1, and JPN1), p-r, FIN. a, d, g, j, m, p are for CD. b, e, h, k, n, q are for UC. c, f, i, l, o, r are for IBD.
Extended Data Fig. 2. Index variants in the 16 new IBD EAS loci.
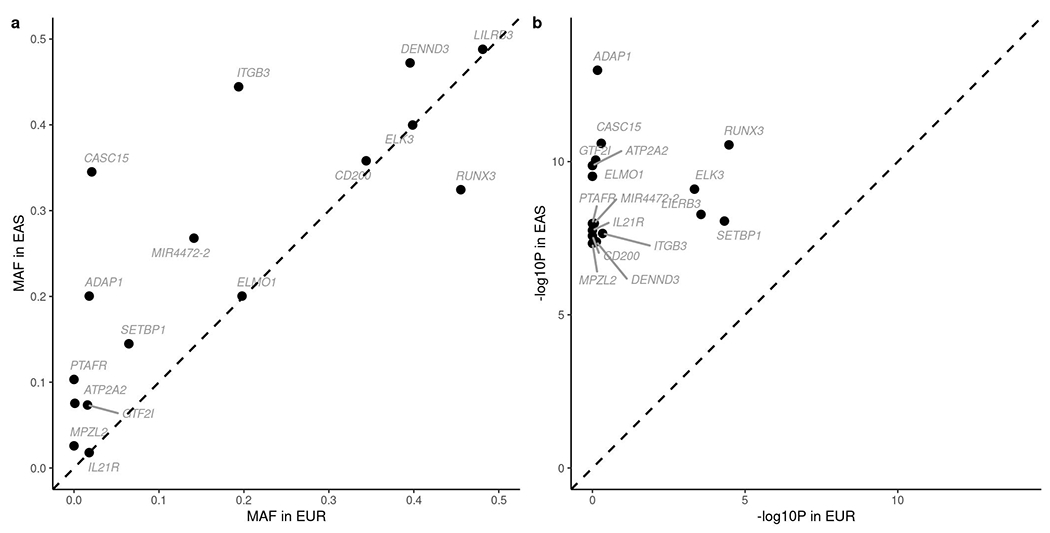
a, Minor allele frequency (MAF) taken from 1000 Genomes EAS and EUR reference panels respectively. b, P-value in respective studies.
Extended Data Fig. 3. Comparison between the fixed-effect (FE) meta-analysis and MANTRA.
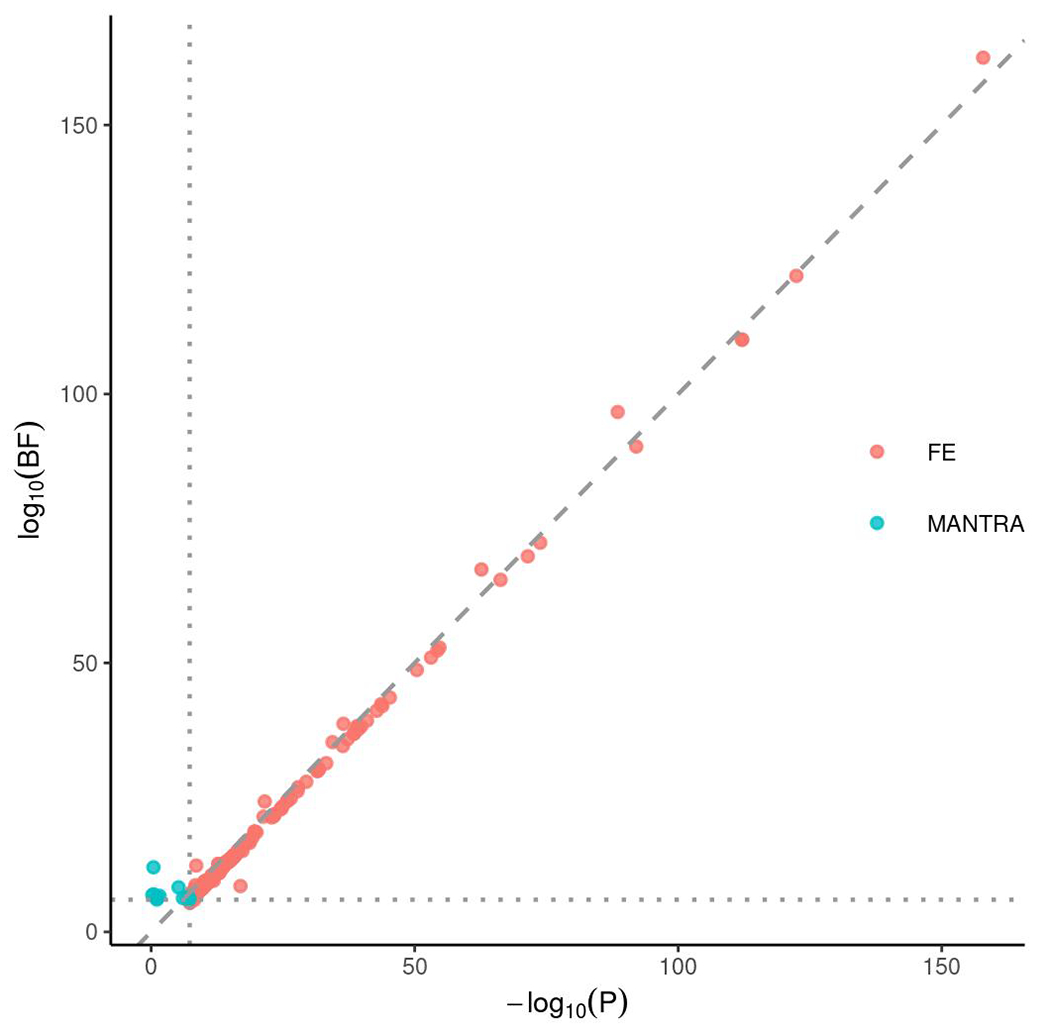
Index variants in loci identified by either FE or MANTRA were plotted. For FE, we used genome-wide significance threshold of 5 × 10−8, and for MANTRA, we used the Bayes Factor threshold of 106, plotted as the vertical and horizontal lines respectively. P: P-value from FE. BF: Bayes factor from MANTRA.
Extended Data Fig. 4. IBD gene network.
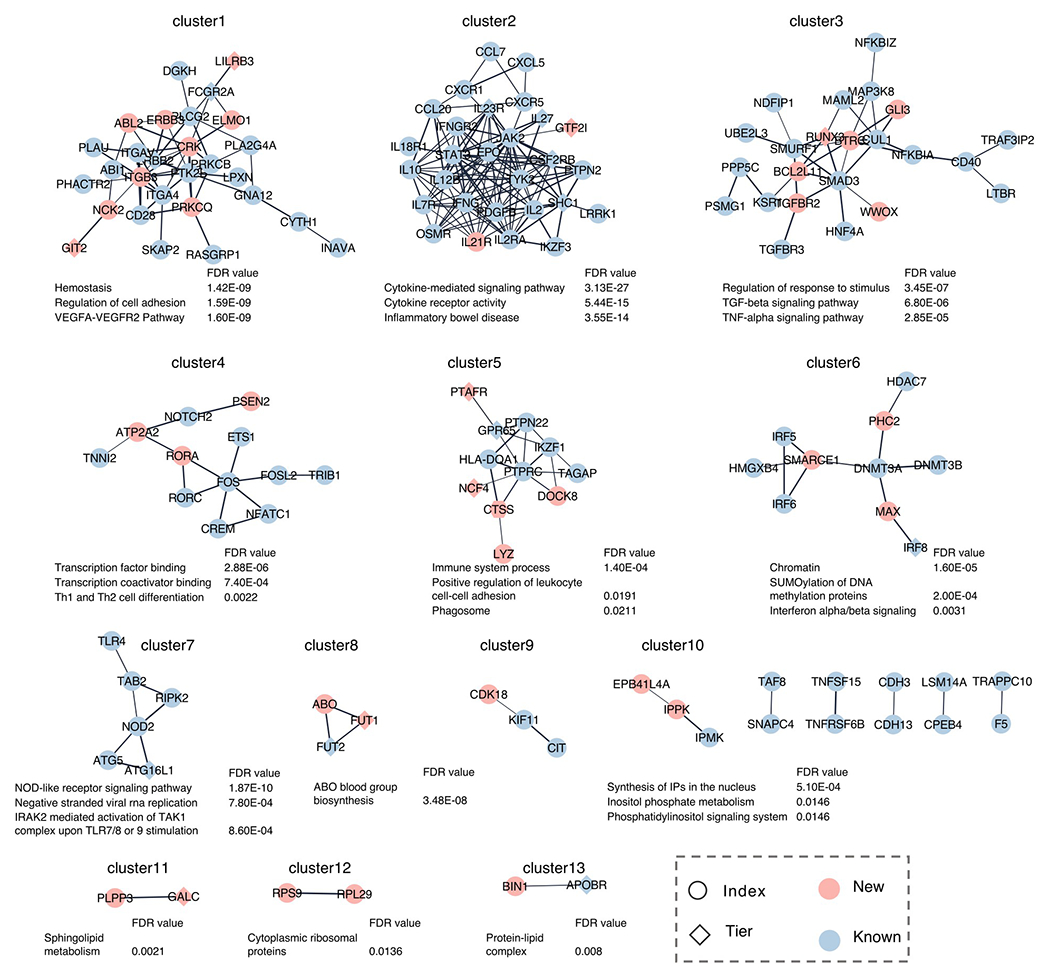
IBD gene network was created using the STRING functional protein association networks and clustered using Community Clustering Glay (Methods). For clusters with more than two genes or with new IBD genes, top three significantly enriched pathways were shown if false-discovery rate (FDR) < 0.05. New: nearest genes to the index variants in new IBD loci or new genes in Table 2 (boldfaced); Known: nearest genes to the index variants in known IBD loci; Index: nearest genes to the index variants in IBD loci except for those in Table 2; Tier: genes in Table 2.
Extended Data Fig. 5. Comparative genetic architecture within EAS.
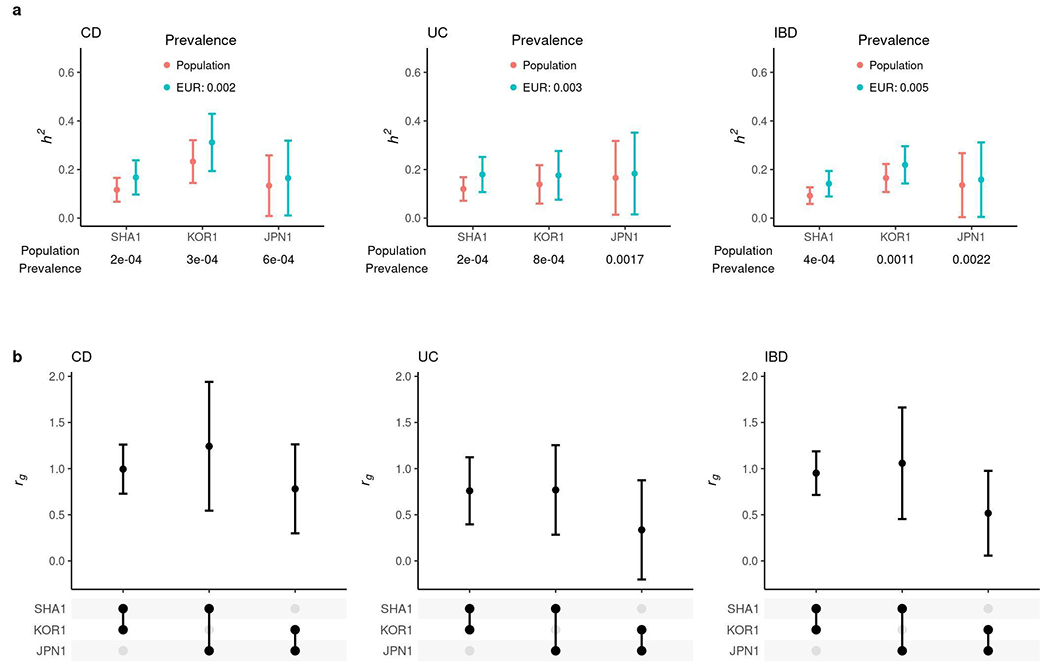
a, SNP-based heritability in the liability scale with the prevalence in its respective population or the European population. b, Genetic correlation (rg). In a and b, the sample sizes used to derive SHA1, KOR1 and JPN1 h2 and their rg were 8,831, 6,038 and 2,624 for CD, and 8,679, 5,988 and 2,803 for UC, respectively. Results are plotted as mean value ± 95% confidence interval (error bar).
Extended Data Fig. 6. Quantile-Quantile plots for the heterogeneity test within EAS.
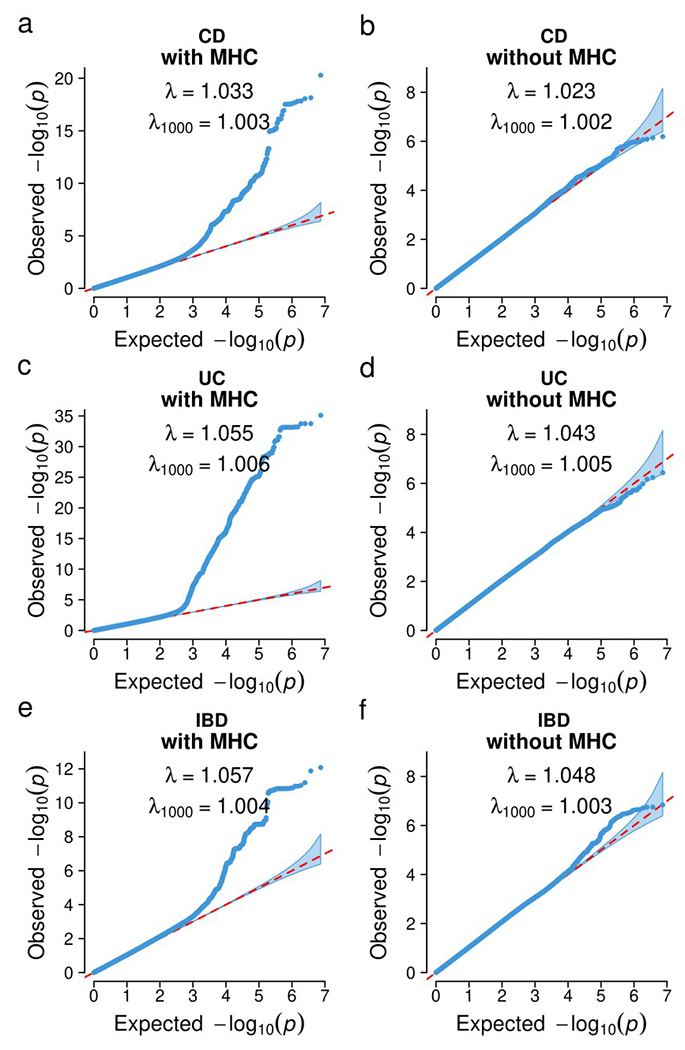
a, b, CD. c, d, UC. e, f, IBD. a, c, e, Genome-wide variants including the MHC locus. b, d, f, Genome-wide variants excluding the MHC locus. Cochran’s Q-test, two-sided, was used for the heterogeneity test. The dots indicate variants. Shaded area indicates the 95% confidence interval under the null distribution.
Extended Data Fig. 7. Enrichment of squared genetic correlation stratified across genomic annotations.
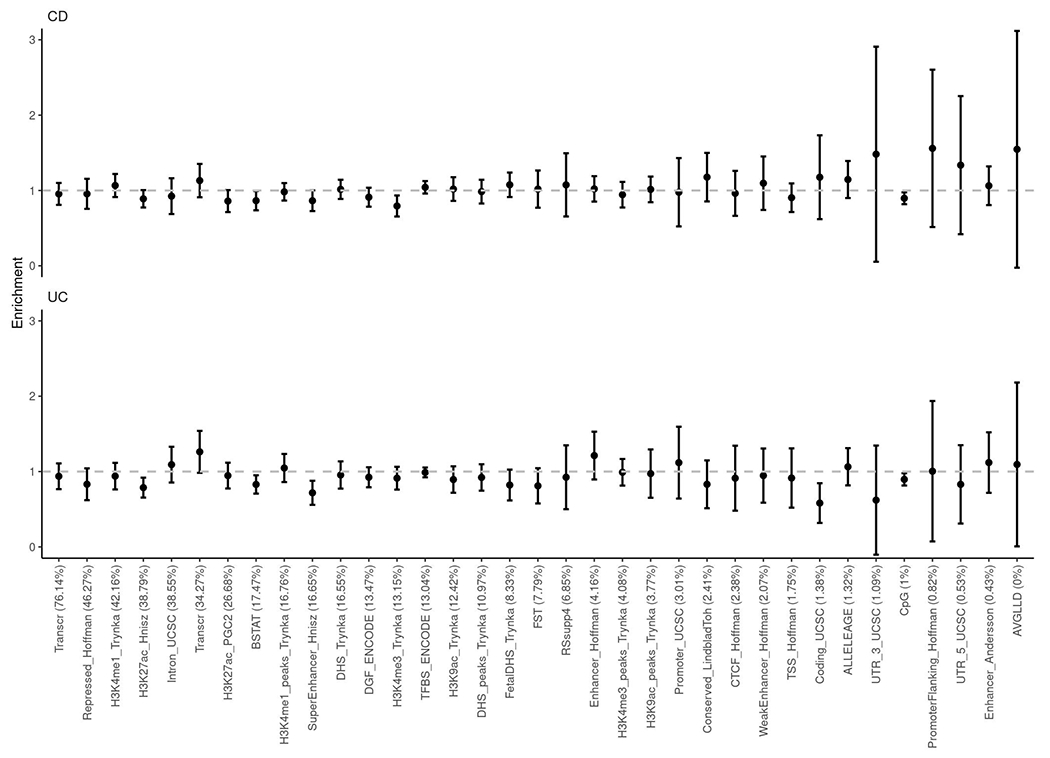
No significant enrichment or depletion (deviation from 1) was observed after Bonferroni corrections. Results are plotted as mean value ± 95% confidence interval before multiple testing corrections (error bar).
Extended Data Fig. 8. Variance explained for IBD associations across EUR and EAS.
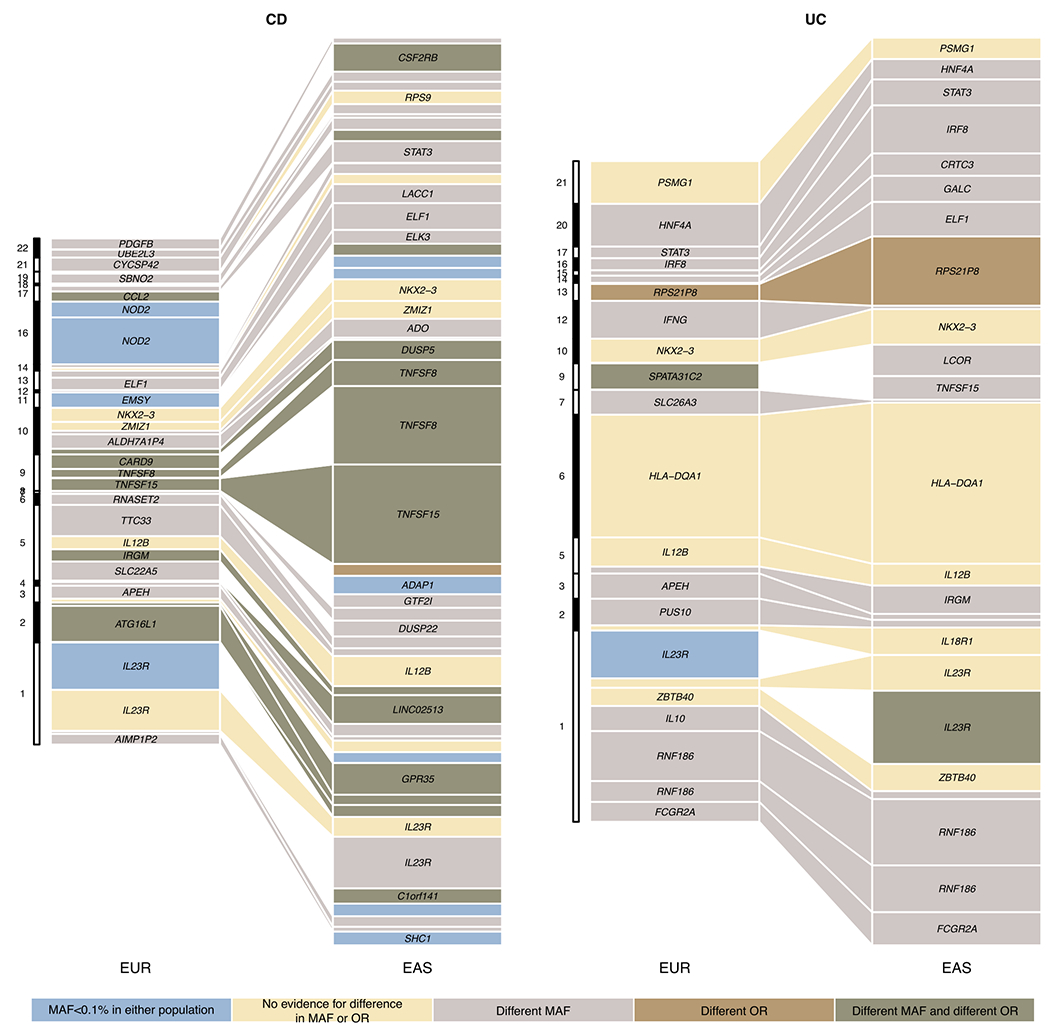
We included all loci from Supplementary Table 8. For loci with fine-mapping analyses performed, we used the conditional OR (using COJO, Methods) for variants with the highest PIP in each credible set to account for multiple independent associations. We took fine-mapping results from ref 12 for EUR and from this study for EAS. For loci with no fine-mapping results, we used the index variant (variant with the most significant P value) as the proxy for the loci. We only plotted associations that have variance explained greater than 0.3% in either EAS or EUR. Different MAF is defined as Fst > 0.01, and different OR is defined as heterogeneity test P value < 0.05 after Bonferroni correction. Because the heterogeneity test was corrected using a higher multiple testing burden, the significance for a handful of loci, e.g., RNF186, can be different from Figure 3c. Nearest genes to the associations were used as labels for associations when the text space is available.
Extended Data Fig. 9. Difference between variance explained for CD and UC across EUR and EAS.
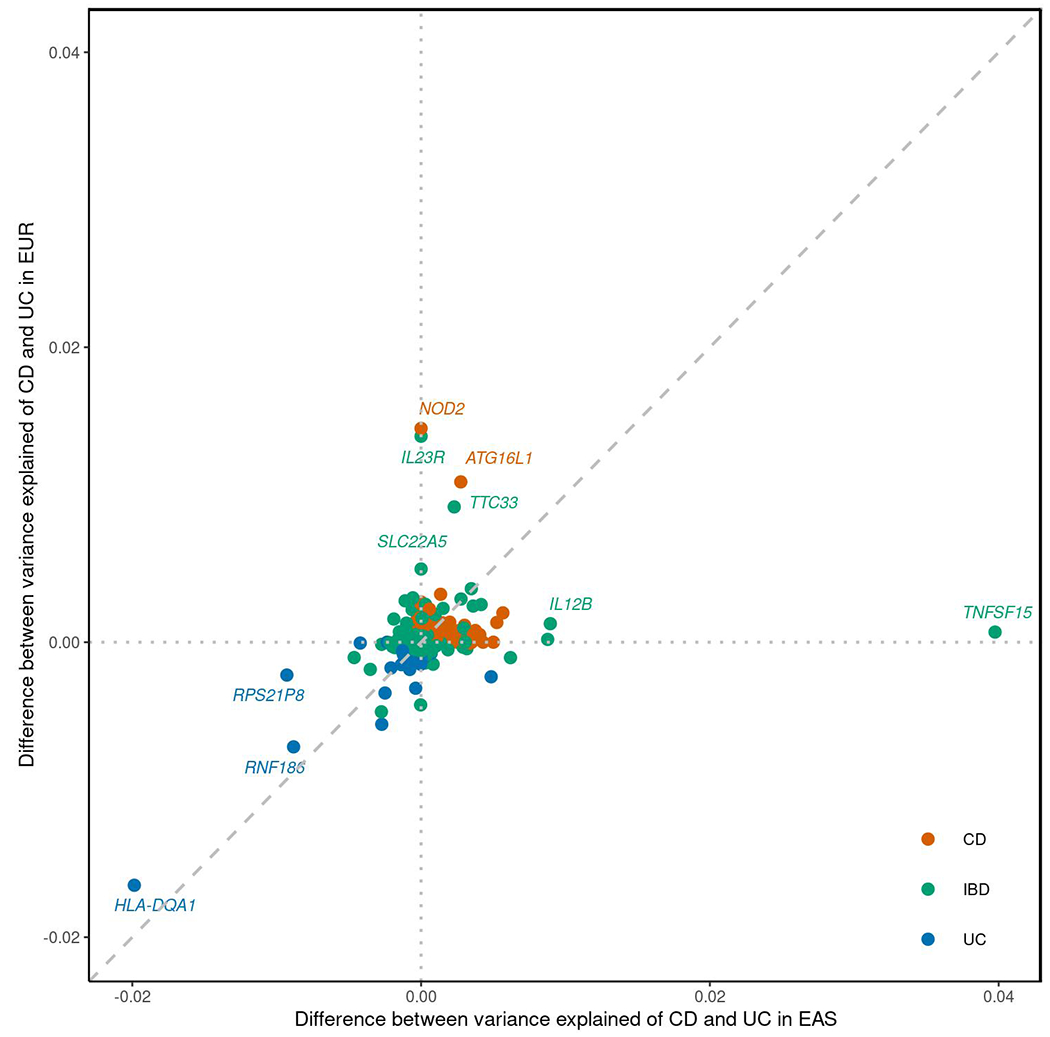
Index variants from Supplementary Table 8 were plotted. Difference between variance explained was calculated as variance explained of CD - variance explained of UC.
Extended Data Fig. 10. Polygenic risk prediction on Chinese, Korean and Japanese subjects.
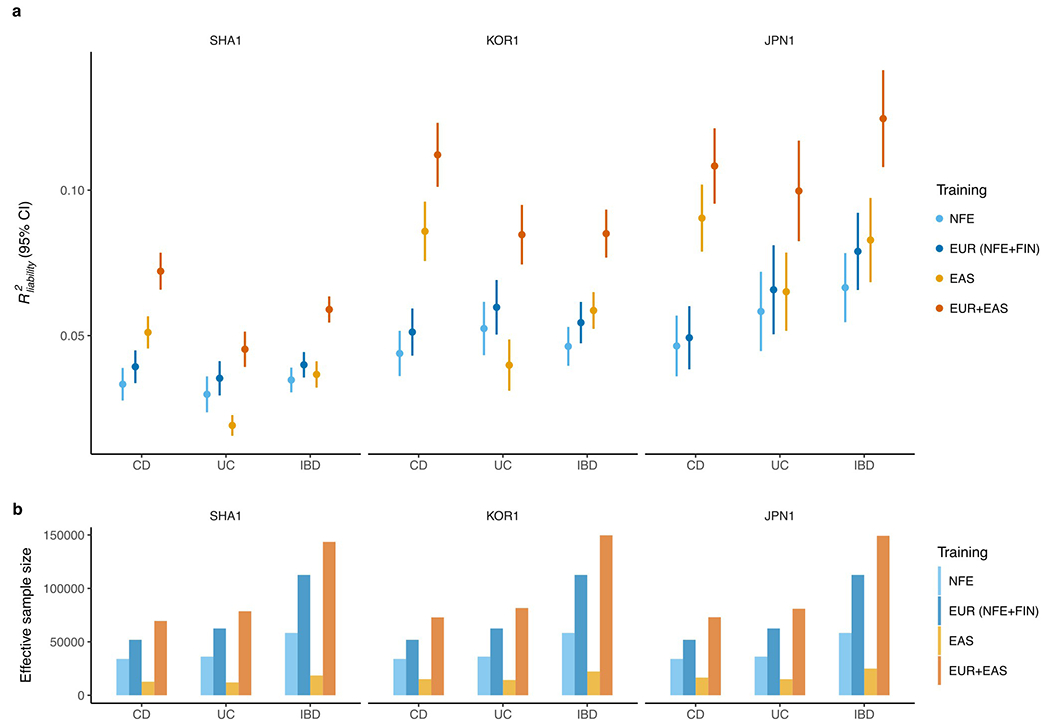
a, Leave-one-country-out strategy was performed to test the performance of PRS on SHA1 (Chinese), KOR1 (Korean), and JPN1 (Japanese) subjects, respectively. The prediction accuracy was measured as R2 on the liability scale using the population prevalence (Methods). For the testing cohort, we randomly split subjects into validation and testing 100 times (Methods). All other EAS cohorts were used as discovery. Results are plotted as mean value ± 95% confidence interval of R2 across the 100 replicates (error bar). b, Effective sample size of training datasets, calculated as 4/(1/ncase+1/ncontrol).
Supplementary Material
Supplementary tables 1-13
Manhattan plot for IBD genetic associations.
Regional association plot for IBD-associated loci in EAS
Forest plot for index variants in IBD-associated loci
Forest plot for putative causal variants identified in EAS and in EUR
Supplementary Note including discussions in the Major histocompatibility complex (MHC) and IBD genes of interest.
ACKNOWLEDGEMENTS
Z.L. acknowledges support from the National Natural Science Foundation of China (91942312, 81630017). H.H. acknowledges support from NIDDK K01DK114379, NIDDK R01DK129364 and the Stanley Center for Psychiatric Research. M.L. acknowledges support from the National Natural Science Foundation of China (81870389, 82070565). Y. Kakuta and Y. Kinouchi acknowledge support from JSPS KAKENHI (21K07884, 21K07955), the Japan Agency for Medical Research and Development (AMED) (JP18kk0305002), and Labour Sciences Research Grants for Research on Intractable Diseases from the Ministry of Health, Labour, and Welfare of Japan. Y. Kakuta, Y. Kawai, K.T. and M.N. acknowledge support from AMED (JP19km0405501). K.S. acknowledges support from the National Research Foundation of Korea (2017R1A2A1A05001119, 2020R1A2C2003275). D.M. acknowledges the Leona M. and Harry B. Helmsley Charitable Trust and NIDDK U01DK062413. K.T. and M.N. acknowledge support from AMED (JP19km0405205). Part of the computations on JPN1 was performed on the NIG supercomputer at the ROIS National Institute of Genetics. Computations on SHA1 were performed in a super computing environment at the Digital Health China Technologies Corp. Ltd.. We want to acknowledge the participants and investigators of FinnGen study. The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme Corp, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, Novartis AG, and Boehringer Ingelheim. The following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). Finnish Biobank Cooperative-FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious® services (https://site.fingenious.fi/en/) managed by FINBB.
CONSORTIUM AUTHOR LISTS AND AFFILIATIONS
FinnGen
Mark J. Daly2,21,22
A list of members is available as Supplementary Table 13.
International Inflammatory Bowel Disease Genetics Consortium
Maria Abreu25, Jean-Paul Achkar26, Vibeke Andersen27, Charles Bernstein28, Steven Brant29, Luis Bujanda30, Siew Chien Ng31, Judy Cho20, Mark J. Daly2,21,22, Lee A. Denson32, Richard H. Duerr33, Lynnette R. Ferguson34, Denis Franchimont35, Andre Franke36, Richard Gearry37, Hakon Hakonarson38, Jonas Halfvarson39, Caren Heller40, Hailiang Huang2,3,24, Antonio Julià41, Judith Kelsen38, Hamed Khalili42, Subramaniam Kugathasan43, Juozas Kupcinskas44, Anna Latiano45, Edouard Louis46, Reza Malekzadeh47, Jacob Mccauley48, Dermot P. B. McGovern14, Christopher Moran49, David Okou43, Tim Orchard50, Aarno Palotie2,3,21, Miles Parkes51, Joel Pekow52, Uroš Potočnik53, Graham Radford-Smith54, John Rioux55, Gerhard Rogler56, Bruce Sands57, Mark Silverberg58, Harry Sokol59, Séverine Vermeire60, Rinse K. Weersma61, Ramnik Xavier62
25Division of Gastroenterology, Department of Medicine, Leonard M. Miller School of Medicine, University of Miami, Miami, FL, USA. 26Cleveland Clinic, Cleveland, OH, USA. 27Focused Research Unit for Molecular Diagnostic and Clinical Research, Hospital of Southern Jutland, Aabenraa, Denmark. 28University of Manitoba IBD Clinical and Research Centre, Winnipeg, Manitoba, Canada. 29Rutgers University, New Brunswick and Piscataway, NJ, USA. 30Osakidetza-Basque Health Service, Bilbao, Spain. 31The Chinese University of Hong Kong, Hong Kong, China. 32Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA. 33Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA. 34The University of Auckland, Auckland, New Zealand. 35Department of Gastroenterology, Erasme Hospital, Free University of Brussels, Brussels, Belgium. 36Institute of Clinical Molecular Biology (IKMB), Christian-Albrechts-University of Kiel, Kiel, Germany. 37Department of Medicine, University of Otago, Christchurch, New Zealand. 38Children’s Hospital of Philadelphia, Philadelphia, PA, USA. 39Department of Gastroenterology, Faculty of Medicine and Health, Örebro University, Örebro, Sweden. 40Crohn’s & Colitis Foundation, New York, NY, USA. 41Rheumatology Research Group, Vall d’Hebron University Hospital, Barcelona, Spain. 42Massachusetts General Hospital Gastroenterology Unit, Boston, MA, USA. 43Emory University, Atlanta, GA, USA. 44Department of Gastroenterology, Lithuanian University of Health Sciences, Kaunas, Lithuania. 45Fondazione IRCCS Casa Sollievo della Sofferenza, Gastroenterology Unit, San Giovanni Rotondo, Foggia, Italy. 46University of Liège, ULG, Liège, Belgium. 47DDRI, Tehran University of Medical Sciences, Tehran, Iran. 48John P. Hussman Institute for Human Genomics, Leonard M. Miller School of Medicine, University of Miami, Miami, FL, USA. 49MassGeneral Hospital for Children, Boston, MA, USA. 50Imperial College London, London, UK. 51Addenborookes Hospital, Cambridge, UK. 52University of Chicago, Chicago, IL, USA. 53University of Maribor, Faculty of Medicine, Center for Human Molecular Genetics and Pharmacogenomics, Maribor, Slovenia. 54QIMR Berghofer MRI, Herston, Australia. 55Montreal Heart Institute, Research Center, Montreal, QC, Canada. 56Department of Gastroenterology and Hepatology, University Hospital Zurich, Zurich, Switzerland. 57Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 58Mount Sinai Hospital, Toronto, ON, Canada. 59St Antoine Hospital, APHP, Paris, France. 60Department of Gastroenterology - University hospitals Leuven, Leuven, Belgium. 61Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, Groningen, The Netherlands. 62The Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Chinese Inflammatory Bowel Disease Genetics Consortium
Naizhong Hu63, Qian Cao64, Yufang Wang65, Yinglei Miao66, Hongjie Zhang67, Xiaoping Lv68, Xiang Gao69, Hu Zhang65, Jingling Su70, Baisui Feng71, Ye Zhao72, Liangru Zhu73, Yan Chen74, Lanxiang Zhu75, Chunxiao Chen76, Yali Wang77, Yingde Wang78, Zhi Pang79, Yingxuan Chen80, Xiaolan Zhang81, Hui Li82, Qin Yu83, Mei Ye84, Sumin Zhang85, Wen Tang86, Mei Wang87, Xiaocang Cao88, Ruixin Zhu89, Guangxi Zhou90, Zhaolian Bian91, Xiaofeng Guo92, Xiaoli Wu93, Jinchun Liu94, Wei Xu95, Yuqin Li96, Qin Guo97, Zhiguo Guo98, Mingsong Li5, Zhanju Liu1
63Department of Gastroenterology, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui province, China. 64Department of Gastroenterology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang province, China. 65Department of Gastroenterology, West China Hospital, Sichuan University, Chengdu, Sichuan Province, China. 66Department of Gastroenterology, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan province, China. 67Department of Gastroenterology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu province, China. 68Department of Gastroenterology, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, China. 69Department of Gastroenterology, the Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong province, China. 70Department of Gastroenterology, Affiliated Zhongshan Hospital of Xiamen University, Xiamen, Fujian province, China. 71Department of Gastroenterology, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan province, China. 72Department of Gastroenterology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan province, China. 73Department of Gastroenterology, Union Hospital, Huazhong University of Science and Technology, Wuhan, Hubei province, China. 74Department of Gastroenterology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang province, China. 75Department of Gastroenterology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu province, China. 76Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang province, China. 77Department of Gastroenterology, Datong Third People’s Hospital, Datong, Shanxi province, China. 78Department of Gastroenterology, the First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning province, China. 79Department of Gastroenterology, the Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou, Jiangsu province, China. 80Department of Gastroenterology and Hepatology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China. 81Department of Gastroenterology, The East Branch of the Second Hospital of Hebei Medical University, Shijiazhuang, Hebei province, China. 82Department of Gastroenterology, The Second Hospital of Harbin Medical University, Harbin, Heilongjiang province, China. 83Department of Gastroenterology, Tongji Hospital, Huazhong University of Science and Technology, Wuhan, Hubei province, China. 84Department of Gastroenterology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei province, China. 85Department of Colorectal Surgery, Nanjing Hospital of TCM Affiliated to Nanjing University of Chinese Medicine, Nanjing, Jiangsu province, China. 86Department of Gastroenterology, The Second Hospital of Soochow University, Suzhou, Jiangsu province, China. 87Department of Gastroenterology, Affiliated Hospital of Yangzhou University, Yangzhou, Jiangsu province, China. 88Department of Gastroenterology, General Hospital, Tianjin Medical University, Tianjin, China. 89Department of Bioinformatics, School of Life Sciences and Technology, Tongji University, Shanghai, China. 90Department of Gastroenterology, Affiliated Hospital of Jining Medical University, Jining, Shandong province, China. 91Department of Gastroenterology, Nantong Third People’s Hospital, Nantong University, Nantong, Jiangsu province, China. 92Department of Gastroenterology, Shanxi Provincial People’s Hospital, Taiyuan, Shanxi province, China. 93Department of Gastroenterology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang province, China. 94Department of Gastroenterology, First Hospital of Shanxi Medical University, Taiyuan, Shanxi province, China. 95Department of Colorectal Surgery, The Affiliated Changzhou Second People’s Hospital of Nanjing Medical University, Changzhou, Jiangsu province, China. 96Department of Gastroenterology, Bethune First Affiliated Hospital of Jilin University, Changchun, Jilin province, China. 97Department of Gastroenterology, Third Xiangya Hospital of Central South University, Changsha, Hunan province, China. 98Department of Gastroenterology, Suzhou Hospital of Anhui Medical University, Suzhou, Anhui province, China.
Footnotes
COMPETING INTERESTS
W.S. and C.S. are employees of Digital Health China Technologies Corp. Ltd.. M.J.D. is a founder of Maze Therapeutics. D.P.B.M. has received consultancy fees from Prometheus Biosciences, Prometheus Laboratories, Takeda, Gilead, Pfizer. Stock - Prometheus Biosciences. B.D.Y. has served on advisory boards for AbbVie Korea, Celltrion, Daewoong Pharma, Ferring Korea, Janssen Korea, Pfizer Korea, and Takeda Korea; has received research grants from Celltrion and Pfizer Korea; has received consulting fees from Chong Kun Dang Pharm., CJ Red BIO, Cornerstones Health, Daewoong Pharma, IQVIA, Kangstem Biotech, Korea United Pharm. Inc., Medtronic Korea, NanoEntek, and Takeda; and has received speaking fees from AbbVie Korea, Celltrion, Ferring Korea, IQVIA, Janssen Korea, Pfizer Korea, Takeda, and Takeda Korea. H.H. received consultancy fees from Ono Pharmaceutical and honorarium from Xian Janssen Pharmaceutical. The remaining authors declare no competing interests.
CODE AVAILABILITY
Computer code relating to this study includes:
RICOPILI v2019_Jun_25.001: https://sites.google.com/a/broadinstitute.org/ricopili
EIGENSTRAT v6.1.4: PCA, https://github.com/DReichLab/EIG/tree/master/EIGENSTRAT
bcftools v1.11: http://samtools.github.io/bcftools/bcftools.html
LDSC v1.0.1: https://github.com/bulik/ldsc
S-LDXR v0.3-beta: https://huwenboshi.github.io/s-ldxr
HRC-1000G-check-bim v4.3.0: https://www.well.ox.ac.uk/~wrayner/tools/HRC-1000G-check-bim-v4.3.0.zip
VcfCooker v1.1.1: https://genome.sph.umich.edu/wiki/VcfCooker
Eagle2 v2.4.1: https://alkesgroup.broadinstitute.org/Eagle/
Minimac4 v1.0.0: https://genome.sph.umich.edu/wiki/Minimac4
apt software v2.10.2.2: https://www.thermofisher.com/us/en/home/life-science/microarray-analysis/microarray-analysis-partners-programs/affymetrix-developers-network/affymetrix-power-tools.html
SNPolisher v3.0: https://downloads.thermofisher.com/SNPolisher_3.0.zip
BEAGLE v5.1: https://faculty.washington.edu/browning/beagle/b5_1.html
PLINK2 v2.00a3.6: https://www.cog-genomics.org/plink/2.0
METAL v2011-03-25: https://genome.sph.umich.edu/wiki/METAL
MANTRA v1: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3460225/
COJO v1.92.2beta: https://yanglab.westlake.edu.cn/software/gcta/#COJO
PRS-CS v1.0.0: https://github.com/getian107/PRScs
PRS-CSx v1.0.0: https://github.com/getian107/PRScsx
Python implementation for SuSiE: https://github.com/getian107/SuSiEx
FinnGen QC and Association analysis: https://finngen.gitbook.io/documentation/methods/genotype-imputation/genotype-data
FinnGen GWAS: https://finngen.gitbook.io/documentation/methods/phewas
IEU open GWAS project: https://gwas.mrcieu.ac.uk/phewas/
VEP v104.3: https://useast.ensembl.org/info/docs/tools/vep/index.html
Cytoscape v3.9.1: https://cytoscape.org/
DATA AVAILABILITY
CaVEMaN and DAP-G GTEx v8 Fine-Mapping cis-eQTL Data was retrieved from https://gtexportal.org/home/datasets#filesetFilesDiv15. 1000 Genomes Project Phase 3 is available from https://www.internationalgenome.org/category/phase-3/. TOPMed reference panel R2 is available from https://imputation.biodatacatalyst.nhlbi.nih.gov/#!. Human Genome Diversity Project is available from https://www.internationalgenome.org/data-portal/data-collection/hgdp. Simons Genome Diversity Project is available from https://www.simonsfoundation.org/simons-genome-diversity-project/. Korean Personal Genome Diversity Project is available from http://opengenome.net/Main_Page. NBDC human database (accession ID: JGAS000114) is available from https://humandbs.biosciencedbc.jp/en/. STRING functional protein association networks are available from https://string-db.org/. NFE summary statistics are from ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/. FIN summary statistics are from FinnGen R7, https://www.finngen.fi/en/access_results. PRS weights and genome-wide summary statistics for the meta-analyzed EAS samples, and across all study samples (EAS and EUR) can be downloaded from https://www.ibdgenetics.org. Individual-level genotype data for EAS samples are available upon request: SHA1, Z.L. (zhanjuliu@tongji.edu.cn); KOR1, K.S. (kysong@amc.seoul.kr); JPN1, Y. Kakuta (ykakuta@med.tohoku.ac.jp) and; ICH1, IIBDGC (ibdgc-dcc@mssm.edu). Access to individual-level genotypes from samples recruited within mainland China are subject to the policies and approvals from the Human Genetic Resource Administration, Ministry of Science and Technology of the People’s Republic of China.
REFERENCES
- 1.Inflammatory bowel disease. in Fast Facts About GI and Liver Diseases for Nurses (Springer Publishing Company, 2016). [Google Scholar]
- 2.GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol 5, 17–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M’Koma AE Inflammatory bowel disease: an expanding global health problem. Clin. Med. Insights Gastroenterol 6, 33–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung S et al. Identification of three novel susceptibility loci for inflammatory bowel disease in Koreans in an extended genome-wide association study. J. Crohns. Colitis 15, 1898–1907 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Yang S-K et al. Identification of loci at 1q21 and 16q23 that affect susceptibility to inflammatory bowel disease in Koreans. Gastroenterology 151, 1096–1099.e4 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Brant SR et al. Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology 152, 206–217.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyette P et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet 47, 172–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleynen I et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 387, 156–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam M et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51, 1670–1678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang CS et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun 6, 10206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugot JP et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Ogura Y et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurki MI et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sazonovs A et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet 54, 1275–1283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai M et al. Meta-analysis fine-mapping is often miscalibrated at single-variant resolution. Cell Genom 2, 100210 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivas MA et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet 43, 1066–1073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivas MA et al. A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat. Commun 7, 12342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lassen KG et al. Genetic coding variant in GPR65 alters lysosomal pH and links lysosomal dysfunction with colitis risk. Immunity 44, 1392–1405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavoie S et al. The Crohn’s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife 8, e39982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varma M et al. Cell type- and stimulation-dependent transcriptional programs regulated by Atg16L1 and its Crohn’s disease risk variant T300A. J. Immunol 205, 414–424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris AP Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol 35, 809–822 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Festen EAM et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut 58, 799–804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki CA et al. Transforming growth factor beta (TGF-beta) and autoimmunity. Autoimmun. Rev 4, 450–459 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Ihara S, Hirata Y & Koike K TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol 52, 777–787 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Kiryluk K et al. GWAS defines pathogenic signaling pathways and prioritizes drug targets for IgA nephropathy. bioRxiv. Preprint at doi: 10.1101/2021.11.19.21265383 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Ge T, Chen C-Y, Ni Y, Feng Y-CA & Smoller JW Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun 10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammer T & Langholz E The epidemiology of inflammatory bowel disease: balance between East and West? A narrative review. Dig. Med. Res 3, 48–48 (2020). [Google Scholar]
- 33.Ruan Y et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet 54, 573–580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asano K et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet 41, 1325–1329 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki K et al. A genome-wide association study identifies 2 susceptibility loci for Crohn’s disease in a Japanese population. Gastroenterology 144, 781–788 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Yang S-K et al. Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm. Bowel Dis 19, 954–966 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Yang S-K et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63, 80–87 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Fuyuno Y et al. Genetic characteristics of inflammatory bowel disease in a Japanese population. J. Gastroenterol 51, 672–681 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Yang S-K et al. Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm. Bowel Dis 21, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H-S et al. X chromosome-wide association study identifies a susceptibility locus for inflammatory bowel disease in Koreans. J. Crohns. Colitis 11, 820–830 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Kakuta Y et al. A genome-wide association study identifying RAP1A as a novel susceptibility gene for Crohn’s disease in Japanese individuals. J. Crohns. Colitis 13, 648–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto D et al. Genetic analysis of ulcerative colitis in Japanese individuals using population-specific SNP array. Inflamm. Bowel Dis 26, 1177–1187 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Ye BD et al. Identification of ten additional susceptibility loci for ulcerative colitis through Immunochip analysis in Koreans. Inflamm. Bowel Dis 22, 13–19 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Kanai M et al. Insights from complex trait fine-mapping across diverse populations. bioRxiv. Preprint at doi: 10.1101/2021.09.03.21262975 (2021). [DOI] [Google Scholar]
- 45.Ye BD et al. Association of FUT2 and ABO with Crohn’s disease in Koreans. J. Gastroenterol. Hepatol 35, 104–109 (2020). [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 46.Magro F et al. European Crohn’s and Colitis Organisation [ECCO]. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J. Crohns. Colitis 11, 649–670 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Gomollón F et al. 3rd European Evidence-based Consensus on the Diagnosis and Management of Crohn’s Disease 2016: Part 1: Diagnosis and Medical Management. J. Crohns. Colitis 11, 3–25 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Sturm A et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 2: IBD scores and general principles and technical aspects. J. Crohns. Colitis 13, 273–284 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Maaser C et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J. Crohns. Colitis 13, 144–164 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Kakuta Y et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: a multicenter study. J. Gastroenterol 53, 1065–1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakuta Y et al. Crohn’s disease and early exposure to thiopurines are independent risk factors for mosaic chromosomal alterations in patients with inflammatory bowel diseases. J. Crohns. Colitis 16, 643–655 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Cortes A & Brown MA Promise and pitfalls of the Immunochip. Arthritis Res. Ther 13, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taliun D et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawai Y et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J. Hum. Genet 60, 581–587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G, Sarkar A, Carbonetto P & Stephens M A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Series B Stat. Methodol 82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bulik-Sullivan B et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SH, Wray NR, Goddard ME & Visscher PM Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi H et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat. Commun 12, 1098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park SH et al. A 30-year trend analysis in the epidemiology of inflammatory bowel disease in the Songpa-Kangdong District of Seoul, Korea in 1986–2015. J. Crohns. Colitis 13, 1410–1417 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Szklarczyk D et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables 1-13
Manhattan plot for IBD genetic associations.
Regional association plot for IBD-associated loci in EAS
Forest plot for index variants in IBD-associated loci
Forest plot for putative causal variants identified in EAS and in EUR
Supplementary Note including discussions in the Major histocompatibility complex (MHC) and IBD genes of interest.
Data Availability Statement
CaVEMaN and DAP-G GTEx v8 Fine-Mapping cis-eQTL Data was retrieved from https://gtexportal.org/home/datasets#filesetFilesDiv15. 1000 Genomes Project Phase 3 is available from https://www.internationalgenome.org/category/phase-3/. TOPMed reference panel R2 is available from https://imputation.biodatacatalyst.nhlbi.nih.gov/#!. Human Genome Diversity Project is available from https://www.internationalgenome.org/data-portal/data-collection/hgdp. Simons Genome Diversity Project is available from https://www.simonsfoundation.org/simons-genome-diversity-project/. Korean Personal Genome Diversity Project is available from http://opengenome.net/Main_Page. NBDC human database (accession ID: JGAS000114) is available from https://humandbs.biosciencedbc.jp/en/. STRING functional protein association networks are available from https://string-db.org/. NFE summary statistics are from ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/. FIN summary statistics are from FinnGen R7, https://www.finngen.fi/en/access_results. PRS weights and genome-wide summary statistics for the meta-analyzed EAS samples, and across all study samples (EAS and EUR) can be downloaded from https://www.ibdgenetics.org. Individual-level genotype data for EAS samples are available upon request: SHA1, Z.L. (zhanjuliu@tongji.edu.cn); KOR1, K.S. (kysong@amc.seoul.kr); JPN1, Y. Kakuta (ykakuta@med.tohoku.ac.jp) and; ICH1, IIBDGC (ibdgc-dcc@mssm.edu). Access to individual-level genotypes from samples recruited within mainland China are subject to the policies and approvals from the Human Genetic Resource Administration, Ministry of Science and Technology of the People’s Republic of China.


