Abstract
Vestimentiferan tubeworms are representative members of deep-sea chemosynthetic ecosystems. In this study, we developed a draft genome and gene models and performed genomic and transcriptomic analyses of Lamellibrachia satsuma, the only vestimentiferan reported from the euphotic zone. The quality of the genome assembly and gene models is comparable to or higher than those of previously reported vestimentiferan tubeworms. Tissue-specific transcriptome sequencing revealed that Toll-like receptor genes and lineage-specific expanded bacteriolytic enzyme genes are highly expressed in the obturacular and vestimental regions, respectively, suggesting the importance of these tissues in defense against pathogens. On the other hand, globin subunit genes are expressed almost exclusively in the trunk region, supporting the hypothesis that the trophosome is the site of haemoglobin biosynthesis. Vestimentiferan-specific expanded gene families included chitinases, ion channels, and C-type lectins, suggesting the importance of these functions for vestimentiferans. C-type lectins in the trunk region, in particular, may be involved in recognition of pathogens, or in interactions between tubeworms and symbiotic bacteria. Our genomic and transcriptomic analyses enhance understanding of molecular mechanisms underlying the unique lifestyle of vestimentiferan tubeworms, particularly their obligate mutualism with chemosynthetic bacteria.
Keywords: tubeworm, genome sequencing, transcriptomic analysis, gene duplication, chemosynthetic symbiosis
1. Introduction
Since the discovery of symbiosis between chemosynthetic bacteria and a tubeworm lacking a digestive tract (Riftia pachyptila), scientists have investigated chemosynthetic symbioses in various marine environments, ranging from hydrothermal vents to shallow-water coastal sediments.1 Tube-dwelling annelids (Siboglinidae) are the best-studied members of deep-sea chemosynthetic ecosystems, and of this family’s four main lineages, Frenulata, Vestimentifera, Sclerolinum, and Osedax, vestimentiferans are typically found in hydrothermal vent and hydrocarbon seep areas.2 Adult vestimentiferan tubeworms lack mouths and digestive systems and depend on chemoautotrophic or methanotrophic endosymbionts for their nutritional demands.3 Symbiotic bacteria are acquired from the surrounding environment by horizontal transmission.4 In a cold seep vestimentiferan tubeworm, Lamellibrachia satsuma, γ- and ε-Proteobacteria have been found in the trophosome, the organ specialized for bacterial symbiosis.5
Bodies of vestimentiferans can be divided into four main regions: an obturacular region, a vestimental region, a trunk region, and an opisthosoma (Fig. 1B).2,9 The obturacular region is composed of the obturaculum and branchial plume, which is the primary site of inorganic compound absorption and gas exchange. This region is in direct contact with seawater, while other parts of the body are enclosed in a chitinous tube, which defines the unique appearance of tubeworms.10 The vestimental region, from which the name Vestimentifera is derived, is a highly muscular region that encloses a heart, brain, and gonopore.11,12 The trophosome, a highly vascularized organ that occupies most of the trunk region, harbours symbiotic bacteria in bacteriocytes.13–15 The opisthosoma exhibits most of the features shared with other annelids, including muscular septae and segmentally arranged chitinous chaetae.9
Sulfide, O2, and CO2, required for chemosynthesis by symbiotic bacteria, are taken up by the branchial plume, transported to the trophosome, and supplied to chemosynthetic symbionts in bacteriocytes. Vestimentiferans inhabiting cold seeps can also acquire sulfide via the ‘root’, an extension of the posterior part of the body.16,17 Extracellular hemoglobins (Hbs) reversibly bind oxygen and sulfide at two sites, and transport them through the vascular blood and coelomic fluid.18,19 Hbs may be involved in protection of cells against sulfide toxicity, by binding H2S with high affinity,19,20 thereby preventing inhibition of cytochrome c oxidase.
While most vestimentiferan tubeworms inhabit the deep sea, only Lamellibrachia satsuma has been reported from the euphotic zone (at a depth of 82 to 110 m).21,22 Thus, this species is relatively easy to collect. In addition, L. satsuma can be maintained under laboratory conditions for long periods, provided with sodium sulfide (Na2S) or a whale vertebra as a source of energy (Fig. 1A).21,23,24 Spawning and early larval development have also been observed using individuals kept in aquaria.21,23,24 These features render L. satsuma suitable as a model system for tubeworm research. However, a draft sequence of its nuclear genome has been lacking, while the complete mitochondrial genome and the γ-proteobacterial symbiont genome have already been reported.25,26
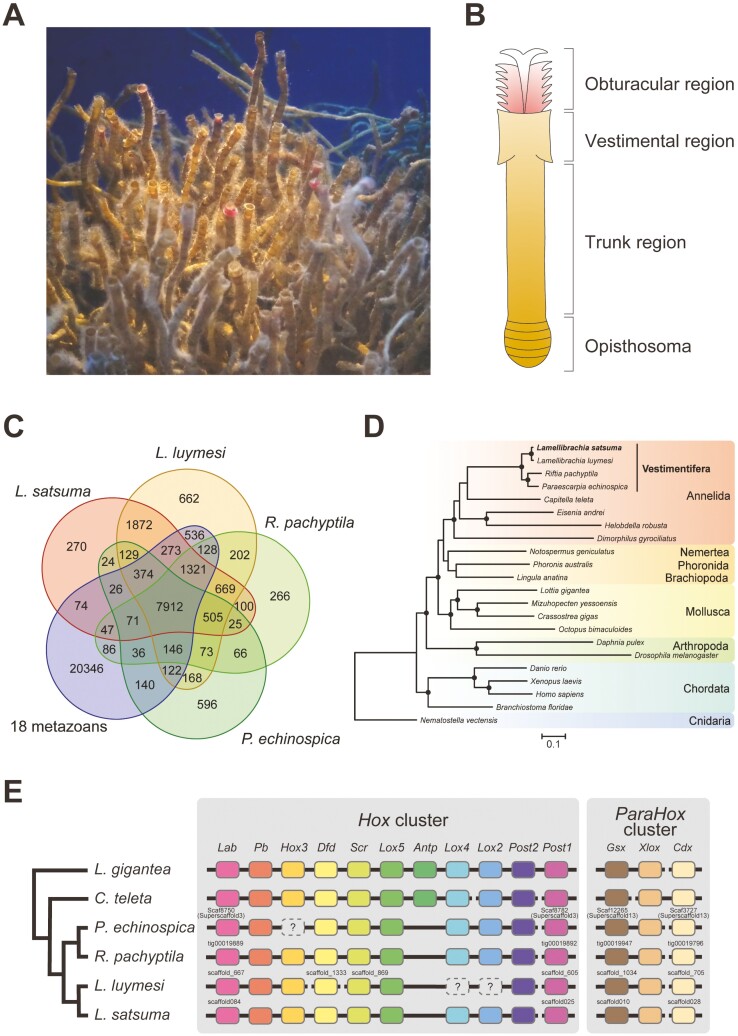
Figure 1.
Genome sequencing of Lamellibrachia satsuma. (A) L. satsuma maintained at Kagoshima City Aquarium, Kagoshima, Japan. (B) Schematic drawing of the body of a vestimentiferan. The chitinous tube that covers the body is omitted for clarity. (C) Venn diagram of shared gene families (OGs) among four vestimentiferans and 18 non-vestimentiferan metazoans. (D) Phylogenetic relationships among four vestimentiferans and 18 non-vestimentiferan metazoans. Concatenated amino acid sequences of 30 single-copy OGs shared by all 22 species were aligned, gap-trimmed, and used for construction of a maximum likelihood phylogenetic tree. The phylogenetic tree constructed with 163 single-copy OGs from 15 lophotrochozoans and two arthropods is also shown in Supplementary Fig. S1. Circles on branches indicate bootstrap values higher than 80%. (E) Schematic representation of Hox and ParaHox cluster organization of four vestimentiferans, C. teleta, and L. gigantea. The last common ancestor of molluscs and annelids is estimated to have possessed 11 Hox genes, as seen in L. gigantea.6 The cladogram on the left is based on phylogenetic relationships inferred from the present study. Hox3 in P. echinospica, Lox4, and Lox2 in L. luymesi were identified in one previous study,7 while not in another previous study8 or in this study.
In this study, we report a high-quality draft genome of Lamellibrachia satsuma, collected from shallow water in Kagoshima Bay, Japan. In order to reveal the evolutionary history of genes involved in the unique vestimentiferan lifestyle, we performed comparative genomic analyses using 21 metazoan genomes, including three vestimentiferan tubeworms, Lamellibrachia luymesi, Paraescarpia echinospica, and Riftia pachyptila.7,8,27 Transcriptomic analyses using three regions of the body of L. satsuma were also conducted to understand functions of these genes. These analyses provide insights that will help to explain unique strategies and evolutionary history of L. satsuma and the Vestimentifera.
2. Materials and methods
2.1. Sample collection, DNA extraction, and genome sequencing
Tubeworms were collected with the Nanseimaru at Kagoshima Bay, Kagoshima, Japan (31_39.756ʹN, 130_48.050ʹE), kept in a tank, and immediately transferred to Kagoshima City Aquarium (25 June 2018). Chitinous tubes were removed, and then the obturacular region, vestimental region, and trunk region were cut into approximately 1 cm pieces. The male gonad with sperm, was isolated from male specimens. Samples were immediately stored at −80℃ until DNA and RNA extraction.
To obtain highly concentrated DNA with little contamination from symbionts, we isolated high-molecular-weight DNA from frozen male gonads, according to the Bionano Prep Cell Culture DNA Isolation Protocol (BioNano Genomics). This isolated, high-molecular weight DNA was used to construct a sequencing library with Ligation Sequencing Kit (SQK-LSK109; Oxford Nanopore Technologies) according to the manufacturer’s instructions. The library was sequenced with PromethION (Oxford Nanopore Technologies) using R9.4.1 flow cells (Oxford Nanopore Technologies). The same DNA was also sequenced using a NovaSeq (250 bp, paired-end) (Illumina).
2.2. Genome assembly
We obtained 54.0 Gb of PromethION raw sequencing data, but only 38.3 Gb, comprising sequences >10 kb, were used for genome assembly. PromethION sequence reads longer than 10 kbp were first assembled using NECAT v.0.0128 with default options. Possible diploid scaffolds were removed with Purge Haplotigs v.1.1.129 and merged with HaploMerger230 using default parameters. Further scaffolding using the raw PromethION data was performed with LINKS v.1.8.531 with a kmer size of 25. Finally, assembled sequences were polished using the raw PromethION data and paired-end Illumina shotgun reads with HyPo v.1.0.3.32
To identify possible symbiotic bacterial scaffolds, all scaffolds were searched using BLASTN (E-value cut off: 1e−5) against the dataset in the Bacteria division of the DDBJ ALL database, and complete genomes of mitochondria and γ-proteobacterial endosymbionts isolated from L. satsuma collected from Kagoshima Bay.25,26 Sixteen relatively short scaffolds with ~0 E-value and ~100% query coverage were concluded to be endosymbiont contaminants and were removed from the draft genome assembly. We assessed completeness of the draft genome with BUSCO ver. 5.3.2,33 using the metazoa_odb10 dataset (954 genes).
2.3. RNA extraction, transcriptome sequencing, gene prediction, and annotation
Total RNA was extracted from the obturacular, vestimental, and trunk regions of three specimens using TRIzol reagent (Thermo Fisher Scientific) and cleaned with an RNeasy Mini Kit (QIAGEN). An mRNA sequencing library was prepared using an MGIEasy RNA Directional Library Prep Set (MGI). The resulting dsDNA library was circularized with an MGIEasy Circularization Kit (MGI), and then DNA nanoballs were prepared with a DNBSEQ-G400RS High-throughput Sequencing Kit (MGI). 2 × 100 bp paired-end sequencing of each library was performed using DNBSEQ-G400 (MGI). Low-quality reads (quality score < 20 and length < 25 bp) and sequence adaptors were trimmed using CUTADAPT v.1.18.34
For gene prediction from the assembled scaffolds, repetitive elements in the assembled scaffolds were identified de novo with RepeatScout v.1.0.635 and RepeatMasker v.4.1.0 (http://www.repeatmasker.org). Repetitive elements were masked based on length (>50 bp) and occurrence (more than 10 times). Gene prediction was first executed with the BRAKER pipeline v2.1.2,36 with AUGUSTUS v.3.3.3 as in Yoshioka et al.37 RNA-seq reads were aligned to the genome sequence with HISAT v2.1.0.38 Then, alignment information was used for BRAKER gene prediction with options ‘UTR = on’, ‘softmasking’, and ‘AUGUSTUS_ab_initio’. To improve gene prediction, we further performed genome-guided transcriptome assembly using StringTie39 with an option ‘-m 500’. During read alignments, we used soft-masked repeats for genome-guided transcriptome assembly and hard-masked repeats for BRAKER gene prediction. Finally, genes that were absent in AUGUSTUS prediction, but present in predictions from genome-guided transcriptome assembly or AUGUSTUS ab initio were added to the gene predictions from AUGUSTUS using GffCompare.40 We named the L. satsuma gene models using the pattern, Lsat_s[scaffold number].g[number of genes in the scaffold]. For example, ‘Lsat_s006.g0545’ is located in scaffold 6 and is the 545th gene in the scaffold. The longest transcript variants of each gene were selected and translated into protein sequences with TransDecoder (https://github.com/TransDecoder/TransDecoder). We assessed completeness of gene models with BUSCO ver. 5.3.233 using the metazoa_odb10 dataset (954 genes).
The proteome was annotated with BLASTP (E-value cut off: 1e−5) against the UniProt/SwissProt database.41 Domain structures and transmembrane regions of proteins were analyzed with InterProScan ver. 5.56-89.0.42 and DeepTMHMM,43 respectively.
2.4. Clustering of orthologous genes and phylogenetic analysis
In addition to the L. satsuma gene models, we used publicly available gene models of 21 metazoans including three vestimentiferans, four non-vestimentiferan annelids, a nemertean, a phoronid, a brachiopod, four molluscs, two arthropods, four chordates, and a cnidarian (Supplementary Table S1). The longest transcript variants of each gene were selected and used for the following analyses. The completeness of gene models was assessed with BUSCO ver. 5.3.233 using the metazoa_odb10 dataset (954 genes). Clustering of orthogroups (OGs) was performed using OrthoFinder ver. 2.5.4,44 and OGs were considered gene families in this study.
For phylogenetic analysis of metazoan genomes, we used 30 OGs assigned as single copy in all of the above metazoan genomes. All amino acid sequences belonging to the same gene family were aligned using MAFFT ver. 7.310 with ‘-auto’ option,45 and all gaps in alignments were removed using TrimAL ver. 1.446 with the ‘-nogaps’ option. All sequences from the same species were concatenated, and then a maximum likelihood analysis of concatenated sequences (6,751 amino acid sequences in length) was performed using RaxML ver. 8.2.12 with 100 bootstrap replicates and the ‘PROTOGAMMAAUTO’ option.47
For phylogenetic analysis of each gene, amino acid sequences were aligned using MAFFT v.7.310 with ‘-auto’ option,45 and gaps were removed using TrimAL v.1.446 with the ‘-gappyout’ option. Trimmed sequences composed only of gaps were removed from the following analyses. Poorly aligned sequences were also removed manually. Then, maximum likelihood analyses of aligned sequences were performed using RaxML v.8.2.12 with 100 bootstrap replicates and the ‘PROTOGAMMAAUTO’ option.47
2.5 Identification of lineage-specific expanded or contracted gene families
In order to identify lineage-specific expanded gene families, we compared gene numbers in each OG of four vestimentiferans and four non-vestimentiferan annelids (Capitella teleta, Helobdella robusta, Eisenia andrei, and Dimorphilus gyrociliatus). We then conducted molecular phylogenetic analysis of genes within candidate OGs to estimate their evolutionary origin.
Only OGs that were shared by all eight species were used for the gene number comparison, and genes associated with transposable elements were excluded from the analysis. We performed Fisher’s exact test (‘average gene number in one OG in lineage A’/‘average gene number in the other OGs in lineage A’ versus the ‘average gene number in one OG in lineage B’/‘average gene number in the other OGs in lineage B’) in R v.4.1.3,48 after rounding all elements to integers. To identify candidate OGs specifically expanded in a specific lineage (lineage A), we selected gene families with the criteria that gene numbers of lineage A should be larger than those of lineage B and the P-value in Fisher’s exact test should be <0.05. We then performed molecular phylogenetic analysis as described above.
OGs specifically contracted in a specific lineage (lineage A) were also selected in the same way, but with the criteria that gene numbers of lineage A should be smaller than those of lineage B and the P-value in Fisher’s exact test should be <0.05.
2.6 RNA-seq gene expression and enrichment analyses
Cleaned RNA-seq reads obtained from three specimens were mapped to L. satsuma gene models using bwa-mem2 v.2.2.1.49 Expression levels were quantified using Salmon v.1.8.0.50 Mapping counts were normalized by the trimmed mean of M values (TMM) method, and then converted to counts per million (CPM) using EdgeR v.3.36.051,52 in R v.4.1.3.48 Before statistical testing, we removed genes with expression levels below 1 TMM-normalized CPM in at least half the RNA-Seq samples. Then TMM-normalized CPMs in each region were compared pairwise with the other two regions to identify DEGs. Obtained P-values were adjusted using the Benjamini–Hochberg method in EdgeR. When the gene expression level of the region is significantly higher (false discovery rate; FDR < 0.01) than the other two regions, genes were considered region-specific DEGs. Expression levels of each gene (average transcripts per million (TPMs)) were visualized using ComplexHeatmap v.2.13.1.53 in R. Genes with average TPMs >1 were considered expressed.
Functional enrichment analysis for UniProt keywords and Gene Ontology terms using region-specific DEGs was performed on the web platform Database for Annotation, Visualization and Integrated Discovery (DAVID) 202154,55 with default settings. UniProt IDs of BLAST top hit for L. satsuma gene models were used as the background dataset, and those for region-specific DEGs were analyzed. Gene models that had no homologies with sequences in the SwissProt database were excluded from the analysis.
3. Results and discussion
3.1. Genome assembly and gene prediction of Lamellibrachia satsuma
After removing possible contaminants originating from symbiotic bacteria, we obtained a draft genome assembly of approximately 736 Mbp with an 8.1 Mbp N50 size and 306 scaffold sequences (Table 1). Benchmarking Universal Single-Copy Orthologs (BUSCO) analyses showed that the draft genome recovered 95.5% of the highly conserved single-copy orthologs in metazoan species, which is higher than other vestimentiferan draft genomes (L. luymesi, P. echinospica, and R. pachyptila) (Table 1). We predicted 27,979 protein-coding genes, which is comparable to the draft genome of the giant tubeworm, R. pachyptila (Table 1).
Table 1.
Statistics of genome assembly and gene annotation of vestimentiferans
| Lamellibrachia satsuma (this study) | Lamellibrachia luymesi 27 | Riftia pachyptila 8 | Paraescarpia echinospica 7 | |
|---|---|---|---|---|
| Total size (bases) | 735,529,629 | 687,711,696 | 560,783,187 | 1,090,967,472 |
| Number of scaffolds | 306 | 11,871 | 447 | 7,389 (14 pseudo-chromosomes + 7375 contigs) |
| N50 scaffold size (bases) | 8,074,470 | 372,990 | 2,870,320 | 67,235,296 |
| GC content (%) | 40.2 | 40.2 | 40.9 | 40.6 |
| Number of protein-coding genes | 27,979 | 38,998 | 25,984 | 22,642 |
| BUSCO completeness for genome assembly (%) | 95.5 [S: 95.3, D: 0.2] F: 2.7, M: 1.8 |
91.8 [S: 91.1, D: 0.7] F: 5.3, M: 2.9 |
94.7 [S: 94.4, D: 0.3] F: 3.4, M: 1.9 |
94.1 [S: 92.2, D: 1.9] F: 3.7, M: 2.2 |
| BUSCO completeness for gene models (%) | 96.2 [S: 95.6, D: 0.6] F: 1.2, M: 2.6 |
94.0 [S: 93.3, D: 0.7] F: 4.5, M: 1.5 |
93.4 [S: 92.9, D: 0.5] F: 3.4, M: 3.2 |
81.4 [S: 79.4, D: 2.0] F: 7.0, M: 11.6 |
Only the longest transcript variant of each gene was used for analysis with BUSCO v.5.3.2.
S: complete and single-copy BUSCOs; D: complete and duplicated BUSCOs; F: fragmented BUSCOs; M: missing BUSCOs.
3.2 Comparative genomics among metazoan genomes
Clustering of orthologous groups (OGs) was performed with OrthoFinder v.2.5.4 using gene models of four tubeworms, including L. satsuma, four non-vestimentiferan annelids, a nemertean, a phoronid, a brachiopod, three molluscs, four chordates, and a cnidarian (Supplementary Table S1). We identified 38,051 gene families, of which 505 gene families were shared exclusively by the four vestimentiferans, so these were considered Vestimentifera-specific gene families (Fig. 1C). These gene families may have been instrumental in the evolutionary history of vestimentiferans. Two Lamellibrachia species, L. satsuma, and L. luymesi, shared 1,872 gene families exclusively, and these may have lineage-specific functions.
We performed molecular phylogenetic analysis using concatenated amino acid sequences of 30 single-copy OGs shared by all 22 species. This analysis showed that these four vestimentiferans form a cluster, and that the genus Lamellibrachia is a sister to the clade that includes Riftia and Paraescarpia, as in a previous study using mitochondrial genomes56 (Fig. 1D). Analyses using 163 single-copy OGs from 15 lophotrochozoans and two arthropods might show more accurate phylogenetic relationships between six phyla, since most clades were supported by 100% bootstrap values. (Supplementary Fig. S1).
Hox genes are clustered on metazoan chromosomes and function as transcriptional regulators that guide axial patterning during development. The ParaHox gene cluster is a paralog of the Hox gene cluster and is also involved in metazoan development.57,58 In the L. satsuma and Riftia genomes, we identified 10 of 11 Hox genes that were present in the last common ancestor of the Mollusca and Annelida (Fig. 1E and Supplementary Fig. S2A).6 All four vestimentiferan genomes lacked Antp, which is hypothesized to have been possessed by the molluscan-annelid common ancestor,6 suggesting the loss of this gene in the common ancestor of vestimentiferans or siboglinids. In the L. satsuma and Riftia genomes, Hox genes except for Post1 are arranged in the same order as in the molluscan-annelid common ancestor. Post1 is reportedly separated from the main Hox cluster by a large distance in the same pseudochromosome in the Paraescarpia genome,7 suggesting a conserved arrangement among vestimentiferan genomes. We also identified three ParaHox genes in all vestimentiferan genomes that were present in the molluscan-annelid common ancestor (Fig. 1E and Supplementary Fig. S2B). As with Post1 in the Hox cluster, Cdx was separated by a large distance (approximately 41 Mbp) from other ParaHox genes in the same pseudochromosome of Paraescarpia, whereas they were located in different scaffolds in the L. satsuma, L. luymesi, and Riftia genomes. Collectively, unlike tapeworms, which also lack digestive tracts, and which are reported to have lost all ParaHox and many Hox genes,59 vestimentiferan tubeworms possess almost completely conserved Hox and ParaHox clusters. However, the loss of Antp in the common ancestor may be responsible for limited segmentation in the posterior region of juveniles, as discussed in a previous study using the P. echinospica genome.7
3.3 Tissue-specific expressed genes
In order to reveal transcriptomic differences between the obturacular, vestimental, and trunk regions of L. satsuma, we performed tissue-specific RNA-seq analysis using three specimens. An average of approximately 17 million RNA-seq reads per sample was retained after quality control (Supplementary Table S2). We compared gene expression levels of the three regions and identified 429, 219, and 637 DEGs specific to the obturacular, vestimental, and trunk regions, respectively (Supplementary Table S9). Among these, 59.6%, 50.9%, and 64.7% had homologies with sequences in the SwissProt database (Supplementary Table S9).
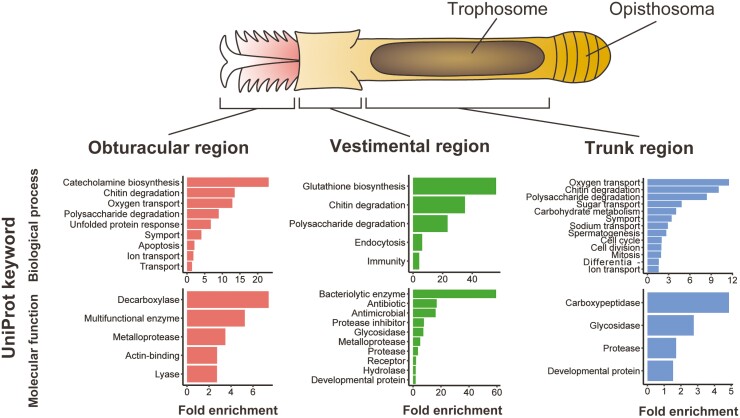
Functional enrichment analysis, based on UniProt keywords, revealed transcriptomic characteristics of the three main regions of the vestimentiferan (Fig. 2). Genes involved in transport, i.e., oxygen transport, ion transport, and symport, were upregulated in the obturacular region, suggesting their roles in acquiring substances from the environment. While the rest of the vestimentiferan body is covered with a chitinous tube, the obturacular region is in direct contact with the surrounding water and is exposed to environmental stresses, including water temperature fluctuations and toxic chemicals. Enrichment of genes related to unfolded protein response and apoptosis suggests their involvement in stress responses to extreme environmental conditions (Fig. 2; see also Supplementary Fig. S3). This result is consistent with a previous study using the Riftia transcriptome, in which apoptosis-related genes are highly expressed in the plume.8 Genes related to metabolism of proteins and peptides, i.e., carboxypeptidase and protease, were enriched among trunk-specific upregulated genes (Fig. 2; see also Supplementary Fig. S5). One of these genes was cathepsin, which is the best-known protease involved in protein degradation in lysosomes.60 Previous studies have shown that several cathepsins are highly expressed in trophosomes of the vestimentiferans, L. luymesi, Paraescarpia, and Riftia at the level of the transcriptome or proteome, suggesting a common route of lysosomal digestion of symbiotic bacteria,7,8,27,61 and our results also support this hypothesis. Enrichment of genes related to transport in this region suggests their involvement in nutrient exchange between host and symbiont (Fig. 2; see also Supplementary Fig. S5). Genes related to spermatogenesis and cell division were upregulated in the trunk region (Fig. 2), possibly because this region contains gonads, though most of this region is occupied by the trophosome.12
Figure 2.
Transcriptomic profiles of body regions of L. satsuma. The schematic drawing at the top shows the basic body structure of a ventimentiferan. The graph at the bottom shows results of the functional enrichment analysis of region-specific DEGs based on UniProt keywords. Keywords with P-values < 0.05 are shown.
Interestingly, obturacular, vestimental, and trunk region-specific DEGs each included different chitinases (Fig. 2). These genes may be involved in reconstruction of chitinous tubes that cover most of vestimentiferan bodies. In addition, oxygen transport-related genes, specifically globins and a linker chain, were upregulated in the trunk region (Fig. 2, see below).
3.4 Innate immunity
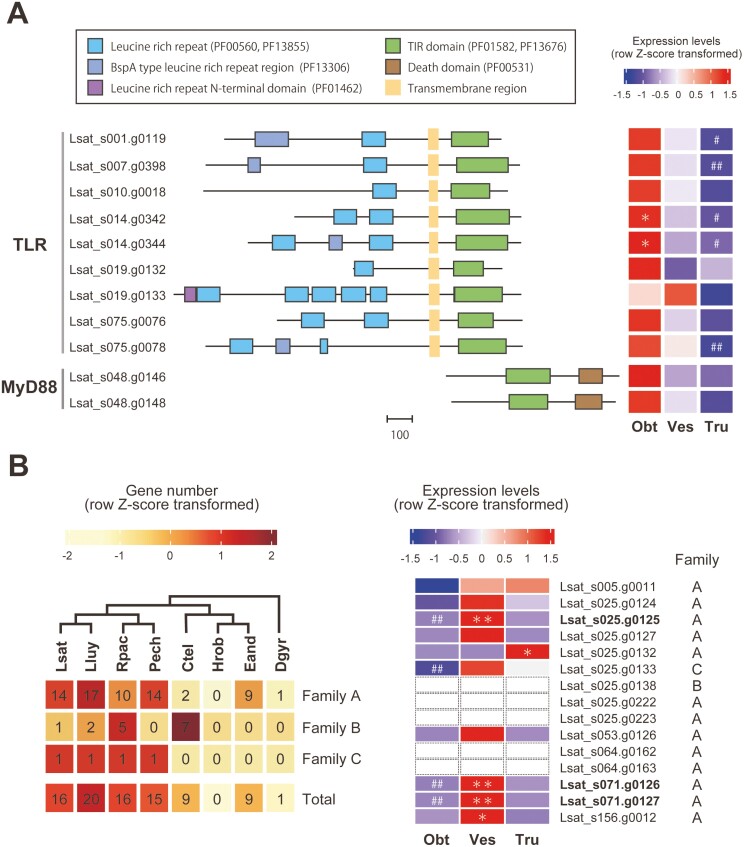
Genes related to antimicrobial responses (‘bacteriolytic enzyme’, ‘antibiotic’, and ‘antimicrobial’ in Fig. 2) were enriched among vestimental region-specific DEGs (Fig. 2). Actually, DEGs characterized by these terms exclusively comprised lysozyme genes (see also Supplementary Fig. S4). Lysozyme genes tended to be highly expressed in the vestimental region, including some genes not identified as DEGs (Fig. 3B and Supplementary Fig. S7), suggesting that they may contribute to innate immune defense against non-symbiotic bacteria in this region. Our analysis also suggests that lysozyme genes possessing a destabilase domain (PF05497), which is found in some invertebrate-type lysozymes,62,63 were expanded by tandem duplication in each lineage of vestimentiferans (Fig. 3B and Supplementary Fig. S8).
Figure 3.
Toll-like receptor, MyD88, and lysozyme genes in the L. satsuma genome. (A) TLR and MyD88. Domain organizations are shown on the left. The scale bar indicates the number of amino acid residues. A heat map of relative gene expression levels (row Z-score transformed) is shown on the right. (B) Lysozymes. Gene numbers of OGs containing putative lysozyme genes (family A, B, and C) and their totals are shown on the left. The cladogram at the top is based on phylogenetic relationships inferred from the present study (Supplementary Fig. S1). All L. satsuma genes in these OGs were annotated as lysozymes by a BLAST search against the SwissProt database, and all except one, possessed a destabilase domain (PF05497). Heat map of relative expression levels (row Z-score transformed) of lysozyme genes with a destabilase domain (PF05497), including three vestimentum-specific DEGs are shown on the right. Expression of Lsat_s025.g0138, 0222, g0223, Lsat_s064.g0162, g0163 were not detected in any of the three regions. Lsat, L. satsuma; Lluy, L. luymesi; Rpac, R. pachyptila; Pech, P. echinospica; Ctel, C. teleta; Hrob, H. robusta; Eand, E. andrei; Dgyr, D. gyrociliatus; Obt, obturacular region; Ves, vestimental region; Tru, trunk region; * or **, genes with expression levels significantly higher in one region than other regions (FDR < 0.05 or 0.01, respectively); # or ##, genes with expression levels significantly lower in a specific region than in other regions (FDR < 0.05 or 0.01, respectively).
Pattern recognition receptors (PRRs) contribute to innate immune system control in both vertebrates and invertebrates. They detect microbe-associated molecular patterns, conserved molecular structures specific to microbes, and trigger host immune responses via activation of complex signalling pathways.64,65 The best-known examples of PRRs are Toll-like receptors (TLRs), transmembrane proteins containing leucine-rich repeats, transmembrane domains, and intracellular Toll-interleukin 1 receptor (TIR) domains.66 They are expressed on cell surfaces or in intracellular vesicles of immune cells and recognize mainly microbial membrane components and microbial nucleic acids, respectively.66 In L. luymesi, TLRs and innate immunity are hypothesized to be involved in endosymbiont acquisition and selective tolerance.27
We performed genomic analysis to identify TLR genes in the L. satsuma genome and compared gene expression levels in the three regions. Our analysis revealed that there are at least nine TLR genes in the L. satsuma genome with different domain structures, and six of them are arranged in three clusters in the genome (Fig. 3A). There is a clear tendency for most TLR genes to be highly expressed in the obturacular region and downregulated in the trunk region. Only one gene tended to be highly expressed in the vestimental region, though the difference was not significant (Fig. 3A and Supplementary Fig. S6). In addition, we identified two copies of MyD88 with TIR and death domains, the essential adaptor molecule for TLRs.66 As with TLRs, MyD88 tended to be highly expressed in the obturacular region and downregulated in the trunk region (Fig. 3A and Supplementary Fig. S6). These results are consistent with previous studies reporting that TLRs are highly expressed in the plume and vestimentum of Paraescarpia and Riftia,7,8,67 and suggest that non-self-recognition and innate immune responses via TLRs are active in the obturacular region, the interface with the environment, yet suppressed in the trunk region. Collectively, pathogen recognition and innate immune responses are presumably active in the obturacular and vestimental regions, which are susceptible to pathogens, yet suppressed in the trunk region, which contains trophosomes. This system is reasonable to protect symbiotic bacteria from the host immune system.
3.4 Sulfide and oxygen transport-related genes
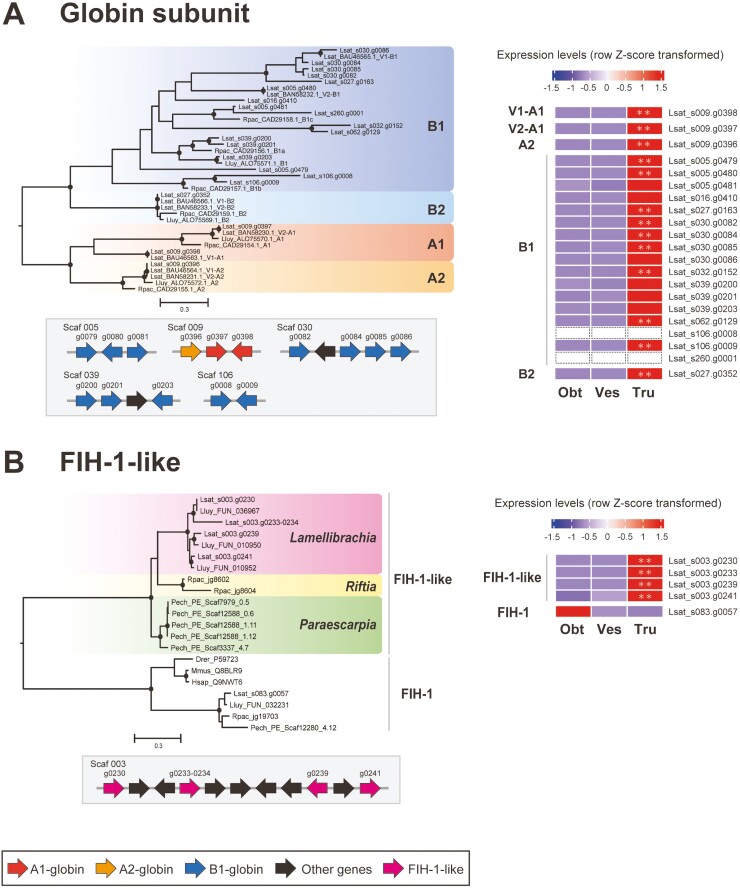
Extracellular hemoglobins (Hbs) function in oxygen and sulfide transport in vestimentiferans by reversibly binding to these molecules.18,19 Siboglinids possess three types of extracellular Hbs, V1, V2, and C1,68 containing four types of globin subunits, A1, A2, B1, and B2. In this study, globin subunit genes were analyzed in the same way as Hox and ParaHox genes using L. satsuma (BAU46563.1 – BAU46566.1, BAN58230.1 – BAN58233.1), L. luymesi,69 and R. pachyptila70 globin genes retrieved from NCBI GenBank as references. We identified 21 globin subunit genes in the L. satsuma genome, all of which contain the globin domain (PF00042). As in previous studies using the L. luymesi, Riftia, and Paraescarpia genomes,7,8,27 we found an expansion of B1-globin subunits. 17 copies of globin genes were placed in the B1-globin clade, while two copies of A1 and a single copy of A2 and B2-globins were identified (Fig. 4A). Fifteen of 21 globin genes were estimated to have been expanded by tandem duplication (Fig. 4A). We also performed molecular phylogenetic analysis using globin genes from three vestimentiferan genomes identified in previous studies.7,8,27 These results supported the hypothesis that expansion of B1-globins occurred at the base of the vestimentiferan lineage (Supplementary Fig. S9A).8 In addition, it suggested parallel gene duplication events after divergence of the Lamellibrachia, Riftia, and Paraescarpia lineages and between L. satsuma and L. luymesi, consistent with a previous study using only two lineages (Supplementary Fig. S9A).7 Gene duplication has contributed to evolutionary acquisition of new gene functions, and species-specific gene duplication can lead to species-specific features.71–73 The complicated evolutionary history of B1-globin genes suggests the importance of gene duplication and diversification in adaptation of each lineage to its habitat. While B1-globins formed nine clades, A1-globins were divided into two clades, supported by bootstrap values of 98 and 99%, respectively. These clades contained one copy of each of the Hb genes of the four vestimentiferans, and the reference sequence of the A1-globin gene of L. satsuma V1 and V2 Hb, respectively (Supplementary Fig. S9A). These results suggest that two A1-globins are used differently, and these clades correspond to A1-globins contained in V1 and V2 Hb.
Figure 4.
Molecular phylogenetic tree, arrangement in the genome, and expression levels of globin, FIH-1, and FIH-1-like genes. (A) Globin subunit genes. The maximum likelihood phylogenetic tree was constructed using aligned and gap-trimmed amino acid sequences. A molecular phylogenetic tree of globin genes identified in gene models of four vestimentiferans is also shown in Supplementary Fig. S9A. For genes retrieved from NCBI GenBank, taxonomy, accession number, and definition are shown. Circles on branches indicate bootstrap values higher than 80%. Arrows indicate transcription directions of the genes. The heat map shows relative gene expression levels (row Z-score transformed). Expression of two B1-globin genes (Lsat_s106.g0008 and Lsat_s260.g0001) was not detected in any of the three regions. (B) FIH-1-like genes. The maximum likelihood phylogenetic tree was constructed using the aligned and gap-trimmed amino acid sequences. For genes retrieved from UniProt/SwissProt database, taxonomy and accession numbers are shown. The heat map shows relative gene expression levels (row Z-score transformed). Lsat, L. satsuma; Lluy, L. luymesi; Rpac, R. pachyptila; Pech, P. echinospica; Drer, Danio rerio; Mmus, Mus musculus; Hsap; Homo sapiens; A1-B2, globin A1-B2 chain (subunit); V1, V2, V1 and V2 haemoglobin; Obt, obturacular region; Ves, vestimental region; Tru, trunk region; **, region-specific DEGs (FDR < 0.01).
Our RNA-seq analysis confirmed the expression of 19 globin genes, except for two B1-globin genes (Fig. 4A and Supplementary Fig. S10). All 19 globin genes showed a tendency to be highly expressed in the trunk region (Fig. 4A and Supplementary Fig. S10). In addition, upregulation of 14 of the 19 globin genes in the trunk region was supported by FDR < 0.01 (Fig. 4A). Functional enrichment analysis also indicated that genes involved in oxygen transport, i.e., globin genes, were enriched among trunk region-specific DEGs (Fig. 2; see also Supplementary Fig. S5). We also compared expression levels of putative heme biosynthesis enzymes between the three regions. All eight heme biosynthesis enzymes were expressed in the trunk region, and four of them tended to be highly expressed compared with other regions, though the difference was not statistically significant (Supplementary Fig. S9B). In vestimentiferans, it has been assumed that Hbs are synthesized in the intravasal (heart) body, located in the dorsal vessel that starts in the anterior vestimental region and continues through the trunk region, into the opisthosoma.74 On the other hand, it was hypothesized based on transcriptomic analyses that the trophosome is the site of haematopoiesis.8 Previous studies have shown that globins are abundant in the plume and trophosome of Riftia at the proteomic level, and that of Paraescarpia at the transcriptomic level.7,61 Our results are consistent with these previous reports, while the exact location of Hb biosynthesis in the trunk region is unclear. Hbs are presumably synthesized in the trophosome or other tissues in the trunk region and transported to other regions, particularly to tissues where oxygen uptake occurs.
Five hundred and five Vestimentifera-specific gene families (Fig. 1C) also included an interesting gene family, hypoxia-inducible factor 1-alpha inhibitor (factor inhibiting HIF-1 alpha; FIH-1)-like genes, which may be involved in control of haemoglobin biosynthesis or oxygen transport in the trophosome. This gene family contained four, three, two, and five genes in L. satsuma, L. luymesi, Riftia, and Paraescarpia, and they showed sequence similarities to mammalian FIH-1. Our molecular phylogenetic analysis suggests that each of the four vestimentiferans possesses one copy of FIH-1 genes, in addition to multi-copy FIH-1-like genes. FIH-1-like genes may have been expanded by parallel gene duplication events after the diversification of Lamellibrachia, Riftia, and Paraescarpia (Fig. 4B). The arrangement of FIH-1-like genes in the L. satsuma genome indicates that they were expanded by tandem duplication (Fig. 4B). All L. satsuma FIH-1-like genes are trunk-region specific DEGs, while the FIH-1 gene tends to be highly expressed in the obturacular region (Fig. 4B and Supplementary Fig. S11). A comparative proteomic analysis revealed that proteins annotated as FIH-1, which may belong to the Vestimentifera-specific FIH-1-like gene family, are detected almost exclusively in the trophosome of Riftia.61 Taken together, FIH-1-like genes may have Vestimentifera-specific and trunk-specific functions.
Hypoxia-inducible factor 1 (HIF-1) is a transcriptional activator that serves in regulation of oxygen homeostasis in metazoans.75,76 HIF-1 mediates adaptive responses to low oxygen levels, including erythropoiesis, angiogenesis, and metabolic reprogramming in mammals. HIF-1 is composed of an oxygen-dependently regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit.75 Reactive oxygen species generated as a result of oxygen deficiency are believed to increase stability of HIF-1α.75 On the other hand, FIH-1 inhibits the transactivation function of HIF-1 under normal oxygen concentrations.77 In vestimentiferan tubeworms, oxygen and sulfide are transported by Hbs into the trophosome, which is highly vascularized and may be the site of haemoglobin biosynthesis. Previous studies revealed that genes involved in antioxidative stress responses are highly expressed in the trophosome, indicating high oxidative stress in this tissue.8,61 Assuming that the Vestimentifera-specific FIH-1-like gene family has a molecular function similar to that of mammalian FIH-1, this family may be involved in regulation of haematopoiesis, angiogenesis, and metabolism in the trophosome in response to oxygen concentration or oxidative stress.
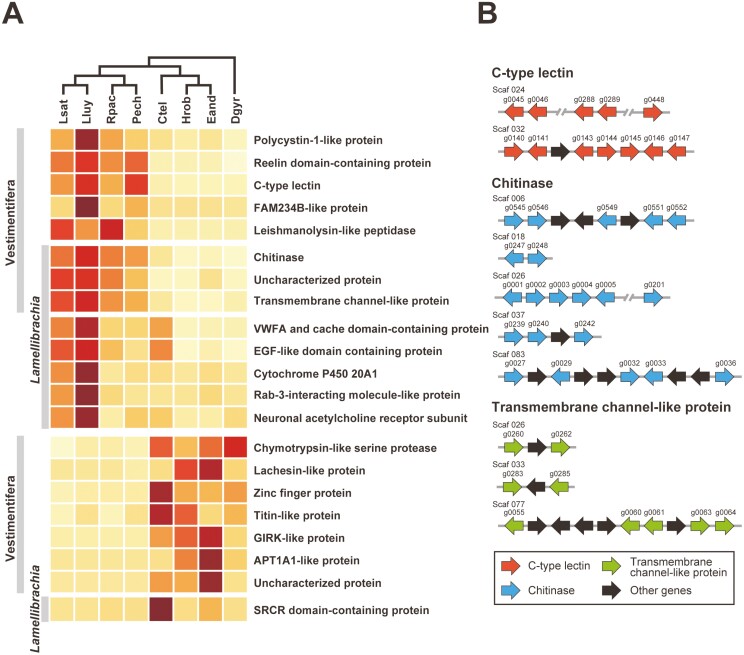
3.5 Lineage-specific expanded and contracted gene families
We identified eight Vestimentifera-specific and eight Lamellibrachia-specific expanded gene family candidates by comparing OGs of four vestimentiferans and four non-vestimentiferan annelids (Fig. 5A). Among them, chitinase, TMC, and an uncharacterized protein family were common to the two (Fig. 5A). Chitinase is a hydrolytic enzyme that degrades glycosidic bonds in chitin. In insects, the exoskeleton, the peritrophic matrix, and gut linings contain chitin as major structural components, and chitinase genes are thought to have been expanded by tandem duplication.78 Previous studies using vestimentiferan genomes also pointed out the importance of the gene repertoire related to chitin metabolism.7,8 Our comparative genomic analysis suggests an expansion of the chitinase family in the Vestimentifera and Lamellibrachia lineages (Fig. 5A and Supplementary Fig. S12A). This gene family contains 26 L. satsuma genes, all of which possess glycosyl hydrolase family 18 domains (PF00704). Some of them are estimated to have been expanded by multiple tandem duplication events (Fig. 5B and Supplementary Fig. S12A). In addition, some genes show tissue-specific expression in different regions (Supplementary Fig. S12B). In insects, chitinase genes show different developmental patterns and tissue specificity, and are believed to be involved in survival, moulting, or development.79 Chitinases in vestimentiferans may also be involved in development or growth by degrading components of chitinous tubes.
Figure 5.
Lineage-specific expanded and contracted gene family candidates and their arrangement in the L. satsuma genome. (A) Gene numbers (row Z-score transformed) of Vestimentifera and Lamellibrachia-specific expanded and contracted gene family candidates. The cladogram on the top is based on phylogenetic relationships inferred from the present study (Supplementary Fig. S1). The top part of the heatmap shows Vestmentifera or Lamellibrachia-specific expanded gene family candidates. The bottom part shows Vestimentifera or Lamellibrachia-specific contracted gene family candidates. (B) Arrangement of genes belonging to the chitinase, TMC, and C-type lectin families in the L. satsuma genome. Arrows indicate transcription directions of genes. Lsat, L. satsuma; Lluy, L. luymesi; Rpac, R. pachyptila; Pech, P. echinospica; Ctel, C. teleta; Hrob, H. robusta; Eand, E. andrei, Dgyr; D. gyrociliatus.
Transmembrane channel-like (TMC) proteins are involved in a wide range of functions in many species, such as chemosensation, hearing, and egg laying.80 The TMC family, specifically expanded in the Vestimentifera and Lamellibrachia, contains 13 L. satsuma genes, 12 of which possess a TMC domain (PF07810) (Fig. 5A and Supplementary Fig. S13A). Some of these genes are tandemly located in three clusters in the L. satsuma genome, suggesting multiple and independent tandem duplication events in the Lamellibrachia lineage (Fig. 5B and Supplementary Fig. S13A). Our RNA-seq analysis revealed that seven and three genes in this family tend to be highly expressed in the obturacular and trunk regions, respectively (Supplementary Fig. S13B), suggesting that they may be involved in chemosensation and transmembrane transport between the plume and surrounding water, or in interactions between host and symbionts.
Comparative genomic analysis revealed expansion of the C-type lectin (CTL) family by tandem duplication events in the Vestimentifera lineage (Fig. 5A and B, and Supplementary Fig. S14A). C-type lectin receptors are a group of PRRs involved in animal innate immune defense, which recognizes specific carbohydrates in a Ca2+-dependent manner. In many invertebrate species, including hydrothermal vent shrimps, corals, sponges, and marine nematodes, lectins are likely involved in symbiont recognition.81–84 Our phylogenetic analysis revealed that this gene family contains 17 L. satsuma genes, 7 of which possess lectin C-type domains (PF00059). In contrast to TLRs, most of which are highly expressed in the obturacular region and downregulated in the trunk region, some genes in the CTL family are highly expressed in the vestimental or trunk region (Supplementary Fig. S14B). These CTLs may participate in pathogen recognition in the trunk region, and more specifically, in the trophosome, where few TLR-expressing innate immune cells reside. Furthermore, these CTLs are possibly involved in acquisition of specific symbiotic bacteria, or in maintenance of symbiotic relationships.
We also identified seven candidate OGs of Vestimentifera-specific contracted gene families and one candidate OG of Lamellibrachia-specific contracted gene family. Candidate OGs of Vestimentifera-specific contracted gene families contained a chymotrypsin-like serine protease family (Fig. 5A). Serine proteases have a wide range of functions such as food digestion, polypeptide metabolism, and immune response.85 Our molecular phylogenetic analysis using four annelids, including two vestimentiferans, indicated that tandem duplication events have occurred in each lineage (Supplementary Fig. S15A). Thus, it is more likely that Vestimentifera lineage experienced slower diversification of the chymotrypsin-like serine protease family compared to other annelids, rather than a contraction of this gene family. This may be due to their unique body structure lacking a digestive tract, or to features of the immune system, discussed above. Titin is a protein involved in contraction of muscle tissues.86 Vestimentiferans exhibit a smaller number of titin-like protein genes than other annelids, which may be possibly due to their sessile lifestyle and nutritional dependence on symbionts. (Fig. 5A, Supplementary Fig. S15B).
4. Conclusions
We reported a high-quality draft genome and tissue-specific transcriptome data of a vestimentiferan tubeworm, Lamellibrachia satsuma. While the L. satsuma genome contains nearly complete Hox and ParaHox clusters, Antp was lost in the common ancestor of vestimentiferans. Our transcriptomic analysis suggests that TLR genes and lineage-specifically expanded lysozyme genes are highly expressed in the obturacular and vestimental regions, whereas innate immune defense is suppressed in the trophosome, which harbours symbiotic bacteria. On the other hand, lineage-specifically expanded C-type lectins, some of which are highly expressed in the trunk region, may be involved in distinguishing symbionts from pathogens, or in host-symbiont interactions. Globin subunits are highly expressed in the trunk region, supporting the hypothesis that the trophosome has a haematopoietic function. FIH-1-like gene family, a Vestimentifera-specific gene family first identified in this study, may have functions related to regulation of haematopoiesis, angiogenesis, and metabolism in the trophosome. In addition to C-type lectins, we also identified Vestimentifera or Lamellibrachia-specific expanded genes such as chitinases and ion channels, suggesting that these genes may contribute to establishment of unique biological characters of tubeworms, such as symbioses with bacteria and tube development. Our study illuminates not only the molecular and evolutionary bases of the unique vestimentiferan lifestyle, but also how animals adapt to deep-sea chemosynthetic ecosystems.
Supplementary Material
Acknowledgements
We thank Prof. Hiroshi Miyake (Kitasato University) for helping dissect Lamellibrachia satsuma tubeworms and for tissue identification. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.FundingThis study was supported in part by JSPS KAKENHI grants (20H03235 and 20K21860 for CS). Conflict of interestsThe authors declare that they have no conflict of interest.
Contributor Information
Taiga Uchida, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba, Japan.
Yuki Yoshioka, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba, Japan; Marine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, Japan.
Yu Yoshida, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba, Japan.
Manabu Fujie, DNA Sequencing Section (SQC), Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, Japan.
Ayuta Yamaki, Kagoshima City Aquarium, Kagoshima, Japan; Enoshima Aquarium, Fujisawa, Kanagawa, Japan.
Akira Sasaki, Kagoshima City Aquarium, Kagoshima, Japan.
Koji Inoue, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba, Japan.
Chuya Shinzato, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba, Japan.
Author contributions
CS and AY conceptualized genome sequencing of Lamellibrachia satsuma, and CS and KI commenced the genome project. AY and AS performed animal collection, and CS and KI performed sample collection. MF prepared sequencing libraries and produced sequencing data. CS assembled the genome, and YYoshioka and YYoshida performed gene prediction. TU performed comparative genomics and transcriptome analyses with support from YYoshioka. TU wrote the main manuscript and CS supervised the project. All authors reviewed and approved the final version of the manuscript.
Data availability
All ONT and illumina reads are available under DRA accession number DRA015634 with the BioProject accession ID PRJDB14199 and BioSample accession IDs SAMD00529701-SAMD00529710. Genome assemblies are available from accession numbers BSQZ01000001-BSQZ01000306.
References
- 1. Dubilier, N., Bergin, C., and Lott, C.. 2008, Symbiotic diversity in marine animals: the art of harnessing chemosynthesis, Nat. Rev. Microbiol., 6, 725–40. [DOI] [PubMed] [Google Scholar]
- 2. Hilário, A., Capa, M., Dahlgren, T.G., et al. 2011, New Perspectives on the Ecology and Evolution of Siboglinid Tubeworms, PLoS One, 6, e16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavanaugh, C. M., McKiness, Z. P., Newton, I. L. G., and Stewart, F. J.. 2006, Marine chemosynthetic symbioses. The Prokaryotes. Springer New York: New York, NY, pp. 475–507. [Google Scholar]
- 4. Nussbaumer, A.D., Fisher, C.R., and Bright, M.. 2006, Horizontal endosymbiont transmission in hydrothermal vent tubeworms, Nature, 441, 345–8. [DOI] [PubMed] [Google Scholar]
- 5. Patra, A.K., Cho, H.H., Kwon, Y.M., et al. 2016, Phylogenetic relationship between symbionts of tubeworm Lamellibrachia satsuma and the sediment microbial community in Kagoshima Bay, Ocean Sci. J., 51, 317–32. [Google Scholar]
- 6. Simakov, O., Marletaz, F., Cho, S.-J., et al. 2013, Insights into bilaterian evolution from three spiralian genomes, Nature, 493, 526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun, Y., Sun, J., Yang, Y., et al. 2021, Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica, Mol. Biol. Evol., 38, 4116–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Oliveira, A.L, Mitchell, J., Girguis, P., and Bright, M.. 2022, novel insights on obligate symbiont lifestyle and adaptation to chemosynthetic environment as revealed by the giant tubeworm genome, Mol. Biol. Evol., 39, msab347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Southward, E.C., Schulze, A., and Gardiner, S.L.. 2005, Pogonophora (Annelida): form and function, Hydrobiologia, 535–536, 227–51. [Google Scholar]
- 10. Minic, Z. and Hervé, G.. 2004, Biochemical and enzymological aspects of the symbiosis between the deep-sea tubeworm Riftia pachyptila and its bacterial endosymbiont, Eur. J. Biochem., 271, 3093–102. [DOI] [PubMed] [Google Scholar]
- 11. Schulze, A. 2003, Phylogeny of Vestimentifera (Siboglinidae, Annelida) inferred from morphology, Zool. Scr., 32, 321–42. [Google Scholar]
- 12. Jones, M.L. 1981, Riftia pachyptila Jones: observations on the vestimentiferan worm from the galápagos rift, Science, 213, 333–6. [DOI] [PubMed] [Google Scholar]
- 13. Cavanaugh, C.M., Gardiner, S.L., Jones, M.L., Jannasch, H.W., and Waterbury, J.B.. 1981, Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts, Science, 213, 340–2. [DOI] [PubMed] [Google Scholar]
- 14. Felbeck, H. 1981, Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera), Science, 213, 336–8. [DOI] [PubMed] [Google Scholar]
- 15. Hand, S.C. 1987, Trophosome ultrastructure and the characterization of isolated bacteriocytes from invertebrate-sulfur bacteria symbioses, Biol. Bull., 173, 260–76. [DOI] [PubMed] [Google Scholar]
- 16. Freytag, J.K., Girguis, P.R., Bergquist, D.C, Andras, J.P., Childress, J.J., and Fisher, C.R.. 2001, A paradox resolved: sulfide acquisition by roots of seep tubeworms sustains net chemoautotrophy, Proc. Natl. Acad. Sci., 98, 13408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Julian, D., Gaill, F., Wood, E., Arp, A.J., and Fisher, C.R.. 1999, Roots as a site of hydrogen sulfide uptake in the hydrocarbon seep vestimentiferan Lamellibrachia sp, J. Exp. Biol., 202, 2245–57. [DOI] [PubMed] [Google Scholar]
- 18. Zal, F., Suzuki, T., Kawasaki, Y., Childress, J.J., Lallier, F.H., and Toulmond, A.. 1997, Primary structure of the common polypeptide chainb from the multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila: an insight on the sulfide binding-site, Proteins, 29, 562–74. [PubMed] [Google Scholar]
- 19. Zal, F., Leize, E., Lallier, F.H., Toulmond, A., Van Dorsselaer, A., and Childress, J.J.. 1998, S-Sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins, Proc. Natl. Acad. Sci., 95, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powell, M.A. and Somero, G.N.. 1983, Blood components prevent sulfide poisoning of respiration of the hydrothermal vent tube worm Riftia pachyptila, Science, 219, 297–9. [DOI] [PubMed] [Google Scholar]
- 21. Miura, T., Tsukahara, J., and Hashimoto, J.. 1997, Lamellibrachia satsuma, a new species of Vestimentiferan worms (Annelida: Pogonophora) from a shallow hydrothermal vent in Kagoshima Bay, Japan, Proc. Biol. Soc. Wash., 110, 447–56. [Google Scholar]
- 22. Hashimoto, J., Miura, T., Fujikura, K., and Ossaka, J.. 1993, Discovery of vestimentiferan tubeworms in the euphotic zone, Zool. Sci., 10, 1063–7. [Google Scholar]
- 23. Miyake, H., Tsukahara, J., Hashimoto, J., Uematsu, K., and Maruyama, T.. 2006, Rearing and observation methods of vestimentiferan tubeworm and its early development at atmospheric pressure, Cah. Biol. Mar., 47, 471–5. [Google Scholar]
- 24. Shinozaki, A., Kawato, M., Noda, C., et al. 2010, Reproduction of the vestimentiferan tubeworm Lamellibrachia satsuma inhabiting a whale vertebra in an aquarium, Cah. Biol. Mar., 51, 467–73. [Google Scholar]
- 25. Patra, A.K., Kwon, Y.M., and Yang, Y.. 2022, Complete gammaproteobacterial endosymbiont genome assembly from a seep tubeworm Lamellibrachia satsuma, J. Microbiol., 60, 916–27. [DOI] [PubMed] [Google Scholar]
- 26. Patra, A.K., Kwon, Y.M., Kang, S.G., Fujiwara, Y., and Kim, S.-J.. 2016, The complete mitochondrial genome sequence of the tubeworm Lamellibrachia satsuma and structural conservation in the mitochondrial genome control regions of Order Sabellida, Mar. Genomics, 26, 63–71. [DOI] [PubMed] [Google Scholar]
- 27. Li, Y., Tassia, M.G., Waits, D.S., Bogantes, V.E., David, K.T., and Halanych, K.M.. 2019, Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi, BMC Biol., 17, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen, Y., Nie, F., Xie, S.-Q., et al. 2021, Efficient assembly of nanopore reads via highly accurate and intact error correction, Nat. Commun., 12, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roach, M.J., Schmidt, S.A., and Borneman, A.R.. 2018, Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies, BMC Bioinf., 19, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang, S., Kang, M., and Xu, A.. 2017, HaploMerger2: rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly, Bioinformatics, 33, 2577–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warren, R.L., Yang, C., Vandervalk, B.P., et al. 2015, LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads, GigaScience, 4, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kundu, R., Casey, J., and Sung, W.-K.. 2019, HyPo: super fast & accurate polisher for long read genome assemblies, bioRxiv, 2019.12.19.882506. [Google Scholar]
- 33. Manni, M., Berkeley, M.R., Seppey, M., Simão, F.A., and Zdobnov, E.M.. 2021, BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes, Mol. Biol. Evol., 38, 4647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin, M. 2011, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal, 17, 10–2. [Google Scholar]
- 35. Price, A.L., Jones, N.C., and Pevzner, P.A.. 2005, De novo identification of repeat families in large genomes, Bioinformatics, 21, i351–8. [DOI] [PubMed] [Google Scholar]
- 36. Brůna, T., Hoff, K.J., Lomsadze, A., Stanke, M., and Borodovsky, M.. 2021, BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database, NAR Genom. Bioinform., 3, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshioka, Y., Suzuki, G., Zayasu, Y., Yamashita, H., and Shinzato, C.. 2022, Comparative genomics highlight the importance of lineage-specific gene families in evolutionary divergence of the coral genus, Montipora, BMC Ecol. Evol., 22, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim, D., Langmead, B., and Salzberg, S.L.. 2015, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods, 12, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pertea, M., Pertea, G.M., Antonescu, C.M., Chang, T.-C., Mendell, J.T., and Salzberg, S.L.. 2015, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nat. Biotechnol., 33, 290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pertea, G. and Pertea, M.. 2020, GFF utilities: GffRead and GffCompare, F1000Res., 9, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bateman, A., Martin, M.-J., Orchard, S., et al. 2021, UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res., 49, D480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones, P., Binns, D., Chang, H.-Y., et al. 2014, InterProScan 5: genome-scale protein function classification, Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hallgren, J., Tsirigos, K.D., Damgaard Pedersen, M., et al. 2022, DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks, bioRxiv 2022.04.08.487609. [Google Scholar]
- 44. Emms, D.M. and Kelly, S.. 2019, OrthoFinder: phylogenetic orthology inference for comparative genomics, Genome Biol., 20, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katoh, K. and Standley, D.M.. 2013, MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol., 30, 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Capella-Gutiérrez, S., Silla-Martínez, J.M., and Gabaldón, T.. 2009, trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses, Bioinformatics, 25, 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stamatakis, A. 2014, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics, 30, 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. R Core Team. 2022, R: A language and environment for statistical computing. R Found. Stat. Comput: Vienna, Austria. [Google Scholar]
- 49. Vasimuddin, M., Misra, S., Li, H., and Aluru, S.. 2019, Efficient architecture-aware acceleration of BWA-MEM for multicore systems. 2019 IEEE Int. Parallel Distrib. Process. Symp., Rio de Janeiro, Brazil. 314–24. [Google Scholar]
- 50. Patro, R., Duggal, G., Love, M.I, Irizarry, R.A., and Kingsford, C.. 2017, Salmon provides fast and bias-aware quantification of transcript expression, Nat. Methods, 14, 417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson, M.D., McCarthy, D.J., and Smyth, G.K.. 2010, edgeR: a bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCarthy, D.J., Chen, Y., and Smyth, G.K.. 2012, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation, Nucleic Acids Res., 40, 4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gu, Z., Eils, R., and Schlesner, M.. 2016, Complex heatmaps reveal patterns and correlations in multidimensional genomic data, Bioinformatics, 32, 2847–9. [DOI] [PubMed] [Google Scholar]
- 54. Sherman, B.T., Hao, M., Qiu, J., et al. 2022, DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update), Nucleic Acids Res., 50, W216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang, D.W., Sherman, B.T., and Lempicki, R.A.. 2009, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 56. Sun, Y., Liang, Q., Sun, J., et al. 2018, The mitochondrial genome of the deep-sea tubeworm Paraescarpia echinospica (Siboglinidae, Annelida) and its phylogenetic implications, Mitochondrial DNA B Resour., 3, 131–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Biscotti, M.A., Canapa, A., Forconi, M., and Barucca, M.. 2014, Hox and ParaHox genes: a review on molluscs, Genesis, 52, 935–45. [DOI] [PubMed] [Google Scholar]
- 58. Brooke, N.M., Garcia-Fernàndez, J., and Holland, P.W.H.. 1998, The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster, Nature, 392, 920–2. [DOI] [PubMed] [Google Scholar]
- 59. Tsai, I.J., Zarowiecki, M., Holroyd, N., et al. ; Taenia solium Genome Consortium. 2013, The genomes of four tapeworm species reveal adaptations to parasitism, Nature, 496, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Appelqvist, H., Wäster, P., Kågedal, K., and Öllinger, K.. 2013, The lysosome: From waste bag to potential therapeutic target, J. Mol. Cell. Biol., 5, 214–26. [DOI] [PubMed] [Google Scholar]
- 61. Hinzke, T., Kleiner, M., Breusing, C., et al. 2019, Host-Microbe Interactions in the Chemosynthetic Riftia pachyptila Symbiosis, Mbio, 10, e0224–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zavalova, L., Lukyanov, S., Baskova, I., et al. 1996, Genes from the medicinal leech (Hirudo medicinalis) coding for unusual enzymes that specifically cleave endo-ɛ(γ-Glu)-Lys isopeptide bonds and help to dissolve blood clots., Mol. Gen. Genet., 253, 20–5. [DOI] [PubMed] [Google Scholar]
- 63. Ren, Q., Qi, Y.-L., Hui, K.-M., Zhang, Z., Zhang, C.-Y., and Wang, W.. 2012, Four invertebrate-type lysozyme genes from triangle-shell pearl mussel (Hyriopsis cumingii), Fish Shellfish Immunol., 33, 909–15. [DOI] [PubMed] [Google Scholar]
- 64. Kumar, H., Kawai, T., and Akira, S.. 2011, Pathogen Recognition by the Innate Immune System, Int. Rev. Immunol., 30, 16–34. [DOI] [PubMed] [Google Scholar]
- 65. Janeway, C.A. and Medzhitov, R.. 2002, Innate Immune Recognition, Annu. Rev. Immunol., 20, 197–216. [DOI] [PubMed] [Google Scholar]
- 66. Kawai, T. and Akira, S.. 2010, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors, Nat. Immunol., 11, 373–84. [DOI] [PubMed] [Google Scholar]
- 67. Yang, Y., Sun, J., Sun, Y., et al. 2020, Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts, ISME J., 14, 135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zal, F., Lallier, F.H., Wall, J.S., Vinogradov, S.N., and Toulmond, A.. 1996, The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila, J. Biol. Chem., 271, 8869–74. [DOI] [PubMed] [Google Scholar]
- 69. Waits, D.S., Santos, S.R., Thornhill, D.J., Li, Y., and Halanych, K.M.. 2016, evolution of sulfur binding by hemoglobin in Siboglinidae (Annelida) with special reference to bone-eating worms, Osedax, J. Mol. Evol., 82, 219–29. [DOI] [PubMed] [Google Scholar]
- 70. Bailly, X., Jollivet, D., Vanin, S., et al. 2002, Evolution of the sulfide-binding function within the globin multigenic family of the deep-sea hydrothermal vent tubeworm Riftia pachyptila, Mol. Biol. Evol., 19, 1421–33. [DOI] [PubMed] [Google Scholar]
- 71. Ohno, S. 1970, Evolution by Gene Duplication. Springer Berlin Heidelberg: Berlin, Heidelberg. [Google Scholar]
- 72. Zhang, J. 2003, Evolution by gene duplication: an update, Trends Ecol. Evol., 18, 292–8. [Google Scholar]
- 73. Conant, G.C. and Wolfe, K.H.. 2008, Turning a hobby into a job: How duplicated genes find new functions, Nat. Rev. Genet., 9, 938–50. [DOI] [PubMed] [Google Scholar]
- 74. Schulze, A. 2002, Histological and ultrastructural characterization of the intravasal body in Vestimentifera (Siboglinidae, Polychaeta, Annelida), Cah. Biol. Mar., 43, 355–8. [Google Scholar]
- 75. Semenza, G.L. 2009, Regulation of oxygen homeostasis by hypoxia-inducible factor 1, Physiology, 24, 97–106. [DOI] [PubMed] [Google Scholar]
- 76. Wang, L., Cui, S., Ma, L., Kong, L., and Geng, X.. 2015, Current advances in the novel functions of hypoxia-inducible factor and prolyl hydroxylase in invertebrates, Insect. Mol. Biol., 24, 634–48. [DOI] [PubMed] [Google Scholar]
- 77. Semenza, G.L. 2003, Targeting HIF-1 for cancer therapy, Nat. Rev. Cancer, 3, 721–32. [DOI] [PubMed] [Google Scholar]
- 78. Dixit, R., Arakane, Y., Specht, C.A., et al. 2008, Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects, Insect Biochem. Mol. Biol., 38, 440–51. [DOI] [PubMed] [Google Scholar]
- 79. Arakane, Y. and Muthukrishnan, S.. 2010, Insect chitinase and chitinase-like proteins, Cell. Mol. Life Sci., 67, 201–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yue, X., Sheng, Y., Kang, L., and Xiao, R.. 2019, Distinct functions of TMC channels: a comparative overview, Cell. Mol. Life Sci., 76, 4221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu, X.-L., Ye, S., Cheng, C.-Y., et al. 2019, Identification and characterization of a symbiotic agglutination-related C-type lectin from the hydrothermal vent shrimp Rimicaris exoculata, Fish Shellfish Immunol., 92, 1–10. [DOI] [PubMed] [Google Scholar]
- 82. Weis, V.M. 2019, Cell biology of coral symbiosis: foundational study can inform solutions to the coral reef crisis, Integr. Comp. Biol., 59, 845–55. [DOI] [PubMed] [Google Scholar]
- 83. Müller, W.E., Zahn, R.K., Kurelec, B., Lucu, C., Müller, I., and Uhlenbruck, G.. 1981, Lectin, a possible basis for symbiosis between bacteria and sponges, J. Bacteriol., 145, 548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bulgheresi, S., Schabussova, I., Chen, T., Mullin, N.P., Maizels, R.M., and Ott, J.A.. 2006, A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode, Appl. Environ. Microbiol., 72, 2950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Di Cera, E. 2009, Serine proteases, IUBMB Life, 61, 510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tskhovrebova, L. and Trinick, J.. 2003, Titin: properties and family relationships, Nat. Rev. Mol. Cell Biol., 4, 679–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ONT and illumina reads are available under DRA accession number DRA015634 with the BioProject accession ID PRJDB14199 and BioSample accession IDs SAMD00529701-SAMD00529710. Genome assemblies are available from accession numbers BSQZ01000001-BSQZ01000306.