Promiscuous recognition of MR1 by canonical MAIT TCRs endowed with dual reactivity to both microbial and self-antigens enables MAIT cell responses in the absence of microbial infection.
Abstract
Mucosal-associated invariant T (MAIT) cells use canonical semi-invariant T cell receptors (TCR) to recognize microbial riboflavin precursors displayed by the antigen-presenting molecule MR1. The extent of MAIT TCR crossreactivity toward physiological, microbially unrelated antigens remains underexplored. We describe MAIT TCRs endowed with MR1-dependent reactivity to tumor and healthy cells in the absence of microbial metabolites. MAIT cells bearing TCRs crossreactive toward self are rare but commonly found within healthy donors and display T-helper-like functions in vitro. Experiments with MR1-tetramers loaded with distinct ligands revealed significant crossreactivity among MAIT TCRs both ex vivo and upon in vitro expansion. A canonical MAIT TCR was selected on the basis of extremely promiscuous MR1 recognition. Structural and molecular dynamic analyses associated promiscuity to unique TCRβ-chain features that were enriched within self-reactive MAIT cells of healthy individuals. Thus, self-reactive recognition of MR1 represents a functionally relevant indication of MAIT TCR crossreactivity, suggesting a potentially broader role of MAIT cells in immune homeostasis and diseases, beyond microbial immunosurveillance.
Introduction
MR1 is a non-polymorphic MHC class I–like molecule that presents small metabolites to T cells. In humans, the MR1-restricted T cell pool consists of two populations, mucosal-associated invariant T (MAIT) cells (Porcelli et al., 1993; Treiner et al., 2003) and MR1T cells (Harriff et al., 2018; Lepore et al., 2017), which significantly differ in antigen (Ag) specificity and TCR repertoire. While MR1T cells react to not yet identified self and potential tumor-associated Ags (Harriff et al., 2018; Lepore et al., 2017) and display polyclonal TCRs, MAIT cells recognize microbial metabolites and are uniquely defined by an almost invariant TCRα chain paired with a restricted TCRβ chain repertoire (Lepore et al., 2014; Porcelli et al., 1993; Tilloy et al., 1999). The canonical MAIT TCRα is made by the TRAV1-2 variable gene rearranged with either the TRAJ33, TRAJ12, or TRAJ20 gene segments and with remarkably limited junctional variability (Lepore et al., 2014; Porcelli et al., 1993; Tilloy et al., 1999). Other MAIT cell defining features include high expression of CD161, CD26 (Sharma et al., 2015), and IL-18Rα; expression of the transcription factor PLZF; and reactivity to microbial Ags (Le Bourhis et al., 2010), although very rare CD161− MAIT cells have also been described (Koay et al., 2019). Microbial Ags include the potent pyrimidine agonists 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU) and 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU), intermediates of riboflavin biosynthesis that convert to 6,7-dimethyl-8-ribityllumazine (RL-6,7-diMe) and 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH), which also stimulate MAIT cells, although with lower potency. 5-OE-RU and 5-OP-RU are captured by MR1 via a Schiff base with MR1 residue K43, whereas the RL-6,7-diMe and RL-6-Me-7-OH metabolites do not form a Schiff base (Corbett et al., 2014; Kjer-Nielsen et al., 2012). Thus, MR1 binds distinct microbial Ags that show different stimulatory capacities (Schmaler et al., 2018).
Several MAIT TCR-MR1 crystal structures demonstrated a highly conserved binding mode that is similar to the conformation adopted by canonical HLA class I– and HLA class II–restricted TCRs (Awad et al., 2020; Corbett et al., 2014; Eckle et al., 2014; Patel et al., 2013). This binding mode positions the invariant MAIT TCRα orthogonally above the Ag, establishing a conserved and essential hydrogen bond with the ribityl moiety of the riboflavin-related agonists via the residue Y95α (encoded by the TRAJ gene segments; Corbett et al., 2014). Despite the conserved mode of MR1 binding of the invariant TCR Vα, the TCRβ chain has an important role in influencing Ag recognition and magnitude of the T cell activation (Eckle et al., 2014; Gherardin et al., 2016).
Although MR1 is ubiquitously expressed, the majority of MR1 molecules remain unfolded within the ER until an abundance of Ags is available for loading (McWilliam et al., 2016). MR1 stabilization is achieved through neutralization of K43 within the Ag binding pocket, allowing egress to the surface. However, small quantities of MR1 are commonly expressed on the cell surface, consistent with the recognition of different cell types such as dendritic cells, epithelial cells, and cancer cells by MR1T cell clones that do not react to microbial Ags (Lepore et al., 2017). This implies a wider role for MR1-restricted T cells in immunity (Crowther et al., 2020; Lepore et al., 2017). Recently, additional MAIT cell agonists beyond 5-OP-RU have been identified (Harriff et al., 2018; Keller et al., 2017) and MAIT cells can distinguish between different Ags through their TCR Vβ-chain, indicating the MAIT TCR repertoire may be shaped by available Ags (Gherardin et al., 2016; Gold et al., 2014; Howson et al., 2018; Lopez-Sagaseta et al., 2013).
The reactivity of MAIT cells toward self has not been systematically investigated since initial experiments, in which mouse MAIT cells were stimulated by uninfected fibroblasts and B cells (Huang et al., 2008). Recent studies extended these observations to humans using MR1 tetramers and structural analyses and suggested that MAIT TCRs can be crossreactive toward other MR1 ligands, including some common drugs and potential self-Ags (Gherardin et al., 2016; Huang et al., 2008; Keller et al., 2017; Young et al., 2013). Further reports on MAIT cells have indicated that while bacterial colonization of the gut is required for their thymic selection and peripheral expansion, MAIT cells are still present in the periphery of germ-free mice, albeit at a much lower frequency (Koay et al., 2019; Legoux et al., 2019), implying microbial-independent thymic selection. Consistently, MAIT cell alterations in inflammatory diseases with no direct microbial etiology, such as diabetes, multiple sclerosis, and obesity, have been reported (Croxford et al., 2006; Miyazaki et al., 2011; Rouxel et al., 2018; Toubal et al., 2020). Furthermore, activation of MAIT cells upon challenge with SARS-CoV-2 infected macrophages has been observed (Flament et al., 2021). Overall, these data suggest that some MAIT cells could become activated in a microbial-independent manner.
Here, we set out to define the extent and mechanisms of MR1-mediated MAIT cell self-reactivity. We demonstrate that microbial-independent MR1 reactivity of a canonical MAIT cell subset is commonly observed in healthy individuals. Furthermore, we describe a novel mode of promiscuous MR1 recognition by a MAIT TCR, associated with a CDR3β-motif enriched in circulating self-reactive MAIT cells.
Results
A subset of MAIT cells responds in vitro and ex vivo to stimulation by MR1-overexpressing tumor cells
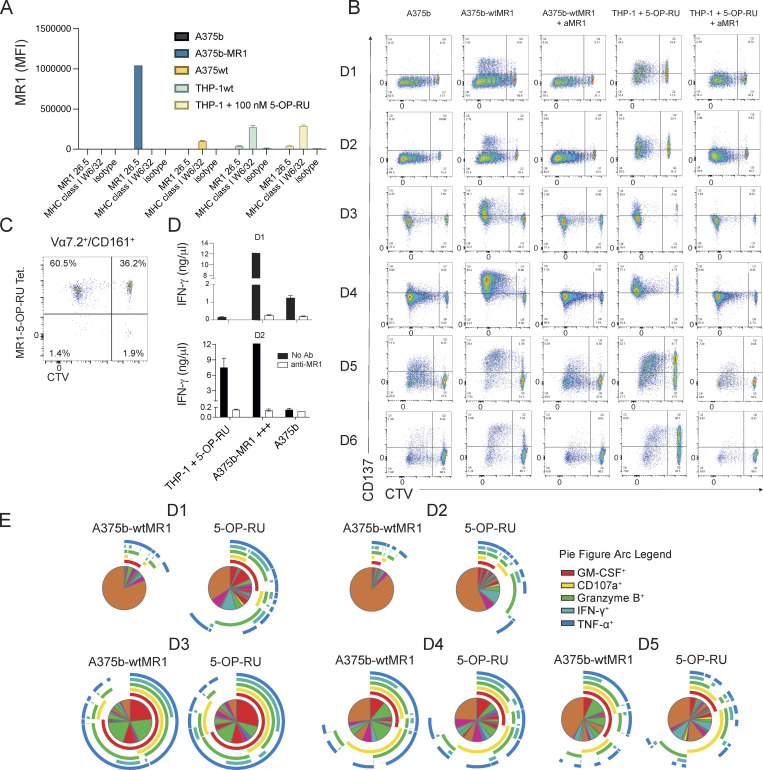
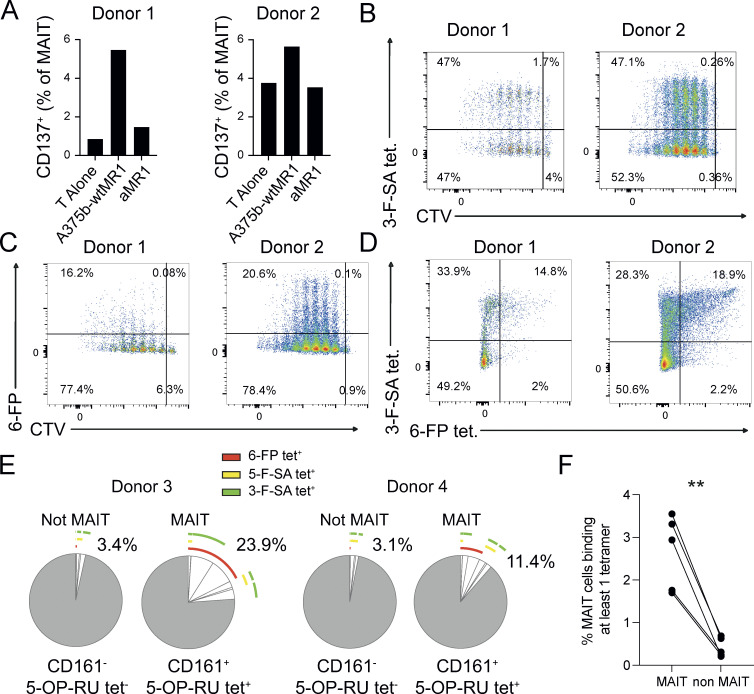
We asked whether canonical MAIT cells that recognize microbial metabolites could also respond to MR1-mediated stimulation in the absence of microbial ligands. Purified, circulating Vα7.2+ T cells from healthy donors were cultured with β2-microglobulin (B2M)-deficient A375 melanoma cells (A375b) engineered to display high levels of surface wild-type MR1 (A375b-wtMR1; Fig. S1 A). We selected MR1-overexpressing A375 cells as APCs because of their capacity to broadly stimulate MAIT and MR1T cells through an efficient presentation of both microbial and self-Ags, respectively (Lepore et al., 2017). Freshly isolated T cells were stimulated with A375b-wtMR1 cells, and proliferative responses as well as MR1-dependent activation upon rechallenge were analyzed. Most of the proliferating Vα7.2+/CD161+ MAIT cells upregulated the activation marker CD137 following rechallenge with A375b-wtMR1 cells, contrary to non-proliferating cells that remained unresponsive (Fig. 1, A and B; and Fig. S1 B). Activation was prevented by anti-MR1 mAb and did not occur when the cultures were rechallenged with A375b cells that lack wtMR1 on their surface (Fig. 1, A and B; and Fig. S1 B). Both proliferating and non-proliferating MAIT cells recognized 5-OP-RU–pulsed THP-1 cells in an MR1-dependent fashion (Fig. 1 A and Fig. S1 B) and bound 5-OP-RU–loaded MR1 tetramers (Fig. S1 C). Of note, a proportion of proliferating non-MAIT cells (defined as Vα7.2+ and CD161−) exhibited an MR1-dependent response to A375b-wtMR1 cells (Fig. 1 A), representing MR1T cells that do not recognize microbial Ags and express distinct TCRs as previously reported (Lepore et al., 2017).
Figure S1.
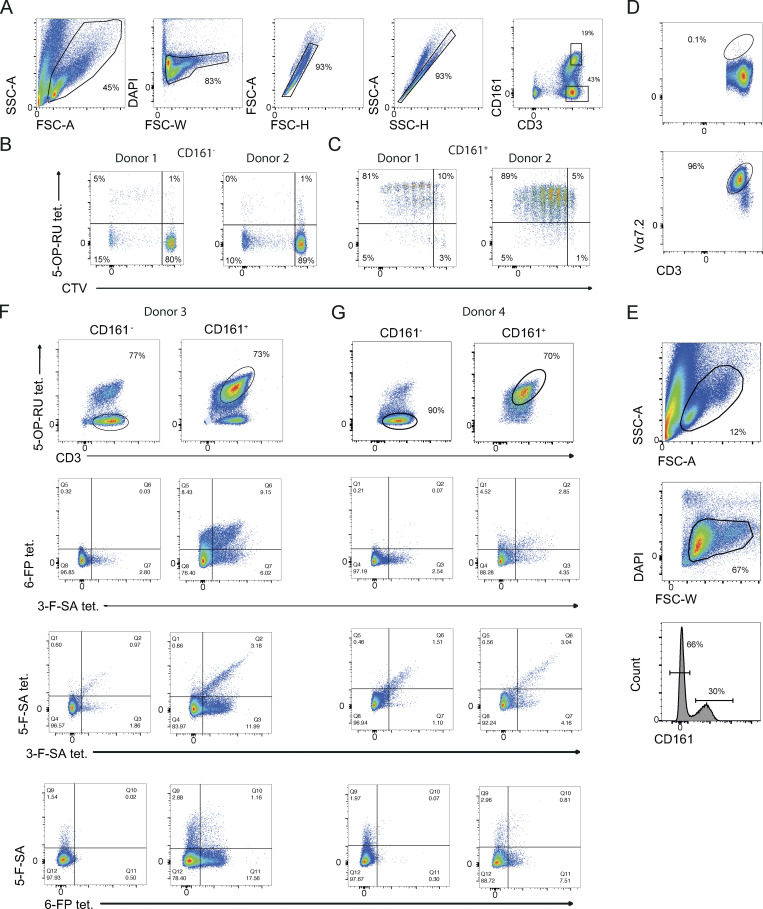
Reactivity and function of self-reactive MAIT cell lines. (A) Expression of MR1 on cell lines used in this study. In the condition with 5-OP-RU, a 6-h incubation time was used. (B) Activation of self-reactive MAIT cells after proliferation with the indicated conditions from six individual donors. (C) Percentage of MR1-5-OP-RU tetramer+ cells within MAIT cells proliferating (CTV dull) and not (CTV bright) after stimulation with A375b-MR1 cells. (D) IFN-γ release by MAIT cells in the cultures illustrated in A (black bars) + aMR1 mAb (white bars). Concentrations are expressed as mean +SD. Data obtained from two donors. (E) Frequency of MAIT cells that secrete combination of the indicated cytokines.
Figure 1.
Self-reactivity and polyfunctionality of circulating MAIT cells from healthy donors. (A) CD137 expression by autoreactive MAIT cells expanded for 10 d. Proliferating and not proliferating MAIT (top row) or non-MAIT (bottom row) cells following stimulation with the indicated APCs ± anti-MR1 mAb (aMR1). MAIT cells (Vα7.2+/CD161+) proliferative status is revealed by CTV emission. Plots are representative of results obtained with six donors. (B) Summary of MAIT cell CD137 expression on proliferating cells (CTV dull) after rechallenge with the indicated condition (numbers as in panel A). Data were obtained from six donors. Statistical significance was determined using a one-way ANOVA with Friedman test, * P ≤ 0.05. (C) Effect of aMR1 mAb on surface expression of the indicated activation markers on CTV-dull MAIT cells stimulated with 5-OP-RU–pulsed THP-1 cells (top row) or with A375b-wtMR1 cells without exogenous antigens (bottom row). Median fluorescence intensity (MFI) is indicated ± aMR1 mAb. Data obtained from five donors. Statistical significance was determined using Student’s t test, * P ≤ 0.05, ** P ≤ 0.01. (D) Vα7.2 surface expression on MAIT cells stimulated with 5-OP-RU–pulsed THP-1 cells (top row) or with A375b-wtMR1 cells without exogenous antigens (bottom row). MFI is indicated ± aMR1 mAb. Data obtained from five donors. Statistical significance was determined using Student’s t test, * P ≤ 0.05. (E) Percentage of ex vivo MAIT cells from healthy donors double positive for CD137 and CD69 after overnight co-culture with A375b-MR1 cells ± aMR1 mAb. Stimulation with 5-OP-RU was used as positive control with the scale on the right-hand y-axis (green). Cells were pregated as CD3+/CD26+Vα7.2+/CD161+. Data are a summary of all five donors tested. Statistical significance was determined using Student’s t test, * P ≤ 0.05. (F and G) Average frequency of cells expressing one or more of the indicated activation-associated molecules within self-reactive MAIT cell lines stimulated with (F) A375b-wtMR1 cells or (G) 5-OP-RU–loaded THP-1 cells. Pie segments indicate cells positive for any combination of the indicated cytokines or activation markers. Pie arcs indicate the cytokine positivity of each segment. Data is averaged from five donors. Source data are available for this figure: SourceData F1.
The dual reactivity of proliferating Vα7.2+/CD161+ MAIT cells in the presence and absence of microbial-Ags was further confirmed by the detection of MR1-dependent IFN-γ release (Fig. S1 D) and upregulation of additional T cell activation markers in response to both 5-OP-RU–pulsed THP-1 and A375b-wtMR1 cells (Fig. 1 C). Notably, both stimuli also induced an MR1-dependent decrease of TCR surface levels, as detected with anti-Vα7.2 mAb staining, suggestive of activation-induced TCR downregulation (Fig. 1 D).
We next asked whether self-reactive MAIT cells could also be identified ex vivo following short-term stimulation. A small but detectable fraction of freshly purified MAIT cells upregulated CD137 and CD69 after overnight culture with A375b-MR1 cells (mean 1.1%, Fig. 1 E) in the absence of microbial Ags. As expected, a large majority of MAIT cells responded instead to 5-OP-RU (Fig. 1 E). Anti-MR1 blocking mAb prevented MAIT cell activation in the control culture (mean 0.1%). Remarkably, ex vivo self-reactive MR1-restricted responses were consistently detected in circulating MAIT cells from all the seven donors investigated.
Finally, we compared the functional response of in vitro expanded self-reactive MAIT cells to either 5-OP-RU or A375b-wtMR1 cells and did not observe major qualitative differences between the two stimuli (Fig. 1, F and G; and Fig. S1 E).
Taken together, these data indicated that MAIT cells endowed with dual reactivity to microbial Ags and cell-endogenous MR1–Ag complexes are present in circulating T cells of healthy donors as they expand in vitro and are detectable ex vivo following co-culture with MR1-overexpressing tumor cells. Furthermore, microbial and cell-endogenous stimulation elicited a comparable effector program within in vitro–expanded self-reactive MAIT cells.
Self-reactive MAIT cells can recognize physiological levels of MR1 in healthy cells and display T-helper-like functions in vitro
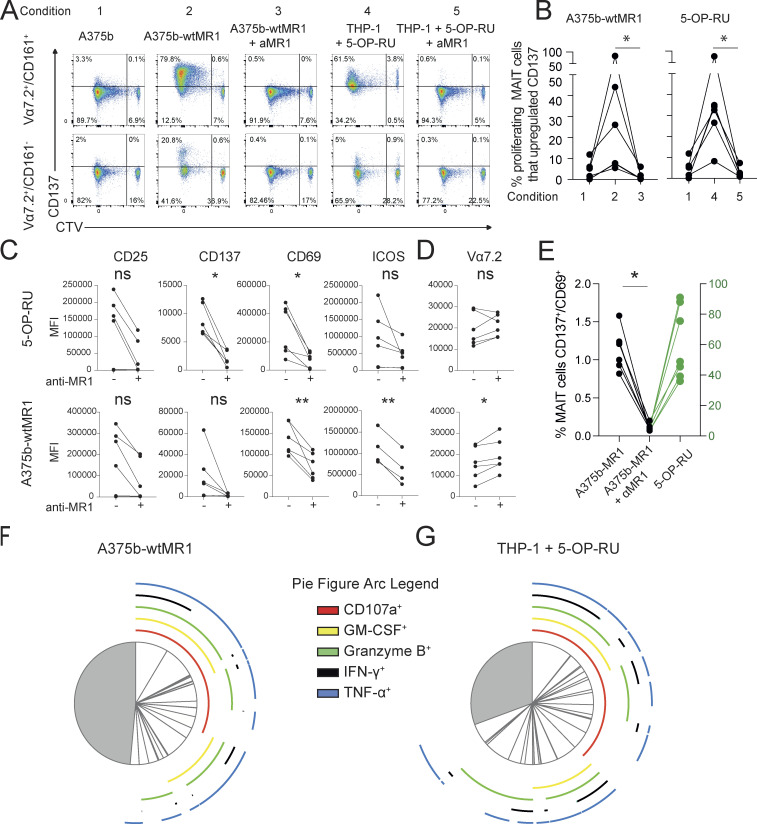
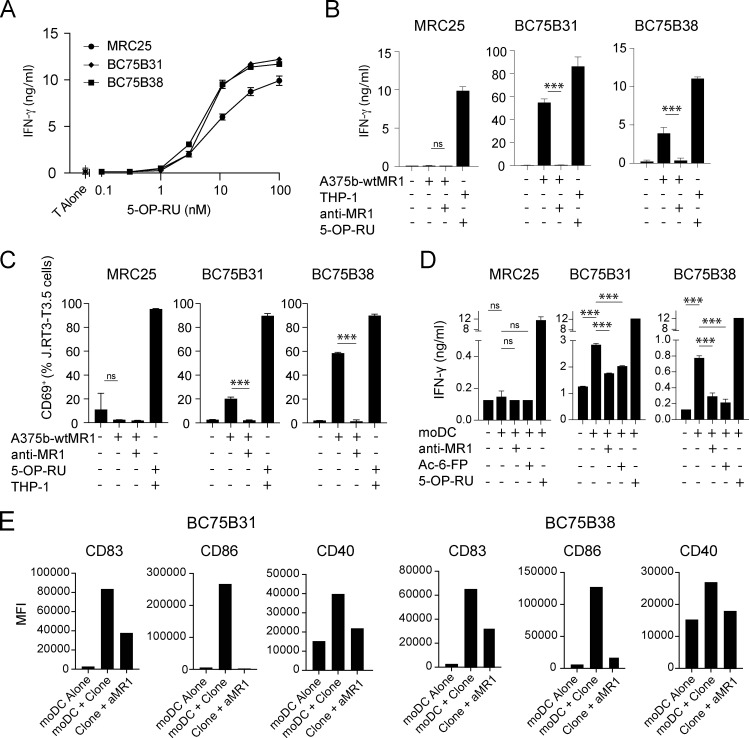
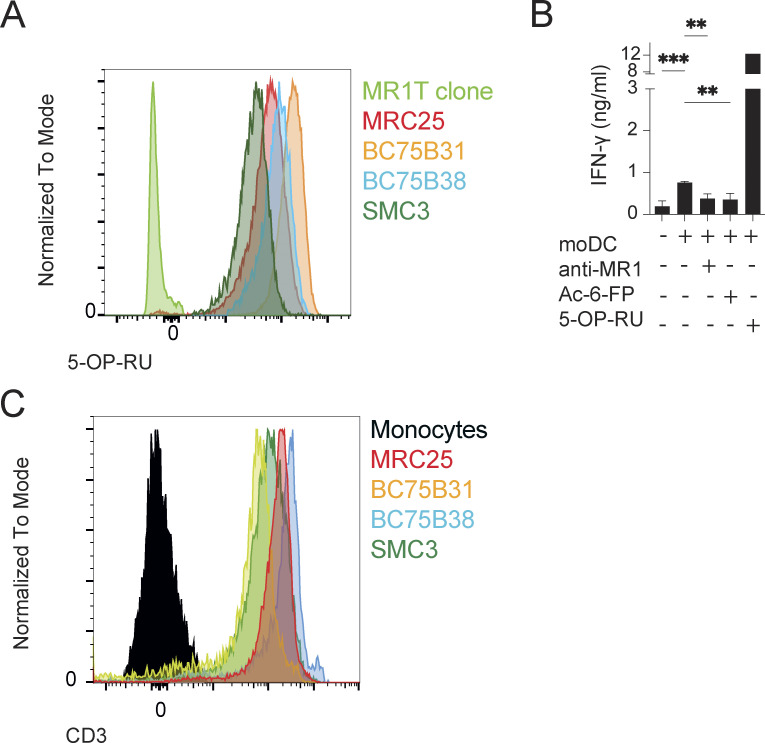
To validate the reactivity and further investigate the function of self-reactive MAIT cells, we interrogated T cell clones. Each clone displayed phenotypic and functional features of canonical MAIT cells, including expression of the classical invariant TCRα chain (Table 1), dose-dependent IFN-γ release by 5-OP-RU stimulation (Fig. 2 A), and binding to 5-OP-RU–loaded MR1 tetramers (Fig. S2 A). When challenged with A375b-wtMR1 cells in the absence of foreign Ags, the clones BC75B31 and BC75B38 released IFN-γ in an MR1-dependent manner (Fig. 2 B). TCR transfer experiments confirmed self-reactive recognition of A375b-wtMR1 (Fig. 2 C). These results indicated that MAIT clones BC75B31 and BC75B38 possess TCRs endowed with dual, MR1-dependent reactivity to microbial and cell-endogenous stimulation. We, therefore, selected these two clones as models to investigate whether MAIT cell self-reactivity also occurs toward healthy cells, expressing physiological MR1 levels on the cell surface. We focused on monocyte-derived dendritic cells (moDCs) and asked whether the low constitutive levels of MR1 they express (Lepore et al., 2017) could be sufficient to drive immunologically relevant recognition by self-reactive MAIT cells. Coculture experiments revealed that moDCs were able to support MR1-dependent IFN-γ release by the BC75B31 and BC75B38 self-reactive MAIT clones, whereas they failed to stimulate a control non-self-reactive MAIT clone (MRC25; Fig. 2 D). A third clone (SMC3) weakly recognized moDCs (Fig. S2 B). Notably, MR1-dependent interaction between the MAIT clones and moDCs resulted in the upregulation of markers associated with DC maturation and licensing, including CD83, CD86, and CD40 (Fig. 2 E). These in vitro results suggested a potential role of self-reactive MAIT cells in modulating DC function.
Table 1.
TCR sequences used in this manuscript
| TCRα | ||||||||
| Clone name | TRAV | TRAJ | CDR3 | |||||
| MRC25 | TRAV1-2 | TRAJ33 | CAVVDSNYQLI | |||||
| BC75B31 | TRAV1-2 | TRAJ33 | CAVMDSNYQLI | |||||
| BC75B38 | TRAV1-2 | TRAJ33 | CAVMDSNYQLI | |||||
| SMC3 | TRAV1-2 | TRAJ33 | CASMDSNYQLI | |||||
| E8 | TRAV1-2 | TRAJ33 | CAVKDSNYQLIW | |||||
| AF-7 | TRAV1-2 | TRAJ33 | CAFLDSNYQLIW | |||||
| TCRβ | ||||||||
| Clone name | TRBV | TRBD | TRBJ | CDR3 | ||||
| MRC25 | TRBV6-1 | TRBD2 a 01 | TRBJ2-7 | CASRLMSGSSYEQYF | ||||
| BC75B31 | TRBV4-2 | TRBD1 a 01 | TRBJ2-1 | CASSHGSTGAYNEQFF | ||||
| BC75B38 | TRBV4-3 | TRBD2 a 02 | TRBJ2-1 | CASSQDPSGSYNEQFF | ||||
| SMC3 | TRBV20-1 | TRBD2 a 01 | TRBJ2-3 | CSAKVTSGQHQGTTDTQYF | ||||
| E8 | TRBV6-1/5 | - | TRBJ1-6 | CASSNREYSPLHF | ||||
| AF-7 | TRBV6-1 | TRBD2 a 01 | TRBJ2-2 | CASSVWTGEGSGELFF | ||||
| 393a | TRBV6-4 | - | TRBJ2-3 | CASSDREADTQYF | ||||
Paired with TCRα chain from the clone SMC3.
Figure 2.
Self-reactivity and T-helper-like functions of MAIT cell clones. (A) Release of IFN-γ by three MAIT cell clones after coculture with 1 × 105 THP-1 cells and indicated concentrations of the microbial Ag 5-OP-RU. IFN-γ is reported as mean ± SD of triplicate cultures. The data are representative of three independent experiments. (B) Release of IFN-γ by three MAIT cell clones in A in response to A375b-wtMR1 cells ± aMR1 mAb. 5-OP-RU–pulsed THP-1 cells were used as a positive control. IFN-γ release is shown as mean ± SD of triplicate cultures. The data is representative of three independent experiments, *** P ≤ 0.001. (C) Percentage of J.RT3-T3.5 cells expressing surface CD69 after incubation with A375b-wtMR1 cells ± aMR1 mAb. A375b-wtMR1 cells pulsed with 5-OP-RU were used as a positive control. J.RT3-T3.5 cell lines express the TCR of the indicated MAIT cell clones. The data are representative of three independent experiments, *** P ≤ 0.001. (D) Release of IFN-γ by three MAIT cell clones stimulated with moDCs ± 5-OP-RU ± aMR1 mAb or ± Ac-6-FP. moDCs pulsed with 5-OP-RU were used as a positive control. IFN-γ release is mean ± SD of triplicate cultures. The data are representative of three independent experiments, *** P ≤ 0.001. (E) Expression levels of the surface maturation markers CD83, CD86, and CD40 on moDCs after overnight coculture with the BC75B31 (left panels) and BC75B38 (right panels) MAIT cell clones. The data are representative of two independent experiments. Statistical significance in all cases was determined using one-way ANOVA with Dunn multiple comparison test.
Figure S2.
Self-reactive MAIT cell phenotype. (A) All MAIT cell clones stain brightly with the 5-OP-RU tetramer. (B) Activation of the MAIT clone SMC3 with moDCs plus either anti-MR1 blocking mAbs or Ac-6-FP or 5-OP-RU. Statistical significance was determined using one-way ANOVA with Dunn multiple comparison test. The data are representative of two independent experiments, ** P ≤ 0.01, *** P ≤ 0.001. (C) CD3 MFI of each clone in panel A.
Canonical MAIT TCRs display various degrees of crossreactive MR1 recognition
Combined microbial and self-reactivity of canonical MAIT cells is an indication of TCR crossreactivity toward different MR1-presented ligands. To further investigate the extent of MAIT TCR crossreactivity, four different approaches were used. In the first, bulk MAIT cell lines were generated from two donors after stimulation with the microbial agonist 5-OP-RU. A fraction of 5-OP-RU–expanded MAIT cells displayed self-reactivity upon challenge with A375b-MR1 cells, as detected by CD137 upregulation (Fig. 3 A). This activation was prevented by anti-MR1 blocking mAb (Fig. 3 A), thus further indicating that this group of MAIT cells was able to recognize both endogenous MR1–ligand complexes displayed by A375b-wtMR1 cells as well as exogenous 5-OP-RU presented by MR1. These experiments also suggested that self-reactive MAIT cells may accumulate in vivo following any microbial infection that drives significant MAIT cell expansion.
Figure 3.
Crossreactivity of circulating MAIT cells from healthy donors. (A) Percentage of CD137+ MAIT cells following activation by A375b-MR1 cells ± aMR1 mAb. MAIT cell lines were previously generated from two donors by in vitro expansion with 5-OP-RU. (B) Plots of MAIT cell lines stained with MR1-3-F-SA tetramer vs. CTV. Cells were pregated on CD161+ cells. Data were obtained from a total of two donors. (C) Representative plots of MAIT cell lines stained with MR1-6-FP tetramer vs. CTV. Cells were pre-gated on CD161+ cells. Data was obtained from a total of two donors. (D) Populations of MAIT cells that are double positive for MR1-3-F-SA and MR1-6-FP tetramers in the same two donors (B–D). (E) Frequency of MR1-5-OP-RU, -6-FP, -3-F-SA, or -5-F-SA MR1 tetramer+ cells from two additional MAIT cell lines derived from the peripheral blood of donors 3 and 4. Pie segments indicate cells positive for any combination of the four tetramer sets. Pie arcs indicate the tetramer positivity of each segment. Percentages indicate the total number of cells positive for at least one tetramer. (F) Percentage of ex vivo MAIT cells from healthy donors stained with at least one of three tetramers: CD8-null MR1-3-F-SA, -5-F-SA, or -6-FP. MAIT cells were pregated on live CD3+/Vα7.2+/CD161+/CD26+ cells. Non-MAIT cells were pregated on live CD3+/Vα7.2− cells. Statistical significance was determined using Student’s t test, ** P ≤ 0.01. Source data are available for this figure: SourceData F3.
The second assay consisted of staining the same MAIT cell lines with MR1-tetramers loaded with either 5-OP-RU, 3-formylsalycilic acid (3-F-SA), or 6-formylpterin (6-FP). All MAIT cells (CD3+CD161high) bound to 5-OP-RU tetramers (Fig. S3, A and C), as expected. A very small fraction of non-classical MAIT cells (CD3+CD161−) also reacted to 5-OP-RU tetramers (Fig. S3, A and B), representing 5-OP-RU–reactive MR1-restricted T cells lacking the canonical MAIT TCR, as previously reported (Gherardin et al., 2018; Gold et al., 2010). Surprisingly, up to 47.1% of MAIT cells were stained by 3-F-SA tetramers (Fig. 3 B), and up to 29.6% by 6-FP tetramers (Fig. 3 C). In addition, a significant number of MAIT cells (∼15 and ∼19% in two donors, respectively) even bound to all three tetramers (Fig. 3 D).
Figure S3.
Gating strategies for identifying bona fide MAIT cells. (A) Gating strategy for identifying MAIT (CD161+) and non-MAIT cells (CD161−) after enrichment for TCR Vα7.2+ T cells, linked to Fig. 3, B–D. (B) Example plots of CD161-fraction that does not contain many 5-OP-RU expanded T cells or T cells that bound to the MR1-5-OP-RU tetramer. (C) Example plots showing that CD161 hi cells within the line simultaneously proliferated when stimulated with 5-OP-RU and bound to the MR1-5-OP-RU tetramer. (D) Example of enrichment of MAIT cells with Vα7.2 mAb. Upper panel shows depleted fraction and lower panel shows enriched fraction. (E) Gating strategy for the identification of MAIT and non-MAIT cells displayed in Fig. 3 E. (F and G) Plots showing populations of MAITs or non-MAITs with different combinations of tetramers.
In the third experiment, MAIT cell lines generated from additional two donors by enrichment of TCR Vα7.2+ cells and subsequent expansion with PHA stimulation were challenged with four sets of MR1 tetramers loaded with either 5-OP-RU, 6-FP, 5-F-SA, and 3-F-SA (Fig. S3, D–G). MAIT cells were identified as CD161+ CD3+ MR1-5-OP-RU tetramer+ (Fig. S3, E–G) whereas CD161− CD3+ MR1-5-OP-RU tetramer− cells represented control non-MAIT cells (Fig. S3, E–G). Within the MAIT group, 23.9% (donor 3) and 11.4% (donor 4) of cells bound to at least one of the other three tetramers bearing different ligands (Fig. 3 E; and Fig. S3, F and G). Distinct patterns of tetramer crossreactivity were observed, with some cells binding to all four tetramer sets (Fig. 3 E; and Fig. S3, F and G). In contrast, non-MAIT cells infrequently reacted to any of the tetramers (3.4%, donor 3 and 3.1%, donor 4; (Fig. 3 E; and Fig. S3, F and G).
Finally, we estimated the proportion of crossreactive MAIT cells able to bind MR1 tetramers loaded with non-microbial Ags directly ex vivo (Fig. 3 F). As previous studies have reported that CD8 has an important impact on MR1-dependent MAIT cell reactivity to microbial metabolites due to its ability to bind to MR1 (Souter et al., 2022), we used the same mutated MR1 tetramers devoid of the CD8 binding capacity (CD8-null). With these reagents, we could strictly assess TCR-dependent binding and rule out any potential contribution of CD8. Significantly more MAIT cells bound to at least one of the three CD8-null tetramers loaded with 6-FP, 5-F-SA, or 3-F-SA ligands (mean 2.7% of MAIT cells) as compared with the control non-MAIT cell group (0.4% of non-MAIT; Fig. 3 F).
Taken together, these data sets indicated that a proportion of MAIT cells feature different degrees of promiscuous TCR interaction with MR1 complexes presenting non-microbial ligands, thus suggesting that TCR crossreactivity is not uncommon within MAIT cells.
Extreme promiscuity in MR1 recognition by a canonical MAIT TCR
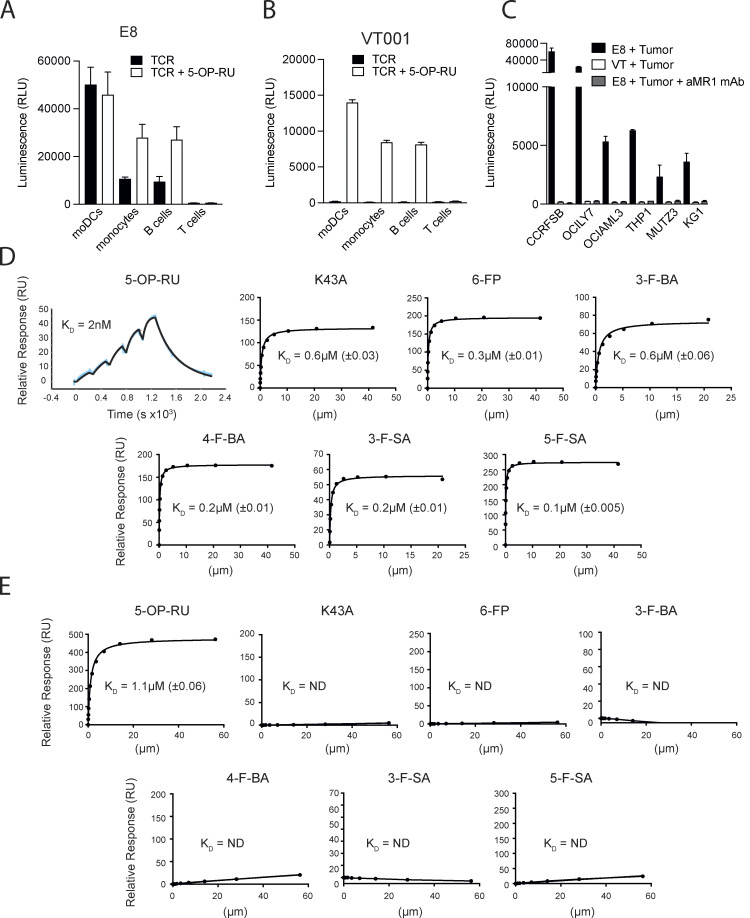
The significant extent of promiscuity in MR1 recognition we observed among MAIT cells prompted us to investigate to what extent unique TCRβ chains contribute to MR1 crossreactivity. For this purpose, a MAIT TCR phage library was generated using a canonical MAIT TCRα chain complemented with random TCRβ chains. When phages were screened for binding to MR1-K43A monomers, a canonical MAIT TCR called E8 was isolated expressing the TRAV1-2-TRAJ33 and a chimera of TRBV6-1/TRBV6-5 gene pairs (Table 1). The E8 TCR displayed MR1-dependent self-reactivity toward healthy moDCs, monocytes, and B cells, but not T cells when transduced into NFAT-Luciferase TCR-null B2M knock-out Jurkat cells (Jurkat-E8; Fig. 4 A). The microbial Ag 5-OP-RU increased their reactivity to monocytes and B cells but did not enhance the already strong Jurkat-E8 cell response to moDCs (Fig. 4 A). A control, non-self-reactive MAIT TCR VT001 (TRAV1-2-TRAJ33/TRBV6-2 gene pairs) also expressed in NFAT-Luciferase TCR-null B2M knock-out Jurkat cells (Jurkat-VT001) did not respond to these APCs unless they were pulsed with 5-OP-RU (Fig. 4 B). In addition, Jurkat-E8, but not Jurkat-VT001, cells reacted to multiple tumor cell lines of different tissue origin in MR1-dependent manner and in the absence of any microbial Ag (Fig. 4 C). Taken together, these data indicated a dual reactivity of the E8 TCR, exemplified by recognition of microbial metabolites and self-reactivity to multiple MR1-expressing cell types.
Figure 4.
Broad self-reactivity and promiscuous recognition of MR1 ligands by the E8 TCR. (A) Recognition of primary immune cells in the absence (black bars) or presence (white bars) of 5-OP-RU by E8 TCR transduced NFAT-Luciferase TCR-null B2M knock-out Jurkat cells. (B) Recognition of primary immune cells in the absence (black bars) or presence (white bars) of 5-OP-RU by VT001 TCR-transduced NFAT-Luciferase TCR-null B2M knock-out Jurkat cells. (C) Recognition of lymphoma cell lines by NFAT-Luciferase TCR-null B2M knock-out Jurkat cells expressing the E8 TCR (black bars), the VT001 TCR (white bars), or the E8 TCR in the presence of blocking aMR1 mAb (gray bars). (A–C) Luminescence measured following NFAT-driven luciferase activity is shown as the cumulative relative luminescence units (RLU) data from three experiments with mean ± SD of duplicate cultures. (D) Binding affinities, as measured by surface plasmon resonance, of the E8 TCR interacting with wildtype MR1 refolded with the indicated range of MR1 ligands, and the empty MR1-K43A mutant. Dissociation constant values (KD) are indicated ± standard error. >150 μM: the measured KD of the TCR MR1 interaction >150 μM and therefore is unlikely to elicit a MAIT cell response. The very high binding affinity of the E8 TCR to MR1 5-OP-RU was measured using the BIAcore8K using single-cycle kinetic analysis. The remaining measurements were performed on a BiacoreT200 and the KDs were calculated using steady-state analysis. (E) Binding affinities, as measured by surface plasmon resonance, of the control AF-7 TCR interacting with wildtype MR1 refolded with the indicated range of MR1 ligands, and MR1-K43A. KD are indicated ± standard error. >150 μM: the measured KD of the TCR MR1 interaction was >150 μM and therefore is unlikely generate a MAIT cell response. Source data are available for this figure: SourceData F4.
We next expressed the E8 TCR in a soluble format and further assessed the extent of cross-reactivity by surface plasmon resonance using MR1 monomers loaded with 5-OP-RU or a range of nonmicrobial ligands, including 6-FP, 3-F-SA, 5-F-SA, 3-formylbenzoic acid (3-F-BA), and 4-formylbenzoic acid (4-F-BA). We also included the empty MR1-K43A mutant of MR1. A previously characterized and non-crossreactive MAIT TCR AF-7 (Eckle et al., 2014) was used as a reference (Table 1). The E8 TCR bound to all MR1–ligand complexes with low nanomolar to low micromolar affinities (KD range = 0.002–0.6 µM), with MR1–5-OP-RU being the strongest affinity ligand (Fig. 4 D). In contrast, the AF-7 TCR bound to MR1-5-OP-RU with KD = 1.1 µM, but not to other immobilized MR1–ligand complexes (Fig. 4 E). Thus, the E8 TCR is an extremely promiscuous classical MAIT TCR that displays self-reactivity toward healthy and tumor cells and binding to a range of soluble MR1–ligand complexes.
Structural basis of promiscuous MR1 recognition by the E8 TCR
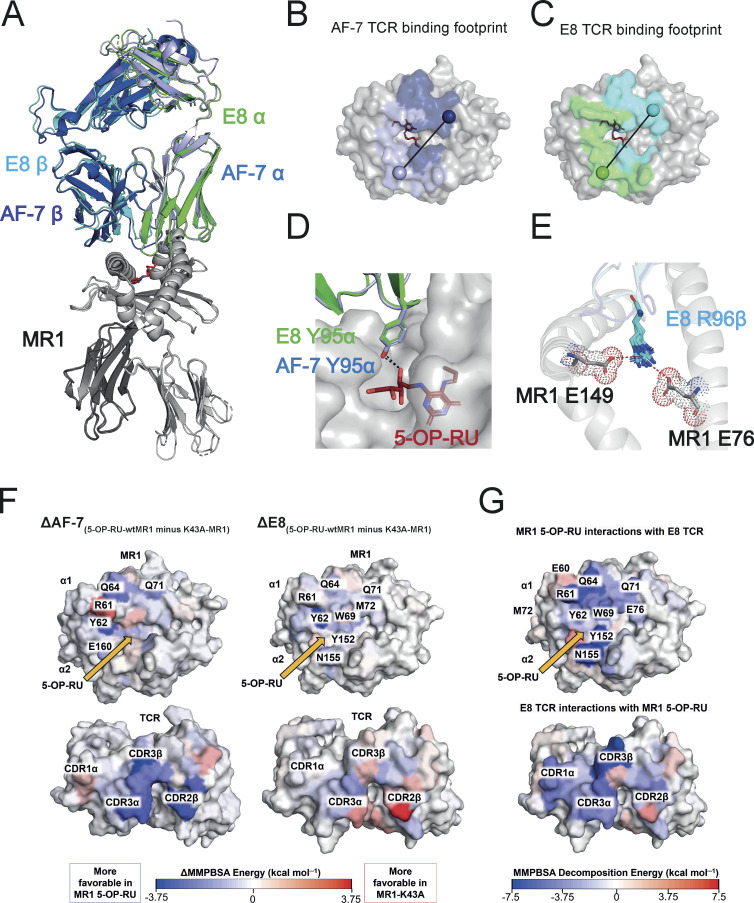
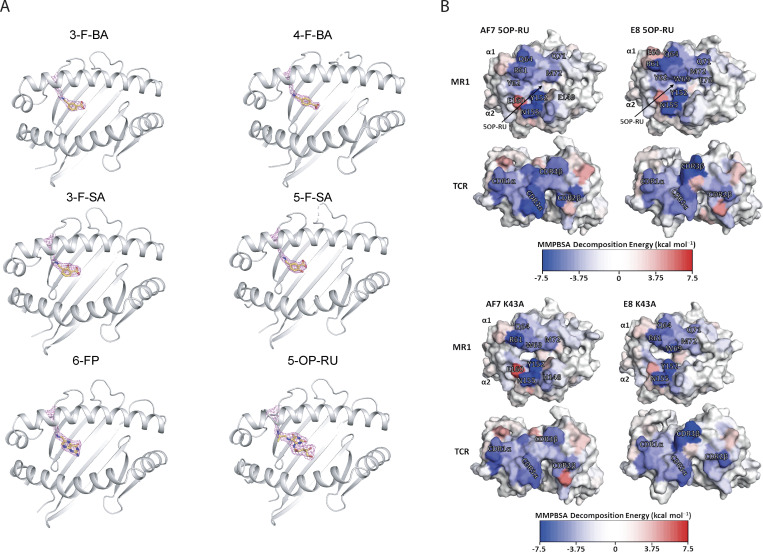
To understand the molecular basis of the broad E8 TCR reactivity, we solved the crystal structures of the E8 TCR in complex with MR1-5-OP-RU, -6-FP, -3-F-SA, -5-F-SA, -3-F-BA, -4-F-BA, and empty MR1-K43A between 1.84 and 2.4 Å resolution (Fig. 5 A, Fig. S4 A, and Table S1). The structures of the E8 TCR were aligned to the structure of the classical MAIT TCR AF-7 (PDB: 6PUC) bound to MR1-5-OP-RU (Awad et al., 2020; Fig. 5 A). The E8 and AF-7 TCRs bound to MR1 in a very similar mode (Fig. 5, A–C). They possess the same chain pairing and virtually identical amino acid sequences in their complementarity-determining region (CDR) loops (Table 1) that adopted identical positions and made a very similar network of contacts with both the MR1 surface and 5-OP-RU (Fig. 5, A–D and Table S2). Like the AF-7 TCR, the E8 TCR contacted the ribityl chain of 5-OP-RU with the Y95α residue in the CDR3α loop (Fig. 5 D and Table S2). The only difference in contacts with MR1-5-OP-RU between the E8 TCR and the AF-7 MAIT TCR was mediated by the E8 TCR residue R96β in the CDR3β loop, which made additional salt bridges with MR1 residues E76 and E149 (Fig. 5 E and Table S2). The structures of the E8 TCR in complex with MR1-5-OP-RU, -K43A, -6-FP, -3-F-SA, -5-F-SA, -3-F-BA, and -4-F-BA revealed an almost identical network of contacts (Table S3). No ligands except 5-OP-RU contributed to TCR binding, providing a possible explanation for the stronger affinity and greater potency of this TCR for MR1–5-OP-RU complexes (Table S3). Together these results suggest that the TCR residue R96β drives broad recognition of MR1–ligand complexes by forming salt bridges to the MR1 heavy chain.
Figure 5.
Structural and energetic basis of promiscuous recognition of MR1 by the E8 TCR. (A) The structures of the E8 TCR (TRAV in green, TRBV in cyan) bound to MR1 loaded (in gray) with 5-OP-RU (shown as red sticks) aligned to the AF-7 TCR (TRAV in light blue and TRBV in dark Blue) bound to MR1 5-OP-RU (PDB 6PUC; Awad et al., 2020). (B) Surface map of the MR1 binding footprint of the AF-7 TCR (α in light blue and β in dark blue) as in Awad et al. (2020). A vector is drawn connecting the disulfide in the α chain variable domain (light blue sphere) to the disulfide in the β chain variable domain (dark blue sphere). (C) Surface map of the MR1 binding footprint of the E8 TCR (α in green and β in cyan). A vector is drawn connecting the disulfide in the α chain variable domain (green sphere) to the disulfide in the β chain variable domain (cyan sphere). (D) The structures of the AF-7 CDR3α Y95 residue light (blue sticks) and E8 CDR3α Y95 residue (green sticks) showing polar interaction (dotted line) with 5-OP-RU (red sticks) bound to MR1 (gray; Awad et al., 2020). (E) Superimposed structures of the CDR3β R96 residue (cyan sticks) in E8 TCRs that form salt bridges to the residues E76 and E149 (gray sticks) in MR1 loaded with ligands (5-OP-RU, 6-FP, 3-F-SA, 5-F-SA, 3-F-BA, and 4-F-BA). (F) Calculated per-residue differences (5-OP-RU-wtMR1 minus K43A-MR1) in the binding free energy for both the AF-7 and E8 TCRs with (5-OP-RU-wtMR1) and without (K43A-MR1) 5-OP-RU bound to MR1. A blue residue is more favorable in the 5-OP-RU form, while a red residue is more favorable in the MR1-K43A form. Yellow arrows indicate the position of 5-OP-RU. (G) Calculated per-residue contributions to the binding free energy for the E8 TCR–MR1 complex with 5-OP-RU bound. The MR1 and TCR molecules are shown as surfaces and color mapped according to their MMPBSA calculated per residue decomposition energies. Color mapping goes from blue (favorable binding) to white (neutral) to red (unfavorable binding) as indicated by the color bar.
Figure S4.
Crystal structure images. (A) Electron density of ligand pockets in the E8 TCR-MR1 crystal structures. MR1 (gray) K43 residue and the covalently bound ligands are shown in sticks. (B) Calculated per-residue contributions to the binding free energy for both the E8 and AF-7 TCRs in complex with MR1 5-OP-RU and the empty MR1 K43A (and therefore in the absence of 5-OP-RU). The MR1 and TCR molecules are shown as surface and color mapped according to their MMPBSA calculated per-residue decomposition energies. Color mapping goes from blue (favorable binding) to white (neutral) to red (unfavorable binding) as indicated by the color bar. Equivalent plots for both TCR–MR1 complexes with 5-OP-RU bound.
To further explore the hypothesis that the salt bridge interaction network mediated by E8 TCR residue R96β was important for the broad recognition of multiple MR1 ligands, MR1 residues E76 and E149 were mutated to Q76 and Q149. E (Glu) and Q (Gln) have very similar physiochemical properties, but Q lacks the anionic carboxylate group that is required to form a salt bridge. Thus, we reasoned that these mutations would abrogate the salt bridge interaction whilst maintaining the overall binding mode of the E8 TCR, and likely still enable some interaction with MR1 residues 76 and 149. The mutated form of MR1 (MR1 E76Q E149Q) was refolded in complex with 5-OP-RU, 6-FP, and 5-FSA, and the soluble protein was used to investigate the binding affinity of the E8 TCR. In addition, we measured the binding affinities to a version of the E8 TCR engrafted with TRBV6-1 (to control for the chimeric output of the phage display) and the canonical AF7 TCR (Table 2). In line with the structural analysis, the binding affinity of the AF7 TCR was not substantially affected by the MR1 mutations: we observed a KD = 0.6 µM for MR1 E76Q E149Q in complex with 5-OP-RU compared with a KD = 1 µM for MR1-5-OP-RU. In contrast, the binding affinity of the E8 TCR to MR1 E76Q E149Q was substantially reduced compared with its binding affinity to MR1 wildtype bound to all ligands tested (eight- to 10-fold reduction in binding affinity; Table 2 and Fig. 4 source data file). Similar observations were made with the E8 TRBV6-1 TCR, with an eightfold reduction in binding affinity for MR1 E76Q E149Q. Previous studies have shown that affinity reductions within this range can abrogate MAIT cell recognition of MR1 ligands (Patel et al., 2013), indicating that the salt bridge interactions mediated by R96β are likely to be central to biologically relevant recognition of MR1 by the E8 TCR. Altogether, these data support the structural analysis and demonstrate that the salt bridge interaction between E8 TCR residue R96β and MR1 residues E76 and E149 plays a central role in crossligand recognition.
Table 2.
Effects of the MR1 E76Q E149Q salt bridge mutations on the binding affinities of AF-7 TCR, E8 TCR, and E8 TRBV6-1 TCR
| MR1 Wt | MR1 E76Q E149Q | |||||
|---|---|---|---|---|---|---|
| TCR | 5-OP-RU | 6-FP | 5-FSA | 5-OP-RU | 6-FP | 5-FSA |
| AF-7 | 1 ± 0.02 μM | >150 μM | >150 μM | 0.6 ± 0.03 μM | >150 μM | >150 μM |
| E8 | 1.3 ± 0.1 nM | 0.6 ± 0.07 μM | 0.2 ± 0.03 μM | 11.2 ± 1.5 nM | 3.5 ± 0.2 μM | 1.8 ± 0.2 μM |
| E8 TRBV6-1 | 80.3 ± 8.3 nM | 10.8 ± 0.7 μM | 6.9 ± 0.9 μM | 0.6 ± 0.05 μM | 42.1 ± 2.6 μM | 32.1 ± 3.7 μM |
Binding affinities, as measured by surface plasmon resonance, of the AF-7, E8 TCR, and E8 TRBV6-1 TCR interacting with wildtype MR1 and MR1 E76Q E149Q refolded with 5-OP-RU, 6-FP, and 5-FSA. >150 μM: the measured KD of the TCR MR1 interaction was >150 μM and therefore is unlikely to generate a MAIT cell response. KD are indicated with the standard error in brackets. The binding affinity of the E8 TCR to MR1 5-OP-RU and MR1 E76Q E149Q 5-OP-RU was measured using the BIAcore8K using single-cycle kinetic analysis. The remining measurements were completed on a BiacoreT200 and the KDs were calculated using steady-state analysis.
Energetic basis of promiscuous MR1 recognition by the E8 TCR
To further explore the basis for the promiscuous behavior of the E8 TCR, we performed molecular dynamics (MD) simulations and binding free energy calculations. These were made on both the AF-7 and E8 TCR bound to wtMR1 loaded with 5-OP-RU and to empty MR1-K43A. The MD simulations allowed us to sample the conformational space available to each complex, and snapshots from these simulations were used to calculate the binding free energy using the molecular mechanics Poisson–Boltzmann surface area (MMPBSA) approach (Genheden and Ryde, 2015; Miller et al., 2012). We utilized two different MMPBSA protocols to predict ΔΔG (Table 3 and Table S4). For the AF-7 TCR–MR1 complex with and without 5-OP-RU bound, we obtained ΔΔG values of −19.6 and −8.7 kcal mol─1 respectively, with the 5-OP-RU bound complex consistently predicted to have a substantially higher affinity (Table 2). In contrast, using the same two protocols for the E8 TCR, we obtained ΔΔG values of +2.8 and −0.9 kcal mol─1 (Table 3), demonstrating our calculations can reproduce the ligand-dependent behavior of AF-7 TCR and the promiscuous behavior of E8 TCR.
Table 3.
Calculated binding free energy differences between the AF7 and E8 TCR
| MMPBSA protocol 1 | MMPBSA protocol 2 | ||||
|---|---|---|---|---|---|
| TCR | MR1 Complex | ΔG (kcal mol−1) | ΔΔG(5-OP-RU-K43A) (kcal mol−1) |
ΔG (kcal mol−1) | ΔΔG(5-OP-RU-K43A) (kcal mol−1) |
| AF-7 | Wt-5-OP-RU | −8.6 ± 3.2 | −19.6a | −28.9 ± 1.1 | −8.7a |
| Empty (K43A) | 11.1 ± 5.3 | −20.2 ± 3.3 | |||
| E8 | Wt-5-OP-RU | −0.8 ± 4.1 | 2.8 | −24 ± 1.6 | −0.9a |
| Empty (K43A) | −3.6 ± 3.8 | −23.1 ± 1.9 | |||
A negative ΔΔG means the 5-OP-RU bound form is more favorable than the MR1-K43A form. Errors are presented as the SD from the five replicas. MMPBSA Protocols 1 and 2 differ by the choice of internal protein dielectric constant setting. For Protocol 1, the internal dielectric constant is set to 1, for Protocol 2, the internal dielectric constant is set to 4.
The MMPBSA approach also allows the decomposition of calculated binding free energy differences into per-residue contributions, which we and others have successfully utilized to identify the key residues and interactions across the binding interface that drive affinity (Crean et al., 2020; Holland et al., 2020; Xiao et al., 2019; Zoete et al., 2010). Thus, we used the MMPBSA approach to understand why the AF-7 and the E8 TCRs have differing ligand dependencies. To this end, the per-residue binding contributions of each TCR to MR1 loaded with and without 5-OP-RU were investigated (Fig. 5 F and Fig. S4 B). For the AF-7 TCR–MR1 complex, several MR1 residues (Y62, Q64, and Q71) and TCR residues on the CDR3α loop (S93α, N94α, and Y95α), on the CDR2β loop (Y48β and D56β), and on the CDR3β loop (T97β and E99β) showed a substantial loss in binding energy in the absence of 5-OP-RU (Fig. 5 F, left panels). This was consistent with previous literature that identified the Y95α residue as a key driver of the AF-7 TCRs MR1 5-OP-RU specificity (Eckle et al., 2014). In contrast, for the E8 TCR–MR1 complex, only two residues (Y62 on MR1 and D56β on the CDR2β loop) across the entire binding interface showed large differences with and without 5-OP-RU bound (Fig. 5 F right panels). Irrespective of 5-OP-RU, the preservation of the binding energy “footprint” is consistent with our structural and Surface Plasmon Resonance (SPR) data on the E8 TCR. Further analysis of the most favorable contributions of the E8 TCR revealed they were within the CDR3β loop (particularly residues R96β and Y98β; Fig. 5 G and Fig. S4 B). For both complexes, MR1 residues Q64, Y152, and N155 provided particularly strong interactions (Fig. 5 G and Fig. S4 B). However, the E8 TCR–MR1 complex was more focused toward MR1 residues located on the α1-helix (R61, W69, and E76; Fig. 5 G and Fig. S4 B).
In summary, our MD analysis revealed that the CDR3β loop of the E8 TCR acts as an anchor, helping to lock in place the remainder of the binding interface independent of the ligand loaded. This is in agreement with structural data which showed the R96β in the E8 CDR3β loop likely forms a salt bridge with the MR1 E76 and E149 residues and is the key driver of promiscuous MR1 binding.
Self-reactive MAIT TCRs can use E8-like CDR3βs
Next, we asked whether a portion of self-reactive MAIT cell TCRβ chains contain E8-like features that could account for the self-recognition of MR1. According to the structural analysis, we reasoned the position of R96β within the CDR3β loop structure would be most influenced by TRBV usage (TRBV6 gene) and CDR3β length of 13 amino acids (CDR3L13). These three features were the basis to search for TCRs with similar motifs.
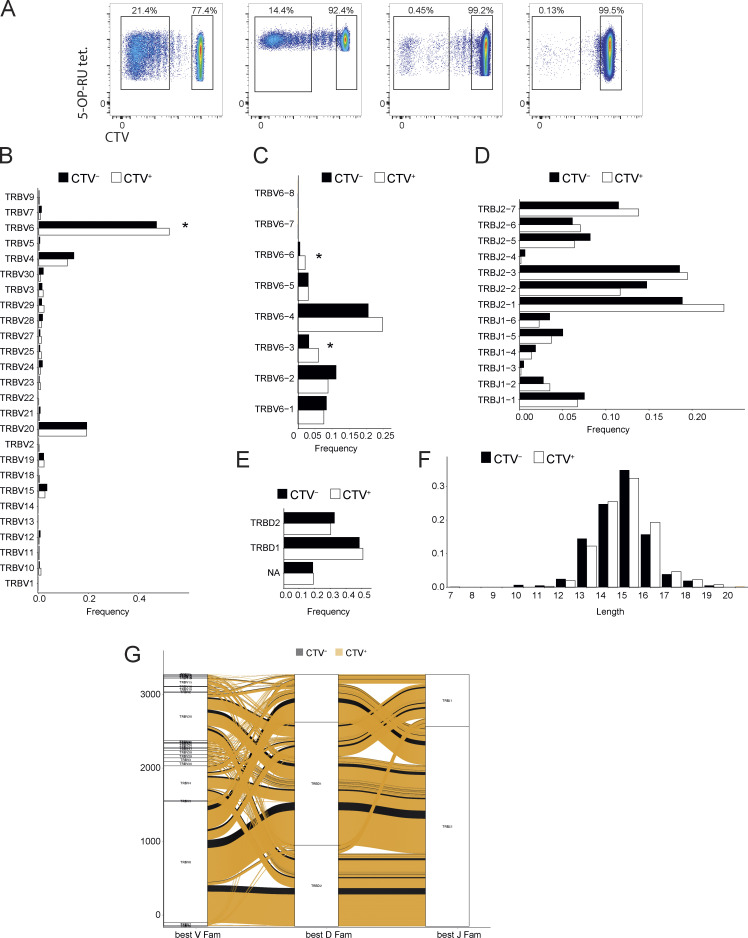
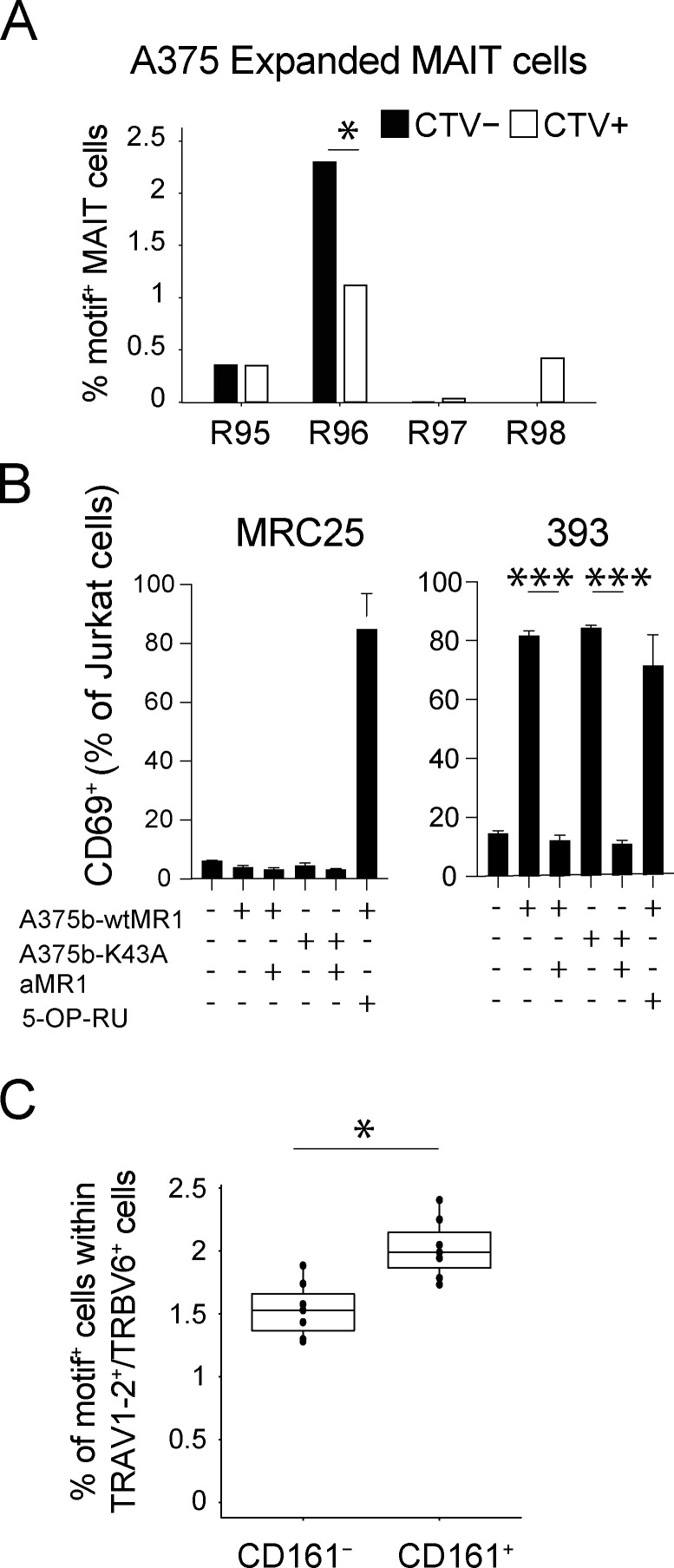
We initially addressed whether this motif was increased in in vitro–expanded autoreactive MAIT cells. Proliferating and non-proliferating MAIT cells stimulated with A375b-wtMR1 cells (from four donors) were sorted into two pools and their TCRβ chains sequenced (Fig. S5 A). A significant increase in the number of TCRβ sequences concomitantly expressing the TRBV6 gene, CDR3L13, and R96β was observed within self-reactive proliferating MAIT cells as compared with non-self-reactive non-proliferating MAIT cells (P = 0.04; Fig. 6 A). Importantly, the frequency of TCRs also co-expressing TRBV6 and CDR3L13 but displaying the Arg in a distinct position nearby the CDR3β residue 96 (R95β, R97β or R98β) was not significantly different between the two MAIT cell groups (Fig. 6 A), indicating that the E8-like motif is enriched within the TCRβ repertoire of healthy donors’ circulating self-reactive MAIT cells.
Figure S5.
Data relating to TCR usage. (A) Sorted 5-OP-RU tetramer+ cell populations that were used for bulk β-chain sequencing. (B) TRBV gene usage of CTV+ or CTV− MAIT cells. Statistical significance was determined using Fisher’s exact test, * P ≤ 0.05. (C) TRBV6 gene usage of CTV+ or CTV− cells. Statistical significance was determined using Fisher’s exact test, * P ≤ 0.05. (D) TRBJ6 usage of CTV+ or CTV− MAIT cells. (E) TRBD gene usage of CTV+ or CTV− MAIT cells. (F) Distribution of CDR3 lengths within either CTV+ or CTV− populations. (G) Alluvial plot or rearranged TCR genes in either CTV+ or CTV− MAIT cells.
Figure 6.
Enrichment of R96 in self-reactive MAIT cells. (A) Frequencies of TRBV6+, CDR3L13+ MAIT cells with either R95, R96, R97, or R98 motif within either self-reactive, proliferated MAIT cells (CTV−) or non-self-reactive, non-proliferated MAIT cells (CTV+). Statistical significance was determined using Fisher’s exact test, * P ≤ 0.05. (B) Activation of J.RT3-T3.5 cells transduced with 393 TRBV (a MAIT TRBV bearing the E8-like motif) or with the control MRC25 TCRBV gene. Percentage of CD69+ cells after co-culture with the indicated APC ± anti-MR1 mAb is illustrated. 5-OP-RU–pulsed THP-1 cells were used as a positive control. Data are representative of three individual experiments each performed in triplicate. Statistical significance was determined using Student’s t test, *** P ≤ 0.001. (C) Frequency of TRBV sequences with the E8-like motif within ex vivo MAIT cells (TRAV1-2+/TRBV6+, and CD161+) or non-MAIT cells (TRAV1-2+/TRBV6+, and CD161−) sorted from the periphery of seven healthy donors (Lepore et al., 2014). Statistical significance was determined using Wilcoxon signed-rank test, * P ≤ 0.05.
To functionally validate these findings, a TCRβ chain bearing this motif was co-expressed with a canonical MAIT TCRα chain (clone SMC3) in J.RT3-T3.5 cells (TCR 393). The expression of this hybrid MAIT TCR pair enabled MR1-dependent self-reactivity toward A375b-wtMR1 cells, in addition to conferring the expected canonical reactivity to 5-OP-RU (Fig. 6 B).
We then performed additional analysis of both proliferating and non-proliferating cells to investigate other unique sequences enriched in self-reactive MAIT cells. TRBV gene usage was biased toward TRBV6, TRBV20, and TRBV4 gene families, which are also the most frequently used in the classical MAIT TCRs (Treiner et al., 2003). In proliferating cells, TRBV6 was significantly over-represented compared with non-proliferating cells (P = 0.03; Fig. S5 B), with a significantly increased use of TRBV6-3 (P = 0.013) and TRBV6-6 (P = 0.013) genes (Fig. S5 C). No significant difference among proliferating and non-proliferating cells was found in the usages of TRBJ (Fig. S5 D) and TRBD (Fig. S5 E) genes or in the lengths of CDR3β (Fig. S5 F). In addition, no significant differences were seen when combinations of genes were analyzed Fig. S5 G). Thus, the motif outlined by the E8 TCR and represented by the combination of TRBV6 and CDR3L13 with R96 might be used to identify and track a population of bona fide self-reactive MAIT cells.
To validate the presence of MAIT cells displaying this motif, we searched within previously acquired TCRβ-chain datasets from healthy donor’s circulating TRAV1-2+/TRBV6+/CD161+ (MAIT cells) and TRAV1-2+/TRBV6+/CD161− (non-MAIT; Lepore et al., 2014). In seven different donors, the motif was observed in 1.73–2.4% of MAIT TCR transcripts (median 1.99%; Fig. 6 C). In contrast, the same motif was detected in 1.28–1.88% (median 1.52%) of TCR transcripts from Τ cells expressing Vα7.2+ and lacking CD161 (Fig. 6 C). Taken together, these findings suggested that in healthy individuals, a small fraction of MAIT cells express the E8-like TRBV6, CDR3L13-R96β motif, which we found associated with self-reactive recognition of MR1.
Discussion
Here, we describe a population of crossreactive human MAIT cells that are activated upon recognition of MR1 in the absence of microbial Ags. The functional responses we investigated did not qualitatively diverge from those induced by the microbial Ag 5-OP-RU and non-transformed target cells expressing low, physiological levels of MR1 were sufficient for productive stimulation of tested clones. Our data revealed that self-reactive recognition of MR1 is a feature of some MAIT cell TCRs, enabling recognition of both tumor and healthy cells. We estimated that self-reactive MAIT cells are rare within circulating T cells of healthy individuals, and whether their frequency is increased and/or their function altered in patients with autoimmunity, inflammatory diseases, and cancer deserves appropriate clinical investigation.
As self-reactive MAIT cells are present in healthy individuals, an important question arises regarding their regulation in vivo. MAIT cells exhibit an effector memory phenotype seemingly due to the abundance of microbial Ags that leads to continual stimulation (Legoux et al., 2019; Seach et al., 2013). Such frequent stimulation promotes the expression of several regulatory molecules, including natural killer inhibitory receptors and immune checkpoint controls (McMahon and Raulet, 2001) that may allow self-reactivity of certain MAIT cells only in circumstances when they are not engaged. The regulation of MR1 levels on APCs could also play a role, only enabling MAIT cell stimulation when enough stimulatory self-Ags are available for loading or when the “correct” Ags for self-reactive MAIT cells are present. Another possibility supported by our in vitro data suggests that self-reactive MAIT cells can expand during microbial infections. Thus, after peripheral expansion by abundant microbial Ags, rare high-affinity self-reactive MAIT TCRs might increase in frequency and subsequently respond to MR1-mediated stimulation in non-infectious settings. Finally, MAIT cell self-reactivity could also be promoted by high SYK Tyrosine kinase expression, which facilitates productive TCR signaling as shown with recognition of CD1d-self lipids by self-reactive human invariant natural killer T cells (iNKT) cells (Perroteau et al., 2020).
Self-reactive MR1 recognition is a clear indication of MAIT TCR crossreactivity, which we found to be represented at different degrees within MAIT cells, thus confirming and extending previous results (Gherardin et al., 2016; Keller et al., 2017). A significant fraction of both in vitro expanded and ex vivo analyzed 5-OP-RU–reactive MAIT TCRs appear to be promiscuous and able to bind MR1 tetramers loaded with distinct non-microbial ligands. Of note, a gradient of promiscuity is observed, where some MAIT TCRs can bind either one, two, or three different MR1 tetramers. An elegant study recently reported a key role of CD8 in enhancing MAIT cell response to 5-OP-RU via lateral binding to MR1 (Souter et al., 2022). The same study also indicated the CD8–MR1 interaction as crucial for the recognition of the weak folate Ags (e.g., 6-FP) by MR1-restricted T cells. Thus, both MR1- and conventional MHC-I–restricted T cells use the CD8 coreceptor to amplify TCR-depended responses, with the greatest impact in the case of low-affinity interactions with Ags (Laugel et al., 2011). Our experiments with both CD8-enabled and CD8-disabled MR1 tetramers, while supporting these findings, also highlight the presence of crossreactive MAIT TCRs that are less influenced by CD8 for the binding of MR1–ligand complexes. Accordingly, CD8-negative MAIT cells are commonly found in both humans and mice, albeit at much lower frequencies (Gherardin et al., 2018; Martin et al., 2009).
An important consideration relates to the non-microbial Ags we and others used to load MR1 tetramers. To our knowledge, these are all physiologically uncommon small molecules that are buried within the MR1 pocket, and therefore not easily accessible for direct contact with TCRs. Thus, a hypothesis that deserves further investigation is whether this type of crossreactive recognition relies on particular ligand-dependent conformations of MR1 sensed by specific TCRs rather than direct TCR–ligand interactions, as already observed within the CD1 system (Cotton et al., 2018). In addition, unique TCR features could also enable broad productive interactions with distinct MR1–ligand complexes as exemplified by the extremely promiscuous E8 TCR mode of MR1 recognition. Here, classical MAIT TCR primary interactions with both 5-OP-RU and MR1 led to robust cell activation, as previously described (Corbett et al., 2014). In addition, key interactions between the CDR3β loop and the MR1 α-helices enabled recognition of a variety of MR1–Ag complexes and contributed to the high affinity for 5-OP-RU. The E8 TCR residue R96β forms a double salt bridge with MR1 residues E76 and E149, allowing productive recognition of MR1 without direct ligand interaction or further stabilizing the complex in the presence of 5-OP-RU. Thus, the two MR1 residues behave as a tweezer, anchoring the TCR residue R96β. MD simulations supported the importance of R96β-mediated interaction in enabling promiscuous and 5-OP-RU–enhanced MR1 recognition compared with the canonical MAIT TCR, AF-7 (Eckle et al., 2014). In addition, biophysical analysis, using MR1 with conservative mutations at residues E76 and E149 to disrupt the salt bridge, confirmed the central role of R96β in driving biologically relevant Ag cross-recognition, and the MD simulations recapitulated the binding affinities for the TCRs to different tested ligands and demonstrated a major energetic role mediated by the E8 TCR CDR3β residue R96.
Our findings fit with the current view of MAIT TCR–MR1–Ag interaction. An elegant study using a series of Ag analogs revealed the importance of a network of polar interactions between TCR, MR1, and Ag, called the interaction triad, which is critical for MAIT cell activation (Awad et al., 2020). Major contributions of different residues within the CDR3β loop were revealed, in some instances directly interacting with the MR1-bound Ag. Another study assigned a degree of autoreactivity to the M33.64 MAIT TCR in which two residues in the CDR3β loop (Thr100β and Asn99β) pinched residue Glu149 of MR1, forming a stable bond (Gherardin et al., 2016). Instead, in the case of the E8 TCR described here, two MR1 residues (E76 and E149) clamp the R96β within the CDR3β. These different modes of binding underline a variety of mechanisms enabling CDR3β-mediated MAIT cell autoreactivity, further describing the unusual plasticity of this interaction. The role of the TCRβ chain in modulating and fine-tuning the response of invariant TCRs to Ag in the context of non-polymorphic molecules has been previously highlighted within the CD1d/semi-invariant iNKT TCR system (Mallevaey et al., 2011). In that study, a hydrophobic motif promoting association with CD1d was found in the CDR3β loops of TCR from self-reactive iNKT cells. This motif facilitated the iNKT response to a broad range of CD1d-restricted self-Ags. It was also described that by transferring this CDR3 β loops into the Vβ6 chain, the resulting TCR started to interact with self-CD1d tetramers. Analogous findings related to the contribution of TCRβ in fine-tuning responses were also subsequently reported for MAIT TCR recognition of the 5-OP-RU Ag (Eckle et al., 2014; Narayanan et al., 2020).
Thus, the immune system deploys distinct mechanisms to extend its breadth of Ag recognition in the case of non-polymorphic Ag-presenting molecules and maximize its ability to interact with these targets.
Last, but not least, the potential Ags involved in the self-reactivity of some MAIT TCRs to both tumor and healthy cells deserve further consideration. While we cannot exclude that some Ag recognition is shared between these cell types, we anticipate that promiscuous recognition of different Ags plays a large role. Some outstanding questions remain, including the identity of recognized Ags, their abundance, expression in healthy vs. tumor cells, and their impact on the immune function of MAIT cells.
In conclusion, our data indicate that a discrete population of MAIT cells is endowed with MR1-dependent self-reactivity toward tumor and healthy cells. In addition, our work provides compelling evidence that TCR crossreactivity is not infrequent within MAIT cells and is characterized by degrees of promiscuity toward distinct MR1–Ag complexes. The self-reactive recognition of MR1 by MAIT cells may have important physiological and immunological implications. Within T cells restricted to non-polymorphic Ag-presenting molecules, self- and tumor-reactivity are also observed toward CD1a, CD1b, CD1c, and CD1d molecules (Bagchi et al., 2017; Bendelac et al., 1995; de Jong et al., 2010; Lepore et al., 2014; Porcelli et al., 1989). On one hand, these autoreactive T cells might participate in inflammatory and autoimmune diseases sustained by unbalanced immune homeostasis and/or play a role in cancer surveillance/progression. On the other hand, the consistent presence of these self-reactive cells in healthy individuals and their T-helper-like properties in vitro suggest they may also have regulatory and/or homeostatic functions, as previously proposed for other non-MHC restricted T cell populations such as iNKT cells, canonical non-self-reactive MAIT cells, and MR1T cells (Cerundolo et al., 2009; Lepore et al., 2017; Salio et al., 2017). Future studies in different cohorts of patients will address the immunological roles of promiscuous Ag recognition by MAIT TCRs.
Materials and methods
Study design
The objectives of this study were to identify whether MAIT cells possess reactivity beyond that toward microbial Ags. To enable these goals, we designed and performed experiments in cellular immunology, protein biochemistry, and crystallography. The number of independent experiments is outlined in the figure legends, where applicable.
Study approval
All human blood samples collected at Immunocore were processed in accordance with the guidelines of Immunocore’s Human Tissue Act compliance team, to conform to the United Kingdom Human Tissue Act 2004, (under ethical approval license IMCres02). Blood specimens obtained from the University Hospital Basel were approved by the local ethical review board (Ethics Committee North-West & Central Switzerland, EKNZ 2017-01888).
Cell lines and primary T cells
The following cell lines were obtained from American Type Culture Collection: A375 (human melanoma), THP-1 (myelomonocytic leukemia), and J.RT3-T3.5 (TCRβ-deficient T cell leukemia). The HEK 293 cell line was obtained from the Leibniz-Institute DSMZ-German Collection of Microorganisms and Cell Cultures. J.RT3-T3.5 were engineered to lack endogenous TCRα protein and express an NFAT-driven luciferase reporter. NFAT-Luciferase TCR-null B2M knock-out Jurkat cells were developed from Parental NFAT-Luciferase Jurkat cells from Promega (part no.: J133A). All cells were routinely tested for the absence of mycoplasma contamination. None of the cell lines used in this study are present in the database of commonly misidentified cell lines. Cell lines were not authenticated. All primary cell lines and clones used in this study were isolated from peripheral blood mononuclear cells (PBMCs) obtained using Lymphoprep (Stemcell Technologies) from the blood of blood bank donors and maintained in culture as previously described (Lepore et al., 2014). Briefly, MAIT cell lines were generated by magnetic bead enrichment using biotinylated anti-Vα7.2 mAb (Clone 3C10; BioLegend) or specific expansion using 5-OP-RU. Enriched MAIT cells were prelabeled with Cell Trace Violet (CTV) according to manufacturer instructions and then cultured with irradiated A375b-wtMR1 cells for the indicated number of days in a 1:1 ratio. Human rIL-2 (5 IU/ml; Peprotech) was added at day 5 and thereafter every 2 d. Cells were washed and rechallenged as indicated (ratio 2:1) in the presence or absence of purified anti-MR1 mAb (20 µg/ml, Ultra-LEAF Purified Clone 26.5; BioLegend). From these lines, self-reactive MAIT clones were derived by limiting dilution in the presence of PHA (1 µg/ml), human rIL-2 (100 U/ml), and irradiated PBMC (5 × 105 cells/ml), and screened for reactivity toward indicated cell. CD14+ monocytes were isolated from PBMCs by positive selection using magnetic beads (Stemcell Technologies) and cultured in the presence of 25 ng/ml GM-CSF and 20 ng/ml IL-4 (both from Peprotech). Activation of monocytes was achieved by treating them with 50 ng/ml LPS (from Escherichia coli O111:B4; Sigma-Aldrich) overnight.
T cell activation
T cell clones (5 × 104 cells/well unless otherwise indicated) were cocultured with indicated target cells (5 × 104 cells/well) in 130 μl total volume in triplicates for 18 h. In some experiments, anti-MR1 mAb (20 µg/ml, Ultra-LEAF Purified Clone 26.5; BioLegend) was added and incubated for 30 min prior to the addition of T cells. In other experiments, APCs were pulsed for 2 h at 37°C with indicated concentrations of Ags or freshly prepared 5-OP-RU as described in Schmaler et al. (2018). J.RT3-T3.5 activation assays were performed in a 1:1 ratio with the indicated APC for 18 h. Cells were then either harvested and stained for surface CD69 upregulation or luciferase was measured using Bio-Glo (Promega).
TCR gene transfer
Total RNA was extracted from snap-frozen cell pellets from each clone. SMARTer RACE 5'/3′ kit (Takara) was used for cDNA synthesis and generation of TCR transcripts. Functional TCRα and β chains were identified by sequencing and analysis using the ImMunoGeneTics information system (http://www.imgt.org). The TCRα and β sequences were either synthesized at Integrated DNA Technologies (TCR 393) or amplified from cDNA with gene-specific primers (TCRs BC75B31, BC75B38, and MRC25) containing cloning adaptors. In both cases, the insert was cloned by In-Fusion HD (Takara) to a lentiviral vector for cotransfection of HEK 293 T LX cells. The endotoxin-free vectors were cotransfected together with the lentivirus packaging plasmids pMD2.G, pMDLg/pRRE, and pRSV-REV (all from Addgene) to HEK 293 T LX cells with Metafectene PRO reagent from Biontex. Lentiviral supernatants of the corresponding TCRα and β sequences were combined and used to transduce J.RT3-T3.5 cells overnight. TCR-expressing J.RT3-T3.5 cells were sorted for CD3 expression before functional analysis. For the experiments described in Fig. 4, TCR genes were transduced in NFAT-Luciferase TCR-null B2MKO Jurkat cells developed from the parental NFAT-Luciferase Jurkat line (Promega, part no. J133A).
Flow cytometry
When staining with MR1 tetramers (20 μg/ml) or anti-human Vα7.2 (2.5 µg/ml Clone 3C10; BioLegend), the cells were pretreated for 30 min at 37°C with 50 nM dasatinib (Sigma-Aldrich) in PBS. All mAb for staining was titrated on appropriate cells before use. Tetramers were added first for 20 min at room temperature and anti-human mAb were added for a further 20 min in PBS with dasatinib: mAb specific for CD3 (Clone UCHT1), CD4 (Clone OKT4), CD8 (Clone RPA-T8), CD161 (Clone HP-3G19), and for activation markers CD137 (Clone 4B4-1), CD69 (Clone FN50), CD25 (Clone BC96), and ICOS (Clone DX29), all from BioLegend. DAPI was used to exclude dead cells. Doublets were excluded by forward scatter-area (FSC-A), forward scatter-width (FSC-W), side scatter-area (SSC-A), and side scatter-height (SSC-H).
Intracellular cytokine staining was performed by the addition of Brefeldin A (5 μg/ml; BioLegend), monensin (2 μM; BioLegend), and 4 μg/ml anti-human CD107a mAb (Clone H43A; BioLegend) 1 h after coculture with indicated APCs. Cells were then harvested and treated with fixable LIVE/DEAD BLUE stain (Thermo Fisher Scientific) for 20 min followed by anti-CD137, CD161, and CD3 mAb before fixation and permeabilization (Buffers from BioLegend). After permeabilization the following mAb specific for intracellular cytokines were added for 40 min on ice: IFN-γ (Clone 4S-B3), TNF-α (Clone MAb.11), GM-CSF (Clone BVD2-21C11), IL-17a (Clone BL168), IL-13 (Clone JES10-5A2), Granzyme B (Clone QA16A02), all from BioLegend. All cells were acquired on a Fortessa (BD) or Aurora spectral analyzer (Cytek) and analyzed using FlowJo v10 software (LLC).
Cytokine analysis
The following human cytokines were assessed by ELISA as previously described (Lepore et al., 2017): human IFN-γ (capture MD-1 mAb; revealing biotinylated 4S.B3 mAb; BioLegend) and human IL-13 (capture clone JES10-5A2; revealing biotinylated clone SB126d 1090; SouthernBiotech).
Identification of VT001 MAIT TCR
TCR was identified from a MAIT T cell clone generated from normal human PBMC as previously described (Lepore et al., 2014, 2017). Briefly, MAIT T cells (CD3+ CD161+ TRAV1-2+ Vδ2−) were sorted by flow cytometry (BD FACS Aria) and expanded on PHA, IL-2, and irradiated allogenic PBMCs to establish a T cell line from which clones were subsequently generated by limiting dilution. Individual clones were assessed for CD161, TRAV1-2, and CD137 expression by flow cytometry following overnight co-culture with 5-OP-RU–loaded THP-1 cells. Positive clones were selected for TCR gene sequencing. Briefly, this involves first-strand cDNA generation and universal amplification using SmartSeq2 chemistry (Picelli et al., 2014), followed by targeted amplification of TCR chains and MiSeq Next Generation Sequencing. Sequencing data were analyzed using a bespoke bioinformatics pipeline (unpublished).
Identification of E8 TCR
TCRs were isolated from phage libraries based on healthy donor MAIT cell TCR repertoire. TCR isolation to produce E8 TCR has been described previously (Li et al., 2005; Liddy et al., 2012). In brief, phage display panning was performed using the MR1-K43A to select an MR1-specific TCR.
Construct design, protein expression, and purification
The sequences coding for the TCR chains were cloned into the pGMT7 vector. TCR constructs were designed to include the variable and constant domains of both α and β chains with an engineered interchain disulfide bond as previously described (Boulter et al., 2003). The sequences coding for wtMR1, MR1-K43A, MR1-E76Q-E149Q, and B2M were cloned into the pET23d vector. CD8-null MR1 coding sequence was generated by introducing the mutations Q223A and E224K as previously described (Souter et al., 2022). The proteins were expressed in the BL21 (DE3) Rosetta pLysS strain (Novagen), refolded from inclusion bodies, and purified as previously described (Garboczi et al., 1992; Boulter et al., 2003; Reantragoon et al., 2013). For SPR measurements, a C-terminal AVI-tag was added to the wtMR1, MR1-K43A, and MR1-E76Q-E149Q constructs and biotinylated after purification using the Avidity Bir A Biotinylation kit, then purified again using a size exclusion column to remove the biotin and Bir A.
SPR single-cycle kinetic analysis
Purified TCRs, MR1-K43A, MR1-E76Q-E149Q, and wtMR1 loaded with 6-FP (Schircks Laboratories), 3-F-BA (Fluorochem), 4-F-BA (Sigma Aldrich), 3-F-SA (Thermo Fisher Scientific Acros Organics), and 5-F-SA (Fluorochem) were subjected to SPR analysis using a BIAcoreT200 using steady-state affinity analysis. The curves were fitted and calculations were completed in GraphPad Prism v9 software. Purified E8 TCR and MR1 (WT and E76Q-E149Q mutant) loaded with 5-OP-RU were subjected to SPR analysis using a BIAcore8K using single cycle kinetic analysis, which was completed with Biacore Insight Evaluation software.
Crystallization and protein structure determination
E8 TCR and MR1 molecules in 10 mM Tris, pH 8.0, 100 mM NaCl were mixed in equimolar ratio and concentrated to 8–10 mg/ml. Sitting drops were set up containing 150 nl of protein solution and 150 nl of reservoir solution in MRC crystallization plates using the Gryphon robot (ART Robbins) and incubated at 20°C. For every E8 TCR-MR1 sample, crystals appeared in many different crystallization conditions. Crystals were cryoprotected using reservoir solution containing 30% ethylene glycol and flash-cooled in liquid N2. Diffraction data were collected at beamlines I03 and I04 at the Diamond Light Source, UK. Datasets used for structure solutions were collected from crystals grown in the following crystallization conditions:
E8-MR1-5-OP-RU: 0.2 M potassium thiocyanate, 0.1 M Bis-Tris propane, pH 8.5, and 20% wt/vol PEG 3350;
E8-MR1-K43A: 0.2 M magnesium chloride hexahydrate, 0.1 M Tris, pH 8.5, and 15% wt/vol PEG 4000;
E8-MR1-6-FP: 0.2 M sodium iodide, 0.1 M Bis-Tris propane, pH 8.5, and 20% wt/vol PEG 3350;
E8-MR1-3-F-SA: 0.2 M sodium malonate dibasic monohydrate, 0.1 M Bis-Tris propane, pH 8.5, and 20% wt/vol PEG 3350;
E8-MR1-5-F-SA: 0.2 M sodium fluoride, 0.1 M Bis-Tris propane, pH 8.5, and 20% wt/vol PEG 3350;
E8-MR1-3-F-BA: 0.1 M magnesium acetate tetrahydrate, 0.1 M MOPS, pH 7.5, and 12% wt/vol PEG 8000;
E8-MR1-4-F-BA: 0.1 M Hepes, pH 7.5, 20% wt/vol PEG 4000, and 15% Glycerol.
The diffraction data were integrated and scaled using the xia2 (Winter et al., 2013) automated processing pipeline using XDS (Kabsch, 2010) and XSCALE. The E8 TCR–MR1–5-OP-RU complex structure was solved by molecular replacement using MR1 and TCR coordinates from PDB 4PJA as the search models in Phaser (McCoy et al., 2007) within the CCP4 suite (Winn et al., 2011). The model was built using iterative cycles of manual model building in COOT (Emsley et al., 2010) and refinement using Refmac (Murshudov et al., 2011). The ligand restraints for refinement were generated using AceDRG (Long et al., 2017). All other E8–MR1 complex structures were solved using the E8 TCR–MR1–5-OP-RU structure (with the ligand removed) as the search model for molecular replacement in Phaser. Model building and refinement processes for these complexes were carried out as explained for the E8 TCR–MR1–5-OP-RU complex structure. The stereochemical properties and validation of the models were assessed using PDB-REDO (Joosten et al., 2012) and MolProbity (Williams et al., 2018). Buried surface area and TCR docking geometry statistics based on those described previously (Rudolph et al., 2006) were generated using Molecular Operating Environment (Chemical Computing Group; Molecular Operating Environment, 2022) The structural figures were generated using Pymol (Schrödinger). The diffraction data were integrated and scaled using the xia2 (Winter et al., 2013) automated processing pipeline using XDS (Kabsch, 2010) and XSCALE. The E8 TCR–MR1–5-OPRU complex structure was solved by molecular replacement using MR1 and TCR coordinates from PDB 4PJA as the search models in Phaser (McCoy et al., 2007) within the CCP4 suite (Winn et al., 2011). The model was built using iterative cycles of manual model building in COOT (Emsley et al., 2010) and refinement using Refmac (Murshudov et al., 2011). The ligand restraints for refinement were generated using AceDRG (Long et al., 2017). All other E8–MR1 complex structures were solved using the E8 TCR–MR1–5-OP-RU structure (with the ligand removed) as the search model for molecular replacement in Phaser. Model building and refinement processes for these complexes were carried out as explained for the E8 TCR–MR1–5-OP-RU complex structure. The stereochemical properties and validation of the models were assessed using PDB-REDO (Joosten et al., 2012) and MolProbity (Williams et al., 2018). Buried surface area and TCR docking geometry statistics based on those described previously (Rudolph et al., 2006) were generated using Molecular Operating Environment (Chemical Computing Group; Molecular Operating Environment, 2022). The structural figures were generated using Pymol (Schrödinger).
MD simulations and MMPBSA calculations
MD simulations were prepared and performed using the Amber18 software package (Song et al., 2019). In all cases, x-ray crystal structures were used as the starting point for simulations. Following structure preparation (His tautomerization states assignments, Asn/Gln flips, protonation states set for an effective pH of 7), each structure was solvated in octahedral water box with all crystallographic water molecules retained. His tautomerization states assignments are provided in Table S4. The Amber ff14SB (Maier et al., 2015) force field and TIP3P water model were used to describe protein and water molecules, respectively. For simulations with 5-OP-RU covalently bound to K43, a custom residue was built using a combination of ff14SB (Maier et al., 2015) for the lysine unit and GAFF2 (Wang et al., 2004) for the connection and 5-OP-RU unit (with HF/6-31G(d) RESP fitted charges obtained using the R.E.D. server [Vanquelef et al., 2011]). To prepare each system for production MD simulations, a standard procedure of energy minimization, heating, and equilibration was performed. Production MD simulations were performed in the NPT ensemble at 300 K and 1 atm using a 2 fs time step (with the SHAKE algorithm applied). Production MD simulations were 100 ns long, and for each system, five replicas were performed, with subsequent simulation analysis performed using CPPTRAJ (Roe and Cheatham, 2013). MMPBSA calculations were performed using MMPBSA.py.MPI (Miller et al., 2012) using snapshots from the previously described MD simulations as input (snapshots were taken every 50 ps from 5 × 100 ns runs, so 10,000 snapshots per complex in total). The implicit salt concentration was set to 150 mM. Calculated binding free energies were decomposed to their per residue contributions using an internal dielectric constant of 1 (i.e., Protocol 1 in Table S4).
System preparation
All simulations were performed starting from crystal structures. For simulations of AF7 MR1 with 5-OP-RU bound, we used PDB 6PUC. For simulations of AF7 K43A without 5-OP-RU, we manually removed the 5-OP-RU unit and mutated K43 to alanine (as no structure exists for this mutant). For simulations of E8 MR1 with 5-OP-RU bound, we used PDB 7ZT2 (generated in this study). For simulations of E8 K43A without 5-OP-RU, we used PDB 7ZT3 (generated in this study). For the structure of AF7 MR1 with 5-OP-RU (PDB 6PUC), we performed simulations using chains A, B, G, and H. Missing residues in chain A 190–195 were added using chain C of 6PUC (Awad et al., 2020) as the template. For the structure of E8 MR1 K43A (produced in this study), chain A has missing residues 189–196. We used the structure of E8 MR1 with 5-OP-RU (also produced in this study) as the template for these residues. Optimal histidine tautomerization states (see Table S4) and asparagine and glutamine side chain orientations were determined using MolProbity (Williams et al., 2018), and all residues, except for His90 on MR1, were simulated in their standard protonation states based on pKa calculations performed using PROPKA 3.1 (for a target pH of 7). His90 on MR1 was simulated as positively charged based on the PROPKA prediction and manual inspection. Further, the tautomerization state of His58 on MR1 was manually assigned to be singly protonated on its Nε2 atom, with this atom coordinating the Schiff base nitrogen on K43–5-OP-RU, which acts as a hydrogen bond acceptor. Then structures were solvated in an octahedral water box, (with all crystallographic water molecules kept). The system box size was set such that no solute atom was within 10 Å of the box boundary. To ensure the total system charge was 0, sodium or chloride ions were added as necessary.
Parametrization of 5-OP-RU
The K43 residue on MR1 which is covalently bound (through a Schiff base) to 5-OP-RU was parameterized for MD simulations as a single (non-standard) amino acid. Here, we followed the recommended procedure as detailed in full here: https://upjv.q4md-forcefieldtools.org/Tutorial/Tutorial-4.php#16. First, we extracted the structure of K43-5-OP-RU from PDB 6PUC (Awad et al., 2020) alongside residues Q42 and E44, which were manually chemically modified to acetylate and amidate the backbone of K43. The structure was then optimized with QM (B3LYP/6-31G(d,p), tight convergence criteria) using Gaussian16 (Frisch et al., 2016). The resulting structure was then submitted to the R.E.D. Server (Vanquelef et al., 2011) for the partial charge calculation (with HF/6-31G(d) RESP fitted charges obtained). For these partial charge calculations, we charge-constrained the acetate and amidate groups to each have a total charge of 0. Atom type definitions for all atoms from the lysine unit with the exception of the side-chain nitrogen were described using the Amber ff14SB (Maier et al., 2015) force field, while atom types for the lysine side-chain nitrogen and the 5-OP-RU unit were described with GAFF2 (Wang et al., 2004) atom types. For bond, angles, dihedrals, and improper torsion terms that contained a mixture of ff14SB and GAFF2 atom types, terms were taken from the GAFF2 library by analogy. Complete parameters alongside an exemplar tleap script to use said parameters are deposited on zenodo (https://zenodo.org/record/6651550).
MD simulations
The structure equilibration procedure was used to prepare all systems simulated for production MD simulations in the NPT ensemble at 300 K and 1 atm. All dynamic steps applied the SHAKE algorithm to constrain all bonds containing a hydrogen atom. Replica simulations were initiated from the second heating step of the following protocol (with each replica therefore assigned different random velocity vectors at this stage). Simulations performed in the NVT ensemble used Langevin temperature control (with a collision frequency of 1 ps−1) and used a simulation time step of 1 fs. Simulations performed in the NPT ensemble used Langevin temperature control (collision frequency of 1 ps−1) and a Berendsen barostat (1 ps pressure relaxation time).
The equilibration protocol is as follows: first, hydrogens atoms and solvent molecules were energy minimized (using 500 steps of steepest descent followed by 500 steps of conjugate gradient minimization). To prevent the movement of non-hydrogen and non-solvent atoms during the minimization, 10 kcal mol−1 Å−1 positional restraints were used to keep all heavy atoms fixed. Then the solvent was heated rapidly from 50 to 300 K (NVT ensemble, 1 fs time step) over the course of 200 ps, with the previously described restraints still maintained. The positional restraints were then replaced with 5 kcal mol−1 Å−1 positional restraints on only the Cα carbon atoms of each residue and subjected to another round of energy minimization (500 steps of steepest descent followed by 500 steps of conjugate gradient). Retaining these positional restraints, the system was heated from 25 to 300 K over the course of 50 ps (NVT ensemble, 1 fs time step). Simulations were then performed in the NPT ensemble (1 atm, 300 K, 2 fs time step) by first gradually reducing the 5 kcal mol−1 Å−1 Cα carbon restraints over the course of 50 ps. This was done by reducing the restraint weight by 1 kcal mol−1 Å−1 every 10 ps. A final 1-ns-long MD simulation with no restraints placed on the system was then performed, with the final structure produced after this run used as the starting point for production MD simulations.
Informatics analysis of TCRs
Raw sequencing data was demultiplexed using Cutadapt v3.5 and the quality of the reads was checked using FastQC v0.11.9. MiXCR v3.0.13 (Bolotin et al., 2015) was used on the demultiplexed data to align the reads and assemble them into clonotypes, groups of reads sharing the same CDR3 sequence. No further trimming was performed as MiXCR by default handles reads containing low-quality nucleotides, performs PCR error correction, and conserves only clonotypes that would present a productive TCR. The output tables of MiXCR were loaded into R, and sequences corroborated by only one read were filtered from the dataset unless they were present in both non-proliferating and proliferating subsets. Exploration of the Vβ, Jβ, and CDR3β lengths as well as the analysis of the sequences carrying Arginine in positions 5, 6, 7, or 8 of the CDR3β was performed via custom R scripts. Statistical significance was determined using Fisher’s exact test and correcting for multiple testing through the Benjamini-Hochberg method for contingency tables or using the Wilcoxon signed-rank test.
Statistical analysis
Cytokine secretion and luciferase assays as well as flow cytometry data were analyzed for normality using Shapiro–Wilk normality test. The appropriate statistical test is indicated in the figure legends and performed using Prism 9, GraphPad software. For SPICE analysis, multiple comparison Student’s t test was performed automatically using the freely available software SPICE 6.
Online supplemental material
Fig. S1 shows functional data relating to Fig. 1. Fig. S2 shows additional clone data. Fig. S3 shows gating strategy. Fig. S4 shows crystal structure images. Fig. S5 shows data relating to TCR usage. Tables S1, S2, S3, S4, and S5 show additional statistics of crystal structures.
Supplementary Material
shows data collection and refinement statistics.
shows contacts between MR1-5-OP-RU and TCRs.
shows contacts between the E8 TCR and MR1 with different ligands.
shows histidine tautomerization states used for simulations of the E8 TCR and AF7 TCR in complex with MR1.
shows duplicate Biacore data for E8 TCR and AF-7 TCR binding to MR1 loaded with an array of ligands and the empty MR1 K43A mutant.
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
Acknowledgments
The authors would like to thank Diamond Light Source for beam time and the staff for assistance with crystal testing and data collection, and Mariolina Salio for critical reading of the manuscript. This research made use of the Balena High-Performance Computing Service at the University of Bath. The MR1 tetramer technology was developed jointly by James McCluskey, Jamie Rossjohn, and David Fairlie, and 5-OP-RU- and 6-FP-hMR1 tetramers were produced by the National Institutes of Health Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
This work received funding from the Swiss National Foundation 310030-173240 and 310030B-192828, Krebsliga BeiderBasel KLbB-4779-02-2019, Swiss Cancer League KFS-4707-02-2019, Department of Biosystems Science and Engineering at Eidgenössische Technische Hochschule Zürich PMB-02-17 (all to G. De Libero), and from the University of Basel 3MM1055 to A. Chancellor. R.M. Crean’s studentship is funded by the Engineering and Physical Sciences Research Council. M.W. an der Kamp is a Biotechnology and Biological Services Research Council David Phillips Fellow (BB/M026280/1).
Author contributions: A. Chancellor, R. Simmons, R.C. Khanolkar, V. Nosi, A. Beshirova, G. Berloffa, R. Colombo, V. Karuppiah, J.M. Paruppiah, V. Tubb, H. Ghadbane, R.J. Suckling, K. Page, R.M. Crean, A. Vacchini, C. De Gregorio, V. Schaefer, D. Constantin, T. Gligoris, A. Lloyd, M. Hock, V. Srikannathasan, R.A. Robinson, G.S. Besra, and M.W. an der Kamp performed experiments, analyzed data, and provided critical inputs to the manuscript. R. Calogero. and L. Mori discussed the data and revised the manuscript. A. Chancellor, D.K. Cole, G. De Libero, and M. Lepore conceived and directed the project. A. Chancellor, D.K. Cole, G. De Libero, and M. Lepore wrote the manuscript. All authors reviewed and commented on the manuscript.
Data availability
The crystal structures of E8 TCR and MR1–ligand complexes have been deposited with the PDB under Accessions # 7ZT2, 7ZT3, 7ZT4, 7ZT5, 7ZT7, 7ZT8, and 7ZT9. T cell clones are available from G. De Libero under a material transfer agreement with the University of Basel. New TCR sequences in this manuscript are available from G. De Libero under a material transfer agreement with the University of Basel. All other data are available in the article itself and its supplementary materials.
References
- Awad, W., Ler G.J.M., Xu W., Keller A.N., Mak J.Y.W., Lim X.Y., Liu L., Eckle S.B.G., Le Nours J., McCluskey J., et al. 2020. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat. Immunol. 21:400–411. 10.1038/s41590-020-0616-6 [DOI] [PubMed] [Google Scholar]
- Bagchi, S., He Y., Zhang H., Cao L., Van Rhijn I., Moody D.B., Gudjonsson J.E., and Wang C.R.. 2017. CD1b-autoreactive T cells contribute to hyperlipidemia-induced skin inflammation in mice. J. Clin. Invest. 127:2339–2352. 10.1172/JCI92217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac, A., Lantz O., Quimby M.E., Yewdell J.W., Bennink J.R., and Brutkiewicz R.R.. 1995. CD1 recognition by mouse NK1+ T lymphocytes. Science. 268:863–865. 10.1126/science.7538697 [DOI] [PubMed] [Google Scholar]
- Bolotin, D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., and Chudakov D.M.. 2015. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods. 12:380–381. 10.1038/nmeth.3364 [DOI] [PubMed] [Google Scholar]
- Boulter, J.M., Glick M., Todorov P.T., Baston E., Sami M., Rizkallah P., and Jakobsen B.K.. 2003. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 16:707–711. 10.1093/protein/gzg087 [DOI] [PubMed] [Google Scholar]
- Cerundolo, V., Silk J.D., Masri S.H., and Salio M.. 2009. Harnessing invariant NKT cells in vaccination strategies. Nat. Rev. Immunol. 9:28–38. 10.1038/nri2451 [DOI] [PubMed] [Google Scholar]
- Corbett, A.J., Eckle S.B., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 509:361–365. 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- Cotton, R.N., Shahine A., Rossjohn J., and Moody D.B.. 2018. Lipids hide or step aside for CD1-autoreactive T cell receptors. Curr. Opin. Immunol. 52:93–99. 10.1016/j.coi.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crean, R.M., MacLachlan B.J., Madura F., Whalley T., Rizkallah P.J., Holland C.J., McMurran C., Harper S., Godkin A., Sewell A.K., et al. 2020. Molecular rules underpinning enhanced affinity binding of human T cell receptors engineered for immunotherapy. Mol. Ther. Oncolytics. 18:443–456. 10.1016/j.omto.2020.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther, M.D., Dolton G., Legut M., Caillaud M.E., Lloyd A., Attaf M., Galloway S.A.E., Rius C., Farrell C.P., Szomolay B., et al. 2020. Author Correction: Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 21:695. 10.1038/s41590-020-0640-6 [DOI] [PubMed] [Google Scholar]
- Croxford, J.L., Miyake S., Huang Y.Y., Shimamura M., and Yamamura T.. 2006. Invariant V(alpha)19i T cells regulate autoimmune inflammation. Nat. Immunol. 7:987–994. 10.1038/ni1370 [DOI] [PubMed] [Google Scholar]
- de Jong, A., Peña-Cruz V., Cheng T.Y., Clark R.A., Van Rhijn I., and Moody D.B.. 2010. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat. Immunol. 11:1102–1109. 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle, S.B., Birkinshaw R.W., Kostenko L., Corbett A.J., McWilliam H.E., Reantragoon R., Chen Z., Gherardin N.A., Beddoe T., Liu L., et al. 2014. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211:1585–1600. 10.1084/jem.20140484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flament, H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., Rousseau C., Soulard P., Gouda Z., Cagninacci L., et al. 2021. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 22:322–335. 10.1038/s41590-021-00870-z [DOI] [PubMed] [Google Scholar]
- Frisch, M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., et al. 2016. Gaussian 16 Rev. C.01. In Wallingford, CT. [Google Scholar]
- Garboczi, D.N., Hung D.T., and Wiley D.C.. 1992. HLA-A2-peptide complexes: Refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA. 89:3429–3433. 10.1073/pnas.89.8.3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., and Ryde U.. 2015. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10:449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardin, N.A., Keller A.N., Woolley R.E., Le Nours J., Ritchie D.S., Neeson P.J., Birkinshaw R.W., Eckle S.B.G., Waddington J.N., Liu L., et al. 2016. Diversity of T Cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. 44:32–45. 10.1016/j.immuni.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Gherardin, N.A., Souter M.N., Koay H.F., Mangas K.M., Seemann T., Stinear T.P., Eckle S.B., Berzins S.P., d’Udekem Y., Konstantinov I.E., et al. 2018. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol. Cell Biol. 96:507–525. 10.1111/imcb.12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, M.C., Cerri S., Smyk-Pearson S., Cansler M.E., Vogt T.M., Delepine J., Winata E., Swarbrick G.M., Chua W.J., Yu Y.Y., et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8:e1000407. 10.1371/journal.pbio.1000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, M.C., McLaren J.E., Reistetter J.A., Smyk-Pearson S., Ladell K., Swarbrick G.M., Yu Y.Y., Hansen T.H., Lund O., Nielsen M., et al. 2014. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211:1601–1610. 10.1084/jem.20140507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriff, M.J., McMurtrey C., Froyd C.A., Jin H., Cansler M., Null M., Worley A., Meermeier E.W., Swarbrick G., Nilsen A., et al. 2018. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol. 3:eaao2556. 10.1126/sciimmunol.aao2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, C.J., Crean R.M., Pentier J.M., de Wet B., Lloyd A., Srikannathasan V., Lissin N., Lloyd K.A., Blicher T.H., Conroy P.J., et al. 2020. Specificity of bispecific T cell receptors and antibodies targeting peptide-HLA. J. Clin. Invest. 130:2673–2688. 10.1172/JCI130562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson, L.J., Napolitani G., Shepherd D., Ghadbane H., Kurupati P., Preciado-Llanes L., Rei M., Dobinson H.C., Gibani M.M., Teng K.W.W., et al. 2018. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella Paratyphi A. Nat. Commun. 9:253. 10.1038/s41467-017-02540-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Gilfillan S., Kim S., Thompson B., Wang X., Sant A.J., Fremont D.H., Lantz O., and Hansen T.H.. 2008. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205:1201–1211. 10.1084/jem.20072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, R.P., Joosten K., Murshudov G.N., and Perrakis A.. 2012. PDB_REDO: Constructive validation, more than just looking for errors. Acta Crystallogr. D Biol. Crystallogr. 68:484–496. 10.1107/S0907444911054515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. 2010. Xds. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, A.N., Eckle S.B., Xu W., Liu L., Hughes V.A., Mak J.Y., Meehan B.S., Pediongco T., Birkinshaw R.W., Chen Z., et al. 2017. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 18:402–411. 10.1038/ni.3679 [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491:717–723. 10.1038/nature11605 [DOI] [PubMed] [Google Scholar]
- Koay, H.F., Gherardin N.A., Xu C., Seneviratna R., Zhao Z., Chen Z., Fairlie D.P., McCluskey J., Pellicci D.G., Uldrich A.P., and Godfrey D.I.. 2019. Diverse MR1-restricted T cells in mice and humans. Nat. Commun. 10:2243. 10.1038/s41467-019-10198-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugel, B., Cole D.K., Clement M., Wooldridge L., Price D.A., and Sewell A.K.. 2011. The multiple roles of the CD8 coreceptor in T cell biology: Opportunities for the selective modulation of self-reactive cytotoxic T cells. J. Leukoc. Biol. 90:1089–1099. 10.1189/jlb.0611316 [DOI] [PubMed] [Google Scholar]
- Le Bourhis, L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11:701–708. 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- Legoux, F., Bellet D., Daviaud C., El Morr Y., Darbois A., Niort K., Procopio E., Salou M., Gilet J., Ryffel B., et al. 2019. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 366:494–499. 10.1126/science.aaw2719 [DOI] [PubMed] [Google Scholar]
- Lepore, M., Kalinichenko A., Colone A., Paleja B., Singhal A., Tschumi A., Lee B., Poidinger M., Zolezzi F., Quagliata L., et al. 2014. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat. Commun. 5:3866. 10.1038/ncomms4866 [DOI] [PubMed] [Google Scholar]
- Lepore, M., Kalinichenko A., Calogero S., Kumar P., Paleja B., Schmaler M., Narang V., Zolezzi F., Poidinger M., Mori L., and De Libero G.. 2017. Correction: Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. 6:e29743. 10.7554/eLife.29743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Moysey R., Molloy P.E., Vuidepot A.L., Mahon T., Baston E., Dunn S., Liddy N., Jacob J., Jakobsen B.K., and Boulter J.M.. 2005. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat. Biotechnol. 23:349–354. 10.1038/nbt1070 [DOI] [PubMed] [Google Scholar]
- Liddy, N., Bossi G., Adams K.J., Lissina A., Mahon T.M., Hassan N.J., Gavarret J., Bianchi F.C., Pumphrey N.J., Ladell K., et al. 2012. Monoclonal TCR-redirected tumor cell killing. Nat. Med. 18:980–987. 10.1038/nm.2764 [DOI] [PubMed] [Google Scholar]
- López-Sagaseta, J., Dulberger C.L., McFedries A., Cushman M., Saghatelian A., and Adams E.J.. 2013. MAIT recognition of a stimulatory bacterial antigen bound to MR1. J. Immunol. 191:5268–5277. 10.4049/jimmunol.1301958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, F., Nicholls R.A., Emsley P., Graǽulis S., Merkys A., Vaitkus A., and Murshudov G.N.. 2017. AceDRG: A stereochemical description generator for ligands. Acta Crystallogr. D Struct. Biol. 73:112–122. 10.1107/S2059798317000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., and Simmerling C.. 2015. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 11:3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey, T., Clarke A.J., Scott-Browne J.P., Young M.H., Roisman L.C., Pellicci D.G., Patel O., Vivian J.P., Matsuda J.L., McCluskey J., et al. 2011. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 34:315–326. 10.1016/j.immuni.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. 2009. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7:e54. 10.1371/journal.pbio.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, C.W., and Raulet D.H.. 2001. Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol. 13:465–470. 10.1016/S0952-7915(00)00242-9 [DOI] [PubMed] [Google Scholar]
- McWilliam, H.E., Eckle S.B., Theodossis A., Liu L., Chen Z., Wubben J.M., Fairlie D.P., Strugnell R.A., Mintern J.D., McCluskey J., et al. 2016. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 17:531–537. 10.1038/ni.3416 [DOI] [PubMed] [Google Scholar]
- Miyazaki, Y., Miyake S., Chiba A., Lantz O., and Yamamura T.. 2011. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int. Immunol. 23:529–535. 10.1093/intimm/dxr047 [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE) . 2022. C.C.G.U., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7 [Google Scholar]
- Murshudov, G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., and Vagin A.A.. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67:355–367. 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, G.A., McLaren J.E., Meermeier E.W., Ladell K., Swarbrick G.M., Price D.A., Tran J.G., Worley A.H., Vogt T., Wong E.B., and Lewinsohn D.M.. 2020. The MAIT TCRβ chain contributes to discrimination of microbial ligand. Immunol. Cell Biol. 98:770–781. 10.1111/imcb.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, O., Kjer-Nielsen L., Le Nours J., Eckle S.B., Birkinshaw R., Beddoe T., Corbett A.J., Liu L., Miles J.J., Meehan B., et al. 2013. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4:2142. 10.1038/ncomms3142 [DOI] [PubMed] [Google Scholar]
- Perroteau, J., Navet B., Devilder M.C., Hesnard L., Scotet E., Gapin L., Saulquin X., and Gautreau-Rolland L.. 2020. Contribution of the SYK Tyrosine kinase expression to human iNKT self-reactivity. Eur. J. Immunol. 50:1454–1467. 10.1002/eji.201948416 [DOI] [PubMed] [Google Scholar]
- Picelli, S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., and Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Porcelli, S., Brenner M.B., Greenstein J.L., Balk S.P., Terhorst C., and Bleicher P.A.. 1989. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 341:447–450. 10.1038/341447a0 [DOI] [PubMed] [Google Scholar]
- Porcelli, S., Yockey C.E., Brenner M.B., and Balk S.P.. 1993. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J. Exp. Med. 178:1–16. 10.1084/jem.178.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon, R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z., Eckle S.B.G., Uldrich A.P., Birkinshaw R.W., Patel O., et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210:2305–2320. 10.1084/jem.20130958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, D.R., and Cheatham T.E. III. 2013. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 9:3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Rouxel, O., DaSilva J., Beaudoin L., Nel I., Tard C., Cagninacci L., Kiaf B., Oshima M., Diedisheim M., Salou M., et al. 2018. Author Correction: Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat. Immunol. 19:1035. 10.1038/s41590-017-0023-9 [DOI] [PubMed] [Google Scholar]
- Rudolph, M.G., Stanfield R.L., and Wilson I.A.. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466. 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Salio, M., Gasser O., Gonzalez-Lopez C., Martens A., Veerapen N., Gileadi U., Verter J.G., Napolitani G., Anderson R., Painter G., et al. 2017. Activation of human mucosal-associated invariant T cells induces CD40L-dependent maturation of monocyte-derived and primary dendritic cells. J. Immunol. 199:2631–2638. 10.4049/jimmunol.1700615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaler, M., Colone A., Spagnuolo J., Zimmermann M., Lepore M., Kalinichenko A., Bhatia S., Cottier F., Rutishauser T., Pavelka N., et al. 2018. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol. 11:1060–1070. 10.1038/s41385-018-0020-9 [DOI] [PubMed] [Google Scholar]
- Seach, N., Guerri L., Le Bourhis L., Mburu Y., Cui Y., Bessoles S., Soudais C., and Lantz O.. 2013. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol. 191:6002–6009. 10.4049/jimmunol.1301212 [DOI] [PubMed] [Google Scholar]
- Sharma, P.K., Wong E.B., Napier R.J., Bishai W.R., Ndung’u T., Kasprowicz V.O., Lewinsohn D.A., Lewinsohn D.M., and Gold M.C.. 2015. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology. 145:443–453. 10.1111/imm.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L.F., Lee T.S., Zhu C., York D.M., and Merz K.M. Jr. 2019. Using AMBER18 for relative free energy calculations. J. Chem. Inf. Model. 59:3128–3135. 10.1021/acs.jcim.9b00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter, M.N.T., Awad W., Li S., Pediongco T.J., Meehan B.S., Meehan L.J., Tian Z., Zhao Z., Wang H., Nelson A., et al. 2022. CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells. J. Exp. Med. 219:e20210828. 10.1084/jem.20210828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B.R., III, McGee T.D. Jr, Swails J.M., Homeyer N., Gohlke H., and Roitberg A.E.. 2012. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 8:3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Tilloy, F., Treiner E., Park S.H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., and Lantz O.. 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189:1907–1921. 10.1084/jem.189.12.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubal, A., Kiaf B., Beaudoin L., Cagninacci L., Rhimi M., Fruchet B., da Silva J., Corbett A.J., Simoni Y., Lantz O., et al. 2020. Mucosal-associated invariant T cells promote inflammation and intestinal dysbiosis leading to metabolic dysfunction during obesity. Nat. Commun. 11:3755. 10.1038/s41467-020-17307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner, E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., and Lantz O.. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 422:164–169. 10.1038/nature01433 [DOI] [PubMed] [Google Scholar]
- Vanquelef, E., Simon S., Marquant G., Garcia E., Klimerak G., Delepine J.C., Cieplak P., and Dupradeau F.Y.. 2011. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 39:W511–W517. 10.1093/nar/gkr288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wolf R.M., Caldwell J.W., Kollman P.A., and Case D.A.. 2004. Development and testing of a general amber force field. J. Comput. Chem. 25:1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Williams, C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. 2018. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27:293–315. 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, G., Lobley C.M., and Prince S.M.. 2013. Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69:1260–1273. 10.1107/S0907444913015308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z., Cong Y., Huang K., Zhong S., Zhang J.Z.H., and Duan L.. 2019. Drug-resistance mechanisms of three mutations in anaplastic lymphoma kinase against two inhibitors based on MM/PBSA combined with interaction entropy. Phys. Chem. Chem. Phys. 21:20951–20964. 10.1039/C9CP02851J [DOI] [PubMed] [Google Scholar]
- Young, M.H., U’Ren L., Huang S., Mallevaey T., Scott-Browne J., Crawford F., Lantz O., Hansen T.H., Kappler J., Marrack P., and Gapin L.. 2013. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS One. 8:e53789. 10.1371/journal.pone.0053789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Irving M.B., and Michielin O.. 2010. MM-GBSA binding free energy decomposition and T cell receptor engineering. J. Mol. Recognit. 23:142–152. 10.1002/jmr.1005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows data collection and refinement statistics.
shows contacts between MR1-5-OP-RU and TCRs.
shows contacts between the E8 TCR and MR1 with different ligands.
shows histidine tautomerization states used for simulations of the E8 TCR and AF7 TCR in complex with MR1.
shows duplicate Biacore data for E8 TCR and AF-7 TCR binding to MR1 loaded with an array of ligands and the empty MR1 K43A mutant.
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
Data Availability Statement
The crystal structures of E8 TCR and MR1–ligand complexes have been deposited with the PDB under Accessions # 7ZT2, 7ZT3, 7ZT4, 7ZT5, 7ZT7, 7ZT8, and 7ZT9. T cell clones are available from G. De Libero under a material transfer agreement with the University of Basel. New TCR sequences in this manuscript are available from G. De Libero under a material transfer agreement with the University of Basel. All other data are available in the article itself and its supplementary materials.