Abstract
Mitochondrial DNA (mtDNA) is a small, circular, double-stranded DNA inherited from the mother during fertilization. Evolutionary evidence supported by the endosymbiotic theory identifies mitochondria as an organelle that could have descended from prokaryotes. This may be the reason for the independent function and inheritance pattern shown by mtDNA. The unstable nature of mtDNA due to the lack of protective histones, and effective repair systems make it more vulnerable to mutations. The mtDNA and its mutations could be maternally inherited thereby predisposing the offspring to various cancers like breast and ovarian cancers among others. Although mitochondria are considered heteroplasmic wherein variations among the multiple mtDNA genomes are noticed, mothers can have mitochondrial populations that are homoplasmic for a given mitochondrial mutation. Homoplasmic mitochondrial mutations may be transmitted to all maternal offspring. However, due to the complex interplay between the mitochondrial and nuclear genomes, it is often difficult to predict disease outcomes, even with homoplasmic mitochondrial populations. Heteroplasmic mtDNA mutations can be maternally inherited, but the proportion of mutated alleles differs markedly between offspring within one generation. This led to the genetic bottleneck hypothesis, explaining the rapid changes in allele frequency witnessed during the transmission of mtDNA from one generation to the next. Although a physical reduction in mtDNA has been demonstrated in several species, a comprehensive understanding of the molecular mechanisms is yet to be demonstrated. Despite initially thought to be limited to the germline, there is evidence that blockages exist in different cell types during development, perhaps explaining why different tissues in the same organism contain different levels of mutated mtDNA. In this review, we comprehensively discuss the potential mechanisms through which mtDNA undergoes mutations and the maternal mode of transmission that contributes to the development of tumors, especially breast and ovarian cancers.
Keywords: cancer, inheritance, mtdna, endosymbiotic theory, mitochondrial dna
Introduction and background
Mitochondrial deoxyribonucleic acid (mtDNA) is a small circular DNA found within mitochondria present in the cytoplasm of a cell. This DNA is supplementary to the nucleic acid material found in the nucleus of each cell. The mtDNA codes for 37 genes that promote the proper functioning of some cells. The mitochondria synthesize adenosine triphosphate (ATP) through oxidative phosphorylation and encode information for the synthesis of enzymes, transfer ribonucleic acid (tRNA), and ribosomal RNA (rRNA) [1]. Disorders of mtDNA and mutations in its genes can predispose to health problems like age-related hearing loss, diabetes, and brain, heart, and liver failure, among other conditions [2]. Moreover, mtDNA and its associated mitochondrial disorders can predispose people to different types of cancers including lymphomas, leukemias, and breast, intestine, liver, and kidney tumors, among others [1, 3]. However, the mechanisms behind carcinogenesis are not yet adequately elaborated.
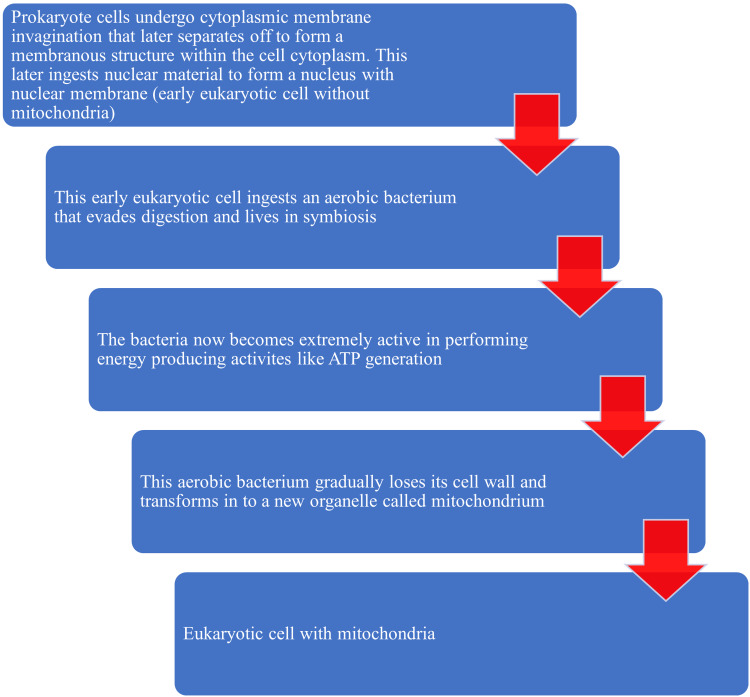
From the results of evolutionary and genetic studies, mtDNA has been shown to be inherited from the mother during fertilization [4]. Additionally, studies have observed some important characteristics of mtDNA like high mutation rates, increased copy numbers in a cell, and unable to undergo recombination [5-7]. According to endosymbiotic theory, the origin of eukaryotic cells has been tracked back to the prokaryotes. It was Lynn Margulis who proposed the endosymbiotic theory in the 1960s [8]. This theory puts forth a gradual process that occurs over a long period of time for the development of eukaryotic cells from the prokaryotes [9]. The mitochondria, plastids, and chloroplasts could have descended from free-living prokaryotes [9]. The endosymbiotic theory presumes that complex multicellular organisms including humans have a small mitochondrial genome that is being generated and transferred through evolution, probably from the prokaryotes [10]. It has been hypothesized that some of the organelles of eukaryotic cells could have developed from the prokaryotes like bacteria. The early eukaryotic cells lacked mitochondria, but during evolution, these cells may have ingested the aerobic bacteria that had started living in symbiosis. During the course of evolution, the ingested aerobic bacteria lost their cell walls and may have developed into mitochondria [11]. The diagrammatic representation of the proposed evolution of mtDNA is shown in Figure 1.
Figure 1. Endosymbiont theory depicting the evolution of mitochondria from prokaryotes.
Note: This figure has been created by the authors
ATP, adenosine triphosphate
Review
Human mtDNA contains 16,569 base pairs and encodes 13 proteins. The mtDNA has a heavy strand and a light strand wherein the heavy strand is guanine rich and encodes 12 subunits. The light strand is rich in cytosine and encodes one subunit that performs oxidative phosphorylation. The regulation of the entire mtDNA genome is at the site of origin of replication of these strands [12]. The hyper diversity is the reason for the high mutational rate seen in mtDNA and vice versa. This hyper-diverse mtDNA causes unusual variations in DNA sequence, promoting coding variations, gene rearrangements, recombination, and finally high mtDNA substitution rates [13]. The initial transcription of mtDNA takes place on a displacement loop (D-loop) that has unusual bases like dihydro uracil, and hence it is the more susceptible region to mutations [14-15].
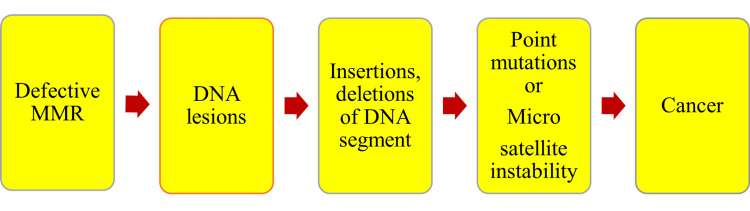
Generally, the hypervariable regions help in the anchoring of proteins to the membrane, protein-protein interactions, and protein signaling not specific to mtDNA. These regions have variations in tandem repeats seen mostly on the D-loop of mtDNA. Tandem repeats are frequently observed sequences on DNA. They are more prone to mispairing of bases known as ‘slipped strand mispairing.’ This may be the reason for the inappropriate mismatch repair (MMR) seen in mtDNA [16]. It was assumed that the eukaryotic mtDNA has an inefficient repair system but recent identification of the mutHLS (consisting of mutH, mutL, and mutS mutation detection and repair proteins) system has confirmed the potential role of mutHLS system in the repair process. Within the mtDNA, mutHLS repairs mismatch, strand break, insertion, and deletion of bases repair through endonuclease activity [17-18].
Less efficient MMR is seen in mtDNA compared to nuclear DNA [19]. Therefore, mtDNA could suffer from lesions, undergo deletions and insertions, and develop mutations that could predispose to carcinogenesis. The identification of mismatches and initiation of repair is very much required for genome stability. Defective MMR leads to the loss of gene activities. This further leads to microsatellite instability, elevates spontaneous mutation rate, and accelerates tumor development and progression as shown in Figure 2.
Figure 2. Effects of mismatch repair on mitochondrial DNA mutations and cancer.
Note: This figure has been created by the authors
MMR, mismatch repair; DNA, deoxyribonucleic acid
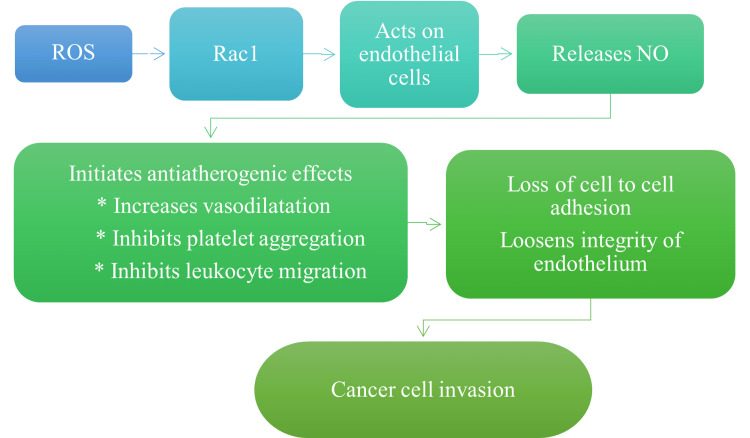
Another mechanism by which the defects in mtDNA can cause carcinogenesis is the generation of reactive oxygen species (ROS) during the process of oxidative phosphorylation [20-21]. Production of ROS by the mitochondria further leads to alterations in cellular vitality and metabolite concentrations. In this process, the ROS stimulates the Rac1 (Ras-related C3 botulinum toxin substrate 1) protein which acts on the vascular endothelial cells. This causes the release of nitric oxide (NO) that results in antiatherogenic effects like vasodilatation and inhibition of platelet and leukocyte aggregation. Moreover, this process results in the loss of cell-to-cell adhesion and loosens the endothelial integrity, and transforms the normal cells into cancer cells. The method of ROS-dependent carcinogenesis is shown in Figure 3.
Figure 3. Role of ROS in the development of cancer.
Note: This figure has been created by the authors
ROS, reactive oxygen species; Rac1, Ras-related C3 botulinum toxin substrate 1; NO, nitric oxide
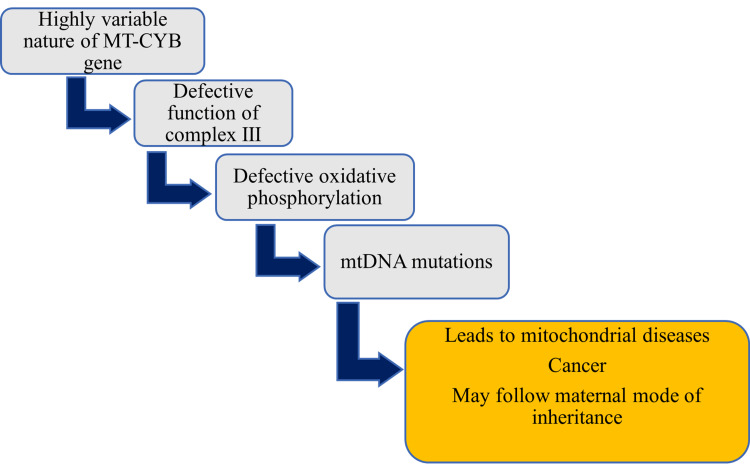
The nuclear genome supplies protein for mitochondrial replication and gene expression. The mutations to these nuclear genomes not only alter mitochondrial function but also influences carcinogenesis and progression. It was found that the D-loop region and mitochondrial cytochrome b (MT-CYB) gene are specific sites of mtDNA where mutations are common [22-23]. The D-loop of mtDNA is required for transcription and gene expression. The strand/primer for initiation of transcription provides an additional strand that makes it a triple-strand DNA structure. This makes mtDNA more susceptible to the mutation of somatic cells that invariably lead to cancers [24]. This influences copy number of cells (variations in a repeated sequence of genes) as well as the regulation of mitochondria.
The MT-CYB gene provides the necessary information for the synthesis of cytochrome b protein of complex III involved in oxidative phosphorylation [20, 23]. This highly variable nature of the MT-CYB gene is also the reason for the development of cancers. It was identified that oral cancers are primarily due to mtDNA mutations [23]. But it is unclear whether it is D-loop region or MT-CYB gene mutations that lead to pathological consequences in cancer. Hence it was suggested that mtDNA could be used as a biomarker for the early identification of cancers [23]. The role of the MT-CYB gene in the development of mitochondrial diseases and cancers is shown in Figure 4.
Figure 4. Role of MT-CYB gene in the development of mitochondrial diseases and cancer.
Note: This figure has been created by the authors
MT-CYB, mitochondrial cytochrome b; mtDNA, mitochondrial DNA
mtDNA and maternal inheritance
The mitochondrial genome is independent of its inheritance pattern compared to chromosomal inheritance. Recent research had identified that paternal mtDNA is eliminated through various mechanisms which include mitophagy, and autophagy-independent degradation, among others. In mitophagy, aged and dysfunctional mitochondria are removed by lysosomal activity and/or proteasomal pathways at the embryonal stage [25-26]. Animal studies have suggested that the paternal mitochondria and the mtDNA could be eliminated by autophagosome-based sperm organelle mechanisms [27-29]. Other mechanisms like mitochondrial endonuclease-mediated elimination of paternal mtDNA have also been implicated in the maternal inheritance of mtDNA [30].
This enables uniparental inheritance which in turn keeps deleterious mutations of DNA genes at their location whilst restricting their spread [31-32]. Mitochondrial DNA (mtDNA) also contributes to the inheritance of cancer-susceptible alleles, loss of heterozygosity (two different genes) of inherited and mutated susceptible genes, and inactivation of tumor suppressor genes by deletion, insertion, or gene rearrangement. Additionally, biallelic inactivation (recessive), and heteroplasmy of mtDNA hold both abnormal and normal mitochondrial genomes due to unequal segregation [33-34]. The heteroplasmic nature of mtDNA leads to a genetic bottleneck which predicts that little mtDNA is transferred to the next generation limiting genetic variations in the family [35-36].
In a recent observation, it has been identified that the mitochondria meet the endoplasmic reticulum (ER) for the exchange of nutrients like calcium and glucose. Additionally, ER has been noted to assist in mitochondrial fusion and fission. Dysplasia of mitochondria in the neurons was found to predispose children to hereditary conditions like spastic paraplegias [37-38].
Influence of mtDNA in ovarian cancer and breast cancer
Current evidence suggests that there is an increasing frequency of hereditary breast and ovarian cancers [39]. Additionally, there is an increased awareness of gene mutations that could predispose to the familial transfer of cancers [40]. However, there is a scarcity of such information regarding the role of mtDNA and its influence on the development of cancers. The D-loop of mtDNA is more prone to mutations in breast and ovarian cancers. The mitochondrial microsatellite instability (mtMSI) and variations in CA (cytosine, adenine) repeats were also observed in ovarian and breast carcinoma. The inefficient MMR after malignant transformation may contribute to mtMSI. The mtMSI is observed in mononucleotide repeats at nucleotide positions 303-309. This region is the site where replication primer binds, making a defective DNA repair system. In a previous study, it was found that homo polymorphic nucleotide (variant forms of specific nucleotide sequence) tracts of mtDNA are more prone to errors due to the low frameshift (addition/deletion of bases) mutations. Moreover, the fidelity of DNA polymerase gamma (PLOG) that carries out the mtDNA replication process is lower than that found in the nucleus and is not severely affected [41-46].
In a recent report, it was noticed that mitochondria may play a significant role in the development, metastasis, chemotherapy resistance, and treatment of ovarian cancers [47]. The mitochondrial ribosomal proteins (MRPs) including the MRPL15 were found to be associated with the development of ovarian cancer. Moreover, this protein was found to be efficient in predicting the prognosis [48]. Damage to the mitochondria present in the ovarian germ and somatic cells may lead to the transfer of mitochondrial non-coding RNAs (ncRNAs) into the nucleus. This could predispose to early ovarian aging and resultant adverse effects including hormonal imbalances and tumorigenesis [49].
The abnormal transmission of mtDNA into the nuclear DNA was found to correlate with the development of breast cancer [50]. An association of mitochondrial D310 mutation was found to correlate with the development of various solid organ cancers including breast cancer [51].
Current research perspectives
Two different mtDNA variants/mutations were identified wherein the mutations that cause the formation of tumors are called inducers and those mtDNA mutations that facilitate the survival of tumor cells are called adaptors. The mtDNA mutations may be of three types based on their origins including those which are inherited and undergo familial transfer, mutations that happen within the cell, and ancient but transferable mtDNA variations [52]. Recent research had also suggested that better mitochondrial health can be instrumental in preventing replication errors and regulating apoptotic activities [53].
Studies have noted that mtDNA mutations, dysregulation of mtDNA, and disturbances in the communication between nuclear DNA and mtDNA may affect cellular health and in turn, predispose to carcinogenesis [54]. A recent hypothesis also suggested that the mitochondria may under the influence of microbial pathogens can transform themselves into super-power cells or immortal cancer cells that enable tumor formation [55]. In view of the potential role of mtDNA in aging and diseases including cancers, the role of gene editing in therapeutic management was being explored [56].
The aldehyde dehydrogenase 2 (ALDH2), a mitochondrial enzyme was found to regulate acetaldehyde toxicity within the cell and was suggested as a potential biomarker for diagnosis, metastasis, and prognosis of cancer. Additionally, abnormalities in ALDH2 and associated gene polymorphisms (single nucleotide polymorphisms-rs671) were noted to influence tumorigenesis and therefore should be considered as potential therapeutic targets [57].
Drugs like nilotinib, salinomycin, tigecycline, and eupatilin among others targeting mitochondrial dysfunctional pathways are being explored for their role in the treatment and management of ovarian cancer [58]. Given that the functional characteristics of somatic mtDNA are not sufficiently elucidated, it is impractical to understand the variations and mutations of mtDNA and its relationship with cancers [59-61].
Conclusions
The mtDNA is inherited uniparentally probably owing to the loss of paternal mtDNA during fertilization. The unique nature of mtDNA to synthesize its proteins and its independent inheritance patterns makes it a strong maternal factor. The unstable nature of replication machinery with a defective repair system makes mtDNA more prone to mutations and tumorigenesis. The inheritance patterns seen in ovarian and breast cancers are solely dependent on the frequency of mtDNA mutations and their ability to utilize different repair mechanisms in various tumor cells. Therefore, it is extremely essential for the researchers and the physicians involved in cancer diagnosis, treatment, and management to improve the understanding of the potential role of mtDNA in the development, diagnosis, prevention, and cure of cancers.
The authors have declared that no competing interests exist.
References
- 1.Mitochondrial DNA. [ Feb; 2023 ]. 2020. https://medlineplus.gov/genetics/chromosome/mitochondrial-dna/ https://medlineplus.gov/genetics/chromosome/mitochondrial-dna/
- 2.The mitochondrion: a perpetrator of acquired hearing loss. Böttger EC, Schacht J. Hear Res. 2013;303:12–19. doi: 10.1016/j.heares.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The role of mitochondria in carcinogenesis. Kozakiewicz P, Grzybowska-Szatkowska L, Ciesielka M, et al. Int J Mol Sci. 2021;22:5100. doi: 10.3390/ijms22105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maternal inheritance of mammalian mitochondrial DNA. Hutchison CA 3rd, Newbold JE, Potter SS, et al. Nature. 1974;251:536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- 5.Maternal ancestry and population history from whole mitochondrial genomes. Kivisild T. Investig Genet. 2015;6:3. doi: 10.1186/s13323-015-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Wei W, Schon KR, Elgar G, et al. Nature. 2022;611:105–114. doi: 10.1038/s41586-022-05288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitochondrial DNA and human evolution. Pakendorf B, Stoneking M. Annu Rev Genomics Hum Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- 8.On the origin of mitosing cells. Sagan L. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 9.Lynn Margulis and the origin of the eukaryotes. Cornish-Bowden A. J Theor Biol. 2017;434:1. doi: 10.1016/j.jtbi.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 10.Sequence and organization of the human mitochondrial genome. Anderson S, Bankier AT, Barrell BG, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 11.The endosymbiotic theory. [ Apr; 2023 ]. 2022. https://bio.libretexts.org/Bookshelves/Microbiology/Microbiology_(Kaiser)/Unit_4%3A_Eukaryotic_Microorganisms_and_Viruses/07%3A_The_Eukaryotic_Cell/7.8%3A_The_Endosymbiotic_Theory pp. 3–2023.https://bio.libretexts.org/Bookshelves/Microbiology/Microbiology_(Kaiser)/Unit_4%3A_Eukaryotic_Microorganisms_and_Viruses/07%3A_The_Eukaryotic_Cell/7.8%3A_The_Endosymbiotic_Theory
- 12.Relation between mitochondrial DNA hyperdiversity, mutation rate and mitochondrial genome evolution in Melarhaphe neritoides (Gastropoda: Littorinidae) and other Caenogastropoda. Fourdrilis S, de Frias Martins AM, Backeljau T. Sci Rep. 2018;8:17964. doi: 10.1038/s41598-018-36428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The mitochondrial genome: structure, transcription, translation and replication. Taanman JW. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 14.Clinical significance of the D-loop gene mutation in mitochondrial DNA in laryngeal cancer. Wang L, Cheng HX, Zhou YH, et al. Onco Targets Ther. 2021;14:3461–3466. doi: 10.2147/OTT.S304836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molecular origins of rapid and continuous morphological evolution. Fondon JW 3rd, Garner HR. Proc Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Why do mammalian mitochondria possess a mismatch repair activity? Mason PA, Lightowlers RN. FEBS Lett. 2003;554:6–9. doi: 10.1016/s0014-5793(03)01169-4. [DOI] [PubMed] [Google Scholar]
- 17.DNA mismatch repair and its many roles in eukaryotic cells. Liu D, Keijzers G, Rasmussen LJ. Mutat Res Rev Mutat Res. 2017;773:174–187. doi: 10.1016/j.mrrev.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Repair of mtDNA in vertebrates. Bogenhagen DF. Am J Hum Genet. 1999;64:1276–1281. doi: 10.1086/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitochondria and mitochondrial ROS in cancer: novel targets for anticancer therapy. Yang Y, Karakhanova S, Hartwig W, et al. J Cell Physiol. 2016;231:2570–2581. doi: 10.1002/jcp.25349. [DOI] [PubMed] [Google Scholar]
- 20.Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Dasgupta S, Hoque MO, Upadhyay S, et al. Cancer Res. 2008;68:700–706. doi: 10.1158/0008-5472.CAN-07-5532. [DOI] [PubMed] [Google Scholar]
- 21.Number of somatic mutations in the mitochondrial D-loop region indicates poor prognosis in breast cancer, independent of TP53 mutation. Kuo SJ, Chen M, Ma GC, et al. Cancer Genet Cytogenet. 2010;201:94–101. doi: 10.1016/j.cancergencyto.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 22.A neonatal polyvisceral failure linked to a de novo homoplasmic mutation in the mitochondrially encoded cytochrome b gene. Fragaki K, Procaccio V, Bannwarth S, et al. Mitochondrion. 2009;9:346–352. doi: 10.1016/j.mito.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Validation of next-generation sequencing of entire mitochondrial genomes and the diversity of mitochondrial DNA mutations in oral squamous cell carcinoma. Kloss-Brandstätter A, Weissensteiner H, Erhart G, et al. PLoS One. 2015;10:0. doi: 10.1371/journal.pone.0135643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitochondria and the hallmarks of cancer. Giampazolias E, Tait SW. FEBS J. 2016;283:803–814. doi: 10.1111/febs.13603. [DOI] [PubMed] [Google Scholar]
- 25.Degradation of paternal mitochondria via mitophagy. Sasaki T, Sato M. Biochim Biophys Acta Gen Subj. 2021;1865:129886. doi: 10.1016/j.bbagen.2021.129886. [DOI] [PubMed] [Google Scholar]
- 26.Allophagy, or how the embryo eliminates mitochondria and other paternal organelles (Article in French) Rawi SA, Galy V. Med Sci (Paris) 2012;28:343–346. doi: 10.1051/medsci/2012284003. [DOI] [PubMed] [Google Scholar]
- 27.Autophagosomal sperm organelle clearance and mtDNA inheritance in C. elegans. Merlet J, Rubio-Peña K, Al Rawi S, et al. Adv Anat Embryol Cell Biol. 2019;231:1–23. doi: 10.1007/102_2018_1. [DOI] [PubMed] [Google Scholar]
- 28.Multiple roles of endocytosis and autophagy in intracellular remodeling during oocyte-to-embryo transition. Sato K. Proc Jpn Acad Ser B Phys Biol Sci. 2022;98:207–221. doi: 10.2183/pjab.98.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post-fertilisation sperm mitophagy: the tale of mitochondrial eve and Steve. Sutovsky P, Song WH. Reprod Fertil Dev. 2017;30:56–63. doi: 10.1071/RD17364. [DOI] [PubMed] [Google Scholar]
- 30.Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Zhou Q, Li H, Li H, et al. Science. 2016;353:394–399. doi: 10.1126/science.aaf4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitochondrial dynamics in health and disease. Yapa NM, Lisnyak V, Reljic B, et al. FEBS Lett. 2021;595:1184–1204. doi: 10.1002/1873-3468.14077. [DOI] [PubMed] [Google Scholar]
- 32.Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Sato M, Sato K. Autophagy. 2012;8:424–425. doi: 10.4161/auto.19243. [DOI] [PubMed] [Google Scholar]
- 33.Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. Passamonti M, Ghiselli F. DNA Cell Biol. 2009;28:79–89. doi: 10.1089/dna.2008.0807. [DOI] [PubMed] [Google Scholar]
- 34.Two genetic hits (more or less) to cancer. Knudson AG. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 35.Biparental inheritance of mitochondrial DNA in humans. Luo S, Valencia CA, Zhang J, et al. Proc Natl Acad Sci USA. 2018;115:13039–13044. doi: 10.1073/pnas.1810946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A genetic bottleneck of mitochondrial DNA during human lymphocyte development. Tang Z, Lu Z, Chen B, et al. Mol Biol Evol. 2022;39:0. doi: 10.1093/molbev/msac090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reorganization, specialization, and degradation of oocyte maternal components for early development. Satouh Y, Sato K. Reprod Med Biol. 2023;22:0. doi: 10.1002/rmb2.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipids in the physiopathology of hereditary spastic paraplegias. Darios F, Mochel F, Stevanin G. Front Neurosci. 2020;14:74. doi: 10.3389/fnins.2020.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hereditary breast and ovarian cancer (HBOC): review of its molecular characteristics, screening, treatment, and prognosis. Yoshida R. Breast Cancer. 2021;28:1167–1180. doi: 10.1007/s12282-020-01148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Implementation of multigene panel testing for breast and ovarian cancer in South Africa: a step towards excellence in oncology for the public sector. van der Merwe NC, Ntaita KS, Stofberg H, et al. Front Oncol. 2022;12:938561. doi: 10.3389/fonc.2022.938561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The mitochondrial DNA genetic bottleneck: inheritance and beyond. Zhang H, Burr SP, Chinnery PF. Essays Biochem. 2018;62:225–234. doi: 10.1042/EBC20170096. [DOI] [PubMed] [Google Scholar]
- 42.Frequent mutations in the mitochondrial control region DNA in breast tissue. Rosson D, Keshgegian AA. Cancer Lett. 2004;215:89–94. doi: 10.1016/j.canlet.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 43.Ovarian cancer: a landscape of mitochondria with emphasis on mitochondrial dynamics. De Rasmo D, Cormio A, Cormio G, et al. Int J Mol Sci. 2023;24:1224. doi: 10.3390/ijms24021224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Research highlights on contributions of mitochondrial DNA microsatellite instability in solid cancers - an overview. Yusoff AA, Radzak SM, Khair SZ. Contemp Oncol (Pozn) 2022;26:8–26. doi: 10.5114/wo.2022.115674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. Longley MJ, Nguyen D, Kunkel TA, et al. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 46.A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. Lee YS, Johnson KA, Molineux IJ, et al. J Biol Chem. 2010;285:28105–28116. doi: 10.1074/jbc.M110.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitochondrial integration and ovarian cancer chemotherapy resistance. Shen L, Xia M, Zhang Y, et al. Exp Cell Res. 2021;401:112549. doi: 10.1016/j.yexcr.2021.112549. [DOI] [PubMed] [Google Scholar]
- 48.MRPL15 is a novel prognostic biomarker and therapeutic target for epithelial ovarian cancer. Xu H, Zou R, Li F, et al. Cancer Med. 2021;10:3655–3673. doi: 10.1002/cam4.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovarian aging: role of pituitary-ovarian axis hormones and ncRNAs in regulating ovarian mitochondrial activity. Colella M, Cuomo D, Peluso T, et al. Front Endocrinol (Lausanne) 2021;12:791071. doi: 10.3389/fendo.2021.791071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Ju YS, Tubio JM, Mifsud W, et al. Genome Res. 2015;25:814–824. doi: 10.1101/gr.190470.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitochondrial D310 mutation as clonal marker for solid tumors. Geurts-Giele WR, Gathier GH, Atmodimedjo PN, et al. Virchows Arch. 2015;467:595–602. doi: 10.1007/s00428-015-1817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitochondrial DNA variation and cancer. Kopinski PK, Singh LN, Zhang S, et al. Nat Rev Cancer. 2021;21:431–445. doi: 10.1038/s41568-021-00358-w. [DOI] [PubMed] [Google Scholar]
- 53.Metabolic health, mitochondrial fitness, physical activity, and cancer. Clemente-Suárez VJ, Martín-Rodríguez A, Redondo-Flórez L, et al. Cancers (Basel) 2023;15:814. doi: 10.3390/cancers15030814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Functional role of mitochondrial DNA in cancer progression. Lin YH, Lim SN, Chen CY, et al. Int J Mol Sci. 2022;23:1659. doi: 10.3390/ijms23031659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.From pathogens to cancer: are cancer cells evolved mitochondrial super cells? Balzanelli MG, Distratis P, Lazzaro R, et al. Diagnostics (Basel) 2023;13:813. doi: 10.3390/diagnostics13040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The mitochondrial genome in aging and disease and the future of mitochondrial therapeutics. Saravanan S, Lewis CJ, Dixit B, et al. Biomedicines. 2022;10:490. doi: 10.3390/biomedicines10020490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The role of ALDH2 in tumorigenesis and tumor progression: targeting ALDH2 as a potential cancer treatment. Zhang H, Fu L. Acta Pharm Sin B. 2021;11:1400–1411. doi: 10.1016/j.apsb.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitochondrial dysfunction pathway alterations offer potential biomarkers and therapeutic targets for ovarian cancer. Shen L, Zhan X. Oxid Med Cell Longev. 2022;2022:5634724. doi: 10.1155/2022/5634724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitochondrial DNA: the overlooked oncogenome? Gammage PA, Frezza C. BMC Biol. 2019;17:53. doi: 10.1186/s12915-019-0668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitochondrial DNA is a major source of driver mutations in cancer. Kim M, Mahmood M, Reznik E, et al. Trends Cancer. 2022;8:1046–1059. doi: 10.1016/j.trecan.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tumour mitochondrial DNA mutations drive aerobic glycolysis to enhance checkpoint blockade. Mahmood M, Liu EM, Shergold AL, et al. bioRxiv. 2023 doi: 10.1038/s43018-023-00721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]