Abstract
Parasites sometimes expand their host range and cause new disease aetiologies. Genetic changes can then occur due to host-specific adaptive alterations, particularly when parasites cross between evolutionarily distant hosts. Characterizing genetic variation in Cryptosporidium from humans and other animals may have important implications for understanding disease dynamics and transmission. We analyse sequences from four loci (gp60, HSP-70, COWP and actin) representing multiple Cryptosporidium species reported in humans. We predicted low genetic diversity in species that present unusual human infections due to founder events and bottlenecks. High genetic diversity was observed in isolates from humans of Cryptosporidium meleagridis, Cryptosporidium cuniculus, Cryptosporidium hominis and Cryptosporidium parvum. A deviation of expected values of neutrality using Tajima's D was observed in C. cuniculus and C. meleagridis. The high genetic diversity in C. meleagridis and C. cuniculus did not match our expectations but deviations from neutrality indicate a recent decrease in genetic variability through a population bottleneck after an expansion event. Cryptosporidium hominis was also found with a significant Tajima's D positive value likely caused by recent population expansion of unusual genotypes in humans. These insights indicate that changes in genetic diversity can help us to understand host-parasite adaptation and evolution.
Key words: Cryptosporidiosis, host shift, neutrality, population genetics
Introduction
Understanding the complex dynamics of host shifting in parasites is vital for the effective development of controls to prevent their transmission to people. Cross-species transmission can be influenced by rates of host–parasite contact, host immune response and parasite adaptation (Lloyd-Smith et al., 2009). For many microorganisms, genetic adaptation is often proposed to be the key driver of successful emergence into a new host species, but it is difficult to distinguish which processes generate genetic change (Pepin et al., 2010; Chabas et al., 2018). The genetic variability of parasites in new hosts may provide some insights into the evolutionary processes driving adaptation (Penczykowski et al., 2016). Comparative analysis of parasite genetic diversity within and among hosts could help to clarify transmission-evolution dynamics, as well as their associated risk for host shifts and outbreaks among people. Distinguishing changes in genetic patterns is fundamental for monitoring pathogen evolution, adaptation to new hosts, identification of factors driving cross-species transmission and, ultimately, the reduction of risk to public health.
Cryptosporidium is one of the most common enteric parasites in humans. This parasite causes infant deaths from diarrhoea, as well as disease in wildlife and domestic animals (Kotloff et al., 2013; Striepen, 2013). Most of the approximately 38 species of Cryptosporidium and several related genotypes are adapted to different vertebrate hosts, which limits between-species transmission (Xiao and Feng, 2008; Garcia-R and Hayman, 2016, 2017; Garcia-R et al., 2017; Feng et al., 2018). Humans are the major host of Cryptosporidium hominis and Cryptosporidium viatorum (Ryan et al., 2014), but C. hominis is reported in other mammal species (Widmer et al., 2020), whereas no alternative animal reservoir has been identified for C. viatorum (Elwin et al., 2012; but see Koehler et al., 2018). Nevertheless, about 17 Cryptosporidium species with diverse animals as main hosts are now found in humans at different notification levels. For instance, cattle are an important reservoir of C. parvum, but this species is reported in a significant proportion of human Cryptosporidium infections (Feng et al., 2018). Likewise, Cryptosporidium meleagridis, C. cuniculus, C. tyzzeri and C. ubiquitum (previously known as the cervine genotype) primarily parasitize birds, rabbits, rodents and ruminants, respectively. These species have recently been found in gastroenteritis cases of humans, but are less commonly reported (Chalmers et al., 2009a; Bouzid et al., 2013; Rašková et al., 2013; Li et al., 2014). This apparent expansion of host range demonstrates the plasticity of these species and their ability to adapt to new niches.
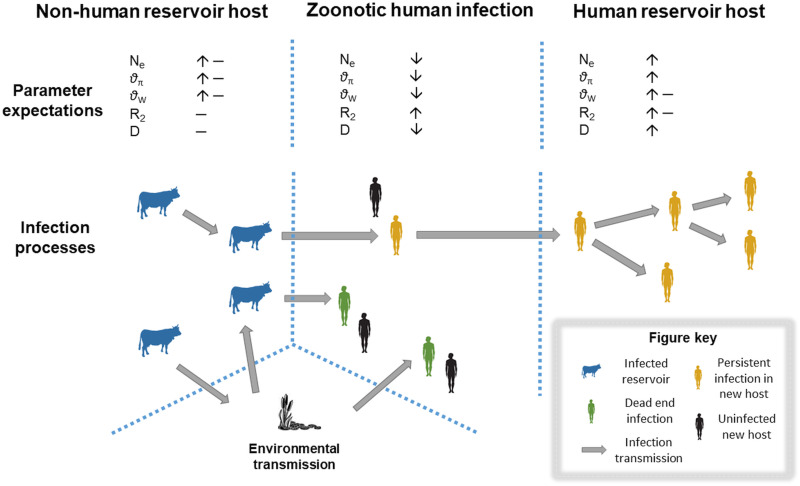
The change to new environments (here, new hosts) must be associated with traits that influence parasite fitness and hence cause signatures in the genetic diversity of the parasites (Barrett et al., 2008). Changes in parasite gene frequencies can be estimated by comparison of genetic variation in the known reservoir host source (where the infection is maintained and perpetuates) and the new host (Fig. 1). The comparison of quantitative genetic differences can help to untangle the processes involved in parasite–host dynamics, and the rates and barriers parasites need to overcome to enable successful cross-species transmissions (Geoghegan et al., 2017). While low genetic diversity in the parasite that colonizes a host does not necessarily imply that an episode of shifting has occurred, it is strongly indicative of genetic factors affecting adaptation and highlights the importance of comparative studies across multiple populations and species (Fig. 1).
Fig. 1.
Population and genetic parameter expectations during Cryptosporidium host shifting. Key transmission pathways show non-human reservoirs (blue) infecting each other and the environment through oocyst shedding. Cross-species transmission and host shifting occur through direct and indirect transmission (e.g. via the environment), mostly leading to dead-end infections (green). Subsequent establishment in new hosts with possible adaptation through ongoing transmission may occur (yellow). Transmission between individuals of the new host is necessary to establish long-term associations that characterize a successful host shift, in contrast to occasional spillover pathogen infections in the new host. The expected ranges (e.g. ↑ = high, ↓ = low, – = no change, ↑– = high or no change) for population genetic parameters are shown above each scenario; Ne – effective population size; θπ – nucleotide diversity; θW – Watterson's theta; R2 – Ramos-Onsins and Rozas' population growth test; and D – Tajima's D.
Cryptosporidium species in a large, well-mixed host population might have high genetic diversity, with their transmission dynamics stable over time (Fig. 1). However, during a parasite population expansion and founder event, a large chain of cascading effects may influence these parameters (Longdon et al., 2014). Infectious doses for humans can be as few as 10 infectious oocysts, each with four diploid sporozoites within the oocysts. Such population bottlenecks typically promote loss of genetic diversity in the parasite population of the new host and generate among-population genetic differences. Upon infection, however, populations may expand massively, with infected hosts excreting up to 6 × 107 oocysts per gram of feces (Uga et al., 2000) and up to 1010 oocysts per day for up to 2 weeks while infected (Meinhardt et al., 1996). The parasite population will reach a recovery point after the colonization event if the host population is large and interconnected (Fig. 1). Although host species differ in many traits, including population size, distribution and resistance, we predict that there is a relationship between the dynamics of host jumps and genetic changes in parasite populations of Cryptosporidium that records important clues to their transmission into new hosts.
Here, we use a population-based study to infer patterns of genetic diversity from host shifting in Cryptosporidium parasites. We study Cryptosporidium genetic variation across host species using publicly available data to discern different likely outcomes of host–parasite dynamics and the cause-effect relationship of Cryptosporidium parasites that host shift. Our aim is to help understand host–parasite shifts, such as for human cryptosporidiosis, by addressing the following questions: can general patterns of changes in genetic diversity be detected in the genes of parasites that host switch? Are genetic diversity measures lower in Cryptosporidium species recently reported in humans? How is the genetic diversity of Cryptosporidium species distributed across different animal hosts?
Methods
Data collection
We use data from previously published sequences of Cryptosporidium species found in GenBank (see Table S1 for details). Here, we defined populations and genetic diversity as genetic variants on six evolutionary lineages of Cryptosporidium (C. hominis, C. parvum, C. meleagridis, C. cuniculus, C. tyzzeri and C. ubiquitum) detected in both humans and other vertebrates around the world. Nucleotide sequences of interest were downloaded using Geneious v.10.2 (Kearse et al., 2012) with a search strategy comprised of fields including species names [‘Organism’] and gene names [‘All Fields’ or ‘Protein Name’]. The 60-kDa glycoprotein (gp60), heat shock protein (HSP-70), oocyst wall protein (COWP) and actin gene (actin) are the most frequent markers used for detection and identification of Cryptosporidium, and we, therefore, focus on these four genes here. Among these markers, gp60 is the only gene considered to be under selection (Widmer, 2009; Abal-Fabeiro et al., 2013) and its adaptive evolution plays a powerful role in pathogen entrenchment across species boundaries. This marker has, however, been readily and extensively adopted as a key component for molecular epidemiological investigations because of its high reliability in the characterization of genotypes and detection of variants within populations (Garcia-R and Hayman, 2017; Garcia-R et al., 2020).
Very short sequences (<200 bp) or sequences overlapping only a short section of the overall alignment were discarded (see Supporting Information). Related meta-information (host and isolation/source) was appended to the final dataset and sequences were organized by species, gene, host and source (feces-only to ensure that samples were obtained only from the specific host). Hosts were chosen according to common names when the scientific name was not available and when the names indicate that sequences are from the given host species. In population genetic studies, assigning individuals to populations may be challenging if there is incomplete knowledge of the sampling design. Due to the low number of samples from geographically and taxonomically distant hosts, we compared the data obtained from humans to a combined dataset of all non-human hosts. The reason for this approach is to test for evidence of diversity estimates supporting our expectations (Fig. 1).
Analyses
Nucleotide diversity (θπ) and Watterson's theta (θW) within hosts were estimated for sequences of Cryptosporidium species associated with humans and other hosts using DnaSP v.5.0 (Librado and Rozas, 2009). The Tajima's D statistic (Tajima, 1989) was used to test the null hypothesis of neutrality and demographic growth was assessed using the R2 test (Ramos-Onsins and Rozas, 2002), both using DnaSP v.5.0. Sensitivity analyses were performed to test the effect of sample size and the number of segregating sites (Subramanian, 2016). We advocate for subsampling the data to ascertain the robustness of the summary statistics using a rarefaction approach for our most data-rich sample, the gp60 gene, which displays a high mutation rate and high level of polymorphisms (Strong et al., 2000).
Results
Sequences used in this study were submitted to GenBank between 2002 and 2017, with the exception of a few COWP gene sequences isolated from humans in previous years. This is important due to nomenclature changes in Cryptosporidium. For instance, C. hominis was referred to as C. parvum genotypes H or 1 before 2002, which can create confusion when assigning sequences to species and influence the results of genetic diversity analyses. In total, we compiled 697 sequences of the gp60 gene from C. cuniculus, C. hominis, C. meleagridis, C. parvum, C. tyzzeri and C. ubiquitum that infect humans, as well as isolates from a range of different hosts (462 sequences from humans and 235 sequences from other animals); 13 sequences of the HSP-70 gene from C. hominis (11 sequences from humans and two sequences from other animals); eight sequences of the actin gene from C. parvum (five sequences from humans and three sequences from other animals); and 46 sequences of the COWP gene from C. parvum (18 sequences from humans and 28 sequences from other animals) (Table S1).
Genetic diversity estimates, including standard deviations, are provided in Table S2. We performed sensitivity analyses and did not find a significant difference between data partitions as a result of reductions in either sample size or the number of segregating sites on θπ and θW (Fig. 2A and B), showing that the estimates of these summaries are robust across a range of sample sizes in this instance. We confirmed that gp60 has the highest variation, but that genetic diversity estimates are not statistically significant among other genes and hosts (Fig. 2C and D).
Fig. 2.
Rarefaction analysis of the stability of genetic diversity measures (θπ and θW) from publicly available data in Cryptosporidium. Outputs derived as a function of downsampling individuals (A) or segregating sites (B) in a number of gp60 sequences. Nucleotide diversity (θπ) and Watterson's estimator (θW), upper and lower respectively, by host (C) and genes (D).
Population parameter estimates yield a high θπ for the gp60 gene in humans infected with C. meleagridis and C. cuniculus in comparison to their other hosts (Fig. 3A and Table S2). In contrast, C. hominis presents high θπ values in its major host group (humans and non-human primates; Fig. 3B) suggesting a bottleneck and likely a dead-end infection in domesticated animals. The more ubiquitous C. parvum has low values in its major host source (cattle) but similar values across several other hosts, even though they cover greater taxonomic distances (Fig. 3C). The intermediate levels across the board of hosts might suggest a sustained infection across a broad host range. The same pattern was found in C. meleagridis and C. tyzzeri with their likely host groups (birds and rodents, respectively) presenting low θπ values (Table S2). Evidence of distortion in the frequency of polymorphism distributions (Tajima's D) was found in two species of Cryptosporidium that have been recently reported in humans (C. meleagridis and C. cuniculus), as well as the type human species (C. hominis), at the gp60 gene (Table 1).
Fig. 3.
Population estimates of Cryptosporidium species infecting humans and other hosts. (A) Estimates of nucleotide diversity (θπ) using the gp60 gene for six Cryptosporidium species infecting humans (red); and variation in nucleotide diversity (θπ) in gp60 in C. hominis (B) and C. parvum (C) among other hosts (blue).
Table 1.
Estimates of nucleotide diversity (θπ), Watterson's estimator (θW), Tajima's D and Ramos-Onsins and Rozas' population growth test (R2) using gp60 and COWP genes for Cryptosporidium species infecting humans and other animal hosts (n ⩾ 4, the minimum number of sequences required to compute these tests). *P < 0.05, ***P < 0.001.
| Gene | Species | Host | θπ | s.d. | θW | s.d. | D | R2 |
|---|---|---|---|---|---|---|---|---|
| gp60 | C. cuniculus | Others | 0.081 | 0.033 | 0.076 | 0.006 | 0.33 | 0.19 |
| Humans | 0.113 | 0.013 | 0.065 | 0.005 | 2.99*** | 0.24 | ||
| C. hominis | Others | 0.1405 | 0.033 | 0.16 | 0.013 | −0.48 | 0.11 | |
| Humans | 0.158 | 0.003 | 0.084 | 0.006 | 2.76* | 0.16 | ||
| C. meleagridis | Others | 0.075 | 0.015 | 0.067 | 0.006 | 0.58 | 0.21 | |
| Humans | 0.19 | 0.033 | 0.123 | 0.011 | 2.23* | 0.21 | ||
| C. parvum | Others | 0.073 | 0.005 | 0.054 | 0.004 | 1.13 | 0.12 | |
| Humans | 0.091 | 0.0064 | 0.085 | 0.0046 | 0.26 | 0.09 | ||
| C. ubiquitum | Others | 0.099 | 0.018 | 0.09 | 0.006 | 0.56 | 0.18 | |
| Humans | 0.053 | 0.01 | 0.046 | 0.0045 | 0.70 | 0.19 | ||
| COWP | C. parvum | Others | 0.0011 | 0.0008 | 0.00414 | 0.0015 | −1.15 | 0.19 |
| Humans | 0.017 | 0.0081 | 0.035 | 0.0045 | −2.08* | 0.16 |
Discussion
Understanding the factors that influence the success of Cryptosporidium to colonize new hosts can help to explain host shifting in humans. The putative main hosts of C. meleagridis (birds), C. cuniculus (rabbits) and C. parvum (cattle) were found to harbour low genetic diversity at the gp60 gene compared to the diversity found in humans. Greater θπ of these parasites in humans did not meet our expectations of high diversity within major hosts and low diversity in new ones (here, humans; Fig. 1). Nonetheless, significant Tajima's D positive values in C. meleagridis and C. cuniculus in humans at the gp60 gene might indicate that these lineages have recently decreased their level of genetic variability through a population bottleneck caused by recent colonization of a new host. These species have lately expanded their range to humans and caused outbreaks worldwide (Chalmers et al., 2009b, 2011; Koehler et al., 2014). An increased number of observed polymorphisms at the gp60 gene must be the result of advantageous adaptations during colonization of the new host and subsequent ‘arms race’, indicating that these parasites are either human-adapted or often cause infection in people (Akiyoshi et al., 2003; Feng et al., 2018).
The genetic diversity of C. hominis observed in people matches our expectations for the reservoir host–parasite relationship. There are genetically diverse C. hominis genotypes identified in humans causing unusual infections (Lebbad et al., 2013, 2018), as well as in other hosts (Plutzer and Karanis, 2009; Feng et al., 2018). This might indicate a predictable relationship between genetic diversity and taxonomic distance of primary and secondary hosts (i.e. similar parasite diversity in closely related hosts). High variable genotypes lead to an increase in genetic polymorphisms that cause a deviation from the theoretical expectation of neutrality. Differences in genetic diversity could be due to higher rates of adaptation in lineages with large population sizes (Feng et al., 2018) that allow the parasite to respond to novel selection pressures from the host(s).
Despite the high genetic diversity in Cryptosporidium species that infect humans, the range of potential evolutionary processes in host–parasite interactions needs to be better understood (Barrett et al., 2008; Longdon et al., 2014). We aim to draw inferences about patterns of host–parasite associations and evolutionary history of episodic host shifting or adaptation within this system (Fig. 1). Nonetheless, we acknowledge that information on the timing of shifting or details of the cross-species transmission events is currently limited. The precise effects of host shifting in Cryptosporidium populations could depend on many factors including the life cycle, population subdivision, sampling strategy (markers under selection pressure or mixed geographical origin of the samples), reproductive mode, genetic recombination, host phylogeny and host resistance to parasites (Lively, 2010; Wang et al., 2014), among others.
Sampling strategies play a key role in observed population summary statistics and we need to study the actual evolutionary units in time and space. Unfortunately, publicly available data are invariably found in scattered locations due to piecemeal submission of sequences to genetic databases and different questions and research designs from the original studies. The scale at which Cryptosporidium species adapt to new hosts might depend on the properties and dynamics of the given system. For instance, a species with a time-dependent association for host adaptation and large geographic and host ranges may be more likely to adapt through multiple, geographically-restricted mutations than by global sweeps of local and host-restricted species (Leffler et al., 2012). Studies at multiple scales are required to tease these processes apart, but clearly defining and delimiting the population being studied is essential for accurate estimates of host shifting.
We previously showed that departures from neutrality in Cryptosporidium from humans in New Zealand could be detected through changing epidemiological patterns (Garcia-R and Hayman, 2017). This means that analysing disease patterns by fine-scale surveillance is likely a more powerful way to infer parasite dynamics. With the advent of cheap and rapid sequencing of whole genomes, we expect that understanding genetic diversity and population structure will provide explanatory power to identify the risk of the spread of Cryptosporidium species and ecological fitting into humans (Nader et al., 2019). Ideally, studies using genomes and fine-scale longitudinal sampling before and after the putative host shifting event will help to identify the patterns in genetic diversity variation caused by parasite evolutionary processes, which can, in turn, be used to predict novel host shifts from genetic data alone.
Acknowledgements
Matt Knox and four anonymous reviewers provided helpful comments that improved this manuscript.
Financial support
JCG-R and DTSH thank the New Zealand Ministry of Health for funding. DTSH acknowledges funding from the Royal Society Te Apārangi Rutherford Discovery Fellowship (MAU1701). The funders played no part in the design of this study.
Ethical standards
Not Applicable.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020001493.
click here to view supplementary material
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Abal-Fabeiro JL, Maside X, Bello X, Llovo J and Bartolomé C (2013) Multilocus patterns of genetic variation across Cryptosporidium species suggest balancing selection at the gp60 locus. Molecular Ecology 22, 4723–4732. [DOI] [PubMed] [Google Scholar]
- Akiyoshi DE, Dilo J, Pearson C, Chapman S, Tumwine J and Tzipori S (2003) Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infection and Immunity 71, 1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ and Linde CC (2008) Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends in Ecology & Evolution 23, 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid M, Hunter PR, Chalmers RM and Tyler KM (2013) Cryptosporidium Pathogenicity and virulence. Clinical Microbiology Reviews 26, 115–134. doi: 10.1128/cmr.00076-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabas H, Lion S, Nicot A, Meaden S, van Houte S, Moineau S, Wahl LM, Westra ER and Gandon S (2018) Evolutionary emergence of infectious diseases in heterogeneous host populations. PLoS Biology 16, e2006738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R, Robinson G, Elwin K, Hadfield S, Xiao L, Ryan U, Modha D and Mallaghan C (2009a) Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerging Infectious Diseases 15, 829–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers RM, Elwin K, Thomas AL, Guy EC and Mason B (2009b) Long-term Cryptosporidium Typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Eurosurveillance 14, e19086. [DOI] [PubMed] [Google Scholar]
- Chalmers RM, Elwin K, Hadfield SJ and Robinson G (2011) Sporadic human cryptosporidiosis caused by Cryptosporidium Cuniculus, United Kingdom, 2007–2008. Emerging Infectious Disease Journal 17, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwin K, Hadfield SJ, Robinson G, Crouch ND and Chalmers RM (2012) Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. International Journal for Parasitology 42, 675–682. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ryan UM and Xiao L (2018) Genetic diversity and population structure of Cryptosporidium. Trends in Parasitology 34, 997–1011. [DOI] [PubMed] [Google Scholar]
- Garcia-R JC and Hayman DTS (2016) Origin of a major infectious disease in vertebrates: The timing of Cryptosporidium evolution and its hosts. Parasitology 143, 1683–1690. [DOI] [PubMed] [Google Scholar]
- Garcia-R JC and Hayman DTS (2017) Evolutionary processes in populations of Cryptosporidium Inferred from gp60 sequence data. Parasitology Research 116, 1855–1861. [DOI] [PubMed] [Google Scholar]
- Garcia-R JC, French N, Pita A, Velathanthiri N, Shrestha R and Hayman D (2017) Local and global genetic diversity of protozoan parasites: spatial distribution of Cryptosporidium and Giardia Genotypes. PLOS Neglected Tropical Diseases 11, e0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-R JC, Pita AB, Velathanthiri N, French NP and Hayman DTS (2020) Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitolology Research 119, 2317–2326. [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Duchêne S and Holmes EC (2017) Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathogens 13, e1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M and Sturrock S (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler AV, Whipp MJ, Haydon SR and Gasser RB (2014) Cryptosporidium Cuniculus – new records in human and kangaroo in Australia. Parasites & Vectors 7, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler AV, Wang T, Haydon SR and Gasser RB (2018) Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus – An emerging zoonotic pathogen? International Journal for Parasitology 7, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque ASG, Zaidi AKM, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM and Levine MM (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. The Lancet 382, 209–222. [DOI] [PubMed] [Google Scholar]
- Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B and Axén C (2013) Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology 140, 1735–1740. [DOI] [PubMed] [Google Scholar]
- Lebbad M, Winiecka-Krusnell J, Insulander M and Beser J (2018) Molecular characterization and epidemiological investigation of Cryptosporidium hominis IkA18G1 and C. hominis monkey genotype IiA17, two unusual subtypes diagnosed in Swedish patients. Experimental Parasitology 188, 50–57. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A, Andolfatto P and Przeworski M (2012) Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biology 10, e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D and Feng Y (2014) Subtyping Cryptosporidium Ubiquitum, a zoonotic pathogen emerging in humans. Emerging Infectious Diseases 20, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P and Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lively CM (2010) The effect of host genetic diversity on disease spread. The American Naturalist 175, E149–E152. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JRC, Dobson AP, Hudson PJ and Grenfell BT (2009) Epidemic dynamics at the human-animal interface. Science (New York, N.Y.) 326, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Brockhurst MA, Russell CA, Welch JJ and Jiggins FM (2014) The evolution and genetics of virus host shifts. PLoS Pathogens 10, e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt PL, Casemore DP and Miller KB (1996) Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiologic Reviews 18, 118–136. [DOI] [PubMed] [Google Scholar]
- Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, Chalmers RM, Hunter PR, van Oosterhout C and Tyler KM (2019) Evolutionary genomics of anthroponosis in Cryptosporidium. Nature Microbiology 4, 826–836. [DOI] [PubMed] [Google Scholar]
- Penczykowski RM, Laine A-L and Koskella B (2016) Understanding the ecology and evolution of host–parasite interactions across scales. Evolutionary Applications 9, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, Lass S, Pulliam JRC, Read AF and Lloyd-Smith JO (2010) Identifying genetic markers of adaptation for surveillance of viral host jumps. Nature Reviews Microbiology 8, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutzer J and Karanis P (2009) Genetic polymorphism in Cryptosporidium species: an update. Veterinary Parasitology 165, 187–199. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins SE and Rozas J (2002) Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution 19, 2092–2100. [DOI] [PubMed] [Google Scholar]
- Rašková V, Květoňová D, Sak B, McEvoy J, Edwinson A, Stenger B and Kváč M (2013) Human cryptosporidiosis caused by Cryptosporidium Tyzzeri and C. parvum Isolates presumably transmitted from wild mice. Journal of Clinical Microbiology 51, 360–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U, Fayer R and Xiao L (2014) Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141, 1667–1685. [DOI] [PubMed] [Google Scholar]
- Striepen B (2013) Time to tackle cryptosporidiosis. Nature 503, 189–191. [DOI] [PubMed] [Google Scholar]
- Strong WB, Gut J and Nelson RG (2000) Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infection and Immunity 68, 4117–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S (2016) The effects of sample size on population genomic analyses – implications for the tests of neutrality. BMC Genomics 17, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga S, Matsuo J, Kono E, Kimura K, Inoue M, Rai SK and Ono K (2000) Prevalence of Cryptosporidium parvum Infection and pattern of oocyst shedding in calves in Japan. Veterinary Parasitology 94, 27–32. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang L, Axen C, Bjorkman C, Jian F, Amer S, Liu A, Feng Y, Li G, Lv C, Zhao Z, Qi M, Dong H, Wang H, Sun Y, Ning C and Xiao LH (2014) Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Scientific Reports 4, srep04208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer G (2009) Meta-analysis of a polymorphic surface glycoprotein of the parasitic protozoa Cryptosporidium parvum And Cryptosporidium hominis. Epidemiology and Infection 137, 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer G, Köster PC and Carmena D (2020) Cryptosporidium hominis infections in non-human animal species: revisiting the concept of host specificity. International Journal for Parasitology 50, 253–262. [DOI] [PubMed] [Google Scholar]
- Xiao L and Feng Y (2008) Zoonotic cryptosporidiosis. FEMS Immunology & Medical Microbiology 52, 309–323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182020001493.
click here to view supplementary material