Abstract
Cancer-associated fibroblasts (CAFs) can promote tumor growth, metastasis and therapeutic resistance in esophageal squamous cell carcinoma (ESCC), but the mechanisms of action remain elusive. Our objective was to identify secreted factor(s) that mediate the communication between CAFs and ESCC tumor cells with the aim of identifying potential druggable targets. Through unbiased cytokine arrays, we have identified CC motif chemokine ligand 5 (CCL5) as a secreted factor that is increased upon co-culture of ESCC cells and CAFs, which we replicated in esophageal adenocarcinoma (EAC) with CAFs. Loss of tumor-cell derived CCL5 reduces ESCC cell proliferation in vitro and in vivo and we propose this is mediated, in part, by a reduction in ERK1/2 signaling. Loss of tumor-derived CCL5 reduces the percentage of CAFs recruited to xenograft tumors in vivo. CCL5 is a ligand for the CC motif receptor 5 (CCR5), for which a clinically approved inhibitor exists, namely Maraviroc. Maraviroc treatment reduced tumor volume, CAF recruitment and ERK1/2 signaling in vivo, thus, mimicking the effects observed with genetic loss of CCL5. High CCL5 or CCR5 expression is associated with worse prognosis in low grade esophageal carcinomas.
Implications:
These data highlight the role of CCL5 in tumorigenesis and the therapeutic potential of targeting the CCL5-CCR5 axis in ESCC.
Keywords: Esophageal cancer, cancer-associated fibroblasts, CCL5, Maraviroc
Introduction
Esophageal carcinomas, represented by two major subtypes - Esophageal squamous cell carcinoma (ESCC) and Esophageal adenocarcinoma (EAC), account for 1% of new cancer cases each year in the USA (1). ESCC is the predominant subtype in Asia, eastern Europe and Africa with EAC more prevalent in western Europe and USA. This geographic distribution can be, in part, explained by the major risk factors for each subtype with tobacco, alcohol consumption and deficiency of minerals/vitamins associated with ESCC, while obesity and gastroesophageal reflux disease (GERD) are the predominant risk factors for EAC (2). Both subtypes are highly aggressive with a proclivity for metastasis, and thus have very poor prognoses with 5-year survival rates of 15–20% (1). The tumor microenvironment (TME) is known to drive tumorigenesis, metastatic colonization and therapeutic resistance, and as such, there is a pressing need to understand how components of the TME influence cancer cell behavior (3). Cancer associated fibroblasts (CAFs), a prominent cell type in the TME, have been shown to promote tumor cell growth, invasion, metastasis and therapeutic resistance in esophageal carcinomas (4) (and other cancers), yet the underlying mechanisms of action remain elusive (5). Understanding how fibroblasts are recruited and transition into pro-tumorigenic CAFs, and determining how CAFs communicate with tumor cells to promote tumorigenesis, are crucial for the future development of targeted therapeutic approaches.
CAFs can influence tumorigenesis by remodeling the extra-cellular matrix (ECM) and secretion of growth factors and immunomodulatory effectors including a variety of cytokines and chemokines (5). These secreted factors can directly act on tumor cells through paracrine signaling or act on other “non-tumor” cells such as immune cells to alter the TME (6). To address which secreted factors are involved in fibroblast-tumor cell communication, our lab completed unbiased cytokine arrays from fibroblast-ESCC cell co-cultures and identified a restricted number of cytokines increased upon co-culture (7). This work identified IL-6 as a mediator of fibroblast-ESCC cell communication and we demonstrated that targeting IL-6 in ESCC has therapeutic potential (7). Those cytokine arrays also highlighted another inflammatory chemokine that was upregulated upon co-culture of ESCC tumor cells and fibroblasts, namely the C-C motif chemokine ligand 5 (CCL5). CCL5 belongs to the CC subfamily of chemokines, which comprises 27 ligands and 10 receptors, and can act through C-C motif chemokine receptors 1, 3 and 5 (CCR1, CCR3 and CCR5) (8). The CCL5-CCR5 axis has been demonstrated to exhibit both pro and anti-cancer effects (9). It is noteworthy that other ligands can activate CCR5 including CCL3, CCL4 and CCL8, which adds further complexity when investigating the role of the CCL5-CCR5 axis in cancer (9). Herein, we demonstrate that CCL5 expression is elevated in ESCC and EAC tissues and upon co-culture of tumor cells with CAFs. We show that CCL5 is expressed predominantly by tumor cells and, using both in vitro and in vivo models, demonstrate that CCL5 loss reduces tumor cell proliferation and CAF recruitment. Thus, CCL5 promotes tumorigenesis through both CAF-dependent and independent mechanisms. Importantly, we illustrate that targeted inhibition of CCR5, by the clinically approved inhibitor, Maraviroc, reduces tumor growth and CAF recruitment in xenograft models. These data suggest that targeting the CCL5-CCR5 axis may have therapeutic potential in ESCC.
Materials and Methods
Cell culture
Esophageal squamous cell carcinoma cell lines; HCE-7, TE-1 and TE-11, which have been previously described (10–12), and esophageal adenocarcinoma cell line; OE-33 (#96070808, Millipore Sigma) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS (HyClone) and 1% penicillin/streptomycin (Fisher Scientific). Fetal esophageal fibroblasts (FEF3) and ESCC-1F fibroblast cell lines have been previously described (7) and were maintained in DMEM containing 15% FBS (HyClone) and 1% penicillin/streptomycin. The EAC-1F fibroblast cell line was generated from an esophageal adenocarcinoma tissue biopsy by dispase/trypsin digestion, established and expanded in Advanced DMEM:F12 culture medium containing 15% FBS (HyClone) and 1% penicillin/streptomycin, and after passage 2 were maintained in DMEM containing 15% FBS (HyClone) and 1% penicillin/streptomycin. Short tandem repeat (STR) analysis (ATCC) was performed on cell lines to validate cell line authenticity and cells were regularly tested for mycoplasma contamination with Mycoalert detection kit (Lonza). To maintain experimental consistency, and minimize the risk of genetic drift, we only used early passage cells (<20 passages) for experiments and routinely assessed cell phenotype, growth rates, viability and expression of common cell lineage markers.
Generation of CCL5−/− cell lines using CRISPR/Cas9
To generate CCL5 knockout cell lines, we used nucleofection to introduce two single guide RNAs and Cas9 ribonucleoprotein. All nucleofection reagents and guide RNAs were purchased from Synthego. The CCL5 knockout reaction mixture was composed of sgCCL5-1 [GACUCACACGACUGCUGGGU], sgCCL5-2 [GGUGUGGUGUCCGAGGAAUC], Cas9 ribonucleoprotein, nucleofection supplement and nucleofector solution. The Cas9 control reaction mixture was prepared as above minus the single guide RNAs. Nucleofection was completed using the Lonza P3 primary Cell Nucleofection kit and Amaxa 4D Nucleofector (Lonza). Following nucleofection, cells were sorted into 96 well plates, coated in 1% gelatin and containing conditioned medium from ESCC cells with an additional 20% FCS, by the Flow Cytometry Core (Columbia University). Plates were incubated for 2–4 weeks before colonies were expanded and the presence of indels was identified by DNA sequencing. DNA was extracted from 96 well plates using QuickExtract DNA extraction solution (Epicentre) and PCRs performed using 2x DreamTaq PCR master mix (ThermoFisher Scientific), CCL5 forward primer: CATGGAGCCTGAGCTTTGGA and CCL5 reverse primer: TTGGAGTGGGCACGATTCTC. DNA sequencing was performed by Genewiz (Azenta Life Sciences) and alignments completed using Benchling software (www.benchling.com).
3D tumoroid cultures
For 3D tumoroid cultures, adherent 2D cell lines were trypsinized and counted with 2000 cells seeded in 30 μl of Matrigel (Corning) per well of a 24-well plate. Plates were incubated at 37ºC for 30 mins to allow Matrigel to solidify then 500 μl of complete culture medium was added to each well. Culture medium was exchanged at day 4 and 7. Tumoroids were imaged and analyzed (counts and area) using the Celigo Image Cytometer (Nexcelom) at day 7 and 10.
Colony formation assays
For colony formation assays, 10,000 cells were seeded in complete culture medium in a 6 well plate and incubated at 37ºC for 10 days. Cells were fixed in 4% paraformaldehyde (ThermoFisher Scientific), washed in PBS then incubated in 0.5% crystal violet (Sigma) solution for 20 mins at room temperature. Plates are washed thoroughly with water and left to dry before imaging with the Chemidoc imaging system (Bio-Rad). Following imaging, 100% methanol was added to each well and plates were incubated for 20 mins on a rocker before transferring the methanol solution to triplicate wells of a 96 well plate. The absorbance was measured at 570 nm and the fold-change between samples and control cells was quantified.
ELISA
Conditioned medium was collected from mono- and co-cultures of either 2D adherent cells or 3D tumoroids cultures. CCL5 secretion levels were measured using the Human RANTES (CCL5) ELISA kit (RayBiotech) following manufacturer’s instructions.
Antibodies
Antibodies recognizing p-ERK1/2 (T202/Y204) (#4370), ERK1/2 (#4695), p-STAT3 (Y705) (#9145), p-STAT3 (S727) (#9134), STAT3 (#9139), Cyclin D1 (#2978) and β-actin (#4967) were purchased from Cell Signaling Technology. Additional antibodies used were GAPDH (#ab8245, Abcam), CCL5 (#ab189841, Abcam) and α-smooth muscle actin (#ab5694, Abcam). Secondary antibodies used were IRDye 800CW Goat anti-Rabbit IgG secondary antibody (#926–32211, LI-COR), IRDye 680RD Goat anti-Mouse IgG secondary antibody (#926–68070, LI-COR), Goat anti-Rabbit IgG Texas Red secondary antibody (#T-2767, ThermoFisher Scientific), Goat anti-mouse IgG Alexa Fluor 488 secondary antibody (#A-11001, ThermoFisher Scientific) and Goat anti-rabbit IgG (H+L) biotinylated secondary antibody (#BA-1000, Vector Labs).
Immunoblotting
Cells were washed and scraped in ice-cold PBS then pelleted at 1800rpm for 5 min at 4°C. Cell pellets were resuspended in an appropriate volume (50–150 ul) of cell lysis buffer [20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% [vol/vol] Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1× complete EDTA-free protease inhibitor cocktail [Roche]). Lysates were incubated on ice for 45 min and vortexed regularly before clarifying at 13,000rpm for 20 min at 4°C. Protein lysates were quantified using Bio-Rad protein assay and adjusted to 0.5–2 μg/μl solutions using 4x Laemmli sample buffer (Bio-Rad). Samples (10–30 μg) were loaded into 4–15% mini-protean TGX precast gels (Bio-Rad) and transferred to 0.2 μm nitrocellulose membrane using Trans-blot turbo mini transfer packs (Bio-Rad). Following transfers, membranes were washed in PBS + 0.1% tween20 then blocked in 5% milk in PBS for 60 mins. Membranes were incubated in primary antibody for 16 hours at 4ºC then secondary antibody for 60 mins at room temperature with antibodies diluted in 5% bovine serum albumin in PBS + 0.1% tween20. Membranes were washed in PBS + 0.1% tween20 between incubations and prior to imaging on Chemidoc imaging system (Bio-rad).
Immunofluorescence
Cells were seeded on coverslips and grown to desired confluency prior to fixing in 4% paraformaldehyde (ThermoFisher Scientific) for 15 mins. Cells were permeabilized by incubating in 0.2% triton x/PBS solution for 10 mins then blocked in 5% bovine serum albumin (Sigma) in PBS + 0.1% tween20 (Sigma) solution for 60 mins with washes in PBS between incubations. Cells were incubated in primary antibody (diluted at 1:100–1:500) for 16 hours at 4ºC then secondary antibody (diluted at 1:500) for 1 hour at room temperature. Antibodies were diluted in 0.5% bovine serum albumin (Sigma) in PBS + 0.1% tween20 (Sigma) solution with washes in PBS + 0.1% tween20 solution between incubations. Slides were mounted using Vectashield Hardset anti-fade mounting medium containing DAPI (Vector Labs) and sealed with clear nail polish. Images were obtained using a Keyence BZ-X800 (Keyence).
Quantitative real-time PCR (qRT-PCR)
RNA was extracted using the RNeasy mini kit (Qiagen) and concentrations were measured using a NanoDrop Lite Spectrophotometer (ThermoFisher Scientific). Synthesis of cDNA was completed using 1 μg of RNA and the iScript cDNA synthesis kit (Bio-Rad). Each qRT-PCR reaction was completed in triplicate with the reaction mixture: 2 μM forward primer, 2 μM reverse primer, 50% SYBR green PCR master mix (Applied Biosystems) and 4 μl diluted cDNA (1:5) in a final volume of 20 μl in 96 well PCR plates. Reactions were completed using the 7500 Real-Time PCR system (ThermoFisher Scientific) with a 60ºC annealing temperature for 40 cycles. Primers used are detailed in Table 1. Relative expression was analyzed using the comparative Ct method (ΔΔCt) with samples compared to the endogenous control gene; GAPDH. Following qRT-PCR, 1 μl of TriTrack DNA loading dye (Fisher Scientific) was added to 5 μl of sample and run on a 2% agarose gel containing SYBR safe DNA gel stain (ThermoFisher Scientific). Agarose gels were imaged on the Chemidoc imaging system (Bio-Rad).
Table 1: List of primers used for qRT-PCR.
Primer sequences were designed on Benchling (www.benchling.com) and specificity verified with Primer Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
| Gene | Forward primer | Reverse Primer |
|---|---|---|
| CCL1 | AATACCAGCTCCATCTGCTCCAA | GAACCCATCCAACTGTGTCCAAG |
| CCL2 | AGGTGACTGGGGCATTGAT | GCCTCCAGCATGAAAGTCTC |
| CCL3 | ATGCAGGTCTCCACTGCTGCCCTT | GCACTCAGCTCCAGGTCGCTGACAT |
| CCL4 | GCAAGTCTGTGCTGATCCCA | GCGGAGAGGAGTCCTGAGTA |
| CCL5 | ATCCTCATTGCTACTGCCCTC | GCCACTGGTGTAGAAATACTCC |
| CCL8 | GCCTGCTGCTCATGGCAGCC | GCACAGACCTCCTTGCCCCG |
| CCR1 | CCTGCTGACGATTGACAGGTA | TTGGAAAAGTATAAGCCTGGCAT |
| CCR2 | GCCTTTTTCACATAGCTCTTGGC | AGGAGTCCTTGTGTAGTCACTTT |
| CCR3 | ATACCAGAGCACTGATGGCC | GATGTTGGTCATAATTCGGAGCC |
| CCR4 | ATGAACCCCACGGATATAGCA | CACCACAGAATTTCCAAGCAGA |
| CCR5 | AGGGCTGTGAGGCTTATCTT | CACCTGCATAGCTTGGTCCA |
| CCND1 | GCCACTGGTGTAGAAATACTCC | GCGCCGCAGGCTTGACTCCAGAA |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
Histology
Clinical materials were obtained from patients following written informed consent in accordance with recognized ethical guidelines (Declaration of Helsinki) and approved by an Institutional Review Board. ESCC tissue samples were obtained as biopsies from Kagoshima University Hospital as previously described (13). EAC tissue samples were obtained as biopsies from Dr. Kenneth K. Wang, M.D at the Mayo Clinic (Rochester, MN; IRB protocol 15–009292).
Immunohistochemistry
Tissue samples were fixed in 10% neutral buffered formalin for 24 hours at 4ºC, washed in PBS twice and transferred to 70% ethanol. Tissue was embedded in paraffin and sectioned at 5 μm thickness by the Molecular Pathology Shared Resource at Columbia University. Slides were de-waxed and rehydrated by repeated washes in 100% xylene then decreasing concentrations of ethanol. Antigen retrieval was completed by incubating slides in boiling 10 mM Citric acid buffer (pH=6) for 30 min. Slides were blocked in 3% H2O2 (Sigma), Avidin (Sigma), Biotin (Sigma) and Starting Block T20 (ThermoFisher Scientific) for 15 mins each with washes in PBS + 0.1% tween20 between incubations. Slides were incubated in primary antibody (1:100 – 1:500) for 16 hours at 4ºC then biotinylated secondary antibody (1:200) for 30 min at 37ºC. Antibodies were diluted in 1% Bovine serum albumin in PBS + 0.3% triton x with washes in PBS + 0.1% tween20 between incubations. Slides were incubated with Vectastain elite ABC HRP kit (Vector Labs) for 30 min at 37ºC before visualization was completed using the DAB/Ni peroxidase substrate kit (Vector Labs). Slides were counterstained in Haematoxylin and dehydrated by repeated washes in increasing concentrations of ethanol then 100% xylene. Slides were mounted using Cytoseal. Imaging of brightfield images was completed using Keyence BZ-X800 (Keyence).
Animal models
Animal studies have been approved by Institutional Animal Care and Use committee (IACUC) at the University of Pennsylvania and Columbia University. For subcutaneous xenograft models, 5 × 106 TE11 cells −/+ 1 × 106 FEF3 fibroblasts were suspended in 50 μl Matrigel and implanted into both flanks of athymic nude mice (Taconic). Prior to implantation, cells were tested for mycoplasma contamination and incubated for 48 hours in antibiotic free cell culture medium. For drug treatments, Maraviroc (SelleckChem) or DMSO was given at 10 mg/kg intraperitoneally daily. For patient derived xenografts (PDX), 1 mm ESCC tumor fragments were placed in the subcutaneous flank incisions and incisions closed by suture in anesthetized mice. Mice were housed for 4–8 weeks with tumor volume measured as [length x width2]. Following conclusion of xenograft growth studies, mice were culled and the tumors were dissected and processed for immunohistochemical analysis.
Statistical analysis
Histological scorings for CCL5 expression (Fig. 1B) and cell type specific p-ERK1/2 expression (Fig. 4D) were performed by Dr. Andres J. Klein-Szanto, pathologist, blinded to the identity of the samples. Staining for α-SMA (Fig. 2G & 4C) and p-ERK1/2 (Fig. 3G & S4F) were quantified as the percentage of staining positivity using Image J software (https://imagej.nih.gov/ij/). Significance has been determined using the unpaired student’s t-test with p-value denoted by asterisk. *<0.05, **<0.01, ***<0.001, ****<0.0001. Kaplan-Meier survival curves (Fig. 5) were generated using data from the TCGA database accessed via UCSC Xena (14). The mean of the whole sample was denoted as the cutoff value, thus an expression above the mean would be categorized as high and those values below the mean categorized as low expression. Significance for survival curves was determined by the log-rank test. Cox proportional hazards analysis was performed to determine the effect of multiple variables on survival. These statistical analyses and corresponding figures were generated with GraphPad Prism (www.graphpad.com).
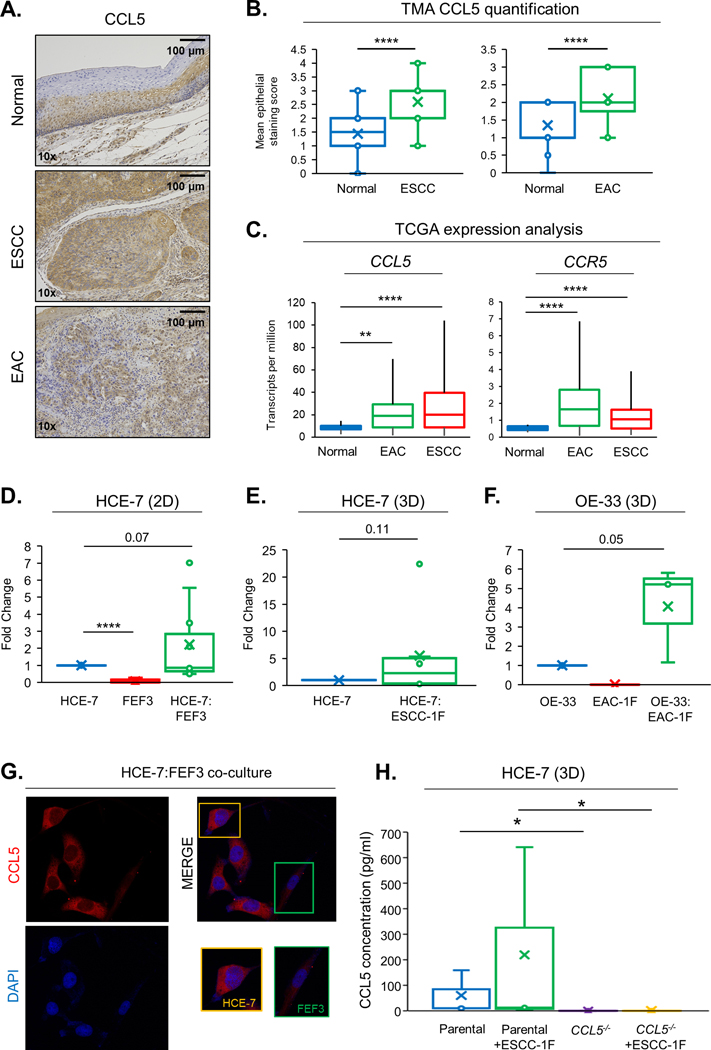
Fig. 1: CCL5 expression is elevated in esophageal carcinoma and co-culture with CAFs induces CCL5 secretion.
(A) Representative images of normal esophagus, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) stained with CCL5 antibody and counterstained with hematoxylin. (B) Quantification of CCL5 staining in epithelial cells from normal esophagus (n=56) vs ESCC (n=116) and normal esophagus (n=20) vs EAC (n=18) from tissue microarrays (TMA). (C) Analysis of CCL5 and CCR5 transcript expression from The Cancer Genome Atlas (TCGA) comparing normal esophagus (n=11), ESCC (n=95) and EAC (n=89). (D) CCL5 secretion from 2D cell cultures; HCE-7 only, FEF3 only and HCE-7:FEF3 co-cultures. (E) CCL5 secretion from 3D ESCC tumoroid cultures; HCE-7 only and HCE-7:ESCC-1F co-cultures. (F) CCL5 secretion from 3D EAC tumoroid cultures; OE33 only, EAC-1F only and OE33:EAC-1F. CCL5 secretion quantification in D-F are represented as fold change compared to tumor cell monoculture. (G) Immunofluorescence of HCE-7:FEF3 2D co-cultures stained with CCL5 antibody (top left panel) and DAPI (bottom left panel). Merged images (right panels) and zoomed images of HCE-7 and FEF3 cells. Images obtained with 20x microscope objective. (H) CCL5 secretion from 3D tumoroid cultures; HCE-7 parental, HCE-7 parental:ESCC-1F co-culture, HCE7 CCL5−/− and HCE-7 CCL5−/−:ESCC-1F co-culture presented as fold-change compared to HCE-7 parental cells. All CCL5 secretion data is representative of at least 3 biological replicates. Significance has been determined using the unpaired student’s t-test with p-value denoted by asterisk. *<0.05, **<0.01, ***<0.001, ****<0.0001.
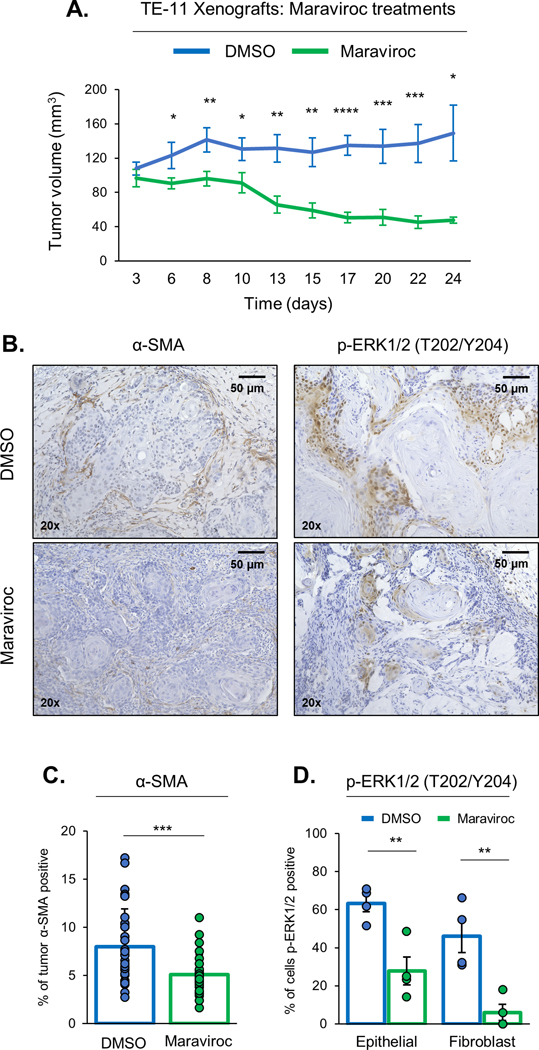
Fig. 4: Targeted CCR5 inhibition replicates CCL5 loss with reduced tumor growth and CAF recruitment in vivo.
(A) Tumor volume of Xenografts generated from co-culture of TE-11 and ESCC-1-F cells then treated with either DMSO or (10 mg/kg interperitoneally daily) Maraviroc. Tumor volume is measure over 24 days (n=10 tumors from 5 mice per cohort). (B) Representative images of Xenografts, generated from TE-11 and ESCC-1-F cells then treated with either DMSO or Maraviroc, stained with either α-SMA or p-ERK1/2 (T202/Y204) and counterstained with hematoxylin. (C) Quantification of α-SMA positivity illustrated as the percentage of tumor area expressing α-SMA. (D) Quantification of p-ERK1/2 (T202/Y204) positivity illustrated as the percentage of cells, separated into either epithelial or fibroblast lineages, for p-ERK1/2 (T202/Y204). Significance has been determined using the unpaired student’s t-test with p-value denoted by asterisk. *<0.05, **<0.01, ***<0.001, ****<0.0001.
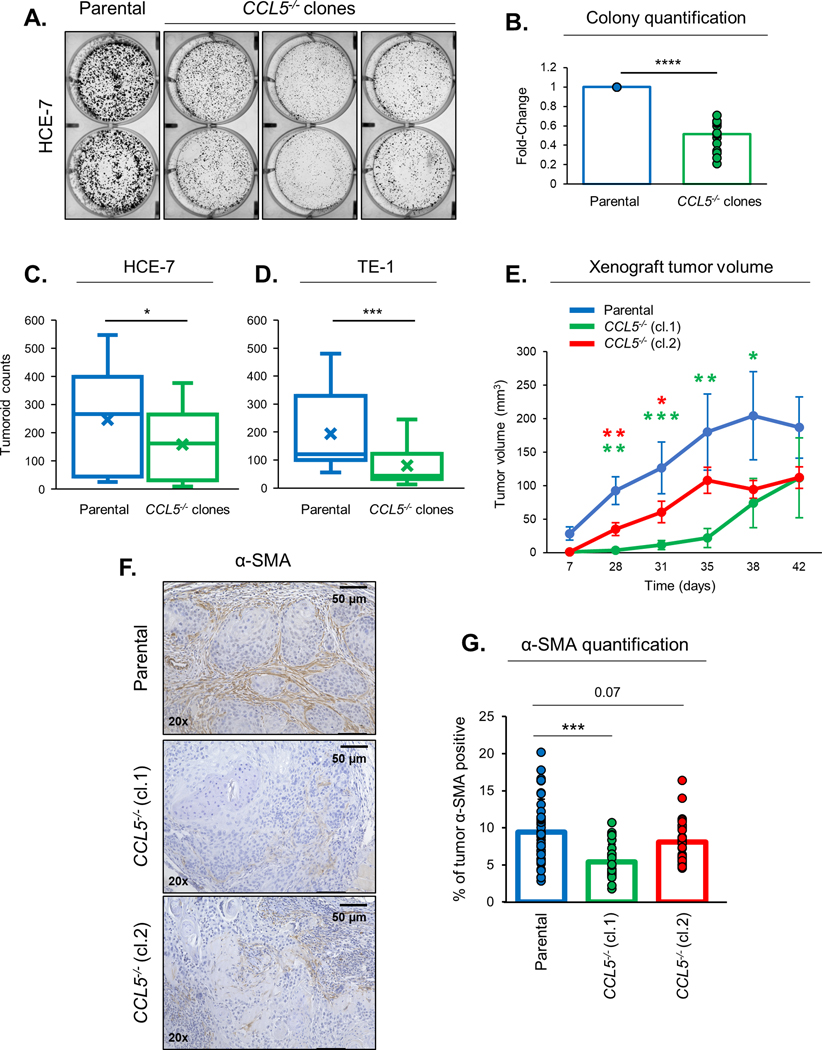
Fig. 2: Loss of tumor-derived CCL5 reduces tumor cell proliferation and CAF recruitment.
(A) Representative images of colony formation assay of HCE-7 parental and CCL5−/− clones in duplicate wells. (B) Quantification of colony formation assays of HCE-7 parental and CCL5−/− clones represented as fold-change compared to parental. (C) Number of tumoroids per well at day 10 in HCE-7 parental and CCL5−/− clones. (D) Number of tumoroids per well at day 10 in TE-1 parental and CCL5−/− clones. (E) Quantification of tumor volumes in Xenografts generated from TE-11, TE-11 CCL5−/− (cl.1) and TE-11 CCL5−/− (cl.2) cell lines with tumors measured over 42 days (n=6 per cohort). (F) Representative images of Xenografts generated from TE-11, TE-11 CCL5−/− (cl.1) and TE-11 CCL5−/− (cl.2) cell lines and stained with α-smooth muscle actin (α-SMA) and counterstained with hematoxylin. (G) Quantification of α-SMA positivity illustrated as the percentage of tumor area expressing α-SMA (n=6 per cohort). Colony formation assay and tumoroid count data are representative of at least 3 biological replicates. Significance has been determined using the unpaired student’s t-test with p-value denoted by asterisk. *<0.05, **<0.01, ***<0.001, ****<0.0001.
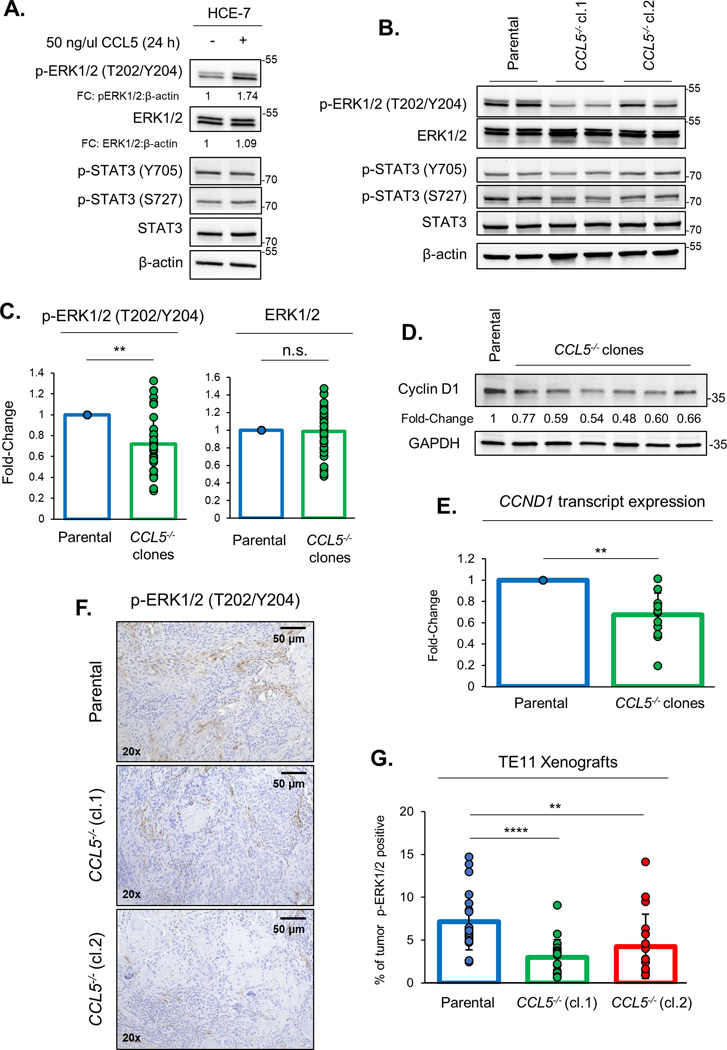
Fig. 3: Loss of tumor-derived CCL5 reduces ERK1/2 signaling.
(A) Immunoblotting of HCE-7 cells treated with 50 ng/μl CCL5 for 24 hours. Quantification of phosphorylated ERK1/2 (T202/Y204) and ERK1/2 protein levels were normalized to β-actin and displayed as fold-change (FC) below corresponding blot. (B) Immunoblotting of HCE-7 parental, HCE-7 CCL5−/− (cl.1) and HCE-7 CCL5−/− (cl.2) cell lines in duplicate. (C) Quantification of phosphorylated ERK1/2 (T202/Y204) and ERK1/2 protein levels were normalized to β-actin and displayed as fold-change as a bar graph with overlay of individual experiments. Results from all HCE-7 CCL5−/− clones were combined and compared to HCE-7 parental cells. (D) Immunoblotting of HCE-7 parental and CCL5−/− clones. Quantification of cyclin D1 protein levels were normalized to GAPDH and displayed as fold-change below corresponding blot. All immunoblotting is representative of a minimum of three biological replicates. (E) CCND1 mRNA transcript levels were quantified by qRT-PCR and normalized to house-keeping gene, GAPDH. Transcript levels are displayed as fold-change compared to HCE-7 parental cells. (F) Representative images of Xenografts generated from TE-11, TE-11 CCL5−/− (cl.1) and TE-11 CCL5−/− (cl.2) cell lines and stained with p-ERK1/2 (T202/Y204) and counterstained with hematoxylin. (G) Quantification of p-ERK1/2 (T202/Y204) positivity illustrated as the percentage of tumor area expressing p-ERK1/2 (T202/Y204) (n=6 per cohort). Significance has been determined using the unpaired student’s t-test with p-value denoted by asterisk. *<0.05, **<0.01, ***<0.001, ****<0.0001.
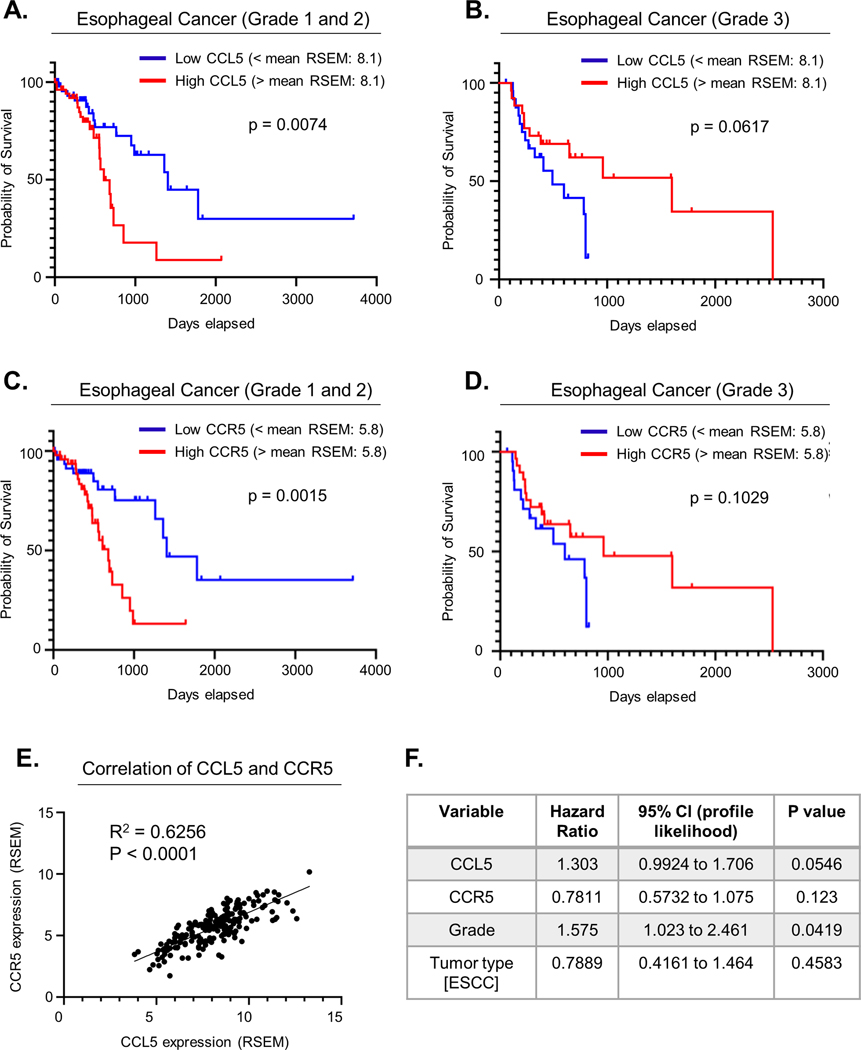
Fig. 5: High CCL5 and CCR5 expression in low grade esophageal cancers is associated with poor prognosis.
(A) Kaplan-Meier survival curve of the effect of CCL5 expression on esophageal cancer patient overall survival with grade 1 (well differentiated) + 2 (moderately differentiated) tumors. (B) Kaplan-Meier survival curve of the effect of CCL5 expression on esophageal cancer patient overall survival with grade 3 (poorly differentiated) tumors. (C) Kaplan-Meier survival curve of the effect of CCR5 expression on dataset analyzed for A. (D) Kaplan-Meier survival curve of the effect of CCR5 expression on dataset analyzed for B. The cut-off used to determine low and high expression was the mean RNA expression of the whole sample by RSEM (CCL5=8.1, CCR5=5.8). (E) Correlation of CCL5 expression and CCR5 expression in esophageal cancers. (F) Multivariable analysis on the association between CCL5 expression, CCR5 expression, tumor grade and tumor type (ESCC vs. EAC) on esophageal cancer patient survival. RSEM = RNA-Seq by Expectation Maximization.
Data Availability
Data sharing not applicable to this article as no datasets were generated and only publicly available datasets were analyzed during the current study.
Results
CCL5 expression is elevated in esophageal carcinomas and co-culture with CAFs induces CCL5 secretion.
Previous cytokine array data indicated that CCL5 secretion is increased upon co-culture of esophageal carcinoma cells with fetal esophageal fibroblast cells (7). We assessed normal esophagus epithelium, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) tissues for CCL5 protein expression by immunohistochemistry (Fig. 1A) and identified significantly higher CCL5 expression in esophageal carcinomas (Fig. 1B). We corroborated these data independently using the TCGA (The Cancer Genome Atlas) database with both ESCC and EAC tissues demonstrating higher CCL5 levels, and its receptor, CCR5, compared to normal esophageal epithelium (Fig. 1C). CCL5 secretion levels was assessed upon co-culture of an ESCC cell line (HCE-7) with either fetal esophageal fibroblasts (FEF3) in a 2D monolayer cell culture or CAFs derived from ESCC (ESCC-1F) in a 3D tumoroid model system (Fig. 1D&E). CCL5 levels in the conditioned growth medium were increased in the co-cultures compared to mono-cultures, as assessed by ELISA (Fig. 1D&E). Similar results were observed with co-culture of OE-33 (EAC) cells and CAFs derived from EAC (EAC-1F), with increased CCL5 secretion in 3D tumoroid models (Fig. 1F). Data from these ELISAs (Fig. 1D-F) indicate that fibroblasts in monoculture do not secrete CCL5 and that only tumor cell monocultures and co-culture of tumor cells with fibroblasts result in secretion of CCL5. Immunofluorescence staining of these co-cultures (HCE-7, an ESCC cell line, and FEF3, a fibroblast cell line) with CCL5 antibody confirmed that HCE-7 cells express higher CCL5 levels with minimal expression in FEF3 cells (Fig. 1G). Therefore, we hypothesized that tumor cells are the predominant source of CCL5 within these co-culture experiments, and as such, we used CRISPR/Cas9 technology to generate CCL5 null cell lines in three ESCC tumor cell lines; HCE-7, TE-1 and TE-11. We confirmed CCL5 loss in these cells by assessing CCL5 mRNA expression, CCL5 secretion and DNA sequencing (Fig. S1). Interestingly, the supernatants of CCL5 null cell lines co-cultured with fibroblasts (ESCC-1F) in 3D tumoroid models have no detectable CCL5 expression (Fig. 1H). This underscores that tumor-derived CCL5 is required for the increased CCL5 secretion detected in tumor cells/fibroblast cells co-cultures.
Loss of tumor-derived CCL5 reduces tumor cell proliferation and CAF recruitment.
When generating CCL5−/− cell lines, we observed a consistent reduction in cell growth rates in our knockout cells. We characterized the effect of CCL5 loss on tumor cell proliferation using 2D colony formation assays (Fig. 2A and S2A). HCE-7 and TE-1 CCL5−/− clones exhibit a significant reduction in cell proliferation compared to parental cells in colony formation assays (Fig. 2A&B and S2A&B). To confirm that the changes in growth rates were caused by loss of CCL5 and not a result of clonal variation during selection or a consequence of transfections with Cas9, we also preformed colony formation assays on parental, Cas9 transfected pools and CCL5−/− pools (Fig. S2C&D). As expected, CCL5−/− pools had reduced cell growth compared to either parental or Cas9 transfected cells (Fig. 2C&D). This reduction in cell proliferation is maintained in 3D tumoroid cultures with significantly lower number of tumoroids formed from CCL5−/− clones compared to parental cells (Fig. 2C&D). TE-11 cells, a commonly studied ESCC cell line, has been previously published for the establishment of xenografts in nude mice (7). We established xenografts from TE-11 parental, TE-11 CCL5−/− (cl.1) and TE-11 CCL5−/− (cl.2) cells, and monitored tumor growth over 42 days. Xenografts generated from TE-11 CCL5−/− cells grow significantly slower over the first 30 days and continued a trend of smaller tumors for the duration of this experiment (Fig. 2E). CCL5 is an established chemokine so we hypothesized that the loss of tumor-derived CCL5 may remove a signal that is involved in CAF recruitment. To assess CAF recruitment, we stained xenografts generated from TE-11 parental, TE-11 CCL5−/− (cl.1) or TE-11 CCL5−/− (cl.2) cells with α-smooth muscle actin (α-SMA), which is a marker of CAFs (15) (Fig. 2F). Xenografts generated from TE-11 CCL5−/− cells had reduced percentage of α-SMA positive cells, consistent with a reduction in CAFs in the TME (Fig. 2G).
Loss of tumor-derived CCL5 reduces ERK1/2 signaling.
ERK1/2 and STAT3 signaling have been reported as downstream effectors of the CCL5-CCR5 axis (9). Phosphorylation of ERK1/2 and STAT3 are essential for the activation of these signaling pathways (16,17). We assessed activation of both pathways in HCE-7 cells following treatment with recombinant CCL5 protein (Fig. 3A). CCL5 treatment increased the levels of phosphorylated ERK1/2 (T202/Y204) but the phosphorylation status of STAT3 was unaffected (Fig. 3A). The treatment of HCE-7 and TE-1 cell lines with recombinant CCL5 protein induces robust phosphorylation of ERK1/2 within 30 mins (Fig. S3A). HCE-7 CCL5−/− clones have reduced levels of p-ERK1/2 (T202/Y204) compared to parental cells but total levels of ERK1/2 protein are unaffected (Fig. 3B&C). There are no consistent changes to p-STAT3 (Y705) or p-STAT3 (S727) protein levels (Fig. 3B). Cyclin D1, an ERK1/2 target gene, has key roles in cell cycle progression and cell proliferation (18). Therefore, we assessed levels of cyclin D1 protein and found a consistent reduction in the levels of cyclin D1 in HCE-7 CCL5−/− clones compared to HCE-7 parental cells (Fig. 3D). As an ERK1/2 target gene, we assessed the levels of cyclin D1 mRNA (CCND1) transcripts and observed that CCL5−/− clones have significantly lower CCND1 transcript levels compared to parental cells (Fig. 3E). This reduction in Cyclin D1 was replicated in TE-1 CCL5−/− cells, but only a slight reduction in total ERK1/2 protein levels were detected in these cell lines (Fig. S3B&C). We also compared pERK1/2(T202/Y204) expression levels from parental, Cas9 transfected and CCL5−/− pools (Fig. S3D). Expression levels were comparable between parental and Cas9 transfected cells and reduced in CCL5−/− pools (Fig. S3D). To assess ERK1/2 activity in vivo, we stained xenografts generated from TE-11 parental, TE-11 CCL5−/− (cl.1) or TE-11 CCL5−/− (cl.2) cells with p-ERK1/2 (T202/Y204) antibody (Fig. 3F). Xenografts generated from TE-11 CCL5−/− clones had reduced p-ERK1/2 expression compared to those derived from TE-11 parental cells (Fig. 3G), thus confirming that loss of CCL5 from ESCC cell lines reduces phosphorylation of ERK1/2 both in vitro and in vivo.
Targeted CCR5 inhibition replicates CCL5 loss with reduced tumor growth and CAF recruitment in vivo.
Targeting a secreted ligand may be replete with technical challenges, whereas, targeting a receptor can be possible with small molecule inhibitor or an antibody-based approach. CCL5 can bind multiple receptors including CCR1, CCR3 and CCR5 (19). Conversely, multiple ligands can activate these receptors, specifically CCL3, CCL4, CCL5 and CCL8 can all bind to CCR5 (8). It is important to note that our cell lines express multiple CC ligands and receptors (Fig. S4A). CCL5 binds CCR5 with high affinity and the CCL5-CCR5 axis has been demonstrated in multiple cancers (9). Maraviroc is a CCR5 inhibitor which is clinically approved for the treatment of human immunodeficiency virus (HIV) infections (20). Despite the presence of multiple CC-receptors, maraviroc has been shown to be highly specific for CCR5 inhibition (21,22). To assess if CCR5 inhibition could replicate the cell proliferation defects observed with loss of CCL5, we preformed colony formation assays using parental HCE-7 and TE-1 cells. Treatment with Maraviroc slightly, albeit significantly, reduced cell proliferation in HCE-7 and TE-1 cells (Fig. S4B). To assess this effect of Maraviroc on cell proliferation in vivo, we generated xenografts from TE-11 and ESCC-1F co-cultures, then treated mice with either DMSO or Maraviroc (Fig. 4A). Maraviroc treatment significantly reduced tumor growth over 35 days of growth (Fig. 4A). We replicated these findings in a patient-derived xenograft (PDX) model where ESCC tumor fragments were used to generate xenografts prior to treatment with either DMSO or Maraviroc (Fig. S4C). We assessed the effect of Maraviroc on CAF infiltration by staining TE-11 xenografts with α-SMA and identified a significant reduction in α-SMA positive cells in Maraviroc treated xenografts (Fig. 4B&C). Since our studies with CCL5 knockout cell lines indicate that ERK1/2 signaling is reduced upon loss of CCL5, we tested if inhibition of CCR5 with Maraviroc could also reduce ERK1/2 signaling both in vitro and in vivo. Maraviroc treatment slightly reduced levels of p-ERK1/2 (T202/Y204) and Cyclin D1 in HCE-7 cells in vitro (Fig. S4D). Interestingly, this reduction in p-ERK1/2 (T202/Y204) levels was more robust in our in vivo model systems. Maraviroc treatment significantly reduced the percentage of cells, both epithelial and fibroblast cells, positive for p-ERK1/2 (T202/Y204) in xenografts generated from TE-11 and ESCC-1F cell lines (Fig. 4B&D). This reduction in p-ERK1/2 (T202/Y204) protein levels was also observed in Maraviroc treated PDX tumors (Fig. S4E&F). These findings indicate that Maraviroc treatment can replicate the reduction in cell proliferation, CAF infiltration and ERK1/2 signaling that was identified with CCL5 loss.
High CCL5 and CCR5 expression in low-grade esophageal cancers is associated with poor prognosis.
To determine if the expression of CCL5 is associated with patient outcome, we interrogated the TCGA survival data. Categorization of esophageal cancers (includes both ESCC and EAC) into specific tumor grades, reveals a significant effect of CCL5 and CCR5 expression on patient prognosis. Patients with high CCL5 expression and grade 1 (well differentiated) or 2 (moderately differentiated) esophageal cancers have significantly reduced overall survival compared to those patients with low CCL5 expression (Fig. 5A). This effect is reversed paradoxically in grade 3 (poorly differentiated) esophageal cancers with high CCL5 expression associated with increased survival rates, albeit this does not reach statistical significance (Fig. 5B). These results were replicated with CCR5 expression profiles (Fig. 5C, D). As expected from the Kaplan-Meier curves, CCL5 and CCR5 expression are positively correlated in esophageal cancers (Fig. 5E). Multivariable analysis of the esophageal cancer dataset reveal that tumor grade is an important determinant for survival outcomes (P=0.0419) with CCL5 expression almost reaching significance (P=0.0546) (Fig. 5F). Tumor type (ESCC vs. EAC) and CCR5 expression were not significantly associated with esophageal cancer survival (Fig. 5F). These data suggest that targeting the CCL5-CCR5 axis in well or moderately-differentiated esophageal cancers may be therapeutically relevant, a novel conceptual framework for these cancers.
Discussion
As the pro-tumorigenic roles of CAFs are identified and explored, there is increasing evidence that targeting CAFs may have therapeutic benefit (23). CAFs not only promote tumor growth, but also shelter tumor cells from conventional anti-cancer therapies such as chemotherapy and radiotherapy (5). Understanding how CAFs are recruited and reprogrammed by tumors and how CAFs communicate with tumor cells to promote tumorigenesis are critical to develop effective anti-CAF specific therapeutic strategies. For our studies we focused on the communication between CAFs and tumor cells derived from ESCC, as ESCC has a particularly poor prognosis that is a result of early metastases and therapeutic resistance (24). We hypothesized that CAFs are partly responsible for these ESCC characteristics. Cell-cell communication is complex and we predict that multiple cytokines, chemokines and growth factors are involved in the communication between CAFs and tumor cells. Indeed, we have demonstrated previously that the cytokine IL-6 has crucial roles in CAF-tumor interactions (7). Here we characterize how tumor-derived CCL5 expression is increased upon co-culture with CAFs and that loss of tumor-derived CCL5 reduces tumor cell growth in vitro and in vivo. It is important to note that loss of tumor-derived CCL5 reduces tumor cell growth in mono-cultures, indicating that this reduction is caused by tumor cell intrinsic effects of CCL5 and independent of CAFs. This tumor cell intrinsic effect of CCL5 signaling has been observed independently in different ESCC cell lines with knockdown of CCL5, through siRNA transfections, reducing basal cell growth (25). Importantly, we also detect a reduction in CAF recruitment in in vivo xenografts generated by CCL5−/− cells. Recently, the CCL5-CCR5 signaling axis has been shown to recruit fibroblasts in colorectal cancer with knockdown of either CCL5 or CCR5 thereby reducing the ability of colorectal cancer cell lines to recruit CAFs in vitro (26). These independent findings in other systems are corroborative of our findings and therefore, we propose that CCL5 has both tumor intrinsic and paracrine signaling effects in ESCC.
The CCL5-CCR5 axis has been reported to induce several downstream signaling pathways including STAT3, ERK1/2, PI3K and NF-κB (9). In our studies we observed consistent alterations in ERK1/2 signaling, thereby indicating that this may be the preferred pathway affected in our in vitro and in vivo conditions. Indeed, we provide evidence that targeting of the CCL5-CCR5 axis with Maraviroc, a clinically approved CCR5 inhibitor, reduces tumor growth, ERK1/2 signaling and CAF recruitment in vivo. These data represent independent corroboration with experiments using CCL5−/− cells. Therefore, we show that targeting of the CCL5-CCR5 axis by either genetic manipulation or chemical inhibition results in reduced tumor growth and CAF recruitment. It is noteworthy that the reduction in tumor cell growth and ERK1/2 signaling are modest in vitro compared to the more compelling reduction observed in vivo upon either genetic or chemical inhibition of the CCL5-CCR5 signaling pathway. We propose these differences can be explained by the absence of CAFs in the in vitro experiments. CAFs promote tumor growth through multiple mechanisms, including re-modeling of the extracellular matrix, promotion of angiogenesis and remodeling of tumor cell metabolism in response to limited nutrients or hypoxia (27). Specifically, co-injection of CAFs with cancer cell lines in subcutaneous xenografts models has been shown to promote primary tumor growth (28). Taken together, we hypothesize that the difference in magnitude between in vitro and in vivo tumor growth results can be explained by the importance of CAFs for tumor progression in vivo.
We noted that CCL5 expression is higher in esophageal cancer tissues compared to normal esophageal epithelium and revealed an association between CCL5 expression and prognosis that was dependent upon tumor grade, indicating that stratification of patients by tumor grade could be considered for future clinical trials. We speculate that these results may be due to alterations in the immune-microenvironment between tumor grades and between high and low CCL5 expressing tumors. CCL5 expression has been associated with the presence of regulatory T cells (Tregs), tumor-infiltrating lymphocytes and natural killer cells in renal clear cell cancer, breast cancer and melanoma respectively (29–31). Interestingly, an increase in immune cell infiltration has been observed in late stage and less-differentiated breast cancers, bladder cancers and oral squamous cell carcinomas (32,33). A recent publication revelated that less-differentiated ESCC tumors have increased infiltration of B cell and Tregs compared to well-differentiated tumors (34). These data indicate that the CCL5-CCR5 axis may have protean roles including promotion of tumor growth, CAF recruitment and immune cell infiltration.
We demonstrate that CCL5 and CCR5 expression is correlated positively in esophageal cancers. Our studies support the premise that tumor cells are the predominant source of CCL5 and CCR5. Esophageal cancers, with high or low CCL5 expression, may trigger complex interactions in the microenvironment that influences patient outcomes. Future studies should interrogate how the CCL5-CCR5 signaling axis regulates immune cell infiltration in ESCC and how this varies between tumor grades. Our findings offer new conceptual perspectives to investigate the use of Maraviroc with current treatment of ESCC and EAC in the neoadjuvant or adjuvant setting.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grants 5P30CA013696 and 5P01CA098101 (AKR, JAD, HN, KJD, AJKS, QT). The authors thank the following shared resources through the Herbert Irving Comprehensive Cancer Center: Biostatistics, Confocal and Specialized Microscopy, Database, Genetically Engineered Mouse Models, and Molecular Pathology.
Financial support
This work was supported by NIH/NCI grants 5P30CA013696 and 5P01CA098101.
Footnotes
Conflicts of interest
The authors declare no potential conflicts of interest
References
- 1.Cancer of the Esophagus - Cancer Stat Facts [Internet]. SEER. [cited 2022 Feb 9]. Available from: https://seer.cancer.gov/statfacts/html/esoph.html [Google Scholar]
- 2.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014. Dec 25;371(26):2499–509. [DOI] [PubMed] [Google Scholar]
- 3.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017. May 1;7(5):1016–36. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Zhang G, Wang J, Wang L, Huang X, Cheng Y. The role of cancer-associated fibroblasts in esophageal cancer. J Transl Med. 2016. Jan 29;14(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020. Mar;20(3):174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021. Oct 11;20(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakasheva TA, Lin EW, Tang Q, Qiao E, Waldron TJ, Soni M, et al. IL-6 mediates cross-talk between tumor cells and activated fibroblasts in the tumor microenvironment. Cancer Res. 2018. Sep 1;78(17):4957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska-Bosiacka I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int J Mol Sci. 2020. Jan;21(20):7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldinucci D, Borghese C, Casagrande N. The CCL5/CCR5 Axis in Cancer Progression. Cancers. 2020. Jul 2;12(7):1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banks-Schlegel SP, Quintero J. Growth and Differentiation of Human Esophageal Carcinoma Cell Lines. 1986;46:9. [PubMed] [Google Scholar]
- 11.Nishihira T, Kasai M, Mori S, Watanabe T, Kuriya Y, Suda M, et al. Characteristics of two cell lines (TE-1 and TE-2) derived from human squamous cell carcinoma of the esophagus. Gan. 1979. Oct;70(5):575–84. [PubMed] [Google Scholar]
- 12.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119(8):441–9. [DOI] [PubMed] [Google Scholar]
- 13.Natsuizaka M, Kinugasa H, Kagawa S, Whelan KA, Naganuma S, Subramanian H, et al. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res. 2014. Jan 15;4(1):29–41. [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020. Jun;38(6):675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han C, Liu T, Yin R. Biomarkers for cancer-associated fibroblasts. Biomark Res. 2020. Nov 11;8(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK signalling pathway and tumorigenesis (Review). Exp Ther Med. 2020. Mar 1;19(3):1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014. Nov;14(11):736–46. [DOI] [PubMed] [Google Scholar]
- 18.Lavoie JN, L’Allemain G, Brunet A, Müller R, Pouysségur J. Cyclin D1 Expression Is Regulated Positively by the p42/p44MAPK and Negatively by the p38/HOGMAPK Pathway*. J Biol Chem. 1996. Aug 23;271(34):20608–16. [DOI] [PubMed] [Google Scholar]
- 19.Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hébert CA. Distinct but Overlapping Epitopes for the Interaction of a CC-Chemokine with CCR1, CCR3, and CCR5. Biochemistry. 1997. Aug 1;36(32):9642–8. [DOI] [PubMed] [Google Scholar]
- 20.Woollard SM, Kanmogne GD. Maraviroc: a review of its use in HIV infection and beyond. Drug Des Devel Ther. 2015;9:5447–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, et al. Maraviroc (UK-427,857), a Potent, Orally Bioavailable, and Selective Small-Molecule Inhibitor of Chemokine Receptor CCR5 with Broad-Spectrum Anti-Human Immunodeficiency Virus Type 1 Activity. Antimicrob Agents Chemother. 2005. Nov;49(11):4721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, et al. Structure of the CCR5 Chemokine Receptor–HIV Entry Inhibitor Maraviroc Complex. Science. 2013. Sep 20;341(6152):1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saw PE, Chen J, Song E. Targeting CAFs to overcome anticancer therapeutic resistance. Trends Cancer. 2022. Jul 1;8(7):527–55. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Liu X, Cao S, Dong X, Rao S, Cai K. Understanding Esophageal Cancer: The Challenges and Opportunities for the Next Decade. Front Oncol. 2020. Sep 10;10:1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu YC, Shen YC, Chang JWC, Hsieh JJ, Chu Y, Wang CH. Autocrine CCL5 promotes tumor progression in esophageal squamous cell carcinoma in vitro. Cytokine. 2018. Oct 1;110:94–103. [DOI] [PubMed] [Google Scholar]
- 26.Gao LF, Zhong Y, Long T, Wang X, Zhu JX, Wang XY, et al. Tumor bud-derived CCL5 recruits fibroblasts and promotes colorectal cancer progression via CCR5-SLC25A24 signaling. J Exp Clin Cancer Res CR. 2022. Mar 3;41(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirri P, Chiarugi P. Cancer-associated-fibroblasts and tumour cells: a diabolic liaison driving cancer progression. Cancer Metastasis Rev. 2012. Jun 1;31(1):195–208. [DOI] [PubMed] [Google Scholar]
- 28.Linxweiler J, Hajili T, Körbel C, Berchem C, Zeuschner P, Müller A, et al. Cancer-associated fibroblasts stimulate primary tumor growth and metastatic spread in an orthotopic prostate cancer xenograft model. Sci Rep. 2020. Jul 28;10(1):12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai S, Wu Y, Yan Y, Kang H, Zhang J, Ma W, et al. The effect of CCL5 on the immune cells infiltration and the prognosis of patients with kidney renal clear cell carcinoma. Int J Med Sci. 2020. Oct 18;17(18):2917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araujo JM, Gomez AC, Aguilar A, Salgado R, Balko JM, Bravo L, et al. Effect of CCL5 expression in the recruitment of immune cells in triple negative breast cancer. Sci Rep. 2018. Mar 20;8(1):4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mgrditchian T, Arakelian T, Paggetti J, Noman MZ, Viry E, Moussay E, et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A. 2017. Oct 31;114(44):E9271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q wen, Liu L, Gong C yang, Shi H shan, Zeng Y hui, Wang X ze, et al. Prognostic Significance of Tumor-Associated Macrophages in Solid Tumor: A Meta-Analysis of the Literature. PLoS ONE. 2012. Dec 28;7(12):e50946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaafar NM, Osman TAH, Ahmed IA, Elsheikh M, Dongre H, Jacobsen MR, et al. Characterization of immune cell infiltrate in tumor stroma and epithelial compartments in oral squamous cell carcinomas of Sudanese patients. Clin Exp Dent Res. 2022;8(1):130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu G, Chen L, Wu S, Feng Y, Lin T. Comprehensive Analysis of Tumor-Infiltrating Immune Cells and Relevant Therapeutic Strategy in Esophageal Cancer. Dis Markers. 2020. May 11;2020:8974793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated and only publicly available datasets were analyzed during the current study.