Significance
Understanding the processes responsible for the striking plant diversity found in tropical forests has been a constant preoccupation of ecology and evolutionary biology. Recent studies have proposed a role for introgressive gene flow. To test the prevalence of introgression in a high-diversity clade of trees with a specialized pollination system, we built a global phylogenomic framework of figs (Ficus), a keystone species across tropical forests and partners in a celebrated pollination mutualism. Our results based on 1,858 genes for 520 species of figs and relatives found limited introgression in the nuclear genome despite widespread cytoplasm transfer, consistent with phylogenetically stable lineages despite occasional hybridization. A well-resolved phylogenomic framework for figs provides an important tool for classification and comparative evolutionary studies.
Keywords: Ficus, hybridization, Moraceae, phylogenomics
Abstract
Studies investigating the evolution of flowering plants have long focused on isolating mechanisms such as pollinator specificity. Some recent studies have proposed a role for introgressive hybridization between species, recognizing that isolating processes such as pollinator specialization may not be complete barriers to hybridization. Occasional hybridization may therefore lead to distinct yet reproductively connected lineages. We investigate the balance between introgression and reproductive isolation in a diverse clade using a densely sampled phylogenomic study of fig trees (Ficus, Moraceae). Codiversification with specialized pollinating wasps (Agaonidae) is recognized as a major engine of fig diversity, leading to about 850 species. Nevertheless, some studies have focused on the importance of hybridization in Ficus, highlighting the consequences of pollinator sharing. Here, we employ dense taxon sampling (520 species) throughout Moraceae and 1,751 loci to investigate phylogenetic relationships and the prevalence of introgression among species throughout the history of Ficus. We present a well-resolved phylogenomic backbone for Ficus, providing a solid foundation for an updated classification. Our results paint a picture of phylogenetically stable evolution within lineages punctuated by occasional local introgression events likely mediated by local pollinator sharing, illustrated by clear cases of cytoplasmic introgression that have been nearly drowned out of the nuclear genome through subsequent lineage fidelity. The phylogenetic history of figs thus highlights that while hybridization is an important process in plant evolution, the mere ability of species to hybridize locally does not necessarily translate into ongoing introgression between distant lineages, particularly in the presence of obligate plant–pollinator relationships.
Interrogating the processes that gave rise to and continue to maintain the splendid plant diversity found in tropical forests has been a constant preoccupation of natural history and evolutionary biology (1). Recent studies have increasingly proposed a role for introgressive gene flow between species, made possible by high numbers of closely related sympatric species, as a feature of tropical biodiversity (2, 3). Yet whether diversity facilitates or results from introgression is difficult to determine (4). Introgression may increase genetic diversity within species (3) and has the potential to move potentially adaptive alleles among them (5), though at the risk of genetic homogenization (6). Factors limiting interspecific hybridization such as pollinator specialization, temporal separation of flowering times, and pollen incompatibility are important balancing mechanisms allowing species coexistence. There is, however, confusion in the tropical plant literature between hybridization, which provides the opportunity for genetic introgression, and introgression, the incorporation of genetic material from one species into a lineage after the initial hybrids and first-generation backcrosses (7). Hybridization may in fact lead to only rare introgression in lineages with strong isolating mechanisms such as high pollinator specificity. Nevertheless, these rare events have the potential to compound, imprinting ancient introgression on global clades. Exemplary model systems with a robust phylogenetic and morphological framework can provide a better understanding of the relative importance of these processes.
The global lineage of fig trees (Ficus, Moraceae) presents a model system for dissecting the history of introgression in the face of strong isolating mechanisms. A keystone genus of tropical forests, a partner in a celebrated pollination mutualism, a shade tree to the Buddha during his enlightenment, the Egyptian tree of life, one of the Seven Species of the Hebrew Bible, and the namesake of a sura in the Qur’an, Ficus has few peers in its combination of ecological and cultural importance. This diverse genus of about 850 species consists of lineages that are largely biogeographically confined, with many sympatric species in forests throughout the tropics (8, 9). Scholars have long recognized coevolution with specialized pollinating wasps (Agaonidae) as a major engine of fig diversity (10, 11), as pollinator specificity can serve as a powerful isolating mechanism, limiting hybridization (12, 13). Yet fig biologists have long recognized occasional pollinator sharing and host switching, either inferred from comparisons of fig and wasp phylogenetic trees or observed directly in some closely related species (7, 14). Some studies have therefore proposed that genetic introgression facilitated initially by incomplete pollinator specificity could play a major role in the evolution of Ficus (14, 15). These two processes, introgression via nonabsolute pollinator specificity and multiple species coexistence facilitated by pollinator specialization, may balance one another to help maintain the high diversity of figs.
The outsized importance of Ficus is matched by the challenges it has posed for classification (16), leading E.D. Merrill to question the sanity of taxonomists who choose to work on Ficus (8). Despite decades of global phylogenetic studies (10, 11, 16–18), variable sampling schemes, lack of phylogenetic resolution, and conflicting nuclear and organelle topologies (15, 19) have hindered conclusive resolution of major questions in the evolutionary history of Ficus. Based on the most complete phylogenetic study of PCR-amplified loci, including six gene regions for 307 species of Ficus, Clement et al. (16) took a major step toward reconciling molecular and traditional classification, establishing informal clade names and providing a tractable framework for future revisionary work. While recent studies largely agree on similar section-level clades, backbone relationships remain uncertain and subject to substantial disagreement. For example, analyses disagree on whether the Neotropical sect. Pharmacosycea is sister to all other figs, whether the monoecious stranglers form a clade, and whether the dioecious species form a clade (10, 11, 15, 16, 19, 20).
Despite the reproductive isolation expected in a group with high pollinator specificity, there is an emerging consensus that hybridization does occur between closely related Ficus species, further complicating the taxonomy of the genus (7, 21, 22). Hybridization may have played an important role in the evolution of Ficus, though only limited published evidence suggests that hybridization results in nuclear gene introgression in the genus (14, 15). The case for whether genetic introgression among Ficus species plays a prominent and ongoing role in the evolution of the genus, contributing to codiversification with pollinating wasps, is still open (10). Indeed, the extent of interspecific gene flow in the genus remains uncertain, and even dramatic plastome–nuclear discordance may represent occasional localized gene flow rather than widespread introgression (23). This is partially because no phylogenomic study exploring reticulate relationships has yet employed dense and widespread taxon sampling comparable to the well-sampled studies based on few loci (16). In addition, the evolution and diversification of Ficus must be understood in the evolutionary and comparative context of the Moraceae family more broadly (16, 20, 24–27).
This study documents the outcome of an ancient balance between introgression and strong reproductive isolation in one of the world’s largest angiosperm genera. We provide the most densely sampled and data-rich phylogenomic reconstruction of Ficus to date, contextualized within comprehensive phylogenetic sampling across Moraceae. We use our phylogeny to investigate two alternatives: whether the phylogeny of Ficus is network-like, with ongoing introgression blurring boundaries between lineages; or more tree-like, with pulses of introgression punctuating evolution within phylogenetically stable lineages. Analyses employed 1,751 nuclear loci for 235 species of Ficus and 285 other Moraceae, representing all genera, with species-level sampling within most of them, including divergence time estimations and ancestral-range reconstructions. Our analyses also provide a well-supported framework for further work on the classification, origin, and dispersal of figs.
Results
Nonitalicized informal clade names follow the system established by Clement et al. (16), except that we now consider Mixtiflores to include the Urostigma clade as shown by Rasplus et al. (11) and this study. Formal taxonomic names follow Berg and Corner (8), as updated by Pederneiras et al. (28). Additional proposed changes by Zhang et al. (29) are noted when relevant but are not used throughout the paper as some of them are not compatible with our results. Synonyms appear in parentheses when the latter are in more common use than the legitimate names. A summary of relevant names appears in Table 1.
Table 1.
Left: Clade names according to Clement et al. (16), as modified here. Middle: Corresponding formal taxonomic names and ranks according to Berg and Corner (8), as updated by Pederneiras et al. (28). Right: Taxonomic names and ranks according to Zhang et al. (29), to the extent they differ
| Clade | Berg and Corner, as updated by Pederneiras et al. | Zhang et al., if different |
|---|---|---|
| Pharmacosycea | Subg. Pharmacosycea, sect. Pharmacosycea | – |
| Mixtiflores | Subg. Spherosuke (=Urostigma) | – |
| Urostigma | Subsect. Urostigma | – |
| Conosycea | Subsect. Cordifoliae (=Conosycea) | – |
| Malvanthera | Sect. Malvanthera | – |
| Madagascar clade | [not recognized] | [not recognized] |
| Galoglychia | Sect. Platyphyllae | – |
| Americanae | Sect. Americanae | – |
| Oreosycea | Subg. Pharmacosycea, sect. Oreosycea | – |
| Caricae | Subg. Ficus, subsect. Ficus | Subg. Ficus in total |
| Sycidium | Subg. Terega (=Sycidium) | – |
| Sycomorus | Subg. Sycomorus | – |
| Frutescentiae | Subg. Ficus sect. Frutescentieae | Subg. Synoecia sect. Plagiostigma in part |
| Eriosycea | Subg. Ficus sect. Eriosycea | Subg. Synoecia sect. Apiosycea in part |
| Synoecia | Subg. Synoecia | Subg. Synoecia in part |
Phylogenetic Trees.
We conducted phylogenetic analyses in four categories: 1) All-Ficus nuclear species tree: a coalescent-based species tree based on 1,858 gene trees for 362 samples (232 Ficus species and 5 outgroups, Fig. 1) from samples enriched for nuclear targets (30, 31); 2) Plastome: a maximum-likelihood tree based on whole-chloroplast sequences for 180 taxa, Fig. 2 and SI Appendix, Fig. S2); 3) Introgression: phylogenetic network analyses under maximum pseudo-likelihood and a related analysis of gene tree conflict using rooted triplets (Figs. 2 and 3 and SI Appendix, Figs. S2–S4) based on 1,223 gene trees; and 4) Moraceae: a whole-family tree, time-calibrated under penalized likelihood, based on 528 samples and 724 genes (including 231 Ficus species, with replicates deduplicated, Fig. 4). All samples with successful technical replicates (successful recovery of at least 1,000 loci for both replicates) were sister to one another in the phylogenetic analyses.
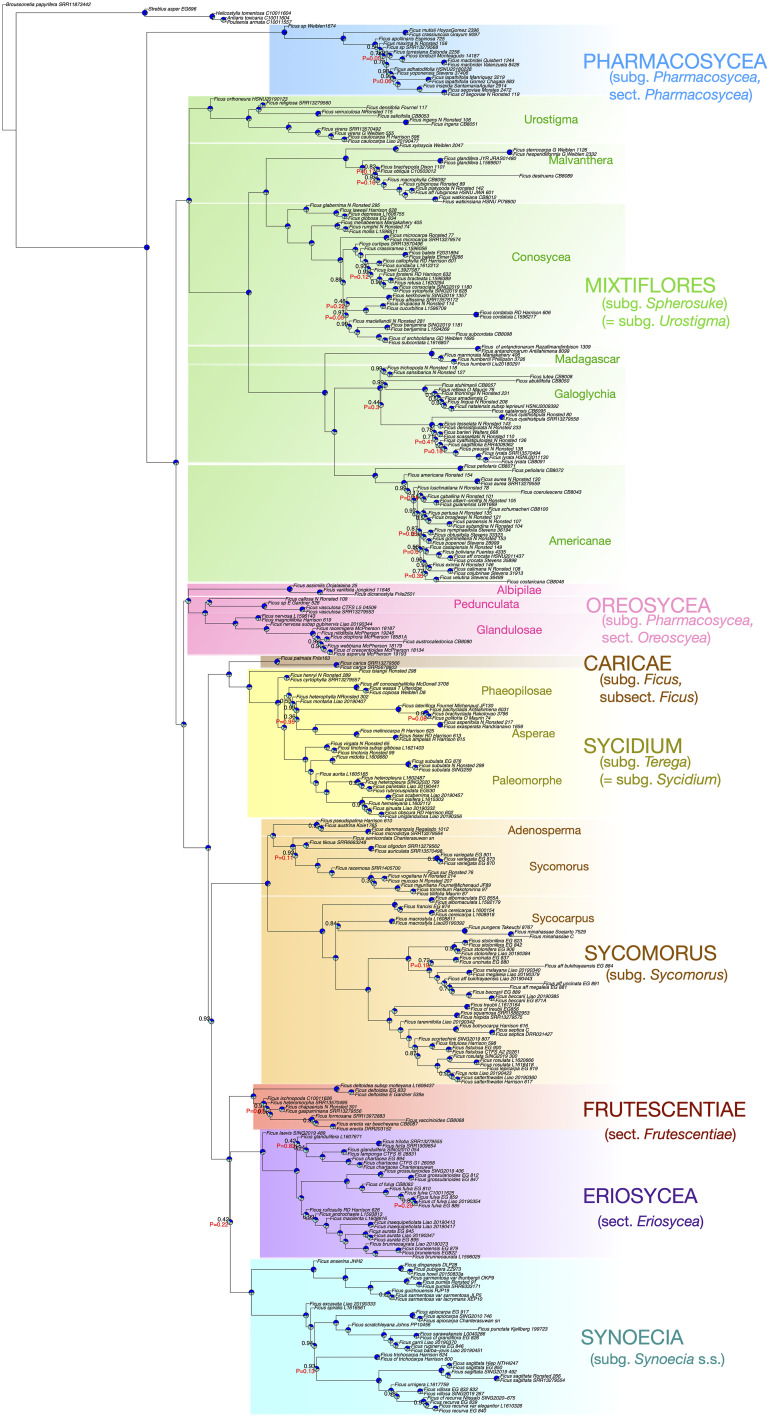
Fig. 1.
All-Ficus tree. ASTRAL species tree based on 1,751 nuclear genes. Node labels denote local posterior probability (LPP) and P-value for the polytomy test. Unlabeled nodes have LPP = 1.0 and P < 0.05. Pie charts denote the quartet frequency of the main topology (blue), the second-most frequent topology (light blue), and the third-most frequent topology (gray).
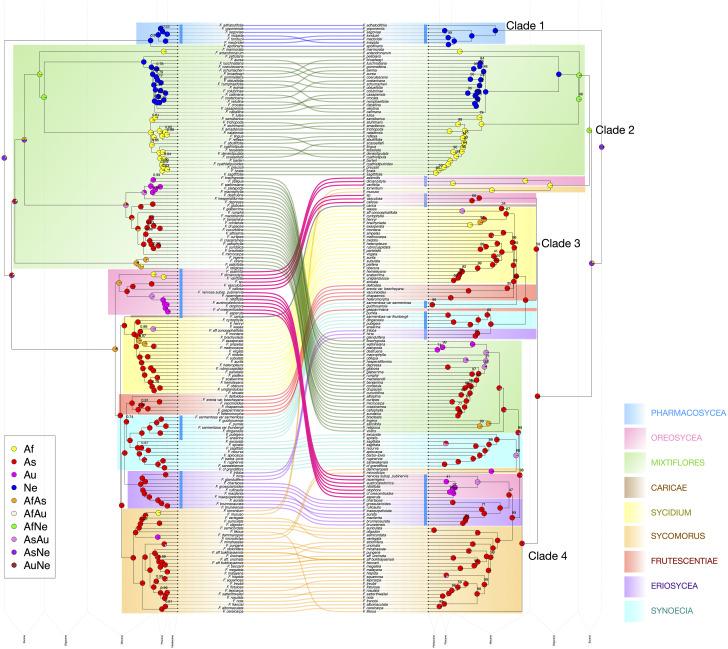
Fig. 2.
Cyto-nuclear discordance. (Left) Time-calibrated nuclear species tree (pruned from Fig. 4), nodes labeled with posterior probability (unlabeled nodes have LPP = 1.0). (Right) Time-calibrated chloroplast tree, nodes labeled with bootstrap support (unlabeled nodes have bootstrap = 100%). Pie charts at nodes on both trees represent inferred ancestral ranges. Clade colors match those in Fig. 1. Blue bars next to tip labels denote taxa pollinated by members of subfamily Blastophaginae.
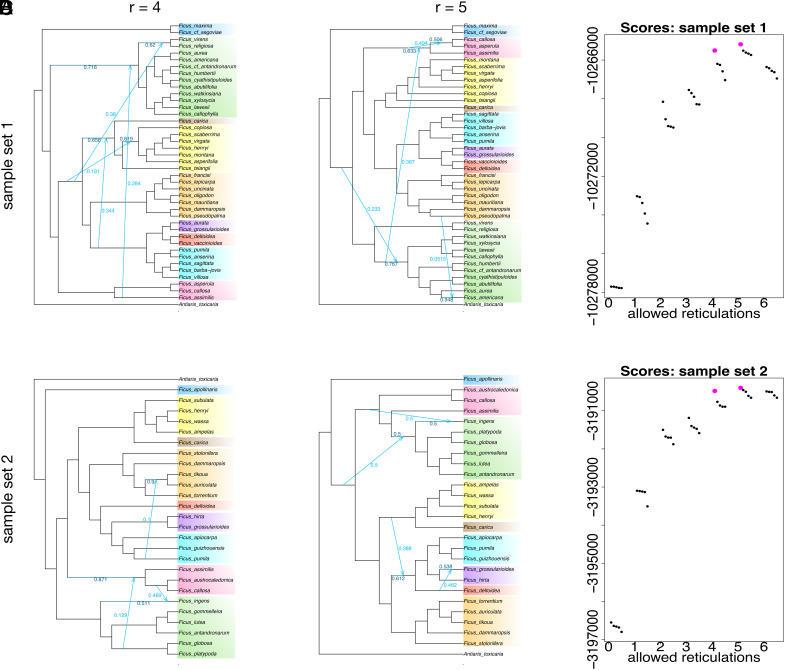
Fig. 3.
The best maximum-pseudo-likelihood networks allowing a maximum of 4 (left column) and 5 (middle column) reticulations, plus associated plots of network scores (right column) for sample sets 1 (top row) and 2 (bottom row) based on 1,223 nuclear gene trees. Clade colors match those in Fig. 1. Inheritance probabilities appear in light blue for reticulated (minor) edges and in dark blue for the corresponding branches on the major (bifurcating) tree.
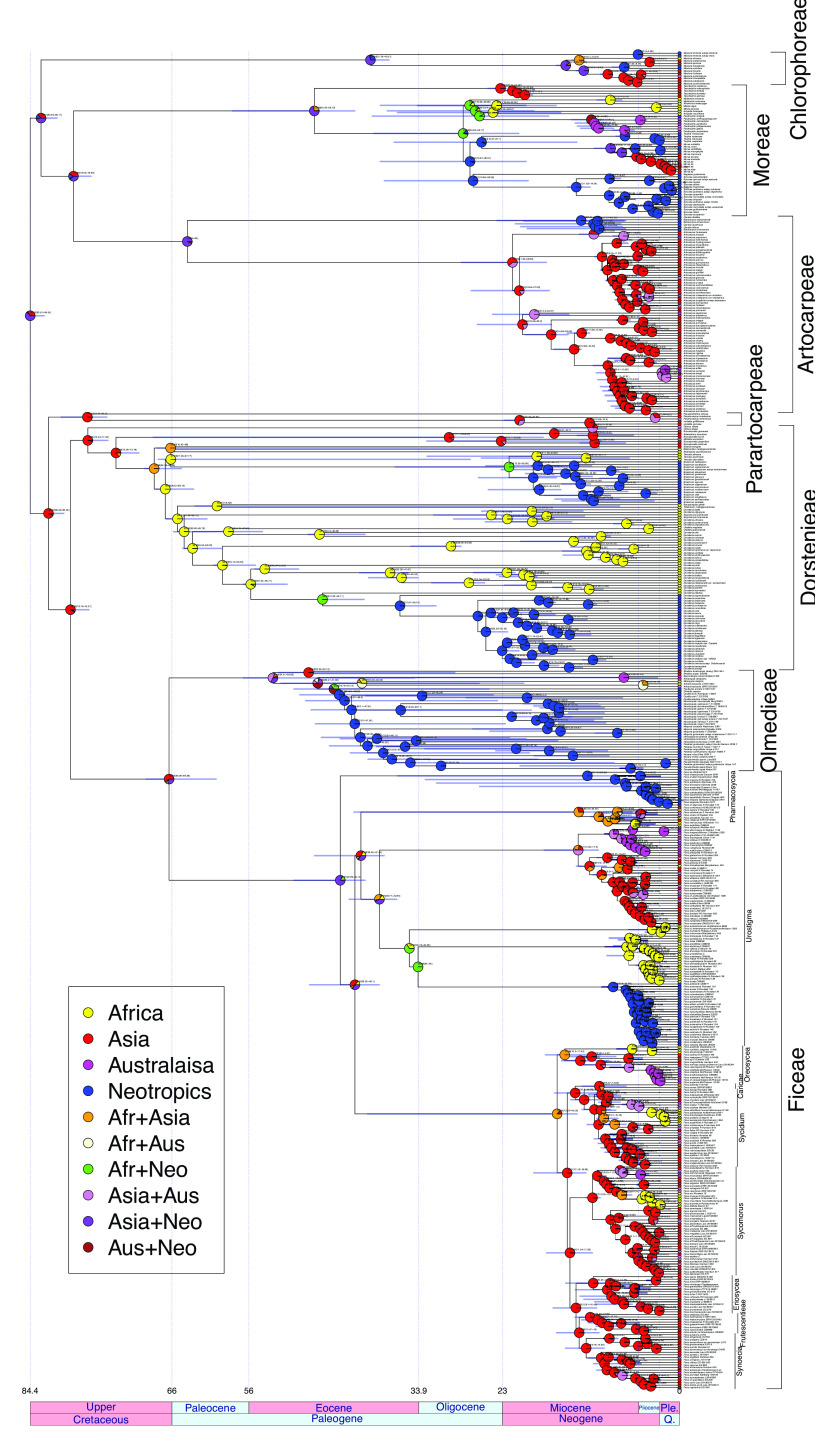
Fig. 4.
Time-calibrated Moraceae tree, showing inferred ancestral ranges at the nodes. Textual node labels indicate node ages and 95% HPD, in Ma.
Nuclear Species Trees.
The All-Ficus tree (Fig. 1) was based on ca. 666.7 billion nuclear quartet trees, and the final normalized quartet score was 0.77. All subgenera recognized by Berg and Corner (8) were monophyletic except Pharmacosycea and Ficus, the latter corresponding instead to Zhang et al.’s reduced circumscription (29), which comprises only the Caricae clade. The monoecious Neotropical Pharmacosycea clade (sect. Pharmacosycea) was sister to the entire rest of the genus, while the monoecious Paleotropical Oreosycea (sect. Oreosycea) was sister to a clade containing all of the dioecious figs: the Caricae (subsect. Ficus) + Sycidium (subg. Terega = subg. Sycidium), subg. Synoecia s.l. sensu Zhang [comprising the Eriosycea (sect. Eriosycea), Frutescentiae (sect. Frutescentiae), and Synoecia (subg. Synoecia s.str.) clades], and Sycomorus (subg. Sycomorus, which also contains a few monoecious figs) clades. Within the Synoecia clade, Frutescentiae was sister to Eriosycea and Synoecia, although with low support (LPP = 0.42) and without rejecting the polytomy hypothesis (P = 0.22). The monophyletic monoecious Mixtiflores (subg. Spherosuke = subg. Urostigma), containing the Urostigma (subsect. Urostigma), Conosycea (sect. Cordifoliae = sect. Conosycea), Malvanthera (sect. Malvanthera), Americanae (sect. Americanae), and Galoglychia (sect. Platyphyllae = sect. Galoglychia) clades, was sister to all of Ficus except Pharmacosycea. Notable within that clade was a previously unrecognized section-level clade of Madagascan species sister to Americanae (Neotropics) and Galoglychia (Africa). The backbone did not change in a more conservative analysis retaining only the least-saturated 50% of genes, those whose GC content did not differ substantially between Pharmacosycea and the rest of Ficus, and gene tree splits with at least 50% bootstrap support. A species tree based on 16,449 genes assembled from publicly available whole-genome reads for 26 samples also had the same backbone, save for a transposition in the positions of Eriosycea and Frutescentiae and a single rearrangement within Conosycea (SI Appendix, Fig. S1).
Chloroplast Tree.
The plastome tree (Fig. 2 and SI Appendix, Fig. S2) revealed four main lineages. One of these lineages (Clade I) was strictly associated with Neotropical Pharmacosycea and was sister to the other three clades. Species belonging to Oreosycea appear in each of these other clades, always sister to most other species within each. Clade II is largely African and Neotropical, comprising a clade of African Oreosycea and Sycomorus, sister to half of Mixtiflores (Madagascar, Galoglychia, and Americanae). Clade III contains three Asian Oreosycea, sister to a mixed assemblage of several dioecious clades (Cariceae, Frutescentiae, and parts of Eriosycea, Sycidium, and Synoecia). Clade IV consists of Ficus tikoua (Sycomorus) and F. albipila (Oreosycea) in a grade, followed by two subclades—one containing the rest of Mixtiflores along with Malesian and Papuasian Synoecia and Sycomorus species and the other containing Australasian Oreosycea plus most of Eriosycea and Sycomorus.
Analyses of the nuclear and chloroplast datasets together revealed substantial cyto-nuclear discordance. Scoring the plastome tree using a pruned set of nuclear gene trees resulted in a low normalized quartet score of 0.61, indicating that nearly 40% of nuclear gene tree quartets were not reflected in the plastome tree. Similarly, coalescent-based simulations starting from the nuclear species tree revealed that the plastome tree contained dramatically more extra lineages (52 clades not appearing in nuclear species tree) than would be expected for a maternally inherited organellar genome (simulated plastome trees had a median of 6 clades not appearing in the nuclear species tree) and instead fell within the expected range of extra lineages present in simulated nuclear gene trees under incomplete lineage sorting (SI Appendix, Fig. S3).
Introgression.
We investigated introgression using maximum pseudo-likelihood networks as well as an analysis of rooted triplet trees.
For network analyses based on 1,223 gene trees, two strategies were employed—one with 27 samples chosen to reflect cyto-nuclear discordance and another with 42 samples representing major nuclear lineages. For both schemes, the optimal networks had four or five reticulations, depending on whether a slope heuristic or BIC was used to select the model (Fig. 3 and SI Appendix, Table S2). All four optimal networks showed reticulation between Oreosycea and Mixtiflores (and Urostigma in particular), reflecting competing topologies placing Oreosycea either sister to the dioecious clade (Fig. 3 A–C) or with the Mixtiflores stranglers (Fig. 3D). These reticulations reflected a substantial signal in the nuclear gene trees, with inheritance probabilities (the proportional genomic contribution to the reticulated edge) mostly above 0.28, and in one network (Fig. 3D) as high as 0.5, reflecting equally strong signals for the major tree and the reticulated edges. Another recurring theme (Fig. 3 A and D) was reticulation along the backbone of the dioecious clade reflecting conflict as to whether Caricae+Sycidium or Sycomorus was sister to the rest of the clade, again reflecting a substantial signal in the gene trees (inheritance probability >0.3 for the reticulated edge in both cases). In one case (Fig. 3C), a weak signal of reticulation (inheritance probability 0.1) was detected between Synoecia (root climbers) and the sublineage of Sycomorus containing F. tikoua, the only member of its clade with a rampant habit. In the best network based on 13,587 genes from whole-genome data for 24 samples (SI Appendix, Fig. S4), the strongest reticulation signal was between Urostigma and the single Oreosycea sample included in that analysis (inheritance probability for the minor edge of 0.383); the other inferred reticulations were based on comparatively weak signals, with inheritance probabilities all below 0.16 (i.e., above 0.84 for the major tree). All networks, scores, and information criteria appear in SI Appendix.
In the triplet analysis of 42 samples (SI Appendix, Fig. S5), patterns of gene tree discordance followed the same themes as those reconstructed under maximum pseudo-likelihood, suggesting reticulation linking Oreosycea to Mixtiflores and the clade containing all of the dioecious lineages (Caricae, Sycidium, Sycomorus, Frutescentiae, Eriosycea, and Synoecia). The pattern was stronger for Ficus assimilis (Albipilae) than for the other two samples of Oreosycea (Glandulosae and Pedunculata), but similar patterns were also observed for the other samples, so the introgression may be shared by all of the samples.
Family-Wide Analysis.
The Moraceae tree (Fig. 4), inferred from 724 genes selected for clock-like behavior (determined by low root-to-tip variance, low site saturation, and concordance with the species tree topology), recovered well-supported clades for each of the seven tribes, sorted into two sister subfamilial clades. The first comprised Chlorophoreae, which was sister to Artocarpeae + Moreae. The second contained Parartocarpeae sister to Dorstenieae+Olmedieae+Ficeae, the latter two sister to one another. Within Olmedieae, the wind-pollinated Streblus was sister to the rest of Olmedieae, and the wind-pollinated Olmedia was sister to the entire (largely insect pollinated) Neotropical clade save for the morphologically distinctive Poulsenia. The Ficus backbone was the same as in the All-Ficus tree described above, including Pharmacosycea as sister to the rest of Ficus, save for a transposition in the positions of Eriosycea and Frutescentiae, mirroring the topology recovered by the whole-genome Ficus tree. The crown and stem ages of Ficus were 44.4 and 63.8 Ma, respectively. The ancestral area reconstruction on the same tree (Fig. 4 and SI Appendix, Fig. S10) reconstructed an ancestral stem range for Ficus of Asia, expanding at the crown to Asia+Neotropics. The same Asia+Neotropics pattern was recovered for three other tribes in the family: Moreae, Artocarpeae, and Chlorophoreae. The crown and stem range for all Ficus except for Pharmacosycea was Asia alone, with subsequent dispersals to other regions. The most likely range for the crown node of Moraceae was also Asia+Neotropics. An analysis of speciation rates recovered seven rate shifts in Moraceae, five of them in Ficus, in Oreosycea+dioecious figs, Conosycea, Malvanthera, Galoglychia, and Americanae, with the highest rate in the latter (SI Appendix, Fig. S7).
Discussion
This study presents the most densely sampled phylogenomic reconstruction of Ficus to date. It is a major advance in untangling deep phylogenetic relationships and clarifying the contribution of introgression to the evolution of Ficus. While previous studies have pointed either to tight codiversification with wasps (10, 32) or to widespread hybridization (14, 15), our results show that both processes are at play: Ficus reflects a history of lineage stability punctuated by introgression events among ancestral lineages. These largely relate to the backbone topology and center around Oreosycea in both the nuclear and chloroplast analyses (Figs. 2 and 3 and SI Appendix, Figs. S2–S9). Traces of deep introgression of Oreosycea cytoplasm into most other lineages in the chloroplast tree largely did not mirror conflicts relating to that lineage in the nuclear tree, which suggests a history of localized introgression in deep time followed by lineage fidelity rather than ongoing introgression between lineages. This finding confirms a basic scenario of coevolution within major fig and wasp lineages, with major introgression events limited to stem lineages likely predating the diversification of extant figs. This is consistent with evidence from population-level studies concluding that while figs can sometimes hybridize readily, species nevertheless usually remain distinct (7, 33).
Echoes of Introgression in Cyto-Nuclear Discordance.
We recovered substantial cyto-nuclear discordance, supporting the findings of Bruun-Lund et al. (19). The chloroplast tree was dramatically more different in topology than would be expected under simple incomplete lineage sorting (SI Appendix, Fig. S3), providing compelling evidence of ancient introgression among major lineages, largely centered around the Oreosycea clade, reflecting both geographic affinity and pollinator alliances (Fig. 2 and SI Appendix, Fig. S2). Yet the particular relationships evident in the chloroplast tree are largely not reflected in phylogenetic networks based on nuclear genes. Combined with the high normalized quartet scores in the nuclear analyses (>0.75)—slightly higher than those previously calculated for Artocarpus (34)—these dynamics suggest occasional rather than sustained introgression through evolutionary time, with the chloroplast tree preserving echoes of events that have largely been drowned out by the intralineage fidelity characterizing the nuclear genome. The Oreosycea clade appears in three out of four chloroplast lineages, usually sister to geographically proximate taxa. This likely represents local gene flow mediated by pollinator host shifting or sharing, perhaps (as discussed below) driven by isolation of individual fig trees in climatically unstable areas. Some African Sycomorus species belong to the same highly supported chloroplast lineage (SI Appendix, Fig. S2, Clade 2) as some African species of Oreosycea; however, this affinity is not supported by the nuclear gene trees, which support an unambiguously monophyletic Sycomorus (LPP = 1.0, P < 0.05). In two chloroplast lineages, the transfer of Oreosycea plastome was probably mediated by members of the Blastophaginae (35), which pollinate both Oreosycea and some of the species associated by their chloroplast lineages (SI Appendix, Fig. S2, Clades 3 and 4 in particular). The Blastophagineae also link Oreosycea and Pharmacosycea—the two lineages that traditionally comprise the (paraphyletic) subgenus Pharmacosycea. Another member of Moraceae, the broad Streblus as defined by Corner, forms a morphological but nonmonophyletic alliance dispersed throughout the phylogeny that may be described as morphological relics of an ancestral taxon (27, 36). The same might be said of the two sections of subg. Pharmacosycea, preserving ancestral characters and occupying sister positions either to the entire genus or to subclades in both the nuclear and chloroplast trees.
The ancient introgressions inferred here are consistent with patterns observed in extant figs. Deep introgression events appear to be limited to certain lineages (primarily Mixtiflores and Oreosycea), and hybridization among extant figs is also apparently unevenly distributed among clades, with many reports focusing on the Neotropical and Afrotropical monoecious figs (10, 14, 33, 37), although hybridization between closely related dioecious figs has also been reported (38–40). This variation in effective gene flow could reflect variation in pollinator specialization (39) or genetic distance among species within clades. Two processes may facilitate the breakdown in pollinator exclusivity necessary for hybridization to occur. Biogeographic processes may lead to secondary contact between recently diverged species or even bring together previously separated species that had never evolved barriers to hybridization. Current hybridization in Panamanian figs (14, 33) might involve this dynamic, facilitated by the recent rise of the Isthmus of Panama. A recent study found ongoing hybridization and historical introgression between two closely related fig species, which nevertheless remain essentially distinct (33), mirroring on a small scale the broader dynamic observed in deep time in this study: stability of lineages despite occasional hybridization. A second kind of breakdown of pollinator exclusivity has been documented when a fig species is isolated and the normal pollinator is absent (13, 41). Historical introgressions may result from the same processes, with isolation perhaps caused by climatic or environmental changes; indeed, many extant Oreosycea and Urostigma species display intermittent growth, a helpful adaptation to environmental fluctuations in seasonally dry forests where some of these species are native. Yet even in isolation, figs often maintain fidelity to their usual pollinators (12, 13), contributing to the episodic nature of introgression in figs. This second pattern mirrors recent findings in the oak family (Fagaceae) in which hybridization at the base of the tree involved lineages that no longer hybridize but that exhibited historical sympatry (at continental levels), while introgression among extant species may reflect adaptive introgression or simply porous barriers between recently diverged species (42, 43). Ficus, like oaks, may be a syngameon reflecting an equilibrium between adaptive introgression and the maintenance of genomically distinct species (5), and occasional introgression involving stress-adapted species (like certain Oreosycea) may contribute to the survival and evolution of other species. This idea might be tested in the future by combining our phylogenetic results with current and past climatic data.
Classification of Figs.
We highlight two noteworthy taxonomic findings. The first is the existence of a section-level clade that is endemic to Madagascar, containing at least three species. The persistence of this lineage fits into a pattern of unique lineages diversifying in Madagascar, highlighting the geographic and evolutionary isolation of the island (44–46). Further study of these taxa may provide insights into the origins of the African and Neotropical stranglers (Galoglychia and Americanae), which have high rates of speciation (SI Appendix, Fig. S7) (47). This finding also highlights the importance of dense taxon sampling, as we had no a priori expectation that these species would form a distinct clade. Second, we note the monophyly of subg. Spherosuke (=Urostigma, the Mixtiflores clade), a subject of conflicting past studies (11, 16). While its monophyly is highly supported in the nuclear tree (LPP = 1.0, P < 0.05), substantial gene tree conflict exists (evidenced by the quartet score of less than 0.5), and all inferred networks showed introgression involving at least part of the subgenus, which appears as two distinct chloroplast lineages.
The well-resolved phylogenetic tree presented here provides a basis for establishing an updated higher classification of Ficus. While the backbone topology here differs in some ways from that recovered by other phylogenomic studies (11, 15), those studies (and for the most part Clement et al. (16) as well) agree on the major clades (Fig. 1). The informal clade names proposed by Clement et al. (16), with the addition of Urostigma to the Mixtiflores clade, already provide a framework for fig classification. Building upon that framework as well as the emerging consensus as to the major clades, the formal taxa could easily be updated with a few mere changes in rank (e.g., elevating sect. Oreosycea to subgenus level) and circumscription (perhaps expanding the circumscription of subg. Ficus).
Biogeography.
Our time-calibrated Moraceae tree reconstructed a Paleocene stem (63.8 Ma, 95% HPD 60.3 to 69.4) and Eocene crown for Ficus (44.4 Ma, 95% HPD 40.9 to 50.1), close to the ages in the study by Zhang et al. (20) (Fig. 4 and SI Appendix, Fig. S6). Given the paucity of verifiable Ficus fossils and the possibility of deep coalescence in gene trees, these dates might best be treated as minimum ages. The biogeographic analysis recovered an Asian stem node with two subsequent dispersals to the Neotropics, consistent with previous studies finding an Asian origin for Ficus (10, 48). The Asia-to-Asia+Neotropics pattern appears at the root of four out of seven tribes in Moraceae, reflecting a recurring phylogenetic pattern within Moraceae of largely Asian clades sister to largely Neotropical clades.
Moraceae were present in boreotropical North America during the Eocene (49), and while Ficus itself has not yet been recorded, the presence of the family raises the possibility that the habitat might have been suitable for the genus. Dispersal from Eurasia via North America to the Neotropics, previously proposed for Artocarpeae (50), is therefore plausible. An alternative possibility is dispersal via Africa, as Moraceae were also present in the late Cretaceous of Egypt (51), or even via subtropical Antarctica during the Eocene. Any of these scenarios would likely have involved substantial extinction of stem lineages due to glaciation in North America or Antarctica, or due to desertification in Africa. Other dynamics observed in our analyses hint at a role for extinct or unknown lineages. For example, the chloroplast affinity between some Australasian Oreosycea (ser. Glandulosae) and Eriosycea likely originated in a hybridization event between an extinct ancestor of both either in Asia or on a drifting India. Likewise, the Madagascar clade, sister to African and Neotropical stranglers, may represent a link between Asian and Neotropical+African Mixtiflores. Long-distance dispersal also cannot be discounted, particularly as both bats and birds are major dispersers of figs (52). The long Ficus stem coupled with the long distance between Asia and the Neotropics strongly suggests the involvement of extinct stem taxa (sometimes called “ghost lineages”) (53) in the evolution of Ficus. The older ages of internal nodes in the chloroplast tree (Fig. 2) as well as the generally older node ages in the most recent phylogenetic study of fig wasps provide additional circumstantial evidence of ghost lineages. The biogeographic interplay between the fig and wasp phylogenies will be a major area of study as detailed and well-supported phylogenetic reconstructions for both figs and wasps come into focus.
The analyses here paint a picture of more or less isolated evolution within phylogenetically stable lineages punctuated by occasional local introgression events likely mediated by local pollinator sharing. While the obligate mutualism between figs and fig wasps promotes lineage fidelity, it is not ironclad. Local gene flow—both within extant clades and among stem lineages predating extant diversity—plays a recurring role in the evolution of figs, perhaps allowing transfer of beneficial genes between species. By the same token, the fig–wasp mutualism appears to play a role in limiting introgression, resulting in a fundamentally tree-like pattern of evolution despite occasional introgression and promoting divergence of species by preventing genetic homogenization. The phylogenetic history of figs thus highlights that while hybridization is an important process in plant evolution, the mere ability of species to hybridize locally does not necessarily translate into ongoing introgression between lineages, particularly in the presence of obligate plant–pollinator relationships. As patterns in the evolution of tropical biodiversity continue to come into focus, researchers must continue to consider the balance between hybridization and the processes that limit it.
Materials and Methods
Taxon Sampling.
The complete study included a total of 627 samples, 245 species of Ficus (330 samples), 284 other Moraceae, and 13 outgroups (in Urticaceae and Cannabaceae) (SI Appendix, Table S1). In Ficus, sampling represented all major lineages and about 30% of the recognized species. In the rest of Moraceae, sampling represented all genera, mostly with species-level sampling (exceptions include Dorstenia, with ca. 50% sampling, and the tribe Olmedieae, with ca. 60% sampling). Data sources included 330 samples newly sequenced for this study and 256 samples from previous studies by the authors (27, 34, 54, 55), supplemented with samples from the NCBI Sequence Read Archive, and outgroups from the Plant and Fungal Tree of Life project (PAFTOL). New samples were collected during fieldwork in Australia, Brunei, Cameroon, Madagascar, Papua New Guinea, Philippines, Reunion, Sabah and Sarawak in Malaysia, Thailand, Yunnan, South China, Ryukyu Islands, from curated living collections of botanic gardens (Bergen Botanical Garden, Fairchild Tropical Botanical Garden, National Tropical Botanical Garden, US Department of Agriculture Subtropical Horticulture Research Station, Xishuangbanna Tropical Botanical Garden), and from herbarium specimens (C, F, K, L, MO, SING).
Sample Preparation and Sequencing.
Approximately 1 cm2 of leaf tissue was ground using a FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA), and DNA was extracted using either the Qiagen DNeasy Plant Mini kit (Qiagen, Hilden, German) or a modified CTAB protocol (56), with the incubation periods for lysis and precipitation in the latter sometimes extended to overnight or longer for herbarium specimens. DNA was quantified using a Qubit fluorometer and fragmented to a target mean size of 550 bp. Due to the scope of this collaboration, library preparation took place in more than one laboratory. Therefore, for some samples, Illumina TruSeq–compatible libraries were prepared using the NEB DNA Ultra 2 kit (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s protocol, except that most pre-PCR steps were carried out in half-volumes to save reagent costs. For other samples, similar libraries were prepared using the blunt-end single-tube protocol “BEST” (57) with an additional column-cleaning step (58). All newly prepared libraries were enriched for 1,315 single-copy nuclear genes (“Ficus1315”) identified by Bruun-Lund (31), and a majority were also enriched for the 333 Moraceae loci (“Moraceae333”) identified by Gardner et al. (30). Enrichment was carried out using a custom myBaits kit (Daicel Arbor Biosciences, Ann Arbor, MI, USA) following the manufacturer’s protocol, except that the RNA baits were diluted 1:1 with nuclease-free water. A subset of samples were subjected to technical replicates to verify the reliability and repeatability of laboratory processes.
Datasets.
We assembled four datasets:
All-Ficus: 1,751 genes for 330 Ficus samples (not including 29 technical replicates) plus five outgroups in the sister tribe, rooted with Broussonetia papyrifera.
WGS-Ficus: 16,449 genes for 26 Ficus samples, plus two outgroups.
Plastome: Whole chloroplasts for 219 Ficus samples.
Dated-Moraceae: 1,751 genes for 528 samples spanning the whole Moraceae, including one sample per species in Ficus.
Sequence Assembly and Alignment.
For the All-Ficus samples, we trimmed reads using Trimmomatic (ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 HEADCROP:3 LEADING:30 TRAILING:25 SLIDINGWINDOW:4:25 MINLEN:20) (59). The trimmed reads were assembled using HybPiper 1.2, which produces gene-by-gene, de novo assemblies guided by a set of reference CDS sequences (60). Assemblies were carried out using default parameters; the reference file consisted of the predicted CDS for each target gene in the Moraceae333 and Ficus1315 sets as well as sequences for the Angiosperms353 genes (61), some of which were recoverable from off-target reads, for a total of 1,751 genes. Subsequent analyses used the default HybPiper output, which is the predicted CDS for each gene. For each gene, sequences were filtered to remove those whose length was less than 100 bp or 25% of the average length of that gene, and samples with less than 50 genes remaining after filtering were discarded. The filtered sequences were aligned with MAFFT 7.450 (62), and sites with over 75% gaps were removed using TrimAl (63). An initial set of gene trees was estimated using FastTree 2.1.10 (64), and sequences corresponding to outlier branches were removed from the alignments using TreeShrink in “all-genes” mode. These cleaned alignments were used for all subsequent sequence analyses.
Phylogenomic Analyses.
Gene trees were generated using IQTree 2.0.3 (65) under the best-fit model for each gene as determined by Bayesian Information Criterion, and node support was calculated using 1,000 ultrafast bootstrap replicates. After collapsing nodes with less than 30% support using TreeCollapseCL 3.0 (66), the gene trees were used to estimate a species tree using ASTRAL-III 5.7.1 (67). Node support was estimated using LPP, a metric based on quartet scores that represents gene tree concordance. We also carried out a polytomy test in ASTRAL (−t 10) to investigate whether the polytomy hypothesis could be rejected for each node.
Because questions have been raised in previous studies about long branch attraction caused by site saturation affecting the rooting of Ficus (11), we reran ASTRAL on a conservative selection of genes filtered for saturation and GC-content heterogeneity using genesortR (68). First, we sorted genes by saturation level (69) and discarded the highest 50%; we then calculated GC content for Pharmacosycea and for the rest of Ficus and retained only those loci for which GC in both groups was within one SD of the mean GC in Ficus. We also applied a stricter filter to the gene trees, retaining only those splits with at least 50% bootstrap support.
Whole-Genome Phylogenetic Analysis.
This analysis contained 30 species, including the 15 analyzed by Wang et al. (15). Samples were trimmed and assembled as described above and then assembled with HybPiper 1.2 under default parameters, using the complete set of predicted CDS from the Ficus microcarpa genome (70) as a reference. We discarded all sequences for any gene that triggered a paralog warning in HybPiper for any sample. Within each locus, sequences less than 50% of the average length or 500 bp were discarded, and loci with an average length of less than 750 bp or containing fewer than 10 samples were also discarded. Alignment and tree inference for the 16,449 genes passing these filters then proceeded as outlined above.
Chloroplast Phylogeny.
For each sample, reads were mapped to the Ficus carica chloroplast genome (GenBank accession number KY635880.1) using BWA (71). Variants were called using SAMtools, and a consensus sequence was generated using the mpileup command (72). To ensure the reliability of the alignments, indels were not included in consensus sequences, as we found that these introduced errors into alignments, particularly in areas with lower depth. Samples with more than 50% undetermined bases across the entire plastome were discarded, and the remaining were concatenated into an alignment (no separate aligning step was necessary as the consensus sequences did not contain indels), which was visually inspected for artifacts in AliView. A sequence for Ficus albipila from Bruun-Lund et al. (19) was added to the alignment. A maximum-likelihood tree based on the unpartitioned alignment was inferred with RAxML-ng under the “GTRCAT” model, with 1,000 bootstrap replicates. We also scored nodes with quartet scores based on the nuclear gene trees using the “−t 1” option in ASTRAL. The chloroplast tree was time calibrated and biogeographic areas were reconstructed following the methods outlined for the Moraceae tree below. The chloroplast tree was compared to the nuclear tree using the cophylo command in Phytools (73) in R after pruning F. albipila, which was not present in the nuclear dataset. To test whether the differences between the nuclear and chloroplast trees were consistent with incomplete lineage sorting or would be better explained by introgression, 1,000 chloroplast trees were simulated as described in ref. 42. The ASTRAL tree, with branches scaled by a factor of 4 to reflect the maternal inheritance of the chloroplast, was used as a starting point. The number of extra lineages in the actual chloroplast tree as well as in the simulated trees compared to the ASTRAL tree (i.e., the number of splits occurring in the chloroplast trees but not in the ASTRAL tree) was counted using the “DeepCoalCount_tree” command in PhyloNet 3.8.0 (74).
Analyses of Hybridization.
We inferred phylogenetic networks based on two sample sets assembled from the 1,751 nuclear gene trees: 1) 41 Ficus samples representing the major lineages observed in the nuclear species tree, and 2) 27 Ficus samples representing the major-lineage discordance between the nuclear and chloroplast trees. We also inferred a network based on ca. 13,586 trees for 26 samples assembled from whole-genome sequences. Using the Phyx package, all gene trees were rooted using Antiaris toxicaria, and those lacking the outgroup were discarded. We inferred networks under maximum-pseudo-likelihood using PhyloNet (74), ignoring splits with less than 30% bootstrap support and otherwise using default settings, returning the best five inferred networks. We ran the analyses seven times, allowing 0 to 6 reticulations, and chose the best network using a slope heuristic (75) and BIC.
We also examined localized gene tree conflict directly by testing whether any sample had a conflicting position in the gene trees. To do this, we reduced all rooted gene trees to sample triplets. For each triplet, we counted the number of times that each of the three possible topologies [A, (B, C); B, (A, C); C, (A, B)] occurred. Topologies with bootstrap support lower than 30% were ignored. When there are two topologies that are more common than the third, this could indicate topological conflict caused by hybridization. The direction of the gene flow can be estimated by counting which topology was underrepresented. Using this method, introgression events can only be detected if both parent lineages and their hybrid are included in the sampling. If only one topology is present in more than a third of gene trees, there is likely a single strongly supported phylogenetic pattern with little conflicting data. If all topologies are equally common, it could be due to poor resolution at the node observed, for instance due to short time between divergence events or due to continuous gene flow (for instance, due to conspecificity). We ran 100 bootstrap replicates of all topology frequencies to derive a SD for them. We then calculated the occurrence of the second-most common topology for each triplet (minus the SD from the bootstrap replicates). If these were more than 1/3rd of the total number of trees, we considered that result as support that the sample being tested might be of hybrid origin. The values were visually observed in a heat map generated for each of the samples and each of the possible parent pairs, arranged in the order of a consensus phylogeny. All possible introgression events were then mapped to a phylogeny, showing the most likely direction of gene flow. Finally, the output was critically assessed to estimate the nodes at which the introgression events took place. In the case of more complex reticulation events, the introgression events could not be placed unambiguously.
Divergence Times and Biogeography.
For reconstruction of historical biogeography, we first inferred a time-calibrated tree on an expanded set of taxa including species-level sampling for most of Moraceae plus select outgroups in Urticaceae, rooted with Trema orientale. Outgroups and other Moraceae were drawn from previous studies focusing on various clades in the family (27, 34, 54, 55, 76), supplemented by 30 samples newly sequenced for this study, mostly from Olmedieae, and publicly available reads from the NCBI Sequence Read Archive (SI Appendix, Table S1). For Moraceae samples outside Ficeae and Olmedieae, sequences mostly consisted of the Moraceae333 loci, with some Angiosperms353 loci assembled from off-target reads, whereas Urticaceae samples were based on the Angiosperms353 loci.
Sequences were aligned and trimmed by tribe within Moraceae, and outgroup sequences were aligned and trimmed together. These alignments were merged with MAFFT using the “--merge” option and further pruned using the FastTree/TreeShrink method described above. Gene trees and an ASTRAL species tree were inferred as described above. We also used genesortR (68) to sort genes by clock-like behavior, determined by low root-to-tip variance, low site saturation, and concordance with the species tree topology. To generate branch lengths proportional to substitutions, we selected 724 genes from this sorted list and generated a partitioned supermatrix, which was used in RAxML-ng (under GTRCAT) to infer branch lengths on the ASTRAL tree. We accounted for uncertainty in branch lengths by repeating the branch-length calculation on a set of 100 jackknife replicates. For each replicate, 50% of loci were dropped, but to prevent taxa from dropping out, we sampled the three overlapping sets of loci separately: Angiosperms 353, Moraceae333, and Ficus1315. Resampling of locus names was carried out in R, and assembly of the jackknife supermatrices was carried out using fasta_merge.py from HybPiper. Before time calibration, the furthest outgroup, Trema orientale, was pruned from the trees.
The main tree and the jackknife trees were time-calibrated under penalized likelihood using treePL (77), with eight fossil constraints (SI Appendix, Table S3) following the workflow described by Maurin (78). The Eocene fossil achene of Ficus lucidus was used to constrain the stem node of Ficus to a minimum of 56 Ma (79), and a fossil fig wasp (80, 81) was used to constrain the stem of Galoglychia to a minimum age of 34 Ma. Finally, the stem nodes of sections Pharmacosycea and Americanae were constrained to a minimum of 16 Ma based on fossil agaonid wasps (82). The ages of the Moraceae crown and stem were bounded by the 95% HPD interval from Zhang et al. (20).
Four biogeographic areas were coded following previous studies (10, 83): 1) Neotropics (Southern and Central America), 2) Afrotropics (Africa and Madagascar), 3) Australasia (including Australia, New Caledonia, New Guinea, and islands east of Lydekker’s line and the Moluccas), and 4) Asia (Asia including three species which are also present in the Palearctic) (SI Appendix, Table S1). Ancestral area reconstruction was carried out under the DEC model as implemented in BioGeoBears (84). We allowed up to two simultaneous areas per node, as several extant taxa occur in two areas. Allowing simultaneous areas on such a large tree was made computationally feasible by parallelizing the DEC analysis on 32 threads.
Finally, we reconstructed diversification rates using BAMM (85, 86) on the time-calibrated tree. Priors were optimized using BAMMtools (86), and less-than-complete sampling of certain lineages was specified using a sample probability table. The analysis was run for 100,000,000 generations, and the first 10% were discarded as burn-in based on a visual inspection of a graphical output of the results.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the late C.C. Berg, D. Dixon, E. Jousselin, O. Maurin, C. Michenaud, and O. Ryding for providing material or assistance in obtaining material for this study; Fairchild Tropical Botanical Garden, Montgomery Botanical Center, and the Ornamental program at the USDA-ARS Subtropical Horticulture Research Station for access to living collections; C. Hansen for help in sample preparation; and Postar Miun, Jeisin Jumian, Markus Gubilil, S. Brono, Aloyzius Laim, Salang anak Nyegang, Saini anak Rosalin, Jugah anak Tagi, Raihan Rashida binti Ruslan, and Norazira binti Ramnor for assistance in the field. We thank the Sabah Biodiversity Centre (permit nos. JKM/MBS.1000-2/2 JLD.9 (26) and JKM/MBS.1000-2/2 JLD.9 (27) and export license no. JKM/MBS.1000-2/3 JLD.4 (15)); Chief Conservator of Forests and Deputy Chief Conservator of Forests (Research & Development), Sabah Forestry Department; and Sabah Parks for permission to conduct field research in Sabah, and the Sarawak Forest Department for permission to conduct field research in Sarawak (permit nos. NPW.907.4.4(JLD.14)-137 and (245)JHS/NCCD/600-7/2/107/JLD.2 and park permit nos. WL69/2017 and WL124/2019). This research was part of the PhD work of SBL under the supervision of N.R., F.K., and W.L.C. and of S.B.-L. supervised by A.L.H. This research was funded by a grant from the Danish Council for Independent Research—Natural Sciences (#7014-00205) to N.R., S.B.-L., and F.K.; a grant from the U.S. NSF to E.M.G. (DBI 1711391); a grant from Guangzhou Collaborative Innovation Center on Science-tech of Ecology and Landscape to Y.-Q.P.; a fellowship from the National Parks Board of Singapore to E.M.G.; a scholarship from the China Scholarship Council (201806140098) to S.L.; and a grant from the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, E–26/202.277/2019, E–26/202.278/2019) to L.C.P.
Author contributions
E.M.G., S.B.-L., A.L.H., G.K., C.B., and N.R. designed research; E.M.G., S.B.-L, M.N., B.C., W.L.C., C.G., R.D.H., A.L.H., M.H., G.K., F.K., S.L., L.C.P., Y.-Q.P., J.T.P., Q.P., A.S.A.P., J.S., S.J.S., E.V., G.D.W., N.J.C.Z., Q.Z., Z.Z., C.B., and N.R. performed research; S.B.-L. and M.N. contributed new reagents/analytic tools; E.M.G., M.N., F.K., Q.P., and J.-Y.R. analyzed data; and E.M.G., S.B.-L., M.N., A.L.H., F.K., J.-Y.R., C.B., and N.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Raw reads have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA956524 (87). Bait sequences, alignments, trees, and analysis scripts have been deposited in the Dryad Data Repository at https://doi.org/10.5061/dryad.x0k6djhqq (88).
Supporting Information
References
- 1.Richardson J. E., Pennington R. T., Editorial: Origin of tropical diversity: From clades to communities. Front. Genet. 7, 186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larson D. A., Vargas O. M., Vicentini A., Dick C. W., Admixture may be extensive among hyperdominant Amazon rainforest tree species. New Phytol. 232, 2520–2534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linan A. G., et al. , Interspecific hybridization and island colonization history, not rarity, most strongly affect the genetic diversity in Diospyros a clade of Mascarene-endemic trees. J. Hered. 113, 336–352 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Mitchell N., Whitney K. D., Limited evidence for a positive relationship between hybridization and diversification across seed plant families. Evolution 75, 1966–1982 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Suarez-Gonzalez A., Lexer C., Cronk Q. C. B., Adaptive introgression: A plant perspective. Biol. Lett. 14, 20170688 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon C. H., Lerdau M., Variable mating behaviors and the maintenance of tropical biodiversity. Front. Genet. 6, 183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Z.-D., Kobmoo N., Cruaud A., Kjellberg F., Genetic structure and hybridization in the species group of Ficus auriculata: Can closely related sympatric Ficus species retain their genetic identity while sharing pollinators? Mol. Ecol. 23, 3538–3550 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Berg C. C., Corner E. J. H., Ficus. Flora Malesiana Ser. 1, 17 (2005). [Google Scholar]
- 9.Harrison R. D., Figs and the diversity of tropical rainforests. BioScience 55, 1053–1064 (2005). [Google Scholar]
- 10.Cruaud A., et al. , An extreme case of plant-insect codiversification: Figs and fig-pollinating wasps. Syst. Biol. 61, 1029–1047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasplus J., et al. , Exploring systematic biases, rooting methods and morphological evidence to unravel the evolutionary history of the genus Ficus (Moraceae). Cladistics 37, 402–422 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Harrison R. D., et al. , Pollination of Ficus elastica: India rubber re-establishes sexual reproduction in Singapore. Sci. Rep. 7, 11616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison R. D., Maintenance of specificity in an isolated fig. Biotropica 39, 275–277 (2007). [Google Scholar]
- 14.Machado C. A., Robbins N., Gilbert M. T. P., Herre E. A., Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proc. Natl. Acad. Sci. U.S.A. 102 Suppl, 6558–6565 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., et al. , Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nat. Commun. 12, 718 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clement W. L., et al. , Evolution and classification of figs (Ficus, Moraceae) and their close relatives (Castilleae) united by involucral bracts. Bot. J. Linn. Soc. 193, 316–339 (2020). [Google Scholar]
- 17.Weiblen G. D., Phylogenetic relationships of functionally dioecious Ficus (Moraceae) based on ribosomal DNA sequences and morphology. Am. J. Bot. 87, 1342–1357 (2000). [PubMed] [Google Scholar]
- 18.Jousselin E., Rasplus J.-Y., Kjellberg F., Convergence and coevolution in a mutualism: Evidence from a molecular phylogeny of Ficus. Evolution 57, 1255–1269 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Bruun-Lund S., Clement W. L., Kjellberg F., Rønsted N., First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Mol. Phylogenet. Evol. 109, 93–104 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q., Onstein R. E., Little S. A., Sauquet H., Estimating divergence times and ancestral breeding systems in Ficus and Moraceae. Ann. Bot. 123, 191–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai L., Hayakawa H., Fukuda T., Yokoyama J., A breakdown of obligate mutualism on a small island: An interspecific hybridization between closely related fig species (Ficus pumila and Ficus thunbergii) in Western Japan. Am. J. Plant Sci. 6, 126–131 (2015). [Google Scholar]
- 22.Costa P. C., et al. , The phylogeography of two disjunct Neotropical Ficus (Moraceae) species reveals contrasted histories between the Amazon and the Atlantic Forests. Bot. J. Linn. Soc. 185, 272–289 (2017). [Google Scholar]
- 23.Petit R. J., Excoffier L., Gene flow and species delimitation. Trends Ecol. Evol. 24, 386–393 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Datwyler S. L., Weiblen G. D., On the origin of the fig: Phylogenetic relationships of Moraceae from NDHF sequences. Am. J. Bot. 91, 767–777 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Zerega N. J. C., Clement W. L., Datwyler S. L., Weiblen G. D., Biogeography and divergence times in the mulberry family (Moraceae). Mol. Phylogenet. Evol. 37, 402–416 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Clement W. L., Weiblen G. D., Morphological evolution in the mulberry family (Moraceae). Syst. Bot. 34, 530–552 (2009). [Google Scholar]
- 27.Gardner E. M., et al. , Repeated parallel losses of inflexed stamens in Moraceae: Phylogenomics and generic revision of the tribe Moreae and the reinstatement of the tribe Olmedieae (Moraceae). Taxon 70, 946–988 (2021). [Google Scholar]
- 28.Pederneiras L. C., Carauta J. P. P., Neto S. R., Vidal de Mansano F., An overview of the infrageneric nomenclature of Ficus (Moraceae). Taxon 64, 589–594 (2015). [Google Scholar]
- 29.Zhang Z., Wang X.-M., Liao S., Zhang J.-H., Li H.-Q., Phylogenetic reconstruction of Ficus subg. Synoecia and its allies (Moraceae), with implications on the origin of the climbing habit. Taxon 69, 927–945 (2020). [Google Scholar]
- 30.Gardner E. M., Johnson M. G., Ragone D., Wickett N. J., Zerega N. J. C., Low-coverage, whole-genome sequencing of Artocarpus camansi (Moraceae) for phylogenetic marker development and gene discovery. Appl. Plant Sci. 4, 1600017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruun-Lund S., “Darwin’s abominable mystery–and the evolution and diversification of Ficus L”, Dissertation (PhD), (University of Copenhagen, Copenhagen, Denmark, 2019), 21 March 2023. [Google Scholar]
- 32.Rønsted N., et al. , 60 million years of co-divergence in the fig-wasp symbiosis. Proc. Biol. Sci. 272, 2593–2599 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satler J. D., et al. , Pollinator and host sharing lead to hybridization and introgression in Panamanian free-standing figs, but not in their pollinator wasps. Ecol. Evol. 13, e9673 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner E. M., et al. , Paralogs and off-target sequences improve phylogenetic resolution in a densely sampled study of the breadfruit genus (Artocarpus, Moraceae). Syst. Biol. 70, 558–575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruaud A., et al. , Laying the foundations for a new classification of Agaonidae (Hymenoptera: Chalcidoidea), a multilocus phylogenetic approach. Cladistics 26, 359–387 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Corner E. J. H., The evolution of Streblus Lour. (Moraceae): With a new species of sect. Bleekrodea. Phytomorphology 25, 1–12 (1975). [Google Scholar]
- 37.a Cornille, et al. , Floral volatiles, pollinator sharing and diversification in the fig-wasp mutualism: insights from Ficus natalensis, and its two wasp pollinators (South Africa). Proc. Biol. Sci. 279, 1731–1739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moe A. M., Rossi D. R., Weiblen G. D., Pollinator sharing in dioecious figs (Ficus: Moraceae). Biol. J. Linn. Soc. 103, 546–558 (2011). [Google Scholar]
- 39.Su Z.-H., et al. , Pollinator sharing, copollination, and speciation by host shifting among six closely related dioecious fig species. Commun. Biol. 5, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wachi N., Kusumi J., Tzeng H.-Y., Su Z.-H., Genome-wide sequence data suggest the possibility of pollinator sharing by host shift in dioecious figs (Moraceae, Ficus). Mol. Ecol. 25, 5732–5746 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Ware A. B., Compton S. G., Breakdown of pollinator specificity in an African fig tree. Biotropica 24, 544–549 (1992). [Google Scholar]
- 42.Zhou B.-F., et al. , Phylogenomic analyses highlight innovation and introgression in the continental radiations of Fagaceae across the Northern Hemisphere. Nat. Commun. 13, 1320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S., et al. , Phylogenomic analyses reveal widespread gene flow during the early radiation of oaks and relatives (Fagaceae: Quercoideae). bioRxiv [Preprint] (2023). 10.1101/2023.04.25.538215 (Accessed 16 May 2023). [DOI]
- 44.Vences M., Wollenberg K. C., Vieites D. R., Lees D. C., Madagascar as a model region of species diversification. Trends Ecol. Evol. 24, 456–465 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Federman S., et al. , The biogeographic origin of a radiation of trees in Madagascar: Implications for the assembly of a tropical forest biome. BMC Evol. Biol. 15, 216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonelli A., et al. , Madagascar’s extraordinary biodiversity: Evolution, distribution, and use. Science 378, eabf0869 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Bruun-Lund S., Verstraete B., Kjellberg F., Rønsted N., Rush hour at the Museum—Diversification patterns provide new clues for the success of figs (Ficus L., Moraceae). Acta Oecologica 90, 4–11 (2018). [Google Scholar]
- 48.Machado A. F. P., Rønsted N., Bruun-Lund S., Pereira R. A. S., Paganucci de Queiroz L., Atlantic forests to the all Americas: Biogeographical history and divergence times of Neotropical Ficus (Moraceae). Mol. Phylogenet. Evol. 122, 46–58 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Boonchai N., Manchester S. R., Wheeler E. A., Welkoetoxylon multiseriatum: Fossil moraceous wood from the Eocene Green River Formation, Wyoming, U.S.A. IAWA J. 36, 158–166 (2015). [Google Scholar]
- 50.Williams E. W., et al. , Out of Borneo: Biogeography, phylogeny, and divergence date estimates of Artocarpus (Moraceae). Ann. Bot. 119, 611–627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Noamani Z. M., Ficoxylon fusiforme (Moraceae), a new species from the Upper Cretaceous Nubian Sandstone in Southern Egypt. Egypt. J. Bot. 62, 31–44 (2021). [Google Scholar]
- 52.Lomascolo S. B., Levey D. J., Kimball R. T., Bolker B. M., Alborn H. T., Dispersers shape fruit diversity in Ficus (Moraceae). Proc. Natl. Acad. Sci. U.S.A. 107, 14668–14672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tricou T., Tannier E., de Vienne D. M., Ghost lineages highly influence the interpretation of introgression tests. Syst. Biol. 71, 1147–1158 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q., Gardner E., Zerega N., Sauquet H., Long-distance dispersal shaped the diversity of tribe Dorstenieae (Moraceae). bioRxiv [Preprint] (2019). 10.1101/531855. [DOI]
- 55.Zerega N. J. C., Gardner E. M., Delimitation of the new tribe Parartocarpeae (Moraceae) is supported by a 333-gene phylogeny and resolves tribal level Moraceae taxonomy. Phytotaxa 388, 253–265 (2019). [Google Scholar]
- 56.Doyle J. J., Doyle J. L., A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987). [Google Scholar]
- 57.Carøe C., et al. , Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410–419 (2018). [Google Scholar]
- 58.Allentoft M. E., et al. , Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinforma. Oxf. Engl. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson M. G., et al. , HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4, 1600016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson M. G., et al. , A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-Medoids clustering. Syst. Biol. 68, 594–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price M. N., Dehal P. S., Arkin A. P., Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodcroft E., TreeCollapserCL4: Removing Doubt from Your Trees! Collapse Trees by Bootstrap (2013) (29 December 2022).
- 67.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 19, 15–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mongiardino Koch N., Phylogenomic subsampling and the search for phylogenetically reliable loci. Mol. Biol. Evol. 38, 4025–4038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nosenko T., et al. , Deep metazoan phylogeny: When different genes tell different stories. Mol. Phylogenet. Evol. 67, 223–233 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Zhang X., et al. , Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell 183, 875–889.e17 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., et al. , The sequence alignment/map format and SAMtools. Bioinforma. Oxf. Engl. 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 74.Wen D., Yu Y., Zhu J., Nakhleh L., Inferring phylogenetic networks using PhyloNet. Syst. Biol. 67, 735–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osuna-Mascaró C., et al. , Hybridization and introgression are prevalent in Southern European Erysimum (Brassicaceae) species. Ann. Bot. 131, 171–184 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gardner E. M., et al. , Phylogenomics of Brosimum (Moraceae) and allied genera, including a revised subgeneric system. Taxon 70, 778–792 (2021). [Google Scholar]
- 77.Smith S. A., O’Meara B. C., treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Maurin K. J. L., An empirical guide for producing a dated phylogeny with treePL in a maximum likelihood framework. arXiv [Preprint] (2020). 10.48550/arXiv.2008.07054. (Accessed 30 August 2022). [DOI]
- 79.Collinson M. E., “The fossil history of the Moraceae, Urticaceae (including Cecropiaceae), and Cannabaceae” in Evolution, systematics, and fossil history of the Hamamelidae, Crane P. R., Blackmore S., Eds. (Clarendon Press, Oxford, 1989), vol. 2. pp. 319–339. [Google Scholar]
- 80.Compton S. G., et al. , Ancient fig wasps indicate at least 34 Myr of stasis in their mutualism with fig trees. Biol. Lett. 6, 838–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Antropov A. V., et al. , The wasps, bees and ants (Insecta: Vespida=Hymenoptera) from the Insect Limestone (Late Eocene) of the Isle of Wight, UK. Earth Environ. Sci. Trans. R. Soc. Edinb. 104, 335–446 (2013). [Google Scholar]
- 82.Pederneiras L. C., Gaglioti A. L., Romaniuc-Neto S., Mansano V. d. F., The role of biogeographical barriers and bridges in determining divergent lineages in Ficus (Moraceae). Bot. J. Linn. Soc. 187, 594–613 (2018). [Google Scholar]
- 83.Lopez-Vaamonde C., et al. , Molecular dating and biogeography of fig-pollinating wasps. Mol. Phylogenet. Evol. 52, 715–726 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Matzke N. J., BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts (R Package Version 02, 2013).
- 85.Rabosky D. L., Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9, e89543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rabosky D. L., et al. , BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707 (2014). [Google Scholar]
- 87.Gardner E. M., et al. , BioProject PRJNA956524: Phylogenomics of Ficus -- target enriched raw sequence reads. National Center for Biotechnology Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA956524. Deposited 23 May 2023.
- 88.Gardner, et al. , Data for: Echoes of ancient introgression punctuate stable genomic lineages in the evolution of figs. Dryad. 10.5061/dryad.x0k6djhqq. Deposited 13 June 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Raw reads have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA956524 (87). Bait sequences, alignments, trees, and analysis scripts have been deposited in the Dryad Data Repository at https://doi.org/10.5061/dryad.x0k6djhqq (88).