Abstract
Progress in cell purification technology is critical to increase the availability of viable cells for therapeutic, diagnostic, and research applications. A variety of techniques are now available for cell separation, ranging from non-affinity methods such as density gradient centrifugation, dielectrophoresis, and filtration, to affinity methods such as chromatography, two-phase partitioning, and magnetic-/fluorescence-assisted cell sorting. For clinical and analytical procedures that require highly purified cells, the choice of cell purification method is crucial, since every method offers a different balance between yield, purity, and bioactivity of the cell product. For most applications, the requisite purity is only achievable through affinity methods, owing to the high target specificity that they grant. In this review, we discuss past and current methods for developing cell-targeting affinity ligands and their application in cell purification, along with the benefits and challenges associated with different purification formats. We further present new technologies, like stimuli-responsive ligands and parallelized microfluidic devices, towards improving the viability and throughput of cell products for tissue engineering and regenerative medicine. Our comparative analysis provides guidance in the multifarious landscape of cell separation techniques and highlights new technologies that are poised to play a key role in the future of cell purification in clinical settings and the biotech industry.
Keywords: cell purification, immunoaffinity, MACS, FACS, microfluidics
1. Introduction
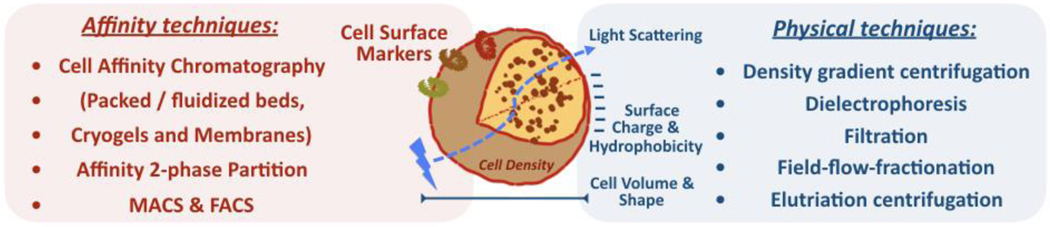
The ability to sort cells into distinct, mono-disperse populations is crucial to advance our knowledge of specific phenotypes, and explore their potential in tissue engineering and regenerative medicine [1, 2]. Efficient cell separation is therefore paramount in a multitude of fields, including personalized cell therapy [3–6], organ recellularization [7–11], diagnostics and disease monitoring [12–17], drug discovery [18–22], and basic cell biology [23–25]. To meet the growing demand for highly pure cell products, there has been considerable effort to develop efficient and high-throughput separation methods. As a result, a multitude of techniques have emerged, which are classified into separations by (i) physical characteristics (i.e., cell volume and shape, density, and light scatter properties or fluorescence), (ii) surface properties (i.e., electrical charges, hydrophobicity, etc.) and cell constituents (i.e., such as nucleic acids, enzymes and other proteins), and (iii) adherence/affinity features [26–29] (Figure 1).
Figure 1.
Cell properties and corresponding purification techniques.
When supplying cells for therapeutic applications, separation technologies must meet analytical benchmarks and regulatory compliance [30–32]. Consistency in product quality, in terms of cell viability and phenotype purity, is highly controlled to ensure product efficacy and patient safety [33–35]. The presence of adventitious agents is also rigorously monitored, and all processing steps must be compatible with sterility requirements [33, 36, 37].
Affinity-based separations have emerged as the main technology for cell isolation, as they meet the demand for high yield and purity, together with scalability and sterility [27, 38]. After three decades of developments, however, a systematic review is needed to recapitulate the diversity and complexity of affinity-based cell separation technologies and guide new users through the selection of appropriate purification methods. To this end, we present a comprehensive survey of affinity-based methods for cell purification, including traditional chromatographic techniques to more recent, non-chromatographic or pseudo-chromatographic systems (Figure 2, Table 1). These methods employ a variety of biorecognition agents for capture, ranging from traditional protein ligands to synthetic binders. Through this comparison, we also aim to identify emerging opportunities for improving the manufacturing of cells for tissue engineering and regenerative medicine.
Figure 2.
Cell purification technologies.
Table 1.
Comparison of physical (non-affinity) and affinity-based cell separation techniques.
| Method | mechanism | Target cells | Advantages |
|---|---|---|---|
| Physical (non-affinity) methods | |||
| Density gradient Centrifugation | Cells migrate through a vertical density gradient (aqueous solutions of biopolymers) during centrifugation and collect in the region where the local density corresponds to their own. | Human mesenchymal stem cells, hematopoietic stem and progenitor cells, blood cells (erythrocyte, platelets, granulocytes, lymphocytes, monocytes), circulating tumor cells, sperm cells, and neurons. Rat pancreatic islets. | Label-free technology, which enables processing large volumes in short process times and concentrating the cell product; high viability of the cell product; reproducible results; facile scale up; commercially available equipment. |
| Dielectrophoresis | Cells placed in a gradient electric field act as induced dipoles and migrate at: different rates based on their size and dielectric properties as well as the dielectric properties of the medium. | Hematopoietic stem and progenitor cells, leukocytes (B and T-lymphocytes, monocytes, and granulocytes), circulating tumor cells, astrocyte and neuron-biased cells, neural stem and progenitor cells. Isolation of pathogenic bacteria from blood. Fractionation of viable vs. non-viable cells (yeast and mammalian cells). | Label-free, continuous technology that enables sorting cells based on viability without dilution, thereby reducing sample volumes; short processing time and high sensitivity; can be integrated with microfluidic devices. |
| Field flow fractionation | A cell suspension is flown through a channel where a field (e.g., crossflow, sedimentation, and electrical) is applied perpendicular to the direction of flow enabling separation based on mobility differences. | Pathogenic bacteria, yeast cell subpopulations. Human blood cells (erythrocyte, platelets, leukocytes), cancer cells, neurons from cerebral cortices, embryonic stem cells, mesenchymal stem cells, hematopoietic stem cells, electroporated vs. non-electroporated cells, and cells undergoing apoptosis. | Label-free, continuous technology that grants high viability and bioactivity of the cell product, under short process time; reduced sample volumes; high reproducibility. |
| Filtration | Cells are captured non-specifically on the surface of a material with controlled porosity based on physical properties such as cell diameter (volume) and aspect ratio. Generally used as preparative tool for further purification. | Blood cells (leukocytes, erythrocytes), hematopoietic stem and progenitor cells, adipose-derived stem cells, mesenchymal stem cells, circulating tumor cells. Bacterial and mammalian (e.g., Chinese Hamster Ovary (CHO) cells) cells for metabolomics preparation. | High-throughput, simple, and scalable technology, which can be integrated into a microfluid device platform. |
| Elutriation Centrifugation | Cells are separated based on their sedimentation velocity. | Blood cells (granulocytes, lymphocytes, monocytes, platelets), macrophages, Kupffer cells from liver, mast cells, hepatocytes, sperm cells (rats and human), separation into age-related fractions (yeast, erythrocytes, prostate and ovarian cancer cells from tumors, and hematopoietic stem and progenitor cells. | Rapid processing of large volumes of cells featuring a wide range of sizes; applicability at low temperatures to impede cell activation; high recovery and viability of the cell product. |
2. Cells of interest
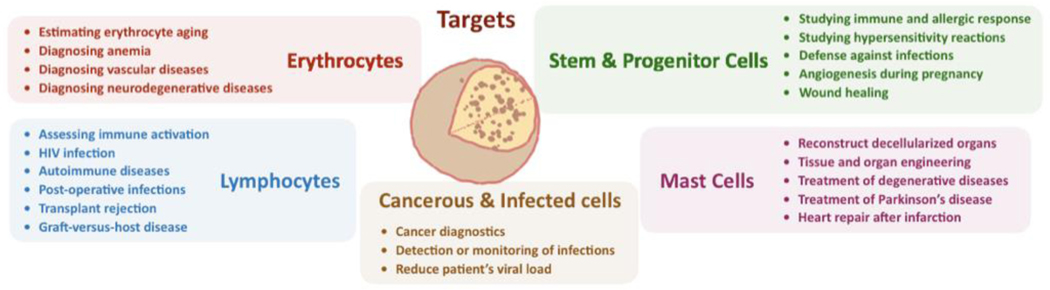
A list of clinically relevant cell products is provided in Figure 3. The isolation of erythrocytes is a prerequisite for estimating erythrocyte aging [39] and diagnosing conditions such as anemia[40] as well as vascular [41] and neurodegenerative diseases (Alzheimer’s and Parkinson’s) [12, 42]. Similarly, the isolation of lymphocytes is needed when assessing immune activation [13, 14], and as such, these cells are valuable in diagnosing or studying HIV infections [43], autoimmune diseases [44], post-operative infections [45], transplant rejection [46], and graft-versus-host disease (GvHD) [47, 48]. Mast cells (MCs) also represent a relevant class of targets, especially for studying innate immune response, as their specific role in vivo is still unclear; while often associated with allergic response, specifically anaphylaxis, and hypersensitivity reactions [49, 50], MCs have also been found to have significant roles in a host’s defense against infections [51–53], angiogenesis during pregnancy [54], wound healing [55, 56], and autoimmune diseases [57]. Obtaining pure mast cell isolates has the potential to greatly improve our knowledge of disease mechanisms through the study of mast cell activation and immune response stimulation [58, 59]. Stem and progenitor cells are key ingredients in regenerative medicine and developmental biology, where they are used to reconstruct decellularized organs or to seed scaffolds for tissue and organ engineering [7, 9, 60]. For these reasons, stem cells have shown promise to help relieve the shortage of transplant organs [61, 62], to treat a number of conditions including macular degeneration [63] and Parkinson’s disease [64, 65], or as a therapy to repopulate heart tissue after myocardial infarction [66, 67]. The isolation of stem cells involves an additional challenge compared to common cell purification, as undifferentiated cells must be removed prior to implantation to reduce the risk of teratoma formation [68, 69]; with an average of 107 – 109 cells being required for a transplant [70, 71], even a 0.1% impurity level can result in a load of 106 undifferentiated cells and teratoma formation [72, 73]. Several technologies have been developed for stem cell purification based on cell phenotype, including density-gradient separation [74, 75], fluorescence-activated cell sorting (FACS) [73, 76, 77], and metabolic selection [78]. Improved cell purification techniques would also be beneficial to detect and monitor circulating tumor cells [79, 80] and pathogen infections [81]. It is in fact particularly difficult to isolate circulating tumor cells due to their rarity (~ 1 circulating tumor cell per 108 red blood cells [82]). Additionally, cell separation techniques have been used to remove virus-infected cells from a patient to reduce a their overall viral load, as shown with malaria and hepatitis C [83–87]. Improved pathogen infection detection is not only beneficial for human related infections like those caused by HIV [88, 89], but also for the monitoring of food-related pathogens [90–92].
Figure 3.
Cell targets and their diagnostic or therapeutic applications.
3. Non-affinity methods
Outside of affinity-based methods, cell separation is typically based on the physical properties of cells [93, 94]. These methods include density gradient centrifugation [95–100], dielectrophoresis [101–105], field-flow fractionation [102, 106–111], filtration [112–118], and elutriation centrifugation [119–123]. While useful for primary enrichment, these methods lack the specificity and resolution to achieve the levels of purity required for therapeutic and analytical applications [93, 94, 124, 125], and typically afford low yield for rare cell types [107, 126]. To overcome these limitations, affinity-based methods have been implemented to improve recovery and purity [29, 127–131]. These rely on the specific recognition and binding of a cell surface target by a complementary molecule, called ligand, immobilized on a suitable carrier or surface [132, 133]. Protein ligands, especially antibodies, are currently the major workhorse in affinity-based cell purification, owing to their high capture strength and selectivity [134–137]. Biological ligands, however, are expensive and often suffer from low biochemical stability. Furthermore, their strong binding often makes the elution of cells challenging [133, 138, 139]. Thus, improvements in affinity methods are needed to enable therapeutic and analytical approaches that rely on consistent and cost-effective cell purification. The main channels of innovation are (i) the identification of cost-effective synthetic affinity or pseudo-affinity ligands for replacing biological ligands, (ii) the development of purification formats that improve upon classic chromatography originally designed for protein purification, and (iii) the determination of unique surface receptors on the target cells that are appropriate for use as affinity targets to ensure high phenotypical purity of the cell product.
4. Conventional Affinity Ligand Formats and Selection for Cell Separation
In cell separations, ligands bind proteins that are ideally unique or overexpressed on the cell membrane of the population of interest. Three ligand families are currently the most employed in cell separation: antibodies, proteins, and lectins. More recently, however, synthetic ligands have emerged as promising and cost-effective alternatives. In this section, conventional ligands used for cell separation are described as well as the desired ligand properties to achieve successful cell purification.
4.1. Cell properties that determine the outcome of affinity-based cell purification.
Cell sorting relies on the identification of a target receptor on either the cell phenotype of interest (positive cell enrichment) or the background cells (negative cell enrichment) [13, 14, 140, 141], based on the target’s abundance on the cell surface, the heterogeneity of cell population, and the requirements for the final cell product [142–144]. Typically, each receptor forms only ~ 0.01% of the total membrane protein content [145, 146], although proteins considered of “low-abundance” can be considerably less, as occurs on T or B lymphocytes [147]. The difficulty in identifying unique biomarkers for a target cell phenotype complicates the separation process and renders the assessment of cell product purity challenging [140, 148–150]. Cell surface receptors may vary among donors and different tissues isolated from an individual donor [151–153]. This heterogeneity complicates the selection of target cell markers for affinity purification and has slowed considerably the study of certain cell classes. This has particularly been the case for mast-cells [51, 53, 56], whose purification by affinity is predominantly based on CD117 (c-Kit) targeting, although this receptor is not specific to mast cells and is present in many stem cell phenotypes [154]. For target cells featuring a particularly low surface density of unique receptors, negative enrichment is the preferred strategy [13, 155–157]. When low expression level is combined with low target cell abundance, microfluidic devices integrating negative selection strategies and physical separation methods (e.g., fluid, electric, or magnetic field) represent the technology of choice [156, 158–160].
Additional considerations when selecting the target receptor come from the biochemical effects that occur upon receptor binding. External cell receptors are inherently connected to cell metabolism, and ligand/receptor interactions can trigger undesired events such as internalization of the receptor, metabolic alteration, and even differentiation, in the case of stem cells [151, 152, 161]. Metabolic changes caused by affinity binding have been observed on mast cells enriched by targeting c-kit and FcεRI; while utilized for the positive selection of mast cells, these markers are crucial in IgE activation and are likely to impact cellular metabolism [58, 162].
Ligand selection must also take into account both kinetic (kon and koff) and thermodynamic (KD) binding parameters [163–166]. Binding strength (KD) is crucial to ensure product purity, and, in the case of positive selection, to ensure that the target cells can be eluted from the affinity adsorbent. High-affinity ligands (low KD), while binding target cells specifically, make cell elution difficult, whereas low-affinity ligands (high KD), while allowing for easier elution, may not provide sufficient throughput. Thus, an ideal affinity ligand offers a balance between specific binding and effective elution [133]. Furthermore, quantifying cell adsorption in terms of KD only is not accurate, due to multi-point interactions between a target cell and multiple immobilized ligands known as avidity. Cell size, aspect ratio, and receptor density can be used to estimate the number of interactions per cell, and select an appropriate ligand density for a given value of KD [167, 168]. Finally, cell elution conditions are also crucial, as they strongly affect the viability of the recovered cells [138]. Elution can be achieved (i) non-specifically, by manipulating the salt concentration, pH, or temperature, or (ii) specifically, by using eluents that inhibit the ligand-cell interactions [139, 140]. Non-specific methods can damage the cells, while specific methods tend to be expensive. To overcome these issues, specific elution methods using multivalent competitive inhibitors have been presented, which have shown increased cell recovery compared to monovalent inhibitors [139, 169].
4.2. Antibodies.
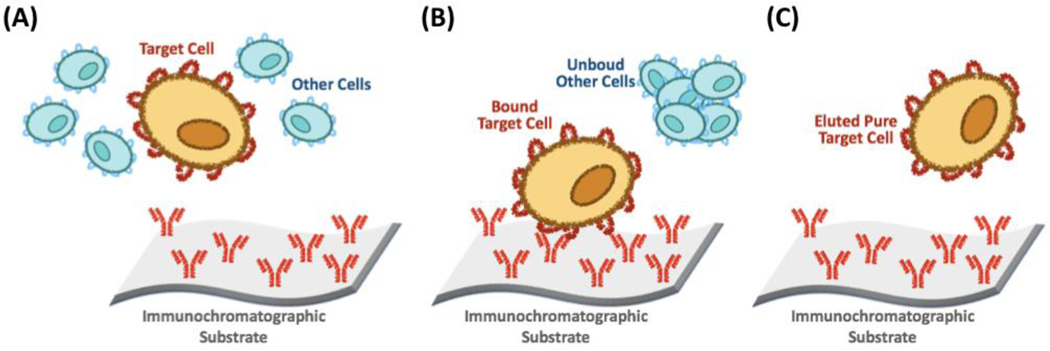
“Immunoaffinity”, i.e. the use of antibodies as affinity ligands (Figure 4), has been widely applied for cell purification, owing to the antibodies’ binding selectivity and ability to operate effectively under physiological conditions [93, 150, 170–175]. Following the seminal work by Peterson [176] and Wigzell [177, 178], immunoaffinity has been employed to purify a wide variety of cells, including pathogenic bacteria [179], lymphocytes [180–185], mast and inflammatory cells [186, 187], neural cells [188, 189], and stem cells [190–192]. Recently, antibody fragments, such as Fabs and scFv, have been utilized as ligands in lieu of whole antibodies, as they possess the same binding activity while being produced more affordably [193, 194]. The strength of the interaction between the antibody and target protein, however, requires harsh elution conditions that may impact the cells’ viability. To address this issue, elution strategies have included competitive elution [181, 195, 196] and cleavable linkers [197, 198].
Figure 4.
Cell immunoaffinity chromatography. (A) Contacting a mixture of cells with the affinity substrate (e.g., an immunoaffinity adsorbent); (B) Removing the unbound cells by washing; (C) Eluting the target cell.
4.3. Protein A/G.
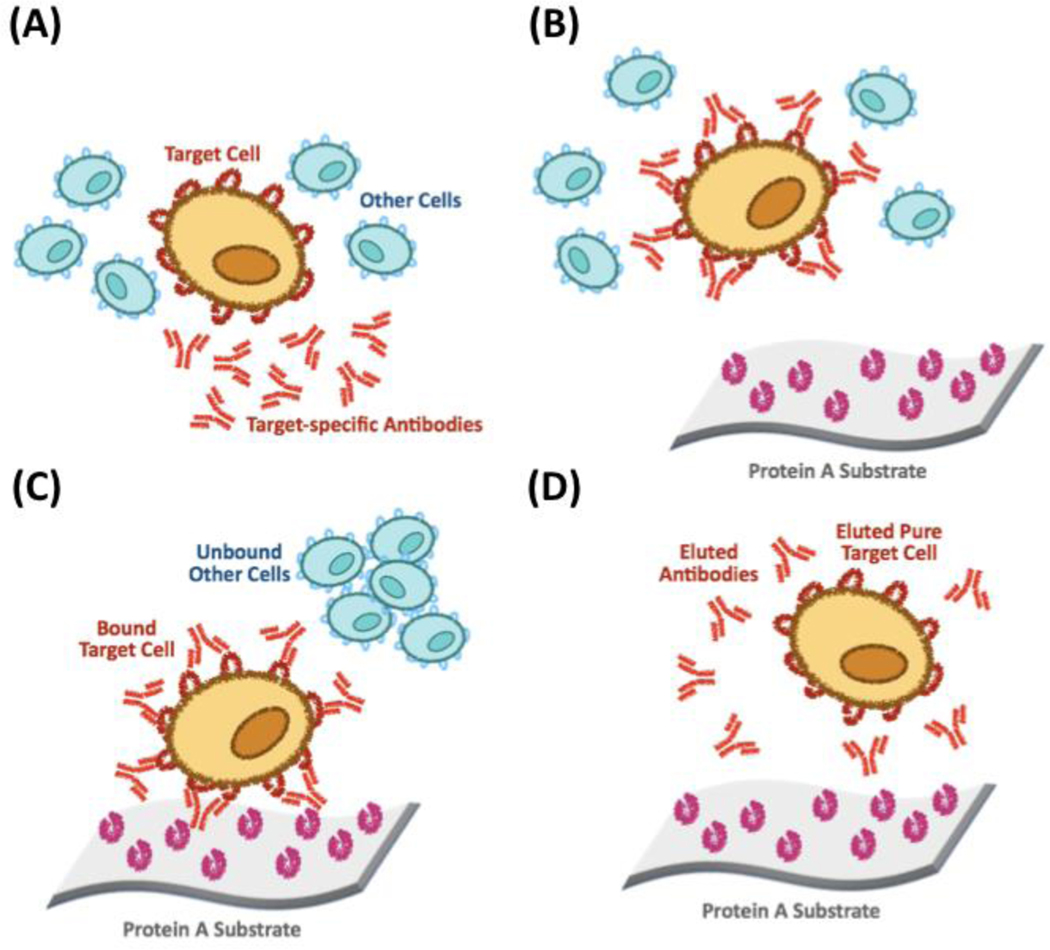
Another antibody-based method for cell isolation relies on Protein A and Protein G, two antibody-binding proteins expressed respectively by Staphylococcus aureus and group C and G Streptococcal bacteria [199]. In Protein A/G-based methods, a cell mixture is incubated with a receptor-specific antibody and passed through a Protein A/G-linked adsorbent [200–205], where the antibody-labeled cells are selectively retained (Figure 5). As the binding to Protein A/G is less impacted by steric hindrance than binding to immobilized antibodies directly, this variant of immunoaffinity cell chromatography is more efficient, and has been demonstrated in different formats, such as rosetting [206, 207] and solid-phase chromatography [201, 208].
Figure 5.
Protein A-based cell affinity chromatography. (A) Mixing the target cell and other cells with target-specific antibodies (e.g., an immunoaffinity adsorbent); (B) Removing the unbound cells by washing; (C) Eluting the target cell.
4.4. Protein and synthetic antigens.
Antigens represent a broad class of ligands ranging from proteins to small synthetic molecules [93, 171, 209–211]. The use of antigenic ligands for purifying white cells has been pioneered by Wigzell et al., who isolated immunized mouse lymph node cells using glass and plastic beads functionalized with human serum albumin, bovine serum albumin, and ovalbumin with yields between 60–95%, but poor enrichment (2.5-fold) [212]. Later work on lymphocytes has utilized enzyme-substrate interactions to isolate lymphocytes raised against enzyme antigens [213]. To purify enzyme-binding lymphocytes, Deluca et al. contacted white cells with the antigen enzyme and then exposed the solution to beads decorated with the enzyme’s substrate to specifically capture enzyme-bound lymphocytes [210].
Synthetic antigens represent the first use of synthetic ligands for cell purification [214–217]. Truffa-Bachi et al. utilized haptens as antigens to stimulate an immune response, and subsequently as immobilized ligands to isolate white cells with anti-hapten activity [214, 215]. This method addresses two main difficulties encountered in affinity-based capture, namely (i) non-specific binding of non-target cells and (ii) detaching cells from the adsorbent without impacting their viability. In this context, Haas et al. utilized a gelatin matrix containing dinitrophenyl as a ligand for adsorbing mouse spleen cells, demonstrating that 30-fold enrichment and high viability could be achieved by melting the gelatin, providing for a gentle elution strategy [217].
4.5. Lectins.
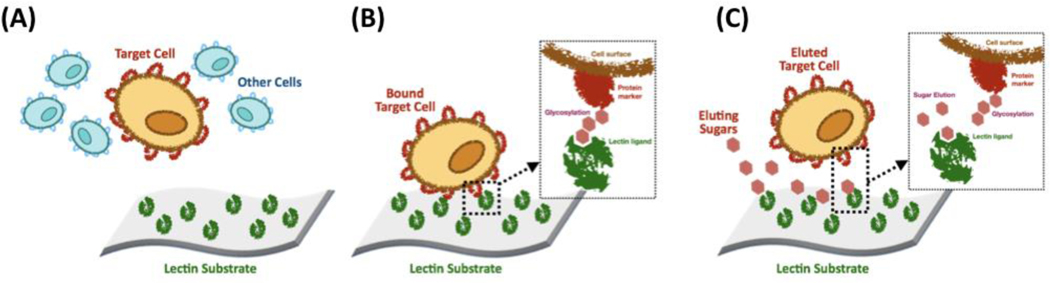
Lectins recognize specific carbohydrate sequences on glycoprotein cell surface markers and have been widely utilized for cell fractionation (Figure 6) [218]. Herz et al. have used soybean agglutinin as a ligand to isolate T lymphocytes from peripheral blood for use in the prevention of graft vs. host disease in bone marrow transplants [219]. Hellström et al. have shown how helix pomatia A hemagglutinin can bind T cells treated with neuraminidase by targeting surface carbohydrates [220]; because only a small fraction of B cells interact with helix pomatia hemagglutinin, this method represents an efficient strategy to separate T cells from B cells [221]. This work shows how lectins enable highly specific cell fractionation as they target post-translational modifications; helix pomatia hemagglutinin, in fact, is selective for human T cells over many B cells since T cells express proteins with unique post-translational modifications [222]. Another major advantage of lectins is that cell elution can be triggered by mono- and disaccharides, which are harmless to cells [171]. In one instance, though, the elution of mouse thymocytes from concanavalin A was accomplished by cleaving the mercury-sulfur bond conjugating the lectin ligands from the chromatographic substrate using a short thiol, affording quantitative recovery and high cell viability [223].
Figure 6.
Lectin-based cell affinity chromatography. (A) Contacting a mixture of cells with the lectin substrate; (B) Removing the unbound cells by washing; (C) Eluting the target cells using a mixture of sugars.
5. Formats of affinity-based cell separation
The principles of affinity purification have been applied in different ways for cell capture, depending on the source fluids and the required throughput [26, 100, 140, 150]. Cell-binding ligands have been immobilized onto solid substrates (chromatographic-like methods) [128] or polymer carriers (pseudo-/non-chromatographic method) [224] as well as magnetic particles (MACS) [225] and fluorescent markers (FACS) [226] that enable separation by an electromagnetic field. More recently, affinity ligands have been displayed on the channels of microfluidic devices. This latest frontier of cell separation offers higher resolution and holds great promise to expedite the clinical implementation of cellular therapies relying on rare cell types.
5.1. Rosetting.
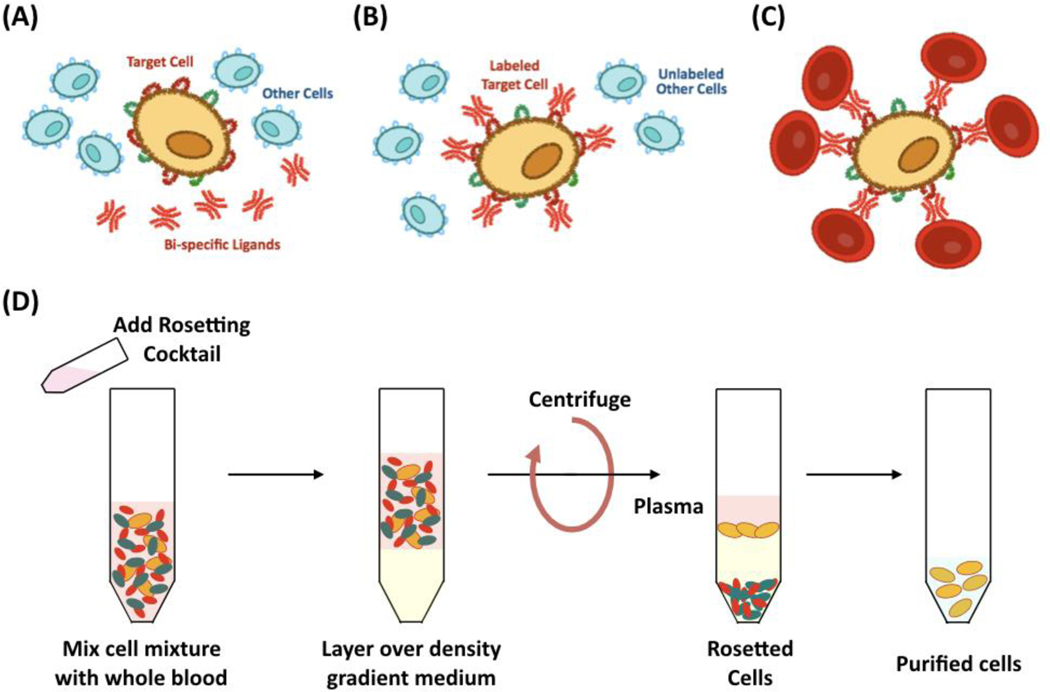
Rosetting was the first isolation method to combine affinity with traditional density-gradient separation methods [150, 227–229]. In this technique, antigen-specific cells are incubated with antigen-coated erythrocytes, with which they form aggregates, called “rosettes”, that are separated from non-rosetted cells by gradient centrifugation (Figure 7) [230]. Rosetting was first utilized to separate two mouse immune cell populations using sheep red blood cells [231]. Further work demonstrated that greater quantities and purities of rosette-forming antigen-specific cells could be obtained through avidin-biotin affinity [207], gradient density centrifugation [18], and in combination with magnetic fields [232]. Rosetting is now routinely employed for purifying B and T lymphocytes and stem cells [233], with commercial products such as the RosetteSep™ kit from StemCell, which offer good recovery and purity.
Figure 7.
Cell rosetting technique. (A) Contacting the target cell and other cells with bispecific (target cell and red blood cells) ligands resulting in (B) the formation of a complex; (C) Incubating the tagged target cells with red blood cells; (C) Procedure of cell purification by rosetting.
5.2. Chromatography.
Besides recovery and purity, other parameters, such as scalability and capacity, are critical to extend cell separation processes to clinical and commercial applications [128, 234]. In this context, cell affinity chromatography (CAC) shows great promise as a scalable technology [235], given its successful use in industrial protein purification [236, 237]. In CAC, cells are injected into a column packed with a porous material functionalized with affinity ligands. Target cells are retained by affinity on the chromatographic medium, while other components flow through (Figure 4). While similar to traditional protein chromatography, CAC faces unique challenges, due to the major differences between cells and proteins: cells are large, sensitive to shear stress [204], and possess a low diffusivity, which results in the need for convective transport to achieve sufficient interaction with the affinity surface [238–240]. Most importantly, high binding avidity requires harsh elution conditions to elute the cells from the chromatographic substrate [204, 241–243]. These challenges have highlighted the need for matrices that are tailored for chromatographic cell separations.
Computational modeling has been utilized to simulate cell interactions with affinity surfaces and guide the design of CAC substrates. Hammer et al. modeled the receptor-mediated adhesion of cells to ligand-decorated surfaces [238] and found that adhesion mainly depends on (i) the cell receptor-ligand interaction, such as the bond formation rate (kon) and strength (KD), and (ii) the fluid mechanical force, receptor mobility, and contact area [244–250]. The model predicts two regimes governing CAC, i.e. a rate-controlled high-affinity regime and a low-affinity regime. Additional studies have expanded on CAC modeling [251] by implementing advanced analytical [252–254] and numerical [244, 255–257] approaches, understanding the effect of contact time and presence of inhibitors on cell adhesion [258], evaluating the effect of cell deformability on adhesion to surfaces [259, 260], and observing cell binding in microfluidic channels [261, 262]. A model based on “cell rolling” behavior, inspired by leukocytes rolling against blood vessel walls [263, 264], was designed to increase the likelihood of ligand-receptor interactions [143, 145, 256, 265–268], reduce residence times, and secure the binding of cells with low surface marker density.
In place of traditional chromatographic substrates, alternatives such as fluidized bed CAC, cryogel CAC, and microfluidic CAC, have been proposed [234].
5.3. Fluidized Beds.
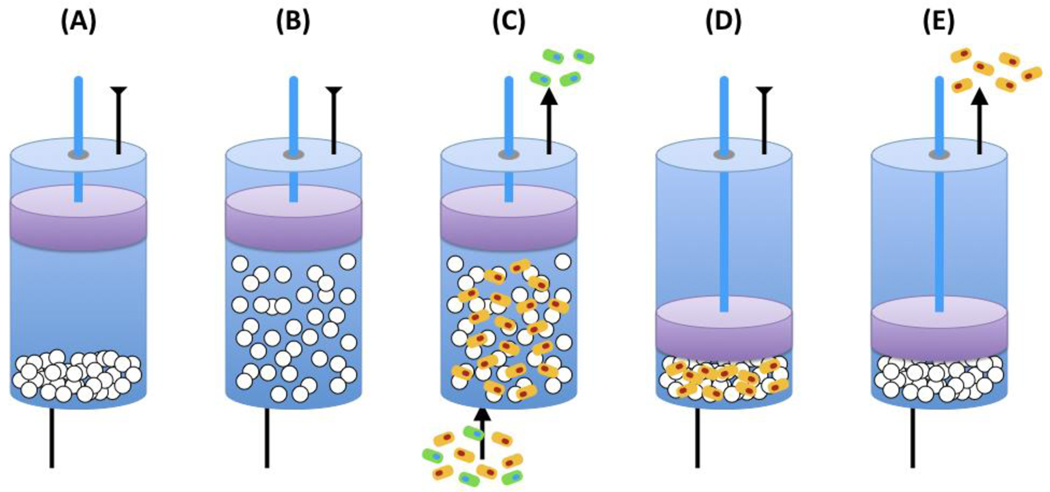
Fluidized bed, or expanded-bed, affinity adsorption is frequently used to harvest from crude feedstocks [269]. A fluidized bed is comprised of porous particles coated with cell-binding affinity ligands that are agitated by an upward flow of fluid containing the target cells (Figure 8). The advantages of this technique over traditional CAC are (i) improved mass transfer and (ii) large inter-particle volume, and (iii) high surface area [221, 270–272]. In one study, perfluorocarbon-based beads functionalized with lectin Concanavalin A were utilized to capture Saccaromyces cervisiae cells [271, 273]. The rapid adsorption kinetics enabled the capture of up to 6.8.109 cells/mL, although elution was hindered by the “avidity” effect; to facilitate elution, ion-exchange groups were used in lieu of Concanavalin A [270, 271]. Fluidized bed separation was also utilized to isolate monocytes labeled with biotinylated antibodies from human peripheral blood using streptavidin beads [272]; cells were eluted using mechanical shear to a purity of 90%, yield of 77%, and viability of greater than 65%. While promising, fluidized beds suffer from limitations such as shear stress on cells, the need for large columns, long equilibration times, non-specific capture by the adsorbent base material, limited flow velocities, disengagement of absorbed cells from ligands [270], and fouling of the beads [274].
Figure 8.
Cell purification by expanded bed chromatography. (A) Filling the column with beads; (B) Expanding the beads; (C) Loading the cell mixture; (D) Compacting and washing the beads; (E) Eluting the target cells
5.4. Cryogels.
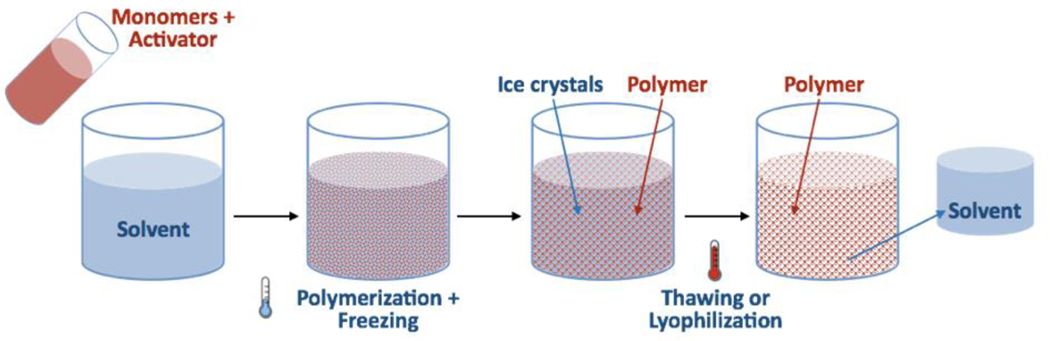
Another alternative to traditional CAC is represented by monolithic cryogels [128, 234, 275, 276]. Cryogel matrices are prepared by gelation or polymerization at sub-zero temperatures to create a continuous macroporous structure that enables cell suspensions to flow through [275] (Figure 9). While initially designed for the separation of proteins [277], oligonucleotides [278], and plasmids [279], cryogels have been shown to be ideal for the purification of viruses [280], cell organelles [281], and whole cells [234] owing to their (i) uniform and highly interconnected pores [128, 234], (ii) high channel width (>30 μm) that provides for efficient transport of cells between 2 and 15 μm [128, 282], (iii) efficient ligand conjugation [204, 205, 283], and (iv) high elasticity and hydrophilicity, which is particularly suited for mammalian cells [234, 284]. Finally, cryogels are attractive for large scale manufacturing as they exhibit high storage stability and have an extended life cycle [205, 285]. The two main approaches for cell capture using cryogels are mechanical entrapment in the cryogel matrix and ligand-mediated binding [140, 275, 286, 287]. Ligands are conjugated to the cryogel either during or after cryogenic pore formation [281, 288]. A wide array of ligand formats [234, 285, 288] including antibodies [204, 205, 247, 289, 290], proteins [128, 204, 205, 286, 291], lectins, and synthetic ligands [290, 292–294] have been incorporated into cryogels for the separation of lymphocyte cells [128, 204, 285], myeloid cells [276, 295], microbial cells like Staphylococcus aureus [290], Escherichia coli [286, 291, 296], Bacillus halodurans [297], and yeast cells [286, 291]. Elution from cryogels can be achieved by traditional methods, as well as by elastic deformation and thermally-induced shrinkage of the matrix to ensure viability of the recovered cell product [291].
Figure 9.
Process of cryogel production.
6. Pseudo-chromatographic systems
6.1. Gel Affinity Separation.
Among polymer-based media, gels are particularly attractive as single-use adsorbents for cell purification, as they can be disintegrated thermally or enzymatically to release viable cells [298–305]. Haas and Layton developed antigen-coated gelatin layers to separate spleen cells with a 30-fold enrichment [217]; the bound lymphocytes were recovered by melting the gelatin. Because cell recovery could only be performed below gelatin’s melting temperature, Maoz et al. modified this process by including matrix-specific enzymes (i.e., collagenase) [303]; less than 5% of non-specific cells bound the gel, and non-adherent cells had significantly lower cytotoxicity than the bound cells, indicating that this method can specifically isolate functional T cells. Bröcker et al. developed antigen-functionalized gelatin for purifying T cells with up to 100-fold enrichment and purity of 80–90% [304]. Similarly, Webb et al. used an anti-mouse IgG for the selective capture of B cells [302]; on average 250 cells/mm2 attached to the immobilized antibody and the B cells had a minimum viability of 60%.
6.2. Fiber-based affinity separations.
Arrays of parallel hollow fibers have gained popularity as substrates for affinity purification of cells [306–313]. Fibers introduce a new component in cell adhesion, represented by the fiber’s cross-section geometry and flexibility [309, 312]. Fiber-based adsorbents are also attractive as they can be regenerated by washing at high shear [308, 312] and can be manufactured affordably at large-scale. The first use of fibers for cell isolation was published by Edelman et al. [306], who described the isolation of spleen cells from mice, immunized against Dnp38-bovine IgG, using nylon fibers coated with Dnp38-BSA, tosyl30-BSA, and BSA antigens. The cells were detached mechanically, chemically, or competitively by incubation with inhibitors. While the eluted cells were up to 90% viable, significant non-specific binding occurred, which limited purity to 63–88%. To increase specificity, several authors have coupled antibodies and antigens to the luminal surface of cellulose hollow fiber modules. Pope et al. covalently attached goat anti-mouse antibodies to cellulose fibers to capture CD4+ lymphocytes, resulting in 63–99.9% depletion of the CD4+ cells from the starting population [314]. Similarly, Nordon et al. covalently coupled an anti-CD34 antibody directly to the luminal surface of their system’s fibers to enrich CD34+ cells from mononuclear cells at 94% purity and 61% yield [307]. Other groups have developed fusion proteins comprising an antibody-binding domain and a fiber-binding domain for mediating the adhesion of antibody-labeled cells onto the fibers. Specifically, Craig et al. developed a fusion protein (“protein LG”) that captured more than 90% of the antibody-labeled CD34+ cells onto a cellulose fiber module [311]. The use of these chimeric proteins helps overcome problems associated with ligand conjugation to hollow fibers such as low yield, random orientation, and structural alterations or degradation caused by the conjugation chemistry [311]. Hollow fiber systems enable the implementation of unconventional elution techniques. For example, bound cells can be fractionated into populations with varying binding strength by adjusting the flow rate (shear elution) [312]. Bound cells can also be eluted if labile links (e.g., a disulfide bond) are included between the ligands and fibers.
6.3. Affinity Membranes.
Membranes are suitable substrates for affinity cell separations, as the balance between trans-membrane flux and fluid velocity parallel to the surface can be easily controlled to optimize adsorption and elution [138, 315]. Additionally, the pore size of membranes and surface shear can be varied to minimize concentration polarization and fouling, which is advantageous when processing high-density cell suspensions. In an early example, Mandrusov et al. used a cellophane dialysis membrane functionalized with goat anti-mouse immunoglobulin to purify mouse B-lymphocytes [316]: cells were eluted with a low pH buffer by trans-membrane diffusion, while a shear-producing flow was applied to promote detachment of the cells from the membrane and neutralization of the acidic environment. Feeding the elution buffer on the membrane side opposite to the bound cells afforded a 100% yield and 60% viability, indicating that a trans-membrane pH gradient is needed to elute cells effectively without decreasing cell viability.
Affinity membranes enable cell separation processes that employ bubble-induced cell detachment [317]. This technique is attractive as cells can be removed from adsorption surfaces without excessive dilution. Wang et al. utilized this method with tubular capillaries coated with antibodies to purify specific blood cell populations [127], obtaining 85.7% yield, 97.6% purity, and 85.8% viability of CD4+ cells isolated from blood samples. Specifically, > 90% of cells detached by bubble-induced elution, whereas compression and flow-induced elution resulted in 40–80% and 10–40% of cell detachment, respectively [317].
Thermo-responsive polymers have also been integrated in membranes to improve elution. Specifically, a poly(N-isopropylacrylamide)-grafted polypropylene (PNIPAAm-g-PP) membrane functionalized with monoclonal antibody ligands was developed for purifying CD80+ cells [318]. PNIPAAm displays a thermo-responsive phase transition at 32°C, where it switches from a hydrophilic to a hydrophobic state. At 37°C, antibody ligands adhere to the PNIPAAm-g-PP membranes by hydrophobic interaction, enabling the affinity capture of CD80+ cells; at 4°C, the IgG ligands detach from the PNIPAAm coating, thereby releasing the cells. The recovered cells were enriched from a 1:1 cell suspension to 72%, proving to be the first case of affinity-based capture of cells where temperature is used for cell elution. In a similar work, anti-CD34 antibodies were adsorbed onto a PNIPAAm-g-PP membrane and utilized to enrich CD34+ cells. The CD34+ cell concentration was increased from 50% in the feedstock to 85% in the eluate, and 95% of the recovered cells were viable [319].
7. Non-chromatographic affinity purification methods
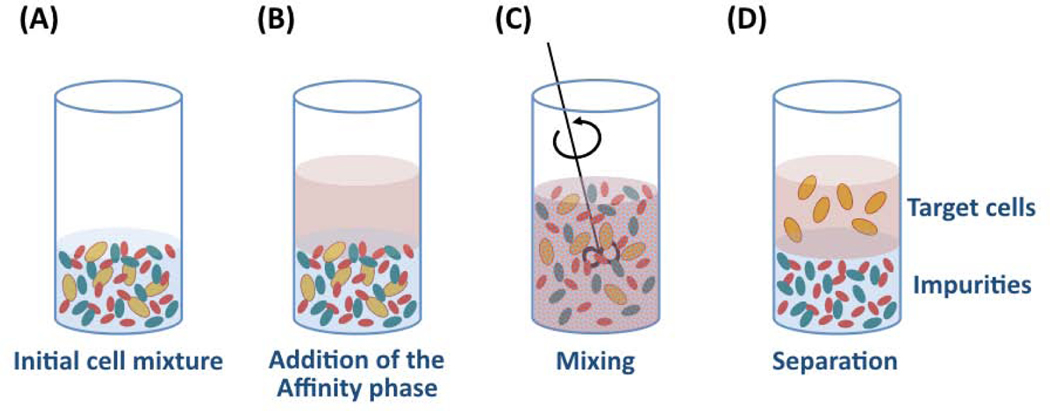
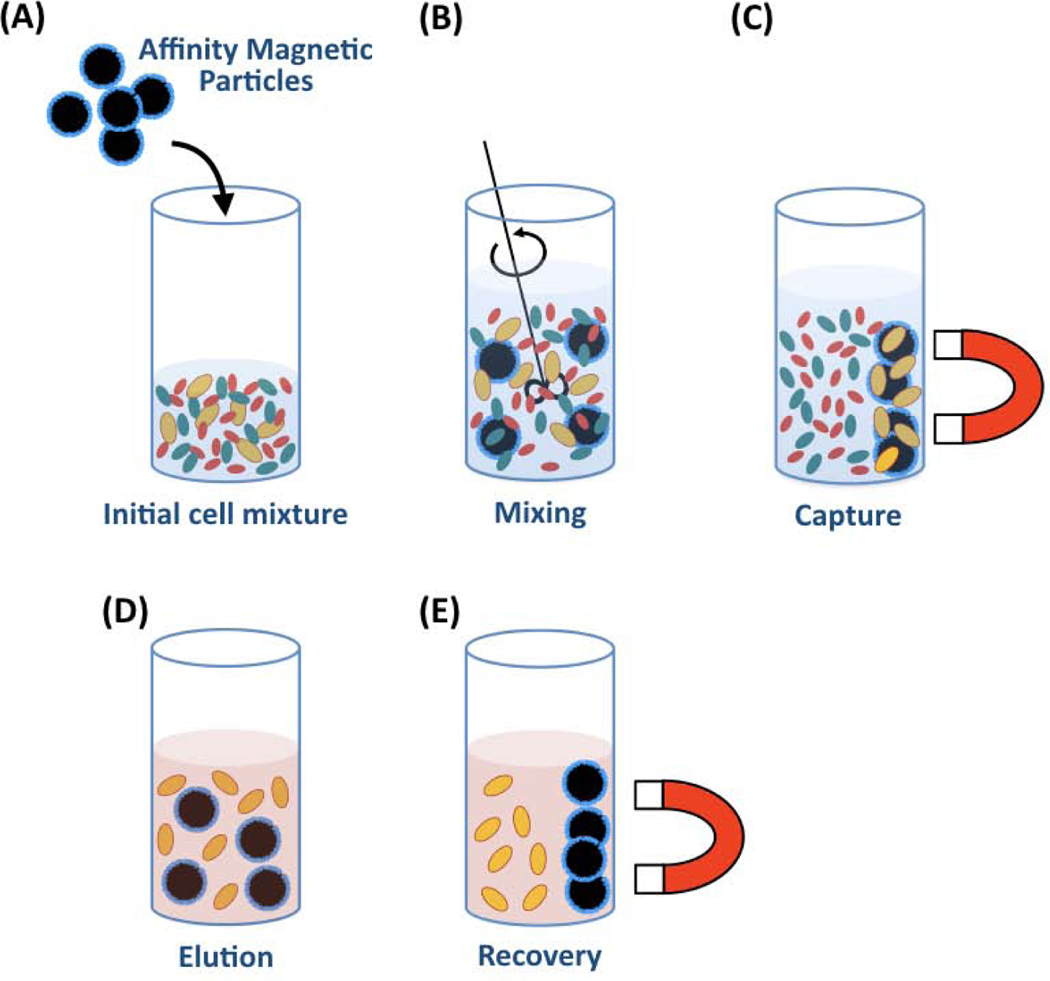
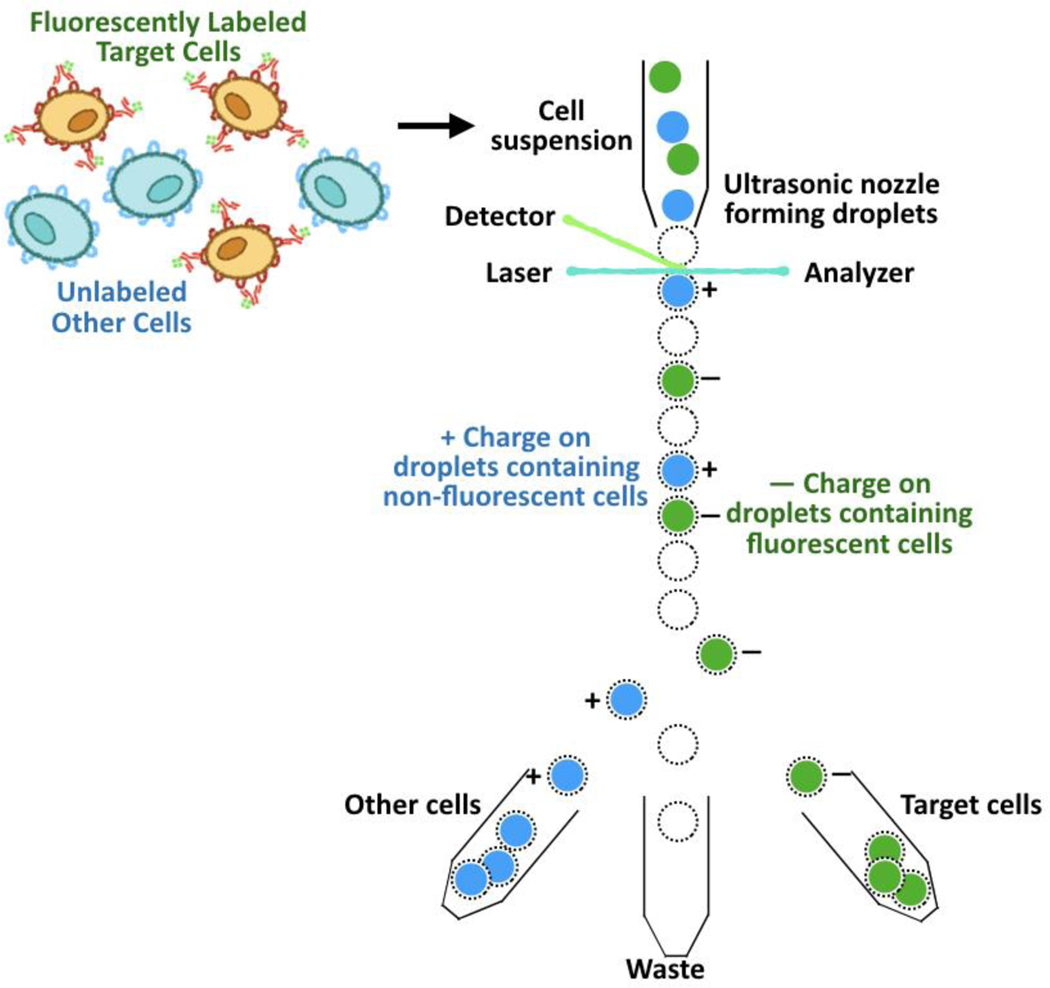
A variety of non-chromatographic techniques have been developed, such as two-phase separations, magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS) [148]. Two-phase separations employ polymeric materials often labeled with affinity ligands to drive the selective migration of cells into an aqueous phase (Figure 10). In MACS and FACS, the target cells are tagged with labeled affinity ligands that enable separation; magnetic labels are used in MACS (Figure 11), while fluorescent labels are used in FACS (Figure 12). While MACS and two-phase separations isolate cells into bulk groups, FACS is unique in its ability to analyze and sort single cells, allowing for more precise cell separation.
Figure 10.
Cell purification by affinity-based aqueous two-phase partition. (A) Suspending the cell mixture; (B) Adding the affinity-polymer forming the second phase; (C) Mixing the two phases; (D) Allowing the two phases to separate and recovering the target cells in the top affinity phase.
Figure 11.
Cell purification by MACS. (A) Contacting the cell mixture with ligand-functionalized magnetic beads; (B) suspending the magnetic beads in the cell mixture; (C) Applying a magnetic field to isolate the magnetized target cells and remove all unbound cells; (D) Resuspend and wash the magnetized target cells; (E) Elute the target cells from the magnetic beads.
Figure 12.
Cell purification by FACS.
7.1. Affinity two-phase partitioning.
Affinity two-phase partitioning is a powerful preparative method for cells, cell membranes and organelles, and viruses [140, 320–329]. Aqueous two-phase systems (ATPS) form when two polymers added to a water solution produce two non-miscible liquid layers, across which other components in solution migrate based on their differential affinity towards the polymers (Figure 10). To improve the selectivity of cell migration, affinity ligands such as lectins, antibodies, and receptor-specific molecules have been conjugated to the phase-forming polymers [330, 331]. Polyethylene glycol (PEG) and dextran are the most commonly utilized polymers for ATPS, with PEG being used as the ligand carrier and the dextran-rich phase acting as the receptacle for the bulk contaminants [325, 332–334]. Monoclonal antibodies coupled to PEG have been utilized for separating human red blood cells from sheep and rabbit blood cells, resulting in up to 92% partitioning of the human red blood cells to the top phase [321, 335, 336]. Antibody-PEG conjugates have also been used to purify hybridoma 16–3F cells from their parental NS-1 cell line, resulting in 24% recovery and 80% purity [332].
Two-phase affinity partitioning has flourished with the introduction of stimuli-responsive polymers. Kumar et al. have utilized PNIPAM decorated with antibody ligands to separate CD34+ human acute myeloid leukemia KG-1 cells from Jurkat cells (immortalized human T lymphocytes) [337]. While more than 80% of the KG-1 cells were partitioned to the top phase, a small contamination of Jurkat cells was observed; however, incomplete recovery of the conjugates lowered the yield of KG-1 cells to 75% during subsequent use. In addition to antibodies, cell separation by two-phase partitioning has also been demonstrated with other ligands, such as transferrin [323], synthetic dyes [338–342], and immobilized metals [333, 343].
Owing to its biocompatibility, mild operating conditions, and scalability, ATPS is regarded as a high-potential technology for the recovery of cell targets for which the minimization of mechanical stimuli is critical (e.g., stem cells and neurons) [327, 328]. Sousa et al. used PEG800-dextran functionalized with anti-CD34 antibodies to separate and recover CD34+ stem cells from umbilical cord blood [325]. The CD34+ cells were enriched from a starting population of ~ 0.2% CD34+ cells to ~ 42% in the final population, and recovered with 81–95% yield; in contrast, with PEG alone, a cell enrichment of 13% and 2.3% recovery was achieved. Using a three-polymer (PEG, ficoll, dextran) system and an anti-CD133 antibody, González-González et al. recovered CD133+ stem cells from umbilical cord blood with a final recovery of 62% and 98% viability [328].
A significant limitation of two-phase separations is the recurring presence of impurities in the top (product) phase. Accordingly, separations using ATPS generally result in lower purity than what is achieved using chromatographic technologies. Further work to improve ATPS separations, especially by increasing the partition preference of antibodies to a top clean phase, is considered a worthwhile effort to achieve a truly scalable process for cell separation.
7.2. Magnetic-activated cell sorting (MACS).
MACS is a relatively recent cell separation technology that employs affinity ligands conjugated to magnetic particles comprising an iron core coated by a hydrophilic shell to reduce non-specific binding (Figure 11) [344]. Upon incubating ligand-coated particles with a cell mixture, a magnetic field is applied to separate the target cells bound to the magnetic particles from the unbound cells [345]. Pioneered by Zborowski and co-workers [142, 346–350], MACS is now recognized for its speed of separation, with rates in the range of ~1011 cells/hour [100]. Recent developments enable simultaneous separation of multiple cell types using magnetic field gradients [348, 351] or by combination with microfluidic devices [352, 353]. The predominant MACS format is antibody-based, where target cells are either directly bound onto antibody-coated beads [225, 347], or are labeled in solution with a primary antibody and subsequently captured onto beads coated with a secondary antibody [354, 355].
Ligand immobilization techniques are highly dependent on the nature of the ligand. Methods for antibody immobilization to magnetic particles include covalent binding [356, 357], streptavidin-mediated immobilization (specific for biotinylated antibodies) [358, 359], Protein A-/G-mediated immobilization [360], conjugation to boronic acid or hydrazinyl groups [360, 361], and oligo-dT coating [362]. Besides antibodies [225, 345, 363–365], other affinity ligands have been successfully utilized in MACS [366–375]. Herr et al. utilized DNA aptamers to capture acute leukemia cells from complex mixtures with a 40% recovery [371]. Magnetic nanoparticles coated with bis-Zn-DPA, a synthetic ligand that binds Gram-positive and Gram-negative bacteria, have been utilized for separating Escherichia coli from blood with complete bacterial clearance in two separation cycles [375]. This work has also demonstrated that nanoparticles outperform micrometer-scale particles in terms of binding capacity and kinetics, and separation output.
The increasing relevance of immunomagnetic separation technology is demonstrated by the recent FDA approval of the CellSearch system, which can isolate circulating tumor cells using a epithelial cell-adhesion molecule (EpCAM) antibody [376–378].
7.3. Fluorescent-Activated Cell Sorting (FACS).
In FACS, fluorescently-tagged ligands are utilized to individually sort cells using fluorescence and light scattering [379–381]. When injected into the sorter, the stream of cells tagged with fluorescent ligands is broken into droplets that contain a single cell; each droplet passes through an illumination detection zone, and a charge is placed onto any cell that meets the separation criteria. As the charged droplets fall through electrostatic deflecting plates, they are sorted into different containers based on their charge (Figure 12). FACS has been extensively utilized for sorting therapeutic cell products, especially stem cells [29, 31, 382–384] and blood cells [385, 386]. FACS has gained popularity as it provides highly pure (>95%) cell populations and can sort at the single cell level due to the high sensitivity of fluorescence detection [100, 387]. FACS also allows population-averaged single cell data, as it can be used to efficiently perform high throughput cell sorting and counting [387]. Recent advances in fluorescent dyes and laser detectors allow researchers to simultaneously track multiple cell parameters [388, 389]. On the other hand, FACS requires the use of expensive equipment and suffers from limited throughput (~ 107 cells/hour) and long processing times (3–6 hours), which prevents its use in large scale manufacturing of therapeutic cells [380].
Elements of MACS and FACS sorting can be combined in a method known as “ratcheting cytometry” to perform multicomponent purifications of specific subpopulations [390]. This method is frequently used for continuous and quantitative purification of T cell subsets for cell therapy manufacturing. Specifically, T cells from apheresis or peripheral blood mononuclear cell samples are magnetically labeled using magnetic particles featuring different iron oxide content and size, and antibody functionalization. As magnetic particles travel differently within the sorting cartridge based on their magnetization and size, cells specifically bound to a magnetic particle population can be isolated from other cells in the mixture [390]. Ratcheting cytometry also enables sorting cells based on differential levels of antigen, as this determines the number of magnetic particles bound to a cell. This method has been used to simultaneously isolate CD4 and CD8 T cells from a sample via labeling with antigen-specific magnetic particles [391].
8. Emerging Trends
Cell purification technology is rapidly evolving, owing to the introduction of target-specific biorecognition moieties for capture (i.e., biological and synthetic ligands) and isolation formats (e.g., microfluidic devices).
8.1. Microfluidic devices for cell separation.
The latest frontier of CAC is represented by microfluidic devices that comprise sub-millimeter channels coated with affinity ligands (M-CAC) [129, 131, 159, 392–396]. The high surface-area-to-volume ratio of microfluidic channels, enhanced by micro-fabricated structures with complex geometry, has enabled the capture of cells at extremely low concentrations by M-CACs [261, 392, 397–399]. M-CAC systems have been utilized to separate T- and B-lymphocytes at high purity (> 97%) from mixed suspensions [127, 175, 400]. To ensure binding specificity, the channels are often grafted with hydrophilic polymer brushes (e.g., PEG) or coated with hydrogels (e.g., alginate) functionalized with antibody ligands [401, 402]. Chang et al. have developed a M-CAC system coated with E-selectin IgG molecules to separate HL-60 and U-937 myeloid cells with purity greater than 70% and 200-fold enrichment [262]. M-CAC also enables the sorting and capture of multiple cell types from a complex mixture. Li et al. incorporated a pneumatic-actuated control layer into an affinity separation layer to create different antibody-coated regions within the same channel [129]. Ramos cells were flown through anti-CD19- and anti-CD71-coated regions, with the anti-CD19 region having a capture density 2.44-fold higher than the anti-CD71 region. The authors also coated a second channel with two different antibodies targeting either Ramos or HuT 78 cells, allowing specific retention of the cells in their complementary region at greater than 90% purity. Lastly, a four-region antibody-coated device was developed for the simultaneous capture of three different cell lines in a single channel, thereby enabling multiple cell sorting.
Microfluidic devices coated with antibodies against specific cell markers have gathered considerable interest as tools for detecting rare tumor circulating cells (CTCs) [397, 403–405]. Isolating CTCs from the bloodstream enables the detection, characterization, and monitoring of non-hematological cancers [406], but is made extremely challenging by their low concentration (1 −100 CTCs per mL of blood [82, 376–378, 407, 408]). Researchers have shown that microfluidic devices (CTC-Chip) containing an array of microposts functionalized with epithelial cell adhesion molecules (EpCAM) can capture CTCs [397]. Many attributes of the device have been explored to enhance CTC enrichment. Gleghorn et al. described how the geometry of the microposts can enhance CTC enrichment [398]. CTC-ligand binding has also been improved by introducing a high-throughput microfluidic device called “HB-Chip”, which mixes the blood cells by generating micro-vortices that increase the interactions between the target CTCs and the antibody-coated channels [406]. To increase binding sensitivity, Myung et al. developed high-avidity ligands by conjugating multiple EpCAM ligands to dendimers [409]. The combination of multivalent binding and cell rolling in the channels mediated by E-selectin granted high sensitivity and specificity towards CTCs. This work has led to a device, commercialized by Biocept, which employs streptavidin-coated microposts to capture CTCs tagged with biotinylated antibodies, followed by fluorescent microscopy-based detection and in situ cytogenic interrogation [399, 410]. Another approach for the isolation of CTCs is represented by negative enrichment (or negative selection) using microfluidic technologies [411], which takes advantage of the physical and biochemical properties of cells [412]. Unlike hematopoietic cells, which display the cell surface markers CD15 (granulocytes), CD66b (granulocytes) and CD45 (leukocytes), CTCs are CD15/45-negative. Accordingly, negative enrichment technologies feature microfluidic channels, nanoparticles, and micro-scale adsorbents functionalized with anti-CD15, anti-CD66b and, most commonly, anti-CD45 antibodies [155–157]. The affinity-based selection alone, however, is often not sufficient to achieve the desired enrichment factor, and must be complemented by size exclusion-based or fluid dynamic-based separation techniques [157, 158, 160, 413, 414].
Lastly, emerging technologies for droplet-based single cell analyses are flooding the contemporary literature landscape. While there is significant focus on droplet barcoding for single cell sequencing and transcriptomics [415–418], some efforts are aimed at employing droplet-based technologies for human cell isolation, sorting, and studying biomolecular interactions [419–425]. An in-depth review of droplet-based cell analyses was recently provided by Huck et al. [426]. Briefly, the formation of water-in-oil plug flow in microfluidics can generate picoliter-sized droplets for carrying cells or other biomolecular residents, and these droplets can be generated in a highly repetitive and chemically-defined manner [427–430]. When employed for cell isolation and sorting, the physico-chemical properties of the droplet can be tuned to promote interaction with specific surface features of the microfluidic device, resulting in droplet isolation and sorting [431–435]; for example, the interfacial tension of the droplet can be made sensitive to pH causing the droplets to interact with the microfluidic channel’s surface [422]. The physico-chemical properties of the resident cell can also be made responsive, so that the droplet can be sorted via imaging and fluorescence-activated techniques [436–440].
In regard to using these techniques for studying affinity-based chemistries, droplet microfluidics have been employed to screen drug and antibody binding by generating sub-nanoliter reactors [441–444]. In one example, hybridoma cells secreting antibodies were individually co-encapsulated with a target cell in nanodroplets to select hybridoma clones expressing antibodies featuring affinity for the target cell [440]. While there have been limited studies of strictly affinity-based sorting via droplet microfluidics, based on the aforementioned examples there is an emerging lane of study for using droplet microfluidics for therapeutic antibody discovery, especially since the single-cell droplet approach is amenable to use with primary human plasma cells, which secrete antibodies.
8.2. Synthetic Ligands.
A significant barrier to improving the affordability of cell products is represented by the cost of biological ligands [133]. While highly selective, proteins and antibodies are biochemically labile [445], and a complex engineering process is required to discover viable ligands [446]. Further, they are generally characterized by high binding strength, which can trigger undesired intracellular signaling cascades upon binding and even cell death [447–449]. To overcome these limitations, synthetic ligands have been proposed to maintain targeted affinity while lowering binding strength to facilitate cell elution. In addition, synthetic ligands are biochemically stable and can be synthesized affordably at large scale.
Hormones are the first small molecules to ever be utilized as affinity ligands [450]. In particular, histamine [451, 452], catecholamines [453], and prostaglandins [454, 455] coupled to Sepharose beads have been used to separate 19S and 7S plaque-forming cells from the total spleen leukocyte population. The hormone-based adsorbents were able to capture 56% and 84% of the 19S and 7S plaque-forming cells respectively. Similarly, glycans (e.g., mannose) immobilized on Dowex resins have been utilized to separate E. coli K12 and Campylobacter jejuni NCTC 11168 cells with high yield (94–96%) and selectivity [456].
Recent advances in selection technology have spurred the use of synthetic ligands with engineered affinity and selectivity for any target cell [133, 457–459]. Aptamers and peptides represent the main classes of synthetic ligands [449, 460–470]. Aptamers consist of single-stranded DNA or RNA molecules and have seen rising popularity in cell purification [469]. The development of these ligands is supported by a high-throughput screening method known as “systematic evolution of ligands by exponential enrichment” (SELEX) [464, 471–474]. Xu et al. have selectively captured three leukemia cell lines (CCL-119, Ramos cells, and Toledo cells) using a microfluidic device coated with cell line-specific aptamers, with up to 136-fold enrichment [475]. Aptamers have also been demonstrated as cell capture ligands in more traditional pseudo-chromatography applications, as described by Zhang et al., where aptamers coupled to a hydrogel bound and eluted target cells with a resulting viability of ~ 99% [476]. Aptamers have also been successfully used in MACS applications [371, 372, 475, 477–480], microfluidic devices, and hydrogels, with reported capture efficiencies and cell purities at > 80% [131, 403, 481–483]. These case studies showcase the value of aptamers as cell capturing ligands. Nonetheless, some improvements are still needed, such as increasing binding selectivity to improve capture [484], tuning the binding strength to facilitate cell elution [485], and addressing safety concerns by implementing rigorous tests of biocompatibility [486].
Peptides have also emerged as robust and cost-effective alternatives to protein ligands [133]. Over the past two decades a number of selection techniques have been developed, ranging from the screening of biological and synthetic libraries in liquid or solid phase to in silico approaches, such as computational design and machine learning. This has resulted in a myriad of peptides targeting analytical and medically relevant target cells. Veleva et al. identified an angiogenic tumor-binding peptide via in vitro enrichment of a peptide library against peripheral blood outgrown endothelial cells followed by in vivo screening of the enriched library to identify tumor-binding peptides [487]. Similarly, Oyama and coworkers identified peptides from a phage library to bind human lung cancer cell lines and noted that the selected peptides were specific towards the target cancer cells without negative selection [461, 466]. In 2008, Choi et al. identified a Raji cell-targeting peptide as a model for Burkitt lymphoma cells with seemingly high specificity for these cells as determined by lack of binding to normal, non-cancerous B cells, peripheral blood cells, or other leukemia cells [488]. Wang et al. also utilized phage display to identify affinity peptides for imaging detection of human colorectal cancer cells (Caco-2); the specificity of the peptide was confirmed using negative-control cell lines HEK293, SGC-7901, and SMMC-7721 [489]. Peptide ligands have also been employed in a number of cell adhesion applications, which are of primary interest for cell separations. De et al. demonstrated the use of peptides in pathogen removal applications by isolating pancreatic beta-cells infected by Mycoplasma arginii from healthy cells, showcasing a 10-fold reduction in the number of infected cells [490]. Success in separating different phenotypes of primary cells has also been shown in multiple cases by microcontact printing of tetrameric peptides in microfluidic devices. These have been used for the separation of osteoblasts from fibroblasts by Hasenbein and coworkers [491], or the fractionation and characterization of different human cell phenotypes by Murthy and coworkers [492–494].Peptide ligands have been discovered for a number of cell surface markers that identify analytically or therapeutically relevant cells, including CD-34 [495], CD-133 [496, 497], CD-38 [498], VCAM-1 [499–502], and Flt-3 [503, 504].
Large peptides, developed from non-antibody scaffolds, also represent a viable alternative for cell separation purposes. Our group has identified the first non-antibody binders for CD-117 by screening a yeast-display scaffold library against magnetized yeast cells expressing the extracellular domain of CD-117 [505]. Two nanobody mutants were identified with good affinity (i.e., 131 and 204 nM) for CD-117. While binding of these mutants for CD-117 was only confirmed for yeast displayed CD-117, a combination of these ligands would likely enable the purification of phenotypically pure cells such as endothelial stem and progenitor cells (ESCs, HSC), and hematopoietic stem and progenitor cells (EPCs, HPCs) [506–517]. Additionally, the mid-nanomolar affinities of the nanobody ligands promote gentler compared to antibody ligands.
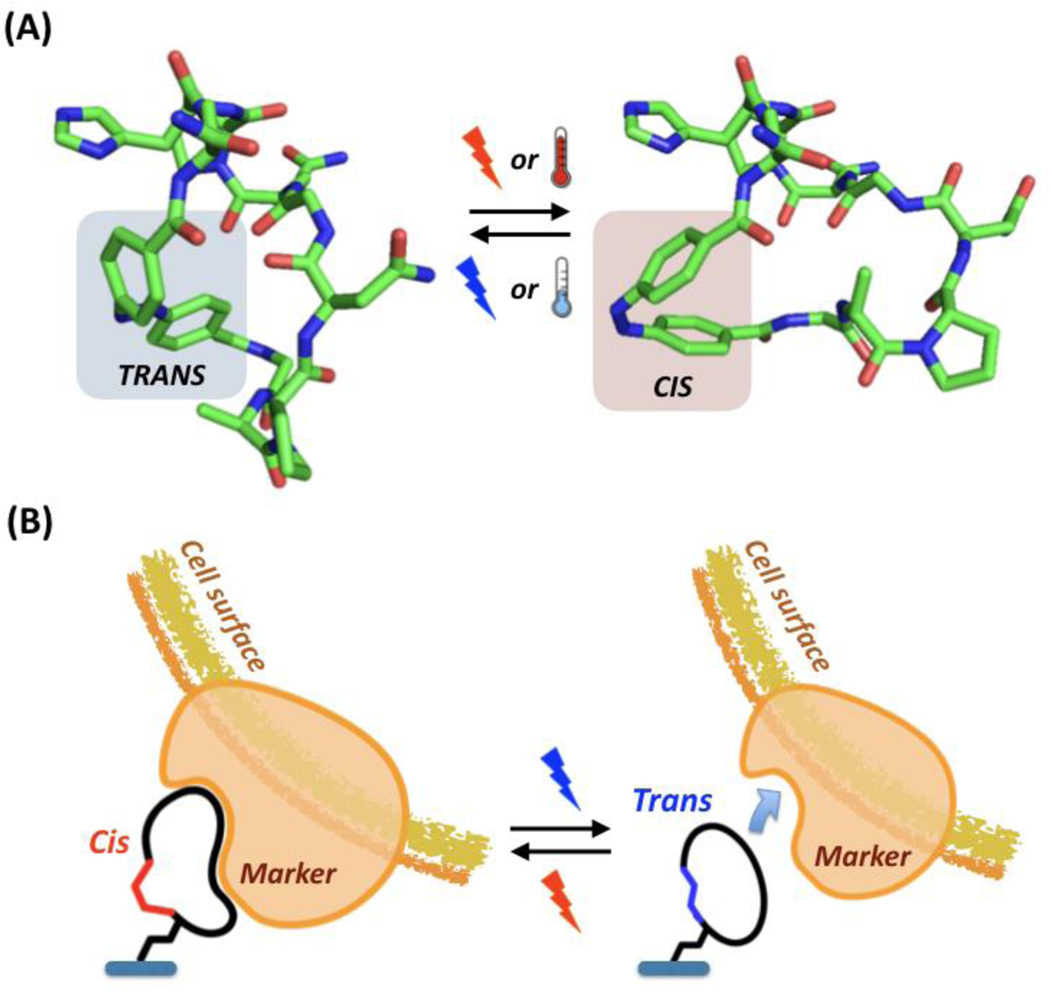
Peptide ligands can also be engineered to enable the control of cell binding and release upon exposure of biocompatible stimuli. To this end, stimuli-responsive monomers can be incorporated into the amino acid sequence, allowing the peptide to reversibly switch between a binding and a non-binding mode upon cooling, or exposure to light or a magnetic field (Figure 13). Our group, for example, has developed VCAM1-binding azobenzene-cyclized peptides for the light-controlled labeling of endothelial progenitor cells [518]. Upon exposure to light, the ligands undergo a remarkable ~ 1300-fold variation in binding strength, which enables selective and stable light-controlled labeling of cells. Notably, modified azobenzene linkers have been engineered to photo-switch in different wavelength windows, namely red, green, and blue (RGB) light [519–524]. Therefore, a combination of peptide ligands targeting different cell markers, whose binding/release is triggered at different wavelengths, could be used to produce and dynamically modify patterns of cells on solid substrates by exposure to sequences of red, green, or blue photo-patterns, for example using liquid crystal display light-emitting diodes (LCD-LED) arrays.
Figure 13.
Cell purification using stimuli-responsive peptide ligands. (A) Reversible photo- or thermo-switching of an azobenzene-cyclized peptide; (B) Principle of cell capture and release using stimuli-controlled cyclic peptides.
9. Conclusions.
Cell separation technologies have progressed steadily to meet the demands for basic research, diagnostic, and therapeutic applications, resulting in cell isolation methods that are more efficient, scalable, and dependable. Affinity-based approaches are now the most utilized, owing to their ability to achieve high purity. A wide variety of affinity-based approaches are available, ranging from traditional chromatographic to pseudo- and non-chromatographic systems. Each system has advantages and disadvantages, as outlined in this work, which must be carefully considered when choosing a cell separation method. Microfluidic technologies represent the next frontier of cell manufacturing as they offer the capacity to perform multiple functions (mixing, counting, lysis, single cell analysis, etc.) in a single device. Advances in parallelization and scale-up hold great promise to overcome the low throughput of current devices and enable processing of large sample volumes. Further, the ability to integrate post-sorting molecular, cellular, and functional characterization furthers the appeal of using microfluidic devices for cell separation.
On the biorecognition front, affinity-based separations are shifting from protein and antibody ligands towards synthetic ligands. Biological ligands, in fact, while highly specific, are limited by their high cost and exceedingly strong binding. Synthetic ligands, on the other hand, can be synthesized affordably, at large scale, and with no batch-to-batch variability. The need to develop gentle cell elution conditions has stimulated the development of stimuli-responsive ligands, such as photo-switchable peptides, whose binding activity can be controlled by exposure to biocompatible stimuli. In this regard, further progress in the fields of in vitro and in silico selection methods is needed to expand the portfolio of peptides and aptamers with tailored affinity and binding mechanism for cellsurface markers. Further studies – both experimental and modeling – are also needed to understand the balance between affinity, multivalent binding due to expression level of surface markers, in order to optimize the balance between efficient cell capture and elution, ultimately enabling high recovery and bioactivity.
Currently, the major challenge for cell therapies and related clinical applications resides in achieving rapid, efficient, and affordable separation while minimizing costs and attaining the required purity, yield, and functionality of the cellular product. Membrane-based separations show exceptional potential in large-scale production, particularly in combination with cell-specific biorecognition moieties that ensure high recovery, purity, and bioactivity of the cell product. Owing to their high pore diameter and porosity, in fact, membranes enable processing high volumes of cell suspensions at high flow rates, thereby increasing throughput and minimizing processing time, which aids in maintaining the viability of the cell product. On the front of basic cellular research and personalized medicine, the continued identification of highly specific markers defining cell populations [525], combined with the advancements in integrating physical and affinity-based strategies in miniaturized devices, will be critical for the fruition of patient-specific cellular therapies.
Statement of significance.
Technologies for cell purification have served science, medicine, and industrial biotechnology and biomanufacturing for decades. This review presents a comprehensive survey of this field by highlighting the scope and relevance of all known methods for cell isolation, old and new alike. The first section covers the main classes of target cells and compares traditional non-affinity and affinity-based purification techniques, focusing on established ligands and chromatographic formats. The second section presents an excursus of affinity-based pseudo-chromatographic and non-chromatographic technologies, especially focusing on magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). Finally, the third section presents an overview of new technologies and emerging trends, highlighting how the progress in chemical, material, and microfluidic sciences has opened new exciting avenues towards high-throughput and high-purity cell isolation processes. This review is designed to guide scientists and engineers in their choice of suitable cell purification techniques for research or bioprocessing needs.
Acknowledgments.
This work was partially funded by a grant from the National Science Foundation (CBET 1511227 and 1743404). KB kindly acknowledges support from an NSF Graduate Research Fellowship and a National Institute of Health Molecular Biotechnology Training Fellowship (NIH T32 GM008776).
Footnotes
Declaration of Interest Statement For:
No personal or financial interests exist.
Conflict of interest. The authors have no conflict of interest to acknowledge.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Food D. Administration, Guidance for industry: guidance for human somatic cell therapy and gene therapy, Fed Reg 36413 (1998). [Google Scholar]
- [2].Halme DG, Kessler DA, FDA regulation of stem-cell–based therapies, Mass Medical Soc, 2006. [DOI] [PubMed] [Google Scholar]
- [3].Food H. Drug Administration, Eligibility determination for donors of human cells, tissues, and cellular and tissue-based products. Final rule, Federal register 69(101) (2004) 29785. [PubMed] [Google Scholar]
- [4].Regulation E, 1394/2007 on advanced therapy medicinal products and amending Directive 2001/83, EC and Regulation (EC) No 726 (2004). [Google Scholar]
- [5].George B, Regulations and guidelines governing stem cell based products: clinical considerations, Perspectives in clinical research 2(3) (2011) 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ratcliffe E, Thomas RJ, Williams DJ, Current understanding and challenges in bioprocessing of stem cell-based therapies for regenerative medicine, British medical bulletin (2011) ldr037. [DOI] [PubMed] [Google Scholar]
- [7].Badylak SF, Taylor D, Uygun K, Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds, Annual review of biomedical engineering 13 (2011) 27–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bijonowski BM, Miller WM, Wertheim JA, Bioreactor design for perfusion-based, highly vascularized organ regeneration, Current opinion in chemical engineering 2(1) (2013) 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Robertson MJ, Dries-Devlin JL, Kren SM, Burchfield JS, Taylor DA, Optimizing recellularization of whole decellularized heart extracellular matrix, PloS one 9(2) (2014) e90406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gilpin SE, Ott HC, Using nature’s platform to engineer bio-artificial lungs, Annals of the American Thoracic Society 12(Supplement 1) (2015) S45–S49. [DOI] [PubMed] [Google Scholar]
- [11].Zhou Q, Li L, Li J, Stem cells with decellularized liver scaffolds in liver regeneration and their potential clinical applications, Liver International 35(3) (2015) 687–694. [DOI] [PubMed] [Google Scholar]
- [12].Bosman G, Bartholomeus I, De Man A, Van Kalmthout P, De Grip W, Erythrocyte membrane characteristics indicate abnormal cellular aging in patients with Alzheimer’s disease, Neurobiology of aging 12(1) (1991) 13–18. [DOI] [PubMed] [Google Scholar]
- [13].Nossal GJ, Negative selection of lymphocytes, cell 76(2) (1994) 229–239. [DOI] [PubMed] [Google Scholar]
- [14].von Boehmer H, Positive selection of lymphocytes, Cell 76(2) (1994) 219–228. [DOI] [PubMed] [Google Scholar]
- [15].Rawstron A, Villamor N, Ritgen M, Böttcher S, Ghia P, Zehnder J, Lozanski G, Colomer D, Moreno C, Geuna M, International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia, Leukemia 21(5) (2007) 956–964. [DOI] [PubMed] [Google Scholar]
- [16].Ozcan A, Demirci U, Ultra wide-field lens-free monitoring of cells on-chip, Lab on a Chip 8(1) (2008) 98–106. [DOI] [PubMed] [Google Scholar]
- [17].Gorges TM, Pantel K, Circulating tumor cells as therapy-related biomarkers in cancer patients, Cancer Immunology, Immunotherapy 62(5) (2013) 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kodituwakku AP, Jessup C, Zola H, Roberton DM, Isolation of antigen-specific B cells, Immunology and cell biology 81(3) (2003) 163–170. [DOI] [PubMed] [Google Scholar]
- [19].Butcher EC, Can cell systems biology rescue drug discovery?, Nature Reviews Drug Discovery 4(6) (2005) 461–467. [DOI] [PubMed] [Google Scholar]
- [20].Folkman J, Angiogenesis: an organizing principle for drug discovery?, Nature reviews Drug discovery 6(4) (2007) 273–286. [DOI] [PubMed] [Google Scholar]
- [21].Zhou B-BS, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB, Tumour-initiating cells: challenges and opportunities for anticancer drug discovery, Nature reviews Drug discovery 8(10) (2009) 806–823. [DOI] [PubMed] [Google Scholar]
- [22].Grskovic M, Javaherian A, Strulovici B, Daley GQ, Induced pluripotent stem cells—opportunities for disease modelling and drug discovery, Nature reviews Drug discovery 10(12) (2011) 915–929. [DOI] [PubMed] [Google Scholar]
- [23].Gee A, Durett A, Cell sorting for therapeutic applications--points to consider, Cytotherapy 4(1) (2002) 91–92. [DOI] [PubMed] [Google Scholar]
- [24].Shulman LP, Fetal cells in maternal blood, Current women’s health reports 3(1) (2003) 47–54. [PubMed] [Google Scholar]
- [25].Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT, Marker-specific sorting of rare cells using dielectrophoresis, Proceedings of the National Academy of Sciences of the United States of America 102(44) (2005) 15757–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kumar RK, Lykke A, Cell separation: a review, Pathology 16(1) (1984) 53–62. [DOI] [PubMed] [Google Scholar]
- [27].Ibrahim SF, van den Engh G, High-speed cell sorting: fundamentals and recent advances, Current opinion in biotechnology 14(1) (2003) 5–12. [DOI] [PubMed] [Google Scholar]
- [28].Kamihira M, Kumar A, Development of separation technique for stem cells, Cell Separation (2007) 173–193. [DOI] [PubMed] [Google Scholar]
- [29].Diogo MM, da Silva CL, Cabral J, Separation technologies for stem cell bioprocessing, Biotechnology and bioengineering 109(11) (2012) 2699–2709. [DOI] [PubMed] [Google Scholar]
- [30].Burger S, Current regulatory issues in cell and tissue therapy, Cytotherapy 5(4) (2003) 289–298. [DOI] [PubMed] [Google Scholar]
- [31].McIntyre CA, Flyg BT, Fong TC, Fluorescence-activated cell sorting for CGMP processing of therapeutic cells, BioProcess International 8(6) (2010) 44–53. [Google Scholar]
- [32].Brandenberger R, Burger S, Campbell A, Fong T, Lapinskas E, Rowley JA, Cell therapy bioprocessing, BioProcess Int 9(Suppl 1) (2011) 30–37. [Google Scholar]
- [33].Goldring CE, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, French N, Hanley NA, Kelly L, Kitteringham NR, Assessing the safety of stem cell therapeutics, Cell stem cell 8(6) (2011) 618–628. [DOI] [PubMed] [Google Scholar]
- [34].Jung Y, Bauer G, Nolta JA, Concise review: induced pluripotent stem cell‐derived mesenchymal stem cells: progress toward safe clinical products, Stem cells 30(1) (2012) 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Okano H, Nakamura M, Yoshida K, Okada Y, Tsuji O, Nori S, Ikeda E, Yamanaka S, Miura K, Steps toward safe cell therapy using induced pluripotent stem cells, Circulation research 112(3) (2013) 523–533. [DOI] [PubMed] [Google Scholar]
- [36].Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Characterization of human embryonic stem cell lines by the International Stem Cell Initiative, Nature biotechnology 25(7) (2007) 803–816. [DOI] [PubMed] [Google Scholar]
- [37].Segers VF, Lee RT, Stem-cell therapy for cardiac disease, Nature 451(7181) (2008) 937–942. [DOI] [PubMed] [Google Scholar]
- [38].Bowen WS, Development and characterisation of affinity devices for cell detection and separation, © William Bowen, 2015. [Google Scholar]
- [39].Bosman G, Werre J, Willekens F, Novotný V, Erythrocyte ageing in vivo and in vitro: structural aspects and implications for transfusion, Transfusion Medicine 18(6) (2008) 335–347. [DOI] [PubMed] [Google Scholar]
- [40].Taimeh Z, Koene RJ, Furne J, Singal A, Eckman PM, Levitt MD, Pritzker MR, Erythrocyte aging as a mechanism of anemia and a biomarker of device thrombosis in continuous-flow left ventricular assist devices, The Journal of Heart and Lung Transplantation 36(6) (2017) 625–632. [DOI] [PubMed] [Google Scholar]
- [41].Dinkla S, Erythrocyte aging and disease A tale of membranes and microparticles, Ned Tijdschr Klin Chem Labgeneesk 41(3) (2016) 217–228. [Google Scholar]
- [42].De Franceschi L, Olivieri O, Corrocher R, Erythrocyte aging in neurodegenerative disorders, Cellular and molecular biology (Noisy-le-Grand, France) 50(2) (2004) 179–185. [PubMed] [Google Scholar]
- [43].Moir S, Fauci AS, B cells in HIV infection and disease, Nature Reviews Immunology 9(4) (2009) 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hampe CS, B cells in autoimmune diseases, Scientifica 2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Edwards MR, Sultan P, Del Arroyo AG, Whittle J, Karmali SN, Moonesinghe SR, Haddad FS, Mythen MG, Singer M, Ackland GL, Metabolic dysfunction in lymphocytes promotes postoperative morbidity, Clinical Science 129(5) (2015) 423–437. [DOI] [PubMed] [Google Scholar]
- [46].Lian CG, Bueno EM, Granter SR, Laga AC, Saavedra AP, Lin WM, Susa JS, Zhan Q, Chandraker AK, Tullius SG, Biomarker evaluation of face transplant rejection: association of donor T cells with target cell injury, Modern Pathology 27(6) (2014) 788–799. [DOI] [PubMed] [Google Scholar]
- [47].Krensky AM, Weiss A, Crabtree G, Davis MM, Parham P, T-lymphocyte-antigen interactions in transplant rejection, New England Journal of Medicine 322(8) (1990) 510–517. [DOI] [PubMed] [Google Scholar]
- [48].Finke J, Schmoor C, Bethge WA, Ottinger H, Stelljes M, Volin L, Heim D, Bertz H, Grishina O, Socie G, Long-term outcomes after standard graft-versus-host disease prophylaxis with or without anti-human-T-lymphocyte immunoglobulin in haemopoietic cell transplantation from matched unrelated donors: final results of a randomised controlled trial, The Lancet Haematology 4(6) (2017) e293–e301. [DOI] [PubMed] [Google Scholar]
- [49].Galli SJ, Tsai M, IgE and mast cells in allergic disease, Nature medicine 18(5) (2012) 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Silver R, Curley JP, Mast cells on the mind: new insights and opportunities, Trends in neurosciences 36(9) (2013) 513–521. [DOI] [PubMed] [Google Scholar]
- [51].Galli SJ, Wedemeyer J, Tsai M, Analyzing the roles of mast cells and basophils in host defense and other biological responses, International journal of hematology 75(4) (2002) 363–369. [DOI] [PubMed] [Google Scholar]
- [52].Ramos L, Peña G, Cai B, Deitch E, Ulloa L, Mast cell stabilization improves survival by preventing apoptosis in sepsis, The Journal of Immunology 185(1) (2010) 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Urb M, Sheppard DC, The role of mast cells in the defence against pathogens, PLoS pathogens 8(4) (2012) e1002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Norrby K, Mast cells and angiogenesis, Apmis 110(5) (2002) 355–371. [DOI] [PubMed] [Google Scholar]
- [55].Bischoff SC, Schwengberg S, Raab R, Manns MP, Functional properties of human intestinal mast cells cultured in a new culture system: enhancement of IgE receptor-dependent mediator release and response to stem cell factor, The Journal of Immunology 159(11) (1997) 5560–5567. [PubMed] [Google Scholar]
- [56].Douaiher J, Succar J, Lancerotto L, Gurish MF, Orgill DP, Hamilton MJ, Krilis SA, Stevens RL, Development of mast cells and importance of their tryptase and chymase serine proteases in inflammation and wound healing, Advances in immunology 122 (2014) 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Benoist C, Mathis D, Mast cells in autoimmune disease, Nature 420(6917) (2002) 875–878. [DOI] [PubMed] [Google Scholar]
- [58].Gibbs BF, Ennis M, Isolation and purification of human mast cells and basophils, Human Airway Inflammation: Sampling Techniques and Analytical Protocols (2001) 161–176. [DOI] [PubMed] [Google Scholar]
- [59].Sellge G, Bischoff SC, Isolation, culture, and characterization of intestinal mast cells, Mast Cells: Methods and Protocols (2005) 123–138. [DOI] [PubMed] [Google Scholar]
- [60].Wang Y, Zhao Z, Ren Z, Zhao B, Zhang L, Chen J, Xu W, Lu S, Zhao Q, Peng J, Recellularized nerve allografts with differentiated mesenchymal stem cells promote peripheral nerve regeneration, Neuroscience Letters 514(1) (2012) 96–101. [DOI] [PubMed] [Google Scholar]
- [61].Rafii S, Lyden D, Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration, Nature medicine 9(6) (2003) 702–712. [DOI] [PubMed] [Google Scholar]
- [62].Bonandrini B, Figliuzzi M, Papadimou E, Morigi M, Perico N, Casiraghi F, Sangalli F, Conti S, Benigni A, Remuzzi A, Recellularization of well-preserved acellular kidney scaffold using embryonic stem cells, Tissue Engineering Part A 20(9–10) (2014) 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R, Embryonic stem cell trials for macular degeneration: a preliminary report, The Lancet 379(9817) (2012) 713–720. [DOI] [PubMed] [Google Scholar]
- [64].Brederlau A, Correia AS, Anisimov SV, Elmi M, Paul G, Roybon L, Morizane A, Bergquist F, Riebe I, Nannmark U, Transplantation of human embryonic stem cell‐derived cells to a rat model of Parkinson’s disease: effect of in vitro differentiation on graft survival and teratoma formation, Stem cells 24(6) (2006) 1433–1440. [DOI] [PubMed] [Google Scholar]
- [65].Gonzalez R, Garitaonandia I, Crain A, Poustovoitov M, Abramihina T, Noskov A, Jiang C, Morey R, Laurent LC, Elsworth JD, Proof of concept studies exploring the safety and functional activity of human parthenogenetic-derived neural stem cells for the treatment of Parkinson’s disease, Cell transplantation 24(4) (2015) 681–690. [DOI] [PubMed] [Google Scholar]
- [66].Lalit PA, Hei DJ, Raval AN, Kamp TJ, Induced Pluripotent Stem Cells for Post–Myocardial Infarction Repair, Circulation research 114(8) (2014) 1328–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict CL, Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction, Circulation research (2015) CIRCRESAHA. 115.305990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Baum CM, Weissman IL, Tsukamoto AS, Buckle A-M, Peault B, Isolation of a candidate human hematopoietic stem-cell population, Proceedings of the National Academy of Sciences 89(7) (1992) 2804–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schriebl K, Lim S, Choo A, Tscheliessnig A, Jungbauer A, Stem cell separation: a bottleneck in stem cell therapy, Biotechnology journal 5(1) (2010) 50–61. [DOI] [PubMed] [Google Scholar]
- [70].Jillella AP, Ustun C, What is the optimum number of CD34+ peripheral blood stem cells for an autologous transplant?, Stem cells and development 13(6) (2004) 598–606. [DOI] [PubMed] [Google Scholar]
- [71].Klimanskaya I, Rosenthal N, Lanza R, Derive and conquer: sourcing and differentiating stem cells for therapeutic applications, Nature reviews Drug discovery 7(2) (2008) 131–142. [DOI] [PubMed] [Google Scholar]
- [72].Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS, Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation, Glia 49(3) (2005) 385–396. [DOI] [PubMed] [Google Scholar]
- [73].Doi D, Samata B, Katsukawa M, Kikuchi T, Morizane A, Ono Y, Sekiguchi K, Nakagawa M, Parmar M, Takahashi J, Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation, Stem cell reports 2(3) (2014) 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Juopperi TA, Schuler W, Yuan X, Collector MI, Dang CV, Sharkis SJ, Isolation of bone marrow–derived stem cells using density-gradient separation, Experimental hematology 35(2) (2007) 335–341. [DOI] [PubMed] [Google Scholar]
- [75].Yamamoto Y, Itoh S, Yamauchi Y, Matsushita K, Ikeda S, Naruse H, Hayashi M, Density Gradient Centrifugation for the Isolation of Cells of Multiple Lineages, Journal of cellular biochemistry 116(12) (2015) 2709–2714. [DOI] [PubMed] [Google Scholar]
- [76].Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK, Bartlett PF, Purification of a pluripotent neural stem cell from the adult mouse brain, Nature 412(6848) (2001) 736–739. [DOI] [PubMed] [Google Scholar]
- [77].Philipp S, Oberg HH, Janssen O, Leippe M, Gelhaus C, Isolation of erythrocytes infected with viable early stages of Plasmodium falciparum by flow cytometry, Cytometry Part A 81(12) (2012) 1048–1054. [DOI] [PubMed] [Google Scholar]
- [78].Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes, Cell stem cell 12(1) (2013) 127–137. [DOI] [PubMed] [Google Scholar]
- [79].Ferreira MM, Ramani VC, Jeffrey SS , Circulating tumor cell technologies, Molecular oncology 10(3) (2016) 374–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, Telekes A, Technologies for circulating tumor cell separation from whole blood, Journal of hematology & oncology 12(1) (2019) 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lu HD, Yang SS, Wilson BK, McManus SA, Chen CV-H, Prud’homme RK, Nanoparticle targeting of Gram-positive and Gram-negative bacteria for magnetic-based separations of bacterial pathogens, Applied Nanoscience 7(3–4) (2017) 83–93. [Google Scholar]
- [82].Nelson NJ, Circulating tumor cells: will they be clinically useful?, Oxford University Press, 2010. [DOI] [PubMed] [Google Scholar]
- [83].Kim J, Massoudi M, Antaki JF, Gandini A, Removal of malaria-infected red blood cells using magnetic cell separators: A computational study, Applied mathematics and computation 218(12) (2012) 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gandini A, Weinstein R, Sawh R.-p., Parks D, Blood purification method and apparatus for the treatment of malaria, Google Patents, 2013. [Google Scholar]
- [85].Nam J, Huang H, Lim H, Lim C, Shin S, Magnetic separation of malaria-infected red blood cells in various developmental stages, Analytical chemistry 85(15) (2013) 7316–7323. [DOI] [PubMed] [Google Scholar]
- [86].Deshpande A, Kalgutkar S, Udani S, Red cell exchange using cell separator (therapeutic erythrocytapheresis) in two children with acute severe malaria, JOURNAL-ASSOCIATION OF PHYSICIANS OF INDIA 51 (2003) 925–926. [PubMed] [Google Scholar]
- [87].Martin AB, Wu W-T, Kameneva MV, Antaki JF, Development of a high-throughput magnetic separation device for malaria-infected erythrocytes, Annals of biomedical engineering 45(12) (2017) 2888–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Stevenson M, Stanwick T, Dempsey M, Lamonica C, HIV-1 replication is controlled at the level of T cell activation and proviral integration, The EMBO journal 9(5) (1990) 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Collins DP, Luebering BJ, Shaut DM, T‐lymphocyte functionality assessed by analysis of cytokine receptor expression, intracellular cytokine expression, and femtomolar detection of cytokine secretion by quantitative flow cytometry, Cytometry Part A 33(2) (1998) 249–255. [DOI] [PubMed] [Google Scholar]
- [90].El-Boubbou K, Gruden C, Huang X, Magnetic glyco-nanoparticles: a unique tool for rapid pathogen detection, decontamination, and strain differentiation, Journal of the American Chemical Society 129(44) (2007) 13392–13393. [DOI] [PubMed] [Google Scholar]
- [91].Cai G, Wang S, Zheng L, Lin J, A fluidic device for immunomagnetic separation of foodborne bacteria using self-assembled magnetic nanoparticle chains, Micromachines 9(12) (2018) 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Matta LL, Alocilja EC, Carbohydrate Ligands on Magnetic Nanoparticles for Centrifuge-Free Extraction of Pathogenic Contaminants in Pasteurized Milk, Journal of food protection 81(12) (2018) 1941–1949. [DOI] [PubMed] [Google Scholar]
- [93].Dainiak M, Kumar A, Galaev I, Mattiasson B, Methods in cell separations, Cell Separation (2007) 1–18. [DOI] [PubMed] [Google Scholar]
- [94].Pretlow TG, Pretlow TP, Cell separation: methods and selected applications, Academic Press; 2014. [Google Scholar]
- [95].Brakke MK, Density gradient centrifugation: a new separation technique1, Journal of the American Chemical Society 73(4) (1951) 1847–1848. [Google Scholar]
- [96].Brewer GJ, Torricelli JR, Isolation and culture of adult neurons and neurospheres, Nature protocols 2(6) (2007) 1490–1498. [DOI] [PubMed] [Google Scholar]
- [97].Sims NR, Anderson MF, Isolation of mitochondria from rat brain using Percoll density gradient centrifugation, Nature protocols 3(7) (2008) 1228–1239. [DOI] [PubMed] [Google Scholar]
- [98].Chang Y, Hsieh P-H, Chao CC, The efficiency of Percoll and Ficoll density gradient media in the isolation of marrow derived human mesenchymal stem cells with osteogenic potential, Chang Gung Med J 32(3) (2009) 264–275. [PubMed] [Google Scholar]
- [99].Fuss IJ, Kanof ME, Smith PD, Zola H, Isolation of whole mononuclear cells from peripheral blood and cord blood, Wiley Online Library; 2009. [DOI] [PubMed] [Google Scholar]
- [100].Zhu B, Murthy SK, Stem cell separation technologies, Current opinion in chemical engineering 2(1) (2013) 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huang Y, Yang J, Wang X-B, Becker FF, Gascoyne PR, The removal of human breast cancer cells from hematopoietic CD34+ stem cells by dielectrophoretic field-flow-fractionation, Journal of hematotherapy & stem cell research 8(5) (1999) 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang X-B, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PR, Cell separation by dielectrophoretic field-flow-fractionation, Analytical Chemistry 72(4) (2000) 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Elitas M, Dhar N, Schneider K, Valero A, Braschler T, McKinney J, Renaud P, Dielectrophoresis as a single cell characterization method for bacteria, Biomedical Physics & Engineering Express 3(1) (2017) 015005. [Google Scholar]
- [104].Fernandez RE, Rohani A, Farmehini V, Swami NS, Microbial analysis in dielectrophoretic microfluidic systems, Analytica chimica acta 966 (2017) 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang J, Song Z, Liu Q, Song Y, Recent Advances in Dielectrophoresis‐Based Cell Viability Assessment, Electrophoresis (2019). [DOI] [PubMed] [Google Scholar]
- [106].Reschiglian P, Zattoni A, Roda B, Michelini E, Roda A, Field-flow fractionation and biotechnology, TRENDS in Biotechnology 23(9) (2005) 475–483. [DOI] [PubMed] [Google Scholar]
- [107].Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PR, Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation, Lab on a Chip 8(8) (2008) 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Roda B, Lanzoni G, Alviano F, Zattoni A, Costa R, Di Carlo A, Marchionni C, Franchina M, Ricci F, Tazzari PL, A novel stem cell tag-less sorting method, Stem Cell Reviews and Reports 5(4) (2009) 420–427. [DOI] [PubMed] [Google Scholar]
- [109].Roda B, Reschiglian P, Alviano F, Lanzoni G, Bagnara GP, Ricci F, Buzzi M, Tazzari PL, Pagliaro P, Michelini E, Gravitational field-flow fractionation of human hemopoietic stem cells, Journal of Chromatography A 1216(52) (2009) 9081–9087. [DOI] [PubMed] [Google Scholar]
- [110].Roda B, Zattoni A, Reschiglian P, Moon MH, Mirasoli M, Michelini E, Roda A, Field-flow fractionation in bioanalysis: a review of recent trends, Analytica chimica acta 635(2) (2009) 132–143. [DOI] [PubMed] [Google Scholar]
- [111].Chianea T, Assidjo N, Cardot P, Sedimentation field-flow-fractionation: emergence of a new cell separation methodology, Talanta 51(5) (2000) 835–847. [DOI] [PubMed] [Google Scholar]
- [112].Choi S, Song S, Choi C, Park J-K, Continuous blood cell separation by hydrophoretic filtration, Lab on a Chip 7(11) (2007) 1532–1538. [DOI] [PubMed] [Google Scholar]
- [113].Chen X, Liu CC, Li H, Microfluidic chip for blood cell separation and collection based on crossflow filtration, Sensors and Actuators B: Chemical 130(1) (2008) 216–221. [Google Scholar]
- [114].Li J, Liu C, Dai X, Chen H, Liang Y, Sun H, Tian H, Ding X, PMMA microfluidic devices with three-dimensional features for blood cell filtration, Journal of Micromechanics and Microengineering 18(9) (2008) 095021. [Google Scholar]
- [115].McFaul SM, Lin BK, Ma H, Cell separation based on size and deformability using microfluidic funnel ratchets, Lab on a chip 12(13) (2012) 2369–2376. [DOI] [PubMed] [Google Scholar]
- [116].Cunha B, Peixoto C, Silva MM, Carrondo MJ, Serra M, Alves PM, Filtration methodologies for the clarification and concentration of human mesenchymal stem cells, Journal of membrane science 478 (2015) 117–129. [Google Scholar]
- [117].Bordag N, Janakiraman V, Nachtigall J, Maldonado SG, Bethan B, Laine J-P, Fux E, Fast filtration of bacterial or mammalian suspension cell cultures for optimal metabolomics results, PloS one 11(7) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ribeiro-Samy S, Oliveira MI, Pereira-Veiga T, Muinelo-Romay L, Carvalho S, Gaspar J, Freitas PP, López-López R, Costa C, Diéguez L, Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients, Scientific reports 9(1) (2019) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Binda E, Erhart D, Schenk M, Zufferey C, Renzulli P, Mueller C, Quantitative isolation of mouse and human intestinal intraepithelial lymphocytes by elutriation centrifugation, Journal of immunological methods 344(1) (2009) 26–34. [DOI] [PubMed] [Google Scholar]
- [120].Grosse J, Meier K, Bauer TJ, Eilles C, Grimm D, Cell separation by countercurrent centrifugal elutriation: recent developments, Preparative Biochemistry and Biotechnology 42(3) (2012) 217–233. [DOI] [PubMed] [Google Scholar]
- [121].Stroncek DF, Fellowes V, Pham C, Khuu H, Fowler DH, Wood LV, Sabatino M, Counter-flow elutriation of clinical peripheral blood mononuclear cell concentrates for the production of dendritic and T cell therapies, Journal of translational medicine 12(1) (2014) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bauer J, Advances in cell separation: recent developments in counterflow centrifugal elutriation and continuous flow cell separation, Journal of Chromatography B: Biomedical Sciences and Applications 722(1–2) (1999) 55–69. [DOI] [PubMed] [Google Scholar]
- [123].Lasch J, Küllertz G, Opalka JR, Separation of erythrocytes into age-related fractions by density or size? Counterflow centrifugation, Clinical chemistry and laboratory medicine 38(7) (2000) 629–632. [DOI] [PubMed] [Google Scholar]
- [124].Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W, Amini H, Di Carlo D, Label-free cell separation and sorting in microfluidic systems, Analytical and bioanalytical chemistry 397(8) (2010) 3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yeo T, Tan SJ, Lim CL, Lau DPX, Chua YW, Krisna SS, Iyer G, San Tan G, Lim TKH, Tan DS, Microfluidic enrichment for the single cell analysis of circulating tumor cells, Scientific reports 6 (2016) 22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Pösel C, Möller K, Fröhlich W, Schulz I, Boltze J, Wagner D-C, Density gradient centrifugation compromises bone marrow mononuclear cell yield, PloS one 7(12) (2012) e50293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang K, Marshall MK, Garza G, Pappas D, Open-tubular capillary cell affinity chromatography: single and tandem blood cell separation, Analytical chemistry 80(6) (2008) 2118–2124. [DOI] [PubMed] [Google Scholar]
- [128].Kumar A, Srivastava A, Cell separation using cryogel-based affinity chromatography, Nature protocols 5(11) (2010) 1737–1747. [DOI] [PubMed] [Google Scholar]
- [129].Li P, Gao Y, Pappas D, Multiparameter cell affinity chromatography: separation and analysis in a single microfluidic channel, Analytical chemistry 84(19) (2012) 8140–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ, Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes, Methods 56(2) (2012) 293–304. [DOI] [PubMed] [Google Scholar]
- [131].Gao Y, Li W, Pappas D, Recent advances in microfluidic cell separations, Analyst 138(17) (2013) 4714–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Labrou N, Clonis Y, The affinity technology in downstream processing, Journal of biotechnology 36(2) (1994) 95–119. [DOI] [PubMed] [Google Scholar]
- [133].Menegatti S, Naik AD, Carbonell RG, The hidden potential of small synthetic molecules and peptides as affinity ligands for bioseparations, Pharmaceutical Bioprocessing 1(5) (2013) 467–485. [Google Scholar]
- [134].Rodrigues G, Fernandes T, Rodrigues C, Diogo M, Cabral J, Enrichment and Separation Technologies for Stem Cell-Based Therapies, Stem Cell Manufacturing (2016) 199. [Google Scholar]
- [135].Hoeve MA, De Sousa PA, Willoughby NA, Challenges of Scale-up of Cell Separation and Purification Techniques, Bioprocessing for Cell-Based Therapies (2017) 127. [Google Scholar]
- [136].Rahmanian N, Bozorgmehr M, Torabi M, Akbari A, Zarnani A-H, Cell separation: Potentials and pitfalls, Preparative Biochemistry and Biotechnology 47(1) (2017) 38–51. [DOI] [PubMed] [Google Scholar]
- [137].Wu J, Chen Q, Lin J-M, Microfluidic technologies in cell isolation and analysis for biomedical applications, Analyst 142(3) (2017) 421–441. [DOI] [PubMed] [Google Scholar]
- [138].Hubble J, Affinity cell separations: problems and prospects, Trends in Biotechnology 15(7) (1997) 249–255. [Google Scholar]
- [139].Cao X, Eisenthal R, Hubble J, Detachment strategies for affinity-adsorbed cells, Enzyme and microbial technology 31(1) (2002) 153–160. [Google Scholar]
- [140].Kumar A, Galaev IY, Mattiasson B, Cell Separation: Fundamentals, Analytical and Preparative Methods, Springer Berlin Heidelberg; 2010. [Google Scholar]
- [141].Rawal S, Yang Y-P, Cote R, Agarwal A, Identification and Quantitation of Circulating Tumor Cells, Annual Review of Analytical Chemistry (0) (2017). [DOI] [PubMed] [Google Scholar]
- [142].Chalmers JJ, Zborowski M, Moore L, Mandal S, Fang B, Sun L, Theoretical analysis of cell separation based on cell surface marker density, Biotechnology and bioengineering 59(1) (1998) 10–20. [PubMed] [Google Scholar]
- [143].Mahara A, Yamaoka T, Antibody‐immobilized column for quick cell separation based on cell rolling, Biotechnology progress 26(2) (2010) 441–447. [DOI] [PubMed] [Google Scholar]
- [144].Lakschevitz FS, Hassanpour S, Rubin A, Fine N, Sun C, Glogauer M, Identification of neutrophil surface marker changes in health and inflammation using high-throughput screening flow cytometry, Experimental cell research 342(2) (2016) 200–209. [DOI] [PubMed] [Google Scholar]
- [145].Yamaoka T, Mahara A, Cell rolling column in purification and differentiation analysis of stem cells, Reactive and Functional Polymers 71(3) (2011) 362–366. [Google Scholar]
- [146].Cahall CF, Lilly JL, Hirschowitz EA, Berron BJ, A Quantitative Perspective on Surface Marker Selection for the Isolation of Functional Tumor Cells, Breast cancer: basic and clinical research 9(Suppl 1) (2015) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Mavrangelos C, Swart B, Nobbs S, Nicholson IC, Macardle PJ, Zola H, Detection of low-abundance membrane markers by immunofluorescence—a comparison of alternative high-sensitivity methods and reagents, Journal of immunological methods 289(1) (2004) 169–178. [DOI] [PubMed] [Google Scholar]
- [148].Mattanovich D, Borth N, Applications of cell sorting in biotechnology, Microbial cell factories 5(1) (2006) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Will B, Steidl U, Multi-parameter fluorescence-activated cell sorting and analysis of stem and progenitor cells in myeloid malignancies, Best Practice & Research Clinical Haematology 23(3) (2010) 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tomlinson MJ, Tomlinson S, Yang XB, Kirkham J, Cell separation: Terminology and practical considerations, Journal of tissue engineering 4 (2013) 2041731412472690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Raj A, van Oudenaarden A, Nature, nurture, or chance: stochastic gene expression and its consequences, Cell 135(2) (2008) 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Cahan P, Daley GQ, Origins and implications of pluripotent stem cell variability and heterogeneity, Nature reviews Molecular cell biology 14(6) (2013) 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oleś AK, Araúzo-Bravo MJ, Saitou M, Hadjantonakis A-K, Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages, Nature cell biology 16(1) (2014) 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Hong S-H, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M, Cell fate potential of human pluripotent stem cells is encoded by histone modifications, Cell stem cell 9(1) (2011) 24–36. [DOI] [PubMed] [Google Scholar]
- [155].Lee TY, Hyun K-A, Kim S-I, Jung H-I, An integrated microfluidic chip for one-step isolation of circulating tumor cells, Sensors and Actuators B: Chemical 238 (2017) 1144–1150. [Google Scholar]
- [156].Chen C-L, Chen K-C, Pan Y-C, Lee T-P, Hsiung L-C, Lin C-M, Chen C-Y, Lin C-H, Chiang B-L, Wo AM, Separation and detection of rare cells in a microfluidic disk via negative selection, Lab on a Chip 11(3) (2011) 474–483. [DOI] [PubMed] [Google Scholar]
- [157].Hyun K-A, Jung H-I, Advances and critical concerns with the microfluidic enrichments of circulating tumor cells, Lab on a Chip 14(1) (2014) 45–56. [DOI] [PubMed] [Google Scholar]
- [158].Bu J, Kang Y-T, Kim YJ, Cho Y-H, Chang HJ, Kim H, Moon B-I, Kim HG, Dual-patterned immunofiltration (DIF) device for the rapid efficient negative selection of heterogeneous circulating tumor cells, Lab on a Chip 16(24) (2016) 4759–4769. [DOI] [PubMed] [Google Scholar]
- [159].Li P, Gao Y, Pappas D, Negative enrichment of target cells by microfluidic affinity chromatography, Analytical chemistry 83(20) (2011) 7863–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sajay BNG, Chang C-P, Ahmad H, Khuntontong P, Wong CC, Wang Z, Puiu PD, Soo R, Rahman ARA, Microfluidic platform for negative enrichment of circulating tumor cells, Biomed. Microdevices 16(4) (2014) 537–548. [DOI] [PubMed] [Google Scholar]
- [161].Masson K, Heiss E, Band H, Rönnstrand L, Direct binding of Cbl to Tyr568 and Tyr936 of the stem cell factor receptor/c-Kit is required for ligand-induced ubiquitination, internalization and degradation, Biochemical journal 399(1) (2006) 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Krishnaswamy G, Ajitawi O, Chi DS, The human mast cell: an overview, Mast Cells: Methods and Protocols (2005) 13–34. [DOI] [PubMed] [Google Scholar]
- [163].Liu Y, Tang X, Liu F, K.a. Li, Selection of ligands for affinity chromatography using quartz crystal biosensor, Analytical chemistry 77(13) (2005) 4248–4256. [DOI] [PubMed] [Google Scholar]
- [164].Drabovich A, Selection of Affinity Ligands Using Kinetic Capillary Electrophoresis, York University; 2008. [Google Scholar]
- [165].Bergsdorf C, Ottl J, Affinity-based screening techniques: their impact and benefit to increase the number of high quality leads, Expert opinion on drug discovery 5(11) (2010) 1095–1107. [DOI] [PubMed] [Google Scholar]
- [166].Wei Y, Wesson PJ, Kourkine I, Grzybowski BA, Measurement of Protein− Ligand Binding Constants from Reaction-Diffusion Concentration Profiles, Analytical chemistry 82(21) (2010) 8780–8784. [DOI] [PubMed] [Google Scholar]
- [167].Mammen M, Choi S-K, Whitesides GM, Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors, Angewandte Chemie International Edition 37(20) (1998) 2754–2794. [DOI] [PubMed] [Google Scholar]
- [168].Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ, Designing a polyvalent inhibitor of anthrax toxin, Nature biotechnology 19(10) (2001) 958–961. [DOI] [PubMed] [Google Scholar]
- [169].Adler P, Wood SJ, Lee YC, Lee RT, Petri WA, Schnaar RL, High affinity binding of the Entamoeba histolytica lectin to polyvalent N-acetylgalactosaminides, Journal of biological chemistry 270(10) (1995) 5164–5171. [DOI] [PubMed] [Google Scholar]
- [170].Brown G, Greaves MF, Cell surface markers for human T and B lymphocytes, European journal of immunology 4(4) (1974) 302–310. [DOI] [PubMed] [Google Scholar]
- [171].Sharma SK, Mahendroo P, Affinity chromatography of cells and cell membranes, Journal of Chromatography A 184(4) (1980) 471–499. [DOI] [PubMed] [Google Scholar]
- [172].Basch RS, Berman JW, Lakow E, Cell separation using positive immunoselective techniques, Journal of immunological methods 56(3) (1983) 269–280. [DOI] [PubMed] [Google Scholar]
- [173].Tadikonda KR, Davis RH, Cell separations using targeted monoclonal antibodies against overproduced surface proteins, Applied biochemistry and biotechnology 45(1) (1994) 233–244. [DOI] [PubMed] [Google Scholar]
- [174].Gomez SM, Choy G, Kabir N, Leonard EF, Capture of Rare Cells in Suspension with Antibody‐Coated Polystyrene Beads, Biotechnology progress 15(2) (1999) 238–244. [DOI] [PubMed] [Google Scholar]
- [175].Sin A, Murthy SK, Revzin A, Tompkins RG, Toner M, Enrichment using antibody‐coated microfluidic chambers in shear flow: Model mixtures of human lymphocytes, Biotechnology and bioengineering 91(7) (2005) 816–826. [DOI] [PubMed] [Google Scholar]
- [176].Evans WH, Mage MG, Peterson EA, A Method for the Immunoadsorption of Cells to an Antibodycoated Polyurethane Foam, The Journal of Immunology 102(4) (1969) 899–907. [PubMed] [Google Scholar]
- [177].Wigzell H, Sundqvist K, Yoshida TO, Separation of Cells According to Surface Antigens by the Use of Antibody‐Coated Columns. Fractionation of Cells Carrying Immunoglobulins and Blood Group Antigen, Scandinavian journal of immunology 1(1) (1972) 75–87. [DOI] [PubMed] [Google Scholar]
- [178].Binz H, Wigzell H, Shared idiotypic determinants on B and T lymphocytes reactive against the same antigenic determinants. III. Physical fractionation of specific immunocompetent T lymphocytes by affinity chromatography using anti-idiotypic antibodies, Journal of Experimental Medicine 142(5) (1975) 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Brewster JD, Isolation and concentration of Salmonellae with an immunoaffinity column, Journal of microbiological methods 55(1) (2003) 287–293. [DOI] [PubMed] [Google Scholar]
- [180].Berenson RJ, Levitt LJ, Levy R, Miller RA, CELLULAR IMMUNOABSORPTION USING MONOCLONAL ANTIBODIES: ELECTIVE REMOVAL OF T CELLS FROM PERIPHERAL BLOOD AND BONE MARROW, Transplantation 38(2) (1984) 136–142. [PubMed] [Google Scholar]
- [181].Hubbard RA, Schluter SF, Marchalonis JJ, [14] Separation of lymphoid cells using immunoadsorbent affinity chromatography, Methods in enzymology 108 (1984) 139–148. [DOI] [PubMed] [Google Scholar]
- [182].Uchiyama T, Hori T, Tsudo M, Wano Y, Umadome H, Tamori S, Yodoi J, Maeda M, Sawami H, Uchino H, Interleukin-2 receptor (Tac antigen) expressed on adult T cell leukemia cells, Journal of Clinical Investigation 76(2) (1985) 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Braun RW, Kümel G, [70] Separation of T cell subpopulations by monoclonal antibodies and affinity chromatography, Methods in enzymology 121 (1986) 737–748. [DOI] [PubMed] [Google Scholar]
- [184].Kataoka K, Sakurai Y, Hanai T, Maruyama A, Tsuruta T, Immunoaffinity chromatography of lymphocyte subpopulations using tert-amine derived matrices with adsorbed antibodies, Biomaterials 9(3) (1988) 218–224. [DOI] [PubMed] [Google Scholar]
- [185].Thomas TE, Lansdorp PM, Immunoadsorption of T cells onto glass beads using tetramolecular complexes of monoclonal antibodies, Journal of immunological methods 112(2) (1988) 219–226. [DOI] [PubMed] [Google Scholar]
- [186].Schulman ES, MAcGLASHAN DW, Peters SP, Schleimer R, Newball H, Lichtenstein L, Human lung mast cells: purification and characterization, The Journal of Immunology 129(6) (1982) 2662–2667. [PubMed] [Google Scholar]
- [187].Van Overveld F, Bruijnzeel P, Kreukniet J, Raaijmakers J, Terpstra G, The isolation of inflammatory cells from normal human lung tissue, International journal of tissue reactions 9(1) (1986) 61–67. [PubMed] [Google Scholar]
- [188].Au AM-J, Varon S, Neural cell sequestration on immunoaffinity columns, Experimental cell research 120(2) (1979) 269–276. [DOI] [PubMed] [Google Scholar]
- [189].Chatterjee D, Mandal C, Sarkar P, Development and characterization of five monoclonal antibodies against neuronal cell surface antigens—Evaluation of their use in cell separation by affinity chromatography, Journal of neuroimmunology 15(3) (1987) 251–262. [DOI] [PubMed] [Google Scholar]
- [190].Körbling M, Drach J, Champlin R, Engel H, Huynh L, Kleine H, Berenson R, Deisseroth A, Andreeff M, Large-scale preparation of highly purified, frozen/thawed CD34+, HLA-DR-hematopoietic progenitor cells by sequential immunoadsorption (CEPRATE SC) and fluorescence-activated cell sorting: implications for gene transduction and/or transplantation, Bone marrow transplantation 13(5) (1994) 649–654. [PubMed] [Google Scholar]
- [191].Wognum AW, Eaves AC, Thomas TE, Identification and isolation of hematopoietic stem cells, Archives of medical research 34(6) (2003) 461–475. [DOI] [PubMed] [Google Scholar]
- [192].Mahara A, Yamaoka T, Continuous separation of cells of high osteoblastic differentiation potential from mesenchymal stem cells on an antibody-immobilized column, Biomaterials 31(14) (2010) 4231–4237. [DOI] [PubMed] [Google Scholar]
- [193].Manderino GL, Gooch GT, Stavitsky AB, Preparation, characterization, and functions of rabbit lymph node cell populations: I. Preparation of KLH primed T and B memory cells with anti-Fab′ affinity columns, Cellular immunology 41(2) (1978) 264–275. [DOI] [PubMed] [Google Scholar]
- [194].Romain PL, Schlossman SF, [15] Use of anti-fab columns for the isolation of human lymphocyte populations, Methods in enzymology 108 (1984) 148–153. [DOI] [PubMed] [Google Scholar]
- [195].Braun R, Teute H, Kirchner H, Munk K, Rapid separation of T cell subpopulations with monoclonal antibodies and affinity chromatography, Journal of immunological methods 54(2) (1982) 251–258. [DOI] [PubMed] [Google Scholar]
- [196].Firat H, Giarratana M, Kobari L, Poloni A, Bouchet S, Labopin M, Gorin N, Douay L, Comparison of CD34+ bone marrow cells purified by immunomagnetic and immunoadsorption cell separation techniques, Bone marrow transplantation 21(9) (1998) 933–938. [DOI] [PubMed] [Google Scholar]
- [197].Thomas D, Phillips B, The separation of human B lymphocytes on a digestible immunoadsorbent column, European journal of immunology 3(11) (1973) 740–742. [DOI] [PubMed] [Google Scholar]
- [198].Thomas TE, Sutherland HJ, Lansdorp PM, Specific binding and release of cells from beads using cleavable tetrameric antibody complexes, Journal of immunological methods 120(2) (1989) 221–231. [DOI] [PubMed] [Google Scholar]
- [199].Zettlitz KA, Protein A/G Chromatography, Antibody Engineering (2010) 531–535. [Google Scholar]
- [200].Nash A, Separation of lymphocyte sub-populations using antibodies attached to staphylococcal protein A-coated surfaces, Journal of immunological methods 12(1–2) (1976) 149–161. [DOI] [PubMed] [Google Scholar]
- [201].Ghetie V, Mota G, Sjöquist J, Separation of cells by affinity chromatography on SpA-Sepharose 6MB, Journal of immunological methods 21(1–2) (1978) 133–141. [DOI] [PubMed] [Google Scholar]
- [202].Kretser T, Bodmer J, Bodmer W, The separation of cell populations using monoclonal antibodies attached to sepharose, HLA 16(4) (1980) 317–325. [DOI] [PubMed] [Google Scholar]
- [203].Ghetie V, Sjöquist J, [13] Separation of cells by affinity chromatography on protein A gels, Methods in enzymology 108 (1984) 132–138. [DOI] [PubMed] [Google Scholar]
- [204].Kumar A, Plieva FM, Galaev IY, Mattiasson B, Affinity fractionation of lymphocytes using a monolithic cryogel, Journal of immunological methods 283(1) (2003) 185–194. [DOI] [PubMed] [Google Scholar]
- [205].Kumar A, Rodríguez‐Caballero A, Plieva FM, Galaev IY, Nandakumar KS, Kamihira M, Holmdahl R, Orfao A, Mattiasson B, Affinity binding of cells to cryogel adsorbents with immobilized specific ligands: effect of ligand coupling and matrix architecture, Journal of Molecular Recognition 18(1) (2005) 84–93. [DOI] [PubMed] [Google Scholar]
- [206].Ghetie V, Stålenheim G, Sjöquist J, Cell Separation by Staphylococcal Protein A‐ Coated Erythrocytes, Scandinavian journal of immunology 4(5) (1975) 471–477. [DOI] [PubMed] [Google Scholar]
- [207].Levitt D, Danen R, Separation of lymphocyte subpopulations using biotin-avidin erythrocyte rosettes, Journal of immunological methods 89(2) (1986) 207–211. [DOI] [PubMed] [Google Scholar]
- [208].Duffey PS, Drouillard DL, Barbe CP, Lymphocyte sorting on albuminated CIBA blue dextran-staphylococcal protein A-conjugated sepharose 6MB affinity columns, Journal of immunological methods 45(2) (1981) 137–151. [DOI] [PubMed] [Google Scholar]
- [209].Wigzell H, Huber C, Schirrmacher V, Affinity fractionation of lymphoid cells according to type and function, Haematologia 6(3) (1972) 369–375. [PubMed] [Google Scholar]
- [210].DeLuca D, Yowell RL, Miller A, The specific separation of enzyme antigen-binding cells by substrate affinity chromatography, Microenvironmental Aspects of Immunity, Springer; 1973, pp. 693–698. [DOI] [PubMed] [Google Scholar]
- [211].MaClennan I, Gray D, Antigen‐driven selection of virgin and memory B cells, Immunological reviews 91(1) (1986) 61–86. [DOI] [PubMed] [Google Scholar]
- [212].Wigzell H, Andersson B, Cell separation on antigen-coated columns, Journal of Experimental Medicine 129(1) (1969) 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Carvajal RE, Porras FM, Garcia-Carreño F, Separation and isolation of lymphocyte subpopulations of different affinity for the antigen, Immunology letters 8(2) (1984) 101–106. [DOI] [PubMed] [Google Scholar]
- [214].Truffa-Bachi P, Wofsy L, Specific separation of cells on affinity columns, Proceedings of the National Academy of Sciences 66(3) (1970) 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Wofsy L, Kimura J, Truffa-Bachi P, Cell separation on affinity columns: the preparation of pure populations of anti-hapten specific lymphocytes, The Journal of Immunology 107(3) (1971) 725–729. [PubMed] [Google Scholar]
- [216].Henry C, Kimura J, Wofsy L, Cell separation on affinity columns: the isolation of immunospecific precursor cells from unimmunized mice, Proceedings of the National Academy of Sciences 69(1) (1972) 34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Haas W, Schrader J, Szenberg A, A new, simple method for the preparation of lymphocytes bearing specific receptors, European journal of immunology 4(8) (1974) 565–570. [DOI] [PubMed] [Google Scholar]
- [218].Kataoka K, Sakurai Y, Tsuruta T, Affinity Selection of Cells on Solid-Phase Matrices with Immobilized Proteins, ACS Publications; 1987. [Google Scholar]
- [219].Hertz C, Graves D, Lauffenburger D, Serota F, Use of cell affinity chromatography for separation of lymphocyte subpopulations, Biotechnology and bioengineering 27(5) (1985) 603–612. [DOI] [PubMed] [Google Scholar]
- [220].Hellström U, Hammarström M-L, Hammarström S, Perlmann P, [16] Fractionation of human lymphocytes on Helix pomatia a hemagglutinin-sepharose and wheat germ agglutinin-sepharose, Methods in enzymology 108 (1984) 153–168. [DOI] [PubMed] [Google Scholar]
- [221].Putnam DD, Namasivayam V, Burns MA, Cell affinity separations using magnetically stabilized fluidized beds: Erythrocyte subpopulation fractionation utilizing a lectin‐magnetite support, Biotechnology and bioengineering 81(6) (2003) 650–665. [DOI] [PubMed] [Google Scholar]
- [222].Whitehurst C, Day N, Gengozian N, A method of purifying feline T lymphocytes from peripheral blood using the plant lectin from Pisum sativum, Journal of immunological methods 175(2) (1994) 189–199. [DOI] [PubMed] [Google Scholar]
- [223].Bonnafous J-C, Dornand J, Favero J, Boschetti E, Mani J-C, Cell affinity chromatography with ligands immobilized through cleavable mercury-sulfur bonds, Journal of immunological methods 58(1–2) (1983) 93–107. [DOI] [PubMed] [Google Scholar]
- [224].Walter H, Partitioning in aqueous two–phase system: theory, methods, uses, and applications to biotechnology, Elsevier; 2012. [Google Scholar]
- [225].Thiel A, Scheffold A, Radbruch A, Immunomagnetic cell sorting—pushing the limits, Immunotechnology 4(2) (1998) 89–96. [DOI] [PubMed] [Google Scholar]
- [226].Basu S, Campbell HM, Dittel BN, Ray A, Purification of specific cell population by fluorescence activated cell sorting (FACS), Journal of visualized experiments: JoVE (41) (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Inaba K, Swiggard WJ, Steinman RM, Romani N, Schuler G, Brinster C, Isolation of dendritic cells, Current protocols in immunology (2009) 3.7.1–3.7.19. [DOI] [PubMed] [Google Scholar]
- [228].Kanof ME, Isolation of T cells using rosetting procedures, Current protocols in immunology 112(1) (2016) 7.2. 1–7.2. 5. [DOI] [PubMed] [Google Scholar]
- [229].Tondreau T, Lagneaux L, Dejeneffe M, Delforge A, Massy M, Mortier C, Bron D, Isolation of BM mesenchymal stem cells by plastic adhesion or negative selection: phenotype, proliferation kinetics and differentiation potential, Cytotherapy 6(4) (2004) 372–379. [DOI] [PubMed] [Google Scholar]
- [230].Reinherz EL, Kung PC, Goldstein G, Schlossman SF, Separation of functional subsets of human T cells by a monoclonal antibody, Proceedings of the National Academy of Sciences 76(8) (1979) 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Brody T, Identification of two cell populations required for mouse immunocompetence, The Journal of Immunology 105(1) (1970) 126–138. [PubMed] [Google Scholar]
- [232].Owen CS, Moore E, High gradient magnetic separation of rosette-forming cells, Cell Biochemistry and Biophysics 3(2) (1981) 141–153. [DOI] [PubMed] [Google Scholar]
- [233].Myers CD, Sanders VM, Vitetta ES, Isolation of antigen-binding virgin and memory B cells, Journal of immunological methods 92(1) (1986) 45–57. [DOI] [PubMed] [Google Scholar]
- [234].Dainiak MB, Galaev IY, Kumar A, Plieva FM, Mattiasson B, Chromatography of living cells using supermacroporous hydrogels, cryogels, Cell Separation, Springer; 2007, pp. 101–127. [DOI] [PubMed] [Google Scholar]
- [235].Mohr F, Przibilla S, Leonhardt F, Stemberger C, Dreher S, Müller TR, Fräßle SP, Schmidt GP, Kiene M-L, Stadler H, Efficient immunoaffinity chromatography of lymphocytes directly from whole blood, Scientific reports 8(1) (2018) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Wilchek M, Chaiken I , An overview of affinity chromatography, Affinity Chromatography: Methods and Protocols (2000) 1–6. [DOI] [PubMed] [Google Scholar]
- [237].Ayyar BV, Arora S, Murphy C, O’Kennedy R, Affinity chromatography as a tool for antibody purification, Methods 56(2) (2012) 116–129. [DOI] [PubMed] [Google Scholar]
- [238].Hammer DA, Lauffenburger DA, A dynamical model for receptor-mediated cell adhesion to surfaces, Biophysical journal 52(3) (1987) 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Hammer DA, Lauffenburger DA, A dynamical model for receptor-mediated cell adhesion to surfaces in viscous shear flow, Cell Biochemistry and Biophysics 14(2) (1989) 139–173. [DOI] [PubMed] [Google Scholar]
- [240].Korn C, Schwarz U, Efficiency of initiating cell adhesion in hydrodynamic flow, Physical review letters 97(13) (2006) 138103. [DOI] [PubMed] [Google Scholar]
- [241].Vauquelin G, Charlton SJ, Exploring avidity: understanding the potential gains in functional affinity and target residence time of bivalent and heterobivalent ligands, British journal of pharmacology 168(8) (2013) 1771–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Vauquelin G, Bricca G, Van Liefde I, Avidity and positive allosteric modulation/cooperativity act hand in hand to increase the residence time of bivalent receptor ligands, Fundamental & clinical pharmacology 28(5) (2014) 530–543. [DOI] [PubMed] [Google Scholar]
- [243].Nauerth M, Wing K, Korner H, Busch D, Relevance of the T cell Receptor-Ligand Avidity for Immunity to Infection, J Microb Biochem Technol 8 (2016) 131–135. [Google Scholar]
- [244].Saterbak A, Kuo SC, Lauffenburger DA, Heterogeneity and probabilistic binding contributions to receptor-mediated cell detachment kinetics, Biophysical journal 65(1) (1993) 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Hubble J, Eisenthal R, Whish WJ, A model for the initial phase of cell/surface interactions based on ligand binding phenomena, Biochemical Journal 311(3) (1995) 917–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Hubble J, Ming F, Eisenthal R, Whish W, Progressive detachment of cells from surfaces: A consequence of heterogeneous ligand populations or a multi-site binding equilibrium?, Journal of theoretical biology 182(2) (1996) 169–171. [DOI] [PubMed] [Google Scholar]
- [247].Ming F, Whish WJ, Hubble J, Eisenthal R, Estimation of parameters for cell-surface interactions: maximum binding force and detachment constant, Enzyme and microbial technology 22(2) (1998) 94–99. [Google Scholar]
- [248].Ming F, Eisenthal R, Whish WJ, Hubble J, The kinetics of affinity-mediated cell-surface attachment, Enzyme and microbial technology 26(2) (2000) 216–221. [DOI] [PubMed] [Google Scholar]
- [249].Tees DF, Chang K-C, Rodgers SD, Hammer DA, Simulation of cell adhesion to bioreactive surfaces in shear: The effect of cell size, Industrial & engineering chemistry research 41(3) (2002) 486–493. [Google Scholar]
- [250].Caputo KE, Lee D, King MR, Hammer DA, Adhesive dynamics simulations of the shear threshold effect for leukocytes, Biophysical journal 92(3) (2007) 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Hammer DA, Linderman JJ, Graves DJ, Lauffenburger DA, Affinity chromatography for cell separation: mathematical model and experimental analysis, Biotechnology progress 3(3) (1987) 189–204. [Google Scholar]
- [252].Hammer DA, Lauffenburger DA, Effects of nonspecific cell/surface interactions on cell affinity chromatographic separations, MRS Online Proceedings Library Archive 110 (1987). [Google Scholar]
- [253].Dembo M, On peeling an adherent cell from a surface, Lectures on Mathematics in the life sciences 24 (1994) 51–77. [Google Scholar]
- [254].Orsello CE, Lauffenburger DA, Hammer DA, Molecular properties in cell adhesion: a physical and engineering perspective, TRENDS in Biotechnology 19(8) (2001) 310–316. [DOI] [PubMed] [Google Scholar]
- [255].Hubble J, Monte Carlo simulation of biospecific interactions between cells and surfaces, Chemical engineering science 58(19) (2003) 4465–4474. [Google Scholar]
- [256].N’dri N, Shyy W, Tran-Son-Tay R, Computational modeling of cell adhesion and movement using a continuum-kinetics approach, Biophysical journal 85(4) (2003) 2273–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Hubble J, Affinity adsorption of cells to surfaces and strategies for cell detachment, Cell Separation (2007) 75–99. [DOI] [PubMed] [Google Scholar]
- [258].Lam A, Cao X, Eisenthal R, Hubble J, Effect of contact time and inhibitor concentration on the affinity mediated adsorption of cells to surfaces, Enzyme and microbial technology 29(1) (2001) 28–33. [DOI] [PubMed] [Google Scholar]
- [259].Khismatullin DB, Truskey GA, A 3D numerical study of the effect of channel height on leukocyte deformation and adhesion in parallel-plate flow chambers, Microvascular research 68(3) (2004) 188–202. [DOI] [PubMed] [Google Scholar]
- [260].Khismatullin DB, Truskey GA, Three-dimensional numerical simulation of receptor-mediated leukocyte adhesion to surfaces: Effects of cell deformability and viscoelasticity, Physics of Fluids 17(3) (2005) 031505. [Google Scholar]
- [261].Murthy SK, Sin A, Tompkins RG, Toner M, Effect of flow and surface conditions on human lymphocyte isolation using microfluidic chambers, Langmuir 20(26) (2004) 11649–11655. [DOI] [PubMed] [Google Scholar]
- [262].Chang WC, Lee LP, Liepmann D, Biomimetic technique for adhesion-based collection and separation of cells in a microfluidic channel, Lab on a Chip 5(1) (2005) 64–73. [DOI] [PubMed] [Google Scholar]
- [263].Greenberg AW, Hammer DA, Cell separation mediated by differential rolling adhesion, Biotechnology and bioengineering 73(2) (2001) 111–124. [DOI] [PubMed] [Google Scholar]
- [264].Karnik R, Hong S, Zhang H, Mei Y, Anderson DG, Karp JM, Langer R, Nanomechanical control of cell rolling in two dimensions through surface patterning of receptors, Nano letters 8(4) (2008) 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Hammer DA, Apte SM, Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion, Biophysical journal 63(1) (1992) 35–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Chang K-C, Tees DF, Hammer DA, The state diagram for cell adhesion under flow: leukocyte rolling and firm adhesion, Proceedings of the National Academy of Sciences 97(21) (2000) 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Dong C, Lei XX, Biomechanics of cell rolling: shear flow, cell-surface adhesion, and cell deformability, Journal of biomechanics 33(1) (2000) 35–43. [DOI] [PubMed] [Google Scholar]
- [268].King MR, Hammer DA, Multiparticle adhesive dynamics. Interactions between stably rolling cells, Biophysical journal 81(2) (2001) 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Mattiasson B, Expanded bed chromatography, Springer Science & Business Media; 2013. [Google Scholar]
- [270].Ujam L, Clemmitt R, Chase H, Cell separation by expanded bed adsorption: use of ion exchange chromatography for the separation of E. coli and S. cerevisiae, Bioprocess and Biosystems Engineering 23(3) (2000) 245–250. [Google Scholar]
- [271].Clemmitt R, Chase H, Impact of operating variables on the expanded bed adsorption of Saccharomyces cerevisiae cells using a concanavalin A derivatized perfluorocarbon, Biotechnology and bioengineering 82(5) (2003) 506–516. [DOI] [PubMed] [Google Scholar]
- [272].Ujam L, Clemmitt R, Clarke S, Brooks R, Rushton N, Chase H, Isolation of monocytes from human peripheral blood using immuno‐affinity expanded‐bed adsorption, Biotechnology and bioengineering 83(5) (2003) 554–566. [DOI] [PubMed] [Google Scholar]
- [273].McCreath GE, Chase HA, Affinity adsorption of Saccharomyces cerevisiae on concanavalin a perflurocarbon emulsions, Journal of Molecular Recognition 9(5‐6) (1996) 607–616. [DOI] [PubMed] [Google Scholar]
- [274].Lin DQ, Kula MR, Liten A, Thömmes J, Stability of expanded beds during the application of crude feedstock, Biotechnology and bioengineering 81(1) (2003) 21–26. [DOI] [PubMed] [Google Scholar]
- [275].Plieva FM, Galaev IY, Noppe W, Mattiasson B, Cryogel applications in microbiology, Trends in microbiology 16(11) (2008) 543–551. [DOI] [PubMed] [Google Scholar]
- [276].Dainiak MB, Kumar A, Galaev IY, Mattiasson B, Cryogels as matrices for cell separation and cell cultivation, Macroporous Polymers: Production Properties and Biotechnological/Biomedical Applications (2009) 363. [Google Scholar]
- [277].Singh NK, Dsouza RN, Grasselli M, Fernández-Lahore M, High capacity cryogel-type adsorbents for protein purification, Journal of Chromatography A 1355 (2014) 143–148. [DOI] [PubMed] [Google Scholar]
- [278].Emin Çorman M, Bereli N, Özkara S, Uzun L, Denizli A, Hydrophobic cryogels for DNA adsorption: Effect of embedding of monosize microbeads into cryogel network on their adsorptive performances, Biomedical Chromatography 27(11) (2013) 1524–1531. [DOI] [PubMed] [Google Scholar]
- [279].Çimen D, Yılmaz F, Perçin I, Türkmen D, Denizli A, Dye affinity cryogels for plasmid DNA purification, Materials Science and Engineering: C 56 (2015) 318–324. [DOI] [PubMed] [Google Scholar]
- [280].Williams SL, Eccleston ME, Slater NK, Affinity capture of a biotinylated retrovirus on macroporous monolithic adsorbents: Towards a rapid single‐step purification process, Biotechnology and bioengineering 89(7) (2005) 783–787. [DOI] [PubMed] [Google Scholar]
- [281].Lozinsky VI, Galaev IY, Plieva FM, Savina IN, Jungvid H, Mattiasson B, Polymeric cryogels as promising materials of biotechnological interest, TRENDS in Biotechnology 21(10) (2003) 445–451. [DOI] [PubMed] [Google Scholar]
- [282].Jungbauer A, Hahn R, Polymethacrylate monoliths for preparative and industrial separation of biomolecular assemblies, Journal of chromatography A 1184(1) (2008) 62–79. [DOI] [PubMed] [Google Scholar]
- [283].Arrua RD, Alvarez Igarzabal CI, Macroporous monolithic supports for affinity chromatography, J. Sep. Sci 34(16‐17) (2011) 1974–1987. [DOI] [PubMed] [Google Scholar]
- [284].Jain E, Kumar A, Disposable polymeric cryogel bioreactor matrix for therapeutic protein production, Nature protocols 8(5) (2013) 821–835. [DOI] [PubMed] [Google Scholar]
- [285].Kumar A, Bhardwaj A, Methods in cell separation for biomedical application: cryogels as a new tool, Biomedical materials 3(3) (2008) 034008. [DOI] [PubMed] [Google Scholar]
- [286].Dainiak MB, Galaev IY, Mattiasson B, Affinity cryogel monoliths for screening for optimal separation conditions and chromatographic separation of cells, Journal of Chromatography A 1123(2) (2006) 145–150. [DOI] [PubMed] [Google Scholar]
- [287].Podgornik A, Krajnc NL, Application of monoliths for bioparticle isolation, J. Sep. Sci 35(22) (2012) 3059–3072. [DOI] [PubMed] [Google Scholar]
- [288].Lozinsky VI, Plieva FM, Galaev IY, Mattiasson B, The potential of polymeric cryogels in bioseparation, Bioseparation 10(4) (2001) 163–188. [DOI] [PubMed] [Google Scholar]
- [289].Dainiak MB, Kumar A, Galaev IY, Mattiasson B, Detachment of affinity-captured bioparticles by elastic deformation of a macroporous hydrogel, Proceedings of the National Academy of Sciences of the United States of America 103(4) (2006) 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Ott S, Niessner R, Seidel M, Preparation of epoxy‐based macroporous monolithic columns for the fast and efficient immunofiltration of Staphylococcus aureus, Journal of separation science 34(16‐17) (2011) 2181–2192. [DOI] [PubMed] [Google Scholar]
- [291].Galaev IY, Dainiak MB, Plieva F, Mattiasson B, Effect of matrix elasticity on affinity binding and release of bioparticles. Elution of bound cells by temperature-induced shrinkage of the smart macroporous hydrogel, Langmuir 23(1) (2007) 35–40. [DOI] [PubMed] [Google Scholar]
- [292].Arvidsson P, Plieva FM, Savina IN, Lozinsky VI, Fexby S, Bülow L, Galaev IY, Mattiasson B, Chromatography of microbial cells using continuous supermacroporous affinity and ion-exchange columns, Journal of Chromatography A 977(1) (2002) 27–38. [DOI] [PubMed] [Google Scholar]
- [293].Dainiak MB, Plieva FM, Galaev IY, Hatti‐Kaul R, Mattiasson B, Cell chromatography: Separation of different microbial cells using IMAC supermacroporous monolithic columns, Biotechnology progress 21(2) (2005) 644–649. [DOI] [PubMed] [Google Scholar]
- [294].Srivastava A, Shakya AK, Kumar A, Boronate affinity chromatography of cells and biomacromolecules using cryogel matrices, Enzyme and microbial technology 51(6) (2012) 373–381. [DOI] [PubMed] [Google Scholar]
- [295].Srivastava A, Singh S, Kumar A, 16 Supermacroporous Functional Cryogel Stationary Matrices for Efficient Cell Separation, Supermacroporous Cryogels: Biomedical and Biotechnological Applications (2016) 445. [Google Scholar]
- [296].Plieva FM, Savina IN, Deraz S, Andersson J, Galaev IY, Mattiasson B, Characterization of supermacroporous monolithic polyacrylamide based matrices designed for chromatography of bioparticles, Journal of Chromatography B 807(1) (2004) 129–137. [DOI] [PubMed] [Google Scholar]
- [297].Szczęsna-Antczak M, Galas E, Bacillus subtilis cells immobilised in PVA-cryogels, Biomolecular Engineering 17(2) (2001) 55–63. [DOI] [PubMed] [Google Scholar]
- [298].Gold EF, Kleinman R, Ben-Efraim S, A heat-digestible cell-immunoadsorbent made by coupling hapten to gelatin, Journal of immunological methods 6(1–2) (1974) 31–37. [DOI] [PubMed] [Google Scholar]
- [299].Kedar E, de Landazuri MO, Bonavida B, Cellular immunoadsorbents: a simplified technique for separation of lymphoid cell populations, The Journal of Immunology 112(3) (1974) 1231–1243. [PubMed] [Google Scholar]
- [300].Haas W, Separation of antigen-specific lymphocytes. II. Enrichment of hapten-specific antibody-forming cell precursors, Journal of Experimental Medicine 141(5) (1975) 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Haas W, Layton JE, Separation of antigen-specific lymphocytes. I. Enrichment of antigen-binding cells, Journal of Experimental Medicine 141(5) (1975) 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Webb C, Teitelbaum D, Rauch H, Maoz A, Arnon R, Fuchs S, Fractionation of functional lymphocytes sensitized to basic encephalitogen on derivatized collagen and gelatin gels, The Journal of Immunology 114(5) (1975) 1469–1472. [PubMed] [Google Scholar]
- [303].Maoz A, Shellam G, Fractionation of cytotoxic cells from tumour-immune rats on derivatized collagen gels, Journal of immunological methods 12(1–2) (1976) 125–130. [DOI] [PubMed] [Google Scholar]
- [304].Bröcker EB, Sorg C, Specific separation of cytotoxic T lymphocytes on immunoadsorptive films, Journal of immunological methods 14(3–4) (1977) 333–342. [DOI] [PubMed] [Google Scholar]
- [305].Nossal G, Pike BL, Improved procedures for the fractionation and in vitro stimulation of hapten-specific B lymphocytes, The Journal of Immunology 120(1) (1978) 145–150. [PubMed] [Google Scholar]
- [306].Edelman G, Rutishauser U, Millette C, Cell fractionation and arrangement on fibers, beads, and surfaces, Proceedings of the National Academy of Sciences 68(9) (1971) 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Nordon RE, Haylock DN, Gaudry L, Schindhelm K, Hollow-fibre affinity cell separation system for CD34+ cell enrichment, Cytometry 24(4) (1996) 340–347. [DOI] [PubMed] [Google Scholar]
- [308].Nordon RE, Schindhelm K, Design of hollow fiber modules for uniform shear elution affinity cell separation, Artificial organs 21(2) (1997) 107–115. [DOI] [PubMed] [Google Scholar]
- [309].Orsello C, Lauffenburger D, Colton C, Characterization of cell detachment from hollow fiber affinity membranes, Biomedical sciences instrumentation 35 (1998) 315–320. [PubMed] [Google Scholar]
- [310].Nordon RE, Shu A, Camacho F, Milthorpe BK, Hollow‐fiber assay for ligand‐ mediated cell adhesion, Cytometry Part A 57(1) (2004) 39–44. [DOI] [PubMed] [Google Scholar]
- [311].Craig SJ, Shu A, Xu Y, Foong FC, Nordon R, Chimeric protein for selective cell attachment onto cellulosic substrates, Protein Engineering, Design & Selection 20(5) (2007) 235–241. [DOI] [PubMed] [Google Scholar]
- [312].Nordon R, Craig S, Hollow-fibre affinity cell separation, Cell Separation (2007) 129–150. [DOI] [PubMed] [Google Scholar]
- [313].Xu G, Tan Y, Xu T, Yin D, Wang M, Shen M, Chen X, Shi X, Zhu X, Hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for cancer cell capture and culture, Biomaterials science 5(4) (2017) 752–761. [DOI] [PubMed] [Google Scholar]
- [314].Pope NM, Kulcinski DL, Hardwick A, Chang YA, New application of silane coupling agents for covalently binding antibodies to glass and cellulose solid supports, Bioconjugate chemistry 4(2) (1993) 166–171. [DOI] [PubMed] [Google Scholar]
- [315].Saranya R, Murugan R, Hegde M, Doyle J, Babu R, Affinity membranes for capture of cells and biological substances, Filtering Media by Electrospinning, Springer; 2018, pp. 175–195. [Google Scholar]
- [316].Mandrusov E, Houng A, Klein E, Leonard EF, Membrane-based cell affinity chromatography to retrieve viable cells, Biotechnology progress 11(2) (1995) 208–213. [DOI] [PubMed] [Google Scholar]
- [317].Barkley S, Johnson H, Eisenthal R, Hubble J, Bubble‐induced detachment of affinity‐adsorbed erythrocytes, Biotechnology and applied biochemistry 40(2) (2004) 145–149. [DOI] [PubMed] [Google Scholar]
- [318].Okamura A, Itayagoshi M, Hagiwara T, Yamaguchi M, Kanamori T, Shinbo T, Wang P-C, Poly (N-isopropylacrylamide)-graft-polypropylene membranes containing adsorbed antibody for cell separation, Biomaterials 26(11) (2005) 1287–1292. [DOI] [PubMed] [Google Scholar]
- [319].Okamura A, Hagiwara T, Yamagami S, Yamaguchi M, Shinbo T, Kanamori T, Kondo S, Miwa K, Itagaki I, Effective cell separation utilizing poly (N-isopropylacrylamide)-grafted polypropylene membrane containing adsorbed antibody, Journal of bioscience and bioengineering 105(3) (2008) 221–225. [DOI] [PubMed] [Google Scholar]
- [320].Albertsson P-A, Separation of cell organelles and membrane vesicles by phase partition, Progress in clinical and biological research 270 (1988) 227. [PubMed] [Google Scholar]
- [321].Karr LJ, Van Alstine JM, Snyder RS, Shafer SG, Harris JM, Cell separation by immunoaffinity partitioning with polyethylene glycol-modified protein A in aqueous polymer two-phase systems, Journal of Chromatography A 442 (1988) 219–227. [DOI] [PubMed] [Google Scholar]
- [322].Delgado C, Anderson RJ, Francis GE, Fisher D, Separation of cell mixtures by immunoaffinity cell partitioning: strategies for low abundance cells, Analytical biochemistry 192(2) (1991) 322–328. [DOI] [PubMed] [Google Scholar]
- [323].Delgado C, Sancho P, Medieta J, Luque J, Ligand-receptor interactions in affinity cell partitioning: studies with transferrin covalently linked to monomethoxypoly (ethylene glycol) and rat reticulocytes, Journal of Chromatography A 594(1–2) (1992) 97–103. [DOI] [PubMed] [Google Scholar]
- [324].Rosa PA, Azevedo AM, Aires-Barros MR, Application of central composite design to the optimisation of aqueous two-phase extraction of human antibodies, Journal of Chromatography A 1141(1) (2007) 50–60. [DOI] [PubMed] [Google Scholar]
- [325].Sousa AF, Andrade PZ, Pirzgalska RM, Galhoz TM, Azevedo AM, Da Silva CL, Raquel Aires-Barros M, Cabral JM, A novel method for human hematopoietic stem/progenitor cell isolation from umbilical cord blood based on immunoaffinity aqueous two-phase partitioning, Biotechnology letters 33(12) (2011) 2373–2377. [DOI] [PubMed] [Google Scholar]
- [326].González-González M, Rito-Palomares M, Aqueous two-phase systems strategies to establish novel bioprocesses for stem cells recovery, Critical reviews in biotechnology 34(4) (2014) 318–327. [DOI] [PubMed] [Google Scholar]
- [327].González‐González M, Rito‐Palomares M, Méndez Quintero O, Partition behavior of CD133+ stem cells from human umbilical cord blood in aqueous two‐phase systems: In route to establish novel stem cell primary recovery strategies, Biotechnology progress 30(3) (2014) 700–707. [DOI] [PubMed] [Google Scholar]
- [328].González‐González M, Rito‐Palomares M, Application of affinity aqueous two‐phase systems for the fractionation of CD133+ stem cells from human umbilical cord blood, Journal of Molecular Recognition 28(3) (2015) 142–147. [DOI] [PubMed] [Google Scholar]
- [329].González-González M, Willson RC, Rito-Palomares M, Elimination of contaminants from cell preparations using aqueous two-phase partitioning, Separation and Purification Technology 158 (2016) 103–107. [Google Scholar]
- [330].Brandt S, Goffe R, Kessler S, O’Connor J, Zale S, Klein E, Affinity Partitioning in Aqueous Two-Phase Systems, (2000). [Google Scholar]
- [331].Kumar A, Kamihira M, Mattiasson B, Two-phase affinity partitioning of animal cells: implications of multipoint interactions, Methods for affinity-based separations of enzymes and proteins (2002) 163–180. [Google Scholar]
- [332].Hamamoto R, Kamihira M, Iijima S, Specific separation of animal cells using aqueous two-phase systems, Journal of fermentation and bioengineering 82(1) (1996) 73–76. [Google Scholar]
- [333].Laboureau E, Capiod J, Dessaint C, Prin L, Vijayalakshmi M, Study of human cord blood lymphocytes by immobilized metal ion affinity partitioning, Journal of Chromatography B: Biomedical Sciences and Applications 680(1) (1996) 189–195. [DOI] [PubMed] [Google Scholar]
- [334].Cabral J, Cell partitioning in aqueous two-phase polymer systems, Cell separation (2007) 151–171. [DOI] [PubMed] [Google Scholar]
- [335].Karr LJ, Shafer SG, Harris JM, Van Alstine JM, Snyder RS, Immuno-affinity partition of cells in aqueous polymer two-phase systems, Journal of Chromatography A 354 (1986) 269–282. [DOI] [PubMed] [Google Scholar]
- [336].Sharp KA, Yalpani M, Howard SJ, Brooks DE, Synthesis and application of a poly (ethylene glycol)-antibody affinity ligand for cell separations in aqueous polymer two-phase systems, Analytical biochemistry 154(1) (1986) 110–117. [DOI] [PubMed] [Google Scholar]
- [337].Kumar A, Kamihira M, Galaev IY, Mattiasson B, Iijima S, Type‐specific separation of animal cells in aqueous two‐phase systems using antibody conjugates with temperature‐sensitive polymers, Biotechnology and bioengineering 75(5) (2001) 570–580. [DOI] [PubMed] [Google Scholar]
- [338].Muiño BM, Cebrian PJ, Olde B, Johansson G, Effect of dextran-and poly (ethylene glycol)-bound procion yellow HE-3G on the partition of membranes from calf brain synaptosomes within an aqueous two-phase system, Journal of chromatography 358(1) (1986) 147–158. [DOI] [PubMed] [Google Scholar]
- [339].Blanco MTM, Cebrian JA, Olde B, Johansson G, Subfractions of membranes from calf brain synaptosomes obtained and studied by liquid—liquid partitioning, Journal of Chromatography A 547 (1991) 79–87. [DOI] [PubMed] [Google Scholar]
- [340].Pérez JC, Blanco MTM, Johansson G, Heterogeneity of synaptosomal membrane preparations from different regions of calf brain studied by partitioning and counter-current distribution, International journal of biochemistry 23(12) (1991) 1491–1495. [DOI] [PubMed] [Google Scholar]
- [341].Zijlstra G, Michielsen M, De Gooijer C, Van der Pol L, Tramper J, Separation of hybridoma cells from their IgG product using aqueous two-phase systems, Bioseparation 6(4) (1995) 201–210. [PubMed] [Google Scholar]
- [342].Zijlstra G, Michielsen M, De Gooijer C, Van der Pol L, Tramper J, IgG and hybridoma partitioning in aqueous two-phase systems containing a dye-ligand, Bioseparation 7(2) (1998) 117–126. [DOI] [PubMed] [Google Scholar]
- [343].Nanak E, Vijayalakshmi M, Chadha K, Segregation of normal and pathological human red blood cells, lymphocytes and fibroblasts by immobilized metal‐ion affinity partitioning, Journal of Molecular Recognition 8(1‐2) (1995) 77–84. [DOI] [PubMed] [Google Scholar]
- [344].Ugelstad J, Berge A, Ellingsen T, Aune O, Kilaas L, Nilsen T, Schmid R, Stenstad P, Funderud S, Kvalheim G, Monosized magnetic particles and their use in selective cell separation, Macromolecular Symposia, Wiley Online Library, 1988, pp. 177–211. [Google Scholar]
- [345].Schmitz B, Radbruch A, Kümmel T, Wickenhauser C, Korb H, Hansmann M, Thiele J, Fischer R, Magnetic activated cell sorting (MACS)—a new immunomagnetic method for megakaryocytic cell isolation: comparison of different separation techniques, European journal of haematology 52(5) (1994) 267–275. [DOI] [PubMed] [Google Scholar]
- [346].Zborowski M, Fuh CB, Green R, Sun L, Chalmers JJ, Analytical magnetapheresis of ferritin-labeled lymphocytes, Analytical chemistry 67(20) (1995) 3702–3712. [DOI] [PubMed] [Google Scholar]
- [347].Sun L, Zborowski M, Moore LR, Chalmers JJ, Continuous, flow‐through immunomagnetic cell sorting in a quadrupole field, Cytometry Part A 33(4) (1998) 469–475. [DOI] [PubMed] [Google Scholar]
- [348].Chalmers JJ, Zhao Y, Nakamura M, Melnik K, Lasky L, Moore L, Zborowski M, An instrument to determine the magnetophoretic mobility of labeled, biological cells and paramagnetic particles, Journal of Magnetism and Magnetic Materials 194(1) (1999) 231–241. [Google Scholar]
- [349].McCloskey KE, Chalmers JJ, Zborowski M, Magnetophoretic mobilities correlate to antibody binding capacities, Cytometry Part A 40(4) (2000) 307–315. [DOI] [PubMed] [Google Scholar]
- [350].McCloskey KE, Chalmers JJ, Zborowski M, Magnetic cell separation: characterization of magnetophoretic mobility, Analytical chemistry 75(24) (2003) 6868–6874. [DOI] [PubMed] [Google Scholar]
- [351].Chalmers JJ, Zborowski M, Sun L, Moore L, Flow through, immunomagnetic cell separation, Biotechnology progress 14(1) (1998) 141–148. [DOI] [PubMed] [Google Scholar]
- [352].Adams JD, Kim U, Soh HT, Multitarget magnetic activated cell sorter, Proceedings of the National Academy of Sciences 105(47) (2008) 18165–18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Huang N-T, Hwong Y-J, Lai RL, A microfluidic microwell device for immunomagnetic single-cell trapping, Microfluidics and Nanofluidics 22(2) (2018) 16. [Google Scholar]
- [354].Islam D, Lindberg A, Detection of Shigella dysenteriae type 1 and Shigella flexneri in feces by immunomagnetic isolation and polymerase chain reaction, Journal of clinical microbiology 30(11) (1992) 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Evaristo C, Steinbrück P, Pankratz J, Yu Z, Dose C, REAlease™ Immunomagnetic Separation Technology with reversible labeling for positive selection of leukocytes, Am Assoc Immnol, 2018. [Google Scholar]
- [356].Mazurek GH, Reddy V, Murphy D, Ansari T, Detection of Mycobacterium tuberculosis in cerebrospinal fluid following immunomagnetic enrichment, Journal of clinical microbiology 34(2) (1996) 450–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Zhou X, Luo B, Kang K, Ma S, Sun X, Lan F, Yi Q, Wu Y, Multifunctional luminescent immuno-magnetic nanoparticles: toward fast, efficient, cell-friendly capture and recovery of circulating tumor cells, Journal of Materials Chemistry B 7(3) (2019) 393–400. [DOI] [PubMed] [Google Scholar]
- [358].Deng MQ, Cliver DO, Mariam TW, Immunomagnetic capture PCR to detect viable Cryptosporidium parvum oocysts from environmental samples, Applied and environmental microbiology 63(8) (1997) 3134–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Rao L, Meng QF, Huang Q, Wang Z, Yu GT, Li A, Ma W, Zhang N, Guo SS, Zhao XZ, Platelet–Leukocyte Hybrid Membrane‐Coated Immunomagnetic Beads for Highly Efficient and Highly Specific Isolation of Circulating Tumor Cells, Advanced Functional Materials 28(34) (2018) 1803531. [Google Scholar]
- [360].Shinkai M, Suzuki M, Iijima S, Kobayashi T, Antibody‐conjugated magnetoliposomes for targeting cancer cells and their application in hyperthermia, Biotechnology and applied biochemistry 21(2) (1995) 125–137. [DOI] [PubMed] [Google Scholar]
- [361].Dhadge VL, Hussain A, Azevedo AM, Aires-Barros R, Roque AC, Boronic acid-modified magnetic materials for antibody purification, Journal of The Royal Society Interface 11(91) (2014) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Scouten WH, Konecny P, Reversible immobilization of antibodies on magnetic beads, Analytical biochemistry 205(2) (1992) 313–318. [DOI] [PubMed] [Google Scholar]
- [363].Benez A, Geiselhart A, Handgretinger R, Schiebel U, Fierlbeck G, Detection of circulating melanoma cells by immunomagnetic cell sorting, Journal of clinical laboratory analysis 13(5) (1999) 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [364].Grunewald S, Paasch U, Glander H-J, Enrichment of non–apoptotic human spermatozoa after cryopreservation by immunomagnetic cell sorting, Cell and tissue banking 2(3) (2001) 127–133. [DOI] [PubMed] [Google Scholar]
- [365].Pimpha N, Chaleawlert-umpon S, Chruewkamlow N, Kasinrerk W, Preparation of anti-CD4 monoclonal antibody-conjugated magnetic poly (glycidyl methacrylate) particles and their application on CD4+ lymphocyte separation, Talanta 84(1) (2011) 89–97. [DOI] [PubMed] [Google Scholar]
- [366].Jackson C, Garbett P, Nissen B, Schrieber L, Binding of human endothelium to Ulex europaeus I-coated Dynabeads: application to the isolation of microvascular endothelium, Journal of cell science 96(2) (1990) 257–262. [DOI] [PubMed] [Google Scholar]
- [367].Rye PD, Sweet and sticky: carbohydrate-coated magnetic beads, Nature Biotechnology 14(2) (1996) 155–157. [Google Scholar]
- [368].Yang LJ-S, Zeller CB, Schnaar RL, Detection and isolation of lectin-transfected COS cells based on cell adhesion to immobilized glycosphingolipids, Analytical biochemistry 236(1) (1996) 161–167. [DOI] [PubMed] [Google Scholar]
- [369].Rye PD, Bovin NV, Selection of carbohydrate-binding cell phenotypes using oligosaccharide-coated magnetic particles, Glycobiology 7(2) (1997) 179–182. [DOI] [PubMed] [Google Scholar]
- [370].Collins BE, Yang L, Schnaar RL, Lectin-mediated cell adhesion to immobilized glycosphingolipids, Methods in enzymology 312 (1999) 438–446. [DOI] [PubMed] [Google Scholar]
- [371].Herr JK, Smith JE, Medley CD, Shangguan D, Tan W, Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells, Analytical Chemistry 78(9) (2006) 2918–2924. [DOI] [PubMed] [Google Scholar]
- [372].Smith JE, Medley CD, Tang Z, Shangguan D, Lofton C, Tan W, Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells, Analytical Chemistry 79(8) (2007) 3075–3082. [DOI] [PubMed] [Google Scholar]
- [373].Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W, Selection of aptamers for molecular recognition and characterization of cancer cells, Analytical chemistry 79(13) (2007) 4900–4907. [DOI] [PubMed] [Google Scholar]
- [374].Dong X, Zheng Y, Huang Y, Chen X, Jing X, Synthesis and characterization of multifunctional poly (glycidyl methacrylate) microspheres and their use in cell separation, Analytical biochemistry 405(2) (2010) 207–212. [DOI] [PubMed] [Google Scholar]
- [375].Lee J-J, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA, Tsui JH, Kohane DS, Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood, Nano letters 14(1) (2013) 1–5. [DOI] [PubMed] [Google Scholar]
- [376].Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system, Clinical cancer research 13(3) (2007) 920–928. [DOI] [PubMed] [Google Scholar]
- [377].Adams DL, Stefansson S, Haudenschild C, Martin SS, Charpentier M, Chumsri S, Cristofanilli M, Tang CM, Alpaugh RK, Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the cellsearch® CTC test, Cytometry Part A 87(2) (2015) 137–144. [DOI] [PubMed] [Google Scholar]
- [378].Beije N, Jager A, Sleijfer S, Circulating tumor cell enumeration by the CellSearch system: the clinician’s guide to breast cancer treatment?, Cancer treatment reviews 41(2) (2015) 144–150. [DOI] [PubMed] [Google Scholar]
- [379].Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, HERzENbERG LA, The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford, Clinical chemistry 48(10) (2002) 1819–1827. [PubMed] [Google Scholar]
- [380].González‐González M, Vázquez‐Villegas P, García‐Salinas C, Rito‐Palomares M, Current strategies and challenges for the purification of stem cells, Journal of Chemical Technology and Biotechnology 87(1) (2012) 2–10. [Google Scholar]
- [381].Liu L, Cheung TH, Charville GW, Rando TA, Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting, Nature protocols 10(10) (2015) 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [382].Pruszak J, Sonntag KC, Aung MH, Sanchez‐Pernaute R, Isacson O, Markers and methods for cell sorting of human embryonic stem cell‐derived neural cell populations, Stem cells 25(9) (2007) 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [383].Fong CY, Peh GS, Gauthaman K, Bongso A, Separation of SSEA-4 and TRA-1–60 labelled undifferentiated human embryonic stem cells from a heterogeneous cell population using magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS), Stem Cell Reviews and Reports 5(1) (2009) 72–80. [DOI] [PubMed] [Google Scholar]
- [384].Lee D-C, Hsu Y-C, Chung Y-F, Hsiao C-Y, Chen S-L, Chen M-S, Lin H-K, Chiu M, Isolation of neural stem/progenitor cells by using EGF/FGF1 and FGF1B promoter-driven green fluorescence from embryonic and adult mouse brains, Molecular and Cellular Neuroscience 41(3) (2009) 348–363. [DOI] [PubMed] [Google Scholar]
- [385].Weissman IL, Shizuru JA, The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases, Blood 112(9) (2008) 3543–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [386].Chen Y, Chung AJ, Wu TH, Teitell MA, Di Carlo D, Chiou PY, Pulsed laser activated cell sorting with three dimensional sheathless inertial focusing, Small 10(9) (2014) 1746–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Bhagat AAS, Bow H, Hou HW, Tan SJ, Han J, Lim CT, Microfluidics for cell separation, Medical and Biological Engineering and Computing 48(10) (2010) 999–1014. [DOI] [PubMed] [Google Scholar]
- [388].De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M, 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity, Nature medicine 7(2) (2001) 245–248. [DOI] [PubMed] [Google Scholar]
- [389].Perfetto SP, Chattopadhyay PK, Roederer M, Seventeen-colour flow cytometry: unravelling the immune system, Nature Reviews Immunology 4(8) (2004) 648–655. [DOI] [PubMed] [Google Scholar]
- [390].Murray C, Pao E, Tseng P, Aftab S, Kulkarni R, Rettig M, Di Carlo D, Quantitative magnetic separation of particles and cells using gradient magnetic ratcheting, Small 12(14) (2016) 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Murray C, Pao E, Jann A, Park DE, Di Carlo D, Continuous and Quantitative Purification of T-Cell Subsets for Cell Therapy Manufacturing Using Magnetic Ratcheting Cytometry, SLAS TECHNOLOGY: Translating Life Sciences Innovation 23(4) (2018) 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [392].Li P, Tian Y, Pappas D, Comparison of inlet geometry in microfluidic cell affinity chromatography, Analytical chemistry 83(3) (2011) 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [393].Chen J, Li J, Sun Y, Microfluidic approaches for cancer cell detection, characterization, and separation, Lab on a Chip 12(10) (2012) 1753–1767. [DOI] [PubMed] [Google Scholar]
- [394].Gao Y, Li W, Zhang Y, Pappas D, Cell Affinity Separations on Microfluidic Devices, Affinity Chromatography: Methods and Protocols (2015) 55–65. [DOI] [PubMed] [Google Scholar]
- [395].Zhang Y, Li W, Zhou Y, Johnson A, Venable A, Hassan A, Griswold J, Pappas D, Detection of sepsis in patient blood samples using CD64 expression in a microfluidic cell separation device, Analyst 143(1) (2018) 241–249. [DOI] [PubMed] [Google Scholar]
- [396].Zhang M, Xu B, Siehr A, Shen W, Efficient release of immunocaptured cells using coiled-coils in a microfluidic device, RSC Advances 9(50) (2019) 29182–29189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Isolation of rare circulating tumour cells in cancer patients by microchip technology, Nature 450(7173) (2007) 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ, Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody, Lab on a chip 10(1) (2010) 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [399].Arya SK, Lim B, Rahman ARA, Enrichment, detection and clinical significance of circulating tumor cells, Lab on a Chip 13(11) (2013) 1995–2027. [DOI] [PubMed] [Google Scholar]
- [400].Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M, A microfluidic device for practical label-free CD4+ T cell counting of HIV-infected subjects, Lab on a Chip 7(2) (2007) 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Hatch A, Hansmann G, Murthy SK, Engineered alginate hydrogels for effective microfluidic capture and release of endothelial progenitor cells from whole blood, Langmuir 27(7) (2011) 4257–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M, Biopolymer system for cell recovery from microfluidic cell capture devices, Analytical chemistry 84(8) (2012) 3682–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [403].Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W, Enrichment of cancer cells using aptamers immobilized on a microfluidic channel, Analytical chemistry 81(3) (2008) 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ, Rare cell capture in microfluidic devices, Chemical engineering science 66(7) (2011) 1508–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [405].Yu M, Stott S, Toner M, Maheswaran S, Haber DA, Circulating tumor cells: approaches to isolation and characterization, The Journal of cell biology 192(3) (2011) 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Isolation of circulating tumor cells using a microvortex-generating herringbone-chip, Proceedings of the National Academy of Sciences 107(43) (2010) 18392–18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Ciurte A, Selicean C, Soritau O, Buiga R, Automatic detection of circulating tumor cells in darkfield microscopic images of unstained blood using boosting techniques, PloS one 13(12) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [408].Shen Z, Wu A, Chen X, Current detection technologies for circulating tumor cells, Chemical Society Reviews 46(8) (2017) 2038–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [409].Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S, Dendrimer‐Mediated Multivalent Binding for the Enhanced Capture of Tumor Cells, Angewandte Chemie International Edition 50(49) (2011) 11769–11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [410].Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ, Detection and isolation of circulating tumor cells: principles and methods, Biotechnology advances 31(7) (2013) 1063–1084. [DOI] [PubMed] [Google Scholar]
- [411].Lustberg M, Jatana KR, Zborowski M, Chalmers JJ, Emerging technologies for CTC detection based on depletion of normal cells, Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer, Springer; 2012, pp. 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [412].Toss A, Mu Z, Fernandez S, Cristofanilli M, CTC enumeration and characterization: moving toward personalized medicine, Annals of translational medicine 2(11) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [413].Kang H, Kim J, Cho H, Han K-H, Evaluation of Positive and Negative Methods for Isolation of Circulating Tumor Cells by Lateral Magnetophoresis, Micromachines 10(6) (2019) 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].Wang X, Sun L, Zhang H, Wei L, Qu W, Zeng Z, Liu Y, Zhu Z, Microfluidic chip combined with magnetic-activated cell sorting technology for tumor antigen-independent sorting of circulating hepatocellular carcinoma cells, PeerJ 7 (2019) e6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [415].Mereu E, Lafzi A, Moutinho C, Ziegenhain C, McCarthy DJ, Álvarez-Varela A, Batlle E, Grün D, Lau JK, Boutet SC, Benchmarking single-cell RNA-sequencing protocols for cell atlas projects, Nature Biotechnology (2020) 1–9. [DOI] [PubMed] [Google Scholar]
- [416].Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Multiplexed droplet single-cell RNA-sequencing using natural genetic variation, Nature biotechnology 36(1) (2018) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [417].Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L, Single-cell barcoding and sequencing using droplet microfluidics, Nature protocols 12(1) (2017) 44. [DOI] [PubMed] [Google Scholar]
- [418].Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells, Cell 161(5) (2015) 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [419].Francz B, Ungai-Salánki R, Sautner É, Horvath R, Szabó B, Subnanoliter precision piezo pipette for single-cell isolation and droplet printing, Microfluidics and Nanofluidics 24(2) (2020) 12. [Google Scholar]
- [420].Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML, Droplet microfluidic technology for single-cell high-throughput screening, Proceedings of the National Academy of Sciences 106(34) (2009) 14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Yuan Y, Brouchon J, Calvo-Calle M, Xia J, Zhang X, Sun L, Ye F, Weitz DA, Heyman JA, Droplet encapsulation improves accuracy of immune cell cytokine capture assays, Lab on a Chip (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [422].Pan CW, Horvath DG, Braza S, Moore T, Lynch A, Feit C, Abbyad P, Sorting by interfacial tension (SIFT): label-free selection of live cells based on single-cell metabolism, Lab on a Chip 19(8) (2019) 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [423].Vallejo D, Nikoomanzar A, Paegel BM, Chaput JC, Fluorescence-activated droplet sorting for single-cell directed evolution, ACS synthetic biology 8(6) (2019) 1430–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].Witek M, Freed I, Soper SA, Cell Separations and Sorting, Analytical Chemistry (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Shields CW IV, Reyes CD, López GP, Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation, Lab on a Chip 15(5) (2015) 1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [426].Matuła K, Rivello F, Huck WT, Single‐Cell Analysis Using Droplet Microfluidics, Advanced Biosystems 4(1) (2020) 1900188. [DOI] [PubMed] [Google Scholar]
- [427].Doufène K, Tourné-Péteilh C, Etienne P, Aubert-Pouëssel A, Microfluidic Systems for Droplet Generation in Aqueous Continuous Phases: A Focus Review, Langmuir 35(39) (2019) 12597–12612. [DOI] [PubMed] [Google Scholar]
- [428].Zhu P, Wang L, Passive and active droplet generation with microfluidics: a review, Lab on a Chip 17(1) (2017) 34–75. [DOI] [PubMed] [Google Scholar]
- [429].Teh S-Y, Lin R, Hung L-H, Lee AP, Droplet microfluidics, Lab on a Chip 8(2) (2008) 198–220. [DOI] [PubMed] [Google Scholar]
- [430].Rosenfeld L, Lin T, Derda R, Tang SK, Review and analysis of performance metrics of droplet microfluidics systems, Microfluidics and nanofluidics 16(5) (2014) 921–939. [Google Scholar]
- [431].Xi H-D, Zheng H, Guo W, Gañán-Calvo AM, Ai Y, Tsao C-W, Zhou J, Li W, Huang Y, Nguyen N-T, Active droplet sorting in microfluidics: a review, Lab on a Chip 17(5) (2017) 751–771. [DOI] [PubMed] [Google Scholar]
- [432].Sajeesh P, Sen AK, Particle separation and sorting in microfluidic devices: a review, Microfluidics and nanofluidics 17(1) (2014) 1–52. [Google Scholar]
- [433].Tan Y-C, Ho YL, Lee AP, Microfluidic sorting of droplets by size, Microfluidics and Nanofluidics 4(4) (2008) 343. [Google Scholar]
- [434].Tan Y-C, Fisher JS, Lee AI, Cristini V, Lee AP, Design of microfluidic channel geometries for the control of droplet volume, chemical concentration, and sorting, Lab on a Chip 4(4) (2004) 292–298. [DOI] [PubMed] [Google Scholar]
- [435].Niu X, Zhang M, Peng S, Wen W, Sheng P, Real-time detection, control, and sorting of microfluidic droplets, Biomicrofluidics 1(4) (2007) 044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [436].Hasan S, Geissler D, Wink K, Hagen A, Heiland JJ, Belder D, Fluorescence lifetime-activated droplet sorting in microfluidic chip systems, Lab on a Chip 19(3) (2019) 403–409. [DOI] [PubMed] [Google Scholar]
- [437].Sciambi A, Abate AR, Accurate microfluidic sorting of droplets at 30 kHz, Lab on a Chip 15(1) (2015) 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [438].Baret J-C, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity, Lab on a Chip 9(13) (2009) 1850–1858. [DOI] [PubMed] [Google Scholar]
- [439].Lagus TP, Edd JF, A review of the theory, methods and recent applications of high-throughput single-cell droplet microfluidics, J. Phys. D: Appl. Phys 46(11) (2013) 114005. [Google Scholar]
- [440].Shembekar N, Hu H, Eustace D, Merten CA, Single-cell droplet microfluidic screening for antibodies specifically binding to target cells, Cell reports 22(8) (2018) 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [441].Yelleswarapu V, Buser JR, Haber M, Baron J, Inapuri E, Issadore D, Mobile platform for rapid sub–picogram-per-milliliter, multiplexed, digital droplet detection of proteins, Proceedings of the National Academy of Sciences 116(10) (2019) 4489–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [442].Shojaeian M, Lehr F-X, Göringer HU, Hardt S, On-demand production of femtoliter drops in microchannels and their use as biological reaction compartments, Analytical chemistry 91(5) (2019) 3484–3491. [DOI] [PubMed] [Google Scholar]
- [443].Sinha N, Subedi N, Tel J, Integrating immunology and microfluidics for single immune cell analysis, Frontiers in immunology 9 (2018) 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [444].Sánchez Barea J, Lee J, Kang D-K, Recent Advances in Droplet-based Microfluidic Technologies for Biochemistry and Molecular Biology, Micromachines 10(6) (2019) 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [445].Linhult M, Gulich S, Hober S, Affinity ligands for industrial protein purification, Protein and peptide letters 12(4) (2005) 305–310. [DOI] [PubMed] [Google Scholar]
- [446].Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L, A recurring theme in protein engineering: the design, stability and folding of repeat proteins, Current opinion in structural biology 15(4) (2005) 464–471. [DOI] [PubMed] [Google Scholar]
- [447].Stroumpoulis D, Zhang H, Rubalcava L, Gliem J, Tirrell M, Cell adhesion and growth to peptide-patterned supported lipid membranes, Langmuir 23(7) (2007) 3849–3856. [DOI] [PubMed] [Google Scholar]
- [448].Kaur G, Wang C, Sun J, Wang Q, The synergistic effects of multivalent ligand display and nanotopography on osteogenic differentiation of rat bone marrow stem cells, Biomaterials 31(22) (2010) 5813–5824. [DOI] [PubMed] [Google Scholar]
- [449].Zhang D, Peptide microarrays for the discovery of cell-ligand interactions that direct cell state, University of Illinois at Urbana-Champaign, 2016. [Google Scholar]
- [450].Melmon KL, Weinstein Y, Shearer G, Bourne HR, Bauminger S, Separation of specific antibody-forming mouse cells by their adherence to insolubilized endogenous hormones, Journal of Clinical Investigation 53(1) (1974) 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [451].Shearer G, Weinstein Y, Melmon KL, Enhancement of immune response potential of mouse lymphoid cells fractionated over insolubilized conjugated histamine columns, The Journal of Immunology 113(2) (1974) 597–607. [PubMed] [Google Scholar]
- [452].Weinstein Y, Melmon K, Control of immune responses by cyclic AMP and lymphocytes that adhere to histamine columns, Immunological communications 5(5) (1976) 401–416. [DOI] [PubMed] [Google Scholar]
- [453].Shearer G, Weinstein Y, Melmon K, Bourne H, Separation of leukocytes by their amine receptors: subsequent immunologic functions, Cyclic AMP, cell growth, and the immune response, Springer; 1974, pp. 135–146. [Google Scholar]
- [454].Melmon K, Weinstein Y, Shearer G, Bourne H, Leukocyte separation on the basis of their receptors for biogenic amines and prostaglandins: relation of the receptor to antibody formation, Cyclic AMP, Cell Growth, and the Immune Response, Springer; 1974, pp. 114–134. [Google Scholar]
- [455].SHEARER G, BOURNE H, LEUKOCYTE SEPARATION ON THE BASIS OF THEIR RECEPTORS FOR BIOGENIC AMINES AND PROSTAGLANDINS: RELATION OF THE RECEPTOR TO ANTIBODY FORMATION, Cyclic AMP, Cell Growth, and the Immune Response: Proceedings of the Symposium Held at Marco Island, Florida January 8– 10, 1973, Springer Science & Business Media, 2013, p. 114. [Google Scholar]
- [456].Van Tassell ML, Price NP, Miller MJ, Glycan-specific whole cell affinity chromatography: A versatile microbial adhesion platform, MethodsX 1 (2014) 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [457].Krag DN, Shukla GS, Shen G-P, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C, Selection of tumor-binding ligands in cancer patients with phage display libraries, Cancer research 66(15) (2006) 7724–7733. [DOI] [PubMed] [Google Scholar]
- [458].Stoltenburg R, Reinemann C, Strehlitz B, SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands, Biomolecular engineering 24(4) (2007) 381–403. [DOI] [PubMed] [Google Scholar]
- [459].Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB, Synthetic ligands discovered by in vitro selection, Journal of the American Chemical Society 129(43) (2007) 13137–13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [460].Gohlke H, Klebe G, Approaches to the description and prediction of the binding affinity of small‐molecule ligands to macromolecular receptors, Angewandte Chemie International Edition 41(15) (2002) 2644–2676. [DOI] [PubMed] [Google Scholar]
- [461].Oyama T, Sykes KF, Samli KN, Minna JD, Johnston SA, Brown KC, Isolation of lung tumor specific peptides from a random peptide library: generation of diagnostic and cell-targeting reagents, Cancer letters 202(2) (2003) 219–230. [DOI] [PubMed] [Google Scholar]
- [462].McGuire MJ, Samli KN, Johnston SA, Brown KC, In vitro selection of a peptide with high selectivity for cardiomyocytes in vivo, Journal of molecular biology 342(1) (2004) 171–182. [DOI] [PubMed] [Google Scholar]
- [463].Askoxylakis V, Zitzmann S, Mier W, Graham K, Krämer S, von Wegner F, Fink RH, Schwab M, Eisenhut M, Haberkorn U, Preclinical evaluation of the breast cancer cell-binding peptide, p160, Clinical cancer research 11(18) (2005) 6705–6712. [DOI] [PubMed] [Google Scholar]
- [464].Guo K, Wendel HP, Scheideler L, Ziemer G, Scheule AM, Aptamer‐based capture molecules as a novel coating strategy to promote cell adhesion, Journal of cellular and molecular medicine 9(3) (2005) 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [465].McGuire MJ, Samli KN, Chang Y-C, Brown KC, Novel ligands for cancer diagnosis: selection of peptide ligands for identification and isolation of B-cell lymphomas, Experimental hematology 34(4) (2006) 443–452. [DOI] [PubMed] [Google Scholar]
- [466].Oyama T, Rombel IT, Samli KN, Zhou X, Brown KC, Isolation of multiple cell-binding ligands from different phage displayed-peptide libraries, Biosensors and Bioelectronics 21(10) (2006) 1867–1875. [DOI] [PubMed] [Google Scholar]
- [467].Lo A, Lin C-T, Wu H-C, Hepatocellular carcinoma cell-specific peptide ligand for targeted drug delivery, Molecular cancer therapeutics 7(3) (2008) 579–589. [DOI] [PubMed] [Google Scholar]
- [468].McGuire MJ, Li S, Brown KC, Biopanning of phage displayed peptide libraries for the isolation of cell-specific ligands, Biosensors and Biodetection: Methods and Protocols: Electrochemical and Mechanical Detectors, Lateral Flow and Ligands for Biosensors (2009) 291–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [469].Nery AA, Wrenger C, Ulrich H, Recognition of biomarkers and cell‐specific molecular signatures: Aptamers as capture agents, Journal of separation science 32(10) (2009) 1523–1530. [DOI] [PubMed] [Google Scholar]
- [470].Svensen N, Díaz-Mochón JJ, Bradley M, Encoded peptide libraries and the discovery of new cell binding ligands, Chemical Communications 47(27) (2011) 7638–7640. [DOI] [PubMed] [Google Scholar]
- [471].Sefah K, Shangguan D, Xiong X, O’donoghue MB, Tan W, Development of DNA aptamers using Cell-SELEX, Nature protocols 5(6) (2010) 1169–1185. [DOI] [PubMed] [Google Scholar]
- [472].Ohuchi S, Cell-SELEX technology, BioResearch open access 1(6) (2012) 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [473].Moon J, Kim G, Park SB, Lim J, Mo C, Comparison of whole-cell SELEX methods for the identification of Staphylococcus aureus-specific DNA aptamers, Sensors 15(4) (2015) 8884–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [474].Ganji A, Varasteh A, Sankian M, Aptamers: new arrows to target dendritic cells, Journal of drug targeting 24(1) (2016) 1–12. [DOI] [PubMed] [Google Scholar]
- [475].Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W, Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells, Analytical chemistry 81(17) (2009) 7436–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [476].Zhang Z, Chen N, Li S, Battig MR, Wang Y, Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences, Journal of the American Chemical Society 134(38) (2012) 15716–15719. [DOI] [PubMed] [Google Scholar]
- [477].Pappas D, Wang K, Cellular separations: a review of new challenges in analytical chemistry, analytica chimica acta 601(1) (2007) 26–35. [DOI] [PubMed] [Google Scholar]
- [478].Chen Y, Tyagi D, Lyu M, Carrier AJ, Nganou C, Youden B, Wang W, Cui S, Servos M, Oakes K, Regenerative NanoOctopus Based on Multivalent-Aptamer-Functionalized Magnetic Microparticles for Effective Cell Capture in Whole Blood, Analytical chemistry 91(6) (2019) 4017–4022. [DOI] [PubMed] [Google Scholar]
- [479].Li Z, Wang G, Shen Y, Guo N, Ma N, DNA‐templated magnetic nanoparticle‐ quantum dot polymers for ultrasensitive capture and detection of circulating tumor cells, Advanced Functional Materials 28(14) (2018) 1707152. [Google Scholar]
- [480].Feng J, Dai Z, Tian X, Jiang X, Detection of Listeria monocytogenes based on combined aptamers magnetic capture and loop-mediated isothermal amplification, Food Control 85 (2018) 443–452. [Google Scholar]
- [481].Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH, Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device, Analytical chemistry 84(9) (2012) 4199–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [482].Zhou Q, Liu Y, Shin D-S, Silangcruz J, Tuleuova N, Revzin A, Aptamer-containing surfaces for selective capture of CD4 expressing cells, Langmuir 28(34) (2012) 12544–12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [483].Liu L, Yang K, Gao H, Li X, Chen Y, Zhang L, Peng X, Zhang Y, Artificial antibody with site-enhanced multivalent aptamers for specific capture of circulating tumor cells, Analytical chemistry 91(4) (2019) 2591–2594. [DOI] [PubMed] [Google Scholar]
- [484].Zhu Q, Liu G, Kai M, DNA aptamers in the diagnosis and treatment of human diseases, Molecules 20(12) (2015) 20979–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [485].Yoon JW, Jang IH, Heo SC, Kwon YW, Choi EJ, Bae K-H, Suh D-S, Kim S-C, Han S, Haam S, Isolation of Foreign Material-Free Endothelial Progenitor Cells Using CD31 Aptamer and Therapeutic Application for Ischemic Injury, PloS one 10(7) (2015) e0131785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [486].Keefe AD, Pai S, Ellington A, Aptamers as therapeutics, Nature reviews Drug discovery 9(7) (2010) 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [487].Veleva AN, Nepal DB, Frederick CB, Schwab J, Lockyer P, Yuan H, Lalush DS, Patterson C, Efficient in vivo selection of a novel tumor-associated peptide from a phage display library, Molecules 16(1) (2011) 900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [488].Choi JH, Lee WK, Han SH, Ha S, Ahn SM, Kang JS, Choi YJ, Yun C-H, Identification and characterization of nonapeptide targeting a human B cell lymphoma, Raji, International immunopharmacology 8(6) (2008) 852–858. [DOI] [PubMed] [Google Scholar]
- [489].Wang H, Ma C, Li R, Guo Y, He Y, Wang X, Chen Y, Hou Y, Selection and characterization of colorectal cancer cell-specific peptides, Biotechnology letters 35(5) (2013) 671–677. [DOI] [PubMed] [Google Scholar]
- [490].De J, Chang Y-C, Samli KN, Schisler JC, Newgard CB, Johnston SA, Brown KC, Isolation of a Mycoplasma-specific binding peptide from an unbiased phage-displayed peptide library, Molecular Biosystems 1(2) (2005) 149–157. [DOI] [PubMed] [Google Scholar]
- [491].Hasenbein M, Andersen T, Bizios R, Micropatterned surfaces modified with select peptides promote exclusive interactions with osteoblasts, Biomaterials 23(19) (2002) 3937–3942. [DOI] [PubMed] [Google Scholar]
- [492].Plouffe BD, Njoka DN, Harris J, Liao J, Horick NK, Radisic M, Murthy SK, Peptide-mediated selective adhesion of smooth muscle and endothelial cells in microfluidic shear flow, Langmuir 23(9) (2007) 5050–5055. [DOI] [PubMed] [Google Scholar]
- [493].Plouffe BD, Radisic M, Murthy SK, Microfluidic depletion of endothelial cells, smooth muscle cells, and fibroblasts from heterogeneous suspensions, Lab on a Chip 8(3) (2008) 462–472. [DOI] [PubMed] [Google Scholar]
- [494].Vickers DA, Murthy SK, Receptor expression changes as a basis for endothelial cell identification using microfluidic channels, Lab on a Chip 10(18) (2010) 2380–2386. [DOI] [PubMed] [Google Scholar]
- [495].Nielsen JS, McNagny KM, Novel functions of the CD34 family, Journal of cell science 121(22) (2008) 3683–3692. [DOI] [PubMed] [Google Scholar]
- [496].Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW, AC133, a novel marker for human hematopoietic stem and progenitor cells, Blood 90(12) (1997) 5002–5012. [PubMed] [Google Scholar]
- [497].Sun J, Zhang C, Liu G, Liu H, Zhou C, Lu Y, Zhou C, Yuan L, Li X, A novel mouse CD133 binding-peptide screened by phage display inhibits cancer cell motility in vitro, Clinical & experimental metastasis 29(3) (2012) 185–196. [DOI] [PubMed] [Google Scholar]
- [498].Mehta K, Shahid U, Malavasi F, Human CD38, a cell-surface protein with multiple functions, The FASEB Journal 10(12) (1996) 1408–1417. [DOI] [PubMed] [Google Scholar]
- [499].Ulyanova T, Scott LM, Priestley GV, Jiang Y, Nakamoto B, Koni PA, Papayannopoulou T, VCAM-1 expression in adult hematopoietic and nonhematopoietic cells is controlled by tissue-inductive signals and reflects their developmental origin, Blood 106(1) (2005) 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [500].Shukla S, Langley MA, Singh J, Edgar JM, Mohtashami M, Zúñiga-Pflücker JC, Zandstra PW, Progenitor T-cell differentiation from hematopoietic stem cells using Delta-like-4 and VCAM-1, Nature Methods (2017). [DOI] [PubMed] [Google Scholar]
- [501].Leung K, VHPKQHRGGSKGC-liquid perfluorocarbon nanoparticles, National Center for Biotechnology Information (US), Bethesda (MD), 2004. [PubMed] [Google Scholar]
- [502].Xu W, Zhang S, Zhou Q, Chen W, VHPKQHR peptide modified magnetic mesoporous nanoparticles for MRI detection of atherosclerosis lesions, Artificial cells, nanomedicine, and biotechnology 47(1) (2019) 2440–2448. [DOI] [PubMed] [Google Scholar]
- [503].Lyman S, Brasel K, Rousseau A, Williams D, The flt3 ligand: a hematopoietic stem cell factor whose activities are distinct from steel factor, Stem cells (Dayton, Ohio) 12 (1993) 99–107; discussion 108–10. [PubMed] [Google Scholar]
- [504].Tima S, Okonogi S, Ampasavate C, Pickens C, Berkland C, Anuchapreeda S, Development and characterization of FLT3-specific curcumin-loaded polymeric micelles as a drug delivery system for treating FLT3-overexpressing leukemic cells, Journal of pharmaceutical sciences 105(12) (2016) 3645–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [505].Bacon K, Burroughs M, Blain A, Menegatti S, Rao BM, Screening yeast display libraries against magnetized yeast cell targets enables efficient isolation of membrane protein binders, ACS combinatorial science 21(12) (2019) 817–832. [DOI] [PubMed] [Google Scholar]
- [506].Terstappen L, Huang S, Analysis of bone marrow stem cell, Blood cells 20(1) (1993) 45–61; discussion 61–3. [PubMed] [Google Scholar]
- [507].Kondo M, Weissman IL, Akashi K, Identification of clonogenic common lymphoid progenitors in mouse bone marrow, Cell 91(5) (1997) 661–672. [DOI] [PubMed] [Google Scholar]
- [508].Bhatia M, Bonnet D, Murdoch B, Gan OI, Dick JE, A newly discovered class of human hematopoietic cells with SCID-repopulating activity, Nature medicine 4(9) (1998) 1038–1045. [DOI] [PubMed] [Google Scholar]
- [509].Guo Y, Lübbert M, Engelhardt M, CD34− hematopoietic stem cells: current concepts and controversies, Stem cells 21(1) (2003) 15–20. [DOI] [PubMed] [Google Scholar]
- [510].Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG, Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo, Journal of Experimental Medicine 198(2) (2003) 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [511].Lai AY, Kondo M, Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors, Journal of Experimental Medicine 203(8) (2006) 1867–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [512].Broxmeyer HE, Umbilical cord transplantation: epilogue, Seminars in hematology, Elsevier, 2010, pp. 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [513].Seita J, Weissman IL, Hematopoietic stem cell: self‐renewal versus differentiation, Wiley Interdisciplinary Reviews: Systems Biology and Medicine 2(6) (2010) 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [514].Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC, All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells, Cell stem cell 9(1) (2011) 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [515].Boyer SW, Beaudin AE, Forsberg EC, Mapping differentiation pathways from hematopoietic stem cells using Flk2/Flt3 lineage tracing, Cell Cycle 11(17) (2012) 3180–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [516].Eaves CJ, Hematopoietic stem cells: concepts, definitions, and the new reality, Blood 125(17) (2015) 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [517].Volpe G, Clarke M, Garcìa P, Walton DS, Vegiopoulos A, Del Pozzo W, O’Neill LP, Frampton J, Dumon S, Regulation of the Flt3 Gene in haematopoietic stem and early progenitor cells, PloS one 10(9) (2015) e0138257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [518].Day K, Schneible JD, Young AT, Pozdin VA, Gaffney LS, Prodromou R, Freytes DO, Daniele M, Menegatti S, Cell Labeling with VCAM1-specific Light-Responsive Peptides, Submitted to Biomaterials Science (2020). [Google Scholar]
- [519].Zebger I, Rutloh M, Hoffmann U, Stumpe J, Siesler H, Hvilsted S, Photoorientation of a liquid crystalline polyester with azobenzene side groups. 1. Effects of irradiation with linearly polarized blue light, The Journal of Physical Chemistry A 106(14) (2002) 3454–3462. [Google Scholar]
- [520].Sadovski O, Beharry AA, Zhang F, Woolley GA, Spectral tuning of azobenzene photoswitches for biological applications, Angewandte Chemie International Edition 48(8) (2009) 1484–1486. [DOI] [PubMed] [Google Scholar]
- [521].Beharry AA, Sadovski O, Woolley GA, Azobenzene photoswitching without ultraviolet light, Journal of the American Chemical Society 133(49) (2011) 19684–19687. [DOI] [PubMed] [Google Scholar]
- [522].Samanta S, Beharry AA, Sadovski O, McCormick TM, Babalhavaeji A, Tropepe V, Woolley GA, Photoswitching azo compounds in vivo with red light, Journal of the American Chemical Society 135(26) (2013) 9777–9784. [DOI] [PubMed] [Google Scholar]
- [523].Zhang L, Zhang H, Gao F, Peng H, Ruan Y, Xu Y, Weng W, Host–guest interaction between fluoro-substituted azobenzene derivative and cyclodextrins, RSC Advances 5(16) (2015) 12007–12014. [Google Scholar]
- [524].Hansen MJ, Lerch MM, Szymanski W, Feringa BL, Direct and Versatile Synthesis of Red‐Shifted Azobenzenes, Angewandte Chemie 128(43) (2016) 13712–13716. [DOI] [PubMed] [Google Scholar]
- [525].Ravenhill BJ, Soday L, Houghton J, Antrobus R, Weekes MP, Comprehensive cell surface proteomics defines markers of classical, intermediate and non-classical monocytes, Scientific reports 10(1) (2020) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]