Keywords: AKI, human genetics, molecular genetics
Abstract
Key Points
Two genetic variants in the DISP1-TLR5 gene locus were associated with risk of AKI.
DISP1 and TLR5 were differentially regulated in kidney biopsy tissue from patients with AKI compared with no AKI.
Background
Although common genetic risks for CKD are well established, genetic factors influencing risk for AKI in hospitalized patients are poorly understood.
Methods
We conducted a genome-wide association study in 1369 participants in the Assessment, Serial Evaluation, and Subsequent Sequelae of AKI Study; a multiethnic population of hospitalized participants with and without AKI matched on demographics, comorbidities, and kidney function before hospitalization. We then completed functional annotation of top-performing variants for AKI using single-cell RNA sequencing data from kidney biopsies in 12 patients with AKI and 18 healthy living donors from the Kidney Precision Medicine Project.
Results
No genome-wide significant associations with AKI risk were found in Assessment, Serial Evaluation, and Subsequent Sequelae of AKI (P < 5×10−8). The top two variants with the strongest association with AKI mapped to the dispatched resistance-nodulation-division (RND) transporter family member 1 (DISP1) gene and toll-like receptor 5 (TLR5) gene locus, rs17538288 (odds ratio, 1.55; 95% confidence interval, 1.32 to 182; P = 9.47×10−8) and rs7546189 (odds ratio, 1.53; 95% confidence interval, 1.30 to 1.81; P = 4.60×10−7). In comparison with kidney tissue from healthy living donors, kidney biopsies in patients with AKI showed differential DISP1 expression in proximal tubular epithelial cells (adjusted P = 3.9×10−2) and thick ascending limb of the loop of Henle (adjusted P = 8.7×10−3) and differential TLR5 gene expression in thick ascending limb of the loop of Henle (adjusted P = 4.9×10−30).
Conclusions
AKI is a heterogeneous clinical syndrome with various underlying risk factors, etiologies, and pathophysiology that may limit the identification of genetic variants. Although no variants reached genome-wide significance, we report two variants in the intergenic region between DISP1 and TLR5, suggesting this region as a novel risk for AKI susceptibility.
Introduction
AKI affects approximately 20% of hospitalized patients,1 and 40% of patients admitted to the intensive care unit2 and contributes significantly to poor long-term outcomes.3–5 Despite this public health effect, no effective pharmacotherapy exists for AKI.6 An approach identifying genetic risk factors for AKI could enhance efforts to develop novel preventive and therapeutic strategies. A genetic predisposition for AKI has been suggested by previous candidate gene studies and two prior genome-wide association studies (GWAS).7,8 However, limitations in power due to sample size and analyses completed in predominantly European ancestry (EA) populations may have restricted identification of genetic variants for the development of AKI.9,10 Another limitation of conducting genetic association studies for AKI in hospitalized cohorts is the issue of competing risk.11 Genetic cohorts of hospitalized patients, particularly critically ill patients, can experience in-hospital mortality of 20%–60%8,12 which could prevent the occurrence of organ failure outcomes, such as the development of AKI, that require some period of observation to ascertain. A final limitation is the difficulties in obtaining an outpatient baseline serum creatinine for accurate diagnosis of AKI.13,14 The Kidney Disease Improving Global Outcomes (KDIGO) consensus group defines AKI as an increase in serum creatinine of ≥0.3 mg/dl from the baseline within a 48-hour period or an increase in serum creatinine of ≥50% from the baseline within 7 days.15 This definition relies on an adequate baseline or outpatient measure of kidney function before hospitalization, which is often lacking in hospitalized populations.
To account for issues of timing, severity of AKI, and competing risks, we prospectively enrolled a multiethnic population for genotyping in the Assessment, Serial Evaluation, and Subsequent Sequelae of AKI (ASSESS-AKI) Study.16–18 All ASSESS-AKI participants had a baseline outpatient serum creatinine before hospitalization, and AKI cases were matched to non-AKI controls on the basis of baseline demographics and comorbidities. In addition, all participants enrolled in ASSESS-AKI had to survive the index hospitalization. Thus, ASSESS-AKI improves the precision in the AKI phenotype and minimizes the competing risk of early death making it a unique cohort to study genetic risk factors for the development of AKI. Given that multiethnic cohorts with genomic data similar to ASSESS-AKI currently do not exist we sought other ways to functionally validate our findings. We identified the precise cell types that express candidate genes associated with AKI in ASSESS-AKI and the degree to which these genes were differentially expressed in AKI using a single-cell Ribonucleic acid sequencing (RNAseq) dataset from AKI biopsy tissue obtained through the Kidney Precision Medicine Project (KPMP).19 Some of the results of these studies have been previously reported in abstract form.20
Methods
Study Populations
We performed a GWAS of AKI in 1369 adults in ASSESS-AKI who consented to genetic analyses and for whom genotyping passed quality measures. The full ASSESS-AKI cohort included 1538 hospitalized adults who did or did not experience an episode of AKI and survived to complete a study visit 90 days after discharge.16,21 Adults with AKI were matched to hospitalized adults without AKI on the basis of site, preadmission CKD status, age, prior cardiovascular disease, diabetes mellitus, and preadmission eGFR using the Chronic Kidney Disease Epidemiology Collaboration equation. This cohort is racially/ethnically diverse, with representation from non-Hispanic White, Hispanic/Latino, East Asian, and African American race/ethnicity groups. All study procedures were approved by the local Institutional Review Boards.
AKI Definition
In ASSESS-AKI, AKI cases were defined using modified KDIGO criteria on the basis of an increase of ≥50% or ≥0.3 mg/dl in serum creatinine above a baseline serum creatinine value obtained at an outpatient, nonemergency department measurement between 7 and 365 days before the index admission.21 Urine output was not used to define AKI.
Genotyping and Imputation
In ASSESS-AKI, genotyping was performed in one batch, using the Illumina Multiethnic Global Array platform. Samples with a call rate ≥97% were included in the analyses. No participants were removed for genotyping quality. Two participants were removed for discordance between genetic sex and self-reported sex. Another participant from a pair of participants identified with cryptic relatedness was removed. Genome-wide variant imputation was conducted with EAGLE v2.322 phasing on the Michigan Imputation Server23 using the Haplotype Reference Consortium r1.1 2016.24 We included genetic variants or single-nucleotide polymorphisms (variants) with an imputation quality r2 >0.3, Hardy–Weinberg Equilibrium P > 10−6 in the EA participants, and a minor allele frequency >1% for a final variant count of 8,669,569.
Two variants with the strongest associations were then selected for direct genotyping using RT-PCR methods. Direct genotyping of rs17538288 and rs7546189 was performed with TaqMan assays using the QuantStudio 7 Real-time PCR System (Thermo Fisher). Lymphoblastoid cell lines derived from participants in the 1000 Genomes Project were used as controls. We achieved a genotype call rate of 98.7% (1471/1490) for rs17538288 and 100% (1456/1456) for rs7546189, with complete agreement between duplicates, and expected genotypes for the 1000 genomes samples. The results did not deviate significantly from Hardy–Weinberg Equilibrium.
Statistical Analysis
Patient demographic variables are reported as either mean and standard deviation or median and quartiles. The primary end point for our GWAS was the development of AKI. Odds ratios (ORs) are reported with 95% confidence intervals (CIs). We tested in logistic regression models for AKI using an additive genetic model with the imputed variants while conditioning on the following covariates: participant's age, sex, diabetes, hypertension, study site enrollment, and ten principal components (PCs) of ancestry. In a sensitivity analysis, we tested whether the top variants were associated with severity of kidney injury modeling AKI status as an ordinal variable (0=no AKI, 1=KDIGO Stage 1 AKI, 2=KDIGO Stage 2, and 3=KDIGO Stage 3). Additional post hoc regressions were performed with R statistical software to evaluate the extent to which associations between variants and AKI at a particular locus were independent. We also performed compound heterozygosity analysis.25 In the post hoc tests, the RT-PCR genotype results of the two most associated variants were used to improve genotype accuracy over the imputation-based genotype GWAS of AKI. We additionally rephased these directly genotyped variants using EAGLE (v2.4.1).23 GWAS association tests were completed using PLINK 1.90.26 We performed both an all ancestry GWAS and an analysis limited to participants of EA. Ten PCs of ancestry were generated with the SNPRelate R package v3.7 at an LD pruning threshold of 0.3.27 These PCs were adjusted to control for population stratification. EA was identified on visual inspection of the plotted clusters and selection of the Europeans in the region PC1 <0 and PC2 <0.012.
In Silico Analysis
We sought evidence for differential expression of dispatched RND transporter family member 1 (DISP1) and toll-like receptor 5 (TLR5) using single-cell RNAseq data from the KPMP.28 Single-cell RNAseq was performed in kidney biopsy tissue from 12 patients with AKI and living donor biopsy tissue from 18 healthy donors. Living donor biopsies were obtained before perfusion and before placement in the recipient. The characteristics of the living donor cohort have been described previously.29 Methods for tissue processing and RNAseq analyses have been previously published.30 We compared DISP1 and TLR5 expression between AKI and no AKI biopsy tissue on the basis of a Bonferroni-corrected P value adjusted for all the features in the analysis. We also tested whether the variants identified in the intergenic region between DISP1 and TLR5 were expression quantitative trait loci (eQTL) in the genotype-tissue Expression (GTEx) Portal.31 In GTEx, we queried data from 73 participants for whom the results from kidney tissue existed. We also queried the NephQTL database, which includes 187 human kidney samples from patients with nephrotic syndrome in the Nephrotic Syndrome Study Network cohort.32 We looked for overlap between the variants with the strongest associations and epigenetic regulatory annotations in the University of California Santa Cruz (UCSC) Genome Browser.33
Results
Characteristics of Patient Populations
Figure 1 presents the selection of the analytic cohort. We identified 637 cases and 732 controls from the ASSESS-AKI population that consented to genome-wide genotyping. Table 1 summarizes the characteristics of the AKI and non-AKI participants. The participants with AKI had higher rates of inpatient incident dialysis (3.6% versus 0%) and lower eGFR at 3 months (66.7 versus 73.9 ml/min per 1.73 m2). Principal Components Analysis (PCA) analysis identified 500 cases and 612 controls as EA and 137 cases and 120 controls who did not cluster with EA participants. See Supplemental Figure 1 for a plot of the PCA and self-reported ancestry.
Figure 1.
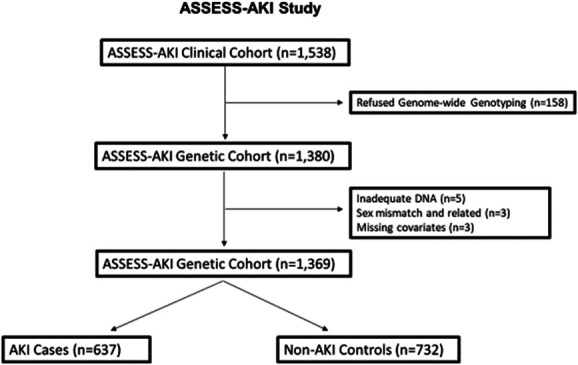
Patient flow. One thousand three hundred and sixty-nine participants were included in ASSESS-AKI genetic cohort with 637 AKI cases and 732 non-AKI controls. ASSESS-AKI, Assessment, Serial Evaluation, and Subsequent Sequelae of AKI.
Table 1.
Patient characteristics in Assessment, Serial Evaluation, and Subsequent Sequelae of AKI cohort
| Characteristics | AKI (n=637) | Non-AKI (n=732) |
|---|---|---|
| Center, n (%) | ||
| Vanderbilt University | 173 (27) | 199 (27) |
| Kaiser permanente, California | 133 (21) | 181 (25) |
| Yale | 133 (21) | 139 (19) |
| University of Washington, Seattle | 198 (31) | 213 (29) |
| Female, n (%) | 209 (33) | 308 (42) |
| Age, yr, mean (SD) | 68.5 (13.1) | 70.0 (13.2) |
| Race, n (%) | ||
| White | 512 (80) | 619 (84) |
| Black | 87 (14) | 71 (10) |
| Other | 38 (6) | 42 (6) |
| Comorbidities, n (%) | ||
| Hypertension | 499 (78) | 501 (68) |
| CKD | 245 (39) | 275 (38) |
| Diabetes | 301 (49) | 228 (33) |
| AKI Risk Factor, n (%) | ||
| Major surgical procedure | 255 (40) | 380 (52) |
| Acute heart failure | 52 (8.2) | 14 (1.9) |
| Acute myocardial infarction | 26 (4.1) | 19 (2.6) |
| Sepsis | 107 (17) | 26 (4) |
| Treated in ICU during index admission, n (%) | 466 (73) | 442 (60) |
| Baseline eGFR, ml/min per 1.73 m2, mean (SD) | 67.9 (25.7) | 71.7 (24.4) |
| KDIGO AKI Stage, n (%) | ||
| Stage 1 | 461 (72.3) | — |
| Stage 2 | 95 (14.9) | — |
| Stage 3 | 81 (12.7) | — |
| Outcomes | ||
| Acute dialysis, n (%) | 23 (3.6) | 0 (0) |
| 3-mo eGFR, ml/min per 1.73 m2, mean (SD) | 66.7 (26.9) | 73.9 (24.4) |
ICU, intensive care unit; KDIGO, Kidney Disease Improving Global Outcomes.
GWAS Analysis in ASSESS-AKI
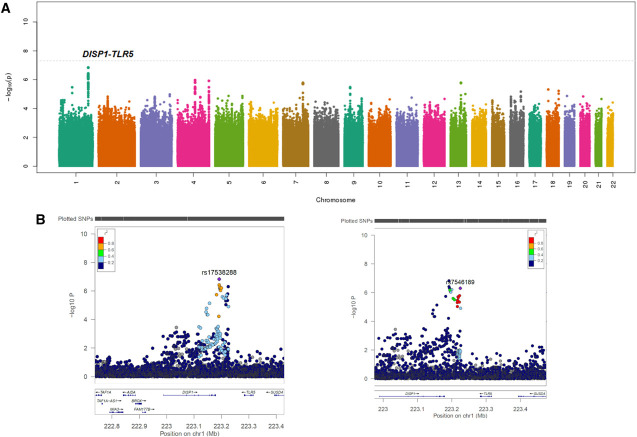
Association testing revealed no variants that satisfied the Bonferroni-corrected genome-wide threshold of P < 5×10−8. The lambda genomic inflation factor on the basis of a median chi-square was estimated at approximately 1.01 in the all ancestry and the EA-stratified analyses, indicating negligible variation in population structure between cases and controls. The quantile–quantile plot showed overall adherence to expected P values (Supplemental Figure 2). Association testing demonstrated 45 variants at nine loci for AKI with P < 5×10−6 in the full cohort with all ancestries (Figure 2A for Manhattan Plot and Supplemental Excel File 1). Our P value threshold for suggestive association was selected based on prior research that a threshold in this range might allow discovery of disease causing variants in smaller sample sizes, while limiting the number of false positive results.34,35 Representative variants from each loci were selected based on lowest P value and independence of linkage disequilibrium (LD) (r2<0.3). Of note, there were two variants selected in the intergenic region between DISP1 and TLR5 genes that showed strong association with AKI and low LD (r2=0.24 and D′=0.68 in all ancestry and r2=0.22 and D′=0.59 in EA). Of the nine top variants at nine different loci, five of the variants were associated with an increased risk of AKI, while four of the nine variants were associated with a decreased risk (Table 2). All nine of the variants were also associated with the severity of AKI (Supplemental Table 1). We also evaluated the association of the top two variants with Stage 1 AKI to no AKI and severe AKI (Stages 2 and 3) compared with no AKI. We found that both the top two variants were nominally associated with severity of AKI (Supplemental Table 2).
Figure 2.
Manhattan plots generated from genome-wide association study analysis of the AKI cases and controls. Plots include (A) association results for imputed single-nucleotide polymorphisms (variants) with AKI in the discovery cohort. (B) Genomic locus for the DISP1-TLR5 top hits, rs17538288 and rs7546189 at higher magnification. LD, r2, is indicated relative to the variant with lowest P value (purple marker) by colored scale. Notice that the top two DISP1-TLR5 hits are not in LD with each other (r2=0.24). Genes located near each locus are displayed in the bottom panels. DISP1, dispatched resistance-nodulation-division (RND) transporter family member 1; LD, linkage disequilibrium; TLR5, toll-like receptor 5.
Table 2.
Summary of suggestive imputed variant associations with AKI in Assessment, Serial Evaluation, and Subsequent Sequelae of AKI for joint all ancestry and stratified to European American analysis
| Variant | Chr | Position | Gene | MA | MAF | EA MAF | Genotyped | Imputation Quality (r2) | All Ancestry OR (95% CI) | All Ancestry Pa |
EA OR (95% CI) |
EA Pa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17538288 | 1 | 223192017 | DISP1-TLR5 | A | 0.439 | 0.429 | Imputed | 0.97 | 1.54 (1.31 to 1.81) | 1.47E-07 | 1.50 (1.25 to 1.79) | 8.49E-06 |
| rs7546189 | 1 | 223224864 | DISP1-TLR5 | T | 0.347 | 0.349 | Imputed | 0.96 | 1.54 (1.3 to 1.82) | 4.85E-07 | 1.62 (1.35 to 1.96) | 3.39E-07 |
| rs80052123 | 1 | 89493224 | GBP3 (5′) | A | 0.143 | 0.152 | Direct | 1.0 | 1.71 (1.36 to 2.15) | 3.41E-06 | 1.67 (1.31 to 2.13) | 3.95E-05 |
| rs6533107 | 4 | 104706914 | TACR3 5′ | A | 0.352 | 0.374 | Imputed | 0.97 | 0.66 (0.56 to 0.78) | 1.09E-06 | 0.65 (0.54 to 0.78) | 4.38E-06 |
| rs9998646 | 4 | 188592759 | LINC02492 | C | 0.441 | 0.447 | Imputed | 0.99 | 0.68 (0.59 to 0.80) | 1.22E-06 | 0.67 (0.56 to 0.79) | 4.10E-06 |
| rs72607731 | 7 | 124916848 | LOC101928283 | T | 0.237 | 0.259 | Imputed | 0.97 | 1.59 (1.32 to 1.92) | 1.65E-06 | 1.53 (1.25 to 1.88) | 3.12E-05 |
| rs1368999 | 9 | 28879870 | LINGO2 | C | 0.492 | 0.54 | Imputed | 0.98 | 0.67 (0.57 to 0.80) | 3.32E-06 | 0.63 (0.53 to 0.76) | 6.10E-07 |
| rs4414368 | 13 | 83974202 | Gene Desert | T | 0.178 | 0.123 | Imputed | 0.99 | 0.58 (0.46 to 0.72) | 1.64E-06 | 0.62 (0.47 to 0.82) | 0.0006643 |
| rs9945894 | 18 | 9083721 | NDUFV2 5′ | C | 0.077 | 0.075 | Imputed | 0.98 | 2.03 (1.50 to 2.75) | 4.83E-06 | 1.90 (1.36 to 2.65) | 0.000163 |
variant, single-nucleotide polymorphism; Chr, chromosome; MA, minor allele; MAF, minor allele frequency; EA, European American; OR, odds ratio; CI, confidence interval; DISP1, dispatched resistance-nodulation-division (RND) transporter family member 1; TLR5, toll-like receptor 5.
P value and odds ratio after adjustment for patient's age, sex, diabetes, hypertension, enrollment site, and first ten principal components of ancestry in cases and controls.
The variant demonstrating the strongest association with AKI was rs17538288 which is a noncoding intergenic variant between DISP1 and TLR5 genes. The minor allele of this variant is associated with an increased risk of AKI (OR, 1.54; 95% CI, 1.30 to 1.81; P = 1.47×10−7). The next strongest association was observed with rs7546189, which is also in the intergenic region between DISP1 and TLR5 and is 33 kilobases from rs17538288. Variant rs7546189 is associated with an increased risk of AKI (OR, 1.54; 95% CI, 1.3 to 1.82; P = 4.85×10−7). When we restricted participants to EA only (500 cases and 612 controls), rs17538288 (OR, 1.50; 95% CI, 1.25 to 1.79; P = 8.48×10−6) and rs7546189 (OR, 1.62; 95% CI, 1.35 to 1.96; P = 3.39×10−7) remained strongly associated with AKI. In the non-EA participants (137 cases, 120 controls), rs17538288 remained nominally significant (OR, 2.06; 95% CI, 1.34 to 3.22; P = 0.0012) and rs7546189 did not (OR, 1.29; 95% CI, 0.81 to 1.87; P = 0.33).
DISP1-TLR5 Variants Direct Genotyping Concordance with Imputation
Both top variants were imputed on the Multiethnic Global Array chip, and we directly genotyped rs17538288 and rs7546189 via Taqman-based RT-PCR to validate the imputation results. Direct genotypes of rs17538288 were highly concordant with the imputed results (1314/1353; approximately 97.12% concordance). Direct genotypes of rs7546189 were also highly concordant with the imputed genotypes (1289/1334; approximately 96.62% concordance). We updated the discordant imputed genotypes with the RT-PCR results for post hoc analyses. The resulting association statistics improved slightly for rs17538288 (OR, 1.55; 95% CI, 1.32 to 1.82; P = 9.47×10−8) and remained similar for rs7546189 (OR, 1.53; 95% CI, 1.30 to 1.81; P = 4.60×10−7).
Compound Heterozygosity and Variant Association Independence Analysis for Haplotypes Near the DISP1-TLR5 Genes
To identify the allele burden association of these two variants (rs17538288 and rs7546189), phased haplotype analysis showed that all four possible two-variant allele haplotype combinations are commonly present (Supplemental Tables 3 and 4). Owing to this, we tested whether the combination of these two variants in a compound heterozygosity association analysis strengthens estimates for AKI. Considering the directly genotyped data, we found that logistic regression tests modeling the combined burden of minor alleles for each variant showed stronger associations for AKI (P value = 1.77×10−9) than each variant independently (Table 3). In addition, testing the residuals of one variant's association with AKI with the other variant led both variants to remain significant (P values = 0.001 and 0.0002) (Supplemental Figure 3 and Supplemental Table 5). We also evaluated the association of these two variants with the risk of AKI stratified by whether patients had surgery as their primary risk factor for AKI. We found that both variants maintained nominal significance for the outcome of AKI stratified by surgery (Supplemental Table 6).
Table 3.
Total burden model of minor alleles for dispatched RND transporter family member 1 toll-like receptor 5 variants, rs17538288 >A and rs7546189 >T.
| Number of Minor Alleles, (Number of Patients) | 0 (n=318) | 1 (n=318) | 2 (n=449) | 3 (n=178) | 4 (n=100) | Risk of AKI (OR, 95% CI) | P Value |
|---|---|---|---|---|---|---|---|
| AKI, n (%) | 119 (37%) | 136 (43%) | 213 (47%) | 102 (57%) | 67 (67%) | 1.34 (1.22 to 1.48) | 1.77×10−9 |
| No AKI, n (%) | 205 (63%) | 182 (57%) | 236 (53%) | 76 (43%) | 33 (33%) | — | — |
Zero designates homozygous major allele for both variants, and four designates homozygous minor allele for both variant. RND, resistance-nodulation-division; OR, odds ratio; CI, confidence interval.
Functional Annotation of Top-Performing Variants from ASSESS
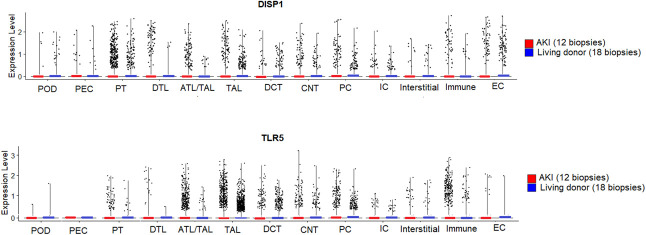
Demographics and kidney function for the cohort of healthy living donor (n=18) and AKI (n=12) participants can be found in the supplement (Supplemental Table 7). Examination of single-cell RNA sequencing analyses from kidney biopsies completed in hospitalized patients with AKI through the KPMP19 initiative demonstrated that DISP1 and TLR5 were expressed in multiple kidney cell types, including proximal tubular epithelial cells and thick ascending limb of the loop of Henle (TAL) (Figure 3). In comparison with kidney tissue from healthy living donor, AKI biopsies showed increased DISP1 expression in proximal tubular epithelial cells, the primary site of injury in AKI (adjusted P = 3.9×10−2) and TAL (adjusted P = 8.7×10−3) (Supplemental Table 7). In addition, AKI biopsies showed decreased expression of TLR5 in TAL (adjusted P = 4.9×10−30) (Supplemental Table 8).
Figure 3.
Single-cell RNA sequencing in living donor (LD) (n=18) and AKI (n=12) kidney biopsy specimens demonstrates DISP1 and TLR5 gene expression in multiple cell lines. DISP1 gene expression is significantly upregulated in PT (adjusted P = 3.9×10−2) and TAL cell types (adjusted P = 8.7×10−3) in AKI compared with no AKI, while TLR5 expression is downregulated in TAL cell types (adjusted P = 4.9×10−30) in AKI compared with no AKI. ATL, ascending thin limb of the loop of Henle; CNT, connecting tubule; DCT, distal convoluted tubule; DISP1, dispatched RND transporter family member 1; DTL, descending thin limb of the loop of Henle; EC, endothelial cells; IC, intercalated cells; PC, principal cells; POD, podocytes; PT, proximal tubular epithelial cells and 1–3 segments; TAL, thick ascending limb of the loop of Henle; TLR5, toll-like receptor 5.
Examination of the GTEx and NephQTL also revealed that the two variants, rs17538288 and rs7546189, were eQTLs for DISP1 gene expression in cardiac tissue (GTEx, P < 0.0001, normalized effect size=0.15 and 0.16, respectively) and decreased HHIPL2 expression in tubulointerstitium kidney tissue (NephQTL, rs17538288, P = 0.048, β=−0.24). The minor allele of each variant was associated with increased DISP1 expression. Increased DISP1 expression was associated with AKI in the KPMP single-cell RNAseq data. By contrast, we found that the minor allele of both variants was associated (P < 3×10−8, normalized effect size=−0.24 and −0.29, respectively) with decreased TLR5 expression in GTEx in cardiac tissue. NephQTL did not show eQTL evidence for either variant with DISP1 or TLR5 in N=166 tubulointerstitium kidney tissue participants. Decreased TLR5 expression was associated with AKI in the single-cell RNAseq KPMP analyses (Supplemental Figures 4 and 5). Further inspection of variants in the UCSC Genome Browser33 revealed epigenetic regulatory functional annotations in multiple tissues and cell lines. The results can be found in the Supplemental Online text (Supplemental Figure 6 and Supplemental Table 9).
Replication of Variants from Two Prior AKI GWAS Studies
Review of two previous reported GWAS of AKI7,8 failed to reveal any shared variants or genetic loci between the literature and the top-performing variants in ASSESS-AKI. None of the previously reported 22 variants were associated with the development of AKI in ASSESS-AKI (nominal P < 0.05) with a similar direction of effect. Because the prior two AKI GWAS studies restricted to persons of EA, we tested the association of prior variants with AKI in persons of EA in ASSESS-AKI and were again unable to detect these previously reported associations (Supplemental Table 10).
Discussion
We identified nine variants with associations at a level suggestive of being susceptibility loci for AKI risk. The two variants most strongly associated with AKI mapped to the DISP1-TLR5 locus. Both variants were associated with an increased risk of AKI that reached genome-wide significance in a post hoc compound heterozygous analysis. Of note, the minor alleles of both variants were eQTLs for DISP1, with the minor allele of each variant associated with increased DISP1 expression in GTEx. Increased DISP1 gene expression was present in kidney biopsies from patients with AKI compared with healthy donors. The minor alleles of both variants were also eQTLs for TLR5 and were associated with reduced TLR5 expression in GTEx. Reduced TLR5 gene expression was present in kidney biopsies from patients with AKI compared with healthy donors. Thus, the direction of variant-modulated gene expression and the relationship to risk for AKI may be in opposite directions for the two proximal genes with increased DISP1 expression and decreased TLR5 expression associated with risk for AKI.
The gene DISP1 is involved in sonic hedgehog signaling, and microdeletions of this gene have been associated with developmental delay and dysmorphic features.36 In experimental models, sonic hedgehog signaling can accelerate fibrosis postkidney injury and contribute to the development of CKD after AKI.37,38 The gene TLR5 plays a fundamental role in pathogen recognition and activation of innate immune responses, and TLR5 is highly expressed in the kidneys. Investigators demonstrated that a TLR5 agonist attenuated kidney injury in a murine model of radiation-induced renal failure and ischemia/reperfusion-induced renal failure through decreased accumulation of reactive oxygen species.39,40 This background knowledge supports that the variants, rs17538288 and rs7546189, may influence the risk of AKI through DISP1 and/or TLR5 signaling. Because these two variants have low correlation and because the minor allele frequency is approximately 10% different, we completed compound heterozygosity analyses. In these analyses, we demonstrated that the combination of variants strengthens the association with AKI. GTEx data show increased expression of DISP1 in cardiac tissue and decreased expression of TLR5 in cardiac tissue in association with rs17538288 and rs7546189. The KPMP single-cell RNAseq results additionally support a differential role for DISP1 and TLR5 with risk for AKI. In kidney biopsies from patients with AKI, DISP1 expression was significantly upregulated, whereas TLR5 expression was reduced compared with healthy donor biopsies. We hypothesize that upregulation of DISP1 may lead to hedgehog-mediated effects on kidney tubule regeneration and healing and that downregulation of TLR5 leads to augmented kidney injury. We also found that the variants identified were distinct from previously reported variants in the TLR5 gene that were associated with pneumonia susceptibility.41–43
Participants in the ASSESS-AKI Study comprise a multiethnic population, and our primary discovery analysis was completed in all persons who agreed to genetic testing in ASSESS-AKI. In sensitivity analyses, we evaluated differences in variant AKI risk on the basis of ancestry. Compared with the all ancestry analysis, we found that rs17538288 had an attenuated risk for AKI in the EA-only population, while the risk of AKI was higher for rs7546189 in the EA-only population. These findings suggest that genetic ancestry may influence the risk of AKI, a complex polygenic trait.44 Further studies are necessary to determine whether ancestry is a surrogate for unmeasured or unknown environmental exposures that may influence the risk of AKI.45–47
Two prior AKI GWAS studies have been published that have only included patients of EA and mostly postcardiac surgery.7,8 We were unable to replicate prior published variants associated with AKI, and this is a challenge in the field.48,49 Potential reasons for the lack of replication may be due to the heterogeneity in the AKI clinical syndrome and lack of power due to small sample size.50,51 Combining patients with different underlying risk, pathophysiology, and outcomes may dilute the signal to identify individual variants. For example, ASSESS-AKI included a general hospitalized population with a distribution of various AKI risk factors, such as sepsis, surgery, and medications.
Strengths of ASSESS-AKI include the enrollment across multiple centers, the inclusion of participants with diverse ancestry, and the prospective matched cohort study design of the facilitated study of genomic risk of AKI independent of several matching covariates. Second, ASSESS-AKI included a general hospital population at risk for AKI with a mandated baseline serum creatinine measurement before hospitalization. Often in hospitalized patients, an outpatient baseline serum creatinine value is lacking, and thus, the severity and timing of AKI are unknown. Misclassification of AKI status due to a lack of a baseline serum creatinine could dilute potential statistical signals. For example, if a patient carrying a high-risk genetic variant presents to the hospital with AKI but is unrecognized due to a lack of an outpatient baseline, then this patient may be classified as a control, attenuating any potential association signal.
We acknowledge a number of limitations. The main limitation is the sample size, limiting the power to detect variants of small effect sizes or lower population frequencies. Another limitation is that we lacked a second cohort with similar inclusion criteria and genotyping to test replication of top variants.
In summary, we describe the results of a GWAS of AKI with case–control matched participants designed to reduce heterogeneity. No variants reached genome-wide significance. We report two novel variants adjacent to the DISP1 and TLR5 genes suggestively associated with AKI susceptibility. Both variants warrant further study on the basis of post hoc independence tests, lack of LD, and the increased strength of association when these two DISP1-TLR5 regulatory variants are considered as compound heterozygotes. Independent evaluation of NephQTL, GTEx, and KPMP single-cell RNAseq data of kidney biopsies supports the interpretation of these variants as being involved in the etiology of AKI. Other independent studies should aim to validate our findings, including functional studies to evaluate the AKI loci reported.
Disclosures
P.K. Bhatraju reports the following: Research Funding: Roche Diagnostics. V.M. Chinchilli reports the following: Advisory or Leadership Role: Allergan; Astra-Zeneca; Biohaven; Jannsen; Regeneron; Sanofi. S.G. Coca reports the following: Consultancy: 3ive, Axon, Bayer, Boehringer Ingelheim, Nuwellis, Renalytix, Reprieve Cardiovascular, Takeda, Vifor; Ownership Interest: pulseData, Renalytix; Research Funding: ProKidney, Renal Research Institute, Renalytix, XORTX; Patents or Royalties: Renalytix; and Other Interests or Relationships: Associate Editor for Kidney360, Editorial Boards of JASN, CJASN, Kidney International. A.X. Garg reports the following: Research Funding: Astellas, Baxter; Advisory or Leadership Role: Currently on the Editorial Boards of Kidney Int and AJKD; and Other Interests or Relationships: Served as the Medical Lead Role to Improve Access to Kidney Transplantation and Living Kidney Donation for the Ontario Renal Network (government funded agency located within Ontario Health). This position ended October 2022. A.S. Go reports the following: Research Funding: Amarin Pharmaceuticals, Bristol MeyersSquibb, CSL Behring, Janssen Research and Development, Novartis, Pfizer. J. Himmelfarb reports the following: Consultancy: Maze Therapeutics; Ownership Interest: Kuleana Technology, Inc.; Research Funding: Aurinia Pharmaceuticals; Honoraria: various academic institutions for invited lectures; Patents or Royalties: Patents held and patents pending owned by the University of Washington; Advisory or Leadership Role: CJASN (Ed board); BMC Medicine (Ed. Board), Nature Reviews Nephrology (Advisory Board); and Other Interests or Relationships: Research grant support from Northwest Kidney Centers; Founder, CEO, President and equity holder, Kuleana Technology, Inc. T.A. Ikizler reports the following: Consultancy: Fresenius-Kabi, Nestle; Research Funding: NIDDK; Veterans Affairs; Honoraria: Fresenius-Kabi, Nestle; and Advisory or Leadership Role: Kidney International. G.P. Jarvik reports the following: Ownership Interest: literally hundreds managed by a professional manager and not chosen by me or my spouse; and Advisory or Leadership Role: I was the 2022 Past President of the American Society of Human Genetics. J.S. Kaufman reports the following: Consultancy: National Kidney Foundation; Otsuka Pharmaceutical; Ownership Interest: Amgen; Advisory or Leadership Role: Digestive and Kidney Diseases, National Institute of Diabetes, paid Steering Committee Chair; and Other Interests or Relationships: Associate Editor, American Journal of Kidney Disease. P.L. Kimmel reports the following: Employer: National Institute of Diabetes and Digestive Kidney Diseases (NIDDK); Ownership Interest: As a Federal Employee at NIDDK, my holdings are reviewed each year for potential conflict of interest. At this time my only stock holding related in any fashion to health care is CVS; Patents or Royalties: Elsevier: Royalties for co-editing Chronic Renal Disease and Psychosocial Aspects of Chronic Kidney Disease; Advisory or Leadership Role: Unpaid member of Board of Directors of Academy of Medicine of Washington, DC; and Other Interests or Relationships: Co-Editor, Chronic Renal Disease Academic Press; Co-Editor, Psychosocial Aspects of Chronic Kidney Disease. Academic Press; Royalties. K. Kiryluk reports the following: Consultancy: Calvariate, HiBio; and Research Funding: Aevi Genomics, AstraZeneca, Bioporto, Vanda, Visterra. S. Menez reports the following: Research Funding: RenalytixAI. A.S. Naik reports the following: Consultancy: CareDX—External Advisor; and Honoraria: CareDx. P.M. Palevsky reports the following: Consultancy: Janssen Research & Development, LLC; and Advisory or Leadership Role: National Kidney Foundation: Past President, Member, Scientific Advisory Board; Renal Physicians Association: Member, Quality, Safety and Accountability Committee; Quality Insights Renal Network 4: Chair, Medical Review Board; UpToDate: Section Editor, RAcute Kidney Injury; Journal of Intensive Care Medicine: Member, Editorial Board. M.R. Palmer reports the following: Employer: Natera, Inc.; and Ownership Interest: Natera, Inc. C.R. Parikh reports the following: Consultancy: Genfit Biopharmaceutical Company; Ownership Interest: Renaltix AI Research Funding: National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); National Heart, Lung and Blood Institute (NHLBI); and Advisory or Leadership Role: Genfit Biopharmaceutical Company; Renalytix. J.A. Schaub reports the following: Consultancy: Cook Biotech (Spouse); Nuvira (Spouse); Uptodate (Spouse). E.D. Siew reports the following: Ownership Interest: Amazon stock, Apple stock; Patents or Royalties: Author for UptoDate (royalties) and Advisory or Leadership Role: Editorial board of CJASN. A. Srivastava reports the following: Consultancy: CVS Caremark; Tate & Latham (Medicolegal consulting); and Honoraria: AstraZeneca; Bayer; FNIH; Horizon Therapeutics PLC. I.B. Stanaway reports the following: Ownership Interest: I have an LLC, Byrell Systems, registered in Washington State that is solely owned by me, Dr. Ian Byrell Stanaway.; and Other Interests or Relationships: ASN Member. F.P. Wilson reports the following: Consultancy: Translational Catalyst, LLC; Ownership Interest: Owner of Efference, LLC; Research Funding: Amgen; Boeringher-Ingelheim; Vifor, Whoop; Advisory or Leadership Role: Editorial Board—American Journal of Kidney Disease; Editorial Board—Clinical Journal of the American Society of Nephrology; and Other Interests or Relationships: Medical columnist-Medscape. L.B. Ware reports the following: Consultancy: Akebia, Global Blood Therapeutics, Santhera; Ownership Interest: Virtuoso Surgical; and Research Funding: Boehringer Ingelheim; Genentech. M.M. Wurfel reports the following: Consultancy: Roche Diagnostics; and Research Funding: Roche. All remaining authors have nothing to disclose.
Supplementary Material
Acknowledgments
Work was performed at the University of Washington, Seattle, Washington. The funding sources had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
J.H. and M.M.W. contributed equally to this work.
Funding
This study was supported by the supplemental American Recovery and Reinvestment Act funds and research grants U01DK082223, U01DK082185, U01DK082192, U01DK082183, U01DK084012 and R01DK098233 from the NIDDK of the National Institutes of Health (NIH), US Department of Health and Human Services. Dr. Bhatraju is supported by grant K23DK116967 from the NIH. Dr. Coca is also supported by grants R01DK106085 and U01DK106962 from the NIH. Dr Parikh is also supported by grant R01HL085757 from the NIH. Dr Garg is also supported by the Dr. Adam Linton Chair in Kidney Health Analytics and a Clinician Investigator Award from the Canadian Institutes of Health Research. Drs. Hsu, Go, and Liu are also supported by grants R01DK101507, U01DK060902, R01DK101507, K24DK113381, and K24DK92291 from the NIH. Dr. Siew is also supported by grant 5K23DK088964 from the NIH. Dr. Himmelfarb is also supported by grants U2C DK114886, UG3TR002158, and U01DK099923 from the NIH. Dr. Menez is supported by grant K23DK128538 from the NIH. Dr. Palevsky is supported by grant 1UH3 DK114861 from the NIH. Dr. Naik is supported by K23DK125529. Dr. Kiryluk is supported by NIH grants R01-LM013061, 2U01-HG008680, and RC2-DK116690.
The KPMP is funded by the following grants from the NIDDK: U2C DK114886, UH3DK114861, UH3DK114866, UH3DK114870, UH3DK114908, UH3DK114915, UH3DK114926, UH3DK114907, UH3DK114920, UH3DK114923, UH3DK114933, and UH3DK114937.
Author Contributions
Conceptualization: Pavan K. Bhatraju, Gail P. Jarvik, Jonathan Himmelfarb, Mark M. Wurfel.
Formal analysis: Pavan K. Bhatraju, Ian B. Stanaway, Melody R. Palmer, Rajasree Menon, Jennifer A. Schaub.
Funding acquisition: Steve G. Coca, Alan S. Go, T. Alp Ikizler, James S. Kaufman, Paul L. Kimmel, Jonathan Himmelfarb, Mark M. Wurfel.
Investigation: All authors
Methodology: Pavan K. Bhatraju, Ian B. Stanaway, Melody R. Palmer, Rajasree Menon, Jennifer A. Schaub, Gail P. Jarvik, Sana S. Sakr, Jonathan Himmelfarb, Mark M. Wurfel
Software: Ian B. Stanaway, Melody R. Palmer, Rajasree Menon
Validation: Krzysztof Kiryluk, Steven Menez, Abhijit S. Naik, Paul M. Palevsky, Anand Srivastava, F. Perry Wilson, Jonathan Himmelfarb.
Visualization: Ian B. Stanaway, Melody R. Palmer, Rajasree Menon
Writing - original draft: Pavan K. Bhatraju.
Writing - review & editing: All authors.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/KN9/A363.
Additional eMethods and eResults.
Supplemental Table 1. Ordinal Regression for KDIGO AKI Severity.
Supplemental Table 2. Association of top two variants and severity of AKI comparing Stage 1 AKI with no AKI and comparing Stage 2 and 3 AKI with no AKI.
Supplemental Table 3. Top Two SNPs adjacent DISP1-TLR5, Linkage Disequilibrium Information.
Supplemental Table 4. Case–control counts by DISP1-TLR5 variants.
Supplemental Table 5. DISP1-TLR5 Top Two Variants Post Hoc Regression Tests and Condition Adjusted Results with updated RT-PCR validation genotypes.
Supplemental Table 6. Demographics and kidney function in living donor and AKI participants who underwent kidney biopsy for single-cell RNAseq analyses.
Supplemental Table 7. Differential DISP1 gene expression between living donor and AKI biopsies.
Supplemental Table 8. Differential TLR5 gene expression between living donor and AKI biopsies.
Supplemental Table 9. Epigenetic Regulatory Information and Cell types from the UCSC Browser.
Supplemental Table 10. European American ASSESS-AKI GWAS Evaluation of SNPs of interest from two prior AKI GWAS.
Supplemental File 1. The 45 associated SNPs with p<5x10−6: ASSESS_AKI_GWAS_regression_results.xlsx.
Supplemental File 2. GTEx: Top-Performing SNPs from ASSESS-AKI: GTEx_AKI_GWAS_hits_eQTLs.xlsx.
Supplemental File 3. NephQTL: Top-Performing SNPs from ASSESS-AKI: NephQTL_AKI_GWAS_hits_eQTLs.xlsx.
Supplemental Figure 1. PCA plot in ASSESS.
Supplemental Figure 2. Quantile–quantile AKI GWAS plot in ASSESS.
Supplemental Figure 3. Barplots of the top two DISP1-TLR5 SNPs.
Supplemental Figure 4. DISP1-TLR5 SNPs GTEx gene expression.
Supplemental Figure 5. DISP1 rs17538288 HHIPL2 NephQTL gene expression.
Supplemental Figure 6. UCSC Genome Browser Screenshot of DISP1-TLR5 locus with the GeneHancer GH01J223018 epigenetic regulatory site annotated.
References
- 1.Wang HE, Muntner P, Chertow GM, Warnock DG. Acute kidney injury and mortality in hospitalized patients. Am J Nephrol. 2012;35(4):349–355. doi: 10.1159/000337487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clermont G, Acker CG, Angus DC, Sirio CA, Pinsky MR, Johnson JP. Renal failure in the ICU: comparison of the impact of acute renal failure and end-stage renal disease on ICU outcomes. Kidney Int. 2002;62(3):986–996. doi: 10.1046/j.1523-1755.2002.00509.x [DOI] [PubMed] [Google Scholar]
- 3.Ishani A, Xue JL, Himmelfarb J, Eggers PW, Kimmel PL, Molitoris BA. Acute kidney injury increases risk of ESRD among elderly. J Am Soc Nephrol. 2009;20(1):223–228. doi: 10.1681/ASN.2007080837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coca SG, Yusuf B, Shlipak MG, Garg AX, Parikh CR. Long-term risk of mortality and other adverse outcomes after acute kidney injury: a systematic review and meta-analysis. Am J Kidney Dis. 2009;53(6):961–973. doi: 10.1053/j.ajkd.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371(1):58–66. doi: 10.1056/nejmra1214243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellomo R, Kellum JA, Ronco C. Acute kidney injury. Lancet. 2012;380(9843):756–766. doi: 10.1016/s0140-6736(11)61454-2 [DOI] [PubMed] [Google Scholar]
- 7.Stafford-Smith M, Li Y-J, Mathew JP, Li Y-W, Ji Y, Phillips-Bute BG. Genome-wide association study of acute kidney injury after coronary bypass graft surgery identifies susceptibility loci. Kidney Int. 2015;88(4):823–832. doi: 10.1038/ki.2015.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Lu Q, Cheng Y, Belcher JM, Siew ED, Leaf DE. A genome-wide association study to identify single-nucleotide polymorphisms for acute kidney injury. Am J Respir Crit Care Med. 2017;195(4):482–490. doi: 10.1164/rccm.201603-0518oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. doi: 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. Plos Biol. 2013;11(9):e1001661. doi: 10.1371/journal.pbio.1001661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noordzij M, Leffondré K, van Stralen KJ, Zoccali C, Dekker FW, Jager KJ. When do we need competing risks methods for survival analysis in nephrology? Nephrol Dial Transplant. 2013;28(11):2670–2677. doi: 10.1093/ndt/gft355 [DOI] [PubMed] [Google Scholar]
- 12.Reilly JP, Wang F, Jones TK, Palakshappa JA, Anderson BJ, Shashaty MGS. Plasma angiopoietin-2 as a potential causal marker in sepsis-associated ARDS development: evidence from Mendelian randomization and mediation analysis. Intensive Care Med. 2018;44(11):1849–1858. doi: 10.1007/s00134-018-5328-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilander LM, Vaara ST, Kaunisto MA, Pettilä V, The Finnaki Study Group. Common inflammation-related candidate gene variants and acute kidney injury in 2647 critically ill Finnish patients. J Clin Med. 2019;8(3):342. doi: 10.3390/jcm8030342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siew ED, Matheny ME, Ikizler TA, Lewis JB, Miller RA, Waitman LR. Commonly used surrogates for baseline renal function affect the classification and prognosis of acute kidney injury. Kidney Int. 2010;77(6):536–542. doi: 10.1038/ki.2009.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison SN, Levin A, Moss AH, Jha V, Brown EA, Brennan F. Executive summary of the KDIGO controversies conference on supportive care in chronic kidney disease: developing a roadmap to improving quality care. Kidney Int. 2015;88(3):447–459. doi: 10.1038/ki.2015.110 [DOI] [PubMed] [Google Scholar]
- 16.Go AS, Parikh CR, Ikizler TA, Coca S, Siew ED, Chinchilli VM. The assessment, serial evaluation, and subsequent sequelae of acute kidney injury (ASSESS-AKI) study: design and methods. BMC Nephrol. 2010;11(1):22. doi: 10.1186/1471-2369-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu C, Chinchilli VM, Coca S, Devarajan P, Ghahramani N, Go AS. Post–acute kidney injury proteinuria and subsequent kidney disease progression: the assessment, serial evaluation, and subsequent sequelae in acute kidney injury (ASSESS-AKI) study. JAMA Intern Med. 2020;180(3):402–410. doi: 10.1001/jamainternmed.2019.6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatraju PK, Zelnick LR, Chinchilli VM, Moledina DG, Coca SG, Parikh CR. Association between early recovery of kidney function after acute kidney injury and long-term clinical outcomes. JAMA Netw Open. 2020;3(4):e202682. doi: 10.1001/jamanetworkopen.2020.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer IH, Alpers CE, Azeloglu EU, Balis UGJ, Barasch JM, Barisoni L. Rationale and design of the kidney precision medicine project. Kidney Int. 2021;99(3):498–510. doi: 10.1016/j.kint.2020.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatraju PK Akilesh S Hsu C-Y, et al. Genome-wide association study for AKI in the ASSESS-AKI study. J Am Soc Nephrol Abstr Suppl. 2019;30:463. https://www.asn-online.org/education/kidneyweek/archives/KW19Abstracts.pdf [Google Scholar]
- 21.Ikizler TA Parikh CR Himmelfarb J, et al. A prospective cohort study that examined acute kidney injury and kidney outcomes, cardiovascular events and death informs on long-term clinical outcomes. Kidney Int. 2020;99(2):456–465. doi: 10.1016/j.kint.2020.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh P-R, Danecek P, Palamara PF, Fuchsberger C, A Reshef Y, K Finucane H. Reference-based phasing using the haplotype reference consortium panel. Nat Genet. 2016;48(11):1443–1448. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy S Das S Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong K, Zhu G, Jing X, Hendriks AEJ, Drop SLS, Ikram MA. Genome-wide compound heterozygote analysis highlights alleles associated with adult height in Europeans. Hum Genet. 2017;136(11-12):1407–1417. doi: 10.1007/s00439-017-1842-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidney Precision Medicine Project | NIDDK [Internet]. National Institute of Diabetes and Digestive and Kidney Diseases. 2022. https://www.niddk.nih.gov/research-funding/research-programs/kidney-precision-medicine-project-kpmp [Google Scholar]
- 29.Menon R, Otto EA, Berthier CC, Nair V, Farkash EA, Hodgin JB. Glomerular endothelial cell-podocyte stresses and crosstalk in structurally normal kidney transplants. Kidney Int. 2022;101(4):779–792. doi: 10.1016/j.kint.2021.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menon R, Otto EA, Sealfon R, Nair V, Wong AK, Theesfeld CL. SARS-CoV-2 receptor networks in diabetic and COVID-19 associated kidney disease. Kidney Int. 2020;98(6):1502–1518. doi: 10.1016/j.kint.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonsdale J Thomas J Salvatore M, et al. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillies CE, Putler R, Menon R, Otto E, Yasutake K, Nair V. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am J Hum Genet. 2018;103(2):232–244. doi: 10.1016/j.ajhg.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CM, Barber GP, Casper J, Clawson H, Diekhans M, Gonzalez JN. UCSC Genome Browser enters 20th year. Nucleic Acids Res. 2020;48(D1):D756–D761. doi: 10.1093/nar/gkz1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32(2):179–185. doi: 10.1002/gepi.20292 [DOI] [PubMed] [Google Scholar]
- 35.Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15(5):335–346. doi: 10.1038/nrg3706 [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld JA, Lacassie Y, El-Khechen D, Escobar LF, Reggin J, Heuer C. New cases and refinement of the critical region in the 1q41q42 microdeletion syndrome. Eur J Med Genet. 2011;54(1):42–49. doi: 10.1016/j.ejmg.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 37.Kramann R, Fleig SV, Schneider RK, Fabian SL, DiRocco DP, Maarouf O. Pharmacological GLI2 inhibition prevents myofibroblast cell-cycle progression and reduces kidney fibrosis. J Clin Invest. 2015;125(8):2935–2951. doi: 10.1172/jci74929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauhauser AA, Ren C, Lu D, Li B, Zhu J, McEnery K. Hedgehog signaling indirectly affects tubular cell survival after obstructive kidney injury. Am J Physiol Renal Physiol. 2015;309(9):F770–F778. doi: 10.1152/ajprenal.00232.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burdelya LG, Krivokrysenko VI, Tallant TC, Strom E, Gleiberman AS, Gupta D. An agonist of toll-like receptor 5 has radioprotective activity in mouse and primate models. Science. 2008;320(5873):226–230. doi: 10.1126/science.1154986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuzawa N, Petro M, Baldwin WM, Gudkov AV, Fairchild RL. A TLR5 agonist inhibits acute renal ischemic failure. J Immunol. 2011;187(7):3831–3839. doi: 10.4049/jimmunol.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. J Exp Med. 2003;198(10):1563–1572. doi: 10.1084/jem.20031220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West TE, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Myers ND. Impaired TLR5 functionality is associated with survival in melioidosis. J Immunol. 2013;190(7):3373–3379. doi: 10.4049/jimmunol.1202974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaichana P, Chantratita N, Brod F, Koosakulnirand S, Jenjaroen K, Chumseng S. A nonsense mutation in TLR5 is associated with survival and reduced IL-10 and TNF-α levels in human melioidosis. Plos Negl Trop Dis. 2017;11(5):e0005587. doi: 10.1371/journal.pntd.0005587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park DS, Eskin I, Kang EY, Gamazon ER, Eng C, Gignoux CR. An ancestry based approach for detecting interactions. Genet Epidemiol. 2018;42(1):49–63. doi: 10.1002/gepi.22087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhry S, Burchard EG, Borrell LN, Tang H, Gomez I, Naqvi M. Ancestry-environment interactions and asthma risk among Puerto Ricans. Am J Respir Crit Care Med. 2006;174(10):1088–1093. doi: 10.1164/rccm.200605-596oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348(12):1170–1175. doi: 10.1056/nejmsb025007 [DOI] [PubMed] [Google Scholar]
- 47.Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121(5):565–575. doi: 10.1007/s00439-007-0350-2 [DOI] [PubMed] [Google Scholar]
- 48.Vilander LM, Kaunisto MA, Pettilä V. Genetic predisposition to acute kidney injury—a systematic review. BMC Nephrol. 2015;16(1):197. doi: 10.1186/s12882-015-0190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renken IJE Vilander LM Kaunisto MA, et al. No association between genetic loci near IRF2 and TBX1 and acute kidney injury in the critically ill. Am J Respir Crit Care Med. 2019;201(1):109–111. doi: 10.1164/rccm.201903-0633LE [DOI] [PubMed] [Google Scholar]
- 50.Bhatraju PK, Zelnick LR, Herting J, Katz R, Mikacenic C, Kosamo S. Identification of acute kidney injury subphenotypes with differing molecular signatures and responses to vasopressin therapy. Am J Respir Crit Care Med. 2019;199(7):863–872. doi: 10.1164/rccm.201807-1346oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiersema R, Jukarainen S, Vaara ST, Poukkanen M, Lakkisto P, Wong H. Two subphenotypes of septic acute kidney injury are associated with different 90-day mortality and renal recovery. Crit Care. 2020;24(1):150. doi: 10.1186/s13054-020-02866-x [DOI] [PMC free article] [PubMed] [Google Scholar]