Abstract
The publication of the complete sequence of Helicobacter pylori 26695 in 1997 and more recently that of strain J99 has provided new insight into the biology of this organism. In this review, we attempt to analyze and interpret the information provided by sequence annotations and to compare these data with those provided by experimental analyses. After a brief description of the general features of the genomes of the two sequenced strains, the principal metabolic pathways are analyzed. In particular, the enzymes encoded by H. pylori involved in fermentative and oxidative metabolism, lipopolysaccharide biosynthesis, nucleotide biosynthesis, aerobic and anaerobic respiration, and iron and nitrogen assimilation are described, and the areas of controversy between the experimental data and those provided by the sequence annotation are discussed. The role of urease, particularly in pH homeostasis, and other specialized mechanisms developed by the bacterium to maintain its internal pH are also considered. The replicational, transcriptional, and translational apparatuses are reviewed, as is the regulatory network. The numerous findings on the metabolism of the bacteria and the paucity of gene expression regulation systems are indicative of the high level of adaptation to the human gastric environment. Arguments in favor of the diversity of H. pylori and molecular data reflecting possible mechanisms involved in this diversity are presented. Finally, we compare the numerous experimental data on the colonization factors and those provided from the genome sequence annotation, in particular for genes involved in motility and adherence of the bacterium to the gastric tissue.
A new era in genome science started with the publication of the complete genome of Haemophilus influenzae (82). Eleven other prokaryotic genomes, including two strains of the gastric pathogen Helicobacter pylori (3, 280), and the eukaryotic genome of Saccharomyces cerevisiae (102) have been sequenced, allowing for genome comparisons. Microbial genomics is providing increasing and useful information on fundamental and applied microbiology, taxonomy, molecular biology, microbial ecology, and medical, veterinary, and agricultural microbiology.
Complete genome sequence information has extended our knowledge of bacterial genomic systems, showing that the organization of orthologous genes on the chromosome is not conserved during evolution and that the genome itself is quite complex and flexible. The advent of these sequences has afforded novel insights into the molecular mechanisms of genetic change and adaptive mutations characteristic of bacteria. The construction of putative minimal gene sets sufficient for sustaining cellular life has become a more realistic possibility through comparison of bacterial genomes.
Genome sequence data also are utilized to obtain a physiological integration of phenotypic characteristics of an organism based on functional identification of genes and network reconstruction of pathways and functional units. A current and widely used method for functional analysis is based on searching for similarities in databases (34). Mapping out metabolic and regulatory networks, signal transduction, etc., follows the functional identification of genes. In practice, these are iterative processes: when a pathway is reconstructed incompletely from the predicted genes, the functional identification of genes has to be reviewed, and either genes are reassigned to the missing steps or a new set of genes encoding alternative proteins that can fill the missing functions is found. In bacterial genomes in which the operon structure is relatively well conserved, the clustering of genes can act as a good indicator for reconstruction of functional units, e.g. the aerobic respiratory chain. Through physiological integration, the results of functional identification of genes and network reconstruction are assembled into coherent wholes describing overall phenotypic features of the organism, e.g., aspects of its physiology, adaptation to an ecological niche, and pathogenicity. Physiological integration also serves to explain characteristics of the phenotype not yet fully understood and predicts properties which had not been observed experimentally.
The problems encountered in functional identification, network reconstruction, and physiological integration of organisms are being solved as a result of the growing number of complete genomes sequenced and the efforts to develop new databases and computational algorithms and techniques. Nevertheless, at present genome data on their own are not sufficient even for complete functional identification of genes. Thus, knowledge of biochemical pathways, mechanisms of gene expression, genetics, etc., must be used to interpret the results of sequence similarity searches performed on complete genomes and to place all this information into a coherent set that provides insight into the physiology of a microorganism. It is not surprising that the best-annotated genome is that of Escherichia coli, which is in fact the best-studied organism.
H. pylori is a gram-negative bacterium that is associated with gastric inflammation (290) and peptic ulcer disease (179) and is a risk factor for gastric cancer (83) and non-Hodgkin’s lymphomas of the stomach (230). Almost half of the world’s population harbors an H. pylori infection, with the nature and severity of the disease depending on both host characteristics and environmental factors. There is a considerable body of evidence indicating that the bacterial genotype is also an important factor (201, 203) in the nature of the disease. Sequencing of the genome of H. pylori 26695 has provided insights into its biology, and the recently published comparison of this genome to that of strain J99 has served to deepen our knowledge of the diversity of this bacterium.
Unless explicitly mentioned, we discuss the published genome of strain 26695, whose genes are numbered and preceded by “HP.” After a brief description of the general features of the genome, the links between experimental data and the information provided by sequence annotations are reviewed within the framework of functional identification, network reconstruction, and physiological integration. The review is divided into five areas: metabolism; replication, transcription, and translation; regulation of gene expression; diversity; and colonization factors.
GENERAL FEATURES OF THE GENOME
The genomes of H. pylori 26695 and J99 were sequenced from circular chromosomes containing 1,667,867 and 1,643,831 bp, respectively. These sizes are similar to that of Haemophilus influenzae and about one-third that of E. coli (30, 82).
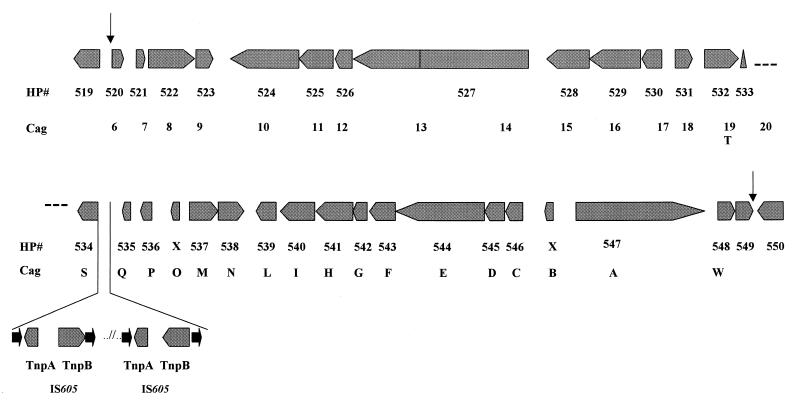
The average G+C content is 39%, but five regions in the genome of strain 26695 (nine in strain J99) have a distinct G+C composition (3, 280). Region 2 (35% G+C) of strain 26695 is the cag pathogenicity island associated with production of the CagA antigen and upregulation of interleukin 8 (42). The other four regions have not been yet characterized experimentally. Regions 1 and 3 (33% G+C) contain copies of the insertion sequence IS605, 5S rRNA genes, and a 521-bp repeat. In addition, region 1 contains the virB4 gene, which encodes the protein involved in the transfer of the T-DNA in Agrobacterium tumefaciens and in the secretion of the Bordetella pertussis toxin (294). Region 4 (43% G+C) contains fused rpoB and rpoC genes encoding the β and β′ subunits of the RNA polymerase. The fusA gene, which codes for the translation elongation factor EF-G, is also associated with this region. Finally, region 5 (33% G+C) contains two restriction/modification systems.
Eight repeat families, with a sequence identity greater than 97% have been found in the H. pylori DNA. They are localized either in intergenic regions (one family) or within coding sequences, where they may represent gene duplications.
Tomb et al. (280) identified 1,590 open reading frames (ORFs), which represent 91% of the H. pylori chromosome of strain 26695, and Alm et al. (3) identified 1,495 ORFs, which represent 90.8% of the chromosome of strain J99. No genome-wide strand bias was observed. The noncoding regions of strain 26695 (9%) are divided into three classes. Intergenic sequences represent 6% of the noncoding regions, while noncoding repeats account for 2.3% and stable RNA accounts for 0.7%. Among the 1,590 ORFs, 1,091 were found to have counterparts in other organisms, allowing putative biological roles to be assigned to them, although not all have orthologues of known function. The 499 ORFs which exhibited no database matches may be considered at this time to be specific to H. pylori. In the reannotation of this genome carried out by Alm et al. (3), the number of ORFs in strain 26695 is 1,552, of which 1,185 have orthologues found in other species, 367 are H. pylori specific, and 69 are specific to strain 26695. The proportion of orphan genes in H. pylori is similar to that found in other bacteria sequenced to date. However, it ought to be kept in mind that orthologous genes of some of these ORFs will probably be identified when further whole genomes are completely sequenced.
METABOLISM
A better understanding of the biochemistry of H. pylori is of fundamental interest to microbiology and also could help in developing new anti-H. pylori therapies. In this section, the principal metabolic pathways of the bacterium are reviewed by comparing the experimental data with those derived from the whole genome sequence (280).
Glucose Metabolism
Soon after its discovery, H. pylori was classified as a Campylobacter species and, like other members of this genus, was reported to be unable to catabolize carbohydrates. Several studies of the physiology of the bacterium (39, 43, 181, 183, 185, 187, 188) have provided evidence that it can metabolize glucose by both oxidative and fermentative pathways, although it is an obligate microaerophile. Moreover, glucose appears to be the only carbohydrate utilized by the bacterium (188). More recently, the whole-genome analysis of H. pylori has supported these findings (280).
Glucose is imported into the cells by a permease which is specific for d-glucose and galactose. This transporter is sodium dependent and is unaffected by inhibitors known to affect other bacterial glucose permeases (39, 181). Analysis of the genome established the presence of the GluP glucose/galactose transporter, and no other saccharide permease has been identified (280). As suggested by Mendz et al. (188), intracellular phosphorylation of glucose is performed by a glucokinase rather than a hexokinase, and there is no evidence for the glucose phosphotransferase system involving the phosphorylation of an E-III enzyme. In agreement with these data, the analysis of the H. pylori genome shows its capacity to code for a glucokinase. The HP1103 gene has 59.5% similarity to the glk gene of E. coli, and genes encoding either a phosphotransferase system or a less specific hexokinase were not found by Tomb et al. (280). This feature could explain the limited range of carbohydrates used by H. pylori. Glucose utilization shows biphasic characteristics, with a slow initial period followed by faster catabolism. The rates of decline of glucose levels in both phases depend on the growth conditions of the bacteria, suggesting that this metabolite is not a preferred energy substrate but can be used when other sources of energy have been exhausted (185, 188). It would be of interest to attempt to relate the characteristics of glucose uptake and catabolism to the properties of the specific proteins encoded in the chromosome.
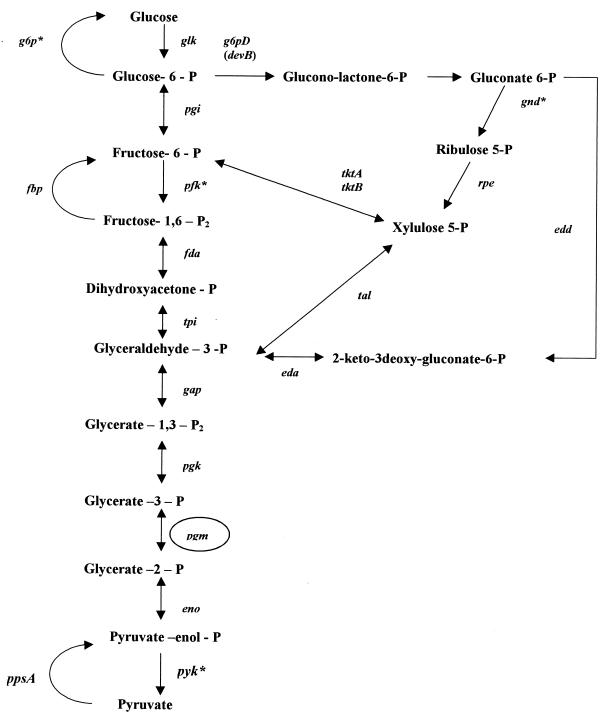
Mendz and Hazell (183) demonstrated the presence of enzymatic activities which are part of the oxidative and nonoxidative steps of the pentose-phosphate pathway in H. pylori. This pathway is an efficient mechanism to provide NADPH and NADH for reductive biosynthesis and C5 phosphorylated carbohydrates essential for nucleotide synthesis (Fig. 1). The genes HP1386, HP1102, HP1101, HP1495, HP1088, and HP0354, orthologous to rpe, devB, g6pD, tal, tktA, and tktB, respectively, encoding enzymes of the pentose phosphate pathway, were identified in the genome, but no sequence similarity to the 6-phosphogluconate dehydrogenase gene, a key enzyme of the pathway, was found in the H. pylori DNA. This suggests the existence of a protein with a similar function to and different characteristics from other 6-phosphogluconate dehydrogenases.
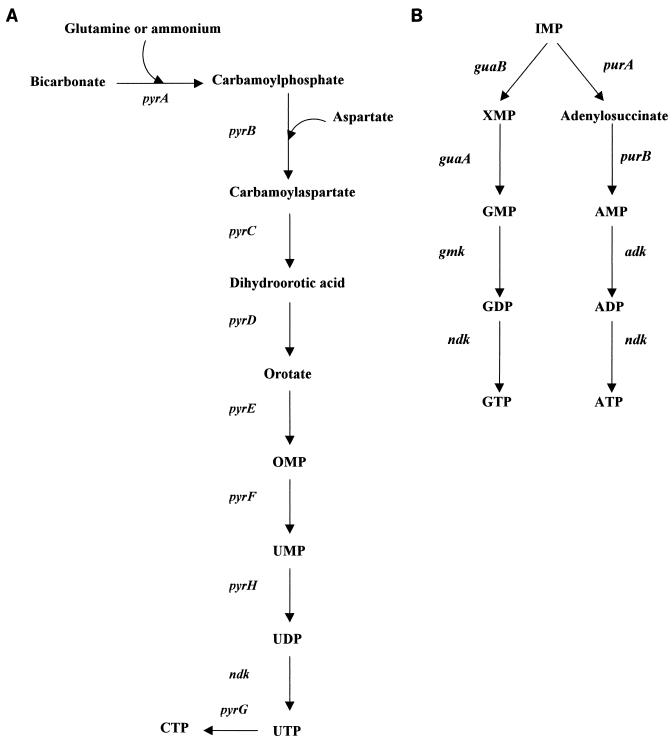
FIG. 1.
Glycolysis, gluconeogenesis, pentose phosphate, and Entner-Doudoroff pathways. Glycolysis: glk, glucokinase; pgi, phosphoglucose isomerase; pfk, phosphofructokinase; fda, fructose-1,6-bisphosphate aldolase; tpi, triose-phosphate isomerase; gap, glyceraldehyde-3-phosphate dehydrogenase; pgk, phosphoglycerate kinase; pgm, phosphoglycerate mutase; eno, enolase; pyk, pyruvate kinase. Gluconeogenesis: the same enzymes as in glycolysis but with unidirectional steps, i.e., ppsA, phosphoenol pyruvate synthase; fbp, fructose-1,6 bisphosphatase; and g6p, glucose-6 phosphatase. Pentose phosphate: g6pD (devB), glucose-6-phosphate dehydrogenase; lactonase; gnd, 6-phosphogluconate dehydrogenase; rpe, d-ribulose-5-phosphate 3 epimerase; tal, transaldolase; tkt, transketolase. Entner-Doudoroff: edd, 6-phosphosgluconate dehydratase; eda, 2-keto-3-deoxy-6-phosphogluconate aldolase. Asterisks denote enzymes for which no gene was identified in the H. pylori sequence. The circle denotes an enzyme whose enzymatic activity has not been observed but whose corresponding gene was identified.
Alternatively, glucose-6-phosphate can be utilized by the Entner-Doudoroff pathway, regarded as an alternative to glycolysis (43, 187). The specific steps of this pathway consist of two reactions: a dehydratase-catalyzed formation of 2-keto-3-deoxygluconate-6-phosphate from gluconate-6-phosphate and an aldolase-catalyzed production of pyruvate and glyceraldehyde-3-phosphate (Fig. 1). These activities were found in H. pylori lysates (187), and ORFs HP1099 and HP1100, with homologies to the 2-keto-3-deoxy-6-phosphogluconate aldolase (eda) gene and the 6-phosphogluconate dehydratase (edd) gene from E. coli, respectively, were identified by Tomb et al. (280). The Entner-Doudoroff pathway is inducible in E. coli and is rarely employed by wild-type strains (85); however, it appears to be constitutive in H. pylori. Although its potential energy yield is less than that of glycolysis, it offers the possibility of metabolizing aldonic acids.
In 1996, Hoffman et al. (121) reported glycolytic activity and gluconeogenesis in H. pylori. The two pathways have seven common reversible reactions and are distinguished by three opposed irreversible steps (Fig. 1). There has been controversy about the existence of glycolysis since Mendz et al. (187) and Chalk et al. (43) were not able to detect some of the enzyme activities of this pathway. In particular, phosphoglycerate mutase activity has not been observed, although a gene (HP0974) coding for a protein with 44.6% similarity to the pgm gene product has been identified. Analysis of the H. pylori genome suggests that other genes coding for the enzymes of the glycolytic pathway are present, with the important exceptions of phosphofructokinase, which affects the irreversible phosphorylation of fructose-6-phosphate to fructose-1,6-biphosphate in glycolysis and of pyruvate kinase. Tomb et al. (280) identified a gene (HP1385) coding for a putative fructose-1,6-bisphosphatase which catalyzes the opposed irreversible reaction in gluconeogenesis and also found a gene (HP0121) encoding phosphoenolpyruvate synthase, an enzyme complex responsible for another of the irreversible reactions in gluconeogenesis. However, a gene coding for glucose-6-phosphatase, the enzyme involved in the third irreversible reaction of gluconeogenesis, has not been identified, and only a gene (HP1103) coding for glucokinase, which catalyzes the opposed reaction of glycolysis, was identified. Thus, in the case of glycolysis and gluconeogenesis, deductions from the H. pylori genome analysis have been useful in confirming some experimental data and at the same time have shown the need for further verification of the presence of these pathways.
Pyruvate Metabolism
Pyruvate is an end product of both the glycolytic and Entner-Doudoroff pathways. The metabolic fate of pyruvate in H. pylori was investigated by several groups. In one study, pyruvate was metabolized to lactate, ethanol, and acetate under anaerobic conditions whereas the major product of pyruvate under aerobic conditions was acetate (43). In another study (190), cells incubated with pyruvate under microaerobic conditions yielded lactate, acetate, formate, succinate, and alanine. The formation of succinate suggests the incorporation of pyruvate into the Krebs cycle, and the presence of alanine supports the view that pyruvate could play an important role in biosynthetic processes. The formation of lactate, ethanol, and acetate suggests the use of pyruvate in fermentative metabolism. In agreement with this observation, Tomb et al. (280) found ORFs HP1222 and HP0357 with similarities to the H. influenzae genes encoding d-lactate dehydrogenase, necessary to convert pyruvate to lactate, and short-chain alcohol dehydrogenase, respectively. The corresponding enzymes catalyze reactions occurring in conjunction with the oxidation of NADH. Formation of acetate from pyruvate requires three steps: the oxidative decarboxylation of pyruvate to produce acetyl coenzyme A (acetyl-CoA) and formate, the formation of acetyl-phosphate, and its dephosphorylation to acetate. H. pylori ORFs corresponding to genes coding for enzymes responsible for the last two steps in E. coli were identified by using sequence similarities: HP0904 to the phosphate acetyltransferase pta gene, and HP0903 to the acetate kinase ackA gene.
To enter the Krebs cycle, as well as to form acetate, pyruvate must be converted to acetyl-CoA. Hughes et al. (126) demonstrated that oxidative decarboxylation of pyruvate is carried out in H. pylori by a pyruvate:acceptor oxidoreductase (POR), instead of the aerobic pyruvate dehydrogenase (AceEF) or the strictly anaerobic pyruvate-formate lyase (Pfl) associated with mixed-acid fermentation. Flavodoxin is believed to be the in vivo electron acceptor for POR. This type of oxidoreductase is found commonly in obligate anaerobes, such as Clostridium spp. The pyruvate-flavodoxin oxidoreductase of H. pylori is composed of four subunits and is related to pyruvate-ferrodoxin oxidoreductases previously detected only in hyperthermophilic organisms (126). In the H. pylori genome sequence, genes HP1108 to HP1111 have similarities to the POR genes of the hyperthermophilic archaeon Pyrococcus furiosus and those of the hyperthermophilic bacterium Thermotoga maritima, depending on the subunit considered. Neither aceEF nor pfl genes were identified (280). The genome analysis of H. pylori supports the experimental data, particularly its utilization of pyruvate in fermentative metabolism. The biochemical characterization of the pyruvate-flavodoxin oxidoreductase and the data provided by the analysis of the genome data provide insight into the microaerophily of the bacterium.
The Krebs Cycle and Related Enzymes
A function of the tricarboxylic acid cycle is the oxidation of acetyl units to produce CO2 and the generation of reduced nucleotides useful for reductive biosynthesis or for storage of energy in the form of ATP. The cycle also provides precursors required for biosynthesis, e.g., oxaloacetate, succinyl-CoA, and α-ketoglutarate.
Elements of the Krebs cycle are present in H. pylori (121, 188, 237, 267). The absence of the α-ketoglutarate dehydrogenase complex was reported by Hoffman et al. (121) and confirmed by other groups (52, 237), and Pitson et al. (237) did not find succinyl-CoA synthetase in the bacterium. Accordingly, genes encoding the two subunits of the α-ketoglutarate dehydrogenase complex (sucA and sucB) or the subunits of the succinyl-CoA synthetase (sucC and sucD) were not identified (280).
Hoffman et al. (121) and Pitson et al. (237) reported the existence of α-ketoglutarate:acceptor oxidoreductase (OOR) which catalyzes the conversion of α-ketoglutarate to succinate. Hughes et al. (126) purified the OOR enzyme and showed that it was composed of four heterogeneous subunits, with significant sequence similarity to archaeal enzymes. Unlike POR, OOR was unable to use a previously identified flavodoxin (FldA) as an electron acceptor. The analysis performed by Tomb et al. (280) yielded the ORFs HP0589, HP0590, and HP0591, which had similarities to the ferrodoxin oxidoreductase genes korA, korB, and korG from Methanococcus jannaschii and which presumably could encode three of the subunits of the H. pylori OOR enzyme. However, the amino acid sequences of H. pylori OorA and OorB determined by Hughes et al. (126) are more closely related to those of the two-subunit POR of the aerobic halophile Halobacterium halobium. The last subunit, OorD, appears to be a 10-kDa integral ferredoxin-like protein. Like OOR, POR is oxygen labile, and both are likely to be major factors in the requirement of H. pylori for low oxygen tensions.
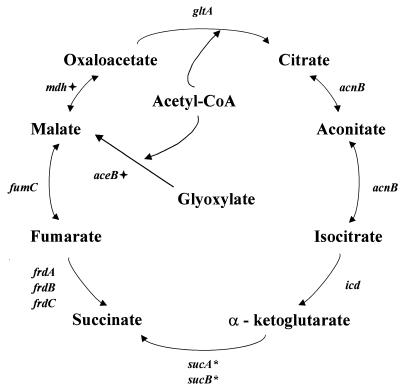
In H. pylori, the Krebs cycle appears to be a branched noncyclic pathway, in which the dicarboxylic acid arm proceeds reductively from oxaloacetate through malate and fumarate to succinate and the tricarboxylic acid arm operates oxidatively from oxaloacetate through citrate and isocitrate to α-ketoglutarate (114, 188, 237) (Fig. 2). The discoveries of genes encoding citrate synthase (gltA, HP0026), aconitase (acnB, HP0779), isocitrate dehydrogenase (icd, HP0027), fumarase (fumC, HP1325), and fumarate reductase (frdABC, HP0191 to HP0193) and the absence of ORFs with similarity to the genes encoding α-ketoglutarate dehydrogenase support this hypothesis (280). Malate dehydrogenase and malate synthase activities have been measured in H. pylori (237), but no genes encoding these enzymes have been identified, suggesting that they are likely to have an amino acid sequence quite different from the others so far determined. These examples illustrate the caution that must be exercised when predicting metabolic and functional characteristics of organisms based solely on genomic analysis.
FIG. 2.
Dicarboxylic and tricarboxylic acid branches of the noncyclic Krebs cycle. gltA, citrate synthase; acnB, aconitase; icd, isocitrate dehydrogenase; sucAB, α-ketoglutarate dehydrogenase; frdABC, fumarate reductase; fumC, fumarase; mdh, malate dehydrogenase; aceB, malate synthase. Asterisks denote enzymes for which no genes were identified in the H. pylori sequence; crosses denote enzymes whose enzymatic activities were observed but the corresponding genes were not identified. Reprinted from reference 237 with permission of the publisher.
An interesting feature of the reductive branch is that fumarate, via fumarate reductase, may act as the terminal electron acceptor in anaerobic respiration. Mendz et al. (189) investigated the properties of the H. pylori enzyme activity in vivo and established that it is a potential therapeutic target against the bacterium. In E. coli, fumarate reductase is made up of four subunits, i.e., a flavoprotein, an iron-sulfur protein, and two membrane anchor proteins (12); and in Wolinella succinogenes, fumarate reductase consists of three subunits (151). Ge et al. (92) cloned and characterized the H. pylori fumarate reductase operon, which is composed of three structural genes coding for subunits highly similar to those of W. succinogenes.
The putative anaerobic respiration with fumarate as the acceptor and the presence of pyruvate-flavodoxin and α-ketoglutarate:acceptor oxidoreductases support the existence of anaerobic metabolism in H. pylori, even though oxygen is required for growth, and contribute to the characterization of the microaerophilic phenotype of the bacterium.
Concerning the Krebs cycle, some features deduced from the genome are in agreement with experimental data, but other deductions need further experimental verification. Conversely, the metabolic data suggest the need for more genomic analyses of the bacterium.
Amino Acid Metabolism
Amino acids are potential sources of carbon, nitrogen, and energy. The development of a defined medium for growing H. pylori and the subsequent determination of the amino acid requirements of the bacterium were important steps in understanding its metabolism (215, 246). All the strains tested required arginine, histidine, isoleucine, leucine, methionine, phenylalanine, and valine. Some of them also required alanine (8 of 10 strains), and serine was also needed for 5 of them. Analyses of genome molecular data support these experimental results.
The requirement for arginine and histidine in the growth medium may be explained by the fact that no protein involved in the arginine or histidine biosynthetic pathways is encoded by the H. pylori chromosome, except glutamate dehydrogenase (GdhA), which catalyzes the synthesis of glutamate from α-ketoglutarate (98, 244, 295). ORF HP0380 has 72.7% similarity to E. coli gdhA.
Aspartate synthesis is a key step in the syntheses of several other amino acids. Aspartate is formed from oxaloacetate by transamination with glutamate as the amino donor (244), and the syntheses of methionine, threonine, and isoleucine are linked to that of aspartate (107, 231). The genes HP1189, HP1229, HP0822, HP1050, and HP0098 in the H. pylori DNA sequence are similar to asd, lysC, metL, thrB, and thrC, respectively, which encode enzymes involved in the threonine synthesis pathway. However, the genes encoding threonine deaminase and acetohydroxy acid synthase, which are necessary to form isoleucine, are lacking; thus, this amino acid is required for growth. Furthermore, a requirement for valine would follow from the absence of the gene which encodes acetohydroxy acid synthase, the first enzyme of the valine biosynthesis pathway (284). In the specific pathway leading to methionine synthesis, only the metB gene (HP0106), encoding cystathionine γ-synthase, seems to be present, which would explain the need for this amino acid. The absence of the leuA gene, encoding isopropyl malate dehydrogenase, is consistent with the need for leucine, which cannot be synthesized from α-ketoglutarate (284).
Chorismate, a precursor of aromatic amino acids, is formed from phosphoenol pyruvate and erythrose 4-phosphate by means of seven enzymes whose genes are present in H. pylori DNA (238). The synthesis of tyrosine and phenylalanine from chorismate requires three enzymes, one of which (tyrosine aminotransferase) is common to both pathways. The tyrB gene, encoding tyrosine aminotransferase, was not identified from the nucleotide sequence, but its function can be supplemented by the branched-amino-acid aminotransferase enzyme, whose gene, ilvE (HP1468), is found in the chromosomal DNA of H. pylori. Since the tyrA gene (HP1380), encoding chorismate mutase-prephenate dehydrogenase, is also present in the genome, the bacterium can synthesize tyrosine. However, H. pylori is not able to synthesize phenylalanine, because the pheA gene, encoding chorismate mutase-prephenate dehydratase, is absent from the chromosome. The five enzymes specific for the biosynthesis of tryptophan have been identified in the genome of H. pylori, and the six sequential genes HP1277 to HP1282 encoding them have a similar organization to the corresponding genes of E. coli, which form the trpEDCBA operon.
The genome sequence of H. pylori suggests that other amino acids are synthesized via their conventional pathways (149, 156, 231, 238, 244, 269, 284).
Although Reynolds and Penn (246) observed that alanine enhances bacterial growth in the presence of glucose, Mendz and Hazell (182) demonstrated that H. pylori grows well in a glucose-free medium supplemented with arginine, aspartate, asparagine, glutamate, glutamine, and serine as sole substrates. The principal metabolic products observed from amino acid catabolism were acetate, formate, succinate, and lactate. These results showed that carbohydrates could be removed from media in which amino acids constituted the basic nutrients. Moreover, glucose added to growth media composed of a mixture of amino acids is not utilized until the other metabolites are significantly depleted (185, 188). These results are contrary to the conclusion of Tomb et al. (280) from genomic analysis that the glycolysis-gluconeogenesis metabolic axis constitutes the backbone of energy production in the bacterium.
Nonetheless, other comparisons between experimental results and molecular data from the genome show good correlations, even before some pathways have been completely elucidated. For example, the catabolism of alanine, arginine, aspartate, asparagine, glutamate, glutamine, and serine is dependent on the presence of several enzymes, whose genes have been identified. Indeed, the genes coding for alanine dehydrogenase (ald, HP1398), aspartate-ammonia lyase (aspA, HP0649), l-asparaginase II (asnB, HP0723), glutamate dehydrogenase (gdhA, HP0380), and l-serine deaminase (sda, HP0132) were found by sequence similarities.
The conversion of arginine to urea and ornithine described by Mendz and Hazell (186) is very interesting because it offers some insight into the nitrogen metabolism of H. pylori. The authors inferred the presence of an arginase necessary to catalyze this reaction, and the kinetic parameters, specificity, effects of cations, pH profile, and inhibition of this enzyme activity were studied in vivo and in situ (123, 191). In agreement with these results, Tomb et al. (280) identified ORF HP1399, with similarity to the rocF gene from Bacillus subtilis encoding arginase, and ORF HP1017, with similarity to the rocE gene encoding an arginine transporter. Moreover, an aliphatic amidase which hydrolyzes short-chain amides to produce carboxylic acid and ammonia has been identified in H. pylori (261). The gene encoding this enzyme was characterized and found to have high similarity (75%) to the amiE gene from Pseudomonas aeruginosa. This aliphatic amidase activity has been previously found only in environmental bacteria. These observations led to the suggestion that the amiE gene was acquired by horizontal transfer. Two paralogous genes, HP0294 and HP1238, seem to be present in H. pylori DNA.
Analysis of the genome sequence supports the experimental data concerning the synthesis pathways of the amino acids and explains the in vitro requirement for some of them by the lack of genes encoding enzymes involved in those synthesis pathways. However, more experiments are needed to fully understand amino acid catabolism in the bacterium, and these investigations should be guided by the genomic data.
Lipid Metabolism
Lipids could be used as a source of carbon and energy, and phospholipids are a potential source of phosphate. Little information is available concerning the metabolism of fatty acids and phospholipids in H. pylori, but a detailed analysis of its DNA sequence has increased our understanding of its lipid metabolism.
In E. coli, fatty acid degradation occurs by cyclic β-oxidation and thiolytic cleavage, yielding acetyl-CoA (62). The first step of the degradation is the activation of the free fatty acids to an acyl-CoA thioester by acyl-CoA synthetase. This β-oxidation requires four enzymes: acetyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and 3-ketoacyl-CoA thiolase. Only the orthologous genes HP1045 and HP0690, corresponding to acoE encoding acyl-CoA synthetase and fadA encoding the 3-ketoacyl-CoA thiolase, respectively, have been identified by genome sequence analysis.
Concerning phospholipid degradation, several studies have reported phospholipase activities in H. pylori. Various strains have been shown to express phospholipase A1 (160), A2 (227), or C (33, 291); however, in the genome, only ORF HP0499 showed similarity to phospholipase A1 gene from E. coli. It has to be concluded that the other genes may not have sufficient sequence similarity to be identified as such.
The biosynthesis of lipids and phospholipids by H. pylori is an area of investigation which requires further basic research. In particular, the biosynthetic pathways of fatty acids have not been completely elucidated, although some features have been established. The mechanism of fatty acid synthesis is conserved in prokaryotes and eukaryotes and proceeds in two stages, initiation and cyclic elongation (63).
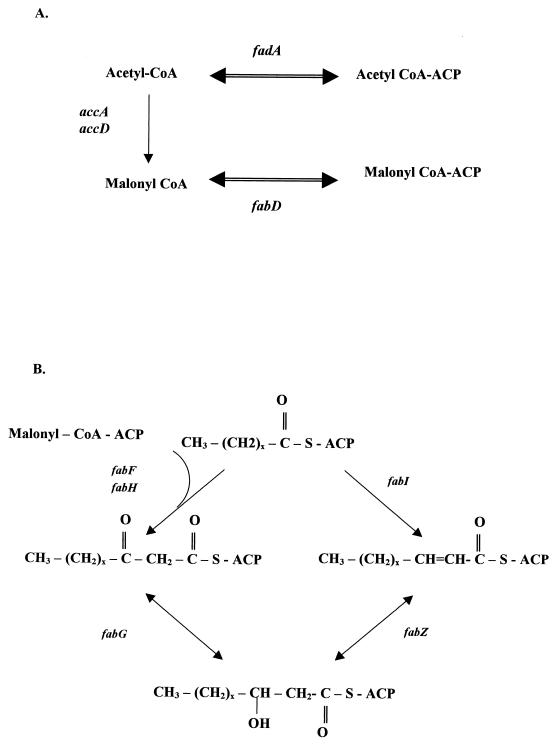
The first committed step in fatty acid biosynthesis is the formation of malonyl-CoA from acetyl-CoA catalyzed by acetyl carboxylase (Fig. 3). The prokaryotic form of this enzyme, exemplified by the E. coli enzyme, consists of a complex of three individual proteins: biotin carboxyl carrier protein, biotin carboxylase, and carboxyltransferase (α and β subunits). The enzyme complex of H. pylori has been characterized by Burns et al. (37), and the genes encoding the proteins in the complex, HP0370 (accC), HP0371 (fabE), HP0557 (accA), and HP0950 (accD), have been found in H. pylori DNA. The next steps in the initiation of fatty acid biosynthesis involve the attachment of the acyl carrier protein (ACP) to the acetyl and malonyl moieties. The genes encoding ACP and the two subunits of the holo-ACP synthase, HP0559, HP0962 (acpP), and HP0808 (acpS), respectively, are also present in the genome. The transfer reactions of CoA-bearing acyl chains to ACP are catalyzed by acetyl-CoA:ACP transacylase and malonyl-CoA:ACP transacylase (Fig. 3A). The gene encoding acetyl-CoA:ACP transacylase was not identified in the H. pylori genome. However, this function could be performed by a thiolase product of HP0690 (fadA). Malonyl-CoA is converted to malonyl-CoA:ACP by the malonyl-CoA:ACP transacylase encoded by HP0090 (fabD).
FIG. 3.
Initiation (A) and elongation (B) phases of fatty acid biosynthesis. Initiation: accA, carboxyltransferase (α subunit); accD, carboxyltransferase (β subunit); fadA, thiolase; fabD, malonyl-CoA:ACP transacylase. Elongation: fabF, 3-ketoacyl-ACP synthase; fabH, 3-ketoacyl-ACP synthase; fabI, enoyl-ACP reductase; fabG, 3-ketoacyl-ACP-reductase; fabZ, (3R)-hydroxymyristoyl-ACP dehydratase.
Five genes, whose corresponding proteins are involved in elongation reactions of fatty acid synthesis, are present in the DNA sequence (Fig. 3B): HP0558 (fabF) and HP0202 (fabH), encoding 3-ketoacyl-ACP synthases; HP0561 (fabG), encoding 3-ketoacyl-ACP reductase; HP0195 (fabI), encoding enoyl-ACP reductase; and HP1376 (fabZ), encoding (3R)-hydroxymyristoyl-ACP dehydratase. In E. coli, a specific dehydrase enzyme, 3-hydroxydecanoyl-ACP dehydrase, catalyzes a key reaction at the point at which the biosyntheses of unsaturated and saturated fatty acids diverge (31). The fabZ gene might function in a similar manner in H. pylori, since the major fatty acids in the bacterium are tetradecanoic acid (14:0) and 19-carbon cyclopropane (19:0). Detection of the ORF HP0416, with similarity to the cfa gene from E. coli, explains the presence of cyclopropane fatty acids in H. pylori, in common with many other bacteria (103, 104).
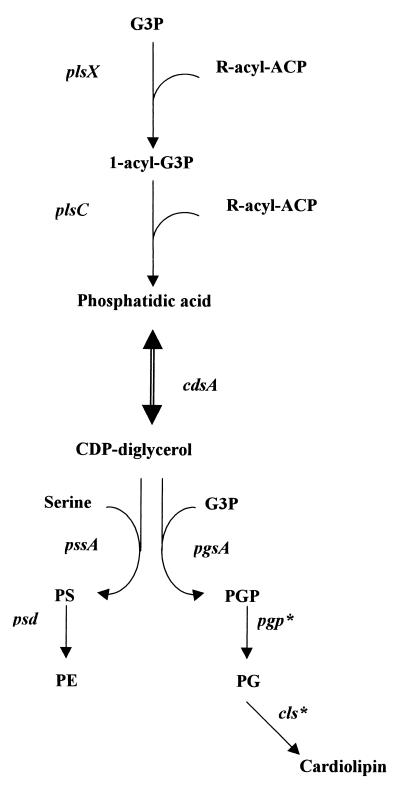
There is only a small proportion of neutral lipids in the lipid composition of H. pylori. The most abundant phospholipids are phosphatidylethanolamine (PE), cardiolipin, and phosphatidylglycerol (PG); this composition is similar to that in many other gram-negative bacteria (119). The biosynthesis pathway for phospholipids utilizes sn-glycerol-3-phosphate (Glp), which can also serve as a substrate for glycolysis or gluconeogenesis in prokaryotes. The key phospholipid synthetic intermediate, phosphatidic acid, is formed in two steps (Fig. 4). Glp is acylated first by the glycerol-3-phosphate acyltransferase, using the acyl-ACP products of fatty acid synthesis, and then a second fatty acid is added by a 1-acyl-Glp acyltransferase to form phosphatidic acid (251). Both enzymes seem to be present in H. pylori, since the corresponding genes, HP0201 (plsX) and HP1348 (plsC), respectively, have been identified. Phosphatidic acid is converted to CDP-diglycerol. This reaction is catalyzed by CDP-diglycerol synthase. CDP-diglycerol reacts with serine to form phosphatidylserine (PS) or with Glp to form phosphatidylglycerolphosphate (PGP). The decarboxylation of PS by PS decarboxylase yields PE, and the dephosphorylation of PGP by PGP phosphatase yields PG. Analysis of the H. pylori DNA sequence led to the identification of three ORFs, HP0215, HP1016, and HP1357, with similarity to the cdsA, pgsA, and psd genes, respectively, of E. coli, whose products are required for the enzymatic reactions listed above (Fig. 4). The PS synthase and its corresponding gene (pssA) have been described by Ge and Taylor (93). However, no sequence similarity to a gene encoding PGP phosphatase, which allows the synthesis of PG, or with the gene encoding the cardiolipin synthase was identified in H. pylori.
FIG. 4.
Synthesis of phosphatidic acid, PS, PE, PGP, PG, and cardiolipin. plsX, glyceraldehyde-3-phosphate (G3P) acyltransferase; plsC, 1-acyl-G1P acyltransferase; cdsA, CDP-diglycerol synthase; pssA, PS synthase; psd, PS decarboxylase; pgsA, PGP synthase; pgp, PGP phosphatase; cls, cardiolipin synthase. Asterisks denote enzymes for which no genes were identified in the H. pylori sequence.
A unique characteristic of the lipid composition of this bacterium is the presence of three cholesteryl glucosides synthesized de novo, which account for about 25% (wt/wt) of the total lipids (110, 119). Cholesteryl glucosides are synthesized in plants and fungi but are rare in higher animals and bacteria. The cholesteryl glucosides found in H. pylori have a glycosidic linkage, which is unusual for natural glycosides, and one has a phosphate-linked glycoside that has not yet been found in prokaryotes or eukaryotes (119). UDP-glucose:sterol glucosyltransferase is the enzyme catalyzing the synthesis of cholesteryl glucosides in plants and fungi. No ORF has been found in the genome of H. pylori to account for this enzymatic activity.
The results of many of these analyses have not yet been backed up by experimental results. Elucidation of the role of a number of different genes identified in the lipid metabolism of the bacterium must await further study.
Lipopolysaccharide Biosynthesis
In general, the surface-exposed lipopolysaccharide (LPS) molecules play an important role in the interaction between gram-negative bacteria and their hosts. They are efficient immunomodulators and potent stimulators of the immune system (208). LPS is composed of three parts: the hydrophobic lipid A, the hydrophilic O-antigen polysaccharide region, and the core polysaccharide region, which connects the other two.
The lipid A portion is the component responsible for the immunological and endotoxic properties. Several groups have studied the relationship between the lipid A structure, LPS endotoxic properties, and immunobiological activities (14, 143, 209, 272); the H. pylori LPS is remarkable in its low toxicity. The polysaccharide moiety of H. pylori LPS contains Lewisx and Lewisy antigenic motifs that mimic Lewis antigens present on parietal cells of the human gastric mucosa (6, 7, 259).
At least 27 genes likely to be involved in LPS biosynthesis have been found in H. pylori (280). In contrast to the situation in other bacteria, LPS biosynthesis genes in H. pylori are scattered throughout the genome, not clustered at one locus. A homologue of GDP-d-mannose dehydratase from Vibrio cholerae and homologues of galactosyltransferases from Klebsiella pneumoniae have been suggested to be involved in O-antigen synthesis. The genes HP0379 and HP0651, coding for two 1,3-fucosyltransferases, have been identified; these transferases catalyze 1,3-glycosidic linkages implicated in the expression of sialyl-Lewisx antigens in humans. DNA motifs evoking phase variation have been found near the 5′ end of the 1,3-fucosyltransferase genes. The relevance of these motifs in vivo is examined in more detail below in the section on the diversity of H. pylori. Berg et al. (22) pointed out the presence of gene HP0326, an orthologue of neuA, that codes for CMP-N-acetylneuraminic acid synthetase. This enzyme is the glycosyl donor of sialyl groups and thus could be involved in the sialyl-Lewisx antigen synthesis which has been reported in some strains (296). ORFs similar to genes encoding lipid A biosynthetic enzymes, namely, HP1375 for UDP-N-acetylglucosamine acyltransferase (lpxA), HP0867 for lipid A disaccharide synthetase (lpxB), HP0196 for UDP-3-O-(3-hydroxymyristoyl)glucosamine-N-acyltransferase (lpxD), and HP1052 for UDP-3-O-acyl-N-acetylglucosamine deacetylase (envA), were found in the H. pylori sequence. Orthologues of genes involved in core region synthesis were also identified: HP0858 (rfaE), HP1191 (rfaF), HP0859 (rfaD), and HP0279 (rfaC) for the heptose region of the inner core; HP0003 (kdsA), HP0230 (kdsB), and HP1386 (rpe) for the 3-deoxy-d-mannooctulosonic acid (KDO) residue region of the inner core; and HP0159, HP0208, HP1416 (three copies of rfaJ), HP1166 (pgi), and HP0646 (galU) for the outer core. KDO transferase, which catalyzes the attachment of KDO groups to lipid A, was also found to be encoded by the genes HP0742, HP1166, and HP0646, which are orthologues of kdsA. The identification of HP0159, HP0208, and HP1416 as homologues of the 1,2-glucosyltransferase gene (rfaJ) is confusing, since no 1,2-linked glucose has been found in H. pylori LPS (22). The authors suggested that these genes may encode 1,4-galactosyltransferase and/or 1,3-N-acetylglucosaminyltransferase functions, for which no orthologous genes have been found in the H. pylori sequence. The gene encoding 1,2-fucosyltransferase is also missing; it seems to be truncated in the genome of the sequenced strain (22).
In conclusion, genomic analysis allowed the identification of genes involved in LPS synthesis. However, in a few cases, further information is required to elucidate the relationship between functions deduced from the sequence analysis and those determined experimentally.
Nucleotide Biosynthesis
Purines and pyrimidines are essential for the synthesis of nucleoside triphosphates (NTPs), which are precursors of nucleic acids. rNMPs, from which dNMPs are derived, may be synthesized de novo from simple precursors or formed via salvage pathways. 5-Phospho-α-d-ribosyl-1-pyrophosphate (PRPP) is synthesized from ATP and ribose 5-phosphate by the action of phosphoribosyl pyrophosphate synthetase. This enzyme is encoded by the HP0742 (prsA) gene in H. pylori. The ribose-5-phosphate moiety of nucleotides is derived from PRPP in de novo synthesis and in some salvage pathways. Nucleoside polyphosphates are formed by successive phosphorylations of their monophosphate counterparts.
De novo synthesis of UTP and CTP.
The pathway responsible for the de novo synthesis of UTP and CTP, two precursors of RNA, comprises nine enzymes (Fig. 5A). Carbamoylphosphate is a substrate for the first reaction of the pathway and is formed by a synthetase which catalyzes the reaction from glutamine or ammonium, bicarbonate, and ATP. Analysis of the H. pylori DNA sequence identified two ORFs, HP1237 and HP0919, with similarities to the pyrAa gene from Salmonella cholerasuis and the pyrAb gene from Bacillus caldolyticus (280), which encode carbamoyl-phosphate synthetases.
FIG. 5.
De novo synthesis of UTP and CTP (A) and ATP and GTP (B). (A) pyrA, carbamoyl-phosphate synthase; pyrB, aspartate transcarbamoylase; pyrC, dihydroorotase; pyrD, dihydroorotase dehydrogenase; pyrE, orotate phosphoribosyltransferase; pyrF, OMP decarboxylase; pyrH, UMP kinase; ndk, nucleoside diphosphokinase; pyrG, CTP synthetase. (B) guaB, IMP dehydrogenase; guaA, GMP synthase; gmk, GMP kinase; purA, adenylosuccinate synthetase; purB, adenylosuccinate lyase; adk, adenylate kinase.
In the first reaction committed solely to pyrimidine biosynthesis, carbamoyl phosphate is converted to carbamoyl aspartate by its condensation with the amino group of aspartate in a reaction catalyzed by aspartate carbamoyl transferase. In E. coli, this enzyme is a dodecamer composed of two catalytic trimers (c3) and three regulatory dimers (r2). The gene for the catalytic chain, pyrB, and the gene for the regulatory chain, pyrI, are organized in an operon transcribed from B to I (218). ORF HP1084 in H. pylori DNA is similar to the pyrB gene from B. subtilis, but no gene orthologous to pyrI seems to be present. Burns et al. (38) characterized the kinetic and regulatory properties of H. pylori aspartate carbamoyl transferase and suggested that it could be grouped as a class A enzyme on the basis of several specific features of its regulation.
Orotate is formed by the action of dihydroorotase (pyrC) and dihydroorotate dehydrogenase (pyrD). Orotate phosphoribosyltransferase (pyrE) catalyzes the transfer of a ribose-5-phosphate moiety from PRPP to orotic acid, yielding the nucleotide orotidylic acid, which is subsequently decarboxylated to UMP by the action of the orotidine-5′-phosphate decarboxylase. H. pylori has the coding capacity for these four enzymes, whose activities in cells and lysates were reported by Mendz et al. (192). HP0266 and HP0581 are similar to the pyrC gene from Pseudomonas putida, HP1011 is similar to the pyrD gene from H. influenzae, HP1257 is similar to the pyrE gene from Thermus aquaticus, and HP0005 is similar to the pyrF gene from B. subtilis. The last steps in the generation of UTP involve phosphorylation of UMP to UDP and UDP to UTP by the sequential action of UMP kinase and nucleoside diphosphokinase, two enzymes whose activities have been observed in H. pylori (192) and which appear to be encoded by HP0777 and HP0198, orthologues of the pyrH and ndk genes, respectively.
Finally, amination of UTP by CTP synthase converts it to CTP. This enzyme activity was measured in bacterial lysates (192), and HP0349 is similar to the pyrG gene from H. influenzae.
Thus, the enzyme activities of the de novo pyrimidine synthesis pathway of H. pylori have been identified in situ, and putative genes coding for the corresponding enzymes have been found in its genome.
De novo synthesis of ATP and GTP.
Inosinate (IMP) is the first purine ribonucleotide formed de novo in the pathway (Fig. 5B). AMP and GMP are synthesized from IMP via separate branches of the pathway. From these monophosphates, ATP and GTP are generated by phosphorylations, with the intermediate formation of ADP and GDP (218).
IMP is formed in 10 enzymatic reactions by stepwise additions of functional groups to PRPP. In the H. pylori genome, only genes HP1218 and HP1112 were found to have similarities to genes coding for enzymes required for IMP synthesis: HP1218 was similar to purD, encoding glycinamide ribonucleotide synthetase, and HP1112 was similar to purB, encoding the bifunctional enzyme adenylosuccinate lyase, which also has a function in the synthesis of AMP from IMP. Although eight of the enzymes have not been identified in the genome, Mendz et al. (194) demonstrated full growth of the bacterium in the absence of purine bases, nucleosides, and nucleotides, indicating that H. pylori is able to synthesize purines de novo to meet its requirements. These results emphasize the need to compare the conclusions of genomic analysis with metabolic data in the quest to understand the physiology of the bacterium.
The conversion of IMP to GTP or ATP requires four enzymes for each branch of the pathway, and the genes coding for these enzymes seem to be present in H. pylori DNA. The genes involved in GTP synthesis are HP0829 (guaB) for IMP dehydrogenase, HP0409 (guaA) for GMP synthetase, and HP0321 (gmk) for 5′-guanylate kinase; and the genes involved in ATP synthesis are HP0255 (purA) for adenylosuccinate synthetase, HP1112 (purB) for adenylosuccinate lyase, and HP0618 (adk) adenylate kinase. GDP and most NDPs are converted to NTPs through the action of NDP kinase with ATP as the phosphate donor. HP0198, with high similarity to the ndk gene encoding this enzyme in Myxococcus xanthus, was found in the H. pylori genome. The presence of the genes of both pathways would suggest that these are the routes used by the bacterium to synthesize ATP and GTP from inosinic acid. However, no guanylate kinase activity is observed in bacterial lysates, and H. pylori adenylate kinase is able to phosphorylate AMP with GTP or ITP as phosphate donors, with production of GDP and IDP, respectively (193). These results suggest that the syntheses of GTP and ITP may follow alternative pathways and raise the possibility that the guaB and guaA genes are not expressed ordinarily. This matter can be resolved by direct experimental confirmation of the presence of the corresponding enzyme activities.
Nucleotide salvage pathways.
Salvage pathways enable cells to scavenge preformed nucleic bases and nucleosides for nucleotide synthesis or to reutilize bases and nucleosides produced endogenously as a result of nucleotide turnover, allowing circumvention of de novo pathways. Utilization of preformed pyrimidine bases is limited in H. pylori, with significant uptake of orotate and a lesser incorporation of uracil into the cells (192), but no gene similar to uraA, which encodes a cytoplasmic membrane protein required for uracil uptake in E. coli, was found in the H. pylori genome. Phosphoribosyltransferases are important for the salvage of free nucleic bases for biosynthesis, and enzyme activities for both orotate (OPRTase) and uracil (UPRTase) were observed in situ in H. pylori (192). In E. coli, the upp gene, coding for UPRTase, is the first gene of a bicistronic operon, with uraA as the second gene. In H. pylori, the gene HP1257, with similarity to pyrE coding for T. aquaticus OPRTase (part of the de novo pathway described above), was identified but no gene orthologous to upp was found. HP1180 has similarity to nupC, coding for a pyrimidine nucleoside transport protein in B. subtilis; however, the bacterium incorporates only very small quantities of uridine (192). No other genes orthologous to those encoding enzymes required for pyrimidine salvage in E. coli, such as deoA and tdk, coding for thymidine phosphorylase and thymidine kinase, respectively, or udp and udk, coding for uridine phosphorylase and uridine kinase, respectively, have been identified in H. pylori DNA.
In contrast, purine base salvage pathways are very active in H. pylori (Fig. 6). Adenine, guanine, and to a lesser extent hypoxanthine are incorporated by the bacterium at significant rates (193), but adenine transport systems like the high-affinity (purP) or low-affinity adenine transporters in E. coli or the transporters for guanine or hypoxanthine encoded by the pbuG gene of B. subtilis have not been identified yet in the H. pylori genome.
FIG. 6.
Purine salvage pathways. deoD, purine nucleoside phosphorylase; apt, adenine phosphoribosyltransferase; gpt, xanthine/guanine phosphoribosyltransferase. Circles denote enzymes whose enzymatic activities have not been observed but whose corresponding genes were identified. Dashed arrows show steps given in detail in Fig. 5B.
In E. coli, Salmonella typhimurium, and B. subtilis, transport of exogenous purine bases is tightly coupled to the metabolic processes that convert them to nucleotides and is controlled by the nucleotide pools in such a way that there is no intracellular accumulation of free bases (221, 307). Purine phosphoribosyltransferase activities converting adenine, guanine, and hypoxanthine to the corresponding NMP have been measured in H. pylori (193). The highest rates were measured for APRTase and GPRTase, converting adenine and guanine to AMP and GMP, respectively. The genes HP0572, orthologous to apt, encoding adenine phosphoribosyltransferase in E. coli, and HP0735, orthologous to gpt, encoding a xanthine/guanine phosphoribosyltransferase (XGPRTase) in H. influenzae, were identified in H. pylori DNA. The rate measured for H. pylori HPRTase activity converting hypoxanthine to IMP was significantly lower than those for APRTase and GPRTase (193), and no orthologous gene for a specific HPRTase was found in the H. pylori genome. In E. coli and S. typhimurium, the gpt and hpt genes encode GPRTase and HPRTase, respectively. However, in H. influenzae, hypoxanthine, xanthine, and guanine are substrates for XGPRTase to synthesize IMP, XMP, and GMP, respectively; and in B. subtilis, the same transferase uses guanine or hypoxanthine (221). Thus, it is possible that the enzyme encoded by the gpt gene in H. pylori is able to utilize both guanine and hypoxanthine. Interestingly, there is a relationship between the uptake of adenine, guanine, and hypoxanthine and the activities of the corresponding phosphoribosyltransferases in H. pylori, suggesting that uptake mechanisms are under the same metabolic controls as the salvage biosynthesis pathways, similarly to what occurs in E. coli, S. typhimurium, and B. subtilis. The significant amounts of purine bases incorporated by H. pylori and the relatively high activities measured for the transferases indicate that the bacterium can salvage purines efficiently via this pathway.
A route for the synthesis of IMP or GMP from inosine or guanosine, respectively, involves the phosphorylation of the corresponding nucleoside. In E. coli and S. typhimurium, the enzyme catalyzing this step is guanosine kinase encoded by gsk. The absence of guanosine kinase activity (193) and of a gene coding for it indicate that this route is not present in H. pylori. In E. coli and S. typhimurium, a major pathway for the salvage of purine nucleosides is their degradation to the corresponding base by a purine-nucleoside phosphorylase encoded by deoD followed by conversion to an NMP by the appropriate phosphoribosyltransferases (307). No purine-nucleoside phosphorylase activity was detected in H. pylori (193), although a gene, HP1178, orthologous to deoD was identified in its genome. This discrepancy may be explained by the relatively high adenine and guanine nucleosidase activities observed in H. pylori cell extracts (193), which could substitute for purine-nucleoside phosphorylase in the hydrolysis of the nucleosides and may have masked a weak phosphorylase activity, since the method used to assess the presence of the latter enzyme measures its activity at the same time as those of the nucleosidases. Alternatively, deoD may not be expressed under the bacterial growth conditions used in that study. In E. coli and S. typhimurium, the synthesis of purine-nucleoside phosphorylase is induced by purine nucleosides in the growth medium with concomitant suppression of the contributions of the de novo pathway to the purine nucleotide pool (307).
It is of interest that considerable purine nucleoside phosphotransferase activities were measured in H. pylori cell extracts (193). These enzymes phosphorylate adenosine or guanosine to AMP or GMP, respectively, and may constitute an alternative nucleoside salvage pathway not found in E. coli, S. typhimurium, or B. subtilis. No genes encoding these enzymes were identified in the H. pylori genome.
Adenine and guanine nucleotides can be interconverted through the common precursor IMP. These conversions serve to balance both types of nucleotides in the cellular pool, and they play an important role when the bacterium can salvage nucleobases in its habitat (307). Adenine nucleotides can be converted to guanine nucleotides via two pathways for which orthologous genes have not been found in H. pylori DNA. GMP is converted to IMP by the action of GMP reductase, which seems to be encoded in H. pylori DNA by the gene HP0854, a guaC orthologue.
Owing to the limited number of studies performed on nucleotide salvage pathways in H. pylori (192, 193), the present genomic analysis will be useful to future investigations in this field. Concerning pyrimidine salvage pathways, genes involved in these pathways in other organisms such as B. subtilis or E. coli are absent from the H. pylori genome, and the experimental data confirm this finding. Further experiments on the purine salvage pathways to support or disprove the conclusions deduced from the genomic analysis are required.
Deoxyribonucleotide biosynthesis.
In cells, the ratio of RNA to DNA is between 5:1 and 10:1; thus, most of the carbon which flows through the nucleotide biosynthesis pathways is directed to the rNTP pools. Nonetheless, the relatively small fraction that is directed to synthesizing dNTPs is of fundamental importance for the life of the cell, since dNTPs are used almost exclusively in the biosynthesis of DNA. In most organisms, the first step specific for deoxyribonucleotide synthesis is catalyzed by rNDP reductase (rNDPase), which converts all four rNDPs to the corresponding 2′-deoxyribonucleoside diphosphates. In H. pylori, the ORFs HP0680 and HP0364 have similarities to the nrdA gene from E. coli, encoding the α subunit of this enzyme, and the nrdB gene from Synechocystis sp., encoding the β subunit, respectively.
Thymine nucleotides are deoxy compounds with no ribonucleotide counterparts and therefore cannot arise simply through the action of rNDPase. The biosynthesis of dTTP occurs partly from the dUDP produced by rNDPase and partly from deoxycytidine nucleotides. In E. coli, dTTP synthesis requires four additional steps catalyzed by dUTPase (dUTP→dUMP) encoded by dut, thymidylate synthase (dUMP→dTMP) encoded by thyA, dTMP kinase (dTMP→dTDP) encoded by tmk, and nucleoside diphosphokinase (dTDP→dTTP) encoded by ndk. In H. pylori DNA, the genes HP0865, HP1474, and HP0198 are orthologous to dut, tmk, and ndk, respectively; but no gene orthologous to thyA was found (Fig. 7). The dTTP salvage pathway described in E. coli (218) requires the presence of uridine phosphorylase or thymidine phosphorylase and thymidine kinase, encoded by udp, deoA, and tdk genes, respectively. No ORFs with similarity to these genes were found in H. pylori. Alternatively, dTTP may be provided by the action of exodeoxyribonuclease on DNA, releasing NMPs, which can be converted to triphosphates as described above. The HP1526 gene is similar to the lexA gene encoding this enzyme in B. subtilis.
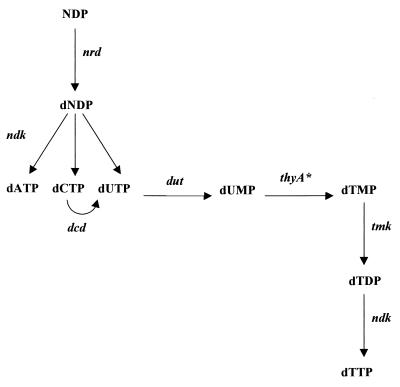
FIG. 7.
Deoxyribonucleotide biosynthesis. nrd, ribonucleoside diphosphate reductase; ndk, nucleoside diphosphokinase; dcd, dCTP deaminase; dut, deoxyuridinetriphosphatase; thyA, thymidylate synthase; tmk, thymidylate kinase. The asterisk denotes an enzyme for which no corresponding gene was identified. N can be A, C, and U.
ADP, GDP, and UDP are substrates in the synthesis of the corresponding NTPs, but CDP is not. It must be derived either from CTP by the action of nucleoside diphosphokinase or from CMP produced as a result of phospholipid synthesis (CDP-glycerides). CDP can also be provided by RNA degradation, and a polynucleotide phosphorylase enzyme seems to be encoded in H. pylori by gene HP1213, an orthologue of the pnp gene of H. influenzae. Once formed, dATP, dGDP, and dCDP are converted directly to the corresponding triphosphates by nucleoside diphosphokinase. dUTP can be also generated by the deamination of dCTP by the action of deoxycytidine triphosphate deaminase, a product of the dcd gene. HP0372, an orthologue of the dcd gene from H. influenzae, was found in H. pylori DNA (Fig. 7).
As noted above, the finding of an ORF similar to the deoD gene from E. coli encoding purine nucleoside phosphorylase suggests that deoxyadenosine, inosine, deoxyinosine, guanosine, and deoxyguanosine could be utilized as precursors for nucleic acid biosynthesis, although these enzymatic activities have not been observed in cell lysates (193).
In conclusion, H. pylori seems to be able to synthesize de novo many of the pyrimidine nucleotides and to have a limited utilization of the pyrimidine salvage pathways. The bacterium shows a greater capacity to salvage preformed purines, but at the same time it is able to grow and proliferate, synthesizing de novo purine nucleotides. The data from molecular analyses are not in complete agreement with the enzymatic activities detected in H. pylori by Mendz et al. (192, 193) and require further experimental verification.
Respiratory Chains
Many substrate oxidations are exploited by cells to generate metabolic energy. A central goal of respiration is to obtain reducing power to energize proton-translocating systems. Bacterial respiratory chains have a modular character, comprising dehydrogenase complexes, quinone pools, and terminal oxidoreductases (95). The terminal respiratory acceptor can be oxygen (aerobic respiration) or other substrates (anaerobic respiration). Besides their bioenergetic function (generation of proton motive force), respiratory systems also participate in the maintenance of intracellular redox balance (regeneration of NAD+) and the control of dioxygen concentration (scavenging of oxygen).
H. pylori has genes coding for proteins involved in both types of respiration. The genes for an NADH-quinone oxidoreductase respiratory chain have been found in an operon coding for the NDH-1 complex, HP1260 to HP1263 and HP1266 to HP1273. In bacteria, this complex is made up of 14 protein subunits whose genes are arranged in the same order with few absences in different NDH-1 operons (302). The H. pylori NDH-1 operon has genes encoding 12 subunits of the complex, and they are in the same order as their homologues in other bacterial NDH-1 operons, but it lacks the genes coding for two subunits of the complex which are involved in binding and oxidizing NADH. These genes are not found anywhere else in the H. pylori genome. In place of the two missing genes, the operon has HP1264 and HP1265, which lack binding motifs for NADH, flavin mononucleotide, and an FeS cluster, suggesting that H. pylori NDH-1 is a quinone reductase and a proton pump but not an NADH dehydrogenase (80). In the H. pylori genome, there is no gene encoding the NDH-2 complex, another type of bacterial NADH-quinone reductase which serves as an entry point to the respiratory chain of electrons donated by NADH but which is not a coupling site and does not translocate protons across the membrane (95). Since NADPH oxidase activity has been observed in membrane preparations (44) and crude bacterial supernatants (264), the hypothesis that NADPH was the electron donor for the H. pylori NDH-1 complex was investigated by Finel (80), who concluded that it was unlikely.
The ORFs HP0265, HP0378, HP0147, HP0144, HP0145, HP1539, HP1538, HP1540, HP0146, HP1461, and HP1227 are similar to genes encoding enzymes required for the synthesis of cytochrome c, cytochrome bc1 complex, and cbb3-type cytochrome c oxidase. In agreement with this observation, Marcelli et al. (170) reported that H. pylori cells and membranes contain b- and c-type cytochromes but not terminal oxidases of the a or d type. Also, the quinol oxidases, cb-type and cbb3-type cytochrome oxidases, appear to act as a terminal oxidases in the respiratory chain of H. pylori (1, 211).
In addition to the NADH-ubiquinone oxidoreductase complex, three other respiratory electron-generating dehydrogenases have been identified in the genome of H. pylori. The cytoplasmic membrane Ni hydrogenase of W. succinogenes consists of three polypeptides, HydA, HydB, and HydC, and contains cytochrome b. The hydrogenase genes are arranged in the order hydA, hydB, and hydC, with the transcription start site in front of hydA. An intergenic region of 69 nucleotides separates hydC from at least two more ORFs of unknown function (68). The sequential genes HP0631, HP0632, and HP0633 of the H. pylori genome are orthologous to the hydABC operon and are followed by ORF HP0634, which is similar to the W. succinogenes hydD, suggesting that H. pylori encodes a quinone-reactive Ni/Fe-hydrogenase. In the chromosome of Rhodobacter capsulatus B10, there is a sequence of 20 hup and hyp genes encoding hydrogenase-specific products which bear significant structural identity to the hydrogenase gene products from other bacteria (50). In the genome of H. pylori, the five ORFs HP0869, HP0900, HP0899, HP0898, and HP0047 are orthologues of the hyp genes, whose proteins are involved in hydrogenase expression and synthesis in several bacteria. In particular, ORF HP0048 has similarity to the hypF gene of R. capsulatus, whose product, HypF, is involved in the regulation of hydrogenase synthesis (49). Maier et al. (166) observed hydrogen uptake hydrogenase activity coupled to whole-cell respiration and identified Ni-Fe hydrogenase activity, which was shown to be subject to anaerobic activation.
H. pylori HP0961 and HP0666 seem to encode two distinct sn-glycerol-3-phosphate dehydrogenases, a so-called aerobic enzyme and an anaerobic enzyme. These enzymes serve as electron donors by converting glycerol-3-phosphate to dihydroxyacetone phosphate. In E. coli, the level of each enzyme is modulated by the growth conditions (32). A putative d-lactate dehydrogenase encoded by HP1222 (dld), which may serve as an electron donor, was found in H. pylori. d-Amino acid dehydrogenases are also potential electron donors, and the gene HP0943, an orthologue of dadA, which encodes the smaller subunit of the d-amino acid dehydrogenase in E. coli, is present in the H. pylori sequence; however, a gene for the larger subunit was not found. Recently, Evans et al. (76) identified a 27-kDa hydrophobic protein whose N-terminal sequence is similar to the β subunit of the formate dehydrogenase of Desulfovibrio vulgaris. The genes encoding the α and β subunits of this enzyme were not found by Tomb et al. (280), but the gene for the γ subunit was located. In D. vulgaris, formate dehydrogenase and cytochrome c553 form an oxidoreduction complex. In H. pylori, the polypeptide encoded by the ORF HP1227 was identified as an orthologue of cytochrome c553 from D. vulgaris, but Evans et al. (76) were not able to detect enzymatic activity of H. pylori formate dehydrogenase, leaving its contribution to energy production uncertain.
Other components that function as oxidoreduction mediators among the donor and acceptor complexes are quinones and b-type cytochromes (see above). Quinones are thought to function as mobile carriers which deliver reducing equivalents from dehydrogenases to terminal oxidoreductases in respiratory chains. Marcelli et al. (170) showed that the major isoprenoid quinone in H. pylori is menaquinone-6, with traces of menaquinone-4. The midpoint potential of the menaquinone/menaquinol couple in the membrane is about 190 mV lower than for the ubiquinone/ubiquinol couple; thus, menaquinones are better suited for a respiratory chain with lower potential electron acceptors; this could be a reason why these naphthoquinones are more commonly used for anaerobic respiration (95). The presence of menaquinones in H. pylori leads to the assumption that the anaerobic respiratory chain is used more often than the aerobic chain (275). This conclusion, however, must be drawn cautiously, because the specificity of a particular quinone for a given respiratory chain may also reflect particular structural features of quinones (95). Present in H. pylori genome are the genes HP0929 and HP0240, orthologues of the H. influenzae ispA gene, encoding geranyl pyrophosphate synthetase, and the E. coli ispB gene, encoding octaprenyl pyrophosphate synthetase, respectively. These are two enzymes of the biosynthesis pathway of the octaprenyl side chain of ubiquinones. HP1360 is an orthologue of the E. coli ubiA gene, which is involved in the second step of ubiquinone biosynthesis.
Fumarate may serve as an electron acceptor in anaerobic respiration. The existence of a fumarate transport system and an active fumarate catabolism via fumarate reductase in H. pylori was demonstrated in situ (184, 189, 191). The presence of fumarate reductase activity suggests the possibility of ATP generation via anaerobic respiration in the bacterium, in a manner similar to that in other anaerobic or facultative bacteria. The cloning and functional characterization of the H. pylori fumarate reductase operon showed that the FrdA and FrdB subunits are the catalytic dimers and that the FrdC subunit serves as a membrane anchor. This protein has a striking degree of sequence identity to W. succinogenes FrdC, which is a cytochrome b with two heme groups (25, 92). It has been proposed that the hydrogenase and fumarate reductase of H. pylori may be two components of an anaerobic respiratory chain which utilizes fumarate as the terminal electron acceptor (111).
The ability of H. pylori to use other terminal electron acceptors for anaerobic respiration seems to be limited, since no orthologues of genes coding for reductases of trimethylamine N-oxide, dimethyl sulfoxide, tetrathionate, or sulfite have been found in H. pylori. HP0642 and HP0954 exhibit similarity to a putative NAD(P)H nitroreductase gene from H. influenzae, and Ziebarth et al. (308) suggested that H. pylori possesses nitrate reductase and/or nitrosating enzymes such as cytochrome cd-1 nitrite reductase. Goodwin et al. (105) have shown that mutation in the rdxA gene encoding an oxygen-insensitive NADPH nitroreductase is associated with metronidazole resistance. HP1572 is a homologue of dniR, whose product in E. coli has a positive regulatory action inducing the synthesis of the nitrite reductase (cytochrome c552) when nitrate is used as a terminal electron acceptor in anaerobic respiration (138). Interestingly, there is no evidence of nitrate reductase activity in H. pylori or of a homologue of nitrite reductase gene in the genome. However, the HP0313 gene is an orthologue of B. subtilis narK, which encodes a nitrite extrusion protein, thus providing a mechanism for exporting nitrite from H. pylori cells.
The conundrum of the presence of an operative aerobic respiratory chain in H. pylori, together with the anaerobic respiration used by the bacterium at the low oxygen tensions, has not been resolved. A complete understanding of the physiology of this microaerophile requires the solution of this apparent paradox.
Catalase, Superoxide Dismutase, and Alkylhydroperoxide Reductase
There appear to be three principal mechanisms which enable H. pylori to resist oxidative damage, and they are catalyzed by the enzymes superoxide dismutase, catalase, and alkylhydroperoxide reductase (Ahp) (112, 167, 224, 233, 266). Inflammation within the gastric mucosa leads to an increase in toxic oxygen metabolites (66, 213). The superoxide anion, a highly reactive oxygen species formed as part of the oxidative burst of polymorphonuclear leukocytes, is dismutated to H2O2 by superoxide dismutase (207). Hydrogen peroxide is in turn converted to oxygen and water by catalase. Alkylhydroperoxide reductase catalyzes the reduction of alkyl hydroperoxide to the corresponding alcohol (233). In most bacteria, alkylhydroperoxide reductase is a two-component system consisting of the proteins AhpF and AhpC. AhpC is responsible for the peroxide reductase activity, while the accessory flavoenzyme, AhpF, possesses NADH or NADPH oxidase activity. In S. typhimurium, NADH oxidase or NADH oxidase-like activity coupled to AhpC is sufficient to generate active alkylhydroperoxide reductase (219, 220). The H. pylori gene HP1563 (tsaA) is an orthologue of ahpC (240, 241), but a homologue of ahpF was not identified in the H. pylori genome. However, there is evidence of the presence of NADH oxidase activity (264) in the bacterium. Catalase appears to be expressed in the cytoplasm and the periplasmic space and at the cell surface (112, 236, 242). It is a “classical” catalase, lacking peroxidase activity. These catalases usually are found in mammalian cells and contain a heme prosthetic group and NADPH binding activity (81). Examination of the H. pylori catalase gene katA (HP0875) product reveals a GXGXXG motif commonly associated with NADH rather than NADPH binding proteins (167). The H. pylori ORF HP0485 has similarity to the catalase-like gene cysR of Synechococcus spp., but at present its function is unclear.
Genomic and experimental data concerning enzymes involved in protection against oxidative damage agree, but the functions of some genes identified by sequence analysis with potential involvement in defence mechanisms require further elucidation.
Nitrogen Sources
Nitrogen is essential for the growth of all living organisms. Genome analysis indicates that H. pylori is able to use several substrates, including urea, ammonia, and some amino acids, as nitrogen sources (182, 280). Ammonia can be produced by the activity of urease (293), which allows access to urea nitrogen in the form of ammonium ions. The availability of energy and nitrogen determines the participation of glutamine synthetase (GSase), glutamate synthase (GOGATase), and glutamate dehydrogenase (GDHase) in ammonia assimilation. In energy-poor, nitrogen-rich (ammonia-containing) media, GDHase is important for glutamate synthesis and ammonia assimilation; in energy-rich, nitrogen-rich (ammonia-containing) media, GDHase levels are high and ammonia can be assimilated via GSase (244). The presence of the HP0512 gene in the H. pylori genome, similar to the glnA gene encoding GSase in E. coli, shows that assimilation of ammonia can be achieved via this route by converting glutamate to glutamine. This hypothesis was confirmed by the results obtained by Garner et al. (91). The failure to detect a gene orthologous to gltB, which encodes GOGATase in E. coli, suggests that α-ketoglutarate is transformed into glutamate by GDHase, whose gene, HP0380, an orthologue of gdhA, is in the H. pylori sequence (Fig. 8). The presence of glnA and gdhA and the absence of gltB suggest that H. pylori is adapted to an ammonia-rich environment. This appears to be the type of environment of the bacterium, whose metabolic machinery is capable of fast deaminations of several amino acids with concomitant generation of ammonium, as described above for amino acid metabolism (182). In addition to asnB, aspA, and sda, coding for various amino acid deaminases, H. pylori has the dadA gene, encoding d-amino acid dehydrogenase. As previously mentioned under “Amino acid metabolism,” there is also evidence for the presence of an aliphatic amidase (261), which catalyzes the degradation of amides supplying a nitrogen source by ammonia production.
FIG. 8.
Ammonia assimilatory cycle in E. coli and in H. pylori.
The analysis of the H. pylori genome sequence identified genes potentially involved in nitrogen assimilation. Although these genes reflect environmental conditions and metabolic characteristics of the bacterium, the way in which they actually function in vivo must be confirmed by experimental data.
Iron Acquisition
Iron is the fourth most abundant element in nature and is very important for biological systems, which require iron only at micromolar concentrations for growth (216). Under aerobic conditions, the oxidation state is Fe(III), which at neutral pH forms insoluble hydrated oxide polymers. Under anaerobic conditions, the predominant iron form is Fe(II), which is relatively soluble. Thus, in an oxidizing atmosphere, organisms have to develop efficient systems for iron assimilation. Presumably, these systems were developed after basic biochemical pathways had arisen, and this situation is reflected in their great diversity at the biochemical level (70). Given the multiple roles of iron-containing proteins, the potential toxicity of iron, and the desirability of keeping stores of iron, it is expected that iron assimilation would be tightly controlled by organisms (70). H. pylori, like other bacteria, requires iron-scavenging systems to survive in its environment. The iron-scavenging and regulation systems in the bacterium are summarized in Table 1.
TABLE 1.
Iron-scavenging and regulation systems
| System | Protein | HP no. | Reference |
|---|---|---|---|
| Storage | Ferritin Pfr | HP0653 | 21 |
| NapA | HP0243 | 20 | |
| Siderophore | FecEa | HP0888 | |
| FecDa | HP0889 | ||
| FecAa | HP0686 | 128 | |
| HP0807 | |||
| HP1400 | |||
| TonB | HP1341 | 288 | |
| ExbB | HP1130 | ||
| HP1139 | |||
| HP1445 | |||
| ExbD | HP1129 | ||
| HP1340 | |||
| HP1446 | |||
| CeuE | HP1561 | ||
| Ferrous iron uptake | FeoBb | HP0687 | |
| Regulation | FrpB | HP0876 | 300 |
| HP0915 | |||
| HP0916 | |||
| HP1512 | |||
| HP0590 | |||
| Fur | HP1027 | 19 | |
| Histidine kinase regulator | HP1043 |
FecR, FecI, FecB, and FecC seem not to be encoded by H. pylori.
The feoA gene was not identified in the H. pylori sequence.
Frazier et al. (87) and later Evans et al. (76) characterized a nonheme cytoplasmic iron-containing ferritin used for storage of iron (Pfr), which forms paracrystalline inclusions. Studies of the function of the pfr gene (HP0653) indicate that a nonheme-iron ferritin is involved in the formation of iron-containing subcellular structures and contributes to the resistance of H. pylori to the metal. Evidence of an interaction of ferritin with iron-dependent regulation mechanisms has also been obtained (20, 21). In addition to the ferritin Pfr mentioned above, H. pylori contains a bacterioferritin possibly involved in the storage of residual iron, encoded by the napA (HP0243) gene.
There has, however, been some controversy about the existence in H. pylori of siderophores, small iron-chelating molecules synthesized and released into the environment to scavenge iron. Husson et al. (127) did not detect siderophores. These authors suggested that the bacterium possesses a lactoferrin receptor, which would allow it to fix iron directly, and identified and characterized lactoferrin-binding proteins (67). In contrast, Illingworth et al. (128) reported the presence of siderophores produced by the bacterium. The results of H. pylori sequence analysis indicate the existence of a system for iron uptake, analogous to the siderophore-mediated ferric citrate (fec) uptake system of E. coli (268). The ORFs HP0888, HP0889, HP0686, HP0807, and HP1400 are similar to the genes belonging to the fec locus of E. coli, but they are not organized in a single operon. It should also be noted that neither the sensor FecR and regulatory FecI proteins nor the FecB dicitrate-binding and FecC transport proteins appear to be encoded by the H. pylori DNA (280). The sequence HP1341, orthologous to the tonB gene in P. aeruginosa encoding the siderophore-mediated iron transport protein TonB, was also found in the H. pylori genome. TonB is an essential component in iron-siderophore uptake in bacteria, apparently functioning as an energy transducer in coupling the energized state of the cytoplasmic membrane to open the outer membrane channel for iron dicitrate (241). In H. pylori, there are three pairs of sequential genes, HP1129-HP1130, HP1139-HP1140, and HP1145-HP1146, orthologues of the exbB and exbD genes of E. coli and H. influenzae, which code for the TonB accessory proteins, ExbB and ExbD, which have been characterized by Velayudhan et al. (288).
HP1561 and HP1562 are similar to the ceuE gene from Campylobacter coli, which is part of the Campylobacter enterochelin uptake operon (ceuBCDE), encoding components of a periplasmic binding protein-dependent transport system for the uptake of the ferric siderophore enterochelin in C. coli (248). Another system for iron uptake present in H. pylori appears similar to the ferrous (feo) iron uptake system present in E. coli. The feoAB two-gene operon encodes proteins which contribute significantly to the iron supply of E. coli cells under anaerobic conditions (139). The feoB-like gene HP0687, coding for a putative cytoplasmic protein with homology to ATPase, was identified in H. pylori, whereas no feoA orthologue seems to be present. Worst et al. (299) suggested the involvement of the ribBA-mediated riboflavin production in the feo-like system by the reduction of ferric iron to ferrous iron. HP0804 is similar to ribA and ribB, encoding GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase of Bacillus amyloliquefaciens. Thus, H. pylori appears to be able to assimilate ferrous iron.
The identification of HP0786, HP0915, HP0916, and HP1512, with sequence similarities to the frpB gene (Fe-regulated protein B) of Neisseria meningitidis, encoding iron-regulated outer membrane proteins (OMPs), suggests the presence of another system for iron uptake in H. pylori. This protein is homologous to several TonB-dependent outer membrane receptors of E. coli (23). The identification of iron-regulated OMP genes in H. pylori proteins is in agreement with the observations by Worst et al. (300) of three heme binding iron-repressible OMPs that might be involved in the uptake of heme from the host.
The ferric uptake regulator (Fur) is the main protein affecting the expression of iron-regulated genes. This protein is known to function in the presence of iron as a repressor of iron-controlled genes (13), and it exerts its regulatory activity by way of Fur binding boxes within the promoter region of selected genes. HP1027 encodes a homologue of the fur gene and was characterized by Bereswill et al. (19). The highest degree of homology was observed for the Fur protein from Campylobacter jejuni. The repressor activity in H. pylori depends on addition of iron to the medium, indicating that iron acts as a corepressor, as with the Fur proteins from other bacteria. Consensus sequences for Fur binding boxes were found upstream of the homologues of the two fecA, three frnB, and one fur genes of H. pylori (280). A Fur-binding box was also found upstream of the catalase katA gene of the bacterium by Odenbreit et al. (224) and was confirmed by Manos et al. (167). This is similar to the situation in C. jejuni, where a Fur binding box is also located upstream of the catalase gene (106).
The putative product of HP1043 is a histidine kinase regulator which has similarity to the ferric uptake PopP response regulator of Rhizobium leguminosarum, required for high-affinity iron acquisition in this bacterium (303). However, the greatest similarity of R. leguminosarum PopP is to E. coli PhoP, which, in E. coli and S. typhimurium, appears to regulate survival under extremely acidic conditions and may play a role in pathogenesis by increasing viability in the stomach (263).
This summary emphasizes the complexity of iron acquisition by H. pylori. The genome sequence will facilitate experimental molecular genetic and biochemical analyses that will address (i) the significance of the redundancy of these iron-scavenging systems, (ii) whether they are present in all H. pylori strains, (iii) whether their expression pattern is altered during infection, (iv) whether regulation of iron uptake is related to the survival of the bacteria under extremely acidic conditions, and (v) whether gastric and nongastric Helicobacter species possess the same iron-scavenging systems.
Urease and Nickel Uptake
H. pylori produces high levels of urease, which makes up about 6% of the total bacterial protein (153). This enzyme breaks down urea into ammonia and carbon dioxide, providing an acid-neutralizing cloud of ammonia that could protect the bacterium from gastric acidity (172). It has become increasingly clear that this is not the only function of urease in the physiology of H. pylori (114). The importance of this enzyme in bacterial colonization has been stated by others (153, 202, 207), and the present discussion is limited to the synthesis of urease and its relationship to nickel uptake.
H. pylori urease has unusual characteristics compared to those of other bacterial species. First, the enzyme is found in the cytoplasm as well as on the bacterial surface (69, 236). Second, it has two optimal pHs, one of which is acidic (78), and it displays higher substrate affinity than other bacterial ureases (204). Third, it is composed of only two subunits, UreA and UreB (46, 125), whereas other bacterial ureases contain three subunits (205).
The synthesis of active urease in H. pylori requires the expression not only of the ureA and ureB structural genes but also of four accessory genes, ureE, ureF, ureG, and ureH, which are necessary to incorporate nickel ions into the apoenzyme (65). The enzymatically active urease is a metalloenzyme, containing nickel ions. A fifth putative accessory gene, designated ureI, encodes a protein which is essential for the survival of H. pylori in the stomachs of mice used as a model of human infection, and it has been proposed that UreI spans the cytoplasmic membrane to facilitate the export of ammonium ions (262).
Other gene products are necessary for full urease activity. Mobley et al. (205) isolated the gene encoding the nickel transporter NixA, and another study (16) demonstrated that this gene is needed for the production of active urease. The structure and function of the nickel transport protein are under investigation (89). The ORF HP1576 encodes a protein similar to an H. influenzae ATP binding cassette (ABC) transporter, ATP binding protein (280). This H. pylori protein also has significant homology (56% amino acid sequence identity) to NikD, an ATP binding protein component of an ATP-dependent nickel transport system in E. coli that, together with NixA, is required for the production of a catalytically active urease (115).
The HP0011 gene encodes the heat shock protein HspA, which is also required for full urease activity (140). The C-terminal domain of this chaperonin exhibits a high affinity for nickel, and it was proposed that the role of HspA may be either to increase nickel incorporation or to stabilize the nickel in urease (153). In addition, a second system for nickel uptake exists in the form of a P-type ATPase transition metal ion uptake system with affinity for Ni2+, Cu2+, and Co2+ (180). Some of the mutants lacking this pump were negative for urease activity, but most of them remained positive.
Gilbert et al. (97) characterized a histidine-rich protein, Hpn, which strongly binds Ni2+ and Zn2+, and Tomb et al. (280) identified the HP1427 gene, encoding this protein. An Hpn-negative, isogenic H. pylori strain, generated by hpn gene deletion and grown on blood agar, had the same urease activity as a wild-type cell. The features of Hpn suggest that it acts as a Ni2+ storage system.
Urease and other proteins necessary for its full expression, particularly those involved in nickel acquisition, as well as their corresponding genes, have been extensively studied. The molecular data deduced from genomic analysis support the experimental findings.
pH Regulation and Responses to Acidic Conditions
The conditions of the ecological niche of H. pylori are dominated by the gastric H+, K+-ATPase, which secretes acid into the lumen of the stomach. Accordingly, the bacterium must have developed physiological strategies to survive in this environment. In vitro, its optimal growth is between pH 6.0 and 7.0, with no growth at all at pH below 4.5 to 5.0 or above 8.0 (210). In the presence of urea, it can survive in vitro at pHs as low as 3.0 (48). The urease is found in the cytoplasm and on the surface of H. pylori, both in vivo and in stationary-phase cultures. The ability of H. pylori to survive exposure to low pH is likely to depend on a combination of cytoplasmic and surface-associated urease activities (150). Glucose metabolism can take place in H. pylori at environmental pHs between 3.5 and 8.6, and cytoplasmic urease activity allows metabolism in the pH range from 2.5 to 4.0 by maintaining the periplasmic pH at 6.2 (245). On the other hand, surface-associated urease decreases H. pylori survival at neutral pH (196). Low environmental pH reduces urease activity as well as synthesis of nascent urease, catalase, and, presumably, many other proteins. This suggests that H. pylori is not acidophilic, although it tolerates short-term exposure to low pH (15). The bacterium is most probably a neutrophile that has adapted itself to the acidic environment of the stomach and can be classified as an acid-tolerant neutrophile.
In addition to the production of urease, H. pylori has developed other mechanisms of pH homeostasis. In many bacteria, the H+-translocating F1F0-ATPase is an important enzyme for regulating intracellular pH or synthesizing ATP. A transmembrane proton gradient or proton motive force (PMF) is formed by reversal of the F1F0-ATPase. In respiring cells, passage of electrons through an electron transfer chain to suitable electron acceptors is coupled to the extrusion of protons and the creation of a transmembrane electrochemical gradient of protons (137). The basic strategy of H. pylori is to maintain the PMF by adjusting the potential difference across the cytoplasmic membrane to compensate for the changes in pH gradient (196, 252). This PMF is used to convert energy into ATP through the F1F0-ATPase; thus, the PMF may be kept at a high enough level to allow ATP synthesis over a wide range of external pHs. The genes encoding orthologues of the α, β, γ, δ, and ɛ subunits of the F1 segment, responsible for the ATP synthesis and hydrolysis, are located in the sequential ORFs HP1134, HP1132, HP1135, HP1131, and HP1133, respectively. These ORFs are followed by HP1136 and HP1137, two genes coding for the b and b′ subunits, respectively, of the F0 segment involved in the H+ permeability pathway. The genes coding for subunits a and c of F0, HP0828 and HP1212, respectively, are found elsewhere in the H. pylori DNA. The distribution of the genes encoding the F1F0-ATPase reflects the more highly conserved structure of the F1 knob subunits and the more variable composition of the F0 base. This enzyme has been extensively analysed by McGowan et al. (174).
The bacterium can create a positive-inside membrane potential by either concentrating cations or pumping out anions. Tomb et al. (280) suggested that the first strategy is more likely since no clear mechanism for anion efflux has been identified. To date, genes coding for only three anion transporters have been found: HP0473 to HP0475, encoding the ABC transporter ModABC involved in the uptake of molybdenum; HP1491, encoding the phosphate permease; and HP0313, encoding the nitrite permease. However, Tomb et al. (280) pointed out that anion conductances associated with other proteins have not been determined, especially for the multidrug resistance (MDR)-like transporters.
Three proton-translocating P-type ATPases have been identified in H. pylori: ATPase-439 (CadA), ATPase-948 (CopA), and ATPase-115 (156, 166, 167). Initially, it was thought that these ATPases played a role in pH regulation by extruding protons from the cytoplasm, but Tomb et al. (280) suggested that they are more closely related to divalent cation transporters. There is agreement that they are involved in importing divalent cations and eliminating toxic metals rather than in pH regulation. HP1552 and HP1183 are similar to genes coding for the Na+/H+ antiporters in E. coli and in Enterococcus hirae, respectively. These H+-coupled ion transport systems are responsible for controlling the flow of ions into and out of the cell and also would play a role in pH regulation.
Basic amines, products of amino acid decarboxylases, provide an important protective mechanism for bacteria living under low-pH conditions, and they may be involved in pH homeostasis in H. pylori. The HP0422 gene is similar to speA, encoding arginine decarboxylase in E. coli. This enzyme may play a role in regulating pH by utilizing products of protein biodegradation (225).
Urease plays an important role in acid resistance in H. pylori, but there are indications that the bacterium has developed other specialized intrinsic defenses. In a study by Bijlsma et al. (24), urease-positive UV mutants grew identically to wild-type strains on pH 7 plates but did not grow on pH 5 plates. Complementary studies with mutant and wild-type strains suggested that in H. pylori a urease-independent acid resistance system, probably dependent on the expression of more than one gene, is involved in growth at low pH. Novel proteins are produced when H. pylori cells are subjected to a pH shift from 7 to 3, and the gene encoding a P-type ATPase that may catalyze NH4+-H+ exchange across the cytoplasmic membrane has been cloned (175). In another study, exposure of H. pylori to an acidic pH induced the production of a neutrophile-activating protein and alkyl hydroperoxide reductase, although no change in the level of transcription of the genes HP1183 or HP1563 was detected (173). A possible explanation is that the effect of acid on these proteins reflects a redistribution of their localization, an alteration in their physical properties, or changes in the levels of their translation or degradation. Investigations of urease-independent mechanisms of resistance to acid stress by H. pylori has led to the identification of genes whose expression is induced after exposure to acidic pH. The HP0045 gene encodes a predicted 34.8-kDa protein (WbcJ) homologous to known bacterial O-antigen biosynthesis proteins involved in the conversion of GDP-mannose to GDP-fucose. An isogenic wbcJ-null mutant strain failed to express O-antigen and Lewisx or Lewisy determinants and was more sensitive to acid stress than was the wild-type strain. Qualitative differences in LPS profiles were observed between H. pylori cells grown at pH 5 and pH 7, suggesting that H. pylori may alter its LPS structure in response to acidic pH. This may be an important adaptation facilitating H. pylori colonization of the acidic gastric environment (176).
The nucleotide excision repair (NER) pathway contributes to the repair of DNA damage, and HP1114 (uvrB) is a putative NER gene in H. pylori. An isogenic H. pylori UvrB-negative mutant constructed by inserting a kanamycin resistance cassette into uvrB showed a greatly increased sensitivity to the DNA-damaging agents methyl methanesulfonate and UV radiation, as is the case with UvrB-negative mutants of other bacterial species. Low pH was significantly more lethal to the uvrB mutant than to the wild-type strain, suggesting that the H. pylori NER pathway is involved in the repair of acid-induced DNA damage (279).
Finally, pathogenic bacteria can respond to chemical and physicochemical signals by induction or repression of appropriate genes. The pH of a host microenvironment appears to be one of these signals (225). At low pH, H. pylori may change the expression of some virulence genes, outer membrane proteins, or sensor-regulator proteins. For example, H. pylori cells of cagA+ strains grown at pH 6 for 48 h, which induces maximal CagA expression, were significantly more susceptible to pH 3 than were wild-type cagA strains or isogenic cagA+ knockouts. These data suggest that H. pylori possesses a differential acid susceptibility that may contribute to preferential colonization of particular H. pylori strains in specific mucosal layer niches (142).
Virulence factors in enterobacteria are coordinately regulated by a variety of environmental signals (198). A growing number of acid-inducible virulence factors have been identified in E. coli, S. typhimurium, and other enterobacteria (263). ToxR is a transmembrane DNA binding protein that regulates the expression of cholera toxin, pilus, and outer membrane proteins in V. cholerae. Some environmental signals such as osmolarity and pH are sensed by ToxR, which couples them to the expression of toxR-regulated proteins (197). The tagE gene is located within a cluster of genes required for efficient intestinal colonization and is ToxR activated (147). In H. pylori, ORFs HP1543 and HP1544 are two orthologues of tagE. If their products are similar to TagE, it would be of considerable interest to determine whether their expression is also pH regulated.
Experimental data show that several mechanisms have been developed by H. pylori to regulate internal pH and respond to acidic conditions. Data obtained from analysis of the genome agree with these observations and suggest areas for investigating further the acid resistance of the bacterium.
REPLICATION, TRANSCRIPTION, AND TRANSLATION
Replication
Tomb et al. (280) found no typical eubacterial origin of replication, and so they arbitrarily designated bp 1 at the beginning of a repeat which produces translational stop codons in the three reading frames. However, the HP1529 gene was identified an orthologue of dnaA from B. subtilis, whose product is the chromosomal replication initiator protein which binds to the replication origin (DnaA boxes) (195). RNA polymerase is involved in the initiation of DNA replication by promoting a transcriptional event which precedes actual initiation (171). No sequence similarity was found to the gene coding for the DnaC protein, which also acts in the initiation of the DNA replication at oriC and is essential for the viability of E. coli (195). No ORF similar to the dnaT gene which is involved in the termination of DNA replication was identified. Interestingly, dnaC and dnaT genes are transcribed in a single operon in E. coli. Nevertheless, the HP1362 gene is an orthologue of the gene coding for DnaB, a replicative helicase, which interacts with DnaC to form the prepriming complex; and the HP0387 gene has sequence similarity to the priA gene from H. influenzae, encoding a primosomal protein replication factor which seems to act as a helicase within the primosome. Sequence comparisons allowed Tomb et al. (280) to identify other genes encoding proteins that participate directly in the propagation of a replication fork: HP1245, an orthologue of ssb, whose product SSB binds to single stranded DNA; HP0012, an orthologue of dnaG, encoding the primase DnaG, whose functional role is to synthesize the RNA primers needed for initiation of the lagging-strand Okazaki fragments; HP1460, HP1387, HP0500, HP1231, and HP0717, orthologues of dnaE, dnaQ, dnaN, holB, and dnaX, respectively, encoding the subunits of DNA polymerase III holoenzyme, which is the replicative DNA polymerase; HP1470, an orthologue of polA, which codes for DNA polymerase I, associated with nick translation activity in discontinuous DNA replication; and HP0615, an orthologue of lig, whose product is the DNA ligase necessary to seal DNA nicks. Genes HP0701 and HP0501 are orthologues of gyrA and gyrB, encoding the subunits of the DNA gyrase which plays a crucial role in initiation, propagation, and termination of replication by introducing negative supercoils into chromosomal DNA. Moore et al. (206) had sequenced the gyrA gene by characterizing ciprofloxacin-resistant mutants of H. pylori. Orthologues of genes encoding proteins that may play an auxiliary role in the replication of the chromosome in E. coli were identified in H. pylori: HP0911 to rep, encoding the Rep protein; HP1478 to uvrD, coding for DNA helicase II; HP0661 and HP1323 to rnhA and rnhB, whose products are RNase H and RNase HII, respectively; HP0116, HP0440, and ORF02428 to three copies of topA, encoding DNA topoisomerase I; HP1332, HP0109, HP0110, HP0011, and HP0010 to dnaJ, dnaK, grpE, groES, and groEL, respectively, coding for the heat shock proteins DnaJ, DnaK, GrpE, GroES, and GroEL; and HP0083 to hup, encoding the histone-like DNA binding protein HU.
Experimental data concerning the replication of H. pylori chromosome are scarce. The genomic analysis revealed surprising findings, in particular in the initiation of replication, and provided sound foundations for achieving a complete understanding of this process in H. pylori.
Transcription
The ORFs HP1293, HP1198, and ORF01039 were found to be similar to rpoA of B. subtilis, rpoB of E. coli, and rpoC of H. influenzae, respectively, encoding the subunits of RNA polymerase. As pointed out by Tomb et al. (280), the genes coding for the β and β′ subunits are fused in H. pylori, while they are contiguous but separate in other prokaryotes studied (306). Tomb et al. (280) identified also orthologues of genes encoding several transcription factors including three transcription initiation factors in H. pylori DNA. Sigma- factors promote attachment of the RNA polymerase to specific initiation sites and are released afterward. HP0088 is an orthologue of the rpoD gene from H. influenzae, which encodes the RNA polymerase sigma 70 factor, generally the primary sigma factor in gram-negative bacteria. ORF HP0714 is similar to the rpoN gene from B. subtilis. This gene codes for the sigma 54 factor, which is used by the promoter of the minor flagellin flaB gene (reference 134 and see below). The alternative transcription initiation factor, sigma F, also is encoded by H. pylori DNA. This sigma factor is specific for the flagellar operon in E. coli. HP0866 is an orthologue of greA from E. coli, which encodes a factor necessary for efficient RNA polymerase transcription elongation beyond template-encoded arresting sites. HP1514, HP1203, and HP0550 have similarities to genes coding for the transcription termination factors NusA, NusG, and Rho, respectively. The NusG protein acts as a component of the transcription complex and interacts with the termination factor Rho and RNA polymerase. NusA participates also in the transcription termination process (247). An ORF similar to the antitermination factor NusB also was identified in the H. pylori genome. Regulation of transcription is discussed later in the review (see “Regulation of gene expression”). Genomic analysis suggests that the transcription process in H. pylori is very similar to that of other gram-negative bacteria. This information requires experimental confirmation.
Translation
The translational apparatus is a major component of the machinery to express the genetic information of the cell. In addition to ribosomes, tRNA, and mRNA, it includes numerous ligands, monovalent and divalent ions, nucleotides, and proteins, such as tRNA-modifying enzymes, aminoacyl-tRNA synthetases, and proteins transiently associated with the ribosomes, e.g., initiation, elongation, and release factors.
H. pylori contains two separate sets of 23S-5S-16S rRNA genes in addition to one orphan 5S rRNA gene and one structural RNA (ssrD) gene (280). A total of 36 tRNA genes were identified in H. pylori. They are organized into seven clusters plus 12 single genes. Orthologues of genes encoding nine tRNA-modifying enzymes were found (Table 2). However, no ORF similar to the rnd, rph, and rnpB genes, whose products are involved in tRNA maturation, were identified. A search for tRNA synthetase genes in the H. pylori genome identified all the expected genes, except glutaminyl-tRNA synthetase (glnS) and asparaginyl-tRNA synthetase (ansS). In other bacteria, a single glutamyl-tRNA synthetase aminoacylates both tRNAGlu and tRNAGln with glutamate. The formation of glutaminyl-tRNA may be accomplished by amidation of glutamate to glutamine in a reaction that is functionally analogous to that catalyzed by glutamine synthetase (270). This transamidation may also occur in H. pylori. However, as pointed out by Tomb et al. (280), in H. pylori the product of the second glutamyl-tRNA synthetase genes HP0643 and HP0476 (gltX) may perform the function of glutaminyl-tRNA synthetase. The lack of the asparaginyl-tRNA synthetase gene is, at this time, unique in whole-genome-sequenced bacteria. Even Mycoplasma genitalium, which is thought to contain the smallest genome for a self-replicating organism, has the coding capacity for this enzyme (86). Tomb et al. (280) reported that a transamidation process from Asp-tRNAAsn to Asn-tRNAAsn may operate in H. pylori, as described for the archeon Haloferax volcanii (64). Alm et al. (3) have proposed an alternative mechanism involving HP0830, HP0658, and HP0975, homologous to the gatABC genes of B. subtilis, whose products amidate glutamate-charged tRNAs to make glutamine-charged tRNAs. The products of these H. pylori genes also would be responsible for the amidation of aspartate-charged tRNAs.
TABLE 2.
tRNA-modifying enzymes and related genes
| HP no. | Gene | Protein |
|---|---|---|
| HP1141 | fmt | Methionyl-tRNA formyltransferase |
| HP1497 | pth | Peptidyl-tRNA hydrolase |
| HP0361 | hisT | Pseudouridylate synthase I |
| HP1148 | rnpA | Protein component of ribonuclease P |
| HP1062 | queA | S-Adenosylmethionine:tRNA ribosyltransferase isomerase |
| HP1513 | selA | Selenocystein synthase |
| HP1148 | trmD | tRNA (guanine-N1) methyltransferase |
| HP1415 | miaA | tRNA delta (Z)-isopentenyl pyrophosphate transferase |
| HP0281 | tgt | tRNA-guanine transglycosylase |
The mature, 70S ribosome particle can be dissociated into two subunits, a small 30S subunit and a large 50S subunit. A total of 21 ribosomal proteins constituting the 30S subunit are encoded by H. pylori (S1 to S21), and 31 orthologues of the ribosomal proteins of the 50S subunit were found (L1 to L7, L9 to L11, L13 to L24, L27 to L29, and L31 to L36), as expected. HP1298, HP1048 and HP0124, with similarities to the infA, infB, and infC genes, respectively, from Bacillus spp., were found in H. pylori DNA. The respective gene products interact and are involved in the formation of the initiation complex of 70S ribosomes with initiator fMet-tRNAfMet, mRNA, and GTP. Four peptide chain elongation factors are encoded by H. pylori: EF-G (HP1995, fusA), EF-P (HP0177, efp), EF-Ts (HP0177, tsf), and EF-Tu (HP1205, tufB). Orthologues of genes coding for the three release factors RF-1, RF-2, and RRF are also found in H. pylori DNA (HP0077, HP0171, and HP1256, respectively).
In conclusion, most of the translational apparatus in H. pylori is similar to that of other bacteria, but the absence of two tRNA synthetases has led to competing explanations of the mechanisms of amidation of aspartate-charged tRNA and formation of glutamine-tRNA. Experimental data are required to identify these mechanisms.
REGULATION OF GENE EXPRESSION
In response to environmental stimuli, bacteria must be able to regulate the expression of their genes. A search for regulatory proteins in the H. pylori genome indicated that it encodes few such proteins in comparison with E. coli. This finding may be indicative of the high level of adaptation to the human gastric environment, the lack of competition from other microorganisms, and the capacity of the bacterium to be transmitted efficiently from person to person, particularly in childhood (200). Examples of a number of important established or putative regulatory factors encoded by the H. pylori genome are discussed below. In most cases, no experimental data are available, and experiments are necessary to establish the existence of regulatory mechanisms in vivo.
Helix-Turn-Helix
Only three predicted proteins containing a helix-turn-helix motif known to allow DNA recognition of transcriptional factors were identified (280). These three predicted proteins consist of two encoded by ORFs HP1124 and HP1349, with no database match, and the putative heat shock protein HspR, encoded by HP1025, which interacts specifically with the dnaK (chaperone) promoter region in Streptomyces coelicolor (36) (Table 2).
Heat Shock and Oxidative Stress
A surprising finding in the genome of H. pylori was the absence of homologues to the transcription regulatory sigma factors 32 (heat shock) and ςS (stress and stationary phase) (280). The absence of these factors does not indicate an absence of heat shock proteins in the bacterium. Indeed, homologues of genes encoding the heat shock proteins discussed above and listed in Table 3 were identified in the genome regulated by a housekeeping ς70 like-sigma factor (2, 17, 274, 280).
TABLE 3.
Proteins involved in gene expression regulation
| Protein | HP no. | Putative function |
|---|---|---|
| HP1124a | ||
| HP1349a | ||
| HspR | HP1025 | Heat shock protein |
| GroEL | HP0010 | Chaperone and heat shock protein |
| GroES | HP0011 | Cochaperone |
| DnaK | HP0109 | Cochaperone and heat shock protein 70 |
| DnaJ | HP1332 | Cochaperone and heat shock protein |
| GrpE | HP0110 | Cochaperone and heat shock protein |
| SpoT | HP0775 | Control of ppGpp synthesis |
| GppA | HP0278 | Control of pppGpp synthesis |
| CstA | HP1168 | Carbon starvation |
| LytB | HP0400 | Penicillin tolerance |
| NdK | HP0198 | De novo synthesis of nucleotide |
| Signal-transducing proteins | ||
| VanRB | HP1365 | Transcriptional activator |
| FleR | HP0703 | Transcriptional activator |
| PopP | HP1043 | Transcriptional regulator |
| CiaR | HP1043 | Transcriptional regulator |
| HypF | HP0048 | Transcriptional regulator |
| TenA | HP1287 | Transcriptional activator |
| NifR3 | HP0727 | Transcriptional activator |
| AtoS | HP0244 | Histidine kinase sensor protein |
| Phor | HP0164 | Signal-transducing protein kinase |
| RprX | HP1364 | Signal-transducing protein |
The products are identified as regulatory proteins by conserved motifs.
The induction of an inflammatory response by H. pylori infection leads to increased potential for oxidative damage to the bacterium. As discussed below, H. pylori has the enzymatic capacity to deal with such oxidative stress. Despite this, no orthologues of the genes encoding the oxidative stress regulator components of OxyR, SoxR, SoxS, or SOS systems have been found in H. pylori DNA.
Stringent Response
Orthologues of genes coding for other proteins without the helix-turn-helix motif but with putative regulatory function were identified in H. pylori (Table 3). Guanosine-3′-diphosphate-5′-diphosphate (ppGpp) is a mediator of the stringent response that coordinates a variety of cellular activities in response to changes in nutritional status (40). The major trigger for the stringent response is a decrease in the intracellular concentration of aminoacyl-tRNA. In bacteria such as E. coli, the synthesis of ppGpp is controlled by two gene products, ppGpp synthetase I (relA) and a bifunctional ppGpp 3′-pyrophosphohydrolase/ppGpp synthetase II (spoT) (116, 301). ppGpp synthetase I, also known as stringent factor, is classified as a ribosome-dependent enzyme, while the SpoT product is classified as a ribosome-independent enzyme.
No gene similar to the relA gene encoding ppGpp synthetase I was found in the H. pylori genome. However, HP0775 is an orthologue of spoT. Indeed, HP0198, HP0278, HP1168, and HP0400 were identified as orthologues of ndk, gppA, cstA, and lytB, coding for NDP kinase, guanosine pentaphosphate (pppGpp) phosphohydrolase, carbon starvation protein A, and penicillin tolerance protein, respectively. The H. influenzae product of lytB plays a role in penicillin tolerance and also in the stringent response. While some regulatory functions relate to nitrogen starvation, the carbon starvation protein A is required for peptide utilization when available carbon is limited.
In addition to ppGpp, the nucleotide pppGpp is involved in the control of the stringent response and may be more important in bacteria such as Staphylococcus spp. than is ppGpp (41). Thus, the presence of orthologues of spoT and gppA in the H. pylori genome indicates that both ppGpp and pppGpp may be synthesized in the bacterium, and hence they are likely to be key elements of the stringent response in H. pylori.
Histidine Kinase Sensors
Members of the two-component transcriptional regulator family, either the histidine kinase sensor (four orthologues) or the DNA binding response regulator (seven orthologues), were identified in H. pylori (Table 3). HP0703 is an orthologue of the gene encoding the FleR response regulator. FleR is a flagellar synthesis transcriptional activator in P. aeruginosa and is homologous to other regulatory proteins that bind to specific upstream activating elements to enhance transcription of genes with ς54 promoters (249). In E. coli, ς54 is also an important regulator in situations of nitrogen (ammonia) limitation.
Unlike other bacterial flagellar filaments, which are composed of a single protein, H. pylori flagellar filaments are composed of polymers of two subunits: the major flagellin, FlaA, and the minor flagellin, FlaB. The flaA gene is expressed from a ς28-like promoter, whereas flaB is expressed from ς54. Tomb et al. (280) reported the presence of these two types of promoter elements upstream of several flagellar genes, suggesting a complex transcriptional regulation of the flagellar regulon.
HP0244, encoding a putative histidine kinase sensor protein, is a homologue of atoS, encoding the AtoS sensor protein. AtoS activates AtoC in E. coli and RprX in Bacteroides fragilis by phosphorylation. In E. coli, AtoC is a positive ς54 RNA polymerase transcriptional activator regulating short-chain fatty acid metabolism (131).
The information provided by the genome about regulatory networks in H. pylori allows some speculation about its possible modes of transmission. Although H. pylori appears to be transmitted from person to person (200), experimental data report the presence of H. pylori in the mouth, feces, and dental plaque (178, 265), and environmental reservoirs also have been implicated in the transmission of the bacterium. As a consequence, one aspect of the physiology of H. pylori which has generated considerable controversy is the biological relevance of coccoidal forms of the bacterium. The controversy is illustrated by Kursters et al. (152), who suggested that the coccoidal form of H. pylori is the morphologic manifestation of bacterial cell death, while on the other hand, Cole et al. (51) claimed that H. pylori exists both as an actively dividing spiral form and a nonculturable but viable metabolizing coccoidal form. Based on the limited number of regulatory networks of the bacterium, it seems unlikely that the coccoidal forms correspond to an adaptative form for survival in the environment. Indeed, if they are involved in survival under unfavorable conditions, one would expect to find an active switch in the metabolism of the bacterium regulated by homologues of the heat shock or stress and stationary-phase transcription regulatory sigma factors ς32 and ςS. The presence of such switching is not supported by the molecular data on the regulatory networks present in H. pylori.
The paucity of regulatory functions in H. pylori may reflect a tight adaptation of this organism to the restricted ecological niche of the human stomach. However, these conclusions are not final and would benefit from further investigation into gene expression in the stomach. In particular, H. pylori may regulate gene expression by methylation, as suggested by the presence of nine type II methyltransferase genes conserved between strains 26695 and J99 and the absence of the restriction subunit partners (3).
DIVERSITY
H. pylori is associated with diverse pathologies of varying severity, such as chronic gastritis, peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma, and gastric carcinoma. The occurrence of such diverse diseases can be due to host genetic factors together with environmental factors including diet, but it may also depend on specific properties of the pathogen (203). For example, a person carrying a strain with the cag pathogenicity island (cag+), s1a (vacA), and iceA1 genes (detailed below) has a higher risk of developing duodenal ulceration than a person infected by a strain harboring different alleles of vacA and iceA and no cag pathogenicity island (27). Also, preliminary studies have shown that the risk of esophageal and gastric cardia adenocarcinoma is decreased in persons infected with a cag+ strain (45). It is therefore important to evaluate the heterogeneity of H. pylori strains within the population.
The genetic diversity of H. pylori has been the focus of many investigations. It can be analyzed at two different levels: the genomic variation between strains originating from different individuals, and the variations in populations within an individual host. Numerous studies based on plasmid profiles (228, 260), restriction fragment length polymorphism (RFLP) analysis of chromosomal DNA (120, 276) or specific loci (10, 90, 141, 243, 286, 297), and repetitive extragenic palindromic PCR (99) have shown that there are substantial levels of variation among natural isolates of H. pylori. The general impression has been that this pathogen may be more diverse than most other microorganisms. Go et al. (101) estimated the genetic diversity of H. pylori isolates by examination of allelic variation in six genes encoding metabolic housekeeping enzymes by using multilocus enzyme electrophoresis. They showed that the mean diversity exceeds the level of diversity recorded in virtually all other bacterial species studied by the same technique. Similarly, based on multilocus enzyme electrophoresis data generated from 23 isolates using 16 enzyme loci, Hazell et al. (111) suggested that in view of the genetic diversity found, isolates may be more appropriately termed as belonging to a species complex than to a single species. Using randomly amplified polymorphic DNA-PCR and DNA fingerprinting, van der Ende et al. (285) showed that strains from unrelated infected patients had unique fingerprints whereas strains isolated from family members had very similar although not identical patterns. These results imply that differences observed between the strains infecting individual family members occurred after the primary infection. They also suggest that the evolution of H. pylori is ongoing and continues to result in highly diverse strains.
On the other hand, the opinion of Hancock et al. (109), who have sequenced the J99 strain, is that H. pylori is not as diverse as may be thought. The authors of a recent study (3) comparing the genome sequences from strains 26695 and J99 conclude that the overall organization, gene order, and predicted proteomes of both strains are quite similar and that an overestimation of the true extent of genetic variation in H. pylori has occurred probably through the use of lower-resolution techniques, such as pulsed-field gel electrophoresis and RFLP.
Different mechanisms, such as point mutations or mosaicism within a conserved gene, nonconservation of some genes, chromosomal rearrangements, variation in a number of intragenic cassettes, and extragenic elements, could contribute to the genetic diversity. We illustrate such mechanisms by providing several examples and then draw together the genetic possibilities relevant to H. pylori.
Point Mutations within a Conserved Gene
The first mechanism through which genetic diversity within the species may occur is the point mutation; i.e., only a single base pair of DNA is changed. Examination of the heterogeneity results from RFLP analysis indicates that much of this phenomenon results from variation in a single base pair. Kansau et al. (141) showed that the primary sequence of a 210-bp region of the glmM (previously ureC) gene was different for each of 29 strains. The same region of the glmM gene (294 bp) was sequenced for 46 strains isolated from unrelated patients by Courcoux et al. (53). The sequences obtained were again unique for each strain. The polymorphism observed by this technique was correlated with that deduced from restriction endonuclease analysis. Alm et al. (3) hybridized in silico the genome of strain 26695 with NotI fragments from strain J99 used as probes and observed fragments with different sizes from those obtained with the genome of strain J99. The difference in sizes in the fragments from strain 26695 and J99 were due mainly to silent nucleotide variations within genes; i.e., no polymorphism was detected at the amino acid level.
Examination of the various sequences shows that many of the point mutations are silent. The mutations have accumulated in the neutral part of the codon (third position). However, nonsynonymous substitutions also occur. In contrast, for the so-called cytotoxin-associated gene (cagA), which is not conserved in all strains, the ratio between the mutations accumulated in the first and second positions and those in the third position of the codon is approximately 1.1:0.9 (55), which suggests that the selective pressure on H. pylori genes is not uniform.
Another example is illustrated by the characterization of the allelic diversity of the iceA gene, a putative restriction enzyme which appears to be induced when H. pylori encounters epithelial cells (232). Sequencing of iceA genes from seven clinical isolates showed that two major allelic types, iceA1 and iceA2, could be defined.
Genotypic and Phenotypic Variation in the vacA Gene
The mechanisms involved in phenotypic and antigenic variation of H. pylori can be divided into three types: gene mosaic organization, intragenic recombination, and “on-off” switching owing to DNA slippage or repeat motif insertion within a coding sequence.
Some H. pylori strains produce a cytotoxin that causes vacuolation in cultured epithelial cells (158). These strains are isolated more frequently from patients with peptic ulcer disease than from patients without it (79, 277). Antibodies to this protein were detected in patients infected with toxin-producing strains, demonstrating the vacuolating cytotoxin production in vivo (157).
Several groups have cloned and sequenced the gene encoding the cytotoxin designated vacA (60, 235, 255, 278). The corresponding vacuolating cytotoxin (VacA) is produced by approximately 50% of H. pylori strains (58), and there is a heterogeneity in the level of vacA transcription among these strains (84), but the vacA gene encoding the cytotoxin was present in all strains tested (60, 235). Inactivation of vacA abolishes the cytotoxicity in cells cultured in vitro (96).
Atherton et al. (10) characterized the vacA gene from 59 different H. pylori isolates and demonstrated the presence of a mosaic structure. The vacA gene contains both conserved regions and regions of diversity. The gene segment encoding the C-terminus of the protoxin, and the segment encoding the region near the amino terminus appear to be conserved in all isolates. However, there is sufficient diversity in the mid-region of the gene to define at least four allelic types, designated m1, m2, m1-like (m1*), and m1*-m2, and at least three different families of vacA signal sequences, designated s1a, s1b, and s2, can be defined, indicating the sequence diversity between strains in this region (Fig. 9). The existence of different s genotypes and m genotypes of vacA yields the possibility of multiple combinations (10, 229). Interestingly, variance in the s and m genotypes appears to be correlated with different levels of production of the VacA cytotoxin and hence may be a marker for differences in the virulence potential of strains (11).
FIG. 9.
Diversity of the VacA protein.
Recently, van Doorn et al. (287) reported a novel vacA subtype, designated s1c, in isolates from East Asia, with three different allelic forms, m1, m2a, and m2b, in the m region. Strobel et al. (271) have also claimed that further variants of the m1 genotype of vacA exist and have proposed an additional designation, m1a. In a study conducted in Germany, H. pylori strains of the vacA-s1 genotype were isolated from 96% of patients with peptic ulcer disease. The vacA-s2 genotype was present in only 4% of these patients but in 31% of the patients with non-ulcer dyspepsia. In contrast, Japanese strains are highly homologous, with more than 96% having the vacA-s1a/m1 genotype. In Japanese strains, there appears to be no association between the specific vacA types and the level of in vitro cytotoxin activity or clinical disease. Thus cagA+, vacA-s1a/m1-type isolates are common in Japan, and the genotype does not predict disease outcome (129). Furthermore, there are regional variations within populations in Europe, with, for instance, the s1b genotype being common in the Portuguese population, while the Dutch typically harbor the s1a genotype of vacA (287).
Such genotypic variation in vacA raises questions about the mechanism by which it may be accomplished. Tomb et al. (280) observed three vacA-related genes, HP0289, HP0922, and HP0610, in the genome of H. pylori (Fig. 10). However, the encoded putative proteins were predicted not to be secreted toxins (8). The same observation has been made by Alm et al. (3). Nonetheless, the presence of vacA-related genes led Tomb et al. (280) to the suggestion that intragenic recombination may occur between these genes to generate new VacA variants.
FIG. 10.
Conserved domains between VacA and related proteins encoded by H. pylori 26695. The different shadings represent regions of similarity. aa, amino acids.
cag Locus
Production of the vacuolating cytotoxin is associated with the presence of a 120- to 140-kDa antigenic protein (CagA) that is expressed in 60 to 80% of H. pylori strains (4, 59, 61). The nucleotide sequence of the cytotoxin-associated gene A cagA has been determined (56, 281), and its disruption does not abolish cytotoxin activity, despite the association of the two gene products (282). The cagA gene is a marker for a large, 40-kb, locus containing over 40 genes, which has been termed the cag pathogenicity island (42) (Fig. 11). Table 4 shows the putative proteins encoded by this cag pathogenicity island. Several characteristics of the cag locus include the presence of short repeated sequences and insertion sequences and a G+C content (35%) which is lower than that of the genome (39%) (280). The cag locus confers inflammation-inducing activity by contributing to the stimulation of interleukin-8 production by epithelial cells (61, 153, 283). This pathogenicity island is also thought to encode a type IV secretion system that allows surface expression of proteins interacting with epithelial cells (9). The implications of the cag pathogenicity island and the vacuolating cytotoxin in the pathogenicity of H. pylori have been extensively reviewed (29, 57, 73, 153, 155, 177, 178, 207) and are not developed here.
FIG. 11.
Schematic organization of the cag pathogenicity island. The orientations of the genes are indicated.
TABLE 4.
Putative proteins encoded by the cag pathogenicity island
| HP no. | Homologue of gene product |
|---|---|
| HP0520 | NSHa |
| HP0521 | NSH |
| HP0522 | Intracellular hyaluronic acid binding protein, Homo sapiens |
| HP0523 | NSH |
| HP0524 | VirD4, T-DNA transfer protein, A. tumefaciens |
| HP0525 | PtlH, pertussis toxin liberation, B. pertussis |
| HP0526 | NSH |
| HP0527 | VirB10, T-DNA transfer protein, A. tumefaciens |
| HP0528 | VirB9, T-DNA transfer protein, A. tumefaciens |
| HP0529 | NSH |
| HP0530 | NSH |
| HP0531 | NSH |
| HP0532 | NSH |
| HP0534 | NSH |
| HP0535 | NSH |
| HP0536 | NSH |
| HP0537 | NSH |
| HP0538 | NSH |
| HP0539 | HP0541 |
| HP0540 | NSH |
| HP0541 | HP0539 |
| HP0542 | NSH |
| HP0543 | NSH |
| HP0544 | PicB, H. pylori; VirB4, pertussis toxin secretion protein, B. pertussis |
| HP0545 | NSH |
| HP0546 | NSH |
| HP0547 | CagA, H. pylori; surface-located membrane protein Lmp3, M. hominis |
| DNA helicase, M. jannaschii |
NSH, no significant homology.
The cag locus provides an extraordinary illustration of multiple mechanisms by which diversity may occur. Firstly, it was acquired most probably by horizontal transfer, integrated into the glutamate racemase gene, and evolved by chromosomal rearrangements and acquisition of insertion sequence elements (42). The molecular characterization of the cagA gene within the locus provides an example of heterogeneity by variation in a number of intragenic cassettes. This was the first H. pylori gene described not to be conserved in all strains (56, 281). In 1988, Apel et al. (4) showed that 80% of the H. pylori-infected patients presented antibodies directed against a 120-kDa protein. The protein was subsequently named CagA, and isolates which fail to express the CagA protein lack the cagA gene (56, 281). Examination of the cagA structure indicates that a number of DNA repeats are present. These intragenic cassettes, whose sizes range up to 102 bp, are present in as many as five copies and account for the size variation of the CagA protein (26).
In addition to variations in cagA, Censini et al. (42) observed three variations in the cag locus: (i) the presence of the cag locus containing the insertion sequence IS605; (ii) the cag locus together with IS605, but with the former containing gene deletions and disruption in its continuity; and (iii) absence of cag and IS605. These observations suggest that the cag locus was acquired originally as a single unit and that the insertion of IS605 occurred more recently, allowing chromosomal rearrangements by recombinant events, which could generate the great variability observed within the intermediate-type lineage.
Four other regions of H. pylori 26695 and eight regions in H. pylori J99 are of particular interest due to their distinct G+C content, and they might also have been acquired by horizontal transfer. An examination of their distribution in different H. pylori strains would provide information about their contribution to H. pylori diversity.
Mosaic Organization and Phase Variation
After examination of the complete genome sequence of H. pylori 26695, Tomb et al. (280) suggested the possibility of a mosaic organization in certain genes different from that observed for the vacA gene. They reported the presence of a large family of OMPs, including two adhesins and four porins, with all of the 32 members of this family having a similar domain at the amino terminus and one of seven domains at the carboxy terminus. Moreover, 11 of these OMPs had extensive similarity over their entire length. Given the large number of sequence-related genes encoding putative surface-exposed proteins, the potential exists for extensive recombination leading to mosaic organization. Tomb et al. (280) suggest that this organization could be the basis for antigenic variation in H. pylori and an effective mechanism for host immune response evasion, similar to that seen in M. genitalium (234).
Antigenic variation could result from gene mosaic organization, but it could also result from genetic regulation at the transcription level. Such transcriptional controls have been documented by the presence of oligonucleotide repeats in promoter regions (304). Stretches of repeats have been identified in potential promoter regions of 18 genes, including eight OMP genes and α-1,3-fucosyltransferase genes, which are involved in LPS synthesis (see above). The synthesis of functional versus nonfunctional proteins may depend on the number and length of such repeats generating on-off switching. This on-off switching occurs by DNA slippage during replication. Recently, Appelmelk et al. (5) described phase variation in Lewis and non-Lewis LPS serotypes due to on-off switching of the α-1,3-fucosyltransferase genes.
Llver et al. (163) identified another mechanism of phase variation in H. pylori. In one isolate, two alleles of the babA gene encoding an adhesin were present (see below). Their sequences were identical except for an insert of a 10-bp repeat motif in the region encoding the signal peptide sequence. This resulted in the creation of a translational initiation codon that was functional in babA2, expressing the adhesin, whereas the babA1 gene was silent. The authors observed frequent deletions of the repeat motif and conversion of the silent babA1 gene, suggesting the presence of hot spots for phase variation within the bab gene family.
The comparison of genes whose expression may be regulated by slipped-strand repair between strains 26695 and J99 led Alm et al. (3) to conclude that some genes may be differently expressed in the two strains.
Genomic analyses and sequence comparison of the two strains have provided examples of genes whose expression may be regulated. Identification of other genes subject to transcriptional control will serve to identify the mechanisms of gene expression present in H. pylori.
Extragenic Elements
Several studies have indicated that 35 to 75% of H. pylori strains possess plasmids, a fraction of whose sequences appears to be related (118, 145, 199). Plasmid acquisition should be a means of diversity for H. pylori strains. Go et al. (100) reported finding repetitive DNA elements in chromosomal DNA which were also present in plasmids, suggesting the possibility of recombination events between the bacterial chromosome and plasmids.
Insertion sequences may also contribute to heterogeneity, as was illustrated above for the cag locus. The complete genome sequencing of H. pylori 26695 and J99 revealed that two distinct insertion sequences are present (3, 280). Five full-length copies of IS605 and two copies of a novel IS606 are inserted into the chromosomal DNA of strain 26695, whereas no complete IS605 and one IS606 are present in the genome of strain J99. Moreover, there are eight partial copies of IS605 and two partial copies of IS606 in strain 26695 and five and four, respectively, in strain J99. Recent studies showed that IS605 and IS606 were found in approximately one-third of H. pylori strains (124, 144).
Molecular and Sequencing Data Reflecting Possible Mechanisms Involved in Diversity
H. pylori strains seem to belong to a very diverse group. Point mutations and similar phenomena contribute to increased diversification, and these events may have led to the birth of a quasispecies, a cluster of closely related organisms forming a species complex (111), although it should be noted that Alm et al. (3) do not appear to agree with this conclusion. This diversity could be considered a form of genetic drift (28). Based on the coding capacity for methyltransferases, DNA glycosylases, and MutS and UvrD proteins involved in error-free and error-prone repairs, the sequence data obtained from the study of Tomb et al. (280) suggest that H. pylori is able to perform mismatch repairs. However, an SOS system appears to be absent. From these observations, it may be concluded that point mutations may account for only a minor level of strain diversity.
Following the discovery of the cag pathogenicity island, horizontal transmission of foreign DNA fragments has been suggested as a source of strain diversity. However, it seems unlikely that this is the most prominent mechanism through which diversity occurs. Indeed, the success of such DNA acquisition depends first on the transfer of DNA and second on its integration into the H. pylori genome by a recombination event. Twenty-three methyltransferase homologues are present in the H. pylori genome, with at least seven being adenine specific and four being cytosine specific (280). Further analysis revealed three type I restriction modification systems, 14 type II systems, and at least two type III systems (250, 254). Hence, the defense system, whose aim is to degrade foreign DNA, is well developed in the bacterium. Although proteins involved in the natural competence for DNA transformation have been identified and characterized, such as those encoded by the comB locus (122), suggesting that recombinations may occur, the efficiency of a successful foreign DNA transfer might be low owing to the existence of numerous restriction modification systems in H. pylori.
In contrast, recombination events within and between strains may lead to genetic shifts which could be responsible for changes of greater magnitude. Observations by Jiang et al. (132) showing that the physical maps of unrelated H. pylori strains are distinct from one another, are in agreement with this concept. Moreover, two other events could increase the probability of occurrence of homologous recombinations: (i) the presence of multiple organisms within a host, and (ii) the natural competence of H. pylori (136, 289), which may lead to homologous recombination phenomena, producing chimeras which could be termed genetic shifts. The RecBCD pathway, which is the major pathway for recombination in wild-type E. coli cells (148), seems to be absent. However, like the mollicute Spiroplasma citri, H. pylori might perform homologous recombination by another pathway such as the RecF pathway (168, 169). In E. coli, this pathway generally depends on the RecA, RecJ, RecN, RecR, RecG, and RuvABC proteins, and the genes HP0153, HP0348, HP1393, HP0925, HP1523, HP0883, HP1059, and HP0877, respectively, orthologues of those encoding these proteins, are present in H. pylori chromosomal DNA. Moreover, the RecA protein has been characterized by Schmitt et al. (256). Thus, a system for homologous recombination appears to be present in H. pylori (280).
Other biochemical functions, such as single-stranded-DNA binding and helicase, may be provided by conserved hypothetical proteins with homology to proteins with similar functions, such as the proteins encoded by HP1553, HP1245, HP0911.
In conclusion, H. pylori has the coding capacity for several mechanisms contributing to diversity, and there is evidence that H. pylori strains are highly diverse (although there is no consensus on this point) and that bacterial populations in the stomach are varied and undergo continuous change. In vivo and in vitro studies of the genes that may be involved in generating diversity will provide a better understanding of the heterogeneity of bacterial populations that must be taken into account in the development of diseases linked to this infection.
COLONIZATION FACTORS
Motility
The motility of H. pylori is essential for colonization. It allows the bacteria to spread through the viscous mucus covering the epithelial cells of the gastric mucosa (113). Eaton (71, 72) demonstrated that nonmotile variants have greater difficulty colonizing gnotobiotic piglets than do motile strains. The studies of Johensans et al. (133) and Haas et al. (108) have confirmed the role of the flagella and the resulting motility in H. pylori pathogenicity.
H. pylori possesses five to seven unipolar flagella. A particular property of these flagella is the presence of a sheath covering the flagellar filament. This sheath is composed of a double layer of phospholipids and is thought to protect the flagella from the gastric acidity, which otherwise would depolymerize flagellar filaments (94). Recently, Jones et al. (135) showed the existence of a flagellar sheath protein identical to the HpaA protein, which has been reported to be an N-acetylneuraminyllactose binding a hemagglutinin (75).
Unlike other bacterial flagellar filaments, which are composed of a single protein, H. pylori flagellar filaments are made of polymers of two subunits, the major flagellin, FlaA, and the minor flagellin, FlaB. The genes encoding these proteins were characterized by Leying et al. (159) and Suerbaum et al. (273). The experimental results obtained by Johensans et al. (134) suggest that the relative composition of the flagellar filaments may vary and adapt to environmental conditions.
H. pylori genome analysis suggests the existence of at least 40 proteins involved in the regulation, secretion, and assembly of the flagellar structure. A gene (flgE) encoding a flagellar hook has been identified (226). The hook connects the filament to the cell and is believed to act as a flexible coupling between filament and cell. Based on sequence similarity, two genes, HP1119 and HP0752, similar to flgK and fliD, encoding hook accessory proteins, have been identified, as well as the HP0907 gene, an orthologue of flgD, encoding a hook assembly protein. The hook is connected to a structural complex known as the basal body, which is embedded in the cell surface. Several proteins are associated with the basal body. Among them are the flagellar basal-body L-, M-, and P-ring proteins encoded by HP0325, HP0351 and HP0246, orthologues of flgH, fliF, and flgI, respectively. The ORFs HP1557, HP1558, HP0770 and HP1092, and HP1585 are similar to the fliE, flgC, flhB, and flgG genes encoding other flagellar basal-body proteins. Although the basal body is a major component of the bacterial flagellum, it does not contain several proteins that are known to be important for motor function. These are the MotA and MotB integral proteins necessary for motor rotation but not for flagellar assembly or for motor switching (165). HP0815 and HP0816 are orthologues of motA and motB from B. subtilis. The proteins encoded by HP0352, HP0584, HP1031, and HP1030, orthologues of the fliG, fliN, fliM, and fliY genes, respectively, may be involved in the rotation of the motor and its switching. The switching complex may provide the interface between the energy-transducing mechanism and the sensory information from the chemotaxis system.
In addition to the transcriptional regulation of the flagellar regulon mentioned above, other mechanisms seem to be involved in the control of expression of flagellar genes. For example, HP0753 and HP1035 have similarities to the fliS and flhF genes of B. subtilis, respectively. The FliS protein is implicated in the negative control of flagellar gene expression, and the FlhF protein is a GTP-binding protein. Schmitz et al. (257) characterized the flbA gene, which encodes a protein with similarity to the InvA, LcrD, and FlbF proteins involved in the regulation of motility or virulence.
Some genes required for flagellum assembly have similarities to those involved in the type III secretion systems (55). The proteins forming the P and L rings cleave signal peptides and are exported through the sec-dependent pathway, whereas the external flagellar components are exported through a specialized pathway encoded by flagellar genes. The flagellar export pathway is assembled from the genes HP0353, HP1420, HP0684 and HP0685, HP1041, HP0770, HP1419, and HPO173 orthologues of the genes encoding FliH, FliI, FliP, FlhA, FlhB, FliQ, and FliR, respectively.
Based on molecular data obtained by genome sequencing, studies can be carried out to identify flagellar gene functions and the control of expression of flagellar and motility genes.
H. pylori has developed metabolic and physiological properties that enable it to survive and grow in a particular ecological niche. Nonetheless, there are further potential advantages to be able to migrate to the most favorable environment available. Thus, chemotaxis may be crucial for gastric colonization. Tomb et al. (280) identified several putative chemotaxis genes in the H. pylori genome, HP0019, HP0082, HP0099, HP0103, HP0391, HP0393, HP0616, and HP1067. Worku et al. (298) confirmed that H. pylori demonstrates plasma chemotaxis, which is mediated, in particular, through the amino acids glutamine, histidine, lysine, and alanine. Similarly, the studies of Yoshiyama et al. (305) and Nakamura et al. (212) lead to the conclusion that movement of H. pylori in the viscous mucin layer is enhanced by chemotactic activity toward urea and bicarbonate.
Together with the genes involved in flagellar structure and assembly, motile and chemotactic activities require genes encoding receptors and transducers and genes involved in intermediate signal processing (164). Many of the genes that function in the primary reception process also function in transport. This is the case for genes encoding periplasmic binding proteins for dipeptides. In this category, the genes encoding dipeptide ABC transporter ATP binding proteins in the H. pylori sequence and their orthologues are HP0301 to dppD from B. subtilis, and HP0302 and HP0298 to dppF and dppA from E. coli; the genes encoding dipeptide ABC transporter permease proteins and their orthologues are HP0299 to dppB, and HP0300 to dppC from E. coli. Methyl-accepting chemotaxis proteins, which are primary receptors for amino acids and secondary receptors for other attractants (164), are also encoded by H. pylori. HP0082, HP0099, and HP010 have similarities to the tlpC, tlpA, and tlpB genes, respectively, that encode methyl-accepting chemotaxis proteins in B. subtilis. Genes involved in communication between receptors-transducers and the motors were also found in H. pylori. Jackson et al. (130) identified homologues of the genes encoding CheA and CheY, which are the major regulators of chemotaxis in bacteria. More recently, CheY was shown to belong to a stress-responsive operon (17). Moreover, Pittman et al. (239) sequenced three genes in H. pylori DNA: one encoding chemotaxis homologues of CheV from B. subtilis, a second encoding a bifunctional CheA-CheY fusion protein (CheF) similar to FRZE of M. xanthus, and a third encoding the enterobacterial CheW protein. Similarly to CheF, the B. subtilis CheV protein contains an amino-terminal domain homologous to CheW linked to a response regulator domain of the CheY family, suggesting that either or both of these functions are duplicated (88).
The components of the motility and chemotaxis machinery of H. pylori are similar to that found in other gram-negative bacteria, with the exception of the two constituent subunits of the flagella. Identification of attractants and repellents would serve to determine the function of chemotaxis and motility genes and the regulation of the system, as well as the role of chemotaxis in colonization by H. pylori.
Adhesins
Although the majority of H. pylori organisms in infected patients are free living in the mucus layer (155), a proportion appear to adhere to the epithelial cells of the gastric tissue (117, 154, 217). Adhesins have been identified either directly or by characterizing cellular receptors (47).
HpaA binds an N-acetylneuraminyllactose-type receptor, and was the first protein described as an H. pylori adhesin (74). However, Jones et al. (135) in 1997 showed that this protein is not an adhesin, at least for AGS cells, but is a flagellar sheath protein. Regarding the H. pylori DNA sequence, Tomb et al. (280) identified 19 other lipoproteins, some of which are likely to contribute to the adherence capacity of the bacterium. In 1990, Fauchère and Blaser (77) showed that surface components of H. pylori cells mediate their adherence to HeLa cell membranes.
Another category of putative adhesins is the OMP family, represented in H. pylori by 32 members (280). Three adhesins described in H. pylori belong to the large OMP family (35, 222, 223). They are AlpA and AlpB, which may act as adhesins and recognize different receptors on the cell surface, and the BabA adhesin, which mediates attachment to the Lewisb antigen (163). The authors of this study suggested that other members of the closely related OMP family may also act as adhesins. Recently, Namavar et al. (214) identified a 16-kDa surface protein that adhered to oligosaccharide ligands such as sulfated Lewisa antigens present on mucin glycoproteins. Tomb et al. (280) pointed out that the OMP genes have highly homologous domains at the 5′ and 3′ ends. This feature offers the possibility of recombination and hence an increase in antigenic variation (see above).
Lingwood et al. (161, 162) described a 63-kDa exoenzyme S-type molecule which binds a PE-type glycerolipid isolated from human gastric cells. The relevance of H. pylori binding to such molecules in vivo has not yet been studied.
Considering the adhesins previously described and the large family of lipoproteins and OMPs identified (280), it appears that H. pylori could use several adherence mechanisms for successful attachment to epithelial cells. Gene disruption experiments should provide data useful to evaluate the role of OMP in adherence.
Other Enzymes
Some enzymes produced by H. pylori have been proposed to be indirectly responsible for mucosal damage (178). Courillon-Mallet et al. (54) showed that H. pylori produces a histamine N-methyltransferase which generates N-methyl histamine, an agonist of H3 histamine receptors, but no orthologous gene was found in H. pylori DNA.
The HP0357 ORF has similarity to the gene encoding alcohol dehydrogenase, which may produce acetaldehyde in an excess of ethanol (253).
H. pylori also exhibits hemolytic activity (18, 258, 292). The gene encoding hemolysin was identified, as well as the HP1086 and HP1490 genes, orthologues of tly, encoding Serpulina hyodysenteriae hemolysin (280). HP0599 is a homologue of hylB which codes for the hemolysin secretion protein precursor of V. cholerae, and the H. pylori protein may form a pore through which hemolysin is exported.
Experimental results are not completely in agreement with the genome sequence analysis, particularly for the histamine N-methyltransferase, and reverse genetic experiments would serve to elucidate some of these discrepancies.
CONCLUSIONS
A total of 1,590 ORFs were identified in the genome of strain 26695, and 1,091 showed similarities to genes contained in the databases. Some of these genes encode proteins with known roles, but others have no assigned functions. Regarding the former, it should be kept in mind that sequence similarities provide predictions of functions but do not prove them and also that a protein whose function has been determined in one organism may play different or additional roles in another organism (8). Nonetheless, the analysis supports the conclusions that the organization of orthologous genes in the bacterial chromosome is not conserved during evolution and that the genome is more complex and flexible than was hitherto thought (146). For example, the location of rRNA genes in H. pylori is not contiguous, at variance with most other bacteria (3, 280).
Comparing the genomes of strains 26695 and J99, Alm et al. (3) suggested that there is a low level of evolutionary divergence. Among the orphan ORFs, for which no orthologue was found in either strain sequenced, 69 and 56 are unique to strains 26695 and J99, respectively (3, 280). Genes orthologous to these may be found as the number of sequenced genomes increases, but the fact that they represent intraspecific differences suggests that there is a core of genes specific to H. pylori. Possible clues to virulence that would have been obtained by comparing the DNA sequences of pathogenic and nonpathogenic strains are not available, since both strains came from patients with gastric diseases, although the diseases were different.
Enzymes represent about 20% of the genes in bacteria, and their corresponding genes constitute the best annotated aspect of genomes. The reason is that metabolic pathways are both highly conserved and the most extensively studied biochemically (34). H. pylori is no exception; although its biochemistry is not well understood and has unique characteristics, the contribution of the analysis of the genome to understanding its metabolic pathways is comparatively much greater than, for example, the contribution to understanding its regulatory pathways, which are more divergent between organisms and for which there is limited biochemical knowledge. The deductions about H. pylori glutaminyl-tRNA synthetase and asparaginyl-tRNA synthetase illustrate this point. The genes coding for these enzymes were not found in the genomes of strains 26695 or J99, but experimental work showed that neither glutamine nor asparagine is essential to grow the bacterium in vitro. The presence in the genomes of the other amino acid-tRNA synthetase genes and the conservation of these synthetases in evolutionary history made it unlikely that the missing genes remained unidentified in the genomes. Consequently, explanations based on identified genes are given in the annotation of both genomes, including alternative mechanisms found in an archeon and another bacterium (3, 280).
Since H. pylori belongs to an ancient branch of the Proteobacteria, its genome may shed some light on the evolution of metabolic pathways in organisms. It is worth noting that many similarities to archeal instead of bacterial enzymes were found; also, genes encoding enzymes which are part of operons in other proteobacteria are found at different loci in the H. pylori genome. In contrast, these characteristics make network reconstruction more difficult. For example, at variance with other bacteria, the genes coding for the enzymes of the de novo pyrimidine nucleotide synthesis pathway do not form an operon in H. pylori, and only two genes, found at different loci in the genome, have similarities to genes encoding enzymes of the de novo purine nucleotide synthesis, although experimental evidence shows that the bacterium can synthesize purine nucleotides de novo. Both the lack of similarities to other enzymes of de novo purine synthesis and the absence of an operon structure will make it more difficult to reconstruct, in the genome of H. pylori, the pathway of de novo purine biosynthesis.
Analysis of the H. pylori genome has provided insights into some aspects of the physiology of the microorganism, including its adaptation to a restricted ecological niche and possible mechanisms that induce antigenic variation within the H. pylori population. The latter aspect is just beginning to be investigated in depth (26, 73). It would be useful to know if such antigenic variation allows H. pylori to evade the host immune response and if it plays a role in the persistence of the organism in the stomach. From this perspective, the family of OMPs which contains a large number of paralogues (32) is of particular interest, and it should be noted that two of inversions-transpositions required to align the genomes of strains 26695 and J99 are associated with genes encoding members of this family (3).
Gene knockout experiments are used widely to investigate the role of proteins in the physiology of H. pylori. Construction of isogenic mutants in which specific genes are inactivated has aided our understanding of the roles of various gene products. The complete genome sequence and the functional predictions for approximately two-thirds of the genes add potential to the physiological integration of phenotypic characteristics of the bacterium. Now it will be possible to disrupt individual genes of interest whose presence in the genome had not been established previously, as well as groups of genes in a specific functional category or a cluster of paralogues.
ACKNOWLEDGMENTS
We thank Hilde De Reuse (Laboratoire de Bactériologie des Muqueuses, Institut Pasteur, Paris, France) for her comments and suggestions.
A.M. and F.M. thank the Conseil Régional d’Aquitaine for support. G.L.M. and S.L.H. are grateful for the support of the Australian Research Council and the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Alderson J, Clayton C L, Kelly D J. Investigations into the aerobic respiratory chain of Helicobacter pylori. Gut. 1997;41:A7. . (Abstract.) [Google Scholar]
- 2.Allan E, Mullany P, Tabaqchali S. Construction and characterization of a Helicobacter pylori clpB mutant and role of the gene in stress response. J Bacteriol. 1998;180:426–429. doi: 10.1128/jb.180.2.426-429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, de Jonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Apel I, Jacobs E, Kist M, Bredt W. Antibody response of patients against a 120 kDa surface protein of Campylobacter pylori. Zentbl Bakteriol Mikrobiol Hyg Ser A. 1988;268:271–276. doi: 10.1016/s0176-6724(88)80012-9. [DOI] [PubMed] [Google Scholar]
- 5.Appelmelk B J, Shiberu B, Trinks C, Tapsi N, Zheng P Y, Verboom T, Maaskant J, Hokke C H, Schiphorst W E C M, Blanchard D, Simoonssmit I M, Vandeneijnden D H, Vandenbrouckegrauls C M J E. Phase variation in Helicobacter pylori lipopolysaccharide. Infect Immun. 1998;66:70–76. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appelmelk B J, Simoonssmit I, Negrini R, Moran A P, Aspinall G O, Forte J G, Devries T, Quan H, Verboom T, Maaskant J J, Ghiara P, Kuipers E J, Bloemena E, Tadema T M, Townsend R R, Tyagarajan K, Crothers J M, Monteiro M A, Savio A, Degraaff J. Potential role of molecular mimicry between Helicobacter pylori lipopolysaccharide and host Lewis blood group antigens in autoimmunity. Infect Immun. 1996;64:2031–2040. doi: 10.1128/iai.64.6.2031-2040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aspinall G O, Monteiro M A, Pang H, Walsh E J, Moran A P. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen chain and core oligosaccharide regions. Biochemistry. 1996;35:2489–2497. doi: 10.1021/bi951852s. [DOI] [PubMed] [Google Scholar]
- 8.Atherton J. Helicobacter pylori unmasked—the complete genome sequence. Eur J Gastroenterol Hepatol. 1997;9:1137–1140. [PubMed] [Google Scholar]
- 9.Atherton J, Covacci A. Pathogenic properties of Helicobacter pylori. Curr Opin Gastroenterol. 1997;13:20–24. [Google Scholar]
- 10.Atherton J C, Cao P, Peek R M, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori—association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 11.Atherton J C, Peek R M, Tham K T, Cover T L, Blaser M J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 12.Bachmann B J. Linkage map of Escherichia coli K12, edition 7. Microbiol Rev. 1983;47:519–522. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker P J, Hraba T, Taylor C E, Stashak P W, Fauntleroy M B, Zähringer U, Takayama K, Sievert T R, Hronowski X, Cotter R J. Molecular structures that influence the immunomodulatory properties of the lipid A inner core region oligosaccharides of bacterial lipopolysaccharides. Infect Immun. 1994;62:2257–2269. doi: 10.1128/iai.62.6.2257-2269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauerfeind P, Garner R, Dunn B E, Mobley H L T. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauerfeind P, Garner R M, Mobley H L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bereswill S, Fassbinder F, Volzing C, Covacci A, Haas R, Kist M. Hemolytic properties and riboflavin synthesis of Helicobacter pylori: cloning and functional characterization of the ribBA gene encoding GTP-cyclohydrolase II that confers hemolytic activity to Escherichia coli. Med Microbiol Immunol. 1998;186:177–187. doi: 10.1007/s004300050062. [DOI] [PubMed] [Google Scholar]
- 19.Bereswill S, Lichte F, Vey T, Fassbinder F, Kist M. Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol Lett. 1998;159:193–200. doi: 10.1111/j.1574-6968.1998.tb12860.x. [DOI] [PubMed] [Google Scholar]
- 20.Bereswill S, Waidner U, Fassbinder F, Hantke K, Kist M. The Helicobacter pylori pfr-gene encoding ferritin inactivates the iron-dependent fur-regulon in Escherichia coli. Gut. 1997;41:A7. . (Abstract.) [Google Scholar]
- 21.Bereswill S, Waidner U, Odenbreit S, Lichte F, Fassbinder F, Hantke K, Bode G, Kist M. Structural functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology. 1998;144:2505–2516. doi: 10.1099/00221287-144-9-2505. [DOI] [PubMed] [Google Scholar]
- 22.Berg D E, Hoffman P, Appelmeck B, Kursters J. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Genet. 1997;13:468–474. doi: 10.1016/s0966-842x(97)01164-5. [DOI] [PubMed] [Google Scholar]
- 23.Beucher M, Sparling P F. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated outer membrane protein of Neisseria gonorrhea. J Bacteriol. 1995;177:2041–2049. doi: 10.1128/jb.177.8.2041-2049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bijlsma J J E, Gerrits M M, Imamdi R, Vandenbroucke-Grauls C M J E, Kusters J G. Urease-positive, acid-sensitive mutants of Helicobacter pylori: urease-independent acid resistance involved in growth at low pH. FEMS Microbiol Lett. 1998;167:309–313. doi: 10.1111/j.1574-6968.1998.tb13244.x. [DOI] [PubMed] [Google Scholar]
- 25.Birkholz S, Knipp U, Lemma E, Kroger A, Opferkuch W. Fumarate reductase: an immunogenic protein. J Med Microbiol. 1994;41:56–62. doi: 10.1099/00222615-41-1-56. [DOI] [PubMed] [Google Scholar]
- 26.Blaser M J. Genetic bases for heterogeneity of Helicobacter pylori. In: Hunt R H, Tytgat G N J, editors. Helicobacter pylori—basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 33–39. [Google Scholar]
- 27.Blaser M J. Helicobacter pylori and gastric diseases. Clin Rev. 1998;316:1507–1510. doi: 10.1136/bmj.316.7143.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaser M J. Heterogeneity of Helicobacter pylori. Eur J Gastroenterol Hepatol. 1997;9:S3–S7. doi: 10.1007/978-94-009-1792-7_3. [DOI] [PubMed] [Google Scholar]
- 29.Blaser M J, Perez-perez G I, Kleanous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 30.Blattner F R, Plunkett I G, Bloch C A, Perna N T, Burland V, Riley M, Glasner C-V J J, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 31.Bloch K. β-Hydroxydecanoylthioester dehydrase. In: Boyer P D, editor. The enzymes. New York, N.Y: Academic Press, Inc.; 1970. pp. 441–464. [Google Scholar]
- 32.Böck A, Sawers G. Fermentation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 262–282. [Google Scholar]
- 33.Bode G, Song Q, Barth R, Adler G. Phospholipase C activity of Helicobacter pylori is not associated with the prevalence of the cagA gene. Gut. 1997;41:A14. doi: 10.1046/j.1365-2362.2001.00814.x. . (Abstract.) [DOI] [PubMed] [Google Scholar]
- 34.Bono H, Ogata H, Goto S, Kanehisa M. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998;8:203–210. doi: 10.1101/gr.8.3.203. [DOI] [PubMed] [Google Scholar]
- 35.Boren T, Falk P, Roth K A, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium. Proc Natl Acad Sci USA. 1993;90:2035–2039. doi: 10.1073/pnas.90.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bucca G, Ferina G, Puglia A M, Smith C P. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol Microbiol. 1995;17:663–674. doi: 10.1111/j.1365-2958.1995.mmi_17040663.x. [DOI] [PubMed] [Google Scholar]
- 37.Burns B P, Hazell S L, Mendz G L. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology. 1995;141:3113–3118. doi: 10.1099/13500872-141-12-3113. [DOI] [PubMed] [Google Scholar]
- 38.Burns B P, Hazell S L, Mendz G L. Regulatory characteristics of a class A aspartate carbomoyl transferase from Helicobacter pylori. Gut. 1997;41:A21. . (Abstract.) [Google Scholar]
- 39.Burns B P, Mendz G L, Hazell S L. Characterization of the glucose transport in Helicobacter pylori. Acta Gastroenterol Belg. 1993;56:44. [Google Scholar]
- 40.Cashel M, Gentry D R, Herdanez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 41.Cassels R, Oliva B, Knowels D. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in Staphylococci. J Bacteriol. 1995;177:5161–5165. doi: 10.1128/jb.177.17.5161-5165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Censini S, Lange C, Xiang Z Y, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalk P A, Roberts A D, Blows W M. Metabolism of pyruvate and glucose by intact cells of Helicobacter pylori studied by C-13 NMR spectroscopy. Microbiology. 1994;140:2085–2092. doi: 10.1099/13500872-140-8-2085. [DOI] [PubMed] [Google Scholar]
- 44.Chang H T, Marcelli S W, Daivson A A, Chalk P A, Poole R K, Miles R J. Kinetics of substrate oxidation by whole cells and cell membranes of Helicobacter pylori. FEMS Microbiol Lett. 1995;129:33–38. doi: 10.1016/0378-1097(95)00130-W. [DOI] [PubMed] [Google Scholar]
- 45.Chow W-H, Blaser M J, Blot W J. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 1998;58:588–590. [PubMed] [Google Scholar]
- 46.Clayton C L, Pallen M J, Kleanthous H, Tabaqchali S. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res. 1990;18:362. doi: 10.1093/nar/18.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clyne M, Drumm B. Adherence of Helicobacter pylori to gastric mucosa. Can J Gastroenterol. 1997;11:243–248. doi: 10.1155/1997/149734. [DOI] [PubMed] [Google Scholar]
- 48.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in presence of urea. Infect Immun. 1995;63:1669–1673. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colbeau A, Elsen S, Tomiyama M, Zorin N A, Dimon B, Vignais P M. Rhodobacter capsulatus HypF is involved in regulation of hydrogenase synthesis through the HupUV proteins. Eur J Biochem. 1998;251:65–71. doi: 10.1046/j.1432-1327.1998.2510065.x. [DOI] [PubMed] [Google Scholar]
- 50.Colbeau A, Richaud P, Caballero F J, Elster C, Delphin C, Smith R L, Chabert J, Vignais P M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993;8:15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 51.Cole S P, Cirillo D, Kagnoff M F, Guiney D G, Eckmann L. Coccoid and spiral Helicobacter pylori differ in their abilities to adhere to gastric epithelial cells and induce interleukin-8 secretion. Infect Immun. 1997;65:843–846. doi: 10.1128/iai.65.2.843-846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corthesy-Theulaz I E, Bergonzelli G E, Henry H, Bachmann D, Schorderet D F, Blum A L, Ornston L N. Cloning and characterization of Helicobacter pylori succinyl CoA:acetoacetate CoA-transferase, a novel prokaryotic member of the CoA-transferase family. J Biol Chem. 1997;272:25659–25667. doi: 10.1074/jbc.272.41.25659. [DOI] [PubMed] [Google Scholar]
- 53.Courcoux P, Freland C, Piemond Y, Fauchère J L, Labigne A. Polymerase chain reaction and direct DNA sequencing as a method for distinguishing between different strains of Helicobacter pylori. Rev Esp Enferm Dig. 1990;78:29. [Google Scholar]
- 54.Courillon-Mallet A, Launet J M, Roucayrol A M, Callebert J, Edmond J P, Tabuteau F, Cattan D. Helicobacter pylori infection: physiopathologic implication of N-α-methyl histamine. Gastroenterology. 1995;108:959–966. doi: 10.1016/0016-5085(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 55.Covacci A. Mobilis in mobile: unexpected flexibility and quantum leaps in the Helicobacter pylori genome. In: Hunt R H, Tytgat G N J, editors. Helicobacter pylori—basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 40–49. [Google Scholar]
- 56.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128 kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 58.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 59.Cover T L, Dooley C P, Blaser M J. Characterization of human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect Immun. 1990;58:603–610. doi: 10.1128/iai.58.3.603-610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cover T L, Tumuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 61.Crabtree J E, Figura N, Taylor J D, Bugnoli M, Armellini D, Tomkins D S. Expression of 120-kilodalton protein and cytotoxicity among Helicobacter pylori. J Clin Pathol. 1992;45:733–736. doi: 10.1136/jcp.45.8.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cronan J E. Two-carbons and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 343–357. [Google Scholar]
- 63.Cronan J E, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 64.Curnow A W, Ibba M, Soll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 65.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;74:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davies G R, Banatvala N, Collins C E, Sheaff M T, Abdi Y, Clements L, Rampton D S. Relationship between infective load of Helicobacter pylori and reactive oxygen metabolite production in antral mucosa. Scand J Gastroenterol. 1994;29:419–424. doi: 10.3109/00365529409096832. [DOI] [PubMed] [Google Scholar]
- 67.Dhaenens L, Szczebara F, Husson M O. Identification, characterization, and immunogenicity of the lactoferrin-binding protein from Helicobacter pylori. Infect Immun. 1997;65:514–518. doi: 10.1128/iai.65.2.514-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dross L J, Geisler V, Lenger R, Theis F, Krafft T, Fahrenholz F, Kojro E, Duchene A, Tripier D, Juvenal K. The quinone-reactive, Ni/Fe-hydrogenase of Wolinella succinogenes. Eur J Biochem. 1992;206:93–102. doi: 10.1111/j.1432-1033.1992.tb16905.x. [DOI] [PubMed] [Google Scholar]
- 69.Dunn B E, Vakil N B, Schneider B G, Miller M M, Zitzer J B, Peutz T, Phadnis S H. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun. 1997;65:1181–1188. doi: 10.1128/iai.65.4.1181-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Earhart C F. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1075–1090. [Google Scholar]
- 71.Eaton K A. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect Immun. 1989;59:2470–2475. doi: 10.1128/iai.57.4.1119-1125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eaton K A. Motility as a factor in the colonization of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 73.Eck M, Schmausser B, Haas R, Greiner A, Czub S, Muller Hermelink H K. MALT-type lymphoma of the stomach is associated with Helicobacter pylori strains expressing the CagA protein. Gastroenterology. 1997;112:1482–1486. doi: 10.1016/s0016-5085(97)70028-3. [DOI] [PubMed] [Google Scholar]
- 74.Evans D G, Evans D J J, Moulds J J, Graham D Y. N-Acetylneuraminyl-lactose-binding fibrillar hemagglutinin of Campylobacter pylori: a putative colonization factor antigen. Infect Immun. 1988;56:2896–2906. doi: 10.1128/iai.56.11.2896-2906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans D G, Karjalainen R K, Evans D J J, Graham D Y, Lee C H. Cloning, nucleotide sequence, and expression of a gene encoding an adhesin subunit protein of Helicobacter pylori. J Bacteriol. 1993;175:674–683. doi: 10.1128/jb.175.3.674-683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans D J J, Evans D G, Lampert H C, Nakano H. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis, and Treponema pallidum, by analysis sequence. Gene. 1995;153:123–127. doi: 10.1016/0378-1119(94)00774-m. [DOI] [PubMed] [Google Scholar]
- 77.Fauchère J L, Blaser M. Adherence of Helicobacter pylori cells and their surface components to HeLa cell membranes. Microb Pathog. 1990;9:427–439. doi: 10.1016/0882-4010(90)90061-t. [DOI] [PubMed] [Google Scholar]
- 78.Ferrero R L, Lee A. The importance of urease in acid protection for the gastric colonising bacteria Helicobacter pylori and Helicobacter felis sp. nov. Microbiol Ecol Health. 1991;4:121–134. [Google Scholar]
- 79.Figura N, Guglielmetti P, Rossolini A, Barben A, Cusi G, Musmanno R, Russi M, Quaranta S. Cytotoxin production by Campylobacter pylori strains isolated from patients with peptic ulcers and from patients with chronic gastritis only. J Clin Microbiol. 1989;27:225–226. doi: 10.1128/jcm.27.1.225-226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finel M. Does NADH play a central role in energy metabolism in Helicobacter pylori? Trends Biochem Sci. 1998;23:412–414. doi: 10.1016/s0968-0004(98)01276-6. [DOI] [PubMed] [Google Scholar]
- 81.Fita I, Rossman M G. The NADPH binding site on beef liver catalase. Proc Natl Acad Sci USA. 1985;82:1604–1608. doi: 10.1073/pnas.82.6.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudeck D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 83.Forman D, Webb P, Parsonnet J. H. pylori and gastric cancer. Lancet. 1994;34:243–244. [PubMed] [Google Scholar]
- 84.Forsyth M H, Atherton J C, Blaser M J, Cover T L. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–3094. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fraenkel D G, Levisohn S R. Glucose, and gluconate metabolism in an Escherichia coli mutant lacking phosphoglucose isomerase. J Bacteriol. 1967;93:1571–1578. doi: 10.1128/jb.93.5.1571-1578.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrman J, Nguyen D, Utterback R T, Saudeck D M, Phillips C A, Merrick J M, Tomb J F, Dougherty B A, Bott K F, Hu P C, Lucier T S, Peterson S N, Smith H O, Hutchinson III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 87.Frazier B A, Pgeifer J D, Russel D G, Klak P, Olsen A N, Hammar M, Westblom T U, Normark S T. Paracrystalline inclusions of a novel ferritin containing non-heme iron, produced by the human gastric pathogen Helicobacter pylori: evidence for a third class of ferritins. J Bacteriol. 1993;175:966–972. doi: 10.1128/jb.175.4.966-972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fredrick K L, Helmann J D. Dual chemotaxis signaling pathways in Bacillus subtilis: a sigma D-dependent gene encodes a novel protein with both CheW and CheY homologous domain. J Bacteriol. 1994;176:2727–2735. doi: 10.1128/jb.176.9.2727-2735.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fulkerson J F, Garner R M, Mobley H L T. Structure and function of the NixA nickel transport protein of Helicobacter pylori. Gut. 1997;41:A107. . (Abstract.) [Google Scholar]
- 90.Garcia-Arata M I, Canton R, de Rafael L, Boixeda D, Martin de Argila C, Gisberg J P, Moreno L, Baquero F. Epidemiological usefulness of the polymorphism in the gene ureC of Helicobacter pylori. Gut. 1997;41:A41. . (Abstract.) [Google Scholar]
- 91.Garner R M, Fulkerson J, Mobley H L T. Helicobacter pylori glutamine synthetase lacks features associated with transcriptional and posttranslational regulation. Infect Immun. 1998;66:1839–1847. doi: 10.1128/iai.66.5.1839-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge Z M, Jiang Q, Kalisiak M S, Taylor D E. Cloning and functional characterization of Helicobacter pylori fumarate reductase operon comprising three structural genes coding for subunits C, A and B. Gene. 1997;204:227–234. doi: 10.1016/s0378-1119(97)00550-7. [DOI] [PubMed] [Google Scholar]
- 93.Ge Z M, Taylor D E. The Helicobacter pylori gene encoding phosphatidylserine synthase: sequence, expression, and insertional mutagenesis. J Bacteriol. 1997;179:4970–4976. doi: 10.1128/jb.179.16.4970-4976.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Geis G, Suerbaum S, Forsthoff B, Leying B, Opferkuch W. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J Med Microbiol. 1993;38:371–377. doi: 10.1099/00222615-38-5-371. [DOI] [PubMed] [Google Scholar]
- 95.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 217–261. [Google Scholar]
- 96.Ghiara P, Marchetti M, Blaser M J, Tummuru M K R, Cover T L, Segal E D, Tompkins L S, Rappuoli R. Role of the Helicobacter pylori virulence factors vacuolating cytotoxin, CagA, and urease in a mouse model of disease. Infect Immun. 1995;63:4154–4160. doi: 10.1128/iai.63.10.4154-4160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gilbert J V, Ramakrishna J, Sunderman F W, Wright A, Plaut A G. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect Immun. 1995;63:2682–2688. doi: 10.1128/iai.63.7.2682-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glansdorff N. Biosynthesis of arginine and polyamines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 408–433. [Google Scholar]
- 99.Go M F, Chan K Y, Versalovic J, Koeuth T, Graham D Y, Lupski J R. Cluster analysis of Helicobacter pylori genomic DNA fingerprints suggests gastroduodenal disease-specific associations. Scand J Gastroenterol. 1995;30:640–646. doi: 10.3109/00365529509096306. [DOI] [PubMed] [Google Scholar]
- 100.Go M F, Cissell L, Graham D Y. Helicobacter pylori plasmids: epidemiology in different geographical regions. In: Hunt R H, Tytgat G N J, editors. Helicobacter pylori—basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. p. 4. [Google Scholar]
- 101.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goffeau A, Barrell B G, Bussey H. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 103.Goldfine H. Comparative aspects of bacterial lipids. Adv Microbiol Physiol. 1972;8:1–58. doi: 10.1016/s0065-2911(08)60187-3. [DOI] [PubMed] [Google Scholar]
- 104.Goldfine H. Lipids of prokaryotes—structure and distribution. Curr Top Membr Transp. 1982;17:2–44. [Google Scholar]
- 105.Goodwin A, Kersulyte D, Sisson G, van Zanten S J O V, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 106.Grant K A, Park S F. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology. 1995;141:1369–1376. doi: 10.1099/13500872-141-6-1369. [DOI] [PubMed] [Google Scholar]
- 107.Greene R C. Biosynthesis of methionine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 542–560. [Google Scholar]
- 108.Haas R, Meyer T, van Putten J. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 109.Hancock R E W, Alm R, Bina J, Trust T. Helicobacter pylori: a surprisingly conserved bacterium. Nat Biotechnol. 1998;16:216–217. doi: 10.1038/nbt0398-216. [DOI] [PubMed] [Google Scholar]
- 110.Haque M, Hirai Y, Yokota K, Oguma K. Lipid profiles of Helicobacter pylori and Helicobacter mustelae grown in serum-supplemented and serum-free media. Acta Med Okayama. 1995;49:205–211. doi: 10.18926/AMO/30380. [DOI] [PubMed] [Google Scholar]
- 111.Hazell S L, Andrews R H, Mitchell H M, Daskalopoulous G. Genetic relationship among isolates of Helicobacter pylori: evidence for the existence of a Helicobacter pylori species-complex. FEMS Microbiol Lett. 1997;150:27–32. doi: 10.1111/j.1574-6968.1997.tb10345.x. [DOI] [PubMed] [Google Scholar]
- 112.Hazell S L, Evans D J J, Graham D Y. Helicobacter pylori catalase. J Gen Microbiol. 1991;137:57–61. doi: 10.1099/00221287-137-1-57. [DOI] [PubMed] [Google Scholar]
- 113.Hazell S L, Lee A, Hennessy W. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 114.Hazell S L, Mendz G L. How Helicobacter pylori works: an overview of the metabolism of Helicobacter pylori. Helicobacter. 1997;2:1–12. doi: 10.1111/j.1523-5378.1997.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 115.Hendricks J K, Mobley H L T. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hernandez V J, Bremer H. E. coli ppGpp synthetase II activity requires SpoT. J Biol Chem. 1991;266:5991–5999. [PubMed] [Google Scholar]
- 117.Hessey S J, Spencer J, Wyatt J I, Sobala G, Rathbone B J, Axon A T R, Dixon M F. Bacterial adhesion and disease activity in Helicobacter associated chronic gastritis. Gut. 1990;31:134–138. doi: 10.1136/gut.31.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heuermann D, Haas R. Genetic organization of a small cryptic plasmid of Helicobacter pylori. Gene. 1995;165:17–24. doi: 10.1016/0378-1119(95)00469-m. [DOI] [PubMed] [Google Scholar]
- 119.Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J Bacteriol. 1995;177:5327–5333. doi: 10.1128/jb.177.18.5327-5333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirschl A M, Richter M, Makristathis A, Prückl P, Willinger B, Schütze K, Rotter M L. Single and multiple strain colonization in patients with Helicobacter pylori-associated gastritis: detection by macrorestriction DNA analysis. J Infect Dis. 1994;170:473–475. doi: 10.1093/infdis/170.2.473. [DOI] [PubMed] [Google Scholar]
- 121.Hoffman P S, Goodwin A, Johnsen J, Magee K, Veldhuyzen S J O, Vanzanten S J O V. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J Bacteriol. 1996;178:4822–4829. doi: 10.1128/jb.178.16.4822-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hofreuter D, Odenbreit S, Henke G, Haas R. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;28:1027–1038. doi: 10.1046/j.1365-2958.1998.00879.x. [DOI] [PubMed] [Google Scholar]
- 123.Holmes E M, Mendz G L. Metabolic fate of arginine, an essential amino acid requirement for Helicobacter pylori. In: Lastovica E J, Newell D G, Lastovica E E, editors. Campylobacter, Helicobacter, and related organisms. Cape Town, South Africa: Institute of Child Health; 1998. pp. 193–196. [Google Scholar]
- 124.Höök-Nikanne J, Berg D, Peek R, Kersulyte D, Tummuru M, Blaser M. DNA sequence conservation and diversity in transposable element IS605 of Helicobacter pylori. Helicobacter. 1998;3:79–85. doi: 10.1111/j.1523-5378.1998.08011.x. [DOI] [PubMed] [Google Scholar]
- 125.Hu L, Foxall P, Russell R, Mobley H. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect Immun. 1992;60:2657–2666. doi: 10.1128/iai.60.7.2657-2666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hughes N J, Clayton C L, Chalk P A, Kelly D J. The porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J Bacteriol. 1998;180:1119–1128. doi: 10.1128/jb.180.5.1119-1128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Husson A M, Legrand D, Spik G, Leclerc H. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect Immun. 1993;61:2694–2697. doi: 10.1128/iai.61.6.2694-2697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Illingworth D S, Walter K S, Griffiths P L, Barclay R. Siderophore production and iron-regulated envelope proteins of Helicobacter pylori. Zentbl Bakteriol Int Med Microbiol. 1993;280:113–119. [PubMed] [Google Scholar]
- 129.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jackson C J, Kelly D J, Clayton C L. The cloning and characterization of chemotaxis genes in Helicobacter pylori. Gut. 1995;37:A71. . (Abstract.) [Google Scholar]
- 131.Jenkins L S, Nunn W D. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol. 1997;169:2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 133.Johensans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johensans C, Labigne A, Suerbaum S. Reporter gene analyses show that expression of both H. pylori flagellins is dependent on the growth phase. Gut. 1995;41:A246. . (Abstract.) [Google Scholar]
- 135.Jones A C, Logan R P H, Foynes S, Cockayne A, Wren B W, Penn C W. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesin for AGS cells. J Bacteriol. 1997;179:5643–5647. doi: 10.1128/jb.179.17.5643-5647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients-from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 137.Kadner R J. Cytoplasmic membrane. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 58–87. [Google Scholar]
- 138.Kajie S, Ideta R, Yamato I, Anraku Y. Molecular cloning and DNA sequence of dniR, a gene affecting anaerobic expression of the Escherichia coli hexaheme nitrite reductase. FEMS Microbiol Lett. 1991;67:205–211. doi: 10.1016/0378-1097(91)90355-e. [DOI] [PubMed] [Google Scholar]
- 139.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kansau I, Guillain F, Thiberge J M, Labigne A. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA) Mol Microbiol. 1996;22:1013–1023. doi: 10.1046/j.1365-2958.1996.01536.x. [DOI] [PubMed] [Google Scholar]
- 141.Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, Braimi N, Dupont C, Labigne A. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol. 1996;147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 142.Karita M, Blaser M J. Acid-tolerance response in Helicobacter pylori and differences between cagA(+) and cagA(−) strains. J Infect Dis. 1998;178:213–219. doi: 10.1086/515606. [DOI] [PubMed] [Google Scholar]
- 143.Kashihara W, Ogawa T, Suda Y, Hayashi T, Kusumoto S, Tamura T, Shimoyama T. Chemical structure and immunobiological activities of lipid A from Helicobacter pylori LPS in comparison with Porphyromonas gingivalis and Escherichia coli lipids As. Gut. 1997;41:A8. doi: 10.1016/s0264-410x(97)00102-3. . (Abstract.) [DOI] [PubMed] [Google Scholar]
- 144.Kersulyte D, Akopyants N, Clifton S, Roe B, Berg D. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene. 1998;223:175–186. doi: 10.1016/s0378-1119(98)00164-4. [DOI] [PubMed] [Google Scholar]
- 145.Kleanthous H, Clayton C L, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-negative bacteria. Mol Microbiol. 1991;5:2377–2389. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 146.Kolsto A B. Dynamic bacterial genome organization. Mol Microbiol. 1997;24:241–248. doi: 10.1046/j.1365-2958.1997.3501715.x. [DOI] [PubMed] [Google Scholar]
- 147.Kovach M E, Hughes K J, Everiss K D, Peterson K M. Identification of a ToxR-activated gene, tagE, that lies within the accessory colonization factor gene cluster of Vibrio cholerae. Gene. 1994;148:91–95. doi: 10.1016/0378-1119(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 148.Kowalczykowski S C, Dixon D A, Eggleston A K, Lander S D, Rebrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 514–523. [Google Scholar]
- 150.Krishnamurthy P, Parlow M, Zitzer J B, Vakil N B, Mobley H L T, Levy M, Phadnis S H, Dunn B E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect Immun. 1998;66:5060–5066. doi: 10.1128/iai.66.11.5060-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kruger A, Geisler V, Lemma E, Theis F, Lenger R. Bacterial fumarate respiration. Arch Microbiol. 1992;158:311–314. [Google Scholar]
- 152.Kursters J G, Gerrits M M, VanStrijp J A G, Vandenbroucke-Grauls C M J E. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun. 1997;65:3672–3679. doi: 10.1128/iai.65.9.3672-3679.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Labigne A, de Reuse H. Determinants of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 154.Lee A, Fox J, Hazell S. Pathogenicity of Helicobacter pylori: a perspective. Infect Immun. 1993;61:1601–1610. doi: 10.1128/iai.61.5.1601-1610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee A, Mitchell H. Basic bacteriology of H. pylori colonization factors. In: Hunt R H, Tytgat G N T, editors. Basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 59–72. [Google Scholar]
- 156.Leisinger T. Biosynthesis of proline. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 434–441. [Google Scholar]
- 157.Leunk H, Fergusson M A, Morgan D R, Low D E, Simor A E. Antibody to cytotoxin in infection by Helicobacter pylori. J Clin Microbiol. 1990;6:1181–1184. doi: 10.1128/jcm.28.6.1181-1184.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leunk R D, Johnson P T, David B C, Kraft W J, Morgan D R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 159.Leying H, Suerbaum S, Geis G, Hass R. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol Microbiol. 1992;6:2863–2874. doi: 10.1111/j.1365-2958.1992.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 160.Lichtenberger L M, Hazell S L, Romero J J, Graham D Y. Helicobacter pylori hydrolysis of artificial phospholipid monolayers: insight into a potential mechanism of mucosal injury. Gastroenterology. 1990;98:A78. . (Abstract.) [Google Scholar]
- 161.Lingwood C A, Hueska M, Kuskis A. The glycerolipid receptor of Helicobacter pylori (and exoenzyme S) is phosphatidylethanolamine. Infect Immun. 1992;60:2470–2474. doi: 10.1128/iai.60.6.2470-2474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lingwood C A, Wasfy G, Han H, Huesca M. Receptor affinity purification of a lipid-binding adhesion from Helicobacter pylori. Infect Immun. 1993;61:2474–2478. doi: 10.1128/iai.61.6.2474-2478.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Llver D, Arnqvist A, Ögren J, Frick I M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 164.Macnab R M. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 165.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 166.Maier R J, Fu C, Gilbert J, Moshiri F, Olson J, Plaut A G. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- 167.Manos J, Kolesnikow T, Hazell S L. An investigation of the molecular basis of the spontaneous occurrence of a catalase-negative phenotype in Helicobacter pylori. Helicobacter. 1998;4:1–7. doi: 10.1046/j.1523-5378.1998.08030.x. [DOI] [PubMed] [Google Scholar]
- 168.Marais A, Bové J, Renaudin J. Characterization of the recA gene regions of Spiroplasma citri and Spiroplasma melliferum. J Bacteriol. 1996;178:7003–7009. doi: 10.1128/jb.178.23.7003-7009.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Marais A, Bové J, Renaudin J. Spiroplasma citri virus SpV1-derived cloning vector: Deletion formation by illegitimate and homologous recombination in a spiroplasmal host strain which probably lacks a functional recA gene. J Bacteriol. 1996;178:862–870. doi: 10.1128/jb.178.3.862-870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Marcelli S W, Chang H T, Chapman T, Chalk P A, Miles R J, Poole R K. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol Lett. 1996;138:59–64. doi: 10.1111/j.1574-6968.1996.tb08135.x. [DOI] [PubMed] [Google Scholar]
- 171.Marians K J. Replication fork propagation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 749–763. [Google Scholar]
- 172.Marshall B J, Barret L, Prakash C, McCallum R, Guerrant R. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology. 1990;99:269–276. doi: 10.1016/0016-5085(90)90957-3. [DOI] [PubMed] [Google Scholar]
- 173.McGowan C, Necheva A, Cover T, Blaser M. Acid-induced expression of oxidative stress protein homologs in H. pylori. Gut. 1997;41:A18. [Google Scholar]
- 174.McGowan C C, Cover T L, Blaser M J. Analysis of F1F0-ATPase from Helicobacter pylori. Infect Immun. 1997;65:2640–2647. doi: 10.1128/iai.65.7.2640-2647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McGowan C C, Cover T L, Blaser M J. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology. 1996;110:926–938. doi: 10.1053/gast.1996.v110.pm8608904. [DOI] [PubMed] [Google Scholar]
- 176.McGowan C C, Necheva A, Thompson S A, Cover T L, Blaser M J. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 177.McGuigan J. Helicobacter pylori: the versatile pathogen. Dig Dis Sci. 1996;14:289–303. doi: 10.1159/000171560. [DOI] [PubMed] [Google Scholar]
- 178.Mégraud F. Helicobacter pylori: bacterial factors and interaction with the epithelial cells. Yale J Biol Med. 1996;69:35–37. [PMC free article] [PubMed] [Google Scholar]
- 179.Mégraud F, Lamouliatte H. Helicobacter pylori and duodenal ulcer. Dig Dis Sci. 1992;37:769–772. doi: 10.1007/BF01296437. [DOI] [PubMed] [Google Scholar]
- 180.Melchers K, Weitzenegger T, Buhmann A, Steinhilber W, Sachs G, Schafer K P. Cloning and membrane topology of a P-type ATPase from Helicobacter pylori. J Biol Chem. 1996;271:446–457. doi: 10.1074/jbc.271.1.446. [DOI] [PubMed] [Google Scholar]
- 181.Mendz G L, Burns B P, Hazell S L. The glucose transporters of Helicobacter pylori. Biochim Biophys Acta. 1995;1244:269–276. doi: 10.1016/0304-4165(95)00018-7. [DOI] [PubMed] [Google Scholar]
- 182.Mendz G L, Hazell S L. Aminoacid utilization by Helicobacter pylori. Int J Biochem Cell Biol. 1995;27:1085–1093. doi: 10.1016/1357-2725(95)00069-2. [DOI] [PubMed] [Google Scholar]
- 183.Mendz G L, Hazell S L. Evidence for a pentose phosphate pathway in Helicobacter pylori. FEMS Microbiol Lett. 1991;84:331–336. [Google Scholar]
- 184.Mendz G L, Hazell S L. Fumarate catabolism in Helicobacter pylori. Biochem Mol Biol Int. 1993;31:325–332. [PubMed] [Google Scholar]
- 185.Mendz G L, Hazell S L. Glucose metabolism by Helicobacter pylori. Microbiology. 1994;140:2179–2180. [Google Scholar]
- 186.Mendz G L, Hazell S L. The urea cycle of Helicobacter pylori. Microbiology. 1996;142:2959–2967. doi: 10.1099/13500872-142-10-2959. [DOI] [PubMed] [Google Scholar]
- 187.Mendz G L, Hazell S L, Burns B P. The Entner-Doudoroff pathway in Helicobacter pylori. Arch Biochem Biophys. 1994;312:349–356. doi: 10.1006/abbi.1994.1319. [DOI] [PubMed] [Google Scholar]
- 188.Mendz G L, Hazell S L, Burns B P. Glucose utilization and lactate production by Helicobacter pylori. J Gen Microbiol. 1993;139:3023–3028. doi: 10.1099/00221287-139-12-3023. [DOI] [PubMed] [Google Scholar]
- 189.Mendz G L, Hazell S L, Srinivasan S. Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori. Arch Biochem Biophys. 1995;321:153–159. doi: 10.1006/abbi.1995.1380. [DOI] [PubMed] [Google Scholar]
- 190.Mendz G L, Hazell S L, Vangorkom L. Pyruvate metabolism in Helicobacter pylori. Arch Microbiol. 1994;162:187–192. doi: 10.1007/BF00314473. [DOI] [PubMed] [Google Scholar]
- 191.Mendz G L, Holmes E M, Ferrero R L. In situ characterization of Helicobacter pylori arginase. Biochim Biophys Acta. 1998;1388:465–477. doi: 10.1016/s0167-4838(98)00207-6. [DOI] [PubMed] [Google Scholar]
- 192.Mendz G L, Jimenez B M, Hazell S L, Gero A M, O’Sullivan W J. De novo synthesis of pyrimidine nucleotides by Helicobacter pylori. J Appl Bacteriol. 1994;77:1–8. doi: 10.1111/j.1365-2672.1994.tb03036.x. [DOI] [PubMed] [Google Scholar]
- 193.Mendz G L, Jimenez B M, Hazell S L, Gero A M, O’Sullivan W J. Salvage synthesis of purine nucleotides by Helicobacter pylori. J Appl Bacteriol. 1994;77:674–681. doi: 10.1111/j.1365-2672.1994.tb02818.x. [DOI] [PubMed] [Google Scholar]
- 194.Mendz G L, Shepley A J, Hazell S L, Smith M A. Purine metabolism and the microaerophily of Helicobacter pylori. Arch Microbiol. 1997;168:448–456. doi: 10.1007/s002030050521. [DOI] [PubMed] [Google Scholar]
- 195.Messer W, Weigel C. Initiation of chromosome. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1579–1601. [Google Scholar]
- 196.Meyer-Rosberg K, Scott D R, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111:886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 197.Miller J F, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miller J F, Mekalanos J J, Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 199.Minnis J A, Taylor T E, Knesek J E, Peterson W L, McIntire S A. Characterization of a 3.5-kbp plasmid from Helicobacter pylori. Plasmid. 1995;34:22–36. doi: 10.1006/plas.1995.1030. [DOI] [PubMed] [Google Scholar]
- 200.Mitchell H M, Li Y Y, Hu P J, Liu Q, Chen M, Du G G, Wang Z J, Lee A, Hazell S L. Epidemiology of Helicobacter pylori in Southern China: identification of early childhood as the critical period for acquisition. J Infect Dis. 1992;166:149–153. doi: 10.1093/infdis/166.1.149. [DOI] [PubMed] [Google Scholar]
- 201.Mobley H. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am J Med. 1996;100:2S–11S. doi: 10.1016/s0002-9343(96)80223-3. [DOI] [PubMed] [Google Scholar]
- 202.Mobley H, Hu L, Foxall P. Helicobacter pylori urease: properties and role in pathogenesis. Scand J Gastroenterol. 1991;187:39–46. [PubMed] [Google Scholar]
- 203.Mobley H L T. Helicobacter pylori factors associated with disease development. Gastroenterology. 1997;113:S21–S28. doi: 10.1016/s0016-5085(97)80006-6. [DOI] [PubMed] [Google Scholar]
- 204.Mobley H L T, Foxall P A. H. pylori urease. In: Hunt R H, Tytgat G N T, editors. Helicobacter pylori—basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 41–58. [Google Scholar]
- 205.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Moore R A, Beckthold B, Wong S, Kureishi A, Bryan L E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:107–111. doi: 10.1128/aac.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Moran A P. Pathogenic properties of Helicobacter pylori. Scand J Gastroenterol. 1996;31:22–31. [PubMed] [Google Scholar]
- 208.Moran A P. Structure-bioactivity relationships of bacterial endotoxins. J Toxicol Toxin Rev. 1995;14:47–83. [Google Scholar]
- 209.Moran A P, Lindner B, Walsh E J. Structural characterization of the lipid a component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J Bacteriol. 1997;179:6453–6463. doi: 10.1128/jb.179.20.6453-6463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Morgan D R, Freedman F, Depew C E, Kraft W G. Growth of Campylobacter in liquid media. J Clin Microbiol. 1987;25:2123–2125. doi: 10.1128/jcm.25.11.2123-2125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nagata K, Tsukita S, Tamura T, Sone N. A cb-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology. 1996;142:1757–1763. doi: 10.1099/13500872-142-7-1757. [DOI] [PubMed] [Google Scholar]
- 212.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. Urease plays an important role in the chemotactic motility of Helicobacter pylori in aviscous environment. Infect Immun. 1998;66:4832–4837. doi: 10.1128/iai.66.10.4832-4837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nalini S, Ramakrishna B S, Mohanty A, Balasubramanian K A. Hydroxyl radical formation in human gastric juice. J Gastroenterol Hepatol. 1992;7:497–501. doi: 10.1111/j.1440-1746.1992.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 214.Namavar F, Sparrius M, Veerman E C I, Appelmelk B J, Vandenbroucke-Grauls C M. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect Immun. 1998;66:444–447. doi: 10.1128/iai.66.2.444-447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nedenskov P. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol. 1994;60:3450–3453. doi: 10.1128/aem.60.9.3450-3453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Neidlands J B. Microbial envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- 217.Neman-Simha V, Mégraud F. In vitro model for Campylobacter pylori adherence properties. Infect Immun. 1988;56:3329–3333. doi: 10.1128/iai.56.12.3329-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Neuhard J, Kelln R A. Biosynthesis and conversion of pyrimidines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 580–599. [Google Scholar]
- 219.Niimura Y, Massey V. Reaction mechanism of Amphibacillus xylanus NADH oxidase alkylhydroperoxide reductase flavoprotein. J Biol Chem. 1996;271:30459–30464. doi: 10.1074/jbc.271.48.30459. [DOI] [PubMed] [Google Scholar]
- 220.Niimura Y, Poole L B, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22 kDa protein component. J Biol Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 221.Nygaard P. Purine and pyrimide salvage pathways. In: Sonenshein A L, editor. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 359–378. [Google Scholar]
- 222.Odenbreit S, Till M, Haas R. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol Microbiol. 1996;20:361–373. doi: 10.1111/j.1365-2958.1996.tb02623.x. [DOI] [PubMed] [Google Scholar]
- 223.Odenbreit S, Till M, Hofreuter D, Haas R. Outer membrane proteins AlpA and AlpB are involved in H. pylori binding to epithelial cells. Gut. 1997;41:A107. [Google Scholar]
- 224.Odenbreit S, Wieland B, Haas R. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J Bacteriol. 1996;178:6960–6967. doi: 10.1128/jb.178.23.6960-6967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Olson E R. Influence of pH on bacterial gene expression. Mol Microbiol. 1993;8:5–14. doi: 10.1111/j.1365-2958.1993.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 226.O’Toole P W, Kostrzynska M, Trust T J. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol. 1994;14:691–703. doi: 10.1111/j.1365-2958.1994.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 227.Ottlecz A, Romero J J, Hazell S L, Graham D Y, Lichtenberger L M. Phospholipase activity of Helicobacter pylori and its inhibition by bismuth salts. Biochem Biophys Res Commun. 1993;38:2071–2080. doi: 10.1007/BF01297087. [DOI] [PubMed] [Google Scholar]
- 228.Oudbier J H, Langenberg W, Rauws E A J, Bruin-Moch C. Genotypical variation of Campylobacter pylori from gastric mucosa. J Clin Microbiol. 1990;28:559–565. doi: 10.1128/jcm.28.3.559-565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pan Z-J, Berg D E, van der Hulst R W M, Su W-W, Raudonikiene A, Xiao S-D, Dankert J, Tytgat G N J, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 230.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 231.Patte J C. Biosynthesis of threonine and lysine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 528–541. [Google Scholar]
- 232.Peek R M, Thompson S A, Atherton J C, Blaser M J, Miller G G. Expression of iceA, a novel ulcer associated Helicobacter pylori gene is induced by contact with gastric epithelial cells and is associated with enhanced mucosal IL-8. Gut. 1996;39:A71. [Google Scholar]
- 233.Pesci E, Pickett C. Genetic organization and enzymatic activity of a superoxide dismutase from the microaerophilic human pathogen, Helicobacter pylori. Gene. 1994;143:111–116. doi: 10.1016/0378-1119(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 234.Peterson S N, Bailey C C, Jensen J S, Borre M B, King E S, Bott K F, Hutchinson C A. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc Natl Acad Sci USA. 1995;92:11829–11833. doi: 10.1073/pnas.92.25.11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Phadnis S H, Ilver D, Janzon L, Normark S, Westblom T U. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pitson S M, Mendz G L, Srinivasan S, Hazell S L. The tricarboxylic acid cycle of Helicobacter pylori. Eur J Biochem. 1999;260:258–267. doi: 10.1046/j.1432-1327.1999.00153.x. [DOI] [PubMed] [Google Scholar]
- 238.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 239.Pittman M S, Jackson C J, Kelly D J, Clayton C L. Novel chemotaxis like genes in Helicobacter pylori: characterization of CheY and CheF, a homologue of the myxobacterial FRZE protein. Gut. 1997;41:A15. . (Abstract.) [Google Scholar]
- 240.Poole K, Zhao Q, Neshat S, Heinrichs D E, Dean C R. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology. 1996;146:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 241.Poole L B. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AHPF and AHPC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 242.Radcliff F J, Hazell S L, Kolesnikow T, Doidge C, Lee A. Catalase, a novel antigen for Helicobacter pylori vaccination. Infect Immun. 1997;65:4668–4674. doi: 10.1128/iai.65.11.4668-4674.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rautelin H, Tee W, Seppala K, Kosumen T U. Ribotyping patterns and emergence of metronidazole resistance in paired clinical samples of Helicobacter pylori. J Clin Microbiol. 1994;32:1079–1082. doi: 10.1128/jcm.32.4.1079-1082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 391–407. [Google Scholar]
- 245.Rektorschek M, Weeks D, Sachs G, Melchers K. Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology. 1998;115:628–641. doi: 10.1016/s0016-5085(98)70142-8. [DOI] [PubMed] [Google Scholar]
- 246.Reynolds D J, Penn C W. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirement. Microbiology. 1994;140:2649–2656. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- 247.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 822–848. [Google Scholar]
- 248.Richardson P T, Park S F. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology. 1995;141:3181–3191. doi: 10.1099/13500872-141-12-3181. [DOI] [PubMed] [Google Scholar]
- 249.Ritchings B, Almira E, Lory S, Ramphal R. Cloning and phenotypic characterization of the fleS and fleR, new response regulators in Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Roberts R J, Macelis D. Rebase-restriction enzymes and methylases. Nucleic Acids Res. 1998;26:338–350. doi: 10.1093/nar/26.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rock C O, Cronan J J E. Regulation of bacterial membrane lipids synthesis. Curr Top Membr Transp. 1982;17:209–233. [Google Scholar]
- 252.Sachs G, Meyer-Rosberg K, Scott D R, Melchers K. Acid, protons and Helicobacter pylori. Yale J Biol Med. 1996;69:301–316. [PMC free article] [PubMed] [Google Scholar]
- 253.Salmela K S, Roine R P, Koivisto T, Höök-Nikanne J, Kosunen T U, Salaspuro M. Characteristics of Helicobacter pylori alcohol dehydrogenase. Gastroenterology. 1993;105:325–330. doi: 10.1016/0016-5085(93)90704-g. [DOI] [PubMed] [Google Scholar]
- 254.Saunders N J, Peden J F, Hood D W, Moxon E R. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 255.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 256.Schmitt W, Odenbreit S, Heuermann D, Haas R. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol Gen Genet. 1995;248:563–572. doi: 10.1007/BF02423452. [DOI] [PubMed] [Google Scholar]
- 257.Schmitz A, Josenhans C, Suerbaum S. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J Bacteriol. 1997;179:987–997. doi: 10.1128/jb.179.4.987-997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Segal E D, Tompkins L S. Identification and characterization of a Helicobacter pylori hemolysin. Infect Agents Dis. 1993;2:178–182. [PubMed] [Google Scholar]
- 259.Sherbrune R, Taylor D E. Helicobacter pylori expresses a complex surface carbohydrate, Lewis X. Infect Immun. 1995;63:4564–4565. doi: 10.1128/iai.63.12.4564-4568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Simor A E, Shames B, Drumm B, Sherman P, Low D E, Penner J L. Typing of Campylobacter pylori by bacterial DNA restriction endonuclease analysis and determination of plasmid profile. J Clin Microbiol. 1990;28:83–86. doi: 10.1128/jcm.28.1.83-86.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–998. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 262.Skouloubris S, Thiberge J M, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1539–1549. [Google Scholar]
- 264.Smith M A, Edwards D I. Oxygen scavenging, NADH oxidase and metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1997;39:347–353. doi: 10.1093/jac/39.3.347. [DOI] [PubMed] [Google Scholar]
- 265.Song Q, Haller B, Schmid R, Adler G, Bode G. PCR detection of Helicobacter pylori in the dental plaque with different primers. Gut. 1997;41:A38. . (Abstract.) [Google Scholar]
- 266.Spiegelhalder C, Gerstenecker B, Kersten A, Schiltz E, Kist M. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect Immun. 1993;61:5315–5325. doi: 10.1128/iai.61.12.5315-5325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Srinivasan S, Mendz G L, Hazell S L. The Krebs’ cycle of Helicobacter pylori. Aust Microbiol. 1994;15:A97. [Google Scholar]
- 268.Staudenmaier H, Van Hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Stauffer G V. Biosynthesis of serine and glycine and one-carbon units. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 506–513. [Google Scholar]
- 270.Strauch M A, Zalkin H, Aronson A I. Characterization of the glutamyl-tRNA (Gln)-to-glutaminyl-tRNA (Gln) amidotransferase reaction of Bacillus subtilis. J Bacteriol. 1988;170:916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Strobel S, Bereswill S, Balig P, Allgaier P, Sonntag H-G, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–1289. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Suda Y, Ogawa T, Kashihara W, Oikawa M, Shimoyama T, Hayashi T, Tamura T, Kusumoto S. Chemical structure of lipid A from Helicobacter pylori strain 206-1 lipopolysaccharide. J Biochem. 1997;121:1129–1133. doi: 10.1093/oxfordjournals.jbchem.a021705. [DOI] [PubMed] [Google Scholar]
- 273.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Suerbaum S, Thilberg J M, Kansau I, Ferrero R L, Labigne A. Helicobacter pylori hspA-hspB heat shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol Microbiol. 1994;14:959–974. doi: 10.1111/j.1365-2958.1994.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 275.Tatusov R L, Mushegian A R, Bork P, Brown N P, Hayes W S, Borodovsky M, Rudd K E, Koonin E V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 276.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Tee W, Lambert J R, Dwyer B. Cytotoxin production by Helicobacter pylori from patients with upper gastrointestinal tract disease. J Clin Microbiol. 1995;33:1203–1205. doi: 10.1128/jcm.33.5.1203-1205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Telford J L, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Thompson S A, Latch R L, Blaser M J. Molecular characterization of the Helicobacter pylori uvrB gene. Gene. 1998;209:113–122. doi: 10.1016/s0378-1119(98)00028-6. [DOI] [PubMed] [Google Scholar]
- 280.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleishmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbuch J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalk H G, Glodek A, McKenney K, Fitzgerald L M, Lee M, Adams M D, Hickey E K, Berg D E, Gocayne I D, Fujii C, Bwoman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 281.Tummuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Tummuru M K R, Cover T L, Blaser M J. Mutations in the cytotoxin associated cagA gene does not affect the vacuolating cytotoxin activity of Helicobacter pylori. Infect Immun. 1994;62:2609–2613. doi: 10.1128/iai.62.6.2609-2613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tummuru M K R, Sharma S A, Blaser M J. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:867–876. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 284.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 442–457. [Google Scholar]
- 285.van der Ende A, Rauws E A J, Feller M, Mulder C J J, Tytgat G N J, Dankert J. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]
- 286.van Doorn L J, Figueiredo C, Carneiro F, Sanna R, Pena S, Midolo P, Blaser M J. Worldwide heterogeneity of the Helicobacter pylori vacA gene. Gut. 1997;41:A34. . (Abstract.) [Google Scholar]
- 287.van Doorn L-J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K W, Atherton J C, Blaser M J, Quint W G V. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Velayudhan J, Hughes N J, Andrews S C, Clayton C L, Kelly D J. Iron acquisition in H. pylori—characterization of tbpA, fecA, feoB, and exbB homologues. Gut. 1997;41:A109. . (Abstract.) [Google Scholar]
- 289.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 290.Warren J R, Marshall B. Unified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 291.Weitkamp H J H, Perez-Perez G I, Bode G, Malfertheiner P, Blaser M J. Identification and characterization of Helicobacter pylori phospholipase C activity. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;280:11–27. doi: 10.1016/s0934-8840(11)80937-0. [DOI] [PubMed] [Google Scholar]
- 292.Wetherall B L, McDonald P J, Johnson A M. Partial characterization of a cell-free hemolytic factor produced by Helicobacter pylori. FEMS Microbiol Immunol. 1992;4:123–128. doi: 10.1111/j.1574-6968.1992.tb04978.x. [DOI] [PubMed] [Google Scholar]
- 293.Williams C L, Preston T, Hossack M, Slater C, Mccoll K E L. Helicobacter pylori utilises urea for amino acid synthesis. FEMS Immunol Med Microbiol. 1996;13:87–94. doi: 10.1111/j.1574-695X.1996.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 294.Winans S C, Burns D L, Christie P J. Adaptation of conjugal system for the export of pathogenic macromolecules. Trends Microbiol. 1996;13:87–94. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Winkler M E. Biosynthesis of histidine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 485–505. [Google Scholar]
- 296.Wirth H P, Yang M Q, Karita M, Blaser M J. Expression of the human cell surface glycoconjugates Lewis X and Lewis Y by Helicobacter pylori isolates is related to cagA status. Infect Immun. 1996;64:4598–4605. doi: 10.1128/iai.64.11.4598-4605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wolle K, Leodolter A, Malfertheiner P, Kanig W. Genomic diversity of urease gene among Helicobacter pylori strains isolated from one geographical area. Gut. 1997;41:A40. . (Abstract.) [Google Scholar]
- 298.Worku M, Sidebotham R L, Wren B W, Karim Q N. Chemotaxis of H. pylori in presence of human plasma. Gut. 1997;41:A25. . (Abstract.) [Google Scholar]
- 299.Worst D J, Gerrits M M, Vandenbroucke-Grauls C M J E, Kusters J G. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J Bacteriol. 1998;180:1473–1479. doi: 10.1128/jb.180.6.1473-1479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Worst D J, Otto B R, Degraaff J. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect Immun. 1995;63:4161–4165. doi: 10.1128/iai.63.10.4161-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Xiao H, Kalman M, Ikhihara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bipyrophosphate synthetic activity in relA null mutants can be eliminated by spoT null mutation. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 302.Yagi T, Di Bernardo S, Matsuno-Yagi A. Procaryotic complex I (NDH-1), an overview. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 303.Yeoman K H, Delgado M J, Wexler M, Downie J A, Johnston A W. High affinity iron acquisition in Rhizobium leguminosarum requires the cycHJKL operon and the feuPQ gene products, which belong to the family of two component transcriptional regulators. Microbiology. 1997;143:127–134. doi: 10.1099/00221287-143-1-127. [DOI] [PubMed] [Google Scholar]
- 304.Yogev D, Rosengarten R, Watson-McKnown R, Wise K. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yoshiyama H, Nakamura H, Mizote T, Nakazawa T. Chemotaxis of Helicobacter pylori in a viscous environment is urease dependent. Gut. 1997;41:A23. . (Abstract.) [Google Scholar]
- 306.Zakharova N, Hoffman P, Berg D E, Severinov K. The largest subunits of RNA polymerase from gastric helicobacters are tethered. J Biol Chem. 1998;273:19371–19374. doi: 10.1074/jbc.273.31.19371. [DOI] [PubMed] [Google Scholar]
- 307.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 561–579. [Google Scholar]
- 308.Ziebarth D, Spiegelhalder B, Bartsch H. N-nitrosation of medicinal drugs catalysed by bacteria from human saliva and gastro-intestinal tract, including Helicobacter pylori. Carcinogenesis. 1997;18:383–389. doi: 10.1093/carcin/18.2.383. [DOI] [PubMed] [Google Scholar]