Abstract
Serum induces Candida albicans to make a rapid morphological change from the yeast cell form to hyphae. Contrary to the previous reports, we found that serum albumin does not play a critical role in this morphological change. Instead, a filtrate (molecular mass, <1 kDa) devoid of serum albumin induces hyphae. To study genes controlling this response, we have isolated the RAS1 gene from C. albicans by complementation. The Candida Ras1 protein, like Ras1 and Ras2 of Saccharomyces cerevisiae, has a long C-terminal extension. Although RAS1 appears to be the only RAS gene present in the C. albicans genome, strains homozygous for a deletion of RAS1 (ras1-2/ras1-3) are viable. The Candida ras1-2/ras1-3 mutant fails to form germ tubes and hyphae in response to serum or to a serum filtrate but does form pseudohyphae. Moreover, strains expressing the dominant active RAS1V13 allele manifest enhanced hyphal growth, whereas those expressing a dominant negative RAS1A16 allele show reduced hyphal growth. These data show that low-molecular-weight molecules in serum induce hyphal differentiation in C. albicans through a Ras-mediated signal transduction pathway.
The ability to switch from a cellular yeast form to a filamentous form is characteristic of many fungi. In Saccharomyces cerevisiae, the switch from yeast cells to pseudohyphae is signaled by nitrogen starvation and appears to be under the dual control of both the mitogen-activated protein (MAP) kinase and A kinase pathways (20, 23, 29, 30). There is considerable evidence that the RAS2 gene acts upstream of both signaling pathways. Deletion of RAS2 reduces filamentous growth, whereas the dominant active allele of RAS2 enhances pseudohyphal growth (12, 30).
In Candida albicans (unlike S. cerevisiae) serum induces the switch from yeast to a variety of filamentous forms (germ tubes, pseudohyphae, and hyphae). This morphological change is thought to be essential for virulence (1). Serum appears to induce the morphologic change in Candida via several pathways (24). This conclusion is based on the response of mutants to serum. Single mutants in either CPH1 (a STE12 orthologue) or EFG1 can still be induced to form hyphae by serum; only a strain containing two mutations, the double mutant cph1 efg1, fails to show vigorous serum induction of hyphae from yeast cells (21). This observation led to the proposal that serum triggers multiple signaling pathways in Candida, any one of which was capable of inducing the yeast to hyphal conversion.
The complexity of serum has hampered the identification of a single component that is responsible for its ability to induce the morphological change in Candida. Previous investigations had shown that the serum inducing factor(s) was heat stable and nondialyzable (5, 28) and that the peak of serum induction activity comigrated with the serum albumin fraction in purification by gel filtration (2). However, several observations argue against albumin itself as the inducing factor. Although some commercially purified serum albumins have the ability to induce hyphae, other preparations of albumin do not. Moreover, an active serum albumin fraction, when hydrolyzed to its constituent amino acids, retained its inductive activity (6, 7). These studies are consistent with the possibility that it is not albumin that evokes the morphologic change but rather components that copurify with it.
We provide here evidence that serum albumin does not play a key role in serum induction. Serum from a mutant rat strain that is completely devoid of albumin induces Candida to form germ tubes and hyphae as effectively as serum from a normal rat that has albumin. Furthermore, a low-molecular-weight serum filtrate (molecular mass, <1 kDa) induces hyphae with the same potency as does serum containing albumin. We show that the Candida RAS1 gene is essential for hyphal differentiation in response to this low-molecular-weight serum factor. A dominant active RAS1 allele can bypass the serum requirement for hyphal growth. Thus, the RAS1 gene plays a key role in regulating the serum response in C. albicans.
MATERIALS AND METHODS
Screen for the Candida RAS1 gene.
The strain ASY240 (ras1 ras2 ade2 ade3 trp1 [Table 1]) provided a convenient colony color system to screen for Candida RAS genes. Colonies of strains carrying the ade2 mutation are red, whereas colonies of an ade2 ade3 strain are white and require adenine and histidine because the ade3 mutation blocks accumulation of the red pigment and results in both an adenine and histidine requirement (15). ASY240 contains the human v-RAS gene and the yeast TRP1 and ADE3 genes on an unstable ARS plasmid (pAS1076). The human v-RAS gene is required for the growth of ASY240 (yeast ras1 ras2 strains are inviable), and the ADE3 gene complements the ade3 defect permitting the formation of the red pigment. Thus, ASY240 grows into homogeneous red colonies on plates containing low amounts of histidine and adenine.
TABLE 1.
Strains, primers, and plasmids used in this study
| Strain, primer, or plasmid | Genotype, sequence, or description |
|---|---|
| S. cerevisiae ASY240 | ura3-1 leu2-3 112 his3-11,15 trp1Δ63 ade2-1 ade3::hisG |
| ras1::HIS3 ras2::LEU2 (pAS1076) | |
| C. albicans | |
| RAS1/ras1-1 | RAS1/ras1Δ::hisG-URA3-hisG |
| RAS1/ras1-2 | RAS1/ras1Δ::hisG |
| ras1-2/ras1-3 | ras1Δ::hisG/ras1Δ::hph-URA3-hph |
| ras1-2/ras1-4 | ras1Δ::hisG/ras1Δ::hph |
| ras1-2/ras1-4/RAS1 | ras1Δ::hisG/ras1Δ::hph/ade2::RAS1-URA3 |
| RAS1/RAS1/M-RAS1 | RAS1/RAS1/ade2::Mal-RAS1-URA3 |
| RAS1/RAS1/M-RAS1V13 | RAS1/RAS1/ade2::Mal-RAS1V13-URA3 |
| RAS1/RAS1/M-RAS1A16 | RAS1/RAS1/ade2::Mal-RAS1A16-URA3 |
| Primers | |
| QF61 (hph gene) | 5′-GGATCGATATGAAAAAGCCTGAACTCACCG-3′ |
| QF62 (hph gene) | 5′-GGATCGATCTATTCCTTTGCCCTCGG-3′ |
| CaRAS1-NF (5′RAS1) | 5′-GGTTAATGAATGAATGGCTTGC-3′ |
| QF92 (5′RAS1) | 5′-CGGGATCCGTATATGTGTGGATATGAATTTG-3′ |
| QF93 (3′RAS1) | 5′-CGGGATCCCCTATCCAATTGATTGTTTCC-3′ |
| QF94 (3′RAS1) | 5′-CTCCAGAAAGATCATTTGGTG-3′ |
| QF91 (RAS1) | 5′-TAAGCTTCATACCATGTTGAGAGAATAT-3′ |
| QF87 (RAS1V13) | 5′-TTGTTGTTGGAGGTGTTGGTGTTGGTAAAT-3′ |
| QF88 (RAS1V13) | 5′-ATTTACCAACACCAACACCTCCAACAACAA-3′ |
| QF89 (RAS1A16) | 5′-GAGGTGGTGGTGTTGCTAAATCCGCTTTAAC-3′ |
| QF90 (RAS1A16) | 5′-GTTAAAGCGGATTTAGCAACACCACCACCTC-3′ |
| Plasmids | |
| pAS1076 | Human v-RAS gene under Adh promoter in pACYC184 with TRP1-ARS1 from YRP7 |
| R2Cla | RAS1 ClaI fragment in pBluescript II KS(+) |
| pQF137 | RAS1 deletion construct with hisG-URA3-hisG cassette |
| pQF138 | RAS1 deletion construct with hph-URA3-hph cassette |
| BES116 | ADE2-URA3-ADE2 AscI fragment in pBluescript II KS(+) |
| BES119 | ADE2-URA3-pMAL2-ADE2 AscI fragment in pBluescript II KS(+) |
| pQF119 | RAS1 in BES116 |
| pQF144.1 | RAS1 in BES119 |
| pQF145.2 | RAS1V13 in BES119 |
| pQF146.1 | RAS1A16 in BES119 |
The Candida RAS gene was cloned by transforming a C. albicans genomic library (19) on a plasmid lacking ADE3 into ASY240. The resulting Saccharomyces transformants were screened for the appearance of white colonies. The presumption was that the white colonies were those that could lose the resident plasmid (pAS1076) containing the human v-RAS gene because they contained a Candida RAS gene capable of suppressing the ras1 ras2 defect (16). The transformants were selected at 30°C for 4 days on a synthetic complete medium (SC) designed to highlight the color difference (it lacks uracil [32] and contains 0.1 mM adenine and 0.1 mM histidine). White colonies were single colony purified on standard SC-Ura plates and replicated to SC-Trp and SC-His plates. Plasmids were isolated from colonies that were white on low-adenine-histidine plates (and tested as Trp− His−, indicating loss of plasmid pAS1076) and transformed into Escherichia coli. Purified plasmids were retransformed into ASY240 to verify the phenotype. The RAS1 ClaI fragment was subcloned into pRS202 (Fink lab plasmid collection), resulting in plasmid R2cla, and sequenced.
Novel hph-URA3-hph disruption cassette.
The creation of null mutations in Candida requires two sequential knockouts because Candida is diploid. To facilitate this process, we created a new construct, the hph-URA3-hph disruption cassette (see Results). The E. coli hph gene was PCR amplified by using primers QF61, QF62, and B690 (Fink lab plasmid collection, hph gene clone) as the template, cloned into pCR2.1 (TA Cloning Kit; Invitrogen), resulting in pQF48.7 and pQF48.17, with the insert at opposite orientations in the vector respectively. The HindIII-XbaI fragment of hph from pQF48.7, the XbaI-ScaI fragment of Candida URA3 from pQF65.2 and the EcoRV-BamHI fragment of hph from pQF48.17 were cloned into the pBluescript II KS(+) HindIII-BamHI site, yielding pQF86. The hph-URA3-hph cassette (3.4 kb) can be released by BamHI digestion.
C. albicans growth and transformation.
The media for growth experiments was yeast extract-peptone-dextrose medium (YPD) with 2% glucose at room temperature and YPD with 4% glucose at 37°C. At 37°C Candida cells form hyphae that interfere with the measurement of growth by optical density (OD). The addition of 4% glucose at 37°C inhibits hyphal growth and permits an accurate measurement of the growth rate at this temperature.
Cells were grown in YPD at room temperature to late log phase (OD600 of ca. 5). The standard S. cerevisiae lithium acetate transformation procedure was followed for transformation of C. albicans.
Deletion of RAS1.
5′-RAS1 was PCR amplified by using QF92 and CaRAS-NF as primers and R2cla as the template. The product was cloned into pCR2.1, resulting in plasmid pQF121. 3′-RAS1 was PCR amplified with QF93 and QF94 as primers and R2cla as the template. The product was cloned into pCR2.1, resulting in plasmid pQF122. The SpeI-BamHI fragment from pQF121, and the BamHI-PstI fragment from pQF122 were inserted at the SpeI/PstI site in pBluescript II KS(+), resulting in pQF124. The RAS1 deletion constructs, pQF137 and pQF138, were generated by inserting the hisG-URA3-hisG (10) cassette and the Hph-URA3-Hph cassette, respectively, at the BamHI site in pQF124. The heterozygous deletion strain (RAS1/ras1-1) was obtained by transforming C. albicans CAI4 strain (10) with the PstI/NotI fragment of pQF137. The homozygous deletion strain (ras1-2/ras1-3) was obtained by transforming the heterozygous deletion strain (RAS1/ras1-2) with the SacI/ClaI fragment of pQF138.
Constructs for ectopic expression of RAS1 in Candida.
A PvuII fragment containing a multiple cloning site and flanking sequences was removed from pBluescript II KS (Stratagene) and replaced by an AscI site. A 2.2-kb Candida ADE2 gene containing SpeI fragment was cloned into the new AscI site, and then the Candida URA3 gene on a 1.8 kb BamHI/XbaI fragment was used to replace a 0.6-kb BamHI/XbaI fragment within ADE2. Next, the multiple cloning site of pBluescript II KS resulting from BssHII digestion was cloned into the XbaI site flanking the URA3 gene. These constructions resulted in plasmid BES1116. Finally, a 0.5-kb fragment of the Candida MAL2 gene promoter (up to but not including a start site) was cloned at SacI/EcoRV within the multiple cloning site. This resulted in plasmid BES119.
The ClaI fragment of R2cla was cloned into the BES116 ClaI site, resulting in pQF119: the RAS1 gene and URA3 marker were flanked by the ADE2 5′- and 3′-end sequences. The AscI fragment of pQF119 was transformed into the homozygous deletion strain (ras1-2/ras1-4), and Ura+ colonies were selected.
Site-directed mutagenesis.
The wild-type RAS1 open reading frame (ORF) was PCR amplified with QF91 and T3 as primers and R2cla as the template. The product was cloned into pCR2.1 to form plasmid pQF120.1. The RAS1V13 mutant was generated by the DpnI method (Stratagene QuickChange Site-Directed Mutagenesis Kit, catalog number 200518) by using primers QF87 and QF88 with pQF120.1 as the template to form plasmid pQF139.2. Similarly, the RAS1A16 plasmid was generated by using QF89 and QF90 as primers to form pQF140.1. The constructs were confirmed by sequencing. The RAS1, RAS1V13, and RAS1A16 ORFs were released from the respective plasmids by HindIII/XhoI digestion and cloned into the plasmid BES119 to form pQF144.1, pQF145.2, and pQF146.1, respectively. The AscI fragments of these plasmids were used to transform C. albicans. The transformants containing these constructs have RAS1 alleles that are inducible when the strain is grown on maltose.
Induction of germ tubes and hyphae.
C. albicans SC5314, the Ura+ ancestor of CAI4 (10) was used to follow the formation of germ tubes and hyphae from yeast form cells. Portions (5 μl) of log-phase cells (OD600 of ca. 1) were mixed in 500 μl (105/ml) 50 mM potassium phosphate (pH 6)–10% serum or serum filtrate and incubated at 37°C. At various times after incubation the morphology of the cells was monitored under the microscope.
The serum filtrate was obtained by dialysis serum against 50 mM potassium phosphate at pH 6 by using a Spectra/Por regenerated cellulose dialysis membrane (CMS 265-015).
To determine colony and cell morphology on solid media, we streaked the cells on agar plates containing serum (5%) and incubated them at 37°C overnight. In other experiments, cells were streaked on SC-Ura sucrose (2%)–50 mM succinic acid (pH 5) plates and incubated at 30°C for a week.
Heat shock sensitivity and glycogen content.
Cells growing in liquid SC-Ura sucrose medium to early stationary phase (OD600 of ca. 10 to 20) were incubated at 50°C for 10 min, diluted fivefold, spotted onto YPD plates, and incubated at 30°C for 2 days. Plates were photographed, and colonies from the last dilution were counted.
Different strains were patched either on synthetic complete medium without uracil plates with sucrose as the sole carbon source or on YPD plates, incubated at 30°C for 2 days, stained with iodine vapor, and photographed.
Nucleotide sequence accession number.
Sequence data described in this report have been submitted to GenBank under accession no. AF177670.
RESULTS
Albumin is not the component of serum that induces hyphal growth.
Serum is a potent and medically relevant inducer of C. albicans hyphal growth. Although Candida forms germ tubes and hyphae in response to diverse external treatments, some strains form germ tubes and hyphae only after exposure to serum (27), and some treatments will induce only stationary-phase cells (3), but not exponentially growing cells, to form germ tubes and hyphae. Previous studies indicated that serum albumin is the component of serum that mediates this induction (2). However, we and others have found that not all commercially available purified serum albumin induces germ tubes and hyphae in Candida (2).
Four lines of evidence suggest that it is not the albumin in serum that causes the morphological induction. First, commercially purified serum albumin is not as effective as unfractionated serum in inducing the morphological change. At equal concentrations of protein (Fig. 1A), serum is at least 50-fold more effective than commercially purified serum albumin in inducing both germ tubes and hyphae. Second, filtration of serum through a molecular-weight cutoff membrane (of ca. 1 kDa) revealed that almost all of the inductive activity resides in the small molecules that flow through the membrane and not in the albumin, which is retained (data not shown). Third, purified recombinant human albumin from Pichia pastoris was not a potent inducer of filamentation in Candida. At concentrations where commercial purified serum albumin (purified from serum) produced complete conversion of yeast to filaments, the recombinant human albumin failed to induce any morphological change in Candida. Tenfold-higher concentrations of recombinant human albumin failed to induce hyphae but were capable of inducing pseudohyphae (Fig. 1A). Fourth, serum obtained from a rat mutant lacking albumin was as potent in inducing filaments as that containing albumin (Fig. 1B). The Nagase analbuminemic rat (NAR) reduces the albumin concentration in serum at least 7,000-fold compared to wild-type rat serum (SD8W) (14, 31). Serial dilution experiments show that 0.6% solutions of either wild-type or mutant sera induced 100% germ tubes and hyphae, 0.3% solutions of either induced 50% germ tubes and hyphae in 2 h, and 0.15% solutions of either failed to induce germ tubes and hyphae even after 24 h. Since serum albumin constitutes about 40% of serum protein, the protein concentration of NAR serum is about two-thirds that of the wild-type rat (SD8W) serum. These data show that serum from the mutant lacking albumin is as potent as serum from the wild type in inducing germ tubes and hyphae.
FIG. 1.
Serum albumin is not the component of serum that induces germ tubes and hyphae. (A) Germ tubes and hyphae are induced by different concentrations of bovine serum and bovine serum albumin (BSA). Serum contains 40 mg of albumin per ml. Therefore, 0.5% serum contains 0.2 mg of albumin and 0.2% serum contains 0.08 mg of albumin. The recombinant human serum albumin (rHSA) only induced pseudohyphae at a high concentration. (B1) NAR serum induces germ tubes (2 h) and hyphae (24 h) as effectively as the wild-type rat (SD8W) serum. (B2) Sodium dodecyl sulfate-polyacrylamide gel. Lanes: 1, molecular weight marker; 2, 14.5 μg of purified bovine serum albumin; 3, 0.5 μl of SD8W serum; 4, 0.5 μl of NAR serum.
C. albicans RAS1 gene.
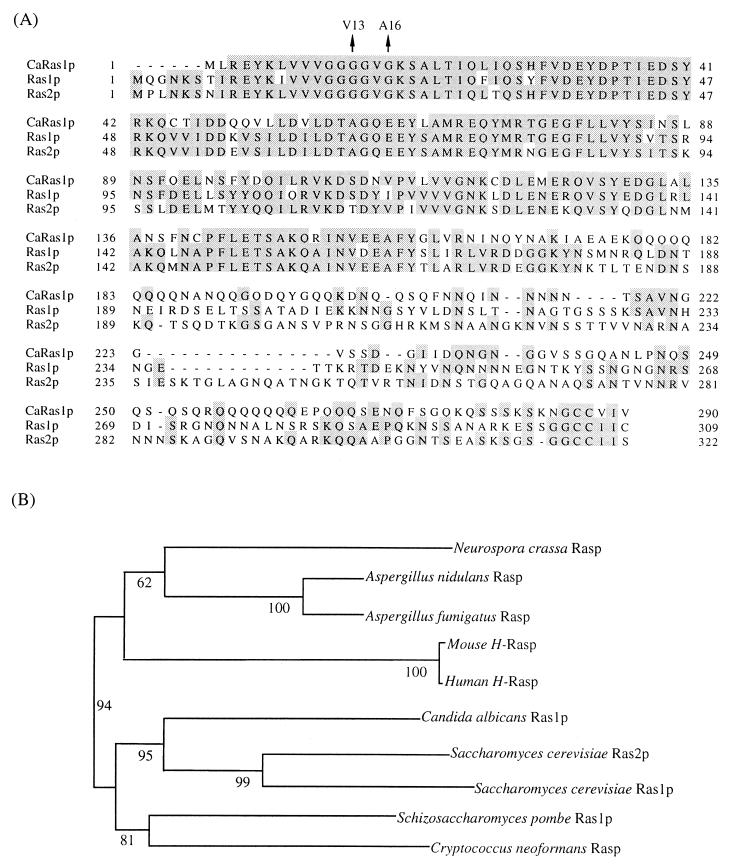
The C. albicans RAS1 gene was cloned from a genomic library (19) by its ability to suppress the viability defect of an S. cerevisiae ras1 ras2 strain (see Materials and Methods). The ORF on the suppressing plasmid is homologous to RAS genes in S. cerevisiae, other fungi, and mammals. The predicted Ras protein encoded by the C. albicans RAS1 gene was aligned with proteins encoded by the S. cerevisiae RAS1 and RAS2, human RAS gene, and RAS genes from other fungi (Fig. 2A). The C. albicans RAS1 protein sequence is most closely related to those predicted for the S. cerevisiae RAS1 and RAS2 genes (Fig. 2B). The Candida Ras1p has an extension in the C terminus typical of the Saccharomyces sequence but is 6-amino-acid residues shorter at the N terminus than the Saccharomyces sequences. The C-terminal extension is glutamine-rich and has no significant homology to the Saccharomyces sequence or to any other sequence in the database. Low-stringency Southern analysis suggests that there is only one RAS gene in Candida genome (Fig. 3B).
FIG. 2.
The C. albicans Ras protein. (A) Sequence alignment of C. albicans Ras1, S. cerevisiae Ras1 and Ras2 proteins. Identical residues are shaded. Amino acids changed in different Ras proteins are indicated by arrows. (B) Phylogenetic tree of Ras proteins from different organisms. The 3′-end sequences of S. cerevisiae Ras1 and Ras2 and C. albicans Ras1 proteins were truncated to create the tree by using the CLUSTALX program. The phylogenetic tree was made with 1,000 bootstrap resamplings. The numbers are bootstrap percentages for each branch point.
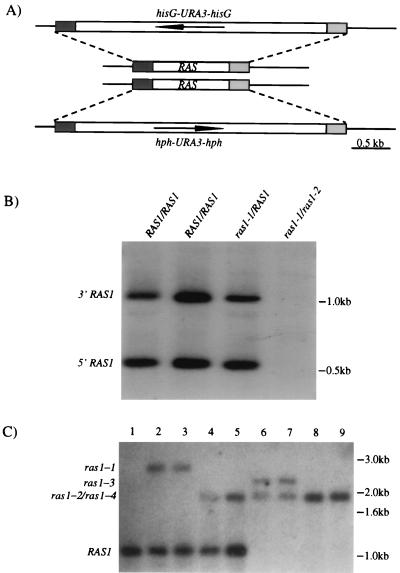
FIG. 3.
Deletion of the RAS1 gene in C. albicans. (A) The RAS1 ORF (0.9-kb), hisG-URA3-hisG (4.0-kb), and hph-URA3-hph (3.4-kb) cassettes are represented by open boxes. The arrows in the boxes indicate the direction of the transcription of URA3. 5′- and 3′-end sequences of RAS1 are represented by shaded boxes. (B) Low-stringency Southern blot analysis of RAS genes in C. albicans. Candida genomic DNAs from different strains were digested with SspI, run on 1% agarose gel, and transferred to the Hybond+ membrane. The membrane was hybridized at 65°C overnight and washed at 60°C in 0.5 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The probe is RAS1 ORF. (C) Southern blot analysis of RAS1 deletion. Genomic DNAs were digested with SspI. The probe is the RAS1 3′ sequence. Lanes: 1, wild-type strain; 2 and 3, RAS1/ras1-1; 4 and 5, RAS1/ras1-2; 6 and 7, ras1-2/ras1-3; 8 and 9, ras1-2/ras1-4.
Disruption of C. albicans RAS1 gene.
To test whether the RAS1 gene plays a role in the response of Candida to serum, we deleted the RAS1 gene in C. albicans. A strain heterozygous for the RAS1 deletion was constructed by using a hisG-URA3-hisG cassette (ras1-1) (10). Segregants that had lost the URA3 marker (ras1-2) were then isolated from this strain. The segregants were shown to be heterozygous for the ras1-2 deletion both by PCR and by Southern analysis (Fig. 3C).
Disruption of the remaining RAS1 gene in the heterozygote proved difficult when using transformation with a plasmid containing the standard hisG-URA3-hisG cassette. This plasmid integrates at high frequency into the ras1-2 deletion allele rather than the RAS1 allele presumably because of the hisG homology between the hisG region on the plasmid and the hisG in the ras1-2 gene on the chromosome. To avoid this problem, we constructed a novel disruption cassette hph-URA3-hph in which the hisG segment was replaced by the E. coli hygromycin gene (hph; see Materials and Methods). When this hygromycin construct (ras1-3) was used, approximately 10% of the transformants had the construct integrated in the intact RAS1 allele in the heterozygote (as shown by PCR and Southern analysis [Fig. 3C]). Two independent strains homozygous for the ras1 deletion (ras1-2/ras1-3) each derived from an independently constructed RAS1/ras1 heterozygote were analyzed and tested for all the phenotypes described in the present study.
Strains homozygous for the deletion of ras1 (ras1-2/ras1-3) are viable but grow more slowly than the progenitor RAS1/RAS1 strain. The doubling times for ras1-2/ras1-3, RAS1/ras1-1, and RAS1/RAS1 at 23°C are 3, 1.5, and 1 h, respectively, and at 37°C are 1.5, 1.16, and 1 h, respectively.
Strains lacking the RAS1 gene have a severe defect in the response to serum. After 24 h, the homozygous ras1-2/ras1-3 mutant failed to form either germ tubes or true hyphae in response to serum or the serum filtrate at pH 6 and 37°C. By contrast, strains heterozygous for the deletion RAS1/ras1-1 form wild type-like germ tubes after 2 h and hyphae after 7 h (Fig. 4A). On an agar plate containing serum, the wild-type (RAS1/RAS1) strain and the heterozygous deletion (RAS1/ras1-1) strain form hyphae, whereas the homozygous deletion (ras1-2/ras1-3) strain forms round colonies without protruding hyphae (Fig. 4B). A ras1-2/ras1-4 strain transformed with the RAS1 gene (ras1-2/ras1-4/RAS1) appears to be identical to wild-type strains with respect to growth rate and the ability to form germ tubes and hyphae both on plates and in liquid serum-containing medium.
FIG. 4.
The homozygous ras1-2/ras1-3 deletion strain was defective for both germ tubes and hyphal formation. (A) Liquid medium assay. Log-phase cells (105/ml) were induced by 10% serum in 50 mM potassium phosphate (pH 6) at 37°C. (B) Solid medium assay. The agar plate contains 5% serum. Cells were streaked onto the plate and incubated at 37°C overnight.
The ras1-2/ras1-3 mutant responds slightly differently to unfractionated serum as compared with the serum filtrate. After 24 h of incubation, the ras1-2/ras1-3 mutant formed some pseudohyphae when exposed to serum but remained as yeast forms when exposed to the serum filtrate. These data could be interpreted to mean that there is some large molecule in the prefractionated serum (and absent from the filtrate) that is capable of inducing pseudohyphae. However, this hypothetical molecule is not very potent and fails to promote conversion of the ras1-2/ras1-3 mutant to hyphae.
Dominant RAS1 mutations affect filamentous growth.
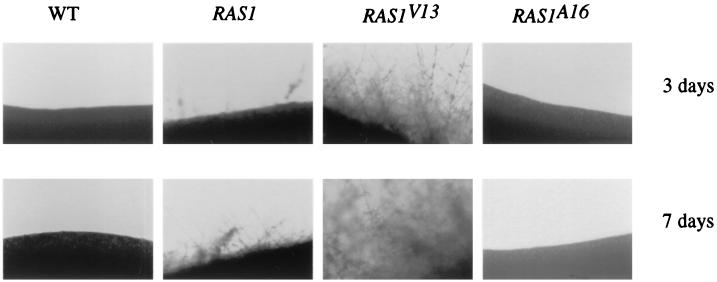
Dominant-active (RAS1V13) and dominant-negative (RAS1A16) alleles (Fig. 2A) of the Candida RAS1 gene were generated based on the amino acid substitutions observed for similar mutations in S. cerevisiae. Plasmids were constructed in which each of these alleles, RAS1, RAS1V13, or RAS1A16, was expressed from the inducible maltose 2 promoter (referred to as M-RAS1) (11), and each of these constructs was introduced into a RAS1/RAS1 strain by transformation. On glucose medium, where the maltose promoter is repressed, all three strains formed hyphae after 1 week of incubation. On sucrose medium at 30°C, where the MAL2 promoter is induced, the wild-type RAS1/RAS1 strain began to form short hyphae after 7 days, whereas the RAS1/RAS1/M-RAS1 strain formed longer and more abundant hyphae during the same time period. By contrast, the RAS1/RAS1/M-RAS1V13 strain produces abundant hyphae after only 3 days of incubation, and the RAS1/RAS1/M-RASA16 strain failed to form hyphae even after 2 weeks of incubation (Fig. 5).
FIG. 5.
The dominant-active allele RAS1V13 enhances filamentous growth, and the dominant-negative allele RAS1A16 suppresses filamentous growth. Cells were streaked onto a SC-Ura–sucrose (2%) plate containing 50 mM succinate at pH 5. The strain carrying the RAS1/RAS1/M-RAS1V13 (the RAS1V13 allele under control of the maltose 2 promoter) formed hyphae at the edge of the colonies after incubation at 30°C for 3 days, whereas RAS1/RAS1/M-RAS1 (a strain containing wild-type RAS1 allele under the maltose promoter) started to form hyphae after 7 days. The wild-type strain formed shorter and fewer hyphae after 7 days than did RAS1/RAS1/M-RAS1. The strain carrying the RAS1A16 allele did not form hyphae even after 2 weeks.
Candida ras1 mutants have phenotypes similar to those of ras2 Saccharomyces mutants.
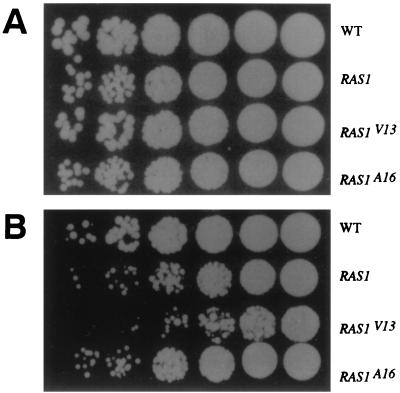
We examined C. albicans strains containing the ras1 mutations to determine whether they had phenotypes in common with Saccharomyces containing comparable ras2 mutations. Strains of S. cerevisiae containing the dominant active allele RAS2V19 are more sensitive to heat-shock than are wild-type strains and fail to accumulate glycogen. The Candida strains were heat shocked at 50°C for 10 min and then plated at 30°C for 2 days. The Candida RAS1/RAS1/M-RASV13 strain was 10- to 20-fold more sensitive to heat shock than the wild type strain; the strain RAS1/RAS1/M-RAS1 was twofold more sensitive. There was no difference in the heat shock sensitivity of the wild-type strain and the RAS1/RAS1/M-RAS1A16 strain (Fig. 6).
FIG. 6.
The strain carrying the dominant active allele RAS1V13 was more sensitive to heat shock. The same number of cells were either left untreated (A) or heat shocked at 50°C for 10 min (B). Each spot represents the culture diluted successively by fivefold (from right to left) on a YPD plate. The plates were incubated at 30°C for 2 days.
Colonies of the Candida wild-type strain stain brown with iodine vapor, whereas those of the Candida RAS1/RAS1/M-RAS1V13 strain stain yellow instead of brown (data not shown). This difference in staining suggests that strains containing the Candida M-RAS1V13 mutation do not accumulate as much glycogen as do the wild-type strains. No difference was observed between the intensity of staining of the wild-type strain (RAS1/RAS1) and either the Candida RAS1/RAS1/M-RASA16 or the ras1-2/ras1-3 strains.
DISCUSSION
The change in morphology in response to serum is a characteristic signature of C. albicans. When incubated with serum at 37°C and neutral pH, C. albicans undergoes a morphological change to form germ tubes and hyphae. Our data strongly suggest that serum albumin is not the key component in serum that induces this response. Serum from the NAR induces germ tubes and hyphae of C. albicans as effectively as the wild-type rat serum. Serum dilution experiments failed to detect a difference between these two sera in their ability to induce filamentation despite the fact that the quantitative difference in albumin concentration between these two rat sera is at least 7,000-fold (14, 31).
Although some samples of serum albumin induce germ tubes and hyphae, none of the samples is as potent as serum. It is likely that the inductive ability resides in other molecules that copurify with the albumin. Consistent with this idea, we found that small molecules filtered out of the serum induced filamentation in Candida. However, we were unable to purify a single small molecule that was responsible for the inductive activity, which raises the possibility that more than one component is required to induce this rapid morphological change. Previous studies have shown that proline (8) or a mixture of amino acids (6, 18) can induce germ tubes and hyphae in C. albicans. Under the conditions described here, an amino acid mixture (reconstituted to equal the known concentrations in serum) is not as effective as the serum filtrate in inducing germ tubes and hyphae.
Our finding that the Candida RAS1 is essential for the morphological change induced by serum is consistent with observations on this morphological change in other fungi. For example, in S. cerevisiae the RAS2/cyclic AMP (cAMP) pathway is one component in the regulation of filamentous growth. Moreover, deletion of RAS2 reduces filamentous growth (17), and elevated levels of cytoplasmic cAMP enhance filamentous growth (22). Both the phosphodiesterase (pde2) mutant and the dominant-active allele of RAS2 (RAS2V19) enhance filamentous growth (12). Recent studies show that of the three cAMP-dependent protein kinases (A kinase) in S. cerevisiae only one, Tpk2, is required for filamentous growth (29). The cAMP pathway has also been implicated in the morphogenesis of other fungi. In the corn pathogen Ustilago maydis and in the rice pathogen Magnaporthe grisea, the cAMP-dependent protein kinase catalytic subunit is required for both morphogenesis and virulence (9, 13, 25, 34). RAS activity also modulates many developmental decisions in Aspergillus nidulans (33).
The C. albicans Ras1 protein is most homologous to the S. cerevisiae Ras1p and Ras2p. The Candida Ras1p is the only other known Ras protein that has a C-terminal extension like that found in the S. cerevisiae Ras proteins. This similarity is reflected in the comparable phenotypes of the Candida and Saccharomyces ras mutants. Strains overexpressing activated RAS are more sensitive to heat shock and accumulate less carbohydrates than Ras+ strains. In Saccharomyces the ras2 deletion has reduced filamentation and in Candida the ras1/ras1 strain is defective in the formation of germ tubes and hyphae.
Despite these similarities, the ras mutants in the two organisms show a number of differences. The Saccharomyces ras2 deletion accumulates more carbohydrates than Ras+ strains, whereas the Candida ras1-2/ras1-3 deletion strain does not. Moreover, deletion of RAS1 in C. albicans has a more severe defect on filamentous growth than deletion of RAS2 in S. cerevisiae. Ras function in Saccharomyces is essential, whereas it does not appear to be in Candida. Although we uncovered only a single RAS gene in Candida, it is possible that there is another RAS gene with insufficient homology to have yielded a signal in the Southern blot analysis and able to supply Ras function for viability.
Experiments in S. cerevisiae suggest that RAS2 acts upstream of both the STE12 MAP kinase pathway (26, 30) and PHD1 pathways. The phenotype of the ras1-2/ras1-3 mutant in C. albicans is also consistent with RAS1 acting upstream of these pathways (CPH1 = STE12 and EFG1 = PHD1 pathways). Although the single cph1 mutant and the efg1 mutant are defective in filament formation on some media, each of these single mutant strains can be induced to form germ tubes and hyphae by serum. However, the cph1 efg1 double mutant, like the ras1-2/ras1-3 mutant, is defective in serum induction. Thus, the phenotypes of the mutants are compatible with a model in which RAS transmits a signal to two parallel pathways, the MAP kinase pathway and the PHD1 pathway, each of which activates the downstream targets required for the formation of germ tubes and hyphae.
Several observations suggest that this linear model may not be an adequate representation of the network controlling the morphological change from yeast cells to filamentous cells. Although neither the cph1 efg1 double mutant or the ras1 mutant make hyphae and pseudohyphae in response to serum, both strains form filaments after growth on rich medium (YPD) at room temperature for a week. Since neither the ras mutant or the cph1 efg1 double mutant block filament production under all environmental conditions, there must be additional routes to activate the downstream functions responsible for the morphological change. The multiple control of the switch from one cell type to another is not surprising because filament formation in C. albicans is induced by an enormous diversity of external cues (4, 8). The unraveling of this network of signaling pathways and their interactions may require not only traditional mutant analysis but also the application of novel technologies that await the completion of the Candida genome.
ACKNOWLEDGMENTS
We thank Mike Lorenz, Hiten Madhani, and Julia Koehler for helpful comments on the manuscript; Amir Sherman for providing strains; and Seymour Packman and Hiroky Kodama for providing the NAR and wild-type rat sera. We also thank Fran Lewitter for helping create the phylogenetic relationships.
This work supported in part by a Damon Runyon Postdoctoral Fellowship DRG-1418 (Q.F.), National Institute of Health postdoctoral fellowship K08 AI01484-02 (E.S.), National Research Service Award F32GM19181-02 (B.G.), and National Institute of Health grant GM40266 (G.R.F.). G.R.F. is American Cancer Society Professor of Genetics.
REFERENCES
- 1.Anderson M L, Odds F C. Adherence of Candida albicans to vaginal epithelia: significance of morphological form and effect of ketoconazole. Mykosen. 1985;28:531–540. doi: 10.1111/j.1439-0507.1985.tb02083.x. [DOI] [PubMed] [Google Scholar]
- 2.Barlow A J, Aldersley T, Chattaway F W. Factors present in serum and seminal plasma which promote germ-tube formation and mycelial growth of Candida albicans. J Gen Microbiol. 1974;82(Pt. 2):261–272. doi: 10.1099/00221287-82-2-261. [DOI] [PubMed] [Google Scholar]
- 3.Bell W M, Chaffin W L. Effect of yeast growth conditions on yeast-mycelial transition in Candida albicans. Mycopathologia. 1983;84:41–44. doi: 10.1007/BF00436995. [DOI] [PubMed] [Google Scholar]
- 4.Berardinelli S, Opheim D J. New germ tube induction medium for the identification of Candida albicans. J Clin Microbiol. 1985;22:861–862. doi: 10.1128/jcm.22.5.861-862.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley H R, Van uden N. The identification of Candida albicans within two hours by the use of an egg white slide preparation. Sabouraudia. 1963;2:205–208. [Google Scholar]
- 6.Chattaway F W, O’Reilly J, Barlow A J, Aldersley T. Induction of the mycelial form of Candida albicans by hydrolysates of peptides from seminal plasma. J Gen Microbiol. 1976;96:317–322. doi: 10.1099/00221287-96-2-317. [DOI] [PubMed] [Google Scholar]
- 7.Chattaway F W, Wheeler P R, O’Reilly J. Purification and properties of peptides which induce germination of blastospores of Candida albicans. J Gen Microbiol. 1980;120:431–437. doi: 10.1099/00221287-120-2-431. [DOI] [PubMed] [Google Scholar]
- 8.Dabrowa N, Taxer S S, Howard D H. Germination of Candida albicans induced by proline. Infect Immun. 1976;13:830–835. doi: 10.1128/iai.13.3.830-835.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durrenberger F, Wong K, Kronstad J W. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc Natl Acad Sci USA. 1998;95:5684–5689. doi: 10.1073/pnas.95.10.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geber A, Williamson P R, Rex J H, Sweeney E C, Bennett J E. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol. 1992;174:6992–6996. doi: 10.1128/jb.174.21.6992-6996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 13.Gold S, Duncan G, Barrett K, Kronstad J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994;8:2805–2816. doi: 10.1101/gad.8.23.2805. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko T, Shima H, Esumi H, Ochiai M, Nagase S, Sugimura T, Nagao M. Marked increases of two kinds of two-exon-skipped albumin mRNAs with aging and their further increase by treatment with 3′-methyl-4-dimethylaminoazobenzene in Nagase analbuminemic rats. Proc Natl Acad Sci USA. 1991;88:2707–2711. doi: 10.1073/pnas.88.7.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- 16.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubler E, Mosch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 18.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. . (Erratum, 267:5194, 1995.) [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 21.Lo H J, Kohler J R, Di Domenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 24.Madhani H D, Fink G R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell T K, Dean R A. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7:1869–1878. doi: 10.1105/tpc.7.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosch H U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogletree F F, Abdelal A T, Ahearn D G. Germ-tube formation by atypical strains of Candida albicans. Antonie Leeuwenhoek. 1978;44:15–24. doi: 10.1007/BF00400073. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds R, Braude A I. The filament inducing property of blood for Candida albicans: its nature and significance. Clin Res Proc. 1956;4:40. [Google Scholar]
- 29.Robertson L S, Fink G R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rupp S, Summers E, Lo H J, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalaby F, Shafritz D A. Exon skipping during splicing of albumin mRNA precursors in Nagase analbuminemic rats. Proc Natl Acad Sci USA. 1990;87:2652–2656. doi: 10.1073/pnas.87.7.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman F, Fink G R, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 33.Som T, Kolaparthi V S. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol Cell Biol. 1994;14:5333–5348. doi: 10.1128/mcb.14.8.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J R, Hamer J E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]