Summary
Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine encephalitis virus-like particles (VLPs), a regimen that protects against aerosol challenge with all three viruses. Single- and triple-virus specific antibodies were isolated, and we identified 21 unique binding groups. Cryo-EM structures revealed that broad VLP binding inversely correlated with sequence and conformational variability. One triple-specific antibody, SKT05, bound proximal to the fusion peptide and neutralized all three Env-pseudotyped encephalitic alphaviruses by using different symmetry elements for recognition across VLPs. Neutralization in other assays (e.g., chimeric Sindbis virus) yielded variable results. SKT05 bound backbone atoms of sequence-diverse residues, enabling broad recognition despite sequence variability; accordingly, SKT05 protected mice against Venezuelan equine encephalitis virus, chikungunya virus, and Ross River virus challenges. Thus, a single vaccine-elicited antibody can protect in vivo against a broad range of alphaviruses.
Keywords: alphavirus, broadly neutralizing antibody, cryo-EM, in vivo challenge, vaccine
In Brief:
Vaccine-elicited antibodies in non-human primates show a wide range of specificities against encephalitic and arthritogenic alphaviruses, providing insights into both antiviral immunity and vaccination strategy for alphavirus-driven diseases.
Graphical Abstract
INTRODUCTION
Alphaviruses are enveloped, positive-sense, single-stranded RNA viruses in the family Togaviridae with substantial impact on animal and human health. Transmitted by mosquitos, alphavirus infection in humans result in a spectrum of disease that can be categorized as either arthritogenic or encephalitic. Arthritogenic alphaviruses, such as chikungunya virus (CHIKV), Ross River virus (RRV), Mayaro virus (MAYV), or o’nyong’nyong virus (ONNV), most often cause acute and chronic musculoskeletal disease. While rarely fatal, symptoms include fever, rash, and myalgia; incapacitating arthralgia and myalgia can persist for months to years after infection.1–3 In contrast, infection with encephalitic alphaviruses, including western, eastern, or Venezuelan equine encephalitis virus (W/E/V/EEV), can vary from asymptomatic or acute febrile illness to severe neurological impairment and death.4,5 Eastern EEV (EEEV) is the most virulent, with a human case fatality rate estimated at 30–75% in symptomatic individuals with neuro-invasive disease.4,6,7 While Venezuelan EEV (VEEV) is the least lethal, with death occurring in <1% of cases, it may be the most important in terms of morbidity.8–11 Unlike WEEV and EEEV, for which humans represent dead-end hosts, outbreaks of VEEV can become self-sustaining as virus can replicate to levels sufficient for domestic/urban transmission between humans.5,12 Moreover, most infections with VEEV are apparent and present abruptly with debilitating symptoms like those caused by arthritogenic alphaviruses.7,11,13 Though outbreaks of alphavirus infection are often geographically isolated by vector and host reservoir availability, social and environmental factors leading to the spread of mosquito vectors into new geographic areas is cause for concern.14,15
The alphavirus genome encodes four non-structural proteins (nsP1, nsP2, nsP3, and nsP4) and six structural proteins (capsid, E3, E2, 6K, TF, and E1).16 On the surface of a mature virion, heterodimers of the envelope proteins, E1 and E2, assemble into 80 trimeric spikes that are arranged with T = 4 icosahedral symmetry.17–19 At the apex of the trimer host receptors engage clefts formed by adjacent E1/E2 heterodimers, with contacts that usually involved both E1 and E2, while viral entry occurs via the hydrophobic fusion loops located within the E1 protein.16,20–22 While the fusion loop is theoretically an appealing target for broadly reactive α-EEV monoclonal antibodies (mAbs), it is occluded by the E2 protein and thought to be primarily accessible following a conformation change when exposed to low pH within endosomes.23–25 Several studies highlight the importance of antibody-mediated protection against alphavirus infection, although only recently have antibodies been identified with breadth against multiple alphaviruses.26–35 These antibodies, from convalescent infected subjects, targeted a conserved, cryptic E1 epitope proximal to and within the fusion peptide and were shown to protect mice against multiple alphaviruses despite poor in vitro neutralization activity.24,26 While these antibodies may prove important as prophylactic or post-exposure therapies, a vaccine candidate that elicits such antibodies remains to be demonstrated.
Multiple vaccine and therapeutic candidates have been explored to combat alphavirus infections, yet none have been approved for public use in the United States.36–43 All three EEVs, and potentially CHIKV, can be transmitted via aerosol exposure.11,44 Accordingly, the EEVs are considered NIAID Category B priority pathogens with EEEV and VEEV being USDA/CDC Select Agents due to their potential as bioterrorism agents.45,46 Investigational vaccines against each EEV have been used by the U.S. military since the 1970s for at-risk laboratory and military personnel, but have resulted in poor immunogenicity, limited longevity, and substantial side effects.42,47,48 Previously, we showed that a trivalent vaccine consisting of WEEV, EEEV, and VEEV (WEVEEV) virus-like particles (VLPs) demonstrated good immunogenicity in nonhuman primates (NHP) and protected them against aerosol challenge with all three EEVs.37 The mechanism of protection was likely humoral, as sera from immunized NHP protected mice against lethal challenge; however, it is not known if protection arose from multiple single-specific antibodies, or one or more multi-specific antibodies.
Here, we demonstrate an approach for the identification of EEV-specific B cells in vaccinated NHPs, in which the same VLPs used for immunization were also individually labeled with distinct fluorophores and used as probes. The trivalent vaccine elicited a wide range of specificities, including antibodies that target all three EEVs. We mapped mAbs to the surface of the encephalitic alphaviruses and determined their neutralization capacities. We defined one group of triple-specific mAbs that both bound and neutralized all three EEV pseudoviruses and showed that two of these triple-specific mAbs, SKT05 and SKT20, protect mice in vivo from lethal challenge with VEEV. With cryo-EM, we identified multiple binding epitopes on the surface of the WEVEEV VLPs and highlighted the structural components underlying the breadth observed by triple-specific mAbs. We demonstrated that the breadth of triple-specific mAbs extends to arthritogenic alphaviruses, as they bound to the surface of infected cells, and that SKT05 protected mice in vivo from challenge with CHIKV and RRV. Finally, we humanized SKT05 and showed equivalent antiviral activity, providing a proof of principle for using vaccinated NHP to generate reagents on a clinical development path.
RESULTS
Trivalent VLP immunization elicits single-specific and triple-specific α-EEV mAbs in NHP
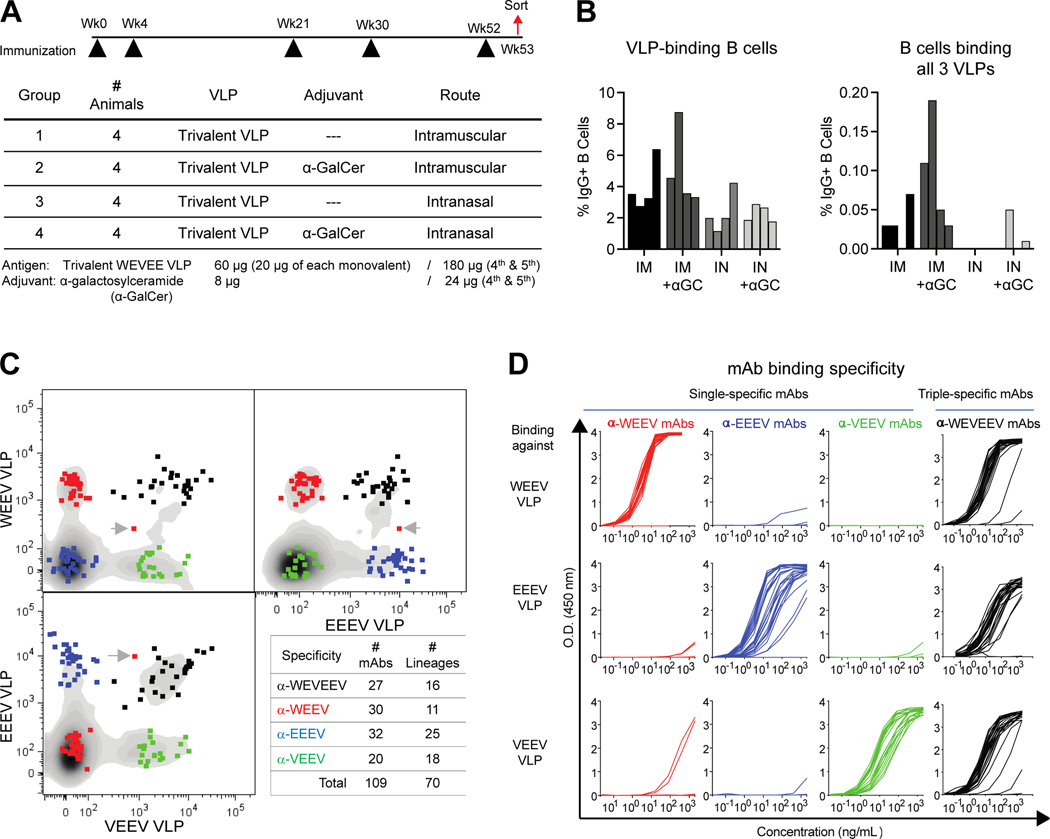
Cynomolgus macaques (n=16) received a vaccine composed of WEEV, EEEV, and VEEV VLPs. Immunizations were administered intramuscularly (IM) or intranasally (IN) five times over 52 weeks, with or without alpha-galactosylceramide (α-GalCer) as an adjuvant (Figure 1A). Systemic and mucosal immune responses were evaluated in serum and bronchoalveolar lavage (BAL) fluid, respectively. After one month, IM immunization induced higher systemic binding IgG responses against all EEVs compared to IN immunization, while addition of α-GalCer adjuvant only generated increased systemic IgG responses in IN-vaccinated animals (Figure S1A). IgG levels in BAL fluid were low, and contamination with small amounts of blood during sampling greatly affected the quantitation of bona fide mucosal antibody responses. Since IgA-dimers do not exist in blood, we quantitated total secretory IgA dimer levels in BAL to assess mucosal humoral immune responses (Figure S1B). There was a trend that IN immunization with adjuvant generated greater IgA dimer response than IM immunization, but responses were variable.
Figure 1. Trivalent VLP immunization elicits single-specific and triple-specific α-EEV mAbs in NHP.
(A) Cynomolgus macaque immunization regimen. (B) Percent of CD19+IgG+ B cells binding at least one VLP probe (left), and those binding all three VLP probes simultaneously (right). (C) Single B cells positive for binding only a single VLP (WEEV: red; EEEV: blue; VEEV: green) or all three VLPs (WEVEEV: black) were sorted into wells. A gray arrow denotes a doublet sort, comprising two B cells in one well (only one sequence was generated). Lineages were defined as having a unique V-gene allele, CDR3 sequence, and CDR3 length. (D) ELISA binding curves of single-specific and triple-specific ⍺-EEV mAbs to WEEV VLPs (top), EEEV VLPs (middle), and VEEV VLPs (bottom). Data are representative of two to three independent experiments. See also Figure S1 and Tables S1–S2.
One week after the 5th immunization, we isolated PBMC and evaluated binding to individual EEV VLPs, each labeled with distinct fluorescent lipophilic membrane staining dye (Figure S1C). These labeled VLPs functioned well as probes for antigen-specific B cell sorting (Figure 1B and 1C), and we identified B cells with surface IgG that putatively bound one or more VLPs. The animal with the greatest number of triple-specific B cells was selected for EEV-specific single B cell sorting, and heavy and light chain immunoglobulins were sequenced and synthesized. In total, we expressed 109 α-EEV mAbs comprising 70 lineages, with a range of single- and triple-specific binding (Figure 1C, Table S1).
Our sorting strategy was confirmed by testing the 109 α-EEV mAbs for binding to each VLP by ELISA (Figure 1D and Table S2). For the α-WEEV mAbs, which were selected positively for binding the WEEV VLP and negatively for not binding EEEV and VEEV VLPs, all (30/30) bound the WEEV VLP, and only two showed very low-level cross-reactivity for binding to VEEV VLP. Similarly, all α-VEEV mAbs (20/20) bound only the VEEV VLP with no cross-reactivity, and 27/32 α-EEEV mAbs bound only the EEEV VLP while 5/32 failed to bind any VLP. For putatively triple-specific mAbs (α-WEVEEV), 26/27 bound all three VLPs.
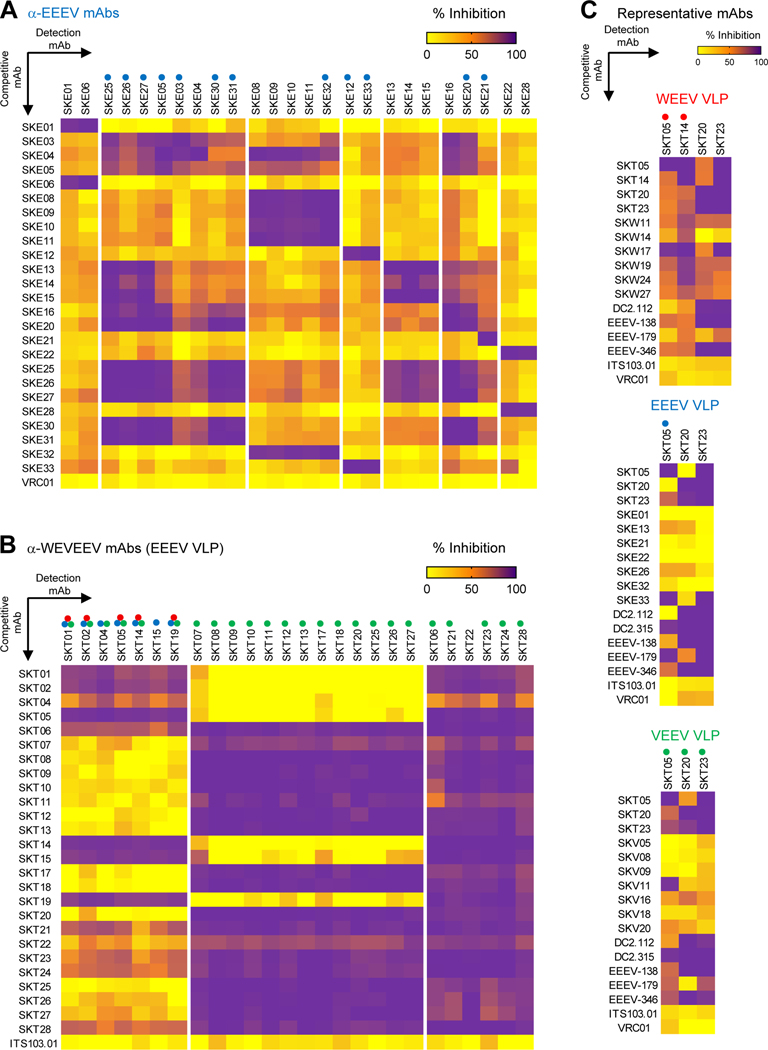
Competition ELISAs identify distinct binding groups of single-specific and triple-specific α-EEV mAbs
To estimate the number of major antigenic sites recognized by single- and triple-specific α-EEV mAbs, we performed competitive-binding ELISAs with each VLP (Figure 2 and Figure S2). For single-specific mAbs, hierarchical clustering identified seven competition groups for α-EEEV mAbs (Figure 2A), six for α-WEEV mAbs (Figure S2A), and seven for α-VEEV (Figure S2B). For triple-specific mAbs (α-WEVEEV), hierarchical clustering identified three competition groups for the EEEV VLP (Figure 2B) and VEEV VLP (Figure S2D) and four for the WEEV VLP (Figure S2C). One mAb was chosen as a representative from each competition group of single-specific and triple-specific mAbs. We tested single-specific mAbs, as well as previously published broadly reactive α-EEV mAbs (DC2.112, DC2.315, EEEV-138, EEEV-179, and EEEV-346) as competitors against the representative triple-specific mAbs for each VLP (Figure 2C).24,26 One group from each of the single-specific mAbs was found to block at least one triple-specific mAb. Thus, while the latter represent broadly reactive epitopes, there are also strain-specific mAbs against those epitopes. We show that most of the previously published broadly reactive α-EEV mAbs fall into one of two triple-specific competition groups: those represented by SKT20 or SKT23. Only EEEV-179 appears to share a competition group with SKT05 when tested on the EEEV but not WEEV VLP. In total, our mAbs map to 21 regions on the surfaces of WEVEE VLPs.
Figure 2. Competition ELISAs identify distinct binding groups of single-specific and triple-specific α-EEV mAbs.
Competitive binding ELISAs were performed to identify overlap in EEEV VLP surface binding areas within (A) ⍺-EEEV mAbs and (B) ⍺-WEVEEV mAbs. Data were hierarchically clustered to determine competition groups for each VLP and are representative of at least two independent experiments (C) Representative single-specific mAbs, as well as previously published broadly reactive ⍺-EEV mAbs (DC2.112, DC2.315, EEEV-138, EEEV-179, and EEEV-346),24,26 were competed against representative triple-specific mAbs. Heat maps display percent inhibition ranging from yellow (minimal competition) to orange (moderate competition) to purple (maximal competition). Negative control mAbs include either the human ⍺-HIV mAb VRC0160 or the NHP ⍺-SIV mAb ITS103.01.62 Colored circles above mAbs along x-axis represent the EEV pseudovirus that was neutralized (red: WEEV; blue: EEEV; green: VEEV). Data are representative of two independent experiments. See also Figures S2–S3, and Table S2.
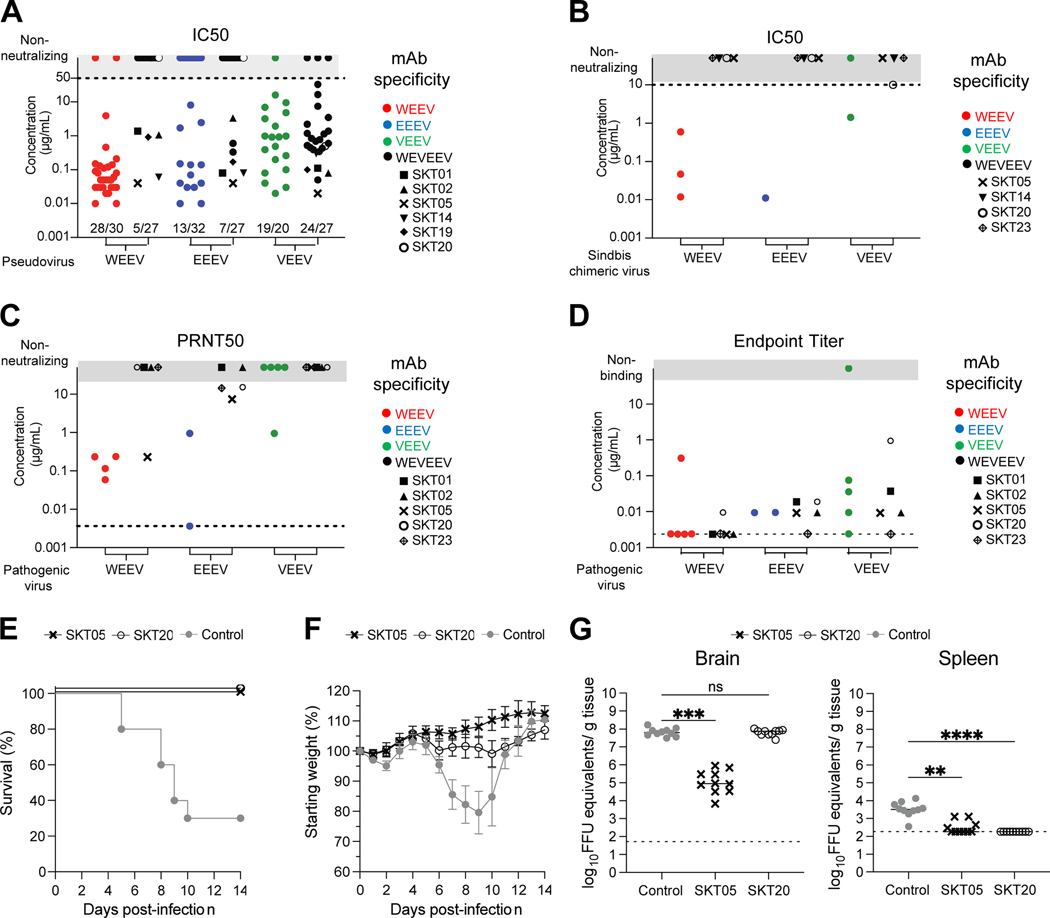
Vaccine-elicited α-EEV mAbs bind, neutralize, and protect against encephalitic alphavirus challenge in vivo
To evaluate the neutralization activity of single-specific and triple-specific α-EEV mAbs we used Env-pseudotyped lentiviral reporter viruses for WEEV, EEEV, and VEEV (Figure 3A and Table S2). For α-WEEV mAbs, 27 reached a half maximal inhibitory concentration (IC50) of less than 1 μg/mL against the WEEV pseudovirus; of those, 13 were particularly potent with 80% inhibitory concentrations (IC80) below 1 μg/mL. Fewer α-EEEV mAbs were neutralizing, with 4 reaching IC80 values below 1 μg/mL. 19/20 α-VEEV mAbs neutralized VEEV pseudovirus, with varying potency. Six α-VEEV mAbs neutralized with IC80 below 1 μg/mL. We also tested the triple-specific mAbs for neutralization of each EEV pseudovirus. Similar to the α-VEEV mAbs, nearly all (24/27) triple-specific mAbs neutralized VEEV pseudovirus with varying potency. While neutralization of WEEV and EEEV pseudovirus was less common, those that did neutralize at least one (i.e., SKT01, SKT02, SKT04, SKT05, SKT14, SKT15, and SKT19) did so with IC50 titers that ranged from 40 ng/mL to 3.3 μg/mL. Of these, we identified 5 (SKT01, SKT02, SKT05, SKT14, and SKT19) that neutralized all three EEV pseudoviruses. One triple-specific mAb (SKT05) exhibited exceptional potency against all three EEV pseudoviruses, with IC80 values of 70 ng/mL (WEEV), 390 ng/mL (EEEV), and 60 ng/mL (VEEV).
Figure 3. Vaccine-elicited ⍺-EEV mAbs bind, neutralize, and protect against encephalitic alphavirus challenge in vivo.
(A) Neutralization IC50 values for single- and triple-specific ⍺-EEV mAbs against WEEV, EEEV, and VEEV Env-pseudotyped lentiviral reporter viruses. Fractions above x-axis indicate number of neutralizing mAbs out of total tested. Data are representative of at least two independent experiments performed in triplicate. (B) IC50 values for select ⍺-EEV mAbs against SINV-chimeric viruses. Data are representative of two independent experiments. (C) PRNT50 values for select ⍺-EEV mAbs against pathogenic WEEV (Fleming strain), EEEV (FL93–939 strain), and VEEV (TrD strain). Data are shown from one experiment after determining the appropriate starting dilution. (D) ELISA endpoint binding titers for select single-specific and triple-specific ⍺-EEV mAbs against WEEV (CBA87 strain), EEEV (FL93–939 strain), and VEEV (TrD strain). Data are shown from one experiment after determining the appropriate starting dilution. In the event an endpoint titer was not identified, results are reported as half of the lowest binding titer tested and are indicated along the dotted line. (E-G) VEEV (TC-83) challenge outcome in mice (n=10/group) that received SKT05, SKT20, or a NHP ⍺-SIV mAb as a control one day prior to inoculation. Data are representative of two independent experiments. (E) Survival rate analysis and (F) change in relative weight in mice for 14 days after inoculation. (G) Viral load was determined in the brain and spleen 5 days after inoculation. Statistical significance related to viral RNA was determined by Kruskal-Wallis test (**p< 0.0021, ***p< 0.0002, ****p< 0.0001). The dotted line indicates the limit of detection for viral RNA analysis. Colors represent mAb specificity while select triple-specific mAbs are shown as unique black symbols. Only select triple-specific mAbs were assessed in B-D, all of which had unique black symbols. See also Figure S2 and Table S2.
We evaluated the neutralization activity of select mAbs using Sindbis (SINV) chimeric viruses that express the nonstructural proteins of SINV and the structural proteins of WEEV, EEEV, or VEEV (Figure 3B). 5/6 single-specific mAbs neutralized with IC50 values that ranged from 10 ng/mL to 1.4 μg/mL. In contrast, of the triple-specific mAbs, only SKT20 weakly neutralized SINV-VEEV, while SKT05, SKT14, and SKT23 did not neutralize any chimeric virus. We also evaluated the neutralization activity of select mAbs against pathogenic WEEV, EEEV, and VEEV (Figure 3C). Some single-specific mAb neutralized the cognate viruses VEEV (strain TrD), WEEV (strain Fleming) and EEEV (strain FL93–939) potently. Neutralization activity was weak by the triple-specific mAbs, although SKT05 did display reduced plaque sizes that could reflect antiviral activity. Of note, both SKT20 and SKT23 neutralized pathogenic EEEV despite not neutralizing EEEV pseudovirus. The variability of neutralization activity in distinct in vitro assays has been published.
Thus, in addition to testing neutralization, we also tested select mAbs for binding to pathogenic WEEV, EEEV, and VEEV (Figure 2D). When tested against WEEV (strain CBA87), α-WEEV and α-WEVEEV mAbs were particularly potent with most endpoint titers <10 ng/mL. Endpoint titers for α-EEEV and α-WEVEEV mAbs binding to EEEV (strain FL93–939) were ~10 ng/mL. We observed the largest variability in binding to pathogenic VEEV (strain TrD); 4/5 α-VEEV mAbs bound with endpoint titers ranging from <10 ng/mL to 100 ng/mL, while α-WEVEEV mAbs ranged from <10 ng/mL to 1 μg/mL. Overall, these are exceptionally potent binding mAbs with a range of epitope and strain specificities.
We selected two triple-specific mAbs from separate competition groups to determine whether protective effects could be conferred in vivo following encephalitic alphavirus challenge. We administered SKT05 or SKT20 to six-week-old C3H-HeN mice one day prior to intranasal inoculation with 107 FFU of VEEV TC-83 (approximately 1 LD50). The animals were followed for 14 days post-infection (dpi). SKT05 or SKT20 treatment resulted in 100% survival following lethal challenge (Figure 3E). Control mAb-treated mice exhibited substantial weight loss, as expected in this challenge model; weights of SKT20-treated mice remained relatively unchanged over the same time, while weights increased normally in SKT05-treated mice (Figure 3F). An additional set of mice were sacrificed at 5 dpi and viral load was assessed in the brain and spleen. Mice administered SKT05, but not SKT20, had significantly reduced viral titers in the brain compared to a control mAb (Figure 3G, left). In the spleen, both SKT05 and SKT20 administration prior to challenge resulted in significantly reduced viral titers compared to a control mAb. Thus, SKT05 and, to a lesser extent SKT20, provide protective benefits against encephalitic alphavirus challenge when administered prophylactically. These results also confirm the limited utility of in vitro assays as predictors of in vivo protection.
Broadly neutralizing α-EEV mAbs are best elicited following three immunizations
To better understand the impact of immunization timing on elicitation of broadly neutralizing α-EEV mAbs, we performed next generation sequencing (NGS) on PBMC isolated at various timepoints after the first four immunizations of the animal from which we isolated broadly neutralizing antibodies. In total, 31 out of 70 mAb lineages were detected with heavy chain NGS. After the first two immunizations, we detected at most only one new lineage of each specificity. In contrast, we detected the greatest number of new lineages appearing at week 23, two weeks after the third immunization (Figure S3A); a similar trend was observed when we assessed the total number of transcripts from all lineages of each specificity, normalized to the total number of transcripts at each time point (Figure S3B). The lineage representing three of our five broadly neutralizing triple-specific mAbs (SKT01, SKT02, SKT05) was first detected between week 23 and 32. Interestingly, at week 21 we first detected a lineage that was shared by one triple-specific mAb (SKT19) and one α-EEEV mAb (SKE12), suggesting that ongoing somatic hypermutation might guide the immune response to greater breadth.
Consistent with these NHP data, we did not detect triple-specific α-EEV mAbs capable of neutralizing all three EEV pseudoviruses from two subjects enrolled in a human phase 1 clinical trial evaluating the safety and immunogenicity of trivalent WEVEEV VLP immunization;49 these subjects received only two doses of VLPs. Two subjects were chosen for EEV-specific single B cell sorting based on neutralizing antibody titers against pathogenic WEEV, EEEV, and VEEV from sera collected two weeks after the final immunization. Both subjects were administered 60 μg of WEVEEV VLPs over eight weeks (Figure S3C). In total, we identified only six triple-specific B cells from both participants and expressed the mAbs from these cells. Four of them bound all three VLPs (hSKT1001, hSKT1002, hSKT1004, hSKT1006), while hSKT1003 bound only VEEV VLP; and hSKT1005 did not bind any VLP (Figure S3D, left).
We tested human triple-specific mAbs for EEV pseudovirus neutralization (Figure S3D, right). Despite positive binding to all three VLPs, none neutralized WEEV or EEEV pseudovirus. Instead, these four mAbs, as well as the one that only bound the VEEV VLP, neutralized only VEEV pseudovirus. We performed competitive-binding ELISAs with each human triple-specific mAb as competitors against representative NHP mAbs (Figure S3E). As predicted from the neutralization profiles, most of the human α-WEVEEV mAbs were in the SKT20 competition group that neutralized only VEEV pseudovirus. However, when they were tested for binding to the surface of live cells infected with SINV-WEEV, SINV-EEEV, or SINV-VEEV, many bound at comparable levels as SKT05 (Figure S3F). Accordingly, these data along with the NHP NGS data suggest that three or more immunizations may be required to elicit high levels of SKT05-like broadly neutralizing α-EEV mAbs, though protective effects conferred by fewer immunizations may still be a possibility in the absence of broad neutralization.
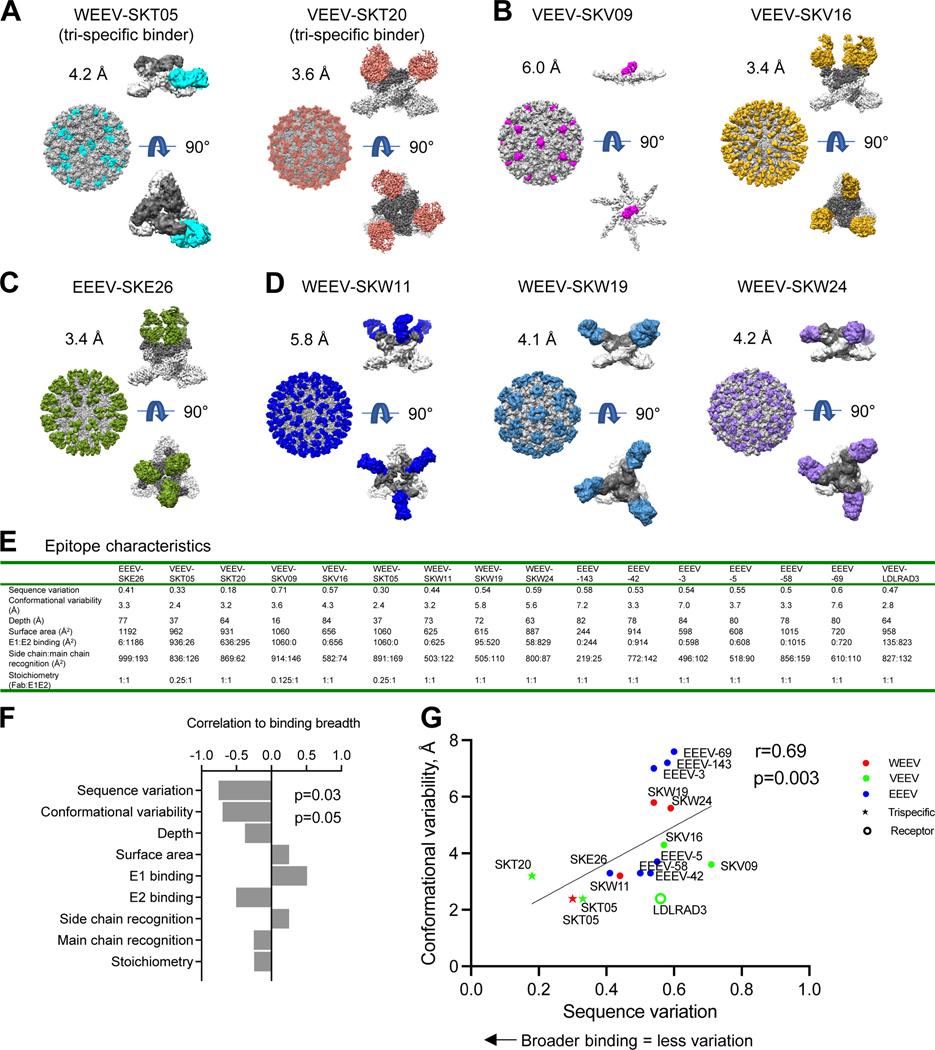
Cryo-EM structures of neutralizing antibodies with VLPs reveal that sequence variation and conformation variability inversely correlate with broad recognition
To gain insight into the broad recognition and protective activity by SKT05 and SKT20, we determined cryo-EM structures of antibody Fab-VLP complexes. While structures of ligand-free VEEV and EEEV VLPs have been determined at resolutions of 3.5 Å and 4.2 Å, respectively,22,27 the ligand-free structure of WEEV VLP had not been defined. We collected single-particle cryo-EM data on a Titan Krios and determined the structure of ligand-free WEEV VLP at 3.4 Å resolution (Figure S4A, Table S3, and Data S1). Like other alphaviruses,19,50 the WEEV VLP comprised a T=4 icosahedron, with 240 copies of each structural protein assembled into 80 trimers. The extracted E1E2 trimer showed moderate structural variability, with an average backbone root-mean-square deviation (RMSD) between WEEV and VEEV of 2.1 Å and between WEEV and EEEV of 2.6 Å (Figure S4B). We calculated sequence variation as a buried surface area (BSA)-weighted average of normalized Shannon entropy, based on WEEV, VEEV, EEEV, CHIKV, RRV, MAYV, and ONNV sequences (Figure S4C). Overall, sequence variation tended to be greater with E2 and on the outer surface of the VLP – and to correlate with conformational variability. The fusion peptide region was highly conserved, though structurally variable (Figure S4D).
There were too many mAbs to characterize Fab-VLP complexes for all representative mAbs. To create an appropriate matrix of information by which to understand broad binding, we chose a subset of antibodies for structural analysis, including two triple-specific antibodies, SKT05 and SKT20, and supplemented these with single-specific antibodies, each from a different competition group and against a different target VLP (Figure 4A–4D, Table S3, and Data S1).
Figure 4. Cryo-EM structures of neutralizing antibodies with VLPs reveal that sequence variation and conformation variability inversely correlate with broad recognition.
(A-D) Cryo-EM structures of select (A) triple-specific antibodies, (B) α-VEEV antibodies, (C) α-EEEV antibodies, or (D) α-WEEV antibodies. Each antibody neutralized at least one pseudovirus. For each antibody-VLP complex, the entire complex is shown with VLP, with VLP in gray and Fabs colored. The E1E2 spike with bound Fab is also shown E1 in light gray, E2 in dark gray, and Fab colored; a side view is shown at top, and under it, a 90° rotation viewing down the spike molecular 3-fold axis (note that for VEEV-SKV09, this Fab binds directly to E1 at the 2-fold axis of the VLP, and this interaction is shown from side and along 2-fold axis). (E) Epitope characteristics are calculated for new antibodies and EEEV-143 (PDB 6xob), EEEV-42 (PDB 6mui), EEEV-3 (PDB 6mw9), EEEV-69 (PDB 6mwx), EEEV-58 (PDB 6mwv), EEEV-5 (PDB 6mwc), and LDLRAD3 (PDB 7ffn). (F) Bar graph displaying correlation of binding breadth and epitope properties. Sequence variation was calculated as a BSA-weighted average of normalized entropy. Conformational variability was calculated as a BSA-weighted average RMSD. (G) Epitope sequence and conformational variability plotted for alphavirus antibodies and receptor. Color identifies VLP with which the antibody complex structure was solved. See also Figures S4–S5 and Data S1.
With VEEV, we determined complex structures with the triple-specific mAb SKT20 as well as the VEEV-neutralizing single-specific mAbs SKV09 and SKV16. SKT20 recognized the conserved fusion peptide in domain II of the E1 protein (Figure 4A, Table S3, and Data S1). SKV09 bound E1 at the VLP pseudo-6-fold axis with a restricted stoichiometry of only 0.375 Fabs per trimer (Figure 4B, Table S3, and Data S1). By contrast, SKV16 and SKT20 both recognized the E1E2 trimer, with stoichiometries of three Fabs per trimer; SKV16 bound specifically to domain B of E2 (Figure 4B, Table S3, and Data S1). With EEEV, we determined the cryo-EM structure at 3.5 Å resolution for the antigen-binding fragment of the EEEV-neutralizing mAb SKE26, which bound primarily to the A domain of the E2 protein (Figure 4C, Table S3, and Data S1).
With WEEV, we determined complex structure with WEEV-neutralizing single-specific mAbs SKW11, SKW19 and SKW24, as well as the triple-specific mAb SKT05. SKT05 (Figure 4A, Table S3, and Data S1) showed almost lateral recognition, binding domain II of the E1 protein, proximal to the fusion peptide. Another notable feature of SKT05 was its restricted stoichiometry, with only a single Fab binding per E1E2 trimer. We developed a refinement algorithm, selecting and grouping only the appropriate, Fab-bound E1E2-protomers in the reconstruction density, which improved resolution from 5.7 Å to 4.2 Å. All three single-specific mAbs bound the E2 protein. SKW11 primarily bound the β-ribbon of the E2 protein with a ~45-degree approach angle (Figure 4D, Table S3, and Data S1). Both SKW19 and SKW24 primarily bound the B domain of the E2 protein, with SKW19 (Figure 4D, Table S3, and Data S1) binding at a more equatorial approach angle compared to SKW24 (Figure 4D, Table S3, and Data S1) that was lateral in its binding angle. All three recognized trimers with a stoichiometry of three Fabs per E1E2 trimer.
To provide insight into the relationship between recognized epitope and neutralization breadth, we assessed epitope characteristics (Figure 4E and Table S4) and correlated these quantitatively with binding breadth. Recognition of E1 positively correlated with binding breadth, whereas recognition of E2 inversely correlated, although these correlations did not reach statistical significance. Other epitope characteristics such as depth, surface area, side-chain recognition, and main-chain recognition showed less significant associations. However, two epitope characteristics showed significant correlation with binding breadth: sequence variation and conformational variability (Figure 4F and Figure S5). Analysis of all α-EEV antibody-VLP structures in the PDB for structural and sequence variation revealed high correlation between these two epitope features (R=0.69, P=0.03). For this analysis, we included not only the WEEV-SKT05 structure at 4.2 Å resolution (Figure 4A and Table S3), but also added the structure of VEEV-SKT05, at 3.5 Å resolution (Table S3, and Data S1). Independent of the virus VLP used for structure determination, the epitopes for two triple-specific binders, SKT05 and SKT20, showed the lowest overall sequence and conformational variation (Figure 4G). Overall, the collection of structures of neutralizing single-specific and triple-specific antibodies showed broad recognition to correlate inversely with two epitope features: sequence variation and conformational variability.
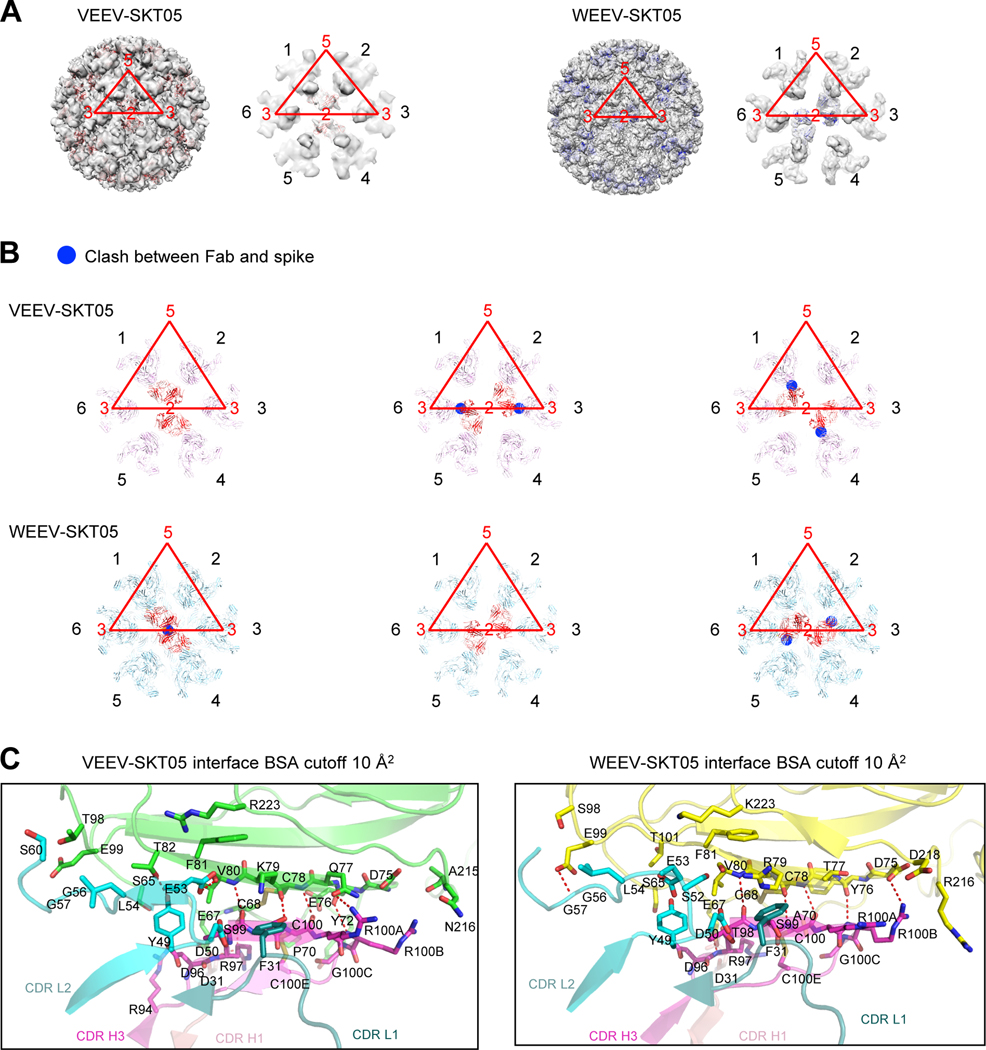
SKT05 recognizes different symmetry-related protomers in VEEV and WEEV VLPs
In addition to differences in overall resolution, the structures of SKT05 with WEEV (4.2 Å) and with VEEV (3.5 Å) revealed SKT05 to recognize different symmetry-defined protomers in VEEV and WEEV VLPs (Figures 5A and 5B). Two SKT05 Fabs bound at the central two-fold axis of the VLP, where three binding orientations were possible. In both VEEV and WEEV, only one of the three potential binding orientations was used, and it was different for the two VLPs. In both cases, modeling indicated SKT05 to clash at the two orientations that were not used for each VLP (Figure 5B).
Figure 5. SKT05 utilizes different symmetry to bind VEEV and WEEV.
(A) Overall view of the reconstruction density for VEEV and WEEV VLPs with bound SKT05 Fab and close-up view of icosahedral 2-fold axis surrounded with six E1:E2 spikes (labeled 1–6 in black font, with VLP symmetry axes labeled in red font, and the icosahedral asymmetric unit also shown in red). SKT05 Fabs are shown as red and blue ribbons, respectively. SKT05 bound to spikes labeled 1 and 4 in VEEV and to spikes labeled 2 and 5 in WEEV. (B) VEEV-SKT05 and WEEV-SKT05 complexes docked into spike reconstruction density surrounding 2-fold axis. Clashes are highlighted with blue circles and were observed between SKT05 and nearest polypeptide chains when VEEV-SKT05 was docked to spike pairs 2,5 and 6,3, and when WEEV-SKT05 was docked to spike pairs 1,4 and 6,3. (C) Details of interactions between VEEV and WEEV VLPs with SKT05 are shown in ribbons, with all interactions with a BSA larger than 10 Å2 shown in sticks. Left panel: interactions of VEEV E1 (green) with CDR-H1, CDR-H3, CDR-L1 and CDR-L2 of SKT05. Right panel: interactions of WEEV E1 (yellow) with CDR-H1, CDR-H3, CDR-L1 and CDR-L2 of SKT05. See also Table S3 and Data S1.
Comparison of the recognition by SKT05 in VEEV and WEEV revealed changes in orientations of side chains, often related to differences in the sequences of the epitopes on VEEV and WEEV (Figure 5C, Table S5). Overall, the two structures were highly similar, with only 0.8 Å RMSD in epitope and 0.6 Å RMSD in paratope. The differences in SKT05 recognition of VEEV and WEEV protomers likely explains difference in competition between SKT05 and other antibodies, which differed depending on the VLP analyzed. For example, the triple-specific mAbs that neutralized all three pseudoviruses all shared the same competition group for the EEEV and VEEV VLP (Figures 2B and S2D), yet SKT05 was not blocked by these mAbs when competition was assessed with the WEEV VLP (Figure S2C). Moreover, mAbs in the SKT23 competition group identified with the VEEV VLP blocked all other mAbs from binding, while this was not the case with the WEEV VLP. Of note, SKT20 clustered into the same competition group regardless of VLP. Collectively these data demonstrate that SKT05 recognizes symmetry-related protomers of VEEV and WEEV differently, and that this difference enables antibodies that bind to the same region on the E1E2 trimer to not compete in the context of the full VLP.
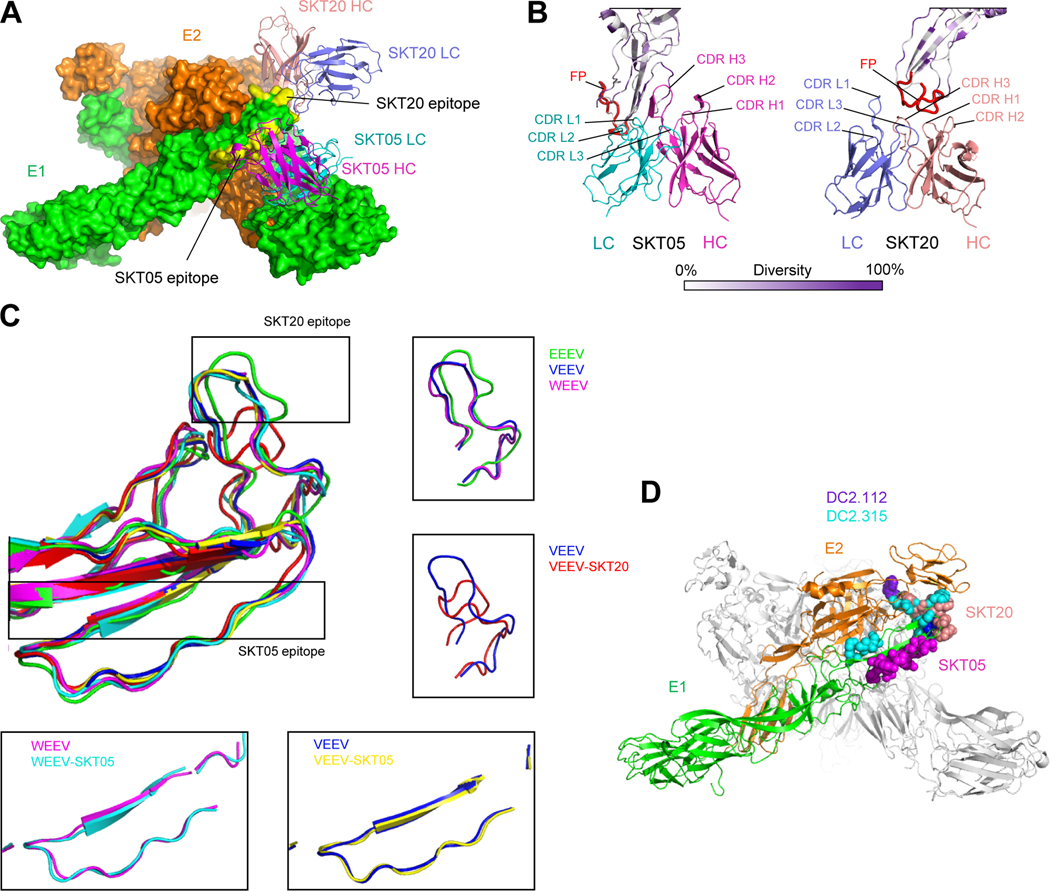
Mechanisms enabling broad recognition by SKT05 and SKT20
Triple-specific mAbs fell into 3–4 competition groups: for EEEV and VEEV, those that competed with SKT05, those that competed with SKT20, and those that competed with both (Figures 2B and S2D); for WEEV, SKT05 was in a competition group of its own (Figure S2C). Thus, we would expect all triple-specific mAbs to recognize regions defined by these two antibodies. Consistent with the competition data, SKT05 and SKT20 bound epitopes that were proximal, but not overlapping (Figure 6A). SKT05 used complementarity-determining regions (CDRs) L1, L2 and H3 regions to bind an epitope immediately N terminal to the fusion peptide, whereas SKT20 utilized all CDRs except L2 to bind directly to the fusion peptide (Figure 6B).
Figure 6. Structural details of SKT05 and SKT20 broad recognition.
(A) Overall view of the WEEV spike bound by SKT05 and SKT20. Molecular surfaces are shown for the E1:E2 trimer (colored green and orange, respectively), with SKT05 and SKT20 variable domains displayed in ribbons and their epitopes highlighted in yellow. (B) Close-up view of SKT20 and SKT05 in contact with WEEV E1 glycoprotein drawn in ribbons and colored by sequence diversity according to the white-to-purple key, with two conformations of the fusion peptide highlighted in red. (C) Superposition of E1 glycoproteins for EEEV, VEEV, WEEV, and their SKT05 and SKT20-bound complexes. In free EEEV, VEEV, and WEEV VLPs, E1 glycoproteins have a similar conformation for the fusion peptides. Binding of SKT05 does not affect the conformation of E1, whereas binding of SKT20 results in a dramatic change in the conformation of the fusion peptide. (D) View of the WEEV spike, displayed in ribbons representation, with one E1:E2 protomer colored green and orange, respectively, and the two other protomers colored medium and light gray. Epitopes for SKT05, SKT20, DC2.112 and DC2.315 are shown in sphere representation, and colored magenta, orange, purple, and cyan, respectively. See also Data S1.
The SKT20 epitope was almost 100% conserved within EEVs, with sequence conservation of the epitope providing a clear mechanism for its broad recognition. The SKT05 epitope, in contrast, involved a mixture of conserved residues (mostly recognized by the light chain) and variable residue (mostly recognized by the heavy chain) (Figure 5C, Table S5). The variable residues recognized by the heavy chain, however, involved backbone contacts not dependent on sequence conservation. Thus, SKT05 utilized a more complex mechanism of binding, with recognition of conserved residues intermixed with recognition of backbone for variable residues.
We also examined the conformational change induced in the VLPs by SKT05 and SKT20 (Figure 6C). Some variation in the fusion loop was observed between EEEV, VEEV and WEEV (RMSD of 1.6 Å), but a much larger change in fusion loop conformation was induced by SKT20 (RMSD of 4.3 Å). By contrast, binding of SKT05 induced negligible backbone movements (RSMD of 0.5 Å).
Lastly, we examined how the SKT05 epitope compared with broadly reactive DC2.112 and DC2.315 antibodies.26 We mapped the location of residues identified by mutational analysis to impact binding of DC2.112 and DC2.315; residues impacting DC2.112 and DC2.315 antibody recognition overlapped with the SKT20 epitope, but not with the SKT05 epitope (Figure 6D). Thus, broad recognition by SKT05 and SKT20 appears to involve different mechanisms. For SKT05, broad recognition involved binding at a conformationally conserved but somewhat sequence-variable epitope, utilizing backbone recognition. For SKT20, broad recognition involved binding at a conformationally divergent but highly sequence-conserved epitope.
SKT05 recognizes and protects in vivo against arthritogenic alphaviruses
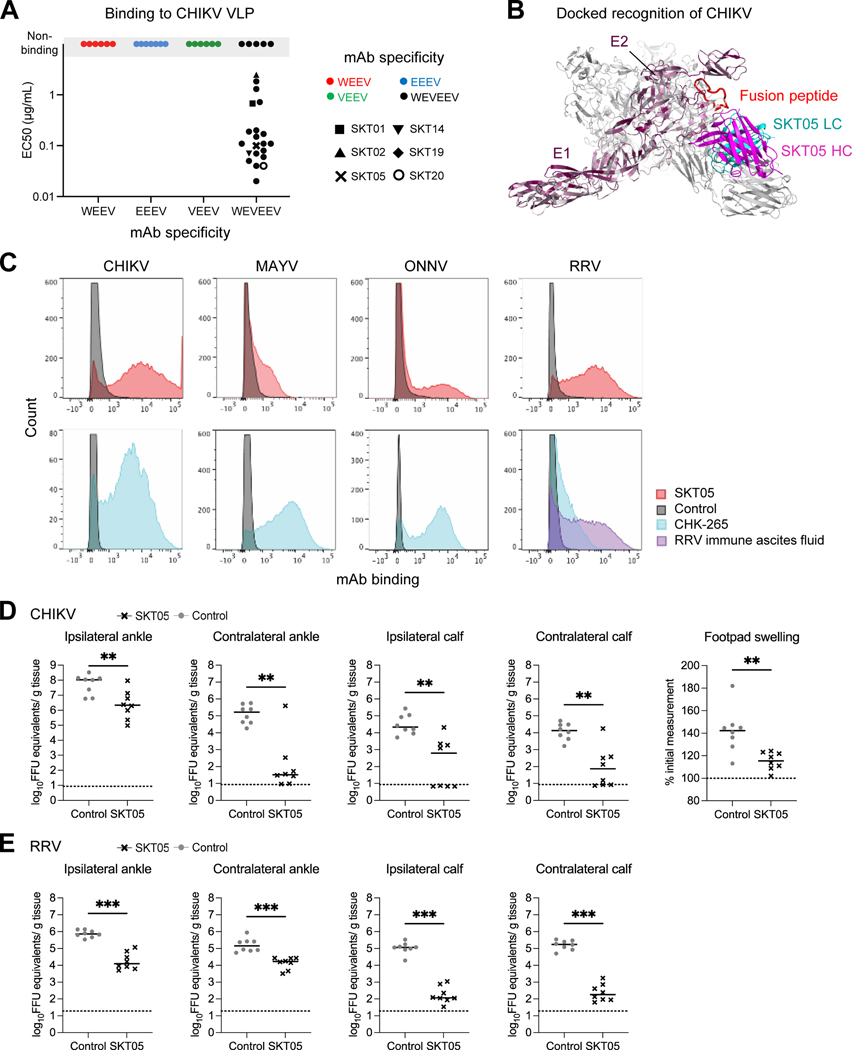
Due to the unique structural characteristics revealed by cryo-EM, we hypothesized that our triple-specific α-EEV mAbs might also bind additional alphaviruses. We first assessed this possibility by testing binding to CHIKV VLPs; representative single-specific mAbs and all triple-specific mAbs were tested for binding by ELISA (Figure 7A). None of the single-specific mAbs bound CHIKV VLP. In contrast, 21 triple-specific mAbs bound CHIKV VLP with EC50s ranging from 20 ng/mL to 2.49 μg/mL (median 110 ng/mL). Among the more potent triple-specific mAbs that bound CHIKV VLP, SKT05 had an EC50 value of 100 ng/mL. These results were substantiated by a docked structure of SKT05 Fab in complex with CHIKV VLP, which revealed compatible binding to those regions identified for WEEV and VEEV (Figure 7B). Indeed, arthritogenic and encephalitic alphaviruses share greater sequence identity and similarity within the E1 protein than the E2 protein.26 However, SKT05 failed to neutralize CHIKV, MAYV, ONNV, and RRV (Figure S6A). Thus, we tested binding of select triple-specific mAbs to live cells infected with CHIKV, MAYV, ONNV, and RRV. Notably, SKT05 bound CHIKV-, RRV-, and ONNV-infected cells, with binding to MAYV-infected cells also being observed but to a lesser extent (Figure 7C). A similar trend was observed with the other broadly neutralizing triple-specific mAbs; SKT20 bound CHIKV- and MAYV-infected cells with limited binding to RRV- and ONNV-infected cells (Figure S6B).
Figure 7. SKT05 broadly recognizes and protects in vivo against arthritogenic alphaviruses.
(A) Representative single-specific mAbs, and all triple-specific mAbs, were tested for binding to chikungunya VLP. Symbol colors represent mAb specificity to the WEEV VLP (red), EEEV VLP (blue), VEEV VLP (green), or all three VLPs (black). Select triple-specific antibodies are shown as unique symbols due to their binding and neutralization profiles. Data are representative of at least two independent experiments. (B) Dock model of SKT05 and CHIKV VLP. (C) Binding to live cells infected with CHIKV, MAYV, ONNV, or RRV by SKT05 (red), CHK-265 (blue), RRV immune ascites fluid (purple), or a control mAb (gray) was determined by flow cytometry. Data are representative of three independent experiments performed in duplicate. (D-E) Arthritogenic alphavirus challenge outcome in mice (n=8/group) that received SKT05 (black) or the NHP α-SIV mAb ITS103.01 as a control (gray) one day prior to inoculation. Data are representative of two independent experiments. Viral load was determined in indicated tissues 3 days following inoculation with (D) CHIKV or (E) RRV. Footpad swelling (width x height) in the ipsilateral foot was measured prior to and 3 days following CHIKV inoculation. Statistical significance related to viral RNA was determined by a Mann-Whitney test and by unpaired t-test for footpad swelling (*p < 0.05, **p < 0.01, ***p < 0.001). The dotted line indicates the limit of detection for viral RNA analysis and the baseline foot measurement for foot swelling. See also Figures S6–S7.
Given the cross-reactive binding to arthritogenic alphaviruses we determined whether SKT05 could confer protective effects in vivo. We administered SKT05 to four-week-old C57BL/6J mice one day prior to subcutaneous challenge with 103 FFUs of CHIKV or RRV. Three days after challenge, mice were euthanized, and tissues were harvested to assess viral load. For CHIKV-infected mice, administration of SKT05 reduced footpad swelling and viral burden in all tissues assessed when compared to a control mAb (Figure 7D and Figure S6C). Similarly, SKT05-treated mice also significantly reduced viral load in all tissues assessed following RRV challenge (Figure 7E and Figure S6D). Thus, when administered prophylactically, SKT05 protects not only against encephalitic alphavirus challenge, but also provides protective benefits against arthritogenic alphavirus challenge.
The close phylogenetic relationship between NHPs and humans provides an opportunity to modify NHP mAbs to resemble those expressed by human germline genes with minimal mutations (Figure S7A) as potential clinically useful drugs. Accordingly, we identified 6 amino acid framework positions of SKT05 that differed from the closest human germline sequence and created 64 SKT05 variants with different degrees of humanization (all combinations of the 6 mutations) (Figure S7B). Of these, 14 bound all three EEV VLPs with EC50 and EC80 values comparable to native SKT05 (Figure S7C). The same 14 SKT05 variants neutralized EEV pseudoviruses with IC50 and IC80 values comparable to SKT05 (Figure S7D). We selected 10 of these SKT05 variants that most closely resembled the human germline gene to test for binding to live cells infected with CHIKV, MAYV, ONNV, and RRV. Similar to SKT05, we observed binding predominantly to cells infected with CHIKV, RRV, and ONNV, while binding to MAYV appeared to be slightly improved (Figure S7E). Thus, we identified several partially humanized SKT05 variants that retain binding and neutralization characteristics of SKT05 and may prove valuable in a clinical setting.
DISCUSSION
In this study, we describe the isolation and structural characterization of α-EEV mAbs with protective effects against both encephalitic and arthritogenic alphaviruses from trivalent VLP-immunized macaques. In total, we identified 109 α-EEV mAbs that (by experimental design) recognized either a single VLP, or all three VLPs. Competitive binding ELISAs revealed 6–7 single-specific competition groups for each virus and 3–4 triple-specific competition groups, and cryo-EM structural analysis of 6 single-specific mAbs and 2 triple-specific antibodies revealed that broad recognition inversely correlated with sequence and conformational variability. Two triple-specific mAbs, SKT05 and SKT20, bound epitopes proximal or including the highly conserved fusion peptide. Despite binding all three viruses at an epitope within the fusion peptide that is almost 100% conserved, SKT20 only neutralized VEEV pseudovirus. In contrast, SKT05 bound proximal to the fusion peptide and potently neutralized all three EEV pseudoviruses. While the SKT05 epitope contained a mixture of both conserved and variable residues, variable residues were recognized through backbone contacts not dependent on sequence conservation. Because of the high reliance of backbone interactions, it is less likely that a virus could select for mutations rendering it resistant to SKT05 neutralization.
Historically, the isolation of anti-alphavirus mAbs has relied on sampling symptomatic individuals in regions following an outbreak. While elicitation of protective mAbs against individual alphaviruses has been reported following vaccination, the development of a vaccine or monoclonal antibody capable of providing protection against both classes of alphaviruses remains highly desirable. Previously, we demonstrated that vaccination with trivalent WEVEEV VLPs protects NHP from aerosol challenge with all three encephalitic alphaviruses, and that this protection was humoral.37 Using the same immunogens, we show here the elicitation of many α-EEV mAbs targeting single or multiple encephalitic alphaviruses. When administered prophylactically, two of these vaccine-elicited triple-specific antibodies, SKT05 and SKT20, provide protection against encephalitic alphavirus challenge. Furthermore, SKT05 also provides protection from arthritogenic alphavirus challenge. Notably, these immunogens have also been assessed in a phase 1 clinical trial and shown to be immunogenic and well tolerated.49 Four weeks after the second immunization, 76% of participants developed neutralizing antibody titers against all three encephalitic alphaviruses. Notably, the triple-specific mAbs isolated from two vaccinated subjects shared a competition group with SKT20 that, despite binding all three EEV VLPs, only neutralized the VEEV pseudovirus. Given the large increase in the elicitation of triple-specific mAbs following a third (perhaps delayed) immunization in NHP, it is likely that the best clinical results will be achieved with a similar delayed-boost regimen.
SKT05 potently neutralized all three pseudotyped EEVs, however, this was not recapitulated with other in vitro assays such as the SINV-EEV chimeric neutralization. SKT05 only moderately neutralized pathogenic WEEV and EEEV and failed to neutralize the pathogenic VEEV and all the chimeric viruses. Since the pseudotyped EEVs lack the alphavirus capsid protein, the particles may not display the structural glycoproteins in the same organization as observed on authentic alphaviruses. The variation in neutralization potency of SKT05 between the chimeric and pathogenic viruses was unexpected and may relate to the assay used for analysis (i.e., FRNT for chimeric viruses and PRNT for pathogenic viruses). For poorly neutralizing mAbs, small foci that are detectable by antigen staining (FRNT) may not produce visible plaques (PRNT) and thus result in slightly different outcomes between the assays. Future studies are needed to sort out the differences between the assays to define the antiviral activity of SKT05 and to determine if a particular assay shows a stronger correlation with in vivo protection. Recently, Kim et. al.24,26 and Williamson et. al.24,26 both reported pan-protective anti-alphavirus mAbs that were fusion-peptide directed yet lacked neutralizing activity. Indeed, SKT05 showed in vivo activity against RRV and CHIKV though it did not neutralize these viruses in vitro. Previous studies have shown a necessary role of Fc-effector functions for mAb efficacy against arthritogenic alphaviruses through Fc-FcγR engagement on monocytes, which may be a potential mechanism of protection for SKT05 in these models.26,51,52 However, all of our mAbs have identical IgG Fc regions by design.
While SKT05 and SKT20 weakly or failed to neutralize the chimeric or pathogenic VEEV, SKT05 and SKT20 protected mice from weight loss and death during VEEV challenge. Interestingly, only SKT05 reduced viral burden in the brain. This suggests that SKT05 and SKT20 may have distinct mechanisms of protection. In this mouse model of VEEV infection, high viral load in the brain does not always correlate with lethality. Other factors such as increased blood brain barrier permeability and induction of cytokines, particularly IFN-γ, can lead to encephalitis and death.53,54 SKT05 may be more efficient at reducing infection of the olfactory neuroepithelium through egress blockade or Fc-mediated effector functions and preventing central nervous system (CNS) infection, while SKT20 may be more primed to alter the inflammation that follows viral penetration of the blood brain barrier and replication in the CNS.55,56 However, the different neutralization and binding activity of SKT05 and SKT20 to VEEV may be associated with the in vivo protection and should be considered during future studies.
A central finding of our study is the definition of the SKT05-binding site of vulnerability, which, unlike the site recognized by other pan-alphavirus mAbs, is accessible to antibody on the fully assembled VLP. The E1 binding site contains some sequence variability, and we show how SKT05 employs a binding mode that surmounts this by recognizing backbone atoms of sequence variable residues. Such backbone-recognition has been observed in other instances of conserved recognition of sequence variable regions, such as with antibody CR8069 and CR8071 that recognize the divergent hemagglutinin head57 and with the CD4 receptor recognizing a region of the HIV-1 envelope that is only partially sequence conserved.58 Fascinatingly, similar to these examples, SKT05 also uses intermolecular β-sheet interaction to facilitate breadth, which in this case extends beyond encephalitic alphaviruses to arthritogenic alphaviruses. This mode of binding, which makes viral resistance more difficult to achieve, provides additional impetus for the clinical development of SKT05.
Limitations of the study
It is likely that mAbs targeting other epitopes are present in the other vaccinated animals and that we have not saturated the epitope definition. Similarly, we sorted from the two clinical trial participants at the only available timepoints, which limited the antigen-specific cells recovered. While these participants were chosen based on neutralizing sera titers against virulent strains of EEVs, it is possible that additional triple-specific mAbs were present in other participants. Although hierarchal clustering identified multiple competition groups for single- and triple-specific mAbs, only one representative mAb from each group was tested in subsequent competition binding ELISAs and only a subset were subject to cryo-EM; additional epitopes and/or neutralizing mAbs may exist. The limited PBMC availability at early time points during the immunization phase of the study led to only 31/70 lineages detected prior to week 53. While the relative frequency of the potent broadly neutralizing mAbs clearly increased following 3rd or subsequent immunizations, additional studies would provide insight into the timing of maturation and the full range of epitopes targeted following trivalent WEVEE vaccination.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mario Roederer (roederer@nih.gov).
Materials availability
Materials described in this paper are available from the Lead Contact for distribution under an NIH Material Transfer Agreement.
Data and code availability
Data.
All data reported in this paper will be shared by the Lead Contact upon request.
Sequencing data were deposited in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA951758.
The atomic models and cryo-EM reconstructions and generated during this study are available at Protein Data Bank (PDB, https://www.rcsb.org) and Electron Microscopy Data Bank (EMDB, https://www.ebi.ac.uk/emdb/) under the accession codes EMDB: 28117; EMDB: 27757, PDB: 8DWO; EMDB: 27389, PDB: 8DEC; EMDB: 27391, PDB: 8DEE; EMDB: 28119; EMDB: 27722, PDB: 8DUL; EMDB: 28118; EMDB: 27723, PDB: 8DUN; EMDB: 28115; EMDB: 27390, PDB: 8DED; EMDB: 28116; EMDB: 27392, PDB: 8DEF; EMDB: 28056; EMDB: 28058, PDB: 8EEU; EMDB: 28187; EMDB: 27395, PDB: 8DEQ; EMDB: 28188; EMDB: 27396, 8DER; EMDB: 28059; EMDB: 28060, PDB: 8EEV.
Additional information can be obtained from the Lead Contact upon request.
Code.
This paper does not report original code.
Additional Information.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contract upon request.
Experimental model and subject details
Animals and study design
Sixteen Chinese-origin cynomolgus macaques (Macaca fascicularis), aged 8–10 years at time of immunization, split evenly by sex, were used for immunogenicity experiments (four groups of four animals); we did not have the power to distinguish the immunogenicity by sex. All in vivo procedures were carried out in accordance to institutional, local, state, and national guidelines and laws governing research in animals including the Animal Welfare Act. The animal protocols “VRC ASP 15–539” and the procedures were reviewed and approved by the Animal Care and Use Committee (ACUC) of both the Vaccine Research Center (in accordance to all the NIH policy and guidelines) as well as the Institutional Animal Care and Use Committee (IACUC) of Bioqual, Inc. where non-human primates were housed for the duration of the study. Bioqual Inc., and the NIH are both accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALACi) and are in full compliance with the Animal Welfare Act and Public Health Service Policy on Humane Care and Use of Laboratory Animals. In accordance to the institutional policies of both institutions, all compatible non-human primates are always pair-housed, and single housing is only permissible when scientifically justified or for veterinary medical reasons, and for the shortest duration possible.
Non-human primates were housed in appropriately sized caging according to the Guide for the Care and Use of Laboratory Animals, 8th ed.59, and supplemented with a variety of enrichment toys, treats, fresh produce, and foraging devices. Water was offered ad libitum and monkeys were fed primate biscuits (Monkey Diet, 5038, Lab diet, St. Louis, MO) twice daily. Animal holding rooms were maintained on a 12-hour light/dark cycle, room temperature of 60–70F, and relative humidity between 30 to 70% as standard practice.
Male C57BL/6J mice (Jackson Laboratory) were used at 4 weeks of age for the CHIKV and RRV challenges. Female C3H/HeN (Charles River Laboratories) were used at 6 weeks of age for the VEEV challenge. Mice were housed in individually ventilated cages with a maximum of 5 mice per cage under barrier and specific-pathogen-free (SPF) conditions. The mice were maintained on a 14h/10h light/dark cycle. The temperature and relative humidity were maintained at 20 – 24°C and 30 – 70%, respectively. Sterile cages contained wood chip bedding (Beta Chip, Animal Specialties and Provisions) and nestlets that were changed weekly. Autoclaved tap water (ABSL2) or acidified water (ABSL3) in sterile bottles and verified 75 IF/Auto Ext Mod Diet (LabDiet) was provided ad libitum and changed weekly. All materials were autoclaved prior to use. If mice were unable to reach food or water due to illness, DietGel Recovery (Clear H2) was provided as a supplement. The studies were conducted in compliance with NIAID ACUC under protocol LVD 6E and are compliant with the recommendations outlined in the Guide for the Care and Use of Laboratory Animals. The SKT05 and SKT20 in vivo efficacy studies were performed in established mouse models for alphavirus infection. For this reason, 4-week-old male mice were used for CHIKV or RRV challenge and 6-week-old female mice were used for VEEV challenge.26,53,54,60–62 However, alphavirus infection in either sex has been shown to result in similar disease manifestation, such as foot swelling, weight loss, and mortality, and/or viral burden.30,63,64 In this study, we did not evaluate the impact of sex on the efficacy of SKT05 and SKT20. We anticipate similar protection would be observed in both sexes, but additional studies will be necessary to confirm these results.
VRC 313 sample collection. Detailed information regarding the age/developmental stage, sex, gender, ancestry, race, ethnicity, and socioeconomic status of participants can be found in Coates et al.49 The study was sponsored by the VRC, NIAID, NIH and conducted at the Hope Clinic of the Emory Vaccine Center at Emory University in Atlanta, GA. The clinical trial protocol was reviewed and approved by the Emory University Institutional Review Board (IRB). All subjects met protocol eligibility criteria and agreed to participate in the study by signing the Emory IRB approved informed consent. Research studies with these samples were conducted by protecting the rights and privacy of the study participants. The study protocol and informed consent form and are available on the ClinicalTrials.gov registry.
Cell lines and primary culture
Vero cells (female) and BHK21 cells (male) were cultured in DMEM supplemented with 5% heat-inactivated (HI)-FBS at 37°C with 5% CO2. 293A cells (female) and 293T cells (female) were cultured in DMEM supplemented with 10% HI-FBS and 1% penicillin/streptomycin (p/s) at 37°C with 10% CO2. Vero76 cells (female) were maintained in minimum essential medium (MEM) supplemented with 10% HI-FBS and 1% p/s at 37°C with 5% CO2. Our cell lines have not been authenticated.
Virus Stocks
Chikungunya virus (CHIKV; strains 181/25 and AF15561) and Ross River virus (RRV; strain T48) were produced from infectious cDNA clones as previously described and passaged once in Vero or BHK cells, respectively.65,66 Mayaro virus (MAYV; strain BeH407) and o’nyong’nyong virus (ONNV; strain MP30) were generously provided by Michael Diamond (Washington University in St. Louis) and propagated in Vero cells. Venezuelan equine encephalitis virus (VEEV; strain TC-83) was passaged on Vero cells. Previously described chimeric viruses encoding the nonstructural proteins of Sindbis virus (strain TR339) and the structural proteins of WEEV (strain CBA87; SINV-WEEV), EEEV (strain FL93–939; SINV-EEEV), and VEEV (strain TrD; SINV-VEEV) were generously provided by Dr. William Klimstra (University of Pittsburgh) and passaged on Vero cells.34,67 All virus experiments were performed in approved biosafety level facilities. VEEV-TC83 vaccine strain from a vial of human use vaccine (Lot 4 Run 2) produced by the National Drug Company was passaged once on Vero76 cells. VEEV-Trinidad donkey, EEEV- FL93–939, and WEEV-Fleming strains produced in Vero76 cells were used for the PRNTs. Sucrose purified VEEV-Trinidad donkey, EEEV- FL93–939, and WEEV-CBA87 strains produced in Vero76 cells were used for the ELISAs.
METHOD DETAILS
Production of virus-like particles (VLPs)
The vectors that express each VLP (C-E3-E2–6K-E1) with capsid (C) and envelope glycoproteins of western, eastern, or Venezuelan equine encephalitis viruses (W/E/V/EEV), as well as chikungunya virus (CHIKV), were constructed as described previously.37,41 The structural genes are from WEEV CBA87, EEEV PE-6, VEEV TC-83, and CHIKV 37997 (GenBank accession numbers: ABD98014.1, AAU95735.1, AAB02517.1, and EU224270, respectively). VLPs were produced by transfecting human embryonic kidney cell line 293-derived suspension cell line 293F by 293fectin transfection reagent according to manufacturer’s instructions. This was followed by purification through a series of steps including centrifugation, filtration, ultrafiltration and diafiltration, chromatography, and sterile filtration as described previously.68 The VLPs were quantitated by BCA protein assay.
Labeling and titration of virus-like particle
Three separate lipophilic membrane staining dyes were used with EEV VLPs: PKH26, PKH67, and CellVue Claret. All labeling was carried out according to manufacturer’s instructions. For PKH26, PKH67, and CellVue Claret, 1 μL of each dye was diluted with 1 mL of diluent C. 1 mL of the diluted dye was added to 200μg of VLP in 500μL PBS, pH 7.4. The mixture was incubated at room temperature for 3 minutes and the labeling was stopped by adding 4.5 mL of 1% BSA in PBS, pH 7.4, mixing by pipetting, and incubating at room temperature for one minute. Free dyes from the labeled VLPs were removed by Amicon 100 KDa centrifugal filter. Theoretical VLP concentrations were calculated using the initial amount of VLP and the volume after free dye removal assuming a 90% recovery. BCA protein quantitation of labeled VLPs was unsuccessful, possibly due to the labeled dye colors. Thus, labeled VLPs were titrated before use to provide appropriate separation of positive and negative VLP+ populations. For titrations, 25μL of α-mouse Igκ compensation beads and 0.1 μg of mouse α-EEEV mAb were incubated at room temperature for 15 minutes and washed with 4% heat-inactivated newborn calf serum (HINCS) in RPMI. The serially diluted labeled VLPs were added to the beads/mAb complex and incubated at room temperature for 15 minutes and washed with 4% HINCS in RPMI. The complexes were analyzed by LSRFortessa X-50 Cell Analyzer (BD Biosciences).
Single cell sorting
To isolate EEV-specific B cells by indexed single cell sorting, cryopreserved PBMCs were thawed and stained as follows; PBMCs were stained with LIVE/DEAD fixable aqua or blue dead cell stain at room temperature in dark for 15 minutes and washed. For macaque PBMC, cells were stained with a panel of WEEV VLP-CellVue Claret, EEEV VLP-PKH67, and VEEV VLP-PKH26 and fluorescently labeled antibodies for CD19 (clone J3–119) and IgG (clone G18–145) at room temperature in dark for 15 minutes and washed three times. For human PBMC, cells were stained with the same labeled VLPs as macaques as well as fluorescently labeled antibodies for CD19 (clone HIB19), CD20 (Clone 2H7), IgM (clone MHM-88), IgG (clone G18–145) at room temperature in dark for 15 minutes and washed three times. Indexed single cell sorting of macaque PBMC was performed by a modified 3-laser BD FACSAria cell sorter, or a FACSymphony S6 cell sorter for bulk B cell sorting, using the FACSDiva software as previously described.69 The cells were sorted into the 96 well plate containing RNase OUT, 5X First Strand Buffer, DTT, and IgePAL.
RT-PCR, cloning, and expression of immunoglobulin genes
The genes of IgG heavy and light chains were amplified by reverse transcription-PCR (RT-PCR) from indexed single sorted B cells in 96 well plate and cloned into eukaryotic expression vector as described previously.69 Briefly, the B cell RNA was reversely transcribed by Superscript III with random hexamer. The IgH, IgLκ, and IgLλ variable region genes were amplified independently by nested PCR from the cDNA as a template. The PCR was carried out in 96 well plate by HotStarTaq Plus DNA Polymerase kit with forward and reverse primer mixtures as described previously (Table S6).70−71The amplified DNA was analyzed on 1%-agarose gel and positive reactions were directly sequenced. The variable regions gene were codon optimized for human cell expression, synthesized (GenScript, Synbio Technologies), and cloned into CMVR expression vectors between a murine Ig leader (GenBank DQ407610) and the constant regions of rhesus macaques IgG1 (GenBank AAF14058), IgLκ (GenBank AAD02577), or IgLλ (GenBank ADX62855), as previously described.70 The paired heavy and light chain plasmids were co-transfected to Expi293 cells by Expifectamine 293 transfection kit. Full length IgG were purified by rProtein A Sepharose Fast Flow antibody purification resin.
Biotinylation of mAbs
Antibody (200 μg) was buffer exchanged into 0.1M bicarbonate buffer, pH 8.4, using Zeba spin desalting columns following the manufacturers recommended protocol. 10 mg EZ-Link NHS-Biotin N-Hydroxysuccinimidobiotin was dissolved in 1mL dry DMSO and was added to the buffer exchanged antibodies at a concentration of 8μL per milligram of antibody. The antibody-biotin mixture was placed on a rotator at room temperature and incubated for 2 hours. After incubation, biotinylated antibodies were buffer exchanged with Zeba spin desalting columns into Tris NaCl pH 8.2 (10mM Trizma, 150mM NaCl, 0.1% NaN3, pH 8.2).
Production of pseudotyped lentiviral reporter(s)
EEV Env-pseudotyped lentiviral reporters were produced from the recombinant lentiviral vectors expressing the structural proteins from WEEV, EEEV, and VEEV (CBA87, PE-6, and TC-83 strain, respectively) and encoding a luciferase reporter gene as previously described.37 Briefly, 293T cells were transfected with 500 ng of WEEV, EEEV, or VEEV envelope plasmid, 7 μg of a transducing vector encoding a luciferase reporter gene under the control of a cytomegalovirus (CMV) promoter (pHR’ CMV-luciferase plasmid), and 7 μg of a packaging plasmid that expresses all HIV-1 structural proteins except envelope (pCMVΔR8.2). Culture medium was completely changed after an overnight Fugene 6 transfection (Promega) followed by harvesting supernatants 48 hours later. During harvesting, supernatants were filtered through a 0.45 mm syringe filter, then stored in aliquots and frozen at −80°C.
Neutralization of WEEV, EEEV, and VEEV Env-pseudotyped lentiviral vectors/reporters
Neutralization assays were similarly performed as described previously.37 Briefly, one day before infection 104 cells/well of 293A cells in Minimum Essential Media (MEM) supplemented with 5% FBS and 1% penicillin-streptomycin (M5 media) were seeded into tissue culture treated black & white 96-well isoplate. Titrated amounts of pseudotyped lentiviral reporters (with p24 levels of about 50 ng/ml) were incubated with 4-fold serially diluted mAbs from 50μg/mL or 10μg/mL at 37 °C in a 5% CO2 incubator for 45 minutes. Next, 50μL/well of the virus:mAb solution were added to 293A cells and incubated at 37 °C in a 5% CO2 incubator for 2 hours. After incubation, 100 μL/well of M5 media was added to the plate. The α-SIV mAb ITS103.0172 served as a negative control. After 24 hours of incubation, cells were lysed using cell lysis buffer. Luciferase activity was measured using the Micro-Beta JET after incubation with a luciferase assay reagent according to the manufacturer’s protocol. All experiments were performed in triplicate. Inhibition values were calculated as follows: inhibition (%) = [1 – [luciferase activity (cps) in pseudotyped lentiviral reporter–infected cells incubated with the indicated dilutions of mAb] / [luciferase activity (cps) in pseudotyped lentiviral reporter–infected cells incubated with the same dilutions of control mAb] X 100. Calculations for the 50, 80, and 90% inhibitory concentrations were made with GraphPad Prism.
Animals and immunizations
Chinese-origin cynomolgus macaques (Macaca fascicularis) were used for all experiments, of which all were reviewed and approved by the Institutional Animal Care and Use Committees (IACUCs) of the VRC/NIAID/NIH. All animals were housed and cared for in accordance with local, state, federal, and institutional policies in facilities accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC) and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011). Hematology parameters were evaluated from whole blood using a hematology analyzer. Alphavirus-naïve, mixed gender Chinese cynomolgus macaques (>3 kg; 4.20 to 11.75 kg) were randomized by gender and body weight and intramuscularly (IM) or intranasally (IN) administered with 60μg of trivalent VLPs (20 μg each of the monovalent VLPs) with or without 8 μg of α-galactosylceramide (α-GalCer) three times at weeks 0, 4 and 21 followed by 180 μg of trivalent VLPs with or without 24 μg of α-GalCer at weeks 30 and 52. At one week after the 5th immunization, PBMCs were isolated and stained for single B cell sorting.
Enzyme-Linked Immunosorbent Assay (ELISA)
The binding of mAbs to VLPs was examined as follows: 96 well ELISA plates were coated with 2 μg/ml of VLP in PBS, pH 7.4, incubated at 4 °C overnight, and blocked with PBS containing 5% skim milk and 2% BSA (blocking buffer) at room temperature for one hour. Each mAb was serially diluted with 5% skim milk and 2% BSA in PBS with 0.05% Tween-20 (PBST) dilution buffer and was added to the plate and incubated at room temperature for one hour. After washing, horseradish peroxidase (HRP)-conjugated Fcγ-specific goat α-monkey IgG, goat α-rhesus IgG (H+L), or mouse α-human IgG Fc was added and incubated at room temperature for one hour. Tetramethylbenzidine (TMB) substrate was added to each well, and the reaction was stopped after 10 minutes by adding 1 M H2SO4. The absorbance was measured at 450 nm. Half-maximal effective concentration (EC50) was calculated by computer-assisted nonlinear fitting using GraphPad Prism. Serum IgG levels in the plasma of immunized animals were calculated as stated above. Total secretory IgA dimer amount in bronchoalveolar lavage (BAL) fluid was quantitated and certain range of total IgA dimer was run against EEV VLPs. Briefly, 2μg/mL of mouse α-rhesus J chain mAb (Nonhuman Primate Reagent Resource, NHPRR; clone CA1L_33e1_Ala3) in carbonate-bicarbonate buffer, pH 9.4 (Sigma) was added to 96 well ELISA plates and incubated at 4ºC overnight. After blocking, 50-fold diluted BAL fluid was added to the plate and incubated at room temperature for an hour. Serially diluted rhesus IgA dimer (NHPRR; clone b12rAid; 200~0.3 ng/mL) was used as a reference standard. After washing, HRP-conjugated Fc-specific goat α-monkey IgA was added and incubated at room temperature for an hour. Tetramethylbenzidine substrate was added to each well, and the reaction was stopped and absorbance measured as above. Total IgA amount was calculated from the standard curve with rhesus IgA dimer (SoftMax Pro, Molecular Devices). Then, 2-fold serially diluted BAL fluids corresponding to 50~1.5625 ng/mL of total IgA dimer were added to VLP-coated plates blocked with blocking buffer, and the plates were incubated at room temperature for one hour. The following procedures with HRP-conjugated Fc-specific goat α-monkey IgA, TMB, and stop solution were carried out as described above. AUC was calculated using GraphPad Prism. Competitive binding ELISAs were similarly performed as stated above with the following adjustments. After blocking and washing VLP-coated plates, 10 μg/mL of individual mAbs were added and incubated at room temperature for 30 minutes. Titrated amounts of biotinylated mAbs were then added, without a wash in between, and incubated at room temperature for one hour. After washing, streptavidin-HRP was added and incubated for one hour followed by development of ELISA plates as described above. To assess binding to virulent strains of WEEV, EEEV, and VEEV indirect ELISAs were performed as follows: 96-well high-binding plates were coated overnight at 4°C with 3 μg/mL sucrose-purified virus in PBS. Next, coated plates were fixed with 10% formalin for 1 hour. After the fixative was removed, plates were washed 3x with PBS + 0.02% Tween-20 and blocked with Neptune buffer + 3% normal goat serum at ambient temperature for 5 hours. After blocking, plates were washed 3x with PBS + 0.02% Tween-20. Samples were diluted two-fold in blocking buffer from a known starting protein concentration for the IgG and added to the plate in duplicate. Plates were incubated overnight at 4°C. Following incubation, plates were washed 3x with PBS + 0.02% Tween-20, a secondary α-human H+L horseradish peroxidase-conjugated antibody diluted in blocking buffer was added, and plates were be incubated for 1 hour at ambient temperature. Next, plates were washed 3x with PBS + 0.02% Tween-20, TMB substrate was added, and plates were incubated for ~5 minutes at ambient temperature. Finally, the reaction was stopped and absorbance was read using a Spectramax M5 instrument set at 450 nm. End titer was determined by the last dilution that was greater than the negative control + 3 standard deviations of the background wells. End titer was reported as the reciprocal dilutions as μg/mL.
Immunoglobulin repertoire library preparation and next generation sequencing
RNA was isolated using Qiagen RNeasy kits. Reverse transcription (RT) was performed using Clontech SMARTer cDNA template switching: 5’ CDS oligo(dT) (12 mM) was added to RNA and incubated at at 72C for 3 minutes and 4C for at least 1 minutes. The RT mastermix (5x RT Buffer (250 mM Tris-HCl (pH 8.3), 375 mM KCl, 30 mM MgCl2), Dithiothreitol, (DTT 20 mM), dNTP Mix (10 mM), RNAse Out (40U/mL), SMARTer II A Oligo (12 mM), Superscript II RT (200U/mL)) was added to the reaction and incubated at 42C for 90 minutes and 70C for 10 minutes. First-strand cDNA was purified using AMPure XP beads. Following RT, two PCR rounds were carried out to generate immunoglobulin amplicon libraries compatible with Illumina sequencing. All oligos were ordered from Integrated DNA Technologies. Separate reactions were carried out for IgM and IgG heavy chains; the primer sets did not allow us to distinguish between the subtypes of IgG. The first PCR amplification was carried out using KAPA Real-Time Library Amplification Kit. cDNA was combined with master mix (2X KAPA PCR Master Mix, 12 mM mL 5PIIA and 5 mL IgM/IgG/IgK/IgL Constant Primer (2 mM)). The amplification was monitored using real-time PCR and was stopped during the exponential phase. The amplified products were again purified using AMPure XP beads. A second round of PCR amplification was carried out for addition of barcodes and Illumina adapter sequences: master mix (2X KAPA PCR Master Mix 2x, Nuclease-free water), 10 mM of P5_Seq BC_XX 5PIIA, 10 mM of P7_ i7_XX IgM/IgG/IgK/IgL and were combined with amplified Immunglobulin from the first round PCR and amplified using real-time PCR monitoring. The P5_Seq BC_XX 5PIIA primers contain a randomized stretch of four to eight nucleotides to increase diversity. This was followed by purification with AMPure XP beads. A final PCR step was performed for addition of remaining Illumina adapters by mixing master mix (2X KAPA PCR Master Mix, 10 mM P5_Graft P5_seq, Nuclease-free water), 10 mM of P7_ i7_XX IgM/IgG/IgK/IgL oligo and amplified products from the previous PCR step followed by purification with AMPure XP beads. The quality of library was assessed using Agilent Bioanalyzer. The amplicon libraries were pooled and sequenced on either an Illumina HiSeq 2500 in rapid run mode or an Illumina MiSeq as 2×300 paired-end runs. The accession number for the NGS data reported in this paper is BioProject: PRJNA951758.
BCR sequence analysis
All sequences from each locus (IgH, IgK, IgL) were individually pooled and clustered at 99% identity using vsearch v2.8.1 (https://doi.org/10.7717/peerj.2584). Centroids from clusters with at least three reads were submitted to partis (commit fcd5ea6) (https://doi.org/10.1371/journal.pcbi.1007133) for germline inference. For IgH, the starting database was the Macaca fascicularis reference set from KIMDB v1.1 (https://pubmed.ncbi.nlm.nih.gov/33484642/); for IgK and IgL the Macaca mulatta reference sets from IMGT (https://doi.org/10.1093/nar/gku1056) were used due to the lack of publicly available Macaca fasciuclaris references at these loci. Orgdbstats (https://github.com/airr-community/ogrdbstats) was used to review the results of the germline inference, and those with low apparent usage were removed. Finally, sequences were annotated based on the inferred personalized gene database using SONAR v4.2 (https://doi.org/10.3389/fimmu.2016.00372). Clonal relatives of experimental mAbs were identified based on V gene match and CDR H3 nucleotide identity using SONAR.
Cryo-EM structure determination
Virus-like particles at a concentration of 2 mg/ml were mixed with Fab fragments at a molar ratio of one expected E1-E2 binding site per one Fab fragment, and the mixture briefly incubated at room temperature. Immediately before vitrification, Quantifoil R 1.2/1.3 gold grids were glow-discharged using a PELCO easiGlow glow-discharger (air pressure: 0.39 mBar, current: 20 mA, duration: 30 s). Cryo-EM specimens were prepared with an FEI Vitrobot Mark IV plunger using the following parameters: chamber temperature of 4°C, chamber humidity of 95%, and drop volume of 2.7 μl. Specimen screening was performed on a Thermo Scientific Talos F200C electron microscope equipped with a Falcon 3EC direct electron detector (DED) and a side-entry holder.
For VEEV-SKT05, the dataset was collected using SerialEM73 at NICE cryo-EM facility on an FEI Titan Krios electron microscope equipped with a Gatan K2 summit DED operated in the super-resolution mode (pixel size before binning: 0.528 Å) (Table S3). For VEEV-SKT20 and WEEV VLP, datasets were collected on the same microscope equipped with a Gatan K3 DED operated in the super-resolution mode (pixel size before binning: 0.41 Å) and with a Gatan BioQuantum energy filter (Table S3). Cryo-EM datasets for VEEV-SKV09, VEEV-SKV33, WEEV-SKT05, WEEV-SKW11, WEEV-SKW19, WEEV-SKW24 and EEEV-SKE26 complexes were collected at the Columbia Cryo-EM Facility on a Titan Krios electron microscope equipped with a Gatan K3 DED and a BioQuantum energy filter using Leginon74 (Table S3).
Single particle analysis of the VEEV-SKT05 complex dataset was performed using the RELION 3.1 pipeline,75 with patch-based movie frame alignment by MotionCor276 and contrast transfer function (CTF) parameters determination by GCTF.77 Particle picking was performed using the template-free Laplacian-Gaussian filter-based approach. Particles were extracted into 920-pixel boxes and subjected to rounds of 2D and 3D classification, with a previously determined low-resolution map of VEEV37 as the initial 3D reference. A 4.1-Å map of the VLP-Fab complex was obtained after iterative rounds of Bayesian polishing and per-particle CTF parameters refinement, with icosahedral symmetry enforced. Examination of this map revealed a one-Fabto-one-VLP-spike binding stoichiometry. To improve the resolution of the spike-antibody interface, a mask encompassing one VLP spike with one bound Fab was created in UCSF Chimera78 and used for partial signal subtraction after symmetry expansion. The signal-subtracted particles were extracted in 296-pixel boxes and subjected to 3D refinement with limited angular search (--sigma_ang = 5). Local 3D classification without particle alignment was then used to isolate particles with well-resolved Fab fragments. This was followed by another round of auto-refinement, which produced the final map with a resolution of 3.5 Å. Local resolution was determined with ResMap 1.1.4.79 To obtain a more detailed version of the experimental map, DeepEMhancer80 was used for automatic post-processing.
Single particle analysis of the VEEV-SKT20 dataset followed the approach developed for the VEEV-SKT05 complex. Examination of the 4.7-Å map of the icosahedral VLP-Fab complex revealed three low-occupancy Fab fragments bound to each viral spike protein. To isolate particles with all three Fabs present after symmetry expansion and partial signal subtraction, a mask was created encompassing each of the three Fabs individually. 3D classification without image alignment was then performed within each of these masks separately, producing three particle subsets with well-defined individual Fabs. The output _data.star files from the three 3D classification jobs were then parsed to identify particles that were present in all three files. Auto-refinement of this subset of 28,235 particles produced a structure with strong density for all three Fab fragments. C3 symmetry was imposed before the final refinement, which produced a 3.6-Å resolution map. DeepEMhancer was used for map post-processing.
For WEEV VLP, WEEV-SKW19, WEEV-SKW24, WEEV-SKT05, WEEV-SKW11, EEEV-SKE26, VEEV-SKV09 and VEEV-SKV33 complexes, datasets were processed using cryoSPARC v3.3.1.81 Movies were aligned and dose-weighted using patch motion correction, and the micrograph contrast transfer function (CTF) parameters were estimated using patch CTF estimation. Particles were picked using the blob picker and subjected to 2D classification with selection of best classes. Ab-initio, homogeneous, non-uniform and 3D refinement jobs were run with I symmetry imposed. For WEEV VLP, WEEV-SKW19, WEEV-SKW24, WEEV-SKT05, WEEV-SKW11 and EEEV-SKE26 complexes, particles were then symmetry expanded and local refinement was performed using a mask covering the region of spike protruding from the VLP membrane and the Fab. For VEEV-SKV09 and VEEV-SKV33 complexes, particle sets were transferred to RELION using the pyem script (https://github.com/asarnow/pyem). This was followed by expansion of the icosahedral symmetry and signal subtraction. For VEEV-SKV33, the map was refined using RELION and sharpened using deepEMhancer. For VEEV-SKV09, particles were subjected to multiple rounds of masked 3D classification and transferred back to cryoSPARC, followed by local refinement.
To obtain atomic models of complexes, homology models of the Fab fragments were generated with the SWISS-MODEL server82 (VEEV-SKT05 and VEEV-SKT20), AlphaFold283 (SKW19, SKW24, SKE26), Alphafold multimer84 (SKV09 and SKV33) or the SAbPred server85 (SKW11). For VEEV- and EEEV-containing complexes, previously deposited VEEV (PDB 7FFE)22 and EEEV (PDB 6XO4)27 structures, respectively, were used as starting models. Initial atomic models for WEEV structural proteins were generated with AlphaFold2 and superimposed with their VEEV counterparts using Coot.86–88 The initial models were docked into corresponding cryo-EM maps using UCSF Chimera, except for the VEEV-SKV09 and VEEV-SKV33 complexes, for which model fitting was performed using ISOLDE 89 integrated into ChimeraX.90 Atomic models were refined by alternating rounds of model building in Coot and real-space refinement in Phenix.91 The cryo-EM map of WEEV-SKT05 complex did not allow visualization of side chains; the model was refined based on the high homology with the VEEV-SKT05 complex structure. The map of the WEEV-SKW11 complex allowed unequivocal identification of the Fab SKW11 heavy and light chain positions but not visualization of side chains; therefore, the SKW11 Fab model was refined as poly-alanine. Structure validation was performed with Molprobity92,93 and the PDB validation server. The analysis of VLP-Fab interfaces was done with PISA.94 Summaries of model refinement statistics and quality assessment for cryo-EM reconstructions are shown in Table S3 and Data S1.
Mouse challenge experiments
Experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health after approval by the NIAID DIR Animal Care and Use Committee.
VEEV challenge:
SKT05, SKT20, or ITS.103 control mAb (200 μg in PBS) was administered intraperitoneally one day prior to virus challenge in 6-week-old female C3H/HeN mice purchased from Charles River Laboratories. Mice were inoculated intranasally with 107 FFUs of VEEV (strain TC-83) in PBS (40 μl; 20 μl per nare) under anesthesia with 2,2,2-Tribromoethanol. Mice were weighed daily and humanely euthanized when ≥ 25% of their starting weight was lost or reach other endpoint criteria. A subset of mice was euthanized at 5 dpi, extensively perfused with PBS, and indicated tissues were harvested. Viral burden was determined by qRT-PCR.
CHIKV and RRV challenge:
SKT05 or ITS103.01 control mAb (200 μg in PBS) was administered intraperitoneally one day prior to virus challenge in 4-week-old male C57BL/6J mice purchased from the Jackson Laboratory. Mice were inoculated subcutaneously in the left rear footpad with 103 FFUs of RRV (strain T48) or CHIKV (strain AF15561) in Hank’s Balanced Salt Solution (HBSS) supplemented 1% HI-FBS under isoflurane anesthesia. For CHIKV-infected mice, swelling of the ipsilateral foot was measured (width x height) prior to infection and on the day of harvest using digital calipers. Mice were euthanized at 3 dpi, extensively perfused with PBS, and indicated tissues were harvested. Viral burden was determined by RT-qPCR.
RNA extraction and RT-qPCR analysis
Perfused tissues were homogenized in 1 mL of DMEM supplemented with 2% HI-FBS, 10 mM HEPES, and 100 U/mL of penicillin and streptomycin using Zirconia/Silica Beads in a MagNa Lyser for 60 seconds at 6,000 rpm. The homogenate was clarified by centrifugation at 10,000 rpm for 5 minutes. For RRV- or VEEV-infected tissue, RNA was extracted from clarified homogenate using the Kingfisher Duo Prime with the MagMAX-96 Viral RNA isolation kit following the manufacturer’s instructions. For CHIKV-infected tissue, RNA was extracted using the RNeasy mini kit following the manufacturer’s instructions.
To determine viral burden, equal quantities of RNA were added to the TaqMan fast virus 1-step master mix with RRV nsp3 specific primers/ probe (Forward: 5’-GTGTTCTCCGGAGGTAAAGATAG-3’, Reverse: 5’-TCGCGGCAATAGATGACTAC-3’, Probe: 5’-/56-FAM/ACCTGTTTA/ZEN/CCGCAATGGACACCA/3IABkFQ/-3’), CHIKV E1 specific primers/probe (Forward: 5’-TCGACGCGCCATCTTTAA-3’, Reverse: 5’- ATCGAATGCACCGCACACT-3’, Probe: 5’-/56-FAM/ACCAGCCTG/ZEN/CACCCACTCCTCAGAC/3IABkFQ/-3’), or VEEV nsp3 specific primers/probe (Forward: 5’-CCATATACTGCAGGGACAAGAA-3’, Reverse: 5’-CACTGAAGAGTCGTCGGATATG-3’, Probe: 5’-56’FAM/ATGACTCTC/ZEN/AAGGAAGCAGTGGCT/3IABkFQ/-3’) and run on a QuantStudio 3 Real-Time PCR System.30,95,96 Viral RNA isolated from RRV, CHIKV, or VEEV stock was used to generate a standard curve to determine FFU equivalents. All samples were normalized to gram of tissue or mL of serum.
mAb binding to surface of live alphavirus-infected cells
Vero cells were inoculated with ONNV (strain MP30), CHIKV (strain 181/25), RRV (strain T48), or MAYV (strain BeH407) at an MOI of 1 for 19 h or with SINV/VEEV, SINV/EEEV, or SINV/WEEV at an MOI of 0.5 for 21 h in DMEM supplemented with 10 mM HEPES, 100 U/mL penicillin and streptomycin, and 2% HI-FBS. Cells were rinsed with PBS, trypsinized, and centrifuged at 1500 rpm for 5 minutes. Cells were transferred to a 96 well U-bottom plate and rinsed with FACS buffer (PBS with 1% HI-FBS). Infected cells were incubated with indicated mAb (10 μg/mL) or 1:2000 RRV immune ascites fluid in FACS Buffer for 1 hour at 4°C. The cells were then rinsed twice with FACS buffer and incubated with 1:2000 AF647-conjugated goat α-mouse or α-human IgG for 1 hour at 4°C. Cells were rinsed with FACS buffer then fixed with 4% PFA in PBS for 10 minutes at 4°C. The cells were washed and resuspended in FACS buffer then run on a LSR Fortessa cell analyzer. Negative gates were set based on a control mAb (α-West Nile virus mAb E60 or α-SIV mAb ITS103.01) The data were analyzed using FlowJo software version 10.8.1. Median Fluorescence Intensity (MFI) was calculated for the Alexa-647 axis.
Focus reduction neutralization tests (FRNT)
FRNT assays were performed as previously described.29 Briefly, mAbs were serially diluted in DMEM supplemented with 2% HI-FBS, 10 mM HEPES, and 100 U/mL penicillin and streptomycin then incubated with 102 FFUs of indicated virus for 1 hour at 37°C. The mAb-virus complexes were added to Vero cells then incubated for 1.5 hours. The cells were overlaid with MEM and 1% methylcellulose supplemented with 2% HI-FBS, HEPES buffer, penicillin and streptomycin and incubated for between 18–22 hours. The cells were fixed with 1% PFA in PBS for 1 hour followed by washing with PBS. The cells were stained with a 500 ng/mL of indicated mAb or 1:4000 dilution of ascites fluid CHIKV (CHK-11), MAYV (CHK-48), ONNV (CHK-11), RRV (mouse α-RRV ascites fluid, SINV/VEEV (SKV09), SINV/EEEV (SKE26), or SINV/WEEV (SKW11) in permeabilization buffer (PBS with 0.1% saponin and 0.1% BSA) for 2 hours at room temperature. The cells were washed with PBS + 0.05% Tween-20 then incubated with a 1:5000 dilution of a peroxidase-conjugated α-mouse IgG or α-monkey IgG diluted in permeabilization buffer for 1 hour. Plates were developed with True Blue Peroxidase substrate then foci were counted on ImmunoSpot plate reader. Wells containing mAb were compared to wells with no mAb to determine percent inhibition. The IC50 value was calculated using non-linear regression with the top and bottom constrained to 100 and 0, respectively.
Plaque Reduction Neutralization Test (PRNT)
Individual antibodies were diluted in MEM with 2% heat inactivated fetal bovine serum (HI-FBS), 1% HEPES, and 2% Penicillin/Streptomycin (10,000 IU/mL, 10,000 IU/mL) (Pen/Strep) and then serially diluted 1:2. Virus stocks were diluted to a concentration of 2.0 × 103 PFU/ml and added 1:1 to diluted samples or the media-only control wells. Samples were incubated overnight at 4°C. Six-well plates of Vero 76 cells were infected with 0.1 mL of each serial dilution per well in duplicate and plates were incubated at 37°C for 1h. After 1h incubation, cells were overlaid with 0.6% agarose in Basal Medium Eagle (BME) with 10% HI-FBS, and 2% Pen/Strep, and incubated for ~24h at 37°C, 5% CO2. A second overlay containing 0.6% agarose in BME with 10% HI-FBS, 2% Pen/Strep, and 5% of total volume neutral red vital stain was added to wells and further incubated for 18–28h for visualization of plaques. Plaques were counted following incubation with stain overlay. The virus only control was counted, and the percent neutralization of the diluted sera was determined. A virus specific hyperimmune mouse serum was utilized as a positive control. Normal human sera were utilized as a negative control.
Isothermal titration calorimetry (ITC)
Binding experiments by ITC were performed at 25 or 37°C using a VP-ITC microcalorimeter from MicroCal⁄Malvern Instruments (Northampton, MA, USA). The VLPs and the antibodies were dissolved in PBS, pH 7.4, except in the titration of VEEV with SKT05 which also included Tween-20 at a concentration of 0.05%. The solution containing either of the antibodies at 0.9 – 1.0 mg/mL (12 – 14 μM antigen binding sites) was added in 7-μL aliquots to the stirred calorimetric cell (~1.4 mL) containing the VLP solution. The VLP solutions were prepared at 0.2 – 0.3 mg/mL, except in the VEEV-SKT05 experiment where VEEV was prepared at 0.9 mg/mL. All reagents were exhaustively dialyzed prior to the experiments using dialysis membranes having a cutoff of 100 kDa. The heat evolved upon each injection was obtained from the integral of the calorimetric signal and the heat associated with binding was obtained after subtraction of the heat of dilution. The enthalpy change, ΔH, the association constant, Ka (the dissociation constant, Kd =1/Ka) and the stoichiometry, N, were obtained by nonlinear regression of the data to a single-site binding model using Origin with a fitting function made inhouse. Gibbs energy, ΔG, was calculated from the binding affinity using ΔG = -RTlnKa, (R = 1.987 cal/(K × mol)) and T is the absolute temperature in kelvin). The entropy contribution to Gibbs energy, -TΔS, was calculated from the relation ΔG = ΔH -TΔS. The results were normalized per mole of antigen binding sites and the stoichiometry, N, denotes the number of antigen binding sites per E1:E2 monomer.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was assigned when P values were < 0.05 using GraphPad Prism Version 9.0. Foot swelling was compared using an unpaired t-test. For tissue viral titers, a Mann-Whitney test was used for the arthritogenic alphavirus challenge and a Kruskal-Wallis with a Dunn’s post-test comparing mAb treatment to the control mAb was used for the VEEV challenge. Statistical details of experiments can also be found in the figures, figure legends, and results.
Supplementary Material
Figure S1. Trivalent VLP immunization elicits systemic and mucosal humoral immune responses, related to Figures 1–3.
(A-B) Systemic IgG responses in plasma (A) and mucosal IgA dimer responses in BAL (B) were measured by ELISA plates coated with WEEV VLP (left), EEEV VLP (middle), or VEEV VLP (right). Endpoint titers (IgG) and AUC (IgA) are shown as mean ± SEM from four animals per group (top) and from individual animals per group (below). The lower limit of detection is indicated by the black line; red arrows indicate the first four immunizations. (C) Representative flow cytometry gating scheme for PBMC stained with labeled VLP probes and fluorophore labeled α-CD19 and α-IgG mAbs one week after the 5th immunization.
Figure S2. Competition ELISAs identify distinct binding groups of single-specific and triple-specific α-EEV mAbs, related to Figure 2.
(A-D) Competitive binding ELISAs were performed to identify overlap in VLP surface binding areas within ⍺-WEEV mAbs (A), ⍺-VEEV mAbs (B), and ⍺-WEVEEV mAbs using the WEEV VLP (C) and VEEV VLP (D). Competitive mAbs were pre-incubated on VLP-bound plates at a concentration of 10 μg/mL for 30 minutes prior to the addition of titrated biotinylated single-specific (A-B) or triple-specific (C-D) mAbs. Data was subject to hierarchical clustering to determine competition groups. Heat maps display percent inhibition ranging from yellow (minimal competition) to orange (moderate competition) to purple (substantial competition). Colored circles above mAbs on x-axis represent the EEV pseudovirus that was neutralized (red: WEEV; blue: EEEV; green: VEEV). Data are representative of at least two independent experiments.
Figure S3. Memory B cells from broadly neutralizing lineages are rare, related to Figure 2.
(A) Most ⍺-EEV lineages are first observed after three immunizations with WEVEEV VLPs. B cells were sorted from PBMC of the animal with the highest percentage of B cells binding all three VLPs at various time points and bulk IgSeq was conducted to determine timing of lineage development. Assessed time points are denoted by different colors and WEEV endpoint titers for each time point are included in the legend. Despite a similar number of PBMC used at each time point, the number of new triple-specific lineages detected differed. (B) There was a pronounced increase in transcript prevalence following third and fourth immunizations. Transcripts from single- and triple-specific ⍺-EEV mAb lineages are displayed by color as fractions of total sequences observed at different time points following bulk IgSeq. (C) Subjects in clinical trial VRC 313 were immunized intramuscularly with or without alum adjuvant twice, eight weeks apart, with 20 μg of each monovalent WEEV, EEEV, and VEEV VLPs. Six total B cells from two subjects, binding all three VLPs simultaneously, were sorted and assessed for binding and neutralization. (D) ELISA binding curves (left) and pseudovirus neutralization curves (right) of triple-specific ⍺-EEV human mAbs against WEE VLP (top), EEE VLP (middle), and VEE VLP (bottom). Data are displayed as nonlinear fit of log transformed data and representative of two independent experiments. (E) Triple-specific ⍺-EEV human mAbs competed with SKT20, but not SKT05, for binding to VLP. Competition binding ELISA assays were performed with triple-specific human mAbs and representative triple-specific NHP mAbs. Heat maps display percent inhibition where colors range from yellow (minimal competition) to orange (moderate competition) to purple (substantial competition). Data are representative of two independent experiments. (F) Triple-specific human mAbs were tested alongside SKT05 and one single-specific mAb from each VLP for binding to cells infected with SINV-WEEV (top), SINV-EEEV (middle), and SINV-VEEV (bottom). The NHP α-SIV mAb ITS103.01 was used as a negative control. Data are representative of two independent experiments.
Figure S4. Cryo-EM structure of WEEV defines WEVEEV surface diversity, related to Figure 4.
(A) Cryo-EM structure of WEEV at 3.4 Å resolution, shown with previously determined VEEV (PDB 7ffe) and EEEV (PDB 6xo4) structures, which were colored radially like WEEV structure except the outer color being cyan or blue for VEEV or EEEV, respectively. (B) Conformational variability was calculated for each residue of WEEV as average distance between WEEV and VEEV (PDB 7ffe), EEEV (PDB 6xo4). E1 is colored green, E2 – orange. (C) Sequence variation was calculated as BSA weighted average of normalized Shannon’s entropy based on WEEV, VEEV, EEEV, CHIKV, RRV, MAYV, ONNV sequences and mapped on to WEEV. (D) Sequence and conformational variability are plotted for each residue. SKT05/SKT20 epitopes as well as virus receptors LDLRAD3 and MXRA8 epitopes are highlighted.
Figure S5. WEVEEV conformational and sequence variation, related to Figures 4–5.
(A) Sequence variation is plotted against conformational variability for each residue. (B) The agreement between conformational and sequence variation is plotted for each residue as difference between fractions of conformational and sequence variations as follows: absolute value of (sequence variation-conformational variability/5.0). Sequence variation is calculated as normalized entropy and values range from 0 to 1. Conformational variability values larger than 5 Å were replaced with 5.0; the fraction of conformational variability is calculated by normalizing all values by 5.0. (C) RMSD for fusion peptides when E1 proteins were superimposed by Cα atoms. (D) Data from calorimetric titrations of VLPs with different antibodies. (E-F) Thermodynamics of mAb binding to WEEV, EEEV, and VEEV VLPs.
Figure S6. Triple-specific α-EEV mAbs do not neutralize arthritogenic alphaviruses, but bind infected cells and protect in vivo, related to Figure 7.
(A) In vitro neutralization curves for SKT05 against chikungunya virus (CHIKV), Mayaro virus (MAYV), o’nyong’nyong virus (ONNV), and Ross River virus (RRV). The broadly reactive mouse mAb CHK-265 was used as a positive control, while the α-West Nile virus mAb E60 and α-SIV mAb ITS103.01 were used as mouse and NHP IgG negative controls, respectively. Data represents the mean ± SD for two independent experiments performed in triplicate. (B) Broadly neutralizing triple-specific mAbs, as well as broadly binding SKT20 mAb, were tested for binding to cells infected with CHIKV, MAYV, ONNV, and RRV. Vero cells were infected with CHIKV, MAYV, ONNV, or RRV (MOI of 1 for each) for 19hr followed by staining with the indicated mAbs (red). Cells were analyzed by flow cytometry and negative gates were set based on a control mAb (gray). Fluorescence intensity is shown along the x-axis and cell count is shown along the y-axis. Data represent three independent experiments performed in duplicate. (C-D) Four-week-old male C57BL/6J mice (n=8; 2 independent experiments) were administered 200 μg of SKT05 or ITS103.01 (control) one day prior to subcutaneous inoculation with 103 FFU of CHIKV (B) or RRV (C) in the left rear footpad. Viral load was determined in indicated tissues 3 days following inoculation. Statistical significance related to viral RNA was determined by a Mann-Whitney test (*p < 0.05, **p < 0.01, ***p < 0.001). The dotted line indicates the limit of detection for viral RNA analysis.
Figure S7. Humanized SKT05 variants display similar binding and neutralization profiles as SKT05, related to Figure 7.
(A) Strategy for humanization of SKT05. We determined the closest human germline gene to the NHP SKT05 sequence and identified differing framework positions. Structurally important amino acids were not altered, as well as amino acids that were common between NHP and humans. (B) Substitution frequency of SKT05 amino acids compared to most closely related human alleles. Magenta color represents framework positions that were altered. (C) Variable binding affinity of humanized SKT05 variants. We performed ELISA assays on 64 humanized SKT05 variants to determine binding to WEEV VLP (left), EEEV VLP (middle), and VEEV VLP (right). Binding titers of the humanized SKT05 variants with comparable binding affinities to all three VLPs are shown to the right as EC50 and EC80 plots. Binding titers for SKT05 are shown as a red symbol for comparison. Binding curves are displayed as nonlinear fit of log transformed data. Data for the 64 humanized variants represent one experiment. The group of best binding mAbs were selected for further analysis. (D) A total of 14 humanized SKT05 variants with similar binding affinities to all three VLPs as SKT05 were tested for pseudovirus neutralization against WEEV (left), EEEV (middle), and VEEV (right). Neutralization IC50 and IC80 titers are shown to the right with SKT05 shown in red for comparison. Neutralization curves are shown as log transformed nonlinear fit of percent inhibition. Data are representative of two independent experiments performed in triplicate. (E) Ten humanized SKT05 variants were tested for binding to live cells infected with chikungunya virus (CHIKV), Mayaro virus (MAYV), o’nyong’nyong virus (ONNV), or Ross River virus (RRV). Vero cells were infected with CHIKV, MAYV, ONNV, or RRV (MOI of 1 for each) or left uninfected for 16hr followed by staining with a humanized SKT05 variant mAb (red), CHK-265 (blue), or RRV immune ascites fluid (purple). Cells were analyzed by flow cytometry and negative gates were set based on a control mAb (gray). Median fluorescence intensity (MFI) was calculated based on the total distribution of cells. Data are representative of three independent experiments performed in duplicate.
Table S6. Rhesus and human primers for the amplification and sequencing of heavy, kappa, and lambda chain genes from single cells, related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Single-specific and triple-specific macaque antibodies (expi293F-produced IgG1) | This paper | Table S1 |
| ITS103.01 | NIH Vaccine Research Center | Welles et. al.72 |
| VRC01 | NIH Vaccine Research Center | Xu et. al.69 |
| WEEV VLP-CellVue Claret | This paper | N/A |
| EEEV VLP-PKH67 | This paper | N/A |
| VEEV VLP-PKH26 | This paper | N/A |
| ECD Mouse anti-human CD19 (clone J3–119) | Beckman Coulter | Cat#IM2708U; RRID:AB_130854 |
| PE/Dazzle 594 Mouse anti-human CD19 (clone HIB19) | BioLegend | Cat#302251; RRID:AB_2563559 |
| PE-Cy5 Mouse anti-human IgG (clone G18–145) | BD Biosciences | Cat#551497; RRID:AB_394220 |
| Brilliant Violet 421 Mouse anti-human IgG (clone G18-145) | BD Biosciences | Cat#562581; RRID:AB_2737665 |
| Brilliant Violet 570 Mouse anti-human IgM (clone MHM-88) | BioLegend | Cat#314517: RRID:AB_10913816 |
| Alexa Fluor 700 Mouse anti-human CD20 (clone 2H7) | BioLegend | Cat#302322; RRID:AB_493753 |
| Mouse anti-human anti-EEEV antibody (clone 1A4B.6) | Millipore | Cat#MAB8754; RRID:AB_95408 |
| Anti-Mouse Igκ beads | BD Biosciences | Cat#552843; RRID:AB_10051478 |
| Goat anti-monkey IgG (Fc-specific)-HRP | Nordic MUbio | Cat#GAMon/IgG(Fc) /PO |
| Goat anti-rhesus IgG (H+L)-HRP | SouthernBiotech | Cat#6200–05; RRID:AB_2796268 |
| Mouse anti-human IgG-HRP | SouthernBiotech | Cat#9040–05; RRID:AB_2687484 |
| Mouse anti-rhesus J chain monoclonal antibody (clone CA1L_33e1_Ala3) | Nonhuman Primate Reagent Resource | RRID:AB_2819358 |
| Rhesus IgA dimer (clone b12rAid) | Nonhuman Primate Reagent Resource | No longer available |
| Anti-human IgG (H+L) antibody, peroxidase-labeled | SeraCare | Cat#5450–0009 |
| High Sensitivity Streptavidin-HRP | Thermo Scientific | Cat#21130 |
| Peroxidase Labeled Goat anti-mouse IgG (H+L) | SeraCare | Cat#5450–0011; RRID:AB_2687537 |
| HRP Labeled Mouse anti-monkey IgG | Southern Biotech | Cat#4700–05; RRID:AB_2796069 |
| Anti-Mouse IgG2a Isotype Control | BioXcell | Cat#BE0085; RRID:AB_1107771 |
| E60 IgG | BioXcell | Cat#BE0325; RRID:AB_2819052 |
| Anti-RRV Ascites Fluid | ATCC | Cat#V-559–701-562 |
| Alexa-647 fluorophore goat anti-mouse | ThermoFisher | Cat#A-21235; RRID:AB_2535804 |
| Alexa-647 fluorophore anti-human | Southern Biotech | Cat#2040–31; RRID:AB_2795651 |
| CHK-265 IgG | Pal et al.97 | N/A |
| CHK-11 IgG | Pal et al.97 | N/A |
| CHK-48 IgG | Pal et al.97 | N/A |
| EEEV-138 | James E. Crowe Jr. | Williamson et al.24 |
| EEEV-179 | James E. Crowe Jr. | Williamson et al.24 |
| EEEV-346 | James E. Crowe Jr. | Williamson et al.24 |
| DC2.112 | Kim et al.26 | GenBank Accession: MZ417554 & MZ417555 |
| DC2.315 | Kim et al.26 | GenBank Accession: MZ417556 & MZ417557 |
| Bacterial and virus strains | ||
| Chikungunya virus (strain 181/25) | Mainou et al.98 | MW473668.1 |
| Chikungunya virus (strain AF15561) | Ashbrook et al.99 | EF452493.1 |
| Ross River virus (strain T-48) | Kuhn et al.100 | GQ433359.1 |
| Mayaro virus (strain BeH407) | Fox et al.29 | MK573238.1 |
| O’nyong’nyong virus (strain MP30) | Fox et al.29 | N/A |
| Venezuelan equine encephalitis virus (strain TC-83) | The National Drug Company |
GenBank#L01443.1 |
| Chimeric SINV-WEEV | Ma et al.67 | N/A |
| Chimeric SINV-EEEV | Kim et al.34 | N/A |
| Chimeric SINV-VEEV | Ma et al.67 | N/A |
| WEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al.37 |
| EEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al.37 |
| VEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al.37 |
| CHIKV virus-like particles (VLPs) | NIH Vaccine Research Center | Akahata et al.41 |
| Env-pseudotyped lentiviral reporter virus | NIH Vaccine Research Center | Ko et al.37 |
| Biological samples | ||
| Normal human sera | Lonza | Cat#14–50014 |
| VEEV hyperimmune mouse sera | ATCC | Cat#VR-1250AF |
| EEEV hyperimmune mouse sera | ATCC | Cat#VR-1242AF |
| WEEV hyperimmune mouse sera | ATCC | Cat#VR-1251AF |
| VRC313 Clinical Trial Participant Sera | Coates et al.49 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Expi293™ Expression Medium | ThermoFisher/Gibco | Cat#A1435101 |
| Opti-MEM™ I Reduced Serum Medium | ThermoFisher/Gibco | Cat#31985070 |
| Minimal Essential Medium (MEM) | ThermoFisher/Gibco | Cat#11095080 |
| Dulbecco’s Modified Eagle Medium (DMEM) | ThermoFisher/Gibco | Cat#12430054 |
| RPMI 1640 custom (w/ HEPES, NaCl, w/o biotin, L-glutamine, phenol red, riboflavin, NaHCO3) | ThermoFisher/Gibco | Cat#A14576DK (custom) |
| Basal Medium Eagle | Quality Biological | Cat#112–339-131 |
| Neptune Buffer | Immunochemistry technologies | Cat#64 |
| Diluent C for general membrane labeling | Sigma-Aldrich | Cat#CGLDIL-6X10ML |
| Amicon Ultra-15 centrifugal filter, 100KDa | Millipore | Cat#UFC910024 |
| Brilliant Stain Buffer Plus | BD Biosciences | Cat#566385 |
| Zeba Spin Desalting Columns, 40K MWCO | Thermo Fisher Scientific |
Cat#87767 |
| Luciferase Cell Culture Lysis 5X Reagent | Promega | Cat#E1531 |
| Superscript III Reverse Transcriptase | ThermoFisher | Cat#18080085 |
| rProtein A Sepharose | Sigma Aldrich | Cat#GE17–1279-03 |
| AMPure XP beads | Beckman Coulter | Cat#A63882 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405nm excitation | Invitrogen | Cat#L34966 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit, for UV excitation | Invitrogen | Cat#L23105 |
| PKH26 membrane staining dye | Sigma-Aldrich | Cat#MINI26–1KT |
| PKH67 membrane staining dye | Sigma-Aldrich | Cat#MINI67–1KT |
| CellVue Claret membrane staining dye | Sigma-Aldrich | Cat#MINCLARET1KT |
| EZ-Link NHS-biotin | ThermoFisher | Cat#20217 |
| Critical commercial assays | ||
| ExpiFectamine™ 293 Transfection Kit | ThermoFisher/Gibco | Cat#A14525 |
| Pierce BCA Protein Assay Kit | Thermo Scientific | Cat#23225 |
| Luciferase Assay System | Promega | Cat#E1501 |
| KAPA HiFi HotStart Real-Time Library Amplification Kit | Roche | Cat#7959028001 |
| MagMAX™−96 Viral RNA Isolation Kit | ThermoFisher | Cat# AMB18365 |
| TaqMan™ Fast Virus 1-Step Master Mix | ThermoFisher | Cat# 4444434 |
| RNeasy Mini Kit | Qiagen | Cat# 74106 |
| HotStar Taq Plus DNA polymerase kit | Qiagen | Cat#203605 |
| Deposited data | ||
| Cryo-EM Structure: SKE26-EEEV VLP | This Paper | EMDB: 28117 |
| Cryo-EM Structure: SKE26-EEEV Spike | This Paper | EMDB: 27757; PDB: 8DWO |
| Cryo-EM Structure: WEEV VLP | This Paper | EMDB: 27389; PDB: 8DEC |
| Cryo-EM Structure: WEEV Spike | This Paper | EMDB: 27391; PDB: 8DEE |
| Cryo-EM Structure: SKT05-WEEV VLP | This Paper | EMDB: 28119 |
| Cryo-EM Structure: SKT05-WEEV Spike | This Paper | EMDB: 27722; PDB: 8DUL |
| Cryo-EM Structure: SKW11-WEEV VLP | This Paper | EMDB: 28118 |
| Cryo-EM Structure: SKW11-WEEV Spike | This Paper | EMDB: 27723; PDB: 8DUN |
| Cryo-EM Structure: SKW19-WEEV VLP | This Paper | EMDB: 28115 |
| Cryo-EM Structure: SKW19-WEEV Spike | This Paper | EMDB: 27390; PDB: 8DED |
| Cryo-EM Structure: SKW24-WEEV VLP | This Paper | EMDB: 28116 |
| Cryo-EM Structure: SKW24-WEEV Spike | This Paper | EMDB: 27392; PDB: 8DEF |
| Cryo-EM Structure: SKT05-VEEV VLP | This Paper | EMDB: 28056 |
| Cryo-EM Structure: SKT05-VEEV Spike | This Paper | EMDB: 28058; PDB: 8EEU |
| Cryo-EM Structure: SKV09-VEEV VLP | This Paper | EMDB: 28187 |
| Cryo-EM Structure: SKV09-VEEV Spike | This Paper | EMDB: 27395: PDB: 8DEQ |
| Cryo-EM Structure: SKV16-VEEV VLP | This Paper | EMDB: 28188 |
| Cryo-EM Structure: SKV16-VEEV Spike | This Paper | EMDB: 27396; 8DER |
| Cryo-EM Structure: SKT20-VEEV VLP | This Paper | EMDB: 28059 |
| Cryo-EM Structure: SKT20-VEEV Spike | This Paper | EMDB: 28060; PDB: 8EEV |
| Heavy chain NGS sequence data related to immunization timing | NCBI Sequence Read Archive | Bioproject: PRJNA951758 |
| Experimental models: Cell lines | ||
| Hamster: BHK-21 | ATCC | Cat#CCL-10; RRID:CVCL_1915 |
| Monkey: Vero | ATCC | Cat#CCL-81; RRID:CVCL_0059 |
| Human: Vero 76 | ATCC | Cat#CRL-1587; RRID:CVCL_0603 |
| Human: 293A | ThermoFisher | Cat#R70507; RRID:CVCL_6910 |
| Human: 293T | ATCC | Cat#CRL-3216; RRID:CVCL_006 |
| Human: Expi293F | ThermoFisher/Gibco | Cat#A14527; RRID:CVCL_D615 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#664 |
| Mouse: C3H/HeN | Charles River Laboratory | Cat#025 |
| Macaques: Chinese origin cynomolgus | Vaccine Research Center, NIH | N/A |
| Oligonucleotides | ||
| Rhesus primers for single cell amplification and sequencing of heavy, kappa, and lambda genes | Gorman et. al.70 | Table S6 |
| Human primers for single cell amplification and sequencing of heavy, kappa, and lambda genes | Wang et al.71 | Table S6 |
| 1st round bulk BCR seq IgM Constant primer | Krebs et al.101 | AGGAGACGAGGGGGA AAAGGGTTGGGGCGG ATG |
| 1st round bulk BCR seq IgG Constant primer | Krebs et al.101 | GCCAGGGGGAAGACC GATGGGCCCTTGGTG GA |
| 1st round bulk BCR seq Igκ Constant primer | Krebs et al.101 | GCGGGAAGATGAAGA CAGATGGTGCAGCCA CAG |
| 1st round bulk BCR seq Igλ Constant primer | Krebs et al.101 | GGCCTTGTTGGCTTG AAGCTCCTCAGAGGA GGG |
| P5_Seq BC_xx 5PIIA | Krebs et al.101 | CACGACGCTCTTCCG ATCTNNNNNNNNNNN NAAGCAGTGGTATCA ACGCAGAGT |
| P7 i7_xx IgG | Krebs et al.101 | CAAGCAGAAGACGGC ATACGAGATNNNNNN NNGCCAGGGGGAAGA CCGATGGGCCCTTGG TGGA |
| P7 i7_xx Igλ | Krebs et al.101 | CAAGCAGAAGACGGC ATACGAGATNNNNNN NNGGCCTTGTTGGCT TGAAGCTCCTCAGAG GAGGG |
| P7 i7_xx Igκ | Krebs et al.101 | CAAGCAGAAGACGGC ATACGAGATNNNNNN NNGCGGGAAGATGAA GACAGATGGTGCAGC CACAG |
| RRV nsp3 specific primers/probe Forward primer: 5’-GTGTTCTCCGGAGGTAAAGATAG-3’ Reverse primer: 5’-TCGCGGCAATAGATGACTAC-3’ Probe: 5’-/56-FAM/ACCTGTTTA/ZEN/CCG CAATGGACACCA/3IABkFQ/−3’ |
IDT | Fox et al.30 |
| CHIKV E1 specific primers/probe Forward primer: 5’-TCGACGCGCCATCTTTAA-3’ Reverse primer: 5’- ATCGAATGCACCGCACACT-3’ Probe: 5’-/56-FAM/ ACCAGCCTG/ZEN/CACCC ACTCCTCAGAC/3IABkFQ/−3’ |
IDT | Zhang et al.95 |
| VEEV nsp3 specific primers/probe Forward primer: 5’-CCATATACTGCAGGGACAAGAA-3’ Reverse primer: 5’-CACTGAAGAGTCGTCGGATATG-3’, Probe: 5’−56’FAM/ATGACTCTC/ZEN/AAGGAAG CAGTGGCT/3IABkFQ/−3’ |
IDT | Bakovic et al.96 |
| Recombinant DNA | ||
| pVRC8400 vector | NIH Vaccine Research Center | Gorman et. al.70 |
| Software and algorithms | ||
| GraphPad Prism v9.0 | GraphPad | RRID:SCR_002798 |
| cryoSPARC v3.3 | Punjani et al.81 | https://cryosparc.com |
| Relion v3.1 | Scheres75 | https://www3.mrclmb.cam.ac.uk/relion/index.php/Main_Page |
| Alphafold2 | Mirdita et al.83 | https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb |
| Phenix v1.20 | Liebschner et al.91 | http://www.phenixonline.org/ |
| CCP4i v7.1 | Winn et al.102 | https://www.ccp4.ac.uk/ |
| Coot v0.9.7 | Emsley and Cowtan86 | https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | Davis et al.93 Chen et al.92 | http://molprobity.biochem.duke.edu/ |
| PDBePISA v1.52 | Krissinel and Henrick94 | https://www.ebi.ac.uk/msd-srv/prot_int/cgibin/piserver |
| Pymol v1.8.6 | Schrödinger, Inc. | https://pymol.org/2/ |
| Chimera v1.14 | Pettersen et al.78 | http://www.cgl.ucsf.edu/chimera/about.html |
| SerialEM | Mastronarde73 | https://www.nexperion.net/serialem |
| Leginon | Carragher et al.74 | https://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage |
| MotionCor2 | Zheng et al.76 | https://emcore.ucsf.edu/ucsf-software |
| GCTF | Zhang77 | https://www2.mrclmb.cam.ac.uk/download/gctf/ |
| ResMap 1.1.4 | Kucukelbir et al.79 | https://resmap.sourceforge.net/ |
| DeepEMhancer | Sanchez-Garcia et al.80 |
https://github.com/rsanchezgarc/deepEMhancer |
| SWISS-MODEL | Waterhouse et al.82 | https://swissmodel.expasy.org/ |
| Alphafold multimer | Jumper et al.84 | https://cosmiccryoem.org/tools/alphafoldmultimer/ |
| SAbPred server | Dunbar et al.85 | https://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/ |
| ISOLDE | Croll89 | https://isolde.cimr.cam.ac.uk/ |
| Flowjo | BD Biosciences | RRID:SCR_008520 |
| FACS Diva | BD Biosciences | RRID:SCR_001456 |
| BioTek Gen5 Software v3.10 | BioTek | RRID:SCR_017317 |
| JMP v.16.2.0 | SAS Institute Inc. | RRID:SCR_014242 |
| Other | ||
| BioTek 405 TS washer | BioTek | RRID:SCR_019725 |
| BioTek Epoch Microplate Spectrophotometer | BioTek | RRID:SCR_019741 |
| LSR Fortessa X-50 Cell Analyzer | BD Biosciences | RRID:SCR_019602 |
| FACS Aria II Cell Sorter | BD Biosciences | RRID:SCR_018934 |
| FACSymphony S6 Cell Sorter | BD Biosciences | Cat#662937–00382906629371 |
| MicroBeta2 Microplate Counter | PerkinElmer | Cat#2450–0120 |
| Illumina MiSeq Next Generation Sequencer | Illumina | RRID:SCR_016379 |
| Illumina HiSeq 2500 | Illumina | RRID:SCR_016383 |
| VP-ITC microcalorimeter | MicroCal⁄Malvern Instruments | N/A |
| Immunospot Plate Reader | Cellular Technology International, Inc. | Cat#S6MACRO |
| SpectraMax M5 | Molecular Devices | RRID:SCR_020300 |
| MagNA Lyser | Roche | RRID:SCR_020506 |
| QuantStudio 3 Real-Time PCR System | Applied Biosystems | RRID:SCR_018712 |
| Traceable digital calipers | Thomas Scientific | Cat# 1235C55 |
| KingFisher™ Duo Prime Purification System | Thermo Scientific | Cat# 5400110 |
Highlights:
Trivalent EEV VLP vaccine elicits antibodies with broad alphavirus activity
mAb SKT05 protects against arthritogenic and encephalitic alphavirus infection
Backbone contacts of sequence-diverse residues enables broad SKT05 recognition
Vaccinated macaques can be used to generate clinically relevant information
Acknowledgements
We thank the study team and the study participants for their contribution to the VRC313 clinical trial. For clinical support, we thank members of the Hope Clinic of the Emory Vaccine Center, and the VRC313 clinical trials team from the Vaccine Research Center. For manufacturing support, we thank the Vaccine Research Center Production Program Laboratory, operated by the Vaccine Clinical Materials Program, Leidos Biomedical Research, Inc. (Frederick, MD). We also thank Ena Tully for experimental assistance with cryo-EM studies, and Jonathan Stuckey for assistance with figure editing and submission. Additionally, we thank Ms. Allison Zeher and Dr. Rick Huang for help with cryo-EM data collection; this work used the NCI/NICE Cryo-EM Facility. We thank James Crowe for providing EEEV-138, EEEV-179, and EEEV-346 IgG, Michael Diamond for providing MAYV and ONNV, and William Klimstra for providing the SINV-EEV chimeric viruses. This study used the Office of Cyber Infrastructure and Computational Biology High Performance Computing cluster at the National Institute of Allergy and Infectious Diseases (NIAID), NIH. This work was supported by the Intramural Research Programs of the Vaccine Research Center and the Division of Intramural Research, NIAID, NIH. This work was funded in part by the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001 and by Federal funds from the National Cancer Institute, NIH, under Contract No. 75N910D00024.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of Interests
NIH has submitted a provisional patent application for select antibodies described in this manuscript on which M.S.S., S.Y.K., R.V., P.D.K., and M.R. are co-inventors. The other authors declare no competing interests.
ADDITIONAL RESOURCES
VRC/NIAID/NIH protocol VRC 313: ClinicalTrials.gov; registry number: NCT03879603
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fox JM, and Diamond MS (2016). Immune-Mediated Protection and Pathogenesis of Chikungunya Virus. J Immunol 197, 4210–4218. 10.4049/jimmunol.1601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaid A., Burt FJ., Liu X., Poo YS., Zandi K., Suhrbier A., Weaver SC., Texeira MM., and Mahalingam S. (2021). Arthritogenic alphaviruses: epidemiological and clinical perspective on emerging arboviruses. Lancet Infect Dis 21, e123–e133. 10.1016/S1473-3099(20)30491-6. [DOI] [PubMed] [Google Scholar]
- 3.Silva LA, and Dermody TS (2017). Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest 127, 737–749. 10.1172/JCI84417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronca SE, Dineley KT, and Paessler S. (2016). Neurological Sequelae Resulting from Encephalitic Alphavirus Infection. Front Microbiol 7, 959. 10.3389/fmicb.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arechiga-Ceballos N, and Aguilar-Setien A. (2015). Alphaviral equine encephalomyelitis (Eastern, Western and Venezuelan). Rev Sci Tech 34, 491–501. 10.20506/rst.34.2.2374. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong PM, and Andreadis TG (2013). Eastern equine encephalitis virus--old enemy, new threat. N Engl J Med 368, 1670–1673. 10.1056/NEJMp1213696. [DOI] [PubMed] [Google Scholar]
- 7.Stromberg ZR, Fischer W, Bradfute SB, Kubicek-Sutherland JZ, and Hraber P. (2020). Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines (Basel) 8. 10.3390/vaccines8020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman-Teran C, Calderon-Rangel A, Rodriguez-Morales A, and Mattar S. (2020). Venezuelan equine encephalitis virus: the problem is not over for tropical America. Ann Clin Microbiol Antimicrob 19, 19. 10.1186/s12941-020-00360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi SL, Russell-Lodrigue KE, Plante KS, Bergren NA, Gorchakov R, Roy CJ, and Weaver SC (2020). Rationally Attenuated Vaccines for Venezuelan Equine Encephalitis Protect Against Epidemic Strains with a Single Dose. Vaccines (Basel) 8. 10.3390/vaccines8030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusnak JM, Dupuy LC, Niemuth NA, Glenn AM, and Ward LA (2018). Comparison of Aerosol- and Percutaneous-acquired Venezuelan Equine Encephalitis in Humans and Nonhuman Primates for Suitability in Predicting Clinical Efficacy under the Animal Rule. Comp Med 68, 380–395. 10.30802/AALAS-CM-18-000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steele KE, and Twenhafel NA (2010). REVIEW PAPER: pathology of animal models of alphavirus encephalitis. Veterinary pathology 47, 790–805. 10.1177/0300985810372508. [DOI] [PubMed] [Google Scholar]
- 12.Weaver SC, and Barrett AD (2004). Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2, 789–801. 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weaver SC, Ferro C, Barrera R, Boshell J, and Navarro JC (2004). Venezuelan equine encephalitis. Annual review of entomology 49, 141–174. 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- 14.Tsetsarkin KA, Vanlandingham DL, McGee CE, and Higgs S. (2007). A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3, e201. 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver SC, Winegar R, Manger ID, and Forrester NL (2012). Alphaviruses: population genetics and determinants of emergence. Antiviral Res 94, 242–257. 10.1016/j.antiviral.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes AC, Basore K, Fremont DH, and Diamond MS (2020). A molecular understanding of alphavirus entry. PLoS Pathog 16, e1008876. 10.1371/journal.ppat.1008876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel RH., Provencher SW., von Bonsdorff CH., Adrian M., and Dubochet J. (1986). Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature 320, 533–535. 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- 18.von Bonsdorff CH, and Harrison SC (1975). Sindbis virus glycoproteins form a regular icosahedral surface lattice. J Virol 16, 141–145. 10.1128/JVI.16.1.141-145.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basore K, Kim AS, Nelson CA, Zhang R, Smith BK, Uranga C, Vang L, Cheng M, Gross ML, Smith J, et al. (2019). Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor. Cell 177, 1725–1737 e1716. 10.1016/j.cell.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M, Chanel-Vos C, and Liao M. (2010). Alphavirus Entry and Membrane Fusion. Viruses 2, 796–825. 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, Rimkunas R, Fong RH, Lin H, Poddar S, et al. (2018). Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574. 10.1038/s41586-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma B, Huang C, Ma J, Xiang Y, and Zhang X. (2021). Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3. Nature 598, 677–681. 10.1038/s41586-021-03909-1. [DOI] [PubMed] [Google Scholar]
- 23.Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, and Rey FA (2004). Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325. 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 24.Williamson LE, Reeder KM, Bailey K, Tran MH, Roy V, Fouch ME, Kose N, Trivette A, Nargi RS, Winkler ES, et al. (2021). Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress. Cell 184, 4430–4446 e4422. 10.1016/j.cell.2021.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, and Rey FA (2010). Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712. 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 26.Kim AS, Kafai NM, Winkler ES, Gilliland TC Jr., Cottle EL, Earnest JT, Jethva PN, Kaplonek P, Shah AP, Fong RH, et al. (2021). Pan-protective anti-alphavirus human antibodies target a conserved E1 protein epitope. Cell 184, 4414–4429 e4419. 10.1016/j.cell.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson LE, Gilliland T Jr., Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, et al. (2020). Human Antibodies Protect against Aerosolized Eastern Equine Encephalitis Virus Infection. Cell 183, 1884–1900 e1823. 10.1016/j.cell.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin J, and Simmons G. (2019). Antiviral Functions of Monoclonal Antibodies against Chikungunya Virus. Viruses 11. 10.3390/v11040305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MKS, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. (2015). Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107. 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox JM, Huang L, Tahan S, Powell LA, Crowe JE Jr., Wang D, and Diamond MS (2020). A cross-reactive antibody protects against Ross River virus musculoskeletal disease despite rapid neutralization escape in mice. PLoS Pathog 16, e1008743. 10.1371/journal.ppat.1008743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rico AB, Phillips AT, Schountz T, Jarvis DL, Tjalkens RB, Powers AM, and Olson KE (2016). Venezuelan and western equine encephalitis virus E1 liposome antigen nucleic acid complexes protect mice from lethal challenge with multiple alphaviruses. Virology 499, 30–39. 10.1016/j.virol.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke CW., Froude JW., Rossi F., White CE., Moye CL., Ennis J., Pitt ML., Streatfield S., Jones RM., Musiychuk K., et al. (2019). Therapeutic monoclonal antibody treatment protects nonhuman primates from severe Venezuelan equine encephalitis virus disease after aerosol exposure. PLoS Pathog 15, e1008157. 10.1371/journal.ppat.1008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke CW, Froude JW, Miethe S, Hulseweh B, Hust M, and Glass PJ (2018). Human-Like Neutralizing Antibodies Protect Mice from Aerosol Exposure with Western Equine Encephalitis Virus. Viruses 10. 10.3390/v10040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim AS, Austin SK, Gardner CL, Zuiani A, Reed DS, Trobaugh DW, Sun C, Basore K, Williamson LE, Crowe JE Jr., et al. (2019). Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat Microbiol 4, 187–197. 10.1038/s41564-018-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swayze RD, Bhogal HS, Barabe ND, McLaws LJ, and Wu JQ (2011). Envelope protein E1 as vaccine target for western equine encephalitis virus. Vaccine 29, 813–820. [DOI] [PubMed] [Google Scholar]
- 36.Dupuy LC, Richards MJ, Livingston BD, Hannaman D, and Schmaljohn CS (2018). A Multiagent Alphavirus DNA Vaccine Delivered by Intramuscular Electroporation Elicits Robust and Durable Virus-Specific Immune Responses in Mice and Rabbits and Completely Protects Mice against Lethal Venezuelan, Western, and Eastern Equine Encephalitis Virus Aerosol Challenges. J Immunol Res 2018, 8521060. 10.1155/2018/8521060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko SY, Akahata W, Yang ES, Kong WP, Burke CW, Honnold SP, Nichols DK, Huang YS, Schieber GL, Carlton K, et al. (2019). A virus-like particle vaccine prevents equine encephalitis virus infection in nonhuman primates. Sci Transl Med 11. 10.1126/scitranslmed.aav3113. [DOI] [PubMed] [Google Scholar]
- 38.Suschak JJ, Bixler SL, Badger CV, Spik KW, Kwilas SA, Rossi FD, Twenhafel N, Adams ML, Shoemaker CJ, Spiegel E, and Hooper JW (2022). A DNA vaccine targeting VEE virus delivered by needle-free jet-injection protects macaques against aerosol challenge. NPJ Vaccines 7, 46. 10.1038/s41541-022-00469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke CW, Erwin-Cohen RA, Goodson AI, Wilhelmsen C, Edmundson JA, White CE, and Glass PJ (2022). Efficacy of Western, Eastern, and Venezuelan Equine Encephalitis (WEVEE) Virus-Replicon Particle (VRP) Vaccine against WEEV in a Non-Human Primate Animal Model. Viruses 14. 10.3390/v14071502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carossino M, Thiry E, de la GA, and Barrandeguy ME (2014). Novel vaccination approaches against equine alphavirus encephalitides. Vaccine 32, 311–319. [DOI] [PubMed] [Google Scholar]
- 41.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, and Nabel GJ (2010). A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat.Med. 16, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pittman PR, Makuch RS, Mangiafico JA, Cannon TL, Gibbs PH, and Peters CJ (1996). Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 14, 337–343. [DOI] [PubMed] [Google Scholar]
- 43.Reed DS., Glass PJ., Bakken RR., Barth JF., Lind CM., da Silva L., Hart MK., Rayner J., Alterson K., Custer M., et al. (2014). Combined alphavirus replicon particle vaccine induces durable and cross-protective immune responses against equine encephalitis viruses. J Virol 88, 12077–12086. 10.1128/JVI.01406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirimotich CM, Vela EM, Garver J, Barnewall RE, Miller BD, Meister GT, and Rogers JV (2017). Chikungunya virus infection in Cynomolgus macaques following Intradermal and aerosol exposure. Virol J 14, 135. 10.1186/s12985-017-0804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidwell RW, and Smee DF (2003). Viruses of the Bunya- and Togaviridae families: potential as bioterrorism agents and means of control. Antiviral Res 57, 101–111. 10.1016/s0166-3542(02)00203–6. [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (2022). HHS/USDA Select Agents and Toxins List. https://www.selectagents.gov/sat/list.htm. [Google Scholar]
- 47.Pittman PR, Liu CT, Cannon TL, Mangiafico JA, and Gibbs PH (2009). Immune interference after sequential alphavirus vaccine vaccinations. Vaccine 27, 4879–4882. 10.1016/j.vaccine.2009.02.090. [DOI] [PubMed] [Google Scholar]
- 48.Reisler RB, Gibbs PH, Danner DK, and Boudreau EF (2012). Immune interference in the setting of same-day administration of two similar inactivated alphavirus vaccines: eastern equine and western equine encephalitis. Vaccine 30, 7271–7277. [DOI] [PubMed] [Google Scholar]
- 49.Coates EE, Edupuganti S, Chen GL, Happe M, Strom L, Widge A, Florez MB, Cox JH, Gordon I, Plummer S, et al. (2022). Safety and immunogenicity of a trivalent virus-like particle vaccine against western, eastern, and Venezuelan equine encephalitis viruses: a phase 1, open-label, dose-escalation, randomised clinical trial. Lancet Infect Dis. 10.1016/S1473-3099(22)00052–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Jose J, Xiang Y, Kuhn RJ, and Rossmann MG (2010). Structural changes of envelope proteins during alphavirus fusion. Nature 468, 705–708. 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fox JM, Roy V, Gunn BM, Huang L, Edeling MA, Mack M, Fremont DH, Doranz BJ, Johnson S, Alter G, and Diamond MS (2019). Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcgammaR interaction on monocytes. Sci Immunol 4. 10.1126/sciimmunol.aav5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Earnest JT, Holmes AC, Basore K, Mack M, Fremont DH, and Diamond MS (2021). The mechanistic basis of protection by non-neutralizing anti-alphavirus antibodies. Cell Rep 35, 108962. 10.1016/j.celrep.2021.108962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollidge BS, Cohen CA, Akuoku Frimpong J, Badger CV, Dye JM, and Schmaljohn CS (2021). Toll-like receptor 4 mediates blood-brain barrier permeability and disease in C3H mice during Venezuelan equine encephalitis virus infection. Virulence 12, 430–443. 10.1080/21505594.2020.1870834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julander JG, Skirpstunas R, Siddharthan V, Shafer K, Hoopes JD, Smee DF, and Morrey JD (2008). C3H/HeN mouse model for the evaluation of antiviral agents for the treatment of Venezuelan equine encephalitis virus infection. Antiviral Res 78, 230–241. 10.1016/j.antiviral.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cain MD, Salimi H, Gong Y, Yang L, Hamilton SL, Heffernan JR, Hou J, Miller MJ, and Klein RS (2017). Virus entry and replication in the brain precedes blood-brain barrier disruption during intranasal alphavirus infection. J Neuroimmunol 308, 118–130. 10.1016/j.jneuroim.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steele KE, Davis KJ, Stephan K, Kell W, Vogel P, and Hart MK (1998). Comparative neurovirulence and tissue tropism of wild-type and attenuated strains of Venezuelan equine encephalitis virus administered by aerosol in C3H/HeN and BALB/c mice. Veterinary pathology 35, 386–397. 10.1177/030098589803500508. [DOI] [PubMed] [Google Scholar]
- 57.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. <j/>(2011).. In Guide for the Care and Use of Laboratory Animals. 10.17226/12910. [DOI] [Google Scholar]
- 60.Powell LA, Miller A, Fox JM, Kose N, Klose T, Kim AS, Bombardi R, Tennekoon RN, Dharshan de Silva A, Carnahan RH, et al. (2020). Human mAbs Broadly Protect against Arthritogenic Alphaviruses by Recognizing Conserved Elements of the Mxra8 Receptor-Binding Site. Cell Host Microbe 28, 699–711 e697. 10.1016/j.chom.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malonis RJ., Earnest JT., Ki AS., Angeliadis M., Holtsberg FW., Aman MJ., Jangra RK., Chandran K., Daily JP., Diamond MS., et al. (2021). Near-germline human monoclonal antibodies neutralize and protect against multiple arthritogenic alphaviruses. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2100104118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpentier KS, Davenport BJ, Haist KC, McCarthy MK, May NA, Robison A, Ruckert C, Ebel GD, and Morrison TE (2019). Discrete viral E2 lysine residues and scavenger receptor MARCO are required for clearance of circulating alphaviruses. Elife 8. 10.7554/eLife.49163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bocan TM, Stafford RG, Brown JL, Akuoku Frimpong J, Basuli F, Hollidge BS, Zhang X, Raju N, Swenson RE, and Smith DR (2019). Characterization of Brain Inflammation, Apoptosis, Hypoxia, Blood-Brain Barrier Integrity and Metabolism in Venezuelan Equine Encephalitis Virus (VEEV TC-83) Exposed Mice by In Vivo Positron Emission Tomography Imaging. Viruses 11. 10.3390/v11111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim AS, Zimmerman O, Fox JM, Nelson CA, Basore K, Zhang R, Durnell L, Desai C, Bullock C, Deem SL, et al. (2020). An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis. Cell Host Microbe 27, 428–440 e429. 10.1016/j.chom.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morrison TE, Whitmore AC, Shabman RS, Lidbury BA, Mahalingam S, and Heise MT (2006). Characterization of Ross River virus tropism and virus-induced inflammation in a mouse model of viral arthritis and myositis. J Virol 80, 737–749. 10.1128/JVI.80.2.737-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, and Vanlandingham DL (2006). Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis 6, 325–337. 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- 67.Ma H, Kim AS, Kafai NM, Earnest JT, Shah AP, Case JB, Basore K, Gilliland TC, Sun C, Nelson CA, et al. (2020). LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314. 10.1038/s41586-020-2915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang LJ, Dowd KA, Mendoza FH, Saunders JG, Sitar S, Plummer SH, Yamshchikov G, Sarwar UN, Hu Z, Enama ME, et al. (2014). Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet 384, 2046–2052. 10.1016/S0140-6736(14)61185-5. [DOI] [PubMed] [Google Scholar]
- 69.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gorman J, Mason RD, Nettey L, Cavett N, Chuang GY, Peng D, Tsybovsky Y, Verardi R, Nguyen R, Ambrozak D, et al. (2019). Isolation and Structure of an Antibody that Fully Neutralizes Isolate SIVmac239 Reveals Functional Similarity of SIV and HIV Glycan Shields. Immunity 51, 724–734 e724. 10.1016/j.immuni.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 71.Wang LT., Pereira LS., Flores-Garcia Y., O’Connor J., Flyn BJ., Scho A., Hurlburt NK., Dillon M., Yang ASP., Fabra-Garcia A., et al. (2020). A Potent Anti-Malarial Human Monoclonal Antibody Targets Circumsporozoite Protein Minor Repeats and Neutralizes Sporozoites in the Liver. Immunity 53, 733–744 e738. 10.1016/j.immuni.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welles HC, King HAD, Nettey L, Cavett N, Gorman J, Zhou T, Tsybovsky Y, Du R, Song K, Nguyen R, et al. (2022). Broad coverage of neutralization-resistant SIV strains by second-generation SIV-specific antibodies targeting the region involved in binding CD4. PLoS Pathog 18, e1010574. 10.1371/journal.ppat.1010574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, and Reilein A. (2000). Leginon: an automated system for acquisition of images from vitreous ice specimens. J Struct Biol 132, 33–45. 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- 75.Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang K. (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 79.Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, and Vargas J. (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874. 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 82.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M. (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunbar J, Krawczyk K, Leem J, Marks C, Nowak J, Regep C, Georges G, Kelm S, Popovic B, and Deane CM (2016). SAbPred: a structure-based antibody prediction server. Nucleic Acids Res 44, W474–478. 10.1093/nar/gkw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 87.Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krissinel E, and Henrick K. (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60, 2256–2268. 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 89.Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530. 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis IW, Murray LW, Richardson JS, and Richardson DC (2004). MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–619. 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krissinel E, and Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 95.Zhang R, Earnest JT, Kim AS, Winkler ES, Desai P, Adams LJ, Hu G, Bullock C, Gold B, Cherry S, and Diamond MS (2019). Expression of the Mxra8 Receptor Promotes Alphavirus Infection and Pathogenesis in Mice and Drosophila. Cell Rep 28, 2647–2658 e2645. 10.1016/j.celrep.2019.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bakovic A, Bhalla N, Kortchak S, Sun C, Zhou W, Ahmed A, Risner K, Klimstra WB, and Narayanan A. (2020). Venezuelan Equine Encephalitis Virus nsP3 Phosphorylation Can Be Mediated by IKKbeta Kinase Activity and Abrogation of Phosphorylation Inhibits Negative-Strand Synthesis. Viruses 12. 10.3390/v12091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, et al. (2013). Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog 9, e1003312. 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mainou BA, Zamora PF, Ashbrook AW, Dorset DC, Kim KS, and Dermody TS (2013). Reovirus cell entry requires functional microtubules. mBio 4. 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ashbrook AW., Burrack KS., Silva LA., Montgomery SA., Heise MT., Morrison TE., and Dermody TS. (2014). Residue 82 of the Chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J Virol 88, 12180–12192. 10.1128/JVI.01672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuhn RJ, Niesters HG, Hong Z, and Strauss JH (1991). Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182, 430–441. 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 101.Krebs SJ, Kwon YD, Schramm CA, Law WH, Donofrio G, Zhou KH, Gift S, Dussupt V, Georgiev IS, Schatzle S, et al. (2019). Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual. Immunity 50, 677–691 e613. 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Trivalent VLP immunization elicits systemic and mucosal humoral immune responses, related to Figures 1–3.
(A-B) Systemic IgG responses in plasma (A) and mucosal IgA dimer responses in BAL (B) were measured by ELISA plates coated with WEEV VLP (left), EEEV VLP (middle), or VEEV VLP (right). Endpoint titers (IgG) and AUC (IgA) are shown as mean ± SEM from four animals per group (top) and from individual animals per group (below). The lower limit of detection is indicated by the black line; red arrows indicate the first four immunizations. (C) Representative flow cytometry gating scheme for PBMC stained with labeled VLP probes and fluorophore labeled α-CD19 and α-IgG mAbs one week after the 5th immunization.
Figure S2. Competition ELISAs identify distinct binding groups of single-specific and triple-specific α-EEV mAbs, related to Figure 2.
(A-D) Competitive binding ELISAs were performed to identify overlap in VLP surface binding areas within ⍺-WEEV mAbs (A), ⍺-VEEV mAbs (B), and ⍺-WEVEEV mAbs using the WEEV VLP (C) and VEEV VLP (D). Competitive mAbs were pre-incubated on VLP-bound plates at a concentration of 10 μg/mL for 30 minutes prior to the addition of titrated biotinylated single-specific (A-B) or triple-specific (C-D) mAbs. Data was subject to hierarchical clustering to determine competition groups. Heat maps display percent inhibition ranging from yellow (minimal competition) to orange (moderate competition) to purple (substantial competition). Colored circles above mAbs on x-axis represent the EEV pseudovirus that was neutralized (red: WEEV; blue: EEEV; green: VEEV). Data are representative of at least two independent experiments.
Figure S3. Memory B cells from broadly neutralizing lineages are rare, related to Figure 2.
(A) Most ⍺-EEV lineages are first observed after three immunizations with WEVEEV VLPs. B cells were sorted from PBMC of the animal with the highest percentage of B cells binding all three VLPs at various time points and bulk IgSeq was conducted to determine timing of lineage development. Assessed time points are denoted by different colors and WEEV endpoint titers for each time point are included in the legend. Despite a similar number of PBMC used at each time point, the number of new triple-specific lineages detected differed. (B) There was a pronounced increase in transcript prevalence following third and fourth immunizations. Transcripts from single- and triple-specific ⍺-EEV mAb lineages are displayed by color as fractions of total sequences observed at different time points following bulk IgSeq. (C) Subjects in clinical trial VRC 313 were immunized intramuscularly with or without alum adjuvant twice, eight weeks apart, with 20 μg of each monovalent WEEV, EEEV, and VEEV VLPs. Six total B cells from two subjects, binding all three VLPs simultaneously, were sorted and assessed for binding and neutralization. (D) ELISA binding curves (left) and pseudovirus neutralization curves (right) of triple-specific ⍺-EEV human mAbs against WEE VLP (top), EEE VLP (middle), and VEE VLP (bottom). Data are displayed as nonlinear fit of log transformed data and representative of two independent experiments. (E) Triple-specific ⍺-EEV human mAbs competed with SKT20, but not SKT05, for binding to VLP. Competition binding ELISA assays were performed with triple-specific human mAbs and representative triple-specific NHP mAbs. Heat maps display percent inhibition where colors range from yellow (minimal competition) to orange (moderate competition) to purple (substantial competition). Data are representative of two independent experiments. (F) Triple-specific human mAbs were tested alongside SKT05 and one single-specific mAb from each VLP for binding to cells infected with SINV-WEEV (top), SINV-EEEV (middle), and SINV-VEEV (bottom). The NHP α-SIV mAb ITS103.01 was used as a negative control. Data are representative of two independent experiments.
Figure S4. Cryo-EM structure of WEEV defines WEVEEV surface diversity, related to Figure 4.
(A) Cryo-EM structure of WEEV at 3.4 Å resolution, shown with previously determined VEEV (PDB 7ffe) and EEEV (PDB 6xo4) structures, which were colored radially like WEEV structure except the outer color being cyan or blue for VEEV or EEEV, respectively. (B) Conformational variability was calculated for each residue of WEEV as average distance between WEEV and VEEV (PDB 7ffe), EEEV (PDB 6xo4). E1 is colored green, E2 – orange. (C) Sequence variation was calculated as BSA weighted average of normalized Shannon’s entropy based on WEEV, VEEV, EEEV, CHIKV, RRV, MAYV, ONNV sequences and mapped on to WEEV. (D) Sequence and conformational variability are plotted for each residue. SKT05/SKT20 epitopes as well as virus receptors LDLRAD3 and MXRA8 epitopes are highlighted.
Figure S5. WEVEEV conformational and sequence variation, related to Figures 4–5.
(A) Sequence variation is plotted against conformational variability for each residue. (B) The agreement between conformational and sequence variation is plotted for each residue as difference between fractions of conformational and sequence variations as follows: absolute value of (sequence variation-conformational variability/5.0). Sequence variation is calculated as normalized entropy and values range from 0 to 1. Conformational variability values larger than 5 Å were replaced with 5.0; the fraction of conformational variability is calculated by normalizing all values by 5.0. (C) RMSD for fusion peptides when E1 proteins were superimposed by Cα atoms. (D) Data from calorimetric titrations of VLPs with different antibodies. (E-F) Thermodynamics of mAb binding to WEEV, EEEV, and VEEV VLPs.
Figure S6. Triple-specific α-EEV mAbs do not neutralize arthritogenic alphaviruses, but bind infected cells and protect in vivo, related to Figure 7.
(A) In vitro neutralization curves for SKT05 against chikungunya virus (CHIKV), Mayaro virus (MAYV), o’nyong’nyong virus (ONNV), and Ross River virus (RRV). The broadly reactive mouse mAb CHK-265 was used as a positive control, while the α-West Nile virus mAb E60 and α-SIV mAb ITS103.01 were used as mouse and NHP IgG negative controls, respectively. Data represents the mean ± SD for two independent experiments performed in triplicate. (B) Broadly neutralizing triple-specific mAbs, as well as broadly binding SKT20 mAb, were tested for binding to cells infected with CHIKV, MAYV, ONNV, and RRV. Vero cells were infected with CHIKV, MAYV, ONNV, or RRV (MOI of 1 for each) for 19hr followed by staining with the indicated mAbs (red). Cells were analyzed by flow cytometry and negative gates were set based on a control mAb (gray). Fluorescence intensity is shown along the x-axis and cell count is shown along the y-axis. Data represent three independent experiments performed in duplicate. (C-D) Four-week-old male C57BL/6J mice (n=8; 2 independent experiments) were administered 200 μg of SKT05 or ITS103.01 (control) one day prior to subcutaneous inoculation with 103 FFU of CHIKV (B) or RRV (C) in the left rear footpad. Viral load was determined in indicated tissues 3 days following inoculation. Statistical significance related to viral RNA was determined by a Mann-Whitney test (*p < 0.05, **p < 0.01, ***p < 0.001). The dotted line indicates the limit of detection for viral RNA analysis.
Figure S7. Humanized SKT05 variants display similar binding and neutralization profiles as SKT05, related to Figure 7.
(A) Strategy for humanization of SKT05. We determined the closest human germline gene to the NHP SKT05 sequence and identified differing framework positions. Structurally important amino acids were not altered, as well as amino acids that were common between NHP and humans. (B) Substitution frequency of SKT05 amino acids compared to most closely related human alleles. Magenta color represents framework positions that were altered. (C) Variable binding affinity of humanized SKT05 variants. We performed ELISA assays on 64 humanized SKT05 variants to determine binding to WEEV VLP (left), EEEV VLP (middle), and VEEV VLP (right). Binding titers of the humanized SKT05 variants with comparable binding affinities to all three VLPs are shown to the right as EC50 and EC80 plots. Binding titers for SKT05 are shown as a red symbol for comparison. Binding curves are displayed as nonlinear fit of log transformed data. Data for the 64 humanized variants represent one experiment. The group of best binding mAbs were selected for further analysis. (D) A total of 14 humanized SKT05 variants with similar binding affinities to all three VLPs as SKT05 were tested for pseudovirus neutralization against WEEV (left), EEEV (middle), and VEEV (right). Neutralization IC50 and IC80 titers are shown to the right with SKT05 shown in red for comparison. Neutralization curves are shown as log transformed nonlinear fit of percent inhibition. Data are representative of two independent experiments performed in triplicate. (E) Ten humanized SKT05 variants were tested for binding to live cells infected with chikungunya virus (CHIKV), Mayaro virus (MAYV), o’nyong’nyong virus (ONNV), or Ross River virus (RRV). Vero cells were infected with CHIKV, MAYV, ONNV, or RRV (MOI of 1 for each) or left uninfected for 16hr followed by staining with a humanized SKT05 variant mAb (red), CHK-265 (blue), or RRV immune ascites fluid (purple). Cells were analyzed by flow cytometry and negative gates were set based on a control mAb (gray). Median fluorescence intensity (MFI) was calculated based on the total distribution of cells. Data are representative of three independent experiments performed in duplicate.
Table S6. Rhesus and human primers for the amplification and sequencing of heavy, kappa, and lambda chain genes from single cells, related to Figure 1.
Data Availability Statement
Data.
All data reported in this paper will be shared by the Lead Contact upon request.
Sequencing data were deposited in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA951758.
The atomic models and cryo-EM reconstructions and generated during this study are available at Protein Data Bank (PDB, https://www.rcsb.org) and Electron Microscopy Data Bank (EMDB, https://www.ebi.ac.uk/emdb/) under the accession codes EMDB: 28117; EMDB: 27757, PDB: 8DWO; EMDB: 27389, PDB: 8DEC; EMDB: 27391, PDB: 8DEE; EMDB: 28119; EMDB: 27722, PDB: 8DUL; EMDB: 28118; EMDB: 27723, PDB: 8DUN; EMDB: 28115; EMDB: 27390, PDB: 8DED; EMDB: 28116; EMDB: 27392, PDB: 8DEF; EMDB: 28056; EMDB: 28058, PDB: 8EEU; EMDB: 28187; EMDB: 27395, PDB: 8DEQ; EMDB: 28188; EMDB: 27396, 8DER; EMDB: 28059; EMDB: 28060, PDB: 8EEV.
Additional information can be obtained from the Lead Contact upon request.
Code.
This paper does not report original code.
Additional Information.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contract upon request.