Abstract
A Gram-staining-negative, oxidase-positive, strictly aerobic rod-shaped bacterium, designated strain PT1T, was isolated from the laboratory-reared larvae of the sea cucumber Apostichopus japonicus. A phylogenetic analysis based on the 16S rRNA gene nucleotide sequences revealed that PT1T was closely related to Neptuniibacter marinus ATR 1.1T (= CECT 8938T = DSM 100783T) and Neptuniibacter caesariensis MED92T (= CECT 7075T = CCUG 52065T) showing 98.2% and 98.1% sequence similarity, respectively. However, the average nucleotide identity (ANI) and in silico DNA-DNA hybridization (DDH) values among these three strains were 72.0%-74.8% and 18.3%-19.5% among related Neptuniibacter species, which were below 95% and 70%, respectively, confirming the novel status of PT1T. The average amino acid identity (AAI) values of PT1T showing 74–77% among those strains indicated PT1T is a new species in the genus Neptuniibacter. Based on the genome-based taxonomic approach, Neptuniibacter victor sp. nov. is proposed for PT1T. The type strain is PT1T (JCM 35563T = LMG 32868T).
Introduction
The genus Neptuniibacter, belonging to the family Oceanospirillaceae, was first proposed by Arahal et al. (2007) with Neptuniibacter caesariensis, which was isolated from surface water from the Eastern Mediterranean [1]. Currently, four species including N. caesariensis have been described in this genus: Neptuniibacter halophilus, isolated from a salt field in southern Taiwan [2], Neptuniibacter marinus and Neptuniibacter pectenicola, both from a scallop hatchery in Norway [3]. Members of the genus Neptuniibacter have been isolated from seawater and/or salt water, and the genus is characterized as being Gram-staining negative, motile, oxidase-positive, strictly aerobic rods requiring NaCl and/or sea salts [1–3].
In the process of collecting reference genomes to understand the structure, function, and dynamics of the sea cucumber microbiota [4–6], the strain PT1T was isolated from the larvae of Apostichopus japonicus [4]. The strain PT1T possesses similar sequences to those of the key Amplicon Sequence Variant (ASV0001-0004), which significantly increased its abundance during the pentactula and juvenile stages found in meta16S analyses of the early developmental stages in sea cucumber [4]. The strain PT1T could have previously unknown interactions with the host sea cucumber. Moreover, PT1T is the only isolate identified to the genus Neptuniibacter among 237 isolates obtained from the early life stage of A. japonicus [4]. Due to the absence of comprehensive studies on the genomic characterization of the genus Neptuniibacter and fewer animal-associated Neptuniibacter isolates, genome fundamentals can contribute not only to host-microbes interaction studies but also to genome taxonomy. Here we report the genome characterization of the newly described Neptuniibacter sp. strain PT1T, and propose it as Neptuniibacter victor sp. nov. based on the genome taxonomy.
Materials and methods
Bacterial strains and phenotyping
The strain PT1T was isolated from the pentactula larvae of A. japonicus reared in a laboratory aquarium in July 2019 [4]. In brief, larvae were collected using 40 μm nylon mesh (FALCON Cell Strainer, Durham, USA), washed once with sterilized seawater, and then manually homogenized in 1 mL filter-sterilized natural seawater for 60 seconds. Ten-fold serial dilutions of the homogenate were cultured on 1/5 strength ZoBell 2216E agar plates [4]. Bacterial colonies were purified using the same agar plates. After purification, the isolate was stored at -80°C suspended in glycerol supplemented 1/5 strength ZoBell 2216E broth. N. halophilus LMG 25378T, N. caesariensis CECT 7075T, N. marinus CECT 8938T, N. pectenicola CECT 8936T were used for genomic and phenotypic comparisons against the strain PT1T. All strains were cultured on Marine agar 2216 (BD, Franklin Lakes, New Jersey, USA). The phenotypic characteristics were determined according to previously described methods and Traitar approach [5–10]. Motility was observed under a microscope using cells suspended in droplets of sterilized 75% artificial seawater [5, 6]. API20 NE (bioMérieux, Craponne, France) was also used to examine phenotypes according to Chen et al. [2].
Due to difficulties in the growth of PT1T in manually prepared marine Oxidative/Fermentative (OF), a salt requirement test, and synthetic basal marine media for evaluating carbon assimilations, we also used the in silico phenotyping tool, Traitar ver. 1.1.2 (Microbial Trait Analyzer) [11], instead of the experimental approach to compare predicted phenotypes based on genome sequences among Neptuniibacter species. This software is capable of predicting 67 phenotypic traits using Prodigal for gene prediction and Pfam family for annotation [11]. The software uses two prediction models, the phypat model (which predicts the presence/absence of proteins found in the phenotype of 234 bacterial species) and a combination of phypat+PGL models (which uses the information of phypat combined with the information of the acquisition or loss of protein families and phenotypes through evolution), to determine the phenotypic characteristics [11].
Whole genome sequencing
Genomic DNAs of PT1T and N. halophilus were extracted from cells grown in Marine Broth 2216 using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Genome sequencing was performed using both Oxford Nanopore Technology (ONT) MinION and Illumina MiSeq platforms. For the ONT sequencing, the library was prepared using Rapid Barcoding Sequence kit SQK-RBK004 (Oxford Nanopore Technologies, Oxford, UK) according to the standard protocol provided by the manufacturer. The library was loaded into a flow cell (FLO-MIN 106), and a 48-hour sequencing run with MinKNOW ver. 3.6.0 software was performed. Basecalling was carried out using Guppy ver. 4.4.1 (Oxford Nanopore Technologies). For Illumina sequencing, a 300 bp paired-end library was prepared using the NEBNext Ultra II FS DNA Library Prep Kit. The ONT and Illumina reads were assembled using Unicycler ver. 0.4.8 [12]. After checking quality value (QV) of these Illumina reads over 27 with no adaptor and barcode sequences assessed using FastQC program ver. 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), these reads were used for genome assembly without preprocessing. Genome sequences of N. caesariensis, N. marinus, and N. pectenicola were retrieved from the NCBI database; the accession numbers are GCF_000153345.1, GCF_001597725.1, and GCF_001597735.1 [1, 3]. The whole genome sequences were annotated with DDBJ Fast Annotation and Submission Tool (DFAST) [13]. The complete genome sequences of PT1T and N. halophilus LMG 25378T acquired in this study were deposited in GenBank/EMBL/DDBJ under accession number AP025763 and AP027292-AP027293, respectively.
Molecular phylogenetic analysis based on 16S rRNA gene nucleotide sequences
The full-length 16S rRNA gene sequence of strain PT1T was obtained from the complete genome sequence. The 16S rRNA gene nucleotide sequences of the type strains of the genus Neptuniibacter and other Oceanospirillaceae species were retrieved from NCBI databases. Sequences were aligned using MEGA X ver. 11.0.11 [14]. A phylogenetic model test and reconstruction of maximum likelihood (ML) tree were performed using the MEGA X ver. 11.0.11 [14, 15]. ML tree was reconstructed with 1,000 bootstrap replications using Kimura 2-parameter (K2) with gamma distribution (+G) and invariant site (+I) model. In addition, nucleotide similarities among strains were also calculated using the MEGA X software [14].
Overall genome relatedness indices (OGRIs)
Overall genome relatedness indices (OGRIs) were calculated to determine the novelty of PT1T using same approach by Yamano et al. [5, 6]. Average nucleotide identities (ANIs) were calculated using the Orthologous Average Nucleotide Identity Tool (OrthoANI) software ver. 0.93.1 [16] using genomes of the PT1T and previously described Neptuniibacter type strains. In silico DNA-DNA hybridization (DDH) values were calculated using Genome-to-Genome Distance Calculator (GGDC) ver. 2.1 with formula 2, being the most robust against incomplete genomes [17]. Average amino acid identities (AAIs) were calculated and compared between PT1T and Neptuniibacter species (Table 1) using an enveomics toolbox [18].
Table 1. List of genomes used in this study.
| Species | Strain | RefSeq/GenBank accession | Size (bp) | No. Of contigs | G+C content | MLSA | AAI |
|---|---|---|---|---|---|---|---|
| Neptuniibacter victor | PT1 T | AP025763 (in this study) | 3,952,146 | 1 | 44.2% | + | + |
| Neptuniibacter halophilus | LMG 25378 T | AP027292-AP027293 (in this study) | 4,113,600 | 2 | 53.3% | + | + |
| Neptuniibacter caesariensis | MED92 T | GCF_000153345.1 | 3,924,755 | 6 | 46.0% | + | + |
| Neptuniibacter marinus | ATR 1.1 T | GCF_001597735.1 | 3,454,191 | 117 | 42.8% | + | + |
| Neptuniibacter pectenicola | LFT 1.8 T | GCF_001597725.1 | 3,631,894 | 203 | 45.7% | + | + |
| Aliamphritea ceti | RA1 T | AP025282 | 5,213,210 | 1 | 47.1% | + | - |
| Aliamphritea hakodatensis | PT3 T | AP025281 | 5,208,344 | 1 | 52.2% | + | - |
| Aliamphritea spongicola | MEBiC05461 T | AP025283 | 4,964,135 | 167 | 51.5% | + | - |
| Amphritea balenae | JAMM1525 T | GCF_014646975.1 | 4,671,747 | 18 | 47.7% | + | - |
| Amphritea pacifica | ZJ14W T | GCF_016924145.1 | 4,702,884 | 282 | 51.2% | + | - |
| Amphritea atlantica | M41 T | AP025284 | 4,804,482 | 1 | 51.1% | + | - |
| Amphritea japonica | JAMM1866 T | AP025761-AP025762 | 3,880,036 | 2 | 47.6% | + | - |
| Amphritea opalescens | ANRC-JH13 T | GCF_003957515.1 | 4,124,452 | 46 | 48.5% | + | - |
| Neptunomonas antarctica | S3-22 T | GCF_900156635.1 | 4,569,005 | 25 | 45.7% | + | - |
| Neptunomonas phycophila | Scap09 | GCF_013394205.1 | 3,976,465 | 1 | 45.5% | + | - |
| Marinomonas arctica | BSI20414 | GCF_014623465.1 | 4,540,024 | 1 | 44.5% | + | - |
| Marinomonas mediterranea | MMB-1 T | GCF_000192865.1 | 4,684,316 | 1 | 44.1% | + | - |
| Marinomonas posidonica | IVIA-Po-181 T | GCF_000214215.1 | 3,899,940 | 1 | 44.3% | + | - |
| Oceanospirillum beijerinckii | NBRC 15445 T | GCF_000422425.1 | 5,325,321 | 99 | 47.8% | + | - |
| Oceanospirillum maris | ATCC 27509 T | GCF_000422865.1 | 3,709,807 | 44 | 45.9% | + | - |
| Oceanospirillum multiglobuliferum | NBRC 13614 T | GCF_900167095.1 | 3,512,709 | 46 | 45.4% | + | - |
| Marinobacterium aestuarii | ST58-10 T | GCF_001651805.1 | 5,191,608 | 1 | 58.8% | + | - |
| Marinobacterium litorale | DSM 23545 T | GCF_000428985.1 | 4,380,400 | 64 | 56.4% | + | - |
| Marinobacterium mangrovicola | DSM 27697 T | GCF_004339595.1 | 4,979,947 | 15 | 57.1% | + | - |
| Nitrincola lacisaponensis | 4CA T | GCF_000691225.1 | 3,412,133 | 43 | 52.1% | + | - |
| Nitrincola tapanii | MEB 193 T | GCF_008368715.1 | 2,793,747 | 19 | 50.8% | + | - |
| Nitrincola tibetensis | xg18 T | GCF_003284585.1 | 4,001,852 | 54 | 46.1% | + | - |
+: used, -: not used.
Multilocus sequence analysis (MLSA)
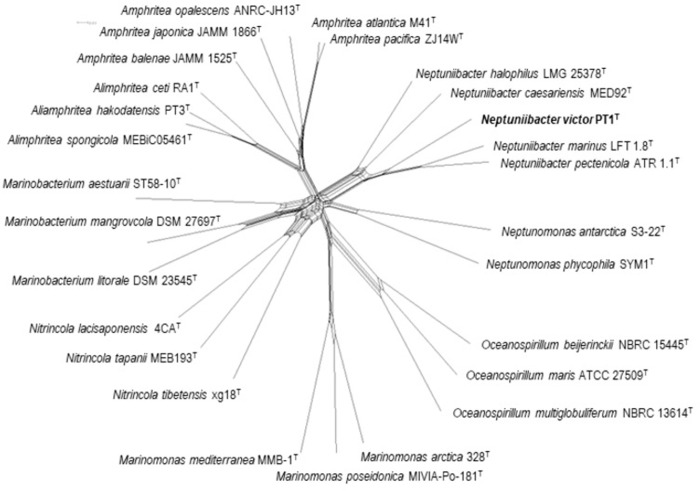
MLSA including concatenation of sequences and phylogenetic network reconstruction were performed using SplitsTree ver. 4.16.2 with the almost same setting [5–10, 19]. The sequences of four protein-coding genes (mreB, recA, rpoA, and topA) were obtained from the genome sequences of PT1T, N. halophilus LMG 25378T, N. caesariensis CECT 7075T, N. marinus CECT 8938T, N. pectenicola CECT 8936T and other related Oceanospirillaceae species (Table 1). These sequences were aligned using Clustal X ver. 2.1 [20]. Regions used for the network reconstruction in Fig 2 were 1–995, 1–1,041, 1–981, and 1–2,648 for mreB, recA, rpoA, and topA, as PT1T nucleotide sequence positions (GenBank accession number AP025763), respectively.
Fig 2. MLSA network of PT1T and related Oceanospirillaceae.
A list of strains and their assembly accession is provided in Table 1. Sequence regions used for reconstructing the network were 1–995 on mreB, 1–1,041 on recA, 1–981 on rpoA, and 1–2,648 on topA. The clade which PT1T belonged was robust with supported by 100% bootstrap.
Pan and core genome analyses
A total of five genomes, including two newly obtained in this study (PT1T and N. halophilus LMG 25378T) and three retrieved from the NCBI database (N. caesariensis, N. marinus, N. pectenicola) were used for pan- and core-genome analyses using the program anvi’o ver. 7 [21] based on previous studies [5, 6, 10, 22], with minor modifications. Briefly, contig databases of each genome were constructed by fasta files (anvi-gen-contigs-database) and decorated with hits from HMM models (anvi-run-hmms). Subsequently, functions were annotated for genes in a contigs database (anvi-run-ncbi-cogs). KEGG annotation was also performed (anvi-run-kegg-kofams). The storage database was generated (anvi-gen-genomes-storage) using all contigs databases and pangenome analysis was performed (anvi-pan-genome). The results were displayed (anvi-display-pan) and adjusted manually.
In silico chemical taxonomy
The genes encoding key enzymes and proteins for the synthesis of fatty acids (Fas), polar lipids, and isoprenoid quinones were retrieved from the genome sequences of PT1T and four previously described Neptuniibacter species using in silico MolecularCloning ver. 7 (In Silico Biology, Yokohama, Japan) based on the same approach of Yamano et al. [5, 6]. The structure and distribution of the genes were also compared using in silico MolecularCloning ver. 7. The 3D-structure of FA desaturase gene products found in Neptuniibacter strains was predicted using Phyre2 ver. 2 [23] with the same approach described previously [5, 6].
Results and discussion
Molecular phylogenetic analysis based on 16S rRNA gene nucleotide sequences
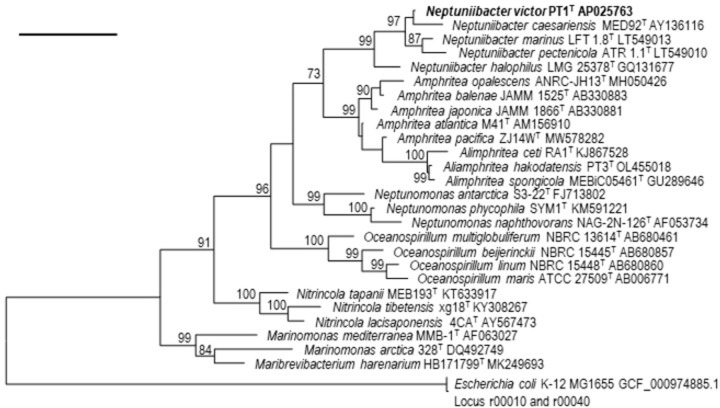
Phylogenetic analysis based on 16S rRNA gene nucleotide sequences showed that the strain PT1T was affiliated to the members of the genus Neptuniibacter showing 96.7–98.2% sequence similarities, which are below the proposed threshold range for species boundary at 98.7% [24, 25]. The strain showed high sequence similarities of 98.2% with N. marinus. The tree also revealed that the PT1T formed a robust monophyletic cluster with N. caesariensis, N. halophilus, N. marinus, and N. pectenicola within the genus Neptuniibacter (Fig 1).
Fig 1. ML tree based on 16S rRNA gene nucleotide sequences of strain PT1T and related type strains.
The ML tree was reconstructed using the K2+G+I model. Numbers shown on branches are bootstrap values (%) based on 1,000 replicates (>70%). A total of 1,284 bp was compared (162–1,446 position in PT1T, GenBank accession number AP025763). 16S rRNA gene nucleotide sequences loci r00010 and r00040 under GenBank accession number GCF_000974885.1 were used as an outgroup to generate this rooted tree. Bar, 0.05 substitutions per nucleotide position.
Genomic features and overall genome relatedness indices (OGRIs)
The ANI values of the PT1T against N. halophilus, N. caesariensis, N. marinus, and N. pectenicola were 72.0%, 72.4%, 74.8%, and 74.7%, respectively (S1 Fig), which are below the species boundary threshold of 95% proposed in previous studies [26]. The DDH values of PT1T against those species were 18.7%, 18.3%, 19.5%, and 19.1%, respectively, which were also below the species delineation threshold (70%). ANI and in silico DDH revealed PT1T as a novel species.
The AAI values of PT1T for against N. halophilus, N. caesariensis, N. marinus, and N. pectenicola were 75.8%, 74.8%, 77.8%, and 77.7%, respectively, which are below the 95% species boundary and above the 64–67% genus boundary, indicating that they are new species within the genus (S2 Fig) [5, 6, 8, 27].
Multilocus sequence analysis (MLSA)
MLSA network showed that PT1T is likely to be monophyletic with previously described Neptuniibacter species but conspecifically separate from them. This result supports the proposal that PT1T is a novel species in the genus Neptuniibacter (Fig 2).
Experimental phenotypic characterization and in silico phenotyping
Several experimental phenotypic characterization tests revealed that PT1T shares several characteristics with Neptuniibacter spp. including being positive for oxidase and growth at 15°C and 30°C [1–3]. PT1T could be differentiated from Neptuniibacter spp. by growth at 4°C, 10°C, and 37°C, catalase reaction, hydrolysis of starch and DNA, nitrate reduction, and utilization of glucose, and DL-malic acid (Table 2).
Table 2. Phenotypic characteristics of PT1T and Neptuniibacter strains.
| Characteristics | N. victor sp. nov. PT1T | N. halophilusT | N. caesariensisT | N. marinusT | N. pectenicolaT |
|---|---|---|---|---|---|
| Growth at | |||||
| 4°C | - | - | - | + | + |
| 10°C | + | - | + | + | + |
| 37°C | - | + | + | - | + |
| 45°C | - | - | - | - | - |
| Catalase | - | - | + | - | + |
| Production of | |||||
| amylase | - | - | - | + | - |
| DNase | w | w | - | nd | nd |
| Nitrate reduction | + | - | - | - | - |
| Utilization of | |||||
| D-glucose | + | - | - | - | - |
| DL-malic acid | - | - | + | + | + |
Phenotypic data for Neptuniibacter are from this and previous studies [1–3]. All strains are positive in growth at 15–30°C, oxidase test. All strains are negative in growth at 45°C, indole production, utilization of L-arabinose, D-mannitol, N-acetyl-D-glucosamine, maltose, and hydrolysis of Tween 20 and 80, and gelatin.
+: present; -: lack; w: weak reaction; nd: not determined.
Unfortunately, PT1T was unable to be grown in manually prepared media commonly used for OF, salt requirement, and carbon assimilation tests [5–8, 10], so we also use in silico phenotyping approach using Traitar software for predicting the phenotype. Before predicting PT1T phenotype, the accuracy of Traitar phenotyping was evaluated to be 81.5% in average using phenotypic characterization from four described Neptuniibacter species (N. halophilus, N. caesariensis, N. marinus, and N. pectenicola) with 40–47 traits compared. The Traitar prediction showed that PT1T was differentiated by nine traits among Neptuniibacter species: utilization of acetate (accuracy 50%), cellobiose (accuracy 100%), maltose (accuracy 75%) and mucate (not determined), hydrolysis of casein (accuracy 50%), production of alkaline phosphatase (accuracy 75%), indole (accuracy 75%), lysine decarboxylase (not determined), and growth at 42°C (accuracy 25%) (S3 Fig). However, no maltose utilization and indole production were observed using the API20NE.
Pan and core genomes and its ecogenomic interpretation
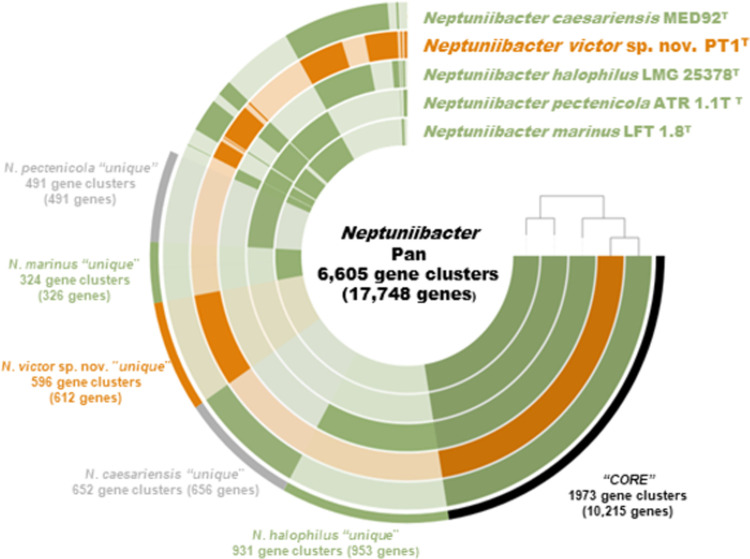
The pangenome of Neptuniibacter species consists of 6,605 gene clusters (17,748 genes) (Fig 3). Genes were classified into Core, present in all strains, and Unique in individual species. Core consisted of 1,973 gene clusters (10,215 genes). COG categories such as J (translation), K (transcription) and E (amino acid transport) were abundant in this bin. Unique in PT1T consists of 596 gene clusters (612 genes). COG categories such as E (amino acid transport system) and I (lipid transport and metabolism) were included in this bin. Genes related to nitrogen metabolism were also detected in Unique in PT1T.
Fig 3. Anvi’o representation of the pangenome of strain PT1T and Neptuniibacter.
Layers represent each genome, and the darker areas represent the occurrence of gene clusters. The outer colored bars indicate the “Core” or “Unique” bins.
Interestingly, PT1T possessed a 45 kb gene island encoding a type VI secretion system (T6SS) and genes (narGHJI and norBC) responsible for performing several steps of denitrification, nitrate and nitric oxide reductions. These genes were grouped into the PT1-Unique genes by pangenomic analysis. Recently, T6SS has been identified as having a role in killing phenotypes of Vibrio fischeri to establish spatial separation in different crypts of a light organ in the host Euprymna scolopes squid [28, 29]. NarGHI is typically identified as a dissimilatory nitrate reductase with a role in nitrate respiration under not only anaerobic but also aerobic conditions [30, 31]. The presence of formate dehydrogenase genes suggests that formate could be served as an electron donor for nitrate respiration in PT1T [30]. Nitrite is further metabolized by a nitrate reductase NirBD to ammonia in the PT1T. The assimilatory nitrate reduction pathway using NasAC and NirA has also been predicted in this strain [32]. Detoxification of nitric oxide (NO), which is a multifunctional immune trigger involving antimicrobial activity, using a nitric oxide reductase, NorBC, has been known as a function in sustaining host-microbes interactions in a wide variety of animals and plants [33]. As PT1 possesses similar sequences to those of the key abundant ASVs associated with the pentactula larvae of A. japonicus, the T6SS with nitrate respiration and NO detoxification genotypes of the PT1T could contribute to competitive dominance in the early life of the benthic host animal. Genes responsible for poly-β-hydroxybutyrate biosynthesis were found in the PT1T genome [34], but the effects of the PT1T on promoting growth in sea cucumber A. japonicus have yet to be observed (data not shown).
In silico chemical taxonomy
As fatty acids, polar lipids, and isoprenoid quinones are common subjects for chemical taxonomic analyses, in silico chemical taxonomy among the Neptuniibacter species including the newly described species of PT1T based on genome sequences, as an alternative to the traditional chemical taxonomy, was performed (S1 Table).
Comparative genome analyses among described Neptuniibacter species referring reported cellular fatty acids of Neptuniibacter species, which are mainly linear, mono-unsaturated or saturated consisting of C16:0, C16:1 and C18:1, with a small amount of C10:0 3-OH (S1 Table) [1–3], reconstructed the basic type II fatty acid biosynthesis (FAS II) pathway driven by fabABFDGIVYZ, which is very similar to that of E. coli [35] (S4–S6 Figs). In particular, long-chain saturated fatty acid (C16:0) is one of the main features of Neptuniibacter fatty acids, comprising approximately 15–26% of the total (S1 Table). C16:0 is one of the main products of the FAS II pathway, suggesting PT1T could also produce C16:0 based on genome comparisons (S6 Fig). In Neptunibacter species, C16:1 and C18:1 together make up over 60% of the total fatty acids (S1 Table), so monounsaturated fatty acids are also major features of the fatty acid profile in PT1T. Monounsaturated fatty acids can be produced through ω7 monounsaturated fatty acid synthesis initiated by isomerization of trans-2-decenoyl-ACP into cis-3-decenoyl-ACP by fabA gene product (S4 Fig). After elongation by fabB gene product, the acyl chain is returned to the FAS II pathway and goes through further elongation, producing C16:1ω7c and C18:1ω7c [5, 6, 36]. The genome comparisons predicted that all strains including PT1T are capable of producing C16:1ω7c and C18:1ω7c by fabA and fabB gene products. 3-hydroxylated FAs, which are the primary fatty acids in lipid A as well as in ornithine-containing lipids, could be supplied by the FAS II pathway since 3-hydroxy-acyl-ACP is known to be normally intermediated in the FAS II elongation cycle [5, 6, 36]. Since no genes responsible for the synthesis of ornithine-containing lipids were found in genomes of PT1T or any other species in either genus, it is likely that 3-hydroxylated FAs in PT1T originate in lipid A [5, 6].
A comparative genome survey of the genes responsible for the FAS II pathway on the PT1T genome reveals the presence of a core gene set, and genomic structures of FAS II core genes are likely to be retained between described Neptuniibacter species (S5 and S6 Figs), which could lead to the conclusion that the novel strain is capable of producing similar FA profiles, mainly consisting of C16:0, C16:1ω7c, and C18:1ω7c. PT1T is also potentially capable of making C10:0 3-OH which is commonly found in this genus because of the presence of the lpxA gene. lpxA is responsible for the incorporation of 3-hydroxyacyl to UDP-N-acetyl-α-D-glucosamine, which is a primary reaction to the biosynthesis of lipid A [37].
Comparative analysis among Neptuniibacter species including PT1T also revealed a complete gene set for the production of PG and PE; plsX, plsY, plsC, cdsA, pssA, psd, pgsA and pgpA (S5 and S6 Figs), which indicate that PT1T produces PG and PE as major polar lipids, similar to other Neptuniibacter species.
The only respiratory quinone reported from previously described Neptuniibacter species is ubiquinone-8 (Q-8) (S1 Table). Biosynthesis of Q-8 consists of nine steps, and Ubi proteins are involved in each reaction; side chain synthesis by ispB gene product, core biosynthetic pathway by ubiC, ubiA, ubiD, ubiX, ubiI, ubiG, ubiH, ubiE, and ubiF, and accessory hypothetical functions by three genes ubiB, ubiJ, and ubiK (S7 Fig) [5, 6, 38]. The set of genes was identified in PT1T genome, of which results strongly suggest that the predominant ubiquinone of PT1T is Q-8 (S7 Fig).
Conclusions
A combination of modern genome taxonomic studies including in silico chemical taxonomy revealed that strain PT1T is a new species in the genus Neptuniibacter. The name Neptuniibacter victor sp. nov. (PT1T = JCM 35563T = LMG 32868T) is proposed.
Description of Neptuniibacter victor sp. nov.
Neptuniibacter victor (vic’tor. L. masc. n. victor, the winner, referring to the predicted ecophysiology of this bacterium possessing T6SS in sea cucumber aquaria, where the bacterium was isolated).
Gram-negative, motile and aerobic rod. Colonies on Marine Agar (BD) are white and 1.0–3.0 mm in diameter after culture for 3 days. No pigmentation and bioluminescence are observed. Growth occurs at 15°C-30°C. Positive for oxidase. Weakly positive for DNase. Negative for growth on a manually prepare basal seawater medium, catalase, hydrolyses of starch, agar, Tween 20 and 80, and gelatin. The newly described species is characterized by positive for nitrate to nitrite, and utilization of D-glucose, using API20NE. The DNA G+C content is 44.2% and the genome size is 3.95 Mb.
The type strain PT1T (= JCM 35563T = LMG 32868T) was isolated from a pentactula larvae of A. japonicus reared in a laboratory aquarium at Hokkaido University, Hakodate, Japan. The GenBank accession number for the 16S rRNA gene sequence of the type strain is LC716006. The complete genome sequence of the strain is deposited in the DDBJ/ENA/GenBank under the accession number AP025763.
Supporting information
PG, phosphatidylglycerol; PE, phosphatidylethanolamine; DPG, diphosphatidylglycerol; PL, phospholipids; AL, aminolipid; PN, phosphoaminolipid; nd: not determined.
(PDF)
(PDF)
Reference genomes were retrieved from NCBI database.
(PDF)
0: negative, 1:phypat positive, 2: phypat+PGL positive, 3: both predictions positive.
(PDF)
Predicted fatty acid synthetic pathway in Neptuniibacter. ACP: acyl-carrier protein; accABCD: acetyl-CoA carboxylase complex; fabA: 3-hydroxyacyl-ACP dehydrase/trans-2-decanoyl-ACP isomerase; fabB: 3-ketoacyl-ACP synthase Ⅰ; fabD: malonyl-CoA: ACP transacylase; fabF: 3-ketoacyl-ACP synthase Ⅱ; fabG: 3-ketoacyl-ACP reductase; fabI: enoyl-ACP reductase Ⅰ; fabV: enoyl-ACP reductase; fabY: 3-ketoacyl-ACP synthase; fabZ: 3-hydroxyacyl-ACP dehydratase; lpxA: glucosamine N-acyltransferase.
(PDF)
(PDF)
Protein/enzyme name each gene is coding: fabA: 3-hydroxyacyl-ACP dehydrase/trans-2-decanoyl-ACP isomerase; fabB: 3-ketoacyl-ACP synthase Ⅰ; fabD: malonyl-CoA: ACP transacylase; fabF: 3-ketoacyl-ACP synthase Ⅱ; fabG: 3-ketoacyl-ACP reductase; fabI: enoyl-ACP reductase Ⅰ; fabV: enoyl-ACP reductase; fabY: 3-ketoacyl-ACP synthase; fabZ: 3-hydroxyacyl-ACP dehydratase; acpP: Acyl-carrier protein; plsX: FA/phospholipid synthesis protein; lpxAD: glucosamine N-acyltransferase; lpxB: lipid-A-disaccharide synthase.
(PDF)
(PDF)
Acknowledgments
We gratefully thank for Professor (Emeritus) Aharon Oren, The Hebrew University of Jerusalem, for his advice on bacterial names.
Data Availability
The GenBank accession number for the 16S rRNA gene sequence of the type strain PT1 is LC716006. The whole genome sequence of the PT1 and Neptuniibacter halophilus LMG 25378T have been deposited to DDBJ/ENA/GenBank under the accession numbers AP025763, and AP027292-AP027293. Raw reads used for those genome assembly have been also deposited at GenBank under accession number DRA015857.
Funding Statement
This study was supported by Kaken 19K22262. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arahal DR, Lekunberri I, Gonzalez JM, Pascual J, Pujalte MJ, Pedros-Alio C, et al. Neptuniibacter caesariensis gen. nov., sp. nov., a novel marine genome-sequenced gammaproteobacterium. Int J Syst Evol Microbiol. 2007; 57:1000–1006. doi: 10.1099/ijs.0.64524-0 [DOI] [PubMed] [Google Scholar]
- 2.Chen MH, Sheu SY, Chiu TF, Chen WM. Neptuniibacter halophilus sp. nov., isolated from a salt pan, and emended description of the genus Neptuniibacter. Int J Syst Evol Microbiol. 2012; 62:1104–1109. doi: 10.1099/ijs.0.030379-0 [DOI] [PubMed] [Google Scholar]
- 3.Dieguez AL, Balboa S, Magnesen T, Romalde JL. Neptuniibacter pectenicola sp. nov. and Neptuniibacter marinus sp. nov., two novel species isolated from a Great scallop (Pecten maximus) hatchery in Norway and emended description of the genus Neptuniibacter. Syst App Microbiol. 2017; 40:80–85. doi: 10.1016/j.syapm.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Sakai Y, Mino S, Sawabe T. Characterization of microbiomes associated with the early life stages of sea cucumber Apostichopus japonicus Selenka. Front Mar Sci. 2022; 9, 37. doi: 10.3389/fmars.2022.801344 [DOI] [Google Scholar]
- 5.Yamano R, Yu J, Jiang C, Haditomo AHC, Mino S, Sakai Y, et al. Taxonomic revision of the genus Amphritea supported by genomic and in silico chemotaxonomic analyses, and the proposal of Aliamphritea gen. nov. PLoS ONE. 2022; 17: e0271174. doi: 10.1371/journal.pone.0271174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamano R, Yu J, Haditomo AHC, Jiang C, Mino S, Romalde JL, et al. Genome taxonomy of the genus Thalassotalea and proposal of Thalassotalea hakodatensis sp. nov. isolated from sea cucumber larvae. PLoS ONE. 2023; 18: e0286693. doi: 10.1371/journal.pone.0286693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Saari N, Gao F, Amin AKMR, Sato K, Sato K, Mino S, et al. Advanced microbial taxonomy combined with genome-based-approaches reveals that Vibrio astriarenae sp. nov., an agarolytic marine bacterium, forms a new clade in Vibrionaceae. PLoS ONE. 2015; 10: e0136279. doi: 10.1371/journal.pone.0136279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amin AKMR, Tanaka M, Al-Saari N, Feng G, Mino S, Ogura Y, et al. Thaumasiovibrio occultus gen. nov. sp. nov. and Thaumasiovibrio subtropicus sp. nov. within the family Vibrionaceae, isolated from coral reef seawater off Ishigaki Island, Japan. Syst Appl Microbiol. 2017; 40:290–296. doi: 10.1016/j.syapm.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 9.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AKMR, Mino S, et al. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol. 2013; 4:414. doi: 10.3389/fmicb.2013.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Hongyu B, Jiang C, Mino S, Meirelles PM, Thompson F, et al. Vibrio taketomensis sp. nov. by genome taxonomy. Syst Appl Microbiol. 2020; 43:126048. doi: 10.1016/j.syapm.2019.126048 [DOI] [PubMed] [Google Scholar]
- 11.Weimann A, Mooren K, Frank J, Pope PB, Bremges A, McHardy AC. From genomes to phenotypes: traitar, the microbial trait analyzer. mSystems. 2016; 1, e00101–e00116. doi: 10.1128/mSystems.00101-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017; 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018; 34:1037–1039. doi: 10.1093/bioinformatics/btx713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018; 35:1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 2020; 33:1870–1874. doi: 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee I, Kim YO, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2015; 66:1100–1103. doi: 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 17.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013; 14:60. doi: 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. 2016; 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1 [DOI] [Google Scholar]
- 19.Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006; 23:254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed]
- 21.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2021; 6:3–6. doi: 10.1038/s41564-020-00834-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018; 6:e4320. doi: 10.7717/peerj.4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015; 10:845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarza P, Yilmaz P, Pruesse E. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014; 12:635–645. doi: 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014; 64:346–351. doi: 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- 26.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018; 68:461–466. doi: 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- 27.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol Res. 2005; 6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spearea L, Cecereb AG, Guckesb KR, Smitha S, Wollenbergc MS, Mandeld MJ, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Aca Sci USA. 2018; 115: E8528–E8537. doi: 10.1073/pnas.1808302115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bongrand C, Ruby EG. The impact of Vibrio fischeri strain variation on host colonization. Curr Opin Microbiol. 2019; 50:15–19. doi: 10.1016/j.mib.2019.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JA, Richardson DJ. Respiration of nitrate and nitrite. EcoSal Plus. 3.2.5, doi: 10.1128/ecosal.3.2.5 [DOI] [PubMed] [Google Scholar]
- 31.Malm S, Tiffert Y, Micklinghoff J, Schultze S, Joost I, Weber I, et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009; 155:1332–1339. [DOI] [PubMed] [Google Scholar]
- 32.Moreno-Vivian C, Cabello P, Martinez-Luque M, Blasco L, Castillo R. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 181:6573–6584. doi: 10.1128/JB.181.21.6573-6584.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Ruby EG. The roles of NO in microbial symbioses. Cellular Microbiol. 2011; 13:518–526. doi: 10.1111/j.1462-5822.2011.01576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki Y, Meirelles PM, Mino S, Suda W, Oshima K, Hattori M, et al. Individual Apostichopus japonicus fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci Rep. 2016; 6:21631. doi: 10.1038/srep21631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janßen HJ, Steinbüchel A. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels. 2014; 7:7. doi: 10.1186/1754-6834-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Lara IM, Geiger O. Formation of fatty acids. In: Timmis KN. Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag Berlin Heidelberg. 2010; pp. 386–393. doi: 10.1007/978-3-540-77587-4_26 [DOI] [Google Scholar]
- 37.Coleman J, Raetz CR. First committed step of lipid A biosynthesis in Escherichia coli: sequence of the lpxA gene. J Bacteriol. 1988; 170:1268–74. doi: 10.1128/jb.170.3.1268-1274.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abby SS, Kazemzadeh K, Vragniau C, Pelosi L, Perrel F. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim Biophys Acta Bioenerg. 2020; 1861:11. doi: 10.1016/j.bbabio.2020.148259 [DOI] [PubMed] [Google Scholar]