Summary
Background
Chronic active lesions (CAL) in multiple sclerosis (MS) have been observed even in patients taking high-efficacy disease-modifying therapy, including B-cell depletion. Given that CAL are a major determinant of clinical progression, including progression independent of relapse activity (PIRA), understanding the predicted activity and real-world effects of targeting specific lymphocyte populations is critical for designing next-generation treatments to mitigate chronic inflammation in MS.
Methods
We analyzed published lymphocyte single-cell transcriptomes from MS lesions and bioinformatically predicted the effects of depleting lymphocyte subpopulations (including CD20 B-cells) from CAL via gene-regulatory-network machine-learning analysis. Motivated by the results, we performed in vivo MRI assessment of PRL changes in 72 adults with MS, 46 treated with anti-CD20 antibodies and 26 untreated, over ∼2 years.
Findings
Although only 4.3% of lymphocytes in CAL were CD20 B-cells, their depletion is predicted to affect microglial genes involved in iron/heme metabolism, hypoxia, and antigen presentation. In vivo, tracking 202 PRL (150 treated) and 175 non-PRL (124 treated), none of the treated paramagnetic rims disappeared at follow-up, nor was there a treatment effect on PRL for lesion volume, magnetic susceptibility, or T1 time. PIRA occurred in 20% of treated patients, more frequently in those with ≥4 PRL (p = 0.027).
Interpretation
Despite predicted effects on microglia-mediated inflammatory networks in CAL and iron metabolism, anti-CD20 therapies do not fully resolve PRL after 2-year MRI follow up. Limited tissue turnover of B-cells, inefficient passage of anti-CD20 antibodies across the blood–brain-barrier, and a paucity of B-cells in CAL could explain our findings.
Funding
Intramural Research Program of NINDS, NIH; NINDS grants R01NS082347 and R01NS082347; Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; Cariplo Foundation (grant #1677), FRRB Early Career Award (grant #1750327); Fund for Scientific Research (FNRS).
Keywords: Susceptibility-based MRI, Paramagnetic rims, Anti-CD20 treatment, Machine learning, Single cell RNA sequencing
Research in context.
Evidence before this study
B-cells are crucial in multiple sclerosis (MS) pathogenesis. B-cell depletion with anti-CD20 antibodies is a successful treatment strategy with profound suppression of clinical relapses and radiological acute inflammatory lesions. This treatment approach has also been approved for some progressive MS cases. Nonetheless, re-analysis of clinical trials has highlighted that anti-CD20 agents do not prevent clinical progression independent of relapse activity (PIRA), a process that might be associated with both smoldering inflammation and relentless neurodegeneration in MS tissue.
Added value of this study
This study addresses two critical questions for MS pathophysiology and treatment: (1) Which lymphocyte types (including CD20 B-cells) are important to sustain microglia/dendritic cell-mediated chronic inflammation at the edge of chronic active MS lesions? (2) Can we target this smoldering unresolved inflammatory process using anti-CD20 therapy? We find that, although they are relatively sparse, CD20 B-cells have the potential to modulate the microglia-mediated inflammatory network at the chronic active lesion, as their in-silico depletion is predicted to affect genes highly expressed in microglia and dendritic cells together with their inflammatory signaling pathways. Despite this predicted effect, an in vivo longitudinal MRI study showed that anti-CD20 B-cell antibody does not resolve chronic active MRI lesions over a median of 2 years of follow-up. Consistently, in this cohort of patients, PIRA events (disability worsening independent of relapse activity) over the follow up were more common in individuals featuring chronic inflammatory MRI activity (≥4 paramagnetic rim brain lesions, or PRL).
Implications of all the available evidence
Our study fills the gap between the potential promising role of anti-CD20 therapies on chronic active MS inflammatory lesions and the limited effect of these drugs on clinical PIRA. In particular, our MRI data suggest that peripheral B-cell depletion does not sufficiently reduce the activity of iron-laden microglia at the edge of chronic active lesions. In these lesions, smoldering inflammatory demyelination and axonal injury are compartmentalized behind a virtually closed blood–brain barrier. Limited tissue turnover of CD20 B-cells, inefficient passage of anti-CD20 antibodies across the blood–brain-barrier, and a paucity of B-cells in chronic active lesions could explain our findings.
Introduction
MS is a chronic immune-mediated disease in which central nervous system (CNS)-reactive T- and B-lymphocytes play a critical pathogenic role. Unlike the relatively stereotyped processes that lead to multifocal inflammatory demyelinating lesions and clinical relapse in so-called “active” MS, the pathogenesis of progressive MS (and of PIRA in particular) is likely pleotropic.1 Among the relevant processes, chronic compartmentalized tissue inflammation, which is most prominent in chronic active/smoldering white matter (WM) lesions and in the leptomeninges, is recognized to play a critical role.2
The surprising efficacy of B-cell depletion treatment in MS with anti-CD20 antibodies has led to reconsideration of the B-cell lineage contribution in MS pathogenesis. A variety of mechanisms have been considered, including immunoglobulin production, antigen presentation, secretion of cytokines and other inflammatory molecules3; B cells also harbor certain viral pathogens.4 Anti-CD20 antibodies deplete various stages of maturing B-cells, thereby affecting B-cell–T-cell interactions and pro-inflammatory myeloid cell responses.5 Randomized clinical trials have consistently shown dramatic reduction of new lesion formation and annualized relapse rate in relapsing-remitting MS.6,7 Ocrelizumab, a humanized anti-CD20 antibody, is also licensed for primary progressive MS, and both ocrelizumab and ofatumumab are licensed for secondary progressive MS with active MRI.8,9 Nonetheless, clinical progression independent of relapse activity (termed “PIRA”) has been identified in MS patients treated with highly effective treatments, including ocrelizumab.10
To understand the potential impact of lymphocyte depletion or manipulation in the CNS, we analyzed in detail the distribution of lymphocyte subpopulations in MS tissue according to different pathological stages by reanalyzing published single-nucleus transcriptome profiles from 3 studies.11, 12, 13 Using computational and machine learning techniques on the immune gene regulatory network at the chronic active lesion (CAL) edge, we predicted the effects of specific lymphocyte subpopulation depletion strategies, including of CD20 B-cells, on arresting chronic inflammation in MS lesions.
As our in silico analysis suggested that CD20 B-cell depletion would have profound effects on such lesions, we analyzed data from clinical cohorts followed at 4 academic medical centers to investigate in vivo the effect of anti-CD20 antibody therapy on paramagnetic rim lesions (PRL), also known as iron rim lesions (IRL),14 a subset of CAL that can be detected in vivo using susceptibility-sensitive MRI. It is now well established that the burden of CAL strongly correlates with disease severity and progression.15,16 Since recent longitudinal MRI studies have reported that the paramagnetic rim is a dynamic biomarker of perilesional chronic inflammation and tends to fade on MRI over a median of 7 years,11,17 we specifically evaluated whether the paramagnetic rim of PRL disappeared in anti-CD20 treated patients over the follow up, and we quantified longitudinal PRL evolution using quantitative susceptibility mapping (QSM) for estimating lesion magnetic susceptibility (paramagnetic rim and myelination status), T1 mapping for estimating lesion microstructural damage (both myelination status and axonal loss), and lesion volume for estimating lesion expansion or shrinkage over time.
Methods
Single-cell lymphocyte phenotyping in MS brain tissue
We downloaded the raw snRNA-seq data or the gene expression matrix available from 3 single-nucleus RNA sequencing (snRNAseq) studies,11, 12, 13 in which nuclei were extracted from MS autopsy snap-frozen brain tissue classified according to the presence of lesions at different pathological stages, such as active, chronic active, chronic inactive, lesion core, and periplaque WM, and from non-neurological control brain tissue. To have a representative but non-comprehensive comparison between brain vs blood/CSF lymphocyte distribution, a fourth available single-cell RNA sequencing (scRNAseq) dataset was also downloaded and re-analyzed.18 The datasets were initially analyzed individually using a bioinformatic pipeline (Seurat v4 R-based package) previously described in detail.11
For each dataset, UMAP plots were generated, and cell-type identification was performed using known lineage markers. A subset of the nuclei belonging to the immune cell clusters was performed using the “subset” function in Seurat. To refine annotation of the different lymphocyte subpopulations, the newly obtained Seurat object (one for each dataset) was used as input into the Azimuth computational platform (https://azimuth.hubmapconsortium.org) and mapped onto the provided multimodal single cell reference atlas (RNA and protein) of circulating immune mononuclear cells (n = 161,764).19 Only those lymphocytes with high mapping score >0.7 (reflecting the confidence associated with a specific annotation) and/or prediction score >0.7 (reflecting how well represented the cell is by the reference atlas) were retained for further analysis. The mapping and prediction scores provided by Azimuth range from 0 to 1, and a threshold of 0.7 was chosen based on the fact that we are mapping nuclei transcriptomes onto an atlas derived from whole cells.
In silico depletion of selected lymphocyte subpopulations and virtual knockout of the BTK gene at the chronic active lesion edge
The immune gene regulatory network (GRN) was generated starting from the immune cell snRNAseq dataset from chronic active lesion edge samples,11 and included 6266 genes. The simulated effect of depleting sparse lymphocyte subpopulations (MS4A1 (CD20)-expressing B-cells, plasmablasts, and T-cell subtypes, separately), on the original GRN, was gauged using scTenifoldNet, a machine learning tool for comparative single-cell network analysis implemented in an R package (https://github.com/cailab-tamu/scTenifoldNet).20 Briefly, the scTenifoldNet workflow takes two snRNA-seq expression matrices as inputs. The analysis aims to identify genes whose transcriptional regulation is shifted between the two conditions. The whole workflow consists of five steps: cell subsampling, network construction, network denoising, manifold alignment, and module detection. The final output is a table of genes with associated Euclidean distance computed between the coordinates of the same gene in both datasets. To help contextualize the results, we also simulated the analysis by comparing the original dataset against itself. As expected, no significant distance was produced. Using the same dataset,11 we also simulated the knockout of the BTK gene, known to be expressed by both microglia and B-cell lineage cells,21 and a similar workflow was implemented using scTenifoldKnk (https://github.com/cailab-tamu/scTenifoldKnk).20
A principal components analysis of the simulated distances from comparing the original immune GRN and the same network after removing specific cell populations or genes was computed. The Euclidean distance between the different comparisons was plotted as a color-coded heatmap. Functional enrichment analysis of gene sets was performed on the subset of simulated significant perturbed genes (p < 0.05) using Enrichr (KEGG Human and MsigDB Hallmarks annotations),22, 23, 24 and the top 20 terms were evaluated for biological interpretation.
Longitudinal clinical-MRI study
Study participants
Imaging, laboratory, and clinical data were prospectively collected under institutional review board-approved protocols in adults with MS from 4 academic research hospitals: the NIH Clinical Center (Bethesda, MD, USA), the Johns Hopkins University Hospital (Baltimore, MD, USA), and the Erasme and Saint Luc University Hospitals (Brussels, Belgium). These protocols allow exploratory analysis and pooling of collected data. Clinical data were obtained from experienced MS clinicians at baseline and at each MRI follow-up. Inclusion criteria for the current retrospective analysis were the following: age 18, diagnosis of MS based on the 2017 McDonald criteria, and availability of a high-resolution susceptibility-based MRI scans acquired longitudinally either before and after anti-CD20 treatment or in the absence of disease-modifying treatment (DMT). Since previous studies have shown that PRL are present in patients receiving DMTs,15,16 washout of a prior DMT was not required here as an inclusion criteria. For the untreated cohort, the baseline MRI scan was the first scan with available high-resolution susceptibility-based MRI, whereas in the anti-CD20 cohort the baseline scan was the latest MRI before treatment administration.
Progression independent of relapse activity (PIRA) was defined as an increase in the EDSS score confirmed at least after six months and occurring in the absence of clinical relapses, of ≥1.5 points if baseline EDSS was 0, ≥1.0 point if baseline EDSS was 1.0–5.5, or ≥0.5 points if baseline EDSS was ≥6.0.25
MRI acquisition and analysis
MRI studies were performed on five 3-T MRI scannerss: 3 Philips Intera or Ingenia scanners (Philips Medical Systems, The Netherlands), 1 S Skyra scanner (Siemens AG, Germany) and 1 General Electric Signa Premier (GE Healthcare, Waukesha, Wisconsin, USA).
-
•
On all scanners, submillimeter isotropic 3D T2∗-weighted segmented echo-planar-imaging (EPI),24, 25, 26 providing magnitude and phase images (from which QSM were derived), was acquired before or during intravenous injection of a single dose (0.1 mmol/kg) of gadobutrol (Gadavist; Bayer Healthcare, Leverkusen, Germany). The same 3D T2∗-EPI sequence16,26 was adapted and optimized to the different MRI scanners with minimal parameter modification (sequence parameters are listed in Supplementary Table S1).
-
•
Additional routine MRI images acquired for clinical use, including 3D T2-FLAIR images and postcontrast T1-weighted sequences, were not standardized across the scanners.
-
•
On the 3T Siemens Skyra and GE Premier (NIH and CUSL, respectively), a 3D MP2RAGE providing T1w images and estimated T1 maps, was acquired (sequence parameters are listed in Supplementary Table S2).
PRL assessment
In each individual and for each MRI timepoint, the number of supratentorial chronic non-enhancing PRL on unwrapped filtered phase images was determined by consensus of 2 raters (PM and MA) blinded to all patient information, including treatment status. A chronic non-enhancing MS lesion was defined as a PRL if it demonstrated a paramagnetic rim on unwrapped filtered phase images,27 and an isointense/slightly paramagnetic core when compared to the extralesional WM. Specific rim features include: (1) colocalization with the edge of an MS lesion on T2-FLAIR or T1-weighted images; (2) visibility on at least two planes for 3D images; and (3) coverage of at least 2/3 of the lesion WM edge on the slice of maximum visibility. 3D T2-FLAIR images were rigidly coregistered to the high-resolution 3D EPI scan. PRL and comparable non-PRL were manually segmented on coregistered T2-FLAIR images. Whenever possible, at least 1 PRL and 1 non-PRL (the latter selected in the contralateral WM) were segmented for each participant.
Longitudinal lesion volume, susceptibility, and T1 times
All included patients underwent longitudinal 3T MRI scans. QSM images were reconstructed from the 3D EPI magnitude and phase images using a MATLAB toolbox (JHU/KKI_QSM_Toolbox_v3.0).28 After selecting the magnitude and raw phase images as input, the following parameters available in the toolbox menu were implemented: phase unwrapping method: Laplacian; threshold for the brain mask = 0.5; background removal: V-SHARP, and iTKD to calculate susceptibility. T1 maps were estimated from the 3D MP2RAGE sequence. For each segmented PRL and non-PRL, longitudinal volume, susceptibility, and, when MP2RAGE was available, T1 times, were determined. 3D T2-FLAIR and 3D MP2RAGE images were rigidly coregistered to the high-resolution 3D EPI scan for lesion volume and T1 time assessment. QSM images were reconstructed from the 3D EPI magnitude and phase images and, thus, naturally coregistered to the 3D EPI scan. Similarly to previous studies,29,30 by segmenting the whole lesion area (vs only the lesion edge), we expected to capture on QSM and T1 maps not only the paramagnetic changes at the edge of PRL, but also the overall tissue microstructural damage, including demyelination and axon loss, occurring within the lesion core.
Statistics
Demographic, clinical, and MRI differences were assessed with ANOVA and Tukey's post-hoc multiple-comparison test, or with Fisher's exact or chi-squared test, as appropriate. Differential abundance analysis of lymphocyte subpopulations in different pathological conditions was performed using propeller,31 a statistical tool designed to take into account the sample variability in single cell genomics data. The association between treatment status and log-lesion volume, susceptibility, and T1 time changes (dependent variables) over time was tested in 3 distinct multivariable linear mixed models with random lesion and participant effect. Lesion category (PRL vs non-PRL), treatment status (treated vs untreated with anti-CD20 therapy), and time (time post baseline MRI acquisition) were considered as fixed effect variables, with interaction between lesion category and time and between lesion category and treatment status. The mixed-model analysis was restricted to a maximum of 4 years MRI follow up (excluded 24/1885 datapoints) to avoid results driven by few datapoints with longer follow up. As a sensitivity analysis, (1) the 3 models were re-run including the clinical-MRI center as fixed effect, and (2) the 3 models were also re-run separately for PRL only and non-PRL only, with and without clinical-MRI center as fixed effect. Statistical power considerations were based on previous calculations on a 7T longitudinal dataset suggesting that in a 1-year trial, 16 patients per arm (treated vs untreated, with a total of 112 PRLs) would be sufficient to detect a 10% treatment effect (fading or disappeared paramagnetic rim) with 80% power.11
Ethics
Study procedures received approval from an ethical standards committee on human experimentation in each of the four academic research hospitals (NIH IRB 89N0045; Johns Hopkins IRB CIR00086166; Brussels Saint-Luc IRB B4032020000104; Brussels Erasme IRB 8406201628764), and written informed consent was obtained from all participants prior to participation.
Role of funders
This work was supported by the Intramural Program of NINDS of National Institutes of Health, by the National Institutes of Health (R01NS082347 and R01NS082347), by the Fund for Scientific Research (FRS, FNRS; grant #40008331), by the Cariplo Foundation (grant #1677) and by the FRRB Early Career Award (grant #1750327). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov) and the research cluster of IRCSS San Raffaele Hospital, Milan, Italy. The funding sources had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Few CD20 B-cells are seen at the chronic active lesion edge
To assess B-cell subtype presence and overall lymphocyte distribution in MS lesions at different histopathological stages (active, chronic active, chronic inactive, lesion core, and periplaque WM), we re-analyzed 3 available snRNAseq brain datasets11, 12, 13 encompassing a total of 123,193 nuclei from 21 progressive MS cases (10 women, mean age 46.7 years, age range 34–58) and 17 non-neurological controls (6 women, mean age 55 years, age range 34–82).
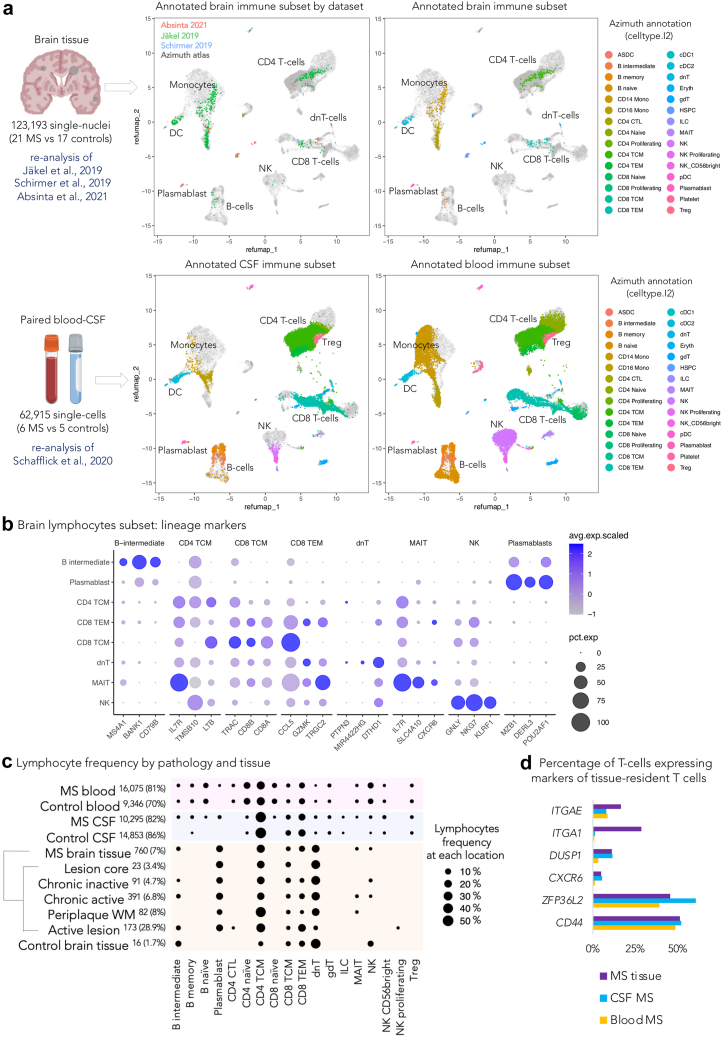
We identified a total of 776 lymphocytes from 10,913 immune cells (7.1%), of which 391 were from chronic active lesions (391 of 5743 immune cells, 6.8%). As expected, the highest frequency of lymphocytes was identified in active lesions (173 of 599 immune cells, 28.9%), while the fewest were in control brain tissue (16 of 921 immune cells, 1.7%). Since the low number of lymphocytes in MS tissue prevented any reliable unsupervised subclustering analysis, lymphocyte identity was refined by implementing a multimodal reference atlas derived from >160,000 circulating mononuclear immune cells.19 Fig. 1 shows the refined annotation of the immune cell subsets from the MS brain, CSF, and blood onto the reference atlas (Fig. 1a) as well as lineage marker genes of the different brain population subtypes (Fig. 1b).
Fig. 1.
Single-cell transcriptome lymphocyte phenotyping reveals few CD20 B-cells in MS brain tissue. (a) Refined annotation of lymphocyte subsets (color-coded) in the brain, CSF, and blood from the re-analysis of available single nucleus or single cell transcriptomic datasets11, 12, 13,18 using the Azimuth reference atlas (gray dots).19 (b) Dot plot depicting selected lineage genes for each brain lymphocyte subpopulation. Dot size corresponds to the percentage of nuclei expressing the gene in each cluster, and color represents the average expression level as shown in the legend. (c) Dot plot depicting the percentage of lymphocyte subsets in brain tissue, CSF, and blood from MS cases vs non-neurological controls. Dot size corresponds to the percentage of lymphocytes as shown in the legend. Data were filtered to show only lymphoid populations with prevalence of >0.1% to preserve readability. The total number of lymphocytes and their percentage relative to mononuclear immune cells (in brackets) are reported for each location/tissue. Activated T-cells (CD4 TCM, CD8 TEM, and dnT) and plasmablasts are preferentially represented in MS brain tissue. The lymphoid profile in CSF and blood is enriched by additional lymphoid populations, including naïve and memory B-cells as well as naïve T-cells. Circulating plasmablasts are seen almost exclusively in CSF and blood from MS, but not non-neurological controls. (d) Bar graph showing the percentages of T-cells expressing gene markers of resident memory T-cells in the 3 compartments (brain vs CSF vs blood) in MS vs non-neurological controls. Abbreviations: MS: Multiple sclerosis; CSF: Cerebrospinal fluid; WM: White matter; CTL: Cytotoxic T cells; TCM: Central memory T cells; TEM: Effector memory T cells; dnT: CD4- CD8-double negative T cells; gdT: Gamma delta T cells; ILC: Innate lymphoid cells; MAIT: Mucosal-associated invariant T cells; T cell NK: natural killer; Treg: Regulatory T cells.
In chronic active lesions, B-cells expressing CD20 (MS4A1 gene) accounted for only 4.3% of all lymphocytes (a total of 17 intermediate B-cells), while a relatively high proportion of terminally differentiated B-cells were seen as antibody-producing plasmablasts (20.5% of lymphocytes). The remaining 74.2% of lymphocytes at this site were activated T cells (CD3+CD4-CD8-double negative T cells [dnT], 33.8%; CD4 T memory cells, 23.5%; CD8 T effector, 10.2%; CD8 T memory cells, 4.9%; mucosal-associated invariant T cell [MAIT], 1.8%) and natural killer (NK) cells (1%).
Overall, except for active lesions, which had additional populations (such as CD4 T effector lymphocytes and proliferating NK cells), differences between pathologically defined chronic active and chronic inactive lesion stages related to the total number of lymphocytes (mean lymphocytes/sample 32 and 10, respectively, t-test p = 0.03) rather than to the different proportions of lymphoid subtypes (ANOVA false discovery rate [FDR] >0.05 for all the lymphocyte subsets, Fig. 1c, Supplementary Table S3a). Scarce MAIT cells (<10 cells) were seen only in chronic active lesions and periplaque, although this result did not reach statistical significance (Supplementary Table S3). No innate lymphoid cells (ILC) or naïve lymphocytes were identified in the brain tissue, and plasmablasts were seen exclusively in the MS brain (vs control brain). Statistically significant expansion of intermediate B cells, plasmablasts and NK cells was seen in the CSF from MS vs control samples (Supplementary Table S3b); while no significant differences were seen in the blood for all lymphocyte subtypes (Supplementary Table S3c).
We then looked specifically for genes known to be expressed in tissue resident memory T-cells, a category of long-lived, non-circulating lymphocytes not depicted by the implemented immune cell reference atlas but recognized as relevant in autoimmune diseases.32, 33, 34 Of interest, among those genes, ITGA1 (CD49a binding type IV collagen) was expressed by 28.5% of T-cells within the brain but was not expressed in the T-cells from CSF or blood (Fig. 1d). In line with a recent study,35 within the MS lesions (regardless of the pathological stage) and MS CSF, we found a higher proportion of T-cells expressing markers of tissue resident memory cells than in the MS blood, such as CXCR6, ZFP36L2 and DUSP1 (Fig. 1d), or markers of interaction with the extracellular matrix, such as CD44 (receptor for hyaluronan; 51.2% of tissue T cells), and ITGAE (or CD103 binding E-cadherin; 16.4% of tissue T-cells, ∼2-time more frequent than in the MS blood).
Predicted effect of in silico depletion of lymphocyte subpopulations on immune gene regulatory networks in chronic active MS lesions
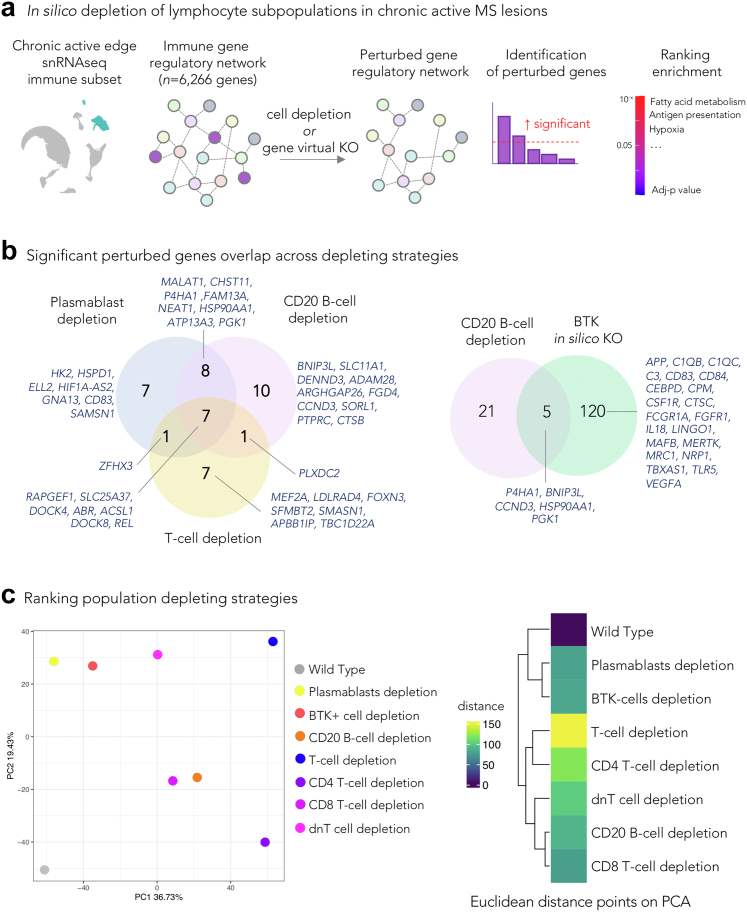
We then assessed whether an efficient CD20 B-cell depletion within the brain tissue could, at least in principle, affect communication within the complex network of immune cells at the chronic active lesion edge.11 To this end, from the immune cell snRNAseq dataset of the chronic active lesion edge samples,11 we generated multiple immune gene regulatory network (GRN) datasets by in silico removal of specific subsets of lymphocytes (CD20 B-cells, plasmablasts, and T-cell subtypes, separately), or by performing virtual knockout of the Bruton tyrosine kinase (BTK) gene, expressed by both microglia and B-cell lineage. Fig. 2a shows the in silico analysis overview that is fully detailed in the Methods.
Fig. 2.
Prediction of the effects of depleting lymphocyte subpopulations in chronic active lesions. (a) Schematic workflow: starting from a snRNAseq dataset of the chronic active MS lesion edge, immune GRNs were computed and compared with GRN versions depleted of specific cell populations or of a gene of interest, using a machine learning tool for comparative single-cell network analysis (scTenifoldNet20 for comparing the effect of cell removal and scTenifoldKnk21 for the virtual knockout). The simulated significantly differentially perturbed features (genes) in the GRN comparison were analyzed for functional enrichment analysis. (b) Venn diagrams showing overlaps of significantly affected genes by in silico lymphocyte or BTK depletion. Most of these genes are known to regulate microglia or dendritic cell activities. (c) PCA plot and heatmap of the simulated distances from comparing the GRN of the original immune dataset vs the same dataset after removing specific cell populations. Wild type is the product of comparing the original dataset with itself using scTenifoldNet. This comparison serves as a reference for a null difference. The result suggests that the removal of T cells (followed by CD4 T-cells) should have a larger impact on the immune GRN at the chronic active lesion edge than removing other lymphocytes subpopulations or inhibiting BTK. The value in the tile of the heatmap is the Euclidean distance among the different comparisons; all distances are relative to the wild type comparison. The result supports the one presented in the PCA. Abbreviations: GRN: Gene regulatory network; BTK: Bruton's tyrosine kinase; dnT: CD4- CD8-double negative T cells; PCA: Principal component analysis.
In all virtual lymphocyte-depleting conditions, most of the significantly affected genes were markers of microglia and dendritic cells (common genes across depletion strategies: RAPGEF1, SLC25A37, DOCK4, ABR, ACSL1, DOCK8, REL), supporting the critical role of lymphocytes in amplifying the inflammatory response of microglia/dendritic cells in tissue (Fig. 2b for overlapping genes). CD20 B-cell depletion was predicted to specifically perturb genes involved in iron/heme metabolism (BNIP3L, SLC25A37, CCND3, CTSB), mitotic spindle (ABR, FGD4, DOCK4), hypoxia (BNIP3L, P4HA1, PGK1), and antigen presentation (HSP90AA1, CTSB).
Additional enriched terms were identified when plasmablasts or T-cell subtypes were selectively depleted, including fatty acid biosynthesis and degradation, ferroptosis, TNF via NFKB signaling, PPAR pathway, and RAP1 pathway. Interestingly, BTK virtual knockout had the highest number of perturbed genes (n = 125) and was associated with enriched terms including angiogenesis, mTORC1 signaling (relevant for microglia priming),36 complement (C1QB, C1QC, C3) and coagulation cascade, fatty acid metabolism, and hypoxia. Supplementary Figure S1 shows the pathway analysis for all the perturbed conditions.
When all the virtual conditions were directly compared to one another and to the basal condition, T-cell depletion (followed by CD4 T-cell depletion only) most strongly affected the immune GRN at the chronic active edge, explaining more than 37% of the total variance (Fig. 2c).
No PRL changes, but PIRA events in anti-CD20 antibody treated patients
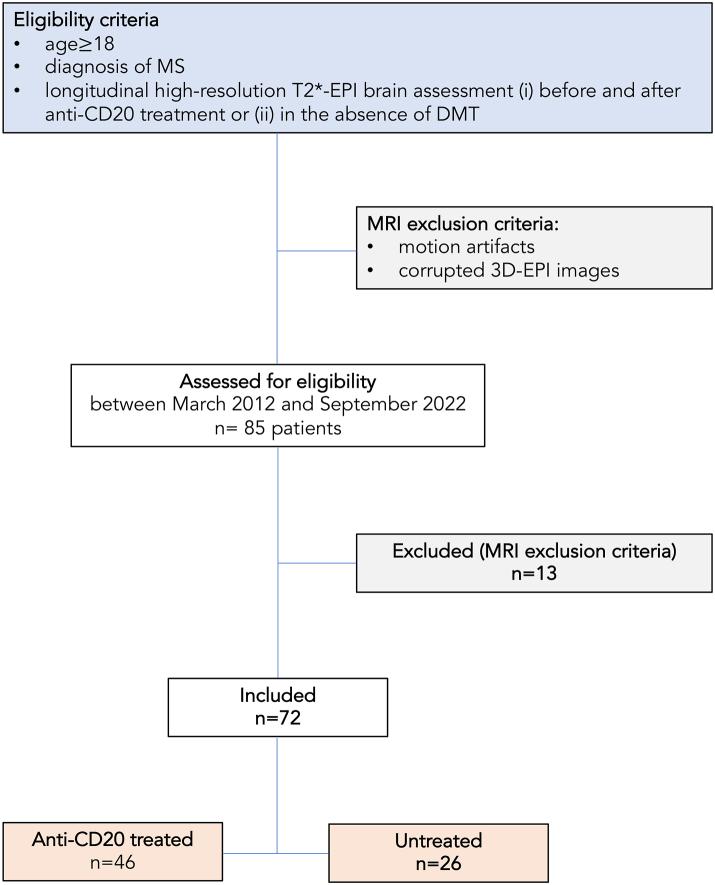
In the multicenter clinical/MRI study, from March 2012 to September 2022, we retrospectively analyzed 3T MRI scans that included high-resolution susceptibility sensitive images in 72 adults with MS (30 with relapsing-remitting and 42 with primary or secondary progressive MS) who were followed at 4 academic centers. Of these, 46 were studied before and after anti-CD20 monoclonal antibody treatment (45 treated with ocrelizumab and one with rituximab), while 26 were untreated for the duration of follow-up (Fig. 3). Demographic and clinical data are provided in Table 1 and Supplementary Table S4.
Fig. 3.
Flow chart summarizing patients' progress through the study. Abbreviations: MS: multiple sclerosis; high-resolution T2∗-EPI: submillimeter isotropic 3D T2∗-weighted segmented echo-planar-imaging; MRI: magnetic resonance imaging; DMT: Disease modifying treatment; Anti-CD20 treated/Untreated: Patients treated/untreated with anti-CD20 antibody therapy.
Table 1.
MS cohort characteristics by treatment category.
| Treatment category | Treated with anti-CD20 monoclonal antibody | Untreated | Statistical analysis |
|---|---|---|---|
| # | 46 (64%) | 26 (36%) | – |
| Demographic and clinical characteristics | |||
| Clinical phenotype | RRMS 17 (37%) PMS 29 (63%) |
RRMS 13 (50%) PMS 13 (50%) |
p = 0.32a n.s. |
| Sex (female, %) | 23/46 (50%) | 21/26 (81%) | p = 0.01a |
| Mean age, years (SD) | 42 (11) | 55 (10) | p < 0.0001b |
| Mean disease duration, years (SD) | 10 (9) | 15 (11) | p = 0.03b |
| # patients with previous DMTe | 33/46 (72%) | 9/26 (35%) | p = 0.002a |
| Median EDSS at baseline (range) | RRMS 1.5 (0–5.5) PMS 5.5 (2–8) |
RRMS 1.5 (0–6.5) PMS 4.5 (2.5–7) |
p = 0.18b n.s. p = 0.76b n.s. |
| Median EDSS last-MRI fu. (range) | RRMS 1.5 (1–5.5) PMS 5.5 (2–8) |
RRMS 1.5 (0–6.5) PMS 4 (1.5–6.5) |
p = 0.23b n.s. p = 0.72b n.s. |
| # patients with PIRA | 9/46 (20%) | 2/26 (8%) | |
| Mean follow-up time, months (range)d | 23 (8–40) | 31 (6–83) | p = 0.9b n.s. |
| Mean number of treatment infusions (SD)c | 4 () | – | |
| # patients with 1 PRL (%) | 34/46 (74%) | 11/26 (42%) | p = 0.01a |
Abbreviations: RRMS: Relapsing-remitting MS; PMS: Progressive MS (including primary and secondary progressive MS patients); DMT: Disease modifying treatment; EDSS: Expanded Disability Status Scale; MRI: Magnetic resonance imaging; PIRA: Progression independent of relapse activity; PRL: Paramagnetic rim lesion; QSM: Quantitative susceptibility mapping; n.s.: Not significant; SD: Standard deviation.
Fisher exact test.
Mann Whitney test.
Mean number of treatment infusions: the first 2 infusion (normally 15 days apart) were counted as 1.
Average post-treatment (for anti-CD20 treated patients) or post baseline MRI acquisition (for untreated patients) follow-up time.
Number of patients having received another DMT before inclusion in the study (e.g. before the baseline MRI acquisition).
The median time between baseline and treatment start was 3 months (range, −27 to 0 months). The average post-treatment follow-up time was 22 months (range, 7–40 months). The average number of treatment infusions was 4 (range, 2–6) at last MRI follow-up; the first two infusions (normally 15 days apart) were counted as one. The median number of MRI timepoints per patient was 2 (range, 2–5).
PRL assessment
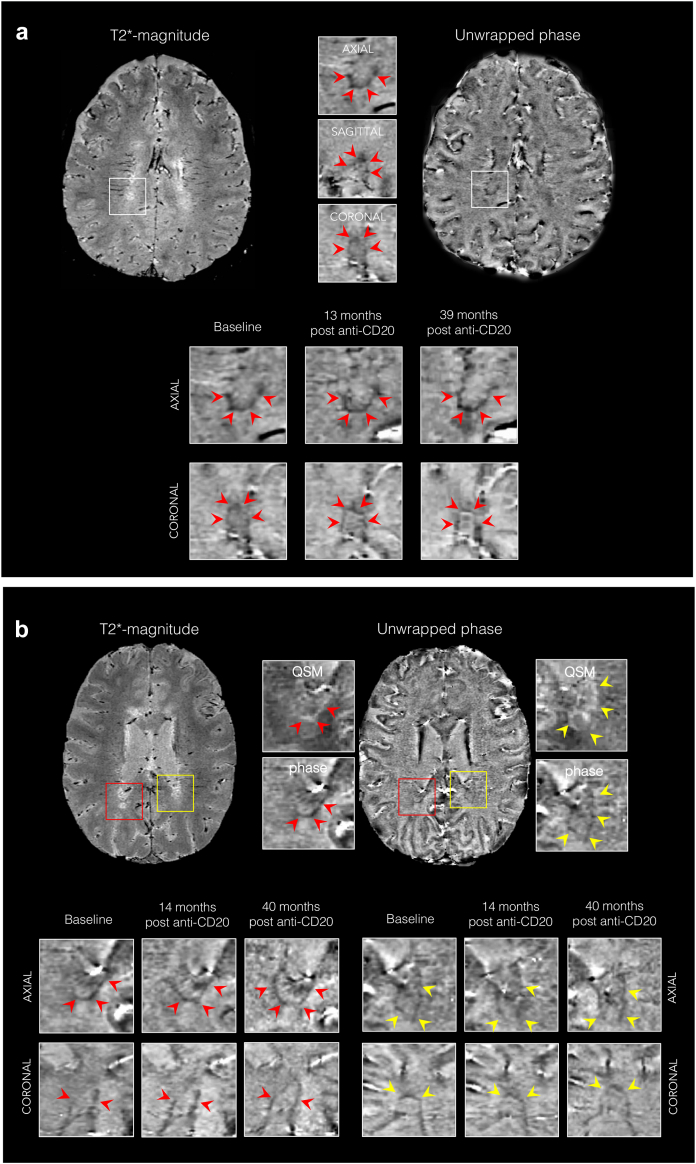
A total of 377 lesions were analyzed: 202 PRL (150 treated and 52 untreated) and 175 non-PRL (124 treated and 51 untreated). Of the 150 treated PRL, 72 (48%) and 36 (24%) were, respectively, followed-up for more than 24 and 36 months after treatment administration. Fig. 4 shows examples of the evolution of treated PRL over 39 and 40 months.
Fig. 4.
Lack of evolution of paramagnetic rim lesions after anti-CD20 treatment. Representative 3D T2∗-weighted magnitude and unwrapped phase MRI images showing paramagnetic rim lesions (PRL) with persistent paramagnetic rims in (a) a 31-year-old with PPMS followed for 39 months, and (b) a 32-year-old with PPMS followed for 40 months, after treatment initiation. In (b), red and yellow arrows delineate two distinct PRL. Abbreviations: MRI, Magnetic resonance imaging; PPMS, Primary progressive MS; PRL, Paramagnetic rim lesions; QSM, Quantitative susceptibility mapping; anti-CD20, anti-CD20 treatment.
Of the 202 segmented PRL, in only one PRL did the paramagnetic rim disappear during the longitudinal MRI follow-up. This PRL, which was untreated, was already barely visible on the 14-month follow-up scan and was not visible on the 38, 51, and 63 month-follow-up scans (Supplementary Figure S2).
Thirteen gadolinium-enhancing lesions were observed on the baseline MRI scan (12 belonging to later-treated and 1 to untreated individuals). None of these 13 gadolinium-enhancing lesions at baseline corresponded to a persistent PRL on the follow-up MRIs. No additional gadolinium-enhancing lesions were detected at follow-up.
Nine of 46 (20%) treated individuals experienced PIRA, compared to 2 of 26 (7.7%) untreated (Table 1).25 When applying a clinically meaningful PRL count threshold,11,15,16,37 PIRA occurred more frequently in individuals with ≥4 PRL on the baseline brain MRI scan than in those with fewer or no PRL (7 of 11 PIRA patients had ≥4 PRL vs 15 of 61 non-PIRA had ≥4 PRL; Fisher exact test, p = 0.027).
Longitudinal lesion volume
A longitudinal multivariable linear mixed model with random lesion and participant effect of the average log-lesion volume showed that PRL were bigger (estimate 0.85 mm3; p < 0.0001; Supplementary Figure S3a) than non-PRL. However, the model did not find a significant treatment effect on either PRL or non-PRL average log-lesion volume (p = 0.68) [Fig. 5a].
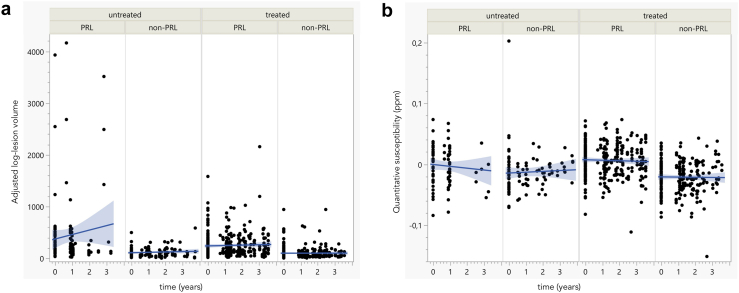
Fig. 5.
Lack of change in lesion volume and susceptibility in paramagnetic vs non-paramagnetic rim lesions. Paramagnetic rim lesions (PRL) and non-PRL average adjusted log-lesion volume over time (a) and magnetic susceptibility, derived from quantitative susceptibility mapping (QSM) (b) in treated and untreated cases. The average log-lesion volume and susceptibility were higher in PRL when compared with non-PRL (multivariable linear mixed model, p < 0.0001). There was no significant treatment effect for PRL or non-PRL average adjusted log-lesion volume (multivariable linear mixed model, p = 0.68). Average susceptibility slightly decreased over time in both treated and untreated non-PRL (multivariable linear mixed model, p < 0.0001 and p = 0.006, respectively) but not in treated and untreated PRL (multivariable linear mixed model, p = 0.18). Abbreviations: PRL, paramagnetic rim lesions; treated/untreated, treated/untreated with anti-CD20 antibody therapy; ppm, parts per million.
Longitudinal magnetic susceptibility
The multivariable linear mixed model with random lesion and participant effect found that lesion quantitative susceptibility was higher in PRL when compared with non-PRL (estimate 0.03 ppm; p < 0.0001; Supplementary Figure S3b). A significant decrease in magnetic susceptibility over time was not found in treated PRL (estimate +0.005 ppm), however it was found in untreated PRL (estimate −0.005 ppm), and in treated/untreated non-PRL (estimate −0.02 and −0.01 ppm, respectively; p = 0.0002) [Fig. 5b]. These results are consistent with the persistency of treated PRL at follow up.
Longitudinal T1 times
For a subset of 20 individuals (14 treated and 6 untreated), T1 lesion values were obtained from the 3D MP2RAGE T1 maps. The same model found higher average T1 times in PRL when compared to non-PRL (estimate 191.56 ms; p < 0.0001; Supplementary Figure S3c). A minimal decrease of T1 values over time was found independently of lesion type (estimate −0.12 ms/months; p = 0.001). However, no treatment effect was found for either PRL or non-PRL (p = 0.39; Supplementary Figure S4).
Sensitivity analysis
No significant center-specific effect affected the results of these 3 independent multivariate analyses (Supplementary Table S5). When the 3 models were run separately for PRL and non-PRL, the results did not change significantly, except for a treatment effect on PRL volume that did not survive to the correction for MRI-clinical center fixed effect (Supplementary Table S5).
Discussion
Halting chronic inflammation is expected to have prognostic relevance in MS, as patients featuring chronic active MS lesions on brain MRI have higher motor/cognitive disability and poor long-term prognosis.15,16,38, 39, 40, 41, 42 In this context, it is critical to identify strategies to modulate chronic inflammation that can be directly tested using longitudinal imaging biomarkers, such as the resolution of paramagnetic rims.
In a previous snRNAseq study,11 the reconstructed glial interactome highlighted the importance of a lymphocyte-microglia-astrocyte axis in sustaining chronic inflammation at the lesion edge.11 To disrupt the vicious circle of chronic inflammation, one possible option is to target reactive glial cells (specifically microglia or astrocytes) or their crosstalk, while another option is to target infiltrated lymphocytes that are still present, albeit sparsely, at the chronic active lesion edge. While there is growing interest in targeting glial activity, this work focused on the latter strategy, in particular on testing the potential ability of B-cell depleting therapy to reduce the compartmentalized chronic inflammation that characterizes chronic active MS lesions.
First, we analyzed whether CD20 B-cells are present in chronic active lesions and their potential role in sustaining chronic inflammation at the lesion edge. Leveraging data from 3 published snRNAseq datasets,11, 12, 13 we delineated different lymphoid subpopulations, annotating them using a multimodal atlas (transcriptome and proteome) of circulating mononuclear immune cells. We found that in chronic active lesions, plasmablasts outnumber CD20 B-cells, the latter accounting for only the 4.3% of all lymphocytes. Activated T cells (CD4 TCM, CD8 TEM and dnT) were more frequent, accounting for 74.2% of all lymphocytes. Among them, few MAIT was seen in chronic active lesions, their periplaque and in CSF from MS samples only; although their role is still elusive, MAIT compartmentalization within the inflamed organ target is seen in other chronic inflammatory diseases, including MS.43, 44, 45 Interestingly, differences between different lesion chronic stages (i.e., chronic active vs chronic inactive lesions) were mainly related to the total number of lymphocytes in tissue rather than to different proportions of their lymphoid subtypes, suggesting that these pathological stages are part of a spectrum rather than fully separate pathological outcomes.
In this context, we interrogated the potential role of lymphocyte subsets, including CD20 B-cells, in sustaining the microglia/dendritic cell-mediated inflammatory network at the chronic active lesion edge. We implemented machine-learning techniques applied to the snRNAseq data to first reconstruct the immune gene regulatory network and compare it with simulated versions depleted of specific lymphocyte subsets or with in silico of the BTK gene knockout (simulating the effect of BTK-inhibitors). Our analysis predicts that depletion of CD20 B-cells in chronic active lesions would affect genes highly expressed in microglia and dendritic cells as well as signaling pathways involved in iron/heme metabolism, mitotic spindle, hypoxia, and antigen presentation. All these processes are known to be highly relevant to MS pathophysiology, especially at the chronic active lesion edge.11 Interestingly, when in silico depletion CD20 B-cells, plasmablasts, and T-cell subtypes were compared,21 T-cell depletion, followed by CD4 T cell depletion only, had the most impact on the immune gene regulatory network at the chronic active edge. This analysis suggests the potential effects of targeted lymphocyte depletion strategies at the chronic edge and helps to rank hypotheses to be further tested, including in in vivo MRI-based clinical trials that use the resolving paramagnetic rim lesion biomarker.
Based on this premise, we tested in vivo the longitudinal effect on PRL of treatment with anti-CD20 monoclonal antibodies, compared to no treatment, over a mean follow-up of two years (range, 8–40 months). Importantly, the same 3T MRI susceptibility sequence was implemented in all centers from which images were aggregated, and MRI data analysis was centralized and blinded to treatment. We found that the paramagnetic rim did not disappear in any of the PRL studied during follow-up in individuals treated with anti-CD20 therapy. Importantly, although the average post-treatment follow-up was about 2 years (a timeframe typical of phase 3 MS clinical trials), ∼50% of the PRL were followed-up for more than 2 years, providing additional time for potential PRL resolution. However, the only PRL in which the paramagnetic rim resolved belonged to an untreated individual (Supplementary Figure S2).
While these results suggest, in agreement with previous studies,11,17 that chronic WM inflammation is a longstanding process lasting many years, it also answers the question of whether anti-CD20 antibodies can promptly and efficiently resolve the chronic inflammatory process ongoing at the edge of PRL. Considering that PRL tend to remain stable or expand over time, unlike the typical shrinkage seen in non-PRL,14,15,46 and that PRL are associated with more severe tissue damage,17,47, 48, 49 we analyzed the longitudinal lesion volume, susceptibility, and T1 time in PRL vs non-PRL. Consistent with previous data,14,15,17,46, 47, 48, 49 we found that PRL were bigger and featured higher susceptibility and T1 times when compared to non-PRL, confirming their destructive phenotype. However, we did not observe a significant treatment effect on any of these measures. We also confirmed that no significant center-specific effect affected the results of the multivariate analysis. Despite the lack of modulation of the paramagnetic rim, in agreement with previous studies,6,8 we confirmed that the anti-CD20 antibody treatment is a successful treatment strategy to suppress active inflammation (no additional gadolinium-enhancing lesions were detected during the follow-up).
A previous study analyzed the effect of ocrelizumab on slowly expanding lesions (SEL), which identify a subset of chronic lesions with radial expansion on longitudinal T2 and T1-weighted MRI scans and correlate with MS disease severity.50 That study found a significant, albeit modest,51 treatment effect on SEL in terms of T1-lesion volume decrease and normalized T1-weighted intensity increase over the 2.5 years post-treatment follow-up. However, regardless of the treatment, normalized T1-weighted intensity was found to decrease, and T1-lesion volume to increase, in both SEL and non-SEL during follow-up. We did not analyze SEL in the current dataset, in part to avoid potential complications related to protocol variation over time and center. Thus, interpreting the results of Elliott et al.50 with respect to those of our study must be approached cautiously. Nonetheless, one should consider that PRL and SEL only show partial correspondence, and that, distinct from lesion volume changes, the paramagnetic rim is a direct marker of the presence of iron-containing inflammatory cells at the lesion edge.51, 52, 53 Interestingly, there is some evidence that SEL with paramagnetic rims are the most severe subset of chronic lesions in terms of tissue damage,54 and it is possible that these lesions are less likely to be altered by B-cell depletion.
In agreement with a recent re-analysis of ocrelizumab phase 3 clinical trials,10 we found that ∼20% of the anti-CD20 treated MS cases experienced disability worsening independent of relapse activity by the end of the study. Of relevance, in our study, PIRA was more common in individuals with ≥4 brain PRL, supporting the critical contribution of chronic inflammation to MS disease progression, a process that we show does not fully resolve with administration of anti-CD20 monoclonal antibody therapies.
Taken together, our MRI data suggest that peripheral B-cell depletion, although efficient in preventing new lesion formation, does not sufficiently impact the tissue inflammatory milieu to the point of reducing the activity of iron-laden microglia at the edge of chronic active lesions over a median follow-up of 2 years. In these lesions, the smoldering inflammatory demyelination and axonal injury is compartmentalized behind a virtually closed blood–brain barrier. Although we cannot directly estimate the rate of ongoing lymphocyte infiltration into tissue, using snRNAseq data, we recognized upregulation of markers of tissue-resident memory T-cells (CD69, CD44, ITGA1, and ITGAE), which are long-lived, non-recirculating lymphocytes that can respond quickly to the presence of their cognate antigen and, potentially, sustain tissue damage in autoimmune diseases.32,33
This study has some limitations. Although no significant treatment effect was found on persistence of the PRL rim, volume, susceptibility, or T1 times, larger cohorts and longer post-treatment follow-up are required to further elucidate the effect of anti-CD20 antibody therapy in chronic active lesions, as paramagnetic rims has been shown to fade or to disappear over almost a decade (median of 7 years).11,17,49,55 Moreover, although large-scale longitudinal studies investigating the impact of DMTs on chronic active lesions are still lacking, preliminary evidence from the literature suggest some potential effect of DMTs on evolving tissue damage within PRL.30,56 Recognizing that dissecting the relative contribution of myelin and iron to the average magnetic susceptibility within MS lesions using longitudinal QSM can be challenging, we additionally measured quantitative longitudinal T1 times within lesions, a strategy which could more reliably depict myelin status.29 Additionally, although lesion volumes were carefully delineated manually during the longitudinal follow-up, we did not perform a comprehensive SEL analysis. Finally, our in silico analysis should be considered to be hypothesis-generating rather than replacing mechanistic studies.
In conclusion, although CD20 B-cells are likely involved in sustaining the microglia/dendritic cell-mediated inflammatory network at the chronic active MS lesion edge, anti-CD20 antibody therapies do not resolve PRL over an average of 2 years of MRI follow up. The limited turnover/tissue infiltration of circulating anti-CD20 B-cells (which are the main target of anti-CD20 treatments),32 the inefficient passage of anti-CD20 antibodies across the blood–brain barrier,3 and the paucity of CD20 B-cells in chronic active lesions could help to explain our results. Future prospective multicenter trials that marshal optimized susceptibility-MRI sequences for PRL detection should further investigate the longer-term effect of anti-CD20 treatments on chronic inflammatory MS lesions. Finally, budding from our in silico analysis, future therapeutic strategies could focus on depleting or modulating specific subsets of lymphocytes in the MS brain.
Contributors
PM, AS, PAC, VVP, DSR, WAM, GK, VL, GP, SES, TD contributed to data acquisition.
PM, CVB, EP, AS, MW, AS, XL, MA analyzed the data.
CB, PM, EP performed the statistical analysis.
PM, DSR, MA drafted and revised the manuscript.
PM, EP, MA drafted the figures.
PM, MA verified the underlying data reported in the manuscript.
All authors approved the submitted the final draft of the manuscript.
Data sharing statement
Qualified researchers can access the individual patient data of this study upon reasonable request and material transfer agreement between institutes.
Raw data from single-cell genomics analyzed in this work are available at: Absinta et al., Nature 2021 accession number GSE180759; Schafflick et al., Nature Comm 2020 accession number GSE138266; Schirmer et al., Nature 2091 accession number PRJNA544731; Jäkel et al., Nature 2019 accession number GSE118257; Hao et al., Cell 2021 accession number GSE164378.
Source code used in the paper is available at https://github.com/AbsintaLab/EBIOM23.git.
Declaration of interests
PM received consultancy honoraria from Sanofi and Biogen and, research funding from Biogen.
VVP received support for attending meetings and/or travel, support for participating in Advisory Board and consulting fees (the latters payed to the institution Cliniques Universitaires Saint-Luc) from Roche, Biogen, Sanofi, Merck Healthcare KGaA (Darmstadt, Germany), Bristol Meyer Squibb, Janssen, Almirall, Alexion and Novartis Pharma.
PAC Is PI on grants to JHU from Genentech and previously Principia; he has received consulting honoraria for serving on SABs for Nervgen, Idorsia, Biogen, Vaccitech, and Lilly.
DSR has received research funding from Abata Therapeutics, Sanofi-Genzyme, and Vertex Pharmaceuticals.
MA received consultancy honoraria from Abata Therapeutics, Biogen, Sanofi-Genzyme and GSK.
Acknowledgements
The authors thank the study participants; the neuroimmunology clinics in each center for recruiting and evaluating the patients and for coordinating the scans; the NIH/NINDS Functional Magnetic Resonance Imaging Facility, Blake Dewey (Johns Hopkins University), Julie Absil (Université Libre de Bruxelles), and Amin Ben Ayad (Université Catholique de Louvain) for assistance with 3T MRI scan acquisition and analysis. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov) and the research cluster of San Raffale Hospital.
PM research activity is supported by the Fund for Scientific Research (F.R.S, FNRS; grant #40008331), Cliniques universitaires Saint-Luc “Fonds de Recherche Clinique” and Biogen.
CVB is supported by the “FSR grant” of the UCLouvain.
EP is supported by FRRB Early Career Award (grant #1750327 to MA).
MW is supported by the “Swiss Government Excellence Scholarship” (grant #2021.0087).
XL is supported by the National Institutes of Health (NIH/NIBIB, P41EB031771).
PAC is supported by the National Institutes of Health (R01NS082347 and R01NS082347).
DSR is supported by the Intramural Research Program of NINDS/NIH and by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
MA is supported by the Conrad N. Hilton Foundation (Marylin Hilton Bridging Award for Physician-Scientists, grant #17313), the International Progressive MS alliance (21NS037), the Roche Foundation for Independent Research, the Cariplo Foundation (grant #1677), the FRRB Early Career Award (grant #1750327), and the National MS Society (NMSS RFA-2203-39325).
Study approval: Study procedures received approval from an ethical standards committee on human experimentation in each of the four academic research hospitals, and written informed consent was obtained from all participants prior to participation.
Footnotes
Appendix A
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104701.
Contributor Information
Pietro Maggi, Email: pietro.maggi@uclouvain.be.
Martina Absinta, Email: absinta.martina@hsr.it.
Appendix A. Supplementary data
Selective lymphocyte-depletion affects genes involved in microglia/dendritic cell activity. Functional enrichment analysis of gene sets performed on the subset of simulated significant perturbed genes (p < 0.05) using Enrichr (KEGG Human and MsigDB Hallmarks annotations).
A disappearing paramagnetic rim during longitudinal MRI follow-up in an untreated individual. The rim of this paramagnetic rim lesion (PRL), which belonged to an untreated 62-year-old with RRMS, was already barely visible on the 14-month follow-up scan and clearly not visible on the 38-, 51-, and 63-month follow-up scans. Abbreviations: MRI, magnetic resonance imaging; RRMS, relapsing remitting MS; PRL, paramagnetic rim lesion.
Overall lesion volume, susceptibility and T1 relaxation changes differences in paramagnetic vs non-paramagnetic rim lesions across all time points. The mean [SD] (a) log-lesion volume (294 [418]; 104 [111]), (b) susceptibility (0,004 [0,028]; −0,019 [0,027]) and (c) T1 times (1456 [268]; 1198 [240]) across all time points were higher in PRL when compared with non-PRL. In (d-f), log-lesion volume (435 [719]; 94 [90] and 249 [240]; 118 [63]), susceptibility (−0,002 [0,032]; −0,013 [0,033] and 0,006 [0,026]; −0,021 [0,025]) and T1 times (1161 [107]; 1119 [107] and 1472 [265]; 1220 [262]) across all time points are shown in untreated and treated patients, respectively. Please refer to multivariable linear mixed model in the manuscript Results for estimates and statistical significance. Abbreviations: PRL, Paramagnetic rim lesions; ppm, parts per million; ms, Milliseconds.
No treatment effect in PRL vs non-PRL by assessing T1 relaxation changes. The average T1 times were higher in PRL when compared with non-PRL (multivariable linear mixed model, p < 0.0001), and average T1 time decreased over time in all lesion types (p = 0.001). However, there was no significant treatment effect for either PRL or non-PRL average T1 time evolution (multivariable linear mixed model, p = 0.39). Abbreviations: treated/untreated with anti-CD20 antibody therapy; PRL, Paramagnetic rim lesions; ms, Milliseconds.
References
- 1.Kuhlmann T., Moccia M., Coetzee T., et al. Multiple sclerosis progression: time for a new mechanism-driven framework. Lancet Neurol. 2023;22(1):78–88. doi: 10.1016/S1474-4422(22)00289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Absinta M., Lassmann H., Trapp B.D. Mechanisms underlying progression in multiple sclerosis. Curr Opin Neurol. 2020;33(3):277–285. doi: 10.1097/WCO.0000000000000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cencioni M.T., Mattoscio M., Magliozzi R., Bar-Or A., Muraro P.A. B cells in multiple sclerosis — from targeted depletion to immune reconstitution therapies. Nat Rev Neurol. 2021;17(7):399–414. doi: 10.1038/s41582-021-00498-5. [DOI] [PubMed] [Google Scholar]
- 4.Bjornevik K., Cortese M., Healy B.C., et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375(6578):296–301. doi: 10.1126/science.abj8222. [DOI] [PubMed] [Google Scholar]
- 5.Myhr K.M., Torkildsen Ø., Lossius A., Bø L., Holmøy T. B cell depletion in the treatment of multiple sclerosis. Expert Opin Biol Ther. 2019;19(3):261–271. doi: 10.1080/14712598.2019.1568407. [DOI] [PubMed] [Google Scholar]
- 6.Hauser S.L., Bar-Or A., Comi G., et al. Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. N Engl J Med. 2017;376(3):221–234. doi: 10.1056/NEJMoa1601277. [DOI] [PubMed] [Google Scholar]
- 7.Hauser S.L., Waubant E., Arnold D.L., et al. B-cell depletion with rituximab in relapsing–remitting multiple sclerosis. N Engl J Med. 2008;358(7):676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 8.Montalban X., Hauser S.L., Kappos L., et al. Ocrelizumab versus placebo in primary progressive multiple sclerosis. N Engl J Med. 2017;376(3):209–220. doi: 10.1056/NEJMoa1606468. [DOI] [PubMed] [Google Scholar]
- 9.Kang C., Blair H.A. Ofatumumab: a review in relapsing forms of multiple sclerosis. Drugs. 2022;82(1):55–62. doi: 10.1007/s40265-021-01650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kappos L., Wolinsky J.S., Giovannoni G., et al. Contribution of relapse-independent progression vs relapse-associated worsening to overall confirmed disability accumulation in typical relapsing multiple sclerosis in a pooled analysis of 2 randomized clinical trials. JAMA Neurol. 2020;77(9):1132. doi: 10.1001/jamaneurol.2020.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Absinta M., Maric D., Gharagozloo M., et al. A lymphocyte–microglia–astrocyte axis in chronic active multiple sclerosis. Nature. 2021;597:709. doi: 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jäkel S., Agirre E., Mendanha Falcão A., et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566(7745):543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schirmer L., Velmeshev D., Holmqvist S., et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573(7772):75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dal-Bianco A., Grabner G., Kronnerwetter C., et al. Slow expansion of multiple sclerosis iron rim lesions: pathology and 7 T magnetic resonance imaging. Acta Neuropathol. 2017;133(1):25–42. doi: 10.1007/s00401-016-1636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Absinta M., Sati P., Masuzzo F., et al. Association of chronic active multiple sclerosis lesions with disability in vivo. JAMA Neurol. 2019;76:1474. doi: 10.1001/jamaneurol.2019.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maggi P., Sati P., Nair G., et al. Paramagnetic rim lesions are specific to multiple sclerosis: an international multicenter 3T MRI study. Ann Neurol. 2020;88(5):1034–1042. doi: 10.1002/ana.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dal-Bianco A., Grabner G., Kronnerwetter C., et al. Long-term evolution of multiple sclerosis iron rim lesions in 7 T MRI. Brain. 2021;144:awaa436. doi: 10.1093/brain/awaa436. [DOI] [PubMed] [Google Scholar]
- 18.Schafflick D., Xu C.A., Hartlehnert M., et al. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat Commun. 2020;11(1):247. doi: 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osorio D., Zhong Y., Li G., Huang J.Z., Cai J.J. scTenifoldNet: a machine learning workflow for constructing and comparing transcriptome-wide gene regulatory networks from single-cell data. Patterns. 2020;1(9) doi: 10.1016/j.patter.2020.100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correale J. BTK inhibitors as potential therapies for multiple sclerosis. Lancet Neurol. 2021;20(9):689–691. doi: 10.1016/S1474-4422(21)00250-7. [DOI] [PubMed] [Google Scholar]
- 22.Chen E.Y., Tan C.M., Kou Y., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuleshov M.V., Jones M.R., Rouillard A.D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z., Bailey A., Kuleshov M.V., et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1(3) doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kappos L., Butzkueven H., Wiendl H., et al. Greater sensitivity to multiple sclerosis disability worsening and progression events using a roving versus a fixed reference value in a prospective cohort study. Mult Scler J. 2018;24(7):963–973. doi: 10.1177/1352458517709619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sati P., Thomasson D., Li N., et al. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Mult Scler J. 2014;20(11):1464–1470. doi: 10.1177/1352458514525868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Absinta M., Sati P., Gaitan M.I., et al. Seven-tesla phase imaging of acute multiple sclerosis lesions: a new window into the inflammatory process. Ann Neurol. 2013;74(5):669–678. doi: 10.1002/ana.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Chen L., Kutten K., et al. Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility. Neuroimage. 2019;191:337–349. doi: 10.1016/j.neuroimage.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolb H., Absinta M., Beck E.S., et al. 7T MRI differentiates remyelinated from demyelinated multiple sclerosis lesions. Ann Neurol. 2021;90(4):612–626. doi: 10.1002/ana.26194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinger N., Ponath G., Sweeney E., et al. Dimethyl fumarate reduces inflammation in chronic active multiple sclerosis lesions. Neurol Neuroimmunol Neuroinflamm. 2022;9(2):e1138. doi: 10.1212/NXI.0000000000001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phipson B., Sim C.B., Porrello E.R., Hewitt A.W., Powell J., Oshlack A. propeller: testing for differences in cell type proportions in single cell data. Bioinformatics. 2022;38(20):4720–4726. doi: 10.1093/bioinformatics/btac582. Mathelier A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machado-Santos J., Saji E., Tröscher A.R., et al. The compartmentalized inflammatory response in the multiple sclerosis brain is composed of tissue-resident CD8+ T lymphocytes and B cells. Brain. 2018;141(7):2066–2082. doi: 10.1093/brain/awy151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fransen N.L., Hsiao C.C., van der Poel M., et al. Tissue-resident memory T cells invade the brain parenchyma in multiple sclerosis white matter lesions. Brain. 2020;143(6):1714–1730. doi: 10.1093/brain/awaa117. [DOI] [PubMed] [Google Scholar]
- 34.Wu H., Liao W., Li Q., et al. Pathogenic role of tissue-resident memory T cells in autoimmune diseases. Autoimmun Rev. 2018;17(9):906–911. doi: 10.1016/j.autrev.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Ostkamp P., Deffner M., Schulte-Mecklenbeck A., et al. A single-cell analysis framework allows for characterization of CSF leukocytes and their tissue of origin in multiple sclerosis. Sci Transl Med. 2022;14(673) doi: 10.1126/scitranslmed.adc9778. [DOI] [PubMed] [Google Scholar]
- 36.Keane L., Antignano I., Riechers S.P., et al. mTOR-dependent translation amplifies microglia priming in aging mice. J Clin Invest. 2021;131(1) doi: 10.1172/JCI132727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maggi P., Kuhle J., Schädelin S., et al. Chronic white matter inflammation and serum neurofilament levels in multiple sclerosis. Neurology. 2021;97(6):e543–e553. doi: 10.1212/WNL.0000000000012326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcille M., Hurtado Rúa S., Tyshkov C., et al. Disease correlates of rim lesions on quantitative susceptibility mapping in multiple sclerosis. Sci Rep. 2022;12(1):4411. doi: 10.1038/s41598-022-08477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood H. Slowly expanding lesions are linked to multiple sclerosis progression. Nat Rev Neurol. 2022;18(5):252. doi: 10.1038/s41582-022-00654-5. [DOI] [PubMed] [Google Scholar]
- 40.Calvi A., Carrasco F.P., Tur C., et al. Association of slowly expanding lesions on MRI with disability in people with secondary progressive multiple sclerosis. Neurology. 2022;98(17):e1783–e1793. doi: 10.1212/WNL.0000000000200144. [DOI] [PubMed] [Google Scholar]
- 41.Preziosa P., Pagani E., Meani A., et al. Slowly expanding lesions predict 9-year multiple sclerosis disease progression. Neurol Neuroimmunol Neuroinflamm. 2022;9(2):e1139. doi: 10.1212/NXI.0000000000001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altokhis A.I., Hibbert A.M., Allen C.M., et al. Longitudinal clinical study of patients with iron rim lesions in multiple sclerosis. Mult Scler J. 2022;28 doi: 10.1177/13524585221114750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carnero Contentti E., Farez M.F., Correale J. Mucosal-associated invariant T cell features and TCR repertoire characteristics during the course of multiple sclerosis. Front Immunol. 2019;10:2690. doi: 10.3389/fimmu.2019.02690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazaki Y., Miyake S., Chiba A., Lantz O., Yamamura T. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int Immunol. 2011;23(9):529–535. doi: 10.1093/intimm/dxr047. [DOI] [PubMed] [Google Scholar]
- 45.Willing A., Leach O.A., Ufer F., et al. CD8 + MAIT cells infiltrate into the CNS and alterations in their blood frequencies correlate with IL-18 serum levels in multiple sclerosis: clinical immunology. Eur J Immunol. 2014;44(10):3119–3128. doi: 10.1002/eji.201344160. [DOI] [PubMed] [Google Scholar]
- 46.Elliott C., Wolinsky J.S., Hauser S.L., et al. Slowly expanding/evolving lesions as a magnetic resonance imaging marker of chronic active multiple sclerosis lesions. Mult Scler J. 2019;25(14):1915–1925. doi: 10.1177/1352458518814117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Absinta M., Sati P., Schindler M., et al. Persistent 7-tesla phase rim predicts poor outcome in new multiple sclerosis patient lesions. J Clin Invest. 2016;126(7):2597–2609. doi: 10.1172/JCI86198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahmanzadeh R., Lu P.J., Barakovic M., et al. Myelin and axon pathology in multiple sclerosis assessed by myelin water and multi-shell diffusion imaging. Brain. 2021;144:awab088. doi: 10.1093/brain/awab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S., Nguyen T.D., Hurtado Rúa S.M., et al. Quantitative susceptibility mapping of time-dependent susceptibility changes in multiple sclerosis lesions. AJNR Am J Neuroradiol. 2019;40(6):987–993. doi: 10.3174/ajnr.A6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott C., Belachew S., Wolinsky J.S., et al. Chronic white matter lesion activity predicts clinical progression in primary progressive multiple sclerosis. Brain. 2019;142(9):2787–2799. doi: 10.1093/brain/awz212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold D.L., Belachew S., Gafson A.R., Gaetano L., Bernasconi C., Elliott C. Slowly expanding lesions are a marker of progressive MS – No. Mult Scler J. 2021;27(11):1681–1683. doi: 10.1177/13524585211017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvi A., Clarke M.A., Prados F., et al. Relationship between paramagnetic rim lesions and slowly expanding lesions in multiple sclerosis. Mult Scler J. 2022;29 doi: 10.1177/13524585221141964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elliott C., Rudko D.A., Arnold D.L., et al. Lesion-level correspondence and longitudinal properties of paramagnetic rim and slowly expanding lesions in multiple sclerosis. Mult Scler J. 2023;29 doi: 10.1177/13524585231162262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elliott C., Belachew S., Fisher E., et al. MRI characteristics of chronic MS lesions by phase rim detection and/or slowly expanding properties (4101) Neurology. 2021;96(15 Supplement):4101. [Google Scholar]
- 55.Weber C.E., Wittayer M., Kraemer M., et al. Long-term dynamics of multiple sclerosis iron rim lesions. Mult Scler Relat Disord. 2022;57 doi: 10.1016/j.msard.2021.103340. [DOI] [PubMed] [Google Scholar]
- 56.Eisele P., Wittayer M., Weber C.E., Platten M., Schirmer L., Gass A. Impact of disease-modifying therapies on evolving tissue damage in iron rim multiple sclerosis lesions. Mult Scler J. 2022;28(14):2294–2298. doi: 10.1177/13524585221106338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selective lymphocyte-depletion affects genes involved in microglia/dendritic cell activity. Functional enrichment analysis of gene sets performed on the subset of simulated significant perturbed genes (p < 0.05) using Enrichr (KEGG Human and MsigDB Hallmarks annotations).
A disappearing paramagnetic rim during longitudinal MRI follow-up in an untreated individual. The rim of this paramagnetic rim lesion (PRL), which belonged to an untreated 62-year-old with RRMS, was already barely visible on the 14-month follow-up scan and clearly not visible on the 38-, 51-, and 63-month follow-up scans. Abbreviations: MRI, magnetic resonance imaging; RRMS, relapsing remitting MS; PRL, paramagnetic rim lesion.
Overall lesion volume, susceptibility and T1 relaxation changes differences in paramagnetic vs non-paramagnetic rim lesions across all time points. The mean [SD] (a) log-lesion volume (294 [418]; 104 [111]), (b) susceptibility (0,004 [0,028]; −0,019 [0,027]) and (c) T1 times (1456 [268]; 1198 [240]) across all time points were higher in PRL when compared with non-PRL. In (d-f), log-lesion volume (435 [719]; 94 [90] and 249 [240]; 118 [63]), susceptibility (−0,002 [0,032]; −0,013 [0,033] and 0,006 [0,026]; −0,021 [0,025]) and T1 times (1161 [107]; 1119 [107] and 1472 [265]; 1220 [262]) across all time points are shown in untreated and treated patients, respectively. Please refer to multivariable linear mixed model in the manuscript Results for estimates and statistical significance. Abbreviations: PRL, Paramagnetic rim lesions; ppm, parts per million; ms, Milliseconds.
No treatment effect in PRL vs non-PRL by assessing T1 relaxation changes. The average T1 times were higher in PRL when compared with non-PRL (multivariable linear mixed model, p < 0.0001), and average T1 time decreased over time in all lesion types (p = 0.001). However, there was no significant treatment effect for either PRL or non-PRL average T1 time evolution (multivariable linear mixed model, p = 0.39). Abbreviations: treated/untreated with anti-CD20 antibody therapy; PRL, Paramagnetic rim lesions; ms, Milliseconds.