Abstract
In humans, methionine derived from dietary proteins is necessary for cellular homeostasis and regeneration of sulfur containing pathways, which produce inorganic sulfur species (ISS) along with essential organic sulfur compounds (OSC). In recent years, inorganic sulfur species have gained attention as key players in the crosstalk of human health and the gut microbiome. Endogenously, ISS includes hydrogen sulfide (H2S), sulfite (SO32−), thiosulfate (S2O32−), and sulfate (SO42−), which are produced by enzymes in the transsulfuration and sulfur oxidation pathways. Additionally, sulfate-reducing bacteria (SRB) in the gut lumen are notable H2S producers which can contribute to the ISS pools of the human host. In this review, we will focus on the systemic effects of sulfur in biological pathways, describe the contrasting mechanisms of sulfurylation versus phosphorylation on the hydroxyl of serine/threonine and tyrosine residues of proteins in post-translational modifications, and the role of the gut microbiome in human sulfur metabolism.
1. Introduction
Sulfur is among the most biologically abundant elements in the human body with functions including cellular signaling, detoxification of free radicals, structural support, and assisting in energy production [[1], [2], [3], [4], [5]]. Sulfur pools in the human body are vast and include both inorganic sulfur species (ISS) as well as critical organic sulfur-containing compounds (OSCs). In humans, sulfur pools (ISS and OSC) are maintained mainly through dietary intake of methionine along with adequate B-vitamins (B1, B2, B3, B5, B6, B7, B9, B12), and trace elements zinc, nickel, molybdenum, cobalt, potassium, magnesium, iron, calcium, and sodium (Table 1) [6,7]. Methionine is an essential regulator of human sulfur pools through its direct regeneration of universal methyl donor s-adenosyl-l-methionine (SAM) and homocysteine, which replenish cysteine concentrations during the fed-state through the transsulfuration pathway [4]. While cysteine is considered a conditionally essential amino acid, it is the rate-limiting amino acid in the synthesis of glutathione, coenzyme A, iron-clusters, taurine, and tertiary proteins, all of which are considered OSCs because they contain sulfur [4]. As a product of sulfur pathways, ISS are commonly produced and include hydrogen sulfide (H2S), sulfite (SO32−), thiosulfate (S2O32−), and sulfate (SO42−). Additionally, microbiota residing in the human gut-microbiome are major contributors and regulators of sulfur pools in the body. Through the sulfate reduction pathways of sulfur-reducing bacteria energy is not only extracted for the microorganism, but hydrogen sulfide is produced. In general, microbiota found in the gut-microbiome favor ISS reduction while human metabolic processes favor ISS oxidation to sulfate for its activation to 3′-phosphoadenosine 5′-phosphate (PAPS) and numerous sulfonation/sulfurylation reactions [[8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18]]. The purpose of this review, is to focus on the systemic effects of sulfur (ISS and OSC) in biological pathways and the role of the gut microbiome in human sulfur metabolism.
Table 1.
Enzymes in sulfur metabolism pathways described in this review. Additional information obtained from The Human Protein Atlas.
| ID | GENE | NAME | CHROMOSOME | EC | COFACTOR/COENZYME | TISSUE RNA EXPRESSION (nTPM) |
|---|---|---|---|---|---|---|
| ENSG00000181915 | ADO | 2-aminoethanethiol dioxygenase | 10 | 1.13.11.19 | Nickel (Ni2+) Fe2+ | Testes - 40 |
| Brain - 30 | ||||||
| Tongue - 20 | ||||||
| ENSG00000101444 | AHCY (SAHH) | Adenosylhomocysteinase | 20 | 3.3.1.1 | NAD + open, NADH closed | Liver - 180, Kidney (Proximal Tubule) - 160, Pancreas - 160, most tissue |
| ENSG00000119689 | α-KGDHC - DLST | α-ketoglutarate dehydrogenase complex | 14 | 1.2.4.2 | TPP (vitamin B1) Mg2+ | Tongue - 230 |
| Muscle - 150 | ||||||
| Kidney - 100 | ||||||
| ENSG00000145692 | BHMT | betaine-homocysteine S-methyltransferase | 5 | 2.1.1.5 | Zinc (Zn2+) | Liver (hepatocytes) - 1150 |
| Potassium (K+) | Kidney (proxima tubule) - 770 | |||||
| ENSG00000132840 | BHMT2 | betaine-homocysteine S-methyltransferase 2 | 5 | 2.1.1.5 | Zinc (Zn2+) | Liver - 430 |
| Potassium (K+) | Kidney - 333 | |||||
| ENSG00000160200 | CBS | Cystathionine beta-synthase | 21 | 4.2.1.22 | Heme (Fe2+) | Liver - 175 |
| Pyridoxal-5-phosphate (PLP) | Pancreas - 100 | |||||
| ENSG00000129596 | CDO1 | Cysteine dioxygenase type 1 | 5 | 1.13.11.20 | Iron (II) | Liver - 500 |
| Adipose - 120 | ||||||
| ENSG00000016391 | CHDH | Choline dehydrogenase | 3 | 1.1.99.1 | FAD+ | Liver - 40 |
| Kidney - 40 | ||||||
| ENSG00000068120 | COASY | Coenzyme A synthase | 17 | PPAT - 2.7.7.3 | ATP | Liver - 70 |
| DPCK - 2.7.1.24 | Magnesium (Mg2+) | Adrenals - 60 | ||||
| ENSG00000139631 | CSAD | Cysteine sulfinic acid decarboxylase | 12 | 4.1.1.29 | Heme (Fe2+) | Liver - 100 |
| Pyridoxal-5-phosphate (PLP) | Adipose - 70 | |||||
| ENSG00000116761 | CTH - CSE - CGL | Cystathionine gamma-lyase | 1 | 4.4.1.1 | Heme (Fe2+) | Liver - 120 |
| Pyridoxal-5-phosphate (PLP) | Ovary - 35 | |||||
| ENSG00000228716 | DHFR | Dihydrofolate reductase 1 | 5 | 1.5.1.3 | NADP+ | Liver - 60 |
| Thymus - 60 | ||||||
| Bone Marrow - 60 | ||||||
| Tonsils/lymph nodes - 40 | ||||||
| ENSG00000178700 | DHFR2 | Dihydrofolate reductase 2 | 3 | 1.5.1.3 | NADP+ | Ovary - 10 |
| Endometrium - 8 | ||||||
| Liver - 7 | ||||||
| ENSG00000102967 | DHODH | Dihydroorotate dehydrogenase (quinone) | 16 | 1.3.5.2 | FAD+ | Liver (hepatocytes) - 100 |
| ENSG00000132837 | DMGDH | Dimethylglycine dehydrogenase | 5 | 1.5.99.2 | FAD+ | Liver - 300 |
| kidney - 170 | ||||||
| ENSG00000105755 | ETHE1/PDO | “ethylmalonic encephalopathy 1 protein” and “per sulfide dioxygenase" | 19 | 1.13.11.18 | Iron (Fe2+) | Colon - 260 |
| Rectum - 230 | ||||||
| ENSG00000010932 | FMO1 | Flavin containing dimethylaniline monoxygenase 1 | 1 | 1.14.13.8 | FAD+ | Kidney (Proximal Tubule) - 200 |
| NADP+ | ||||||
| ENSG00000094963 | FMO2 | Flavin containing dimethylaniline monoxygenase 2 | 1 | 1.14.13.8 | FAD + NADPH Magnesium (Mg2+) | Lung - 100 |
| Adipose - 70 | ||||||
| Esophagus - 70 | ||||||
| Muscle - 60 | ||||||
| ENSG00000007933 | FMO3 | Flavin containing dimethylaniline monoxygenase 3 | 1 | 1.14.13.8 | FAD+ | Liver - 1120 |
| NADP+ | ||||||
| ENSG00000076258 | FMO4 | Flavin containing dimethylaniline monoxygenase 4 | 1 | 1.14.13.8 | FAD+ | Liver - 70 |
| NADP+ | Kideny - 50 | |||||
| ENSG00000131781 | FMO5 | Flavin containing dimethylaniline monoxygenase 5 | 1 | 1.14.13.8 | FAD+ | Liver - 700 |
| NADP+ | ||||||
| ENSG00000165060 | FXN | Frataxin | 9 | 1.16.3.1 | Iron (Fe2+) | Liver - 46 |
| Bone Marrow - 30 | ||||||
| Muscle - 25 | ||||||
| ENSG00000001084 | y-GCS - GCLC | Glutamate-cysteine ligase | 6 | 6.3.2.2 | ATP Magnesium (Mg2+) | Liver - 150 |
| Bladder - 60 | ||||||
| Fallopian Tubes - 60 | ||||||
| ENSG00000120053 | GOT1 - CAT1 - AST1 | Aspartate aminotransferase 1 | 10 | 2.6.1.3 | Pyridoxal 5′-phosphate (PLP) | Tongue - 1010 |
| Heart - 560 | ||||||
| Muscle - 500 | ||||||
| Liver - 370 | ||||||
| Kidney - 140 | ||||||
| ENSG00000125166 | GOT2 - CAT2 - AST2 | Aspartate aminotransferase 2 | 16 | 2.6.1.3 | Pyridoxal 5′-phosphate (PLP) | Tongue - 730 |
| Skeletal muscle - 700 | ||||||
| Liver - 320 | ||||||
| Heart - 310 | ||||||
| ENSG00000100983 | GSS | Glutathione synthetase | 20 | 6.3.2.3 | ATP Magnesium (Mg2+) | Testis - 70 |
| Liver - 45 | ||||||
| Kidney - 45 | ||||||
| GI - 40 | ||||||
| Skin - 30 | ||||||
| Brain - 20 | ||||||
| ENSG00000151224 | MAT1A | Methionine adenosyltransferase 1A | 10 | 2.5.1.6 | ATP | Liver (hepatocytes) - 1400 |
| Magnessium (Mg2+) | ||||||
| Potassium (K+) | ||||||
| ENSG00000168906 | MAT2A | Methionine adenosyltransferase 2A | 2 | 2.5.1.6 | ATP | Pancreas - 900 |
| Magnessium (Mg2+) | Thyroid - 400 | |||||
| Potassium (K+) | Brain White Matter - 300 | |||||
| ENSG00000038274 | MAT2B | Methionine adenosyltransferase 2B | 5 | 2.5.1.7 | NADP+ | Nonspecific - 100 (Kidney, duodenum, lymph) |
| ENSG00000128309 | MPST, TST2 | Mercaptopyruvate sulfurtransferase | 22 | 2.8.1.2 | Zinc (Zn2+) | Liver - 400 |
| GI TRACT - 100 | ||||||
| ENSG00000177000 | MTHFR | Methylenetetrahydrofolate reductase | 1 | 1.5.1.20 | FAD+ | Epidydemis - 60 |
| NADP+ | Bone - 30 | |||||
| ENSG00000116984 | MTR | Methionine synthase | 1 | 2.1.1.13 | B12 | Parathyroid - 30 |
| Zinc (Zn2+) | Muscle - 20 | |||||
| Cobalt (Co3+) | Cardiomyocytes - 20 | |||||
| ENSG00000124275 | MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 5 | 2.1.1.3 | FAD+, NADP+ | Parathyroid - 20 |
| Tongue 20 | ||||||
| Muscle - 16 | ||||||
| ENSG00000152782 | PANK1 | Pantothenate kinase 1 | 10 | 2.7.1.33 | ATP Magnesium (Mg2+) | Liver - 110 |
| Kidney - 40 | ||||||
| Tongue - 30 | ||||||
| GI-tract - 20 | ||||||
| ENSG00000125779 | PANK2 | Pantothenate kinase 2 | 20 | 2.7.1.33 | ATP Magnesium (Mg2+) | Testis - 25 |
| Tonsils - 20 | ||||||
| Bone Marrow - 20 | ||||||
| Esophagus - 20 | ||||||
| ENSG00000120137 | PANK3 | Pantothenate kinase 3 | 5 | 2.7.1.33 | ATP Magnesium (Mg2+) | Liver - 40 |
| GI Tract - 30 | ||||||
| Breast - 25 | ||||||
| ENSG00000157881 | PANK4 | Pantothenate kinase 4 | 1 | 2.7.1.33 | ATP, Maganase (Mn2+), Nickel (Ni2+), Cobalt (Co2+) | Skeletal Muscle - 35 |
| Heart - 20 | ||||||
| Tongue - 20 | ||||||
| ENSG00000138801 | PAPSS1 | 3′-phosphoadenosine 5′-phosphosulfate synthase 1 | 4 | ATPS - 2.7.7.4, APSK - 2.7.1.25 | ATP, Magnesium (Mg2+) | Brain - 65 |
| Salvilary gland - 65 | ||||||
| Stomach - 60 | ||||||
| Skin - 50 | ||||||
| ENSG00000198682 | PAPSS2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | 10 | ATPS - 2.7.7.4 , APSK - 2.7.1.25 | ATP, Magnesium (Mg2+) | Adrenal - 135 |
| Liver - 80 | ||||||
| Colon - 80 | ||||||
| Lung - 70 | ||||||
| Placenta - 60 | ||||||
| ENSG00000138621 | PPCDC | Propionyl-CoA decarboxylase, alpha subunit | 15 | 4.1.1.36 | Coenzyme A, FMN | Parathyroid - 20 |
| Bone Marrow - 20 | ||||||
| Adrenal - 20 | ||||||
| Esophagus - 20 | ||||||
| ENSG00000127125 | PPCS | Propionyl-CoA synthase | 1 | 6.3.2.51 | B5, ATP, Magnesium (Mg2+) | Liver - 100 |
| Kidney - 90 | ||||||
| Pancreas - 70 | ||||||
| Testis - 70 | ||||||
| ENSG00000123453 | SARDH | Sarcosine dehydratase | 9 | 1.5.8.3 | FAD+, Pyridoxal-5-phosphate (PLP) | Liver - 115 |
| Pancreas - 40 | ||||||
| Testis - 40 | ||||||
| ENSG00000073578 | SDHA | Succinate dehydrogenase complex, subunit A | 5 | 1.3.5.1 | FAD+ | Heart - 600 |
| Skeletal muscle - 480 | ||||||
| Liver - 250 | ||||||
| ENSG00000117118 | SDHB | Succinate dehydrogenase complex, subunit B | 1 | 1.3.5.1 | Iron–Sulfur [Fe–S] | Tongue - 580 |
| Skeletal muscle - 500 | ||||||
| Liver - 310 | ||||||
| ENSG00000143252 | SDHC | Succinate dehydrogenase complex, subunit C | 1 | 1.3.5.1 | Iron–Sulfur [Fe–S] | Liver - 300 |
| Tongue - 280 | ||||||
| Kidney - 280 | ||||||
| Skeletal muscle - 280 | ||||||
| ENSG00000204370 | SDHD | Succinate dehydrogenase complex, subunit D | 11 | 1.3.5.1 | Iron–Sulfur [Fe–S], Heme | Tongue - 450 |
| Liver - 350 | ||||||
| Skeletal muscle - 330 | ||||||
| Kidney - 320 | ||||||
| ENSG00000176974 | SHMT1 | Serine hydroxymethyltransferase 1 | 17 | 2.1.2.1 | Pyridoxal-5-phosphate (PLP) | Liver- 400 |
| Kidney - 170 | ||||||
| ENSG00000182199 | SHMT2 | Serine hydroxymethyltransferase 2 | 12 | 2.1.2.1 | Pyridoxal-5-phosphate (PLP) | Liver - 180 |
| Pancreas - 60 | ||||||
| ENSG00000081800 | SLC13A1 | Solute carrier family 13 member A1 | 7 | NA | Sodium (Na+) | Kidney (Proximal Tubule) - 80 |
| ENSG00000145217 | SLC26A1 | Solute carrier family 26 member 1 | 4 | NA | NA | Liver - 18 |
| Adrenals - 10 | ||||||
| ENSG00000155850 | SLC26A2 - DTDST | diastrophic dysplasia sulfate transporter | 5 | NA | NA | Colon - 210 |
| Rectum - 200 | ||||||
| ENSG00000137767 | SQOR | Sulfide quinone oxidoreductase | 15 | 1.8.5.8 | FAD+ | Colon - 210 |
| Rectum - 180 | ||||||
| Muscle - 130 | ||||||
| Esophagus - 110 | ||||||
| ENSG00000196502 | SULT1A1 | Sulfotransferase family 1A member 1 | 16 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Liver - 250 |
| Duodenum - 140 | ||||||
| Small intestines - 100 | ||||||
| Adrenals - 100 | ||||||
| ENSG00000197165 | SULT1A2 | Sulfotransferase family 1A member 2 | 16 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) Calcium (Ca2+) | Liver - 50 |
| Duodenum - 50 | ||||||
| Small intestine - 50 | ||||||
| ENSG00000261052 | SULT1A3 | Sulfotransferase family 1A member 3 | 16 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Duodenum - 240 |
| Small Intestine - 140 | ||||||
| BRAIN - 50 | ||||||
| ENSG00000213648 | SULT1A4 | Sulfotransferase family 1A member 4 | 16 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Small Intestine - 70 |
| Colon - 50 | ||||||
| Adipose - 30 | ||||||
| Breast - 30 | ||||||
| ENSG00000173597 | SULT1B1 | Sulfotransferase family 1B member 1 | 4 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Duodenum - 100 |
| Small Intestine - 100 | ||||||
| ENSG00000198203 | SULT1C2 | Sulfotransferase family 1C member 2 | 2 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Stomach (gastric mucus cells) - 170 |
| Kidney - 70 | ||||||
| ENSG00000196228 | SULT1C3 | Sulfotransferase family 1C member 3 | 2 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Breast - 370 |
| ENSG00000198075 | SULT1C4 | Sulfotransferase family 1C member 4 | 2 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Brain - 30 |
| Gall bladder - 30 | ||||||
| Ovary - 30, | ||||||
| ENSG00000109193 | SULT1E1 | Sulfotransferase family 1E member 1 | 4 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) Sodium (Na+) | Liver - 40 |
| Duodenum - 30 | ||||||
| Vagina - 30 | ||||||
| Skin - 20 | ||||||
| ENSG00000105398 | SULT2A1 | Sulfotransferase family 2A member 1 | 19 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Liver - 1080 |
| Duodenum - 240 | ||||||
| Small intestine 100 | ||||||
| Duodenum - 60 | ||||||
| Parathyroid - 40 | ||||||
| ENSG00000088002 | SULT2B1a | Sulfotransferase family 2B member 1a | 19 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) Sodium (Na+) | Esophogus - 300 |
| Skin (karatinocytes), 200 | ||||||
| Vagina 150 | ||||||
| Cervix - 70 | ||||||
| Salivary glands - 50 | ||||||
| ENSG00000088002 | SULT2B1b | Sulfotransferase family 2B member 1b | 19 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) Sodium (Na+) | Esophogus - 300 |
| Skin (karatinocytes) - 200 | ||||||
| Vagina 150 | ||||||
| Cervix - 70 | ||||||
| ENSG00000130540 | SULT4A1 | Sulfotransferase family 4A member 1 | 22 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Brain (cerebral cortex) - 100 |
| Brain (white matter) - 30 | ||||||
| ENSG00000138068 | SULT6B1 | Sulfotransferase family 6B member 1 | 2 | 2.8.2.1 | 3-phospho-5-adenosyl sulfate (PAPS) | Testes - 5 |
| Brain - 3 | ||||||
| ENSG00000139531 | SUOX | Sulfite oxidase | 12 | 1.8.3.1 | Molybdenum (Mo) Cobalt (Co) Heme (Fe2+) | Liver - 70 |
| Parathyroid - 50 | ||||||
| Kidney - 40 | ||||||
| Muscle - 40 | ||||||
| ENSG00000138336 | TET1 | Ten-eleven translocation methylcytosine deoxigenase 1 | 10 | NA | Zinc (Zn2+) Iron (Fe2+) | Brain - 5 |
| ENSG00000168769 | TET2 | Ten-eleven translocation methylcytosine deoxigenase 2 | 4 | NA | Zinc (Zn2+) Iron (Fe2+) | Bone marrow - 25 |
| Skin - 10 | ||||||
| ENSG00000187605 | TET3 | Ten-eleven translocation methylcytosine deoxigenase 3 | 2 | NA | Zinc (Zn2+) Iron (Fe2+) | Bone marrow - 20 |
| Skin - 15 | ||||||
| Brain - 13 | ||||||
| ENSG00000169902 | TPST1 | Tyrosylprotein sulfotransferase 1 | 7 | 2.8.2.20 | NA | Liver - 50 |
| Gallbladder – 40 | ||||||
| Urinary bladder - 40 | ||||||
| ENSG00000128294 | TPST2 | Tyrosylprotein sulfotransferase 2 | 7 | 2.8.2.20 | NA | Pancreas - 400 |
| Epididymis - 100 | ||||||
| Liver - 70 | ||||||
| ENSG00000176890 | TS/TYMS | Thymidylate synthase | 18 | 2.1.1.45 | Tetrahydrofolate (THF) | Thymus - 115 |
| Bone - 90 | ||||||
| Tonsil - 45 | ||||||
| lymph - 40 | ||||||
| ENSG00000128311 | TST | Thiosulfate sulfurtransferase | 22 | 2.8.1.1 | Zinc (Zn2+) | Liver - 560 |
| GI Tract - 150 | ||||||
| Adrenals - 150 | ||||||
| ENSG00000112299 | VNN1 | Vanin 1 | 6 | 3.5.1.92 | NA | Liver – 220 |
| Duodenum 140 | ||||||
| Gallbladder - 90 | ||||||
| ENSG00000112303 | VNN2 | Vanin 2 | 6 | 3.5.1.92 | NA | Spleen – 60 |
| Tonsils & Lymph nodes – 40 | ||||||
| Bone Marrow - 30 | ||||||
| ENSG00000093134 | VNN3 | Vanin 3 | 6 | 3.5.1.92 | NA | Liver – 35 |
| Bone Marrow - 35 |
2. Inorganic sulfur species – hydrogen sulfide (H2S)
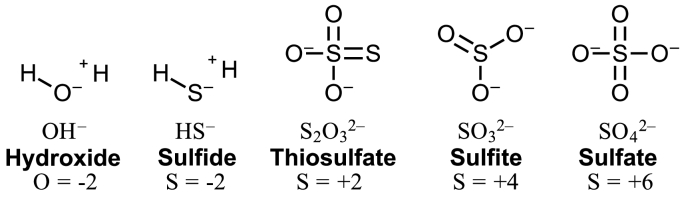
Among the most remarkable distinctions between sulfur and oxygen is the range of accessible oxidation states available for sulfur biology [19,20] (Fig. 1). The oxidation states of inorganic sulfur species found in the human body include hydrogen sulfide (H2S) at −2, thiosulfate (S2O32−) at +2, sulfite (SO32−) at +4, and sulfate (SO42−) at +6. Compared to oxygen, sulfur contains an available d-orbital for bond formation, has a larger radius, and is less electronegative (2.58 for sulfur, compared to oxygen at 3.44 on the Pauling scale). While the redox potential for H2O reduction is strongly positive (+0.82 V), the redox potential for H2S is more negative (−0.27 V). This redox potential of H2S is in the same range as NADH and FADH2 (−0.32 V and −0.20 V, respectively), which likely contributes to its ability to carry electrons and detoxify free radicals [1]. Additionally, H2S has a pKa1 of close to 7, which is near the physiological pH, which is significant as fluctuating H2S and HS− anions have different physiological functions and properties allowing sulfur compounds to determine the cellular redox potential, regulate metabolic pathways, and react to oxidative stress [21]. Notably, the ionized HS− is more reactive than the un-ionized H2S and exerts its biological effects through binding electrophiles in a process known as sulfhydration or attaching to other reactive sulfur species (thiols) to form disulfide bonds in a process known as persulfhydration. These post-translational processes, sulfhydration and persulfidation, also apply to thiol containing molecules (R–SH) such as cysteine and are regarded as fundamental molecular mechanisms for sensing/regulating oxidative stress [[22], [23], [24]].
Fig. 1.
2-D molecular structure of hydroxide anion (left) and inorganic sulfur species (sulfide, thiosulfate, sulfite, and sulfate) at the physiological plasma pH of about 7.4.
In healthy humans, the plasma baseline levels of H2S have been reported to lie in the range of 34 μM–274 μM; however, this does not specify the concentration of ionized to un-ionized H2S forms [25,26]. By assuming the plasma pH is about 7.4, the ionized HS− is the predominate form at 80% of the concentration, while the un-ionized H2S exists at 20%. This is important for two major reasons; first, HS− anions are biologically more reactive, attaching to proteins found on platelets which inhibit aggregation and also attaching to potassium-ATP channels causing vasodilation [2,19,[27], [28], [29], [30], [31]]. Secondly, since the ionized HS− form carries an overall negative charge, it requires facilitated diffusion and therefore specific transporters, such as anion exchange protein AE1 present on erythrocytes to enter cells [29]. On the other hand, the 20% H2S at the physiological plasma pH of 7.4 is more hydrophobic and penetrates the lipid bilayer of the cell membrane through simple diffusion. Inside the cell, where the pH of the cytosol is closer to a neutral 7, un-ionized H2S and ionized HS− anions would become roughly equal in concentration. In the basic environment of the mitochondria, where the pH can reach above 8, the concentration of the HS− anion again predominates at more than 90% of the sulfide concentration. Lastly, in a more acidic environment such as the lysosomes with a pH below 5, nearly 99% of un-ionized H2S predominates [21].
In recent years, accumulating evidence continues to show a key reciprocal relationship between hydrogen sulfide and oxygen in regulating metabolism and electron flow within the mitochondria [3,[32], [33], [34]]. More specifically, Kumar et al. and others have shown that under conditions of excess HS− concentrations, oxidative phosphorylation is inhibited by HS− binding to cytochrome a3 (cytochrome oxidase) of mitochondrial complex IV in the electron transport chain. Inhibition of Complex IV by HS− anions results in increased electrons at complex II, III, and the Coenzyme Q (CoQH2) pools. Although electrons readily escape as free radicals, the majority of electrons are shunted in a reverse flow through complex II to the sulfide quinone oxidoreductase (SQOR) reducing fumarate to succinate along with thiosulfate or oxidized glutathione (GSSH), depending on sulfur acceptor availability [1,3,20,[32], [33], [34], [35], [36], [37]]. A sulfide-driven metabolic flux may be a protective mechanism used in mammalian cells to avoid free radical damage during conditions of low pH, high proliferation, or hypoxia. In hypoxia, succinate accumulation is observed, and restoration of Complex II activity through pure oxygen reperfusion induces rapid succinate oxidation met with an already abundant CoQH2 pool causing electrons to leak as reactive oxygen species (ROS) and free radical tissue damage to occur [36]. To avoid tissue damage in reperfusion cases according to these mechanisms, hypothermic therapy and gentle rewarming of organs mixed with oxygen and sulfide monitoring may ameliorate the events and significantly reduce succinate-driven ROS production [37] (Fig. 2).
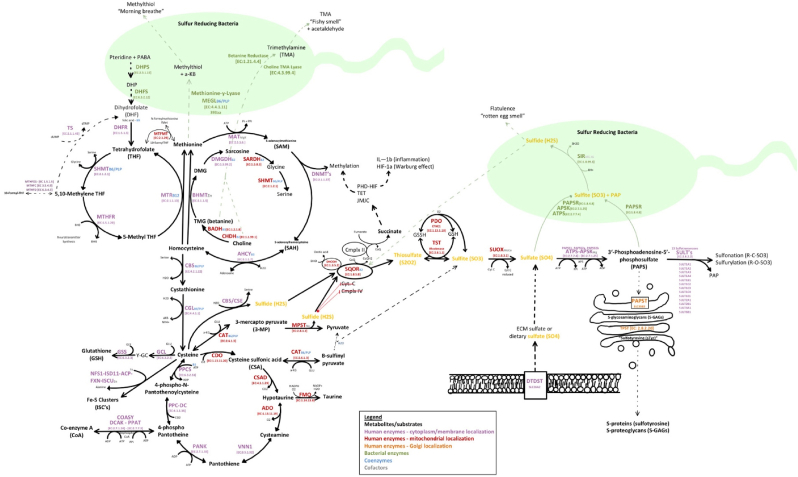
Fig. 2.
–In humans, sulfur pathways require sufficient intake of methionine along with adequate B-vitamins (B2, B3, B5, B6, B7, B9, B12), and trace elements zinc, nickel, molybdenum, cobalt, potassium, magnesium, iron, calcium, and sodium (Table 1) [4,6,7,19,68,71,77,174]. Methionine is essential as it can regenerate critical organic sulfur compounds, including the universal methyl donor s-adenosyl-l-methionine (SAM), homocysteine, cysteine, glutathione (GSH), iron-clusters (Fe–S), coenzyme A (CoA), hypotaurine, and taurine [4]. As a by-product of transsulfuration pathways, inorganic sulfur species (ISS) are produced. Specifically, the Cystathionine β-synthase (CBS), cystathionine γ-lyase (CGL), and 3-mercaptopyruvate sulfurtransferase (MST) enzymes produce endogenous H2S, mitochondrial SQOR, ETHE1/PDO, and TST enzymes produce SO32− and S2O32−, and mitochondrial SUOX produces endogenous SO42. Additionally, extracellular sulfate can also be transported into cells by SLC26A1 or SLC26A2 (DTDST) sulfate/chloride antiporters [63]. The oxidation of inorganic sulfur species (H2S, SO32− and S2O32−) then occurs step-wise in the mitochondria where thiosulfate and GSSH are oxidized by either iron-dependent persulfide dioxygenase (PDO/ETHE1) or by thiosulfate sulfur transferase (TST/rhodanese) to produce sulfite (SO32−) and regenerate reduced glutathione (GSH) [40,42,43,45,47,49,175]. The final and most oxidized inorganic sulfur species produced from sulfite (S = +4), is inorganic sulfate (SO42−) (S = +6), by molybdenum-dependent sulfite oxidase (SOUX), which resides in the intermembrane space of the mitochondria [54]. Sulfate is readily used as a substrate for sulfonation/sulfurylation reactions after activation to 3′-phosphoadenosine-5′-phosphosulfate (PAPS) by PAPS synthase [13,14].
3. Inorganic sulfur species – thiosulfate (S2O32−)
Thiosulfate has been shown to possesses antioxidant, anti-inflammatory, and antihypertensive properties [20,[38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50]]. Clinically, low concentrations of thiosulfate have been shown to reduce hypertension, left ventricular hypertrophy, fibrosis, and even rescue a failing heart [42]. Currently, sodium thiosulfate is an FDA approved medicine used for treating acute cyanide poisoning, carbon monoxide toxicity, and calcific uremic arteriolopathy found in dialysis patients [38,40,[42], [43], [44],48,51]. Systemically, thiosulfate (S2O32−) mainly serves as a superoxide scavenger to be excreted in the urine but can also be a major sulfur source for sulfonation/sulfurylation pathways [42] (Fig. 1).
Thiosulfate and sulfite are produced within the mitochondria of various tissues by the thiosulfate sulfurtransferase (TST) using inorganic sulfide anions (HS−) and glutathione-persulfide substrates [49]. For thiosulfate to be produced, a sulfhydryl group is donated to glutathione by sulfur-donating enzymes such as the membrane-bound sulfide quinone oxidoreductase (SQOR). TST can then use the sulfhydryl group from glutathione-persulfide to produce thiosulfate using an additional inorganic sulfide during sulfur oxidation [49]. Thiosulfate (S = +2) can be excreted into the plasma and or further oxidized to sulfite (S = +4) for sulfate-PAPS production (Fig. 1, Fig. 2).
4. Inorganic sulfur species – sulfite (SO32−)
Inorganic sulfite (SO32−) and sulfur dioxide (SO2) have both shown to cause a wide-range of allergic reactions and inflammatory symptoms at high concentrations [52]. While both sulfur dioxide (S = +4) and inorganic sulfite (S = +4) share similar sulfur oxidation states, sulfur dioxide has been described to exist as an aerosolized pollutant whereas inorganic sulfite is a dissolved ISS in human plasma [52]. Notably, mast cells and basophils have been reported to be sulfite-sensitive and induce histamine degranulation independent of calcium or IgE cross-linking upon exposure [53]. This means, inhalation of high concentrations of sulfur dioxide pollutants, ingestion of high volume sodium sulfites found in food additives, or dysfunctional sulfur oxidation pathways resulting in higher plasma sulfite concentrations may all be risk factors for anaphylaxis, especially in sulfite-sensitive patients (Fig. 1).
Under physiological conditions, sulfite (S = +4) is endogenously produced in the mitochondria and then further oxidized to inorganic sulfate (SO42−) (S = +6), by molybdenum-dependent sulfite oxidase (SOUX), which resides in the intermembrane space of the mitochondria [54]. Since SOUX is the rate-limiting enzyme for sulfite oxidation, single nucleotide polymorphisms (SNPs) or molybdenum deficiencies effecting enzymatic function can cause a wide range of sulfite sensitivities due to increased plasma sulfite (Fig. 1, Fig. 2).
5. Inorganic sulfate (SO42−) & 3′-phosphoadenosine-5′-phosphosulfate (PAPS)
In humans, inorganic sulfate is a vital ISS that is produced through sulfite oxidation by either molybdenum-dependent sulfite oxidase (SOUX) residing in the intermembrane space of the mitochondria or by the moonlighting functions of endothelial nitric oxide synthase (eNOS) [15,54,55]. Inorganic sulfate can also be consumed through diet or produced as a degradative product of the extracellular matrix by sulfatase enzymes [[56], [57], [58], [59]].
In humans, inorganic sulfate is activated through ATP and magnesium-dependent PAPS synthase (PAPSS1 or PAPSS2a/b) and then incorporated into biomolecules such as glycoproteins, glycosaminoglycans (GAGS), cholesterols, or even proteins in sulfonation/sulfurylation reactions by several sulfotransferase enzymes (SULTs/TPSTs). Inorganic sulfate levels in the plasma are retained by reabsorption of the filtered sulfate in the renal proximal tubule which requires the SLC13A1 (Solute Carrier Family 13 Member 1) which is a sodium-sulfate cotransporter (NaS1) located in the apical brush-border membrane and together with the SLC26A1-encoded sulfate-anion exchange transporter in the contra luminal basolateral membrane, allows for sulfate re-entry into blood circulation [[60], [61], [62]]. Of these two transport proteins, SLC13A1 Na-sulfate cotransport is believed to be the rate-limiting step in plasma sulfate retention and loss of this transporter may deplete the circulating sulfate pools and negatively affect general sulfonation/sulfurylation capacity [[60], [61], [62]]. When in circulation, inorganic sulfate can be transported into cells by SLC26A1 or SLC26A2 (DTDST) sulfate/chloride antiporters (Fig. 2) [63].
Upon entering the cell, inorganic sulfate is activated/adenylated using ATP to form adenosine phosphosulfate (APS) by ATP sulfurylase and then phosphorylated to form PAPS by the magnesium-dependent APS kinase activity [13,14]. Notably, the ATP sulfurylase and APS kinase activities are fused into a single polypeptide known as PAPSS which forms the universal sulfuryl donor. In humans there are two major isoforms PAPSS1 and PAPSS2 with a 77% identity in amino acid sequence. In addition, there is a splice variant PAPSS2b which contains an extra five amino acid sequence GMALP [13,14]. As the rate-limiting enzymes in the sulfurylation pathway, PAPSS supplies the sulfuryl group from PAPS to several different sulfotransferase (SULT) enzymes for sulfonation/sulfurylation reactions [64]. Among the thousands of end-products in sulfonation/sulfurylation, cholesterol sulfate (Ch-S), complex glycosaminoglycans (GAGS), and sulfotyrosine (proteins) are arguably the most vital for cellular homeostasis.
6. Organic sulfur compounds – methionine: folate-dependent and independent cycles
Methionine is an essential regulator of cellular homeostasis and sulfur pools through its direct regeneration of universal methyl donor s-adenosyl-l-methionine (SAM) and homocysteine, which replenish cysteine concentrations during the fed-state through the transsulfuration pathway [4]. The magnesium-dependent methionine adenosyltransferase (MAT1A or MAT2A/B) produces SAM from methionine using ATP. SAM is the universal methyl group donor used by nearly all methyltransferase reactions in every cellular compartment, second only to ATP in the number of biological reactions that use it [65]. When the methyl group is transferred from SAM, the remaining molecule is s-adenosyl-l-homocysteine (SAH) which functions as a feedback inhibitor of SAM-dependent methyltransferases. SAH can then be reversibly hydrolyzed to produce homocysteine and adenosine by the B3/NAD+-dependent SAH hydrolase (SAHH; AHCY) [66]. Notably, AHCY is the only reversible enzyme in the methionine cycle and as such is a key regulator of the SAM to SAH molar ratio, commonly referred to as the “methylation index” or “indicator of methylation capacity.” [67] When SAH and homocysteine concentrations increase faster than methionine regeneration, SAM utilizing enzymes are inhibited by SAH and the SAM concentrations then act in a feed-forward mechanism to allosterically activate the B6-dependent cystathionine beta-synthase (CBS) which metabolizes homocysteine through the transsulfuration pathway and allows the methionine cycle to continue [68,69]. While mammals assimilate methionine primarily through the diet, methionine is also regenerated from homocysteine by the folate cycle which utilizes the zinc and B12-dependent methionine synthase (MS; MTR) and or the zinc-dependent betaine-homocysteine S-methyltransferase (BHMT) with betaine (trimethylglycine; TMG) being the one-carbon donor produced from choline in the mitochondria of the liver and the kidneys [[70], [71], [72]]. It has been reported that ∼50% of methionine regeneration occurs through the folate cycle and ∼50% of methionine regeneration occurs by zinc-dependent BHMT (Fig. 2, Fig. 3) [71,72].
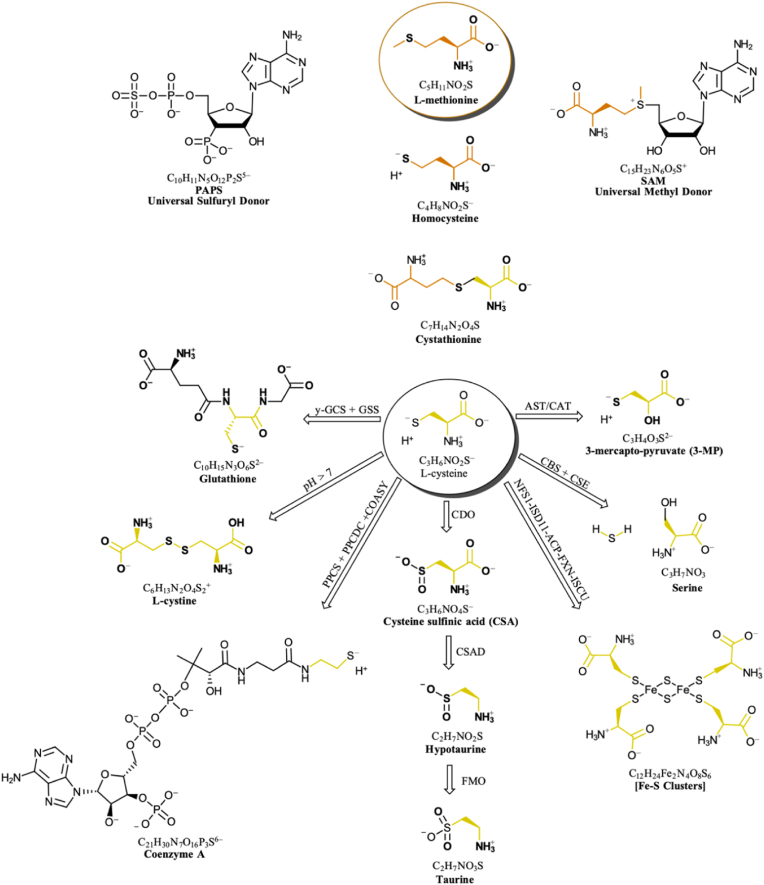
Fig. 3.
Organic sulfur compounds (OSC's) in human metabolism described in this review.
7. Transsulfuration – homocysteine to cysteine
In order to maintain cellular homeostasis and cysteine concentrations, dietary intake of methionine is vital because it replenishes cysteine through the transsulfuration pathway [73]. Transsulfuration from homocysteine to cysteine occurs by two steps; first, through a condensation reaction combining homocysteine with a serine amino acid by the enzyme CBS, which is the rate-limiting step of the transsulfuration pathway [74]. Secondly, cystathionine is hydrolyzed by cystathionine gamma lyase (CGL; CSE) into cysteine producing alpha keto-butyrate and ammonia by-products. Studies have shown, SAM allosterically activates CBS and inhibits of methylenetetrahydrofolate reductase (MTHFR) increasing the irreversible reaction of homocysteine to cysteine [21,[74], [75], [76], [77], [78], [79]] (Fig. 2).
8. Organic sulfur compounds – cysteine to glutathione (GSH), coenzyme A, taurine etc
While cysteine and the disulfide form cystine are relatively insoluble and toxic at high concentrations [80], cysteine is a crucial OSC and intermediate in the synthesis of proteins [81], coenzyme A [[82], [83], [84], [85], [86]], taurine [[87], [88], [89], [90]], iron-sulfur clusters [[91], [92], [93]], zinc-finger complexes/metalloproteins [94,95], and glutathione (GSH or γ-glutamyl-cysteinyl-glycine) [71,77,78,80,92,[96], [97], [98], [99], [100], [101], [102]]. Intracellular cysteine is maintained in the 80–100 μM range in most tissues except in the kidney, where its concentration is only ∼1 mM [58]. In the mitochondria, cysteine can be oxidized to hypotaurine and taurine [81,103,104], used by cysteine desulfurase (NFS1) to generate iron–sulfur (Fe–S) clusters [[91], [92], [93]], or function in sulfhydration and persulfidation reactions [22,23,30,105]. In the cytosol under conditions of abundant ATP concentrations and pantothenate (B5), cysteine is combined with B5 through a series of reactions that utilize three ATP molecules to form coenzyme A [[82], [83], [84], [85], [86]] (Fig. 2).
Under conditions of increased oxidative stress, cysteine concentrations are shifted toward producing glutathione (GSH; γ-glutamyl-cysteinyl-glycine), which represents the most abundant low molecular weight thiol present in the human body with an intracellular concentration of 1–10 mM [71]. GSH is among the most important thiols in the human body because of its ability to scavenge free radicals and regulate superoxides [16,106]. Low GSH levels are associated with rheumatoid arthritis, Alzheimer's, Parkinson's, cirrhosis, human immunodeficiency virus (HIV), asthma, cystic fibrosis, diabetes, hemorrhagic strokes, atherosclerosis, and benign cancers [71]. GSH is synthesized from the precursor of three common amino acids; cysteine, glutamate, and glycine by two ATP-dependent reactions catalyzed by γ-glutamylcysteine synthetase (GCS; GCL) and by GSH synthetase (GSS), respectively [98]. While cysteine is the rate-limiting substrate in GSH synthesis [71,[96], [97], [98], [99],107,108], the concentration of GSH is also enzymatically regulated. Specifically, GSH concentrations are increased by upregulation of the GCS enzyme through the nuclear erythroid factor 2-related factor 2 (NRF2) pathway and GSH concentrations are decreased by the γ-glutamyltranspeptidase (GGT) enzyme present on the external surface of certain cell types [71,77,78,96,[109], [110], [111]] (Table 1 and Fig. 3).
Most prominently, cysteine oxidation to taurine accounts for around 50% of cysteine pool utilization in most tissues [112]. Taurine serves various cellular functions including osmotic pressure regulation, cellular membrane stabilization, calcium signaling regulation, mitochondrial biogenesis upregulation, neurotransmitter inhibition through weak binding of the GABA-a receptor, conjugation of bile salts, and as a radical scavenger [87,88,[112], [113], [114]]. The process of cysteine oxidation to taurine occurs by two main pathways; first, through the oxidation of cysteine to form cysteine sulfonic acid (CSA) by the mitochondrial enzyme cysteine dioxygenase (CDO), followed by the decarboxylation to hypotaurine by cysteine sulfonic acid decarboxylase (CSAD) and an oxidation to taurine by the flavin-containing monooxygenase (FMO) enzyme [90]. Secondly, taurine can be formed through the breakdown of coenzyme A [103,104] (Fig. 2, Fig. 3).
9. Sulfur in post-translational modifications (S-PTMs)
Among the mechanisms for cellular signaling and sensing in both eukaryotic and prokaryotic cells, post-translational modifications (PTMs) are an abundant group of mechanisms where biochemical alterations significantly affect the diversity of protein localization, stability, and function (Fig. 2). In contrast to the better-known system of phosphorylation/dephosphorylation, the PTM system of sulfurylation is relatively less understood. Upon phosphorylation the recipient molecule receives two negative charges, whereas the recipient molecule upon sulfurylation receives one negative charge. Aside from this crucial difference, the bond angle, bond distance and the chemical natures are not very different at the macroscopic levels. Yet the charge differences alone can make some uniqueness to the sets of molecules that are being either phosphorylated or sulfurylated [115,116]. From the global analysis it is apparent that many eukaryotic proteins/enzymes are hormonally regulated by phosphorylation/dephosphorylation mechanisms, and small molecules (primary metabolites) are phosphorylated to keep the phosphorylated compounds trapped inside the cell. On the contrary, it is tempting to speculate that molecules that are sulfurylated are mostly secondary metabolites in nature and are typically directed outside of the cell although not exclusively. Some examples of sulfurylated macromolecules (>10,000 Da) include proteoglycans (glycosaminoglycans), cholecystokinin (CCK), thyroglobulin (TG), and glycoprotein receptors (TSHr, LH/hCGr, FSHr) [[117], [118], [119]]. Some small molecules that get sulfurylated include dopamine, estrogen, pregnenolone, glycan moiety of high endothelial venule, and selectin proteins [[119], [120], [121]]. Importantly, the recipient amino acid residues of the proteins that are often phosphorylated are serine, threonine and less frequently tyrosine. In contrast, the peptide/protein residue that is sulfurylated is only tyrosine. Since sulfurylated serine or threonine have not been discovered thus far in humans, we speculate that phosphorylation happens uniquely to serine/threonine residues of proteins (Venkatachalam unpublished) whereas tyrosine residues of peptide/proteins can be either phosphorylated or sulfurylated [120]. Thus, in broad terms clear evolutionary differences between sulfuryl versus phosphoryl transfer processes and their specificities must be understood for targeted therapies.
10. Sulfotyrosine
Of the potential post-translational modifications (PTMs) that can occur on a tyrosine residue, sulfurylation (R1-O-(SO32−)) of tyrosine is the most abundant PTM and is vital for the biological activity of many proteins directed out of the cell [122]. With a pKa near 1.5, the sulfuryl group of the sulfotryrosine remains fully ionized at any pH found in the biological system and therefore can serve to increase the plasma solubility and half-life of proteins, influence binding interactions, and even regulate the peptide/protein activities [122]. Sulfurylation of tyrosine occurs in the Golgi apparatus where the pH is more acidic (pH 6.0–6.8) and is directed by two protein-tyrosine sulfotransferases (TPST1/TPST2), each with different specificities and expression patterns [117,119] (Fig. 2). The catalyzed reaction involves the transfer of a sulfuryl group from the 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to the hydroxyl group of tyrosine residues which are surrounded by clusters of acidic residues, glutamate and aspartate [119,121,123].
11. Sulfated Glycosaminoglycans (S-GAGs)
Sulfated Glycosaminoglycans (S-GAGs) are highly sulfated macromolecules that are found on the surface of every cell and in the extracellular matrix (ECM) throughout the human body [124,125]. S-GAGs are a diverse class of long, linear, and heterogeneous polysaccharides characterized by disaccharide repeats composed of alternating units of uronic acid and amino-sugar, forming chains that range from 1 to 25,000 disaccharide units [124,125]. Heparin (HP), heparan sulfate (HS), dermatan sulfate (DS), chondroitin sulfate (CS), and keratan sulfate (KS) are all S-GAGs that play an important role in cell-cell communications, structural support, cell adhesion and signaling [46,[124], [125], [126], [127], [128], [129], [130]].
The biosynthesis of S-GAGs begins in the cytoplasm with the formation of a tetrasaccharide linkage to the core proteins, catalyzed by the sequential actions of four glycosyltransferases, which adds one xylose, two galactose, and one glucuronic acid residues (Xyl-gal-gal-glucA). These blocks are then transported into the Golgi lumen through a transmembrane antiporter, where the polymerization and sulfurylation of HP, HS, DS, CS, and KS occurs followed by anterograde vesicle transport to the outer cell membrane [110,111,124,125,129,131]. The sulfurylation of HP and HS typically occurs at positions 2, 3, and 6 of the glucuronic or iduronic acids. The sulfurylation of DS and CS typically occur at positions 4 and 6 of the N-acetylgalactosamine (GalNAc) residues. Lastly, sulfurylation of KS occurs at position 6 of both the N-acetylglucosamine and galactose [122,124].
Inversely, the degradation of the S-GAGs occurs both extracellularly and within the lysosomes [124]. Within the lysosomes, acid hydrolases degrade S-GAGs by the sequential removal of monosaccharides followed by the removal of the sulfur groups; sulfonate/sulfamate groups (R–NH–SO32-) and sulfuryl/sulfate groups (R-O-SO32-) [110,111,131]. If any of the 12 enzymes participating in S-GAG degradation malfunctions, the lysosomal accumulation of the substrates result in disorders known as mucopolysaccharidoses (MPS) [57,110,111,131]. Overall, S-GAGs are essential in providing structural support and negative charge to cells, and are particularly abundant in the cardiovascular endothelium and the glycocalyx, a protective barrier for epithelial cells in the gut [125,132] (Fig. 2).
12. Sulfur homeostasis in the gut microbiome and crosstalk
The human gut is home to a diverse community of microorganisms, including bacteria, viruses, fungi, and protozoa. It is estimated that the gut microbiome contains tens of thousands of different species of microorganisms, with the majority being bacteria [11,[133], [134], [135], [136]] (Table 2). The dominant gut microbial phyla are Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, with the two phyla Bacteroidetes and Firmicutes representing 90% of gut microbiota [137]. Bacteroidetes consists of a diverse group of bacteria, the predominant genera being Bacteroides and Prevotella. Firmicutes also consists of a diverse group of bacteria, the predominant genera being Clostridium, Staphylococcus, Lactobacillus, and Bacillus [137]. The Actinobacteriaphylum is proportionally less abundant and mainly represented by the Bifidobacterium genus. Overall, it is estimated that the gut microbiome contains 39 trillion microbial cells compared to 37 trillion human cells [138].
Table 2.
The dominant human gut microbial phyla are Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, with the two phyla Bacteroidetes and Firmicutes representing 90% of gut microbiota [137]. It is estimated that the gut microbiome contains 39 trillion microbial cells compared to 37 trillion human cells [138]. Additional enzyme information was obtained from KEGG pathways.
| PHYLUM | SUMMARY | COMMON BACTERIA | BACTERIAL SULFUR REDUCING ENZYMES |
|---|---|---|---|
| Bacteroides | Gram Negative (−) Anaerobic Rod-shaped |
Alistipes (−) Rod Bacteroides cellulosilyticus(−) Rod Bacteroides dorei(−) Rod Bacteroides fragilis(−) Rod Bacteroides ovatus(−) Rod Bacteroides vulgatus(−) Rod Bacteroides xylanisolvens(−) Rod Faecalibacterium prausnitzii(−) Rod Oscillibacter (−) Rod Parabacteroides(−) Rod Porphyromonas gingivalis(−) Rod Prevotella (−) Rod |
3-mercaptopyruvate sulfurtransferase (3-MST; sseA) PAPS Reductase (PAPSR/CysH) Desulfhydrase enzymes Sulfite Reductase (CysJ/CysI; SIR) O-acetylserine(thiol)lyas (OASS) enzymes. ArylSulfatases Cystathionine-gamma-synthase (metB) Cystathionine-gamma-lyase (mccB) Cystiene synthase (cysK, cysM) Desulfurylases (dcyD, malY, metC) Cysteine desulfhydrases (dcyD/CDS, CdsH, LCD/DES1) Methionine Gamma Lyase (MGL) GlycoSulfatase |
| Firmicutes | Gram Positive (+) Aerobic/Anaerobic/Facultative Rod/Coccus-shaped |
Bacillus anthrax (+) Rod Staphylococcus (+) Coccus Streptococcus (+) Coccus Clostridium difficile(+) Rod Clostridium perfinges(+) Rod Clostridium tetanus(+) Rod Lactobacillus(+) Rod Roseburia(+) Rod Fusobacterium (−) Spindle rod Eubacteria (+) Rod Haemophilis influenzae(−) Rod Listeria(+) Rod |
3-mercaptopyruvate sulfurtransferase (3-MST; sseA) PAPS Reductase (PAPSR/CysH) Desulfhydrase enzymes Sulfite Reductase (CysJ/CysI; SIR) O-acetylserine(thiol)lyas (OASS) enzymes. ArylSulfatase Cystathionine-gamma-synthase (metB) Cystathionine-gamma-lyase (mccB) Cystiene synthase (cysK, cysM) Desulfurylases (dcyD, malY, metC) Cysteine desulfhydrases (dcyD/CDS, CdsH, LCD/DES1) Methionine Gamma Lyase (MGL) |
| Proteobacteria | Gram Negative (−) Anaerobic/Facultative Rod-shaped |
Desulfovibrio (−) Rod Corynebacterium diphtheria(+) Rod Escherichia Coli(−) Rod Klebsiella(−) Rod Proteus (−) Rod Salmonella (−) Rod Shigella (−) Rod Vibrio cholerae(−) Rod Campylobacter jejuni(−) Spiral rod Helicobacter pylori(−) Spiral rod |
3-mercaptopyruvate sulfurtransferase (3-MST; sseA) PAPS Reductase (PAPSR/CysH) Desulfhydrase enzymes Sulfite Reductase (CysJ/CysI; SIR) O-acetylserine(thiol)lyas (OASS) enzymes. ArylSulfatases Cystathionine-gamma-synthase (metB) Cystathionine-gamma-lyase (mccB) Cystiene synthase (cysK, cysM) Desulfurylases (dcyD, malY, metC) Cysteine desulfhydrases (dcyD/CDS, CdsH, LCD/DES1) Methionine Gamma Lyase (MGL) GlycoSulfatase |
| Actinomycetota (Actinobacteria) | Gram Positive (+) Aerobic/Anaerobic Rod/Coccus/Branching |
Mycobacterium Leprae (+) Rod Mycobacterium Tuberculosis(+) Rod Nocardia (+) Rod Rhodococcus (+) Coccus/Branching Streptomyces (+) Coccus/Branching Actinomyces(+) Branching rod Bifidobacterium (+) Branching rod Collinsella (+) Cocci/Branching |
3-mercaptopyruvate sulfurtransferase (3-MST; sseA) PAPS Reductase (PAPSR/CysH) Desulfhydrase enzymes Sulfite Reductase (CysJ/CysI; SIR) O-acetylserine(thiol)lyas (OASS) enzymes. ArylSulfatases Cystathionine-gamma-synthase (metB) Cystathionine-gamma-lyase (mccB) Cystiene synthase (cysK, cysM) Desulfurylases (dcyD, malY, metC) Cysteine desulfhydrases (dcyD/CDS, CdsH, LCD/DES1) Methionine Gamma Lyase (MGL) |
In general, microbiota found in a healthy human gut form colonies and biofilms on the mucus layers of the host intestinal tissues which is spatially segregated and maintained by the secretion of thrombin from the epithelium [139]. In homeostasis, the mucus layers and thrombin keep the biofilms at bay, through proteolytic degradation of its matrix-associated proteins [139]. In the small intestine, antimicrobial peptides, IgA, IL-33, IL-10 and transforming growth factor-β (TGFβ), as well as cells such as CD103+ dendritic cells and regulatory T-cells are produced to also maintain gut homeostasis while the large intestine relies on a thick continuous mucus layer to compartmentalize the microbiome [32,[140], [141], [142], [143], [144]]. It is worth noting that bacterial composition and colonization is dynamic, not only diverse between individuals, but also in a single individual as trillions of microorganisms are constantly exposed to gut-environment changes (pH, temperature, etc.), diet-choices by the host, and other microbes. Among the diversity of the trillions of microorganisms, there are numerous metabolic processes that allow microbes to utilize elements and metabolites. Apart from sulfur, nitrogen, carbon, and phosphorus are heavily involved in the metabolic flux and contribute to the complexity of the overall gut-microbiome metabolism and its homeostasis.
Due to the dynamic complexity of the gut-microbiome, the mechanisms by which bacteria interact with the human host is complex and multifaceted. Some major mechanisms by which bacteria interact with the human host is through fermenting and producing metabolites such as short-chain fatty acids, like acetate, propionate, and butyrate, as well as more complex metabolites such as the hormones serotonin (5-HT) [145,146], cholecystokinin (CCK), gastric inhibitory peptide (GIP) and glucagon-like peptide 1 (GLP-1) [147], and various vitamins [147]. The metabolites can then affect the human host through direct metabolite PTM or indirect metabolic pathways, including chromatin regulation [10,[148], [149], [150]], chemical regulation of the gut–brain axis [16,135], and contributions to the immunomodulatory functions of the host [143,144].
Among the various metabolites and necessary elements, sulfur plays a crucial role in shaping the overall composition and contributions of the gut microbiome. Studies have reported that in healthy humans, concentrations of H2S were highest in the colon and commonly ranged from 0.3 to 3.4 mM [[151], [152], [153], [154]]. Low physiological H2S levels exert anti-inflammatory benefits, stabilize mucus layers in the gut, prevent fragmentation and adherence of the microbiota biofilm to the epithelium, inhibit the release of invasive pathogens, and help resolve inflammation and tissue injury [19,27,28,35,153,155,156]. In contrast, at excessive concentrations in the gut, H2S can cause mucus disruption, inflammation, and can then diffuse into the bloodstream where it has been reported to enter red blood cells and bind hemoglobin (Hb), methemoglobin (metHb), and glutathione disrupting systemic redox homeostasis, oxygen utilization, and free radical scavenging [157,158].
While oxygen is the final electron acceptor in oxidative phosphorylation for human metabolism, many microbes utilize ISS or OSC's rather than oxygen as electron acceptors for energy producing pathways. Sulfur-reducing bacteria (SRBs) such as those residing in the human gut, for example, rely on metabolizing OSCs through assimilatory pathways and ISS through dissimilatory pathways for energy which require sulfur species to be electron acceptors and produce H2S or reduced sulfur species as final products [[9], [10], [11]]. Two key enzymes able to complete the final step of sulfite reduction ubiquitous among microbes are the dissimilatory sulfite reductase (Dsr) and anaerobic sulfite reductase (Asr) enzymes [113]. While studies have mainly focused on bacteria harboring Dsr enzymes, Asr may be a more important contributor of sulfate reduction in the human gut [113].
Cowley et al. describes the diversity and distribution of sulfur enzymes within the gut microbiome with great detail [113]. Although the assimilatory metabolism of OSC's like methionine by methionine gamma lyase (mgl) and 3′-phosphoadenosine 5′-phosphate by PAPS Reductase (PAPSR), and sulfite by sulfite reductase (SIR) are common within the microbiome, cysteine-metabolizing enzymes are by far the most expressed and highly conserved genes across nearly all microbiota residing in the human gut-microbiome [113]. Genes associated with cysteine metabolism include cysteine synthase (cysK, cysM), cysteine desulfurase (iscS, sufS), cystathionine-γ-synthase (metB), desulfuryalases (dcyD, malY, metC, and mgl) as well as the three human orthologs cystathionine-β-synthase (CBS), cystathionine-γ-lyase (CSE/mccB), and 3-mercaptopyruvate sulfurtransferase (3MST) [113]. Additionally, cysteine desulfhydrase (dcyD/CDS, CdsH, LCD/DES1) are enzymes that produces H2S from cysteine and have been described in diverse intestinal pathogens such as Salmonella typhi [80,159], Mycobacterium tuberculosis, [31,80,159] Helicobacter pylori [160], and the protozoan Leishmania major, [161] as well as various oral microorganisms, including Streptococcus, Prevotella, and Fusobacterium [109,159,[161], [162], [163], [164], [165], [166], [167]].
In addition to cysteine metabolizing enzymes, arylsulfatases, glycosulfatases, and O-acetyl-l-serine sulfhydrylase (OASS) are enzymes that produce reduced sulfur products. Arylsulfatases are involved in the degradation of sulfated compounds and have been purified from Clostridium perfringens [57,58]. Glycosulfatase enzymes are also involved in the degradation of sulfated compounds and have been described primarily in the intestinal Prevotella and Bacteroides fragilis and have also been identified in the stomach Helicobacter pylori [160]. Conversely, the prevalent O-acetyl-l-serine sulfhydrylase (OASS) and homocysteine synthase mediate the microbial synthesis of cysteine from O-acetyl-l-serine and sulfide in bacteria and archaea [168].
The detailed mechanisms of the cross-talk between the gut microbiome and the human host are complex, dynamic, and still to be fully determined. While the mutually beneficial relationship of the host provides an environment and nutrients for the bacteria to thrive, the bacteria contribute to various aspects of human health. The functional contributions of the gut microbiome to host health are immense [8,12,16,135]. To name a few, microbiota maintain gut-barrier function in the mucus layers of the GI tract [139,153,[169], [170], [171], [172], [173]], support gut motility [145,148], secrete hormones like serotonin (5-HT) [145,146], cholecystokinin (CCK), gastric inhibitory peptide (GIP) and glucagon-like peptide 1 (GLP-1) [147], regulate chromatin [10,[148], [149], [150]], contribute to the gut–brain axis [16,135], and contribute to immunomodulatory functions of the host [143,144] (Table 2).
13. Conclusion
Sulfur pools in the human body are vast and include both inorganic sulfur species (ISS) as well as critical organic sulfur-containing compounds (OSCs). Endogenously produced ISS includes hydrogen sulfide (H2S), sulfite (SO32−), thiosulfate (S2O32−), and sulfate (SO42−). Endogenous OSCs include methionine, cysteine, universal methyl donor s-adenosyl-l-methionine (SAM), homocysteine, cystathionine, cysteine, glutathione (GSH), coenzyme A (CoA), hypotaurine, taurine, and others [4]. Through this review, we have explored the systemic effects of sulfur in biochemical pathways, including mechanisms of regulation by post-translational modifications, and explored the role of the gut microbiome in human sulfur metabolism. While this review has attempted to be thorough, the growing body of literature on the interconnectedness of sulfur pathways and crosstalk with gut microbiota, as well as, human brain sulfurylation are vast warranting further investigation and an up to date review. In future studies, we feel it is important to characterize rate-limiting enzymes including MTHFR, CBS, GCL, PAPSS, eNOS, SUOX, SQOR, and others within the sulfur pathways as they have implications for human health and disease. By investigating specific rate-limiting enzymes and sulfur pathway variations, we can begin to create pharmacological agents to treat associated diseases.
Declaration of competing interest
This is to attest that we do not have or claim “conflict of interest”.
Data availability
Data will be made available on request.
References
- 1.Bouillaud F. Sulfide oxidation evidences the immediate cellular response to a decrease in the mitochondrial ATP/O2 ratio. Biomolecules. 2022;12(3):361. doi: 10.3390/biom12030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kabil O., Banerjee R. Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 2010;285(29):21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar R., Landry A.P., Guha A., et al. A redox cycle with complex II prioritizes sulfide quinone oxidoreductase-dependent H2S oxidation. J. Biol. Chem. 2022;298(1) doi: 10.1016/j.jbc.2021.101435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller C.G., Schmidt E.E. Sulfur metabolism under stress. Antioxidants Redox Signal. 2020;33(16):1158–1173. doi: 10.1089/ars.2020.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson K.R. A case for hydrogen sulfide metabolism as an oxygen sensing mechanism. Antioxidants. 2021;10(11):1650. doi: 10.3390/antiox10111650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller C.G., Holmgren A., Arnér E.S.J., Schmidt E.E. NADPH-dependent and -independent disulfide reductase systems. Free Radic. Biol. Med. 2018;127:248–261. doi: 10.1016/j.freeradbiomed.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stipanuk M.H. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 2004;24(1):539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 8.Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16(8):461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 9.de Groot P., Scheithauer T., Bakker G.J., et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. 2020;69(3):502–512. doi: 10.1136/gutjnl-2019-318320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donohoe D.R., Holley D., Collins L.B., et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4(12):1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donohoe D.R., Garge N., Zhang X., et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metabol. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 13.Venkatachalam K.V., Fuda H., Koonin E.V., Strott C.A. Site-selected mutagenesis of a conserved nucleotide binding HXGH motif located in the ATP sulfurylase domain of human bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase. J. Biol. Chem. 1999;274(5):2601–2604. doi: 10.1074/jbc.274.5.2601. [DOI] [PubMed] [Google Scholar]
- 14.Venkatachalam K.V. Human 3′-phosphoadenosine 5′-phosphosulfate (PAPS) synthase: biochemistry, molecular biology and genetic deficiency. IUBMB Life. 2003;55(1):1–11. doi: 10.1080/1521654031000072148. [DOI] [PubMed] [Google Scholar]
- 15.Cooper I.D., Crofts C.A.P., DiNicolantonio J.J., et al. Relationships between hyperinsulinaemia, magnesium, vitamin D, thrombosis and COVID-19: rationale for clinical management. Open Heart. 2020;7(2) doi: 10.1136/openhrt-2020-001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makki K., Deehan E.C., Walter J., Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Scheithauer T.P.M., Rampanelli E., Nieuwdorp M., et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.571731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabil O., Vitvitsky V., Banerjee R. Sulfur as a signaling nutrient through hydrogen sulfide. Annu. Rev. Nutr. 2014;34(1):171–205. doi: 10.1146/annurev-nutr-071813-105654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishanina T.V., Libiad M., Banerjee R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 2015;11(7):457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bearden S.E., Beard R.S., Pfau J.C. Extracellular transsulfuration generates hydrogen sulfide from homocysteine and protects endothelium from redox stress. Am. J. Physiol. Heart Circ. Physiol. 2010;299(5):H1568–H1576. doi: 10.1152/ajpheart.00555.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul B.D., Snyder S.H. H2S: a novel gasotransmitter that signals by sulfhydration. Trends Biochem. Sci. 2015;40(11):687–700. doi: 10.1016/j.tibs.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., Du J., Tang C., Huang Y., Jin H. H2S-Induced sulfhydration: biological function and detection methodology. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang G., Zhao K., Ju Y., et al. Hydrogen sulfide protects against cellular senescence via S -sulfhydration of Keap1 and activation of Nrf2. Antioxidants Redox Signal. 2013;18(15):1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]
- 25.Karunya R., Jayaprakash K.S., Gaikwad R., et al. Rapid measurement of hydrogen sulphide in human blood plasma using a microfluidic method. Sci. Rep. 2019;9(1):3258. doi: 10.1038/s41598-019-39389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furne J., Saeed A., Levitt M.D. Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;295(5):R1479–R1485. doi: 10.1152/ajpregu.90566.2008. [DOI] [PubMed] [Google Scholar]
- 27.Zaorska E., Tomasova L., Koszelewski D., Ostaszewski R., Ufnal M. Hydrogen sulfide in pharmacotherapy, beyond the hydrogen sulfide-donors. Biomolecules. 2020;10(2):323. doi: 10.3390/biom10020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streeter E., Ng H.H., Hart J.L. Hydrogen sulfide as a vasculoprotective factor. Med. Gas Res. 2013;3(1):9. doi: 10.1186/2045-9912-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Łowicka E., Bełtowski J. Hydrogen sulfide (H2S) - the third gas of interest for pharmacologists. Pharmacol. Rep. 2007;59(1):4–24. [PubMed] [Google Scholar]
- 30.Sun H.J., Wu Z.Y., Nie X.W., Bian J.S. Role of hydrogen sulfide and polysulfides in neurological diseases: focus on protein S-persulfidation. Curr. Neuropharmacol. 2021;19(6):868–884. doi: 10.2174/1570159X18666200905143550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saini V., Chinta K.C., Reddy V.P., et al. Hydrogen sulfide stimulates Mycobacterium tuberculosis respiration, growth and pathogenesis. Nat. Commun. 2020;11(1):557. doi: 10.1038/s41467-019-14132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landry A.P., Ballou D.P., Banerjee R. H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase. J. Biol. Chem. 2017;292(28):11641–11649. doi: 10.1074/jbc.M117.788547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry A.P., Ballou D.P., Banerjee R. Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. Chembiochem. 2021;22(6):949–960. doi: 10.1002/cbic.202000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libiad M., Yadav P.K., Vitvitsky V., Martinov M., Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 2014;289(45):30901–30910. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szabo C., Ransy C., Módis K., et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 2014;171(8):2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chouchani E.T., Pell V.R., Gaude E., et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmann J., Otarashvili G., Meszaros A., et al. Restoring mitochondrial function while avoiding redox stress: the key to preventing ischemia/reperfusion injury in machine perfused liver grafts? Int. J. Mol. Sci. 2020;21(9):3132. doi: 10.3390/ijms21093132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder P.M., Frenay A.R.S., Koning A.M., et al. Sodium thiosulfate attenuates angiotensin II-induced hypertension, proteinuria and renal damage11These authors contributed equally to this manuscript. Nitric Oxide. 2014;42:87–98. doi: 10.1016/j.niox.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Song K., Wang F., Li Q., et al. Hydrogen sulfide inhibits the renal fibrosis of obstructive nephropathy. Kidney Int. 2014;85(6):1318–1329. doi: 10.1038/ki.2013.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bijarnia R.K., Bachtler M., Chandak P.G., van Goor H., Pasch A. Sodium thiosulfate ameliorates oxidative stress and preserves renal function in hyperoxaluric rats. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0124881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng T., Zhuo L., Wang Y., et al. Systematic review of sodium thiosulfate in treating calciphylaxis in chronic kidney disease patients. Nephrology. 2018;23(7):669–675. doi: 10.1111/nep.13081. [DOI] [PubMed] [Google Scholar]
- 42.Sen U., Vacek T.P., Hughes W.M., et al. Cardioprotective role of sodium thiosulfate on chronic heart failure by modulating endogenous H2S generation. Pharmacology. 2008;82(3):201–213. doi: 10.1159/000156486. [DOI] [PubMed] [Google Scholar]
- 43.Bebarta V.S., Brittain M., Chan A., et al. Sodium nitrite and sodium thiosulfate are effective against acute cyanide poisoning when administered by intramuscular injection. Ann. Emerg. Med. 2017;69(6):718–725.e4. doi: 10.1016/j.annemergmed.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen I.T.N., Wiggenhauser L.M., Bulthuis M., et al. Cardiac protection by oral sodium thiosulfate in a rat model of L-NNA-induced heart disease. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.650968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furne J., Springfield J., Koenig T., DeMaster E., Levitt M.D. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem. Pharmacol. 2001;62(2):255–259. doi: 10.1016/S0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 46.Chwatko G., Forma E., Wilkosz J., et al. Thiosulfate in urine as a facilitator in the diagnosis of prostate cancer for patients with prostate-specific antigen less or equal 10 ng/mL. Clin. Chem. Lab. Med. 2013;51(9) doi: 10.1515/cclm-2013-0069. [DOI] [PubMed] [Google Scholar]
- 47.Kruithof P.D., Lunev S., Aguilar Lozano S.P., et al. Unraveling the role of thiosulfate sulfurtransferase in metabolic diseases. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(6) doi: 10.1016/j.bbadis.2020.165716. [DOI] [PubMed] [Google Scholar]
- 48.Strazzula L., Nigwekar S.U., Steele D., et al. Intralesional sodium thiosulfate for the treatment of calciphylaxis. JAMA Dermatol. 2013;149(8):946. doi: 10.1001/jamadermatol.2013.4565. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M.Y., Dugbartey G.J., Juriasingani S., Sener A. Hydrogen sulfide metabolite, sodium thiosulfate: clinical applications and underlying molecular mechanisms. Int. J. Mol. Sci. 2021;22(12):6452. doi: 10.3390/ijms22126452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roorda M., Miljkovic J.L., van Goor H., Henning R.H., Bouma H.R. Spatiotemporal regulation of hydrogen sulfide signaling in the kidney. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsang R.Y., Al-Fayea T., Au H.J. Cisplatin overdose. Drug Saf. 2009;32(12):1109–1122. doi: 10.2165/11316640-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 52.Meng Z., Qin G., Zhang B., et al. Oxidative damage of sulfur dioxide inhalation on lungs and hearts of mice. Environ. Res. 2003;93(3):285–292. doi: 10.1016/S0013-9351(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 53.Liu M., Lu J., Chen Y., et al. Sodium sulfite-induced mast cell pyroptosis and degranulation. J. Agric. Food Chem. 2021;69(27):7755–7764. doi: 10.1021/acs.jafc.1c02436. [DOI] [PubMed] [Google Scholar]
- 54.Johnson-Winters K., Tollin G., Enemark J.H. Elucidating the catalytic mechanism of sulfite oxidizing enzymes using structural, spectroscopic, and kinetic analyses. Biochemistry. 2010;49(34):7242–7254. doi: 10.1021/bi1008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seneff S., Lauritzen A., Davidson R., Lentz-Marino L. Is endothelial nitric oxide synthase a moonlighting protein whose day job is cholesterol sulfate synthesis? Implications for cholesterol transport, diabetes and cardiovascular disease. Entropy. 2012;14(12):2492–2530. doi: 10.3390/e14122492. [DOI] [Google Scholar]
- 56.Sardiello M., Annunziata I., Roma G., Ballabio A. Sulfatases and sulfatase modifying factors: an exclusive and promiscuous relationship. Hum. Mol. Genet. 2005;14(21):3203–3217. doi: 10.1093/hmg/ddi351. [DOI] [PubMed] [Google Scholar]
- 57.Verheyen S., Blatterer J., Speicher M.R., et al. Novel subtype of mucopolysaccharidosis caused by arylsulfatase K (ARSK) deficiency. J. Med. Genet. 2022;59(10):957–964. doi: 10.1136/jmedgenet-2021-108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huijghebaert S.M., Mertens J.A., Eyssen H.J. Isolation of a bile salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl. Environ. Microbiol. 1982;43(1):185–192. doi: 10.1128/aem.43.1.185-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Agostini A.I., Dong J.C., de Vantéry Arrighi C., et al. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J. Biol. Chem. 2008;283(42):28115–28124. doi: 10.1074/jbc.M805338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee A., Beck L., Markovich D. The human renal sodium sulfate cotransporter (SLC13A1; hNaSi-1) cDNA and gene: organization, chromosomal localization, and functional characterization. Genomics. 2000;70(3):354–363. doi: 10.1006/geno.2000.6404. [DOI] [PubMed] [Google Scholar]
- 61.Markovich D. Na+-sulfate cotransporter SLC13A1. Pflügers Archiv. 2014;466(1):131–137. doi: 10.1007/s00424-013-1388-8. [DOI] [PubMed] [Google Scholar]
- 62.van de Kamp J.M., Bökenkamp A., Smith D.E.C., et al. Biallelic variants in the <scp> SLC13A1 </scp> sulfate transporter gene cause hyposulfatemia with a mild spondylo‐epi‐metaphyseal dysplasia. Clin. Genet. 2023;103(1):45–52. doi: 10.1111/cge.14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forlino A. A diastrophic dysplasia sulfate transporter (SLC26A2) mutant mouse: morphological and biochemical characterization of the resulting chondrodysplasia phenotype. Hum. Mol. Genet. 2005;14(6):859–871. doi: 10.1093/hmg/ddi079. [DOI] [PubMed] [Google Scholar]
- 64.Riches Z., Stanley E.L., Bloomer J.C., Coughtrie M.W.H. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie.”. Drug Metabol. Dispos. 2009;37(11):2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eustáquio A.S., Härle J., Noel J.P., Moore B.S. S-Adenosyl-L-Methionine Hydrolase (Adenosine-forming), a conserved bacterial and archeal protein related to SAM-dependent halogenases. Chembiochem. 2008;9(14):2215–2219. doi: 10.1002/cbic.200800341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nilsson R., Nicolaidou V., Koufaris C. Mitochondrial MTHFD isozymes display distinct expression, regulation, and association with cancer. Gene. 2019;716 doi: 10.1016/j.gene.2019.144032. [DOI] [PubMed] [Google Scholar]
- 67.Xiao J., You Y., Chen X., et al. Higher S-adenosylhomocysteine and lower ratio of S-adenosylmethionine to S-adenosylhomocysteine were more closely associated with increased risk of subclinical atherosclerosis than homocysteine. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.918698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pascale R.M., Peitta G., Simile M.M., Feo F. Alterations of methionine metabolism as potential targets for the prevention and therapy of hepatocellular carcinoma. Medicina. 2019;55(6):296. doi: 10.3390/medicina55060296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kulkarni A., Dangat K., Kale A., Sable P., Chavan-Gautam P., Joshi S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in wistar rats. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teng Y.W., Mehedint M.G., Garrow T.A., Zeisel S.H. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 2011;286(42):36258–36267. doi: 10.1074/jbc.M111.265348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masella Roberta, Mazza Giuseppe (Joe) John Wiley & Sons; 2009. Glutathione and Sulfur Amino Acids in Human Health and Disease. [Google Scholar]
- 72.Nazki F.H., Sameer A.S., Ganaie B.A. Folate: metabolism, genes, polymorphisms and the associated diseases. Gene. 2014;533(1):11–20. doi: 10.1016/j.gene.2013.09.063. [DOI] [PubMed] [Google Scholar]
- 73.Cooper A.J.L. Biochemistry of sulfur-containing amino acids. Annu. Rev. Biochem. 1983;52(1):187–222. doi: 10.1146/annurev.bi.52.070183.001155. [DOI] [PubMed] [Google Scholar]
- 74.Zuhra K., Augsburger F., Majtan T., Szabo C. Cystathionine-β-synthase: molecular regulation and pharmacological inhibition. Biomolecules. 2020;10(5):697. doi: 10.3390/biom10050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Settembre C., Di Malta C., Polito V.A., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garg S.K., Yan Z., Vitvitsky V., Banerjee R. Differential dependence on cysteine from transsulfuration versus transport during T cell activation. Antioxidants Redox Signal. 2011;15(1):39–47. doi: 10.1089/ars.2010.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mosharov E., Cranford M.R., Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39(42):13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 78.Vitvitsky V., Thomas M., Ghorpade A., Gendelman H.E., Banerjee R. A functional transsulfuration pathway in the brain links to glutathione homeostasis. J. Biol. Chem. 2006;281(47):35785–35793. doi: 10.1074/jbc.M602799200. [DOI] [PubMed] [Google Scholar]
- 79.Sbodio J.I., Snyder S.H., Paul B.D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 2019;176(4):583–593. doi: 10.1111/bph.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Go Y.M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J.H., Ling Y.Q., Fan J.J., Zhang X.P., Cui S. Expression of cysteine sulfinate decarboxylase (CSD) in male reproductive organs of mice. Histochem. Cell Biol. 2006;125(6):607–613. doi: 10.1007/s00418-005-0095-8. [DOI] [PubMed] [Google Scholar]
- 82.Badgley M.A., Kremer D.M., Maurer H.C., et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368(6486):85–89. doi: 10.1126/science.aaw9872. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gout I. Coenzyme A, protein CoAlation and redox regulation in mammalian cells. Biochem. Soc. Trans. 2018;46(3):721–728. doi: 10.1042/BST20170506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robishaw J.D., Neely J.R. Coenzyme A metabolism. Am. J. Physiol. Endocrinol. Metabol. 1985;248(1):E1–E9. doi: 10.1152/ajpendo.1985.248.1.E1. [DOI] [PubMed] [Google Scholar]
- 85.Theodoulou F.L., Sibon O.C.M., Jackowski S., Gout I. Coenzyme A and its derivatives: renaissance of a textbook classic. Biochem. Soc. Trans. 2014;42(4):1025–1032. doi: 10.1042/BST20140176. [DOI] [PubMed] [Google Scholar]
- 86.Matye D., Gunewardena S., Chen J., et al. TFEB regulates sulfur amino acid and coenzyme A metabolism to support hepatic metabolic adaptation and redox homeostasis. Nat. Commun. 2022;13(1):5696. doi: 10.1038/s41467-022-33465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hussy N., Deleuze C., Desarménien M.G., Moos F.C. Osmotic regulation of neuronal activity: a new role for taurine and glial cells in a hypothalamic neuroendocrine structure. Prog. Neurobiol. 2000;62(2):113–134. doi: 10.1016/S0301-0082(99)00071-4. [DOI] [PubMed] [Google Scholar]
- 88.Albrecht J., Schousboe A. Taurine interaction with neurotransmitter receptors in the CNS: an update. Neurochem. Res. 2005;30(12):1615–1621. doi: 10.1007/s11064-005-8986-6. [DOI] [PubMed] [Google Scholar]
- 89.Kevresan S., Kuhajda K., Kandrac J., Fawcett J.P., Mikov M. Biosynthesis of bile acids in mammalian liver. Eur. J. Drug Metab. Pharmacokinet. 2006;31(3):145–156. doi: 10.1007/BF03190711. [DOI] [PubMed] [Google Scholar]
- 90.Stipanuk M.H., Dominy J.E., Lee J.I., Coloso R.M. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 2006;136(6):1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 91.Ferecatu I., Canal F., Fabbri L., Mazure N.M., Bouton C., Golinelli-Cohen M.P. Dysfunction in the mitochondrial Fe-S assembly machinery leads to formation of the chemoresistant truncated VDAC1 isoform without HIF-1α activation. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poltorack C.D., Dixon S.J. Understanding the role of cysteine in ferroptosis: progress & paradoxes. FEBS J. 2022;289(2):374–385. doi: 10.1111/febs.15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Melber A., Winge D.R. 2018. Steps toward Understanding Mitochondrial Fe/S Cluster Biogenesis; pp. 265–292. [DOI] [PubMed] [Google Scholar]
- 94.Pace N., Weerapana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014;4(2):419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Foster M., Samman S. Zinc and redox signaling: perturbations associated with cardiovascular disease and diabetes mellitus. Antioxidants Redox Signal. 2010;13(10):1549–1573. doi: 10.1089/ars.2010.3111. [DOI] [PubMed] [Google Scholar]
- 96.Lu S.C. Regulation of glutathione synthesis. Mol. Aspect. Med. 2009;30(1–2):42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dickinson D.A., Forman H.J. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 2002;64(5–6):1019–1026. doi: 10.1016/S0006-2952(02)01172-3. [DOI] [PubMed] [Google Scholar]
- 98.Vitvitsky V., Mosharov E., Tritt M., Ataullakhanov F., Banerjee R. Redox regulation of homocysteine-dependent glutathione synthesis. Redox Rep. 2003;8(1):57–63. doi: 10.1179/135100003125001260. [DOI] [PubMed] [Google Scholar]
- 99.Griffith O.W. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 1999;27(9–10):922–935. doi: 10.1016/S0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 100.Sekhar R.V., Patel S.G., Guthikonda A.P., et al. Deficient synthesis of glutathione underlies oxidative stress in aging and can be corrected by dietary cysteine and glycine supplementation. Am. J. Clin. Nutr. 2011;94(3):847–853. doi: 10.3945/ajcn.110.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Combs J.A., DeNicola G.M. The non-essential amino acid cysteine becomes essential for tumor proliferation and survival. Cancers. 2019;11(5):678. doi: 10.3390/cancers11050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allocati N., Masulli M., Di Ilio C., Federici L. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7(1):8. doi: 10.1038/s41389-017-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao P., Yang C., Nesvick C.L., et al. Hypotaurine evokes a malignant phenotype in glioma through aberrant hypoxic signaling. Oncotarget. 2016;7(12):15200–15214. doi: 10.18632/oncotarget.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ueki I., Stipanuk M.H. 3T3-L1 adipocytes and rat adipose tissue have a high capacity for taurine synthesis by the cysteine dioxygenase/cysteinesulfinate decarboxylase and cysteamine dioxygenase pathways. J. Nutr. 2009;139(2):207–214. doi: 10.3945/jn.108.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ida T., Sawa T., Ihara H., et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA. 2014;111(21):7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bannai S., Sato H., Ishii T., Sugita Y. Induction of cystine transport activity in human fibroblasts by oxygen. J. Biol. Chem. 1989;264(31):18480–18484. doi: 10.1016/S0021-9258(18)51491-5. [DOI] [PubMed] [Google Scholar]
- 107.Bhatt A., Parsi M.A., Stevens T., et al. Volatile organic compounds in plasma for the diagnosis of esophageal adenocarcinoma: a pilot study. Gastrointest. Endosc. 2016;84(4):597–603. doi: 10.1016/j.gie.2015.11.031. [DOI] [PubMed] [Google Scholar]
- 108.Dickinson D.A., Moellering D.R., Iles K.E., et al. Cytoprotection against oxidative stress and the regulation of glutathione synthesis. Biol. Chem. 2003;384(4) doi: 10.1515/BC.2003.061. [DOI] [PubMed] [Google Scholar]
- 109.Castellarin M., Warren R.L., Freeman J.D., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Celik B., Tomatsu S.C., Tomatsu S., Khan S.A. Epidemiology of mucopolysaccharidoses update. Diagnostics. 2021;11(2):273. doi: 10.3390/diagnostics11020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khan S.A., Peracha H., Ballhausen D., et al. Epidemiology of mucopolysaccharidoses. Mol. Genet. Metabol. 2017;121(3):227–240. doi: 10.1016/j.ymgme.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kurtz J.A., VanDusseldorp T.A., Doyle J.A., Otis J.S. Taurine in sports and exercise. J Int Soc Sports Nutr. 2021;18(1) doi: 10.1186/s12970-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolf P.G., Cowley E.S., Breister A., et al. Diversity and distribution of sulfur metabolic genes in the human gut microbiome and their association with colorectal cancer. Microbiome. 2022;10(1):64. doi: 10.1186/s40168-022-01242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grimble R.F. The effects of sulfur amino acid intake on immune function in humans. J. Nutr. 2006;136(6):1660S–1665S. doi: 10.1093/jn/136.6.1660S. [DOI] [PubMed] [Google Scholar]
- 115.Venkatachalam K. Biochemical sulfuryl group transfer from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) versus phosphoryl transfer from ATP: what can Be learnt? Biochem. Physiol. Open Access. 2016;1(5) doi: 10.4172/2168-9652.1000192. [DOI] [Google Scholar]
- 116.Venkatachalam K. Novel cancer therapy: targeting methionine metabolism. Faseb. J. 2015;29(S1) doi: 10.1096/fasebj.29.1_supplement.897.32. [DOI] [Google Scholar]
- 117.Yang Y.S., Wang C.C., Chen B.H., Hou Y.H., Hung K.S., Mao Y.C. Tyrosine sulfation as a protein post-translational modification. Molecules. 2015;20(2):2138–2164. doi: 10.3390/molecules20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stone M.J., Chuang S., Hou X., Shoham M., Zhu J.Z. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. N. Biotech. 2009;25(5):299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 119.Hoffhines A.J., Damoc E., Bridges K.G., Leary J.A., Moore K.L. Detection and purification of tyrosine-sulfated proteins using a novel anti-sulfotyrosine monoclonal antibody. J. Biol. Chem. 2006;281(49):37877–37887. doi: 10.1074/jbc.M609398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gamage N., Barnett A., Hempel N., et al. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006;90(1):5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 121.Stewart V., Ronald P.C. Sulfotyrosine residues: interaction specificity determinants for extracellular protein–protein interactions. J. Biol. Chem. 2022;298(8) doi: 10.1016/j.jbc.2022.102232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Strott C.A. Sulfonation and molecular action. Endocr. Rev. 2002;23(5):703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 123.Niu H., Li M.Y., Wu X yuan, Long Y.T. Directly mapping the O-sulfation sites within CCR2 via an engineered nanopore. Biophys. J. 2023;122(3):319a. doi: 10.1016/j.bpj.2022.11.1789. [DOI] [Google Scholar]
- 124.Gandhi N.S., Mancera R.L. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 2008;72(6):455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 125.Solakyildirim K. Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques. Anal. Bioanal. Chem. 2019;411(17):3731–3741. doi: 10.1007/s00216-019-01722-4. [DOI] [PubMed] [Google Scholar]
- 126.Kumar S., Huang J., Cushnir J.R., Španěl P., Smith D., Hanna G.B. Selected ion flow tube-MS analysis of headspace vapor from gastric content for the diagnosis of gastro-esophageal cancer. Anal. Chem. 2012;84(21):9550–9557. doi: 10.1021/ac302409a. [DOI] [PubMed] [Google Scholar]
- 127.Yamagishi K., Onuma K., Chiba Y., et al. Generation of gaseous sulfur-containing compounds in tumour tissue and suppression of gas diffusion as an antitumour treatment. Gut. 2012;61(4):554–561. doi: 10.1136/gutjnl-2011-300721. [DOI] [PubMed] [Google Scholar]
- 128.Higashiyama S., Abraham J., Klagsbrun M. Heparin-binding EGF-like growth factor stimulation of smooth muscle cell migration: dependence on interactions with cell surface heparan sulfate. JCB (J. Cell Biol.) 1993;122(4):933–940. doi: 10.1083/jcb.122.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Villanueva I., Gladem S.K., Kessler J., Bryant S.J. Dynamic loading stimulates chondrocyte biosynthesis when encapsulated in charged hydrogels prepared from poly(ethylene glycol) and chondroitin sulfate. Matrix Biol. 2010;29(1):51–62. doi: 10.1016/j.matbio.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moran A.P., Gupta A., Joshi L. Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut. 2011;60(10):1412–1425. doi: 10.1136/gut.2010.212704. [DOI] [PubMed] [Google Scholar]
- 131.Filocamo M., Tomanin R., Bertola F., Morrone A. Biochemical and molecular analysis in mucopolysaccharidoses: what a paediatrician must know. Ital. J. Pediatr. 2018;44(S2):129. doi: 10.1186/s13052-018-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Woods E.C., Kai F., Barnes J.M., et al. A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. Elife. 2017;6 doi: 10.7554/eLife.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davis L.M.G., Martínez I., Walter J., Goin C., Hutkins R.W. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.De Angelis M., Piccolo M., Vannini L., et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Vadder F., Kovatcheva-Datchary P., Goncalves D., et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 136.Wolf P.G., Cowley E.S., Breister A., et al. Diversity and distribution of sulfur metabolic genes in the human gut microbiome and their association with colorectal cancer. Microbiome. 2022;10(1):64. doi: 10.1186/s40168-022-01242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arumugam M., Raes J., Pelletier E., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8) doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johansson M.E.V., Sjövall H., Hansson G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013;10(6):352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gharagozloo M., Gris K.V., Mahvelati T., Amrani A., Lukens J.R., Gris D. NLR-dependent regulation of inflammation in multiple sclerosis. Front. Immunol. 2018;8 doi: 10.3389/fimmu.2017.02012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lemire P., Robertson S.J., Maughan H., et al. The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep. 2017;21(13):3653–3661. doi: 10.1016/j.celrep.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 142.Mamantopoulos M., Ronchi F., Van Hauwermeiren F., et al. Nlrp6- and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity. 2017;47(2):339–348.e4. doi: 10.1016/j.immuni.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 143.Smith P.M., Howitt M.R., Panikov N., et al. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bachem A., Makhlouf C., Binger K.J., et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity. 2019;51(2):285–297.e5. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 145.Fukumoto S., Tatewaki M., Yamada T., et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;284(5):R1269–R1276. doi: 10.1152/ajpregu.00442.2002. [DOI] [PubMed] [Google Scholar]
- 146.Yano J.M., Yu K., Donaldson G.P., et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin A.M., Sun E.W., Rogers G.B., Keating D.J. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front. Physiol. 2019;10 doi: 10.3389/fphys.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Soret R., Chevalier J., De Coppet P., et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138(5):1772–1782.e4. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 149.Kaiko G.E., Ryu S.H., Koues O.I., et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165(7):1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krautkramer K.A., Kreznar J.H., Romano K.A., et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell. 2016;64(5):982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Attene-Ramos M.S., Nava G.M., Muellner M.G., Wagner E.D., Plewa M.J., Gaskins H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 2010 doi: 10.1002/em.20546. NA-NA. [DOI] [PubMed] [Google Scholar]
- 152.Kushkevych I., Dordević D., Kollar P., Vítězová M., Drago L. Hydrogen sulfide as a toxic product in the small–large intestine Axis and its role in IBD development. J. Clin. Med. 2019;8(7):1054. doi: 10.3390/jcm8071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ijssennagger N., van der Meer R., van Mil S.W.C. Sulfide as a mucus barrier-breaker in inflammatory bowel disease? Trends Mol. Med. 2016;22(3):190–199. doi: 10.1016/j.molmed.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 154.Dordević D., Jančíková S., Vítězová M., Kushkevych I. Hydrogen sulfide toxicity in the gut environment: meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes. J. Adv. Res. 2021;27:55–69. doi: 10.1016/j.jare.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wallace J.L., Wang R. Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat. Rev. Drug Discov. 2015;14(5):329–345. doi: 10.1038/nrd4433. [DOI] [PubMed] [Google Scholar]
- 156.Bos E.M., van Goor H., Joles J.A., Whiteman M., Leuvenink H.G.D. Hydrogen sulfide: physiological properties and therapeutic potential in ischaemia. Br. J. Pharmacol. 2015;172(6):1479–1493. doi: 10.1111/bph.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Klingerman C.M., Trushin N., Prokopczyk B., Haouzi P. H 2 S concentrations in the arterial blood during H 2 S administration in relation to its toxicity and effects on breathing. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;305(6):R630–R638. doi: 10.1152/ajpregu.00218.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bianco C.L., Savitsky A., Feelisch M., Cortese-Krott M.M. Investigations on the role of hemoglobin in sulfide metabolism by intact human red blood cells. Biochem. Pharmacol. 2018;149:163–173. doi: 10.1016/j.bcp.2018.01.045. [DOI] [PubMed] [Google Scholar]
- 159.Kumagai H., Choi Y.J., Sejima S., Yamada H. Formation of cysteine desulfhydrase by bacteria. Agric. Biol. Chem. 1975;39(2):387–392. doi: 10.1271/bbb1961.39.387. [DOI] [Google Scholar]
- 160.Slomiany B.L., Murty V.L., Piotrowski J., Grabska M., Slomiany A. Glycosulfatase activity of H. pylori toward human gastric mucin: effect of sucralfate. Am. J. Gastroenterol. 1992;87(9):1132–1137. [PubMed] [Google Scholar]
- 161.Marciano D., Santana M., Mantilla B.S., Silber A.M., Marino-Buslje C., Nowicki C. Biochemical characterization of serine acetyltransferase and cysteine desulfhydrase from Leishmania major. Mol. Biochem. Parasitol. 2010;173(2):170–174. doi: 10.1016/j.molbiopara.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 162.Takahashi Y., Yoshida A., Nagata E., et al. Streptococcus anginosusl-cysteine desulfhydrase gene expression is associated with abscess formation in BALB/c mice. Mol Oral Microbiol. 2011;26(3):221–227. doi: 10.1111/j.2041-1014.2010.00599.x. [DOI] [PubMed] [Google Scholar]
- 163.Yoshida Y., Ito S., Kamo M., et al. Production of hydrogen sulfide by two enzymes associated with biosynthesis of homocysteine and lanthionine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2010;156(7):2260–2269. doi: 10.1099/mic.0.039180-0. [DOI] [PubMed] [Google Scholar]
- 164.Dharmani P., Strauss J., Ambrose C., Allen-Vercoe E., Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect. Immun. 2011;79(7):2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Strauss J., Kaplan G.G., Beck P.L., et al. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17(9):1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 166.Metaxas M.A., Delwiche E.A. THE <scp>l</scp> -CYSTEINE DESULFHYDRASE OF ESCHERICHIA COLI. J. Bacteriol. 1955;70(6):735–737. doi: 10.1128/jb.70.6.735-737.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kostic A.D., Gevers D., Pedamallu C.S., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Borup B., Ferry J.G. O-acetylserine sulfhydrylase from methanosarcina thermophila. J. Bacteriol. 2000;182(1):45–50. doi: 10.1128/JB.182.1.45-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Desai M.S., Seekatz A.M., Koropatkin N.M., et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schroeder B.O. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf). 2019;7(1):3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lewis K., Lutgendorff F., Phan V., Söderholm J.D., Sherman P.M., McKay D.M. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm. Bowel Dis. 2010;16(7):1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 172.Martens E.C., Chiang H.C., Gordon J.I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Peng L., Li Z.R., Green R.S., Holzmanr I.R., Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in caco-2 cell monolayers. J. Nutr. 2009;139(9):1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cooper A.J.L. Biochemistry of sulfur-containing amino acids. Annu. Rev. Biochem. 1983;52(1):187–222. doi: 10.1146/annurev.bi.52.070183.001155. [DOI] [PubMed] [Google Scholar]
- 175.Tiranti V., Viscomi C., Hildebrandt T., et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 2009;15(2):200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.