Abstract
The skeletal muscle provides a very permissive physiological environment for adeno-associated virus (AAV) type 2-mediated gene transfer. We have studied the early steps leading to the establishment of permanent transgene expression, after injection of recombinant AAV (rAAV) particles in the quadriceps muscle of mice. The animals received an rAAV encoding a secreted protein, murine erythropoietin (mEpo), under the control of the human cytomegalovirus major immediate-early promoter and were sacrificed between 1 and 60 days after injection. The measurement of plasma Epo levels and of hematocrits indicated a progressive increase of transgene expression over the first 2 weeks, followed by a stabilization at maximal plateau values. The rAAV sequences were analyzed by Southern blotting following neutral or alkaline gel electrophoresis of total DNA from injected muscles. While a high number of rAAV sequences were detected during the first 5 days following the injection, only a few percent of these sequences was retained in the animals analyzed after 2 weeks, in which transgene expression was maximal. Double-stranded DNA molecules resulting from de novo second-strand synthesis were detected as early as day 1, indicating that this crucial step of AAV-mediated gene transfer is readily accomplished in the muscle. The templates driving stable gene expression at later time points are low in copy number and structured as high-molecular-weight concatemers or interlocked circles. The presence of the circular form of the rAAV genomes at early time points suggests that the molecular transformations involved in the formation of stable concatemers may involve a rolling-circle type of DNA replication.
Human adeno-associated virus (AAV) type 2 is a nonpathogenic parvovirus, whose 4.7-kb genome consists in two 145-nucleotide inverted terminal repeats (ITRs) and two genes, rep, encoding several proteins involved in the virus life cycle, and cap, encoding structural virion proteins (4). In infected cells, the incoming AAV single-stranded DNA (ssDNA) is converted into a double-stranded (ds) transcriptional template. Further steps in the productive viral replication require functions provided by a helper adenovirus or herpesvirus. In the absence of helper functions, the AAV dsDNA is able to integrate at specific sites into the human genome, most frequently on chromosome 19q (21). It then can persist indefinitely in a latent state until mobilized and rescued by a helper virus superinfection. This site-specific integration involves the Rep 68/78 proteins (37), which are thought to initiate the process by bridging genomic and viral sequences and nicking one strand in the ds AAV genome. Integration per se probably involves cellular recombination pathways. The absence of known pathogenicity together with this capacity for integrating in the host genome has rendered recombinant AAV (rAAV) vectors attractive for gene therapy prospects. The ITRs from the AAV genome are the only viral sequences required in cis to generate rAAV vectors. Recombinant constructs containing two ITRs bracketing a gene expression cassette of up to 4.5 kb are converted into an ssDNA vector genome and packaged into AAV particles, in the presence of the AAV rep and cap gene products and helper functions from an adenovirus (32).
The terminally differentiated and postmitotic muscle cells provide a highly permissive environment for rAAV-mediated gene transfer, and long-term expression in animal experiments has been obtained previously (7, 13, 15, 18, 25, 34, 38). The mechanism by which rAAV genomes become stabilized in the muscle fiber nuclei is still unclear. This establishment phase may take several weeks, as suggested by the progressive increase in gene expression that takes place following gene transfer (14, 18, 26, 33, 34). A consequence of this peculiar mode of expression may be the weak and transient immune response observed even with transgenes whose products are known to be strongly immunogenic in the context of an adenoviral vector (16).
Here, we have sought to characterize the status of vector DNA during this establishment phase. We have injected an rAAV encoding the murine erythropoietin (mEpo) into the muscles of mice and analyzed both transgene expression and rAAV DNA status at different time points. Our analysis indicates that a series of molecular transformations of rAAV DNA occur over the first month following gene transfer. The nature of the molecular intermediates identified here provides clues for the definition of a maintenance mechanism for the rAAV genome in the transduced muscle.
MATERIALS AND METHODS
rAAV vector production.
pSSV9-MD-mEPO was described previously (34). It contains the mEpo cDNA under the control of the cytomegalovirus (CMV) immediate-early promoter and intron 2 and 3′ untranslated region from the human β-globin gene (Fig. 1). rAAV AAV-MD-mEpo was prepared as described elsewhere (35). The titers of the rAAV preparations were determined by dot blot hybridization (1012 physical particles/ml) and by limiting-dilution infections. The stocks were determined to be free of detectable contamination by adenovirus and by replication-competent AAV (less than 1 wild-type AAV/108 rAAVs).
FIG. 1.
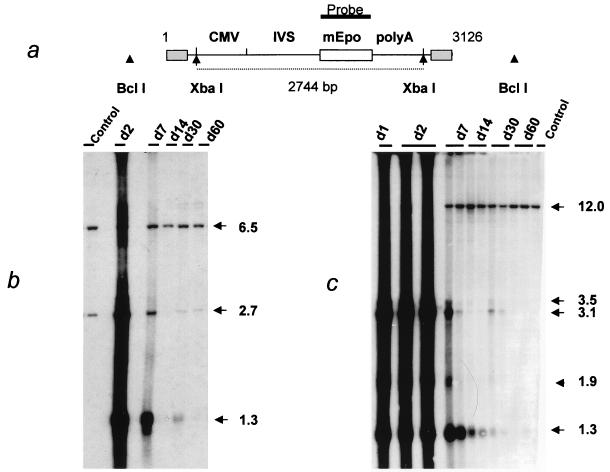
Southern blot analysis of the AAV-MD-mEpo AAV genomes in injected muscles. (a) Positions of restriction sites and probe (Epo cDNA) in AAV-MD-mEpo. Digestion with XbaI releases a 2.7-kb DNA fragment internal to the vector. BclI does not cut in the vector. AAV ITRs are represented as shaded boxes. CMV, immediate-early promoter from the human CMV; IVS, second intron from the human β-globin gene; polyA, untranslated region from the human β-globin gene. (b) Time course detection of AAV-MD-mEpo DNA: Southern blot analysis of XbaI-digested total DNA (4 μg) from injected muscles of animals sacrificed on day (d) 2 to day 60 after injection (experiment 1). Sacrifice dates are indicated above the lanes. Control, DNA from a noninjected muscle (4 μg) spiked with plasmid pSSV9-MD-mEpo (0.5 copy/haploid genome) and cut by XbaI. Apparent sizes of the hybridizing bands are indicated in kilobases on the right of the autoradiogram. The mEpo cDNA probe detects an endogeneous 6.5-kb fragment in the mouse genome. (c) Southern blot analysis of BclI-digested genomic DNA (8 μg) from injected muscles (day [d] 1 to day 60 after injection). Control, BclI-digested DNA from a noninjected muscle (8 μg). Apparent sizes (kilobases) of the different fragments are indicated on the right of the gel. The mEpo cDNA probe detects an endogenous 12-kb fragment in the mouse genome.
Animal studies.
Eight-week-old BALB/c mice were anesthetized with sodium barbitol (95 mg/kg of body weight) and injected in the quadriceps muscle with a single dose of 30 μl (2.7 × 1010 particles) of AAV-MD-mEpo. Two animals were sacrificed 1, 2, 7, 14, 30, and 60 days following injection (experiment 1), and three animals were sacrificed 1, 2, 3, 4, 5, 6, and 7 days following injection (experiment 2). For each animal, both quadriceps were taken and frozen in liquid nitrogen. Blood samples were collected into EDTA-coated tubes, by punction of the retroorbital sinus, with heparin-coated capillaries. Detection of mEpo was carried out on 20 μl of plasma in a total volume of 200 μl by a sandwich radioimmunoassay (I-125 EPO COATRIA; BioMerieux, Marcy l’Etoile, France). Each measurement was carried out in duplicate. For each assay, a standard curve was established by measuring the binding of total activity of standards included in the kit, ranging from 5 to 900 mU of Epo per ml. Since we used 10-fold-diluted samples and the first point of the standard curve is at 5 mU/ml, only values greater than 50 mU/ml were considered accurate. Transgene expression was also indirectly estimated by measuring the hematocrit (reflecting the level of circulating erythrocytes) by a microhematocrit method (20).
DNA analysis.
Total genomic DNA was isolated from injected quadriceps or control quadriceps by a standard procedure (3). Briefly, the muscle was frozen in liquid nitrogen, crushed with a mortar and pestle, and digested for 18 h at 50°C in a solution containing 100 mM NaCl, 10 mM Tris-HCl (pH 8), 25 mM EDTA (pH 8), and 0.5% sodium dodecyl sulfate, and 0.1 mg of proteinase K per ml. After two phenol-chloroform-isoamyl alcohol extractions and one chloroform extraction, the DNA was ethanol precipitated for 18 h at −20°C in 3.5 M ammonium acetate and resuspended in a solution consisting of 10 mM Tris-HCl (pH 8) and 0.1 mM EDTA. Standard Southern blot analysis was performed as follows. Four or eight micrograms of DNA was digested with restriction endonuclease for 16 h, electrophoresed through an 0.8% agarose gel, transferred onto a nylon membrane, and hybridized to a radiolabeled mEpo cDNA probe or to a probe complementary to the CMV immediate-early promoter (XbaI/HindIII restriction fragment of pSSV9-MD-mEPO). Filters were exposed for 1 to 7 days and analyzed on a Storm scanner (Molecular Dynamics). The ratio of viral genomes per haploid host genome was assessed on XbaI blots with the ImageQuant program by measuring the ratio of the hybridization signal from the vector internal 2.7-kb fragment to the hybridization signal from the endogenous Epo gene.
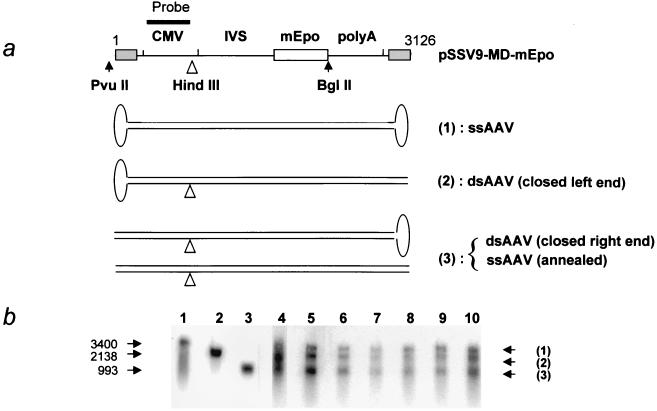
293 cells (American Type Culture Collection) were coinfected with AAV-MD-mEpo (multiplicity of infection of 5) and an adenovirus with E1 deleted (Ad dl324) (multiplicity of infection of 5). Cells were harvested 48 h after infection and lysed for total DNA extraction by a standard procedure (3). Active rAAV replication in these cells was assessed by Southern blot detection of ds species after nondenaturing gel electrophoresis. Alkaline gel electrophoresis was performed on 4 μg of HindIII-digested total DNA either from AAV-MD-Epo-injected muscles or from AAV-MD-Epo-infected 293 cells, as a control. Samples were loaded on a 25-cm-long denaturing gel as described elsewhere (31) and run for 4 h at 50 V. After alkali blotting, the filter was hybridized with the CMV promoter probe. We used PvuII-BglII- or PvuII-HindIII-purified restriction fragments from pSSV9-MD-mEPO as size markers (2,138- and 993-base-long molecules, respectively). DNA extracted from a purified preparation of an rAAV Neo green fluorescent protein (3,400 bases) (40) was used as a size marker for the full-length rAAV ssDNA.
RESULTS
An rAAV vector encoding mEpo (AAV-MD-mEpo) (34) was injected as a single intramuscular dose (2.7 × 1010 particles) into two groups of adult immunocompetent mice. The first group (n = 12) was studied over a 60-day period, and the second group (n = 21) was used to examine the events taking place during the first week following gene transfer. Blood was collected at the time of sacrifice, and the transgene activity was documented, either directly by measuring serum Epo levels or indirectly by assessing the hematocrit. Genomic DNA was prepared from the injected muscle, and the vector sequences were analyzed by Southern blotting.
In the first experimental group, an increase in mEpo plasma levels (Table 1) was first detected on day 7 (165-mU/ml average) and reached a plateau on day 14 (490-mU/ml average). This represented a 20-fold increase compared to the baseline value in noninjected animals. As a consequence of these elevated Epo levels, the hematocrit of animals sacrificed after day 7 was high, with values around 85% reached on day 30. The analysis of the second group indicated that the Epo transgene expression can be detected with confidence (i.e., Epo levels over 50 mU/ml [see Materials and Methods]) as early as day 4, with an effect on the hematocrit on day 6 (Table 2). Altogether, these measurements showed that following rAAV injection in the muscle, there was a progressive transgene expression over the first 2 weeks, before maximal stable values were reached.
TABLE 1.
Transgene expression and number of rAAV copies in injected animals (experiment 1)
| Days postinjection | Hematocrit (%) | Epo level (mU/ml) | No. of rAAV mEpo DNA copies/haploid genome |
|---|---|---|---|
| 1 | 50 | 13 | 45 |
| 48 | 31 | <0.1 | |
| 2 | 46 | 35 | 54 |
| 47 | 14 | 44 | |
| 7 | 56 | 208 | 2.1 |
| 55 | 122 | 0.3 | |
| 14 | 75 | 535 | 0.3 |
| 72 | 445 | 0.3 | |
| 30 | 85 | 330 | 0.2 |
| 83 | 374 | <0.14 | |
| 60 | 81 | 103 | <0.1 |
| 87 | 321 | 0.35 |
TABLE 2.
Transgene expression and number of rAAV copies in injected animals (experiment 2)
| Days postinjection | Hematocrit (%) | Epo level (mU/ml) | No. of rAAV mEpo DNA copies/haploid genome |
|---|---|---|---|
| 1 | 46 | 11 | 16 |
| 50 | 15 | 33 | |
| 50 | 22 | 12 | |
| 2 | 46 | 10 | 19 |
| 47 | 24 | 14 | |
| 49 | 19 | 11 | |
| 3 | 48 | 23 | 4 |
| 48 | 44 | 4.5 | |
| 9 | 50 | 33 | |
| 4 | 48 | 88 | 3 |
| 47 | 25 | 4.5 | |
| 46 | 85 | 7 | |
| 5 | 48 | 80 | 0.8 |
| 48 | 83 | <0.1 | |
| 52 | 110 | 2 | |
| 6 | 55 | 190 | 0.2 |
| 54 | 185 | <0.1 | |
| 54 | 119 | 0.6 | |
| 7 | 56 | 153 | 0.5 |
| 57 | 258 | 0.5 | |
| 54 | 216 | 0.5 |
The progressive appearance of transgene expression in rAAV-transduced tissues may be related to the kinetics of synthesis of ds forms of the vector DNA, suitable for transcription. In order to examine the formation of ds transcription templates, total DNA was extracted from the injected muscles and analyzed by Southern blotting. Digestion with XbaI releases a 2.7-kb DNA fragment internal to the vector (Fig. 1a). The intensity of this fragment is a measure of the total amount of rAAV dsDNA in the muscle DNA preparation. Digestion with BclI, an enzyme that does not cut within the rAAV genome, was used to analyze the status of the input vector DNA at different time points (Fig. 1c). At days 1 and 2, a strong signal with an apparent size of 1.3 kb was present in both XbaI and BclI digests. Since this band was not affected by any restriction enzyme used (including NheI and BglII [see Fig. 4b]), we attributed it to the input ss rAAV genome, according to previous reports (7, 13, 24, 34). This signal progressively decreased thereafter and was almost undetectable on day 60. Unexpectedly, the 2.7-kb XbaI fragment indicative of the presence of ds rAAV genomes was detected from day 1. It persisted up to 60 days (Fig. 1b), and there was an inverse relationship between its intensity and the amount of transgene expression. The XbaI analysis was used to quantify the amount of ds rAAV genome per haploid mouse genome (Table 1). On day 7, only 1 to 3% of the signal detected at day 2 remained (approximately 0.8 versus 28 copies/haploid genome). On day 60, the rAAV internal fragment was detectable at a level of 0.3 copy/haploid genome. Digestion with BclI resulted in a signal at 3.1 kb, the expected size for the linear ds monomer. This band was intense on days 1 and 2, decreased until day 30, and was no longer detected in animals sacrificed at day 60 (Fig. 1c), even when blots were overexposed (data not shown). Two additional bands at 3.5 and 1.9 kb were also detected from day 1 to day 30 and were no longer seen on day 60. These three forms of the rAAV genome were further analyzed in the second series of animals (see below). These data indicated that the ds rAAV genomes detected in the XbaI digest on day 60 had been converted to higher-molecular-weight forms that were not resolved in the BclI digest. The low intensity of the rAAV-specific hybridization signal (0.3 copy/genome) precluded the detection of a smear, corresponding to randomly integrated vector sequences, or to large concatemers of the rAAV genome. When a single-cut enzyme (BglII) was used on DNA obtained on day 30, only a 3.1-kb monomer-size band was observed, indicating that most vector genomes were structured either as head-to-tail repeats or possibly as complexes of interlocked circles (Fig. 2, lane 3).
FIG. 4.
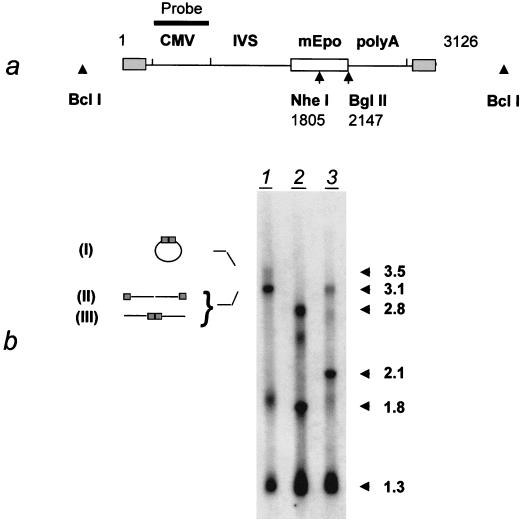
Southern blot analysis of rAAV DNA species in an animal sacrificed 1 day after injection. (a) Positions of restriction sites and probe (CMV promoter) in MD-AAV-mEpo (see legend to Fig. 1a). IVS, second intron from the human β-globin gene. (b) Four micrograms of total DNA was digested by BclI (lane 1), BglII (lane 3), or both BglII and NheI (lane 2). The diagram on the left indicates the deduced structure of the 3.5- and 3.1-kb DNA species: circular monomeric form (I), ds monomeric form (linear) (II), or linearized circular form and/or digested head-to-tail concatemer (III). The shaded boxes indicate the ITRs.
FIG. 2.
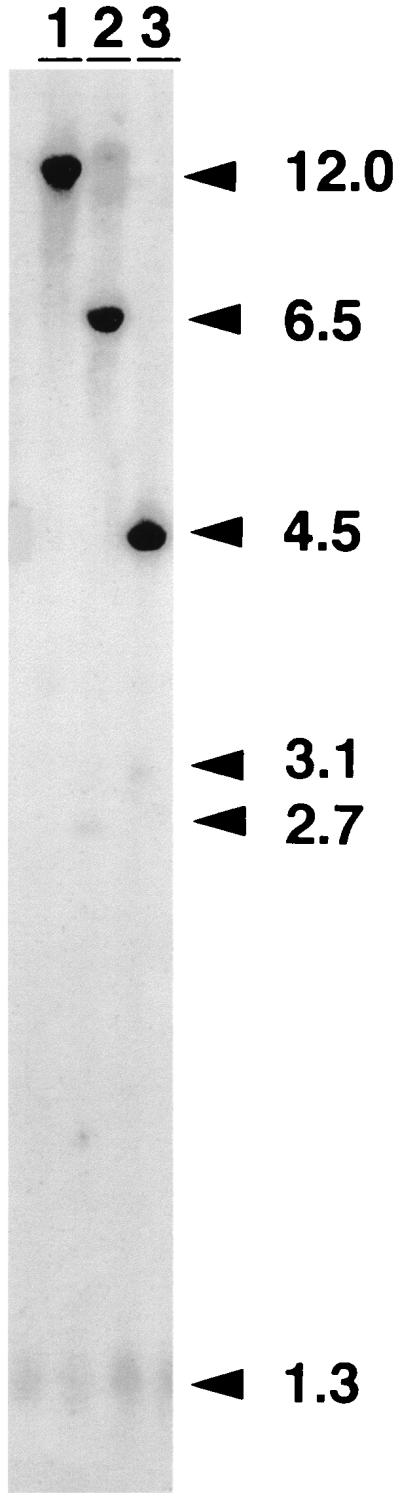
Detection of head-to-tail tandem by Southern blot analysis (mEpo cDNA probe). Each lane contains 8 μg of total DNA from an injected muscle (day 30 after injection) digested with BclI (lane 1), XbaI (lane 2), or BglII (lane 3). The mEpo cDNA probe detects in each lane an endogenous fragment (12, 6.5, and 4.5 kb, respectively). Numbers at right indicate size in kilobases.
The same analysis of genomic DNA prepared from rAAV-transduced muscles was performed on the second group of animals sacrificed between days 1 and 7. The same profiles as in experiment 1 were obtained when the DNA was digested by XbaI or BclI (data not shown). A very intense signal corresponding to over 20 copies of the 3.1-kb monomer per haploid genome was detected in samples from animals sacrificed between days 1 and 4 (Table 2). A 10-fold-weaker signal was present at day 4, a situation similar to the one observed at day 7 in the previous experiment. In order to verify whether the ds monomers arose from second-strand synthesis or from the annealing of complementary positive- and negative-strand ssDNA equally present in the AAV population (5), we performed alkaline gel electrophoresis of HindIII-digested DNA from injected quadriceps of five animals sacrificed 1 (n = 2), 2 (n = 1), and 3 (n = 2) days after injection.
As depicted in Fig. 3a, HindIII cuts at position 993 in the rAAV genome. After digestion, alkaline gel separation, and blot hybridization with a CMV promoter probe, the ds forms resulting from complementary-strand reannealing or from negative-strand synthesis (closed right end, Fig. 3a) are expected to yield the same band as the control pSSV9-MD-Epo plasmid cut with PvuII and HindIII (Fig. 3b, lane 3). In contrast, the ds species resulting from positive-strand synthesis and containing a closed terminal hairpin (closed left end, Fig. 3a) are expected to yield the same band as the control pSSV9-MD-Epo plasmid cut with PvuII and HindIII (Fig. 3b, lane 3). In contrast, the ds species resulting from positive-strand synthesis and containing a closed terminal hairpin (closed left end, Fig. 3a) are expected to be retarded on the alkaline gel. Both bands were indeed detected in a control DNA sample prepared from 293 cells coinfected with AAV-MD-mEpo and an E1-deleted adenovirus which contained newly synthesized ds forms (Fig. 3b, lane 4). The third, slowly migrating band corresponded to the input ss genome (Fig. 3b, lane 1). Figure 3b shows that these three molecular species were detected in all samples (lanes 5 to 10). In particular, the presence of the middle band indicated that a fraction of the ds monomers observed as early as day 1 arose from second-strand synthesis.
FIG. 3.
Southern blot analysis of the vector DNA after alkaline gel electrophoresis. (a) Positions of restriction sites and probe (CMV promoter) in pSSV9-MD-mEpo. The different species expected to be detected in the analysis are represented below. Three different fragments are expected upon HindIII digestion of these forms: (1) undigested ssDNA, (2) fragment from closed-left-end ds species (ds forms resulting from the synthesis of positive strand), and (3) fragment either from reannealed ds species or from closed-right-end species (ds forms resulting from synthesis of negative strand). IVS, second intron from the human β-globin gene. (b) Southern blot following alkaline gel electrophoresis of HindIII-digested total muscle DNA (4 μg) from animals sacrificed on day 1 (lanes 5 and 6 correspond to two different DNA samples from the same injected muscle split prior to the extraction process, and lane 7 corresponds to a second animal), day 2 (one animal, lane 8), and day 3 (two animals, lanes 9 and 10). Lane 4 contains DNA from 293 cells coinfected with AAV-MD-mEpo and a helper adenovirus as a control for second-strand synthesis. Lanes 1 to 3 contain the following size markers: purified DNA from rAAV green fluorescent protein Neo particles (lane 1), PvuII/BglII fragment from pSSV9-MD-mEpo (lane 2), and PvuII/HindIII fragment of pSSV9-MD-mEpo (lane 3). Sizes in bases are indicated on the left.
As mentioned above, two additional, rAAV-specific bands with apparent sizes of 1.9 and 3.5 kb were detected in early samples treated with BclI. In order to elucidate the nature of the 3.5-kb band, the total DNA from an animal sacrificed 1 day after injection was analyzed with BglII and NheI, two enzymes that cut the vector DNA once (Fig. 4a). Digestion with BglII (Fig. 4b, lane 3) yielded the expected 2.1-kb fragment from the linear monomer and eliminated the 3.5-kb species present in the BclI digest (Fig. 4b, lane 1). The signal remaining at 3.1 kb in the BglII digest was eliminated in the double digest with BglII and NheI (Fig. 4b, lane 2), in which a 2.8-kb fragment appeared. The presence of this fragment indicated that the 3.1-kb band in the BglII digest was a permuted form of the monomeric genome. Such a permutation can originate from either head-to-tail concatemers or circular forms of the genome. The fact that the 3.5-kb band disappeared in the BglII digest suggests that this band represents such circular forms.
The data shown in Fig. 4 also indicated that the 1.9-kb band was partially resistant to digestion by BglII (Fig. 4b). This suggests the presence of several comigrating molecular species. They possibly correspond to partially ds forms, produced by pausing of the second-strand DNA synthesis: depending on which strand is used as the DNA polymerase template, pauses will produce intermediates whose ds portion may or may not contain a BglII site. Further experiments will be needed to validate this hypothesis.
DISCUSSION
Permissiveness for rAAV-mediated gene transfer varies broadly among different tissues or cell types. For instance, differentiated tissues such as brain (22), retina (14), liver (19, 33), and skeletal muscle (7, 13, 15, 18, 25, 34, 38) are naturally permissive, whereas primary cell cultures or bone-marrow-derived cells (27) have been more difficult to transduce. Susceptibility to rAAV transduction may in some cases simply be related to the efficiency of vector entry. In this respect, cell surface levels of heparan sulfate, which acts as an attachment receptor, have been shown to be important (36). Gene expression following transduction is also highly dependent on the presence of the appropriate cellular environment. The cellular DNA synthesis machinery is needed to convert the vector ssDNA into a ds form that can serve as a transcription template (11, 12). Functions expressed by a coinfecting helper adenovirus, as well as various genotoxic treatments (1, 2, 29), are known to enhance this process. Cellular permissiveness has been correlated elsewhere with the phosphorylation status of a protein able to bind the D sequence on the AAV genome (28). Here, we have undertaken a time course analysis of transgene expression and vector genome status in the permissive context of the skeletal muscle. We observe that, following injection, high levels of rAAV genomes are found in total DNA preparations from days 1 to 5. The input ssDNA is retained for several days, and in some animals, it is still detectable at day 60. It is possible that this vector DNA comes from particles trapped by the heparan sulfate in the extracellular matrix surrounding muscle fibers (8) and that these vector particles continuously initiate transduction events over several days.
Our measurements indicate that transgene expression can be detected as early as day 4. This is consistent with the efficient conversion of the input ssDNA into dsDNA that we observed in DNA samples analyzed between days 1 and 3. Yet, most of the vector DNA originally present ends up being eliminated. Around day 5, the number of rAAV genome copies is dramatically reduced, and only 1 to 3% of the original amount of genetic material is retained after day 7. Unexpectedly, the clearance of most of the vector DNA is concomitant with the elevation of expression levels. Therefore, only a few genomes are stabilized and established as transcriptionally active. Monomers are not detectable on day 60, and our analysis suggests that most of the remaining genomes exist as high-molecular-weight species. These can be either head-to-tail concatemers as reported previously (7, 13, 34, 38) or, alternatively, interlocked circular forms of the genome.
The formation of high-molecular-weight structures appears to be key in the establishment of a stable genetic modification, but the underlying mechanism remains unclear. It is interesting to note that multimers which can be directly synthesized from a linear ss AAV template with the ITR priming structure are expected to be in an inverted configuration (head to head or tail to tail) (39). The predominance of direct repeats suggests that another mechanism is actually involved in concatemer formation. Here, we observe the presence of circular monomers which are formed very early after transduction and progressively disappear thereafter. This is in agreement with a recent report where Duan et al. describe the rescue of monomeric circular forms of the AAV genome from muscles injected with an rAAV (10). Interestingly, such circles could be used as templates for the formation of head-to-tail arrays of the rAAV genome by a rolling-circle replicative mechanism similar to the one described elsewhere for herpesviruses (6). Rolling-circle replication could be initiated through the interaction of a cruciform structure formed by the ITR and a cellular endonuclease (39).
Whether these high-molecular-weight forms are integrated into the host genome or are episomal remains to be elucidated. The integration of rAAV, mostly as single copies, has been directly documented in cell culture experiments, upon selection of the transduced cells (9, 17, 23, 30). Miao et al. have reported that, following rAAV transduction of the liver, multiple copies of rAAV genomes are associated with high-molecular-weight DNA in the megabase range, again as head-to-tail concatemers (24). A direct association of the vector sequences with the chromosomal DNA was suggested by in situ hybridization on metaphase chromosomes from explanted hepatocytes (24). However, direct evidence for integration of the rAAV genome following in vivo administration, such as the identification of junctions between rAAV and the host DNA, is still lacking. It remains possible that, given the appropriate cellular milieu, the multimerized vector DNA tightly associates with the chromosome and is constantly replicated, thereby escaping degradation within the nucleus.
ACKNOWLEDGMENTS
We thank Anne Marie Douar and Antoine Kichler for discussions and critical reading of the manuscript and Melinda Van Roey for animal care.
This work was supported by the Association Française contre les Myopathies.
REFERENCES
- 1.Alexander I E, Russell D W, Miller A D. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J Virol. 1994;68:8282–8287. doi: 10.1128/jvi.68.12.8282-8287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander I E, Russell D W, Spence A M, Miller A D. Effects of gamma irradiation on the transduction of dividing and nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum Gene Ther. 1996;7:841–850. doi: 10.1089/hum.1996.7.7-841. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]
- 4.Berns K I. Parvoviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 1743–1764. [Google Scholar]
- 5.Berns K I, Rose J A. Evidence for a single-stranded adenovirus-associated virus genome: isolation and separation of complementary single strands. J Virol. 1970;5:693–699. doi: 10.1128/jvi.5.6.693-699.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 7.Clark K R, Sferra T J, Johnson P R. Recombinant adeno-associated viral vectors mediate long-term transgene expression in muscle. Hum Gene Ther. 1997;8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 8.Cullen M J, Landon D N. The normal ultrastructure of skeletal muscle. In: Walton J, Karpati G, Hilton-Jones D, editors. Disorders of voluntary muscle. 6th ed. New York, N.Y: Churchill Livingstone; 1994. pp. 87–131. [Google Scholar]
- 9.Duan D, Fisher K J, Burda J F, Engelhardt J F. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 1997;48:41–56. doi: 10.1016/s0168-1702(96)01425-6. [DOI] [PubMed] [Google Scholar]
- 10.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher K J, Gao G P, Weitzman M D, De Matteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 14.Flannery J G, Zolotukhin S, Vaquero M I, LaVail M M, Muzyczka N, Hauswirth W W. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:6916–6921. doi: 10.1073/pnas.94.13.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzog R W, Hagstrom J N, Kung S H, Tai S J, Wilson J M, Fisher K J, High K A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jooss K, Yang Y, Fisher K J, Wilson J M. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol. 1998;72:4212–4223. doi: 10.1128/jvi.72.5.4212-4223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearns W G, Afione S A, Fulmer S B, Pang M C, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 18.Kessler P D, Podsakoff G M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeberl D D, Alexander I E, Halbert C L, Russell D W, Miller A D. Persistent expression of human clotting factor IX from mouse liver after intravenous injection of adeno-associated virus vectors. Proc Natl Acad Sci USA. 1997;94:1426–1431. doi: 10.1073/pnas.94.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koepke J A. Practical laboratory hematology. New York, N.Y: Churchill Livingstone; 1991. pp. 112–114. [Google Scholar]
- 21.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandel R J, Spratt S K, Snyder R O, Leff S E. Midbrain injection of recombinant adeno-associated virus encoding rat glial cell line-derived neurotrophic factor protects nigral neurons in a progressive 6-hydroxydopamine-induced degeneration model of Parkinson’s disease in rats. Proc Natl Acad Sci USA. 1997;94:14083–14088. doi: 10.1073/pnas.94.25.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao C H, Snyder R O, Schowalter D B, Patijn G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. [DOI] [PubMed] [Google Scholar]
- 25.Monahan P E, Samulski R J, Tazelaar J, Xiao X, Nichols T C, Bellinger D A, Read M S, Walsh C E. Direct intramuscular injection with recombinant AAV vectors results in sustained expression in a dog model of hemophilia. Gene Ther. 1998;5:40–49. doi: 10.1038/sj.gt.3300548. [DOI] [PubMed] [Google Scholar]
- 26.Murphy J E, Zhou S, Giese K, Williams L T, Escobedo J A, Dwarki V J. Long-term correction of obesity and diabetes in genetically obese mice by a single intramuscular injection of recombinant adeno-associated virus encoding mouse leptin. Proc Natl Acad Sci USA. 1997;94:13921–13926. doi: 10.1073/pnas.94.25.13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnazhagan S, Mukherjee P, Wang X S, Qing K, Kube D M, Mah C, Kurpad C, Yoder M C, Srou E F, Srivastava A. Adeno-associated virus type 2-mediated transduction in primary human bone marrow-derived CD34+ hematopoietic progenitor cells: donor variation and correlation of transgene expression with cellular differentiation. J Virol. 1997;71:8262–8267. doi: 10.1128/jvi.71.11.8262-8267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qing K, Khuntirat B, Mah C, Kube D M, Wang X S, Ponnazhagan S, Zhou S, Dwarki V J, Yoder M C, Srivastava A. Adeno-associated virus type 2-mediated gene transfer: correlation of tyrosine phosphorylation of the cellular single-stranded D sequence-binding protein with transgene expression in human cells in vitro and murine tissues in vivo. J Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell D W, Alexander I E, Miller A D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutledge E A, Russell D W. Adeno-associated virus vector junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder R O, Miao C H, Patijn G A, Spratt S K, Danos O, Nagy D, Gown A M, Winther B, Meuse L, Cohen L K, Thompson A R, Kay M A. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 34.Snyder R O, Spratt S K, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 35.Snyder R O, Xiao X, Samulski R J. Vectors for gene therapy: production of recombinant adeno-associated viral vectors. In: Dracopoli N, et al., editors. Current protocols in human genetics. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 12.1.1–12.1.24. [Google Scholar]
- 36.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X, Xiao W, Li J, Samulski R J. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]