ABSTRACT
The global prevalence of infections caused by extended-spectrum β-lactamase-producing Enterobacterales (ESBL-E) is increasing, and for Escherichia coli, observations indicate that this is partly driven by community-onset cases. The ESBL-E population structure in the community is scarcely described, and data on risk factors for carriage are conflicting. Here, we report the prevalence and population structure of fecal ESBL-producing E. coli and Klebsiella pneumoniae (ESBL-Ec/Kp) in a general adult population, examine risk factors, and compare carriage isolates with contemporary clinical isolates. Fecal samples obtained from 4,999 participants (54% women) ≥40 years in the seventh survey of the population-based Tromsø Study, Norway (2015, 2016), were screened for ESBL-Ec/Kp. In addition, we included 118 ESBL-Ec clinical isolates from the Norwegian surveillance program in 2014. All isolates were whole-genome sequenced. Risk factors associated with carriage were analyzed using multivariable logistic regression. ESBL-Ec gastrointestinal carriage prevalence was 3.3% [95% confidence interval (CI) 2.8%–3.9%, no sex difference] and 0.08% (0.02%–0.20%) for ESBL-Kp. For ESBL-Ec, travel to Asia was the only independent risk factor (adjusted odds ratio 3.46, 95% CI 2.18–5.49). E. coli ST131 was most prevalent in both collections. However, the ST131 proportion was significantly lower in carriage (24%) versus clinical isolates (58%, P < 0.001). Carriage isolates were genetically more diverse with a higher proportion of phylogroup A (26%) than clinical isolates (5%, P < 0.001), indicating that ESBL gene acquisition occurs in a variety of E. coli lineages colonizing the gut. STs commonly related to extraintestinal infections were more frequent in clinical isolates also carrying a higher prevalence of antimicrobial resistance, which could indicate clone-associated pathogenicity.
IMPORTANCE
ESBL-Ec and ESBL-Kp are major pathogens in the global burden of antimicrobial resistance. However, there is a gap in knowledge concerning the bacterial population structure of human ESBL-Ec/Kp carriage isolates in the community. We have examined ESBL-Ec/Kp isolates from a population-based study and compared these to contemporary clinical isolates. The large genetic diversity of carriage isolates indicates frequent ESBL gene acquisition, while those causing invasive infections are more clone dependent and associated with a higher prevalence of antibiotic resistance. The knowledge of factors associated with ESBL carriage helps to identify patients at risk to combat the spread of resistant bacteria within the healthcare system. Particularly, previous travel to Asia stands out as a major risk factor for carriage and should be considered in selecting empirical antibiotic treatment in critically ill patients.
KEYWORDS: Escherichia coli, Klebsiella pneumoniae, extended-spectrum β-lactamase, antimicrobial resistance, carriage, risk factors, general population, bacterial genomics
INTRODUCTION
Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli (ESBL-Ec) and Klebsiella pneumoniae (ESBL-Kp) are major contributors to the global burden of disease due to the antibiotic-resistant bacteria (1). The prevalence of infections caused by ESBL-Ec or ESBL-Kp is increasing, even in countries with low antibiotic use and availability on prescription only, like Norway (2). The mortality of invasive infections with ESBL-producing Enterobacterales (ESBL-E) ranges from 10% to 35%, depending on bacterial species, host factors, severity of disease, and initial antibiotic therapy (3, 4). Consequently, ESBL-Ec/Kp are high-priority pathogens when developing new drugs to combat the threat from antibiotic-resistant bacteria (5).
ESBL-Ec/Kp colonization precedes invasive infections (6 - 8), and recent reviews report an increasing global prevalence of human ESBL-E carriage in the community, with an eightfold rise over the past two decades (9, 10). The highest prevalence is detected in Asian and African regions (20%–70%) and the lowest in Europe and the Americas (<10%) (9, 10). A study from the USA shows that the increased incidence of ESBL-Ec infections was driven by an increase in community-onset cases (11), and human-to-human transmission was the attributable source to 60% of community-acquired gastrointestinal ESBL-Ec carriage in the Netherlands (12).
Several risk factors for ESBL-E gastrointestinal carriage have been described. However, these have, with a few exceptions (13, 14), mainly been investigated in small and/or selected study populations, such as international travelers (15), patients with gastroenteritis (16), patients recruited by general practitioners (17), persons with recent healthcare contact (18), pregnant women (19), children (20), or persons living in a livestock-dense area (21). Many studies have identified international travel as a risk factor for ESBL acquisition, while the significance of sex, age, antibiotic or proton pump inhibitor use, hospitalization, and diet are conflicting (13 - 17 - 18 - 22 - 24 - 24). Factors associated with the carriage of pathogenic bacteria are also shown to overlap. For instance, factors associated with Kp gastrointestinal carriage (25) overlap with those associated with ESBL-Ec carriage (15, 22, 26, 27). Additionally, an association between ESBL-E carriage and vancomycin-resistant enterococci (VRE) has been described previously (7, 28), and also that VRE colonization is significantly associated with Kp colonization among intensive care unit patients (29).
We have comprehensive knowledge of the prevalence and population structure of clinical ESBL-E isolates showing a dominance of CTX-M-group ESBL enzymes and the association with specific extraintestinal pathogenic E. coli (ExPEC) and multidrug-resistant K. pneumoniae high-risk clones (30 - 33). Large-scale genomic studies have identified specific subclades of E. coli sequence type (ST) 131 and K. pneumoniae ST307 as major contributors to the increasing prevalence of ESBL infections (30, 32, 34, 35). Community-based studies from the Netherlands and Sweden also observed a predominance of ST131, but a high genetic diversity within the ESBL-Ec population (13, 14).
Improved knowledge of risk factors for ESBL-E carriage and their population structure in the general human population may provide information for risk stratification and targeted infection control measures. In addition, studies show that it is important to consider ESBL-E carriage in critically ill septic patients with respect to the choice of empirical antibiotic treatment since ESBL-Ec/Kp colonization can precede invasive infections and carriage or non-carriage status may guide empirical antibiotic treatment (36 - 38). The aims of this study were to examine the prevalence of, and risk factors associated with, ESBL-Ec/Kp gastrointestinal carriage in a general adult population in Norway and to compare the ESBL-Ec population structure with a national collection of clinical isolates.
MATERIALS AND METHODS
Study population and design
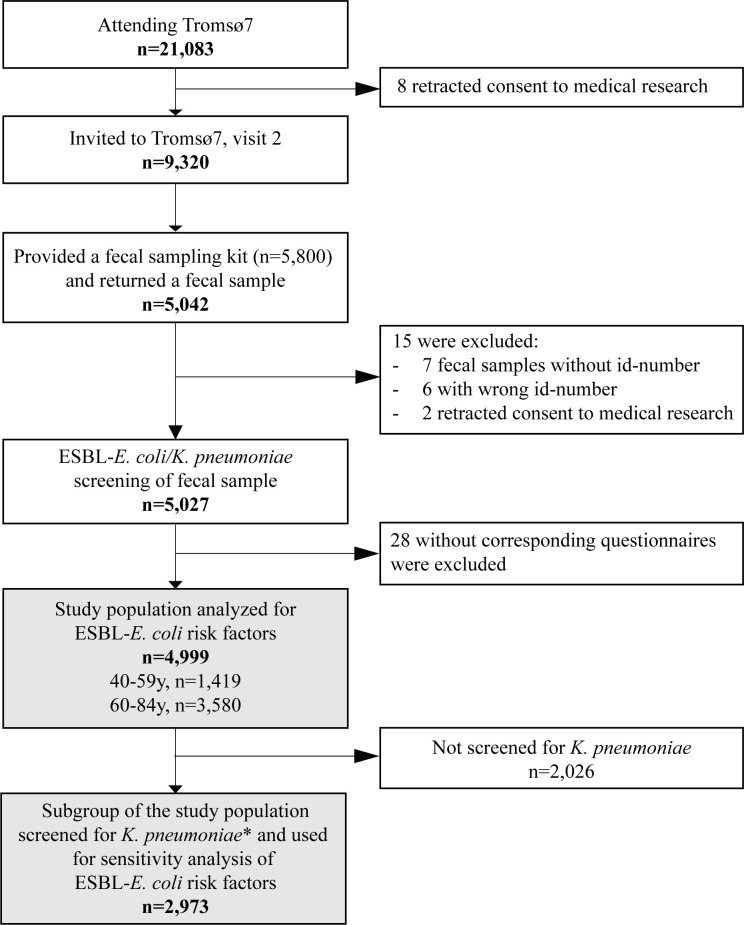
Our study sample was drawn from Tromsø7, the last of seven cross-sectional health surveys conducted between 1974 and 2016 in Tromsø municipality, Norway (https://uit.no/research/tromsostudy). Tromsø is representative of a Northern European, urban population (39). Tromsø7 (March 2015–October 2016) included questionnaires and two clinical visits (https://uit.no/research/tromsostudy/project?pid=708909). Unique national identity numbers from the official population registry were used to invite all citizens ≥40 years (n = 32,591). Sixty-five percent (n = 21,083, 11,074 women) attended the first clinical visit in the study (Fig. 1).
Fig 1.
Flow diagram of the study population, Tromsø7, 2015–2016. *Subgroup of the study population screened for K. pneumoniae carriage as previously described (25).
A selection of 9,320 persons attending the first visit was invited for a second visit, also including 3,154 former Tromsø Study participants not already included in the random selection process, which was required for other clinical research purposes. From March 2015 to March 2016, 5,800 participants at the first visit were consecutively provided a fecal self-sampling kit. Participants collected fecal material using nylon-flocked ESwab 490CE.A (Copan, Brescia, Italy). In total, 87% (n = 5,042) returned a sample either at the second visit or by mail to the laboratory.
All 5,042 fecal samples were screened for the presence of ESBL-producing Ec and Kp via selective culture (see below). All participants completed two self-administered structured questionnaires on sociodemographics, smoking, alcohol use, hospitalization, drug use, and travel abroad. We excluded 13 participants with the wrong or missing sample identification numbers, two retracting consent to medical research, and 28 with incomplete questionnaires for a final study population of 4,999 participants (Fig. 1). We analyzed the association between ESBL-Ec gastrointestinal carriage and different risk factors in 4,999 participants (Fig. 1). Next, we conducted a sensitivity analysis studying the association between Kp gastrointestinal carriage and ESBL-Ec carriage among 2,973 participants additionally screened for Kp in our previous study (25), irrespective of resistance (Fig. 1).
Isolation of ESBL-producing E. coli and K. pneumoniae
We added 200 µL of 85% glycerol to the ESwab tubes on arrival at the local microbiological laboratory and stored the samples at −80°C. From the thawed media, 100 µL were plated onto CHROMagar ESBL (CHROMagar, Paris, France) and incubated for 48 hours at 37°C. Pink, purple, and blue colonies suspected of being ESBL-producing Ec or Klebsiella spp. were identified using mass spectrometry (matrix-assisted laser desorption ionization-time of flight [MALDI-TOF]; Bruker Daltonics, Bremen, Germany). The first colony identified as either E. coli, K. pneumoniae, or Klebsiella variicola from each sample was kept and further analyzed. All samples were plated on cysteine lactose electrolyte deficient agar (MAST Group, Bootle, UK) to assess the growth of fecal flora and validity of the samples.
K. pneumoniae isolation
The screening strategy and isolation procedure for Kp gastrointestinal carriage of 2,973 participants in Tromsø7 are described in detail elsewhere (25). Briefly, we plated and screened the fecal samples onto the selective SCAI (Simons citrate agar with inositol; both Sigma-Aldrich, Darmstadt, Germany) medium and identified suspected colonies using MALDI-TOF.
Antimicrobial susceptibility testing and phenotypic ESBL identification
Susceptibility testing was performed according to the European Commitee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution method for carriage isolates and disc diffusion method (40) for the clinical isolates, and both were interpreted using the EUCAST 2023 clinical breakpoint table (https://eucast.org/). For the confirmation of ESBL-producing Ec and Kp, we followed the EUCAST algorithm for the phenotypic detection of ESBLs using the BD BBL combination disk test (Becton Dickinson and Company, Sparks, NV, USA).
Genomic sequencing and bioinformatic analysis
Genomic DNA from all 166 ESBL-Ec of the Tromsø7 collection was extracted with the MagNA Pure 96 system (Roche Applied Science, Mannheim, Germany), and sequencing libraries were prepared according to the Nextera Flex sample preparation protocol (Illumina, San Diego, CA, USA). Samples were sequenced on the Illumina MiSeq platform to generate 300-bp paired-end reads. All reads were trimmed with TrimGalore v0.6.4 and assembled with Unicycler v0.4.8 including SPAdes v3.13.0 (41 - 43). STs were assigned using the multilocus sequence type (MLST) software E. coli scheme v2.19.0 and Enterobase (44 - 46). AMRFinderPlus v3.10.16 was used to determine resistance genes among the Ec isolates (47). Plasmid replicons were identified using Abricate v1.0.1 including the PlasmidFinder 2021-March-27 database (48, 49). Phylogroup assignment was based on ClermonTyping v20.03 (March 2020) (50). The fimH type was identified using FimTyper v1.0 and BLAST+ v2.12.0 (51, 52). Regarding the Klebsiella isolates, Kleborate v2.0.0 was used to determine species identification, ST, and acquired genes encoding virulence or antibiotic resistance (53, 54).
The clinical ESBL-producing Ec included 118 isolates out of 123 representing all ESBL-Ec isolates collected in 2014 (55), as part of the yearly Norwegian Surveillance Program of Antimicrobial Resistance (NORM). The surveillance program included all clinical microbiological laboratories in Norway. The sampling period was 6 months and 2 days for blood and urine isolates, respectively. Genome sequencing of five isolates was unsuccessful. Before sequencing, the NORM 2014 isolates were stored at −80°C and then sent to GATC Biotech AG (part of Eurofins Genomics/Eurofins Scientific) in Germany for DNA isolation and WGS. Raw reads were trimmed using Trimmomatic v0.39 and assembled with SPAdes v3.15.0 (56, 57). Contigs shorter than 200 bp were discarded. The genomic data were analyzed as described above.
Phylogenetic and population structure analysis of ESBL-E. coli
We used Prokka v.1.14.6 (58) to annotate genomes and snippy v.4.6.0 (59) to map the sequence reads to the ST131 Ec EC958 chromosome (HG941718.1) (https://www.ncbi.nlm.nih.gov/nuccore/HG941718.1) to create the core genome alignment with sufficient resolution to differentiate between the ST131 C1 and C2 subclades. We used snp-dists v.0.8.2 (https://github.com/tseemann/snp-dists/) to create the single nucleotide polymorphism (SNP) distance matrix from the core genome alignment. We used a 17 SNP cutoff to define the similarity between two genomes and to identify probable ESBL-Ec transmission events among cases (60). To assess the phylogenetic relatedness, we used the core SNP alignment to infer a maximum-likelihood tree using RAxML v.8.2.8 with the GTR + Gamma rate model and 100 rapid bootstraps visualized in iTol (v6.5.2) (61, 62). ST131 subclades were determined based on subclade-specific SNPs and fimH alleles, and subclade membership was corrected when assignment based on the SNP profile of sporadic isolates did not fit with the phylogenetic distribution of clades (30, 35, 51, 63).
Statistical analysis
Our primary analysis was a multivariable logistic regression model, with the outcome variable ESBL-Ec gastrointestinal carriage using SPSS v.26.0 (SPSS, Inc., Chicago, IL, USA). We analyzed factors associated with ESBL-Ec gastrointestinal carriage among 4,999 participants (Table 1). Both the primary (Table 1) and the sensitivity analysis (Table S2) were multivariable logistic regression analyses. Explanatory variables were selected with the help of a directed acyclic graph constructed using DAGitty v3.0 (Fig. S3 and S4) (64). All explanatory variables were kept in the fully adjusted models. Multicollinearity between the entered variables was assessed by calculating the variance inflation factor (VIF) and tolerance statistic. Multicollinearity was not a problem with VIF >10 and tolerance statistic <0.2 (65). The strength of the associations was examined by calculating adjusted odds ratios (AORs) with 95% confidence interval (CI). Two-sided P-values <0.05 were considered statistically significant. The prevalence of ST131 among carrier and clinical isolates was compared by calculating the OR with 95% CI using logistic regression in SPSS. The comparison of ESBL-Ec and phenotypic resistance proportions were assessed using χ2 test.
TABLE 1.
ESBL-producing E. coli gastrointestinal carriage and associated factors among 4,999 participants in Tromsø7 d
| Characteristics | % (ESBL-E. coli) | n (ESBL-E. coli) | N | AOR | 95% CI | P-value |
|---|---|---|---|---|---|---|
| Age (years) | 0.232 | |||||
| 40–49 | 3.6 | 22 | 605 | 1.00 | ||
| 50–59 | 4.1 | 33 | 814 | 1.15 | 0.66–2.01 | |
| 60–69 | 3.2 | 68 | 2,128 | 0.81 | 0.49–1.33 | |
| 70–84 | 3.0 | 43 | 1,452 | 0.72 | 0.42–1.26 | |
| Hospitalization past 12 mo | 0.125 | |||||
| No | 3.3 | 142 | 4,344 | 1.00 | ||
| Yes | 4.0 | 24 | 593 | 1.44 | 0.90–2.28 | |
| Antibiotic use past 14 days a | 0.269 | |||||
| No | 3.3 | 158 | 4,832 | 1.00 | ||
| Yes | 5.2 | 8 | 155 | 1.56 | 0.71–3.43 | |
| Acid-suppressive medication past 4 wk | 0.949 | |||||
| Not used | 3.3 | 123 | 3,762 | 1.00 | ||
| ≤Weekly | 3.2 | 13 | 403 | 1.02 | 0.57–1.83 | |
| Every week, but not daily | 3.5 | 9 | 260 | 1.10 | 0.55–2.20 | |
| Daily | 3.7 | 11 | 297 | 1.20 | 0.64–2.27 | |
| Travel abroad past 12 mo b | <0.001 | |||||
| No | 2.5 | 53 | 2,145 | 1.00 | ||
| Other regions (excluding Asia) | 3.2 | 70 | 2,214 | 1.36 | 0.93–1.98 | |
| Asia exclusively or Asia + other regions | 8.2 | 38 | 462 | 3.46 | 2.18–5.49 | |
| Traveler’s diarrhea past 12 mo c | 0.970 | |||||
| No | 3.3 | 156 | 4,724 | 1.00 | ||
| Yes | 4.8 | 8 | 166 | 1.02 | 0.48–2.16 |
Have you taken any antibiotics (tablets or oral suspensions, nasal ointments, eye drops, or eye ointment) during the past 14 days?
Traveled outside the Nordic countries >1-week duration in the past 12 months.
For each travel abroad the past 12 months, the participants were asked if they had experienced diarrhea in connection with the travel.
ESBL, extended-spectrum β-lactamase; N, denominator; AOR, adjusted odds ratio; CI, confidence interval. AOR adjusted for age, hospitalization in the past 12 months, antibiotic use in the past 14 days, acid suppressive medication in the past 4 weeks, travel abroad in the past 12 months, and traveler’s diarrhea in the past 12 months.The multivariable model includes 4,491 participants with complete information on all variables.
RESULTS
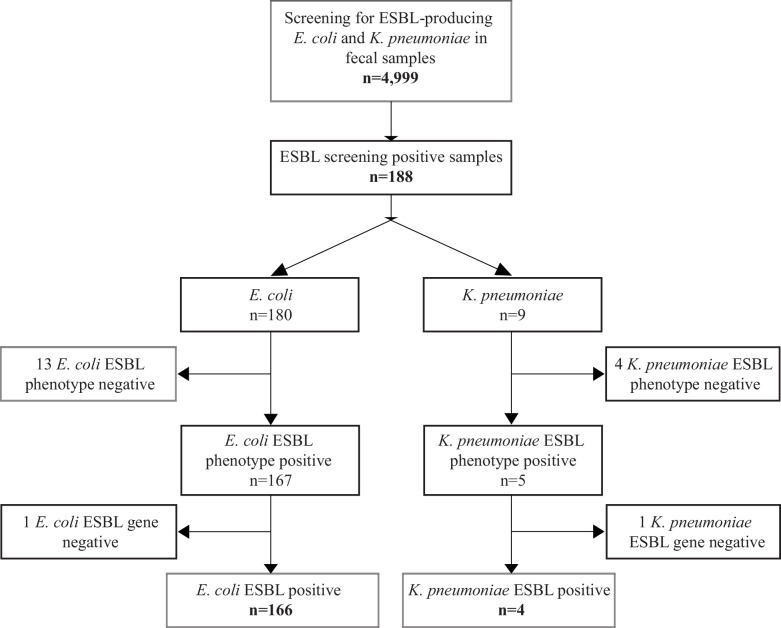
We detected gastrointestinal carriage of putative ESBL-Ec/Kp in 188 of 4,999 randomly selected participants who provided fecal samples in the seventh survey of the population-based Tromsø study (Tromsø7) (39) (Table S1; Fig. 2). Overall, 87% of participants receiving a sampling kit returned a fecal sample. In total, 180 Ec and 9 Kp putative ESBL-positive isolates were isolated from the 188 ESBL screening-positive fecal samples. Both ESBL-Ec and ESBL-Kp were detected in one sample.
Fig 2.
Flowchart and results of fecal sample screening for gastrointestinal carriage of ESBL-producing E. coli and K. pneumoniae in 4,999 participants in Tromsø7, 2015–2016.
Phenotypic and genotypic analyses showed that 14 putative ESBL-Ec isolates were either phenotypically and/or genotypically ESBL-negative. Of these, one isolate harbored plasmid-mediated AmpC (blaCMY-2). After exclusion of the ESBL-negative isolates, the prevalence of ESBL-Ec gastrointestinal carriage was 3.3% (95% CI 2.8%–3.9%, 166 of 4,999 participants): 3.1% (2.5%–3.9%) in women and 3.5% (2.8%–4.4%) in men.
Among the nine screening ESBL-positive Kp isolates, one harbored blaCMY-2 and four did not express an ESBL phenotype. Consequently, four samples were considered positive for ESBL-producing Kp, corresponding to a prevalence of 0.08% (0.02%–0.20%), all among male participants.
Factors associated with gastrointestinal carriage of ESBL-E. coli
We analyzed data from 4,999 participants (Table S1). Median age was 65 years (interquartile range 58–70 years, no sex difference) of the whole study population. In multivariable logistic regression analyses adjusted for all explanatory variables, only travel to Asia in the past 12 months was associated with ESBL-Ec gastrointestinal carriage with an AOR of 3.46 (2.18–5.49) (Table 1). Among participants reporting hospitalization in the past year, or recent use of antibiotics or acid-suppressive medication, we observed a non-significant increase in the prevalence of ESBL-Ec.
Considering the overlapping risk factors between ESBL-E and Kp gastrointestinal carriage identified in the literature (15, 22, 26, 27) and in our previous study (25), we conducted a sensitivity analysis with Kp carriage as a risk factor in a subgroup of 2,973 participants (of the total 4,999 participants in this current study) previously screened for Kp (Table S2). In this model including Kp carriage, AOR was 1.65 (0.98–2.77), indicating a possible association between Kp and ESBL-Ec carriage (Table S2). However, no significant differences in the estimates of the risk factors were observed compared to the model without Kp carriage conducted on the whole study population (Table 1; Table S2), indicating the validity of our primary model.
Comparative analysis of ESBL-E. coli carriage and clinical isolates
To explore the population structure and genomic characteristics of ESBL-Ec in community carriage, we whole-genome sequenced (WGS) all 166 isolates. Furthermore, we sequenced a contemporary national collection of 118 clinical ESBL-Ec isolates (NORM 2014) for comparative analysis (Fig. 3; Table S3).
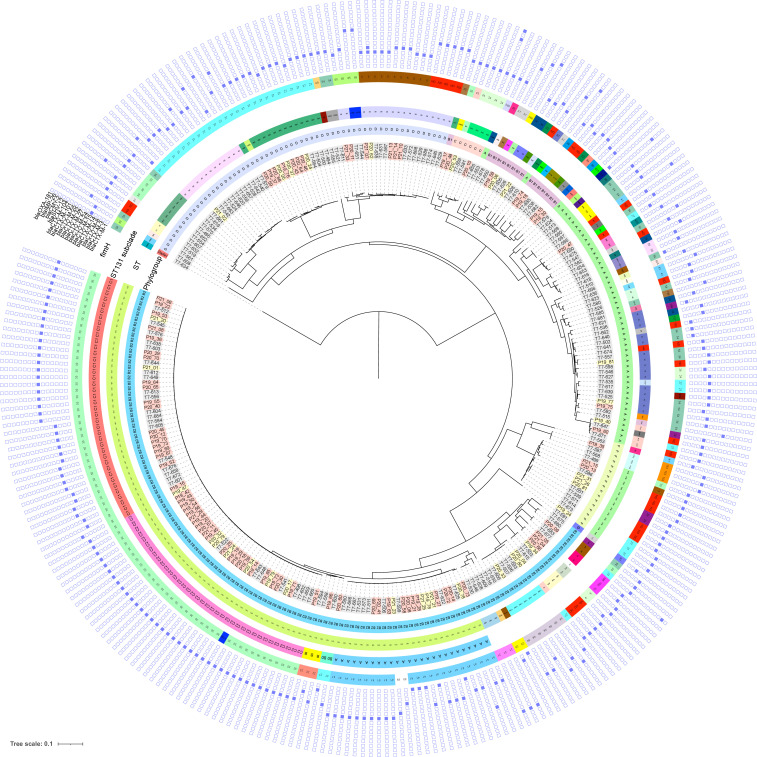
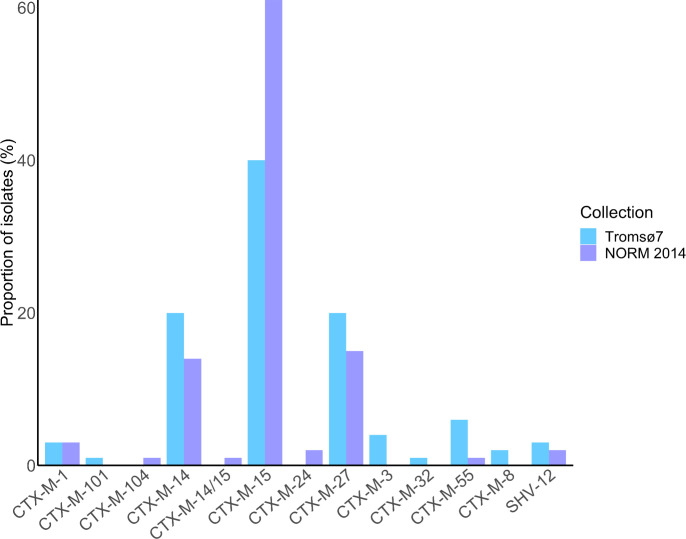
Fig 3.
Maximum-likelihood phylogenetic tree based on core genome alignment of the genomes of ESBL-E. coli carriage isolates from Tromsø7 (labeled gray, n = 166) and clinical isolates from NORM 2014 (blood isolates labeled with red and urine isolates with yellow, n = 118). The innermost ring illustrates phylogroups, followed by a ring with sequence types (STs), a ring with ST131 subclades, and a ring with fimH types (ND, not detected). The heatmap shows the presence (blue color) or absence (white) of ESBL gene variants and other relevant β-lactamases.
At the phylogroup level, 52.4% of the ESBL-Ec carriage isolates belonged to either phylogroup A (26.5%) or D (25.9%), and 33.7% belonged to phylogroup B2 (Fig. 3 and 4 ; Table S3). This contrasts with the clinical isolates where 64.4% (P < 0.001) of the isolates belonged to phylogroup B2 and only 20.4% (P < 0.001) belonged to phylogroup A (5.1%) and D (15.3%) combined.
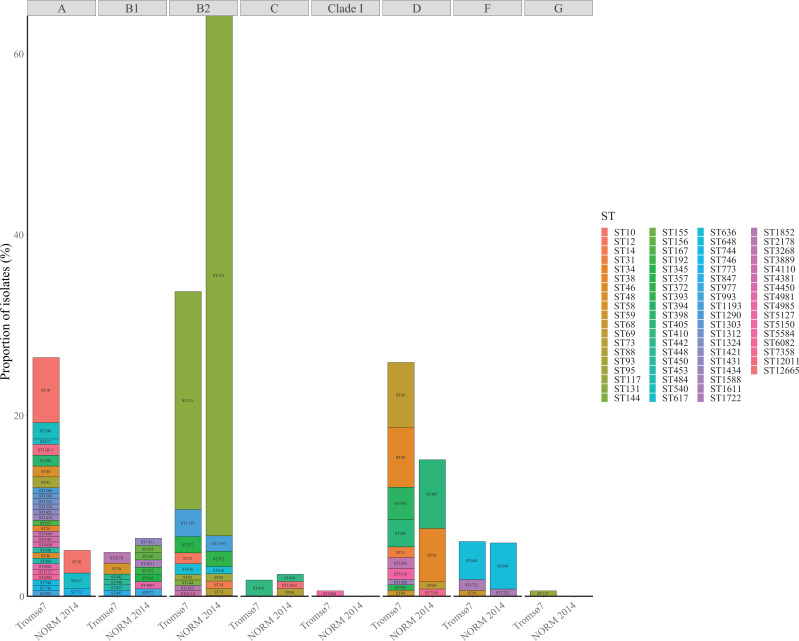
Fig 4.
Phylogroup (labeled on the top) and sequence type (ST) distribution of ESBL-E. coli carriage isolates from Tromsø7 (n = 166) and clinical isolates from NORM 2014 (n = 118).
Carriage isolates had higher ST diversity ( Fig. 3 and 4; Table S3) with Simpson’s diversity index of 92.4% compared to clinical isolates (65.9%, P < 0.001). We identified 58 different STs among the carriage isolates while the clinical isolates included 27 STs (Fig. 3 and 4; Table S3). ST131 (phylogroup B2) was the dominant ST in both collections, however, more prevalent among clinical isolates (57.6%, n = 68) than carriage isolates (24.1%, n = 40, P < 0.001). Comparison of the prevalence of ESBL-producing ST131 colonization versus infection in our study results in a crude odds ratio (OR) for infection of 4.3 (2.57–7.13, P < 0.001).
Within ST131, the multidrug-resistant subclades C1 (alternatively referred to as H30-R) and C2 (H30-Rx) accounted for 67.5% in carriage and 73.5% in clinical isolates (Fig. 3; Fig. S1 and Table S3). The subclade C1 (47.5%, n = 19) was the most prevalent among carriage isolates and C2 (42.6%, n = 29) among clinical isolates. The difference in the proportion of C2 between clinical and carriage isolates (20.0%, n = 8, P = 0.017) was statistically significant. For the overall ESBL-Ec population, crude OR for infection among C2 was 6.44 (2.82–14.68, P < 0.001) (Table S4).
The proportion of subclades A, B, and B0, less associated with antibiotic resistance, was not different between ST131 carriage isolates (32.5%) and clinical isolates (26.5%, P = 0.508) (Fig. S1). However, with regard to the overall ESBL-Ec population, subclade A was associated with infection (carriage 6.0% versus clinical 13.6%, OR 2.45, 1.07–5.60, P = 0.034) (Table S4).
The most prevalent STs in carriage isolates following ST131 were ST10 (7.2%, n = 12, phylogroup A), ST69 (7.2%, n = 12), and ST38 (6.6%, n = 11, both phylogroup D) compared to ST405 (7.6%, n = 9), ST38 (5.9%, n = 7, both phylogroup D), and ST648 (5.1%, n = 6, phylogroup F) in clinical isolates (Fig. 3 and 4; Table S3). We did not identify the common ExPEC lineage ST73 (30, 34) among the carriage isolates but found one ST95 (30, 34) (0.6%) and five isolates of the emerging ST1193 (66, 67) (3.0%).
Using an SNP cutoff of ≤17 (60), we detected 2 putative clusters (Table S5) among 5 of 284 isolates. All five were carriage isolates. One ST357 cluster (6–9 SNP differences) consisted of three isolates and the other cluster of two ST131 isolates (4 SNP differences). We detected no clusters among the clinical isolates.
Antimicrobial resistance and plasmid replicon content
CTX-M enzymes accounted for at least 97.0% of the ESBL phenotypes in both collections (Fig. 3 and 5 ; Table S3). The most prevalent ESBL genes among both carriage and clinical isolates were blaCTX-M-15 (40.4%, n = 67 versus 61.0%, n = 72, P = 0.001), blaCTX-M-14 (20.5%, n = 34 versus 14.4%, n = 17, P = 0.188), and blaCTX-M-27 (19.9%, n = 33 versus 15.3%, n = 18, P = 0.321). One clinical isolate of ST38 harbored both blaCTX-M-15 and blaCTX-M-14. blaCTX-M-3, blaCTX-M-8, blaCTX-M-32, and blaCTX-M-101 were exclusively detected in carriage isolates, whereas blaCTX-M-24 and blaCTX-M-104 were present only in clinical isolates. The remaining ESBL producers contained blaSHV-12 in 3.0% of the carriage and in 1.7% of the clinical isolates. blaTEM ESBL genes were not detected.
Fig 5.
ESBL gene prevalence in ESBL-E. coli carriage isolates from the Tromsø7 study (n = 166) and clinical isolates from NORM 2014 (n = 118).
The clinical isolates showed an overall higher proportion of phenotypic resistance compared to the carriage isolates (Table 2; Table S3). Only 2.4% of carriage isolates were phenotypically resistant to piperacillin–tazobactam compared to 31.8% (P < 0.001) of clinical isolates. We also detected a high prevalence of phenotypic co-resistance to non-β-lactam antibiotics, such as gentamicin, ciprofloxacin, and trimethoprim–sulfamethoxazole, both in clinical and carriage isolates. Isolates co-resistant to all these three antibiotic classes accounted for 10.2% (17/166) of carriage and 33.1% (39/118, P < 0.001) of clinical isolates.
TABLE 2.
Susceptibility profile of ESBL-E. coli carriage isolates from Tromsø7 (n = 166) and clinical isolates from NORM 2014 (n = 118) d
| Carriage | Clinical | |||||
|---|---|---|---|---|---|---|
| %S | %I | %R | %S | %I | %R | |
| Amoxicillin–clavulanic acid i.v. | 57.2 | – | 42.8 | 25.4 | – | 74.6 |
| Amoxicillin–clavulanic acid a | 90.4 | – | 9.6 | 54.2 | – | 45.8 |
| Piperacillin–tazobactam | 97.6 | – | 2.4 | 68.2 b | – | 31.8 b |
| Cefuroxime a | 0.6 | – | 99.4 | 1.7 | – | 98.3 |
| Ceftazidime | 20.5 | 32.5 | 47.0 | 10.2 | 11.0 | 78.8 |
| Cefotaxime | 0.0 | 2.4 | 97.6 | 0.8 | 0.8 | 98.4 |
| Cefepime | 7.8 | 54.8 | 37.3 | 4.7 b | 5.9 b | 89.4 b |
| Ceftazidime–avibactam | 100.0 | – | 0.0 | NA | NA | NA |
| Ertapenem | 100.0 | – | 0.0 | NA | NA | NA |
| Meropenem | 100.0 | 0.0 | 0.0 | 99.2 | 0.8 | 0.0 |
| Aztreonam | 5.4 | 23.5 | 71.1 | NA | NA | NA |
| Amikacin | 100.0 | – | 0.0 | NA | NA | NA |
| Gentamicin | 80.1 | – | 19.9 | 48.3 | – | 51.7 |
| Tobramycin | 76.5 | – | 23.5 | NA | NA | NA |
| Ciprofloxacin | 49.4 | 13.9 | 36.7 | 14.4 | 8.5 | 77.1 |
| Trimethoprim–sulfamethoxazole | 48.8 | 0.6 | 50.6 | 27.1 | 0.0 | 72.9 |
| Nitrofurantoin | 99.4 | – | 0.6 | 93.9 c | – | 6.1 c |
| Fosfomycin | 99.4 | – | 0.6 | NA | NA | NA |
| Fosfomycin a | 96.4 | – | 3.6 | NA | NA | NA |
| Colistin | 98.8 | – | 1.2 | NA | NA | NA |
| Tigecycline | 100.0 | – | 0.0 | 100.0 b | – | 0.0 b |
Breakpoints for uncomplicated urinary tract infections.
Available for blood culture isolates only (n = 85).
Available for urine culture isolates only (n = 33).
S, susceptible; I, susceptible, increased exposure; R, resistant; i.v., intravenous; –, no I category; NA, not available.
Phenotypic ciprofloxacin resistance was different for ST131 (70.0%) versus non-ST131 (26.2%, P < 0.001) among carriage isolates in contrast to the clinical isolates (Table S6). Two carriage isolates of ST484 and ST1324 (both phylogroup A) were phenotypically resistant to colistin and harbored mcr-1.1 and mcr-3.5, respectively. No isolates expressed clinical resistance against carbapenems. However, two clinical isolates of ST95 (phylogroup B2) and ST410 (phylogroup C) harbored blaIMP-26 and blaOXA-181, respectively. All isolates were susceptible to tigecycline.
Despite the overall higher proportion of phenotypic resistance among clinical isolates, the average number of different plasmid replicon-types per isolate (both 3.1/isolate) did not differ between the carriage and clinical collections. We identified 43 and 39 different plasmid replicon types among 97.1% (162/166) carriage and 95.0% (112/118) clinical isolates (Table S3; Fig. S2). The most prevalent were IncFIB(AP001918) (carriage 22.7% versus clinical 22.3%) followed by Col156 (carriage 11.1% versus clinical 11.5%) and IncFIA (carriage 10.9% versus clinical 17.7%, P = 0.101). Replicon type Col156 and IncFIA were mainly detected in globally disseminated ExPEC clones (ST131, ST1193, ST648, ST69, ST405), whereas IncFIB(AP001918) was additionally frequently spread among other STs (Table S3).
ESBL-K. pneumoniae carriage isolates
We detected only four ESBL-Kp, each of a different ST (ST29, ST211, ST261, and ST2459) harboring blaCTX-M-15 (n = 2), blaCTX-M-14 (n = 1), or blaSHV-12 (n = 1). None of the isolates were genomically assigned with a virulence score >1 using Kleborate (53).
DISCUSSION
Our study contributes to the knowledge of prevalence of, and factors associated with, ESBL-Ec/Kp gastrointestinal carriage in a general adult population and the bacterial population structure of carriage isolates. The comparison to a contemporary collection of clinical ESBL-Ec isolates revealed differences in the population structure and the prevalence of phenotypic resistance between carriage and clinical isolates. Travel to Asia was identified as a major risk for ESBL-Ec gastrointestinal carriage.
An ESBL-Ec carriage prevalence of 3.3% (2.8%–3.9%) is lower but comparable to previous community-based data from Europe including Sweden 4.4% (3.5%–5.3%, n = 2,134; data collected 2012–2013) (13), the Netherlands 4.5% (3.9%–5.1%, n = 4,177; 2014–2016) (14), and a Norwegian study 4.9% (2.7%–8.1%, n = 284; 2014–2016) (17) using similar screening approaches.
We identified significant differences in the ESBL-Ec population structure between the community and clinical isolates. The globally disseminated phylogroup B2 clone ST131 has been identified as a key contributor to the increase in ESBL prevalence (68) and in a longitudinal study of E. coli bloodstream isolates we identified ST131 to be the single largest contributor to the increase in the prevalence of ESBL-Ec in Norway (30). We also observed the predominance of ST131 in both of our collections. However, the proportion is significantly lower in carriage isolates due to lower numbers of the multidrug-resistant subclade C2. Moreover, the carriage population had a higher proportion of phylogroup A, associated with asymptomatic intestinal carriage in humans (69, 70), and a significantly greater ST diversity overall, compared to the clinical isolates. These observations indicate that the acquisition of ESBL genes frequently occurs in a variety of E. coli lineages colonizing the gut. However, there are differences in the colonization potential of E. coli lineages and the risk of invasive infection by ESBL-Ec which seems to be clone dependent (68, 71). The higher odds for the infection that we detected for ST131 is similar to that of the Swedish study (AOR 3.4, 1.8–6.4) indicating a higher pathogenicity potential of ST131 compared to commensal E. coli lineages of phylogroup A, such as ST10 (13). Moreover, we found that ST131 subclade A, previously reported with less resistance, and the multidrug-resistant subclade C2 had higher odds for infection, and this may contribute to the sustained establishment of these subclades among bloodstream infections in Norway (30).
Assuming a 100% colonization rate of E. coli, the large proportion of STs notorious as common causes of extraintestinal clinical infections (e.g., ST131, ST405, ST38, and ST648) (30, 32, 33) could at least partly explain the higher prevalence of ESBL among E. coli causing bloodstream infections in Norway (5.8% in 2016) (72) compared to the carriage prevalence of 3.3% identified here. We also observed emerging clones such as ST1193, which appears to have disseminated rapidly worldwide over the last decade (66, 67). The low prevalence of ESBL-Kp gastrointestinal carriage (0.08%) is consistent with previous community-based reports (9, 14, 17).
The identification of clusters with closely related isolates could indicate putative clonal spread. This could include within-household and social network transmission or nosocomial spread. However, we did not have access to epidemiological data to examine this further.
In line with other studies, we found a strong association between ESBL-Ec carriage and travel to Asian regions (13 - 17 - 22 - 22). This supports the current patient screening recommendations for ESBL-producing Gram-negative bacteria after a hospital stay abroad in the past year before hospital admission in Norway (73). In contrast to a Swedish and a Dutch study (15, 74), we did not identify travelers’ diarrhea as a risk factor for ESBL-Ec carriage. However, our study was not designed to specifically investigate international travelers but rather focused on risk factors in the general adult population.
We found no association between ESBL-Ec gut carriage and factors such as hospitalization, antibiotic use, and acid-suppressive medication, and conflicting results have been detected in previous studies (22, 75). Hospitalization as a risk factor has mainly been reported in studies investigating patients with ESBL-E infections (23, 76). In line with most studies that assessed risk factors regarding ESBL-E carriage in individuals in the community, we did not identify hospitalization as an independent risk factor (13, 22, 74). This may be due to the increased ESBL prevalence not only in hospitals but also in the community over the last decades and implies that boundaries have become blurred between those two settings (9, 22, 77, 78).
The non-significant effect of antibiotic use is mainly due to the limitations of the drug variable in our study which is based on self-reported data, including topical antibiotics and covers only the last 2 weeks before self-sampling. As antibiotic use has been found as a risk factor for resistance in many other studies (14, 15, 22), we cannot rule out that antibiotic use plays a role in ESBL-E carriage. There are reports identifying an association between the use of gastric acid–suppressive medication and intestinal colonization or infections with ESBL-E (18, 21, 23, 26, 27, 76). However, a Dutch study comparable to ours did not find an association between proton pump inhibitor use and ESBL-E carriage in the overall analysis (14).
Interestingly, we found a possible association between Kp and ESBL-Ec carriage. This may support the previously described link between ESBL-E, VRE (7, 28), and Kp colonization among intensive care unit patients (29). These associations warrant further investigations to assess if a common set of risk factors for carriage of different clinically important pathogens can be identified.
Decolonization of ESBL-E gastrointestinal carriage has been investigated as a possible strategy to reduce the risk of infection and transmission. However, decolonization is not recommended due to insufficient evidence (79). Our findings could also be considered in designing future decolonization strategies.
An important strength of our study is the non-selective recruitment from the official population registry, and the high participation (87%) compared to 18.3% and 18.8% in comprehensive studies from the Netherlands (14) and Sweden (13), respectively. It is a limitation that we only captured the general population 40 years and older. However, other studies have not found an association between age and ESBL-E carriage (13, 14). If age is associated with specific Ec STs, this might potentially bias the ST-specific estimates. Moreover, more extensive data on drug use would have strengthened the analyses. Additionally, the genomic diversity of carriage isolates is likely to be underestimated due to the isolation and sequencing of only one colony per fecal sample. A previous nationwide genomic study on E. coli causing bloodstream infections detected no discernible spatiotemporal spread or phylogenetic structure within Norway (30), indicating limited bias in comparing a local and national collection of isolates. The short-read data did not allow us to investigate differences in plasmid population structure between carriage and clinical isolates beyond the level of replicon types.
Conclusions
The prevalence of ESBL-Ec carriage in a general adult urban Norwegian population was low reflecting the relatively low prevalence of ESBL-Ec in clinical isolates. Travel to Asia was the only independent risk factor for ESBL-Ec carriage and should be considered in terms of screening recommendations before hospital admission. The differences in ESBL-Ec populations between carriage and clinical isolates indicating a higher risk of infection dependent on the ESBL-Ec clone support the integration of genomics in risk assessments.
ACKNOWLEDGMENTS
We are grateful for technical assistance from Bjørg Haldorsen, Bettina Aasnæs, and Ellen Josefsen in organizing the collection of fecal samples in the laboratory; Eva Bernhoff and Ragna-Johanne Bakksjø for performing WGS; Marit Andrea Klokkhammer Hetland for the initial sequence analysis of carriage isolates; and Rod Wolstenholme for figure editing. We thank Sigma2, the national provider of e-infrastructure, for access to High-Performance Computing and large-scale data storage (Project ID: NN9794K).
This study was supported by grants from the Northern Norway Regional Health Authority (HNF1415-18) and the Trond Mohn Foundation (TMF2019TMT03). The funders of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Collaborators forming the Norwegian E. coli ESBL Study Group include Nina Handal (Akershus University Hospital), Elisabeth Sirnes (Førde Hospital), Liv Jorunn Hafne (Haugesund Hospital), Paul Christoffer Lindemann (Haukeland University Hospital), Lovise Marie Norgaard (Innlandet Hospital), Kyriakos Zaragkoulias (Levanger Hospital), Einar Nilsen (Molde and Ålesund Hospital), Hege Elisabeth Larsen (Nordland Hospital), Marcela Pino (Oslo University Hospital, Rikshospitalet), Nils Olav Hermansen (Oslo University Hospital, Ullevål), Iren H. Löhr (Stavanger University Hospital), Aleksandra Jakovljev (St. Olav University Hospital), Ståle Tofteland (Sørlandet Hospital), Kristina Papp (Unilabs Telelab), Gunnar Skov Simonsen (University Hospital of North Norway), Åshild Marvik (Vestfold Hospital), Nadine Durema Pullar (Vestre Viken, Bærum Hospital), Roar Magne Bævre-Jensen (Vestre Viken, Drammen Hospital), and Anita Kanestrøm (Østfold Hospital).
Ø.S. conceptualized and acquired funding for the study in collaboration with A.S., K.G., and I.H.L. Ø.S. was responsible for organizing the collection of fecal samples. L.L.E.A. did the screening of fecal samples and phenotypic testing. N.R., K.S., L.S., and K.G. conceptualized and conducted the epidemiological analysis of risk factors. I.H.L. organized W.G.S. of carriage isolates. G.S.S. and the Norwegian E. coli ESBL Study Group provided data and clinical isolates. D.J.B., A.K.P., N.R., and Ø.S. analyzed the WGS data. N.R., Ø.S., K.S., and K.G. prepared first manuscript draft. All authors contributed to review and editing of the manuscript and approved the final version.
All authors report no conflicts of interest.
Contributor Information
Niclas Raffelsberger, Email: niclas.raffelsberger@unn.no.
Mariana Castanheira, JMI Laboratories, North Liberty, Iowa city, Iowa, USA .
Collaborators: Nina Handal, Liv Jorunn Hafne, Paul Christoffer Lindemann, Lovise Marie Norgaard, Kyriakos Zaragkoulias, Einar Nilsen, Hege Elisabeth Larsen, Marcela Pino, Nils Olav Hermansen, Iren H. Löhr, Aleksandra Jakovljev, Ståle Tofteland, Kristina Papp, Gunnar Skov Simonsen, Åshild Marvik, Nadine Durema Pullar, Roar Magne Bævre-Jensen, and Anita Kanestrøm
ETHICS APPROVAL
The study was approved by the Regional Committee for Medical and Health Research Ethics, North Norway (REC North reference: 2016/1788 and 2014/940) and the Data Protection Officer at University Hospital of North Norway (reference: 2019/4264). The study complied with the Declaration of Helsinki. All participants in Tromsø7 signed an informed consent form prior to participation.
DATA AVAILABILITY
Bacterial genome data (raw Illumina reads) are publicly available in National Center for Biotechnology Information under BioProject PRJEB53319 (NORM 2014 collection, https://www.ebi.ac.uk/ena/browser/view/PRJEB53319) and PRJEB57251 (Tromsø7 collection, https://www.ebi.ac.uk/ena/browser/view/PRJEB57251). This study is based on data owned by a third party (The Tromsø Study, Department of Community Medicine, UiT The Arctic University of Norway, Norway). Confidentiality requirements according to the Norwegian law prevent sharing of individual patient level data in public repositories. Application of legal basis and exemption from professional secrecy requirements for the use of personal health data in research may be sent to a regional committee for medical and health research ethics (https://rekportalen.no/). The authors gained access to the data through the Tromsø Study’s application process. Guidelines on how to access the data are available at the website https://uit.no/research/tromsostudy. All inquiries about the Tromsø Study should be sent by e-mail to tromsous@ism.uit.no. All the questionnaire variables are published in the NESSTAR program system, and results can be viewed online: http://tromsoundersokelsen.uit.no/tromso/.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00025-23.
ST131 clade distribution among ESBL-E. coli carriage isolates from Tromsø7 (n=40) and clinical isolates from NORM 2014 (n=68).
Replicon type distribution among ESBL-E. coli from carriage isolates from Tromsø7 (n=166) and clinical isolates from NORM 2014 (n=118).
Directed acyclic graph (DAG) illustrating the causal relationship between ESBL-E. coli (ESBL-Ec) gastrointestinal carriage (outcome) and relevant covariates among 4,999 participants in Tromsø7. The variable ‘drug use’ includes antibiotic use past 14 days and acid suppressive medication past four weeks. To set up the most plausible causal relationship between covariates and the outcome, we first searched the literature for relevant factors associated with ESBL-E. coli gastrointestinal carriage. Thereafter, the DAG guided the selection of the multivariable logistic regression model, which was adjusted for the minimal sufficient adjustment set comprising age, drug use, hospitalization, travel abroad, and traveler`s diarrhea (i.e. the variables constituting a confounding pathway). Controlling for the minimal sufficient adjustment set warrants that confounding paths are blocked in order to minimize bias of the causal relationship. The model was not adjusted for sex due to no statistically significant sex difference in prevalence of ESBL-E. coli carriage and because it does not constitute biasing paths after adjustment. For the variables sex and alcohol consumption, a direct effect on ESBL-E. coli carriage is not described or investigated, however they are known to affect the microbiota composition. Hence, intestinal microbiota was included as an unobserved mediator of the causal relationship.
Directed acyclic graph (DAG) illustrating the causal relationship between K. pneumoniae species complex (Kp) gastrointestinal carriage (exposure), ESBL-E. coli (ESBL-Ec) gastrointestinal carriage (outcome), and relevant covariates among 2,973 participants in Tromsø7. The variable ‘drug use’ includes antibiotic use past 14 days and acid suppressive medication past four weeks. K. pneumoniae is a common cause of healthcare associated infections often combined with antimicrobial resistance. In the literature and our previous study (Raffelsberger N, Hetland MAK, Svendsen K, et al. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes. 2021 Jan-Dec;13(1):1939599.), we identified several factors associated with K. pneumoniae gastrointestinal carriage, which are overlapping with those associated with ESBL-E. coli carriage. To capture the relevance of K. pneumoniae carriage as an exposure, we included this variable into the causal relationship with ESBL-E. coli carriage. The DAG was used for the selection of variables for the multivariable logistic regression model, to conceptualize confounding and to identify the minimal sufficient adjustment set. Although sex and alcohol consumption represent ancestors of exposure and outcome, the adjustment for age, drug use, hospitalization, travel abroad, and traveler`s diarrhea controls for relevant confounders and blocks biasing paths. The absence of red arrows in the DAG implies that there are no open unadjusted confounding pathways.
Characteristics of the study population.
Sensitivity analysis.
Genome characteristics of ESBL-E. coli from carriage isolates from Tromsø7 (n=166) and clinical isolates from NORM 2014 (n=118) isolates.
Comparison of ESBL-E. coli ST131 subclade prevalence.
SNP distances among two putative ESBL-producing E. coli clusters.
Susceptibility profile of carriage and clinical isolates of ESBL-E. coli ST131 and non-ST131.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Antimicrobial Resistance C. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. NORM/NORM-VET 2021 . 2022. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromsø/Oslo/Ås; ISSN: 1502-2307 (print)/1890- 9965 (electronic).
- 3. Schwaber MJ, Carmeli Y. 2007. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J Antimicrob Chemother 60:913–920. doi: 10.1093/jac/dkm318 [DOI] [PubMed] [Google Scholar]
- 4. Scheuerman O, Schechner V, Carmeli Y, Gutiérrez-Gutiérrez B, Calbo E, Almirante B, Viale P-L, Oliver A, Ruiz-Garbajosa P, Gasch O, Gozalo M, Pitout J, Akova M, Peña C, Molina J, Hernández-Torres A, Venditti M, Prim N, Origüen J, Bou G, Tacconelli E, Tumbarello M, Hamprecht A, Karaiskos I, de la Calle C, Pérez F, Schwaber MJ, Bermejo J, Lowman W, Hsueh P-R, Navarro-San Francisco C, Bonomo RA, Paterson DL, Pascual A, Rodríguez-Baño J, REIPI/ESGBIS/INCREMENT investigators . 2018. Comparison of predictors and mortality between bloodstream infections caused by ESBL-producing Escherichia coli and ESBL-producing Klebsiella pneumoniae. Infect Control Hosp Epidemiol 39:660–667. doi: 10.1017/ice.2018.63 [DOI] [PubMed] [Google Scholar]
- 5. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Aboderin AO, Al-Abri SS, Awang Jalil N, Benzonana N, Bhattacharya S, Brink AJ, Burkert FR, Cars O, Cornaglia G, Dyar OJ, Friedrich AW, Gales AC, Gandra S, Giske CG, Goff DA, Goossens H, Gottlieb T, Guzman Blanco M, Hryniewicz W, Kattula D, Jinks T, Kanj SS, Kerr L, Kieny M-P, Kim YS, Kozlov RS, Labarca J, Laxminarayan R, Leder K, Leibovici L, Levy-Hara G, Littman J, Malhotra-Kumar S, Manchanda V, Moja L, Ndoye B, Pan A, Paterson DL, Paul M, Qiu H, Ramon-Pardo P, Rodríguez-Baño J, Sanguinetti M, Sengupta S, Sharland M, Si-Mehand M, Silver LL, Song W, Steinbakk M, Thomsen J, Thwaites GE, van der Meer JW, Van Kinh N, Vega S, Villegas MV, Wechsler-Fördös A, Wertheim HFL, Wesangula E, Woodford N, Yilmaz FO, Zorzet A. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 6. Prevel R, Boyer A, M’Zali F, Cockenpot T, Lasheras A, Dubois V, Gruson D. 2019. Extended spectrum beta-lactamase producing Enterobacterales faecal carriage in a medical intensive care unit: low rates of cross-transmission and infection. Antimicrob Resist Infect Control 8:112. doi: 10.1186/s13756-019-0572-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wyres KL, Hawkey J, Mirčeta M, Judd LM, Wick RR, Gorrie CL, Pratt NF, Garlick JS, Watson KM, Pilcher DV, McGloughlin SA, Abbott IJ, Macesic N, Spelman DW, Jenney AWJ, Holt KE. 2021. Genomic surveillance of antimicrobial resistant bacterial colonisation and infection in intensive care patients. BMC Infect Dis 21:683. doi: 10.1186/s12879-021-06386-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denkel LA, Maechler F, Schwab F, Kola A, Weber A, Gastmeier P, Pfäfflin F, Weber S, Werner G, Pfeifer Y, Pietsch M, Leistner R. 2020. Infections caused by extended-spectrum β-lactamase-producing enterobacterales after rectal colonization with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin Microbiol Infect 26:1046–1051. doi: 10.1016/j.cmi.2019.11.025 [DOI] [PubMed] [Google Scholar]
- 9. Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bezabih YM, Sabiiti W, Alamneh E, Bezabih A, Peterson GM, Bezabhe WM, Roujeinikova A. 2021. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother 76:22–29. doi: 10.1093/jac/dkaa399 [DOI] [PubMed] [Google Scholar]
- 11. Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, McCarthy N, Paul P, McDonald LC, Kallen A, Fiore A, Craig M, Baggs J. 2020. Multidrug-resistant bacterial Infections in U.S. hospitalized patients, 2012-2017. N Engl J Med 382:1309–1319. doi: 10.1056/NEJMoa1914433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mughini-Gras L, Dorado-García A, van Duijkeren E, van den Bunt G, Dierikx CM, Bonten MJM, Bootsma MCJ, Schmitt H, Hald T, Evers EG, de Koeijer A, van Pelt W, Franz E, Mevius DJ, Heederik DJJ, ESBL Attribution Consortium . 2019. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Health 3:e357–e369. doi: 10.1016/S2542-5196(19)30130-5 [DOI] [PubMed] [Google Scholar]
- 13. Ny S, Löfmark S, Börjesson S, Englund S, Ringman M, Bergström J, Nauclér P, Giske CG, Byfors S. 2017. Community carriage of ESBL-producing Escherichia coli is associated with strains of low pathogenicity: a swedish nationwide study. J Antimicrob Chemother 72:582–588. doi: 10.1093/jac/dkw419 [DOI] [PubMed] [Google Scholar]
- 14. van den Bunt G, van Pelt W, Hidalgo L, Scharringa J, de Greeff SC, Schürch AC, Mughini-Gras L, Bonten MJM, Fluit AC. 2019. Prevalence, risk factors and genetic characterisation of extended-spectrum beta-lactamase and carbapenemase-producing enterobacteriaceae (ESBL-E and CPE): a community-based cross-sectional study, the Netherlands, 2014 to 2016. Euro Surveill 24:1800594. doi: 10.2807/1560-7917.ES.2019.24.41.1800594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, Grobusch MP, Lashof AMO, Molhoek N, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. 2017. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 17:78–85. doi: 10.1016/S1473-3099(16)30319-X [DOI] [PubMed] [Google Scholar]
- 16. Jørgensen SB, Samuelsen O, Sundsfjord A, Bhatti SA, Jørgensen I, Sivapathasundaram T, Leegaard TM. 2014. High prevalence of faecal carriage of ESBL-producing Enterobacteriaceae in Norwegian patients with gastroenteritis. Scand J Infect Dis 46:462–465. doi: 10.3109/00365548.2014.896031 [DOI] [PubMed] [Google Scholar]
- 17. Ulstad CR, Solheim M, Berg S, Lindbæk M, Dahle UR, Wester AL. 2016. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia Coli and Klebsiella spp. in healthy people in Norway. Antimicrob Resist Infect Control 5:57. doi: 10.1186/s13756-016-0156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kohler P, Seiffert SN, Kessler S, Rettenmund G, Lemmenmeier E, Qalla Widmer L, Nolte O, Seth-Smith HMB, Albrich WC, Babouee Flury B, Gardiol C, Harbarth S, Münzer T, Schlegel M, Petignat C, Egli A, Héquet D. 2022. Molecular epidemiology and risk factors for extended-spectrum β-lactamase-producing Enterobacterales in long-term care residents. J Am Med Dir Assoc 23:475–481. doi: 10.1016/j.jamda.2021.06.030 [DOI] [PubMed] [Google Scholar]
- 19. Rettedal S, Löhr IH, Bernhoff E, Natås OB, Sundsfjord A, Øymar K. 2015. Extended-spectrum β-lactamase-producing Enterobacteriaceae among pregnant women in Norway: prevalence and maternal-neonatal transmission. J Perinatol 35:907–912. doi: 10.1038/jp.2015.82 [DOI] [PubMed] [Google Scholar]
- 20. Birgy A, Cohen R, Levy C, Bidet P, Courroux C, Benani M, Thollot F, Bingen E. 2012. Community faecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in French children. BMC Infect Dis 12:315. doi: 10.1186/1471-2334-12-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wielders CCH, van Hoek A, Hengeveld PD, Veenman C, Dierikx CM, Zomer TP, Smit LAM, van der Hoek W, Heederik DJ, de Greeff SC, Maassen CBM, van Duijkeren E. 2017. Extended-spectrum β-Lactamase- and pAmpC-producing Enterobacteriaceae among the general population in a livestock-dense area. Clin Microbiol Infect 23:120. doi: 10.1016/j.cmi.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 22. Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. 2016. Fecal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis 63:310–318. doi: 10.1093/cid/ciw283 [DOI] [PubMed] [Google Scholar]
- 23. Richelsen R, Smit J, Laxsen Anru P, Schønheyder HC, Nielsen H. 2021. Risk factors of community-onset extended-spectrum β-lactamase Escherichia coli and Klebsiella pneumoniae bacteraemia: an 11-year population-based case-control-control study in Denmark. Clin Microbiol Infect 27:871–877. doi: 10.1016/j.cmi.2020.08.004 [DOI] [PubMed] [Google Scholar]
- 24. Leistner R, Meyer E, Gastmeier P, Pfeifer Y, Eller C, Dem P, Schwab F. 2013. Risk factors associated with the community-acquired colonization of extended-spectrum beta-lactamase (ESBL) positive Escherichia coli. an exploratory case-control study. PLoS One 8:e74323. doi: 10.1371/journal.pone.0074323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raffelsberger N, Hetland MAK, Svendsen K, Småbrekke L, Löhr IH, Andreassen LLE, Brisse S, Holt KE, Sundsfjord A, Samuelsen Ø, Gravningen K. 2021. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes 13:1939599. doi: 10.1080/19490976.2021.1939599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Willems RPJ, van Dijk K, Ket JCF, Vandenbroucke-Grauls C. 2020. Evaluation of the association between gastric acid suppression and risk of intestinal colonization with multidrug-resistant microorganisms: a systematic review and meta-analysis. JAMA Intern Med 180:561–571. doi: 10.1001/jamainternmed.2020.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reuland EA, Al Naiemi N, Kaiser AM, Heck M, Kluytmans J, Savelkoul PHM, Elders PJM, Vandenbroucke-Grauls C. 2016. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother 71:1076–1082. doi: 10.1093/jac/dkv441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy P, Malczynski M, Obias A, Reiner S, Jin N, Huang J, Noskin GA, Zembower T. 2007. Screening for extended-spectrum β-Lactamase-producing Enterobacteriaceae among high-risk patients and rates of subsequent bacteremia. Clin Infect Dis 45:846–852. doi: 10.1086/521260 [DOI] [PubMed] [Google Scholar]
- 29. Collingwood A, Blostein F, Seekatz AM, Wobus CE, Woods RJ, Foxman B, Bachman MA. 2020. Epidemiological and microbiome associations between Klebsiella pneumoniae and vancomycin-resistant Enterococcus colonization in intensive care unit patients. Open Forum Infect Dis 7:faa012. doi: 10.1093/ofid/ofaa012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gladstone RA, McNally A, Pöntinen AK, Tonkin-Hill G, Lees JA, Skytén K, Cléon F, Christensen MOK, Haldorsen BC, Bye KK, Gammelsrud KW, Hjetland R, Kümmel A, Larsen HE, Lindemann PC, Löhr IH, Marvik Å, Nilsen E, Noer MT, Simonsen GS, Steinbakk M, Tofteland S, Vattøy M, Bentley SD, Croucher NJ, Parkhill J, Johnsen PJ, Samuelsen Ø, Corander J. 2021. Emergence and dissemination of antimicrobial resistance in Escherichia coli causing bloodstream infections in Norway in 2002–17: a nationwide, longitudinal, microbial population genomic study. Lancet Microbe 2:e331–e341. doi: 10.1016/S2666-5247(21)00031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fostervold A, Hetland MAK, Bakksjø R, Bernhoff E, Holt KE, Samuelsen Ø, Simonsen GS, Sundsfjord A, Wyres KL, Löhr IH, Norwegian Study Group on Klebsiella pneumoniae . 2022. A nationwide genomic study of clinical Klebsiella pneumoniae in Norway 2001-2015: introduction and spread of ESBL facilitated by CG15 and CG307. J Antimicrob Chemother 77:665–674. doi: 10.1093/jac/dkab463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunn SJ, Connor C, McNally A. 2019. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: the complexity of clones and plasmids. Curr Opin Microbiol 51:51–56. doi: 10.1016/j.mib.2019.06.004 [DOI] [PubMed] [Google Scholar]
- 33. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. 2019. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev 32:e00135-18. doi: 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, Peacock SJ, Parkhill J. 2017. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res 27:1437–1449. doi: 10.1101/gr.216606.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goodman KE, Lessler J, Cosgrove SE, Harris AD, Lautenbach E, Han JH, Milstone AM, Massey CJ, Tamma PD. 2016. A clinical decision tree to predict whether a bacteremic patient is infected with an extended-spectrum β-lactamase–producing organism. Clin Infect Dis 63:896–903. doi: 10.1093/cid/ciw425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de la Court JR, Heijmans J, Huynh J, Sieswerda E, de Jonge NA, van Dijk K, Sigaloff KCE, Schade RP. 2022. Guidance of empirical antimicrobial therapy by surveillance cultures in high-risk neutropenic patients: a retrospective cohort study. Antimicrob Resist Infect Control 11. doi: 10.1186/s13756-022-01198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stupica D, Lusa L, Klevišar MN, Terzić S, Pirš M, Premru MM, Strle F. 2017. Should we consider faecal colonisation with extended-spectrum β-lactamase-producing Enterobacteriaceae in empirical therapy of community-onset sepsis? Int J Antimicrob Agents 50:564–571. doi: 10.1016/j.ijantimicag.2017.06.019 [DOI] [PubMed] [Google Scholar]
- 39. Hopstock LA, Grimsgaard S, Johansen H, Kanstad K, Wilsgaard T, Eggen AE. 2022. The seventh survey of the Tromsø study (Tromsø7) 2015–2016: study design, data collection, attendance, and prevalence of risk factors and disease in a multipurpose population-based health survey. Scand J Public Health 50:919–929. doi: 10.1177/14034948221092294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matuschek E, Brown DFJ, Kahlmeter G. 2014. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect 20:255–266. doi: 10.1111/1469-0691.12373 [DOI] [PubMed] [Google Scholar]
- 41. Krueger FT. 2019. Available from: https://github.com/FelixKrueger/TrimGalore. Retrieved Aug 2019.
- 42. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seemann T . 2021. Mlst github. Available from: https://github.com/tseemann/mlst2021
- 46. Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. 2020. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152. doi: 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. doi: 10.1038/s41598-021-91456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seemann T. 2019. Abricate, github. Available from: https://github.com/tseemann/abricate
- 49. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using Plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. Clermontyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4:e000192. doi: 10.1099/mgen.0.000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen MCF, Lund O, Hansen F, Hammerum AM, Sokurenko E, Hasman H. 2017. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 55:2538–2543. doi: 10.1128/JCM.00737-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Madden T. 2002. The BLAST sequence analysis tool, . In McEntyre J OJ (ed), The NCBI handbook. Bethesda (MD): national center for biotechnology information (US); 2002-Chapter 16. http://www.ncbi.nlm.nih.gov/books/NBK21097/. [Google Scholar]
- 53. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. 2018. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med 10:77. doi: 10.1186/s13073-018-0587-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. NORM/NORM-VET 2014 . 2015. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromsø/Oslo; 2015. ISSN: 1502-2307 (print)/1890- 9965 (electronic).
- 56. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 58. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 59. Seemann T snippy: fast bacterial variant calling from NGS reads. 2015. https://github.com/tseemann/snippy.
- 60. Ludden C, Coll F, Gouliouris T, Restif O, Blane B, Blackwell GA, Kumar N, Naydenova P, Crawley C, Brown NM, Parkhill J, Peacock SJ. 2021. Defining nosocomial transmission of Escherichia Coli and antimicrobial resistance genes: a genomic surveillance study. Lancet Microbe 2:e472–e480. doi: 10.1016/S2666-5247(21)00117-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377–e00413. doi: 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Textor J, van der Zander B, Gilthorpe MS, Liskiewicz M, Ellison GT. 2016. Robust causal inference using directed acyclic graphs: the R package “dagitty.” Int J Epidemiol 45:1887–1894. doi: 10.1093/ije/dyw341 [DOI] [PubMed] [Google Scholar]
- 65. Field A. 2017. Discovering statistics using IBM SPSS statistics. SAGE Publications Ltd. [Google Scholar]
- 66. Johnson JR, Johnston BD, Porter SB, Clabots C, Bender TL, Thuras P, Trott DJ, Cobbold R, Mollinger J, Ferrieri P, Drawz S, Banerjee R, Diekema DJ. 2019. Rapid emergence, subsidence, and molecular detection of Escherichia coli sequence type 1193- fimH64, a new disseminated multidrug-resistant commensal and extraintestinal pathogen . J Clin Microbiol 57:e01664–18. doi: 10.1128/JCM.01664-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tchesnokova V, Radey M, Chattopadhyay S, Larson L, Weaver JL, Kisiela D, Sokurenko EV. 2019. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc Natl Acad Sci U S A 116:14740–14748. doi: 10.1073/pnas.1903002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 70. Denamur E, Clermont O, Bonacorsi S, Gordon D. 2021. The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19:37–54. doi: 10.1038/s41579-020-0416-x [DOI] [PubMed] [Google Scholar]
- 71. Marin J, Clermont O, Royer G, Mercier-Darty M, Decousser JW, Tenaillon O, Denamur E, Blanquart F. 2022. The population genomics of increased virulence and antibiotic resistance in human commensal Escherichia coli over 30 years in France. Appl Environ Microbiol 88:e0066422. doi: 10.1128/aem.00664-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. NORM/NORM-VET . 2017. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromsø/Oslo; ISSN: 1502-2307 (print)/1890- 9965 (electronic).
- 73. Health NIoP. ESBL-holdige gramnegative stavbakterier - smitteverntiltak i helseinstitusjoner. 2015. https://www.fhi.no/sv/forebygging-i-helsetjenesten/smittevern-i-institusjoner/tiltak/esbl-holdige-gramnegative-stavbakte
- 74. Ostholm-Balkhed A, Tärnberg M, Nilsson M, Nilsson LE, Hanberger H, Hällgren A, Travel Study Group of Southeast Sweden . 2013. Travel-associated faecal colonization with ESBL-producing Enterobacteriaceae: incidence and risk factors. J Antimicrob Chemother 68:2144–2153. doi: 10.1093/jac/dkt167 [DOI] [PubMed] [Google Scholar]
- 75. Nicolas-Chanoine M-H, Gruson C, Bialek-Davenet S, Bertrand X, Thomas-Jean F, Bert F, Moyat M, Meiller E, Marcon E, Danchin N, Noussair L, Moreau R, Leflon-Guibout V. 2013. 10-fold increase (2006-11) in the rate of healthy subjects with extended-spectrum β-lactamase-producing Escherichia coli faecal carriage in a parisian check-up centre. J Antimicrob Chemother 68:562–568. doi: 10.1093/jac/dks429 [DOI] [PubMed] [Google Scholar]
- 76. Søgaard M, Heide-Jørgensen U, Vandenbroucke JP, Schønheyder HC, Vandenbroucke-Grauls C. 2017. Risk factors for extended-spectrum β-Lactamase-producing Escherichia coli urinary tract infection in the community in Denmark: a case-control study. Clin Microbiol Infect 23:952–960. doi: 10.1016/j.cmi.2017.03.026 [DOI] [PubMed] [Google Scholar]
- 77. Vihta K-D, Stoesser N, Llewelyn MJ, Quan TP, Davies T, Fawcett NJ, Dunn L, Jeffery K, Butler CC, Hayward G, Andersson M, Morgan M, Oakley S, Mason A, Hopkins S, Wyllie DH, Crook DW, Wilcox MH, Johnson AP, Peto TEA, Walker AS. 2018. Trends over time in Escherichia coli bloodstream infections, urinary tract infections, and antibiotic susceptibilities in Oxfordshire, UK, 1998-2016: a study of electronic health records. Lancet Infect Dis 18:1138–1149. doi: 10.1016/S1473-3099(18)30353-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thaden JT, Fowler VG, Sexton DJ, Anderson DJ. 2016. Increasing incidence of extended-spectrum β-lactamase-producing Escherichia coli in community hospitals throughout the Southeastern United States. Infect Control Hosp Epidemiol 37:49–54. doi: 10.1017/ice.2015.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tacconelli E, Mazzaferri F, de Smet AM, Bragantini D, Eggimann P, Huttner BD, Kuijper EJ, Lucet J-C, Mutters NT, Sanguinetti M, Schwaber MJ, Souli M, Torre-Cisneros J, Price JR, Rodríguez-Baño J. 2019. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin Microbiol Infect 25:807–817. doi: 10.1016/j.cmi.2019.01.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ST131 clade distribution among ESBL-E. coli carriage isolates from Tromsø7 (n=40) and clinical isolates from NORM 2014 (n=68).
Replicon type distribution among ESBL-E. coli from carriage isolates from Tromsø7 (n=166) and clinical isolates from NORM 2014 (n=118).
Directed acyclic graph (DAG) illustrating the causal relationship between ESBL-E. coli (ESBL-Ec) gastrointestinal carriage (outcome) and relevant covariates among 4,999 participants in Tromsø7. The variable ‘drug use’ includes antibiotic use past 14 days and acid suppressive medication past four weeks. To set up the most plausible causal relationship between covariates and the outcome, we first searched the literature for relevant factors associated with ESBL-E. coli gastrointestinal carriage. Thereafter, the DAG guided the selection of the multivariable logistic regression model, which was adjusted for the minimal sufficient adjustment set comprising age, drug use, hospitalization, travel abroad, and traveler`s diarrhea (i.e. the variables constituting a confounding pathway). Controlling for the minimal sufficient adjustment set warrants that confounding paths are blocked in order to minimize bias of the causal relationship. The model was not adjusted for sex due to no statistically significant sex difference in prevalence of ESBL-E. coli carriage and because it does not constitute biasing paths after adjustment. For the variables sex and alcohol consumption, a direct effect on ESBL-E. coli carriage is not described or investigated, however they are known to affect the microbiota composition. Hence, intestinal microbiota was included as an unobserved mediator of the causal relationship.
Directed acyclic graph (DAG) illustrating the causal relationship between K. pneumoniae species complex (Kp) gastrointestinal carriage (exposure), ESBL-E. coli (ESBL-Ec) gastrointestinal carriage (outcome), and relevant covariates among 2,973 participants in Tromsø7. The variable ‘drug use’ includes antibiotic use past 14 days and acid suppressive medication past four weeks. K. pneumoniae is a common cause of healthcare associated infections often combined with antimicrobial resistance. In the literature and our previous study (Raffelsberger N, Hetland MAK, Svendsen K, et al. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes. 2021 Jan-Dec;13(1):1939599.), we identified several factors associated with K. pneumoniae gastrointestinal carriage, which are overlapping with those associated with ESBL-E. coli carriage. To capture the relevance of K. pneumoniae carriage as an exposure, we included this variable into the causal relationship with ESBL-E. coli carriage. The DAG was used for the selection of variables for the multivariable logistic regression model, to conceptualize confounding and to identify the minimal sufficient adjustment set. Although sex and alcohol consumption represent ancestors of exposure and outcome, the adjustment for age, drug use, hospitalization, travel abroad, and traveler`s diarrhea controls for relevant confounders and blocks biasing paths. The absence of red arrows in the DAG implies that there are no open unadjusted confounding pathways.
Characteristics of the study population.
Sensitivity analysis.
Genome characteristics of ESBL-E. coli from carriage isolates from Tromsø7 (n=166) and clinical isolates from NORM 2014 (n=118) isolates.
Comparison of ESBL-E. coli ST131 subclade prevalence.
SNP distances among two putative ESBL-producing E. coli clusters.
Susceptibility profile of carriage and clinical isolates of ESBL-E. coli ST131 and non-ST131.
Data Availability Statement
Bacterial genome data (raw Illumina reads) are publicly available in National Center for Biotechnology Information under BioProject PRJEB53319 (NORM 2014 collection, https://www.ebi.ac.uk/ena/browser/view/PRJEB53319) and PRJEB57251 (Tromsø7 collection, https://www.ebi.ac.uk/ena/browser/view/PRJEB57251). This study is based on data owned by a third party (The Tromsø Study, Department of Community Medicine, UiT The Arctic University of Norway, Norway). Confidentiality requirements according to the Norwegian law prevent sharing of individual patient level data in public repositories. Application of legal basis and exemption from professional secrecy requirements for the use of personal health data in research may be sent to a regional committee for medical and health research ethics (https://rekportalen.no/). The authors gained access to the data through the Tromsø Study’s application process. Guidelines on how to access the data are available at the website https://uit.no/research/tromsostudy. All inquiries about the Tromsø Study should be sent by e-mail to tromsous@ism.uit.no. All the questionnaire variables are published in the NESSTAR program system, and results can be viewed online: http://tromsoundersokelsen.uit.no/tromso/.