Abstract
Prenatal exposure to toxic metals is linked to numerous adverse birth and later-in-life outcomes. These outcomes are tied to disrupted biological processes in fetal-derived tissues including the placenta and umbilical cord yet the precise pathways are understudied in these target tissues. We set out to examine the relationship between metal concentrations in umbilical cord and altered gene expression networks in placental tissue. These novel relationships were investigated in a subset of the Extremely Low Gestational Age Newborn (ELGAN) cohort (n=226). Prenatal exposure to 11 metals/metalloids was measured using inductively coupled plasma tandem-mass spectrometry (ICP-MS/MS) in cord tissue, ensuring passage through the placental barrier. RNA-sequencing was used to quantify >37,000 mRNA transcripts. Differentially expressed genes (DEGs) were identified with respect to each metal. Weighted gene co-expression analysis identified gene networks modulated by metals. Two innovative mixtures modeling techniques, namely principal components analysis and quantile-based g-computation, were employed to identify genes/gene networks associated with multi-metal exposure. Individually, lead was associated with the strongest genomic response of 191 DEGs. Joint lead and cadmium exposure was related to 657 DEGs, including DNA Methyl Transferase 1 (DNMT1). These genes were enriched for the Eukaryotic Initiation Factor 2 (EIF2) pathway. Four gene networks, each containing genes within a Nuclear Factor kappa-light-chain-enhancer of Activated B Cells (NFkB)-mediated network, were significantly increased in average expression level in relation to increases in all metal concentrations. All four of these metal mixture-associated gene networks were negatively correlated with important predictors of neonatal health including birth weight, placenta weight, and fetal growth. Bringing together novel methodologies from epidemiological mixtures analyses and toxicogenomics, applied to a unique cohort of extremely preterm children, the present study highlighted critical genes and pathways in the placenta dysregulated by prenatal metal mixtures. These represent potential mechanisms underlying the developmental origins of metal-induced disease.
Keywords: mixtures, metals, placenta, gene expression, pregnancy, DOHaD
Graphical Abstract
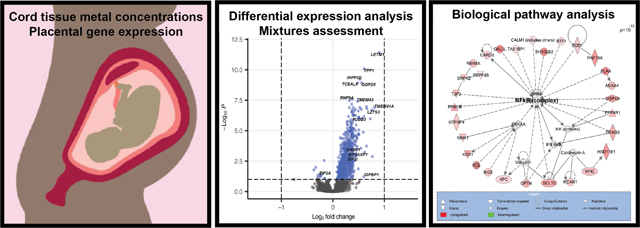
1. Introduction
Exposure to even trace amounts of toxic metals (e.g. cadmium (Cd), lead (Pb), and mercury (Hg)) and metalloids (e.g. arsenic (As)), during pregnancy is a major public health problem worldwide (Michelsen‑Correa et al. 2021). In the United States (U.S), toxic metals are ubiquitous contaminants. For example, among reproductively aged women, inorganic As (iAs) and Cd in urine, and Pb and Hg in blood were detected in 97.8, 87.9, 99.0 and 86.9% of samples (Bulka et al. 2019). Prenatal exposure to metals is associated with acute adverse perinatal outcomes, such as preeclampsia, preterm birth, and low birth weight (Ferguson et al. 2013; Rosen et al. 2018; Rager et al. 2020). Early-life exposure to toxic metals is also implicated in the Developmental Origins of Health and Disease (DOHaD) with long-lasting effects documented on growth and metabolic health (Gardner et al. 2013; Kupsco et al. 2019), immune regulation (Cao et al. 2016; Farzan et al. 2016), and cognitive function (Sanders et al. 2015). Notably, infants born extremely preterm are especially vulnerable to developing many of these later-in-life outcomes (Kuban et al. 2016; Goedicke‑Fritz et al. 2017). Given the prevalence of metal exposure and the severity of the impacts, understanding the underlying biological mechanisms of metal-induced developmental effects remains an urgent public health need.
Dysregulation of critical biological pathways in the placenta, particularly pro-inflammatory pathways, represents one such potential mechanism. The placenta is the master regulator of the intrauterine environment, responsible for the transfer of nutrients, waste, and gases as well as hormonal regulation of the maintenance of pregnancy (Nelson and Myatt 2020). There are numerous, interconnected mechanisms by which trace metals may alter gene expression in the placenta. For instance, some trace metals may alter the conformation of critical proteins involved in transcription or translation, and/or upregulate transcription factors that have cascading effects, or influence epigenetic processes (Blanchard and Cousings 1997; Ryu et al. 2015). Among these mechanisms, metal-responsive placental epigenetic patterning, leading to upregulation/downregulation of key biological pathways is coming to the forefront as a mechanism of interest for both immediate neonatal health as well as DOHaD related outcomes (Marsit 2016). For example, prenatal exposure to As, Cd, Hg, and Pb exposure has been linked to CpG methylation in the placenta in human cohorts (Martin and Fry 2018; Tung et al. 2022). (Payton et al. 2020; Santos et al. 2020). In particular, inflammatory processes, which are in part under epigenetic control, are posited as one of the critical mediating biological mechanisms connecting metal exposure and associated adverse outcomes (Leviton et al. 2015; Ferguson and Chin 2017).
Over the last decade, improved analytical precision and capacity in the generation of high-dimensional molecular data and statistical advancements in bioinformatics have made it more feasible to assess -OMICs data in relation to toxic exposures (Everson and Marsit 2018; Martins et al. 2019). Mirroring the leaps in bioinformatics approaches, the field of environmental epidemiology has made recent strides to overcome statistical barriers to assessing the effects of multiple exposures (Hamra and Buckley 2018; Gibson et al. 2019). This is particularly critical when it comes to assessing the effects of metals because they frequently co-occur, especially in highly contaminated areas (Zota et al. 2011). Moreover, metals may act synergistically or antagonistically with one another to produce different biological effects in mixtures than those for single metals alone (Adebambo et al. 2015; Everson et al. 2017). However, environmental mixtures methods are not often utilized in the toxicogenomics setting, particularly in human cohort studies. This is related to the challenges of high dimensionality that exist both with regards to exposure measures as well as with the -OMICs data collected. Few studies have conducted mixtures-based toxicogenomic assessments in the placenta (Deyssenroth et al. 2018), and no study to our knowledge has done so utilizing cord tissue to measure trace metal exposures. Assessment in this biospecimen ensures the placenta was directly exposed to metals (Rager et al. 2020).
In the present study, we set out to test the hypothesis that inflammation-related genes in the placenta would display altered expression in relation to cord tissue metal concentrations, both individually and in mixtures. Innovatively bringing together data dimension reduction techniques, mixtures modeling and toxicogenomics approaches, we examined the multidimensional relationship between 11 metals/metalloids and 30,000+ genes in placenta in a unique cohort of children born extremely preterm, a population particularly susceptible to the developmental effects of environmental chemicals. Our results confirm metal-associated inflammation-related gene expression in the placenta, thus providing critical information for understanding the role of placental molecular changes in the connection between early-life metal exposure and the developmental origins of metal-associated disease.
2. Materials and Methods
2.1. The ELGAN cohort.
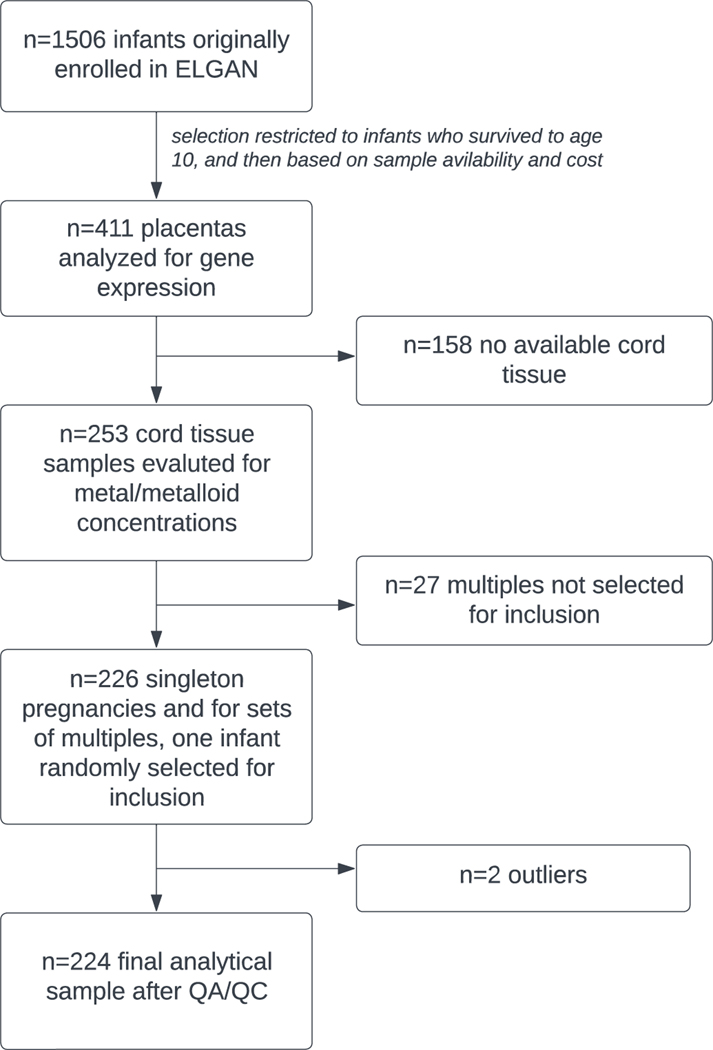
The Extremely Low Gestational Age Newborn (ELGAN) study is an ongoing, multi-site, prospective cohort study originally designed to assess developmental brain abnormalities in children born extremely preterm (O’Shea et al. 2009). Eligible infants were liveborn at less than 28 weeks’ gestation between 2002–2004 at one of the 14 participating sites in 5 different states (North Carolina, Massachusetts, Michigan, Illinois, Connecticut). No specific exclusion criteria were applied and 83% of eligible infants were enrolled. The Institutional Review Board at each site approved the study procedures, and all mothers provided written informed consent. Gestational age was estimated using fetal ultrasound, self-reported last menstrual period, and/or neonatal intensive care unit records. At enrollment, a structured questionnaire collected self-reported information on sociodemographic data and medical information was obtained from medical records. Placental and umbilical cord samples were collected at delivery. Overall, n=1,249 mothers of n=1,506 infants were enrolled and constitute the parent ELGAN study. From this parent study, children were followed prospectively, and among those who survived to age 10, a representative sample of 411 placental samples was selected for the measurement of gene expression. Of these, n=253 also had umbilical cord tissue samples available that were analyzed for a suite of trace metals.
Due to the selection of extremely preterm infants, the ELGAN cohort has a large proportion of multiple births (e.g., twins, triplets). The inclusion of multiple infants born to the same mother would violate the independence of observations assumption required for later statistical modeling given the shared prenatal environment. Therefore, we selected all singletons for inclusion and then randomly selected one infant from each set of multiples to be included in the present study. This filtering removed n=27 subjects, leaving a study population of n=226 (Figure 1).
Figure 1.
Flow diagram illustrating the selection of analytic sample used for this study, a subgroup of the ELGAN study.
2.2. Umbilical cord tissue collection and measurement of trace metal concentrations.
A total of 8 metals (barium, (Ba), Cd, copper (Cu), Pb, manganese (Mn), Hg, strontium (Sr), zinc (Zn)) and 3 metalloids (As, antimony (Sb), selenium (Se)) were targeted for analysis and measured in cord tissue using inductively coupled plasma tandem-mass spectrometry (ICP-MS/MS). Note that while As, Sb, and Se are technically metalloids, with properties of both metals and non-metals, for ease of reading they will henceforth be referred to as part of the collective term “metals.” Within an hour after delivery of the infant, two 1 cm segments of the umbilical cord were collected using sterile technique with a stainless-steel scalpel blade. These segments were placed into separate cryostorage vials and immersed into liquid nitrogen for transport to a −80 °C freezer before being shipped to the Wadsworth Center for analysis. On receipt at the Wadsworth Center, umbilical cord samples were accessioned per standard laboratory procedure and stored at −80 °C pending analysis. Samples were thawed and sectioned into ~0.4 g pieces using high-purity tantalum tools that were fabricated in-house for use in trace element analysis. All umbilical cords were rinsed with double-deionized (DDI) water to remove superficial blood and placed in a 13 mL acid-washed tube. The samples were freeze-dried to constant mass using a slow 5-step program to ensure thorough removal of water content. Each batch of samples was digested in concentrated, double-distilled, HNO3 using a Microwave Assisted Reaction System (MARS 6, Matthews, NC) with closed “XPress” vessels and the “One-Touch Animal Tissue” method. Digests were diluted to ~10 g with DDI water and stored at 4 °C pending analysis. At the time of analysis, tissue digests were further diluted with a reagent containing internal standards with a final HNO3 concentration ~10% (v/v). A method was developed and optimized to analyze the cord tissue samples on an Agilent 8900 ICP-MS/MS equipped with a SPS 4 autosampler and an Octopole Reaction System (ORS) with axial acceleration technology. A collision cell gas (He) was used to reduce polyatomic interferences on 65Cu, 66Zn, 78Se, and 88Sr. The reaction gas O2 was used to mitigate interferences on 55Mn, 75As, 111Co, 121Sb, 202Hg. In O2 gas mode the elements As and Sb were monitored as 75As16O and 121Sb16O, respectively, i.e., mass shifted. Pb was monitored as Σ206, 207, 208Pb, to account for the natural variation in the relative abundance of stable isotopes for Pb. The final optimized method was validated using two Standard Reference Materials (SRM) from the National Institute of Standards and Technology (NIST): NIST SRM 1577b Bovine Liver and NIST SRM 1577c Bovine Liver. Two other SRMs, NBS 1577 Bovine Liver and SRM 8414 Bovine Muscle, were used for additional quality control. The SRMs were freeze-dried, digested, and analyzed alongside the umbilical cord samples. In addition, recovery values for sample spikes (typically within ±20%), duplicates (typically agreement to within ±20%), blanks, and calibration curve robustness were all carefully monitored throughout the study. The method limit of detection (LOD) for each of the 11 trace metals measured was: As (0.42 ng/g), Ba (8.5 ng/g), Cd (0.32 ng/g), Cu (0.074 μg/g), Hg (0.79 ng/g), Mn (0.010 μg/g), Pb (2.6 ng/g), Sb (0.88 ng/g), Se (0.10 μg/g), Sr (0.039 μg/g), Zn (1.4 μg/g). All samples had detectable levels for Mn, Cu, Zn, As, Se, Sr, Ba, and Pb. Detection frequency for Sb, Hg, and Cd was 94, 97, and 98%, respectively, for the 253 cord tissue samples analyzed. Samples at or below the LOD were imputed as , as is commonly implemented in environmental chemistry data (EPA 2000).
2.3. Placental tissue collection and measurement of gene expression.
Methods for the collection of placental tissue samples and assessment of placental gene expression within the ELGAN cohort have been described in detail elsewhere (Payton et al. 2020). In summary, <1 g placental biopsy samples were removed from the base of the chorion and stored in sterile 2 mL cryovials in a −80 °C freezer. Segments (0.2 g) were extracted using a sterile dermal curette and were then washed in 1x PBS (Fisher Scientific, Waltham, MA) to reduce any potential blood contamination, snap frozen in homogenization tubes and placed on dry ice. A sterile stainless-steel bead (Qiagen, Germantown, MD) in RLT + lysis buffer (Qiagen) with the TissueLyserII instrument (Qiagen) was used to homogenize the tissue segments, which were then stored at 80 °C until nucleic acid extraction. The AllPrep DNA/RNA/miRNA Universal kit (Qiagen) was used to extract RNA molecules 18 nucleotides and greater. RNA quality was determined using LabChip (Perkin Elmer, Waltham, MA) to generate RNA integrity numbers (RIN). Genome-wide mRNA expression was determined using QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen, Vienna, Austria) and RNA-sequencing libraries were pooled and sequenced (single-end 50 bp) on one lane of the Illumina Hiseq 2500 (Illumina, San Diego, CA). Libraries were prepared by automation on Sciclone G3 (Perkin Elmer, Waltham, MA) to avoid potential batch to batch artifacts. The counts of sequencing reads were aligned to the GENCODE database (v.30) and organized using Salmon (v0.11.3) to give a total of n=37,268 unique human RNA transcripts (Harrow et al. 2012; Patro et al. 2017).
2.4. Statistical analysis.
2.4.1. Metals data descriptive statistics.
All statistical analysis was conducted in R (v4.0.2) (R Core Team 2020). Descriptive statistics, both overall and stratified by key sociodemographic variables, were calculated to describe the distributions of metals. Shapiro-Wilks tests were used to test for normality in the cord metals data. Since none of the metals were normally distributed, Spearman rank correlation was used to evaluate pairwise correlations between the metals.
2.4.2. Placental mRNA data processing.
Prior to modeling, count data were first filtered to exclude universally lowly expressed transcripts, requiring that > 25% of the samples be expressed at signals above the overall median signal intensity (Payton et al. 2020). This resulted in a total of n=11,402 mRNA transcripts included in analyses. QA/QC was conducted on the count data using both calculation and visualization of principal components via the prcomp function and hierarchical clustering, including calculation of distance metrics and visualization, using the hclust function in the stats R package (v4.0.2) (R Core Team 2020). Two samples were deemed to be outliers and were thus removed, resulting in a final analytic sample of n=224 (Figure 1). Count data were normalized utilizing the DESeq2 R package (v1.30.1) using median signal intensity (Love et al. 2014). The SVA R package (v 3.38.0) was used to account for potential batch effects and sources of sample heterogeneity (e.g. cell type proportions) with control probes empirically estimated using default parameters (Leek et al. 2012, 2020; Leek 2014). Two surrogate variables were calculated and included as covariates in all subsequent models (Figure S1, Additional File 1).
2.4.3. Covariate selection.
Covariates were chosen a priori based on a directed acyclic graph approach. Selected covariates included maternal pre-pregnancy BMI (underweight, normal, overweight, obese), maternal smoking during pregnancy (yes/no), maternal SES score and infant sex (male/female). Maternal SES score was derived as a summative count of: less than college education, single marital status, eligibility for Supplemental Nutrition Assistance Program (SNAP), and public health insurance, as this measure has previously shown utility for predicting molecular signatures within the placenta in the ELGAN cohort (Santos et al. 2019). Data were missing on seven subjects for the maternal SES score, seven subjects for maternal pre-pregnancy BMI, and four subjects for maternal smoking during pregnancy. Random forest modeling was utilized to impute covariates where they were missing utilizing the missForest R package (v1.4)(Stekhoven and Bühlmann 2012; Stekhoven 2013). The out-of-bag imputation error value for the normalized root mean squared error was 0.006, indicating good performance of the random forest imputation (Stekhoven and Bühlmann 2012).
2.4.4. Evaluation of metals/metal-mixtures to single-gene expression.
Using the DESeq2 method, negative binomial generalized linear models were used to identify genes with differential expression based on individual metal concentrations (at or above median versus below median concentration) (Love et al. 2014). To account for multiple testing, p-values were adjusted using the Benjamini and Hochberg (BH) procedure (Benjamini and Hochberg 1995).
In addition to analyses of individual metals, two different approaches were utilized within the DESeq2 framework to evaluate whether individual genes were differentially expressed in relation to metal mixtures. First, a principal components analysis (PCA) approach was used and second, an a priori toxic/essential metals grouping approach was used. PCA produces linear combinations (principal components, “PC”) of the input dataset (in this case, 11 metals) to describe as much of the information as possible in fewer variables (Jolliffe and Cadima 2016). Each PC has loading values for each of the metals. A higher loading value means that the metal contributes more to the variability represented in that PC, and a positive loading means that metal’s variability is positively correlated with that PC. PCA was conducted using the prcomp function in the stats R package (R Core Team 2020). The PCs were used as independent variables in models fit using DESeq2 (Love et al. 2014). The first four PCs were evaluated; however, only the first two PCs (PC1 and PC2) were ultimately included in the final results as PC3 was associated with only two genes and PC4 was not associated with any gene.
Following the PCA-based analysis, two different groupings of metals based on a priori knowledge of their toxicity were generated. The toxic metal index was the sum of the z-score scaled concentrations of As, Ba, Cd, Hg, Pb, Mn, Sb, and Sr. The essential metal index was the sum of the z-score scaled concentrations of Cu, Se, and Zn. These two index variables were used as independent variables in models fit using DESeq2 (Love et al. 2014). In the DESeq2-based analyses, differentially expressed genes were defined as those with BH-adjusted p-value<0.1.
2.4.5. Examination of the relationship between metals/metal-mixtures and co-expressed gene networks.
To derive the co-expressed gene networks, we utilized WGCNA within the WGCNA R package (v1.70–3) (Langfelder and Horvath 2008). WGCNA is a technique used to identify modules of genes with highly correlated expression and to describe these clusters by calculating module eigengenes, representing the first PC of each module (Langfelder and Horvath 2008). The application of WGCNA by our team has been described in detail elsewhere (Rager et al. 2017, 2021). The minimum module size was set to 30, as is suggested in in the WGCNA tutorial to encourage larger module sizes (Langfelder 2014).
First, to assess the relationship between co-expressed gene networks and single-metal exposure, linear regression models with the eigengene as the dependent variable and each metal as the independent variable were fit. The metal variables were quantized so that the resulting beta estimates represented the expected change in the eigengene value for a quartile increase in the metal concentration.
To evaluate the relationship of the co-expressed gene networks with metal mixtures, two approaches were utilized. First, for comparison to the single-gene analysis, PCs, described above, were used as independent variables in linear regression models with the eigengene as the dependent variable. Second, quantile-based g-computation was employed utilizing the qgcomp R package (v2.8.0) (Keil et al. 2020; Keil 2021). Quantile-based g-computation is a recently developed statistical method that employs a generalized-linear-model based implementation of g-computation (Keil et al. 2020). It provides estimates of the effect of simultaneously increasing all exposures (in this case, 11 metals) within a mixture by one quartile, which we refer to as the “overall mixture” association. It also calculates a weighted index of the component exposures. The components of the index can have negative or positive weights (that sum to 1) which represent proportional contributions to negative or positive partial effects from an individual metal (Keil et al. 2020). To emulate the a priori defined toxic index and essential index approach used in the single gene analysis, described above, we also fit models within the quantile-based g-computation framework using only the toxic metals (“toxic only mixture”), and then only essential metals (“essential only mixtures”). Therefore, the association for the toxic only mixture/essential only mixture can be interpreted as the expected change in the module eigengene value when all toxic metals/essential metals are increased by one quartile. Metalmixtures significantly associated with eigengene values were defined as those with model pvalues <0.05.
2.4.6. Biological pathway analysis.
Canonical pathway and network enrichment analyses were carried out utilizing the Ingenuity Knowledge Database (Ingenuity Pathway Analysis, Qiagen, Redwood City, USA). For any metal with differentially expressed genes identified, all differentially expressed genes were used as input data. For any co-expressed gene network associated with a metal mixture, all genes within the network were used as input data. Over-represented canonical pathways were defined as those containing more genes than expected by random chance, as based on q-value calculated from a right-tailed Fisher’s Exact Test (significance defined at p<0.05). Networks were constructed based on known protein-protein interactions and other molecular interactions and were ranked based on right-tailed Fisher’s Exact test p-values, indicating the likelihood of observing a network containing at least the same number of proteins encoded by differentially expressed genes/gene networks by chance in comparison to random selections of other genes within the genome.
2.4.7. Correlation between metal mixture-associated genes/gene networks and neonatal phenotypes.
In order to understand the potential clinical implications of the metal mixture-associated genes and gene networks, we evaluated correlations with four critical phenotypes indicative of neonatal health: placental weight (grams), birth weight (grams), fetal growth and gestational age (days). Fetal growth was calculated as the birth weight for gestational age, z score transformed among all ELGAN participants. Genes were assessed for correlation with phenotypes if they were significantly differentially expressed in relation to a PC. Normalized counts derived from DESeq2 were used in the correlation analysis. Gene networks were assessed for correlation if they were significantly associated with the metal mixture in the quantilebased g-computation analysis and PC-based analysis. For the gene networks, the module eigengene value was used to correlate with the phenotype values. Spearman rank correlation was used given the non-normality of the module eigengene data with a significance threshold of p<0.05.
2.5. Data Availability.
Code and data used in this analysis are publicly available from the UNC Superfund Research Program GitHub page (https://github.com/UNCSRP/Metal-mixtures-modeling-BW-gene-networks-PTB-placentas). mRNA count data are publicly available from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository under GEO series GSE154829.
Results
3.1. Study population characteristics.
General characteristics of the ELGAN participants included in the present study are provided in Table 1. Within this group, 59.3% of the mothers self-identified as White, 29.6% as Black. The majority of participants were non-Hispanic. Most mothers did not smoke during pregnancy (91.2%). The majority of the mothers were healthy weight (51.8%), with only 6.6% of mothers being underweight and the remaining 41.6% being overweight or obese. The majority of mothers were also either married or cohabitating (77.45%), had some college education or more (58.85%), did not use public insurance (66.4%), and were not eligible for SNAP (88.1%). The n=226 cohort used in this study did not vary significantly in the proportions of any key clinical or demographic variables from the cohort in which placental gene expression is characterized (n=411) or the parent ELGAN cohort (n=1506) (Table S1, Additional File 2).
Table 1.
Socio-demographic characteristics of the ELGAN sub-cohort analyzed in this study (n=226).
| N (%) | |
|---|---|
| Maternal race | |
| White | 134 (59.3) |
| Black | 68 (30.1) |
| Other | 21 (9.3) |
| Missing | 3 (1.3) |
| Maternal ethnicity | |
| Non-Hispanic | 207 (91.6) |
| Hispanic | 19 (8.4) |
| Maternal age | |
| <21 | 25 (11.1) |
| 21–35 | 155 (68.6) |
| >35 | 46 (20.4) |
| Maternal BMI a | |
| Underweight (<18.5) | 15 (6.6) |
| Normal weight (18.5 =< BMI <25) | 117 (51.8) |
| Overweight (15.0 =< BMI <30) | 41 (18.1) |
| Obese (>30) | 53 (23.5) |
| Maternal smoking while pregnant a | |
| No | 206 (91.2) |
| Yes | 20 (8.8) |
| Maternal marital status | |
| Married or cohabitating | 175 (77.4) |
| Single | 51 (22.6) |
| Maternal education level | |
| Less than college education | 88 (38.9) |
| Some college education or more | 133 (58.8) |
| Missing | 5 (2.2) |
| Maternal public insurance use | |
| No | 150 (66.4) |
| Yes | 73 (32.3) |
| Missing | 3 (1.3) |
| Maternal eligibility for SNAP | |
| No | 199 (88.1) |
| Yes | 24 (10.6) |
| Missing | 3 (1.3) |
| Infant sex | |
| Male | 123 (54.4) |
| Female | 103 (45.6) |
| Singleton pregnancy | |
| No | 70 (31) |
| Yes | 156 (69) |
| Gestational age in weeks, (median (min, max)) | 26.3 (23, 27.9) |
based on imputed values following random forest modeling imputation. Pre-imputation, there was data missing on seven subjects for the maternal SES score, seven subjects for maternal pre-pregnancy BMI, and four subjects for maternal smoking during pregnancy.
Overall distributions of metal levels in umbilical cord tissue are detailed in Table 2. Metal levels varied by socio-demographic variables in bivariate assessments (Table S2, Additional File 2). As, Cd, Pb, and Zn varied significantly by maternal race with As and Zn displaying the highest levels in White mothers and Pb and Cd having the highest levels in mothers who self-identified as Asian, Native American, Mixed race or Other (grouped together as “Other” due to small sample size). As, Ba, Cd, Hg, Pb, Sr, and Zn concentrations all significantly increased with increasing maternal age. Cd levels were higher in mothers who smoked, although this difference was not statistically significant. Zn significantly varied by maternal pre-pregnancy BMI with lower concentrations at extremes (ie. underweight or obese). No significant differences in metal concentrations were observed by fetal sex or maternal SES score, and only Pb levels differed between singleton and multiple pregnancies.
Table 2.
Distributions of the concentrations of the 11 metals measured in cord tissue among the samples used in this analysis (n=226).
| Metal | Units | Median | Interquartile Range | 25th percentile | 75th percentile | Minimum | Maximum |
|---|---|---|---|---|---|---|---|
| As | ng/g | 4.59 | 4.10 | 3.30 | 7.40 | 1.28 | 70.65 |
| Ba | ng/g | 81.05 | 73.35 | 55.95 | 129.30 | 17.50 | 3131.90 |
| Cd | ng/g | 1.22 | 2.13 | 0.73 | 2.86 | <0.32 | 4057.21 |
| Cu | μg/g | 3.54 | 1.46 | 3.01 | 4.47 | 1.85 | 14.31 |
| Hg | ng/g | 7.72 | 12.03 | 3.62 | 15.65 | <0.79 | 96.36 |
| Mn | μg/g | 0.34 | 0.12 | 0.29 | 0.42 | 0.10 | 1.70 |
| Pb | ng/g | 15.75 | 20.55 | 9.20 | 29.75 | <2.6 | 708.70 |
| Sb | ng/g | 3.24 | 3.90 | 1.93 | 5.83 | <0.88 | 360.93 |
| Se | μg/g | 0.86 | 0.19 | 0.78 | 0.97 | 0.44 | 1.98 |
| Sr | μg/g | 0.55 | 0.44 | 0.39 | 0.83 | 0.17 | 3.33 |
| Zn | μg/g | 59.40 | 18.38 | 52.20 | 70.58 | 29.90 | 487.50 |
3.2. Correlations between cord metals.
Many metal pairs were significantly correlated (Table S3, Additional File 2; Figure S2, Additional File 1). Specifically, 38 pairings (69% of 55 pairings) were significantly correlated (p<0.05) in a Spearman correlations test. Nearly all of these significant correlations were positively correlated, potentially indicative of shared sources of exposure. Only one pairing was negatively correlated: Se and Sb (Spearman rank: −0.170, p=0.01).
3.3. Single metal and metal-mixture associations with individual genes.
In the single metal analysis, across the transcriptome, Pb, Hg, and Mn demonstrated the greatest genomic response with 191, 95, and 37 significantly differentially expressed genes (DEGs), respectively (Table 3; Table S4, Additional File 2; Figure S3, Additional File 1). For each of these, the majority of DEGs demonstrated decreased expression with increasing concentrations of the metal, defined as genes demonstrating a log2(Fold Change)<0. Ba, Cd, Cu, and As all had fewer than 15 DEGs and null associations were observed for Sb, Se, Sr, and Zn.
Table 3.
Summary of differential expression analyses in relation to individual metals and mixtures, PC1, and PC2.
| Metal | Total number of DEGs | Number of up-regulated DEGs | Number of down-regulated DEGs |
|---|---|---|---|
| Arsenic | 1 | 0 | 1 |
| Barium | 12 | 5 | 7 |
| Cadmium | 2 | 1 | 1 |
| Copper | 2 | 1 | 1 |
| Mercury | 95 | 5 | 90 |
| Manganese | 37 | 6 | 31 |
| Lead | 191 | 45 | 146 |
| Antimony | 0 | 0 | 0 |
| Selenium | 0 | 0 | 0 |
| Strontium | 0 | 0 | 0 |
| Zinc | 0 | 0 | 0 |
| PC1 | 40 | 29 | 11 |
| PC2 | 657 | 646 | 11 |
| Toxic index | 112 | 88 | 24 |
| Essential index | 62 | 56 | 6 |
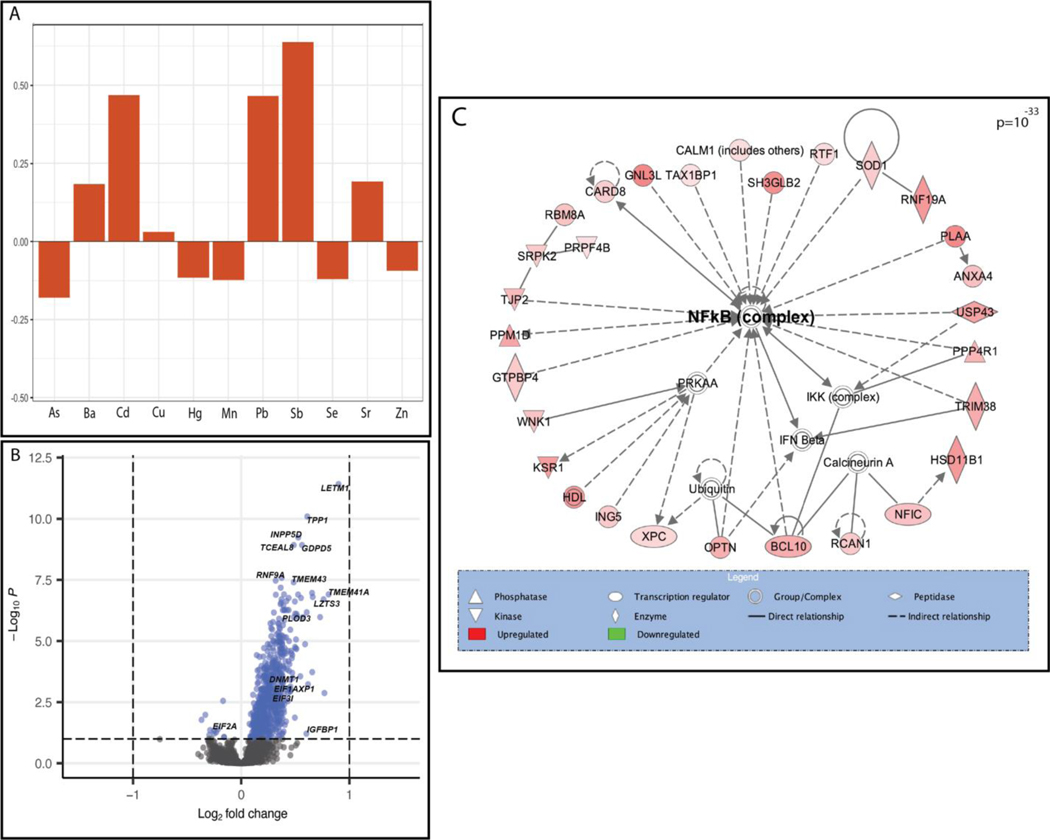
We then examined the relationship between individual genes and metal mixtures using a principal components (PC)-based approach and an a priori groupings approach. Two PCs were identified that captured the variability in the levels of the 11 metals. PC1 displayed a profile distinguished through an inverse association with only Cd and highest positive loadings for Cu, Mn, Se, and Zn, indicative of a metal profile predominated by high levels of essential metals (Mn being essential at low doses). PC1 was associated with 40 DEGs (Table 3). Notably, this number of genes exceeded the findings for some of the single metals alone. PC2 displayed a profile with more variability of the direction of loadings across metals, with several essential metals, including Mn, Se, and Zn all having negative loadings and the strongest positive loadings being for Cd, Pb, and Sb, all toxic metals; thus, representative of a profile of high toxic metals and low essential metals (Table S6, Additional File 2; Figure 2A). In comparison to the results for the single metals and PC1, PC2 demonstrated a substantially larger genomic response (16.4 times as many genes as PC1), with 657 associated DEGs (Table 2; Table S4, Additional File 2; Figure 2B). Among all DEGs identified in response to PC1 or PC2, 94% were differentially expressed in relation to PC2.
Figure 2.
The multi-metal PC (PC2) was associated with the expression of NF-kB enriched genes. (A) Plot demonstrating the individual loading values of metals onto the principal component 2 (PC2) eigenvector, (B) Volcano plot demonstrating differentially expressed (BH p-value <0.1) genes in relation to the multi-metal PC, adjusted for maternal pre-pregnancy BMI, maternal smoking, maternal SES score and infant sex, with select significant genes highlighted, (C) a significantly enriched biological network identified in pathway analysis of genes that were differentially expressed in response to the multi-metal PC.
Notable genes that were differentially expressed in relation to the PC2 included DNA Methyltransferase 1 (DNMT1) (log2FC= 0.228, BH-adjusted p-value <0.001), Insulin Like Growth Factor Binding Protein-1 (IGFBP1) (log2FC= 0.602, BH-adjusted p-value= 0.06), Insulin Like Growth Factor-2 Binding Protein-1 (IGF2BP1) (log2FC= 0.092, BH-adjusted p-value =0.07), INSIGF2 readthrough (INS-IGF2) (log2FC= 0.083, BH-adjusted p-value =0.05), Eukaryotic Translation Initiation Factor 2A (EIF2A) (log2FC= −0.249, BH-adjusted p-value= 0.06), and Eukaryotic Translation Initiation Factor 3 Subunit I (EIF3I) (log2FC= 0.341, BH-adjusted p-value= 0.002) (Table S4, Additional File 2; Figure 2). Through pathway analysis of PC2 associated DEGs, Eukaryotic Initiation Factor 2 (EIF2) Signaling (p=6.20×10−09) was identified as the topmost enriched canonical pathway and Gene Expression was identified as the topmost enriched molecular and cellular function (p=3.48×10−14). Furthermore, a notable significant biological network among these genes included Nuclear Factor kappa-light-chain-enhancer of Activated B Cells (NF-kB) as a central node (Figure 2C). Among the notable genes listed above, INS-IGF2 and IGF2BP1 were both significantly negatively correlated with birth weight (INS-IGF2: −0.29, p=1.05×10−5; IGF2BP1: −0.23, p=4.72×10−4); fetal growth (INS-IGF2: −0.19, p=5.14×10−3; IGF2BP1: −0.16, p=1.77×10−2), and placenta weight (INS-IGF2: −0.14, p=3.9×10−2; IGF2BP1: 0.16, p=2.02×10−2), (Table S5, Additional File 2). INS-IGF2 was also negatively correlated with gestational age (−0.21, p=1.81×10−3), but IGF2BP1 was not (Table S5, Additional File 2). EIF3I was negatively correlated with birth weight (−0.24, p=3.05×10−4), gestational age (−0.16, p=1.66×10−2) and fetal growth (−0.19, p=5.07×10−3) and EIF2A was negatively correlated with birth weight (−0.17, p=1.23×10−2) (Table S5, Additional File 2). Overall, of the 657 DEGs associated with PC2, 505 were significantly correlated with birth weight, 322 with fetal growth and 213 with placenta weight (Table S5, Additional File 2).
We also assessed genome-wide individual gene expression in relation to two a priori defined indices: a toxic metal index and an essential metal index. When examining the genes that were differentially expressed in response to these indices, 35.6% of them were differentially expressed in relation to the essential metals index (62 DEGs) and the remaining 64.4% were differentially expressed in relation to the toxic metal index (112 DEGs). This mirrors the findings from the PC-based analysis, in which the predominantly essential metal loaded PC1 had a weaker response than the predominately toxic metal loaded PC2. However, overall, the toxic metals or essential metals only indices appeared to underestimate the genomic response to metal mixtures in comparison to the combined toxic and essential metal profiles captured by the PCs. For example, PC2 was associated with 5.87 times as many genes as the toxic metal index. All DEGs for single metal and multi-metal-based analyses are detailed in Table S4, Additional File 2.
3.4. Single metal and metal-mixture associations with co-expressed gene networks.
We are mindful that genes operate as co-expressed networks rather than as independent units and for this reason, we utilized WGCNA to identify a total of 25 co-expressed gene networks, which ranged from including 3249 to 38 genes (Table S7, Additional File 2). Table S8 (Additional File 2) details the gene to module assignments. These co-expressed gene networks were used to calculate module eigengenes (ME) which were assessed for associations with single metals and metal mixtures using two approaches, a principal components-based approach (evaluating the association with PC1 and PC2) and quantile-based g-computation approach.
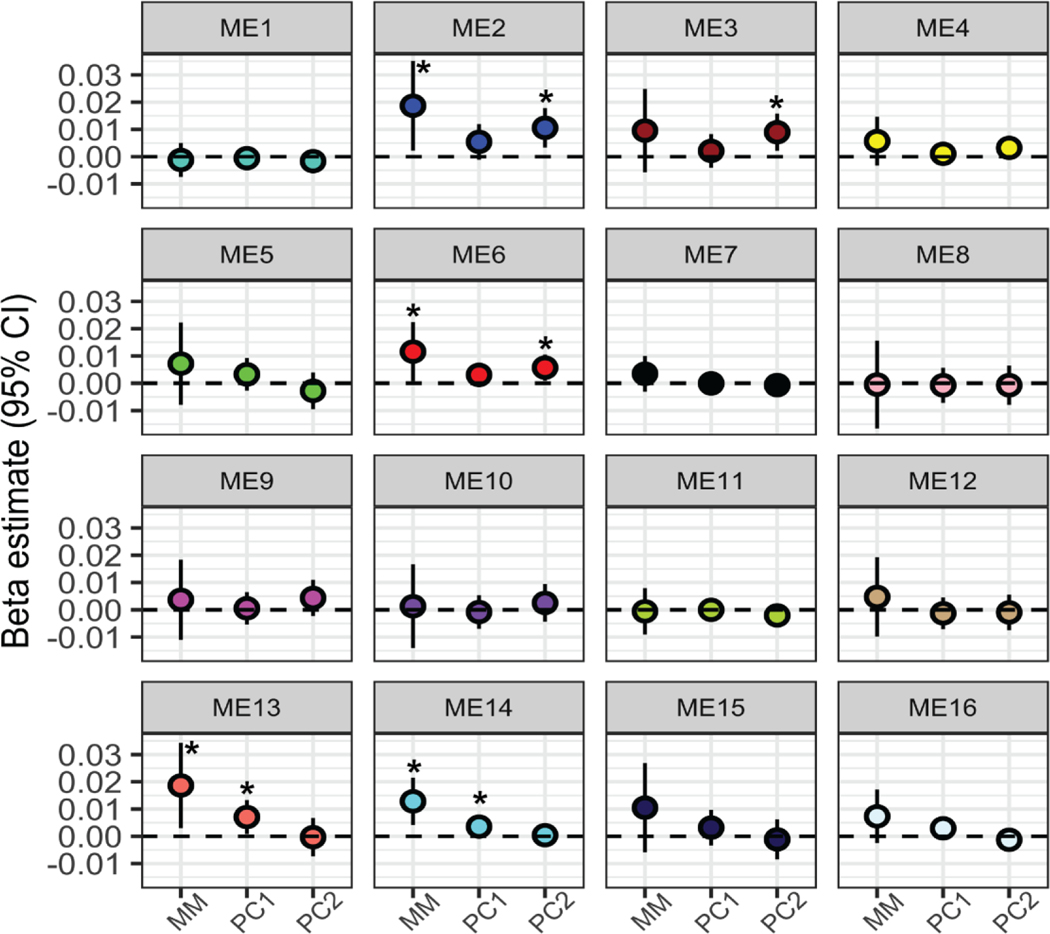
Overall, we identified more significant changes in co-expressed gene networks when evaluating the association with the metal mixture compared to single metals alone. Regarding associations with single metals, Pb and Sb were significantly associated with the expression of ME23; however, no other metals were associated with any gene networks (Table S9, Additional File 2; Figure S4, Additional File 1). In contrast, four gene networks were identified as having significantly associated expression levels in relation to metal mixtures. Specifically, ME2, ME6, ME13 and ME14 were all significantly associated with the overall mixture that included all 11 metals in the quantile-based g-computation analysis (Table S9, Additional File 2; Figure 3). In other words, when all 11 metal concentrations increased by a quartile, the average gene expression level of each of these co-expressed gene networks increased. In addition, ME14 and ME22 were significantly associated with the essential only mixture (Table S9, Additional File 2). No gene networks were associated with the toxic only mixture.
Figure 3.
Metal mixture associations with placental co-expressed gene networks. Beta estimates correspond to the expected average change in each module eigengene (ME) value for each placental co-expressed gene network with (1) a one quartile increase in the entire metal mixture (MM), (2) one unit increase in principal component 1 (PC1), and (3) one unit increase in principal component 2 (PC2). All estimates are adjusted for maternal pre-pregnancy BMI, maternal smoking, maternal SES score and infant sex. *= significant estimates (p<0.05).
The overall mixture associations with ME2, ME6, ME13 and ME14 can be broken down into the partial positive and negative contributions by the individual metal components of the mixture. In each of these cases, the mixture association is positive overall, with an increase by one quartile of all metals corresponding to an overall increase in gene expression (Figure S5, Additional File 1). For ME2 and ME6, As, Hg, and Ba contribute negatively to this effect, acting in opposition to the overall association’s direction (Figure S5, Additional File 1). For ME13, As, Pb, and Ba contribute negatively to the overall effect, with Cd, Cu, and Mn contributing in the same direction as the overall effect (Figure S5, Additional File 1). For ME14, Ba, Zn, and Mn contribute in the opposing direction to the overall effect (Figure S5, Additional File 1).
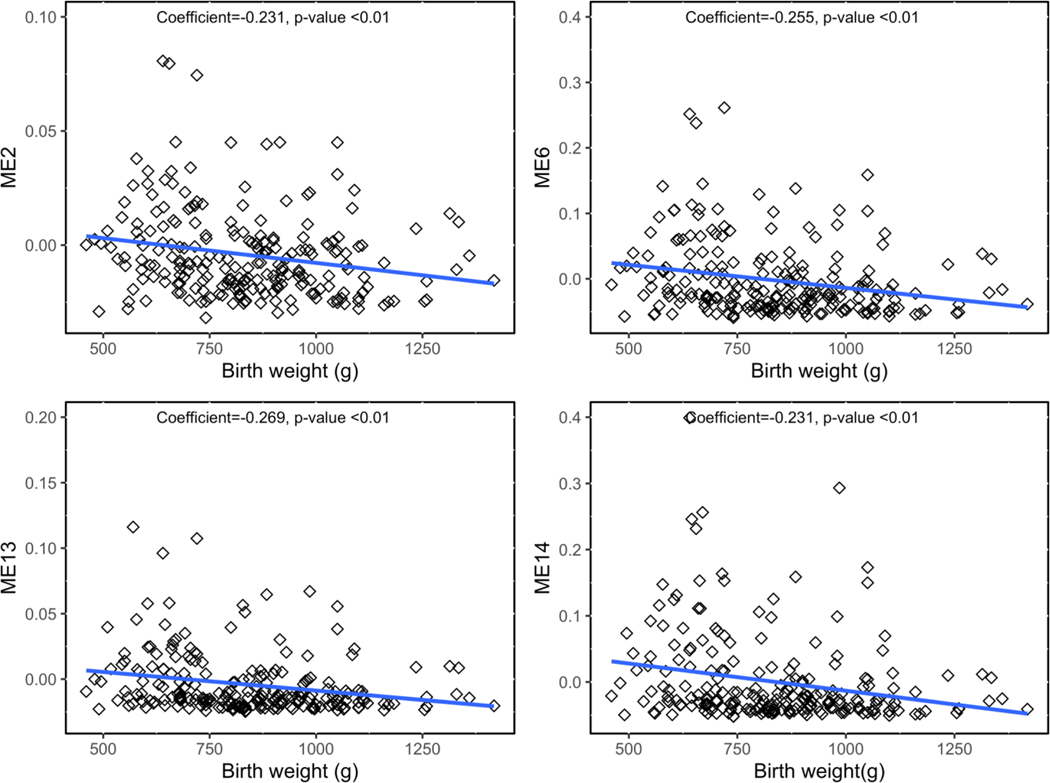
When utilizing PC1 and PC2 in relation to the gene networks, PC1 was associated with ME14 and ME13 and PC2 was associated with ME2 and ME6 (Table S9, Additional File 2; Figure 3). Therefore, across two different methods, four gene networks were associated with metal mixtures (ME2, ME6, ME13, and ME14) and are thus termed metal mixture associated gene networks. Notably, ME2 contained DNMT1, and one of the topmost enriched molecular and cellular functions among ME2 genes was Gene Expression (p=1.38×10−5). Moreover, for ME6 the topmost enriched canonical pathway was EIF2 signaling (p=2.07×10−6), mirroring the findings from the PCA for individual gene expression. Strikingly, across all metal mixture-associated gene networks, genes within the networks contained significant sub-networks with NF-kB as a central node, similar to the enriched biological network in genes responsive to PC2 (Figure S6, Additional File 1). All of the metal mixture-associated gene networks were significantly negatively correlated with birth weight and fetal growth (Spearman rank correlation p<0.05) (Table S10, Additional File 2). Three of the networks, ME2, ME6, and ME14 were also significantly negatively correlated with placenta weight, and two were significantly negatively correlated with gestational age (Table S10, Additional File 2; Figure S7, Additional File 1). The strongest correlations were observed for birthweight, shown in Figure 4.
Figure 4.
Spearman correlations between four metal mixture-associated gene networks and birth weight. Spearman’s rho and p-value are listed on each plot.
4. Discussion
Exposure to trace metals during pregnancy is associated with pregnancy complications, adverse neonatal outcomes, and later-in-life disease (Rager et al. 2020). The adverse health effects tied to metal exposure are tightly linked to the placenta, where dysregulated expression of genes, particularly those involved in inflammatory processes, represents a potential underlying mechanism (Marsit 2016; Bommarito et al. 2017). Few studies have compared trace metals exposure with placental gene expression directly in this target biospecimen. In the present study, we integrated chemical measurements in umbilical cord and placental transcriptomic data within a subsample of the ELGAN cohort to examine the multidimensional relationship between >37,000 placental transcripts and 11 metals/metalloids. Mixtures modeling approaches identified the altered expression of genes involved in epigenetic processing (DNMT1) as well as inflammation-related pathways including EIF2 and NF-kB signaling. These pathways represent key targets for future intervention.
One particularly striking finding in this study was that DNMT1 was upregulated in relation to PC2, characterized by high levels of Pb, Cd, and Sb. This gene was also a member of one of the metal mixture-associated gene networks, ME2, which was functionally enriched for gene expression processes. Interestingly, in the only other human cohort study that has evaluated metal mixtures and placental genome-wide gene expression, a gene network enriched for gene expression processes was also associated with multi-metal exposure (Deyssenroth et al. 2018). DNMT1 is responsible for maintaining and repairing established DNA methylation, an epigenetic process that is critical for healthy development of the fetus and placenta (Bianco‑Miotto et al. 2016). While individual metals have been associated with altered DNA methylation in the placenta, few studies have assessed metal mixtures (Marsit 2016; Bommarito et al. 2017). A recent study that assessed maternal blood levels of metal mixtures and newborn cord CpG methylation found a suggestive interaction between As and Hg (Weyde et al. 2021). This type of synergistic effect of multi-metal exposure on gene expression is supported by toxicologic in vitro studies assessing metal mixtures in placenta (Adebambo et al. 2015). The upregulation of DNMT1 found in this study provides evidence for the hypothesis that metal-responsive gene expression is partly mediated through epigenetic mechanisms. This activation of DNMT1 may also have implications for DOHaD, as epigenetic alterations are believed to be a major mechanism driving early life influences on later life health (Marsit 2016). In fact, there is a growing literature demonstrating the role of DNA methylation as a mediator that transduces the effects of metals into neonatal and developmental disease (Maccani et al. 2015; Bozack et al. 2018). Therefore, our results and others point to a potential role for DNMT1 as an epigenetic master regulator of gene expression modulation in the placenta in relation to metal mixtures.
The mixtures approach also identified increased expression of IGFBP1, IGF2BP1, and INS-IGF2, genes that are implicated in inflammation regulation, in relation to PC2 (Lee et al. 1997; Leviton et al. 2019). High levels of IGFBP1 or IGF2BP1 in the placenta may reduce levels of insulin like growth factor −1 and −2 (IGF1 and IGF2), important fetal and placental growth factors, leading to reduced growth. Indeed, we found that IGF2BP1 and INS-IGF2 were negatively correlated with birth weight, fetal growth, and placental weight. Furthermore, neonatal IGFBP1 protein concentrations have been tied in the ELGAN cohort to severe fetal growth restriction, medically indicated preterm delivery as well as elevated inflammation in the neonate (Leviton et al. 2019). Thus, IGFBP1, IGF2BP1, and INS-IGF2 may represent critical biomarkers of placental and/or neonatal inflammatory response in response to metal exposure and may act as a mediating mechanism between metal exposure and birthweight/fetal growth.
Two biological pathways were identified among the metal mixture-responsive gene networks, namely the NF-kB signaling pathway and the EIF2 signaling pathway. NF-kB is a master regulator of inflammation that controls over 400 inflammation- and apoptosis-related genes. This pathway plays critical roles during the implantation, maintenance of pregnancy, and labor (Sakowicz 2018). The upregulation of NF-kB may also represent an underlying mechanism driving more global gene expression changes through its signaling cascade. We have previously identified the NF-kB signaling pathway as induced in newborns of As-exposed mothers (Fry et al. 2007). Relevant to this cohort of extremely preterm infants, the maintenance of pregnancy depends on the inhibition of NF-kB and dysregulation of NF-kB during pregnancy may be implicated in pregnancy complications, such as intrauterine growth restriction, preeclampsia and preterm birth (Aban et al. 2004; Lindström and Bennett 2005; Sakowicz 2018). Moreover, NF-kB has been identified as a critical master regulator of the relationship between neonatal inflammation and early-life brain damage that can lead to later-in-life impaired neurocognition among those born extremely preterm, thus providing a potential link to DOHaD outcomes (Leviton et al. 2015).
The results also highlighted genes that are part of the EIF2 signaling pathway in relation to multi-metal exposure. EIF2 signaling pathway is one of the main ways in which cells regulate translation initiation in response to stress (Shrestha et al. 2012). Importantly, EIF2 signaling has been shown to be downregulated in response to toxic metal exposure in vitro, corresponding with our finding of downregulated expression of EIF2A with respect to the multi-metal PC (Shrestha et al. 2012). Importantly, alterations in EIF2 phosphorylation have been linked to placental inflammation, reduced cellular proliferation (required for healthy placentation) and intrauterine growth restriction and preeclampsia (Yung et al. 2008; Gaccioli et al. 2013). Thus, the finding of dysregulation of EIF2 and NF-kB signaling in response to prenatal metal mixtures has implications for both perinatal and developmental outcomes.
While this study is among the first to identify metals-mixture associated transcriptomic changes in the placenta, it is not without limitations. First, the ELGAN cohort is comprised of infants born extremely preterm. While this is a critical population to study given their vulnerability to environmentally-induced disease, the findings may not be generalizable to children born at term. Second, cord tissue is a powerful yet underutilized biospecimen for assessing prenatal chemical exposures making it challenging to make comparisons of the exposure levels recorded in this study to other populations. Nevertheless, from studies that have assessed cord tissue levels of trace metals, the levels in ELGAN are somewhat comparable, although our sample did have lower Pb and higher Zn levels overall compared to the literature (Sakamoto et al. 2013; Ni et al. 2018). Third, the ELGAN study did not record data on maternal diet during pregnancy which could represent a source of exposure to both metals and critical nutrients, particularly for certain food groups, such as seafood. Therefore, lack of nutritional data could represent a source of residual confounding. Lastly, selection bias is a possibility within this study given that a subsample of the overall parent cohort was used herein. Only infants who survived to age 10 have been included in the subsample used in this study, thus selecting for infants with perhaps less extreme pathologies.
5. Conclusions
In the present study, we used novel mixtures-based methodologies and toxicogenomics approaches to identify critical metal mixture-associated genes and biological pathways in the placenta. Future research will investigate the relationships among these metals, placental pathways identified herein and health effects of the children in this prospective cohort, for whom there is now data up to 18 years of age. Of particular relevance to the DOHaD framework, the findings of disrupted genes and biological pathways involved in epigenetic processing (DNMT1) and inflammation (EIF2-signalling, NF-kB signaling and IGFBP1/IGF2BP1) provide plausible underlying mechanisms connecting metals and adverse perinatal and later-in-life outcomes.
Supplementary Material
Highlights.
37,000+ placental genes evaluated in response to 11 cord tissue metal concentrations
Pb was associated with strongest genomic response in single-metals analysis
Pro-inflammatory pathways, EIF2- and NF-kB, enriched in response to metal mixtures
Expression of DNMT1, IGF2BP1, IGFBP1 was associated with multi-metal exposure
Metal mixture-associated gene networks were correlated with birth weight
Funding.
This work was funded in part by (1) Office of the Director, National Institutes of Health (OD) and the National Institute of Environmental Health Sciences (NIEHS), grant number 1U2CES026542-03 to the Wadsworth Center (PJP); (2) Office of the Director, National Institutes of Health (OD) and the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD), grant numbers 4UH3OD023348-03 and 5R01HD092374-04 (TMO, RCF).
Ethics approval and consent to participate.
Each of the 14 participating sites’ Institutional Review Boards approved the study procedures and all mothers provided written informed consent for the ELGAN study.
Abbreviations
- BH
Benjamini-Hochberg
- DEG(s)
differentially expressed gene(s)
- DNMT1
DNA Methyl Transferase 1
- EIF2(A)
Eukaryotic Initiation Factor 2 (A)
- IGFBP1
Insulin Like Growth Factor Binding Protein 1
- NF-kB
Nuclear Factor kappa-light-chain-enhancer of Activated B Cells (“Nuclear Factor kappa B”)
- ME
module eigengene
- MM
metal mixture
- WGCNA
weighted gene expression analysis
- PC(A)
principal component (analysis)
Footnotes
Competing interests. The authors declare that they have no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability of data and materials.
Datasets generated in the study supporting the conclusions of this article are included within the article and its additional files.
Bibliography
- Aban M, Cinel L, Arslan M, Dilek U, Kaplanoglu M, Arpaci R, et al. Expression of nuclear factor-kappa B and placental apoptosis in pregnancies complicated with intrauterine growth restriction and preeclampsia: an immunohistochemical study. Tohoku J Exp Med. 2004. Nov;204(3):195–202. [DOI] [PubMed] [Google Scholar]
- Adebambo OA, Ray PD, Shea D, Fry RC. Toxicological responses of environmental mixtures: Environmental metal mixtures display synergistic induction of metal-responsive and oxidative stress genes in placental cells. Toxicol Appl Pharmacol. 2015. Dec 15;289(3):534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995. Jan;57(1):289–300. [Google Scholar]
- Bianco-Miotto T, Mayne BT, Buckberry S, Breen J, Rodriguez Lopez CM, Roberts CT. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction. 2016. Jul;152(1):R23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard R, Cousings R. Emerging Technologies for Nutrition Research: Potential for Assessing Military Performance Capability. National Academies Press (US; ); 1997. [PubMed] [Google Scholar]
- Bommarito PA, Martin E, Fry RC. Effects of prenatal exposure to endocrine disruptors and toxic metals on the fetal epigenome. Epigenomics. 2017. Mar;9(3):333–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozack AK, Cardenas A, Quamruzzaman Q, Rahman M, Mostofa G, Christiani DC, et al. DNA methylation in cord blood as mediator of the association between prenatal arsenic exposure and gestational age. Epigenetics. 2018. Oct 11;13(9):923–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulka CM, Bommarito PA, Fry RC. Predictors of toxic metal exposures among US women of reproductive age. J Expo Sci Environ Epidemiol. 2019. Sep;29(5):597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Xu X, Hylkema MN, Zeng EY, Sly PD, Suk WA, et al. Early-life Exposure to Widespread Environmental Toxicants and Health Risk: A Focus on the Immune and Respiratory Systems. Ann Glob Health. 2016. Feb;82(1):119–31. [DOI] [PubMed] [Google Scholar]
- Deyssenroth MA, Gennings C, Liu SH, Peng S, Hao K, Lambertini L, et al. Intrauterine multi-metal exposure is associated with reduced fetal growth through modulation of the placental gene network. Environ Int. 2018. Nov;120:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. EPA: Guidance for DataQuality Assessment. Section 4.7. Values below detection limits [Internet]. 2000. [cited 2022 Oct 26]. Available from: https://www.epa.gov/sites/default/files/2015-06/documents/g9-final.pdf [Google Scholar]
- Everson TM, Kappil M, Hao K, Jackson BP, Punshon T, Karagas MR, et al. Maternal exposure to selenium and cadmium, fetal growth, and placental expression of steroidogenic and apoptotic genes. Environ Res. 2017. Oct;158:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson TM, Marsit CJ. Integrating -Omics Approaches into Human Population-Based Studies of Prenatal and Early-Life Exposures. Curr Environ Health Rep. 2018. Sep;5(3):328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan SF, Li Z, Korrick SA, Spiegelman D, Enelow R, Nadeau K, et al. Infant infections and respiratory symptoms in relation to in utero arsenic exposure in a U.S. cohort. Environ Health Perspect. 2016. Jun;124(6):840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KK, Chin HB. Environmental chemicals and preterm birth: Biological mechanisms and the state of the science. Curr Epidemiol Rep. 2017. Mar;4(1):56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KK, O’Neill MS, Meeker JD. Environmental contaminant exposures and preterm birth: a comprehensive review. J Toxicol Environ Health B Crit Rev. 2013;16(2):69–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, et al. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007. Nov;3(11):e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaccioli F, White V, Capobianco E, Powell TL, Jawerbaum A, Jansson T. Maternal overweight induced by a diet with high content of saturated fat activates placental mTOR and eIF2alpha signaling and increases fetal growth in rats. Biol Reprod. 2013. Oct 24;89(4):96. [DOI] [PubMed] [Google Scholar]
- Gardner RM, Kippler M, Tofail F, Bottai M, Hamadani J, Grandér M, et al. Environmental exposure to metals and children’s growth to age 5 years: a prospective cohort study. Am J Epidemiol. 2013. Jun 15;177(12):1356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EA, Goldsmith J, Kioumourtzoglou M-A. Complex mixtures, complex analyses: an emphasis on interpretable results. Curr Environ Health Rep. 2019;6(2):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedicke-Fritz S, Härtel C, Krasteva-Christ G, Kopp MV, Meyer S, Zemlin M. Preterm Birth Affects the Risk of Developing Immune-Mediated Diseases. Front Immunol. 2017. Oct 9;8:1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra GB, Buckley JP. Environmental exposure mixtures: questions and methods to address them. Curr Epidemiol Rep. 2018. Jun;5(2):160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012. Sep;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Transact A Math Phys Eng Sci. 2016. Apr 13;374(2065):20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil A. qgcomp: Quantile G-Computation. R package; 2021. [Google Scholar]
- Keil AP, Buckley JP, O’Brien KM, Ferguson KK, Zhao S, White AJ. A Quantile-Based Computation Approach to Addressing the Effects of Exposure Mixtures. Environ Health Perspect. 2020. Apr 7;128(4):47004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban KCK, Joseph RM, O’Shea TM, Allred EN, Heeren T, Douglass L, et al. Girls and Boys Born before 28 Weeks Gestation: Risks of Cognitive, Behavioral, and Neurologic Outcomes at Age 10 Years. J Pediatr. 2016. Jun;173:69–75.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsco A, Kioumourtzoglou M-A, Just AC, Amarasiriwardena C, Estrada-Gutierrez G, Cantoral A, et al. Prenatal metal concentrations and childhood cardiometabolic risk using bayesian kernel machine regression to assess mixture and interaction effects. Epidemiology. 2019. Mar;30(2):263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008. Dec 29;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P. Tutorials for WGCNA R package [Internet]. 2014. [cited 2021 Oct 19]. Available from: https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/ [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Zhang Y, et al. sva: Surrogate Variable Analysis. R package; 2020. [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012. Mar 15;28(6):882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014. Dec 1;42(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PD, Giudice LC, Conover CA, Powell DR. Insulin-like growth factor binding protein 1: recent findings and new directions. Proc Soc Exp Biol Med. 1997. Dec;216(3):319–57. [DOI] [PubMed] [Google Scholar]
- Leviton A, Allred EN, Fichorova RN, VanderVeen DK, O’Shea TM, Kuban K, et al. Early Postnatal IGF-1 and IGFBP-1 Blood Levels in Extremely Preterm Infants: Relationships with Indicators of Placental Insufficiency and with Systemic Inflammation. Am J Perinatol. 2019. Dec;36(14):1442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton A, Gressens P, Wolkenhauer O, Dammann O. Systems approach to the study of brain damage in the very preterm newborn. Front Syst Neurosci. 2015. Apr 14;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström TM, Bennett PR. The role of nuclear factor kappa B in human labour. Reproduction. 2005. Nov;130(5):569–81. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani JZJ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, et al. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect. 2015. Jul;123(7):723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ. Placental epigenetics in children’s environmental health. Semin Reprod Med. 2016. Jan;34(1):36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Dreij K, Costa PM. The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures. Int J Environ Res Public Health. 2019. Nov 26;16(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EM, Fry RC. Environmental influences on the epigenome: Exposure-associated DNA methylation in human populations. Annu Rev Public Health. 2018. Apr 1;39:309–33. [DOI] [PubMed] [Google Scholar]
- Michelsen-Correa S, Martin CF, Kirk AB. Evaluation of Fetal Exposures to Metals and Metalloids through Meconium Analyses: A Review. Int J Environ Res Public Health. 2021. Feb 18;18(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DM, Myatt L. The human placenta in health and disease. Obstet Gynecol Clin North Am. 2020. Mar;47(1):xv–xviii. [DOI] [PubMed] [Google Scholar]
- Ni W, Yang W, Yu J, Li Z, Jin L, Liu J, et al. Umbilical cord concentrations of selected heavy metals and risk for orofacial clefts. Environ Sci Technol. 2018. Sep 18;52(18):10787–95. [DOI] [PubMed] [Google Scholar]
- O’Shea TM, Allred EN, Dammann O, Hirtz D, Kuban KCK, Paneth N, et al. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum Dev. 2009. Nov;85(11):719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017. Apr;14(4):417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton A, Clark J, Eaves L, Santos HP, Smeester L, Bangma JT, et al. Placental genomic and epigenomic signatures associated with infant birth weight highlight mechanisms involved in collagen and growth factor signaling. Reprod Toxicol. 2020. Jul 25;96:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Auerbach SS, Chappell GA, Martin E, Thompson CM, Fry RC. Benchmark dose modeling estimates of the concentrations of inorganic arsenic that induce changes to the neonatal transcriptome, proteome, and epigenome in a pregnancy cohort. Chem Res Toxicol. 2017. Oct 16;30(10):1911–20. [DOI] [PubMed] [Google Scholar]
- Rager JE, Bangma J, Carberry C, Chao A, Grossman J, Lu K, et al. Review of the environmental prenatal exposome and its relationship to maternal and fetal health. Reprod Toxicol. 2020. Dec;98:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Clark J, Eaves LA, Avula V, Niehoff NM, Kim YH, et al. Mixtures modeling identifies chemical inducers versus repressors of toxicity associated with wildfire smoke. Sci Total Environ. 2021. Jun 25;775:145759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- Rosen EM, Muñoz MI, McElrath T, Cantonwine DE, Ferguson KK. Environmental contaminants and preeclampsia: a systematic literature review. J Toxicol Environ Health B Crit Rev. 2018. Dec 24;21(5):291–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H-W, Lee DH, Won H-R, Kim KH, Seong YJ, Kwon SH. Influence of toxicologically relevant metals on human epigenetic regulation. Toxicol Res. 2015. Mar;31(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Yasutake A, Domingo JL, Chan HM, Kubota M, Murata K. Relationships between trace element concentrations in chorionic tissue of placenta and umbilical cord tissue: potential use as indicators for prenatal exposure. Environ Int. 2013. Oct;60:106–11. [DOI] [PubMed] [Google Scholar]
- Sakowicz A. The role of NFκB in the three stages of pregnancy - implantation, maintenance, and labour: a review article. BJOG. 2018. Oct;125(11):1379–87. [DOI] [PubMed] [Google Scholar]
- Sanders AP, Claus Henn B, Wright RO. Perinatal and childhood exposure to cadmium, manganese, and metal mixtures and effects on cognition and behavior: A review of recent literature. Curr Environ Health Rep. 2015. Sep;2(3):284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HP, Bhattacharya A, Joseph RM, Smeester L, Kuban KCK, Marsit CJ, et al. Evidence for the placenta-brain axis: multi-omic kernel aggregation predicts intellectual and social impairment in children born extremely preterm. Mol Autism. 2020. Dec 11;11(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HP, Bhattacharya A, Martin EM, Addo K, Psioda M, Smeester L, et al. Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics. 2019. Aug;14(8):751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA, Schesser K. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J Biol Chem. 2012. Aug 17;287(34):28738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekhoven DJ, Bühlmann P. MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012. Jan 1;28(1):112–8. [DOI] [PubMed] [Google Scholar]
- Stekhoven DJ. missForest: Nonparametric Missing Value Imputation using Random Forest. R package; 2013. [Google Scholar]
- Tung PW, Kennedy EM, Burt A, Hermetz K, Karagas M, Marsit CJ. Prenatal lead (Pb) exposure is associated with differential placental DNA methylation and hydroxymethylation in a human population. Epigenetics. 2022. Sep 23;1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyde KVF, Olsen A-K, Duale N, Kamstra JH, Skogheim TS, Caspersen IH, et al. Gestational blood levels of toxic metal and essential element mixtures and associations with global DNA methylation in pregnant women and their infants. Sci Total Environ. 2021. Sep 15;787:147621. [DOI] [PubMed] [Google Scholar]
- Yung H, Calabrese S, Hynx D, Hemmings BA, Cetin I, Charnock-Jones DS, et al. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol. 2008. Aug;173(2):451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zota AR, Schaider LA, Ettinger AS, Wright RO, Shine JP, Spengler JD. Metal sources and exposures in the homes of young children living near a mining-impacted Superfund site. J Expo Sci Environ Epidemiol. 2011. Oct;21(5):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and data used in this analysis are publicly available from the UNC Superfund Research Program GitHub page (https://github.com/UNCSRP/Metal-mixtures-modeling-BW-gene-networks-PTB-placentas). mRNA count data are publicly available from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository under GEO series GSE154829.
Datasets generated in the study supporting the conclusions of this article are included within the article and its additional files.