Abstract
Purpose:
This research investigates the association between benzodiazepines (BZD) and cancer patient survival outcomes, the pancreatic cancer tumor microenvironment, and cancer-associated fibroblast (CAF) signaling.
Experimental Design:
Multivariate Cox regression modeling was used to retrospectively measure associations between Roswell Park cancer patient survival outcomes and BZD prescription records. IHC, H&E, Masson's trichrome, RNAscope, and RNA sequencing were used to evaluate the impact of lorazepam (LOR) on the murine PDAC tumor microenvironment. ELISA and qPCR were used to determine the impact of BZDs on IL6 expression or secretion by human-immortalized pancreatic CAFs. PRESTO-Tango assays, reanalysis of PDAC single-cell sequencing/TCGA data sets, and GPR68 CRISPRi knockdown CAFs were used to determine the impact of BZDs on GPR68 signaling.
Results:
LOR is associated with worse progression-free survival (PFS), whereas alprazolam (ALP) is associated with improved PFS, in pancreatic cancer patients receiving chemotherapy. LOR promotes desmoplasia (fibrosis and extracellular matrix protein deposition), inflammatory signaling, and ischemic necrosis. GPR68 is preferentially expressed on human PDAC CAFs, and n-unsubstituted BZDs, such as LOR, significantly increase IL6 expression and secretion in CAFs in a pH and GPR68-dependent manner. Conversely, ALP and other GPR68 n-substituted BZDs decrease IL6 in human CAFs in a pH and GPR68-independent manner. Across many cancer types, LOR is associated with worse survival outcomes relative to ALP and patients not receiving BZDs.
Conclusions:
We demonstrate that LOR stimulates fibrosis and inflammatory signaling, promotes desmoplasia and ischemic necrosis, and is associated with decreased pancreatic cancer patient survival.
Translational Relevance.
Benzodiazepines (BZD) are commonly prescribed to cancer patients to treat anxiety, insomnia, and chemotherapy-induced nausea. We are the first group to identify an association between BZDs and survival outcomes in early- and late-stage disease across multiple cancer types. In pancreatic cancer patients receiving chemotherapy, we found that lorazepam (LOR) correlates with worsened progression-free survival outcomes, although alprazolam (ALP) is associated with improved PFS outcomes. We then provide a potential mechanistic explanation for these clinical observations. Ultimately, this research supports the need to perform prospective clinical trials to determine how different BZDs impact survival across multiple cancer types.
Introduction
Pancreatic cancer is a recalcitrant disease with the poorest five-year survival rate (12%) relative to all cancers assessed by the American Cancer Society from 2012 to 2018 (1). In the United States, pancreatic cancer is projected to be the second leading cause of cancer-related death by 2030, despite accounting for only ∼3% of all estimated new cancer cases (2). Over 90% of patients with pancreatic cancer present with pancreatic adenocarcinoma (PDAC), which is associated with the worst clinical outcomes (3). This disease is often lethal because patients present with nonspecific symptoms such as weight loss, abdominal pain, and fatigue, and are consequently diagnosed at late stages. Complete surgical resection is the only curative therapy. However, at diagnosis, only 20% of patients are surgical candidates (4).
A unique feature further driving this deadly disease is the presence of a dense, desmoplastic (fibrotic) stroma that impedes drug delivery. The PDAC tumor microenvironment (TME), which is composed of cancer-associated fibroblasts (CAF), immune cells, and extracellular matrix (ECM) proteins, can comprise up to 90% of the tumor volume and plays important roles in PDAC development, progression, and therapeutic resistance (5). CAFs are plastic, highly heterogeneous cells, with both tumor-promoting and tumor-restraining roles (6). The two most well-characterized CAF subtypes are myofibroblastic CAFs (myCAF) and inflammatory CAFs (iCAF; ref. 7). myCAFs preferentially express α-SMA and are thought to be tumor restraining. iCAFs secrete high levels of inflammatory cytokines, most notably interleukin-6 (IL6), and are thought to be protumorigenic due to the fact IL6 is associated with worse survival outcomes (8). CAFs influence tumor cell growth, angiogenesis, metastasis, ECM remodeling, and immune cell signaling and function by secreting ECM proteins, growth factors, chemokines, and cytokines (6). Therefore, understanding how CAFs develop, undergo subtype switching, and interact with tumor and immune cells, subsequently modulating therapy response, is fundamental to improving PDAC patient survival.
The role of palliative care medicine in influencing the TME and cancer patient outcomes is also vitally important. Cancer is a devastating diagnosis, associated with emotional distress, anxiety, and depression (9). Harsh surgical, radiologic, and chemotherapeutic interventions can induce numerous side effects, including nausea, anxiety, fatigue, and insomnia (10). To combat these cancer-associated effects, patients are frequently prescribed an array of palliative care drugs such as aspirin, cannabinoids, antihistamines, selective serotonin reuptake inhibitors, opioids, and benzodiazepines (BZD). There is a growing appreciation that many commonly prescribed drugs can either positively or negatively impact cancer risk, tumor progression, and chemotherapeutic efficacy (11). Many of these interactions are being tested experimentally, providing insight into clinical observations, and opening new avenues to improve patient outcomes. This is a highly significant problem due to the vast majority of patients who are taking these medications, and our general lack of knowledge regarding their impact on the cancer phenotype (11).
In this study, we report the novel discovery that lorazepam (LOR, Ativan) and alprazolam (ALP, Xanax), BZDs frequently prescribed to cancer patients to treat anxiety, affect patient survival outcomes across the cancer spectrum. We use a combination of in vivo and in vitro models to mechanistically determine the effects of LOR and ALP on the PDAC TME. Specifically, we find that LOR promotes IL6 secretion from CAFs and drives ischemic necrosis and desmoplasia in mouse models of PDAC. To our knowledge, this is the first study to demonstrate that the commonly prescribed BZD lorazepam modifies the TME and has potential clinical implications when prescribing BZDs to cancer patients.
Materials and Methods
Benzodiazepine prescription frequency
We used Roswell Park Comprehensive Cancer Center's web-based tool, nSight, which allows users to explore and analyze clinical data. We compared BZD prescription records (alprazolam, lorazepam, chlordiazepoxide, clobazam, clonazepam, clorazepate, diazepam, estazolam, flurazepam, midazolam, oxazepam, temazepam, and triazolam) in Roswell Park patients with primary cancers of the prostate, pancreas, ovary, kidney, head and neck, corpus uteri, colon, breast, brain, and those with invasive nevi/melanomas. Pan-cancer analysis assessed all Roswell Park patients. Patients with multiple primary cancers were excluded. The data were acquired on February 3, 2023.
Pancreatic cancer epidemiology study
This study was conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule) and received approval from an institutional review board (Study ID: BDR 125720). Due to the retrospective, blinded nature of this analysis, written informed consent from patients was not required. This study assesses the association between BZD prescription records (diazepam, lorazepam, alprazolam, temazepam, clonazepam, nordiazepam, and oxazepam) and the survival outcomes of Roswell Park pancreatic cancer patients treated with chemotherapy from 2004 to 2020. Patients who did not receive chemotherapy (n = 4) or had clinical stage 0 disease (n = 2) were removed. Patient characteristics were summarized by BZD use (overall and by type; Supplementary Tables S1–S3) using the mean, median, and standard deviation for quantitative variables; and using frequencies and relative frequencies for categorical variables. Comparisons were made using the Mann–Whitney U or Kruskal–Wallis tests for quantitative variables, and Fisher exact or Chi-square tests for categorical variables. The time-to-event outcomes were summarized by group using standard Kaplan–Meier methods, where the 1/3-year rates and medians were estimated with 95% confidence intervals. Associations were evaluated using the log-rank test. Overall survival (OS) is defined as the time from first chemotherapy until death due to any cause or last follow-up. Disease-specific survival is defined as the time from chemotherapy until death due to cancer or last follow-up. Progression-free survival (PFS) is only defined in those who were disease-free (i.e., nonpersistent disease), and is the time from chemotherapy until recurrence, death from disease, or last follow-up. Disease-free survival (DFS) is defined as the time from chemotherapy until persistent disease, recurrence, death from disease, or last follow-up. To account for potential imbalances in patient demographic and clinical characteristics, multivariable Cox regression models were used to evaluate the association between group (i.e., BZD usage) and the survival outcomes while adjusting for age, sex, race, clinical stage, additional treatments, and progressive disease [for OS and disease-specific survival (DSS) only]. Hazard ratios for BZD, with 95% confidence intervals, were obtained from model estimates. All analyses were conducted in SAS v9.4 at a significance level of 0.05. Tx: Surgery refers to any cancer-related surgical procedure including surgical biopsies and partial resections.
Murine experiments
Mice were housed and maintained in the Lab Animal Shared Resource or the Translational Imaging Shared Resource at Roswell Park Comprehensive Cancer Center. All experiments were conducted under IACUC protocol #1381M.
LSL-KrasG12D/+; LSL-trp53r172h/+; pdx-1-cre (KPC) subcutaneous syngeneic allograft long-term study
A subcutaneously passaged KPC002 allograft derived from a female KPC mouse was stored in freezing media (50% RPMI, 40% FBS, and 10% DMSO) in liquid nitrogen. The p3 allograft tissue was passaged once in strain-matched C57BL/6 female mice by dipping the tumor tissue (2–3 mm in size) in Corning Matrigel (cat. #356231) and implanting the tissue bilaterally into the flank of each mouse. The tumor tissue was harvested 2 weeks later. ∼0.55 mm3 tumor pieces were implanted into the left flank of 24 C57BL/6 female mice. When the tumors reached 100 to 200 mm3, the mice were enrolled in the study. Each mouse was treated with 0.5 mg/kg lorazepam or DMSO control [0.25% DMSO in a sodium chloride solution (0.9%), Sigma,cat. #S8776] daily by intraperitoneal (i.p.) injection. A 50 μg/mL lorazepam was prepared fresh by diluting a 20 mg/mL stock of lorazepam (Sigma-Aldrich, cat. #L1764) or alprazolam (Sigma-Aldrich, cat. #A8800) dissolved in DMSO in a sodium chloride solution (0.9%; Sigma, cat. #S8776) and each mouse received 0.01 mL/g. Mice were weighed daily, and tumor growth was measured biweekly using Fisherbrand Traceable Digital Calipers (0–150 mm). When the tumors measured 2,000 mm3 or other endpoint criteria were reached, the mice were sacrificed 2 hours after drug administration.
KPC subcutaneous syngeneic allograft short-term study
A subcutaneously passaged KPC002 allograft derived from a female KPC mouse was stored in freezing media (50% RPMI, 40% FBS, and 10% DMSO) in liquid nitrogen. The p2 allograft tissue was passaged once in strain-matched C57BL/6 female mice by dipping the tumor tissue (2–3 mm in size) in Corning Matrigel (cat. #356231) and implanting the tissue bilaterally into the flank of each mouse. The tumor tissue was harvested 2 weeks later. ∼0.55 mm3 tumor pieces were implanted into the left flank of 20 C57BL/6 female mice. When the tumors reached 100 to 200 mm3, the mice were enrolled in the study. Each mouse was treated daily with 0.5 mg/kg lorazepam or DMSO control (0.2% DMSO in a sodium chloride solution (0.9%; Sigma, cat. #S8776) by i.p. injection. A 50 μg/mL lorazepam was prepared fresh by diluting a 25 mg/mL stock of lorazepam (Sigma-Aldrich, L1764, LOT#035F0115) dissolved in DMSO in a sodium chloride solution (0.9%; Sigma, cat. #S8776) and each mouse received 0.01 mL/g. Mice were weighed daily, and tumor growth was measured daily using Fisherbrand Traceable Digital Calipers (0–150 mm). After 1 or 2 weeks, the mice were sacrificed 2 hours after drug administration.
LC-MS analysis of subcutaneous syngeneic KPC allograft tumors
141.9 to 255.6 mg mouse tumor pieces (2-week timepoint, 2 hours post-dosing) were snap frozen in homogenizing tubes and stored at −80˚C. Prior to analysis, the tumors were homogenized in 25% methanol. Calibrators, quality controls, plasma blanks, and study samples were thawed and vortexed for 5 to 10 seconds. To separate 1.5 mL microcentrifuge tubes, 50 μL of spiking solution was added to 50 μL of blank plasma for calibrators A-I and quality controls. 50 μL of 50% methanol was added to 50 μL plasma blank with internal standard, plasma blank, reagent blank (water), and study samples. 200 μL of WIS was added to each sample (or 100% methanol to plasma blank and reagent blank) using a repeater pipet and vortexed for ∼10 seconds. Samples were allowed to digest for 10 minutes in the refrigerator or on wet ice. Samples were vortexed for ∼10 seconds and centrifuged at 13,500 rpm for 10 minutes at 4°C. 150 μL of each sample was transferred to the autosampler vial and 5.00 μL was injected into the LC-MS/MS (Sciex 5500 QTrap) system.
H&E
Freshly isolated tumors were fixed in 10% neutral buffered formalin solution (Sigma-Aldrich; cat. #HT501128) for 24 hours prior to processing. Tumor processing was performed in the Experimental Tumor Model (ETM) Shared Resource using a HistoCore Arcadia H (Leica) embedder and sliced in 5-μm sections using an RM2235 (Leica) microtome. FFPE unstained slides were rehydrated as follows: xylene: 5 minutes (repeat 3 times), 100% ethanol: 10 minutes, 95% ethanol: 10 minutes (repeat twice), 70% ethanol: 10 minutes, distilled water 5 minutes. The slides were then placed in hematoxylin for 2 minutes, rinsed with cold running tap water for ∼3 min, dipped twice in 1% acid alcohol, rinsed with cold running tap water until tissue turned blue color. Next, the slides were placed in 95% ethanol for 3 minutes, eosin for 30 seconds, dipped in 95% ethanol 4–5 times, and dehydrated as follows: 95% ethanol: 3 minutes, 100% ethanol: 3 minutes, xylene: 3 minutes (repeat twice), xylene: 5 minutes. Slides were dried briefly and cover-slipped using Poly-Mount.
Ischemic necrosis quantification
H&E slides were imaged using the ScanScope XT System, and necrotic area relative to total area was determined in a blinded manner by a PDAC pathologist.
Masson's trichrome
Freshly isolated tumors were fixed in 10% neutral buffered formalin solution (Sigma-Aldrich, cat. #HT501128) for 24 hours prior to processing. Tumor processing was performed in the ETM Shared Resource using a HistoCore Arcadia H (Leica) embedder and sliced in 5-μm sections using an RM2235 (Leica) microtome. Tissue was rehydrated as follows: xylene: 3 minutes (repeat 3 times), 100% ethanol: 3 minutes (repeat three times), 95% ethanol: 3 minutes, 70% ethanol: 3 minutes, deionized water: 5 minutes. The Abcam trichrome stain kit (ab150686) was then used according to the manufacturer's instructions. For step 5.9, the slides were rinsed in distilled water for 2 minutes, and in step 5.12 the slides were rinsed in distilled water for 30 seconds. The slides were dehydrated as follows: 95% ethanol: 3 minutes (repeat twice), 100% ethanol: 3 minutes (repeat twice), and xylene: 5 minutes (repeat three times). The slides were dried briefly and cover-slipped using Poly-Mount.
Immunohistochemistry
Freshly isolated tumors were fixed in 10% neutral buffered formalin solution (Sigma-Aldrich, cat. #HT501128) for 24 hours prior to processing. All IHC processing and staining was performed in the ETM Shared Resource using an AutoStainer Plus (Dako) using the antibodies alpha-smooth muscle actin (Sigma-Aldrich, cat. # A5228, RRID: AB_262054), vimentin (Cell Signaling Technology, cat. # 5741, RRID: AB_10695459), cytokeratin-19 (Abcam, cat. # ab15463, RRID: AB_2281021), and Ki67 (Abcam, cat. # ab15580, RRID: AB_443209), except phospho-STAT3 (Cell Signaling Technology, cat. #9145S).
Phospho-STAT3 IHC was performed manually. The FFPE slides were deparaffinized and hydrated. For antigen retrieval, the slides were placed in slide chambers containing pH 6.0 1× Antigen Unmasking Solution, Citric Acid Based (Vector Laboratories, cat. #H-3300) in the 2100 Retriever (Aptum Biologics Ltd) and the antigen-unmasking cycle was run according to the manufacturer's instructions, and allowed to cool overnight. Slides were washed 3× with 1× PBS, and then endogenous peroxidase activity was blocked for 10 minutes with 3% lab-grade hydrogen peroxide (Fisher Science Education, cat. # S25359). Slides were washed 1× with PBS (0.1% Tween), blocked for 20 minutes with normal goat serum (2.5%), and incubated overnight at 4˚C with anti-phospho-STAT3 (1:250 dilution) using the ImmPRESS HRP Goat Anti-Rabbit IgG Polymer Detection Kit, Peroxidase (Vector Laboratories, cat. #MP-7451-15). The following day the slides were washed with PBS (0.1% Tween) for 5 minutes, incubated with ImmPRESS Polymer reagent for 30 minutes, washed 2× with PBS (0.1% Tween; 5 minutes), and incubated for 1 minute with DAB Substrate Kit, Peroxidase (HRP; Vector Laboratories, cat. #SK-4100) according to the manufacturer's instructions. Phospho-STAT3 was imaged in a blinded manner, and the area fraction was quantified using ImageJ (ImageJ, RRID: SCR_003070) HDab color deconvolution plugin (Color 2/Color 1) × 100 (Threshold 0.162).
Second harmonic generation (SHG) of polarized light detection and analysis
As previously reported (12), imaging of SHG signal from collagen bundles was performed with a Leica SP8 DIVE confocal/multiphoton microscope system (Leica Microsystems, Inc.), using a 25× HC FLUOTAR L 25×/0.95NA W VISIR water-immersion objective. H&E-stained specimens were excited at 850 nm using an IR laser Chameleon Vision II (Coherent Inc.), and blackguard SHG emitted signal was collected using a nondescanned detector configured to record wavelengths between 410 and 440 nm. Under pathologist supervision, two different areas containing tumor and stromal tissue were selected from three different animals of each cohort. Using the automated Leica Application Suite X 3.5.5 software, 2–4 regions of interest (ROI) from each area were set up for SHG signal collection using identical settings and recorded as monochromatic, 16-bit image stacks of 5-μm depth (Z total distance). Image processing and digital analyses were conducted via FIJI (ImageJ 1.52p, RRID: SCR_003070; Fiji, RRID: SCR_002285) software (13). Raw image stack files were tridimensionally reconstituted as two-dimensional maximal projection 16-bit images. For all images, signal-to-noise identical thresholds were set. Resultant SHG-positive-signal pixels were used to calculate integrated densities (e.g., SHG signal/SHG area). SHG-integrated density data were standardized to the mean value obtained from the vehicle cohort. Results represent SHG arbitrary units compared with control tissues. Additionally, CT-FIRE (V2.0 Beta; https://eliceirilab.org/software/ctfire/) software (14) was used for individual collagen fiber (SHG-positive) architecture analyses. Following the pipeline described by the authors in the provided manual document, SHG images were loaded in batches organized by cohorts. Using similar settings for both groups, single collagen fibers were analyzed for length, width, and straightness. A threshold for fibers with a minimum of 10 μm length was set to reduce error from smaller objects detected. Readouts were plotted in graphs, expressed in micron units for length, width parameters, and arbitrary units for fiber straightness.
KPC short-term lorazepam study
Male and female autochthonous LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) mice (n = 2–3/arm) with a C57BL/6 background were enrolled when their tumors reached 100 to 150 mm3, as measured by MRI (Translational Imaging Shared Resource, Roswell Park). All experimental MRI studies used a 4.7T MR scanner (Roswell Park) dedicated to preclinical research. Baseline MRI scans were acquired prior to treatment. Each KPC mouse was treated daily with 0.5 mg/kg lorazepam or DMSO control [0.2% DMSO in a sodium chloride solution (0.9%); Sigma, cat. #S8776; by i.p. injection. A 50 μg/mL lorazepam was prepared fresh by diluting a 25 mg/mL stock of lorazepam (Sigma-Aldrich, L1764) dissolved in DMSO in a sodium chloride solution (0.9%; Sigma, cat. #S8776), and each mouse received 0.01 mL/g. Mice were weighed daily and were monitored for hunching, anemia, labored breathing, and decreased activity. Follow-up MR imaging was performed at 1 and 2 weeks to assess tumor growth. Multislice high-resolution T2-weighted images were acquired for visualization of tumor extent in vivo.
Magnetic resonance imaging
Experimental magnetic resonance imaging (MRI) studies were carried out using a 4.7T horizontal bore MR scanner (GE NMR Instruments) incorporating AVANCE digital electronics, a removable gradient coil insert (G060, Bruker Medical Inc.) and a custom-designed 35-mm radiofrequency transmit–receive coil. Induction and maintenance of anesthesia for imaging were accomplished using Isoflurane inhalation (2%–3% in oxygen; Abbott Laboratories). Anesthetized autochthonous KPC mice were placed on a sled equipped with temperature and positioned within the magnet. To visualize autochthonous KPC tumor growth and quantify tumor burden, multislice T2-weighted (T2W) spin echo images were acquired on the coronal and axial planes using previously described protocols (15, 16). Additionally, T1-weighted images were acquired before and after gadolinium contrast to assess vascular response to lorazepam treatment. T1-relaxation rate measurements (R1) were obtained using a saturation recovery fast spin echo (FSE) sequence as described previously (17, 18). Following image acquisition, the data sets were transferred to a processing workstation and processed using the medical imaging software Analyze (version 10; AnalyzeDirect). R1 maps were calculated on a pixel-by-pixel basis and tumor ΔR1 (ΔR1 = R1Post Gd − R1PreGd) values were calculated from the pre- and post-contrast images as an indirect measure of tumor perfusion. A two-tailed unpaired t test was used to compare differences between control and treatment groups.
RNA sequencing of subcutaneous syngeneic KPC allograft tumors
Heat maps were generated using a regularized-log transformation (DSEQ2-implement) from raw counts. Each individual gene is row normalized to highlight and examine the differentially expressed genes. Pheatmap package (v1.0.12) from R was used to produce all DE-related heatmaps. As previously described (Venkat and Feigin. BioRxiv, 2021), gene set enrichment analysis and Enrichr were used to perform pathway analysis using the MSigDB hallmark, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome Gene sets (refs. 19, 20; Edward, Subramanian). Enrichment of the input genes (LOR/VEH) in Enrichr was computed using the Fisher exact test, and P values were adjusted using the Benjamini–Hochberg correction (FDR < 0.01).
RNAscope multiplex fluorescent detection with immunofluorescence
Tumor processing was performed in the ETM Shared Resource using a HistoCore Arcadia H (Leica) embedder and sliced in 5-μm sections using an RM2235 (Leica) microtome. Chosen slides were warmed at 65°C for 60 minutes, cooled for 10 minutes, deparaffinized with xylene for 2 × 5 minutes, dehydrated in 100% ethanol for 2 × 1 minute, and washed with 0.1% Tween-20 RNAse-free 1× phosphate-buffered saline (PBST) three times. RNAscope Multiplex Fluorescent Detection was performed according to modified instructions provided by the Pasca Di Magliano lab (21). Briefly, slides were incubated with hydrogen peroxide (H2O2) for 10 minutes at room temperature followed by target retrieval at 98°C for 15 minutes. Slides were then blocked with Codetection antibody diluent for 30 minutes and incubated with Primary Antibody solutions overnight at 4°C.
The following day, tissue sections were fixed with formalin, treated with Protease Plus Reagent for 11 minutes at 40°C, and washed with PBST three times. RNAscope probes (if any) were then added for a 2-hour incubation at 40°C. Following two washes with RNAscope wash buffer at each step, the signal for each of the probes was amplified with AMP 1, 2, and 3 reagents, horseradish peroxidase, and tyramide signal amplification kit at 40°C. Slides were then stained with DAPI for 15 minutes at room temperature and incubated with appropriate secondary antibody solution for 45 minutes at room temperature before being mounted with ProLong Diamond Antifade. Probe and antibody information can be found in Supplementary Table S4.
RNAscope imaging analysis
Images were obtained using confocal microscopy and exported as multiple-image LIFs for analysis in HALO-v3.5 software (Indica Labs). For each slide, five representative confocal microscopy images were obtained, totaling 10 images. Images were imported directly into the HALO software for analysis. Images were analyzed with HALO image analysis software (Indica Labs) using the Indica Labs FISH-IF module. After cells were detected based on nuclear recognition (DAPI stain), the fluorescence intensity of the cytoplasmic areas of each cell was measured. A mean intensity threshold above the background was used to determine positivity for each fluorochrome within the cytoplasm, thereby defining cells as either positive or negative for each marker. The positive cell data were then used to define colocalized populations. The percentage of αSMA, Il6, and/or Gpr68-positive cells were calculated by fluorescence-positive cell counts, divided by total DAPI-positive nuclei. The number of cells was quantified by the HALO programming system and recorded. Percent positive cell values were imported into Excel (Microsoft) for graphing and statistical analysis. Statistics: two-tailed unpaired t test.
Re-analysis of single-cell sequencing data
Peng and colleagues (22) data set was processed and analyzed as described in Venkat and Feigin (BioRxiv, 2021). In brief, single-cell RNA-seq FASTQ files of human PDAC tumors (n = 24) and normal human pancreata (n = 11) were downloaded from the Genome Sequence Archive [Accession: CRA001160, Bioproject (NCBI BioProject, RRID: SCR_004801): PRJCA001063]. Files were aligned to the hg19 genome with Cell Ranger 3.1.0 using standard parameters (23). Twenty-one of the human PDAC tumors and all 11 normal human pancreata have proper chemistry and alignment and were used for downstream analyses. Annotated cells with 200 to 6,000 genes/cell (upper limit to exclude possible doublets) were filtered to remove cells with >10% mitochondrial counts and genes occurring in <3 cells, yielding a final count of 10,345 normal pancreas cells and 22,053 PDAC cells. Analyses were carried out in R 4.0.4. Differentially expressed genes between the subclusters were identified using the FindMarkers function in Seurat4 (24).
Steele and colleagues (25) and Kemp and colleagues (26) data sets were processed as previously described. In brief, h5 files were imported into R, and processed with the Seurat package (24, 27). Data were normalized and integrated for batch correction. PCA clustering and UMAP visualization were performed to generate unbiased clusters. Populations were labeled based on established lineage markers (25, 26). Feature plots or Dot plots were generated to visualize specific gene-expression profiles.
GPR68 correlation analysis
cBioPortal was used to assess GPR68 correlation with CAF and epithelial markers in the pancreatic adenocarcinoma (TCGA and Pan-Cancer Atlas) data set (n = 175 patients/samples).
Measuring pH of murine pancreas and pancreatic tumors
The fabrication of H+-sensitive microelectrodes and their use for measuring pH was performed as described in detail by Lee and colleagues (28). In brief, borosilicate glass (no. BF200-156-10, Sutter Instrument) is pulled to a fine tip (∼1 megaohm resistance) using a model P-1000 micropipette puller (Sutter Instrument). To create an electrode that monitors the H+-sensitive electrical potential, VH, the tip of one electrode is filled with H+-selective ionophore cocktail B (Sigma-Aldrich) and backfilled with a solution: 40 mmol/L K2HPO4, 15 mmol/L NaCl, pH 7.0. To monitor the reference electrical potential, Vref, a second microelectrode is filled with 3 M KCl. The true H+-selective signal is the subtracted signal (VH-Vref), acquired using an HiZ-223 dual channel electrometer (Warner Instruments) and digitized using a Digidata 1550 unit. The signal is converted to pH by a three-point calibration at pH 6.0, pH 7.5, and pH 8.0 using custom software (Courtesy of Dale Huffman and Walter Boron at Case Western Reserve University). The electrical potential of the fluid in the measurement chamber (PBS pH 7.50) is maintained at 0 mV using a bath clamp (no. 725I, Warner Instruments). Pancreatic tissue was sectioned into a 5-mm thick slice to allow submersion in the bath and was impaled with the Vref and VH electrodes. Vref did not deviate from 0 mV, demonstrating electrode placement in the extracellular milieu, whereas the measured pH dropped rapidly to a new level that plateaued after 5 minutes.
Cell culture
Human immortalized CAF (C7-TA-PSC) cells were a gift from Dr. Edna Cukierman (Fox Chase Cancer Center). HTLA cells were a gift from Dr. Brian Roth (University of North Carolina). All cell lines were routinely tested for mycoplasma at the end of each experiment. The C7-TA-PSC cells were last tested on June 27, 2022, using the Genome Modulation Services Shared Resource. The HTLA cells were last tested on May 3, 2019, using the Lonza MycoAlert Plus Mycoplasma Detection Kit (VWR; cat. #75860-358). All experiments were performed with cells with a total passage number of 5–18. The cell lines were not authenticated.
Acidic media preparation
All acidic media preparation was based on a protocol by Dr. Tonio Pera (Thomas Jefferson University).
HTLA media
Following the instructions for powdered DMEM (Sigma-Aldrich, cat. #D5030), when the media were fully dissolved 10% FBS, 12.5 mL 1 M HEPES, 2 μg/mL puromycin, 100 μg/mL hygromycin B, 1 mmol/L sodium pyruvate, and 1% P/S were added to the media. The media were aliquoted into separate beakers and were adjusted to the appropriate pH using 10 N HCl/NaOH. pH was measured with a VWR Traceable pH/ORP meter (10539-802). Media were sterile filtered with a 0.22-μm pore size SteriCup (MilliporeSigma SteriCup Quick Release-GV Vacuum Filtration System, 500 mL, Fisher Scientific, cat. #S2GVU05RE).
CAF media
Following the instructions for powdered DMEM (Sigma-Aldrich D5030), when the media were fully dissolved 10% FBS, 12.5 mL 1 M HEPES, 1 mmol/L sodium pyruvate, and 1% P/S were added to the media. The media were aliquoted into separate beakers and were adjusted to the appropriate pH using 10 N HCl/NaOH. pH was measured with a VWR Traceable pH/ORP meter (cat. #10539-802). Media were sterile filtered with a 0.22-μm pore size SteriCup (MilliporeSigma SteriCup Quick Release-GV Vacuum Filtration System, 500 mL, Fisher Scientific, S2GVU05RE).
PRESTO-Tango protocol
HTLA cells were maintained in DMEM supplemented with 10% FBS, 2 μg/mL puromycin, 100 μg/mL hygromycin B, and 1% P/S at 37°C in a 5% CO2 incubator. For acidic pH studies 37°C, a 0% CO2 incubator was used (see Acidic media preparation). For transfection, 400,000 HTLA cells/well were plated in a 6-well dish. The next day, Lipofectamine 3000 (L3000008, Thermo Scientific) was used according to the manufacturer's instructions to transfect 500 ng GPR68-Tango (Addgene, cat. #66371) construct per well. The transfection reagent remained on the cells overnight. Three wells were not transfected to serve as a negative control. On day 3, the cells were replated in a white flat-bottom polystyrene TC-treated Corning 384-well plate (8,000 cells/well). A Bio-Rad TC-20 automated cell counter was used to count the cells. On day 4, the Tecan D300e digital drug dispenser was used to plate the desired drug concentrations using 10 mmol/L drug stocks resuspended in DMSO. DMSO concentration was normalized. On day 5, the luminescence of each well was measured using the Promega Bright-Glo Luciferase Assay System (cat. #E2610) according to the manufacturer's instructions.
Western blot
Protein lysis was performed following the Silva and colleagues (29) rapid extraction method for mammalian cell culture. Proteins were transferred to nitrocellulose membranes (0.2 μm, Bio-Rad, cat. #1620112) at a constant voltage of 100 V for 70 minutes at 4°C using Mini Trans-Blot Cell (Bio-Rad). Membranes were blocked in TBS-T (Tris-buffered saline (TBS) with 0.5% v/v TWEEN-20, Sigma-Aldrich) and 5% w/v nonfat dry milk (Blotting-Grade Blocker, Bio-Rad, cat. #1706404). Primary antibodies were diluted in 5% milk in TBS-T and incubated overnight at 4°C phospho-CREB (Ser133) (87G3) monoclonal anti-rabbit antibody (Cell Signaling Technology, cat. #9198S, 1:1,000 dilution), GAPDH anti-mouse monoclonal antibody (Proteintech, cat. #60004-1-Ig, 1:20,000 dilution). Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,000 Donkey anti-rabbit; Fisher Scientific; cat. #45-000-682, or 1:2,000 Goat anti-mouse (Sigma-Aldrich; cat. # A4416, RRID: AB_258167) for 45 to 90 minutes at room temperature. Pierce ECL Western Blotting Substrate (Thermo Scientific, cat. #32106) was used for chemiluminescent detection. Signals were visualized and imaged using the ChemiDoc XRS+ System and Image Lab Software (Bio-Rad).
qPCR
Cells were washed once with ice-cold PBS and then lysed and homogenized in TRIzol reagent according to the manufacturer's protocol. RNA was isolated and DNase I treated using a Direct-zol RNA miniprep kit (Zymo Research) according to the manufacturer's protocol. RNA concentration and purity were measured using a Thermo Scientific NanoDrop 8000 Spectrophotometer. Any RNA with an A260/280 ratio below 1.9 or an A260/230 ratio below 1.9 were excluded from the analysis. RNA was aliquoted and stored at −80°C. 300–900 ng RNA was converted to 20 μL cDNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. The cDNA was diluted with nuclease-free water (∼15 ng/μL) and the qPCR was performed in 10 μL reactions using iTaq Universal SYBR green Supermix according to the manufacturer's instructions using 0.5 μL primer and 1 μL cDNA per reaction. Thermal cycling was performed using a Bio-Rad CFX Connect Realtime System. All primers were Bio-Rad PrimePCR SYBR Green Assay primers. Gene-expression analysis was performed using the ΔΔCt method.
CRISPRi GPR68 knockdown cell generation
The knockdown cells were generated according to a modified protocol from Francescone and colleagues (30). The following GPR68 CRISPRi gRNA sequences were used (gRNA sequences were selected from the top guide RNA sequences for GPR68 as determined by Horlbeck and colleagues; ref. 31):
1.1 Caccgggagggagagctgggatcg
1.2 Aaaccgatcccagctctccctccc
Generation of lentiviral vectors
Designed guide sequences (Integrated DNA Technologies) were cloned into the lentiviral vector CRISPRi-Puro (gifted from the Cukierman Lab: modified from Addgene Plasmid #71236 to contain a “stuffer” to promote gRNA cloning efficiency). 8 μg CRISPRi-Puro plasmid was linearized and dephosphorylated with 2 μL BSMBI enzyme and 5 μL NE buffer 3.1 diluted in distilled water for a final volume of 50 μL. The mixture was placed into Eppendorf Thermomixer C (55°C, 300 rpm, 3 hours) and then 1 μL of CIP was added and incubated for 1 hour (55°C, 300 rpm). After linearization, the digested plasmid was loaded into an agarose gel (0.6%) and the higher molecular weight band was gel purified using an Invitrogen PureLink Quick Gel Extraction Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The guide RNA oligos were phosphorylated and annealed: 1 μL Oligo 1 (100 μmol/L), 1 μL Oligo 2 (100 μmol/L), 1 μL 10× T4 ligation buffer (NEB), 6.5 μL ddH2O, and 0.5 μL T4 PNK (10 μL total volume). The phosphorylation/annealing mixture was placed into the Bio-Rad T100 Thermocycler: 37°C (30 min), 95°C (5 min), then ramped down to 25°C at 5°C/min, then diluted 1:200 with ddH2O. The annealed and phosphorylated guide sequences were ligated into the linearized and dephosphorylated CRISPRi-Puro plasmid as follows: 25 ng linearized CRISPRi-Puro plasmid, 1 μL 1:200 annealed guides, 1 μL 10× T4 ligase buffer, and 1 μL T4 ligase (10 μL total volume) was incubated at room temperature for 30 minutes. 3 μL of the ligation reaction was transformed into 25 μL of Stbl3 competent cells (NEB) by keeping the mixture on ice for 10 minutes, heat shocking at 42°C for ∼1 minute, placing on ice for 10 minutes, adding 100 μL sterile LB to each tube, and incubating for 30 minutes in the Eppendorf Thermomixer C (37°C, 300 rpm). The entire mixture was plated on LB-AMP plates (100 μg/mL Ampicillin), 2–3 colonies from each plate were miniprepped using the Thermo Scientific GeneJet miniprep kit according to the manufacturer's instructions. The plasmid DNA was sequenced by Eurofins Genomics using the hU6-F primer: GAGGGCCTATTTCCCATGATT. Lentiviruses were produced as follows: Day 1: Transfect 293T cells (∼75% confluent, 10 cm plate, 6 mL fresh complete media) with 2 μg of the CRISPRi-Puro plasmid containing the appropriate guide (and CRISPRi-Puro uncut as a control), 1.5 μg psPAX2 (RRID: Addgene_12260), and 0.5 μg pMD2.G using Lipofectamine 3000 according to the manufacturer's instructions. Day 2: Gently add 4 mL fresh complete media to each plate and incubate for 24 hours. Day 3: Collect virus and replace it with 10 mL fresh media, filter (0.45 μm), aliquot, and store at −80°C. Day 4: Collect virus, filter (0.45 μm), aliquot, and store at −80°C.
Lentiviral reverse transduction (based on addgene protocol)
60,000 C7-TA-PSC cells per mL of media containing 10 μg/mL polybrene were prepared. Lentiviral media were rapidly thawed, diluted, and mixed with 60,000 cells in 1 mL of media. The virus was left on the cells for 48 hours and then replaced with fresh complete media. A no virus control was made for selection purposes. 72 hours after the reverse transduction, puromycin selection was performed (2 μg/mL).
Human IL6 ELISA
For the GPR68 overexpression ELISA, day 1: 1 mL of media containing 28,000 C7-TA-PSC immortalized human CAFs was plated into each well of a 12-well plate. Day 2: Wells were transfected with 125 ng GPR68 cDNA or a no DNA control using Lipofectamine 3000 according to the manufacturer's instructions for a 12-well plate. Day 3: 20 μmol/L benzodiazepine/DMSO control was bulk prepared in pH 6.8 media and 1 mL per well was added (24 hours timepoint), 6 hours timepoint wells received pH 6.8 media, the plate was kept in the 37°C, 0% CO2 incubator. Day 4: 20 μmol/L benzodiazepine/DMSO control were bulk prepared in pH 6.8 media and 1 mL per well was added to the 6 hours timepoint wells, the plate was kept in the 37°C, 0% CO2 incubator. The media were collected from the wells, centrifuged at 1,000 rpm, 4°C, 3 minutes, and the supernatant was transferred to freshly labeled tubes. 100 μL of each sample, as well as 100 μL of each standard (0–1,000 pg/mL, prepared according to the manufacturer's instructions for cell culture supernatants), was plated into the wells of the ELISA test strips and incubated overnight, 4°C, with gentle rocking (Sigma-Aldrich, RAB0306, Human IL6 ELISA Kit). Day 5: Finished ELISA according to the manufacturer's instructions. For the ELISAs without GPR68 expression 45,000 to 50,000 C7-TA-PSC immortalized human CAF cells per well were plated in 12-well plates, 20 μmol/L BZDs were added on day 2, 24 hours later, the conditioned media were collected and centrifuged, as described above. Statistics: One or two-way ANOVA with Bonferroni multiple comparisons or Holm–Šídák multiple comparisons test, respectively.
Pan-cancer epidemiology study
This study was conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule) and received approval from an institutional review board (Study ID: BDR 161522). Due to the retrospective, blinded nature of this analysis, written informed consent from patients was not required. All statistics were performed using SAS version 9.4 (SAS Institute Inc.; Statistical Analysis System, RRID: SCR_008567). All analyses were performed within the disease site (brain, breast, corpus uteri, head and neck, melanoma, kidney, ovary, pancreas, colon, and prostate). Only patients with a diagnostic date starting in the year 2000 were used for this analysis. Within the disease site, patient characteristics were summarized by cohort (LOR, ALP, No Benzo). Frequencies and relative frequencies were provided for categorical variables and compared using Chi-square test. P values were provided. The overall and PFS summaries were summarized by cohort using standard Kaplan–Meier methods. The median survival rate, Kaplan–Meier curves, and log-rank P values were provided. Time to progression was calculated from “recurrence days from Dx” if recurrence occurred. Otherwise, OS time was used. Multivariate Cox regression modeling was performed to measure associations between survival outcomes and cohort. Models were adjusted for sex (where applicable), clinical grade, and clinical stage. Hazard ratios and corresponding 95% confidence intervals were provided for individual LOR and ALP groups, with “No Benzo” as the referent group. Type 3 test was used, and an overall P value measuring the association between survival and cohort was provided.
Statistical analysis
Statistics were performed in GraphPad Prism 9.3.1 (RRID: SCR_002798). Unless otherwise noted, P < 0.05 was considered statistically significant. All statistical methods and P values are described in the figure legends. Asterisks on the graphs denote statistically significant differences: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data availability
The RNA sequencing data reported in this paper have been deposited in the GEO repository under accession number GSE237649.
Results
Lorazepam is associated with poor survival outcomes in pancreatic cancer patients
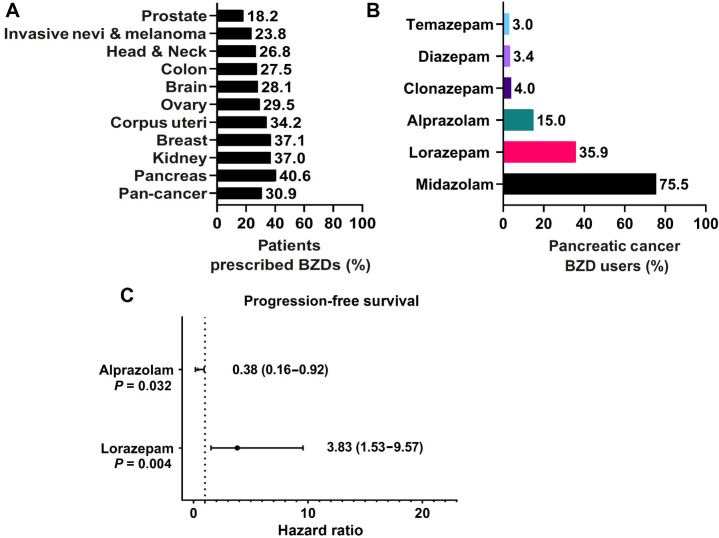
To determine how frequently benzodiazepines (BZD) are prescribed to cancer patients, we broadly examined BZD use in Roswell Park Comprehensive Cancer Center patients. We specifically assessed patients with primary cancers of the prostate, pancreas, ovary, kidney, head and neck, corpus uteri, colon, breast, and brain, and those with invasive nevi/melanoma. Across all cancer types, 30.9% of patients had a record of BZD usage (Fig. 1A). Female patients had an equal or higher record of BZD prescriptions relative to males (34.2% vs. 27.4%) across all cancer types (Supplementary Fig. S1A). Pancreatic cancer patients had the highest record of BZD usage, with 40.6% of patients prescribed at least one BZD (Fig. 1A). Due to the high frequency of BZD use, we assessed the impact of BZDs on pancreatic cancer patient survival outcomes. We first evaluated how BZD prescription records correlated with survival outcomes in Roswell Park pancreatic cancer patients treated with chemotherapy from 2004 to 2020. Pancreatic cancer patients with a BZD prescription record had no significant difference in progression-free survival (PFS; Supplementary Fig. S1B) but were associated with significantly improved DSS relative to those without prescription records of BZDs (Supplementary Fig. S1C). Improved DSS can be partially attributed to imbalances in patient demographic and clinical characteristics; patients prescribed BZDs were significantly more likely to be white, younger, and were less likely to receive radiotherapy or surgery compared with non-BZD users (Supplementary Table S1). Therefore, we performed covariate-adjusted analyses to account for age, sex, race, clinical stage, additional treatments, and progressive disease. With these factors considered, DSS was significantly improved in patients prescribed BZDs (HR: 0.70; 0.60–0.82; Supplementary Fig. S1D).
Figure 1.
Lorazepam is associated with poor survival outcomes in pancreatic cancer patients. A, Percentage of Roswell Park patients with a prescription record of benzodiazepines (BZDs) by cancer type. B, Percentage of pancreatic cancer patients prescribed BZDs who are receiving the top six most commonly prescribed BZDs. C, Covariate-adjusted analysis evaluating the impact of lorazepam (n = 40) or alprazolam (n = 27) prescription records on pancreatic cancer patient PFS, accounting for age, sex, race, clinical stage, additional treatments, and progressive disease relative to no lorazepam (n = 29) or no alprazolam (n = 42). Pan-cancer analysis refers to the combined average of all cancer types in the nSight database. Statistics: To account for potential imbalances in patient demographic and clinical characteristics, multivariable Cox regression models were used to evaluate the association between group (i.e., BZD usage) and the survival outcomes while adjusting for age, sex, race, clinical stage, and additional treatments. Hazard ratios for BZD, with 95% confidence intervals, were obtained from model estimates. All analyses were conducted in SAS v9.4 at a significance level of 0.05.
We then sought to investigate if any specific commonly prescribed BZDs were associated with significant differences in survival. The most commonly prescribed BZD in pancreatic cancer, and all other cancer types with the exception of brain cancer, was midazolam, a short-acting (half-life 2–5 hours) agent often used as a sedative prior to surgery or medical procedures (Supplementary Fig. S1E; ref. 32). The intermediate-acting (half-life 6–24 hours) BZDs lorazepam (LOR) and alprazolam (ALP) were the second and third most commonly prescribed BZDs to pancreatic cancer patients, respectively (Fig. 1B). LOR and ALP are frequently prescribed to pancreatic cancer patients to treat anxiety and anticipatory nausea prior to chemotherapy (10, 33). Due to the frequency of use and the longer-acting effect of LOR and ALP relative to midazolam, we assessed the impact of LOR and ALP on pancreatic cancer patient survival outcomes (Supplementary Tables S2 and S3). We performed covariate-adjusted analyses to account for age, sex, race, clinical stage, and additional treatments (Supplementary Table S5). Strikingly, LOR was associated with significantly worse PFS (HR: 3.83; 1.53–9.57) relative to patients not prescribed LOR (Fig. 1C). In contrast, ALP was associated with significantly improved PFS (HR: 0.38; 0.16–0.92) relative to patients not prescribed ALP (Fig. 1C). Collectively, we find that BZDs are commonly prescribed to pancreatic cancer patients. Importantly, specific BZD choice is associated with positive (ALP) or negative (LOR) survival outcomes.
Lorazepam promotes ischemic necrosis and desmoplasia in murine PDAC tumors
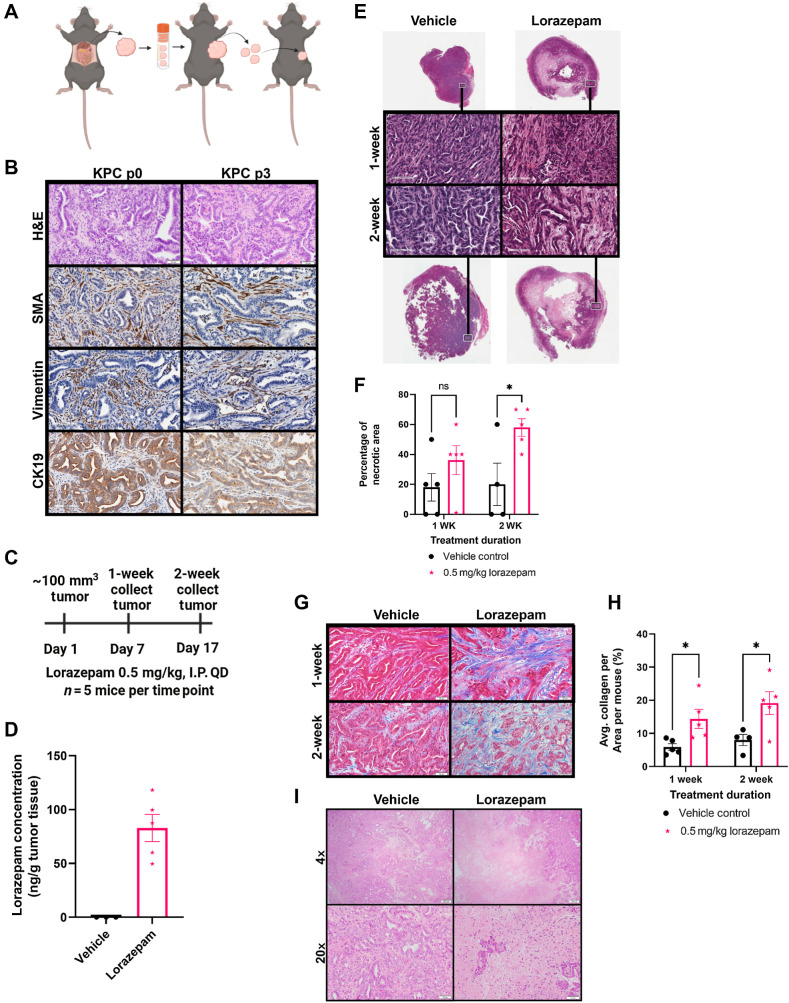
Due to the differential effect of LOR and ALP on pancreatic cancer patient survival, we sought to characterize how these BZDs impact the growth and histology of murine pancreatic ductal adenocarcinoma (PDAC), the most common and deadly form of pancreatic cancer. We subcutaneously implanted LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) tumor pieces into strain-matched, immunocompetent C57BL/6 mice (Fig. 2A). Our model accurately recapitulated the histology of the autochthonous KPC spontaneous tumor as demonstrated by H&E staining (Fig. 2B). The stromal compartment was maintained as indicated by α-SMA and vimentin staining, and the epithelial compartment was well-differentiated as evidenced by CK19 staining (Fig. 2B). To elucidate the effect of LOR and ALP on tumor growth, we treated C57BL/6 mice bearing KPC subcutaneous syngeneic allograft tumors with 0.5 mg/kg LOR or ALP daily until the tumors reached 2,000 mm3 or the mice reached endpoint criteria (Supplementary Fig. S2A). All the mice used in this study were female to match the sex of the syngeneic allograft tumor, and there were no significant differences in the age, weight, and enrollment tumor size of the mice (Supplementary Fig. S2B–S2D). We did not observe significant differences in tumor growth or survival of the mice (Supplementary Fig. S2E–S2G). However, upon histologic examination, we observed the presence of ischemic necrosis in tumors from LOR-treated mice (Supplementary Fig. S2H and S2I). Next, we examined collagen deposition and found a significant increase upon BZD treatment, which was again most striking in the LOR-treated mice (Supplementary Fig. S2J and S2K). This experiment suggested that LOR may remodel the PDAC TME.
Figure 2.
Lorazepam promotes ischemic necrosis and desmoplasia in murine PDAC tumors. A, Schematic of subcutaneous LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) syngeneic allograft model generation. B, Comparison (top to bottom) of H&E (20×), α-SMA IHC (20×), vimentin IHC (20×), and CK19 IHC (20×) in the KPC spontaneous tumor (left) and the p3 KPC syngeneic allograft derived from the KPC spontaneous tumor (right). C, Experimental schematic of short-term LOR (n = 5/arm) or vehicle treatment (n = 4–5/arm). D, Scatter plot with bar (mean with SEM) of LOR concentration per mouse quantified by liquid chromatography–mass spectroscopy (LC-MS) in the 2-week LOR (n = 5) or vehicle (n = 3) treated subcutaneous KPC syngeneic allograft tumors collected 2 hours post-dosing. E, Representative Aperio scanned H&E section of 1-week (top) and 2-week (bottom) vehicle (left) and LOR (right) treated mice, representative zoomed-in 20× images (black and white box) of 1-week (second row) and 2-week (third row) vehicle (left) and LOR (right) treated mice. F, Quantification of the percentage of necrotic area per slide. G, Representative 20× Masson's trichrome images of 1-week (top) and 2-week (bottom) treated mice. H, Quantification of the percentage of collagen per area. ImageJ (ImageJ, RRID: SCR_003070) color deconvolution plugin was used to quantify collagen area/20× field of 5 randomly selected images per mouse in a blinded manner. I, Representative 4× (top) and 20× (bottom) H&E image of KPC spontaneous tumors treated with 0.5 mg/kg vehicle (left) or LOR (right) for 2 weeks (n = 2–3/arm). Statistics: Groups were compared by mixed-effects analysis with Bonferroni's multiple comparison test, black = vehicle, pink = 0.5 mg/kg LOR.
To more definitively assess the impact of LOR on the TME, we performed a short-term treatment study. When the syngeneic subcutaneous allograft tumors reached 100 mm3, we treated the mice daily for 1 week or 2 weeks with 0.5 mg/kg LOR or vehicle (Fig. 2C). As noted in the previous study, all of the mice were female, and there were no significant differences in murine age, weight, and enrollment tumor size (Supplementary Fig. S2L–S2N). To ensure therapeutic relevance, our dosing scheme was based on previous murine studies assessing the anxiolytic impact of LOR (34). We performed pharmacokinetic studies on endpoint tumors and found LOR concentrations of 49.6–118 ng/g, 2 hours post-dosing (Fig. 2D). These concentrations were comparable to those observed in the brains of male CD-1 mice 1 hour post-intraperitoneal injection with 0.1–0.3 mg/kg LOR, supporting that the drug deposited in the tumor tissue at therapeutically relevant quantities (35). We performed H&E staining to identify histologic changes resulting from LOR treatment. Control tumors were differentiated with a well-defined stromal compartment (Fig. 2E). In contrast, LOR-treated tumors were more poorly differentiated, had increased stromal area, and had a significant increase in ischemic necrosis in the center of the tumors (Fig. 2E and F). LOR treatment did not impact endpoint tumor weight or tumor volume, supporting that increasing levels of necrosis was independent of tumor size (Supplementary Fig. S2O and S2P). Tumor size was likely maintained by the presence of rapidly proliferating tumor cells on the leading edge of the LOR-treated tumors, as indicated by Ki67 staining (Supplementary Fig. S2Q). Strikingly, we observed significant increases in collagen deposition at the 1- and 2-week time points (Fig. 2G and H), indicating that LOR treatment increases desmoplasia. We did not observe any significant changes in collagen fiber integrated density, length, width, or straightness by second harmonic generation imaging (Supplementary Fig. S2R–S2U). Therefore, LOR promotes collagen deposition but not collagen remodeling. Next, we sought to extend these findings to the spontaneous KPC model. We treated KPC mice bearing 100 mm3 tumors daily with LOR (0.5 mg/kg) or vehicle for 2 weeks. Consistent with the transplant model, LOR treatment resulted in ischemic necrosis in KPC mice and did not influence the tumor growth kinetics (Fig. 2I; Supplementary Fig. S2V). However, T1-weighted contrast-enhanced MRI revealed a significant reduction in tumor perfusion following LOR treatment (Supplementary Fig. S2W and S2X). Poor perfusion is associated with desmoplasia and impairment of chemotherapeutic drug delivery in PDAC (36). Aggregately, these results support that LOR promotes desmoplasia within the PDAC tumor microenvironment.
Lorazepam promotes inflammatory response and extracellular matrix signature in PDAC tumors
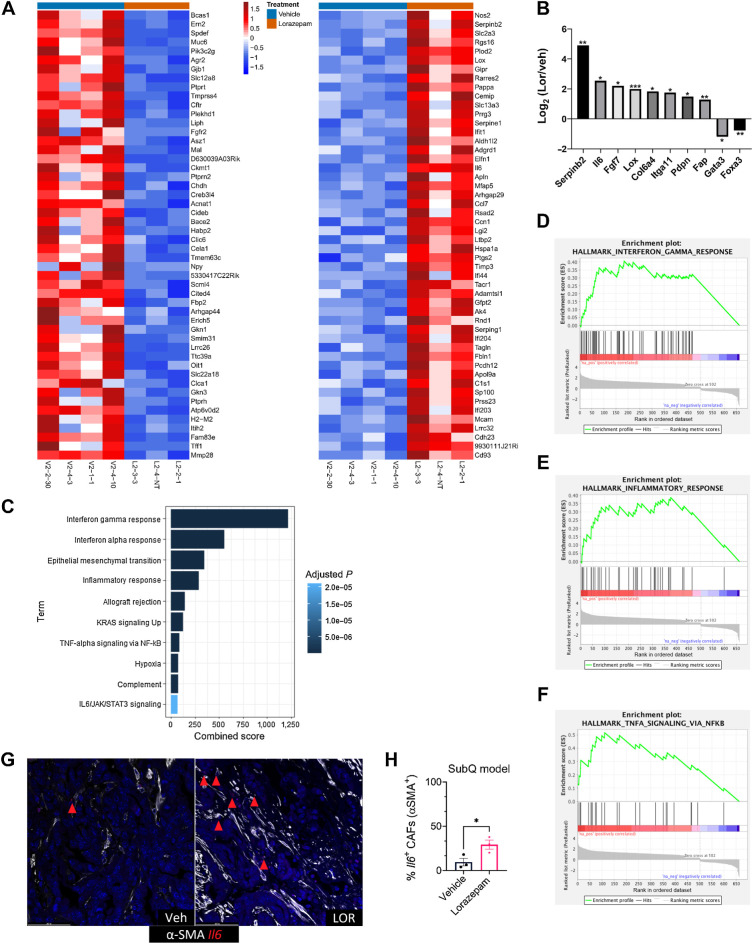
To assess transcriptional changes associated with LOR treatment, we performed RNA sequencing on the 2-week vehicle and LOR-treated subcutaneous syngeneic allograft tumors (Fig. 3A). There were 370 significantly upregulated genes and 617 significantly downregulated genes associated with LOR treatment. Consistent with increased stromal area and desmoplasia, we found a significant upregulation of extracellular matrix (ECM)-related genes, including Serpinb2, Il6, Fgf7, Lox, Col6a4, Iga11, Pdpn, and Fap, in the LOR-treated tumors (Fig. 3A and B). We also observed a significant downregulation of the epithelial-related genes Muc5ac and Gata3 (Fig. 3A and B). We performed pathway analysis to assess the top signaling pathways enriched upon LOR treatment. Among the top 10 upregulated KEGG pathways were interferon gamma response, interferon alpha response, epithelial–mesenchymal transition, TNF-alpha signaling via NF-κB, hypoxia, complement, and IL6/JAK/STAT3 signaling (Fig. 3C–F). These pathways, and IL6, are highly enriched in the proinflammatory iCAF subpopulation (ref. 7; Venkat and Feigin. BioRxiv, 2021). Although IL6 has been reported to be associated with iCAFs, recent work has highlighted the extreme heterogeneity of CAF subtypes in the PDAC TME, and IL6 is broadly expressed across multiple CAF subpopulations in murine PDAC models (6, 7, 21, 37, 38). Therefore, we determined if the LOR-induced IL6 was produced in CAFs. To determine if upregulated IL6 mRNA expression was produced by CAFs, we used RNAscope to perform RNA in situ hybridization (ISH) using Il6 and Acta2 probes. We found that LOR was associated with a significantly higher number of IL6-positive CAFs in both the KPC syngeneic and KPC spontaneous models (Fig. 3G and H; Supplementary Fig. S3A and S3B). LOR treatment also increased IL6 secretion in PDGFRβ+ CAFs in the subcutaneous syngeneic allograft model (Supplementary Fig. S3C). These results indicate that LOR increases inflammatory signaling by CAFs and ECM-related gene expression in murine models of PDAC.
Figure 3.
Lorazepam promotes inflammatory response and extracellular matrix signature in PDAC tumors. A, Heat map of top 50 downregulated (left) and upregulated (right) genes in the 2-week LOR-treated (orange bar) subcutaneously implanted KPC tumors relative to the vehicle-treated (blue bar) tumors. B, Differentially expressed extracellular matrix–related genes and epithelial genes in the 2-week LOR-treated mice relative to the vehicle-treated mice. Statistics: adjusted P-value of log2 fold change of LOR/VEH. C, Enrichr combined scores of the top 10 enriched KEGG terms in the 2-week LOR-treated tumors relative to vehicle. D–F, Enrichment plots of (D) Hallmark_Interferon_Gamma_Response (adjusted P = 2.23E−36) and (E) Hallmark_Inflammatory Response (adjusted P = 1.98E−16), and (F) Hallmark_TNFA_Signaling_via_NFKB (adjusted P = 5.57E−08). G, Representative 40 P = RNAscope images of IL6+/SMA+ cells in the 2-week treated vehicle (left) and LOR-treated subcutaneously implanted KPC tumors (n = 3/arm). H, Quantification of G.
GPR68 is preferentially expressed on human PDAC CAFs
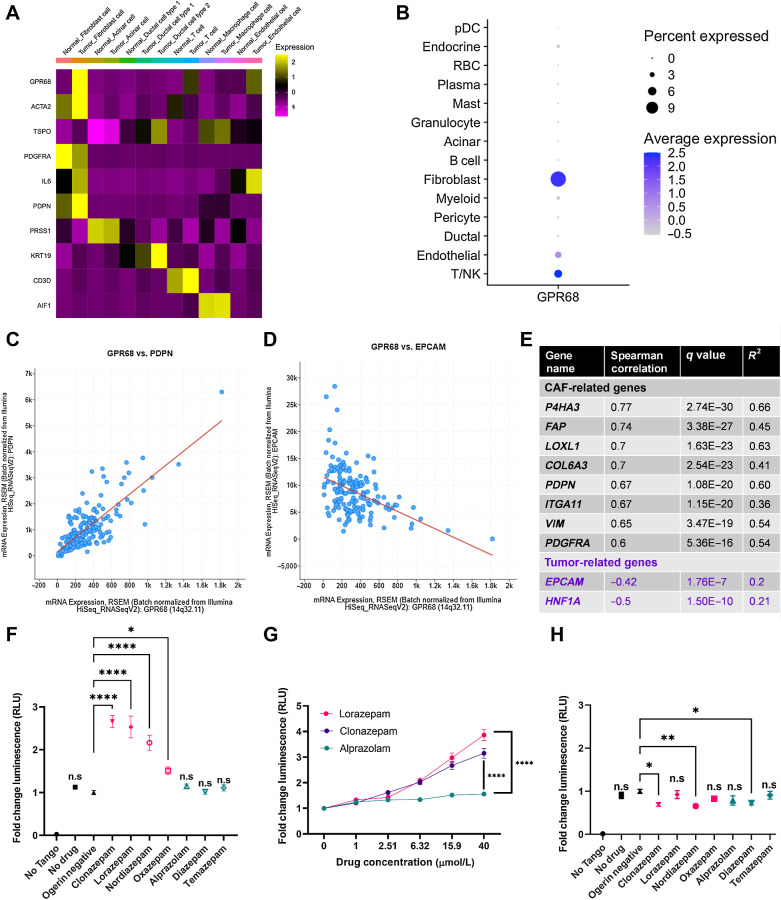
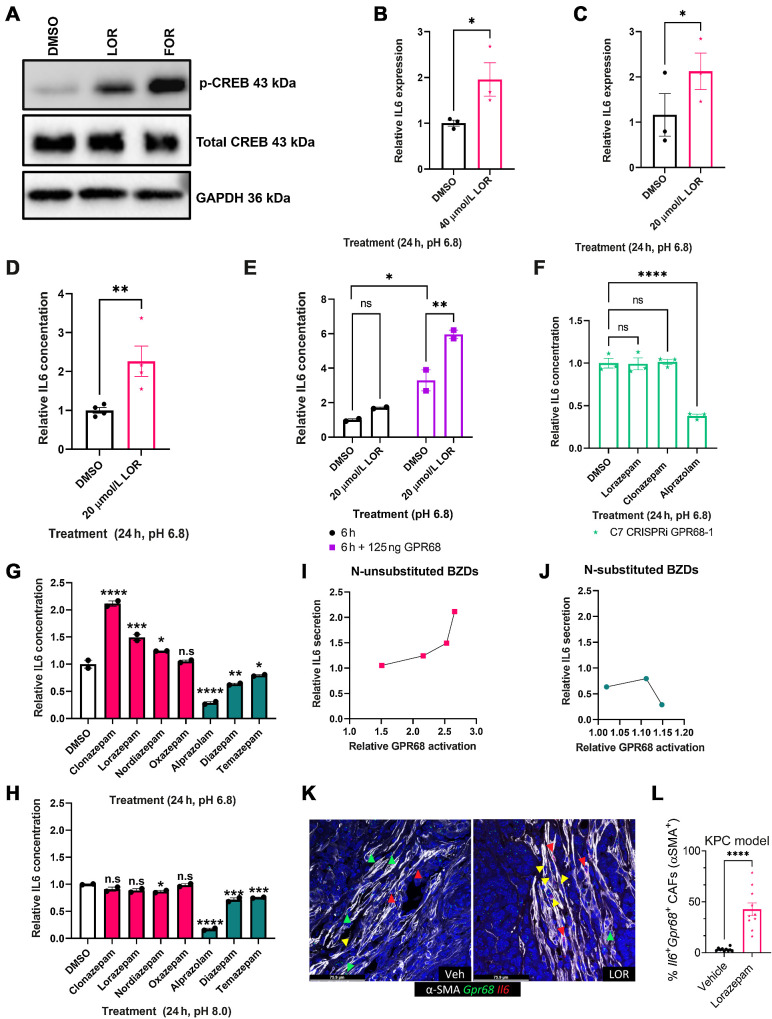
We next sought to determine the molecular mechanism by which LOR regulates IL6 production. First, we assessed the expression of common BZD targets in PDAC tumors, including the pentameric GABA-A receptors, the proton-sensing G-protein coupled receptor (GPCR) GPR68, and the translocator protein (TSPO, also known as the peripheral benzodiazepine receptor). We reprocessed human PDAC single-cell sequencing data from Peng and colleagues (22) and found that PDAC CAFs preferentially express Gpr68 and the GABA-A receptor subunits Gabra1, Gabrb2, Gabrg2, and Gabrr1 (Fig. 4A; Supplementary Fig. S4A).
Figure 4.
N-unsubstituted benzodiazepines potentiate activation of GPR68, a receptor preferentially expressed on human PDAC CAFs. A, Heat map of GPR68 and TSPO expression by cell type from the Peng et al (22) human pancreatic ductal adenocarcinoma tumor single-cell sequencing data set. Yellow represents upregulated gene expression relative to other cell types within a row. B, Dot plot visualization of GPR68 gene expression level (color intensity) and frequency (size of dot) in different cell populations of human PDAC samples from Steele et al (25). C and D, Correlation plot of (C) GPR68 and PDPN, and (D) GPR68 and EPCAM in the human PDAC Pan-Cancer Atlas (TCGA data set). E, Summary table of the Spearman correlation of CAF-related genes with GPR68 in the human PDAC Pan-Cancer Atlas (TCGA data set). F–H, PRESTO-Tango Assay for GPR68 activation (F) pH 6.8 BZD screen; (G) pH 6.8 dose-response curve for LOR, CLZ, and ALP; and (H) pH 7.4 BZD screen. Each plot represents the normalized average of 2–3 biological replicates. Statistics: BZD screens were analyzed by ordinary one-way ANOVA with Dunnett multiple comparison test, and dose–response curves were analyzed by two-way ANOVA with Holm–Šídák multiple comparisons test.
We chose to focus on GPR68, an acid-sensing receptor, for two reasons. First, activation of GPR68 in pancreatic CAFs is known to upregulate IL6 secretion under acidic conditions (39). Second, n-unsubstituted BZDs (Supplementary Fig. S4B), such as LOR and clonazepam (CLZ), are strong positive allosteric modulators of GPR68, meaning they potentiate GPR68 activation only under acidic conditions. Conversely, n-substituted BZDs, including ALP, do not activate GPR68 (Supplementary Fig. S4B; ref. 40). Therefore, we hypothesized that LOR increases inflammatory signaling by promoting GPR68 activation in CAFs. To further support that GPR68 is preferentially expressed in CAFs, we reprocessed human PDAC single-cell sequencing data from Steele and colleagues (25). As observed in the Peng and colleagues data set, GPR68 was most highly expressed in human PDAC CAFs (Fig. 4B). Furthermore, there is a strong, significant positive correlation between GPR68 and CAF-related genes, such as podoplanin (PDPN), and a strong, significant negative correlation between GPR68 and epithelial-related genes, such as epithelial cellular adhesion molecule (EPCAM) in the human PDAC Pan-Cancer Atlas TCGA data set (Fig. 4C–E). To ensure that murine PDAC CAFs also express Gpr68, we reanalyzed single-cell sequencing data from Kemp and colleagues (26). Similar to the human PDAC data set, Gpr68 was preferentially expressed on KPC tumor fibroblasts, T cells, and endothelial cells (Supplementary Fig. S4C). We confirmed that both SMA+ and PDGFRβ+ murine CAFs express Gpr68 by performing RNA ISH (Supplementary Fig. S4D and S4E). In addition to being expressed on CAFs, reanalysis of the CAF cluster in the human PDAC single-cell sequencing by Steele and colleagues (25) indicated that Gpr68 is not highly expressed on pericytes [RGS5 (regulator of G-protein signaling 5) marker], supporting that it is a fibroblast-specific marker (Supplementary Fig. S4F–S4H). To determine the relationship between GPR68 expression and PDAC progression, we reanalyzed GPR68 expression by disease stage in the human PDAC single-cell sequencing by Steele and colleagues (25). GPR68 was not expressed strongly in the normal human pancreas but was expressed in PDAC primary tumors and PDAC metastases, supporting its likely role in disease pathogenesis (Supplementary Fig. S4I–S4K).
N-unsubstituted benzodiazepines potentiate activation of GPR68
To identify which commonly prescribed BZDs were the strongest GPR68 activators, we performed PRESTO-Tango assays at pH 6.8, the optimal pH for GPR68 activation. This luciferase-based assay measures GPCR activity in a G-protein–independent manner. We found that at pH 6.8, the n-unsubstituted BZDs (LOR, CLZ, nordiazepam, and oxazepam) promoted GPR68 activation. In contrast, the n-substituted BZDs (ALP, diazepam, and temazepam) did not promote GPR68 activation (Fig. 4F). GPR68 activation by the n-unsubstituted BZDs LOR and CLZ was dose-dependent at pH 6.8, whereas the n-substituted BZD ALP did not activate GPR68 at any dose (Fig. 4G). When we rescreened the BZDs at pH 7.4 (a pH where GPR68 is not active), there was no significant increase in GPR68 activation by any BZD, supporting that n-unsubstituted BZDs are positive allosteric modulators of GPR68 (Fig. 4H).
Next, we sought to determine if murine PDAC tumors had a pH in the relevant range to support GPR68 activation. We assessed the pH of orthotopically implanted syngeneic KPC tumors (n = 2), adjacent normal pancreas from the orthotopic model (n = 1), bilaterally implanted subcutaneous KPC tumors (n = 4), and the corresponding pancreata of the subcutaneously implanted tumors (n = 2) using an H+ sensitive microelectrode. In the subcutaneous model, the normal pancreata had an average pH of 6.9568 ± 0.1559. The tumors (weighing 0.985 g, 0.331 g, 0.214 g, and 0.078 g) were significantly more acidic, with an average pH of 6.7270 ± 0.2292 (Supplementary Fig. S4L–S4N). Additionally, the subcutaneous tumors were well-differentiated with a clearly defined stromal compartment (Supplementary Fig. S4O). For the orthotopic model, the adjacent normal pancreas had a pH of 6.8833 (Supplementary Fig. S4P). Similar to the subcutaneous tumors, the orthotopic tumors (weighing 1.448 g and 1.713 g) were significantly more acidic than the normal pancreas with a pH of 6.6056 ± 0.2313 and were well-differentiated with a well-defined stromal compartment (Supplementary Fig. S4Q–S4S). Taken together, these results support that GPR68, a receptor preferentially expressed on PDAC CAFs, is activated by n-unsubstituted BZDs under acidic conditions present in the PDAC TME.
Lorazepam promotes IL6 secretion by human PDAC CAFs in a GPR68-dependent manner
Insel and colleagues (41) previously established that GPR68 activation in human CAFs increases IL6 secretion in a cAMP–PKA–pCREB-dependent manner. We hypothesized that n-unsubstituted BZDs, including LOR, would increase IL6 expression in CAFs in a GPR68-dependent and pH-dependent manner. First, we treated immortalized human CAFs with LOR for 3 hours at pH 6.8, and observed a significant increase in phospho-CREB (p-CREB) protein levels by Western blot (Fig. 5A). Next, we assessed the role of LOR in regulating IL6 expression. To determine if LOR modulated Il6 mRNA expression, we treated immortalized human CAFs with LOR at pH 6.8 and performed qPCR. LOR significantly increased Il6 expression at 24 hours (Fig. 5B). Similarly, Il6 mRNA expression was significantly increased upon LOR treatment in human primary pancreatic CAFs (Fig. 5C). Il6 mRNA expression was also significantly upregulated in the LOR-treated KPC syngeneic allograft tumors at the 2-week timepoint (Fig. 3B). Next, we performed an IL6 ELISA which revealed that 24 hours LOR treatment significantly increased IL6 protein secretion in immortalized human CAFs at pH 6.8 (Fig. 5D). Then, we evaluated whether GPR68 overexpression would promote even higher levels of IL6 secretion. GPR68 overexpression in human immortalized CAFs significantly increased IL6 secretion by LOR (Fig. 5E). In fact, 24 hours LOR treatment of human immortalized CAFs with GPR68 overexpression produced such high levels of IL6 that the readings were too high to register (data not shown). To determine if LOR-mediated IL6 secretion by CAFs was GPR68-dependent, we knocked down GPR68 in human immortalized CAFs using CRISPRi (Supplementary Fig. S5A). As expected, GPR68 knockdown potently decreased IL6 levels (Supplementary Fig. S5B). We then treated the control and GPR68 knockdown CAFs with LOR, CLZ, ALP, or DMSO at pH 6.8. GPR68 knockdown prevented LOR and CLZ from increasing IL6 secretion at pH 6.8 (Fig. 5F). To determine if all GPR68 activator BZDs increase IL6 secretion, we treated immortalized human CAFs with a panel of the most commonly prescribed BZDs at pH 6.8 and pH 8.0 for 24 hours, collected the conditioned media, and performed an IL6 ELISA. At pH 6.8, n-unsubstituted BZDs (GPR68 activators) significantly increased IL6 secretion (Fig. 5G). Unexpectedly, n-substituted BZDs (nonactivators) significantly decreased IL6 secretion (Fig. 5G). When we performed the ELISA at pH 8.0, there was no significant increase in IL6 secretion by the n-unsubstituted BZDs. This supports the contention that n-unsubstituted BZDs promote IL6 secretion through GPR68 in CAFs (Fig. 5H). In contrast, at pH 8.0, n-substituted BZDs continued to significantly decrease IL6 secretion, suggesting that this is occurring in a GPR68-independent manner (Fig. 5H). In fact, ALP still potently decreased IL6 in the presence of GPR68 knockdown (Fig. 5F). We compared GPR68 activation by PRESTO-Tango with the ability of each BZD to increase IL6 levels to further establish GPR68 dependence. We found that there was a direct correlation between the degree of GPR68 activation and the increase in IL6 secretion by n-unsubstituted BZDs (Fig. 5I). There was no correlation between decreased IL6 secretion and GPR68 activation by the n-substituted BZDs (Fig. 5J). To determine the relationship between GPR68 and IL6 in vivo, we performed RNA ISH using Il6, Gpr68, and Acta2 probes. In KPC tumors, LOR treatment significantly increased the number of triple-positive (Il6+/Gpr68+/Acta2+) cells, supporting that GPR68 increases IL6 secretion by CAFs in vivo (Fig. 5K and L). To begin elucidating the effects of IL6 secretion in vivo, we performed phospho-STAT3 IHC on the 2-week treated subcutaneous KPC syngeneic allograft tumors. We observed an increase in phospho-STAT3 with LOR treatment, supporting that IL6 secretion increases phospho-STAT3 signaling (Supplementary Fig. S5C and S5D). In summary, these results indicate that BZDs differentially affect IL6 secretion based on the structure of the BZD. N-unsubstituted BZDs promote IL6 secretion under acidic conditions in a GPR68-dependent manner whereas n-substituted BZDs decrease IL6 secretion in a pH and GPR68-independent manner.
Figure 5.
Lorazepam increases IL6 secretion by human PDAC CAFs in a GPR68-dependent manner. A, Western blot of immortalized human PDAC CAFs treated with LOR or forskolin (positive control) at pH 6.8 for 3 hours. B,Il6 qPCR of immortalized human PDAC CAFs treated with 40 μmol/L LOR at pH 6.8 for 24 hours. C,Il6 qPCR of primary human PDAC CAFs treated with 20 μmol/L LOR at pH 6.8 for 24 hours. D, IL6 ELISA of conditioned media from immortalized human PDAC CAFs treated with BZDs (20 μmol/L) or DMSO control for 24 hours at pH 6.8. E, IL6 ELISA of conditioned media from immortalized human PDAC CAFs treated with 20 μmol/L LOR or DMSO control for 6 hours in the presence or absence of GPR68 overexpression. F, IL6 ELISA of GPR68 knockdown immortalized human PDAC CAFs treated with LOR, CLZ, ALP, or DMSO control for 24 hours at pH 6.8. G–H, IL6 ELISA of conditioned media from immortalized human PDAC CAFs treated with BZDs (20 μmol/L) or DMSO control for 24 hours at (G) pH 6.8 or (H) pH 8.0. Pink represents n-unsubstituted BZDs, teal represents n-substituted BZDs. I and J, Correlation plot of relative GPR68 activation of each BZD by PRESTO-Tango relative to IL6 secretion by IL6 ELISA for (I) n-unsubstituted BZDs and (J) n-substituted BZDs at pH 6.8. K, Representative 40× RNAscope images of IL6+/GPR68+/SMA+ cells in the 2-week treated vehicle (left) and LOR-treated KPC tumors. L, Quantification of K. All experiments are representative of 2–4 biological replicates. Statistics: To analyze two groups, paired/unpaired one-tailed t tests were performed. For the analysis of multiple groups, we performed ordinary one-way ANOVA with Bonferroni multiple comparison test. In the case of multiple groups with two independent variables, groups were compared by two-way ANOVA with Holm–Šídák multiple comparisons test.
Lorazepam is associated with worse patient survival across multiple cancer types
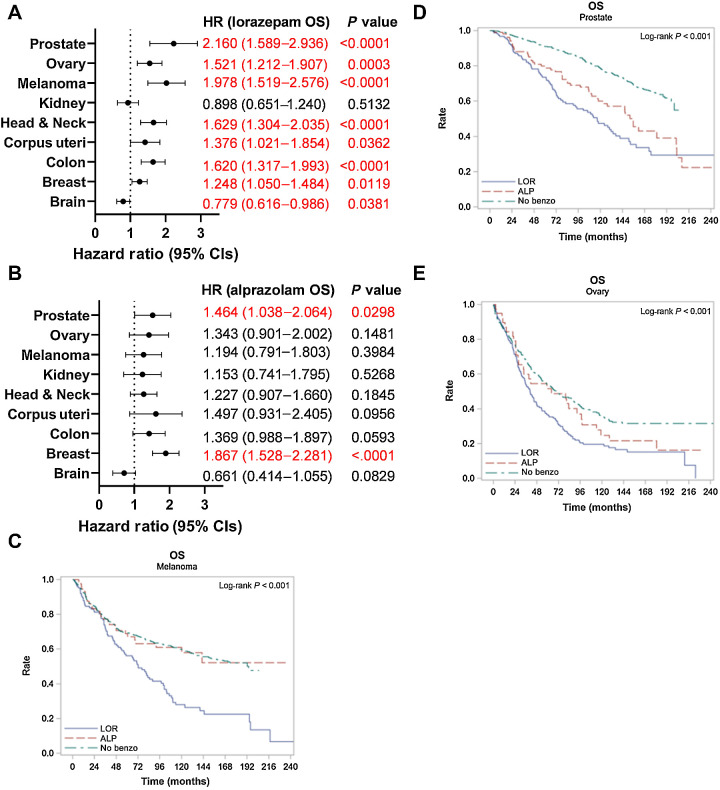
Based on the differential effect of BZDs on IL6 secretion by CAFs (Fig. 5), and the established role of IL6 in promoting worse clinical outcomes (42–44), we compared OS differences in Roswell Park patients (2000–2022) prescribed LOR or ALP relative to patients with no record of BZDs treated for primary cancers of the brain (Supplementary Table S6), breast (Supplementary Table S7), corpus uteri (Supplementary Table S8), head and neck (Supplementary Table S9), skin (Supplementary Table S10), kidney (Supplementary Table S11), ovary (Supplementary Table S12), colon (Supplementary Table S13), and prostate (Supplementary Table S14). LOR and ALP are commonly prescribed to patients with these cancer types (Supplementary Fig. S6A and S6B). We calculated hazard ratios accounting for sex (where applicable), clinical grade, and clinical stage. LOR was associated with significantly worse OS and PFS in prostate cancer [HR OS: 2.160 (1.589, 2.936), HR PFS: 1.899 (1.433, 2.517)], ovarian cancer [HR OS: 1.521 (1.212, 1.907), HR PFS: 1.464 (1.174, 1.826)], invasive nevi/melanoma [HR OS: 1.978 (1.519, 2.576), HR PFS: 2.195 (1.699, 2.835)], head and neck cancer [HR OS: 1.629 (1.304, 2.035), HR PFS: 1.635 (1.313, 2.036)], colon cancer [HR OS: 1.620 (1.317, 1.993), HR PFS: 1.782 (1.457, 2.179)], uterine cancer [HR OS: 1.376 (1.021, 1.854), and breast cancer [HR OS: 1.248 (1.050, 1.484), HR PFS: 1.345 (1.138, 1.591)] relative to patients not prescribed BZDs (Fig. 6A; Supplementary Fig. S6C). In contrast, ALP was infrequently associated with significant differences in survival outcomes, with the exception of hormonal cancers where there was significantly worse OS and PFS in breast cancer [HR OS: 1.867 (1.528, 2.281), HR PFS: 1.850 (1.523, 2.248)], worse OS in prostate cancer [HR OS: 1.464 (1.038, 2.064)], and worse PFS in uterine cancer patients [HR PFS: 1.668 (1.051, 2.646); Fig. 6B; Supplementary Fig. S6D]. Intriguingly, LOR was associated with significantly improved OS in patients with brain cancer (Fig. 6A). LOR and ALP did not correlate with altered survival outcomes in kidney cancer (Fig. 6A and B; Supplementary Table S11). The Kaplan–Meier curves for OS and PFS for melanoma (Fig. 6C; Supplementary Fig. S6E), prostate cancer (Fig. 6D; Supplementary Fig. S6F), and ovarian cancer (Fig. 6E; Supplementary Fig. S6G) clearly demonstrate that LOR correlates with worse survival outcomes relative to patients prescribed ALP or those with no record of BZD use. Overall, we find that LOR is associated with poor survival outcomes across multiple cancer types.
Figure 6.
Lorazepam is associated with worse patient survival across multiple cancer types. A and B, Association between prescription or infusion records of (A) LOR or (B) ALP and OS by cancer type in Roswell Park patients with a diagnostic date from 2000 to 2022; significant values are highlighted in red. C–E, Kaplan–Meier curve comparing OS in Roswell Park patients with prescription or infusion records of LOR or ALP, or those with no history of BZD use treated for primary (C) invasive nevi or melanoma, (D) prostate cancer, or (E) ovarian cancer. Statistics: Multivariate Cox regression modeling was performed to measure associations between survival outcomes and cohort. Models were adjusted for sex (where applicable), clinical grade, and clinical stage. HR and corresponding 95% CIs were provided for individual LOR and ALP groups, with “No Benzo” as the referent group. Type 3 test was used, and an overall P value measuring the association between survival and cohort was provided. CI, confidence interval; HR, hazard ratio; OS, overall survival.
Discussion
We provide evidence that the commonly prescribed anti-anxiety drug LOR promotes desmoplasia in the PDAC tumor microenvironment (Figs. 2 and 3), increases IL6 secretion by CAFs (Fig. 5), and is associated with poor cancer patient survival outcomes (Figs. 1 and 6). Retrospective epidemiologic studies found that LOR was associated with worse PFS, whereas ALP was associated with improved PFS in pancreatic cancer patients (Fig. 1). LOR promotes desmoplasia (Fig. 2), inflammatory signaling (Fig. 3), IL6 expression in CAFs (Figs. 3 and 5) and ischemic necrosis in murine PDAC models (Fig. 2). LOR is likely promoting inflammatory signaling and IL6 secretion by CAFs through activation of GPR68. GPR68 is preferentially expressed on human PDAC CAFs and n-unsubstituted BZDs significantly increase GPR68 activation under acidic conditions (Fig. 4). LOR increases IL6 expression and secretion in human immortalized CAFs in a pH and GPR68-dependent manner (Fig. 5). Conversely, ALP and other GPR68 nonactivator BZDs decrease IL6 in human immortalized CAFs in a pH and GPR68-independent manner (Fig. 5). We propose that LOR stimulates fibrosis and inflammatory signaling, promoting desmoplasia and ischemic necrosis, phenotypes associated with chemoresistance, subsequently decreasing pancreatic cancer patient survival (36, 37, 45). Across many cancer types, LOR is associated with worse survival outcomes, supporting a protumorigenic role (Fig. 6).
In the context of cancer, BZDs are commonly used in palliative care (46). High usage of BZDs is concerning because many epidemiologic studies have found that BZDs increase the risk of cancer (47–52). However, few experimental studies have been performed to mechanistically link BZDs to increased cancer risk. Studies in mice and rats have shown that diazepam and oxazepam can spontaneously induce liver cancer and clobazam can induce thyroid cancer (53–55). These studies support that BZD use may promote cancer development, but no study has definitively addressed the association between BZDs and human cancer progression.
To our knowledge, our research is the first retrospective cohort study to assess the association between BZDs and cancer patient survival, accounting for potential confounding variables, including disease stage (Fig. 6). We are also the first to perform a comprehensive analysis regarding the association between BZDs and pancreatic cancer survival outcomes (Fig. 1). Previously, O'Donnell and colleagues (56) performed a systematic review to determine the relationship between BZDs and cancer patient survival. Their cohort was primarily late-stage cancer patients receiving the short-acting sedative BZD, midazolam. Unsurprisingly, they did not observe significant survival differences.
Experimentally, few studies have quantified the effect of commonly prescribed BZDs on cancer progression and the TME. Oshima and colleagues studied the impact of the short-acting BZD midazolam on LSL-KrasG12D/+; Trp53flox/flox; Pdx-1cre/+ (KPPC) mice (57). They found that midazolam slowed tumor growth/proliferation, decreased inflammatory cytokine production (including IL6), and reduced the number of α-SMA+ cells. Our studies are the first to test physiologically relevant doses of LOR in immunocompetent cancer models with intact stroma (Fig. 2). Fafalios and colleagues (58) found that LOR decreased prostate cancer cell growth in vivo. Their study used very high LOR concentrations (40 mg/kg) and differences in tumor growth were only observed at very large tumor volumes in immunocompromised mice. Previous studies in rats injected intravenously with W-256 carcinosarcoma cells indicate that ALP inhibits lung metastases in a central BZD receptor–dependent manner (59). Additionally, ALP, LOR, and CLZ enhance or suppress immune function in cancer and noncancer settings (60–64). We are the first to comprehensively assess the impact of commonly prescribed BZDs on IL6 signaling by CAFs (Fig. 5).
IL6 plays important roles in pancreatic cancer development and progression (65). Inhibition of IL6 improves the efficacy of PD-L1 immunotherapy in mouse models (66). Conversely, high IL6 levels are associated with lower survival and decreased gemcitabine efficacy in PDAC patients (8). We show that there is a strong association between BZDs and survival outcomes in PDAC patients receiving chemotherapy. Additional epidemiology studies should be performed to determine if BZDs are associated with altered survival outcome in cancer patients receiving immunotherapy drugs.
IL6 is also associated with a subset of pancreatic CAFs known as inflammatory CAFs or iCAFs, characterized by high expression of inflammatory cytokines (37). Due to the protumorigenic nature of IL6, this subtype is presumed to be associated with poor survival outcomes relative to myofibroblastic CAFs (myCAF), which are characterized by high levels of alpha-smooth muscle actin (α-SMA; ref. 7). Interestingly, pathway analysis of our LOR-treated tumors overlapped significantly with iCAF-related signaling pathways (Fig. 3C–F), supporting that LOR may increase the level of iCAFs (ref. 7; Venkat and Feigin. BioRxiv, 2021). It is well established that CAF subtypes are plastic (37). We identify a significant increase in IL6+/SMA+ cell populations in murine PDAC tumors, suggesting that LOR may promote CAF subtype plasticity (Fig. 3G and H, Supplementary Fig. S3A and S3B).
An off-target effect of n-unsubstituted BZDs is positive allosteric modulation of GPR68 (Fig. 4). GPR68 activation increases IL6 and IL8 in various cell types (67–70). Our studies are the first to determine how BZDs influence GPR68 signaling in pancreatic CAFs. To our knowledge, we are also the first to determine the pH of subcutaneous and orthotopically implanted KPC tumors using a microelectrode pH meter. Our findings were consistent with High and colleagues' (71) measurement of the pH of murine pancreatic tumors from cerulein-treated K-rasLSL.G12D/+; Pdx-1-Cre (KC) mice using acidoCEST magnetic resonance imaging (MRI). Acidic pH can alter the TME by modulating enzymatic function, as well as by promoting epithelial-to-mesenchymal transition, metastasis, and T-cell anergy (72–75), common features of pancreatic cancer. To ensure that pancreatic cancer research is physiologically relevant, it is vital that in vitro models accurately mimic the acidic pH conditions observed in vivo.
In addition to impacting inflammation, GPR68 regulates fibrosis and mechanosensing, important factors in promoting pancreatic cancer development and progression (39, 76–78). An unbiased screen revealed GPR68 as a fibroblast-specific drug target in colon cancer (67). Knockdown of GPR68 in bone marrow–derived mesenchymal stem cells (BMSC), which can become CAFs, slowed tumor growth when subcutaneously coinjected with tumor cells into nude mice, further supporting the CAF-specific importance of GPR68 in cancer (79). Additionally, mechanosensing and acid-sensing are vital to fibrosis and cancer cell survival. GPR68 senses and responds to membrane stretch and shear stress, regulating blood vessel dilation/remodeling (78, 80, 81). Wei and colleagues (78) proposed that GPR68 will likely be a potential drug target for solid cancers and fibrotic diseases, thus the role of GPR68 in pancreatic cancer, which is very fibrotic, is highly relevant.
In summary, we have interrogated the role of BZDs on the PDAC TME and patient survival. We made the significant, novel discovery that certain types of BZDs may negatively affect cancer patient survival, whereas others may be beneficial. Due to the frequency that BZDs are prescribed, this is an issue that could affect a large percentage of cancer patients. Performing prospective clinical trials and additional experimental studies to determine whether BZDs affect therapeutic efficacy is vital. Additionally, this research provides a platform to guide others interested in determining how commonly prescribed drugs influence the tumor microenvironment via on-target or off-target mechanisms.
Our studies have a number of limitations that need to be addressed before any clinical recommendations can be made regarding the use of BZDs in cancer patients. Although our BZD dosing strategies in vivo were designed based on previous BZD studies to assess anxiety in mice, the treatment regimen does not completely replicate BZD use in humans due to differences in drug formulation, drug metabolism, and differences in route of administration (82–84). Additionally, BZD dosage differs based on the indication of use (83). Although we focused on the anxiolytic role of BZDs, our epidemiology study did not address the indication of use as a confounding variable. We were also unable to quantify relative BZD usage in our patient cohorts and therefore could not identify if the survival differences were dose-dependent. In our analysis, we accounted for comorbid conditions, which is important because some subsets of BZDs may induce pancreatitis, particularly in the context of acute BZD poisoning in women (85, 86). Information on confounding variables was not available for all patients. However, missing information is assumed to be random (no association with patient characteristics or outcomes), thus any multivariable analyses are not biased by the exclusion of subjects with missing data. Our epidemiology studies only included pancreatic cancer patients receiving chemotherapy, whereas the mice did not receive chemotherapy. The phenotypes (increased collagen, inflammatory signaling, IL6, upregulated EMT/hypoxia pathways, etc.) we observed in our KPC syngeneic subcutaneous allograft model are commonly associated with more aggressive, chemoresistant tumors, supporting that LOR may be promoting worse survival outcomes by impacting chemotherapeutic efficacy (36, 37, 45). Our murine studies also used tumor volume as a survival endpoint, although humans usually die from metastatic disease (87). Although our murine subcutaneous model had a similar histology to human PDAC tumors, the fibroblasts are mostly derived from the skin which may not recapitulate spontaneous tumors (88–90). To establish that LOR-induced IL6 secretion is promoting worse clinical outcomes, future work is needed to determine whether IL6 inhibition/ablation, GPR68 inhibition/ablation, or JAK/STAT inhibitors decrease the phenotypic effect of LOR treatment in vivo.
Supplementary Material
Supplementary Figure S1. BZD prescriptions are associated with improved disease-specific pancreatic cancer patient survival
Supplementary Figure S2. LOR and ALP promote collagen deposition but do not impact tumor growth
Supplementary Figure S3. LOR treatment promotes IL-6 expression in authochthonous KPC CAFs in vivo
Supplementary Figure S4. KPC subcutaneous syngeneic allograft tumors are acidic and express GPR68.
Supplementary Figure S5. LOR treatment increases STAT3 phosphorylation
Supplementary Figure S6. LOR is associated with worse progression-free survival across multiple cancer types.
Supplementary Table S1. Pancreatic cancer patient characteristics by benzodiazepine prescription records.
Supplementary Table S2. Pancreatic cancer patient characteristics by lorazepam prescription records
Supplementary Table S3. Pancreatic cancer patient characteristics by alprazolam prescription records
Supplementary Table S4. RNAscope Antibodies and Probes
Supplementary Table S5. Pancreatic Cancer Time-to-Event Outcomes: Multivariate Summaries.
Supplementary Table S6. Brain cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S7. Breast cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S8. Corpus uterine cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S9. Head and neck cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S10. Invasive nevi and melanoma patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S11. Renal cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S12. Ovarian cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S13. Colon cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S14. Prostate cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Acknowledgments
We thank Dr. Ralph Francescone for providing advice and reagents for generating the CRISPRi knockdown cell lines, Dr. Agnieszka Witkiewicz for providing human primary stellate cells, Michael Habitzruther and Steve Turowski for technical assistance performing KPC murine imaging, Dr. Tonio Pera for protocols to prepare acidic pH media, and Richard A. Pasternack for technical support with pH measurements. Research reported in this publication was supported by the NCI of the NIH under Award Number F31CA260942 and seed funding from the Roswell Park Alliance Foundation. NCI grant P30CA016056 supported the use of Roswell Park Comprehensive Cancer Center's Pathology Network; Biomedical Research Informatics; Biostatistics and Statistical Genomics; Genome Modulation Services; Translational Imaging; Experimental Tumor Models; Laboratory Animal; Drug Discovery Core; and Bioanalytics, Metabolomics, and Pharmacokinetics Shared Resources. Additional support from NIH (R01EY028580, to M.D. Parker) and the 5th AHEPA Cancer Research Foundation Inc, as well as R01CA269660, S10ODO23666 and P30CA06927 (Core Microscopy Facility) to J. Franco-Barraza and E. Cukierman. C. Fountzilas receives funding from the National Comprehensive Cancer Network Foundation, National Comprehensive Cancer Network Oncology Research Program, Taiho Oncology, and Pfizer Inc (all to Institute).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
A.C. Cornwell reports grants from the National Cancer Institute during the conduct of the study. C. Fountzilas reports grants and nonfinancial support from Taiho Oncology and Pfizer outside the submitted work. M.E. Feigin reports grants from the NIH during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
A.C. Cornwell: Conceptualization, investigation, writing–original draft, writing–review and editing. A.A. Tisdale: Investigation. S. Venkat: Formal analysis. K.E. Maraszek: Investigation. A.A. Alahmari: Investigation. A. George: Formal analysis. K. Attwood: Formal analysis. M. George: Investigation. D. Rempinski: Investigation. J. Franco-Barraza: Investigation. M. Seshadri: Formal analysis. M.D. Parker: Formal analysis, investigation, methodology. E. Cortes Gomez: Formal analysis. C. Fountzilas: Formal analysis. E. Cukierman: Formal analysis. N.G. Steele: Formal analysis, supervision, investigation. M.E. Feigin: Resources, formal analysis, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. SEER*Explorer: an interactive website for SEER cancer statistics, National Cancer Institute [Internet]. 2022. Available from:https://seer.cancer.gov/statistics-network/explorer/application.html.
- 2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 3. Sarantis P, Koustas E, Papadimitropoulou A, Papavassiliou AG, Karamouzis MV. Pancreatic ductal adenocarcinoma: treatment hurdles, tumor microenvironment and immunotherapy. World J Gastrointest Oncol 2020;12:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu YA, Oba A, Lin R, Watanabe S, Meguid C, Schulick RD, et al. Selecting surgical candidates with locally advanced pancreatic cancer: a review for modern pancreatology. J Gastrointest Oncol 2021;12:2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res 2008;68:918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitsunaga S, Ikeda M, Shimizu S, Ohno I, Furuse J, Inagaki M, et al. Serum levels of IL-6 and IL-1β can predict the efficacy of gemcitabine in patients with advanced pancreatic cancer. Br J Cancer 2013;108:2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller K, Massie MJ. Depression and anxiety. Cancer J 2006;12:388–97. [DOI] [PubMed] [Google Scholar]
- 10. Bektay MY, İzzettin FV. Oncology pharmacy practice: the clinical pharmacist's perspective 2021.
- 11. Cornwell AC, Feigin ME. Unintended effects of GPCR-targeted drugs on the cancer phenotype. Trends Pharmacol Sci 2020;41:1006–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruggeri JM, Franco-Barraza J, Sohail A, Zhang Y, Long D, di Magliano MP, et al. Discoidin domain receptor 1 (DDR1) is necessary for tissue homeostasis in pancreatic injury and pathogenesis of pancreatic ductal adenocarcinoma. Am J Pathol 2020;190:1735–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bredfeldt JS, Liu Y, Pehlke CA, Conklin MW, Szulczewski JM, Inman DR, et al. Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J Biomed Opt 2014;19:016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma R, Buitrago S, Pitoniak R, Gibbs JF, Curtin L, Seshadri M, et al. Influence of the implantation site on the sensitivity of patient pancreatic tumor xenografts to Apo2L/TRAIL therapy. Pancreas 2014;43:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito F, Camoriano M, Seshadri M, Evans SS, Kane JM, Skitzki JJ. Water: a simple solution for tumor spillage. Ann Surg Oncol 2011;18:2357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalmuk J, Folaron M, Buchinger J, Pili R, Seshadri M. Multimodal imaging guided preclinical trials of vascular targeting in prostate cancer. Oncotarget 2015;6:24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seshadri M, Spernyak JA, Mazurchuk R, Camacho SH, Oseroff AR, Cheney RT, et al. Tumor vascular response to photodynamic therapy and the antivascular agent 5, 6-dimethylxanthenone-4-acetic acid: implications for combination therapy. Clin Cancer Res 2005;11:4241–50. [DOI] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf 2013;14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steele NG, Biffi G, Kemp SB, Zhang Y, Drouillard D, Syu L, et al. Inhibition of hedgehog signaling alters fibroblast composition in pancreatic CancerHedgehog signaling in pancreatic cancer. Clin Cancer Res 2021;27:2023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R. et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A. et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L. et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nature Cancer 2020;1:1097–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kemp SB, Steele NG, Carpenter ES, Donahue KL, Bushnell GG, Morris AH. et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci Alliance 2021;4:e202000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM. et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S-K, Boron WF, Parker MD. Monitoring ion activities in and around cells using ion-selective liquid-membrane microelectrodes. Sensors 2013;13:984–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silva RC, Castilho BA, Sattlegger E. A Rapid Extraction Method for mammalian cell cultures, suitable for quantitative immunoblotting analysis of proteins, including phosphorylated GCN2 and eIF2a. MethodsX 2018;5:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Francescone R, Vendramini-Costa DB, Franco-Barraza J, Wagner J, Muir A, Lau AN. et al. Netrin G1 promotes pancreatic tumorigenesis through cancer-associated fibroblast–driven nutritional support and immunosuppression. Cancer Discov 2021;11:446–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 2016;5:e19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tradounsky G. Seizures in palliative care. Can Fam Physician 2013;59:951–5. [PMC free article] [PubMed] [Google Scholar]
- 33. Howard P, Twycross R, Shuster J, Mihalyo M, WA. Benzodiazepines. J Pain Symptom Manage 2014;47:955–64. [DOI] [PubMed] [Google Scholar]
- 34. Tang X, Yang L, Fishback NF, Sanford LD. Differential effects of lorazepam on sleep and activity in C57BL/6J and BALB/cJ strain mice. J Sleep Res 2009;18:365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller LG, Greenblatt DJ, Paul SM, Shader RI. Benzodiazepine receptor occupancy in vivo: correlation with brain concentrations and pharmacodynamic actions. J Pharmacol Exp Ther 1987;240:516–22. [PubMed] [Google Scholar]
- 36. Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinomapathway antagonism shapes CAF heterogeneity in PDAC. Cancer Discov 2019;9:282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019;9:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wiley SZ, Sriram K, Liang W, Chang SE, French R, McCann T, et al. GPR68, a proton-sensing GPCR, mediates interaction of cancer-associated fibroblasts and cancer cells. FASEB J 2018;32:1170–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang X-P, Karpiak J, Kroeze WK, Zhu H, Chen X, Moy SS, et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 2015;527:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Insel PA, Sriram K, Wiley SZ, Wilderman A, Katakia T, McCann T, et al. GPCRomics: GPCR expression in cancer cells and tumors identifies new, potential biomarkers and therapeutic targets. Front Pharmacol 2018;9:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Michalaki V, Syrigos K, Charles P, Waxman J. Serum levels of IL-6 and TNF-α correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer 2004;90:2312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lane D, Matte I, Rancourt C, Piché A. Prognostic significance of IL-6 and IL-8 ascites levels in ovarian cancer patients. BMC Cancer 2011;11:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duffy SA, Taylor JM, Terrell JE, Islam M, Li Y, Fowler KE, et al. Interleukin-6 predicts recurrence and survival among head and neck cancer patients. Cancer 2008;113:750–7. [DOI] [PubMed] [Google Scholar]
- 45. Dauer P, Nomura A, Saluja A, Banerjee S. Microenvironment in determining chemo-resistance in pancreatic cancer: neighborhood matters. Pancreatology 2017;17:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stark DPH, House A. Anxiety in cancer patients. Br J Cancer 2000;83:1261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kripke DF, Langer RD, Kline LE. Hypnotics' association with mortality or cancer: a matched cohort study. BMJ open 2012;2:e000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kao C-H, Sun L-M, Su K-P, Chang S-N, Sung F-C, Muo C-H, et al. Benzodiazepine use possibly increases cancer risk: a population-based retrospective cohort study in Taiwan. J Clin Psychiatry 2012;73:e555–e60. [DOI] [PubMed] [Google Scholar]
- 49. Iqbal U, Nguyen P-A, Syed-Abdul S, Yang H-C, Huang C-W, Jian W-S, et al. Is long-term use of benzodiazepine a risk for cancer? Medicine (Baltimore) 2015;94:e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harlow BL, Cramer DW. Self-reported use of antidepressants or benzodiazepine tranquilizers and risk of epithelial ovarian cancer: evidence from two combined case-control studies (Massachusetts, United States). Cancer Causes Control 1995;6:130–4. [DOI] [PubMed] [Google Scholar]
- 51. Pottegård A, Friis S, Andersen M, Hallas J. Use of benzodiazepines or benzodiazepine related drugs and the risk of cancer: a population-based case-control study. Br J Clin Pharmacol 2013;75:1356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim HB, Myung SK, Park YC, Park B. Use of benzodiazepine and risk of cancer: a meta-analysis of observational studies. Int J Cancer 2017;140:513–25. [DOI] [PubMed] [Google Scholar]
- 53. Diwan BA, Rice JM, Ward JM. Tumor-promoting activity of benzodiazepine tranquilizers, diazepam and oxazepam, in mouse liver. Carcinogenesis 1986;7:789–94. [DOI] [PubMed] [Google Scholar]
- 54. Miyawaki I, Moriyasu M, Funabashi H, Yasuba M, Matsuoka N. Mechanism of clobazam-induced thyroidal oncogenesis in male rats. Toxicol Lett 2003;145:291–301. [DOI] [PubMed] [Google Scholar]
- 55. Fox K, Lahcen R. Liver-cell adenomas and peliosis hepatis in mice associated with oxazepam. Res Commun Chem Pathol Pharmacol 1974;8:481–8. [PubMed] [Google Scholar]
- 56. O'Donnell SB, Nicholson MK, Boland JW. The association between benzodiazepines and survival in patients with cancer: a systematic review. J Pain Symptom Manage 2019;57:999–1008. [DOI] [PubMed] [Google Scholar]
- 57. Oshima Y, Sano M, Kajiwara I, Ichimaru Y, Itaya T, Kuramochi T, et al. Midazolam exhibits antitumour and anti-inflammatory effects in a mouse model of pancreatic ductal adenocarcinoma. Br J Anaesth 2022;128:679–90. [DOI] [PubMed] [Google Scholar]
- 58. Fafalios A, Akhavan A, Parwani AV, Bies RR, McHugh KJ, Pflug BR. Translocator protein blockade reduces prostate tumor growth. Clin Cancer Res 2009;15:6177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freiregarabal M, Nuneziglesias M, Balboa J, Fernandezrial J, Garciavallejo L, Gonzalezpatino E, et al. Inhibitory effects of alprazolam on the enhancement of lung metastases induced by operative stress in rats. Int J Oncol 1993;3:513–7. [DOI] [PubMed] [Google Scholar]
- 60. Fride E, Skolnick P, Arora PK. Immunoenhancing effects of alprazolam in mice. Life Sci 1990;47:2409–20. [DOI] [PubMed] [Google Scholar]
- 61. Freire-Garabal M, Núñez MJ, Balboa J, Fernández-Rial J, Vallejo LG, González-Bahillo J, et al. Effects of alprazolam on cellular immune response to surgical stress in mice. Cancer Lett 1993;73:155–60. [DOI] [PubMed] [Google Scholar]
- 62. Elmesallamy G, Abass MA, Atta A, Refat NA. Differential effects of alprazolam and clonazepam on the immune system and blood vessels of non-stressed and stressed adult male albino rats. Mansoura J Forensic Med Clin Toxicol 2011;19:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Covelli V, Maffione AB, Greco B, Cannuscio B, Calvello R, Jirillo E. In vivo effects of alprazolam and lorazepam on the immune response in patients with migraine without aura. Immunopharmacol Immunotoxicol 1993;15:415–28. [DOI] [PubMed] [Google Scholar]
- 64. Ramirez K, Niraula A, Sheridan JF. GABAergic modulation with classical benzodiazepines prevent stress-induced neuro-immune dysregulation and behavioral alterations. Brain Behav Immun 2016;51:154–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Y, Yan W, Collins MA, Bednar F, Rakshit S, Zetter BR, et al. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res 2013;73:6359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mace TA, Shakya R, Pitarresi JR, Swanson B, McQuinn CW, Loftus S, et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut 2018;67:320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Horman SR, To J, Lamb J, Zoll JH, Leonetti N, Tu B. et al. Functional profiling of microtumors to identify cancer associated fibroblast-derived drug targets. Oncotarget 2017;8:99913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chandra V, Karamitri A, Richards P, Cormier F, Ramond C, Jockers Ret al. Extracellular acidification stimulates GPR68 mediated IL-8 production in human pancreatic β cells. Sci Rep 2016;6:25765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ichimonji I, Tomura H, Mogi C, Sato K, Aoki H, Hisada T, et al. Extracellular acidification stimulates IL-6 production and Ca2+ mobilization through proton-sensing OGR1 receptors in human airway smooth muscle cells. Am J Physiol Lung Cell Molec Physiol 2010;299:L567–L77. [DOI] [PubMed] [Google Scholar]
- 70. Horiguchi K, Higuchi M, Yoshida S, Nakakura T, Tateno K, Hasegawa R, et al. Proton receptor GPR68 expression in dendritic-cell-like S100β-positive cells of rat anterior pituitary gland: GPR68 induces interleukin-6 gene expression in extracellular acidification. Cell Tissue Res 2014;358:515–25. [DOI] [PubMed] [Google Scholar]
- 71. High RA, Randtke EA, Jones KM, Lindeman LR, Ma JC, Zhang S, et al. Extracellular acidosis differentiates pancreatitis and pancreatic cancer in mouse models using acidoCEST MRI. Neoplasia 2019;21:1085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu S, Zhou H-Y, Deng S-C, Deng S-J, He C, Li X, et al. ASIC1 and ASIC3 contribute to acidity-induced EMT of pancreatic cancer through activating Ca2+/RhoA pathway. Cell Death Dis 2017;8:e2806–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lardner A. The effects of extracellular pH on immune function. J Leukocyte Biol 2001;69:522–30. [PubMed] [Google Scholar]
- 74. Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, et al. editors. Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol; 2017;43:74–89. [DOI] [PubMed] [Google Scholar]
- 75. Swietach P, Vaughan-Jones RD, Harris AL, Hulikova A. The chemistry, physiology and pathology of pH in cancer. Philos Trans R Soc Lond B Biol Sci 2014;369:20130099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hutter S, van Haaften WT, Hünerwadel A, Baebler K, Herfarth N, Raselli T, et al. Intestinal activation of pH-sensing receptor OGR1 [GPR68] contributes to fibrogenesis. J Crohn's Colitis 2018;12:1348–58. [DOI] [PubMed] [Google Scholar]
- 77. Matsuzaki S, Ishizuka T, Yamada H, Kamide Y, Hisada T, Ichimonji I, et al. Extracellular acidification induces connective tissue growth factor production through proton-sensing receptor OGR1 in human airway smooth muscle cells. Biochem Biophys Res Commun 2011;413:499–503. [DOI] [PubMed] [Google Scholar]
- 78. Wei W-C, Bianchi F, Wang Y-K, Tang M-J, Ye H, Glitsch MD. Coincidence detection of membrane stretch and extracellular pH by the proton-sensing receptor OGR1 (GPR68). Curr Biol 2018;28:3815–23. e4. [DOI] [PubMed] [Google Scholar]
- 79. Zhu H, Guo S, Zhang Y, Yin J, Yin W, Tao S, et al. Proton-sensing GPCR-YAP signalling promotes cancer-associated fibroblast activation of mesenchymal stem cells. Int J Bio Sci 2016;12:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martin AL, Steurer MA, Aronstam RS. Constitutive activity among orphan class-A G protein coupled receptors. PLoS One 2015;10:e0138463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu J, Mathur J, Vessières E, Hammack S, Nonomura K, Favre J, et al. GPR68 senses flow and is essential for vascular physiology. Cell 2018;173:762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Elliott H. Metabolism of lorazepam. Br J Anaesth 1976;48:1017–23. [DOI] [PubMed] [Google Scholar]
- 83. Griffin CE, Kaye AM, Bueno FR, Kaye AD. Benzodiazepine pharmacology and central nervous system–mediated effects. Ochsner J 2013;13:214–23. [PMC free article] [PubMed] [Google Scholar]
- 84. Deguchi T, Watanabe N, Kurihara A, Igeta K, Ikenaga H, Fusegawa K, et al. Human pharmacokinetic prediction of UDP-glucuronosyltransferase substrates with an animal scale-up approach. Drug Metab Dispos 2011;39:820–9. [DOI] [PubMed] [Google Scholar]
- 85. Liaw G-W, Hung D-Z, Chen W-K, Lin C-L, Lin I-C, Kao C-H. Relationship between acute benzodiazepine poisoning and acute pancreatitis risk: a population-based cohort study. Medicine (Baltimore) 2015;94:e2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Abed A, Minaiyan M, Safaei A, Taheri D. Effect of diazepam on severity of acute pancreatitis: possible involvement of peripheral benzodiazepine receptors ISRN Gastroenterol 2013;2013:484128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chuong MD, Herrera R, Ucar A, Aparo S, De Zarraga F, Asbun H, et al. Causes of death among patients with initially inoperable pancreas cancer after induction chemotherapy and ablative 5-fraction stereotactic magnetic resonance image guided adaptive radiation therapy. Adv Radiat Oncol 2023;8:101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li J, Qian W, Qin T, Xiao Y, Cheng L, Cao J, et al. Mouse-derived allografts: a complementary model to the KPC mice on researching pancreatic cancer in vivo. Comput Struct Biotechnol J 2019;17:498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Erstad DJ, Sojoodi M, Taylor MS, Ghoshal S, Razavi AA, Graham-O'Regan KA, et al. Orthotopic and heterotopic murine models of pancreatic cancer and their different responses to FOLFIRINOX chemotherapy. Disease Models Mech 2018;11:dmm034793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Arina A, Idel C, Hyjek EM, Alegre M-L, Wang Y, Bindokas VP, et al. Tumor-associated fibroblasts predominantly come from local and not circulating precursors. Proc Natl Acad Sci 2016;113:7551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. BZD prescriptions are associated with improved disease-specific pancreatic cancer patient survival
Supplementary Figure S2. LOR and ALP promote collagen deposition but do not impact tumor growth
Supplementary Figure S3. LOR treatment promotes IL-6 expression in authochthonous KPC CAFs in vivo
Supplementary Figure S4. KPC subcutaneous syngeneic allograft tumors are acidic and express GPR68.
Supplementary Figure S5. LOR treatment increases STAT3 phosphorylation
Supplementary Figure S6. LOR is associated with worse progression-free survival across multiple cancer types.
Supplementary Table S1. Pancreatic cancer patient characteristics by benzodiazepine prescription records.
Supplementary Table S2. Pancreatic cancer patient characteristics by lorazepam prescription records
Supplementary Table S3. Pancreatic cancer patient characteristics by alprazolam prescription records
Supplementary Table S4. RNAscope Antibodies and Probes
Supplementary Table S5. Pancreatic Cancer Time-to-Event Outcomes: Multivariate Summaries.
Supplementary Table S6. Brain cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S7. Breast cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S8. Corpus uterine cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S9. Head and neck cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S10. Invasive nevi and melanoma patient characteristics, median survival, and Multivariate Cox Regression Modeling
Supplementary Table S11. Renal cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S12. Ovarian cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S13. Colon cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling.
Supplementary Table S14. Prostate cancer patient characteristics, median survival, and Multivariate Cox Regression Modeling
Data Availability Statement
The RNA sequencing data reported in this paper have been deposited in the GEO repository under accession number GSE237649.