Abstract
Adeno-associated virus (AAV) is a powerful gene therapy vector that has been used in several FDA-approved therapies as well as in multiple clinical trials. This vector has high therapeutic versatility with the ability to deliver genetic payloads to a variety of human tissue types, yet there is currently a lack of transgene expression control once the virus is administered. There are also times when transgene expression is too low for the desired therapeutic outcome, necessitating high viral dose administration resulting in possible immunological complications. Herein, we validate a chemically controllable AAV transgene expression technology in vitro that utilizes bifunctional molecules known as chemical epigenetic modifiers (CEMs). These compounds employ endogenous epigenetic machinery to specifically enhance transgene expression of episomal DNA. A recombinant AAV (rAAV) was designed to both deliver the reporter transgene as well as deliver a synthetic zinc finger (ZFs) protein fused to FK506 binding protein (FKBP). These synthetic ZFs target a DNA-binding array sequence upstream of the promoter expressing the AAV transgene to specifically enhance AAV transgene expression in the presence of a CEM. The transcriptional activating compound CEM87 functions by recruiting the epigenetic transcription activator bromodomain-containing protein 4 (BRD4), increasing AAV transgene activity up to fivefold in a dose-dependent manner in HEK293T cells. The highest levels of transgene product activity are seen 24 h following CEM87 treatment. Additionally, the CEM87-mediated enhancement of different transgene products with either Luciferase or green fluorescent protein (GFP) was observed in multiple cell lines and enhancement of transgene expression was capsid serotype independent. The impact of CEM87 activity can be disrupted through drug removal or chemical recruitment site competition with FK506, thus demonstrating the reversibility of the impact of CEM87 on transgene expression. Collectively, this chemically controllable rAAV transgene technology provides temporal gene expression control that could increase the safety and efficiency of AAV-based research and therapies.
Keywords: AAV, gene regulation, AAV transgene expression, zinc finger proteins, chemical biology
INTRODUCTION
Adeno-associated virus (AAV) is a small single-stranded DNA parvovirus that is considered defective for autonomous replication and is considered nonpathogenic.1 In the 1980s, it was demonstrated that the AAV coding sequences could be replaced with transgenic DNA and packaged into the protein capsid, termed recombinant AAV (rAAV).2,3 Since then, rAAV has been developed as a promising episomal gene delivery vector highlighted by the FDA's approval of a handful of rAAV drugs to date for ocular, neurological, and hematological disorders with more than 100 ongoing rAAV clinical trials for diverse diseases.4–6
While rAAV is a popular choice for gene therapies because of its broad spectrum of tissue tropism and its lack of pathogenicity, an issue facing rAAV therapies is insufficient rAAV transgene expression upon delivery to induce the desired therapeutic effect.2 Additionally, there is currently no clinically applicable technology available that controls transgene expression postinfection partially due to the size constraints of the rAAV vectors. The chromatinization of the AAV genome in vitro and in vivo provides a route to control AAV transgene expression with host epigenetic machinery.7,8 Here, we introduce a chemically controlled method to regulate rAAV gene expression in vitro through the utilization of endogenous transcriptional machinery.
We have previously shown that endogenous epigenetic machinery can be harnessed to control both exogeneous and endogenous gene expression in vitro by using bifunctional molecules known as chemical epigenetic modifiers (CEMs) along with DNA-binding proteins.9–11 CEM compounds are made of a FK506 molecule with a polyethylene glycol (PEG) linker connecting a compound that can recruit endogenous epigenetic machinery, when coupled with a protein targeting system we can regulate the gene of interest.
CEM87 is the lead-activating compound composed of FK506 linked with a PEG linker to iBET762, which is known to bind bromodomain-containing proteins (BRD) BRD2, BRD3, and BRD4.12 Through chromatin immunoprecipitation (ChIP) experiments, we have previously determined that CEM87 recruits BRD4 to targeted gene loci.10 BRD4 is an epigenetic protein whose presence on gene loci is associated with chromatin decompaction and subsequent gene expression through acetylation of histones and recruitment of other transcription factors.13 Utilizing CEM87 to recruit BRD4 to gene loci allows for targeted enhancement of gene expression.
Chemical-induced proximity (CIP) is a chemical biology strategy that uses chemical ligands to bring different proteins physically closer, thus allowing temporal control of a diverse set of biological processes including the regulation of transcription by recruitment of chromatin regulators to gene promoters.14 Recent advances have coupled chromatin regulators, including those tethered through CIP technology, with DNA-targeting system such as nuclease-deficient CRISPR Cas Protein 9 (dCas9) to epigenetically control target genes.15–19 Building on this work, our lab previously combined CEM compounds along with dCas9, F506 binding protein (FKBP), and small guide RNAs (sgRNAs) to target-specific DNA sequences to regulate gene expression.
While the utilization of dCas9, FKBP, sgRNA, and CEM compounds (dCas9-CEM) was able to significantly regulate gene expression, there are benefits to utilizing other DNA-binding proteins rather than dCas9. The current dCas9-CEM technology requires multiple vector delivery with lentivirus because of the large plasmid payload requiring separate plasmids for dCas9 and sgRNAs. An alternative to dCas9 that requires less payload space are zinc finger (ZFs) proteins. ZFs are small eukaryotic proteins that exhibit DNA-binding specificity and are present in many endogenous human transcription factors.20 ZF engineering allows multiple zinc fingers to be linked together in an array that targets larger DNA sequences.21 Engineered zinc finger arrays have previously been linked to nucleases to enable gene editing and to effector proteins to enable transcriptional regulation.22–27
This inspired the design of a ZF array linked to FKBP to allow for ZF array mediated control of gene expression with CEMs (ZF-CEM). Creation of a rAAV episome that is controllable with ZF-CEM technology requires the use of a ZF array that has a binding preference for a synthetic DNA-binding array rather than endogenous DNA. Zinc finger homeodomain 1 (ZFHD1) was chosen because it has previously been verified and used within the literature as a ZF array linked to effector domains to regulate gene expression.28,29
Herein we describe the use of ZF-CEM technology to regulate the expression of AAV transgenes in vitro. The collective results highlight that rAAV transgene expression is not maximal in tested contexts and demonstrate that the AAV ZF-CEM system can specifically enhance episomal transgene production. These data shed insight into targeted epigenetic regulation of transduced rAAV genomes and warrant further testing in disease contexts toward future strategies for enhanced and potentially safer gene therapies.
MATERIALS AND METHODS
Plasmid design
+ZF-FKBP and −ZF-FKBP plasmids with AAV2 and eight compatible ITRs were adapted from Hirsch et al.30 Renilla luciferase plasmid in a lentiviral backbone plasmid was adapted from Hathaway et al.29 and Toyama et al.31 All plasmid sequences were verified with sanger sequencing. Single-stranded AAV plasmids had their inverted terminal repeats verified through restriction enzyme digestion (Supplementary Fig. S1) using XmaI (R0180L; New England Biolabs). Additional plasmid design information can be found in the Supplementary Data.
Cell culture
HEK293, HEK293T, and HEK293T Lenti-X (Clontech) cells were cultured with high glucose Dulbecco's Modified Eagle's Medium (DMEM) (10-013-CV; Corning). The media was supplemented with 0.1% 55 mM of 2-mercaptoethanol, 0.9% 100 × non-essential amino acids solution (11140-050; Gibco), 0.9% 10 mM N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (25-060-CI; Corning), 1% penicillin-streptomycin (15140-122; Gibco), and 10% FBS (S11550; Atlantic Biologicals). HCT116 cells and U2OS cells were cultured in McCoy's 5A (10-050-CV; Corning), 1% penicillin-streptomycin (15140-122; Gibco), and 10% FBS (S11550; Atlantic Biologicals). Every 3–5 days the cells were passaged. Cells were maintained at 20–90% confluency in an incubator at 37°C and 5% CO2.
Lenti-virus infection
HEK293 Lenti-X cells at a low passage (10–13) were transfected with polyethlenimine (PEI; 23966-1; Polysciences) containing 18 μg of EF1α-Renilla plasmid containing puromycin resistance, 13.5 μg of Gag-Pol plasmid (#12260; Addgene) and 4.5 μg of VSV-G envelope protein plasmid (12259; Addgene). Media was changed after 16 h, and lentivirus was harvested from cells 48 h postmedia change. Lentivirus containing EF1α-Renilla was then added to HEK293T cells (passage 35–45), HCT116 cells (passage 33–34), and U2OS cells (passage 130–135). HEK293T EF1α-Renilla and HCT116 EF1α-Renilla were continually cultured under puromycin selection at a concentration of 1.5 μg/mL. U2OS EF1α-Renilla cells were continually cultured with 2 μg/mL of puromycin.
AAV2 production
AAV2 was packaged according to methods previously published.32 In 15 cm plates, HEK293 cells at 70% confluency were transfected with PEI containing 10 μg XR2,32 12 μg XX680,32 and 9 μg of plasmid to be packaged. After 3–5 days posttransfection, cells were harvested, lysed through sonication, and purified through a CsCl gradient. Titer of AAV was determined through qPCR with Roche FastStart Universal SYBR Green Master (Rox) (4913850001; Sigma) on the QuantStudio 6 Flex (Thermo Scientific). Further information relating to the titering of and actual titers of all viruses in the manuscript can be found in the Supplementary Data.
Dual luciferase assay
Cells stably expressing EF1α-Renilla were either transfected with PEI or transduced with AAV2 in biological triplicate. For transfection experiments, cells were transfected with a ratio of 1:3 of DNA (in μg) to PEI (in μL) in minimal DMEM media (10-013-CV; Corning). The media was changed 16 h later, and CEM87 was added. For AAV2 infection experiments, cells were transduced with AAV2 the day after cells were split. Twenty-four hours after infection, CEM87 was added to the media. Unless otherwise stated, 48 h post CEM87 addition to media in both transfection and infection experiments, the dual luciferase assay was run using Dual Luciferase Reagent Assay kit (E1980; Promega). PheraStar FS (BMG Labtech) was used to analyze changes in luminescence. Firefly luciferase luminescence was normalized to Renilla luciferase luminescence. Fold change in luminescence was determined by dividing the normalized luminescence of the treated cell replicate by the normalized luminescence of the 0 nM CEM87 cell replicate. Raw data for all experiments available upon request.
Luciferase assay
Cells were plated in a 24-well plate in biological triplicate. After 16 h, HEK293 cells were transfected with a ratio of 1:3 of DNA (in μg) to PEI (in μL) in minimal DMEM media (10-013-CV; Corning). AAV8 +ZF-FKBP was added 8 h later. Twenty-four hours after AAV8 infection, CEM87 was added to the media. Twenty-four hours post CEM87 addition to media, the luciferase assay was run using Luciferase Assay Reagent (Promega). PheraStar was used to analyze changes in luminescence. The gain was set to 3,600 for all AAV8 infection experiments. Total protein levels were determined by Bradford Assay (23200; Thermo Scientific). The GloMax Discovery (Promega) was used to determine the relative absorbances from the Bradford Assay. Firefly luciferase luminescence was normalized to Total Protein levels. Fold change in luminescence was determined by dividing the normalized luminescence of the treated cell replicate by the normalized luminescence of the 0 nM CEM87 cell replicate. Raw data for all experiments available upon request.
Flow cytometry
Cells were plated in a 48-well plate in biological triplicate. The next day, the cells were infected with AAV2 +ZF-FKBP_GFP. After 24 h, the cells were treated with CEM87. Flow cytometry was performed using Attune NxT 24 h post CEM treatment to detect for changes in fluorescence. Live cells were gated for using FSC-Area and SCC-Area. Single cells were gated for using SCC-Height and SCC-Area. GFP was detected with a BL1 laser and gated for on a histogram of BL1-Area and count. Raw data for all experiments available upon request.
Western blot
HEK293T EF1α-Renilla cells were either transfected with +ZF-FKBP plasmid or transduced with AAV2 +ZF-FKBP. After 24 h, the cells were treated with CEM87. Forty-eight hours post CEM87 treatment, total protein was taken from cells transfected or transduced with +ZF-FKBP plasmids and quantified using the Bradford Assay (23200; Thermo Scientific). The GloMax Discovery (Promega) was used to determine the relative absorbances from the Bradford Assay. Fifteen micrograms per lane of protein were ran on 4–20% Mini Protean TGX Gels (4561096; BioRad) then transferred to PVDF membrane. Membranes were imaged by infrared fluorescence (Li-Cor Biosciences). Antibodies for FKBP12 (ab2918; ABCAM) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (ab8245; ABCAM) were used.
Microscope images
Images were taken with Olympus IX83 Microscope after CEM87 treatment of HEK293T cells that had been transduced with either AAV2 +ZF-FKBP or AAV2 +ZF-FKBP_GFP. Images were taken at a magnification of either 10 × or 40 × .
Cell titer glo assay
HEK293T cells were plated in a 96-well plate in biological triplicate. Some cells were then infected with AAV2 +ZF-FKBP and treated with various concentrations of CEM87, while other cells were just treated with various concentrations of CEM87. After 24 h, Promega's Cell Titer Glo Assay (G7570) was used to determine cell viability. Raw data for all experiments available upon request.
Statistical analysis
Figures were made and statistically analyzed using GraphPad Prism v.9 (GraphPad Software). Statistical significance was when the p-value compared to 0 nM CEM87 was determined to be ≤0.05. Additional information is available in the Supplementary Data.
RESULTS
Establishing ZF-based CEM technology in a tissue culture model system
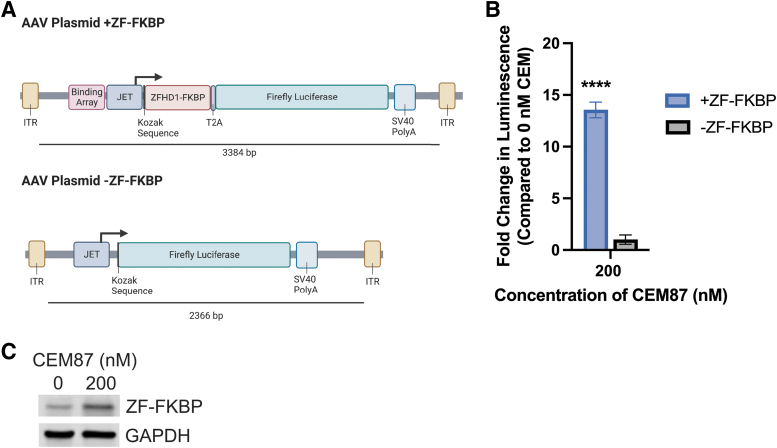
To determine if CEM87 could be used in conjunction with ZFHD1 fused to FKBP (ZF-FKBP) to control transgene expression, HEK293T cells that stably express EF1α upstream of Renilla luciferase (EF1α-Renilla) as a luminescence control were transfected with AAV plasmids (Fig. 1A). Renilla luciferase was used as an internal luciferase normalization control. The luminescence from the firefly luciferase expressed from the AAV plasmid transgene can be normalized to the luminescence of the stable Renilla luciferase in the cells.33 One of the plasmids contains 12 repeats of a ZFHD1 DNA-binding domain upstream of a JET promoter upstream of a ZF-FKBP with a T2A peptide sequence followed by a firefly luciferase gene (+ZF-FKBP).34 This portion of the cassette, which is required for CEM87-mediated control of transgene expression, takes up 1,306 bp of vector space. The control plasmid contains a JET promoter upstream of the firefly luciferase gene (−ZF-FKBP). CEM87 was added to the cells 16 h post-transfection. After 48 h, the relative luminescence of the cells was determined (Fig. 1B; Supplementary Fig. S2).
Figure 1.
ZF-CEM technology design and transgene induction following transfection. (A) AAV +ZF-FKBP and AAV −ZF-FKBP plasmid cassettes. (B) HEK293T cells were transfected with the plasmids depicted in A. Cells were treated with CEM87 and after 48 h, the change in luciferase luminescence was determined. At 200 nM, CEM87 was able to significantly increase luciferase transgene expression when cells were transfected with plasmids containing +ZF-FKBP. (C) Western blot of HEK293T cells transfected with +ZF-FKBP plasmid and then treated with CEM87. Western blot is detecting ZF-FKBP fusion protein with anti-FKBP primary antibody. p Value parameters: ****p ≤ 0.0001. AAV, adeno-associated virus; FKBP, FK506 binding protein; ITR, inverted terminal repeat; ZF-FKBP, ZFHD1 fused to FKBP.
There was a statistically significant 14-fold increase in relative luminescence when 200 nM of CEM87 was added to the cells transfected with +ZF-FKBP plasmids. The cells that were transfected with −ZF-FKBP plasmid did not have a statistically significant change in luminescence when CEM87 was added. These results indicate that ZF-FKBP can be used with CEM87 to increase expression of a gene in a plasmid.
Western blotting was performed on HEK293T EF1α-Renilla cells that were transfected with +ZF-FKBP plasmid and treated with CEM87. An increase in ZF-FKBP fusion protein is observed in cells transfected with +ZF-FKBP plasmid and treated with 200 nM CEM87 compared to transfected cells treated with 0 nM CEM87 (Fig. 1C; Supplementary Fig. S3). The observed increase in ZF-FKBP fusion protein in cells transfected with +ZF-FKBP plasmid supports that CEM87 treatment increases both ZF-FKBP levels and luciferase protein levels.
Characterization of CEM87 control of AAV transgene in different cell lines
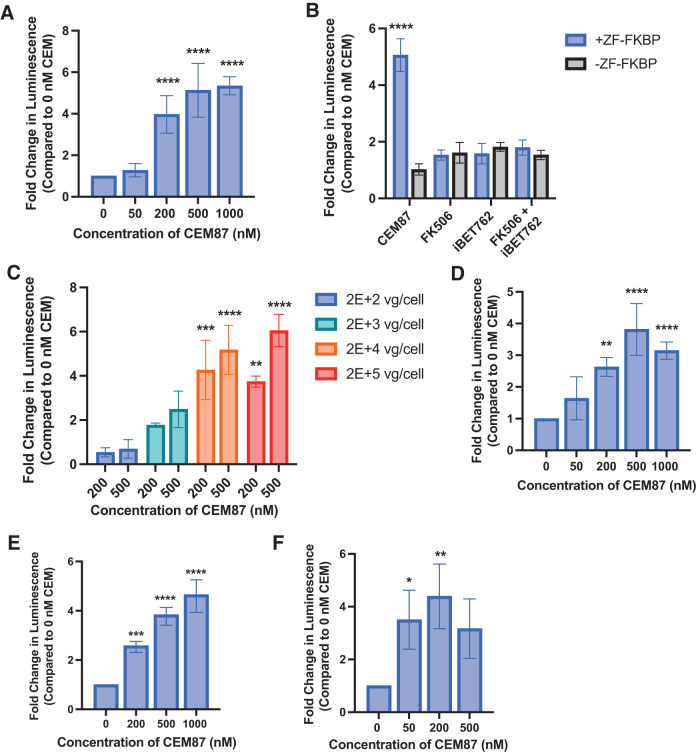
Next, we wanted to determine if ZFHD1 fused to FKBP could be used with CEM87 to increase expression of a transgene within an AAV episome. HEK293T EF1α-Renilla cells were transduced with AAV2 +ZF-FKBP at a multiplicity of infection (MOI) of 2E+4 vg/cell. After 24 h, various concentrations of CEM87 were added to the cells, and after 48 h, the relative luminescence was determined (Fig. 2A; Supplementary Fig. S4). Cells that had been treated with AAV2 +ZF-FKBP and CEM87 at concentrations between 200 and 1,000 nM had a statistically significant fourfold to fivefold increases in relative luminescence compared to cells that had been transduced with the same virus and were treated with 0 nM of CEM87. These results indicate that CEM87 can increase expression of the transgene-derived luciferase activity in a dose-dependent manner in vitro.
Figure 2.
CEM87-mediated induction of firefly luciferase transgene expression following AAV vector transduction. (A) Dose-dependent impact of CEM87 treatment on transgene expression following HEK293T cells transduction (2E+4 vg/cell). (B) The effect of CEM87 on luciferase transgene expression compared to the impact of its individual chemical parts. (C) Transgene product activity in HEK293 cells treated with 2E+4 and 2E+5 vg/cell at CEM87 concentrations of 200 and 500 nM. (D) Fold change in luminescence in HCT116 cells infected with AAV2 +ZF-FKBP then treated with 0–1,000 nM of CEM87. (E) Fold change in luminescence in U2-OS cells infected with AAV2 +ZF-FKBP then treated with 0–1,000 nM of CEM87. (F) Fold change in luminescence in HEK293 cells transfected with a plasmid encoding E4orf6, infected with AAV8 +ZF-FKBP, and treated 0–500 nM of CEM87. p Value parameters: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Microscope images were taken at the time of infection, at the time of CEM87 addition, and 24 h after CEM87 addition. No observable toxicity and/or changes in cell morphology were noted among the treatment groups (Supplementary Fig. S5). To quantitatively determine if AAV2 and CEM87 or CEM87 alone impact cell viability, HEK293T cells were either infected and treated with various doses of CEM87, or just treated with various doses of CEM87. Promega's Cell Titer Glo reagent was used to determine the impact of these conditions on HEK293T cell viability. Based on the data, neither AAV2 and CEM87 or just CEM87 alone impact HEK293T cell viability at any concentration of CEM87 (Supplementary Fig. S6).
To confirm that the impact of CEM87 on transgene expression is due to the bifunctional design of the compound rather than any one of its key moieties alone, HEK293T EF1α-Renilla cells were treated with 200 nM of CEM87, FK506, iBET762, or a combination of FK506 and iBET762. A fivefold change in luciferase activity was only seen in cells transduced with AAV2 +ZF-FKBP and treated with CEM87 (Fig. 2B; Supplementary Fig. S7). These results indicate that the effects on gene expression seen with CEM87 treatment are due to the entire compound rather than one of the individual parts.
All previous experiments were conducted with an MOI of 2E+4 vg/cell. While the data do indicate CEM87 can control luciferase activity at 200 nM with an approximate fivefold change, we transduced HEK293T EF1α-Renilla cells to test a range MOIs from 2E+2 to 2E+5 vg/cell to determine what impact MOI has on CEM87 ability to control AAV2 transgene expression. At MOIs of 2E+4 and 2E+5 vg/cell, CEM87 at 200 and 500 nM significantly increases luciferase activity between fivefold and sixfold (Fig. 2C; Supplementary Fig. S8). At lower MOIs of 2E+2l and 2E+3 vg/cell, CEM87 at 200 and 500 nM can increase luciferase activity but not in a significant manner. Based on these results, we see a substantial fold-change in luminesce at a CEM87 concentration of 200 nM and at a MOI of 2E+4 vg/cell and use these conditions in subsequent work.
To determine whether CEM87 can enhance AAV transgene production in cell lines beyond HEK293T, HCT116 EF1α-Renilla cells and U2OS EF1α-Renilla cells were transduced with AAV2 +ZF-FKBP at an MOI of 2E+4 vg/cell. After 24 h, CEM87 was able to increase the fold change in luminescence compared to no CEM87 treatment in a statistically significant manner at 200 nM and higher doses in both HCT116 and U2OS cell lines. In the HCT116 cell line, the highest fold change of approximately fivefold was achieved at 500 nM of CEM87 (Fig. 2D; Supplementary Fig. S9). We hypothesize that the slight decrease in expression at 1,000 nM is due to the hook effect, where excess bifunctional compound blocks each target site instead of bridging them causing nonproductive events. In the U2OS cell line, the highest fold increase in luminescence of fivefold was seen also at 500 nM (Fig. 2E; Supplementary Fig. S10). These data indicate CEM87 can chemically control AAV2 transgene expression in a dose-dependent manner in different cell lines.
AAV8 +ZF-FKBP vectors were produced, and gene expression control was explored in vitro to determine if ZF-CEM technology can regulate expression of transgenes delivered by a different AAV serotype. To enhance AAV8 vector transduction, HEK293 cells were first transfected with a plasmid containing the Adenovirus gene encoding E4orf6, a protein that enhances AAV transduction in conjunction with E1B-55kD (the gene of which is an integrant in 293 cells).35,36 AAV8 +ZF-FKBP was then added to cells at an MOI of 2E+4 vg/cell. After 24 h, CEM87 was added to the cells at various concentrations. After an additional 24 h, relative luminescence was determined. CEM87 was able to increase luciferase activity at concentrations of 50 and 200 nM to threefold and fourfold, respectively (Fig. 2F; Supplementary Fig. S11). These results indicate that CEM87 can increase expression of transgenes delivered by both AAV2 and AAV8 serotypes.
Characterization of versatility of CEM-mediated AAV control in different cell lines and with an alternate AAV capsid
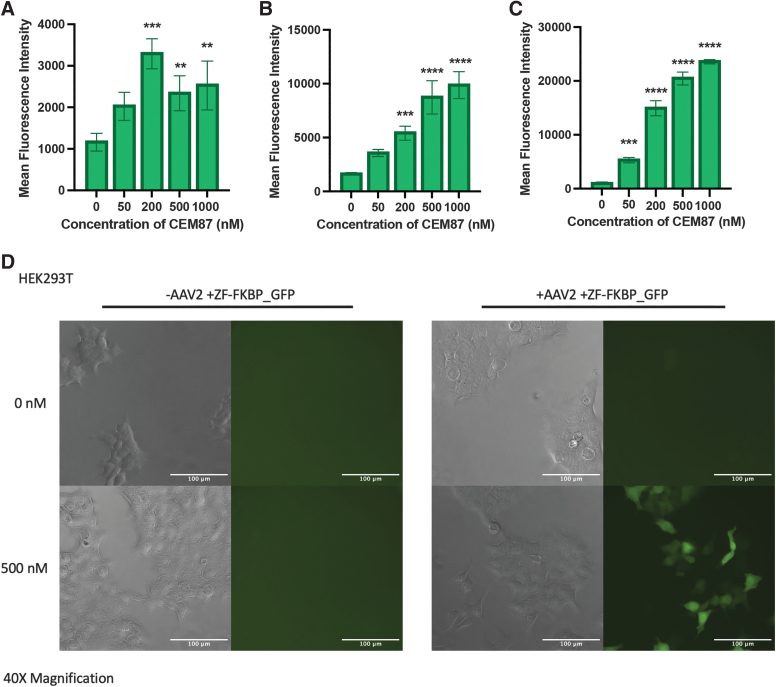
An analogous version of AAV2 +ZF-FKBP was made that would deliver a GFP gene rather than a firefly luciferase gene (AAV2 +ZF-FKBP_GFP) to determine if CEM87 can control the expression of different transgenes. HCT116, U2OS, and HEK293T cells were infected with 2E+4 vg/cell of AAV2 +ZF-FKBP_GFP. The cells were treated with various concentrations of CEM87. After 24 h, flow cytometry was used to determine the mean fluorescence of the cells.
HCT116 cells had an increase in mean fluorescence intensity of approximately threefold when treated with 200 nM of CEM87 and approximately twofold when treated with 500 and 1,000 nM of CEM87 (Fig. 3A; Supplementary Figs. S12 and S13). In U2OS cells, the highest increases in mean fluorescence intensity were fivefold and sixfold when cells were treated with 500 and 1,000 nM of CEM87, respectively (Fig. 3B; Supplementary Figs. S14 and S15). HEK293T cells had increases in mean fluorescence intensity of 17- and 20-fold when treated with 500 and 1,000 nM of CEM87, respectively (Fig. 3C; Supplementary Fig. S16). When imaged, the increase in GFP is apparent when cells are treated with 500 nM of CEM87 compared to cells that only received 0 nM (Fig. 3D). These data indicate the CEM87 can achieve dose-dependent chemical control of expression of different AAV transgenes in various cell lines and demonstrate that some cell lines are more responsive than others.
Figure 3.
CEM87-mediated induction of GFP transgene expression following AAV vector transduction. (A) Mean fluorescence intensity of HCT116 cell infected with AAV2 +ZF-FKBP_GFP then treated with 0–1,000 nM of CEM87. (B) Mean fluorescence intensity of U2-OS cells infected with AAV2 +ZF-FKBP_GFP then treated with 0–1,000 nM of CEM87. (C) Mean fluorescence intensity of HEK293T cells infected with AAV2 +ZF-FKBP_GFP then treated with 0–1,000 nM of CEM87. (D) Phase and fluorescent images of HEK293T cells either infected with AAV2 +ZF-FKBP_GFP or no virus and then treated with 0 or 500 nM of CEM87. Images were taken at 40 × magnification. p Value parameters: **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. GFP, green fluorescent protein; ZF, zinc finger protein.
Determining the impact of CEM87 over time and the reversibility of CEM transcriptional control
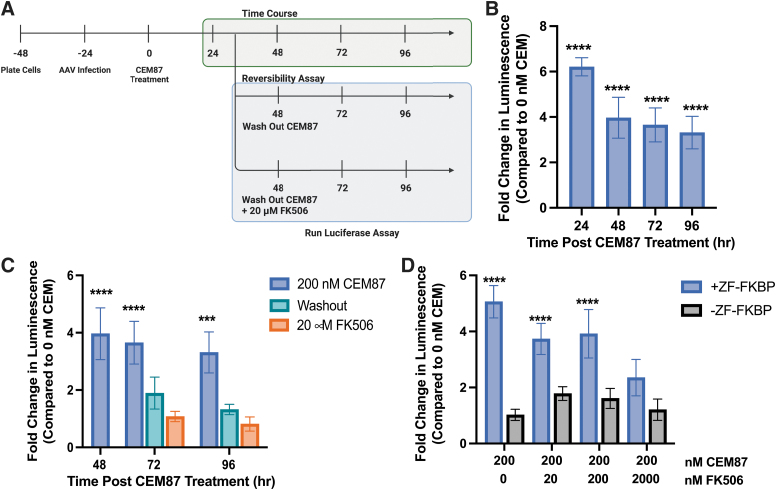
To better understand how quickly CEM87 begins to increase relative luminescence and at what time the largest increase in luminescence is observed, changes in luminescence were measured at multiple time points after initial CEM87 treatment. HEK293T EF1α-Renilla cells were transduced with AAV2 +ZF-FKBP at an MOI of 2E+4 vg/cell. Cells were treated with 200 nM of CEM87 24 h postinfection. Relative luminescence was measured between 24 and 96 h post-CEM87 treatment and the fold change in relative luminescence in the presence of 200 nM CEM87 was determined (Fig. 4A). After 24 h, cells that had been transduced with AAV2 +ZF-FKBP and CEM87 treatment had the highest fold change at sixfold. The fold change begins to decrease after 48 h of chemical treatment to fourfold, with additional small decreases in activity at 72 and 96 h (Fig. 4B; Supplementary Fig. S17).
Figure 4.
Kinetic effect of CEM87 on transgene expression and FK506-reversibility of the transgene expression enhancement. (A) Diagram of time course and reversibility of CEM87 experiments in HEK293T cells. (B) Impact of CEM87 on AAV transgene activity over time after a single dose at time 0 h. (C) AAV transgene activity following CEM87 removal and the addition of FK506. (D) Dose-dependent effect of FK506 on chemical competition with CEM87 to suppress an CEM87-mediated increase in AAV transgene activity. p Value parameters: ***p ≤ 0.001, ****p ≤ 0.0001.
To investigate the ability to reverse the transgene expression impacts of CEM87, CEM87 washout, or chemical competition with FK506 was tested in HEK293T EF1α-Renilla cells after transduction with AAV2 +ZF-FKBP. Washing away CEM87 with fresh medium or replacement with medium containing 100 × FK506 (20 μM) began 48 h after CEM87 treatment. Within 24 h, luciferase activity in both the washout group and washout +FK506 group were reduced to similar levels as the no CEM87 treatment group (Fig. 4C; Supplementary Fig. S18). The fold changes in luminescence were determined not to be statistically significant. This was also the case 48 h after media wash out and chemical competition with 20 μM FK506. While 100 × more FK506 coupled with CEM87 washout was able to reverse the transcriptional impacts of CEM87, less FK506 might be able to chemically compete with CEM87. To test this, HEK293T EF1α-Renilla cells were transduced with AAV2 +ZF-FKBP or −ZF-FKBP and then treated with CEM87 coupled with various concentrations of FK506. At 10 × more, 2 μM FK506 can compete with CEM87 and reduce luciferase activity (Fig. 4D; Supplementary Fig. S19). These results indicate that CEM87 effects on luciferase activity are reversible within 24 h when the compound is removed from the cells and that 2 μM of FK506 can compete with CEM87.
DISCUSSION
Here, we present a novel solution that has broad potential utility to control gene therapy vectors postadministration with value both in the research laboratory and potential future clinical value. Gene therapies have tremendous potential, yet the ability to control the transgene payload is currently limited primarily to a fixed vector design and choice of a gene promoter. Once the viral vector is administered, there are no approved methods of controlling the gene expression off the AAV cassette. Furthermore, transgene expression tends to diminish over time lowering the long-term efficacy of treatment. Currently, there are no methods to boost expression other than redosing with AAV, which potentially presents complications like immunogenicity. Here, we present a platform capable of substantially enhancing AAV transgene expression posttransduction in multiple human cells. We demonstrate for the first time that a bifunctional chemical that tethers transcriptional activators to the AAV transgene can significantly increase AAV transgene expression after initial transduction in vitro.
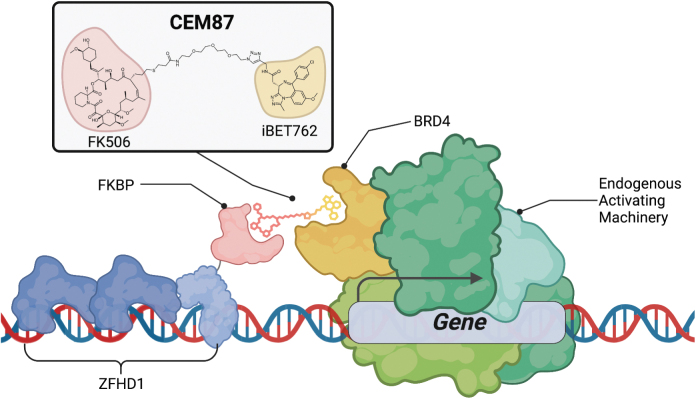
Based on our previous work with CEM technology coupled with CRISPR-based approaches to regulate gene expression, we hypothesized that a we could adapt this platform to efficiently control AAV transgenes.9–11 It is known that rAAV DNA forms double-stranded circular monomers or concatemers that largely persist as extrachromosomal species.37,38 Previous reports using histone deacetylase inhibitors demonstrated enhanced rAAV expression suggesting that the AAV episomes persist in a chromatin-like state in a repressed conformation.39 Furthermore, a recent study revealed that NP220 in concert with the human silencing hub complex represses expression of rAAV genomes.40 These studies demonstrate that rAAV genomes are epigenetically repressed to some extent and that repression can be relieved pharmacologically. This provided some of the rationale for the hypothesis that the CEM-mediated recruitment of BRD4 would enhance transgene expression when ZFHD1 is used to target DNA upstream of the promoter and transgene (Fig. 5A).
Figure 5.
Zinc finger-CEM induction model. This technology utilizes ZFHD1 as the ZF-FKBP. ZF-FKBP binds to the specific DNA binding domain sequence upstream of the promoter and gene. When treated with CEM87, the FK506 moiety binds to FKBP and the iBET762 moiety has affinity to BRD4 which is an endogenous transcriptional activator. BRD4, bromodomain-containing protein 4; CEM, chemical epigenetic modifier; ZF-FKBP, ZF array fused to FKBP; ZFHD1, zinc finger homeodomain 1.
In this work, we demonstrate that ZF-CEM technology was able to chemically control expression of different AAV transgenes in a dose-dependent manner in three different cell types. CEM87 was able to chemically control expression of transgenes delivered by two different serotypes, AAV2 and AAV8. Importantly, the increase in transgene expression that CEM87 causes can be reversed with chemical competition using FK506 demonstrating a method to reduce transgene enhancement. Impacts of CEM87 on gene and transgene expression do vary depending on whether the gene is on a plasmid or a transgene on an episome; the different fold increases seen between transfection of plasmid data and transduced transgenes makes this evident. The difference in fold increase between the luciferase data and GFP data also indicates that the impact CEM87 can have on transgene expression is specific to the transgene being delivered. HEK293T cells transduced with +ZF-FKBP_GFP had higher fold changes in mean fluorescence intensity. Even though this is not directly comparable to the luciferase data seen in the HEK293T cells, the differences in fold change could be due to various reasons including the fact that GFP is a smaller gene to transcribe than firefly luciferase. While the gene expression enhancements seen in this work were exciting, we plan to further explore the role of vector design, serotype compatibility, and application of the approach in more diverse tissues.
ZF-CEM technology provides a chemically responsive method to control AAV transgene expression posttransduction using chemical biology tools previously used to explore chromatin biology. This new application of epigenome editing tools for the gene therapy field allows for a way to directly edit the AAV epigenome to control transgene expression. While this technology is promising, there are some limitations. CEM87 is reliant on BRD4 availability in the cell, so this technology may not work as well in cell types with reduced or missing BRD4. To address this limitation, future work will examine other CEMs that target different epigenetic machineries to expand the library of compounds that can be used with AAV ZF-CEM control technology. Further investigation is also required to determine if the in vitro abilities of this technology translate to in vivo applications.
The first iteration of this technology provides a way for the first time to chemically control AAV transgene expression through direct targeting of epigenetic machinery at the start of transcription. ZF-CEM technology is versatile and can be applied to different AAV serotypes as well as cell lines if they are transducible by AAV and express the epigenetic proteins targeted by CEMs. This approach alone has value in biomedical research as different levels of gene expression can be driven off a single viral cassette when investigating gene replacement in cells. We hope that CEM-mediated control of AAV expression may also be applied to human disease models that could benefit from the ability to enhance AAV transgene expression after infection to a desired gene dose range.
ACKNOWLEDGMENTS
We thank the Hathaway laboratory as well as the lab of Dr. Matt Hirsch for stimulating discussions of this work and experimental advice. The authors would like to thank Brain Goltz and the UNC CRISPR Screening Facility for set up and access to the PheraStar FS luminometer used for all luciferase experiments. We would also like to thank Dr. Daniel Crona and his lab for the use of the GloMax Discovery plate reader. We would like to acknowledge the UNC Vector Core for packaging AAV2 +ZF-FKBP and AAV8 +ZF-FKBP viruses. We also thank UNC Lineberger Comprehensive Cancer Center for use of the Li-Cor for western blot imaging. Diagrams for figures were made on BioRender.com. The UNC Flow Cytometry Core allowed access to Attune NxT for all flow experiments. Special thanks to Brian Anderson, Zachary Davis-Gilbert, Kareem Galal, and the UNC Structural Genomics Consortium for additional CEM87 compound characterization.
Authors' Contributions
J.D.U., S.R.W., L.S., J.J., and N.A.H. designed the research. J.D.U., S.R.W., L.S., and A.A.G. conducted the experiments. X.Y. synthesized the compounds. J.D.U., S.R.W., L.S., and N.A.H. wrote the manuscript with input from all authors.
Supplementary Material
AUTHOR DISCLOSURE
N.A.H. is the founder and a shareholder of Epigenos Biosciences, Inc., which is commercially advancing chemical epigenetic modifier technology. J.D.U., L.S., N.A.H., and M.L.H. have filed a provisional patent application based on the results described in this manuscript. J.J. is a cofounder and equity shareholder in Cullgen, Inc., a scientific cofounder and scientific advisory board member of Onsero Therapeutics, Inc., and a consultant for Cullgen, Inc., EpiCypher, Inc., and Accent Therapeutics, Inc. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, Inc., Cullgen, Inc., and Cullinan Oncology, Inc.
FUNDING INFORMATION
Funding for this work was received from the U.S. National Institutes of Health grants R01GM118653 (to N.A.H.), R35GM148365 (N.A.H.) T32GM135122 (to J.D.U.), T32GM12274 (to S.R.W.), R41EY033603 (to N.A.H.), as well as R21AI164214 (to N.A.H.). This work utilized the NMR Spectrometer Systems at Mount Sinai acquired with funding from the NIH's SIG grants 1S10OD025132 and 1S10OD028504.
SUPPLEMENTARY MATERIAL
REFERENCES
- 1. Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science 1965;149:754–756. [DOI] [PubMed] [Google Scholar]
- 2. Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: Transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA 1984;81:6466–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tratschin JD, West MH, Sandbank T, et al. A human parvovirus, adeno-associated virus, as a eucaryotic vector: Transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol Cell Biol 1984;4:2072–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berns KI, Muzyczka N. AAV: An overview of unanswered questions. Hum Gene Ther 2017;28:308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Z, Anselmo AC, Mitragotri S. Viral vector-based gene therapies in the clinic. Bioeng Transl Med 2022;7:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, et al. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther 2021;29:464–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marcus-Sekura CJ, Carter BJ. Chromatin-like structure of adeno-associated virus DNA in infected cells. J Virol 1983;48:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Penaud-Budloo M, Le Guiner C, Nowrouzi A, et al. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol 2008;82:7875–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butler KV, Chiarella AM, Jin J, et al. Targeted gene repression using novel bifunctional molecules to harness endogenous histone deacetylation activity. ACS Synth Biol 2018;7:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiarella AM, Butler KV, Gryder BE, et al. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat Biotechnol 2020;38:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu D, Foley CA, Birla SV, et al. Bioorthogonal chemical epigenetic modifiers enable dose-dependent CRISPR targeted gene activation in mammalian cells. ACS Synth Biol 2022;11:1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung CW, Coste H, White JH, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem 2011;54:3827–3838. [DOI] [PubMed] [Google Scholar]
- 13. Devaiah BN, Case-Borden C, Gegonne A, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 2016;23:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanton BZ, Chory EJ, Crabtree GR. Chemically induced proximity in biology and medicine. Science 2018;(80-.):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Braun SMG, Kirkland JG, Chory EJ, et al. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat Commun 2017;8:560–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y, Xiong X, Wong S, et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods 2016;13:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura M, Gao Y, Dominguez AA, et al. CRISPR technologies for precise epigenome editing. Nat Cell Biol 2021;23:11–22. [DOI] [PubMed] [Google Scholar]
- 18. Chiarella AM, Lu D, Hathaway NA. Epigenetic control of a local chromatin landscape. Int J Mol Sci 2020;21:6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gjaltema RAF, Rots MG. Advances of epigenetic editing. Curr Opin Chem Biol 2020;57:75–81. [DOI] [PubMed] [Google Scholar]
- 20. Brayer KJ, Segal DJ. Keep your fingers off my DNA: Protein–protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys 2008;50:111–131. [DOI] [PubMed] [Google Scholar]
- 21. Gersbach CA, Gaj T, Barbas CF. Synthetic zinc finger proteins: The advent of targeted gene regulation and genome modification technologies. Acc Chem Res 2014;47:2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 2009;27:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Straimer J, Lee MC, Lee AH, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat Methods 2012;9:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beerli RR, Segal DJ, Dreier B, et al. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA 1998;95:14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mussolino C, Sanges D, Marrocco E, et al. Zinc-finger-based transcriptional repression of rhodopsin in a model of dominant retinitis pigmentosa. EMBO Mol Med 2011;3:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nomura W, Barbas CF. In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc 2007;129:8676–8677. [DOI] [PubMed] [Google Scholar]
- 27. Beerli RR, Dreier B, Barbas CF. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA 2000;97:1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pollock R, Giel M, Linher K, et al. Regulation of endogenous gene expression with a small-molecule dimerizer. Nat Biotechnol 2002;20:729–733. [DOI] [PubMed] [Google Scholar]
- 29. Hathaway NA, Bell O, Hodges C, et al. Dynamics and memory of heterochromatin in living cells. Cell 2012;149:1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hirsch ML, Li C, Bellon I, et al. Oversized AAV transductifon is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol Ther 2013;21:2205–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Toyama EQ, Herzig S, Courchet J, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016;351:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc 2006;1:1412–1428. [DOI] [PubMed] [Google Scholar]
- 33. Schagat TPA, Paguio A, Kopish K. Normalizing genetic reporter assays approaches and considerations for increasing consistency and statistical significance. Cell Notes 2007;17:9–12. [Google Scholar]
- 34. Tornøe J, Kusk P, Johansen TE, et al. Generation of a synthetic mammalian promoter library by modification of sequences spacing transcription factor binding sites. Gene 2002;297:21–32. [DOI] [PubMed] [Google Scholar]
- 35. Lentz TB, Samulski RJ. Insight into the mechanism of inhibition of adeno-associated virus by the Mre11/Rad50/Nbs1 complex. J Virol 2015;89:181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song L, Samulski RJ, Hirsch ML. Adeno-associated virus vector mobilization, risk versus reality. Hum Gene Ther 2020;31:1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi VW, Samulski RJ, McCarty DM. Effects of adeno-associated virus DNA hairpin structure on recombination. J Virol 2005;79:6801–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakai H, Yant SR, Storm TA, et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol 2001;75:6969–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okada T, Uchibori R, Iwata-Okada M, et al. A histone deacetylase inhibitor enhances recombinant adeno-associated virus-mediated gene expression in tumor cells. Mol Ther 2006;13:738–746. [DOI] [PubMed] [Google Scholar]
- 40. Das A, Vijayan M, Walton EM, et al. Epigenetic silencing of recombinant adeno-associated virus genomes by NP220 and the HUSH complex. J Virol 2022;96:e0203921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.