Abstract
Purpose:
Advances in genomic research have facilitated rare disease diagnosis for thousands of individuals. Unfortunately, the benefits of advanced genetic diagnostic technology are not distributed equitably among the population, as has been seen in many other healthcare contexts. Quantifying and describing inequities in genetic diagnostic yield is inherently challenging due to barriers to both clinical and research genetic testing. We therefore present an implementation protocol developed to expand access to our rare disease genomic research study and to further understand existing inequities.
Methods and Findings:
The Rare Genomes Project (RGP) at the Broad Institute of MIT and Harvard offers research genome sequencing to individuals with rare disease who remain genetically undiagnosed through direct interaction with the individual or family. This presents an opportunity for diagnosis beyond the clinical context, thus eliminating many barriers to access. An initial goal of RGP was to equalize access to genomic sequencing by decoupling testing access from proximity to a major medical center and physician referral. However, study participants over the initial three years of this project were predominantly white and well-resourced. To further understand and address the lack of diversity within RGP, we developed a novel protocol embedded within the larger RGP study, in an approach informed by an implementation science framework. The aims of this protocol are 1) to diversify recruitment and enrollment within RGP, 2) understand the process and context of implementing genomic medicine for rare disease diagnosis, and 3) investigate the value of a diagnosis for underserved populations.
Implications:
Improved understanding of existing inequities and potential strategies to address them are needed in order to advance equity in rare disease genetic diagnosis and research. In addition to the moral imperative of equity in genomic medicine, this approach is also critical in order to fully understand the genomic underpinnings of rare disease.
Keywords: genetics, genomics, rare disease, implementation, disparities, inequities
Introduction
Remarkable advances in genomic sequencing have transformed rare disease diagnosis (1, 2).. The ability to search for pathogenic variants across the genome have led to the identification of a molecular diagnosis for many individuals and the discovery of thousands of disease genes (1, 2). This has a profound and multifaceted impact on these individuals and their families, in addition to broader implications for the larger rare disease community towards understanding the genomic landscape of these conditions. At the level of the patient, the psychosocial challenges of remaining undiagnosed may be alleviated by a molecular diagnosis, allowing for increased understanding, feelings of empowerment and control, and connection with other people with the same condition for further support (3, 4). Unfortunately, the benefits of these advances in genomic technology are not distributed equitably, as has been seen in many other aspects of healthcare (5, 6). Multiple prior studies of genomic sequencing, its diagnostic yield, and its clinical and psychosocial impact involve primarily white participants with a relatively high socioeconomic status (6–8). There are many structural and systemic barriers to a genetic diagnosis among diverse populations (9, 10), including failure to suspect or recognize genetic disorders in non-white individuals (11), decreased referral rates for clinical genetic testing (12), insurance challenges (13), difficulty interpreting test results given lack of ancestry diversity in the reference genome (14), and low referral rates to rare disease research programs when standard clinical evaluation is unsuccessful (15).
These inequities in access to genomic sequencing are not the result of lower incidence of genetic disorders in non-white populations, but highlight systemic and structural barriers in access to appropriate health services. Indeed, quantifying and defining inequities in genetic diagnostic yield has also been challenging, particularly because a genetic diagnostic evaluation is never even initiated for many individuals with limited access to healthcare, as we have previously demonstrated (12). This may result in fewer diagnoses identified in minoritized populations (16, 17), a longer time to diagnosis (18), and a lack of diversity in genomic research studies (6, 19). In addition, the features leading to effective implementation of genomic medicine for underserved communities remain poorly understood (20). This, in turn, impedes optimal development of genomic medicine programs and perpetuates inequities.
The Rare Genomes Project (RGP) is an ongoing study offering research genome sequencing (GS) that is free to participants for whom the clinical route to testing has been unsuccessful. A founding goal of RGP was to equalize access to GS by decoupling testing access from proximity to a major medical center and physician referral and by removing cost and insurance coverage as barriers to access. However, over the first three years of RGP, the study participants who have self-reported race and other sociodemographic features were overwhelmingly white (>90%), highly-educated (>90% of adults with some college attendance or higher) and well-resourced, as evident from their self-reported household income (>60% reporting annual income >$90,000/year and only 5% below the federal poverty line) and testing history prior to RGP enrollment. Of all 2,712 applications to RGP since its inception, 1,482 applicants have reported having genetic testing, 328 reported having no genetic testing, and 268 were uncertain if genetic testing has been done; of those with prior testing, 423 reported having prior exome sequencing, and 406 reported having prior GS. Although insurance is not a barrier in research, the process required to enroll in a research study, including RGP, and have genomic sequencing performed is still fraught with multiple barriers to access. A recent analysis of individuals with rare disease who declined participation in the Undiagnosed Diseases Network identified geographic and other logistical barriers to participation, such as the need to take time off from work, and highlighted the emotional exhaustion of families dealing with complex medical conditions that may limit their enthusiasm for research enrollment (21); these barriers may be further magnified for minoritized individuals facing of multiple sources of disenfranchisement. In addition, a problematic legacy of racism and misconduct within genetics and genomics in particular impedes the trustworthiness of genomic research, particularly for minoritized populations (9).
Diverse and population-representative genomic studies that include individuals from multiple ancestries will ideally augment the ability to identify molecular diagnoses for individuals with rare disease, as has been seen already in research on complex traits (22). As such, research GS with a specific focus on broad and diverse enrollment presents a unique opportunity not only to increase genetic diagnosis for under-represented individuals by potentially eliminating access barriers, but also to generate data to improve diagnostic likelihood for all. We therefore present our protocol to implement and study the impact of approaches to address the systemic and structural barriers to genetic testing within the ongoing RGP study, with the goals of diversifying the study population, understanding current barriers to access, and evaluating the priorities and values of historically medically-underserved communities and the existing inequities perpetuated by structural barriers. In this context, we consider the term medically-underserved to encompass people whose access to genomic research has historically been limited due to structural racism, language barriers, economic challenges, or other socio-demographic features.
Methods
Overview of Study Design
The protocol presented within this manuscript represents a novel initiative embedded within the parent RGP study, which was launched in 2017. The experience over the first three years of the RGP study (2017–2020) informed the development of the present protocol, officially launched in 2022, that layers in additional outreach and enrollment support for historically medically-underserved populations, with additional outcomes measures included (as outlined below). This novel protocol was developed with input from key stakeholders across multiple domains, including clinicians, genomic researchers, genetic counselors, and individuals with rare disease or their family members, including partnerships with many patient advocacy organizations. Stakeholder input was ascertained via a variety of approaches, including conversations with clinicians, genetic counselors, and genomic researchers serving medically-underserved populations regarding their perceived barriers to rare disease research enrollment. In addition, the entire research team reviewed and provided feedback on the study design, with particular attention paid to insight from the research staff who directly interface with applicants and participants. Input from individuals with rare disease was also ascertained verbally, through conversation with families who had successfully enrolled in the RGP study.
The three primary aims of this protocol are: 1) to diversify RGP enrollment, 2) understand the process and context of implementing genomic medicine for rare disease diagnosis, and 3) investigate the value of a diagnosis for underserved populations. Inclusion criteria include the presence of a condition that is likely to be monogenic, the ability to provide samples for genetic testing, and living within the United States, in addition to meeting at least one of our criteria of historic underrepresentation (Table 1). Exclusion criteria include a confirmed genetic diagnosis or a condition unlikely to be monogenic. In addition to genomic data (first aim), we collect and analyze data regarding the implementation of our process to diversify RGP enrollment (second aim) and survey data regarding diagnostic impact for participants (third aim).
Table 1.
Underrepresentation in the Rare Genomes Project cohort.
| Category | Proportion prior to the current study |
|---|---|
| Non-white race | 8.0% |
| Hispanic or Latinx ethnicity | 8.5% |
| Limited English proficiency | 0.0%a |
| Household income under the federal poverty line | 5.0% |
| Parental education high school level or less | 3.8% |
| Primary residence in a non-metropolitan area | 15% |
English proficiency was one of the original inclusion criteria for this study
The Rare Genomes Project
Overview:
The typical process of RGP enrollment begins with the affected individual or their parent/guardian self-referring through the application form on our website (Figure 1), often connecting to us via social media or at the recommendation of another individual, patient advocacy organization, or provider who knows about the project. Participants can access additional information by reviewing information on the website, including a brief participation overview video, and by visiting the RGP Facebook page that is maintained by study staff. An internal application review process is then performed by two clinicians (genetic counselors, medical geneticists, or relevant disease-area specialists such as neurologists) to determine the eligibility of these applicants for enrollment. For cases that are accepted, RGP study staff contact the affected individual or parent/guardian to explain the study and obtain informed consent for enrollment in the RGP study protocol from affected individuals and their family members. Finally, blood samples from the affected individuals and biologic parents, when available, are obtained for GS and analysis. If a likely diagnosis is found, the causal variant(s) are confirmed in a CLIA-certified lab (cost covered by the study) and returned through the participant’s local physician. This study framework (including the components below) was maintained for our novel protocol, with additional supports added.
Figure 1. The Rare Genomes Project homepage.
From this website, participants may click a link to self-apply or apply on behalf of their child for consideration of study enrollment.
Consent:
Eligible participants are contacted via phone or email to schedule a consent appointment, which is typically 30–45 minutes in length and held either by phone or video conferencing.
Data and sample collection:
Once enrolled, a blood draw collection kit is sent to the participant’s mailing address to be used at a local phlebotomy site. Medical records are also collected after enrollment to be used at time of analysis of the data.
Sequencing and analysis:
GS is performed by the Genomics Platform at the Broad Institute and analyzed by a team within the Broad Institute Center for Mendelian Genomics.
Reporting and disclosure:
Diagnostic variants and candidates of high clinical suspicion that are approved by RGP leadership (which includes board-certified clinical geneticists and molecular diagnosticians) undergo CLIA-certified confirmation and are returned to the families in coordination with their local provider. If families do not have a local provider comfortable with returning these results, a genetic counseling telemedicine visit is offered. Variant classifications are shared in ClinVar, and de-identified genomic data from all cases will be shared with other researchers through the National Human Genome Research Institute’s AnVIL.
Population and recruitment
The targeted enrollment for this prospective cohort study, based upon the existing RGP framework, is 50 participants who are medically-underserved and currently underrepresented in the RGP cohort as well as in prior studies (23, 24). Specifically, we employ the following criteria: non-white race, Hispanic or Latinx ethnicity, limited English proficiency, household income under the federal poverty line, parental education high school level or less, and primary residence in a non-metropolitan area (Table 1). These criteria are identified in potential participants in multiple ways: RGP applicants are asked to self-identify their race and ethnicity at the time of initial application, or referring clinicians may identify the race and ethnicity for the individuals whom they refer; enrolled participants also provide additional demographic data via our baseline RGP survey (which participants complete online, on paper, or via interview).
To expand our outreach efforts, we work closely with clinicians who care for a large population of medically-underserved individuals, in addition to community partnerships. A short physician referral form that typically takes under 5 minutes to complete has been created for physicians to refer patients who would typically face barriers to participation through the online RGP application process. Additional outreach efforts include establishing relationships with rare disease organizations that focus on increasing access to genomic medicine for minoritized communities.
Implementation of enhanced recruitment and enrollment support
The remote nature of RGP makes participating in the study feasible for those who have access to online resources such as a computer or smartphone, along with internet or cellular data access. However, since flexibility within the RGP study design is needed for medically-underserved populations, we now address multiple barriers to enrollment, including limited English proficiency, difficulty accessing online resources, lack of transportation, and inability to communicate with RGP study staff during normal working hours. These barriers are addressed by implementing the following modifications (Figure 2): (1) Outreach to providers to identify individuals with rare diseases who are unable to access clinical genetic testing. (2) Application assistance by RGP staff for any participants who have difficulty accessing online resources, rather than relying on self-referral via our website. The online application may be completed over the phone with the participant and a Spanish-speaking RGP coordinator or an interpreter for other languages. Alongside this, RGP staff are available outside normal working hours to accommodate participants who need assistance during evenings and weekends; this is important to increase access across all areas of employment, particularly shift workers, who comprise up to a quarter of the U.S. workforce (25). (3) Increased access to interpreter services and translated study materials. While the RGP study initially launched in English and required English language proficiency as an original inclusion criteria, the protocol has been amended to eliminate the English language requirement. The website, recruitment materials, and consent forms have been fully translated into Spanish, and enrollment in other languages is possible through interpreters and short form consent. (4) Mobile phlebotomy services for participants unable to travel to a clinical lab to provide a sample for sequencing (our current phlebotomy partner requires blood draws by 1pm, presenting a potential barrier to access) (5) Finally, as certain families may experience medical mistrust and/or have ambivalence about genetic testing in general, we continue to discuss questions and concerns with prospective participants, which will also inform our process. We anticipate that these strategies will allow us to increase RGP access for medically-underserved populations that are currently underrepresented in our study cohort and in genomic medicine studies in general.
Figure 2. Rare Genomics Project enrollment process, barriers, and proposed solutions.
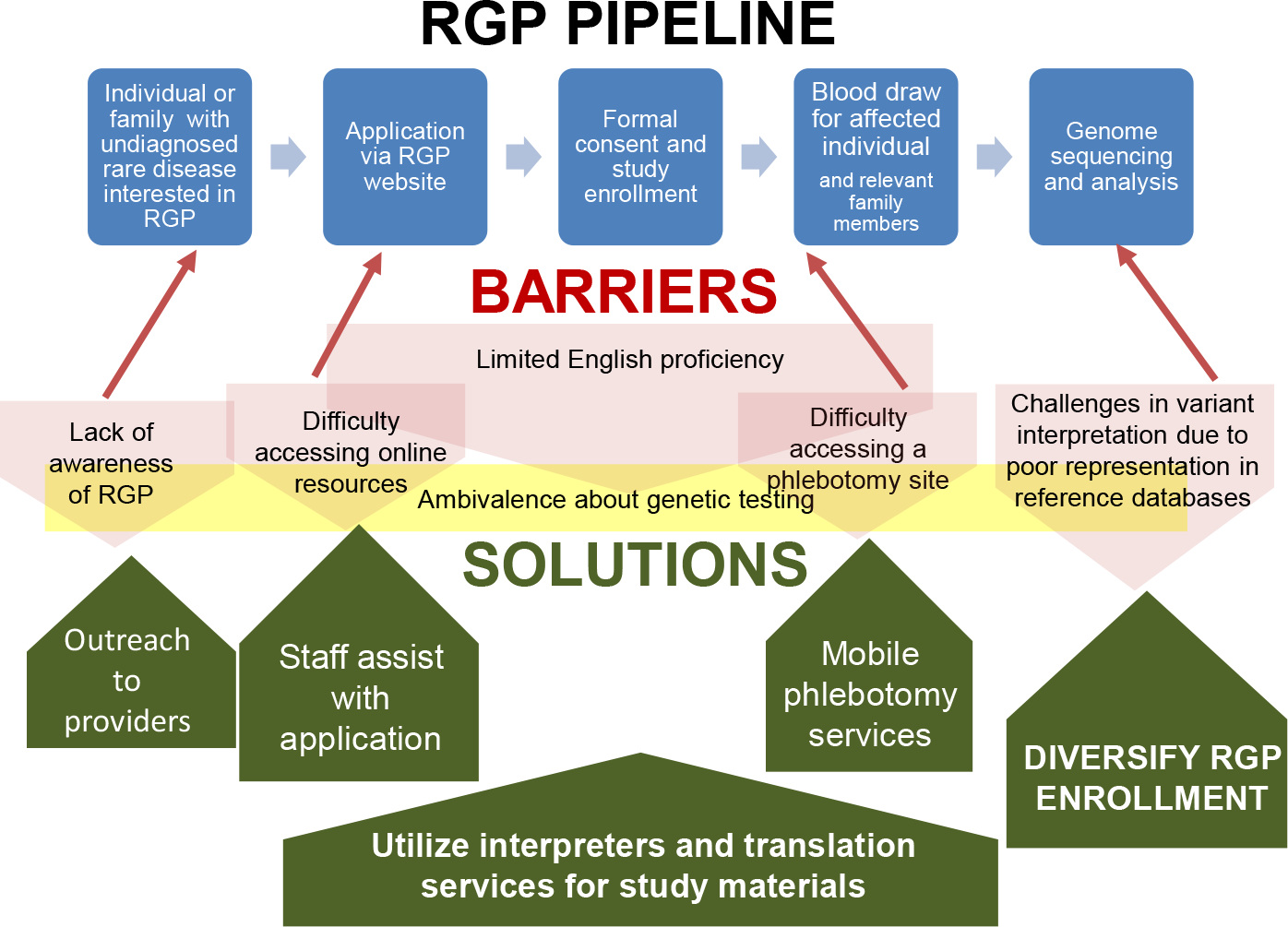
The original process is represented by the blue squares, barriers by the red and yellow shaded shapes, and solutions by the green arrows.
Evaluation of the intervention
To evaluate the process and context of implementing genomic medicine for rare disease diagnosis in diverse populations, we employ a mixed-methods approach informed by Consolidated Framework for Implementation Research (CFIR) constructs as portrayed in a recent taxonomy (26, 27), used to design our outcome measures in order to generate valid empirical data. The implementation outcomes of adoption (measured at the level of the provider) and acceptability (measured at the level of the consumer, in this case, the study participant) are evaluated in this project, where adoption refers to the uptake of the intervention and acceptability refers to how agreeable the intervention is to the participant (26). The referent for these outcomes (the item whose acceptability and adoption is being assessed) will be the RGP process from enrollment to sequencing, analysis, and return of results (Figure 3).
Figure 3. The Rare Genomes Project process, interventions to improve diversity, and implementation outcomes measures.
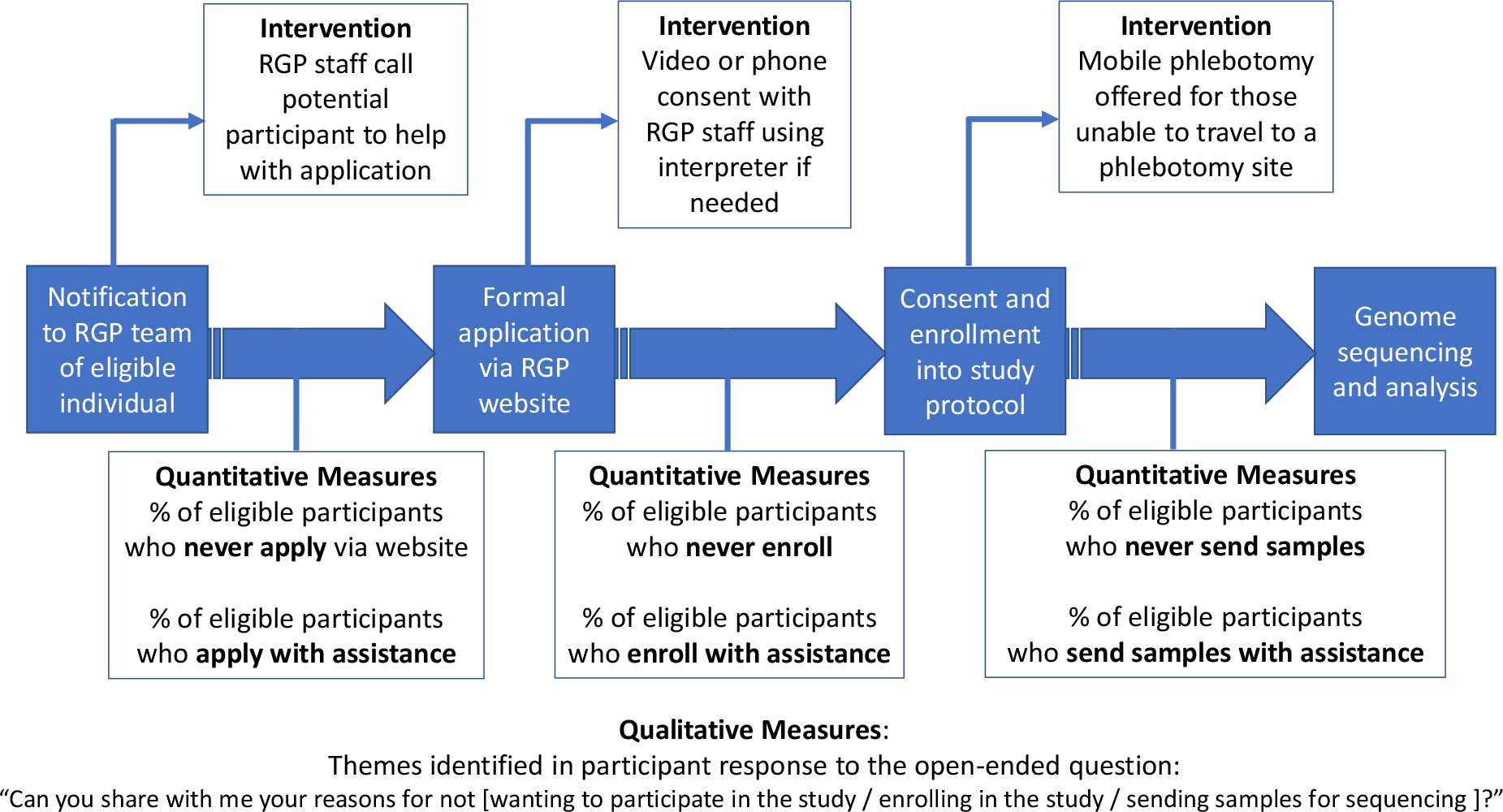
Interventions are presented in text boxes above the current workflow (in blue), and outcomes measures are presented in text boxes below.
We employ a mixed-methods approach, in a convergent design, to evaluate participant features related to these outcomes of adoption and acceptability and the association with completion of the RGP process (Figure 3). Our quantitative measures describe the proportions of individuals who are able to progress through the various stages of the study. For the qualitative component of this protocol, RGP study staff contact prospective participants who have been referred but do not apply via our website, who apply but do not complete enrollment, or who have enrolled but do not provide samples for sequencing (see below). As the study progresses, we will use this information to address unforeseen barriers that arise.
Referral/application stage: We note the number of individuals who complete this process with assistance or who do not proceed further. Those who elect not to proceed further will be offered the opportunity to share their experience in an open-ended manner.
Enrollment stage: We note the number of prospective participants who decline enrollment and those who require the assistance of interpreter services to enroll. Those who decline enrollment will be asked to share their reasoning as above.
Sample collection stage: If a sample has not been received three months after enrollment, we contact the participant or family to determine whether mobile phlebotomy would be useful. If the participant would like mobile phlebotomy services, we arrange for this and contact participants again if samples have not been received three months after mobile phlebotomy is offered. We note the number of participants who require mobile phlebotomy to provide samples and those who never provide samples. Those who decline to provide samples at any time will be asked to share their reasoning as above.
In this way, adoption is assessed by determining the proportion of referrals/applicants that do not enroll in the study or enroll but do not submit samples for sequencing within the first six months after enrollment. Acceptability is assessed by evaluating the reasons that potential participants do not proceed from referral to receipt of samples for GS and the motivations for pursuing a diagnosis.
Understanding priorities of study participants
To evaluate the priorities, values, and impact of a diagnosis for underrepresented individuals and families, we developed baseline and follow-up survey tools (Supplement) that address multiple domains of diagnostic impact and are informed by CFIR constructs (Table 2). This survey has been in use within RGP for the past year and has been well-received by study participants, with 62% completing the baseline survey and 89% completing follow-up surveys. Surveys are administered to enrolled participants or their parents/guardians and are available in English and Spanish. For additional languages, we offer verbal administration of our surveys using interpreter services. RGP participants or their parents/guardians are also asked to complete the Perceived Stress Scale (PSS), a validated instrument that is available in multiple languages (28). The PSS is a 10-item survey that asks participants to respond on a 5-point Likert scale from “never” to “very often” regarding certain thoughts and feelings over the past month. A total score is calculated, with lower scores reflecting less stress, and comparisons may be made to population averages.
Table 2.
Domains included in study surveys at baseline and follow-up
| Variable Domains | Baseline | Follow-up | CFIR Construct |
|---|---|---|---|
| Participant Characteristics | Personal attributes | ||
| Socio-demographicsa | X | ||
| Utility of Genetic Diagnosis | Knowledge and Beliefs about the Intervention | ||
| Overall importance | X | X | |
| Understanding | X | X | |
| Responsibility | X | X | |
| Worry | X | X | |
| Preparation for the future | X | X | |
| Better treatment | X | X | |
| Social support | X | X | |
| Indirect healthcare costs | X | X | |
| Decision to have more children | X | X | |
| Planning for next pregnancy | X | X | |
| Perceived Stress Scale | X | X | Personal Attributes |
including race, ethnicity, household income, education level
The RGP baseline survey and PSS are offered upon enrollment, and the follow-up survey and PSS are repeated at 3- and 12-months post diagnosis (if one is found via GS) to evaluate changes in priorities, values and stress level. Surveys may be completed online via REDCap (29), on paper via mail, or administered verbally via an interview for those who are unable to access the online or mailed version.
Analysis
Genomic analysis:
We analyze the GS data to identify molecular diagnoses under the assumptions that the affected individual has a severe, rare, Mendelian condition. Our interpretation of gene-disease association is guided by the Clinical Genomic Resource (ClinGen) framework (30) and variants are classified according to criteria defined by the American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) (31). We return pathogenic, likely pathogenic, and variants of uncertain significance (VUS) of high interest in established disease genes, along with VUSs of high interest in candidate disease genes. We consider a case “solved” if a variant is clinically validated and the local physician, if involved, interprets the variant as causal.
Analysis of diagnostic utility:
Future analyses will compare the diagnostic yield in our target cohort of 50 RGP participants who qualify as underserved or historically underrepresented (based upon our six criteria outlined in Table 1) to the yield in the larger RGP cohort. Additional analyses will include a) the proportion of diagnoses requiring GS (versus those that could have been found using other common clinical tests such as chromosomal microarray, karyotype, or targeted gene panel), b) the proportion of diagnoses involving previously-known versus novel disease genes and previously-known versus novel variants, and c) the phenotypic characteristics of each population. Phenotypes are characterized using the Human Phenotype Ontology (HPO) (32), and we will evaluate for differences in number and types of HPO terms associated with the underserved and non-underserved cohorts.
Analysis of intervention success:
At the conclusion of this study, we will compare the overall proportions of RGP participants across the entire study period who fall into our six measured socio-demographic categories both before and after the implementation of the described measures. This will indicate whether or not we have achieved our goal of increasing RGP diversity. Subsequent analyses will focus on the process and context of our implementation measures, evaluating the implementation outcomes of adoption and acceptability of GS for a diverse cohort of individuals with rare disease (Table 3) as previously outlined, using descriptive statistics as well as an mixed deductive and inductive analysis of qualitative data in an approach based in grounded theory.
Table 3.
Implementation Outcomes
| Domain | Quantitative Measures | Qualitative Measures |
|---|---|---|
| Adoption | Proportion of referred participants who fully enroll Proportion of enrolled participants who are sequenced Proportion of participants belonging to at least one underserved category |
|
| Acceptability | Differences in motivations for pursuing GS and in perceived impact of genetic diagnosis | Reasons not to participate and free-text responses on RGP survey |
Analysis of psychosocial impact:
The primary outcome identified from survey data is the perceived overall importance of a genetic diagnosis, measured on a 5-point Likert scale (“not at all important” to “extremely important”). Secondary outcomes include other domains of diagnostic utility, also measured on a 5-point Likert scale, in addition to the mean perceived stress score determined by the 10-item PSS (28). We will determine the effects of participant characteristics on the primary outcome by performing a multiple linear regression analysis with perceived importance of a genetic diagnosis as the dependent variable and socio-demographic features as covariates. We will test variables individually and construct regression models to evaluate the interaction between these and other covariates such as clinical features of the affected child. For our secondary outcomes, we will similarly evaluate the influence of sociodemographic features on the various domains of perceived diagnostic utility and on the perceived stress score. Additionally, we will evaluate the impact of a diagnosis by comparing the domains of diagnostic utility and perceived stress at baseline to these same features both immediately (3 months) and longer-term (12 months) post-diagnosis for those who receive a molecular genetic diagnosis.
Anticipated Results:
The first goal of this study is to increase genetic diagnoses for medically-underserved populations through a personalized, supportive approach to participation and targeted outreach to populations currently underrepresented in RGP. Thus, a clear benchmark for success for this study will be an increase in the racial, ethnic, and socioeconomic diversity of the RGP study cohort. Our framework conceptualizes underrepresentation as a result of structural and systemic discrimination. As such, we hypothesize that there is genetic basis of molecular underdiagnosis in underrepresented populations. In fact, we hypothesize that the diagnostic yield for our diverse and previously-underrepresented cohort will be higher than the diagnostic yield for the remainder of the RGP participants due to referral bias: we have previously demonstrated in the outpatient pediatric genetics setting that the diagnostic yield for families with limited English proficiency was higher than the diagnostic yield for English-speaking families, attributable to more severe phenotypes seen in the patients with limited English proficiency who were able to access our clinic despite the language barriers (12).
A second goal is to evaluate the process and context of implementing genomic medicine for rare disease diagnosis in diverse populations. A crucial outcome from this study will therefore be an analysis of the process involved in successful rare disease research enrollment, sample acquisition, analysis, and return of results for underserved populations, reflected in our defined outcomes of adoption and acceptability. We hypothesize that our results will provide new insight into barriers to and facilitators of rare disease genomic research that will inform future approaches.
Our final goal is to evaluate the priorities, values and impact of a diagnosis for underrepresented individuals and families. We will describe the motivations for seeking a diagnosis and diagnostic impact (both psychosocial and related to healthcare utilization) in this cohort of 50 individuals with comparison to our overall RGP study population. We hypothesize that the current overrepresentation of white, well-resourced participants is the result of access barriers rather than differences in the perceived benefit of a genetic diagnosis, though prior research suggests that perceived utility of genetic testing may vary by cultural context (33).
Discussion:
The Rare Genomes Project is an innovative study related to the ability for prospective participants to self-refer and participate from anywhere in the United States without receiving care at a major academic medical center. In addition, diagnostic findings, as well as promising candidate disease genes, that are identified through GS are returned to RGP participants in coordination with their local medical care team, with support from study genetic counselors. The goal of RGP is to thus empower families with genomic understanding of their own rare conditions. However, due to lack of diversity in genomic research studies for individuals with rare disease, the optimal approach for implementation of genomic medicine, particularly for medically-underserved and minoritized populations, remains unknown and under-realized (34). We therefore present our approach to devote resources that facilitate access for these populations to our ongoing research study, while concurrently analyzing not only the diagnostic, clinical, and psychosocial impact of GS but also the approach itself, via our implementation outcomes.
Like many research studies, RGP was originally designed to be accessible by any person. However, structural barriers including, but not limited to, the need to navigate online resources and speak English likely contributed to the inequitable access to RGP and ultimately led to the lack of diversity within the study, as prior research has demonstrated that language barriers as well as dependence upon referrals from subspecialists, such as clinical geneticists, obstructs access to rare disease research (15). This protocol details our approach to increase our outreach and engagement, including liaisons with clinicians in the community, dedicated support for participants unable to participate online, mobile phlebotomy for those with transportation barriers, expansion of research team availability to accommodate those with long working hours or unable to access medical care during typical business hours, and expanded use of interpreter services. Through this implementation process, we hope to improve the participant experience and make our study accessible to those who would not have otherwise been able to enroll in RGP.
One potential limitation that is not directly addressed in our study design relates to issues of a patient-provider trust. This issue has been extensively studied, particularly in genetic research, and is a major barrier to enrollment of minoritized racial groups in the United States and elsewhere (35, 36). Because enrollment in RGP provides a service to participants in performing GS and identifying diagnoses that have been difficult for them to access previously, we hope that our approach may begin the process of reparative justice to a therapeutic relationship between research and participant that has been marred by past injustice, disenfranchisement, and medical racism. As RGP staff work closely with the individuals and families to help overcome challenges, we hope to build trust with the participants and improve the experience of the diagnostic odyssey. The design of this study is such that even if participation is not increased significantly, we will generate valuable insight into any shortcomings of our implementation process. Future plans to expand upon this protocol include the incorporation of additional semi-structured interviews with study participants to further understand barriers to and facilitators of access.
Prior research suggests that implementation science has potential to address the barriers and challenges around health inequities in genomic medicine (37). However, prior initiatives related to genomic medicine implementation have focused on individual rather than contextual factors and less than 2% invoked an implementation science framework, without which it is difficult for results to be generalizable and informative; additionally, individual-level interventions to address inequity are often unsuccessful (20). Furthermore, the racial and ethnic composition of study populations are often underreported, limiting the ability to contextualize and generalize study outcomes (20). Finally, prior research related to inequities in genomic medicine have focused primarily on cancer or prenatal genetics and have not been centered around rare disease diagnosis in children and adults (38). Therefore, this protocol presented herein seeks to address an understudied area of rare disease genomic research. The interventions designed for this protocol to enhance enrollment support primarily relate to personnel: increasing availability of research staff by hiring research assistants dedicated to enrollment of participants who face logistical barriers, and adding study team members who speak languages other than English for ease of participant communication. As the use of mobile phlebotomy for select participants amounts to a start-up cost of approximately $5,000 with an additional cost per participant of approximately $100, we feel that our approach to diversifying enrollment is easily portable to other genomic research studies. If successful, our results would inform future study designs and would also support the hiring of additional support staff in clinical practice as well in order to support implementation of genomic medicine for historically underserved populations. Overall, our interventions represent more of a culture shift rather than a resource-intensive approach, as they focus on being deliberate and thoughtful about sociodemographic features and addressing barriers that may result in inequitable access.
Conclusions:
We present our approach to address inequities in access to genomic research for individuals with rare disease. Our plan to measure not only diagnostic yield for diverse populations but also to generate empiric data regarding implementation outcomes is crucial to maximize the power of the conclusions from this project as well as to inform further efforts in the field. Our focus on implementation outcomes rather than solely on clinical outcomes provides valuable insight into the context and process of achieving the desired clinical outcomes and will allow for adjustment of approach if the desired clinical outcomes are not achieved. Even a highly effective clinical intervention such as GS may not be successful in “real world” practice if various implementation factors, such as the acceptability and adoption that we are measuring, are not optimized. As precision medicine continues to rapidly develop, ensuring equitable access to these technological advances is paramount.
Supplementary Material
Highlights.
Genomic research studies present a unique opportunity to identify precise diagnoses for individuals with rare disease
Participation in such studies has been marked by a lack of diversity, particularly related to race, ethnicity, and socioeconomic status, leading to inequity in genomic healthcare
Barriers to participation for minoritized populations are poorly understood
Implementation-focused research presents an opportunity to understand and address these barriers to participation
Acknowledgements:
The authors would like to express their gratitude to all the families who participate in our ongoing research.
Disclosure of Funding Support:
This work was supported by grant number 2020-224274 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, NIH/NHGRI R21HG012397, and U01HG011755 and funding by Illumina. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study sponsors had no role in the study design, collection, analysis, and interpretation of data, writing of the manuscript, or the decision to submit the manuscript for publication.
Footnotes
Declaration of Interest
MHW has served as a consultant to Illumina and Sanofi. HLR has received research funding from Illumina and Microsoft and serves on the scientific advisory board for Genome Medical. This work was supported by grant number 2020-224274 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, NIH/NHGRI R21HG012397, and U01HG011755 and funding by Illumina. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study sponsors had no role in the study design, collection, analysis, and interpretation of data, writing of the manuscript, or the decision to submit the manuscript for publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bamshad MJ, Nickerson DA, Chong JX. Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am J Hum Genet. 2019;105(3):448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posey JE, O’Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Coban Akdemir ZH, et al. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet Med. 2019;21(4):798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spillmann RC, McConkie-Rosell A, Pena L, Jiang YH, Schoch K, Walley N, et al. A window into living with an undiagnosed disease: illness narratives from the Undiagnosed Diseases Network. Orphanet J Rare Dis. 2017;12(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McConkie-Rosell A, Hooper SR, Pena LDM, Schoch K, Spillmann RC, Jiang YH, et al. Psychosocial Profiles of Parents of Children with Undiagnosed Diseases: Managing Well or Just Managing? J Genet Couns. 2018;27(4):935–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert PL, Sisk JE, Howell EA. When does a difference become a disparity? Conceptualizing racial and ethnic disparities in health. Health Aff (Millwood). 2008;27(2):374–82. [DOI] [PubMed] [Google Scholar]
- 6.Hussain SB, Quittner AL, Brown M, Li-Rosi AM. Understanding access to genomics in an ethnically diverse south Florida population: A comparison of demographics in odyssey and rapid whole genome sequencing programs. J Genet Couns. 2020;29(4):553–61. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JS, Robinson JO, Diamond PM, Bharadwaj A, Christensen KD, Lee KB, et al. Patient understanding of, satisfaction with, and perceived utility of whole-genome sequencing: findings from the MedSeq Project. Genet Med. 2018;20(9):1069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walley NM, Pena LDM, Hooper SR, Cope H, Jiang YH, McConkie-Rosell A, et al. Characteristics of undiagnosed diseases network applicants: implications for referring providers. BMC Health Serv Res. 2018;18(1):652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Angelo CS, Hermes A, McMaster CR, Prichep E, Richer E, van der Westhuizen FH, et al. Barriers and Considerations for Diagnosing Rare Diseases in Indigenous Populations. Front Pediatr. 2020;8:579924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraiman YS, Wojcik MH. The influence of social determinants of health on the genetic diagnostic odyssey: who remains undiagnosed, why, and to what effect? Pediatr Res. 2021;89(2):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muenke M, Adeyemo A, Kruszka P. An electronic atlas of human malformation syndromes in diverse populations. Genet Med. 2016;18(11):1085–7. [DOI] [PubMed] [Google Scholar]
- 12.Wojcik MH, Bresnahan M, Del Rosario MC, Ojeda MM, Kritzer A, Fraiman YS. Rare diseases, common barriers: disparities in pediatric clinical genetics outcomes. Pediatr Res. 2022;93(1):110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuter CM, Kohler JN, Bonner D, Zastrow D, Fernandez L, Dries A, et al. Yield of whole exome sequencing in undiagnosed patients facing insurance coverage barriers to genetic testing. J Genet Couns. 2019;28(6):1107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, et al. Genetic Misdiagnoses and the Potential for Health Disparities. N Engl J Med. 2016;375(7):655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young JL, Halley MC, Anguiano B, Fernandez L, Bernstein JA, Wheeler MT, et al. Beyond race: Recruitment of diverse participants in clinical genomics research for rare disease. Front Genet. 2022;13:949422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landry LG, Rehm HL. Association of Racial/Ethnic Categories With the Ability of Genetic Tests to Detect a Cause of Cardiomyopathy. JAMA Cardiol. 2018;3(4):341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonkowsky JL, Wilkes J, Bardsley T, Urbik VM, Stoddard G. Association of Diagnosis of Leukodystrophy With Race and Ethnicity Among Pediatric and Adolescent Patients. JAMA Netw Open. 2018;1(7):e185031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omorodion J, Dowsett L, Clark RD, Fraser J, Abu-El-Haija A, Strong A, et al. Delayed diagnosis and racial bias in children with genetic conditions. Am J Med Genet A. 2022;188(4):1118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odgis JA, Gallagher KM, Suckiel SA, Donohue KE, Ramos MA, Kelly NR, et al. The NYCKidSeq project: study protocol for a randomized controlled trial incorporating genomics into the clinical care of diverse New York City children. Trials. 2021;22(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts MC, Kennedy AE, Chambers DA, Khoury MJ. The current state of implementation science in genomic medicine: opportunities for improvement. Genet Med. 2017;19(8):858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConkie-Rosell A, Spillmann RC, Schoch K, Sullivan JA, Walley N, McDonald M, et al. Unraveling non-participation in genomic research: A complex interplay of barriers, facilitators, and sociocultural factors. J Genet Couns. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins BD, Fischer CG, Polito CA, Maiese DR, Keehn AS, Lyon M, et al. The 2019 US medical genetics workforce: a focus on clinical genetics. Genet Med. 2021;23(8):1458–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiese DR, Keehn A, Lyon M, Flannery D, Watson M, Working Groups of the National Coordinating Center for Seven Regional Genetics Service C. Current conditions in medical genetics practice. Genet Med. 2019;21(8):1874–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.United States Department of Labor: Bureau of Labor Statistics. [Accessed 5 May 2023];Workers on flexible and shift schedules in May 2004. http://www.bls.gov/cps/ [Google Scholar]
- 26.Proctor E, Silmere H, Raghavan R, Hovmand P, Aarons G, Bunger A, et al. Outcomes for implementation research: conceptual distinctions, measurement challenges, and research agenda. Adm Policy Ment Health. 2011;38(2):65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damschroder LJ, Aron DC, Keith RE, Kirsh SR, Alexander JA, Lowery JC. Fostering implementation of health services research findings into practice: a consolidated framework for advancing implementation science. Implement Sci. 2009;4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav. 1983;24(4):385–96. [PubMed] [Google Scholar]
- 29.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42(2):377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100(6):895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine JP, et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halley MC, Young JL, Fernandez L, Kohler JN, Undiagnosed Diseases N, Bernstein JA, et al. Perceived utility and disutility of genomic sequencing for pediatric patients: Perspectives from parents with diverse sociodemographic characteristics. Am J Med Genet A. 2022;188(4):1088–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fatumo S, Chikowore T, Choudhury A, Ayub M, Martin AR, Kuchenbaecker K. A roadmap to increase diversity in genomic studies. Nat Med. 2022;28(2):243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shavers VL, Lynch CF, Burmeister LF. Racial differences in factors that influence the willingness to participate in medical research studies. Ann Epidemiol. 2002;12(4):248–56. [DOI] [PubMed] [Google Scholar]
- 36.Murthy VH, Krumholz HM, Gross CP. Participation in cancer clinical trials: race-, sex-, and age-based disparities. JAMA. 2004;291(22):2720–6. [DOI] [PubMed] [Google Scholar]
- 37.Roberts MC, Mensah GA, Khoury MJ. Leveraging Implementation Science to Address Health Disparities in Genomic Medicine: Examples from the Field. Ethn Dis. 2019;29(Suppl 1):187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Southwick SV, Esch R, Gasser R, Cragun D, Redlinger-Grosse K, Marsalis S, et al. Racial and ethnic differences in genetic counseling experiences and outcomes in the United States: A systematic review. J Genet Couns. 2020;29(2):147–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


