Abstract
Understanding the origins of past and present viral epidemics is critical in preparing for future outbreaks. Many viruses, including SARS-CoV-2, have led to significant consequences not only due to their virulence, but also because we were unprepared for their emergence. We need to learn from large amounts of data accumulated from well-studied, past pandemics and employ modern informatics and therapeutic development technologies to forecast future pandemics and help minimize their potential impacts. While acknowledging the complexity and difficulties associated with establishing reliable outbreak predictions, herein we provide a perspective on the regions of the world that are most likely to be impacted by future outbreaks. We specifically focus on viruses with epidemic potential, namely SARS-CoV-2, MERS-CoV, DENV, ZIKV, MAYV, LASV, noroviruses, influenza, Nipah virus, hantaviruses, Oropouche virus, MARV, and Ebola virus, which all require attention from both the public and scientific community to avoid societal catastrophes like COVID-19. Based on our literature review, data analysis, and outbreak simulations, we posit that these future viral epidemics are unavoidable, but that their societal impacts can be minimized by strategic investment into basic virology research, epidemiological studies of neglected viral diseases, and antiviral drug discovery.
Keywords: viral outbreaks, epidemics, influenza, coronavirus, Mayaro, Oropouche
While future viral outbreaks are unavoidable, we should be able to forecast the time and location of the outbreaks and minimize their potential impacts through consistently funded research in the fields of virology and antiviral drug discovery. Visual representation of countries at risk for originating new viral outbreaks.
Introduction
Despite the rapid development and distribution of vaccines (Excler et al. 2021), the world is still grappling with SARS-CoV-2, which has infected an estimated 761 million people worldwide in 3 years after the pandemic begun. While we continue to grapple with the consequences of this most recent pandemic, it is important to learn from the lessons of history so that we are better prepared for likely unavoidable future viral outbreaks.
Throughout the 20th century, several strains of deadly influenza hit the world. The most severe, the 1918 influenza pandemic, infected 500 million people and killed an estimated 20–50 million people worldwide (CDC 2019a). In the last 40 years, several viruses have caused global epidemics that have substantially affected humankind including (but not limited to) human immunodeficiency virus (HIV), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), Ebola virus, Zika virus (ZIKV), and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Morse et al. 2012). This also includes the currently emerging concerns about the monkeypox virus outbreak, lack of treatments, and its continued spread (Centers for Disease Control and Prevention, CDC 2022a). Factors like expanding mosquito-borne viral diseases and climate change (Reperant and Osterhaus 2017) contribute to sincere concerns about what virus is to come next, and whether we are prepared to prevent future viral outbreaks (Gates 2020). Some groups attribute the increase in viral epidemics to human population growth (Bloom et al. 2017, Spernovasilis et al. 2021, Baker et al. 2021). Historical modeling predicts that there is a 22%–28% chance of a COVID-19 scale viral outbreak again in the next 10 years, and a 47%–57% chance of such an outbreak over the next 25 years, given the yearly probability of 2.5%–3.3% (Agyarko et al. 2023). These estimates emphasize the need for antiviral drug discovery and vaccine development research to lessen the impact of future pandemics.
There is a published history of potential epidemic monitoring well before COVID-19. For example, in 1970, annual global rates of the 1968 Hong Kong influenza were being monitored by many groups (Gill et al. 1971, Mandin et al. 1971, Salim 1971, Gill and Murphy 1972, Jackson et al. 2010) and, in 1999, Sandra Keavey, a practicing physician’s assistant, predicted that the next influenza outbreak would be a pandemic in her published notice to clinicians (Keavey 1999). In 1997, Andrick et al. (1997) discussed the prediction of viral outbreaks as it relates to climate change. In The Coming Plague, Laurie Garrett discussed the epidemiological past and outbreaks of many viruses, including Lassa virus (LASV) and Ebola virus, in addition to their threatened impact on the future of modern society (Garrett 1994). More recent projects such as the Global Virome Project (Carroll et al. 2018) and PREDICT (UC Davis 2009) aimed to analyze all available public health data on every possible zoonotic virus in the hopes of predicting and preparing for the next outbreak. The PREDICT project uncovered nearly 1000 novel viruses but, unfortunately, was shut down just prior to the emergence of the SARS-CoV-2 pandemic (Global Biodefense Staff 2020). Similarly, tools such as HealthMap (https://www.healthmap.org/en/), aggregate all viable resources to locate small-scale, unnoticed outbreaks before they escalate to endemics or pandemics.
It is imperative to develop pre-emptive strategies to deal with new viruses as they emerge to mitigate future outbreaks (Boston’s Children Hospital researchers 2006, Myers et al. 2000). Recently, the Director General of the World Health Organization’s reinforced that humankind must be better prepared for the next pandemic, since outbreaks are “facts of life,” and called on countries to invest more in public health (Reuters Staff 2020). In this perspective, we organize and summarize the various methodologies and outcomes of previous predictions of viral outbreaks in the literature. We aim to identify regions of the world where the next outbreaks are more likely to occur and summarize developments that can help us prepare for such outbreaks.
Below, we consider the factors responsible for the emergence, spread, and eradication of both current and previous viruses including SARS-CoV, MERS-CoV, Ebola virus, ZIKV, SARS-CoV-2, Dengue virus (DENV), Mayaro virus (MAYV), LASV, noroviruses, influenza, Nipah virus, hantaviruses, Oropouche virus (OROV), and Marburg virus (MARV). We collate information about human viral pathogen infection predictions and provide a perspective on the next outbreaks and regions of the world where these viruses are most likely to emerge. Establishing the value and accuracy of outbreak predictions is crucial to minimizing the impact of novel outbreaks given the paucity of available therapeutics for most viruses (Bobrowski et al. 2020). Currently, few approved antiviral drugs are available to treat the viral infections reviewed herein: a handful of drugs are available to treat influenza yet are susceptible to the development of resistance; EUA drugs and vaccines and a few repurposed FDA-approved drugs are available to treat SARS-CoV-2; and there is one antibody treatment against Ebola (US Food and Drug Administration, FDA 2023, National Institute of Allergy and Infectious Diseases, NIAID 2017, Centers for Disease Control and Prevention, CDC 2021a). Although current approaches cannot forecast the exact time and place of the new outbreak, based on systematic literature review, data analysis, and outbreak simulations, we anticipate that the next viral epidemics are unavoidable. We summarize the existing outbreak predictions, current and potential treatments, including both molecular therapeutics and vaccines, and emphasize the importance of continued funding of both virology and antiviral drug discovery research as critical means to minimize the burden of future pandemics on mankind.
A tale of the outbreaks
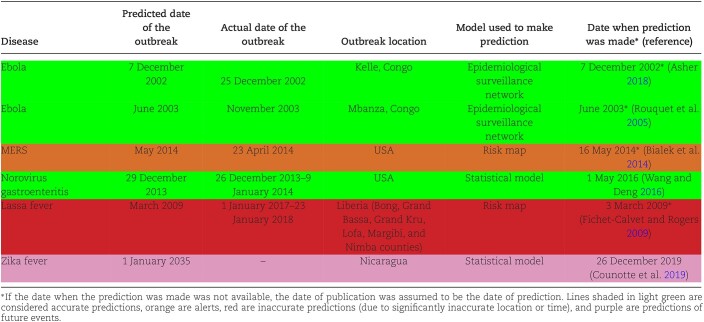
In the discussion that follows, we will reference both the first event and the occurrence of disease cases above average expectancy as an “outbreak.” However, as we analyze the outbreak forecasts retrospectively, it is crucial to understand which event(s) qualify as an outbreak and which do not. For example, the first case of MERS-CoV in the USA occurred on 1 May 2014 (2 months after it was rampant in the Middle East in March 2014), but the outbreak did not begin until 16 May 2014 (Bialek et al. 2014). According to the CDC, an epidemic refers to “an increase, often sudden, in the number of cases of a disease above what is normally expected in that population in that area,” but an outbreak is typically considered an epidemic in a geographically confined area (Centers for Disease Control and Prevention, CDC 2022b). Thus, throughout this paper, we used both terms, i.e. “epidemic” and “outbreak” following these definitions. The use of these definitions in the literature is variable and depends entirely on the severity of the disease, location, frequency, and spread of the virus. Therefore, the term “outbreak” may be frequently used inappropriately or out of context by many of the sources referenced in this review. In some situations, a single case can constitute an outbreak, while in others, thousands of cases should be reported to constitute an outbreak (Association of Professionals in Infection Control and Epidemiology, APIC 2023). Discrepancies like this were evident in our literature search as well, where some viruses had multiple models, some had simulations, and some had no more than observations from past outbreaks. Several groups have tried to develop outbreak prediction models for a variety of viruses, albeit with limited success. In Table 1, the dates and information associated with these predictions are charted. Most of these models proved to be inaccurate or were reported retrospectively. However, some of these efforts can still be useful as warnings and/or observations. For instance, epidemiological surveillance networks monitoring Ebola in the Congo (Rouquet et al. 2005, Asher 2018) and norovirus in the USA (Wang and Deng 2016) resulted in accurate predictions of the outbreak occurrences. These networks’ successes were largely due to their foundations in infection tracing, described below when we discuss Ebola virus and norovirus. Given the last-minute circumstances of the MERS prediction (Bialek et al. 2014), this aligns more to an alert than a prediction, and both the Zika (Counotte et al. 2019) and Lassa fever (Fichet-Calvet and Rogers 2009) predictions were too far removed to determine their accuracy. In all, there is an acute difficulty in accurately assessing when a new outbreak will appear. Furthermore, in some cases (such as with Ebola), even forewarning was not enough to prevent or mitigate an outbreak. Many factors in outbreak models are difficult to assess, especially with little knowledge of viral transmission and pathogenesis; factors such as host–vector transmissibility can be difficult to model due to the vastness of the global virome (Albery et al. 2021). In Table 2, we describe the molecular biology behind each of the viruses, and in the following sections we provide detailed discussions of their outbreak risk based on data and literature analysis. Because different past outbreaks received varying levels of attention and scrutiny, the depth and amount of information discussed below will be different for each virus, with the most elaborate discussion being on the current COVID-19 pandemics.
Table 1.
Summary of predicted outbreaks.
Table 2.
Molecular biology, disease characteristics, and timelines of virus discovery and outbreak.
| Virus | Genome organization | First confirmed isolation | First major outbreak | Latest outbreak |
|---|---|---|---|---|
| SARS-CoV-2 | Spherical, enveloped virus with single-strand, linear, positive-sense RNA genome (∼29.9 Kb) (Naqvi et al. 2020) | December 2019—Wuhan, China (Worobey et al. 2022) | December 2019–January 2020—Wuhan, China (Worobey et al. 2022) | December 2019–January 2020—Wuhan, China (Worobey et al. 2022) |
| SARS-CoV | Spherical, enveloped virus with single-strand, positive-sense RNA genome (∼29.7 Kb) (Xu et al. 2003) | November 2002—Foshan, Guangdong, China (Chinese Law and Government 2014) | February 2003—Guangdong, China (WHO 2009) | January 2004—Guangdong, China (Gralinski and Baric 2015) |
| MERS-CoV | Spherical, enveloped virus with single-strand, positive-sense RNA genome (∼29.9 Kb) (Chu et al. 2014) | June 2012—Jeddah, Saudi Arabia (van Boheemen et al. 2012) | April 2012—Zarqa, Jordan (Hijawi et al. 2013) | Early 2019—Saudi Arabia, occasional cases reported up to March 2022 (World Health Organization) |
| Ebola virus | Filamentous, enveloped virus with single-strand, negative-sense RNA genome (∼19.0 Kb) (Bharat et al. 2012) | June 1976—Nzara, Sudan (Feldmann et al. 2003) | June–August 1976—Sudan and Democratic Republic of the Congo (Feldmann et al. 2003) | June 2021—Guinea (World Health Organization 2021) |
| ZIKV | Spherical, enveloped virus with single-strand, positive-sense RNA genome (∼10.8 Kb) (Wang et al. 2017) | April 1947—Uganda (isolated from monkeys) (Dick et al. 1952) | 1952—Uganda and United Republic of Tanzania (Robinson 2016) | November 2021—Uttar Pradesh, Kerala, and Maharashtra, India (Jha 2021) |
| DENV | Icosohedral, enveloped virus with single-strand, positive-sense RNA genome (∼10.7 Kb) (Kuhn et al. 2002) | August 1942—Nagasaki, Japan (Takasaki 2011) | 1635—Martinique and Guadeloupe, Caribbean (suspected DENV outbreak) (Dick et al. 2012) | Endemic to numerous, mainly tropical, areas with cases still being reported worldwide January–March 2023—Europe, Asia, Africa, and South America (Boston’s Children Hospital researchers 2006, Bolivian hospitals under strain as dengue kills dozens; Associated Press 2023). |
| MAYV | Icosohedral, enveloped virus with single-strand, positive-sense RNA genome (∼10.7 Kb) (Ribeiro-Filho et al. 2021) | August, 1954—Trinidad, Caribbean (Anderson et al. 1957) | August, 1954—Trinidad, Caribbean (Anderson et al. 1957) | October 2020—Cayenne, French Guiana (World Health Organization 2020b) |
| LASV | Round, enveloped virus with two single-strand, ambisense RNA segments (∼3.4 Kb [S] and ∼7.0 Kb [L]) (Hass et al. 2004) | 1969—Lassa, Nigeria (Monath 2019) | 1969—Nigeria (Monath 2019) | Ongoing outbreak (as of June 2022)—Nigeria (Tolu-Kolawole 2022) |
| Norovirus | Icosohedral, nonenveloped virus with single-strand, positive-sense RNA genome (∼7.5 Kb) (Chan et al. 2017) | August 1972—Norwalk, Ohio (Kapikian et al. 1972) | Suspected outbreaks since 1940s, (Kapikian et al. 1972) first confirmed in 1968—Norwalk, Ohio (The inexorable progress of norovirus 2013) | Ongoing outbreak (as of June 2022)—British Colombia, Alberta, Manitoba, and Ontario, Canada (Entis 2022) |
| Influenza virus | Filamentous or spherical, enveloped virus with eight (influenza A and B) or seven (influenza C) single-strand, negative-sense RNA segments (∼2.3, 2.3, 2.2, 1.8, 1.6, 1.4, 1.0, and 0.9 Kb segments in H1N1 influenza A) (Bouvier and Palese 2008) | Isolated from ferrets in 1933—National Institute for Medical Research at Mill Hill, London, England (Smith et al. 1933) | First agreed upon influenza pandemic began in 1729, but reports of outbreaks with influenza-like symptoms date back to 1173 (Potter 2001) | Seasonal influenza cases reported worldwide yearly. Last major global outbreak was during 2009–2010 H1N1 pandemic (Centers for Disease Control and Prevention, CDC 2021b) |
| Nipah virus | Spherical, enveloped virus with single-strand, linear, negative-sense RNA genome (∼18.2Kb) (Eaton et al. 2006) | March 1999—Kampung Sungei Nipah, Malaysia (Chua 2012) | September 1998—Ipoh, Malaysia (Chua 2012) | September 2021—Kerala, India (Yadav et al. 2022) |
| Hanta virus | Spherical, enveloped virus with three single-strand, negative-sense RNA segments (∼1.8–2.1, 3.7–3.8, and 6.5–6.6 Kb segments) (Hepojoki et al. 2012) | March 1978—Seoul, South Korea (Lee et al. 1978) | 1951—Korean Peninsula (Muranyi et al. 2005), but English “sweate” outbreak of 1485 is suspected (Bridson 2001) | May 2022—Palos Blancos, Bolivia (Herriman 2022) |
| OROV | Spherical, enveloped virus with three single-strand, negative-sense RNA segments (∼6.9, 4.4, and 1.0 Kb segments) (da Rosa et al. 2017) | 1955—Vega de Oropouche, Trinidad and Tobago (Anderson et al. 1961; Downs et al. 1961) | 1955—Trinidad and Tobago (Anderson et al. 1961), but first large-scale outbreak in 1961—Belém, Brazil (Azevedo et al. 2007) | August–September 2020—Saül, French Guiana (Organização Mundial da Saúde, World Health Organization 2020a) |
| MARV | Filamentous, enveloped virus with single-strand, negative-sense RNA genome (∼19.1 Kb) (Fujita-Fujiharu et al. 2022) | Pathogen isolated in November 1967—Hamburg, Germany (Slenczka and Klenk 2007) | August 1967—Marburg and Frankfurt, Germany (Slenczka and Klenk 2007) | 23 February 2023—Equatorial Guinea; (Marburg virus disease—Equatorial Guinea; Disease Outbreak News 2023). |
Major viral outbreaks
In the section below, we summarize factors affecting the transmission and outbreak potential, as well as the available models, of SARS-CoV-2, MERS-CoV, DENV, ZIKV, MAYV, LASV, noroviruses, influenza, Nipah virus, hantaviruses, OROV, MARV, and Ebola virus. Brief outbreak histories of each can be found in the Supplementary materials. We acknowledge that not every virus with outbreak potential may be included, but we aimed to include all that are not (i) largely eradicated, (ii) privy to well-accepted preventative treatments, or (iii) vector-borne. The following is a commentary on factors that needed to build successful predictive models.
Coronaviruses
SARS-CoV-2 (2019–2023)
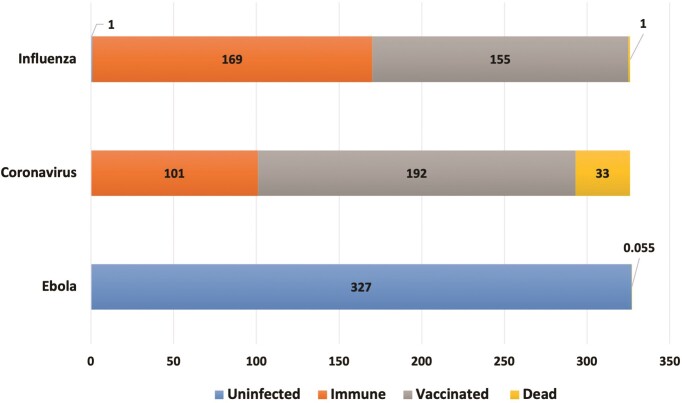
It has been particularly challenging to develop models that capture the effect of increasingly virulent SARS-CoV-2 mutants observed in human populations (Chen et al. 2020, Wang et al. 2020). For example, the B.1.617.2 variant, better known as the Delta variant, is 50% more contagious than the Alpha variant and is 75% more contagious than the original SARS-CoV-2. Thus, Delta also became a prevalent variant for many new COVID-19 cases in 2021 (Katella 2021). The Virus Outbreak Simulator for the USA (https://bioinformaticshome.com/online_software/virus-outbreak/US/index.html#) simulates the spread of select viruses; simulations were available for influenza, coronaviruses, and Ebola (Andrews 2019). We employed this server to run simulations using the settings summarized in Table 3, and the code for the simulation can be found on GitHub (https://github.com/s-andrews/virusbreak). The simulation was set so that the total population was 327 million people and on Day 1, there were 55k carriers of each virus. The coronavirus numbers used by the simulation have been updated daily since December 2019 and were taken from the Institute for Health Metrics and Evaluation (https://covid19.healthdata.org/projections) projections (Institute for Health Metrics and Evaluation 2021).
Table 3.
The input settings and outputs from the Virus Outbreak Simulator. The duration parameter was collected after the simulation ran to completion.
| Parameter | Ebola | Coronavirus | Influenza |
|---|---|---|---|
| Duration (days) | 16 | 126 | 96 |
| Virulence | 2 | 50 | 50 |
| Lethality | 50 | 5 | 0.5 |
| Incubation time | 10 | 5 | 2 |
| Infection time | 5 | 9 | 4 |
| Vaccination (%) | 0 | 60 | 47 |
| Containment | Quarantine and physical distancing | No containment | No containment |
The numeric results of the simulations can be seen in Fig. 1. While vaccinations played a significant role in the number of people predicted to be affected by influenza, only a tiny portion of the population was predicted to die from the infections. Similarly, only a small percentage of the US population was predicted to be infected with Ebola, likely due to the full containment that was in effect for the Ebola simulation. While this severely limited the spread of the infections, all infected persons were predicted to die.
Figure 1.
Number of affected people in the USA based on the simulations of influenza, coronaviruses, and Ebola outbreaks (using https://bioinformaticshome.com/online_software/virus-outbreak/US/index.html#; see text for the input parameters used). All numbers are in millions of people, and the respective colors represent those who would be uninfected, vaccinated (when applicable), immune, and those who would die from the infection.
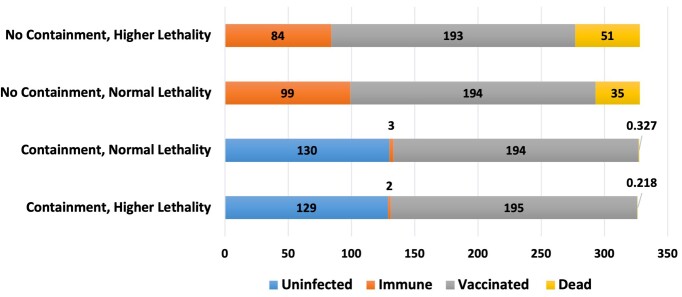
To understand the threat posed by the Delta variant (we launched this study in 2020), we aimed to simulate how it would impact the USA using the simulator under different conditions. Primarily, we varied the lethality of the virus and containment strategies and assumed that 47% of the US population was vaccinated as of 4 July 2021 (Carlsen et al. 2021). We varied the containment, to view the effects of the reopening of the USA and vaccine distribution, and the lethality, to view the differences given the unestablished relationship between the lethality and the roughly 60% increased virulence of the Delta variant (Lovelace Jr. 2021). The input parameters and the results of these simulations are summarized in Table 4 and Fig. 2.
Table 4.
The properties of the simulated Delta variant to hypothesize its effects on a large population, especially comparing lethality and containment efforts.
| Parameter | Containment, higher lethality | Containment, normal lethality | No containment, normal lethality | No containment, higher lethality |
|---|---|---|---|---|
| Duration (days) | 65 | 60 | 124 | 127 |
| Virulence | 80 | 80 | 80 | 80 |
| Lethality | 8 | 5 | 5 | 8 |
| Incubation time | 5 | 5 | 5 | 5 |
| Infection time | 9 | 9 | 9 | 9 |
| Vaccination (%) | 60 | 60 | 60 | 60 |
| Containment | Quarantine and physical distancing | Quarantine and physical distancing | No containment | No containment |
Figure 2.
Number of affected people in the USA based on the simulation of the outbreak of a modified coronavirus emulating the Delta variant of SARS-CoV-2. All numbers are in millions of people, and the respective colors represent those who would be uninfected, vaccinated (when applicable), immune, and those who would die from the infection
These predictions demonstrate how important containment practices like quarantining and social distancing are. Even with increased lethality, containment decreases the number of fatalities by more than 200-fold. Furthermore, increased simulated vaccination significantly decreased the number of deaths (data not shown). Similar studies were run for the Omicron variant. Including the US vaccination status (64%) as of 16 February 2022, we ran the same simulations, only adjusting the settings from our alpha SARS-CoV-2 model by increasing the virulence to 90% and decreasing the lethality to 1%. These percentages were chosen based on the several fold increase in contagiousness (Park 2022) and less than 1% average fatality rate seen in Omicron cases (Arnott 2022). Compared to alpha settings with 64% vaccination, Omicron reduced the duration of the simulated outbreak by 19% and the deaths by 87%. However, with full containment methods both simulated outbreaks ended in 30 days (83% less than the previous alpha simulation) and left the vast majority of the population alive and immune.
Obviously, containment and vaccination are crucial factors in the impacts of SARS-CoV-2 on humanity. Furthermore, the individual specifications of each variant can provide a moving target for therapeutics and models alike. For example, a 3% change in lethality can lead to a 45% increase in deaths when there are no social containment measures. Therefore, rapid changes in containment, vaccination, and viral mutation present major complications to building successful models. Our simulation was limited by using flat vaccination rates as well as poorly defined containment procedures. The flat vaccination rate does not account for those who have been partially vaccinated or any growth in the number of people vaccinated over time. As for the containment, “quarantine” and “physical distancing” are not specific and leave room for interpretation. Notably, IHME COVID-19 data (Institute for Health Metrics and Evaluation 2021) were used to set up the parameters for isolation and social distancing in the simulation. Additionally, the data were collected from a simulator, not real world observations; however, we believe the presentation of the data is highly useful to visualizing the impacts of the parameters discussed. We recognize these limitations and consider it essential to emphasize the importance of interventions and modeling, especially in considering a more lethal virus.
Beyond the current variants of SARS-CoV-2, an impending SARS-CoV-3 should warrant further concern. One study (Wardeh et al. 2021) summarized the data of past transmission cases to highlight the probability of both a novel SARS-CoV outbreak event, as well as novel vector and species transmission. Then, the authors compiled and sequenced viral genomic data to compare it with potential mammalian vectors and generated models predicting each potential coronavirus–mammal association. In doing so, they suggest that dogs, rats, Chinese ferret-badgers, and the Asian palm civet, among more than 100 mammals, should be monitored as major potential host reservoirs for both MERS-CoV and SARS-CoV-2 mutation and transmission (Wardeh et al. 2021). Other studies (Wang et al. 2020, Murray and Piot 2021) have also implied the threat of both continued SARS-CoV-2 and further mutations, but have not built models to predict the future SARS-CoV outbreaks.
MERS-CoV (2012–2014)
Most of the focus on modeling the spread of MERS-CoV revolved around proper data mining and the allocation of patient information. One group of models was based on two key components, the patients’ ages and whether they were symptomatic. These models produced accuracies ranging from 53.6% to 71.58% and suggested older patients being at the highest risk of complications (Al-Turaiki et al. 2016). However, publications describing modeling or forecasting MERS-CoV were extremely limited. We have identified a study describing an epidemiological model that concluded that hospital transmission cases were four times more influential to viral spread than community transmission. Written during the 2014 outbreak, the study suggested efforts to contain the then-current MERS-CoV should be focused on hospitals. They also shared an idea that the weak secondary MERS-CoV transmission removed its “epidemic” status based on the R < 1 where secondary community transmission cases were concerned (Chowell et al. 2014). One group modeled two strains of MERS-CoV to evaluate their epidemic potential and suggest that MERS-CoV had a high risk of developing rapidly in Riyadh and Macca, Saudi Arabia. Through their bilinear, non-monotone, and saturated social behavior models, they retrospectively predicted the number of cases in the 2015–2016 outbreak and used this as their models’ validation. They too emphasized the importance of research given the high mortality rate (35.5%) and complete lack of treatment or prevention (Sardar et al. 2020).
DENV
Many health authorities have taken precautions, like creating clinical networks to monitor DENV based on symptomatology (Gubler 1998). In Australia, predictive models have been used to show correlations between the Southern Oscillation Index (SOI), a measure of sea level air pressure difference between the eastern and western tropical Pacific, and dengue cases. A multivariate Seasonal AutoRegressive Integrated Moving Average model was generated using Queensland Health and Australia’s Bureau of Statistics data from January 1993 and December 2005. It was discovered that dengue fever cases increased as the SOI decreased and temperatures increased; their predictive model values matched observed cases with an error of 1.93% (Hu et al. 2010). Models like these could be pivotal in other at-risk countries if data can be reliably collected. These models also emphasize the importance of including environmental factors in modeling disease outbreaks. This should be of high priority, as there are over 100 countries and 3 billion people worldwide that are at risk of contracting DENV (Centers for Disease Control and Prevention, CDC 2020).
ZIKV
Two major factors have been identified to explain the increasing global spread of the ZIKV: urban transmission and stochastic factors. Experimental studies did not support a genetic variation hypothesis suggesting that an adaptive evolution of ZIKV was responsible for the rapid spread (Gubler et al. 2017). Rather, it is theorized that high levels of viremia in humans facilitates vector-borne transmission (Gubler et al. 2017). Newly infected, immunologically naïve populations were the most likely causes of rapid spread (Gubler et al. 2017), as it is assumed that infected individuals have lifelong immunity. Given the 2015–2016 epidemic timeline, the next outbreak is predicted to occur around 2035 (see Table 1), as this is when the virus would be introduced to naïve populations in the Americas; the chance for outbreaks goes above 50% in 2047 (Counotte et al. 2019). It is thought young women of reproductive age will be at the most risk for ZIKV in the future (Counotte et al. 2019) due to the particularly dangerous threat Zika poses to pregnant persons and their unborn children. However, we could not identify any reliable simulations or models that could confidently predict future ZIKV spread.
MAYV
Lack of an effective vaccine and documented spread to new regions increase the global risk of MAYV outbreaks. Additionally, there has been movement of the outbreaks from primarily South American countries (Caicedo et al. 2021) to North America and Europe (Acosta-Ampudia et al. ), suggesting the potential for rapid globalization. These outbreaks bring mild to moderate febrile illness to the affected countries and are frequently misdiagnosed as other arborviruses early in disease (Acosta-Ampudia et al. ). Currently, vector control (the use of techniques to mitigate the transmitting species, in this case mosquitos) and personal protective measures are the only forms of infection prevention and are relatively ineffective, as the number of new arboviral infections continues to increase (Esposito and Fonseca 2017). One group performed a biomathematical analysis on the epidemiological consequences of MAYV in Colombia (Valencia-Marín et al. 2020). Their model was based on the temperature, migration patterns, rates of development, and the flow of land cargo, all of which contribute to the vector spread and ability to infect the human population. Specifically, this study determined that regions with high rates of land cargo movement and temperatures between 23 and 28°C were most at risk for MAYV spread via Ae. Aegypti. Therefore, they concluded Magdalena, Imerí, and the biogeographical Chocó areas to be at the highest risk for rapid spread of MAYV in Columbia (Valencia-Marín et al. 2020).
LASV
The major risk areas of LASV outbreak are Sierra Leone, Liberia, Guinea, Nigeria, and other countries in the region (Fichet-Calvet and Rogers 2009). This was determined by environmental data and statistical analyses, as these have proven to be the most effective method of producing Lassa fever risk maps (see Table 1). Models were generated using nonlinear maximum likelihood discriminant analysis techniques. Areas of presence and absence of Lassa fever were identified in West Africa, which considers the distribution of the highly populous rodent hosts (Fichet-Calvet and Rogers 2009). The survival of LASV in decreased humidity significantly increases its transmission potential. Rainfall conditions appear to have the most substantial influence, but the temperature has a variable impact, especially in high-risk areas of Western Africa (Fichet-Calvet and Rogers 2009).
Noroviruses
Some norovirus outbreaks can be predicted using factors specific to the host of the norovirus. For example, oyster-borne norovirus can be monitored and predicted using an Artificial Neural Network model called NORF (Wang and Deng 2016). The authors successfully used the NORF model to predict these outbreaks to take place on the Gulf of Mexico using environmental factors such as salinity, water level height, temperature, wind, and rainfall (see Table 1). The model was trained on 14 years of data (from 1994 to 2007) and validated exclusively using seven additional years of data (from 2007 to 2014). The model predicted that an outbreak on the Louisiana coast would occur on 29 December 2013 and the outbreak indeed occurred from 26 December 2013 to 9 January 2014 (Wang and Deng 2016). While this was retrospective in nature, the model was successful in predicting a confirmed outbreak from external data and so other risk assessment models may consider water irrigation of produce (Fiona Barker et al. 2013) to predict future outbreaks. Collaborative tools such as NoroNet, a scientific surveillance collective for norovirus (https://www.rivm.nl/en/noronet), may also help identify outbreaks before they occur. Another model called NOROCAST was created in Japan to predict norovirus genotype and herd immunity; this model found one structural protein in particular impacted herd immunity the most and should be targeted for therapeutics (Suzuki et al. 2019).
Influenza virus
In addition to the four currently approved anti-influenza drugs (Roguski and Fry 2017), annual vaccination against seasonal influenza (both influenza A and B) is available in many countries to protect against the predicted strains of that year and those that are antigenically similar. Unfortunately, it is difficult to predict what strains will emerge, so these vaccines typically have low efficacy due to the high evolutionary rate of influenza viruses (Centers for Disease Control and Prevention). It is believed that the most proactive response to preventing the next influenza resurgence is to survey wild birds and farm swine actively to observe how they interact with domestic animals and humans (Taubenberger and Morens 2010). Antigen shift/drift and flu recombination in intermediate hosts present severe challenges to accurately predicting future outbreaks (Ma et al. 2008, Kim et al. 2018). In this regard, many models integrating past mutation data and other creative tactics, such as local search query data, have been built to predict the next pandemic flu strain (Łuksza and Lässig 2014, Zhang et al. 2019, Yin et al. 2020), all claiming limited success, but acknowledging the stochastic nature of the influenza virus evolution and the plethora of data needed to build models. For instance, Yin et al. (2020) employed their model to predict mutations of influenza A viruses. They concluded that they could predict point mutations at selected residues to provide better insights than current methods, which could prove incredibly useful in vaccine and outbreak prediction development. Many models have also targeted recombination in swine, but to this point all the authors acknowledged that none of the available containment strategies (including vaccination) are sufficient at preventing genetic variation in swine populations infected with influenza (Reynolds et al. 2014, Etbaigha et al. 2018, Li et al. 2022).
Nipah virus
In nature, the primary reservoirs of the Nipah virus are flying foxes, a large bat of the genus Pteropus (Lo Presti et al. 2016). Recent outbreaks in Bangladesh and India trace back to contaminated date palm sap (Ang et al. 2018). An intermediate host that facilitates the transmission of the virus from bats to humans is pigs; the 1999 outbreak in Malaysia resulted in a massive, nationwide culling of pigs to halt the spread of the virus. As they are a frequent host of human-threatening virulent diseases, many studies monitor swine for pathogens hoping to stop the next viral outbreak before it occurs (Ruiz-Fons 2017). Geographical mapping considers climate, longitude, latitude, and previous outbreaks to predict the next outbreak (Peterson 2015), however, we were unable to find any modeling attempts for Nipah virus. Based on what we know about the virus, we hypothesize successful models would take factors such as climate, wildlife monitoring, agricultural factors, and human–host interactions into account.
Hantaviruses
Most approaches to predict hantavirus outbreaks depend on examining the relationships between rodent population growth and hantavirus exposure. These models consider aspects such as random mating and deaths relating to disease or natural causes. Mathematical models have been developed to analyze and interpret the interaction between rodents and hantavirus (Jonsson et al. 2010). These models (SIR and SEIR models) aim to characterize epidemic dynamics and have been useful in predicting possible contagion scenarios (Jonsson et al. 2010). In silico models based on human data are preferred due to the complexities of relating animal models to humans (Safronetz et al. 2013). Other sources posit that adding seasonal interactions with rodents is crucial to models when trying to predict outbreaks (Sauvage et al. 2007). It is also notable that there is increased interaction with rodents as the human population grows and more rodents flock to urban areas due to the destruction of their ecosystems (Neiderud 2015).
OROV
It is predicted that OROV outbreaks and sporadic cases will increase in Brazil and the Amazonian region over time, largely due to recent wildfires and rapid deforestation (de Oliveira Andrade 2019, Tollefson 2020). Because these tropical regions are amenable to arthropod vectors such as mosquitoes, a favored host of this virus, zoonotic arboviruses can cause great harm in these areas. While patients with OROV frequently present with mild symptoms similar to that of other arboviruses many go on to develop meningitis, making outbreaks of the utmost concern (Gutierrez et al. 2019). The densely populated region of southeastern Brazil also has a higher risk of an outbreak due to internal and international migration and tourism (Vasconcelos et al. 2001, Lowe et al. 2013) Evolving environmental, demographic, and social factors make it likely that OROV will spread outside Central and Latin America as the climate change, globalization, and deforestation all continues to increase (Sakkas et al. 2018). Despite our growing knowledge of the epidemiological, clinical, and molecular features of OROV, at this point we do not have sufficient foundations to develop treatments. This is largely due to the nonfatal pathogenesis of OROV infection and its primary involvement with the CNS (da Rosa et al. 2017).
MARV
While the broader epidemiology of MARV is unknown, at this point, nonhuman primates have not proven susceptible to the disease, unlike the Ebola virus (Valentine et al. 2020). Most human outbreaks occurred due to spillover events in caves and mines. But, as is common with other viral hemorrhagic fevers, a failure to rapidly diagnose cases leads to the potential for transmission originating in a hospital or healthcare facility (Pigott et al. 2015). No vaccines or antiviral medications are currently approved for MARV. Predictive models that use transmission maps with calculated uncertainty forecast East Africa as the main region of concern for future outbreaks (Peterson and Samy 2016). The models were built using Maxent ecological niche modeling, however, they are limited by time-averaging the data, as well as access to outdated ecological, viral, and vector data for some of these regions from nearly 20 years ago.
Ebola (1976, 1979, 1994–1997, 2000–2003, 2014–2016, and 2021)
Currently, reports say that 22 countries in Central and West Africa have the potential for zoonotic transmission (Gulland 2014, Pigott et al. 2014). Many forecasting tools, such as stochastic models and Susceptible–Infected–Recovered (SIR) approaches, were used in the wake of the 2014 outbreak and have continued to be tested and optimized (Asher 2018). A total of 6 months after the first reported case, the CDC stated that if nothing changed behaviorally, West Africa could expect 1.4 million cases (Meltzer et al. 2014). Before all of this, in 2004, Leroy et al. (2004) discussed the trend in Ebola-infected apes and how these trends should be surveyed for predictions of human infection. They, and others (Legrand et al. 2007), concur that the virus originated in wildlife, specifically great apes near the Ebola River, who were then handled or hunted by local villages, thus leading to the transmission of the virus (Leroy et al. 2004). In fact, one group claims to have alerted local health authorities to the severe risk of both the December 2002 and November 2003 human outbreaks well before they occurred (see Table 1). They identified these villages as having potential for an outbreak using their Animal Mortality Monitoring Network, an epidemiological surveillance network that was set up to identify infected hosts before they reach the human populations. The two predictions made (based on the data of the five previous outbreaks) were considered accurate, with both occurring in the predicted locations and within a few weeks or a few months later (Rouquet et al. 2005). In 2014, the same group published the approach on how to predict human outbreaks based on ape fecal samples from 2005 and 2007 (Reed et al. 2014). (Kuisma et al. 2019) implemented animal monitoring programs with the government and education programs with the locals in the Democratic Republic of the Congo as an attempt to prevent or lessen the next outbreak, but the system has yet to be tested as there has not been an outbreak in this location since the implementation.
When creating predictive models, the most crucial step is acquiring data properly. One Ebola model uses a mimicking system to test the viability of an outbreak in certain areas synthetically. This system considers different transmission routes such as direct contact, vector-borne, and enteric transmission (Viboud et al. 2018). Variable symptoms should also be noted when improving the accuracy of these models, as more symptoms involved in the model could cause the accuracy to diminish. Constant surveillance and epidemiological characteristics are necessary to define the accuracy of these models (Hart et al. 2019).
Accuracy of outbreak prediction models
A few models were able to predict respective outbreaks within a month’s precision (cf. Table 1). We note that while models offering long-term forecasts were not very successful, short-term models based on environmental changes and/or known wildlife patterns have been more accurate. The two most accurate models detailed in this review were the epidemiological surveying network and statistical models which displayed day level precision. The NORF model (Wang and Deng 2016) was able to use environmental factors to predict when the virus would flourish and transmit to humans through food, whereas the Ebola surveying network managed to alert local governments of human contamination with infected wildlife (Rouquet et al. 2005) with sufficient time prior to both outbreaks. While the success of the governments’ interventions varied, the models made reasonably accurate forecasts. Thus, the surveillance models, especially those monitoring the spread of viruses between humans and animals, have demonstrated their worth and should be researched and supported more widely (PLOS; Wille et al. 2021). This was echoed again recently by researchers who found 35 cases of a henipavirus, named the “Langya” virus, from the last 4 years; they assert the importance of global viral surveillance models given the frequency of human–animal viral transmission (Mallapaty 2022).
Many factors might affect the predictivity of these types of models. Similar models should be considered for future predictive tasks. Due to differing viral evolution rates, distinct spillover events, and specific vector–host interactions, the most significant factors of highly predictive models are commonly virus-specific. However, some of them can be generalized, including epidemiological surveillance measures and monitoring climate change and other forms of human-induced disruptions to the natural order. These environmental factors are frequently indicative of viral spread as they relate to the abundance of the virus or its ability to transmit.
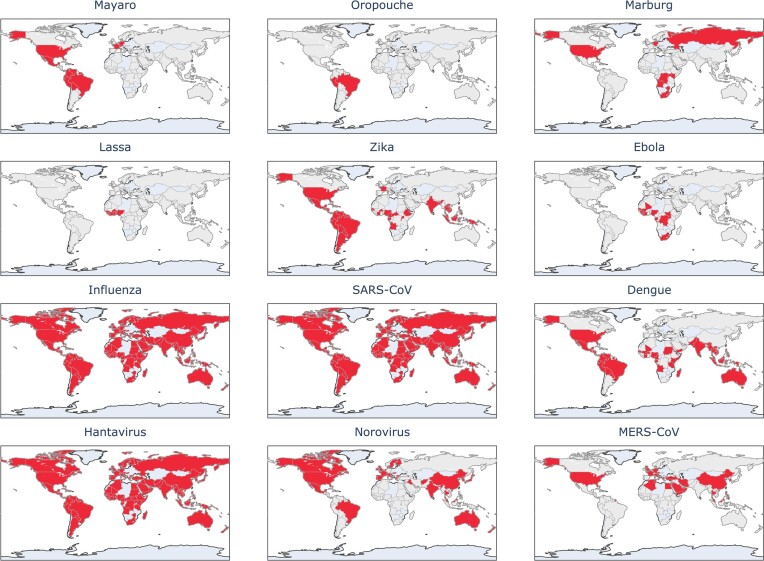
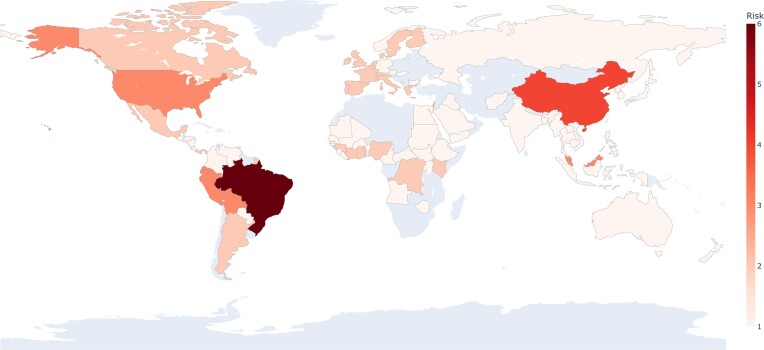
Where possible, these should be considered heavily, especially in areas highlighted in Figs 3 and 4. For both these figures, we compiled information from the literature to identify countries that are within reasonable risk for certain outbreaks. We then generated maps highlighting countries identified in the literature. Neither of these maps should be considered as predictions. Rather, they are observation-based risk maps with no assumed date, exclusively based on literature and prior trends. In Fig. 3, we use a map to visualize areas where outbreaks have occurred (both in origination and spread) to signify the impact of these viruses on the human population. We recognize that this information may be incomplete due to the prerequisite of proper testing and reporting that would allow one to find information on these viruses. Therefore, this knowledge is limited in that not all past outbreaks may be included in any study and may be missing from the data. For updated alerts, websites like healthmap.org (https://www.healthmap.org/en/) provide a map with classified alerts based on literature and news reports. In Fig. 4, we recognize these same shortcomings, especially in considering areas at risk for being the origin of novel outbreaks but aim to emphasize how imminent viral threats are.
Figure 3.
Map highlighting countries that either have experienced or been impacted by outbreaks of the selected viruses (Centers for Disease Control and Prevention, CDC, National Center for Emerging and Zoonotic Infectious Diseases, NCEZID 2023, Verhoef et al. 2015, Sakkas et al. 2018, Acosta-Ampudia et al. 2018b, Memish et al. 2020).
Figure 4.
Visual representation of countries most at risk for originating new viral outbreaks, based on observed trends from previous outbreaks and literature describing those outbreaks. The darker the color and higher the number, the more at risk the country is at for being the epicenter of a novel outbreak. The lack of color in several countries does not imply safety, but rather there is no data or models implying the country to be at risk for the origination of a novel virus.(Centers for Disease Control and Prevention, CDC, Dowdle 1976, Gulland 2014, Lewnard et al. 2014, Clayton 2017, Jiang et al. 2017, Sakkas et al. 2018, de O Mota et al. 2019) This map does not reflect the viral spread that may cover neighbor countries or all the world, depending on the virus and medical counter-measures.
As seen in Fig. 4, the prevalence of outbreak risks, particularly in Brazil, China, and the USA, is clear and should raise concerns over rapid globalization, deforestation, and urbanization. Conversely, there are few parts of the world that are not dealing with the imminent threat of originating a viral outbreak. This further emphasizes the need for collaborative, worldwide research. As we learned with SARS-CoV-2, viral threats can grow quickly and become a concern for the entire world. A hot spot for these threats, Brazil has not only had the highest number of past outbreaks, but it also appears at risk for most future outbreaks. In combination with the tropical climate, deforestation exposes human populations to new species (and therefore new pathogens) by destroying the natural habitats in which reservoir hosts live. As seen with the SARS-CoV-2 pandemic, globalization is integral to spreading an emerging virus (Jeanne et al. 2022). Relatedly, environmental changes and technological advances (such as those contributing to globalization and property development) were also discussed in relation to Zika and OROVs (Vasconcelos et al. 2001, Lowe et al. 2013, Liu-Helmersson et al. 2019). Factors like these should be heavily weighed when considering viral forecasting. There is much evidence connecting increased development and associated loss of biodiversity to an increasing number of disease outbreaks. Deforestation and extinction make pandemics and viral outbreaks more likely, as the species that survive and migrate during severe ecological changes are more likely to host pathogens (e.g. bats, rats, and birds) (Tollefson 2020, Carlson et al. 2022). Globally, it is estimated that sudden (on an ecological timescale) changes in climate and land use will drive new interactions between humans and zoonotic viral host species, resulting in 4000 cross-species transmission events for novel viruses by year 2070. (Carlson et al. 2022) For these and many more reasons, all viruses mentioned here are imminent threats.
It is indisputable that there will be a plethora of viral outbreaks to come. However, learning from the past and using modern technologies, it may be possible to help mitigate the impact of these outbreaks. Whether this comes from predictive models, monitoring of wildlife, or pre-emptive drug discovery and development, we must continue to adapt and learn from what has been successful in the past. Moving forward, models should aim to include as many salient factors as possible and rely heavily on data from previous outbreaks (Drake 2021), as well as animal and viral surveillance wherever possible. The predictive models discussed herein demonstrate that it is possible to predict pandemics with some accuracy before they occur, but it is also important that policy makers and other leaders listen to scientists performing this work and act upon it to prevent massive loss of life based on social measures (masks, distancing, and so on) as well as preventive development of drugs, especially broad-spectrum antivirals, and vaccines. In the past, hesitancy over drug resistant strains has halted development of antivirals (De Clercq 2005); our experience with COVID-19 exemplifies how important it is to continue and broaden this type of research. Although mutations and alternate strains of viruses cannot always be predicted (e.g. influenza virus), scientists’ warnings should be heeded when they are well-founded. As the current practice showed, once the viral outbreaks occur, the best way to contain them is through diligent quarantine, disinfection, and travel restrictions.
It should be noted that new outbreaks have already started to appear in the post-Covid-19 world. In February 2023, an outbreak of MARV was reported in Equatorial Guinea with every known patient dying (Marburg virus disease—Equatorial Guinea; Disease Outbreak News 2023). While the highest probability of outbreak was in Eastern Africa, rather than in Western Africa, it is of note that an outbreak in Africa was expected and predicted. Between 1 January 2023 and 16 March 2023, the USA, parts of Europe and Asia, and much of Africa and South America have all seen DENV outbreaks. Bolivia has reported more than 7000 cases in the first 6 weeks of 2023 Bolivian hospitals under strain as dengue kills dozens; Associated Press 2023). Given the two hot (the average temperature is constantly rising) and rainy years in a row in 2020–2022, we also expected to see a Dengue outbreak in Brazil (which is happening right now) in accord with expectations one could draw from the data in Fig. 4 where we indicated that Brazil is in the most danger for the appearance of new outbreaks. It has already been verified in several studies that the highest incidence of the disease and levels of infestation by Dengue vectors in Brazil coincided with the rainy months, which were also the hottest months of the year in the country (Viana and Ignotti 2013).
While we continue to expect viral outbreaks in the future, we need to admit that there is no blanket approach to predicting these outbreaks. Therefore, we must continue our research into the outbreak prediction, but acknowledge that in all likelihood, we will not precisely know what, when, and where the next viral threat will happen. COVID-19 taught humanity that new enemies are always waiting at the gate and will show no mercy. We have been warned, but now we need to be armed, and thus, antiviral drug discovery and development must be of the highest importance.
Summary and perspectives
Viruses have always been present alongside humankind and new outbreaks are constantly happening. In addition, ecological disruption and climate change make it more likely that new zoonotic viruses will jump over to humans. However, as mentioned in this Perspective, there are many ways we can prevent these outbreaks from turning into mass tragedies like the 1918 flu pandemic and the current COVID-19 pandemic. Such methods include employing data science to predict emerging outbreaks, surveillance of reservoir populations, promoting scientific awareness and literacy, following guidelines set by scientists and officials as to how to prevent infection and spread, providing consistent funding for virology and broad-spectrum antiviral drug discovery research, and ultimately heeding the warnings of the virologists and data scientists that forecast the occurrence of such outbreaks. For instance, where the NOROCAST predictions were heeded, potentially hazardous food was not distributed, and the impending illness was, therefore, prevented (Wang and Deng 2016); on the contrary, the warnings against MERS in the USA were not acted on promptly and severe illness struck those infected in the following months (Bialek et al. 2014, Donnelly et al. 2019).
The coexistence of humans and viruses is an intrinsic aspect of life. Epidemics of various sizes have occurred throughout recorded history and will continue to occur in the future. The constant increase in globalization, encroachment on wildlife, and climate change are all likely to increase the spread of emerging viruses (Baker et al. 2021). Therefore, it is essential to establish a long-standing political and financial investment in virology research to understand the etiology of new viruses and predict geographical areas and circumstances in which outbreaks are more likely to occur. Additionally, ongoing research is necessary to advance vaccine development as well as both broad-spectrum and targeted antiviral treatments. The time a country or community might have to combat a virus may be minimal, making this an urgent task.
Outbreak analytics is an emerging research area focused on employing data science technology and methods to collect, curate, visualize, model, and report outbreaks to inform better and drive proper epidemiological response (Polonsky et al. 2019). However, as discussed in this perspective, current efforts have relied on forecasting new cases likely to arise, spread, and impact an ongoing outbreak. We posit that efforts should be made to employ data science to predict the date and locale that emerging pathogens are more likely to appear to enable pointed research and public health efforts to prevent outbreaks from happening. The rapid and ubiquitous use of smartphones and heavy accumulation of social media data, electronic health records, surveillance and geospatial systems, health sensing systems, online search, and Bluetooth exposure apps (Google 2023) have created an unprecedented technological infrastructure for achieving this goal, similarly to the prediction of other temporal catastrophic events, such as natural disasters (Goswami et al. 2018), terrorism (Ding et al. 2017), and urban pipeline leakage accidents (Qiu et al. 2018). Although the use of health data and phone location continues to raise privacy concerns (Schomakers et al. 2019), progress has been made to develop privacy-preserving technologies to allow health data for contact tracing and epidemiological surveillance and outbreak analytics (Altuwaiyan et al. 2018, Anjum et al. 2018).
Before computational models can be considered a reliable data-driven approach for forecasting emerging pathogens, several steps in the data science of outbreak analytics should be executed. Ideally, data generated by different countries should be shared through government and research institutes, industry, and so on, following the FAIR (findability, accessibility, interoperability, and reusability) data principles (Wilkinson et al. 2016). The logistics underlying building technologies able to predict future outbreaks are complex and will involve the development of both point-of-care data collection, database design, mobile apps, network infrastructure, privacy-preserving technologies, and the development of knowledge graphs algorithms. An epidemiological data model with a defined and well-established ontology needs to be developed to guarantee a good data ecosystem. Further, the contact tracing apps for COVID-19 have faced low efficacy due to the low usage and testing rates (Cebrian 2021, Munzert et al. 2021). These recent studies have shown that data science can be used to better predict and assess the current state of outbreaks. Still, significant political and economic commitment to providing infrastructure for diagnostic testing, collecting, and evaluating the resulting data is necessary.
Interest in viruses should not be a temporal issue, dependent on whether a viral pathogen is currently circulating through the population or not. However, it tends to be in many cases (Bobrowski et al. 2020). Scientific literacy and understanding how viruses and other pathogens cause human disease and spread throughout populations are necessary to promote widespread health. Vaccines are available for viruses that will likely cause outbreaks in the future, such as Ebola virus and influenza virus, and new vaccines for other viruses are on the way. Widespread scientific literacy can facilitate the implementation of mass vaccination campaigns that will prevent outbreaks of these viruses from occurring in the first place.
As seen in the COVID-19 pandemic in certain countries such as the USA, public resistance to health agencies’ guidelines and vaccination has resulted in the accentuated spread of SARS-CoV-2. States with lower levels of mask adherence before the relaxation of CDC guidelines were associated with high COVID-19 case rates in the following month, excluding other factors (Fischer et al. 2021). Regions in the USA that are less likely to mask or get vaccinated against the virus are more likely to be rural regions, which are already more at risk for COVID-19 due to other factors (Centers for Disease Control and Prevention, CDC, Texas A&M University 2021, Callaghan et al. 2021). Relatedly, in many states and presumably beyond, rural counties were associated with higher COVID-19 case rates and mortality rates (Huang et al. 2021). This association between the willingness to follow CDC guidelines and regulations surrounding COVID-19 and the rate or severity of COVID-19 suggests that increased scientific literacy and boosted public awareness campaigns surrounding pandemics and viral diseases might assist in preventing viral spread within the general population.
As highlighted in our recent analysis, the consistent funding for research on HIV/acquired immunodeficiency syndrome has resulted in the development of a plethora of antiviral drugs of different classes that can be used in combination to stave off a disease that was once a death sentence (Bobrowski et al. 2021). This is one of the few examples of a virus that has received consistent attention since it began a pandemic in the 1980s that continues today, and one of the few true success stories in conquering a dangerous viral illness in modern history through antiviral drug development. Having effective treatments for a viral disease, be it preventative care or postsymptomatic treatment, completely changes the course of the epidemic.
For most of the viruses mentioned in this paper there are no available, effective antiviral treatments or vaccines (Table 5). Likewise, there is no consistent funding for these diseases, especially those with high potential to cause disease (such as the Oropouche and MAYVs) but have yet to cause widespread, global outbreaks. Interest in these viruses should not wane past the point where a particular epidemic ends, but more than often, this is the case; as public interest wanes in major epidemic viruses, so does the funding for research into said virus (Bobrowski et al. 2020). Both the rapidity and bulk of immediate responses to temporal viral epidemics are typically insufficient to result in a tangible outcome (i.e. vaccine or antiviral medication) past the point the epidemic has ended. EBOV and ZIKV, both described in this paper as being high-risk viruses for future outbreaks, have seen decreased NIH funding available after their initial outbreaks (2014–2016). This decrease in funding is also associated with a decrease in publications associated with these viruses in PubMed (Bobrowski et al. 2020). Therefore, it is necessary to maintain interest in these viruses past the point at which they cease to be a problem; just because one epidemic has ended does not mean another will not begin soon after, as with the more recent outbreak of EBOV (2018–2020).
Table 5.
Examples of drugs and vaccines that are approved, in development, or may be repurposed against future viral outbreaks.
| Virus | Approved vaccines | Vaccine development | Approved drugs | Repurposed drugs |
|---|---|---|---|---|
| SARS-CoV-2 | Pfizer-BioNTech (Comirnaty), Janssen (J&J), Moderna (Spikevax) (COVID-19 Vaccines, FDA 2022) | Many (Li et al. 2020) | Remdesivir, Paxlovid (EUA), Molnupiravir (EUA) (Melo-Filho et al. 2022) | Galdesivir, Remdesivir, Penciclovir, Disulfiram, Lopinavir, Boceprevir (Melo-Filho et al. 2022) |
| SARS-CoV | Inactivated SARS–CoV vaccine (ISCV), VRC-SRSDNA015-00-VP (Li et al. 2020) | Galdesivir, Remdesivir, Penciclovir, Disulfiram, Lopinavir, Boceprevir (Melo-Filho et al. 2022) | ||
| MERS-CoV | ChAdOx1 (Folegatti et al. 2020) BVRS-GamVac (Study of Safety and Immunogenicity of BVRS-GamVac—ClinicalTrials.gov 2020) MVA-MERS-S_DF1 (Safety and Immunogenicity of the Candidate Vaccine MVA-MERS-S_DF-1 Against MERS—ClinicalTrials.gov 2021) GLS-5300 (INO-4700) (Safety, Tolerability and Immunogenicity of INO-4700 for MERS-CoV in Healthy Volunteers—ClinicalTrials.gov 2022) |
Galdesivir, Ementine, Remdesivir, Disulfiram, Lopinavir, Boceprevir (Melo-Filho et al. 2022) | ||
| Ebola virus | ERVEBO, Zabdeno/Mvabea (World Health Organization 2020c) | ChAd3-EBOZ, Ad5-EBOV, GamEvac-Combi and GamEvacLyo (Woolsey and Geisbert 2021) | Inmazeb, Ebanga (Centers for Disease Control and Prevention, CDC 2021c) | Miglustat, Clomiphene, Toremifene (Yuan 2015) Tilorone, Quinacrine, Pyronaridine (Puhl et al. 2021) |
| ZIKV | rZIKV/D4Δ30–713, ZPIV (NIH Diseases 2018) | Cilexetil, Auranofin, Bortezomib, Dactinomycin, Ivermectin, Mycophenolic Acid, Thioguanine (Loe et al. 2019) |
||
| DENV | Dengvaxia (Deng et al. 2020) | LATV rDEN4∆30, TV003/TV005, S16803, EDIII-P64K, V180 (Deng et al. 2020) | Nelfinavir, Balapiravir, Mycophenolic acid and ribavirin, ZX-2401, Dasatinib, Ivermectin (Botta et al. 2018) | |
| MAYV | ChAdOx1 May (Kroon Campos et al. 2020) | EIDD-1931,Favipiravir, Suramin (Langendries et al. 2021) | ||
| LASV | INO-4500 (ClinicalTrials.gov 2022) | Ribavirin, Favipiravir (Rosenke et al. 2018) | ||
| Norovirus | TAK-214, VXA-NVV-104, Hansenulapolymorpha, Longkoma (Tan 2021) | 2′C-methylcytidine, Ribavirin, 2-thiouridine, 5-nitrocytidine, Suramin, Nitazoxanide (Kaufman et al. 2014) | ||
| Influenza virus | Afluria Quadrivalent, Fluarix Quadrivalent, FluLaval Quadrivalent, Fluzone Quadrivalent, Flucelvax Quadrivalent, Flublok Quadrivalent, FluMist Quadrivalent (Centers for Disease Control and Prevention) | Oseltamivir, Zanamivir, Peramivir, Baloxavir marboxil (Centers for Disease Control and Prevention, CDC 2021c) | Diltiazem (Pizzorno et al. 2019) | |
| Nipah virus | rVSV-ΔG-NiVBG (PHV02) (Foster et al. 2022) | HeV-sG-V (Foster et al. 2022) | ||
| Hanta virus | Hantavax (Liu et al. 2020) | pWRG/HTN-M(x) (Liu et al. 2020) | Ribavirin, Chloroquine (Vergote et al. 2021) | |
| OROV | Favipiravir (Files et al. 2022) | |||
| MARV | VSV-MARV (Marzi et al. 2021) | Tilorone, Quinacrine, Pyronaridine (Puhl et al. 2021) |
For decades, most antiviral research followed the “one bug, one drug” paradigm, but with a recent paradigm shift toward broad spectrum drugs, it is unclear how many existing compounds are active against multiple viruses. An open-access small molecule antiviral compound collection (SMACC) was recently developed (Martin et al. 2023) to support the discovery of broad-spectrum antiviral drug molecules; currently, it contains over 32 500 chemical bioactivity entries for 13 viruses with high pandemic potential. Their analysis revealed several compounds with multiple antiviral activities suggesting the feasibility of broad spectrum antivirals but underscoring the need for systematic efforts toward discovery of such agents, like the Rapidly Emerging Antiviral Drug Development Initiative (READDI) at UNC-Chapel Hill. Current research indicates the conservation of viral proteins (Melo-Filho et al. 2022) or other conserved viral mechanisms, like involvement of common host factors (Kumar et al. 2020), could be the key to broad-spectrum antiviral discovery. Obviously, host targets responsible for viral entry should not be forgotten as well (Hochuli et al. 2022).
Biomedical knowledge mining tools like ROBOKOP (Bizon et al. 2019) (Reasoning Over Biomedical Objects linked in Knowledge-Oriented Pathways) and Chemotext (Capuzzi et al. 2018) have been developed to help elucidate biological pathways underlying compound activity or toxicity. Similar technologies can be leveraged and deployed to mine data to detect emerging pathogens and estimate data of new outbreaks to facilitate rapid responses, such as patient isolation and contact tracing to prevent the spread of the virus. For example, a recent study reported the development of a tool leveraging deep learning and a computer sensor system capable of predicting influenza outbreaks 15 weeks in advance based on and real-time data of flu patterns and symptoms (Al Hossain et al. 2020).
Ultimately, we should focus more on the future, particularly on how the past informs that future. Anyone alive today can say it is infinitely better not to experience a viral pandemic than to live through the associated economic, mental, and personal tragedies associated with it. All the viruses mentioned—SARS-CoV-2, MERS-CoV, DENV, ZIKV, MAYV, LASV, noroviruses, influenza, Nipah virus, hantaviruses, OROV, MARV, and Ebola virus—have epidemic potential and require attention to avoid becoming catastrophes. The available predictive models can advise us on when, where, or what strain of virus may emerge, and more attention should be given to alerting models. Consistent investment in research and public literacy in science is integral to implementing actual policies that can affect individual lives. If public health officials and politicians worldwide heed the warnings of virologists and data scientists who predict and generate data, they could prevent the next viral epidemic and avoid mass morbidity and mortality. History sets a precedent for successes and failures, and the handling of many major pandemics in the past are failures. However, this does not have to be the case in the future. We have been warned of the dangers of currently circulating virus strains that exist and their potential for disease, what remains to be determined is if these dangers will be given proper attention by the scientific community and funding agencies.
Acknowledgments
The UNC team was supported in part by the NIH grants U19AI171292 and R01 AI108197. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. H.J.M. acknowledges financial support from the American Foundation for Pharmaceutical Education's Pre-Doctoral Fellowship. V.A. is currently an employee at Takeda Pharmaceuticals, San Diego, CA.
Contributor Information
Zoe Sessions, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Tesia Bobrowski, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Holli-Joi Martin, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Jon-Michael T Beasley, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Aneri Kothari, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Trevor Phares, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States; School of Chemistry, University of Louisville, 2320 S Brook St, Louisville, KY 40208, United States.
Michael Li, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Vinicius M Alves, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Marcus T Scotti, Department of Pharmaceutical Sciences, Federal University of Paraiba, Campus I Lot. Cidade Universitaria, PB, 58051-900, Brazil.
Nathaniel J Moorman, Department of Microbiology and Immunology, University of North Carolina, 116 Manning Drive, Chapel Hill, NC 27599, United States.
Ralph Baric, Department of Epidemiology, University of North Carolina, 401 Pittsboro St, Chapel Hill, NC 27599, United States.
Alexander Tropsha, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Eugene N Muratov, Laboratory for Molecular Modeling, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina, 301 Pharmacy Ln, Chapel Hill, NC 27599, United States.
Conflict of interest
AT, VA, and ENM are co-founders of Predictive, LLC, which develops computational methodologies and software for toxicity prediction. All other authors declare they have nothing to disclose.
References
- Acosta-Ampudia Y, Monsalve DM, Rodríguez Yet al. Mayaro: an emerging viral threat?. Emerg Microbes Infect. 2018b;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agyarko R, Jamison D, Oppenheim Bet al. What's Next? Predicting The Frequency and Scale of Future Pandemics. Washington: Center for Global Development, 2023. [Google Scholar]
- Al Hossain F, Lover AA, Corey GAet al. FluSense. Proc ACM Interact Mob Wearable Ubiquitous Technol. 2020;4:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Turaiki I, Alshahrani M, Almutairi T. Building predictive models for MERS-CoV infections using data mining techniques. J Infect Publ Health. 2016;9:744–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albery GF, Becker DJ, Brierley Let al. The science of the host–virus network. Nat Microbiol. 2021;6:1483–92. [DOI] [PubMed] [Google Scholar]
- Altuwaiyan T, Hadian M, Liang X. EPIC: efficient privacy-preserving contact tracing for infection detection. In: Proceedings of the 2018 IEEE International Conference on Communications (ICC). Piscataway: IEEE, 2018, 1–6. [Google Scholar]
- Anderson CR, Spense L, Downs WGet al. Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg. 1961;10:574–8. [DOI] [PubMed] [Google Scholar]
- Anderson CR, Wattley GH, Ahin NWet al. Mayaro virus: a new human disease agent. Am J Trop Med Hyg. 1957;6:1012–6. [DOI] [PubMed] [Google Scholar]
- Andrews S. Virus Outbreak Simulator for The US. 2019. https://bioinformaticshome.com/online_software/virus-outbreak/US/index.html
- Andrick B, Clark B, Nygaard Ket al. Infectious disease and climate change: detecting contributing factors and predicting future outbreaks. In: Proceedings of the 1997 IEEE International Geoscience and Remote Sensing Symposium Proceedings. Remote Sensing – A Scientific Vision for Sustainable Development, IGARSS'97. Vol. 4. Piscataway: IEEE, 1997, 1947–9. [Google Scholar]
- Ang BSP, Lim TCC, Wang L. Nipah Virus Infection. J Clin Microbiol. 2018;56:e01875–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum A, Ahmed T, Khan Aet al. Privacy preserving data by conceptualizing smart cities using MIDR-Angelization. Sustain Cities Soc. 2018;40:326–34. [Google Scholar]
- Arnott R. Omicron variant may end up saving lives. 2022. https://www.wsj.com/articles/omicron-variant-may-end-up-saving-lives-infection-antibodies-spread-sick-covid-19-coronavirus-hospitalization-death-vaccine-11641153969 (2 January 2022, date last accessed).
- Asher J. Forecasting Ebola with a regression transmission model. Epidemics. 2018;22:50–55. [DOI] [PubMed] [Google Scholar]
- Associated Press . Bolivian hospitals under strain as dengue kills dozens. New York, 2023. [Google Scholar]
- Association of Professionals in Infection Control and Epidemiology, APIC . Outbreaks, epidemics and pandemics—what you need to know. Arlington, 2023. [Google Scholar]
- Azevedo RDSDS, Nunes MRT, Chiang JOet al. Reemergence of Oropouche fever, Northern Brazil. Emerg Infect Dis. 2007;13:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, Mahmud AS, Miller IFet al. Infectious disease in an era of global change. Nat Rev Microbiol. 2021;20:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TAM, Noda T, Riches JDet al. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA. 2012;109:4275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek SR, Allen D, Alvarado-Ramy Fet al. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities – May 20. Morb Mortal Wkly Rep. 2014;63:431–6. [PMC free article] [PubMed] [Google Scholar]
- Bizon C, Cox S, Balhoff Jet al. ROBOKOP KG and KGB: integrated knowledge graphs from federated sources. J Chem Inf Model. 2019;59:4968–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom DE, Black S, Rappuoli R. Emerging infectious diseases: a proactive approach. Proc Natl Acad Sci USA. 2017;114:4055–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T, Chen L, Eastman RTet al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol Ther. 2021;29:873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T, Melo-Filho CC, Korn Det al. Learning from history: do not flatten the curve of antiviral research!. Drug Discov Tod. 2020;25:1604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston’s Children Hospital Researchers . HealthMap. Boston, 2006. [Google Scholar]
- Botta L, Rivara M, Zuliani Vet al. Drug repurposing approaches to fight Dengue virus infection and related diseases. Front Biosci Landmark. 2018;23:997–1019. [DOI] [PubMed] [Google Scholar]
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26:D49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridson E. The English “sweate” (Sudor Anglicus) and Hantavirus pulmonary syndrome – PubMed. Br J Biomed Sci. 2001;58:1–6. [PubMed] [Google Scholar]
- Caicedo E-Y, Charniga K, Rueda Aet al. The epidemiology of Mayaro virus in the Americas: a systematic review and key parameter estimates for outbreak modelling. PLoS Negl Trop Dis. 2021;15:e0009418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan T, Lueck JA, Trujillo KLet al. Rural and urban differences in COVID-19 prevention behaviors. J Rural Health. 2021;37:287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuzzi SJ, Thornton TE, Liu Ket al. Chemotext: a publicly available web server for mining drug–target–disease relationships in PubMed. J Chem Inf Model. 2018;58:212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen A, Huang P, Levitt Zet al. How is the COVID-19 vaccination campaign going in your state?. 2021. https://www.npr.org/sections/health-shots/2021/01/28/960901166/how-is-the-covid-19-vaccination-campaign-going-in-your-state
- Carlson CJ, Albery GF, Merow Cet al. Climate change increases cross-species viral transmission risk. Nature. 2022;607:1–1. [DOI] [PubMed] [Google Scholar]
- Carroll D, Daszak P, Wolfe NDet al. The Global Virome Project. Science. 2018;359:872–4. [DOI] [PubMed] [Google Scholar]
- CDC . 1918 pandemic (H1N1 virus). 2019a. https://www.cdc.gov/flu/pandemic-resources/1918-pandemic-h1n1.html
- CDC . Principles of Epidemiology in Public Health Practice. 3rd edn. An Introduction to Applied Epidemiology and Biostatistics. 2019b. https://stacks.cdc.gov/view/cdc/6914
- CDC . Zika travel information. https://wwwnc.cdc.gov/travel/page/zika-information
- Cebrian M. The past, present and future of digital contact tracing. Nat Electron. 2021;4:2–4. [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . 2022 U.S. Monkeypox outbreak. 2022a. https://www.cdc.gov/poxvirus/mpox/response/2022/index.html
- Centers for Disease Control and Prevention (CDC) . Dengue around the World. 2020. https://www.cdc.gov/dengue/areaswithrisk/around-the-world.html
- Centers for Disease Control and Prevention (CDC) . Ebola treatments. 2021a. https://www.cdc.gov/vhf/ebola/treatment/index.html#:~:text=Providing%20fluids%20and%20electrolytes%20(body,other%20infections%2C%20if%20they%20occur
- Centers for Disease Control and Prevention (CDC) . Outbreak distribution map, Lassa fever, CDC. https://www.cdc.gov/vhf/lassa/outbreaks/index.html
- Centers for Disease Control and Prevention (CDC) . Rural communities. 2021b. https://www.cdc.gov/nchs/products/databriefs/db447.htm
- Centers for Disease Control and Prevention (CDC) . Ten years of gains: a look back at progress since the 2009 H1N1 pandemic. https://www.cdc.gov/flu/spotlights/2018-2019/decade-since-h1n1-pandemic.html
- Centers for Disease Control and Prevention (CDC) . Vaccine effectiveness: how well do the flu vaccines work?. https://www.cdc.gov/flu/vaccines-work/vaccineeffect.htm
- Centers for Disease Control and Prevention (CDC) . What you should know about flu antiviral drugs. https://www.cdc.gov/flu/treatment/whatyoushould.htm#:~:text=Flu%20antiviral%20drugs%20are%20prescription,from%20a%20health%20care%20provider.
- Centers for Disease Control and Prevention, CDC . Different types of flu vaccines. 2021c. https://www.cdc.gov/flu/prevent/flushot.htm
- Centers for Disease Control and Prevention, CDC . History of Marburg Virus Disease (MVD) outbreaks, Marburg (Marburg Virus Disease). 2022b. https://www.cdc.gov/vhf/marburg/outbreaks/chronology.html
- Chan MCW, Kwan HS, Chan PKS. Structure and genotypes of Noroviruses. The Norovirus. 2017;51–63. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7173473/ [Google Scholar]
- Chen J, Wang R, Wang Met al. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432:5212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Law and Government . A chronicle on the SARS epidemic. Chin Law Govern. 2014;36:12–5. [Google Scholar]
- Chowell G, Blumberg S, Simonsen Let al. Synthesizing data and models for the spread of MERS-CoV, 2013: key role of index cases and hospital transmission. Epidemics. 2014;9:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DKW, Poon LLM, Gomaa MMet al. MERS Coronaviruses in Dromedary Camels, Egypt. Emerg Infect Dis. 2014;20:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB. Introduction: Nipah virus – discovery and origin. Curr Top Microbiol Immunol. 2012;359:1–9. [DOI] [PubMed] [Google Scholar]
- Clayton BA. Nipah virus: transmission of a zoonotic paramyxovirus. Curr Opin Virol. 2017;22:97–104. [DOI] [PubMed] [Google Scholar]
- Counotte MJ, Althaus CL, Low Net al. Impact of age-specific immunity on the timing and burden of the next Zika virus outbreak. PLoS Negl Trop Dis. 2019;13:e0007978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rosa JFT, de Souza WM, de Paula Pinheiro Fet al. Oropouche virus: clinical, epidemiological, and molecular aspects of a neglected Orthobunyavirus. Am J Trop Med Hyg. 2017;96:16–0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Recent highlights in the development of new antiviral drugs. Curr Opin Microbiol. 2005;8:552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De O Mota MT, Avilla CM, Nogueira ML. Mayaro virus: a neglected threat could cause the next worldwide viral epidemic. Fut Virol. 2019;14:375–7. [Google Scholar]
- Oliveira De, Andrade R. Alarming surge in Amazon fires prompts global outcry. Nature. 2019. 10.1038/d41586-019-02537-0 [DOI] [PubMed] [Google Scholar]
- Deng S-Q, Yang X, Wei Yet al. A review on dengue vaccine development. Vaccines. 2020;8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GWA, Kitchen SF, Haddow AJ. Zika Virus (I). Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–20. [DOI] [PubMed] [Google Scholar]
- Ding F, Ge Q, Jiang Det al. Understanding the dynamics of terrorism events with multiple-discipline datasets and machine learning approach. PLoS ONE. 2017;12:e0179057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disease Outbreak News . Marburg virus disease – Equatorial Guinea, 2023.
- Donnelly CA, Malik MR, Elkholy Aet al. Worldwide reduction in MERS cases and deaths since 2016. Emerg Infect Dis. 2019;25:1758–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WR. Marburg virus. Bull Pan Am Health Organ. 1976;10:333–4. [PubMed] [Google Scholar]
- Downs WG, Aitken THG, Anderson CRet al. Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg. 1961;10:574–8. [DOI] [PubMed] [Google Scholar]
- Drake J. Can future epidemics be predicted?. Jersey City: Forbes, 2021. [Google Scholar]
- Eaton BT, Broder CC, Middleton Det al. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entis P. Canadian Norovirus outbreak linked to live spot prawns – eFoodAlert. 2022.
- Esposito DLA, Fonseca BALD. Will Mayaro virus be responsible for the next outbreak of an arthropod-borne virus in Brazil?. Braz J Infect Dis. 2017;21:540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etbaigha F, Willms AR, Poljak Z. A SEIR model of influenza A virus infection and reinfection within a farrow-to-finish swine farm. PLOS ONE. 2018;13:e0202493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excler J-L, Saville M, Berkley Set al. Vaccine development for emerging infectious diseases. Nat Med. 2021;27:591–600. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Jones S, Klenk H-Det al. Ebola virus: from discovery to vaccine. Nat Rev Immunol. 2003;3:677–85. [DOI] [PubMed] [Google Scholar]
- Fichet-Calvet E, Rogers DJ. Risk maps of lassa fever in West Africa. PLoS Negl Trop Dis. 2009;3:e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files MA, Hansen CA, Herrera VCet al. Baseline mapping of Oropouche virology, epidemiology, therapeutics, and vaccine research and development. Npj Vacc. 2022;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiona Barker S, O'toole J, Sinclair MIet al. A probabilistic model of norovirus disease burden associated with greywater irrigation of home-produced lettuce in Melbourne, Australia. Water Res. 2013;47:1421–32. [DOI] [PubMed] [Google Scholar]
- Fischer CB, Adrien N, Silguero JJet al. Mask adherence and rate of COVID-19 across the United States. PLoS ONE. 2021;16:e0249891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti PM, Bittaye M, Flaxman Aet al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect Dis. 2020;20:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Woolsey C, Borisevich Vet al. A recombinant VSV-vectored vaccine rapidly protects nonhuman primates against lethal Nipah virus disease. Proc Natl Acad Sci USA. 2022;119. 10.1073/PNAS.2200065119/SUPPL_FILE/PNAS.2200065119.SD01.XLSX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita-Fujiharu Y, Sugita Y, Takamatsu Yet al. Structural insight into Marburg virus nucleoprotein–RNA complex formation. Nat Commun. 2022;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett L. The Coming Plague: Newly Emerging Diseases in a World Out of Balance. New York: Farrar, Straus and Giroux, 1994. [Google Scholar]
- Gates B. Responding to Covid-19—a once-in-a-century pandemic?. N Engl J Med. 2020;382:NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- Gill PW, Babbage NF, Gunton PEet al. Did the Asian virus protect us from Hong Kong influenza?. Med J Aust. 1971;2:53–4. [DOI] [PubMed] [Google Scholar]
- Gill PW, Murphy AM. The incidence of A2-Hong Kong infections in blood donors during the winter of 1970. Med J Aust. 1972;2:242–6. [PubMed] [Google Scholar]
- Global Biodefense Staff . Shutdown of PREDICT infectious disease program challenged by senators Warren and King. 2020.
- Google . Exposure Notifications: using technology to help public health authorities fight COVID19. 2023.
- Goswami S, Chakraborty S, Ghosh Set al. A review on application of data mining techniques to combat natural disasters. Ain Shams Eng J. 2018;9:365–78. [Google Scholar]
- Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. J Pathol. 2015;235:185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, Vasilakis N, Musso D. History and emergence of Zika virus. J Infect Dis. 2017;216:S860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulland A. Fifteen countries are at risk of Ebola outbreak, says WHO. BMJ. 2014;349:g6305–. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Wise EL, Pullan STet al. Evolutionary dynamics of Oropouche virus in South America. Parrish CR (ed.). J Virol. 2019;94:e01127–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart WS, Hochfilzer LFR, Cunniffe NJet al. Accurate forecasts of the effectiveness of interventions against Ebola may require models that account for variations in symptoms during infection. Epidemics. 2019;29:100371. [DOI] [PubMed] [Google Scholar]
- Hass M, GöLnitz U, MüLler Set al. Replicon system for Lassa virus. J Virol. 2004;78:13793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepojoki J, Strandin T, Lankinen Het al. Hantavirus structure–molecular interactions behind the scene. J Gen Virol. 2012;93:1631–44. 10.1099/vir.0.042218-0 [DOI] [PubMed] [Google Scholar]
- Herriman R. Bolivia: hantavirus health alert issued for Palos Blancos. Outbreak News Today, 2022. [Google Scholar]
- Hijawi B, Abdallat M, Sayaydeh Aet al. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19:S12–8. [PubMed] [Google Scholar]
- Hochuli JE, Jain S, Melo-Filho Cet al. Allosteric binders of ACE2 are promising anti-SARS-CoV-2 agents. ACS Pharmacol Transl Sci. 2022;5:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Clements A, Williams Get al. Dengue fever and El Nino/Southern oscillation in Queensland, Australia: a time series predictive model. Occup Environ Med. 2010;67:307–11. [DOI] [PubMed] [Google Scholar]
- Huang Q, Jackson S, Derakhshan Set al. Urban-rural differences in COVID-19 exposures and outcomes in the South: a preliminary analysis of South Carolina. PLoS ONE. 2021;16:e0246548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute for Health Metrics and Evaluation . COVID-19 projections. Seattle, 2021. [Google Scholar]
- Jackson C, Vynnycky E, Mangtani P. Estimates of the transmissibility of the 1968 (Hong Kong) influenza pandemic: evidence of increased transmissibility between successive waves. Am J Epidemiol. 2010;171:465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanne L, Bourdin S, Nadou Fet al. Economic globalization and the COVID-19 pandemic: global spread and inequalities. GeoJournal. 2022;88:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha DN. Zika: Delhi gets first confirmed Zika case, surveillance up. The Times of India. 2021. [Google Scholar]
- Jiang H, Zheng X, Wang Let al. Hantavirus infection: a global zoonotic challenge. Virol Sin. 2017;32:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CB, Figueiredo LTM, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23:412–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian AZ, Wyatt RG, Dolin Ret al. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katella K. 5 Things To Know About the Delta Variant. New Haven: Yale Medicine, 2021. [Google Scholar]
- Kaufman SS, Green KY, Korba BE. Treatment of norovirus infections: moving antivirals from the bench to the bedside. Antiviral Res. 2014;105:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keavey S. Preparing for the next influenza outbreak – or (inevitably) pandemic. J Am Acad Phys Assist. 1999;12:28. [PubMed] [Google Scholar]
- Kim H, Webster RG, Webby RJ. Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol. 2018;31:174–83. [DOI] [PubMed] [Google Scholar]
- Kroon Campos R, Preciado-Llanes L, Azar SRet al. Adenoviral-vectored Mayaro and Chikungunya virus vaccine candidates afford partial cross-protection from lethal challenge in A129 mouse model. Front Immunol. 2020;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MGet al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuisma E, Olson SH, Cameron KNet al. Correction to ‘Long-term wildlife mortality surveillance in northern Congo: a model for the detection of Ebola virus disease epizootics.’. Phil Trans R Soc B. 2019;374:20190658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Sharma S, Kumar Ret al. Host-directed antiviral therapy. Clin Microbiol Rev. 2020;33. 10.1128/CMR.00168-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langendries L, Abdelnabi R, Neyts Jet al. Repurposing drugs for mayaro virus: identification of eidd-1931, favipiravir and suramin as mayaro virus inhibitors. Microorganisms. 2021;9. 10.3390/MICROORGANISMS9040734/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis. 1978;137:298–308. [DOI] [PubMed] [Google Scholar]
- Legrand J, Grais RF, Boelle PYet al. Understanding the dynamics of Ebola epidemics. Epidemiol Infect. 2007;135:610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Rouquet P, Formenty Pet al. Multiple Ebola virus transmission events and rapid decline of Central African wildlife. Science. 2004;303:387–90. [DOI] [PubMed] [Google Scholar]
- Lewnard JA, Ndeffo Mbah ML, Alfaro-Murillo JAet al. Dynamics and control of Ebola virus transmission in Montserrado, Liberia: a mathematical modelling analysis. Lancet Infect Dis. 2014;14:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Culhane MR, Schroeder DCet al. Vaccination decreases the risk of influenza A virus reassortment but not genetic variation in pigs. Elife. 2022;11:e78618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-D, Chi W-Y, Su J-Het al. Coronavirus vaccine development: from SARS and MERS to COVID-19. J Biomed Sci. 2020a;27:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Ma H, Shu Jet al. Vaccines and therapeutics against hantaviruses. Front Microbiol. 2020;10:2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Helmersson J, Rocklöv J, Sewe Met al. Climate change may enable Aedes aegypti infestation in major European cities by 2100. Environ Res. 2019;172:693–9. [DOI] [PubMed] [Google Scholar]
- Lo Presti A, Cella E, Giovanetti Met al. Origin and evolution of Nipah virus. J Med Virol. 2016;88:380–8. [DOI] [PubMed] [Google Scholar]
- Loe MWC, Lee RCH, Chu JJH. Antiviral activity of the FDA-approved drug candesartan cilexetil against Zika virus infection. Antiviral Res. 2019;172:104637. [DOI] [PubMed] [Google Scholar]
- Lovelace B Jr WHO says delta is the fastest and fittest Covid variant and will ‘pick off’ most vulnerable. Englewood Cliffs: CNBC, 2021. [Google Scholar]
- Lowe R, Bailey TC, Stephenson DBet al. The development of an early warning system for climate-sensitive disease risk with a focus on dengue epidemics in Southeast Brazil. Statist Med. 2013;32:864–83. [DOI] [PubMed] [Google Scholar]
- Łuksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. [DOI] [PubMed] [Google Scholar]
- Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med. 2008;3:158–66. [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. New ‘Langya’ virus identified in China: what scientists know so far. Nature. 2022;608:656–7. [DOI] [PubMed] [Google Scholar]
- Mandin J, Clot J, Delport Het al. Epidemic of A2 Hong-Kong influenza in Montpellier (winter 1969-70). Presse Med. 1971;79:294. [PubMed] [Google Scholar]
- Martin H-J, Melo-Filho CC, Korn Det al. Small Molecule Antiviral Compound Collection (SMACC): a database to support the discovery of broad-spectrum antiviral drug molecules. Antiviral Res. 2023;217:105620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Jankeel A, Menicucci ARet al. Single dose of a VSV-based vaccine rapidly protects macaques from Marburg virus disease. Front Immunol. 2021;12:4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Filho CC, Bobrowski T, Martin H-Jet al. Conserved coronavirus proteins as targets of broad-spectrum antivirals. Antiviral Res. 2022;204:105360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer MI, Atkins CY, Santibanez Set al. Uppl. 2014;63:1–14. [PubMed] [Google Scholar]
- Memish ZA, Perlman S, Van Kerkhove MDet al. Middle East respiratory syndrome. Lancet North Am Ed. 2020;395:1063–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP. A short history of Lassa fever: the first 10–15 years after discovery. Curr Opin Virol. 2019;37:77–83. [DOI] [PubMed] [Google Scholar]
- Morse SS, Mazet JA, Woolhouse Met al. Prediction and prevention of the next pandemic zoonosis. Lancet North Am Ed. 2012;380:1956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzert S, Selb P, Gohdes Aet al. Tracking and promoting the usage of a COVID-19 contact tracing app. Nat Hum Behav. 2021;5:247–55. [DOI] [PubMed] [Google Scholar]
- Muranyi W, Bahr U, Zeier Met al. Hantavirus Infection. J Am Soc Nephrol. 2005;16:3669–79. [DOI] [PubMed] [Google Scholar]
- Murray CJL, Piot P. The potential future of the COVID-19 pandemic. JAMA. 2021;325:1249. [DOI] [PubMed] [Google Scholar]
- Myers MF, Rogers DJ, Cox Jet al. Forecasting disease risk for increased epidemic preparedness in public health. Adv Parasitol. 2000;47:309–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi AAT, Fatima K, Mohammad Tet al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Emerging and Zoonotic Infectious Diseases (NCEZID) . Chronology of Marburg hemorrhagic fever outbreaks. Atlanta, 2023. [Google Scholar]
- National Institute of Allergy and Infectious Diseases (NIAID) . Influenza treatment. Maryland, 2017. [Google Scholar]
- Neiderud C-J. How urbanization affects the epidemiology of emerging infectious diseases. Infect Ecol Epidemiol. 2015;5:27060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH US National Library of Medicine , Safety tolerability and immunogenicity of INO-4700 for MERS-CoV in healthy volunteers – ClinicalTrials.gov, 2022.
- NIH US National Library of Medicine . Safety and immunogenicity of the candidate vaccine MVA-MERS-S_DF-1 against MERS – ClinicalTrials.gov, 2021a.
- NIH: National Institute of Allergy and Infectious Diseases . Zika virus vaccines. Maryland, 2018. [Google Scholar]
- Park A. Omicron could be the beginning of the end of the COVID-19 pandemic. Time, 2022. [Google Scholar]
- Peterson AT, Samy AM. Geographic potential of disease caused by Ebola and Marburg viruses in Africa. Acta Trop. 2016;162:114–24. [DOI] [PubMed] [Google Scholar]
- Peterson AT. Mapping risk of Nipah virus transmission across Asia and across Bangladesh. Asia Pac J Pub Health. 2015;27:NP824–32. [DOI] [PubMed] [Google Scholar]
- Pigott DM, Golding N, Mylne Aet al. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife. 2014;3. 10.7554/eLife.04395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott DM, Golding N, Mylne Aet al. Mapping the zoonotic niche of Marburg virus disease in Africa. Trans R Soc Trop Med Hyg. 2015;109:366–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A, Terrier O, Nicolas De Lamballerie Cet al. Repurposing of drugs as novel influenza inhibitors from clinical gene expression infection signatures. Front Immunol. 2019;10:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonsky JA, Baidjoe A, Kamvar ZNet al. Outbreak analytics: a developing data science for informing the response to emerging pathogens. Phil Trans R Soc B. 2019;374:20180276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CW. A history of influenza. J Appl Microbiol. 2001;91:572–9. [DOI] [PubMed] [Google Scholar]
- Puhl AC, Fritch EJ, Lane TRet al. Repurposing the Ebola and Marburg virus inhibitors Tilorone, Quinacrine, and Pyronaridine: in vitro activity against SARS-CoV-2 and potential mechanisms. ACS Omega. 2021;6:7454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Liang W, Zhang L. Tracing and prediction analysis of an urban pipeline leakage accident based on the catastrophe DBN model. J Nat Gas Sci Eng. 2018;57:339–48. [Google Scholar]
- Reed PE, Mulangu S, Cameron KNet al. A new approach for monitoring Ebolavirus in wild great apes. PLoS Negl Trop Dis. 2014;8:e3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reperant LA, Osterhaus ADME. AIDS, Avian flu, SARS, MERS, Ebola, Zika… what next?. Vaccine. 2017;35:4470–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuters Staff . World must be better prepared for next pandemic, says WHO boss. 2020.
- Reynolds JJH, Torremorell M, Craft ME. Mathematical modeling of influenza A virus dynamics within swine farms and the effects of vaccination. PLOS ONE. 2014;9:e106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Filho HV, Coimbra LD, Cassago Aet al. Cryo-EM structure of the mature and infective Mayaro virus at 4.4 Å resolution reveals features of arthritogenic alphaviruses. Nat Commun. 2021;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JK. Zika virus. JAMA Dermatol. 2016;152:693. [DOI] [PubMed] [Google Scholar]
- Roguski K, Fry A. Infectious agents. In: CDC Yellow Book 2020. Oxford: Oxford University Press, 2017. [Google Scholar]
- Rosenke K, Feldmann H, Westover JBet al. Use of Favipiravir to treat Lassa virus infection in macaques. Emerg Infect Dis. 2018;24:1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquet P, Froment J-M, Bermejo Met al. Wild animal mortality monitoring and human ebola outbreaks, Gabon and Republic of Congo, 2001–2003. Emerg Infect Dis. 2005;11:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Fons F. Review of the current status of relevant zoonotic pathogens in wild swine (Sus scrofa) populations: changes modulating the risk of transmission to humans. Transbound Emerg Dis. 2017;64:68–88. [DOI] [PubMed] [Google Scholar]
- Safronetz D, Geisbert TW, Feldmann H. Animal models for highly pathogenic emerging viruses. Curr Opin Virol. 2013;3:205–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkas H, Bozidis P, Franks Aet al. Oropouche fever: a review. Viruses. 2018;10:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim AR. Hong Kong influenza in the Sudan. Bull World Health Organ. 1971;44:713–6. [PMC free article] [PubMed] [Google Scholar]
- San Martín JL, Brathwaite Dick O, Del Diego Jet al. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg. 2012;87:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar T, Ghosh I, Rodó Xet al. A realistic two-strain model for MERS-CoV infection uncovers the high risk for epidemic propagation. PLoS Negl Trop Dis. 2020;14:e0008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage F, Langlais M, Pontier D. Predicting the emergence of human hantavirus disease using a combination of viral dynamics and rodent demographic patterns. Epidemiol Infect. 2007;135:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomakers E-M, Lidynia C, Ziefle M. Listen to my heart? How privacy concerns shape users’ acceptance of e-health technologies. In: Proceedings of the 2019 International Conference on Wireless and Mobile Computing, Networking and Communications (WiMob). Piscataway: IEEE, 2019, 306–11. [Google Scholar]
- Slenczka W, Klenk HD. Forty years of Marburg virus. J Infect Dis. 2007;196:S131–5. [DOI] [PubMed] [Google Scholar]
- Smith W, Andrewes CH, Laidlaw PP. A virus obtained from influenza patients. Lancet North Am Ed. 1933;222:66–68. [Google Scholar]
- Spernovasilis N, Markaki I, Papadakis Met al. Epidemics and pandemics: is human overpopulation the elephant in the room?. Ethics Med Pub Health. 2021;19:100728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Doan YH, Kimura Het al. Predicting directions of changes in genotype proportions between norovirus seasons in Japan. Front Microbiol. 2019;10. 10.3389/fmicb.2019.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T. Imported dengue fever/dengue hemorrhagic fever cases in Japan. TropMedHealth. 2011;39:S13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. Norovirus vaccines: current clinical development and challenges. Pathogens. 2021;10:1641. 10.3390/PATHOGENS10121641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. Influenza: the once and future pandemic. Pub Health Rep. 2010;125:16–26. [PMC free article] [PubMed] [Google Scholar]
- Texas A&M University . Urban Americans more likely to follow Covid-19 prevention behaviors than rural Americans. College Station, 2021. [Google Scholar]
- The inexorable progress of norovirus . Lancet Infect Dis. 2013;13:97. https://pubmed.ncbi.nlm.nih.gov/23347621/ [DOI] [PubMed] [Google Scholar]
- Tollefson J. Why deforestation and extinctions make pandemics more likely. Nature. 2020;584:175–6. [DOI] [PubMed] [Google Scholar]
- Tolu-Kolawole D. FG deploys response teams as Lassa fever kills 153. PUNCH, 2022. [Google Scholar]
- U.S. Food & Drug Administration (FDA) . Coronavirus (COVID-19) drugs. Maryland, 2023. [Google Scholar]
- Univeristy of California, Davis . PREDICT. Davis, 2009. [Google Scholar]
- US Food and Drug Administration, FDA . COVID-19 vaccines. Maryland, 2022. [Google Scholar]
- US National Library of Medicine . Safety tolerability and immunogenicity of INO-4500 in healthy volunteers – ClinicalTrials.gov, 2020.
- Valencia-Marín BS, Gandica ID, Aguirre-Obando OA. The Mayaro virus and its potential epidemiological consequences in Colombia: an exploratory biomathematics analysis. Parasit Vectors. 2020a;13:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MJ, Ciraola B, Aliota MTet al. No evidence for sylvatic cycles of chikungunya, dengue and Zika viruses in African green monkeys (Chlorocebus aethiops sabaeus) on St. Kitts, West Indies. Parasit Vectors. 2020;13:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boheemen S, De Graaf M, Lauber Cet al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3. 10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos PFC, Costa ZG, Travassos Da Rosa ESet al. Epidemic of jungle yellow fever in Brazil, 2000: implications of climatic alterations in disease spread. J Med Virol. 2001;65:598–604. [PubMed] [Google Scholar]
- Vergote V, Laenen L, Mols Ret al. Chloroquine, an anti-malaria drug as effective prevention for hantavirus infections. Front Cell Infect Microbiol. 2021;11:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef L, Hewitt J, Barclay Let al. Norovirus genotype profiles associated with foodborne transmission, 1999–2012. Emerg Infect Dis. 2015;21:592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana DV, Ignotti E. The ocurrence of dengue and weather changes in Brazil: a systematic review. Rev Bras Epidemiol. 2013;16:240–56. [DOI] [PubMed] [Google Scholar]
- Viboud C, Sun K, Gaffey Ret al. The RAPIDD Ebola forecasting challenge: synthesis and lessons learnt. Epidemics. 2018;22:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Thurmond S, Islas Let al. Zika virus genome biology and molecular pathogenesis. Emerg Microbes Infect. 2017;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Deng Z. Modeling and prediction of oyster norovirus outbreaks along Gulf of Mexico coast. Environ Health Perspect. 2016;124:627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang L, Zhuang H. Profiling and characterization of SARS-CoV-2 mutants’ infectivity and antigenicity. Sig Transduct Target Ther. 2020;5:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardeh M, Baylis M, Blagrove MSC. Predicting mammalian hosts in which novel coronaviruses can be generated. Nat Commun. 2021;12:780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Severe Acute Respiratory Syndrome (SARS) multi-country outbreak - Update 6. Geneva, 2009. [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJet al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Geoghegan JL, Holmes EC. How accurately can we assess zoonotic risk?. PLoS Biol. 2021;19:e3001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C, Geisbert TW. Current state of Ebola virus vaccines: a snapshot. PLoS Pathog. 2021;17:e1010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Oropouche virus disease – French Guiana, France. Geneva, 2020a. [Google Scholar]
- World Health Organization . Mayaro virus disease – French Guiana, France. Geneva, 2020b. [Google Scholar]
- World Health Organization . Ebola virus disease: vaccines. Geneva, 2020c. [Google Scholar]
- World Health Organization . Ebola Outbreak 2021– N'Zerekore, Guinea. Geneva, 2021. [Google Scholar]
- Worobey M, Levy JI, Malpica Serrano Let al. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science. 2022;377:951–9. 10.5281/zenodo.6299600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hu J, Wang Jet al. Genome organization of the SARS-CoV. Genomics Proteomics Bioinformatics. 2003;1:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Sahay RR, Balakrishnan Aet al. Nipah virus outbreak in Kerala State, India amidst of COVID-19 pandemic. Front Pub Health. 2022;10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Luusua E, Dabrowski Jet al. Tempel: time-series mutation prediction of influenza A viruses via attention-based recurrent neural networks. Bioinformatics. 2020;36:2697–704. [DOI] [PubMed] [Google Scholar]
- Yuan S. Possible FDA-approved drugs to treat Ebola virus infection. Infect Dis Poverty. 2015;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yakob L, Bonsall MBet al. Predicting seasonal influenza epidemics using cross-hemisphere influenza surveillance data and local internet query data. Sci Rep. 2019;9:3262. [DOI] [PMC free article] [PubMed] [Google Scholar]