Abstract
Fetal lung fibroblasts contribute dynamic infrastructure for the developing lung. These cells undergo dynamic mechanical transitions, including cyclic stretch and spreading, which are integral to lung growth in utero. We investigated the role of the nuclear envelope protein emerin in cellular responses to these dynamic mechanical transitions. In contrast to control cells, which briskly realigned their nuclei, actin cytoskeleton, and extracellular matrices in response to cyclic stretch, fibroblasts that were acutely downregulated for emerin showed incomplete reorientation of both nuclei and actin cytoskeleton. Emerin-downregulated fibroblasts were also aberrantly circular in contrast to the spindle-shaped controls and exhibited an altered pattern of filamentous actin organization that was disconnected from the nucleus. Emerin knockdown was also associated with reduced myosin light chain phosphorylation during cell spreading. Interestingly, emerin-downregulated fibroblasts also demonstrated reduced fibronectin fibrillogenesis and production. These findings indicate that nuclear-cytoskeletal coupling serves a role in the dynamic regulation of cytoskeletal structure and function and may also impact the transmission of traction force to the extracellular matrix microenvironment.
Significance
Mechanical forces are transmitted to cells via a network of extracellular matrices, integrins, cytoskeleton, and nuclei. Coupling between the nucleus and cytoskeletal actin may be a critical factor driving cellular responses to mechanical stimuli from the microenvironment and for adaptive changes in cellular function during development. Cyclic stretch associated with breathing is important in fetal lung development and adaptation to extrauterine life. Factors that limit frequency or force transduction during fetal lung cycling could result in incomplete lung morphogenesis or maturation. Our investigation of the role of the nuclear envelope protein emerin in fetal lung fibroblast morphology and function suggests that perturbed force transduction at the nuclear-cytoskeletal interface may have a pathogenic role in developmental lung diseases.
Introduction
Mechanical forces contribute to the development of many cells and tissues, including the heart and lungs (1,2). Breathing movements are important for fetal lung development, and begin at approximately 10 weeks of gestation (3). Fetal breathing consists of intermittent excursions during which lung parenchyma undergoes cyclic stretch by diaphragmatic movement and the inflow of amniotic fluid for a few hours of alternating expansion and relaxation each day, and these cycles are separated by long intervals of rest or stasis (4,5). This mechanical cycling is critical for fetal lung growth and maturation to prepare for adaptation to breathing air during extrauterine life (6,7,8). Factors that limit the force or frequency of fetal lung cycling, including abnormal neuromuscular development or injuries to neural structures, result in incomplete lung morphogenesis or maturation (9,10). At the cellular level, cyclic stretch improves lung cell proliferation (11,12), differentiation (13), production of extracellular matrix (ECM) (14,15) and surfactant (16), and vasculogenesis (17,18). Therefore, an understanding of mechanosensing and response to stretch in fetal lung cells may uncover disease mechanisms and potential therapies for developmental lung pathologies.
In tissues under dynamic mechanical loading from environmental movement, forces exerted on cells are transmitted from ECM outside of the cell, via matrix adhesions, integrins, and cytoskeleton, to the nuclear envelope (NE) and nucleus (19,20,21). At the receiving end of this chain of mechanotransduction, the nucleus has a dominant role in overall cellular mechanical response as it is the largest and stiffest organelle and may provide a clutch or anchor for cytoskeletal force (22,23,24). The NE provides the physical connection between the cytoskeleton and the nucleus (25,26), and participates in the mechanosensitive transcriptional regulation and chromatin modification, which in turn alters cell functions (27). LINC (linker of the nucleoskeleton and cytoskeleton) complexes and nuclear lamins are essential for force transmission, cellular reorganization, nuclear rotation, and nuclear shape (28,29,30,31,32), and are also necessary for mechanosensitive transcriptional factor translocation including NF-kB (30) and YAP/TAZ (33), and stretch-induced chromatin changes and gene expression (34).
Emerin is a LEM (LAP2, emerin, MAN1) domain protein that interacts with LINC complexes and localizes predominantly at the inner nuclear membrane (35). Emerin directly binds a chromatin protein barrier-to-autointegration factor and nuclear lamins, together contributing to nuclear lamina networks, which mediate chromatin organization and tethering at the NE (36). Emerin contributes to nuclear structural integrity and responses to stress induced by spatial restriction (35,37,38,39), and has been shown to interact with cytoskeletal microtubules (40), myosin IIB (41), filamentous actin (F-actin) (42), and in vitro myosin-1 (43). Emerin also partners in interactions with LINC complex proteins (44,45,46). In addition to supporting nuclear mechanical integrity and interacting with cytoskeletal and nuclear proteins, emerin modulates the expression of the mechanosensitive genes egr-1, anti-apoptotic gene iex-1, and serum responsive factor-dependent genes that are involved in cell migration, growth, and differentiation (47,48). More recent studies highlighted new roles of emerin in NE protein distribution, cytoskeletal organization, and cell migration. Under biaxial cyclic strain, emerin was detected at the outer nuclear membrane with locally enriched actin polymerization (49). Either upregulation or downregulation of emerin was found to reduce the speed of ameboid migration in melanoma cells (39). Furthermore, RNAi knockdown of emerin was shown to alter the front-back polarization of NE proteins and chromosomes, decrease focal adhesion size, and increase cell migration velocity (50). We interrogated the roles of emerin in nuclear and cytoskeletal responses to mechanical perturbations in the physiological context of cells from a specific mechanically responsive tissue—the human lung.
We used cyclic stretch and cell spreading assays to test the hypothesis that emerin facilitates mechanical responses of fibroblasts during development by coupling the actomyosin cytoskeleton to nuclei. Fetal lung fibroblasts with emerin knockdown (emerin KD) showed multiple phenotypes relevant to lung development, including decreased cell reorientation in response to stretch and loss of F-actin contiguity with the nucleus. Cell size, cell shape, myosin regulatory light chain phosphorylation, and matrix patterning were also affected by emerin KD. These results indicate that nuclear-cytoskeletal interactions may tether actomyosin filaments to the nucleus and provide essential torque points for normal nuclear, cytoskeletal, and matrix responses to mechanical cues during lung development.
Materials and methods
Cell culture
WI-38 human fetal lung fibroblasts (early passage; American Type Culture Collection, Manassas, VA) were maintained in Eagle’s minimal essential medium with Earle’s salts and L-glutamine (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (R&D Systems, Minneapolis, MN), 1 mM sodium pyruvate (Thermo Fisher Scientific), 1% nonessential amino acids (Thermo Fisher Scientific), and 1% antibiotic-antimycotic solution (Thermo Fisher Scientific). Cells were maintained at 37°C and 5% CO2.
Mechanical stretch and cell substrates
Uniaxial stretch was applied with 15% strain at a frequency of 0.5 Hz, unless otherwise specified, by an automated cell stretching system (STREX, model STB-1400, San Diego, CA). Stretch chambers were cleaned twice with decellularization buffer (1% Triton, 1% SDS, 20 mM NH4OH) and washed extensively with deionized water before sterilization with UV irradiation and 70% ethanol. Chambers were then rinsed with phosphate-buffered saline (PBS) three times and coated with 10 μg/mL human plasma fibronectin (Sigma, F1056, St. Louis, MO) in a universal buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.2% sodium azide) at 4°C overnight. Chambers were washed with sterile PBS before cell plating. Before stretch, cells from culture flasks were detached by 0.05% trypsin-EDTA (Thermo Fisher Scientific), resuspended in culture medium, and plated at 25,000 cells/cm2 for a confluent monolayer, or at 5000 cells/cm2 for cell shape and spreading analysis. Cells were incubated for 2 h without stretch to allow initial adhesion, and then subjected to stretch or control treatment for 6 h.
For the stretch experiments (data shown in Figs. 1, 2, 3, S2, and S3), cells were plated on polydimethylsiloxane (PDMS) stretch chambers (STREX, STB-CH-10) that were coated with 10 μg/mL fibronectin. The Young’s modulus of PDMS is ∼3 MPa. For the biochemistry experiments (data shown in Figs. 4 and 5), cells were plated in tissue culture plastic (p35 dish; Corning, Corning, NY) that was coated with 10 μg/mL fibronectin. The Young’s modulus of the plastic is ∼10 GPa. For the cell spreading experiments (immunolabeled images in Figs. 5, S4, and S5), cells were plated on coverslips that were coated with 1% gelatin (bloom 175, Sigma). The Young’s modulus for the glass coverslips is ∼70 GPa.
Figure 1.
Cyclic stretch induces alignment of extracellular matrix, F-actin, and nuclei in fetal lung fibroblasts. (A) A schematic illustration of the experimental setup. WI-38 fibroblasts plated on stretch chambers were subjected to cyclic uniaxial stretch (direction indicated by arrows). Fibronectin (dark gray), F-actin (light gray), and nuclei are highlighted. Top and side views of cells are shown. (B–D) Distribution of fibronectin fibrils (B), F-actin (C), and nuclear (D) orientations at 0, 2, 15, and 20% strain. The angle of 0° is defined as the orientation perpendicular to the axis of stretch. Insets show representative images of the feature under study in grayscale. Images were analyzed using customized scripts (materials and methods). (E) The percentages of objects (fibronectin fibrils, F-actin, and nuclei) within ±15° of 0°, at 0, 2, 15, and 20% strain. Values are shown as mean ± standard error of the mean. Higher strains caused more complete realignment.
Figure 2.
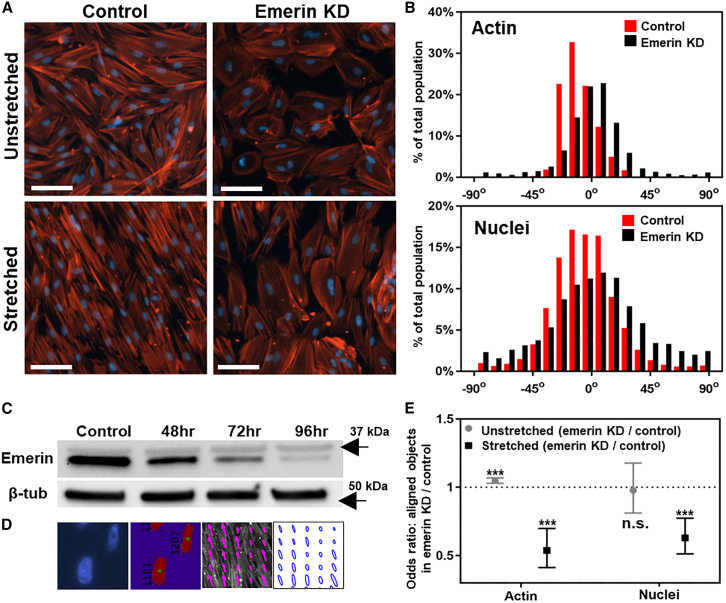
Emerin downregulation reduces alignment of F-actin and nuclei in response to stretch. (A) Epifluorescence images show nuclei and F-actin in control and emerin KD cells, with and without 15% stretch. Scale bars, 100 μm. (B) Distribution of F-actin and nuclear orientation observed in control (red) and emerin KD (black) cells. The angle of 0° is defined as the orientation perpendicular to the axis of stretch. (C) Western blot of emerin protein expression in WI-38 cells at different time points after adenovirally mediated knockdown. Emerin KD was maximal at 4 days after adenoviral transduction. (D) Representative images show the image analysis strategies to quantify the orientation of nuclei and F-actin. Nuclei were recognized as individual objects and orientation was determined by the long axis. For F-actin, each ellipse of persistence represents F-actin pattern in the corresponding tile. The orientation of the long axis represents F-actin orientation. Long and short axis values represent the extent of F-actin persistence parallel or perpendicular to the long axis, respectively. See materials and methods for details. (E) Odds ratios (OR) (dots and squares) and 95% confidence intervals (95% CI; bars) for the orientation of F-actin and nuclei. The OR is defined as the odds of F-actin filaments (or nuclei) within ±15° of the mean orientation in emerin KD compared with the odds in control. For the stretched groups, an OR of 0.63 indicates that the odds of having an emerin KD nucleus within ±15° of the mean orientation is 37% smaller than the odds of having a control nucleus within ±15° of the mean. OR (95% CI) comparing emerin KD with control cells were as follows: F-actin in unstretched condition, 1.05 (1.03–1.07, p < 0.001); F-actin in stretched conditions, 0.54 (0.41–0.70, p < 0.001); nuclei in unstretched condition, 0.97 (0.81–1.18, p = 0.81); nuclei in stretched conditions, 0.63 (0.51–0.77, p < 0.001). ∗∗∗p < 0.001; n.s. denotes that differences are not significant.
Figure 3.
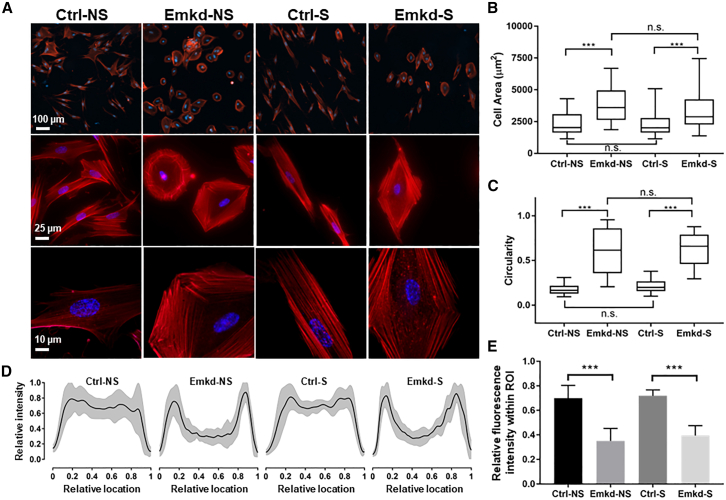
Emerin-downregulated cells are larger, more circular, and have a peripheral F-actin distribution. (A) Epifluorescence images of control (Ctrl) and emerin KD (Emkd) fibroblasts in unstretched (NS) or stretched (S) conditions. The middle and lower rows are shown with 4× and 10× greater magnification (respectively) than the top row (please see scale bars). The F-actin (red, rhodamine-phalloidin) and nuclei (blue, 4′,6-diamidino-2-phenylindole [DAPI]) are shown. (B) Quantitative analysis of cell size shows that Emkd cells are significantly larger than Ctrl in either unstretched or stretched conditions. Boxes indicate the median, first, and third quartiles. Whiskers indicate the 5th and 95th percentiles. Mean cell sizes (95% CI) were as follows (in μm2): Ctrl-NS, 908 (828–988); Emkd-NS, 1475 (1329–1622); Ctrl-S, 936 (815–1057); Emkd-S, 1341 (1277–1404). ∗∗∗p < 0.001; n.s., differences are not significant. (C) Quantitative analysis of cell circularity shows that there is a significant difference in circularity between Ctrl and Emkd in both unstretched and stretched conditions. Boxes indicate the median, first, and third quartiles. Whiskers indicate the 5th and 95th percentiles. Circularity (95% CI) values were as follows: Ctrl-NS, 0.17 (0.15–0.20); Emkd-NS, 0.58 (0.45–0.71); Ctrl-S, 0.22 (0.20–0.24); Emkd-S, 0.63 (0.51–0.76). ∗∗∗p < 0.001; n.s., differences are not significant. (D) F-actin fluorescence intensity distribution over the width of the cells. The y axis shows the normalized F-actin fluorescence intensity. The x axis shows the relative location in the cell normalized by the total width. The black line represents the mean value, and the gray zone represents the standard deviation. See materials and methods for details. (E) Quantitative analysis of the mean F-actin fluorescence intensity at region of interest (ROI): between 20 and 80% of the relative cell width. Bars and whiskers represent mean ± standard deviation. F-actin intensity values within the ROI (mean ± standard deviation) were as follows: Ctrl-NS, 0.70 ± 0.10; Emkd-NS, 0.35 ± 0.10; Ctrl-S, 0.72 ± 0.05; Emkd-S, 0.40 ± 0.08. ∗∗∗p < 0.001. See materials and methods for details.
Figure 4.
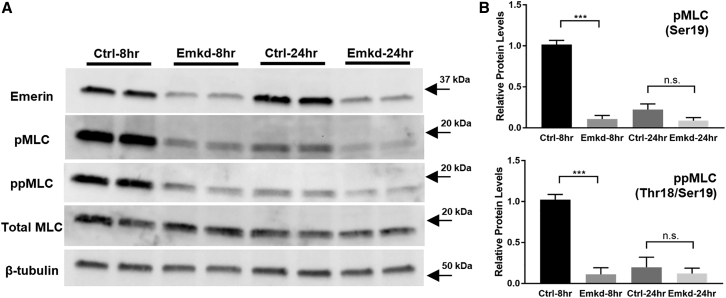
Emerin-downregulated cells exhibit less myosin light chain phosphorylation during cell spreading than controls. (A) Control (Ctrl) and emerin KD (Emkd) fibroblasts were replated and cultured for 8 or 24 h. Immunoblotting was performed to probe monophosphorylation (Ser19; pMLC) and diphosphorylation (Thr18/Ser19; ppMLC) of the myosin light chain. Two technical replicates from each condition are shown. (B) Relative protein levels of pMLC and ppMLC demonstrated a large diminution of both pMLC and ppMLC in Emkd cells at 8 h, and a trend toward a similar pattern at 24 h. Bars and whiskers represent mean ± standard error of the mean based on four biological replicates. Relative pMLC levels were as follows (mean ± standard error of the mean): Ctrl-8hr, 1.02 ± 0.05; Emkd-8hr, 0.11 ± 0.04; Ctrl-24hr, 0.22 ± 0.07; Emkd-24hr, 0.09 ± 0.04. For ppMLC: Ctrl-8hr, 1.02 ± 0.06; Emkd-8hr, 0.11 ± 0.08; Ctrl-24hr, 0.20 ± 0.12; Emkd-24hr, 0.12 ± 0.06.∗∗∗p < 0.001; n.s. denotes that differences are not significant.
Figure 5.
Emerin downregulation decreases fibronectin fibrillogenesis and expression. (A) Epifluorescence images of fibrillogenesis in control (Ctrl) and emerin KD (Emkd) cells on gelatin-coated coverslips for 8 and 24 h. Cells were labeled for F-actin (red, phalloidin), nuclei (blue, DAPI), and fibronectin (grayscale, antibody labeling). Green fluorescence is from GFP that was co-transduced by the adenovirus for emerin KD. Scale bars, 25 μm. (B) Distribution of fibrillogenesis scores in control and emerin KD cells. Higher scores represent more fibrillogenesis (Fig. S6). A score of 5 indicates several continuous fibers over the cell body; a score of 1 indicates only one short fiber at a single edge of a cell. The plot shows the distribution of scores in Ctrl and Emkd at 8 and 24 h. (C) Comparison of the mean (95% CI) of fibrillogenesis scores in the control and emerin KD cells. The mean scores (95% CI) were as follows: Ctrl-8hr, 1.78 (1.56–2.00); Emkd-8hr, 0.50 (0.38–0.63); Ctrl-24hr, 2.71 (2.18–3.24); Emkd-24hr, 0.93 (0.64–1.22). ∗∗∗p < 0.001; n.s. denotes that differences are not significant. Data were analyzed from three biological replicates. (D) Fibronectin protein expression in control and emerin KD cells that were replated and cultured for 24 h after the 4-day knockdown or control treatment. Two technical replicates from each condition are shown. (E) Relative fibronectin protein levels at 24 h. Bars and whiskers represent mean ± standard error of the mean, based on five biological replicates. Relative protein levels were as follows (mean ± standard error of the mean). For fibronectin: Ctrl, 0.97 ± 0.04; Emkd, 0.26 ± 0.10. For emerin: Ctrl, 1.03 ± 0.04; Emkd, 0.15 ± 0.05. ∗∗∗p < 0.001.
Adenoviral constructs
Please refer to the supporting material for details. In brief, Adenoviruses encoding shRNA targeted to green fluorescence protein sequence (Ad.Con-shRNA) and human emerin (Ad.Emerin-shRNA) were generated using a pAd/BLOCK-iT kit (Life Technologies, Carlsbad, CA). For cell transduction, adenoviral vectors for control or emerin KD were added to WI-38 cells at 60–70% confluence. The multiplicity of infection was ∼40. The medium was replaced with fresh medium after 8 h. Cells were used for assays 96 h after transduction.
Antibodies
For immunofluorescence staining, the following antibodies were used at specified concentrations: mouse monoclonal anti-fibronectin (1:100, Millipore Sigma, IST-10, Burlington, MA), mouse monoclonal anti-emerin (1:100, Leica Biosystems, 4G5, Wetzlar, Germany), mouse monoclonal anti-vimentin (1:100, Abcam, RV202), and rabbit polyclonal anti-β-tubulin (1:100, Abcam, ab6046, Cambridge, UK). Secondary antibodies (Jackson ImmunoResearch, West Grove, PA) include: Cy5-conjugated donkey anti-mouse IgG (1:50), Cy5-conjugated donkey anti-rabbit IgG (1:50), and Alexa Fluor 488-conjugated goat anti-mouse IgG (1:100). Rhodamine phalloidin (1:200, Molecular Probes, Eugene, OR) was used to label F-actin. 4′,6-Diamidino-2-phenylindole (1:10,000, Sigma-Aldrich, St. Louis, MO) was used to label nuclei.
For immunoblotting, the following antibodies were used at specified concentrations: mouse monoclonal anti-fibronectin (1:1000, Millipore Sigma, IST-10), mouse monoclonal anti-emerin (1:1000, Leica Biosystems, 4G5), rabbit polyclonal anti-β-tubulin (1:2000, Abcam, ab6046), mouse monoclonal anti-lamin A/C (1:1000, Santa Cruz, sc376248, Santa Cruz, CA); from Cell Signaling Technology, Danvers, MA: rabbit polyclonal anti-phospho-myosin light chain 2 (Ser19) (no. 3671; 1:1000, in 5% cold water fish gelatin in Tris-buffered saline with Tween-20 (TBST): 50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.05% Tween-20), rabbit polyclonal anti-phospho-myosin light chain 2 (Thr18/Ser19) (no. 3674; 1:1000, in 5% cold water fish gelatin in TBST), rabbit polyclonal anti-myosin light chain 2 (no. 3672; 1:1000, in 5% bovine serum albumin in TBST); from Jackson ImmunoResearch, peroxidase-conjugated AffiniPure goat anti-mouse IgG (1:3000), and peroxidase-conjugated AffiniPure goat anti-rabbit (1:3000).
Immunofluorescence staining and imaging
Cells were permeabilized in 0.5% Triton X-100 in 4% paraformaldehyde (Sigma-Aldrich) for 2 min, fixed in 4% paraformaldehyde for 20 min, rinsed twice in PBS, and stored at 4°C. Primary and secondary antibodies were diluted in 0.1% bovine serum albumin in universal buffer. Coverslips were incubated with primary antibodies for 1 h at 37°C, washed in universal buffer three times, incubated with secondary antibodies for 1 h at 37°C, and washed in universal buffer and PBS. Samples were mounted with FluorSave (EMD Millipore, Burlington, MA) on glass slides. Cells were imaged using an inverted Nikon Eclipse TE-200 microscope equipped with epifluorescence, a Rolera EMCCD camera (Q-Imaging), and Volocity software (PerkinElmer, Waltham, MA). All images were acquired at zero strain when the substrates returned to the original unstretched state.
Western blotting
Cells were lysed at the specified time points in lysis buffer containing 0.1% sodium deoxycholate, 0.1% Triton X-100, 2 mM EDTA, 2 mM sodium orthovanadate, 20 μg/mL leupeptin, and 20 μg/mL aprotinin in universal buffer. Protein quantification was performed using Bradford protein assay (Thermo Fisher Scientific). Cell lysates were adjusted to equal protein concentration and reduced in 4× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA) supplemented with 10% β-mercaptoethanol (Sigma-Aldrich). Lysates were denatured for 5 min at 100°C and subjected to SDS-PAGE on 4–15% gradient gels (Bio-Rad Laboratories). Proteins were transferred to nitrocellulose membranes by wet transfer. Membranes were blocked with 5% nonfat milk powder in TBST for 30 min at room temperature. Membranes were probed with primary antibody in blocking buffer at 4°C overnight, washed three times in TBST, and incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature. Pierce enhanced chemiluminescence substrate (Thermo Fisher Scientific) and an Image Lab scanner (Bio-Rad Laboratories) were used to detect signals. Protein densitometry was performed using Image Lab software (Bio-Rad Laboratories).
Finite element analysis
Please refer to supporting material for details. In brief, finite element analysis (FEA) on the deformation of the stretch chamber was performed in Abaqus CAE software (Dassault Systèmes, Waltham, MA). The entire chamber was modeled as a solid 3D object of 10 mm thickness. Extrusion was used to create a thin film of thickness 0.1 mm where cells were attached. Material properties were based on published data for PDMS and ECM (Table S1 in the supporting material) (51,52). A static model and structured hexahedron elements were used. Boundary conditions were based on displacement at the posts where the chambers were pulled.
Image analysis
Please refer to supporting material for details. All customized scripts used in this section are available from the corresponding author. For nuclei alignment, fluorescence images were analyzed in Spyder software (Spyder project contributors) using a Python script developed by the authors. Images were converted to grayscale, and nuclei were identified by adaptive local thresholding. A size filter was applied to remove overlapping nuclei and background noise. The image of each individual nucleus was analyzed and indexed for parameters including major axis orientation, aspect ratio, area, and perimeter. For F-actin and fibronectin alignment and spread, fluorescence images were analyzed in MATLAB software (The MathWorks, Natick, MA) using a script developed by the authors. Each image was divided into 12 × 12 tiles and each tile approximated the size of a single cell. An autocorrelation method was used to describe the pattern of fiber alignment in a tile as an ellipse of persistence with long and short axes.
For cell shape and circularity, images of F-actin in sparsely plated cells were analyzed in ImageJ software using the MorphoLibJ plugin (53). Images were converted to grayscale and thresholded in ImageJ by the Li method to generate binary images. Each cell was labeled and analyzed for cell area, perimeter, and circularity. For F-actin distribution in single cells, images of F-actin in sparsely plated cells were analyzed in ImageJ software using the plot profile function. For each cell, a straight line was drawn across the center of the nucleus along the direction of the short axis and ending at cell boundaries. The F-actin fluorescence intensity profile along this line was normalized and used for analysis.
Statistical analysis
Please refer to supporting material for details. Statistical analysis was performed on data from three or more biological replicates, as detailed in each experiment. For stretch experiments, the conditions were: control unstretched, control stretched, emerin KD unstretched, and emerin KD stretched. For cell spreading experiments, the conditions were: control 8 h, control 24 h, emerin KD 8 h, and emerin KD 24 h.
For nuclear orientation distribution, in each image we assigned a value of 1 if nuclear orientation was within ±15° of the mean orientation, and a value of 0 if it was outside of ±15° of the mean orientation. The values from all biological replicates across experimental conditions were compared by a multilevel logistic regression as odds ratios (ORs) with 95% confidence intervals (95% CI) in STATA software (StataCorp, College Station, TX). The model included two factors (stretch and emerin KD) and their interaction and accounted for the data’s hierarchy level: image per chamber per condition per replicate. The odds are defined as the ratio between the number of nuclei within ±15° of the mean (henceforth named “aligned” nuclei) and the number of nuclei outside of ±15° of the mean (henceforth named “unaligned” nuclei) in one experimental condition. Odds describe the likelihood of having an aligned nucleus in one experimental condition. The OR compares the odds of having an aligned nucleus between two different experimental conditions. For an OR between emerin KD stretched and control stretched, for example, an OR of 1 means that there is no difference between the two groups, whereas an OR of 0.5 means that the odds of having an aligned nucleus in emerin KD stretched is 50% of the odds of having an aligned nucleus in control stretched. Similarly, an OR of 2 means that the odds of having an aligned nucleus in emerin KD stretched is double the odds of having an aligned nucleus in control stretched condition. The distribution of F-actin orientation was analyzed similarly.
For cell area, circularity, and fibronectin score (an ordinal fibrillogenesis scoring system with scores of zero to five indicating no to abundant fibrils, respectively; Fig. S6 in the supporting material), a STATA multilevel regression analysis was used to compare the differences of mean values between conditions with all three replicates combined. The analysis accounted for the data’s hierarchy level: image per chamber or coverslip per condition per replicate. Densitometry data from western blots were analyzed using t-test. Statistical significance was indicated if p < 0.05. Significance levels in the figures are denoted as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Additional details regarding the statistical methodology are included in the supporting material.
Results
Stretch induces alignment of ECM, F-actin, and nuclei in human lung fibroblasts
We plated WI-38 fetal lung fibroblasts on fibronectin-coated stretch chambers, and applied uniaxial cyclic stretch (Fig. 1 A) with a pattern reported for human fetal breathing movements: 15% strain at 0.5 Hz for 6 h (5,54). We mapped the strain distribution in the stretch chamber using FEA simulation and digital image correlation experiment. The results confirmed a uniform strain distribution over the majority of the film and a sinusoidal stretching pattern (Fig. S1 in the supporting material). FEA informs the area with uniform strain distribution, whence the images were sampled. Epifluorescence images of WI-38 cells demonstrated stretch-induced reorientation of three components—nuclei, cytoskeletal F-actin, and fibronectin in the ECM—to nearly 90° relative to the axis of uniaxial stretch (Fig. S2 in the supporting material). To the best of our knowledge, while previous studies reported alignment of cytoskeleton and nuclei (55,56,57), alignment across the entire confluent cell layer and ECM has not been reported. The morphology and orientation of these components are shown as distributions of orientations for fibronectin (Fig. 1 B), F-actin (Fig. 1 C), and nuclei (Fig. 1 D) based on quantitative image analysis.
We further studied the effects of strain values on the alignment by applying 0, 2, 15, and 20% strain to the fetal lung fibroblasts (Fig. 1 B–E). We designated the orientation that was perpendicular to the axis of stretch as 0°. In WI-38 cells subjected to 15 and 20% strain, we detected a threefold increase in the fractions of F-actin, fibronectin, and nuclei that aligned within ±15° of this 0° orientation, compared with those at lower strains (2 and 0%). The collective realignment of nuclei, F-actin, and ECM suggested that these lung fibroblasts respond to cyclic strain of 15% or greater as an integrated network. Subsequently, the roles of the NE in cellular mechanical response, cell shape, cytoskeleton, and ECM were investigated to evaluate the potential role of nuclear-cytoskeletal coupling in coordinating the cellular responses to mechanical deformation in lung fibroblasts.
Emerin downregulation reduces stretch-induced realignment of nuclei and F-actin
We studied the role of nuclear-cytoskeletal coupling in the responses of lung fibroblasts to mechanical stretch by downregulating the expression of the NE protein emerin. We transduced cells with adenoviruses encoding either a control shRNA (Ad.con-shRNA) or an emerin-specific shRNA with green fluorescence protein marker (Ad.Emerin-shRNA) at a multiplicity of infection of ∼40. By immunoblotting, we found that emerin protein expression was reduced to ∼5% compared with control, at 96 h after Ad.Emerin-shRNA delivery (Fig. 2 C), consistent with the 1.5-day half-life that is reported in the literature (58). Therefore, we utilized WI-38 fibroblasts 4 days after delivery of either Ad.con-shRNA or Ad.Emerin-shRNA. After 6 h of 15% strain at 0.5 Hz, control cells showed highly uniform perpendicular realignment of nuclei and F-actin relative to the axis of uniaxial stretch, whereas emerin KD cells showed incomplete reorientation of nuclei and F-actin (Fig. 2 A). Due to imperfect horizontal leveling of the stretch chambers, the alignment of objects showed some offset from an ideal perpendicular direction. We took this into account and subtracted the mean angle from the observed objects to evaluate the width of the distribution from the mean.
We quantified nuclear and F-actin realignment and morphology using customized scripts (Fig. 2 D; materials and methods). Image analysis showed that emerin KD cells had fewer aligned nuclei and less aligned F-actin after stretch, compared with those in control cells (Fig. 2 B). We used the concept of OR (materials and methods) and statistical analysis to quantify the alignment tendency. The OR compares the odds of having an aligned nucleus (within ±15° of the mean) between two different experimental conditions. For nuclear alignment, the OR (95% CI) between emerin KD and control cells was 0.97 (0.81–1.18, p = 0.81) for the unstretched groups, and 0.63 (0.51–0.77, p < 0.001) for the stretched groups (Fig. 2 E). For F-actin alignment, the OR (95% CI) was 1.05 (1.03–1.07, p < 0.001) for the unstretched groups, and 0.54 (0.41–0.70, p < 0.001) for the stretched groups (Fig. 2 E). To interpret the numbers, in the stretched groups the odds of having an aligned nucleus in emerin KD were 37% lower (OR = 0.63) than the odds in control, and the odds of having an aligned F-actin tile in emerin KD were 46% lower (OR = 0.54) than the odds in control. In the unstretched groups, the OR for nuclei alignment between emerin KD and control was not statistically different (OR = 0.97, p = 0.81); the OR for F-actin alignment was statistically significant (OR = 1.05, p < 0.001), but its value close to 1 indicates that it is likely not biologically significant.
We also studied the impact of emerin KD on stretch-induced changes in F-actin pattern by image analysis, where the pattern of F-actin was represented by ellipses of persistence, and the major and minor axis values of the ellipse represented the spread of F-actin (details in supporting material). Stretch increased F-actin anisotropy, with lengthened major axis and shortened minor axis of the representative ellipses in both control and emerin KD fibroblasts (Fig. S3). Emerin KD cells showed larger values of both the major and minor axes compared with the respective measurements in control cells, leading us to investigate the impact of emerin KD on cell shape.
Emerin downregulation changes cell size, circularity, and F-actin organization
To quantitatively characterize morphology and F-actin organization, we treated and stretched the WI-38 cells as described previously and at a lower plating density. Noting that nearly all emerin KD cells had a strikingly round shape, independent of stretch (Fig. 3 A), we examined cells at higher magnification. Emerin KD cells had consistently larger sizes (∼60%) than control cells (Fig. 3 B; p < 0.001 for each). The size was not affected by stretch in either control or emerin KD cells (Fig. 3 B). We also applied a circularity metric and found that emerin KD cells contrasted substantially with the spindle shape of control cells in both unstretched and stretched conditions (Fig. 3 C; p < 0.001 for each).
F-actin was strikingly limited to the cell periphery in emerin KD cells, in stark contrast to control cells (Fig. 3 A). Higher-magnification images confirmed that the perinuclear cell center was often largely devoid of F-actin in emerin KD cells, and that bundles of short F-actin fibers localized near the cell perimeter (Fig. 3 A). These short F-actin filaments were randomly oriented in unstretched emerin KD cells (Fig. 3 A), but formed diamond shapes with the long axis perpendicular to the stretch direction (Fig. 3 A). By a quantitative analysis of F-actin organization (Fig. 3 D), we found uniform fluorescence intensity for F-actin across the entire cell width in the control group, but we documented consistently lower signals near the nucleus in emerin KD cells (p < 0.001; Fig. 3 D and E). We define the perinuclear region as a region of interest (ROI) bounded by margins 20 and 80% of the distance along a line scan of the width of each studied cell. Interestingly, microtubules and intermediate filaments vimentin were present throughout the perinuclear region in both emerin KD and control cells (Figs. S4 and S5 in the supporting material) and were therefore apparently unaffected by emerin downregulation. This suggested a role for emerin and the NE in organization and anchoring F-actin in lung fibroblasts during spreading as well as during stretch.
Emerin downregulation decreases myosin light chain phosphorylation
Alterations of cell shape and F-actin organization in emerin KD cells suggested changes in functional interactions of the actomyosin cytoskeleton with the nucleus. We were motivated to examine myosin activation in the setting of emerin KD by previous evidence of emerin mediation of F-actin binding to the nucleus and of nuclear tethering of actomyosin during force generation (24,43,59). Furthermore, previous work showed that phosphorylation of myosin light chain (MLC) regulated filament formation with a preferential spatial distribution, where monophosphorylated MLC at Ser19 (pMLC) localized to peripheral stress fibers and diphosphorylated MLC at Thr18 and Ser19 (ppMLC) localized to central stress fibers (60). Since the stress fibers are absent in the center region in emerin KD cells, we investigated both pMLC and ppMLC. After 8 or 24 h of cell plating, we collected cell lysates and probed for pMLC, ppMLC, and unmodified total MLC (Fig. 4 A). At 8 h early stages of spreading and adhesion, control cells exhibited robust pMLC and ppMLC. By contrast, pMLC and ppMLC in emerin KD cells were reduced by ∼90% at 8 h (Fig. 4 B; p < 0.001 for each). At 24 h, when cells typically reach a steady state of spreading and adhesion, pMLC and ppMLC decreased in control cells and remained low in emerin KD cells (Fig. 4 B). We concluded that emerin may be required to fully activate or maintain MLC kinase activity during early cell attachment, spreading, and growth.
Emerin downregulation decreases fibronectin fibrillogenesis and protein expression
Matrix patterning is a key component of tissue morphogenesis (61). Fibronectin fibrillogenesis is facilitated by cell shape, traction force, and transmission of force to ECM in an MLC-requiring manner (62,63,64). Perturbations in morphology, cytoskeletal organization, and MLC activity in the emerin KD fibroblasts prompted us to study matrix assembly in these cells. We examined fibronectin fibril formation in control and emerin KD fibroblasts at 8 and 24 h after replating (Fig. 5 A). We developed an ordinal fibrillogenesis scoring system from 0 to 5 (no to abundant fibrils; Fig. S6 in the supporting material). Fibrillogenesis was scored by a single-blind observer (Fig. 5 B). The analysis of the fibrillogenesis (Fig. 5 B) showed that 60% of control cells had scores of 2 and 3 at 8 h, whereas only ∼10% of the emerin KD cells had scores of 2 and ∼60% had scores of 0. At 24 h, about 15% of control cells had elongated fibronectin fibrils spanning the entire cell (scores of 4 and 5), whereas most emerin KD cells exhibited only early fibril formation, and none had scores above 3. Fig. 5 C showed that the mean fibrillogenesis scores were higher for control than for emerin KD cells, at both 8 and 24 h (p < 0.001 for each).
Since emerin and other NE proteins have been implicated in the regulation of chromatin binding, transcription factor activation, and transcription (35,48,65), we also measured fibronectin protein expression in WI-38 fibroblasts with and without emerin KD. We measured total fibronectin protein levels 24 h after replating (Fig. 5 D). Emerin KD cells produced about 75% less fibronectin protein than control (Fig. 5 E; p < 0.01). These data indicate that emerin modulates both fibronectin fibrillogenesis and production.
Discussion
Growing appreciation for the role of the nucleus in mechanical signal transduction during motility, development, mechanical perturbation, and disease has catalyzed work to elucidate the interactional dynamics and unique roles of molecules that mediate nuclear-cytoskeletal linkages in the NE (35,59,66,67). LINC proteins, lamins A/C, and emerin have all been shown to modulate nuclear shape and positioning during cell motility (39,41,50,68). Nesprin and LINC components regulate nuclear movement during retinal development in mammals (59,69). SUN and lamin A impart resilience to nuclear responses and transcriptional modulation during mechanical distortion (27,34,67). Finally, high-impact debilitating diseases including cerebellar ataxia, progeria syndromes, dilated cardiomyopathy, and muscular dystrophies stem from NE dysfunction due to mutations in nesprins 1, 2, and 4, lamin A/C, emerin, and SUN proteins (70).
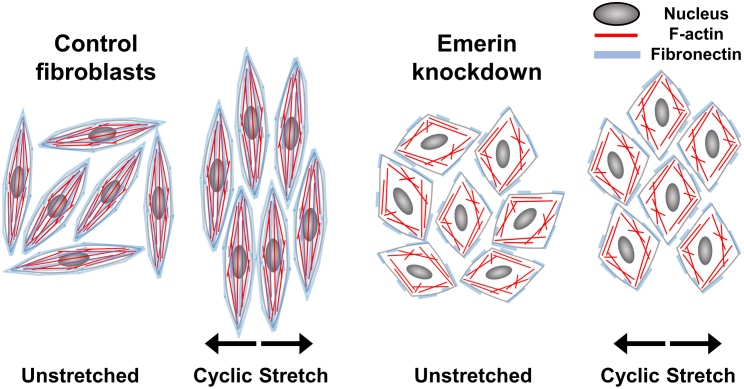
Our work harnesses quantitative imaging and image analysis metrics to provide insights into the role of emerin in nuclear-cytoskeletal coupling during two mechanical transitions—cyclic stretch and spreading. We interrogated the impact of acute emerin KD on mechanical transitions during both cell spreading and cyclic stretch because both are essential components of tissue development. Differences in the microenvironmental mechanics of the two systems used for these two lines of inquiry (standard tissue culture surfaces and PDMS stretch chambers) arise directly from pragmatic aspects of experimental efficiency and are reported. Future work would benefit from a uniform microenvironment for all experiments. For cyclic stretch experiments, 2 h of adhesion and 6 h of stretch were used; for cell spreading, 8 and 24 h were designed to match the stretch experiments and for achieving a stabilized spreading state, respectively. We used shRNA to rapidly reduce emerin protein expression to ∼5% of normal levels within 4 days after treatment. We graphically summarize the consequences of this acute knockdown in Fig. 6. We demonstrated that the odds of stretch-induced realignment with emerin KD were nearly 40% lower for the nucleus (OR = 0.63) and ∼50% lower for F-actin (OR = 0.54) compared with control counterparts. We probed further for alterations in cytoskeletal organization, connectivity, and function that might cause this change in mechanical responsiveness and showed that perinuclear F-actin occupancy fell by over 50% in emerin KD cells, while mean values for cell size were 60% larger and circularity index scores tripled. Further functionally important cytoskeletal aberrations in emerin KD cells included reduction of regulatory MLC phosphorylation by ∼90% during cell adhesion and spreading, and a dramatic decrement in fibronectin fibrillogenesis. Our findings support the hypothesis that emerin has a key functional role in the mechanical continuity between the microfilament cytoskeleton and the NE. Severe acute reduction in emerin availability disconnects the physical basis of mechanical force transduction from polymerized F-actin filaments to the NE, resulting in the loss of a key fulcrum at the center of the cell’s actomyosin network. Consequences for the fetal lung fibroblasts used here include an actual spatial gap between this actomyosin network and the nucleus with resulting circular morphology, decrements in actomyosin force generation as evidenced by lower levels of myosin light chain phosphorylation and less efficient fibronectin fibrillogenesis, and disconnection of coordinated realignment of the nucleus with the ECM and actin cytoskeleton in response to cyclic stretch.
Figure 6.
Featured phenotypes of acute emerin KD in fetal lung fibroblasts. Unstretched control fibroblasts grew in random directions, and after stretch they aligned perpendicularly to the axis of stretch. Actin filaments traversed the nuclear surface and the cells assembled fibronectin in ECM. Emerin KD cells were larger and more circular. Upon stretch, emerin KD cells exhibited less alignment than control cells. In emerin KD cells, actin filaments were disconnected from the nuclei, and cells demonstrated less effective fibronectin assembly. These findings indicate that acute emerin downregulation diminishes nuclear-cytoskeletal coupling and reduces fibronectin assembly.
Emerin is an LEM domain protein that has classically been found to be localized to the inner nuclear membrane, but has been observed at the outer nuclear membrane during nuclear movement and under mechanical strain where it was associated with enriched foci of F-actin polymerization (41,49,71). Furthermore, emerin caps the pointed end and stabilizes actin filaments in vitro and has been shown to facilitate polarization of cytoskeletal F-actin flow and front-rear nuclear polarity during migration of several cell types (39,41,42,50). Recent super-resolution and single-molecule tracking data on the nanoscale organization of emerin in the NE provide detailed insights on its transitions between monomeric and oligomeric forms and emerin scaffolding interactions with barrier-to-autointegration factor, lamin A/C, and LINC complexes (35). These dynamic interactions regulate nuclear shape change in response to mechanical stress—induced in their study by substrates with 5–15 μm wide micropatterns—and include emerin-requiring bridges that span the NE and bind to the actomyosin cytoskeleton (35).
The strikingly atypical fibroblast phenotype that we observe with emerin KD was characterized by rounded cells with a paucity of perinuclear F-actin and poor mechanical responsiveness. These features all suggest reduced nuclear-cytoskeletal connectivity. Precedents for these observations include the perturbed front-rear nuclear polarity, and diffused and disorganized F-actin, matrix adhesions, MTOC, and Golgi distributions in retinal pigment epithelial cells subjected to RNAi for emerin (50). In addition, siRNA-mediated emerin KD in melanoma cells decreased the efficiency of ameboid movement and hindered F-actin-dependent modulation of nuclear stiffness (39). Together with our findings, data from these studies of others suggest that emerin facilitates the generation and adaptive harnessing of force by the actin cytoskeleton. Graham et al. hypothesize that tethering of actomyosin filaments to the nucleus is a critical component of the molecular clutch that improves the efficiency of force transmission from the ECM to the cytoskeleton. Their findings of poor 3D migration and deficient traction force generation by fibroblast and endothelial cell cytoplasts supports this hypothesis (24).
Lemmon and Romer previously proposed a model of cellular mechanotransduction in which both magnitude and direction of traction force at any discrete location in the cell are proportional to the first moment of area about that point in the cell, suggesting that the cytoskeleton provides a cohesive network of connectivity for the cell’s contractile forces (63). The reciprocal relationship between actin recruitment to stress fibers and MLC phosphorylation, and the inefficiency of regulatory MLC that we report here indicate that emerin KD reduces this cytoskeletal network connectivity and resultant force generation (72,73). The reduced traction force at matrix adhesion sites may result in less-efficient exposure of intermolecular fibronectin-to-fibronectin binding sites on the cell surface (74,75). This effect may be exacerbated by the loss of the prototypic fibroblastic spindle shape of the cells that enhances ECM fibril formation, and together these changes may explain the retardation of fibronectin fibrillogenesis that we observe with emerin KD (63,64). In addition, others have observed altered distribution of chromatin and impaired expression of mechanosensitive genes such as egr-1 and iex-1 in the setting of decreased emerin expression (47,50). Subnormal emerin levels and subsequent lower levels of cytoskeletal-nuclear connectivity may contribute to the abnormally low levels of fibronectin protein expression that we observed here.
The prominent phenotype displayed by the human fetal lung fibroblasts with emerin KD in our study is discrepant with both data from murine emerin knockout work (47,76) and with the apparent lack of primary pulmonary dysfunction in patients with X-linked Emery-Dreifuss muscular dystrophy, for whom the loss of emerin function is the primary driver of disease (77). In one previous study (47), emerin-deficient mouse embryonic fibroblasts derived from male emerin hemizygous mice (Emd–/y) show abnormal nuclear shape, increased nuclear cross-sectional area, decreased nuclear shape stability, impaired expression of mechanosensitive genes in response to stretch, but no apparent alteration of F-actin organization. Our study with acute emerin KD by adenoviral transduction in WI-38 human fetal lung fibroblasts shows disconnection of F-actin from nuclei and incomplete reorientation of nuclei and cytoskeleton upon stretch. Functional redundancies in the NE proteins may explain some of these divergent findings, since the time course of full-term embryonic development allows for upregulation of proteins that may compensate for emerin function, whereas our acute emerin KD approach does not (70). In addition, lamina-associated polypeptide 1 (LAP1) may have substantial functional overlap with emerin and murine expression levels are relatively much higher than those in humans (78,79). Future investigations of downstream molecular signaling consequences of emerin KD or emerin mobility within the NE may further clarify the role of emerin partners such as LAP1 and Bcl-2-associated transcription factor 1. Compensatory changes in gene expression of these partners may moderate developmental lung pathology in the setting of congenital emerin dysfunction or absence (80,81).
Conclusion
Work presented here focuses on the functional impact of acute knockdown of the NE protein emerin in human fetal lung fibroblasts during actomyosin-mediated responses to mechanical transitions. Responsiveness to positioning cues, connectivity of the actomyosin network to the nucleus, and activation and harnessing of cell traction force via myosin light chain phosphorylation and matrix fibrillogenesis are all perturbed by acute emerin KD. All of these are essential contributions of lung fibroblasts to tissue morphogenesis. Albeit a part of a multifaceted interrogation of the whole spectrum of emerin-associated proteins, our findings advance an important paradigm that expands perspective on the role of emerin and its binding partners in providing mechanical continuity via a direct linkage between the NE to the cytoskeleton in mechanotransduction in development and disease.
Our investigations of the impact of fetal breathing patterns on cellular organization provide an innovative platform for examining mechanisms and consequences of mechanical responsiveness of the nuclear-cytoskeletal continuum. Mechanical stretch is one facet of signal integration that influences tissue development and growth. Our recent findings on the combinatorial effects of substrate stiffness and geometry on tissue organization pave the way for exciting future investigations (82). Incorporation of micropatterns (Fig. S7) and substrates of various stiffnesses into the stretch platform used here could facilitate the investigation of multimodal signal integration in the developing lung and other mechanosensitive tissues (83).
Author contributions
Q.J., K.L.W., D.H.G., and L.H.R. designed the experiments. Q.J., D.P., and S.L. performed experiments under the supervision of D.H.G. and L.H.R. C.B.T. and S.K. contributed analytic tools. Q.J., H.W.S., and C.B.T. performed data analyses. T.D.N. provided simulation advice. Q.J., K.L.W., D.H.G., and L.H.R. wrote the manuscript with input from all authors.
Acknowledgments
The authors would like to thank Dr. Anil Bhatta and Andrea Wecker for their support with technical and experimental discussions. This work was supported by the National Science Foundation (NSF) under award no. DMR1709349 (to D.H.G. and L.H.R.), by a grant from Kley Dom Biomimetics (to L.H.R. and D.H.G.) and by a StAAR award from the Department of ACCM at the Johns Hopkins University School of Medicine. We would also like to acknowledge partial support for the statistical analysis from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health through grant no. 1UL1TR001079.
Declaration of interests
The authors declare no competing interests.
Editor: Klaus Michael Hahn.
Footnotes
David H. Gracias and Lewis H. Romer contributed equally to this work.
Qianru Jin’s present address is School of Engineering and Applied Sciences, Harvard University, Boston, Massachusetts.
Deepesh Pandey’s present address is Department of Medicine (Cardiology), Medical College of Georgia, Augusta University, Augusta, Georgia.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.05.027.
Supporting citations
Reference (84) appears in the supporting material.
Supporting material
References
- 1.Wozniak M.A., Chen C.S. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mammoto T., Ingber D.E. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vries J.I., Visser G.H., Prechtl H.F. The emergence of fetal behaviour. II. Quantitative aspects. Early Hum. Dev. 1985;12:99–120. doi: 10.1016/0378-3782(85)90174-4. [DOI] [PubMed] [Google Scholar]
- 4.Natale R., Nasello-Paterson C., Connors G. Patterns of fetal breathing activity in the human fetus at 24 to 28 weeks of gestation. Am. J. Obstet. Gynecol. 1988;158:317–321. doi: 10.1016/0002-9378(88)90146-9. [DOI] [PubMed] [Google Scholar]
- 5.Patrick J., Campbell K., et al. Richardson B. Patterns of human fetal breathing during the last 10 weeks of pregnancy. Obstet. Gynecol. 1980;56:24–30. [PubMed] [Google Scholar]
- 6.Kitterman J.A. The effects of mechanical forces on fetal lung growth. Clin. Perinatol. 1996;23:727–740. [PubMed] [Google Scholar]
- 7.Harding R., Hooper S.B. Regulation of lung expansion and lung growth before birth. J. Appl. Physiol. 1996;81:209–224. doi: 10.1152/jappl.1996.81.1.209. [DOI] [PubMed] [Google Scholar]
- 8.Liu M., Post M. Invited review: mechanochemical signal transduction in the fetal lung. J. Appl. Physiol. 2000;89:2078–2084. doi: 10.1152/jappl.2000.89.5.2078. [DOI] [PubMed] [Google Scholar]
- 9.Harding R., Hooper S.B., Han V.K. Abolition of fetal breathing movements by spinal cord transection leads to reductions in fetal lung liquid volume, lung growth, and IGF-II gene expression. Pediatr. Res. 1993;34:148–153. doi: 10.1203/00006450-199308000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Adzick N.S., Harrison M.R., et al. Finkbeiner W. Experimental pulmonary hypoplasia and oligohydramnios: relative contributions of lung fluid and fetal breathing movements. J. Pediatr. Surg. 1984;19:658–665. doi: 10.1016/s0022-3468(84)80349-8. [DOI] [PubMed] [Google Scholar]
- 11.Liu M., Skinner S.J., et al. Post M. Stimulation of fetal rat lung cell proliferation in vitro by mechanical stretch. Am. J. Physiol. 1992;263:L376–L383. doi: 10.1152/ajplung.1992.263.3.L376. [DOI] [PubMed] [Google Scholar]
- 12.Bishop J.E., Mitchell J.J., et al. Low R.B. Cyclic mechanical deformation stimulates human lung fibroblast proliferation and autocrine growth factor activity. Am. J. Respir. Cell Mol. Biol. 1993;9:126–133. doi: 10.1165/ajrcmb/9.2.126. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Maciejewski B.S., et al. Sanchez-Esteban J. Mechanical stretch promotes fetal type II epithelial cell differentiation via shedding of HB-EGF and TGF-α. J. Physiol. 2009;587:1739–1753. doi: 10.1113/jphysiol.2008.163899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mourgeon E., Xu J., et al. Post M. Mechanical strain-induced posttranscriptional regulation of fibronectin production in fetal lung cells. Am. J. Physiol. 1999;277:L142–L149. doi: 10.1152/ajplung.1999.277.1.L142. [DOI] [PubMed] [Google Scholar]
- 15.Breen E.C. Mechanical strain increases type I collagen expression in pulmonary fibroblasts in vitro. J. Appl. Physiol. 2000;88:203–209. doi: 10.1152/jappl.2000.88.1.203. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez J.A., Suzara V.V., Dobbs L.G. Continuous mechanical contraction modulates expression of alveolar epithelial cell phenotype. Am. J. Respir. Cell Mol. Biol. 2003;29:81–87. doi: 10.1165/rcmb.2002-0135OC. [DOI] [PubMed] [Google Scholar]
- 17.Muratore C.S., Nguyen H.T., et al. Wilson J.M. Stretch-induced upregulation of VEGF gene expression in murine pulmonary culture: a role for angiogenesis in lung development. J. Pediatr. Surg. 2000;35:906–912. doi: 10.1053/jpsu.2000.6916. [DOI] [PubMed] [Google Scholar]
- 18.Quinn T.P., Schlueter M., et al. Gutierrez J.A. Cyclic mechanical stretch induces VEGF and FGF-2 expression in pulmonary vascular smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;282:L897–L903. doi: 10.1152/ajplung.00044.2001. [DOI] [PubMed] [Google Scholar]
- 19.Maniotis A.J., Chen C.S., Ingber D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N., Tytell J.D., Ingber D.E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey J.D., Dufresne E.R., Schwartz M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammerding J. Mechanics of the nucleus. Compr. Physiol. 2011;1:783–807. doi: 10.1002/cphy.c100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caille N., Thoumine O., et al. Meister J.-J. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 2002;35:177–187. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 24.Graham D.M., Andersen T., et al. Burridge K. Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J. Cell Biol. 2018;217:895–914. doi: 10.1083/jcb.201706097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luxton G.W.G., Gomes E.R., et al. Gundersen G.G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombardi M.L., Jaalouk D.E., et al. Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer M., Lammerding J. The driving force: nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng. 2019;21:443–468. doi: 10.1146/annurev-bioeng-060418-052139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y.L., Burke B. LINC complexes and nuclear positioning. Semin. Cell Dev. Biol. 2018;82:67–76. doi: 10.1016/j.semcdb.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Chancellor T.J., Lee J., et al. Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 2010;99:115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brosig M., Ferralli J., et al. Chiquet-Ehrismann R. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. Int. J. Biochem. Cell Biol. 2010;42:1717–1728. doi: 10.1016/j.biocel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Anno T., Sakamoto N., Sato M. Role of nesprin-1 in nuclear deformation in endothelial cells under static and uniaxial stretching conditions. Biochem. Biophys. Res. Commun. 2012;424:94–99. doi: 10.1016/j.bbrc.2012.06.073. [DOI] [PubMed] [Google Scholar]
- 32.Kim J.-K., Louhghalam A., et al. Kim D.-H. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat. Commun. 2017;8:2123. doi: 10.1038/s41467-017-02217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elosegui-Artola A., Andreu I., et al. Roca-Cusachs P. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell. 2017;171:1397–1410.e14. doi: 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Tajik A., Zhang Y., et al. Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez A., Bautista M., et al. Pinaud F. Emerin self-assembly and nucleoskeletal coupling regulate nuclear envelope mechanics against stress. J. Cell Sci. 2022;135 doi: 10.1242/jcs.258969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berk J.M., Tifft K.E., Wilson K.L. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowat A.C., Lammerding J., Ipsen J.H. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guilluy C., Osborne L.D., et al. Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavenus S.B., Vosatka K.W., et al. Logue J.S. Emerin regulation of nuclear stiffness is required for fast amoeboid migration in confined environments. J. Cell Sci. 2022;135 doi: 10.1242/jcs.259493. [DOI] [PubMed] [Google Scholar]
- 40.Salpingidou G., Smertenko A., et al. Hutchison C.J. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang W., Folker E.S., et al. Gundersen G.G. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol. Biol. Cell. 2013;24:3869–3880. doi: 10.1091/mbc.E13-06-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holaska J.M., Kowalski A.K., Wilson K.L. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231–E1362. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holaska J.M., Wilson K.L. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q., Ragnauth C.D., et al. Shanahan C.M. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 45.Mislow J.M.K., Holaska J.M., et al. McNally E.M. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 46.Haque F., Mazzeo D., et al. Shackleton S. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lammerding J., Hsiao J., et al. Lee R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho C.Y., Jaalouk D.E., et al. Lammerding J. Lamin A/C and emerin regulate MKL1–SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le H.Q., Ghatak S., et al. Wickström S.A. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 2016;18:864–875. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- 50.Nastały P., Purushothaman D., et al. Maiuri P. Role of the nuclear membrane protein Emerin in front-rear polarity of the nucleus. Nat. Commun. 2020;11:2122. doi: 10.1038/s41467-020-15910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston I.D., McCluskey D.K., et al. Tracey M.C. Mechanical characterization of bulk Sylgard 184 for microfluidics and microengineering. J. Micromech. Microeng. 2014;24 [Google Scholar]
- 52.Soucy P.A., Werbin J., et al. Romer L.H. Microelastic properties of lung cell-derived extracellular matrix. Acta Biomater. 2011;7:96–105. doi: 10.1016/j.actbio.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legland D., Arganda-Carreras I., Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 54.Tschumperlin D.J., Margulies S.S. Alveolar epithelial surface area-volume relationship in isolated rat lungs. J. Appl. Physiol. 1999;86:2026–2033. doi: 10.1152/jappl.1999.86.6.2026. [DOI] [PubMed] [Google Scholar]
- 55.Wang J.H., Goldschmidt-Clermont P., et al. Yin F.C. Specificity of endothelial cell reorientation in response to cyclic mechanical stretching. J. Biomech. 2001;34:1563–1572. doi: 10.1016/s0021-9290(01)00150-6. [DOI] [PubMed] [Google Scholar]
- 56.Morioka M., Parameswaran H., et al. Ito S. Microtubule dynamics regulate cyclic stretch-induced cell alignment in human airway smooth muscle cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greiner A.M., Chen H., et al. Kemkemer R. Cyclic tensile strain controls cell shape and directs actin stress fiber formation and focal adhesion alignment in spreading cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buchwalter A., Schulte R., et al. Hetzer M. Selective clearance of the inner nuclear membrane protein emerin by vesicular transport during ER stress. Elife. 2019;8 doi: 10.7554/eLife.49796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson P.M., Cadot B. Actin on and around the nucleus. Trends Cell Biol. 2021;31:211–223. doi: 10.1016/j.tcb.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 60.Kassianidou E., Hughes J.H., Kumar S. Activation of ROCK and MLCK tunes regional stress fiber formation and mechanics via preferential myosin light chain phosphorylation. Mol. Biol. Cell. 2017;28:3832–3843. doi: 10.1091/mbc.E17-06-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plikus M.V., Wang X., et al. Horsley V. Fibroblasts: origins, definitions, and functions in health and disease. Cell. 2021;184:3852–3872. doi: 10.1016/j.cell.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu J., Doyle A.D., et al. Yamada K.M. Basement membrane regulates fibronectin organization using sliding focal adhesions driven by a contractile winch. Dev. Cell. 2020;52:631–646.e4. doi: 10.1016/j.devcel.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lemmon C.A., Romer L.H. A predictive model of cell traction forces based on cell geometry. Biophys. J. 2010;99:L78–L80. doi: 10.1016/j.bpj.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemmon C.A., Chen C.S., Romer L.H. Cell traction forces direct fibronectin matrix assembly. Biophys. J. 2009;96:729–738. doi: 10.1016/j.bpj.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X., Phua D.Y.Z., et al. Alushin G.M. Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell. 2020;55:468–482.e7. doi: 10.1016/j.devcel.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sankaran J., Uzer G., et al. Rubin J. Gene regulation through dynamic actin control of nuclear structure. Exp. Biol. Med. 2019;244:1345–1353. doi: 10.1177/1535370219850079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Janota C.S., Calero-Cuenca F.J., Gomes E.R. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell Biol. 2020;63:204–211. doi: 10.1016/j.ceb.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Thiam H.R., Vargas P., et al. Piel M. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016;7 doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu J., Lei K., et al. Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum. Mol. Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janin A., Bauer D., et al. Méjat A. Nuclear envelopathies: a complex LINC between nuclear envelope and pathology. Orphanet J. Rare Dis. 2017;12:147. doi: 10.1186/s13023-017-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bautista M., Fernandez A., Pinaud F. A micropatterning strategy to study nuclear mechanotransduction in cells. Micromachines. 2019;10 doi: 10.3390/mi10120810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe T., Hosoya H., Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol. Biol. Cell. 2007;18:605–616. doi: 10.1091/mbc.e06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishimura Y., Shi S., et al. Viasnoff V. Crosstalk between myosin II and formin functions in the regulation of force generation and actomyosin dynamics in stress fibers. Cells Dev. 2021;168 doi: 10.1016/j.cdev.2021.203736. [DOI] [PubMed] [Google Scholar]
- 74.Zhong C., Chrzanowska-Wodnicka M., et al. Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lemmon C.A., Weinberg S.H. Multiple cryptic binding sites are necessary for robust fibronectin assembly: an in silico study. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-18328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hale C.M., Shrestha A.L., et al. Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys. J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gueneau L., Bertrand A.T., et al. Bonne G. Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy. Am. J. Hum. Genet. 2009;85:338–353. doi: 10.1016/j.ajhg.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin J.Y., Méndez-López I., et al. Dauer W.T. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev. Cell. 2013;26:591–603. doi: 10.1016/j.devcel.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y., Shin J.Y., et al. Worman H.J. Postnatal development of mice with combined genetic depletions of lamin A/C, emerin and lamina-associated polypeptide 1. Hum. Mol. Genet. 2019;28:2486–2500. doi: 10.1093/hmg/ddz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Z., Zhu J., et al. Jin X. Function of BCLAF1 in human disease. Oncol. Lett. 2022;23:58. doi: 10.3892/ol.2021.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saunders C.A., Harris N.J., et al. Luxton G.W.G. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol. 2017;216:657–674. doi: 10.1083/jcb.201507113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pahapale G.J., Tao J., et al. Gracias D.H. Directing multicellular organization by varying the aspect ratio of soft hydrogel microwells. Adv. Sci. 2022;9 doi: 10.1002/advs.202104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haniffa M., Taylor D., et al. Human Cell Atlas Developmental Biological Network A roadmap for the human developmental cell atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eberl C. MATLAB Central File Exchange. 2023. Digital image correlation and tracking.https://www.mathworks.com/matlabcentral/fileexchange/12413-digital-image-correlation-and-tracking [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.