Summary
Psyllids, a group of insects that feed on plant sap, have a symbiotic relationship with an endosymbiont called Carsonella. Carsonella synthesizes essential amino acids and vitamins for its psyllid host, but lacks certain genes required for this process, suggesting a compensatory role of psyllid host genes. To investigate this, gene expression was compared between two psyllid species, Bactericera cockerelli and Diaphorina citri, in specialized cells where Carsonella resides (bacteriomes). Collaborative psyllid genes, including horizontally transferred genes, showed patterns of conserved gene expression; however, species-specific patterns were also observed, suggesting differences in the nutritional metabolism between psyllid species. Also, the recycling of nitrogen in bacteriomes may primarily rely on glutamate dehydrogenase (GDH). Additionally, lineage-specific gene clusters were differentially expressed in B. cockerelli and D. citri bacteriomes and are highlighted here. These findings shed light on potential host adaptations for the regulation of this symbiosis due to host, microbiome, and environmental differences.
Subject areas: Microbiology, Phylogenetics, Transcriptomics
Graphical abstract
Highlights
-
•
Psyllids exhibit divergent and conserved gene expression patterns in bacteriomes
-
•
All nutritional HTG copies are upregulated for both psyllid species in bacteriomes
-
•
The recycling of ammonia in bacteriomes is via the GS and GDH pathway
-
•
Sulfur, cysteine, and methionine metabolisms enriched only in D. citri bacteriomes
Microbiology; Phylogenetics; Transcriptomics
Introduction
A multitude of insects have forged mutualistic relationships with microbial symbionts, thereby enhancing their metabolic capabilities and tolerance to diverse environmental conditions. These insects rely on symbionts to supplement their nutrition and aid in the digestion of plant materials, enabling them to thrive in their respective habitats.1 These associations require specific genetic traits in insects, encompassing gene expression patterns in cells hosting symbionts,2 gene duplication,3,4,5 and horizontal gene transfer.6,7,8 These insect traits play a pivotal role in the development and maintenance of obligatory symbiotic mutualisms.
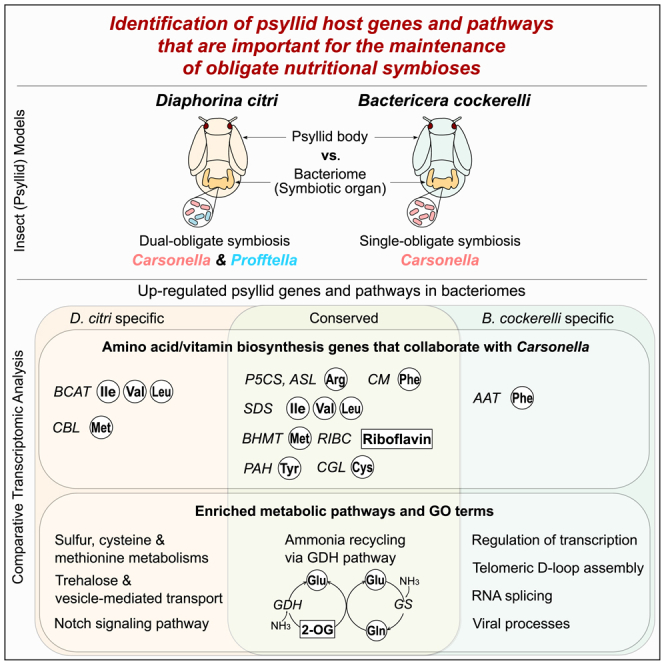
Psyllids, sap-feeding insects in the order Hemiptera, serve as an exemplary illustration of such mutualisms. They uphold an obligate symbiotic relationship with Candidatus Carsonella ruddii (referred to as Carsonella hereafter), which synthesizes vital amino acids that are typically scarce in plant sap.9,10,11,12,13,14 Carsonella resides within specialized cells called bacteriocytes, which are found in the abdominal organ known as the bacteriome.15,16 This long-term, host-restricted relationship leads to continuous gene loss in Carsonella’s genome because of genetic drift.11,14,17,18 Research conducted on Carsonella and the hackberry petiole gall psyllid, Pachypsylla venusta, has unveiled that a distinct subset of psyllid genes is up-regulated in bacteriomes, as compared to other body tissues.7 This finding suggests a potential compensation mechanism for Carsonella’s gene loss and the establishment of integrated host-microbe metabolic pathways, similar to those observed in other hemipteran species.2,6,19 Nonetheless, the extent of functional and expression divergence among these collaborative genes, including horizontally transferred genes (HTGs), remains uncertain across divergent psyllid species.
Recently, chromosomal-level genome assemblies have been successfully obtained for three divergent psyllid species: P. venusta (Carsidaridae),20 the Asian citrus psyllid Diaphorina citri (Psyllidae),21 and the potato/tomato psyllid Bactericera cockerelli (Triozidae).22 Bactericera cockerelli and D. citri diverged from each other approximately 86 million years ago, while their most recent common ancestor separated from P. venusta around 130 million years ago.22 Our previous comparative genomic analysis has revealed that these three psyllid species share notable homologs which could potentially compensate for Carsonella’s incomplete amino acid biosynthesis pathways, specifically in six essential amino acid pathways.22 Remarkably, some of these homologs have been acquired through horizontal gene transfer events from bacteria to an early ancestor of psyllids before the major divergence of the Psylloidea superfamily.7,22 While these HTGs are largely conserved among the three psyllid species, lineage-specific gene duplication events have also been observed.22 Nevertheless, It remains uncertain whether divergent psyllid species exhibit similar expression patterns in bacteriomes for the homologs that are predicted to collaborate with Carsonella in essential amino acid biosynthesis, as the expression patterns of psyllid bacteriomes have only been examined for one psyllid species to date (P. venusta).7
In this study, we present bacteriome and body transcriptomic data for two additional psyllid species D. citri and B. cockerelli. Furthermore, we conduct inter-species comparative transcriptomic analyses, aiming to enhance our understanding of the evolutionary aspects of psyllid host regulation. In comparison to B. cockerelli, D. citri hosts an additional obligate bacterial endosymbiont known as Candidatus Profftella armatura (hereafter referred to as Profftella), which resides in the syncytial region of the bacteriome.23 Profftella is presumed to primarily serve as a defensive symbiont for the psyllid by producing a polyketide toxin called diaphorin, but its genome also contains genes related to the biosynthesis of cysteine, hemolysin, riboflavin, biotin, and carotenoids, which may contribute to D. citri’s nutritional symbiosis.23,24 Both psyllid species also harbor Wolbachia,25,26 one of the most prevalent facultative endosymbionts of arthropods,27 including psyllids.26,28,29,30,31 The impact of Wolbachia on the nutritional metabolism of the psyllid host-symbiont relationship is also currently unknown. Therefore, It is of significant interest to investigate the following questions: 1) To what extent are gene expression patterns conserved between D. citri and B. cockerelli bacteriomes for psyllid homologs that collaborate with Carsonella?, 2) Are there species-specific gene expression patterns within D. citri and B. cockerelli bacteriomes, suggesting divergence in their nutritional symbioses?, and 3) Do psyllid bacteriomes exhibit distinct gene expression patterns for homologs with multiple gene copies, such as the horizontally transferred genes (HTGs) predicted to collaborate with Carsonella? By examining these questions, we can gain deeper insights into the evolution of psyllid host regulation and the intricate dynamics of their nutritional symbioses.
Results
Global differential gene expression in bacteriomes of D. citri and B. cockerelli
To investigate conserved and lineage-specific gene expression patterns related to the nutritional symbiosis in two psyllid species, D. citri and B. cockerelli, we pooled 60 bacteriomes and the rest of the body tissues with three biological replicates each for each species (N = 12 samples; Figure S1; see STAR methods for more detail). For D. citri, we obtained an average of 24,621,735 and 24,442,844 high-quality trimmed reads from three biological replicates of bacteriomes and body tissues, respectively, with an average of 75% and 89% of these reads mapping to the D. citri genome, respectively (Table S1). For B. cockerelli, we obtained an average of 26,365,738 and 27,932,791 trimmed reads from three biological replicates of bacteriomes and body tissues, respectively, with an average of 80% and 76% of these reads mapping to the B. cockerelli genome, respectively (Table S1).
In D. citri, for a total of 23,078 genes, 3,673 (16%) and 4,112 (18%) genes were significantly up- and down-regulated, respectively, in bacteriomes compared to the body tissues (Tables S2A and S3A). In B. cockerelli, for a total of 19,032 genes, 4,589 (24%) and 3,982 (21%) genes were significantly up- and down-regulated, respectively, in bacteriomes compared to the body tissues (Tables S2B and S3B).
Inter-species comparison of one-to-one orthologs
When comparing one-to-one orthologs (N = 5,555) between both psyllid species (Table S4), we identified an overlap of 852 orthologs (15%) that were significantly up-regulated in bacteriomes compared to body for both psyllid species, while 511 and 1,002 orthologs were significantly up-regulated in only D. citri or B. cockerelli, respectively (Figure 1; Table S4). The most represented GO-terms for shared up-regulated one-to-one orthologs include metal ion binding, ubiquitin-dependent protein catabolic process, protein transport, and transmembrane transport. The most represented GO-terms for one-to-one orthologs with species-specific up-regulation patterns include trehalose transport and vesicle-mediated transport for D. citri, and regulation of transcription, RNA splicing and viral processes for B. cockerelli (Table S4). For significantly down-regulated genes in bacteriomes compared to body for both psyllid species there was an overlap of 1,076 orthologs (19%), whereas 364 and 776 orthologs were significantly down-regulated in only D. citri or B. cockerelli, respectively (Figure 1; Table S4). The most represented GO-terms for shared down-regulated one-to-one orthologs include signal transduction, visual perception, integral component of membrane and multicellular organism development. The most represented GO-terms for one-to-one orthologs with species-specific down-regulation include zinc ion binding, response to oxidative stress and Wnt signaling pathway for D. citri and translation, ventral cord development and cell adhesion for B. cockerelli (Table S4).
Figure 1.
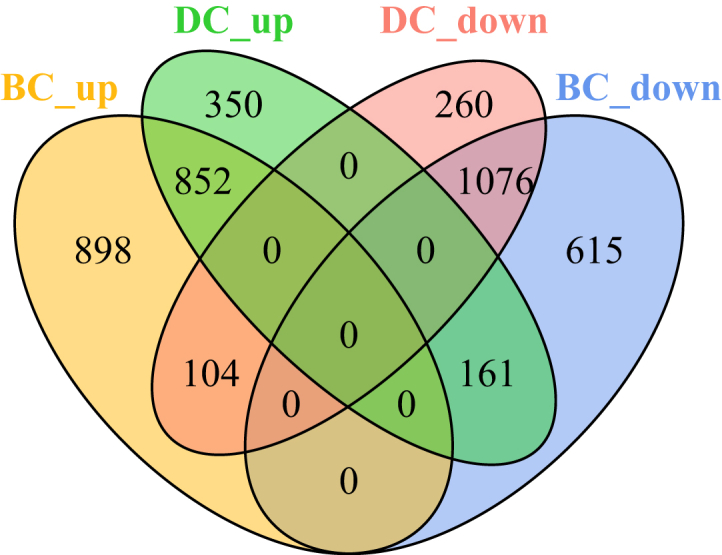
A Venn diagram showing the number of significantly differentially expressed one-to-one orthologs in D. citri and B. cockerelli for bacteriomes compared to body tissues (FDR adjusted p value ≤0.05 and fold change (FC) ≥ 1.5)
Four sets of significantly differentially expressed one-to-one orthologs are represented (from left to right; up-regulated one-to-one orthologs in B. cockerelli (BC_up; outlined in yellow), up-regulated one-to-one orthologs in D. citri (DC_up; outlined in green), down-regulated one-to-one orthologs in D. citri (DC_down; outlined in pink), and down-regulated one-to-one orthologs in B. cockerelli (BC_down; outlined in blue).
To further explore the similarities and differences in gene expression between D. citri and B. cockerelli, we conducted Principal Component Analyses (PCA) on the relative expression of one-to-one orthologs similar to Argandona et al. (2023),32 Georgiadou et al. (2022)33 and Korb et al. (2021).34 Approximately 39% of one-to-one orthologs were significantly differentially expressed in bacteriomes compared to the body tissues for both psyllid species and these genes were examined further using PCA analysis where axis 1 and 2 explained ∼51% and 48% of the variance in the data, respectively. Out of the top 100 orthologs that displayed the highest positive and negative correlations with principal components 1 and 2, 67 of these genes were non-redundant and examined further. These orthologs consist of 43 up-regulated and 24 down-regulated genes in B. cockerelli and 41 up-regulated and 26 down-regulated genes in D. citri (Figure S2; Table S5).
Out of the 67 orthologs, those that are involved in a horizontal gene transfer event and symbiosis include the rRNA methylation gene (RSMJ) which was up-regulated in both species in bacteriomes compared to the body tissues (Tables 2 and S5). Moreover, up-regulated orthologs appear to have two distinct expression profiles with a subset of orthologs having greater (or lower) FC magnitudes in either D. citri or B. cockerelli (Figure S2). For instance, orthologs that were associated with the sulfur compound metabolism and ubiquitin binding showed higher up-regulation profiles in D. citri, while orthologs that were associated with rRNA-methyltransferase activity, trigeminal ganglion development exhibited higher up-regulation profiles in B. cockerelli (Figure S2; Table S5). Down-regulated genes in both species include those associated with thermotaxis, bursicon, and the regulation of sleep (protein quiver/sleepless) (Figure S2; Table S5).
Table 2.
Differential gene expression for HTGs in bacteriomes compared to the body tissues in B. cockerelli and D. citri
| B. cockerellia | LogFC | FDR | D. citri | LogFC | FDR | |
|---|---|---|---|---|---|---|
| Argininosuccinate lyase | ||||||
| ASL-1 | ANN12874 | 3.75 | 4.81E-40 | M8J77_005191 | 3.27 | 2.13E-29 |
| ASL-2a | ANN10361 | 7.63 | 7.48E-93 | M8J77_020699 | 10.07 | 7.17E-93 |
| ASL-2b | ANN20354 | 8.06 | 4.11E-11 | |||
| Chorismate mutase | ||||||
| CM-1 | ANN05927 | 2.63 | 4.89E-11 | M8J77_003746 | 4.26 | 3.79E-65 |
| CM-2 | ANN06704 | 3.07 | 4.02E-41 | M8J77_022915 | 3.98 | 3.37E-45 |
| CM-3 | ANN17115 | 7.403 | 5.66E-40 | M8J77_007440 | 8.84 | 4.20E-96 |
| M8J77_019892 | 8.84 | 4.20E-96 | ||||
| A/G-specific adenine glycosylase | ||||||
| MUTY | ANN05978 | −2.66 | 1.66E-85 | M8J77_002314 | −2.19 | 3.37E-05 |
| AAA-ATPase-like | ||||||
| ORF-1 | ANN01458 | 6.99 | 2.07E-09 | M8J77_017214 | 5.85 | 3.62E-66 |
| ORF-2 | ANN17655 | 6.86 | 5.70E-38 | M8J77_013596 | 7.96 | 5.46E-73 |
| ORF-3a | ANN18155 | 6.957 | 7.57E-42 | M8J77_011284 | 6.90 | 1.64E-75 |
| ORF-3b | ANN18147 | 11.68 | 1.60E-113 | N/A | N/A | N/A |
| Riboflavin synthase | ||||||
| RIBC | ANN14810 | 7.88 | 1.07E-83 | M8J77_002167 | 6.15 | 3.21E-85 |
| 16S rRNA methyltransferase | ||||||
| RSMJ | ANN13802 | 11.70 | 1.44E-86 | M8J77_006534 | 5.87 | 3.27E-46 |
| VOC family protein | ||||||
| YDCJ | ANN12599a | 0.28 | 0.23 | M8J77_004361 | 1.55 | 8.64E-10 |
Homologs in the same row are one-to-one or one to many orthologs. “N/A” indicates no ortholog was identified.
All genes in the table are significantly differentially expressed except for ANN12599.
Species-specific gene expression patterns are prevalent for collaborative genes in the essential amino acid metabolism
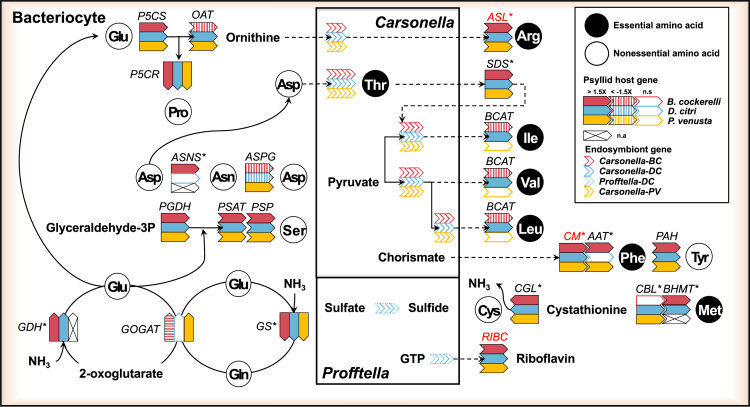
To gain insights into the mechanisms by which divergent psyllid hosts maintain their mutualistic relationship with Carsonella (as well as Profftella), we explored the differential expression patterns of psyllid host genes in the bacteriomes compared to other body tissues. This investigation aimed to shed light on the hypothesized role of certain psyllid genes in collaborating with Carsonella for the biosynthesis of essential amino acids, including arginine, phenylalanine, isoleucine, leucine, valine, and methionine (Figure 2).7,22 Three psyllid homologs hypothesized to be involved in the arginine, isoleucine, and methionine pathway, such as 1-pyrroline-5-carboxylate synthase (P5CS, EC 2.7.2.11/1.2.1.41), L-serine/L-threonine ammonia-lyase (SDS, EC 4.3.1.17/4.3.1.19), and homocysteine S-methyltransferase (BHMT, EC 2.1.1.10), respectively, were all significantly up-regulated in bacteriomes compared to the body tissues (Table 1; Figure 2), suggesting that both psyllid hosts may complement Carsonella’s biosynthesis pathways for arginine, isoleucine, and methionine biosynthesis.
Figure 2.
Differential gene expression of psyllid genes involved in the integrative metabolism with Carsonella and Profftella
Gene expression data are from NCBI SRA: SRA099681 for Pachypsylla venusta, and from this study for B. cockerelli and D. citri. Asterisks indicate more than one homolog is present in B. cockerelli or D. citri. HTGs are shown in red. (>1.5X, significantly up-regulated; <-1.5X, significantly down-regulated; n.s., not significant; n.a. data not available from P. venusta).
Table 1.
Expression of symbiosis-related homologs in bacteriomes compared to the body tissues for D. citri and B. cockerelli
| Name | EC Number | Enzyme | B. cockerelli gene ID | LogFC | FDR | D. citri gene ID | LogFC | FDR |
|---|---|---|---|---|---|---|---|---|
| Collaborative essential amino acid genes | ||||||||
| P5CS | 2.7.2.11/1.2.1.41 | delta-1-pyrroline-5-carboxylate synthetase | ANN07147 | 3.74 | 1.36E-16 | M8J77_013564 | 5.49 | 1.33E-51 |
| OAT | 2.6.1.13 | ornithine—oxo-acid transaminase | ANN07991 | −1.19 | 5.46E-05 | M8J77_012790 | 5.63 | 1.28E-39 |
| BCAT | 2.6.1.42 | branched-chain amino acid aminotransferase | ANN10973 | −0.87 | 0.02 | M8J77_022158 | 0.93 | 0.00 |
| SDS | 4.3.1.17/4.3.1.19 | L-serine/L-threonine ammonia-lyase | ANN22278 | 6.23 | 6.25E-11 | N/A | N/A | N/A |
| N/A | N/A | N/A | M8J77_020702 | 1.04 | 0.01 | |||
| M8J77_019021 | 9.68 | 2.43E-128 | ||||||
| AAT | 2.6.1.1 | ANN17803 | 1.51 | 0.00 | M8J77_007026 | 0.17 | 0.43 | |
| M8J77_014008 | −0.22 | 0.76 | ||||||
| M8J77_009315 | 0.55 | 0.08 | ||||||
| CBL | 4.4.1.13 | cysteine-S-conjugate beta-lyase | ANN16046 | 0.16 | 0.47 | M8J77_020257 | 1.19 | 0.01 |
| M8J77_023431 | 0.14 | 0.57 | ||||||
| ANN16088 | 0.51 | 0.00 | M8J77_004574 | 0.72 | 0.03 | |||
| M8J77_012408 | 0.50 | 0.45 | ||||||
| BHMT | 2.1.1.10 | homocysteine S-methyltransferase | ANN06058 | −1.36 | 9.56E-09 | M8J77_021131 | 0.49 | 0.04 |
| ANN06059 | 3.55 | 1.13E-59 | M8J77_014773 | 5.70 | 1.11E-82 | |||
| Non-essential amino acid psyllid genes | ||||||||
| GOGAT | 1.4.1.13 | Glutamate synthase (NADH) | ANN19375 | −0.75 | 0.02 | M8J77_004711 | 0.57 | 0.02 |
| GDH | 1.4.1.3 | glutamate dehydrogenase | ANN04489 | −1.43 | 5.39E-10 | M8J77_003227 | −0.94 | 3.63E-06 |
| ANN10746 | 7.77 | 0.00 | M8J77_018416 | 5.48 | 0.01 | |||
| GS | 6.3.1.2 | glutamine synthetase | ANN10146 | 1.69 | 8.73E-05 | M8J77_024675 | 3.08 | 4.63E-39 |
| ANN10148 | 1.85 | 2.92E-06 | ||||||
| PAH | 1.14.16.1 | phenylalanine-4-hydroxylase | ANN09753 | 1.92 | 2.03E-06 | M8J77_010704 | 5.54 | 1.51E-55 |
| ASNS | 6.3.5.4 | Asparagine synthase | ANN02335 | 1.75 | 5.62E-12 | N/A | N/A | N/A |
| ANN09881 | 1.68 | 3.20E-26 | M8J77_011885 | −0.17 | 0.47 | |||
| ANN21723 | 1.65 | 3.94E-07 | M8J77_022917 | −0.13 | 0.66 | |||
| ASPG | 3.5.1.1 | L-asparaginase | ANN07324 | −2.71 | 1.29E-12 | M8J77_015442 | −1.54 | 0.00 |
| P5CR | 1.5.1.2 | pyrroline-5-carboxylate reductase | ANN22280 | 2.96 | 4.31E-17 | N/A | N/A | N/A |
| N/A | N/A | N/A | M8J76_017158 | −0.50 | 0.28 | |||
| M8J77_004141 | 5.88 | 6.60E-76 | ||||||
| CGL | 4.4.1.1 | Cystathionine gamma lyase | ANN04485 | 4.96 | 2.95E-38 | M8J77_022568 | 6.66 | 1.26E-85 |
| ANN04499 | 5.27 | 1.64E-34 | N/A | N/A | N/A | |||
| PGDH | 1.1.1.95 | Phosphoglycerate dehydrogenase | ANN19564 | 2.76 | 1.00E-37 | M8J77_016165 | 4.66 | 8.83E-39 |
| PSAT | 2.6.1.52 | Phosphoserine aminotransferase | ANN12533 | 3.35 | 7.73E-82 | M8J77_018069 | 2.95 | 2.71E-29 |
| PSP | 3.1.3.3 | Phosphoserine phosphatase | ANN22282 | 6.18 | 2.97E-29 | N/A | N/A | N/A |
| N/A | N/A | N/A | M8J77_025516 | 6.09 | 4.14E-77 | |||
Homologs in the same row are one-to-one or one to many orthologs. “N/A” indicates no ortholog was identified. Bolded gene IDs indicate that the psyllid gene is significantly differentially expressed in bacteriomes compared to the body tissues.
In contrast, the ortholog ornithine aminotransferase (OAT, EC 2.6.1.13), which provides an alternative pathway to synthesize ornithine via P5CS in the arginine pathway, was significantly up-regulated in D. citri and significantly down-regulated in B. cockerelli (Table 1; Figure 2). Species-specific expression patterns were also observed for other collaborative psyllid homologs in the phenylalanine, methionine, and branched-chain amino acid biosynthesis pathways (e.g., leucine, valine, and isoleucine), suggesting distinct strategies in collaborating with Carsonella to synthesize essential amino acids. For instance, aspartate aminotransferase (AAT, EC 2.6.1.1) was significantly up-regulated in bacteriomes compared to body tissues only in B. cockerelli for phenylalanine synthesis. Cysteine-S-conjugate beta-lyase (CBL, EC 4.4.1.13) was only significantly up-regulated in bacteriomes compared to body tissues in D. citri for the methionine pathway, and branched-chain aminotransferase (BCAT, EC 2.6.1.42) was significantly up- and down-regulated in bacteriomes compared to body tissues in D. citri and B. cockerelli, respectively, for the biosynthesis of leucine, valine, and isoleucine (Table 1; Figure 2).
Expression patterns of psyllid genes in the non-essential amino acid metabolism
The two enzymes, glutamine synthetase (GS, EC 6.3.1.2) and glutamine oxoglutarate aminotransferase (GOGAT, EC 1.4.1.13), are hypothesized to work together in recycling waste ammonia (NH3) in aphid bacteriocytes into glutamine and glutamate, which are used as amino donors for amino acid biosynthesis.2 Recycling of ammonia is crucial for sap-feeding insects due to the limited availability of nitrogen in their diets.35 Here in both B. cockerelli and D. citri, we found all GS orthologs were up-regulated in bacteriomes compared to the body tissues, while species-specific expression patterns for the GOGAT ortholog was observed (Table 1; Figure 2). The GOGAT ortholog in B. cockerelli was significantly down-regulated in bacteriomes compared to the body tissues whereas this ortholog was not significantly differentially expressed in D. citri (Table 1; Figure 2). Interestingly, orthologs for glutamate dehydrogenase (GDH, EC 1.4.1.3), an alternative to the GS/GOGAT pathway for recycling ammonia into glutamate, were significantly up-regulated in bacteriomes compared to body tissues for both psyllid species (Table 1; Figure 2).
The activity of asparaginase (ASPG, EC 3.5.1.1), that converts asparagine to aspartate, has been hypothesized to be a primary source of ammonia for the GS/GOGAT cycle.6,7,36 Here, we found that the ASPG ortholog in both psyllid species were significantly down-regulated in bacteriomes compared to the body tissues (Table 1; Figure 2). However, there were species-specific expression patterns for the biosynthesis of asparagine via asparagine synthase (ASNS, EC 6.3.5.4), where all three homologs in B. cockerelli were significantly up-regulated in bacteriomes compared to the body tissues, however, the two ASNS genes identified in D. citri were not significantly differentially expressed (Table 1; Figure 2).
In regard to other non-essential amino acid biosynthesis pathways, such as tyrosine, cysteine, proline, and serine biosynthesis, we observed that the majority of homologs for both psyllid species were significantly up-regulated in bacteriomes compared to the body tissues. For instance, phenylalanine-4-hydroxylase (PAH, EC 1.14.16.1) in the tyrosine pathway, cystathionine gamma lyase (CGL, EC 4.4.1.1) in the cysteine pathway, which contributes to ammonia production, pyrroline-5-carboxylate reductase (P5CR, EC 1.5.1.2) in the proline pathway, and three genes in the serine pathway such as phosphoglycerate dehydrogenase (PGDH, EC 1.1.1.95), phosphoserine aminotransferase (PSAT, EC 2.6.1.52), and phosphoserine phosphatase (PSP, EC 3.1.3.3), were significantly up-regulated in bacteriomes compared to body tissues for both psyllid species (Table 1; Figure 2). These findings indicate a significant involvement of the psyllid host in the bacteriome of both species, for the biosynthesis of tyrosine, cysteine, proline, and serine.
Horizontal gene transfer events in psyllid genomes may support gene losses in Carsonella
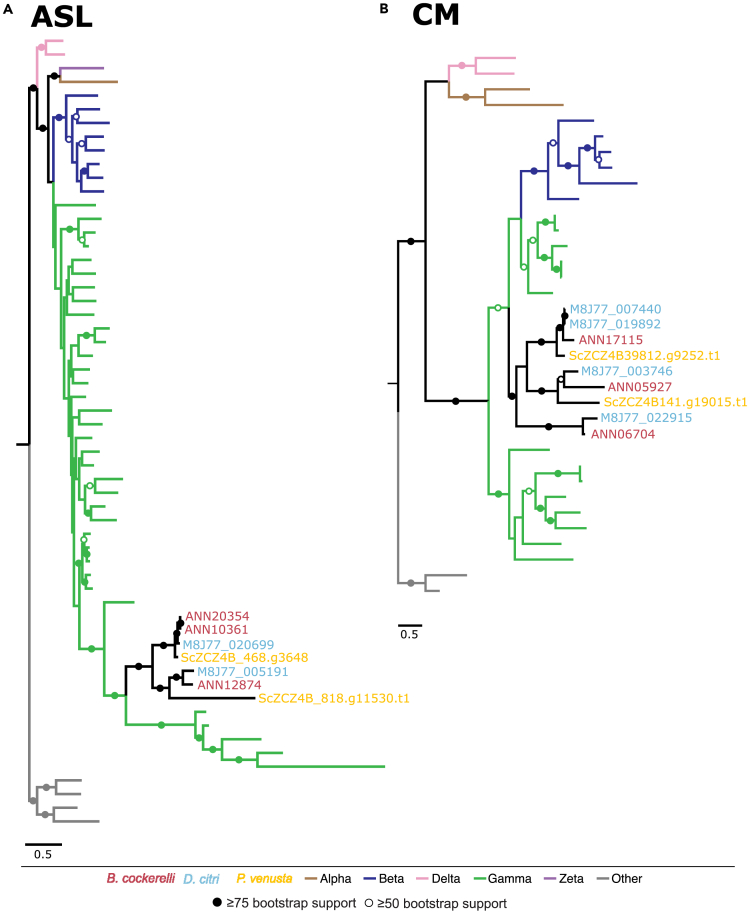
Expression levels of HTGs were examined for differential gene expression in bacteriomes compared to body for both psyllid species B. cockerelli and D. citri (Table 2). Two of these HTGs are predicted to collaborate with Carsonella for the biosynthesis of the essential amino acids, arginine and phenylalanine.7,22 These genes include argininosuccinate lyase (ASL, EC 4.3.2.1), which catalyzes the terminal step in the arginine biosynthesis pathway, and chorismate mutase (CM, EC 5.4.99.5), which is responsible for the conversion of chorismate into an intermediate precursor in the phenylalanine pathway (Figure 2). Phylogenetic analyses carried out in this study reveal distinct patterns within the genomes of B. cockerelli and D. citri. Specifically, we observed the presence of three distinct gene copies of ASL and CM in the genome of B. cockerelli, whereas D. citri’s genome possesses two gene copies of ASL and four gene copies of CM (Figure 3). All copies of ASL and CM in both psyllid species were significantly up-regulated in bacteriomes compared to the body tissues (Table 2; Figure 3), suggesting that all gene copies play important roles in mediating the symbiosis with Carsonella for arginine and phenylalanine biosynthesis.
Figure 3.
Phylogenetic analyses of gene duplications for HTGs in the psyllid genomes of P. venusta (GenBank: GCA_012654025.1), D. citri (GenBank: GCA_024506315.2), and B. cockerelli (GenBank: GCA_024516035.1) for (A) argininosuccinate lyase (ASL) genes and (B) chorismate mutase (CM) genes
Branches are colored according to their bacterial classes within the Proteobacteria, and ranges of bootstrap values are indicated by filled or open circles as indicated in the key.
For the other HTGs in the psyllid genomes, six out of eight HTGs share the same expression pattern for both D. citri and B. cockerelli. These HTGs exhibit significant up-regulation in the bacteriomes compared to the body tissues, indicating their involvement in vital cellular processes. For instance, five HTGs were significantly up-regulated in bacteriomes compared to the body tissues (RIBC, RSMJ, ORF-1, -2, and -3a) for both psyllid species and are predicted to be involved in riboflavin biosynthesis (RIBC), rRNA methylation (RSMJ) and diverse cellular activities (ORFs, AAA-ATPase). One ortholog was significantly down-regulated (MUTY) in bacteriomes compared to body tissues for both psyllid species (Table 2) and is predicted to encode an A/G specific mismatch repair enzyme. Gene expression patterns here indicate that this latter enzyme may play a more important role in body tissues compared to the bacteriome for both psyllid species (Table 2). The HTG YDCJ, a conserved bacterial gene which belongs to the vicinal oxygen chelate (VOC) family, showed species-specific expression where YDCJ was significantly only up-regulated in D. citri bacteriomes (Table 2).
Lineage-specific gene clusters differentially expressed in bacteriomes
To explore the evolutionary divergence and functional specialization of bacteriome expression in D. citri and B. cockerelli, we investigated lineage-specific gene clusters in both psyllid species. These gene clusters are unique to either D. citri or B. cockerelli, encompassing a total of 1,314 clusters comprising 5,150 genes in D. citri and 724 clusters comprising 3,133 genes in B. cockerelli (Table S6). In D. citri, 26 percent of lineage-specific genes (1,331 genes) were significantly differentially expressed, where a total of 525 and 804 genes were significantly up- and down-regulated, respectively, in bacteriomes compared to the body tissues (Table S7). The up-regulated lineage-specific genes in D. citri are primarily associated with nucleus-related activities, transposition and the regulation of the Notch signaling pathway (Table S7). The down-regulated lineage-specific genes in D. citri are primarily associated with DNA integration, RNA-directed DNA polymerase activity and the tachykinin receptor signaling pathway (Table S7).
In B. cockerelli, 24 percent of lineage specific genes (749 genes) were significantly differentially expressed, where a total of 427 and 322 genes were significantly up- and down-regulated, respectively, compared to the body tissues (Table S8). The up-regulated lineage-specific genes in B. cockerelli are primarily associated with protein processing, telomeric D loop disassembly, and transposition (Table S8). The down-regulated lineage-specific genes in B. cockerelli are primarily associated with locomotor rhythm, protein dephosphorylation, positive regulation of transcription, and proteolysis (Table S8).
Additionally, we examined the gene expression patterns of significantly expanding gene families previously identified in B. cockerelli.22 Out of 3,375 significantly expanding genes in 157 clusters, we observed that almost one-third of genes (a total of 1,011 genes) were significantly differentially expressed in bacteriomes relative to the body tissues in B. cockerelli (Table S9). Over 60 percent of these genes in 119 clusters were significantly up-regulated in bacteriomes compared to the body tissues, and the remaining genes in 99 clusters was significantly down-regulated in bacteriomes compared to the body tissues (Table S9). The up-regulated genes are primarily associated with transposons and gene regulation, such as nucleic acid and ATP binding proteins (Table S9A).
KEGG pathway analyses
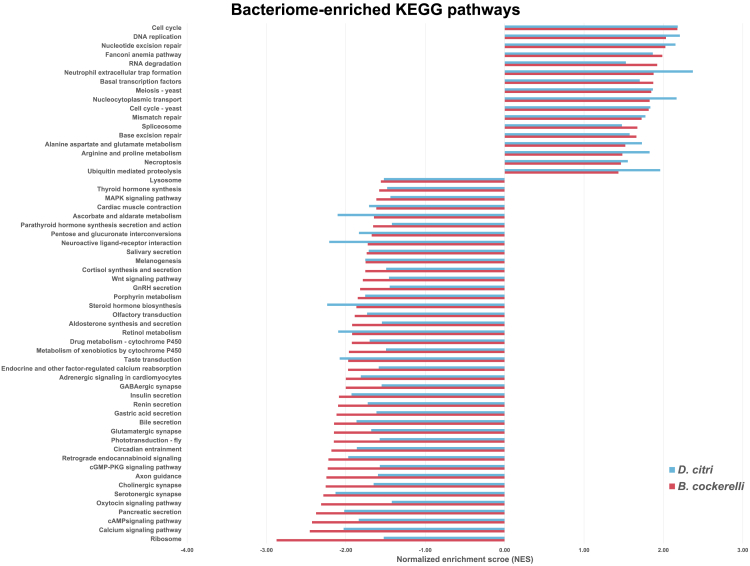
Using GSEA with KEGG pathways, we investigated the enrichment of biological pathways in the bacteriomes compared to body tissues for both species. In the bacteriomes of both psyllid species, we identified 17 and 39 pathways that were significantly positively and negatively enriched in bacteriomes compared to the body tissues, respectively (Figure 4; Table S10). The significantly positively enriched pathways in the bacteriomes include processes such as cell growth and death, transcription, nucleic acid replication and repair, and amino acid metabolisms (Figure 4; Table S10). The positive enrichment of these latter pathways in bacteriomes suggests that bacteriomes are metabolically highly active compared to other body cells, especially within the amino acid metabolism. The significantly negatively enriched pathways in bacteriomes were primarily involved in signaling pathways, lysosomes, and the digestive and endocrine systems (Figure 4; Table S10). These results indicate that genes involved in cell communication, and hormone and enzyme production, such as in the digestive and endocrine systems, are turned off and/or dampened in expression in the bacteriome. Furthermore, each psyllid species also exhibits species-specific pathways that were either positively or negatively enriched in their bacteriomes. For example, 45 and four unique KEGG pathways were significantly positively enriched in D. citri and B. cockerelli bacteriomes compared to the body tissues, respectively, and eight and 60 unique KEGG pathways were significantly negatively enriched in D. citri and B. cockerelli bacteriomes compared to the body tissues, respectively (Table S10). For example, in D. citri the sulfur metabolism and vitamin pathways related to B6 and folate were only significantly positively enriched in D. citri bacteriomes. In B. cockerelli the N-glycan biosynthesis pathways were only significantly positively enriched in B. cockerelli bacteriomes (Table S10). These results suggest that bacteriomes may play species-specific roles in the expression of pathways that are involved in the biosynthesis of sulfur, vitamins, and the modulation of protein folding, stability, and trafficking in D. citri compared to B. cockerelli.
Figure 4.
Gene Set Enrichment Analyses of D. citri and B. cockerelli displaying all KEGG pathways that were significantly enriched in bacteriomes compared to the body tissues based on transcriptomics data
Significance = the normalized p ≤ 0.05 and FDR q ≤ 0.25.
Tissue-specific quantification of Wolbachia in D. citri and B. cockerelli
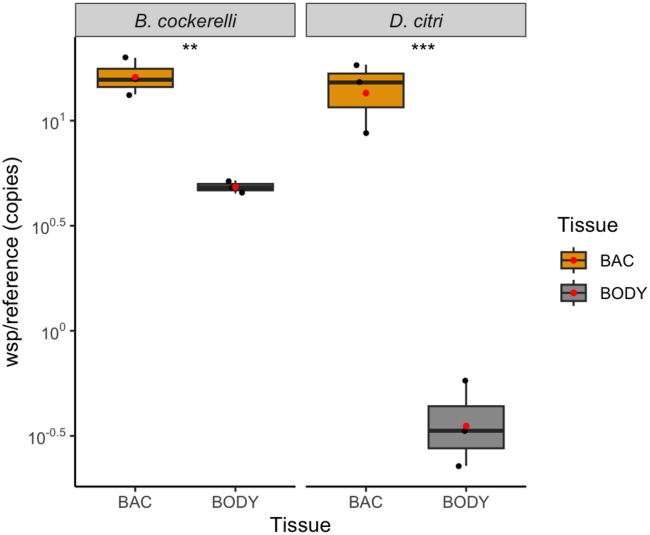
We quantified genome copy numbers of the Wolbachia endosymbiont from both psyllid species in the bacteriome and body tissues to assess how the Wolbachia chromosomal copies fluctuate depending on the tissue type and whether it may directly interact with the integrative metabolism inside of the bacteriome. For both psyllid species, Wolbachia genome copies relative to psyllid genome copies were significantly higher in bacteriomes compared to body tissues (Figure 5). The copy number of the single copy Wolbachia gene (wsp) in B. cockerelli relative to the single copy psyllid gene (FORKHEAD) was 16X ± 3 (mean ± standard deviation, n = 3) higher in bacteriomes and 5X ± 0.3 (n = 3) higher for body tissues (Figure 5). In D. citri, the copy number of the single copy Wolbachia gene (wsp) gene relative to the single copy psyllid gene (ACTB) was 14X ± 5 (mean ± standard deviation, n = 3) higher for bacteriomes and 0.4X lower ±0.2 (n = 3) for body tissues (Figure 5). These findings indicate a potential direct interaction between Wolbachia and both psyllid hosts and Carsonella (and Profftella in D. citri) within the bacteriome, suggesting that Wolbachia could have a substantial impact on the integrated metabolism between Carsonella and the psyllid host.
Figure 5.
Boxplots showing the Wolbachia wsp gene copy numbers in bacteriomes and body tissues for B. cockerelli and D. citri
The wsp gene copy numbers were normalized to the psyllid single copy house-keeping genes encoding FORKHEAD and ACTB in B. cockerelli and D. citri, respectively. All data points of three biological replicates of each tissue type for both psyllid species are presented. Tissues for each biological replicate are pooled tissues from 15 fifth instar nymphs. Red dots and asterisks represent means and significance (p < 0.01 ∗∗, p < 0.001 ∗∗∗) of normalized abundance values using a t-test.
Discussion
Conservation and divergence of gene expression patterns between two psyllid species for the integrated metabolism of Psylloidea and Carsonella
In this study, we observed both conserved and variable gene expression profiles for specialized hemipteran cells (bacteriome) that harbor the nutritional endosymbiont Carsonella for two psyllid species that diverged ∼86 million years ago22 (Figures 1 and 2; Table 1). Drawing on previous transcriptomic and proteomic research conducted in Hemiptera,2,6,7,8,19,36,37,38 we specifically investigated homologs that are postulated to collaborate with the obligate nutritional endosymbiont, Carsonella. Five out of nine enzymes (55%) that are predicted to collaborate in six of Carsonella’s essential amino acid pathways (arginine, isoleucine, valine, leucine, phenylalanine, and methionine) were significantly up-regulated in psyllid bacteriomes compared to body tissues for both B. cockerelli and D. citri (Figure 2). Interestingly, there is a high level of conservation for the differential expression patterns of these collaborative genes in bacteriomes when compared to the more distantly related psyllid species, P. venusta.7 In both P. venusta and D. citri, as well as P. venusta and B. cockerelli, a notable 75% and 62% of these collaborative genes, respectively, exhibited significant up-regulation in bacteriomes compared to body tissues (Figure 2).
Species-specific control of amino acid biosynthesis and its relation to psyllid microbiome differences
Even though the majority of the psyllid’s collaborative genes displayed similar expression patterns in bacteriomes, it is noteworthy that differences in expression patterns were observed between species. Some of these species-specific differences may be attributed to microbiome differences between the psyllid species. For example, a key difference between D. citri and B. cockerelli’s microbiomes is the presence of a co-endosymbiont, Profftella, in D. citri. Here we found that the sulfur metabolism and the cysteine and methionine metabolism were only significantly positively enriched in D. citri bacteriomes, suggesting that Profftella may play a key role in collaborating with D. citri for the biosynthesis of sulfur-containing metabolites (Table S10). Indeed, in D. citri, the enzymatic machinery required for sulfate reduction is exclusively found in Profftella (cysDHIN).24 In contrast Carsonella has completely lost all genes associated with this pathway, and there is no evidence suggesting that these genes have been substituted by any functionally acquired HTGs in the psyllid host.7,22 In the model aphid-Buchnera system, the sulfur assimilation pathway is generally present in Buchnera, and it is primarily inferred to have a role in synthesizing cysteine from serine.39 The origin of cysteine for D. citri remains uncertain as the absence of cysEK in Carsonella hinders the completion of the pathway, making it unclear whether cysteine is synthesized from serine.22 A previous hypothesis put forth in P. venusta7 suggests that cysteine could potentially be synthesized from cystathionine through the activity of cystathionine gamma-lyase (CGL). It is plausible that a similar mechanism may also operate in D. citri. Future studies will be needed to determine the exact role of sulfur assimilation in Profftella and its interactions with D. citri.
Another important species-specific expression difference for collaborative psyllid genes was observed for the enzyme BCAT, which serves as the final step in the biosynthesis of the branched-chain amino acids, isoleucine, valine, and leucine. The expression pattern of the collaborative enzyme BCAT varied among the three divergent psyllid species, where it was up-regulated in bacteriomes in D. citri, down-regulated in B. cockerelli, and non-significantly differentially expressed in P. venusta (Figure 2). One possible explanation for these divergent expression patterns is that, unlike Buchnera in aphids2 and Tremblaya and Moranella in mealybugs,6 all Carsonella genomes still encode ilvE, which potentially has a similar enzyme function as BCAT in these latter insect bacteriomes. Therefore, BCAT may not be essential in some psyllid bacteriomes for this collaborative, redundant function. In D. citri, both Profftella and Wolbachia lack the ilvE gene in their genomes (Table S11).24,40 Therefore, the observed up-regulation of BCAT in D. citri, as compared to B. cockerelli and P. venusta, could potentially be attributed to the presence of Profftella and Wolbachia in D. citri’s bacteriomes. This presence might lead to an increased nutritional demand for essential amino acids, such as leucine, valine, and threonine, consequently relying more on the psyllid host for the regulation of these branched-chain amino acids.
In this study both psyllid species harbor Wolbachia, which is in contrast to the previous analysis of P. venusta.7 Wolbachia is currently categorized in 16–17 distinct monophyletic lineages known as “supergroups” from A to S,41 where supergroups A and B are found only in arthropods.42 The Wolbachia strains discovered in psyllid species to date all belong to supergroup B.43,44 Due to its limited capacity for amino acid biosynthesis, Wolbachia supergroups A and B have been traditionally known to be highly dependent on the amino acid metabolism of its hosts,45,46 while recent studies have provided evidence of evolutionary transitions from facultative to obligate mutualisms in certain members of the Wolbachia F supergroups.47,48 Here higher genome copy counts of Wolbachia were measured in D. citri and B. cockerelli bacteriomes compared to body tissues (Figure 5), suggesting that Wolbachia may have direct metabolic influences on the host and obligate symbiont(s) within bacteriomes for amino acids. A study by Ren et al. (2018)49 corroborates our data here and found that Wolbachia-DC was detected with the highest density in the D. citri bacteriome compared to other body tissues using Fluorescence In Situ Hybridization (FISH) visualization. Hosseinzadeh et al. (2019),50 however, found that Wolbachia-DC was the most abundant in D. citri’s Malpighian tubules among a variety of organs including heads, guts, testes/ovaries and bacteriomes. To our knowledge, it is not known whether Wolbachia has any role in osmoregulation or nitrogen excretion, which are the primary functions of the Malpighian tubules. Based on Wolbachia-DC’s genome, it does not have the enzymatic capabilities required for recycling nitrogen from uric acid,40 which can be found in other insect bacterial endosymbionts.51,52,53
While Wolbachia exhibits genomic hallmarks of being parasitic and reliant on amino acids from its psyllid host and Carsonella, it may also play a role in essential amino acid biosynthesis and exhibit facultative or even mutualistic interactions. For example, cysteine-S-conjugate beta-lyase (CBL; EC 4.4.1.13), is encoded in both D. citri and B. cockerelli and is hypothesized to collaborate with Carsonella in the methionine biosynthesis pathway.7 Interestingly, the two copies of CBL in D. citri exhibited significant up-regulation in bacteriomes, while the CBL copies in B. cockerelli showed no differential regulation in bacteriomes (Table 1). The bacterial enzyme metC (EC 4.4.1.13), a cystathionine β-lyase enzyme in bacteria, is widely conserved among different Wolbachia strains (Table S11)47,54 suggesting that Carsonella might not be entirely dependent on host-encoded CBL for methionine biosynthesis. Instead Wolbachia may help complement the methionine biosynthesis pathway in B. cockerelli. Further functional genomics analyses of Wolbachia in B. cockerelli are needed to further understand this potential metabolic interaction.
Role of the glutamate dehydrogenase pathway in the recycling of waste ammonia in bacteriomes
Sap-feeding insects feed on a diet that is limited in nitrogen, especially essential amino acids.55,56 The recycling of waste ammonia in sap-feeding mutualistic relationships serves as a potential solution to fuel the nitrogen-demanding nature of these symbioses. One enzyme that can upgrade nitrogen from compounds such as ammonia is GS, which is significantly up-regulated in bacteriomes of both D. citri and B. cockerelli here. The GOGAT gene, which works in a cycle with GS to produce glutamate however is down-regulated in B. cockerelli and not statistically differentially expressed in D. citri (Figure 1; Table 1). The GS/GOGAT cycle is known to be an important mechanism in symbiotic relationships between sap-feeding insects and their obligate nutritional symbionts, as highlighted by previous studies on the upgrading of nitrogen from waste ammonia to fuel the amino acid metabolism.2,6,7,19,38,57,58 Interestingly, we identified that glutamate dehydrogenase (GDH) was significantly up-regulated in both D. citri and B. cockerelli bacteriomes, which might be an alternative to GOGAT for recycling ammonia into glutamate (Figure 2). The enzyme GDH is known to play a key role in assimilating waste nitrogen for the production of amino acids in human breast cancer cells where ammonia accumulates rapidly.59 Another recent study also revealed that raised levels of ammonia (hyperammonemia) results in GDH to play an essential role in the recycling of ammonia within non-cancerous brain cells.60 As a result, when ammonia levels rise in bacteriomes, the GS/GOGAT cycle may undergo a shift in its role, favoring GDH for the recycling of ammonia in the biosynthesis of amino acids.
Most studies on hemipteran systems have not reported any involvement of the GDH pathway.2,6,7,38 However, a recent study has found a potential role of the GDH pathway in the glassy-winged sharpshooter (GWSS) symbiosis system.37 In a novel dual obligate symbiosis between Baumannia and Sulcia-GWSS, where the GWSS host has two types of bacteriome tissues for the two endosymbionts, the GDH gene of GWSS is more highly expressed than GOGAT of the GWSS in the yellow bacteriome that harbors both Baumannia and Sulcia-GWSS.37 Future studies are warranted to investigate whether the up-regulation of GDH or the GS/GOGAT cycle in insects for nitrogen upgrading in bacteriomes for amino acid biosynthesis is influenced by the symbiosis, microbiome composition, and/or environmental conditions.
Functional significance of horizontally transferred genes in the amino acid metabolism
In sternorrhynchan insects, a consistent pattern arises with the loss of endosymbiont enzymes involved in the terminal steps of essential amino acid biosynthesis pathways. In response, host genes are hypothesized to step in and bridge these gaps.39 Notably, this pattern extends to the two HTGs in the psyllid’s genome, ASL providing the terminal step for the arginine pathway and CM providing the terminal step for the phenylalanine pathway in psyllids. Here we found the significant up-regulation of these HTGs in bacteriomes for both psyllid species (Figures 2 and 3). This finding aligns with previous research conducted in P. venusta, suggesting a consistent pattern of HTG up-regulation in psyllid bacteriomes when compared to body tissues.7 Despite the potential for subfunctionalization and/or neofunctionalization of the paralogous copies of ASL and CM, all of the copies were significantly up-regulated in the bacteriomes compared to the body tissues in both psyllid species B. cockerelli and D. citri (Table S4.2). This suggests the potential importance of gene dosage for these collaborative enzymes within bacteriomes. Further investigation is needed to determine whether these gene copies exhibit differential expression in response to varying environmental conditions.
This study also identified the HTG RSMJ, a bacterial 16S rRNA methylation gene to have one of the highest relative expression levels of any gene in the bacteriome compared to the body for both psyllid species, as determined by comparative transcriptomics analysis using PCA (Tables 2 and S5). Phylogenetic analysis revealed that the RSMJ gene from the three psyllid species cluster within the ‘class I SAM-dependent methyltransferase’ group within Gammaproteobacteria (e.g., Escherichia coli, Photobacterium spp.)61 (Figure S3). The InterPro GO term (GO:0008990) and Superfamily annotations (IPR029063) provide further evidence for classifying psyllid RSMJ as a SAM-dependent 16S rRNA methyltransferase, however the precise role of this gene in psyllid bacteriomes and the factors contributing to its conserved high expression levels in psyllids remain uncertain without additional functional assays. In E. coli, ten 16S rRNA methylation genes are involved in the modification of the small ribosomal subunit. The gene rsmJ in E. coli is characterized as a methyltransferase specific for the methylation of guanosine in the 1516 position of the 16S rRNA that has a cold sensitive mutant phenotype.62 In bacteria, rRNA methylation serves various purposes, including facilitating rRNA development, enhancing the stability of rRNA configurations, and modifying translation speed, however, alternative roles have been described, such as conferring resistance against aminoglycoside antibiotics derived from actinomycetes.63
Horizontal gene transfer events in psyllids are also hypothesized to play a critical role in B vitamin biosynthesis. Insects require eight B vitamins as coenzymes for essential reactions, but they are unable to produce these vitamins and must obtain them through their diet.64 Some insects can supplement B vitamins using their microbial symbionts,39,65 while some genes in vitamin biosynthesis have been acquired from bacteria by the insect host.6,7 In psyllids, the RIBC gene, also known as riboflavin synthase, has been found to be horizontally transferred to the host genome.7,22 This gene in bacteria, ribC, catalyzes the final step of the pathway, converting 6,7-dimethyl-8-ribityllumazine (DMRL) into riboflavin.66 Our data shows that RIBC was significantly up-regulated in bacteriomes at very high levels in both B. cockerelli and D. citri (LogFC = 7.88 and 6.15, respectively), similar to RIBC in P. venusta.7 However, only D. citri has been reported to possess a complete riboflavin biosynthetic pathway, where the co-obligate endosymbiont Profftella in D. citri retains all the genes in the riboflavin biosynthesis pathway except for the ribC (Figure 2).24 In contrast, the genomes of both the host and Carsonella in B. cockerelli and P. venusta appear to lack the rest of the genes in the riboflavin biosynthesis pathway. Therefore B. cockerelli and P. venusta may rely on their diet for the acquisition of the intermediate DMRL to produce riboflavin.
Regulation of lineage-specific gene clusters and its implications in evolutionary divergence
Our analysis of lineage-specific gene clusters that are unique to either D. citri or B. cockerelli highlights gene clusters that are differentially expressed in bacteriomes compared to body tissues for each species. Notably, the up-regulation of the Notch signaling pathway was prominent among D. citri-specific gene clusters, while no B. cockerelli-specific gene clusters were related to this pathway (Tables S6 and S7). The Notch signaling pathway, particularly in insects, plays crucial roles in body segmentation, proliferation, embryogenesis and cell fate determination.67 In the aphid-Buchnera system, it has been observed that the signaling pathways, such as TGF-beta, Wnt, Hippo, Hedgehog and Notch, are significantly enriched in the 3rd instar bacteriocytes58 and the 4th instar bacteriocytes of low-Buchnera titer genotypes.68 Smith and Moran (2020)68 hypothesize that the up-regulation of signaling pathways such as Notch in bacteriomes may result in an increase in both the number and size of aphid bacteriocytes in response to essential amino acid limitation.69 Hence, the observed up-regulation of lineage specific genes associated with the Notch signaling pathway in D. citri suggests that D. citri may possess distinct developmental processes that necessitate precise regulation to maintain a balance between metabolic production and cell growth. This phenomenon could be attributed to the heightened nutritional requirements imposed by Profftella, which is absent in B. cockerelli.
Our analysis showed that telomeric D loop disassembly was one of the most represented up-regulated GO-terms for B. cockerelli lineage specific clusters, and this GO-term was not found in D. citri specific gene clusters. (Tables S7 and S8). Telomeric D loop disassembly is a critical process involved in chromosome stability, integrity, and maintenance.70 The telomere motif (TTAGG)n is considered canonical for insects,71,72 however, the structure of telomeric repeats can vary depending on the insect group as observed in some coleopteran species.73,74 Interestingly, in a distantly related hemipteran species Myzus persicae, the (TTAGG)n motif has been found to be interspersed with inserted non-LTR retrotransposable elements,75 which could potentially be an intermediate state between the canonical insect telomere and retrotransposon-based ones.76 It is not clear at this point whether B. cockerelli has a canonical or retrotransposon-based (or intermediate) telomeric structure. To date the telomere structure in psyllids has been investigated for only five species within four genera from the families Psyllidae and Aphalaridae,77 and all five species in the study appear to have the (TTAGG)n motif according to FISH visualization. Nevertheless, given that transposition is another highly represented GO-term for B. cockerelli specific gene clusters in bacteriomes, and that B. cockerelli shows a significant increase of transposable elements in its genome,22 the up-regulation of lineage specific genes related to the telomeric D loop assembly in B. cockerelli bacteriomes suggests that there might be a potential association between telomere maintenance and transposable elements that differs from D. citri bacteriomes.
Conclusion
This study found both conserved and variable gene expression profiles for bacteriomes of two divergent psyllid species. Both psyllid species rely on Carsonella for the production of essential amino acids however their regulation of this symbiosis may vary due to host and symbiont genetic factors, differences within their microbiomes, and/or the environment including diet. Future studies aiming to investigate additional psyllid species, as well as considering various factors within species, will provide valuable insights into the evolutionary mechanisms underlying the integrated metabolisms of psyllids. This comprehensive approach will contribute to a deeper understanding of the complex dynamics involved in the evolution of metabolic interactions in psyllids.
Limitations of the study
Functional genetics techniques are still not available in these two psyllid species therefore metabolomic analyses and interpretations here are based on transcriptome data and comparative evolutionary genomics analyses. Due to available insectary and quarantine space, available funding, and timing of life stage dissections of this project, which were conducted around the same time frame to prevent batch effects, only one psyllid genetic line was used per species for this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli K12 JM109 | Promega | Cat#L2005 |
| Biological samples | ||
| Diaphorina citri bacteriome tissues | This study | N/A |
| D. citri body tissues | This study | N/A |
| Bactericera cockerelli bacteriome tissues | This study | N/A |
| B. cockerelli body tissues | This study | N/A |
| Deposited data | ||
| BioProject | This study | NCBI BioProject: PRJNA939696 |
| Raw data | This study | NCBI SRA: SRR23641431 to SRR23641440 |
| Experimental models: Organisms/strains | ||
| D. citri | USA:CA | N/A |
| B. cockerelli | USA:CA | N/A |
| Oligonucleotides | ||
| Dcit-wsp-qF/R | This study | See STAR methods section for sequences |
| Bcoc-wsp-qF/R | This study | See STAR methods section for sequences |
| Recombinant DNA | ||
| pGEM-T-Easy-Dcit-wsp | This study | N/A |
| pGEM-T-Easy-Bcoc-wsp | This study | N/A |
| pGEM-T-Easy-ACTB | This study | N/A |
| pGEM-T-Easy-FORKHEAD | This study | N/A |
| Software and algorithms | ||
| FASTQC | Andrews (2010)81 | v.0.11.8 |
| Trimmomatic | Bolger et al. (2014)82 | v.0.36 |
| HiSAT | Kim et al. (2015) | v.2.1.0 |
| StringTie | Pertea et al. (2015)84 | v.2.2.1 |
| RStudio | R Core Team (2022)85 | v.4.2.0 |
| EdgeR | Robinson et al. (2010)86 | N/A |
| BlastKOALA | Kanehisha et al. (2016) | N/A |
| OrthoVenn2 | Xu et al. (2019)92 | N/A |
| GSEA | Subramanian et al. (2005)88 | v4.0.0 |
| RAxML-HPC BlackBox | Miller et al. (2010)90 | v.8.2.12 |
| Figtree | Rambaut (2018)91 | v.1.4.4 |
| ggplot2 | Wickham (2016)98 | N/A |
| Other | ||
| D. citri reference genome | Carlson et al. (2022)21 | GCA_024506315.2 |
| B. cockerelli reference genome | Kwak et al. (2023)22 | GCA_024516035.1 |
Resource availability
Lead contact
Further information and requests for resources can be directed to and will be fulfilled by the lead contact, Allison K. Hansen (allison.hansen@ucr.edu).
Materials availability
This study did not generate new unique reagents.
Experimental models and study participants
Plant and insect materials
The B. cockerelli line was derived from a wild population in Temecula, California, USA, in August 2019, and is from the same culture that was sequenced in Kwak et al. (2023).22 The established line was maintained on 8-12-week-old Capsicum annuum plants (California Wonder pepper) at 25°C under a 16L:8D light/dark cycle. The Diaphorina citri nymph culture was obtained from the Stouthamer lab at the University of California (UC), Riverside, and was maintained on 6-month-old Murraya koenigii leaves (Curry tree) at 27°C under a 16L:8D light/dark cycle.
For both B. cockerelli and D. citri, 60 bacteriomes and the rest of the body, without the bacteriomes, were dissected from 5th instar nymphs and pooled into two separate tissue samples similar to Hansen & Moran (2011),2 resulting in three biological replicates per species (N = 12 samples). The D. citri and B. cockerelli nymphs were aged for bacteriome dissections according to their morphological characters and developmental periods as described in Hall et al. (2013)79 and Knowlton & Janes (1931),80 respectively. Samples were stored at −80°C in the RNAprotect bacterial reagent (QIAGEN, Germantown, MD, USA) until RNA extraction.
Method details
Total RNA sample preparation and RNA sequencing
Total RNA was purified, DNase 1 treated, and cleaned using Quick-RNA Microprep kit and RNA Clean & Concentrator kit-5 (Zymo research, Irvine, CA, USA) following the manufacturer’s instructions. Purified RNA sample quality and quantity were measured using the Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) at the Institute of Integrative Genome Biology Instrumentation Facilities Services at the University of California, Riverside.
High-quality total RNA (>1 μg) from each pooled bacteriome sample and corresponding body tissue sample was submitted to the DNA Technology Core at the University of California, Davis for library preparation and sequencing. Strand-specific and barcode indexed RNA-seq libraries were generated from 300ng total RNA for each sample. Poly-A enrichment and library prep was done using the Kapa mRNA Stranded library preparation kit (KK8421, Kapa Biosystems, Cape Town, South Africa), following the instructions of the manufacturer. Libraries were amplified with 12 cycles of PCR. The fragment size distribution of the libraries was verified via micro-capillary gel electrophoresis on a Bioanalyzer 2100 (Agilent, Santa Clara, CA). The libraries were quantified by fluorometry on a Qubit fluorometer (LifeTechnologies, Carlsbad, CA) and pooled in equimolar ratios. The pool was quantified by qPCR with a Kapa Library Quant kit (Kapa Biosystems) and sequenced on 1 lane of the Illumina NovaSeq S4 platform (Illumina, San Diego, CA) with paired-end 150bp reads. Reads for all RNA-Seq samples were submitted to the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) under BioProject ID PRJNA939696.
Ortholog analysis and interspecies comparative transcriptomic analysis
Orthologous clusters, lineage-specific clusters, and one-to-one orthologs of D. citri and B. cockerelli were determined using OrthoVenn292 using default settings. To visualize the differentially expressed one-to-one orthologs, a Venn diagram was generated using SeqCode VennPlotter.93 Similar to Georgiadou et al. (2022)33 and Argandona et al. (2023),32 one-to-one orthologs were further examined for inter-species gene expression comparisons. Specifically, the relative magnitude of changes of orthologs in bacteriomes were compared to body tissues for each species using Principal Components Analyses (PCA). To do this, we selected one-to-one orthologs that displayed significant differential gene expression (see above for the thresholds) in both D. citri and B. cockerelli. These orthologs were ranked in descending order of absolute logFC, and each ortholog was then assigned a value of 100 divided by rank, which was then multiplied by the sign of the original logFC. These values were then used as the input for PCA to identify one-to-one orthologs that display the most similar and different gene expression profiles in the bacteriomes compared to the body tissues. The PCA was performed using PC-ORD (version 4.25) 94. For each principal component (axis 1 and 2), we selected the top 50 genes with the highest correlations (the top 25 genes for positive or negative correlations) from the principal components output loading matrix similar to Korb et al. (2021).34 We examined the GO annotations for these top 100 orthologs using OrthoVenn2.92 We generated a heatmap of logFC for one-to-one orthologs using SeqCode Heatmapper.93
Quantitative real-time PCR
Fifth instar nymphs of both species were collected for bacteriome dissections for Wolbachia endosymbiont quantification similar to the RNAseq analysis (above). Bacteriomes and the rest of the body tissues without the bacteriomes were pooled from 15 individuals of both sexes for each biological replicate with a total of three biological replicates per species per tissue type. Genomic DNA was extracted from pooled bacteriome and body tissues using Quick-DNA Microprep Plus Kit (Zymo, Irvine, CA, USA). Samples were homogenized, lysed, and purified following the Solid Tissue Protocol. DNA samples were treated with RNase A (Thermo Scientific, Waltham, MA, USA), and cleaned with the Genomic DNA Clean and Concentrator kit-10 (Zymo research, Irvine, CA, USA). Purified DNA sample quality and quantity were measured via QuickDrop (Molecular Devices, San Jose, CA, USA) and Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA, USA).
Primers for the Wolbachia surface protein (wsp) gene were designed for this analysis using Primer-BLAST in NCBI using GenBank: MN809922.1 for D. citri (Dcit-wsp-qF/R: 5′-TGCTGGAGCTCGTTACTTCG-3’/5′- CAGCTTCTGCACCAACAGTG-3′) and GenBank: KM267307.1 for B. cockerelli (Bcoc-wsp-qF/R: 5′- ATAGCTGCTGGTGGTGCATT-3’/5′- CACCAACACCAACACCAACG-3′). For calibration, the psyllid housekeeping genes encoding ACTB95 and FORKHEAD,96 which are single-copy genes in the D. citri and B. cockerelli genomes, respectively, were also quantified. The PCR products were cloned into the pGEM-T Easy vector (Promega) and amplified in Escherichia coli JM109. Plasmids with inserts were amplified in E. coli and purified using Plasmid Minikit (Invitrogen). Subsequently, Plasmid DNA was quantified using a Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Copy numbers of the plasmid DNA were calculated based on their concentration and molecular weights. For each target gene, 108, 107, 106, 105, 104 and 103 copies/μL of plasmid DNA solutions were freshly prepared for standard samples. The qPCR reactions were run with three technical replicates using iTaq Universal SYBR Green Supermix on a Bio-Rad CFX96 touch machine (Bio-Rad, Hercules, CA, USA), and the conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and the final melt curve for primer specificity from 65°C to 95°C with 0.5°C.
Quantification and statistical analysis
Bioinformatic analysis
Following the same RNAseq pipeline detailed in Pers & Hansen (2021)58 and Argandona et al. (2023),32 raw RNA-seq reads were quality checked and trimmed with FASTQC v.0.11.881 and Trimmomatic v. 0. 3682 with the following parameters: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. The trimmed reads were aligned using HISAT v. 2.1. 083 against the chromosomal assemblies of CRF-CA_Dcit for D. citri (GenBank: GCA_024506315.2)21 and isoF-IL for B. cockerelli (GenBank: GCA_024516035.1).22 The mapped reads for each gene were quantified as raw read counts using StringTie v.2.2.184 using gff files annotated for each psyllid species.21,22 Differential expression of gene transcripts between bacteriome and body samples were determined using R (version 4.2.0)85 using the exact test with EdgeR.86 The exact test was chosen because it has been demonstrated to be more conservative compared to other tests when the number of sample replicates is smaller than five.87 Statistical significance of differentially expressed genes was determined with a false discovery rate (FDR) adjusted p ≤ 0.05 and ≥1.5-fold change of the normalized expression values similar to Argandona et al. (2023).32 In this context, “logFC” indicate log2 fold change between the groups and “logFC” ≥ 0.5849 represents significantly up-regulated genes and “logFC” ≤ −0.5849 represents significantly down-regulated genes. Gene Set Enrichment Analysis (GSEA, v4. 0. 0)88 was used to determine which Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are differently expressed at the normalized p ≤ 0.05 and FDR q ≤ 0.25, as described in Pers & Hansen (2021).58 Annotations for subsets of genes that are related to HTGs and symbiosis genes are obtained from the previous genome analyses,22 and further annotated here for the new genome dataset of D. citri-CRF using NCBI Blast and BlastKOALA.89 Further phylogenetic analyses for three HGTs were performed with RAxML-HPC BlackBox version 8.2.12 on the CIPRES webserver.90 The JTT model was employed, and RAxML was allowed to halt bootstrapping automatically. The resulting bipartition trees generated from RAxML were visualized and exported using Figtree version 1.4.4.91
Wolbachia endosymbiont quantification
The normalized ratio value for Wolbachia genome copies relative to psyllid genome copies was calculated for three biological replicates using the following equation: average Wolbachia DNA single gene copy quantity/average psyllid DNA single gene copy quantity based on the protocol outlined in Bookout et al. (2006).97 Statistical analysis of the copy number variation was conducted using an independent t-test from the ggplot298 package in R version 4.1.285 to determine significant (p < 0.05) differences between tissue types for the normalized ratio values for both psyllid species.
Acknowledgments
We thank the Stouthamer lab for providing Diaphorina citri, Dr. Patrick H. Degnan and Dr. Dohyup Kim for bioinformatic support. The sequencing was carried out at the UC Davis Genome Center DNA Technologies and Expression Analysis Core, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. This work was supported by funding from the National Institute of Food and Agriculture (NIFA), United States Department of Agriculture (USDA) (Award number: 2019-70016-29066), and the Department of Entomology at the University of California, Riverside (UCR).
Author contributions
YK helped set up experiments, conducted the data and bioinformatic analyses and helped write the article. AH helped design the study, conducted data and bioinformatic analyses, and helped write the article.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107930.
Contributor Information
Younghwan Kwak, Email: ykwak@ucmerced.edu.
Allison K. Hansen, Email: allison.hansen@ucr.edu.
Supplemental information
Data and code availability
-
•
Raw RNA-seq data have been deposited at NCBI under BioProject accession number PRJNA939696. Accession numbers are also listed in the key resources table.
-
•
All code used in this paper is from the “Dataset_S9_RNAseq_Code”,58 which is publicly available on figshare: https://doi.org/10.25387/g3.14109851.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen A.K., Moran N.A. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan R.P., Husnik F., Van Leuven J.T., Gilbert D.G., Dávalos L.M., McCutcheon J.P., Wilson A.C.C. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol. Ecol. 2014;23:1608–1623. doi: 10.1111/mec.12627. [DOI] [PubMed] [Google Scholar]
- 4.Price D.R.G., Tibbles K., Shigenobu S., Smertenko A., Russell C.W., Douglas A.E., Fitches E., Gatehouse A.M.R., Gatehouse J.A. Sugar transporters of the major facilitator superfamily in aphids; from gene prediction to functional characterization. Insect Mol. Biol. 2010;19:97–112. doi: 10.1111/j.1365-2583.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- 5.Price D.R.G., Duncan R.P., Shigenobu S., Wilson A.C.C. Genome Expansion and Differential Expression of Amino Acid Transporters at the Aphid/Buchnera Symbiotic Interface. Mol. Biol. Evol. 2011;28:3113–3126. doi: 10.1093/molbev/msr140. [DOI] [PubMed] [Google Scholar]
- 6.Husnik F., Nikoh N., Koga R., Ross L., Duncan R.P., Fujie M., Tanaka M., Satoh N., Bachtrog D., Wilson A.C.C., et al. Horizontal Gene Transfer from Diverse Bacteria to an Insect Genome Enables a Tripartite Nested Mealybug Symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 7.Sloan D.B., Nakabachi A., Richards S., Qu J., Murali S.C., Gibbs R.A., Moran N.A. Parallel Histories of Horizontal Gene Transfer Facilitated Extreme Reduction of Endosymbiont Genomes in Sap-Feeding Insects. Mol. Biol. Evol. 2014;31:857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan J.-B., Chen W., Hasegawa D.K., Simmons A.M., Wintermantel W.M., Ling K.-S., Fei Z., Liu S.-S., Douglas A.E. Metabolic Coevolution in the Bacterial Symbiosis of Whiteflies and Related Plant Sap-Feeding Insects. Genome Biol. Evol. 2015;7:2635–2647. doi: 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thao M.L., Moran N.A., Abbot P., Brennan E.B., Burckhardt D.H., Baumann P. Cospeciation of Psyllids and Their Primary Prokaryotic Endosymbionts. Appl. Environ. Microbiol. 2000;66:2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spaulding A.W., von Dohlen C.D. Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol. Biol. 2001;10:57–67. doi: 10.1046/j.1365-2583.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M. The 160-Kilobase Genome of the Bacterial Endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 12.Moran N.A., Wernegreen J.J. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 2000;15:321–326. doi: 10.1016/S0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 13.Baumann P. Biology of Bacteriocyte-Associated Endosymbionts of Plant Sap-Sucking Insects. Annu. Rev. Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 14.McCutcheon J.P., Moran N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 15.Fukatsu T., Nikoh N. Two Intracellular Symbiotic Bacteria from the Mulberry Psyllid Anomoneura mori (Insecta, Homoptera) Appl. Environ. Microbiol. 1998;64:3599–3606. doi: 10.1128/AEM.64.10.3599-3606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchner P. Interscience Publishers; 1965. Endosymbiosis of Animals with Plant Microorganisms. [Google Scholar]
- 17.Sloan D.B., Moran N.A. Genome Reduction and Co-evolution between the Primary and Secondary Bacterial Symbionts of Psyllids. Mol. Biol. Evol. 2012;29:3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamames J., Gil R., Latorre A., Peretó J., Silva F.J., Moya A. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poliakov A., Russell C.W., Ponnala L., Hoops H.J., Sun Q., Douglas A.E., van Wijk K.J. Large-Scale Label-Free Quantitative Proteomics of the Pea aphid-Buchnera Symbiosis. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.007039. M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Zhang B., Moran N.A. The Aphid X Chromosome Is a Dangerous Place for Functionally Important Genes: Diverse Evolution of Hemipteran Genomes Based on Chromosome-Level Assemblies. Mol. Biol. Evol. 2020;37:2357–2368. doi: 10.1093/molbev/msaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson C.R., ter Horst A.M., Johnston J.S., Henry E., Falk B.W., Kuo Y.-W. High-quality, chromosome-scale genome assemblies: comparisons of three Diaphorina citri (Asian citrus psyllid) geographic populations. DNA Res. 2022;29:dsac027. doi: 10.1093/dnares/dsac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak Y., Argandona J.A., Degnan P.H., Hansen A.K. Chromosomal-level assembly of Bactericera cockerelli reveals rampant gene family expansions impacting genome structure, function and insect-microbe-plant-interactions. Mol. Ecol. Resour. 2023;23:233–252. doi: 10.1111/1755-0998.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakabachi A., Ueoka R., Oshima K., Teta R., Mangoni A., Gurgui M., Oldham N.J., van Echten-Deckert G., Okamura K., Yamamoto K., et al. Defensive Bacteriome Symbiont with a Drastically Reduced Genome. Curr. Biol. 2013;23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 24.Nakabachi A., Piel J., Malenovský I., Hirose Y. Comparative Genomics Underlines Multiple Roles of Profftella, an Obligate Symbiont of Psyllids: Providing Toxins, Vitamins, and Carotenoids. Genome Biol. Evol. 2020;12:1975–1987. doi: 10.1093/gbe/evaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subandiyah S., Nikoh N., Tsuyumu S., Somowiyarjo S., Fukatsu T. Complex Endosymbiotic Microbiota of the Citrus Psyllid Diaphorina citri (Homoptera: Psylloidea) Zool. Sci. (Tokyo) 2000;17:983–989. doi: 10.2108/zsj.17.983. [DOI] [Google Scholar]
- 26.Nachappa P., Levy J., Pierson E., Tamborindeguy C. Diversity of Endosymbionts in the Potato Psyllid, Bactericera cockerelli (Hemiptera: Triozidae), Vector of Zebra Chip Disease of Potato. Curr. Microbiol. 2011;62:1510–1520. doi: 10.1007/s00284-011-9885-5. [DOI] [PubMed] [Google Scholar]
- 27.Stouthamer R., Breeuwer J.A., Hurst G.D. Wolbachia Pipientis: Microbial Manipulator of Arthropod Reproduction. Annu. Rev. Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 28.Cooper W.R., Swisher K.D., Garczynski S.F., Mustafa T., Munyaneza J.E., Horton D.R. Wolbachia Infection Differs Among Divergent Mitochondrial Haplotypes of Bactericera cockerelli (Hemiptera: Triozidae) Ann. Entomol. Soc. Am. 2015;108:137–145. doi: 10.1093/aesa/sau048. [DOI] [Google Scholar]
- 29.Hoffmann M., Coy M.R., Kingdom Gibbard H.N., Pelz-Stelinski K.S. Wolbachia Infection Density in Populations of the Asian Citrus Psyllid (Hemiptera: Liviidae) Environ. Entomol. 2014;43:1215–1222. doi: 10.1603/EN14193. [DOI] [PubMed] [Google Scholar]
- 30.Kwak Y., Sun P., Meduri V.R., Percy D.M., Mauck K.E., Hansen A.K. Uncovering Symbionts Across the Psyllid Tree of Life and the Discovery of a New Liberibacter Species, “Candidatus” Liberibacter capsica. Front. Microbiol. 2021;12:739763. doi: 10.3389/fmicb.2021.739763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Štarhová Serbina L., Gajski D., Malenovský I., Corretto E., Schuler H., Dittmer J. Wolbachia infection dynamics in a natural population of the pear psyllid Cacopsylla pyri (Hemiptera: Psylloidea) across its seasonal generations. Sci. Rep. 2022;12:16502. doi: 10.1038/s41598-022-20968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argandona J.A., Kim D., Hansen A.K. Comparative transcriptomics of aphid species that diverged > 22 MYA reveals genes that are important for the maintenance of their symbiosis. Sci. Rep. 2023;13:5341. doi: 10.1038/s41598-023-32291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgiadou A., Dunican C., Soro-Barrio P., Lee H.J., Kaforou M., Cunnington A.J. Comparative transcriptomic analysis reveals translationally relevant processes in mouse models of malaria. Elife. 2022;11 doi: 10.7554/eLife.70763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korb J., Meusemann K., Aumer D., Bernadou A., Elsner D., Feldmeyer B., Foitzik S., Heinze J., Libbrecht R., Lin S., et al. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376 doi: 10.1098/rstb.2019.0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douglas A.E. Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot. 2006;57:747–754. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- 36.Macdonald S.J., Lin G.G., Russell C.W., Thomas G.H., Douglas A.E. The central role of the host cell in symbiotic nitrogen metabolism. Proc. Biol. Sci. 2012;279:2965–2973. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao M., Bennett G.M. Symbiont replacements reset the co-evolutionary relationship between insects and their heritable bacteria. ISME J. 2020;14:1384–1395. doi: 10.1038/s41396-020-0616-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao M., Yang X., Bennett G.M. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc. Natl. Acad. Sci. USA. 2018;115:E11691–E11700. doi: 10.1073/pnas.1811932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen A.K., Moran N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014;23:1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- 40.Neupane S., Bonilla S.I., Manalo A.M., Pelz-Stelinski K.S. Complete de novo assembly of Wolbachia endosymbiont of Diaphorina citri Kuwayama (Hemiptera: Liviidae) using long-read genome sequencing. Sci. Rep. 2022;12:125. doi: 10.1038/s41598-021-03184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefoulon E., Clark T., Borveto F., Perriat-Sanguinet M., Moulia C., Slatko B.E., Gavotte L. Pseudoscorpion Wolbachia symbionts: diversity and evidence for a new supergroup S. BMC Microbiol. 2020;20:188. doi: 10.1186/s12866-020-01863-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo N., Casiraghi M., Salati E., Bazzocchi C., Bandi C. How Many Wolbachia Supergroups Exist? Mol. Biol. Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- 43.Ou D., Qiu J.-H., Su Z.-Q., Wang L., Qiu B.-L. The phylogeny and distribution of Wolbachia in two pathogen vector insects, Asian citrus psyllid and Longan psyllid. Front. Cell. Infect. Microbiol. 2023;13:1121186. doi: 10.3389/fcimb.2023.1121186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapoval N.A., Nokkala S., Nokkala C., Kuftina G.N., Kuznetsova V.G. The Incidence of Wolbachia Bacterial Endosymbiont in Bisexual and Parthenogenetic Populations of the Psyllid Genus Cacopsylla (Hemiptera, Psylloidea) Insects. 2021;12:853. doi: 10.3390/insects12100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiménez N.E., Gerdtzen Z.P., Olivera-Nappa Á., Salgado J.C., Conca C. A systems biology approach for studying Wolbachia metabolism reveals points of interaction with its host in the context of arboviral infection. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M., Sun L.V., Vamathevan J., Riegler M., Deboy R., Brownlie J.C., McGraw E.A., Martin W., Esser C., Ahmadinejad N., et al. Phylogenomics of the Reproductive Parasite Wolbachia pipientis wMel: A Streamlined Genome Overrun by Mobile Genetic Elements. PLoS Biol. 2004;2:e69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahmood S., Nováková E., Martinů J., Sychra O., Hypša V. Supergroup F Wolbachia with extremely reduced genome: transition to obligate insect symbionts. Microbiome. 2023;11:22. doi: 10.1186/s40168-023-01462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikoh N., Hosokawa T., Moriyama M., Oshima K., Hattori M., Fukatsu T. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA. 2014;111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren S.-L., Li Y.-H., Ou D., Guo Y.-J., Qureshi J.A., Stansly P.A., Qiu B.-L. Localization and dynamics of Wolbachia infection in Asian citrus psyllid Diaphorina citri, the insect vector of the causal pathogens of Huanglongbing. MicrobiologyOpen. 2018;7 doi: 10.1002/mbo3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosseinzadeh S., Shams-Bakhsh M., Mann M., Fattah-Hosseini S., Bagheri A., Mehrabadi M., Heck M. Distribution and Variation of Bacterial Endosymbiont and “Candidatus Liberibacter asiaticus” Titer in the Huanglongbing Insect Vector, Diaphorina citri Kuwayama. Microb. Ecol. 2019;78:206–222. doi: 10.1007/s00248-018-1290-1. [DOI] [PubMed] [Google Scholar]
- 51.Kashima T., Nakamura T., Tojo S. Uric acid recycling in the shield bug, Parastrachia japonensis (Hemiptera: Parastrachiidae), during diapause. J. Insect Physiol. 2006;52:816–825. doi: 10.1016/j.jinsphys.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Sabree Z.L., Kambhampati S., Moran N.A. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. USA. 2009;106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potrikus C.J., Breznak J.A. Gut bacteria recycle uric acid nitrogen in termites: A strategy for nutrient conservation. Proc. Natl. Acad. Sci. USA. 1981;78:4601–4605. doi: 10.1073/pnas.78.7.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown A.M.V., Wasala S.K., Howe D.K., Peetz A.B., Zasada I.A., Denver D.R. Comparative Genomics of Wolbachia–Cardinium Dual Endosymbiosis in a Plant-Parasitic Nematode. Front. Microbiol. 2018;9:2482. doi: 10.3389/fmicb.2018.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandström J., Pettersson J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 1994;40:947–955. doi: 10.1016/0022-1910(94)90133-3. [DOI] [Google Scholar]
- 56.Sandström J.P., Moran N.A. Amino acid budgets in three aphid species using the same host plant. Physiol. Entomol. 2001;26:202–211. doi: 10.1046/j.0307-6962.2001.00235.x. [DOI] [Google Scholar]
- 57.Kim D., Minhas B.F., Li-Byarlay H., Hansen A.K. Key Transport and Ammonia Recycling Genes Involved in Aphid Symbiosis Respond to Host-Plant Specialization. G3 (Bethesda). 2018;8:2433–2443. doi: 10.1534/g3.118.200297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pers D., Hansen A.K. The boom and bust of the aphid’s essential amino acid metabolism across nymphal development. G3 (Bethesda). 2021;11:jkab115. doi: 10.1093/g3journal/jkab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spinelli J.B., Yoon H., Ringel A.E., Jeanfavre S., Clish C.B., Haigis M.C. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017;358:941–946. doi: 10.1126/science.aam9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voss C.M., Arildsen L., Nissen J.D., Waagepetersen H.S., Schousboe A., Maechler P., Ott P., Vilstrup H., Walls A.B. Glutamate Dehydrogenase Is Important for Ammonia Fixation and Amino Acid Homeostasis in Brain During Hyperammonemia. Front. Neurosci. 2021;15:646291. doi: 10.3389/fnins.2021.646291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M., Kong D., Wang Y., Ma Q., Han X., Zhou Y., jiang X., Zhang Y., Ruan Z., Zhang Q. Photobacterium salinisoli sp. nov., isolated from a sulfonylurea herbicide-degrading consortium enriched with saline soil. Int. J. Syst. Evol. Microbiol. 2019;69:3910–3916. doi: 10.1099/ijsem.0.003705. [DOI] [PubMed] [Google Scholar]
- 62.Basturea G.N., Dague D.R., Deutscher M.P., Rudd K.E. YhiQ Is RsmJ, the Methyltransferase Responsible for Methylation of G1516 in 16S rRNA of E. coli. J. Mol. Biol. 2012;415:16–21. doi: 10.1016/j.jmb.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doi Y., Arakawa Y. 16S Ribosomal RNA Methylation: Emerging Resistance Mechanism against Aminoglycosides. Clin. Infect. Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 64.Douglas A.E. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr. Opin. Insect Sci. 2017;23:65–69. doi: 10.1016/j.cois.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Santos-Garcia D., Juravel K., Freilich S., Zchori-Fein E., Latorre A., Moya A., Morin S., Silva F.J. To B or Not to B: Comparative Genomics Suggests Arsenophonus as a Source of B Vitamins in Whiteflies. Front. Microbiol. 2018;9:2254. doi: 10.3389/fmicb.2018.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer M., Bacher A. Biosynthesis of vitamin B2: Structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 2008;474:252–265. doi: 10.1016/j.abb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Tautz D. Segmentation. Dev. Cell. 2004;7:301–312. doi: 10.1016/j.devcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 68.Smith T.E., Moran N.A. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc. Natl. Acad. Sci. USA. 2020;117:2113–2121. doi: 10.1073/pnas.1916748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colella S., Parisot N., Simonet P., Gaget K., Duport G., Baa-Puyoulet P., Rahbé Y., Charles H., Febvay G., Callaerts P., Calevro F. Bacteriocyte Reprogramming to Cope With Nutritional Stress in a Phloem Sap Feeding Hemipteran, the Pea Aphid Acyrthosiphon pisum. Front. Physiol. 2018;9:1498. doi: 10.3389/fphys.2018.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vicari M.R., Bruschi D.P., Cabral-de-Mello D.C., Nogaroto V. Telomere organization and the interstitial telomeric sites involvement in insects and vertebrates chromosome evolution. Genet. Mol. Biol. 2022;45 doi: 10.1590/1678-4685-GMB-2022-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mandrioli M., Monti V., Manicardi G.C. Starting at the end: telomeres and telomerase in arthropods. Biomol. Concepts. 2012;3:465–470. doi: 10.1515/bmc-2012-0008. [DOI] [PubMed] [Google Scholar]
- 72.Traut W., Szczepanowski M., Vítková M., Opitz C., Marec F., Zrzavý J. The telomere repeat motif of basal Metazoa. Chromosome Res. 2007;15:371–382. doi: 10.1007/s10577-007-1132-3. [DOI] [PubMed] [Google Scholar]
- 73.Frydrychová R., Marec F. Repeated Losses of TTAGG Telomere Repeats in Evolution of Beetles (Coleoptera) Genetica. 2002;115:179–187. doi: 10.1023/A:1020175912128. [DOI] [PubMed] [Google Scholar]
- 74.Mravinac B., Meštrović N., Čavrak V.V., Plohl M. TCAGG, an alternative telomeric sequence in insects. Chromosoma. 2011;120:367–376. doi: 10.1007/s00412-011-0317-x. [DOI] [PubMed] [Google Scholar]
- 75.Monti V., Serafini C., Manicardi G.C., Mandrioli M. Characterization of Non-LTR Retrotransposable TRAS Elements in the Aphids Acyrthosiphon pisum and Myzus persicae (Aphididae, Hemiptera) J. Hered. 2013;104:547–553. doi: 10.1093/jhered/est017. [DOI] [PubMed] [Google Scholar]
- 76.Mason J.M., Randall T.A., Capkova Frydrychova R. Telomerase lost? Chromosoma. 2016;125:65–73. doi: 10.1007/s00412-015-0528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maryańska-Nadachowska A., Kuznetsova V.G., Golub N.V., Anokhin B.A. Detection of telomeric sequences and ribosomal RNA genes in holokinetic chromosomes of five jumping plant-lice species: First data on the superfamily Psylloidea (Hemiptera: Sternorrhyncha) EJE (Eur. J. Epidemiol.) 2018;115:632–640. doi: 10.14411/eje.2018.061. [DOI] [Google Scholar]
- 79.Hall D.G., Richardson M.L., Ammar E.-D., Halbert S.E. Asian citrus psyllid, Diaphorina citri, vector of citrus huanglongbing disease. Entomol. Exp. Appl. 2013;146:207–223. doi: 10.1111/eea.12025. [DOI] [Google Scholar]
- 80.Knowlton G.F., Janes M.J. Studies on the Biology of Paratrioza Cockerelli (Sulc) Ann. Entomol. Soc. Am. 1931;24:283–292. doi: 10.1093/aesa/24.2.283. [DOI] [Google Scholar]
- 81.Andrews FastQC: A Quality Control Tool for High Throughput Sequence Dataics - FastQC A Quality Control tool for High Throughput Sequence Data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 82.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.R Core Team . 2022. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 86.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X., Cooper N.G.F., O’Toole T.E., Rouchka E.C. Choice of library size normalization and statistical methods for differential gene expression analysis in balanced two-group comparisons for RNA-seq studies. BMC Genom. 2020;21:75. doi: 10.1186/s12864-020-6502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 90.Miller M.A., Pfeiffer W., Schwartz T. 2010 Gateway Computing Environments Workshop (GCE) IEEE; 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [DOI] [Google Scholar]
- 91.Rambaut A. 2018. FigTree v1.4.4. [Google Scholar]
- 92.Xu L., Dong Z., Fang L., Luo Y., Wei Z., Guo H., Zhang G., Gu Y.Q., Coleman-Derr D., Xia Q., Wang Y. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blanco E., González-Ramírez M., Di Croce L. Productive visualization of high-throughput sequencing data using the SeqCode open portable platform. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-98889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCUNE B. Pc-Ord: An Integrated System for Multivariate Analysis of Ecological Data. Abstr. Bot. 1986;10:221–225. [Google Scholar]
- 95.Tiwari S., Gondhalekar A.D., Mann R.S., Scharf M.E., Stelinski L.L. Characterization of five CYP4 genes from Asian citrus psyllid and their expression levels in Candidatus Liberibacter asiaticus-infected and uninfected psyllids. Insect Mol. Biol. 2011;20:733–744. doi: 10.1111/j.1365-2583.2011.01103.x. [DOI] [PubMed] [Google Scholar]
- 96.Casteel C.L., Hansen A.K., Walling L.L., Paine T.D. Manipulation of Plant Defense Responses by the Tomato Psyllid (Bactericerca cockerelli) and Its Associated Endosymbiont Candidatus Liberibacter Psyllaurous. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bookout A.L., Cummins C.L., Mangelsdorf D.J., Pesola J.M., Kramer M.F. High-Throughput Real-Time Quantitative Reverse Transcription PCR. Curr. Protoc. Mol. Biol. 2006;Chapter 15 doi: 10.1002/0471142727.mb1508s73. Unit 15.8, 8.1-15, 8.28. [DOI] [PubMed] [Google Scholar]
- 98.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw RNA-seq data have been deposited at NCBI under BioProject accession number PRJNA939696. Accession numbers are also listed in the key resources table.
-
•
All code used in this paper is from the “Dataset_S9_RNAseq_Code”,58 which is publicly available on figshare: https://doi.org/10.25387/g3.14109851.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.