Summary
Objective
COVID-19 might cause neuroinflammation in the brain, which could decrease neurocognitive function. We aimed to evaluate the causal associations and genetic overlap between COVID-19 and intelligence.
Methods
We performed Mendelian randomization (MR) analyses to assess potential associations between three COVID-19 outcomes and intelligence (N = 269 867). The COVID phenotypes included severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (N = 2 501 486), hospitalized COVID-19 (N = 1 965 329) and critical COVID-19 (N = 743 167). Genome-wide risk genes were compared between the genome-wide association study (GWAS) datasets on hospitalized COVID-19 and intelligence. In addition, functional pathways were constructed to explore molecular connections between COVID-19 and intelligence.
Results
The MR analyses indicated that genetic liabilities to SARS-CoV-2 infection (odds ratio [OR]: 0.965, 95% confidence interval [CI]: 0.939–0.993) and critical COVID-19 (OR: 0.989, 95% CI: 0.979–0.999) confer causal effects on intelligence. There was suggestive evidence supporting the causal effect of hospitalized COVID-19 on intelligence (OR: 0.988, 95% CI: 0.972–1.003). Hospitalized COVID-19 and intelligence share 10 risk genes within 2 genomic loci, including MAPT and WNT3. Enrichment analysis showed that these genes are functionally connected within distinct subnetworks of 30 phenotypes linked to cognitive decline. The functional pathway revealed that COVID-19-driven pathological changes within the brain and multiple peripheral systems may lead to cognitive impairment.
Conclusions
Our study suggests that COVID-19 may exert a detrimental effect on intelligence. The tau protein and Wnt signaling may mediate the influence of COVID-19 on intelligence.
Introduction
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), known as COVID-19, has an average infection fatality rate of approximately 0.5–2% in most locations worldwide.1 A number of risk or protective factors for COVID-19 outcomes have been reported, including neuropsychiatric diseases.2–12 Meanwhile, a sizable subpopulation of individuals who recovered from acute COVID-19 may suffer from a variety of lingering symptoms, collectively known as long COVID-19.13–17
SARS-CoV-2 can infect cells within the lower respiratory tract (trachea and lungs) and the upper respiratory tract (sinuses, nose and throat),18 in addition to damaging a wide range of human organs and systems, such as the immune system,19 nervous system20 and microvessels.21 Neuropsychiatric manifestations are common among individuals with COVID-19.22 Moreover, it has also been shown that COVID-19 could lead to a loss of 0.2–2% of brain tissue in regions processing the sense of smell and taste, as well as supporting higher functions; these losses are typically more pronounced among older individuals.23 A longitudinal magnetic resonance imaging (MRI) study revealed that individuals who contracted COVID-19 infection, on average, show more pronounced age-associated reductions in brain size and gray matter thickness as well as a larger cognitive decline than controls.24 It was reported that recovered COVID-19 patients have a higher risk of memory decline.25 The neurological sequelae of COVID-19 are associated with increased mental stress and the risks for mental disorders.14,26–29
It is worth mentioning that many of the peripheral pathological changes observed in COVID-19 patients are directly or indirectly linked to cognition.30 Recently, several studies have tested the relationship between COVID-19 and intelligence. In particular, Li et al.8 showed that education may act independently and jointly with intelligence in improving COVID-19 outcomes. Zhu et al.10 suggested a causal genetic linkage between an increased risk of symptomatic COVID-19 and decreased intelligence in children. A significantly increased risk of newly diagnosed Alzheimer’s disease was noted within 360 days after the initial COVID-19 diagnosis in elderly people.31 All of the evidence prompts a detailed evaluation of the relationships between COVID-19 and general intelligence.
Here, we hypothesize that COVID-19 may exert a detrimental effect on intelligence. We sought to evaluate the effects by using the Mendelian randomization (MR) framework applied to genome-wide association study (GWAS) summary results. Using large-scale automated mining of the literature, we also constructed functional pathways connecting COVID-19 and cognitive function.
Methods
GWAS summary datasets
The study utilized publicly available GWAS summary results, with all the participants of European origin. The summary results of the GWAS for intelligence contained 269 867 participants, including those from the UK Biobank (UKB).32 The COVID-19 datasets from the European population were obtained from the COVID-19 HGI GWAS round 7 (release date: 8 April 2022, without the 23andMe cohort).33 To avoid sample overlaps in the MR analysis, we selected the COVID-19 datasets without UKB participants, including hospitalized COVID-19 (40 929 hospitalized cases and 1 924 400 controls), critical COVID-19 (very severe respiratory confirmed 17 472 cases and 725 695 controls) and SARS-CoV-2 infection (143 839 virus-positive cases and 2 357 647 controls). In the identification of overlapping genomic loci between COVID-19 and intelligence, the hospitalized COVID-19 dataset of the European population, including 32 519 hospitalized cases and 2 062 805 controls, was utilized. The latter dataset included the UKB population. The SARS-CoV-2 infection dataset mainly reflects the susceptibility to the virus. The hospitalized COVID-19 and critical COVID-19 datasets characterize the severity of the disease, which we collectively called ‘severe COVID-19’ in this study. Ethical approval had been obtained from each of the original studies.
MR analysis
The analyses were conducted using three complementary methods from TwoSampleMR,34 including weighted median (WM), inverse variance weighted (IVW) and MR-Egger. These models have different assumptions on pleiotropy.35 The IVW model was used as the primary MR method, which assumes an intercept of zero and estimates the causality by a fixed-effect model.36 The WM and MR-Egger models are more sensitive to horizontal pleiotropy but less powerful than IVW. The intercept of the MR-Egger regression was employed to assess the average horizontal pleiotropy.35
For each exposure phenotype, genome-wide significant single-nucleotide polymorphisms (SNPs) (P < 5 × 10−8) were selected as candidate instrumental variables (IVs). Then, these candidate IVs were pruned by a clumping r2 value of 0.001 within a 10-Mb window. The 1000 Genomes Project Phase 3 (EUR) was used as the reference panel.
Shared genomic loci between COVID-19 and intelligence
To identify genetic overlaps between COVID-19 and intelligence, we compared their respective GWAS datasets. For each dataset, we used Functional Mapping and Annotation software to identify LD-independent genomic loci and map SNPs to genes.37 Independent significant SNPs (IndSigSNPs) were identified by their P values (P ≤ 5.0E−08) and their independence from each other (r2 < 0.6). The IndSigSNPs that were in LD with each other within a 500-kb window (r2 < 0.1) were called lead SNPs. For each locus, regional associations were plotted by LocsZoom.38
Protein–protein interaction analysis and pathway construction
The protein-coding genes shared between the sets identified for hospitalized COVID-19 and intelligence were used for the protein–protein interaction (PPI) analysis using STRING v11,39 followed by a subnetwork enrichment analysis (SNEA).40
To explore the molecular network alterations caused by COVID-19 and their influences on intelligence, we constructed functional pathways connecting these two entities using large-scale mining of the literature with Pathway Studio (www.pathwaystudio.com). The following criteria were applied to select the COVID-19-driven cognition/intelligence regulators: (i) the direction of the effect was from COVID-19 to cognition; (ii) exerted changes were in brain regions and other tissues linked to cognition/intelligence; and (iii) the supporting references passed quality control through manual inspection. The relationships that survived the filtering were used to construct the COVID-19-driven signaling pathways that may influence intelligence.
Results
MR analysis
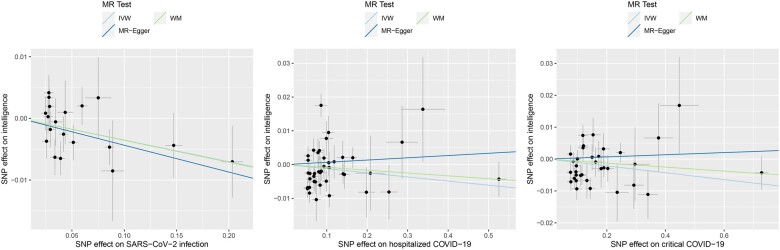
In the MR analysis of the causal effects of the three COVID-19 phenotypes on intelligence, a total of 19, 41 and 34 IVs were extracted for SARS-CoV-2 infection, hospitalized COVID-19 and critical COVID-19, respectively. We found that genetic liabilities to SARS-CoV-2 infection (odds ratio [OR]: 0.965, 95% confidence interval [CI]: 0.939–0.993, P = 0.015) and critical COVID-19 (OR: 0.989, 95% CI: 0.979–0.999, P = 0.036) conferred causal effects on intelligence. There was suggestive evidence supporting the causal effect of hospitalized COVID-19 on intelligence (OR: 0.988, 95% CI: 0.972–1.003, P = 0.127) (Table 1 and Figure 1).
Table 1.
Causal effects of the COVID-19 outcomes on intelligence
| Exposure | Method | b (se) | OR [95%CI] | N_IV | Q_P | I2 | Egger_intercept | P_pleiotropy | P |
|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 infection | IVW | -0.035 (0.014) | 0.965 [0.939-0.993] | 19 | 0.368 | 0.072 | NA | NA | 0.015 |
| SARS-CoV-2 infection | WM | -0.036 (0.019) | 0.965 [0.929-1.002] | 19 | NA | NA | NA | NA | 0.066 |
| SARS-CoV-2 infection | MR Egger | -0.044 (0.026) | 0.957 [0.910-1.007] | 19 | 0.316 | 0.063 | 0.001 | 0.697 | 0.109 |
| Hospitalized COVID-19 | IVW | -0.012 (0.008) | 0.988 [0.972-1.003] | 41 | 2.26E-06 | 0.579 | NA | NA | 0.127 |
| Hospitalized COVID-19 | WM | -0.008 (0.009) | 0.992 [0.975-1.009] | 41 | NA | NA | NA | NA | 0.343 |
| Hospitalized COVID-19 | MR Egger | 0.007 (0.014) | 1.007 [0.979-1.035] | 41 | 8.07E-06 | 0.552 | -0.002 | 0.123 | 0.644 |
| Critical COVID-19 | IVW | -0.011 (0.005) | 0.989 [0.979-0.999] | 34 | 7.36E-03 | 0.411 | NA | NA | 0.036 |
| Critical COVID-19 | WM | -0.006 (0.006) | 0.994 [0.982-1.006] | 34 | NA | NA | NA | NA | 0.306 |
| Critical COVID-19 | MR Egger | 0.003 (0.009) | 1.003 [0.986-1.021] | 34 | 0.022 | 0.342 | -0.003 | 0.061 | 0.702 |
IVW: inverse variance weighted; WM: weighted median; N_IV: number of instrumental variables; Q_P: Cochran’s P value of heterogeneity analysis.
Figure 1.
Causal effects of the COVID-19 outcomes on intelligence. The trait on the x-axis denotes exposure, the trait on the y-axis denotes outcome and each cross point represents an instrumental variant. The lines denote the effect sizes (b) of an exposure on an outcome.
The sensitivity analyses revealed that the directions of causal effect estimates across the methods were largely the same (Table 1 and Figure 1). Notably, tests of MR-Egger regression did not support directional pleiotropy in this MR analysis (MR-Egger intercept < 0.01, P > 0.05). Cochran’s test suggested possible heterogeneity in the hospitalized COVID-19 dataset and the critical COVID-19 dataset.
Shared genomic loci influencing both COVID-19 and intelligence
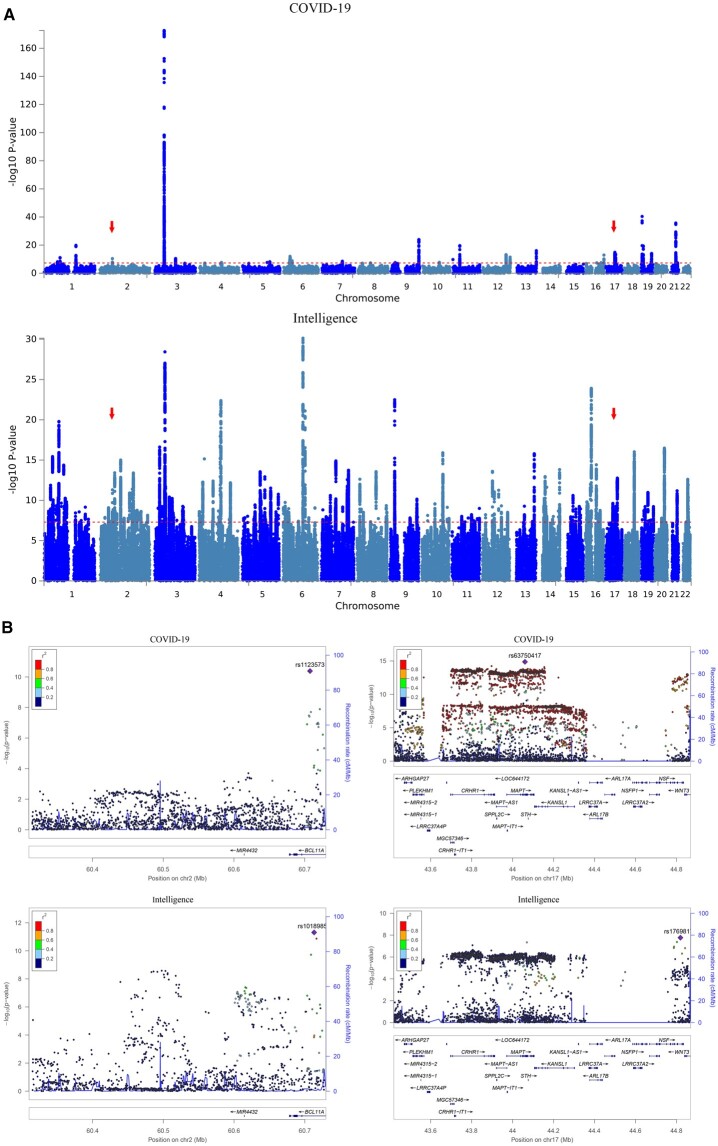
A total of 32 and 203 genomic loci were associated with COVID-19 and intelligence, respectively (Figure 2A and Supplementary Tables S1 and S2). Specifically, we detected two loci overlapping between COVID-19 and intelligence gene sets, including the 2p16.1 locus and the 17q21.31 locus (Table 2 and Figure 2). Ten genes overlapped between COVID-19 and intelligence gene sets included BCL11A, MAPT, KANSL1, ARL17B, NSF, WNT3, LRRC37A, NSFP1, ARL17A and LRRC37A2.
Figure 2.
Overlapping genes between hospitalized COVID-19 and intelligence. (A) Manhattan plot of GWAS results of hospitalized COVID-19 and intelligence. The x-axis is the chromosomal position of SNPs and the y-axis is the significance of the SNPs (−log10P). Each horizontal dashed line denotes the genome-wide significance level of 5E-8. Red arrows indicate the two overlapping genomic loci between COVID-19 hospitalization and intelligence. (B) Two overlapping loci between hospitalized COVID-19 and intelligence. Left is the 2p16.1 locus and right is the 17q21.31 locus in hg19.
Table 2.
Overlapping genomic loci between hospitalized COVID-19 and intelligence
| Trait | SNP | CHR | BP | Start: End | A1/A2 | P | Genes |
|---|---|---|---|---|---|---|---|
| Hospitalized COVID-19 | rs1123573 | 2 | 60707588 | 60705232:60727416 | G/A | 4.13E−11 | CL11A |
| Intelligence | rs10189857 | 2 | 60713235 | 60317457:60726427 | A/G | 4.91E−12 | BCL11A |
| Hospitalized COVID-19 | rs63750417 | 17 | 44060775 | 43422855:44865603 | T/C | 1.37E−15 | ARHGAP27; PLEKHM1; DND1P1; RPS26P8; CRHR1-IT1; CRHR1; MAPT-AS1; SPPL2C; MAPT; STH; KANSL1; KANSL1-AS1; ARL17B; LRRC37A; NSFP1; ARL17A; LRRC37A2; NSF; RPS7P11; WNT3 |
| Intelligence | rs17698176 | 17 | 44819595 | 44040184:44848314 | T/G | 1.70E−08 | MAPT; KANSL1; ARL17B; NSF; WNT3; LRRC37A; NSFP1; ARL17A; LRRC37A2; FAM215B |
CHR, chromosome; BP, base pair.
PPI analysis and SNEA results
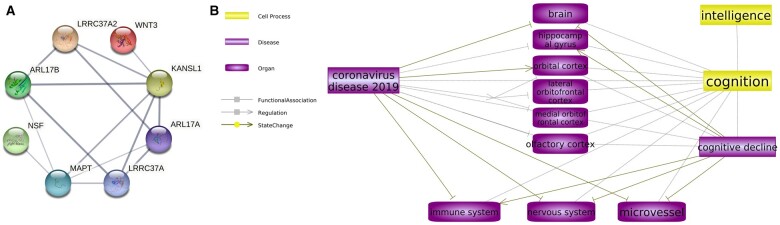
Among the 10 overlapping genes, all except NSFP1 were protein coding. PPI analysis showed that a majority of protein-coding genes formed an interconnected group, with BCL11A remaining an extant entity (Figure 3A).
Figure 3.
PPIs and COVID-19–intelligence connections. (A) PPIs between the shared protein-coding genes. (B) Biological abnormalities induced by COVID-19 at the organ and system levels contribute to the development of intelligence decline. The relation type ‘–|’ represents an inhibition and ‘→’ represents a relationship with no polarity.
The SNEA results showed that 6 out of these 10 genes were enriched within 37 disease-centered subnetworks (P < 0.05, Supplementary Figure S1 and Supplementary Table S3). Interestingly, 30 out of these 37 pathophysiological subnetworks were related to cognitive decline, indicating that these genes may contribute to the impairment of intelligence in a variety of contexts.
Functional pathways connecting COVID-19 and intelligence decline
The analysis of data obtained from structural and functional MRI studies (Supplementary Table S4) allowed the construction of functional pathways that connect COVID-19 with changes in different brain regions. Figure 3B illustrates various noticeable alterations in brain structure resulting from COVID-19, such as decreased gray matter thickness and tissue contrast in the orbitofrontal cortex and parahippocampal gyrus, tissue damage in regions connected to the primary olfactory cortex and a reduction in overall brain size. These brain abnormalities often coincide with the pattern of cognitive decline associated with aging. Some of these changes may be attributed to COVID-19-induced dysfunction of the microvessels, while others could be caused by direct damage to the neuroglial and immune systems. Both of these pathophysiological processes have been linked to impaired cognition. The pathway depicted in Figure 3B provides a potential framework for understanding the possible connection between COVID-19 and cognitive decline at the level of observable traits.
Discussion
In this study, we conducted an MR analysis to explore the potential causality between three forms of COVID-19 and intelligence. Our results showed the causal effects of SARS-CoV-2 infection and critical COVID-19 on intelligence, as well as the possible influence of hospitalized COVID-19 on intelligence, indicating that COVID-19 patients might be at risk of intelligence decline.
Our study shows that the genes located at the 2p16.1 and 17q21.31 regions influence both severe COVID-19 and intelligence. The 2p16.1 locus harbors the single protein-coding gene BCL11A, which plays a vital role in B and T lymphopoiesis41 and defines dendritic cell fate.42 Genetic variation within BCL11A determines residual levels of fetal hemoglobin,43 which may be protective against the symptoms of coronavirus infection.44 In undifferentiated epithelial cells, the product of this gene prevents senescence by accelerating the repair of oxidized DNA.45 During postnatal corticogenesis, BCL11A prevents the death of projection neurons.46 Haploinsufficiency of BCL11A underlines intellectual disability syndrome (IDS) associated with the hereditary persistence of fetal hemoglobin (HbF), also known as Dias-Logan syndrome47 and a chromosome 2p16.1p15 microdeletion syndrome.48 Peculiarly, BCL11A was previously reported as a genome-wide risk gene for COVID-1949 and as a pleiotropic gene for attention-deficit/hyperactivity disorder, autism spectrum disorder and intelligence.50,51 Notably, these three neurodevelopmental features are underpinned by shared genetics.51,52
The 17q21.31 locus contains eight overlapping protein-coding genes, including MAPT, KANSL1, ARL17B, NSF, WNT3, LRRC37A, ARL17A and LRRC37A2. PPI analysis showed that the respective proteins form an interconnected network (Figure 3A), which is functionally linked to a set of diseases associated with cognitive decline (SNEA results). The genes located within the contiguous region were repeatedly identified as contributors to COVID-19 phenotypes. For example, the chromatin modifier gene KANSL1, which is also a risk gene for atrial fibrillation and flutter as well as for pulmonary fibrosis, was identified in studies of genetic associations with severe COVID-19.53,54 The same gene serves as a pathogenic culprit for Koolen De Vries syndrome characterized by intellectual disability accompanied by characteristic facial features and hypotonia,55 a longevity gene56 and a contributor to Alzheimer’s disease phenotypes.57
The MAPT gene encodes the microtubule-associated protein tau. This gene was identified by our previous multi-omics integrative analyses as a contributor to COVID-19.58 A recent study reported that increased levels of tau in the blood, which is possibly due to its excretion by exosomes,55 are associated with fatal outcomes of COVID-19.59 Notably, by adhering to the SARS-CoV-2 S1 receptor-binding domain, tau protein precipitates the aggregation of amyloid-like proteins and promotes neurodegeneration.60MAPT is central to the pathogenesis of multiple neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease and some neuropsychiatric conditions.32,51,61–64 Although it is tempting to establish direct connections between COVID-19 and neurodegeneration through MAPT, it is important to consider that these links could also be indirect. One possible indirect association is the previously documented involvement of MAPT in the phenotypes of aging-promoting interstitial lung disease65 and overall lung function.66 Therefore, further exploration is warranted to fully understand the relationship between COVID-19 and neurodegeneration, taking into account potential indirect pathways involving MAPT.
The study of WNT3 involvement in the intersection of COVID-19 and cognitive phenotypes closely follows that of MAPT. While WNT3 is involved in intelligence and multiple psychiatric conditions,51,67,68 its roles in COVID-19 phenotypes are more elusive and likely defined by indirect relationships with blood–brain barrier permeability.69–71
Colocated genes are rarely separated by recombination and are commonly coregulated. When analysis of coregulation was performed for MAPT-associated gene units, the levels of transcripts produced by LRRC37A2, KANSL1, ARL17B, LRRC37A and ARL17A were found to be affected by the MAPT haplotype in a dose-dependent manner.72 Although each of these genes may have a distinct impact on COVID-19, neurodegenerative phenotypes or both, the existence of embedded coregulation adds complexity to the study of this region. Therefore, it is crucial to conduct functional investigations both in vitro and in model animals to gain a deeper understanding of the interplay between these genes and their roles in the context of COVID-19 and neurodegeneration.
The composed map of the functional pathways (Figure 3B) revealed that COVID-19 influences the structure and function of multiple brain regions, including the hippocampal gyrus, orbitofrontal cortex and olfactory cortex.24,73,74 In both survivors of severe COVID-19 and elderly individuals, the loss of brain tissue may lead to cognitive decline.23,75 The correlations between changes in brain structure and age-related cognitive decline have been extensively documented in the latter group. For instance, among elderly individuals, a notable reduction in the mean volume of the right parahippocampal gyrus corresponds to their cognitive decline.76 The changes in the frontal cortex, especially the orbitofrontal cortex, cingulate cortex and amygdala, are associated with emotional and cognitive impairments.77 The subjective cognitive decline in patients is also connected to significantly reduced activation in the bilateral primary olfactory cortex.78 Moreover, COVID-19 may also lead to dysfunctions in the immune system, the peripheral nervous system and the lining of microvessels,19,21,79 a set of pathological features commonly associated with cognitive decline.80–82 Taken together, the functional pathways presented in Figure 3 may provide some insights into the causal effect of COVID-19 on intellectual impairment.
A limitation of this study is that the sample datasets were derived solely from European populations. To validate the findings, it is necessary to incorporate additional datasets from various population regions.
Conclusions
In summary, our study suggests that COVID-19 may contribute to cognitive impairment. Functional variation within the tau locus and the genes of the Wnt signaling pathway may be relevant to COVID-19 and especially to its neurological sequelae.
Supplementary Material
Acknowledgments
The authors thank all investigators and participants from the COVID-19 Host Genetics Initiative and other groups for sharing these data.
Contributor Information
Hongbao Cao, School of Systems Biology, George Mason University, Manassas, VA 20110, USA.
Ancha Baranova, School of Systems Biology, George Mason University, Manassas, VA 20110, USA; Research Centre for Medical Genetics, Moscow 115478, Russia.
Yuqing Song, Institute of Mental Health, Peking University Sixth Hospital; NHC Key Laboratory of Mental Health (Peking University), National Clinical Research Center for Mental Disorders (Peking University Sixth Hospital), Beijing 100191, China.
Jian-Huan Chen, Laboratory of Genomic and Precision Medicine, Wuxi School of Medicine, Jiangnan University, Wuxi 214122, China.
Fuquan Zhang, Institute of Neuropsychiatry, The Affiliated Brain Hospital of Nanjing Medical University, Nanjing 210029, China; Department of Psychiatry, The Affiliated Brain Hospital of Nanjing Medical University, Nanjing 210029, China.
Supplementary material
Supplementary material is available at QJMED online.
Funding
None to declare.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Hongbao Cao (Formal analysis [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Ancha Baranova (Writing—original draft [equal], Writing—review & editing [equal]), Yuqing Song (Writing—review & editing [supporting]), Jian-Huan Chen (Writing—review & editing [equal]) and Fuquan Zhang (Conceptualization [lead], Data curation [lead], Formal analysis [lead], Resources [lead], Supervision [lead], Validation [lead], Visualization [lead], Writing—original draft [equal], Writing—review & editing [equal])
References
- 1. Levin AT, Hanage WP, Owusu-Boaitey N, Cochran KB, Walsh SP, Meyerowitz-Katz G.. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol 2020; 35:1123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baranova A, Cao H, Teng S, Zhang F.. A phenome-wide investigation of risk factors for severe COVID-19. J Med Virol 2023; 95:e28264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rao S, Baranova A, Cao H, Chen J, Zhang X, Zhang F.. Genetic mechanisms of COVID-19 and its association with smoking and alcohol consumption. Brief Bioinform 2021; 22:bbab284. [DOI] [PubMed] [Google Scholar]
- 4. Cao H, Baranova A, Wei X, Wang C, Zhang F.. Bidirectional causal associations between type 2 diabetes and COVID-19. J Med Virol 2023; 95:e28100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baranova A, Song Y, Cao H, Zhang F.. Causal associations between basal metabolic rate and COVID-19. Diabetes 2023; 72:149–54. [DOI] [PubMed] [Google Scholar]
- 6. Baranova A, Xu Y, Cao H, Zhang F.. Associations between pulse rate and COVID-19. J Med Virol 2023; 95:e28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baranova A, Cao H, Chen J, Zhang F.. Causal association and shared genetics between asthma and COVID-19. Front Immunol 2022; 13:705379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H-Y, Lam K-K, Wong C-K, Chu K-P, Cheung C-L.. Education Attainment, Intelligence and COVID-19: A Mendelian Randomization Study. Journal of Clinical Medicine 2021; 10:4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baranova A, Zhao Y, Cao H, Zhang F.. Causal associations between major depressive disorder and COVID-19. Gen Psychiatr 2023; 36:e101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu G, Zhou S, Xu Y, Gao R, Li H, Su W, et al. Mendelian randomization study on the causal effects of COVID-19 on childhood intelligence. J Med Virol 2022; 94:3233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang F, Baranova A.. Smoking quantitatively increases risk for COVID-19. Eur Respir J 2022; 60:2101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baranova A, Cao H, Zhang F.. Causal associations and shared genetics between hypertension and COVID-19. J Med Virol 2023; 95:e28698. [DOI] [PubMed] [Google Scholar]
- 13. Paterson C, Davis D, Roche M, Bissett B, Roberts C, Turner M, et al. What are the long-term holistic health consequences of COVID-19 among survivors? An umbrella systematic review. J Med Virol 2022; 94:5653–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiabane E, Pain D, Aiello EN, Radici A, Manera MR, Grossi F, et al. Psychiatric symptoms subsequent to COVID-19 and their association with clinical features: a retrospective investigation. Psychiatry Res 2022; 316:114757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baranova A, Cao H, Teng S, Su KP, Zhang F.. Shared genetics and causal associations between COVID-19 and multiple sclerosis. J Med Virol 2023; 95:e28431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murata F, Maeda M, Ishiguro C, Fukuda H.. Acute and delayed psychiatric sequelae among patients hospitalised with COVID-19: a cohort study using LIFE study data. Gen Psychiatr 2022; 35:e100802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai X, Cao X, Jiang Q, Wu B, Lou T, Shao Y, et al. Neurological complications of COVID-19. QJM 2023; 116:161–80. [DOI] [PubMed] [Google Scholar]
- 18. Harrison AG, Lin T, Wang P.. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol 2020; 41:1100–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chao CM, Lai CC, Yu WL.. COVID-19 associated mucormycosis—an emerging threat. J Microbiol Immunol Infect 2022; 55:183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu JM, Tan BH, Wu S, Gui Y, Suo JL, Li YC.. Evidence of central nervous system infection and neuroinvasive routes, as well as neurological involvement, in the lethality of SARS-CoV-2 infection. J Med Virol 2021; 93:1304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haberecker M, Schwarz EI, Steiger P, Frontzek K, Scholkmann F, Zeng X, et al. Autopsy-based pulmonary and vascular pathology: pulmonary endotheliitis and multi-organ involvement in COVID-19 associated deaths. Respiration 2022; 101:155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S.. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol 2020; 77:1018–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee MH, Perl DP, Nair G, Li W, Maric D, Murray H, et al. Microvascular injury in the brains of patients with Covid-19. N Engl J Med 2021; 384:481–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Douaud G, Lee S, Alfaro-Almagro F, Arthofer C, Wang C, McCarthy P, et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature 2022; 604:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Merza MA, Almufty HB, Younis HA, Rasool SO, Mohammed SA.. Memory impairment among recovered COVID-19 patients: the prevalence and risk factors, a retrospective cohort study. J Med Virol 2023; 95:e28459. [DOI] [PubMed] [Google Scholar]
- 26. Fioravanti G, Bocci Benucci S, Prostamo A, Banchi V, Casale S.. Effects of the COVID-19 pandemic on psychological health in a sample of Italian adults: a three-wave longitudinal study. Psychiatry Res 2022; 315:114705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baranova A, Cao H, Zhang F.. Causal effect of COVID-19 on Alzheimer’s disease: a Mendelian randomization study. J Med Virol 2023; 95:e28107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baranova A, Cao H, Zhang F.. Severe COVID-19 increases the risk of schizophrenia. Psychiatry Res 2022; 317:114809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X, Dai Z, Fu J, Si M, Jing S, Wu Y, et al. Suicidal ideation and associated risk factors among COVID-19 patients who recovered from the first wave of the pandemic in Wuhan, China. QJM 2023. 10.1093/qjmed/hcad083. [DOI] [PubMed] [Google Scholar]
- 30. Dissel S, Seugnet L, Thimgan MS, Silverman N, Angadi V, Thacher PV, et al. Differential activation of immune factors in neurons and glia contribute to individual differences in resilience/vulnerability to sleep disruption. Brain Behav Immun 2015; 47:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Davis PB, Volkow ND, Berger NA, Kaelber DC, Xu R.. Association of COVID-19 with new-onset Alzheimer’s disease. J Alzheimers Dis 2022; 89:411–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 2018; 50:912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Initiative C-HG. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet 2020; 28:715–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015; 44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burgess S, Butterworth A, Thompson SG.. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013; 37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe K, Taskesen E, van Bochoven A, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47:D607–D613. D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langlois VS, Martyniuk CJ.. Genome wide analysis of Silurana (Xenopus) tropicalis development reveals dynamic expression using network enrichment analysis. Mech Dev 2013; 130:304–22. [DOI] [PubMed] [Google Scholar]
- 41. Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med 2012; 209:2467–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ippolito GC, Dekker JD, Wang YH, Lee BK, Shaffer AL 3rd, Lin J, et al. Dendritic cell fate is determined by BCL11A. Proc Natl Acad Sci USA 2014; 111:E998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tommassen J, van der Ley P, van Zeijl M, Agterberg M.. Localization of functional domains in E. coli K-12 outer membrane porins. EMBO J 1985; 4:1583–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rawat M, Chandrasekharan P, Hicar MD, Lakshminrusimha S.. COVID-19 in newborns and infants-low risk of severe disease: silver lining or dark cloud? Am J Perinatol 2020; 37:845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vickridge E, Faraco CCF, Tehrani PS, Ramdzan ZM, Rahimian H, Leduy L, et al. The DNA repair function of BCL11A suppresses senescence and promotes continued proliferation of triple-negative breast cancer cells. NAR Cancer 2022; 4:zcac028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wiegreffe C, Wahl T, Joos NS, Bonnefont J, Liu P, Britsch S.. Developmental cell death of cortical projection neurons is controlled by a Bcl11a/Bcl6-dependent pathway. EMBO Rep 2022; 23:e54104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Korenke GC, Schulte B, Biskup S, Neidhardt J, Owczarek-Lipska M.. A novel de novo frameshift mutation in the BCL11A gene in a patient with intellectual disability syndrome and epilepsy. Mol Syndromol 2020; 11:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peter B, Matsushita M, Oda K, Raskind W.. De novo microdeletion of BCL11A is associated with severe speech sound disorder. Am J Med Genet A 2014; 164A:2091–6. [DOI] [PubMed] [Google Scholar]
- 49. Kousathanas A, Pairo-Castineira E, Rawlik K, Stuckey A, Odhams CA, Walker S, et al. ; GenOMICC investigators. Whole genome sequencing reveals host factors underlying critical Covid-19. Nature 2022; 607:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai T, Chen X, Li J, Xiang B, Yang L, Liu Y, et al. Identification of novel mutations in the HbF repressor gene BCL11A in patients with autism and intelligence disabilities. Am J Hematol 2017; 92:E653–6. [DOI] [PubMed] [Google Scholar]
- 51. Rao S, Baranova A, Yao Y, Wang J, Zhang F.. Genetic relationships between attention-deficit/hyperactivity disorder, autism spectrum disorder, and intelligence. Neuropsychobiology 2022; 81:484–96. [DOI] [PubMed] [Google Scholar]
- 52. Baranova A, Wang J, Cao H, Chen JH, Chen J, Chen M, et al. Shared genetics between autism spectrum disorder and attention-deficit/hyperactivity disorder and their association with extraversion. Psychiatry Res 2022; 314:114679. [DOI] [PubMed] [Google Scholar]
- 53. Regan JA, Abdulrahim JW, Bihlmeyer NA, Haynes C, Kwee LC, Patel MR, et al. Phenome-wide association study of severe COVID-19 genetic risk variants. J Am Heart Assoc 2022; 11:e024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Degenhardt F, Ellinghaus D, Juzenas S, Lerga-Jaso J, Wendorff M, Maya-Miles D, et al. Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum Mol Genet 2022; 31:3945–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bigoni S, Marangi G, Frangella S, Panfili A, Ognibene D, Squeo GM, et al. Clinical genetics can solve the pitfalls of genome-wide investigations: lesson from mismapping a loss-of-function variant in KANSL1. Genes (Basel) 2020; 11:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deelen J, Evans DS, Arking DE, Tesi N, Nygaard M, Liu X, et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun 2019; 10:3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gouveia C, Gibbons E, Dehghani N, Eapen J, Guerreiro R, Bras J.. Genome-wide association of polygenic risk extremes for Alzheimer’s disease in the UK Biobank. Sci Rep 2022; 12:8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baranova A, Cao H, Zhang F.. Unraveling risk genes of COVID-19 by multi-omics integrative analyses. Front Med (Lausanne) 2021; 8:738687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De Lorenzo R, Lore NI, Finardi A, Mandelli A, Cirillo DM, Tresoldi C, et al. Blood neurofilament light chain and total tau levels at admission predict death in COVID-19 patients. J Neurol 2021; 268:4436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Idrees D, Kumar V.. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: potential clues to neurodegeneration. Biochem Biophys Res Commun 2021; 554:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. ; 23andMe Research Team. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, et al. ; 23andMe Research Team. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 2017; 49:1511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Witoelar A, Jansen IE, Wang Y, Desikan RS, Gibbs JR, Blauwendraat C, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC), North American Brain Expression Consortium (NABEC), and United Kingdom Brain Expression Consortium (UKBEC) Investigators. Genome-wide pleiotropy between Parkinson disease and autoimmune diseases. JAMA Neurol 2017; 74:780–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang CC, Zhu JX, Wan Y, Tan L, Wang HF, Yu JT, et al. Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget 2017; 8:44994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet 2013; 45:613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet 2019; 104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun 2018; 9:2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang F, Cao H, Baranova A.. Genetic variation mediating neuroticism’s influence on cardiovascular diseases. J Psychopathol Clin Sci 2022; 131:278–86. [DOI] [PubMed] [Google Scholar]
- 69. Pathak GA, Singh K, Miller-Fleming TW, Wendt FR, Ehsan N, Hou K, et al. Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization. Nat Commun 2021; 12:4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Niladhuri SB, Clare GO, Robinson KF, Class J, Almousawi AA, Trevino TN, et al. Age exacerbates SARS-CoV-2-induced blood–brain barrier leakage and neuropsychiatric dysfunction. bioRxiv2022: 2022.06.02.494552.
- 71. Laksitorini MD, Yathindranath V, Xiong W, Hombach-Klonisch S, Miller DW.. Modulation of Wnt/beta-catenin signaling promotes blood-brain barrier phenotype in cultured brain endothelial cells. Sci Rep 2019; 9:19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Koks S, Pfaff AL, Bubb VJ, Quinn JP.. Transcript variants of genes involved in neurodegeneration are differentially regulated by the APOE and MAPT haplotypes. Genes (Basel) 2021; 12:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jeong HG, Ham BJ, Yeo HB, Jung IK, Joe SH.. Gray matter abnormalities in patients with premenstrual dysphoric disorder: an optimized voxel-based morphometry. J Affect Disord 2012; 140:260–7. [DOI] [PubMed] [Google Scholar]
- 74. Bremner JD, Vythilingam M, Vermetten E, Nazeer A, Adil J, Khan S, et al. Reduced volume of orbitofrontal cortex in major depression. Biol Psychiatry 2002; 51:273–9. [DOI] [PubMed] [Google Scholar]
- 75. Heald CJ, Sarma A, Sachs JR, Zapadka ME, Jewett T, Bunch PM.. Practical genetics for the neuroradiologist: adding value in neurogenetic disease. Acad Radiol 2022; 29(Suppl 3):S1–S27. [DOI] [PubMed] [Google Scholar]
- 76. Pantel J, Kratz B, Essig M, Schroder J.. Parahippocampal volume deficits in subjects with aging-associated cognitive decline. Am J Psychiatry 2003; 160:379–82. [DOI] [PubMed] [Google Scholar]
- 77. Sommer M, Hajak G, Dohnel K, Schwerdtner J, Meinhardt J, Muller JL.. Integration of emotion and cognition in patients with psychopathy. Prog Brain Res 2006; 156:457–66. [DOI] [PubMed] [Google Scholar]
- 78. Krstic D, Pfister S, Notter T, Knuesel I.. Decisive role of Reelin signaling during early stages of Alzheimer’s disease. Neuroscience 2013; 246:108–16. [DOI] [PubMed] [Google Scholar]
- 79. Akhmedzhanova LT, Voskresenskaia ON, Isaikin AI, Ermilova EV, Nasonova TI, Chernousov PA, et al. Acute disseminated encephalomyelitis and myelitis associated with new coronavirus infection COVID-19. Case report. Terapevt Arkh 2021; 93:1375–80. [DOI] [PubMed] [Google Scholar]
- 80. Saveanu RV, Mayes T.. Diagnosing depression in congestive heart failure. Heart Fail Clin 2011; 7:75–9. [DOI] [PubMed] [Google Scholar]
- 81. Zhou Y, Yu C, Zheng H, Liu Y, Song M, Qin W, et al. Increased neural resources recruitment in the intrinsic organization in major depression. J Affect Disord 2010; 121:220–30. [DOI] [PubMed] [Google Scholar]
- 82. Greaney JL, Saunders EFH, Santhanam L, Alexander LM.. Oxidative stress contributes to microvascular endothelial dysfunction in men and women with major depressive disorder. Circ Res 2019; 124:564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.