SUMMARY
We conducted high coverage (>30X) whole-genome sequencing of 180 individuals from 12 indigenous African populations. We identify millions of unreported variants, many predicted to be functionally important. We observe that the ancestors of southern African San and central African rainforest hunter-gatherers (RHG) diverged from other populations >200 kya and maintained a large effective population size. We observe evidence for ancient population structure in Africa and for multiple introgression events from “ghost” populations with highly diverged genetic lineages. Although currently geographically isolated, we observe evidence for gene flow between eastern and southern Khoesan-speaking hunter-gatherer populations lasting until ~12 kya. We identify signatures of local adaptation for traits related to skin color, immune response, height, and metabolic processes. We identify a positively selected variant in the lightly pigmented San that influences pigmentation in vitro by regulating the enhancer activity and gene expression of PDPK1.
Graphical Abstract
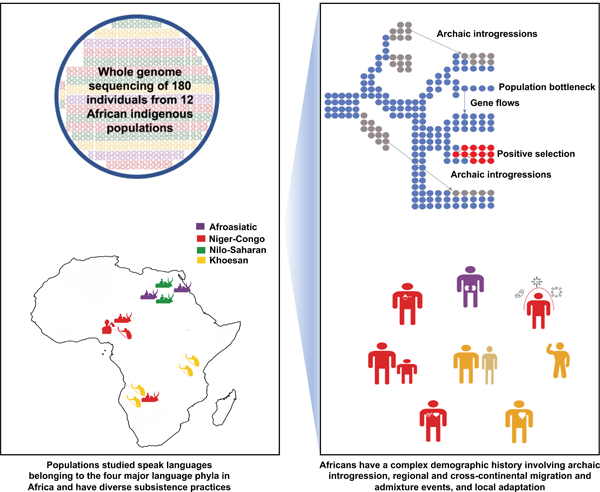
In Brief:
Fan et al. studied the complex demographic history and the genetic basis of local adaptation for traits related to skin color, immune response, and metabolic processes across African populations using high-coverage whole-genome sequencing data of 180 individuals from 12 indigenous African populations.
INTRODUCTION
Africa is the continent where anatomically modern humans originated within the past 300 ky and the source of migration of anatomically modern humans out of Africa within the past 80 ky.1 Africa is also a continent of tremendous cultural, linguistic, phenotypic, and genetic diversity.2,3 More than 2,000 ethnolinguistic groups have been identified in Africa, representing around one-third of the world’s languages.3,4 These languages are classified into four major phyla: Afroasiatic, Nilo-Saharan, Niger-Congo, and Khoesan.5 The Afroasiatic phylum, consisting of ~400 languages, is mainly spoken by agro-pastoralist and agriculturalist populations in northern and eastern Africa. The Nilo-Saharan phylum, comprised of ~206 languages forming ~12 subfamilies, is predominantly spoken by pastoralists in central and eastern Africa. Genetic, linguistic, and archeological data suggest a possible common ancestry of Nilo-Saharan-speaking populations originating near the Ethiopian and Sudanese border within the past 10,500 years.3,6 The Niger-Congo phylum, consisting of ~1,500 sub-languages, is the largest language phylum in Africa.4 The largest subfamily of languages are the Bantu languages, which originated near the border of Cameroon and Nigeria. Bantu-speaking populations used iron tool technology and slash-and-burn agriculture facilitating larger population sizes and migration to eastern and southern Africa beginning ~5 kya (a.k.a, the “Bantu expansion”).7 The Khoesan languages, which are characterized by click consonants, are mainly spoken by the San populations in southern Africa and the Hadza and Sandawe in Tanzania, all of whom currently, or until recently, practice hunting and gathering.5,8 Yet, the San, Hadza, and Sandawe languages are highly divergent and their classification as a single language family remains contentious.9,10 Linguistic studies suggest that the Sandawe language is more similar to that of the southern African San than that of the Hadza.11 Additionally, African populations live in various environments including desert, tropical rainforest, savanna, swamps, and high-altitude mountains, and have adapted to diverse selection pressures such as climate, diet, and pathogen exposure, driving local adaptation.12–14
Despite the essential role that Africa has played in the origin and evolution of anatomically modern humans, Africans are still underrepresented in human genomic studies.15,16 People of African ancestry in the United States have a disproportionately higher burden of common diseases, such as hypertension, diabetes, and kidney failure, likely due to both environmental (including sociodemographic, economic, and health access) and genetic factors.16,17 Therefore, a lack of representation of African populations in genetic research not only hinders our understanding of human evolutionary history, but also limits the development of equitable precision medicine.16
While prior WGS studies in Africa focused on targeted geographic regions18,19 or used 1 - 6 individuals from particular ethnic groups20–25, in this study, we generated high-coverage whole-genome sequencing of 180 individuals from 12 indigenous African populations (15 individuals per population): the Amhara, Dizi, Chabu, and Mursi from Ethiopia, the Hadza and Sandawe from Tanzania, the RHG (Baka and Bagyeli merged into one population), Fulani, and Tikari from Cameroon, and the Herero, Ju|’hoansi and !Xoo (the latter two collectively referred to as “San”) from Botswana (Figure 1A). These populations speak languages encompassing all four African language phyla. The Hadza and San, still practice traditional hunter-gatherer subsistence styles (though the San now receive food subsidies), whereas the Sandawe have adopted agriculture and herding within the past few hundred years.3 The RHG who, based on their short stature, have been referred to as “Pygmies”, have lost their traditional language and now speak Bantu languages.3 Such language replacement also happened to the Fulani, who are traditionally nomadic pastoralists living across a broad range of Africa encompassing the Sudan, Central, and Western Africa.3 The Fulani now speak a Niger-Congo language most similar to languages spoken on the west coast of Africa.26 The Chabu have a census population size of only 1,000-2,000 individuals,27 live in a mountainous region in southwestern Ethiopia, and practice a foraging lifestyle. Their language is considered a ‘language isolate’ and one of the ‘severely endangered languages’ of the world. Linguistic studies suggest that the proto-Chabu language may have originated as an early branch of the Nilo-Saharan phylum.4,28
Figure 1. Geographic locations of the samples and summary of the variants identified in this study.
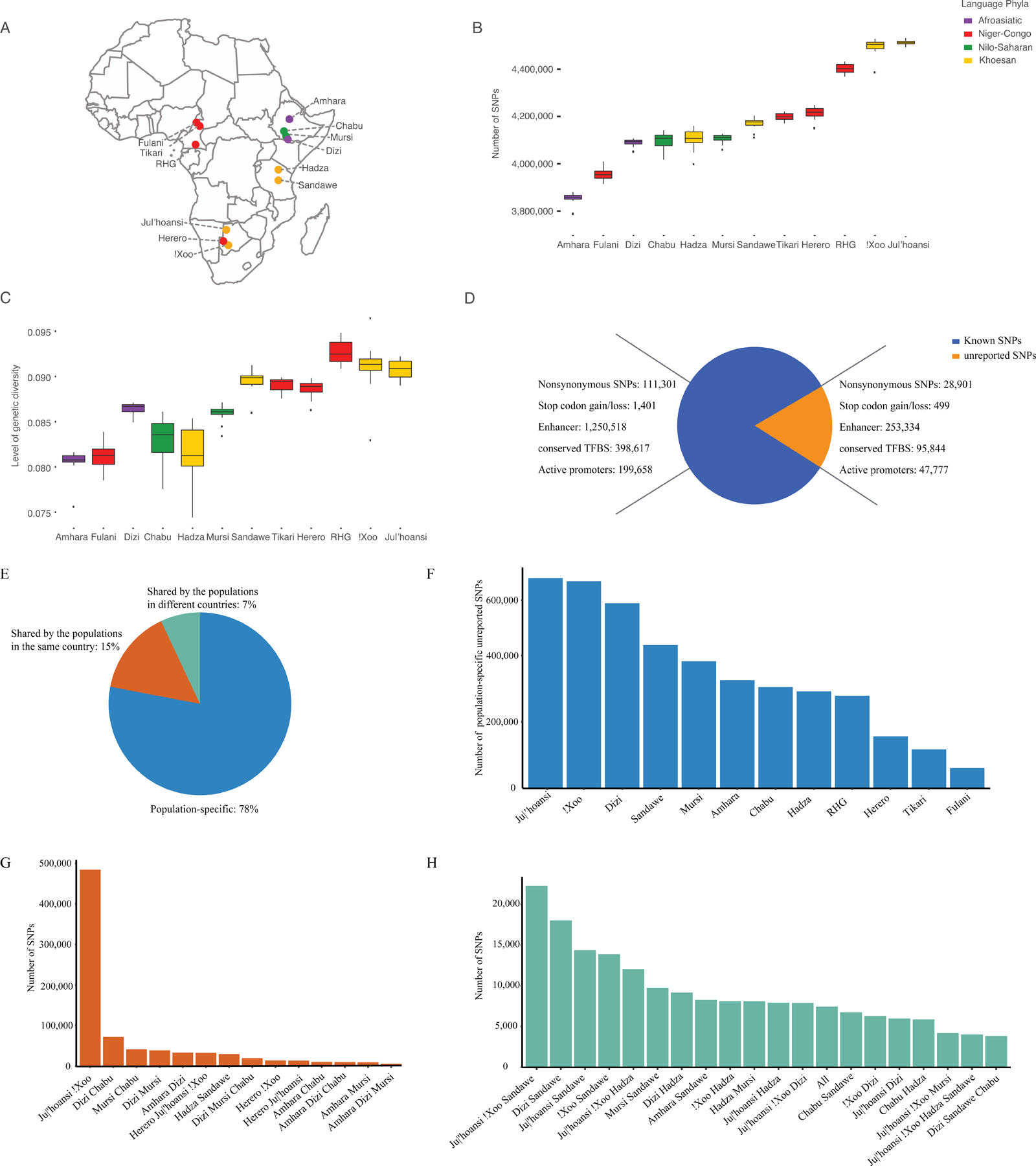
A: Points are populations, with color indicating language classification.
B: Number of SNPs across populations compared to the human reference genome (hg19).
C: Genetic diversity in terms of heterozygosity across populations.
D: Number of unreported and known SNPs and their potentially functional impacts. Here, unreported SNPs were identified by comparison to dbSNP100 (version 155) and gnomAD101 (version 2.1) databases. Annotations of regulatory elements were generated by the Encode project102 based on predicted chromatin state of lymphoblastoid cells from the “GM12878” sample as well as conserved transcription factor binding sites (TFBS). These annotations were downloaded from the UCSC genome browser website.103
E: Pattern of shared unreported SNPs in different populations.
F: Number of population-specific unreported SNPs in each population.
G: Number of unreported SNPs identified in populations in the same country.
H: Number of unreported SNPs identified in populations in different countries. “All” corresponds to SNPs that were shared by all 12 populations. RHG: rainforest hunter-gatherers.
Across these populations, we characterized millions of genomic variants, many of which were predicted to be functional and of potential biomedical relevance. We used multiple approaches to reconstruct the phylogenetic relationship, admixture events, and effective population sizes of these populations. Moreover, we identified population-specific signals of positive selection that may have contributed to local adaptation, and we identified the functional impact of some of these variants on adaptive phenotypes.
Results
We generated high coverage (> 30X) WGS data from 15 individuals per population from 12 African populations (180 individuals total), representing the most diverse genetic ancestries in sub-Saharan Africa based on prior admixture analyses (Figure 1A).3,29 After quality control (STAR Methods), we identified a total of 35,201,568 variants: 32,438,935 single nucleotide polymorphisms (SNPs) and 2,762,633 small insertions and deletions. Further analyses were restricted to 32,044,896 biallelic SNPs. The average number of SNPs varies greatly among populations (Figure 1B). The San and RHG individuals have greatest number of SNPs (Figure 1B) and the highest levels of genetic diversity (Figure 1C), whereas individuals from populations that experienced strong non-African admixture (e.g., Amhara from Ethiopia) or small census sizes (e.g., Hadza or Chabu), carry the fewest SNPs (Figure 1B) and have the lowest genetic diversity (Figure 1C).
We identified 5,344,342 SNPs that are not reported in dbSNP version 155 nor gnomAD version 2.1 (Figure 1D). Around 78% of the unreported SNPs are population-specific, 15% are shared by populations in the same country, and 7% are shared by populations residing in different countries (Figure 1E). Variants at unreported SNPs are significantly rarer than those at previously reported SNPs (Wilcoxon rank sum test, p < 0.001). The Dizi, Ju|’hoansi and !Xoo have the greatest numbers of population-specific unreported variants (Figure 1F), and the Ju|’hoansi and !Xoo shared the greatest number of unreported SNPs among populations in the same country (Figure 1G). Of the unreported variants shared between populations in different countries (Figure 1H), most are shared between the hunter gatherer populations in Southern (Ju|’hoansi and !Xoo) and Eastern (Hadza and Sandawe) Africa and between the Hadza and Sandawe and Ethiopian populations (Amhara, Dizi, Mursi, and Chabu).
Among the unreported SNPs, we identified 28,901 and 499 causing amino acid changes or stop codon gain/loss, respectively, as well as 95,844, 253,334, and 47,777 located in transcription factor binding site regions, enhancers, and active promoter regions, respectively (Figure 1D) based on functional annotations using ANNOVAR. Further, 154 SNPs in our dataset were reported as “Pathogenic” or “Likely Pathogenic” in the ClinVar database (STAR Methods). Of these, 44 are at frequencies higher than 0.05 in at least one of the populations from this study but are either absent or at frequencies lower than 0.01 in non-African populations in gnomAD (Table S1). For example, rs74853476-C is a splice donor variant at dopamine beta-hydroxylase (DBH) associated with orthostatic hypotension 1 in non-African samples.30 While rs74853476-C is rare in all super-populations in gnomAD, it reaches 13% in the Fulani (Figure S1A). Another example consists of three missense mutations (Figures S1B–D), rs139426141-G, rs140482516-T, and rs34097903-A, in Peptidyl Arginine Deiminase 3 (PADI3) reported to associate with central centrifugal cicatricial alopecia in patients of African ancestry.31 Each of these variants is at a high frequency in at least one of the studied populations (Figures S1B–D) but is rare in the non-African super-populations in gnomAD. Thus, a number of variants that are labeled by ClinVar as putatively pathogenic are seen at high frequencies in one or more of our populations and, in fact, may be benign. These observations emphasize a strong need to include ethnically diverse populations in human genetic studies, especially because rarity is used as a criterion for determining a variant’s pathogenicity in clinical studies.16
Phylogenetic relationship of African populations in a worldwide context
After merging our African WGS data with WGS data for Papuans from the Simons Genome Diversity Project (SGDP) and the Northern and Western Europeans from Utah, Tuscans, and Han Chinese in Beijing from the 1000 Genomes Project (1KGP) (STAR Methods), we constructed a neighbor-joining phylogenic tree using MEGA, which neglects migration and recombination. Therefore, admixed populations may cluster near each other. We observed that the Ju|’hoansi and !Xoo have the most basal lineages of all modern humans, followed by the RHG (Figure 2). The remaining populations largely clustered by their current geographical locations with a few exceptions. For example, the Fulani from Cameroon clustered with Afroasiatic-speaking populations in East Africa, suggesting common ancestry with those populations and a language replacement during their migration across the Sahel.3
Figure 2. A neighbor-joining phylogeny of African and representative global individuals based on whole genome sequence data.
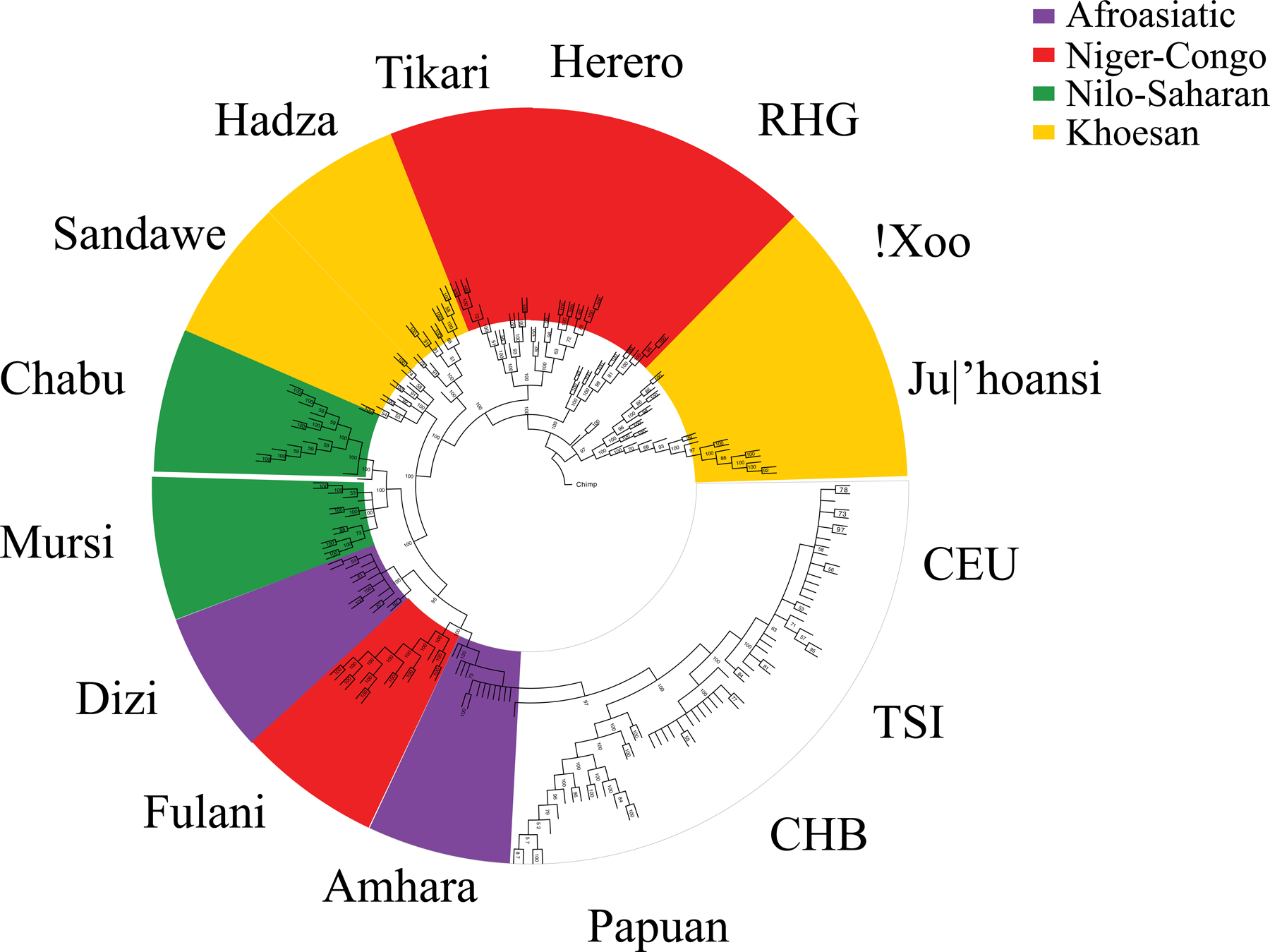
Numbers at each node indicate bootstrap values based on 100 bootstraps. CEU: Northern Europeans from Utah. TSI: Toscani in Italia. CHB: Han Chinese in Beijing, China are from the 1000 Genomes Project.104 Papuan samples were sequenced by the SGDP.105
Further, the Chabu clustered with the Nilo-Saharan-speaking Mursi, consistent with the linguistic classification of the Chabu language.28 The Hadza and Sandawe clustered near each other, though they did not form a monophyletic group, possibly due to strong admixture between the Sandawe and other East African populations (Figures 3E and S2). Consistent with previous studies3,21,32, the Fulani and two Ethiopian Afroasiatic-speaking populations, the Amhara and Dizi, are genetically closest to non-African populations. Yet, a more careful analysis with D-statistics suggests that the out-of-Africa source population was ancestral to all non-RHG, non-San populations in our dataset (Note S1). This suggests that the clustering of non-African populations with the Fulani, Amhara, and Dizi in Figure 2 is due to gene flow from non-Africans into these populations (directly or indirectly), which we confirmed using D-statistics (Note S1).
Figure 3. Population structural analyses based on principal component analysis (PCA) and ADMIXTURE.
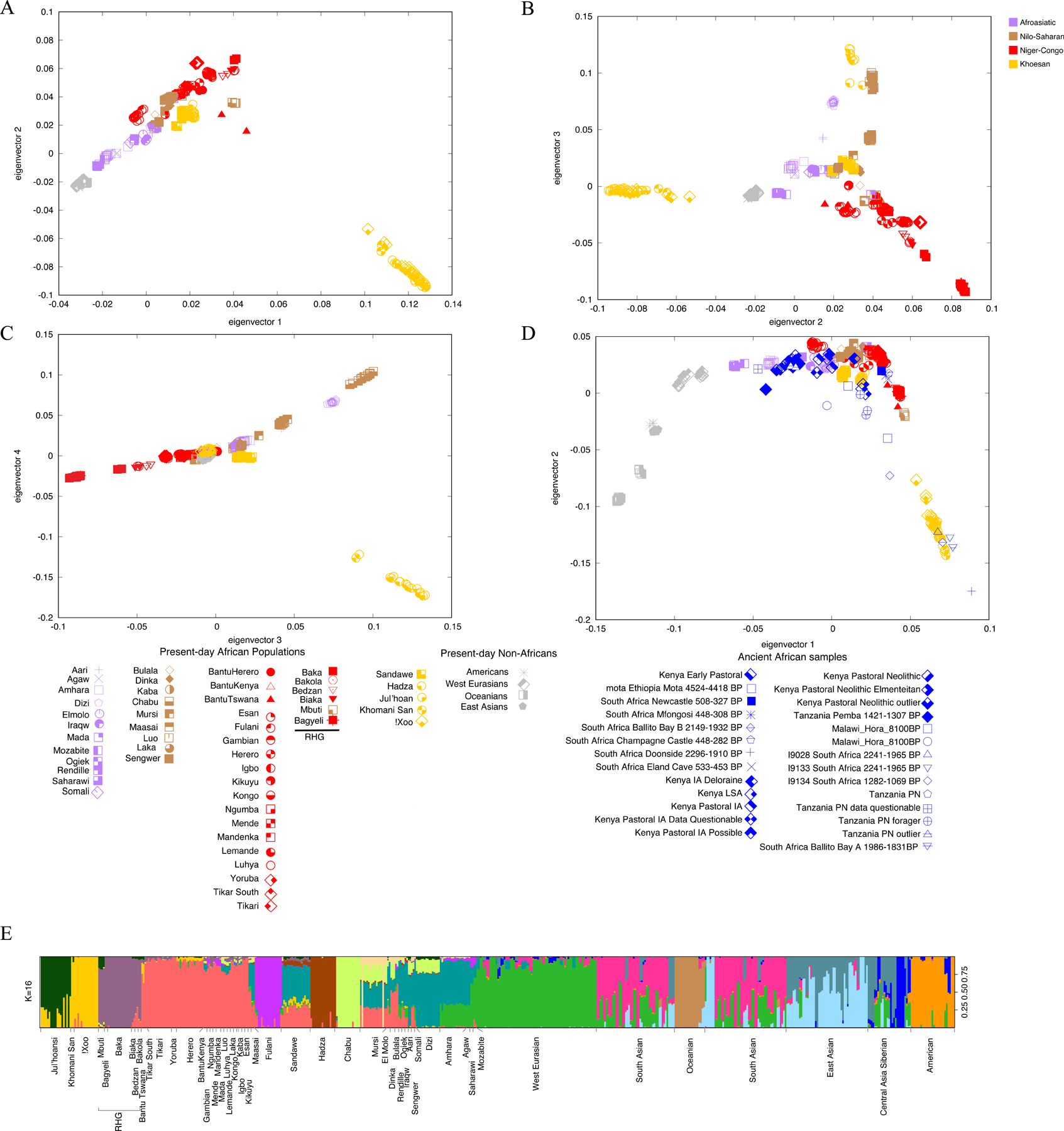
A-C: PCA of modern human populations from the present study with the SGDP.105
D: Projection of ancient samples from previous studies34,93,97,98 onto PCs 1 and 2. Points are individuals and colors indicate language classification (purple Afroasiatic, brown Nilo-Saharan, red Niger-Congo, and yellow Khoesan).
E: ADMIXTURE result for K=16. Bars are individuals and colors indicate ancestry proportions.
Complex demographic history of African populations
Principal component analysis (PCA) of the current dataset merged with a global WGS dataset from the SGDP reveals both continental and population-specific patterns of genetic variation. PC1 separates Africans and non-Africans, with the exception of populations in North Africa and the Middle East, consistent with prior studies (Figure 3A).3,21,33 PC2 distinguishes the San from other Africans (Figure 3A). Subsequent principal components differentiate the Hadza, Chabu, Dizi, and Mursi from other populations along PC3 (Figure 3B), and RHG populations (Baka, Bagyeli, Bakola, Biaka, Bedzan and Mbuti) are distinguished along PC4 (Figure 3C). Including 55 ancient Eastern and Southern African samples dated from 10,000 – 160 BP in the PCA, we observed a wide geographic distribution of Khoesan-related individuals in Africa as previously noted (Figure 3D)34; 15 ancient samples either overlap or fall onto a geographic cline between the present-day Eastern and Southern African Khoesan-speaking hunter-gatherer populations (Figure 3D). For example, Mota from Ethiopia (4524 – 4418 BP) and ancient foragers from Tanzania and Kenya (4080 – 160 BP) overlap in the PCA with the Sandawe and Hadza. Five ancient samples from South Africa (8173 – 1069 BP) either overlap or are close to the present-day southern African San populations, consistent with prior studies.34
ADMIXTURE analysis of the merged dataset separated African and non-African populations at K = 2 (Figure S2). At K = 4, San ancestry (yellow) becomes distinct, which is also common in the RHG, Sandawe and Hadza. At K=7, east African populations (e.g., Hadza, Sandawe, Chabu, Dizi, Amhara, and Mursi) emerged as a cluster (teal). The Fulani formed a distinct cluster at K = 8 (purple). The Hadza emerged as a cluster at K = 10 (brown) and the RHG (dark purple), and Chabu (light green) became distinct clusters at K = 12 (Figure S2). At K = 16 the Ju|’hoansi (dark green) who speak a northern Khoesan language and the !Xoo and Khomani San (yellow) who speak a southern Khoesan language become distinguished (Figure 3E). Additionally, Nilo-Saharan-speaking populations (e.g., the Dinka, Mursi, and Sengwer) became a single cluster (beige) at K = 16. Niger-Congo-related ancestry (red) was inferred to be widely spread across sub-Saharan Africa, but was most common in west and central African Niger-Congo-speaking populations (e.g., Lemande and Tikari) compared to eastern and southern Niger-Congo-speaking populations that have admixed to varying degrees with neighboring populations. The Herero, who speak a Bantu language, have low levels of admixture with the San.3,35 Furthermore, the Sandawe have high levels of Afroasiatic-related (light blue, ~50%) and Niger-Congo-related (red, ~25%) ancestries, but also low levels of ancestries related to the Hadza (brown) and San (yellow/dark green), reflecting shared common ancestry and/or ancient gene flow among southern and eastern African hunter-gatherer populations.
We modeled more complex demographic histories using TreeMix and qpgraph. When no admixture is allowed, the topologies based on qpgraph (Figure 4A) and TreeMix (Figure S3A) are consistent with the topology of the neighbor-joining tree (Figure 2), with the San as an outgroup to all other populations. However, the topologies of qpgraph (Figures 4B and S4) and TreeMix (Figure S3) vary tremendously when allowing admixture among populations. When modeling 10 admixture events, qpgraph estimated that the East African Khoesan populations, the Hadza and Sandawe, respectively derive 71% and 38% ancestry from a population ancestral to the Southern African Khoesan population (consistent with migration events between the Hadza, Sandawe and San inferred from TreeMix with 9 migration events). These populations, particularly the Sandawe (Figure 4B), also derive ancestries from an Afroasiatic-like population, likely reflecting recent Afroasiatic gene flow (Figure 3E), consistent with TreeMix with 4 migration events (Figure S3E). We estimated that the Ethiopian populations (Amhara, Dizi, Mursi, and Chabu) derived 98% and 2% of their ancestries from a population ancestral to the Hadza and a population ancestral to all modern human populations, respectively (Figure 4B). The latter may reflect Neanderthal introgression introduced into Ethiopians indirectly due to high levels of non-African admixture (Figure 3E).36,37 Furthermore, 80% of the Omotic-speaking Dizi ancestry can be traced back to a Chabu-related population and 20% to an Amhara-related population (consistent with TreeMix results with 7 migration events) (Figure S3H). In addition, qpgraph indicates that the RHG derive 37% of their ancestry from a population ancestral to the San and 63% of their ancestry from a Niger-Congo-speaking population (Figure 4B) consistent with high levels of Bantu gene-flow to the RHG.38–40 The relationship of the Tikari and Herero with other populations is complex. They could be modeled as having 23% ancestry related to an archaic population that diverged prior to the divergence of all modern human populations (possibly reflecting introgression from an archaic population into modern populations) and 77% ancestry from a population related to the Nilo-Saharan-speaking Mursi. A similar pattern was observed in the ADMIXTURE analyses at K = 7 to 11 but with much lower inferred Nilo-Saharan-related ancestries in the Tikari and Herero (Figure S2). The TreeMix analyses showed evidence of gene flow between the Mursi and the ancestors of the Tikari and Herero starting at 5 migration events (Figure S3F). The results indicating archaic introgression in a population ancestral to the Bantu-speaking lineage are consistent with previous studies based on ancient African samples which suggested that the West African Niger-Congo-speaking populations carry lineages ancestral to all modern human lineages.41 However, time-resolved demographic history models inferred using alternate methods (described below) suggest that the ancestors of San and RHG may have been the first to split from other modern human lineages.
Figure 4. Demographic history of African populations modeled by qpgraph and momi.
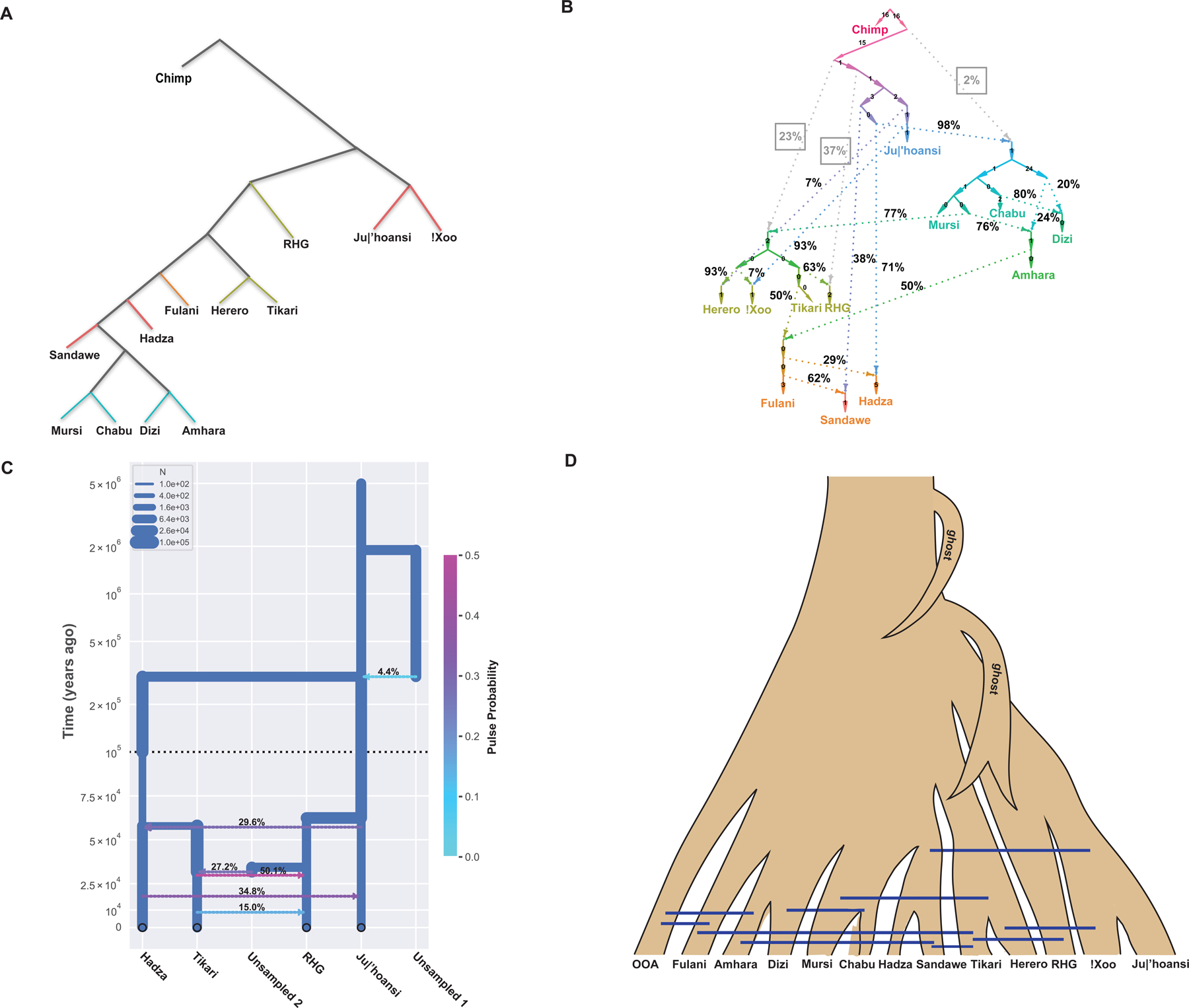
A: Demographic history without admixture inferred by qpgraph.106
B: Demographic history with 10 admixture events inferred by qpgraph.106 Percentages on the dashed lines show ancestry proportions from the two source populations. Numbers on solid lines are inferred drift lengths. The percentages of archaic ancestries are boxed and highlighted in grey.
C: Divergence times and gene flow inferred by momi.107,108 Modeling San and RHG as a sister clade consistently had the highest likelihood compared to other topologies.
D: Summarization of the results of demographic analyses. Blue bars show inferred gene flow among modern human populations. OOA: out of Africa populations. Ghost: inferred introgression from a ghost population. We observe evidence of introgression from a deeply diverged population into the ancestor of all modern human populations. In addition, the Bantu-speaking and RHG populations show some ancestry that is very old, possibly reflecting subsequent introgression with a deeply diverged population.
Consistent with the ADMIXTURE results, TreeMix and qpgraph analyses detected extensive recent gene flow among African populations (Figures 4B and S3–4). For example, the Herero derived 7% of their ancestries from the !Xoo (consistent with TreeMix results with 10 migration events) (Figures 4B and S3K).42–44 The Fulani derived 50% of their ancestry from a population related to the Amhara and 50% from a population related to the Tikari (consistent with TreeMix results with 3 migration events) (Figure S3D). The latter results are consistent with the ADMIXTURE analyses discussed above (Figures 3E and S2) and previous studies based on nuclear genomic variation suggesting that the Fulani share ancestry with Afroasiatic-speaking populations and admixed with Niger-Congo-speaking populations as they migrated across the Sahel.3,21,32,45 Using DATES, which uses the decay of ancestry covariance along the genome to date recent gene flow events, we estimated that the Fulani admixture event occurred 90 ± 40 generations ago (1.4 to 3.8 kya, assuming 29 years per generation), corresponding with later Holocene expansion events of nomadic pastoralists.46,47
Our WGS data also enabled detailed analyses of demographic history using two modeling approaches, MSMC and momi. Because MSMC analyses do not model gene flow, it likely underestimates divergence times in highly admixed populations. We began by investigating the population ancestral to all modern populations. Using momi we compared a model where the populations split from a single panmictic source to a model where the populations split from a structured population. Across all pairs of populations, we inferred that all modern humans descend from deeply structured populations and that they derive approximately 5–15% of their ancestry from a lineage that may have diverged as long ago as 1–3 Mya (Figure 4C), consistent with previous findings suggesting archaic introgression in some African populations.48,49 However, such a model is also consistent with the population ancestral to modern humans being deeply structured.
We next dated the divergence times between modern human populations. To interrogate the oldest population splits we used momi to infer a time-resolved demographic model relating the San (Ju|’hoansi), East African Khoesan (Hadza), RHG (Baka), and Bantu-speaking (Tikari) populations. We tested models with RHG as an outgroup, with the San populations as outgroups, and with the RHG and San as a sister clade derived from a population ancestral to all other populations. The models which had the San and RHG as a sister clade consistently had the highest likelihood (Figure S5) indicating that the oldest split between these populations separated the San and RHG from the Hadza and Tikari as early as 285 kya (Figure 4C). Similarly, when comparing either San or RHG to any other African population, the MSMC CCR does not reach higher than 90% until 150 kya to more than 200 kya (Figure S6). Together, these results indicate that the oldest split separated the San and RHG from all other populations, and this split occurred at least 150 kya and may have occurred as long as 285 kya.
All other pairs of populations were inferred to split more recently, with momi inferring divergence times less than 68 kya and MSMC CCRs reaching 50% before 42 kya (Figures S6D–F). In particular, despite speaking language “isolates” controversially placed within the Khoesan family, we inferred more recent divergence times between the Hadza, Sandawe and non-San/non-RHG populations relative to divergence times between San, RHG and other populations. When comparing the Hadza to non-San/non-RHG populations, momi inferred divergence times 25–60 kya and MSMC inferred a 50% CCR between 29–42 kya (Figure S6G). Similarly, for the Sandawe, momi inferred divergence times between 25–45 kya, and MSMC inferred a 50% CCR between 23–30 kya (Figure S6H). We estimated divergence times between Afroasiatic-speaking and Nilo-Saharan-speaking populations to be around 22–35 kya using momi and MSMC (Figure S6E).
Even within language groups, we observed evidence of ancient population structure. For example, between the Bantu-speaking Tikari and Herero, momi inferred a divergence time of 20 kya and the MSMC CCR reaches 50% at 11 kya (Figure S6I). We estimated that the divergence times between the Khoesan-speaking Ju|’hoansi and !Xoo are 18 kya and 24 kya using momi and MSMC, respectively (Figure S6J), consistent with prior estimates.8 Additionally, the East African Khoesan-speaking Hadza and Sandawe were inferred to have diverged ~23 and 25 kya using MSMC and momi, respectively. The Afroasiatic-speaking Amhara and Dizi were inferred to have diverged 30 kya using momi and 22 kya using MSMC (Figure S6E). Finally, the Nilo-Saharan-speaking Chabu and Mursi were inferred to have diverged 22 kya using momi and 17 kya using MSMC (Figure S6E). All pairwise momi results are presented in Table S2 and are based on the models in Figure S7.
Temporal dynamics of effective population size in Africa
Using PSMC and SMC++, we observed the emergence of effective population size (Ne) differences as early as ~200 kya (Figure 5). From 200–50 kya, the RHG and San have greater Ne compared to other populations (Figure 5A). The Amhara and Dizi have the lowest Ne compared to other African populations (Figure 5A). Four populations, including Hadza, Chabu, Herero, and Fulani, experienced dramatic population size declines 1–10 kya (Figure 5B). In particular, the Ne of both the Hadza and Chabu dropped from ~10,000 to ~200 (Figure 5B), consistent with their current census sizes of ~1,000.
Figure 5. Inferred effective population sizes.
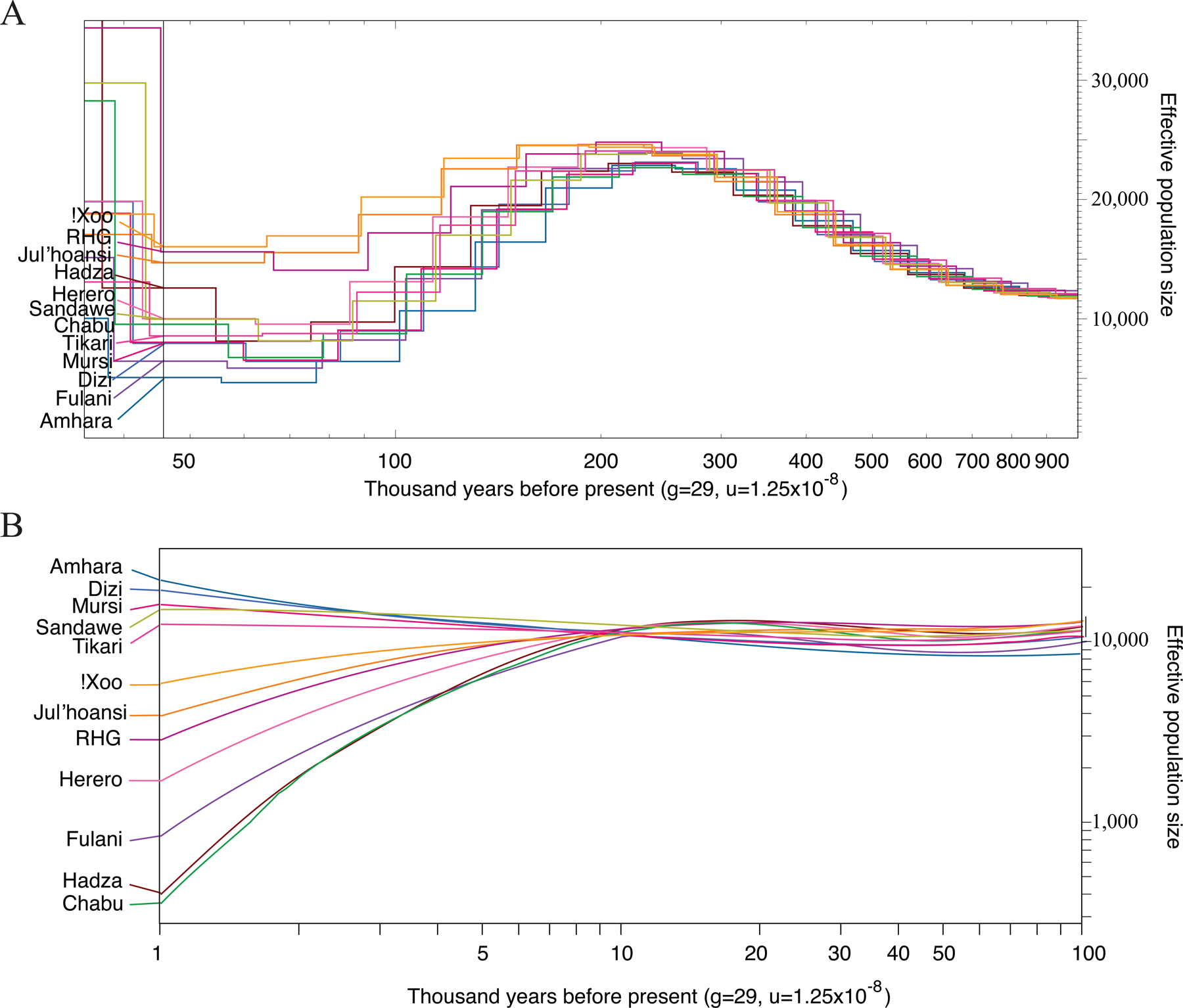
A: the results of PSMC.109 B: the results from SMC++110, plotting effective population size against time, assuming a per-nucleotide, per-generation mutation rate of 1.25 × 10−8 and generation time of 29 years.
Local adaptation in Africans
To identify candidate loci that may play a role in local adaptation to diverse environments and diets, we identified loci that have highly differentiated allele frequencies in each population compared to other African populations using the Di statistic. We calculated Di value for each SNP and defined outliers falling in the 99.9th percentile as Di-SNPs. The functional impact of genes near Di-SNPs was inferred using GREAT (Table S3). We also identified Di-SNPs that overlap GWAS associations from populations with African ancestry using the EBI GWAS catalog and studies using UKBB samples.50,51 We observed evidence for local adaptation for different traits in diverse populations (Figure 6).
Figure 6. Representative phenotypic and physiological traits shaped by positive selection due to local adaptation in African populations.
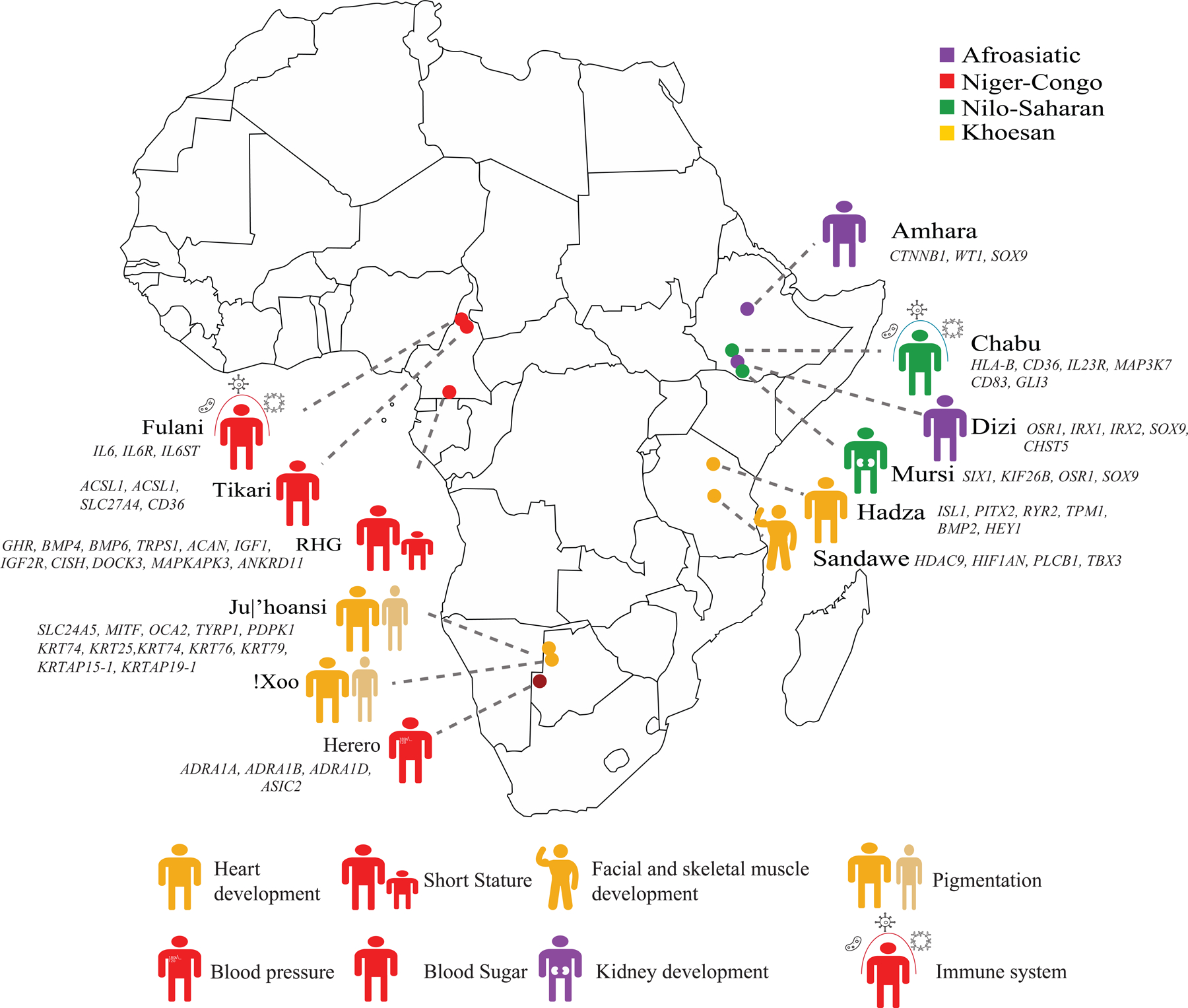
We identified signatures of positive selection in different populations using the di statistic. Representative traits and genes were selected based on functional annotation of outlier SNPs in different populations using GREAT.111
We found that the San, who have lighter skin than other African populations52, have enrichment for Di-SNPs near genes involved in skin pigmentation, including OCA2, TYRP1, SLC24A5, MITF, and other skin phenotypes, including keratin loci (e.g., KRT25, KRT27, and KRT71) (Table S3). Previous studies show that mutations in OCA2, TYRP1, SLC24A5 and MITF can cause ocular albinism type 2, type 3, and type 6, as well as Tietz albinism-deafness syndrome.53,54 In the gene body of OCA2, we identified 112 Di-SNPs, including one synonymous, one nonsynonymous, and 110 intronic mutations. While the nonsynonymous variant (rs1800417) at OCA2 was previously reported to not be associated with skin pigmentation variation in the San55, rs1800404, a synonymous variant in exon 10, associates with skin pigmentation and eye color variation across multiple ethnicities.52,56,57 The light-pigmentation associated allele rs1800404-T, which is a splicing QTL of OCA252,58, is most frequent in the San (83%) compared to all other populations in the present study and gnomAD except for the Finnish population (frequency of 84%;Table S4).
We also observed 22 Di-SNPs in the San within the gene body of PDPK1 (Figure 7A). PDPK1 is an important regulator of melanocyte proliferation and loss of PDPK1 reduces skin pigmentation in mice.59 Interestingly, one Di-SNP, rs77665059, overlaps a melanocyte-specific open chromatin region in the intron of PDPK1 (Figure 7A). The ancestral allele, rs77665059-C, shows higher frequencies in the Ju|’hoansi (0.67) and !Xoo (0.83) compared with other populations (average frequency of 0.14 and 0.03 in the non-San populations of the present study and the global populations in gnomAD, respectively) (Figure 7B). ChIP-seq data revealed that this region is enriched for H3K27ac and H3K4Me1 signals in melanocytes, and binding sites for the transcription factors MITF, SOX10 (involved in melanocyte development and expression of pigmentation genes60,61) and SMARCA4 (chromatin remodeler) (Figure 7A). Based on luciferase expression assays in two melanoma cell lines: MNT-1 (highly pigmented) and WM88 (lightly pigmented). We observed that the ancestral C allele is associated with increased enhancer activity compared to the derived A allele in both cell lines (Figure 7C), consistent with the C allele being associated with lower expression of PDPK1 in fibroblasts in GTEx (Figure 7D).58 Individuals with the C allele have lighter skin pigmentation compared to individuals with the A allele in the San (Figure 7E). Furthermore, CRISPR inhibition of this enhancer indicates significantly reduced expression of PDPK1 and melanin levels in MNT-1 cells (P-value <0.001, one-way ANOVA post hoc test) (Figure 7F). These observations indicate that SNP rs77665059 is within an enhancer active in melanocytes that impacts pigmentation in vitro and may influence skin color in the San by regulating the enhancer activity and gene expression of PDPK1.
Figure 7. rs77665059 affects the enhancer activity of PDPK1 and may contribute to light skin color of the San.
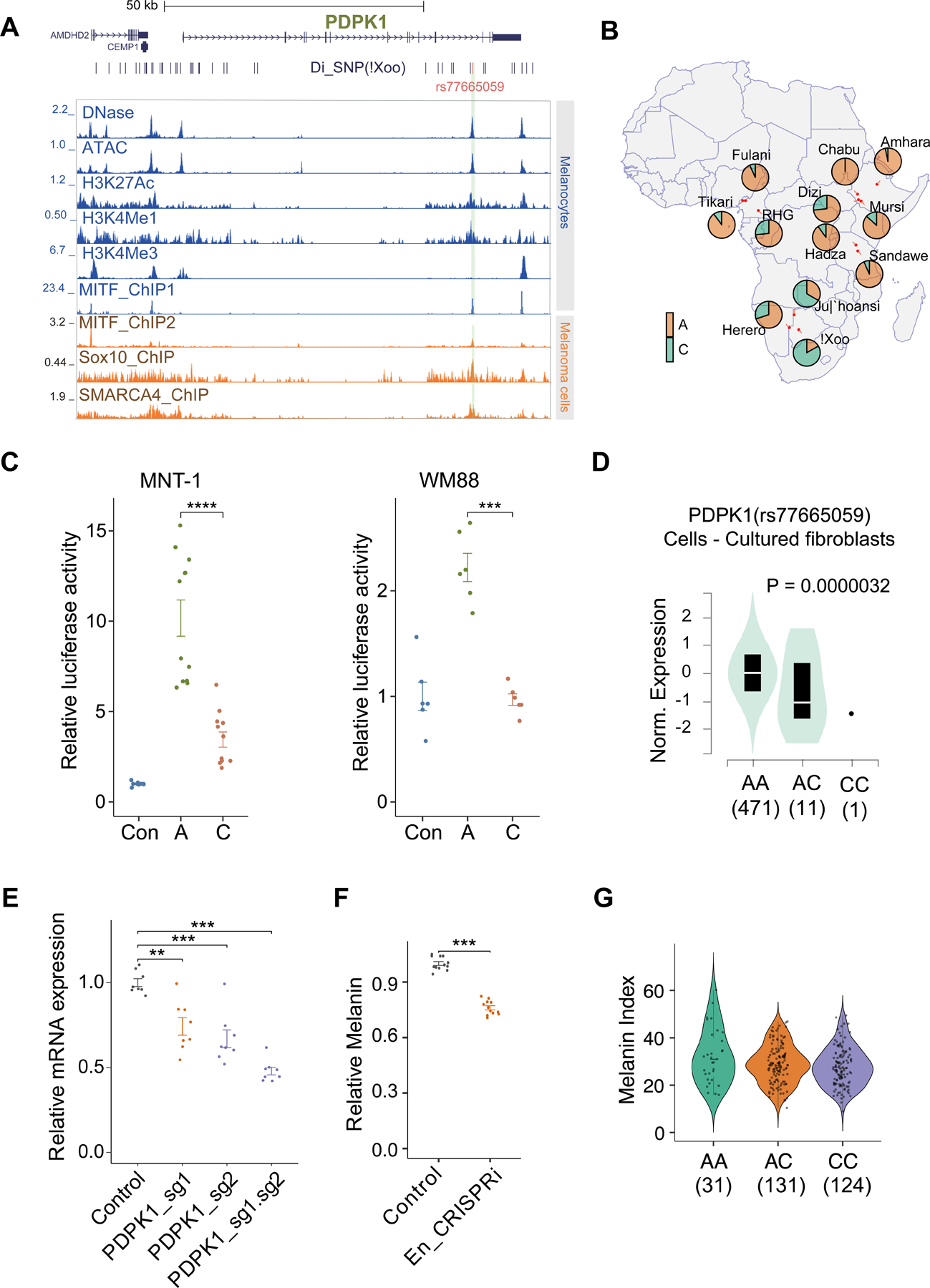
(A) rs77665059 overlaps a melanocyte-specific open chromatin region in the intron of PDPK1.
(B) Allele frequency of rs77665059 in 12 African populations. C is the ancestral allele, highlighted in green.
(C)Luciferase reporter assay of rs77665059 in MNT-1 and WM88 melanoma cells. N=10–12.
(D) rs77665059 is an eQTL of PDPK1 in cultured fibroblast cells. Data from GTEx.
(E) CRISPRi of the enhancer inhibits PDPK1 gene expression.
(F) CRISPRi of the PDPK1 enhancer decreases the melanin level in MNT-1 cells.
(G)Melanin index for different genotypes of rs77665059 in the San. One-way ANOVA with pos hoc tests were used in C, E, and F. **** indicates p < 0.0001, *** indicates p < 0.001.
We also observed enrichment for Di-SNPs in the San near genes involved in hair follicle development and “narrow eye opening” in mice (Table S3). This observation is consistent with descriptions of unique hair follicle morphology (tightly spiraled) and narrow eye morphology in the San.62,63 One SNP of particular interest is a non-synonymous variant, rs111298318, in KRT74. Mutations at KRT74 are known to cause a “wooly hair” phenotype in humans.64 The rs111298318-C variant is at > 0.73 frequency in the San, is <0.05 frequency in other African populations in the present study and is almost absent in non-African super-populations in gnomAD.
In the RHG we found enrichment for Di-SNPs near genes involved in bitter taste receptor activity (e.g., TAS2R1, TAS2R10) and immune response (e.g., HLA-DOA, IL2, and IL4R) consistent with previous studies.23,39 Additionally, we observed enrichment for Di-SNPs near genes involved in bone growth and chondrocyte differentiation (Table S3) including CISH/DOCK3/MAPKAPK3, GHR, IGF1, BMP4, BMP6, ANKRD11, TRPS1, and ACAN23,50,65–69, potentially involved in the short stature of the RHG. Notably, 75 out of 76 Di-SNPs in a 15 Mb region of chromosome 3 (between 45–60 Mb) that were significantly associated with height variation in the RHG23,65 were predicted to be eQTLs of DOCK3 or MAPKAPK3 in GTEx. Further, 312 Di-SNPs (Table S5) were significantly associated with height (P-value < 1e-8) in previous GWAS51,70,71, suggesting that the short stature phenotype in the RHG likely evolved due to positive selection at multiple loci.
We observed an enrichment for Di-SNPs near genes that play a role in immune-related pathways in the Fulani and Chabu (Figure 6 and Table S3). Studies have shown that the Fulani are more resistant to severe malaria relative to other ethnic groups in similar environments.72,73 In the Fulani, we observed significant enrichment for Di-SNPs near genes involved in the “Cellular response to interleukin 6” including IL6, IL6R, and IL6ST (Table S3). Previous studies based on gene expression analysis suggest that genes in the IL6 signaling pathway may play a role in relative resistance to malaria observed in the Fulani.74,75 The rs1889314-A, rs10908834-T, and rs12118634-T alleles of three Di-SNPs are more frequent in the Fulani than other African populations in our study or in the gnomAD database (Table S4) and are significantly associated with increased expression of IL6R compared to the alternative alleles.58
In the Chabu, we observed enrichment for Di-SNPs near genes involved in positive regulation of immune effector processes, positive alpha-beta T cell activation and differentiation (Table S3) reflecting an adaptation to a different environment and different pathogens, compared to the Fulani. We also detected 318 Di-SNPs within or near the MICA locus (±50 kb), including 8 missense mutations (rs1063630, rs1051786, rs1051792, rs1051794, rs1131898, rs1051798, rs1051799, rs61738275). SNPs rs1063630 and rs61738275 are in one LD group (R2 = 1), while the other six SNPs are in a separate LD group (R2 = 1). MICA is a ligand of NKG2D and triggers the cytotoxicity of natural killer cells and CD8 T cells, acting as an important component of the innate immune response.76
In the Hadza, we observed enrichment for Di-SNPs near genes that play a role in pathways related to cardiac function and development, including BMP2, HEY1, MYH6, RYR2, PITX2, and TPM1 (Table S3). Previous studies have shown that genes in cardiac-related pathways are enriched for being targets of positive selection in RHG populations in Africa and Asia.77 The Hadza are one of the few populations globally that continue to practice a traditional hunting and gathering lifestyle and are well-known for the remarkable distance that they travel daily; on average men walk 13 km per day hunting animals and gathering honey, and women walk 8 km per day foraging for plant foods.78 Thus, selection at loci involved in heart development could be adaptive in this population.
The Di-SNPs in the Sandawe are near genes involved in facial and skeletal muscle development, such as regulation of skeletal muscle fiber development, embryonic cranial skeleton morphogenesis, and cranial and craniofacial suture morphogenesis (Table S3). For example, we detected Di-SNPs near MEF2C79, TBX380, and HIF1AN81, which are involved in skeletal muscle development as well as FGFR282, TGFBR283, TBX1584, and TWIST185, which play important roles in cranial development and morphology. The adaptive significance of these loci is unclear.
We observed Di-SNPs in Herero and Tikari at loci that play roles in hypertension, kidney disease, obesity, and diabetes (Table S3), diseases which are relatively common in African Americans compared to other ethnic groups in the U.S..86,87 In the Herero, ontologies such as regulation of systemic arterial blood pressure by baroreceptor feedback, positive regulation of blood pressure by epinephrine-norepinephrine, regulation of systemic arterial blood pressure by norepinephrine-epinephrine, and neurological system process involved in regulation of systemic arterial blood pressure, are significantly enriched for Di-SNPs. A set of 23 Di-SNPs in the Herero were significantly associated with blood pressure traits (e.g., systolic/diastolic blood pressure) in previous GWAS of UKBB samples.50 For example, rs7821832-G is most frequent in the Herero compared to other populations in our study and to populations in gnomAD (Table S4) and is significantly associated with systolic blood pressure (P-value=5.4x10−20) and diastolic blood pressure (P-value= 8.2x10−9) in the UKBB samples.50 In the Tikari, we observed enrichment for Di-SNPs near genes involved in long-chain fatty acid import (Table S3). For example, one of the Di-SNPs, rs2717609-T, is significantly associated with traits such as body fat percentage (P-value=1.1x10−10), whole body fat mass (P-value=4.9x10−10), trunk fat mass (P-value=7.6x10−12), and hip circumference (P-value=5.0x10−9) in a prior GWAS of UKBB samples.50
In the Mursi, Amhara, and Dizi (Table S3), we observed enrichment for genes involved in pathways related to kidney development and morphology which could reflect an adaptation to environments that are often arid, with little access to water. For example, we found that the Di-SNPs rs9823161, rs72841902, and rs4567493 in the Amhara, Dizi, and Mursi are significantly associated with traits related to kidney function in previous GWAS based on multi-ancestry samples.88 rs9823161-A and rs72841902-A are positively associated with estimated glomerular filtration rate, and rs4567493-A is negatively associated with blood urea nitrogen levels.
We also detected loci showing signatures of recent positive selection based on extended haplotype homozygosity using the integrated haplotype score statistic (Table S6). We defined the top 1% of windows with the highest fraction of extreme integrated haplotype scores as outliers and observed some loci that show a shared signature of recent positive selection (Table S6). For example, we observed a shared signature of positive selection at the MHC locus in the Chabu, Mursi and Dizi from Ethiopia (Table S6). We also identified population-specific positive selection signals. For example, genes located in the outlier windows showing strong iHS signals are significantly enriched (FDR adjusted P-value <0.01) in pathways involved in alcohol dehydrogenase activity (e.g., ADH4, ADH5, ADH6, ADH7, and ADH1A) in the Amhara (Table S6), consistent with observations in this population based on SNP array data89, in bitter taste receptor activity (e.g.,TAS2R20, TAS2R30, TAS2R31, TAS2R43,TAS2R46, and TAS2R50) in the Hadza, and in growth hormone receptor binding (e.g., GH1, GH2, CSH1, CSH2, and CSHL1) in the Fulani (Table S6).
Discussion
In this study, we analyzed high-coverage whole-genome sequencing data from 180 individuals from twelve indigenous African populations representing a wider range of cultural, linguistic, phenotypic, and genetic diversity in Africa than previous studies of Africans.3,90 We identified ~5.3 million previously unreported variants, many of which are predicted to be functional. Furthermore, we found that 44 out of 154 “Pathogenic” or “Likely Pathogenic” SNPs are common (frequency > 0.05) in one or more populations in this study but are rare (frequency < 0.01) in non-African populations. These results do not imply that African populations have a high frequency of pathogenic variants but likely reflect that low prevalence of variants is a factor for determining pathogenicity in current clinical studies, and bias toward non-African populations may result in the misclassification of pathogenic variants. These observations emphasize the importance of including ethnically diverse populations and developing unbiased genotyping (e.g., SNP arrays designed for samples of African ancestries) in human genetic studies.16,91
Our study depicts a complex demographic history of African populations, consisting of ancient population divergence, regional and cross-continental migration, and admixture events (Figures 3 and 4B–D). Although phylogenetic analyses indicate that the San descend from a population ancestral to all other modern humans, demographic modeling using momi, allowing for changes in effective population size and migration between populations, consistently supports a model in which the RHG and San form a sister clade deriving from a population ancestral to all other modern human populations. We find similar effective population sizes of the San and RHG from 50–200 kya (Figure 5), consistent with shared common ancestry. Similarly, ADMIXTURE analysis identifies shared ancestry between the San and RHG, particularly at low K values (Figure S2). On the other hand, qpgraph suggests that the RHG and Bantu populations derive a substantial portion of their ancestry from a population that is an outgroup to all modern populations (Figure 4B). One possibility to explain these observations is a model with multiple introgression events between a deeply diverged population (diverged >1 – 3 Mya) with the ancestors of all modern humans (Figure 4C) and, more recently, with the ancestors of the RHG and Bantu populations (Figure 4D), consistent with previous reports of archaic introgression in African populations.23,24,34,41. However, these results could also be explained if the lineages related to modern-day African populations were part of a deeply structured ancestral population (a “multiregional” model of modern human origins in Africa which could have been facilitated by gene flow between structured populations). Sequencing ancient DNA from archaic hominid fossils in Africa, if it becomes feasible, may provide more direct evidence of archaic admixture in Africa as has been the case for Neanderthal and Denisovan introgression in non-Africans.36,92
Thus, the early demographic history of the lineages leading to modern humans is complex with multiple episodes of gene flow between modern human lineages and possibly with other hominid lineages. When accounting for gene flow, we estimated that the deepest divergence among modern humans dates back to 285 kya, which is consistent with the estimates based on ancient African samples41,93 and fossil records in Africa.1 Without accounting for gene flow, however, our estimates from MSMC analyses are much more recent (~100–150 kya) but still quite deep. We also show that populations speaking all major language families diverged tens of thousands of years ago, consistent with long term population structure both within and between populations speaking languages from different phyla.34,94,95
Although their languages are highly divergent and their classification into the Khoesan phylum is still contentious, our analyses based on qpgraph, TreeMix, and momi identified signals of ancient gene flow between currently geographically isolated Khoesan-speaking hunter gatherer populations, the Hadza and Sandawe in East Africa and the Ju|’hoansi and !Xoo who currently reside in southern Africa, as recent as within the last 12 ky. Evidence based on mtDNA and autosomal data from modern and ancient samples34,95–98 suggests the present-day San may have originated in Eastern Africa, then migrated into southern Africa and that there could have been a broader distribution of Khoesan-speaking populations in Africa. Therefore, there could have been continuous gene flow between the Khoesan-speaking populations in Eastern and Southern Africa over long periods of time. In addition, we observed that Niger-Congo-related ancestries are highest in the Niger-Congo-speaking populations in West and Central Africa (e.g., Tikari), but are slightly lower in the Herero of Botswana, reflecting an origin of Bantu speaking populations in West and Central Africa with the past 5 ky and more recent migration of the Herero into southern Africa in the past 1 ky and subsequent admixture with Khoesan-speaking populations such as the !Xoo. We also observed Bantu ancestry in the Sandawe and !Xoo, reflecting admixture of Bantu-speaking people with indigenous populations as they spread throughout Africa. Consistent with the linguistic and archeologic record, we observe evidence for migration and gene flow of Nilo-Saharan and Afro-Asiatic speaking populations from a homeland in present day Sudan/Ethiopia southward into Kenya and Tanzania (Figure 3). The local indigenous hunter gatherer populations were either assimilated or forced to move into harsh habitats leading to severe decreases in effective population size in the Hadza23 and Chabu27 but not the Sandawe, who assimilated with the neighboring Cushitic and Bantu speaking populations, resulting in high levels of gene flow, adoption of agro-pastoralism, and population growth. We also observed decreases in effective population size in the Fulani (consistent with a study based on mtDNA markers99) and Herero. German colonial soldiers nearly exterminated the Herero people of Namibia in the past 100 years, which likely explains the bottleneck in that population.
We identified loci that may play a role in phenotypic and physiological adaptation to diverse environments, diets, and pathogens across African populations. Some of these loci may affect disease susceptibility in current populations living in more urban environments. Combining in silico and in vitro data, we show that one of the Di-SNPs, rs77665059, may play a role in light skin color of the San by regulating expression of PDPK1, which could be adaptive in this population living relatively far from the equator. With ongoing deep phenotyping of global populations based on multiple “-omics” data and the advances of in vitro and in vivo technologies, we expect the functions of adaptive variants in more human populations will be characterized in the future.21 The identification of genetic variants that differ in frequency in ethnically diverse populations is a complementary approach to GWAS for identifying functionally important variation, particularly in cases where that variation is strongly correlated with ancestry and where GWAS may have limited power due to small sample sizes and/or variants that are close to fixation in particular populations.
Limitations of the study
There are still some ambiguities in our inferences of African demographic history because we can only model simple demographic histories whereas the real demographic histories are likely to be much more complex. Additionally, given 15 samples per population, we may be underpowered to detect all loci that are under selection. Moreover, we may be missing some rare, but functionally important SNPs as well as SNPs that may be specific to populations from regions not well represented in the current study such as western and northern Africa. To deepen our understanding of complex evolutionary history of Africans, we must develop more efficient computational methods, include more indigenous populations and ancient samples at broad geographic and temporal scales, and integrate genomic data with paleobiological, archeological and linguistic data. Additional genomic data modalities, such as long read sequencing to uncover structural variants, may illuminate additional forms of genetic variation beyond SNPs and small insertions and deletions.
STAR+METHODS
Resource availability
Lead contact
Further information and requests for resources and information should be directed to and will be fulfilled by the Lead Contact, Dr. Sarah A. Tishkoff (tishkoff@pennmedicine.upenn.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The SNP data are publicly available through the dbGAP database (accession number: phs003096.v1.p1). Links to the software and algorithms in the present study were listed in the key resources table.
Key resources table
Experiment model and subject details
Before sample collection, permits were received from the Ministry of Health and National Committee of Ethics in Cameroon, COSTECH, NIMR in Dar es Salaam, Tanzania, the University of Addis Ababa and the Federal Democratic Republic of Ethiopia Ministry of Science and Technology National Health Research Ethics Review Committee; the University of Botswana and the Ministry of Health in Gaborone, Botswana. We obtained Informed consent from all research participants. In addition, appropriate IRB approval was obtained from the University of Pennsylvania. We merged the Baka and Bagyeli into one population, RHG. All the samples were males and > 18 years old.
Method details
DNA Sequencing
We conducted whole genome sequencing of 180 individuals (fifteen unrelated samples per population) at high coverage (on average >30X) using Illumina HiSeq X Ten platform. All samples were processed using the same PCR-free library preparation and the same sequencing protocol, which reduces the potential for PCR bias and also minimizes artifactual differences caused by sample preparation. The samples were sequenced using paired-end sequencing with 150 bp at each end and a 350 bp insertion size.
Curation of Sequencing Data and short variant calling
We trimmed the sequencing adapters using trimadap (https://github.com/lh3/trimadap) and masked optical duplicate reads using SAMBLASTER119 (version 0.1.22). The reads were mapped to the decoy version of the human reference genome (hs37d5) with bwa mem mode (version 0.7.10).116 SAMtools version 1.4130 was used to sort and index the mapping results. We also filtered out the reads with mapping quality < 20 using SAMtools. We conducted variant calling using Haplotypecaller module in GATK Toolkit (version nightly-2016–09-26-gfade77f)117 following the best practice guidance of germline short variant discovery. In addition, we used a prior of (0.4995, 0.001, 0.4995) for the homozygous to reference, heterozygote, and homozygous non-reference allele were used in the Bayesian SNP calling step to generate reference-bias free genotypes following the recommendations of the Simons Genome Diversity Project (SGDP).105 The sample-level variant calling results were stored in intermediate files with genomic variant calling format (gVCF) that contain a record for every position of the examined regions in the genome. We then merged the sample-level gVCF files using CombineGVCFs module. We performed joint genotyping using GenotypeGVCFs module, which generated one quality score for each variant site based on the inferred genotype likelihood across all the samples. We filtered the variants using a two-fold filtering strategy. First, SNPs from the 1000 Genomes project Phase 3104, Illumina Omini 5M SNP array52, and the HapMap project were used as the truth data in the GATK variant quality score recalibration (VQSR) step. For the INDEL VQSR, we used the curated genotypes from Mills et al131 as the training dataset. We obtained 33,360,065 SNPs and 2,762,633 Indels after VQSR. Second, we further excluded the variants (28 SNPs and 0 InDels) that locate in the potentially duplicated regions identified by Delly (version 0.7.6)120, and are in the low complexity regions of the human reference genome (921,130 SNPs and 0 InDels).132 Finally, we obtained 35,201,568 variants, consisting of 32,438,935 SNPs and 2,762,633 InDels. We note that no variant violates HWE (p-value <1E-6) when calculating HWE for each population using Plink.133
SNP annotation
We used ANNOVAR version 2018–04-16 to annotate the bi-allelic SNPs. The novel SNPs are identified based on the comparisons to the variants in dbSNP100 (version 155) and gnomAD (version 2.1) databases.101 The functional impacts of the SNPs in the coding regions were predicted based on RefSeq annotation of hg19.134 We intersected the SNPs in our dataset with the annotations of transcription factor binding sites (TFBS) and predicted chromatin state segmentations of GM12878 generated by the ENCODE project102. We also intersected our SNPs with the variants in the ClinVar database (as of 2021–05-01).135 We reported the variants that were only labelled as “Pathogenic” or “Likely Pathogenic” in the Clinvar database.
Merging with the SGDP data
We first removed SNPs in linkage disequilibrium (LD) using Plink version v1.90b3j133 with parameters --indep-pairwise 50 10 0.1. The pruned data were recruited as query to extract the genotype information of 251 and 93 non-African and African samples, respectively, from the Simons Genome Diversity Project (SGDP)21,105 using cTools (https://github.com/DReichLab/cTools). We used VCFtools129 version 0.1.17 to merge the variants in our and SGDP datasets. A total of 12,443,243 SNPs were used in the ADMIXTURE and principal components analysis (PCA).
Quantification and statistical analysis
ADMIXTURE and PCA
The merged data were used as input for ADMIXTURE version 1.3.0.118 The number of ancestral groups (K) was set from 2 to 16. We conducted 10 runs at each K value using the default parameters to avoid local optima and merged the results of the 10 runs at each K value using CLUMPP version 1.1.2.121 We performed PCA of our dataset with the global samples of the SGDP using smartpca in the EIGENSOFT toolkit version 6.0.1.114,115
Incorporation of ancient African DNA samples
We repeated the PCA incorporating genotypes from 55 ancient African samples from four studies.34,93,97,98 The genotype information of Prendergast et al were obtained from the authors directly. For the other three studies, we downloaded the bam files and conducted SNP calling using apulldown.py (https://github.com/mathii/gdc3/blob/master/apulldown.py), which conducts haploid calling for ancient samples. 345,065 transversion SNPs from the ancient samples were merged with our dataset. Using the lsq mode in smartpca114, we projected the ancient samples to present-day populations.
Phylogenetic relationship inference
We first extracted the orthologous base pairs in the chimpanzee genome from the alignment of human and chimpanzee genomes generated by the Ensembl database. 136 Using chimpanzee as outgroup, we inferred the phylogenetic relationship of African and non-African populations with the neighbor-joining method in MEGA version 11.122 We evaluated the robustness of the phylogeny using 100 bootstraps. We used Figtree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) to visualize the results from MEGA.
Effective population size and divergence time inference
We estimated effective population sizes using the pairwise sequentially Markovian coalescent (PSMC) version 0.6.4-r49 with default parameters.109 Since the PSMC model is only capable of inferring the effective population size > 60 kya109, we also used the SMC++ model version 1.11.1 with default parameters110 to infer the recent effective population sizes of African populations. To convert from generations to years, we assumed a generation time of 29 years and 1.25×10−8 mutations per site per year.
D-statistic analysis of the relationship of African and non-African populations
We used D statistics to identify the potential out-of-Africa source population(s) and waves of admixture between African and non-Africans using D-statistics. D-statistics measure excess allele sharing between populations and are computed for a set of four populations. For populations 1, 2, 3, and 4 D(1, 2; 3, 4) is computed as (p(ABBA) - p(BABA))/(p(ABBA) + p(BABA)) where p(ABBA) represents the probability that for a randomly chosen biallelic site a randomly chosen individual from population 1 and a randomly chose individual from population 4 have the same allele (which we call A) while a randomly chosen individual chosen from population 2 and a randomly chosen individual from population 3 have the other allele (which we call B). Similarly, p(BABA) represents the probability that for a randomly chosen biallelic site randomly chosen individuals from populations 1 and 3 have the same allele (B) and randomly chosen individuals from populations 2 and 4 have the same allele (A). If the four populations are related by an unrooted tree such that populations 1 and 2 are sister taxa and populations 3 and 4 are sister taxa, then both ABBA and BABA sites are discordant with the tree. That is, the individuals that share alleles are not from sister taxa, and hence the mutation must have either arisen independently in two different populations or have occurred in the population ancestral to all four populations. Since both ABBA and BABA sites are discordant, they should be approximately equally likely, and so D(1, 2; 3, 4) is approximately zero. In the case that populations 1 and 2 are not sister taxa, then population 1 must either be more closely related to population 3 than population 4, in which case p(BABA) will be larger than p(ABBA), or vice-versa, in which case p(ABBA) will be larger than p(BABA). In either case, D(1, 2; 3, 4) will differ significantly from zero. Therefore, we can interpret D-statistics that are close to zero as being consistent with populations 1 and 2 being sister taxa and populations 3 and 3 being sister taxa. We merged our dataset with the SGDP data21,105 and restricted our analyses to biallelic SNPs. All D-statistics106 were computed using admixtools2 version 2.0.0 (https://uqrmaie1.github.io/admixtools/index.html), using the commands “extract_f2” with the options “minmaf=0.05” and “maxmiss=0.01” to precompute f2 statistics and using the “qpdstat” command to compute D-statistics. To obtain genetic distances between markers, we used the pyrho YRI recombination map, which was inferred to be a population-specific recombination map for the Yoruba in Ibadan Nigeria (YRI) as a proxy for the recombination rates in the present samples.137
Demographic inference based on momi
We performed two main types of analyses in momi.107,108 In one set of analyses, we considered “generic” models and fit these models to many sets of populations. These analyses used all 15 individuals from each population. We considered four different generic models (Figure S7).
Model 1 has two populations that start as a single population, and then population 2 splits from population 1 at an inferred divergence time. The ancestral population has some size, that changes 100kya to the present-day size of population 1 and population 2 has a constant size from the time of divergence onward. The divergence time and the three populations sizes are all inferred from the data.
Model 2 is identical to Model 1 in the recent past but at some time pre-dating the split of the two present-day populations, an unsampled “ghost” population splits from the ancestral population and at some point after this split but prior to the divergence of the two modern populations there is a pulse of gene flow from the ghost population into the population ancestral to the two modern populations. In this model, we infer all of the same parameters as in the first model but, additionally, the divergence time of the ghost population, the size of the ghost population, and the timing and amount of pulse admixture from the ghost population into the ancestral population. Both Model 1 and Model 2 were fit using each possible pair of populations as the first and second population.
Model 3 was designed to account for pervasive gene flow between RHG and Tikari when computing divergence times between RHG and other populations. In this model, RHG and population 1 each diverge from an ancestral population at their own inferred divergence time and having their own population sizes after divergence. At an inferred time there is a pulse admixture of an inferred strength from the Tikari into RHG. The ancestral population has some size, which changes to the present-day size of the Tikari 100kya. In this model we infer the two divergence times, the four population sizes (three present day, one ancient), and the timing and strength of the pulse admixture event.
Model 4 is identical to Model 3 but with a ghost population added in the same way as going from Model 1 to Model 2. Models 3 and 4 were fit by including each non-Tikari, non-RHG population with Tikari and RHG populations.
In another set of analyses, we fit more complex models to specific sets of populations. Due to the complexity of these models, these were fit using only two arbitrarily chosen individuals per population. These models were initially based on qpgraph results or known historical events (e.g., the Bantu expansion). Using goodness-of-fit criteria from momi (f2 and identity-by-state) additional admixture events, population size changes, or unsampled populations were added to the model. To investigate the deepest splits between populations, we considered models with Ju|’hoansi, RHG, Tikari, and Hadza and explored models where either RHG was the outgroup, Ju|’hoansi was the outgroup, or RHG and Ju|’hoansi were sister groups. Keeping this aspect of the tree topology fixed, we tried several different demographic models by adding admixture events to try to find a sensible model with that topology that produced a good likelihood and also had good goodness-of-fit.
In all cases, models were initialized randomly several times and re-optimized to avoid getting stuck in local optima. Both sets of analyses determined the derived allele based on the ancestral alleles provided by the 1000 Genomes Project104, although results were qualitatively similar when using the “folded” frequency spectrum obtained using the momi function “fold()”.
Demographic inference using qpgraph
We used the f2 statistics computed using admixtools2 (https://uqrmaie1.github.io/admixtools/articles/admixtools.html) as above (Out-of-Africa Source Population) as the input to qpgraph as implemented in admixtools2. In particular, we used the “find_graphs” function in admixtools2. This performs an automated search similar to simulated annealing to find the best fitting admixture graph with a given number of admixture events, but can get stuck in local optima. To this end, we ran “find_graphs” 20 times per number of admixture events and stored the best fitting graph. We used the parameters “stop_gen=1000” and “numgraphs=25”, which determine the extent of the search for the optimal graph. For the graph with no admixture events, we initialized each search randomly. For graphs with one or more admixture graphs we initialized the search at the best graph we found with one fewer admixture event. In all qpgraph analyses we used chimpanzee as an outgroup.
DATES analysis
The Fulani show clear signatures of being admixed with some ancestry similar to the Amhara and some ancestry similar to the Tikari. To date the timing of this admixture, we used the software DATES124. DATES uses the covariance of ancestry as a function of genetic distance between markers to estimate a time of admixture. We obtained the genetic distances between markers as described above in “Out-of-Africa Source Population”. We ran DATES version 753 using the parameters “binsize: 0.001”, “maxdis: 1.0”, “qbin: 10”, “afffit: yes”, and “lovalfit: 0.45”.
Divergence time estimates based on MSMC
The SNPs in our dataset were phased with SHAPEIT version 2.r837138 using the haplotypes of African populations in the 1000 Genomes Project phase 3104 as the reference panel (with parameters --no-mcmc, --input-ref, --include-grp AFR, -- effective-size 17469, -window 0.5). The heterozygous sites that were not reported in the 1000 Genomes Project were kept as unphased. We used a mutation rate 2 μ =2.5 × 10−8 mutations per nucleotide per generation and generation time g = 29 years in the MSMC analysis.
Identification of signatures of positive selection
We employed the di statistic139 to identify signals of positive selection in different populations. di statistics normalizes the Fst values between populations and identifies the most differential variants in each population.
where Fst (i,j) is the Fst value of an SNP site between populations, E[Fst(i, j)] and sd[Fst(i, j)] is the average and standard deviation of Fst value between populations. Here, we defined outliers falling in the 99.9th percentile of the empirical distribution of Di values as Di-SNPs. We functionally annotated the outlier SNPs using the Genomic Regions Enrichment of Annotations Tool (GREAT) version 3.0.111 We first run di using all 15 populations. Due to the recent divergence between Ju|’hoansi and !Xoo, we also performed di analysis using the merged Ju|’hoansi and !Xoo as a single group against other populations.
We also used integrated haplotype score (iHS) statistics140 to detect recent hard selection sweeps. The SNPs were phased with Shapeit4126 version 4.1.3 using the haplotypes of the 1000 Genomes project104 as a reference panel. We calculated iHS for every SNP with minor allele frequency > 5% within each population using selscan version 2.0.0127 with default parameters. The unstandardized integrated haplotype scores were normalized in frequency bins across the genome using norm module in selscan. We partitioned the genome to 100 kb non-overlapping windows. The top 1% of windows with the highest fraction of extreme integrated haplotype scores were defined as outliers.140 Using the genes located in the outlier windows in each population as query, we conducted GO enrichment tests using DAVID, an online functional annotation tool.128 We reported the GO ontologies with an FDR adjusted P-value <0.05.
Functional analyses of rs77665059 at the PDPK1 locus
Cell culture
MNT-1 cells (ATCC, #CRL-3450), were obtained from Dr. Michael S. Marks at Children’s Hospital of Philadelphia Research Institute, were grown in DMEM (Gibco, #11965084) supplemented with 20% Fetal Bovine Serum (FBS), 1% GlutaMAX (Gibco, #35050061), 1% NEAA (Gibco, #10370021),1% penicillin/streptomycin (Gibco, #15140122), and 10% AIM-V (Gibco, # 12055–091). Cells were transfected using Lipofectamine 3000 Transfection Reagent (Invitrogen, #L3000150).
WM88, melanocytic patient-derived melanoma tumor cell line, was obtained from Dr. Ashani Weeraratna at Wistar institute, Philadelphia, PA, were cultured in Tumor Specialized medium (80% MCDB153, 20% Leibovitz’s L-15, supplemented with 2% fetal bovine serum (FBS) and 1.68 mM CaCl2) at 37°C with 5% CO2 in a humidified incubator.
Luciferase reporter assay
The MNT-1 and WM88 cell lines were used for luciferase reporter assays. The cells were plated in 24-well plates at 0.1M per well, and 500 ng firefly luciferase plasmid, 20 ng pRL Renilla luciferase plasmid (Promega, # E2231) and 1.5 µL Lipofectamine 3000 (Invitrogen, #L3000150) were added to each well. 36 hours post transfection, luciferase activity was determined using the Dual-Luciferase Assay kit (Promega, # E1910) according to manufacturer instructions. The luminescence signal was detected in a white 96-well plate using SpectraMax i3x Multi-Mode Microplate Reader. The reporter gene activity of firefly luciferase was normalized to that of Renilla luciferase to determine the activity of functional elements.
Plasmid cloning
For the luciferase assay, human enhancer elements were cloned using genomic DNA extracted from MNT-1 cells. The amplified enhancer fragments were sequenced and ligated to PGL4.23 vector (Promega, #E8411) using Gibson assembly (NEB, #E2621). Candidate functional SNPs were introduced by mutated primers.
For CRISPR inhibition experiments, sgRNAs were designed using IDT (https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM) or CRISPOR (http://crispor.tefor.net/) and cloned into a pLKO5.sgRNA.EFS.GFP (Addgene, #57822) vector using FastDigest BsmBI (Fermentas) following the protocol (https://media.addgene.org/data/plasmids/52/52961/52961-attachment_B3xTwla0bkYD.pdf).
CRISPR mediated inhibition
To perform enhancer CRISPR inhibition, we first constructed MNT-1 cells stably expressing dCas9-KRAB-MeCP2 (Addgene, #110821). We produced dCas9-KRAB-MeCP2 (CRISPRi) lentiviruses following the published protocol141. Then, MNT-1 cells were infected with each virus with 8 µg/mL Polybrene (Sigma CatNo.H9268). For CRISPR inhibition of the enhancer, we cultured MNT-1-dCas9-KRAB-Mecp2 cells in 24-well plates at a density of 0.05M per well and cultured for 24h. We changed to fresh medium with 8 µg/mL Polybrene (MNT-1 cells) before infection. We added PLKO5-sgRNA (target to enhancer) virus at ~10 MOI, centrifuged at 1000g for 30min at 32 °C. 24hrs post infection, we replaced the medium using medium with Blasticidin (5 µg/mL), and changed the medium every 24hrs. 5 days after infection, we harvested the cells for total RNA extraction or melanin assay.
RT-qPCR
Total RNA was purified from all the cultured cells (CRISPR KO, CRISPR inhibition, Overexpression) using Direct-zol RNA Miniprep Kits (Zymo, R2052) following manufacturer’s instruction, and concentration was determined by a Nanodrop.
For RT-qPCR, 200–500 ng RNA was used for reverse transcription using M-MLV Reverse Transcriptase (Promega, # M1701) and Random Primer Mix (NEB, S1330). qPCR was conducted using Luna Universal qPCR Master Mix (NEB, M3003) on a QuantStudio 6 Flex Real-Time PCR machine.
Melanin Assay
MNT-1 cells were washed with PBS twice and detached by 0.25% trypsin. The cells were pelleted at 300g for 3 mins and supernatant was remove gently. The cell pellet was washed once with PBS and lysed in 200μL lysis buffer (50 mM Tris-HCl, pH 7.4, 2 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol) per million cells and vortexed 3 times, every 5 minutes. We centrifuged the lysate at 12,000g, 10 min, 4°C. We used 50 μL supernatant for protein quantification (BCA assay, Thermo Scientific, #23225). Then, 150 μL 2X Protease Lysis buffer (20mM Tris (pH 8), 200mM NaCl, 50mM EDTA, 1% SDS, 0.5 mg/mL protease K) was added to the 150 uL cell lysate to digest the pellet. We rotated at 65 °C for 5 hours and spun at 12,000g for 10 min at room temperature to collect melanin. We dissolved the melanin pellets in 0.45 mL 2M NaOH/20%DMSO and then incubated at 60°C for 30min with 850 rpm shaking. Once the melanin has fully dissolved (if not, we performed sonication for 5 mins), we vortexed to mix and read the absorbance at 450 nm. If necessary, we diluted the melanin with the buffer (2M NaOH/20%DMSO) so that the reading is less than 0.35.
Melanocyte epigenomic data
The chromatin accessibility data and ChIP-seq data of melanocytes or melanoma cells are collected from the Cistrome and ENCODE databases. The melanocyte data include DNase-Seq (Citrome: #41038), ATAC-Seq (Citrome: #79019), H3K27Ac ChIP-Seq (Citrome: #39849), H3K4Me1 ChIP-Seq (Citrome: #85888), H3K4Me3 ChIP-Seq (Citrome: #34310), MITF ChIP-Seq (Citrome: #42176). The melanoma (501-MEL) data include SOX10 ChIP-Seq (Citrome: #52549), MITF ChIP-Seq (Citrome: #52398), SMARCA4 ChIP-Seq (Citrome: #52555). The ENCODE cell line DNase data are from http://hgdownload.soe.ucsc.edu/gbdb/hg38/bbi/wgEncodeRegDnase/SNP frequency plots were plotted using R packages ("ggplot2", "ggrepel", "ggspatial", "sf", "scatterpie","tidyverse","data.table") in R version 4.1.1.
Supplementary Material
Supplementary Note. D-statistic analysis of African and non-African populations, related to STAR methods.
Figure S1. Frequencies of four annotated “Pathogenic” variants, related to Figure 1.
A: rs74853476-C is a splice donor mutation and was linked to Orthostatic hypotension 1. We found that the frequency of rs74853476-C reaches 13% in the Fulani but is nearly absent in other African populations in the current study and in populations from the gnomAD project. The X- and Y-axis shows the population name and the frequency of rs74853476-C of each population respectively.
B-D: rs139426141-G, rs140482516-T, rs34097903-A are three nonsynonymous mutations that were reported to associate with central centrifugal cicatricial alopecia in individuals of African ancestry. We found that the frequencies of these three mutations are > 5% in the African populations in our current study but are rare in the populations from the gnomAD database. The X- and Y-axis shows the population name and the frequencies of rs139426141-G, rs140482516-T, rs34097903-A in each population, respectively.
AFR, AMR, EAS, NFE, FIN, ASJ: samples of African, American, East Asian, European (non-Finnish), European, and Ashkenazi Jewish ancestry.
OTH: Others
We used K from 2 to 16 including the populations in the present study and SGDP. The SGDP dataset includes 251 and 108 non-African and African samples, respectively. We used a total of 12,443,243 SNPs in the ADMIXTURE analyses.
Figure S3. Treemix results when allowing number of migrations to range from 0 to 11, related to Figure 4.
Figure S4. qpgraph results when allowing number of migrations to range from 0 to 11, related to Figure 4.
We found that model (C) which had the San and RHG as a sister clade consistently had the highest likelihood when comparing the models using the RHG (A) or Ju|’hoansi (B) as outgroups, respectively.
A: divergence time between Ju|’hoansi and non-Khoesan-speaking, non-RHG populations. B: divergence time between !Xoo and non-Khoesan-speaking, non-RHG populations. C: divergence time between RHG and other populations. D: divergence time between NC- and NS-speaking populations. E: divergence time between NS- and AA-speaking populations. F: divergence time between AA- and NC-speaking populations. G: divergence time between the Hadza and AA-speaking populations. H: divergence time between the Sandawe and AA-speaking populations. I. divergence time between two NC-speaking populations, Herero and Tikari. J: divergence time between Ju|’hoansi and !Xoo. X- and Y-axis is the inferred divergence time and relative cross coalescence rate, respectively.
Model 1 has two populations sharing a common ancestor. We assumed a constant population size after the population split (upper boundary 100 kya) and inferred the divergence time and population sizes of populations 1 and 2 and the ancestral population. Model 2 assumes an unsampled “ghost” population splits from the ancestral population of populations 1 and 2 as well as a pulse of gene flow from the ghost population into the population ancestral to the two present-day populations. We inferred the same parameters as in model 1, but also the divergence time and the population size of the ghost population and the amount of pulse admixture from the ghost population into the ancestral population. Both Model 1 and Model 2 were fit using each possible pair of populations as the first and second population. We used models 3 and 4 to infer the divergence times between RHG and each non-Tikari, non-RHG population, due to strong gene flow between the RHG and Tikari. In model 3, RHG and population 1 each diverge from an ancestral population at their own inferred divergence time and having their own population sizes after divergence. At an inferred time, there is a pulse admixture of an inferred strength from the Tikari into RHG. The ancestral population has some size, which changes to the present-day size of the Tikari 100 kya. In this model, we infer the two divergence times, the four population sizes (three present day, one ancient), and the timing and strength of the pulse admixture event. Model 4 is identical to Model 3 but with a ghost population added in the same way as going from Model 1 to Model 2.
Table S1. 44 SNPs that were reported as "Pathogenic or likely Pathogenic" in the Clinvar database but were with non-reference allele frequency > 5% in at least one population in the present study, related to Figure 1.
Table S2. momi results based on pairwise comparison, related to Figure 4.
Table S3. GO enrichment results based on the Di-SNPs of different populations using GREAT, related to Figure 6.
Table S4. Frequencies of select Di-SNPs in the gnomAD database, related to Figure 6.
Table S5. 312 Di-SNPs in the RHG that are significant in previous GWAS of height in modern humans, related to Figure 6
Table S6. GO enrichment test results of the genes that were located in the outlier windows based on iHS analyses, related to Figure 6.
Highlights.
Whole genome sequencing of 180 Africans identifies millions of unreported variants
Complex African demographic history with ancient structure, admixture, and introgression
Southern and Central African hunter-gatherers share a unique ancient common ancestry
Signatures of local adaptation for skin color, immune response, height, and metabolism
Acknowledgments
We thank Huiru Sun and Shuhang Li for careful reading of the manuscript, Iain Mathieson at the University of Pennsylvania for discussing the analyses of ancient samples and participants who donated samples. We thank Michael S. Marks at Children’s Hospital of Philadelphia Research Institute and Ashani Weeraratna at Wistar institute for providing melanoma tumor cell lines. Research supported in part by NIH grants 1R35GM134957, R01AR076241, and ADA 1–19-VSN-02 (to S.A.T.), R35-GM134922 (to Y.S.S.) and the Penn Skin Biology and Diseases Resource-based Center, funded by NIH/NIAMS grant P30-AR069589 and the University of Pennsylvania Perelman School of Medicine. S.F. is supported by grants from the National Key R&D Program of China (2020YFE0201600 and 2021YFC2500202), National Natural Science Foundation of China (Grant No. 31970563), the 111 Project (B13016), and Shanghai Municipal Science and Technology (Grant No. 2017SHZDZX01, Grant No. 19410741100).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online
References
- 1.Hublin J-J, Ben-Ncer A, Bailey SE, Freidline SE, Neubauer S, Skinner MM, Bergmann I, Le Cabec A, Benazzi S, Harvati K, and Gunz P (2017). New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546, 289–292. 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 2.Beltrame MH, Rubel MA, and Tishkoff SA (2016). Inferences of African evolutionary history from genomic data. Curr. Opin. Genet. Dev 41, 159–166. 10.1016/j.gde.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. (2009). The genetic structure and history of Africans and African Americans. Science 324, 1035–1044. 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blench R (2006). Archaeology, Language, and the African Past (Rowman Altamira; ). [Google Scholar]
- 5.Heine B, and Nurse D (2000). African Languages: An Introduction (Cambridge University Press; ). [Google Scholar]
- 6.Ehret C (1983) Population Movement and Culture Contact in the Southern Sudan, c. 3000 BC to AD 1000. In Culture History in the Southern Sudan, Mack J and Robertshaw P, ed., (Memoire 8. Nairobi: British Institute in Eastern Africa; ) pp. 19–48. [Google Scholar]
- 7.Diamond J, and Bellwood P (2003). Farmers and their languages: the first expansions. Science 300, 597–603. 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 8.Pickrell JK, Patterson N, Barbieri C, Berthold F, Gerlach L, Güldemann T, Kure B, Mpoloka SW, Nakagawa H, Naumann C, et al. (2012). The genetic prehistory of southern Africa. Nat. Commun 3, 1143. 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elderkin ED (1982). SUGIA, Sprache und Geschichte in Afrika (Buske H.).
- 10.Schladt M (1998). Language, Identity, and Conceptualization Among the Khoisan (Köppe R).
- 11.Güldemann T, and Stoneking M (2008). A Historical Appraisal of Clicks: A Linguistic and Genetic Population Perspective. Annu. Rev. Anthropol 37, 93–109. 10.1146/annurev.anthro.37.081407.085109. [DOI] [Google Scholar]
- 12.Campbell MC, Hirbo JB, Townsend JP, and Tishkoff SA (2014). The peopling of the African continent and the diaspora into the new world. Curr. Opin. Genet. Dev 29, 120–132. 10.1016/j.gde.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell MC, and Tishkoff SA (2010). The evolution of human genetic and phenotypic variation in Africa. Curr. Biol 20, R166–173. 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan S, Hansen MEB, Lo Y, and Tishkoff SA (2016). Going global by adapting local: A review of recent human adaptation. Science 354, 54–59. 10.1126/science.aaf5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popejoy AB, and Fullerton SM (2016). Genomics is failing on diversity. Nature 538, 161–164. 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirugo G, Williams SM, and Tishkoff SA (2019). The Missing Diversity in Human Genetic Studies. Cell 177, 1080. 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, et al. (2021). Heart disease and stroke statistics-2021 update: A report from the American Heart Association. Circulation 143, e254–e743. 10.1161/CIR.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 18.Choudhury A, Aron S, Botigue LR, Sengupta D, Botha G, Bensellak T, Wells G, Kumuthini J, Shriner D, Fakim YJ, et al. (2020). High-depth African genomes inform human migration and health. Nature 586, 741–748. 10.1038/s41586-020-2859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurdasani D, Carstensen T, Fatumo S, Chen G, Franklin CS, Prado-Martinez J, Bouman H, Abascal F, Haber M, Tachmazidou I, et al. (2019). Uganda genome resource enables insights into population history and henomic discovery in Africa. Cell 179, 984–1002 e1036. 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhury A, Ramsay M, Hazelhurst S, Aron S, Bardien S, Botha G, Chimusa ER, Christoffels A, Gamieldien J, Sefid-Dashti MJ, et al. (2017). Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat. Commun 8, 2062. 10.1038/s41467-017-00663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan S, Kelly DE, Beltrame MH, Hansen MEB, Mallick S, Ranciaro A, Hirbo J, Thompson S, Beggs W, Nyambo T, et al. (2019). African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations. Genome Biol 20, 82. 10.1186/s13059-019-1679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HL, Ratan A, Perry GH, Montenegro A, Miller W, and Schuster SC (2014). Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat. Commun 5, 5692. 10.1038/ncomms6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo J-M, Lema G, Fu W, Nyambo TB, Rebbeck TR, et al. (2012). Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell 150, 457–469. 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorente-Galdos B, Lao O, Serra-Vidal G, Santpere G, Kuderna LFK, Arauna LR, Fadhlaoui-Zid K, Pimenoff VN, Soodyall H, Zalloua P, et al. (2019). Whole-genome sequence analysis of a Pan African set of samples reveals archaic gene flow from an extinct basal population of modern humans into sub-Saharan populations. Genome Biol 20, 77. 10.1186/s13059-019-1684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. (2010). Complete Khoisan and Bantu genomes from southern Africa. Nature 463, 943–947. 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff HE (2019). The Cambridge Handbook of African Linguistics (Cambridge University Press; ). [Google Scholar]
- 27.Gopalan S, Berl REW, Myrick JW, Garfield ZH, Reynolds AW, Bafens BK, Belbin G, Mastoras M, Williams C, Daya M, et al. (2022). Hunter-gatherer genomes reveal diverse demographic trajectories during the rise of farming in Eastern Africa. Curr. Biol 32, 1–9. 10.1016/j.cub.2022.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blench R, and Spriggs M (2003). Archaeology and language II: archaeological data and linguistic hypotheses (Routledge; ). [Google Scholar]
- 29.Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, and Tishkoff SA (2012). Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol 13, R1. 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold AC, Garland EM, Celedonio JE, Raj SR, Abumrad NN, Biaggioni I, Robertson D, Luther JM, and Shibao CA (2017). Hyperinsulinemia and insulin resistance in dopamine beta-hydroxylase deficiency. J. Clin. Endocrinol. Metab 102, 10–14. 10.1210/jc.2016-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malki L, Sarig O, Romano MT, Mechin MC, Peled A, Pavlovsky M, Warshauer E, Samuelov L, Uwakwe L, Briskin V, et al. (2019). Variant PADI3 in central centrifugal cicatricial alopecia. N. Engl. J. Med 380, 833–841. 10.1056/NEJMoa1816614. [DOI] [PubMed] [Google Scholar]
- 32.Vicente M, Priehodova E, Diallo I, Podgorna E, Poloni ES, Cerny V, and Schlebusch CM (2019). Population history and genetic adaptation of the Fulani nomads: inferences from genome-wide data and the lactase persistence trait. BMC Genomics 20, 915. 10.1186/s12864-019-6296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra-Vidal G, Lucas-Sanchez M, Fadhlaoui-Zid K, Bekada A, Zalloua P, and Comas D (2019). Heterogeneity in palaeolithic population continuity and neolithic expansion in North Africa. Curr. Biol 29, 3953–3959 e3954. 10.1016/j.cub.2019.09.050. [DOI] [PubMed] [Google Scholar]
- 34.Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, et al. (2017). Reconstructing prehistoric african population structure. Cell 171, 59–71.e21. 10.1016/j.cell.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. (2012). Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338, 374–379. 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49. 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Lachance J, Tishkoff SA, Hey J, and Xing J (2013). Apparent variation in Neanderthal admixture among African populations is consistent with gene flow from Non-African populations. Genome. Biol. Evol 5, 2075–2081. 10.1093/gbe/evt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh P, Veeramah KR, Lachance J, Tishkoff SA, Wall JD, Hammer MF, and Gutenkunst RN (2016). Whole-genome sequence analyses of Western Central African Pygmy hunter-gatherers reveal a complex demographic history and identify candidate genes under positive natural selection. Genome Res 26, 279–290. 10.1101/gr.192971.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry GH, Foll M, Grenier J-C, Patin E, Nédélec Y, Pacis A, Barakatt M, Gravel S, Zhou X, Nsobya SL, et al. (2014). Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers. Proc. Natl. Acad. Sci. USA 111, E3596–3603. 10.1073/pnas.1402875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintana-Murci L, Quach H, Harmant C, Luca F, Massonnet B, Patin E, Sica L, Mouguiama-Daouda P, Comas D, Tzur S, et al. (2008). Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc. Natl. Acad. Sci. USA 105, 1596–1601. 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipson M, Ribot I, Mallick S, Rohland N, Olalde I, Adamski N, Broomandkhoshbacht N, Lawson AM, Lopez S, Oppenheimer J, et al. (2020). Ancient West African foragers in the context of African population history. Nature 577, 665–670. 10.1038/s41586-020-1929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehret C (2001). Bantu expansions: re-envisioning a central problem of early African history. Int. J. Afr. Hist 34, 5–41. doi. 10.2307/3097285. [DOI] [Google Scholar]
- 43.Pakendorf B, de Filippo C, and Bostoen K (2011). Molecular perspectives on the Bantu expansion: a synthesis. Lang. Dyn. Chang 1, 50–88. 10.1163/221058211X570349. [DOI] [Google Scholar]
- 44.Phillipson DW (2005). African Archaeology (Cambridge University Press; ). [Google Scholar]
- 45.Hassan HY, Underhill PA, Cavalli-Sforza LL, and Ibrahim ME (2008). Y-hromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history. Am. J. Phys. Anthropol 137, 316–323. 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- 46.Blench R (1999). The westward wanderings of Cushitic pastoralists: explorations in the prehistory of Central Africa In ĽHomme et l'animal dans le bassin du lac Tchad Paris, B.J. Baroin C, ed. pp. 39–80.
- 47.Ehret C (2001). An African classical age: eastern and southern Africa in world history, 1000 BC to AD 400 (Rutgers University Press; ). [Google Scholar]
- 48.Durvasula A, and Sankararaman S (2020). Recovering signals of ghost archaic introgression in African populations. Sci. Adv 6, eaax5097. 10.1126/sciadv.aax5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ragsdale AP, and Gravel S (2019). Models of archaic admixture and recent history from two-locus statistics. PLoS Genet 15, e1008204. 10.1371/journal.pgen.1008204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canela-Xandri O, Rawlik K, and Tenesa A (2018). An atlas of genetic associations in UK Biobank. Nat. Genet 50, 1593–1599. 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh PR, Kichaev G, Gazal S, Schoech AP, and Price AL (2018). Mixed-model association for biobank-scale datasets. Nat. Genet 50, 906–908. 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. (2017). Loci associated with skin pigmentation identified in African populations. Science 358, eaan8433. 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gronskov K, Ek J, and Brondum-Nielsen K (2007). Oculocutaneous albinism. Orphanet. J. Rare. Dis 2, 43. 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tietz W (1963). A syndrome of deaf-mutism associated with albinism showing dominant autosomal inheritance. Am. J. Hum. Genet 15, 259–264. [PMC free article] [PubMed] [Google Scholar]
- 55.Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Moller M, Sandhu MS, et al. (2017). An unexpectedly complex architecture for skin pigmentation in Africans. Cell 171, 1340–1353 e1314. 10.1016/j.cell.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adhikari K, Mendoza-Revilla J, Sohail A, Fuentes-Guajardo M, Lampert J, Chacon-Duque JC, Hurtado M, Villegas V, Granja V, Acuna-Alonzo V, et al. (2019). A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat. Commun 10, 358. 10.1038/s41467-018-08147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lona-Durazo F, Hernandez-Pacheco N, Fan S, Zhang T, Choi J, Kovacs MA, Loftus SK, Le P, Edwards M, Fortes-Lima CA, et al. (2019). Meta-analysis of GWA studies provides new insights on the genetic architecture of skin pigmentation in recently admixed populations. BMC Genet 20, 59. 10.1186/s12863-019-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium GTEx (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585. 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scortegagna M, Ruller C, Feng Y, Lazova R, Kluger H, Li JL, De SK, Rickert R, Pellecchia M, Bosenberg M, and Ronai ZA (2014). Genetic inactivation or pharmacological inhibition of Pdk1 delays development and inhibits metastasis of Braf(V600E)::Pten(-/-) melanoma. Oncogene 33, 4330–4339. 10.1038/onc.2013.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris ML, Baxter LL, Loftus SK, and Pavan WJ (2010). Sox proteins in melanocyte development and melanoma. Pigment. Cell. Melanoma. Res 23, 496–513. 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy C, Khaled M, and Fisher DE (2006). MITF: master regulator of melanocyte development and melanoma oncogene. Trends. Mol. Med 12, 406–414. 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Ribot I (2004). Differentiation of modern sub-Saharan African populations: craniometric interpretations in relation to geography and history. Bull. Mem. Soc. Anthropol. Paris 16 (3–4), 143–165 [Google Scholar]
- 63.Tobias PV, and Biesele M (1978). The Bushmen: San hunters and herders of southern Africa (Human & Rousseau; ). [Google Scholar]
- 64.Shimomura Y, Wajid M, Petukhova L, Kurban M, and Christiano AM (2010). Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am. J. Hum. Genet 86, 632–638. 10.1016/j.ajhg.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarvis JP, Scheinfeldt LB, Soi S, Lambert C, Omberg L, Ferwerda B, Froment A, Bodo J-M, Beggs W, Hoffman G, et al. (2012). Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet 8, e1002641. 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. (2010). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838. 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suemoto H, Muragaki Y, Nishioka K, Sato M, Ooshima A, Itoh S, Hatamura I, Ozaki M, Braun A, Gustafsson E, and Fassler R (2007). Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev. Biol 312, 572–581. 10.1016/j.ydbio.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Chen G, Deng C, and Li YP (2012). TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci 8, 272–288. 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sirmaci A, Spiliopoulos M, Brancati F, Powell E, Duman D, Abrams A, Bademci G, Agolini E, Guo S, Konuk B, et al. (2011). Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am. J. Hum. Genet 89, 289–294. 10.1016/j.ajhg.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet 46, 1173–1186. 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM, and Consortium G (2018). Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet 27, 3641–3649. 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Modiano D, Petrarca V, Sirima BS, Nebie I, Diallo D, Esposito F, and Coluzzi M (1996). Different response to Plasmodium falciparum malaria in west African sympatric ethnic groups. Proc. Natl. Acad. Sci. USA 93, 13206–13211. 10.1073/pnas.93.23.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seck MC, Thwing J, Badiane AS, Rogier E, Fall FB, Ndiaye PI, Diongue K, Mbow M, Ndiaye M, Diallo MA, et al. (2020). Analysis of anti-Plasmodium IgG profiles among Fulani nomadic pastoralists in northern Senegal to assess malaria exposure. Malar. J 19, 15. 10.1186/s12936-020-3114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bostrom S, Giusti P, Arama C, Persson JO, Dara V, Traore B, Dolo A, Doumbo O, and Troye-Blomberg M (2012). Changes in the levels of cytokines, chemokines and malaria-specific antibodies in response to Plasmodium falciparum infection in children living in sympatry in Mali. Malar. J 11, 109. 10.1186/1475-2875-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quin JE, Bujila I, Cherif M, Sanou GS, Qu Y, Vafa Homann M, Rolicka A, Sirima SB, O'Connell MA, Lennartsson A, et al. (2017). Major transcriptional changes observed in the Fulani, an ethnic group less susceptible to malaria. Elife 6, e29156. 10.7554/eLife.29156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xing S, and Ferrari de Andrade L (2020). NKG2D and MICA/B shedding: a ťag game' between NK cells and malignant cells. Clin. Transl. Immunology 9, e1230. 10.1002/cti2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergey CM, Lopez M, Harrison GF, Patin E, Cohen JA, Quintana-Murci L, Barreiro LB, and Perry GH (2018). Polygenic adaptation and convergent evolution on growth and cardiac genetic pathways in African and Asian rainforest hunter-gatherers. Proc. Natl. Acad. Sci. USA 115, E11256–E11263. 10.1073/pnas.1812135115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wood BM, Harris JA, Raichlen DA, Pontzer H, Sayre K, Sancilio A, Berbesque C, Crittenden AN, Mabulla A, McElreath R, et al. (2021). Gendered movement ecology and landscape use in Hadza hunter-gatherers. Nat. Hum. Behav 5, 436–446. 10.1038/s41562-020-01002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, and Olson EN (2014). Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. USA 111, 4109–4114. 10.1073/pnas.1401732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colasanto MP, Eyal S, Mohassel P, Bamshad M, Bonnemann CG, Zelzer E, Moon AM, and Kardon G (2016). Development of a subset of forelimb muscles and their attachment sites requires the ulnar-mammary syndrome gene Tbx3. Dis. Model. Mech 9, 1257–1269. 10.1242/dmm.025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang X, Yang S, Wang C, and Kuang S (2017). The hypoxia-inducible factors HIF1alpha and HIF2alpha are dispensable for embryonic muscle development but essential for postnatal muscle regeneration. J. Biol. Chem 292, 5981–5991. 10.1074/jbc.M116.756312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iseki S, Wilkie AO, and Morriss-Kay GM (1999). Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development 126, 5611–5620. 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- 83.Seo HS, and Serra R (2009). Tgfbr2 is required for development of the skull vault. Dev. Biol 334, 481–490. 10.1016/j.ydbio.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singh MK, Petry M, Haenig B, Lescher B, Leitges M, and Kispert A (2005). The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev 122, 131–144. 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 85.Bildsoe H, Fan X, Wilkie EE, Ashoti A, Jones VJ, Power M, Qin J, Wang J, Tam PPL, and Loebel DAF (2016). Transcriptional targets of TWIST1 in the cranial mesoderm regulate cell-matrix interactions and mesenchyme maintenance. Dev. Biol 418, 189–203. 10.1016/j.ydbio.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 86.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, and Rotimi C (2009). A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet 5, e1000564. 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marshall MC Jr. (2005). Diabetes in African Americans. Postgrad. Med. J 81, 734–740. 10.1136/pgmj.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, Tin A, Wang L, Chu AY, Hoppmann A, et al. (2019). A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet 51, 957–972. 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McQuillan MA, Ranciaro A, Hansen MEB, Fan S, Beggs W, Belay G, Woldemeskel D, and Tishkoff SA (2022). Signatures of convergent evolution and natural selection at the alcohol dehydrogenase gene region are correlated with agriculture in ethnically diverse Africans. Mol. Biol. Evol 39. 10.1093/molbev/msac183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scheinfeldt LB, Soi S, Lambert C, Ko WY, Coulibaly A, Ranciaro A, Thompson S, Hirbo J, Beggs W, Ibrahim M, et al. (2019). Genomic evidence for shared common ancestry of East African hunting-gathering populations and insights into local adaptation. Proc. Natl. Acad. Sci. USA 116(10), 4166–4175. 10.1073/pnas.1817678116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, and Kohane IS (2016). Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375, 655–665. 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. (2012). A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226. 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schlebusch CM, Malmström H, Günther T, Sjödin P, Coutinho A, Edlund H, Munters AR, Vicente M, Steyn M, Soodyall H, et al. (2017). Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358(6363), 652–655. 10.1126/science.aao6266. [DOI] [PubMed] [Google Scholar]
- 94.Lipson M, Sawchuk EA, Thompson JC, Oppenheimer J, Tryon CA, Ranhorn KL, de Luna KM, Sirak KA, Olalde I, Ambrose SH, et al. (2022). Ancient DNA and deep population structure in sub-Saharan African foragers. Nature 603, 290–296. 10.1038/s41586-022-04430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wohns AW, Wong Y, Jeffery B, Akbari A, Mallick S, Pinhasi R, Patterson N, Reich D, Kelleher J, and McVean G (2022). A unified genealogy of modern and ancient genomes. Science 375, eabi8264. 10.1126/science.abi8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonder MK, Mortensen HM, Reed FA, de Sousa A, and Tishkoff SA (2007). Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol 24, 757–768. 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 97.Prendergast ME, Lipson M, Sawchuk EA, Olalde I, Ogola CA, Rohland N, Sirak KA, Adamski N, Bernardos R, Broomandkhoshbacht N, et al. (2019). Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science 365, eaaw6275. 10.1126/science.aaw6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gallego Llorente M, Jones ER, Eriksson A, Siska V, Arthur KW, Arthur JW, Curtis MC, Stock JT, Coltorti M, Pieruccini P, et al. (2015). Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820–822. 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 99.Černý V, Pereira L, Musilová E, Kujanová M, Vašíková A, Blasi P, Garofalo L, Soares P, Diallo I, Brdička R, and Novelletto A (2011). Genetic structure of pastoral and farmer populations in the African Sahel. Mol. Biol. Evol 28, 2491–2500. 10.1093/molbev/msr067. [DOI] [PubMed] [Google Scholar]
- 100.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, and Sirotkin K (2001). dbSNP: the NCBI database of genetic variation. Nucleic. Acids. Res 29, 308–311. 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Encode Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. (2016). The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206. 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, and Reich D (2012). Ancient admixture in human history. Genetics 192, 1065–1093. 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kamm JA, Terhorst J, and Song YS (2017). Efficient computation of the joint sample frequency spectra for multiple populations. J. Comput. Graph. Stat 26, 182–194. 10.1080/10618600.2016.1159212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kamm J, Terhorst J, Durbin R, and Song YS (2020). Efficiently inferring the demographic history of many populations with allele count data. J. Am. Stat. Assoc 115, 1472–1487. 10.1080/01621459.2019.1635482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, and Durbin R (2011). Inference of human population history from individual whole-genome sequences. Nature 475, 493–496. 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Terhorst J, Kamm JA, and Song YS (2017). Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet 49, 303–309. 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, and Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang K, Li M, and Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids. Res 38, e164. 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Patterson N, Price AL, and Reich D (2006). Population structure and eigenanalysis. PLoS Genet 2, e190. 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, and Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38, 904–909. 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 116.Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, and DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alexander DH, Novembre J, and Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–1664. 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Faust GG, and Hall IM (2014). SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505. 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, and Korbel JO (2012). DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339. 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jakobsson M, and Rosenberg NA (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 122.Tamura K, Stecher G, and Kumar S (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol 38, 3022–3027. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schiffels S, and Durbin R (2014). Inferring human population size and separation history from multiple genome sequences. Nat. Genet 46, 919–925. 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, et al. (2019). The formation of human populations in South and Central Asia. Science 365, eaat7487. 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pickrell JK, and Pritchard JK (2012). Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8, e1002967. 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Delaneau O, Zagury JF, Robinson MR, Marchini JL, and Dermitzakis ET (2019). Accurate, scalable and integrative haplotype estimation. Nat. Commun 10, 5436. 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szpiech ZA, and Hernandez RD (2014). selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol 31, 2824–2827. 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, and Chang W (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic. Acids. Res 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed]
- 129.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mills RE, Pittard WS, Mullaney JM, Farooq U, Creasy TH, Mahurkar AA, Kemeza DM, Strassler DS, Ponting CP, Webber C, and Devine SE (2011). Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res 21, 830–839. 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H (2014). Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851. 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, and Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.OĽeary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic. Acids. Res 44, D733–745. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic. Acids. Res 46, D1062–D1067. 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. (2022). Ensembl 2022. Nucleic. Acids. Res 50, D988–D995. 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spence JP, and Song YS (2019). Inference and analysis of population-specific fine-scale recombination maps across 26 diverse human populations. Sci. Adv 5, eaaw9206. 10.1126/sciadv.aaw9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Delaneau O, Zagury JF, and Marchini J (2013). Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6. 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 139.Akey JM, Ruhe AL, Akey DT, Wong AK, Connelly CF, Madeoy J, Nicholas TJ, and Neff MW (2010). Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. USA 107, 1160–1165. 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Voight BF, Kudaravalli S, Wen X, and Pritchard JK (2006). A map of recent positive selection in the human genome. PLoS Biol 4, e72. 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gordon MG, Inoue F, Martin B, Schubach M, Agarwal V, Whalen S, Feng S, Zhao J, Ashuach T, Ziffra R, et al. (2020). lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nat. Protoc 15, 2387–2412. 10.1038/s41596-020-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note. D-statistic analysis of African and non-African populations, related to STAR methods.
Figure S1. Frequencies of four annotated “Pathogenic” variants, related to Figure 1.
A: rs74853476-C is a splice donor mutation and was linked to Orthostatic hypotension 1. We found that the frequency of rs74853476-C reaches 13% in the Fulani but is nearly absent in other African populations in the current study and in populations from the gnomAD project. The X- and Y-axis shows the population name and the frequency of rs74853476-C of each population respectively.
B-D: rs139426141-G, rs140482516-T, rs34097903-A are three nonsynonymous mutations that were reported to associate with central centrifugal cicatricial alopecia in individuals of African ancestry. We found that the frequencies of these three mutations are > 5% in the African populations in our current study but are rare in the populations from the gnomAD database. The X- and Y-axis shows the population name and the frequencies of rs139426141-G, rs140482516-T, rs34097903-A in each population, respectively.
AFR, AMR, EAS, NFE, FIN, ASJ: samples of African, American, East Asian, European (non-Finnish), European, and Ashkenazi Jewish ancestry.
OTH: Others
We used K from 2 to 16 including the populations in the present study and SGDP. The SGDP dataset includes 251 and 108 non-African and African samples, respectively. We used a total of 12,443,243 SNPs in the ADMIXTURE analyses.
Figure S3. Treemix results when allowing number of migrations to range from 0 to 11, related to Figure 4.
Figure S4. qpgraph results when allowing number of migrations to range from 0 to 11, related to Figure 4.
We found that model (C) which had the San and RHG as a sister clade consistently had the highest likelihood when comparing the models using the RHG (A) or Ju|’hoansi (B) as outgroups, respectively.
A: divergence time between Ju|’hoansi and non-Khoesan-speaking, non-RHG populations. B: divergence time between !Xoo and non-Khoesan-speaking, non-RHG populations. C: divergence time between RHG and other populations. D: divergence time between NC- and NS-speaking populations. E: divergence time between NS- and AA-speaking populations. F: divergence time between AA- and NC-speaking populations. G: divergence time between the Hadza and AA-speaking populations. H: divergence time between the Sandawe and AA-speaking populations. I. divergence time between two NC-speaking populations, Herero and Tikari. J: divergence time between Ju|’hoansi and !Xoo. X- and Y-axis is the inferred divergence time and relative cross coalescence rate, respectively.
Model 1 has two populations sharing a common ancestor. We assumed a constant population size after the population split (upper boundary 100 kya) and inferred the divergence time and population sizes of populations 1 and 2 and the ancestral population. Model 2 assumes an unsampled “ghost” population splits from the ancestral population of populations 1 and 2 as well as a pulse of gene flow from the ghost population into the population ancestral to the two present-day populations. We inferred the same parameters as in model 1, but also the divergence time and the population size of the ghost population and the amount of pulse admixture from the ghost population into the ancestral population. Both Model 1 and Model 2 were fit using each possible pair of populations as the first and second population. We used models 3 and 4 to infer the divergence times between RHG and each non-Tikari, non-RHG population, due to strong gene flow between the RHG and Tikari. In model 3, RHG and population 1 each diverge from an ancestral population at their own inferred divergence time and having their own population sizes after divergence. At an inferred time, there is a pulse admixture of an inferred strength from the Tikari into RHG. The ancestral population has some size, which changes to the present-day size of the Tikari 100 kya. In this model, we infer the two divergence times, the four population sizes (three present day, one ancient), and the timing and strength of the pulse admixture event. Model 4 is identical to Model 3 but with a ghost population added in the same way as going from Model 1 to Model 2.
Table S1. 44 SNPs that were reported as "Pathogenic or likely Pathogenic" in the Clinvar database but were with non-reference allele frequency > 5% in at least one population in the present study, related to Figure 1.
Table S2. momi results based on pairwise comparison, related to Figure 4.
Table S3. GO enrichment results based on the Di-SNPs of different populations using GREAT, related to Figure 6.
Table S4. Frequencies of select Di-SNPs in the gnomAD database, related to Figure 6.
Table S5. 312 Di-SNPs in the RHG that are significant in previous GWAS of height in modern humans, related to Figure 6
Table S6. GO enrichment test results of the genes that were located in the outlier windows based on iHS analyses, related to Figure 6.
Data Availability Statement
The SNP data are publicly available through the dbGAP database (accession number: phs003096.v1.p1). Links to the software and algorithms in the present study were listed in the key resources table.
Key resources table


