Abstract
Advances in genomics have enabled the development of polygenic scores (PGS), sometimes called polygenic risk scores, in the context of multifactorial diseases and disorders such as cancer, cardiovascular disease, and schizophrenia. PGS estimate an individual’s genetic predisposition, as compared to other members of a population, for conditions which are influenced by both genetic and environmental factors. There is significant interest in using genetic risk prediction afforded through PGS in public health, clinical care, and research settings, yet many acknowledge the need to thoughtfully consider and address ethical, legal, and social implications (ELSI). To contribute to this effort, this paper reports on a narrative review of the literature, with the aim of identifying and categorizing ELSI relating to genetic risk prediction in the context of multifactorial disease, which have been raised by scholars in the field. Ninety-two articles, spanning from 1977 to 2021, met the inclusion criteria for this study. Identified ELSI included potential benefits, challenges and risks that focused on concerns about interpretation and use, and ethical obligations to maximize benefits, minimize risks, promote justice, and support autonomy. This research will support geneticists, clinicians, genetic counselors, patients, patient advocates, and policymakers in recognizing and addressing ethical concerns associated with PGS; it will also guide future empirical and normative research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12687-022-00625-9.
Keywords: Polygenic risk score, Polygenic score, Ethics, Multifactorial disease, Multifactorial inheritance, Genetic testing
Introduction
Ethical, legal, and social implications (ELSI) of genetic tests in the context of monogenic (Mendelian) disorders, in which rare variants in a particular gene underlie a disease, have received significant attention and analysis (Berliner 2014; Nussbaum et al. 2015), predominantly using the four principles framework (Beauchamp and Childress 2013). Autonomy-related issues include privacy and shared decision-making about undergoing genetic testing (Stiles and Appelbaum 2019). Regarding beneficence and non-maleficence, the pros and cons of testing are context-dependent: there is potential for anxiety, stigma, guilt, and discrimination, but results may guide care. However, utility of testing is not limited to clinical actionability, as it may enrich self or family knowledge, inform reproductive decision making, or enable connections to supportive communities (Grosse and Khoury 2006). Justice concerns include equitable access (Delikurt et al. 2015) and appropriate allocation of resources to address social as well as genetic/biological determinants of health (Artiga and Hinton 2018).
Although ELSI analysis has predominantly focused on monogenic disorders, most human traits including disease susceptibility are not only polygenic (arising from many genes) but also multifactorial, meaning that the probability and strength (expressivity) of the phenotype are influenced by variation in many genes as well as environmental and lifestyle factors. ELSI related to genetic testing in the context of common multifactorial diseases such as diabetes and cardiovascular disease are less developed, but recent advances in polygenic risk prediction make research in this area increasingly important (National Human Genome Research Institute 2020a; Polygenic Risk Score Task Force of the International Common Disease Alliance 2021).
Polygenic risk prediction relies on genome-wide association studies (GWAS), which identify associations between single nucleotide polymorphisms (SNPs, variants at specific positions in the genome), and human diseases or traits (McCarthy et al. 2008; National Human Genome Research Institute 2020b). To conduct GWAS, investigators leverage the availability of large cohorts of genotyped individuals where phenotypes for particular quantitative or dichotomous traits (cases and controls) are known. Early GWAS research was disappointing, as effect sizes of identified SNPs did not match expected disease heritability; this became known as the “missing heritability” problem (Crouch and Bodmer 2020; McCarthy et al. 2008; Torkamani et al. 2018). However, as sample sizes and statistical power increased, more variants with small (even nonsignificant) effect sizes were identified, which, when combined, made genetic risk prediction for multifactorial traits feasible (Crouch and Bodmer 2020; Khoury 2003; Pharoah et al. 2002; Purcell et al. 2009; Ronald 2020; Yang et al. 2010).
Two 2018 papers intensified the enthusiasm for polygenic scores (PGS), also known as polygenic risk scores (PRS), for major common diseases (Inouye et al. 2018; Khera et al. 2018). PGS “aggregate the contribution of an individual’s germline genome into a single number proportional to the risk for a given disease”(Lambert et al. 2019). Since then, many publications have reported on PGS for a variety of multifactorial diseases and phenotypes (Cattarinussi et al. 2022; Dixon et al. 2022; Fusar-Poli et al. 2022; Klarin and Natarajan 2022; Meerman et al. 2022), evidencing significant interest in this approach and reflecting its accessibility in terms of cost (Lambert et al. 2019; Murray et al. 2021; Polygenic Risk Score Task Force of the International Common Disease Alliance 2021).
There is considerable hope for the potential of genetic risk prediction for multifactorial disease, specifically PGS, to improve clinical care and public health (National Human Genome Institute 2020a) and a recognized need to thoughtfully consider and address the associated ELSI (Polygenic Risk Score Task Force of the International Common Disease Alliance 2021). To contribute to this effort, this paper reports on a narrative review of the literature, with the aim of identifying and categorizing ELSI raised by scholars related to genetic risk prediction in the context of multifactorial disease, predominantly PGS. This research will be useful to researchers, healthcare professionals, patients, advocates, and policymakers in framing and addressing ELSI associated with PGS, as well as to guide future research.
Methods
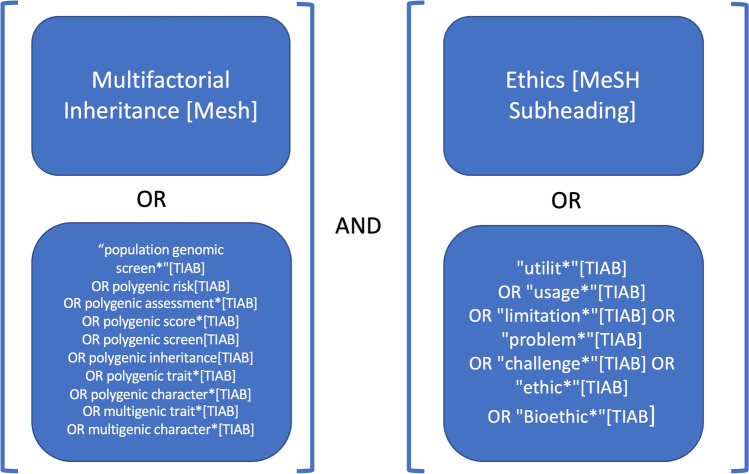
PubMed and PsychInfo databases were searched in January 2022 using a combination of Medical Subject Headings [MeSH] and title/abstract terms [tiab] on multifactorial inheritance and ethics (see Fig. 1). Using this strategy, 1258 unique articles were identified. Titles and abstracts were screened for selection. To be included, articles should have relevance to polygenic scores or genetic risk prediction of multifactorial human disease, and mentioned ELSI. Articles not written in English, only addressing scientific issues, or focused on agriculture, model organisms, forensic testing, or behavioral genetics (e.g., educational attainment) were excluded.
Fig. 1.
Complete literature search strategy. MeSH stands for Medical Subject Headings and TIAB stands for TItle/ABstract
Articles meeting the inclusion criteria (see Supplementary File 1) were read and highlighted, and a reflexive thematic analysis was conducted (Braun and Clarke 2006, 2014, 2021). Representative quotes addressing ELSI (data) were extracted from articles and iteratively organized into categories using Microsoft Word and Excel. The four principles (Beauchamp and Childress 2013)and Ethical Requirements for Clinical Research (Emanuel et al. 2000) were employed as deductive ELSI frameworks, but categories were also identified inductively. Finally, data were coded in Excel with a basic codebook representing 11 identified high-level themes. More than half of the quotes received two or more codes. Due to space constraints, ELSI specific to reproductive uses (one identified theme) will not be reported in this manuscript.
Results
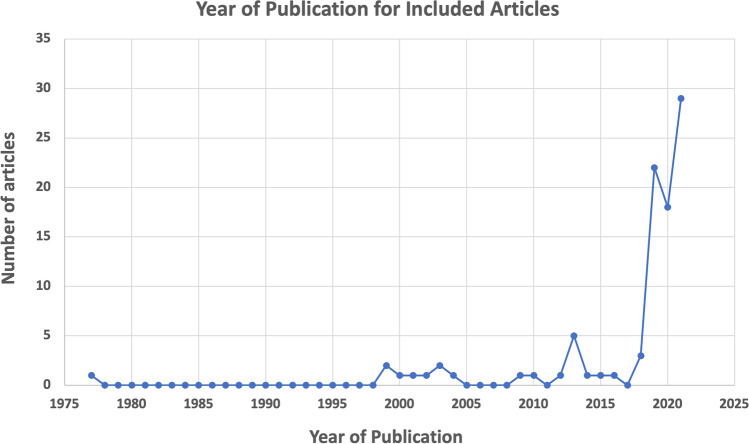
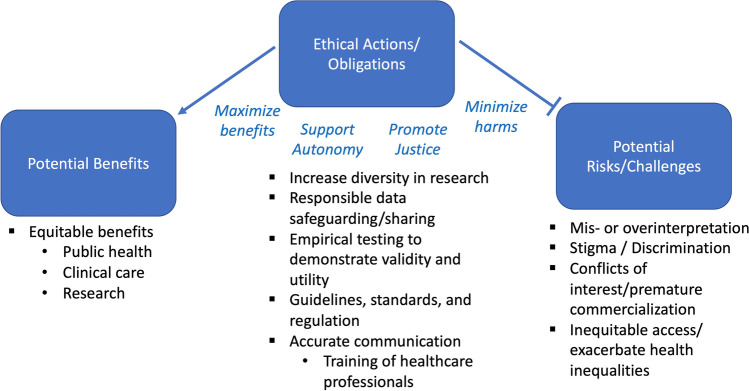
Ninety-two articles spanning 1977–2021 were reviewed and thematically analyzed (Braun and Clarke 2006, 2014, 2021) for ELSI issues related to genetic risk prediction in the context of multifactorial disease (see Fig. 2). Earlier papers were more theoretical, but more recent papers in the set focus on PGS, which has received considerable attention since 2018. Many articles addressed a specific therapeutic area (e.g., psychiatry, cardiovascular disease, and cancer). Most ELSI concerns apply across therapeutic areas, but psychiatric and reproductive applications were viewed as particularly sensitive. Ten identified ELSI themes of polygenic risk prediction, discussed below, are organized into three groups: potential benefits; challenges and risks; and ethical actions and obligations (see Fig. 3).
Fig. 2.
Year of publication for articles meeting the inclusion criteria
Fig. 3.
Ten identified ELSI themes for genetic risk prediction for multifactorial disease, i.e., PGS, fall into three groups: potential benefits, potential risks and challenges, and ethical actions and obligations. ELSI specific to reproductive uses was also identified as a theme but is not reported in this manuscript. The four classic bioethics principles--beneficence, autonomy, justice and non-maleficence (Beauchamp and Childress 2013)--are represented with words in blue text
Potential benefits
Potential benefits of genetic risk prediction for multifactorial disease, i.e., PGS, were identified as a prominent ELSI theme in the literature. PGS may improve healthcare in three different settings: public health (population screening for early detection of disease risk), clinical care (guiding individual care), and research (e.g., testing investigational drugs on patients with high genetic risk) (Yanes et al. 2020a). In public health, determination of genetic risk for multifactorial disease may guide adjustments to screening prioritization or protocols, which may optimize resource allocation and/or allow for earlier diagnosis or identification of individuals with high genetic predisposition. Earlier diagnosis may increase opportunities to prevent or delay multifactorial disease via lifestyle changes, prophylactic treatments, or avoidance of risk-increasing environmental factors (Briggs and Slade 2019; Chaudhari et al. 2020; Driver et al. 2020; Ganna et al. 2013; Hall et al. 2004; Lambert et al. 2019; Palk et al. 2019; Pashayan et al. 2013; Rubin and Glusman 2019; Yanes et al. 2020b). As compared with other clinical risk factors, an advantage of PGS, which provides an estimate of germline (inherited) genetic-based risk, is that it can be assessed very early in human life (Torkamani et al. 2018). Also, significantly larger numbers of people with high genetic risk for multifactorial disease can potentially be identified with PGS than with monogenic testing (Lewis and Vassos 2020; Palk et al. 2019; Tellier et al. 2021; Zeinomar and Chung 2021).
Other possible benefits of PGS are to improve individual clinical care by clarifying diagnosis, prognosis, or guiding treatments (Duncan et al. 2019; Lambert et al. 2019; Lewis and Vassos 2020; Murray et al. 2021; Zeinomar and Chung 2021). For example, addition of polygenic risk may refine risk for individuals undergoing testing for monogenic risk variants. Interestingly, PGS may provide insights into genetic overlap as well as differentiation between phenotypes and diseases (DiBlasi et al. 2021; Fullerton and Nurnberger 2019; Martin et al. 2019a; Ronald 2020; Yanes et al. 2020a). PGS may also be useful in clinical research (Slunecka et al. 2021; Weinberger 2019; Zhou et al. 2021); for example, efficacy of drugs can be specifically evaluated in cohorts of patients with high genetic risk (Manrique de Lara et al. 2019; Martin et al. 2019a).
The predictive power of PGS is also expected to improve over time due to increasing size of GWAS datasets and methodological improvements (Karavani et al. 2019; Lambert et al. 2019; Martin et al. 2019b; Ronald 2020). To increase predictive power, models may combine PGS with risk from rare variants typically not included (Briggs and Slade 2019; Choi et al. 2020; Fabbri and Serretti 2020; Fullerton and Nurnberger 2019; Lambert et al. 2019; Rashkin et al. 2019; Yanes et al. 2020a) or other factors like health conditions or family history (Lambert et al. 2019; Polygenic Risk Score Task Force of the International Common Disease Alliance 2021; Ronald 2020).
Potential risks and challenges
Four identified themes, potential risks and challenges of PGS, express concerns about interpretation and use: the potential for mis- or over-interpretation, stigma and discrimination, conflicts of interest and premature commercialization, and inequitable access to potential benefits.
Potential for mis- or overinterpretation
A significant challenge for PGS is the potential for mis- or over-interpretation by healthcare providers, patients, and consumers (Torkamani et al. 2018), which may lead to inappropriate actions such as overdiagnosis or overtreatment (Lewis and Green 2021). General understanding of genetics may not translate into an ability to interpret PGS, the simplicity of which, provided as numbers or percentiles, belies its complexity (Driver et al. 2020). Studies have demonstrated that individuals struggle with probability literacy and risk numeracy, and that recipients of health information understand absolute risks better than relative risks (Peay 2020; Slunecka et al. 2021). Also, inappropriate expectations “for distinctly separated phenotypic subpopulations” may be anticipated from a monogenic paradigm (Ozdemir et al. 2009). PGS do not perfectly differentiate individuals with and without disease: the mean score for cases will be higher than for controls (Pashayan et al. 2013), but cases and controls can have low and high scores, respectively (Fullerton and Nurnberger 2019; Lambert et al. 2019). Individuals with extremely high PGS are at higher genetic risk for a disease, but if the absolute risk for the disease is very low, this increased risk may not be clinically meaningful (Rosenberg et al. 2019).
Another article astutely points out that genetic testing for complex diseases raises “fundamental questions of what should be considered a ‘genetic’ disorder in the first place, and what the psychological, social, ethical, and legal implications are of labeling a common condition like heart disease as ‘genetic’” (Andrews and Zuiker 2003). PGS may actually change how we perceive multifactorial disease; instead of a binary state (someone has a disease or doesn’t), it could be viewed as more of a continuum (Lewis and Green 2021). After learning about genetic predisposition for multifactorial disease, people may inappropriately discount environmental and lifestyle influences on diseases (Driver et al. 2020; Peay 2020; Warren 2018).
The potential for gene-environment interactions may also be overlooked or underappreciated. Although PGS intends to measure genetic contributions to a particular phenotype, some variant-associated effects may be conferred by gene-environment correlations (Ronald 2020). Although controlling for and separating genetic and environmental effects in PGS is a “long-studied issue… it is still not a ‘solved’ problem in genetic studies… [and] care is needed to ensure this powerful tool is applied appropriately” (Blanc and Berg 2020). Appreciation for the limitations of polygenic scores is also required when interpreting phenotype differences across populations: “genetic contributions to traits, as estimated by polygenic scores, combine with environmental contributions so that differences among populations in trait distributions need not reflect corresponding differences in genetic propensity” (Rosenberg et al. 2019).
Overinterpretation is also possible (Curtis 2019). Some caution that even though PGS “demonstrate the importance of genetic variation in the etiology of the disorders,” this does not validate “the value of the risk score proposal in disease prediction (i.e., screening)” (Wald and Old 2019). Although moderate relative risks (e.g., three- to sixfold) “can have considerable significance in determining causes of disease… estimates of the relative risk between a disease marker and a disease have to be extremely high for the risk factor to merit consideration as a worthwhile screening test” (Wald and Old 2019). In these authors’ estimation, PGS will not meet this requirement, and they warn: “it is important that the potential applications of genomic medicine are not compromised by raising unrealistic expectations in medical screening” (Wald and Old 2019).
Potential for stigma and discrimination
Several articles note the connection between the potential for misunderstanding PGS and possible downstream negative consequences (Andrews and Zuiker 2003; Fabbri and Serretti 2020; Galton and Ferns 1999; Ikeda et al. 2021; Palk et al. 2019; Saya et al. 2021; Torkamani et al. 2018). While some advocate that PGS should be treated like other non-genetic laboratory tests and biomarkers (Andrews and Zuiker 2003; Saya et al. 2021), others worry that genetic information in the context of multifactorial disease may elicit stigma or discrimination (Chowdhury et al. 2013; Ikeda et al. 2021; Kious et al. 2021). This potential is particularly acute in certain therapeutic areas, such as mental health, as biogenetic explanations and the de-emphasis of social determinants may be associated with lower social acceptance for individuals with mental health disorders (Palk et al. 2019). More research is needed to evaluate these issues (Driver et al. 2020; Rashkin et al. 2019), including “the impact of PRSs on knowledge, self-concept, symptom burden, and treatment adherence for affected individuals. For at-risk individuals, studies may evaluate knowledge and risk perception, the positive and negative psychological and social impact of learning the risk information, and any resulting behavior changes for participants” (Peay 2020).
Potential conflicts of interest/premature commercialization
Scholars have long expressed concerns about premature commercialization and potential conflicts of interest (Motulsky 2002).The potential benefits of identifying those at high genetic risk for common diseases “have fallen on fertile ground among politicians, healthcare providers, and the general public, particularly in light of the increasing costs of healthcare in developed societies” (Hall et al. 2004). It is important to recognize that interpretation of genomic data, including “genotype associations with multifactorial phenotypes,” rests with humans who may have particular values, biases, associations, interests or even conflicts that impact their analysis (Ozdemir et al. 2009).
Despite the uncertain clinical value of PGS at the present time (Curtis 2019; Parens et al. 2020; Rosenberg et al. 2019; Wald and Old 2019), PGS are increasingly becoming available through direct-to-consumer (DTC) testing companies, including in sensitive and controversial contexts, such as psychiatric conditions and preimplantation genetic testing of embryos after in vitro fertilization (Docherty et al. 2021; Lewis and Vassos 2020; Motulsky 2002; Rashkin et al. 2019; Treff et al. 2019; Turley et al. 2021). Companies offering genetic testing “with varying clinical utility” may give “consumers an expanded sense of agency and autonomy around their genetic information” (Rashkin et al. 2019). However, while DTC companies and even healthcare systems may argue that individuals have a right to learn about what is currently known about their (or their future child’s) genomic liability, some voice strong concern about the potential for significant harm (Docherty et al. 2021; Fabbri and Serretti 2020; Parens et al. 2020).
Concerns about equitable access
Notwithstanding its many potential benefits, many articles voice concern that PGS may exacerbate health inequities: current algorithms have a varying accuracy across different population groups due to the Eurocentric bias in genetic databases (Briggs and Slade 2019; Cavazos and Witte 2021; Chowdhury et al. 2013; Dikilitas et al. 2020; Fernandez-Rhodes et al. 2020; Lambert et al. 2019; Martin et al. 2019b; Palk et al. 2019; Slunecka et al. 2021; Warren 2018; Yanes et al. 2020a; Zhou et al. 2021). Indeed, one group “consider[s] the consistent observation that [PGS] are currently of far greater predictive value in individuals of recent European descent than in others to be the major ethical and scientific challenge surrounding clinical translation and, at present, the most critical limitation to genetics in precision medicine.” (Martin et al. 2019b). Some commercial tests for polygenic risk are restricted by ancestry (Lewis and Green 2021).
Ethical actions and obligations
The remaining identified themes convey ethical obligations and actions to maximize benefits, minimize harm, promote justice, and support autonomy (Beauchamp and Childress 2013) in the context of genetic risk prediction for multifactorial disease: increasing diversity in research, responsible data safeguarding and sharing, empirical demonstration of validity and utility, clear and accurate communication, and the development of guidelines, standards, and possible regulations. These themes are further discussed below, with representative quotes provided in Table 1.
Table 1.
Illustrative quotes related to identified themes that express ethical actions and obligations to maximize benefits, minimize risks, promote justice, and support autonomy (Beauchamp and Childress 2013). Note that many of the quotes include the abbreviation PRS for polygenic risk score (citations within quotes are omitted)
| Theme | Illustrative quote |
|---|---|
| Need for increased diversity in genetic research | “We discuss the important ethical, legal, and social implications of increasing ancestral diversity in genetic studies of cardiometabolic disease and the challenges that arise from the (1) lack of diversity in current reference populations and available analytic samples and the (2) unequal generation of health-associated genomic data and their prediction accuracies” (Fernandez-Rhodes et al. 2020) |
| “The lack of representative GWAS has been recognized as a key obstacle in the project of Precision Medicine. Initiatives like the All of Us study, the Clinical Sequencing Evidence-Generating Research Consortium (CSER), the Human Genome Reference Program (HGRP), the PRS Diversity Consortium, and others have been designed to address both the underlying science and clinical translation, as well as longstanding debates about the role of race in medicine, genetics, and genomics” (James et al. 2021) | |
| Need for responsible data safeguarding and sharing | “In addition, many of the approaches used in research (e.g., anonymization, de-identification) are not applicable to genetic information because the genome is the ultimate identifier. Thus there is a requirement for additional strategies that preserve the privacy of genomic data while not compromising the accuracy of the results” (McLaren et al. 2016) |
| “In particular, disease risk prediction based on patient characteristics, including PRSs, has the potential to be used against the patient, and thus tight regulation is needed. To balance the advantage of advancing healthcare using large-scale EHR data and potential concerns of privacy violations, more up-to-date regulatory measures are needed to match the pace of technological development” (Li et al. 2020) | |
| “Barriers to such data-sharing include non-harmonized or unclear data protection laws and data localization requirements, which can preclude the creation of large representative datasets. Legal doctrines including collection limitation and data minimization, purpose limitation, and strict interpretations of consent requirements and anonymization requirements, all common to data protection law, can impede the collection of rich datasets and the efficient sharing thereof” (Knoppers et al. 2021) | |
| Empirical demonstration of validity and utility | “For both sets of evidence, assumptions on effectiveness and cost-effectiveness based on results of modeling must be backed by evidence of scientific validity and clinical utility from systematic empirical research such as pilot studies or clinical trials” (Chowdhury et al. 2013) |
| “PRSs have strong face validity; they intuitively seem to make sense, but this apparent face validity is not enough. More comparative research is needed to investigate the construct, content, and criterion validity of PRS, to explore alternative ways of quantifying polygenic risk, and to rigorously compare new and current methods” (Janssens 2019) | |
| “There remains a gap in evidence from prospective observational studies or treatment trials regarding the appropriate placement of PRS in risk assessment and lipid treatment decisions relative to information on rare monogenic gene variants, particularly in multiethnic populations” (O'Donnell 2020) | |
| “Poorly designed and/or described studies call into question the validity of some PRS to predict their target outcome, and relatively few studies have externally benchmarked multiple scores’ performance” (Wand et al. 2021) | |
| Need for accurate communication | “This conceptual transfer from monogenic disorders to polygenic disease is quite inappropriate, because polygenic disease involves the co-inheritance of several genetic determinants that usually have to interact with environmental factors before the disease becomes manifest. The genetic determinants for a phenotype can be variable and they may interact with different ways; some of the genetic factors can even be protective for the occurrence of the disease” (Galton and Ferns 1999) |
| “We argue that particular attention should be paid to the difficulties associated with the communication and interpretation of results. This would be due, in part, to the fact that, given the etiological complexity of psychiatric disorders, a PRS in the top percentile would be an indicator of risk, not a definitive prognosis. For this reason, nuance and skill would be required in articulating and ensuring correct understanding (both of counsellors and patients) of ‘complex’ risk. While the difficulties associated with feedback of complex genetic risk are not necessarily unique to PRS, they nevertheless warrant consideration given its recency” (Palk et al. 2019) | |
| Need for guidelines, standards, and possible regulation | “Another measure to build public trust and sustainability of omics fields could be legislative initiatives to create a multidisciplinary oversight body, at arm’s length from conflicts of interests, to carry out independent, impartial, and transparent innovation analyses and prospective technology assessment” (Ozdemir et al. 2009) |
| “As GWAS sample sizes increase and PRS become more powerful, they are set to play a role in research and personalized medicine. However, despite the growing application and importance of PRS, there are limited guidelines for performing PRS analyses, which can lead to inconsistency between studies and misinterpretation of results” (Choi et al. 2020) | |
| “Although we have provided explicit recommendations on how to acknowledge study design limitations and their effects on the interpretation and generalizability of a PRS, future research should attempt to establish best practices to guide the field” (Wand et al. 2021) | |
| "Stricter controls need to be put in place to regulate companies offering direct-to-consumer genetic tests” (Manrique de Lara et al. 2019) |
Need for increased diversity in genetic research
According to the PRS Task Force, “responsible use” of a PRS is achieved when “there are clear benefits that outweigh risks, and where effort is taken toward a goal of equitable benefit for all” (Polygenic Risk Score Task Force of the International Common Disease Alliance 2021). To ensure equitable access to PGS, many recommend increased representation and diversity in genetic research (Cavazos and Witte 2021; Chowdhury et al. 2013; Dikilitas et al. 2020; Duncan et al. 2019; Durvasula and Lohmueller 2021; Fernandez-Rhodes et al. 2020; James et al. 2021; Knoppers et al. 2021; Lambert et al. 2019; Manrique de Lara et al. 2019; Martin et al. 2019b; Mudd-Martin et al. 2021; Murray et al. 2021; Palk et al. 2019; Rubin and Glusman 2019; Slunecka et al. 2021; Yanes et al. 2020a; Zhou et al. 2021). Aside from ensuring that PGS realize “comparable performance across sub-populations and across human genetic diversity,” it is also important to make sure that there is equitable access to risk-stratified care and follow-up (Knoppers et al. 2021). There is also concern about how to ethically use race, ancestry, and ethnicity in PGS reporting (Fernandez-Rhodes et al. 2020; James et al. 2021; Lewis and Green 2021; Mudd-Martin et al. 2021).
Need for data safeguarding and sharing
Related to concerns about maximizing the accuracy and applicability of PGS and diversifying genetic research is an expressed need for responsible data sharing and safeguarding in the context of genomic data (Andrews and Zuiker 2003; Briggs and Slade 2019; Daniels et al. 2021; Li et al. 2020; McLaren et al. 2016; Pashayan et al. 2013). The potential for stigma and/or discrimination necessitates special attention to “how and when genetic samples and data are acquired, stored, and used” (Chowdhury et al. 2013). However, barriers to data sharing limit the power of PGS (Knoppers et al. 2021). One organization, the Global Alliance for Genomics and Health, has developed policies to guide ethical sharing of genomic and clinical data (Mudd-Martin et al. 2021).
Empirical demonstration of validity and utility
Benefit/risk analysis of PGS will depend on the exact context: how the score was developed, its predictive power in a given population for a specific disease, and what decisions it is intended to inform (Kotze et al. 2015; Lewis and Vassos 2020; Yanes et al. 2020a). Ultimately, clinical or implementation research must empirically determine whether specific PGS or models incorporating PGS improve outcomes (Choi et al. 2020; Chowdhury et al. 2013; Duncan et al. 2019; O'Donnell 2020), including whether risk results motivate behavior change, which is not clear (Driver et al. 2020; Yanes et al. 2020a). Studies must determine the clinical utility of specific PGS for different uses; some trials are already underway (Lambert et al. 2019; Polygenic Risk Score Task Force of the International Common Disease Alliance 2021; Yanes et al. 2020a).
Need for accurate communication
Clear and accurate communication of the benefits and limitations of PGS will be critical to its ethical implementation (Ronald 2020). Indeed, one study found that patients did not question the quality or utility of genomic information about polygenic risk for melanoma (Smit et al. 2021). Although some individuals are interested in genomic tests “to be empowered with personal risk information” (Saya et al. 2021), many people are more interested in genetic information if it can guide actions (Driver et al. 2020). Notwithstanding individuals’ desire for genetic information, it “is not automatically empowering” and “if the results are not carefully communicated, patients may be confused about their impact, and unsure of what steps to take next” (Kious et al. 2021).
Further, to mitigate the potential for mis- or overinterpretation of PGS, many articles stress the need for training healthcare professionals who will encounter and interpret PGS and counsel patients (Andrews and Zuiker 2003; Briggs and Slade 2019; Chowdhury et al. 2013; Kotze et al. 2015; Lea 2003; Lewis and Green 2021; McLaren et al. 2016; Motulsky 2002; Palk et al. 2019; Pashayan et al. 2013; Pashayan and Pharoah 2012; Rashkin et al. 2019; Torkamani et al. 2018; Vassy et al. 2018). For example, there are concerns about genetic determinism: “without appropriate communication of the uncertainty around [PGS], large-scale deployment…could potentially reinforce and amplify false genetic-determinism attitudes” (Polygenic Risk Score Task Force of the International Common Disease Alliance 2021). One study concluded that return of polygenic risk results was feasible in a primary care setting (Saya et al. 2021); however, the physicians had received genomics training so the results may not generalize to all settings.
Also acknowledged is that responsibility for accurate communication is shared and should involve engagement of patient and public groups and investment in genetic counseling (Ronald 2020) as well as responsible dissemination of the science by investigators and journalists (Rosenberg et al. 2019). To support individuals’ decisions, it is important to set policies around what information should be provided (Pashayan et al. 2013), including to family members, (Lewis and Green 2021; Zhou et al. 2021), although this may not be as critical for polygenic prediction as with genetic testing for single-gene disorders (Briggs and Slade 2019; Rashkin et al. 2019), as complex trait theory suggests that most cases will arise in individuals without family history of the disease (Lambert et al. 2019). Other issues related to communication that were raised in the literature include communication of genetic overlap between different phenotypes and diseases, particularly in psychiatric settings (DiBlasi et al. 2021; Driver et al. 2020), communication of results that have been changed or updated over time (Briggs and Slade 2019; Knoppers et al. 2021; Pashayan and Pharoah 2012; Slunecka et al. 2021), and the appropriateness of returning PGS results to minors or vulnerable adults (Docherty et al. 2021; Lewis and Green 2021; Manrique de Lara et al. 2019; Palk et al. 2019).
Guidelines, standards, and regulation
Standards and guidelines are needed as they are critical for quality control with PGS (Choi et al. 2020). Since different research groups use different methods to develop PGS, results can diverge: “we believe this lack of consistency to be a prime concern for the PRS field, and additional resources, such as a centralized public database of published polygenic scores, are necessary to increase PRS comparability and evaluation and thus improve their potential for translation” (Lambert et al. 2019). Problems with PGS include overfitting caused by using the same dataset for generating and training (Yanes et al. 2020a), or errors caused by misclassifying a phenotype or treatment of missing information (Li et al. 2020). Inaccuracies may have significant downstream repercussions, especially if commercialization of tests and/or health policy is based on biased or substandard studies (Ozdemir et al. 2009). However, guidelines for PGS will depend on the specific disease, as well as the availability of GWAS summary data for discovery and target populations (Yanes et al. 2020a). In response to the need to standardize development and reporting, and enable evaluation, the PGS Catalog (www.pgscatalog.org), an open database of PGSs, was developed as a resource for the community (Yanes et al. 2020a). The “Polygenic Risk Score Reporting Standards,” a framework defining minimal information needed to interpret and evaluate PGS, was put forward in 2021 (Wand et al. 2021). Guidance and frameworks around best practices for communication of polygenic risk are also recommended (Lewis and Green 2021; Ozdemir et al. 2009; Slunecka et al. 2021; Tabor et al. 2014; Turley et al. 2021; Vassy et al. 2018; Yanes et al. 2020a).
Regulation of PGS also needs consideration: too much could delay availability and limit access, but too little could jeopardize safe and appropriate use (Knoppers et al. 2021). To counter potential misuse of polygenic risk prediction, some call for stronger legal protections against genetic discrimination (Torkamani et al. 2018) and privacy regulations (Li et al. 2020). An article by Docherty et al. on PGS for suicide prediction includes a robust discussion of FDA regulation of DTC genetic tests (Docherty et al. 2021). Risks of DTC genetic tests include false positives, false negatives, and errors in interpretation (Docherty et al. 2021). The authors believe the current regulatory environment enables “oversimplification and exaggeration of research results for marketing purposes” and provision of genetic tests “without demonstration of clinical validity” (Docherty et al. 2021). “At a minimum, companies offering DTC genetic testing for polygenic risk should publish guidelines for interpreting their results that, in layperson terms, acknowledge a lack of clinical utility,” the authors write. “Again, however, the current failure of DTC companies to do so may be difficult to remedy without regulatory changes” (Docherty et al. 2021). Appropriate governance and regulation of PGS in the context of embryo screening is another noteworthy concern (Karavani et al. 2019; Lázaro-Muñoz et al. 2021; Munday and Savulescu 2021; Turley et al. 2021).
Discussion
Many ELSI concerns related to genetic risk prediction for multifactorial disease were raised in the 92 articles included in this study, which spanned from 1977 to 2021 (see Fig. 2). There is significant enthusiasm for the potential of polygenic risk prediction to improve healthcare of many multifactorial diseases by facilitating screening through risk stratification (public health), contributing to diagnosis and/or prognosis or guiding care by informing therapy or approach (clinical care), and enhancing research. However, scholars are also concerned about potential risks and challenges of PGS, including the potential for mis- or overinterpretation, stigma and discrimination, premature commercialization, and inequitable access to benefits. In order to realize the potential of PGS in an ethically and socially responsible manner, researchers and developers are obligated to take steps that will maximize benefits, minimize harm, promote justice, and support autonomy (Beauchamp and Childress 2013). These actions include diversifying research, safeguarding and sharing data responsibly, empirical testing of PGS validity and utility, accurately communicating the meaning and limitations of PGS, and developing appropriate guidelines, standards, and regulations.
Concerns about informed consent, which dominate the ethics literature for genetic testing for monogenic disorders, were not as prominent in the context of genetic risk prediction for multifactorial disease. Although reasons for this are unclear, it could be because PGS are only emerging in clinical and direct-to-consumer settings, are perceived as less specifically actionable than monogenic tests (e.g., they do not provide clarity on disease mechanism), and/or have less significance for family members. Though informed consent was not a dominant theme, autonomy was a significant focus in the literature, as it relates to accurate understanding of PGS. Indeed, apart from the issue that PGS will not be equally accurate across different population groups and therefore may have the unintended effect of contributing to health inequities, the major concern about PGS emerging from this review relates to possible misinterpretation, misrepresentation, or misuse. Appropriate interpretation and use align with the ethical principles of beneficence, nonmaleficence, and justice (Beauchamp and Childress 2013).
Based on the findings from this review, it will be critical to support unbiased assessment and communication of the benefits and risks of PGS. Many stress the need for education of healthcare professionals who will assist patients with understanding and interpreting PGS results, but given potential conflicts of interest, it is unclear what entity could or should take responsibility for this formidable obligation, to what extent those developing or commercializing PGS should be involved, and what oversight mechanisms are needed. Important questions that need consideration include: who should conduct risk/benefit analysis and determine the appropriate validation, interpretation, and use of PGS? What role should pharmaceutical companies and genetic testing companies play in educating health professionals and consumers about PGS? Especially given commercial interest, what standards should be imposed and what is the role for regulation, if any? To ensure the ethical translation of polygenic risk prediction from research into healthcare, stakeholders must carefully consider these critical questions.
A notable limitation of this narrative review is that it was conducted by a single author. As thematic analysis is inherently a qualitative, subjective endeavor (Braun and Clarke 2021), important articles or ELSI themes may have been overlooked.
Conclusion
This narrative review confirmed widespread interest in the potential for polygenic risk prediction such as PGS to improve public health, clinical care, and research in the context of multifactorial disease. However, to ensure ethical translation of PGS, many potential risks and challenges must be addressed, including the potential for mis- or overinterpretation, stigma and discrimination, and conflicts of interest and premature commercialization. Scholars also voice significant concern about equitable access to benefits. Ethical actions and obligations expressed in the literature, which relate to maximizing benefits, minimizing harm, promoting justice, and supporting autonomy, include increasing diversity in research, responsible data safeguarding and sharing, the need for empirical determination of validity and utility, the need for clear and accurate communication; and development of guidelines, standards, and possible regulations. This research should support the ethical research, development, and translation of polygenic risk prediction.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Thank you to Tim Roberts, NYU librarian, for assistance with the literature search strategy, and to Dr. Aravinda Chakravarti and Dr. Herb Leventer, who provided helpful comments on earlier drafts of this article. Thank you to the anonymous reviewers for their suggestions, which improved the manuscript.
Author contributions
CRC conceived the idea for this work, reviewed and analyzed the articles, and drafted and edited the manuscript.
Declarations
Competing interests
No competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andrews L, Zuiker ES. Ethical, legal, and social issues in genetic testing for complex genetic diseases. Valparaiso Univ Law Rev. 2003;37:793–829. [PubMed] [Google Scholar]
- Artiga S, Hinton E (2018) Beyond health care: the role of social determinants in promoting health and health equity (issue brief). Henry J. Kaiser Family Foundation (KFF) From https://www.kff.org/racial-equity-and-health-policy/issue-brief/beyond-health-care-the-role-of-social-determinants-in-promoting-health-and-health-equity/
- Beauchamp TL, Childress JF (2013) Principles of biomedical ethics, 7th edn. Oxford University Press, New York
- Berliner J (ed) (2014) Ethical dilemmas in genetics and genetic counseling: principles through case scenarios, Oxford University Press, New York
- Blanc J, Berg JJ (2020) How well can we separate genetics from the environment? Elife 9. 10.7554/eLife.64948 [DOI] [PMC free article] [PubMed]
- Braun V, Clarke V. Using thematic analysis in psychology. Qual Res Psychol. 2006;3:77–101. doi: 10.1191/1478088706qp063oa. [DOI] [Google Scholar]
- Braun V, Clarke V. What can “thematic analysis” offer health and wellbeing researchers? Int J Qual Stud Health Well Being. 2014;9:26152. doi: 10.3402/qhw.v9.26152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Clarke V. One size fits all? What counts as quality practice in (reflexive) thematic analysis? Qual Res Psychol. 2021;18:328–352. doi: 10.1080/14780887.2020.1769238. [DOI] [Google Scholar]
- Briggs S, Slade I. Evaluating the integration of genomics into cancer screening programmes: challenges and opportunities. Curr Genet Med Rep. 2019;7:63–74. doi: 10.1007/s40142-019-00162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattarinussi G, Delvecchio G, Sambataro F et al (2022) The effect of polygenic risk scores for major depressive disorder, bipolar disorder and schizophrenia on morphological brain measures: a systematic review of the evidence. J Affect Disord 310:213–222. 10.1016/j.jad.2022.05.007 [DOI] [PubMed]
- Cavazos TB, Witte JS (2021) Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. HGG Adv 2 . 10.1016/j.xhgg.2020.100017 [DOI] [PMC free article] [PubMed]
- Chaudhari BP, Manickam K, McBride KL. A pediatric perspective on genomics and prevention in the twenty-first century. Pediatr Res. 2020;87:338–344. doi: 10.1038/s41390-019-0597-z. [DOI] [PubMed] [Google Scholar]
- Choi SW, Mak TS, O'Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Dent T, Pashayan N, et al. Incorporating genomics into breast and prostate cancer screening: assessing the implications Genetics in medicine : official journal of the American College of Medical. Genetics. 2013;15:423–432. doi: 10.1038/gim.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch DJM, Bodmer WF. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc Natl Acad Sci U S A. 2020;117:18924–18933. doi: 10.1073/pnas.2005634117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D. Clinical relevance of genome-wide polygenic score may be less than claimed. Ann Hum Genet. 2019;83:274–277. doi: 10.1111/ahg.12302. [DOI] [PubMed] [Google Scholar]
- Daniels H, Jones KH, Heys S, et al. Exploring the use of genomic and routinely collected data: narrative literature review and interview study. J Med Internet Res. 2021;23:e15739. doi: 10.2196/15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delikurt T, Williamson GR, Anastasiadou V, et al. A systematic review of factors that act as barriers to patient referral to genetic services. Eur J Hum Genet. 2015;23:739–745. doi: 10.1038/ejhg.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBlasi E, Kang J, Docherty AR. Genetic contributions to suicidal thoughts and behaviors. Psychol Med. 2021;51:2148–2155. doi: 10.1017/s0033291721001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikilitas O, Schaid DJ, Kosel ML, et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am J Hum Genet. 2020;106:707–716. doi: 10.1016/j.ajhg.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P, Keeney E, Taylor JC et al (2022) Can polygenic risk scores contribute to cost-effective cancer screening? A systematic review genetics in medicine. Genet Med 24:1604–1617. 10.1016/j.gim.2022.04.020 [DOI] [PMC free article] [PubMed]
- Docherty A, Kious B, Brown T, et al. Ethical concerns relating to genetic risk scores for suicide. Am J Med Genet B Neuropsychiatr Genet. 2021;186:433–444. doi: 10.1002/ajmg.b.32871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver MN, Kuo SI, Dick DM. Genetic feedback for psychiatric conditions: where are we now and where are we going. Am J Med Genet B Neuropsychiatr Genet. 2020;183:423–432. doi: 10.1002/ajmg.b.32815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Lohmueller KE. Negative selection on complex traits limits phenotype prediction accuracy between populations. Am J Hum Genet. 2021;108:620–631. doi: 10.1016/j.ajhg.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel EJ, Wendler D, Grady C. What makes clinical research ethical? JAMA. 2000;283:2701–2711. doi: 10.1001/jama.283.20.2701. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Serretti A. Genetics of Treatment outcomes in major depressive disorder: present and future clinical psychopharmacology and neuroscience : the official scientific journal of the Korean College of. Neuropsychopharmacology. 2020;18:1–9. doi: 10.9758/cpn.2020.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Rhodes L, Young KL, Lilly AG, et al. Importance of genetic studies of cardiometabolic disease in diverse populations. Circ Res. 2020;126:1816–1840. doi: 10.1161/circresaha.120.315893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton JM, Nurnberger JI (2019) Polygenic risk scores in psychiatry: will they be useful for clinicians? F1000Res 8. 10.12688/f1000research.18491.1 [DOI] [PMC free article] [PubMed]
- Fusar-Poli L, Rutten BP, van Os J et al (2022) Polygenic risk scores for predicting outcomes and treatment response in psychiatry: hope or hype? Int Rev Psychiatry. 10.1080/09540261.2022.2101352 [DOI] [PubMed]
- Galton DJ, Ferns GA. Genetic markers to predict polygenic disease: a new problem for social genetics. Qjm. 1999;92:223–232. doi: 10.1093/qjmed/92.4.223. [DOI] [PubMed] [Google Scholar]
- Ganna A, Magnusson PK, Pedersen NL, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33:2267–2272. doi: 10.1161/atvbaha.113.301218. [DOI] [PubMed] [Google Scholar]
- Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–450. doi: 10.1097/01.gim.0000227935.26763.c6. [DOI] [PubMed] [Google Scholar]
- Hall WD, Morley KI, Lucke JC (2004) The prediction of disease risk in genomic medicine EMBO Rep 5 Spec No:S22–26 . 10.1038/sj.embor.7400224 [DOI] [PMC free article] [PubMed]
- Ikeda M, Saito T, Kanazawa T, et al. Polygenic risk score as clinical utility in psychiatry: a clinical viewpoint. J Hum Genet. 2021;66:53–60. doi: 10.1038/s10038-020-0814-y. [DOI] [PubMed] [Google Scholar]
- Inouye M, Abraham G, Nelson CP, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JE, Riddle L, Koenig BA, et al. The limits of personalization in precision medicine: polygenic risk scores and racial categorization in a precision breast cancer screening trial. PLoS ONE. 2021;16:e0258571. doi: 10.1371/journal.pone.0258571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens A. Validity of polygenic risk scores: are we measuring what we think we are? Hum Mol Genet. 2019;28:R143–r150. doi: 10.1093/hmg/ddz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavani E, Zuk O, Zeevi D, et al. Screening human embryos for polygenic traits has limited utility. Cell. 2019;179(1424–1435):e1428. doi: 10.1016/j.cell.2019.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ. Genetics and genomics in practice: the continuum from genetic disease to genetic information in health and disease. Genet Med. 2003;5:261–268. doi: 10.1097/01.GIM.0000076977.90682.A5. [DOI] [PubMed] [Google Scholar]
- Kious BM, Docherty AR, Botkin JR, et al. Ethical and public health implications of genetic testing for suicide risk: family and survivor perspectives Genetics in medicine : official journal of the American College of Medical. Genetics. 2021;23:289–297. doi: 10.1038/s41436-020-00982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarin D, Natarajan P. Clinical utility of polygenic risk scores for coronary artery disease. Nat Rev Cardiol. 2022;19:291–301. doi: 10.1038/s41569-021-00638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppers BM, Bernier A, Granados Moreno P et al. (2021) Of Screening, stratification, and scores. J Personalized Med 11 . 10.3390/jpm11080736 [DOI] [PMC free article] [PubMed]
- Kotze MJ, Luckhoff HK, Peeters AV, et al. Genomic Medicine and Risk Prediction across the Disease Spectrum. Crit Rev Clin Lab Sci. 2015;52:120–137. doi: 10.3109/10408363.2014.997930. [DOI] [PubMed] [Google Scholar]
- Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019;28:R133–r142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- Lázaro-Muñoz G, Pereira S, Carmi S, et al. Screening embryos for polygenic conditions and traits: ethical considerations for an emerging technology. Genet Med. 2021;23:432–434. doi: 10.1038/s41436-020-01019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea DH. How genetics changes daily practice. Nurs Manage. 2003;34:19–24. doi: 10.1097/00006247-200311000-00010. [DOI] [PubMed] [Google Scholar]
- Lewis ACF, Green RC. Polygenic risk scores in the clinic: new perspectives needed on familiar ethical issues. Genome Med. 2021;13:14. doi: 10.1186/s13073-021-00829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Chen Y, Ritchie MD, et al. Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet. 2020;21:493–502. doi: 10.1038/s41576-020-0224-1. [DOI] [PubMed] [Google Scholar]
- Manrique de Lara A, Soto-Gomez L, Nunez-Acosta E, et al. Ethical issues in susceptibility genetic testing for late-onset neurodegenerative diseases. Am J Med Genet B Neuropsychiatr Genet. 2019;180:609–621. doi: 10.1002/ajmg.b.32699. [DOI] [PubMed] [Google Scholar]
- Martin AR, Daly MJ, Robinson EB, et al. Predicting polygenic risk of psychiatric disorders. Biol Psychiatry. 2019;86:97–109. doi: 10.1016/j.biopsych.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McLaren PJ, Raisaro JL, Aouri M, et al. Privacy-preserving genomic testing in the clinic: a model using HIV treatment Genetics in medicine : official journal of the American College of Medical. Genetics. 2016;18:814–822. doi: 10.1038/gim.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerman JJ, Janzing JG, Ter Hark SE et al. (2022) The potential of polygenic risk scores to predict antidepressant treatment response in major depression: a systematic review. J Affect Disord 304:1–11. 10.1016/j.jad.2022.02.015 [DOI] [PubMed]
- Motulsky A. From pharmacogenetics and ecogenetics to pharmacogenomics. Med Secoli. 2002;14:683–705. [PubMed] [Google Scholar]
- Mudd-Martin G, Cirino AL, Barcelona V, et al. Considerations for cardiovascular genetic and genomic research with marginalized racial and ethnic groups and indigenous peoples: a scientific statement from the american heart association. Circ Genom Precis Med. 2021;14:e000084. doi: 10.1161/hcg.0000000000000084. [DOI] [PubMed] [Google Scholar]
- Munday S, Savulescu J. Three models for the regulation of polygenic scores in reproduction. J Med Ethics. 2021;47:e91. doi: 10.1136/medethics-2020-106588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray GK, Lin T, Austin J, et al. Could polygenic risk scores be useful in psychiatry?: a review. JAMA Psychiatry. 2021;78:210–219. doi: 10.1001/jamapsychiatry.2020.3042. [DOI] [PubMed] [Google Scholar]
- National Human Genome Research Institute (2020a) Polygenic risk scores. https://www.genome.gov/Health/Genomics-and-Medicine/Polygenic-risk-scores. Accessed 11 Oct 2022
- National Human Genome Research Institute (2020b) Genome-wide association studies fact sheet. https://www.genome.gov/about-genomics/fact-sheets/Genome-Wide-Association-Studies-Fact-Sheet. Accessed 6 July 2022
- Nussbaum R, McInnes RR, Willard HF (2015) Chapter 19: Ethical and social issues in genetics and genomics. In: Thompson & Thompson genetics in medicine, 8th edn. Elsevier inc., p 383–390
- O'Donnell CJ. Opportunities and challenges for polygenic risk scores in prognostication and prevention of cardiovascular disease. JAMA Cardiol. 2020;5:399–400. doi: 10.1001/jamacardio.2019.6232. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Suarez-Kurtz G, Stenne R, et al. Risk assessment and communication tools for genotype associations with multifactorial phenotypes: the concept of “edge effect” and cultivating an ethical bridge between omics innovations and society. Omics. 2009;13:43–61. doi: 10.1089/omi.2009.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palk AC, Dalvie S, de Vries J, et al. Potential use of clinical polygenic risk scores in psychiatry - ethical implications and communicating high polygenic risk. Philos Ethics Humanit Med: PEHM. 2019;14:4. doi: 10.1186/s13010-019-0073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parens E, Matthews L, Appelbaum PS. Polygenic risk scores, prediction of psychiatric disorders, and the health of all of us. Lancet Psychiatry. 2020;7:481. doi: 10.1016/s2215-0366(20)30185-1. [DOI] [PubMed] [Google Scholar]
- Pashayan N, Hall A, Chowdhury S, et al. Public health genomics and personalized prevention: lessons from the COGS project. J Intern Med. 2013;274:451–456. doi: 10.1111/joim.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashayan N, Pharoah P. Population-Based Screening in the Era of Genomics. Per Med. 2012;9:451–455. doi: 10.2217/pme.12.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peay HL (2020) Genetic risk assessment in psychiatry. Cold Spring Harb Perspect Med 10. 10.1101/cshperspect.a036616 [DOI] [PMC free article] [PubMed]
- Pharoah PDP, Antoniou A, Bobrow M, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Polygenic Risk Score Task Force of the International Common Disease Alliance Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat Med. 2021;27:1876–1884. doi: 10.1038/s41591-021-01549-6. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkin MD, Bowes J, Dunaway K, et al. Genetic counseling, 2030: an on-demand service tailored to the needs of a price conscious, genetically literate, and busy world. J Genet Couns. 2019;28:456–465. doi: 10.1002/jgc4.1123. [DOI] [PubMed] [Google Scholar]
- Ronald A. Editorial: polygenic scores in child and adolescent psychiatry - strengths, weaknesses, opportunities and threats. J Child Psychol Psychiatry. 2020;61:519–521. doi: 10.1111/jcpp.13246. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Edge MD, Pritchard JK, et al. Interpreting polygenic scores, polygenic adaptation, and human phenotypic differences. Evol Med Public Health. 2019;2019:26–34. doi: 10.1093/emph/eoy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin IR, Glusman G (2019) Opportunities and challenges in interpreting and sharing personal genomes. Genes 10. 10.3390/genes10090643 [DOI] [PMC free article] [PubMed]
- Saya S, McIntosh JG, Winship IM, et al. Informed choice and attitudes regarding a genomic test to predict risk of colorectal cancer in general practice. Patient Educ Couns. 2021 doi: 10.1016/j.pec.2021.08.008. [DOI] [PubMed] [Google Scholar]
- Slunecka JL, van der Zee MD, Beck JJ, et al. Implementation and implications for polygenic risk scores in healthcare. Hum Genomics. 2021;15:46. doi: 10.1186/s40246-021-00339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AK, Reyes-Marcelino G, Keogh L, et al. ‘There is a lot of good in knowing, but there is also a lot of downs’: public views on ethical considerations in population genomic screening. J Med Ethics. 2021;47:e28–e28. doi: 10.1136/medethics-2019-105934. [DOI] [PubMed] [Google Scholar]
- Stiles D, Appelbaum PS. Cases in precision medicine: concerns about privacy and discrimination after genomic sequencing. Ann Intern Med. 2019;170:717–721. doi: 10.7326/M18-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor HK, Auer PL, Jamal SM, et al. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet. 2014;95:183–193. doi: 10.1016/j.ajhg.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier L, Eccles J, Treff NR et al (2021) Embryo screening for polygenic disease risk: recent advances and ethical considerations Genes (Basel) 12. 10.3390/genes12081105 [DOI] [PMC free article] [PubMed]
- Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- Treff NR, Eccles J, Lello L, et al. Utility and First Clinical Application of Screening Embryos for Polygenic Disease Risk Reduction. Front Endocrinol. 2019;10:845. doi: 10.3389/fendo.2019.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley P, Meyer MN, Wang N, et al. Problems with using polygenic scores to select embryos. N Engl J Med. 2021;385:78–86. doi: 10.1056/NEJMsr2105065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassy JL, Davis JK, Kirby C, et al. How primary care providers talk to patients about genome sequencing results: risk, rationale, and recommendation. J Gen Intern Med. 2018;33:877–885. doi: 10.1007/s11606-017-4295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald NJ, Old R. The illusion of polygenic disease risk prediction. Genet Med. 2019;21:1705–1707. doi: 10.1038/s41436-018-0418-5. [DOI] [PubMed] [Google Scholar]
- Wand H, Lambert SA, Tamburro C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021;591:211–219. doi: 10.1038/s41586-021-03243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M. The approach to predictive medicine that is taking genomics research by storm. Nature. 2018;562:181–183. doi: 10.1038/d41586-018-06956-3. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Thinking about schizophrenia in an era of genomic medicine. Am J Psychiatry. 2019;176:12–20. doi: 10.1176/appi.ajp.2018.18111275. [DOI] [PubMed] [Google Scholar]
- Yanes T, McInerney-Leo AM, Law MH, et al. The emerging field of polygenic risk scores and perspective for use in clinical care. Hum Mol Genet. 2020;29:R165–R176. doi: 10.1093/hmg/ddaa136. [DOI] [PubMed] [Google Scholar]
- Yanes T, Young MA, Meiser B, et al. Clinical applications of polygenic breast cancer risk: a critical review and perspectives of an emerging field. Breast Cancer Res. 2020;22:21. doi: 10.1186/s13058-020-01260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeinomar N, Chung WK. Cases in precision, medicine: the role of polygenic risk scores in breast cancer risk assessment. Ann Intern Med. 2021;174:408–412. doi: 10.7326/m20-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li YYT, Fu AKY, et al. Polygenic Score Models for Alzheimer’s Disease: from Research to Clinical Applications. Front Neurosci. 2021;15:650220. doi: 10.3389/fnins.2021.650220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.