Abstract
Various cellular sources hamper interpretation of positron emission tomography (PET) biomarkers in the tumor microenvironment (TME). We developed an approach of immunomagnetic cell sorting after in vivo radiotracer injection (scRadiotracing) with three-dimensional (3D) histology to dissect the cellular allocation of PET signals in the TME. In mice with implanted glioblastoma, translocator protein (TSPO) radiotracer uptake per tumor cell was higher compared to tumor-associated microglia/macrophages (TAMs), validated by protein levels. Translation of in vitro scRadiotracing to patients with glioma immediately after tumor resection confirmed higher single-cell TSPO tracer uptake of tumor cells compared to immune cells. Across species, cellular radiotracer uptake explained the heterogeneity of individual TSPO-PET signals. In consideration of cellular tracer uptake and cell type abundance, tumor cells were the main contributor to TSPO enrichment in glioblastoma; however, proteomics identified potential PET targets highly specific for TAMs. Combining cellular tracer uptake measures with 3D histology facilitates precise allocation of PET signals and serves to validate emerging novel TAM-specific radioligands.
The assessment of single-cell tracer uptake and 3D histology advances development of TAM-specific PET biomarkers in glioblastoma.
INTRODUCTION
The tumor microenvironment (TME) emerged as an acknowledged key component in cancer biology and treatment (1). In particular, novel immunotherapies against targets of the TME facilitate potentiation of host antitumor immune responses (2). However, despite the need to monitor novel anticancer therapeutics in vivo, specific biomarkers for immune cells of the TME are still limited (3). Recent efforts originated in radiolabeled antibodies for positron emission tomography (PET) that specifically target immune cells (4), but the cell type heterogeneity of the TME hampers cellular allocation and interpretation of these biomarkers. Furthermore, antibody-based PET biomarkers have unpredictable penetrance through the blood-brain barrier due to variable disruption in presence of brain tumors, neurodegeneration, and neuroinflammatory conditions. Thus, their use is limited in brain malignancies, such as glioblastoma, which represents the most common malignant primary brain tumor with a very poor prognosis (5). In this disease, 18-kDa translocator protein (TSPO)–PET showed opportunities to assess myeloid cells of the TME as important contributors to immune evasion of glioblastoma (6). However, the detailed sources of TSPO and other TME biomarkers in glioma have not yet been thoroughly elucidated, which again hampers interpretation and understanding of individual PET images. In glioma, tumor-associated microglia and macrophages (TAMs) show elevated expression of TSPO in vitro (7) and in vivo (6), acting as a potential signal target of TSPO ligands. However, this finding is a matter of debate since in high-grade tumors, TSPO was found to be highly overexpressed by tumor cells as well (8–10).
To overcome the limitations of previous immunohistochemistry to PET comparisons, we aimed to resolve the complex sources of TME biomarkers in glioma at cellular resolution. We therefore established and validated an approach of immunomagnetic cell sorting (MACS) after radiotracer injection [single-cell radiotracing (scRadiotracing)] (11) and exploited this technique in a mouse model of experimental glioblastoma. TSPO tracer uptake was measured in single tumor cells and CD11b+ immune cells of a SB28 glioblastoma mouse model, validated against TSPO protein levels as determined by proteomics as a proof of concept. Furthermore, we investigated associations between single-cell TSPO tracer uptake and tumor heterogeneity in TSPO-PET. To bridge the gap between implantation of murine tumors and spontaneous glioma development in the human brain, we transferred TSPO scRadiotracing to human samples and determined proportions of TSPO tracer uptake in vitro in tumor and immune cells of patients with high-grade and low-grade glioma immediately after surgery. As a second major innovation, three-dimensional (3D) histology via tissue clearing and light sheet microscopy was integrated to acknowledge PET signals as a composite of cell type abundance and single-cell tracer uptake. Last, the full proteome of isolated SB28 tumor cells and TAMs was analyzed for identification of potential TAM-specific TME radioligand targets with superior binding capacity when compared to TSPO.
RESULTS
Single-cell tracer uptake measures of microglia as calculated by scRadiotracing correspond to PET signal changes upon microglia depletion
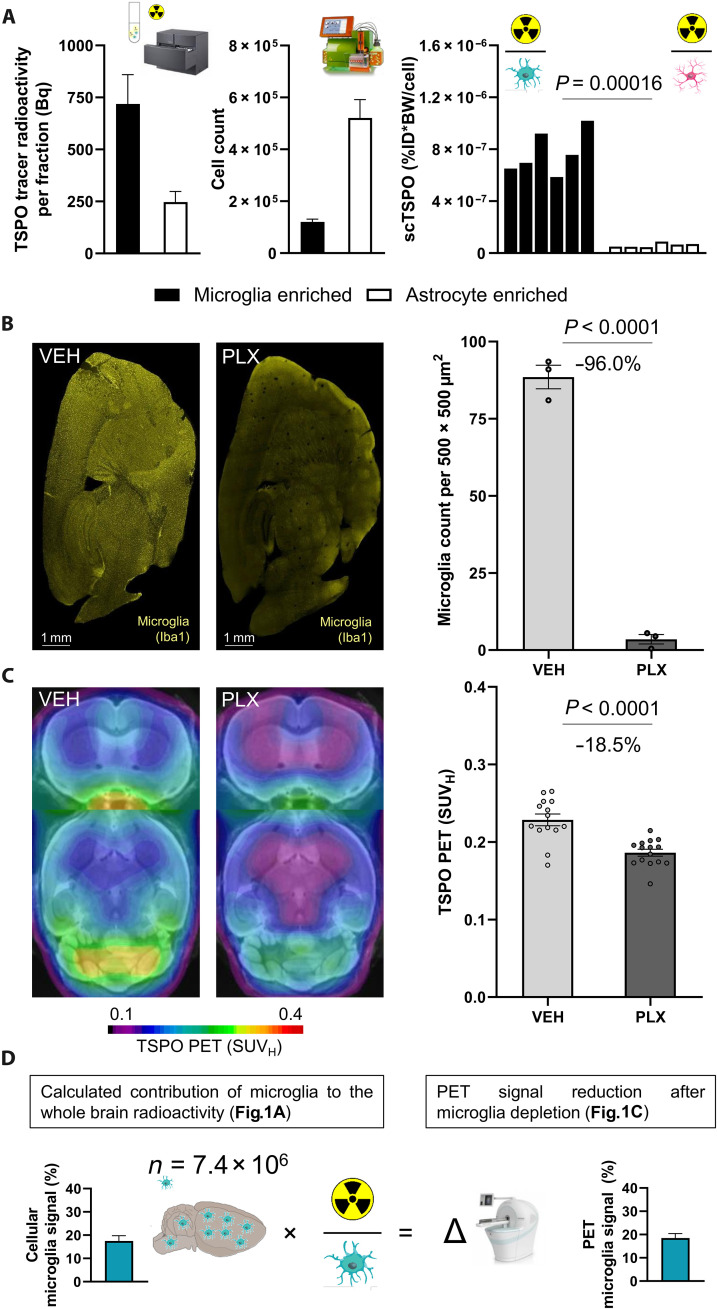
Immune PET biomarkers such as TSPO have the potential to monitor TAMs of the TME, but the target signal of currently available TSPO radioligands is challenged by several cellular sources. To validate scRadiotracing for the TSPO tracer [18F]GE-180, we compared the magnitude of the microglia-specific TSPO-PET signal with the recovered radioactivity in isolated microglia of untreated mice. First, we investigated the allocation of the TSPO tracer in the healthy mouse brain. Isolated CD11b+ microglia (1.2 × 105 ± 0.1 × 105 cells) of untreated mice revealed 7.7 × 10−7 ± 0.7 × 10−7 percentage radioactivity [normalized to injected dose (ID) and body weight (BW), %ID*BW] per single cell, whereas enriched anti-astrocyte cell surface antigen-2 (ACSA2)+ astrocytes (5.2 × 105 ± 0.7 × 105 cells) indicated 12.5-fold lower radioactivity per single cell (6.2 × 10−8 ± 0.7 × 10−8 %ID*BW; P = 0.00016; Fig. 1A), speaking for strong specificity of the tracer for microglia over astrocytes. Next, extrapolation by published microglia cell numbers (6% of all brain cells, 7.63 × 106 microglia cells) from dedicated studies (12), whole-brain radioactivity (0.19 to 0.30 MBq) and %brain dose per single microglia cell (1.8 × 10−6 to 3.6 × 10−6) were used to calculate the contribution of the microglia population to the radioactivity in the whole brain (17.5% ± 2.2%).
Fig. 1. TSPO-PET signal reduction in the whole brain after microglia depletion corresponds to the whole-brain signal attributable to tracer uptake in microglia.
(A) Components of scRadiotracing after in vivo tracer injection. Upon in vivo tracer injection and magnetic cell separation, cell pellets were analyzed by a high-sensitive gamma counter to measure the radioactivity (in becquerels) in the sample (left) (n = 6, means ± SEM) and by flow cytometry to determine the cell count (middle) (n = 6, means ± SEM). After calculation of radioactivity per single cell [single-cell TSPO tracer (scTSPO), normalized to ID and BW] in each sample, untreated mice showed significantly higher TSPO tracer uptake in microglia when compared to astrocytes (right) (each bar represents a single animal; n = 6, paired t test). (B) Representative sagittal Iba1 immunohistochemistry sections of vehicle treatment (VEH; n = 3) and PLX5622 (PLX)–treated (n = 3) mice and quantification of stained Iba1 area (%). Unpaired t test, means ± SEM. (C) Group average TSPO-PET images of healthy mice upon a magnetic resonance imaging (MRI) template after VEH (n = 14) or CSF1R inhibition (PLX5622; microglia depletion, n = 15) indicated a distinct signal reduction in the whole brain of PLX5622-treated animals (−18.5%). Unpaired t test, means ± SEM. (D) Extrapolation of the signal attributable to microglia using published cell numbers (76) and tracer uptake per single microglia cell as calculated by scRadiotracing. The calculated 17.5% contribution of microglia to the whole-brain TSPO-PET signal corresponded to the observed TSPO-PET signal reduction after microglia depletion (18.5%). Means ± SEM.
To quantify the contribution of microglia to the overall TSPO-PET signal, we used colony stimulating factor–1 receptor (CSF1R) inhibition via PLX5622 and hereby depleted 96% of all microglia in the mouse brain (Fig. 1B). As a result of microglia depletion, the TSPO-PET signal (Fig. 1C) in mice at 10.1 ± 2.1 months of age (13, 14) showed 18.5% lower heart-normalized standardized uptake values (SUVH) when compared to vehicle controls (0.19 ± 0.01 versus 0.23 ± 0.01; P < 0.0001). This decrease corresponded to the contribution of the microglia cell population extrapolated after scRadiotracing (P = 0.770; Fig. 1D). Thus, TSPO scRadiotracing provided a nearly complete recovery of the microglia-specific PET signal in healthy mice and suggested capability of the method to be used for tracer allocation in cells of the TME.
TSPO tracer uptake of tumor cells and TAMs reflects cellular TSPO protein levels in the SB28 glioblastoma mouse model
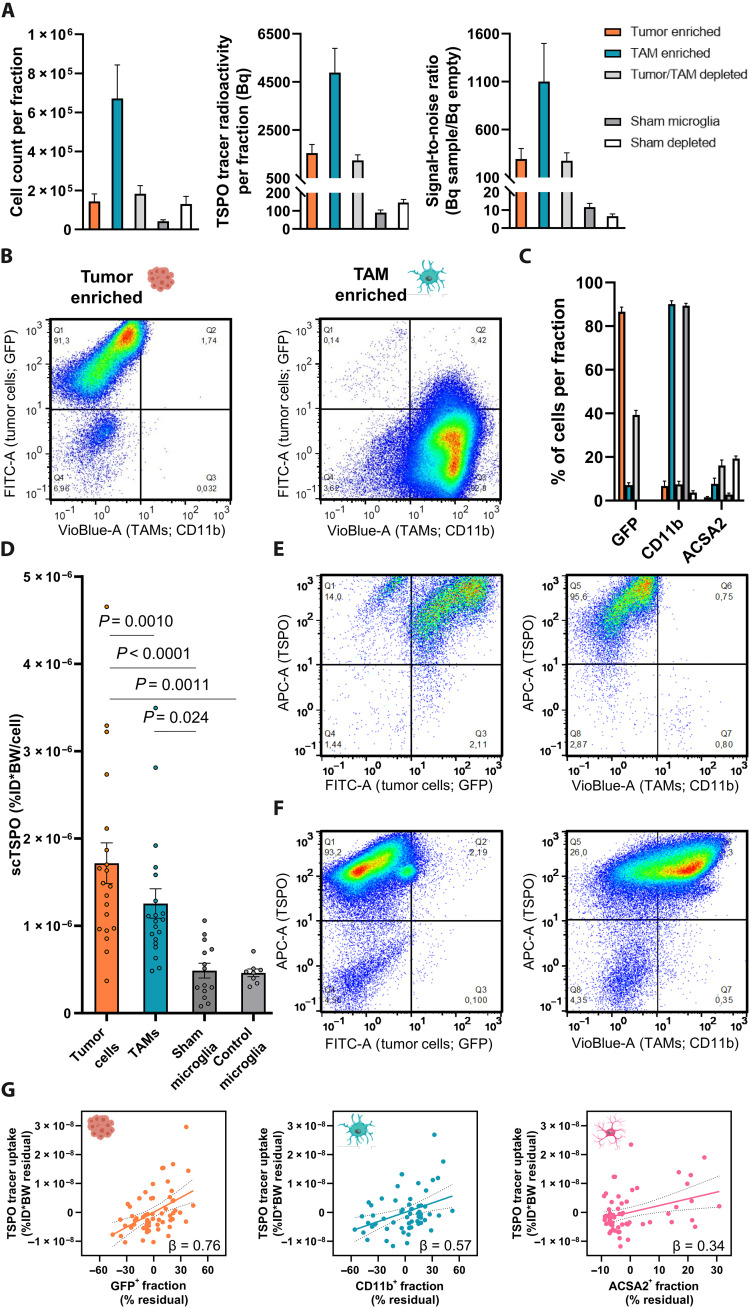
In the next step, we applied scRadiotracing to an experimental orthotopic glioblastoma model (SB28) that closely reflects the human TME (15) to investigate the contribution of tumor cells and TAMs to the TSPO-PET signal (Fig. 2, A to C). Using MACS, CD11b+ immune cells were isolated from tumor and sham-injected brains (TAMs or sham microglia), and tumor samples underwent subsequent tumor cell isolation. Residual cell pellets were analyzed as depleted fractions. We extracted 1.4 × 105 ± 0.4 × 105 green fluorescent protein (GFP)–positive tumor cells and 6.7 × 105 ± 1.7 × 105 CD11b+ TAMs from tumor mice and 4.2 × 104 ± 0.8 × 104 CD11b+ microglia from sham mice (Fig. 2A) in enriched fractions at high purity as determined by flow cytometry (tumor cells, 87% ± 2%; TAMs, 90% ± 1%; sham microglia, 89% ± 1%; Fig. 2, B to D). Signal-to-noise ratios for radioactivity detection were consistently ≥2 (Fig. 2A).
Fig. 2. scRadiotracing in the SB28 glioblastoma mouse model allows differential assessment of TSPO tracer uptake in tumor cells and TAMs.
(A) Acquired components of scRadiotracing after in vivo tracer injection in SB28 glioblastoma (n = 20) and sham (n = 14) mice. Using MACS, tumor cells (GFP+) and CD11b+ TAMs from SB28-bearing mice and CD11b+ microglia from sham-injected mice were enriched, thus leaving the respective residual–depleted cell fractions. Absolute cell numbers (left) and measured radioactivity (middle) resulted in satisfying signal-to-noise ratios (right) for all enriched and depleted fractions investigated. Means ± SEM. (B) MACS of tumor cells and TAMs led to >90% purity in enriched fractions as confirmed by flow cytometry. (C) Distribution of tumor cells (GFP), TAMs (CD11b), and astrocytes (ACSA2) in enriched and depleted fractions of SB28 glioblastoma (n = 20) and sham (n = 14) mice. Means ± SEM. (D) Comparison of the scTSPO uptake of isolated tumor cells (n = 20) and TAMs (n = 20) in SB28 glioblastoma mice as well as microglia of sham (n = 14) and untreated control (n = 8) mice. Paired t test for tumor cells versus TAMs, one-way ANOVA for all other comparisons, means ± SEM. (E and F) TSPO costaining in flow cytometry shows that nearly all tumor cells (GFP) (E) and TAMs (CD11b) (F) were also positive for TSPO. Notably, the minor population of CD11b− cells in the TAM-enriched fraction did not show positivity for TSPO (F, lower left quadrant), confirming the specificity of the TSPO costaining. Pooled data from n = 3 tumors. (G) Regression model including the enriched and depleted fractions of SB28 mice indicated highest contribution of tumor cells (left) to the radioactivity in the sample, followed by TAMs (middle) and astrocytes (right). Linear regression, β = standardized regression coefficient. n = 60 samples. Error bands represent 95% confidence interval.
Subsequently, single-cell tracer uptake of tumor cells and TAMs was quantified to decipher cellular contributions to the TSPO tracer uptake in glioblastoma. Both single SB28 tumor cells [3.53-fold, 1.7 × 10−6 ± 0.2 × 10−6 %ID*BW; P < 0.0001, one-way analysis of variance (ANOVA); Fig. 2D] and TAMs (2.58-fold, 1.3 × 10−6 ± 0.2 × 10−6 %ID*BW; P = 0.024, one-way ANOVA; Fig. 2D) indicated higher TSPO tracer uptake when compared to sham microglia (4.9 × 10−7 ± 0.8 × 10−7 %ID*BW). We also tested whether sham injections had an impact on TSPO tracer uptake of microglia and did not find any remarkable difference in these cells when compared to control microglia of untreated age-matched mice (P = 0.999, one-way ANOVA; Fig. 2D). As a proof of specificity, we injected excessive cold GE-180 before application of [18F]GE-180 and found nearly complete blocking of TSPO tracer uptake in SB28 tumor cells (−98%; P = 0.012), TAMs (−97%; P = 0.012), and sham microglia (−93%; P = 0.029; fig. S1A).
In the direct comparison of tumor cells and TAMs, we noted 1.37-fold higher TSPO tracer uptake of single SB28 tumor cells when compared to TAMs (P = 0.0010, paired t test), indicating a dual cellular allocation of the tracer with even slightly higher single-cell uptake of SB28 tumor cells when compared to the immune cell target population. Highly congruent to scRadiotracing, proteomics revealed increased TSPO protein levels in SB28 tumor cells [2.98-fold, false discovery rate (FDR)–corrected P = 0.015] and TAMs (2.04-fold, P = 0.0016) when compared to control microglia. Although not reaching significance after correction for multiple comparisons, higher TSPO tracer uptake of SB28 tumor cells over TAMs was also reflected by higher TSPO protein abundance (1.46-fold, P = 0.357; fig. S1B). We additionally proved the presence of the tracer target in SB28 tumor and immune cells by revealing that the vast majority of isolated CD11b+ and GFP+ cells showed TSPO colabeling in flow cytometry (Fig. 2, E and F), which is in accordance to the previously reported colocalization of TSPO and GFP/CD11b in immunohistochemistry (16). Validation experiments with isotype controls and unstained samples showed excellent specificity of CD11b and TSPO detection by flow cytometry in stained TAM and SB28 tumor cell–enriched samples (fig. S2, A and B). Detection of GFP was highly specific against tumor-free brain of sham animals (fig. S2B).
To determine the contributions of the distinct cell types with TSPO expression to the radioactivity in the cell pellets, we calculated a linear regression with normalized radioactivity per cell count as dependent variable and cellular proportions (GFP+ tumor cells, CD11b+ TAMs, and ACSA2+ astrocytes) of all enriched and depleted fractions as predictors. The regression model significantly explained the variance in single-cell TSPO tracer uptake (F = 6.2, P = 0.0011, R2 = 0.25, R2adj = 0.21). Tumor cells showed highest contribution to the single-cell TSPO tracer uptake (β = 0.76, P = 0.00017), followed by TAMs (β = 0.57, P = 0.0032) and astrocytes (β = 0.34, P = 0.0074) (Fig. 2G). A global model using measured radioactivity, total cell count, and cell proportions indicated that 76% of the variance in radioactivity measures could be explained by the cellular component of tumor cells, TAMs, and astrocytes (F = 46.6, P < 0.0001, R2 = 0.772, R2adj = 0.756). To question the performed calculations in depth, we correlated the radioactivity measured in the depleted tumor fractions, containing 39% ± 2% GFP+, 7% ± 1% CD11b+, and 16% ± 3% ACSA2+ cells, with estimated summed up radioactivity by single-cell tracer uptake and cell count (R = 0.74, P = 0.0002).
In summary, the established methodology accurately determines radiotracer uptake of single cells, which closely reflects relative protein abundance in respective cell types. The cellular allocation of TSPO as a biomarker consists of overexpression in both tumor and immune cells in SB28 tumors.
Single-cell tracer uptake explains interindividual PET heterogeneity and reveals dominant association of tumor cells with TSPO-PET signals
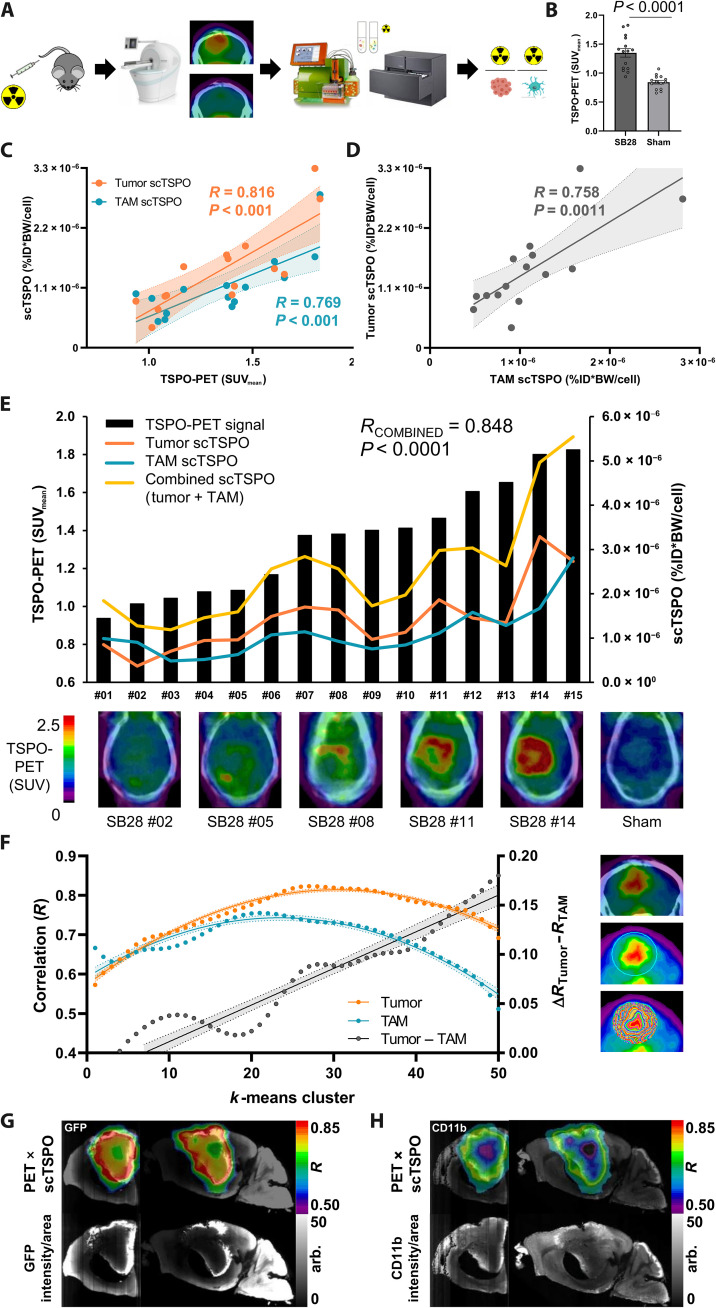
To bridge the gap between cellular tracer uptake and tumor imaging, we added PET to the experimental workflow of scRadiotracing in SB28 glioblastoma mice (Fig. 3A). TSPO-PET indicated heterogeneity of tumor signals 2.5 weeks after surgery (range of SUVmean, 0.937 to 1.825), which were strongly elevated when compared to sham injection (SUVmean, 0.842 ± 0.032; P < 0.0001; Fig. 3B). We correlated the individual single-cell TSPO tracer uptake of SB28 tumor cells and TAMs with the TSPO-PET signal in the lesion and observed similar degrees of strong association for both cell types (Fig. 3C). We also found an intercorrelation between single-cell tracer uptake of tumor cells and TAMs (R = 0.758, P = 0.0011), which indicated bidirectional dependence of TSPO enrichment in tumor and immune cells (Fig. 3D). Supporting these data, we found positive correlations between TSPO-PET and TSPO immunohistochemistry in both SB28 tumor cells and myeloid cells (fig. S3). A combined vector of single-cell TSPO tracer uptake of both tumor cells and TAMs strongly correlated with the magnitude of the tumor TSPO-PET signal (R = 0.848, P < 0.0001; Fig. 3E). A regression model of TSPO tracer uptake of tumor cells and TAMs, however, demonstrated that only tumor cells contributed significantly to the TSPO-PET signal (tumor cells: β = 0.55, P = 0.038; TAMs: β = 0.35, P = 0.157).
Fig. 3. Correlation of single-cell TSPO tracer uptake of tumor cells and TAMs with TSPO-PET indicates association of TSPO-PET heterogeneity with single-cell tracer enrichment.
(A) Schematic illustration of the scRadiotracing workflow. (B) TSPO-PET imaging indicated significantly higher lesion site signals in SB28 tumor mice (n = 15) than in sham (n = 14) animals at day 18 after inoculation. Unpaired t test, means ± SEM. (C) Correlation between scRadiotracing and TSPO-PET showed a strong dependence of PET signals from both tumor and TAM scTSPO uptake (normalized to ID and BW) in SB28 mice. N = 15, R = Pearson’s coefficient of correlation. Error bands represent 95% confidence interval. (D) Intercorrelation of scTSPO of tumor cells and TAMs in the SB28 glioblastoma mouse model. N = 15, R = Pearson’s coefficient of correlation. Error band represents 95% confidence interval. (E) Association between combined scTSPO and heterogeneous TSPO-PET signals. Black bars symbolize the individual TSPO-PET signal for all SB28 mice investigated (n = 15). Curves represent scTSPO of tumor cells (orange) and TAMs (blue) and a combined vector of cellular tracer uptake (yellow) for each individual animal. Axial sections of TSPO-PET images upon the individual contrast-enhanced computed tomography (CT) illustrate interindividual TSPO-PET signal heterogeneity of SB28 tumors. R = Pearson’s coefficient of correlation. (F) Visualization of the regional k-means cluster analysis. A sphere was placed over the entire signal enhancement in TSPO-PET, which was followed by application of k-means clustering. This resulted in 50 volumes of interest (VOIs) defining 50 intratumoral regions of increasing signal intensity (images on the right). (G and H) Strong regional agreement between tumor and TAM cell density (GFP/CD11b, light sheet microscopy) and regions with high scTSPO-to-PET associations. Coronal and sagittal slices show projections of correlation coefficients (R) onto k-means cluster VOIs of an individual mouse. arb., arbitrary units.
Going into more detail, we interrogated the regional heterogeneity of tumor TSPO-PET signals and correlated single-cell TSPO enrichment with the regional PET signal magnitude. Therefore, we performed a cluster-based analysis (k-means clustering, 50 clusters defining intratumoral regions of increasing signal intensity) of TSPO-PET in the tumor lesion (Fig. 3F) and correlated single-cell tracer uptake with the PET signal in respective clusters. The association between single-cell TSPO enrichment and TSPO-PET signal intensity in the cluster followed an inverted U-shape function for tumor cells and TAMs, with higher agreement between tumor cell TSPO and regional TSPO-PET signal when compared to TAM TSPO and regional TSPO-PET signal (Fig. 3, F to H). Notably, the association between TSPO-PET and single-cell tracer uptake was increasingly dominated by tumor cell TSPO enrichment as a linear function of cluster hierarchy (y = 0.004x − 0.026; R = 0.947; P < 0.0001; Fig. 3, F to H). This indicates that, in particular, regions with high TSPO-PET signal intensities were characterized by strong dependence from single–tumor cell TSPO. Thus, the combination of PET and scRadiotracing identified a strong association between PET signals and cellular tracer uptake, which was characterized by a dominant dependency of TSPO-PET signals from tumor cells when compared to immune cells.
To transfer our methodology to a different glioblastoma model, we performed TSPO-PET imaging and scRadiotracing in a pilot cohort of GL261 tumor mice, implanted in mice with wild-type and nude (Fox1nu) background (fig. S4). Keeping potential differences in the time courses of GL261 and SB28 mice in mind, we observed similar cellular TSPO tracer uptake of tumor cells and TAMs of GL261 mice with wild-type and Fox1nu background compared to SB28 implanted in wild-type mice. Noteworthy, the very low cell count of TAMs in Fox1nu mice was associated with low TSPO-PET signals.
In vitro scRadiotracing in human glioma underlines translational value of the methodology and confirms higher TSPO tracer uptake of tumor cells when compared to TAMs
To test for a potential translational value of single-cell tracer uptake measures in human glioblastoma, we applied in vitro scRadiotracing in a cohort of patients with high-grade and low-grade glioma that underwent biopsy or tumor resection (Table 1 and table S1). Tissue samples were investigated immediately after surgery applying in vitro [18F]GE-180 incubation of individual single-cell suspensions and subsequent cell sorting (Fig. 4A). The rationale was to check for discrepancies of cellular tracer binding between species.
Table 1. Characteristics of the human glioma cohort.
rs6971, TSPO polymorphism rs6971 (77); LAB, low-affinity binding status; MAB, medium-affinity binding status; HAB, high-affinity binding status; Dx, diagnosis; IDH, isocitrate dehydrogenase mutation; m, male; f, female; y, years. See table S1 for supplementary data.
| ID | Age (y) | Sex | Dx | Disease duration (months) | WHO grade | rs6971 | IDH | Sample acquisition | Previous therapy | [18F]GE-180 TSPO-PET |
|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 36.0 | m | Astrocytoma | 5.6 | 2 | HAB | + | Resection | − | + |
| #2 | 59.0 | f | Oligodendroglioma | 13.6 | 2 | MAB | + | Resection | + | + |
| #3 | 50.6 | f | Glioblastoma | 0.9 | 4 | MAB | − | Resection | + | + |
| #4 | 46.9 | f | Oligodendroglioma | 0.3 | 2 | HAB | + | Resection | − | + |
| #5 | 75.7 | f | Glioblastoma | 0.2 | 4 | MAB | − | Biopsy | − | + |
| #6 | 69.1 | f | Glioblastoma | 1.0 | 4 | LAB | − | Resection | + | − |
| #7 | 64.3 | f | Oligodendroglioma | 17.8 | 2 | − | + | Biopsy | + | − |
| #8 | 59.3 | m | Astrocytoma | 2.6 | 2 | HAB | + | Biopsy | + | + |
| #9 | 36.4 | f | Oligodendroglioma | 1.1 | 2 | HAB | + | Biopsy | − | + |
| #10 | 44.0 | m | Oligodendroglioma | 12.3 | 3 | MAB | + | Biopsy | + | + |
| #11 | 72.3 | f | Glioblastoma | 0.1 | 4 | − | − | Resection | − | − |
| #12 | 39.7 | f | Astrocytoma | 4.5 | 2 | − | + | Biopsy | − | − |
| #13 | 27.2 | f | Astrocytoma | 0.7 | 2 | − | + | Biopsy | − | − |
| #14 | 68.7 | m | Glioblastoma | 0.1 | 4 | LAB | − | Resection | − | + |
| #15 | 66.8 | f | Oligodendroglioma | 25.9 | 2 | − | + | Resection | + | − |
| #16 | 58.5 | m | Astrocytoma | 20.8 | 3 | − | + | Biopsy | + | + |
| #17 | 52.8 | f | Oligodendroglioma | 20.9 | 3 | HAB | + | Biopsy | + | + |
| #18 | 49.6 | m | Oligodendroglioma | 1.2 | 3 | HAB | + | Biopsy | + | + |
| #19 | 57.9 | m | Astrocytoma | 12.9 | 3 | MAB | + | Biopsy | + | − |
| #20 | 72.0 | m | Glioblastoma | 1.4 | 4 | HAB | − | Resection | − | + |
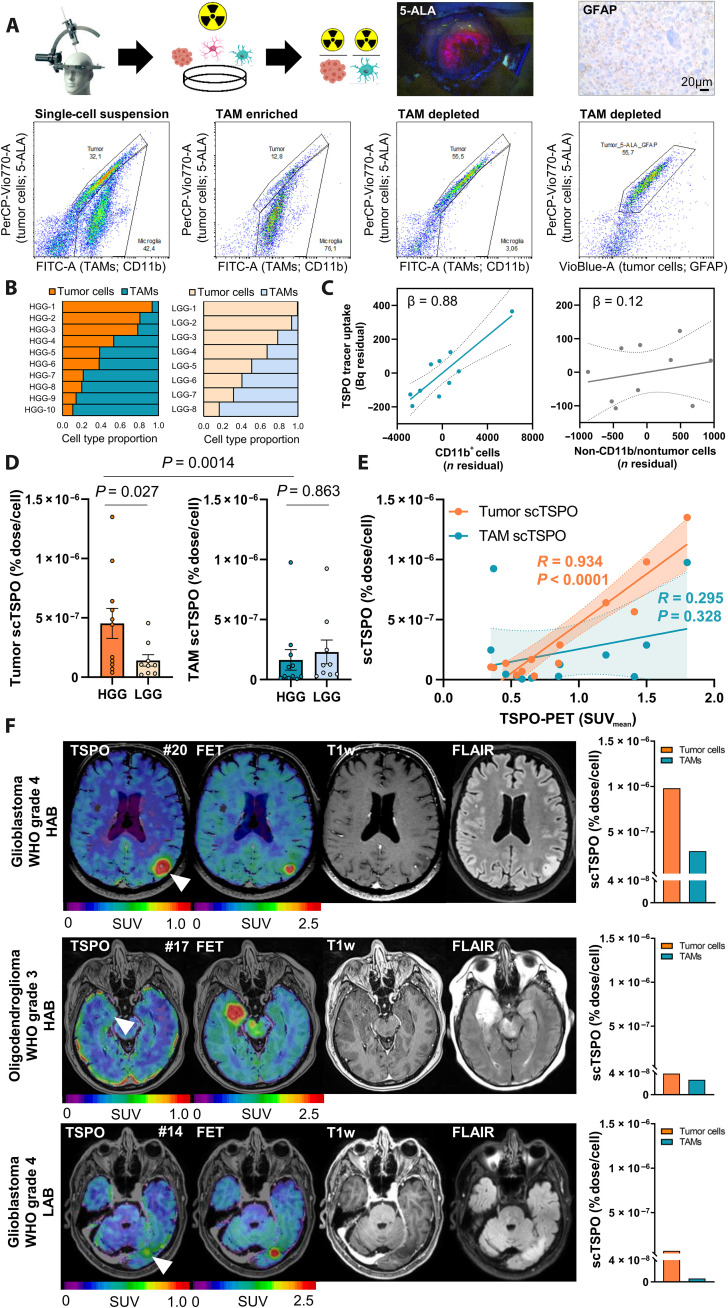
Fig. 4. Translation of scRadiotracing to human glioma samples.
(A) Schematic illustration of the workflow and the gating strategy in human glioma samples (n = 20). The single-cell suspension (left) was separated into TAM-enriched (CD11b+; second from left) and tumor-enriched (third from left) fractions. Tumor cells were defined via glial fibrillary acidic protein (GFAP) or 5-aminolevulinic acid (5-ALA) after confirmation of 5-ALA positivity during surgery or after confirmation of GFAP positivity during neuropathological workup. 5-ALA+ cells colocalized with GFAP+ cells (right). (B) Relative distribution of tumor cells and TAMs in the single-cell suspension of human high-grade glioma (HGG; n = 10) and low-grade glioma (LGG; n = 8) samples. The first two patients did not receive an analysis of the single-cell suspension. (C) Strong contribution of TAMs (CD11b+ cells) but not of CD11b−/nontumor cells to the measured activity (n = 10 biopsy samples). Linear regression, β = standardized regression coefficient. Error bands represent 95% confidence interval. (D) Comparison of scTSPO uptake of tumor cells and TAMs in samples of human HGG (n = 11) and LGG (n = 9) by a multivariate model including tumor grade, age, and sex. Means ± SEM. (E) Correlation of TSPO-PET signals with scTSPO of tumor cells and TAMs. N = 13, R = Pearson’s coefficient of correlation. Error bands represent 95% confidence interval. (F) Three patient examples with similar signals in amino acid [O-(2-[18F]fluoroethyl)-l-tyrosine (FET)] PET and only little contrast enhancement in MRI. The patient with high tumoral TSPO-PET signal [top row, glioblastoma, World Health Organization (WHO) grade-4, high-affinity binding status (HAB), patient #20] showed distinctly more scTSPO compared to the patients with only faint (middle row, oligodendroglioma, WHO grade-3, HAB, patient #17) or low [bottom row, glioblastoma, WHO grade-4, low-affinity binding status (LAB), patient #14] tumoral signal in TSPO-PET.
scRadiotracing resulted in enriched fractions with visually well discernible populations of tumor cells and TAMs (fig. S5). High-grade glioma indicated similar heterogeneity of tumor cell–to–TAM ratios in the single-cell suspension when compared to low-grade glioma (Fig. 4B). As the purity of tumor- and TAM-enriched fractions was limited in human samples, negligibility of TSPO tracer uptake of nontumor/non-TAM cells was proven by showing that only the number of isolated TAMs correlated with measured tracer signal in the TAM-enriched cell pellet (β = 0.875, P = 0.0021), whereas nontumor/non-TAM cells did not contribute to the overall magnitude of radioactivity in the same sample (β = 0.118, P = 0.542; Fig. 4C). Single-cell TSPO enrichment in TAMs was similar between patients with high-grade and low-grade glioma (1.6 × 10−7 ± 0.9 × 10−7 %ID versus 2.3 × 10−7 ± 1.0 × 10−7 %ID; P = 0.863), without significant explanation by age (P = 0.541) or sex (P = 0.869). Contrary, TSPO enrichment of tumor cells was higher in patients with high-grade glioma (4.5 × 10−7 ± 1.3 × 10−7 %ID) when compared to patients with low-grade glioma (1.4 × 10−7 ± 0.5 × 10−7 %ID; P = 0.027; Fig. 4D), again without significant explanation by age (P = 0.192) or sex (P = 0.799). Within the group of patients with high-grade glioma, TSPO enrichment of tumor cells was stronger when compared to TAMs (P = 0.0014; Fig. 4D). Contrary, TSPO enrichment of tumor cells showed no significant difference when compared to TAMs within the group of patients with low-grade glioma. We questioned whether previous therapy constitutes a potential confounder of single-cell tracer uptake, but we did not find any significant impact of a previous therapy (radiation and/or chemotherapy, 11 of 20) or resection (9 of 20) on TSPO enrichment of single tumor cells or TAMs (all P > 0.05). Noteworthy, in a small subset of patients, a head-to-head comparison of tissue samples acquired at primary diagnosis and recurrence indicated higher percentages of infiltrating myeloid cells in the recurrent situation compared to primary diagnosis, and we observed a higher degree of TSPO positivity in infiltrating myeloid cells compared to resident myeloid cells (fig. S6). Thus, these preliminary data suggest that a higher share of the TSPO-PET signal in the recurrent situation derives from myeloid cells, enhanced by the stronger TSPO expression of increased infiltrating macrophages.
In accordance with the combination of PET and scRadiotracing in the SB28 mouse model, we correlated single-cell TSPO enrichment with TSPO-PET imaging, which was performed in a subset of patients before surgery. TSPO enrichment of tumor cells (R = 0.934, P < 0.0001) but not TSPO enrichment of TAMs (R = 0.295, P = 0.328) was associated with the lesion signal in TSPO-PET (Fig. 4E). Individual patients with varying TSPO-PET signal intensities but similar O-(2-[18F]fluoroethyl)-l-tyrosine ([18F]FET) and contrast-enhanced magnetic resonance imaging (MRI) findings showed a strong agreement between lesion signals and corresponding tumor cell TSPO enrichment (Fig. 4F). Immunofluorescence TSPO positivity of the sections matched with high TSPO tracer uptake of single cells in a patient with glioblastoma (#20) and explained the distinct difference of the TSPO-PET signal compared to a patient with oligodendroglioma (#17). Both patients were characterized by similar immunofluorescence-derived cell density. The lower TSPO-PET signal in the second patient with glioblastoma (#14) most likely derived from a lower cell density in combination with low-affinity binding, as also indicated by the lower TSPO tracer uptake of single tumor cells and TAMs (fig. S6). Together, the dominance of tumor cell radiotracer uptake and the association between scRadiotracing and PET in human tissue samples closely reflected our findings in SB28 mice. Therefore, in vitro scRadiotracing provides an opportunity to efficiently check for consistency in cellular target binding between murine and human samples.
The triangle of PET, scRadiotracing, and 3D histology dissects cellular sources of regional PET signals and pinpoints dominant contribution of tumor cells to the TSPO-PET signal
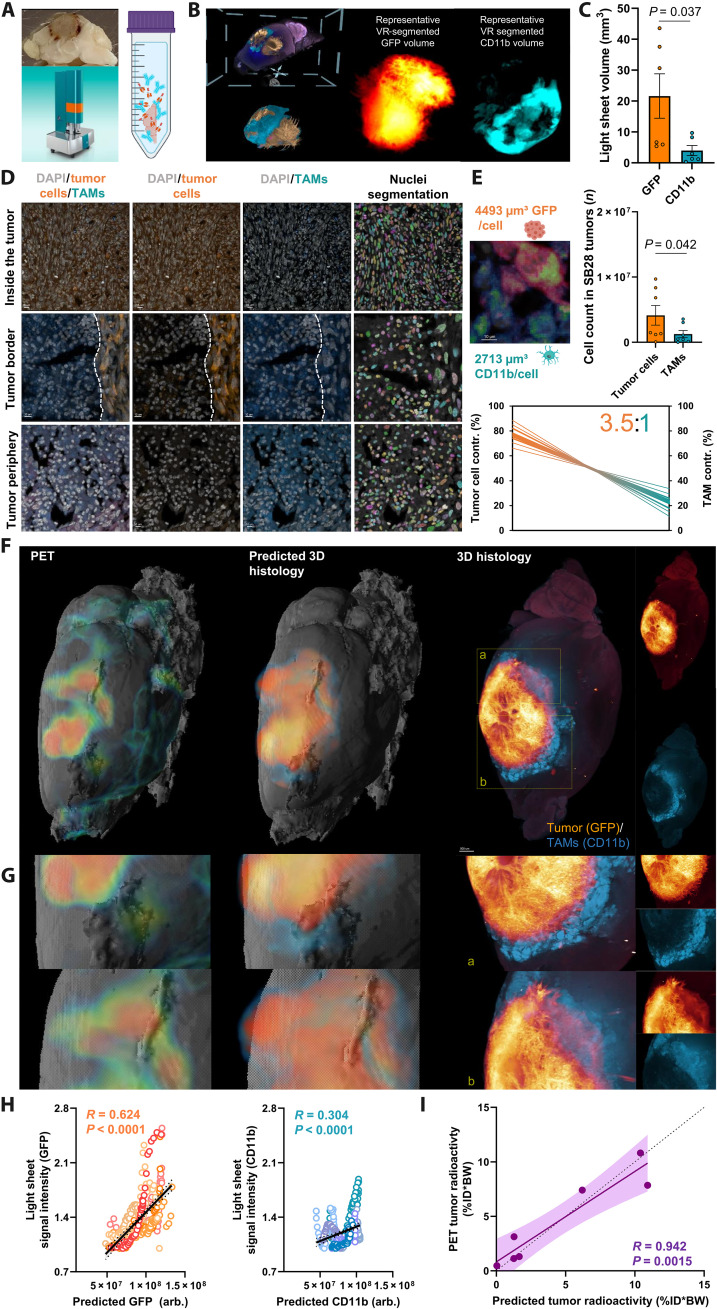
Since PET tracer signals are a product of cellular tracer uptake and regional cell type abundance, we used 3D histology via tissue clearing and light sheet microscopy (Fig. 5A) to determine absolute and relative cell numbers of GFP+ tumor cells and CD11b+ immune cells throughout entire SB28 tumors in the intact ipsilateral hemispheres. GFP+ volumes were 5.5-fold larger when compared to CD11b+ volumes (18.6 ± 6.8 mm3 versus 3.4 ± 1.5 mm3; P = 0.037; Fig. 5, B and C). Confocal microscopy revealed a GFP+ volume of 4493 μm3 per SB28 tumor cell and a CD11b+ volume of 2713 μm3 per TAM (Fig. 5D), resulting in higher cell count of SB28 tumor cells when compared to TAMs in individual SB28 lesions (Fig. 5E). Considering both single-cell tracer uptake and cell distributions of tumor cells and TAMs, we calculated a tumor cell–to–TAM contribution to the TSPO-PET signals in SB28 lesions of 3.5:1 (Fig. 5E).
Fig. 5. Integrated analysis of regional PET signals, cellular tracer uptake, and 3D histology.
(A) Tissue clearing with a modified version of 3DISCO (65) and light sheet florescent microscope imaging. (B) Representative masks of tumor cells and TAMs in the virtual reality (VR) annotation tool. This process determined the occupied volume of GFP+ tumor cells and CD11b+ TAMs within the whole tumor using segmentation via syGlass (69). (C) Quantitative comparison of GFP+ and CD11b+ volumes within individual SB28 tumors (n = 7, paired t test). (D) Confocal microscopy determined the average GFP+ volume per single tumor cell and the average CD11b+ volume per single TAM. Nuclei segmentation using cellpose (68) determined cell numbers per section (right column). (E) Quantitative comparison of the absolute tumor and TAM cell count (n = 7, paired t test) and contribution of tumor cells and TAMs to the overall TSPO-PET signal (3.5:1). Thick lines indicate the mean of scTSPO × top/bottom standard error of the mean (SEM) band of tumor volumes; thin lines indicate top/bottom SEM band of scTSPO × top/bottom SEM band of tumor volumes. (F) Regional TSPO-PET signals (left) were combined with single-cell tracer uptake values of tumor cells and TAMs to predict cell type abundance within individual SB28 tumors (middle) with validation by light sheet microscopy (right). All mice are illustrated in fig. S7. (G) Representative magnified areas within the SB28 tumor underline the regional agreement between predicted and standard of truth 3D histology. (H) Correlation of predicted and standard of truth 3D histology within k-means clusters (Fig. 3). n = 350 single regions from n = 7 tumors. R = Pearson’s coefficient of correlation. Error bands represent 95% confidence interval. (I) Prediction of radioactivity measured in PET by cellular abundance and cellular tracer uptake. R = Pearson’s coefficient of correlation. Error bands represent 95% confidence interval. Dashed line represents line of identity (y = x).
Next, PET, scRadiotracing, and 3D histology were used as an integrated concept. The topology of 3D histology could be predicted by combining lesion PET signals with single-cell tracer values (Fig. 5, F and G, and fig. S7). For this purpose, we used the regional association between single-cell tracer uptake of SB28 tumor cells and TAMs with TSPO-PET (i.e., correlation coefficients per cluster; Fig. 3F) as a cluster-based PET weighting factor. k-means cluster PET signals of all mice that received TSPO-PET imaging and 3D histology were multiplied with their respective correlation coefficients and divided by single-cell tracer uptake of both tumor cells and TAMs (Fig. 2D) to predict individual regional abundance (i.e., light sheet signal intensity) for each cell type (Fig. 5, F to H). Predicted and standard of truth 3D histology were strongly correlated (tumor cells: R = 0.624, P < 0.0001; TAMs: R = 0.304, P < 0.0001; Fig. 5H and fig. S7). Hot spots of GFP positivity were colocalized with regions of high TSPO-PET signal intensity (Fig. 5G), fitting to the strong association between tumor cell TSPO and regional TSPO-PET (Fig. 3F). The radioactivity in individual tumors as measured by PET was precisely predicted by aggregated single-cell tracer uptake and cell count of tumor cells and TAMs (Fig. 5I). In summary, exploiting the established methodological combination of PET, scRadiotracing, and 3D histology enables to pinpoint complex PET signals to their detailed cellular origins.
Proteomics identifies potential TAM-specific radiotracer targets with superior binding capacity when compared to TSPO
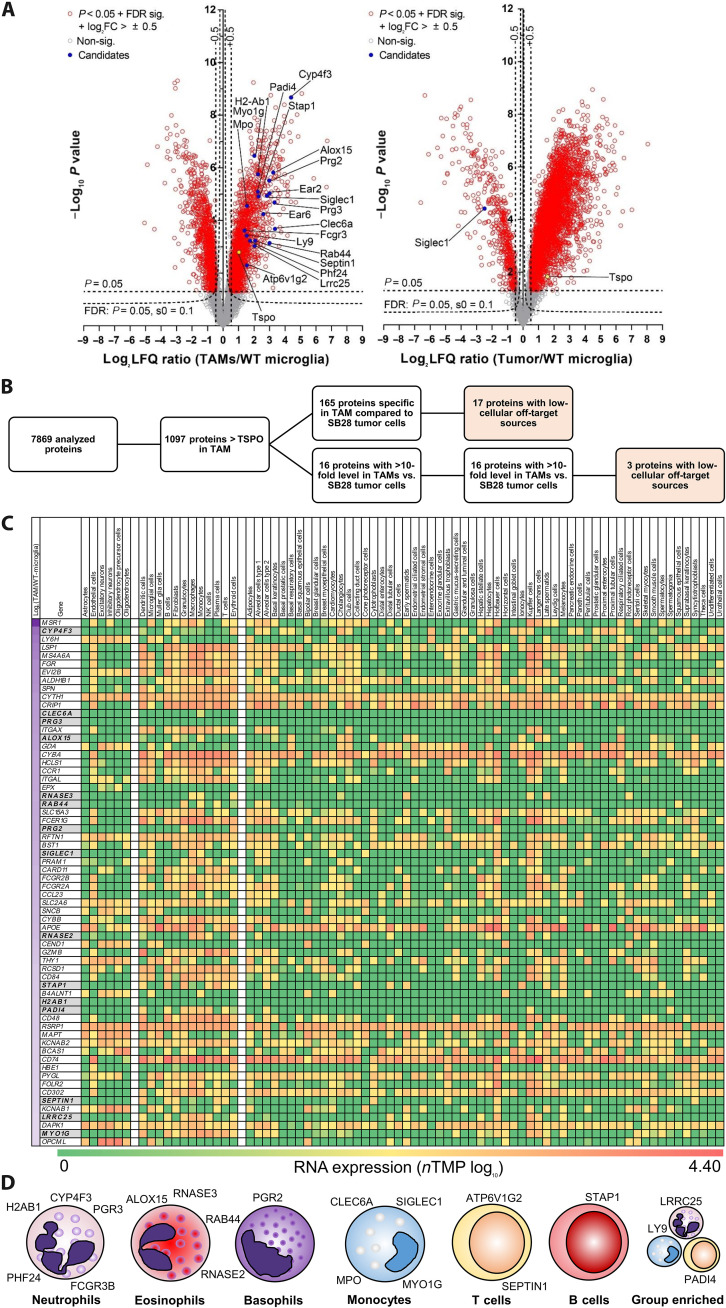
Since tumor cells dominate the TSPO-PET signal, the development of novel PET radiotracers with higher specificity for TAMs could provide added value to glioblastoma diagnostics. We therefore generated murine proteome data to identify potential TAM-specific radiotracer targets. Starting from a total of 7869 proteins relatively quantified between TAMs and microglia of healthy control mice, 1097 showed higher fold changes than TSPO (Fig. 6, A and B). A total of 165 of these proteins could not be quantified in SB28 tumor cells, and additional 16 proteins showed >10-fold higher levels in TAMs when compared to SB28 tumor cells (Fig. 6B), thus providing sufficient specificity for TAM over tumor cell expression.
Fig. 6. Proteome analysis of the SB28 TME identifies TAM-specific radiotracer targets for glioblastoma.
(A) Volcano plot representation of the different protein levels in isolated TAMs (n = 5) and isolated SB28 tumors cells (n = 3) in comparison with control microglia isolated from age-matched mice (WT-microglia; n = 6). TSPO (yellow) showed intermediate elevation of protein levels in TAMs and SB28 tumor cells. A total of 1098 of 7869 analyzed proteins indicated higher protein levels in TAMs than did TSPO. FC, fold change. (B) Selection process of potential radiotracer targets for specific detection of TAMs over SB28 tumor cells. A total of 165 of the identified proteins in TAMs were not detected in SB28 tumor cells, and another 16 proteins had >10-fold higher levels in TAMs compared to SB28 tumor cells. A total of 20 proteins showed low-cellular off-target sources and were identified as potential TAM radiotracer targets. (C) Heatmap screening for TAM specificity of the 181 proteins of interest. The Human Protein Atlas (17) was used to determine RNA expression levels of all proteins of interest in (i) resident off-target cells of the brain (left cell type columns), (ii) resident and infiltrating cells in the presence of glioblastoma (middle cell type columns), and (iii) off-target cells of the organism (right cell type columns). Genes are sorted by protein level differences in TAMs compared to control microglia of untreated mice (top to bottom; log2 label-free quantification ratio; first column). Sixty-four proteins with highest elevation [log2 (TAMs/WT-microglia) > 2; left column] are illustrated in (C) and all proteins are provided in fig. S8. Proteins of interest are highlighted in gray. (D) Immune cell expression cluster allocation of proteins of interest. Single-cell RNA data of the Human Protein Atlas were used to categorize the final set of identified proteins.
To translate these findings from mouse to human, we used the Human Protein Atlas (17) to identify expression of respective human gene homologs in off-target cells in the brain as an index of TAM specificity (see first set of columns in Fig. 6C and fig. S8). Physiological expression in resident and invading cells of the TME (see second set of columns in Fig. 6C and fig. S8) was characterized. In a next step, we ensured low target expression in other cells in the organism to prevent peripheral radiotracer absorbance since this may cause a sink effect and high radiation exposure (see third set of columns in Fig. 6C and fig. S8). This selection process resulted in 20 proteins with highly suitable characteristics as specific immune PET tracer targets (Fig. 6D), which were ultimately categorized into their immune cell expression cluster (Fig. 6D). CYP4F3, H2AB1, PGR3, FCGR3B, and PHF24 were allocated to the neutrophil expression cluster. ALOX15 and RNASE3 were associated with the eosinophil expression cluster, and PGR2 was localized in the basophil expression cluster, while both expression clusters were shared for RAB44 and RNASE2. An association with the monocyte expression cluster was identified for CLEC6A, MPO, MYO1G, and SIGLEC1. Few identified genes belonged to B cell (ATP6V1G2 and SEPTIN1) and T cell (STAP1) expression clusters, and group enrichment was found for LRRC25, LY9, and PADI4 (Fig. 6D). High protein abundance, which allows sufficient PET signal-to-noise ratios, was observed for ALOX15, PRG2, SIGLEC1, MYO1G, and MPO [label-free quantification (LFQ) intensity >18; fig. S9A]. To account for potential down regulation due to immunosuppression, we also considered the entire fraction of TAM-selective proteins identified in TAMs and microglia of healthy control mice as extended targets of interest (10.5%; fig. S9B). These proteins were screened for targets with scientific relevance in several areas of research and we identified NLRP3, CSF1R, and TREM2 as rising candidate proteins with a broad range of possible applications (fig. S9C). Radiolabeled ligands with binding affinity to these proteins may re-enter the workflow of scRadiotracing, 3D histology, and human translation to test for their cellular specificity in vivo.
DISCUSSION
Our study determined in vivo radiotracer uptake in the complex TME of glioblastoma at cellular resolution. After validation of scRadiotracing upon TSPO tracer injection, we deciphered the cellular sources of TSPO-PET signals in experimental glioblastoma and patients with glioma. The established methodology accurately determined higher radiotracer uptake of single glioblastoma tumor cells when compared to immune cells, which closely reflected protein levels of respective cell types. The combination of PET and single-cell tracer uptake served to predict 3D histology. Ultimately, a detailed proteome analysis facilitated identification of TAM-specific radiotracer targets beyond TSPO.
As a prerequisite of the main analysis, a methodological validation of TSPO radiotracer measures in single cells had to be addressed. We applied a microglia depletion paradigm (18) to compare the signal reduction observed by in vivo [18F]GE-180 TSPO-PET of mice after CSF1R inhibition with the total radioactivity attributable to the microglia cell population after scRadiotracing. Here, we confirmed that scRadiotracing was able to closely recover the tracer signal reduction observed in PET (Fig. 1). By CSF1R inhibition with PLX5622, microglia cells can be depleted up to 99%, which may be maintained under continuous treatment for up to 2 months (19). CSF1R inhibition likely has only minor effect on the TSPO gene expression of nonmyeloid central nervous system cells in wild-type mice (20), but we note that some downstream effect on other cell types cannot be fully excluded since the number of TSPO-positive neurons decreased upon CSF1R inhibition in a mouse model of prion-induced chronic neurodegeneration (21). Still, we claim that the reduction of the TSPO-PET signal derived from the nearly complete depletion of microglia after 7 weeks of PLX5622 treatment. Furthermore, we observed 12-fold higher single-cell TSPO tracer uptake of microglia when compared to astrocytes, pinpointing specificity of [18F]GE-180 to microglia in untreated mice. In addition, the high congruence between tracer signal in the microglia population and PET signal reductions after microglia depletion fits to previous investigations that outlined a high correlation between [18F]GE-180 TSPO-PET and the microglia markers Iba1 and CD68 in immunohistochemistry (22). With regard to the choice of the radiotracer, we note that other frequently used TSPO radioligands such as [18F]DPA-714, [11C]PBR28, or [11C]PK11195 may show better or worse uptake characteristics compared to [18F]GE-180 (23). Although direct comparisons of several TSPO ligands would be desirable in future studies, the short half-life of carbon-11–labeled TSPO tracers likely prevents the assessment of their cellular uptake via scRadiotracing.
We were able to further validate scRadiotracing in tumor cells and TAMs of the experimental SB28 glioblastoma model by quantification of relative cellular protein levels of TSPO in respective cell populations using proteomics. The application of proteomics was particularly important since transcriptomics does not necessarily reflect the amount of protein that can be targeted by the radiotracer (24). We also found that the interindividual heterogeneity of SB28 and human tumor PET signals was correlated with the magnitude of tracer uptake per each single cell, which implies that the PET signal is mainly driven by cellular uptake and not by confounding alterations of the blood-brain barrier (25) or off-target binding (26). Our observations of heterogeneous single-cell tracer uptake fit to the biological variability of TSPO that has been outlined not only in glioblastoma (7, 27) but also in neurodegenerative diseases (28, 29). Ultimately, the combination of PET and scRadiotracing with 3D histology confirmed the methodological capability to quantify cellular sources of in vivo PET signals. In this regard, PET tracer signals are not only a function of cellular uptake but also a product of cell type abundance and single-cell tracer uptake, making the combination of scRadiotracing and 3D assessment of specific cell counts (30), a major achievement of the current work.
In this study, we used TSPO as an established PET biomarker in neurological and neuro-oncological diseases with controversially discussed cellular signal sources. Although several correlative investigations questioned the cellular sources of TSPO tracer signals in glioma (6, 31), direct measures of radiotracer allocation have not yet been performed, and contributions of tumor and immune cells to the PET signal remained unclear. Notably, not all immune subsets of glioblastoma express TSPO, but TSPO is dominantly expressed by resident microglia (32) and infiltrating TAMs (6). Recent data showed that TSPO expression of glioblastoma cells is also involved in T cell–mediated cytotoxicity (33), which indicates that TSPO can also have an indirect impact on immune cells in the TME of glioblastoma. In this regard, T cells are a numerically small subset of immune cells in glioblastoma that typically do not express TSPO. Thus, it needs to be considered that T cells will not relevantly contribute to the immune cell–driven TSPO-PET signal in glioblastoma, although T cells constitute a major effector of immunotherapy (34). Our data indicated higher enrichment of [18F]GE-180 in SB28 tumor cells when compared to TAMs after in vivo tracer administration, and similar results were obtained in human high-grade glioma cells when compared to TAMs of the same tumor after in vitro tracer incubation. These findings were supported by stronger agreement of tumor TSPO-PET signals with individual tumor cell tracer uptake in mice and humans. 3D histology in SB28 mice moreover showed higher abundance of tumor cells when compared to TAMs, pinpointing the dominance of tumor cells to SB28 tumor TSPO-PET signals. Still, potential discrepancies between murine and human glioblastoma (e.g., inoculation versus spontaneous tumor growth) need to be considered. Although the SB28 model is reported to closely mimic the human TME (15) and our translational data indicate similar results regarding single-cell TSPO enrichment across species, the small sample size of the human analysis in respect to large heterogeneity in diagnoses and previous therapy strategies remains to be acknowledged. Further investigations in predefined subgroups with larger sample sizes may aid to disentangle and differentiate therapy-related effects on TSPO expression in cells of the human TME, i.e., after radiation therapy (35). The assessment of TSPO expression in specific immune cell subsets of glioblastoma could still be biologically very meaningful, even when tumor cells quantitatively dominate the TSPO-PET signal. Furthermore, a pilot experiment with nude GL261-implanted mice indicated that strong changes in TAM cell numbers may also have a considerable impact on TSPO-PET signals. Distinct differences in immune phenotypes of available syngeneic glioblastoma mouse models can also likely shift the proportions of cellular contributions to PET signals (36), which calls for comparative studies using TSPO-PET or novel immune PET tracers across various cell lines.
A high contribution of tumor cells to the TSPO-PET signal of glioblastoma is in agreement with preclinical investigations that used TSPO-PET and autoradiography after implantation of GL261 cells into a full TSPO knockout (31, 37). However, several preclinical studies have revealed a distinct colocalization of TSPO and Iba1/CD68 in immunohistochemistry in different glioblastoma mouse models and concluded that TAMs contribute substantially to the TSPO-PET signal (7–10). While some of these studies have already drawn a qualitative comparison between immunohistochemistry and TSPO-PET quantification by various ligands (8–10), our study provides direct evidence for simultaneous allocation of a TSPO-PET tracer in both cell types. Future research should also compare single-cell TSPO enrichment across the broad spectrum of different glioblastoma models (i.e., GL261) and primary human glioblastoma cell lines.
Various signal sources with a considerable impact on the TSPO-PET signal in the TME call for detailed interrogation of TSPO as a glioblastoma imaging biomarker. First, lacking specificity of TSPO-PET signals for TAMs in glioblastoma hampers the biomarker’s application as a mere immune index. However, since TSPO-PET signals are associated with survival times in patients with primary and recurrent glioma (38, 39), TSPO-PET still adds valuable biomarker features to the toolbox of glioblastoma imaging assessments (40). These quantitative TSPO-PET data even exceeded the prognostic value of gene expression levels (41), which could be related to the direct quantification of TSPO receptor abundance instead of indirect mRNA measures. In general, PET imaging is increasingly used for the assessment of patients with brain tumors, in addition to conventional MRI and provides clinically valuable information (40). As radiolabeled glucose ([18F]FDG) shows limited sensitivity for glioma tissue, radiolabeled amino acids such as [18F]FET are the tracer of choice for the delineation of tumor volumes for treatment planning, response assessment, and differentiation between tumor recurrence and posttherapeutic changes (40). However, in recent years, innovative PET tracers for improved brain tumor characterization, e.g., targeting tumor-associated immune cells, have gained increasing interest (42). TSPO-PET is one of these promising investigational candidates, but we note that the TSPO polymorphism in humans, which is associated with differential binding of TSPO ligands including PET tracers, may limit broader use outside university hospitals. Nevertheless, the pilot dataset of in vitro scRadiotracing in patients with glioma already showed feasibility even with small amounts of tissue obtained from biopsy. Thus, the current results should encourage to investigate the separate prognostic value of single-cell TSPO enrichment in tumor and immune cells since such data would potentially result in different treatment strategies. Distinct binding of radioligands to different cells of the TME will also affect personalized medicine, including direct targeting of various tumors with radiotheranostics.
Beyond TSPO, TAM-specific radioligands for assessment of immune cells in the TME could facilitate monitoring of novel immunotherapies (43). We used our murine bulk proteomics dataset to classify TSPO within the spectrum of potential radiotracer targets. In this regard, we note that single-cell profiling approaches (44, 45) are more comprehensive to understand potential immune cell radiotracer targets at a granular level. Contrary, our bulk proteome analysis of one murine cell line only identified targets that are enriched across the whole CD11b+ population in contrast to tumor cell–enriched samples. Nevertheless, this proof of concept allowed a direct comparison of protein levels and cellular tracer binding in the same samples, which can act as a benchmark for identification of novel immune radiotracer targets. Since TSPO was only moderately elevated in TAMs and surpassed by the TSPO expression in tumor cells, there is a major need for further improvement, identification, and development of TAM-specific radiotracers. The identified targets need to be further evaluated in terms of ligand accessibility and stable target expression and screened for available crystal structures or known binders that aid generation of lead molecules for radiotracer development. Several of the identified proteins matched with signature genes of immunosuppressive [i.e., ALOX15 (46)] TAM phenotypes or predicted therapy response upon checkpoint inhibition [i.e., LY9 (47) or MPO (48)], thus indicating their potential as biomarkers of immunomodulation in cancer. Noteworthy, several rising immune targets with relevance not only in glioblastoma but also in other areas of research, such as NLRP3 (49), CSF1R (50), and TREM2 (51), were also located within the small proportion of TAM-selective proteins, allowing broad application of an emerging radiotracer. scRadiotracing will provide the opportunity for precise determination of the in vivo specificity of potential radioligands to immune targets in their developmental process despite the complexity of the TME. We note that future studies including scRadiotracing in glioblastoma should also focus on immune cell and TAM subsets to allow development of improved imaging approaches of the complex TME. Single-cell profiling approaches will provide the opportunity to perform preselection of immune subsets before cell sorting with specific antibodies.
In a nutshell, the established methodology yields robust and reliable quantification of cellular radiotracer uptake in mice and patients and will serve to disentangle cellular sources of PET signals of established or newly developed tracers. This is of particular importance since many potential radiotracer targets are shared between tumor cells, TME cells, and resident cells of the brain (52).
MATERIALS AND METHODS
Study design
The primary goal of the study is to resolve PET tracer signals at cellular resolution. We challenged the methodology by targeting TSPO as a biomarker with complex cellular sources in glioblastoma. For this purpose, a combination of radiotracer injection and MACS was applied [scRadiotracing (11)]. To validate the workflow, the MACS data of microglia in untreated mice were compared to microglia depletion experiments using CSF1R inhibition in combination with [18F]GE-180 TSPO-PET. In mice with orthotopic implanted tumor (SB28; 2.5 weeks after inoculation) or sham injection, the brain was removed 75 min after [18F]GE-180 injection, and whole-brain activity was measured. The tumor or the injection site was dissected, followed by radioactivity measurement and dissociation of the tumor tissue into a single-cell suspension. Tumor cells (negative selection) and TAMs (CD11b) were isolated via magnetic cell sorting using specific antibodies. The radioactivity in the cell-specific pellets was measured by a highly sensitive gamma counter. To determine the absolute cell number in each pellet and the proportions (i.e., purity) of the different cell populations, flow cytometry was performed subsequent to cell sorting. TSPO tracer uptake was determined per single cell and compared to TSPO protein levels, whereas contributions of the different cell populations to the total activity were calculated by multiple regression. The methodology was translated to human resection and biopsy samples of patients with high-grade and low-grade glioma, and in vitro scRadiotracing was performed to determine proportions of tumor and immune cell tracer uptake. To acknowledge PET signals as product of cellular uptake and cellular abundance, 3D histology via light sheet microscopy was performed in perfused mouse brains to obtain absolute cell counts and relative quantitative proportions of glioblastoma and immune cells within the tumor. Then, an integrated model of PET, single-cell tracer uptake, and 3D histology was used to disentangle regional PET signals by their distinct cellular components. Last, TSPO protein levels were characterized within the whole spectrum of the tumor and immune cell proteome, with the goal to determine TAM-specific radiotracer targets. The local ethics committee of the LMU medical faculty gave permission to perform the human study (Institutional Review Board 601-16 and 17-457, updated protocol 23-0339). All patients signed an informed consent form.
Animals
All animal experiments were performed in compliance with the National Guidelines for Animal Protection, Germany and with the approval of the regional animal committee (Government of Upper Bavaria) and overseen by a veterinarian. All animals were housed in a temperature- and humidity-controlled environment with a 12-hour light-dark cycle, with free access to food (Ssniff, Soest, Germany) and water.
For scRadiotracing experiments, 8-week-old C57BL/6 mice were purchased from Charles River (Sulzfeld, Germany) and acclimated for at least 1 week. At day 0, the mice were randomized inoculated with 100,000 SB28-GFP cells suspended in 2 μl of Dulbecco’s modified Eagle’s medium (DMEM) (Merck, Darmstadt, Germany) (glioblastoma mice, n = 27) or 2 μl of saline (sham mice, n = 14). Additional n = 8 mice received no treatment before scRadiotracing, serving as control animals. For inoculation, mice were anesthetized with intraperitoneal injections of 10% ketamine (100 mg/kg) and 2% xylazine (10 mg/kg) in 0.9% NaCl. Anesthetized mice were immobilized and mounted onto a stereotactic head holder (David Kopf Instruments, Tujunga, CA, USA) in the flat-skull position. After surface disinfection, the skin of the skull was dissected with a scalpel blade. The scull was carefully drilled with a micromotor high-speed drill (Stoelting Co., Wood Dale, IL, USA) 2-mm posterior and 1-mm left of the bregma. By stereotactic injection, 1 × 105 cells were applied with a 10-μl Hamilton syringe (Hamilton, Bonaduz, Switzerland) at a depth of 2 mm below the drill hole. Cells were slowly injected within 1 min, and after a settling period of another 2 min, the needle was removed in 1-mm steps per minute. After that, the wound was closed by suturing. Mice were checked daily for tumor-related symptoms and euthanized when tumor burden (i.e., appearance, coordinative deficits, and motor symptoms) reached stop criteria (not reached in any animal). On the last day of the experiment, the mice were injected with 15 ± 1 MBq of [18F]GE-180 into the tail vein before cervical dislocation and brain extraction at 75 min after injection (n = 33). N = 15 of these glioblastoma and all sham mice received TSPO-PET imaging directly before brain extraction. Additional n = 7 glioblastoma mice received TSPO-PET imaging and perfusion with 4% paraformaldehyde (PFA) before 3D histology by light sheet microscopy. Another n = 14 C57BL/6 mice were used for extraction of naïve control microglia (n = 8 scRadiotracing and n = 6 proteomics). N = 3 glioblastoma mice and n = 3 sham mice received injection of 0.345 μmol of cold GE-180 (25,000-fold excess), 20 min before injection of [18F]GE-180 and subsequent scRadiotracing. N = 7 glioblastoma mice (d17) received scRadiotracing with splitting of tumor cell–enriched and TAM-enriched samples into unstained processing and processing of all variants of isotype control combinations. N = 7 glioblastoma mice received TSPO-PET imaging at day 18 and perfusion with 4% PFA at day 19 before immunohistochemistry for Iba1 and TSPO. As a pilot experiment, 100,000 GL261–blue fluorescent protein cells were inoculated in one wild-type mouse and two Fox1nu mice, with TSPO-PET and scRadiotracing at day 19.
For validation of scRadiotracing as a reliable method to determine TSPO-PET signal sources at cellular resolution, we analyzed combined data of pharmacological depletion of microglia by CSF1R inhibition (53), which was performed in C57BL/6 mice of two datasets (13, 14) by PLX5622 [1200 parts per million (ppm), n = 15] and vehicle controls (n = 14), aged 10.1 ± 2.1 months. The TSPO-PET scan was performed in the last week of 7 weeks of treatment. All sample sizes were determined by previous PET data using G*power.
Cell culture
SB28-GFP cells (54) were cultured in DMEM containing minimum essential medium nonessential amino acids (1×), 1% penicillin-streptomycin solution (Thermo Fisher Scientific, Waltham, MA, USA), and 10% fetal bovine serum (Biochrome, Berlin, Germany). Cell cultures were maintained in the incubator at 37°C in humidified and 5% CO2-conditioned atmosphere. Cells were passaged when the cell density in the flask reached 80% confluence.
Radiosynthesis
Automated production of [18F]GE-180 was performed on a FASTlab synthesizer with single-use disposable cassettes. The prefilled precursor vial was assembled on the cassette, and the cassette was mounted on the synthesizer according to the setup instructions. The FASTlab control software prompts were followed to run the cassette test and to start the synthesis. No carrier–added 18F-fluoride was produced via 18O(p, n)18F reaction by proton irradiation of 18O-enriched water and delivered to the 18F incoming reservoir. The fully automated manufacturing process consists of the following steps: trapping of 18F-fluoride on a quaternary methyl ammonium cartridge, elution using Kryptofix222, potassium hydrogen carbonate, water and acetonitrile, azeotropic drying of 18F-fluoride at 120°C for 9 min, labeling of the precursor in MeCN at 100°C for 6 min, dilution of the crude product with water, tC18 cartridge–based purification by use of 20 ml of 40% (v/v) ethanol and 11.5 ml of 35% (v/v) ethanol, elution of the product with 3.5 ml of 55% (v/v) ethanol, and final formulation with phosphate buffer. Radiochemical yield 39 ± 7% (n = 16) non d. c., synthesis time 43 min, radiochemical purity ≥ 98%. Specific activity was 3285 ± 1000 GBq/μmol (range, 1698 to 4898 GBq/μmol) at the end of the synthesis.
Immunomagnetic cell separation
MACS was performed as described previously (13) with slight modifications for additional tumor cell isolation. Detailed descriptions of brain dissociation and isolation of different cell types were as follows.
Mouse brain dissociation
Adult mouse brains were removed after cervical dislocation at 75 min after injection and stored in cold Dulbecco’s phosphate-buffered saline (D-PBS). The brains were cut into small pieces and dissociated using gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec, 130-096-427) in combination with different dissociation kits according to the manufacturer’s instructions. Adult brain dissociation kit, mouse and rat (Miltenyi Biotec, 130-107-677), was used for adult mouse brain dissociation of untreated mice and sham mice. Tumor dissociation kit (Miltenyi Biotec, 130-096-730) was used for dissociation of tumor tissue. The dissociated cell suspension was applied to prewet 70-μm cell strainer (Miltenyi Biotec, 130-110-916). The cell pellet was resuspended using cold D-PBS and cold debris removal solution. Cold D-PBS was gently overlaid on the cell suspension and centrifuged at 4°C and 3000g for 10 min with acceleration at 9 and deceleration at 5. The two top phases were removed entirely. The cell pellets were collected. Cell pellets were resuspended with 1 ml of cold red blood cell removal solution, followed by 10-min incubation. Cell pellets were collected for further applications.
Isolation of tumor cells
Tumor cell isolation kit, mouse (Miltenyi Biotec, 130-110-187), was used according to the manufacturer’s instructions. The prepared cell pellets were resuspended in 80 μl of D-PBS–0.5% bovine serum albumin (BSA) buffer per 107 total cells including red blood cells. Twenty microliters of nontumor cell depletion cocktail was added, and the suspension was incubated at 4°C in the dark for 15 min. The volume was adjusted to 500 μl per total 107 cells with D-PBS–0.5% BSA buffer before proceeding to magnetic separation. The prewet LS columns (Miltenyi Biotec, 130-042-401) were placed at QuadroMACS Separator (Miltenyi Biotec, 130-090-976). The cell suspensions were applied onto the column. The columns were washed with 2 × 1 ml of D-PBS–0.5% BSA buffer. The flow-through containing the unlabeled cells was collected as the tumor cell–enriched fractions. The columns were removed from the magnetic field, and the nontumor cells were flashed out using 3 ml of D-PBS–0.5% BSA buffer.
Isolation of TAMs and microglia
TAMs (glioblastoma mice) or microglia (sham-injected or untreated control mice) were isolated from animals using CD11b MicroBeads, human and mouse (Miltenyi Biotec, 130-049-601), and a MACS separation system (Miltenyi Biotec) as described previously (55, 56). For murine samples, the prepared cell pellets were resuspended in 90 μl of D-PBS–0.5% BSA buffer per 107 total cells. Ten microliters of CD11b MicroBeads per 107 total cells were added and incubated for 15 min in the dark at 4°C. Human samples were resuspended in 80 μl of D-PBS–0.5% BSA buffer per 107 total cells, 20 μl of CD11b MicroBeads per 107 total cells were added, and cells were incubated for 15 min in the dark at 4°C. Cells were washed by adding 1 to 2 ml of buffer per 107 cells and centrifuged at 300g for 10 min. The cell pellets were resuspended in 500 μl of D-PBS–0.5% BSA. The prewet LS columns (Miltenyi Biotec, 130-042-401) were placed onto a QuadroMACS Separator (Miltenyi Biotec, 130-090-976). The cell suspensions were applied onto the column. The columns were washed with 3 × 3 ml of D-PBS–0.5% BSA buffer. The flow-through containing the unlabeled cells was collected as the microglia-depleted fractions. The columns were removed from the magnetic field, and microglia were flashed out using 5 ml of D-PBS–0.5% BSA buffer.
Isolation of astrocytes
Adult brain dissociation kit, mouse and rat (Miltenyi Biotec, 130-107-677), was used according to the manufacturer’s instructions. The prepared cell pellets were resuspended in 80 μl of AstroMACS separation buffer (Miltenyi Biotec, 130-117-336) per 107 total cells. Ten microliters of Fc receptor blocking reagent was added and incubated for 10 min in the dark at 4°C. Ten microliters of anti-ACSA2 MicroBeads were added and incubated for 15 min in the dark at 4°C. Cells were washed by adding 1 ml of AstroMACS separation buffer and centrifuged at 300g for 5 min. Cell pellets were resuspended in 500 μl of AstroMACS separation buffer. The prewet MS columns (Miltenyi Biotec, 130-042-201) were placed at OctoMACS Separator (Miltenyi Biotec, 130-042-109). The cell suspensions were applied onto the column, followed by washing with 3 × 500 μl of AstroMACS separation buffer. The flow-through was collected containing nonastrocytic cells as an astrocyte-depleted fraction. The columns were removed from the magnetic field, and the astrocytes were flashed out using 1 ml of AstroMACS separation buffer.
Gamma emission measurements
Radioactivity concentrations of cell pellets were measured in a gamma counter (Hidex AMG Automatic Gamma Counter, Mainz Germany), cross-calibrated to the activity in the whole brain, with decay correction to time of tracer injection for final activity calculations.
Flow cytometry
Flow cytometry staining was performed at 4°C. After gamma emission measurement, the cell suspension was centrifuged at 400g for 5 min, and the supernatant was aspirated completely. The cell pellet was then resuspended in 100 μl of cold D-PBS containing fluorochrome-conjugated antibodies recognizing mouse CD11b and ACSA2 (Miltenyi Biotec, 130-113-810 and 130-116-247) in a 1:100 dilution and incubated for 10 min at 4°C in the dark. Samples were washed with 2 ml of D-PBS and centrifuged for 5 min at 400g. Last, cell pellets were resuspended in 500 μl of D-PBS, and samples were immediately used for flow cytometry using a MACSQuant analyzer. Precision Count Beads (BioLegend, 424902) were added for counting the absolute number of cells for the samples of n = 9 mice measured with a BD LSRFortessa cell analyzer (BD Biosciences, Franklin Lakes, NJ, USA). Acquired data included absolute cell numbers and purity of GFP+, CD11b+, and ACSA2+ cells in each sample.
TSPO costaining was performed for tumor- and CD11b-enriched cell fractions of n = 3 SB28 mice. To this end, cells were permeabilized after initial flow cytometry using Inside Stain Kit (Miltenyi Biotec, 130-090-477) according to the manufacturer’s instructions. Cell pellets were resuspended in 250 μl of cold D-PBS, 250 μl of Inside Fix Solution was added, and cells were incubated for 20 min in the dark at 4°C and then centrifuged at 300g for 5 min. Complete aspiration of supernatant was followed by washing with 1 ml of cold D-PBS and another centrifugation step at 300g for 5 min. Cell pellets were washed with 1 ml of Inside Perm Solution and centrifuged at 300g for 5 min. Supernatant was aspirated, cells were resuspended in 47 μl of Inside Perm Solution and stained with 3 μl of anti-PBR antibody [EPR5384] (Abcam, ab199836). After 10 min of incubation, cells were washed by adding 1 ml of Inside Perm Solution, centrifuged at 300g for 5 min, and resuspended in 300 μl of cold D-PBS for flow cytometry analysis.
Validation experiments with isotype controls and unstained samples were performed using n = 7 glioblastoma mice at day 17 after inoculation. Isotype controls as recommended by the manufacturers were used for CD11b and TSPO: REA control antibody, human immunoglobulin G (IgG1) (Miltenyi Biotec, 130-113-454); rabbit IgG, monoclonal [EPR25A]–isotype control (ab199093). SB28 tumor cell–enriched samples and TAM-enriched samples were split up into unstained (10%), CD11b/TSPO (22.5%), IgG1/TSPO (22.5%), CD11b/IgG (22.5%), and IgG1/IgG (22.5%) before flow cytometry. All other pellet processing was performed as described above.
For subsequent proteome analysis (see below), cell pellets of n = 3 tumor-enriched and n = 5 CD11b-enriched fractions of SB28 tumor samples were stored at −80° together with n = 6 CD11b-enriched fractions (i.e., control microglia) derived from an independent cohort of age- and sex-matched untreated control mice.
Calculation of single-cell TSPO tracer signal
Measured radioactivity (in becquerels) of cell pellets was divided by the specific cell number in the pellet resulting in calculated radioactivity per cell. Radioactivity per cell was normalized by injected radioactivity and BW (i.e., %ID*BW). Published cell numbers of microglia (12) were used to extrapolate the whole-brain radioactivity located in the microglia fraction of untreated mice. For tumor probes, cell numbers (tumor cells and TAMs) determined by light sheet microscopy were multiplied with single-cell %ID*BW as an estimate of cell fraction–specific contributions to the PET signal.
Human samples and in vitro scRadiotracing
Human tumor tissue samples were acquired during neurosurgical biopsy or open resection and stored in tissue storage solution (Miltenyi Biotec, 130-100-008) for 2 to 30 hours until further processing for scRadiotracing. Details on the patient cohort are provided in Results (Table 1). Tissue was manually cut into smaller pieces if necessary and dissociated using tumor dissociation kit, human (Miltenyi Biotec, 130-095-929). Removal of debris and red blood cells was performed for tumor resections as described above. Single-cell suspensions were incubated in vitro at a concentration of 56 ± 8 MBq of [18F]GE-180 in 1 ml for 30 min, then washed twice with 3 ml of cold D-PBS, and centrifuged at 400g for 5 min. Cell pellets were resuspended in 100 μl of cold D-PBS and stained with fluorochrome-conjugated antibodies recognizing human CD11b and glial fibrillary acidic protein (GFAP; Miltenyi Biotec, 130-113-810 and 130-123-846) as described above. Gamma counter measurement and flow cytometry analysis of the stained single-cell suspension were performed using 10% of the probe after resuspension in 500 μl. Subsequently, TAMs were isolated using CD11b microbeads (Miltenyi Biotec, 130-093-634), followed by further gamma counter measurement and flow cytometry. Proportions and cell count of TAMs in the resulting cell pellets were determined using CD11b. For tumor cells, a step-wise validation of the gating strategy was performed. Detection of GFAP+ cells was used for n = 6 samples where GFAP positivity was validated by staining during neuropathological workup of the same tumors. N = 4 patients received 5-aminolevulinic acid (5-ALA) guided surgery and differentiation of tumor cell populations by 5-ALA positivity in flow cytometry (57). These n = 10 samples were used for cross-validation of autofluorescence-based gating (excitation wavelength of 488 nm and emission wavelengths of 655 to 730 nm) (58), which was applied in another n = 10 samples without GFAP or 5-ALA positivity.
For samples containing very high proportions of TAMs, in the first step, the radioactivity per single TAM cell was calculated using the well-purified CD11b+ TAM–enriched fraction (see gating and analysis strategy A in fig. S5). Since the tumor cells in the depleted fraction were not purified to a satisfactory degree (<70%), the radioactivity attributable to the fraction of TAM cells was subtracted from the total activity of the depleted fraction, and the remaining radioactivity was divided by the total cell number of tumor cells in the depleted fraction for calculation of radioactivity per single tumor cell. For samples containing very low concentrations of TAMs, in the first step, the activity per single tumor cell was calculated in the CD11− fraction (see gating and analysis strategy B in fig. S5). Since TAMs in the CD11b+ fractions were not purified to a satisfactory degree (<70%), the radioactivity attributable to tumor cells was subtracted from the measured radioactivity in the TAM-enriched fraction, and the remaining radioactivity was divided by the total cell number of CD11b+ cells in the TAM-enriched fraction to obtain radioactivity per TAM. Negligibility of TSPO tracer uptake by non-TAM and nontumor cells for the overall radioactivity in the cell pellets was confirmed by a regression model of cell numbers and gamma emission in TAM-enriched fractions of all biopsy samples.
Small-animal PET/computed tomography
All small-animal PET procedures followed an established standardized protocol for radiochemistry, acquisition, and postprocessing (59, 60). For tumor and sham mice, [18F]GE-180 TSPO small-animal PET (12 ± 1 MBq, n = 36) recordings with an emission window of 0 to 60 min after injection were obtained to measure cerebral TSPO expression before sorting using a Mediso PET/computed tomography (CT) system (Mediso, Budapest, Hungary). A contrast enhanced CT was performed before the PET scan. For the mice of the depletion experiment, a static [18F]GE-180 emission (13 ± 2 MBq) was recorded between 60 and 90 min after injection using a harmonized Siemens Inveon DPET (Siemens, Munich, Germany). All small-animal PET experiments were performed with isoflurane anesthesia (1.5% at time of tracer injection and during imaging; delivery 3.5 liters/min).
All analyses were performed by PMOD (V3.5, PMOD Technologies, Basel, Switzerland) using CT (tumor and sham mice) and tracer-specific templates (depletion experiment) for spatial normalization (59). For tumor mice, a 40- to 60-min frame was analyzed and normalization of activity was performed by SUV. The average TSPO-PET SUV was obtained from a spherical volume of interest at the tumor site as the primary PET readout (16). Within the tumor volume of interest, we performed a 50-step k-means clustering using the PMOD segmentation tool (10 iterations) to delineate individual clusters of tumor tracer uptake in each tumor. For correlation analysis with light-sheet microscopy, a 50% threshold was applied before clustering to avoid artificial associations by inclusion of necrotic parts of the tumor. For the depletion experiment, normalization of injected activity was performed by the previously validated myocardium correction method (61), and PET estimates deriving from a whole-brain volume of interest (59) were extracted.
Correlation between small-animal TSPO-PET and TSPO immunohistochemistry
Immunohistochemistry was performed to assess the TSPO expression levels in the tumor core, the tumor boarder, and the infiltration zone. N = 7 SB28 glioblastoma mice perfused at day 19 after inoculation that received a TSPO-PET on day 18 were used for this analysis. To this end, PFA-fixed 80-μm-thick coronal brain sections were incubated overnight at 4°C in PBS with 5% normal goat serum and 0.5% Triton X-100 containing guinea pig monoclonal anti-Iba1 primary antibody (1:500; Synaptic Systems GmbH, Göttingen, Germany, 234308) and rabbit monoclonal anti-TSPO primary antibody (1:500; EPR5384, Abcam, Cambridge, UK, ab109497). Afterward, slices were washed three times with PBS supplemented with 0.5% Triton X-100, and, subsequently, slices were incubated for 2 hours at room temperature (RT) with a suitable secondary antibody. Imaging was performed on a confocal microscope (LSM 780 Axio invers, Carl Zeiss AG, Jena, Germany) with a ×20 objective in two coronal sections. Images contained a volume of 0.002 mm3. Target areas were selected on the basis of the tumor anatomy and consisted of the tumor core, the tumor boarder, and the infiltration zone. Images were processed with the ZEN 3.1 software, and image analysis was performed with Fiji/ImageJ by quantifying the area with a signal over a certain threshold for GFP, Iba1, and TSPO. Furthermore, TSPO area% was obtained in thresholded GFP+ and Iba1+ areas. TSPO-PET SUVmean values were determined from matching areas (1-mm3 spheres) and correlated with immunohistochemistry.
Multiplexed immunofluorescence and image analysis
A small subset of patients, including patient #3 (recurrence), patient #6 (primary and recurrence), and patient #20 (primary and recurrence), were analyzed for the comparison of infiltrating and resident myeloid cells via immunofluorescence. Formalin-fixed and paraffin-embedded (FFPE) tissue sections were deparaffinized and rehydrated. Antigen retrieval was performed using a pressure cooker and 1× citrate buffer, pH 6.0. Subsequently, sections were stained using the Opal Polaris 7 Color Manual IHC Detection Kit (Akoya Biosciences). Image analysis was performed by using inForm analysis software (Akoya Biosciences) and the open-source software for digital pathology image analysis QuPath. The following primary antibodies were used at a 1:100 dilution: CD11b recombinant rabbit monoclonal antibody clone JU93-81 (Thermo Fisher Scientific, MA5-32793), Purified anti-human CD162 antibody clone KPL-1 (BioLegend, 328802), and TSPO polyclonal antibody (Thermo Fisher Scientific, PA5-75544).
Multiplex Opal immunofluorescence staining and counting
Sections of patients #14, #17, and #20 underwent a second workflow of immunofluorescence to correlate TSPO-PET signals, scRadiotracing, and histology. For fluorescent, 3-plex staining of TSPO/CD11b/4′,6-diamidino-2-phenylindole (DAPI) and p53/TSPO/DAPI the Opal system (Akoya Biosciences) were used. TSPO were stained with the recombinant anti-PBR antibody [EPR5384] (Abcam, ab109497; 1:5000), CD11b with a monoclonal CD11b antibody [C67F154] (eBioscience, 14-0196-82; 1:500) and p53 with a monoclonal p53 antibody [DO-7] (Dako, M7001; 1:50). Heat-induced antigen retrieval for TSPO and CD11b was performed in 10 mM citrate buffer (pH 6.0) for p53 in 1 mM tris/EDTA buffer (pH 9.0). Briefly, paraffin sections (3 μm) were baked (50°C), deparaffinized, and rehydrated. Antigen retrieval for the first target stain was performed for 30 min. After washing (PBS with 0.05% Tween 20), sections were blocked for 10 min with kit-provided endogenous enzyme block (Dako, K4065) following a 1% BSA incubation for 10 min (RT). Incubation with primary antibodies was performed (TSPO, 1 hour at RT; p53, overnight at 4°C). After washing, sections were labeled with Opal polymer horseradish peroxidase Ms + Rb (10 min at RT). After additional washing, sections were incubated with Opal Dye working solution (10 min at RT). After another washing step, the second target stain was performed like the first one (CD11b, overnight at 4°C; TSPO, 1 hour at RT). Last, another antigen retrieval step in 10 mM citrate buffer (pH 6.0) was followed by nuclear counterstaining using spectral DAPI solution [TBST (1 droplet/ml) (pH 7.5) and 0.05% Tween 20] for 5 min at RT. Last, slides were coverslipped using Aquatex (Merck KGaA, 1.08562.0050), and JPEG images (400×) were counted within Fiji (62) with the Cell Counter plugin (https://biii.eu/cell-counter-imagej).
Human PET/CT
TSPO-PET was available for 13 of 20 (65%) patients who underwent scRadiotracing. All human TSPO-PET scans were performed on a Biograph 64 PET/CT scanner (Siemens, Erlangen, Germany). Tracer production and image acquisition were performed as described previously (63). Approximately 180 MBq of [18F]GE-180 was injected as an intravenous bolus, and summation images 60 to 80 min after injection were used for image analysis using a Hermes workstation (Hermes Medical Solutions, Stockholm, Sweden). MRI sequences included gadolinium-enhanced T1- and T2-weighted images and were used for anatomical mapping of stereotactic biopsy coordinates and resected tumor mass within the TSPO-PET images using Brainlab Elements (Brainlab AG, Munich, Germany). For correlation analysis between scRadiotracing and TSPO-PET imaging, the mean SUV was obtained from a 0.2-cm3 (25 voxels) volume of interest using the sample coordinates.
Amino acid PET was available for a subset of the investigated patients (80%) and served for characterization of vital tumor tissue. Approximately 180 MBq of [18F]FET was injected, and summation images 20 to 40 min after injection were analyzed. Tumor uptake was assessed as maximum SUV (SUVmax). As described previously (64), the mean background activity was defined as the mean activity of at least six crescent-shaped cortical areas in the healthy contralateral side, and SUVmean and SUVmax were divided by the mean background activity to obtain mean and maximal tumor-to-background ratios (TBRmean and TBRmax). The biological tumor volume was semiautomatically delineated using the standard 1.6 × background activity as threshold.
Neuropathological analysis
Five-micrometer-thick sections of FFPE tumor tissue were routinely stained with hematoxylin and eosin. In addition, immunohistochemical stainings using antibodies against GFAP (polyclonal; Agilent Technologies, Santa Clara, CA, USA), MAP2 (HM-2; Merck, Darmstadt, Germany), isocitrate dehydrogenase mutation 1 (IDH1) (R132H) (QM002; Quartett GmbH, Potsdam, Germany), ATRX (BSB-108; Bio SB, Goleta, CA, USA), and Ki67 (MIB1; Agilent Technologies, Santa Clara, CA, USA) were routinely performed according to standard protocols.
Transcardial perfusion, immunohistochemistry, and tissue clearing
Mice that were intended for 3D histology were intraperitoneally injected with 10% ketamine (100 mg/kg) and 2% xylazine (10 mg/kg) in 0.9% NaCl. After expiration of the pedal reflex, intracardial perfusion was performed with 0.1 M PBS (10 U/ml; Ratiopharm) for 6 min, followed by the administration of 4% PFA in 0.1 M PBS (pH 7.4) (Morphisto, 11762.01000) for another 6 min. Afterward, the brain was removed and postfixed by 4% PFA for 6 to 12 hours at 4°C and washed with 0.1 M PBS, and stored in 0.1 M PBS.
Samples were subjected to a modified version of vDISCO (65) and SHANEL (66) protocols. Samples were decolorized with 25% CUBIC solution for 1 day. An extra permeabilization step was performed using SHANEL reagents: 10% CHAPS with 25% N-methyldiethanolamine in distilled H2O at 37°C overnight to access the dense glioblastoma. Then, samples were further permeabilized with vDISCO permeabilization solution with additional 10% 2-hydroxypropyl-β-cyclodextrin overnight at 37°C. The next day, antibody labeling and boosting step was performed with the addition of CD11b (1:50; Miltenyi Biotec, 130-110-611) and GFP-nanobooster LOT 102 (1:500; Chromotek, gba647n). The samples were incubated in 37°C for 9 days. After labeling, samples were washed with vDISCO washing solution for 3 hours, and 3DISCO clearing (67) with 50-70-90-100-100% tetrahydrofuran (THF) (Sigma-Aldrich, 186562), 1 hour each, and dichloromethane (Sigma-Aldrich, 270997), 30 min, was performed. Last, samples were placed in BABB (2:1; benzyl benzoate and benzyl alcohol) (Sigma-Aldrich, 24122 and W213802) solution indefinitely for refractive index matching and light sheet fluorescent imaging.
Light sheet fluorescent microscopy
Ultramicroscope Blaze (Miltenyi Biotec) was used to acquire single plane illumination (light sheet) image stacks. Filters used were excitation of 470/40 nm, emission of 535/50 nm; excitation of 545/25 nm, emission of 605/70 nm; and excitation of 560/30 nm, emission of 609/54 nm for autofluorescence, CD11b signal, and cancer cells, respectively. For all scans, ×1.66 magnification, 1 × 2 tiling, 30% overlap, 120-ms exposure time, 100% light sheet width, ~7-μm laser sheet thickness, ~0.31 numerical aperture (NA), and 5-μm z-step were used. To prevent signal saturation, laser power was adjusted per each sample.
Cryosectioning and immunofluorescence staining
To determine the absolute cell numbers within tumors, after acquiring the light sheet images of brains, tissue was rehydrated for sectioning. After sequential incubation in THF, samples were washed in PBS and placed in 30% sucrose solution for dehydration overnight. The next morning, samples were embedded in optimal cutting temperature and frozen. Fifteen-micrometer sagittal sections were acquired using a cryostat (CM3050S, 665 Leica, Wetzlar, Germany). Tissue sections blocked 0.2% Triton X-100, 10% dimethyl sulfoxide, and 10% goat serum in 0.1 M PBS for 1 hour, stained CD11b (1:50) and GFP-nanobooster LOT 102 (1:500) in 0.2% Tween 20, 5% dimethyl sulfoxide, 5% goat serum, and 0.001% heparin in 0.1 M PBS, and washed with 0.2% Tween 20 and 0.001% heparin in 0.1 M PBS for 3 × 2 min.
Laser scanning confocal microscopy
Leica SP8 was used for laser scanning confocal microscopy. Images were acquired with ×40 magnification acquired as 8-bits with HCPL APO CS2 40×/1.30 NA HCPL APO CS2 40×/1.30 NA or a HCPL APO CORR CS2 63×/1.30 NA objective, 1024 × 1024 resolution, 200 Hz, at 20° to 25°C. Tile scans were obtained with z-step size of 1 μm to allow 3D reconstruction of tissue for a depth of 15 μm.
Image processing
Tiles from each brain were stitched as described previously (65). Stitched images were saved in TIFF format for further processing. Volumes of segmented CD11b positivity per TAM and segmented GFP positivity per tumor cell were obtained via confocal images to allow calculation of absolute cell numbers in 3D histology (see Eq. 1). Regions of interest with predominant CD11b (n = 5) and cancer cell (n = 5) signal were acquired, and DAPI nuclear stains were used to count cells per region of interest (ROI). To segment single nuclei from cancer cells and CD11b+ cells for estimation of absolute cell numbers, cellpose (68) was used. The percentage of fluorescence+ nuclei/cells was determined via colocalization of nuclei with adjacent CD11b and GFP positivity. Volumes of CD11b and GFP positivity were obtained via assessment of segmented areas per channel (50% maximum threshold) for each single layer after applying a Gaussian filter of 6 mm. To account for positive areas not belonging to captured nuclei (i.e., at the edge of cells), we determined the area of CD11b and GFP positivity remote from nuclei (manual threshold visually matching the cell borders). To account for caption of variable profiles in single cells, we assumed a Gaussian distribution of modeled percentage areas in a fluorescence positive sphere (i.e., cell) with an internal fluorescence negative sphere (i.e., nucleus), calculating a correction factor of 1.044. Areas per cell were then extrapolated to volumes per cell using spherical assumption.
| (1) |
To calculate the volume of tumor cells and TAMs within the tumor in 3D histology, two channels were separately segmented using syGlass virtual reality tool (69). The TIFF stack was imported into the syGlass software where the stack is rendered as a 3D image in a virtual space. The ROI tool was used to annotate the signal of interest with adjustable thresholding in the 3D space to get the best signal-to-noise ratio. The annotation mask was exported as TIFFs for further downstream processing. On the basis of the virtual reality–aided manual annotations of tumor and TAM regions, areas were calculated as the sum of the marked mask voxels. Light sheet fluorescence microscopy’s resolution of 19.58 mm3 per voxel was used to compute the total volume in mm3. The percentage of TAMs within the tumor was obtained by first filtering the annotated TAM mask with the tumor area by dot multiplication between the two masks. Then, the number of obtained voxels was divided by the total amount of tumor voxels. Last, the PET signal of the tumor was compared with the aggregated recovered signals of tumor cells and TAMs (see Eq. 2).
| (2) |
Proteomics
Isolated TAMs (n = 5) and tumor cells (n = 3), as well as control microglia isolated from untreated control mice (n = 6), were prepared for mass spectrometry–based label-free protein quantification. The cell pellets were lysed in 50 μl of radioimmunoprecipitation assay lysis buffer [50 mM tris, 150 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, and 0.1% (w/v) SDS (pH 8.0)] at 4°C with intermediate vortexing. DNA was disrupted by ultrasonication (M220 Focused-ultrasonicator, Covaris, Woburn, MA, USA). The samples were centrifuged for 5 min at 16,000g and 4°C to remove cell debris and undissolved material. The supernatants were transferred to protein LoBind tubes (Eppendorf, Hamburg, Germany). Proteins were reduced at 37°C for 30 min with 15 mM dithiothreitol (DTT), followed by cysteine alkylation with 60 mM iodoacetamide (IAA) for 30 min at 20°C. Excess of IAA was removed by adding DTT. Detergent removal and subsequent digestion with 0.2 μg of LysC and 0.2 μg of trypsin (Promega, Walldorf, Germany) was performed using the single-pot, solid phase–enhanced sample preparation as previously described (70). After vacuum centrifugation, peptides were dissolved in 20 μl of 0.1% formic acid. The peptide amount of tumor cell samples was estimated using a fluorescence-based protein assay (Qubit Protein assay, Thermo Fisher Scientific, Waltham, MA, USA).
For mass spectrometry, the injected volume was adjusted on the basis of the counted cell numbers to analyze equal cell numbers with a maximum injection amount of 400 ng of peptides. The peptides were separated on a nanoElute nanoHPLC system (Bruker, Bremen, Germany) using a 5-mm trapping column (Thermo Fisher Scientific, Waltham, MA, USA) and an in-house packed C18 analytical column (15 cm × 75 μm ID, ReproSil-Pur 120 C18-AQ, 1.9 μm, Dr. Maisch GmbH). Peptides were separated with a binary gradient of water and acetonitrile (B) containing 0.1% formic acid at a flow rate of 300 nl/min (0 min, 2% B; 2 min, 5% B; 70 min, 24% B; 85 min, 35% B; 90 min, 60% B) and a column temperature of 50°C.
The nanoHPLC was online coupled to a TimsTOF pro mass spectrometer (Bruker, Bremen, Germany) with a CaptiveSpray ion source (Bruker, Bremen, Germany). A data-independent acquisition parallel accumulation–serial fragmentation (PASEF) method for spectrum acquisition was used. Briefly, ion accumulation and separation using trapped ion mobility spectrometry (TIMS) was set to a ramp time of 100 ms. One scan cycle included one TIMS full MS scan and with 26 windows with a width of 27 mass/charge ratio (m/z) covering a m/z range of 350 to 1002 m/z. Two windows were recorded per PASEF scan. This resulted in a cycle time of 1.4 s.
The data analysis and protein LFQ was performed with the software DIA-NN (71) version 1.8. The data were analyzed using a spectral library including 13,912 protein groups and 208,218 precursors in 174,909 elution groups generated with the same samples and additional murine microglia samples using DIA-NN searching against a one protein per gene database from Mus musculus (download date: 2022-01-25, 21,994 entries). Trypsin was defined as protease, and two missed cleavages were allowed. Oxidation of methionines and acetylation of protein N termini were defined as variable modifications, whereas carbamidomethylation of cysteines was defined as fixed modification. The precursor and fragment ion m/z ranges were limited from 350 to 1002 and 200 to 1700, respectively. Precursor charge states of 2 to 4 were considered. The mass accuracy for peptides and peptide fragments was set to 15 and 20 ppm, respectively. An FDR threshold of 1% was applied for peptide and protein identifications. Data normalization was disabled to obtain quantification values, which reflect the protein amounts per cell.
Ratios of TSPO protein expression between SB28 tumor cells, TAMs, and control microglia were compared against corresponding ratios obtained by scRadiotracing. Furthermore, all proteins were analyzed regarding their SB28 tumor cell and TAM expression levels relative to control microglia. Potential TAM-specific radiotracer targets were identified in a multistep process. First, all proteins with higher levels in TAMs (TAM–to–control microglia ratio) when compared to TSPO were selected. Second, among these, we identified proteins that were not present in SB28 tumor cells or showed high specificity in TAMs over SB28 tumor cells (TAM–to–tumor cell ratios of ≥10). Third, to further interrogate specificity, the Human Protein Atlas (17) was used to analyze single-cell RNA expression levels of brain and nonbrain cells for all identified target proteins that could serve for subsequent development of TAM-specific radioligands. Fourth, the predominant cell type and the functional immune cell cluster of the identified proteins for radiotracer development were determined by the Human Protein Atlas. To account for potential down-regulation in late stage SB28 tumors due to immunosuppression, TAM-selective proteins were also screened for important immune targets with lower expression compared to TSPO. Proteins were ranked based on the number of references linked at the UniProt library (72) as a surrogate of scientific relevance not only in glioblastoma but all areas of research and screened for targets of interest.
Statistics
Statistical analyses were performed with GraphPad Prism (V9, San Diego, CA, USA) and SPSS (V26, IBM, Armonk, NY, USA). A significance threshold of P < 0.05 was considered as significant for all experiments.
Validation of TSPO scRadiotracing
Radioactivity per cell was compared between microglia and astrocytes by a paired Student’s t test. In the depletion experiment, TSPO-PET measures of the whole brain were compared between treatment and vehicle by an unpaired Student’s t test. Iba1 immunoreactivity was compared between treatment and vehicle groups by an unpaired Student’s t test. The decrease in the TSPO-PET signal in the treatment group was considered as microglia bound radioactivity in the brain and compared to extrapolated radioactivity of the microglia cell population as assessed by scRadiotracing using a Student’s t test.
scRadiotracing in SB28 glioblastoma mice
Radioactivity per cell (%ID*BW normalized) was compared between different cell populations (tumor cells, TAMs, sham microglia, and control microglia) by a one-way ANOVA with Tukey post hoc test or by a paired Student’s t test for populations that were present in the same animals. Multiple regression was used to determine contributions of tumor cells (GFP+), microglia (CD11b+), and astrocytes (ACSA2+) to the radioactivity in the cell pellet. As a validation, measured radioactivity in the depleted fraction of the tumor was correlated with predicted radioactivity by cell count and single-cell tracer uptake (Pearson’s coefficient of correlation).
Association between scRadiotracing and PET imaging
Lesion site SUV was compared between SB28 and sham mice by an unpaired Student’s t test. Single-cell tracer uptake (tumor cells, TAMs, and a summed vector) was correlated with the tumor PET signal and between cell types using Pearson’s coefficient of correlation, and we applied a regression model with both cell types as predictors and the PET signal as dependent variable. PET cluster values of the tumor segmentation were correlated with single-cell tracer uptake and plotted as a function of cluster grade, after determining the best curve fit (linear, quadratic, and exponential) by Akaike information criteria. The difference between the cluster agreement of tumor cells and TAMs was calculated to determine dominant contribution of cell types and likewise plotted as a function of cluster grade.
Human in vitro scRadiotracing
A regression model with cell count of TAMs and non-TAM/nontumor cells as predictors and pellet radioactivity as dependent variable was applied to test for significant contribution of non-TAM/nontumor cells to the total radioactivity. Tumor grade (high versus low), age, and sex were tested as predictors of single-cell tracer uptake by a multivariate regression model. Within the groups of high-grade and low-grade glioma, single-cell tracer uptake of tumor cells and TAMs was compared by a paired Student’s t test. Single-cell tracer uptake (tumor cells and TAMs) was correlated with the tumor PET signal using Pearson’s coefficient of correlation.
Integrated model of PET, scRadiotracing, and 3D histology
Comparisons of GFP+ and CD11b+ volumes and cell counts in SB28 tumors were performed using a paired Student’s t test. The following analysis was performed using the 50 cluster regions of interest per individual mouse. TSPO-PET SUVs were multiplied with the regional correlation coefficient of either tumor or TAM single-cell tracer uptake correlations with PET. The individual single-cell tracer uptake was estimated according to TSPO-PET SUV of the tumor (Fig. 3B) and applied to predict light sheet signal intensity per cluster as a surrogate for cell density. This was performed separately for tumor cells and TAMs. A linear regression was performed to test for the agreement between predicted and standard of truth cell density per region. Last, a combined tumor cell and TAM model was used to predict absolute radioactivity in SB28 tumors as measured by means of TSPO-PET. To this end, the cell count of both tumor cells and TAMs in seven SB28 tumors was multiplied with the individual single-cell tracer uptake (as estimated according to TSPO-PET SUV of the tumor) and aggregated before correlation with equally normalized (%ID*BW) PET radioactivity.
Proteomics
The statistical data analysis of the DIA-NN output was performed with the software Perseus version 1.6.14.0 (73). Only LFQ intensities from protein groups with at least two peptides were considered. The protein LFQ intensities were log2-transformed. A one-way ANOVA test was applied to determine statistically significant differences between the means of the three groups. Afterward, individual Student’s t tests were applied to evaluate proteins with a significantly different abundance between the TAMs, tumor cells, and control microglia. In addition, a permutation-based FDR estimation was used with a FDR of 5% at s0 = 0.1 as threshold (74). All numeric values are reported as average group values ± SEM unless otherwise indicated.
Acknowledgments
We thank R. Oos and G. Palumbo for excellent technical support during PET imaging.
Funding: This project was partly funded by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) (FOR 2858 project numbers 421887978 and 422188432 and Research Training Group GRK 2274). N.L.A. is supported by a research grant of the Else Kröner-Fresenius-Stiftung. M.B. and S.F.L. were funded by the DFG under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy—ID 390857198). S.V.K. was supported by the Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed) and the Friedrich-Baur-Stiftung.
Author contributions: Conceptualization: L.M.B., S.V.K., J.B., L.v.B., N.L.A., and M.B. Methodology: L.M.B., S.V.K., Z.I.K., J.B., S.A.M., S.U., L.Hoeh., I.H., L.H.K., S.T.K., P.Be., A.Z., and M.B. Software: L.M.B., Z.I.K., S.Q., J.B., S.A.M., S.U., L.Hoeh., I.H., L.G., L.H.K., A.Z., N.L.A., and M.B. Validation: L.M.B., S.V.K., S.L., N.L.A., and M.B. Formal analysis: L.M.B., S.V.K., Z.I.K., S.A.M., L.H.K., S.T.K., L.Hoer., and M.B. Investigation: L.M.B., S.V.K., Z.I.K., A.B., J.L., S.A.M., S.U., L.Hoeh., I.H., K.W.-M., V.C.R., L.H.K., S.T.K., P.Be., H.E.P., M.A., N.B., L.Hoer., R.S., D.M., and M.B. Resources: L.M.B., Z.I.K., S.Q., A.B., J.L., J.B., S.A.M., S.U., L.Hoeh., I.H., K.W.-M., V.C.R., L.G., L.H.K., S.T.K., H.E.P., A.Z., N.B., L.Hoer., R.S., D.M., S.L., N.L.A., and M.B. Data curation: L.M.B., S.V.K., Z.I.K., S.Q., S.A.M., S.U., L.Hoeh., I.H., V.C.R., L.H.K., A.Z., N.L.A., and M.B. Writing—original draft: L.M.B. Writing—review and editing: A.H., P.Ba., M.J.R., S.L., S.Z., J.H., S.F.L., A.E., J.C.T., L.v.B., N.L.A., and M.B. Visualization: L.M.B., Z.I.K., A.B., J.L., S.A.M., N.L.A., and M.B. Supervision: A.H., K.W.-M., M.A., P.Ba., M.J.R., S.L., S.Z., J.H., S.F.L., A.E., J.C.T., L.v.B., N.L.A., and M.B. Project administration: L.M.B., N.L.A., and M.B. Funding acquisition: P.Ba., M.J.R., S.L., S.Z., J.H., S.F.L., A.E., J.C.T., L.v.B., N.L.A., and M.B.
Competing interests: N.L.A. and M.B. are members of the Neuroimaging Committee of the EANM. J.C.T. has a paid consultancy relationship with AAA-Novartis and received research grants from Novocure and Munich Surgical Imaging and a speaker honorarium from Seagen. N.L.A. received funding from Novocure. M.B. received speaker honoraria from Roche, GE Healthcare and Life Molecular Imaging and is an advisor of Life Molecular Imaging. V.C.R. received speaker honoraria from Novocure. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions of Figs. 1 to 5 are present in the paper and/or the Supplementary Materials. Proteomics source data of Fig. 6 are available as extended data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://www.proteomexchange.org/) via the PRIDE (75) partner repository with the dataset identifier PXD044033. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Jin M.-Z., Jin W.-L., The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 5, 166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitt J. M., Marabelle A., Eggermont A., Soria J. C., Kroemer G., Zitvogel L., Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 27, 1482–1492 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Fernandes B., Feltes P. K., Luft C., Nazario L. R., Jeckel C. M. M., Antunes I. F., Elsinga P. H., de Vries E. F. J., Potential PET tracers for imaging of tumor-associated macrophages. EJNMMI Radiopharm. Chem. 7, 11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kist de Ruijter L., van de Donk P. P., Hooiveld-Noeken J. S., Giesen D., Elias S. G., Lub-de Hooge M. N., Oosting S. F., Jalving M., Timens W., Brouwers A. H., Kwee T. C., Gietema J. A., Fehrmann R. S. N., Fine B. M., Sanabria Bohórquez S. M., Yadav M., Koeppen H., Jing J., Guelman S., Lin M. T., Mamounas M. J., Eastham J. R., Kimes P. K., Williams S. P., Ungewickell A., de Groot D. J. A., de Vries E. G. E., Whole-body CD8+ T cell visualization before and during cancer immunotherapy: A phase 1/2 trial. Nat. Med. 28, 2601–2610 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low J. T., Ostrom Q. T., Cioffi G., Neff C., Waite K. A., Kruchko C., Barnholtz-Sloan J. S., Primary brain and other central nervous system tumors in the United States (2014-2018): A summary of the CBTRUS statistical report for clinicians. Neurooncol. Pract. 9, 165–182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinnhardt B., Müther M., Roll W., Backhaus P., Jeibmann A., Foray C., Barca C., Döring C., Tavitian B., Dollé F., Weckesser M., Winkeler A., Hermann S., Wagner S., Wiendl H., Stummer W., Jacobs A. H., Schäfers M., Grauer O. M., TSPO imaging-guided characterization of the immunosuppressive myeloid tumor microenvironment in patients with malignant glioma. Neuro. Oncol. 22, 1030–1043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai L., Kirchleitner S. V., Zhao D., Li M., Tonn J. C., Glass R., Kalin R. E., Glioblastoma exhibits inter-individual heterogeneity of TSPO and LAT1 expression in neoplastic and parenchymal cells. Int. J. Mol. Sci. 21, 612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinnhardt B., Pigeon H., Theze B., Viel T., Wachsmuth L., Fricke I. B., Schelhaas S., Honold L., Schwegmann K., Wagner S., Faust A., Faber C., Kuhlmann M. T., Hermann S., Schafers M., Winkeler A., Jacobs A. H., Combined PET imaging of the inflammatory tumor microenvironment identifies margins of unique radiotracer uptake. Cancer Res. 77, 1831–1841 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Buck J. R., McKinley E. T., Fu A., Abel T. W., Thompson R. C., Chambless L., Watchmaker J. M., Harty J. P., Cooper M. K., Manning H. C., Preclinical TSPO ligand PET to visualize human glioma xenotransplants: A preliminary study. PLOS ONE 10, e0141659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pigeon H., Pérès E. A., Truillet C., Jego B., Boumezbeur F., Caillé F., Zinnhardt B., Jacobs A. H., Le Bihan D., Winkeler A., TSPO-PET and diffusion-weighted MRI for imaging a mouse model of infiltrative human glioma. Neuro Oncol. 21, 755–764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartos L. M., Kunte S. T., Beumers P., Xiang X., Wind K., Ziegler S., Bartenstein P., Choi H., Lee D. S., Haass C., von Baumgarten L., Tahirovic S., Albert N. L., Lindner S., Brendel M., Single-cell radiotracer allocation via immunomagnetic sorting to disentangle PET signals at cellular resolution. J. Nucl. Med. 63, 1459–1462 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos Santos S. E., Medeiros M., Porfirio J., Tavares W., Pessoa L., Grinberg L., Leite R. E. P., Ferretti-Rebustini R. E. L., Suemoto C. K., Filho W. J., Noctor S. C., Sherwood C. C., Kaas J. H., Manger P. R., Herculano-Houzel S., Similar microglial cell densities across brain structures and mammalian species: Implications for brain tissue function. J. Neurosci. 40, 4622–4643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang X., Wind K., Wiedemann T., Blume T., Shi Y., Briel N., Beyer L., Biechele G., Eckenweber F., Zatcepin A., Lammich S., Ribicic S., Tahirovic S., Willem M., Deussing M., Palleis C., Rauchmann B. S., Gildehaus F. J., Lindner S., Spitz C., Franzmeier N., Baumann K., Rominger A., Bartenstein P., Ziegler S., Drzezga A., Respondek G., Buerger K., Perneczky R., Levin J., Hoglinger G. U., Herms J., Haass C., Brendel M., Microglial activation states drive glucose uptake and FDG-PET alterations in neurodegenerative diseases. Sci. Transl. Med. 13, eabe5640 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Shi Y., Cui M., Ochs K., Brendel M., Strubing F. L., Briel N., Eckenweber F., Zou C., Banati R. B., Liu G. J., Middleton R. J., Rupprecht R., Rudolph U., Zeilhofer H. U., Rammes G., Herms J., Dorostkar M. M., Long-term diazepam treatment enhances microglial spine engulfment and impairs cognitive performance via the mitochondrial 18 kDa translocator protein (TSPO). Nat. Neurosci. 25, 317–329 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Genoud V., Marinari E., Nikolaev S. I., Castle J. C., Bukur V., Dietrich P.-Y., Okada H., Walker P. R., Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology 7, e1501137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartos L. M., Kirchleitner S. V., Blobner J., Wind K., Kunze L. H., Holzgreve A., Gold L., Zatcepin A., Kolabas Z. I., Ulukaya S., Weidner L., Quach S., Messerer D., Bartenstein P., Tonn J. C., Riemenschneider M. J., Ziegler S., von Baumgarten L., Albert N. L., Brendel M., 18 kDa translocator protein positron emission tomography facilitates early and robust tumor detection in the immunocompetent SB28 glioblastoma mouse model. Front. Med. 9, 992993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F., Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Dagher N. N., Najafi A. R., Kayala K. M. N., Elmore M. R., White T. E., Medeiros R., West B. L., Green K. N., Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflammation 12, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya M. M., Green K. N., Allen B. D., Najafi A. R., Syage A., Minasyan H., Le M. T., Kawashita T., Giedzinski E., Parihar V. K., West B. L., Baulch J. E., Limoli C. L., Elimination of microglia improves cognitive function following cranial irradiation. Sci. Rep. 6, 31545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmore M. R., Najafi A. R., Koike M. A., Dagher N. N., Spangenberg E. E., Rice R. A., Kitazawa M., Matusow B., Nguyen H., West B. L., Green K. N., Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicente-Rodríguez M., Mancuso R., Peris-Yague A., Simmons C.; NIMA Consortium, Gómez-Nicola D., Perry V. H., Turkheimer F., Lovestone S., Parker C. A., Cash D., Pharmacological modulation of TSPO in microglia/macrophages and neurons in a chronic neurodegenerative model of prion disease. J. Neuroinflammation 20, 92 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parhizkar S., Arzberger T., Brendel M., Kleinberger G., Deussing M., Focke C., Nuscher B., Xiong M., Ghasemigharagoz A., Katzmarski N., Krasemann S., Lichtenthaler S. F., Muller S. A., Colombo A., Monasor L. S., Tahirovic S., Herms J., Willem M., Pettkus N., Butovsky O., Bartenstein P., Edbauer D., Rominger A., Erturk A., Grathwohl S. A., Neher J. J., Holtzman D. M., Meyer-Luehmann M., Haass C., Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci. 22, 191–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cumming P., Burgher B., Patkar O., Breakspear M., Vasdev N., Thomas P., Liu G. J., Banati R., Sifting through the surfeit of neuroinflammation tracers. J. Cereb. Blood Flow Metab. 38, 204–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenbaum D., Colangelo C., Williams K., Gerstein M., Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanotti-Fregonara P., Pascual B., Rizzo G., Yu M., Pal N., Beers D., Carter R., Appel S. H., Atassi N., Masdeu J. C., Head-to-head comparison of 11C-PBR28 and 18F-GE180 for quantification of the translocator protein in the human brain. J. Nucl. Med. 59, 1260–1266 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Wimberley C., Lavisse S., Brulon V., Peyronneau M. A., Leroy C., Bodini B., Remy P., Stankoff B., Buvat I., Bottlaender M., Impact of endothelial 18-kDa translocator protein on the quantification of 18F-DPA-714. J. Nucl. Med. 59, 307–314 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Foray C., Barca C., Winkeler A., Wagner S., Hermann S., Schäfers M., Grauer O. M., Zinnhardt B., Jacobs A. H., Interrogating glioma-associated microglia and macrophage dynamics under CSF-1R therapy with multitracer in vivo PET/MRI. J. Nucl. Med. 63, 1386–1393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palleis C., Sauerbeck J., Beyer L., Harris S., Schmitt J., Morenas-Rodriguez E., Finze A., Nitschmann A., Ruch-Rubinstein F., Eckenweber F., Biechele G., Blume T., Shi Y., Weidinger E., Prix C., Botzel K., Danek A., Rauchmann B. S., Stocklein S., Lindner S., Unterrainer M., Albert N. L., Wetzel C., Rupprecht R., Rominger A., Bartenstein P., Herms J., Perneczky R., Haass C., Levin J., Hoglinger G. U., Brendel M., In vivo assessment of neuroinflammation in 4‐repeat tauopathies. Mov. Disord. 36, 883–894 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Pascoal T. A., Benedet A. L., Ashton N. J., Kang M. S., Therriault J., Chamoun M., Savard M., Lussier F. Z., Tissot C., Karikari T. K., Ottoy J., Mathotaarachchi S., Stevenson J., Massarweh G., Scholl M., de Leon M. J., Soucy J. P., Edison P., Blennow K., Zetterberg H., Gauthier S., Rosa-Neto P., Microglial activation and tau propagate jointly across Braak stages. Nat. Med. 27, 1592–1599 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Albert-Smet I., Marcos-Vidal A., Vaquero J. J., Desco M., Muñoz-Barrutia A., Ripoll J., Applications of light-sheet microscopy in microdevices. Front. Neuroanat. 13, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banati R. B., Wilcox P., Xu R., Yin G., Si E., Son E. T., Shimizu M., Holsinger R. M. D., Parmar A., Zahra D., Arthur A., Middleton R. J., Liu G. J., Charil A., Graeber M. B., Selective, high-contrast detection of syngeneic glioblastoma in vivo. Sci. Rep. 10, 9968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao R., Pan R., Shang C., Li X., Cheng J., Xu J., Li Y., Translocator protein 18 kDa (TSPO) deficiency inhibits microglial activation and impairs mitochondrial function. Front. Pharmacol. 11, 986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menevse A. N., Ammer L. M., Vollmann-Zwerenz A., Kupczyk M., Lorenz J., Weidner L., Hussein A., Sax J., Mühlbauer J., Heuschneider N., Rohrmus C., Mai L. S., Jachnik B., Stamova S., Volpin V., Durst F. C., Sorrentino A., Xydia M., Milenkovic V. M., Bader S., Braun F. K., Wetzel C., Albert N. L., Tonn J. C., Bartenstein P., Proescholdt M., Schmidt N. O., Linker R. A., Riemenschneider M. J., Beckhove P., Hau P., TSPO acts as an immune resistance gene involved in the T cell mediated immune control of glioblastoma. Acta Neuropathol. Commun. 11, 75 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathewson N. D., Ashenberg O., Tirosh I., Gritsch S., Perez E. M., Marx S., Jerby-Arnon L., Chanoch-Myers R., Hara T., Richman A. R., Ito Y., Pyrdol J., Friedrich M., Schumann K., Poitras M. J., Gokhale P. C., Gonzalez Castro L. N., Shore M. E., Hebert C. M., Shaw B., Cahill H. L., Drummond M., Zhang W., Olawoyin O., Wakimoto H., Rozenblatt-Rosen O., Brastianos P. K., Liu X. S., Jones P. S., Cahill D. P., Frosch M. P., Louis D. N., Freeman G. J., Ligon K. L., Marson A., Chiocca E. A., Reardon D. A., Regev A., Suvà M. L., Wucherpfennig K. W., Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184, 1281–1298.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betlazar C., Middleton R. J., Howell N., Storer B., Davis E., Davies J., Banati R., Liu G. J., Mitochondrial translocator protein (TSPO) expression in the brain after whole body gamma irradiation. Front. Cell Dev. Biol. 9, 715444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalsa J. K., Cheng N., Keegan J., Chaudry A., Driver J., Bi W. L., Lederer J., Shah K., Immune phenotyping of diverse syngeneic murine brain tumors identifies immunologically distinct types. Nat. Commun. 11, 3912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banati R. B., Middleton R. J., Chan R., Hatty C. R., Kam W. W., Quin C., Graeber M. B., Parmar A., Zahra D., Callaghan P., Fok S., Howell N. R., Gregoire M., Szabo A., Pham T., Davis E., Liu G.-J., Positron emission tomography and functional characterization of a complete PBR/TSPO knockout. Nat. Commun. 5, 5452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quach S., Holzgreve A., Kaiser L., Unterrainer M., Dekorsy F. J., Nelwan D. V., Bartos L. M., Kirchleitner S. V., Weller J., Weidner L., Niyazi M., Ruf V. C., Herms J., Stöcklein S., Wetzel C., Riemenschneider M. J., Baumgarten L. V., Thon N., Brendel M., Rupprecht R., Bartenstein P., Tonn J.-C., Albert N. L., TSPO PET signal using [18F]GE180 is associated with survival in recurrent gliomas. Eur. J. Nucl. Med. Mol. Imaging 50, 859–869 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albert N. L., Nelwan D. V., Fleischmann D. F., Quach S., von Rohr K., Kaiser L., Teske N., Unterrainer L. M., Bartos L. M., Ruf V. C., Brendel M., Riemenschneider M. J., Wetzel C., Herms J., Rupprecht R., Thon N., Tonn J. C., Belka C., Bartenstein P., von Baumgarten L., Niyazi M., Unterrainer M., Holzgreve A., Prognostic value of TSPO PET before radiotherapy in newly diagnosed IDH-wild-type glioblastoma.J. Nucl. Med., jnumed.122.265247, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law I., Albert N. L., Arbizu J., Boellaard R., Drzezga A., Galldiks N., la Fougere C., Langen K. J., Lopci E., Lowe V., McConathy J., Quick H. H., Sattler B., Schuster D. M., Tonn J. C., Weller M., Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F]FDG: Version 1.0. Eur. J. Nucl. Med. Mol. Imaging 46, 540–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Ren P., Zeng Z., Ma L., Li Y., Zhang H., Guo W., Inhibition of translocator protein 18 kDa suppressed the progression of glioma via the ELAV-like RNA-binding protein 1/MAPK-activated protein kinase 3 axis. Bioengineered 13, 7457–7470 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galldiks N., Langen K. J., Albert N. L., Law I., Kim M. M., Villanueva-Meyer J. E., Soffietti R., Wen P. Y., Weller M., Tonn J. C., Investigational PET tracers in neuro-oncology-What’s on the horizon? A report of the PET/RANO group. Neuro Oncol. 24, 1815–1826 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon I., Amann M., Goubier A., Arce Vargas F., Zervas D., Qing C., Henry J. Y., Ghorani E., Akarca A. U., Marafioti T., Śledzińska A., Werner Sunderland M., Franz Demane D., Clancy J. R., Georgiou A., Salimu J., Merchiers P., Brown M. A., Flury R., Eckmann J., Murgia C., Sam J., Jacobsen B., Marrer-Berger E., Boetsch C., Belli S., Leibrock L., Benz J., Koll H., Sutmuller R., Peggs K. S., Quezada S. A., CD25-Treg-depleting antibodies preserving IL-2 signaling on effector T cells enhance effector activation and antitumor immunity. Cancer 1, 1153–1166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landry A. P., Balas M., Alli S., Spears J., Zador Z., Distinct regional ontogeny and activation of tumor associated macrophages in human glioblastoma. Sci. Rep. 10, 19542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pombo Antunes A. R., Scheyltjens I., Lodi F., Messiaen J., Antoranz A., Duerinck J., Kancheva D., Martens L., De Vlaminck K., Van Hove H., Kjølner Hansen S. S., Bosisio F. M., Van der Borght K., De Vleeschouwer S., Sciot R., Bouwens L., Verfaillie M., Vandamme N., Vandenbroucke R. E., De Wever O., Saeys Y., Guilliams M., Gysemans C., Neyns B., De Smet F., Lambrechts D., Van Ginderachter J. A., Movahedi K., Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 24, 595–610 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Wang Y.-C., Wang X., Yu J., Ma F., Li Z., Zhou Y., Zeng S., Ma X., Li Y.-R., Neal A., Huang J., To A., Clarke N., Memarzadeh S., Pellegrini M., Yang L., Targeting monoamine oxidase A-regulated tumor-associated macrophage polarization for cancer immunotherapy. Nat. Commun. 12, 3530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sade-Feldman M., Yizhak K., Bjorgaard S. L., Ray J. P., de Boer C. G., Jenkins R. W., Lieb D. J., Chen J. H., Frederick D. T., Barzily-Rokni M., Freeman S. S., Reuben A., Hoover P. J., Villani A.-C., Ivanova E., Portell A., Lizotte P. H., Aref A. R., Eliane J.-P., Hammond M. R., Vitzthum H., Blackmon S. M., Li B., Gopalakrishnan V., Reddy S. M., Cooper Z. A., Paweletz C. P., Barbie D. A., Stemmer-Rachamimov A., Flaherty K. T., Wargo J. A., Boland G. M., Sullivan R. J., Getz G., Hacohen N., Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martín-Ruiz A., Fiuza-Luces C., Martínez-Martínez E., Arias C. F., Gutiérrez L., Ramírez M., Martín-Acosta P., Coronado M. J., Lucia A., Provencio M., Effects of anti-PD-1 immunotherapy on tumor regression: Insights from a patient-derived xenograft model. Sci. Rep. 10, 7078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue L., Lu B., Gao B., Shi Y., Xu J., Yang R., Xu B., Ding P., NLRP3 promotes glioma cell proliferation and invasion via the interleukin-1β/NF-κB p65 signals. Oncol. Res. 27, 557–564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akkari L., Bowman R. L., Tessier J., Klemm F., Handgraaf S. M., de Groot M., Quail D. F., Tillard L., Gadiot J., Huse J. T., Brandsma D., Westerga J., Watts C., Joyce J. A., Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med. 12, eaaw7843 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Sun R., Han R., McCornack C., Khan S., Tabor G. T., Chen Y., Hou J., Jiang H., Schoch K. M., Mao D. D., Cleary R., Yang A., Liu Q., Luo J., Petti A., Miller T. M., Ulrich J. D., Holtzman D. M., Kim A. H., TREM2 inhibition triggers antitumor cell activity of myeloid cells in glioblastoma. Sci. Adv. 9, eade3559 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couturier C. P., Ayyadhury S., Le P. U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., Lee C., Wang Y. C. D., Wee Yong V., Guiot M.-C., Najafabadi H., Misic B., Antel J., Bourque G., Ragoussis J., Petrecca K., Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 11, 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spangenberg E., Severson P. L., Hohsfield L. A., Crapser J., Zhang J., Burton E. A., Zhang Y., Spevak W., Lin J., Phan N. Y., Habets G., Rymar A., Tsang G., Walters J., Nespi M., Singh P., Broome S., Ibrahim P., Zhang C., Bollag G., West B. L., Green K. N., Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10, 3758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haddad A. F., Young J. S., Amara D., Berger M. S., Raleigh D. R., Aghi M. K., Butowski N. A., Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neurooncol. Adv. 3, vdab100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colombo A. V., Sadler R. K., Llovera G., Singh V., Roth S., Heindl S., Sebastian Monasor L., Verhoeven A., Peters F., Parhizkar S., Kamp F., Gomez de Aguero M., MacPherson A. J., Winkler E., Herms J., Benakis C., Dichgans M., Steiner H., Giera M., Haass C., Tahirovic S., Liesz A., Microbiota-derived short chain fatty acids modulate microglia and promote Aβ plaque deposition. eLife 10, e59826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang X., Piers T. M., Wefers B., Zhu K., Mallach A., Brunner B., Kleinberger G., Song W., Colonna M., Herms J., Wurst W., Pocock J. M., Haass C., The Trem2 R47H Alzheimer’s risk variant impairs splicing and reduces Trem2 mRNA and protein in mice but not in humans. Mol. Neurodegener. 13, 49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith S. J., Rowlinson J., Estevez-Cebrero M., Onion D., Ritchie A., Clarke P., Wood K., Diksin M., Lourdusamy A., Grundy R. G., Rahman R., Metabolism-based isolation of invasive glioblastoma cells with specific gene signatures and tumorigenic potential. Neurooncol. Adv. 2, vdaa087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black D., Kaneko S., Walke A., König S., Stummer W., Suero Molina E., Characterization of autofluorescence and quantitative protoporphyrin IX biomarkers for optical spectroscopy-guided glioma surgery. Sci. Rep. 11, 20009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brendel M., Probst F., Jaworska A., Overhoff F., Korzhova V., Albert N. L., Beck R., Lindner S., Gildehaus F. J., Baumann K., Bartenstein P., Kleinberger G., Haass C., Herms J., Rominger A., Glial activation and glucose metabolism in a transgenic amyloid mouse model: A triple-tracer PET study. J. Nucl. Med. 57, 954–960 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Overhoff F., Brendel M., Jaworska A., Korzhova V., Delker A., Probst F., Focke C., Gildehaus F.-J., Carlsen J., Baumann K., Haass C., Bartenstein P., Herms J., Rominger A., Automated spatial brain normalization and hindbrain white matter reference tissue give improved [18F]-florbetaben PET quantitation in Alzheimer’s model mice. Front. Neurosci. 10, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deussing M., Blume T., Vomacka L., Mahler C., Focke C., Todica A., Unterrainer M., Albert N. L., Lindner S., von Ungern-Sternberg B., Baumann K., Zwergal A., Bartenstein P., Herms J., Rominger A., Brendel M., Coupling between physiological TSPO expression in brain and myocardium allows stabilization of late-phase cerebral [18F]GE180 PET quantification. Neuroimage 165, 83–91 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Unterrainer M., Fleischmann D. F., Vettermann F., Ruf V., Kaiser L., Nelwan D., Lindner S., Brendel M., Wenter V., Stocklein S., Herms J., Milenkovic V. M., Rupprecht R., Tonn J. C., Belka C., Bartenstein P., Niyazi M., Albert N. L., TSPO PET, tumour grading and molecular genetics in histologically verified glioma: A correlative 18F-GE-180 PET study. Eur. J. Nucl. Med. Mol. Imaging 47, 1368–1380 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Unterrainer M., Vettermann F., Brendel M., Holzgreve A., Lifschitz M., Zahringer M., Suchorska B., Wenter V., Illigens B. M., Bartenstein P., Albert N. L., Towards standardization of 18F-FET PET imaging: Do we need a consistent method of background activity assessment? EJNMMI Res. 7, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai R., Pan C., Ghasemigharagoz A., Todorov M. I., Forstera B., Zhao S., Bhatia H. S., Parra-Damas A., Mrowka L., Theodorou D., Rempfler M., Xavier A. L. R., Kress B. T., Benakis C., Steinke H., Liebscher S., Bechmann I., Liesz A., Menze B., Kerschensteiner M., Nedergaard M., Erturk A., Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao S., Todorov M. I., Cai R., Maskari R. A., Steinke H., Kemter E., Mai H., Rong Z., Warmer M., Stanic K., Schoppe O., Paetzold J. C., Gesierich B., Wong M. N., Huber T. B., Duering M., Bruns O. T., Menze B., Lipfert J., Puelles V. G., Wolf E., Bechmann I., Erturk A., Cellular and molecular probing of intact human organs. Cell 180, 796–812.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erturk A., Becker K., Jahrling N., Mauch C. P., Hojer C. D., Egen J. G., Hellal F., Bradke F., Sheng M., Dodt H. U., Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 69.S. Pidhorskyi, M. Morehead, Q. Jones, G. Spirou, G. Doretto, syGlass: Interactive exploration of multidimensional images using virtual reality Head-mounted displays. arXiv:1804.08197 [cs.GR] (2018).
- 70.Hughes C. S., Moggridge S., Muller T., Sorensen P. H., Morin G. B., Krijgsveld J., Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Demichev V., Messner C. B., Vernardis S. I., Lilley K. S., Ralser M., DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.The UniProt Consortium , UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., Cox J., The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Tusher V. G., Tibshirani R., Chu G., Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D. J., Prakash A., Frericks-Zipper A., Eisenacher M., Walzer M., Wang S., Brazma A., Vizcaíno J. A., The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herculano-Houzel S., The human brain in numbers: A linearly scaled-up primate brain. Front. Hum. Neurosci. 3, 31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berroterán-Infante N., Tadić M., Hacker M., Wadsak W., Mitterhauser M., Binding affinity of some endogenous and synthetic TSPO ligands regarding the rs6971 polymorphism. Int. J. Mol. Sci. 20, 563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1
Legend for data S1
Data S1