Abstract
Normal human cells in culture enter replicative senescence after a finite number of population doublings. The exact molecular mechanisms triggering the growth arrest are poorly understood. A recent report on the disappearance of the G-rich 3′ telomeric overhang in senescent cells led to the hypothesis that loss of the 3′ G-rich overhang is the molecular signal that triggers senescence. Here, we describe a quantitative assay to measure the length of the G-rich 3′ telomeric overhangs from cultured cells. Using both this assay and the conventional nondenaturing hybridization assay for measuring G-rich overhangs, we show that normal human fibroblasts can maintain their overhangs at senescence. Furthermore, cells do not lose their overhangs when they bypass senescence after the inactivation of p53 and Rb. We thus conclude that a global reduction in overhang length is not the molecular signal that triggers replicative senescence.
Telomeres are distinct DNA-protein structures that protect eukaryotic chromosome ends from degradation and inappropriate recombination or fusions. Human telomeric DNA is composed of many kilobases of (TTAGGG)n repetitive hexamers followed by a single-stranded overhang at the 3′ end of the G-rich strand. Telomere ends are prevented from being recognized as damaged DNA by a variety of protection mechanisms (3, 6, 12, 36). The 3′ overhangs may be protected by single-stranded DNA binding proteins, such as Cdc13 in Saccharomyces cerevisiae (26, 34), TEBP (telomere end binding protein) in the ciliate Oxytricha nova (16, 38, 39), or Pot1 (2, 10, 25, 28). The 3′ overhang can also be tucked into the duplex telomeric DNA region by telomere binding proteins and form a special chromatin structure, the t-loop, that presumably helps shield telomere ends from recognition as double-stranded breaks (13, 17, 33).
In cultured normal human cells, telomeres progressively shorten with each cell division. As a consequence, cells enter replicative senescence (mortality stage 1 [M1]) when at least some telomeres become critically short (5, 18, 41, 48, 52). Senescence can be overcome by inactivation of the tumor suppressor p53 and retinoblastoma (Rb) proteins, resulting in a limited extension of life span before reaching crisis (M2), where most cells eventually undergo apoptosis (41).
The molecular mechanisms underlying replicative senescence are not fully understood. It has long been thought that replicative senescence in human cells is telomere length regulated. It has been proposed that disruption of telomere end capping (initiated by “too-short” telomeres or other mechanisms) can cause senescence. This is based on the observation that ectopic expression of a dominant-negative version of TRF2, a telomeric DNA binding protein, induces rapid loss of 3′ overhangs, end-to-end chromosome fusions, and senescence-like growth arrest without detectable loss of double-stranded telomere DNA (43). It was later discovered that the loss of 3′ overhangs induced by a dominant-negative TRF2 was the consequence of cleavage of the overhang at the D-loop by ERCC1/XPF endonuclease rather than erosion of overhangs per se (51).
A second hypothesis arose from the observation that the 3′ G-rich overhang is eroded at senescence (42). Those authors proposed that loss of the 3′ G-rich overhang is the molecular signal that triggers senescence (42). However, it is unclear whether the observed loss of overhangs is a primary event or a secondary consequence of short telomeres producing DNA damage signals when approaching senescence. These signals might then induce the end-processing events that cause overhang shortening.
We wished to understand the role of telomeric overhang dynamics in replicative senescence. To date, there is no reliable quantitative method to measure the length of telomeric overhangs in human cells. Both the conventional nondenaturing hybridization assay (30) and the newly developed telomere oligonucleotide ligation assay (T-OLA) (9) only determine the relative but not absolute length of telomeric overhangs. The electron microscopy method for measuring overhang length (21) requires large amounts of DNA (hundreds of micrograms of DNA). We thus developed a novel biochemical technique to determine telomeric overhang length. We observed that normal human fibroblasts could maintain their telomeric overhangs at senescence. We also found that cells that have escaped senescence through inactivation of p53 and Rb by overexpressing human papillomavirus type 16 E6 and E7 gene products (HPV E6/E7) maintained their overhangs during their extended life span, whereas cells overexpressing simian virus 40 large T-antigen (SV40 L-Tg) partially shortened their overhangs in culture. We also confirmed our results by using a nondenaturing hybridization assay. Our results strongly suggest that overhang loss is not the trigger of replicative senescence.
MATERIALS AND METHODS
Assay for measuring the size of telomeric overhangs.
Five micrograms of total DNA was resuspended in 15 μl of gp32 protection buffer (10 mM HEPES [pH 7.5], 100 mM LiCl, 2.5 mM MgCl2, 5 mM CaCl2; Li+ rather than Na+ or K+ salts was used to minimize potential G-quartet formation [47], while HEPES was used rather than Tris to avoid amino groups that would interfere with subsequent cross-linking steps) and was treated with 1 μg of RNase A with or without Exo I for 30 min at 37°C. The DNA was then treated with T4 gene protein 32 (10 pmol/μl; Roche) and glutathione S-transferase (GST)-UP1 (0.6 μg/μl; see below) for 1 h at room temperature (RT), followed by cross-linking with 0.025% glutaraldehyde for 15 min at RT. Cross-linking was stopped by adding Tris (pH 7.5) to 30 mM for 15 min at RT. The unprotected DNA was digested by DNase I (0.0025 U/μl; Roche) for 30 min at 37°C, which was then inactivated at 80°C for 30 min. Protease K (1 μg/μl; Roche) and sodium dodecyl sulfate (SDS; 0.5%) were added, and samples were incubated at 55°C for 16 h to digest proteins and reverse cross-linking. The high-specific-activity C-rich probe (5 fmol; see below) was added for hybridization to the G-rich overhang overnight at RT. Samples were analyzed the next day on 0.5× Tris-borate-EDTA (TBE)-6% polyacrylamide gels at 24 V/cm at 4°C to prevent melting of the annealed overhangs during electrophoresis. Gels were dried on Hybond N+ membrane (Amersham), exposed to a phosphorimager screen, and quantified by ImageQuant software (Amersham). To measure the size of the 3′ G-rich overhang, we used Exo I-treated sample as background and subtracted its signal from that of the untreated sample at each measured size interval. The weighted mean size was calculated using the following formula: Σ(ODi)/Σ(ODi/Li), where ODi is the phosphorimager output (signal intensity) and Li is the length in nucleotides of the DNA at position i, using the range of the molecular weight marker standards (see below). Results shown are representative of at least three independent experiments. A mixture of all size standards, both untreated and those subjected to the protection assay (coated, cross-linked, and digested with DNase), was included in every experiment to verify consistent protection. Although experiments with individual size standards suggested that the measured sizes using the overhang protection assay were approximately 65% of actual sizes (see Fig. 2B, below), the weighted mean size of the mixture of untreated size standards did not decrease following coating, cross-linking, and digestion with DNase. We thus did not adjust sizes by a correction factor but based sizes on the weighted means by using the bands from the (TTAGGG)n markers as size standards.
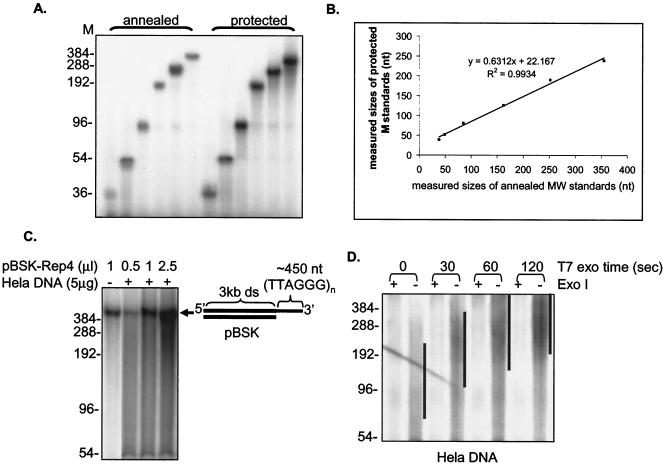
FIG. 2.
Validation of the overhang protection assay. (A) The overhang protection assay was performed on single-stranded oligonucleotides containing (TTAGGG)n repeats (n = 6, 9, 16, 32, 48, and 64). Oligonucleotides were either annealed directly to the C-rich probe (annealed) or underwent the overhang protection assay (protected). Weighted mean sizes were calculated after quantitating the signals in each lane with ImageQuant software between molecular size (M) marker positions of 36 to 384 bp. (B) Results of the assay in panel A are plotted. (C) The overhang protection assay was performed on the model template pBSK-Rep4, which contained an ∼450-nt 3′ (TTAGGG)n overhang after 3 kb of double-stranded pBSK vector sequences. Prior to the assay, a different amount of pBSK-Rep4 was mixed with or without HeLa genomic DNA. (D) Whole genomic DNA from HeLa cells was digested with T7 gene 6 exonuclease (a 5′-to-3′ exonuclease) for the indicated times and then treated with or without Exo I before being analyzed in the overhang protection assay. Black lines indicate positions of the bulk of the overhang DNA.
Expression and purification of GST-UP1.
The GST-UP1 protein (24) was expressed in Escherichia coli and affinity purified on glutathione-Sepharose (Amersham) following the manufacturer's instructions. The purified protein was >95% homogeneous as tested by SDS-polyacrylamide gel electrophoresis.
Synthesis of high-specific-activity C-rich probe.
Synthesis of a high-specific-activity (CCCTAA)3 probe has been described elsewhere (20). Briefly, a uracil-containing G-rich template was annealed to a short oligonucleotide which was extended by the Klenow fragment of DNA polymerase (Invitrogen) in the presence of [α-32P]dCTP (29). Six [32P]dCTP were incorporated per molecule, which gave a much stronger signal than end-labeled probe. Single-stranded probe was obtained by treating the extended probe with uracil deglycosylase to remove the template oligonucleotide.
Construction of model template.
pBSK-Rep4 is a plasmid containing an ∼450-nucleotide (nt) (TTAGGG)n sequence in a pBlueScript vector (49). Single-stranded circular DNA from both the pBSK-Rep4-containing plus strand and a pBSK vector-only minus strand was made through phage packaging. Equal amounts were annealed in 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, and 50 mM NaCl at 75°C for 5 min and slowly cooled to RT to create circular DNA containing double-stranded pBSK sequences with an ∼450-nt single-stranded TTAGGG loop. This product was then digested at the 3′ end of the telomeric repeats with SmaI and gel purified to create an artificial 3′ G-rich overhang.
Creation of G-rich telomeric repeat molecular marker size standards.
(TTAGGG)n repeat sequences annealed to the high-specific-activity C-rich probe migrated differently from the commonly used DNA molecular weight standards on nondenaturing polyacrylamide gel. We thus created specific TTAGGG size standards for this study. Briefly, 20 pmol of 32P-end-labeled (TTAGGG)16 oligo was annealed to 6 pmol of single-stranded plasmid containing 300 nt of (CCCTAA)n telomeric repeats in 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 50 mM KCl, and 1 mM β-mercaptoethanol. The annealed product was then diluted fourfold in T4 ligase buffer and ligated overnight at RT by using T4 ligase (0.3 U/μl; Invitrogen). After ethanol precipitation, the pellet was dissolved in H2O, melted for 3 min at 97°C, quick-chilled, and run on a 6% native polyacrylamide gel. The large 3-kb single-stranded plasmid stayed near the origin, allowing easy separation of the single-stranded ligation products. The gel-purified individual ligation products were macerated by spinning through a 22-gauge needle hole in a 0.5-ml tube and eluted overnight in 10 mM HEPES (pH 7.5) and 1 mM EDTA at RT.
Nondenaturing hybridization assay.
Five micrograms of undigested genomic DNA was diluted in 50 μl of gp32 protection buffer and hybridized with 5 fmol of high-specific-activity C-rich probe at RT overnight. Samples were size fractionated on a 0.7% agarose gel in 0.5× TBE at 8 V/cm and 4°C for 4 h. The gel was then denatured in 0.5 M NaOH-1.5 M NaCl at 4°C for 40 min to denature the annealed probe and the DNA (the probe did not diffuse significantly during this denaturation step). After washing in water for 15 min at 4°C, the denatured gel was dried on Hybond N+ membrane (Amersham) at 55°C. All the C-rich probe that had hybridized to the single-stranded overhangs was transferred onto the membrane after drying. The dried gel was peeled off the membrane and neutralized in 1.5 M NaCl-0.5 M Tris-HCl (pH 8.0) for 30 min and then hybridized to an Alu repeat probe (5′-GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCA-3′) at 42°C overnight. The gel was then washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min followed by two washes with 0.1× SSC-0.1% SDS for 10 min each. Both the membrane and gel were exposed to a PhosphorImager screen (Amersham). Relative amounts of overhangs were calculated by normalizing signals from the membrane (representing overhang signals) with the Alu repeat signal (representing total genomic DNA).
Enzymatic reactions.
To remove the 3′ G-rich overhang, total DNA was treated with the 3′-to-5′ exonuclease E. coli Exo I (0.3 U/μg of DNA; Amersham) in 15 μl of gp32 protection buffer supplemented with β-mercaptoethanol (20 mM) at 37°C for 1 h to overnight. To generate longer 3′ overhangs, total DNA was treated with the 5′-to-3′ T7 gene 6 exonuclease (2 U/μg of DNA; Amersham) in 40 mM Tris-HCl (pH 7.5)-20 mM MgCl2-50 mM NaCl at 37°C for the indicated times and stopped with 25 mM EDTA (pH 8.0).
Cells and culture conditions.
All cells were cultured at 37°C under 5% CO2 in a 4:1 mixture of Dulbecco's modified Eagle's medium and medium 199 supplemented with 10% cosmic calf serum (HyClone, Logan, Utah) and 50 μg of gentamicin (Sigma). Cells were trypsinized at RT, and Sylvania Gold fluorescent bulbs were used for both culture room and laminar flow hood lighting. Cells were considered senescent after failure to increase cell number in 4 weeks. Detection of senescence-associated β-galactosidase activity was performed as described previously (14).
Extopic expression of hTERT, SV40 L-Tg, pLXSN, and HPV E6, E7, and E6/E7 by using retroviruses was described previously (7, 32). The SV40 L-Tg vector used did not express small t antigen.
TRAP assay.
Nonradioactive telomeric repeat amplification protocol (TRAP) analysis was performed as described previously (7).
RESULTS
Measurement of length of G-rich overhangs by telomere overhang protection assay.
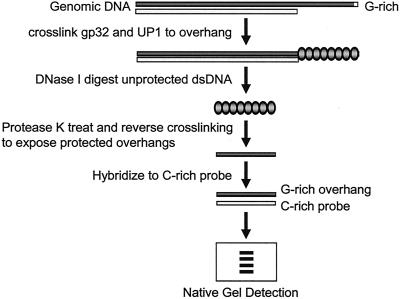
We first developed a method for quantitatively measuring the length of telomeric G-rich overhangs (Fig. 1). Genomic DNA was incubated with both T4 gene 32 protein (gp32) and UP1 to coat the G-rich overhangs. gp32 binds cooperatively to single-stranded DNA independent of DNA sequence (40). UP1 is the proteolytic fragment of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and binds to single-stranded (TTAGGG)n repeats in vitro (24). Using a model template containing an ∼450-nt 3′ single-stranded (TTAGGG)n overhang (see below), we found that addition of UP1 increased the protection of long G-rich overhangs (see Fig. S1A in the supplemental material), possibly due to the unfolding of potential quadruplex structures in the G-rich sequences by UP1 (15). Addition of more gp32 protein alone did not produce the same increase of the overhang protection but rather caused a shift towards increased size under the experimental conditions (see Fig. S1B in the supplemental material). These proteins were then glutaraldehyde cross-linked, and the uncoated double-stranded DNA was digested by DNase I treatment. After reversal of the cross-linking and removal of proteins with protease K, the single-stranded overhangs were hybridized to a high-specific-activity [32P](CCCTAA)3 probe and subjected to nondenaturing polyacrylamide gel analysis.
FIG. 1.
Outline for measuring the size of telomeric 3′ G-rich overhang using the overhang protection assay. Total genomic DNA is incubated with T4 gp32 and GST-UP1, which bind to single-stranded 3′ G-rich overhangs. After the proteins are cross-linked to DNA, unprotected double-stranded DNA is removed by DNase I digestion. After heat inactivation of the DNase I, cross-linking is reversed and proteins are digested to make the protected overhangs available for hybridization to 32P-labeled C-rich probe. Overhangs are finally size fractionated on a native polyacrylamide gel.
The overhang measurement technique was validated on single-stranded (TTAGGG)n oligonucleotides (Fig. 2A). The (TTAGGG)n size standards, ranging from 36 to 384 nt, were subjected to the overhang protection assay. As a comparison, standards were also directly annealed to the [32P](CCCTAA)3 probe. As shown in Fig. 2B, the measured mean sizes of each signal after the overhang protection assay were comparable to the input sizes of each oligonucleotide annealed directly to the probe. There was a minor smear to lower molecular weight in the protected fragments, indicating that although the protection was good, it was not absolute. Since larger overhangs hybridized to more probe and thus had a larger signal per molecule, size quantification involved normalizing signals by molecular weight (see below). In the absence of gp32 and UP1, no signals were detected (see Fig. S2 in the supplemental material), indicating that DNase I digestion was complete and protein protection was required for overhang measurement.
We next tested this method on a substrate mimicking a real telomere, a linear DNA molecule containing 3 kb of double-stranded pBlueScript vector sequence with ∼450 nt of single-stranded (TTAGGG)n repeats at the 3′ end (pBSK-Rep4). The overhang size measured by the overhang protection assay produced a fragment of the expected size (Fig. 2C). Moreover, we were able to detect this fragment after mixing pBSK-Rep4 with HeLa genomic DNA (Fig. 2C), suggesting that the smear contributed by the HeLa DNA represented a diversity of sizes in the G-rich overhangs in genomic DNA.
We then applied the overhang protection assay to DNA isolated from HeLa cells. Using whole genomic DNA, we detected the expected smear of signals instead of discrete bands (Fig. 2D, no T7 exonuclease). Because this smear could be due to single-stranded gaps, loops, or incomplete digestion of double-stranded DNA, we treated paired aliquots with the 3′-to-5′ enzyme Exo I to remove the 3′ overhang prior to adding gp32 and UP1. The difference in signals between the presence or absence of Exo I represented the specific contribution of the G-rich 3′ overhang. The resulting smear represented the heterogeneity of telomeric overhang length in human cells, consistent with previous results from electron microscopy showing that human cells have overhangs ranging from 50 to ∼400 nt long (49). Progressive resection of the C-rich strand with T7 gene 6 exonuclease (a 5′-to-3′ exonuclease) prior to the overhang protection assay greatly increased the overhang sizes (Fig. 2D). When the protected overhangs were hybridized to a high-specific-activity G-rich probe, no signals were observed (see Fig. S2 in the supplemental material), indicating that we could not detect the presence of any C-rich overhangs and were specifically measuring C-rich overhangs. In the absence of gp32 and UP1 protein, no signal above background was detected (Fig. S2 in the supplemental material). Taken together, we conclude that the overhang protection assay measures the mean size of telomeric overhangs from cultured cells.
Quantitation of mean overhang sizes from the overhang protection assay.
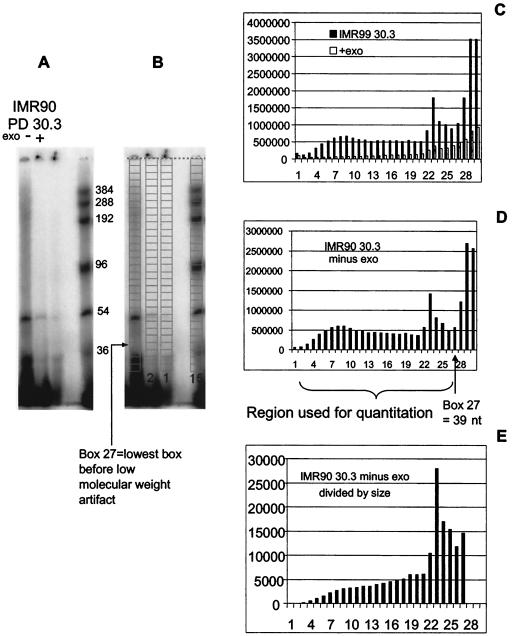
Figure 3 shows an example of how we quantitated the mean overhang size from a broad smear of signals. A gel of the protected overhangs (Fig. 3A) was first overlaid in ImageQuant with a grid of 30 boxes (Fig. 3B). The box position of each of the protected standards (right lane in Fig. 3B) was used to calculate the actual molecular weight represented at the midpoint of each box. The volume (signal intensity) of each box was determined by ImageQuant and imported into an Excel spread sheet. A bar graph of the signal intensities for both the overhang signal (IMR90 PD 30) and the background control (digested with Exo I to remove the overhang) is shown in Fig. 3C. The intensity was corrected by subtracting the background (with Exo) signal for each location (box) from the overhang signal, shown in Fig. 3D. There was a low-molecular-weight artifact in boxes 28 to 30 of the IMR90 PD 30. After eliminating these boxes, the signals from the remaining boxes were divided by the size in nucleotides calculated for each box (since longer telomeres hybridize to more probe), and these values are shown in Fig. 3E. The values of intensity shown in Fig. 3D correspond to the term ODi, and the values divided by size shown in Fig. 3E correspond to the term ODi/Li in the formula Σ(ODi)/Σ(ODi/Li) used to calculate the final weighted mean size of the overhang. The program used for calculating the weighted mean size of overhang is the same as that for calculating the weighted mean size of telomere restriction fragments and is available at the website http://www.swmed.edu/home_pages/cellbio/shay-wright/research/1UTSWTelorunforweb.xls.
FIG. 3.
Quantitation of mean overhang sizes from the overhang protection assay. The gel (A) was first overlaid with a grid of boxes (B) and then quantitated (C). The values after subtracting the background (D) correspond to the term ODi, while the values after dividing the signal by the size in nucleotides (E) correspond to the signal ODi/Li in the formula Σ(ODi)/Σ(ODi/Li) used to calculate the final mean sizes. See text for details.
We would like to point out that the measurements in the overhang protection assay below 45 nt are difficult to reproducibly quantitate. When we treated genomic DNA with T7 exonuclease prior to the overhang protection assay, we found some signals below 45 nt remained unchanged (see Fig. S3 in the supplemental material). Furthermore, based upon the signals obtained from the nondenaturing hybridization assay of measuring telomeric overhangs, the intensity of the signal, particularly below ∼35 nt, was often >10-fold greater than could be explained by authentic telomeric overhangs. This signal was Exo I sensitive and might represent some cross-hybridization of the probe to single-stranded material, including small RNase A-resistant fragments present in the genomic DNA preparations. We therefore did not include signals below 45 nt in the quantitative determination of mean overhang length. Based on the distribution of signals, this probably had little effect on mean sizes greater than 100 nt, but it would have progressively greater impact on accuracy as the mean size becomes smaller, as a greater fraction of the total signal potentially falls below this 45-nt cutoff.
Senescent cells maintain their 3′ G-rich overhangs.
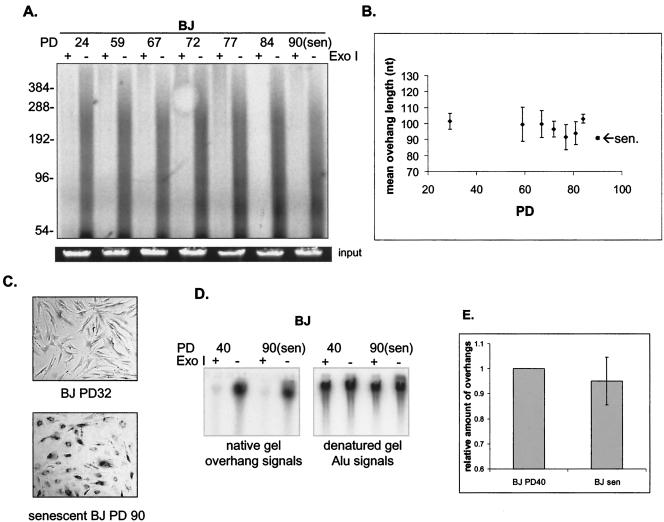
Telomeres in senescent normal human fibroblasts have been reported to lose 60 to 80% of their 3′ overhangs (42). We reexamined this conclusion by using the overhang protection assay to analyze both BJ foreskin and IMR90 lung fibroblasts. We found minimal (∼10%) overhang loss in senescent BJ cells (Fig. 4A and B) and no overhang loss in senescent IMR90 cells (Fig. 5A and B). Although it appears that senescent BJ cellslost some of their long overhangs (>300 nt) (Fig. 4A), the longest overhangs quantitatively contributed very little to the weighted mean sizes of overhangs because larger overhangs hybridized to more probes. We confirmed these were senescent cells by lack of proliferation for 4 weeks and by detecting the presence of senescence-associated β-galactosidase activity (Fig. 4C). To determine whether the length of overhangs changed prior to senescence, we collected DNA from BJ cells every 5 to 10 population doublings (PD) for the last 30 doublings prior to senescence. To minimize the stress, cells were continuously passaged without freezing and thawing and culture media were changed at least once a week. We found that overhang length remained unchanged prior to senescence (Fig. 4A and B).
FIG. 4.
BJ cells maintain their telomeric 3′ overhangs at senescence. (A) Overhang protection assay of DNA from BJ fibroblasts at different PD up to senescence (sen). DNA was treated with or without Exo I before being analyzed in the overhang protection assay. To verify approximately equal amounts of input DNA, 1/30 of each sample was run on a 0.7% agarose gel and visualized with ethidium bromide (input). (B) Weighted mean sizes of the overhangs were quantified and are plotted. Results shown are representative of at least three independent experiments. Error bars represent one standard deviation. Cells were cultured in three different growth series until senescence, and DNA from these cells was used for the overhang protection assay. (C) Senescence-associated β-galactosidase staining of BJ fibroblasts at PD 32 and at senescence. (D) Nondenaturing hybridization analysis of telomeric restriction fragments from BJ fibroblasts at PD 40 and 90 (senescence). Undigested genomic DNA was hybridized to 32P-labeled C-rich high-specificity probe and then gel fractionated in 0.5× TBE. After the gel was denatured, overhang signals were transferred to a nylon membrane and exposed to a phosphorimager screen (left panel). The denatured gel was neutralized and rehybridized to the Alu repeat probe (right panel). (E) Relative amounts of overhangs were calculated by normalizing signals from the membrane (overhang signals) by the Alu repeat signal (representing total genomic DNA) and plotted. Results shown are representative of three independent experiments. Error bars represent one standard deviation.
FIG. 5.
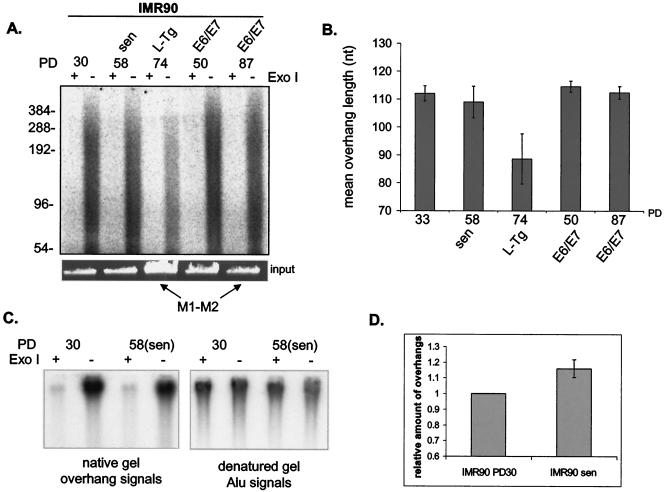
IMR90 cells maintain their telomeric 3′ overhangs at senescence. (A) Overhang protection analysis of DNA from IMR90 fetal lung fibroblasts at different PD up to senescence (sen) and from IMR90 cells expressing SV40 L-Tg or HPV E6 and E7 (E6/E7). Cells that are in the extended life span through expression of L-Tg (PD 74) or E6/E7 (PD 87) are indicated as M1-M2. DNA was treated with or without Exo I before being analyzed in the overhang protection assay. To verify approximately equal amounts of input DNA, 1/30 of each sample was run on a 0.7% agarose gel and visualized with ethidium bromide (input). (B) Weighted mean sizes of overhangs were quantified and plotted. Results shown are representative of at least three independent experiments. Error bars represent one standard deviation. (C) Nondenaturing hybridization analysis of telomeric restriction fragments from young (PD 30) and senescent (PD 58) IMR90 fetal lung fibroblasts. Undigested genomic DNA was hybridized to 32P-labeled C-rich high-specificity probe and then gel fractionated in 0.5× TBE. After the gel was denatured, overhang signals were transferred to a nylon membrane and exposed to a phosphorimager screen (left panel). The denatured gel was then neutralized and rehybridized to the Alu repeat probe (right panel). (D) Relative amounts of overhangs were calculated by normalizing signals from the membrane (overhang signals) to the Alu repeat signal (representing total genomic DNA) and plotted. Results shown are representative of three independent experiments. Error bars represent one standard deviation.
We next used the nondenaturing hybridization assay to confirm that senescent BJ and IMR90 cells maintained their overhangs. DNA was first hybridized to a C-rich probe under nondenaturing conditions to allow the probe to hybridize to the telomeric overhangs. Undigested whole genomic DNA was used to get a compact signal for easy quantitation. After gel fractionation, we denatured the gel under conditions that retained the annealed probe inside the gel. The gel was then dried on a membrane to transfer the overhang-hybridized probe onto the membrane. The total amount of signal on the membrane represented the overhang signal. The same gel was then hybridized to an Alu repeat probe. Overhang signals were normalized to the total amount of genomic DNA determined from signals hybridized to the Alu repeat probe. We treated paired aliquots with Exo I to remove the 3′ overhang, and the difference in signals with or without Exo I represented the specific contribution of the G-rich 3′ overhangs (Fig. 4D and 5C). Results showed that neither senescent BJ nor senescent IMR90 lost a substantial amount of their overhangs (Fig. 4E and 5D), consistent with results from the overhang protection assay. We therefore found no evidence that global overhang loss occurred or triggered replicative senescence.
Cells bypassing M1 maintain their overhang length.
We next examined the overhang length in cells bypassing senescence due to the expression of HPV E6 and E7. The E6 and E7 gene products cause degradation of p53 and hyperphosphorylated Rb, respectively, thus allowing cells to bypass senescence (19, 46). During their extended life span, both BJ and IMR90 cells infected with E6/E7 showed no decrease in overhang length (Fig. 5B and 6C).
FIG. 6.
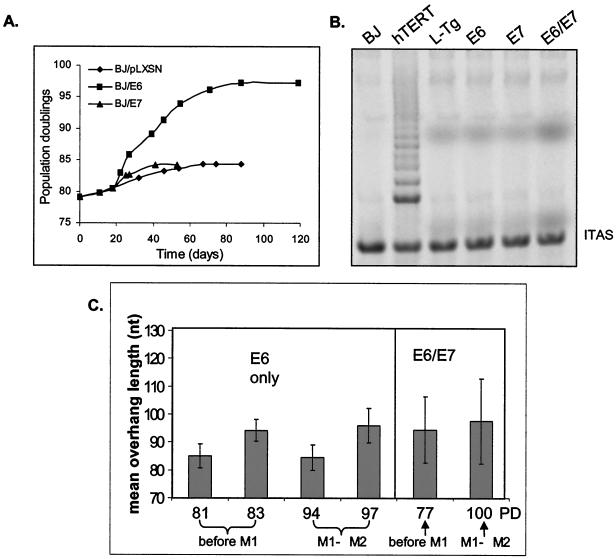
Cells maintain their 3′ overhangs during their extended life span. (A) Growth curve of BJ fibroblasts ectopically expressing HPV E6 only, E7 only, or pLXSN (vector-only control). BJs were infected at PD 78 with retrovirus containing E6 (filled squares), E7 (filled triangles), or vector only (diamonds). After selection, cells were cultured until no cell number increase in 4 weeks was observed. (B) TRAP analysis of BJ cells ectopically expressing E6, E7, E6 and E7, L-Tg, or hTERT. Nonradioactive TRAP assays were performed by using whole-cell lysates from 2,500 cells. ITAS represents the 36-bp internal TRAP assay standards. (C) Weighted mean sizes of overhangs from the overhang protection assay from BJ fibroblasts ectopically expressing E6 or E6 and E7 (E6/E7) at different PD. Results are representative of at least three independent experiments. Error bars represent one standard deviation.
We then tested cells infected with HPV16 E6 only and E7 only. BJ cells were infected with retrovirus containing E6, E7, E6 plus E7, or empty vector. Ectopic expression of E6 extended life span for about 13 PD beyond normal senescence, and cells eventually growth arrested. Cells expressing E7 growth arrested at approximately the same PD as the control cells infected with vector control (Fig. 5A). Although expression of E6 is able to activate telomerase in epithelial cells and primary keratinocytes (1, 23, 44), telomerase activity remained undetectable in BJ/E6, BJ/E7, or BJ/E6/E7 cells (Fig. 6B).
Cells expressing E6 only and cells expressing both E6 and E7 showed overhangs with lengths similar to those of normal cells before and after bypassing M1 (Fig. 6C). Cells expressing only E7 showed no reduction in overhang length at senescence compared to proliferating cells (data not shown), supporting our conclusion that a reduction of overhang length is not the cause of replicative senescence. The behavior of cells expressing HPV oncoproteins showed that there is no association of short overhangs with the extended life span between M1 and M2.
Cells expressing SV40 L-Tg gradually lose some of their overhangs during passaging.
We also examined the overhang length in cells bypassing senescence due to the expression of SV40 L-Tg. In contrast to cells expressing HPV E6/E7, L-Tg-infected BJ and IMR90 cells exhibited about a 20 to 30% overhang shortening during their extended life span (Fig. 5B and 7B).
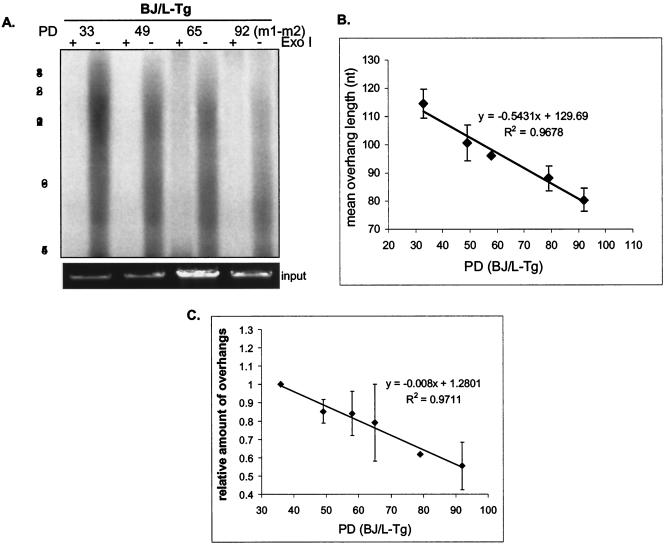
FIG. 7.
Loss of telomeric overhangs in cells expressing SV40 L-Tg, as shown in the overhang protection analysis of DNA from BJ fibroblasts infected with retrovirus containing SV40 L-Tg at different PD. Cells bypassed senescence at approximately PD 90. DNA was treated with or without Exo I before being analyzed in the overhang protection assay. To verify approximately equal amounts of input DNA, 1/30 of each sample was run on a 0.7% agarose gel and visualized with ethidium bromide (input). (B) Weighted mean sizes of overhangs were quantified and are plotted. Results shown are representative of at least three independent experiments. Error bars represent one standard deviation. (C) Relative amount of overhangs of BJ/L-Tg cells measured by nondenaturing hybridization assay. Undigested whole genomic DNA from BJ at PD 40 and at senescence was first hybridized to the C-rich telomeric probe and then gel fractionated. After the gel was denatured, overhang signals were transferred to a membrane and the gel was rehybridized to the Alu repeat probe. Relative amounts of overhangs were calculated by normalizing signals from the membrane (overhang signals) by the Alu repeat signal (representing total genomic DNA). Results shown are representative of three independent experiments. Error bars represent one standard deviation.
To test whether overhang shortening in L-Tg-infected cells occurred prior to M1, we infected young BJ cells at PD 27 with L-Tg. The young BJ/L-Tg cells displayed similar overhang sizes to the uninfected control. However, their overhangs shortened as these cells were passaged (Fig. 7A and B). The overhangs continued to shorten after cells bypassed senescence (PD 92) (Fig. 7A and B). No change in the rate of decrease was seen throughout the culture period. We also used the nondenaturing hybridization assay and confirmed the results from the overhang protection assay (Fig. 7C). At present, we do not know the reasons why overhang sizes are reduced in L-Tg-expressing cells. The overhang loss measured by nondenaturing hybridization was greater than that measured by the overhang protection assay (40 versus 30% at PD 92), indicating that we underestimated the contribution of the shortest overhangs (below our 45-nt cutoff) induced by L-Tg at higher PD when we calculated the mean overhang size using the overhang protection assay.
DISCUSSION
A 60 to 80% loss of the G-rich telomeric 3′ overhangs has been claimed to be the proximate cause of replicative senescence (42). Using both the overhang protection and the nondenaturing hybridization assays, we failed to find evidence consistent with this conclusion. Overhang length was not appreciably shorter at senescence in either BJ foreskin or IMR90 fetal lung fibroblasts. BJ cells were also used in the above study, while IMR90 fetal lung fibroblasts are similar to the MRC5 fetal lung fibroblasts that were used. As in the previous report, we did observe overhang shortening in cells expressing SV40 L-Tg. However, with the HPV oncoproteins E6 and E7 we did not find overhang shortening either before or during the period of extended life span.
Using a combination of immunofluorescence and fluorescence in situ hybridization analyses, our group has recently demonstrated that in normal human fibroblasts (BJ) it is specifically 10% of telomeres with the shortest ends that cause senescence, with only a few telomeres initiating end associations in any given cell (52). The senescence-associated DNA damage γ-H2AX/53BP1 foci (11) colocalized with these shortest telomeres but not to the longer telomeres (52). These results are essentially incompatible with a global change in overhang length being the cause of replicative senescence. One should not observe a 60 to 80% reduction in overhangs when using an assay that measures all 92 ends if only a few ends are limiting for growth. Thus, our present conclusion that global overhang loss is not the cause of replicative senescence is further supported by a totally different analysis.
It is a formal possibility that only the shortest telomeres responsible for signaling replicative senescence lose their overhangs. If so, it indicates that processing of these shortest telomere ends may be different from that of the rest of the telomere ends. Cells must be able to distinguish these shortest telomere ends from others, perhaps thorough recruiting or modifying DNA damage response factors at these ends. At present, there is no available method to measure the overhang length at each individual telomere end. Future study is needed to determine the length of overhangs on these shortest telomeres.
The difference in results cannot be explained solely on the basis of different methods of analysis. The original report claiming overhang loss as the cause of senescence confirmed their observations by also conducting hybridization to overhangs in native (nondenaturing) gels (42). We have used this same technique, and we found that the overhangs did not decrease in size (Fig. 4E and 5D). We speculate that the most likely explanation lies in different culture conditions that produce different baseline levels of stress or DNA damage. Some types of stress and DNA damage have been shown to induce a shortening of overhangs (42), and a reduction in overhang size with ongoing culture has also been reported by Keys et al. (22). Because of our interest in the long-term cultivation of normal diploid cells, we have optimized culture conditions as much as possible. In addition to carefully screening different serum lots, our culture facility contains longer-wavelength gold lighting to prevent the formation of toxic products in tissue culture media produced by standard fluorescent lighting (4, 45). In addition, since trypsinization at 37°C has been shown to be toxic (31), all trypsinizations were performed at RT. It is possible that these or other differences in culture technique produce a lower level of stress or damage in our cells that prevents the overhangs from shortening. Irrespectively, the presence of normal replicative aging in the absence of significant overhang shortening establishes that global overhang shortening cannot have a causal relationship with this phenomenon.
Differences between our overhang protection assay and the T-OLA could also contribute to the differences observed. The T-OLA requires the uninterrupted ligation of multiple short oligonucleotides hybridized to the G-rich overhang in order to provide an estimate of the size of the overhang (9). Electron microscopic studies have shown that human telomeric overhangs are quite heterogeneous in length, ranging from 50 to at least 400 nt (21, 49). The T-OLA technique thus attempts to analyze a heterogeneously sized target with a technique that produces a heterogeneously sized product even from a unique model target. In addition, anything that prevents a continuous gap-free alignment of oligonucleotides would prevent the production of large ligated products. The G-rich sequence has the ability to form unique secondary structures called G-quadruplexes (50). The presence of small “G-knots” anywhere in the overhang could potentially interrupt the continuous alignment of adjacent nucleotides and could contribute to a decreased size of T-OLA products. It is unknown whether such G-knots are present in normal telomeres or if their abundance changes under different conditions such as stress or senescence. The increased protection we observed using UP1, which has at least some capacity to unwind G-quartets under physiological conditions (15, 24), suggests that some such structures might be present in overhangs in isolated genomic DNA.
Although signals below 45 nt are excluded from quantitation in the overhang protection assay, we do not believe this limitation affects our conclusions that overhang loss is not a molecular trigger for replicative senescence. The previous report using T-OLA (42) also had a detection cutoff at 48 nt, because the probe used was 24 nt and the minimal ligation product was 48 nt long. The overhang protection assay described for the first time in this report, which measures the absolute length of overhangs and provides information about overhang size distribution, provides a useful complement to the conventional nondenaturing hybridization assay.
We observed a gradual decrease of overhang sizes in SV40 L-Tg-expressing cells but not in HPV E6/E7-overexpressing cells. At present, we have no adequate explanation for this phenomenon, although it is known that L-Tg and E6/E7 block p53 and Rb pathways through different mechanisms (19). The effect of L-Tg cannot be direct, since there was initially no change in overhang length and the reduction was progressive over the next 60 PD (Fig. 7). Given that some forms of damage induce overhang shortening (42), we speculate that differences in the mechanisms by which HPV E6/E7 and L-Tg affect stress and damage signal processing are responsible for the observed difference.
It has long been hypothesized that telomere overhangs are generated by the combined effects of end replication problem and telomere end processing. The conventional DNA replication machinery will generate a 3′ G-rich overhang at the end of the lagging-strand daughter, while the end of the leading-strand daughter will initially be blunt (27, 35). Both ends can be further trimmed by hypothetical nucleases to generate longer overhangs (8, 37). This hypothesis is supported by the observations that overhangs are detected at both ends of chromosomes in ciliates and human cells. Our result demonstrating that human telomeres have long 3′ overhangs is consistent with previous findings from other assays (29, 49). It will be important to pinpoint the factors and/or nucleases responsible for overhang creation and processing that contribute to the telomere shortening rate (21).
Supplementary Material
Acknowledgments
We are grateful to B. Chabot (Université de Sherbrooke, Québec, Canada) for the GST-UP1 construct. We also thank anonymous reviewers for their helpful comments on the manuscript. We thank O. Bechter and N. Forsyth for discussion and critical comments on the manuscript and W. Walker and J. Sherrell for technical assistance.
This work was supported by a Ruth L. Kirschstein National Research Service Award Individual Fellowship (W.C.) and NIH AG01228. W.E.W. is an Ellison Foundation Senior Scholar.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baege, A. C., A. Berger, R. Schlegel, and T. Veldman. 2002. Cervical epithelial cells transduced with the papillomavirus E6/E7 oncogenes maintain stable levels of oncoprotein expression but exhibit progressive, major increases in hTERT gene expression and telomerase activity. Am. J. Pathol. 160:1251-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, M. O., and N. A. Sharkey. 1977. Mutagenicity and toxicity of visible fluorescent light to cultured mammalian cells. Nature 266:724-726. [DOI] [PubMed] [Google Scholar]
- 5.Campisi, J., S. H. Kim, C. S. Lim, and M. Rubio. 2001. Cellular senescence, cancer and aging: the telomere connection. Exp. Gerontol. 36:1619-1637. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes, R. B., and V. Lundblad. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14:351-356. [DOI] [PubMed] [Google Scholar]
- 7.Chai, W., L. P. Ford, L. Lenertz, W. E. Wright, and J. W. Shay. 2002. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277:47242-47247. [DOI] [PubMed] [Google Scholar]
- 8.Chakhparonian, M., and R. J. Wellinger. 2003. Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet. 19:439-446. [DOI] [PubMed] [Google Scholar]
- 9.Cimino-Reale, G., E. Pascale, E. Battiloro, G. Starace, R. Verna, and E. D'Ambrosio. 2001. The length of telomeric G-rich strand 3′-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 29:E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colgin, L. M., K. Baran, P. Baumann, T. R. Cech, and R. R. Reddel. 2003. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13:942-946. [DOI] [PubMed] [Google Scholar]
- 11.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194-198. [DOI] [PubMed] [Google Scholar]
- 12.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 13.de Lange, T. 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5:323-329. [DOI] [PubMed] [Google Scholar]
- 14.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda, H., M. Katahira, N. Tsuchiya, Y. Enokizono, T. Sugimura, M. Nagao, and H. Nakagama. 2002. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc. Natl. Acad. Sci. USA 99:12685-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., and V. A. Zakian. 1986. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47:195-205. [DOI] [PubMed] [Google Scholar]
- 17.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 18.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 19.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24:159-169. [DOI] [PubMed] [Google Scholar]
- 20.Herbert, B., J. W. Shay, and W. E. Wright. 2003. Analysis of telomeres and telomerase, p. 18.6.6. In Current protocols in cell biology. Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 21.Huffman, K. E., S. D. Levene, V. M. Tesmer, J. W. Shay, and W. E. Wright. 2000. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem. 275:19719-19722. [DOI] [PubMed] [Google Scholar]
- 22.Keys, B., V. Serra, G. Saretzki, and T. Von Zglinicki. 2004. Telomere shortening in human fibroblasts is not dependent on the size of the telomeric-3′-overhang. Aging Cell 3:103-109. [DOI] [PubMed] [Google Scholar]
- 23.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 24.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 25.Lei, M., P. Baumann, and T. R. Cech. 2002. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41:14560-14568. [DOI] [PubMed] [Google Scholar]
- 26.Lin, J. J., and V. A. Zakian. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93:13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingner, J., J. P. Cooper, and T. R. Cech. 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269:1533-1534. [DOI] [PubMed] [Google Scholar]
- 28.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 29.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 30.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeehan, W. L. 1977. The effect of temperature during trypsin treatment on viability and multiplication potential of single normal human and chicken fibroblasts. Cell. Biol. Int. Rep. 1:335-343. [DOI] [PubMed] [Google Scholar]
- 32.Morales, C. P., S. E. Holt, M. Ouellette, K. J. Kaur, Y. Yan, K. S. Wilson, M. A. White, W. E. Wright, and J. W. Shay. 1999. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 21:115-118. [DOI] [PubMed] [Google Scholar]
- 33.Munoz-Jordan, J. L., G. A. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 35.Olovnikov, A. M. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41:181-190. [DOI] [PubMed] [Google Scholar]
- 36.Price, C. 1999. Telomeres. Capping off the ends. Nature 397:213-214. [DOI] [PubMed] [Google Scholar]
- 37.Price, C. M. 1997. Synthesis of the telomeric C-strand. A review. Biochemistry (Moscow) 62:1216-1223. [PubMed] [Google Scholar]
- 38.Price, C. M., and T. R. Cech. 1989. Properties of the telomeric DNA-binding protein from Oxytricha nova. Biochemistry 28:769-774. [DOI] [PubMed] [Google Scholar]
- 39.Price, C. M., and T. R. Cech. 1987. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev. 1:783-793. [DOI] [PubMed] [Google Scholar]
- 40.Shamoo, Y., A. M. Friedman, M. R. Parsons, W. H. Konigsberg, and T. A. Steitz. 1995. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature 376:362-366. [DOI] [PubMed] [Google Scholar]
- 41.Shay, J. W., and W. E. Wright. 2001. Telomeres and telomerase: implications for cancer and aging. Radiat. Res. 155:188-193. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, S. A., I. Ben-Porath, V. J. Carey, B. F. O'Connor, W. C. Hahn, and R. A. Weinberg. 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33:492-496. [DOI] [PubMed] [Google Scholar]
- 43.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 44.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, R. J., and B. R. Nixon. 1978. Identification of hydrogen peroxide as a photoproduct toxic to human cells in tissue-culture medium irradiated with “daylight” fluorescent light. In Vitro 14:715-722. [DOI] [PubMed] [Google Scholar]
- 46.White, A. E., E. M. Livanos, and T. D. Tlsty. 1994. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 8:666-677. [DOI] [PubMed] [Google Scholar]
- 47.Williamson, J. R., M. K. Raghuraman, and T. R. Cech. 1989. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59:871-880. [DOI] [PubMed] [Google Scholar]
- 48.Wright, W. E., O. M. Pereira-Smith, and J. W. Shay. 1989. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol. Cell. Biol. 9:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zahler, A. M., J. R. Williamson, T. R. Cech, and D. M. Prescott. 1991. Inhibition of telomerase by G-quartet DNA structures. Nature 350:718-720. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12:1489-1498. [DOI] [PubMed] [Google Scholar]
- 52.Zou, Y., A. Sfeir, J. W. Shay, and W. E. Wright. 2004. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell 15:3709-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.