Abstract
Class Ia phosphoinositide 3-kinases (PI3Ks) are heterodimers of p110 catalytic and p85 regulatory subunits that mediate a variety of cellular responses to growth and differentiation factors. Although embryonic development is not impaired in mice lacking all isoforms of the p85α gene (p85α−/− p55α−/− p50α−/−) or in mice lacking the p85β gene (p85β−/−) (D. A. Fruman, F. Mauvais-Jarvis, D. A. Pollard, C. M. Yballe, D. Brazil, R. T. Bronson, C. R. Kahn, and L. C. Cantley, Nat Genet. 26:379-382, 2000; K. Ueki, C. M. Yballe, S. M. Brachmann, D. Vicent, J. M. Watt, C. R. Kahn, and L. C. Cantley, Proc. Natl. Acad. Sci. USA 99:419-424, 2002), we show here that loss of both genes results in lethality at embryonic day 12.5 (E12.5). The phenotypes of these embryos, including subepidermal blebs flanking the neural tube at E8 and bleeding into the blebs during the turning process, are similar to defects observed in platelet-derived growth factor receptor α null (PDGFRα−/−) mice (P. Soriano, Development 124:2691-2700, 1997), suggesting that PI3K is an essential mediator of PDGFRα signaling at this developmental stage. p85α−/− p55α+/+ p50α+/+ p85β−/− mice had similar but less severe defects, indicating that p85α and p85β have a critical and redundant function in development. Mouse embryo fibroblasts deficient in all p85α and p85β gene products (p85α−/− p55α−/− p50α−/− p85β−/−) are defective in PDGF-induced membrane ruffling. Overexpression of the Rac-specific GDP-GTP exchange factor Vav2 or reintroduction of p85α or p85β rescues the membrane ruffling defect. Surprisingly, reintroduction of p50α also restored PDGF-dependent membrane ruffling. These results indicate that class Ia PI3K is critical for PDGF-dependent actin rearrangement but that the SH3 domain and the Rho/Rac/Cdc42-interacting domain of p85, which lacks p50α, are not required for this response.
Hormone and growth factor stimulation of phosphoinositide 3-kinases (PI3Ks) results in production of lipid second messengers at the plasma membrane that initiate a complex network of signaling pathways that have been implicated in cell growth, cell survival, cell proliferation, and cell migration (7, 12, 38, 44). Disregulation of the PI3K pathway by loss of PTEN, the phosphatase that degrades phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3), or by activating mutations in the p110α catalytic subunit of PI3K is one of the most frequent events in human cancers (9, 27, 33, 42). However, the role of individual isoforms of PI3K in development and in signaling is poorly understood.
Mammalian PI3Ks can be grouped into three major classes, based on primary sequence, mechanism of regulation and substrate specificity. Class Ia PI3Ks are heterodimers consisting of a catalytic subunit (p110) and a regulatory subunit (p85). These enzymes are thought to provide the major source of PI-3,4,5-P3 in vivo upon activation of receptor type protein-tyrosine kinases. Multiple isoforms of class Ia PI3K exist in mammals (14). Three different genes encode class Ia catalytic subunits, termed p110α, p110β, and p110δ. Three genes have also been described for the associating regulatory subunit. Two genes encode isoforms of 85 kDa, termed p85α (Pik3r1) and p85β (Pik3r2). The Pik3r1 gene also produces two major alternative transcripts encoding the smaller proteins p55α and p50α. These shorter isoforms include the two C-terminal Src-homology 2 (SH2) domains and the p110 interacting domain but lack the N-terminal SH3 domain, proline-rich regions, and a domain that mediates interaction with Rho/Rac/Cdc42 family members. A third gene encodes p55γ (or p55PIK), a protein with similar structure to p55α. The p85α and p85β proteins are ubiquitously expressed, whereas the smaller isoforms are distributed in a tissue-specific manner.
Activation of PI3K by the platelet-derived growth factor (PDGF) receptor (PDGFR) has been extensively studied. PDGF binding to the receptor activates autophosphorylation at sites (Tyr 740 and Tyr 751) that mediate binding to the SH2 domains of the p85 regulatory subunit of PI3K (21). This interaction recruits PI3K to the plasma membrane and turns up its catalytic activity for local production of PI-3,4,5-P3. PI-3,4,5-P3 acts as a second messenger that recruits and activates intracellular signaling molecules such as the Ser/Thr protein kinase AKT/PKB, Tec family tyrosine kinases, and Arf exchange factors. In vitro studies using PI3K binding mutants of the PDGFRβ have indicated that PI3K is responsible for PDGF-induced cell proliferation, cell survival, and migration (2, 20, 24, 41). However, these mutations have also been shown to disrupt association with other signaling molecules (30). Experiments using dominant-negative forms of PI3K and drug inhibitors have supported a role for PI3K in PDGF-dependent cell ruffling and chemotaxis (24, 34, 45, 46). But these studies are complicated by the ability of dominant-negative forms of PI3K to compete with other signaling proteins for binding to the PDGFR and by the ability of existing PI3K inhibitors to inhibit a variety of lipid kinases and protein kinases. Thus, to evaluate the role of PI3K isoforms in PDGF signaling during development and in isolated cells, it is critical to delete specific genes.
We show here that targeted disruption of all p85α and p85β gene products (p85α−/− p55α−/− p50α−/− p85β−/−) causes death by embryonic day 12.5 (E12.5), accompanied by similar defects to those seen in PDGFRα null mice. We observed similar phenotypes in p85α−/− p55α+/+ p50α+/+ p85β−/− mouse embryos and in p110α−/− mouse embryos. We conclude that during development, p110α fulfills unique functions downstream of the PDGFR α that cannot be mediated by p110β. We also conclude that the shorter alternative splice forms of p85α cannot fully compensate for p85α and p85β at this stage of development.
We also show that mouse embryonic fibroblasts (MEFs) lacking all isoforms of p85α and p85β are impaired in PDGF-induced membrane ruffling and that this defect can be explained by a defect in PDGF-induced Rac activation, since overexpression of the Rac-specific GDP/GTP exchange factor (GEF) Vav2 can restore the defects in membrane ruffling. Reintroduction of either p85α or p85β can also restore the PDGF-dependent membrane ruffling. Surprisingly, the p50α protein can also restore PDGF-dependent ruffling. These results indicate that the SH3 domain, Pro-rich domains, and Rho/Rac/CDC42 binding domains in the N terminus of p85α and p85β are not required for PDGF-dependent remodeling of the actin cytoskeleton.
MATERIALS AND METHODS
Targeted disruption of the Pik3r2 gene and generation of mice lacking p85β.
A mouse p85β cDNA (gift from Sebastian Pons) was used to screen a genomic DNA library from the 129Sv strain in LambdaFIXII (provided by F. Alt, Children′s Hospital, Boston, Mass.). Genomic clones where obtained and mapped by restriction digest, Southern blotting, and sequencing. We designed a 2-kb targeting construct containing a pgk-neomycin resistance cassette to replace a 1.8-kb region of Pik3r2 which included 5′-untranslated sequence, the start codon, and the first exon. The pLNTK vector carrying the p85β homologous arms surrounding the neomycin replacement cassette contained the thymidylate kinase (TK) gene peripheral to these segments. The mouse embryonic stem (ES) cell line TC-1 (strain 129SvEv) was electroporated (2 × 10e7 cells) with 20 μg of linearized DNA and subjected to positive selection with G418 (0.4 mg/ml) and negative selection with ganciclovir (1 μM). One clone of 450 screened contained a heterozygous disruption of the Pik3r2 gene was identified by Southern blot analysis with probes flanking the 5′ homology region and a probe internal to the 3′ homology region. The presence of a single integration event was verified by hybridization with a Neo probe. We obtained one ES clone with a correctly targeted p85β allele and injected it into C57BL/6 blastocysts (Beth Israel Deaconess Medical Center Transgenic Facility). Highly chimeric mice were bred with C57BL/6 females to obtain mice with heterozygous loss of p85β. The offspring of p85β+/− intercrosses were born with expected Mendelian ratios of 1:2:1 (wild-type/heterozygous/homozygous ratio), revealing that the p85β gene product is not required during embryonic development. The mutation was subsequently maintained and studied in a mixed 129SvEv × C57BL/6 background.
p85α+/− p55α+/− p50α+/− mice were crossed with p85β−/− mice to obtain double heterozygous mice. p85α+/− p55α+/− p50α+/− p85β+/− mice were crossed with p85β−/− mice to generate p85α+/− p55α+/− p50α+/− p85β−/−, which were viable and fertile. Intercrosses of these mice were performed to generate embryos.
p85α−/− p55α+/+ p50α+/+ mice, lacking only the p85α isoform and retaining p55α and p50α, were purchased from Taconic Farms. These animals, in the C57BL/6 background, were crossed with p85β−/− mice (129SvEv × C57BL/6) under a research crossbreeding license. The resulting double heterozygous mice were crossed with p85β−/− mice to generate p85α+/− p55α+/+ p50α+/+ p85β−/−, which were viable and fertile. Intercrosses of these mice were performed to generate embryos.
Southern blot analysis.
Recombinant ES cells were screened by Southern blot (ZetaProbe GT membranes; Bio-Rad) of EcoRI-digested genomic DNA with a BamHI-digested, 32P-labeled probe upstream of the 5′ homologous region and confirmed with a SacI/EcoRI-digested 32P-labeled probe inside the 3′ homologous region. The 5′ probe produced a band at about 9 kb representing the wild-type allele and a 6-kb band representing the recombinant allele. The 3′ probe produced a 9-kb wild-type band and a 3-kb recombinant band. The presence of a single integration event was verified by hybridization to genomic DNA digested with either EcoRI or BamHI with a 32P-labeled probe to the neomycin resistance gene by Southern blotting.
Genotyping of p85β-deficient mice.
Mice were genotyped by PCR of crude tail genomic DNA (digested with modified Gitschier's buffer) with three primers, P1 (5′-GTCGCCTGTGACTTCTGGAAGT-3′), P2 (5′-GCATCCAGCCCACATTGTGT-3′), and P3 (5′-TGTTAAGAAGGGTGAGAACAGAGTACC-3′). The PCR volume used was 16 μl containing 0.33 μM (each) P2 and P3 and 0.76 μM P1, 0.1 μl of Hot Start Taq polymerase, and 1× accompanying buffer and MgCl2 (QIAGEN). A 295-bp product of P1 and P2 represented the endogenous allele, and a 340-bp product of P1 and P3 represented the recombinant allele, visualized on a 2% 1× TAE (40 mM Tris, 0.02 N glacial acetic acid, 1 mM EDTA [pH 8])-agarose gel by ethidium bromide staining.
The offspring (and embryos) of crosses between double mutant p85α+/− p55α+/− p50α+/− and p85β−/− mice or p85α+/− p55α+/+ p50α+/+ and p85β−/− mice were genotyped for p85α as described by Fruman et al. (13) (p85α+/− p55α+/− p50α+/−) or Terauchi et al. (37) (p85α+/− p55α+/+ p50α+/+) and for p85β as described above.
Histopathology in p85α−/− p55α−/− p50α−/− p85β−/− embryos.
E8.5 to 12.5 embryos (129 × BL6) were separated from their yolk sacs, which were then used for genotyping by PCR. The specimens were fixed in Bouin′s solution or Formalin for 4 h or overnight and then transferred into 70% ethanol. Sagittal sections were stained by standard hematoxylin and eosin (H&E) staining procedure.
Tissue cell culture.
MEFs were generated from embryos at various developmental stages (E8.5 for p85α−/− p55α−/− p50α−/− p85β−/− and littermate controls and E11.5 for p85α−/− p55α+/+ p50α+/+ p85β−/− and littermate controls). The embryos were removed from their yolk sac and transferred onto gelatinized 12-well tissue culture dishes containing 15% fetal calf serum-Dulbecco's modified Eagle's medium (FCS/DMEM). The cells were trypsinized and expanded very slowly (1:2 or 1:3). The MEFs were immortalized with a replication-defective retrovirus expressing SV40 large antigen (gift from D. Livingston). The cells were used for experiments when they achieved a stable growth rate in growth media containing 15% FCS.
Antibodies and inhibitors.
Mouse monoclonal anti-p55γ antibody was a generous gift from M. White. Anti-p110α monoclonal antibody (P94520) was purchased from Transduction Laboratories. Rabbit polyclonal anti-p110α antibody (SC-7174) was purchased from Santa Cruz. Rabbit polyclonal anti-p110β antibody (SC-602) was purchased from Santa Cruz. Mouse monoclonal antiphosphotyrosine antibody (4G10) was a gift from T. Roberts. Rabbit polyclonal anti-Akt antibody (9272), rabbit polyclonal anti-mitogen-activated protein kinase (anti-MAPK) antibody (9102), or rabbit polyclonal antibodies for the phosphorylated proteins (9271L and 9101S) were purchased from Cell Signaling Technology, Beverly, Mass. Mouse monoclonal anti-T7 antibody was purchased from Novagen. PDGF-BB was purchased from Austral Biologicals. PDGF-AA was purchased from Sigma. IGF-1 was purchased from Austral Biologicals. Epidermal growth factor (EGF) was purchased from Upstate Technologies. PS, PI, and PI-4,5-P2 were purchased from Avanti Polar Lipids. PI-4-P was purchased from Sigma. Wortmannin was purchased from Sigma. LY294002 was purchased from Calbiochem.
Immunoblot.
Subconfluent cells were starved for 24 h and then stimulated with various concentrations of growth factors for the indicated times (with or without pretreatment with 100 nM wortmannin for 20 to 30 min). Cells were washed three times with ice-cold phosphate-buffered saline (PBS) and then lysed in lysis buffer (25 mM Tris [pH 7.2], 137 mM NaCl, 10% glycerol, 1% NP-40, 25 mM NaF, 10 mM sodium pyrophosphate, 1 mM EDTA, containing fresh protease-phosphatase inhibitors) for 10 min at 4°C. The clarified lysates were standardized by Bradford assay and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and probed with various antibodies according to the manufacturers' manuals.
PI3K assay.
Equal amounts of clarified cell lysate were immunoprecipitated with anti-p85pan, anti-p110α, anti-p110β, or 4G10 antisera. The immunoprecipitates (IP) were washed two times with lysis buffer and twice with TNE (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA). The PI3K assay was performed by adding 20 μl of HEPES (100 mM, pH 7), 20 μl of lipids (67 μM lipid substrate, 133 μM phosphoserine), and 10 μl of ATP mix (70 mM HEPES [pH 7], 50 mM MgCl2, 0. 5 mM ATP, [γ-32P]ATP at 10 μCi/assay) to 50 μl of IP. The reactions were performed at room temperature and stopped after 10 min by adding 25 μl of 5 M HCl. The lipids were extracted with 160 μl of CHCl3-methanol (MeOH) (1:1). The phosphorylated lipids were spotted on a thin-layer chromatography plate and separated overnight with 1-propanol-2 M acetic acid (65:35). The radioactivity was visualized by PhosphorImager (Molecular Dynamics).
HPLC analysis.
Subconfluent cells were starved overnight and incubated for 45 min in phosphate-free medium and then labeled for 4 h with 32P (2 mCi/10-cm plate). The cells were stimulated with 10 ng of PDGF per ml for 5 min, with or without pretreatment with 100 nM wortmannin for 20 to 30 min. The reaction was stopped with ice-cold PBS. The cells were lysed in 400 μl of MeOH and 400 μl of 1 M HCl. The lipids were extracted with 400 μl of CHCl3 and then with 400 μl of MeOH-0.1 M EDTA. Lipids were deacylated with 1 ml of methylamine reagent (26.8 ml of 40% methylamine in H2O, 16 ml of H2O, 45.7 ml of methanol, 11.4 ml of n-butanol) for 50 min at 53°C. Deacylated lipids were dried and resuspended in 600 μl of H2O and extracted with 600 μl of n-butanol-petroleum ether-ethyl formate (20:4:1). Then the lipids were mixed with 3H-labeled standards and analyzed by anion-exchange high-performance liquid chromatography (HPLC) with a Partisphere SAX column (Whatman) as described previously (L.A. Serunian and L. C. Cantley).
Rac activation assay.
The levels of Rac GTP were measured by affinity precipitation with glutathione S-transferase (GST)-CRIB (Cdc42 and Rac interactive region) of PAK65 (28), as previously described (19).
Time-lapse ruffling.
Subconfluent cells plated on coverslips were starved overnight up to 48 h. The coverslips were then placed into a heated device (37°C) on an inverted microscope (DIAPHOT 300; Nikon). Pictures were taken every 15 s, and movies were analyzed with Image Pro Plus. Basal ruffling was assessed for 3 min, and then PDGF-BB or insulin-like growth factor 1 (IGF-1) was added to a final concentration of 50 ng/ml or 20 nM, respectively. Ruffling activity was monitored for about 15 to 60 min, and then 100 nM wortmannin was added to determine PI3K-dependent ruffling. The movies were analyzed for the percentage of cells forming ruffles after growth factor stimulation.
Phalloidin staining of fixed ruffles.
Cells on coverslips were prepared as just described above. The subconfluent, resting cells were stimulated with 50 ng of PDGF-BB per ml for 15 min and then washed with CBS (10 mM morpholineethanesulfonic acid [MES; pH 6.1], 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 10 to 11% sucrose). The cells were fixed in CBS containing 3.7% paraformaldehyde for 20 min and then permeabilized for 10 to 20 min in CBS with 0.1% Triton X-100. The actin cytoskeleton was stained during the permeabilization process with rhodamine-coupled phalloidin. The coverslips were washed once in CBS and then mounted and analyzed under the microscope for the percentage of cells exhibiting lamellipodium formation. In order to stain for T7-tagged Vav2, cells were lysed 24 h after transfection (with or without pretreatment with 100 nM wortmannin for 30 min), fixed, and permeabilized as described above. The cells were blocked in Tris-buffered saline (TBS) containing 5% horse serum and 1% bovine serum albumin (BSA) for 15 min. The primary anti-T7 antibody was incubated overnight at 4°C, and the secondary fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody was incubated for 30 min at room temperature in TBS containing 1% BSA.
Retroviral addback.
p85α−/− p55α−/− p50α−/− p85β−/− MEFs were infected with p85α and p85β or p50α in murine stem cell virus (MSCV)-internal ribosomal entry site (IRES) green fluorescent protein (GFP) (pMIG) or pMIG alone (constructs gift from D. Fruman). The GFP-expressing cells were separated via fluorescence-activated cell sorting (FACS; BIDMC, Boston, Mass.).
Transient transfections.
T7-tagged Vav2 (gift from C. Carpenter) or HA-p55γ (gift from M. White) was introduced into p85α−/− p55α−/− p50α−/− p85β−/− cells by transient transfection with the Lipofectamine Plus protocol according to the manufacturer′s manual.
RESULTS
Embryonic death of mice lacking all gene products of p85α and p85β.
The early embryonic lethality of mice lacking PI3K catalytic isoforms p110α or p110β demonstrated an important nonredundant function of these two catalytic isoforms of class Ia PI3K in murine development (3, 4). In contrast, mice lacking all splice forms of p85α survive to birth, and although most die within a week, occasionally these mice survive to become adults (13). Mice lacking p85α but expressing the shorter alternative splice forms of this gene, p55α and p50α are fully viable (37). Although we have not previously published our strategy for generating p85β−/− mice (presented here in detail in Materials and Methods), we have reported that these mice are viable and have increased insulin sensitivity (40). Adult male mice lacking p85β have a statistically significant 9% decrease in body weight (data not shown). In order to determine whether the viability of mice lacking individual p85 genes is due to redundant functions, we generated mice with a combined loss of all p85α and p85β gene products. No mice deficient in both p85α (and its smaller variants) and p85β were born, demonstrating the requirement for these class Ia PI3K regulatory subunits in development.
In order to determine at which embryonic stage the mutant mice died, timed pregnancies were set up and embryos were analyzed at various stages. Expected Mendelian ratios of viable embryos lacking p85α, p55α, p50α and p85β were detected until E11.5. However, the mutant embryos exhibited severe developmental defects (Table 1). All mutant embryos displayed one or more subepidermal blebs (detachment of the most outer epithelial cell layer) flanking the neural tube at a time just before the embryos turn to achieve the fetal position (E8 to 8.5) (Fig. 1A). In a few mutant embryos, a dilated pericardium or twining was observed. After the embryos had turned about their anterior-posterior axis (E8.5 to 9.5) many of the blebs were filled with blood (Fig. 1B, C, and E). At later stages (E10 to 11.5), blebs and other facial abnormalities (e.g., clefted face) were detected on the head (Fig. 1D). Some of the mutant embryos had a dilated, wavy neural tube and exhibited multiple hemorrhages in the head region and branchial arches. At E12.5, p85α−/− p55α−/− p50α−/− p85β−/− embryos were pale and exhibited no heartbeat. The mutant embryos were delayed in their development and therefore smaller than their littermates. These observations are remarkably similar to defects observed in PDGFRα−/− mice (36). This correlation suggests that class Ia PI3K isoforms are required for PDGFRα-dependent development at E8 to 12. Histological sections of p85α−/− p55α−/− p50α−/− p85β−/− and control embryos at E8 to 11.5 were analyzed to elucidate defects of the mutant mice on the cellular level. The sections of the mutant mice at E8.5 showed detachment of the outer epithelial cell layer (Fig. 1E). Sections between E9.5 and 11.5 revealed wavy neural tubes, disturbed somites as consequences of epidermal blebbing, increased mesenchyme, and hemorrhaging to various degrees (data not shown).
TABLE 1.
p85 is required for proper embryonic development
| Stage | Viability of intercross offspringa
|
|||||
|---|---|---|---|---|---|---|
| α−/− β−/−
|
α+/− β−/−
|
α+/+ β−/−
|
||||
| No. | Phenotype | No. | Phenotype | No. | Phenotype | |
| E8-9 | 9 | All embryos have subepidermal blebs flanking neural tube. Some of the blebs are filled with blood. | 6 | Normal | 5 | Normal |
| E9.5-11.5 | 12 (of those, 2 dead) | All embryos have subepidermal blebs, mostly flanking neural tube and some on branchial arches or head. Some of these structures are filled with blood. Some embryos also exhibit a wavy neural tube. | 27 | Normal | 15 | Normal |
| E11.5-12.5 | 14 (of those, 10 dead) | Alive embryos have severe facial abnormalities, such as clefted face, blebbing, and hemorrhaging. A few embryos exhibit blebs on the trunk and a wavy neural tube. | 33 | Normal | 15 | Normal |
| >E12.5 | 10 (of those, 9 dead) | Alive embryo is developmentally delayed, with a small bloody dot in the trunk. | 30 | Normal | 12 | Normal |
Shown is the viability of offspring from p85α+/− p55α+/− p50α+/− p85β−/− intercrosses. Alive embryos deficient in p85α−/− p55α−/− p50α−/− p85β−/− were detected in the expected Mendelian ratios until E11.5. Ten mutant embryos were found after E12.5. However, nine of those were already dead or resorbing. The phenotypes of p85α−/− p55α−/− p50α−/− p85β−/− embryos are also shown. The defects of the mutant embryos included subepidermal blebbing, hemorrhaging, and waviness of the neural tube. Facial abnormalities occurred after the appearance of blebs in the trunk.
FIG. 1.
Deficiency in class Ia PI3K causes subepidermal blebbing. (A) Subepidermal blebbing in unturned E8 p85α−/− p55α−/− p50α−/− p85β−/− embryo. Shown are one unturned control littermate embryo (left) and one unturned p85α−/− p55α−/− p50α−/− p85β−/− embryo (right) with a subepidermal bleb on the trunk. (B and C) Blood-filled bleb in turned E9.5 p85α−/− p55α−/− p50α−/− p85β−/− embryos. Shown are one control littermate embryo (left) and two p85α−/− p85β−/− embryos (middle and right). Both embryos lacking p85α gene products and p85β display blood-filled blebs flanking their neural tube. (D) Facial abnormalities in E11.5 p85α−/− p55α−/− p50α−/− p85β−/− embryo. Shown are one control littermate embryo (left) and a p85α−/− p55α−/− p50α−/− p85β−/− embryo (right) which has a non-blood-filled subepidermal structure on the head. (E) Blood-filled, subepidermal bleb in turned E9 p85α−/− p55α−/− p50α−/− p85β−/− embryo. Shown are one control littermate embryo (left) and one p85α−/− p55α−/− p50α−/− p85β−/− embryo (right). Embryos were dissected at E9 and embedded in paraffin, and sagittal sections were stained with H&E to study their cell sructures.
Partial redundancy of the smaller PI3K regulatory isoforms, p55α and p50α, in embryonic development.
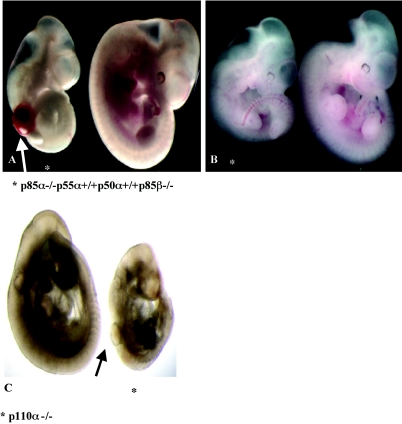
Next, we investigated whether intact expression of the smaller PI3K regulatory isoforms p55α and p50α (which do not contain the N-terminal domains of 85-kDa isoforms) can rescue the defects seen in p85α−/− p55α−/− p50α−/− p85β−/− embryos. To generate mice that lack both 85-kDa isoforms but express the p55α and p50α isoforms, p85α−/− mice—which have the first exon of p85α deleted but retain exons that allow expression of the p55α and p50α isoform (37)—were crossed with p85β−/− mice. Viable p85α−/− p55α+/+ p50α+/+ p85β−/− embryos were detected until E12.5, but similarly to p85α−/− p55α−/− p50α−/− p85β−/− mice, p85α−/− p55α+/+ p50α+/+ p85β−/− mice exhibited blood-filled blebs flanking their neural tube, hemorrhaging, wavy neural tubes, and facial abnormalities (Fig. 2A and B) and the embryos were delayed in their development (Fig. 2A). However, the phenotype of p85α−/− p55α+/+ p50α+/+ p85β−/− embryos was less severe (fewer embryos exhibited blebs) than that of p85α−/− p55α−/− p50α−/− p85β−/− embryos, and p85α−/− p55α+/+ p50α+/+ p85β−/− mice survived until E13. These data demonstrate that intact expression of p55α and p50α together can only partially compensate for loss of PI3K 85-kDa isoforms during embryonic development.
FIG. 2.
Partial rescue in p85α−/− p55α+/+ p50α+/+ p85β−/− embryos. (A) Blood-filled bleb in turned E10.5 p85α−/− p55α+/+ p50α+/+ p85β−/− embryo. Shown are one control littermate embryo (right) and one p85α−/− p55α+/+ p50α+/+ p85β−/− embryo (left). Embryos lacking both 85-kDa isoforms (p85α and p85β) display a blood-filled bleb flanking their neural tube. The mutant embryo is smaller than the control littermate. (B) Minor defects in turned E10.5 p85α−/− p55α+/+ p50α+/+ p85β−/− embryo. Shown are one control littermate embryo (right) and one p85α−/− p55α+/+ p50α+/+ p85β−/− embryo (left). The embryo lacking both 85-kDa isoforms (p85α and p85β) is smaller than the control littermate but exhibits no blebs. (C) Subepidermal bleb in turned E10.5 p110α−/− embryo. Shown are one control littermate embryo (left) and one p110α−/− embryo (right). The embryo lacking p110α displays a subepidermal bleb flanking the neural tube. The mutant embryo is smaller than the control littermate.
Developmental abnormalities in p110α−/− mice are similar to those due to combined loss of p85α and p85β.
Next, we compared the phenotype of p110α−/− mice to that of mice lacking p85α (with or without intact expression of p55α and p50α) and p85β. In contrast to p110β null mice, which die at <E3.5 (3), p110α null mice survive until E10.5 (4) and therefore overlap temporally with the time frame of the phenotype observed in p85α−/− p55α−/− p50α−/− p85β−/− mice. We intercrossed p110α+/− mice and analyzed timed pregnancies up to E10.5. Extensive hemorrhaging and proliferative defects have been described previously for mice with targeted disruption of p110α. Interestingly, some of these mutant embryos have subepidermal blebs (frequently filled with blood) in the same position as that observed in the p85α−/− p55α−/− p50α−/− p85β−/− mice (Fig. 2C). The frequency of this and additional defects are described in detail in the publication by Bi et al. (4). Thus, p110α and either p85α or p85β appear to be essential for PDGFRα-dependent developmental events between E8 and E11.
MEFs lacking p85α, p55α, p50α, and p85β fail to proliferate unless immortalized with SV40.
Mice deficient in all gene products of p85α and p85β (p85α−/− p55α−/− p50α−/− p85β−/−) die during early embryonic development (described above). Therefore, the role of PI3K isoforms was studied in ex vivo experiments. MEFs from p85α−/− p55α−/− p50α−/− p85β−/− embryos (E8.5) and littermate control p85α+/− p55α+/− p50α+/− p85β−/− embryos were generated. While the control primary cells could be expanded, the p85α−/− p55α−/− p50α−/− p85β−/− primary cells stopped proliferating after about five passages and adopted a senescence-like morphology. In order to establish cell lines, the primary pools were immortalized by retroviral expression of the SV40 large T antigen. Three independent cell pools derived from p85α−/− p55α−/− p50α−/− p85β−/− embryos (named M1, M2, and M3) and one control cell pool derived from a p85α+/− p55α+/− p50α+/− p85β−/− littermate embryo (named C) were established. Western blot analysis of normalized total cell lysates showed complete loss of p85α and p85β in the mutant cells as expected (Fig. 3A, upper panel). The smaller isoforms, p55α and p50α, were not detected in either control or mutant fibroblasts in general. Surprisingly, p55γ, which is primarily expressed in brain (31), was substantially upregulated in the mutant cell pools (Fig. 3A, lower panel).
FIG. 3.
Loss of all p85α and p85β gene products is associated with increased expression of p55γ but reduced overall PI3K activity. (A) p85 protein levels. HA-p55γ was transfected into a mutant cell pool (M1). The next day, growing MEFs of the indicated genotypes were lysed and equal amounts of protein were subjected to SDS-PAGE and immunoblotted with anti-p85pan (upper panel) or anti-p55γ antisera (lower panel). Panel A shows one out of three independent experiments with similar results. (B) p85-associated PI3K activity. Exponentially growing MEFs of the indicated genotypes were lysed, and equal amounts of protein were immunoprecipitated with anti-p85pan antibody. The immunoprecipitates were subjected to in vitro PI 3 -kinase assay with phosphoinositol (PI) as a substrate. The left panel in panel B shows a representative radiograph with one mutant cell pool (M2) and one control cell pool (C). A duplicate set was pretreated with 100 nM wortmannin (WM) for 20 min to inhibit the reaction. The right panel in panel B shows a quantification of three independent experiments described in the left panel, using three different mutant cell pools (M) and one control cell pool (C). Results are means ± standard error of the relative PI3K activity. (C) p110α protein levels. Exponentially growing MEFs of the indicated genotypes were lysed, and equal amounts of protein were immunoprecipitated with an antibody specific for p110α. The immunoprecipitates were subjected to SDS-PAGE and probed with anti-p110α antibody. Equal loading was verified by detecting similar amounts of Erk on the same membrane(data not shown). Panel 3C shows a representative immunoblot with one control pool (C) and two mutant pools (M1 and M3). (D) p110α-associated PI3K activity. Exponentially growing MEFs of the indicated genotypes were lysed, and equal amounts of protein were immunoprecipitated with anti-p110α antibody. The immunoprecipitates were subjected to in vitro PI3K assay with PI as a substrate. The left panel in panel D shows a representative radiograph on one control pool (C) and one mutant pool (M2). The right panel in panel D shows a quantification of three independent experiments described in the left panel, using three mutant pools (M) and one control pool (C). Results are means ± standard error of the relative PI3K activity.
Further assessment of class Ia PI3K in the cells was made by measuring PI3K activity in anti-p85 immunoprecipitates from control cells and the three mutant cell lines M1, M2, and M3. The antibody used for this immunoprecipitation (anti-p85pan) was raised against the N-terminal SH2 domain of p85α, which is retained in p55α and p50α and highly conserved in p85β and p55γ, allowing for cross-recognition of all isoforms by the antiserum. The amount of PI3K activity in the immunoprecipitates varied between the three mutant pools and was about 5 to 20% of the activity observed in the control cell pool (Fig. 3B). This residual activity is apparently due to the up-regulated p55γ in these cells (Fig. 3A). The Western blot does not convincingly reveal p55γ protein in the M2 cells, consistent with the very low PI3K activity (anti-85pan- and anti-p110-associated PI3K activities [see below] are much lower in the M2 than in M1 and M3 cell pools). Therefore, M2 cells have much less p55γ protein than M1 and M3 cells.
As described previously, p110α stability is enhanced by its association with the regulatory subunit (48). Therefore, p110α protein levels and kinase activity were assessed on immunoprecipitates of mutant and control cell pools with anti-p110α antibodies. The immunoprecipitates were then either subjected to SDS-PAGE and Western blot analysis with an anti-p110α antibody, or the immunoprecipitates were subjected to in vitro PI3K assays. Consistent with greatly reduced p85 levels, the protein level of p110α was significantly decreased as well (Fig. 3C). The p110α activity was diminished to about 10 to 40% in the three different mutant cell pools compared to that in control cells as judged by in vitro PI3K assays (Fig. 3D).
p85α−/− p55α−/− p50α−/− p85β−/− MEFs show defective PI3K activation upon stimulation with PDGF.
In order to assess the contribution of the residual class Ia PI3K (p55γ and associated p110 isoforms) in PDGF-stimulated PI3K activity, the amount of PI3K activity recruited to Tyr-phosphorylated proteins after PDGF-BB treatment was determined. Subconfluent serum-starved MEFs were stimulated for 5 min with 10 ng of PDGF-BB per ml with or without wortmannin (100 nM) pretreatment. The cells were lysed, immunoprecipitated with antiphosphotyrosine antibody, and then subjected to in vitro PI3K assays. Only about 10 to 15% of the PI3K activity recruited in control cells was detected in the mutant cells, and as expected, this activity could be completely inhibited with wortmannin pretreatment (Fig. 4A). We also analyzed the generation of class Ia PI3K lipid products in intact cells after PDGF-BB treatment. Consistent with the in vitro data, PDGF-BB stimulated production of both PI-3,4-P2 and PI-3,4,5-P3 in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs but the increases in these lipids were only 20 and 30%, respectively, of the increases observed in control cells (Fig. 4B). As expected, pretreatment with 100 nM wortmannin completely blocked production of both lipids.
FIG. 4.
Loss of all p85α and p85β gene products results in greatly reduced PDGF-stimulated PI3K activity in vitro and in vivo. (A) In vitro generation of PI3K products. Growing MEFs of the indicated genotypes were starved and then stimulated for 5 min with 10 ng of PDGF per ml (with or without pretreatment with 100 nM WM for 20 min). Equal amounts of protein were immunoprecipitated with antiphosphotyrosine (anti-PY) (4G10), and then an in vitro PI3K assay was performed. The left panel shows a representative experiment with one control clone (C) and one mutant clone (M1). The right panel shows the fold increase in PDGF-stimulated PI3K activity of three mutant pools in comparison to one control pool (result of three independent experiments). Results are means ± standard error of the relative PI3K activity. (B) In vivo generation of PI3K products. Growing MEFs of the indicated genotypes were starved, metabolically labeled with 32PO4, and then stimulated for 5 min with 10 ng of PDGF per ml (with or without pretreatment with 100 nM wortmannin for 20 min). Lipids were extracted and analyzed by HPLC. The panel shows one representative experiment out of two independent experiments with similar results.
p85α−/− p55α−/− p50α−/− p85β−/− MEFS show defective PDGF-dependent phosphorylation of Akt and MAPK.
Phosphorylation of Akt on residue Ser 473 reflects Akt activity and has been shown to correlate with PI3K activation. Consistent with the decreased PDGF-stimulated PI-3,4-P2 and PI-3,4,5-P3 production in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs, there was also less PDGF-dependent phosphorylation of Akt on serine 473 when either subsaturating (1 ng/ml) or saturating (10 ng/ml) concentrations of PDGF-BB were used. Also the duration of Akt phosphorylation was reduced in p85α−/− p55α−/− p50α−/− p85β−/− MEFs (Fig. 5A). IGF-1- and EGF-dependent Akt activations were also reduced in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs (Fig. 5B).
FIG. 5.
Defective signaling in p85α−/− p55α−/− p50α−/− p85β−/− MEFs. (A and B) p85 is required for activation of Akt. MEFs with the indicated genotypes were starved overnight and then stimulated with 1 to 10 ng of PDGF per ml for 7 to 60 min (A) or saturating amounts of PDGF (10 ng/ml), IGF-1 (20 nM), or EGF (100 ng/ml) for 5 min (B). Lysates were resolved by SDS-PAGE and immunoblotted with anti-Akt and anti-phospho-Akt antibodies. Blots are representative of at least three independent experiments for each condition. (C) p85 is required for PDGF-induced activation of Erk. MEFs with indicated genotypes were starved overnight and then stimulated with 1, 3, or 10 ng of PDGF per ml for 5 min. Lysates were resolved by SDS-PAGE and immunoblotted with anti-Erk and anti-phospho-Erk antibodies. Note that the antibody that recognizes total Erk preferentially blots the nonphosphorylated Erk, so increased phosphorylation correlates with decreased blotting of total Erk. A second loading control for this experiment was added, a blot for total Akt. Blots are representative of at least three independent experiments. (D) Normal PDGF-dependent tyrosine phosphorylation of the PDGF receptor in mutant cell pools. MEFs with indicated genotypes were starved overnight and then stimulated with 10 or 20 ng of PDGF per ml for the indicated times. Lysates were resolved by SDS-PAGE and immunoblotted with anti-PDGF receptor and anti-Akt antibodies. The activation of these proteins wase assessed by antiphosphotyrosine (anti-PY) or phospho-specific antibodies. The blots are representative of three independent experiments.
We also investigated PDGF-dependent Erk activation in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs. We found that Erk phosphorylation was impaired in the mutant cells when a low level of PDGF (1 ng/ml) was used (Fig. 5C). However, saturating amounts of PDGF-BB (3 to 10 ng/ml) led to normal Erk1/2 phosphorylation (Fig. 5C). PDGF-dependent phosphorylation of the PDGFR on tyrosine residues was not significantly different between the control and mutant cell lines (Fig. 5D).
MEFs lacking all isoforms of p85α and p85β exhibit defects in PDGF-dependent membrane ruffling.
PI3K has been implicated in PDGF-dependent membrane ruffling and lamellipodium formation based on studies with receptor mutations, dominant-negative forms of PI3K, and drugs (8, 24, 34, 45, 46). Consistent with these results, we found that while PDGF stimulated ruffling and lamellipodium formation in control MEFs, it failed to do so in the p85α−/− p55α−/− p50α−/− p85α−/− MEFs (Fig. 6A).
FIG. 6.
P85 is required for PDGF-induced actin rearrangements. (A) P85 is required for PDGF-induced lamellipodium formation. MEFs with indicated genotypes were starved and then stimulated with 50 ng of PDGF per ml for 10 min (with or without pretreatment with 100 nM wortmannin [WM] for 20 min). The cells were fixed and actin structures were stained with rhodamine-phalloidin. The panels are representative of at least three independent experiments. (B) p85 is required for PDGF-induced activation of Rac. MEFs with the indicated genotypes were transfected with HA-Rac. The cells were starved overnight and then stimulated for 3 min with 30 ng of PDGF per ml. Equal amounts of lysates were subjected to in vitro binding assays with the immobilized CRIB domain of PAK65. Bound proteins were resolved by SDS-PAGE and immunoblotted with anti-Rac antibodies (Rac GTP). The levels of total Rac (Rac) and p85 are shown below. Equal stimulation was assessed by Erk activation. Blots are representative of at least three independent experiments. While 65% of the control MEFs exhibited PDGF-induced ruffling, 0% of the mutant MEFs ruffled upon PDGF treatment. (C) Wild-type Vav2 overexpression leads to membrane ruffling in the absensce of PI3K signaling. Control and mutant MEFs were transfected with T7-tagged wild-type Vav2. The transfected cells were grown for 24 h. Then the cells were fixed (with or without pretreatment with 100 nM wortmannin for 30 min), and the actin structures were stained with rhodamine-phalloidin. Vav2-expressing cells were identified with anti-T7 antibody staining. About 90% of either control or mutant MEFs (with or without wortmannin) that were overexpressing Vav2 exhibited lamellipodium formation. (D) Retroviral restoration of p85 isoforms. Mutant (M1) cell pools were infected with retroviral pMIG constructs to restore expression of p85α, p85β, or p50α similarly to the control (C) cell pool. The infected cells were lysed, and the protein levels were examined by Western blot analysis of the lysates, or, alternatively, p85/p110 complexes were immunoprecipitated with the anti-p85pan antibody and then examined by Western blot analysis. Blots are representative of at least three independent experiments. (E) The Rho-GAP domain of p85 is not necessary for PDGF-induced membrane ruffling. Various p85 isoforms mediate PDGF- or IGF-1-induced membrane ruffling. Mutant (p85α−/− p55α−/− p50α−/− p85β−/−) cell pools were infected with retroviral pMIG constructs to restore expression of p85α, p85β, or p50α similarly to the control (p85α+/− p55α+/− p50α+/− p85β−/−) cell pool. Mutant and control cells were plated on coverslips, starved overnight, and then stimulated with either 50 ng of PDGF-BB per ml or 20 nM IGF-1. Pictures were taken every 15 s for 2 h. The number of cells exhibiting circular ruffles were counted and expressed as a percentage of total cells. The data represent the means of at least three independent experiments ± standard error.
It has been shown that Rac is a crucial mediator of PDGF-BB-induced ruffling and lamellipodium formation downstream of PI3K (16). Thus, we investigated Rac activation in the control and mutant cells. We assessed the activation state of Rac by using the CRIB domain of PAK65 to pull down GTP-Rac from serum-starved or PDGF-stimulated cells. PDGF-dependent activation of Rac was observed in the control MEFs but was impaired in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs (Fig. 6B). As an internal control, PDGF-dependent Erk phosphorylation was similar in the two cell types, as expected for saturating levels of PDGF.
We also investigated the possibility that restoring Rac activation by overexpressing the Rac-GEF Vav2 would restore lamellipodium formation in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs. The mutant cell pool was transfected with Vav2. The cells were fixed and the actin structures were stained with rhodamine phalloidin. Interestingly, overexpression of Vav2 circumvented the block in ruffling in p85α−/− p55α−/− p50α−/− p85α−/− MEFs and caused lamellipodium formation even in the presence of 100 nM wortmannin (Fig. 6C). These findings indicate that the impaired Rac activation is the major cause of the ruffling defects in p85α−/− p55α−/− p50α−/− p85α−/− MEFs and that overexpression of Vav2 circumvents the requirement of PI3K for membrane ruffling.
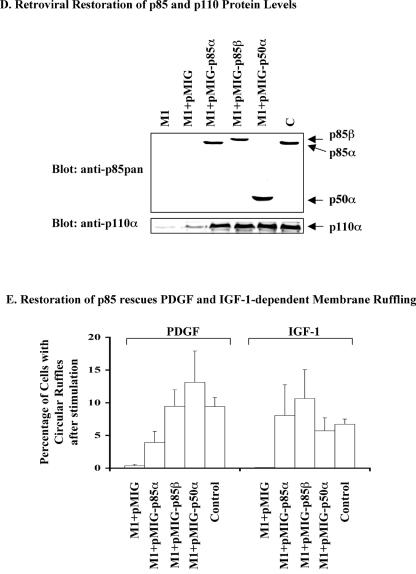
Previous studies using general PI3K inhibitors, dominant-negative p85, or receptors lacking the p85 binding sites did not address whether specific PI3K isoforms are required for cellular responses. The failure of the p85α−/− p55α−/− p50α−/− p85β−/− MEFs to ruffle in response to PDGF, despite a significant but reduced PDGF-dependent stimulation of PI-3,4-P2 and PI-3,4,5-P3 production and activation of Akt, could be explained by a failure to reach the signal threshold for actin rearrangement. Alternatively, the remaining regulatory isoform (p55γ) might lack critical domains (SH3, Pro-rich or Rho/Rac/CDC42-interacting domains) needed for this response. To assess the capability of the various p85 isoforms to mediate PDGF-induced ruffling, the expression of p85α, p85β, or p50α was restored in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs. To do so, the mutant cell pool was infected with retroviral constructs for p85α, p85β, or p50α. The infected cells were sorted by FACS analysis, since the retrovirus introduced an additional GFP that could be used as a marker for infection.
The infected cell pools were first analyzed for expression of p85 and p110 isoforms by Western blot analysis. The levels of expression of the p85α, p85β, and p50α proteins following retroviral infection and sorting of infected cells were in a range similar to the expression of p85α in the control cells based on blotting with the anti-p85pan antibody (Fig. 6D, upper panel). (This antibody detects p85α somewhat better than p85β, implying that p85β expression is slightly higher than the other isoforms.) In agreement with results in Fig. 3C, p110α levels are reduced in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs compared to control MEFs and expression of each of the regulatory subunit isoforms restored the p110α protein to a level comparable to that observed in the control MEFs (Fig. 6D, lower panel).
Next, the formation of circular ruffles in response to PDGF or IGF-1 was assessed by time-lapse microscopy of live cells. Retroviral reintroduction of p85α or p85β into p85α−/− p55α−/− p50α−/− p85β−/− MEFs rescued the ruffling defect (Fig. 6E). This finding confirms that the impaired membrane ruffling in p85α−/− p55α−/− p50α−/− p85β−/− MEFs is due to absence of these genes and not another undefined defect in the cell lines. Surprisingly, the expression of the smaller p85α isoform, p50α, which lacks the SH3 domain, Pro-rich regions, and Rho/Rac/Cdc42 binding domain, was also sufficient to rescue both PDGF- and IGF-1-dependent membrane ruffling (Fig. 6E). These results indicate that the failure of the p85α−/− p55α−/− p50α−/− p85β−/− MEFs to ruffle is due to a failure to reach a threshold of PI3K signaling rather than absence of the SH3 domain, Pro-rich regions, and Rho/Rac/Cdc42 binding domain in the p55γ regulatory subunit.
DISCUSSION
We report here that p85α−/− p55α−/− p50α−/− p85β−/− mice have an embryonic lethal phenotype and exhibit defects similar to those seen in PDGFRα-deficient mice and p110α−/− mice. We also show that MEFs derived from p85α−/− p55α−/− p50α−/− p85β−/− mice have impaired PDGF-dependent signaling, including a defect in Rac activation and in membrane ruffling. Reintroduction of p85α or p85β restored the PDGF-dependent ruffling. Surprisingly, the p50α isoform, which lacks the SH3 domain, Pro-rich regions, and Rho/Rac/CDC42 binding domain common to p85α and p85β, also restored PDGF-dependent ruffling. Expression of the Rac-GEF Vav2 in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs also restored membrane ruffling. These findings suggest that PDGF-induced membrane ruffling is dependent on the ability of class Ia PI3K to promote GTP exchange on Rac (by activating Rac-specific GEFs), but does not require the direct interaction of Rho/Rac/CDC42 members with the Rho-GAP homology domain of p85.
Previous studies have shown that targeted disruption of genes for PDGF or PDGFR family members cause early embryonic death, accompanied by severe developmental abnormalities (6, 25, 26, 35, 36). In vitro studies with PI3K inhibitors, PDGFR mutants, and dominant-negative forms of class Ia PI3K have suggested that PI3K mediates PDGF-induced cellular events, such as proliferation, cell survival, and cell migration. Since PI3K is thought to be a major target of the PDGFR, it was surprising that mice with tyrosine-to-phenylalanine mutations in the PI3K binding sites of the PDGFRβ were viable and had only minor defects in their capillary pressure (17). However, interpretation of these results is complicated by the fact that multiple adapter proteins can potentially mediate PI3K activation independent of direct binding to receptors. We show here that mice deficient in p85α p55α p50α p85β have very similar developmental defects to mice deficient in PDGFRα (36). This finding points to a major role of class Ia PI3K downstream of the PDGFRα in vivo. Mice deficient in p110α also exhibit similar defects, as do mice lacking p85α and p85β but capable of expressing the p55α and p50α alternative splice forms of p85α. These results indicate that during embryonic development stages E8 to E12, the PDGFRα requires full-length p85 isoforms (either p85α or p85β) and requires p110α. The shorter isoforms of p85 (p50α, p55α, and p55γ) cannot fully substitute for p85 isoforms during development, either because of inappropriate expression or lack of essential domains. Also, p110β and p110δ cannot substitute for p110α during development. Since PDGFRα−/− mice survive longer than mice lacking p85α p55α p50α p85β, during development, PI3K must be an important mediator of additional receptors other than the PDGFRα.
The p110 catalytic subunits are thermally unstable when not bound to a p85 regulatory subunit (48), and the finding of reduced p110α expression in double-mutant embryos is consistent with our previous observation that both p110α and p110β protein levels are decreased in tissues from p85α−/− mice (13). In light of the previous observation that p110β−/− embryos die before E3.5, we were initially surprised that p85α−/− p55α−/− p50α−/− p85β−/− embryos survive until E12.5. Consistent with the importance of regulatory subunits for stability of p110 subunits, we found that p110α levels and total PI3K activity are decreased dramatically in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs and that the level of p110α is restored when p85 isoforms are added back. However, there was a significant amount of p110α protein and class Ia PI3K activity remaining in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs, and this could be explained by the up-regulation of p55γ that we observe in these MEFs compared to control MEFs. An up-regulation of p55γ during development of the p85α−/− p55α−/− p50α−/− p85β−/− embryos might provide sufficient stability of p110β to avoid early lethality, and this would also explain why the p85α−/− p55α−/− p50α−/− p85β−/− MEFs have elevated levels of p55γ. Unfortunately available antibodies against p55γ and p110β are not sufficiently sensitive to detect these proteins in either wild-type or p85α−/− p55α−/− p50α−/− p85β−/− E12 embryos by in situ hybridization.
MEFs derived from mutant p85α−/− p55α−/− p50α−/− p85β−/− embryos proliferated in culture for no more than five passages before they adopted a typical senescent morphology. This finding is in agreement with reports that PI3K inhibitors can induce a senescence-like phenotype in wild-type MEFs that is associated with up-regulation of p27 (Kip1) (10). By retroviral introduction of the SV40 large T antigen, we could rescue the mutant MEFs and establish immortalized, stable cell lines. As indicated above, the SV40-immortalized p85α−/− p55α−/− p50α−/− p85β−/− MEFs have elevated p55γ compared to SV40-immortalized control MEFs. Whether the elevation of p55γ preexisted in the MEFs from these embryos or was selected for during immortalization is not clear, but it is likely that this increase is important for the growth and survival of the MEFs. Interestingly, there is recent evidence that p55γ can interact with the retinoblastoma tumor suppressor protein (Rb) via an N-terminal sequence common to the p55γ and p55α proteins but absent in other isoforms (47). Rb is a cellular target of the SV40 large T antigen and a crucial regulator of the G1/S transition in mammalian cells (for review, see reference 15).
In vitro kinase assays on antiphosphotyrosine immunoprecipitates after PDGF stimulation, along with in vivo 32Pi labeling, showed substantially reduced, albeit still detectable PI3K activity in the mutant cell pools in comparison to control cell pools in vitro and in vivo. This activity could be completely inhibited by treating cells with wortmannin. Though equally sensitive to wortmannin as class Ia PI3K, class II PI3K C2β can utilize PI and PI4P but not PI-4,5-P2 (1). Therefore we conclude, that in the mutant cell pools the PDGF-BB-induced PIP3 production is most likely attributed to p55γ/p110, whereas the PDGF-BB-induced PI-3,4-P2 generation might be attributed to p55γ/p110 as well as class II PI3K C2β.
Short-term and long-term stimulation with subsaturating as well as saturating doses of PDGF-BB both resulted in diminished Akt phosphorylation in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs compared to that in control MEFs. Reintroduction of p85α, p85β, or p50α restored this defect (data not shown). It was surprising, however, that despite the substantial loss of PI3K isoforms, the reduction in Akt phosphorylation was relatively mild when saturating doses of PDGF were added. This might be explained by the amplification of signal at the level of PI3K due to the relative large amount of PI-3,4,5-P3 produced by each PI3K molecule.
In contrast to Akt, Erk phosphorylation is not PI3K dependent at high doses of PDGF-BB. However, at lower doses of PDGF-BB (such as 1 ng/ml), Erk phosphorylation is also diminished in mutant cells compared to that in control cells. This finding is in agreement with previous results that the PI3K dependency of Erk activation inversely correlates with the strength of stimulus (11).
PDGF-BB-stimulated GTP loading of Rac is greatly diminished in p85α−/− p55α−/− p50α−/− p85β−/− cells. Since Rac is a crucial mediator of PDGF-induced membrane ruffling, it was not surprising that the mutant cell pools showed impaired PDGF-dependent ruffling. The p85α−/− p55α−/− p50α−/− p85β−/− MEFs showed a total absence of actin ruffles upon PDGF-BB treatment. These findings suggest that class Ia PI3K is a necessary mediator of PDGF-BB-induced membrane ruffling. The p85α−/− p55α−/− p50α−/− p85β−/− fibroblasts also fail to ruffle upon IGF-1 stimulation. This finding is in agreement with reports that IGF-1-induced ruffling is completely blocked by wortmannin treatment or overexpression of dominant-negative p85 (23). Facial abnormalities, such as those seen in the p85α−/− p55α−/− p50α−/− p85β−/− mice and PDGFRα−/− mice, are often attributed to the failure of neural crest cells to migrate from the somites to the facial regions. Therefore, the defect in PDGF-dependent actin remodeling seen in the p85α−/− p55α−/− p50α−/− p85β−/− MEFs could explain a defect in cell migration. It is possible that the smaller isoforms p55α and p50α might be able to mediate PDGF-induced actin rearrangements in vitro but cannot fulfill their role in vivo since they might not be expressed in the crucial cell types. Alternatively, PI3K is implicated in many cellular responses (cell proliferation, cell growth, and cell survival), and it is likely that the SH3, proline-rich regions, and/or Rho-GAP domain of p85 contributes to one or more of these events.
In order to test the hypothesis that impaired Rac activation is the cause of the inability of the p85α−/− p55α−/− p50α−/− p85β−/− cell pools to ruffle, Rac activation was driven by overexpressing the Rac-specific GEF Vav2. Vav2 overexpression in the mutant cells rescued the ruffling defect even in the presence of wortmannin. Vav2 overexpression in NIH 3T3 fibroblasts has been shown to result in Rac activation, as judged by increased Rac-GTP levels and lamellipodium formation (29). These data show that PI3K is not necessary for Vav2-induced lamellipodium formation.
There is evidence that PI3K can function downstream of Rac (18, 22, 32). The GTP-bound form of Rac can interact with the Rho-GAP domain of p85 (5, 39, 49). Since the p85 Rho-GAP domain lacks GAP activity, this interaction might play a role in recruitment of PI3K to regions of membranes where Rac or Cdc42 is activated (5, 39, 49). By acting both upstream and downstream of Rac, class Ia PI3K could be in a positive feedback loop to concentrate Rac signaling (and actin rearrangement) at specific locations in the cell membrane (43). However, our results show clearly that the small p85α isoform p50α is able to mediate PDGF- or IGF-1-induced membrane ruffling. Therefore, the interaction between Rac-GTP or Cdc42-GTP with the Rho-GAP domain of p85α/β is not essential for PDGF- or IGF-1-induced membrane ruffling in these cells under our experimental conditions.
Acknowledgments
S.M.B was supported by a scholarship from Boehringer Ingelheim Fonds. D.A.F was supported in part by fellowships from the Damon Runyon Cancer Research Foundation and the Leukemia and Lymphoma Society. S.M.T. was supported by the Leukemia and Lymphoma Society. This work was supported by NIH grants GM41890 and PO1-CA089021 to L.C.C, AI50831 to D.A.F, and CA75621 to S.M.T.
We thank Morris White for sharing the anti-p55γ antisera and Benjamin Neel for anti-PDGFR antibody. We also thank John Watt, Monica Kosmatka, Nicole Logsdon, and Nikki Madson for maintaining the mouse colonies.
REFERENCES
- 1.Arcaro, A., S. Volinia, M. J. Zvelebil, R. Stein, S. J. Watton, M. J. Layton, I. Gout, K. Ahmadi, J. Downward, and M. D. Waterfield. 1998. Human phosphoinositide 3-kinase C2beta, the role of calcium and the C2 domain in enzyme activity. J. Biol. Chem. 273:33082-33090. [DOI] [PubMed] [Google Scholar]
- 2.Bazenet, C. E., and A. Kazlauskas. 1994. The PDGF receptor alpha subunit activates p21ras and triggers DNA synthesis without interacting with rasGAP. Oncogene 9:517-525. [PubMed] [Google Scholar]
- 3.Bi, L., I. Okabe, D. J. Bernard, and R. L. Nussbaum. 2002. Early embryonic lethality in mice deficient in the p110beta catalytic subunit of PI 3-kinase. Mamm. Genome 13:169-172. [DOI] [PubMed] [Google Scholar]
- 4.Bi, L., I. Okabe, D. J. Bernard, A. Wynshaw-Boris, and R. L. Nussbaum. 1999. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J. Biol. Chem. 274:10963-10968. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch, G. M., C. J. Vlahos, Y. Wang, U. G. Knaus, and A. E. Traynor-Kaplan. 1996. Rac GTPase interacts specifically with phosphatidylinositol 3-kinase. Biochem. J. 315:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostrom, H., K. Willetts, M. Pekny, P. Leveen, P. Lindahl, H. Hedstrand, M. Pekna, M. Hellstrom, S. Gebre-Medhin, M. Schalling, M. Nilsson, S. Kurland, J. Tornell, J. K. Heath, and C. Betsholtz. 1996. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85:863-873. [DOI] [PubMed] [Google Scholar]
- 7.Cantley, L. C. 2002. The phosphoinositide 3-kinase pathway. Science 296:1655-1657. [DOI] [PubMed] [Google Scholar]
- 8.Cantley, L. C., K. R. Auger, C. Carpenter, B. Duckworth, A. Graziani, R. Kapeller, and S. Soltoff. 1991. Oncogenes and signal transduction. Cell 64:281-302. [DOI] [PubMed] [Google Scholar]
- 9.Cantley, L. C., and B. G. Neel. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 96:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado, M., R. H. Medema, I. Garcia-Cao, M. L. Dubuisson, M. Barradas, J. Glassford, C. Rivas, B. M. Burgering, M. Serrano, and E. W. Lam. 2000. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J. Biol. Chem. 275:21960-21968. [DOI] [PubMed] [Google Scholar]
- 11.Duckworth, B. C., and L. C. Cantley. 1997. Conditional inhibition of the mitogen-activated protein kinase cascade by wortmannin. Dependence on signal strength. J. Biol. Chem. 272:27665-27670. [DOI] [PubMed] [Google Scholar]
- 12.Franke, T. F., C. P. Hornik, L. Segev, G. A. Shostak, and C. Sugimoto. 2003. PI3K/Akt and apoptosis: size matters. Oncogene 22:8983-8998. [DOI] [PubMed] [Google Scholar]
- 13.Fruman, D. A., F. Mauvais-Jarvis, D. A. Pollard, C. M. Yballe, D. Brazil, R. T. Bronson, C. R. Kahn, and L. C. Cantley. 2000. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat. Genet. 26:379-382. [DOI] [PubMed] [Google Scholar]
- 14.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 15.Harbour, J. W., and D. C. Dean. 2000. Rb function in cell-cycle regulation and apoptosis. Nat. Cell Biol. 2:E65-E67. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins, P. T., A. Eguinoa, R. G. Qiu, D. Stokoe, F. T. Cooke, R. Walters, S. Wennstrom, L. Claesson-Welsh, T. Evans, M. Symons et al. 1995. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr. Biol. 5:393-403. [DOI] [PubMed] [Google Scholar]
- 17.Heuchel, R., A. Berg, M. Tallquist, K. Ahlen, R. K. Reed, K. Rubin, L. Claesson-Welsh, C. H. Heldin, and P. Soriano. 1999. Platelet-derived growth factor beta receptor regulates interstitial fluid homeostasis through phosphatidylinositol-3′ kinase signaling. Proc. Natl. Acad. Sci. USA 96:11410-11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inabe, K., M. Ishiai, A. M. Scharenberg, N. Freshney, J. Downward, and T. Kurosaki. 2002. Vav3 modulates B cell receptor responses by regulating phosphoinositide 3-kinase activation. J. Exp. Med. 195:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innocenti, M., E. Frittoli, I. Ponzanelli, J. R. Falck, S. M. Brachmann, P. P. Di Fiore, and G. Scita. 2003. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joly, M., A. Kazlauskas, F. S. Fay, and S. Corvera. 1994. Disruption of PDGF receptor trafficking by mutation of its PI-3 kinase binding sites. Science 263:684-687. [DOI] [PubMed] [Google Scholar]
- 21.Kazlauskas, A., and J. A. Cooper. 1990. Phosphorylation of the PDGF receptor beta subunit creates a tight binding site for phosphatidylinositol 3 kinase. EMBO J. 9:3279-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keely, P. J., J. K. Westwick, I. P. Whitehead, C. J. Der, and L. V. Parise. 1997. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature 390:632-636. [DOI] [PubMed] [Google Scholar]
- 23.Kotani, K., K. Yonezawa, K. Hara, H. Ueda, Y. Kitamura, H. Sakaue, A. Ando, A. Chavanieu, B. Calas, F. Grigorescu et al. 1994. Involvement of phosphoinositide 3-kinase in insulin- or IGF-1-induced membrane ruffling. EMBO J 13:2313-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundra, V., J. A. Escobedo, A. Kazlauskas, H. K. Kim, S. G. Rhee, L. T. Williams, and B. R. Zetter. 1994. Regulation of chemotaxis by the platelet-derived growth factor receptor-beta. Nature 367:474-476. [DOI] [PubMed] [Google Scholar]
- 25.Leveen, P., M. Pekny, S. Gebre-Medhin, B. Swolin, E. Larsson, and C. Betsholtz. 1994. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 8:1875-1887. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, P., M. Hellstrom, M. Kalen, L. Karlsson, M. Pekny, M. Pekna, P. Soriano, and C. Betsholtz. 1998. Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development 125:3313-3322. [DOI] [PubMed] [Google Scholar]
- 27.Luo, J., B. D. Manning, and L. C. Cantley. 2003. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4:257-262. [DOI] [PubMed] [Google Scholar]
- 28.Manser, E., T. Leung, H. Salihuddin, Z. S. Zhao, and L. Lim. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367:40-46. [DOI] [PubMed] [Google Scholar]
- 29.Marignani, P. A., and C. L. Carpenter. 2001. Vav2 is required for cell spreading. J. Cell Biol. 154:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura, R., W. Li, A. Kashishian, A. Mondino, M. Zhou, J. Cooper, and J. Schlessinger. 1993. Two signaling molecules share a phosphotyrosine-containing binding site in the platelet-derived growth factor receptor. Mol. Cell. Biol. 13:6889-6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pons, S., T. Asano, E. Glasheen, M. Miralpeix, Y. Zhang, T. L. Fisher, M. G. Myers, Jr., X. J. Sun, and M. F. White. 1995. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 15:4453-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachdev, P., L. Zeng, and L. H. Wang. 2002. Distinct role of phosphatidylinositol 3-kinase and Rho family GTPases in Vav3-induced cell transformation, cell motility, and morphological changes. J. Biol. Chem. 277:17638-17648. [DOI] [PubMed] [Google Scholar]
- 33.Samuels, Y., Z. Wang, A. Bardelli, N. Silliman, J. Ptak, S. Szabo, H. Yan, A. Gazdar, S. M. Powell, G. J. Riggins, J. K. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304:554. [DOI] [PubMed] [Google Scholar]
- 34.Severinsson, L., B. Ek, K. Mellstrom, L. Claesson-Welsh, and C. H. Heldin. 1990. Deletion of the kinase insert sequence of the platelet-derived growth factor β-receptor affects receptor kinase activity and signal transduction. Mol. Cell. Biol. 10:801-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano, P. 1994. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 8:1888-1896. [DOI] [PubMed] [Google Scholar]
- 36.Soriano, P. 1997. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124:2691-2700. [DOI] [PubMed] [Google Scholar]
- 37.Terauchi, Y., Y. Tsuji, S. Satoh, H. Minoura, K. Murakami, A. Okuno, K. Inukai, T. Asano, Y. Kaburagi, K. Ueki, H. Nakajima, T. Hanafusa, Y. Matsuzawa, H. Sekihara, Y. Yin, J. C. Barrett, H. Oda, T. Ishikawa, Y. Akanuma, I. Komuro, M. Suzuki, K. Yamamura, T. Kodama, H. Suzuki, T. Kadowaki, et al. 1999. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat. Genet. 21:230-235. [DOI] [PubMed] [Google Scholar]
- 38.Toker, A., and L. C. Cantley. 1997. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387:673-676. [DOI] [PubMed] [Google Scholar]
- 39.Tolias, K. F., L. C. Cantley, and C. L. Carpenter. 1995. Rho family GTPases bind to phosphoinositide kinases. J. Biol. Chem. 270:17656-17659. [DOI] [PubMed] [Google Scholar]
- 40.Ueki, K., C. M. Yballe, S. M. Brachmann, D. Vicent, J. M. Watt, C. R. Kahn, and L. C. Cantley. 2002. Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 99:419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valius, M., and A. Kazlauskas. 1993. Phospholipase C-gamma 1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor's mitogenic signal. Cell 73:321-334. [DOI] [PubMed] [Google Scholar]
- 42.Vivanco, I., and C. L. Sawyers. 2002. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2:489-501. [DOI] [PubMed] [Google Scholar]
- 43.Weiner, O. D., P. O. Neilsen, G. D. Prestwich, M. W. Kirschner, L. C. Cantley, and H. R. Bourne. 2002. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch, H. C., W. J. Coadwell, L. R. Stephens, and P. T. Hawkins. 2003. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 546:93-97. [DOI] [PubMed] [Google Scholar]
- 45.Wennstrom, S., P. Hawkins, F. Cooke, K. Hara, K. Yonezawa, M. Kasuga, T. Jackson, W. L. Claesson, and L. Stephens. 1994. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr. Biol. 4:385-393. [DOI] [PubMed] [Google Scholar]
- 46.Wennstrom, S., A. Siegbahn, K. Yokote, A. K. Arvidsson, C. H. Heldin, S. Mori, and W. L. Claesson. 1994. Membrane ruffling and chemotaxis transduced by the PDGF beta-receptor require the binding site for phosphatidylinositol 3′ kinase. Oncogene 9:651-660. [PubMed] [Google Scholar]
- 47.Xia, X., A. Cheng, D. Akinmade, and A. W. Hamburger. 2003. The N-terminal 24 amino acids of the p55 gamma regulatory subunit of phosphoinositide 3-kinase binds Rb and induces cell cycle arrest. Mol. Cell. Biol. 23:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, J., Y. Zhang, J. McIlroy, T. Rordorf-Nikolic, G. A. Orr, and J. M. Backer. 1998. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 18:1379-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, Y., S. Bagrodia, and R. A. Cerione. 1994. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J. Biol. Chem. 269:18727-18730. [PubMed] [Google Scholar]