Targeting the PRMT5/MTA complex selectively inhibits growth of MTAP-deleted preclinical cancer models and demonstrates preliminary clinical activity in MTAP-deleted cancers.
Abstract
Previous studies implicated protein arginine methyltransferase 5 (PRMT5) as a synthetic lethal target for MTAP-deleted (MTAP del) cancers; however, the pharmacologic characterization of small-molecule inhibitors that recapitulate the synthetic lethal phenotype has not been described. MRTX1719 selectively inhibited PRMT5 in the presence of MTA, which is elevated in MTAP del cancers, and inhibited PRMT5-dependent activity and cell viability with >70-fold selectivity in HCT116 MTAP del compared with HCT116 MTAP wild-type (WT) cells. MRTX1719 demonstrated dose-dependent antitumor activity and inhibition of PRMT5-dependent SDMA modification in MTAP del tumors. In contrast, MRTX1719 demonstrated minimal effects on SDMA and viability in MTAP WT tumor xenografts or hematopoietic cells. MRTX1719 demonstrated marked antitumor activity across a panel of xenograft models at well-tolerated doses. Early signs of clinical activity were observed including objective responses in patients with MTAP del melanoma, gallbladder adenocarcinoma, mesothelioma, non–small cell lung cancer, and malignant peripheral nerve sheath tumors from the phase I/II study.
Significance:
PRMT5 was identified as a synthetic lethal target for MTAP del cancers; however, previous PRMT5 inhibitors do not selectively target this genotype. The differentiated binding mode of MRTX1719 leverages the elevated MTA in MTAP del cancers and represents a promising therapy for the ∼10% of patients with cancer with this biomarker.
See related commentary by Mulvaney, p. 2310.
This article is featured in Selected Articles from This Issue, p. 2293
INTRODUCTION
Tumor suppressor genes lost via biallelic deletions cannot be directly targeted per se; however, cancers with tumor suppressor alterations may exhibit vulnerabilities that can be exploited for the treatment of cancer. Functional genomic screens in molecularly annotated cancer cell lines have cataloged the dependencies of thousands of genes and are particularly useful for the identification of long-sought synthetic lethal targets and exploitable vulnerabilities for tumor suppressor–altered states (1, 2). Homozygous deletion of the CDKN2A region including methylthioadenosine phosphorylase (MTAP) is frequently observed in cancer, as it is an especially efficient mechanism for removing multiple tumor suppressor transcripts, including CDKN2A (encoding p16Ink4a and p14ARF) and CDKN2B (encoding p15Ink4b; ref. 3).
Protein arginine methyltransferase 5 (PRMT5) is a methylosome protein 50 (MEP50) cofactor-dependent methyltransferase that utilizes the universal methyl donor S-adenosyl methionine (SAM) to add symmetric dimethylarginine (SDMA) posttranslational modifications to target proteins involved in a variety of essential cellular functions including RNA splicing, transcription, and translation (4–6). PRMT5 was identified as a synthetic lethal target for cancers harboring homozygous deletion of the MTAP gene (hereafter denoted MTAP del cancers), which is adjacent to and codeleted with the most frequently deleted tumor suppressor gene, CDKN2A (7–9). MTAP is deleted in a high percentage of several cancers, including non–small cell lung cancer (NSCLC), mesothelioma, pancreatic cancer, glioblastoma, head and neck cancer, esophageal cancer, bladder cancer, and malignant peripheral nerve sheath tumors (MPNST). MTAP catalyzes the phosphorolysis of 5′-deoxy-5′-methylthioadenosine (MTA) to adenine and 5-methylthio-D-ribose-1-phosphate and is required for the methionine and adenosine salvage pathways (10). MTAP loss leads to the accumulation of MTA, which was biochemically determined to compete with SAM for binding to PRMT5 and is a moderately potent and selective inhibitor of PRMT5-dependent deposition of SDMA on key protein substrates (7–9). This creates a state whereby MTAP del cancer cells are particularly vulnerable to further inhibition of PRMT5. Importantly, the synthetic lethality effect was observed only in screens that used short hairpin RNA (shRNA) technology, which reduces, but does not eliminate, PRMT5 activity. Thus, a small-molecule inhibitor that mechanistically exploits the partially inhibited state of PRMT5 was predicted to recapitulate the selective synthetic lethality observed in MTAP-deleted cells in shRNA screens. The synthetic lethal effect was not observed in single-guide RNA (sgRNA) screens that knocked out all PRMT5 activity, and, in fact, sgRNA screens further suggested that PRMT5 is an essential gene for all cells. Notably, previously identified small-molecule PRMT5 inhibitors described to date bind either apo or SAM-bound PRMT5 (11, 12), do not bind PRMT5 complexed with MTA, and do not selectively impact the viability of MTAP del cell lines. These first-generation PRMT5 inhibitors also exhibit mechanism-based toxicities, including neutropenia, thrombocytopenia, and anemia, which likely limit the ability to reach the dose and exposure levels that achieve the requisite (near complete) target inhibition necessary to drive tumor regression in patients (13, 14).
A small molecule that selectively binds and stabilizes the catalytically inactive PRMT5/MTA complex is predicted to further inhibit the residual PRMT5 activity in MTAP del tumors and may represent a synthetic lethal–based precision medicine for patients with these cancers. MRTX1719 was identified through structure-based drug design as a potent small-molecule inhibitor of the PRMT5/MTA complex (15). In the present studies, we characterize its mechanism of action for selective inhibition of PRMT5-dependent cellular function and demonstrate its selective antitumor activity in MTAP del cancer models and patients.
RESULTS
PRMT5-Dependent SDMA Is Reduced in MTAP-Nonexpressing Human Tumors Compared with MTAP-Expressing Tumors across Multiple Tumor Types
Human cancers harboring MTAP gene deletions are anticipated to exhibit increased levels of MTA, which would, in turn, compete with SAM and partially inhibit PRMT5-dependent methyltransferase activity. To directly investigate whether MTAP del human tumors exhibit reduced PRMT5 activity, MTAP protein expression and SDMA protein modification levels were specifically evaluated in MTAP del versus MTAP wild-type (WT) tumor cells. MTAP and SDMA IHC was performed on a panel of non–small cell lung (adenocarcinoma and squamous cell carcinoma) cancer, bladder cancer, head and neck cancer, mesothelioma, and pancreatic cancer tissue microarrays (TMA; Supplementary Materials and Methods). For each tumor type, the SDMA IHC H-score was determined for a similar number of cases without detectable MTAP protein expression and with MTAP expression. SDMA modification in neoplastic cells from MTAP-nonexpressing tumors was reduced by ∼50% compared with MTAP expressing tumors across tumor types (Supplementary Fig. S1). These data indicate that human cancers that do not express MTAP protein exhibit an apparent reduction of PRMT5 activity relative to MTAP-expressing tumors. Because PRMT5 is a cell-essential gene, these data suggest that MTAP del neoplastic cells may be differentially susceptible to tumor cell–selective inhibition of PRMT5.
Identification and In Vitro Characterization of MRTX1719
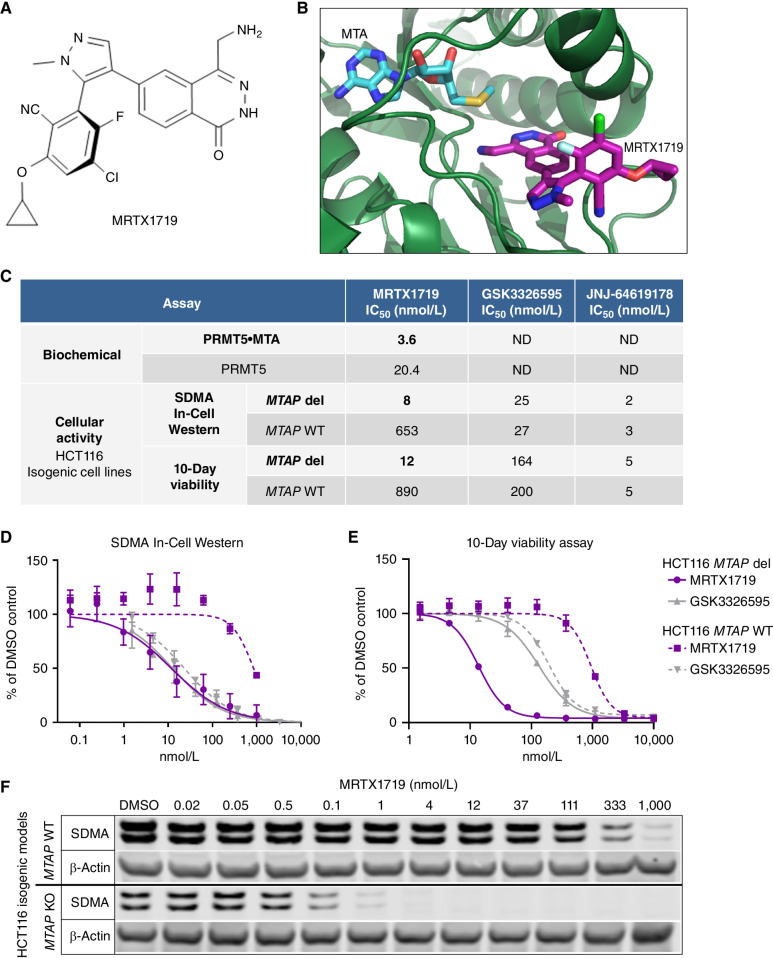
To identify compounds that selectively bound to the PRMT5/MTA complex, a screen was performed using a surface plasmon resonance (SPR) strategy whereby PRMT5/MEP50 was screened for small-molecule fragment binding in the presence of 20 μmol/L MTA and counter-screened against both apo-PRMT5/MEP50 and PRMT5/MEP50 in the presence of 20 μmol/L SAM (15). A 4-(aminomethyl)phthalazine-1(2H)-one fragment hit that selectively bound the PRMT5/MEP50 complex in the presence of MTA was identified. This initial fragment was elaborated, and a lead series was expanded further utilizing a structure-based drug discovery approach leading to the identification of MRTX1719 (Fig. 1A). MRTX1719 binding to the MTA-bound PRMT5/MEP50 complex was confirmed by X-ray crystallography (Fig. 1B) and is further described in Smith and colleagues (15).
Figure 1.
MRTX1719 binds PRMT5/MTA and selectively inhibits MTAP del cancer cells with reduced activity in MTAP WT cells. A, Chemical structure of MRTX1719. B, X-ray crystal structure of MRTX1719 cocomplexed with PRMT5/MEP50 and MTA (right; Protein Data Bank code 7S1S; ref. 14). C, MRTX1719 was run in a PRMT5/MEP50 biochemical assay that measures the activity of the complex in the absence and presence of MTA at an approximate IC50 concentration of MTA in the assay (2 μmol/L). MRTX1719, GSK3326595, and JNJ-64619178 were run in SDMA In-Cell Western (SYM11 antibody; C and D) and 10-day viability assays (C and E) in MTAP del and WT HCT116 cell lines. ND, not determined. F, Cropped SDMA Western blot showing protein bands that correlate with the molecular weight of SmD3 in MTAP del and WT HCT116 cell lines following 4 days of treatment with a range of concentrations of MRTX1719, with β-actin run as a loading control. KO, knockout.
MRTX1719 was evaluated in biochemical assays that measure the inhibition of the PRMT5/MEP50 complex methyltransferase activity using a histone 4 peptide substrate. The assay was performed with and without MTA to determine the IC50 in conditions intended to model elevated MTA levels present in MTAP del tumor cells compared with MTAP WT cells. The average IC50 value in the PRMT5 biochemical assay without MTA added was 20.4 nmol/L, and the average IC50 value with MTA present was 3.6 nmol/L (Fig. 1C). Furthermore, by SPR, MRTX1719 demonstrated ∼70-fold increased binding to the MTA/PRMT5 complex versus SAM-bound PRMT5 (KD = 0.14 pmol/L vs. 9.4 pmol/L; ref. 15). These data demonstrate that MRTX1719 selectively binds to the PRMT5/MEP50/MTA complex and exhibits increased potency against PRMT5 in the presence of MTA. In addition, MRTX1719 was evaluated in a biochemical assay format amenable to testing whether an inhibitor is peptide substrate competitive. The IC50 of MRTX1719 determined in the presence of 0.25 μmol/L MTA with increasing histone 4 peptide substrate concentration exhibited decreased potency, indicating the mode of inhibition is substrate competitive similar to the binding mode of GSK3326595 (Supplementary Materials and Methods; Supplementary Fig. S2A; ref. 12).
MRTX1719 was then evaluated in cellular mechanistic and viability assays using the human colorectal cancer HCT116 cell line, which is MTAP WT, and an isogenic paired cell line that utilized a CRISPR-based strategy to knock out the MTAP gene (MTAP del; Supplementary Materials and Methods). MRTX1719 inhibited PRMT5-dependent SDMA protein modification in MTAP del HCT116 cells with an IC50 value of 8 nmol/L and in parental HCT116 cells with an IC50 value of 653 nmol/L (Fig. 1C and 1D). Additionally, MRTX1719 inhibited the viability of MTAP del HCT116 cells with an IC50 value of 12 nmol/L and parental HCT116 cells with an IC50 value of 890 nmol/L using a 10-day viability assay format (Fig. 1C and E). Thus, MRTX1719 demonstrated greater than 70-fold selectivity for inhibiting both PRMT5-mediated SDMA protein modification and viability in MTAP del compared with MTAP WT HCT116 cancer cells in vitro. A second isogenic MTAP del and WT pair was generated using the human pancreatic cancer cell line PK-1, and similar potency and selectivity in SDMA and viability assays were observed (Supplementary Fig. S2B and S2C). In contrast, GSK3326595 and JNJ-64619178 have been previously described as SAM-uncompetitive and SAM-competitive PRMT5 inhibitors, respectively (11, 12), and demonstrated a lack of selectivity between MTAP del and WT cell lines in both SDMA and viability assays (Fig. 1C–E; Supplementary S2C).
MRTX1719 was also evaluated for modulation of SDMA levels over a longer time course of 4 days in MTAP del and WT HCT116 cells. A concentration-dependent reduction of PRMT5-mediated SDMA protein modification was observed that was highly selective for the MTAP del cell line (Fig. 1F). SDMA levels corresponding to the SmD3 protein molecular weight were undetectable at concentrations greater than 1 nmol/L in MTAP del cells but were still present at 1 μmol/L in MTAP WT cells, confirming MRTX1719 is highly selective for inhibition of PRMT5 activity in MTAP del versus MTAP WT cells.
As MRTX1719 exhibits a unique binding mode, it was evaluated for PRMT5 target occupancy kinetics and duration of target inhibition in biochemical and cellular assay formats, respectively. MRTX1719 demonstrated a long dissociation half-life from MTA-bound PRMT5 (∼14 days) as determined by SPR (15). To explore the impact of a long dissociation half-life in intact cells, MTAP del HCT116 cells were incubated with a range of concentrations of MRTX1719 for 3 hours, 72 hours, or 3 hours followed by washing the cells with PBS and incubating for an additional 72 hours in media without inhibitor (a 72-hour washout) and subsequently evaluated in the SDMA assay. Treatment of MTAP del cells for 3 hours followed by immediate evaluation of SDMA levels did not result in any effect (Supplementary Fig. S2D), as a reduction in SDMA levels following PRMT5 inhibition is known to require a longer time frame (11). In contrast, treatment of MTAP del HCT116 cells for 3 hours followed by a 72-hour washout resulted in a concentration-dependent reduction of SDMA with an IC50 of 44 nmol/L compared with 3.4 nmol/L in 72-hour treated cells, suggesting MRTX1719 binds the PRMT5/MTA complex tightly and exhibits a durable pharmacodynamic effect in cells (Supplementary Fig. S2D). Furthermore, MRTX1719 demonstrated a comparable increase in potency and extended target occupancy in MTAP WT HCT116 cells incubated with 20 μmol/L MTA compared with MTAP WT HCT116 parental cells incubated without MTA, confirming that the elevated MTA in MTAP del cells contributes significantly to increased potency and long target occupancy of MRTX1719 in MTAP del cells (Supplementary Fig. S2E and S2F).
To further characterize the specificity of MRTX1719 binding to the PRMT5/MTA complex, thermal shift experiments were conducted and compared with first-generation PRMT5 inhibitors (Supplementary Materials and Methods). After a 15-minute incubation, MRTX1719 demonstrated complete binding to MTA-bound PRMT5, as evidenced by the clear, single peak increase (right shift) in the melting temperature trace, whereas MRTX1719 exhibited only partial binding to SAM-bound PRMT5 at this time point (Supplementary Fig. S3A and S3B). After a 4-hour incubation, MRTX1719 binding to SAM-bound PRMT5 was still incomplete, whereas complete binding of MRTX1719 to MTA-bound PRMT5 was maintained. These data suggest that MRTX1719 binds both MTA-bound and SAM-bound PRMT5; however, binding to SAM-bound PRMT5 does not reach saturation in the thermal shift experiment and is incomplete. Moreover, based on the melting temperature shift, the ternary complex of MRTX1719–MTA–PRMT5 displayed increased stability compared with the ternary complex of MRTX1719–SAM–PRMT5 as well as MRTX1719—PRMT5. In comparison, GSK3326595 demonstrated high-affinity binding only to SAM-bound PRMT5, as would be expected for a SAM-uncompetitive PRMT5 inhibitor, whereas JNJ-64619178 demonstrated high-affinity binding to apo, SAM, and MTA-bound PRMT5 as would be expected for a potent SAM-competitive PRMT5 inhibitor. These data confirm MRTX1719 binds tightly and preferentially to the PRMT5/MTA complex and exhibits a differentiated binding mechanism compared with first-generation PRMT5 inhibitors.
MRTX1719 Demonstrates Selective and Dose-Dependent Reduction of PRMT5-Dependent SDMA and Tumor Growth in MTAP Del Tumor Models while Sparing MTAP WT Tissue
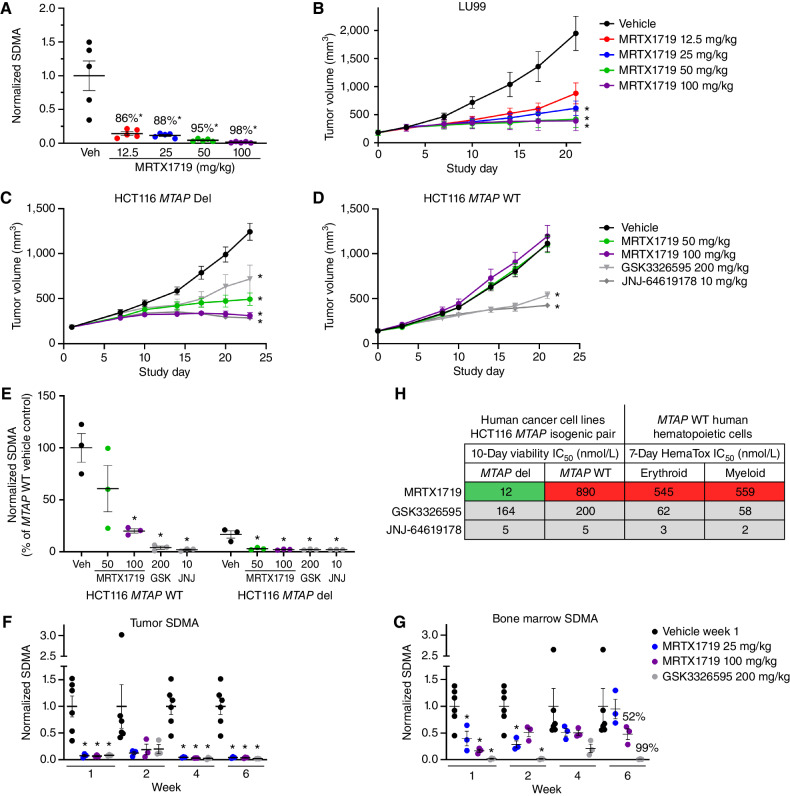
To evaluate target pharmacodynamics and antitumor activity in vivo, MRTX1719 was administered orally once daily for 22 days to immunocompromised mice bearing human MTAP del LU99 lung tumor xenografts and demonstrated dose-dependent inhibition of PRMT5-dependent SDMA modification (Fig. 2A). Furthermore, MRTX1719 demonstrated dose-dependent tumor growth inhibition of MTAP del LU99 tumor xenografts established in immunocompromised mice (Fig. 2B). Oral daily (q.d.) administration at 50 and 100 mg/kg resulted in similar antitumor activity—approximate tumor stasis—and correlated with 95% or greater SDMA reduction, indicating that a near-maximal pharmacodynamic target inhibition and antitumor efficacy are achieved in this dose range in mice. All dose levels of MRTX1719 up to 100 mg/kg q.d. were well tolerated with no overt signs of toxicity or evidence of body weight loss (Supplementary Fig. S4A). The SAM-competitive PRMT5 inhibitor JNJ-64619178 was evaluated in the LU99 model and demonstrated antitumor activity that correlated with strong tumor SDMA reduction (Supplementary Fig. S4B and S4C). However, at these doses, strong SDMA modulation was also observed in the bone marrow, illustrating on-target liabilities previously associated with non–MTA-cooperative PRMT5 inhibitors.
Figure 2.
MRTX1719 exhibits selective, dose-dependent inhibition of PRMT5-dependent SDMA modification in MTAP del tumor xenografts in vivo. A, MRTX1719 was administered daily via daily oral gavage for 22 days to immunocompromised mice bearing LU99 tumor xenografts (average initial tumor volume ∼180 mm3) at 12.5, 25, 50, and 100 mg/kg. Tumors were collected 4 hours after dose, and SDMA was analyzed by immunoblot and quantified with densitometry. Average normalized SDMA values were divided by the average value in vehicle-treated tumors to calculate percent inhibition. Data shown represent the average of 2 to 3 tumors per treatment group ± SEM. Statistics were determined using a two-tailed Student t test with significance indicated (*, P < 0.05). Veh, vehicle. B, MRTX1719 was administered at the indicated doses via daily oral gavage to mice bearing established LU99 cell line–derived tumor xenografts as in A. Data, mean tumor volume ± SEM. Statistics were determined using a two-way ANOVA with significance indicated (*, P < 0.05). A and B were previously published in Smith et al. (14). C and D, MRTX1719, GSK3326595, or JNJ-64619178 was administered via daily oral gavage at the doses indicated to mice bearing established HCT116 MTAP del (C) or MTAP WT (D) cell line–derived tumor xenografts. Dosing was initiated when tumors were ∼150 mm3. Data, mean tumor volume ± SEM. Statistics were determined using a two-way ANOVA with significance indicated (*, P < 0.05). E, Tumors from the HCT116 MTAP WT or HCT116 MTAP del cell line–derived xenografts in C and D were collected 4 hours after dose, and SDMA was analyzed by immunoblot. Data shown represent the average of 3 tumors per treatment group ± SEM. Statistics were determined using a two-tailed Student t test with significance indicated (*, P < 0.05). GSK, GSK3326595; JNJ, JNJ-64619178. F and G, MRTX1719 and GSK3326595 were administered via daily oral gavage to immunocompromised mice bearing established LU99 xenograft tumors at the indicated doses for the indicated number of weeks. Tumor (F) and bone marrow (G) were collected 4 hours after last dose, and SDMA was analyzed by Western blot densitometry. Representative SDMA bands were analyzed from 3 to 5 tumor lysates and bone marrow lysates from 3 to 5 mice per treatment group, and data are shown as mean normalized SDMA levels ± SEM. Statistics were determined using a two-tailed Student t test with significance indicated (*, P < 0.05). H, MRTX1719, JNJ-64619178, and GSK3326595 were run in human erythroid and myeloid 7-day HemaTox assays (STEMCELL Technologies) and were compared with IC50 values from 10-day HCT116 MTAP del and MTAP WT cell line viability assays.
Daily (q.d.), twice-a-day (b.i.d.), and every-other-day (Q2D) dosing schedules were evaluated to determine the optimal dosing schedule. Total dose-matched q.d. and b.i.d. dosing resulted in comparable antitumor activity at the 50 mg/kg b.i.d. and 100 mg/kg q.d. as well as the 25 mg/kg b.i.d. versus 50 mg/kg q.d. dose groups (Supplementary Fig. S4D). In contrast, Q2D dosing at 100 or 200 mg/kg demonstrated reduced antitumor activity compared with total dose-matched 50 or 100 mg/kg q.d. dosing. These data demonstrate q.d. dosing is optimal for achieving maximal antitumor activity in preclinical models.
The selective impact of MRTX1719 on SDMA and antitumor efficacy in MTAP del tumors in vivo was evaluated utilizing the isogenic paired MTAP ± HCT116 model system. MRTX1719 administered orally at 50 and 100 mg/kg q.d. demonstrated tumor growth inhibition of HCT116 MTAP del tumor xenografts; however, no effect on tumor growth was observed at these dose levels in the isogenic HCT116 MTAP WT tumor xenograft model (Fig. 2C and D). In contrast, GSK3326595 and JNJ-64619178 demonstrated similar antitumor activity in both the MTAP del and WT xenograft models.
In addition, MRTX1719 demonstrated a greater magnitude of inhibition of PRMT5-dependent SDMA modification in MTAP del compared with MTAP WT HCT116 xenograft tumors (Fig. 2E). In contrast, GSK3326595 and JNJ-64619178 demonstrated similar reduction in SDMA in both HCT116 MTAP genotypes. In summary, MRTX1719 consistently resulted in near-complete reduction of PRMT5-dependent SDMA selectively in MTAP del tumor cells, with reduced SDMA reduction in MTAP WT normal cells, which was associated with significant antitumor activity at doses that were well tolerated.
To evaluate SDMA inhibition kinetics and subsequent mechanistic impact of MRTX1719 in vivo, mice bearing LU99 xenograft tumors were treated over a time course, tumors were collected 4 hours after last dose, and immunoblot analyses were performed. SDMA levels were unchanged after one dose, partially reduced after 3 days of dosing, and maximally inhibited following 14 or 21 days of dosing (Supplementary Fig. S4E). Levels of phosphorylated Rb (phospho-Ser 807/811) were reduced with similar kinetics relative to SDMA and were nearly undetectable after 14 and 21 days of administration. The cell-cycle inhibitory proteins p21 and p27 were also increased following 3 days of dosing, with p21 remaining elevated and p27 levels recovering after 14 and 21 days of dosing. Cleaved caspase-7 and cleaved PARP were also elevated following 14 and 21 days of dosing, providing evidence of apoptosis coincident with maximal PRMT5 pathway modulation. Finally, MRTX1719 was administered daily at 100 mg/kg to mice bearing LU99 xenograft tumors for 21 days, and tumors were collected 4, 24, 72, and 120 hours after the last dose. SDMA levels were still 95% and 69% reduced relative to vehicle-treated tumors in treated tumors harvested 72 and 120 hours after the last dose, respectively (Supplementary Fig. S4F), demonstrating that MRTX1719 exhibits a durable pharmacodynamic effect for several days after cessation of therapy in vivo. These data demonstrate that maximal inhibition of PRMT5-mediated SDMA modification requires several days of administration in vivo, which, in turn, coincides with subsequent modulation of key proliferation and apoptosis pathways in mediating the antitumor activity of MRTX1719.
Based on the lack of significant activity of MRTX1719 in the MTAP WT tumor model, studies were extended to also evaluate its effects in mouse Mtap WT tissue. To assess the relative effects of MRTX1719 on SDMA levels in normal tissue, a time-course analysis was conducted in MTAP del tumors and Mtap WT bone marrow from immunocompromised mice bearing LU99 tumor xenografts treated with MRTX1719 or GSK3326595. MRTX1719 and GSK3326595 both demonstrated marked inhibition of PRMT5-mediated SDMA modification in LU99 tumors following 1, 2, 4, or 6 weeks of daily dosing (25 or 100 mg/kg MRTX1719 and 200 mg/kg GSK3326595; Fig. 2F; see also Supplementary Fig. S4C). In contrast, MRTX1719 treatment exhibited comparatively less reduction of bone marrow SDMA levels (52% SDMA reduction), whereas GSK3326595 treatment resulted in a pronounced reduction of both bone marrow and tumor SDMA levels (99% SDMA reduction in bone marrow; Fig. 2G). Finally, in a 28-day repeat dose study, female and male CD-1 mice dosed with MRTX1719 at 150 mg/kg q.d. daily exhibited minimal changes in hematologic parameters (Supplementary Table S1).
To evaluate the effect of PRMT5 inhibitors on normal human hematopoietic cell viability, MRTX1719, GSK3326595, and JNJ-64619178 were evaluated in 7-day erythroid and myeloid cell HemaTox viability assays (STEMCELL Technologies). Similar to the differential effect in HCT116 MTAP WT tumor cells, MRTX1719 was ∼50-fold less potent in HemaTox viability assays compared with its activity in the HCT116 MTAP del cell line viability assay (Fig. 2H). In contrast, GSK3326595 and JNJ-64619178 demonstrated ∼2- to 3-fold increased potency in HemaTox viability assays compared with their activity in either HCT116 MTAP WT or MTAP del cell line viability assays. Together, these data demonstrate that MRTX1719 selectively affects the viability of MTAP del cancer cells, with significantly reduced activity in mouse or human bone marrow, and, furthermore, highlight the significantly improved selectivity margin of MRTX1719 relative to previously described PRMT5 inhibitors.
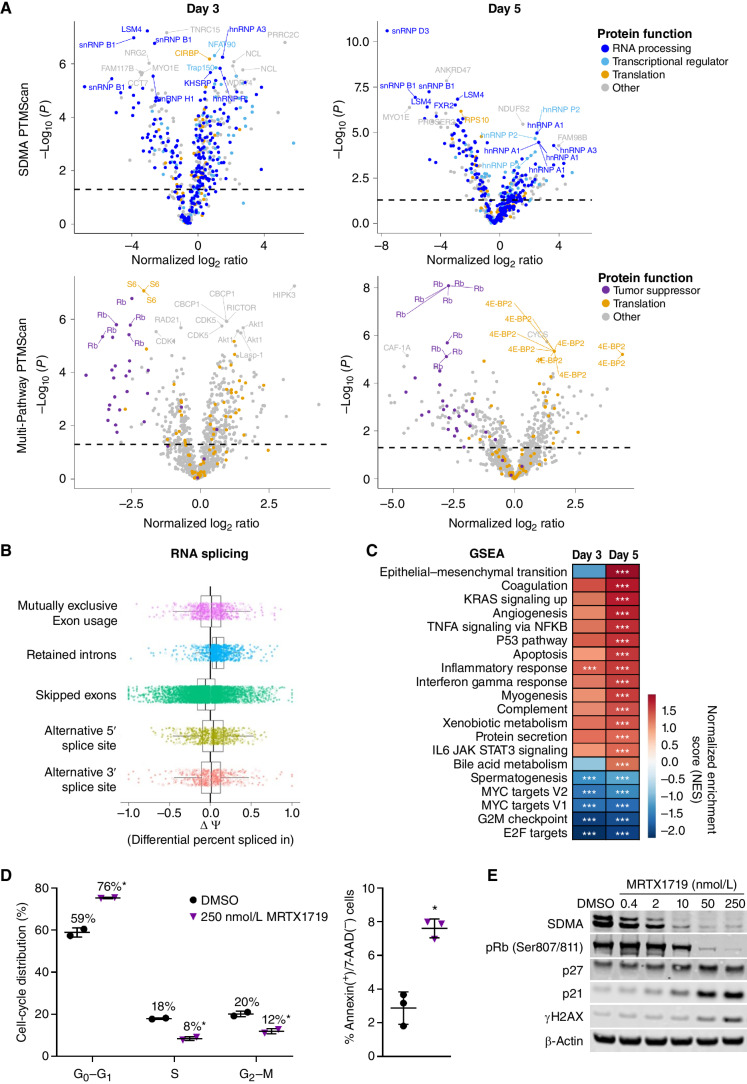
MRTX1719 Inhibits Critical PRMT5-Dependent Molecular and Cellular Processes in MTAP Del Cells
To explore the molecular and cellular consequences of PRMT5 inhibition in MTAP del cancer cells, differentially abundant posttranslational protein modifications were measured in cell lysates from LU99 cells (MTAP del) treated with DMSO versus 1 μmol/L MRTX1719 for 3 or 5 days using LC-MS/MS following antibody enrichment using an SDMA motif antibody or the Multi-Pathway Enrichment Kit, which includes anti–phospho-site antibodies (PTMScan, Cell Signaling Technology; Supplementary Table S2). The most differentially abundant posttranslationally modified proteins following 3 or 5 days of treatment with MRTX1719 included components of the spliceosome (snRNP D3, snRNP B1, and LSM4), transcriptional proteins (S6 and RPS10), and the cell-cycle regulators Rb and CDK1 (Fig. 3A; Supplementary Table S2). Symmetric dimethylarginine modification of spliceosome components has been previously demonstrated to be essential for spliceosome function (16, 17). RNA sequencing (RNA-seq) was also performed on LU99 cells treated as above followed by analyses of aberrant splicing and differential expression. Transcripts with retained introns were increased in MRTX1719-treated versus DMSO-treated LU99 cells (Fig. 3B), which is a previously described mechanism following PRMT5 inhibition (18, 19). Gene set enrichment analysis (GSEA) demonstrated that p53, apoptosis, and immune-related hallmark pathways were significantly upregulated, whereas MYC-related and cell cycle–related pathways were significantly downregulated after 3- or 5-day treatment with MRTX1719 (Fig. 3C). Finally, cell-cycle analysis demonstrated treatment with MRTX1719 led to a marked reduction in the percentage of cells in S-phase and G2–M, an increase in the percentage of cells in G0–G1, and an approximately 3-fold increase in cells positive for the early apoptotic marker Annexin V (Fig. 3D). Protein analyses of lysates from LU99 cells treated with MRTX1719 further demonstrated that maximal or near-maximal reduction of PRMT5-dependent SDMA modification coincided with decreased pRb (Ser807/811) as well as increased p27, p21, and γH2AX (Fig. 3E). These data are in agreement with previous mechanistic studies designed to evaluate the consequences of PRMT5 inhibition (12, 18–20). Collectively, these data demonstrate that MRTX1719 exhibits selective on-target activity in MTAP del cells and alters PRMT5-regulated molecular and cellular processes essential for cell proliferation and viability.
Figure 3.
MRTX1719 treatment disrupts PRMT5-regulated processes in MTAP del cancer cells. A, Volcano plots of posttranslational modifications of trypsin-digested peptides from LU99 cells treated for 3 or 5 days with 1 μmol/L MRTX1719 vs. DMSO as measured by LC-MS/MS following antibody enrichment using the SDMA PTMScan kit (top) or the Multi-Pathway PTMScan kit (bottom) that includes phosphorylation (Cell Signaling Technology). B, RNA-seq data were generated from LU99 cells treated with MRTX1719 or DMSO for 3 days, and transcripts were analyzed for alternative RNA splicing using rMATS. Transcripts with retained introns were increased in MRTX1719-treated vs. DMSO-treated cells. C, GSEA was performed on RNA-seq data from 3- and 5-day MRTX1719-treated LU99 cells compared with DMSO-treated. ***, FDR <0.25. D, LU99 cells were treated with 250 nmol/L MRTX1719 for 3 days and analyzed on a Guava flow cytometer. Cell-cycle distribution analysis (left) was performed, and apoptotic cells were analyzed for surface Annexin V–positive, 7-AAD–negative staining (right). Summary data from 2 or 3 independent experiments are presented as the mean ± SD. Statistics were determined using a two-tailed Student t test with significance indicated (*, P < 0.05). E, Cell lysates from LU99 cells treated for 3 days with MRTX1719 and analyzed for selected protein markers by Western blot.
MRTX1719 Demonstrates Broad-Spectrum Activity against MTAP Del Cancer Cell Lines from a Variety of Tumor Types
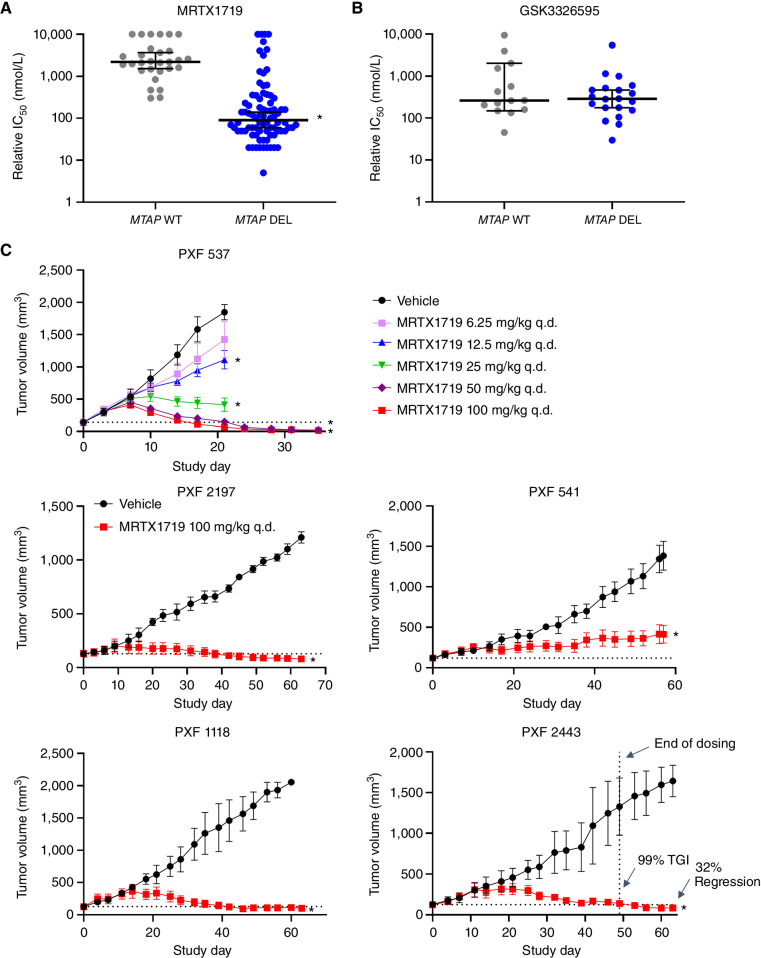
To further investigate the selectivity of MRTX1719 for reducing the viability of MTAP del tumor cells, it was evaluated across a panel of 70 MTAP del and 26 MTAP WT human cancer cell lines in 5-day CellTiter-Glo (CTG) viability assays. GSK3326595 was also evaluated in CTG assays in a subset of cell lines. MRTX1719 selectively reduced the viability of MTAP del cell lines with a minimal effect on MTAP WT cells, whereas GSK3326595 demonstrated similar activity across cell lines independent of MTAP genotype (Fig. 4A and B; Supplementary Table S3). The median IC50 value for MRTX1719 in the MTAP del cell lines was 90 nmol/L, whereas the median IC50 of MRTX1719 in the MTAP WT cell lines was 2.2 μmol/L. Of note, a 5-day viability assay was utilized, as it was amenable to screening an expanded panel of cell lines; however, IC50 values were 3- to 4-fold lower when using a 10-day viability assay (e.g., MRTX1719 LU99 5-day viability IC50: 72 nmol/L; 10-day viability IC50: 20 nmol/L), suggesting the cellular potency of MRTX1719 is increased with a longer duration of PRMT5 inhibition.
Figure 4.
MRTX1719 selectively inhibits MTAP del cancer cell line growth in vitro and in vivo. A and B, MRTX1719 in vitro activity across a panel of MTAP WT and MTAP del cell line models (5-day viability assay, Crown Biosciences). Dot plots showing median with 95% confidence intervals of IC50 values for MRTX1719 (A) and GSK3326595 (GSK-595; B) in MTAP WT and MTAP del cell line models. A smaller cohort of models was tested with GSK-595. Median values: MRTX1719-MTAP del: IC50 = 90 nmol/L; MTAP WT: IC50 = 2.2 mmol/L; GSK-595-MTAP del: IC50 = 262 nmol/L; MTAP WT: IC50 = 286 nmol/L. Statistics were determined using a two-tailed Student t test with significance indicated (*, P < 0.05). C, MRTX1719 was administered orally at the doses indicated to five mesothelioma PDX models. n = 3–5 mice per treatment group. Data, average tumor volume ± SEM. Statistics were determined using a two-way ANOVA with significance indicated (*, P < 0.05). TGI, tumor growth inhibition.
MRTX1719 Maintains Activity against MTAP Del Cells When Cocultured with MTAP WT Fibroblasts
Although the selective effect of MRTX1719 on MTAP del tumor cell viability is anticipated to be cell autonomous, a recent study reported MTA does not accumulate in human MTAP del glioblastoma tissue and further reported MTAP WT cells are able to metabolize elevated MTA secreted by MTAP del tumor cells in vitro, potentially offsetting an MTA-dependent synthetic lethal vulnerability (21). To determine whether MTAP WT fibroblasts/stromal cells can impair the activity of MRTX1719 in selectively targeting MTAP del cancer cell viability, LU99 cells were labeled with luciferase and were cocultured with and without the MRC-5 human lung fibroblast cell line, and cell viability was evaluated (Supplementary Materials and Methods). MRTX1719 IC50 values were similar for LU99 cells cultured with and without an equal number of MRC-5 cells in viability assays (Supplementary Fig. S5A). Viability was also independently assessed for each cell line without admixing, and it was confirmed that MRTX1719 selectively reduced viability in the LU99 cell line, but not MRC-5 cells (Supplementary Fig. S5B). These data suggest MTAP WT admixed human fibroblasts do not impact the antitumor activity of MRTX1719 against MTAP del cancer cells in vitro.
MRTX1719 Demonstrates Antitumor Activity and Tumor Regression across a Variety of Cell Line– and Patient-Derived Tumor Xenograft Models, Including Mesothelioma Models
Cell lines derived from NSCLC, pancreatic cancer, and mesothelioma were among the most sensitive cell lines to MRTX1719 cell viability screens (Supplementary Fig. S6A). MRTX1719 was evaluated in ∼3-week efficacy studies in selected MTAP del cell line–derived xenograft (CDX) and patient-derived xenograft (PDX) models. Notable antitumor activity was observed in pancreatic, lung, esophageal, cholangiocarcinoma, and gastric cancer models, including tumor regression in a subset of models (Supplementary Fig. S6B). MTAP del is commonly observed in patients with mesotheliomas (∼40%), a disease with limited effective therapeutic options and approved targeted therapies. Therefore, MRTX1719 was evaluated in five MTAP del mesothelioma PDX models. MRTX1719 administered orally q.d. at 100 mg/kg demonstrated marked antitumor activity in all models tested, including regression in four out of five models (Fig. 4C). In the PXF 537 model, treatment at 50 or 100 mg/kg dose levels led to near-complete regression. In the four PDX models that exhibited regressions, it was notable that tumor growth in MRTX1719-treated mice was comparable to vehicle-treated mice for the first 5 to 10 days before tumor growth slowed and began to regress around day 20 in some models, suggesting sustained PRMT5 inhibition over several weeks is required to observe a maximal antitumor effect.
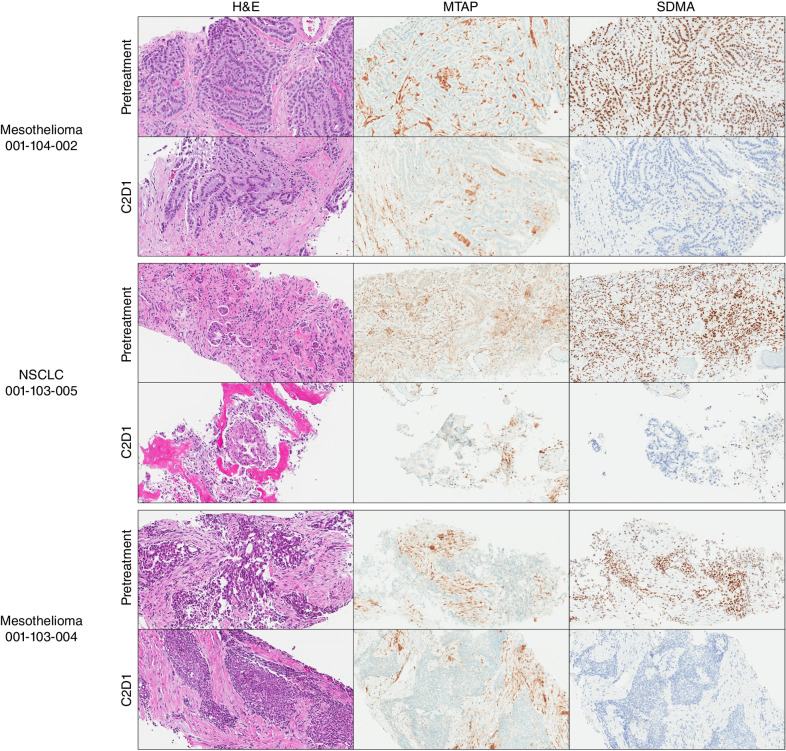
MRTX1719 Treatment of Patients with MTAP Del Cancers Demonstrated Marked Reduction of PRMT5-Dependent SDMA in Repeat Tumor Biopsies
MRTX1719 is currently in a first-in-human, phase I/II clinical trial (study 1719-001, NCT05245500) in patients with solid tumors harboring MTAP gene deletions. To evaluate the clinical pharmacodynamics of MRTX1719 in patients with MTAP del cancers, pretreatment and cycle 2 day 1 biopsies were obtained from three patients orally administered 200 mg q.d. and analyzed for MTAP and SDMA protein expression by IHC. SDMA expression was completely extinguished in the neoplastic cells of all three tumor biopsies, with baseline H-scores of 240, 250, and 280 determined in the three pretreatment biopsies, whereas SDMA was undetectable (H-score = 0) in all three cycle 2 day 1 biopsies (Fig. 5; Supplementary Fig. S7A). The percentage of SDMA-positive stromal cells was also evaluated and was 100% in all three pretreatment biopsies. In cycle 2 day 1 biopsies, the percentage of SDMA-positive stromal cells was 90%, 50%, and 0%. Importantly, dose-limiting hematopoietic-based toxicities similar to those reported for non-MTAP del-selective PRMT5 inhibitors, have not been observed at any dose level evaluated to date (up to 800 mg q.d.; Supplementary Table S4). These data suggest MRTX1719 completely inhibits PRMT5 activity in MTAP del neoplastic tumor cells in patients administered MRTX1719 200 mg q.d. without observation of mechanism-based or dose-limiting toxicities.
Figure 5.
PRMT5-mediated SDMA modification in MTAP del patient tumor biopsies is extinguished following daily oral administration of 200 mg MRTX1719. Pretreatment and C2D1 tumor biopsies were stained for MTAP and SDMA protein expression by IHC. Adjacent slides were also stained with hematoxylin and eosin (H&E). MTAP and SDMA H-scores were determined by a pathologist and scored in Supplementary Fig. S7A.
MRTX1719 Demonstrates RECIST Responses in Patients with MTAP Del Cancer
MRTX1719 has been evaluated in patients with solid tumors harboring MTAP deletions at the starting dose level of 50 mg followed by 100% dose escalations to 100, 200, 400, and 800 mg dose levels administered once daily orally over 21-day cycles. Dose escalation is based on the modified toxicity probability interval-2 (mTPI-2) methodology with planned cohorts of at least three patients at each dose level (22). Patients were evaluated for clinical activity in accordance with RECIST v1.1 (23). MRTX1719 was well tolerated with no dose-limiting toxicities observed at dose levels up to 400 mg q.d. (800 mg q.d. under evaluation). Six individual case narratives for patients treated in dose escalation cohorts are included below. These case narratives for responding patients were selected from a total of 18 patients who were evaluable for clinical response as of June 13, 2023, at dose levels in the therapeutic range of 100 mg q.d. or greater; one of the 18 patients had stable disease with a decrease in tumor as of June 13, 2023, and achieved a response shortly after (June 19, 2023), which was confirmed (August 1, 2023) and is included in the case narratives.
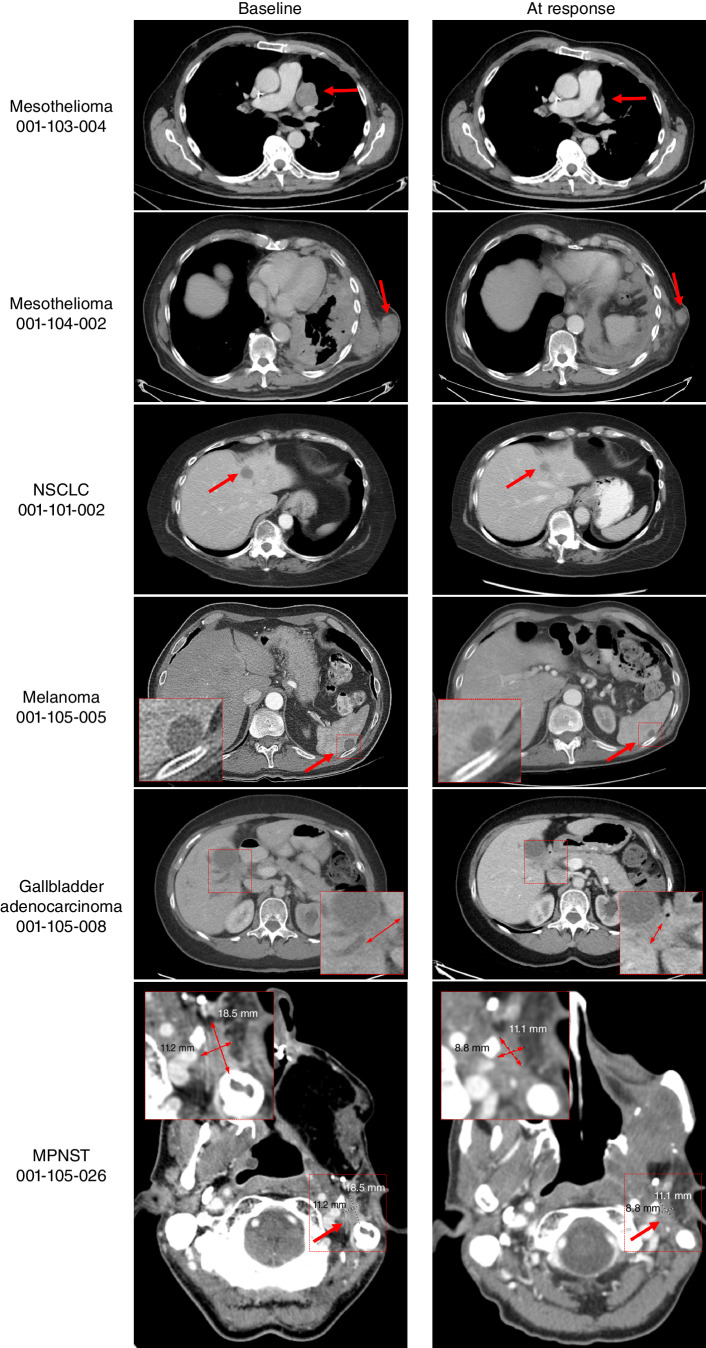
A 70-year-old male diagnosed with epithelioid malignant mesothelioma, previously treated with pleurectomy/decortication, cisplatin/pemetrexed chemotherapy, and immunotherapy with ipilimumab and nivolumab, was enrolled into the 1719-001 clinical trial 2 years after diagnosis for asymptomatic progressive pleural disease. Next-generation sequencing (NGS) demonstrated homozygous MTAP–CDKN2A–CDKN2B deletion and homozygous BAP1 deletion. At the time of enrollment, five pleural rind lesions were identified as target lesions, with a maximum diameter of 2.4 cm. The patient was administered MRTX1719 200 mg q.d. p.o. (per os, or by mouth) on a 21-day cycle, which he tolerated well. After 5 cycles, lipase and amylase blood levels were increased without symptoms, and MRTX1719 was held for 3 weeks, after which they returned to normal, and MRTX1719 was resumed at 100 mg q.d. p.o. for cycle 7 day 1. Asymptomatic elevations in lipase and amylase recurred, and therapy is currently being held. The first on-study disease assessment at cycle 2 day 20 demonstrated a reduction in the size of all five target lesions, with an overall 33.3% reduction by RECIST. Subsequent disease assessments at cycle 4 day 20 and cycle 6 day 21 showed continued RECIST-defined partial response with 50.1% and 56.1% reduction in target lesions, respectively, despite the dose interruption (Fig. 6). The patient remains on study through cycle 8.
Figure 6.
Activity of MRTX1719 in patients with MTAP del cancers. Mesothelioma patient 001-103-004: Pretreatment and follow-up (FU) C6D20 scans, indicating 56% reduction of target lesions, including the pleural disease shown here. The patient continues on study. Mesothelioma patient 001-104-002: Activity of MRTX1719 in patient epithelioid mesothelioma with MTAP deletion. Pretreatment and FU 3 scans, indicating 30% reduction of target lesions, including soft tissue lesions shown here. The patient continues on study. NSCLC patient 001-101-002: Baseline: dominant mass left hepatic dome measuring 29 × 19 mm, previous 20 × 19 mm. Stable other tiny scattered hypodense hepatic lesions too small to accurately characterize. C5D1: left hepatic dome metastasis, measuring 17 × 17 mm. Stable and mild decreasing size of hepatic metastases. The patient continues on study. Melanoma patient 001-105-005: Baseline and cycle 9 scans of a patient with MTAP del melanoma. Partial response was confirmed at cycle 9, and the patient continues on study. Gallbladder adenocarcinoma patient 001-105-008: Pretreatment and cycle 9 scans of a patient with treatment-refractory MTAP loss gallbladder indicating 43% reduction of target lesions. The patient continues on study. MPNST patient 001-105-026: Patient received MRTX1719 800 mg q.d. Restaging after 4 cycles shows a partial response with a 38.9% decrease of target lesions. The patient continues on study.
An 81-year-old male diagnosed with unresectable epithelioid malignant mesothelioma previously treated with carboplatin/pemetrexed and an investigational WT1 vaccine plus nivolumab was enrolled in the 1719-001 clinical trial. NGS demonstrated homozygous MTAP deletion and a PIK3CA mutation. At the time of enrollment, metastatic disease identified as target lesions included a 2.2-cm mediastinal lymph node, a 2.2-cm retrocrural lymph node, and a 4.7-cm soft tissue lesion. The patient was administered MRTX1719 200 mg q.d. p.o. on a 21-day cycle and demonstrated clinical improvement with respect to pain and swelling at sites of soft tissue lesions. MRTX1719 was interrupted due to grade 3 diarrhea during cycle 5, which resolved after treatment interruption of approximately 2 weeks. MRTX1719 was resumed at 100 mg q.d. p.o. without recurrence of diarrhea. The first on-study disease assessment at cycle 2 day 20 demonstrated stable disease with a 13% reduction of target lesions. The subsequent disease assessment at cycle 4 day 20 achieved RECIST-defined partial response with a 30.7% reduction of target lesions, and despite dose interruption, disease assessment on cycle 6 day 9 showed ongoing confirmed partial response (Fig. 6). The patient remains on study treatment through cycle 8.
A 61-year-old female with NSCLC harboring EGFR mutation and both EGFR and MET gene amplification with progressive disease after treatment with osimertinib was enrolled in the 1719-001 clinical trial. NGS demonstrated homozygous MTAP deletion. At the time of enrollment, she had metastatic disease to the left hepatic dome (2.9-cm target lesion) and inferomedial lateral segment of the left hepatic lobe (nontarget lesion) of the liver. The patient was administered MRTX1719 at 400 mg q.d. p.o. on a 21-day cycle and tolerated the treatment well. The first on-study disease assessment at cycle 2 day 20 demonstrated a RECIST-defined partial response with a 41% reduction of the target lesion and was confirmed at cycle 5 day 1 (Fig. 6). The patient remains on study treatment through cycle 6.
A 73-year-old male diagnosed with BRAF WT, stage IV melanoma with prior disease interventions including adjuvant nivolumab, ipilimumab, and nivolumab following metastatic recurrence, and palliative radiation to head and neck lymph nodes and brain metastasis was enrolled in the 1719-001 clinical trial. Targeted NGS demonstrated a homozygous CDKN2A/B/MTAP deletion and TERT mutation. The patient had multiple subcutaneous nodules in the chest and abdomen, a nodule in the nondependent portion of the gallbladder, as well as numerous bilateral pulmonary nodules and a splenic lesion. During treatment with ipilimumab and nivolumab, some of these lesions had initial enlargement and subsequent stabilization, but the splenic lesion increased in size over several scans, so the splenic lesion (2.0 cm) was considered a target lesion for the trial. The patient was administered MRTX1719 100 mg q.d. p.o. on a 21-day cycle and a slow decrease in all lesions, including a RECIST-defined partial response of 30% reduction of the splenic lesion that was observed at cycle 7 day 1. Subsequent disease assessments at cycle 9 day 1 and cycle 14 day 1 demonstrated continued response, with a 40% reduction of the splenic lesion in the latest assessment (Fig. 6). The patient remains on study treatment through cycle 15 after a dose interruption due to fatigue.
A 74-year-old female presented with metastatic adenocarcinoma with mucinous features including pelvic implants affecting the colon, omentum, and left ovary (CK7 positive, CDX2 weakly positive, CK20 and SATB2 negative) and a gallbladder thickening and adjacent soft mass. She was diagnosed with metastatic gallbladder adenocarcinoma with low tumor mutation burden that harbored ARID1A deletion (exons 6–20) and rearrangement, CDKN2A/B and MTAP homozygous deletion, PIK3R1 N564D, and NBN P81fs*23 in mutational analysis. The patient was enrolled in the 1719-001 clinical trial with progressive disease after treatment with cisplatin and gemcitabine. At the time of enrollment, she had recurrent disease in the gallbladder (3.2-cm target lesion) and extensive metastases involving the peritoneum. The patient was administered MRTX1719 100 mg q.d. p.o. on a 21-day cycle. The patient tolerated treatment well but experienced an episode of bowel obstruction due to peritoneal disease during the first cycle. She demonstrated clinical improvement with no further episodes of bowel obstruction. The patient had an initially increasing tumor marker that later decreased (Supplementary Fig. S7B), and a gradual decrease in the gallbladder lesion that achieved a RECIST-defined partial response with a 43% reduction at cycle 8 day 12 and was confirmed at cycle 10 day 12 (Fig. 6). The patient's dose was escalated to 200 mg q.d. p.o. during cycle 10 and she remains on study treatment through cycle 13.
A 45-year-old male was diagnosed with MPNST of the left maxillary sinus (IHC negative for AE1/AE3, CAM 5.2, CK903, SMA, desmin, myogenin, CD34, and ALK with a WT pattern of p53 expression, retained INI1 and BRG1, and an elevated Ki-67 index) in 2017. Upon diagnosis, the patient was treated with up-front surgery and radiation. He had recurrent disease in 11/2019 and was treated with surgery followed by ifosfamide/etoposide with concurrent radiation and, upon progression, with MAI (mesna, doxorubicin, ifosfamide chemotherapy) and a trial with pembrolizumab plus APG-115 (MDM2 inhibitor). At the time of enrollment in the 1719-001 trial, he had biopsy-proven recurrence in the left masticator space and was using a PEG (percutaneous endoscopic gastrostomy) tube for nutrition. Genetic analysis shows NF1 splice-site 1063 mutated, CDKN2A/B/MTAP loss. The patient received MRTX1719 800 mg q.d. p.o. on a 21-day cycle and tolerated treatment well with only grade 1 treatment-related adverse events of hypercalcemia, hypophosphatemia, and neutropenia that resolved without intervention or dose modification. The first restaging scan after 2 cycles showed stable disease with a 16.7% decrease of target lesions. Restaging after 4 cycles demonstrated an unconfirmed partial response with a 38.9% decrease of target lesions (Fig. 6). The patient remains on study treatment.
DISCUSSION
Following the identification of PRMT5 as a synthetic lethal target for the treatment of MTAP del cancers (7–9), several crucial challenges needed to be addressed in order to develop a small-molecule–targeting strategy to exploit these observations and to bring this therapeutic approach to the clinic. The strategy identified to surmount these challenges was to develop an inhibitor that selectively binds to the PRMT5/MTA complex. This strategy exploits the elevated MTA concentrations present in MTAP del cells and turns this altered metabolic state into a therapeutic vulnerability to selectively kill MTAP del neoplastic tumor cells. The discovery of an orally bioavailable small molecule capable of selective binding to PRMT5 when bound to MTA was accomplished using a fragment screen against MTA-soaked PRMT5/MEP50, followed by iterative structure-supported drug optimization (15). Although small molecules capable of binding PRMT5 when bound to SAM had been described (12), the development of a small molecule that selectively binds to an enzyme when complexed with a substrate-competitive metabolite, which then locks that complex in a catalytically inactive state with a biochemically determined residence time on the order of days, is largely unprecedented. It is important to reiterate that previously developed PRMT5 inhibitors do not bind the PRMT5/MTA complex and therefore do not exhibit selectivity for MTAP del cancer cells. Because PRMT5 is required for the viability of all cells, and hematopoietic cells in particular, these inhibitors do not appear to have a sufficiently wide therapeutic index and have shown limited clinical utility to date (24, 25). Given the prevalence of the MTAP deletion in ∼10% of all cancers, this differentiated drug discovery approach may be relevant for a very large patient population, the vast majority of which have high unmet medical need.
In addition, the degree of selectivity required for a small molecule to preferentially bind PRMT5/MTA relative to PRMT5 ± SAM and to selectively inhibit PRMT5 in MTAP del cells was unknown. In the original shRNA screens used, the concentrations of SAM-bound, MTA-bound, and apo-PRMT5 were presumably reduced to an equivalent degree across the cancer cell lines evaluated. These initial functional genomics studies suggest that a compound with equivalent binding affinity to MTA-bound and SAM-bound PRMT5 should be sufficient to recapitulate the original synthetic lethal phenotype observed. However, given the need to achieve complete or near-complete inhibition of PRMT5 in MTAP del tumor cells to achieve maximal antitumor activity in preclinical studies, a compound that maximizes PRMT5 inhibition in MTAP del cells while sparing normal cells is anticipated to exhibit a sufficiently wide therapeutic index to avoid mechanism-based toxicity observed with first-generation SAM-competitive/cooperative PRMT5 inhibitors. Modeling a ∼70-fold selectivity index from the cellular assays is anticipated to maximize target inhibition (>95%–99% reduction of SDMA in MTAP del tumor cells) and antitumor activity while sparing inhibition of PRMT5 in MTAP WT tissues (estimated to be <∼75% reduction of SDMA in nontumor cells; see ref. 13) to avoid on-target PRMT5-mediated toxicities, including anemia, thrombocytopenia, and neutropenia.
One of the most critical questions following the discovery of PRMT5 as a synthetic lethal target in MTAP del cancers from shRNA screens was whether selective inhibition of PRMT5 in MTAP del tumors could elicit objective response and marked reduction of tumor burden at tolerated dose levels. The inhibition of a protein target implicated in the regulation of a cell-essential epigenetic pathway in a genetically defined patient population is largely unprecedented. The antitumor activity of MRTX1719, including regression, across lung cancer, pancreatic cancer, and mesothelioma CDX or PDX models was encouraging preclinically. The partial responses to MRTX1719 across several cancer types demonstrate that objective tumor regression is achievable with this approach and provide initial proof of concept for this therapeutic strategy in MTAP del cancers. Whether distinct tumor types ultimately respond better to MRTX1719 and whether additional biomarkers can be identified that correlate with increased response represent major questions moving forward in this field.
The complete inhibition of PRMT5-dependent SDMA in the neoplastic cells from three MTAP del tumors from patients treated with 200 mg MRTX1719 provides early clinical proof of mechanism for this therapeutic strategy and demonstrates that complete inhibition of SDMA in MTAP del tumor cells, as assessed by an SDMA IHC assay, is achievable. Moreover, the six confirmed objective responses observed in the phase I study demonstrate that the ability of MRTX1719 to achieve apparent complete SDMA inhibition at well-tolerated doses translates to objective tumor response, a phenomenon that was infrequently observed for first-generation PRMT5 inhibitors (14). Importantly, none of the patients treated with MRTX1719 experienced dose-limiting adverse events associated with non–MTAP del–selective PRMT5 inhibitors, such as thrombocytopenia, anemia, or neutropenia, at dose levels up to 400 mg q.d. This suggests that a PRMT5 inhibition threshold associated with a functional impact on MTAP WT normal cell populations was not exceeded in these patients. These data suggest that MRTX1719 is well positioned to test the MTAP del synthetic lethal hypothesis by maximizing the anticipated therapeutic index via sparing MTAP WT normal cells and enabling maximal, if not complete, inhibition of PRMT5 in MTAP del tumors.
Given the unique mechanism of action of MRTX1719, the relatively delayed kinetics of the pharmacodynamic and antitumor response in nonclinical models and patients was noteworthy. These observations likely have implications for the present dataset and for the clinical development of MRTX1719. Most targeted therapies are designed to block oncogenic driver pathways, resulting in rapid target inhibition (within hours). Rapid target inhibition kinetics of oncogenic drivers exhibit a near-instantaneous impact on cell proliferation and survival pathways and subsequent initiation of apoptotic pathways, often leading to objective tumor regression (e.g., RECIST response) after only weeks of therapy. In contrast to oncogene-directed therapies, MRTX1719 targets an epigenetic pathway that is crucial for basic cellular functions that, when inhibited, may result in differential and potentially slower acting tumor response kinetics. Our data demonstrate that complete or near-complete reduction of PRMT5-dependent SDMA was required for maximal antitumor activity. Maximal reduction of SDMA required treatment with MRTX1719 for several days in vitro and 2 weeks in vivo. Secondary effects of MRTX1719 treatment on cellular function, including disrupted RNA splicing as well as modulation of key cell-cycle regulators and apoptotic proteins, were then observed subsequent to maximal SDMA modulation. PRMT5-dependent mechanistic effects impacting cell viability, growth, proliferation, and survival were thus observed within weeks of treatment initiation as opposed to hours. These findings likely provide insight into the delayed onset of the tumor response in the nonclinical models, which also required several days or weeks until the maximal antitumor response was observed. In the MRTX1719 clinical trial, four patients were on therapy for several cycles (21 days each) before experiencing an objective response (i.e., by RECIST 1.1 criteria) with their tumors slowly decreasing in size over the entire course of their treatment. These findings suggest that tumor response may continue to deepen over time, and it will be of interest to continue to evaluate tumor response kinetics following treatment with MRTX1719 across a broader patient population with longer patient follow-up. In addition, the duration of treatment will be of major importance, as CDKN2A/MTAP deletion is an early driver event that is clonal in most cancers and mechanisms of resistance to MRTX1719 are unknown and may be more elusive.
The preclinical and early clinical proof-of-concept data provided herein support the adoption of synthetic lethal strategies selectively targeting PRMT5 and the development of MRTX1719 in cancers harboring MTAP deletions. The partial responses observed in six patients with advanced cancer demonstrate that MRTX1719 may represent a new therapeutic strategy for the treatment of multiple cancer types that are in dire need of additional therapies. MTAP gene deletions encompass several cancer types for which few, if any, biomarker-guided targeted therapies currently exist. The detailed pharmacologic and mechanistic characterization of MRTX1719 demonstrates that blocking PRMT5 complexed with MTA is a rational approach for targeting the large and diverse population of patients with cancer with this previously untargetable genetic alteration.
METHODS
Chemical Compounds
MRTX1719 was synthesized at Wuxi AppTec (lot numbers EW14002-1215-p1, EW14002-1236-P1, EW24742-131-P1, EW24742-138-P1, EW24742-154-p1, EW28124-11-P1, EW4257-2945-p1, EW8101-1782-P2, and EW8101-1848-P1; average purity 97.3%). JNJ-64619178 (HY-101564) and GSK3326595 (HY-101563) were purchased from MedChemExpress. All compounds in powder form were stored desiccated at room temperature and protected from light. For in vitro experiments, all compounds were formulated in 100% DMSO, aliquoted, and stored at −20°C.
Biochemical Assay
PRMT5 biochemical studies were conducted using the FlashPlate system at Reaction Biology Corporation using the PMRT5/MEP50 complex (#HMT-22-148: Human recombinant PRMT5 [GenBank Accession No NM_006109; amino acids 2–637 (end)], with N-terminal Flag-Tag and Human recombinant MEP50 [Gene GenBank Accession No. NM_024102; amino acids 2–342 (end)], with N-terminal His tag, coexpressed in Sf9 cell expression system). The FlashPlate substrate was a Biotin-Histone H4 (1–15) peptide (Rockland #000-006-K75). The methyl donor was S-Adenosyl-L-[methyl-3H]methionine (PerkinElmer #NET155H001MC). The reaction buffer was 50 mmol/L Tris-HCl (pH 8.5), 0.002% Tween20, 0.005% bovine serum albumin (BSA), 1 mmol/L TCEP, and 1% dimethyl sulfoxide (DMSO).
The final reaction conditions consisted of 3 nmol/L PRMT5/MEP50, 40 nmol/L Histone H4 (1–15)-Biotin peptide, and 1 μmol/L SAM. Substrate was prepared in a reaction buffer. Enzyme was delivered into the substrate solution at 40 μL/well. Compounds were delivered in DMSO into the reaction mixture and preincubated for 20 minutes at room temperature. Tritiated SAM was delivered into the reaction mixture to initiate the reaction at 10 μL/well, and the reaction was incubated for 2 hours at room temperature. To stop the reaction, 10 μL/well of 600 μmol/L cold SAM was delivered, and 50 μL/well of stopped reaction was transferred into the FlashPlate and incubated for 1 hour at room temperature. Plates were washed with PBS and counted on a TopCount HTS Microplate Scintillation and Luminescence Counter (Packard Instrument Company). A 10-dose IC50 with 3-fold serial dilution starting at 1 μmol/L was run in the presence or absence of 2 μmol/L MTA.
Raw data were converted to percentage of enzyme activity, transformed data were curve fit, and IC50 values were calculated using GraphPad [Prism; Sigmoidal dose–response (variable slope), bottom = 0, top <120].
Cell Lines
HCT116 parental cells were obtained from the American Type Culture Collection (CCL-247, RRID: CVCL_0291). The LU99 cell line was obtained from RIKEN BioResource Research Center (RCB1900, RRID: CVCL_3015). All cell lines were cultured in their recommended media and purchased between 2015 and 2021. Banked cell stocks were short tandem repeat authenticated and IMPACT tested at IDEXX. Revived cells were thawed and scaled up with minimal passaging for in vivo studies. Revived cells used for in vitro studies were not in culture longer than 8 weeks. All cell lines in culture were tested monthly using MycoAlert (#LT07-318) purchased from Lonza.
In-Cell Western Assay
Anti-dimethyl-Arginine, symmetric (SYM11, #07-413, RRID: AB_310595) was purchased from Sigma-Aldrich. IRDye 800CW goat anti-mouse (#926-32210, RRID: AB_621842) was purchased from LI-COR. Methanol-free formaldehyde (#28906) and DRAQ5 (#62251) were obtained from Thermo Scientific.
To prepare assay plates for the SYM11 In-Cell Western assay, cells were trypsinized and resuspended in fresh media, and viable cells were counted utilizing a Cellometer Mini (Nexcelom) and trypan blue exclusion. Cells were diluted in complete growth media and seeded at 2,000 cells/well in 96-well, black-walled, clear bottom assay plates. PBS was added to the outer wells of each 96-well plate. Cells were incubated for 24 hours at 37°C 5% CO2 in 100% humidity to adhere prior to treatment. MRTX1719 was serially diluted (1:4) in DMSO. A 10× dosing plate was prepared from the DMSO serial dilutions using an intermediate dilution (1:100) into complete growth media. Cells were dosed with 10 μL of the 10× intermediate drug dilutions added to the 96-well cell plates. Six DMSO vehicles and six positive control (200 nmol/L JNJ-64619178) wells were included on each assay plate. After 4-day drug treatment, cells were fixed by adding freshly prepared 4% formaldehyde to each well and incubated for 20 minutes at room temperature. Formaldehyde was removed, and cells were permeabilized with ice-cold methanol for 10 minutes overnight at −20°C. Following permeabilization, methanol was removed from each well and Odyssey blocking buffer (#927-50000; LI-COR) with 0.05% Tween-20 was added to each well and incubated for 1 hour at room temperature. Blocking buffer was removed, and the SYM11 primary antibody (1:500 in blocking buffer) was added and incubated overnight at 4°C on a rocking platform. Plates were then washed 3 times with PBS + 0.1%Tween 20 (PBST) and incubated with LI-COR IRDye 800CW secondary antibody (1:800) and DRAQ5 (1:10,000) in blocking buffer for 2 hours at room temperature. Plates were washed 3 times with PBST and then imaged using the LI-COR Odyssey CLx Imaging system set to Plate Acquisition format for both the 700- and 800-nm wavelength channels to measure the signal intensity from each well for the DRAQ5 and goat anti-rabbit IRDye 800CW secondary antibody, respectively. Signal outputs were exported to Excel for analysis. To quantify the fluorescence intensity for each selected well in the In-Cell Western, IRDye 800CW secondary antibody–alone control wells were subtracted from the SDMA signal (800 nm) to remove the background signal. The background-subtracted SDMA signal was divided by the DRAQ5 signal (700 nm) to normalize for cell number. The average normalized SDMA signal from the JNJ-64619178 control wells was subtracted from the normalized SDMA signal. Percent inhibition values were calculated by dividing normalized values from each treated well by the average of the normalized values in the vehicle-treated wells and multiplying by 100. The percentage of vehicle control values was plotted as log(inhibitor) vs. response – Variable slope (four parameters) for curve fitting and IC50 value determination in GraphPad Prism.
10-Day Viability Assay
To prepare assay plates for viability assays, cells were trypsinized and resuspended in fresh media, and viable cells were counted utilizing a Cellometer Mini and trypan blue exclusion. Cells were diluted in complete growth media and seeded at 250 cells/well in 96-well, clear round-bottom plates (#3358 Corning). PBS was added to the outer wells of each 96-well plate. After seeding, cell plates were incubated for 24 hours at 37°C 5% CO2 in 100% humidity to adhere prior to treatment. The following day, MRTX1719 was serially diluted (1:3) in 100% DMSO, and a 10× dosing plate was prepared from the DMSO serial dilutions using an intermediate dilution (1:100) into complete growth media. Cells were dosed with 10 μL of the 10× intermediate drug dilutions added to the 96-well cell plates. Six vehicle (DMSO) control wells were included on each assay plate. After drug treatment for 5 days, cells were rinsed with PBS, trypsinized, and split 1:20 into a new 96-well plate containing fresh media and 1× drug with a final volume of 100 μL per well. After 5 additional days of treatment, cell plates were equilibrated to room temperature, 30 μL of CTG Luminescent Cell Viability Assay (#G7573, Promega) was added to each well, plates were covered in aluminum foil to protect from light, and incubated at room temperature for 30 minutes on a microtiter plate shaker and day-10 luminescence readings were collected using a CLARIOstar microplate reader (BMG Labtech). Percent inhibition values were calculated by dividing relative luminescence unit (RLU) values from each treated well by the average RLU values in the vehicle-treated wells and multiplying by 100. The percentage of vehicle control values was plotted as log(inhibitor) vs. response – Variable slope (four parameters) for curve fitting and IC50 value determination in GraphPad Prism.
5-Day Viability Assay
A panel of MTAP WT and MTAP del human cancer cell lines was screened in a 5-day viability assay. Optimized cell numbers were plated for each cell line and incubated overnight to adhere prior to treatment. Baseline CTG readings were taken on the day of dosing and subtracted from day-5 CTG values prior to IC50 calculation, as described for the 10-day viability assay. Statistical significance was determined using a two-tailed Student t test comparing IC50 values in MTAP WT and MTAP del cancer cell lines assuming equal variance.
Immunoblotting
Pierce radioimmunoprecipitation assay (RIPA) lysis buffer (#89901), 10× HALT (#PI78443), and phenylmethanesulfonyl fluoride (PMSF; #ICN19538105) were purchased from Thermo Fisher. Sodium orthovanadate (#S6508-10G) was purchased from Sigma-Aldrich. The following antibodies were used at the indicated dilution: SDMA (1:1000; Cell Signaling Technology #13222; RRID: AB_2714013), β-Actin (1:2,000; Abcam #ab8226; RRID: AB_306371), phospho-Rb (Ser807/811; 1:500; Cell Signaling Technology #8516; RRID: AB_11178658), p21 (1:500; Cell Signaling Technology #2947; RRID: AB_823586), p27 (1:500; Cell Signaling Technology #3686; RRID: AB_2077850), phospho–Histone H2A.X (1:500; Cell Signaling Technology #2577; RRID: AB_2118010), IRDye 680RD goat anti-rabbit (1:10,000; LI-COR #926-68071; RRID: AB_10956166), IRDye 800CW goat anti-mouse (1:10,000; LI-COR #926-32210; RRID: AB_621842), and goat anti-rabbit IgG (H+L)–HRP conjugate (1:3,000; Bio-Rad #1706515; RRID: AB_11125142).
In studies in which tumors were collected 4 hours after last dose, mice were humanely sacrificed and tumors were surgically removed, cut into fragments, and transferred to a 2 mL screw cap tube containing ceramic beads (Lysing Matrix A, #6910100; MP Biomedicals) and immediately snap frozen in liquid nitrogen. Frozen tumor fragments were stored at −80°C.
In studies in which end-of-study bone marrow was collected 4 hours after last dose, mice were humanely sacrificed and one femur was surgically removed. The top of the femur was cut off, placed upside down in a 0.2-mL Eppendorf tube with a small hole cut into the bottom end, placed inside a 1.5 mL Eppendorf tube, and centrifuged for 30 seconds at 4,000 RPM and 4°C. The femurs were flushed with 1 mL of cold PBS and collected into the 1.5-mL Eppendorf tubes containing the pelleted bone marrow. The pellet was gently disrupted, washed with ice-cold PBS, and centrifuged again for 15 seconds at >10,0000 RPM and 4°C. The PBS wash was aspirated before snap freezing the pelleted bone marrow in liquid nitrogen and stored at −80°C until further processing was performed.
An equal volume of ice-cold RIPA lysis buffer supplemented with 1× HALT, 1 mmol/L PMSF, and 1 mmol/L sodium orthovanadate was added (∼300 to 650 μL), and tumors were homogenized using the MP FastPrep-24 homogenizer (MP Biomedicals) with high-speed shaking 3 to 5 times for 20 seconds while keeping the tumor lysate on ice between cycles. After homogenization, tubes were spun at 15,000 RPM for 10 minutes at 4°C, and supernatant was collected.
Mouse bone marrow pellets were lysed with an equal volume of RIPA lysis buffer supplemented with 1× HALT, 1 mmol/L PMSF, and 1 mmol/L sodium orthovanadate on ice with gentle agitation. Following complete lysis, tubes were spun at 15,000 RPM for 10 minutes at 4°C, and supernatant was collected. Protein concentrations of each lysate sample were determined using a Pierce BCA protein assay kit (#PI23227; Thermo Fisher) per the manufacturer's instructions.
Approximately 30 μg of total protein was added to 4× sample loading buffer (#161-0791; Bio-Rad) and 10× reducing agent (#NP0009; Invitrogen). Samples were boiled for 5 minutes. Processed samples were loaded onto either 12% Bis-Tris 26-well or 18-well gels (#3450119/#3450118; Bio-Rad) using MOPS running buffer (#161-0788; Bio-Rad) or NuPAGE 10% or 4%–12% Bis-Tris Midi Protein 20-well gels (WG1202BOX/WG1402BOX; Invitrogen) using NuPAGE MES SDS 20× Running Buffer (NP0002; Invitrogen). Proteins were transferred from the gels to a nitrocellulose membrane using the iBlot 2 Dry Blotting System (#IB23001; Thermo Fisher) and run at 20 volts for 1 minute, 23 volts for 4 minutes, and 25 volts for 2 minutes.
Membranes were blocked with LI-COR Intercept TBS Blocking Buffer (#927-60001; Thermo Fisher) for 1 hour at room temperature on a rocking platform. Primary antibodies were diluted in LI-COR Blocking Buffer and incubated overnight at 4°C on a rocking platform. Membranes were then washed 3 times for 10 minutes with Tris-buffered saline-Tween 20 (TBS-T) and incubated with LI-COR IRDye secondary antibodies for 1 hour at room temperature. Membranes were washed 3 times for 10 minutes with TBS-T.
Images were acquired from probed nitrocellulose membranes using the LI-COR Odyssey CLx Imaging System (LI-COR) set to the AutoScan channel for both the 700- and 800-nm wavelength channels to measure the signal intensity from the IRDye 680RD goat anti-rabbit and IRDye 800CW goat anti-mouse secondary antibodies, respectively. Images were imported into LI-COR's Image Studio software version 4.0 and then .tif files were exported for annotation. To quantify the pixel intensity for each selected protein band, the Add Rectangle tool in the image viewer was used to identify a consistently sized area of interest for each band of a given target protein as well as a representative background region of the immunoblot. The signal output column from the software subtracts background pixel intensity and is used to determine the target pixel intensity for each protein band. This corrected signal intensity was determined for each target protein of interest, and data were exported to Excel. The target protein normalization of each sample was determined by dividing the signal output of the target protein by the signal output of the loading control protein. Each target protein was also averaged within each vehicle or treatment group. The vehicle value was normalized to 1 by dividing all average values by the vehicle value, and standard deviation was calculated from the normalized values. Percent inhibition of normalized SDMA in MRTX1719-treated tumors compared with vehicle-treated control tumors was calculated by dividing the average MRTX1719-treated normalized SDMA signal by the average vehicle-treated normalized SDMA signal and multiplying by 100. GraphPad Prism 8 was used to graph the data.
Statistical significance of SDMA densitometry in tumor and bone marrow following drug treatment was determined using a two-tailed Student t test equation in Microsoft Excel comparing normalized SDMA of vehicle samples to treated samples assuming equal variance.
HemaTox Erythroid and Myeloid Differentiation Assessment
At the time of assay setup, one prequalified lot of frozen bone marrow CD34+ cells was thawed and viability assessed. CD34+ cells were seeded in 96-well culture plates at 1,000 to 3,000 cells/well in the presence of a test or control article. Control cultures containing an equivalent amount of solvent (solvent control), but no test or control article, were also initiated. Three replicates were initiated for each condition. The culture plates were incubated at 37°C, 5% CO2 for 7 days. After the appropriate incubation period, a portion of the cells from each well was stained to evaluate live erythroid cells (CD71 and CD235A/Glycophorin A) or live myeloid cells (CD15 and CD13) by flow cytometric analysis. The total number of live erythroid (CD71+CD235A−, CD71−CD235A+, and CD71+CD235A+) or myeloid (CD13+CD15−, CD13−CD15+, and CD13+CD15+) cells in the test and control article–treated cultures was compared with the solvent control cultures to determine the percent of control growth and the IC50 values for each compound.
PTMScan
In vitro LU99-treated cells for PTMScan analysis were generated in 150-mm dishes with cell densities that ensured subconfluent plates following treatment. Cells were treated for 3 or 5 days with DMSO or MRTX1719 (1 μmol/L) and collected in urea lysis buffer following the manufacturer's recommended protocol (Cell Signaling Technology). PTM enrichment and analysis were performed by Cell Signaling Technology (Supplementary Table S2).
Cell-Cycle Assay
For cell seeding, LU99 cells were seeded in 10-cm dishes at 6e5 cells/plate. After seeding, cell plates were incubated overnight to adhere prior to treatment. Cells were treated for 3 days and then trypsinized and counted. Cells were fixed and stained following the Luminex Guava Cell Cycle Reagent (#4500-0220) manufacturer's protocol. Cells were fixed in a 15-mL conical tube by rinsing cells with PBS and then resuspending cells in 1 mL of ice-cold 70% ethanol per 1e6 cells while vortexing at medium speed. For cell staining, 200 μL of the fixed cell suspension was moved to a 96-well, round-bottom plate and rinsed with 200 μL of PBS twice prior to staining with 200 μL of room temperature Guava Cell Cycle Reagent. Cells were incubated at room temperature in the dark for 30 minutes prior to running samples on the Guava EasyCyte System. For each sample, 5,000 events were acquired and GuavaSoft Software was used to analyze the data. The stained negative control (DMSO-treated) sample was used to set the gate for the assay. Statistical significance was determined using a two-tailed Student t test equation in Microsoft Excel comparing vehicle samples to treated samples assuming equal variance for indicated cell-cycle populations.
Annexin V Apoptosis Assay
LU99 cells were seeded in 6-well plates. DMSO-treated wells were seeded at 3e4 cells/well, and drug-treated wells were seeded at 5e4 cells/well. After seeding, cell plates were incubated overnight to adhere prior to treatment. Cells were treated for 3 days and then trypsinized, counted, and normalized to 4e5 cells/mL. Following cell preparation, cells were stained following Luminex Guava Nexin Reagent (#4500-0450) manufacturer's protocols. The cell suspension (100 μL) was added to a 96-well plate along with 100 μL of room temperature Nexin Reagent. Cells were incubated at room temperature in the dark for 20 minutes prior to running samples on the Guava EasyCyte System. For each sample, 5,000 events were acquired and GuavaSoft Software was used to analyze the data. The stained negative control (DMSO- treated) sample was used to set the gate by adjusting the quadrant markers for the Annexin V–PE versus 7-AAD dot plot and applied to the rest of the samples. Statistical significance was determined using a two-tailed Student t test equation in Microsoft Excel comparing vehicle samples to treated samples assuming equal variance.
RNA-seq
LU99 cells were treated with DMSO or MRTX1719 (1 μmol/L) for 3 or 5 days. Cells were rinsed with ice-cold PBS, and cell pellets were snap frozen. RNA extraction and RNA sequencing were performed by Genewiz, and data files were transferred to Monoceros Biosciences LLC for bioinformatic analysis.
RNA-seq Preprocessing
The reads were trimmed, and low-quality reads were removed using Trimgalore er.0.6.3 dev (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) with the “paired” parameter and length of 151 bps. Trimmed fastq sequences were aligned to the human reference genome (GRCh38; ref. 26) using STAR aligner v2.7.1a (27), with the produced bam files sorted by coordinate by using option “–outSAMtype BAM SortedByCoordinate.” Raw read gene counts were obtained using STAR aligner with options “–quantMode GeneCounts” and “–sjdbGTFfile,” with gene models in GTF format obtained from EnsEMBL release 83. Alignment QC (quality control) and read mapping statistics were obtained from Picard v2.20.3 tools using the function “CollectMultipleMetrics” (http://broadinstitute.github.io/picard/).
RNA-seq Data Analysis
Raw gene counts were used for quality control and transformed to counts per million (CPM) for downstream differential expression analysis. The “prcomp” function in the stats R library was applied for the principal component analysis (PCA). One DMSO day-5 sample was determined to be an outlier based on a distinct distribution of GC content and expression distribution compared with all other samples and was removed from downstream analyses. After removing the outlier, all samples clustered according to treatment in the PCA and the DMSO vehicles were selected as controls. Differential expression analysis was performed with limma-trend (version 3.40.6; ref. 28) in R (version 3.6.1). Very lowly expressed genes with a mean of Log2(CPM) smaller than −2 were eliminated. GSEA (gsea v2.2) and the Molecular Signature Database (MSigDB; msigdb_v7.04) were used for gene enrichment analysis (29).
Differential Splicing Analysis
RNA sequence alignment files (BAM) were used as input to rMATS version 4.1.0. rMATS is a computational tool designed to identify differential alternative splicing events across samples being compared (30). A custom set of scripts was used to automate the execution of rMATS and the summarization of the results in Excel files. Five types of alternative splicing events were identified by rMATS: alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), mutually exclusive exon usage (MXE), retained intron (RI), and skipped exons (SE). A summary sheet highlighting genes affected by 1 or many differential splicing events was generated. Additional sheets with detailed information for each of the alternative splicing event identified by rMATS was also generated as part of the analysis workflow (http://rnaseq-mats.sourceforge.net).
Custom R code using the ggplot graph library was used to generate additional visualizations from the analysis results. A box plot showing the ΔΨ (differential percent spliced in) for all events grouped by the splice event type was generated to highlight the enrichment of any event type in any of the groups being compared. A stacked column plot was also generated to visualize the total number of alternative spliced events identified in each of the comparison groups.
In Vivo Studies
All mouse studies were approved by the Institutional Animal Care and Use Committee based on guidelines from the NIH (Explora protocol EB17-010-049). Mice were maintained under pathogen-free conditions, and food and water were provided ad libitum.
Six- to 8-week-old female Hsd:Athymic Nude-Foxn1nu mice (Envigo) were injected subcutaneously with tumor cells in 100 μL of PBS and Matrigel matrix (Corning #356237) in the right hind flank of each mouse with 5e6 cells (LU99) or 1e6 cells (HCT116 parental and HCT116 MTAP del) 50:50 cells:Matrigel. Mouse health was monitored daily, and caliper measurements began when tumors were palpable. Tumor volume measurements were determined utilizing the formula 0.5 × L × W2, in which L refers to the length and W refers to the width of each tumor. When tumors reached the desired average study start tumor volume (LU99: 179 mm3 or 116 mm3; HCT116 MTAP WT: 140 mm3; HCT116 MTAP del: 185 mm3), mice were randomized into treatment groups. MRTX1719 was formulated in 0.5% methylcellulose (4,000 cps) + 0.2% Tween80 in water once per week and stored at room temperature protected from light. GSK3326595 was formulated in 0.5% methylcellulose (4,000 cps) + 0.2% Tween80 in water once per week and stored at 4°C protected from light. JNJ-64619178 was formulated in 20% HP-β-CD in water once per week and stored at room temperature protected from light. Mice were orally administered vehicle, MRTX1719, GSK3326595, or JNJ-64619178 at the indicated doses and schedules. Mice were monitored daily, with tumor volumes and body weights measured 2 or 3 times per week.
The PXF 537, PXF 2197, PXF 1118, PXF 541, and PXF 2442 mesothelioma PDX model experiments were conducted at Charles River with similar study designs to the LU99 and HCT116 tumor models. Briefly, NMRI nu/nu mice were inoculated subcutaneously with a 2- to 3-mm3 PDX fragment and randomized for treatment initiation when the mean tumor volumes reached ∼120 to 140 mm3. Mice were treated with vehicle or MRTX1719 (n = 3 per group) at the indicated dose and duration.
Percent tumor growth inhibition (% TGI) was calculated using the following formula: [1 − (final drug-treated tumor volume – initial drug-treated tumor volume)/(final vehicle-treated tumor volume – initial vehicle-treated tumor volume)] × 100.
Human Tumor IHC
IHC was performed on formalin-fixed, paraffin-embedded pretreatment and cycle 2 day 1 biopsies, sectioned at 4-μm thickness onto positively charged glass slides. Serial tissue sections were stained for MTAP with an anti-MTAP antibody (Abcam #ab55517; RRID:AB_944282) or SDMA using a rabbit MultiMab SDMA antibody (Cell Signaling Technology #13222, RRID: AB_2714013) and automated detection on a Dako Link 48 autostainer at Mosaic Laboratories.
MTAP and SDMA staining was evaluated by a pathologist with a semiquantitative scale, and the percentage of neoplastic, stromal, and other cell type staining at each of the following four intensity levels was recorded: 0 (unstained), 1+ (weak staining), 2+ (moderate staining), and 3+ (strong staining). An H-score was calculated based on the summation of the product of the percent of cells stained at each intensity using the equation: (3 × % of “3+” cells) + (2 × % of “2+” cells) + (1 × % of “1+” cells).
Statistical Analysis
The statistical significance of TGI was determined using two-way ANOVA calculated in GraphPad Prism 8 (GraphPad). Significance was noted in tumor growth plots with “*” for Dunnett multiple comparisons test with an adjusted P < 0.05 between vehicle and treated tumor volume measurements on the last day of treatment. Statistical significance of pharmacodynamic (in vitro and in vivo) and in vitro functional effects was determined using a two-tailed Student t test equation comparing vehicle samples to treated samples assuming equal variance.
Data Availability
The RNA-seq data generated in this study are publicly available in the Gene Expression Omnibus (GEO) at GSE240145.
Supplementary Material
Materials and methods related to supplemental data
Hematological parameters in CD-1 mice treated with MRTX11719
PTMScan data tables
5-Day in vitro viability data
Treatment-related treatment-emergent adverse events by preferred term and maximum grade safety population
SDMA and MTAP IHC TMA data
Supplemental biochemical and cellular characterization of MRTX1719
Thermal shift PRMT5 inhibitor data
Supplemental in vivo pharmacodynamic characterization of MRTX1719
Co-culture experiments with MTAP-expressing fibroblasts
Supplemental in vitro and in vivo efficacy of MRTX1719
Supplemental pharmacodynamic effects of MRTX1719 in patients
Acknowledgments
The authors thank Reaction Biology Corporation and BPS Biosciences for biochemical assay support, Crown Biosciences and Charles River Laboratories for mouse tumor xenograft studies, STEMCELL Technologies for performing the HemaTox assays, Cell Signaling Technology for PTMScan SDMA and PTMScan Multi-Pathway Enrichment data, Mosaic (CellCarta) for the SDMA and MTAP IHC, and all patients participating in the 1719-001 clinical trial. Mirati Therapeutics provided all of the funding for this study. Funding from Mirati Therapeutics was not provided through a grant.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
V. Bowcut reports other support from Mirati Therapeutics, Inc. during the conduct of the study; other support from Mirati Therapeutics, Inc. outside the submitted work; and a patent for combination therapies using PRMT5 inhibitors pending. L. Vegar reports personal fees and other support from Mirati Therapeutics, Inc. during the conduct of the study, as well as personal fees and other support from Mirati Therapeutics, Inc. outside the submitted work. A. Blaj reports personal fees from Mirati Therapeutics, Inc. during the conduct of the study. J. Rodon reports nonfinancial support and reasonable reimbursement for travel from the European Society for Medical Oncology; consulting and travel fees from Peptomyc, Kelun Pharmaceuticals/Klus Pharma, Ellipses Pharma, Molecular Partners, and IOnctura (including serving on the scientific advisory board); consulting fees from Vall d'Hebron Institute of Oncology/Ministero De Empleo Y Seguridad Social, Chinese University of Hong Kong, Boxer Capital, LLC, and Tang Advisors, LLC; research funding from Blueprint Medicines, Black Diamond Therapeutics, Merck Sharp & Dohme, Hummingbird, Yingli, and Vall d'Hebron Institute of Oncology/Cancer Core Europe; and serving as investigator in clinical trials with Novartis, Spectrum Pharmaceuticals, Symphogen, BioAlta, Pfizer, GenMab, CytomX, Kelun-Biotech, Takeda/Millenium, GSK, Taiho, Roche Pharmaceuticals, Hummingbird, Yingli, Bicycle Therapeutics, Merus, Curis, Bayer, AadiBioscience, Nuvation, ForeBio, BioMed Valley Discoveries, Loxo Oncology, Hutchinson MediPharma, Cellestia, Deciphera, Ideaya, Amgen, Tango Therapeutics, Mirati Therapeutics, Inc., Linnaeus Therapeutics, and Cancer Core Europe. M. Offin is an uncompensated scientific advisory board member for the Mesothelioma Applied Research Foundation, has received the LUNGevity Foundation Career Development Award, and within the last 36 months has received an honorarium from Pfizer, Jazz Pharmaceuticals, and Astro. K.C. Arbour reports other support from Mirati Therapeutics, Inc. during the conduct of the study, as well as personal fees from Amgen, Novartis, AstraZeneca, G1 Therapeutics, and Sanofi/Genzyme, and other support from Revolution Medicines and Genentech outside the submitted work. M.L. Johnson reports grants from Mirati Therapeutics, Inc. during the conduct of the study, as well as grants and other support from AbbVie, Amgen, Apexigen, Arcus Biosciences, AstraZeneca, Black Diamond, Boehringer Ingelheim, Calithera, Daiichi Sankyo, Genentech/Roche, GenMab, Genocea Biosciences, GSK, Gritstone Oncology, Ideaya Biosciences, Mirati Therapeutics, Inc., Merck, Janssen, Immunocore, Novartis, Regeneron Pharmaceuticals, Revolution Medicines, Sanofi, Takeda Pharmaceuticals, and Turning Point Therapeutics, grants from Acerta, Adaptimmune, Array BioPharma, Artios Pharma, Atreca, BeiGene, BerGenBio, BioAtla, Bristol Myers Squibb, Carisma Therapeutics, Checkpoint Therapeutics, City of Hope National Medical Center, Corvus Pharmaceuticals, Curis, CytomX, Dracen Pharmaceuticals, Dynavax, Lilly, Elicio Therapeutics, EMD Serono, EQRx, Erasca, Exelixis, Fate Therapeutics, Guardant Health, Harpoon, Helsinn Healthcare SA, Hengrui Therapeutics, Hutchinson MediPharma, IGM Biosciences, Immunitas Therapeutics, Incyte, Jounce Therapeutics, Kadmon Pharmaceuticals, Kartos Therapeutics, Loxo Oncology, Lycera, Memorial Sloan Kettering, Merus, Mythic Therapeutics, NeoImmune Tech, Neovia Oncology, Numab Therapeutics, Nuvalent, OncoMed Pharmaceuticals, Palleon Pharmaceuticals, Pfizer, PMV Pharmaceuticals, Rain Therapeutics, RasCal Therapeutics, Relay Therapeutics, Ribon Therapeutics, Rubius Therapeutics, Seven and Eight Biopharmaceuticals/Birdie Biopharmaceuticals, Shattuck Labs, Silicon Therapeutics, Stem CentRx, Syndax Pharmaceuticals, Tarveda, TCR2 Therapeutics, Tempest Therapeutics, Tizona Therapeutics, Tmunity Therapeutics, University of Michigan, Vyriad, WindMIL Therapeutics, and Y-mAbs Therapeutics, and other support from Arrivent, Astellas, EcoR1, Gilead Sciences, iTeos, Jazz Pharmaceuticals, Molecular Axiom, Normunity, Oncorus, Pyramid Biosciences, Seagen, Synthekine, and VBL Therapeutics outside the submitted work. P.A. Jänne reports personal fees from Mirati Therapeutics, Inc. during the conduct of the study; grants and personal fees from AstraZeneca and Boehringer Ingelheim, personal fees from Pfizer, Roche/Genentech, Chugai Pharmaceuticals, Eli Lilly, SFJ Pharmaceuticals, Voronoi, Daiichi Sankyo, Biocartis, Novartis, Sanofi, Takeda Oncology, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Eisai, Allorion Therapeutics, Accutar Biotech, AbbVie, Monte Rosa, Scorpion Therapeutics, Merus, Frontier Medicines, Hongyun Biotechnology, and Duality Biologics, and grants from PUMA, Astellas, and Revolution Medicines outside the submitted work; and a patent for EGFR mutations issued and licensed to Labcorp. C.L. Haddox reports other support from Mirati Therapeutics, Inc. during the conduct of the study. K.P. Papadopoulos reports other support from Mirati Therapeutics, Inc. during the conduct of the study, as well as other support from AbbVie, ADC Therapeutics, Amgen, Anheart Therapeutics, Bayer, Daiichi Sankyo, AstraZeneca, F-Star, Incyte, Jounce, Lilly/Loxo, Merck, Mersana, Pfizer, Regeneron, Revolution Medicines, Syros Pharma, Tempest Therapeutics, Treadwell Therapeutics, CytomX Therapeutics, Kezar, Monte Rosa, and Storm outside the submitted work. J.T. Henry reports full or part-time employment with Sarah Cannon Research Institute; stocks/shares in HCA; and his institution (Sarah Cannon Research Institute) receives research funding from the following companies: Abbiscko Therapeutics, ABl Bio, Accutar Biotech, ADC Therapeutics, Agenus, Aileron Therapeutics, Amgen, Artios, AstraZeneca, Bicycle Therapeutics, BioAlta, BioInvent Pharma, Biosplice Therapeutics, Black Diamond Therapeutics, Boehringer, Cyteir, Daiichi Sankyo, Eli Lilly, Epizyme, Erasca, Exelixis, FujiFilm, GSK, Hutchison MediPharma, ICON plc, IGM Biosciences, Immunogen, Ingelheim, Jacobio Pharmaceuticals, Jounce Pharma, Jubilant Therapeutics, Loxo Oncology, Merck & Co, Metabomed, Mirati Therapeutics, Inc., Molecular Templates, Navire Pharma, Nikang Pharmaceuticals, Oncorus, Poseida, Prelude Therapeutics, PureTech, Pyramid, Rascal Therapeutics, Regeneron, Relay Therapeutics, Rgenix, Ribon Therapeutics, Sapience, Sarah Cannon Development Innovations, Sarah Cannon Research Institute, Seagen, Simcha Therapeutics, Siranomics, Stingthera, Synthorx Inc., Takeda Pharmaceuticals, Tallac Therapeutics, Tarveda, Teneothree, Tesaro, Turning Point Pharma, Xencor, and Tango Pharmaceuticals. K. Leventakos reports other support from Mirati Therapeutics, Inc. during the conduct of the study, as well as other support from Amgen, Boehringer Ingelheim Pharmaceuticals, AstraZeneca, Janssen, Jazz Pharmaceuticals, Mirati Therapeutics, Inc., Regeneron, Takeda, Targeted Oncology, OncLive State of the Summit, MJH Life Sciences, AstraZeneca, and Mirati Therapeutics, Inc. outside the submitted work. J.G. Christensen reports other support from Mirati Therapeutics, Inc. during the conduct of the study; other support from Tango Therapeutics outside the submitted work; and patent 20220331323 pending and patent 20220331324 pending. R. Shazer reports other support from Mirati Therapeutics, Inc. during the conduct of the study. P. Olson reports other support from Mirati Therapeutics, Inc. during the conduct of the study, as well as other support from Pfizer and Tango Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
L.D. Engstrom: Conceptualization, resources, formal analysis, supervision, validation, investigation, visualization, methodology, project administration, writing–review and editing. R. Aranda: Resources, formal analysis, investigation, methodology. L. Waters: Resources. K. Moya: Resources. V. Bowcut: Resources. L. Vegar: Resources, investigation. D. Trinh: Resources. A. Hebbert: Resources. C.R. Smith: Supervision, project administration. S. Kulyk: Supervision. J.D. Lawson: Resources, software, supervision. L. He: Resources, software, formal analysis. L.D. Hover: Resources, software, formal analysis. J. Fernandez-Banet: Supervision. J. Hallin: Resources, supervision, methodology. D. Vanderpool: Conceptualization, supervision. D.M. Briere: Conceptualization, supervision, project administration. A. Blaj: Resources, investigation, writing–original draft. M.A. Marx: Supervision, project administration. J. Rodon: Resources, investigation, visualization, methodology, writing–original draft. M. Offin: Resources, investigation, visualization, methodology. K.C. Arbour: Resources, investigation, visualization, methodology, writing–original draft. M.L. Johnson: Resources, investigation, visualization, methodology, writing–original draft. D.J. Kwiatkowski: Resources, investigation, visualization, methodology, writing–original draft. P.A. Jänne: Resources, investigation, visualization, methodology, writing–original draft. C.L. Haddox: Resources, investigation, visualization, methodology, writing–original draft. K.P. Papadopoulos: Resources, investigation, visualization, methodology, writing–original draft. J.T. Henry: Resources, investigation, visualization, writing–original draft. K. Leventakos: Resources, investigation, visualization, methodology. J.G. Christensen: Conceptualization, resources, supervision, project administration. R. Shazer: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. P. Olson: Conceptualization, resources, data curation, formal analysis, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat Rev Drug Discov 2020;19:23–38. [DOI] [PubMed] [Google Scholar]
- 2. Muller FL, Aquilanti EA, DePinho RA. Collateral lethality: a new therapeutic strategy in oncology. Trends Cancer 2015;1:161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gu F, Pfeiffer RM, Bhattacharjee S, Han SS, Taylor PR, Berndt S, et al. Common genetic variants in the 9p21 region and their associations with multiple tumours. Br J Cancer 2013;108:1378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell 2017;65:8–24. [DOI] [PubMed] [Google Scholar]
- 5. Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress 2020;4:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O'Keefe M, et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell 2021;81:3481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 2016;351:1214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep 2016;15:574–87. [DOI] [PubMed] [Google Scholar]
- 9. Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 2016;351:1208–13. [DOI] [PubMed] [Google Scholar]
- 10. Appleby TC, Erion MD, Ealick SE. The structure of human 5′-deoxy-5′-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis. Structure 1999;7:629–41. [DOI] [PubMed] [Google Scholar]
- 11. Brehmer D, Beke L, Wu T, Millar HJ, Moy C, Sun W, et al. Discovery and pharmacological characterization of JNJ-64619178, a novel small molecule inhibitor of PRMT5 with potent anti-tumor activity. Mol Cancer Ther 2021;20:2317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 2015;11:432–7. [DOI] [PubMed] [Google Scholar]
- 13. Feustel K, Falchook GS. Protein arginine methyltransferase 5 (PRMT5) inhibitors in oncology clinical trials: a review. J Immunother Precis Oncol 2022;5:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vieito M, Moreno V, Spreafico A, Brana I, Wang JS, Preis M, et al. Phase 1 study of JNJ-64619178, a protein arginine methyltransferase 5 inhibitor, in advanced solid tumors. Clin Cancer Res 2023; 29:3592–602. [DOI] [PubMed] [Google Scholar]
- 15. Smith CR, Aranda R, Bobinski TP, Briere DM, Burns AC, Christensen JG, et al. Fragment-based discovery of MRTX1719, a synthetic lethal inhibitor of the PRMT5*MTA complex for the treatment of MTAP-deleted cancers. J Med Chem 2022;65:1749–66. [DOI] [PubMed] [Google Scholar]
- 16. Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, et al. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol 2002;159:957–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 2001;7:1531–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, et al. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell 2017;32:411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamard PJ, Santiago GE, Liu F, Karl DL, Martinez C, Man N, et al. PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep 2018;24:2643–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerhart SV, Kellner WA, Thompson C, Pappalardi MB, Zhang XP, Montes de Oca R, et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep 2018;8:9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barekatain Y, Ackroyd JJ, Yan VC, Khadka S, Wang L, Chen KC, et al. Homozygous MTAP deletion in primary human glioblastoma is not associated with elevation of methylthioadenosine. Nat Commun 2021;12:4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan F, Mandrekar SJ, Yuan Y. Keyboard: a novel Bayesian toxicity probability interval design for phase I clinical trials. Clin Cancer Res 2017;23:3994–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 24. Liu F, Cheng G, Hamard PJ, Greenblatt S, Wang L, Man N, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J Clin Invest 2015;125:3532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 2009;16:304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 2017;27:849–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci U S A 2014;111:E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods related to supplemental data
Hematological parameters in CD-1 mice treated with MRTX11719
PTMScan data tables
5-Day in vitro viability data
Treatment-related treatment-emergent adverse events by preferred term and maximum grade safety population
SDMA and MTAP IHC TMA data
Supplemental biochemical and cellular characterization of MRTX1719
Thermal shift PRMT5 inhibitor data
Supplemental in vivo pharmacodynamic characterization of MRTX1719
Co-culture experiments with MTAP-expressing fibroblasts
Supplemental in vitro and in vivo efficacy of MRTX1719
Supplemental pharmacodynamic effects of MRTX1719 in patients
Data Availability Statement
The RNA-seq data generated in this study are publicly available in the Gene Expression Omnibus (GEO) at GSE240145.