Abstract
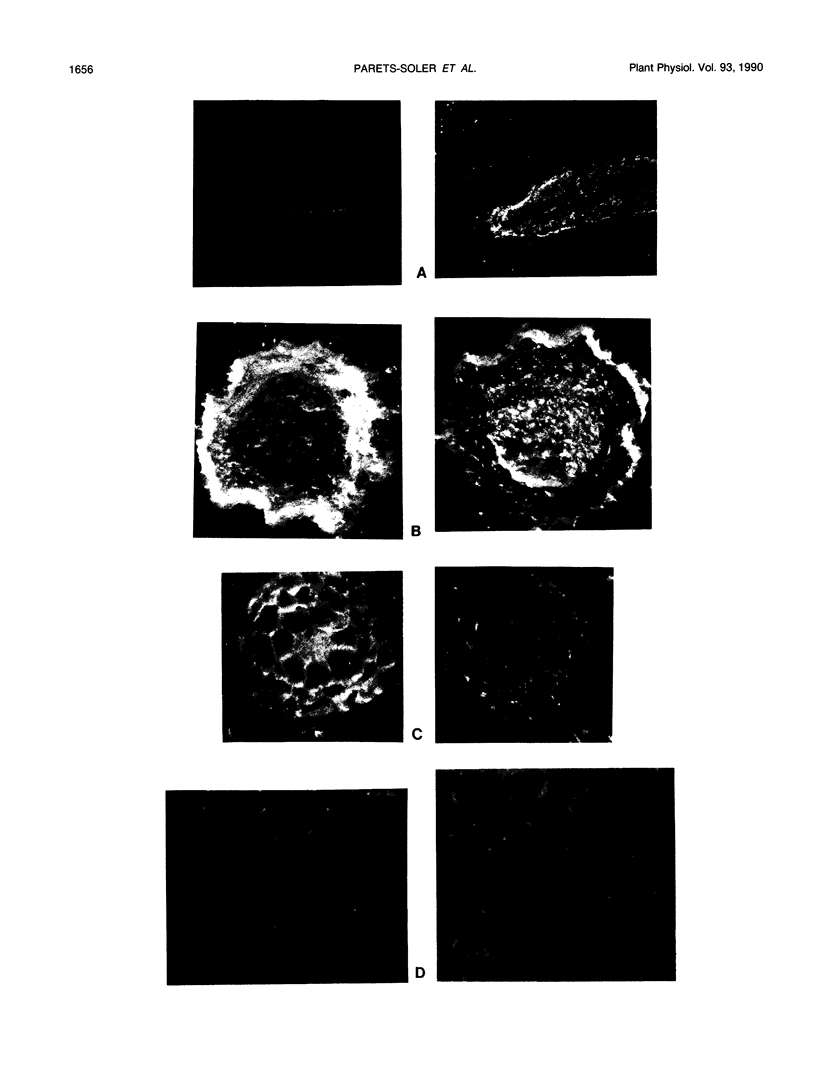
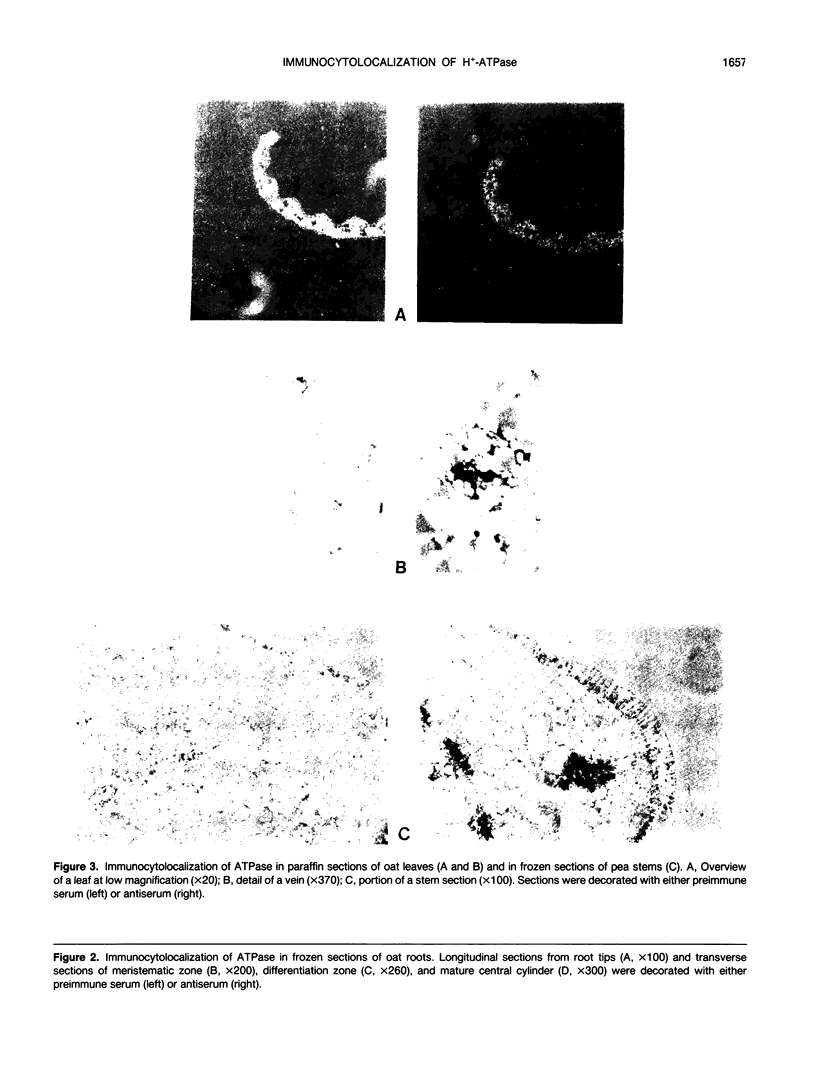
The localization of plasma membrane H+-ATPase has been studied at the optical microscope level utilizing frozen and paraffin sections of Avena sativa and Pisum sativum, specific anti-ATPase polyclonal antibody, and second antibody coupled to alkaline phosphatase. In leaves and stems the ATPase is concentrated at the phloem, supporting the notion that it generates the driving force for phloem loading. In roots the ATPase is concentrated at both the periphery (rootcap and epidermis) and at the central cylinder, including endodermis and vascular cells. This supports a `two-pump' mechanism for ion absorption, involving active uptake at the epidermis, symplast transport across the cortex, and active efflux at the xylem. The low ATPase content of root meristem and elongation zone may explain the observed transorgan H+ currents, which leave nongrowing parts and enter growing tips.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Boutry M., Michelet B., Goffeau A. Molecular cloning of a family of plant genes encoding a protein homologous to plasma membrane H+-translocating ATPases. Biochem Biophys Res Commun. 1989 Jul 31;162(2):567–574. doi: 10.1016/0006-291x(89)92348-6. [DOI] [PubMed] [Google Scholar]
- Gilder J., Cronshaw J. A biochemical and cytochemical study of adenosine triphosphatase activity in the phloem of Nicotiana tabacum. J Cell Biol. 1974 Jan;60(1):221–235. doi: 10.1083/jcb.60.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G., Simons K., Warren G., Tokuyasu K. T. Immunoelectron microscopy using thin, frozen sections: application to studies of the intracellular transport of Semliki Forest virus spike glycoproteins. Methods Enzymol. 1983;96:466–485. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- Hanson J. B. Application of the chemiosmotic hypothesis to ion transport across the root. Plant Physiol. 1978 Sep;62(3):402–405. doi: 10.1104/pp.62.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. F., Surowy T. K., Sussman M. R. Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump (H+-ATPase) of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1234–1238. doi: 10.1073/pnas.86.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. B., Sussman M. R., Mierzwa R. J., Evert R. F. Cytochemical localization of ATPase activity in oat roots localizes a plasma membrane-associated soluble phosphatase, not the proton pump. Plant Physiol. 1988 Mar;86(3):841–847. doi: 10.1104/pp.86.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J. A., Metzler M. C., Nelson T. The argentia mutation delays normal development of photosynthetic cell-types in Zea mays. Dev Biol. 1987 Jul;122(1):243–255. doi: 10.1016/0012-1606(87)90349-6. [DOI] [PubMed] [Google Scholar]
- Pardo J. M., Serrano R. Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J Biol Chem. 1989 May 25;264(15):8557–8562. [PubMed] [Google Scholar]
- Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- Stanley K. K., Luzio J. P. Construction of a new family of high efficiency bacterial expression vectors: identification of cDNA clones coding for human liver proteins. EMBO J. 1984 Jun;3(6):1429–1434. doi: 10.1002/j.1460-2075.1984.tb01988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenseel M. H., Dorn A., Jaffe L. F. Natural H Currents Traverse Growing Roots and Root Hairs of Barley (Hordeum vulgare L.). Plant Physiol. 1979 Sep;64(3):512–518. doi: 10.1104/pp.64.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter-Sluiter E., Läuchli A., Kramer D. Cytochemical Localization of K-stimulated Adenosine Triphosphatase Activity in Xylem Parenchyma Cells of Barley Roots. Plant Physiol. 1977 Dec;60(6):923–927. doi: 10.1104/pp.60.6.923. [DOI] [PMC free article] [PubMed] [Google Scholar]