Abstract
Helicobacter pylori infection is a major risk factor for the development of gastric cancer. The bacteria reside in close proximity to gastric surface mucous as well as stem and progenitor cells. Here, we take advantage of wild-type and genetically engineered murine gastric organoids and organoid-derived monolayers to study the cellular targets of H. pylori–induced DNA damage and replication stress and to explore possible interactions with preexisting gastric cancer driver mutations. We find using alkaline comet assay, single-molecule DNA fiber assays, and immunofluorescence microscopy of DNA repair foci that H. pylori induces transcription-dependent DNA damage in actively replicating, Leucine-rich-repeat containing G-Protein-Coupled Receptor 5 (Lgr5)–positive antral stem and progenitor cells and their Troy-positive corpus counterparts, but not in other gastric epithelial lineages. Infection-dependent DNA damage is aggravated by Apc inactivation, but not by Trp53 or Smad4 loss, or Erbb2 overexpression. Our data suggest that H. pylori induces DNA damage in stem and progenitor cells, especially in settings of hyperproliferation due to constitutively active Wnt signaling.
Stem cells sustain DNA damage upon exposure to Helicobacter pylori that is aggravated by constitutively active Wnt signaling.
INTRODUCTION
Chronic colonization of mucosal surfaces by bacterial pathogens and by certain constituents of a normal gastrointestinal microbiota is increasingly recognized as an early driver of human gastrointestinal carcinogenesis. Examples include Helicobacter pylori colonization of the gastric mucosa leading to gastric adenocarcinoma (1), genotoxin-producing Salmonella enterica associated with gallbladder (2, 3) and colorectal carcinoma (4), and colibactin-producing Escherichia coli and colorectal carcinoma (5–7). All three bacterial species share the ability to damage their target cells’ DNA either directly by the production of genotoxins such as colibactin and typhoid toxin (6–9) or by promoting DNA damage via the induction of transcription/replication conflicts (TRCs) (10). Additional bacterial pathogens, particularly Chlamydia trachomatis, also compromise their host cells’ genomic integrity and are therefore considered candidate bacterial carcinogens (11). The link to cancer is best documented for H. pylori and gastric cancer (12), the third most common cause of cancer-related deaths (13). Chronic infection with H. pylori is considered the main risk factor by far for gastric cancer (12, 14, 15), and eradication therapy has proven an effective prevention strategy in particularly high-risk geographical areas of the world (16, 17). Eradication of H. pylori clearly reduces the risk of progression from gastric cancer precursor lesions (such as chronic or atrophic gastritis) to gastric cancer (16) and may even be beneficial once in situ carcinomas (i.e., limited to the gastric mucosa and submucosa) have been diagnosed (18).
The genetic makeup of the infecting H. pylori strain has emerged as an important determinant of gastric cancer risk. Strains harboring the Cag pathogenicity island, which encodes a type IV secretion system (T4SS) (19), are more tightly associated with gastric cancer than strains lacking the ability to assemble a functional Cag pathogenicity island–encoded T4SS and to deliver the main protein effector of the T4SS, CagA (20). T4SS-positive strains inject not only CagA but also an intermediate of inner core lipopolysaccharide (LPS) biosynthesis, adenosine 5′-diphosphate-β-d-manno-heptose (β-ADP-heptose), into the cytoplasm of target cells, where it binds to an innate immune sensor, the alpha kinase 1 (ALPK1) (21–23). Binding of β-ADP-heptose to ALPK1 stimulates its kinase domain to phosphorylate and activate TRAF interacting protein with forkhead associated domain (TIFA) (24), which forms large complexes called TIFAsomes (21). H. pylori mutants that lack the ability to produce β-ADP-heptose are incapable of activating the ALPK1/TIFA pathway (21–23). Activation of the ALPK1/TIFA signaling axis leads to nuclear factor κB (NF-κB) activation and the subsequent production of proinflammatory cytokines and other NF-κB target gene products (21–23). We have shown previously that activation of the β-ADP-heptose/ALPK1/TIFA/NF-κB signaling axis results in DNA double-strand break formation in S phase cells at genomic sites where transcription and replication machineries collide (10). H. pylori–induced DNA damage is further aggravated by the co-occurring T4SS-driven delivery of the bacterial oncoprotein CagA, which, by interacting with the Polarity-regulating kinase partitioning-defective 1b (PAR1b) inhibits PAR1b-mediated breast cancer 1 (BRCA1) phosphorylation and nuclear translocation and thereby disables error-free homologous recombination–mediated DNA repair (25). Inherited pathogenic variants affecting any one of four homologous recombination repair factors, i.e., BRCA1, BRCA2, ATM, and PALB2, predispose the carriers to a 45% lifetime risk of developing gastric cancer only if the carrier is infected with H. pylori (26). Here, we use murine gastric organoids to investigate in a relevant model system to which gastric lineages H. pylori adheres and in which lineages it preferentially induces DNA damage. We further use genetically modified organoids to explore possible synergies of early gastric cancer driver mutations and H. pylori in promoting DNA damage and replication stress in gastric organoid–derived epithelial cells.
RESULTS
H. pylori induces DNA damage and replication stress in murine gastric organoid cells
To address whether H. pylori exposure affects the genomic integrity of murine gastric organoid cells, we developed a protocol that allows us to cultivate antrum or corpus gland–derived organoids within a time frame of 1 week (fig. S1A). Our culture conditions preserve to some extent the diversity of gastric antral and corpus lineages (mucous pit, mucous neck, chief, and stem/progenitor cells), both when organoids are cultured as three-dimensional (3D) structures and when seeded in 2D for subsequent procedures (fig. S1, B to D). One exception is the corpus-specific parietal cell lineage, for which signature transcripts are present in the 3D organoids but lost upon seeding in 2D (fig. S1D). Single-cell RNA sequencing (scRNA-seq) of 2D cultures segregated seven main clusters—which we identified in accordance with existing literature on human gastric organoid scRNA-seq data (27), murine gastric and small intestinal gland scRNA-seq data (28–30), and the mouse cell atlas database (31)—as mature mucous pit cells (cluster 1), immature mucous pit cells (2), isthmus stem and progenitor cells (3), and basal stem and progenitor cells deriving from Leucine-rich-repeat containing G-Protein-Coupled Receptor 5 (Lgr5)–positive precursors (cluster 4) and mucous neck/chief cells (cluster 5; Fig. 1A). Two additional clusters were numerically less important (<1% of all cells) and proved difficult to annotate. Antral and corpus gland–derived 2D organoid cultures were quite similar with respect to the relative representation of the five major clusters (Fig. 1B); most clusters (with the exception of cluster 5) were preserved also during infection with H. pylori (Fig. 1B). Marker transcripts of the mature pit cells of cluster 1 included Muc5ac, Gkn1, and Gkn2 (encoding the pit cell mucin Muc5ac and the gastrokines 1 and 2); typical markers of the immature pit cells of cluster 2 were Tff1 as well as Psca and Mucl3 (encoding the trefoil factor 1, prostate stem cell antigen, and Mucin-like 3; Fig. 1C and fig. S1E) (27, 28, 30). Isthmus stem and progenitor cells of cluster 3 were characterized by expression of Mki67, Stmn1, and Top2a (encoding murine Ki67, stathmin1, and topoisomerase 2a; and the expression of Birc5, Cdk1, and Ube2c; Fig. 1C and fig. S1E) (27, 28, 30), whereas cluster 4 (tentatively annotated as basal stem cell–derived cluster) was positive for Cth, Gas5, Zfas1, and numerous ribosomal protein transcripts (Fig. 1C and fig. S1E) (29). Cluster 5 was the least well defined and showed evidence of expression of not only various neck and chief cell transcripts but also of transcripts that could not readily be assigned to published gastric epithelial cell profiles (Cd44 and Vil1; Fig. 1C and fig. S1E).
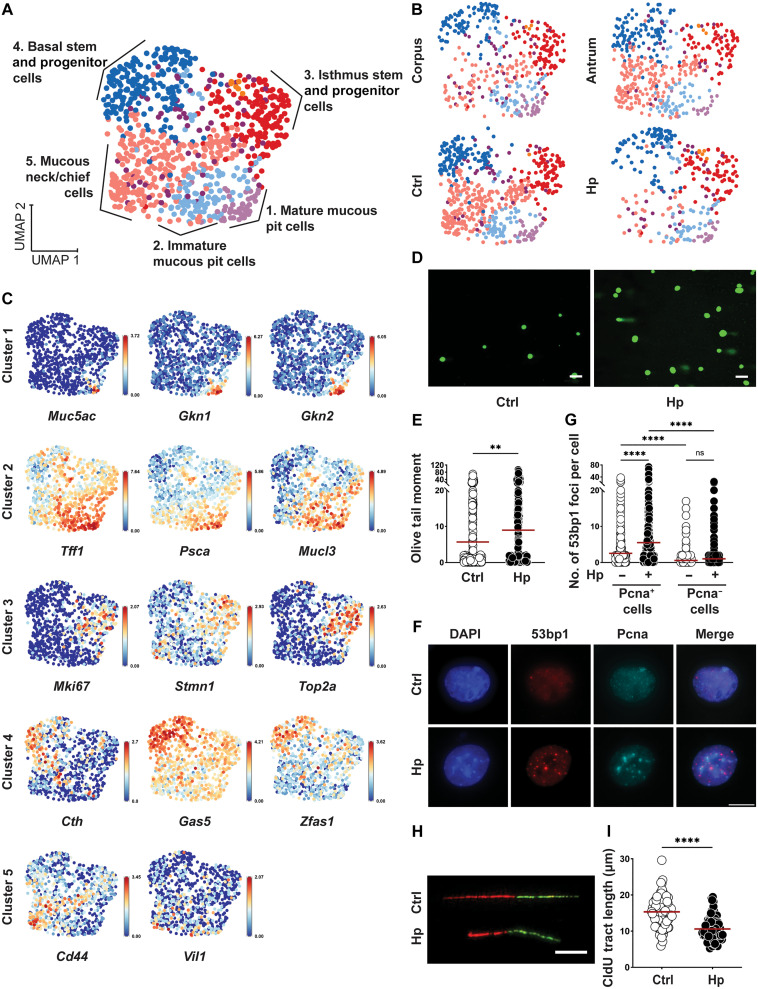
Fig. 1. Exposure to H. pylori induces DNA damage and replication stress in murine gastric organoid cells.
(A to C) Antrum and corpus-derived organoids were dissociated, seeded in 2D, and infected with H. pylori (Hp) G27 for 6 hours. Cells were labeled with antibody-conjugated hashtags and subjected to scRNA-seq. Unsupervised clustering using Seurat identified five main clusters; the indicated cluster identities were assigned on the basis of their expression levels of cell type–specific markers. Uniform Manifold Approximation and Projection (UMAP) visualizations in (A) and (B) show the combined data points from all four conditions (A) and separated by site and infection status (B); the expression of 14 select cluster marker transcripts is projected on top of the UMAP visualizations in (C) (data were normalized for the expression of the gene within the dataset). (D to I) Antrum-derived organoid cells seeded in 2D were infected with H. pylori strain G27 [multiplicity of infection (MOI) of 50] for 6 hours; cells were harvested and analyzed for the appearance of DNA damage by alkaline comet assay [(D) and (E)] and by 53bp1/Pcna-specific immunofluorescence microscopy (F and G); replication stress was quantified by DNA fiber assay [(H) and (I)]. Representative images of comets, 53bp1 foci formation and DNA fibers are shown in (D), (F), and (H). Scale bars, 10 μm. Data in (E), (G), and (I) are representative of at least three independent experiments; ~150 cells per condition are shown in (E), ~1000 cells per condition are shown in (G), and ~100 fibers per condition are shown in (I). Horizontal lines represent means. P values were calculated using Student’s t test; **P < 0.01; ****P < 0.001. ns, not significant.
Several transcripts that we expected to find on the basis of positive reverse transcription polymerase chain reaction (RT-PCR) and immunofluorescence microscopy results (fig. S1, B to D) were notably absent in our scRNA-seq data; examples include Muc6, Lgr5, and Tnfrsf19 encoding Troy, all of which were readily detectable at the protein level but absent in our scRNA-seq data. Having established and characterized murine organoid cultures in 2D and, less extensively, in 3D, we asked whether their exposure to live H. pylori would induce DNA damage as determined by alkaline comet assay. Organoids that were microinjected with H. pylori (fig. S1, F and G) or cultured as monolayers in 2D and infected with H. pylori (Fig. 1, D and E) showed extensive DNA damage in the form of comet tails. DNA damage and replication stress were further also observed in the form of p53-binding protein 1 (53bp1) foci formation and shortening of DNA fibers (Fig. 1, F to I); DNA damage occurred upon exposure to two different strains of H. pylori (PMSS1 and G27) and was detected in S phase cells positive for proliferating cell nuclear antigen (Pcna), but not in Pcna− cells (Fig. 1, E to I, and fig. S1, F and G). The frequencies of Pcna+ cells did not change upon infection (fig. S1H). Similar levels of H. pylori–induced DNA damage were detected in antral and corpus-derived organoid cells (fig. S1, I and J); antral 2D cultures are therefore shown representatively for both sites for all subsequent experiments, unless otherwise indicated.
H. pylori–induced DNA damage depends on a functional T4SS/β-ADP-heptose/ALPK1 axis and active replication
Having shown previously for a human gastric cancer cell line that H. pylori–induced DNA damage depends on a functional T4SS, as well as the RfaE enzyme and thus the ability of H. pylori to synthesize the LPS biosynthetic intermediate β-ADP-heptose (10), we infected murine organoid cells with mutants lacking RfaE or CagE, the latter being incapable of assembling a functional T4SS (32). Neither mutant induced DNA damage as judged by alkaline comet assay (fig. S2, A and B). Conversely, the exogenous addition of synthetic β-ADP-heptose was as efficient as live infection at inducing DNA damage, as determined by comet assay (Fig. 2, A and B) and 53bp1 staining (Fig. 2, C and D).
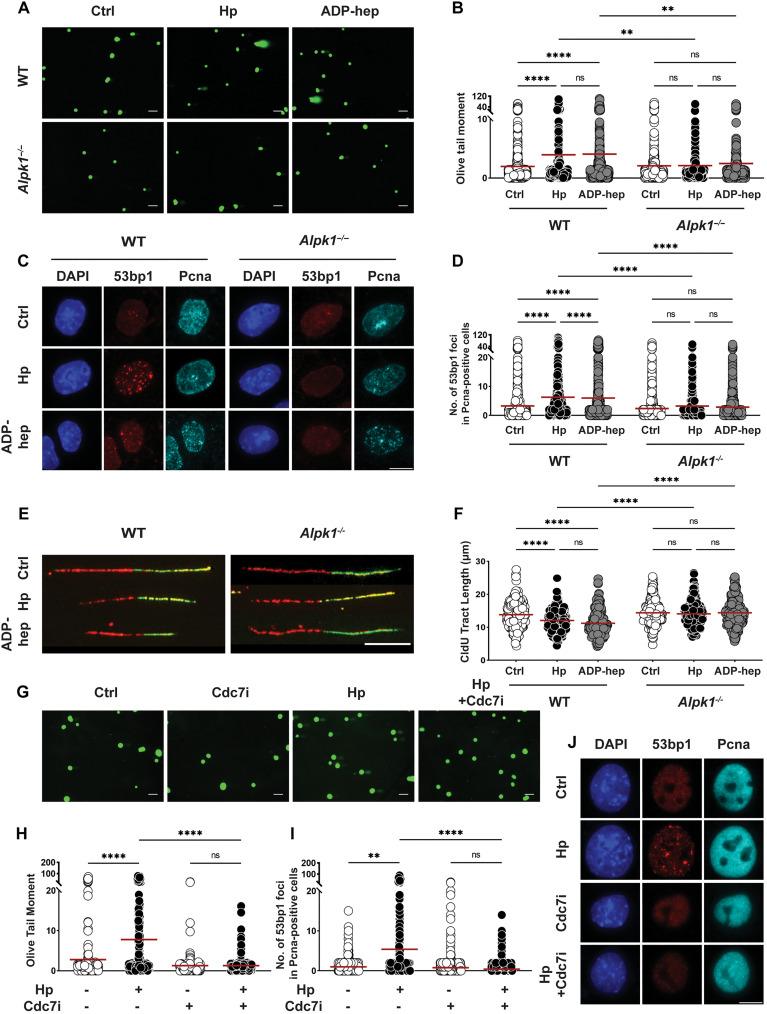
Fig. 2. H. pylori–induced DNA damage and replication stress is dependent on Alpk1 signaling and unimpaired progression through S phase.
(A to F) Antrum-derived organoid cells seeded in 2D were infected with H. pylori strain G27 (MOI of 50) for 6 hours. Cells were harvested and analyzed for the appearance of DNA damage by alkaline comet assay [(A) and (B)] and by 53bp1/Pcna-specific immunofluorescence microscopy [(C) and (D)]; replication stress was quantified by DNA fiber assay [(E) and (F)]. Representative images of comets, 53bp1 foci formation, and DNA fibers are shown in (A), (C), and (E) (scale bars, 10 μm); pooled data from three independent experiments are shown in (B), (D), and (F) [~300 cells are shown per condition in (B), ~1500 cells per condition in (D), and 300 fibers per condition in (F)]. (G to J) Antrum-derived organoid cells seeded in 2D were infected with H. pylori strain G27 (MOI of 50) for 6 hours, in the presence or absence of the Cdc7 inhibitor XL413 at 0.2 μM final concentration. Cells were harvested and analyzed by alkaline comet assay [(G) and (H)] and by 53bp1/Pcna-specific immunofluorescence microscopy [(I) and (J)]. Data are from one experiment and representative of two independently conducted ones. A total of ~150 cells and ~1000 cells per condition are shown in (H) and (I), respectively. Horizontal lines represent means. P values were calculated using one-way analysis of variance (ANOVA); **P < 0.01; ****P < 0.001.
53bp1 foci indicative of DNA damage were observed in Pcna+ cells in S phase (Fig. 2, C and D). The genetic ablation of Alpk1, a cytoplasmic serine/threonine kinase that binds β-ADP-heptose and activates NF-κB by triggering TIFAsome formation (24), abrogated the DNA damage induced by H. pylori and β-ADP-heptose (Fig. 2, A to D). We have demonstrated earlier using transformed cell lines that H. pylori–induced DNA damage is a consequence of transcription/replication conflicts (TRCs) in actively cycling cells in S phase (10) and indeed detect evidence of RNA:DNA hybrids (R-loops) indicative of these TRCs also in H. pylori–exposed organoid cells (by staining with the RNA:DNA hybrid–specific antibody S9.6; fig. S2, C and D). Blockade of NF-κB–dependent transcription downstream of Alpk1 using the NF-κB inhibitor BAY-11 prevented DNA damage induced by H. pylori (fig. S2E). Consistent with the observed R-loop formation and TRCs, H. pylori–induced organoid cells are subject to replication stress as evidenced by shortening of DNA fibers; replication stress is also observed upon exposure to synthetic β-ADP-heptose and depends on Alpk1 (Fig. 2, E and F). To address whether the observed DNA damage indeed depends on active replication, we blocked the activity of the CDC7 kinase with the specific inhibitor XL413. Exposure to this inhibitor completely prevented comet and 53bp1 foci formation upon H. pylori infection (Fig. 2, G to J) and reduced the number of evaluable fibers by >70%; the length of the remaining fibers was reduced even in the absence of infection and could not be reduced further by infection (fig. S2F). The combined results indicate that H. pylori infection compromises the genomic integrity of primary gastric epithelial cells in a manner that depends on its ability to produce and deliver β-ADP-heptose and to cause TRCs through activation of the Alpk1/NF-κB signaling pathway.
Gastric stem and progenitor cells are the main targets of H. pylori–induced DNA damage
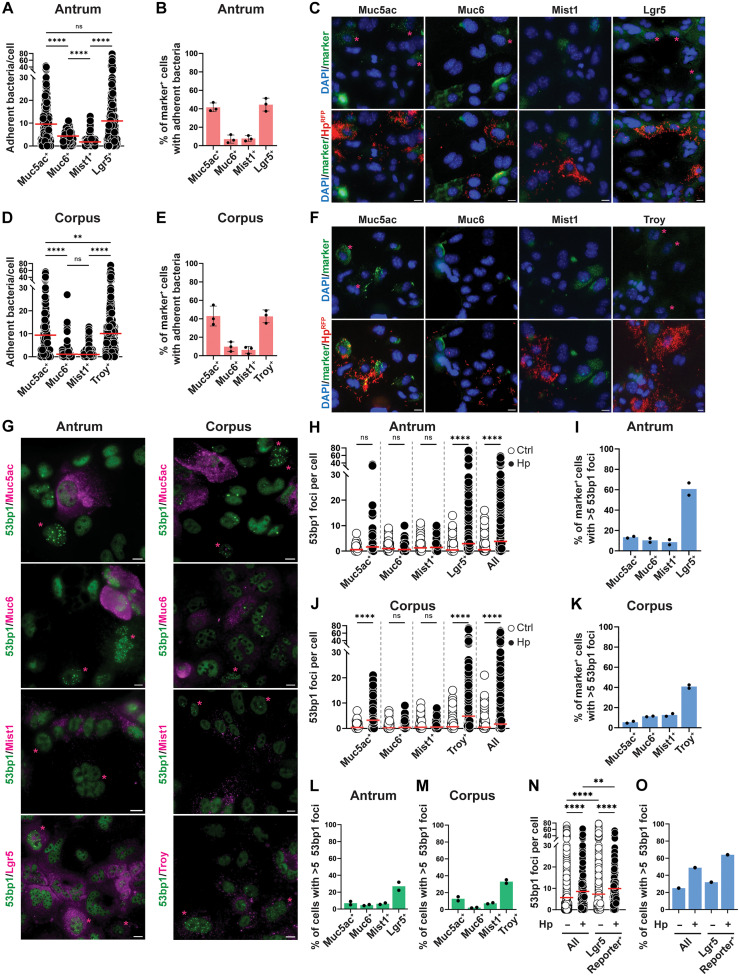
The presence of diverse gastric cell lineages in our organoid cell monolayers allowed us to investigate the preferred target cells of H. pylori. To this end, we took advantage of an H. pylori strain that was engineered to express red fluorescent protein (RFP) (33) and used it in combination with immunofluorescence staining for gastric lineage markers, followed by automated image analysis. Gastric antral and corpus organoid cell lineages were identified by their Muc5ac and Muc6 expression as mucous pit and mucous neck cells, respectively, by their Mist expression as chief cells, and by their Troy (corpus) and Lgr5 (antrum) expression as stem and progenitor cells. Expression of the lineage markers was mutually exclusive (fig. S3A) with less than 5% of cells estimated to be positive for more than one marker. H. pylori was found to adhere preferentially to Muc5ac-positive mucous pit, but not to Muc6-positive mucous neck or Mist-positive chief cells (Fig. 3, A to F). Unexpectedly, a large fraction of RFP+ H. pylori was found in tight contact with Troy+ and Lgr5+ stem and progenitor cells in corpus- and antrum-derived organoid cultures, respectively (Fig. 3, A to F). We found, on average, more bacteria attached to Muc5ac+ mucous pit and to Troy/Lgr5+ stem and progenitor cells than to the other cell lineages (Fig. 3, A to D, and high-magnification images in fig. S3B), and the fraction of cells with attached bacteria was higher for mucous and stem/progenitor cells than for the other lineages (Fig. 3, B and E). The preference for mucous pit and stem/progenitor cells was confirmed also at a lower multiplicity of infection (MOI) (of 10; fig. S3, C to H). We next asked which of the lineages sustained DNA damage, quantified microscopically as 53bp1+ foci formation. DNA damage was mostly observed in Troy+ and Lgr5+ stem cells in corpus- and antrum-derived organoid cultures, respectively; Muc5ac-expressing mucous pit cells sustained modest levels of DNA damage, and no damage was observed in mucous neck or chief cells (Fig. 3, G to M). We observed, on average, more 53bp1 foci per cell in the stem/progenitor compartment than in the other lineages (Fig. 3, H and J), and the fraction of cells with DNA damage (>5 foci per cell) was also highest in this compartment (Fig. 3, I and K). Among all cells exhibiting DNA damage in our antrum and corpus cultures, the stem/progenitor compartment made up the largest fraction (Fig. 3, L and M), despite contributing only marginally more cells to the overall pool than the differentiated lineages (fig. S3I). Given that H. pylori–induced DNA damage occurs in Pcna+ cells, we asked whether more stem and progenitor cells might be undergoing active replication at any given time than cells belonging to the differentiated lineages. This was indeed the case, as 50 to 60% of Lgr5/Troy+ cells, but only 10 to 25% of differentiated lineage-positive cells, stained positive for Pcna (fig. S3J). As many commercially available Lgr5-directed antibodies (and antibody lots) are unspecific, we used a recently generated transgenic Lgr5 reporter line (34, 35) to validate our antibody and to assess DNA damage in Lgr5-mOrange reporter cells. In this mouse line, the introduction of the transgene affects neither the expression nor the function of endogenous Lgr5 (34, 35). More than 90% of Lgr5-mOrange reporter cells were stained by our antibody, and 66% of cells stained by the antibody also expressed the reporter (fig. S3, K and L). Lgr5-mOrange reporter cells exhibited a higher baseline DNA damage as detected by 53bp1 foci than the total cell population and showed a further increase in DNA damage upon H. pylori infection (Fig. 3, N and O). The susceptibility of Lgr5-mOrange reporter cells to DNA damage could be linked to a high proportion of cells in S phase (fig. S3M), confirming the results obtained for antibody-stained Lgr5+ cells.
Fig. 3. H. pylori–induced DNA damage occurs in gastric stem and progenitor cells.
(A to F) Antrum-derived [(A) to (C)] or corpus-derived [(D) to (F)] organoid cells seeded in 2D were infected with RFP-expressing H. pylori PMSS1 (MOI of 50) for 6 hours. Cells were stained for the indicated lineage markers and subjected to quantitative automated microscopy. The number of adherent RFP+ bacteria per cell is shown for each lineage in (A) and (D); the fraction of cells per lineage with at least 10 adherent bacteria is shown in (B) and (E). Representative images are shown in (C) and (F), with lineage marker–positive cells in green (indicated by asterisks). (G to M) Antrum-derived [(G) to (I) and (L)] or corpus-derived [(G), (J), (K), and (M)] organoid cells were seeded and infected as described in (A) to (F). Cells were stained for the indicated lineage markers and for 53bp1. The number of 53bp1 foci per cell is shown for each lineage in (H) and (J); the fraction of cells per lineage with five or more 53bp1 foci is shown in (I) and (K). The fraction of lineage marker–positive cells among all cells with five or more 53bp1 foci is shown in (L) and (M). (N and O) Antrum-derived organoid cells from an Lgr5-mOrange reporter mouse were seeded and infected as described in (A) to (F). Cells were stained with an RFP/mOrange-specific antibody and for 53bp1; data are plotted as described above. Data are pooled from two [(A), (D), and (H) to (M)] and three [(B) and (E)] identically designed experiments or are from a single experiment [(N) and (O)]; 200 to 1000 cells per condition were analyzed. Red horizontal lines represent means. Means ± SD are shown alongside the individual data points in (B) and (E); means and individual data points are shown in (I) and (K) to (M). Scale bars, 10 μm throughout. P values were calculated using one-way ANOVA; **P < 0.01; ****P < 0.001.
Last, we set out to determine in which of the lineages the exposure to H. pylori would induce a transcriptional response. To this end, infected and control antrum and corpus 2D monolayers were subjected to RNA sequencing. Unsupervised hierarchical clustering and principal components analysis (PCA) revealed the site (antrum versus corpus) to be the main driver of sample segregation, followed by H. pylori exposure (fig. S3, N and O). Gene ontology analysis of the top 117 overlapping differentially up-regulated genes (in both antrum and corpus, log2 fold change > 1, adjusted P < 0.05) revealed pathways such as “response to bacteria,” “leukocyte chemotaxis,” and “inflammatory response” to be induced upon infection (fig. S3P). Superimposing the top 50 most differentially expressed transcripts onto the five gastric epithelial cell clusters shown in Fig. 1 (A to C) revealed that most of the differentially expressed genes mapped to the basal stem and progenitor cell cluster (24 of 50), whereas only very few differentially expressed genes mapped to any of the other clusters (four each to mature and immature pit cells, nine to chief/neck cells, and one to isthmus progenitors); nine induced genes were expressed throughout (representative transcripts shown in fig. S3Q). Together, the data suggest that H. pylori preferentially binds to mucous pit cells and Lgr5+ stem and progenitor cells but induces DNA damage only in the latter compartment.
The mutational inactivation of Apc exacerbates H. pylori–induced DNA damage
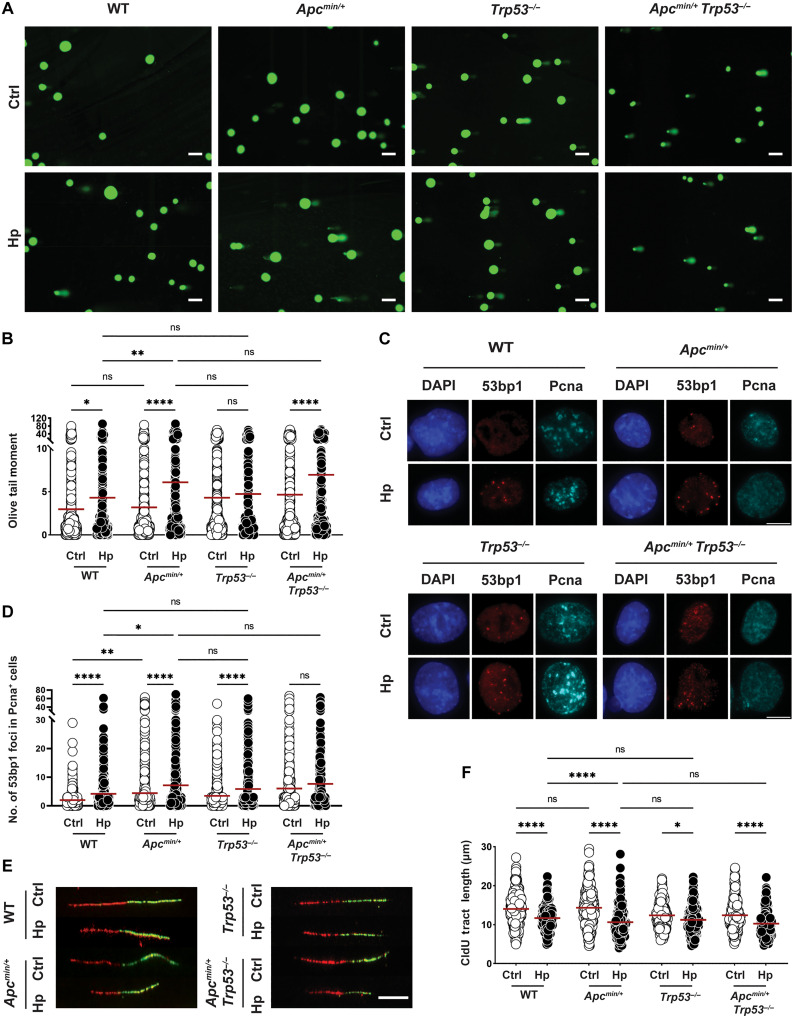
To determine how gastric cancer driver mutations affect H. pylori–induced DNA damage, we chose three tumor suppressor genes and one oncogene, all of which are among the most recurrently mutated genes in gastric cancer, for DNA damage analyses. Mutations in the tumor suppressor genes adenomatous polyposis coli (APC), TP53, and SMAD4 affect roughly 8, 50, and 7% of patients with gastric cancer; TP53 and APC are predominantly mutationally inactivated in the chromosomally instable subtype, whereas SMAD4 mutations affect all four molecular subtypes of gastric cancer (36). The oncogene ERBB2 is subject to both point mutations and gene amplification (affecting 3 and 17% of gastric cancer cases) (36). To address the consequences of Apc truncation and of Trp53 deletion for H. pylori–induced DNA damage and replication stress, we cultured antrum-derived organoids from wild-type (WT), Apcmin/+, Trp53−/−, and Apcmin/+Trp53−/− mice and infected the cells with H. pylori in 2D. H. pylori induced more comets and more 53bp1 foci indicative of DNA damage and shorter fibers in Apcmin/+ relative to WT organoid cells, whereas the loss of Trp53 had no major effect on DNA damage (although the baseline of DNA damage was higher; Fig. 4, A to F). The dual inactivation of Apc and Trp53 did not produce stronger effects than the Apc mutation alone (Fig. 4, A to F). As the main function of P53 is the orchestration of DNA repair, we asked whether Trp53−/− cells would be as capable as WT cells in repairing their H. pylori–induced DNA damage. Both WT and Trp53−/− cells repaired their DNA damage equally efficiently, with comets reduced to baseline after a 24-hour recovery period during which H. pylori was eradicated with antibiotics (fig. S4A). To study how the loss of Smad4 and gain of Erbb2 would affect DNA damage, we genetically engineered WT organoid cells to constitutively express a short hairpin RNA (shRNA) for Smad4 that would down-regulate its expression by greater than 80% (fig. S4B); Erbb2 was overexpressed under the constitutive cytomegalovirus (CMV) promoter (fig. S4C). Smad4 loss, but not Erbb2 overexpression, had modest effects on DNA damage incurred upon H. pylori infection relative to an empty vector control as judged by alkaline comet assay and 53bp1 staining (fig. S4, D to G), despite the fact that Erbb2-overexpressing cells exhibited numerous genomic abnormalities (irrespective of infection; these included micronuclei, bulky chromosome bridges, and misaligned chromosomes; fig. S4, H and I). Given that Apc emerged as the most relevant cancer gene among our small selection of four, we asked whether corpus-derived Apcmin/+ organoid cells would recapitulate the effects observed with the antral Apcmin/+ organoids relative to WT; this was indeed the case for both DNA damage (comet assay) and replication stress (fiber assay) readouts (fig. S4, J to M). Furthermore, administration of β-ADP-heptose recapitulated the elevated DNA damage induced by live infection in Apcmin/+ relative to WT organoid cells of both antrum and corpus origin (fig. S4, J to M). In summary, inactivation of a single allele of Apc by a nonsense mutation at codon 851 synergized with H. pylori infection or β-ADP-heptose exposure in driving DNA damage, whereas the loss of Trp53 or of Smad4 or overexpression of Erbb2 had no effect.
Fig. 4. Apc truncation, but not Trp53 loss, aggravates H. pylori–induced DNA damage.
(A to F) Antrum-derived organoid cells of the indicated genotypes were seeded in 2D and infected with H. pylori strain G27 (MOI of 50) for 6 hours. Cells were harvested and analyzed for the appearance of DNA damage by alkaline comet assay [(A) and (B)] and by 53bp1/Pcna-specific immunofluorescence microscopy [(C) and (D)]; replication stress was quantified by DNA fiber assay [(E) and (F)]. Representative images of comets, 53bp1 foci formation, and DNA fibers are shown in (A) (scale bars, 100 μm), (C) (scale bar, 10 μm), and (E) (scale bar, 10 μm); pooled data from two to four independent experiments are shown in (B) and (F) [~400 to 600 cells are shown per condition in (B), and 300 fibers are shown per condition in (F)]. A representative experiment of two independently conducted ones is shown in (D) (~1000 cells per condition). Red horizontal lines represent means throughout. P values were calculated using one-way ANOVA; *P < 0.05; **P < 0.01; ****P < 0.001.
Apc inactivation results in an expanded pool of actively replicating cells that are susceptible to H. pylori–induced DNA damage
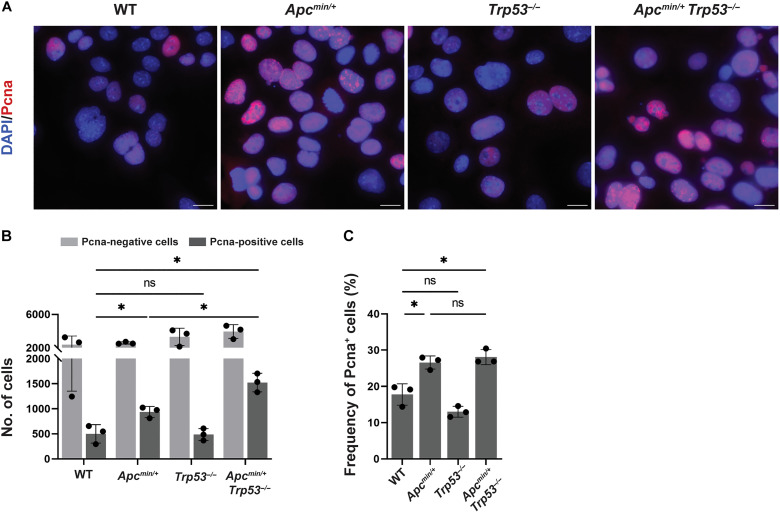
Having shown earlier that both active transcription and active replication are prerequisites of H. pylori–induced DNA damage (Fig. 2, H to J, and fig. S2, E and F), we asked whether Apc inactivation affected the level of DNA damage by enhancing one or the other process. RNA sequencing of antral and corpus-derived organoid cells, infected or not with H. pylori, failed to expose any major differences; WT and Apcmin/+ cells of the same anatomical origin (antrum and corpus) and infection status clustered together tightly, and differences were negligible (fig. S5, A and B). In contrast, when staining for Pcna+ cells, we found that the inactivation of Apc, with and without the additional loss of Trp53, strongly boosted the pool of Pcna+ cells, thereby expanding the potential target cell pool susceptible to H. pylori–induced DNA damage (Fig. 5, A to C). The increase in Pcna+ cells resulting from Apc inactivation coincided with an increase in the Lgr5+ stem and progenitor cell pool in organoids derived from Apcmin/+ relative to WT mice (fig. S5C); this observation is consistent with Lgr5 itself being a target gene of the Wnt signaling pathway and hyperactive Wnt signaling due to Apc inactivation driving stem and progenitor cell expansion (37). The combined data indicate that Apc inactivation synergizes with H. pylori infection to promote potentially carcinogenic DNA damage by increasing the pool of actively proliferating stem and progenitor cells.
Fig. 5. Apc inactivation increases the pool of Pcna+ cells in S phase.
(A to C) Antrum-derived organoid cells of the indicated genotypes were seeded in 2D and stained with a Pcna-specific antibody. Representative images are shown in (A) (scale bars, 10 μm), alongside the absolute numbers of Pcna+ and Pcna− cells in (B), and the frequency of Pcna+ cells among all cells in (C). Three experiments are shown in (B) and (C). Data are plotted as means ± SD. Several thousand cells were analyzed per condition. P values were calculated using one-way ANOVA; *P < 0.05.
DISCUSSION
In earlier work focused on elucidating how H. pylori damages the genome of its target cells, we have mostly relied on transformed or immortalized cell lines as model systems to study DNA damage and the associated replication stress (10, 38, 39). Here, we have used procedures for 2D and 3D growth and infection of murine gastric organoid cultures and have developed genetic modification strategies and downstream readouts for DNA damage and replication stress, to shed light on the preferred target cell type of H. pylori, the cell lineage most susceptible to infection-induced DNA damage, and the interaction of H. pylori with early gastric cancer driver mutations in DNA damage induction. Infection of organoid cells confirmed a key role of active transcription, induced via the previously described H. pylori T4SS/β-ADP-heptose/Alpk1/NF-κB axis (10), in DNA damage and replication stress, and provided more evidence in the form of RNA:DNA hybrids or R-loops (labeled by S9.6 antibody) for the concept that TRCs in actively replicating cells are the main source of H. pylori–induced DNA damage. As a detailed understanding of the cell lineages represented in 2D (and to a lesser extent in 3D) organoid cultures was key to determining the preferred H. pylori target cell in this model system, we used both conventional immunofluorescence and RT-PCR–based strategies and scRNA-seq to characterize the cells in some detail. Immunofluorescence staining and RT-PCR for key markers revealed that Muc5ac+ pit cells, Muc6+ chief cells, and Mist+/pepsinogen C+ chief cells were present in 2D cultures, alongside Lgr5+ or Troy+ stem and progenitor cells. scRNA-seq revealed the presence of five predominant clusters, which, in accordance with published transcriptional profiles of human organoid cultures (27) and of sorted murine and human gastric gland populations (28, 30, 31), could be assigned to specific gastric lineages. We found evidence in our cultures of mature pit cells characterized by their hallmark Muc5ac and gastrokine 1 and 2 expression (Gkn1, Gkn2; cluster 1), of immature pit cells (cluster 2, characterized by Tff1, Psca, and Mucl3), and of isthmus stem and progenitor cells (cluster 3; Stmn1, Mki67, and Top2a). The latter gives rise not only to the former at steady state as judged by lineage tracing but also to Muc6+ mucous neck cells (30); isthmus stem and progenitor cells differentiate as they migrate from the isthmus toward the tip (pit cells) and the neck/base of the gland (neck cells). The two remaining clusters are much less well defined in the literature and are absent from 2D cultures of human organoids (27). Certain overlaps in signature transcripts with previous studies point to cluster 5 as a mixture of neck and chief cells (Cd44 and Vil1; note that several key marker transcripts, such as Muc6 and Pgc encoding pepsinogen C, are notably missing from this list although they are readily amplified by quantitative RT-PCR) (40, 41) and to cluster 4 as the direct progeny of Lgr5-expressing basal stem and progenitor cells as defined by single-cell trancriptomics of small intestinal crypts (29). We believe on the basis of their signature transcripts that clusters 1 and 5 represent terminally differentiated lineages, whereas clusters 3 and 4 both represent stem/progenitor populations; cluster 2, containing immature pit cells, is most likely a transitional cell type on its way to terminal pit cell differentiation. We confirmed the finding made previously by others that parietal cells are difficult to maintain in 2D monolayers (and are therefore absent from the infection system used here); coculturing with immortalized stomach mesenchymal cells can overcome this technical challenge but was not tried here (42).
H. pylori interacts directly with two of the gastric lineages represented in our antral and corpus organoid cultures, the Muc5ac+ pit cells on the one hand, and Lgr5+ or Troy+ stem and progenitor cells on the other. The gastric pit cell is the canonical target cell of H. pylori in vivo and in vitro. Early work has shown that only the pit cell, but not other terminally differentiated lineages (parietal, chief cells), of the gastric mucosa responds to H. pylori infection by mounting a transcriptional response in experimentally infected mice (43); H. pylori was found in close proximity to MUC5AC+ pit cells, but not MUC6+ neck cells, in the gastric pits of human carriers of the infection (44). Human organoid cells cultured in 2D, which feature immature and mature pit cells and their precursors, confirmed that H. pylori adheres to mature pit cells in a manner that depends on urea-driven chemotaxis (27). While we recapitulated the preference for pit cells in our murine organoid model, we additionally detected H. pylori bound to Lgr5+ stem and progenitor cells in antral cultures and to Troy+ stem and progenitor cells in corpus cultures. This observation is in line with the ability of H. pylori to colonize not only the surface epithelium but also the depth of both murine and human gastric glands, where the bacteria can be found in close proximity to Lgr5+ cells in murine antrum and to mitotic progenitor cells in human antrum and corpus (45, 46). This interaction with Lgr5+ cells leads to their T4SS-dependent expansion (hyperplasia) and to the induction of antimicrobial factors (45). Lgr5+ cells mount a robust, R-spondin-3–driven antimicrobial response to H. pylori in vivo, which results in clearance (or at least a reduction) of the bacteria and presumably serves to protect the gland base and its stem cell pool (41). The expression and secretion of antimicrobial factors by gland base cells and their expansion are NF-κB dependent (47). Gland base stem cells are the primary cell lineage responding to H. pylori and its ADP-heptose with the production of proinflammatory mediators; this has been definitively shown for antrum stem cells (47) and possibly also applies to corpus basal stem cells. The expansion of Lgr5+ gland base stem cells is not only observed in infected mice (45) but also observed in human carriers of H. pylori (48). These various observations are very well in line with our observation that DNA damage induced by H. pylori in organoid cells preferentially affects Lgr5+/Troy+ stem and progenitor cells and is dependent on both active transcription and replication. A specific inhibitor of NF-κB–dependent transcription or the genetic ablation of the H. pylori innate immune sensor Alpk1, which drives H. pylori–dependent NF-κB activation, both prevented DNA damage in our hands. DNA damage was further observed only in Pcna+ cells in S phase and could be blocked by an inhibitor of Cdc7 kinase that prevents origin firing and thereby entry into S phase. It is interesting to note that a specific type of DNA damage on guanosine that is indicative of oxidative stress (8-hydroxydeoxyguanosine) was detected in non-neoplastic antral Lgr5+ cells in patients with gastric cancer with H. pylori–driven gastritis (48). Lgr5+ stem cells in the corpus of the stomach have been shown to give rise to Kras(G12D)-driven intestinal metaplasia and gastric cancer, and LGR5 expression is seen in human gastric cancers (49); furthermore, in vivo ablation strategies confirm Lgr5+ populations as functional cancer stem cells in orthotopic cancer organoid transplantation models (50). All observations are in line with the notion that H. pylori–induced DNA damage in stem and progenitor cells, if unrepaired, or erroneously repaired, may serve as an early event driving gastric carcinogenesis. Such a setting of erroneous DNA double-strand break repair is constituted by inherited (germline) mutations in the homologous recombination repair genes BRCA1, BRCA2, ATM, and PALB2, all of which have been shown to synergize with H. pylori infection in predisposing carriers of the respective pathogenic variants to an extremely high gastric cancer risk (26).
Having identified stem and progenitor cells as the predominant targets of H. pylori–induced DNA damage in vitro, we asked whether preexisting mutations affecting Trp53, Apc, and the two additional commonly mutated gastric cancer driver genes Smad4 and Erbb2 would affect the extent and repair of DNA damage incurred upon H. pylori exposure. We chose Trp53 because earlier reports had shown somatic TP53 mutations to be fairly common already in noncancerous H. pylori–positive inflamed mucosal samples from patients with gastric cancer but not in H. pylori–negative normal gastric mucosa (51). TP53, APC, and CTNNB1 hotspot mutations, as well as ERBB2 amplifications, were among the earliest somatic gastric cancer mutations found by sequencing of 50 cancer genes in well-differentiated intramucosal gastric cancers obtained by endoscopic submucosal dissection (52). Hereditary pathogenic mutations in SMAD4, APC, and TP53 predispose to gastric cancer; these cause various familial cancer syndromes, i.e., juvenile polyposis syndrome (SMAD4), gastric adenocarcinoma, proximal polyposis and familial adenomatous polyposis syndromes (APC; the latter predisposing to gastric cancer only in Asia), and Li Fraumeni syndrome (TP53) (53, 54). We found only Apc truncation, but not the loss of Smad4 or Trp53, or overexpression of Erbb2 to synergize with H. pylori infection or exogenously added ADP-heptose in causing DNA damage and replication stress. Apc truncation boosts the pool of Pcna+ cells that are susceptible to DNA damage, which likely accounts for the increase in damage observed. The fact that the other three genetic modifications failed to show a phenotype in our acute DNA damage model does not rule out that these alterations interact with H. pylori in some other way in driving gastric carcinogenesis. In summary, our data point to stem and progenitor cells as main targets of H. pylori–induced DNA damage, which is further aggravated by concurrent Apc inactivation.
MATERIALS AND METHODS
Mice and organoid cultures
C57BL/6N-Alpk1em1Fsha/J (strain #: 032561), B6.129S2-Trp53tm1Tyj/J (strain #: 002101), and C57BL/6J-ApcMin/J (strain #: 002020) were obtained from the Jackson laboratory. Composite Trp53−/− Apcmin/+ mice were generated in-house. Lgr5-2A-CreERT2-2A-mOrange2 mice were described previously (34, 35). C57BL/6J WT and mutant mouse lines were maintained in groups of maximum five animals in individually ventilated cages at approved animal facilities of the University of Zürich, with access to food and water ad libitum. Gastric organoid culture conditions were adapted on the basis of previously published protocols (55); briefly, stomachs of 8- to 10-week-old mice were harvested, opened along the greater curvature, and washed in ice-cold Dulbecco’s phosphate-buffered saline (DPBS) (catalog #14190-094, Gibco). Mucus and muscle/serosa layer were removed under a stereomicroscope using forceps; antrum and corpus were separated with a large safety margin, and both tissues were cut into 2- to 5-mm2-sized pieces. Pieces were dissociated in 43.4 mM sucrose (#A2211, 5000, HUBERLAB), 54.9 mM d-sorbitol (#240850, Sigma-Aldrich) in DPBS by 5 to 10 rounds of vigorous pipetting and washing, followed by incubation in 10 mM EDTA (#A1103, 1000, Biochemica) at 4°C for 1 hour and 20 min (rolling shaker). After one more washing step in 43.4 mM sucrose and 54.9 mM d-sorbitol, glands were extracted by placing tissue pieces under a glass slide (#AA00000112E01MNZ10, Epredia) and applying enough pressure to release individual glands; glands were collected in Dulbecco’s modified Eagle’s medium (DMEM), pushed through a 40-μm filter, counted, collected by centrifuging and mixed with Matrigel (#356231, Corning), seeded into 24-well plates (50 μl per well), and cultivated at 37°C in ADMEM [advanced DMEM/F12 (#12634-010, Gibco) + 10 mM penicillin/streptomycin (#167369, Gibco)] supplemented with Wnt-conditioned medium (50%, v/v; supernatants from L Wnt3a cells; CRL-2647, American Type Culture Collection), R-spondin-1 (1 μg/ml; #315-32, PeproTech), N-acetylcysteine (1 mM; #A9165, Sigma-Aldrich), epidermal growth factor (50 ng/ml; #AF-315-09, PeproTech), fibroblast growth factor (100 ng/ml; #100-26, PeproTech), Noggin (100 ng/ml; #250-38, PeproTech), [Leu15]–Gastrin I (10 nM; #G9145, Sigma-Aldrich), N2 (1×; #17502-048, Gibco), B27 (1×; #17504-044, Gibco), Hepes (10 mM; #15630-056, Gibco), GlutaMAX (2 mM; #35050-038, Gibco), and Y27632 (10 μM; only on the seeding day; exclude thereafter, #S1049, Selleckchem). For seeding in 2D, 7-day-old 3D organoids were dissociated in cold DMEM by vigorous pipetting. Organoid fragments were resuspended in TrypLE (#12605028, Gibco) and incubated at 37°C for 10 min. Single-cell suspensions were washed, centrifuged, counted, and seeded at 15,000 cells per well into rat tail collagen (#50201, Ibidi)–coated wells in 2D culture medium [which is identical to 3D medium but lacks Wnt-conditioned medium, R-spondin, penicillin/streptomycin, fibroblast growth factor, Noggin, and N-acetylcysteine and contains 10% fetal bovine serum (#10270-106, Gibco) and 1 μM transforming growth factor–β inhibitor (#SML0788, Sigma-Aldrich)]; cells receive fresh medium after 2 days in culture and are infected with H. pylori after 3 days in culture. H. pylori was grown as described previously (56); bacterial numbers were determined by measuring the optical density at 600 nm, and bacteria were added to cells at an MOI of 50 for 6 hours, unless otherwise specified. Bacteria were killed by addition of tetracycline (1 μg/ml) for 24 hours. The H. pylori strains used in this study were G27 (57), PMSS1 (58), and PMSS1RFP (33). Organoid cells were treated with the NF-κB inhibitor BAY 11-7082 (Sigma-Aldrich) and the Cdck1 inhibitor XL413 at 0.2 μM final concentration. β-ADP-heptose (Invivogen) was added at 0.5 μM final concentration for 6 hours.
Alkaline comet assay
Cells were trypsinized, and 2500 cells per condition were imbedded in 150 μl of 0.8% SeaPlaque low-melting point agarose (Lonza) on two-well comet slides (Trevigen; 60 μl per well) and lysed overnight at 4°C in lysis buffer: 2.5 M NaCl, 100 mM EDTA, and 10 mM tris (pH 10), supplemented with 10% dimethyl sulfoxide and 1% Triton X-100. Slides were washed after overnight lysis and incubated for 40 min in denaturation buffer (300 mM NaOH and 1 mM EDTA), followed by electrophoresis for 20 min at 21 V and 300 mA. Afterward, all slides were washed with PBS, fixed in ice-cold ethanol for 10 min, and dried at 37°C. DNA staining was done with SYBR gold (Thermo Fisher Scientific) for 15 min, followed by washing with PBS and drying at 37°C. Samples were imaged using a Leica DMI 6000 inverted microscope and analyzed using CometScore2.0.
DNA fiber assay
For the analysis of DNA fiber length, cells were sequentially pulse-labeled with 30 mM CldU (#C6891, Sigma-Aldrich) and 250 mM 5′-iododeoxyuridine (IdU) (European Pharmacopoeia) for 30 min each. The cells were collected and resuspended in PBS at 2.5 × 105 cells/ml. The labeled cells were diluted 1:1 (v/v) with unlabeled cells, and 4 μl of cells were mixed with 7.5 μl of lysis buffer [200 mM tris-HCl (pH 7.5), 50 mM EDTA, and 0.5% (w/v) SDS] on a glass slide. After 9 min, the slides were tilted to 15° to 45°, and the resulting DNA spreads were air-dried and fixed in methanol/acetic acid (3:1) solution overnight at 4°. The DNA fibers were denatured with 2.5 M HCl for 60 min, washed several times with PBS to neutralize the pH, and blocked with 0.1% Tween 20 in 2% bovine serum albumin (BSA)/PBS for 40 min. The newly replicated CldU and IdU tracks were labeled for 2.5 hours in the dark, at room temperature, with anti–5-bromo-2′-deoxyuridine antibodies recognizing CldU (rat, ab6326, Abcam; 1:500) and IdU (mouse, 347580 B44, BD Biosciences; 1:100), followed by 1 hour of incubation with secondary antibodies at room temperature in the dark: anti-mouse Alexa Fluor 488 (A11001, Invitrogen; 1:300) and anti-rat Cy3 (712-166-153, Jackson ImmunoResearch Laboratories; 1:150). Fibers were visualized on a Leica DMI 6000 inverted microscope using an HCX Plan APO DIC 63× oil objective [1.4 to 0.6 numerical aperture (NA)] and analyzed using Fiji (59). At least 100 fibers were analyzed per replicate condition.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells grown on autoclaved coverslips and infected with H. pylori for 6 hours. Cells were washed three times with 1× PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. After fixation, cells were washed three times with 1× PBS, followed by a second fixation with ice-cold methanol for 20 min at −20°C. After this second fixation, cells were washed three times with 1× PBS and permeabilized with 0.1% Triton X-100/1× PBS for 20 min at room temperature. After a brief wash, cells were incubated in blocking solution (1% BSA, 100 mM NH4Cl, and 1× PBS) for 30 min at room temperature. Coverslips were then incubated with appropriate primary antibodies diluted in blocking solution for 4 hours at room temperature or overnight at 4°C. The following antibodies and dilutions were used: anti-53bp1 rabbit monoclonal (clone EPR2172, ab175933, Abcam; 1:400), anti-Mucin 5 AC mouse monoclonal (clone 45 M1, ab3649, Abcam; 1:400), anti-Mucin6 rabbit monoclonal (ab192318, Abcam; 1:100), anti-MIST1 mouse monoclonal (clone 6E8/A12/C11P1, NBP2-22478, Novus Biologicals; 1:100), anti-LGR5 mouse monoclonal (clone OTI2A2, MA5-25644, Thermo Fisher Scientific; 1:200), anti-TROY mouse monoclonal (D-4, sc-398526, Santa Cruz Biotechnology; 1:100), anti-PCNA mouse monoclonal (clone PC10, sc-56, Santa Cruz Biotechnology; 1:300), and anti-RFP rabbit polyclonal (600-401-379, Rockland; 1:100). Coverslips were washed three times with 1× PBS and then incubated with secondary antibodies diluted in blocking solution for 1 hour at room temperature in the dark. Secondary antibodies and dilutions were as follows: Alexa Fluor 488 goat anti-mouse immunoglobulin G (IgG) (A110001, Thermo Fisher Scientific; 1:500), Alexa Fluor 488 goat anti-rabbit IgG (A110008, Thermo Fisher Scientific; 1:500), Alexa Fluor 568 goat anti-rabbit (A11011, Thermo Fisher Scientific; 1:500), Alexa Fluor 568 goat anti-mouse (A11004, Thermo Fisher Scientific; 1:500), Alexa Fluor 647 goat anti-rabbit (A21244, Thermo Fisher Scientific; 1:500), and Alexa Fluor 647 goat anti-mouse (A21235, Thermo Fisher Scientific; 1:500). After 3 washes with 1× PBS, coverslips were stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) diluted in distilled H2O for 5 min at room temperature. Coverslips were mounted with Fluoromount-G mounting medium (00-4958-02, Invitrogen). Images were acquired with a Leica DM 6B fluorescent microscope using a 63×/1.4 NA objective with an oil immersion. For the analysis of 53bp1 foci, automated image acquisition was performed on a IX83 microscope (Olympus) equipped with ScanR imaging platform using a 40×/0.9 NA objective. The analysis of acquired images, commonly referred to as a quantitative image–based cytometry, was performed using CellProfiler 4.2.4. (Broad Institute of MIT and Harvard, Cambridge, MA, USA). Nuclei were identified on the basis of the DAPI signal, and the parameters of interest were analyzed for each nuclear object using modules of the CellProfiler 4.2.4. For the quantification of bacteria, the cells were infected with H. pylori genetically tagged with RFP. The bacteria were identified on the basis of the RFP signal and were counted for each cell, which were defined by an enlarging of a nuclear mask using modules of the CellProfiler 4.2.4. Immunofluorescence microscopy of 3D organoids was performed using the antibodies listed above, according to previously published procedures (60); imaging was performed on the inverted confocal laser scanning microscope SP8 (Leica) using a 10×/0.3 NA objective. The images were then processed in Fiji using maximum intensity Z-projection.
Detection of RNA:DNA hybrids with S9.6 antibody
Staining of RNA:DNA hybrids with the anti-RNA:DNA Hybrid [S9.6] antibody was performed using a previously published protocol (61). Briefly, the cells grown on coverslips were fixed with ice-cold methanol for 10 min at 4°C, followed by permeabilization with acetone for 1 min on ice. After fixation and permeabilization, the cells were washed with 1× PBS and 4× saline‑sodium citrate buffer (4× SSC). The cells were incubating in blocking solution (3% BSA, 0.1% Tween 20, and 4× SSC) overnight at 4°C. After a brief wash with blocking solution, the cells were incubated with anti-RNA:DNA Hybrid [S9.6] mouse antibody (ENH001, Kerafast; 1:100) and anti-nucleolin rabbit polyclonal antibody (ab22758, Abcam; 1:1000) diluted in a blocking solution overnight at 4°C and 90 min at room temperature, respectively. After a brief wash with 4× SSC, the coverslips were incubated with secondary antibodies Alexa Fluor 488 goat anti-mouse IgG (A110001, Thermo Fisher Scientific; 1:500) and Alexa Fluor 647 goat anti-rabbit IgG (A21244, Thermo Fisher Scientific; 1:500) diluted in the blocking solution for 90 min at room temperature in dark. Coverslips were then counterstained with DAPI (1 μg/ml) diluted in distilled H2O for 5 min and mounted with Fluoromount-G mounting medium (00-4958-02, Invitrogen). The images were acquired with Leica DM 6B fluorescent microscope using a 63×/1.4 NA objective with an oil immersion. The quantitative image–based cytometry analysis of S9.6 signal was performed using CellProfiler 4.2.4 (Broad Institute of MIT and Harvard, Cambridge, MA, USA). The DAPI signal was used for a segmentation of images to identify individual nuclei. For each nuclear object, the nucleoplasmic signal of S9.6 was measured excluding the nucleolar S9.6 signal.
Engineering of organoid cells
An Erbb2 overexpression plasmid was obtained from Addgene (plasmid #66945); the full-length WT coding sequence was amplified using the primers: forward, CAGATCGCCTGGAGAATT GGCTAGCATCAACAAGTTTGTACAAAAAAAGCAGGCTACCATGATCATCATGGAGCTG; reverse, TCCCCTACCCGGTAGAATTGGATCCAAACCACTTTGTACAAGAAAGCTGGGTTCATACAGGTACATCCAG. The resulting Erbb2 sequence was cloned into the Eco RI and Cla I restriction sites of the pHIV-EF1a-puro-T2A expression vector. For Smad4 knockdown, an shRNA construct (forward, CCGGGCGATTGTGCATTCTCAGGATCTCGAGATCCTGAGAATGCACAATCGCTTTTTG; reverse, AATTCAAAAAGCGATTGTGCATTCTCAGGATCTCGAGATCCTGAGAATGCACAATCGC) was cloned into the Age I and Eco RI restriction sites of the pLKO.1-TRC vector. The following oligos were used for a control shRNA construct: forward, CCGGCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG; reverse, AATTCAAAAACCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG. For packaging, 293 T cells were transfected with 12 μg of transfer plasmid, 6 μg of psPax2, and 3 μg of pCMV-VSVG in serum-free DMEM and polyethylenimine; virus-containing supernatants were collected after 3 days and passed through a 0.45-μm filter. Three volumes of clarified supernatant were combined with one volume of Lenti-X concentrator (631232, Takara), incubated at 4°C overnight, and then centrifuged at 1500g for 45 min at 4°C. Virus pellets were suspended in 250 μl of 3D organoid culture medium plus polybrene (8 μg/ml). For transduction, small clusters of organoid cells were prepared according to a procedure described by Koo et al. (62). Briefly, organoids were broken up by vigorous pipetting (30×), followed by a 5-min incubation in TripLE (#12605028, Gibco) at 37°C. The resulting small clusters were collected by centrifugation, combined with 250 μl of the viral suspension described above, and spinoculated at 600g at 32°C for 60 min. The plate was incubated 6 hours at 37°C; after this time, cells were centrifuged at 1000g for 5 min, resuspended in 100 μl of Matrigel (BD Biosciences), and split it into two wells of a 24-well culture plate. A total of 500 μl of 3D organoid culture medium was added after the Matrigel had polymerized. Puromycin selection was initiated 2 days after transduction (2 μg/ml for 3 days, followed by seven more days with puromycin of 0.6 μg/ml), followed by seeding of organoid cells in 2D for infection.
Bulk and scRNA-seq
Bulk RNA sequencing was performed by the Functional Genomics Center Zürich. RNA was extracted using the QIAGEN RNeasy Mini Kit following the manufacturer’s protocol. Extracted RNA was prepared for sequencing using the Illumina TruSeq mRNA Library assay following the manufacturer’s protocol. Sequencing was performed on the Illumina NovaSeq 6000 using the S1 Reagent Kit v1.5 (100 cycles) as per the manufacturer’s protocol. Demultiplexing was performed using the Illumina bcl2fastq conversion software. Individual library size ranged from 17 million to 42.7 million reads. RNA sequencing analysis was performed using the SUSHI framework (63), which encompassed the following steps: Read quality was inspected using FastQC, and sequencing adaptors removed using fastp; Alignment of the RNA sequencing reads using the STAR aligner (64) and with the Ensembl mouse genome build GRCm39 (release 106) as the reference (65); the counting of gene-level expression values using the “featureCounts” function of the R package Rsubread (66); differential expression using the generalized linear model as implemented by the DESeq2 Bioconductor R package (67), and gene ontology term pathway analysis using the hypergeometric overrepresentation test via the “enrichGO” function of the clusterProfiler Bioconductor R package (68). All R functions were executed on R version 4.2.2 and Bioconductor version 3.16. scRNA-seq was performed by the Functional Genomics Center Zürich. TotalSeq anti-mouse Hashtag reagent [a mixture of two monoclonal antibodies—specific against mouse CD45 and major histocompatibility complex class I—conjugated to the same oligonucleotide was used for multiplex scSeq according to the manufacturer’s instructions (BioLegend)]. Samples were prepared for sequencing using the 10X Genomics with the BioLegend TotalSeq-C Feature Barcoding Single-Cell reagent kit, following the manufacturer’s protocol. Sequencing was performed on the Illumina NovaSeq 6000 (100 cycles). Demultiplexing was performed using the Illumina bcl2fastq conversion software. Gene expression sequencing generated 482 million reads, and feature barcode sequencing 66 million reads. Initial analysis of scRNA-seq data was performed using the SUSHI framework that encompassed the following steps: Read quality was inspected using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/); cell identification, read mapping and gene quantification, and sample demultiplexing were performed using STARsolo for the gene expression data, and 10X Genomics cellRanger version 7.0.0 for the hashtag antibodies data, with the GENCODE mouse genome build GRCm39 release M26 (69) and the Seurat version 4.1.1.9003 (70) Bioconductor R package, was used to demultiplex the cells, identify cell doublets and dropouts based on hashtags, normalize the gene expression counts (log-normalized, scale factor of 10,000), find variable genes (vst, 3000 genes), scale and center the dataset, variance-transform the counts using “SCTransform,” run PCA and Uniform Manifold Approximation and Projection (UMAP) dimensional reduction (1 to 20 dimensions), find neighbors (PCA, 1 to 20 dimensions) and clusters (multiple resolutions from 0.2 to 1, with 0.6 the resolution used), and find all marker genes for each cluster (Wilcox). The code used for these steps is available on the “ezRun” GitHub page (https://github.com/uzh/ezRun/blob/master/R/app-SCFeatBarcoding.R). All R functions were executed on R version 4.2.2 and Bioconductor version 3.16. Bulk RNA sequences are available under the accession number GSE225189 and scRNA sequences under GSE225379.
Statistics
All statistical analysis was performed using GraphPad prism 5.0 software. Student’s t test was used for comparisons between two groups, and one-way analysis of variance (ANOVA) was used for comparisons of more than two groups, followed by Dunn’s multiple comparisons correction. P values are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Acknowledgments
We thank M. Artola-Boran for help in cultivating organoids, K. Bertram for support with cloning strategies, and all members of the Müller laboratory for helpful discussions.
Funding: This work was supported by the Swiss National Science Foundation project grant 310030_192490 (to A.M.), the Comprehensive Cancer Center Zürich project grant (to A.M. and P.L.), the Forschungskredit of the University of Zürich (to J.H. and Z.N.), and the Medical Faculty of the University of Zürich (to A.M.).
Author contributions: Conceptualization: A.M., J.H., and Z.N. Methodology: J.H., Z.N., P.L., G.P., T.V., and K.B. Investigation: J.H., Z.N., P.L., and G.P. Visualization: J.H., Z.N., and P.L. Supervision: A.M. Writing—original draft: A.M. Writing—review and editing: A.M., J.H., Z.N., P.L., G.P., T.V., and K.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.. Bulk and scRNA-seq data are available at GEO under the accession numbers GSE225189 and GSE225379.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.M. Amieva, R. M. Peek Jr., Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 150, 64–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.U. Dutta, P. K. Garg, R. Kumar, R. K. Tandon, Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am. J. Gastroenterol. 95, 784–787 (2000). [DOI] [PubMed] [Google Scholar]
- 3.V. Nagaraja, G. D. Eslick, Systematic review with meta-analysis: The relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment. Pharmacol. Ther. 39, 745–750 (2014). [DOI] [PubMed] [Google Scholar]
- 4.L. Mughini-Gras, M. Schaapveld, J. Kramers, S. Mooij, E. A. Neefjes-Borst, W. v. Pelt, J. Neefjes, Increased colon cancer risk after severe Salmonella infection. PLOS ONE 13, e0189721 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J. C. Arthur, E. Perez-Chanona, M. Mühlbauer, S. Tomkovich, J. M. Uronis, T. J. Fan, B. J. Campbell, T. Abujamel, B. Dogan, A. B. Rogers, J. M. Rhodes, A. Stintzi, K. W. Simpson, J. J. Hansen, T. O. Keku, A. A. Fodor, C. Jobin, Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. J. Dziubanska-Kusibab, H. Berger, F. Battistini, B. A. M. Bouwman, A. Iftekhar, R. Katainen, T. Cajuso, N. Crosetto, M. Orozco, L. A. Aaltonen, T. F. Meyer, Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26, 1063–1069 (2020). [DOI] [PubMed] [Google Scholar]
- 7.C. Pleguezuelos-Manzano, J. Puschhof, A. Rosendahl Huber, A. van Hoeck, H. M. Wood, J. Nomburg, C. Gurjao, F. Manders, G. Dalmasso, P. B. Stege, F. L. Paganelli, M. H. Geurts, J. Beumer, T. Mizutani, Y. Miao, R. van der Linden, S. van der Elst, J. C. Ambrose, P. Arumugam, E. L. Baple, M. Bleda, F. Boardman-Pretty, J. M. Boissiere, C. R. Boustred, H. Brittain, M. J. Caulfield, G. C. Chan, C. E. H. Craig, L. C. Daugherty, A. de Burca, A. Devereau, G. Elgar, R. E. Foulger, T. Fowler, P. Furió-Tarí, J. M. Hackett, D. Halai, A. Hamblin, S. Henderson, J. E. Holman, T. J. P. Hubbard, K. Ibáñez, R. Jackson, L. J. Jones, D. Kasperaviciute, M. Kayikci, L. Lahnstein, L. Lawson, S. E. A. Leigh, I. U. S. Leong, F. J. Lopez, F. Maleady-Crowe, J. Mason, E. M. McDonagh, L. Moutsianas, M. Mueller, N. Murugaesu, A. C. Need, C. A. Odhams, C. Patch, D. Perez-Gil, D. Polychronopoulos, J. Pullinger, T. Rahim, A. Rendon, P. Riesgo-Ferreiro, T. Rogers, M. Ryten, K. Savage, K. Sawant, R. H. Scott, A. Siddiq, A. Sieghart, D. Smedley, K. R. Smith, A. Sosinsky, W. Spooner, H. E. Stevens, A. Stuckey, R. Sultana, E. R. A. Thomas, S. R. Thompson, C. Tregidgo, A. Tucci, E. Walsh, S. A. Watters, M. J. Welland, E. Williams, K. Witkowska, S. M. Wood, M. Zarowiecki, K. C. Garcia, J. Top, R. J. L. Willems, M. Giannakis, R. Bonnet, P. Quirke, M. Meyerson, E. Cuppen, R. van Boxtel, H. Clevers; Genomics England Research Consortium , Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. E. M. Ibler, M. ElGhazaly, K. L. Naylor, N. A. Bulgakova, S. F. el-Khamisy, D. Humphreys, Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection. Nat. Commun. 10, 4040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O. C. B. Martin, A. Bergonzini, M. Lopez Chiloeches, E. Paparouna, D. Butter, S. D. P. Theodorou, M. M. Haykal, E. Boutet-Robinet, T. Tebaldi, A. Wakeham, M. Rhen, V. G. Gorgoulis, T. Mak, I. S. Pateras, T. Frisan, Influence of the microenvironment on modulation of the host response by typhoid toxin. Cell Rep. 35, 108931 (2021). [DOI] [PubMed] [Google Scholar]
- 10.M. Bauer, Z. Nascakova, A. I. Mihai, P. F. Cheng, M. P. Levesque, S. Lampart, R. Hurwitz, L. Pfannkuch, J. Dobrovolna, M. Jacobs, S. Bartfeld, A. Dohlman, X. Shen, A. A. Gall, N. R. Salama, A. Töpfer, A. Weber, T. F. Meyer, P. Janscak, A. Müller, The ALPK1/TIFA/NF-κB axis links a bacterial carcinogen to R-loop-induced replication stress. Nat. Commun. 11, 5117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C. Chumduri, R. K. Gurumurthy, P. K. Zadora, Y. Mi, T. F. Meyer, Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe 13, 746–758 (2013). [DOI] [PubMed] [Google Scholar]
- 12.M. Plummer, S. Franceschi, J. Vignat, D. Forman, C. de Martel, Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 136, 487–490 (2015). [DOI] [PubMed] [Google Scholar]
- 13.F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre, A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 14.J. Parsonnet, G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, R. K. Sibley, Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325, 1127–1131 (1991). [DOI] [PubMed] [Google Scholar]
- 15.J. Parsonnet, D. Vandersteen, J. Goates, R. K. Sibley, J. Pritikin, Y. Chang, Helicobacter pylori infection in intestinal- and diffuse-type gastric adenocarcinomas. J. Natl. Cancer Inst. 83, 640–643 (1991). [DOI] [PubMed] [Google Scholar]
- 16.T. Rokkas, A. Rokka, P. Portincasa, A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann. Gastroenterol. 30, 414–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.I. J. Choi, M. C. Kook, Y. I. Kim, S. J. Cho, J. Y. Lee, C. G. Kim, B. Park, B. H. Nam, Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N. Engl. J. Med. 378, 1085–1095 (2018). [DOI] [PubMed] [Google Scholar]
- 18.I. J. Choi, Y. I. Kim, B. Park, Helicobacter pylori and prevention of gastric cancer. N. Engl. J. Med. 378, 2244–2245 (2018). [DOI] [PubMed] [Google Scholar]
- 19.S. Backert, R. Haas, M. Gerhard, M. Naumann, The Helicobacter pylori type IV secretion system encoded by the cag pathogenicity island: Architecture, function, and signaling. Curr. Top. Microbiol. Immunol. 413, 187–220 (2017). [DOI] [PubMed] [Google Scholar]
- 20.J. Q. Huang, G. F. Zheng, K. Sumanac, E. J. Irvine, R. H. Hunt, Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology 125, 1636–1644 (2003). [DOI] [PubMed] [Google Scholar]
- 21.S. Zimmermann, L. Pfannkuch, M. A. al-Zeer, S. Bartfeld, M. Koch, J. Liu, C. Rechner, M. Soerensen, O. Sokolova, A. Zamyatina, P. Kosma, A. P. Mäurer, F. Glowinski, K. P. Pleissner, M. Schmid, V. Brinkmann, A. Karlas, M. Naumann, M. Rother, N. Machuy, T. F. Meyer, ALPK1- and TIFA-dependent innate immune response triggered by the Helicobacter pylori type IV secretion system. Cell Rep. 20, 2384–2395 (2017). [DOI] [PubMed] [Google Scholar]
- 22.A. Gall, R. G. Gaudet, S. D. Gray-Owen, N. R. Salama, TIFA signaling in gastric epithelial cells initiates the cag type 4 secretion system-dependent innate immune response to Helicobacter pylori infection. mBio 8, e01168-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. C. Stein, E. Faber, S. H. Bats, T. Murillo, Y. Speidel, N. Coombs, C. Josenhans, Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLOS Pathog. 13, e1006514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.P. Zhou, Y. She, N. Dong, P. Li, H. He, A. Borio, Q. Wu, S. Lu, X. Ding, Y. Cao, Y. Xu, W. Gao, M. Dong, J. Ding, D. C. Wang, A. Zamyatina, F. Shao, Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature 561, 122–126 (2018). [DOI] [PubMed] [Google Scholar]
- 25.S. Imai, T. Ooki, N. Murata-Kamiya, D. Komura, K. Tahmina, W. Wu, A. Takahashi-Kanemitsu, C. T. Knight, A. Kunita, N. Suzuki, A. A. del Valle, M. Tsuboi, M. Hata, Y. Hayakawa, N. Ohnishi, K. Ueda, M. Fukayama, T. Ushiku, S. Ishikawa, M. Hatakeyama, Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 29, 941–958.e10 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Y. Usui, Y. Taniyama, M. Endo, Y. N. Koyanagi, Y. Kasugai, I. Oze, H. Ito, I. Imoto, T. Tanaka, M. Tajika, Y. Niwa, Y. Iwasaki, T. Aoi, N. Hakozaki, S. Takata, K. Suzuki, C. Terao, M. Hatakeyama, M. Hirata, K. Sugano, T. Yoshida, Y. Kamatani, H. Nakagawa, K. Matsuda, Y. Murakami, A. B. Spurdle, K. Matsuo, Y. Momozawa, Helicobacter pylori, homologous-recombination genes, and gastric cancer. N. Engl. J. Med. 388, 1181–1190 (2023). [DOI] [PubMed] [Google Scholar]
- 27.C. Aguilar, M. Pauzuolis, M. Pompaiah, E. Vafadarnejad, P. Arampatzi, M. Fischer, D. Narres, M. Neyazi, Ö. Kayisoglu, T. Sell, N. Blüthgen, M. Morkel, A. Wiegering, C. T. Germer, S. Kircher, A. Rosenwald, A. E. Saliba, S. Bartfeld, Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis. Nat. Commun. 13, 5878 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. A. Busslinger, B. L. A. Weusten, A. Bogte, H. Begthel, L. A. A. Brosens, H. Clevers, Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 34, 108819 (2021). [DOI] [PubMed] [Google Scholar]
- 29.A. E. Moor, Y. Harnik, S. Ben-Moshe, E. E. Massasa, M. Rozenberg, R. Eilam, K. Bahar Halpern, S. Itzkovitz, Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156–1167.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 30.S. Han, J. Fink, D. J. Jörg, E. Lee, M. K. Yum, L. Chatzeli, S. R. Merker, M. Josserand, T. Trendafilova, A. Andersson-Rolf, C. Dabrowska, H. Kim, R. Naumann, J. H. Lee, N. Sasaki, R. L. Mort, O. Basak, H. Clevers, D. E. Stange, A. Philpott, J. K. Kim, B. D. Simons, B. K. Koo, Defining the identity and dynamics of adult gastric isthmus stem cells. Cell Stem Cell 25, 342–356.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.X. Han, R. Wang, Y. Zhou, L. Fei, H. Sun, S. Lai, A. Saadatpour, Z. Zhou, H. Chen, F. Ye, D. Huang, Y. Xu, W. Huang, M. Jiang, X. Jiang, J. Mao, Y. Chen, C. Lu, J. Xie, Q. Fang, Y. Wang, R. Yue, T. Li, H. Huang, S. H. Orkin, G. C. Yuan, M. Chen, G. Guo, Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 32.T. L. Cover, D. B. Lacy, M. D. Ohi, The Helicobacter pylori Cag type IV secretion system. Trends Microbiol. 28, 682–695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.I. C. Arnold, X. Zhang, S. Urban, M. Artola-Borán, M. G. Manz, K. M. Ottemann, A. Müller, NLRP3 controls the development of gastrointestinal CD11b+ dendritic cells in the steady state and during chronic bacterial infection. Cell Rep. 21, 3860–3872 (2017). [DOI] [PubMed] [Google Scholar]
- 34.J. Reichmuth, J. vom Berg, M. Brügger, G. Hausmann, T. Valenta, K. Basler, Manipulating the murine Lgr5 locus using a rapid, efficient and flexible CRISPR/Cas9 pipeline. bioRxiv 2020.12.31.424946 [Preprint] (1 January 2021). 10.1101/2020.12.31.424946. [DOI]
- 35.H. Fazilaty, M. D. Brügger, T. Valenta, B. M. Szczerba, L. Berkova, N. Doumpas, G. Hausmann, M. Scharl, K. Basler, Tracing colonic embryonic transcriptional profiles and their reactivation upon intestinal damage. Cell Rep. 36, 109484 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network , Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. van de Wetering, E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, H. Clevers, The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 (2002). [DOI] [PubMed] [Google Scholar]
- 38.I. M. Toller, K. J. Neelsen, M. Steger, M. L. Hartung, M. O. Hottiger, M. Stucki, B. Kalali, M. Gerhard, A. A. Sartori, M. Lopes, A. Müller, Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14944–14949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. L. Hartung, D. C. Gruber, K. N. Koch, L. Grüter, H. Rehrauer, N. Tegtmeyer, S. Backert, A. Müller, H. pylori -induced DNA strand breaks are introduced by nucleotide excision repair endonucleases and promote NF-κB target gene expression. Cell Rep. 13, 70–79 (2015). [DOI] [PubMed] [Google Scholar]
- 40.D. E. Stange, B. K. Koo, M. Huch, G. Sibbel, O. Basak, A. Lyubimova, P. Kujala, S. Bartfeld, J. Koster, J. H. Geahlen, P. J. Peters, J. H. van Es, M. van de Wetering, J. C. Mills, H. Clevers, Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Sigal, M. D. M. Reinés, S. Müllerke, C. Fischer, M. Kapalczynska, H. Berger, E. R. M. Bakker, H. J. Mollenkopf, M. E. Rothenberg, B. Wiedenmann, S. Sauer, T. F. Meyer, R-spondin-3 induces secretory, antimicrobial Lgr5+ cells in the stomach. Nat. Cell Biol. 21, 812–823 (2019). [DOI] [PubMed] [Google Scholar]
- 42.M. A. Schumacher, E. Aihara, R. Feng, A. Engevik, N. F. Shroyer, K. M. Ottemann, R. T. Worrell, M. H. Montrose, R. A. Shivdasani, Y. Zavros, The use of murine-derived fundic organoids in studies of gastric physiology. J. Physiol. 593, 1809–1827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Mueller, D. S. Merrell, J. Grimm, S. Falkow, Profiling of microdissected gastric epithelial cells reveals a cell type-specific response to Helicobacter pylori infection. Gastroenterology 127, 1446–1462 (2004). [DOI] [PubMed] [Google Scholar]
- 44.G. R. Van den Brink, K. M. Tytgat, R. W. Van der Hulst, C. M. Van der Loos, A. W. Einerhand, H. A. Büller, J. Dekker, H. pylori colocalises with MUC5AC in the human stomach. Gut. 46, 601–607 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. Sigal, M. E. Rothenberg, C. Y. Logan, J. Y. Lee, R. W. Honaker, R. L. Cooper, B. Passarelli, M. Camorlinga, D. M. Bouley, G. Alvarez, R. Nusse, J. Torres, M. R. Amieva, Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology 148, 1392–1404.e21 (2015). [DOI] [PubMed] [Google Scholar]
- 46.D. Keilberg, Y. Zavros, B. Shepherd, N. R. Salama, K. M. Ottemann, Spatial and temporal shifts in bacterial biogeography and gland occupation during the development of a chronic infection. mBio 7, e01705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Wizenty, S. Müllerke, M. Kolesnichenko, J. Heuberger, M. Lin, A. S. Fischer, H. J. Mollenkopf, H. Berger, F. Tacke, M. Sigal, Gastric stem cells promote inflammation and gland remodeling in response to Helicobacter pylori via Rspo3-Lgr4 axis. EMBO J. 41, e109996 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Uehara, D. Ma, Y. Yao, J. P. Lynch, K. Morales, A. Ziober, M. Feldman, H. Ota, A. R. Sepulveda, H. pylori infection is associated with DNA damage of Lgr5-positive epithelial stem cells in the stomach of patients with gastric cancer. Dig. Dis. Sci. 58, 140–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Leushacke, S. H. Tan, A. Wong, Y. Swathi, A. Hajamohideen, L. T. Tan, J. Goh, E. Wong, S. L. I. J. Denil, K. Murakami, N. Barker, Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017). [DOI] [PubMed] [Google Scholar]
- 50.A. Fatehullah, Y. Terakado, S. Sagiraju, T. L. Tan, T. Sheng, S. H. Tan, K. Murakami, Y. Swathi, N. Ang, R. Rajarethinam, T. Ming, P. Tan, B. Lee, N. Barker, A tumour-resident Lgr5+ stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 23, 1299–1313 (2021). [DOI] [PubMed] [Google Scholar]
- 51.T. Shimizu, H. Marusawa, Y. Matsumoto, T. Inuzuka, A. Ikeda, Y. Fujii, S. Minamiguchi, S.’. Miyamoto, T. Kou, Y. Sakai, J. E. Crabtree, T. Chiba, Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 147, 407–17.e3 (2014). [DOI] [PubMed] [Google Scholar]
- 52.T. Yoshida, T. Yamaguchi, S. Maekawa, S. Takano, T. Kuno, K. Tanaka, F. Iwamoto, Y. Tsukui, S. Kobayashi, Y. Asakawa, H. Shindo, M. Fukasawa, Y. Nakayama, T. Inoue, T. Uetake, M. Ohtaka, T. Sato, K. Mochizuki, N. Enomoto, Identification of early genetic changes in well-differentiated intramucosal gastric carcinoma by target deep sequencing. Gastric Cancer 22, 742–750 (2019). [DOI] [PubMed] [Google Scholar]
- 53.M. V. Nemtsova, A. I. Kalinkin, E. B. Kuznetsova, I. V. Bure, E. A. Alekseeva, I. I. Bykov, T. V. Khorobrykh, D. S. Mikhaylenko, A. S. Tanas, S. I. Kutsev, D. V. Zaletaev, V. V. Strelnikov, Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci. Rep. 10, 504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.N. Setia, J. W. Clark, D. G. Duda, T. S. Hong, E. L. Kwak, J. T. Mullen, G. Y. Lauwers, Familial gastric cancers. Oncologist 20, 1365–1377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Bartfeld, H. Clevers, Organoids as model for infectious diseases: Culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J. Vis. Exp. 12, 53359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A. Sayi, E. Kohler, I. Hitzler, I. Arnold, R. Schwendener, H. Rehrauer, A. Müller, The CD4+ T cell-mediated IFN-γ response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J. Immunol. 182, 7085–7101 (2009). [DOI] [PubMed] [Google Scholar]
- 57.S. Censini, C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, A. Covacci, Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U.S.A. 93, 14648–14653 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.I. C. Arnold, J. Y. Lee, M. R. Amieva, A. Roers, R. A. Flavell, T. Sparwasser, A. Müller, Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140, 199–209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. F. Dekkers, M. Alieva, L. M. Wellens, H. C. R. Ariese, P. R. Jamieson, A. M. Vonk, G. D. Amatngalim, H. Hu, K. C. Oost, H. J. G. Snippert, J. M. Beekman, E. J. Wehrens, J. E. Visvader, H. Clevers, A. C. Rios, High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019). [DOI] [PubMed] [Google Scholar]
- 61.A. De Magis, S. G. Manzo, M. Russo, J. Marinello, R. Morigi, O. Sordet, G. Capranico, DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 816–825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.B. K. Koo, D. E. Stange, T. Sato, W. Karthaus, H. F. Farin, M. Huch, J. van Es, H. Clevers, Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 9, 81–83 (2011). [DOI] [PubMed] [Google Scholar]
- 63.M. Hatakeyama, L. Opitz, G. Russo, W. Qi, R. Schlapbach, H. Rehrauer, SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinformatics 17, 228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.K. L. Howe, P. Achuthan, J. Allen, J. Allen, J. Alvarez-Jarreta, M. R. Amode, I. M. Armean, A. G. Azov, R. Bennett, J. Bhai, K. Billis, S. Boddu, M. Charkhchi, C. Cummins, L. da Rin Fioretto, C. Davidson, K. Dodiya, B. el Houdaigui, R. Fatima, A. Gall, C. Garcia Giron, T. Grego, C. Guijarro-Clarke, L. Haggerty, A. Hemrom, T. Hourlier, O. G. Izuogu, T. Juettemann, V. Kaikala, M. Kay, I. Lavidas, T. le, D. Lemos, J. Gonzalez Martinez, J. C. Marugán, T. Maurel, A. C. McMahon, S. Mohanan, B. Moore, M. Muffato, D. N. Oheh, D. Paraschas, A. Parker, A. Parton, I. Prosovetskaia, M. P. Sakthivel, A. I. A. Salam, B. M. Schmitt, H. Schuilenburg, D. Sheppard, E. Steed, M. Szpak, M. Szuba, K. Taylor, A. Thormann, G. Threadgold, B. Walts, A. Winterbottom, M. Chakiachvili, A. Chaubal, N. de Silva, B. Flint, A. Frankish, S. E. Hunt, G. R. IIsley, N. Langridge, J. E. Loveland, F. J. Martin, J. M. Mudge, J. Morales, E. Perry, M. Ruffier, J. Tate, D. Thybert, S. J. Trevanion, F. Cunningham, A. D. Yates, D. R. Zerbino, P. Flicek, Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Y. Liao, G. K. Smyth, W. Shi, The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. Frankish, M. Diekhans, I. Jungreis, J. Lagarde, J. E. Loveland, J. M. Mudge, C. Sisu, J. C. Wright, J. Armstrong, I. Barnes, A. Berry, A. Bignell, C. Boix, S. Carbonell Sala, F. Cunningham, T. di Domenico, S. Donaldson, I. T. Fiddes, C. García Girón, J. M. Gonzalez, T. Grego, M. Hardy, T. Hourlier, K. L. Howe, T. Hunt, O. G. Izuogu, R. Johnson, F. J. Martin, L. Martínez, S. Mohanan, P. Muir, F. C. P. Navarro, A. Parker, B. Pei, F. Pozo, F. C. Riera, M. Ruffier, B. M. Schmitt, E. Stapleton, M. M. Suner, I. Sycheva, B. Uszczynska-Ratajczak, M. Y. Wolf, J. Xu, Y. T. Yang, A. Yates, D. Zerbino, Y. Zhang, J. S. Choudhary, M. Gerstein, R. Guigó, T. J. P. Hubbard, M. Kellis, B. Paten, M. L. Tress, P. Flicek, Gencode 2021. Nucleic Acids Res. 49, D916–D923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5