Abstract
The essential oils of peppermint (Mentha x piperita) and spearmint (Mentha spicata) are distinguished by the oxygenation position on the p-menthane ring of the constitutive monoterpenes that is conferred by two regiospecific cytochrome P450 limonene-3- and limonene-6-hydroxylases. Following hydroxylation of limonene, an apparently similar dehydrogenase oxidizes (−)-trans-isopiperitenol to (−)-isopiperitenone in peppermint and (−)-trans-carveol to (−)-carvone in spearmint. Random sequencing of a peppermint oil gland secretory cell cDNA library revealed a large number of clones that specified redox-type enzymes, including dehydrogenases. Full-length dehydrogenase clones were screened by functional expression in Escherichia coli using a recently developed in situ assay. A single full-length acquisition encoding (−)-trans-isopiperitenol dehydrogenase (ISPD) was isolated. The (−)-ISPD cDNA has an open reading frame of 795 bp that encodes a 265-residue enzyme with a calculated molecular mass of 27,191. Nondegenerate primers were designed based on the (−)-trans-ISPD cDNA sequence and employed to screen a spearmint oil gland secretory cell cDNA library from which a 5′-truncated cDNA encoding the spearmint homolog, (−)-trans-carveol-dehydrogenase, was isolated. Reverse transcription-PCR amplification and RACE were used to acquire the remaining 5′-sequence from RNA isolated from oil gland secretory cells of spearmint leaf. The full-length spearmint dehydrogenase shares >99% amino acid identity with its peppermint homolog and both dehydrogenases are capable of utilizing (−)-trans-isopiperitenol and (−)-trans-carveol. These isopiperitenol/carveol dehydrogenases are members of the short-chain dehydrogenase/reductase superfamily and are related to other plant short-chain dehydrogenases/reductases involved in secondary metabolism (lignan biosynthesis), stress responses, and phytosteroid biosynthesis, but they are quite dissimilar (approximately 13% identity) to the monoterpene reductases of mint involved in (−)-menthol biosynthesis. The isolation of the genes specifying redox enzymes of monoterpene biosynthesis in mint indicates that these genes arose from different ancestors and not by simple duplication and differentiation of a common progenitor, as might have been anticipated based on the common reaction chemistry and structural similarity of the substrate monoterpenes.
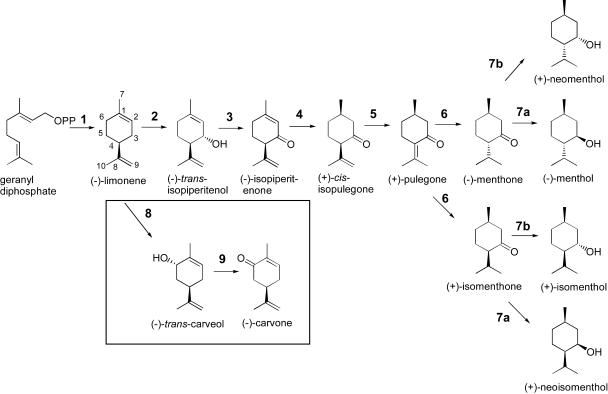
Monoterpenes, the C10 members of the terpenoid family of secondary metabolites, are largely responsible for the fragrance and flavor characteristics of plants and are well known as components of plant essential oils (Parry, 1969). The essential oils of both peppermint (Mentha x piperita), a sterile cross between Mentha spicata and Mentha aquatica (Murray et al., 1972; Harley and Brighton, 1977), and spearmint (Mentha spicata) consist primarily of monoterpenes (Lawrence, 1981) that are distinguished by the position of oxygenation on the p-menthane ring conferred by two distinct regiospecific cytochrome P450 limonene hydroxylases (Fig. 1). Thus, following cyclization of geranyl diphosphate by limonene synthase (Alonso et al., 1992), the common precursor (−)-limonene is oxygenated either by (−)-4S-limonene-3-hydroxylase in peppermint to form (−)-trans-isopiperitenol or (−)-4S-limonene-6-hydroxylase in spearmint to form (−)-trans-carveol (Karp et al., 1990). Whereas the limonene synthase from these 2 plants is very similar (93% identical at the amino acid level; Colby et al., 1993), the 2 hydroxylases exhibit only 70% deduced identity with most of the sequence differences localized to the presumptive active sites (Lupien et al., 1999).
Figure 1.
The principal pathways of monoterpene biosynthesis in peppermint and spearmint. The enzymes indicated are (−)-limonene synthase (1), cytochrome P450 (−)-limonene-3-hydroxylase (2), (−)-trans-isopiperitenol dehydrogenase (3), (−)-isopiperitenone reductase (4), (+)-cis-isopulegone isomerase (5), (+)-pulegone reductase (6), (−)-menthone:(−)-menthol reductase (7a), and (−)-menthone:(+)-neomenthol reductase (7b) in peppermint, and cytochrome P450 (−)-limonene-6-hydroxylase (8) and (−)-trans-carveol dehydrogenase (9) in spearmint (boxed structures). The more commonly used p-menthane monoterpene numbering system is utilized here in which the exocyclic methyl (C7) is appended to C1, the oxygen is at C3 (or C6), and the isopropyl group is attached to C4; thus, (−)-menthol is 1R:3R:4S configured.
In peppermint, several redox enzymes and an isomerase transform the initial hydroxylated product to (−)-menthol (Fig. 1), the principal monoterpene of the commercial essential oil and the component responsible for the familiar cooling sensation of peppermint (Lawrence, 1981). First, a soluble NAD+-dependent dehydrogenase oxidizes the α,β-unsaturated alcohol (−)-trans-isopiperitenol to the corresponding ketone (−)-isopiperitenone. A soluble NADPH dependent, endocyclic double-bond reductase then reduces (−)-isopiperitenone to (+)-cis-isopulegone. The remaining double bond is next moved into conjugation with the carbonyl by an isomerase to produce (+)-pulegone, and a second NADPH-dependent reductase reduces the conjugated double bond to yield both (−)-menthone and (+)-isomenthone. Finally, two NADPH-dependent reductases utilize both (−)-menthone and (+)-isomenthone to complete the reaction sequence. One reductase produces mainly (−)-menthol (from menthone) and (+)-neoisomenthol (from isomenthone), while the other produces primarily (+)-neomenthol (from menthone) and (+)-isomenthol (from isomenthone).
In contrast to the extended pathway in peppermint, the initial hydroxylated product of spearmint, (−)-trans-carveol, is simply oxidized to (−)-carvone, the principal and characteristic component of spearmint oil (Lawrence, 1981). Although the full complement of redox enzymes and the isomerase are present in spearmint (Croteau et al., 1991), (−)-carvone is an inefficient substrate for the initial double-bond reductase, thereby allowing (−)-carvone to accumulate. Thus, the oxidation of the initially formed monoterpenol is the only common redox step in monoterpene metabolism of these two Mentha species. The monoterpenol dehydrogenases of spearmint, peppermint, and scotch spearmint (Mentha x gentiles) all appear capable of utilizing both (−)-trans-carveol and (−)-trans-isopiperitenol as a substrate, an observation perhaps not surprising because spearmint is a parent of both peppermint and scotch spearmint (Harley and Brighton, 1977).
The monoterpene biosynthetic pathways in peppermint and spearmint have been elucidated mainly by time-course studies, feeding experiments with labeled precursors, and characterization of the responsible enzymes in cell-free enzyme systems (for reviews of the pathway, see Croteau, 1987; Wise and Croteau, 1997). Several of the genes encoding early pathway enzymes have been cloned by reverse genetic methods and characterized, including geranyl diphosphate synthase (Burke et al., 1999), limonene synthase (Colby et al., 1993), and the cytochrome P450 limonene-3- and limonene-6-hydroxylases (Lupien et al., 1999).
The remaining pathway genes encoding mainly redox enzymes appeared difficult to approach by reverse genetic methods, and homology-based cloning seemed impractical. Completion of an expressed sequence tag (EST) project based on a cDNA library from peppermint oil gland secretory cells (the site of monoterpene biosynthesis; Lange et al., 2000) coupled with a novel in situ screening assay, employing functional heterologous expression in Escherichia coli (Ringer et al., 2003) provided the impetus for isolating cDNAs encoding these later pathway enzymes. In this article, we describe the isolation and characterization of cDNA clones encoding (−)-trans-isopiperitenol/(−)-trans-carveol dehydrogenase from peppermint and spearmint, report the comparison of the recombinant enzyme and gene with previously described (−)-isopiperitenone reductase and (+)-pulegone reductase (Ringer et al., 2003), and with the menthone reductases described in the accompanying article (Davis et al., 2005), and discuss the implications of these findings for the evolutionary origins of monoterpene biosynthesis in mint.
RESULTS
Isolation of a cDNA Encoding Isopiperitenol Dehydrogenase
Monoterpene biosynthesis in peppermint and spearmint is localized to the specialized secretory cells of epidermal oil glands (Gershenzon et al., 1989; McCaskill et al., 1992), thus making these cells a highly enriched source of transcripts for the relevant biosynthetic enzymes. Random sequencing of a cDNA library constructed from transcripts isolated from peppermint oil gland secretory cells revealed very high abundance of cDNAs encoding previously described enzymes involved in (−)-menthol biosynthesis, as well as a high proportion of cDNAs encoding several groups of putative redox enzymes that could be involved in monoterpene metabolism (Lange et al., 2000). In a previous contribution (Ringer et al., 2003), we described an in situ functional screening strategy whereby groups of these candidate genes were subcloned into the expression vector pSBET and expressed in E. coli BL21(DE3) as host, followed by incubation of the intact transformed bacteria with test substrates, isolation of volatile products by steam distillation, and product analysis by gas chromatography-mass spectrometry (GC-MS). This efficient approach led to the isolation of cDNAs encoding the monoterpene biosynthetic enzymes isopiperitenone reductase and pulegone reductase (Ringer et al., 2003), as well as menthone:neomenthol reductase (Davis et al., 2005; Fig. 1). The identical functional screening approach was employed in an attempt to clone isopiperitenol dehydrogenase (ISPD) from the EST library.
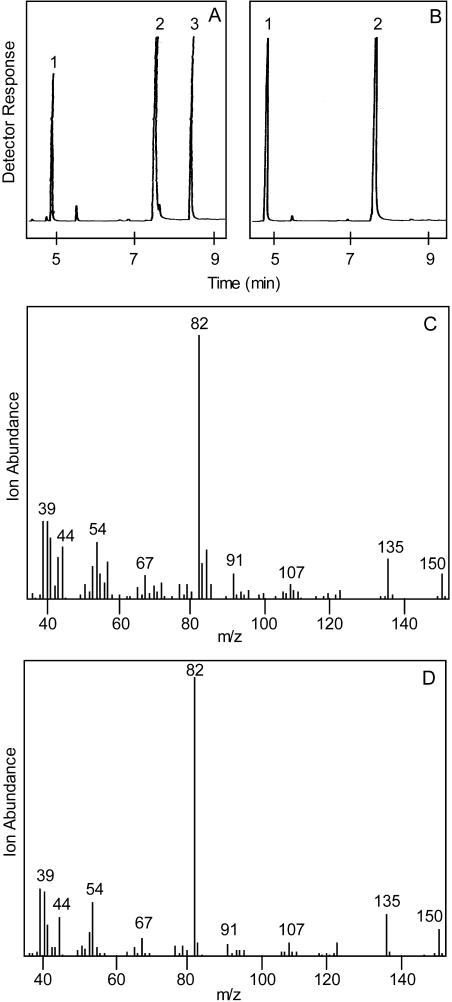
Bioinformatic processing (see “Materials and Methods”) of the secretory cell cDNA library (Lange et al., 2000) yielded approximately 60 ESTs that appeared to encode dehydrogenases and allowed elimination of previously defined dehydrogenases. The remaining ESTs were sorted by fragment assembly into eight unique, putative dehydrogenase acquisitions. In contrast to the reductase groups for which there were usually multiple copies of similar cDNAs (>90% identity) in each group (Ringer et al., 2003), probably indicative of allelic variants in the hexaploid peppermint genome, the putative dehydrogenase clones were much less abundant with two or three acquisitions in fewer overall groups and with several unique sequences as single acquisitions. The functional screening assay of eight candidate genes revealed a single full-length cDNA from the EST library that encoded a dehydrogenase capable of oxidizing (−)-trans-carveol to (−)-carvone. (−)-Carveol was used as substrate in the initial screen, because it is more readily available than (−)-trans-isopiperitenol. Based on previous results (Croteau et al., 1991), the dehydrogenase was presumed to utilize both monoterpenols, and this was subsequently verified by cell-free assay of the recombinant enzyme with NAD+ as cofactor (Figs. 2 and 3). No detectable product was formed in the absence of NAD+ or with boiled enzyme in the presence of this cofactor. Sequencing of the full-length clone revealed an open reading frame of 795 bp encoding a 265-residue enzyme of calculated molecular mass of 27,191.
Figure 2.
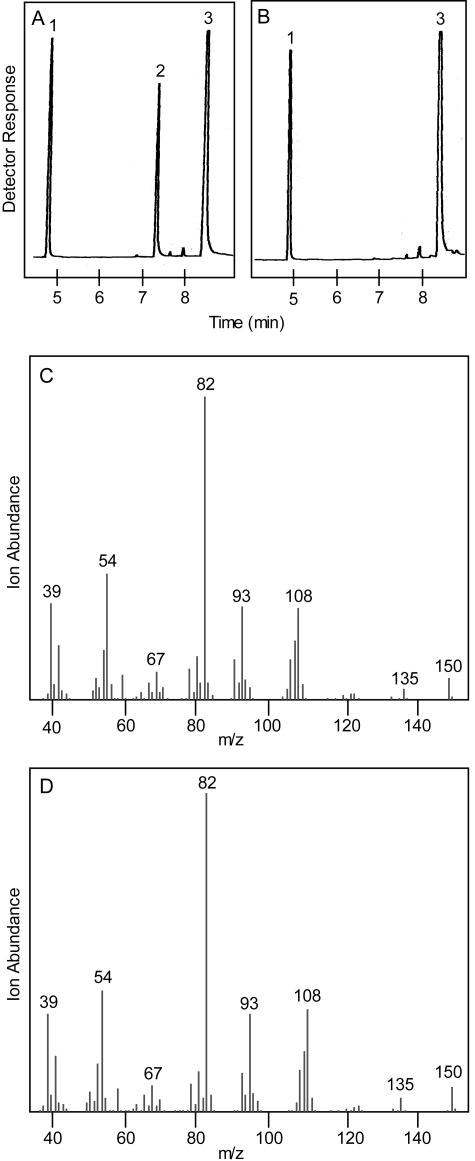
Capillary GC analysis of the products obtained in a standard assay with recombinant (−)-trans-ISPD from peppermint incubated with (−)-trans-carveol and NAD+ (A) and in the same assay without cofactor (NAD+) similarly incubated (B). The peaks indicated are camphor (internal standard; 1), (−)-carvone (2), and (−)-trans-carveol (3). The retention time and mass spectrum (C) of peak 2 are identical to those of authentic (−)-carvone (D).
Figure 3.
Capillary GC analysis of the products obtained in a standard assay with recombinant (−)-trans-ISPD from peppermint incubated with (−)-trans-isopiperitenol and NAD+ (A) and in the same assay without cofactor (NAD+) similarly incubated (B). The peaks indicated are camphor (internal standard; 1), (−)-trans-isopiperitenol (2), and (−)-isopiperitenone (3). The retention time and mass spectrum (C) of peak 3 are identical to those of authentic (−)-isopiperitenone (D).
Immunodetection of Carveol Dehydrogenase
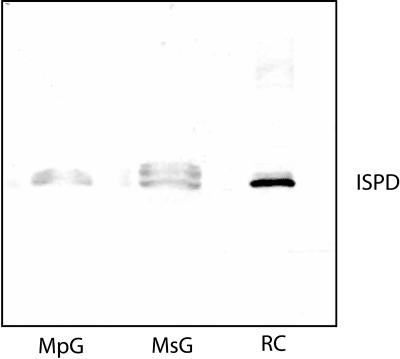
In spite of the significant differences in essential oil chemistry, the observation that both dehydrogenases of peppermint and spearmint appear to utilize (−)-trans-carveol and (−)-trans-isopiperitenol as substrates (Croteau et al., 1991), and the facts that spearmint is a parent of peppermint (Murray et al., 1972; Harley and Brighton, 1977) and that other genes of the two species are very similar (Lupien et al., 1999), suggested that the target monoterpenol dehydrogenases would also be very similar. Polyclonal antibodies prepared against the peppermint ISPD (Turner and Croteau, 2004) were used for immunoblotting of secretory cell extracts. A single band corresponding to ISPD (27 kD) was observed in the peppermint gland secretory cell extract, whereas 3 bands of different sizes (ranging from 27 kD to 30 kD) were visible in the spearmint gland secretory cell extract (Fig. 4). The smallest of the three was equivalent to the ISPD (peppermint) in size. This result suggested that several variants of carveol dehydrogenase may be expressed in spearmint.
Figure 4.
Immunoblot analysis of oil gland secretory cell extracts from peppermint (MpG) and spearmint (MsG), and recombinant (RC) ISPD using polyclonal antibodies raised against ISPD. A protein band equivalent in size to the recombinant ISPD is present in the secretory cell extracts of both plants. Two additional protein bands of larger size are present in the spearmint secretory cell extract.
Homology-Based Cloning of Carveol Dehydrogenase
Because the monoterpenol dehydrogenases appeared to be very similar (despite the two size variants), a homology-based PCR cloning approach was used to isolate the carveol dehydrogenase cDNA from a spearmint oil gland secretory cell library (Lupien et al., 1999). Primers designed to the 5′- and 3′-termini of the peppermint ISPD cDNA amplified what appeared to be a full-length version of the spearmint homolog (several independent clones were sequenced and were 100% identical) with >99% identity to the peppermint dehydrogenase. Additional 5′- and 3′-sequence information was gained with the use of gene-specific and vector-specific primers, and these additional data confirmed the original 3′-coding sequence and demonstrated the 3′-untranslated sequence to be 62% identical with the untranslated region of the peppermint ISPD cDNA (data not shown); however, additional 5′-sequence data showed all clones were truncated (1–7-bp truncations). To obtain the missing 5′-sequence and possibly isolate clones encoding the larger versions of spearmint carveol dehydrogenase detected in the immunoblot, internal primers were designed based on the isopiperitenol/carveol dehydrogenase sequence and single-stranded target cDNA generated from reverse transcription (RT) of RNA isolated and purified from oil gland secretory cells of spearmint leaves were used for 5′-RACE. A pool of 7-, 8-, and 10-d-old spearmint leaves were used for oil gland secretory cell isolation to ensure the presence of transcript for carveol dehydrogenase in immature glands. Because carveol does not accumulate in the essential oil of spearmint, the presence of both limonene (the precursor to carveol) and carvone (the product of the dehydrogenase) in the oil of these leaves (data not shown) indicated that carveol dehydrogenase transcript should be present. The clones obtained from 5′-RACE (approximately 550- and 720-bp fragments) had 100% identity with the cDNA clones obtained from the spearmint cDNA library but with additional 5′-sequence (1–7 bp), including a start codon. With this additional 5′-sequence, the deduced amino acid sequence of the spearmint dehydrogenase retained >99% amino acid identity with the peppermint dehydrogenase. Functional characterization of the recombinant spearmint dehydrogenase demonstrated it was capable of utilizing both monoterpenols as substrates. Using the homology-based PCR approach described above, several attempts were made to isolate a cDNA clone that encoded the presumptive larger versions of the dehydrogenase (Fig. 4), but these attempts were unsuccessful.
Enzyme Characterization
Because the peppermint and spearmint dehydrogenases were capable of utilizing both (−)-trans-carveol and (−)-trans-isopiperitenol and exhibited high amino acid sequence identity (>99% identity), only the ISPD from peppermint was chosen for characterization. This cDNA expressed well in E. coli in operationally soluble form and, following partial purification (approximately 75% purity), kinetic parameters were determined (Table I). At pH 7.5, the Km value for (−)-trans-carveol was 1.8 ± 0.2 μm and for NAD+ was 410 ± 29 μm with a kcat of 0.02 s−1. At pH 10, subsequently determined to be the optimum, kcat increased by 3-fold. The Km value for (−)-trans-isopiperitenol was 72 ± 10 μm with kcat about 10 times less than with (−)-trans-carveol at pH 7.5. The recombinant ISPD was capable of using NADP+ (Vrel approximately 8% at saturation compared to NAD+, measured with (−)-trans-carveol as substrate). Compared to (−)-trans-carveol, other alcohols tested at saturation with NAD+ as cofactor exhibited considerably lower relative velocities: (−)-trans-isopiperitenol (10%), (+)-neomenthol (8%), and (+)-neoisomenthol (4%). Whereas (−)-cis-isopiperitenol was a substrate (Vrel not determined), (−)-cis-carveol, (−)-menthol, and (+)-isomenthol were not detectably oxidized with either NAD+ or NADP+ as cofactor. (−)-Perillyl alcohol (7-hydroxy-limonene) was also not utilized by the peppermint dehydrogenase. ISPD was unable to catalyze the reduction of (−)-isopiperitenone or (−)-carvone to (−)-isopiperitenol or (−)-carveol, respectively, in the presence of NADH at pH 7.5.
Table I.
Kinetic parameters for recombinant ISPD of peppermint
|
Km ± se
|
kcat
|
|||
|---|---|---|---|---|
| (−)-Trans-carveol | (−)-Trans-isopiperitenol | NAD+ | (−)-Trans-carveol | (−)-Trans-isopiperitenol |
| μm | s−1 | |||
| 1.8 ± 0.2 | 72 ± 10 | 410 ± 29 | 0.02a/0.06b | 0.002a |
Assay pH 7.5.
Assay pH 10.
Calibrated size exclusion chromatography (Sephadex 200) of the recombinant ISPD (calculated size 27 kD), with eluant monitoring by activity assay and SDS-PAGE, revealed a broad elution pattern corresponding to dimeric and tetrameric forms (with slightly higher activity and SDS-PAGE band intensity at approximately 108 kD, corresponding to the tetramer). Previous studies with the native ISPD of peppermint had indicated a size of 66 kD (Kjonaas et al., 1985) corresponding to the dimer.
Sequence Analysis
The ISPD cDNA from peppermint (open reading frame of 795 nucleotides) encodes a 265-residue protein of approximately 27 kD that exhibits >99% identity (2 residue differences) to that of the spearmint homolog. Sequence analyses using TargetP V1.0 and Predotar V0.5 gave ambiguous results with regard to the presence of N-terminal subcellular targeting information.
Analysis of the isopiperitenol/carveol dehydrogenase sequence by BLASTP (Altschul et al., 1997) searching at the Biology Workbench site (http://biology.ncsa.uiuc.edu/) and the identification of conserved sequence motifs established the dehydrogenase as a member of the short-chain dehydrogenase/reductase (SDR) superfamily. The isopiperitenol/carveol dehydrogenase shows the closest sequence relationship to other plant dehydrogenases (Figs. 5 and 6), including a stress induced SDR from pea (Pisum sativum; 53% identity, 67% similarity; accession no. AF097651; Brosché and Strid, 1999); Δ5-3β-hydroxysteroid dehydrogenase from Digitalis lanata (48% identity, 61% similarity; accession no. Q93Y47; Finsterbusch et al., 1999); secoisolariciresinol dehydrogenase from Forsythia x intermedia (43% identity, 61% similarity; accession no. AF352735) and Podophyllum peltatum (41% identity, 58% similarity; accession no. AF352734; Xia et al., 2001), and a number of uncharacterized SDRs from tomato, rice, and Arabidopsis (Arabidopsis thaliana).
Figure 5.
Deduced amino acid sequence of ISPD (accession no. AY641428) aligned with the sequences of putative, plant derived SDRs from pea (PsSDR; accession no. AF097651), P. peltatum (PpSDR; accession no. AF352734), and D. lanata (DlSDR; accession no. Q93Y47). The conserved coenzyme binding motif (Motif I), the NAG site (Motif II), and the active site YXXXK element (Motif III) are underlined. The asterisk denotes a conserved Asp indicative of NAD(H) specificity, and the arrow heads indicate the catalytic tetrad. Black boxes indicate identical residues for two or more sequences; gray boxes indicate similar residues. The alignment was created using ClustalX and is displayed using Prettybox (GCG).
Figure 6.
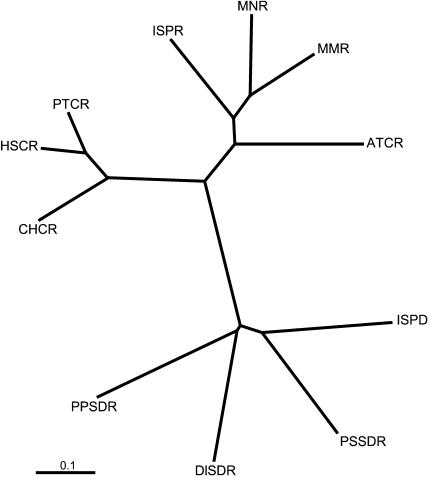
The full-length ISPD and the three related plant SDRs of Figure 5 were aligned with isopiperitenone reductase (ISPR), menthone-menthol reductase (MMR), menthone-neomenthol reductase (MNR), and with closest relative SDRs from human (HSCR; accession no. J04056), pig (PTCR; accession no. M80709), CHO-K1/hamster cells (CHCR; accession no. AB020238), and Arabidopsis (Arabidopsis thaliana; ATCR; accession no. NM_115986) and a phylogenetic tree generated using ClustalX and displayed with Treeview 1.6.6. The reductases form a cluster distinct from the dehydrogenases and are more closely related to mammalian SDRs than to the dehydrogenases. The scale value of 0.1 represents 0.1 amino acid substitutions/site.
The peppermint oil gland EST library contained only 2 representatives of the dehydrogenase (100% identity). In contrast, multiple copies of isopiperitenone reductase (16 representatives) and pulegone reductase (11 representatives) were observed in this library (Ringer et al., 2003).
DISCUSSION
Characterization of ISPD and Carveol Dehydrogenase
Based on several lines of evidence, we anticipated that the ISPD of peppermint and the carveol dehydrogenase of spearmint would be very similar if not identical. Immunoblot analysis of spearmint oil gland extracts with anti-ISPD polyclonal antibodies revealed the presence of three cross-reacting dehydrogenases, one identical in size to peppermint ISPD (Fig. 4). Although a cDNA encoding carveol dehydrogenase of equivalent size and >99% amino acid identity to the peppermint dehydrogenase cDNA was cloned, no longer versions were obtained. It is possible that other (as yet unidentified) genes express long forms of carveol dehydrogenase, but it seems more likely that other proteins are present in oil gland extracts that simply share epitopes with the monoterpenol dehydrogenase and cross react with the antibody. This question will be more fully addressed in future studies directed to expression profiling of these and other biosynthetic enzymes during oil gland development.
The native (−)-ISPD from peppermint leaves (Kjonaas et al., 1985) and the recombinant enzyme are very similar in pH optimum, substrate utilization (with preference for α,β-unsaturated alcohols and the capability of utilizing both trans-carveol and trans-isopiperitenol), and the absolute requirement for oxidized pyridine nucleotide with preference for NAD+ (but at fairly high Km value). Recent immunolocalization (Turner and Croteau, 2004) indicates that this dehydrogenase is localized to mitochondria where NAD(H) concentrations have been reported as 600 μm (Heineke et al., 1991) and as high as 2.4 mm (Wigge et al., 1993). Thus, even with a Km of 410 μm, this enzyme is likely to function at catalytically competent rates in vivo.
The ability of the dehydrogenase to utilize isopiperitenol and carveol is of little consequence in planta because peppermint produces only isopiperitenol and spearmint only carveol (Lawrence, 1981). The higher Km value and slower relative velocity of the peppermint dehydrogenase with isopiperitenol compared to carveol also appears to be insignificant in planta as the rate of conversion is sufficient such that isopiperitenol does not accumulate in peppermint oil (Gershenzon et al., 2000).
Isopiperitenol/Carveol Dehydrogenase Encodes a Member of the SDR Superfamily
Isopiperitenol/carveol dehydrogenase is a member of the ubiquitous SDR superfamily (Jörnvall et al., 1999), a group with at least 3,000 described members (Kallberg et al., 2002) that exhibit diverse substrate utilization. Typical SDRs contain about 250 amino acid residues, but deduced identities among members are quite low; however, the three-dimensional fold of the N-terminal (coenzyme binding) region and several sequence motifs show a high degree of conservation (Jörnvall et al., 1995).
Conserved sequence motifs (Fig. 5) include the GXXXGXG coenzyme-binding motif (motif I; Jörnvall et al., 1995; Kallberg et al., 2002) and a conserved Asp (residue 41 of ISPD) indicative of a preference for NAD(H) over NADP(H) (Kallberg et al., 2002), and the (N)NAG site (motif II; Filling et al., 2002; Kallberg et al., 2002). The YXXXK active site element (motif III) of which the Tyr and Lys form a catalytic tetrad with a Ser (residue 147 in ISPD; Jörnvall et al., 1995) and an Asn (residue 118 in ISPD; Filling et al., 2002) is also highly conserved among SDRs.
The isopiperitenol/carveol dehydrogenase shows significant sequence identity (43%) with secoisolariciresinol dehydrogenase involved in lignan formation, perhaps suggesting the recruitment of dehydrogenases involved in secondary metabolism (phenylpropanoids and terpenoids) from a similar ancestor; however, related enzymes (48%–50% identity) are also involved in phytosterol metabolism and stress induction. In addition to isopiperitenol/carveol dehydrogenase, isopiperitenone reductase (a monoterpene double-bond reductase from peppermint; Ringer et al., 2003) and the menthone:menthol/neomenthol reductases (carbonyl reductases from peppermint; Davis et al., 2005) are members of the SDR superfamily, although they share only approximately 13% identity with the dehydrogenases. Moreover, the reductases are functional monomers and bear an insertion preceding the catalytic Tyr typical of the subclass (Ghosh et al., 2001) unlike isopiperitenol/carveol dehydrogenase and the majority of defined SDRs, which do not bear the insertion and are functional as either homodimers or homotetramers (Jörnvall et al., 1995). Nevertheless, the reductases and the dehydrogenases share common SDR sequence motifs (for sequence alignments, see the accompanying article, Davis et al., 2005).
Generation of a phylogenetic tree (Fig. 6) of the peppermint SDRs and their closest relatives illustrates the proximity of the reductases to an uncharacterized SDR from Arabidopsis and that the reductases are more closely related to certain mammalian SDRs than they are to the dehydrogenase. Whereas the double-bond and carbonyl reductases illustrated could certainly have evolved through gene duplication and differentiation from a proximate SDR ancestor, the dehydrogenase appears to have been recruited from a quite different ancestral source of SDRs. It is notable in this context that (+)-pulegone reductase of peppermint [another monoterpene double-bond reductase related in catalytic function to (−)-isopiperitenone reductase] belongs to the distinct medium chain dehydrogenase/reductase superfamily that is quite dissimilar from, and does not share similar sequence motifs with, the SDRs (Ringer et al., 2003).
The isolation and characterization of the genes and enzymes of monoterpene biosynthesis in mint is important for understanding these pathways and their organization and regulation, and it provides the necessary first step in genetic engineering of these commercial plant species for improved essential oil yield and composition. In the broader context, this work illustrates that the redox steps of the extended monoterpene biosynthetic pathway at focus here (and perhaps other natural product pathways) were derived from an assortment of diverse gene ancestors in primary metabolism and not by duplication and differentiation of a single dehydrogenase/reductase ancestor as might reasonably have been anticipated (Pichersky and Gang, 2000) based on the similarity of substrates and redox chemistry involved. It is clear from the diversity of the redox genes that participate in menthol biosynthesis that homology-based cloning of such genes from other pathways of secondary metabolism may be challenging, and that direct functional screening from enriched cDNA libraries, as described here, may be the most expedient approach to this goal.
MATERIALS AND METHODS
Experimental Materials, Substrates, and Reagents
Peppermint (Mentha x piperita) L. cv Black Mitcham and spearmint (Mentha spicata) plants were propagated and grown in a greenhouse as described in the accompanying article (Davis et al., 2005). (−)-Isopiperitenone was prepared by allylic oxidation of (−)-limonene (Guillon et al., 2000) using chromium hexacarbonyl in acetonitrile, yielding a roughly equal mixture of (−)-carvone and (−)-isopiperitenone. (−)-Isopiperitenone was purified by column chromatography on silica gel by isocratic elution with 27% diethylether in n-hexane. (−)-Isopiperitenol (mixture of cis-and trans-isomers) was prepared by lithium aluminum hydride reduction of (−)-isopiperitenone in anhydrous diethyl ether (Guillon et al., 2000), and the isomers were separated by HPLC on a Machery Nagel Nucleodex β-OH cyclodextrin column (5 μm, 200 × 4 mm) by isocratic elution with 25% acetonitrile in 25 mm ammonium formate, pH 6.0, with elution monitoring at 210 nm. (−)-Carveol (mixture of cis- and trans-isomers) was obtained from Sigma Chemical (St. Louis) and the isomers were separated by HPLC using an Adsorbosphere HS C18 column (250 × 10 mm; Alltech, Deerfield, IL) by isocratic elution with 25% acetonitrile in water with monitoring as before. Other substrates and standards were obtained from Aldrich Chemical (Milwaukee, WI), Haarman and Reimer (Holzminden, Germany), Fluka Chemical (Buchs, Switzerland), or Sigma Chemical.
Cloning and In Situ Assay of (−)-ISPD
ESTs initially annotated as dehydrogenases (Lange et al., 2000) were reevaluated by sequence comparison against the GenBank nonredundant protein database using the BLASTX search algorithm (Altschul et al., 1990). Following elimination of known dehydrogenase gene types (i.e. glyceraldehyde 3-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase), the remaining sequences were aligned using the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, WI; Staden, 1996) into three groups of less than three acquisitions each and five single acquisitions. A full-length representative from each of the three groups and several of the single acquisitions were selected for subcloning, expression in Escherichia coli, and test of function.
Pfu DNA polymerase-based sticky-end PCR (Zeng, 1998) was employed to transfer the original clones from pBluescript SK- (library vector; Lange et al., 2000) into the high-level expression vector pSBET (Schenk et al., 1995) as described previously (Ringer et al., 2003). Briefly, universal reverse primers (Ringer et al., 2003) were designed from the vector sequence to generate a 3′- BamHI overhang for use with any of the putative dehydrogenase library acquisitions. Forward primers were designed from candidate sequences to create a 5′-NdeI overhang at the predicted start codon for each of the putative dehydrogenase clones. Sticky-end forward primers for the cDNA clone encoding the functional ISPD were primer 1, 5′-TATGGCAAGCGTGAAGAAGCTCGCAGGC-3′, and primer 2, 5′-TGGCAAGCGTGAAGAAGCTCGCAGGC-3′. Typical conditions for PCR reactions were 50 to 100 ng DNA template/reaction with 0.2 mm dNTPs, 1 μm of each primer, and 1 unit Pfu polymerase. An MJ thermocycler model PTC-100 (MJ Research, Waltham, MA) was used with a PCR program of initial denaturation (94°C, 1 min), 40 amplification cycles (94°C for 45 s, 50°C for 1 min, and 72°C for 2 min), and final extension at 72°C for 10 min.
pSBET clones were expressed and screened for (−)-ISPD activity using the previously described E. coli in situ assay (Ringer et al., 2003). Thus, pSBET clones were used to transform E. coli BL21(DE3) cells (Invitrogen, Carlsbad, CA), which were grown to A600 of 0.8 to 1.0 at 37°C. Approximately 4 h after induction with 1 mm isopropyl β-d-thiogalactopyranoside, 100 μm (−)-trans-carveol was added (neat) and the mixture was allowed to incubate overnight at 28°C; (−)-carveol is commercially available and was used as test substrate based on previous evidence that this monoterpenol was used by both peppermint and spearmint dehydrogenases (Croteau et al., 1991). At the completion of incubation, cultures were steam distilled-pentane extracted using a Likens-Nickerson apparatus (J&W Scientific, Folsum, CA) that was equipped with standard condenser cooled with chilled glycol. The pentane extract was collected and the substrate and product(s) contained therein were separated by GC using (+)-camphor as internal standard. GC analysis was performed on a Hewlett-Packard 5890 Series II gas chromatograph equipped with a flame ionization detector (at 300°C), 7673A autosampler, and a 30-m × 0.25-mm i.d. capillary column coated with a 0.2-μm film of polyethyleneglycol ester (AT-1000; Alltech, Deerfield, IL) using hydrogen as carrier gas (13 p.s.i. head pressure) with a temperature program of 40°C initial temp, 40°C/min to 50°C, 10°C/min to 180°C, and 40°C/min to 220°C. Products were identified by comparison of retention times and mass spectra to those of authentic standards and were quantified by comparison of the detector response to that of the internal standard. GC-MS analysis was performed on a Hewlett-Packard 5840A-5985B MSD system (at 70 eV) using a ZB5 column (30-m × 0.25-mm i.d., Phenomenex, Torrance, CA), and with a temperature program of initial temp 40°C, 40°C/min to 50°C, and 15°C/min to 305°C. A single acquisition encoding the target dehydrogenase was obtained and fully sequenced. E. coli BL21(DE3) cells with plasmid containing no insert were included as a negative control in these experiments.
Immunoblot Analysis
Equivalent amounts of leaf tissue were harvested from peppermint and spearmint plants and oil gland secretory cells were isolated by an established protocol (Gershenzon et al., 1992). The isolated cells were disrupted by grinding in a mortar with liquid nitrogen and extracted with 50 mm KH2PO4, pH 7.5, containing 10% glycerol, 10 mm sodium sulfite, 1 mm ascorbic acid, 1 mm EDTA, 1 mm dithiothreitol (DTT), and 5 μm each FAD and FMN. The extract was filtered and centrifuged (McConkey et al., 2000) to give the soluble enzyme fraction that was assayed to confirm the presence of dehydrogenase activity. Approximately 0.5 mg of total protein (Bradford, 1976) was separated by SDS-PAGE, blotted to Trans-Blot nitrocellulose membranes according to the manual instructions (Bio-Rad Laboratories, Hercules, CA), and probed with a 1:200 dilution of polyclonal antibodies prepared against the purified isopiperitenol dehydrogenase in rabbits (Turner and Croteau, 2004). Secondary antibody incubation was performed using a 1:3,000 dilution of alkaline phosphatase-conjugated affinipure goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) with nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate staining. Recombinant ISPD was included for size comparison.
Cloning of (−)-Carveol Dehydrogenase
A homology-based PCR cloning strategy, using nondegenerative forward and reverse primers designed from the peppermint dehydrogenase cDNA, was employed. Pfu DNA polymerase amplification, using a spearmint oil gland secretory cell cDNA library (Lupien et al., 1999) as template and a primer set consisting of the gene-specific sticky-end forward primer 1 (described above) and a cDNA-specific reverse primer (5′-TCACTTGGCCACGGCCACGAATGG-3′) based on the last 24 nucleotides of the peppermint dehydrogenase, yielded an amplicon of apparent full length after separation on a 1% agarose gel and ethidium bromide staining. Following excision and gel extraction (Qiagen, Valencia, CA), the PCR product was treated with Taq polymerase (Invitrogen) for addition of the 3′-thymidine overhang, cloned into the pCR2.1 TOPO vector using the TOPO TA cloning kit (Invitrogen), and sequenced. Additional 3′- and 5′- sequence was obtained using forward primer 1 (described above) with a vector-specific reverse primer [M13 forward (−20) primer; 5′-GTAAAACGACGGCCAG-3′], and the peppermint dehydrogenase reverse primer described above with a vector-specific forward primer (M13 reverse primer; 5′-CAGGAAACAGCTATGAC-3′), respectively. The resulting amplicons were cloned as before using the TOPO TA cloning kit and sequenced to provide additional 5′-coding sequence and untranslated 3′-sequence. All amplicons were 5′-truncated with no start codon.
To obtain additional 5′-sequence, RT-PCR amplification and RACE were utilized as described in the accompanying article (Davis et al., 2005). Secretory cells (RNA source) were isolated from equivalent amounts of spearmint leaves of 7, 8, and 10-d-old, using an established protocol (Gershenzon et al., 1992) with a 0.5 mm NaHPO4 and 25 mm MOPSO buffer, pH 6.6, containing 200 mm sorbitol, 10 mm Suc, 1 mm aurintricarboxylic acid, 5 mm thiourea, 2 mm DTT, 1% (w/v) polyvinylpyrrolidone-40, and 0.6% (w/v) methyl cellulose. The intact secretory cells were disrupted by sonication, and RNA was extracted and purified using the Qiagen RNeasy Mini kit. RT of mRNA/single-stranded cDNA synthesis was performed with approximately 1 to 5 μg of the purified RNA as template, an oligo(dT)16-20 primer, and Moloney murine leukemia virus-reverse transcriptase following the manufacturer's instructions (Promega, Madison, WI).
5′-RACE was achieved by homopolymeric tailing of the single-stranded cDNA using terminal transferase (Invitrogen) with dCTP, followed by Taq polymerase mediated PCR amplification of the dC-tailed cDNA with an oligo(dG) forward primer (5′-GGAAACAGCTATGACCATGACGGGIIGGGIIGGGIIGG-3′) and each of three isopiperitenol/carveol dehydrogenase gene-specific internal primers (5′-CGTCCGGAGCCCGATCGTCGC-3′, 5′-CGTCCGGAGCCCGATCGTCGC-3′, and 5′-CTCATCCGAAGCCAGAAA-3′). PCR conditions were similar to those described above with the addition of 1.5 mm Mg2+. PCR amplicons were gel purified, excised, gel extracted, cloned using the TOPO TA cloning kit, and sequenced.
Dehydrogenase Assay
To verify positive results of the in situ assay for the peppermint and spearmint dehydrogenases, cell-free assays with recombinant enzyme were performed as described before (Ringer et al., 2003). E. coli BL21(DE3) cells expressing the relevant pSBET clone were harvested, lysed by sonication and centrifuged to remove cellular debris. Aliquots of the resulting supernatant, containing the soluble enzymes, were analyzed by SDS-PAGE and assayed for dehydrogenase activity. The assay was conducted in 1.5 mL of 25 mm KH2PO4, pH 7.5, containing 10% v/v glycerol, 1 mm DTT, recombinant enzyme and 2 mm NAD+, and was incubated overnight at 31°C with 100 μm (−)-carveol or (−)-isopiperitenol. The mixture was then extracted by vigorous mixing with 0.5 mL pentane; an aliquot of the pentane extract was analyzed by GC and GC-MS as described above.
Dehydrogenase Purification and Characterization
The pSBET clones were sequence verified to ensure that no mutations had been introduced during subcloning, and the recombinant ISPD was prepared at large scale and purified as described previously (Ringer et al., 2003) with minor modifications. Briefly, 1-L cultures were induced with 100 μm isopropyl β-d-thiogalactopyranoside, allowed to incubate for 24 h at 15°C, and the bacteria were harvested by centrifugation, resuspended in a MOPSO buffer system (25 mm MOPSO, pH 7.5, 10% glycerol, and 1 mm DTT), lysed by sonication, and the soluble enzyme fraction prepared as before. This material was combined with 5 mL ceramic hydroxyapatite beads (Bio-Rad) equilibrated with MOPSO buffer and allowed to mix at 4°C for 30 min. Following protein binding, the hydroxyapatite beads were allowed to settle and the supernatant containing the dehydrogenase was immediately applied to a DEAE cellulose (DE52; Whatman International, Maidstone, England) column equilibrated with the same MOPSO buffer to obtain the flow through containing the dehydrogenase. Calibrated SDS-PAGE analysis of ISPD demonstrated purification to approximately 75% by this simple protocol and permitted estimation of protein concentration.
Following establishment of reaction linearity with respect to time and protein concentration, kinetic parameters were determined in potassium phosphate, pH 7.5, containing 10% glycerol and 1 mm DTT, by varying the concentration of substrate [(−)-trans-isopiperitenol or (−)-trans-carveol] or NAD+ while maintaining the other reactant at saturation. All assays were terminated by pentane extraction with (+)-camphor as internal standard and quantification by GC as before. Kinetic constants were determined using nonlinear regression analysis of Michaelis-Menten plots (Enzyme Kinetics Software, Trinity Software, Plymouth, NH) and the results presented are the means of 2 or 3 separate experiments with se < 16% for most experiments.
pH optimum was determined using the phosphate buffer system over a pH range of 5 to 12 at 0.5 pH intervals. Substrate and cofactor specificity was also determined in the same phosphate buffer system, pH 7.5, with 100 μm of the test substrates (−)-trans-carveol, (−)-cis-carveol, (−)-trans-isopiperitenol, (−)-cis-isopiperitenol, (−)-perillyl alcohol, and (+)-neomenthol, (−)-menthol, (+)-isomenthol, and (+)-neoisomenthol, or the corresponding ketones, with 2 mm NAD+ or 2 mm NADH, respectively.
Gel permeation chromatography of the recombinant ISPD was conducted on a calibrated Sephadex 200 (Amersham Pharmacia Biotech, Piscataway, NJ) column, with 50 mm KH2PO4, pH 7.5, containing 2 mm DTT, 100 mm NaCl, and 10% (v/v) glycerol as running buffer at a flow rate of 0.5 mL/min. The 1-mL column fractions were monitored by enzyme assay and SDS-PAGE, and molecular weight was determined by fitting to the Ve/Vo versus molecular weight plot of the standards.
Sequence Analysis
Initial sequence comparisons of the putative dehydrogenase genes from the peppermint oil gland cDNA library (Lange et al., 2000) were performed using the GCG version 10 sequence analysis package (GCG, Madison, WI), and the BLASTX algorithm (Altschul et al., 1990). The BLASTP (Altschul et al., 1997) database search program at the Biology Workbench site (http://biology.ncsa.uiuc.edu/) was used for obtaining related SDR sequences. SDR sequences were aligned, analyzed and displayed using the GCG version 10 sequence analysis package and ClustalX (Thompson et al., 1997). The phylogenetic tree was generated using ClustalX and displayed with Treeview 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). The full-length nucleotide sequence for ISPD was used in a NCBI nucleotide-nucleotide BLAST (Altschul et al., 1997) of the peppermint EST library to determine the abundance of this cDNA. The search for putative subcellular targeting sequences was conducted using TargetP V1.0 (http://www.cbs.dtu.dk/services/TargetP/) and Predotar V0.5 (http://www.inra.fr/predotar/).
Sequence data from this article were obtained from the EMBL/GenBank data libraries under accession numbers AF097651, Q93Y47, AF352734, AF352735, J04056, M80709, AB020238, and NM_115986. The nucleotide sequence reported in this article has been deposited in the GenBank database with the accession number AY641428.
Acknowledgments
We thank Rachael Parkin, Amanda Grimm, and Derek Pouchnik for technical assistance, Julianna Gothard for growing the peppermint and spearmint plants, and Glenn Turner and Charles Burke for the ISPD antibody.
This work was supported by the U.S. Department of Energy, by the Mint Industry Research Council, and by the Washington State University Agricultural Research Center (project 0268).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053298.
References
- Alonso WR, Rajaonarivony JIM, Gershenzon J, Croteau R (1992) Purification of 4S-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha x piperita) and spearmint (M. spicata). J Biol Chem 267: 7582–7587 [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 2: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 200: 248–254 [DOI] [PubMed] [Google Scholar]
- Brosché M, Strid Å (1999) Cloning, expression, and molecular characterization of a small pea gene family regulated by low levels of ultraviolet B radiation and other stresses. Plant Physiol 121: 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CC, Wildung MR, Croteau R (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA 96: 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Alonso WR, Katahira E, McGarvey DJ, Croteau R (1993) 4S-Limonene synthase from the oil glands of spearmint (Mentha spicata): cDNA isolation, characterization and bacterial expression of the catalytically active monoterpene cyclase. J Biol Chem 268: 23016–23024 [PubMed] [Google Scholar]
- Croteau R (1987) Biosynthesis and catabolism of monoterpenoids. Chem Rev 87: 929–954 [Google Scholar]
- Croteau R, Wagschal KC, Karp F, Satterwhite DM, Hyatt DC, Skotland CB (1991) Biochemical characterization of a spearmint mutant that resembles peppermint in monoterpene content. Plant Physiol 96: 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EM, Ringer KL, McConkey ME, Croteau R (2005) Monoterpene metabolism. Cloning, expression, and characterization of menthone reductases from peppermint. Plant Physiol 137: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277: 25677–25684 [DOI] [PubMed] [Google Scholar]
- Finsterbusch A, Lindemann P, Grimm R, Eckerskorn C, Luckner M (1999) Δ5-3β-Hydroxysteroid dehydrogenase from Digitalis lanata Ehrh.: a multifunctional enzyme in steroid metabolism? Planta 209: 478–486 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Maffei M, Croteau R (1989) Biochemical and histochemical localization of monoterpene biosynthesis in the glandular trichomes of spearmint (Mentha spicata). Plant Physiol 89: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, McCaskill D, Rajaonarivony JIM, Mihaliak C, Karp F, Croteau R (1992) Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem 200: 130–138 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, McConkey ME, Croteau R (2000) Regulation of monoterpene accumulation in leaves of peppermint (Mentha x piperita L.). Plant Physiol 122: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Sawicki M, Pletnev V, Erman M, Ohno S, Nakajin S, Duax WL (2001) Porcine carbonyl reductase: structural basis for a functional monomer in short chain dehydrogenases/reductases. J Biol Chem 276: 18457–18463 [DOI] [PubMed] [Google Scholar]
- Guillon J, Rioult JP, Robba M (2000) New synthesis of isopiperitenol, previously isolated from species of Cymbopogon. Flavour Fragr J 15: 223–224 [Google Scholar]
- Harley RM, Brighton CA (1977) Chromosome numbers in the genus Mentha. Bot J Linn Soc 74: 71–96 [Google Scholar]
- Heineke K, Riens B, Grosse H, Hoferichter P, Peter U, Flügge U-i, Heldt HW (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H, Höög J-O, Persson B (1999) SDR and MDR: Completed genome sequences show these families to be large, of old origin, and of complex nature. FEBS Lett 445: 261–264 [DOI] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34: 6003–6013 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDR): coenzyme-based functional assignments in completed genomes. Eur J Biochem 269: 4409–441712230552 [Google Scholar]
- Karp F, Mihaliak CA, Harris JL, Croteau R (1990) Monoterpene biosynthesis: specificity of the hydroxylations of (−)-limonene by enzyme preparations from peppermint (Mentha x piperita), spearmint (Mentha spicata) and perilla (Perilla frutescens) leaves. Arch Biochem Biophys 276: 219–226 [DOI] [PubMed] [Google Scholar]
- Kjonaas RB, Venkatachalam KV, Croteau R (1985) Metabolism of monoterpenes: oxidation of isopiperitenol to isopiperitenone, and subsequent isomerization to piperitenone by soluble enzyme preparations from peppermint (Mentha piperita) leaves. Arch Biochem Biophys 238: 49–69 [DOI] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence BM (1981) Monoterpene interrelationships in the Mentha genus: a biosynthetic discussion. In BD Mookherjee, CJ Mussinan, eds, Essential Oils. Allured Publishing, Wheaton, IL, pp 1–81
- Lupien S, Karp F, Wildung M, Croteau R (1999) Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (−)-4S-limonene-3-hydroxylase and (−)-4S-limonene-6-hydroxylase. Arch Biochem Biophys 368: 181–192 [DOI] [PubMed] [Google Scholar]
- McCaskill D, Gershenzon J, Croteau R (1992) Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187: 445–454 [DOI] [PubMed] [Google Scholar]
- McConkey ME, Gershenzon J, Croteau RB (2000) Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant Physiol 122: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MJ, Lincoln DE, Marble PM (1972) Oil composition of Mentha aquatica x M. spicata F1 hybrids in relation to the origin of M. x piperita. Can J Genet Cytol 14: 13–29 [Google Scholar]
- Parry JW (1969) Spices, Vol I. Chemical Publishing, New York
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–444 [DOI] [PubMed] [Google Scholar]
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau R (2003) Monoterpene double-bond reductases of the (−)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (−)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys 418: 80–92 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss H-H (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19: 196–200 [PubMed] [Google Scholar]
- Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5: 233–241 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GW, Croteau R (2004) Organization of monoterpene biosynthesis in mentha. Immunocytochemical localizations of geranyl diphosphate synthase, limonene-6-hydroxylase, isopiperitenol dehydrogenase, and pulegone reductase. Plant Physiol 136: 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge B, Krömer S, Gardeström P (1993) The redox levels and subcellular distribution of pyridine nucleotides in illuminated barley leaf protoplasts studied by rapid fractionation. Physiol Plant 88: 10–18 [Google Scholar]
- Wise ML, Croteau R (1999) Biosynthesis of monoterpenes. In DE Cane, ed, Comprehensive Natural Products Chemistry, Vol 2, Isoprenoids Including Carotenoids and Steroids. Elsevier, Oxford, pp 97–153
- Xia Z-Q, Costa MA, Pélissier HC, Davin LB, Lewis NG (2001) Secoisolariciresinol dehydrogenase purification, cloning, and functional expression. J Biol Chem 276: 12614–12623 [DOI] [PubMed] [Google Scholar]
- Zeng G (1998) Sticky-end PCR: new method for subcloning. Biotechniques 25: 206–208 [DOI] [PubMed] [Google Scholar]