Abstract
Plant viruses elicit the expression of common sets of genes in susceptible hosts. Studies in Arabidopsis (Arabidopsis thaliana) and tomato (Lycopersicon esculentum) indicate that at least one-third of the genes induced in common by viruses have been previously associated with plant defense and stress responses. The genetic and molecular requirements for the induction of these stress and defense-related genes during compatible host-virus interactions were investigated with a panel of Arabidopsis mutant and transgenic plants defective in one or more defense signaling pathways. pad4, eds5, NahG, npr1, jar1, ein2, sid2, eds1, and wild-type Columbia-0 and Wassilewskija-2 plants were infected with two different viruses, cucumber mosaic virus and oilseed rape mosaic virus. Gene expression was assayed by a high-throughput fiber-optic bead array consisting of 388 genes and by RNA gel blots. These analyses demonstrated that, in compatible host-virus interactions, the expression of the majority of defense-related genes is induced by a salicylic acid-dependent, NPR1-independent signaling pathway with a few notable exceptions that did require NPR1. Interestingly, none of the mutant or transgenic plants showed enhanced susceptibility to either cucumber mosaic virus or oilseed rape mosaic virus based on both symptoms and virus accumulation. This observation is in contrast to the enhanced disease susceptibility phenotypes that these mutations or transgenes confer to some bacterial and fungal pathogens. These experimental results suggest that expression of many defense-related genes in compatible host plants might share components of signaling pathways involved in incompatible host-pathogen interactions, but their increased expression has no negative effect on viral infection.
Viruses are obligate pathogens that use various strategies to coopt cellular resources of compatible host plants to promote their infections. Compatible host-virus interactions result in systemic infections that are typically accompanied by the onset of disease symptoms. By contrast, incompatible interactions result in cessation of virus replication and movement at or near the sites of inoculation. In compatible hosts, viral invasion triggers numerous biochemical and physiological changes in cells, tissues, and even whole plants (Maule et al., 2002). Among these are local and systemic changes in host gene expression. Some local changes occur in the cells where viruses are actively replicating and include both induction and shutoff of host gene expression (Wang and Maule, 1995; Aranda et al., 1996; Escaler et al., 2000; Havelda and Maule, 2000). Other local changes in gene expression can happen in advance of or behind the viral replication front. For example, NADP-dependent malic enzyme was induced in uninfected cells surrounding virus-infected lesions, whereas catalase had altered expression in cells where viral replication had ceased (Havelda and Maule, 2000). These virus-induced modifications in gene expression in cotyledons occur transiently and concomitantly with viral replication as it spreads cell to cell away from the site of inoculation.
These spatial and temporal studies involving in situ hybridization in cotyledons cited above were applied to only a handful of plant genes. To gain a more global perspective on how viruses alter host gene expression patterns, DNA microarrays and other genomics methods are beginning to be applied, albeit with less spatial resolution at this time. cDNA subtraction followed by macroarray analysis identified at least 55 tomato (Lycopersicon esculentum) genes that were differentially expressed after infection with two potato spindle tuber viroid strains. Functional classification of these genes suggested that they are involved in defense/stress response, cell wall structure, chloroplast function, and other diverse cellular processes (Itaya et al., 2002). Expression patterns of Arabidopsis (Arabidopsis thaliana) genes in tobacco mosaic virus (TMV)-infected leaves showed significant alterations in cDNA microarray experiments (Golem and Culver, 2003). Five different positive-stranded RNA viruses elicited the expression of common sets of genes in susceptible Arabidopsis plants in experiments utilizing Arabidopsis GeneChip probe arrays (Whitham et al., 2003). About one-third of the genes induced in common by these viruses were associated with plant defense and stress responses. The association between virus and viroid infections and defense-related gene expression indicates that similar transcriptional responses occur in different hosts in response to these pathogens (Itaya et al., 2002; Whitham, 2004). The time course of gene expression changes occurring in Arabidopsis leaves also revealed that host responses are coordinately regulated during infection by diverse viruses. Moreover, the similar kinetics by which different groups of defense or stress-related genes were induced suggested a common pathway or mechanism leading to their increased expression.
The mechanisms by which plant viruses alter the expression of different groups of host genes are beginning to be deciphered and in part have revealed the underlying causes of virus-induced symptoms (Whitham and Wang, 2004). For example, the processing of micro RNAs can be disrupted by plant viruses leading to ectopic expression of their target genes and symptom-like plant phenotypes (Voinnet, 2001; Mallory et al., 2002; Kasschau et al., 2003; Palatnik et al., 2003; Chapman et al., 2004; Chen et al., 2004; Dunoyer et al., 2004). It is unlikely that disruption of micro RNA processing will explain all of the alterations in plant gene expression and all of the symptoms that occur as a result of viral infections. Of particular interest to us were the mechanisms by which defense-related genes are induced during viral infections and the potential impact of the induction of these genes on the outcome of these interorganismal interactions. The signal transduction pathways controlling plant defense systems are mediated by signaling molecules, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). These molecules are required for expression of defense-related genes and development of resistance phenotypes, and they synergistically and antagonistically influence each other through an intricate network of regulatory interactions (Glazebrook, 2001; Kunkel and Brooks, 2002). Several of the genes that function in SA-, JA-, and ET-dependent defense signaling cascades have been identified and cloned in Arabidopsis (Glazebrook, 2001). In SA signaling, genes such as EDS1 and PAD4 have relatively early signaling functions downstream of R genes but upstream of SA (Zhou et al., 1998; Feys et al., 2001). EDS5 and SID2 are likely to be involved in SA biosynthesis either indirectly or directly (Nawrath and Metraux, 1999; Wildermuth et al., 2001; Nawrath et al., 2002). Along these lines, the NahG transgene, encoding a bacterial salicylate hydroxylase, prevents SA accumulation by degrading it into catechol and thus inhibits many plant defense responses (Gaffney et al., 1993; Delaney et al., 1994). NPR1/NIM1 functions downstream of SA and these other mutants and the encoded protein is stimulated by SA to translocate to the nucleus where it interacts with TGA transcription factors that bind the TGACG motif and that lead to expression of pathogenesis-related (PR) and other defense genes (Kinkema et al., 2000; Fan and Dong, 2002; Mou et al., 2003). In the JA and ET signaling pathways, JAR1 and COI1 are required for perception of JA accumulation and induction of marker genes like PDF1.2 (Pieterse et al., 1998). EIN2 is required for ET perception and also the accumulation of marker genes like PDF1.2 (Pieterse et al., 1998). In general, both JA and ET perception is required for an effective resistance response mediated by these molecules. Loss of either can result in enhanced susceptibility to some pathogens (Thomma et al., 1999; Norman-Setterblad et al., 2000).
Since little is known about the signal transduction pathways that are involved in compatible plant-virus interactions, we were interested to expand our understanding of how the SA and JA/ET signaling pathways influence expression of host genes in susceptible plants and if either of these pathways play significant roles in basal antiviral defenses. To accomplish these objectives, pad4-1, eds5-1, NahG, npr1-1, jar1-1, ein2-1, and wild-type Columbia-0 (Col-0) plants were infected with cucumber mosaic virus strain Y (CMV-Y) and oilseed rape mosaic virus (ORMV). The expression of 388 selected genes, accumulation of viral RNAs, and disease symptoms were monitored. These studies were facilitated by recent development of a high-throughput gene expression profiling system using the cDNA-mediated annealing, selection, extension, and ligation (DASL) assay coupled with universal fiber-optic bead array matrices (Fan et al., 2004; Shou et al., 2004). The custom fiber-optic bead arrays enabled the parallel interrogation of a focused set of 388 genes in 126 samples. Analysis of the resulting data demonstrated that, in compatible host-virus interactions, the expression of the majority of defense-related genes is induced by a SA-dependent, NPR1-independent signaling pathway with a few notable exceptions that did require NPR1. Interestingly, none of the mutant or transgenic plants showed enhanced susceptibility to either CMV-Y or ORMV based on both symptoms and virus accumulation. This observation is in contrast to the enhanced disease susceptibility phenotypes that these mutations or transgenes confer to bacterial and fungal pathogens (e.g. Nawrath and Metraux, 1999; Dewdney et al., 2000; Ton et al., 2002). These experimental results suggest that expression of many defense-related genes in compatible host plants might share a common signaling pathway with incompatible host-pathogen interactions, but their increased expression has no negative effect on viral infections in Arabidopsis.
RESULTS
Expression-Profiling Responses to Viral Infection in Mutant and Wild-Type Plants by the High-Throughput DASL Assay and Fiber-Optic Bead Array Method
Our previous work demonstrated that genes associated with plant defense responses, including PR genes, are induced in compatible Arabidopsis leaves during virus infection (Whitham et al., 2003). The expression profiles of these genes along with other genes that were not obviously related by function suggested coordinated induction in response to diverse RNA viruses despite the lack of specific recognition that would occur in an R-gene-mediated incompatible interaction. The coordinated induction of defense-related genes in compatible host-virus interactions led us to investigate the mechanism(s) underlying their regulation as well as the effects of their induced expression on virus accumulation and symptomatology in Arabidopsis. We hypothesized that the induction of many genes in response to viruses was regulated by a known signaling pathway possibly involving SA, JA, and/or ET as a key signaling molecule. To investigate these possibilities, leaves of pad4-1, eds5-1, npr1-1, jar1-1, and ein2-1 mutants, NahG transgenic, and Col-0 wild-type plants were mock inoculated or inoculated with CMV-Y or ORMV. In each of the 3 independent replicates of this experiment, 4 fully expanded rosette leaves were inoculated on each of 3 plants that were at growth stage 3.00 to 3.50 as defined by (Boyes et al., 2001). The inoculated and mock-inoculated leaves were collected at 2 and 5 days after inoculation (DAI) for RNA extraction. At these time points, the inoculated leaves and upper uninoculated leaves were symptomless on all the plants, as expected.
To interrogate Arabidopsis gene expression in response to ORMV and CMV infections in the defense signaling mutants, we used a recently developed gene expression method called the DASL assay (Fan et al., 2004). In this assay, total RNA is first converted to cDNA using biotinylated oligo(dT) and random hexamers. The biotinylated cDNA is then attached to a streptavidin solid support, and query oligos are annealed to their target sequences in the cDNA. Currently, 3 nonoverlapping sites (probes) are targeted per gene, and up to 500 genes can be assayed in a single DASL reaction (Fan et al., 2004). In this study, the expression of 388 genes was assayed. Most genes were selected for this microarray because they were up- or down-regulated by at least 2-fold in response to viral infections in our previous microarray experiments (Whitham et al., 2003; S. Whitham, unpublished data). The list of genes that were assayed and the sequences of each of their three corresponding unique target sites (probes) are provided in Supplemental Table I along with hybridization and negative controls. Average signal values were computed for each gene by determining the mean signal for the three representative probes. Our previous study showed that three probes per gene lend the assay enough sensitivity and reproducibility for quantitative detection of differential expression (Bibikova et al., 2004; Fan et al., 2004). The query oligos are designed in a way in which they all share common primer landing sites, so that one PCR primer pair is used to amplify all the annealed and amplifiable templates and generate amplicons of similar size (approximately 100 bp). This uniformity minimizes potential bias during amplification of many different targets.
To allow the use of universal microarrays, the query oligos also contain a unique address sequence that is associated with each targeted site. This address sequence allows the amplified product, which is labeled during PCR with a fluorescent primer, to hybridize to a microarray bearing the complementary address sequences. The DASL assay used 100 ng of total RNA to analyze the 388 genes, 5- to 100-fold less than that required by quantitative PCR, which usually takes 2 to 50 ng per reaction (per gene). All these advantages make it an ideal platform for validation of candidate genes or for studying the transcriptional regulation of preselected gene sets derived from initial genome-wide screenings, such as in this study.
The fiber-optic bundles are assembled into an array matrix (Sentrix Array Matrix, Illumina, San Diego) comprising 96 bundles (i.e. arrays) arranged in an 8 × 12 matrix that matches the dimensions of standard microtiter plates. This arrangement allows simultaneous processing of 96 samples at once, which permits rapid analyses of expression patterns in hundreds of samples. The 63 RNA samples (3 treatments × 7 genotypes × 3 biological replicates) from each time point were grouped together on separate 96-well plates (126 total samples) for labeling by the DASL procedure and hybridization to array matrices. For details on labeling, hybridization, and scanning, see “Materials and Methods” (see also Fan et al., 2003). After scanning, background subtraction was performed and then an average signal was computed for each gene based on the signals from the three probes. The background-subtracted data is provided in Supplemental Table II, A and B. The cubic splines method was used to normalize the average signals across 96 arrays within each array matrix (Workman et al., 2002).
A linear mixed-effects model was fit to the data, ANOVA was used to compute an F-statistic, and then P values were assigned to each gene in the normalized data set. We first analyzed the data to identify the genes that had expression patterns that were significantly altered by plant genotype (P < 0.01). This analysis identified 187 and 176 genes from the 2 and 5 DAI data sets, respectively. Extensive overlap between the resulting gene lists was expected, and indeed 133 genes were present at both time points. In total, this analysis indicated that the expression of 230 of the 388 Arabidopsis genes on the array was significantly affected (P < 0.01) by genotype of the plants at one or both time points.
To understand how genotype affected the expression of these 230 genes in the context of virus treatment, their expression profiles were grouped using average linkage-hierarchical clustering, and the output was viewed with the TreeView program (Eisen et al., 1998). This analysis revealed two major clusters of genes with either increased or decreased signals after infection by CMV-Y and/or ORMV in wild-type Col-0 plants (Supplemental Fig. 1). CMV-Y and ORMV elicited similar patterns of gene expression at the 2-DAI time point. However, at 5 DAI, ORMV caused overall greater increases or decreases in the expression of these genes relative to CMV-Y-infected Col-0 leaves. At 5 DAI, the expression of many genes might have been returning to near-basal or marginally induced levels in the CMV-Y infections. These observations are confirmed by calculating the P values for pairwise comparisons of gene expression between ORMV- or CMV-Y-infected Col-0 plants versus mock-infected Col-0 plants. The P values for CMV-Y are typically more significant at 2 DAI than 5 DAI and the P values for ORMV are typically more significant than the P values for CMV, especially at 5 DAI (Table I; Supplemental Table III). The more robust responses to ORMV are consistent with our previous observations using the Arabidopsis 8K GeneChip probe array (Whitham et al., 2003).
Table I.
P values of selected defense-related and genes of unknown function for various pairwise comparisons
| Arabidsopsis Gene Index No. (Gene Name, if Known) GO Biological Process
|
P Values for the Indicated Pairwise Comparisons
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAI | Col-0 CMV/Mc | Col-0 ORMV/M | CMVaein2/Col | ORMVbein2/Col | CMV jar1/Col | ORMV jar1/Col | CMV eds5/Col | ORMV eds5/Col | CMV npr1/Col | ORMV npr1/Col | CMV pad4/Col | ORMV pad4/Col | CMV NahG/Col | ORMV NahG/Col | |
| At3g57260 (BGL2) | 2 | 0.00293 | 0.00026 | 0.69554 | 0.55187 | 0.47836 | 0.20380 | 0.36009 | 0.62654 | 0.76327 | 0.06072 | 0.24481 | 0.43650 | 0.00001 | 0.21890 |
| SAR | 5 | 0.11770 | 0.00006 | 0.03279 | 0.64109 | 0.29773 | 0.83447 | 0.46792 | 0.06539 | 0.99228 | 0.00329 | 0.01514 | 0.09617 | 0.00027 | 2.56E-08 |
| At1g75040 (PR-5) | 2 | 0.02518 | 0.00059 | 0.91343 | 0.80721 | 0.12539 | 0.12424 | 0.53606 | 0.79870 | 0.55590 | 0.14641 | 0.31241 | 0.48899 | 0.00358 | 0.00220 |
| SAR | 5 | 0.06183 | 0.00001 | 0.64229 | 0.81381 | 0.96831 | 0.61953 | 0.06565 | 0.01526 | 0.43174 | 0.00114 | 0.05628 | 0.01513 | 0.01078 | 1.07E-06 |
| At3g48090 (EDS1) | 2 | 0.70794 | 0.06132 | 0.43813 | 0.00762 | 6.12E-07 | 0.14306 | 0.46548 | 0.00230 | 0.18042 | 0.45977 | 0.89478 | 0.00049 | 0.08630 | 0.00023 |
| SAR | 5 | 0.00749 | 0.00001 | 0.61434 | 2.00E-08 | 0.26476 | 0.16881 | 0.49829 | 1.94E-09 | 0.09807 | 1.58E-09 | 0.38694 | 3.29E-09 | 0.12532 | 1.75E-12 |
| At3g52430 (PAD4) | 2 | 0.05172 | 0.00662 | 0.12032 | 0.96954 | 0.05804 | 0.74646 | 0.12313 | 0.32722 | 0.23772 | 0.11497 | 0.00038 | 0.00006 | 0.00065 | 0.00185 |
| SAR | 5 | 0.15884 | 0.00005 | 0.67028 | 0.59636 | 0.77808 | 0.66621 | 0.17272 | 0.00602 | 0.85250 | 0.00852 | 0.00235 | 0.00002 | 0.02028 | 9.35E-07 |
| At2g32680 | 2 | 0.00076 | 0.00107 | 0.06498 | 0.75936 | 0.20693 | 0.67137 | 0.05733 | 0.25382 | 0.82813 | 0.45011 | 0.02929 | 0.05787 | 1.58E-06 | 0.00004 |
| Defense response | 5 | 0.64167 | 0.00001 | 0.31018 | 0.30384 | 0.30642 | 0.87703 | 0.14748 | 0.00017 | 0.58902 | 0.00016 | 0.10547 | 0.15695 | 0.01091 | 1.42E-09 |
| At1g64280 (NPR1) | 2 | 0.02931 | 0.14941 | 0.04772 | 0.48330 | 0.41053 | 0.87900 | 0.22358 | 0.74646 | 0.24572 | 0.71409 | 0.05413 | 0.43202 | 0.05223 | 0.14314 |
| Cell death, response to pathogens | 5 | 0.53514 | 0.01328 | 0.62668 | 0.24988 | 0.55890 | 0.06402 | 0.88187 | 0.15257 | 0.32924 | 0.50428 | 0.88747 | 0.67758 | 0.69616 | 0.00035 |
| At3g20600 (NDR1) | 2 | 0.22816 | 0.07656 | 0.29348 | 0.14915 | 0.26565 | 0.81305 | 0.85986 | 0.81888 | 0.48246 | 0.19499 | 0.12412 | 0.48650 | 0.00001 | 0.00280 |
| Defense response to pathogens | 5 | 0.13025 | 0.01207 | 0.52423 | 0.00515 | 0.09856 | 0.77159 | 0.78413 | 2.83E-06 | 0.67925 | 0.00155 | 0.73835 | 0.04397 | 0.00052 | 7.54E-11 |
| At2g30550 | 2 | 0.00001 | 0.23445 | 0.69564 | 0.19596 | 0.02994 | 0.43996 | 0.00010 | 0.21418 | 0.12239 | 0.08042 | 0.00803 | 0.27363 | 0.00011 | 0.13814 |
| Lipid metabolism | 5 | 0.00431 | 0.06279 | 0.83804 | 0.00925 | 0.09000 | 0.84615 | 0.02187 | 0.04226 | 0.09761 | 0.73349 | 0.25056 | 0.13639 | 0.01809 | 0.02677 |
| At2g30140 | 2 | 0.00036 | 0.00971 | 0.94594 | 0.05704 | 0.71662 | 0.33557 | 0.35557 | 0.33631 | 0.76612 | 0.58959 | 0.84729 | 0.02741 | 0.03842 | 0.67971 |
| Metabolism | 5 | 0.22032 | 0.19854 | 0.73578 | 0.36760 | 0.89673 | 0.43420 | 0.22090 | 0.69527 | 0.20763 | 0.80220 | 0.67370 | 0.99888 | 0.27200 | 0.28996 |
| At1g02920 | 2 | 0.00155 | 0.72964 | 0.02326 | 0.88354 | 0.83160 | 0.95315 | 0.02840 | 0.89054 | 0.96423 | 0.68380 | 0.00035 | 0.31680 | 0.00001 | 0.84785 |
| Toxin catabolism | 5 | 0.00158 | 0.00021 | 0.06149 | 0.99874 | 0.53879 | 0.92116 | 0.07545 | 0.20944 | 0.03195 | 0.12306 | 0.98336 | 0.70865 | 0.00113 | 0.00018 |
| At2g14560 | 2 | 0.33826 | 0.08313 | 0.90294 | 0.36943 | 0.02474 | 0.17793 | 0.13838 | 0.61072 | 0.66108 | 0.61518 | 0.73996 | 0.75525 | 0.00032 | 0.00339 |
| Biological_process unknown | 5 | 0.88802 | 0.00171 | 0.88321 | 0.10113 | 0.77859 | 0.53949 | 0.00084 | 1.59E-09 | 0.02022 | 5.27E-09 | 0.36590 | 0.10736 | 4.57E-06 | 2.81E-12 |
| At2g14610 (PR-1) | 2 | 0.00244 | 0.00247 | 0.80232 | 0.96311 | 0.15094 | 0.03238 | 0.00488 | 0.00001 | 0.00166 | 0.02646 | 0.50670 | 0.22705 | 6.51E-07 | 4.44E-08 |
| Biological_process unknown | 5 | 1.66E-06 | 8.86E-09 | 0.03215 | 0.86817 | 0.00883 | 0.67091 | 4.40E-11 | 1.48E-15 | 4.67E-13 | 1.73E-15 | 0.64820 | 0.00487 | 1.79E-13 | 3.65E-16 |
| At3g51860 (ATCAX3) | 2 | 0.00334 | 0.04204 | 0.11019 | 0.36493 | 0.55912 | 0.19315 | 0.29530 | 0.48882 | 0.10901 | 0.70606 | 0.61733 | 0.02870 | 0.10917 | 0.92285 |
| Biological_process unknown | 5 | 0.12793 | 0.12616 | 0.98971 | 0.89083 | 0.61118 | 0.96456 | 0.52463 | 0.05116 | 0.06454 | 0.40263 | 0.67246 | 0.20691 | 0.15064 | 0.01994 |
| At5g02490 | 2 | 0.01107 | 0.80462 | 0.00565 | 0.89894 | 0.55725 | 0.28988 | 0.03065 | 0.20833 | 0.83310 | 0.90221 | 0.00053 | 0.39852 | 0.00242 | 0.17064 |
| Biological_process unknown | 5 | 0.11085 | 0.00015 | 0.13008 | 0.81845 | 0.67443 | 0.25188 | 0.13764 | 0.29353 | 0.95902 | 0.60313 | 0.25958 | 0.94842 | 0.20957 | 0.23875 |
| At4g39030 (EDS5) | 2 | 0.21962 | 0.00793 | 0.86456 | 0.25233 | 0.00009 | 0.76587 | 0.55231 | 0.17117 | 0.00394 | 0.26546 | 0.20722 | 0.00356 | 0.15244 | 0.42384 |
| SA biosynthesis | 5 | 0.28635 | 0.00039 | 0.70189 | 0.65522 | 0.31164 | 0.79348 | 0.11604 | 0.02345 | 0.35201 | 0.46213 | 0.42840 | 0.17254 | 0.23853 | 0.02771 |
CMV, CMV-Y infected.
ORMV, ORMV infected.
M, Mock inoculated.
Effects of NahG Transgene on Genes Induced by Viral Infection and Affected by Genotype
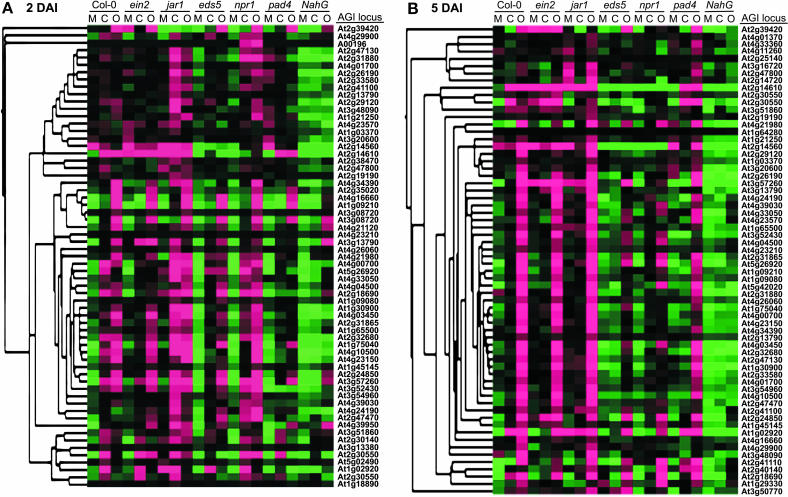
Here, we focus on genes that were induced in response to CMV-Y and/or ORMV at 2 and 5 DAI in wild-type plants and that had expression profiles that were significantly altered in one or more of the mutant genotypes (P ≤ 0.01). Hierarchical clustering of these genes at 2 and 5 DAI is shown in Figure 1, A and B, respectively. Prior to clustering, the expression values of each gene were centered relative to its mean signal among all samples. In Figure 1, the magenta-colored boxes represent expression values in samples that were above the mean, green represents expression values in samples that were below the mean, and black represents samples in which the gene expression level was near the mean. By using this strategy it is possible to visualize relative transcript levels across all samples, because the intensity of the magenta or green correlates to the degree that expression was above or below the mean, respectively. For example, the magenta coloring in the Col-0 CMV-Y and ORMV samples compared to the green in the Col-0 mock illustrates that expression was higher in the virus-infected samples. Thus, the expression of these genes was induced in response to CMV-Y and ORMV. However, their induction was significantly diminished or abolished or in one or more mutant genotypes (specific examples provided in Table I; Supplemental Table III). NahG plants provided the most striking example of this. At both 2 and 5 DAI, expression of these genes was similar in mock-, CMV-Y-, and ORMV-inoculated NahG plants, indicating that there was weak or no induction in response to these viruses, which is in contrast to virus-infected Col-0 leaves (Fig. 1, A and B). Furthermore, these genes typically had lower basal expression levels in the NahG mock-infected plants when compared to the Col-0 wild-type mock-infected plants. Since the accumulation of SA is impaired in the NahG plants, these data suggest that SA is required for the induction of this set of genes during viral infections and for maintenance of their basal expression levels.
Figure 1.
Expression profiles of genes that are induced by CMV-Y and ORMV by an SA-dependent mechanism. This set of genes has expression profiles that were significantly affected by one or more plant genotypes and were induced in response to CMV-Y and ORMV infection in wild-type plants. A, Genes that were induced at 2 DAI. B, Genes that were induced at 5 DAI. The columns represent genotypes and treatments, and the rows represent genes. Each colored box represents the average signal of a gene for three replicates in the indicated sample. The expression data for each gene were mean centered prior to average linkage-hierarchical clustering and the results were displayed using the TreeView program. Magenta indicates samples in which the gene was expressed above the mean for the 21 treatments, black represents samples in which expression was at or near the mean, and green represents expression below the mean. The intensity of color corresponds to the degree to which expression was above or below the mean. M, Mock inoculated; C, CMV-Y infected; O, ORMV infected. Genes are listed by their Arabidopsis gene index designation.
Effects of NahG and Mutants on Basal Expression Levels of Virus-Induced Genes
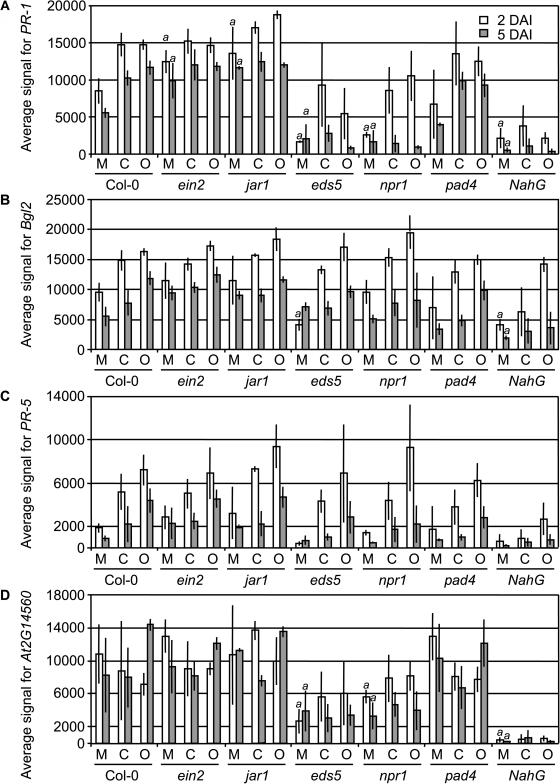
The sensitivity of the DASL assay allowed us to examine the basal expression levels of these genes, which is typically not possible by RNA gel blots or other array-based technologies. To further examine the effects of genotype on basal expression, we plotted the expression values of some selected genes at 2 and 5 DAI (Fig. 2) and determined P values of each gene for pairwise comparisons of the mock-inoculated Col-0 versus each of the mock-inoculated mutants (Supplemental Table IV). The expression of PR-1, Bgl2, and PR-5 was consistently lower in mock-inoculated NahG plants versus mock-inoculated Col-0 plants (Fig. 2). Expression of PR-1 and Bgl2 was significantly lower (P < 0.01) in mock-inoculated NahG plants than in Col-0. PR-5 was consistently lower in NahG plants, but the biological variability did not result in a P < 0.01. As shown in Supplemental Table IV, 27 of these 65 genes have P < 0.01 for the mock-infected NahG versus the Col-0 suggesting that SA is an important component in determining their basal expression levels. Furthermore, PR-1 and another gene, At2g14560, have a strong requirement for EDS5 and NPR1 for their basal accumulation levels (Fig. 2). Thus, SA and other components of defense signaling pathways are not only required in the induction process but function in maintaining some steady-state level of expression.
Figure 2.
Expression of PR-1, Bgl2, PR-5, and At2g14560 in the 7 genotypes after mock inoculation or viral infection. The average signal for each gene was graphed for each sample at 2 and 5 DAI. The vertical lines represent the sd for the three biological replicates, and the “a” above columns represents the mock-inoculated mutant samples that were significantly different from the mock-inoculated Col-0 wild-type samples. M, Mock inoculated; C, CMV-Y infected; O, ORMV infected.
Effects of ein2 and jar1 Mutants on Genes that Are Induced and Regulated by SA
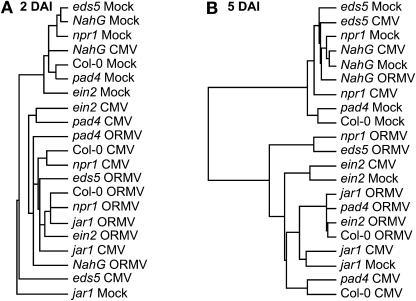
Inspection of Figure 1 reveals some distinct profiles of gene expression among the remaining mutant genotypes. To further examine the relationships of the expression profiles for the different combinations of treatment and genotype, we used average linkage-hierarchical clustering to group the samples at 2 or 5 DAI (Fig. 3). jar1 mock-infected plants do not cluster with other mock-infected plants at 2 or 5 DAI. At 5 DAI, jar1 mock-infected plants are most similar to jar1 plants inoculated with CMV-Y, and ein2 mock-inoculated plants are most similar to ein2 plants inoculated with CMV-Y. This point is illustrated best in Figure 1; here, the ein2-1 and jar1-1 mock-inoculated samples have overall higher basal levels of expression (less intense green) when compared to the Col-0 mock-inoculated sample. This was not the case for mock-inoculated samples of the other mutants. This is also illustrated in Figure 2A, in which the average signal values for PR-1 are plotted. PR-1 was the gene most significantly affected by the jar1 and ein2 mutations. In the mock-infected plants, PR-1 expression is significantly higher compared to wild-type plants (P < 0.01; Supplemental Table IV), and its induced levels in jar1 mutants are significantly higher than virus-infected Col-0 plants (P < 0.01; Table I). Thus, EIN2 and JAR1 repress the expression of PR-1. This observation is consistent with cross talk between the SA and JA/ET pathways that has previously been described in incompatible host-pathogen interactions (Glazebrook, 2001; Kunkel and Brooks, 2002), although it appears some sets of genes, including PR-1, are more strongly affected by cross talk than other sets, including PR-5 and Bgl2.
Figure 3.
Hierarchical clustering of samples at 2 and 5 DAI. Samples were grouped by average linkage-hierarchical cluster using the expression data from the genes listed in Figure 1, A or B.
Effects of eds5, npr1, and pad4 Mutants on Genes That Are Induced and Regulated by SA
At 2 DAI, the CMV-Y- and ORMV-infected npr1-1 samples cluster with their Col-0 counterparts (Fig. 3A), indicating that early expression of these genes is not dependent on NPR1. However, at 5 DAI the CMV-infected npr1-1 sample clusters with NahG and mock-infected samples (Fig. 3B), indicating that sustained induction of these genes in response to CMV-Y requires NPR1. The ORMV-infected npr1-1 sample clusters with the corresponding eds5-1 treatment. As illustrated in Figure 1B, the overall expression of genes in response to ORMV in these 2 mutants is reduced compared with the ORMV-infected Col-0 plants. These results suggest a partial requirement for NPR1 in the virus-induced expression of this set of genes, especially at the 5 DAI time point. pad4-1 mutants cluster with other infected plants at 2 DAI (Fig. 3A) and cluster with their corresponding infected wild-type Col-0 samples at 5 DAI (Fig. 3B). This indicates that PAD4 has less effect on the expression of defense-related genes, although its function is needed for maximal induction in compatible host-virus interactions.
Accumulation of CMV-Y and ORMV in Defense Signaling Mutants
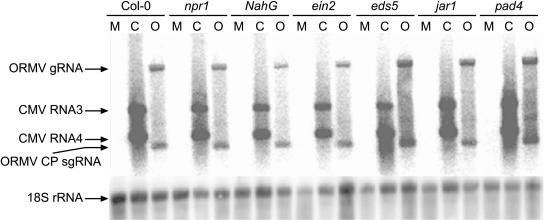
The dramatic effects of an SA-mediated signaling pathway on host gene expression profiles in these compatible host-virus interactions led us to investigate if any of the corresponding signaling mutants had a significant effect on the accumulation of CMV-Y or ORMV. For both viruses, the 2-DAI time point is within the interval when virus accumulation is increasing rapidly with time, and 5 DAI represents a relatively late time point at which the accumulation rate is decreasing. To further investigate the susceptibility of these plants to CMV-Y and ORMV, northern-blot hybridization was used to determine the accumulation of viral RNAs in inoculated leaves of each of the seven Arabidopsis genotypes. Displayed in Figure 4 are data from a single biological replicate illustrating that the accumulation of ORMV and CMV-Y genomic and subgenomic RNAs was not enhanced at 5 DAI in mutant and NahG lines compared to Col-0 wild-type plants. Conclusions from these northern-blot data were supported by quantitative real-time PCR assays performed on the virus-infected mutant and wild-type RNA samples at 2 and 5 DAI. Among the three replicates of this experiment, CMV-Y and ORMV did not accumulate to significantly higher levels in the mutants compared to Col-0 at early or late time points (P > 0.15 for all mutants; Supplemental Table V). In each replicate, a set of mock- and virus-inoculated plants was also saved and monitored for symptoms through 14 DAI. However, no consistent differences in symptoms were observed on the mutants or NahG plants in comparison to wild-type Col-0 plants (data not shown). To ensure that all plants had the expected genotype, we confirmed the sequence of the corresponding mutant alleles by sequencing PCR products amplified from mutant and wild-type genomic DNA samples. Taken together, these results suggested that the basal susceptibility of Arabidopsis to CMV-Y and ORMV was not significantly enhanced when the SA-, JA-, or ET-dependent signaling pathways were disrupted, which is unlike the case for bacterial and fungal pathogens (e.g. Nawrath and Metraux, 1999; Dewdney et al., 2000; Ton et al., 2002).
Figure 4.
Accumulation of CMV-Y and ORMV genomic and subgenomic RNAs in wild-type and mutant plants at 5 DAI. These data suggest that none of the mutants have enhanced susceptibility to CMV-Y or ORMV in comparison to wild-type Col-0 plants. To detect accumulation of viral RNAs, RNA gel blots were hybridized with probes corresponding to the CPs of CMV-Y and ORMV. The CMV-Y CP probe hybridizes to the genomic RNA3 and the subgenomic RNA4, which both contain the CP sequence. The ORMV CP probe hybridizes to the ORMV genomic RNA and the ORMV CP subgenomic RNA as indicated. The Arabidopsis 18S ribosomal RNA was used as a loading control. Data for one biological replicate is presented. M, Mock inoculated; C, CMV-Y inoculated; O, ORMV inoculated; g, genomic, sg, subgenomic.
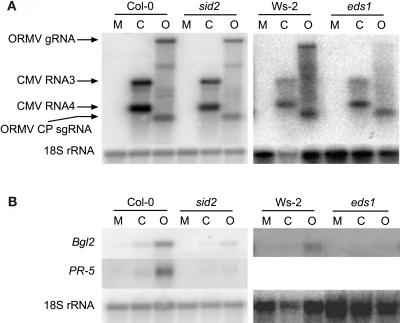
Expression of Defense-Related Genes in sid2 and eds1 Mutants in Response to Virus Infection
To extend our findings and confirm the requirement for SA in the expression of the defense-related genes, we inoculated sid2-1 and eds1-1 mutants with ORMV and CMV. sid2 was of particular interest, because it is deficient in SA biosynthesis (Wildermuth et al., 2001). Total RNA extracted from these plants at 5 DAI was hybridized to coat protein (CP) probes from the two viruses and Bgl2 or PR-5. As with the other mutants, accumulation of these viruses was not enhanced in the mutants compared to wild-type plants (Fig. 5A). Hybridization with the Bgl2 and PR-5 probes demonstrated that induction of these genes was abolished or significantly reduced in infected mutants compared to infected wild-type plants (Fig. 5B). PR-5 expression was not detected by RNA gel blot in the Wassilewskija (Ws)-2 background after virus infection, and thus it was not included for eds1 in Figure 5B. Nevertheless, these data support the conclusion that compatible viruses elicit changes in Arabidopsis gene expression that requires SA and signaling components upstream of it such as EDS1.
Figure 5.
Accumulation of CMV-Y and ORMV and expression of Bgl2 and PR-5 in sid2 mutants. Leaves were harvested from mock-inoculated and virus-inoculated plants at 5 DAI. RNA gel blots containing 2.5 μg of total RNA were hybridized with labeled DNA fragments corresponding to Bgl2, PR-5, CMV CP, or ORMV CP. The Arabidopsis 18S ribosomal RNA was used as a loading control. Data for one biological replicate is presented. M, Mock inoculated; C, CMV-Y inoculated; O, ORMV inoculated; g, genomic, sg, subgenomic.
DISCUSSION
We explored the functions of signal transduction pathways involving SA, JA, and ET in mediating responses to infection in compatible host-virus interactions. This research was aided by an emerging technology, a fiber-optic bead array coupled with the DASL assay (Fan et al., 2004), that allowed us to analyze the expression of a focused set of 388 genes among 7 Arabidopsis genotypes that were mock inoculated or infected by either CMV-Y or ORMV. Most of the 388 genes had been previously identified by a global expression-profiling approach that enabled us to catalog genes that were coordinately up- or down-regulated in response to diverse viral infections in wild-type Col-0 plants (Whitham et al., 2003; S. Whitham, unpublished data). Subsequently, we were interested in establishing the signaling pathways that mediate the altered expression profile of coregulated genes in compatible host-virus interactions. Our strategy is consistent with the systems biology approach described by Katagiri (2003) in which one round of profiling and interpretation leads to subsequent rounds of profiling designed to analyze each interpretation in more detail.
The experimental design allowed for statistical analysis of the bead array data using a linear mixed-effects model as opposed to using a fold change cutoff. The use of a strict fold change cutoff is not appropriate for this gene expression assay or other types of microarray analysis, because they tend to underestimate fold change (Yuen et al., 2002; Fan et al., 2004). Our statistical analysis of the data enabled us to identify genes with expression patterns that were significantly altered in one or more genotypes. Hierarchical clustering enabled us to visualize the relationships between the treatments, genotypes, and time points. Here, we focused on 65 genes, many of which were previously associated with plant defense responses, which were induced in response to CMV-Y and/or ORMV at 2 and/or 5 DAI in wild-type Col-0 plants. In general, ORMV induced the most robust changes in the expression of these genes, which is consistent with our previous observations (Whitham et al., 2003). Induction of these 65 genes was severely compromised in NahG transgenic plants (Fig. 1) and in sid2 mutants (Fig. 5), demonstrating that this response is dependent on SA.
Previous studies have demonstrated that SA synthesis is not induced in tobacco (Nicotiana tabacum) or Arabidopsis in response to compatible viruses. For example, tobacco plants carrying the N gene conferring TMV resistance rapidly accumulate free and conjugated forms of SA following virus infection at temperatures permissive to the resistance response. In contrast, these same N plants do not develop a resistance response at 32°C, allow systemic TMV infection, and do not accumulate additional free or conjugated forms of SA (Malamy et al., 1992). Along a similar line, Arabidopsis plants of the Dijon-17 ecotype that carry the HRT gene required for turnip crinkle virus (TCV) resistance dramatically accumulate SA during the first 3 DAI. In contrast, TCV infection in the Dijon-3 ecotype or other susceptible plants derived from populations segregating for HRT did not accumulate SA any differently than mock-inoculated control plants (Dempsey et al., 1997). Based on these studies, it is unlikely that SA accumulation was induced in response to ORMV or CMV-Y in the susceptible Col-0 or Ws-2 ecotypes. Thus, the basal levels of SA that are present in the leaves of wild-type plants are sufficient to mediate expression of defense-related gene sets in compatible host-virus interactions.
It was also noteworthy that the basal expression levels of many of these defense-related genes were lower in the mock-inoculated NahG plants compared to wild-type Col-0 plants. This observation suggested to us that not only is SA important for induction of the defense-like responses, but in the absence of pathogen attack, SA may sustain basal levels of genes associated with resistance responses and keep the defense system primed. We were able to identify this role for SA, because the sensitivity of the DASL gene expression assay allows detection of low-abundance transcripts that are below the threshold of widely used assays such as RNA gel blots and other microarray techniques (Yeakley et al., 2002; Fan et al., 2004).
In addition to SA, plant responses to pathogen infections are controlled by JA, ET, and several other regulatory genes. A role for JA and ET in the induction of these genes was excluded by using the ein2 and jar1 mutants, which are insensitive to ET and JA, respectively. These mutants are also compromised in their resistance to certain bacterial and fungal pathogens and fail to express defense marker genes such as PDF1.2 (Pieterse et al., 1998; Thomma et al., 1999). Neither the ein2 nor jar1 mutants prevented the induced expression of the SA-dependent genes, and in cases like PR-1, these mutants enhanced the basal and induced expression levels. Thus, JA and ET are not required for induction of these genes and in some cases may be antagonistic to their expression. This observation is consistent with the complex cross talk that occurs between these pathways in which JA and ET antagonize SA-induced expression of genes associated with plant defense responses (Petersen et al., 2000; Kloek et al., 2001; Li et al., 2004). Many other studies have also demonstrated that SA is antagonistic to responses mediated by JA and ET (Clarke et al., 1998, 2000, 2001; Dewdney et al., 2000; Gupta et al., 2000; Jirage et al., 2001). One measure of this is enhanced expression of PDF1.2 in response to pathogen infection in mutants or transgenic plants that do not accumulate SA. In our experiments, there was no significant expression of PDF1.2 in NahG plants or sid2 mutants after virus infection, providing further evidence that the JA and ET defense signaling pathway is not activated in compatible host-virus interactions in Arabidopsis.
Despite the significant negative effects of the defense signaling pathway mutants related to SA on basal expression of and induction of numerous defense-related genes in compatible virus infections, there was not a concomitant enhancement of susceptibility to CMV-Y or ORMV. None of the mutants tested accumulated significantly more viral genomic or subgenomic RNAs than wild-type Col-0 plants (Figs. 4 and 5A; Supplemental Table V) nor did they develop more severe symptoms (data not shown). These results suggest that the components of the SA, JA, and ET signaling pathways that were examined do not confer basal resistance to CMV-Y and ORMV nor do they have roles in symptomatology. We conclude that the lack of functional systemic-acquired resistance (SAR) or induced-systemic resistance pathways does not necessarily confer enhanced susceptibility to infection by ORMV and CMV. This is in contrast to the important role of SA in gene-for-gene types of resistant host-virus interactions that involve the hypersensitive response. For example, the N-, HRT-, and RCY1-mediated resistance responses to TMV, TCV, and CMV-Y, respectively, have an absolute requirement for SA (Friedrich et al., 1995; Kachroo et al., 2000; Takahashi et al., 2002). In the absence of SA, these viruses are not contained by an effective resistance response. In addition, SA treatment of plant tissue or the onset of SAR results in inhibition of viruses (Chivasa et al., 1997; Naylor et al., 1998; Murphy and Carr, 2002). Based on the requirement for SA in these incompatible interactions, it is interesting that depletion of SA does not cause plants to become more susceptible to infection. It is possible that the viruses counteract these defense responses in compatible host-virus interactions similar to the ways that they suppress RNA silencing. Alternatively, gene silencing or other antiviral mechanisms like down-regulation of host mRNA translation could be more limiting to virus replication and movement. In this scenario, the effects of disrupting the SA-mediated defense-like responses could be masked by more robust antiviral defenses. Finally, this ineffective response may simply lack some quality, such as speed and/or magnitude, needed to halt virus infection as occurs in robust R gene-mediated defenses (Tao et al., 2003). Of course, other possibilities cannot be ruled out, but we expect that the described scenarios could be tested with the appropriate combinations of Arabidopsis mutants.
Overall, we propose the following model that incorporates elements from our studies and others to explain the transcriptional behavior of the set of genes investigated here. In the absence of an Avr/R gene interaction, virus invasion is somehow detected and then this information is relayed through PAD4 and EDS1. The response that is initiated requires SA to potentiate and sustain it, but appears to result from both NPR1-independent early events and later events that are both NPR1-dependent and -independent. Basal levels of SA are sufficient to activate defense-associated genes through both NPR1-dependent and NPR1-independent pathways. NPR1, EDS5, SID2, EDS1, PAD4, and SA are required for maximal accumulation of the transcripts. EDS5 and SID2 are likely to be involved via their roles in SA biosynthesis, whereas NPR1, EDS1, and PAD4 are involved in the signaling functions. The existence of the NPR1-independent pathway leading to PR gene expression has been noted by others in host responses to bacterial and fungal pathogens (Glazebrook et al., 1996; Reuber and Ausubel, 1996; Shah et al., 1997; Clarke et al., 2000). In virus-host interactions, the Arabidopsis ecotype Dijon-17 confers resistance to TCV through an undefined SA-dependent pathway that does not involve NPR1, ET, or JA (Kachroo et al., 2000). The genetic and molecular requirements of the responses that were observed in the compatible interactions have similarities to this TCV resistance response. It is possible that components of this signaling pathway function in both incompatible and compatible host-virus interactions. Regardless of how the expression of these defense-related genes is activated, the resulting response has little or no negative effects on viral pathogenesis in compatible Arabidopsis plants (Fig. 4).
The data described here provide insight into a signaling pathway that accounts for one major facet of the gene expression changes elicited during compatible host-virus interactions. Many genes were induced that were not affected by the signal transduction pathways investigated here. This is apparent, because not all the induced genes were significantly affected by one of the plant genotypes that were tested. We also noted that many Arabidopsis genes were down-regulated by virus infection. Although our preliminary analyses suggest that one or more of the genotypes tested might perturb the expression of some of the down-regulated genes, more experiments are needed to clarify the mechanisms involved.
MATERIALS AND METHODS
Preparation of Virions
ORMV and CMV-Y were propagated in tobacco (Nicotiana tabacum) cv SR1 (nn genotype) and tobacco cv Xanthi nc (NN genotype), respectively. The virions were purified as previously described (Chapman, 1998; Roossinck and White, 1998), aliquoted, and stored at −20°C.
Arabidopsis Growth and Virus Inoculation
The pad4-1, npr1-1, eds1-1, eds5-1, ein2-1, sid2-1, and jar1-1 mutants were obtained from the Arabidopsis Biological Resource Center, and the NahG line was obtained from Dr. X. Dong (Duke University). These mutants and transgenic lines have the Col-0 ecotype as their background except for eds1-1, which has Ws-2 as its wild-type progenitor. Appropriate wild-type plants were used as controls in all experiments. Arabidopsis (Arabidopsis thaliana) plants were grown in a growth room set for a 14-h photoperiod and 22°C. Four leaves of 21-d-old plants were labeled with a felt pen and dusted with carborundum. Leaves were rub-inoculated with 10 μL of CMV or ORMV virions that were diluted to 10−1 in 20 mm potassium phosphate buffer, pH 7.2. Plants of each genotype were also mock inoculated with phosphate buffer alone. Leaves from 3 plants were harvested at 2 and 5 DAI, snap frozen in liquid nitrogen, and stored at −80°C. For each treatment and genotype, 3 additional plants were kept for symptom observation until 14 DAI. The three biological replicates of these experiments that were performed were consistent with a randomized complete block design. In total, 126 independent samples were collected for RNA extraction and array analysis.
RNA Extraction
Total RNA was isolated using a modified TRIzol method (38% saturated phenol, pH 4.3, 0.8 m guanidine thiocyanate, 0.4 m ammonium thiocyanate, 0.1 m sodium acetate, and 5% glycerol; Chomczynski and Sacchi, 1987). Leaf tissue was ground in liquid nitrogen, 10 mL of TRIzol solution (preheated to 60°C) was added per 1 g of tissue, and samples were mixed vigorously and incubated at 60°C for 5 min. Next, 2 mL of chloroform was added and samples were vortexed and incubated at room temperature for 5 min. Samples were centrifuged at 15,000g for 15 min at 4°C, and then the upper aqueous layers were collected and transferred to new tubes. RNA was precipitated by adding 0.5 volume of isopropanol and 0.5 volume of 0.8 m sodium citrate/1.2 m sodium chloride and pelleted by centrifugation at 15,000g for 10 min at 4°C. Pellets were washed in ice-cold 75% (v/v) ethanol and centrifuged at 6,000g for 5 min at 4°C. Air-dried RNA pellets were dissolved in 300 μL of diethyl pyrocarbonate-treated water, and the concentration of each sample was determined spectrophotometrically and validated by agarose gel electrophoresis and ethidium bromide staining.
RNA Gel-Blot Analyses
Total RNA, 2.5 μg, was diluted to 11 μL with diethyl pyrocarbonate-water, mixed with 4.5 μL 100 mm sodium phosphate, pH 7.0, 22.5 μL dimethyl sulfoxide, and 6.6 μL 6 m glyoxal (deionized pH > 5), and incubated 1 h at 50°C. Next, 6 μL of glyoxal-loading buffer (10 mm sodium phosphate, pH 7.0, 0.25% [w/v] bromphenol, 0.25% [w/v] xylene cyanol, and 50% [v/v] glycerol) was added to RNA samples. RNA samples were electrophoresed at 4 V cm−1 in a 1% (w/v) agarose gel in 10 mm sodium phosphate running buffer, pH 7.0. Gels were rinsed with distilled water and equilibrated in 10 gel volumes of 20 × SSC (3 m sodium chloride and 0.3 m sodium citrate) for 45 min. RNA was transferred to nylon membranes (Zeta-probe GT, Bio-Rad Laboratories, Hercules, CA), which were washed in 20 mm Tris-HCl, pH 8.0, at 65°C to remove glyoxal after transfer, and UV cross-linked (1,200 Js cm−2).
The following oligonucleotide primer pairs were used to amplify the ORMV and CMV CP genes, BGL2, PDF1.2, and the 18S ribosomal RN:. ORMV CP FP (5′-TCACCCATGGTTTACAACATCACGAGCTCG-3′), ORMV CP RP (5′-CACTTCTAGACTATGTAGCTGGCGCAGTAGCC-3′), CMV CP FP (5′-TCATCCATGGCTTTCCAAGGTACCAGTA-3′), CMV CP RP (5′-CATATCTAGACTAAAGACCGTTAACCACCTGCG-3′), 18S rRNA FP (5′-GACAGACTGAGAGCTCTTTCTTGA-3′), 18S rRNA RP (5′-ACGTAGCTAGTTAGCAGGCTGAG-3′), BGL2 FP (5′-TCAACAGCTATAGCCACTGACAC-3′), BGL2 RP (5′-CTATCACTGGTTCGAGAAAGCTC-3′), PDF1.2 FP (5′-AATGAGCTCTCATGGCTAAGTTTGCTTCC-3′), and PDF1.2 RP (5′-AATCCATGGAATACACACGATTTAGCACC-3′; Penninckx et al., 1996). The Prime-A-Gene labeling system (Promega, Madison, WI) was used to label PCR products with 32P-dCTP for hybridization. Hybridizations and washes were performed as described in the protocol accompanying the Rapid-Hyb buffer (Amersham, Buckinghamshire, UK). Radioactive signals were detected by exposure to x-ray film (Eastman-Kodak, Rochester, NY) or phophorimaging screen (Molecular Dynamics, Sunnyvale, CA). Membranes to be reprobed were stripped by washing twice with 0.1× SSC/0.5% (w/v) SDS at 95°C for 30 min.
Gene Expression Measurements by DASL Assay and Fiber-Optic Bead Array Matrices
A set of 388 genes was selected for microarray analyses based upon previous experiments that identified genes that were up- or down-regulated in response to virus infection using Affymetrix GeneChip probe arrays (Whitham et al., 2003; S. Whitham, unpublished data). A custom oligo pool was made for the 388 genes and was used in conjunction with a universal array matrix (Sentrix Array Matrix, Illumina), comprising 96 arrays arranged in an 8 × 12 matrix that matches the dimensions of standard microtiter plates (Fan et al., 2003). This arrangement allows simultaneous processing of 96 samples using standard robotics. For each of the 388 genes, 3 unique target sites were chosen and corresponding query oligonucleotides were synthesized. A complete list of the genes and their corresponding probes along with negative and hybridization controls are provide in Supplemental Table I. Array manufacturing, sample preparation, and hybridization were performed as previously described (Fan et al., 2003, 2004). Total RNA samples isolated from Col-0, pad4-1, npr1-1, eds5-1, ein2-1, jar1-1, and NahG leaves that were either mock-, CMV-Y-, or ORMV-infected were labeled for hybridization to the bead arrays using the DASL assay (Fan et al., 2004). Briefly, cDNA was synthesized from total RNA (100 ng) with a 20-μL reverse transcription reaction containing a reaction mix (MMC; Illumina) and immobilized onto a solid phase. Gene-specific query oligonucleotides were annealed to the cDNA, ligated, and finally amplified with a Cy-3 labeled universal PCR primer. Labeled PCR products were then hybridized to bead array matrices. Arrays were washed and then imaged using a BeadArray Reader 1000 scanner (Illumina; Barker et al., 2003). Image analysis and data extraction software were as described (Galinsky, 2003). Signal intensity was normalized across each 96-well array matrix using the cubic splines method (Workman et al., 2002). Average signal values were computed for each gene in each sample by determining the mean signal for the three representative probes (Fan et al., 2004).
Statistical Analysis of Bead Array Data
To determine which of the 388 genes monitored by the bead array were most significantly affected by the plant genotype or various pairwise interactions, a linear mixed-effects statistical model was used to perform ANOVA analyses on the normalized gene expression data (Wolfinger et al., 2001). The following linear mixed-model equation was fit to the data sets: yijk = μ + αi +βj + α × βij + Σijk. In this equation, μ represents overall means of genotypes and treatments in our experiments; α represents treatment factors; i = 1 of 3 treatments: mock, CMV, and ORMV; β represents genotype factors; j = 1 of 7 genotypes: col-0, ein2-1, eds5-1, jar1-1, npr1-1, NahG, and pad4-1; α × β represents interaction of α and β; Σ represents error factors; and k represents replicate number.
The SAS program version 9 (SAS Institute, Cary, NC) was used to analyze the microarray data using this mixed-effects model. This program was run independently for the 2-DAI and 5-DAI data sets. The null hypothesis was that there was no difference in gene expression among the genotypes. Genes at or below the P < 0.01 threshold were selected as likely to be reliably influenced by plant genotype. This analysis provided sets of 187 and 176 genes at the 2 -and 5-DAI time points, respectively. Additional pairwise comparisons were performed to examine specific relationships between individual treatments across the genotypes (Supplemental Tables III and IV). To better understand how the expression profiles of the genes were related to each other, these genes were subjected to average linkage-hierarchical clustering using the Cluster program, and the TreeView program was used to display the results (Eisen et al., 1998).
Quantitative Real-Time PCR Analysis of Virus Accumulation
Quantitative real-time reverse transcriptase (RT)-PCR assays of CMV-Y and ORMV were performed using the iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad) on the iQ Real-Time PCR Detection System (Bio-Rad). Each reaction contained 12.5 μL (reaction mix), 5 pmol of forward and reverse primers, 0.5 μL (RT mix), 1.0 ng RNA template, and RNase free water to 25 μL. Samples were incubated for 10 min at 50°C followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Relative quantification was performed using the standard curve method, and all relative virus levels were normalized to the quantity of 18S. Normalized virus quantities were averaged for the three biological replicates and Student's t test was used determine if virus quantities in each mutant were significantly different from virus quantities in wild Col-0 leaves (Supplemental Table V). Primers used for quantitative PCR studies were: 18SF, GACAGACTGAGAGCTCTTTCTTGA; 18SR, ACGTAGCTAGTTAGCAGGCTGAG; ORMVREP1F, GATGCCTATGTGGTGAAGGAATTCAGCG; ORMVREP1R, GCCGGCAAATCCACAAAGTTGAAATCCG; CMVREP1F, AAACGTATTTGGAACATGGCAGGCGG; CMVREP1R, CCACCGACCCGTGGAGAAATGAATG.
Supplementary Material
Acknowledgments
We thank Dan Nettleton for guidance and assistance on the linear mixed-effects statistical analyses. The authors thank Philippe Rigault and Eugene Chudin for their assistance with assay probe design and initial data analysis. We thank Arnold Oliphant, David Barker, and Kan Wang for their support and guidance for this work.
This work was supported by the U.S. Department of Agriculture-National Research Initiative (grant no. 02–35319–12566 to S.A.W.), by the Iowa State University Plant Sciences Institute, and by the Hatch Act and State of Iowa Funds.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056028.
References
- Aranda MA, Escaler M, Wang D, Maule AJ (1996) Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc Natl Acad Sci USA 93: 15289–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJT, Theriault G, Che D, Dickinson T, Shen R, Kain R (2003) Self-assembled random arrays: high performance imaging and genomics applications on high-density microarray platform. Proc SPIE 4966: 1–11 [Google Scholar]
- Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, Wickham E, Atkins D, Barker D, Chee M, et al (2004) Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol 165: 1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SN (1998) Tobamovirus isolation and RNA extraction. In GD Foster, SC Taylor, eds, Plant Virology Protocols: From Virus Isolation to Transgenic Resistance. Humana Press, Totowa, NJ
- Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell 16: 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Naylor M, Carr JP (1997) Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate- phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J 26: 409–420 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T, Uknes S, Vernooji B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al(1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Pathirana MS, Wobbe KK, Klessig DF (1997) Identification of an Arabidopsis locus required for resistance to turnip crinkle virus. Plant J 11: 301–311 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24: 205–218 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaler M, Aranda MA, Thomas CL, Maule AJ (2000) Pea embryonic tissues show common responses to the replication of a wide range of viruses. Virology 267: 318–325 [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al (2003) Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol 68: 69–78 [DOI] [PubMed] [Google Scholar]
- Fan JB, Yeakley JM, Bibikova M, Chudin E, Wickham E, Chen J, Doucet D, Rigault P, Zhang B, Shen R, et al (2004) A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res 14: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich L, Vernooij B, Gaffney T, Morse A, Ryals J (1995) Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol Biol 29: 959–968 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Galinsky VL (2003) Automatic registration of microarray images. II. Hexagonal grid. Bioinformatics 19: 1832–1836 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4: 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golem S, Culver JN (2003) Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana. Mol Plant Microbe Interact 16: 681–688 [DOI] [PubMed] [Google Scholar]
- Gupta V, Willits MG, Glazebrook J (2000) Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol Plant Microbe Interact 13: 503–511 [DOI] [PubMed] [Google Scholar]
- Havelda Z, Maule AJ (2000) Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell 12: 1975–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya A, Matsuda Y, Gonzales RA, Nelson RS, Ding B (2002) Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol Plant Microbe Interact 15: 990–999 [DOI] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26: 395–407 [DOI] [PubMed] [Google Scholar]
- Kachroo P, Yoshioka K, Shah J, Dooner HK, Klessig DF (2000) Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 12: 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Katagiri F (2003) Attacking complex problems with the power of systems biology. Plant Physiol 132: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Hennig J, Klessig DF (1992) Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Bartel D, Vance VB, Bowman LH (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA 99: 15228–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A, Leh V, Lederer C (2002) The dialogue between viruses and hosts in compatible interactions. Curr Opin Plant Biol 5: 279–284 [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Carr JP (2002) Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol 128: 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor M, Murphy AM, Berry JO, Carr JP (1998) Salicylic acid can induce resistance to plant virus movement. Mol Plant Microbe Interact 11: 121–128 [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET (2000) Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol Plant Microbe Interact 13: 430–438 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ, White PS (1998) Cucumovirus isolation and RNA extraction. In GD Foster, SC Taylor, eds, Plant Virology Protocols: From Virus Isolation to Transgenic Resistance. Humana Press, Totowa, NJ
- Shah J, Tsui F, Klessig D (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of tms2 gene. Mol Plant Microbe Interact 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Shou H, Bordallo P, Fan JB, Yeakley JM, Bibikova M, Sheen J, Wang K (2004) Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc Natl Acad Sci USA 101: 3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miller J, Nozaki Y, Takeda M, Shah J, Hase S, Ikegami M, Ehara Y, Dinesh-Kumar SP (2002) RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J 32: 655–667 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, De Vos M, Robben C, Buchala A, Metraux JP, Van Loon LC, Pieterse CM (2002) Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J 29: 11–21 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17: 449–459 [DOI] [PubMed] [Google Scholar]
- Wang D, Maule AJ (1995) Inhibition of host gene expression associated with plant virus replication. Science 267: 229–231 [DOI] [PubMed] [Google Scholar]
- Whitham SA (2004) Viral host genomics. In RM Goodman, ed, Encyclopedia of Plant and Crop Science. Marcel Dekker, New York, pp 1269–1272
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Whitham SA, Wang Y (2004) Roles for host factors in plant viral pathogenicity. Curr Opin Plant Biol 7: 365–371 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3: research0048 [DOI] [PMC free article] [PubMed]
- Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD (2002) Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol 20: 353–358 [DOI] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC (2002) Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res 30: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.