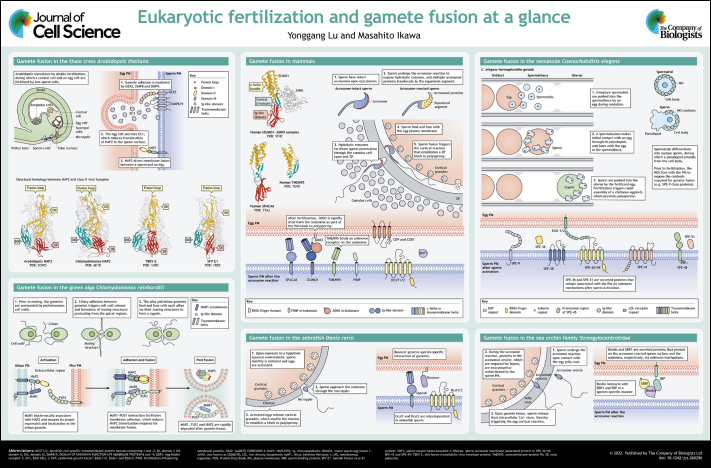
ABSTRACT
In sexually reproducing organisms, the genetic information is transmitted from one generation to the next via the merger of male and female gametes. Gamete fusion is a two-step process involving membrane recognition and apposition through ligand–receptor interactions and lipid mixing mediated by fusion proteins. HAP2 (also known as GCS1) is a bona fide gamete fusogen in flowering plants and protists. In vertebrates, a multitude of surface proteins have been demonstrated to be pivotal for sperm–egg fusion, yet none of them exhibit typical fusogenic features. In this Cell Science at a Glance article and the accompanying poster, we summarize recent advances in the mechanistic understanding of gamete fusion in eukaryotes, with a particular focus on mammalian species.
Keywords: Fertilization, Sperm–egg fusion, Fusogen, Eukaryotes, Mammals
Summary: This Cell Science at a Glance and the accompanying poster summarize recent advances in the mechanistic understanding of gamete fusion in eukaryotes, with a particular focus on mammalian species.
Introduction
In multicellular organisms, the plasma membrane – consisting of a phospholipid bilayer in which proteins, carbohydrates and other substances are embedded – creates a selectively permeable barrier that separates the intracellular compartment of a cell from the external environment. Cells exist as independent entities, each with their own nucleus and cytoplasm, and communicate with each other via the direct transport of small molecules across cell borders, while spontaneous or promiscuous fusion between neighboring cells is prevented by repulsive forces between the juxtaposed lipid membranes (Chen and Olson, 2005).
Cell–cell fusion, however, is a fundamental biological phenomenon in numerous physiological and pathological processes. To overcome the energy barriers between neighboring cells, fusion proteins, or fusogens, are thought to be necessary on one or both fusing membranes (Hernández and Podbilewicz, 2017). Most fusogens tether the two lipid bilayers by inserting a hydrophobic fusion peptide into the target membrane (Hernández and Podbilewicz, 2017; Vance and Lee, 2020). The fusogens then undergo conformational changes in their ectodomains, further bringing the apposed lipid membranes into immediate contact (Chernomordik and Kozlov, 2005). The membrane fusion process then passes through an intermediate state termed hemifusion, during which lipid exchange occurs between the outer leaflets of the two fusing membranes, while the inner leaflets remain separated (Chernomordik and Kozlov, 2005). Upon the merger of the inner leaflets, a nascent fusion pore opens and expands at the hemifusion diaphragm, which eventually permits mixing of the cell contents (Brukman et al., 2019; Hernández and Podbilewicz, 2017; Podbilewicz, 2014).
Somatic cell fusion, such as trophoblast fusion, myoblast fusion and osteoclast fusion, occurs between cells of the same type and culminates in the formation of multinucleated cells. In contrast, gamete fusion involves the joining of two morphologically and genetically distinct cells, as well as their genetic materials. Notwithstanding the diverse fertilization strategies adopted by various species, their gamete fusion processes share distinct similarities. Generally, prior to fusion with eggs, sperm cells are activated by extracellular agonists to display adhesion and fusion proteins on the cell surface (Ikawa et al., 2010; Sprunck et al., 2012). Gamete fusion is then carried out in two steps: recognition and merger of the two plasma membranes, which are mediated by adhesion proteins and fusion proteins, respectively (Bianchi and Wright, 2020; Pinello et al., 2021). Finally, the sperm receptors on the egg surface are subject to degradation to establish a membrane block to polygamy (Bianchi et al., 2014; Liu et al., 2010; Pinello et al., 2021). Despite decades of research on the unique sperm–egg fusion event, the underlying molecular mechanisms, particularly in vertebrates, remain mysterious. In this Cell Science at a Glance article and the accompanying poster, we review the current understanding of gamete fusion in various eukaryotic model organisms and highlight novel interdisciplinary strategies to tackle unsolved questions in the field.
Gamete fusion in flowering plants and protists
In flowering plants, male gametes, which develop in the pollen grain, are non-motile and are transported to the ovary by a pollen tube (Higashiyama and Takeuchi, 2015). Once the tip of the pollen tube reaches the ovule, it ruptures to release two sperm cells for double fertilization – one sperm cell fuses with an egg to form an embryo, while the other one merges with a central cell to produce an endosperm, which provides nutrition for embryonic development (see poster) (Dresselhaus and Franklin-Tong, 2013; Dresselhaus and Snell, 2014; Dresselhaus et al., 2016).
The bona fide gamete fusogen HAPLESS2 (HAP2, also known as GCS1), first identified in the thale cress Arabidopsis thaliana through a pollen-specific transfer DNA (T-DNA) insertional mutagenesis screen (Johnson et al., 2004; Mori et al., 2006; Von Besser et al., 2006), is a male-specific type-I transmembrane glycoprotein bearing three distinct β-sheet-rich regions termed domain I, II and III (DI, DII and DIII; see poster). DIII is associated with the transmembrane domain at the C terminus via a flexible linker, whilst DII contains a fusion loop that projects an amphipathic α-helix at the apical region (Fédry et al., 2018). Prior to gamete interaction in Arabidopsis, the egg cell secretes small cysteine-rich EGG CELL 1 (EC1) proteins, which trigger the approaching sperm to display HAP2 at the cell surface (see poster) (Sprunck et al., 2012). As revealed by a liposome fusion assay, HAP2 induces fusion by inserting its fusion helix into the opposing membrane and undergoes trimerization upon interaction with the lipids (Fédry et al., 2018).
Interestingly, HAP2 is conserved in a range of protists (for example, the unicellular green alga Chlamydomonas reinhardtii, the malaria parasite Plasmodium falciparum, the ciliate Tetrahymena thermophila, and the slime mold Dictyostelium discoideum) and invertebrates (for example, the cnidarian Hydra vulgaris, the sponge Amphimedon queenslandica and the honey bee Apis mellifera) (Mori et al., 2015; Steele and Dana, 2009; Wong and Johnson, 2010). As demonstrated by X-ray crystallography and in silico homology modeling, HAP2 orthologs in Arabidopsis, Chlamydomonas and Tetrahymena share structural similarity with class II viral fusogens (see poster and Box 1) (Fédry et al., 2018, 2017; Feng et al., 2018; Pinello et al., 2017; Valansi et al., 2017).
Box 1. Viral and cell–cell fusogens.
Viral fusogens are subdivided into four classes. Class I fusogens, such as influenza HA2 protein, Ebola virus GP2 protein and human immunodeficiency virus gp41 protein, form trimeric α-helical hairpins with a central coiled-coil structure after fusion (Kielian and Rey, 2006; Podbilewicz, 2014; Vance and Lee, 2020). Class II fusogens, such as tick-borne encephalitis virus envelope protein and Semliki Forest virus E1 protein, contain an elongated ectodomain composed of β-sheets that refolds into a trimer of hairpins (Podbilewicz, 2014; Vance and Lee, 2020). Class III fusogens, such as herpes simplex virus glycoprotein B and vesicular stomatitis virus glycoprotein, contain a trimeric coiled-coil structure and an elongated β-sheet-enriched region that resembles the class I and II fusogens, respectively (Igonet and Rey, 2012; Vance and Lee, 2020). Class IV fusogens, or fusion-associated small transmembrane proteins, are encoded by reoviruses and are characterized by short membrane-disruptive ectodomains (Chan et al., 2021, 2020).
Interestingly, somatic cell–cell fusion proteins show structural and functional similarities to viral fusogens. Syncytins, which mediate trophoblast fusion during mammalian placentation, are retrovirus-derived class I fusogens (Dupressoir et al., 2005; Podbilewicz, 2014). Epithelial cell membrane fusion protein EFF-1 (epithelial fusion failure 1) in C. elegans and gamete fusogen HAP2 in Arabidopsis and Chlamydomonas show structural homology to class II viral fusogens (Fédry et al., 2017; Pérez-Vargas et al., 2014). Distinct from typical class II fusogens, EFF-1 lacks a hydrophobic fusion loop and adopts a homotypic bilateral fusion strategy (Clark, 2018; Zeev-Ben-Mordehai et al., 2014). HAP2 harbors a fusion loop, which significantly varies in structure in Arabidopsis, Chlamydomonas and the parasite Trypanosoma cruzi (Fédry et al., 2018). Strikingly, the Fsx1 (Fusexin1) protein identified in prokaryotic Archaea species has recently been demonstrated to be a bona fide fusogen homologous to HAP2 (Moi et al., 2022), further expanding the class II fusogen superfamily. The molecular function and physiological relevance of Fsx1 in vivo need further investigation.
Membrane recognition and adhesion between gametes, which is usually mediated by extracellular ligand–receptor interactions, is a fundamental prerequisite for fusion to take place. GEX2 (GAMETE EXPRESSED 2) is a sperm-specific protein involved in gamete adhesion in Arabidopsis; sperm cells lacking GEX2 fail to bind to the egg or the central cell in situ (Mori et al., 2014). GEX2 is a type-I transmembrane protein harboring two filamin-like domains that assemble into immunoglobulin (Ig)-like folds composed of multiple antiparallel β-sheets in its extracellular segment (Mori and Igawa, 2014). Similar to GEX2, the tetraspanin proteins DMP8 and DMP9 (DOMAIN OF UNKNOWN FUNCTION 679 MEMBRANE PROTEIN-8 and -9, respectively) exhibit sperm-specific expression and are important for fertilization in Arabidopsis (Cyprys et al., 2019; Takahashi et al., 2018). However, while ablation of DMP8 and DMP9 significantly reduces sperm–egg adhesion, it does not affect sperm–central cell interaction, suggesting that the two gamete fusion events in Arabidopsis might be modulated by independent ligand–receptor pairs (Cyprys et al., 2019). Whether GEX2, DMP8 and DMP9 promote gamete binding through interaction with egg and/or central cell surface receptors, or whether they do so through another mechanism, remains unknown.
In Chlamydomonas, gametes of opposite mating types interact during fertilization, and HAP2, which is expressed in the minus gamete, is indispensable for fusion with the plus gamete (Liu et al., 2008). The cytoplasmic domain of HAP2 is crucial for its localization to an apical membrane projection specialized for fusion, termed the mating structure (Liu et al., 2015). Gamete adhesion in Chlamydomonas is mediated by the direct interaction of FUS1 and MAR1 (Minus Adhesion Receptor 1), which reside at the mating structures of plus and minus gametes, respectively (see poster) (Pinello et al., 2021). FUS1 bears seven Ig-like domains in its extracellular region, structurally resembling the gamete adhesion protein GEX2 in Arabidopsis. MAR1 is a type-I transmembrane protein containing a cysteine-rich growth factor receptor domain and a proline-rich region in its ectodomain. It ensures the proper expression and localization of HAP2 in the minus gamete; in mar1 mutant gametes, the abundance of HAP2 is drastically reduced and its localization is substantially altered (Pinello et al., 2021). During gamete fusion, Chlamydomonas HAP2 interacts with the target membrane via its hydrophobic fusion loops and concomitantly transforms into a stable trimeric conformation (Fédry et al., 2017). FUS1-mediated membrane adhesion is necessary for HAP2 trimerization; the minus gametes fail to form HAP2 trimers when pairing with FUS1-deficient plus gametes (Zhang et al., 2021). After fusion, HAP2, FUS1 and MAR1 are rapidly degraded, suggesting a possible mechanism for the membrane block to polygamy (see poster) (Liu et al., 2010; Pinello et al., 2021).
Gamete fusion in invertebrates
Although HAP2 orthologous gene sequences have been detected in several invertebrate species, it is unclear whether this ancestral fusogen drives gamete fusion in these species.
In the nematode Caenorhabditis elegans, hermaphrodites undergo self-fertilization or pair with male worms to produce self-progeny or outcross progeny, respectively (Mei and Singson, 2021). The maturing oocytes migrate toward the spermatheca, a myoepithelial compartment that stores mature sperm, and undergo fertilization inside the spermatheca (see poster) (Strome, 1986). To acquire fertilization competency, the spherical, non-motile spermatids develop into highly specialized spermatozoa through pseudopod extension, the merger of Golgi-derived membranous organelles (MOs) with the plasma membrane to release or expose the MO contents, and redistribution of essential proteins to the pseudopod and the cell body (see poster) (Karuo et al., 2021; Krauchunas et al., 2018; Singaravelu et al., 2012). During fertilization, an individual spermatozoon is thought to make initial contact with an egg via its pseudopod; however, a detailed model of sperm–egg fusion in C. elegans has not been established.
To date, researchers have reported ten sperm-specific proteins and two egg-specific proteins required for sperm–egg interaction in C. elegans; depletion of any of these proteins causes sterility due to defective gamete fusion (reviewed in Mei and Singson, 2021). The sperm proteins necessary for gamete binding or fusion belong to the SPE-9 class (see poster). SPE-9 and SPE-45 are type-I transmembrane proteins containing ten epidermal growth factor (EGF)-like repeats and an Ig-like domain, respectively (Nishimura et al., 2015; Putiri et al., 2004; Singaravelu et al., 2015; Singson et al., 1998). SPE-42 and SPE-49 – which contain six transmembrane helices, a dendritic cell-specific transmembrane protein (DC-STAMP) domain and a RING-finger domain – are orthologous to Sneaky in the fruit fly Drosophila melanogaster (Kroft et al., 2005; Wilson et al., 2006, 2018, 2011). Mutations of the cysteine residues in the RING-finger domain, which are predicted to coordinate zinc ions, impair sperm–egg interaction (Wilson et al., 2011). SPE-41 (also known as TRP-3) is another six-pass transmembrane protein that functions as a Ca2+ channel in mature spermatozoa (Nishimura and L'Hernault, 2010; Singaravelu et al., 2012). The redistribution of SPE-41 from the MOs to the entire sperm surface is dependent on a tetraspanin, SPE-38, as revealed by a mutagenesis analysis (Singaravelu et al., 2012). SPE-36 (encoded by F40F11.4) and SPE-51 (encoded by T22B11.1) are secreted proteins with an EGF-like and an Ig-like domain, respectively (Krauchunas et al., 2021 preprint; Mei et al., 2021 preprint). SPE-13 and FER-14 are additional sperm membrane proteins of the SPE-9 class, yet their domain composition has not been reported (L'Hernault et al., 1988; Singson, 2001). Most of these SPE-9 class proteins are localized to the MOs in the spermatids and are redistributed to the pseudopods or the entire sperm surface during sperm activation, implying their direct involvement in gamete binding or fusion (Mei and Singson, 2021). Low-density lipoprotein (LDL) receptor repeat-containing EGG-1 and EGG-2 are type-II transmembrane proteins that are expressed on the egg surface and are necessary for fertilization (Kadandale et al., 2005). So far, no sperm–egg ligand–receptor pairs have been identified among these proteins (Mei and Singson, 2021).
In the Strongylocentrotidae family of sea urchins, another invertebrate model system for studying fertilization, gamete recognition is mediated by a secreted protein Bindin and its egg surface receptors, the 350 kDa transmembrane glycoprotein SBP (sperm-binding protein) and the secreted protein EBR1 (egg bindin receptor 1) (Hirohashi and Lennarz, 1998, 2001; Kamei and Glabe, 2003; Ohlendieck et al., 1993; Vacquier, 2021; Wessel et al., 2021a,b). Sperm are triggered by the glycoconjugates in the extracellular coat of the eggs to undergo the acrosome reaction, during which Bindin is translocated from the acrosomal vesicle to the sperm plasma membrane (see poster) (Moy and Vacquier, 1979). The interactions between Bindin and its receptors occur in a species-specific manner (Hirohashi and Lennarz, 2001; Kamei and Glabe, 2003; Vacquier, 2021). So far, only Bindin has been validated to be indispensable for sperm–egg interaction by a knockout study (Wessel et al., 2021a).
Gamete fusion in vertebrates
The house mouse Mus musculus and zebrafish Danio rerio are the primary model organisms for studying fertilization in vertebrates (see poster). Mammals, including humans and mice, have adopted internal fertilization that involves an intricate cascade of molecular processes that alter the male and female gametes, culminating in their fusion (Box 2) (reviewed in Bianchi and Wright, 2016; Deneke and Pauli, 2021; Siu et al., 2021). Distinctly, zebrafish reproduce via external fertilization, during which sperm and eggs are directly released into the water. Zebrafish sperm become motile upon contact with a hypotonic environment, pass through the egg chorion and approach the egg plasma membrane, or the oolemma, through a funnel-shaped canal called the micropyle (Binner et al., 2022; Yanagimachi et al., 2013). Recent knockout studies have unveiled multiple male-specific proteins important for sperm–egg interaction in mice and zebrafish (Binner et al., 2022; Noda et al., 2022). Remarkably, mice lacking any of the essential proteins show normal sperm–oolemma adhesion but defective fusion, whereas the zebrafish mutants exhibit impaired sperm–egg binding.
Box 2. Fertilization in mammals.
After being deposited in the vagina at coitus, sperm travel through the female reproductive tract to reach the fallopian tube (i.e. the oviduct), where fertilization takes place (Suarez and Pacey, 2006). During this long and perilous journey, sperm experience dramatic biochemical and physiological changes, collectively known as capacitation, to gain fertilization competency (Gervasi and Visconti, 2016). As a major hallmark of capacitation, sperm exhibit hyperactivated movement characterized by a high-amplitude, asymmetrical flagellar beating pattern, which contrasts with the relatively low-amplitude, symmetrical beating of non-capacitated spermatozoa (Suarez, 2008). Simultaneously, sperm undergo the acrosome reaction, during which the acrosomal membrane fuses with the sperm plasma membrane at multiple points to expose the acrosomal contents important for fertilization (Okabe, 2018). The essential acrosomal contents include hydrolytic enzymes that allow sperm to loosen and penetrate the structures encapsulating the eggs – the cumulus oophorus and the zona pellucida (ZP), which is a glycoproteinaceous layer surrounding the oolemma – as well as acrosomal proteins that mediate sperm–egg binding and fusion (Ikawa et al., 2010; Khawar et al., 2019).
Upon internalization of a first spermatozoon, cortical granules poised at the egg cortex fuse with the oolemma to release the cortical contents into the perivitelline space, the gap between the oolemma and the ZP (Liu, 2011; Rojas et al., 2021). The released proteases modify ZP proteins, thus inducing ZP hardening that prevents binding and penetration of additional spermatozoa (Fahrenkamp et al., 2020). Meanwhile, the oolemma undergoes structural and molecular alterations involving shedding of sperm-binding receptors, which further establishes the plasma membrane block to polyspermy (Bianchi et al., 2014; Evans, 2020).
IZUMO1 and JUNO
Sperm protein IZUMO1 (Izumo sperm-egg fusion 1) and its oolemma counterpart JUNO (also known as IZUMO1 receptor, or IZUMO1R) form the only ligand–receptor pair known to be pivotal for sperm–egg interaction in mammals (Bianchi et al., 2014; Inoue et al., 2005). Izumo1, but not Juno, is conserved in zebrafish (Binner et al., 2022), but whether it is required for zebrafish fertilization is yet to be explored. Mice lacking IZUMO1 show male-specific sterility; their spermatozoa can elicit an acrosome reaction but fail to fuse with the oolemma (Inoue et al., 2005). IZUMO1 is a type-I transmembrane glycoprotein, which has an ectodomain comprising an N-terminal four-helix bundle (4HB), a central β-hairpin and a C-terminal Ig-like β-sandwich fold (Aydin et al., 2016; Inoue et al., 2005; Ohto et al., 2016). It localizes to the acrosomal membrane before the acrosome reaction and translocates to the equatorial segment or the entire sperm head surface in the acrosome-reacted sperm (Inoue et al., 2005; Satouh et al., 2012). The redistribution of IZUMO1 corroborates previous ultrastructural observations that sperm–oolemma fusion initiates at the equatorial segment of mouse (Nicosia et al., 1977), hamster (Bedford et al., 1979; Huang and Yanagimachi, 1985; Yanagimachi and Noda, 1970) and human sperm (Courtot and Lin-Tong, 1988).
JUNO is a glycosylphosphatidylinositol (GPI)-anchored oolemma protein that adopts a globular architecture stabilized by multiple disulfide bridges (Aydin et al., 2016; Bianchi et al., 2014; Ohto et al., 2016). Female mice deficient in JUNO are infertile, owing to impaired sperm–oolemma fusion (Bianchi et al., 2014). As revealed by X-ray crystallography, the central β-hairpin of IZUMO1 serves as the primary interface for JUNO binding (Aydin et al., 2016; Ohto et al., 2016). In vitro analyses suggest that after the initial interaction with JUNO, IZUMO1 undergoes conformational changes, dimerizes and binds to an unknown receptor, thereby bringing the juxtaposing plasma membranes into closer proximity (Inoue et al., 2015). Upon sperm entry, JUNO is rapidly released from the oolemma as part of the mechanism underlying the plasma membrane block to polyspermy (Bianchi et al., 2014).
IZUMO1 and JUNO do not exhibit structural homology to any known fusogens, nor do they contain a hydrophobic stretch that potentially serves as a fusogenic peptide (Aydin et al., 2016). In addition, they do not show apparent fusogenic activity in vitro; mammalian cultured cells expressing IZUMO1 can adhere to, but not fuse with, cells expressing JUNO or wild-type zona pellucida (ZP)-free eggs (Bianchi et al., 2014; Inoue et al., 2015; Noda et al., 2020). Despite the defective gamete fusion, Izumo1 knockout sperm can bind to wild-type eggs, and wild-type sperm can bind to Juno knockout eggs (Bianchi et al., 2014; Inoue et al., 2005). However, in both cases, binding of acrosome-reacted sperm to the oolemma is significantly reduced (Bianchi et al., 2014; Matsumura et al., 2022). Furthermore, deletion of Izumo1 completely abolishes sperm–oolemma binding in rats, suggesting that the IZUMO1–JUNO interaction underpins sperm–egg adhesion prior to fusion (Matsumura et al., 2022). Contradictory to these observations and assumptions, a recent study has suggested that IZUMO1 can act as a unilateral fusogen in vitro, functioning independently of JUNO and facilitating membrane fusion upon overexpression in baby hamster kidney (BHK) and human embryonic kidney 293 T (HEK293T) cells (Brukman et al., 2022 preprint). This discovery, which goes against the paradigm, requires further confirmation, and the mechanism of IZUMO1-mediated cell–cell fusion needs thorough investigation.
Eggs from the golden hamster Mesocricetus auratus can fuse with spermatozoa from a wide range of species, such as humans, mice and pigs (Yanagimachi, 1981). Consistently, hamster JUNO interacts with human, mouse and pig IZUMO1 (Bianchi and Wright, 2015). Human IZUMO1, however, cannot bind to mouse JUNO in vitro, in agreement with the incompatibility between human sperm and mouse eggs (Bianchi and Wright, 2015). Thus, IZUMO1–JUNO interaction might contribute to species-specific gamete recognition in mammals.
DCST1 and DCST2
Mouse and zebrafish DCST1 and DCST2 (DC-STAMP domain-containing-1 and -2, respectively) are predicted to have five or six transmembrane helices and a RING-finger domain in their C termini, resembling their invertebrate orthologs Sneaky in Drosophila, and SPE-42 and SPE-49 in C. elegans (Inoue et al., 2021; Kroft et al., 2005; Noda et al., 2022; Wilson et al., 2018). Mouse DCST2, like SPACA6 and TMEM95 (which are discussed below), localizes to the sperm acrosomal cap and the equatorial segment before and after the acrosome reaction, respectively (Lamas-Toranzo et al., 2020; Noda et al., 2022, 2020). Zebrafish sperm lack a typical acrosome; Dcst2 localizes to the sperm surface, as indicated by immunocytochemistry (Binner et al., 2022; Noda et al., 2022). Ablation of Dcst1 in zebrafish results in the loss of Dcst2, and vice versa, demonstrating their interdependent relationship (Noda et al., 2022).
SPACA6
SPACA6 (sperm acrosome membrane-associated protein 6), similar to IZUMO1, is a type-I transmembrane protein containing an N-terminal 4HB, a central β-hairpin and an Ig-like β-sandwich in its ectodomain (Aydin et al., 2016; Ohto et al., 2016; Vance et al., 2022). An anti-SPACA6 antibody markedly decreases in vitro fertilization between human sperm and human ZP-free eggs, suggesting the direct involvement of SPACA6 in sperm–egg interaction (Barbaux et al., 2020). Intriguingly, SPACA6 is absent in mouse sperm lacking any of the fusion-related proteins, such as IZUMO1, DCST1 and DCST2 (Inoue et al., 2021; Lu et al., 2022 preprint). In zebrafish, the amount of Dcst2 is significantly decreased in spaca6 knockout sperm (Binner et al., 2022). The interdependent relationships among the fusion-related proteins warrant further investigation.
TMEM95
Similar to IZUMO1 and SPACA6, TMEM95 (transmembrane protein 95) has a helical bundle and a β-hairpin region in its extracellular region but, distinctly, it harbors three α-helices and a coil in its N-terminal helical bundle and lacks an Ig-like fold (Tang et al., 2022). Notwithstanding the structural similarity to IZUMO1, neither TMEM95 nor SPACA6 interact with JUNO in vitro (Tang et al., 2022; Vance et al., 2022). The fragment antigen-binding (Fab) region of an anti-human IZUMO1 monoclonal antibody completely abolishes binding of human sperm to ZP-free golden hamster eggs (Tang et al., 2022). Remarkably, Fab regions of monoclonal antibodies against human TMEM95 do not affect sperm–egg binding but significantly reduce gamete fusion. Furthermore, recombinant human TMEM95 binds hamster oolemma, demonstrating that TMEM95 has a receptor on the egg surface (Tang et al., 2022). Taken together, these findings suggest that TMEM95 and its putative oolemma receptor may have a role distinct from IZUMO1 and JUNO during sperm–egg adhesion and fusion.
FIMP
Mouse Fimp (fertilization-influencing membrane protein) encodes two protein isoforms: a full-length transmembrane form and a secreted form. Male mice carrying a frameshift mutation in Fimp are severely subfertile (Fujihara et al., 2020). Transgenic expression of the mCherry-tagged transmembrane-type FIMP restores the fecundity of Fimp knockout males, suggesting that the secreted isoform is dispensable for sperm–egg fusion (Fujihara et al., 2020). FIMP–mCherry is detected in the equatorial segment of the acrosome-intact sperm and becomes absent in a portion of the acrosome-reacted sperm (Fujihara et al., 2020).
SOF1
SOF1 (sperm-oocyte fusion-required 1, also known as LLCFC1) is a secreted protein that potentially undergoes posttranslational modifications during sperm maturation; it is immunodetected in western blots as a single protein band in lysates of testicular germ cells and as two protein bands in lysates of acrosome-intact sperm. The slower-migrating SOF1 band is lost from lysates of acrosome-reacted sperm, likely as a result of exocytosis during the acrosome reaction (Noda et al., 2020). Whether SOF1 directly or indirectly facilitates gamete fusion remains elusive.
1700029I15Rik
Mouse 1700029I15Rik (also known as C11orf94 or Frey) is a type-II transmembrane protein that is exclusively expressed in the early round spermatids but is absent in the elongated spermatids and mature spermatozoa (Contreras et al., 2022; Lu et al., 2022 preprint). This protein interacts with endoplasmic reticulum (ER) chaperones involved in N-linked glycosylation, protein folding and quality control, and ER–Golgi trafficking (Lu et al., 2022 preprint). Deletion of 1700029I15Rik leads to reduced levels of acrosomal membrane proteins (such as IZUMO1 and SPACA1) in the testes, as well as an absence of SPACA6 in mature spermatozoa, thereby causing defective sperm–egg fusion (Lu et al., 2022 preprint).
CD9 and CD81
CD9 and its paralog CD81 are egg surface proteins important for sperm–egg interaction (Kaji et al., 2000; Miyado et al., 2000; Naour et al., 2000; Rubinstein et al., 2006). Both proteins contain four transmembrane helices flanking a small and a large extracellular loop, assembling into a cone-shaped structure with a central intramembrane cavity (Umeda et al., 2020; Zimmerman et al., 2016). Such a molecular architecture of CD9 creates curvature on the oolemma; eggs lacking CD9 show short and sparse microvilli (Runge et al., 2007; Umeda et al., 2020). Mutations in the large extracellular loop do not alter the abundance of CD9 on the oolemma but do impair sperm–egg fusion (Umeda et al., 2020; Zhu et al., 2002). CD9 also regulates the organization and distribution of sperm receptors, such as JUNO, on the oolemma (Inoue et al., 2020; Jégou et al., 2011). CD81 is immunodetected at the inner surface of the ZP (Ohnami et al., 2012). How this unique subcellular localization correlates with the molecular function of CD81 remains unclear.
Bouncer
In zebrafish, Bouncer is a Ly6/uPAR (lymphocyte antigen-6/urokinase-type plasminogen activator receptor)-family protein that attaches to the oolemma via a GPI anchor (Fujihara et al., 2021; Herberg et al., 2018). Wild-type fish sperm cannot bind the plasma membrane of bouncer knockout eggs, indicating the essential role of Bouncer in sperm–egg interaction. Furthermore, Bouncer governs species-specific gamete recognition in fish; sperm from the medaka Oryzias latipes, but not those from zebrafish, can fertilize bouncer knockout zebrafish eggs expressing a medaka bouncer transgene (Herberg et al., 2018). Interestingly, the homolog of bouncer in mammals, Spaca4, exhibits testis-biased expression and has no role sperm–egg interaction; Spaca4 knockout male mice are subfertile due to defects in sperm ZP binding and penetration (Fujihara et al., 2021).
Perspectives and concluding remarks
Sperm–egg fusion is a highly orchestrated, spatiotemporally dynamic process that requires strict timing and proper interplay of multiple proteins. Despite the enormous biological relevance of gamete fusion in sexually reproducing organisms, our knowledge of the underlying mechanisms remains rudimentary. With the emergence of the CRISPR/Cas9 system, genome editing in vertebrate model species has become rapid and efficient, thus permitting the recent discoveries of numerous sperm proteins required for gamete fusion (Binner et al., 2022; Fujihara et al., 2020; Noda et al., 2020). To acquire an in-depth understanding of the in vivo functions of these proteins, several important questions need to be answered. First, how do these proteins underpin fertilization? A defective gamete fusion phenotype revealed by reverse genetics does not guarantee direct involvement of the protein in the fusion process, such as in the case of 1700029I15Rik (Contreras et al., 2022; Lu et al., 2022 preprint). The features of sperm proteins can be preliminarily investigated using various techniques, including immunocytochemistry and co-immunoprecipitation coupled to mass spectrometry, and their roles in gamete interaction can be subsequently interrogated by fertilization inhibition assays using antibodies or recombinant proteins. To further dissect detailed domain functions, the protein structures can be solved using X-ray crystallography or cryo-electron microscopy, or predicted in silico using homology modeling software such as AlphaFold and RoseTTAFold (Baek et al., 2021; Jumper et al., 2021). Based on the structural characteristics, point mutations can be rationally introduced in vitro (through recombinant expression) or in vivo (using knock-in approaches or transgenics). A second question is whether these sperm proteins have binding partners on the oolemma. To tackle this problem, epitope-tagged recombinant proteins have been demonstrated as sensitive probes to detect weak and transient extracellular ligand–receptor interactions (Bianchi et al., 2014; Tang et al., 2022). How do the ligand–receptor pairs, in addition to IZUMO1–JUNO, contribute to gamete fusion? As proposed previously, during sperm–egg interaction, an array of surface proteins, which have various functions (for example, cell–cell adhesion and intercellular signaling), collectively establish a fusion synapse, which is a specialized area where the membranes are brought into close apposition and a favorable environment is created for membrane fusion (Krauchunas et al., 2016). Finally, does one or more of these proteins act as a gamete fusogen? To examine fusogenic activity, sophisticated cell- or liposome-based membrane fusion assays can be developed (Brukman et al., 2022 preprint; Moi et al., 2022; Valansi et al., 2017). When proteins fail to induce unilateral or homotypic bilateral fusion in vitro, other possibilities should be considered, such as heterotypic bilateral fusion or fusion occurring upon IZUMO1–JUNO-mediated membrane attachment. Given the recent surge of interest in the study of gamete fusion, we anticipate that these mysteries will soon be addressed.
Poster
Panel 1. Gamete fusion in the thale cress Arabidopsis thaliana
Panel 2. Structural homology between HAP2 and class II viral fusogens
Panel 3. Gamete fusion in mammals I
Panel 4. Gamete fusion in mammals II
Panel 5. Gamete fusion in the nematode Caenorhabditis elegans I
Panel 6. Gamete fusion in the nematode Caenorhabditis elegans II
Panel 7. Gamete fusion in the green alga Chlamydomonas reinhardtii
Panel 8. Gamete fusion in the zebrafish Danio rerio
Panel 9. Gamete fusion in the sea urchin family Strongylocentrotidae
Acknowledgements
We thank the members of the Department of Experimental Genome Research for insightful discussions, and we apologize to colleagues whose work could not be cited due to space limitations.
Footnotes
Funding
This work was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP18K16735 and JP22K15103 to Y.L.; JP19H05750, JP21H04753 and JP21H05033 to M.I.); the Takeda Science Foundation (to M.I.); and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01HD087157 and R01HD088412 to M.I.). Deposited in PMC for release after 12 months.
Contributor Information
Yonggang Lu, Email: yonggang-lu@biken.osaka-u.ac.jp.
Masahito Ikawa, Email: ikawa@biken.osaka-u.ac.jp.
Cell science at a glance
Individual poster panels are available for downloading at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.260296#supplementary-data.
References
- Aydin, H., Sultana, A., Li, S., Thavalingam, A. and Lee, J. E. (2016). Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature 534, 562-565. 10.1038/nature18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, M., Dimaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., Wang, J., Cong, Q., Kinch, L. N., Schaeffer, R. D.et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871-876. 10.1126/science.abj8754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaux, S., Ialy-Radio, C., Chalbi, M., Dybal, E., Homps-Legrand, M., Do Cruzeiro, M., Vaiman, D., Wolf, J.-P. and Ziyyat, A. (2020). Sperm SPACA6 protein is required for mammalian Sperm-Egg Adhesion/Fusion. Sci. Rep. 10, 5335. 10.1038/s41598-020-62091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford, J. M., Moore, H. D. M. and Franklin, L. E. (1979). Significance of the equatorial segment of the acrosome of the spermatozoon in eutherian mammals. Exp. Cell Res. 119, 119-126. 10.1016/0014-4827(79)90341-0 [DOI] [PubMed] [Google Scholar]
- Bianchi, E. and Wright, G. J. (2015). Cross-species fertilization: the hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140101. 10.1098/rstb.2014.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, E. and Wright, G. J. (2016). Sperm meets egg: the genetics of mammalian fertilization. Annu. Rev. Genet. 50, 93-111. 10.1146/annurev-genet-121415-121834 [DOI] [PubMed] [Google Scholar]
- Bianchi, E. and Wright, G. J. (2020). Find and fuse: unsolved mysteries in sperm–egg recognition. PLoS Biol. 18, e3000953. 10.1371/journal.pbio.3000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, E., Doe, B., Goulding, D. and Wright, G. J. (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483-487. 10.1038/nature13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binner, M. I., Kogan, A., Panser, K., Schleiffer, A., Deneke, V. E. and Pauli, A. (2022). The sperm protein Spaca6 is essential for fertilization in zebrafish. Front. Cell Dev. Biol. 9, 806982. 10.3389/fcell.2021.806982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukman, N. G., Uygur, B., Podbilewicz, B. and Chernomordik, L. V. (2019). How cells fuse. J. Cell Biol. 218, 1436-1451. 10.1083/jcb.201901017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukman, N. G., Nakajima, K. P., Valansi, C., Flyak, K., Li, X., Higashiyama, T. and Podbilewicz, B. (2022). IZUMO1 is a sperm fusogen. bioRxiv, 2022.02.01.478669. 10.1101/2022.02.01.478669 [DOI] [Google Scholar]
- Chan, K. M. C., Son, S., Schmid, E. M. and Fletcher, D. A. (2020). A viral fusogen hijacks the actin cytoskeleton to drive cell-cell fusion. Elife 9, e51358. 10.7554/eLife.51358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K. M. C., Arthur, A. L., Morstein, J., Jin, M., Bhat, A., Schlesinger, D., Son, S., Stevens, D. A., Drubin, D. G. and Fletcher, D. A. (2021). Evolutionarily related small viral fusogens hijack distinct but modular actin nucleation pathways to drive cell-cell fusion. Proc. Natl Acad. Sci. USA 118, e2007526118. 10.1073/pnas.2007526118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E. H. and Olson, E. N. (2005). Unveiling the mechanisms of cell-cell fusion. Science 308, 369-373. 10.1126/science.1104799 [DOI] [PubMed] [Google Scholar]
- Chernomordik, L. V. and Kozlov, M. M. (2005). Membrane hemifusion: crossing a chasm in two leaps. Cell 123, 375-382. 10.1016/j.cell.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Clark, T. (2018). HAP2/GCS1: mounting evidence of our true biological EVE? PLoS Biol. 16, e3000007. 10.1371/journal.pbio.3000007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras, W., Wiesehöfer, C., Schreier, D., Leinung, N., Peche, P., Wennemuth, G., Gentzel, M., Schröder, B. and Mentrup, T. (2022). C11orf94/Frey is a key regulator for male fertility by controlling Izumo1 complex assembly. Sci. Adv. 8, eabo6049. 10.1126/sciadv.abo6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot, A. M. and Lin-Tong, W. (1988). Initial stages of sperm-egg interaction in a heterospecific system: fate of the post-acrosomal sheath and appearance of a particular material within the oocyte. Hum. Reprod. 3, 651-655. 10.1093/oxfordjournals.humrep.a136761 [DOI] [PubMed] [Google Scholar]
- Cyprys, P., Lindemeier, M. and Sprunck, S. (2019). Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plants 5, 253-257. 10.1038/s41477-019-0382-3 [DOI] [PubMed] [Google Scholar]
- Deneke, V. E. and Pauli, A. (2021). The fertilization enigma: how sperm and egg fuse. Annu. Rev. Cell Dev. Biol. 37, 391-414. 10.1146/annurev-cellbio-120219-021751 [DOI] [PubMed] [Google Scholar]
- Dresselhaus, T. and Franklin-Tong, N. (2013). Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018-1036. 10.1093/mp/sst061 [DOI] [PubMed] [Google Scholar]
- Dresselhaus, T. and Snell, W. J. (2014). Fertilization: a sticky sperm protein in plants. Curr. Biol. 24, R164-R166. 10.1016/j.cub.2013.12.044 [DOI] [PubMed] [Google Scholar]
- Dresselhaus, T., Sprunck, S. and Wessel, G. M. (2016). Fertilization mechanisms in flowering plants. Curr. Biol. 26, R125-R139. 10.1016/j.cub.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir, A., Marceau, G., Vernochet, C., Bénit, L., Kanellopoulos, C., Sapin, V. and Heidmann, T. (2005). Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA 102, 725-730. 10.1073/pnas.0406509102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. P. (2020). Preventing polyspermy in mammalian eggs—Contributions of the membrane block and other mechanisms. Mol. Reprod. Dev. 87, 341-349. 10.1002/mrd.23331 [DOI] [PubMed] [Google Scholar]
- Fahrenkamp, E., Algarra, B. and Jovine, L. (2020). Mammalian egg coat modifications and the block to polyspermy. Mol. Reprod. Dev. 87, 326-340. 10.1002/mrd.23320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry, J., Liu, Y., Péhau-Arnaudet, G., Pei, J., Li, W., Tortorici, M. A., Traincard, F., Meola, A., Bricogne, G., Grishin, N. V.et al. (2017). The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168, 904-915.e10. 10.1016/j.cell.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry, J., Forcina, J., Legrand, P., Péhau-Arnaudet, G., Haouz, A., Johnson, M., Rey, F. A. and Krey, T. (2018). Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol. 16, e2006357. 10.1371/journal.pbio.2006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., Dong, X., Pinello, J., Zhang, J., Lu, C., Iacob, R. E., Engen, J. R., Snell, W. J. and Springer, T. A. (2018). Fusion surface structure, function, and dynamics of gamete fusogen HAP2. Elife 7, e39772. 10.7554/eLife.39772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara, Y., Lu, Y., Noda, T., Oji, A., Larasati, T., Kojima-Kita, K., Yu, Z., Matzuk, R. M., Matzuk, M. M. and Ikawa, M. (2020). Spermatozoa lacking fertilization influencing membrane protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. USA 117, 9393-9400. 10.1073/pnas.1917060117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara, Y., Herberg, S., Blaha, A., Panser, K., Kobayashi, K., Larasati, T., Novatchkova, M., Theussl, H.-C., Olszanska, O., Ikawa, M.et al. (2021). The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization. Proc. Natl. Acad. Sci. USA 118, e2108777118. 10.1073/pnas.2108777118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi, M. G. and Visconti, P. E. (2016). Chang's meaning of capacitation: a molecular perspective. Mol. Reprod. Dev. 83, 860-874. 10.1002/mrd.22663 [DOI] [PubMed] [Google Scholar]
- Herberg, S., Gert, K. R., Schleiffer, A. and Pauli, A. (2018). The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science 361, 1029-1033. 10.1126/science.aat7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, J. M. and Podbilewicz, B. (2017). The hallmarks of cell-cell fusion. Development 144, 4481-4495. 10.1242/dev.155523 [DOI] [PubMed] [Google Scholar]
- Higashiyama, T. and Takeuchi, H. (2015). The mechanism and key molecules involved in pollen tube guidance. Annu. Rev. Plant Biol. 66, 393-413. 10.1146/annurev-arplant-043014-115635 [DOI] [PubMed] [Google Scholar]
- Hirohashi, N. and Lennarz, W. J. (1998). The 350-kDa sea urchin egg receptor for sperm is localized in the vitelline layer. Dev. Biol. 204, 305-315. 10.1006/dbio.1998.9015 [DOI] [PubMed] [Google Scholar]
- Hirohashi, N. and Lennarz, W. J. (2001). Role of a vitelline layer-associated 350 kDa glycoprotein in controlling species-specific gamete interaction in the sea urchin. Dev. Growth Differ. 43, 247-255. 10.1046/j.1440-169x.2001.00571.x [DOI] [PubMed] [Google Scholar]
- Huang, T. T. F., Jr. and Yanagimachi, R. (1985). Inner acrosomal membrane of mammalian spermatozoa: its properties and possible functions in fertilization. Am. J. Anat. 174, 249-268. 10.1002/aja.1001740307 [DOI] [PubMed] [Google Scholar]
- Igonet, S. and Rey, F. A. (2012). SnapShot: viral and eukaryotic protein fusogens. Cell 151, 1634-1634.e1. 10.1016/j.cell.2012.11.041 [DOI] [PubMed] [Google Scholar]
- Ikawa, M., Inoue, N., Benham, A. M. and Okabe, M. (2010). Fertilization: a sperm's journey to and interaction with the oocyte. J. Clin. Invest. 120, 984-994. 10.1172/JCI41585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Ikawa, M., Isotani, A. and Okabe, M. (2005). The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234. 10.1038/nature03362 [DOI] [PubMed] [Google Scholar]
- Inoue, N., Hagihara, Y., Wright, D., Suzuki, T. and Wada, I. (2015). Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm–egg fusion in mice. Nat. Commun. 6, 8858. 10.1038/ncomms9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Saito, T. and Wada, I. (2020). Unveiling a novel function of CD9 in surface compartmentalization of oocytes. Development 147, dev189985. 10.1242/dev.189985 [DOI] [PubMed] [Google Scholar]
- Inoue, N., Hagihara, Y. and Wada, I. (2021). Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. Elife 10, e66313. 10.7554/eLife.66313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou, A., Ziyyat, A., Barraud-Lange, V., Perez, E., Wolf, J. P., Pincet, F. and Gourier, C. (2011). CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc. Natl. Acad. Sci. USA 108, 10946-10951. 10.1073/pnas.1017400108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M. A., Von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., Levin, J. Z. and Preuss, D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971-982. 10.1534/genetics.104.029447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A.et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale, P., Stewart-Michaelis, A., Gordon, S., Rubin, J., Klancer, R., Schweinsberg, P., Grant, B. D. and Singson, A. (2005). The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr. Biol. 15, 2222-2229. 10.1016/j.cub.2005.10.043 [DOI] [PubMed] [Google Scholar]
- Kaji, K., Oda, S., Shikano, T., Ohnuki, T., Uematsu, Y., Sakagami, J., Tada, N., Miyazaki, S. and Kudo, A. (2000). The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 24, 279-282. 10.1038/73502 [DOI] [PubMed] [Google Scholar]
- Kamei, N. and Glabe, C. G. (2003). The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 17, 2502-2507. 10.1101/gad.1133003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuo, Y., Shiraki, R., Yoshida, A., Tsunokawa, R., Nakahara-Yamada, M., Tarui, A., Sato, K., Kawai, K., Omote, M. and Nishimura, H. (2021). Identification and synthesis of DDI-6, a quinolinol analog capable of activating both Caenorhabditis elegans and mouse spermatozoa. Chem. Pharm. Bull. 69, 557-563. 10.1248/cpb.c21-00127 [DOI] [PubMed] [Google Scholar]
- Khawar, M. B., Gao, H. and Li, W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7, 195. 10.3389/fcell.2019.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian, M. and Rey, F. A. (2006). Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67-76. 10.1038/nrmicro1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauchunas, A. R., Marcello, M. R. and Singson, A. (2016). The molecular complexity of fertilization: introducing the concept of a fertilization synapse. Mol. Reprod. Dev. 83, 376-386. 10.1002/mrd.22634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauchunas, A. R., Mendez, E., Ni, J. Z., Druzhinina, M., Mulia, A., Parry, J., Gu, S. G., Stanfield, G. M. and Singson, A. (2018). spe-43 is required for sperm activation in C. elegans. Dev. Biol. 436, 75-83. 10.1016/j.ydbio.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauchunas, A. R., Marcello, M. R., Looper, A. M., Mei, X., Putiri, E., Singaravelu, G., Ahmed, I. I. and Singson, A. (2021). The EGF-motif containing protein SPE-36 is a secreted protein required for sperm function at fertilization in C. elegans. bioRxiv, 2021.07.07.451551. 10.1101/2021.07.07.451551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroft, T. L., Gleason, E. J. and L'Hernault, S. W. (2005). The spe-42 gene is required for sperm–egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev. Biol. 286, 169-181. 10.1016/j.ydbio.2005.07.020 [DOI] [PubMed] [Google Scholar]
- Lamas-Toranzo, I., Hamze, J. G., Bianchi, E., Fernández-Fuertes, B., Pérez-Cerezales, S., Laguna-Barraza, R., Fernández-González, R., Lonergan, P., Gutiérrez-Adán, A., Wright, G. J.et al. (2020). TMEM95 is a sperm membrane protein essential for mammalian fertilization. Elife 9, e53913. 10.7554/eLife.53913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault, S. W., Shakes, D. C. and Ward, S. (1988). Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics 120, 435-452. 10.1093/genetics/120.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. (2011). The biology and dynamics of mammalian cortical granules. Reprod. Biol. Endocrinol. 9, 149. 10.1186/1477-7827-9-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Tewari, R., Ning, J., Blagborough, A. M., Garbom, S., Pei, J., Grishin, N. V., Steele, R. E., Sinden, R. E., Snell, W. J.et al. (2008). The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22, 1051-1068. 10.1101/gad.1656508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Misamore, M. J. and Snell, W. J. (2010). Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137, 1473-1481. 10.1242/dev.044743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Pei, J., Grishin, N. and Snell, W. J. (2015). The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142, 962-971. 10.1242/dev.118844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Shimada, K., Zhang, J., Ogawa, Y., Tang, S., Noda, T., Shibuya, H. and Ikawa, M. (2022). 1700029I15Rik orchestrates the biosynthesis of acrosomal membrane proteins required for sperm–egg fusion. bioRxiv, 2022.04.15.488448. 10.1101/2022.04.15.488448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, T., Noda, T., Satouh, Y., Morohoshi, A., Yuri, S., Ogawa, M., Lu, Y., Isotani, A. and Ikawa, M. (2022). Sperm IZUMO1 is required for binding preceding fusion with Oolemma in mice and rats. Front. Cell Dev. Biol. 9, 810118. 10.3389/fcell.2021.810118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, X. and Singson, A. W. (2021). The molecular underpinnings of fertility: genetic approaches in Caenorhabditis elegans. Adv. Genet. 2, e10034. 10.1002/ggn2.10034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, X., Druzhinina, M., Dharia, S., Krauchunas, A. R., Ni, J., Singaravelu, G., Gu, S. G., Shakes, D. C., Grant, B. D. and Singson, A. W. (2021). SPE-51, a sperm secreted protein with an Immunoglobulin-like domain, is required for sperm-egg fusion in C. elegans. bioRxiv, 2021.07.07.451548. 10.1101/2021.07.07.451548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado, K., Yamada, G., Yamada, S., Hasuwa, H., Nakamura, Y., Ryu, F., Suzuki, K., Kosai, K., Inoue, K., Ogura, A.et al. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321-324. 10.1126/science.287.5451.321 [DOI] [PubMed] [Google Scholar]
- Moi, D., Nishio, S., Li, X., Valansi, C., Langleib, M., Brukman, N. G., Flyak, K., Dessimoz, C., De Sanctis, D., Tunyasuvunakool, K.et al. (2022). Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins. Nat. Commun. 13, 3880. 10.1038/s41467-022-31564-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T. and Igawa, T. (2014). Gamete attachment process revealed in flowering plant fertilization. Plant Signal. Behav. 9, e977715. 10.4161/15592324.2014.977715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Higashiyama, T. and Kuroiwa, T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64-71. 10.1038/ncb1345 [DOI] [PubMed] [Google Scholar]
- Mori, T., Igawa, T., Tamiya, G., Miyagishima, S.-. and Berger, F. (2014). Gamete attachment requires GEX2 for successful fertilization in Arabidopsis. Curr. Biol. 24, 170-175. 10.1016/j.cub.2013.11.030 [DOI] [PubMed] [Google Scholar]
- Mori, T., Kawai-Toyooka, H., Igawa, T. and Nozaki, H. (2015). Gamete dialogs in green lineages. Mol. Plant 8, 1442-1454. 10.1016/j.molp.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Moy, G. W. and Vacquier, V. D. (1979). Chapter 2 immunoperoxidase localization of bindin during the adhesion of sperm to sea urchin eggs. In Current Topics in Developmental Biology, Vol. 13 (ed. Moscona A. A., Monroy A. and Friedlander M.), pp. 31-44. Academic Press. [DOI] [PubMed] [Google Scholar]
- Naour, F. L., Rubinstein, E., Jasmin, C., Prenant, M. and Boucheix, C. (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319-321. 10.1126/science.287.5451.319 [DOI] [PubMed] [Google Scholar]
- Nicosia, S. V., Wolf, D. P. and Inoue, M. (1977). Cortical granule distribution and cell surface characteristics in mouse eggs. Dev. Biol. 57, 56-74. 10.1016/0012-1606(77)90354-2 [DOI] [PubMed] [Google Scholar]
- Nishimura, H. and L'hernault, S. W. (2010). Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev. Dyn. 239, 1502-1514. 10.1002/dvdy.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, H., Tajima, T., Comstra, H. S., Gleason, E. J. and L'hernault, S. W. (2015). The Immunoglobulin-like Gene spe-45 Acts during Fertilization in Caenorhabditis elegans like the Mouse Izumo1 Gene. Curr. Biol. 25, 3225-3231. 10.1016/j.cub.2015.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Lu, Y., Fujihara, Y., Oura, S., Koyano, T., Kobayashi, S., Matzuk, M. M. and Ikawa, M. (2020). Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm–oocyte fusion in mice. Proc. Natl. Acad. Sci. USA 117, 11493-11502. 10.1073/pnas.1922650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Blaha, A., Fujihara, Y., Gert, K. R., Emori, C., Deneke, V. E., Oura, S., Panser, K., Lu, Y., Berent, S.et al. (2022). Sperm membrane proteins DCST1 and DCST2 are required for sperm-egg interaction in mice and fish. Commun. Biol. 5, 332. 10.1038/s42003-022-03289-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendieck, K., Dhume, S. T., Partin, J. S. and Lennarz, W. J. (1993). The sea urchin egg receptor for sperm: isolation and characterization of the intact, biologically active receptor. J. Cell Biol. 122, 887-895. 10.1083/jcb.122.4.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnami, N., Nakamura, A., Miyado, M., Sato, M., Kawano, N., Yoshida, K., Harada, Y., Takezawa, Y., Kanai, S., Ono, C.et al. (2012). CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol. Open 1, 640-647. 10.1242/bio.20121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, U., Ishida, H., Krayukhina, E., Uchiyama, S., Inoue, N. and Shimizu, T. (2016). Structure of IZUMO1–JUNO reveals sperm–oocyte recognition during mammalian fertilization. Nature 534, 566-569. 10.1038/nature18596 [DOI] [PubMed] [Google Scholar]
- Okabe, M. (2018). Sperm–egg interaction and fertilization: past, present, and future. Biol. Reprod. 99, 134-146. 10.1093/biolre/ioy028 [DOI] [PubMed] [Google Scholar]
- Pérez-Vargas, J., Krey, T., Valansi, C., Avinoam, O., Haouz, A., Jamin, M., Raveh-Barak, H., Podbilewicz, B. and Rey, F. A. (2014). Structural basis of eukaryotic cell-cell fusion. Cell 157, 407-419. 10.1016/j.cell.2014.02.020 [DOI] [PubMed] [Google Scholar]
- Pinello, J. F., Lai, A. L., Millet, J. K., Cassidy-Hanley, D., Freed, J. H. and Clark, T. G. (2017). Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr. Biol. 27, 651-660. 10.1016/j.cub.2017.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello, J. F., Liu, Y. and Snell, W. J. (2021). MAR1 links membrane adhesion to membrane merger during cell-cell fusion in Chlamydomonas. Dev. Cell 56, 3380-3392.e9. 10.1016/j.devcel.2021.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz, B. (2014). Virus and Cell Fusion Mechanisms. Annu. Rev. Cell Dev. Biol. 30, 111-139. 10.1146/annurev-cellbio-101512-122422 [DOI] [PubMed] [Google Scholar]
- Putiri, E., Zannoni, S., Kadandale, P. and Singson, A. (2004). Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev. Biol. 272, 448-459. 10.1016/j.ydbio.2004.05.014 [DOI] [PubMed] [Google Scholar]
- Rojas, J., Hinostroza, F., Vergara, S., Pinto-Borguero, I., Aguilera, F., Fuentes, R. and Carvacho, I. (2021). Knockin’ on egg's door: maternal control of egg activation that influences cortical granule exocytosis in animal species. Front. Cell Dev. Biol. 9, 704867. 10.3389/fcell.2021.704867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E., Ziyyat, A., Prenant, M., Wrobel, E., Wolf, J.-P., Levy, S., Le Naour, F. and Boucheix, C. (2006). Reduced fertility of female mice lacking CD81. Dev. Biol. 290, 351-358. 10.1016/j.ydbio.2005.11.031 [DOI] [PubMed] [Google Scholar]
- Runge, K. E., Evans, J. E., He, Z.-Y., Gupta, S., Mcdonald, K. L., Stahlberg, H., Primakoff, P. and Myles, D. G. (2007). Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev. Biol. 304, 317-325. 10.1016/j.ydbio.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Satouh, Y., Inoue, N., Ikawa, M. and Okabe, M. (2012). Visualization of the moment of mouse sperm–egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125, 4985-4990. 10.1242/jcs.100867 [DOI] [PubMed] [Google Scholar]
- Singaravelu, G., Chatterjee, I., Rahimi, S., Druzhinina, M. K., Kang, L., Xu, X. Z. S. and Singson, A. (2012). The sperm surface localization of the TRP-3/SPE-41 Ca2+-permeable channel depends on SPE-38 function in Caenorhabditis elegans. Dev. Biol. 365, 376-383. 10.1016/j.ydbio.2012.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu, G., Rahimi, S., Krauchunas, A., Rizvi, A., Dharia, S., Shakes, D., Smith, H., Golden, A. and Singson, A. (2015). Forward genetics identifies a requirement for the Izumo-like immunoglobulin superfamily spe-45 gene in Caenorhabditis elegans fertilization. Curr. Biol. 25, 3220-3224. 10.1016/j.cub.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson, A. (2001). Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev. Biol. 230, 101-109. 10.1006/dbio.2000.0118 [DOI] [PubMed] [Google Scholar]
- Singson, A., Mercer, K. B. and L'Hernault, S. W. (1998). The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 93, 71-79. 10.1016/S0092-8674(00)81147-2 [DOI] [PubMed] [Google Scholar]
- Siu, K. K., Serrão, V. H. B., Ziyyat, A. and Lee, J. E. (2021). The cell biology of fertilization: gamete attachment and fusion. J. Cell Biol. 220, e202102146. 10.1083/jcb.202102146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck, S., Rademacher, S., Vogler, F., Gheyselinck, J., Grossniklaus, U. and Dresselhaus, T. (2012). Egg cell–secreted EC1 triggers sperm cell activation during double fertilization. Science 338, 1093-1097. 10.1126/science.1223944 [DOI] [PubMed] [Google Scholar]
- Steele, R. E. and Dana, C. E. (2009). Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One 4, e7680. 10.1371/journal.pone.0007680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S. (1986). Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J. Cell Biol. 103, 2241-2252. 10.1083/jcb.103.6.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, S. S. (2008). Control of hyperactivation in sperm. Hum. Reprod. Update 14, 647-657. 10.1093/humupd/dmn029 [DOI] [PubMed] [Google Scholar]
- Suarez, S. S. and Pacey, A. A. (2006). Sperm transport in the female reproductive tract. Hum. Reprod. Update 12, 23-37. 10.1093/humupd/dmi047 [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Mori, T., Ueda, K., Yamada, L., Nagahara, S., Higashiyama, T., Sawada, H. and Igawa, T. (2018). The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145, dev170076. 10.1242/dev.170076 [DOI] [PubMed] [Google Scholar]
- Tang, S., Lu, Y., Skinner, W. M., Sanyal, M., Lishko, P. V., Ikawa, M. and Kim, P. S. (2022). Human sperm TMEM95 binds eggs and facilitates membrane fusion. Proc. Natl. Acad. Sci. USA 119, e2207805119. 10.1073/pnas.2207805119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, R., Satouh, Y., Takemoto, M., Nakada-Nakura, Y., Liu, K., Yokoyama, T., Shirouzu, M., Iwata, S., Nomura, N., Sato, K.et al. (2020). Structural insights into tetraspanin CD9 function. Nat. Commun. 11, 1606. 10.1038/s41467-020-15459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier, V. D. (2021). A protein bridging the gap between sea urchin generations. Proc. Natl. Acad. Sci. USA 118, e2114056118. 10.1073/pnas.2114056118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valansi, C., Moi, D., Leikina, E., Matveev, E., Graña, M., Chernomordik, L. V., Romero, H., Aguilar, P. S. and Podbilewicz, B. (2017). Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 216, 571-581. 10.1083/jcb.201610093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, T. D. R. and Lee, J. E. (2020). Virus and eukaryote fusogen superfamilies. Curr. Biol. 30, R750-R754. 10.1016/j.cub.2020.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, T. D. R., Yip, P., Jiménez, E., Li, S., Gawol, D., Byrnes, J., Usón, I., Ziyyat, A. and Lee, J. E. (2022). SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins. Commun. Biol. 5, 984. 10.1038/s42003-022-03883-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Besser, K., Frank, A. C., Johnson, M. A. and Preuss, D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133, 4761-4769. 10.1242/dev.02683 [DOI] [PubMed] [Google Scholar]
- Wessel, G. M., Wada, Y., Yajima, M. and Kiyomoto, M. (2021a). Bindin is essential for fertilization in the sea urchin. Proc. Natl. Acad. Sci. USA 118, e2109636118. 10.1073/pnas.2109636118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, G. M., Wada, Y., Yajima, M. and Kiyomoto, M. (2021b). Sperm lacking Bindin are infertile but are otherwise indistinguishable from wildtype sperm. Sci. Rep. 11, 21583. 10.1038/s41598-021-00570-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K. L., Fitch, K. R., Bafus, B. T. and Wakimoto, B. T. (2006). Sperm plasma membrane breakdown during Drosophila fertilization requires Sneaky, an acrosomal membrane protein. Development 133, 4871-4879. 10.1242/dev.02671 [DOI] [PubMed] [Google Scholar]
- Wilson, L. D., Sackett, J. M., Mieczkowski, B. D., Richie, A. L., Thoemke, K., Rumbley, J. N. and Kroft, T. L. (2011). Fertilization in C. elegans requires an intact C-terminal RING finger in sperm protein SPE-42. BMC Dev. Biol. 11, 10. 10.1186/1471-213X-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L. D., Obakpolor, O. A., Jones, A. M., Richie, A. L., Mieczkowski, B. D., Fall, G. T., Hall, R. W., Rumbley, J. N. and Kroft, T. L. (2018). The Caenorhabditis elegans spe-49 gene is required for fertilization and encodes a sperm-specific transmembrane protein homologous to SPE-42. Mol. Reprod. Dev. 85, 563-578. 10.1002/mrd.22992 [DOI] [PubMed] [Google Scholar]
- Wong, J. L. and Johnson, M. A. (2010). Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 20, 134-141. 10.1016/j.tcb.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Yanagimachi, R. (1981). Mechanisms of fertilization in mammals. In Fertilization and Embryonic Development In Vitro, (ed. Mastroianni L. and Biggers J. D.), pp. 81-182. Boston, MA: Springer US. [Google Scholar]
- Yanagimachi, R. and Noda, Y. D. (1970). Physiological changes in the postnuclear cap region of mammalian spermatozoa: a necessary preliminary to the membrane fusion between sperm and egg cells. J. Ultrastruct. Res. 31, 486-493. 10.1016/S0022-5320(70)90164-4 [DOI] [PubMed] [Google Scholar]
- Yanagimachi, R., Cherr, G., Matsubara, T., Andoh, T., Harumi, T., Vines, C., Pillai, M., Griffin, F., Matsubara, H., Weatherby, T.et al. (2013). Sperm attractant in the micropyle region of fish and insect eggs. Biol. Reprod. 88, 47. 10.1095/biolreprod.112.105072 [DOI] [PubMed] [Google Scholar]
- Zeev-Ben-Mordehai, T., Vasishtan, D., Siebert, C. A. and Grünewald, K. (2014). The full-length cell–cell fusogen EFF-1 is monomeric and upright on the membrane. Nat. Commun. 5, 3912. 10.1038/ncomms4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Pinello, J. F., Fernández, I., Baquero, E., Fédry, J., Rey, F. A. and Snell, W. J. (2021). Species-specific gamete recognition initiates fusion-driving trimer formation by conserved fusogen HAP2. Nat. Commun. 12, 4380. 10.1038/s41467-021-24613-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G.-Z., Miller, B. J., Boucheix, C., Rubinstein, E., Liu, C. C., Hynes, R. O., Myles, D. G. and Primakoff, P. (2002). Residues SFQ (173-175) in the large extracellular loop of CD9 are required for gamete fusion. Development 129, 1995-2002. 10.1242/dev.129.8.1995 [DOI] [PubMed] [Google Scholar]
- Zimmerman, B., Kelly, B., Mcmillan, B. J., Seegar, T. C. M., Dror, R. O., Kruse, A. C. and Blacklow, S. C. (2016). Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041-1051.e11. 10.1016/j.cell.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.