Abstract
Inhalation is a major exposure route to nanoparticles. Following inhalation, nanoparticles first interact with the lung lining fluid, a complex mixture of proteins, lipids, and mucins. We measure the concentration and composition of lung fluid proteins adsorbed on the surface of titanium dioxide (TiO2) nanoparticles. Using proteomics, we find that lung fluid results in a unique protein corona on the surface of the TiO2 nanoparticles. We then measure the expression of three cytokines (interleukin 6 (IL-6), tumor necrosis factor-alpha (TNF-α), and macrophage inflammatory protein 2 (MIP-2)) associated with lung inflammation. We find that the corona formed from lung fluid leads to elevated expression of these cytokines in comparison to bare TiO2 nanoparticles or coronas formed from serum or albumin. These experiments show that understanding the concentration and composition of the protein corona is essential for understanding the pulmonary response associated with human exposure to nanoparticles.
Graphical Abstract
Introduction
Nanoparticles (NPs) are increasingly used on a large scale in industrial materials and agriculture.1–7 These applications pose the risk of environmental exposure, either directly, as in agriculture, or during the subsequent degradation of nano-containing materials. 8–11 A major concern is inhalation of NPs, especially titanium dioxide (TiO2) NPs,12–16 which are used at very large scale. TiO2 NPs are widely used in the paint, plastics, rubber, adhesives, coatings, and paper industries, and are also a common food coloring.12, 17, 18 The Department of the Interior reported U.S. production of TiO2 at 1.1 million metric tons, valued at $3.2 billion, in 2022.19
In any biological system, proteins adsorb on the surface of NPs forming a protein “corona.”8, 20–26 The specific proteins that adsorb on the NP surface determine the subsequent interactions of the NP with cells and organs including cellular internalization, immune response, biodistribution, and circulation time.22, 24, 25, 27–29 The majority of research on the protein corona, including our own,30–32 has focused on blood serum proteins relevant to nanomedicines. In comparison, inhalation of NPs brings the NPs into contact with the more complex environment of lung lining fluid. The lung has a thin layer of fluid that lines the airspaces. This fluid is a mixture of proteins, lipids, and mucins. The proteins present in lung fluid include those designed for antimicrobial and oxidant defense. Therefore, this lung fluid has important functions in the innate defense mechanisms of the lung.33–35
Our goal was to characterize the concentration and composition of the protein corona formed by bronchoalveolar lung fluid (BALF) and then determine the cellular response to the lung fluid proteins adsorbed on TiO2 NPs. In comparison, previous work has examined a single lung fluid protein, lung surfactant protein A (SPA), isolated from BALF,36, 37 surfactant separated from BALF,38 or BALF from a patient with a pulmonary disease, pulmonary alveolar proteinosis,39 that results in elevated lipids and surfactant proteins in BALF. Our goal was to examine the full BALF mixture obtained from healthy animals. The concentration of the protein corona was measured with a colorimetric assay and the protein composition was determined by gel electrophoresis and proteomics. We found that incubating TiO2 NPs with BALF resulted in a protein corona dominated by albumin, the most abundant protein present in BALF. The production of cytokines (interleukin 6 (IL-6), tumor necrosis factor-alpha (TNF-α), and macrophage inflammatory protein 2 (MIP-2)) by macrophage cells has been associated with lung inflammation.40–42 These cytokines were used as measures of the inflammatory response to the BALF-TiO2 NPs. Coronas formed from bovine serum albumin (BSA) and fetal bovine serum (FBS) were used for comparison. We observed that the BALF corona led to an increased production of pro-inflammatory cytokines in comparison to bare TiO2 NPs and the other protein coronas. We hope our work will help address underlying mechanisms of the observed lung toxicity and inflammation associated with inhalation of TiO2 NPs.43, 44
Materials and Methods
Nanoparticles (NPs) and characterization
TiO2 NPs (#718467, Sigma-Aldrich, Burlington, MA) were used for all experiments. Hydrodynamic diameter, polydispersity index, and zeta potential of the NPs (100 μg/mL in phosphate buffered saline (PBS) diluted 1:100 in ultrapure water) were measured using dynamic light scattering (DLS; Zetasizer, Malvern Instruments, Worcestershire, England). Measurements were carried out in triplicate with three distinct samples. Average and standard deviation are reported for all measurements. Electrophoretic mobility was converted to zeta potential using the Smoluchowski approximation.
Rodent bronchoalveolar lavage (BAL)
C57BL/6 male mice (8–10 weeks) were purchased from Jackson Laboratories (Bar Harbor, ME). All procedures were approved by the Duke University Institutional Animal Care and Use Committee (IACUC) and were performed under an IACUC approved animal protocol (A053–21-03). BAL was performed following a published protocol.45 Prior to BAL, mice were deeply anesthetized with an intraperitoneal injection of ketamine (100 mg/kg), xylazine (100 mg/kg), and saline (0.9%), dosed by weight (350–500 μL). The chest and trachea were dissected to expose the lungs and trachea. Following a nick in the trachea, PE-60 tubing (#9565S30, Thomas Scientific, Swedesboro, NJ) was inserted into the trachea and attached to a 12-inch infusion set (#SV-25BLK, Terumo, Tokyo, Japan), which was connected to a 10 mL syringe held on a ring stand. Lungs were passively filled to 20 cm H2O with PBS to reach total lung capacity. The BALF was then passively drained. The BALF was placed on ice for further processing and for use in NP incubation experiments. BALF used for protein corona formation was pooled from 10–20 mice to reduce mouse-to-mouse variation.
Protein corona formation and quantification
TiO2 NPs (1 mg/mL) were first suspended in PBS and sonicated (5 min, room temperature (RT); #Q700, Qsonica, Newton, CT). A protein corona was then formed by incubating the TiO2 NPs (1 mg/mL) in 10% solutions of FBS (#10437028, Thermo Fisher Scientific, Waltham, MA), BSA (#A2153, Sigma-Aldrich) or BALF diluted in PBS. 10% solutions of FBS, BSA, and BALF correspond to 6, 1.6, and 0.017 mg/mL, respectively. The incubations were performed at RT on an orbital shaker for 30 minutes. The corona formed under these conditions is independent of time (30 min −120 min) and temperature (RT and 37 °C; Fig. S1). To remove unbound proteins, the protein-NP complexes were “washed” with PBS (x3). Each wash step consisted of centrifugation (18,000 rcf, 15 mins), removal of the supernatant, and then resuspension in an equal volume of PBS.
Protein concentration of both the protein corona and unbound protein was measured with a bicinchoninic acid assay (BCA assay; #2260, Thermo Fisher Scientific) according to the manufacturer’s instructions. Washed protein-NP complexes were resuspended in 50 μL of PBS and sonicated (5 min, RT) to aid resuspension. The BCA assay was carried out with proteins present on the NPs (30 min, 37 °C). The protein-NP complexes were then centrifuged (15 min, 18,000 rcf, 4 °C) to separate protein-NPs complexes from the BCA reagent prior to measuring BCA absorption at 562 nm (SpectraMax, iD3, Molecular Devices, San Jose, CA). The TiO2 NP concentration was measured by absorption at 440 nm and comparison with a calibration curve of known concentrations (Fig. S2). Protein concentration is reported as protein relative to NP concentration (μg protein/mg NPs). Removal of free protein was monitored by BCA (Fig. S3), which showed no significant decrease in free protein in the supernatant following additional wash steps. The hard corona is defined as the protein that remains bound to the NPs at this point.
Gel electrophoresis
Gel electrophoresis was used to visualize individual proteins present in the corona of the NPs, as previously described.30 The protein coronas were removed from the NPs by suspending the NPs in loading buffer (Laemmli, #BP-110R; Boston BioProducts, Ashland, MA), incubating for at least 5 min at 95°C, and then loaded onto a gel (tris-glycine sodium dodecyl sulfate (SDS) gel, #4561093, Bio-Rad, Hercules, CA) for SDS-polyacrylamide gel electrophoresis (PAGE; 230 V, 35 min). A 10 to 250 kDa molecular weight marker (Precision Plus Protein Dual Color Standards, #1610374, Bio-Rad) was included. Gels were rinsed by microwaving in deionized water (1 min heat, 1 min rocking at RT, replace water, x3), stained (SimplyBlue Safe Stain, #LC6060, Thermo Fisher) by microwaving until near boiling (1 min), and then rocked for 5 min. Gels were destained in deionized water (10 min, rocking) and NaCl solution (20% w/v, >5 minutes, rocking) and then imaged (PhotoDoc-It, Analytik Jena, Jena, Germany).
Proteomic analysis
Prior to proteomic analysis, samples were digested using a modified S-Trap micro column (Protifi, Farmingdale, NY) protocol, as previously described.30 Proteins were removed from the NP surface by resuspending in 5% SDS buffer, sonicating to reduce aggregates followed by incubating at 95 °C for 15 minutes. Protein concentration was measured with the Pierce 660 nm Protein Assay Reagent (#2260, Thermo Fisher Scientific) with the addition of Ionic Detergent Compatibility Reagent (#22663, Thermo Fisher Scientific) according to the manufacturer’s instructions. A minimum of 5 μg of protein was loaded on each S-Trap. Two modifications were made to the S-trap protocol: dithiothreitol (DTT; #R0861, Thermo Fisher Scientific) and iodoacetamide (IAM; #I1149, Sigma-Aldrich) were used as the reducer (20 mM) and alkylator (40 mM), respectively. DTT and IAM are commonly used for proteomics and are recommended substitutions. Following the completion of the S-trap protocol, the resulting digested proteins were lyophilized and stored at −20 °C until proteomic analysis.
Proteomic analysis was carried out in the Proteomics and Metabolomics Core Facility, part of the Duke Center for Genomics and Computational Biology, as previously described.30, 31 In brief, digested samples were analyzed using LC-MS/MS with ≤ 500 ng of digested protein injected. NanoFlow LC was performed with an ultra-performance liquid chromatography (UPLC, 75 μm × 250 mm, nanoAcquity, Waters Corporation; 400 nL/min) column and a 60 minute total elution time. The column was run with an acetonitrile gradient (5–40%) with 0.1% formic acid. Peptide fragments were analyzed using in-line tandem mass spectrometry (Orbitrap Fusion Lumos, Thermo Fisher).
We analyzed the LC-MS/MS data using MaxQuant (v2.1.0, Max Plank Institute, Munich, Germany), an open-source software designed to analyze mass spectrometry data qualitatively and quantitatively.46, 47 The raw LC-MS/MS spectra were searched, using their integrated Andromeda search engine, against the Swiss-Prot murine canonical protein database from UniProt, accessed on June 6th, 2022. A custom contaminants file was used while searching, which contained a relevant subset of the Common Repository of Adventitious Proteins (cRAP) database.48 For protein and peptide quantification and identification, default MaxQuant parameters were used including a 0.01 false discovery rate, a minimum peptide length of 7 amino acids, a maximum peptide length of 25 amino acids, oxidation and acetyl groups as variable modifications, and carbamidomethyl as a fixed modification. The proteins were quantified by their summed intensity.
Proteomic data was analyzed and filtered in Perseus (v2.0.3.1, Max Plank Institute).49 Proteins were excluded if they were considered contaminants, quality control standards, or were not observed in at least 2 samples. After filtering in Perseus, 112 proteins were observed across the samples. To correct for any change in performance or differences in protein loading, each sample was normalized to itself by dividing by the mean of the interior 80% of the protein intensities. Each sample was scaled to have the same average. We report these values as percent normalized abundance. Fold change for each protein was calculated by taking the log base 2 of the normalized abundance of samples divided by BALF.
The complete lists of proteins are included in the Electronic Supplementary Information (ESI; Table ESI1). In addition, the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD041036 and 10.6019/PXD041036.50
Cell culture, TiO2 NP incubations, and cytokine assays
RAW 264.7 mouse macrophage cells (TIB-71, ATCC, Manassas, VA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, #12100046, Thermo Fisher Scientific) supplemented with 10% FBS (#F4135, Sigma-Aldrich) at 37 °C and 5% CO2. Cells were passaged by scraping (#08100240, Thermo Fisher Scientific) every 4–5 days. Cells were seeded at 250,000 cells/mL in 6-well plates (#353046, Corning, Corning, NY) for gene expression experiments. Cells were seeded and grown overnight in DMEM with 10% FBS, and this media was removed and replaced with DMEM without FBS immediately prior to addition of NPs. Cells were incubated with TiO2 NPs (250 μg/mL) or a PBS vehicle control in serum-free media for 24 h.
Following NP or PBS exposure, cells were processed to extract RNA using the RNeasy Plus Micro Kit (#74034, Qiagen, Hilden Germany). After RNA extraction, RNA content was quantified by Nano Drop (Thermo Fisher Scientific). cDNA H Minus Synthesis Master Mix (#M1681, Thermo Fisher Scientific) was used to generate cDNA. Power Up SYBR Green Master Mix (#A25743, Thermo Fisher Scientific) was used for RT-PCR and reactions were run in QuantStudio 6 (Applied Biosystems, Waltham, MA). The following primer sequences were used: IL-6 Forward TTGGTCCTTAGCCACTCCTTC, IL-6 Reverse TAGTCCTTCCTACCCCAATTTCC; TNF-α Forward CTATGTCTCAGCCTCTTCTC, TNF-α Reverse CATTTGGGAACTTCTCATCC; Cxcl2(MIP-2) Forward GGGTTGACTTCAAGAACATC, CXCL2(MIP-2) Reverse CCTTGCCTTTGTTCAGTATC; 18s Forward TTGACGGAAGGGCACCACCAG, 18s Reverse GCACCACCACCCACGGAATCG. Data collected by QuantStudio 6 was analyzed in Prism (v. 9.5.1, GraphPad Software, San Diego, CA). Cycle threshold (CT) values were normalized to housekeeping gene (18S). Data was expressed as a fold change compared to the control treated groups.
Results and Discussion
NP characterization
The TiO2 NPs used in this study have a primary diameter of ~21 nm, but are observed as fused aggregates by electron microscopy and DLS (Fig. S4).51, 52 The hydrodynamic diameter and zeta potential were measured without sonication (Table 1), to avoid possible disruption of the protein corona.
Table 1.
TiO2 NP hydrodynamic diameter (dh), polydispersity index (PDI), and zeta potential (ZP) in the absence and presence of a protein corona.
| NP and corona | dh (nm) | PDI | ZP (mV) | ΔZP |
|---|---|---|---|---|
|
| ||||
| TiO 2 | 900 ± 410 | 0.40 ± 0.14 | −43 ± 4 | - |
| FBS-TiO 2 | 900 ± 120 | 0.41 ± 0.01 | −23 ± 2 | +20 |
| BSA-TiO 2 | 800 ± 40 | 0.50 ± 0.06 | −32 ± 2 | +11 |
| BALF-TiO 2 | 1200 ± 150 | 0.26 ± 0.08 | −38 ± 3 | +5 |
BALF forms a protein corona on the surface of TiO2 NPs
Protein coronas were formed by incubating TiO2 NPs for 30 minutes at RT with three different protein sources: FBS (6 mg/mL, equivalent to a 10% v/v FBS solution), BSA (1.6 mg/mL), and BALF (0.017 mg/mL). FBS (10%) is a common nutrient source for cell lines, making it relevant to in vitro studies. Murine BALF obtained from mice by lavage was used to model the protein corona formed following inhalation. Albumin is the most abundant protein in both FBS and BALF. The concentration of BSA was chosen to be comparable to the amount of BSA that is found in FBS.30, 53 Unbound proteins were removed from the protein-TiO2 NP suspensions by centrifugation (18,000 rcf, 15 minutes) and resuspension (3x), as described previously (Fig. S3).51, 52, 54, 55
Hydrodynamic diameter and zeta potential of the protein-TiO2 NP complexes were measured using DLS (Table 1). There was no significant change in the diameter of the NPs with the addition of the corona. Previous work examined the coronas formed from porcine BALF on 8 different metal oxide NPs and found that BALF did not lead to a disruption of the TiO2 NP aggregates.36 The zeta potential increased significantly from the bare NPs (−35 ± 3 mV) to the NPs with FBS corona (−23 ± 2 mV; p < 0.05), in agreement with previous studies by our lab in which a FBS corona on TiO2 NPs was observed to have a zeta potential of (−24 ± 2).51, 54 In comparison, there was no significant change in zeta potential with the formation of a BSA (−32 ± 2 mV) or BALF (−38 ± 2 mV) corona. The differences in zeta potential are correlated with the protein corona concentration, as shown using BSA as a representative protein (Fig. S5). In addition, previous work has shown the change in zeta potential is related to the initial charge of the NP and the specific proteins that form the corona.56
The concentration of protein present in the corona is measured by BCA analysis and reported as protein (μg)/NPs (mg), as described in the Materials and Methods (Fig. 1A). The differences in protein corona concentrations (FBS = 53 ± 1.3 μg/mg NP; BSA = 20 ± 1 μg/mg NP, BALF = 1.9 ± 0.3 μg/mg NP) is, at least in part, due to the concentration of the initial protein solution. Using BSA as a representative protein, we found that concentration of the protein corona scaled with the initial concentration of protein used to form the corona (Fig. S6), in agreement with previous work that showed a positive correlation between protein corona concentration and initial protein concentration.30, 57–59 While this may seem intuitive, there are exceptions. For example, a direct comparison of silica NPs (200 nm) and polystyrene NPs (200 nm) incubated with 3%−80% human plasma (1 hr incubation) showed an increasing corona on the polystyrene NPs and a decreasing corona on the silica NPs at increasing plasma concentrations. 57 Previous work with porcine BALF showed relatively low corona concentrations on similar TiO2 NPs following incubation with BALF (7.4 mg/mL, 1 hr, RT).36
Fig. 1.
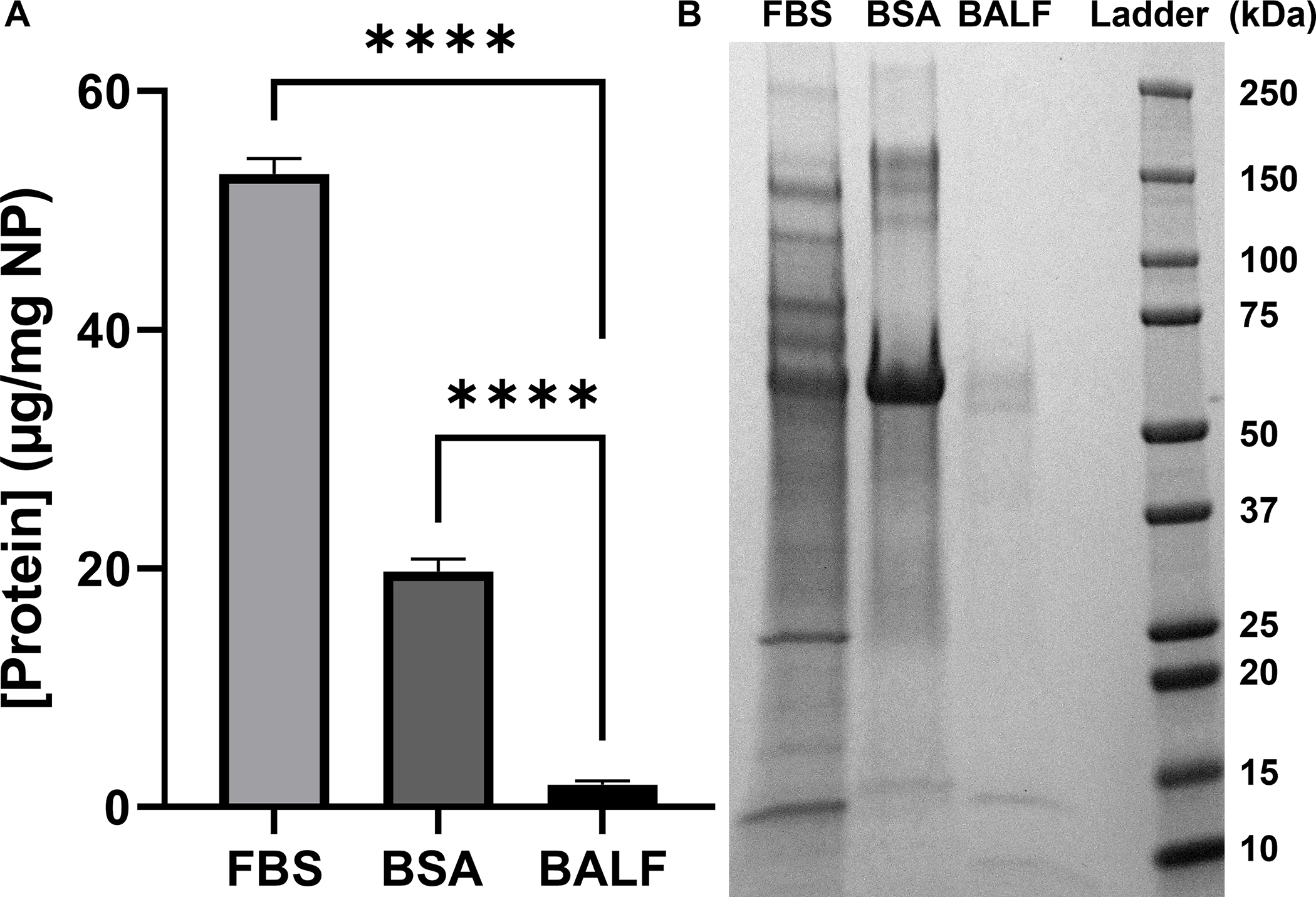
Concentration and composition of proteins adsorbed on TiO2 NPs incubated with FBS, BSA, and BALF. (A) Protein concentration per NP (μg/mg) when (FBS 6 mg/mL; BSA 1.6 mg/mL; BALF 0.017 mg/mL; n=3). The mean protein per NP (μg/mg) is reported with error bars showing standard deviation. Significance was calculated using an ANOVA with a post-hoc Tukey test. ****p<0.0001. (B) Gel electrophoresis of each protein corona.
Composition of the BALF corona
The composition of each protein corona was analyzed using gel electrophoresis (Fig. 1B) and proteomics (Table 1). Gel electrophoresis was in agreement with the protein corona concentration assay (Fig. 1A), with less protein present in the BSA and BALF coronas compared to FBS (Fig. 1B). The gel also shows that the most abundant protein in each of the coronas is albumin (66 kDa). Bands in the BALF corona at ~13 kDa and ~8 kDa were tentatively assigned to uteroglobin (10 kDa) and the monomer of pulmonary surfactant-associated protein B (SPB; 8.7 kDa) based on molecular weight.60
Proteomic analysis of the BALF corona is shown as the relative amount of protein detected (normalized abundance; Table 2) and the amount of protein in the corona relative to the amount in BALF (enrichment, Fig. 2). In general, the normalized abundance shows that the top 15 proteins in a solution of BALF are also present in the corona formed from BALF. For example, albumin is the most abundant protein in the corona formed from BALF (78 ± 2.6%) and in BALF (88.4 ± 4.8%). In comparison, the abundance of uteroglobin in the corona suggests selective adsorption on the TiO2 NP surface. Uteroglobin is the 8th most abundant protein in the corona (0.6 ± 0.3%), while it is the 24th most abundant protein in BALF (0.04 ± 0.07%). Uteroglobin, also known as blastokinin and club-cell secretory protein (CCSP), is an immunomodulatory, anti-inflammatory, and anti-chemotaxis secretoglobin expressed by epithelial cells that interact with external environments. 61
Table 2.
Normalized abundance (%) of the top 15 most abundant corona proteins (n=3). For comparison, the rank order of proteins present in BALF is shown in parentheses. Mean and standard deviation are reported.
| Protein | Protein Corona (%) | BALF (%) |
|---|---|---|
|
| ||
| Albumin | 78 ± 2.6 | 88.4 ± 4.8 (1) |
| Serotransferrin | 6 ± 3.2 | 3.4 ± 1.8 (2) |
| Hemoglobin subunit beta-1 | 4.2 ± 0.7 | 2.2 ± 1.7 (3) |
| Alpha-1-antitrypsin 1–1 | 2 ± 1.2 | 0.9 ± 0.5 (5) |
| Hemoglobin subunit alpha | 2 ± 1.2 | 0.4 ± 0.6 (8) |
| Serine protease inhibitor A3K | 1.0 ± 0.6 | 1.1 ± 0.6 (4) |
| Lysozyme C-2 | 0.7 ± 0.6 | 0.7 ± 0.4 (7) |
| Uteroglobin | 0.6 ± 0.3 | 0.04 ± 0.07 (24) |
| Haptoglobin | 0.6 ± 0.3 | 0.1 ± 0.1 (12) |
| Transthyretin | 0.6 ± 0.2 | 0.08 ± 0.09 (14) |
| Alpha-1-antitrypsin 1–5 | 0.5 ± 0.8 | 0.4 ± 0.2 (10) |
| Hemopexin | 0.5 ± 0.1 | 0.2 ± 0.1 (11) |
| Chitinase-like protein 3 | 0.47 ± 0.05 | 0.7 ± 0.6 (6) |
| Alpha-1-antitrypsin 1–4 | 0.4 ± 0.1 | 0.4 ± 0.2 (9) |
| Peroxiredoxin-6 | 0.2 ± 0.2 | 0.1 ± 0.1 (15) |
Fig. 2.
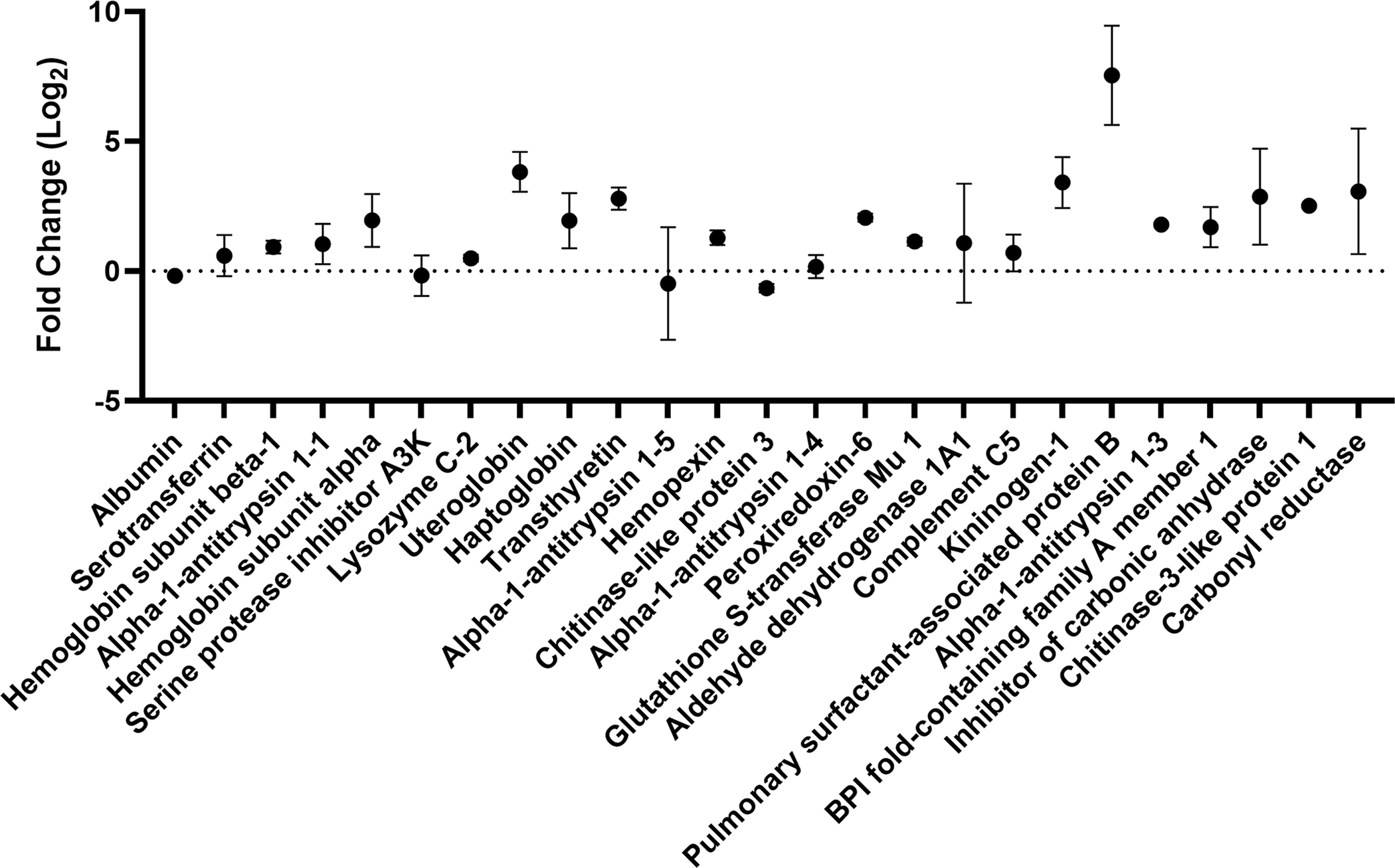
Enrichment (positive fold change) and depletion (negative fold change) of the 25 most abundant proteins present in the BALF protein corona present on TiO2 NPs relative to their abundance in BALF. Fold change, log base 2, with error bars showing standard deviation is plotted for each protein (n=3, n=2 for lysozyme C-2 and peroxiredoxin-6 with outliers removed). Proteins are listed in order of their relative abundance (Table 2).
Enrichment and depletion of a protein in the corona compared to the solution used to form the corona can be visualized in an enrichment plot (Fig. 2). In the enrichment plot, the fold change from serum to corona is displayed as log2 so that an enrichment of 0 is no change, while an enrichment of 1 is a 2-fold enrichment on the corona. Likewise, an enrichment of −1 is a 2-fold depletion in abundance on the corona. This type of plot can be used to determine which proteins are outliers, suggesting they are enriched or depleted in the protein corona. For example, albumin is slightly depleted (−0.18 ± 0.05) in the corona relative to BALF. Of the top 25 most abundant proteins in the protein corona, only SPB is an outlier, as determined by the robust regression and outlier removal (ROUT) method (Q = 0.1%). SPB is the most enriched protein within the top 25 proteins suggesting that it has high affinity for the TiO2 NP surface relative to other proteins in BALF. Previous proteomics experiments with TiO2 NPs (14 nm) incubated with BALF obtained from a human with pulmonary alveolar proteinosis showed binding of SPA, B, and D.39 SPB is a hydrophobic apolipoprotein that is essential for lung function.62 The observed enrichment of SPB is interesting considering molecular dynamics studies that showed anionic NPs were selective for SPB, compared to the similar lung surfactant protein C (SPC) due to a combination of hydrophobics and electrostatics. 63 Although not an outlier, the next most highly enriched protein is uteroglobin (3.8 ± 0.8, corona abundance rank 8). Overall, this enrichment and depletion profile (Fig. 2) shows that there is a selection of specific proteins in the NP corona. Several of these proteins (serotransferrin, uteroglobin, haptoglobin, hemopexin, and chitinase-like protein 3) have immunoregulatory properties suggesting that these proteins adsorbed on the surface of the TiO2 NPs could regulate the inflammation associated with TiO2 lung toxicity in vivo.64–67
BALF corona leads to increased expression of inflammatory cytokines in macrophage cells
To first determine if bare TiO2 NPs elicit an inflammatory response in macrophage cells, cells were incubated with TiO2 NPs for 24 hours. To prevent the formation of a protein corona in situ, serum-free media was used for the incubation. Following the incubation with TiO2 NPs, pro-inflammatory cytokine gene expression (IL-6, TNF-α, and MIP-2) was measured by real time PCR. We observed that bare TiO2 NPs significantly increased the expression of these pro-inflammatory cytokines (Fig. 3). This observation is in agreement with previous work showing TiO2 NPs increased pro-inflammatory cytokines associated with lung inflammation.14, 68, 69
Fig. 3.
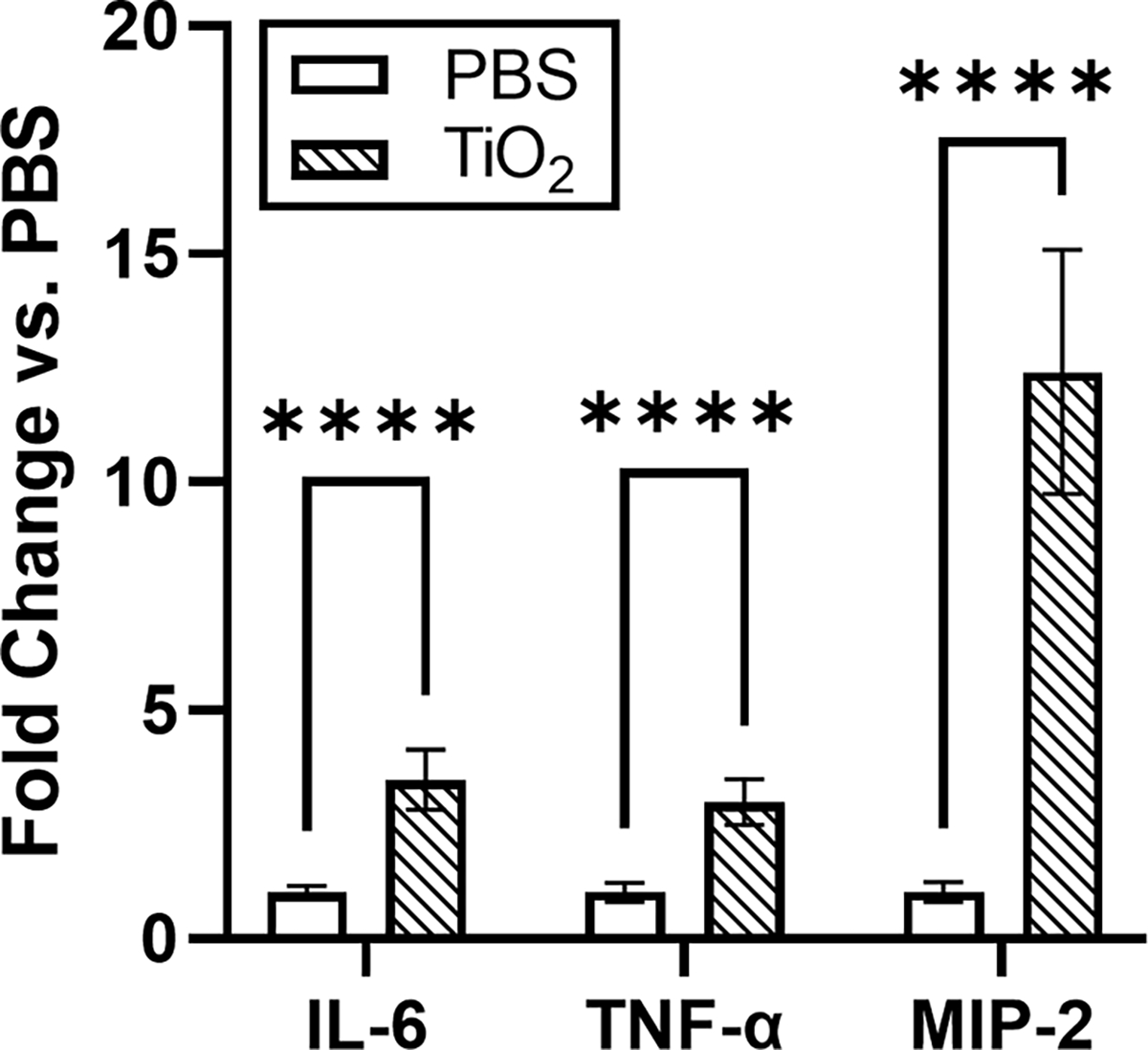
Expression of the pro-inflammatory genes IL-6, TNF-α, and MIP-2 increased in response to bare TiO2 NPs compared to untreated control cells (n=6). Expression change was found to be significant for each gene using unpaired t-tests. **** p<0.0001.
To determine if a BALF corona altered the cellular response to the TiO2 NPs, cells were incubated with BALF-TiO2 NPs and cytokine expression was compared to the response to bare TiO2 NPs (Fig. 3). FBS and BSA coronas were used for comparison as the predominant protein in BALF is albumin. The formation of the protein coronas is described in Materials and Methods. We observed that exposure of macrophage cells with BALF-TiO2 NPs increased the expression of pro-inflammatory cytokines (IL-6, TNF-α and MIP-2; Fig. 4). In comparison, coronas formed from FBS and BSA did not alter the expression of pro-inflammatory cytokines when compared to bare TiO2 NPs. These results suggest that a BALF corona is unique in increasing macrophage expression of pro-inflammatory cytokines and that proteins enriched in the BALF corona (Table 2 and Fig. 2) may drive the enhanced toxicity of these NPs.
Fig. 4.
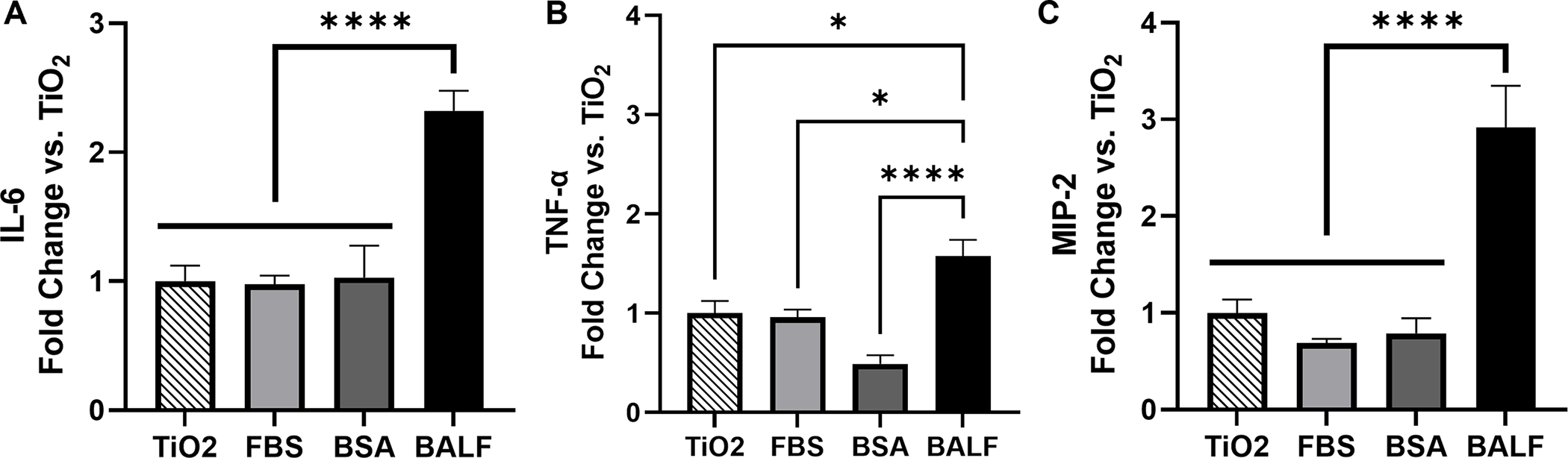
(A) IL-6, (B) TNF-α, and (C) MIP-2 showed elevated expression levels in response to BALF-TiO2 NP compared to FBS- and BSA-TiO2 NPs (n=6). Cytokine expression is reported as fold change relative to bare TiO2 NPs. Significance was determined using a one-way ANOVA with a post hoc Tukey test. Comparisons to the PBS control were also performed (Fig. S7). *p<0.05, ****p<0.0001.
Conclusion
Our studies describe the interaction of TiO2 NPs with BALF and the subsequent response of macrophage cells. We propose that these interactions are critical initial steps in the pulmonary response following the inhalation of these NPs. We find that the proteins present in BALF form a corona on the surface of the TiO2 NPs (Fig. 1). The composition of this corona (Table 1 and Fig. 2) results in a distinct protein-TiO2 NP complex that is associated with an elevated cytokine response in macrophage cells. The incubation of macrophage cells with bare TiO2 NPs leads to increased expression of pro-inflammatory genes, IL-6, TNF-α, and MIP-2 (Fig. 3). A BALF corona further enhances this pro-inflammatory response (Fig. 4). In comparison to FBS and BSA coronas, the BALF corona leads to a much greater response for all three cytokines. It is possible that BALF-specific proteins, other than albumin, are responsible for this increase in cytokine expression either through protein-dependent responses, an increased uptake of NPs, or a combination of both. For example, previous work has measured the uptake of polystyrene NPs (50 nm and 100 nm) by human alveolar lung cells (Type I and II).70 Pre-incubation of these polystyrene NPs with BALF led to increased cellular uptake by Type I cells. Type II cells did not internalize the polystyrene NPs. SPA and surfactant protein D (SPD) were identified on the surface of these polystyrene NPs by western blot. It is not known if SPB was probed. Similarly, work with magnetite NPs (110 nm - 180 nm) showed that adsorption of SPA on the surface of the NPs led to increased uptake by alveolar macrophage cells. 71 Other surfactant proteins were not examined. In comparison, BSA led to decreased uptake. In addition, previous work with polystyrene NPs (100 nm) showed that isolated SPA on the surface of the NPs led to increased uptake of the NPs by macrophage cells,37 suggesting differences in NP uptake could be responsible for the observed difference in cytokine response.
It is important to note the limitations associated with this study. One potential limitation of our studies is that our current work has not addressed the lipid corona that is expected to form following incubation of TiO2 NPs with BALF.72–75 Previous molecular dynamics simulations showed that adsorption of specific proteins (SPA, B, and C) on silver and polystyrene NPs (5 nm and 15 nm, anionic) was determined by the hydrophobicity of the NPs. The lipid composition was insensitive to the type of NP.76 Proteomics experiments with porcine surfactants were in agreement with these simulations (PEG-, PLGA-, and lipid-NPs (~200 nm)).38 We hope future work will probe the lipid composition on these TiO2 NPs. In addition, our experiments use protein coronas formed in situ (RT, 30 min) from isolated BALF. While FBS coronas are insensitive to time (30 min-120 min) and temperature (RT and 37 °C; Fig. S1), it is possible that the BALF corona formed in vivo would result in enrichment of different proteins. Previous experiments using polystyrene NPs with BALF obtained from a human with pulmonary alveolar proteinosis showed that the high abundance proteins present in the corona did not change after 15 min, but the lower abundance proteins varied over a 2 hr period.39 Future studies should address the role of these lower abundance proteins in the cellular response.
In addition to characterizing the BALF corona formed on TiO2 NPs and the resulting cytokine response, we hope these experiments will provide a starting point for future in vivo studies examining the mechanism of the toxicity associated with the inhalation of TiO2 NPs.77, 78 For example, if this enrichment of SPB on the surface of the TiO2 NPs leads to a corresponding depletion of free SPB in the lung there could be a reduced surface tension in the lungs associated with respiratory distress. 79 It is also possible that protein coronas could sequester specific lung lining fluid proteins required for normal host-defense and maintenance of lung homeostasis or the interaction of proteins with the NP surface could activate proteins to make them more pro-inflammatory. These questions will need to be addressed in future studies to define specific effects of the protein corona on the functions of these proteins. Beyond TiO2 NPs, previous work has examined changes in macrophage uptake and cytokine response for diesel exhaust NPs (132 nm) and carbon black NPs (154 nm) with BALF coronas and observed an increase in IL-8 release, suggesting that a BALF corona may be important for a wide range of environmental inhalation exposures.80
Supplementary Material
Environmental significance.
Nanoparticles are used on increasingly large scales in industrial materials and agriculture. These applications pose the risk of environmental exposures either directly, as in agriculture, during manufacturing, or following the degradation of nanoparticle-containing materials. Following this environmental release, humans are exposed to nanoparticles through inhalation. Our studies provide a detailed characterization of the interaction of titanium dioxide (TiO2) nanoparticles with bronchoalveolar lung fluid and the subsequent response of macrophage cells. We propose that the initial interaction of nanoparticles with lung fluid proteins is the critical first step that shapes the pulmonary response to nanoparticles. We hope this research will enable future work reducing the toxicity of nanoparticle exposures.
Acknowledgments
We thank the Duke University School of Medicine for the use of the Proteomics and Metabolomics shared resource, which provided proteomics services, with special thanks to Erik Soderblom and Greg Waitt for technical advice. We thank the National Institutes of Health (NIH 5R21-ES031191) and Duke MEDx for funding.
References
- 1.Mohajerani A, Burnett L, Smith JV, Kurmus H, Milas J, Arulrajah A, Horpibulsuk S and Abdul Kadir A, Nanoparticles in construction materials and other applications, and implications of nanoparticle use, Materials, 2019, 12, 3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giraldo JP and Kruss S, Nanosensors for monitoring plant health, Nat. Nanotechnol., 2023, 18, 107–108. [DOI] [PubMed] [Google Scholar]
- 3.Newkirk GM, de Allende P, Jinkerson RE and Giraldo JP, Nanotechnology approaches for chloroplast biotechnology advancements, Front. Plant Sci, 2021, 12, 691295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu L, Xu M, Wang R, Yin Y, Lynch I and Liu S, The crucial role of environmental coronas in determining the biological effects of engineered nanomaterials, Small, 2020, 16, e2003691. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann T, Lowry GV, Ghoshal S, Tufenkji N, Brambilla D, Dutcher JR, Gilbertson LM, Giraldo JP, Kinsella JM, Landry MP, Lovell W, Naccache R, Paret M, Pedersen JA, Unrine JM, White JC and Wilkinson KJ, Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture, Nat. Food, 2020, 1, 416–425. [Google Scholar]
- 6.Hanus MJ and Harris AT, Nanotechnology innovations for the construction industry, Prog. Mater. Sci, 2013, 58, 1056–1102. [Google Scholar]
- 7.Lee J, Mahendra S and Alvarez PJJ, Nanomaterials in the construction industry: A review of their applications and environmental health and safety considerations, ACS Nano, 2010, 4, 3580–3590. [DOI] [PubMed] [Google Scholar]
- 8.Wheeler KE, Chetwynd AJ, Fahy KM, Hong BS, Tochihuitl JA, Foster LA and Lynch I, Environmental dimensions of the protein corona, Nat. Nanotechnol, 2021, 16, 617–629. [DOI] [PubMed] [Google Scholar]
- 9.Zahra Z, Habib Z, Chung S and Badshah MA, Exposure route of TiO2 NPs from industrial applications to wastewater treatment and their impacts on the agro-environment, Nanomaterials, 2020, 10, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runa S, Hussey M and Payne CK, Nanoparticle–cell interactions: Relevance for public health, J. Phys. Chem. B, 2018, 122, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmudun Nabi M, Wang J, Erfani M, Goharian E and Baalousha M, Urban runoff drives titanium dioxide engineered particle concentrations in urban watersheds: Field measurements, Environ. Sci.: Nano, 2023, DOI: 10.1039/D2EN00826B. [DOI] [Google Scholar]
- 12.Current intelligence bulletin 63: Occupational exposure to titanium dioxide, U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institute for Occupational Safety and Health, 2011. [Google Scholar]
- 13.Sha B, Gao W, Cui X, Wang L and Xu F, The potential health challenges of TiO2 nanomaterials, J. Appl. Toxicol, 2015, 35, 1086–1101. [DOI] [PubMed] [Google Scholar]
- 14.Shi H, Magaye R, Castranova V and Zhao J, Titanium dioxide nanoparticles: A review of current toxicological data, Part. Fibre Toxicol, 2013, 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelclova D, Zdimal V, Fenclova Z, Vlckova S, Turci F, Corazzari I, Kacer P, Schwarz J, Zikova N, Makes O, Syslova K, Komarc M, Belacek J, Navratil T, Machajova M and Zakharov S, Markers of oxidative damage of nucleic acids and proteins among workers exposed to TiO2 (nano) particles, Occup. Environ. Med., 2016, 73, 110–118. [DOI] [PubMed] [Google Scholar]
- 16.Hussain S, Vanoirbeek JAJ and Hoet PHM, Interactions of nanomaterials with the immune system, Wiley Interdiscip. Rev.: Nanomed. and Nanobiotechnol., 2012, 4, 169–183. [DOI] [PubMed] [Google Scholar]
- 17.Weir A, Westerhoff P, Fabricius L, Hristovski K and von Goetz N, Titanium dioxide nanoparticles in food and personal care products, Environ. Sci. Technol, 2012, 46, 2242–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CFR Part 73 listing of color additives exempt from certification, Code of Federal Regulations, Washington DC, 2023. [Google Scholar]
- 19.Survey USG, Mineral commodity summaries 2022, Mineral Commodity Summaries, 2022, DOI: 10.3133/mcs2022. [DOI] [Google Scholar]
- 20.Payne CK, A protein corona primer for physical chemists, J. Chem. Phys, 2019, 151, 130901. [DOI] [PubMed] [Google Scholar]
- 21.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S and Dawson KA, The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century, Adv. Colloid Interface Sci, 2007, 134–135, 167–174. [DOI] [PubMed] [Google Scholar]
- 22.Walczyk D, Bombelli FB, Monopoli MP, Lynch I and Dawson KA, What the cell “sees” in bionanoscience, J. Am. Chem. Soc, 2010, 132, 5761–5768. [DOI] [PubMed] [Google Scholar]
- 23.Tomak A, Cesmeli S, Hanoglu BD, Winkler D and Oksel Karakus C, Nanoparticle-protein corona complex: Understanding multiple interactions between environmental factors, corona formation, and biological activity, Nanotoxicology, 2021, 15, 1331–1357. [DOI] [PubMed] [Google Scholar]
- 24.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DW, Cohen Y, Emili A and Chan WC, Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles, ACS Nano, 2014, 8, 2439–2455. [DOI] [PubMed] [Google Scholar]
- 25.Monopoli MP, Aberg C, Salvati A and Dawson KA, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol, 2012, 7, 779–786. [DOI] [PubMed] [Google Scholar]
- 26.Ke PC, Lin S, Parak WJ, Davis TP and Caruso F, A decade of the protein corona, ACS Nano, 2017, 11, 11773–11776. [DOI] [PubMed] [Google Scholar]
- 27.Kobos L and Shannahan J, Biocorona-induced modifications in engineered nanomaterial-cellular interactions impacting biomedical applications, Wiley Interdiscip. Rev.: Nanomed. and Nanobiotechnol., 2020, 12, e1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamad-Schifferli K, Exploiting the novel properties of protein coronas: Emerging applications in nanomedicine, Nanomedicine (Lond), 2015, 10, 1663–1674. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Hood ED, Nong J, Ding J, Marcos-Contreras OA, Glassman PM, Rubey KM, Zaleski M, Espy CL, Gullipali D, Miwa T, Muzykantov VR, Song WC, Myerson JW and Brenner JS, Combating complement’s deleterious effects on nanomedicine by conjugating complement regulatory proteins to nanoparticles, Adv. Mater, 2022, 34, e2107070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulsen KM and Payne CK, Concentration and composition of the protein corona as a function of incubation time and serum concentration: An automated approach to the protein corona, Anal. Bioanal. Chem, 2022, 414, 7265–7275. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen KM, Pho T, Champion JA and Payne CK, Automation and low-cost proteomics for characterization of the protein corona: Experimental methods for big data, Anal. Bioanal. Chem, 2020, 412, 6543–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleischer CC and Payne CK, Nanoparticle-cell interactions: Molecular structure of the protein corona and cellular outcomes, Acc. Chem. Res, 2014, 47, 2651–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogan MP, Geraghty P, Greene CM, O’Neill SJ, Taggart CC and McElvaney NG, Antimicrobial proteins and polypeptides in pulmonary innate defence, Respir Res, 2006, 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitsett JA and Alenghat T, Respiratory epithelial cells orchestrate pulmonary innate immunity, Nat. Immunol, 2015, 16, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Numata M and Voelker DR, Anti-inflammatory and anti-viral actions of anionic pulmonary surfactant phospholipids, Biochim. Biophys. Acta - Mol. Cell. Biol. Lipids, 2022, 1867, 159139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulze C, Schaefer UF, Ruge CA, Wohlleben W and Lehr C-M, Interaction of metal oxide nanoparticles with lung surfactant protein a, Eur. J. Pharm. Biopharm, 2011, 77, 376–383. [DOI] [PubMed] [Google Scholar]
- 37.McKenzie Z, Kendall M, Mackay RM, Whitwell H, Elgy C, Ding P, Mahajan S, Morgan C, Griffiths M, Clark H and Madsen J, Surfactant protein a (SP-A) inhibits agglomeration and macrophage uptake of toxic amine modified nanoparticles, Nanotoxicology, 2015, 9, 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raesch SS, Tenzer S, Storck W, Rurainski A, Selzer D, Ruge CA, Perez-Gil J, Schaefer UF and Lehr C-M, Proteomic and lipidomic analysis of nanoparticle corona upon contact with lung surfactant reveals differences in protein, but not lipid composition, ACS Nano, 2015, 9, 11872–11885. [DOI] [PubMed] [Google Scholar]
- 39.Whitwell H, Mackay RM, Elgy C, Morgan C, Griffiths M, Clark H, Skipp P and Madsen J, Nanoparticles in the lung and their protein corona: The few proteins that count, Nanotoxicology, 2016, 10, 1385–1394. [DOI] [PubMed] [Google Scholar]
- 40.Arango Duque G and Descoteaux A, Macrophage cytokines: Involvement in immunity and infectious diseases, Front. Immunol, 2014, 5, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussain S, Johnson CG, Sciurba J, Meng X, Stober VP, Liu C, Cyphert-Daly JM, Bulek K, Qian W, Solis A, Sakamachi Y, Trempus CS, Aloor JJ, Gowdy KM, Foster WM, Hollingsworth JW, Tighe RM, Li X, Fessler MB and Garantziotis S, TLR5 participates in the TLR4 receptor complex and promotes myd88-dependent signaling in environmental lung injury, Elife, 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tighe RM, Birukova A, Yaeger MJ, Reece SW and Gowdy KM, Euthanasia- and lavage-mediated effects on bronchoalveolar measures of lung injury and inflammation, Am. J. Respir. Cell Mol. Biol, 2018, 59, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonner James C, Silva Rona M, Taylor Alexia J, Brown Jared M, Hilderbrand Susana C, Castranova V, Porter D, Elder A, Oberdörster G, Harkema Jack R, Bramble Lori A, Kavanagh Terrance J, Botta D, Nel A and Pinkerton Kent E, Interlaboratory evaluation of rodent pulmonary responses to engineered nanomaterials: The niehs nano go consortium, Environ. Health Perspect, 2013, 121, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia T, Hamilton Raymond F, Bonner James C, Crandall Edward D, Elder A, Fazlollahi F, Girtsman Teri A, Kim K, Mitra S, Ntim Susana A, Orr G, Tagmount M, Taylor Alexia J, Telesca D, Tolic A, Vulpe Christopher D, Walker Andrea J, Wang X, Witzmann Frank A, Wu N, Xie Y, Zink Jeffery I, Nel A and Holian A, Interlaboratory evaluation of in vitro cytotoxicity and inflammatory responses to engineered nanomaterials: The niehs nano go consortium, Environ. Health Perspect, 2013, 121, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tighe RM, Birukova A, Yaeger MJ, Reece SW and Gowdy KM, Euthanasia- and lavage-mediated effects on bronchoalveolar measures of lung injury and inflammation, Am. J. Respir. Cell Mol. Biol, 2018, 59, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J and Mann M, Maxquant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol, 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 47.Tyanova S, Temu T and Cox J, The maxquant computational platform for mass spectrometry-based shotgun proteomics, Nat. Protoc, 2016, 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- 48.Crap protein sequences, https://www.thegpm.org/crap/index.html, 2012.
- 49.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M and Cox J, The perseus computational platform for comprehensive analysis of (prote)omics data, Nat. Methods, 2016, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A and Vizcaíno JA, The pride database and related tools and resources in 2019: Improving support for quantification data, Nucleic Acids Res, 2019, 47, D442–d450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jayaram DT and Payne CK, Intracellular generation of superoxide by TiO2 nanoparticles decreases histone deacetylase 9 (hdac9), an epigenetic modifier, Bioconjugate Chem, 2020, 31, 1354–1361. [DOI] [PubMed] [Google Scholar]
- 52.Jayaram DT, Runa S, Kemp ML and Payne CK, Nanoparticle-induced oxidation of corona proteins initiates an oxidative stress response in cells, Nanoscale, 2017, 9, 7595–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martel J, Young D, Young A, Wu C-Y, Chen C-D, Yu J-S and Young JD, Comprehensive proteomic analysis of mineral nanoparticles derived from human body fluids and analyzed by liquid chromatography–tandem mass spectrometry, Anal. Biochem, 2011, 418, 111–125. [DOI] [PubMed] [Google Scholar]
- 54.Runa S, Khanal D, Kemp ML and Payne CK, TiO2 nanoparticles alter the expression of peroxiredoxin antioxidant genes, J. Phys. Chem. C, 2016, 120, 20736–20742. [Google Scholar]
- 55.Runa S, Lakadamyali M, Kemp ML and Payne CK, TiO2 nanoparticle-induced oxidation of the plasma membrane: Importance of the protein corona, J. Phys. Chem. B, 2017, 121, 8619–8625. [DOI] [PubMed] [Google Scholar]
- 56.Fleischer CC and Payne CK, Nanoparticle surface charge mediates the cellular receptors used by protein-nanoparticle complexes, J. Phys. Chem. B, 2012, 116, 8901–8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB and Dawson KA, Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles, J. Am. Chem. Soc, 2011, 133, 2525–2534. [DOI] [PubMed] [Google Scholar]
- 58.Partikel K, Korte R, Mulac D, Humpf HU and Langer K, Serum type and concentration both affect the protein-corona composition of plga nanoparticles, Beilstein J. Nanotechnol., 2019, 10, 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grafe C, Weidner A, Luhe MV, Bergemann C, Schacher FH, Clement JH and Dutz S, Intentional formation of a protein corona on nanoparticles: Serum concentration affects protein corona mass, surface charge, and nanoparticle-cell interaction, Int. J. Biochem. Cell Biol, 2016, 75, 196–202. [DOI] [PubMed] [Google Scholar]
- 60.Courtney Broaddus RJMV, Ernst Joel D., King Talmadge E. Jr, Lazarus Stephen C., Murray John F., Nadel Jay A., Arthur Slutsky, Murray and nadeĺs textbook of respiratory medicine 2 / ed.-in-chief: V. Courtney broaddus, Elsevier Saunders, Philadelphia, Pa., 6. ed edn., 2016. [Google Scholar]
- 61.Mukherjee AB, Zhang Z and Chilton BS, Uteroglobin: A steroid-inducible immunomodulatory protein that founded the secretoglobin superfamily, Endocr. Rev, 2007, 28, 707–725. [DOI] [PubMed] [Google Scholar]
- 62.Ryan MA, Qi X, Serrano AG, Ikegami M, Perez-Gil J, Johansson J and Weaver TE, Mapping and analysis of the lytic and fusogenic domains of surfactant protein B, Biochemistry, 2005, 44, 861–872. [DOI] [PubMed] [Google Scholar]
- 63.Hu G, Jiao B, Shi X, Valle RP, Fan Q and Zuo YY, Physicochemical properties of nanoparticles regulate translocation across pulmonary surfactant monolayer and formation of lipoprotein corona, ACS Nano, 2013, 7, 10525–10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandal AK, Zhang Z, Ray R, Choi MS, Chowdhury B, Pattabiraman N and Mukherjee AB, Uteroglobin represses allergen-induced inflammatory response by blocking pgd2 receptor-mediated functions, J. Exp. Med, 2004, 199, 1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janicova A, Becker N, Xu B, Wutzler S, Vollrath JT, Hildebrand F, Ehnert S, Marzi I, Störmann P and Relja B, Endogenous uteroglobin as intrinsic anti-inflammatory signal modulates monocyte and macrophage subsets distribution upon sepsis induced lung injury, Front. Immunol, 2019, 10, 2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao T, Su Z, Li Y, Zhang X and You Q, Chitinase-3 like-protein-1 function and its role in diseases, Signal Transduct. Target. Ther, 2020, 5, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin T, Sammy F, Yang H, Thundivalappil S, Hellman J, Tracey KJ and Warren HS, Identification of hemopexin as an anti-inflammatory factor that inhibits synergy of hemoglobin with hmgb1 in sterile and infectious inflammation, J. Immunol, 2012, 189, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. L. K, Moon C, Lee HS, Lee HW, Park EM, Kim HS and Castranova, Comparison of the biological activity between ultrafine and fine titanium dioxide particles in raw 264.7 cells associated with oxidative stress, J. Toxicol. Environ. Health Part A, 2008, 71. [DOI] [PubMed] [Google Scholar]
- 69.Miller TJ, Knapton A, Adeyemo OO, Noory LS, Weaver JL, Hanig JP, Honchel R, Zhang J, Espandiari P, Benedick MF, Umbreit TH, Tomazic-Jezic VJ and Sadrieh N, Toxicology of titanium dioxide (TiO2) nanoparticles: In vitro and in vivo evaluation of macrophage uptake of TiO2, The FASEB Journal, 2007, 21, A812–A812. [Google Scholar]
- 70.Thorley AJ, Ruenraroengsak P, Potter TE and Tetley TD, Critical determinants of uptake and translocation of nanoparticles by the human pulmonary alveolar epithelium, ACS Nano, 2014, 8, 11778–11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruge CA, Kirch J, Cañadas O, Schneider M, Perez-Gil J, Schaefer UF, Casals C and Lehr C-M, Uptake of nanoparticles by alveolar macrophages is triggered by surfactant protein a, Nanomedicine, 2011, 7, 690–693. [DOI] [PubMed] [Google Scholar]
- 72.Olenick LL, Troiano JM, Vartanian A, Melby ES, Mensch AC, Zhang L, Hong J, Mesele O, Qiu T, Bozich J, Lohse S, Zhang X, Kuech TR, Millevolte A, Gunsolus I, McGeachy AC, Doğangün M, Li T, Hu D, Walter SR, Mohaimani A, Schmoldt A, Torelli MD, Hurley KR, Dalluge J, Chong G, Feng ZV, Haynes CL, Hamers RJ, Pedersen JA, Cui Q, Hernandez R, Klaper R, Orr G, Murphy CJ and Geiger FM, Lipid corona formation from nanoparticle interactions with bilayers, Chem., 2018, 4, 2709–2723. [Google Scholar]
- 73.Zhang X, Pandiakumar AK, Hamers RJ and Murphy CJ, Quantification of lipid corona formation on colloidal nanoparticles from lipid vesicles, Anal. Chem, 2018, 90, 14387–14394. [DOI] [PubMed] [Google Scholar]
- 74.L. B. G., Capriotti AL, Caracciolo G, Cavaliere C, Cerrato A, Montone CM, Piovesana S, Pozzi D, Quagliarini E and Laganà, A comprehensive analysis of liposomal biomolecular corona upon human plasma incubation: The evolution towards the lipid corona, Talanta, 2020, 209. [DOI] [PubMed] [Google Scholar]
- 75.L. T., Bernfur K, Vilanova M and Cedervall, Understanding the lipid and protein corona formation on different sized polymeric nanoparticles, Sci. Rep, 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Q, Bai X, Hu G and Zuo YY, Unveiling the molecular structure of pulmonary surfactant corona on nanoparticles, ACS Nano, 2017, 11, 6832–6842. [DOI] [PubMed] [Google Scholar]
- 77.Gakis GP, Aviziotis IG and Charitidis CA, Metal and metal oxide nanoparticle toxicity: Moving towards a more holistic structure–activity approach, Environ. Sci.: Nano, 2023, DOI: 10.1039/d2en00897a. [DOI] [PubMed] [Google Scholar]
- 78.Ma Y, Shi J, Zhang Y, Chen Z and Jia G, Titanium dioxide nanoparticles altered the lncrna expression profile in human lung cells, Int. J. Environ. Res. Public Health, 2023, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tredano M, van Elburg RM, Kaspers AG, Zimmermann LJ, Houdayer C, Aymard P, Hull WM, Whitsett JA, Elion J, Griese M and Bahuau M, Compound SFTPB 1549C-->GAA (121ins2) and 457delc heterozygosity in severe congenital lung disease and surfactant protein b (SP-B) deficiency, Hum. Mutat, 1999, 14, 502–509. [DOI] [PubMed] [Google Scholar]
- 80.Shaw CA, Mortimer GM, Deng ZJ, Carter ES, Connell SP, Miller MR, Duffin R, Newby DE, Hadoke PW and Minchin RF, Protein corona formation in bronchoalveolar fluid enhances diesel exhaust nanoparticle uptake and pro-inflammatory responses in macrophages, Nanotoxicology, 2016, 10, 981–991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


