ABSTRACT
Microbial assemblages inhabiting soils sheltered by rocks are widespread in the hyperarid and hypersaline Dalangtan Playa, which could contribute significantly to the ecological processes in this desert ecosystem. Here, we performed geochemical and metagenomic analyses of soils below and beside rocks in the Dalangtan Playa. Fourier transform infrared (FTIR) spectra show more evident signals of organic aromatic molecules in soils below rocks than in nearby bare soils. Soils below rocks harbor distinct microbial communities and much higher biomass and Cyanobacteria abundance in comparison to soils beside rocks. High-quality genomes recovered by hybrid short- and long-read assembly reveal the previously underappreciated contribution of Actinobacteriota in sheltered soils to the primary production of desert ecosystems. In contrast to bare soil communities, the metabolic versatility of sheltered soil communities indicates their critical roles in carbon, nitrogen, and sulfur cycling in the Dalangtan Playa. Trace gases (CO and H2) could be significant energy sources for the respiration and sometimes carbon fixation of both bare and sheltered soil microorganisms in nutrient-deprived environments. The presence of multiple stress response genes against DNA damage, oxidative stress, and osmotic stress, as well as pathways for the biosynthesis and uptake of compatible solutes in genomes across all samples, reveals their capacity for tolerating polyextreme conditions.
IMPORTANCE
The hyperarid Dalangtan Playa in the western Qaidam Basin, northwestern China, is a unique terrestrial analog of Mars. Despite the polyextreme environments of this area, habitats below translucent rocks capable of environmental buffering could serve as refuges for microbial life. In this study, the hybrid assembly of Illumina short reads and Nanopore long reads recovered high-quality and high-continuity genomes, allowing for high-accuracy analysis and a deeper understanding of extremophiles in the sheltered soils of the Dalangtan Playa. Our findings reveal self-supporting and metabolically versatile sheltered soil communities adapted to a hyperarid and hypersaline playa, which provides implications for the search for life signals on Mars.
KEYWORDS: hyperaridity, sheltered soil, metagenome, hybrid assembly, desert ecosystems, adaptive strategies
INTRODUCTION
Deserts or drylands comprise the most extensive terrestrial biomes (1), and microorganisms account for the majority of bioprocesses in desert ecosystems due to the paucity of animals and plants (2). Deserts with polyextreme conditions, such as the well-known Atacama Desert (3, 4) and Antarctic Dry Valleys (5 – 7), provide optimal models for studying the limits of Earth’s life, which by extension could serve as terrestrial analogs to extraterrestrial environments (e.g., Mars) (8, 9). Thus, investigations of the distribution, diversity, and adaptive strategies of extremophiles in extremely arid deserts are fascinating, as they provide unique astrobiological insights into the search for terrestrial-like life beyond Earth (10).
Desert pavements are widespread in Mars-analogous extremely arid areas, and the translucent rocks embedded in regolith commonly serve as refuges for life due to their environmental buffering in coping with ultraviolet (UV) radiation, moisture, and thermal stress (2). The rock substrates contribute to the colonization of lithic microbial communities, which can be categorized into epiliths, endoliths, and hypoliths. Lithic microbial colonization is ubiquitous and frequently reported in worldwide hot and cold deserts, including the Atacama Desert (11 – 14), the Namib Desert (15, 16), the Antarctic Dry Valleys (17 – 21), and the Qinghai-Tibet Plateau (22 – 25). They are often highly specialized communities dominated by Cyanobacteria, acting as important primary producers to support diverse heterotrophic bacteria and archaea (26 – 30). These self-sustaining communities are valuable biomass resources and play critical roles in biogeochemical cycles in oligotrophic deserts (2, 27). However, the majority of previous studies have focused on conspicuous lithic communities, while soil microbial communities sheltered by rocks are still largely unexplored. A previous study on soil microbiomes below the boulders of the Atacama Desert reported abundant ammonia-oxidizing Thaumarchaeota and their potential in nitrogen and carbon cycling (31), indicating the significant yet overlooked roles of sheltered soil microorganisms besides lithic communities in hyperarid desert ecosystems.
The Qaidam Basin in the northern Qinghai-Tibet Plateau is one of the highest (average elevation: ~2,800 m) and driest (average annual precipitation: <45 mm) deserts on Earth with low temperature (average annual temperature: 1–5°C) and high UV radiation (32, 33). The Qaidam Basin is a unique and exemplary terrestrial analog of the Late Noachian-Early Hesperian Mars due to its extreme environments, aqueously altered setting, widespread evaporites, and Mars-like geomorphologies (33 – 35). The regolith microbial communities in Qaidam, dominated by the bacterial phyla Actinobacteria and Proteobacteria, were found to potentially scavenge trace gases (H2 and CO) to acquire energy and develop protein protection and DNA repair mechanisms to cope with harsh conditions (36). The Dalangtan Playa located in the northwest of the Qaidam Basin is one of the most arid regions in the basin (arid index: 0.01–0.05) (37). Sulfate-rich sediments, (per)chlorates, and halites are widespread in the Dalangtan Playa (37, 38). The hyperaridity, high salinity, intense UV radiation, and deprivation of nutrients give rise to the exceptional polyextreme environment of the Dalangtan Playa. To date, only some halotolerant microorganisms, mainly Firmicutes and Actinobacteria, have been isolated and cultured from the Dalangtan Playa sediments (39), while the diversity, metabolic potential, and adaptive strategies of Dalangtan microbial communities remain poorly understood.
Here, we report the diversity and metabolic capabilities of soil microbial communities sheltered by translucent rocks in the Dalangtan Playa and compare them with nearby soil communities beside the rocks. We combined short- and long-read metagenomic assembly to obtain near-complete, high-quality metagenome-assembled genomes (MAGs) of these soil microorganisms and decipher their metabolic and biogeochemical functional trait profiles. In addition to distinct community structures and microbial biogeochemical functions between sheltered and bare soils, stress response genes involved in microbial adaptive strategies to environmental stressors (e.g., radiation, osmotic pressure, and starvation) were further compared. Overall, we unveil the underappreciated but important roles of sheltered soil microbial communities below the rocks, especially Actinobacteriota, in Dalangtan desert ecosystems, which further provides implications for the search for life signals on Mars.
MATERIALS AND METHODS
Sampling procedures and geochemical and mineralogical analyses
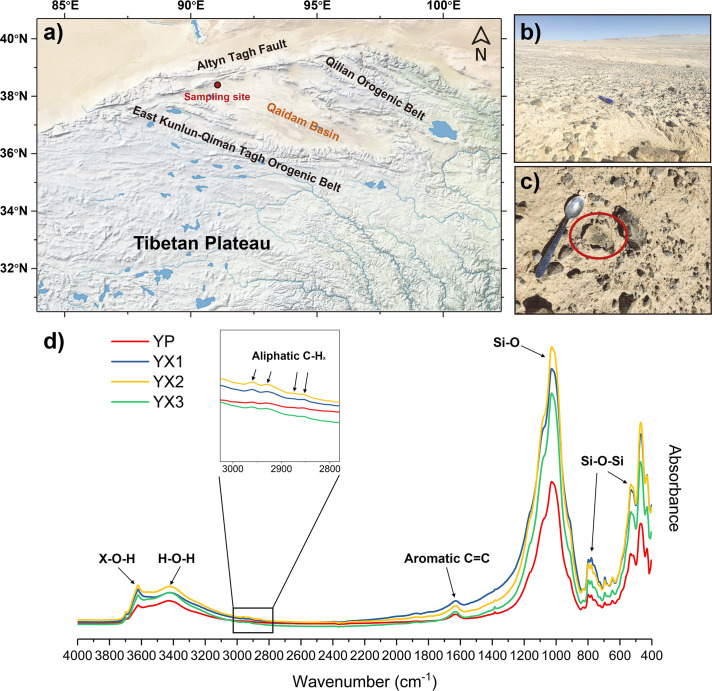
Sheltered and bare soil samples were all collected on 29 June 2021 from a pavement in the Dalangtan Playa of the western Qaidam Basin, China (38°35′44″N, 90°59′6″E) at an elevation of 3,245 m (Fig. 1a). Three sampling sites ~10 m away from each other were chosen to collect soils below (YX) and beside (YP) rocks. Within each sampling field (~2-m radius), subsurface soils (~5-cm depth) under five rocks with visible microbial assemblages were collected and then mixed up (Fig. 1b and c). Due to the low biomass in soils beside rocks, bare soil controls were collected from different sites and mixed up in one sample in order to obtain enough samples for DNA extraction. Finally, four samples, including three soil samples (YX1, YX2, and YX3) below rocks and one bare soil sample (YP) beside rocks, were collected aseptically and stored in sterile 50-mL centrifuge tubes. All samples were immediately delivered to the laboratory with ice packs and stored at −80°C until further processing.
Fig 1.
Location of sampling sites and geochemical characteristics of soils. (a) Sampling site in the Dalangtan Play, western Qaidam Basin (Made with Natural Earth). (b) Image of the desert pavement for sampling. (c) Soil samples with visible green microbial assemblages under translucent rocks. (d) FTIR spectra in the range of 400–4,000 cm−1 of bare soils (YP) beside rocks and soils below rocks (YX1-3). Bands in the region of 2,800–3,000 cm−1 denote different vibrational modes from aliphatic C-H bonds, and peaks at ~1,600 cm−1 denote the vibrations of unsaturated C=C bonds in aromatic materials.
Before geochemical and mineralogical analyses, soils were first pulverized in an agate ball mill cleaned with 75% ethanol before each sample. X-ray diffraction (XRD) analysis was conducted using a D8 Discovery (Bruker, MA, USA) at the Beijing Kuangyan Geoanalysis Laboratory Co. Ltd. with a CuKα (1.5406 Å) radiation source. The XRD patterns were scanned between 2° (2θ) and 70° (2θ) with a step size of 0.02° (2θ) and a scan speed of 0.08° (2θ)/s (40 mA, 40 kV). For other analyses, the powdered samples were then decarbonated with 3 M HCl to remove inorganic carbon and washed with double-distilled H2O. The total organic carbon (TOC) concentrations of these decarbonated soil powders were measured by an Elemental Analyzer ECS 4024 (NC Technologies, Milan, Italy) at the Qingdao Institute of Marine Geology, China Geological Survey, with sediment IVA33802150 as the reference material. A Vertex 70V Fourier transform infrared (FTIR) spectrometer (Bruker, Germany) at the Institute of Geology and Geophysics, Chinese Academy of Sciences, was used to characterize organic matter in soils. After drying at 100°C overnight, decarbonated sample powders were mixed with KBr (dried at 110°C to remove water) and then ground in a mortar to prepare pressed pellets for FTIR measurements. A pure KBr pellet was measured as the background for calibration. Mid-infrared regions in the range of 400–4,000 cm−1 wavenumber were generated with 64 scans per sample and a resolution of 4 cm−1.
Metagenomic DNA extraction and sequencing
Metagenomic DNA was extracted from soil samples with a modified procedure as described previously (36). Briefly, 30 g of soil was ground with liquid nitrogen in a sterilized mortar. Cell lysis and DNA purification were conducted using a PowerMax Soil DNA Isolation Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The supernatant was combined with high-concentration salt solution C4 and filtered through the same silica filter membrane. After the elution step, DNA was precipitated with absolute ethanol, sodium acetate, and glycogen and then washed with 70% ethanol. DNA concentration was measured by a Qubit DNA Assay Kit in a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, USA). The DNA library preparations and Illumina and Nanopore sequencing were conducted at Novogene Bioinformatics Technology Co., Ltd. (Novogene, Beijing, China). All samples were sequenced on the Illumina NovaSeq platform, and 150-bp paired-end reads were generated. However, considering the low DNA yields of bare soil samples (Table 1), only sheltered soil samples were further sequenced using Oxford Nanopore Technologies (ONT).
TABLE 1.
Dalangtan Playa soil sample information and microbial diversity
| Soil type | TOC (%) a | DNA (ng/g) b | ASVs c | Evenness | Shannon | |
|---|---|---|---|---|---|---|
| YX1 | Sheltered | 0.028 | 214 | 1,161 | 0.58 | 5.9 |
| YX2 | Sheltered | 0.063 | 346 | 1,681 | 0.65 | 6.9 |
| YX3 | Sheltered | 0.098 | 388 | 1,513 | 0.58 | 6.2 |
| YP | Bare | 0.022 | 48 | 1,039 | 0.62 | 6.2 |
Total organic carbon (wt%).
Metagenomic DNA yield per gram soil.
Amplicon sequence variants (ASVs) generated by QIIME 2 Deblur.
16S rRNA gene amplification, sequencing, and analysis
Approximately 10 ng of DNA was used for 16S rRNA gene amplification using primers 515F (GTGYCAGCMGCCGCGGTAA) and 806R (GGACTACNVGGGTWTCTAAT) (40) through an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s, and a final extension at 72°C for 10 min. PCR products were monitored by 1% agarose gel electrophoresis and purified by a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). Recovered products were sequenced on an Illumina NovaSeq 6000 platform with 250-bp paired-end reads at Novogene Bioinformatics Technology Co., Ltd. (Novogene, Beijing, China). In order to compare the microbial diversity between sheltered soils and bare soils, three previously investigated bare soil samples (samples 0707, 0711, and 0712) collected from other areas in the northwestern Qaidam Basin were included in the 16S rRNA gene-based analysis (36). QIIME 2 v.2021.11 (41) was used to analyze taxonomic classification. Sequences were trimmed and denoised using the plugin Deblur (42). The representative taxonomic identities were aligned at 99% full-length sequence similarity with pre-trained “Silva 138 99% OTUs from 515F/806R region of sequences” databases using the plugin “feature classifier” (43). Principal coordinate analysis (PCoA) plots of unweighted UniFrac and weighted UniFrac were also generated using QIIME 2.
Metagenomic assembly and binning
Hybrid metagenomic assembly and binning using short- and long-read data from sheltered soil samples were performed with the combination of MetaWRAP (44) and OPERA-MS (45). First, Illumina reads were quality-controlled using MetaWRAP’s “read_qc” module. Short-read assembly was conducted using metaSPAdes (46), and assembled scaffolds ≤2,000 bp were filtered out. Then, short-read assembled results along with short-read Illumina reads and long-read Nanopore reads were set as inputs for hybrid assembly using OPERA-MS with the “no-polishing” option. The binning of hybrid assembled contigs was performed using MetaWRAP’s “Binning” module with MetaBAT2 (47), MaxBin2 (48), and CONCOCT (49). The results of binning were refined using the “Bin_refinement” module. CheckM (50) was used to evaluate the quality of bins, and only high-quality bins (completeness – 5 × contamination > 50%) were kept for further analysis. For the bare soil sample (only Illumina short-read data were obtained), metagenomic assembly and binning were performed using MetaWRAP without the hybrid assembly step.
Phylogenetic analysis
Taxonomic classification of 89 MAGs was assigned using GTDB-Tk (v1.5.1, database release R04-RS89) with the “classify_wf” function (51). Then, we selected reference genomes from the GTDB database (52) based on the taxonomic classification of each genome. Only “GTDB species representatives” were retained. In total, 336 reference genomes were selected and then downloaded from the NCBI database. Phylogenomic analysis of all MAGs was performed using PhyloPhlAn 3.0 (version 3.0.58) (53). The configuration “supermatrix_aa.cfg” and the “phylophlan” database with 400 universal marker genes were used. The maximum likelihood phylogenomic tree of genomes was constructed by RAxML version 8 (54). Reference genomes from the archaeal phylum Thermoproteota were used as an outgroup. Additionally, in order to validate the phylogenomic analysis, we additionally constructed a maximum likelihood phylogenomic tree inferred from an aligned concatenated set of 120 single-copy marker proteins (51). Reference genomes from the bacterial phylum Bacillota were used as an outgroup, and the tree was built using IQ-TREE (v1.6.9) (55) with 1,000 ultrafast bootstraps under the LG+F+R10 model.
Maximum likelihood trees were constructed for group 1 [NiFe]-hydrogenase large subunits (groups 1a, 1b, 1c, 1d, 1e, 1f, 1h, and 1l) and form I carbon monoxide dehydrogenases (CoxL). MAGs from Antarctic Desert soils (n = 451) (56) and other soils across the world (n = 756) (57) were selected as reference genomes because these soil microbial communities have been shown to be capable of consuming atmospheric CO and H2. Amino acid sequences of proteins were retrieved from all MAGs (n = 1,296) using hidden Markov models (HMMs) (58) in METABOLIC software (59). The preliminary classification of group 1 [NiFe]-hydrogenase large subunits was performed using HydDB (60). Protein sequences were aligned using ClustalW in MEGA 11 (61). Maximum likelihood trees were constructed using IQ-TREE (v1.6.9) (55) with 1,000 ultrafast bootstraps under the LG+R9 and LG+F+R10 models for group 1 [NiFe]-hydrogenase large subunits and CoxL, respectively. All trees were visualized and modified using Interactive Tree Of Life (iTOL) v6 (62).
Annotation of metagenomes and MAGs
METABOLIC v4.0 (59) was used to predict the metabolic and functional potential of genomes and metagenomes. Metabolic capabilities of genomes and community-scale functional networks based on the “MW-score” (metabolic weight score) were generated. Compatible solute biosynthesis pathways or transport systems (i.e., ectoine, trehalose, and glycine-betaine) were searched against the “KEGGModuleHit” in METABOLIC output files. Representative proteins involved in the stress response to DNA damage [UvrABC (63), UmuCD (64), MutSL (65), RecA (66), and RadA (67)], osmotic stress (TrkA) (68), oxidative stress (SodF) (69), starvation (Dps) (70), and multiple stresses (SigB) (71) were selected for subsequent analysis. These genes were searched manually through HMMs (58) across all MAGs.
RESULTS AND DISCUSSION
Characterization of sheltered and bare soil samples in the Mars analog Dalangtan Playa
We found abundant rock accumulations on the top of a hill in the Dalangtan Playa (Fig. 1a). Visible microbial assemblages were commonly observed under the translucent rocks, especially quartz stones, embedded in regolith compared to the seemingly lifeless bare soils (Fig. 1b and c). Hypolithic communities beneath rocks have been previously reported in northwestern China’s hyperarid deserts (22, 24, 25). As expected, soil samples sheltered by rocks had a much higher concentration of extracted metagenomic DNA (>200 ng/g) than the bare soil sample (<50 ng/g) (Table 1). Relatively higher TOC concentrations were found in sheltered soils than in bare soils. The range of TOC concentrations (0.022%–0.098%) is broadly consistent with records from previous studies in the hyperarid Qaidam Basin (36, 72). The mineral assemblages of sheltered and bare soils were generally similar and all dominated by silicates, including quartz, chlorite, albite, and illite (Table S1). Calcite, a carbonate mineral, was also abundant in both types of soils. Dolomite (carbonate), orthoclase (silicate), and gypsum (sulfate) were only observed in sheltered soil samples. Interestingly, dolomite was scarce in the YP sample of this study and other surface samples previously reported in the Qaidam Basin (72) but abundant in sheltered soil samples with higher biomass.
FTIR can trace organic matter by distinguishing specific bands in the IR spectra induced by bond vibrations of functional groups in organic materials (73 – 75). In this study, soil organic matter can be inferred from the FTIR spectra of all sheltered and bare soils (Fig. 1d). Except for the distinct IR bands at 900–1,200 cm−1, 430–800 cm−1, and 3,400 cm−1 recording bond vibrations of functional groups from siliceous minerals, all FTIR spectra exhibited bands in the regions of 2,800–3,000 cm−1 (the vibrational modes of aliphatic C-H bonds) and ~1,600 cm−1 (the vibrations of unsaturated C=C bonds in aromatic materials) (76, 77). Additionally, we found more evident peaks of aromatic molecules in sheltered soil samples than in the bare soil sample.
Actinobacteriota-dominated soil microbial communities and distinct taxonomic compositions between sheltered and bare soils
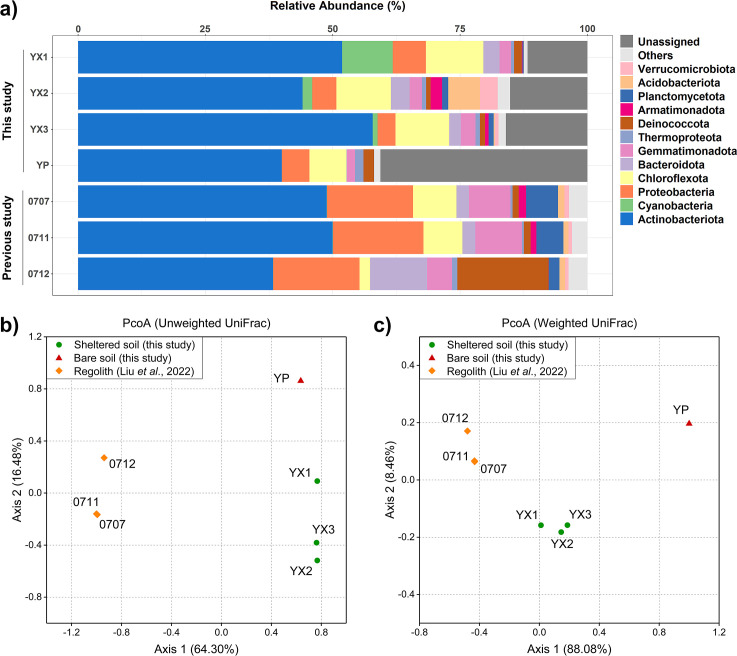
According to 16S rRNA gene sequencing and analysis of four soil samples, we obtained a total of 2,712 amplicon sequence variants (ASVs). Sheltered soils showed a slightly higher number of features than bare soils, while microbial community alpha diversity (Shannon index) and evenness were similar across samples, with no significant difference between sample types (Table 1). Similar microbial evenness and diversity across soils below and beside boulders were also detected in the Atacama Desert (31). All soil microbial communities were dominated by the bacterial phylum Actinobacteriota (39.97%–57.84%), followed by Chloroflexota (7.30%–11.29%) and Proteobacteria (3.52%–6.51%), which is similar to our previously investigated bare regolith samples (samples 0707, 0711, and 0712) collected from different areas in the northwestern Qaidam Basin (Fig. 2a) (36). These microbes, especially Actinobacteriota, are also dominant and prevalent in other hyperarid desert soils (78 – 81) across the world, implying their potentially vital roles in desert ecosystems.
Fig 2.
Microbial communities of sheltered soil and bare soil samples from the Qaidam Basin. (a) Relative abundance of dominated taxa in soil microbial communities according to 16S rRNA gene sequencing. Bare regolith samples 0707, 0711, and 0712 without shields have been previously investigated (36), which were collected in different areas from YX and YP samples in the northwestern Qaidam Basin. (b and c) Principal coordinate analysis (PCoA) ordination plots of all samples based on unweighted (b) and weighted (c) UniFrac.
However, distinct microbial community compositions were found between sheltered (YX) and bare (YP) samples. The bare soil sample had a much higher relative abundance of unassigned features (40.65%) than sheltered soil samples (11.75%–16.02%) (Fig. 2a), suggesting a higher percentage of unclassified microorganisms in Dalangtan air-exposed soils. We identified 16 bacterial phyla and one archaeal phylum (Thermoproteota) from four soil samples. All 17 phyla were detected in sheltered soils except for the phylum Patescibacteria not found in the YX1 sample, while only 14 phyla were detected in the bare soil sample. Notably, sheltered soil samples (0.94%–9.93%) contained a significantly higher abundance of Cyanobacteria than the bare soil samples YP, 0707, 0711, and 0712 (<0.01%) (Fig. 2a). Consistent with the diversity results, PCoA ordination plots based on unweighted UniFrac and weighted UniFrac showed that sheltered soils and bare soils have notably different microbial community compositions (Fig. 2b and c). Given that bare soil samples 0707, 0711, and 0712 were collected from different regions, they also show different microbial communities from YP and YX according to PCoA plots.
High-quality novel genomes recovered from hybrid and short-read metagenomic assembly
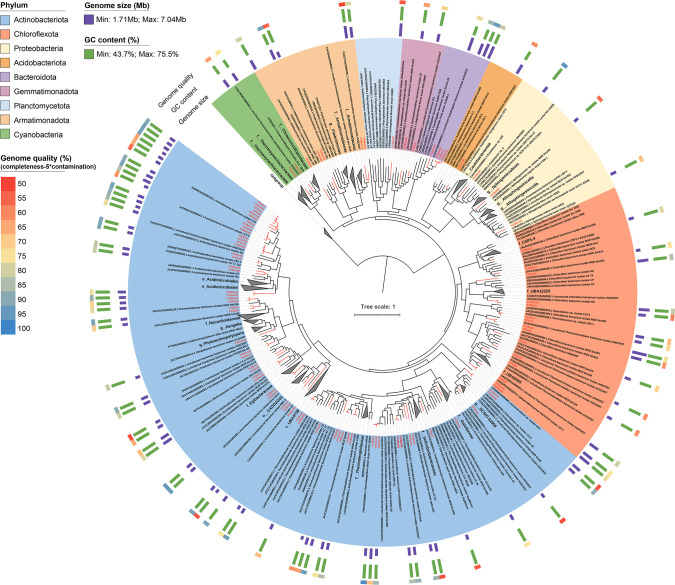
In contrast to the limitations of conventional assembly methods, hybrid assembly combining the accurate but fragmented short reads and error-prone long reads enables the acquirement of much higher-quality and even complete genomes (45, 82 – 84) from metagenomes independent of pure culture methods. In this study, we recovered a total of 112 draft genomes (completeness ≥50% and contamination ≤10%), among which 89 with genome quality ≥50 (defined as completeness – 5 × contamination) (85) were retained and 46 were above the high-quality threshold (completeness ≥90% and contamination ≤5%) (86) (Fig. 3; Table S2). In general, the YX MAGs (YX1, n = 12; YX2, n = 33; and YX3, n = 31) generated by the hybrid assembly of Illumina and Nanopore reads have significantly higher continuity (far fewer gaps) compared to YP MAGs (n = 13) assembled only by short-read Illumina reads. The 89 retained MAGs contained a high average GC content (67.8%), and nearly half of the MAGs (n = 43) had a GC content ≥70%. The genome size of MAGs ranged from 1.71 (YX2bin5, Solirubrobacterales) to 7.04 Mb (YX1bin8, Thermomicrobiales). These high-quality MAGs enable high-accuracy analysis and further understanding of extremophilic microorganisms in the Dalangtan Playa.
Fig 3.
Phylogenomic tree of 89 retained MAGs (genome quality ≥50) and 336 reference genomes using 400 optimized universal marker genes. MAGs reconstructed from this study are marked with red lines. Reference genomes were selected from the GTDB database (GTDB species representatives only).
The GTDB taxonomic classification is congruent with the phylogenomic analysis (Fig. 3; Fig. S1; Table S2); therefore, the GTDB taxonomy is used for genomic classification throughout. According to the GTDB taxonomy, 96.63% of MAGs (n = 83) were unclassified at the species level (57.3% at the genus level, n = 51) against the GTDB database, revealing that a significant amount of microbial “dark matter” (novel MAGs) was detected. The 89 retained MAGs belonged to nine bacterial phyla, including Actinobacteriota (n = 56), Chloroflexota (n = 15), Armatimonadota (n = 5), Bacteroidota (n = 4), Proteobacteria (n = 3), Cyanobacteria (n = 2), Gemmatimonadota (n = 2), Acidobacteriota (n = 1), and Planctomycetota (n = 1) (Fig. 3). In YP samples, only MAGs belonging to the phyla Actinobacteriota (n = 6), Chloroflexota (n = 6), and Gemmatimonadota (n = 1) were recovered. YX1, YX2, and YX3 samples identified four, seven, and seven phyla, respectively. These bacterial phyla were also detected in our previous research on the Qaidam Basin regolith (36). Two Cyanobacteria MAGs (YX2bin18 and YX3bin19) affiliated with the families Chroococcidiopsidaceae and Thermosynechococcaceae, respectively, were found in sheltered soil samples. Except for one genome (YX3bin35) that was unclassified at the order level, the remaining MAGs were distributed into 25 orders, mainly including Acidimicrobiales (n = 20), Solirubrobacterales (n = 16), Thermomicrobiales (n = 8), Euzebyales (n = 4), Propionibacteriales (n = 4), CADDZG01 (n = 4), and Cytophagales (n = 4). Among them, only six orders were identified in YP samples, which were dominated by Thermomicrobiales and Acidimicrobiales, while more taxa at the order level were found in YX samples (8 in YX1, 18 in YX2, and 16 in YX3) (Fig. 3; Table S2).
Diverse metabolic capabilities of microbial communities sheltered by rocks and their potential significant roles in biogeochemical processes
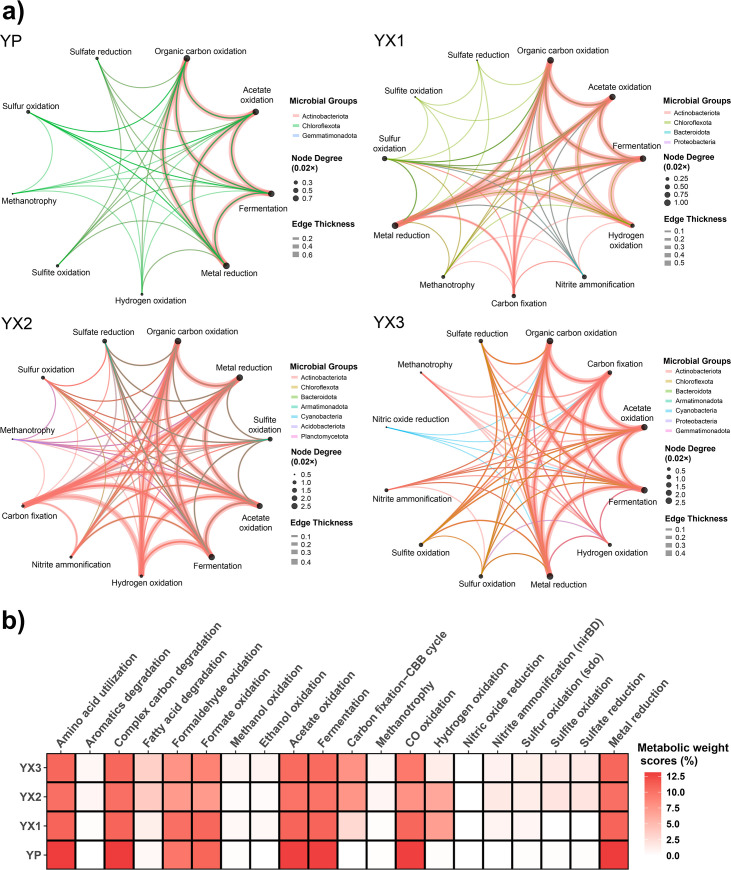
Metabolic and biogeochemical functional trait profile analysis of metagenomes and genomes revealed the potential for organic carbon oxidation, C1 metabolism, acetate oxidation, fermentation, methanotrophy, and metal reduction across all samples (Fig. 4a and b; Table S3). Notably, we also found diverse metabolic capabilities of carbon fixation, hydrogen oxidation, sulfur metabolism, and nitrogen metabolism in microbial communities sheltered below rocks. Compared to bare soil communities, microbial communities with the shelter of rocks had more complex functional networks and developed local self-supporting micro-ecosystems (Fig. 4).
Fig 4.
Metabolic and biogeochemical functional trait profile analysis of metagenomes and genomes. (a) Microbial community functional network of MAGs from the bare soil sample YP and three sheltered soil samples (YX1-3). Nodes represent different biogeochemical processes, and the size of the node was depicted according to the number of connections. Edges connecting two nodes represent functional connections, and edge thickness was determined by the average gene coverage values of the two nodes. Edge colors represent different taxa contributing to biogeochemical processes. (b) Weights of different metabolic functions in four metagenomes. The metabolic weight score (MW-score) was calculated by METABOLIC v4.0 based on gene coverage, which reflects function weights in metagenomes.
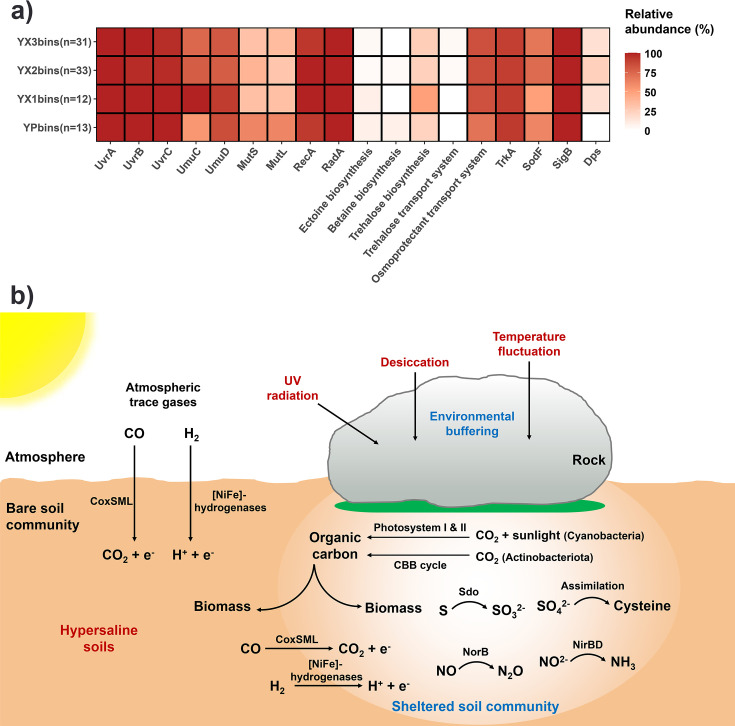
We identified the potential for carbon fixation in sheltered soil communities but not in bare soil communities (Fig. 4a and b). Carbon fixation pathways via the Calvin-Benson-Bassham (CBB) cycle (Form I RuBisCO) were found in Actinobacteriota and Cyanobacteria from YX. Photosynthesis key genes in photosystems I and II (e.g., psaA and psbA) were found in Cyanobacteria genomes. These primary producers could make sheltered soil microbial communities self-sustainable. In oligotrophic deserts that are typical of arid regions, Cyanobacteria are conventionally thought to serve as major primary producers to support other heterotrophs (2, 5). The availability of liquid water is essential for the diversity and primary production of photosynthetic life in deserts (22, 30). Transient hydration events are critical moisture sources to support cyanobacterial phototrophic activity in deserts (87, 88). Carbon fixation by Cyanobacteria in deserts was reported to occur only when local humidity increased, such as fog formation, morning dewfall, and occasional precipitation events (11, 89, 90). We thus argue that the stable environmental buffering and moisture retention provided by rock substrates in the Dalangtan Playa could facilitate Cyanobacteria to use photosystems and the CBB cycle to transduce energy and fix CO2 to organic carbon.
In addition, our (meta)genomic analyses reveal that diverse microbes that dominate in Dalangtan sheltered and bare soils have the potential to use multiple energy sources, including diffusible trace gases (CO and H2) from the atmosphere or deeper anaerobic soil zones, to support aerobic respiration (Fig. 4a and b). Overall, the H2 oxidation pathway was present in all YX samples while restricted to Actinobacteriota, Chloroflexota, and Proteobacteria. Although not abundant in the YP metagenome, group 1 [NiFe]-hydrogenase genes were detected in two MAGs (Thermomicrobiales and Acidimicrobiales) of YP. Abundant genes encoding aerobic carbon monoxide dehydrogenases (coxSML) for CO oxidation were also detected across all samples but restricted within the phyla Actinobacteriota, Chloroflexota, and Gemmatimonadota. It should be noted that the pathway of CO oxidation is more widespread in microbial communities than H2 oxidation according to the metagenomic functional analysis (Fig. 4b).
Besides Cyanobacteria, we highlight the previously underappreciated contribution of Actinobacteriota to chemosynthetic primary production in desert ecosystems (Fig. 4a and b). The high abundance of genes for high-affinity [NiFe]-hydrogenases (group 1) involved in hydrogen (H2) oxidation and carbon monoxide (CO) dehydrogenases (coxSML) in Actinobacteriota in sheltered soils suggests their potential capabilities to make use of atmospheric H2 and CO to support aerobic respiration and carbon fixation. Microbial atmospheric chemosynthesis is a globally widespread phenomenon and contributes greatly to carbon fixation in cold deserts, including deserts in the Qinghai-Tibet Plateau, Antarctica, and Arctic (91). Our results complement the findings by Bay et al. (88) that the primary production role of desert Actinobacteriota, the most abundant bacterial group in biocrusts and topsoils, has been long overlooked. Bay et al. argued that chemosynthetic primary production by Actinobacteriota is likely to be more important than expected in arid soils where photoautotrophs become increasingly sparse. It should be noted that although genes encoding [NiFe]-hydrogenases (group 1) and carbon monoxide dehydrogenases were identified in Actinobacteriota in Dalangtan sheltered soils, Actinobacteriota in bare soils presented no potential for carbon fixation according to our genomic analysis. Moreover, our previous study showed that only a low percentage of Actinobacteriota in the Qaidam Basin regolith harbor key genes for carbon fixation (36). These findings indicate that primary producers, whether chemosynthetic or photosynthetic bacteria, prefer to colonize specific locations with relatively moist and habitable conditions in deserts (as we called “micro-oases”), such as soils sheltered by rocks and wetter biocrusts. Collectively, (meta)genomic data suggest that chemosynthetic Actinobacteriota and photosynthetic Cyanobacteria in desert “micro-oases” both play important roles in contributing to the primary production of organic carbon to support nearby heterotrophic assemblages in desert ecosystems.
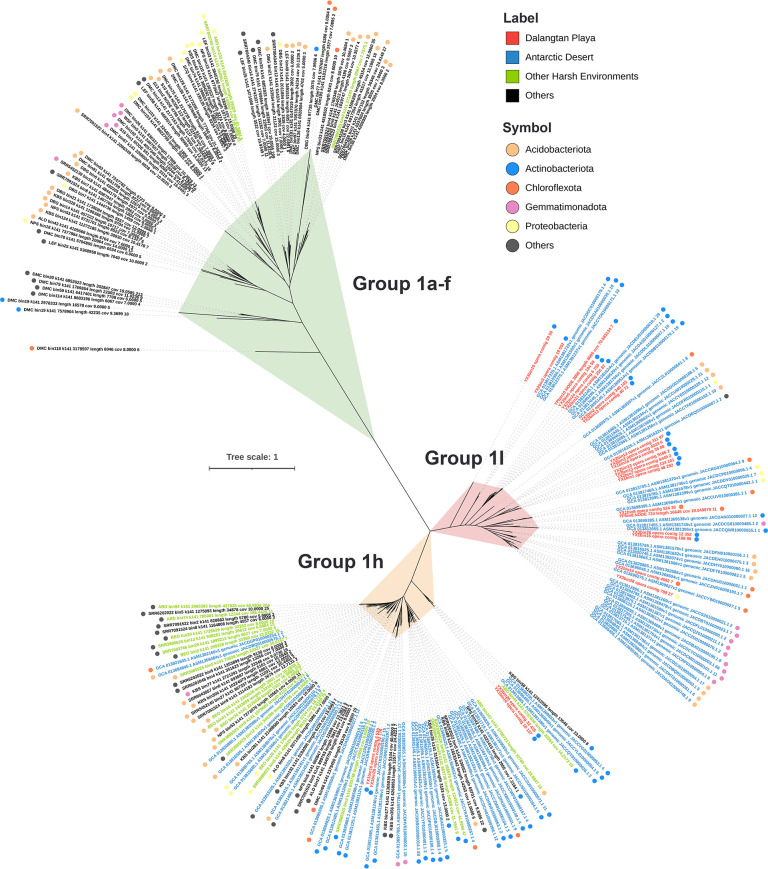
Trace gas consumption (in situ and ex situ) experiments have demonstrated that atmospheric H2 and CO could be scavenged by soil microbes across the world to support aerobic respiration and drive carbon fixation (56, 57, 91 – 94). Many studies have revealed that ubiquitous trace gases CO and H2 are critical to microbial survival and persistence in nutrient-deprived environments (56, 93 – 95), supporting microbial energy acquisition and growth in oligotrophic deserts, such as the Antarctic Dry Valleys (56, 92) and Atacama Desert (31, 96). In this study, we further combined our 89 MAGs with 1,207 MAGs in the studies mentioned above from various soils around the world (56, 57) to gain insight into the phylogenetic diversity of key enzymes involved in H2 and CO oxidation. Far more coxL genes (n = 1,018) than group 1 [NiFe]-hydrogenase genes (n = 216) were detected in 1,296 MAGs (Fig. 5; Fig. S2). In general, group 1 [NiFe]-hydrogenase and CoxL from Dalangtan microorganisms are phylogenetically close to those from the Antarctic Desert, which had experimental evidence to scavenge atmospheric trace gases H2 and CO (56) (Fig. 5; Fig. S2). Notably, the phylogenetic tree shows that group 1l and 1h [NiFe]-hydrogenases are more common in microbes from extreme environments, such as the Dalangtan Playa, the Antarctic Desert (56), and other drylands (57) (Fig. 5). The widespread distribution and predominance of group 1l and 1h [NiFe]-hydrogenases (HylL and HhyL) in extremophiles, particularly group 1l, suggest that these two types of hydrogenases may help cells cope with starvation in harsh conditions by scavenging ambient H2 (97). The CoxL tree shows a broad distribution of this gene in microbes across various environments (Fig. S2), which supports the view that trace gas CO is a widespread energy source for microbial survival (93). Our findings in the Dalangtan Playa consistently indicate that consuming ambient gases could be a widespread and significant survival mechanism for microbial communities in desert ecosystems.
Fig 5.
Maximum likelihood phylogenetic tree of group 1 [NiFe]-hydrogenases identified in 89 MAGs from this study (Dalangtan Playa) and in 1,207 MAGs from previous studies (Antarctic Desert and other soil samples across the world) (56, 57). The tree was constructed using IQ-TREE under the LG+R9 substitution model with 1,000 ultrafast bootstraps.
Representative genes involved in sulfur and nitrogen metabolism were also detected, including sulfur oxidation (sdo), sulfite oxidation and assimilatory sulfate reduction (sat, cysD, cysN, cysNC, and cysH), nitrite ammonification (nirBD), and nitric oxide reduction (norB), in YX metagenomes and MAGs (Fig. 4a and b; Table S3). The representative genes for sulfur metabolism in YX communities were mainly distributed in Actinobacteriota and were also present in Chloroflexota, Acidobacteriota, Bacteroidota, and Proteobacteria. The nirBD gene encoding nitrite reductase was distributed in Actinobacteriota, Chloroflexota, Bacteroidota, and Armatimonadota in YX. The norB gene encoding nitric oxide reductase was only found in Chloroflexota from the YX3 sample. In YP, no genes involved in nitrogen metabolism were detected, and only a small proportion of Chloroflexota and Actinobacteriota had the potential for sulfur metabolism. In line with our previous study on regolith microbiomes (36), we found that microorganisms in Qaidam bare soils have less potential for nitrogen metabolism, although microbial nitrogen cycling has been reported in many other deserts, including the Atacama Desert, the Antarctic Dry Valleys, and the Namib Desert (15, 20, 98). However, in this study, biogeochemical functional analysis reveals that sheltered soil microbial communities, especially Actinobacteriota, are likely important contributors to nitrogen and sulfur cycling in the Dalangtan Playa.
Adaptive strategies of microorganisms to the extreme environments of the Dalangtan Playa
Here, we identified various stress response genes involved in resistance to environmental stressors, such as intense radiation, osmotic stress, oxidative stress, and low nutrient content, across all samples (Fig. 6a). Almost all MAGs harbored different kinds of genes for DNA repair proteins UvrABC (63), MutSL (65), RadA (67), DNA recombinase RecA (66), and/or DNA polymerase UmuCD (64) (Fig. 6a). These DNA repair genes are responsible for the recovery of DNA damage induced by environmental pressure, helping microorganisms cope with intense UV radiation and desiccation (99 – 101). To respond to osmotic stress caused by a high-salt environment, “salt-in” (accumulating ions, mainly K+, in the cytoplasm to achieve osmotic equilibrium with the external environment) and “salt-out” (excluding salts and producing compatible solutes to prevent osmotic pressure) strategies are two common microbial adaptive mechanisms (102, 103). Here, we found different metabolic pathways for the biosynthesis and uptake of compatible solutes, including the biosynthesis of ectoine, betaine, and trehalose and the transport of trehalose and glycine-betaine according to KEGG module analysis (Table S4). Among these functions, glycine-betaine transport and trehalose biosynthesis were the most abundant pathways identified in YX and YP communities, while others were only present in ~4% or even less of total MAGs. In addition to utilizing compatible solutes to cope with osmotic pressure, trkA genes involved in the potassium (K+) uptake system were also present in over 90% of total MAGs. Altogether, soil microbes in the Dalangtan Playa could use salt-in and salt-out approaches to adapt to hypersaline environments. For other stress response genes, sigma factor sigB involved in stress adaptation (71) was present in all MAGs. Over 60% of total MAGs contained genes for superoxide dismutase SodF, which could protect cells from the hazards of reactive oxygen species (ROS) induced by desiccation and radiation (69). Genes encoding DNA starvation/stationary phase protection proteins (dps) that protect DNA from oxidative damage (70) were found in ~20% of YX MAGs. These stress response genes and metabolic versatility could play important roles in microbes below rocks to survive and adapt to the polyextreme environments in the Dalangtan Playa (Fig. 6b). Together with our findings of microbial colonization patterns below rocks and the identification of organic molecules in sheltered soils in the Dalangtan Playa, we argue that rocks are of vital value for life signal exploration on Mars.
Fig 6.
Adaptive strategies and predicted metabolic models of sheltered and bare soil communities in the Dalangtan Playa. (a) Stress response genes and pathways identified in 89 MAGs. The shade of color reflects the relative abundance of MAGs containing the genes. UvrABC, excinuclease ABC subunit; UmuCD, DNA polymerase; MutSL, DNA mismatch repair protein; RecA, recombinase; RadA, DNA repair protein; TrkA, Trk system potassium uptake protein; SodF, superoxide dismutase; SigB, RNA polymerase sigma factor; Dps, DNA starvation/stationary phase protection protein. (b) Predicted conceptual model of metabolic and biogeochemical functions of sheltered soil and bare soil communities based on metagenomes and metagenome-assembled genomes.
ACKNOWLEDGMENTS
We thank Wensi Zhang for his assistance in sample collection and Runjia Ji for her suggestions in genomic analysis. We also thank Xiaoguang Li for his help in the measurement of Fourier transform infrared spectra.
L.L. and W.L. conceived the project, designed the experiments, and wrote the manuscript with input from Y.C., J.S., and Y.P. W.L. supervised the study. L.L. collected the samples and conducted all experiments and data analyses. Y.C. assisted in the sample collection and the interpretation of spectral data.
This work was supported by the National Natural Science Foundation of China (NSFC) (T2225011), the Key Research Program of the Chinese Academy of Sciences (ZDBS-SSW-TLC001), and the Key Research Programs of the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS-201904 and IGGCAS-202102).
Contributor Information
Wei Lin, Email: weilin@mail.iggcas.ac.cn.
John R. Spear, Colorado School of Mines, Golden, Colorado, USA
DATA AVAILABILITY
Sequencing data from this study have been deposited under the NCBI BioProject PRJNA727938. The SRA accession numbers of Nanopore reads, Illumina reads, and 16S rRNA gene sequences are SRR24860171-SRR24860173, SRR24765285-SRR24765288, and SRR24860110-SRR24860113, respectively. The genome accession numbers of reconstructed MAGs are JAUJLY000000000-JAUJPI000000000.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01072-23.
Phylogenomic tree.
Phylogenetic tree of CoxL.
Four supplementary tables.
Supplemental figure legends.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Peel MC, Finlayson BL, McMahon TA. 2007. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci 11:1633–1644. doi: 10.5194/hess-11-1633-2007 [DOI] [Google Scholar]
- 2. Pointing SB, Belnap J. 2012. Microbial colonization and controls in dryland systems. Nat Rev Microbiol 10:551–562. doi: 10.1038/nrmicro2831 [DOI] [PubMed] [Google Scholar]
- 3. Azua-Bustos A, González-Silva C, Fairén AG. 2022. The atacama desert in northern chile as an analog model of Mars. Front Astron Space Sci 8:810426. doi: 10.3389/fspas.2021.810426 [DOI] [Google Scholar]
- 4. Navarro-González R, Rainey FA, Molina P, Bagaley DR, Hollen BJ, de la Rosa J, Small AM, Quinn RC, Grunthaner FJ, Cáceres L, Gomez-Silva B, McKay CP. 2003. Mars-like soils in the Atacama desert, Chile, and the dry limit of microbial life. Science 302:1018–1021. doi: 10.1126/science.1089143 [DOI] [PubMed] [Google Scholar]
- 5. Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of antarctic dry valley soils. Nat Rev Microbiol 8:129–138. doi: 10.1038/nrmicro2281 [DOI] [PubMed] [Google Scholar]
- 6. Heldmann JL, Pollard W, McKay CP, Marinova MM, Davila A, Williams KE, Lacelle D, Andersen DT. 2013. The high elevation dry valleys in Antarctica as analog sites for subsurface ice on Mars. Planetary and Space Science 85:53–58. doi: 10.1016/j.pss.2013.05.019 [DOI] [Google Scholar]
- 7. Cassaro A, Pacelli C, Aureli L, Catanzaro I, Leo P, Onofri S. 2021. Antarctica as a reservoir of planetary analogue environments. Extremophiles 25:437–458. doi: 10.1007/s00792-021-01245-w [DOI] [PubMed] [Google Scholar]
- 8. Fairén AG, Davila AF, Lim D, Bramall N, Bonaccorsi R, Zavaleta J, Uceda ER, Stoker C, Wierzchos J, Dohm JM, Amils R, Andersen D, McKay CP. 2010. Astrobiology through the ages of Mars: the study of terrestrial analogues to understand the habitability of Mars. Astrobiology 10:821–843. doi: 10.1089/ast.2009.0440 [DOI] [PubMed] [Google Scholar]
- 9. Martins Z, Cottin H, Kotler JM, Carrasco N, Cockell CS, de la Torre Noetzel R, Demets R, de Vera J-P, d’Hendecourt L, Ehrenfreund P, Elsaesser A, Foing B, Onofri S, Quinn R, Rabbow E, Rettberg P, Ricco AJ, Slenzka K, Stalport F, ten Kate IL, van Loon JJWA, Westall F. 2017. Earth as a tool for astrobiology-a European perspective. Space Sci Rev 209:43–81. doi: 10.1007/s11214-017-0369-1 [DOI] [Google Scholar]
- 10. Coleine C, Delgado-Baquerizo M. 2022. Unearthing terrestrial extreme microbiomes for searching terrestrial-like life in the solar system. Trends Microbiol 30:1101–1115. doi: 10.1016/j.tim.2022.04.002 [DOI] [PubMed] [Google Scholar]
- 11. Azua-Bustos A, Gonzalez-Silva C, Mancilla RA, Salas L, Gomez-Silva B, McKay CP, Vicuna R.. 2011. Hypolithic cyanobacteria supported mainly by fog in the coastal range of the Atacama Desert. Microb Ecol 61:568-581. doi: 10.1007/s00248-010-9784-5. [DOI] [PubMed] [Google Scholar]
- 12. Crits-Christoph A, Robinson CK, Ma B, Ravel J, Wierzchos J, Ascaso C, Artieda O, Souza-Egipsy V, Casero MC, DiRuggiero J.. 2016. Phylogenetic and functional substrate specificity for endolithic microbial communities in hyper-arid environments. Front Microbiol 7:301. doi: 10.3389/fmicb.2016.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ertekin E, Meslier V, Browning A, Treadgold J, DiRuggiero J.. 2021. Rock structure drives the taxonomic and functional diversity of endolithic microbial communities in extreme environments. Environ Microbiol 23:3937-3956. doi: 10.1111/1462-2920.15287. [DOI] [PubMed] [Google Scholar]
- 14. Wierzchos J, Casero MC, Artieda O, Ascaso C.. 2018. Endolithic microbial habitats as refuges for life in polyextreme environment of the Atacama Desert. Curr Opin Microbiol 43:124-131. doi: 10.1016/j.mib.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 15. Ramond JB, Woodborne S, Hall G, Seely M, Cowan DA.. 2018. Namib Desert primary productivity is driven by cryptic microbial community N-fixation. Sci Rep 8:6921. doi: 10.1038/s41598-018-25078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stomeo F, Valverde A, Pointing SB, McKay CP, Warren-Rhodes KA, Tuffin MI, Seely M, Cowan DA. 2013. Hypolithic and soil microbial community assembly along an aridity gradient in the Namib desert. Extremophiles 17:329–337. doi: 10.1007/s00792-013-0519-7 [DOI] [PubMed] [Google Scholar]
- 17. Friedmann EI. 1982. Endolithic microorganisms in the Antarctic cold desert. Science 215:1045–1053. doi: 10.1126/science.215.4536.1045 [DOI] [PubMed] [Google Scholar]
- 18. Coleine C, Biagioli F, de Vera JP, Onofri S, Selbmann L. 2021. Endolithic microbial composition in Helliwell hills, a newly investigated mars-like area in Antarctica. Environ Microbiol 23:4002–4016. doi: 10.1111/1462-2920.15419 [DOI] [PubMed] [Google Scholar]
- 19. Makhalanyane TP, Valverde A, Birkeland NK, Cary SC, Tuffin IM, Cowan DA. 2013. Evidence for successional development in antarctic hypolithic bacterial communities. ISME J 7:2080–2090. doi: 10.1038/ismej.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mergelov N, Dolgikh A, Shorkunov I, Zazovskaya E, Soina V, Yakushev A, Fedorov-Davydov D, Pryakhin S, Dobryansky A. 2020. Hypolithic communities shape soils and organic matter reservoirs in the ice-free landscapes of East Antarctica. Sci Rep 10:10277. doi: 10.1038/s41598-020-67248-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei STS, Lacap-Bugler DC, Lau MCY, Caruso T, Rao S, de los Rios A, Archer SK, Chiu JMY, Higgins C, Van Nostrand JD, Zhou J, Hopkins DW, Pointing SB. 2016. Taxonomic and functional diversity of soil and hypolithic microbial communities in Miers valley, Mcmurdo dry valleys, Antarctica. Front Microbiol 7:1642. doi: 10.3389/fmicb.2016.01642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pointing SB, Warren-Rhodes KA, Lacap DC, Rhodes KL, McKay CP. 2007. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold Hyperarid deserts. Environ Microbiol 9:414–424. doi: 10.1111/j.1462-2920.2006.01153.x [DOI] [PubMed] [Google Scholar]
- 23. Warren-Rhodes KA, Rhodes KL, Boyle LN, Pointing SB, Chen Y, Liu S, Zhuo P, McKay CP. 2007. Cyanobacterial Ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol 61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x [DOI] [PubMed] [Google Scholar]
- 24. Wong FKY, Lacap DC, Lau MCY, Aitchison JC, Cowan DA, Pointing SB. 2010. Hypolithic microbial community of quartz pavement in the high-altitude tundra of central tibet. Microb Ecol 60:730–739. doi: 10.1007/s00248-010-9653-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu MH, Li T, Zhang GS, Wu FS, Chen T, Zhang BL, Wu XK, Liu GX, Zhang KC, Zhang W. 2023. Seasonal variation of hypolithic microbiomes in the Gobi desert. Microb Ecol 85:1382–1395. doi: 10.1007/s00248-022-02043-3 [DOI] [PubMed] [Google Scholar]
- 26. Lacap-Bugler DC, Lee KK, Archer S, Gillman LN, Lau MCY, Leuzinger S, Lee CK, Maki T, McKay CP, Perrott JK, de Los Rios-Murillo A, Warren-Rhodes KA, Hopkins DW, Pointing SB. 2017. a. Global diversity of desert hypolithic cyanobacteria. Front Microbiol 8:867. doi: 10.3389/fmicb.2017.00867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meslier V, DiRuggiero J. 2019. Endolithic microbial communities as model systems for ecology and astrobiology, p 145–168. In Model Ecosystems in extreme environments [Google Scholar]
- 28. Chan Y, Lacap DC, Lau MCY, Ha KY, Warren-Rhodes KA, Cockell CS, Cowan DA, McKay CP, Pointing SB. 2012. Hypolithic microbial communities: between a rock and a hard place. Environ Microbiol 14:2272–2282. doi: 10.1111/j.1462-2920.2012.02821.x [DOI] [PubMed] [Google Scholar]
- 29. Lacap-Bugler DC, Lee KK, Archer S, Gillman LN, Lau MCY, Leuzinger S, Lee CK, Maki T, McKay CP, Perrott JK, de Los Rios-Murillo A, Warren-Rhodes KA, Hopkins DW, Pointing SB. 2017. b. Global diversity of desert hypolithic cyanobacteria. Front Microbiol 8:867. doi: 10.3389/fmicb.2017.00867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Warren-Rhodes KA, Rhodes KL, Pointing SB, Ewing SA, Lacap DC, Gómez-Silva B, Amundson R, Friedmann EI, McKay CP. 2006. Hypolithic cyanobacteria, dry limit of photosynthesis, and microbial ecology in the hyperarid atacama desert. Microb Ecol 52:389–398. doi: 10.1007/s00248-006-9055-7 [DOI] [PubMed] [Google Scholar]
- 31. Hwang Y, Schulze-Makuch D, Arens FL, Saenz JS, Adam PS, Sager C, Bornemann TLV, Zhao W, Zhang Y, Airo A, Schloter M, Probst AJ. 2021. Leave no stone unturned: individually adapted xerotolerant thaumarchaeota sheltered below the boulders of the Atacama desert Hyperarid core. Microbiome 9:234. doi: 10.1186/s40168-021-01177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng X, Zhang M, Xu Y, Li B.. 2002. Salt lakes of China. Sci Press. [Google Scholar]
- 33. Xiao L, Wang J, Dang Y, Cheng Z, Huang T, Zhao J, Xu Y, Huang J, Xiao Z, Komatsu G. 2017. A new terrestrial analogue site for Mars research: the Qaidam Basin, Tibetan plateau (NW China). Earth Sci Rev 164:84–101. doi: 10.1016/j.earscirev.2016.11.003 [DOI] [Google Scholar]
- 34. Anglés A, Li Y. 2017. The western qaidam basin as a potential martian environmental analogue: an overview. J Geophys Res Planets 122:856–888. doi: 10.1002/2017JE005293 [DOI] [Google Scholar]
- 35. Shen J, Chen Y, Sun Y, Liu L, Pan Y, Lin W. 2022. Detection of biosignatures in terrestrial Mars analogs: strategical and technical assessments. Earth Planet Phys. doi: 10.26464/epp2022042 [DOI] [Google Scholar]
- 36. Liu L, Liu H, Zhang W, Chen Y, Shen J, Li Y, Pan Y, Lin W. 2022. Microbial diversity and adaptive strategies in the mars-like qaidam basin, North Tibetan plateau, China. Environ Microbiol Rep 14:873–885. doi: 10.1111/1758-2229.13111 [DOI] [PubMed] [Google Scholar]
- 37. Kong F, Zheng M, Hu B, Wang A, Ma N, Sobron P. 2018. Dalangtan saline playa in a hyperarid region on Tibet plateau-I: evolution and environments. Astrobiology 18:1243–1253. doi: 10.1089/ast.2018.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kong WG, Zheng MP, Kong FJ, Chen WX. 2014. Sulfate-bearing deposits at dalangtan playa and their implication for the formation and preservation of martian salts. American Mineralogist 99:283–290. doi: 10.2138/am.2014.4594 [DOI] [Google Scholar]
- 39. Huang T, Wang R, Xiao L, Wang H, Martínez JM, Escudero C, Amils R, Cheng Z, Xu Y. 2018. Dalangtan playa (qaidam basin, NW China): Its microbial life and physicochemical characteristics and their astrobiological implications. PLoS One 13:e0200949. doi: 10.1371/journal.pone.0200949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS II, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R, Gilbert JA. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191–16. doi: 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s Q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uritskiy GV, DiRuggiero J, Taylor J. 2018. Metawrap-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. doi: 10.1186/s40168-018-0541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bertrand D, Shaw J, Kalathiyappan M, Ng AHQ, Kumar MS, Li C, Dvornicic M, Soldo JP, Koh JY, Tong C, Ng OT, Barkham T, Young B, Marimuthu K, Chng KR, Sikic M, Nagarajan N. 2019. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat Biotechnol 37:937–944. doi: 10.1038/s41587-019-0191-2 [DOI] [PubMed] [Google Scholar]
- 46. Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. Metabat 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Y-W, Simmons BA, Singer SW. 2016. Maxbin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638 [DOI] [PubMed] [Google Scholar]
- 49. Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103 [DOI] [PubMed] [Google Scholar]
- 50. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. Checkm: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. 2020. GTDB-TK: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parks DH, Chuvochina M, Chaumeil P-A, Rinke C, Mussig AJ, Hugenholtz P. 2020. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol 38:1079–1086. doi: 10.1038/s41587-020-0539-7 [DOI] [PubMed] [Google Scholar]
- 53. Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N. 2020. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using phylophlan 3.0. Nat Commun 11:2500. doi: 10.1038/s41467-020-16366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ortiz M, Leung PM, Shelley G, Jirapanjawat T, Nauer PA, Van Goethem MW, Bay SK, Islam ZF, Jordaan K, Vikram S, Chown SL, Hogg ID, Makhalanyane TP, Grinter R, Cowan DA, Greening C. 2021. Multiple energy sources and metabolic strategies sustain microbial diversity in Antarctic desert soils. Proc Natl Acad Sci U S A 118:e2025322118. doi: 10.1073/pnas.2025322118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bay SK, Dong X, Bradley JA, Leung PM, Grinter R, Jirapanjawat T, Arndt SK, Cook PLM, LaRowe DE, Nauer PA, Chiri E, Greening C. 2021. Trace gas oxidizers are widespread and active members of soil microbial communities. Nat Microbiol 6:246–256. doi: 10.1038/s41564-020-00811-w [DOI] [PubMed] [Google Scholar]
- 58. Johnson LS, Eddy SR, Portugaly E. 2010. Hidden markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou Z, Tran PQ, Breister AM, Liu Y, Kieft K, Cowley ES, Karaoz U, Anantharaman K. 2022. Metabolic: high-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome 10:33. doi: 10.1186/s40168-021-01213-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Søndergaard D, Pedersen CNS, Greening C. 2016. Hyddb: A web tool for hydrogenase classification and analysis. Sci Rep 6:34212. doi: 10.1038/srep34212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tamura K, Stecher G, Kumar S. 2021. Mega11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Letunic I, Bork P. 2019. Interactive tree of life (iTOL) V4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Truglio JJ, Croteau DL, Van Houten B, Kisker C. 2006. Prokaryotic nucleotide excision repair: the uvrabc system. Chem Rev 106:233–252. doi: 10.1021/cr040471u [DOI] [PubMed] [Google Scholar]
- 64. Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. 1999. The mutagenesis protein Umuc is a DNA polymerase activated by Umud', Reca, and SSB and is specialized for translesion replication. J Biol Chem 274:31763–31766. doi: 10.1074/jbc.274.45.31763 [DOI] [PubMed] [Google Scholar]
- 65. Li GM. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res 18:85–98. doi: 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- 66. Bell JC, Kowalczykowski SC. 2016. Reca: regulation and mechanism of a molecular search engine. Trends Biochem Sci 41:491–507. doi: 10.1016/j.tibs.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beam CE, Saveson CJ, Lovett ST. 2002. Role for radA/SMS in recombination intermediate processing in Escherichia coli. J Bacteriol 184:6836–6844. doi: 10.1128/JB.184.24.6836-6844.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dosch DC, Helmer GL, Sutton SH, Salvacion FF, Epstein W. 1991. Genetic analysis of potassium transport Loci in Escherichia coli: evidence for three constitutive systems mediating uptake potassium. J Bacteriol 173:687–696. doi: 10.1128/jb.173.2.687-696.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shirkey B, Kovarcik DP, Wright DJ, Wilmoth G, Prickett TF, Helm RF, Gregory EM, Potts M. 2000. Active Fe-containing superoxide dismutase and abundant sodF mRNA in nostoc commune (Cyanobacteria) after years of Desiccation. J Bacteriol 182:189–197. doi: 10.1128/JB.182.1.189-197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martinez A, Kolter R. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rodriguez Ayala F, Bartolini M, Grau R. 2020. The stress-responsive alternative sigma factor sigb of Bacillus subtilis and its relatives: an old friend with new functions. Front Microbiol 11:1761. doi: 10.3389/fmicb.2020.01761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen Y, Shen J, Liu L, Sun Y, Pan Y, Lin W. 2022. Preservation of organic matter in aqueous deposits and soils across the Mars‐analog qaidam basin, NW China: implications for biosignature detection on Mars. JGR Planets 127. doi: 10.1029/2022JE007418 [DOI] [Google Scholar]
- 73. Marshall CP, Javaux EJ, Knoll AH, Walter MR. 2005. Combined micro-fourier transform infrared (FTIR) spectroscopy and micro-raman spectroscopy of proterozoic acritarchs: a new approach to palaeobiology. Precambrian Research 138:208–224. doi: 10.1016/j.precamres.2005.05.006 [DOI] [Google Scholar]
- 74. Igisu M, Ueno Y, Shimojima M, Nakashima S, Awramik SM, Ohta H, Maruyama S. 2009. Micro-FTIR spectroscopic signatures of bacterial lipids in proterozoic microfossils. Precambrian Research 173:19–26. doi: 10.1016/j.precamres.2009.03.006 [DOI] [Google Scholar]
- 75. Igisu M, Takai K, Ueno Y, Nishizawa M, Nunoura T, Hirai M, Kaneko M, Naraoka H, Shimojima M, Hori K, Nakashima S, Ohta H, Maruyama S, Isozaki Y. 2012. Domain-level identification and quantification of relative prokaryotic cell abundance in microbial communities by micro-FTIR spectroscopy. Environ Microbiol Rep 4:42–49. doi: 10.1111/j.1758-2229.2011.00277.x [DOI] [PubMed] [Google Scholar]
- 76. Chen Y, Sun Y, Liu L, Shen J, Qu Y, Pan Y, Lin WJA. 2023. Biosignatures preserved in carbonate nodules from the Western qaidam basin, NW China: implications for life detection on mars. Astrobiology 23:172–182. doi: 10.1089/ast.2021.0196 [DOI] [PubMed] [Google Scholar]
- 77. Tanykova N, Petrova Y, Kostina J, Kozlova E, Leushina E, Spasennykh MJG. 2021. Study of organic matter of unconventional reservoirs by IR spectroscopy and IR microscopy. Geosciences 11:277. doi: 10.3390/geosciences11070277 [DOI] [Google Scholar]
- 78. Armstrong A, Valverde A, Ramond J-B, Makhalanyane TP, Jansson JK, Hopkins DW, Aspray TJ, Seely M, Trindade MI, Cowan DA. 2016. Temporal dynamics of hot desert microbial communities reveal structural and functional responses to water input. Sci Rep 6:34434. doi: 10.1038/srep34434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Crits-Christoph A, Robinson CK, Barnum T, Fricke WF, Davila AF, Jedynak B, McKay CP, Diruggiero J. 2013. Colonization patterns of soil microbial communities in the Atacama desert. Microbiome 1:28. doi: 10.1186/2049-2618-1-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goordial J, Davila A, Lacelle D, Pollard W, Marinova MM, Greer CW, DiRuggiero J, McKay CP, Whyte LG. 2016. Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J 10:1613–1624. doi: 10.1038/ismej.2015.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Neilson JW, Quade J, Ortiz M, Nelson WM, Legatzki A, Tian F, LaComb M, Betancourt JL, Wing RA, Soderlund CA, Maier RM. 2012. Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama desert, Chile. Extremophiles 16:553–566. doi: 10.1007/s00792-012-0454-z [DOI] [PubMed] [Google Scholar]
- 82. Chen YH, Chiang PW, Rogozin DY, Degermendzhy AG, Chiu HH, Tang SL. 2021. Salvaging high-quality genomes of microbial species from a meromictic Lake using a hybrid sequencing approach. Commun Biol 4:996. doi: 10.1038/s42003-021-02510-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. De Maio N, Shaw LP, Hubbard A, George S, Sanderson ND, Swann J, Wick R, AbuOun M, Stubberfield E, Hoosdally SJ, Crook DW, Peto TEA, Sheppard AE, Bailey MJ, Read DS, Anjum MF, Walker AS, Stoesser N, on behalf of the Rehab C . 2019. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb Genom 5:1–12. doi: 10.1099/mgen.0.000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu L, Wang Y, Che Y, Chen Y, Xia Y, Luo R, Cheng SH, Zheng C, Zhang T. 2020. High-quality bacterial genomes of a partial-nitritation/anammox system by an iterative hybrid assembly method. Microbiome 8:155. doi: 10.1186/s40168-020-00937-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542. doi: 10.1038/s41564-017-0012-7 [DOI] [PubMed] [Google Scholar]
- 86. Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Schriml L, Banfield JF, Hugenholtz P, Woyke T, Genome Standards Consortium . 2017. Minimum information about a single amplified genome (MISAG) and a Metagenome-assembled genome (MIMAG of bacteria and Archaea). Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rajeev L, da Rocha UN, Klitgord N, Luning EG, Fortney J, Axen SD, Shih PM, Bouskill NJ, Bowen BP, Kerfeld CA, Garcia-Pichel F, Brodie EL, Northen TR, Mukhopadhyay A. 2013. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J 7:2178–2191. doi: 10.1038/ismej.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bay SK, Waite DW, Dong X, Gillor O, Chown SL, Hugenholtz P, Greening C. 2021. Chemosynthetic and Photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient. ISME J 15:3339–3356. doi: 10.1038/s41396-021-01001-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kidron GJ, Herrnstadt I, Barzilay E. 2002. The role of dew as a moisture source for sand microbiotic crusts in the Negev desert. J Arid Environ 52:517–533. doi: 10.1006/jare.2002.1014 [DOI] [Google Scholar]
- 90. Rao B, Liu Y, Wang W, Hu C, Dunhai L, Lan S. 2009. Influence of dew on Biomass and Photosystem II activity of Cyanobacterial crusts in the Hopq desert, Northwest China. Soil Biol Biochem 41:2387–2393. doi: 10.1016/j.soilbio.2009.06.005 [DOI] [Google Scholar]
- 91. Ray AE, Zaugg J, Benaud N, Chelliah DS, Bay S, Wong HL, Leung PM, Ji M, Terauds A, Montgomery K, Greening C, Cowan DA, Kong W, Williams TJ, Hugenholtz P, Ferrari BC. 2022. Atmospheric chemosynthesis is phylogenetically and geographically widespread and contributes significantly to carbon fixation throughout cold deserts. ISME J 16:2547–2560. doi: 10.1038/s41396-022-01298-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, Montgomery K, Lines T, Beardall J, van Dorst J, Snape I, Stott MB, Hugenholtz P, Ferrari BC. 2017. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552:400–403. doi: 10.1038/nature25014 [DOI] [PubMed] [Google Scholar]
- 93. Cordero PRF, Bayly K, Man Leung P, Huang C, Islam ZF, Schittenhelm RB, King GM, Greening C. 2019. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J 13:2868–2881. doi: 10.1038/s41396-019-0479-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jordaan K, Lappan R, Dong X, Aitkenhead IJ, Bay SK, Chiri E, Wieler N, Meredith LK, Cowan DA, Chown SL, Greening C. 2020. Hydrogen-oxidizing bacteria are abundant in desert soils and strongly stimulated by hydration. mSystems 5:e01131-20. doi: 10.1128/mSystems.01131-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Greening C, Carere CR, Rushton-Green R, Harold LK, Hards K, Taylor MC, Morales SE, Stott MB, Cook GM. 2015. Persistence of the dominant soil phylum acidobacteria by trace gas scavenging. Proc Natl Acad Sci U S A 112:10497–10502. doi: 10.1073/pnas.1508385112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lynch RC, Darcy JL, Kane NC, Nemergut DR, Schmidt SK. 2014. Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert actinobacteria. Front Microbiol 5:698. doi: 10.3389/fmicb.2014.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shen J, Zerkle AL, Claire MW. 2022. Nitrogen cycling and biosignatures in a hyperarid mars analog environment. Astrobiology 22:127–142. doi: 10.1089/ast.2021.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Humann JL, Ziemkiewicz HT, Yurgel SN, Kahn ML. 2009. Regulatory and DNA repair genes contribute to the desiccation resistance of sinorhizobium meliloti Rm1021. Appl Environ Microbiol 75:446–453. doi: 10.1128/AEM.02207-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mosca C, Rothschild LJ, Napoli A, Ferré F, Pietrosanto M, Fagliarone C, Baqué M, Rabbow E, Rettberg P, Billi D. 2019. Over-expression of UV-damage DNA repair genes and ribonucleic acid persistence contribute to the resilience of dried Biofilms of the desert cyanobacetrium Chroococcidiopsis exposed to mars-like UV flux and long-term desiccation. Front Microbiol 10:2312. doi: 10.3389/fmicb.2019.02312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Timmins J, Moe E. 2016. A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: major findings, functional and mechanistic insight and challenges. Comput Struct Biotechnol J 14:168–176. doi: 10.1016/j.csbj.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gunde-Cimerman N, Plemenitaš A, Oren A. 2018. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev 42:353–375. doi: 10.1093/femsre/fuy009 [DOI] [PubMed] [Google Scholar]
- 103. Merino N, Aronson HS, Bojanova DP, Feyhl-Buska J, Wong ML, Zhang S, Giovannelli D. 2019. Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10:780. doi: 10.3389/fmicb.2019.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenomic tree.
Phylogenetic tree of CoxL.
Four supplementary tables.
Supplemental figure legends.
Data Availability Statement
Sequencing data from this study have been deposited under the NCBI BioProject PRJNA727938. The SRA accession numbers of Nanopore reads, Illumina reads, and 16S rRNA gene sequences are SRR24860171-SRR24860173, SRR24765285-SRR24765288, and SRR24860110-SRR24860113, respectively. The genome accession numbers of reconstructed MAGs are JAUJLY000000000-JAUJPI000000000.