Abstract
Imaging mass spectrometry (IMS) allows for the untargeted mapping of biomolecules directly from tissue sections. This technology is increasingly integrated into biomedical and clinical research environments to supplement traditional microscopy and provide molecular context for tissue imaging. IMS has widespread clinical applicability in the fields of oncology, dermatology, microbiology, and others. This review summarizes the two most widely employed IMS technologies, matrix-assisted laser desorption/ionization (MALDI) and desorption electrospray ionization (DESI), and covers technological advancements, including efforts to increase spatial resolution, specificity, and throughput. We also highlight recent biomedical applications of IMS, primarily focusing on disease diagnosis, classification, and subtyping.
Keywords: Imaging Mass Spectrometry, Clinical Mass Spectrometry, Matrix-Assisted Laser Desorption Ionization, Desorption Electrospray Ionization, Direct Tissue Analysis
1. Introduction
Mass spectrometry (MS) is an invaluable analytical tool for biomedical research and clinical applications due to its high chemical specificity and unparalleled sensitivity.1 When coupled to chromatography, including liquid (LC) and gas chromatography (GC), MS is routinely used to characterize biofluids, such as blood, urine, and plasma.2,3 While these analyses are regarded as the gold standard for guiding modern patient care, they require rigorous sample preparation and pre-treatment, as well as lengthy analysis time. This presents a challenge to accommodate the growing need for real-time assessment of biological specimens in clinical practice. Additionally, traditional chromatography-based MS analyses lack spatial awareness, often critical for tissue-based diagnostics. 2,3
Spatially driven MS technologies, such as imaging mass spectrometry (IMS), can be used to visualize molecular distributions within tissue sections in an untargeted manner.4 Many IMS approaches follow a similar workflow to traditional histology, allowing IMS to be easily integrated into pathology-related research for deeper molecular insight.5 This review summarizes recent advances in IMS instrumentation and methods to increase spatial resolution, specificity, throughput, and analyte coverage. We also highlight current biomedical and clinical applications of IMS, with a primary focus on studies intended to supplement histopathological analyses, including advances in disease diagnosis, classification, and subtyping.
2. Advances in Modern IMS
In recent years, numerous IMS surface sampling technologies have been advanced for improved spatial interrogation of tissue samples (Table I). These technologies differ in instrumental features, such as ionization source and operation principles; however, their workflows remain similar. A typical IMS experiment begins with tissue sectioning and mounting onto a substrate (typically a glass slide) followed by sample preparation, which is unique to each imaging platform and analysis type but can include tissue washing, application of reagents (e.g., enzymes or derivatization reagents), and matrix application. Advancements in IMS technology for biomedical research have focused on improving spatial resolution, increasing throughput, and enhancing molecular coverage. With improved analytical capabilities, parallel efforts to computationally link IMS and histological information have emerged. Herein, recent examples of how machine learning is guiding the translation of IMS technologies into clinical workflows will be highlighted. Finally, while improvements in sample preparation and matrix selection have also been integral to modern IMS,6,7 we will focus on instrumental advances which have improved image quality, reproducibility, and throughput.
Table I.
Overview of IMS Platforms Commonly Employed for Tissue Interrogation
| Ion Source | Matrix | Sample Preparation Time | Source Vacuum Requirements | Spatial Resolution | Molecular Class | Applications | |
|---|---|---|---|---|---|---|---|
| MALDI | Laser | Yes | 1 hour | Intermediate-High Vacuum | <1μm possible,13 10–50 μm standard14,15 | Drugs, metabolites, lipids, proteins | Molecular Profiling, Biomarker Discovery, Disease Classification |
| DESI | ESI | No | 0 min | Ambient | 10 μm possible (nanoDESI), 50–200 μm standard 16,17 | Drugs, metabolites, lipids, proteins | Margin Evaluation, Disease Classification, Biomarker Discovery |
| SIMS | Ion beam | No | 0 min | High Vacuum | 30 nm possible (nanoSIMS), 70 –100 nm standard18,19 | Elements, drugs, metabolites, lipids | Tissue characterization and biomarker discovery, single-cell imaging |
| LAESI | Laser/ ESI | Yes | 30 min | Ambient | 20 μm possible, 200–400 μm standard3,20 | Drugs, metabolites, lipids, proteins | Molecular profiling, biomarker discovery |
| LESA | Micro/ nanoESI | No | 0 min | Ambient | ~100 μm possible (microLESA), >200 μm standard 21,22 | Metabolites, peptides, proteins | Molecular profiling, biomarker discovery |
| MALDESI | Laser/ ESI | Yes | 1 hour | Intermediate- High Vacuum | 50 μm possible, 100–200 μm standard23,24 | Drugs, metabolites, lipids, proteins | Molecular profiling, biomarker discovery |
| LA-ICP | Laser | No | 0 min | Ambient; Vacuum | 2.5 μm possible, 30 μm standard 25 | Elements | Elemental composition of biological samples |
MALDI - Matrix-Assisted Laser Desorption/Ionization; DESI - Desorption Electrospray Ionization; SIMS - Secondary Ionization Mass Spectrometry; LAESI - Laser Ablation Electrospray Ionization; LESA - Liquid Extraction Surface Analysis; MALDESI - Matrix-Assisted Laser Desorption/Ionization Electrospray Ionization; LA-ICP - Laser Ablation Inductively Coupled Plasma
Spatial Resolution and Throughput
Matrix-assisted laser desorption/ionization (MALDI) and desorption electrospray ionization (DESI) IMS have extensively been applied to clinical tissue analysis.8–10 MALDI utilizes a focused UV laser for the desorption/ionization of matrix-coated tissue samples, offering an unparalleled combination of molecular coverage and spatial resolution for IMS enabling analysis of molecular classes ranging from small metabolites and lipids to peptides, glyfvcans, and proteins.10 MALDI IMS can routinely achieve high spatial resolutions, which is crucial for the interrogation of small clinical samples, such as needle biopsies, and for linking IMS data to specific cellular features. MALDI IMS is rapidly approaching single-cell imaging capabilities, with recent publications reporting <10 μm spatial resolutions. Higher spatial resolutions (<1 μm) have been demonstrated with the optimization of custom front-side laser systems11 and the development of transmission geometry12, which redirects the laser path to irradiate the tissue through the back of the slide. The sampled area per pixel decreases with higher spatial resolution, therefore decreasing the number of molecules sampled in a given pixel. To overcome this, instrumental approaches, such as continuous accumulation of selected ions (CASI), which enriches ions in a narrow mass window, and MALDI-2, which uses a second laser for postionization of the MALDI plume, can be used to enhance signal intensity.26 Studies combining transmission geometry and MALDI-2 have achieved imaging at subcellular resolution (<1 μm) and have been applied to study clinically relevant samples.27 DESI deflects a spray of charged droplets onto the tissue surface to extract analytes for detection by mass spectrometry.28 It is appealing for clinical applications because it can utilize nondestructive extraction solvents, such as water, and can be performed under atmospheric pressure without any sample preparation. Solvent-based IMS techniques like DESI require a liquid droplet deposition on the sample, limiting achievable spatial resolution to 50 – 250 μm in most cases.29 Optimization of solvent flow rates, composition, spray-to-sample distance, and other parameters have been explored to improve spatial resolutions.29 In one example, modifications to the DESI sprayer design included the addition of a capillary positioning device, a gas nozzle with an aperture of 400 μm, and a solvent capillary with a tapered tip and a larger outer-to-inner diameter ratio − 360 μm − 40 μm, as compared to 150 μm − 50 μm. 30 They report improved spray focusing and stability, enabling 20 μm spatial resolution imaging. Nano-DESI is a similar liquid extraction-based IMS approach in which analytes are desorbed from the tissue surface via a continuous liquid junction maintained between two capillaries. Ions are subsequently introduced to the mass spectrometer via electrospray ionization.31 This approach has allowed higher spatial resolution and throughput imaging under ambient conditions. Recently, Laskin and coworkers have achieved ~10 μm spatial resolutions with nano-DESI using an improved microfluidic probe, forming a smaller liquid bridge on the tissue’s surface.16
Increased IMS spatial resolution has also motivated development efforts for improved image acquisition speed. Instrument improvements to enhance throughput include modifications to stage mechanics, development of lasers with high repetition rates, implementation of continuous raster sampling for MALDI-TOF instruments, and optimized sample preparation and data generation workflows. Improvements in stage mechanics and laser optics have led to the development of commercial platforms capable of high acquisition rates, such as the RapifleX Tissuetyper (Bruker Daltonics), which utilizes a 10 kHz laser and is capable of acquiring >25 pixels/sec. Modified workflows using this instrument have been demonstrated to approach ~50 pixels/sec.32,33 Others have developed high throughput IMS instruments that utilize continuous raster sampling where the sample stage is moved continuously while sampling with either a laser or liquid droplet. Spatial resolution is defined by the sampling rate, the sample stage velocity, and the distance between line scans. Continuous raster sampling by MALDI has been shown to generate up to 50 pixels/sec.33,34 More recently, a laser scanning technique has been used to achieve 100 pixels/sec.35 Most DESI imaging experiments are performed as continuous raster experiments with data acquisition rates of 30 pixels/sec routinely achievable.36 These technologies and similar advances to increase throughput are critical for large-scale biomedical research projects and for incorporating IMS into clinical settings for real-time tissue interrogation.
Analyte Coverage, Identification, and Quantitation
Molecular coverage, specificity, and quantitation are critical for robust MS analysis. Time-of-flight (TOF) and quadrupole time-of-flight (qTOF) are the most used mass analyzers in IMS today. TOF instruments offer the widest detectable mass range (>100 kDa)37, while qTOF instruments provide resolving powers of ~ 50 000 and <5 ppm mass accuracy.38 Both systems enable high throughput molecular imaging (30–100 pixels/sec). Fourier transform mass spectrometers, such as Fourier transform ion cyclotron resonance (FT-ICR) MS39 and orbitrap MS, offer much higher resolving powers (>100 000) and improved accurate mass (<1ppm) and are increasingly used for clinical sample analysis.3 These platforms are ideal for separating and identifying analytes with similar m/z values; however, they remain costly and have lengthy data acquisition times.40 Alternatively, incorporating orthogonal separation techniques, such as ion mobility spectrometry that provides a gas-phase separation of ions, can increase sensitivity and specificity in IMS experiments.41,42 For example, high-field asymmetric ion mobility spectrometry (FAIMS) has been integrated with DESI IMS to decrease chemical noise and improve signal-to-noise ratios.41,43 Higher resolution ion mobility devices, such as traveling wave ion mobility spectrometry (TWIMS) and trapped ion mobility spectrometry (TIMS), have also been used with imaging mass spectrometry to elucidate isobaric/isomeric species in direct tissue analyses.41,42,44 Furthermore, collision cross section (CCS) values, a descriptor of the structure and three-dimensional conformation of the gas phase ions, can be derived with ion mobility, aiding in more accurate analyte identification. Finally, tandem MS (MSn) can be performed directly on tissue to provide analyte structural information based on detected fragments.45 More advanced fragmentation techniques, such as ultraviolet photodissociation46 and ozone-induced dissociation47,48, can also provide detailed structural information, such as double bond position and stereochemistry, making these techniques more suitable for targeted analyses.45
Although relative quantitation in IMS is routine, absolute quantitation has been a significant challenge in the field of IMS, owing to the high pixel-to-pixel variability, differences in analyte extraction efficiencies, and ion suppression effects.49 Absolute quantitation is most commonly achieved using internal standards against which analyte signals are calibrated.50 Internal standards can be sprayed, spotted, or otherwise deposited onto the tissue surfaces.51 In MALDI IMS workflows, this can be done prior to (or with) the matrix application. The internal standard can also be included in the extraction solvent in DESI IMS applications.52 There are numerous ways of incorporating standards, such as using a single standard and constructing an on-tissue calibration curve; however, mixtures of internal standards are often preferred for quantitation, as they are selected to be structurally similar to various analytes of interest. Alternatively, computational approaches for quantitation have been explored, including calculating matrix correction factors and normalizing against other quantitative approaches.49,52 Despite significant progress, quantitation remains one of the most significant limitations of IMS; a more in-depth exploration of the topic can be found elsewhere.49,52,53
IMS can be used to study a wide range of analytes, including small metabolites, lipids, peptides, glycans, and proteins. MALDI and DESI efficiently ionize metabolites and lipids without significant sample preparation or instrument modifications.54 Untargeted imaging of larger molecules, such as proteins and glycans, is more challenging and requires additional sample preparation steps.55–57 Aberrant protein glycosylation has been linked to numerous health conditions; therefore, characterizing the glycome with spatial specificity is an active area of interest for clinical and pharmaceutical applications alike. This topic is covered in more detail in the following reviews.58,59 Similarly, spatial interrogation of proteins, protein-ligand complexes, proteoforms, and various modifications can provide critical insight into numerous biological processes.60 Intact MS-based protein imaging was first achieved with MALDI IMS.61 This approach offers a broad mass range of approximately 500 Da to over 80 kDa62, depending on the instrument platform. However, MALDI-generated ions have lower charge states (≤3), greatly reducing gas-phase fragmentation efficiency and hindering direct top-down protein identification.60,63 Extraction-based approaches like DESI and nano-DESI generate higher charge-state ions and are particularly well suited for intact protein analysis. In a recent example, nano-DESI was used to map proteins in rat brain tissue at <10μm spatial resolution with oversampling.64
To improve protein coverage in IMS experiments, protein imaging can also be accomplished by bottom-up proteomics following on-tissue digestion.65 In an imaging context, simultaneously achieving robust protein identification, high protein coverage, and high spatial resolutions with either approach remains challenging. Spatial profiling techniques, such as laser capture microdissection (LCM)66 and liquid-extraction surface analysis (LESA)67 can be used to probe specific tissue locations for in-depth protein analyses.63 Once proteins are extracted, the sample can be subjected to either bottom-up or top-down workflows compatible with downstream LC-MS/MS.68 These approaches offer high molecular coverage and robust quantitation but at limited spatial information. More recently, nanodroplet processing in One Pot for Trace Samples (NanoPOTS) has emerged as a technique for working with smaller sample amounts, allowing the mapping of protein abundance with improved sensitivity and at higher spatial resolutions with LCM LC-MS/MS workflows.69 IMS and spatial profiling approaches can be used in parallel to maximize information gleaned from precious tissue samples. Modern multimodal imaging workflows can use IMS data to drive spatial omics (proteomics, glycomics, transcriptomics, etc.) data collection from specific areas of interest to better understand localized molecular biology within tissue microenvironments.
Tissue Processing and Sample Preparation
In addition to instrumental considerations, the information gleaned from a biological system is dictated by the nature of the sample itself and how the specimen is prepared. Surgical samples are used for intraoperative diagnosis, stored for post-surgical analysis, and for developing and validating new diagnostic methods. Common clinical tissue samples include biopsies (needle, wedge), excised tumors, smears, and whole organs.40 Upon excision, samples can be fixed or flash frozen and embedded to preserve tissue morphology and aid in their handling. Both tissue fixation and embedding can affect postoperative IMS analysis and profoundly impact data quality.40,70 For example, fixation materials, such as formalin and paraformaldehyde, alter the chemical composition of the samples through reactions such as cross-linking and denaturing.70 Similarly, common embedding materials, such as paraffin and optimal cutting temperature (OCT) gel, can introduce ion suppression effects in IMS analysis.71 Formalin-fixed paraffin-embedded (FFPE) tissue blocks have long been used for immunohistochemical analysis and are ideal for spatial proteomics and glycomics; however, this embedding material and the washing steps required for deparaffinization significantly hinder lipid and small metabolite detection in MALDI IMS. In addition to the sample procurement and preservation, sample preparations vary by the instrument employed and the target analyte class. These considerations range from the mounting substrate to sample pre-treatment, such as washing, chemical derivatizations, and type of matrix employed, if any.9 For example, protein imaging requires a series of washes to remove salts and lipids that introduce ionization suppression, and some approaches also require on-tissue enzymatic digestion.60 Similarly, glycan imaging requires lengthy sample preparation, including antigen retrieval and enzymatic digestion.58 Comprehensive reviews of sample preparation considerations and their impact on IMS analysis can be found elsewhere.1,6,7,40
3. Multimodal Data Integration and Visualization
Histology uses numerous stains and dyes to reveal the morphological and cellular composition of tissues. While these approaches are highly informative and routinely used by pathologists for diagnosis in clinical settings, they provide only limited molecular information.5,72 For example, immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) are targeted approaches to map the spatial distributions of specific protein or RNA markers in tissue sections.73,74 Untargeted molecular investigations, such as with IMS, can reveal spatially resolved molecular features but do not provide information on cellular and morphological structures. When collectively employed in multimodal imaging experiments, these integrated approaches greatly increase the information obtained from biological systems, enabling molecular histology and furthering next-generation molecular pathology practices.5,72
Correlation of IMS data with stained microscopy can be achieved through computational approaches, such as multimodal registration workflows, as demonstrated by Patterson et al. 75 and Race et al.76 (Figure 1). Patterson et al.75 used autofluorescence microscopy and computational registration to link IMS and microscopy data. An advantage of this approach is the ability to register serial tissue sections that have undergone different analyses with varying spatial resolutions (Figure 1A). Building on image registration, Van de Plas et al.77 demonstrated a predictive imaging modality, termed data-driven image fusion, by combining hematoxylin and eosin (H&E) stained microscopy images with IMS using highly multivariate linear regression (Figure 1B). The resulting model enables the prediction of IMS data to higher spatial resolution (i.e., spatial sharpening) and in tissue areas not analyzed by IMS (i.e., out-of-sample prediction). Race et al.76 combined IMS, histological staining, and deep learning for automatic annotation of histology images and annotation transfer between two modalities. The workflow was demonstrated on data generated from multiple IMS technologies, including MALDI, DESI, and rapid evaporative ionization mass spectrometry (REIMS) (Figure 1C).
Figure 1.
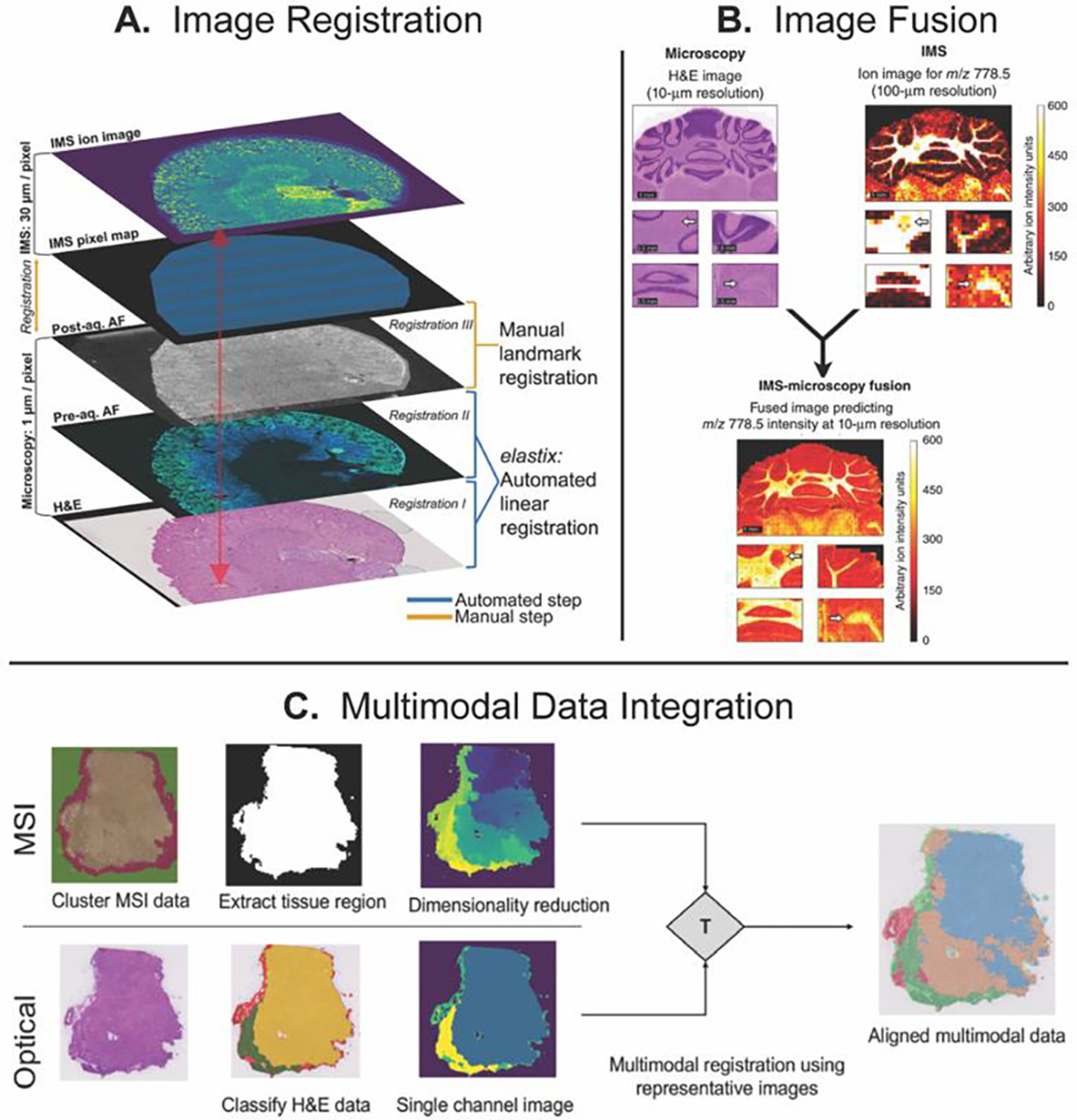
Multimodal image registration workflows by (A) Patterson et al., (B) data-driven image fusion by Van de Plas et al., and (C) Deep-learning image registration by Race et al. Figures adapted from Ref. [75], [76], and [77], with permission from the publishers.
A crucial aspect of obtaining biological insight from multimodal data is the development of computational workflows to interpret large datasets. Classification models require “training” datasets to build models and “testing” datasets to evaluate their performance. The performance is measured by the ability to (1) correctly differentiate disease and healthy cases (accuracy), (2) identify the diseased cases correctly (sensitivity), and (3) determine the healthy cases correctly (specificity), where these characteristics are reported in percentages.78 Pathologists can manually interpret stained sections to annotate important features, such as tumors, tumor margins, and normal tissues. This provides meaningful labels for the training data sets, which can be superimposed onto the molecular data via multimodal image registration.15,79 In the case of biomarker discovery, mass spectra from tissue regions labeled as “tumor” and “control” are compared to reveal mass spectral differences. Once candidate signals (m/z values) are detected, statistical tests, such as Student’s t-test and analysis of variance (ANOVA), are performed to determine if the chemical signal significantly differs between the two groups.5 When building classification models based on multiple mass spectral differences, supervised multivariate analyses, such as least absolute shrinkage and selection operator (LASSO) and partial least squares regression, are performed.80 These approaches are intended to predict tumor class and tissue labels based on training sets. Recently, Tideman et al.81 demonstrated automated biomarker discovery by interpreting classification models constructed from IMS data. This approach offers a spatially specific method for determining important molecular features across large cohorts of biological specimens.
The power of integrating molecular and cellular observations has been demonstrated in numerous applications, including in precision medicine, diagnosis, and biomarker discovery.29,36 For example, recent publications by Schwamborn and coworkers82,83 used molecular information from MALDI IMS to supplement cellular observations for a more comprehensive understanding of pancreatic tumors. Bollwein et al.82 used MALDI IMS and machine learning models to differentiate between pancreatic ductal adenocarcinoma and cholangiocarcinoma tumors, which are extremely difficult to distinguish based on morphology alone. The machine learning models, built on tryptic peptide data from tissue microarrays, correctly classified more than 80% of cases. Similarly, Gonçalves et al.83 identified molecular differences between primary pancreatic ductal adenocarcinoma and distant metastases, the presence of which can affect treatment options for patients. Multiple candidates for protein biomarkers of disease progression and development of distant metastases were identified, and the classification model reported a > 90% accuracy. Both studies showcase the combination of IMS and histological studies for a more comprehensive understanding of disease states.
Building robust prediction models to discern disease states requires significant sample cohorts and rigorous validation with currently employed tissue diagnostic methods. Tissue microarrays and biopsies from large cohorts of diseased and normal patients are ideal for evaluating not only model performance but also the robustness and repeatability of the employed analytical technique. In a recent study, Deininger et al.84 conducted a multi-site study to examine the reproducibility of MALDI imaging techniques for tissue classification. The study developed a standard operating procedure that included histological annotation and an optimized data pre-processing pipeline designed to remove technical variation while retaining biological information. The authors report that using the standard procedure increased classification accuracy and overall robustness in multi-site studies. Similar large-scale studies will be needed to fully incorporate spatially oriented mass spectrometry techniques in the clinical setting.
Unsupervised machine learning strategies can also be employed to extract valuable information from highly dimensional IMS datasets. These strategies are generally explorative and seek to uncover trends and correlations within IMS datasets without a priori information (i.e., labels).85 Unsupervised methods include component analyses and clustering. Briefly, component analyses, such as principal component analysis and non-negative matrix factorization, are intended to reduce multidimensional IMS data and reveal underlying trends in the datasets.5,15,80 Clustering methods, such as hierarchical clustering and k-means, partition pixels based on mass spectral similarities and reveal the molecular ions that drive the resulting groupings.5,80 Clusters are often color-coded and visualized as image segments. These methods are especially powerful when combined with traditional microscopy techniques, as they enable the discovery of molecular profiles with distinct spatial patterns (i.e., segments) that can be linked to specific morphological features.
There are both commercially available and open-source software solutions capable of analyzing and visualizing IMS data. Commercially available IMS software include SCiLS and FlexImaging (Bruker Daltonics), LipostarMSI (Molecular Discovery), and HDI (Waters). Free, open-source IMS processing solutions include MSIReader86, METASPACE87, and numerous R packages specific for IMS analysis (Cardinal88, MALDIquant89, and others90). As IMS workflows evolve to incorporate multimodal imaging data, new software solutions for multimodal data analysis and/or registration have emerged. For example, MZ mine 3 is open-source software that supports hybrid datasets, including LC-MS, ion mobility-MS, and IMS datasets.91 Similarly, several tools for registration and alignment of IMS with other clinically relevant imaging modalities have emerged; these are summarized in recent review papers.92,93
4. Clinical Applications of IMS
Perioperatively, histopathology methods, including frozen section analysis and imprint cytology, provide valuable diagnostic information. These strategies are often time-consuming, require expert pathologists for their interpretation, and may introduce unwanted freezing artifacts.5,94 More advanced analytical approaches, such as IMS, can be integrated to provide complementary information in tumor diagnosis, prognosis, and margin assessment.29 Beyond the clinical lab, IMS has played a vital role in the pharmaceutical field, aiding drug discovery and development. 95,96 Here, we showcase recent biomedical, clinical, and pharmaceutical applications of IMS, with a primary focus on MALDI and DESI. The reader is directed to the following reviews for a more in-depth look into the techniques not covered in this review.8
Matrix-assisted Laser Desorption/Ionization Imaging Mass Spectrometry (MALDI IMS)
MALDI is the most widespread IMS technology and has been used in numerous biomedical and clinical applications, including biomarker discovery, cancer diagnosis, therapeutic testing, and microorganism identification.29 In fact, MALDI MS is routinely employed in clinical microbiology to identify microorganisms in biological isolates, where rapid and precise pathogen identification is critical for selecting appropriate treatment.97 MALDI MS can detect pathogens on a species level within minutes, offering a high-throughput alternative to conventional identification strategies that require lengthy incubation procedures.98 Incorporating the spatial dimension with MALDI IMS offers a unique insight into the molecular species implicated at the host-pathogen interface and molecular alterations that occur during the course of an infection.99,100 Such studies are integral in developing new therapeutics and understanding antibiotic resistance on a molecular level.5
MALDI IMS offers broad analyte coverage and is well-suited for the analysis of large biomolecules, such as proteins and glycans. Because of this, numerous studies have investigated alterations of protein and glycan signatures to identify disease states. For example, Capitoli et al.101 demonstrated MALDI IMS as a complementary tool to cytopathology diagnosis of thyroid nodules based on proteomic signatures. Thyroid fine-needle aspirations were analyzed by MALDI IMS, and the probability of malignancy was mapped on a pixel-by-pixel basis. Although further work is necessary to systematically test these workflows on larger sample cohorts, this introductory study showed promising results in building a MALDI IMS-based proteomic diagnostic tool for thyroid cytopathology. In a more recent example, Ochoa-Rios et al.102 used MALDI IMS to investigate changes in N-glycan composition in mouse and human liver tissues due to non-alcoholic steatohepatitis. N-glycan changes were detected even in early stages of the disease and could be used to identify progression before tissue damage was pathologically visible.
Similarly, recent studies have showcased MALDI IMS as a powerful diagnostic tool for cancer types that are particularly challenging to classify with histopathology alone. For example, Sommella et al.103 used MALDI IMS to investigate salivary tumor tissues (Figure 2A–D). This tumor type’s diverse morphological profile and cell types may not follow traditional observations that allow pathologists to differentiate malignant and benign tumors. Although this tumor type is difficult to characterize with conventional pathology practices, the authors successfully delineated normal and tumor tissue based on metabolomic and lipidomic signatures with a reported 95% accuracy. Janßen et al.104 applied MALDI IMS and computational classification models to differentiate between the two most common non-small cell lung cancer tumor types – adenocarcinoma and squamous cell carcinoma (Figure 3E & 3F). This study exemplifies a successful transfer of classifiers developed on tissue microarrays to whole lung tissue sections, which are generated in routine clinical practices. These and similar studies not only elucidate molecular alteration due to disease but also pave the way for incorporating MALDI IMS in real-time clinical decision making.
Figure 2.
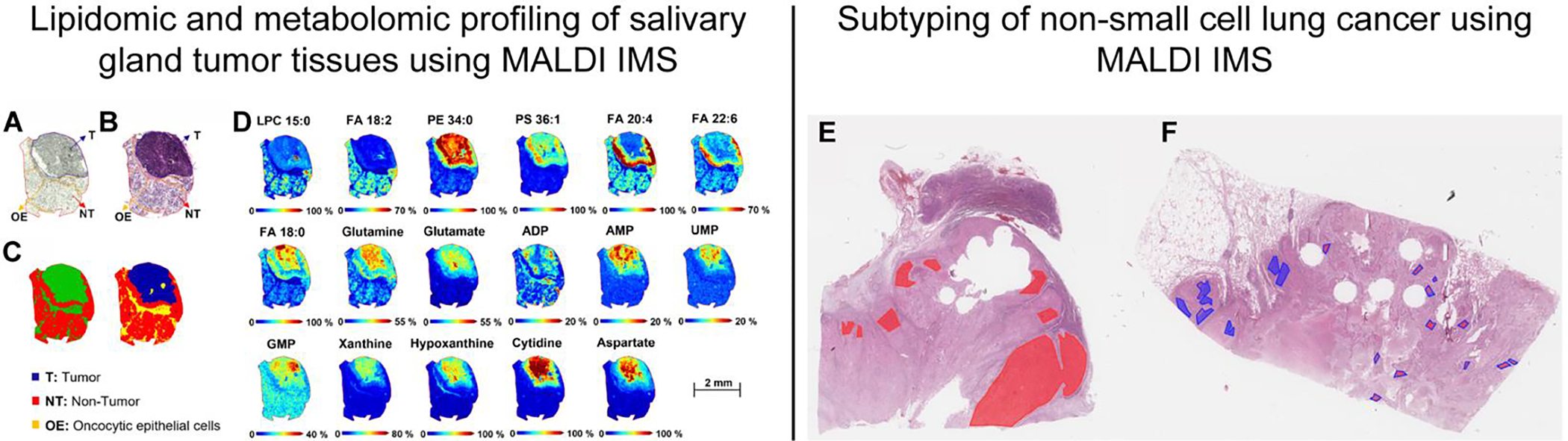
Determining the metabolomic and lipidomic profiles of salivary tumor tissue (A-D) [reference: 103], and subtyping non-small cell lung cancer (E-F) [reference: 104 using MALDI IMS. Annotated optical image (A) and hematoxylin and eosin-stained parotid tissue (B), bisecting K-means segmentation map generated from the MALDI data (C). MALDI IMS images of relevant lipids and metabolites obtained in negative ionization mode, comparing the distribution between tumor and healthy regions of the tissue (D). Annotated (lines) stained sections of adenocarcinoma (E), squamous cell carcinoma (F), and heatmaps (filling) of the neural network classification. Heatmaps show the location of spectra in colors corresponding to their classification: adenocarcinoma –red and squamous cell carcinoma–blue. Figures adapted from Sommella et al. (2022) [reference: 103] and Janßen et al.(2022) [reference: 104] with permissions from the publisher.
Figure 3.
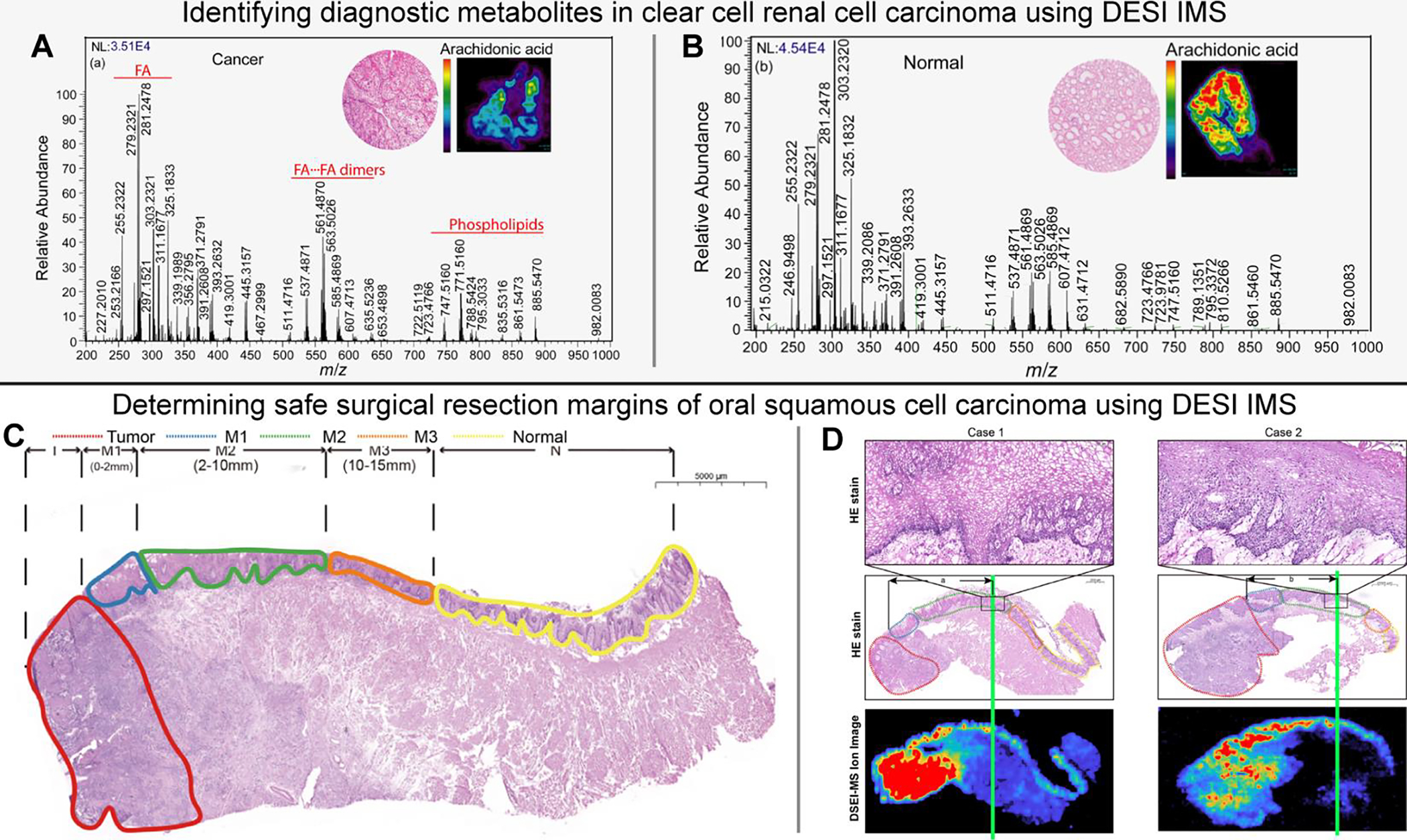
Identifying diagnostic metabolite signatures in clear cell renal cell carcinoma (A-B) [Reference 113] and determining safe surgical resection margins of oral squamous cell carcinoma (C-D) [Reference 114] using DESI IMS. Comparison between average mass spectra of normal (A) and ccRCC tissues (B) (m/z200 –1000), where the inset shows hematoxylin and eosin stains of each as well as ion images of arachidonic acid. Annotated hematoxylin and eosin-stained tissue with oral squamous cell carcinoma including T - Tumor (red), M1- positive margin (blue), M2 - close margin (green), M3 - negative margin (orange), N - normal (yellow) (C). Annotated stains and DESI IMS ion images depicting safe margin distance (green line) as the region where ion intensity decreases/disappears (D). Figures adapted from Vijayalakshmi et al. (2019) [Reference 113] and Yang et al. (2022) [Reference 114] with permissions from the publisher.
Perioperatively, MALDI IMS can provide complementary diagnostic information. A proof-of-concept study by Calligaris et al.105 investigated the use of MALDI IMS for near-real time pituitary tumor detection based on proteomic signatures. The authors demonstrated that MALDI IMS could successfully classify tumors by directly detecting excessive hormonal production from functional pituitary adenomas. Their analyses were performed in under 30 minutes, a timeframe compatible with guiding surgical decision making. As MALDI IMS requires more extensive sample preparation, such as the deposition of the MALDI matrix, this step may lengthen analysis time. New workflows, such as implementing matrix pre-coated slides106 and simplified sample preparation strategies, have been proposed to increase throughput significantly.107
MALDI IMS has also been employed to study drug metabolism, formulation, and delivery, among other pharmaceutical applications.95 In a recent proof-of-concept study, MALDI IMS was used to determine if an inhaled therapy drug can effectively be delivered to the target therapeutic area within human lungs.108 In this study, biopsies were taken from patients with confirmed intestinal lung disease after administering a single dose of nebulized ipratropium bromide. Biopsy sections were analyzed with MALDI IMS and histopathology; the data revealed that the drug co-localized with fibrotic regions in the samples, suggesting that drugs designed to treat pulmonary fibrosis could be administered via inhalation. More examples of MALDI IMS applied to pharmaceutical research and drug development can be found in the highlighted review.95
Desorption Electrospray Ionization Imaging Mass Spectrometry (DESI IMS)
DESI IMS has been increasingly employed in cancer research for diagnosis, subtyping, staging, and surgical margin evaluation.29 Target analyte classes for DESI IMS include metabolites, fatty acids, and lipids; however, modification of spray conditions and extraction solvent compositions have expanded this list to include proteins.109 Robust classification models have been developed for distinguishing gastric cancer110, skin cancer111, breast cancer112, and others28 to guide real-time surgical decision making. Of note, Vijayalakshmi et al.113 used DESI IMS and a LASSO predictive model to identify diagnostic metabolomic signatures in clear cell renal cell carcinoma (ccRCC) from human nephrectomy specimens (Figure 3A & 3B). The authors report that combined indicators (baseline LASSO of differentially expressed metabolites and the ratio of glucose to arachidonic acid) can discriminate between cancer and normal tissue with 85.3% accuracy. In a similar study, Santoro et al.112 used DESI IMS to investigate the metabolomic and lipidomic compositions of molecular subtypes of breast cancer. With similar modeling approaches, the authors could distinguish between invasive breast cancer and adjacent benign tissues, as well as between ductal carcinoma in situ and invasive breast cancer. In another recent study, Yang et al.114 used DESI IMS and machine learning to determine the safe surgical resection distance and margin status of oral squamous cell carcinoma based on diagnostic lipid ions (Figure 3C & 3D). With this method, the authors report an overall prediction accuracy of 92.6%. Moreover, the total data acquisition and analysis procedure can be performed within 30 minutes, which is compatible with perioperative analysis timeframes.
Growing evidence suggests that intraoperative molecular investigation can provide additional information on surgical margins not revealed by histology. For example, in a study of pancreatic cancer margin detection by Eberlin et al.115, several tissue margins diagnosed as non-cancerous by histopathological evaluation were classified as cancerous based on a predictive model built upon metabolomic features. Although the disagreement of classification may be attributed to the error range in the DESI/LASSO analysis, it is possible that molecular markers of pancreatic cancers could be detected prior to morphological alterations that are typically discerned by histopathology.
DESI IMS is perhaps the most promising IMS technique for translation into the surgical suite because it does not require any sample preparation and can be operated under ambient conditions.2,94 DESI IMS offers high throughput analysis in applications where spatial resolution is not critical, such as analyzing surgical swabs and smears. For example, Pirro et al.116 demonstrated DESI IMS for intraoperative diagnosis of glioma from brain tissue swabs from 10 different surgeries. They reported the detection of cancerous tissues with 93% sensitivity and 83% specificity based on lipid and metabolite signals. More recently, Brown et al.117 monitored 2-hydroxyglutarate ion intensities along with lipid and metabolite profile changes for intraoperative glioma diagnosis. The authors report the sensitivity, specificity, and accuracy of predicting disease status as 63%, 83%, and 74%, respectively. Overall, the authors successfully identified isocitrate dehydrogenase mutation status, glioma diagnosis, and intraoperative estimation of tumor cell infiltration.
The advent of nano-DESI has allowed for higher spatial resolutions and enhanced molecular coverage of extraction-based IMS approaches. Thus, nano-DESI is increasingly used in biomedical applications to study a wide range of analytes, including small metabolites, drugs, and proteins.118 In a recent application, nano-DESI was used to examine the localization of diclofenac, a widely used nonsteroidal anti-inflammatory drug, and its metabolites in mouse kidney and liver tissues.119 Nano-DESI has also been applied for high spatial resolution imaging of native proteins in rat kidney120, proteoforms in rat brain64,121 among other applications.54,122 In a recent application to human samples, nano-DESI IMS was used for direct imaging of proteoforms (<70 kDa) in human kidney samples, where clear localizations to distinct anatomical structures and cellular neighborhoods within the major component of the kidney were demonstrated.123 Identification was performed on a serial tissue section by top-down MS. These studies pave the way for future application to molecular tissue mapping, biomarker discovery, and disease diagnostics.
Finally, technological improvements in ambient ionization for clinical applications have led to the development of numerous MS-based surgical devices. For example, the MasSpec Pen is a handheld surgical device that operates via a liquid extraction mechanism.124 The device is intended for in vivo surgical use and has shown promising results in clinical applications. Recently, Zhang et al.125 described the clinical translation of the MasSpec Pen in 100 surgeries performed by seven surgical teams, where tissue analysis was performed during parathyroidectomy, thyroidectomy, breast, and pancreatic surgeries for various clinical indications. Other surgical MS techniques, including laser desorption (SpiderMass), thermal evaporation (REIMS), and solid extraction, have been developed and reviewed in detail elsewhere.10
5. Conclusions
This review highlights significant technological advances in modern imaging mass spectrometry and showcases recent biomedical and clinical applications. In the future, careful validation of experimental methods and classification models across numerous centers is necessary to evaluate the reproducibility, robustness, and applicability of the described workflows. As IMS continues to mature as a field, it is increasingly being used in tandem with other technologies for multimodal tissue interrogation. Multi-institution research consortia, such as the Human BioMolecular Atlas Program (HuBMAP)126, the Human Tumor Atlas Network (HTAN)127, and the Kidney Precision Medicine Project (KPMP)128, represent recent efforts to provide a comprehensive atlas of human tissues using traditional histology methods, fluorescence microscopy, IMS, and other cellular and molecular characterization methods. Ultimately, multimodal imaging strategies are promising for broad biomedical and clinical applications, including disease prediction, diagnosis, guiding treatment, and improving patient care.
HIGHLIGHTS.
Imaging mass spectrometry can advance clinical and biomedical research by providing molecular context to tissue pathology.
Instrumentation developments are improving molecular coverage, image quality, reproducibility, and throughput.
Multimodal data integration and visualization enable novel data mining strategies and are essential for deriving biological insight.
Recent studies showcase the value of MALDI and DESI IMS in clinical tissue interrogation.
Acknowledgements:
This work was funded by grant support provided by the National Institute of Health (NIH) including grants U01 DK133766, U54 DK134302, R01 AG078803, U54 EY032442, R01 AI145992, and R01 AI138581 awarded to J.M.S. K.V.D was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) training grant (T32DK007569-33)
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Buchberger AR; DeLaney K; Johnson J; Li L Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2018, 90 (1), 240–265. 10.1021/ACS.ANALCHEM.7B04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhou X; Zhang W; Ouyang Z Recent Advances in On-Site Mass Spectrometry Analysis for Clinical Applications. Trends Anal. Chem. 2022, 149, 116548. 10.1016/j.trac.2022.116548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Addie RD; Balluff B; Bovée JVMG; Morreau H; McDonnell LA Current State and Future Challenges of Mass Spectrometry Imaging for Clinical Research. Anal. Chem. 2015, 87 (13), 6426–6433. 10.1021/acs.analchem.5b00416. [DOI] [PubMed] [Google Scholar]
- (4).Seeley EH; Schwamborn K; Caprioli RM Imaging of Intact Tissue Sections: Moving beyond the Microscope. J. Biol. Chem. 2011, 286 (29), 25459–25466. 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chung H; Huang P; Chen C; Lee C; Hsu C Next-generation Pathology Practices with Mass Spectrometry Imaging. Mass Spectrom. Rev. 2022, e21795. 10.1002/mas.21795. [DOI] [PubMed] [Google Scholar]
- (6).Ma X; Fernández FM Advances in Mass Spectrometry Imaging for Spatial Cancer Metabolomics. Mass Spectrom. Rev. 2022, e21804. 10.1002/MAS.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhou Q; Fülöp A; Hopf C Recent Developments of Novel Matrices and On-Tissue Chemical Derivatization Reagents for MALDI-MSI. Anal. and Bioanal. Chem. 2021, 413 (10), 2599–2617. 10.1007/S00216-020-03023-7/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).He MJ; Pu W; Wang X; Zhang W; Tang D; Dai Y Comparing DESI-MSI and MALDI-MSI Mediated Spatial Metabolomics and Their Applications in Cancer Studies. Front. Oncol. 2022, 12, 3485. 10.3389/FONC.2022.891018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Maciel LÍL; Bernardo RA; Martins RO; Batista Junior AC; Oliveira JVA; Chaves AR; Vaz BG Desorption Electrospray Ionization and Matrix-Assisted Laser Desorption/Ionization as Imaging Approaches for Biological Samples Analysis. Analytical and Bioanalytical Chemistry 2023 415:18 2023, 415 (18), 4125–4145. 10.1007/S00216-023-04783-8. [DOI] [PubMed] [Google Scholar]
- (10).Zhang J; Sans M; Garza KY; Eberlin LS MASS SPECTROMETRY TECHNOLOGIES TO ADVANCE CARE FOR CANCER PATIENTS IN CLINICAL AND INTRAOPERATIVE USE. Mass Spectrom Rev 2021, 40 (5), 692–720. 10.1002/MAS.21664. [DOI] [PubMed] [Google Scholar]
- (11).Spengler B; Hubert M Scanning Microprobe Matrix-Assisted Laser Desorption Ionization (SMALDI) Mass Spectrometry: Instrumentation for Sub-Micrometer Resolved LDI and MALDI Surface Analysis. J. Am. Soc. Mass Spectrom. 2002, 13 (6), 735–748. 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- (12).Zavalin A; Todd EM; Rawhouser PD; Yang J; Norris JL; Caprioli RM Direct Imaging of Single Cells and Tissue at Sub-Cellular Spatial Resolution Using Transmission Geometry MALDI MS. J. Mass Spectrom. 2012, 47 (11), 1473–1481. 10.1002/JMS.3108. [DOI] [PubMed] [Google Scholar]
- (13).Zavalin A; Todd EM; Rawhouser PD; Yang J; Norris JL; Caprioli RM Direct Imaging of Single Cells and Tissue at Sub-Cellular Spatial Resolution Using Transmission Geometry MALDI MS. J. Mass Spectrom. 2012, 47 (11), 1473–1481. 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- (14).Zhu X; Xu T; Peng C; Wu S Advances in MALDI Mass Spectrometry Imaging Single Cell and Tissues. Front. Chem. 2022, 9, 1076. 10.3389/FCHEM.2021.782432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tuck M; Grélard F; Blanc L; Desbenoit N MALDI-MSI Towards Multimodal Imaging: Challenges and Perspectives. Front. Chem. 2022, 10. 10.3389/FCHEM.2022.904688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yin R; Burnum-Johnson KE; Sun X; Dey SK; Laskin J High Spatial Resolution Imaging of Biological Tissues Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Nat. Protoc. 2019, 14 (12), 3445–3470. 10.1038/s41596-019-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Spruill ML; Maletic-Savatic M; Martin H; Li F; Liu X Spatial Analysis of Drug Absorption, Distribution, Metabolism, and Toxicology Using Mass Spectrometry Imaging. Biochem. Pharmacol. 2022, 201, 115080. 10.1016/j.bcp.2022.115080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Anderton CR; Gamble LJ Secondary Ion Mass Spectrometry Imaging of Tissues, Cells, and Microbial Systems. Micros. Today 2016, 24 (2), 24–31. 10.1017/s1551929516000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Taylor MJ; Lukowski JK; Anderton CR Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J Am Soc Mass Spectrom 2021, 32 (4), 872–894. 10.1021/jasms.0c00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nemes P; Vertes A Laser Ablation Electrospray Ionization for Atmospheric Pressure Molecular Imaging Mass Spectrometry. In Mass Spectrometry Imaging. Methods in Molecular Biology; Rubakhin SS, Sweedler JV, Eds.; Humana Press: Totwa, NJ, 2010; Vol. 656, pp 159–171. [DOI] [PubMed] [Google Scholar]
- (21).Ryan DJ; Patterson NH; Putnam NE; Wilde AD; Weiss A; Perry WJ; Cassat JE; Skaar EP; Caprioli RM; Spraggins JM MicroLESA: Integrating Autofluorescence Microscopy, in Situ Micro-Digestions, and Liquid Extraction Surface Analysis for High Spatial Resolution Targeted Proteomic Studies. Anal Chem 2019, 91 (12), 7578–7585. 10.1021/ACS.ANALCHEM.8B05889/ASSET/IMAGES/LARGE/AC-2018-058898_0004.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang X; Na N; Ouyang J Droplet-Based Extraction Mass Spectrometry. TrAC - Trends in Analytical Chemistry 2021, 143. 10.1016/J.TRAC.2021.116366. [DOI] [Google Scholar]
- (23).Bokhart MT; Manni J; Garrard KP; Ekelöf M; Nazari M; Muddiman DC IR-MALDESI Mass Spectrometry Imaging at 50 Micron Spatial Resolution. J Am Soc Mass Spectrom 2017, 28 (10), 2099–2107. 10.1007/s13361-017-1740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Joignant AN; Bai H; Manni JG; Muddiman DC Improved Spatial Resolution of Infrared Matrix-assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging Using a Reflective Objective. Rapid Communications in Mass Spectrometry 2022, 36 (23). 10.1002/rcm.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Schoeberl A; Gutmann M; Theiner S; Schaier M; Schweikert A; Berger W; Koellensperger G Cisplatin Uptake in Macrophage Subtypes at the Single-Cell Level by LA-ICP-TOFMS Imaging. Anal. Chem. 2021, 93 (49), 16456–16465. 10.1021/acs.analchem.1c03442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Niehaus M; Soltwisch J; Belov ME; Dreisewerd K Transmission-Mode MALDI-2 Mass Spectrometry Imaging of Cells and Tissues at Subcellular Resolution. Nat. Methods 2019, 16 (9), 925–931. 10.1038/s41592-019-0536-2. [DOI] [PubMed] [Google Scholar]
- (27).Dreisewerd K; Bien T; Soltwisch J MALDI-2 and t-MALDI-2 Mass Spectrometry Imaging. Methods Mol. Biol. 2022, 2437, 21–40. 10.1007/978-1-0716-2030-4_2/FIGURES/7. [DOI] [PubMed] [Google Scholar]
- (28).Brown HM; Pirro V; Graham Cooks R From DESI to the MasSpec Pen: Ambient Ionization Mass Spectrometry for Tissue Analysis and Intrasurgical Cancer Diagnosis. Clin. Chem. 2018, 64 (4), 628–630. 10.1373/CLINCHEM.2017.281923. [DOI] [PubMed] [Google Scholar]
- (29).Zhang J; Sans M; Garza KY; Eberlin LS MASS SPECTROMETRY TECHNOLOGIES TO ADVANCE CARE FOR CANCER PATIENTS IN CLINICAL AND INTRAOPERATIVE USE. Mass Spectrom. Rev. 2021, 40 (5), 692–720. 10.1002/MAS.21664. [DOI] [PubMed] [Google Scholar]
- (30).Tillner J; Wu V; Jones EA; Pringle SD; Karancsi T; Dannhorn A; Veselkov K; McKenzie JS; Takats Z Faster, More Reproducible DESI-MS for Biological Tissue Imaging. J. Am. Soc. Mass Spectrom. 2017, 28 (10), 2090–2098. 10.1007/S13361-017-1714-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Roach PJ; Laskin J; Laskin A Nanospray Desorption Electrospray Ionization: An Ambient Method for Liquid-Extraction Surface Sampling in Mass Spectrometry. Analyst 2010, 135 (9), 2233–2236. 10.1039/C0AN00312C. [DOI] [PubMed] [Google Scholar]
- (32).Ogrinc Potočnik N; Porta T; Becker M; Heeren RMA; Ellis SR Use of Advantageous, Volatile Matrices Enabled by next-Generation High-Speed Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Employing a Scanning Laser Beam. Rapid Commun. Mass Spectrom. 2015, 29 (23), 2195–2203. 10.1002/RCM.7379. [DOI] [PubMed] [Google Scholar]
- (33).Spraggins JM; Caprioli RM High-Speed MALDI-TOF Imaging Mass Spectrometry: Rapid Ion Image Acquisition and Considerations for Next Generation Instrumentation. J Am Soc Mass Spectrom 2011, 22 (6), 1022–1031. 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Prentice BM; Chumbley CW; Caprioli RM High-Speed MALDI MS/MS Imaging Mass Spectrometry Using Continuous Raster Sampling. J. Mass Spectrom. 2015, 50 (4), 703–710. 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bednařík A; Machálková M; Moskovets E; Coufalíková K; Krásenský P; Houška P; Kroupa J; Navrátilová J; Šmarda J; Preisler J MALDI MS Imaging at Acquisition Rates Exceeding 100 Pixels per Second. J. Am. Soc. Mass Spec. 2019, 30 (2), 289–298. 10.1007/S13361-018-2078-8. [DOI] [PubMed] [Google Scholar]
- (36).Vaysse PM; Heeren RMA; Porta T; Balluff B Mass Spectrometry Imaging for Clinical Research – Latest Developments, Applications, and Current Limitations. Analyst 2017, 142 (15), 2690–2712. 10.1039/C7AN00565B. [DOI] [PubMed] [Google Scholar]
- (37).Haag AM Mass Analyzers and Mass Spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. 10.1007/978-3-319-41448-5_7. [DOI] [PubMed] [Google Scholar]
- (38).Spraggins JM; Djambazova KV; Rivera ES; Migas LG; Neumann EK; Fuetterer A; Suetering J; Goedecke N; Ly A; Van De Plas R; Caprioli RM High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (TimsTOF) Mass Spectrometry. Anal. Chem. 2019, 91 (22), 14552–14560. 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Marshall AG; Hendrickson CL; Jackson GS FOURIER TRANSFORM ION CYCLOTRON RESONANCE MASS SPECTROMETRY: A PRIMER. Mass Spectrom. Rev. 1998, 17, 1–35. 10.1002/(SICI)1098-2787(1998)17:1. [DOI] [PubMed] [Google Scholar]
- (40).Basu SS; Agar NYR Bringing Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging to the Clinics - ClinicalKey. Clinics in Laboratory Medicine. https://www.clinicalkey.com/#!/content/playContent/1-s2.0-S0272271221000093?returnurl=null&referrer=null (accessed 2022-09-24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sans M; Feider CL; Eberlin LS Advances in Mass Spectrometry Imaging Coupled to Ion Mobility Spectrometry for Enhanced Imaging of Biological Tissues. Curr. Opin. Chem. Biol. 2018, 42, 138–146. 10.1016/j.cbpa.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rivera ES; Djambazova K. v.; Neumann EK; Caprioli RM; Spraggins JM Integrating Ion Mobility and Imaging Mass Spectrometry for Comprehensive Analysis of Biological Tissues: A Brief Review and Perspective. J. Mass Spectrom. 2020, 55 (12), e4614. 10.1002/JMS.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Feider CL; Elizondo N; Eberlin LS Ambient Ionization and FAIMS Mass Spectrometry for Enhanced Imaging of Multiply Charged Molecular Ions in Biological Tissues. Anal. Chem. 2016, 88 (23), 11533–11541. 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Djambazova K; Klein DR; Migas L; Neumann E; Rivera E; van de Plas R; Caprioli RM; Spraggins J Resolving the Complexity of Spatial Lipidomics with MALDI Trapped Ion Mobility Spectrometry. 2020. 10.26434/CHEMRXIV.12331652.V1. [DOI] [PubMed] [Google Scholar]
- (45).Porta Siegel T; Ekroos K; Ellis SR Reshaping Lipid Biochemistry by Pushing Barriers in Structural Lipidomics. Angew. Chem. Int. Ed. 2019, 58 (20), 6492–6501. 10.1002/anie.201812698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Desorption Electrospray Ionization Coupled with Ultraviolet Photodissociation for Characterization of Phospholipid Isomers in Tissue Sections. Anal. Chem. 2018, 90, 10. 10.1021/acs.analchem.8b03026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kozlowski RL; Mitchell TW; Blanksby SJ A Rapid Ambient Ionization-Mass Spectrometry Approach to Monitoring the Relative Abundance of Isomeric Glycerophospholipids. Sci Rep 2015, 5 (1), 1–9. 10.1038/srep09243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Liu X; Jiao B; Cao W; Ma X; Xia Y; Blanksby SJ; Zhang W; Ouyang Z Development of a Miniature Mass Spectrometry System for Point-of-Care Analysis of Lipid Isomers Based on Ozone-Induced Dissociation. Anal. Chem. 2022, 94, 13944–13950. 10.1021/ACS.ANALCHEM.2C03112. [DOI] [PubMed] [Google Scholar]
- (49).Ellis SR; Bruinen AL; Heeren RMA A Critical Evaluation of the Current State-of-the-Art in Quantitative Imaging Mass Spectrometry. Anal Bioanal Chem 2014, 406 (5), 1275–1289. 10.1007/S00216-013-7478-9/FIGURES/5. [DOI] [PubMed] [Google Scholar]
- (50).Dewez F; de Pauw E; Heeren RMA; Balluff B Multilabel Per-Pixel Quantitation in Mass Spectrometry Imaging. Anal Chem 2021, 93 (3), 1393–1400. 10.1021/ACS.ANALCHEM.0C03186/ASSET/IMAGES/LARGE/AC0C03186_0007.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lamont L; Hadavi D; Viehmann B; Flinders B; Heeren RMA; Vreeken RJ; Porta Siegel T Quantitative Mass Spectrometry Imaging of Drugs and Metabolites: A Multiplatform Comparison. Anal Bioanal Chem 2021, 413 (10), 2779–2791. 10.1007/S00216-021-03210-0/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Unsihuay D; Mesa Sanchez D; Laskin J Quantitative Mass Spectrometry Imaging of Biological Systems. 10.1146/annurev-physchem-061020-053416 2021, 72, 307–329. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kertesz V; Cahill JF Spatially Resolved Absolute Quantitation in Thin Tissue by Mass Spectrometry. Anal Bioanal Chem 2021, 413 (10), 2619–2636. 10.1007/S00216-020-02964-3/FIGURES/4. [DOI] [PubMed] [Google Scholar]
- (54).Ma X; Fernández FM Advances in Mass Spectrometry Imaging for Spatial Cancer Metabolomics. Mass Spectrom Rev 2022, e21804. 10.1002/MAS.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Seeley EH; Oppenheimer SR; Mi D; Chaurand P; Caprioli RM Enhancement of Protein Sensitivity for MALDI Imaging Mass Spectrometry after Chemical Treatment of Tissue Sections. J. Am. Soc. Mass Spectrom. 2008, 19 (8), 1069–1077. 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Seeley EH; Caprioli RM Molecular Imaging of Proteins in Tissues by Mass Spectrometry. Proc Natl Acad Sci U S A 2008, 105 (47), 18126–18131. 10.1073/PNAS.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Cornett DS; Reyzer ML; Chaurand P; Caprioli RM MALDI Imaging Mass Spectrometry: Molecular Snapshots of Biochemical Systems. Nat Methods 2007, 4 (10), 828–833. 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- (58).McDowell CT; Lu X; Mehta AS; Angel PM; Drake RR Applications and Continued Evolution of Glycan Imaging Mass Spectrometry. Mass Spectrom Rev 2023, 42 (2), 674–705. 10.1002/MAS.21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Donohoo KB; Wang J; Goli M; Yu A; Peng W; Hakim MA; Mechref Y Advances in Mass Spectrometry-Based Glycomics—An Update Covering the Period 2017–2021. Electrophoresis 2022, 43 (1–2), 119–142. 10.1002/ELPS.202100199. [DOI] [PubMed] [Google Scholar]
- (60).Carver Wong KF; Greatorex RE; Gidman CE; Rahman S; Griffiths RL Surface-Sampling Mass Spectrometry to Study Proteins and Protein Complexes. Essays Biochem 2023, 67 (2), 229–241. 10.1042/EBC20220191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Caprioli RM; Farmer TB; Gile J Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69 (23), 4751–4760. 10.1021/AC970888I. [DOI] [PubMed] [Google Scholar]
- (62).Stoeckli M; Chaurand P; Hallahan DE; Caprioli RM Imaging Mass Spectrometry: A New Technology for the Analysis of Protein Expression in Mammalian Tissues. Nat. Med. 2001, 7 (4), 493–496. 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- (63).Ryan DJ; Spraggins JM; Caprioli RM Protein Identification Strategies in MALDI Imaging Mass Spectrometry: A Brief Review. Curr Opin Chem Biol 2019, 48, 64–72. 10.1016/J.CBPA.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Yang M; Unsihuay D; Hu H; Nguele Meke F; Qu Z; Zhang ZY; Laskin J Nano-DESI Mass Spectrometry Imaging of Proteoforms in Biological Tissues with High Spatial Resolution. Anal Chem 2023, 95 (12), 5214–5222. 10.1021/ACS.ANALCHEM.2C04795/ASSET/IMAGES/LARGE/AC2C04795_0007.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Judd AM; Gutierrez DB; Moore JL; Patterson NH; Yang J; Romer CE; Norris JL; Caprioli RM A Recommended and Verified Procedure for in Situ Tryptic Digestion of Formalin-Fixed Paraffin-Embedded Tissues for Analysis by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry. Journal of Mass Spectrometry 2019, 54 (8), 716–727. 10.1002/JMS.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Datta S; Malhotra L; Dickerson R; Chaffee S; Sen CK; Roy S Laser Capture Microdissection: Big Data from Small Samples. Histol Histopathol 2015, 30 (11), 1255. 10.14670/HH-11-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kertesz V; Van Berkel GJ Fully Automated Liquid Extraction-Based Surface Sampling and Ionization Using a Chip-Based Robotic Nanoelectrospray Platform. Journal of Mass Spectrometry 2010, 45 (3), 252–260. 10.1002/JMS.1709. [DOI] [PubMed] [Google Scholar]
- (68).Mund A; Brunner AD; Mann M Unbiased Spatial Proteomics with Single-Cell Resolution in Tissues. Mol Cell 2022, 82 (12), 2335–2349. 10.1016/J.MOLCEL.2022.05.022. [DOI] [PubMed] [Google Scholar]
- (69).Zhu Y; Piehowski PD; Zhao R; Chen J; Shen Y; Moore RJ; Shukla AK; Petyuk VA; Campbell-Thompson M; Mathews CE; Smith RD; Qian WJ; Kelly RT Nanodroplet Processing Platform for Deep and Quantitative Proteome Profiling of 10–100 Mammalian Cells. Nature Communications 2018 9:1 2018, 9 (1), 1–10. 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Dey P Fixation of Histology Samples: Principles, Methods and Types of Fixatives. Basic and Advanced Laboratory Techniques in Histopathology and Cytology 2018, 3–17. 10.1007/978-981-10-8252-8_1. [DOI] [Google Scholar]
- (71).Truong JXM; Spotbeen X; White J; Swinnen JV; Butler LM; Snel MF; Trim PJ Removal of Optimal Cutting Temperature (O.C.T.) Compound from Embedded Tissue for MALDI Imaging of Lipids. Anal. and Bioanal. Chem. 2021, 413 (10), 2695–2708. 10.1007/S00216-020-03128-Z/FIGURES/6. [DOI] [PubMed] [Google Scholar]
- (72).Porta Siegel T; Hamm G; Bunch J; Cappell J; Fletcher JS; Schwamborn K Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. Springer New York LLC; December 1, 2018, pp 888–901. 10.1007/s11307-018-1267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Khaitan T; Tomar Bhattacharya P PRINCIPLE AND TECHNIQUES OF IMMUNOHISTOCHEMISTRY-A REVIEW. J. Biol. Med. Res 2015, 6 (3), 5204–5210. [Google Scholar]
- (74).Levsky JM; Singer RH Fluorescence in Situ Hybridization: Past, Present and Future. J. Cell Sci. 2003, 116 (14), 2833–2838. 10.1242/JCS.00633. [DOI] [PubMed] [Google Scholar]
- (75).Patterson NH; Tuck M; Van De Plas R; Caprioli RM Advanced Registration and Analysis of MALDI Imaging Mass Spectrometry Measurements through Autofluorescence Microscopy. Anal. Chem. 2018, 90 (21), 12395–12403. 10.1021/ACS.ANALCHEM.8B02884. [DOI] [PubMed] [Google Scholar]
- (76).Race AM; Sutton D; Hamm G; Maglennon G; Morton JP; Strittmatter N; Campbell A; Sansom OJ; Wang Y; Takáts Z; Barry ST; Goodwin RJA; Bunch J Deep Learning-Based Annotation Transfer between Molecular Imaging Modalities: An Automated Workflow for Multimodal Data Integration. Anal. Chem. 2021, 93 (6), 3061–3071. 10.1021/ACS.ANALCHEM.0C02726. [DOI] [PubMed] [Google Scholar]
- (77).Van De Plas R; Yang J; Spraggins J; Caprioli RM Image Fusion of Mass Spectrometry and Microscopy: A Multimodality Paradigm for Molecular Tissue Mapping. Nat. Methods 2015, 12 (4), 366–372. 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Baratloo A; Hosseini M; Negida A; Ashal G. El. Part 1: Simple Definition and Calculation of Accuracy, Sensitivity and Specificity. Emergency 2015, 3 (2), 48–49. [PMC free article] [PubMed] [Google Scholar]
- (79).Neumann EK; Djambazova KV; Caprioli RM; Spraggins JM Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine. J. Am. Soc. Mass Spectrom. 2020, 31 (12), 2401–2415. 10.1021/JASMS.0C00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Verbeeck N; Caprioli RM; Van de Plas R Unsupervised Machine Learning for Exploratory Data Analysis in Imaging Mass Spectrometry. Mass Spectrom. Rev. 2020, 39 (3), 245–291. 10.1002/MAS.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Tideman LEM; Migas LG; Djambazova K. v.; Patterson NH; Caprioli RM; Spraggins JM; van de Plas R Automated Biomarker Candidate Discovery in Imaging Mass Spectrometry Data through Spatially Localized Shapley Additive Explanations. Anal. Chim. Acta 2021, 1177, 338522. 10.1016/J.ACA.2021.338522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Bollwein C; Gonҫalves JPL; Utpatel K; Weichert W; Schwamborn K MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree. Mol. 2022, 27 (11), 3464. 10.3390/MOLECULES27113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gonçalves JPL; Bollwein C; Schlitter AM; Kriegsmann M; Jacob A; Weichert W; Schwamborn K MALDI-MSI: A Powerful Approach to Understand Primary Pancreatic Ductal Adenocarcinoma and Metastases. Mol. 2022, 27 (15), 4811. 10.3390/MOLECULES27154811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Deininger S-O; Bollwein C; Casadonte R; Wandernoth P; Gonçalves JPL; Kriegsmann K; Kriegsmann M; Boskamp T; Kriegsmann J; Weichert W; Schirmacher P; Ly A; Schwamborn K Multicenter Evaluation of Tissue Classification by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2022, 94 (23), 8194–8201. 10.1021/acs.analchem.2c00097. [DOI] [PubMed] [Google Scholar]
- (85).Prasad M; Postma G; Franceschi P; Buydens LMC; Jansen JJ Evaluation and Comparison of Unsupervised Methods for the Extraction of Spatial Patterns from Mass Spectrometry Imaging Data (MSI). Sci. Rep. 2022, 12 (1), 1–12. 10.1038/s41598-022-19365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Bokhart MT; Nazari M; Garrard KP; Muddiman DC MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J Am Soc Mass Spectrom 2018, 29 (1), 8–16. 10.1007/S13361-017-1809-6/ASSET/IMAGES/LARGE/JS8B05659_0005.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Alexandrov T; Ovchinnikova K; Palmer A; Kovalev V; Tarasov A; Stuart L; Nigmetzianov R; Fay D; contributors KM; Gaudin M; Lopez CG; Vetter M; Swales J; Bokhart M; Kompauer M; McKenzie J; Rappez L; Velickovic D; Lavigne R; Zhang G; Thinagaran D; Ruhland E; Sans M; Triana S; Sammour DA; Aboulmagd S; Bagger C; Strittmatter N; Rigopoulos A; Gemperline E; Joensen AM; Geier B; Quiason C; Weaver E; Prasad M; Balluff B; Nagornov K; Li L; Linscheid M; Hopf C; Heintz D; Liebeke M; Spengler B; Boughton B; Janfelt C; Sharma K; Pineau C; Anderton C; Ellis S; Becker M; Pánczél J; Violante G. Da; Muddiman D; Goodwin R; Eberlin L; Takats Z; Shahidi-Latham S METASPACE: A Community-Populated Knowledge Base of Spatial Metabolomes in Health and Disease. bioRxiv 2019, 539478. 10.1101/539478. [DOI] [Google Scholar]
- (88).Bemis KD; Harry A; Eberlin LS; Ferreira C; Van De Ven SM; Mallick P; Stolowitz M; Vitek O Cardinal: An R Package for Statistical Analysis of Mass Spectrometry-Based Imaging Experiments. Bioinformatics 2015, 31 (14), 2418–2420. 10.1093/BIOINFORMATICS/BTV146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Gibb S; Strimmer K MALDIquant: A Versatile R Package for the Analysis of Mass Spectrometry Data. Bioinformatics 2012, 28 (17), 2270–2271. 10.1093/BIOINFORMATICS/BTS447. [DOI] [PubMed] [Google Scholar]
- (90).Zhang Y; Wang X; Li M; Guo T; Zhao Z; Zhang X; Zhang Y; Liu K An Easy-to-Use Graphical User Interface for Mass Spectrometry Imaging Analysis. Int J Mass Spectrom 2023, 492, 117105. 10.1016/J.IJMS.2023.117105. [DOI] [Google Scholar]
- (91).Schmid R; Heuckeroth S; Korf A; Smirnov A; Myers O; Dyrlund TS; Bushuiev R; Murray KJ; Hoffmann N; Lu M; Sarvepalli A; Zhang Z; Fleischauer M; Dührkop K; Wesner M; Hoogstra SJ; Rudt E; Mokshyna O; Brungs C; Ponomarov K; Mutabdžija L; Damiani T; Pudney CJ; Earll M; Helmer PO; Fallon TR; Schulze T; Rivas-Ubach A; Bilbao A; Richter H; Nothias LF; Wang M; Orešič M; Weng JK; Böcker S; Jeibmann A; Hayen H; Karst U; Dorrestein PC; Petras D; Du X; Pluskal T Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3. Nature Biotechnology 2023 41:4 2023, 41 (4), 447–449. 10.1038/s41587-023-01690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Balluff B; Heeren RMA; Race AM An Overview of Image Registration for Aligning Mass Spectrometry Imaging with Clinically Relevant Imaging Modalities. Journal of Mass Spectrometry and Advances in the Clinical Lab 2022, 23, 26–38. 10.1016/J.JMSACL.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Hu H; Laskin J Emerging Computational Methods in Mass Spectrometry Imaging. Advanced Science 2022, 9 (34), 2203339. 10.1002/ADVS.202203339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Santilli AML; Ren K; Oleschuk R; Kaufmann M; Rudan J; Fichtinger G; Mousavi P Application of Intraoperative Mass Spectrometry and Data Analytics for Oncological Margin Detection, A Review. IEEE Trans Biomed Eng 2022. 10.1109/TBME.2021.3139992. [DOI] [PubMed] [Google Scholar]
- (95).Schulz S; Becker M; Groseclose MR; Schadt S; Hopf C Advanced MALDI Mass Spectrometry Imaging in Pharmaceutical Research and Drug Development. Curr Opin Biotechnol 2019, 55, 51–59. 10.1016/J.COPBIO.2018.08.003. [DOI] [PubMed] [Google Scholar]
- (96).Granborg JR; Handler AM; Janfelt C Mass Spectrometry Imaging in Drug Distribution and Drug Metabolism Studies – Principles, Applications and Perspectives. TrAC Trends in Analytical Chemistry 2022, 146, 116482. 10.1016/J.TRAC.2021.116482. [DOI] [Google Scholar]
- (97).Patel R MALDI-TOF MS for the Diagnosis of Infectious Diseases. Clin. Chem. 2015, 61 (1), 100–111. 10.1373/CLINCHEM.2014.221770. [DOI] [PubMed] [Google Scholar]
- (98).Wieser A; Schneider L; Jung J; Schubert S MALDI-TOF MS in Microbiological Diagnostics-Identification of Microorganisms and beyond (Mini Review). Appl. Microbiol. Biotechnol. 2012, 93 (3), 965–974. 10.1007/S00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- (99).Smith AB; Jenior ML; Keenan O; Hart JL; Specker J; Abbas A; Rangel PC; Di C; Green J; Bustin KA; Gaddy JA; Nicholson MR; Laut C; Kelly BJ; Matthews ML; Evans DR; Van Tyne D; Furth EE; Papin JA; Bushman FD; Erlichman J; Baldassano RN; Silverman MA; Dunny GM; Prentice BM; Skaar EP; Zackular JP Enterococci Enhance Clostridioides Difficile Pathogenesis. Nature 2022 611:7937 2022, 611 (7937), 780–786. 10.1038/s41586-022-05438-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Cassat JE; Moore JL; Wilson KJ; Stark Z; Prentice BM; Van De Plas R; Perry WJ; Zhang Y; Virostko J; Colvin DC; Rose KL; Judd AM; Reyzer ML; Spraggins JM; Grunenwald CM; Gore JC; Caprioli RM; Skaar EP Integrated Molecular Imaging Reveals Tissue Heterogeneity Driving Host-Pathogen Interactions. Sci Transl Med 2018, 10 (432). 10.1126/SCITRANSLMED.AAN6361/SUPPL_FILE/AAN6361_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Capitoli G; Piga I; Galimberti S; Leni D; Pincelli AI; Garancini M; Clerici F; Mahajneh A; Brambilla V; Smith A; Magni F; Pagni F MALDI-MSI as a Complementary Diagnostic Tool in Cytopathology: A Pilot Study for the Characterization of Thyroid Nodules. Cancers (Basel) 2019, 11 (9), 1377. 10.3390/CANCERS11091377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Ochoa-Rios S; O’connor IP; Kent LN; Clouse JM; Hadjiyannis Y; Koivisto C; Pecot T; Angel PM; Drake RR; Leone G; Mehta AS; Rockey DC Imaging Mass Spectrometry Reveals Alterations in N-Linked Glycosylation That Are Associated With Histopathological Changes in Nonalcoholic Steatohepatitis in Mouse and Human In Brief. Mol. Cell Proteomics 2022, 21 (5), 100225. 10.1016/j.mcpro.2022.100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Sommella E; Salviati E; Caponigro V; Grimaldi M; Musella S; Bertamino A; Cacace L; Palladino R; Di Mauro G; Marini F; D’ursi AM; Campiglia P MALDI Mass Spectrometry Imaging Highlights Specific Metabolome and Lipidome Profiles in Salivary Gland Tumor Tissues. Metabolites 2022, 12 (6), 530. 10.3390/METABO12060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Janßen C; Boskamp T; Hauberg-Lotte L; Behrmann J; Deininger SO; Kriegsmann M; Kriegsmann K; Steinbuß G; Winter H; Muley T; Casadonte R; Kriegsmann J; Maaß P Robust Subtyping of Non-Small Cell Lung Cancer Whole Sections through MALDI Mass Spectrometry Imaging. Proteom. Clin. Appl. 2022, 16 (4), 2100068. 10.1002/PRCA.202100068. [DOI] [PubMed] [Google Scholar]
- (105).Calligaris D; Feldman DR; Norton I; Olubiyi O; Changelian AN; Machaidze R; Vestal ML; Laws ER; Dunn IF; Santagata S; Agar NYR MALDI Mass Spectrometry Imaging Analysis of Pituitary Adenomas for Near-Real-Time Tumor Delineation. Proc. Natl. Acad. Sci. 2015, 112 (32), 9978–9983. 10.1073/PNAS.1423101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Yang J; Caprioli RM Matrix Pre-Coated Targets for High Throughput MALDI Imaging of Proteins. Journal of Mass Spectrometry 2014, 49 (5), 417–422. 10.1002/JMS.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Basu SS; Regan MS; Randall EC; Abdelmoula WM; Clark AR; Gimenez-Cassina Lopez B; Cornett DS; Haase A; Santagata S; Agar NYR Rapid MALDI Mass Spectrometry Imaging for Surgical Pathology. NPJ Precis Oncol 2019, 3 (17). 10.1038/s41698-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Mikolasch TA; Oballa E; Vahdati-Bolouri M; Jarvis E; Cui Y; Cahn A; Terry RL; Sahota J; Thakrar R; Marshall P; Porter JC Mass Spectrometry Detection of Inhaled Drug in Distal Fibrotic Lung. Respir Res 2022, 23 (1), 118. 10.1186/S12931-022-02026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Garza KY; Feider CL; Klein DR; Rosenberg JA; Brodbelt JS; Eberlin LS Desorption Electrospray Ionization Mass Spectrometry Imaging of Proteins Directly from Biological Tissue Sections. Anal. Chem. 2018, 90 (13), 7785–7789. 10.1021/ACS.ANALCHEM.8B00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Eberlin LS; Tibshirani RJ; Zhang J; Longacre TA; Berry GJ; Bingham DB; Nortone JA; Zare RN; Poultsides GA Molecular Assessment of Surgical-Resection Margins of Gastric Cancer by Mass-Spectrometric Imaging. Proc. Natl. Acad. Sci. 2014, 111 (7), 2436–2441. 10.1073/PNAS.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Margulis K; Chiou AS; Aasi SZ; Tibshirani RJ; Tang JY; Zare RN Distinguishing Malignant from Benign Microscopic Skin Lesions Using Desorption Electrospray Ionization Mass Spectrometry Imaging. Proc. Natl. Acad. Sci. 2018, 115 (25), 6347–6352. 10.1073/PNAS.1803733115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Santoro AL; Drummond RD; Silva IT; Ferreira SS; Juliano L; Vendramini PH; da Costa Lemos MB; Eberlin MN; Andrade VP In Situ Desi-MSI Lipidomic Profiles of Breast Cancer Molecular Subtypes and Precursor Lesions. Cancer Res. 2020, 80 (6), 1246–1257. 10.1158/0008-5472.CAN-18-3574. [DOI] [PubMed] [Google Scholar]
- (113).Vijayalakshmi K; Shankar V; Bain RM; Nolley R; Sonn GA; Kao CS; Zhao H; Tibshirani R; Zare RN; Brooks JD Identification of Diagnostic Metabolic Signatures in Clear Cell Renal Cell Carcinoma Using Mass Spectrometry Imaging. Int J Cancer 2020, 147 (1), 256–265. 10.1002/IJC.32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Yang X; Song X; Zhang X; Shankar V; Wang S; Yang Y; Chen S; Zhang L; Ni Y; Zare RN; Hu Q In Situ DESI-MSI Lipidomic Profiles of Mucosal Margin of Oral Squamous Cell Carcinoma. EBioMedicine 2021, 70, 103529. 10.1016/J.EBIOM.2021.103529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Eberlin LS; Margulis K; Planell-Mendez I; Zare RN; Tibshirani R; Longacre TA; Jalali M; Norton JA; Poultsides GA Pancreatic Cancer Surgical Resection Margins: Molecular Assessment by Mass Spectrometry Imaging. PLoS Med. 2016, 13 (8), e1002108. 10.1371/JOURNAL.PMED.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Pirro V; Alfaro CM; Jarmusch AK; Hattab EM; Cohen-Gadol AA; Cooks RG Intraoperative Assessment of Tumor Margins during Glioma Resection by Desorption Electrospray Ionization-Mass Spectrometry. Proc. Natl. Acad. Sci. 2017, 114 (26), 6700–6705. 10.1073/PNAS.1706459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Brown HM; Alfaro CM; Pirro V; Dey M; Hattab EM; Cohen-Gadol AA; Cooks RG Intraoperative Mass Spectrometry Platform for IDH Mutation Status Prediction, Glioma Diagnosis, and Estimation of Tumor Cell Infiltration. J. Appl. Lab. Med. 2021, 6 (4), 902–916. 10.1093/JALM/JFAA233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Cardoso-Palacios C; Lanekoff I Direct Analysis of Pharmaceutical Drugs Using Nano-DESI MS. J Anal Methods Chem 2016, 2016. 10.1155/2016/3591908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Mesa Sanchez D; Brown HM; Yin R; Chen B; Vavrek M; Cancilla MT; Zhong W; Shyong BJ; Zhang NR; Li F; Laskin J Mass Spectrometry Imaging of Diclofenac and Its Metabolites in Tissues Using Nanospray Desorption Electrospray Ionization. Anal Chim Acta 2022, 1233, 340490. 10.1016/J.ACA.2022.340490. [DOI] [PubMed] [Google Scholar]
- (120).Cooper HJ; Hale OJ Native Mass Spectrometry Imaging of Proteins and Protein Complexes by Nano-Desi. Anal Chem 2021, 93 (10), 4619–4627. 10.1021/ACS.ANALCHEM.0C05277/ASSET/IMAGES/LARGE/AC0C05277_0006.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Yang M; Hu H; Su P; Thomas PM; Camarillo JM; Greer JB; Early BP; Fellers RT; Kelleher NL; Laskin J Proteoform-Selective Imaging of Tissues Using Mass Spectrometry**. Angewandte Chemie International Edition 2022, 61 (29), e202200721. 10.1002/ANIE.202200721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Chung H; Huang P; Chen C; Lee C; Hsu C Next-generation Pathology Practices with Mass Spectrometry Imaging. Mass Spectrom Rev 2022. 10.1002/mas.21795. [DOI] [PubMed] [Google Scholar]
- (123).Su P; McGee JP; Durbin KR; Hollas MAR; Yang M; Neumann EK; Allen JL; Drown BS; Butun FA; Greer JB; Early BP; Fellers RT; Spraggins JM; Laskin J; Camarillo JM; Kafader JO; Kelleher NL Highly Multiplexed, Label-Free Proteoform Imaging Oftissues by Individual Ion Mass Spectrometry. Sci Adv 2022, 8 (32), 9929. 10.1126/SCIADV.ABP9929/SUPPL_FILE/SCIADV.ABP9929_TABLES_S1_TO_S4.ZIP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Zhang J; Rector J; Lin JQ; Young JH; Sans M; Katta N; Giese N; Yu W; Nagi C; Suliburk J; Liu J; Bensussan A; Dehoog RJ; Garza KY; Ludolph B; Sorace AG; Syed A; Zahedivash A; Milner TE; Eberlin LS Nondestructive Tissue Analysis for Ex Vivo and in Vivo Cancer Diagnosis Using a Handheld Mass Spectrometry System. Sci. Trans. Med. 2017, 9 (406), eean3968. 10.1126/scitranslmed.aan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Zhang J; Sans M; DeHoog RJ; Garza KY; King ME; Feider CL; Bensussan A; Keating MF; Lin JQ; Povilaitis SC; Katta N; Milner TE; Yu W; Nagi C; Dhingra S; Pirko C; Brahmbhatt KA; Van Buren G; Carter S; Thompson A; Grogan RH; Suliburk J; Eberlin LS Clinical Translation and Evaluation of a Handheld and Biocompatible Mass Spectrometry Probe for Surgical Use. Clin. Chem. 2021, 67 (9), 1271–1280. 10.1093/CLINCHEM/HVAB098. [DOI] [PubMed] [Google Scholar]
- (126).HuBMAP consortium. The Human Body at Cellular Resolution: The NIH Human Biomolecular Atlas Program. Nature 2019, 574 (7777), 187–192. 10.1038/S41586-019-1629-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Rozenblatt-Rosen O; Regev A; Oberdoerffer P; Nawy T; Hupalowska A; Rood JE; Ashenberg O; Cerami E; Coffey RJ; Demir E; Ding L; Esplin ED; Ford JM; Goecks J; Ghosh S; Gray JW; Guinney J; Hanlon SE; Hughes SK; Hwang ES; Iacobuzio-Donahue CA; Jané-Valbuena J; Johnson BE; Lau KS; Lively T; Mazzilli SA; Pe’er D; Santagata S; Shalek AK; Schapiro D; Snyder MP; Sorger PK; Spira AE; Srivastava S; Tan K; West RB; Williams EH; Aberle D; Achilefu SI; Ademuyiwa FO; Adey AC; Aft RL; Agarwal R; Aguilar RA; Alikarami F; Allaj V; Amos C; Anders RA; Angelo MR; Anton K; Aster JC; Babur O; Bahmani A; Balsubramani A; Barrett D; Beane J; Bender DE; Bernt K; Berry L; Betts CB; Bletz J; Blise K; Boire A; Boland G; Borowsky A; Bosse K; Bott M; Boyden E; Brooks J; Bueno R; Burlingame EA; Cai Q; Campbell J; Caravan W; Chaib H; Chan JM; Chang YH; Chatterjee D; Chaudhary O; Chen AA; Chen B; Chen C; Chen C. hui; Chen F; Chen YA; Chheda MG; Chin K; Chiu R; Chu SK; Chuaqui R; Chun J; Cisneros L; Colditz GA; Cole K; Collins N; Contrepois K; Coussens LM; Creason AL; Crichton D; Curtis C; Davidsen T; Davies SR; de Bruijn I; Dellostritto L; De Marzo A; DeNardo DG; Diep D; Diskin S; Doan X; Drewes J; Dubinett S; Dyer M; Egger J; Eng J; Engelhardt B; Erwin G; Esserman L; Felmeister A; Feiler HS; Fields RC; Fisher S; Flaherty K; Flournoy J; Fortunato A; Frangieh A; Frye JL; Fulton RS; Galipeau D; Gan S; Gao J; Gao L; Gao P; Gao VR; Geiger T; George A; Getz G; Giannakis M; Gibbs DL; Gillanders WE; Goedegebuure SP; Gould A; Gowers K; Greenleaf W; Gresham J; Guerriero JL; Guha TK; Guimaraes AR; Gutman D; Hacohen N; Hanlon S; Hansen CR; Harismendy O; Harris KA; Hata A; Hayashi A; Heiser C; Helvie K; Herndon JM; Hirst G; Hodi F; Hollmann T; Horning A; Hsieh JJ; Hughes S; Huh WJ; Hunger S; Hwang SE; Ijaz H; Izar B; Jacobson CA; Janes S; Jayasinghe RG; Jiang L; Johnson BE; Johnson B; Ju T; Kadara H; Kaestner K; Kagan J; Kalinke L; Keith R; Khan A; Kibbe W; Kim AH; Kim E; Kim J; Kolodzie A; Kopytra M; Kotler E; Krueger R; Krysan K; Kundaje A; Ladabaum U; Lake BB; Lam H; Laquindanum R; Laughney AM; Lee H; Lenburg M; Leonard C; Leshchiner I; Levy R; Li J; Lian CG; Lim KH; Lin JR; Lin Y; Liu Q; Liu R; Longabaugh WJR; Longacre T; Ma CX; Macedonia MC; Madison T; Maher CA; Maitra A; Makinen N; Makowski D; Maley C; Maliga Z; Mallo D; Maris J; Markham N; Marks J; Martinez D; Mashl RJ; Masilionais I; Mason J; Massagué J; Massion P; Mattar M; Mazurchuk R; Mazutis L; McKinley ET; McMichael JF; Merrick D; Meyerson M; Miessner JR; Mills GB; Mills M; Mondal SB; Mori M; Mori Y; Moses E; Mosse Y; Muhlich JL; Murphy GF; Navin NE; Nederlof M; Ness R; Nevins S; Nikolov M; Nirmal AJ; Nolan G; Novikov E; O’Connell B; Offin M; Oh ST; Olson A; Ooms A; Ossandon M; Owzar K; Parmar S; Patel T; Patti GJ; Pe’er I; Peng T; Persson D; Petty M; Pfister H; Polyak K; Pourfarhangi K; Puram SV; Qiu Q; Quintanal-Villalonga Á; Raj A; Ramirez-Solano M; Rashid R; Reeb AN; Reid M; Resnick A; Reynolds SM; Riesterer JL; Rodig S; Roland JT; Rosenfield S; Rotem A; Roy S; Rudin CM; Ryser MD; Santi-Vicini M; Sato K; Schrag D; Schultz N; Sears CL; Sears RC; Sen S; Sen T; Shalek A; Sheng J; Sheng Q; Shoghi KI; Shrubsole MJ; Shyr Y; Sibley AB; Siex K; Simmons AJ; Singer DS; Sivagnanam S; Slyper M; Sokolov A; Song SK; Southard-Smith A; Spira A; Stein J; Storm P; Stover E; Strand SH; Su T; Sudar D; Sullivan R; Surrey L; Suvà M; Terekhanova NV; Ternes L; Thammavong L; Thibault G; Thomas GV; Thorsson V; Todres E; Tran L; Tyler M; Uzun Y; Vachani A; Van Allen E; Vandekar S; Veis DJ; Vigneau S; Vossough A; Waanders A; Wagle N; Wang LB; Wendl MC; West R; Wu C. yun; Wu H; Wu HY; Wyczalkowski MA; Xie Y; Yang X; Yapp C; Yu W; Yuan Y; Zhang D; Zhang K; Zhang M; Zhang N; hang Y; Zhao Y; Zhou DC; Zhou Z; Zhu H; Zhu Q; Zhu X; Zhu Y; Zhuang X The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 2020, 181 (2), 236–249. 10.1016/J.CELL.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).de Boer IH; Alpers CE; Azeloglu EU; Balis UGJ; Barasch JM; Barisoni L; Blank KN; Bomback AS; Brown K; Dagher PC; Dighe AL; Eadon MT; El-Achkar TM; Gaut JP; Hacohen N; He Y; Hodgin JB; Jain S; Kellum JA; Kiryluk K; Knight R; Laszik ZG; Lienczewski C; Mariani LH; McClelland RL; Menez S; Moledina DG; Mooney S; O’Toole JF; Palevsky PM; Parikh CR; Poggio ED; Rosas SE; Rosengart MR; Sarwal MM; Schaub JA; Sedor JR; Sharma K; Steck B; Toto RD; Troyanskaya OG; Tuttle KR; Vazquez MA; Waikar SS; Williams K; Wilson FP; Zhang K; Iyengar R; Kretzler M; Himmelfarb J; Lecker S; Stillman I; Waikar S; Mcmahon G; Weins A; Short S; Hoover P; Aulisio M; Cooperman L; Herlitz L; Poggio E; Jolly S; Appelbaum P; Balderes O; Barasch J; Bomback A; Canetta PA; d’Agati VD; Kudose S; Mehl K; Radhakrishnan J; Weng C; Alexandrov T; Ashkar T; Barwinska D; Dagher P; Dunn K; Eadon M; Ferkowicz M; Kelly K; Sutton T; Winfree S; Parikh C; Rosenberg A; Villalobos P; Malik R; Fine D; Atta M; Monroy Trujillo JM; Slack A; Rosas S; Williams M; Azeloglu E; He C. (John); Hansen J; Parikh S; Rovin B; Anderton C; Pasa-Tolic L; Velickovic D; Lukowski J; Oliver G. (Holt); Ardayfio J; Bebiak J; Campbell T; Campbell C; Hayashi L; Jefferson N; Koewler R; Roberts G; Saul J; Shpigel A; Stutzke EC; Wright L; Miegs L; Pinkeney R; Sealfon R; Troyanskaya O; Tuttle K; Dobi D; Goltsev Y; Lake B; Joanes M; Laszik Z; Schroeder A; Sarwal M; Sigdel T; Balis U; Blanc V; He O; Hodgin J; Mariani L; Menon R; Otto E; Schaub J; Eddy S; Elder M; Hall D; Kruth M; Murugan R; Palevsky P; Randhawa P; Rosengart M; Sims-Lucas S; Stefanick M; Stull S; Tublin M; Alpers C; de Boer I; Dighe A; Mcclelland R; Shankland S; Blank K; Carson J; Dowd F; Drager Z; Park C; Zhang G; Bansal S; Venkatachalam M; Kermani A; Lee S; Lu C; Miller T; Moe O; Park H; Sambandam K; Sanchez F; Torrealba J; Robert T; Vazquez M; Wang N; Vijayan A; Luciano R; Moledina D; Ugochukwu U; Alfano S Rationale and Design of the Kidney Precision Medicine Project. Kidney Int. 2021, 99 (3), 498–510. 10.1016/J.KINT.2020.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]


