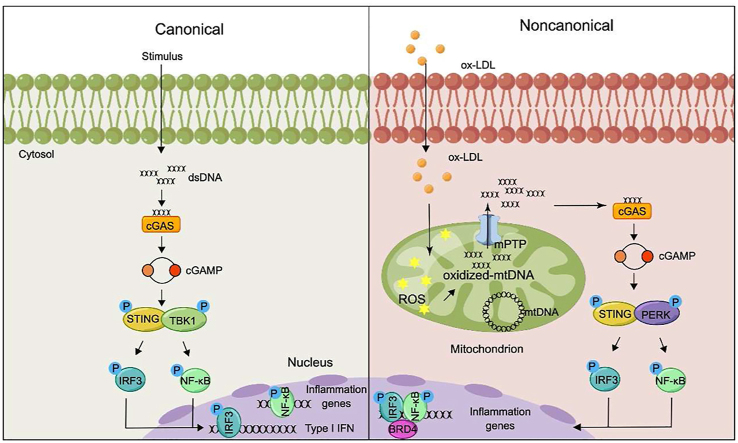
Abstract
Inflammation-driven endothelial dysfunction is the major initiating factor in atherosclerosis, while the underlying mechanism remains elusive. Here, we report that the non-canonical stimulator of interferon genes (STING)–PKR-like ER kinase (PERK) pathway was significantly activated in both human and mice atherosclerotic arteries. Typically, STING activation leads to the activation of interferon regulatory factor 3 (IRF3) and nuclear factor-kappa B (NF-κB)/p65, thereby facilitating IFN signals and inflammation. In contrast, our study reveals the activated non-canonical STING–PERK pathway increases scaffold protein bromodomain protein 4 (BRD4) expression, which encourages the formation of super-enhancers on the proximal promoter regions of the proinflammatory cytokines, thereby enabling the transactivation of these cytokines by integrating activated IRF3 and NF-κB via a condensation process. Endothelium-specific STING and BRD4 deficiency significantly decreased the plaque area and inflammation. Mechanistically, this pathway is triggered by leaked mitochondrial DNA (mtDNA) via mitochondrial permeability transition pore (mPTP), formed by voltage-dependent anion channel 1 (VDAC1) oligomer interaction with oxidized mtDNA upon cholesterol oxidation stimulation. Especially, compared to macrophages, endothelial STING activation plays a more pronounced role in atherosclerosis. We propose a non-canonical STING–PERK pathway-dependent epigenetic paradigm in atherosclerosis that integrates IRF3, NF-κB and BRD4 in inflammatory responses, which provides emerging therapeutic modalities for vascular endothelial dysfunction.
Key words: Endothelial dysfunction, Inflammation, Mitochondrial DNA, STING, ROS, PERK, BRD4, Atherosclerosis
Graphical abstract
Non-canonical STING–PERK pathway dependent epigenetic regulation of vascular endothelial dysfunction promotes atherosclerosis via integrating IRF3, NF-κB and BRD4 in inflammatory response, which shed new light for atherosclerosis therapy.
1. Introduction
Atherosclerosis, a leading cause of cardiovascular morbidity and mortality, emerges as a significant contemporary public health issue1. As a chronic inflammatory disease, endothelial dysfunction is the major initiating factor in the atherogenic process2,3. Therefore, identifying the molecular mechanism underlying endothelial dysfunction is an essential step in developing preventive and therapeutic approaches for atherosclerosis.
The abnormal signal transduction associated with cardiovascular diseases (CVDs) has been linked to inflammatory responses. However, the precise mechanism connecting endothelial dysfunction and inflammation during atherosclerosis remains elusive. Recently, numerous evidence has shown the stimulator of interferon genes (STING), a proinflammatory molecule in the cyclic GMP–AMP (cGAMP) synthase (cGAS)–STING pathway, is a critical signaling molecule in immunity and inflammation4. STING signaling-driven inflammation has been implicated in the pathogenesis of many diseases, such as Parkinson's disease, nonalcoholic steatohepatitis, amyotrophic lateral sclerosis and myocardial infarction5, 6, 7, 8. Shen's group9 has emphasized the critical role of STING in the rupture of aortic aneurysm and dissection, from the aspects of inflammation and destruction. Gain-of-function mutations in STING have also been identified in STING-associated vasculopathy early onset in infancy and familial inflammatory syndrome with lupus-like manifestations10. These findings suggest STING's involvement in vascular disease. However, its role in endothelial dysfunction and atherosclerosis is yet to be determined. By leveraging publicly available microarray data, we found that the expression levels of STING pathway-related genes were elevated in patients with plaques, indicating a potential link between the STING pathway and inflammation in atherosclerosis.
The cGAS–STING pathway is activated by recognizing cytoplasmic foreign double-stranded DNA (dsDNA) from DNA viruses, bacteria, or parasites4,11. Besides, it can also be triggered by self-DNA, including genomic and mitochondrial DNA (mtDNA), to catalyze the synthesis of cGAMP12. Here, we aimed to explore whether dsDNA exists in the cytoplasm of activated endothelium, the source of cytoplasmic DNA, and how dsDNA contributes to endothelial dysfunction and atherosclerosis. The overproduction of cGAMP binds and activates STING to recruit the classical effector tank-binding kinase 1 (TBK1), which mediates the activation of interferon regulatory factor 3 (IRF3) or nuclear factor-kappa B (NF-κB)/p65 to initiate the transcription of type I interferon and proinflammatory cytokines, respectively13. However, some strong phenotypes observed in STING-associated vasculopathy early onset in infancy and lung disease appear to be independent of the classical STING–TBK1–IRF3 axis10,14. Therefore, the role of the non-canonical STING pathway in diseases is still under intensive investigation. Notably, Xu's group15 has recently reported that PKR-like ER kinase (PERK) as the non-canonical effector of STING could be activated to promote senescence and organ fibrosis. In our study, we found that the STING was activated in oxidized cholesterol-challenged endothelial cells without activating TBK1, the canonical effector of STING. These findings prompt us to hypothesize a crucial role of the non-canonical STING pathway in the oxidized cholesterol-induced endothelial injury. Meanwhile, whether STING activates PERK during endothelial dysfunction and atherosclerosis is entirely unknown.
Typically, STING activates IRF3 and NF-κB contributing to IFN signals and inflammation, respectively13. Interestingly, IRF3 colocalizes with NF-κB to the genomic targets to regulate the innate antiviral transcription16. Additionally, genome-bound nuclear NF-κB could form super-enhancers with bromodomain protein 4 (BRD4) to activate inflammatory genes transcription via chromatin remodeling17. BRD4, an important transcriptional regulator, acts as a scaffold for transcription factors via recognizing acetylated histones through bromodomains, to regulate the chromatin landscape and motivate the transactivation of pathologic genes18. Therefore, we propose BRD4 maybe a novel transcriptional regulator in the STING pathway.
Here, we explored the role of the STING pathway in atherosclerosis via human coronary artery endothelial cells (HCAECs) and an endothelium-directed STING or BRD4 knockout mouse on ApoeKO background (ApoeKOStingEC-KO or ApoeKOBrd4EC-KO). Our results revealed that the non-canonical STING-PERK pathway is activated in the aortic endothelium of high-fat diet (HFD)-fed ApoeKO mice by cytoplasmic mtDNA released through mitochondrial permeability transition pore (mPTP) formed by voltage-dependent anion channel 1 (VDAC1). Unlike the canonical mode that STING activates IRF3 and NF-κB contributing to IFN signals and inflammation respectively, the activated STING synergized the interaction between IRF3, NF-κB, and BRD4 in the chromatin microenvironment of regulatory regions of inflammatory cytokines to form super-enhancer via a BRD4-dependent condensation process to precisely orchestrate inflammatory gene expression. Compared to HFD-fed ApoeKOStingWT mice, the ApoeKOStingEC-KO mice presented decreased lipid deposition and inflammation, enhanced plaque stability, and ameliorative endothelium-dependent relaxation ability. These findings have also been validated in clinical samples. Besides, our bone marrow transplantation experiments established that the non-canonical STING–PERK pathway activation in ECs plays a more critical role in atherosclerosis compared to macrophages. We elucidate a novel non-canonical STING–PERK pathway dependent epigenetic paradigm that integrates IRF3, NF-κB and BRD4 in the inflammatory response, which provides emerging therapeutic modalities for ameliorating vascular endothelial dysfunction.
2. Materials and methods
2.1. Cell lines
Human coronary artery endothelial cells (HCAECs) were purchased from ScienCell (USA). HCAECs were cultured in Endothelial Cell Medium (ScienCell, CA, USA) containing 5% fetal bovine serum (FBS) and 1% endothelial cell growth supplement, and 1% penicillin/streptomycin. The HCAECs were treated with oxidative low-density lipoprotein (ox-LDL, 100 μg/mL, 24 h, Peking Union-Biology), lipopolysaccharide (LPS, 1 μg/mL, 24 h, Beyotime), and tumor necrosis factor α (TNFα, 10 ng/mL, 24 h, Beyotime) to establish atherosclerotic endothelial injury. The HCAECs treated with phosphate buffer saline (PBS) were used as the control group. The thiobarbituric acid-reactive substance in ox-LDL is 18 nmol/L malondialdehyde equivalents per milligram of LDL proteins. Mouse aortic endothelial cells (MAECs) were isolated from ApoeKOStingWT and ApoeKOStingEC-KO mice and cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin19. Cells were used for a maximum of six passages. Human monocytic THP-1 cells were cultured in RPMI-1640 medium with 10% FBS supplemented with 20 μg/mL penicillin/streptomycin at 37 °C in a 5% CO2 humidified atmosphere.
2.2. Animal
Sting floxed (Stingfl/fl) mice were generated by Gempharmatech Co., Ltd. (Jiangsu, China). Endothelium-directed Sting knockout hyperlipidemic apolipoprotein e-knockout (ApoeKOStingEC-KO) mice were generated by crossing Stingfl/fl mice with Tek promoter-driven Cre recombinase line (Tek-Cre) on an ApoeKO background. ApoeKO mice without Sting deficiency were employed as control animals (ApoeKOStingWT). ApoeKO mice were purchased from Sippr-BK laboratory animal Co., Ltd. (Shanghai, China). Mice were housed in a specific pathogen-free facility under controlled conditions (temperature, 22 ± 2 °C; relative humidity, 55 ± 15%; noise <60 dB; light/dark cycle, 12 h/12 h) that conform to the Guide for the Care and Use of Laboratory Animals (NIH), and approved by the Animal Care and Use Committee of Nanjing Medical University (IACUC-2105032). An equal number of female and male mice were used for experiments. ApoeKO mice were fed with a normal chow diet (NC) or a HFD (21% fat and 0.15% cholesterol; Xietong Organism lnc., Shanghai, China) for 16 weeks to induce atherosclerosis starting at 8 weeks of age. For the experiment with C-176 (MedChemExpress, NJ, USA), the HFD-induced ApoeKOStingWT mice were treated with a dose of 4 mg/kg intraperitoneally every other day from 8 weeks until 24 weeks. C-176 was dissolved in DMSO at 10 mg/mL and diluted in 5% Tween-80 and 5% PEG-400. For the experiment with VBIT-4 (Selleckchem, USA), the ApoeKOStingWT and ApoeKOStingEC-KO mice were treated with a daily freshly diluted dose of VBIT-4 (20 mg/kg) or vehicle in drinking water from 8 weeks until to 24 weeks. VBIT-4 was freshly dissolved in DMSO and diluted in water (final pH 5.0, DMSO 0.05%). At the end of the experiments, mice were euthanized by inhalation of 2%–3% isoflurane.
Brd4 floxed (Brd4fl/fl) mice were generated by Gempharmatech Co., Ltd. (Jiangsu, China). Endothelium-specific Brd4 knockout hyperlipidemic ApoeKO (ApoeKOBrd4EC-KO) mice were generated by crossing Brd4fl/fl mice with Cdh5 Cre recombinase line (Cdh5-Cre) on an ApoeKO background. ApoeKO mice without Brd4 deficiency were employed as control animals (ApoeKOBrd4WT). The ApoeKOBrd4EC-KO mice (8 weeks old) were injected with endothelial-enhanced adeno-associated virus (AAVendo)-Sting-WT via tail veins leading to the endothelium-specific overexpressing STING. After 4 weeks, the mice were fed with HFD for 16 weeks.
2.3. Bone marrow transplantation
8-week-old recipient mice were maintained on antibiotic water (sulfratrim, 4 μg/mL) for one week followed by lethally irradiated by 9 Gy. On the second day, bone marrow cells were collected by flushing the femurs and tibias of donor mice with sterile PBS. Single-cell suspensions were prepared by passing the cells through a 30 μm nylon gauze. Irradiated recipients were reconstituted with 5 × 106 bone marrow cells by tail vein injection. Mice were maintained on antibiotic water and NC diet for 4 weeks after irradiation. Then, the mice were fed with regular water and HFD for 16 weeks.
2.4. Atherosclerotic lesion analysis
The aortas were perfused with PBS and excised from the proximal aortic arch to the common iliac artery. Then, the connective and adipose tissues were removed from the aorta and the section of the aorta was cut longitudinally and stained with oil red O solution. Finally, the specimens were digitally photographed. The percentage of oil red O-positive area reflecting the extent of lesion development was calculated using Image-Pro Plus software by dividing oil red O-positive areas by the area of the entire aorta.
For histological analysis, the section of aortas was fixed in 4% paraformaldehyde at 4 °C for 24 h, dehydrated in a graded series of alcohol (70%, 80%, 90%, 95% and 100%, each 90 min), embedded in 5-μm thick paraffin. Then, they were stained with hematoxylin & eosin, oil red O, Sirius red and Masson. Images were captured using a light microscope (OLYMPUS BX53; Olympus Corporation) at magnifications, 10 × and 20 ×.
2.5. In vitro permeability assay
The monolayer integrity of HCAECs was assessed by non-fluorescent live cell staining prior to stimulation. Then, the medium was refreshed and appropriate stimuli were added. Next, the endothelial permeability was measured by addition of a FITC-Dextran solution (MedChemExpress, NJ, USA) into the insert and quantification of the permeated FITC-Dextran in the bottom well using a fluorescence plate reader.
2.6. Expression analysis
Total RNA from human HCAECs and indicated mouse arteries was homogenized in Trizol Lysis Reagent (Takara, Japan) according to the manufacturer's instructions and the quality control of samples was carried out by Nanodrop OneC (Thermo Fisher Scientific, Madison, WI, USA). RNA was converted to cDNA using HiScript II Q RT SuperMix for qPCR (Vazyme, Nanjing, China). Quantitative real-time PCRs (qRT-PCRs) were carried out with cDNA in triplicate using AceQ qPCR SYBR Green Master Mix (Vazyme) on QuantStudio 5 Real-Time PCR system (Thermo Fisher Scientific). Relative expression levels of different genes were obtained after normalization with Actin values. Data was analyzed using the 2−ΔΔCT method. The PCR primer sequences are provided in Supporting Information Table S1.
2.7. mtDNA isolation and transfection
The mtDNA from ox-LDL-treated HCAECs was isolated using Mitochondrial DNA isolation kit (Biovision lnc., Mountain view, CA, USA). Following the manufacturer's instructions, 5 × 106 cells were collected by centrifugation at 600 × g for 5 min at 4 °C. Then, the cells were washed with ice-cold PBS and centrifugated at 600 × g for 5 min at 4 °C to remove the supernatant. The cells were resuspended in 1 × cytosol extraction buffer for 10 min on ice. Subsequently, the cells were homogenized by ice-cold tissue grinder and centrifuged at 1200 × g for 10 min at 4 °C to remove the nuclei and intact cells. The supernatant was transferred to a new microcentrifuge tube and centrifuged at 10,000 × g for 30 min at 4 °C. The pellet is resuspended in 1 × cytosol extraction buffer and centrifuged at 10,000 × g for 30 min at 4 °C. The pellet were the mitochondria, which were lysed in mitochondrial lysis buffer for 10 min on ice. The proteins were digested with an enzyme mix (provided in the kit) at 50 °C for at least 2 h. The mtDNA was precipitated with absolute ethanol and stored at –20 °C. For the transfection, the HCAECs were transfected with mtDNA using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer's instructions.
2.8. Mitochondrial DNA-depleted cells
To further confirm the role of redistribution of mtDNA in ox-LDL-treated HCAECs, we constructed mtDNA-depleted HCAECs. The HCAECs were grown in endothelial cell medium supplemented with 50 ng/mL ethidium bromide (EtBr; MedChemExpress, NJ, USA), 100 μg/mL sodium pyruvate, 2 mmol/L l-glutamine, 50 μg/mL uridine and 25 mmol/L HEPES as previously demonstrated20. The depletion was analyzed using qPCR. The primer sequences used are provided in Table S1.
2.9. Quantification of mitochondrial DNA release
After different treatments, the HCAECs (2 × 106) were collected and resuspended in 170 μL digitonin buffer containing 150 mmol/L NaCl, 50 mmol/L HEPES pH 7.4, and 25 μg/mL digitonin (Millipore, USA). Then, they were incubated on a rotator for 10 min at room temperature. After centrifugated at 16,000 × g for 25 min at 4 °C, a 1:20 dilution of the supernatant was used for qPCR. Here, we detected the source of cytoplasmic DNA by nuclei DNA, mitochondrial DNA and ribosomal DNA by corresponding primers provided in Table S1.
2.10. Human specimens
Atherosclerotic and non-atherosclerotic human aorta specimens were obtained from transplant donors at endarterectomy, heart transplantation or coronary artery bypass surgery (Supporting Information Table S2). Written informed consent was obtained from all patients. All procedures involving sampling were performed according to the principles outlined in the Declaration of Helsinki and were approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School (2019-219-01).
2.11. Immunofluorescence analysis
The cells seeded in glass-bottom cell culture dishes were rinsed in PBS followed by fixed in 4% paraformaldehyde and soaked in 0.1% Triton X-100 for 30 min, respectively. After being blocked with 3% bovine serum albumin (BSA) for 1 h, the cells were incubated with specific primary antibodies overnight at 4 °C. For mice, the chest cavity was opened after anesthesia and perfused with 4 °C PBS. Gently stripping away the vascular adventitial fat from the aorta and removing it from the spine subsequently. Then, the heart with the proximal part of the ascending aorta was embedded in OCT and snap frozen at −80 °C. After being sliced into an 8 μm frozen section, the aorta was washed 3 times with PBS and incubated in PBS containing 3% BSA and 0.2% Triton X-100 for 2 h. The slides were incubated with primary antibodies overnight at 4 °C. The cells and slides were then incubated with appropriate TRITC/FITC-conjugated secondary antibodies (diluted in 1% BSA) for 1 h at 37 °C. The cell nuclei were counterstained with DAPI. For en face immunofluorescence staining on the mouse aorta, the thoracic aortas were harvested and fixed in 4% paraformaldehyde. The samples were incubated with primary dsDNA, TOMM20 and intercellular adhesion molecule 1 (ICAM-1) antibody overnight at 4 °C. After washing with PBS, the sample was incubated with appropriate TRITC/FITC-conjugated secondary antibodies (diluted in 1% BSA) for 1 h at 37 °C. The nuclei of aortas were counterstained with DAPI. The sections were viewed under a confocal laser scanning microscope (Oberkochen, Germany).
2.12. Western blotting
HCAECs and isolated arteries from mice or human were lysed on ice in 1 × radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors (Beyotime, Beijing, China). After centrifugation at 4 °C (13,800 × g) for 20 min, protein extract supernatant was collected, followed by denaturalized in 1 × loading buffer at 95 °C for 10 min. Then, samples were separated on SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes (Millipore, USA). Then, the membranes were blocked in 5% skim milk diluted in 1 × Tris-buffered saline containing 0.1% Tween20 (TBST) before overnight incubation at 4 °C with appropriate primary antibodies prepared in 3% BSA (in 1 × TBST) (Supporting Information Table S3). Next, the membranes were washed with 1 × TBST three times before incubation with HRP-conjugated secondary antibodies (prepared in 1 × TBST) for 1 h at room temperature and developed using the ECL detection system (Thermo Scientific Pierce, Life Technologies). Fresh second antibody solutions were used for each experiment. Densitometric analysis of bands of target proteins and loading controls (β-actin) was performed with ImageJ software. Intensity values of bands representing phosphorylated sites of proteins were normalized to the intensity of the band representing total protein. All samples were subjected to SDS-PAGE separation and Western blotting with the indicated antibodies.
2.13. Co-immunoprecipitation
After different treatments, the HCAECs were lysed in lysis buffer (Beyotime, Beijing, China) and placed on a low-speed rotating shaker for 30 min at 4 °C. After centrifuged at 12,000 × g for 10 min, 50 μL of the whole cell lysates were loaded as input sample and the remaining cell lysates were incubated with the indicated antibody with protein A/G agarose beads at 4 °C overnight. The next day, beads were washed with lysis buffer for 5 times and boiled at 95 °C for 10 min. All samples were subjected to SDS-PAGE separation and Western blotting with the indicated antibodies.
2.14. Chromatin immunoprecipitation (ChIP) and Re-chromatin immunoprecipitation (Re-ChIP)
After different treatments, the cells were cross-linked in 1% formaldehyde for 10 min at room temperature and quenched in 0.125 mol/L glycine for 5 min. Then, the cells were collected by centrifugation and sonicated in SDS lysis buffer to generate ∼500 bp chromatin fragments. After centrifugated at 4 °C for 10 min, 20 μL supernatant was used as the input for quantitation. The left supernatant was incubated with the indicated antibody protein A-agarose (Santa, USA) slurry at 4 °C. The Txn stop buffer with 0.4 mg/mL glycogen and proteinase K (0.45 mg/mL, Roche, USA) was used to digest protein in immunoprecipitated DNA at 37 °C for 1 h. The DNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and was used as a template for PCR. For Re-ChIP, the immunocomplex was eluted with elution buffer (1% SDS, 100 mmol/L NaHCO3), diluted with the Re-ChIP buffer (1% Triton X-100, 2 mmol/L EDTA, 150 mmol/L NaCl, 20 mmol/L Tris pH 8.1), which was subjected to immunoprecipitation with H3K27ac antibody. Finally, the enrichment of immunoprecipitated material relative to input with gene-specific primers to the specified regions was determined by qRT-PCR (Supporting Information Table S4).
2.15. Chromatin conformation capture assay-qPCR (3C-qPCR)
After different treatments, the cells were cross-linked in 1% formaldehyde for 10 min at room temperature and quenched in 0.125 mol/L glycine for 5 min. After centrifugation, the cells were lysed in lysis buffer for 10 min on ice. Then, the lysis was centrifuged to get nuclei, which were digested in restriction enzyme buffer containing 0.3% SDS for 1 h at 37 °C. Triton X-100 was added to the buffer for 1 h at 37 °C followed by 400 U XbaI incubation overnight to digest DNA. Next, the nuclei were incubated in 1.6% SDS for 25 min at 65 °C, suspended in ligation buffer containing 1% Triton X-100 for 1 h at 37 °C, then ligated by 100 U ligase for 4 h at 16 °C followed by 30 min at room temperature. The reaction was terminated, and de-crosslinking was performed with PK buffer (5 mmol/L EDTA, pH 8.0; 10 mmol/L Tris-HCl, pH 8.0; 0.5% SDS) at 65 °C overnight. After digested in 300 μg RNase for 45 min at 37 °C, the DNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and amplified by qPCR.
2.16. RNA-seq of HCAECs
Total RNA was isolated and purified using Trizol according to the manufacturer's procedure. The RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was detected by Bio-analyzer 2100 (Agilent, CA, USA) at concentration >50 ng/μL, RIN > 7.0, OD 260/280 > 1.8, and total RNA >1 μg. The detection results were confirmed via electrophoresis with denaturing agarose gel. Poly(A) RNA was purified from 1 μg total RNA using Dynabeads Oligo (dT) 25-61005 (Thermo Fisher, CA, USA) with two rounds of purification and fragmented into small pieces by Magnesium RNA Fragmentation Module (NEB, e6150, USA) at 94 °C for 5–7 min. The RNA fragments were converted to cDNA, which was next used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, USA), RNase H (NEB, USA) and dUTP Solution (Thermo Fisher, USA). The addition of an A-base to the blunt ends of each strand was performed to prepare them for ligation to the indexed adapters. Each adapter contains a T-base overhang for ligating the adapter to the A-tailed DNA fragments. Single- or dual-index adapters were ligated to the fragments and size selection was performed using AMPureXP beads. After the heat-labile UDG enzyme (NEB, USA) treatment of the U-labeled second-stranded DNAs, PCR amplification of the ligated products was carried out under the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 bp (±50 bp). At last, we performed the 2 × 150 bp paired-end sequencing (PE150) on the Illumina Novaseq 6000 (LC-Bio Technology Co., Ltd., Hangzhou, China) following the vendor's recommended protocol.
2.17. RNA-seq analysis
The Cutadapt software was used to remove the reads that contained adapter contamination (command line: ∼cutadapt -a ADAPT1 -A ADAPT2 -o out1.fastq -p out2.fastq in1.fastq in2.fastq -O 5 -m 100). After removing the low-quality and undetermined bases, we employed the HISAT2 software to map reads to the genome (Homo sapiens Ensembl v101, command line: ∼hisat2 -1 R1.fastq.gz -2 R1.fastq.gz -S sample_mapped.sam). The mapped reads of each sample were assembled using StringTie with default parameters (command line: ∼stringtie -p 4 -G genome.gtf -o output.gtf -l sample input.bam). The gffcompare software was used to generate the final transcriptome. Quantification of all transcript abundances denoted as FPKM values (FPKM = [total_exon_fragments/mapped_reads (millions) × exon_length (kb)], command line: ∼stringtie -e -B -p 4 -G merged.gtf -o samples.gtf samples.bam) was performed using ballgown. We identified the differentially expressed genes using the R package named edgeR and set the screening threshold to a P value < 0.05 and |fold change| > 2. Pathway analysis was carried out using the DAVID Gene Functional Classification tool.
2.18. ChIP-seq data and visualization
We obtained the ChIP-seq data from the Gene Expression Omnibus with the identification codes GSE44939 and GSE53998. The IGVTools (version: IGV 2.8.10) was used to visualize these data.
2.19. Microarray data
We obtained the microarray data from the Gene Expression Omnibus with the identification code GSE43292. We presented the gene expression levels in heat maps using the R package named heatmap.
2.20. Assessment of endothelium-dependent relaxation
Endothelium-dependent relaxation was detected by DMT-620M pressure myograph system (Arhus, Denmark). Mouse thoracic aorta was dissected and cut into 2 mm rings. The rings were maintained in oxygenated Kreb's solution at 37 °C and stretched to an optimal baseline tension of 9.8 mN, followed by equilibration for 1 h. Then, the first contraction of the rings was induced by 80 mmol/L KCl and washed with Kreb's solution. Next, norepinephrine (NE, 0.1 μmol/L, MedChemExpress, NJ, USA) was used to produce steady vasoconstriction, and acetylcholine (Ach, 10−9–10−5 mol/L) was added cumulatively to induce endothelium-dependent relaxation.
2.21. Measurement of cytosolic dsDNA concentration in culture HCAECs or aortic tissue
To get the cytosolic dsDNA from cultured HCAECs or aortic tissues, we first used Mitochondria Isolation Kit (Thermo Fisher Scientific, Madison, USA) to remove the mitochondria to avoid the contamination of mtDNA released into the cytosol according to the manufacturer's instructions. Briefly, the pellet from the single-cell suspension of tissues or cells was added Mitochondria Isolation Reagent A and incubated on ice for 2 min after sufficient vortex. Next, the Mitochondria Isolation Reagent B was added and incubated on ice for 5 min, which was vortexed at maximum speed every minute. Then, the tube was added Mitochondria Isolation Reagent C and inverted several times to mix. After centrifugation, the concentration of the final supernatant containing cytosolic dsDNA was measured by PicoGreen Assay Kit (Thermo Fisher Scientific, Madison, USA).
2.22. Mitochondrial stress assay
We examined the mitochondria membrane potential of the cells using a TMRM assay kit (Abcam, Cambridge, UK). According to the instructions, the growth medium was removed from cells after different treatments by aspiration and cells were washed with 1 × PBS. The cells were incubated in TMRM working solution at 15−45 min at 37 °C in the dark. Then, the TMRM working solution was replaced with 1 × Live Cell Imaging Buffer. The fluorescence intensity was measured at 548 nm excitation and 573 nm emission wavelengths by a microplate reader.
2.23. Small interference RNA-mediated knockdown and transfection
Small interference RNA (siRNA; GenePharma, Shanghai, China) mediated knockdown of STING, IRF3, p65 and BRD4 was performed using Opti-MEM and Lipofectamine 3000 (Invitrogen, USA). Scrambled sequences were used as controls. The respective siRNA was diluted in Lipofectamine 3000 and added to HCAECs grown to 30%–50% confluency. The media was changed 6 h after silencing procedures were completed. HCAECs were incubated 24 h prior to assessing the knockdown efficiency via immunoblotting. Transfection of HCAECs with plasmids (or control vectors) for STING was performed using Lipofectamine 3000 for 24 h. The overexpression efficiency was assessed by immunoblotting. The targeted sequences are shown in Supporting Information Table S5.
2.24. Reactive oxygen species (ROS) assay
After different treatments, the level of intracellular ROS was detected by DCFH-DA staining. Firstly, the cells were washed twice with Endothelial Cell Medium. Then, the cells were incubated with DCFH-DA working solution (Beyotime, Beijing, China) at a 1:1000 ratio for 30 min at 37 °C in the dark. Finally, the fluorescent signal of the intracellular ROS was measured by fluorescence microscope at 488/525 nm.
2.25. Monocyte adhesion assay
In adhesion experiments, THP-1 cells, a human monocytic cell line, were labeled with CM-Dil (2.5 μg/mL, Invitrogen, USA) at 37 °C for 10 min as described by the manufacturer. Subsequently, the pre-labeled THP-1 cells were co-cultured with HCAECs with different treatments and allowed to attach at 37 °C for 60 min. Then, the non-adherent THP-1 cells were removed by washing with PBS. The THP-1 cells attached to HCAECs were observed by fluorescence microscope and calculated by ImageJ software.
2.26. Enzyme-linked immunosorbent assay
The levels of cGAMP in cell lysates and aorta tissues from mice and atherosclerotic patients were measured by 2′,3′-cGAMP competitive ELISA Kit (Invitrogen, USA). The aorta tissues from mice and human were grinded into powder in liquid nitrogen. Then, cells and tissues were lysed in lysis buffer on ice. After centrifuged at 21,000 × g for 5 min at 4 °C, the supernatant was collected to detect the cGAMP level according to the manufacturer's instructions. The levels of 8-OHDG (DNA oxidation marker) in cells and tissues from atherosclerosis patients was detected by EpiQuik 8-OHDG DNA Damage Quantification Direct Kit (Epigentek, USA).
2.27. Cytosolic DNA immunoprecipitation and subcellular fractionation
After different treatments, cells were lysed on ice for 10 min with 0.025% w/v digitonin in 20 mmol/L HEPES buffer. Cytosol and membrane fractions were separated by centrifugation (13,000 × g, 5 min, 4 °C). The interaction of VDAC1–mtDNA was detected using an Immunoprecipitation Assay Kit (Millipore, USA) following the manufacturer's instructions with anti-VDAC1 antibody or normal IgG for negative control.
2.28. Statistical analysis
All statistical analyses were performed with SPSS Version 25 software, and graphs were generated using GraphPad Prism 8. Values are presented as mean ± standard error of mean (SEM). Statistical analysis between 2 groups was performed with an unpaired 2-sided Student's t test when the data passed normality and equal variance test, otherwise Man-Whitney U text was used. For the comparisons among more than two groups, one-way ANOVA with the Tukey post hoc test was used to adjust for multiple comparisons. P < 0.05 was considered to be statistically significant. Statistical results and the corresponding methods were presented in the figure legends.
3. Results
3.1. Atherosclerosis-related endothelial dysfunction mediated by oxidized LDL is dependent on the activation of the non-canonical STING–PERK pathway
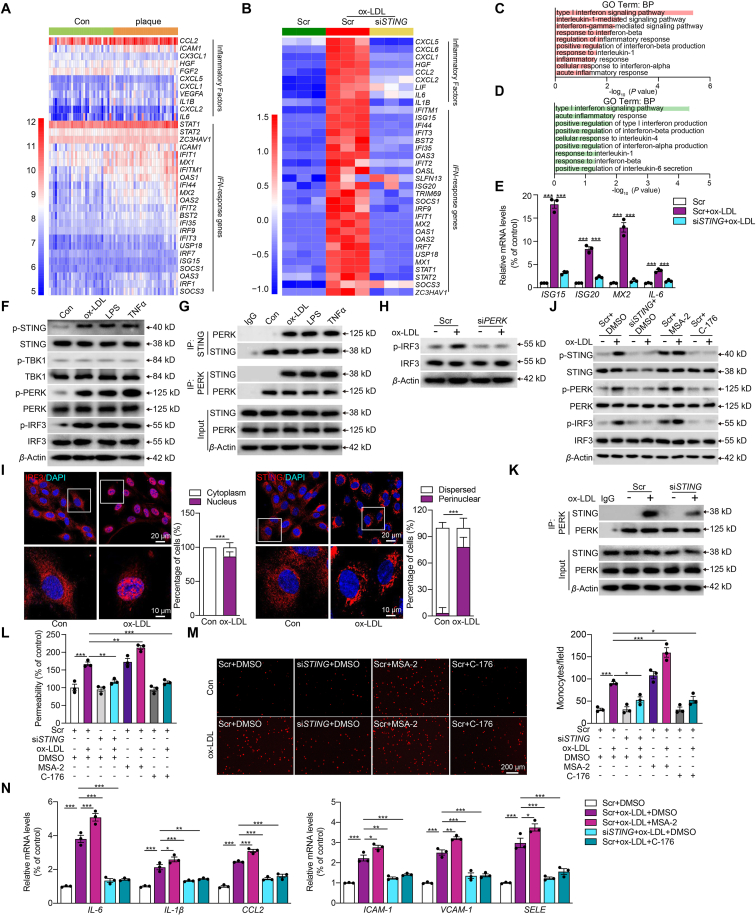
Gene expression patterns of human carotid with or without atheroma plaque have been reported by microarray analysis previously21. We re-analyzed this publicly available microarray data and revealed a pronounced expression of IFN-response and inflammatory genes in patients with plaques, suggesting the involvement of the STING pathway in atherosclerosis (Fig. 1A). To confirm the role of STING in atherosclerosis-related endothelial dysfunction, we profiled the transcriptome changes in HCAECs upon ox-LDL stimulation with and without STING inhibition by siRNA. The efficacy of siRNA-targeted STING inhibition was evaluated by Western blot shown in Supporting Information Fig. S1A. Consistently, inflammatory genes were greatly upregulated in ox-LDL-treated HCAECs, and were mitigated by STING-knockdown (Fig. 1B). Pathway analysis of differentially expressed genes revealed enrichment in biological processes associated with inflammatory response and interferon signaling between scramble (Scr)+ox-LDL and Scr-treated HCAECs (Fig. 1C). Meanwhile, the differentially expressed genes between siSTING + ox-LDL and Scr + ox-LDL groups were also enriched in similar pathways (Fig. 1D). To verify the RNA-seq results, we selected four representative genes for qRT-PCR. The ISG15, ISG20, MX2, and IL-6 were significantly increased upon ox-LDL stimulation and downregulated when STING was knocked down (Fig. 1E).
Figure 1.
Atherosclerosis-related endothelial dysfunction mediated by oxidized LDL is dependent on the activation of the non-canonical STING–PERK pathway. (A, B) Heat map displaying the expression (microarray; log2 RMA signal) of inflammatory factors and IFN-response genes in distant macroscopically intact tissue as control (Con, light green) and atherosclerotic plaque (orange) in (A) and human coronary artery endothelial cells (HCAECs) treated with scramble control (Scr, green), Scr + oxidative low-density lipoprotein (ox-LDL, red), and stimulator of interferon genes siRNA (siSTING)+ox-LDL (yellow) in (B). (C, D) GO biological process enrichment in HCAECs treated with Scr + ox-LDL versus Scr (C) and in HCAECs treated with siSTING + ox-LDL versus Scr + ox-LDL (D) by DAVID analysis. (E) Quantitative real-time PCR (qRT-PCR) analysis of ISG15, ISG20, MX2, and IL-6 mRNA expression in ox-LDL-treated HCAECs combined with siSTING. (F) Western blot analysis of the non-canonical STING–PKR-like ER kinase (PERK) pathway in HCAECs treated with ox-LDL (100 μg/mL), lipopolysaccharide (LPS, 1 μg/mL), or tumor necrosis factor α (TNFα, 10 ng/mL). (G) Co-immunoprecipitation analysis of STING-PERK binding in HCAECs treated with ox-LDL, LPS and TNFα. (H) Western blot analysis of the phosphorylated interferon regulatory factor 3 (p-IRF3) in ox-LDL-treated HCAECs combined with siPERK. (I) Immunofluorescence staining of STING and IRF3 in ox-LDL-treated HCAECs versus control (Scale bar = 20 μm). (J) Western blot analysis of the non-canonical STING–PERK pathway in ox-LDL-treated HCAECs combined with siSTING, STING inhibitor C-176 (0.5 μmol/L) or STING activator MSA-2 (10 μmol/L). (K) Co-immunoprecipitation analysis of STING–PERK binding in ox-LDL-treated HCAECs combined with siSTING. (L, M) Permeability of HCAECs to FITC-Dexstran (L), THP-1 monocytes adhesion (M) in ox-LDL-treated HCAECs combined with siSTING or MSA-2 or C-176. (N) qRT-PCR analysis of adhesions molecules and chemokines mRNA expression in ox-LDL-treated HCAECs combined with siSTING or MSA-2 or C-176. Data are expressed as mean ± SEM, n = 3; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To validate the findings of transcriptome analysis, we next employed Western blot, co-immunoprecipitation and immunofluorescence to corroborate the activation of the STING pathway. As shown in Fig. 1F and Fig. S1B, the protein expression of total STING, PERK and IRF3 had no change as well as the phosphorylated TBK1 usually, whereas the phosphorylation of STING, PERK, and IRF3 increased frequently in HCAECs treated with the risk factors of atherosclerosis including ox-LDL, LPS or TNFα. These suggest the activation of non-canonical STING–PERK pathway in atherosclerosis-related endothelial dysfunction. Additionally, ox-LDL, LPS, and TNFα promoted the binding between STING and its downstream non-canonical effector PERK (Fig. 1G and Fig. S1C). Considering the results Fig. 1F and G and ox-LDL is the classic atherosclerotic stimulator, HCAECs with ox-LDL stimulation were employed as the endothelial dysfunction ex vivo model in the following experiments. Here, we found that PERK deficiency could effectively reverse the activation of IRF3 in ox-LDL-challenged HCAECs (Fig. 1H, Fig. S1D and S1E). ox-LDL induced STING perinuclear re-localization and IRF3 nuclear translocation, indicating the activation of both STING and IRF3 (Fig. 1I). We further confirmed the role of STING in ox-LDL-induced endothelial injury by employing both siRNA and pharmacological approaches. MSA-2 is a STING-specific agonist, and C-176 is a covalent STING antagonist22,23. Activation of STING pathway by ox-LDL was inhibited by siSTING and C-176, as was the interaction between PERK and STING (Fig. 1J and K, Fig. S1F and S1G). Besides, the ox-LDL-induced permeability (Fig. 1L), THP-1 monocytes attachment to HCAECs (Fig. 1M), and the expression of adhesion molecules and chemokines (Fig. 1N) were all reduced by siSTING and C-176. Moreover, STING agonist MSA-2 had similar effects as ox-LDL and partially exacerbated the ox-LDL-induced endothelial injury. These results demonstrate that the non-canonical STING–PERK pathway is crucial in ox-LDL-induced endothelial injury.
3.2. Non-canonical STING–PERK pathway is activated by leaked mtDNA in oxidized LDL-treated endothelial injury
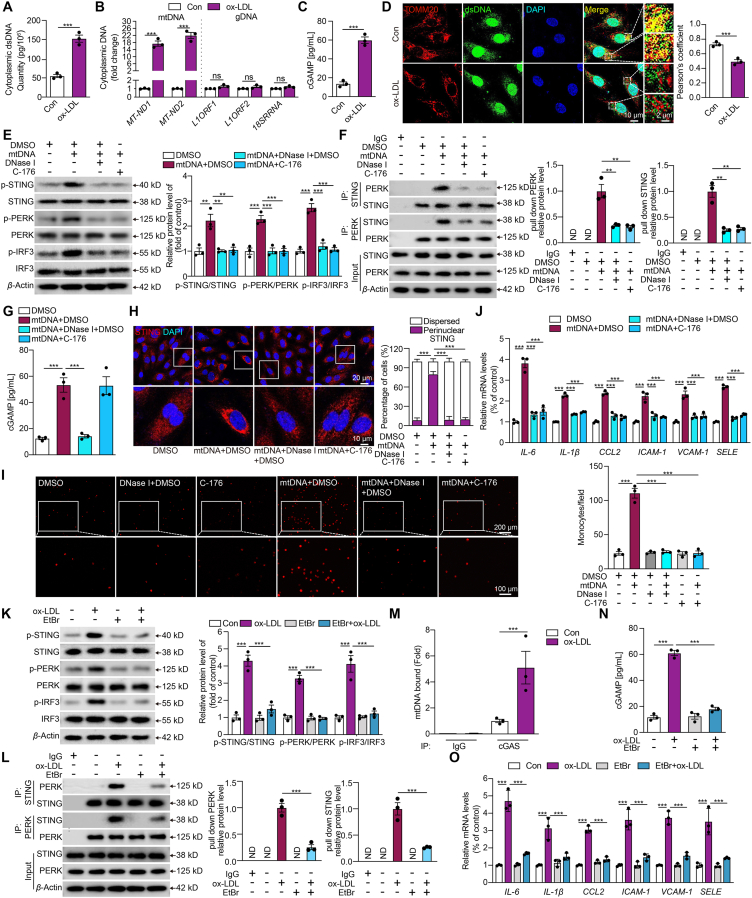
DNA from damaged nuclei or mitochondria leaked into the cytosol is known to promote STING signaling24. Here, we found that the cytosolic dsDNA levels increased markedly in ox-LDL-treated HCAECs compared with the control (Fig. 2A). Then, we used primers specific for mtDNA, nuclear DNA, and ribosome DNA sequences to examine the cytosolic DNA by qPCR analysis. The result showed that the principal source of cytosolic dsDNA in ox-LDL-treated HCAECs was found to be mtDNA (Fig. 2B). Additionally, cGAMP, as a second messenger for foreign DNA sensing and the trigger of STING, demonstrated a substantial increase in ox-LDL-treated HCAECs (Fig. 2C). The co-staining of dsDNA and mitochondria specific maker TOMM20 showed the expected co-localization of dsDNA with mitochondria in HCAECs. In contrast, ox-LDL treatment resulted in a prominent presence of cytosolic dsDNA outside the mitochondria (Fig. 2D). These results suggest that ox-LDL might provoke the release of mtDNA from mitochondria into the cytoplasm, subsequently generating cGAMP in HCAECs.
Figure 2.
Mitochondrial dsDNA leaking mediated the non-canonical STING–PERK pathway activation in the oxidized LDL-induced endothelial injury. (A–D) The cytoplasmic double-stranded DNA (dsDNA) concentration (A), qPCR analysis of cytoplasmic mitochondrial DNA (mtDNA, MT-ND1 and MT-ND2), nuclear LINE1 elements (L1ORF1 and L1ORF2), ribosomal gene (RNA 18S) (B), cyclic GMP-AMP (cGAMP) concentration (C) and immunofluorescence staining of mitochondria by TOMM20 (mitochondria, red) and dsDNA (green) (D) in HCAECs treated with or without ox-LDL (Scale bar = 10 μm). (E–J) Western blot analysis of the non-canonical STING-PERK pathway (E), co-immunoprecipitation analysis of STING-PERK binding (F), cGAMP concentration (G), immunofluorescence staining of STING (H), THP-1 monocytes adhesion (I), and adhesions molecules and chemokines mRNA expression (J) in mtDNA (3 μg/mL)-transfected HCAECs combined with DNase I (1 μg/mL) or C-176. (K–O) Western blot analysis of the non-canonical STING–PERK pathway (K), co-immunoprecipitation analysis of STING-PERK binding (L), cyclic GMP–AMP synthase (cGAS) bound mtDNA by cytosolic DNA immunoprecipitation (M), cGAMP concentration (N) and adhesions molecules and chemokines mRNA expression (O) in ox-LDL-treated HCAECs combined with ethidium bromide (EtBr). Data are expressed as mean ± SEM, n = 3; ∗∗P < 0.01, ∗∗∗P < 0.001; ND, not detected; ns, no significance.
To further confirm that mtDNA was the direct mediator of STING activation in ox-LDL-induced endothelial injury, mtDNA was isolated from ox-LDL-treated HCAECs and then transfected into the naive HCAECs. This intervention mirrored the effects seen in ox-LDL-treated HCAECs as indicated in Fig. 1, including the phosphorylation of STING, PERK, and IRF3 (Fig. 2E), a significant increase in the interaction between PERK and STING (Fig. 2F) and elevated cGAMP generation (Fig. 2G). The perinuclear re-localization of STING (Fig. 2H), nuclear translocation of IRF3 (Supporting Information Fig. S2A), THP-1 cell adhesion (Fig. 2I), and the elevated expression of adhesion molecules and chemokines (Fig. 2J) were observed in mtDNA-transfected HCAECs. When the native HCAECs were transfected with mtDNA pre-digested with DNase I to eliminate mtDNA, or incubated with STING antagonist C-176 after mtDNA transfection, all of the above phenomena were attenuated markedly as shown by Fig. 2E–J.
Additionally, we depleted mtDNA by culturing HCAECs with low dose EtBr and confirmed the depletion efficiency (Fig. S2B). Western bot results showed that mtDNA depletion effectively inhibited the activation of non-canonical STING pathway in ox-LDL-treated HCAECs (Fig. 2K and L). Moreover, an antibody against cGAS pulled down more mtDNA in ox-LDL-treated HCAECs by co-immunoprecipitation (Fig. 2M). mtDNA depletion in HCAECs notably inhibited the ox-LDL-induced increase in cGAMP levels (Fig. 2N) and the upregulated expression of adhesion molecules and chemokines (Fig. 2O). Additionally, the increased activation of the non-canonical STING–PERK pathway, the interaction between PERK and STING, and the expression of adhesion molecules and chemokines in ox-LDL-challenged HCAECs were also inhibited by DNase I (Fig. S2C–S2F). All of these results indicated that the non-canonical STING–PERK pathway is activated by leaked mtDNA in ox-LDL-induced endothelial injury.
3.3. Oxidized LDL causes mtDNA release into the cytoplasm via the VDAC1-dependent mitochondrial permeability transition pore opening
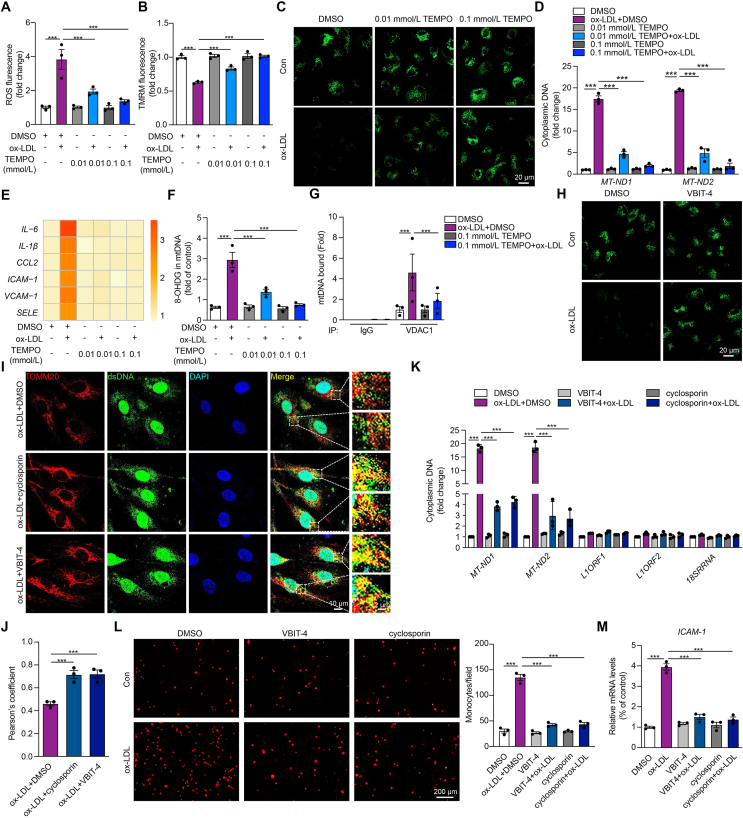
ox-LDL has been documented to cause mitochondria damage and increase intracellular ROS25. Meanwhile, mitochondrial reactive oxygen species (mROS) could trigger mitochondrial stress and the release of mtDNA due to increased mitochondrial outer membrane permeability26. Here, we found the increase of ROS (Fig. 3A), the apparent mitochondrial membrane potential destabilization (Fig. 3B), and the enhanced mPTP opening in ox-LDL-treated HCAECs (Fig. 3C) were reversed by specific mROS scavenger TEMPO in a dose-dependent manner. In addition, TEMPO prevented the elevation of cytosolic mtDNA content and the inflammatory responses in ox-LDL-treated HCAECs (Fig. 3D and E). These observations suggest that mROS may serve as the upstream trigger of mtDNA release through mPTP opening. To test our hypothesis, we first examined the DNA oxidation, as represented by the level of 8-OHDG. As shown in Fig. 3F, the level of 8-OHDG was upregulated in ox-LDL-treated HCAECs, and reversed by TEMPO, indicating significant mtDNA oxidation by mROS. VDAC1, the most abundant VDAC isoforms, has been reported to form the large mitochondrial outer membrane pores under mitochondrial stress via oligomers, which are stabilized by mtDNA through direct interaction26. Therefore, we explored whether VDAC1 could contribute to the release of mtDNA via initiating mPTP opening in ox-LDL-treated HCAECs. As anticipated, co-immunoprecipitation with an antibody against VDAC1 pulled down more mtDNA in ox-LDL-treated HCAECs, while reversed by TEMPO (Fig. 3G). Moreover, the VDAC1 oligomerization inhibitor, VBIT-4, effectively attenuated the mPTP opening in ox-LDL-treated HCAECs (Fig. 3H). Besides, the cytosolic accumulation of mtDNA (Fig. 3I–K), the increased THP-1 cells attachment to HCAECs (Fig. 3L), and the upregulated expression of intercellular adhesion molecule 1 (ICAM-1) (Fig. 3M) caused by ox-LDL were all prevented by mPTP inhibitor cyclosporin or VBIT-4. These data suggest that in ox-LDL-treated HCAECs, the release of mtDNA into the cytoplasm is triggered by mtDNA oxidation, followed by mPTP opening in a VDAC1-dependent manner.
Figure 3.
Oxidized LDL increases 8-OHDG of mtDNA via reactive oxygen species (ROS) and causes mtDNA release into the cytoplasm via voltage-dependent anion channel 1 (VDAC1)-dependent mitochondrial permeability transition pore (mPTP) opening. (A–F) The ROS level stained by DCFH-DA (A), mitochondrial membrane potential stained by TMRM (B), the opening of mPTP stained by MPTP assay kit (C), the expression level of cytoplasmic mtDNA by qPCR (D), heat map results of inflammatory genes (E), and mtDNA oxidation reflected by the level of 8-OHDG (F) in ox-LDL-treated HCAECs combined with mitochondrial ROS scavenger TEMPO at the dose of 0.01 mmol/L and 0.1 mmol/L. (G) Cytosolic DNA immunoprecipitation analysis of the relative amount of mtDNA binding to VDAC1 in ox-LDL-treated HCAECs combined with TEMPO. (H) Representative images of mPTP assay in ox-LDL-treated HCAECs combined with VDAC1 oligomerization inhibitor VBIT-4 (10 μmol/L). (I–M) Immunostaining of TOMM20 (mitochondria, red) and dsDNA (green) (I, J), qPCR analysis of mtDNA, nuclear LINE1 elements or a ribosomal gene (K), attachment of THP-1 monocytes (L), and intercellular adhesion molecule 1 (ICAM-1) mRNA expression (M) in ox-LDL-treated HCAECs combined with mPTP inhibitor cyclosporin and VBIT-4. Data are shown as mean ± SEM, n = 3; ∗∗∗P < 0.001.
3.4. Cooperation among IRF3, p65, and BRD4 in transcriptional regulation of inflammatory genes in oxidized LDL-induced endothelial injury
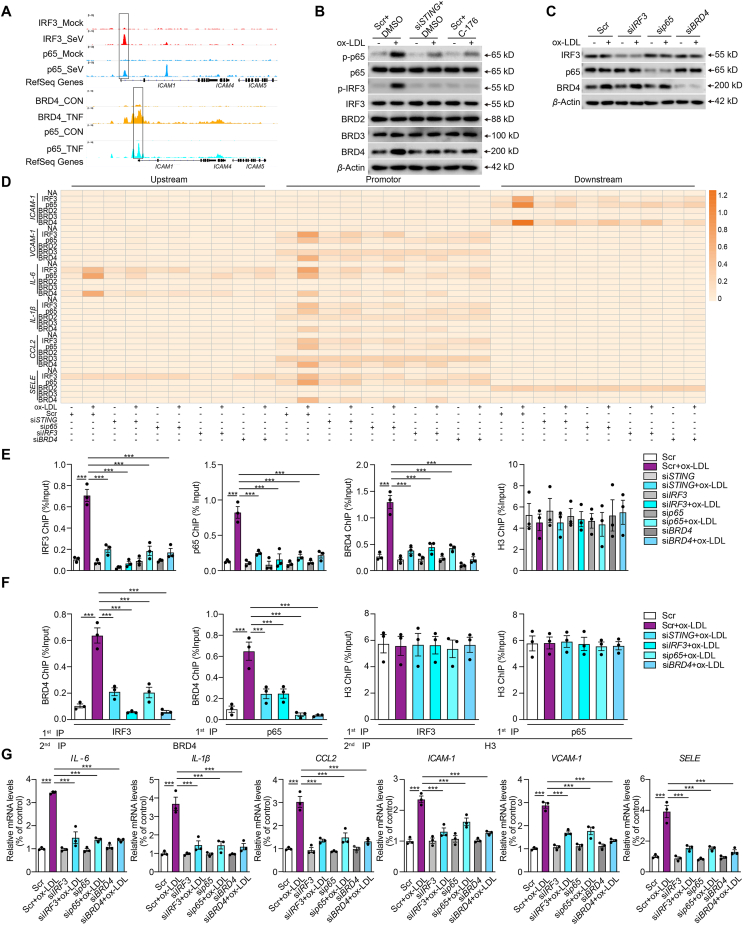
Conventionally, the activated STING mediates the activation of IRF3 or NF-κB to initiate independent transcription of diverse targets27. Interestingly, we discovered that inflammatory stimulation promotes the dynamic colocalization of IRF3, p65, and BRD4 to the proximal promoter region of ICAM-1, as evidenced by chromatin immunoprecipitation sequence results from various studies (Fig. 4A)16,17. We propose that these three proteins may work together to regulate the expression of ICAM-1. Of note, BRD4 was greatly upregulated in ox-LDL-treated HCAECs compared to other BET family members such as BRD2 and BRD3 (Fig. 4B and Supporting Information Fig. S3A). Concurrently, STING inhibition caused by siRNA or pharmacological inhibitor C-176 markedly suppressed the activation of p65 and IRF3, and the expression of BRD4. However, p65, IRF3 and BRD4 had no effect on each other's total protein (Fig. 4C, Fig. S3B and S3C). To further prove our hypothesis, we performed ChIP of the promoter elements of inflammatory genes in ox-LDL-treated HCAECs, employing ChIP to probe for no-antibody control (NA), IRF3, p65, BRD2, BRD3, BRD4, and H3 (Positive control). As expected, there was no significant change in NA, BRD2, and BRD3 enrichments for the specified genes in each treatment group. Interestingly, after ox-LDL treatment, BRD4, IRF3, and p65 were notably recruited to the proximal promoter regions of the inflammatory genes, but disappeared with STING, IRF3, p65, or BRD4 deficiency (Fig. 4D). The control marker H3 was pulled down evenly (Fig. S3D). ChIP data of ICAM-1 in Fig. 4D were replotted into a bar graph in Fig. 4E. Besides, we confirmed that BRD4 and IRF3 occupied the same DNA element of ICAM-1 through a Re-ChIP assay, as did BRD4 and p65 in ox-LDL-treated HCAECs. Such phenomenon was also reversed by the deficiency of STING, IRF3 or p65, suggesting the possibility of a substantial transcriptional complex composed of BRD4, activated p65, and IRF3, regulated by activated STING (Fig. 4F). Finally, the increased expression of inflammatory genes was universally diminished in the presence of STING, IRF3, and p65 deficiency (Fig. 4G). All these results suggest that STING activation regulated the transactivation of inflammatory genes through the synergistic cooperation among BRD4, p65, and IRF3, which is distinct from the canonical mode that STING activates IRF3 and NF-κB, contributing to IFN signals and inflammation, respectively.
Figure 4.
Cooperation of IRF3, NF-κB and BRD4 in the transcription of inflammation genes in oxidized LDL-induced endothelial injury. (A) Tracks of chromatin immunoprecipitation sequencing (ChIP-seq) peaks for IRF3 (red), p65 (blue, upper; cyan, down), and BRD4 (yellow) at the ICAM1 gene loci in control (top) or stimulant-induced inflammation (bottom) samples. (B) Western blot analysis of total and phosphorylated p65 and IRF3 and BRD family in ox-LDL-treated HCAECs combined with siSTING or C-176. (C) Western blot analysis of total and phosphorylated p65 and IRF3 and BRD4 in ox-LDL-treated HCAECs combined with IRF3, p65, or BRD4 siRNA. (D) ChIP analysis of the enrichment of IRF3, p65, BRD2, BRD3, and BRD4 at −1 kb, promoter and +1 kb of inflammatory genes in ox-LDL-treated HACECs combined with STING, p65, IRF3 or BRD4 siRNA. (E, F) The ChIP-qPCR (E) and Re-ChIP (F) analysis of the binding of BRD4, IRF3, p65, and IRF3 on the +1 kb region of ICAM1 in ox-LDL-treated HCAECs combined with STING, p65, IRF3 or BRD4 siRNA. (G) qRT-PCR analysis of adhesions molecules and chemokines mRNA expression in ox-LDL-induced HCAECs combined with STING, p65, IRF3 or BRD4 siRNA. Data are shown as mean ± SEM, n = 3; ∗∗∗P < 0.001.
3.5. STING signal regulates open chromatin state of inflammatory genes in oxidized LDL-treated ECs by a BRD4-dependent super-enhancer with condensation
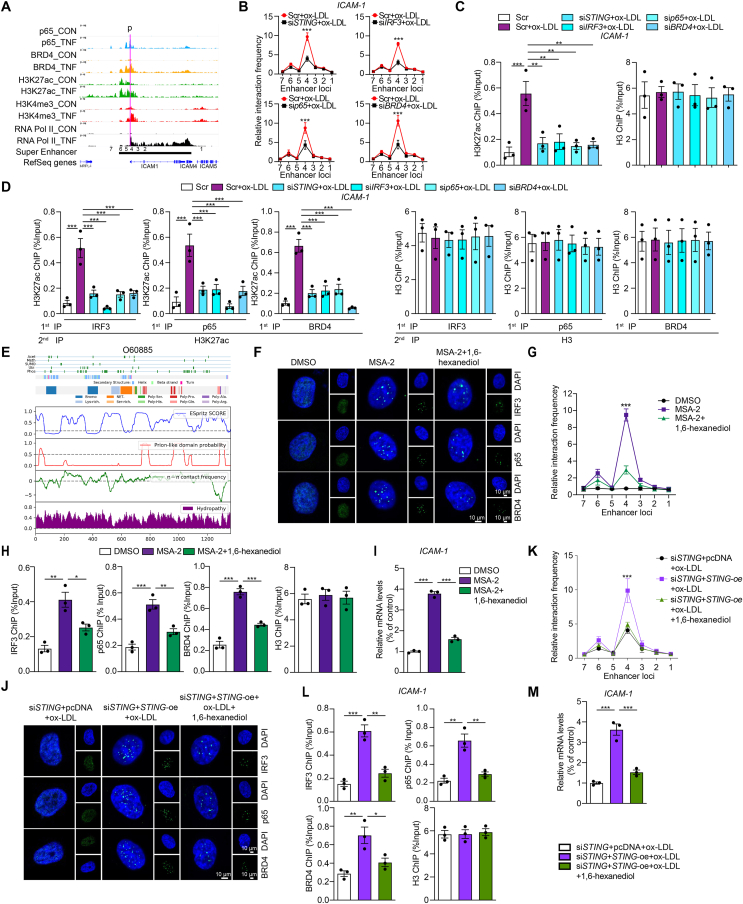
Chromatin immunoprecipitation sequencing results demonstrated that TNFα could promote the recruitment of p65 and BRD4 to the enhancer and proximal promoter region of ICAM-1 gene loci marked by H3K27ac and H3K4me3 (Fig. 5A). To confirm the interaction between ICAM-1 promoter and these potential enhancer clusters, we performed a quantitative analysis of 3C-qPCR. The primers for the promoter and the enhancer locus 4 showed the greatest interaction strength, which were most affected by the deficiency of STING, IRF3, p65, and BRD4 (Fig. 5B). Super-enhancer, strongly enriching for the H3K27ac, BRD4, and the mediator complex1, comprises clusters of transcriptional enhancers28. Furthermore, the ChIP assay showed the +1 kb of ICAM-1 promoter was strongly enriched for H3K27ac, but not with the deficiency of STING, IRF3, p65, or BRD4 (Fig. 5C). Besides, we identified IRF3, p65, BRD4 and H3K27ac occupied the same DNA element of ICAM-1 in ox-LDL-treated HCAECs by Re-ChIP assay (Fig. 5D), suggesting that the enrichment of IRF3, p65 and BRD4 in the super-enhancer is all required for inflammatory genes expression.
Figure 5.
Oxidized LDL regulates the open chromatin state of inflammatory genes in ECs in a BRD4-dependent super-enhancer associated with condensation. (A) ChIP-seq peaks (p65, BRD4, H3K27ac, H3K4me3 and RNA Pol II levels) and putative super-enhancer location around ICAM-1. The putative super-enhancer region is marked as the horizontal line in black. The promoter is marked as the purple line. (B) Quantitative analysis of chromosome conformation capture assays-qPCR (3C-qPCR) analysis of long-distance interactions between ICAM-1 promoter and seven enhancer loci in ox-LDL-treated HACECs combined with STING, IRF3, p65 or BRD4 siRNA (x-axis means seven enhancer loci of ICAM-1). (C, D) The ChIP-qPCR (C) and Re-ChIP (D) analysis of the binding of H3K27ac and BRD4, IRF3, or p65, separately, on the +1 kb region of ICAM1 in ox-LDL-treated HCAECs combined with STING, p65, IRF3 or BRD4 siRNA. The Re-ChIP was performed with 1st round pull-down of antibodies, including IRF3, p65, and BRD4 as well as 2nd round pull-down of H3K27ac antibody. (E) The color-coded schematic representation of the aligned amino acid sequence and corresponding prion-like domain disorder propensity plots for BRD4 using the PLAAC (prion-like amino acid composition) pool. (F–I) Immunofluorescence staining of IRF3 (green), p65 (green), and BRD4 (green) in the nuclear (blue) (F), 3C-qPCR analysis of long-distance interactions between ICAM-1 promoter and seven enhancer loci (the positions of different loci were described as Fig. 5A, x-axis means seven enhancer loci of ICAM-1) (G), ChIP analysis of the enrichment of IRF3, p65, and BRD4 at the super-enhancer region of ICAM-1 (H), and ICAM-1 mRNA level (I) in ox-LDL-treated HCAECs combined with MSA-2 or condensate inhibitor 1,6-hexanediol. (J–M) Immunofluorescence staining of IRF3 (green), p65 (green) and BRD4 (green) in the nuclear (blue) (J), 3C-qPCR analysis of long-distance interactions between ICAM-1 promoter and seven enhancers (x-axis means seven enhancer loci of ICAM-1) (K), ChIP analysis of the enrichment of IRF3, p65 and BRD4 at the super-enhancer region of ICAM-1 (L), and ICAM-1 mRNA level (M) in siSTING- and ox-LDL-treated HCAECs combined with STING overexpression or 1,6-hexanediol. Data are shown as mean ± SEM, n = 3; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Condensation is a characteristic of protein that the low-complexity disordered regions or intrinsically disordered regions aggregate together to form liquid-like condensates. As a hot topic in the field, condensation formation has been found during super-enhancer assembling to compartmentalize and concentrate the transcription apparatus. Such progress was mediated by certain transcriptional factors, for instance, BRD429. The architectural features of BRD4 determined its propensity towards condensate formation (Fig. 5E). After treatment with STING agonist MSA-2, there was a dramatic increase in BRD4 puncta numbers in the nuclei. Notably, p65 and IRF3 presented a similar behavior as BRD4 (Fig. 5F). The 3C-qPCR analysis revealed that the primers for the promoter and the enhancer locus 4 showed the greatest interaction strength upon MSA-2 stimulation (Fig. 5G). Consistently, MSA-2 treatment led to increased binding of IRF3, p65, and BRD4 to the super-enhancer region of ICAM-1 (Fig. 5H) and the upregulated expression of ICAM-1 (Fig. 5I). Similarly, puncta of IRF3, p65, and BRD4 was not observed in ox-LDL-treated STING-deficient HCAECs, while increased with the STING overexpression (Fig. 5J). The extent of STING re-expression in siSTING-treated HCAECs was evaluated by Western blot (Supporting Information Fig. S4). In concordance, the interaction between the promoter and the enhancer locus 4, the binding of IRF3, p65, and BRD4 to the super-enhancer region of ICAM-1, and the mRNA level of ICAM-1 showed the same trends (Fig. 5K–M). All of the above phenomena were reversed by condensate inhibitor 1,6-hexanediol. Thus, these data suggest that the STING signal regulates the open chromatin state of inflammatory genes in ox-LDL-treated HCAECs through a BRD4-dependent super-enhancer with condensation.
3.6. Endothelial deletion of STING reverses high-fat enhanced atherosclerotic lesions in ApoeKO mice
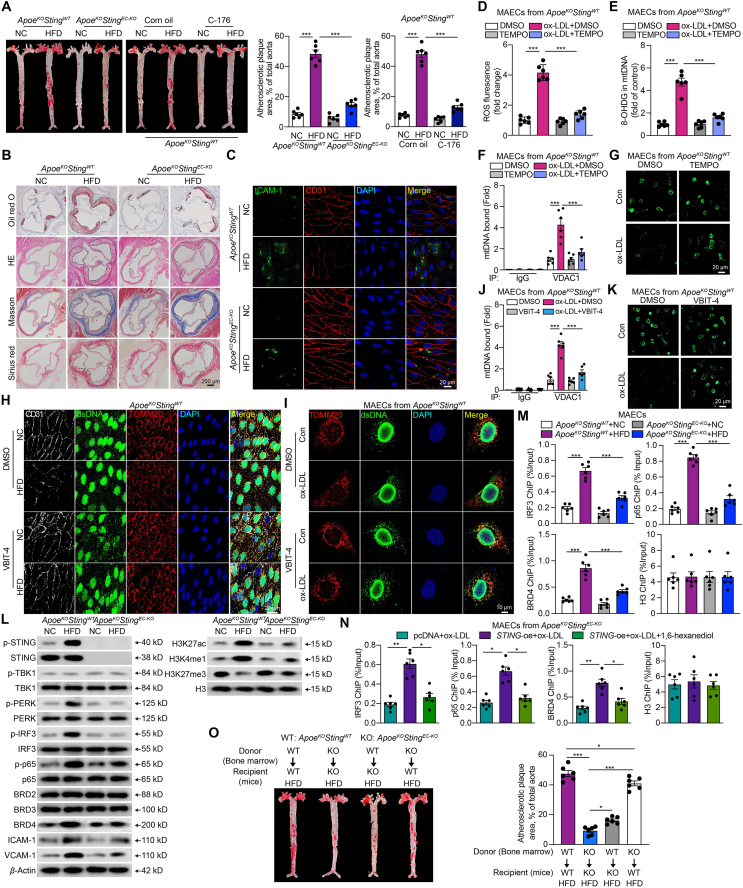
We next evaluated the role of STING in atherosclerosis in vivo. We crossed endothelium-directed Sting knockout mice with ApoeKO to generate a double knockout mouse model (ApoeKOStingEC-KO). Both ApoeKOStingWT and ApoeKOStingEC-KO mice were fed with either a NC or a HFD for 16 weeks to induce atherosclerosis (Supporting Information Fig. S5A). In contrast to HFD-fed ApoeKOStingWT mice, ApoeKOStingEC-KO mice showed decreased atherosclerotic lesions. Meanwhile, pharmacological inactivation of the STING pathway by C-176 has similar effects (Fig. 6A). There was no difference in the atherosclerotic plaque area between females and males in each group (Fig. S5B). Functionally, the ApoeKOStingEC-KO mice showed decreased lipid deposition and necrotic core (Fig. 6B, Fig. S5C and S5D), decreased leucocyte adhesion (Fig. S5E), enhanced plaque stability by histological analysis (Fig. 6B and Fig. S5C), and ameliorative endothelium-dependent vasodilation (Fig. S5F), compared to the ApoeKOStingWT mice. The mRNA levels of adhesions molecules and chemokines, including Il-6, Il-1β, Ccl2, Icam-1, Vcam-1, and Sele, increased in the aortas of HFD-fed ApoeKOStingWT mice and reduced in ApoeKOStingEC-KO mice (Fig. S5G). Consistently, the ICAM-1 immunoreactivity in the endothelium (co-localized with CD31) was significantly increased in the aortas of HFD-fed ApoeKOStingWT mice and was reversed in HFD-fed ApoeKOStingEC-KO mice (Fig. 6C and Fig. S5H). Total cholesterol, low-density lipoprotein cholesterol, triglyceride, and high-density lipoprotein cholesterol levels did not differ between ApoeKOStingWT and ApoeKOStingEC-KO mice fed with HFD (Fig. S5I).
Figure 6.
Endothelial deletion of STING reverses the atherosclerotic lesions in ApoeKOStingWT mice. (A) Representative images of en face aortas stained with oil red O from ApoeKOStingWT and ApoeKOStingEC-KO mice fed with normal chow diet (NC) or high-fat diet (HFD), as well as the ApoeKOStingWT mice fed with NC or HFD combined with STING inhibitor C-176. (B, C) Representative oil red O, hematoxylin and eosin (H&E), Masson and Sirius red staining of the aortic root (B), immunofluorescence staining of CD31 (red) and ICAM-1 (green) (C) in arteries from ApoeKOStingWT and ApoeKOStingEC-KO mice fed with NC or HFD. (D–G) The ROS level (D), mtDNA oxidation (E), VDAC1 bound mtDNA amount by cytosolic DNA immunoprecipitation (F), and the opening of mPTP (G) in mouse aortic endothelial cells (MAECs) derived from ApoeKOStingWT mouse thoracic aorta treated with ox-LDL and 0.1 mmol/L TEMPO. (H) Representative en face aortas immunofluorescence staining of endothelium (CD31), dsDNA and mitochondria (TOMM20) in the arteries from ApoeKOStingWT mice fed with NC or HFD combined with VBIT-4. (I–K) Immunofluorescence staining of dsDNA and mitochondria (TOMM20) (I), VDAC1 bound mtDNA amount by cytosolic DNA immunoprecipitation (J), the opening of mPTP (K) in MAECs derived from ApoeKOStingWT mouse thoracic aorta treated with ox-LDL and VBIT-4. (L, M) The protein levels of non-canonical STING pathway, BRD family, inflammation and super-enhancer specific makers (L), and ChIP analysis of the enrichment of IRF3, p65, and BRD4 at the super-enhancer region of Icam-1 (M) of MAECs derived from the thoracic aorta of ApoeKOStingWT and ApoeKOStingEC-KO mice fed with NC or HFD. (N) ChIP analysis of the enrichment of IRF3, p65, and BRD4 at the super-enhancer region of Icam-1 of ox-LDL-treated MAECs derived from the thoracic aorta of ApoeKOStingEC-KO mice, combined with STING overexpression and 1,6-hexanediol. (O) Representative images of en face aortas stained with oil red O from ApoeKOStingWT and ApoeKOStingEC-KO mice which were transplanted with bone marrow cells from ApoeKOStingWT and ApoeKOStingEC-KO mice. The recipient mice were fed with HFD. Data are shown as mean ± SEM, n = 6; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To substantiate the upstream triggers of STING and the downstream regulatory mechanism we found in HCAECs, we next validated these findings in vivo. Firstly, in ox-LDL-treated MAECs from ApoeKOStingWT, the ROS level and 8-OHDG of mtDNA increased significantly (Fig. 6D and E). Co-immunoprecipitation using an antibody against VDAC1 pulled down more mtDNA, with a rise in mPTP opening in same MAECs (Fig. 6F and G). These changes were reversed by the specific mROS scavenger TEMPO. The concentration of cytoplasmic DNA (Fig. S5J) and the expression of cytoplasmic mtDNA were markedly increased in arteries obtained from HFD-fed ApoeKOStingWT mice (Fig. S5K). As shown in Fig. 6H and Fig. S5L, the cytosolic dsDNA predominantly co-localized with mitochondria in en face aorta samples taken from ApoeKOStingWT fed with NC, but not the case in the HFD group, suggesting a dsDNA leak from mitochondria in the aorta, correlated with enhanced atherosclerotic lesions. This condition could be rescued by VDAC1 oligomerization inhibitor VBIT-4. Consistently, a similar phenomenon was observed in MAECs harvested from ApoeKOStingWT mice (Fig. 6I). The augmented capture of mtDNA by VDAC1 antibody, increased mPTP opening, and the increased phosphorylated STING in ox-LDL-treated MAECs from ApoeKOStingWT were all rescued by VBIT-4 (Fig. 6J and K, Fig. S5M). These data support the idea that STING activation in plaques is triggered by the dsDNA released from the mitochondria via VDAC1.
The relevant proteins in the non-canonical STING-PERK pathway, BRD4 and adhesion proteins, exhibited significant activation or elevation in the aorta from HFD-fed ApoeKOStingWT mice, while decreased in ApoeKOStingEC-KO mice (Fig. 6L and Supporting Information Fig. S5N). ChIP results confirmed the recruitment of BRD4, IRF3, and p65 to the downstream of Icam-1 promoter, dependent on STING activation, as evidenced by the increased enrichment in MAECs from HFD-induced ApoeKOStingWT, whereas it was decreased in ApoeKOStingEC-KO (Fig. 6M). Additionally, the protein levels of super-enhancer specific marker H3K27ac and H3K4me1 were upregulated significantly in MAECs from HFD-fed ApoeKOStingWT mice, accompanied by the downregulated H3K27me3 (Fig. 6L and Fig. S5N). The enrichment of IRF3, p65, and BRD4 on the downstream of Icam-1 promoter was barely detected in ox-LDL-treated MAECs from ApoeKOStingEC-KO mice, but increased with STING overexpression and inhibited by 1,6-hexanediol (Fig. 6N and Fig. S5O). Altogether, these results demonstrated that the activation of non-canonical STING pathway promotes the development of atherosclerosis by exacerbating inflammation, which is mediated by the formation of a super-enhancer complex involving BRD4 and condensate formation.
Given that Tek-cre mice may also induce gene deletion in myeloid cells, we performed bone marrow transplantation to eliminate the effect of STING in myeloid cells on atherosclerosis. The cell transplant efficiency of the irradiation and bone marrow transplant is depicted in Supporting Information Fig. S6A. Remarkably, irradiated ApoeKOStingEC-KO mice subjected to autologous ApoeKOStingEC-KO transplantation led to a significant decrease in plaque area compared with ApoeKOStingWT subjected to autologous ApoeKOStingWT transplantation. However, compared with ApoeKOStingEC-KO subjected to autologous ApoeKOStingEC-KO transplantation, transplantation of ApoeKOStingWT bone marrow cells into ApoeKOStingEC-KO recipient mice slightly increased plaque area, but it did not match the levels seen in of ApoeKOStingWT. Meanwhile, compared with ApoeKOStingWT subjected to autologous ApoeKOStingWT transplantation, transplantation of ApoeKOStingEC-KO bone marrow cells into ApoeKOStingWT recipient mice led to a minor reduction in plaque area, which remained higher than in the ApoeKOStingEC-KO mice subjected to autologous ApoeKOStingEC-KO transplantation (Fig. 6O). The mRNA levels of adhesions molecules and chemokines from the bone marrow transplantation mice arteries had the similar change (Fig. S6B). Our bone marrow transplantation experiment suggests that despite Tek-cre inducing deletion in both myeloid cells and ECs, restoration of macrophage STING could not effectively inhibit the atheroprotective effect achieved through EC-directed STING deletion. These results indicate that the STING in ECs plays a more important role than macrophages in atherosclerosis.
In addition, we also constructed the endothelium-directed Brd4 knockout hyperlipidemic mice (ApoeKOBrd4EC-KO) by crossing Brd4fl/fl mice with Cdh5 promoter-driven Cre recombinase line on an ApoeKO background to explore the functional consequence of IRF3/BRD4/p65 complex formation. Endothelium-specific overexpression of STING was induced in ApoeKOBrd4EC-KO mice through the injection of endothelial-enhanced adeno-associated virus (AAVendo)-Sting-WT via the tail vein. After 4 weeks, the mice were fed with HFD for 16 weeks. The successful construction of the endothelium-specific Brd4 knockout ApoeKO mice was confirmed by qRT-PCR (Supporting Information Fig. S7A), and the efficiency of the endothelium-specific overexpressing STING was also validated (Fig. S7B). Based on the oil red O staining of en face aorta, we found that in contrast to HFD-fed ApoeKOBrd4WT mice, ApoeKOBrd4EC-KO mice showed decreased atherosclerotic lesion. However, there was no difference in the atherosclerotic plaque between ApoeKOBrd4EC-KO mice transfected with AAVendo-Sting-Vector and AAVendo-Sting-WT (Fig. S7C). Besides, the mRNA levels of inflammatory genes decreased in the aortas of HFD-fed ApoeKOBrd4EC-KO mice, which were not affected by AAVendo-Sting-WT (Fig. S7D). These results suggest that BRD4 is the core effector of the IRF3/BRD4/p65 complex responding to the activation of the STING pathway and subsequently triggering the transactivation of inflammatory genes.
3.7. Activation of non-canonical STING-PERK pathway in human atherosclerotic plaques
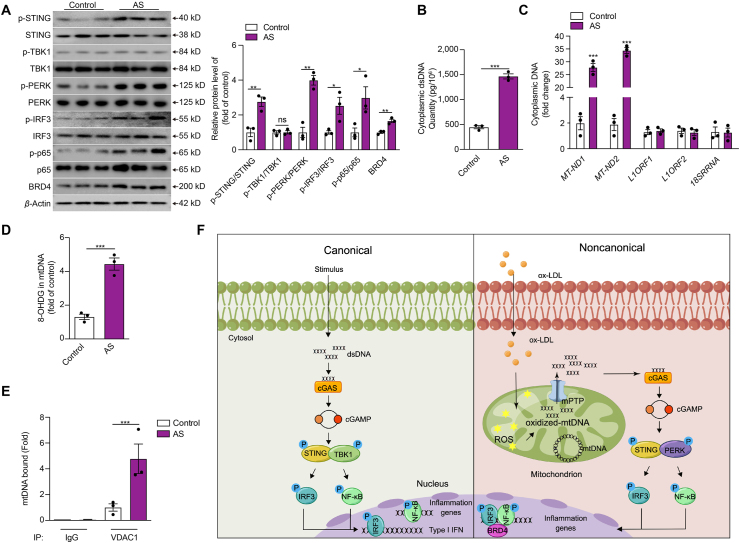
We first examined the activity of the non-canonical STING-PERK pathway and the presence of cytoplasmic DNA in human atherosclerotic tissue. Aortic tissue samples were obtained from atherosclerotic patients (n = 3) and age-matched organ donors (control group, n = 3). We found that the phosphorylated-STING, phosphorylated-PERK, phosphorylated-IRF3, phosphorylated-p65, as well as BRD4 was highly expressed in atherosclerotic plaques compared with those from controls (Fig. 7A). Cytoplasmic dsDNA content, especially the mtDNA, increased dramatically in atherosclerotic plaques compared with those from controls (Fig. 7B and C). Besides, we also confirmed that the oxidation level of mtDNA was increased in atherosclerotic plaques compared with those from controls (Fig. 7D). Meanwhile, co-immunoprecipitation with antibody to VDAC1 pulled down more mtDNA in atherosclerotic plaques than controls (Fig. 7E).
Figure 7.
The activation of the non-canonical STING-PERK pathway in human atherosclerotic plaques. (A–E) The protein level of the non-canonical STING–PERK pathway (A), the cytoplasmic dsDNA concentration (B), the cytoplasmic levels of mtDNA, nuclear LINE1 elements or RNA 18S (C), the mtDNA oxidation (D), mtDNA binding to VDAC1 (E) in the aortic tissues from patients with and without atherosclerosis. (F) The proposed model of non-canonical STING–PERK pathway activation and development of atherosclerosis. ox-LDL damages mitochondria of ECs and increases the generation of ROS, which oxidizes the mtDNA. The oxidized mtDNA interacts with VDAC1 and induces oligomerization of VDAC1 to form mPTP, releasing mtDNA to the cytosol. DNA binds and activates DNA sensor cGAS through conformational changes, producing cGAMP from ATP and GTP to activate the adaptor STING. STING dimerizes and recruits PERK to phosphorylate and activate transcription factors IRF3 and NF-κB. Meanwhile, STING regulates the formation of the transcriptional complex of super-enhancer on the proximal promoter regions of proinflammatory cytokines, including p-IRF3, p-p65, and BRD4 via the condensation characteristic of BRD4 to promote the transactivation of proinflammatory cytokines. Data are shown as mean ± SEM, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001; ns, no significance.
4. Discussion
Traditionally, cytoplasmic dsDNA could activate the classical STING–TBK1–IRF3 axis to participate in the progression of various diseases. Our data identified for the first time that the non-canonical STING–PERK pathway plays a vital role in endothelial dysfunction and atherosclerosis via promoting inflammation. Our results revealed that oxidized mtDNA, resulting from ox-LDL, encourages the release of mtDNA into the cytosol via VDAC1-dependent mPTP opening. Then, the cytoplasmic dsDNA sensor cGAS recognizes mtDNA and catalyzes the synthesis of cGAMP, which activates the non-canonical STING–PERK pathway. Typically, STING activates IRF3 and NF-κB via TBK1 contributing to IFN signals and inflammation respectively. Diversely, in our study, the activated non-canonical STING–PERK pathway integrating IRF3, NF-κB, and BRD4 in the chromatin microenvironment of regulatory regions of inflammatory cytokines to form super-enhancer via a BRD4-dependent condensation process to precisely orchestrate pro-inflammatory gene expression, distinct from the canonical STING–TBK1–IRF3 pathway (Fig. 7F).
Endothelial dysfunction driven by inflammation resulting from the damage of oxidized lipoproteins is considered the major initiating factor in atherosclerosis. Although the STING pathway as a research hotspot in antimicrobial and immunity30, has been reported in myocardial infarction and aortic aneurysm and dissection9,31, the role of the STING pathway in endothelial dysfunction and atherosclerosis has been unclear. Here, we discovered for the first time that the non-canonical STING–PERK pathway is significantly activated in human atherosclerotic tissue and the vascular walls of the HFD-challenged ApoeKO atherosclerosis mouse model. Compared to WT mice, endothelium-directed Sting knockout hyperlipidemic ApoeKO mice displayed a substantial decrease in plaque area and the expression of inflammation genes in the endothelium as well as the ameliorative endothelium-dependent vasodilation, without affecting the plasma lipids content. Genome-wide expression profiling of WT and STING knockdown ECs using RNA-seq also confirmed that pro-inflammatory genes were among the most highly differentially expressed genes after ox-LDL stimulation. Especially, we also demonstrated that compared to macrophages, the non-canonical STING–PERK pathway activation in ECs plays a more crucial role in atherosclerosis by bone marrow transplantation in Tek-cre mice. Together, these data identify that the non-canonical STING–PERK pathway plays a crucial role in atherosclerosis via affecting inflammation other than cholesterol metabolism.
The cGAS–STING pathway is activated by cytoplasmic dsDNA from either mitochondrial or nuclear origin24. Consistent with this concept, we detected a marked increase of cytoplasmic dsDNA content in human atherosclerotic tissue and the vascular wall of HFD-challenged ApoeKO mice. Moreover, the dsDNA sensed by cGAS was mainly the mtDNA rather than nuclear or ribosomal DNA. However, how mtDNA leaked from mitochondria remains unknown. Our study found obvious mitochondrial damage and mPTP opening in ox-LDL-treated HCAECs. As mitochondria are the key sources of ROS, we explored whether the leak of mtDNA was related to mitochondrial ROS. The results showed that the leak of mtDNA into the cytoplasm, mPTP opening, and expression of inflammatory genes challenged with ox-LDL were all reversed by mitochondrial ROS scavenger. VDAC1, the most abundant VDAC isoform, has been reported to form the large mitochondrial outer membrane pores under mitochondrial stress via oligomers, which is stabilized by mtDNA through direct interaction26. Furthermore, we observed a significant degree of mtDNA oxidization and mtDNA interaction with VDAC1, which were also inhibited by mitochondrial ROS scavenger. This suggests that the leak of mtDNA into the cytoplasm in ox-LDL-treated HCAECs is triggered by mtDNA oxidation followed by mPTP opening in a VDAC1-dependent manner. The endothelial injury caused by ox-LDL could also be ameliorated by the potent VDAC1 oligomerization inhibitor VBIT-4, which is consistent with Kim et al.26. These suggest that mitochondria could have attempted to activate the innate immune defenses via mtDNA under oxidized LDL. At the same time, persistent stress would provoke inflammation leading to diseases. Interestingly, we identified for the first time that the inflammatory response relies on the interaction between oxidized mtDNA and VDAC1. Therefore, VDAC1 can also be a therapeutic target for atherosclerosis in addition to STING.
The abnormal activation of the classical STING–TBK1–IRF3 pathway is closely associated with many diseases, such as Aicardi-Goutières syndrome, tumorigenesis and Parkinson's disease32, 33, 34. However, some phenotypes with abnormal STING signaling activation appear to be independent of the classical STING–TBK1–IRF3 axis10,14. PERK is an endoplasmic reticulum stress sensor, regulating the efficiency of ER quality control and mitochondrial homeostasis35. Recently, the role of PERK in regulating STING pathway has received increasing attention. Almeida et al.35 have reported that PERK regulates myeloid cell-induced immunosuppression in cancer by inhibiting STING signaling. Besides, STING could interact with the kinase domain of PERK in the cytoplasmic reticulum membrane through its carboxyl terminal domain localized in the cytoplasm and directly promote its activation, promoting senescence and organ fibrosis15. In our study, for the first time, we identified the crucial function of non-canonical STING–PERK–IRF3 axis in promoting endothelial dysfunction and atherosclerosis. However, the mechanism underlying the activation of PERK by STING needs further investigation.
The microenvironment of the genomic locus plays a crucial role in the fine-tuning of gene expression, including the high level of chromosome architecture and long-distance interaction. NF-κB, as a master regulatory transcription factor, cooperates with chromatin-associated regulatory complexes to drive inflammatory genes transcription in several dominant inflammatory signaling cascades36. IRF3 has been reported to be an essential coactivator for NF-κB-dependent transcriptional responses to virus infection16. Besides, the transcriptional and epigenetic regulator BRD4 could also recruit NF-κB to the proximal promoter regions and transactivates the proinflammatory genes by forming super-enhancers17. Our result showed that STING activation mediates the recruitment of p65, IRF3, and BRD4 simultaneously on the proximal promoter regions of the proinflammatory genes during ox-LDL-induced endothelial injury, such as ICAM-1, to synergistically enhance gene transcription. Moreover, p65, IRF3, and BRD4 co-localized with super-enhancer maker H3K27ac in the proximal promoter region of ICAM-1, and the absence of either component affects the expression of inflammatory genes. This result suggests that STING pathway activation transactivates the expression of inflammatory genes via facilitating the formation of super-enhancer in the proximal promoter regions by synergistic cooperation among BRD4, p65, and IRF3, which is different from the canonical mode that STING activates IRF3 and NF-κB contributing to IFN signals and inflammation respectively. Whereas how the STING pathway regulates the expression and activation of BRD4 in ox-LDL-treated ECs still needs to be further explored.
Interestingly, emerging evidence has reported that the binding factors at super-enhancers condense into membrane-less organelles, resulting in condensate formation from the nucleoplasm, which compartmentalizes and concentrates transcriptional machinery within restricted regions for superior transcriptional output29. Condensation is a process by which a homogeneous liquid solution (or phase) of macromolecular components separates (or demixes) into two distinct phases, which is thought to underlie many biological processes, including cancer, neurodegeneration, and infectious diseases37. However, the role of condensation in CVDs is still unclear. BRD4 is a major tandem-bromodomain-containing transcription regulator and can form phase-separated droplets at the proximal promoter regions of the proinflammatory genes to facilitate the formation of super-enhancer29. Here, we found that the activation of the STING pathway promotes the formation of a super-enhancer complex consisting of IRF3, p65, and BRD4, which could be destructed by condensate inhibitor 1,6-hexanediol. Therefore, we reported a novel condensation-dependent transcriptional regulatory mechanism of the STING pathway, which would benefit the development of therapeutic targets for atherosclerosis and STING related to inflammatory diseases.
Recently, medicines that target the STING pathway have been intensely pursued by the pharmaceutical industry. Preclinical studies in animal models have confirmed that both agonists and antagonists of the STING pathway can influence the outcome across a broad spectrum of diseases38. Here, both in vitro and in vivo data showed that the STING-specific inhibitor C-176 could rescue the inflammatory phenotypes and plaque progression. Moreover, the VDAC1 oligomerization inhibitor VBIT-4 is also beneficial for atherosclerosis by inhibiting mtDNA's release into the cytoplasm in vivo. Besides, the deficiency of IRF3, p65, and BRD4 also significantly inhibited the expression of inflammatory genes. Therefore, our study laid the foundation that the molecules in the upstream and downstream of STING signaling pathway are all potential targets of treatment and clinical drug development for atherosclerosis and CVDs.
5. Conclusions
We identified for the first time that the non-canonical STING–PERK pathway-dependent epigenetic paradigm via integrating IRF3, NF-κB and BRD4 in inflammatory response, which provides emerging therapeutic modalities for ameliorating vascular endothelial dysfunction and atherosclerosis.
Acknowledgments
We thank Yong Ji from the Nanjing Medical University for providing the Tek-Cre mice. This work was supported by the National Nature Science Foundation of China (82270421, 81970428, 31771334, 81800385, 82270484, 81873654, 31800971, and 82170503), and the Major Research Plan of the National Natural Science Foundation of China (91649125). University Natural Science Research of Jiangsu Province (18KJB310008, China), Natural Science Foundation of Jiangsu Province (BK20180684, China). This research was also supported by the program of special professor of Jiangsu Province, the program of the special medical experts of Jiangsu Province and the program of innovation and entrepreneurship team plan of Jiangsu Province, Major project supported by the Basic Science (Natural Science) Foundation of the Jiangsu Higher Education Institutions, Jiangsu Provincial Social Development Project (BE2021749, China).
Author contributions
Xuesong Li, Xiang Chen, Longbin Zheng, Qingguo Li, Yuanqing Gao and Hongshan Chen conceived and designed the study. Xuesong Li, Xiang Chen, Longbin Zheng, Minghong Chen, Yunjia Zhang, Ruigong Zhu and Xian Ji performed the in vitro experiments. Xuan Wu, Jiajing Chen, Quanwen Yin, Jiaming Gu, Xiang Chen, Longbin Zheng, Yunjia Zhang, Minghong Chen, Mengdie Dong performed the in vivo experiments. Bioinformatics analysis was performed by Hong Jiang. All authors analyzed the data. Xuesong Li, Xiang Chen, Longbin Zheng, Xin Tang, Yuanqing Gao, Qingguo Li and Hongshan Chen wrote the manuscript. Yuanqing Gao and Hongshan Chen modified the language of this article. All authors reviewed and approved the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.08.015.
Contributor Information
Qingguo Li, Email: lqg0235062@163.com.
Yuanqing Gao, Email: yuanqinggao@njmu.edu.cn.
Hongshan Chen, Email: hongshanchen@njmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Benjamin E.J., Virani S.S., Callaway C.W., Chamberlain A.M., Chang A.R., Cheng S., et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Xu S., Ilyas I., Little P.J., Li H., Kamato D., Zheng X., et al. Endothelial dysfunction in atherosclerotic cardiovascular diseases and beyond: from mechanism to pharmacotherapies. Pharmacol Rev. 2021;73:924–967. doi: 10.1124/pharmrev.120.000096. [DOI] [PubMed] [Google Scholar]
- 3.Zheng S., Li Z., Song J., Wang P., Xu J., Hu W., et al. Endothelial METRNL determines circulating METRNL level and maintains endothelial function against atherosclerosis. Acta Pharm Sin B. 2023;13:1568–1587. doi: 10.1016/j.apsb.2022.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Bai X.C., Chen Z.J. Structures and mechanisms in the cGAS–STING innate immunity pathway. Immunity. 2020;53:43–53. doi: 10.1016/j.immuni.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Hinkle J.T., Patel J., Panicker N., Karuppagounder S.S., Biswas D., Belingon B., et al. STING mediates neurodegeneration and neuroinflammation in nigrostriatal α-synucleinopathy. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2118819119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King K.R., Aguirre A.D., Ye Y.X., Sun Y., Roh J.D., Ng R.P., Jr., et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med. 2017;23:1481–1487. doi: 10.1038/nm.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C.H., Davidson S., Harapas C.R., Hilton J.B., Mlodzianoski M.J., Laohamonthonkul P., et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183:636–649.e18. doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y., Liu Y., An W., Song J., Zhang Y., Zhao X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J Clin Invest. 2019;129:546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo W., Wang Y., Zhang L., Ren P., Zhang C., Li Y., et al. Critical role of cytosolic DNA and its sensing adaptor STING in aortic degeneration, dissection, and rupture. Circulation. 2020;141:42–66. doi: 10.1161/CIRCULATIONAHA.119.041460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Sanchez G.A.M., et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oduro P.K., Zheng X., Wei J., Yang Y., Wang Y., Zhang H., et al. The cGAS–STING signaling in cardiovascular and metabolic diseases: future novel target option for pharmacotherapy. Acta Pharm Sin B. 2022;12:50–75. doi: 10.1016/j.apsb.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner K.P., Hornung V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat Rev Mol Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Sun L., Chen Z.J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 14.Motwani M., Pawaria S., Bernier J., Moses S., Henry K., Fang T., et al. Hierarchy of clinical manifestations in SAVI N153S and V154M mouse models. Proc Natl Acad Sci U S A. 2019;116:7941–7950. doi: 10.1073/pnas.1818281116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D., Liu Y., Zhu Y., Zhang Q., Guan H., Liu S., et al. A non-canonical cGAS–STING–PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat Cell Biol. 2022;24:766–782. doi: 10.1038/s41556-022-00894-z. [DOI] [PubMed] [Google Scholar]
- 16.Freaney J.E., Kim R., Mandhana R., Horvath C.M. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N., Wu R., Tang D., Kang R. The BET family in immunity and disease. Signal Transduct Targeted Ther. 2021;6:23. doi: 10.1038/s41392-020-00384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y., Lee Y., Cui K., He M., Wang B., Bhattacharjee S., et al. Epsin-mediated degradation of IP3R1 fuels atherosclerosis. Nat Commun. 2020;11:3984. doi: 10.1038/s41467-020-17848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baselet B., Driesen R.B., Coninx E., Belmans N., Sieprath T., Lambrichts I., et al. Rosiglitazone protects endothelial cells from irradiation-induced mitochondrial dysfunction. Front Pharmacol. 2020;11:268. doi: 10.3389/fphar.2020.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayari H., Bricca G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci. 2013;38:311–315. doi: 10.1007/s12038-013-9310-2. [DOI] [PubMed] [Google Scholar]
- 22.Pan B.S., Perera S.A., Piesvaux J.A., Presland J.P., Schroeder G.K., Cumming J.N., et al. An orally available non-nucleotide STING agonist with antitumor activity. Science. 2020;369 doi: 10.1126/science.aba6098. [DOI] [PubMed] [Google Scholar]
- 23.Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z., Zhou H., Ouyang X., Dong Y., Sarapultsev A., Luo S., et al. Multifaceted functions of STING in human health and disease: from molecular mechanism to targeted strategy. Signal Transduct Targeted Ther. 2022;7:394. doi: 10.1038/s41392-022-01252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X., Fu W., Zhou J., Shuai N., Yang Y., Wang B. Oxymatrine attenuates oxidized low-density lipoprotein-induced HUVEC injury by inhibiting NLRP3 inflammasome-mediated pyroptosis via the activation of the SIRT1/Nrf2 signaling pathway. Int J Mol Med. 2021;48:187. doi: 10.3892/ijmm.2021.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J., Gupta R., Blanco L.P., Yang S., Shteinfer-Kuzmine A., Wang K., et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366:1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decout A., Katz J.D., Venkatraman S., Ablasser A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pott S., Lieb J.D. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 29.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M., Jia K., Wang L., Li W., Chen B., Liu Y., et al. Alterations of DNA damage response pathway: biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm Sin B. 2021;11:2983–2994. doi: 10.1016/j.apsb.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu D., Cui Y.X., Wu M.Y., Li L., Su L.N., Lian Z., et al. Cytosolic DNA sensor cGAS plays an essential pathogenetic role in pressure overload-induced heart failure. Am J Physiol Heart Circ Physiol. 2020;318:H1525–H1537. doi: 10.1152/ajpheart.00097.2020. [DOI] [PubMed] [Google Scholar]
- 32.Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M., et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H., Xue X., Panda S., Kawale A., Hooy R.M., Liang F., et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019;38 doi: 10.15252/embj.2019102718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sliter D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D., et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Almeida L.M., Pinho B.R., Duchen M.R., Oliveira J.M. The PERKs of mitochondria protection during stress: insights for PERK modulation in neurodegenerative and metabolic diseases. Biol Rev Camb Phil Soc. 2022;97:1737–1748. doi: 10.1111/brv.12860. [DOI] [PubMed] [Google Scholar]
- 36.Natoli G. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harbor Perspect Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberti S., Dormann D. Liquid–liquid phase separation in disease. Annu Rev Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 38.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS–STING pathway in health and disease. Nat Rev Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.