Abstract
Adeno-associated virus (AAV) is a member of the Parvoviridae, belonging to the Dependovirus genus. Currently, several distinct isolates of AAV are in development for use in human gene therapy applications due to their ability to transduce different target cells. The need to manipulate AAV capsids for specific tissue delivery has generated interest in understanding their capsid structures. The structure of AAV type 4 (AAV4), one of the most antigenically distinct serotypes, was determined to 13-Å resolution by cryo-electron microscopy and image reconstruction. A pseudoatomic model was built for the AAV4 capsid by use of a structure-based sequence alignment of its major capsid protein, VP3, with that of AAV2, to which AAV4 is 58% identical and constrained by its reconstructed density envelope. The model showed variations in the surface loops that may account for the differences in receptor binding and antigenicity between AAV2 and AAV4. The AAV4 capsid surface topology also shows an unpredicted structural similarity to that of Aleutian mink disease virus and human parvovirus B19, autonomous members of the genus, despite limited sequence homology.
Adeno-associated virus (AAV) is a member of the Parvoviridae family (45). AAV virions have a T=1 icosahedral capsid consisting of 60 copies of three related proteins, VP1, VP2, and VP3, at an estimated ratio of 1:1:8, which surrounds a single-stranded DNA genome. These three proteins share a common C-terminal region (the ∼590 amino acids of VP3) but have different amino termini resulting from alternative start codon usage. The entire sequence of VP3 is present in VP2, whose sequence is in turn entirely contained within VP1. The VP3 common region appears to be essential for cell binding and antigenic properties and possibly plays a role in genomic DNA packaging. VP1 of AAV serotype 2 (AAV2) has a unique N-terminal region of ∼130 amino acids, which is important in the viral life cycle after cell binding and entry, and displays a phospholipase A2 function (25) that also appears to be conserved in other serotypes.
The AAVs are distinct from the autonomous parvoviruses by their dependence on a helper virus for replication. These viruses require coinfection with either an adenovirus or a herpesvirus for replicative infection (45). To date, nine distinct serotypes, AAV1-5, AAV7-9, and AAAV, have been cloned from either human or primate samples (5, 7, 10, 23, 41). In addition, a large number of sequences were recently isolated from monkeys (24). There are no diseases associated with AAVs, and their capability of packaging foreign DNA makes them attractive for development as gene therapy vectors (39).
The three-dimensional (3D) structures of several autonomous parvoviruses and that of AAV2 have been determined by X-ray crystallography (1, 2, 4, 37, 53, 61, 68). Lower resolution structures of AAV2 and AAV5 (40, 63), as well as those of the autonomous parvoviruses Aleutian mink disease parvovirus (ADV) and human parvovirus B19 (16, 44), have been mapped by a combination of cryo-electron microscopy (cryo-EM) and pseudoatomic model building. In all of these structures, only the C-terminal common region of VP1/2/3 is observed. The core of the protein is composed of a conserved eight-stranded antiparallel β-barrel motif (3, 14). The majority of the variable surface structure consists of large loops inserted between the strands of the β-barrel. Structural features on the capsid surfaces of these viruses include projections at or surrounding the icosahedral threefold axis and depressions at the twofold and around the fivefold axes (2, 4, 16, 37, 44, 52, 53, 61, 63, 68). A conserved cylindrical channel is present at the icosahedral fivefold axis formed by symmetry-related β-ribbons (3, 67).
Biochemical and molecular characterizations of several of the different serotypes of AAV have indicated that each has unique cell binding characteristics and tissue transduction efficiencies (19, 35, 49, 64). AAV2, the best characterized serotype, has a broad tropism, including tropisms for the eye, central nervous system (CNS), liver, and muscle (21, 42, 54, 66), and has been studied for the treatment of genetic diseases such as cystic fibrosis and alpha 1 antitrypsin deficiency (22, 55, 62). However, vectors derived from other AAV serotypes have been reported to be more efficient at transducing certain cell types than AAV2. AAV1 has demonstrated an improved tropism for striated muscle cells compared to AAV2 (29). AAV3 can transduce hematopoietic cells (28). AAV4, originally isolated from African green monkeys, is able to infect cells from humans and rodents (15). The direct injection of AAV4 into the striata of mice demonstrated a strong tropism for ependymal cells in the CNS (18). In vitro and in vivo experiments with AAV5 have demonstrated improved binding and transduction of airway lung epithelia, muscles, CNS neurons, and the eye compared to AAV2 (18). The more recently identified AAV serotypes, including AAV7 and AAV8, also appear to have unique cell tropisms. For example, AAV8 is reported to efficiently transduce liver cells (23). These differences in cell tropism appear to be due to the ability of the AAV capsids to utilize different cell surface carbohydrates and/or protein receptors for cell binding and entry.
For AAV2, an interaction with heparin sulfate proteoglycans is important for cell binding and transduction (57), although a requirement for human fibroblast growth factor 1 (48) or integrin αvβ5 (56) has been reported. Mutagenesis and the available crystal structure of AAV2 have enabled the identification of the basic residues in VP3 that are responsible for its interaction with heparin (38, 46, 68). The suggested binding site contains a clustering of residues, contributed by icosahedral threefold axis symmetry-related VP3 molecules, that form three basic patches surrounding the icosahedral threefold axes. AAV1, which is ∼83% identical to AAV2 and contains most of the basic amino acids in VP3 that have been identified as being important for heparin binding (but not the critical R585 and R588 residues [38, 46]), does not appear to bind heparin. The carbohydrate moiety recognized by AAV1 for transduction has yet to be determined. The ability to bind heparin is conserved in AAV3, which is ∼87% identical to AAV2, although its affinity is weaker (49). However, AAV3 also lacks R585 and R588. Transduction by AAV4 and AAV5, which are ∼55% identical to AAV2 and to each other, is insensitive to competition with soluble heparin (35, 64). Recent studies with AAV4 and AAV5 indicated that they have hemagglutination activity and require sialic acid for binding and transduction but differ in their specificities (35). AAV4 requires α2,3-O-linked sialic acid and AAV5 requires α2,3-N-linked sialic acid for transduction (35). In addition, the platelet-derived growth factor receptors (PDGFRa and PDGFRb) have been identified as protein receptors for AAV5, and their expression correlates with transduction in vivo (19). The nature of the carbohydrate molecules utilized for cell binding by AAV7-9, AAAV, and the other newly isolated AAVs is yet to be identified.
Unlike heparin sulfate binding, which requires defined basic patches or consensus sequences, to our knowledge there are no consensus amino acid motifs for sialic acid recognition. To explore structural features of the AAV virus capsid responsible for sialic acid binding, we initiated structural studies of AAV4 and AAV5. Our 16-Å-resolution cryo-EM and image reconstruction of AAV5 have been reported elsewhere (63). Here we report a structural determination of intact wild-type (wt) AAV4 particles to a 13-Å resolution by cryo-EM and image reconstruction. Using this reconstruction and a structure-based alignment with the 3D structure of the AAV2 VP3 protein (68), we developed a pseudoatomic model for AAV4. At this medium level of resolution, the AAV4 capsid surface topology clearly highlights differences from those of AAV2 and AAV5 associated with the capsid regions involved in carbohydrate binding and antigenic properties. Interestingly, the overall AAV4 capsid topology is very similar to that of ADV and B19 (1, 16, 44), which are both autonomous parvoviruses and to which AAV4 has very low sequence identity. Inspired by this coincidence, we compared the surface topologies of all available autonomous and dependovirus parvovirus structures at a low resolution and observed that general parvovirus surface topologies are not dictated by tissue tropism, pathogenicity, antigenicity, or specific carbohydrate recognition phenotypes, but might rather be a result of capsid protein translation and maturation characteristics.
MATERIALS AND METHODS
Cells and virus.
Cos cells were maintained as monolayer cultures in D10 medium (Dulbecco's modified Eagle's medium) containing 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 13 U of amphotericin B as recommended by the manufacturer (GIBCO, Gaithersburg, Md.). Wild-type AAV4 was produced and purified as previously described (36).
Cryo-EM and 3D image reconstruction.
Purified wild-type AAV4 particles, at a concentration of 5 × 1013 particles/ml, were quick-frozen on “holey” carbon-coated copper grids as previously described (44). The grids were transferred to the microscope by use of a precooled Gatan cryotransfer holder and were maintained at liquid nitrogen temperatures. Micrographs were recorded under low-dose conditions with a total dose of ∼20.1 e/Å2 at a calibrated magnification of ×47,000 by use of a Philips CM300 FEG transmission cryo-electron microscope operating at 300 kV and Kodak SO-163 film. The images were scanned by use of a Zeiss SCAI scanner with a step size of 7 μm/pixel and then twofold bin averaged to 14 μm/pixels, resulting in a pixel size of 2.98 Å at the specimen. A total of 4,205 particles were boxed, as previously described (44), from 14 micrographs with defocus values ranging from 1- to 3-μm underfocus. The contrast transfer functions for each micrograph were corrected as previously described (11) by use of the ROBEM subroutine of the Purdue suite of EM programs (6). The particle centers and orientations were estimated by the use of EMPFT in the Purdue suite of EM programs (6), with the available low-resolution reconstructed image of ADV (44) used as a starting model. Iterative rounds of image reconstruction (with the Purdue program EM3DR) and model improvement by strict selection of the particles by the EMSEL program based on correlation coefficient calculations were employed to improve these parameters (6). During iterations to improve the cryo-EM map resolution, the overall correlation coefficients and those for each particle image were kept above 0.3. The final density map was calculated from 2,892 particles to a resolution cutoff of 13 Å. The resolution of this map was estimated by dividing the particle images into two sets, generating two reconstructions and comparing the correlation coefficients and phase residuals of the two sets of structure factors as a function of spatial frequency (6). A cryo-EM image of AAV2 was reconstructed to 13-Å resolution from 3,032 particles selected from 14 frozen micrographs of capsids produced by use of an adenovirus expression system (70) as described above for AAV4, but employed an AAV5 reconstructed cryo-EM image (63) as the model for particle center and orientation determination in EMPFT (6).
Generating a pseudoatomic model for the AAV4 capsid.
The amino acid sequences of AAV2 and AAV4 (obtained from the National Center for Biotechnology Information [NCBI] database) were aligned by using the pair-wise option in the program CLUSTALW (v. 1.4) (59), with the following default parameters: gap opening penalty, 10; gap extension penalty, 0.5; and the BLOSUM matrix (30). This alignment procedure was followed by the prediction of the AAV4 secondary structure elements by submission of its sequence to the PSIPRED website (http://bioinf.cs.ucl.ac.uk/psipred/) (33, 43). The atomic coordinates of an AAV2 VP3 monomer (PDB accession no. 1LP3) (68) were rotated from their crystallographic unit cell orientation into a standard orientation by a least-square fit in the interactive graphics program O (34) and then were used to generate a pseudoatomic model of AAV4 based on the sequence alignment. The AAV2 VP3 amino acids were mutated to those of AAV4, coordinates for inserted amino acid sequences were obtained from the O database, and deleted sequences were removed. The model was subjected to energy minimization by use of the CNS program (13). In addition to this interactive model building, the amino acid sequence of AAV4 VP3 was submitted to the online atomic model generator Swiss Model (http://www.expasy.org/) (50), with the AAV2 structure (68) supplied as a template. We had previously used a similar protocol to generate a working model for AAV5 (63). The model generated by Swiss Model (50) was essentially the same as that built interactively. This generated model was used for all comparisons with AAV2, AAV5, and available autonomous parvovirus structures after adjustment to fit the reconstructed cryo-EM density by use of the program O (34).
The Swiss Model coordinates, in a standard orientation, were translated into the reconstructed density map as a rigid body in the program O and then interactively adjusted, within one viral asymmetric unit, to better fit the map. The cryo-EM density map was generated for display in the program O as previously described (11). Due to the medium resolution of the map, to 13 Å, the model was only further fitted into the reconstructed density by adjusting the main chain in accordance with the capsid density envelope. The modified model was energy minimized by use of the CNS program (13). The 60 VP3 icosahedral symmetry equivalent subunits of the virus capsid were generated by matrix multiplication after refinement. The AAV4 capsid pseudoatomic model was used to generate an electron density map for comparison with the cryo-EM density in the OVERLAPMAP subroutine in CCP4 package (17) to obtain a correlation coefficient. An electron density difference map was calculated between the cryo-EM density of the wt AAV4 capsids (containing VP1, VP2, and VP3) and a map generated for the AAV4 VP3 model (for residues 215 to 735 [VP1 numbering]), also by the use of CCP4 package (17). The resultant map was displayed in the program O (34). No attempt was made to model amino acids into this difference density.
Structural alignment of AAV serotypes.
The amino acid sequences of AAV1, -3, AAV6-9 and AAAV (obtained from the NCBI database) were submitted to the online Swiss Model program (http://www.expasy.org/) (50) to generate three-dimensional structures, with the atomic coordinates of AAV2 (PDB accession no. 1LP3) (68) as a template. The resulting models, plus the pseudoatomic models built into the cryo-EM reconstructed densities of AAV4 and AAV5 (63), were superimposed onto the atomic coordinates of AAV2 (68) in the molecular graphics program O by a least-squares algorithm (34) to generate a structural alignment.
Comparison of AAV4 to the amino acid sequences and structures of autonomous parvoviruses.
The amino acid sequence of AAV4 was compared to those of the autonomous parvoviruses ADV, B19, canine parvovirus (CPV), feline panleukopenia virus (FPV), and porcine parvovirus (PPV) (amino acid sequences were obtained from the NCBI database) by the use of CLUSTAL W (59) as described above for AAV2 and AAV4. The pseudoatomic model of AAV4 built into its reconstructed cryo-EM density was superimposed onto the pseudoatomic coordinates of ADV (44) and the atomic coordinates of B19 (37), CPV (67), FPV (51), minute virus of mice (MVM) (4), and PPV (53) (PDB accession no. 1S58, 2CAS, 1C8E, 1MVM, and IK3V) with the program O (34) to obtain a root means squared deviation (RMSD) for their C-α positions, and a percentage of structural similarity was calculated.
Generation of low-resolution surface maps for available parvovirus structures.
Cryo-EM surface images of AAV5, ADV, and B19 were generated from reconstructed densities at 16-, 22-, and 25-Å resolutions, respectively (16, 44, 63) by use of the Purdue suite of EM programs (6). Low-resolution surface maps of B19, CPV, Galleria mellonella densovirus (DNV), FPV, MVM, and PPV were calculated from atomic coordinates obtained by X-ray crystallographic structure determinations (PDB accession numbers given above, plus 1DNV). Low-resolution surface maps of ADV, AAV4, and AAV5 were calculated from pseudoatomic models built into their reconstructed cryo-EM density maps. Structure factors were calculated to 13 Å (to be comparable to the resolution of the AAV4 cryo-EM density map) by use of the CCP4 package (17) after the generation of 60 VP2/3/4 molecules (depending on the virus) by applying icosahedral symmetry to a reference monomer in a standard orientation. A Fourier transform of the structure factors was used to generate low-resolution surface density maps according to a previously described procedure (9).
RESULTS AND DISCUSSION
Capsid surface topology of AAV4.
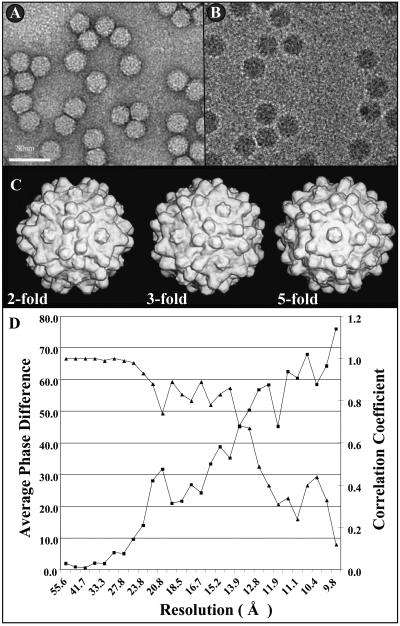
A cryo-EM reconstructed image of AAV4 was obtained from micrographs of frozen wt particles (Fig. 1A and B) propagated in Cos cells and purified by a combination of isopycnic density CsCl gradient ultracentrifugation and molecular weight exclusion filtration (36). A total of 2,892 2D particle projections of known centers and orientations from 14 micrographs were used to reconstruct the final 3D image to a 13-Å resolution (Fig. 1C and D). The AAV4 structure clearly shows the surface features associated with parvovirus capsids, namely, three mound-like protrusions surrounding the icosahedral threefold axes and depressions at the two- and fivefold axes (Fig. 1C and 2). The external diameters of the AAV4 capsid are approximately 220, 230, and 240 Å across the two-, three-, and fivefold icosahedral axes, respectively, and 290 Å across the threefold mounds (Fig. 1C). The peaks of the mounds surrounding the threefold axes in AAV4 are spaced at a distance of ∼26 Å from the exact icosahedral threefold axes.
FIG. 1.
Cryo-EM reconstruction of wild-type AAV4. Negatively stained (A) and frozen (B) micrographs of wt AAV4 particles are shown. The majority of the particles were boxed from carbon film as shown in panel B. Bar = 50 nm. (C) Surface-shaded cryo-EM reconstructed AAV4 images (Purdue image format maps [pifmaps]) viewed down the icosahedral twofold (left), threefold (center), and fivefold (right) axes. The pifmaps were generated from 2D projections by use of the EM3DR subroutine in the Purdue suite of EM programs (6). (D) AAV4 map resolution (Å) determination. The resolution was determined to be where the average phase difference dropped below 50% or the correlation coefficient dropped below 0.5 (6).
FIG. 2.
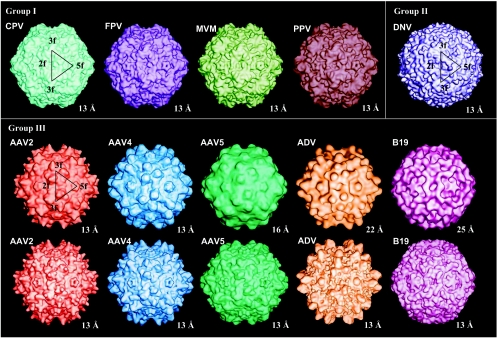
Parvovirus structures at low resolution. A comparison of surface-shaded low-resolution maps of AAV2 (in red), AAV4 (in blue), AAV5 (in dark green), ADV (in orange), B19 (in pink), CPV (in cyan), DNV (in purple), FPV (in magenta), MVM (in green), and PPV (in brown) at the indicated resolution is shown. The surface structure topology groupings (group I to III), as discussed in the text, are given above the respective panels. The 13-Å resolution map images for the group I and II viruses and for AAV2 and B19 in group III (bottom row of group III panel) were generated as previously described from structure factors and phases calculated from atomic coordinates (9). The 13-Å resolution maps for AAV4, AAV5, and ADV (bottom row of group III panel) were generated from pseudoatomic models built into their cryo-EM reconstructed densities. The pifmap images for AAV2, AAV4, AAV5, ADV, and B19 (top row of group III panel) were generated as described for Fig. 1C. The lower-resolution appearance of the reconstructed maps (top row of group III panel) compared to the rendered maps (groups I and II and bottom row of group III) stems from the high thermal motion inherent in cryo-EM data, which tends to decrease the apparent resolution of generated maps. The black triangles on AAV2, CPV, and DNV depict a viral asymmetric unit bound by icosahedral twofold (2f), two threefold (3f), and fivefold (5f) axes. The maps are viewed down an icosahedral twofold axis.
A cryo-EM image reconstruction of AAV2 at ∼13-Å resolution and our published structure of AAV5 (63) at a 16-Å resolution were compared to that of AAV4 (Fig. 2, group III, top row). In addition, low-resolution surface images of AAV2, AAV4, and AAV5, calculated to 13 Å from the atomic model of AAV2 (68) and the pseudoatomic models of AAV4 and AAV5 (63), were compared (Fig. 2, group III, bottom row). As expected for homologous proteins and/or viruses that are ∼55% or more identical (Table 1), the general surface topology of the reconstructed image of the AAV4 capsid is similar to those of AAV2 and AAV5 (Fig. 2, group III). The protrusion that extends out at the fivefold axes is more pronounced in AAV4 than in AAV2 and AAV5. The tips of the threefold mounds are finger-like in AAV2 and round in AAV4 and are larger in these two viruses than in AAV5. The twofold depression is deep in AAV4, shallow in AAV2, and barely discernible in AAV5 (63). Comparisons of the 13-Å-resolution rendered surface images were consistent with those for the cryo-EM reconstructed images, except for the icosahedral twofold axes of AAV4. The twofold axes in AAV4 were still like those of AAV2 as a result of the model (see below).
TABLE 1.
Sequence and structural alignment of parvoviruses with AAV4
| Sequence | % Identity | % Strongly similar | % Weakly similar | % Different | No. of C-α atoms aligned/total (%) | RMDS in C-α (Å) |
|---|---|---|---|---|---|---|
| AAV2a | 58 | 16 | 9 | 17 | 508/520 (98) | 0.5 |
| AAV5a | 52 | 16 | 11 | 21 | 499/520 (96) | 0.6 |
| ADVa | 18 | 18 | 11 | 53 | 377/520 (73) | 1.7 |
| B19a | 25 | 18 | 14 | 43 | 396/520 (76) | 1.8 |
| CPVb | 20 | 21 | 11 | 48 | 380/520 (73) | 1.7 |
| FPVb | 20 | 21 | 11 | 48 | 380/520 (73) | 1.7 |
| MVMb | 18 | 19 | 13 | 50 | 377/520 (73) | 1.7 |
| PPVb | 18 | 20 | 12 | 50 | 375/520 (72) | 1.7 |
| DNVc | 13 | 16 | 12 | 59 |
Group III.
Group I.
Group II.
The AAV4 capsid structure surface topology was also compared to those available for autonomous parvovirus and DNV (52) at 13-Å resolution, calculated from atomic or pseudoatomic models, or at their cryo-EM reconstructed resolution (Fig. 2, groups I, II, and III). The AAV4 capsid topology is surprisingly similar to those reported for ADV (44) and B19 (1, 16, 37), to which its amino acid sequence identities are 18 and 25%, respectively (Table 1). Depressions or valleys are created at the icosahedral threefold axes of the AAVs, ADV, and B19 due to the mounds that surround them (Fig. 2, group III). This threefold topology differs from the pinwheel protrusions at the icosahedral threefold axes in CPV (61), FPV (2), MVM (4), and PPV (53) (Fig. 2, group I). The AAV4, ADV, and B19 twofold depressions are narrower and deeper than those in CPV, FPV, MVM, and PPV, which appear wide and shallow (Fig. 2, group I). The surface depression around the icosahedral fivefold axis was conserved in all group I and III parvovirus structures compared, but it was shallow in AAV5 (Fig. 2). The DNV surface was observed to be drastically different from those of the group I and III viruses, including this fivefold region (Fig. 2).
Pseudoatomic model of the AAV4 capsid protein.
There is no structural information available for the unique region of the VP1 minor coat protein or the region where VP1 overlaps with only VP2 and the extreme N-terminal end of VP3 (where present) for any parvovirus, and as such, these regions were not represented in the pseudoatomic model generated for AAV4. A model of the main-chain atoms of the major coat protein of AAV4, VP3 (residues 215 to 734, based on VP1 numbering), was generated by use of the interactive graphics program O (34) based on the available atomic coordinates of AAV2, guided by an amino acid sequence alignment and the cryo-EM reconstructed density envelope, and by use of the protein model building program Swiss Model (http://www.expasy.org) (50) (Fig. 3 and 4A). The AAV4 VP3 model clearly shows the conservation of the core β-barrel motif (βB-βI in Fig. 5A and 6) that makes up the contiguous shell in all of the parvovirus capsids for which structures are known (1, 2, 4, 37, 44, 52, 53, 61, 63, 68), with no adjustments being required for it to fit into the EM density. This conservation of the β-barrel structural motif was also predicted for the AAV4 VP3 sequence by use of the PSIPRED algorithm (http://bioinf.cs.ucl.ac.uk/psipred) (33, 43). However, differences in the locations of surface loops between the β-strands in AAV4 and AAV2 were clearly evident when the model was placed inside the 13-Å-resolution AAV4 cryo-EM reconstructed density (Fig. 4A).
FIG.3.
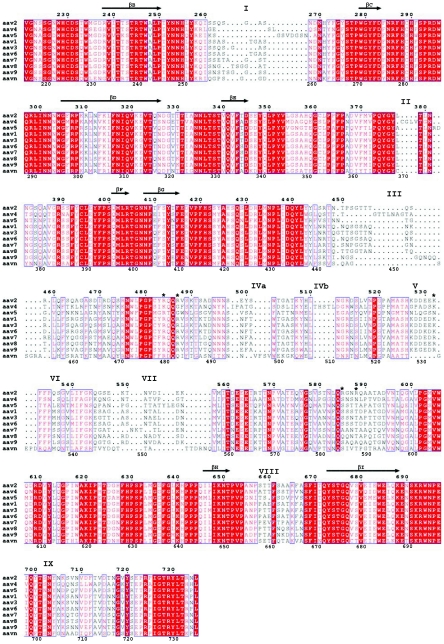
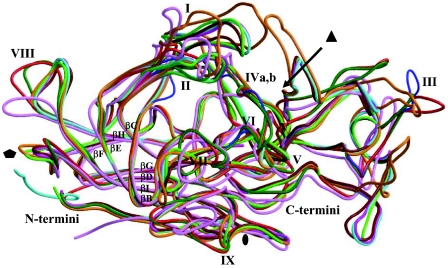
Structural alignment of AAV1-9 and AAAV (AAVN). The alignment was generated based on superimposition of the atomic coordinates of AAV2 (68), the pseudoatomic structures of AAV4 and AAV5 (63) built into their reconstructed cryo-EM densities, and structural models of AAV1, AAV3, AAV6-9, and AAAV generated with the Swiss Model program (50), with AAV2 as a template. The format was output by use of the program ESPript (26). Identical residues are white characters in red boxes; homologous residues are red characters in white boxes; differing residues are black characters. The numbering at the top of the alignment is based on AAV2 VP1 sequence numbers, while that at the bottom is based on AAV4 VP1 sequence numbers. Variable surface loop regions are labeled I to IX. Residues R484, R487, K532, R585, and R588 (AAV2 VP1 numbering), implicated in AAV2 heparin binding (38, 46), are indicated by asterisks. The residues within the eight core β-strands that form the contiguous parvovirus capsid are indicated by arrows above the residues.
FIG. 4.
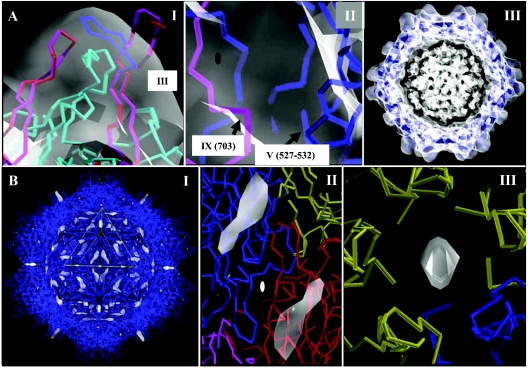
Pseudoatomic model building for the AAV4 capsid. (A) Panel 1 shows the C-α traces of the AAV4 monomer (in magenta) generated by Swiss Model (50) that required adjustment (in blue) in the program O (34) to fit inside the reconstructed density (in transparent white contoured at a σ of 1.0) of the mounds surrounding the icosahedral threefold axes and the starting AAV2 crystal structure model (in red) (68). An intervening loop from a threefold related VP3 subunit is shown in cyan. Variable region III is labeled. Panel II shows C-α traces for loop regions in a reference AAV4 VP3 (in blue) and a twofold related monomer (in magenta), residues 527 to 532 and near 703, respectively, that could not be adjusted to fit into the cryo-EM density envelope (in transparent white contoured at a σ of 1.0) at 13-Å resolution. These regions are labeled V and IX, respectively, as in Fig. 3 to 6. The approximate icosahedral twofold axis is indicated by the filled oval. Panel III shows the C-α traces for 60 AAV4 VP3 monomers (in blue) fitted into the cryo-EM density (in transparent white contoured at a σ of 1.0). The 60 VP3 molecules were generated by matrix multiplication of the coordinates of a reference VP3, in a standard orientation, with 60 icosahedral symmetry operators after model refinement in a crystallography and NMR system (CNS) program (13). The map is sectioned at the center to show the protein envelope. The density in the middle of the particle, at the same contour level as the protein envelope, could not be modeled as a protein or nucleic acid. The view is approximately down an icosahedral twofold axis. (B) Difference map density subtracting the VP3 model from the cryo-EM density. Panel I shows the 60 AAV4 VP3 monomers (in blue) and the positions of difference density (in transparent white) at a contour level of 2.2 σ. A viral asymmetric unit is shown in the yellow triangle, bounded by two threefold axes (bottom right and left edges) and a fivefold axis (top edge), with a twofold axis in the middle. The view, down an icosahedral twofold axis, is rotated 90° compared to the twofold views shown in Fig. 2. Panels II and III show close-up views of the difference densities (in transparent white) viewed down an icosahedral twofold axis (from the interior of the capsid) and in the channel at the icosahedral fivefold axes (viewed from the capsid exterior). The reference monomer is in blue in both panels, the twofold monomer is in red in panel II, and the fivefold monomers are in yellow in panel III.
FIG. 5.
Comparison of AAV2, AAV4, and AAV5. (A) Superimposition of coil representations of the VP3 monomers of AAV2 atomic model coordinates (in red) (68) and of AAV4 (in blue) and AAV5 (in dark green) (63) pseudoatomic models built into the cryo-EM density. Variable surface loop regions (adjusted and unmodified) are labeled I to IX as in Fig. 3. The insert shows a snapshot (from the program O [34]) of the position of the intervening icosahedral threefold related VP3 loop (in cyan) and variable region VI. The residues that form the basic cluster in AAV2 (R484, R487, K532, R585, and R588 [VP1 numbering]) utilized for heparin binding are shown as small red balls and are labeled. The first N-terminal residue modeled (215), the C-terminal residue 734, and β-strands B to I are also labeled. (B) Coil representations of a trimer of AAV2 VP3s (in red, magenta, and cyan), an AAV4 monomer (in blue), and an AAV5 monomer (in dark green), viewed down the icosahedral threefold (solid triangle) axes. The superimposed AAV2, AAV4, and AAV5 monomers are shown rotated ∼90° relative to the view in panel A. The AAV2 basic residues become clustered from the threefold related monomers and are shown in the respective colors of their monomers and outlined in dotted ovals indicated by arrows. The positions of the variable regions (labeled I to IX) are shown as black, dark gray, and gray balls in the red, cyan, and magenta AAV2 monomers, respectively. Panels A and B were generated with the program Bobscript (20). (C) Close-up of the icosahedral threefold (solid triangle) axes of the pifmaps of AAV2 (left), AAV4 (center), and AAV5 (right), showing the local differences in topology of their threefold mounds. The resolution for each map is given on the bottom right-hand side of each panel. Approximate locations for variable regions I to VI are shown on top of one AAV2 monomer and mound. Arrows on the AAV2 surface indicate the approximate locations of the basic residues (shown in A and B) utilized for heparin binding (38, 46). The approximate icosahedral two-, three-, and five-fold axes are indicated by the filled ovals, triangles, and pentagons, respectively, in panels A to C.
FIG. 6.
Superimposition of coil representations of the VP3 monomers of AAV2 (atomic coordinates) (in red), the pseudoatomic models of AAV4 (in blue) and AAV5 (in dark green), as in Fig. 5, the VP2/VP3 monomers of the atomic coordinates of B19 (in pink) (37), CPV (in cyan) (67), FPV (in magenta) (51), MVM (in green) (4), and PPV (in brown) (53), and the VP2 pseudoatomic coordinates of ADV (in orange) (44). Variable surface loop regions labeled I to IX are the same as those in Fig. 3 and 5. The N and C termini of the VPs are indicated. The approximate icosahedral two-, three-, and fivefold axes are indicated by the filled oval, triangle, and pentagon, respectively.
The variable surface loops (Fig. 3 to 5) were interactively adjusted and constrained by the cryo-EM density envelope (where possible) to better represent the AAV4 structure (Fig. 4A). The majority of the AAV2 and AAV4 capsid surface structure differences are located at the base, wall, and top of the mounds surrounding the icosahedral threefold axes (Fig. 4A and 5), even though they are spread throughout the primary sequence (Fig. 3). The insertion of amino acids after residue 262 (between βB and βC) in AAV2 compared to AAV4 (labeled “I” in Fig. 3 and 5) creates a slight protrusion just below the mounds surrounding the threefold axes in AAV2 (Fig. 5C). The insertion of amino acids after residues 369 and 506 in AAV4 (between βE and βF and between βG and βH, respectively) compared to AAV2 (labeled “II” and “IVb” in Fig. 3 and 5) creates an additional density that forms part of the threefold mounds in AAV4 (Fig. 5B and C). The finger-like projection of the AAV2 threefold mounds (40, 68), due to the loop formed between AAV2 residues 451 and 456 (AAV2 VP1 numbering, between βG and βH), was not observed in the AAV4 structure (Fig. 2, group III). Rather, the equivalent AAV4 loop, between residues 445 and 454 (AAV4 VP1 numbering in Fig. 3; labeled “III” in Fig. 3 to 6), had to be modeled folded down towards a loop that intervenes from a threefold icosahedrally related monomer to fit the rounder mounds of AAV4 (Fig. 2, 4A, and 5). This structural difference was also predicted by the Swiss Model program (50), although the loop required further interactive adjustment to fit the AAV4 density (Fig. 4A). There are no equivalent residues to those that form this loop in AAV5 (63), resulting in smaller mounds (Fig. 2, 3, and 5). A single amino acid deletion in AAV5 compared to AAV2 and AAV4 alters the position of the loop region that lies on the surface of the depression at the icosahedral fivefold axes, on top of adjacent fivefold axis-related monomers (residues 646 to 649 in AAV5, 655 to 659 in AAV4, and 656 to 660 in AAV2) (labeled “VIII” in Fig. 3, 5, and 6).
There were some loop variations between AAV4 and AAV2; for example, residues 527 to 532 (between βG and βH) and the area near residue 703 (AAV4 VP1 numbering) (labeled “V” and “IX,” respectively, in Fig. 3 to 6) were outside the cryo-EM envelope, but because of the ambiguity of where to place them in the 13-Å resolution map, the residues were not adjusted from the positions observed in the AAV2 VP3 crystal structure (Fig. 4A, panel II). This major conformational difference was not predicted by the Swiss Model algorithm (Fig. 4A, panel II). Region V is located on the wall of the depression at the icosahedral twofold axes and results from a structural rearrangement, and region IX is very close to the twofold axes (Fig. 5). A surface comparison of the 13-Å-resolution map generated from structural factors and phases calculated for the AAV4 pseudoatomic model with the original cryo-EM reconstruction clearly showed that this loop is incorrectly positioned in the current model (Fig. 2, group III). Region V's rearrangement caused a notable difference in the surface topology of the twofold depressions in AAV2 and AAV4 (Fig. 2, group III, and 5C). The current AAV4 pseudoatomic model superimposes onto the atomic coordinates of AAV2 with an RMSD of 0.5 Å for 508 of 520 aligned Cα positions (Table 1), but the exact locations of various surface loops in AAV4 await a higher-resolution structure.
When the amino acid sequence of AAV4 is compared to that of the autonomous parvoviruses with known structures, it is equally dissimilar to ADV and B19 as to CPV, FPV, MVM, and PPV (Table 1). However, a superimposition of the AAV4 pseudoatomic model onto the coordinates of these viruses shows extensive overlap at the Cα positions (Table 1), with >70% of them being aligned (Fig. 6). A similar percentage for the structural overlap of B19 with the AAV4 model (Table 1) was reported for a comparison of AAV2 and B19 (37). However, the AAV4 model is biased towards the AAV2 structure, and as such, higher-resolution structures of AAV4 (and AAV5) will be required for a more precise comparison to B19, particularly at the threefold mounds, which appear rounder and smaller in B19 (37). The ADV structure is a pseudoatomic model based on CPV, which may account for the similar percentages of structural overlap. Interestingly, the surface loop regions displaying the most variability among the AAVs (labeled “I” to “IX” in Fig. 3 to 6) are also the most variable among all of the known autonomous parvovirus structures and between the autonomous parvoviruses and the AAVs (Fig. 6). The structural homologies thus highlight the conservation of 3D structures among homologous proteins and viruses compared to that of primary sequences and indicate that the observed general surface topology differences (Fig. 2) are mainly due to local conformational alterations of surface loop regions.
An attempt to localize the N-terminal VP1/VP2/VP3 amino acids for AAV4 included the calculation of a difference electron density map by subtracting a map generated with just 60 copies of our VP3 model (residues 215 to 734) from the cryo-EM map (generated from wt capsids that should contain all three VPs) (Fig. 4B). The model map was calculated for all atoms, not just the C-α backbone. A difference map contoured at a sigma level (σ, signal-to-noise ratio) of 2.2 (in a normalized map) showed density extending from the base into the channel at the icosahedral fivefold axes and also extending toward the twofold axes (Fig. 4B). The model of the protein shell had been built at a σ level of 1.0 (Fig. 4A). The density inside the fivefold axis is consistent with the PLA2 role of the VP1 unique region (25, 69) because the channel is the only hole in the mature capsid that can accommodate its externalization for this activity. This channel has also been postulated as the region through which VP2 is externalized for maturation cleavage to VP3 after DNA packaging in several autonomous parvoviruses (4). Thus, the difference density was interpreted as belonging to the N termini of VP1, VP2, and VP3 (Fig. 4B), although no attempt was made to build amino acids into it since there is no obvious connection from the first N-terminal residue in the current model (V215 based on AAV4 VP1 numbering) to the difference densities observed. The observation of two possible positions for this N-terminal region is consistent with the size of the fivefold channel (67), dictating that only one VP chain may pass through it at any one time, and thus the remaining four chains must occupy an internal location.
Density in the interior of the capsid, at a σ level of 1.0, the same as the level used to build the VP3 protein shell fit, could not be modeled (Fig. 4A). This density might represent more unbuilt regions of VP3, VP2, and VP1 or disordered nucleic acids that should be present in the wt AAV4 capsids. Despite this uninterpreted density, a comparison of the electron density map generated for the pseudoatomic capsid model of AAV4 with the cryo-EM reconstructed density, calculated by use of the OVERLAPMAP program in the CCP4 package (17), resulted in a correlation coefficient of 0.8. This high correlation of the model map to the original reconstructed density affirms its applicability. Further confidence in the accuracy of the model also stemmed from the observation that the backbone structures of other parvoviruses that are at least 50% identical are generally superimposable, even at the C-α positions where amino acid types differ (2, 4, 53). This pseudoatomic model of AAV4 provides a means for preliminary comparative analyses with the available structures of AAV2 (68) and AAV5 (63) with regards to possible cell receptor binding sites and antigenic differences.
Comparison of AAV capsid structures and implications for receptor attachment.
Variations in parvovirus capsids occur at surface amino acid positions that control host-specific interactions, such as tissue tropism, pathogenicity, receptor attachment, and antigenicity (3, 31, 32). An amino acid alignment of the VP3 sequence of AAV2 with those of AAV1, AAV3, AAV4, and AAV5 (46) had shown that a number of the basic residues on AAV2 that cluster to confer a heparin binding phenotype were missing in AAV4 and AAV5. In addition, our comparison of the pseudoatomic model generated for AAV5 with the crystal structure of AAV2 had shown that the surface loops of the two serotypes differed on the walls and tops of threefold protrusions that contain the AAV2 basic patch residues (63). Superimposition of the AAV4 VP3 model onto the crystal structure of AAV2 and the pseudoatomic model of AAV5 clearly shows the localization of the majority of the variable regions discussed above (I to IV and VI) on or close to the threefold mounds (Fig. 5). Variable region III (Fig. 3 and 5), plus the loop that contains AAV2 basic residues R585 and R588, surrounds the region containing variable region VI from a threefold symmetry-related monomer to create the threefold mounds (Fig. 5B and C). This threefold monomer would also position regions I, II, IV, and V immediately adjacent to the basic patch of AAV2 (Fig. 5). The basic patch is only formed when the threefold mound-related monomers intertwine, as shown in Fig. 5B, to bring residues R484, R487, and K532 in one monomer close to residues R585 and R588 in an adjacent monomer. Furthermore, residue K532 in variable region V adopts a different conformation in AAV2 compared to that in AAV4 on the wall of the twofold dimple/base of the threefold mounds (Fig. 5B and C). Thus, the unique AAV2, AAV4, and AAV5 capsid topologies at and surrounding their threefold axes (Fig. 5) may control heparin binding phenotypes and may play a role in sialic acid binding, although residues involved in this interaction have yet to be identified.
Sialic acid binding is an activity that has been reported for a number of autonomous parvoviruses, including bovine parvovirus (BPV), CPV, FPV, and MVM (27, 58, 60; A. Lopez-Bueno, N. Bryant, M. Kontou, M.-P. Rubio, R. McKenna, M. Agbandje-McKenna, and J. Almendral, unpublished results). However, while the receptor molecule utilized by CPV, FPV, and AAV5 has been identified (19, 47), the regions of their capsids utilized for receptor recognition have not been unequivocally verified. Mutational and structural studies of CPV suggested that amino acid residues on the wall of the dimple at the icosahedral twofold axis as well as the surface between the three- and fivefold axes control receptor attachment (8, 27, 31, 32, 60). Crystallographic studies of an MVMp-sialic acid complex identified the icosahedral twofold dimple as its sialic acid binding site (Lopez-Bueno et al., unpublished results). For human parvovirus B19, which is known to have hemagglutination activity, by utilizing a non-sialic acid glycolipid, globoside, which is its cellular receptor (12), cryo-EM and image reconstruction showed receptor binding in a depression at its icosahedral threefold axes (16). This location is close to that proposed as being responsible for heparin binding in AAV2 (38, 46). These observations highlight the ability of the parvovirus capsid, regardless of whether it is a dependo- or an autonomous virus, to utilize different surface regions for the recognition of carbohydrate moieties during cell recognition and infection. Thus, the observed surface loop variability between AAV2, -4, and -5 at both the two- and threefold axes may play a role in disparate receptor recognition and attachment. Structural studies of AAV4 and AAV5 in complex with sialic acid molecules are under way to identify their interacting sites.
To probe the general applicability of the observed surface loop variability at the icosahedral twofold axes or surrounding the AAV threefold mounds in other serotypes, we aligned structural models generated for the VP3 amino acid sequences of AAV1, AAV3, AAV6-9, and AAAV (Fig. 3). The loops equivalent to those making up the threefold protrusions in AAV2, -4, and -5 (Fig. 3) are also the most variable in other parvovirus structures (Fig. 6) (3, 14). Interestingly, this variable region spans the center of the primary sequence (residues ∼440 to 600), while residues located at the N and C termini are conserved (Fig. 3). Crystal structures of parvovirus capsids show that the conserved N- and C-terminal regions knit together in β-strands to form the core contiguous capsid, while the variable regions decorate the capsid surface (Fig. 6). Other regions that show the most variability among the AAVs are near residues 260 and 380 (AAV2 VP1 numbering) (labeled I and II in Fig. 3, 5, and 6).
Comparison of AAV capsid structures and implications for antigenic phenotypes.
AAV4 and AAV5 are the most antigenically distant AAV serotypes, with AAV4 being unable to cross-react with antibodies generated to linear AAV2 epitopes, such as the AAV2 B1 site, which cross-reacts with AAV1, AAV3, and AAV5 (65). In addition, the crystal structure of AAV2 located the A20 antigenic epitope (65) specific to this virus on the wall between the two-, three-, and fivefold axes in the AAV2 capsid structure (68). The superimposed VP3s of AAV2, AAV4, and AAV5 (Fig. 5A and B) show that four variable loops (I, II, V, and VI in Fig. 3) contain residues comprising the conformational epitope recognized by the AAV2 A20 antibody (65). Considering that a single amino acid change on a viral surface region is able to eliminate antibody recognition, and given the clustering of local differences on the surface structures of these three AAV capsids, it is not surprising that only AAV2 is recognized by A20. At 13-Å resolution, the rationale for AAV4's unique ability to prevent recognition by the B1 antibody directed to a linear epitope at the extreme C-terminal end of the AAV VP3 protein (65) is not evident.
Significance of parvovirus capsid surface topologies.
A comparison of capsid surface topologies of parvoviruses for which structural information is available (Fig. 2) shows a clear division into three distinct structural groups: group I, comprised of CPV, FPV, MVM, and PPV; group II, comprised of DNV; and group III, comprised of AAV2, AAV4, AAV5, ADV, and B19. DNV (52), currently the sole member of group II, is not discussed in this paper. Group I, distinguished by a single pinwheel protrusion at the icosahedral threefold axes, contributed from three monomers, and a wider twofold depression with small insertions, contains only members of the autonomous parvovirus genus (Fig. 2). Group III, containing both autonomous (ADV and B19) and dependovirus (AAV2, AAV4, and AAV5) parvovirus capsids, has three distinct mounds at a distance of ∼20 to 26 Å from the icosahedral threefold axes and narrower deep twofold axes (except in AAV2 and AAV5) (Fig. 2). Each group III mound is formed by a contribution from two VP monomers (Fig. 4A and 5A and B). The intervening of threefold symmetry-related surface loops is similar in both capsid types, but group I viruses have a closer contact between the six loops at the icosahedral threefold axes, hence the pinwheel arrangement. Group III capsids appear to have depressions at their icosahedral threefold axes due to the surrounding mounds being offset at a distance (Fig. 2). The five-fold channel formed by symmetry-related β-ribbons is conserved in the group I and III capsid structures. In B19 (37), the wall between the twofold and fivefold depressions is more pronounced than in the other group III viruses, appearing at almost the same height as its threefold protrusions, giving the particle a rounder appearance (Fig. 2).
A small number of amino acids generally define differences in tissue tropism, pathogenicity, receptor attachment, and antigenicity phenotypes between highly homologous parvovirus strains and serotypes. The capsid sequences that account for the two- and threefold parvovirus topologies contain the most variable amino acids among the parvoviruses (Fig. 6) (14). However, there are no unifying similarities among the viruses in group I or III with respect to tissue tropism, pathogenicity, or infectious receptor recognition to explain their surface topology similarities. When considering group I capsids, tropism and pathogenicity differences are observed between the highly homologous CPV and FPV (27, 31, 32) as well as MVM and PPV strains (4, 53). The available information shows that the group I viruses utilize different receptors and capsid attachment sites for infectious binding (27, 47; Lopez-Bueno et al., unpublished results), and there is no conformational antigenic cross-reactivity of CPV and FPV with MVM and PPV. For the group III viruses, the AAVs are nonpathogenic, while ADV and B19 can cause lethal diseases. AAV2 and B19 are proposed to bind their infectious receptors, with different terminal carbohydrates, at or close to the icosahedral threefold axes (16, 37, 38, 46), and the carbohydrate binding sites are not known for AAV4, AAV5, and ADV. There is no conformational antigenic cross-reactivity between the AAVs, ADV, and B19. Thus, the observed capsid surface topology groupings are not dictated by similarities in tissue tropism, pathogenicity, antigenicity, or binding of infectious receptors, at least not with respect to the molecule(s) recognized, although similar capsid regions may be utilized for some of these functions.
Our current understanding of parvovirus biology suggests that the common topologies among members of the group I and group III viruses might result from capsid protein translation and posttranslational modification mechanisms. Group I viruses are approximately 50% identical at the amino acid level (except for CPV and FPV, with ∼98% identity), their VP1 and VP2 proteins are translated from the same message from alternative start codons, and VP3 is formed by a posttranslational cleavage of 15 to 18 amino acids from the N terminus of VP2 after DNA packaging. However, this event does not occur in empty particles, which have similar structures to DNA-containing capsids (4). Group III viruses, with ∼50% identity between the AAV capsid sequences and ∼20% identity between ADV and B19, also have a similar protein translation mechanism, since all of their VPs are encoded from alternative start codons. While the AAV capsids are made up of VP1, VP2, and VP3, ADV and B19 do not contain VP3. There is no capsid protein maturation cleavage after the group III capsid proteins are assembled. The true nature of the structural differences between the group I and III capsids at the two- and threefold icosahedral axes remains to be verified, possibly through a comparison of higher-resolution structures, particularly for members of group III, and by probing with molecular biology techniques.
Acknowledgments
We thank Beverly Handelman for technical assistance.
This project was supported by a UF HHMI Pilot program and NIH NHLBI grant P0I HL51811 (M.A.-M. and N.M.), by a Keck Foundation award to the Purdue Structural Biology group for the purchase of a CM300 FEG microscope, a Purdue Reinvestment to the Structural Biology group, and NIH grant GM33050 (T.S.B.), and by NIH intramural support (J.A.C.).
This study is the result of a joint collaboration between J.A.C., T.S.B., and M.A.-M. to define the structure of AAV4.
REFERENCES
- 1.Agbandje, M., S. Kajigaya, R. McKenna, M. G. Rossmann, and N. S. Young. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203:106-115. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje, M., C. R. Parrish, and M. G. Rossmann. 1995. The structure of parvoviruses. Semin. Virol. 6:299-309. [Google Scholar]
- 4.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 5.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 194:754-756. [DOI] [PubMed] [Google Scholar]
- 6.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bantel-Schaal, U., and H. Zur Hausen. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52-63. [DOI] [PubMed] [Google Scholar]
- 8.Barbis, D. P., S. F. Chang, and C. R. Parrish. 1992. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology 191:301-308. [DOI] [PubMed] [Google Scholar]
- 9.Belnap, D. M., A. Kumar, J. T. Folk, T. J. Smith, and T. S. Baker. 1999. Low-resolution density maps from atomic models: how stepping “back” can be a step “forward.” J. Struct. Biol. 125:166-175. [DOI] [PubMed] [Google Scholar]
- 10.Bossis, I., and J. A. Chiorini. 2003. Cloning of an adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J. Virol. 77:6799-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman, V., E. S. Chase, A. W. E. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114-117. [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 14.Chapman, M. S., and M. G. Rossmann. 1993. Structure, sequence, and function correlations among parvoviruses. Virology 194:491-508. [DOI] [PubMed] [Google Scholar]
- 15.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chipman, P. R., M. Agbandje-McKenna, S. Kajigaya, K. E. Brown, N. S. Young, T. S. Baker, and M. G. Rossmann. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. USA 93:7502-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 18.Davidson, B. L., S. C. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Pasquale, G., B. L. Davidson, C. S. Stein, I. Martins, D. Scudiero, A. Monks, and J. A. Chiorini. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9:1306-1312. [DOI] [PubMed] [Google Scholar]
- 20.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132-134. [DOI] [PubMed] [Google Scholar]
- 21.Flannery, J. G., S. Zolotukhin, M. I. Vaquero, M. M. LaVail, N. Muzyczka, and W. W. Hauswirth. 1997. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc. Natl. Acad. Sci. USA 94:6916-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flotte, T. R., and B. J. Carter. 1995. Adeno-associated virus vectors for gene therapy. Gene Ther. 2:357-362. [PubMed] [Google Scholar]
- 23.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 100:6081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 26.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305-358. [DOI] [PubMed] [Google Scholar]
- 27.Govindasamy, L., K. Hueffer, C. R. Parrish, and M. Agbandje-McKenna. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 77:12211-12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handa, A., S. Muramatsu, J. Qiu, H. Mizukami, and K. Brown. 2000. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 81:2077-2084. [DOI] [PubMed] [Google Scholar]
- 29.Hauck, B., and W. Xiao. 2003. Characterization of tissue tropism determinants of adeno-associated virus type I. J. Virol. 77:2768-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77:10099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 34.Jones, T., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 35.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaludov, N., E. Padron, L. Govindasamy, R. McKenna, J. A. Chiorini, and M. Agbandje-McKenna. 2003. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 4. Virology 306:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. USA 101:11628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotin, R. M. 1994. Prospects for the use of adeno-associated virus as a vector for human gene therapy. Hum. Gene Ther. 5:793-801. [DOI] [PubMed] [Google Scholar]
- 40.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor, H. D., and J. L. Melnick. 1966. Small deoxyribonucleic acid-containing viruses (picornavirus group). Nature 210:331-332. [DOI] [PubMed] [Google Scholar]
- 42.McCown, T. J., X. Xiao, J. Li, G. R. Breese, and R. J. Samulski. 1996. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 713:99-107. [DOI] [PubMed] [Google Scholar]
- 43.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 44.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2360. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 46.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, J. S. L., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferring receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a coreceptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 49.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson, A. A., V. Chandrasekar, B. Hebert, G. Sullivan, M. G. Rossmann, and C. R. Parrish. 2000. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300:597-610. [DOI] [PubMed] [Google Scholar]
- 52.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 54.Snyder, R. O., C. H. Miao, G. A. Patijn, S. K. Spratt, O. Danos, D. Nagy, A. M. Gown, B. Winther, L. Meuse, L. K. Cohen, A. R. Thompson, and M. A. Kay. 1997. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat. Genet. 16:270-276. [DOI] [PubMed] [Google Scholar]
- 55.Song, S., M. Morgan, T. Ellis, A. Poirier, K. Chesnut, J. Wang, M. Brantly, N. Muzyczka, B. J. Byrne, M. Atkinson, and T. R. Flotte. 1998. Sustained secretion of human alpha-1-antitrypsin from murine muscle transduced with adeno-associated virus vectors. Proc. Natl. Acad. Sci. USA 95:14384-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Summerford, C. M., J. S. Bartlett, and R. J. Samulski. 1999. A role for integrin αvβ5 in adeno-associated virus type 2 (AAV-2) infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 57.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparin sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thacker, T. C., and F. B. Johnson. 1998. Binding of bovine parvovirus to erythrocyte membrane sialylglycoproteins. J. Gen. Virol. 79:2163-2169. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tresnan, D. B., L. Southard, W. Weichert, J. Y. Sgro, and C. R. Parrish. 1995. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology 211:123-132. [DOI] [PubMed] [Google Scholar]
- 61.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Copans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 62.Wagner, J. A., T. Reynolds, M. L. Moran, R. B. Moss, J. J. Wine, T. R. Flotte, and P. Gardner. 1998. Efficient and persistent gene transfer of AAV-CFTR in maxillary sinus. Lancet 351:1702-1703. [DOI] [PubMed] [Google Scholar]
- 63.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walters, R. W., J. M. Pilewski, J. A. Chiorini, and J. Zabner. 2002. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 277:23709-23713. [DOI] [PubMed] [Google Scholar]
- 65.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao, X., J. Li, and R. J. Samulski. 1996. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 11:8098-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]
- 68.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 70.Zolotukhin, S., M. Porter, I. Zolotukhin, Y. Sakai, S. Loiler, T. J. Fraites, Jr., V. A. Chiodo, T. Phillipsberg, N. Muzyczka, W. W. Hauswirth, T. R. Flotte, B. J. Byrne, and R. O. Snyder. 2002. Production and purification of serotypes 1, 2 and 5 recombinant adeno-associated viral vectors. Methods Enzymol. 28:158-167. [DOI] [PubMed] [Google Scholar]