Abstract
Understanding of tumor biology and identification of effective therapies is lacking for many rare tumors. My Pediatric and Adult Rare Tumor (MyPART) network was established to engage patients, advocates, and researchers and conduct a comprehensive longitudinal Natural History Study of Rare Solid Tumors. Through remote or in-person enrollment at the NIH Clinical Center, participants with rare solid tumors ≥4 weeks old complete standardized medical and family history forms, patient reported outcomes, and provide tumor, blood and/or saliva samples. Medical records are extracted for clinical status and treatment history, and tumors undergo genomic analysis. A total of 200 participants (65% female, 35% male, median age at diagnosis 43 years, range = 2–77) enrolled from 46 U.S. states and nine other countries (46% remote, 55% in-person). Frequent diagnoses were neuroendocrine neoplasms (NEN), adrenocortical carcinomas (ACC), medullary thyroid carcinomas (MTC), succinate dehydrogenase (SDH)-deficient gastrointestinal stromal tumors (sdGIST), and chordomas. At enrollment, median years since diagnosis was 3.5 (range = 0–36.6), 63% participants had metastatic disease and 20% had no evidence of disease. Pathogenic germline and tumor mutations included SDHA/B/C (sdGIST), RET (MTC), TP53 and CTNNB1 (ACC), MEN1 (NEN), and SMARCB1 (poorly-differentiated chordoma). Clinically significant anxiety was observed in 20%–35% of adults. Enrollment of participants and comprehensive data collection were feasible. Remote enrollment was critical during the COVID-19 pandemic. Over 30 patients were enrolled with ACC, NEN, and sdGIST, allowing for clinical/genomic analyses across tumors. Longitudinal follow-up and expansion of cohorts are ongoing to advance understanding of disease course and establish external controls for interventional trials.
Significance:
This study demonstrates that comprehensive, tumor-agnostic data and biospecimen collection is feasible to characterize different rare tumors, and speed progress in research. The findings will be foundational to developing external controls groups for single-arm interventional trials, where randomized control trials cannot be conducted because of small patient populations.
Introduction
Rare tumors and cancers occur in <15 incident cases per 100,000 per year. By this definition in the United States, all pediatric tumors (<20 years) are rare and <13% of tumors diagnosed in adults (≥20 years) are rare (1). Hereafter, we use the term “rare tumors” to include both rare tumors and rare cancers. The overall burden of rare tumors is significant with 27% of U.S. tumor diagnoses being rare. Rare solid tumors are understudied and knowledge of their biology, genomics, clinical course, and psychologic impact is limited. For many rare tumors, no effective therapy exists, and clinical trials are difficult to conduct. Several programs have begun to address obstacles in rare tumor research (2–9), such as designing effective interventional trials (10–13), developing new models (14) and identifying new therapeutic targets (1, 15); however, there remains a large unmet need to study rare tumors comprehensively and longitudinally. My Pediatric and Adult Rare Tumor (MyPART) network (https://www.cancer.gov/pediatric-adult-rare-tumor/) was established at the NCI, Center for Cancer Research (CCR) in response to NCI Cancer MoonshotSM recommendations to increase patient engagement in cancer research (16). MyPART is a team of clinicians and researchers partnering with advocates, patients, and their families to accelerate rare solid tumor research, focusing on patient experience over time through development of the Natural History and Biospecimen Acquisition Study for Children and Adults with Rare Solid Tumors (NHRST; NCT03739827; ref. 17).
Natural history studies are critical to understanding rare tumors. Draft guidance by the FDA provides specifics on leveraging natural history studies in the drug development process for patients with rare diseases, to create external control groups in instances where randomized clinical trials are difficult to conduct due to few enrollees (18). For example, regulatory approval of selumetinib for plexiform neurofibromas (PN) relied on natural history data of PN growth rates to demonstrate that selumetinib significantly altered tumor growth trajectory (19, 20). In addition, the FDA recently described how external controls contributed to regulatory approvals for several targeted therapies (21). The goal of NHRST is to develop deep understanding of the clinical course and pathogenesis of rare solid tumors of unmet need, translating findings to improve outcomes for patients with rare solid tumors. Here we report initial results from baseline evaluation of the first 200 participants.
Materials and Methods
Study Design and Patient Eligibility
This single-institution NHRST study was conducted in accordance with recognized ethical guidelines as per The Belmont Reports and the Department of Health and Human Services Common Rule and was approved by the NIH Institutional Review Board. Written informed consent was obtained from all participants 18 years and older or their legally authorized representative, and from parent(s) of minors. Verbal assent was obtained as appropriate from participants 7–17 years of age. Consent and assent language was approved by the NIH Institutional Review Board. Patients ≥4 weeks old with a rare solid tumor (<15 cases in 100,000 people per year), a genetic cancer predisposition syndrome (GCPS), or their family members were eligible for participation. Of note, the study focuses on enrolling participants with very rare tumors, defined as <2 per million per year, rare tumors that lack clinical trials or standard therapy, tumors that are rare in the pediatric population even if they are common in adults and unusual presentations of rare tumors (2). Participants could enroll remotely (field cohort) or in person at the NIH Clinical Center (NIHCC; clinic cohort). Clinic cohort patients underwent clinical evaluation at NIHCC. The detailed study design, data collection methods and all study forms are provided in Wedekind and colleagues (17).
Recruitment
To increase public awareness, facilitate referrals, and provide educational information, a website was established (www.cancer.gov/mypart) providing information on rare solid tumors, advocacy partnerships, and a lay study description.
Data Collection
NHRST collects all available medical history, including imaging studies and pathology specimens retrospectively and prospectively. Participants (or their caregivers) are asked to complete standardized forms on medical, behavioral, and family tumor history. Follow-up forms are sent to participants yearly in the month of their original consent. Clinicians review yearly medical records and extract data on pathology, presenting symptoms, and treatment by event.
Clinical Evaluations for the Clinic Cohort
For participants coming to NIHCC, clinical phenotyping, clinical laboratory studies, and imaging are conducted, as well as germline genotyping as indicated. A senior genetic counselor (M. Raygada) constructs pedigrees using Progeny Clinical—v.10.6.2.0 (Progeny Genetics LLC. 2019).
Patient Reported Outcomes
The Patient-Reported Outcome Measurement Information System (PROMIS; https://commonfund.nih.gov/promis/tools) is used to collect patient reported outcomes (PRO) from adult/pediatric participants and pediatric caregivers. Encrypted patient responses are converted to standard T-scores through the Assessment Center-Application Programming Interface (https://staging.healthmeasures.net/implement-healthmeasures/administration-platforms/assessment-center-api), such that mean = 50 and SD = 10. Patients are reminded 4 weeks after initial request to complete PROs and data are considered missing if participants do not complete forms within 6 weeks after initial contact. For patients unable to complete forms independently due to illness or preference, PROs may be administered by videoconference by a clinical psychologist (R. Lockridge). Anyone with a depression score >70 receives a follow-up call by a psychologist or member of the behavioral health team.
Data Storage
All standardized forms and PROs are entered into a secure, custom Labmatrix database (Biofortis). Participant and caregiver-reported data are collected electronically using Scribe developed by NCI CCR Center for Information Technology (CIT).
Biospecimen Collection and Analysis
Participants provide a saliva sample and clinic cohort participants provide blood samples. Tumor diagnosis is confirmed by NIHCC pathologists. Patients with sufficient pathology specimens have tumor molecular analysis performed, using either gene panel sequencing of 500+ genes [Trusight Oncology 500 (TSO500), Illumina] plus RNA sequencing (RNA-seq) exome fusion analysis (Supplementary Table S1), tumor-normal matched whole-exome sequencing (T-N WES; available starting in second year of accrual), and/or specific molecular analyses based on tumor type, for example, detection of characteristic gene fusions. The current somatic analysis is limited to TSO500 small nucleotide variants and does not include large gene amplifications/deletions (e.g., brachyury) or fusions. Somatic variants were classified from TSO500 gene panel sequencing data using QIAGEN (RRID:SCR_008539) Clinical Insights Interpret (QCI) by the NIHCC Laboratory of Pathology and compiled for the analysis presented here. Because RNA-seq exome fusion and copy-number variation (CNV) calling was not initially available, fusions and CNVs are not included in this analysis. Participants also get clinical-grade germline sequencing as clinically indicated through commercial vendors (GeneDx or Invitae) or T-N WES (Laboratory of Pathology, CCR, NCI). The current germline analysis was compiled from all available sources of germline genotypes. Germline variants for T-N WES performed at NIHCC were called using the NCI Oncogenomics platform and the ClinOmics analysis pipeline developed by Dr. Javed Khan's group (NCI, CCR). Variants scored as Path/LP by Clinvar or Intervar were selected and those with a variant allele frequency (VAF) of ≥25% were included in the analysis. Pathogenicity was classified as pathogenic (Path), likely pathogenic (LP), and variants of uncertain significance (VUS) while actionability was tiered as 1A, 1B, 2C, 2D, and 3 according to Association for Medical Pathology (AMP)/American Society of Clinical Oncology/College of American Pathologists (CAP) guidelines (22). Participants are provided recommendations about management, treatment options, and relevant available clinical trials, either at NIHCC or outside sites.
Data Analysis
Data linked to the first 200 enrollees were downloaded from Labmatrix, the NIH (BTRIS), NCI Oncogenomics platform, and NIH Integrated Data Analysis Platform (NIDAP). Data were validated, cleaned, and combined using Excel v16 for Mac (Microsoft). Prism 9 for Mac (GraphPad, RRID:SCR_002798) was used to graph data and analyze statistics. SSPS v28 (IBM) was used to analyze PRO results. Data entered as free text were further manually curated after download.
Participant home zip codes were converted to longitude and latitude, using data from U.S. Zip Code Database (https://www.unitedstateszipcodes.org/zip-code-database/) or from web searches for international participants. Distance from the participants home to NIHCC was estimated using the formula D = SQRT[(Latitudehome − LatitideNIHCC)2 + (Longitudehome − LongitudeNIHCC)2] and the distributions for clinic and field groups compared using two-tailed Mann–Whitney test.
Race and ethnicity data were collected through medical records and self-reports. Clinical data were extracted from medical records (treatments received) or recorded when patients came to clinic (performance scores, height, weight, substance use). Body mass index (BMI) [participants ≥20 years (23)] or BMI percentile (participants <20 years old; (https://www.cdc.gov/healthyweight/bmi/widget/calculator.html) were calculated at baseline. U.S. population BMI ranges were taken from the National Health and Nutrition Examination Survey (NHAHES) prepandemic data (24, 25). Occupation, education levels, and cigarette exposure were from self-reports. Drug names were standardized to generic name and classified according to the Cancer Drug Pharmacology Table from the BC Cancer Pharmacology Education Program (26) as cytotoxic chemotherapy, hormonal therapy, immunotherapy, and targeted therapy.
PRO measures of pain, depression, and anxiety were analyzed for adult subjects only. Although pediatric patients (ages 8–17) and their caregivers were also administered PROMIS measures (RRID:SCR_004718), fewer than 10 participant responses were collected out of 29 possible pediatric participants and results were not analyzed at this time. In accordance with PROMIS T-score threshold guidance, participants with T-scores greater than 1 SD above the mean were considered to have clinically significant (CS) levels of pain, depression, and/or anxiety (27, 28).
Data Availability
Data are available through controlled access at dbGAP (RRID:SCR_002709) accession phs003143.v1.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003143.v1.p1).
Results
Enrollment and Participant Demographics
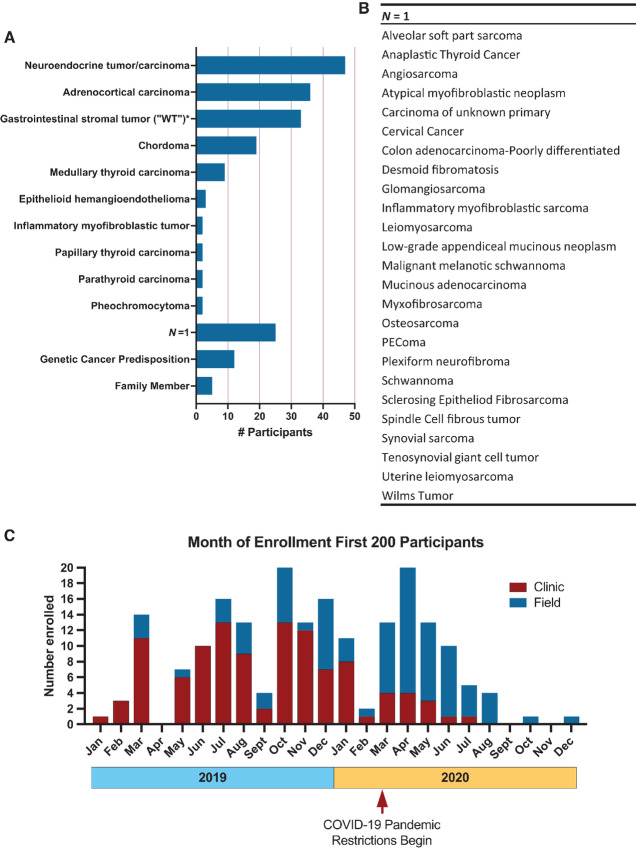
Between January 2019 and December 2020, we enrolled 200 NHRST participants. Three participants withdrew (all unaffected family members of one affected participant) leaving 197 participants for analysis. The most frequent tumor type was NEN (24%), followed by ACC (18%), succinate dehydrogenase (SDH)-deficient gastrointestinal stromal tumor (sdGIST; 17%), chordoma (10%), and MTC (5%; Table 1; Fig. 1A). Participants with GCPS and biological relatives of patients with rare tumors comprise 9% of enrollees. A substantial proportion of participants (13%) were the only enrollee with their tumor type in the first 200 participants (Fig. 1B).
TABLE 1.
Demographics and clinical characteristics of first 200 enrollees showing number of participants in each category
| All | ACC | NEN | MTC | GIST | Chord | Othera | Mut Carr | Fam Memb | |
|---|---|---|---|---|---|---|---|---|---|
| Total subjects | 200 | 36 | 47 | 9 | 33 | 19 | 36 | 12 | 8 |
| Consent withdrawn | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Subjects with data analyzed | 197 | 36 | 47 | 9 | 33 | 19 | 36 | 12 | 5 |
| Sex | |||||||||
| Male | 68 | 5 | 19 | 1 | 14 | 9 | 14 | 2 | 4 |
| Female | 129 | 31 | 28 | 8 | 19 | 10 | 22 | 10 | 1 |
| Age at diagnosis (yrs) | |||||||||
| <18 | 29 | 2 | 1 | 1 | 7 | 11 | 7 | N/A | N/A |
| ≥18, <40 | 54 | 11 | 4 | 6 | 14 | 4 | 15 | N/A | N/A |
| ≥40 | 97 | 23 | 42 | 2 | 12 | 4 | 14 | N/A | N/A |
| Race | |||||||||
| White | 150 | 34 | 36 | 4 | 26 | 7 | 31 | 10 | 2 |
| Black/African American | 9 | 0 | 2 | 2 | 2 | 1 | 2 | 0 | 0 |
| Asian | 9 | 0 | 0 | 1 | 2 | 3 | 2 | 0 | 1 |
| Pacific Islanderb | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Mixed | 10 | 1 | 4 | 1 | 1 | 1 | 0 | 2 | 0 |
| Not reported | 18 | 1 | 5 | 1 | 1 | 7 | 1 | 0 | 2 |
| Enrollment type | |||||||||
| Clinic | 109 | 26 | 20 | 8 | 21 | 10 | 20 | 4 | 0 |
| Field | 88 | 10 | 27 | 1 | 12 | 9 | 16 | 8 | 5 |
| Days dx to enrl Median (range) |
1,311 (0–13,366) |
1,349 (56–8,350) |
1,391 (18–7,035) |
3,057 (481–13,129) |
2,029 (0–13,366) |
882 (22–6,302) |
376 (2–5,727) |
N/A | N/A |
| Disease status at diagnosis | |||||||||
| Localized | 103 | 24 | 15 | 4 | 22 | 15 | 23 | N/A | N/A |
| Metastatic | 73 | 12 | 32 | 5 | 10 | 3 | 11 | N/A | N/A |
| Not available | 4 | 0 | 0 | 0 | 1 | 1 | 2 | N/A | N/A |
| Disease status at enrollment | |||||||||
| NED | 36 | 5 | 8 | 2 | 7 | 7 | 7 | N/A | N/A |
| Localized | 31 | 4 | 3 | 0 | 8 | 6 | 10 | N/A | N/A |
| Metastatic | 113 | 27 | 36 | 7 | 18 | 6 | 19 | N/A | N/A |
| Clinic cohort patients: Performance score – Median, (range, N) | |||||||||
| Karnofsky (≥16yrs) | 90 (50–100, 86) | 90 (70–100, 24) | 90 (60–100, 20) | 90 (80–100, 8) | 90 (70–100, 21) | 80 (70–100, 5) | 95 (50–100, 20) | 90 (80–100, 2) | N/A |
| Lansky (<16 yrs) | 100 (90–100, 5) | 100 (100, 2) | N/A | N/A | N/A | 100 (90–100, 5) | N/A | 100 (100, 2) | N/A |
| Cancer in first-degree relative | |||||||||
| Yes | 93 | 19 | 21 | 5 | 11 | 7 | 13 | 12 | 5 |
| No | 99 | 15 | 25 | 4 | 22 | 12 | 21 | 0 | 0 |
| Not reported | 5 | 2 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
Abbreviations: ACC, adrenocortical carcinoma; Chord, chordoma; dx, diagnosis; enrl, enrollment; Fam Memb, family member; GIST, gastrointestinal stromal tumor; MTC, medullary thyroid carcinoma; Mut Carr, mutation carrier; NED, no evidence of disease; NEN, neuroendocrine neoplasm (includes neuroendocrine carcinoma); yrs, years.
aOther is the combined data for all other tumor types, not ACC, chordoma, GIST, MTC, and NET.
bIncludes Native Hawaiians and Pacific Islanders; American Indian or Alaskan Natives were not identified in the cohort.
FIGURE 1.
A, Number of participants by rare tumor diagnoses (N = 197). B, Diagnoses with only one participant enrolled (N = 1). C, Month of enrollment of 197 participants in 2019 and 2020, relative to the start of COVID pandemic restrictions at NIHCC in March 2020. Clinic cohort patients are shown in red and field cohort patients are in blue.
We enrolled more females (65%), reflecting female predominance in some of the tumor types enrolled, such as ACC and sdGIST (Table 1). Participants with ACC were almost entirely female (86%). We enrolled children, adolescents and young adults (AYA), and older adults for several tumor types from 46 U.S. states, and nine countries across six continents (Supplementary Fig. S1A and S1B); however, particular tumors were more frequently pediatric (e.g., chordoma) or adult (e.g., ACC; Table 1; Supplementary Fig. S2).
We found 14% discrepancy in race between participant-reported data and medical record extraction, with neither source being complete for all participants. Notably, race could be assigned to more participants when self-reported data were combined with medical record data (Supplementary Fig. S3A and S3B), and self-reported data demonstrated minority racial backgrounds within parent ancestry (Supplementary Fig. S3C). Self-reported race was preferentially used if a discrepancy was noted. Table 1 summarizes compiled race information.
Until March 2020, when COVID-19 pandemic restrictions began at NIHCC, most participants were in the clinic group, whereas during pandemic restrictions, most were in the field group due to restrictions on bringing patients to NIHCC (Fig. 1C). Remote participation thus allowed for continued enrollment during the pandemic, although at a reduced capacity, particularly during the last quarter of 2020. In addition, during the fall 2020, there was a pause in enrollment to validate and analyze data. There was no significant difference between field and clinic group in how close they lived to NIHCC (Supplementary Fig. S4A and S4B) over the time of enrollment for the first 200 enrollees. However, during the pandemic the field cohort enrolled from significantly further away (Supplementary Fig. S4C). Form completion by clinic participants was significantly greater than field participants (Supplementary Fig. S5). Overall, the ability to collect data using our forms ranged from 43% for the pain interference index in the field cohort to 98% for medical record extraction.
Most participants did not enroll at the time of diagnosis, but a median of 3.5 years (range = 0–36.3) past initial diagnosis (Supplementary Fig. S6). Analysis of tumor status at diagnosis versus time of enrollment confirmed that most participants had developed metastatic disease by the time of enrollment (Table 1), although 20% of participants had no evidence of disease (NED), highlighting the wide range of disease states of enrolling participants.
Baseline performance scores were relatively high in both pediatric and adult patients (Table 1; Supplementary Fig. S7A). Patients with chordoma reported the highest general health at baseline, with few respondents reporting their health as poor (Supplementary Fig. S7B). Most participants <20 years old were in a healthy weight range (Supplementary Fig. S7C), whereas most adults ≥20 years old were overweight or obese (Supplementary Fig. S7D). However, the proportion of overweight/obese participants was similar to U.S. obesity trends (Supplementary Fig. S7E).
Social/Environmental Variables
The most frequent occupations reported were management, office/administration support, and business/financial operations (Supplementary Fig. S8A) and education data showed that 75% of adults over 21 completed college or graduate school (Supplementary Fig. S8B). Collection of substance use data from clinic evaluations and by self-report of tobacco use and exposure showed low levels of tobacco exposure and recreational drug use (Supplementary Fig. S8C and S8D).
Other Health Issues
Self-reported data of other health issues experienced by participants demonstrated that allergies to medicines and environmental factors were widely reported in all groups (Supplementary Fig. S9). Anxiety and depression were reported in all groups except family members who do not carry a known germline mutation. Hyperthyroidism or hypothyroidism was reported by participants with MTC, ACC, sdGIST, and NEN. Relatively more participants with ACC and NEN reported asthma and high blood pressure. Gastrointestinal disorders, including colitis/Crohn disease/inflammatory bowel disease and gastroesophageal reflux disease were reported by 17% of participants with NEN, with the majority being diagnosed with neuroendocrine tumor (NET) of the gastrointestinal tract. Patients with chordoma reported developmental delays and mental illness, in addition to anxiety and depression. Patients with sdGIST and NEN reported ulcers. For comparison, family members reported common health problems for their age group, such as high cholesterol and diabetes in older individuals.
Prior Tumor-directed Therapy
At initial diagnosis, most rare tumor patients were treated either with surgery alone, or surgery combined with drug treatment (Supplementary Fig. S10A). NEN was the only group with a substantial number of patients receiving medical therapy without surgery at initial diagnosis. Surgery was the most common treatment across tumor types, with 88% of patients undergoing at least one surgery (Supplementary Fig. S10B). Cytotoxic chemotherapy was more common for ACC, whereas targeted therapy was more common in sdGIST, and radiotherapy and hormone therapy were more common in NEN.
PRO Results for Anxiety, Depression, and Pain
PROMIS measures assessing depression, anxiety, and pain interference were completed by 114 adult patients (≥18 years) at baseline. Across patient groups, mean T-scores were within normal limits for depression, anxiety, and pain interference (Table 2). For groups with at least 10 respondents, the percentage of respondents reporting CS anxiety ranged from 20% to 35%. Fewer respondents reported CS depression, with ACC (17%) demonstrating the highest percentage of CS depression. We assessed depression and anxiety scores in participants further out from diagnosis compared with newly diagnosed and show that while anxiety and depression trend lower further from initial diagnosis, there were still participants with CS anxiety and/or depression up to 8 years later (Supplementary Fig. S11). CS pain interference ranged from 14% to 22% of respondents for groups with ≥10 respondents.
TABLE 2.
Adult self-report of PROMIS data
| Total Responses | CSa Responses | |||
|---|---|---|---|---|
| PROMIS Measure/Tumor | n | Mean (SD) | n | % |
| Anxiety | ||||
| ACC | 23 | 55.6 (8.8) | 8 | 34.8 |
| Chordoma | 9 | 55.3 (8.5) | 3 | 33.3 |
| GIST | 20 | 54.9 (7.1) | 4 | 20.0 |
| MTC | 8 | 56.4 (9.2) | 4 | 50.0 |
| NEN | 32 | 55.4 (7.7) | 7 | 21.9 |
| Other | 22 | 50.9 (11.7) | 5 | 22.7 |
| Depression | ||||
| ACC | 23 | 49.7 (9.6) | 4 | 17.4 |
| Chordoma | 9 | 48.5 (9.2) | 1 | 11.1 |
| GIST | 20 | 47.8 (8.0) | 1 | 5.0 |
| MTC | 8 | 49.2 (8.0) | 0 | 0 |
| NEN | 32 | 50.0 (8.6) | 3 | 9.4 |
| Other | 22 | 46.7 (9.7) | 3 | 13.6 |
| Pain interference | ||||
| ACC | 23 | 51.1 (9.8) | 5 | 21.7 |
| Chordoma | 9 | 49.2 (8.7) | 1 | 11.1 |
| GIST | 20 | 51.7 (8.8) | 3 | 15.0 |
| MTC | 8 | 57.4 (6.6) | 3 | 37.5 |
| NEN | 32 | 52.7 (8.6) | 5 | 15.6 |
| Other | 22 | 48.2 (9.8) | 3 | 13.6 |
NOTE: T-scores are compared with a mean of 50, SD of 10.
Abbreviation: CS: clinically significant.
aCS range is ≥60.
Tumors in First-degree Relatives
Using genetic counseling interviews and self-reported family tumor cases, we found that many participants had a first-degree relative with a tumor diagnosis, ranging from 33% to 57% depending on tumor type (Table 1).
Tumor and Germline Mutations
Of 180 participants with rare tumors, 147 had at least one pathology sample available for analysis. For 117 participants, successful panel sequencing was performed on 130 total tumor samples. Of the remaining 30 participants, all available samples failed sequencing for a variety of reasons (e.g., low tumor content or poor DNA quality). Analyzing the age of submitted formalin-fixed paraffin-embedded (FFPE) samples (Supplementary Fig. S12), newer samples tended to be more successful; however, sequencing could be successful in older samples of up to approximately 10 years, and more recent samples could also fail.
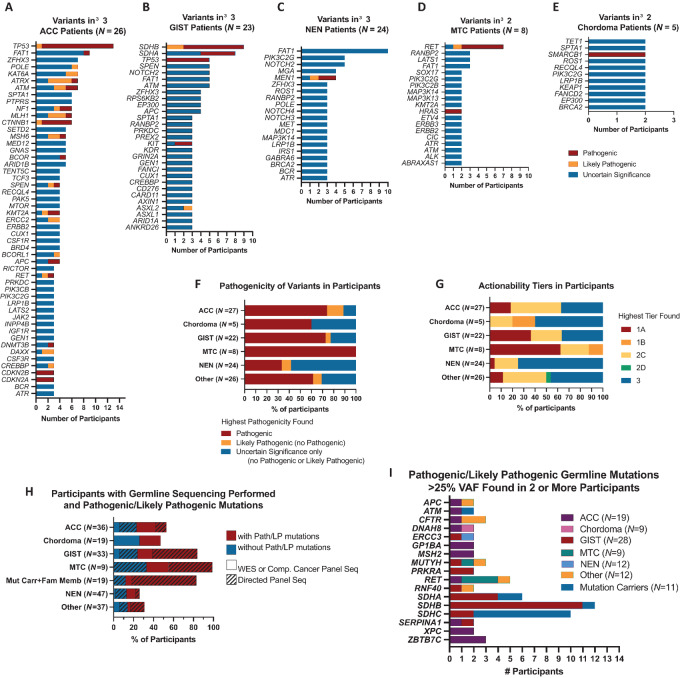
TSO500 assay results for 86 participants in the largest tumor cohorts are shown in Fig 2. In ACCs, Path/LP variants were found in TP53, CTNNB1, ATRX, MLH1, ATM, CDKN2A/CDKN2B, KMT2A, and MSH6 in 3 or more patients (Fig. 2A), but only 63% of patients with ACC had at least one of these mutations and 10 participants had Path/LP variants in genes found in fewer than 3 patients, demonstrating the molecular heterogeneity of this ACC cohort. Most sdGISTs had Path/LP mutations in one of the SDH subunit genes (SDHA N = 4, SDHB N = 8, SDHC N = 2; Fig. 2B) or hypermethylation of the SDHC promoter (epimutants). Consistent with previous reports, only female epimutants were identified (29). NENs were molecularly diverse (Fig. 2C), with Path/LP variants in MEN1 detected in tumors from 3 participants. Two of these patients do not have germline MEN1 mutations. The third has not completed germline testing for MEN1 mutations, to date. Pathogenic TP53 variants were found in 2 NEN participants and all other Path/LP variants detected (N = 14) were found in a single participant. For 14 NEN participants (58%), no Path/LP variants were found. Path/LP variants in RET were found in 6 out of 8 patients with MTC (Fig. 2D), with 3 having germline mutations indicative of MEN type 2 (30). HRAS mutations were found in patients without a RET Path/LP variant. Pathogenic SMARCB1 variants were found in 2 patients with chordoma (Fig. 2E). Overall, 19% of TSO500 genes had a Path/LP variant identified (Supplementary Table S1) and 71% of participants tested had at least one Path/LP variant (Fig. 2F) with the fewest found in NEN. Actionable mutations (Tier 1A/1B/2C/2D) were found in 55% of participants (Fig. 2G).
FIGURE 2.
Variant genes found in 3 or more patients for ACC, N = 27 (A), GIST, N = 22 (B), and NEN, N = 24 (C). Variant genes found in 2 or more patients for MTC, N = 8 (D) and chordoma, N = 5 (E). TSO500 gene panel sequencing identified pathogenic or likely pathogenic mutations for most tumors (F), with NEN and chordoma showing the lowest percentage of pathogenic mutations identified. ACC and MTC tumor analysis identified the highest percentage of pathogenic mutations. G, sdGIST and MTC had the most actionable mutations in Tier 1A. Germline sequencing on participants was performed using directed variant, single gene, or small gene panels (<100 genes; hatched bars) or using comprehensive large gene panels or WES (solid bars; H). The proportion of Path/LP mutations identified (red) varied by tumor type. I, The most common Path/LP germline mutations found were SDHA/B/C, in participants with sdGIST and their family members, and RET, in an ACC, three MTCs, and a pheochromocytoma.
Of the first 200 participants, 102 either had record of previous germline mutation testing or were evaluated for GCPS at NIHCC. Germline mutation testing of participants ranged from examining a specific variant in family members to genome-wide WES (Fig. 2H; Supplementary Table S2). SDHB, SDHC, and SDHA were the most common germline Path/LP variants identified (Fig. 2I), found in participants with sdGIST and their relatives. Path/LP variants in RET were identified in 5 participants with different tumor types. Path/LP variants were also found in 3 participants for CFTR, MUTYH, and ZBTB7C. Of note, six out of 12 GCPS cases were newly diagnosed on NHRST. Of 102 participants with germline data available, 10 had germline VUS identified without any pathogenic germline variant.
N-of-1 Patients
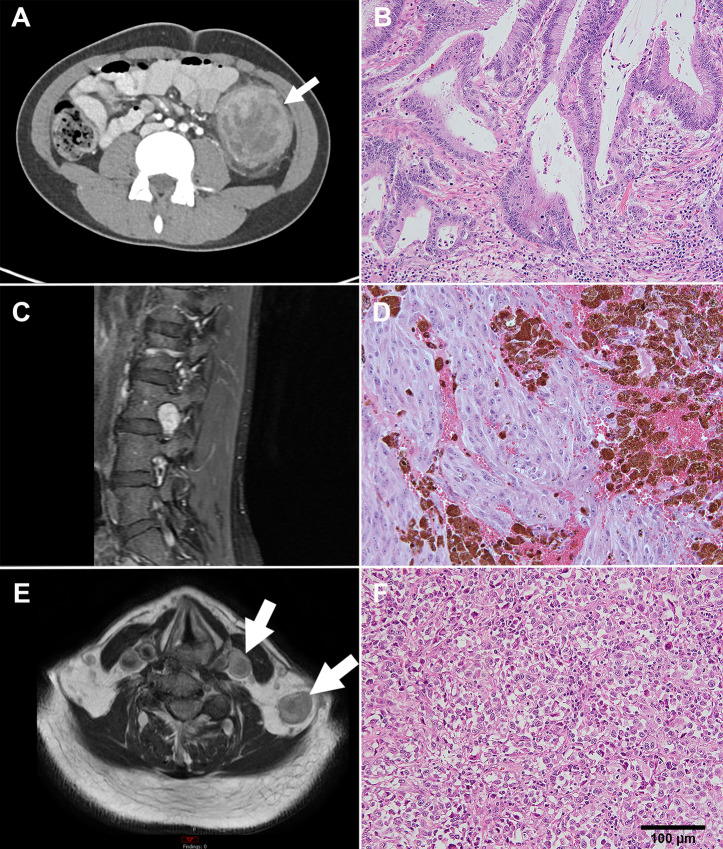
Of 25 participants enrolled with a diagnosis only represented once in our cohort, three are presented in clinical (Fig. 3) and molecular (Fig. 4) detail, representing three age groups enrolled on NHRST (<18, 18–39, and ≥40 years old).
FIGURE 3.
Imaging and histology for 3 highlighted patients. A, Axial CT image of the abdomen showing a mass in the descending colon (arrow) in a 15-year-old male diagnosed with poorly differentiated adenocarcinoma. B, Histology shows high dysplasia and stromal microinvasion. C, MRI of the lumbar spine in a 36-year-old female diagnosed with malignant melanotic schwannoma shows oval-shaped mass in left neural foramen of L3-L4 with avid enhancement on sagittal T1-weighted fat-suppressed image (arrow). Histology (D) shows pigmented spindle and epithelioid cells. E, MRI displays two left supraclavicular masses on T1-weighted image in a 75-year-old male with metastatic poorly differentiated carcinoma of unknown primary in the neck. F, Histology shows epithelioid tumor cells. Scale bar = 100 µm and is the same for B, D, and F.
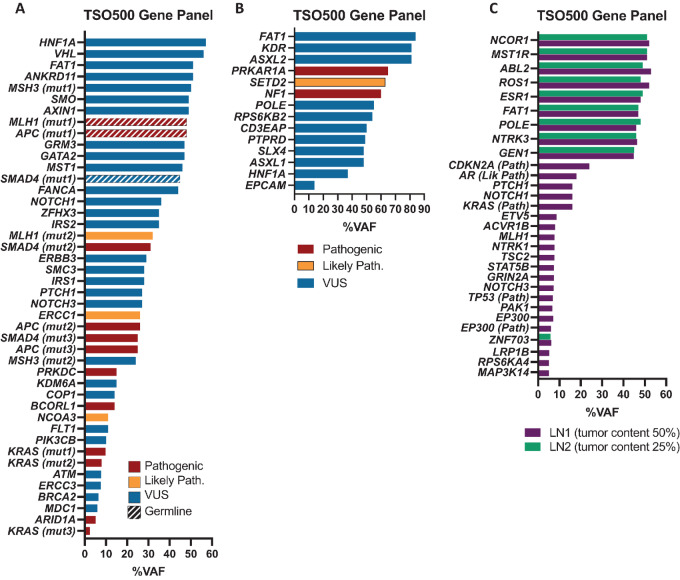
FIGURE 4.
Molecular analysis for 3 highlighted patients. A, Path (red), LP (gold), and VUS (Blue) variants identified by TSO500 in colon adenocarcinoma from 15-year-old male. Pathogenic germline mutations were identified in MLH1 (c.156del p.Glu53Argfs*4) and APC (c.3329C>A, p.Ser1110*) and VUS in SMAD4 (c.947A>G, p.Asn316Ser; hatched bars). B, Variants identified by TSO500 in a malignant melanotic schwannoma from a 36-year-old female, with Path mutations in NF1 (c.276dupA, p.C93fs*14) and PRKAR1A (c.207_208delGA, p.K70fs*11) and LP variant in SETD2 (c.4405dupA, p.M1469fs*6).C, Variants identified in two lymph node metastases collected 1 year apart from a carcinoma of unknown primary in a 75-year-old male (purple collected first, green collected second). VUS with higher percentage VAF (>45%) were identified in both samples (NCOR1, MST1R, ABL2, ROS1, ESR1, FAT1, POLE, NTRK3, and GEN1), as well as low VAF (∼6%) VUS in ZNF703. Pathogenicity of variants is indicated in y-axis label.
A pediatric patient was diagnosed with poorly differentiated adenocarcinoma of the descending colon at age 15. He presented with symptoms of abdominal pain, 30-pound weight loss, and nausea/vomiting. Imaging showed a descending colon mass (Fig. 3A). Pathology showed multiple colonic tubular adenomas, and focal adenocarcinoma (Fig. 3B). The patient was found to have pathogenic germline mutations in MLH1 and APC and VUS (strong evidence of pathogenicity) in SMAD4 (Fig. 4A). Molecular tumor testing (Fig. 4A) revealed pathogenic mutations with low VAF in KRAS, ARID1A, BCORL1, and PRKDC. Low VAF LP variants were found in NCOA3 and ERCC1. Multiple additional mutations were identified in APC, MLH1, and SMAD4. The patient was treated with surgery followed by capecitabine/oxaliplatin for seven cycles and subsequent prophylactic total colectomy. He continues to be followed according to guidelines for Lynch syndrome and familial adenomatous polyposis.
A 36-year-old female presented with back pain and was diagnosed with malignant melanotic schwannoma of the lumbar spine. MRI showed a mass in the left neural foramen of L3-L4 (Fig. 3C). Pathologic examination by IHC showed positive S100, HMB45, Melan-A stains with pigmented spindle and epithelioid cells (Fig. 3D) and psammoma bodies suggestive of Carney Complex (31). Germline mutation of PRKAR1A was negative, although a pathogenic PRKAR1A mutation was identified on tumor sequencing and mosaicism for germline PRKAR1A mutation could not be excluded. In addition, a somatic pathogenic variant in NF1 and LP variant in SETD2 were identified (Fig. 4B). She was treated with surgery, photon radiotherapy, and nivolumab/ipilimumab, but died from disease 2 years after diagnosis.
A 75-year-old male presenting with left neck pain was diagnosed with metastatic poorly differentiated carcinoma of unknown primary. MRI scan showed a left supraclavicular mass (Fig. 3E) and avid 2[18F]fluoro-2-deoxy-D-glucose uptake on PET scan. Biopsy demonstrated a malignant epithelioid tumor (Fig. 3F) positive for keratins (AE1/AE3) and weakly positive for PAX8. Because PAX8 is a common marker of renal cell carcinoma (32), the carcinoma was suspected to be metastatic renal carcinoma; however, no renal primary tumor could be identified. Two metastases to lymph nodes collected 1 year apart were sequenced (Fig. 4C). Path/LP mutations with low VAF were identified in KRAS, TP53, AR, CDKN2A, and EP300 in the first sample, but not the second sample, likely due to lower tumor content of the second sample. VUS with greater percentage VAF (>45%) were identified in both. He was treated with photon radiotherapy and surgical resection and is alive 3 years after diagnosis.
Discussion
We developed the NHRST to advance rare tumor research and ultimately improve patient outcomes through comprehensive study of rare solid tumors while engaging patients, advocates, and researchers. We were able to successfully enroll participants with rare tumors and collect comprehensive standardized clinical information on 98% of participants, self-reported PRO from 74% of participants, and tumor and/or germline biospecimens from 87% of participants. Analysis of the first 197 participants (out of 200 enrolled) of our ambitious study revealed several findings, which serve to inform the future of our study and possibly other rare tumor efforts.
Our study design allowed for enrollment in person at the NIHCC or remotely from the participants’ homes. Study enrollment began the year before the COVID-19 pandemic and remote enrollment allowed continued study participation and study feasibility through the pandemic. This may thus be an efficient approach in future studies.
Despite broad eligibility of any patient with a rare solid tumor, our cohort was not balanced in that we enrolled more participants from the U.S. East Coast closer to the NIHCC in Bethesda, MD, more females than males, and predominately white, non-Hispanic, more highly educated participants. To increase geographic diversity and recruitment of patients from the Southwest and Western United States, we are expanding the study to partner institutions. Several rare tumors in NHRST such as ACC and sdGIST have reported higher incidence in females (29, 33, 34). However, while most of our MTC participants were female (89%), a larger study showed little sex bias in sporadic and hereditary MTC (35). While participants are predominantly White and non-Hispanic, self-reported data on ancestry demonstrated contributions of different races to biospecimen genetic background. We found that reported race and ethnicity in the medical record was inconsistent with self-reported data, which were more detailed, demonstrating the value of self-reporting to reflect race and ethnicity accurately. Our participants showed high levels of education, with management, administrative support, and business/financial operations being the most reported occupations. Previous research has shown that education and income levels can affect diagnosis and progression of disease (36, 37). Recruitment to NHRST relies on referrals by physicians, and on patients finding our study through internet searches or interactions with advocacy groups, which may require higher levels of computer and medical literacy. Sex, race, and ethnicity can impact tumorigenesis and response to treatments both biologically (38) and socioeconomically (39, 40). We recognize that enrollment bias in these factors may limit the relevance of our findings to diverse patient populations and are aiming to develop targeted recruitment strategies for underrepresented populations.
Using a histology-agnostic approach, we enrolled participants with 35 different types of rare solid tumors. NIHCC infrastructure and collaboration among pediatric and adult oncologists facilitated enrollment of both pediatric and adult rare tumor patients, with 34% of participants being AYA, a population of unmet need for whom tumor incidence is increasing (41). We were also able to enroll pediatric participants with tumors diagnosed commonly in adults, but rarely in children, such as colorectal cancer, which is increasing in incidence in younger individuals (42). While any patient with a rare solid tumor was eligible, we enrolled five tumor types (NET, ACC, sdGIST, chordoma, MTC) with 9–47 participants per group. These tumors are the subject of strong research interests at NIHCC, including NIH rare tumor clinics for sdGIST, MTC, and chordoma, and strong collaborations with advocacy (43–45). It is thus feasible to enroll cohorts of patients with specific very rare tumor types. Importantly, most of our participants were not newly diagnosed, but enrolled several years after diagnosis when most participants had metastatic disease. This may reflect the large unmet need for this population where interventional trials are frequently not available and curative approaches are lacking. Recurrent disease after standard upfront therapy may also be the disease status for which development of external controls may be most needed, as novel interventional agents are typically first tested in the relapsed/refractory setting. This highlights the potential of NHRST to illuminate the natural history of recurrent disease and long-term survival and develop robust external controls for intervention trials.
Although we are collecting and extracting retrospective data on prior medical history and treatment, as well as following GCPS for development of tumors, it would be ideal to robustly characterize the clinical course of all very rare tumors of unmet need from diagnosis throughout the disease trajectory. Rapid progress will benefit from dedicated effort by additional “rare tumor champions” for specific rare tumors working with clinicians, researchers, and advocacy to develop additional patient cohorts. NCI's Childhood Cancer Data Initiative (CCDI) in collaboration with the Children's Oncology Group has established the Molecular Characterization Initiative (46), which provides state-of-the-art clinical molecular profiling and collection of clinical information focused on children and young adults with newly diagnosed malignancies including rare malignancies. The NCI CCDI is currently exploring the possibility of a CCDI-coordinated national very rare cancer longitudinal study (46) with participation by multiple sites and consortia.
In addition to information included in the medical record such as imaging, pathology, and medical history, we included PROs and comprehensive, standardized self-reported family and medical history. Although we had significantly fewer respondents completing self-report forms in the field group, we demonstrated feasibility of remote PRO administration to gather data from participants unable to travel due to illness, or other circumstances. PROs can be used to characterize quality of life relative to disease status, effectiveness of therapies, adverse effects of drugs, disease progression (47), and comparing patients with reference populations (e.g., comparing chordoma with other skull-based tumors). Patients with rare and potentially hereditary tumors are at risk for psychologic problems throughout life. Indeed, we found that anxiety and depression can remain CS long after initial diagnosis. Although there are many potential benefits to obtaining PROs from both in-clinic and field cohorts, we also acknowledge several limitations. First, our sample is potentially biased toward those with progressive disease who may be more likely to reach out to the NIH, enroll on the study, and report CS symptoms. Notably, 63% of our cohort had metastatic disease at enrollment. However, because many of our patients were further out from diagnosis at the time of enrollment, the impact of disease status on PROs at the time of diagnosis, is largely unavailable to comment on. In contrast, for field cohort participants, progressive disease may lead to difficulty completing PROs without in-person assistance. Similarly, this study provides insight into depression, pain, and anxiety symptoms at a single timepoint, and does not consider social-emotional history that preceded the cancer diagnosis, or life events that may contribute to elevated mental health symptoms at the time of PRO administration. Although administering PROs remotely is advantageous for patients unable to travel, we note anecdotally that patients completing PROs during visits to NIHCC ask clinical staff for clarification of questions, an opportunity lacking for remote participants. In the case of PROs administered to children, it is unknown how parental interactions in the remote setting might bias how children respond to questions. These confounding factors must be considered when weighing benefits of remote PRO administration in developing external control groups. Future strategies to increase patient engagement and completion of PRO measures, may include offering PROMIS forms in languages other than English and identification of challenges to form completion. In addition, we plan to conduct in-depth, longitudinal analysis of PROs by tumor type to include correlation of PRO data with measures of disease severity.
Broad collection of health-related data allows for comparisons between participants and other patient cohorts or the general population. Although 29% of measured adult participants were obese, this was less than 2017–2018 U.S. population data from NHAHES (42% obese). Our cohort had a bias toward White college-educated women, who typically have lower rates of obesity (48). Therefore, more data and detailed analyses are needed to make BMI comparisons by demographic and tumor type. When looking at participant reported health issues, some were common across all groups (e.g., allergies) likely reflecting the prevalence of these issues in the United States, whereas others were more frequent in certain tumor cohorts. Some health issues associated with specific tumor types likely reflect effects of tumor or treatment on the body (e.g., hypothyroidism or hyperthyroidism resulting from treatment in MTC or high blood pressure due to hormone disruption in ACC), whereas others (e.g., developmental delay in chordoma) could suggest genetic or developmental factors co-occurring with tumor risk or effects of treatment, particularly in developing children.
The molecular drivers of tumorigenesis are unknown for many rare tumors. To identify new targets for therapeutic intervention we analyzed genomic results from tumor biospecimens. TP53 and CTNNB1 are well-established ACC drivers (49–51). Path/LP variants found in ACC are involved in negative regulation of cell cycle and cell-cycle arrest (ATM, ATRX, CDKN2A, CDKN2B, CTNNB1, MSH6, and TP53), chromatin remodeling and organization (ATM, ATRX, CDKN2A, CTNNB1, and KMT2A), DNA repair and response to DNA damage (ATM, ATRX, MLH1, MSH6, and TP53), and intrinsic apoptosis (ATM, MLH1, MSH6, and TP53). While most GIST carry activating mutations in KIT or PDGFRA (52), our study focuses on a rare subset, sdGIST, lacking mutations in KIT or PDGFRA, which are resistant to imatinib and found in younger patients. sdGIST are driven by germline mutations or epigenetic silencing of the SDHC promoter (29, 43). These alterations were also observed in our patient cohort as well as a subset of their relatives. Mutations in RET, found in most MTCs in NHRST, are also found in the GCPS MEN2A and MEN2B, and RET is being targeted for systemic therapies (53–55). Somatic mutations in MEN1 have been previously identified in 44% of pancreatic NETs (56) and were found in 13% of our patients with NEN, including 2 with pancreatic NET. In our cohort, very few driver mutations were identified in chordoma, although amplification of brachyury (57) was not examined and only a small number of biospecimens were available for analysis. Loss of INI1 expression, encoded by SMARCB1, is characteristic of poorly differentiated chordomas more common in children and young adults (56). In addition to the Path/LP variants, many of the VUSs identified in our cohort are in molecular pathways implicated in rare tumors; studies are ongoing for further characterization of all VUSs.
In a study of 1120 pediatric patients (58), Path/LP germline mutations were found in 8.5% of patients and approximately 7%–14% of pediatric tumors are associated with GCPS (59). A survey of 27,999 adults found that 35.6% of respondents reported cancer in a first-degree relative (60). We found that 46% of participants had a first-degree relative with cancer. Of participants with clinically directed germline sequencing, 65% had at least one Path/LP germline mutation (33% of total cohort), likely due to our initial focus on sdGIST and MEN2A/B. Because many patients had targeted sequencing, rather than WES, the number Path/LP mutations present is likely underestimated. Future studies could identify new GCPS in addition to those we focused on.
Challenges
Over the course of enrolling and following the first 200 participants, we identified several challenges in the execution of NHRST. First, the COVID pandemic impeded enrollment, particularly for the clinic group, preventing collection of biospecimens and data from physical exams, imaging, and clinical labs. In contrast, although remote participation had many advantages, we found it was less robust for collecting self-reported data. Second, to take advantage of archival tumor samples for longitudinal analysis, we focused our molecular analysis on FFPE samples. In general, we found that sample quality becomes compromised after 5 years, limiting retrospective analyses. Third, in comparing overlapping self-reported and clinician-collected information, we found discrepancies, primarily due to exact phrasing and interpretation of questions. Finally, we have found that focused effort by clinicians and advocates is required to build and study useful cohorts for rare tumors.
Future Directions
We established a comprehensive, longitudinal natural history study for rare solid tumor types in children and adults, demonstrating how data specific to rare tumor types (e.g., correlated health issues, molecular drivers, and PROs) can be collected in a tumor-agnostic manner. Going forward, we plan to focus on developing interventional trials, increasing representation of minority and underserved populations, building select additional cohorts, and analyzing longitudinal data and comprehensive biospecimen molecular data. Interventional trials could enroll multiple rare tumor types based on mutation status, such as the recently opened trial for SMARCB1/SMARCA4-deficient tumors (NCT05286801). Increasing representation may be facilitated by developing specific partnerships with hospitals, advocacy groups, and professional organizations targeting underrepresented groups. Increasing enrollment of very rare solid tumors affecting children and AYA will be achieved by opening NHRST at other institutions and collaborating with other rare tumor efforts. Longitudinal timepoints will be compared to analyze tumor progression and response to treatment. Collected biospecimens are being comprehensively sequenced to interrogate the transcriptome, genome, and epigenome, in addition to the focused gene panel sequencing described here. As we gather data on NHRST, tumor-specific subprotocols will be developed to collect data tailored for use in interventional trials as external controls. Comprehensive longitudinal data gathered will be shared to facilitate collaborative rare tumor research. All data will be made publicly available per requirement of the NCI Cancer MoonshotSM. Data presented here are being made available through dbGAP and the NCI Cancer Data Service. Additional data from the study that is analyzed in future publications will be released upon validation in NCI-supported databases with controlled access provided by dbGAP.
Supplementary Material
Table S1 shows genes assayed in the TSO500 panel. Table S2 shows the sources of germline data for individual subjects.
Geographic distribution of participants by age group.
Age at diagnosis.
Race and ancestry of participants.
Distribution of enrollees participating remotely (field; red) or traveling to NIHCC (clinic; blue), located in Bethesda, MD.
Protocol form completion.
Time from diagnosis to enrollment by tumor type.
Performance scores, self-reported general health rating, and BMI.
Example of social and environmental characteristics of the NHRST cohort.
Other health issues reported in 3 or more participants out of 115 respondents by tumor type.
Tumor-directed treatments.
Anxiety and depression over time.
Success of TSO500 panel sequencing depending on age of formalin-fixed paraffin-embedded (FFPE) tissue.
Acknowledgments
The authors are thankful to the patients and families for participating in this protocol. We are grateful to members of the NCI CCR Pediatric Oncology Branch for clinical and research support, collaborations, and helpful discussions. We thank the CCR CIT for database support. Palantir Foundry was used in the integration, harmonization, and analysis of a subset of the reported data inside the secure NIDAP. This research was supported by the NCI Cancer MoonshotSM for the My Pediatric and Adult Rare Tumor (MyPART) network (S. Ahmed, M.F. Wedekind, M. Raygada, C. Flowers, B.J. Thomas, D.B. Bernstein, O.B. Kapustina, A.B. Sandler, and A. Jain), by the Intramural Research Program, of the NIH, NCI, Center for Cancer Research (J. Del Rivero, J.W. Glod, M. Miettinen, M. Raffeld, L. Xi, M. Tyagi, J. Kim, K. Aldape, A.A. Malayeri, R.N. Kaplan, B.C. Widemann, and K.M. Reilly) and with federal funds from the NCI, NIH, to Leidos Biomedical Research, Inc, under contract no. 75N91019D00024 (R. Lockridge, T. Allen) and Kelly Government Solutions, under contract no. 75N95021D00012 (C.A. Vivelo).
The following members of the MyPART network, past and present, have contributed to the design, implementation, and/or analysis of the MyPART Natural History and Biospecimen Acquisition Study for Children and Adults with Rare Solid Tumors (NHRST; NCT03739827).
| Name | Affiliations |
|---|---|
| Ken Adalpe | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Shadin Ahmed | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Srivandana Akshintala | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Taryn Allen, | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, Leidos Biomedical Research, Inc. Frederick, MD |
| Fernanda Arnaldez | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Donna Bernstein | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Paula Carter | Blood Processing Core, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Fiona Chalmers | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Michael Davies | Clinical Support Laboratory, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Jaydira del Rivero | Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Sherri DePollar | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Renee Donahue | Center for Immuno-Oncology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| W. Douglas Figg | Genitourinary Malignancies Branch, Blood Processing Core, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Crystal Flowers | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Sarah Fuller | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Yuti Gandhi | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| John W. Glod | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Andrea Gross | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Stephen Hewitt | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Susan Hughes | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Maran Illanchezhian | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Jon Inglefield | Clinical Support Laboratory, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Ashish Jain | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Parthav Jailwala | CCR Collaborative Bioinformatics Resource, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Liny John | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Rosandra Kaplan | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Oxana Kapustina | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Erika Kaschak | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Jason Levine | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Iris Li | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Jung Lim | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Ashkan Malayeri | Department of Radiology and Imaging Sciences, Clinical Center, National Institutes of Health, Bethesda, MD |
| Christian J. Mambrin | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Staci Martin Peron | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Arnulfo Mendoza | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Maria Merino | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Markku Miettinen | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Katie Pendo | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Trung Pham | Blood Processing Core, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Kristen Pike | CLIA Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Mark Raffeld | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Margarita Raygada | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Karlyne M. Reilly | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Robin Lockridge | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD, Leidos Biomedical Research, Inc. Frederick, MD |
| Abby B. Sandler | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Julianne Schultz | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Jeffrey Schlom | Center for Immuno-Oncology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| John (Jack) Shern | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Impana Shetty | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Nurlan Shonkoev | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Hannah Smith | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Mayank Tandon | CCR Collaborative Bioinformatics Resource, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Naomi Taylor | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| BJ Thomas | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Francesco Tomassoni Ardori | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Manoj Tyagi | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Christina Vivelo | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Yanyu Wang | Clinical Support Laboratory, Frederick National Laboratory for Cancer Research, National Cancer Institute, Frederick, MD |
| Mary Frances Wedekind Malone | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Trish Whitcomb | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Brigitte C. Widemann | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Lori Wiener | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Laura Wisch | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Pam Wolters | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Miguel Wolfe | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Liqiang Xi | Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
| Marcia Young | Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD |
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
J.W. Glod reports other from Celgene and Apolomics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
S. Ahmed: Data curation, formal analysis, validation, investigation, visualization, writing-original draft. M.F. Wedekind: Data curation, supervision, validation, methodology, project administration, writing-review and editing. J. Del Rivero: Data curation, validation, methodology, writing-review and editing. M. Raygada: Data curation, formal analysis, validation, investigation, methodology, writing-review and editing. R. Lockridge: Data curation, formal analysis, validation, investigation, visualization, methodology, writing-original draft. J.W. Glod: Data curation, validation, writing-review and editing. C. Flowers: Data curation, validation, writing-review and editing. B.J. Thomas: Data curation, validation, writing-review and editing. D.B. Bernstein: Data curation, validation, writing-review and editing. O.B. Kapustina: Data curation, validation, writing-review and editing. A. Jain: Software, visualization, writing-review and editing. M. Miettinen: Data curation, validation, writing-review and editing. M. Raffeld: Data curation, validation, writing-review and editing. L. Xi: Data curation, validation, writing-review and editing. M. Tyagi: Data curation, software, validation, writing-review and editing. J. Kim: Data curation, validation, writing-review and editing. K. Aldape: Resources, supervision, validation, methodology, writing-review and editing. A.A. Malayeri: Data curation, validation, writing-review and editing. R.N. Kaplan: Data curation, validation, writing-review and editing. T. Allen: Data curation, validation, methodology, writing-review and editing. C.A. Vivelo: Resources, writing-review and editing. A.B. Sandler: Supervision, project administration, writing-review and editing. B.C. Widemann: Conceptualization, resources, data curation, supervision, funding acquisition, validation, methodology, project administration, writing-review and editing. K.M. Reilly: Conceptualization, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-original draft, project administration, writing-review and editing.
References
- 1. Sharifnia T, Hong AL, Painter CA, Boehm JS. Emerging opportunities for target discovery in rare cancers. Cell Chem Biol 2017;24:1075–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrari A, Schneider DT, Bisogno G, Reguerre Y, Godzinski J, Bien E, et al. Facing the challenges of very rare tumors of pediatric age: The European Cooperative Study Group for Pediatric Rare Tumors (EXPeRT) background, goals, and achievements. Pediatr Blood Cancer 2021;68:e28993. [DOI] [PubMed] [Google Scholar]
- 3. Abbas-Aghababazadeh F, Mo Q, Fridley BL. Statistical genomics in rare cancer. Semin Cancer Biol 2020;61:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kee D, Parker C, Bae S, Tucker KM, Harrison M, Tohidi-Esfahani I, et al. CART-WHEEL.org: an ethically approved online database for patient-entered data to facilitate rare cancer research. JCO Clin Cancer Inform 2020;4:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fair D, Potter SL, Venkatramani R. Challenges and solutions to the study of rare childhood tumors. Curr Opin Pediatr 2020;32:7–12. [DOI] [PubMed] [Google Scholar]
- 6. Painter CA, Jain E, Tomson BN, Dunphy M, Stoddard RE, Thomas BS, et al. The Angiosarcoma Project: enabling genomic and clinical discoveries in a rare cancer through patient-partnered research. Nat Med 2020;26:181–7. [DOI] [PubMed] [Google Scholar]
- 7. Blay JY, Coindre JM, Ducimetiere F, Ray-Coquard I. The value of research collaborations and consortia in rare cancers. Lancet Oncol 2016;17:e62–9. [DOI] [PubMed] [Google Scholar]
- 8. Gatta G, Capocaccia R, Botta L, Mallone S, De Angelis R, Ardanaz E, et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet-a population-based study. Lancet Oncol 2017;18:1022–39. [DOI] [PubMed] [Google Scholar]
- 9. Lilly JV, Rokita JL, Mason JL, Patton T, Stefankiewiz S, Higgins D, et al. The children's brain tumor network (CBTN) – accelerating research in pediatric central nervous system tumors through collaboration and open science. Neoplasia 2023;35:100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patel SP, Othus M, Chae YK, Giles FJ, Hansel DE, Singh PP, et al. A phase II Basket Trial of dual anti-CTLA-4 and anti-PD-1 blockade in rare tumors (DART SWOG 1609) in patients with nonpancreatic neuroendocrine tumors. Clin Cancer Res 2020;26:2290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keat N, Law K, Seymour M, Welch J, Trimble T, Lascombe D, et al. International rare cancers initiative. Lancet Oncol 2013;14:109–10. [DOI] [PubMed] [Google Scholar]
- 12. Parsons DW, Janeway KA, Patton DR, Winter CL, Coffey B, Williams PM, et al. Actionable tumor alterations and treatment protocol enrollment of pediatric and young adult patients with refractory cancers in the National Cancer Institute-Children's Oncology Group Pediatric MATCH Trial. J Clin Oncol 2022;40:2224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kline C, Jain P, Kilburn L, Bonner ER, Gupta N, Crawford JR, et al. Upfront biology-guided therapy in diffuse intrinsic pontine glioma: therapeutic, molecular, and biomarker outcomes from PNOC003. Clin Cancer Res 2022;28:3965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rokita JL, Rathi KS, Cardenas MF, Upton KA, Jayaseelan J, Cross KL, et al. Genomic profiling of childhood tumor patient-derived xenograft models to enable rational clinical trial design. Cell Rep 2019;29:1675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singer DS, Jacks T, Jaffee E. A U.S. Cancer Moonshot" to accelerate cancer research. Science 2016;353:1105–6. [DOI] [PubMed] [Google Scholar]
- 17. Wedekind MF, Reilly KM, Rivero JD, Lockridge R, Allen T, Raygada M, et al. NCI intramural program approach to rare tumors: Natural History Study of Rare Solid Tumors in children and adults: a longitudinal, comprehensive data and biospecimen collection protocol. Pediatr Blood Cancer 2023:e30495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Office of the Commissioner Office of Clinical Policy and Programs, Office of Orphan Products Development, Center for Drug Evaluation and Research, Center for Biologics Evaluation and Research. Rare diseases: natural history studies for drug development: draft guidance for industry. March 2019. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/rare-diseases-natural-history-studies-drug-development.
- 19. Gross AM, Wolters PL, Dombi E, Baldwin A, Whitcomb P, Fisher MJ, et al. Selumetinib in children with inoperable plexiform neurofibromas. N Engl J Med 2020;382:1430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gross AM. Using real world data to support regulatory approval of drugs in rare diseases: a review of opportunities, limitations & a case example. Curr Probl Cancer 2021;45:100769. [DOI] [PubMed] [Google Scholar]
- 21. Mishra-Kalyani PS, Amiri Kordestani L, Rivera DR, Singh H, Ibrahim A, DeClaro RA, et al. External control arms in oncology: current use and future directions. Ann Oncol 2022;33:376–83. [DOI] [PubMed] [Google Scholar]
- 22. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW Jr. Body-mass index and mortality in a prospective cohort of U.S. adults. N Engl J Med 1999;341:1097–105. [DOI] [PubMed] [Google Scholar]
- 24. Fryar C, Carroll MD, Afful J. Prevalence of overweight, obesity, and severe obesity among adults aged 20 and over: United States, 1960–1962 through 2017–2018. US Department of Health and Human Services, Centers for Disease Control and Prevention; 2020.
- 25. Stierman B, Afful J, Carroll M, Chen T, Davy O, Fink S, et al. National health and nutrition examination survey 2017–March 2020 prepandemic data files—development of files and prevalence estimates for selected health outcomes. US Department of Health and Human Services, Centers for Disease Control and Prevention 2021. [DOI] [PMC free article] [PubMed]
- 26. Kalyn R. BC Cancer Pharmacy Education Program Cancer Drug Pharmacology Table. Pharmacology_Table.pdf. British Columbia, Canada: BC Cancer; 2018. [Google Scholar]
- 27. Cella D, Choi S, Garcia S, Cook KF, Rosenbloom S, Lai JS, et al. Setting standards for severity of common symptoms in oncology using the PROMIS item banks and expert judgment. Qual Life Res 2014;23:2651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rothrock NE, Amtmann D, Cook KF. Development and validation of an interpretive guide for PROMIS scores. J Patient Rep Outcomes 2020;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Killian JK, Miettinen M, Walker RL, Wang Y, Zhu YJ, Waterfall JJ, et al. Recurrent epimutation of SDHC in gastrointestinal stromal tumors. Sci Transl Med 2014;6:268ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lodish MB, Stratakis CA. RET oncogene in MEN2, MEN2B, MTC and other forms of thyroid cancer. Expert Rev Anticancer Ther 2008;8:625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharath KGG, Shruthi P, Pooja BV, Ravi MR. Trigeminal nerve melanotic schwannoma in carney complex. Indian J Radiol Imaging 2022;32:422–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel SA, Hirosue S, Rodrigues P, Vojtasova E, Richardson EK, Ge J, et al. The renal lineage factor PAX8 controls oncogenic signalling in kidney cancer. Nature 2022;606:999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Audenet F, Mejean A, Chartier-Kastler E, Roupret M. Adrenal tumours are more predominant in females regardless of their histological subtype: a review. World J Urol 2013;31:1037–43. [DOI] [PubMed] [Google Scholar]
- 34. Wang YM, Gu ML, Ji F. Succinate dehydrogenase-deficient gastrointestinal stromal tumors. World J Gastroenterol 2015;21:2303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotwal A, Erickson D, Geske JR, Hay ID, Castro MR. Predicting outcomes in sporadic and hereditary medullary thyroid carcinoma over two decades. Thyroid 2021;31:616–26. [DOI] [PubMed] [Google Scholar]
- 36. Xu L, Wang X, Pan X, Wang X, Wang Q, Wu B, et al. Education level as a predictor of survival in patients with multiple myeloma. BMC Cancer 2020;20:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y, Zhang J, Huang R, Feng WL, Kong YN, Xu F, et al. Influence of occupation and education level on breast cancer stage at diagnosis, and treatment options in China: A nationwide, multicenter 10-year epidemiological study. Medicine 2017;96:e6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pala L, Conforti F. The effect of patient sex on the efficacy and safety of anticancer immunotherapy. Expert Opin Drug Saf 2021;20:1535–44. [DOI] [PubMed] [Google Scholar]
- 39. Mauvais-Jarvis F, Bairey Merz N, Barnes PJ, Brinton RD, Carrero JJ, DeMeo DL, et al. Sex and gender: modifiers of health, disease, and medicine. Lancet 2020;396:565–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hann KEJ, Freeman M, Fraser L, Waller J, Sanderson SC, Rahman B, et al. Awareness, knowledge, perceptions, and attitudes towards genetic testing for cancer risk among ethnic minority groups: a systematic review. BMC Public Health 2017;17:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller KD, Fidler-Benaoudia M, Keegan TH, Hipp HS, Jemal A, Siegel RL. Cancer statistics for adolescents and young adults. CA Cancer J Clin 2020;70:443–59. [DOI] [PubMed] [Google Scholar]
- 42. Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, Siena S. Early-onset colorectal cancer in young individuals. Mol Oncol 2019;13:109–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boikos SA, Pappo AS, Killian JK, LaQuaglia MP, Weldon CB, George S, et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: a report from the National Institutes of Health gastrointestinal stromal tumor clinic. JAMA Oncol 2016;2:922–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weldon CB, Madenci AL, Boikos SA, Janeway KA, George S, von Mehren M, et al. Surgical management of wild-type gastrointestinal stromal tumors: a report from the National Institutes of Health pediatric and wildtype GIST clinic. J Clin Oncol 2017;35:523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. John L, Smith H, Ilanchezhian M, Lockridge R, Reilly KM, Raygada M, et al. The NIH pediatric/young adult chordoma clinic and natural history study: making advances in a very rare tumor. Pediatr Blood Cancer 2023. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Flores-Toro JA, Jagu S, Armstrong GT, Arons DF, Aune GJ, Chanock SJ, et al. The Childhood Cancer Data Initiative: using the power of data to learn from and improve outcomes for every child and young adult with pediatric cancer. J Clin Oncol 2023;41:4045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weldring T, Smith SM. Patient-reported outcomes (PROs) and patient-reported outcome measures (PROMs). Health Serv Insights 2013;6:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogden CL, Fakhouri TH, Carroll MD, Hales CM, Fryar CD, Li X, et al. Prevalence of obesity among adults, by household income and education—United States, 2011–2014. MMW Morb Mortal Wkly Rep 2017;66:1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wasserman JD, Novokmet A, Eichler-Jonsson C, Ribeiro RC, Rodriguez-Galindo C, Zambetti GP, et al. Prevalence and functional consequence of TP53 mutations in pediatric adrenocortical carcinoma: a Children's Oncology Group study. J Clin Oncol 2015;33:602–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu S, Ding G, Zhou Z, Feng C. beta-Catenin-driven adrenocortical carcinoma is characterized with immune exclusion. Onco Targets Ther 2018;11:2029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiorentini C, Grisanti S, Cosentini D, Abate A, Rossini E, Berruti A, et al. Molecular drivers of potential immunotherapy failure in adrenocortical carcinoma. J Oncol 2019;2019:6072863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nannini M, Rizzo A, Indio V, Schipani A, Astolfi A, Pantaleo MA. Targeted therapy in SDH-deficient GIST. Ther Adv Med Oncol 2021;13:17588359211023278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ilanchezhian M, Khan S, Okafor C, Glod J, Del Rivero J. Update on the treatment of medullary thyroid carcinoma in patients with multiple endocrine neoplasia type 2. Horm Metab Res 2020;52:588–97. [DOI] [PubMed] [Google Scholar]
- 54. Okafor C, Hogan J, Raygada M, Thomas BJ, Akshintala S, Glod JW, et al. Update on targeted therapy in medullary thyroid cancer. Front Endocrinol 2021;12:708949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kraft IL, Akshintala S, Zhu Y, Lei H, Derse-Anthony C, Dombi E, et al. Outcomes of children and adolescents with advanced hereditary medullary thyroid carcinoma treated with vandetanib. Clin Cancer Res 2018;24:753–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mobley BC, McKenney JK, Bangs CD, Callahan K, Yeom KW, Schneppenheim R, et al. Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathol 2010;120:745–53. [DOI] [PubMed] [Google Scholar]
- 57. Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, et al. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet 2009;41:1176–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015;373:2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Muskens IS, Zhang C, de Smith AJ, Biegel JA, Walsh KM, Wiemels JL. Germline genetic landscape of pediatric central nervous system tumors. Neuro Oncol 2019;21:1376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kumerow MT, Rodriguez JL, Dai S, Kolor K, Rotunno M, Peipins LA. Prevalence of Americans reporting a family history of cancer indicative of increased cancer risk: Estimates from the 2015 National Health Interview Survey. Prev Med 2022;159:107062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 shows genes assayed in the TSO500 panel. Table S2 shows the sources of germline data for individual subjects.
Geographic distribution of participants by age group.
Age at diagnosis.
Race and ancestry of participants.
Distribution of enrollees participating remotely (field; red) or traveling to NIHCC (clinic; blue), located in Bethesda, MD.
Protocol form completion.
Time from diagnosis to enrollment by tumor type.
Performance scores, self-reported general health rating, and BMI.
Example of social and environmental characteristics of the NHRST cohort.
Other health issues reported in 3 or more participants out of 115 respondents by tumor type.
Tumor-directed treatments.
Anxiety and depression over time.
Success of TSO500 panel sequencing depending on age of formalin-fixed paraffin-embedded (FFPE) tissue.
Data Availability Statement
Data are available through controlled access at dbGAP (RRID:SCR_002709) accession phs003143.v1.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003143.v1.p1).