Abstract
Marine derived cyclic imine toxins, portimine A and B, have attracted extensive attention owing to their intriguing chemical structure and promising anti-cancer therapeutic potential1-4. However, access to large quantities is currently unfeasible and the molecular mechanism behind their potent activity is unknown. To address this, a scalable, concise synthesis of portimines is presented, which benefits from the logic used in two-phase terpenoid synthesis5,6 along with unique tactics such as exploiting ring-chain tautomerization and skeletal reorganization to minimize protecting group chemistry through “self-protection”. Critically, this total synthesis enabled a structural reassignment of portimine B and an in-depth functional evaluation of portimine A, revealing that it induces apoptosis selectively in human cancer cell lines with high potency and is efficacious in vivo in tumor clearance models. Finally, practical access to the portimines and analogs thereof simplified the development of photoaffinity analogs, which were used in chemical proteomic experiments to identify a primary target of portimine A as the 60S ribosomal export protein NMD3.
For decades, cyclic imine (CI) toxins have stimulated extensive interest from the broad scientific community based on their unique and potent bioactivity coupled with their captivating chemical structures1,2. The therapeutic potentials of larger members in this family, such as pinnatoxins7, spirolides8, and gymnodimines9, have been thwarted by their high neurotoxicity in vivo. More compact members of this family were isolated from benthic dinoflagellate Vulcanodinium rugosum in 2013 and 2018, portimine A (PA, 1) and B (PB, 2) respectively (Fig. 1, absolute configuration confirmed in 2019)3,10,11. In sharp contrast to classic CI toxins, preliminary reports indicate that 1 is highly cytotoxic (~3 nM) in several cancer cell lines, while displaying lower acute toxicity in mice, highlighting its potential as a therapeutic agent3,4. Further, recent reports have established portimine A as a selective inducer of apoptosis with minimal necrotic activity4, however, the mechanism of action (MoA) behind its potent activity is unknown. The Achilles heel of such a compound is of course scalable access to its complex architecture. As it is derived from a dinoflagellate in low yield3, chemical synthesis appears to be the only means of realistically procuring such molecules. Even if a bioengineered synthesis could be achieved, semi-synthetic analogs with deep-seated modifications would be unworkable. Featuring a spiro-fused five-membered cyclic imine embedded in a highly oxidized all-carbon tricyclic macrocyclic core, 1 and 2 are formidable targets for synthesis. The unusual peripheral oxidations such as that adjacent to imine carbon (C-5) and neighboring labile medium-sized cyclic ketal add to this challenge. In this work, the first total synthesis of 1 and 2 is presented, which features a carefully choreographed sequence to rapidly build up a minimally oxidized carbon framework followed by strategic oxidations and ring-chain isomerizations that minimize concession steps. In addition, this scaled synthesis enabled elaborated in situ and in vivo investigations as well as illumination of the cellular targets of 1 via chemical proteomics, revealing that 1 targets the 60S ribosomal export protein NMD3, blocks polysome formation, and inhibits protein translation.
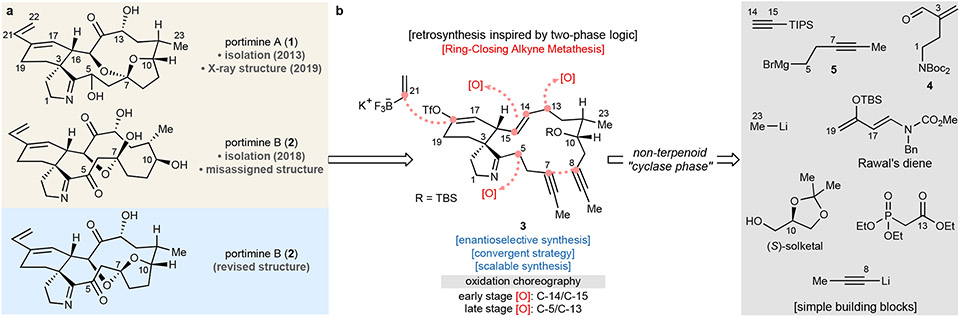
Fig. 1. Retrosynthetic analysis in this work.
Historically, CI toxins have been constructed by patterning the retrosynthetic analysis on the presumed biogenesis12-14, wherein an acyclic structure with maximum functionality is subjected to macrocyclization15-19. This pioneering approach was first accomplished by Kishi et al. in the 1998 total synthesis of pinnatoxin A15. In the case of the portimines, approaches thus far have followed this dogma20-23. The difficulty encountered in these routes points to the challenge of forging key bonds in such a densely functionalized polycyclic alkaloid from an acyclic precursor. From a high level, this scenario is not unlike that encountered in the synthesis of densely functionalized, highly oxidized terpene natural products. In those cases, it has been shown that a two-phase approach to synthesis can be beneficial by building up a minimally oxidized carbon framework followed by strategic late-stage oxidations5,6.
Total synthesis of portimine A and B
By analogy to the logic of two-phase synthesis, a minimally oxidized macrocyclic intermediate to the portimines was targeted with the assumption that a proper choreography of oxidation events would solve both connectivity and stereochemical issues. The only C─O bonds to be installed at the outset were those residing at C-4 (imine carbon) and C-10 (secondary alcohol). A triple bond on C-7/8 would be a surrogate for the eventual C-7 oxidation and, critically, offer a strategic disconnection to the macrocycle using ring-closing alkyne metathesis (RCAM)24. Such a tactic would thereby minimize unstable functional groups and redundant redox manipulations since four key oxidations (C-5, C-13, C-14, C-15) would occur post-macrocyclization. To minimize protecting groups (PGs), the innate reactivity and conformational preferences are utilized via ring-chain reorganization/reconstitution steps. Upon unraveling the macrocycle, the dialkynated precursor 3 traces back to accessible building blocks (Fig. 1). Finally, in order to maximize access to useful analogs, a vinyl triflate was selected as a key functionality to be carried through the entire synthesis.
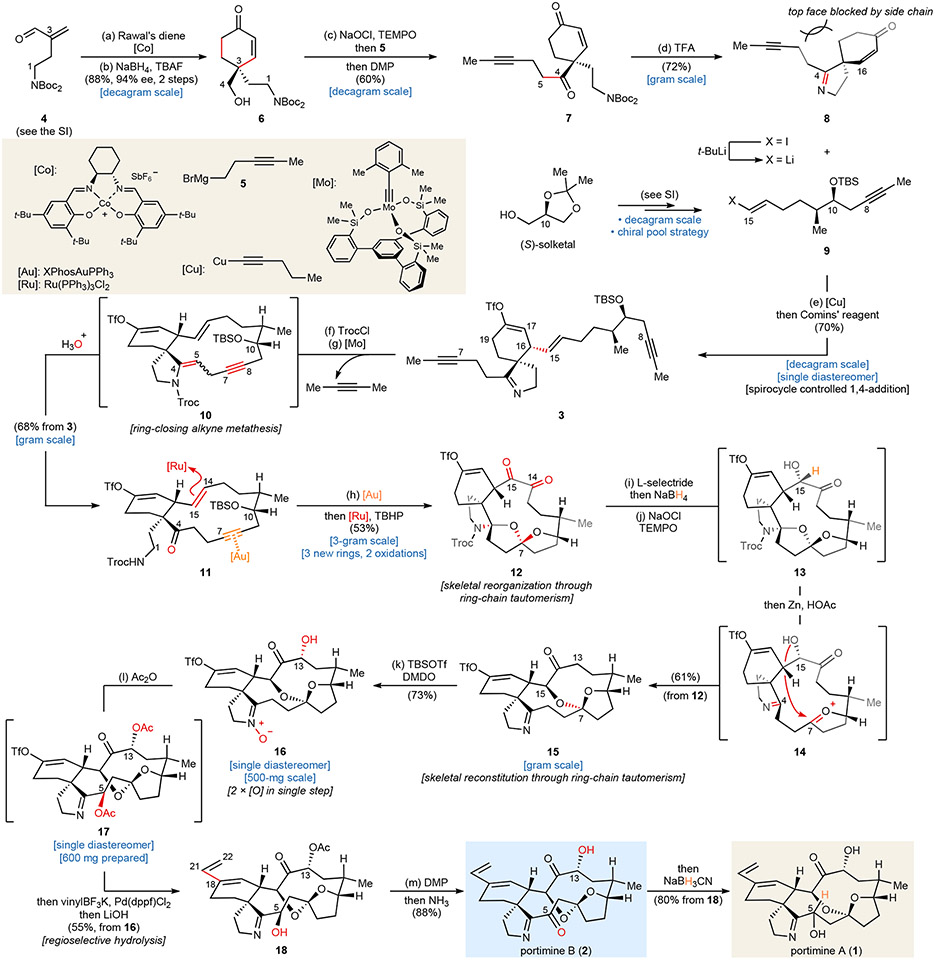
The synthesis is outlined in Fig. 2 and commences with a scalable, asymmetric Diels-Alder cycloaddition25, which established the C-3 chirality. Reduction of C-4, followed by removing the carbamate auxiliary with tetrabutylammonium fluoride (TBAF), afforded 6 in 88% yield, and 94% ee on 50-gram scale. The requisite methyl-capped alkyne side chain can be installed via a sequence including Grignard addition and two oxidations, delivering 7 as the final product in 60% yield after a single purification. Treatment of 7 with trifluoroacetic acid (TFA) in CH2Cl2 afforded spirocyclic imine 8 in 72% yield on gram scale.
Fig. 2. Total synthesis of portimine A and B.
The synthesis of fragment 9 was carried out from inexpensive (S)-solketal (ca. $ 1.1/g, see Supplementary Information for detailed steps). To affix this subunit onto the established chiral spirocyclic core 8, a stereoselective Cu-mediated conjugate addition was applied. The choice of copper (I) reagent26 was crucial since switching to other common copper (I) salts, such as CuI, CuCN, and CuBr, showed little to no observable conversion (Supplementary Table 1). The stereochemical outcome in this step is controlled by the intrinsic configuration of the spiro-cycle, wherein the side chain blocked the top face. Direct treatment of the in situ generated enolate with Comins’ reagent ensured the correct regiochemical olefin outcome and smoothly delivered vinyl triflate 3 as a single product (6.5-g scale), which possessed all skeletal carbon atoms required for 1 and 2.
Attention was then turned to constructing the 14-membered macrocycle in portimines’ skeleton through RCAM, a maneuver that might be derailed by the imine, olefins, or vinyl triflate. Fürstner’s extremely efficient, canopy-shaped catalyst, [Mo] was chosen at this point due to its outstanding functional compatibility and demonstrated robustness27. The pivotal RCAM step could indeed be achieved in 53 – 65% yield when heated 7 with [Mo] in toluene. However, a relatively high catalyst loading (12.5 mol %) was required, presumably due to the basic imine nitrogen. To lower the catalyst loading, the imine was masked with a 2,2,2-trichloroethoxycarbonyl (Troc) group, followed by exposing the formed enamide to 2.0 mol % [Mo]. In this case, formation of macrocycle 10 was completed in one hour. Treating crude 10 with acidic wet methanol liberated the C-4 ketone and deprotected the t-butyldimethylsilyl (TBS) ether on C-10, affording 11 as a white powder (68% overall isolated yield, multigram-scale).
Arrival at macrocycle 11 (the end of the “cyclase phase”) was a milestone since all requisite core C─C bonds were in place to arrive at 1 and 2. All that remained was installation of five oxygen atoms at C-5, C-7, C-13, C-14, and C-15. The “oxidase-phase” commenced with oxidation of C-14/15 since olefin-oxidations are well-known to be compatible with alkynes. Initial attempts using oxidants (i.e., OsO4, m-chloroperoxybenzoic acid (m-CPBA), etc.) delivered undesired stereochemical outcome at C-15 in all cases (see Supplementary information for more details). Therefore, a net six-electron oxidation catalyzed by ruthenium was chosen to arrive at a diketone28, with an eventual strategic reduction to set the desired stereochemistry. Stronger oxidants of this type, however, will not tolerate the presence of an alkyne. For this purpose, an internal protection strategy was designed and achieved upon skeletal reorganization by refluxing 11 and XPhosAuNTf2 (0.8 mol %) in CH2Cl2. The newly-formed tricyclic system reorganized all potentially sensitive sites (nitrogen on C-1, C-4 ketone, C-7-C-8 alkyne, and C-10 alcohol) into their inactive states. Subsequent six-electron oxidation under Ru-catalysis led to diketone 12 in a 53% isolated yield (gram-scale) thereby minimizing reliance on PGs.
With this newly constructed rigid polycyclic system in place, the correct oxidation state and stereochemistry on C-14 and C-15 were installed. Site-specific reduction of C-14 was accomplished using L-selectride, followed by treating the crude material with NaBH4 to afford diol 28 (see Supplementary Information for detailed steps), which possessed the desired stereochemistry on C-15. Subsequently, C-14 was selectively returned to the ketone oxidation state with (2,2,6,6-tetramethylpiperidin-1-yl)oxy (TEMPO)/NaOCl. Other oxidants tested for this step showed poor selectivity (Supplementary Table 3). Remarkably, upon heating crude 13 with zinc powder in acetic acid, the Troc group was dismantled and the polycyclic ring system spontaneously unraveled through ring-chain tautomerization (presumably via 14) to liberate the deoxyportimine triflate 15 in 74% yield on gram-scale.
Only two oxygen atoms, at C-13 and C-5, remained to be installed to complete the synthesis. Both oxidations could be achieved in a single step by treating the crude silyl enol ether of 15 with dimethyldioxirane (DMDO) to afford a nitrone 16 (73% overall yield). Subsequent heating of 16 in the presence of acetic anhydride (Ac2O) and triethylamine (TEA) presumably triggers a Boekelheide type rearrangement29 to deliver a diacetate 17 as a single diastereomer. At this point, the vinyl triflate which had remained a silent observer throughout the synthesis was now called upon to append the final two carbon atoms of 1 and 2. Thus, a Suzuki coupling was chosen to install the exocyclic vinyl group, followed by a regioselective hydrolysis of the C-5 acetate using LiOH to yield monoacetate 18 after a single purification (55% overall yield from 16). To complete the synthesis, 18 was oxidized with Dess-Martin periodinane (DMP), followed by hydrolysis, affording portimine B (PB, 2) in 88% yield. During these studies we suspected that the originally assigned structure of 2 as a ring-opened tautomer was incorrect and this was now confirmed to be the same ring-closed tautomer expressed in 1. To complete the synthesis of portimine A (PA, 1), crude 2 in methanol could be stereoselectively reduced by NaBH3CN in high isolated yield (80%).
Portimine A displays acute toxicity
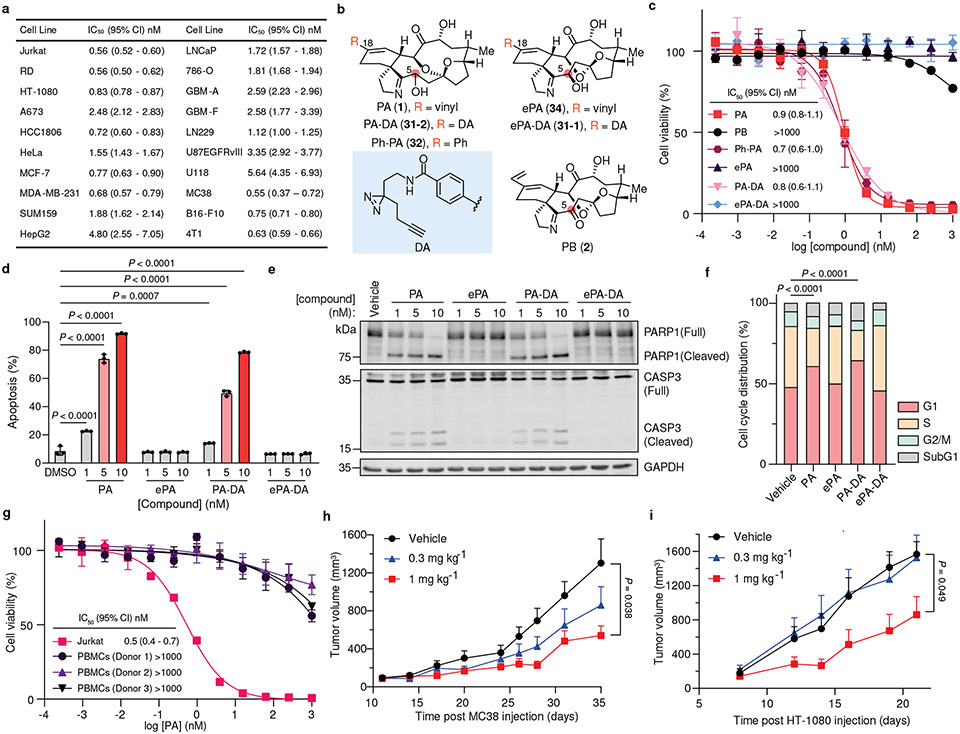
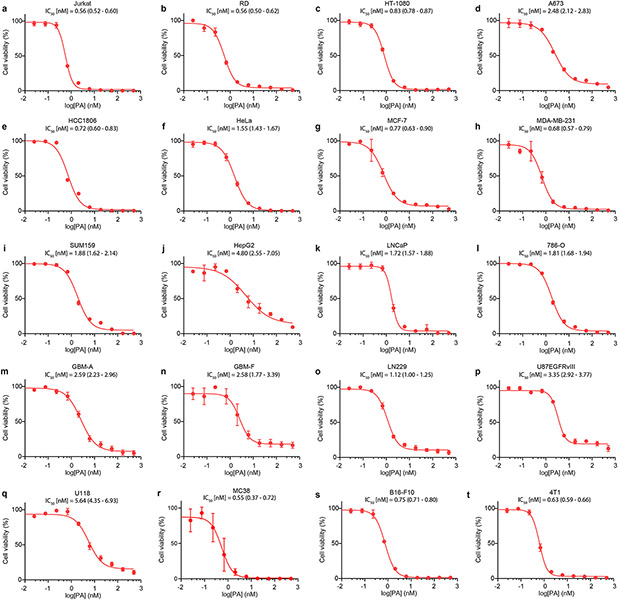
With this scalable synthesis in hand, we first assessed the cytotoxic effects of fully synthetic PA (1) across a broad panel of 20 human and mouse cancer cell lines, ranging from the Jurkat leukemia cell line originally used to characterize PA bioactivity4, to metastatic human fibrosarcoma cells (i.e, HT-1080), triple negative breast cancer (TNBC) cells (i.e., MDA-MB-231) and primary patient-derived glioblastoma brain tumor initiating cell lines that display hallmark features of human brain tumors including capacity to metastasize in orthotopic tumor xenograft models (i.e., GBM-A30). Consistently potent cytotoxic activity was observed across the entire panel of cancer cell lines that were evaluated (IC50 = 0.55 – 5.6 nM, Fig. 3a and Extended Data Fig. 1). In order to understand the mechanism of this activity, we next sought to identify sites that could accommodate a “fully functionalized” retrieval tag to enable chemical proteomic target identification while not interfering with the biological activity of the parent structure31-33. Here, an evaluation of portimine A analogs with modifications at multiple sites (Fig. 3b) in Jurkat cells confirmed that fully-synthetic PB (2) is significantly less toxic compared to PA (1)10, as is epi-portimine A (ePA, 34), the C-5 epimer of PA (1) (Fig. 3c). Taken together, these results suggest that the anti-proliferative activity of PA (1) is dependent on the stereochemistry of C-5, and that ePA (34) could be employed as an inactive control compound for further studies. We also noted that phenyl-derivative Ph-PA (32) showed similar activity to PA (1), indicating the terminal vinyl group (C-21/22) is not essential for the observed biological activity (Fig. 3b-c). Based on these results, we pursued the assembly of a diazirine-alkyne (DA) containing photoaffinity tag at the C-18 position of PA (1) through Suzuki coupling with the key intermediate (compound 21, for detailed synthesis steps, see Supplementary Information). Encouragingly, we observed both the epimeric photoaffinity probes PA-DA (31-2) and ePA-DA (31-1) retained identical activities to their parent analogs, suggesting that they could serve as target identification tools (Fig. 3b-c).
Fig. 3. Portimine A and its functional analogs show selective acute toxicity in cancer cells and in vivo tumor models.
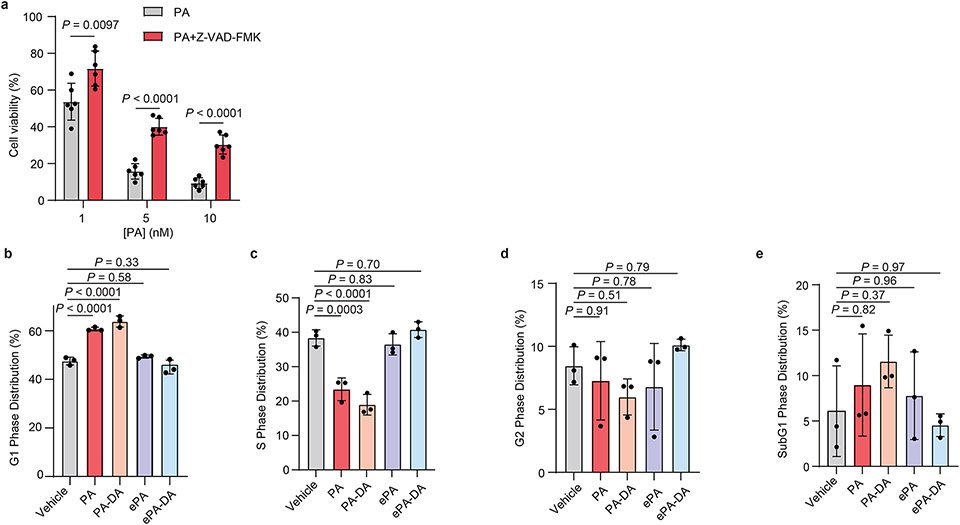
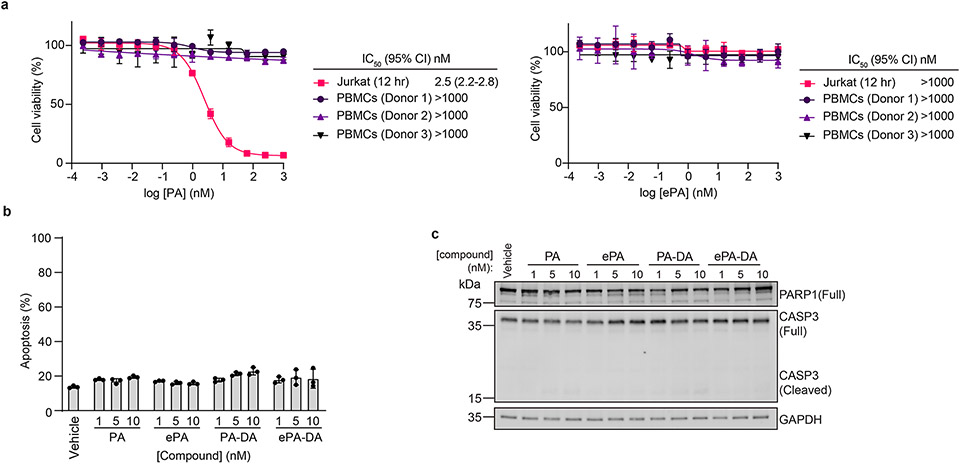
a, Anti-proliferation activity of portimine A (PA) against human and mouse cancer cell lines (see Extended Data Fig. 1 for cell line descriptions). b, Structure of portimine related analogs: phenylportimine A (Ph-PA, 32) and epi-portimine A (ePA, 34), portimine A-diazirine-alkyne (PA-DA, 31-2) and epi-portimine A-diazirine-alkyne (ePA-DA, 31-1). c, Effects of PA and analogs on Jurkat cell viability (24 h; n = 3 biologically replicated experiments; mean ± s.d.). d, FACS analysis of apoptosis using annexin V/ eFluor 780 viability dye after PA and analogs treatment (12 h) in Jurkat cells (n = 3 biologically independent samples; mean ± s.d.). Statistical analysis was performed using one-way ANOVA with multiple comparisons. P-values are shown. e, Immunoblot of caspase-3 and PARP1 after PA and analogs (12 h) treatment in Jurkat cells (n = 3 biologically independent samples). f, PA induce G1-phase arrest. Jurkat cells were treated with portimine A or analogs for 12 h at 1 nM (n = 3 biologically independent samples for each condition). Statistical analysis was performed for G1 phase distribution using one-way ANOVA multiple comparisons. P-values are shown. g, PA displays minimal toxicity in freshly isolated human PBMCs (36 h) from three different donors (n = 3 biologically replicated experiments from each donor; mean ± s.d.). h-i, Impact of PA [0.3 or 1 mg kg−1 intraperitoneally] on h) MC38 or i) HT-1080 tumor growth in WT C57BL/6 mice (n = 10 mice per group). Values indicate mean ± SEM. Mice were euthanized when tumor area exceeded 2000 mm3. Statistical analysis was performed using one-way ANOVA analysis with multiple comparisons. P-values are shown for day35 (MC38) or day21 (HT-1080) by comparing vehicle and 1 mg kg−1 PA treated group. For uncropped western blot images, see Supplementary Fig. 2.
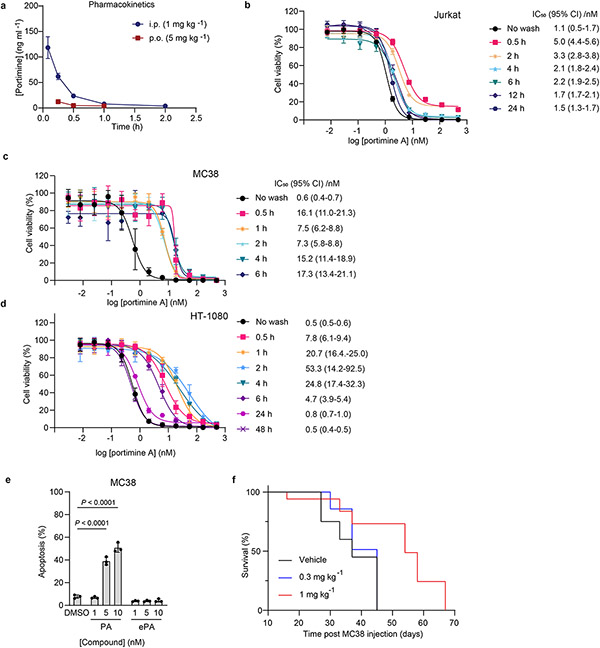
It had previously been reported that isolated PA (1) induces cellular apoptosis in human T cell leukemia and squamous cell carcinoma cell lines, however its molecular target is unknown3,10,4. In line with these previous reports, we observe that fully synthetic PA (1) and PA-DA (31-2) induce apoptosis at low nanomolar concentrations, as evidenced by increased caspase-3 activation, PARP cleavage, and annexin V staining in Jurkat cells, however controls ePA (34) and ePA-DA (31-1) do not (Fig. 3d-e). In addition, the pan-caspase inhibitor Z-VAD-FMK significantly rescued PA-mediated cell death (Extended Data Fig. 2a), consistent with PA effects on viability being caused, at least in part, by apoptosis. Further investigation also revealed that PA (1) and PA-DA (31-2) markedly increased the proportion of Jurkat cells in G1 phases (Fig. 3f and Extended Data Fig. 2b-e), but not in ePA (34) and ePA-DA (31-1) treated cells, indicating PA also induces cell cycle arrest. Notably, PA (1) does not induce apoptosis of isolated human peripheral blood mononuclear cells (PBMCs) and has minimal effects on PBMCs viability (Fig. 3g and Extended Data Fig. 3a-c), suggesting that its toxicity is selective towards rapidly proliferating cells. Considering this observed toxicity and selectivity, we next wondered whether PA (1) would be an effective agent for tumor growth suppression in vivo. Towards this end, preliminary pharmacokinetic evaluation of PA (1) in male C57BL/6J mice revealed a half-life of ~30 minutes, both by intraperitoneal (i.p., 1 mg kg−1) or oral (p.o., 5 mg kg−1) administration (Extended Data Fig. 4a and Supplementary Table 6). Fortunately, subsequent cell washout experiments in Jurkats, as well as in MC38 mouse colon carcinoma and human HT-1080 fibrosarcoma cells, indicated that minimal exposure time to PA (1) is required to maintain impressive potency, with as little as 30 minutes of exposure resulting in IC50 values of 5-16 nM (Extended Data Fig. 4b-d). PA (1) also potently induced apoptosis in MC38 cells, as observed in human cancer cell lines (Extended Data Fig. 4e). Treatment with PA (1) decreased tumor progression compared to vehicle at tolerated dose levels. Specifically, administration of 1 mg kg−1 portimine (i.p., q.d.) resulted in a significant reduction in tumor growth in the context of a syngeneic MC38 mouse colon carcinoma (Fig. 3h), or an HT-1080 human fibrosarcoma tumor xenograft model (Fig. 3i), while also prolonging survival (Extended Data Fig. 4f).
Portimine A targets NMD3
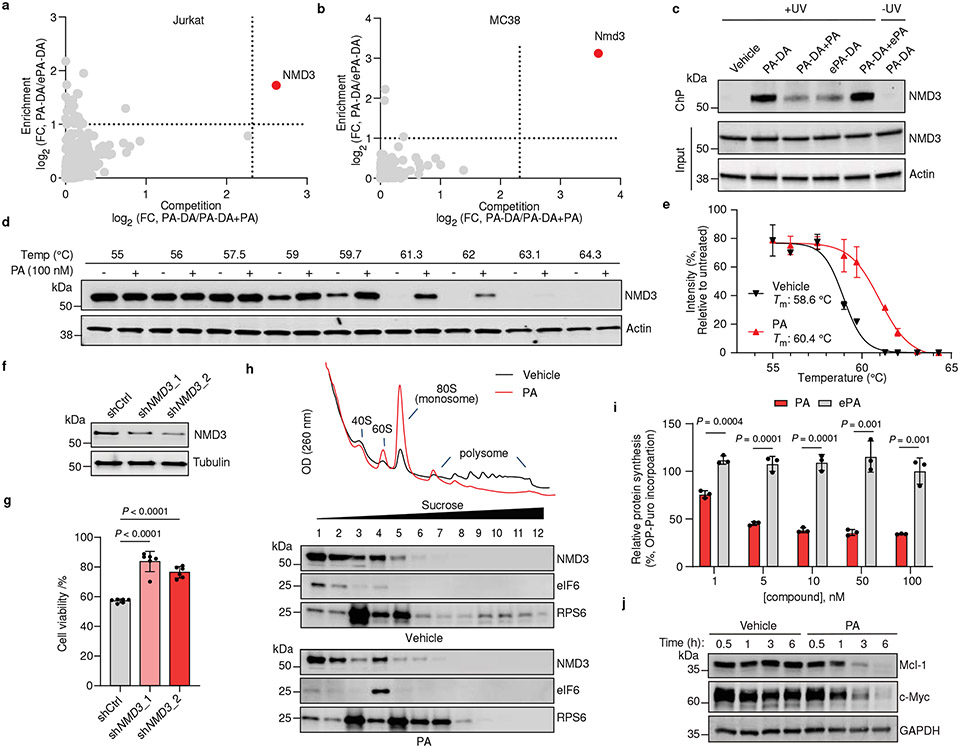
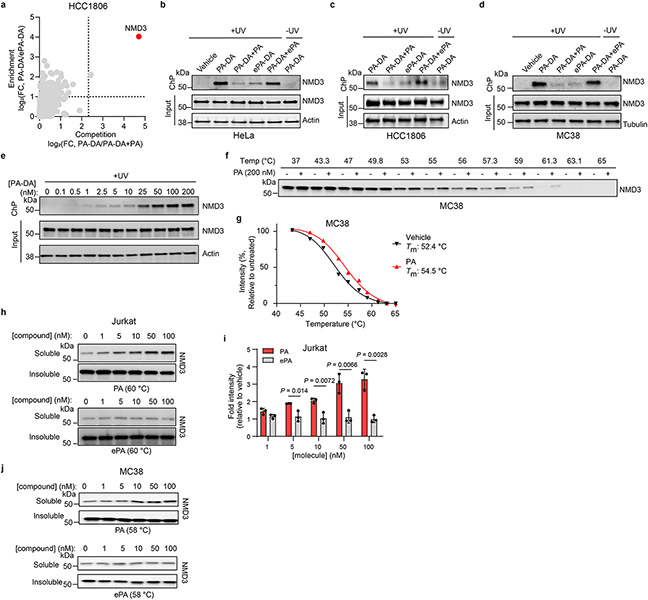
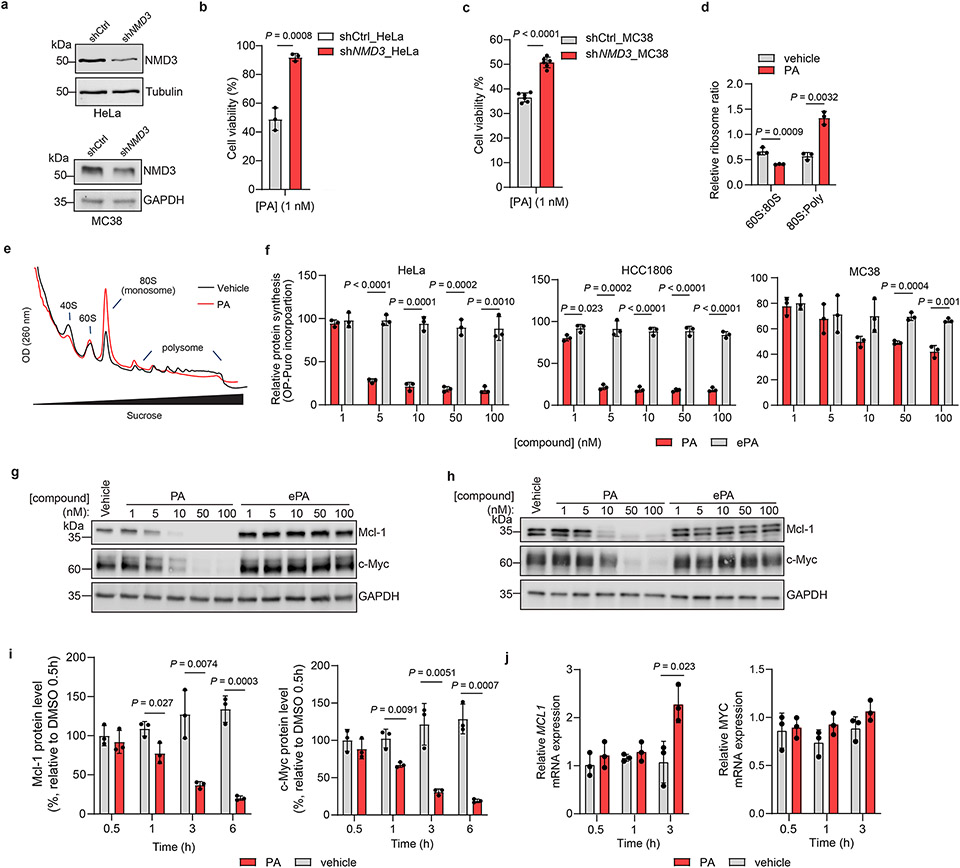
We next pursued identification of the protein targets of PA (1) in cells by tandem mass tag (TMT)-based proteomics32,34,35 in multiple cell lines where PA displays cytotoxicity, including human Jurkat and HCC1806 as well as mouse MC38 cells used in the above in vivo studies. In these experiments, we considered proteins that were substantially enriched (> 2-fold) by PA-DA over ePA-DA (500 nM) and competed (> 5-fold) upon coincubation with excess (8 ×) of PA (1) but not the inactive epimer ePA (34) (Fig. 4a-b and Extended Data Fig. 5a, Supplementary Table 7-9) as pharmacologically-relevant targets of PA (1). We identified only one protein, 60S ribosomal export protein NMD3 that satisfied these criteria in each cell line. We verified this interaction by chemoprecipitation (ChP) and immunoblotting, where labeling of endogenous NMD3 by PA-DA (31-2) is blocked by excess PA (1), but not with excess ePA (34) and little to no labeling occurs in the absence of UV irradiation or with ePA-DA (31-1) (Fig. 4c and Extended Data Fig. 5b-d). In addition, PA-DA labeling of endogenous NMD3 is dose-dependent and occurs at concentrations that mirror its biological activity (Extended Data Fig. 5e). To further confirm a direct interaction between PA (1) and NMD3, we assessed ligand-induced stabilization using a cellular thermal shift assay (CETSA)36. We observed a strong PA-induced stabilization of NMD3 in both human (Jurkat) and mouse (MC38) cell lines (Fig. 4d-e and Extended Data Fig. 5f-g). Additionally, isothermal dose-response fingerprinting (ITDRF) showed that PA stabilizes NMD3 in a concentration range that corresponds to its potent cytotoxic activity, while the inactive epimer ePA had no effect on NMD3 stability (Extended Data Fig. 5h-j). To assess NMD3 as a mechanistically relevant target of PA, we measured compound-induced cytotoxicity in cells in which NMD3 levels were reduced by shRNA. We observed that NMD3-deficient Jurkat, HeLa, and MC38 cells were significantly less sensitive to PA (1), compared to control cells (Fig. 4f-g and Extended Data Fig. 6a-c), confirming that NMD3 is necessary for PA activity.
Fig. 4. Portimine A targets NMD3, prevents polysome formation and inhibits protein translation.
Chemoproteomic profiling of PA in (a) Jurkat and (b) MC38 cells. X-axes correspond to PA-DA (500 nM) enriched proteins competed by PA (8 ×); y-axes correspond to proteins enriched by PA-DA over ePA-DA (500 nM). Designated PA-specific targets in red (competed by active competitor > 5-fold; enriched by PA-DA > 2-fold; and > 4-fold competition difference between PA and ePA). Dotted lines indicate competition (x-axis) and enrichment (y-axis) thresholds. Data presented as mean of biological replicates (n = 2). See Supplementary Tables 7-9 for source data and Methods for additional details. c, Immunoblot of NMD3 engagement by PA-DA (500 nM) co-treated with PA or ePA (8 ×) as well as ePA-DA (500 nM). Representative of three biologically independent samples. d, Immunoblot of NMD3 thermal aggregation (CETSA) in the presence of PA (100 nM). Results representative of two biological replicates. e, Quantitation of the thermal aggregation curves (n = 2 biological replicates; mean ± s.d.). f, Immunoblot of NMD3 in shNMD3 Jurkat cells. g, PA (1 nM, 24 h) has reduced viability effects in Jurkat cells transduced with shRNA targeting NMD3 (n = 6 biological replicates; mean ± s.d.). Statistical analysis performed using unpaired two-tailed Student t-test; P-values are shown. h, Polysome profiling of Jurkat cells treated with PA (50 nM, 6 h). Results representative of three independent experiments. i, Protein synthesis in Jurkat cells inhibited by PA, but not ePA (6 h), as measured by O-propargyl puromycin (OPP) incorporation. Data normalized to vehicle and presented as mean ± s.d. across biological replicates (n = 3). Statistical analysis performed using multiple unpaired two-tailed Student t-test; P-values are shown. j, Immunoblot of Mcl-1 and c-Myc in Jurkat cells treated with PA (10 nM). Results representative of three biological replicates. For uncropped western blot images, see Supplementary Fig. 2.
NMD3 is an adaptor of 60S ribosomal subunit nuclear export and is released upon pre-60S maturation in the cytosol and subsequent polysome formation, though the precise mechanism of this regulation is not fully elucidated37-39. As NMD3 is required for nascent 60S ribosome maturation37, we next examined whether PA (1) affects ribosome assembly. Here, we observed that in both Jurkat and MC38 cells treated with PA (1), but not in those treated with inactive ePA (34), leads to the accumulation of 80S monosomes and disomes as well as the blockade of polysome formation (Fig. 4h and Extended Data Fig. 6d-e). Further, we observed that eukaryotic translation initiation factor 6 (eIF6), an essential factor of 60S maturation and 80S assembly which is reported to be modulated by NMD340-41, is localized in the 60S fraction and decreased in RNA-free fractions in cells treated with PA (1) (Fig. 4h). Given that PA suspends polysome formation, we next assessed whether protein translation is affected42. Indeed, we observe a strong dose-dependent suppression of new protein synthesis in multiple cell lines (Jurkat, HeLa, HCC1806 and MC38) treated with PA, but not the inactive epimer ePA (Fig. 4i and Extended Data Fig. 6f). Translation inhibition is a mechanism shared by several cytotoxic natural products, such as the translation elongation inhibitor homoharingtonine (HHT, omacetaxine mepesuccinate), approved for the treatment of chronic myeloid leukemia (CML)43-44. It is proposed that selective effects of these molecules on cancer cell viability stems from blocking the synthesis of proteins with short half-lives critical for cell growth and maintenance, such as the proto-oncogenic protein c-Myc and anti-apoptotic protein Mcl-143-48. Indeed, we observe a rapid (~3 h), dose- and time-dependent decrease of c-Myc and Mcl-1 in Jurkat and MC38 cells treated with PA, but not ePA. Notably, PA does not impair MYC and MCL1 mRNA levels in this time frame, consistent with PA inhibiting the translation, and not transcription, of these proteins (Fig. 4j and Extended Data Fig. 6g-j).
Discussion
The scalable total synthesis of portimine A (1) and B (2) presented herein benefits from a strategy that is distinct from both biosynthesis and prior approaches. Driven by a desire to avoid an abundance of functional group manipulations, non-strategic redox fluctuations, and PGs, a plan was forged to forego the installation of oxygenation until a late stage. By analogy to two-phase terpenoid synthesis, this required the construction of a minimally decorated carbon skeleton followed by sequential oxidations. In this way, the innate reactivity of a macrocycle could be leveraged to install both the correct oxygenation pattern as well as the stereochemistry. Certain intermediates served to “self-protect” key functional groups and enable the final sequence by strategically timed ring-chain tautomerization events. Finally, an unusually stable vinyl triflate was carried through the majority of the synthesis allowing for flexible diversification of this biologically promising lead compound, enabling the construction of photoaffinity probes for target identification studies. With sizable quantities in hand, we have shown that PA (1) is highly cytotoxic in a panel of 20 cancer cells, but not in human PBMCs, and reduces tumor burden in vivo. Further, chemical proteomic studies revealed NMD3 to be a primary target of PA (1), and to our knowledge, is the first reported small molecule ligand of NMD3. Mechanistically, engagement of NMD3 by PA results in stabilization of 80S, subsequent impairment of polysome formation, and inhibition of protein synthesis, likely leading to cell cycle arrest and apoptosis. We observe that decreasing NMD3 expression reduces PA potency, consistent with a mechanism that requires a direct interaction between NMD3 to achieve functional inhibition. However, the precise molecular details of how binding of PA to NMD3 induces this cascade remain to be fully elucidated and will be subject of future studies. By analogy with other small molecule translation inhibitors, one possible explanation for the potent effects observed in cancer cells, but not PBMCs, could be the result of PA blocking the expression of short-lived proteins critical for proliferation and survival, such as c-Myc and Mcl-1. However, we recognize that PA effects on NMD3 and translation may impact multiple survival pathways and vary based on cell type and associated genetic lesions49. Additional studies should be directed to assess whether malignancies harboring dysregulated expression of such genes display higher vulnerability to PA and whether PA might be synergistic with targeted therapies to achieve higher selectivity and efficacy, as observed with other translation inhibitors50. Overall, this study demonstrates the utility of totally synthetic routes designed with ideality criterion combined with powerful chemical proteomic methods to reveal new potential therapeutic targets51.
Methods
Antibodies and reagents
Antibodies against NMD3 (16060-1-AP), eIF6 (10291-1-AP), and PARP-1 (66520-1-Ig) were purchased from Proteintech. Anti-RPS6 (2217), CASP3 (9662), Mcl-1 (5453), c-Myc (9402) were from Cell Signaling Technology. Rhodamine labeled anti-tubulin (12004165), GAPDH (12004168) and actin (12004164) hFAB were obtained from Bio-Rad.
Biotin-PEG3-Azide (AZ104), o-propargyl puromycin (1407), AZDye 488 Azide (1275), and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA; 1010) were from Click Chemistry Tools. Cycloheximide (C81040) was obtained from RPI Corporation. Sequencing Grade Modified Trypsin (V5111) and CellTiter-Glo Assay (G7570) were purchased from Promega. Pierce Streptavidin Agarose (20353), Pierce High pH Reversed-Phase Fractionation Kit (84868), TMT10plex™ Isobaric Label Reagent Set (90406), TMTpro™ 16plex Label Reagent Set (A44520), Halt Protease Inhibitor (78438), rNaseA (R1253), and RiboLock rNase Inhibitor (EO0381) were all obtained from Thermo Fisher Scientific.
shRNA MISSION RNA interference vectors targeting NMD3 were obtained through Sigma-Aldrich. The targeting sequences of human shNMD3 are as following: shNMD3_1 5’-GATGCTTGAAGACCTTCATAT-3’; shNMD3_2 5’-CTACTATCTTTGCCTCCAGAT-3’; sequence targeting mouse Nmd3: 5’-GATGCTTGAAGACCTTCATAT-3’. Control scramble shRNA was obtained from Addgene (#1864).
Mice
Wild type, immune competent C57BL/6 mice were purchased from The Scripps Research Institute DAR breeding colony. Athymic nude mice (Foxn1nu) were obtained from Jackson Laboratories (strain #: 002019). Animals were handled according to institutional and regulatory guidelines. Mice were maintained in 12 h light-dark cycles at 22 °C with 30-70% humidity and fed a standard irradiated rodent chow diet. Experiments were performed on male and female, seven-week-old mice. Treatment regimens were initiated at the time point corresponding to the formation of palpable tumors. All mice were purchased from The Scripps Research Institute DAR breeding colony or Jackson laboratories. Animals were handled according to institutional and regulatory guidelines, using protocols (number 22-0020-1) approved by the Scripps Research Institutional Animal Care and Use Committee (IACUC).
Cell culture conditions
Jurkat, HCC1806, and HeLa cells were purchased from ATCC. MC38 cells were kindly provided by Dr. Jeff Schlom at National Cancer Institute. Lenti-X™ 293T cells were purchased from Takara bio. Jurkat and HCC1806 cells were cultured in RPMI-1640 supplemented with 10% (v/v) FBS, penicillin-streptomycin and 2 mM L-glutamine. HeLa and Lenti-X™ 293T cells were cultured in DMEM (same supplements as above). MC38 cells were maintained in DMEM supplemented with 10% (v/v) FBS, glutaMAX and 1 × Antibiotic-Antimycotic (Gibco). RD, HT-1080, A673, MCF-7, MDA-MB-231, HepG2, B16-F10 cells were purchased from ATCC, and SUM159 were kindly provided by Laboratory of Dr. Sendurai Mani at Brown University. These cell lines were maintained in DMEM (Corning) supplemented with 10% FBS (Corning) and Anti-anti (Gibco). LNCaP, 786-O, 4T1 cells were obtained from ATCC and maintained in RPMI-1640 (Corning) supplemented with 10% FBS (Corning) and Anti-anti (Gibco). Before the screens, the cell lines were passaged at least twice after thawing. Cultures were confirmed to be free of mycoplasma infection using the MycoAlert Mycoplasma Detection Kit (Lonza). GBM CSCs (GBM-A and GBM-F) were cultured in Neurobasal-A medium (Gibco) supplemented with N2, B27, Glutamax, Anti-Anti (Life Tech), and human FGF (R&D, 20 ng mL−1) and human EGF (R&D, 20 ng mL−1). Tumor samples classified as GBM CSCs were acquired from patients undergoing surgery at the Stanford Medical Center in accordance with Institutional Review Boards at Stanford University and The Scripps Research Institute. Adherent monolayer cultures were maintained in culture dishes coated with poly-d lysine (Sigma, 2 μg mL−1), and laminin (Gibco, 2 μg mL−1). U87EGFRvIII cells were a generous gift from Dr. Paul Mischel (Stanford University) and established by stably expressing EGFRvIII in U87 cells purchased from ATCC and maintained in DMEM containing 10% FBS, as previously described52. LN229 and U118MG cells were purchased from ATCC and maintained in DMEM supplemented with 5% and 10% FBS, respectively. All cell lines were cultured at 37 °C in a humidified 5% CO2 atmosphere.
Lentivirus production and transduction
Lentiviral particles were produced by transfecting Lenti-X™ 293T cells with pMD2.G (Addgene, 12259), psPAX2 (Addgene, 12260), and pLKO.1-puro lentiviral vectors using PEI (Polysciences Inc). Growth medium was changed 24 h later, and viral supernatants were collected 48 and 72 h after transfection and filtered through a 0.2 μm PES membrane. Cells were transduced with viral supernatants and 8 μg ml−1 polybrene, and the viral supernatants were replaced 24 h after infection. Transduced cells were selected with 2 μg ml−1 puromycin for 4 days and then maintained in culture media containing 1 μg ml−1 puromycin.
Isolation of peripheral blood mononuclear cells (PBMCs)
All studies with non-diseased primary human cells were performed on samples obtained from Scripps Research’s Normal Blood Donor Service following protocols approved by The Scripps Research Institute Institutional Review Board (IRB). Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood using Lymphoprep™ (STEMCELL Technologies) gradient and washed twice with FACS buffer (PBS without calcium and magnesium, 2% FBS, and 1 mM EDTA).
Proliferation assay
For SAR experiments, Jurkat cells were plated at 50,000 cells per well in 96-well plates with media containing different concentrations of compounds in triplicate (0.2 pM – 1 μM) for 24 h. For caspase inhibitor rescue experiments, Jurkat cells were treated with media containing different concentrations of portimine A in triplicate (1, 5, 10 nM) with or without Z-VAD-FMK (50 μM, InvivoGen) in six replicates for 24 h. For cell screen experiments, cells were plated at 1,000 cells per well in 384-well plates with fresh media with different concentrations of portimine A in triplicate (8 pM – 500 nM) for 72 h. For Jurkat and PBMCs comparison experiments, cells were plated at 50,000 per well in 96-well plates with fresh media. Cells were then treated with media containing different concentrations of compounds in triplicate (0.2 pM – 1 μM) for 12 or 36 h. All compounds were used as 200 μM DMSO stocks, of which the DMSO content does not exceeded 1% of the total media. The same amount of DMSO was used as a vehicle control. For NMD3 knockdown rescue experiment, control and NMD3 knockdown cells were plated with fresh media without puromycin. Cells were treated with portimine A (1 nM) in biological replicates for 24 h (for Jurkat (n = 6) and HeLa (n = 3) cells) or 48 h (for MC38 cells (n = 6)). Cell proliferation was assayed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) following manufacturer’s guidelines. Data represents mean ± s.d..
Washout experiments
For Jurkat cells, cells were plated at 50,000 cells in 96-well plates. The cells were treated in triplicate with media containing different concentrations of portimine A (0.2 pM – 1 μM) for different periods of incubation time (0.5, 2, 4, 6, 12, 24 and 36 h). The media was removed by centrifugation at 800 × g for 3 min and fresh drug-free media was added. The cells were then allowed to recover until 36 h. MC38 and HT-1080 cells were treated with in triplicate with media containing different concentrations of portimine A (0.003 nM – 500 nM) for different periods of incubation time (0.5, 1, 2, 4, 6, and 72 h). The media was removed for each time point, and fresh drug-free growth media was added. The cells were then allowed to recover until 72 h. Cell viability was measured using CellTiter-Glo reagent. Data represents mean ± s.d.
Pharmacokinetics studies
Male C57BL/6J mice (n = 3 per group, weight 18 – 22 g and age: 6 – 10 weeks) were treated with a solution of portimine (10% DMSO, 10% Solutol, 80% water) at doses of 1 mg kg−1, and 5 mg kg−1 via intraperitoneal (i.p.) and orally (p.o.), respectively. Blood samples were collected at 0.083, 0.25, 0.5, 1, 2, 4, 8, and 24 h after i.p. administration, and at 0.25, 0.5, 1, 2, 4, 8, and 24 h after p.o. administration. All blood samples were transferred into pre-chilled commercial EDTA-K2 tubes and placed on wet ice until centrifugation. Blood samples were processed for plasma by centrifugation at approximately 4 °C, 3,200 × g for 10 min. Plasma was collected respectively and transferred into pre-labeled 96 well plate or polypropylene tubes, quick frozen over dry ice and kept at −60 °C or lower until LC-MS/MS analysis. Then the supernatant was directly injected into an LC-MS/MS-AK_Q-Trap 6500 system for analysis. The pharmacokinetics parameters were calculated by analyzing the compound concentration in plasma samples using the pharmacokinetic software Phoenix WinNonlin 6.3.
Mouse tumor xenograft studies
Seven-week-old female C57BL/6 mice were injected subcutaneously with 5 × 105 MC38 tumor cells and tumor size was measured every other day using a digital caliper. Tumor volume was estimated using the formula: tumor volume = length × width2 /2. On day 12 post tumor cell injection, mice were treated by intraperitoneal injection with portimine A (1 mg kg−1 or 0.3 mg kg−1, resuspended DMSO/Solutol/water 1:1:8, v/v/v) or vehicle (DMSO/Solutol/water 1:1:8, v/v/v) daily for 15 days. Mice were euthanized when tumor area exceeded 2000 mm3. HT-1080 xenograft; Athymic nude mice (Foxn1nu) were obtained from Jackson Laboratories (strain # 002019). Seven-week-old male mice were injected subcutaneously with 5 × 106 HT-1080 tumor cells resuspended in HBSS and tumor size was measured every other day using a digital caliper. Tumor volume was estimated using the formula: tumor volume = length × width2 /2. Beginning on day 8 post tumor cell injection, mice were treated by intraperitoneal injection with portimine A (1 mg kg−1 or 0.3 mg kg−1, resuspended in DMSO/Solutol/water 1:1:8, v/v/v) or vehicle (DMSO/Solutol/water 1:1:8, v/v/v) daily. Mice were euthanized when tumor area exceeded 2000 mm3.
Photoaffinity probe cell treatments
For HCC1806, HeLa and MC38 cells, 15-cm dishes of cells were grown to 80 – 95% confluency. The growth media was aspirated, and the cells were washed with Dulbecco’s phosphate buffered saline (DPBS). For Jurkat cells, cells were separated into 15-cm dishes (30 million per dish) with serum-free medium. The cells were incubated with serum-free medium containing portimine-based probes (PA-DA or ePA-DA, 500 nM) with competitors (PA or ePA, 4 μM) or DMSO for 30 min at 37 °C under a 5% CO2 atmosphere. Treated cells were then irradiated under UV light (365 nm, 20 min), harvested, washed with cold DPBS, and collected into tubes. The cell suspensions were collected and washed through centrifugation (400 × g, 5 min), the pellets were stored at −80 °C until processing. UVP Crosslinker from Analytik Jena is equipped with 5 × Hitachi F8T5 UV bulbs (emission maximum 365 nm).
Preparation of samples for chemoproteomic analysis
Chemoproteomic samples were prepared in biological duplicates for each condition following similar procedures as previously described40. In general, cell pellets were resuspended in 500 μl cold DPBS and lysed by sonication (Branson Sonifier probe, method setting: 15 milliseconds (msec) on, 40 msec off, 15% amplitude, 1 second total on). Protein concentrations were normalized (2 mg ml−1 in 500 μl with ice-cold DPBS) using a DC Protein Assay (Bio-Rad) with absorbance measured using the BMG Labtech CLARIOstar® Microplate Reader following manufacturer’s instructions. Followed by add the “click-chemistry cocktail” to each 500 μl lysate sample: tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 30 μl, 1.7 mM in DMSO/t-BuOH 1:4 v/v), tris(2-carboxyethyl)phosphine (TCEP, 10 μl, 50 mM), biotin-PEG3-azide (5 μl, 100 μM), and CuSO4 (10 μl, 50 mM) and samples were shaken at room temperature (RT) for 1 h. After click reaction, 2.5 ml of ice-cold 4:1 MeOH/CHCl3 solution and 1 ml ice-cold DPBS was added to each sample followed by centrifugation (3,200 × g, 4 °C, 10 min) to precipitate proteins. The precipitated protein was washed with 4:1 MeOH/CHCl3 two more times and resuspended in 6 M urea in DPBS, 500 μl; 10% SDS w/v, 10 μl. Protein reduction was then carried out by the addition of a freshly prepared 1:1 solution (50 μl total) of TCEP (25 μl, 200 mM in DPBS) and K2CO3 (25 μl, 600 mM in DPBS) and incubating the samples for 30 min at 37 °C. Following reduction, proteins were alkylated by the addition of freshly prepared iodoacetamide (70 μl, 400 mM in DPBS) and incubated for 30 min at RT in the dark. After alkylation, 130 μl of 10% SDS was added to samples, and then diluted with 5.5 ml DPBS. 100 μl of 50% slurry of streptavidin agarose slurry (Thermo Fisher Scientific, 20349) was added to each tube and rotated for 90 min at RT for enrichment of biotin-labeled proteins. The beads were pelleted by centrifugation (750 × g, 2 min, 4 °C) and subsequently washed once with 0.2% SDS in DPBS (5 min, 5 ml), twice with DPBS (5 ml), once with water (5 ml), and transferred to LoBind microcentrifuge tubes with 100 mM TEAB (1 ml, pH 8.5).
Streptavidin enriched proteins were digested overnight with trypsin in digestion buffer [100 ml, 100 mM TEAB (pH 8.5), 100 μM CaCl2 at 37 °C. The digest in each condition were taken out to new LoBind tubes, labeled with respective tandem-mass-tags (TMT; Thermo Fisher Scientific) 10plex or 16plex reagents (8 μl, 20 μg μl−1) for 1 h at 37 °C, quenched by hydroxylamine (6 μl, 5% v/v) for additional 15 min, acidified by formic acid (8 μl), and dried through SpeedVac vacuum concentrator (Thermo Fisher Scientific). The dried pellets were dissolved, combined, and proceeded to be fractionated into 12 fractions by Pierce high pH Reversed-Phase Fractionation Kit (Thermo Fisher Scientific, 84868) according to manufacturer’s instructions. The fractions were combined into 6 final fractions in a pairwise fashion, dried, and stored at −80 °C until ready for mass spectrometry analysis.
LC-MS analysis of TMT samples
TMT labeled peptides were resuspended in MS buffer (20 μl, 5% acetonitrile, 0.1 % formic acid in water) prior to LC-MS analysis as previously published methoeds40. In brief, 3 μl of each sample was loaded onto an Acclaim PepMap 100 precolumn (75 μm × 2 mm) and eluted on an Acclaim PepMap RSLC analytical column (75 μm × 15 cm) using an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Buffer A was (0.1% formic acid in H2O) and buffer B (0.1% formic acid in acetonitrile) were used in a 220 min gradient (0.3 ml min−1, 35 °C) of 2% buffer B for 10 min, followed by an incremental increase to 30% buffer B over 192 min, 60% buffer B for additional 5 min, 60 – 95% buffer B for 1 min, and 95% buffer B for 5 min, followed by descent to 2% buffer B for 1 min followed by re-equilibration at 2% buffer B for 6 min. The eluents were analyzed with a Thermo Fisher Scientific Orbitrap Fusion Lumos mass spectrometer with a cycle time of 3 sec and nano-LC electrospray ionization source applied voltage of 2,000 V, the data were collected using Xcalibur (v4.1.50). MS1 spectra were recorded at a scan range was specified from 375 to 1,500 m/z, maximum injection time of 50 msec (dynamic exclusion enabled, repeat count 1, duration 20 sec), the resolution is 120,000 with an automatic gain control (AGC) value of 1 × 106 ions. Peptides selected for MS2 analysis were isolated with the quadrupole (isolation window 1.6 m/z) and fragmented using collision-induced dissociation (CID) with 30 % collision energy, resultant fragments recorded in the ion trap (AGC 1.8 × 104, maximum inject time 120 msec). MS3 spectra were generated through high-energy collision-induced dissociation (HCD, 65% collision energy). Synchronous precursor selection (SPS) was employed to isolate up to 10 MS2 ions for MS3-based quantitation.
Gel-based analysis of endogenous NMD3 engagement in cells
Cells were treated as the “Photoaffinity probe cell treatments” section described. Probe-labeled cell pellets were resuspended in ice-cold DPBS supplemented and 1 ×protease inhibitor (Thermo Fisher Scientific, 78438) and lysed by sonication. Protein concentrations were normalized and click reaction were performed as described in “chemoproteomic preparation” section. In brief, after click reaction, ice-cold MeOH was added to each sample overnight at −20 °C to precipitate proteins. The samples were centrifuged and washed with 4:1 MeOH/CHCl3, resuspended by sonication and washed as above. Pellets were resuspended in freshly prepared 6 M urea solution with 1.7% SDS. Protein pellets were allowed to fully dissolve for 30 minutes at 37 °C. Biotin-abeled proteins were enriched by streptavidin-agarose beads and eluted with 2 × sample buffer with 50 mM biotin for 15 min. Samples were resolved by SDS-PAGE followed by immunoblotting analysis.
Thermo stability experiments
For cellular thermal shift assay (CETSA) and isothermal dose-response fingerprinting (ITDRF) experiments, cells were incubated with DMSO or indicated concentration of compound in fresh media (1.0 × 106 cell ml−1 for Jurkat or 80 % confluency in 15-cm2 plates for MC38 cells) for 30 min at 37 °C. Cells then were harvested, washed with binding buffer (cold DPBS containing DMSO or indicated concentration of compound and protease inhibitor cocktail) once. Resuspend in 1 ml of binding buffer, 50 μl of each aliquot was transferred to PCR tubes and heated at indicated temperatures on Bio-Rad C1000 Thermal Cycler for 3 min, followed by incubate for 3 min at 25 °C. After 3 snap freeze-thaw cycles with liquid nitrogen, lysates were then transferred to 1.5 ml Eppendorf tubes and centrifuged at 100,000 × g for 20 min at 4 °C. Supernatant was transferred to new Eppendorf tubes and add with 4 × SDS sample buffer, boiled for 15 min and submitted to immunoblotting. The remaining insoluble fraction was resuspended in 1 × SDS sample buffer, sonicated, and stored in −80 °C before immunoblotting.
Immunoblots
Cells were lysed by sonication and protein concentrations were normalized as described above. Lysates were then mixed with SDS sample buffer and resolved by SDS-PAGE. Proteins were transferred to PVDF membrane, blocked with 5 % milk in TBST, sequentially incubated with primary and secondary antibody, visualized with Bio-Rad ChemiDoc Imaging System and quantified with Bio-Rad Image Lab software (v6.1.0, Bio-Rad Laboratories, Inc.).
Flow cytometry
For cell cycle analysis, Jurkat cells were fixed in ice-cold 70 % ethanol for 30 min on ice. Fixed cells were then washed twice with PBS and treated with rNaseA (100 μg ml−1 in PBS, Thermo Fisher Scientific, R1253), and propidium iodide solution (50 μg ml−1 in PBS, Thermo Fisher Scientific, P3566) were added sequentially. Cells were stained for 15 minutes at room temperature and immediately analyzed by flow cytometry. For detecting cellular apoptosis, Annexin V and viability staining was performed based on the manufacturer’s instructions with slight modifications. In brief, Jurkat cells or PBMCs were collected and washed twice with cold PBS in 96-well plates, and stained with Fixable Viability Dye eFluor™ 780 (Thermo Fisher Scientific, 65-0865-14) on ice for 20 min. Cells were then washed once with 1% BSA in PBS, once with annexin V binding buffer (10 mM Hepes; pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2) and stained with Annexin V-FITC (Abcam, ab14085) at RT for 10 min in the dark. After staining, cells were washed twice with annexin V binding buffer, and fixed in 1% PFA in annexin V binding buffer for 30 min on ice. Cells were resuspended in binding buffer before analysis. Flow cytometry experiments were performed in the Scripps Research Flow Core using the NovoCyte Analyzers (ACEA Biosciences, Inc.). FACS data was analyzed using the software FlowJo 10.8.0 (BD Biosciences).
O-propargyl-puromycin (OPP) translation inhibition assay
For analysis of protein synthesis, O-propargyl-puromycin (OPP) incorporation assay was performed as previously described49. Briefly, cells were treated with escalating doses of compounds for 6 h (for Jurkat, HeLa, and HCC1806 cells) or 18 h (for MC38 cells) in 96-well plates, and then pulsed with OPP (5 mM final concentration) at 37 °C for 60 minutes. After incubation, cells were harvested, washed with ice-cold PBS, fixed with 1% PFA on ice for 30 min, and then permeabilized with BD perm/wash buffer (BD Biosciences, 554723) at RT for 10 min. Cells were washed with PBS once, and the click reaction mixture (2.5 mM THPTA, 1 mM CuSO4, 2 mM AZDye™ 488 Azide, 5.5 mM sodium ascorbate) were added to each sample to label the alkyne-tagged peptides with fluorescence azide at RT for 30 min. After reaction, the click solution was removed, and the cells were washed with BD perm/wash solution twice, resuspended in FACS buffer and analyzed by flow cytometry (for Jurkat cells) or plate reader (BMG Labtech CLARIOstar® Microplate Reader, for HeLa, HCC1806, and MC38 cells).
Sucrose gradient profiling
Jurkat or MC38 cells (2 × 107 per condition) were treated with DMSO or 50 nM compounds for 6 h. Cells were then treated with 100 μg ml−1 cycloheximide (RPI Corp., C81040) for 5 minutes, collected and washed twice with ice-cold PBS containing cycloheximide, and stored at −80 °C prior to lysis. Frozen cell pellets were thawed and lysed in 200 μl gradient buffer [15 mM Tris-HCl (pH 7.5), 300 mM NaCl, 15 mM MgCl2, 2 mM DTT, 100 μg ml−1 cycloheximide] supplemented with 1% Triton X-100, 0.5 U μl−1 RiboLock rNase Inhibitor (Thermo Fisher Scientific, EO0381), and 1 × protease inhibitor for 15 min on ice. Cells were then centrifuged at 16,000 × g for 7 min at 4 °C to remove the debris. The supernatants were loaded onto a 12 ml 5 – 50% continuous sucrose gradient in gradient buffer and centrifuged in a Beckman SW41 rotor at 38,000 rpm for 2 h at 4 °C. For ribosomal protein distribution analysis, approximately 24 fractions were collected from each sucrose gradient using the Brandel gradient fractionator with monitoring of optical density at 260 nm following instructions. Samples were merged into 12 fractions, mixed with sample buffer, and resolved by SDS-PAGE. Data were analyzed with Microsoft Excel®(v2122) and GraphPad Prism 9.3.0. Quantitation of ribosome ratios were performed by calculating the area below the curve for each fraction using ImageJ (v1.53r17, NIH).
Quantitative PCR
Total RNA for quantitative real-time PCR was extracted using iScript™ RT-qPCR Sample Preparation Reagent (BioRad, 1708898) according to the manufacturer’s protocol. Genomic DNA was removed and cDNA was generated using Maxima H Minus First Strand cDNA Synthesis Kit with dsDNase (K1682, Thermo Fisher Scientific), and RT-qPCR was performed with PowerUp™ SYBR™ Green Master Mix (A25742, Thermo Fisher Scientific) using QuantStudio™ 6 Pro Real-Time PCR System (Thermo Fisher Scientific). The following primers were used: MCL1-F 5’-TGCTTCGGAAACTGGACATCA-3’; MCL1-R 5’-TAGCCACAAAGGCACCAAAAG-3’; MYC-F 5’-CCTGGTGCTCCATGAGGAGAC-3’; MYC-R 5’-CAGACTCTGACCTTTTGCCAGG-3’; GAPDH-F 5’-GTCTCCTCTGACTTCAACAGCG-3’; GAPDH-R 5’-xACCACCCTGTTGCTGTAGCCAA-3’. Results were normalized to expression of GAPDH and 0.5h vehicle-treated group using the ΔΔCt method. Each sample was measured in technical duplicates and each condition was tested with three biological replicates.
Statistics and reproducibility
All immunoblots were performed with a minimum of two biological replicates. All statistical analysis was performed using Microsoft Excel® (v2122) and GraphPad Prism 9.3.0. Proteomic analysis was performed with Proteome Discoverer 2.4 (Thermo Fisher Scientific) and analyzed by previous published methods40. In brief, peptide sequences were identified through the SEQUEST HT algorithm. Fragment tolerances were set to 0.6 Da, and precursor mass tolerances were set to 10 ppm with one missed cleavage site allowed. Oxidation (M, +15.994915) was specified as variable modification while carbamidomethyl (C, +57.02146) and TMT-tag (K and N-terminal, +229.1629 for 10plex, +304.2071 for 16plex) were specified as static modifications. Spectra were searched against the Homo Sapiens proteome database (Uniprot, 2018; 42,358 sequences) or Mus musculus proteome database (Uniprot, 2021; 25,314 sequences) using 1 % false discovery rate (Percolator). MS3 peptides were quantified with a mass tolerance of 20 ppm. Abundances in each channel were normalized to the average of the median signal intensity across all channels. The reported proteins were required to have at least two unique peptides. TMT ratios obtained from Proteome Discoverer were transformed with log2(x), and statistics for calculated ratios were determined by two-tailed t-tests across two biological replicates (significant if P <0.05). Proteomics source data are provided in the Supplementary Tables 7-9.
Extended Data
Extended Data Fig. 1 ∣. Portimine A (PA) inhibits the growth of human and mouse cell lines in the low nanomolar range.
Concentration–response curves for PA and IC50 (95% IC) values in human (a-q) and mouse (r-t) cell lines: a) Jurkat (leukemia). b) RD (rhabdomyosarcoma). c) HT-1080 (fibrosarcoma). d) A673 (Ewings sarcoma). e) HCC1806 (acantholytic squamous cell carcinoma). f) HeLa (cervical adenocarcinoma). gx) MCF-7 (breast adenocarcinoma). h) MDA-MB-231 (breast adenocarcinoma). i) SUM159 (breast adenocarcinoma). j) HepG2 (hepatocellular carcinoma). k) LNCaP (prostate carcinoma). l) 786-O (renal adenocarcinoma). m) GBM-A (glioblastoma). n) GBM-F (glioblastoma). o) LN229 (glioblastoma). p) U87EGFRvIII (glioblastoma). q) U118 (glioblastoma). r) MC38 (colon carcinoma). s) B16-F10 (melanoma). t) 4T1 (mammary carcinoma). Cells were treated with PA at different concentrations for 72 h, followed by the CellTiter-Glo proliferation assay. Data represents mean ± s.d. of three biologically replicated experiments.
Extended Data Fig. 2 ∣. Portimine A triggers apoptosis and cell cycle arrest in Jurkat cells.
(a) PA induced-toxicity (24 h) can be rescued by caspase pan-inhibitor Z-VAD-FMK (50 μM) in Jurkat cells. Data are mean ± s.d. (n = 6 biologically independent samples). Statistical analysis performed using multiple unpaired two-tailed Student t-test; P-values are shown. (b-e) Cell cycle analysis in Jurkat cells. Jurkat cells were treated with 1 nM of indicated compounds for 12 h and propodium iodide used to identify different stages of cells by flow cytometry. Shown are G1 (b), S (c), G2 (d) and SubG1 (e) phase distributions. Data are mean ± s.d of three independent biologically replicated experiments. Statistical analysis was performed using one-way ANOVA analysis with multiple comparisons. P-values are shown.
Extended Data Fig. 3 ∣. Portimine A and analogs do not affect cell viability or induce apoptosis in freshly isolated human PBMCs.
(a) PA displays minimal viability effects on human PBMCs. ePA displays no toxicity to both Jurkat and PBMCs at the concentration range displayed. All presented data as the mean ± s.d. of biological replicated experiments (n = 3). (b) FACS analysis of apoptosis using annexin V/ eFluor 780 viability dye after PA and analogs (12 h) treatment in PBMCs. All data presented as mean ± s.d. of biological replicated experiments (n = 3). (c) Immunoblot of caspase-3 and PARP1 in PBMCs with indicated conditions (n = 3 biologically independent samples). For uncropped western blot images, see Supplementary Fig. 3.
Extended Data Fig. 4 ∣. Portimine A mouse pharmacokinetic properties and fast acting in vitro cell-based target engagement properties based on compound wash-out.
(a) Pharmacokinetic studies of mouse intraperitoneal (i.p.) and oral (p.o.) administration for portimine. Data presented as mean ± s.d. (n = 3 biologically independent samples). (b-d) Washout experiment performed in b) Jurkat c) MC38 and d) HT-1080 cells showed exposure-dependent decrease in IC50, reveals PA has a fast-acting cytotoxicity mechanism. Data presented as mean ± s.d. (n = 3 biologically independent samples). (e) FACS analysis of apoptosis using annexin V/ eFluor 780 viability dye after PA and analogs (24 h) treatment in MC38 cell line. Data presented as mean ± s.d. of biological replicated experiments (n = 3 biologically independent samples). Statistical analysis was performed using one-way ANOVA analysis with multiple comparisons test analysis; P-values are shown. (f) Kaplan-Myer survival curve of WT C57BL/6 MC38 tumor-bearing mice (n = 6 mice per group) after treatment with PA 0.3 or 1 mg kg−1 intraperitoneally.
Extended Data Fig. 5 ∣. Chemical proteomic analysis reveals NMD3 is the target of portimine A.
(a) Chemical proteomic profiling with PA-DA in HCC1806. X-axis corresponds to PA-DA (500 nM) enriched proteins competed by PA (PA, 8 ×); y-axis corresponds to proteins enriched by PA-DA over ePA-DA (500 nM). Designated PA-specific targets in red (competed by active competitor > 5-fold; enriched by PA-DA > 2-fold; and > 4-fold competition difference between PA and ePA). Dotted lines indicate competition (x-axis) and enrichment (y-axis) thresholds. Data presented as mean of biological replicates (n = 2). See Supplementary Tables 7-9 for source data. (b-d) Immunoblot of NMD3 engagement by PA-DA (500 nM) co-treated with PA or ePA (8 ×) as well as by ePA-DA (500 nM) in HeLa (b), HCC1806 (c), and MC38 cells (d). (n = 2 biologically independent samples). (e) NMD3 is engaged by PA-DA in a dose-dependent manner in Jurkat cells. Results are representative of three independent experiments. (f-i) CETSA validation of NMD3 as a target of PA in Jurkat and MC38 cells. (f-g) Immunoblotting and quantitation of NMD3 thermal aggregation curves (mean ± s.d.) in MC38 cells treated with PA. (n = 3 biologically independent samples). (h-i) Dose-response (ITDR) fingerprint of NMD3 stabilization by PA in Jurkat cells and corresponding quantitation. Data presented as mean ± s.d. of biological replicated experiments (n = 3). Statistical analysis performed using multiple unpaired two-tailed Student t-test; P-values are shown. (j) Isothermal dose-response (ITDR) fingerprint of NMD3 stabilization by PA in MC38 cells (n = 3 biologically independent samples). For uncropped western blot images, see Supplementary Fig. 3.
Extended Data Fig. 6 ∣. Portimine A activity dependent on NMD3 and impairs polysome formation.
(a) Immunoblot of NMD3 in HeLa and MC38 cells expressing control or NMD3-specific shRNAs. (b-c) PA has reduced viability effects in Jurkat cells transduced with shRNA targeting NMD3 compared to shCtrl cells in HeLa (b, 1 nM, 24 h) and MC38 (c, 1 nM, 48 h). Data presented as mean ± s.d. of biologically replicated experiments (n = 3 for HeLa, n = 6 for MC38). Statistical analysis was performed using unpaired two-tailed Student t-test, P-values are shown. (d) Quantitation of 60S:80S ratio and 80S:polysome ratio from data shown in Fig. 4f. Presented as mean ± s.d. of biological replicates (n = 3). Statistical analysis performed using unpaired two-tailed Student t-test; P-values are shown. (e) Polysome profiling of MC38 cells treated with PA (50 nM, 6 h). Results are representative of two independent experiments. (f) PA, but not ePA, inhibits new protein synthesis in HeLa, HCC1806, and MC38 cells as determined by O-propargyl puromycin (OPP) incorporation. Results normalized to vehicle and presented as the mean ± s.d. across biologically replicated experiments (n = 3). Statistical analysis was performed using multiple unpaired two-tailed Student t-test. P-values are shown. (g, h) Immunoblot of Mcl-1 and c-Myc after PA and ePA treatment in g) Jurkat and h) MC38 cells. Results are representative of two independent experiments. (i) Quantitation of protein abundances from Fig. 4j. Results presented as mean ± s.d. of biologically replicates (n = 3). Statistical analysis was performed using multiple unpaired two-tailed Student t-test; P-values are shown. (j) MCL1 and MYC mRNA expression assessed by quantitative PCR in Jurkat cells treated with PA (10 nM). Results normalized to vehicle and values indicate mean ± s.d. (n = 3 biologically independent samples). Statistical analysis performed using multiple unpaired two-tailed Student t-test. P-values are shown. For uncropped western blot images, see Supplementary Fig. 3.
Supplementary Material
Acknowledgments
We thank Prof. J. Williamson and Dr. A. Popova (Scripps Research) for their invaluable discussions and generous technical assistance with polysome profiling experiments. We thank Prof. J. Teijaro and D. Lazar for their assistance with PBMC isolation. We are grateful to Dr. D.-H. Huang and Dr. L. Pasternack (Scripps Research) for NMR spectroscopic assistance, Ms. B. Sanchez, Ms. Q. N. Wong, and CoreService Team (Scripps Research) for analytical support. We thank Dr. Eduardo Esquenazi, Tamara Schwent, and Dr. Paige Stout (Sirenas Marine Discovery) for first alerting us to the structure and bioactivity of portimine A, as well as providing NMR spectra of the authentic sample. We thank Prof. A. Fürstner and Dr. J. Hillenbrand (Max-Planck-Institut für Kohlenforschung) for providing RCAM catalysts. Financial support for this work was provided by NIGMS (GM118176).
Footnotes
Competing interests
Scripps Research filed a U.S. patent application on 17 January 2023 covering the chemical structure described in this Article and their use. P.S.B., C.G.P., L.L.L., T. J., W. L., F.M.P. and T.C. are listed as inventors on this patent. The other authors declare no competing financial interest.
Data Availability
The data that support the findings in this work are available within the paper and Supplementary Information (Supplementary text and Extended Data Figs. 1-6, Supplementary Tables 1-9). Uncropped, full western blot images and gels are provided in Supplementary Figure 2-3. All raw proteomics data files have been deposited to the PRIDE53 repository and available under the accession PXD041911 (https://www.ebi.ac.uk/pride/archive/projects/PXD041911). Source data are provided with this paper.
References
- 1.Stivala CE et al. Synthesis and biology of cyclic imine toxins, an emerging class of potent, globally distributed marine toxins. Nat. Prod. Rep 32, 411–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molgo J. et al. Cyclic imine toxins from dinoflagellates: a growing family of potent antagonists of the nicotinic acetylcholine receptors. J. Neurochem 142, 41–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selwood AI et al. Portimine: a bioactive metabolite from the benthic dinoflagellate Vulcanodinium rugosum. Tetrahedron Lett. 54, 4705–4707 (2013). [Google Scholar]
- 4.Cuddihy SL et al. The marine cytotoxin portimine is a potent and selective inducer of apoptosis. Apoptosis 21, 1447–1452 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Jørgensen L. et al. 14-Step Synthesis of (+)-Ingenol from (+)-3-Carene. Science 341, 878–882 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Kanda Y. et al. Two-Phase Synthesis of Taxol. J. Am. Chem. Soc 142, 10526–10533 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munday R, Selwood AI & Rhodes L Acute toxicity of pinnatoxins E, F and G to mice. Toxicon 60, 995–999 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Munday R. et al. Investigations into the toxicology of spirolides, a group of marine phycotoxins. Toxins 4, 1–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munday R. et al. Acute toxicity of gymnodimine to mice. Toxicon 44, 173–178 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Fribley AM et al. Identification of Portimine B, a New Cell Permeable Spiroimine That Induces Apoptosis in Oral Squamous Cell Carcinoma. ACS Med Chem. Lett 10, 175–179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermawan I. et al. Kabirimine, a New Cyclic Imine from an Okinawan Dinoflagellate. Mar. Drugs 17, 353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacKinnon SL et al. Biosynthesis of 13-desmethyl spirolide C by the dinoflagellate Alexandrium ostenfeldii. J. Org. Chem 71, 8724–8731 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kellmann R, Stuken A, Orr RJS, Svendsen HM & Jakobsen KS Biosynthesis and Molecular Genetics of Polyketides in Marine Dinoflagellates. Mar. Drugs 8, 1011–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Wagoner RM, Satake M & Wright JL Polyketide biosynthesis in dinoflagellates: what makes it different? Nat. Prod. Rep 31, 1101–1137 (2014). [DOI] [PubMed] [Google Scholar]
- 15.McCauley JA et al. Total synthesis of pinnatoxin A. J. Am. Chem. Soc 120, 7647–7648 (1998). [Google Scholar]
- 16.Stivala CE & Zakarian A Total synthesis of (+)-pinnatoxin A. J. Am. Chem. Soc 130, 3774–3776 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Nakamura S, Kikitchi F & Hashimoto S Total synthesis of pinnatoxin A. Angew. Chem. Int. Ed 47, 7091–7094 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Araoz R. et al. Total Synthesis of Pinnatoxins A and G and Revision of the Mode of Action of Pinnatoxin A. J. Am. Chem. Soc 133, 10499–10511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong K, Moussa Z, Lee C & Romo D Total Synthesis of the Spirocyclic Imine Marine Toxin (−)-Gymnodimine and an Unnatural C4-Epimer. J. Am. Chem. Soc 133, 19844–19856 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T, Fujiwara K, Kondo Y, Akiba U & Suzuki T Synthesis of the cyclohexene segment of portimine. Tetrahedron Lett. 60, 386–389 (2019). [Google Scholar]
- 21.Aitken HRM, Brimble MA & Furkert DP A Catalytic Asymmetric Ene Reaction for Direct Preparation of alpha-Hydroxy 1,4-Diketones as Intermediates in Natural Product Synthesis. Synlett. 31, 687–690 (2020). [Google Scholar]
- 22.Ding X-B, Aitken HRM, Pearl ES, Furkert DP & Brimble MA Synthesis of the C4-C16 Polyketide Fragment of Portimines A and B. J. Org. Chem 86, 12840–12850 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Li L, El Khoury A, Clement BO, Wu C & Harran PG Asymmetric Organocatalysis Enables Rapid Assembly of Portimine Precursor Chains. Org. Lett 24, 2607–2612 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Fürstner A. Alkyne Metathesis on the Rise. Angew. Chem. Int. Ed 52, 2794–2819 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Iwama T & Rawal VH Design and development of highly effective Lewis acid catalysts for enantioselective Diels-Alder reactions. J. Am. Chem. Soc 124, 5950–5951 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Corey EJ & Beames DJ Mixed Cuprate Reagents of Type R1R2CuLi Which Allow Selective Group Transfer. J. Am. Chem. Soc 94, 7210–7211 (1972). [Google Scholar]
- 27.Hillenbrand J. et al. “"Canopy Catalyst”" for Alkyne Metathesis: Molybdenum Alkylidyne Complexes with a Tripodal Ligand Framework. J. Am. Chem. Soc 142, 11279–11294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S. et al. Ruthenium-Catalyzed Oxidation of Alkenes at Room Temperature: A Practical and Concise Approach to alpha-Diketones. Org. Lett 13, 2274–2277 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Cummins CH & Coates RM Alpha-Oxygenation of Aldehydes and Cyclic-Ketones by Acylation Rearrangement of Nitrones. J. Org. Chem 48, 2070–2076 (1983). [Google Scholar]
- 30.Wurdak H. et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell 6, 37–47 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Li Z. et al. Design and Synthesis of Minimalist Terminal Alkyne-Containing Diazirine Photo-Crosslinkers and Their Incorporation into Kinase Inhibitors for Cell- and Tissue-Based Proteome Profiling. Angew. Chem. Int. Ed 52, 8551–8556 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Parker CG et al. Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell 168, 527–541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker CG et al. Chemical Proteomics Identifies SLC25A20 as a Functional Target of the Ingenol Class of Actinic Keratosis Drugs. ACS Cent. Sci 3, 1276–1285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway LP et al. Evaluation of fully-functionalized diazirine tags for chemical proteomic applications. Chem. Sci 12, 7839–7847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlister GC et al. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal. Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez Molina D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Sengupta J et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell. Biol 189, 1079–1086 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho JH-N, Kallstrom G & Johnson AW Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell. Biol 151, 1057–1066 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trotta CR, Lund E, Kahan L, Johnson AW & Dahlberg JE Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. Embo J. 22, 2841–2851 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceci M. et al. Release of eIF6 (p27(BBP)) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Malyutin AG, Musalgaonkar S, Patchett S, Frank J & Johnson AW Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. Embo J. 36, 854–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koga Y. et al. Discovery of C13-aminobenzoyl cycloheximide derivatives that potently inhibit translation elongation. J. Am. Chem. Soc 143, 13473–13477 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang R. et al. Semisynthetic homoharringtonine induces apoptosis via inhibition of protein synthesis and triggers rapid myeloid cell leukemia-1 down-regulation in myeloid leukemia cells. Mol. Cancer Ther 5, 723–731 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Chen R. et al. Homoharringtonine reduced Mcl-1 expression and induced apoptosis in chronic lymphocytic leukemia. Blood 117, 156–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X. et al. Targeting translation initiation by synthetic rocaglates for treating MYC-driven lymphomas. Leukemia 34, 138–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider-Poetsch T. et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol 6, 209–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manier S. et al. Inhibiting the oncogenic translation program is an effective therapeutic strategy in multiple myeloma. Sci. Transl. Med 9, eaal2668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alinari L. et al. Dual Targeting of the Cyclin/Rb/E2F and Mitochondrial Pathways in Mantle Cell Lymphoma with the Translation Inhibitor Silvestrol. Clin. Cancer Res 18, 4600–4611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindqvist LM et al. Translation inhibitors induce cell death by multiple mechanisms and Mcl-1 reduction is only a minor contributor. Cell Death Dis. 3, e409–e409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen R, Gandhi V & Plunkett W A Sequential Blockade Strategy for the Design of Combination Therapies to Overcome Oncogene Addiction in Chronic Myelogenous Leukemia. Cancer Res. 66, 10959–10966 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Peters DS et al. Ideality in Context: Motivations for Total Synthesis. Acc. Chem. Res 54, 605–617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang MY et al. Mammalian Target of Rapamycin Inhibition Promotes Response to Epidermal Growth Factor Receptor Kinase Inhibitors in PTEN-Deficient and PTEN-Intact Glioblastoma Cells. Cancer Research 66, 7864–7869 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Perez-Riverol Y et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, 543–552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings in this work are available within the paper and Supplementary Information (Supplementary text and Extended Data Figs. 1-6, Supplementary Tables 1-9). Uncropped, full western blot images and gels are provided in Supplementary Figure 2-3. All raw proteomics data files have been deposited to the PRIDE53 repository and available under the accession PXD041911 (https://www.ebi.ac.uk/pride/archive/projects/PXD041911). Source data are provided with this paper.