Abstract
Many biomolecular condensates appear to form via spontaneous or driven processes that have the hallmarks of intracellular phase transitions. This suggests that a common underlying physical framework might govern the formation of functionally and compositionally unrelated biomolecular condensates. Here, we summarize recent work that leverages a stickers-and-spacers framework adapted from the field of associative polymers for understanding how multivalent protein and RNA molecules drive phase transitions that give rise to biomolecular condensates. We provide an overview of the model and its connections to known drivers of biomolecular condensates. We discuss how the valence of stickers impacts the driving forces for condensate formation and elaborate on how stickers can be distinguished from spacers in different contexts. We touch upon the impact of sticker- and spacer-mediated interactions on the rheological properties of condensates and show how the model can be mapped to known drivers of different types of biomolecular condensates.
Keywords: Phase separation, phase transition, biomolecular condensates, stickers-and-spacers
INTRODUCTION
Spatial and temporal organization of cellular matter determines the regulation and control of key cellular processes that lead to nontrivial outcomes such as cell division, differentiation, adhesion, motility, stress response, metabolic control, and cell death (1-7). Cellular matter can be organized into membrane-bound or membraneless organelles. The latter are biomolecular condensates, which are defined as concentrated non-stoichiometric assemblies of biomolecules (8), that can form via spontaneous or driven processes sharing many of the hallmarks of phase transitions (9; 10).
In 1995 Walter and Brooks proposed that “microcompartmentalization”, which refers to the spatial organization of cellular matter, might arise due to phase separation mediated by macromolecular crowding in the cytoplasm (11). A generalization of this idea reemerged following the work of Brangwynne et al. who showed that P-granules in germ cells form via phase separation (12). Since then, there has been a surge of interest in the phenomenon of liquid-liquid phase separation (LLPS) whereby biomolecular condensates form by spontaneous or driven phase separation of macromolecular components from their liquid-like environments into distinct condensates with liquid-like properties (Figure 1) (10; 13-131). The resulting condensates, enriched in specific macromolecules, coexist with the surrounding milieu, which is relatively deficient in the macromolecules of interest. Indeed, there is growing consensus that many, if not all, membraneless biomolecular condensates form via some combination of spontaneous or driven phase separation and percolation (10; 132). Specific types of protein and RNA molecules drive intracellular phase transitions and a defining characteristic of these molecules is the multivalence of interaction domains or motifs (8).
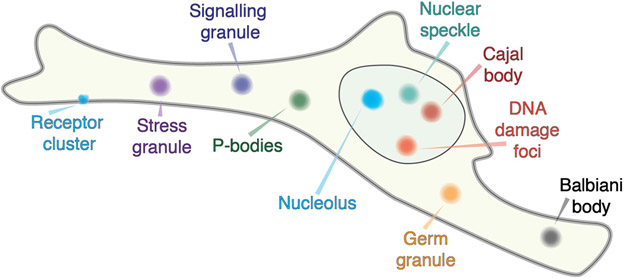
Figure 1: Overview of cellular bodies that are well-described as condensates.
These bodies include large well-studied structures such as the nucleolus, nuclear speckles, and P-bodies, but also smaller assemblies including those such as signaling granules, receptor clusters and DNA damage foci. The size of assemblies in this schematic are not to scale.
Phase separation, especially LLPS, is described using an assortment of analogies to observations made in everyday life. In a two-component system comprising of liquids such as oil and water, phase separation is a demixing process whereby two mutually immiscible liquids form two distinct coexisting phases. Alternatively, a mixture comprising of a water-soluble polymer in an aqueous solvent can separate into a dense polymer-rich phase that coexists with a dilute, polymer-deficient phase. In two-component systems comprising of polymer and solvent we shall use ϕp and ϕs to denote the volume fractions of polymer and solvent, respectively. If the system is closed, ϕp + ϕs = 1, and it follows that ϕs = (1 − ϕp); accordingly, if we set ϕp to be ϕ, then the volume fraction of the polymer becomes the order parameter for describing phase separation. Because the system is closed, ϕ is referred to as a conserved order parameter.
In a binary mixture comprising of a polymer and poor solvent there exists a system-specific concentration threshold designated as the saturation concentration or ϕsat (22; 133; 134) beyond which the system separates into a dense polymer-rich phase that coexists with a dilute phase. The volume fractions of the polymer in the coexisting dense and dilute phases are designated as ϕdilute and ϕdense, respectively, where ϕdilute = ϕsat. For ϕdilute < ϕ < ϕdense, the numbers of the polymer molecules in the two phases are determined by the so-called lever rule: ndilute = ntotal (ϕdense − ϕ)/(ϕdense − ϕdilute) and ndense = ntotal (ϕ − ϕdilute)/(ϕdense − ϕdilute), where ndilute and ndense are respectively the numbers of polymer molecules in the dilute and dense phases, and ntotal is the total number of polymer molecules: ntotal = ndilute + ndense (135).
In mean-field theories for homopolymer solutions, the length (N) of the polymer and the magnitude of the Flory interaction parameter χps, which is positive in a poor solvent (136), will determine the values of ϕdense and ϕdilute. The parameter χps is defined as:
| (1) |
In equation (1), the terms uxy refer to mean-field energies for interactions between species x and y; the subscripts p and s refer to the polymer and solvent, respectively. The algebraic sum of energies is made dimensionless by normalization using the parameter kBT, which quantifies the thermal energy at temperature T; z is a coordination number that represents the average number of nearest-neigbor interactions that each monomeric unit within the polymer can make.
In a good solvent, χps is negative, implying that polymer-solvent interactions are favored over polymer-polymer interactions. As a result, a homogeneous, one-phase mixture is preferred, irrespective of the value of ϕ. Conversely, χps is positive in a poor solvent reflecting the fact that polymer-polymer interactions are favored over polymer-solvent interactions. In a poor solvent, there exists a χps-dependent threshold value of ϕ beyond which the system separates into two coexisting phases; this threshold value is designated as ϕsat. In a theta solvent, also known as an indifferent solvent, χps = 0 and the polymer-solvent, polymer-polymer, and solvent-solvent interactions are perfectly counterbalanced. Accordingly, the entropy of mixing is the only relevant term and this favors the formation of an ideal, one-phase mixture in a theta solvent.
For closed multicomponent systems that comprise of n types of homopolymers plus a solvent, the relevant conserved order parameter is a vector denoted as [ϕ1, ϕ2…ϕn]; here ϕi denotes the volume fraction of the polymer of type i. In a closed system, the volume fraction of the solvent is readily calculated using: . For fixed temperature and pressure, the Gibbs phase rule prescribes that there can be a maximum of n+1 coexisting phases. The determinants of the phase behavior of mixtures comprising of multiple types of homopolymers plus a solvent are components of the vector where each χij is defined as in equation (1); the numeric subscripts correspond to the identities of polymers and the subscript s denotes the solvent. The main upshot is that for aqueous mixtures of homopolymers, the mapping of phase boundaries will require knowledge of the components of the vector either through numerical calculations or suitable measurements (137).
The preceding narrative is inspired by the influential theories of Flory and Huggins (135) and it relies on a purely mean-field description for homopolymers. The vector contains all of the information that is relevant for describing the driving forces for phase separation. It is a measure of solvent quality and the mutual (in)compatibilities of polymers with one another. The compositions of dense phases and the interfacial tensions between pairs of coexisting phases are determined directly by the values of the components in the vector . For homopolymers, all interactions are equivalent and can be used to capture the interplay of polymer and solvent interactions to describe phase behavior.
Going beyond homopolymers:
Homopolymers are poor approximations of most protein and RNA molecules that are drivers of intracellular phase transitions. Protein and RNA molecules are finite-sized heteropolymers of precise molecular weights that comprise of structured domains / motifs, intrinsically disordered regions (IDRs), or some combination of the two. The vector does not capture either the sequence and structural heterogeneities or the hierarchy of anisotropic interactions encoded by the multi-way interplay among heteropolymers and the solvent. Therefore, motivated by modern developments in polymer theories, we propose that protein / RNA molecules that drive intracellular phase transitions are in fact biological instantiations of associative polymers, which were defined by Rubinstein and Dobrynin to be “macromolecules with attractive groups” (138). The attractive groups are distributed across the polymer. Further, the interactions involving these groups can be anisotropic and these interactions include ionic bonds, hydrogen bonds, and interactions mediated by solvents known as gluonic and regulatory solvents (139).
In much the same way that Flory-Huggins theory provides a general framework for describing homopolymeric systems, associative polymers can be described using a stickers-and-spacers model. The groups that participate in attractive interactions are considered as stickers and the parts of the chain that are interspersed between stickers but do not significantly drive attractive interactions are considered as spacers. Non-covalent interactions between stickers within and from different chains will lead to the formation of reversible physical crosslinks (138). Although spacers are not directly involved in these crosslinks, they can have a profound impact on the assembly of associative polymers.
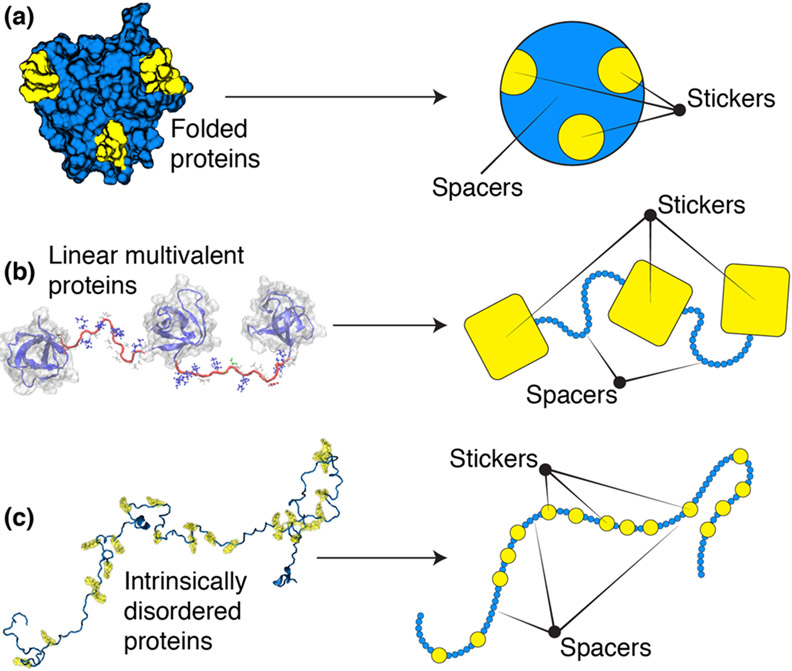
Multivalent protein / RNA molecules may either be branched or linear associative polymers. Therefore, an important question pertains to the molecular identities of stickers in applying the stickers-and-spacers model to biopolymers. In IDRs, stickers are likely to be Short Linear Motifs (SLiMs) that are 1-10 residues in length while spacers are the intervening residues in the IDR (140). Analogously, in unfolded RNA molecules, stickers may be short sequence motifs or even individual nucleotides. Stickers in folded protein domains or structured regions of RNA are surface patches or motifs that emerge from the formation of specific structures. Accordingly, non-sticker regions on the surfaces of folded domains of proteins and disordered loop regions can be considered as spacers. In linear multivalent systems, stickers may be folded binding domains while spacers are the flexible disordered linkers that connect them together. For branched multivalent proteins, disordered regions give these systems a hairy colloidal architecture and the disordered regions, sans the SLiMs, may be thought of as spacers. Figure 2 shows a schematic of different types of multivalent protein and RNA molecules that are mapped onto sticker-and-spacer architectures.
Figure 2: Schematic of different types of stickers and spacers for different systems.
(a) For folded proteins we can map stickers and spacers to the patchy colloid formalism. (b) For linear multivalent proteins we can broadly map stickers as folded binding domains while spacers are flexible linkers that connected domains (c) For intrinsically disordered proteins stickers may be single residues, short linear motifs, or some combination of both.
Importantly, the stickers-and-spacers model does not have restrictions on either the identity of or the resolution at which stickers are defined. Accordingly, the stickers-and-spacers model offers an intuitive and highly generalizable approach for quantitative descriptions of complex biological systems. In the following sections we discuss key details of the stickers-and-spacers formalism for obtaining a thermodynamic description of the phase behavior of associative polymers in solution. We summarize predictions that can be made using this model, cite numerical instantiations of the model, and highlight applications to specific protein and RNA systems.
MEAN-FIELD INCARNATION OF THE STICKERS-AND-SPACERS MODEL
One possible quantitative realization of the stickers-and-spacers framework is an analytical mean-field model for associative polymers. This model rests on the simplifying assumption that the conformational preferences of individual associative polymers are similar in their dense versus dilute phases. This simplification allows us to ignore the possibility that conformational changes might lead to changes in the valence and identities of stickers.
We shall consider a two-component system comprising of associative polymers with multiple interacting stickers interspersed by non-interacting phantom spacers. The model for spacers assumes that the interactions involving spacers (i.e., sticker-spacer, spacer-spacer, and spacer-solvent) counterbalance one another thus making the spacer regions behave like an ideal chain. Physical crosslinks between stickers enable two types of transitions. Aided by the nature of spacers, the physical crosslinks among stickers can lead to a density transition (which is phase separation) whereby above a threshold concentration denoted as csat, the associative polymers in a binary mixture comprising of the polymers and solvent will form a dense phase defined by physically crosslinked stickers that coexists with a dilute phase comprising of minimal inter-sticker crosslinks. The concentrations of associative polymers or stickers in the coexisting dilute and dense phases are denoted respectively as cdilute = csat and cdense. Associative polymers also undergo a networking transition known as percolation. This refers to the topological connectivity among stickers that is engendered by physical crosslinks. Above a concentration threshold cperc known as the gel point or percolation threshold, the system of associative polymers can form a system-spanning network. Phase separation leads to percolation if csat ≤ cperc < cdense. Conversely, phase separation and percolation become decoupled from one another if cdense < cperc (phase separation without percolation) or cperc < csat (percolation without phase separation). For the purpose of developing the mean-field theory, we shall assume that we are operating in the regime where csat ≤ cperc < cdense.
For a system comprising of associative polymers in a solvent, each with n self-interacting stickers (n >> 1), Semenov and Rubinstein (141) showed that the percolation threshold for a system in which stickers are described as phantom chains is estimated as:
| (2) |
Here, n is the apparent valence (number) of stickers and where vb is the volume associated with each inter-sticker crosslink, ε is the effective interaction energy between stickers (ε ≤ 0), kB is the Boltzmann constant, and T is the system temperature. We shall describe the origins of the relationship between cperc and the valence of stickers by summarizing the extension of the theory of Semenov and Rubinstein (141) to the case of an associative polymer that consists of two types of stickers A and B.
An obligate heterotypic-interaction model for stickers and spacers
The theory of Semenov and Rubinstein (141) was extended by Wang et al. (44) to describe the measured variations of csat with the apparent valence (numbers) of Arg and Tyr residues that are considered as the main stickers in FUS / FET family proteins. We follow Wang et al. (44) and impose the constraint that εAA = εBB = 0 and εAB < 0. This implies that the effective attractions arise purely from heterotypic interactions among stickers. Since εAB < 0, it follows that (−εAB/kBT) > 0 and hence we shall write the Boltzmann weight as exp(∣εAB∣/kBT) where the numerator of the exponent refers to the magnitude of the attractive interactions between A-B stickers.
We shall consider a system comprising of N associative polymers in a solvent, each with nA and nB stickers of type A and B, respectively. The spacers between the stickers are inert, phantom chains. The free energy of the system is written as:
| (3) |
The partition function Z can be calculated using the mean-field approach of Semenov and Rubinstein (141) as:
| (4) |
In equation (4), Ω is a combinatorial factor, Npairs is the total number of extant pairs of A-B crosslinks in the system, vb is the bond volume associated with each physical crosslink, and V is the system volume. The combinatorial factor is computed as:
| (5) |
Substitution of (5) into (4) and (3) leads to:
| (6) |
Minimizing this free energy with respect to Npairs leads to:
| (7) |
Here, the attractive volume . Solving the quadratic equation yields the following expression for Npairs that minimizes F:
| (8) |
A total of Npairs stickers of type A and Npairs stickers of type B will participate in Npairs crosslinks. Accordingly, the fraction of interacting stickers (or crosslinks) in each chain will be:
| (9) |
Replacing Npairs in equation (8) with p leads to:
| (10) |
Here, c is the polymer concentration N/V. If we set the strengths of sticker-sticker interactions such that λc << 1 (weak interactions), then:
| (11) |
Based on Flory-Stockmayer theory (142; 143), we know that at the percolation threshold cperc the value of p designated as pperc is:
| (12) |
Substituting (12) into the left-hand side of (11) and setting c = cperc on the right-hand side of (11) leads to the following estimate for cperc, which is valid for nA + nB >> 1:
| (13)(44) |
Extending the obligate heterotypic model to include homotypic sticker interactions
Using a similar extension of the mean-field model developed by Prusty et al. (144) who applied the model to a system comprising of distinct polymers with A- and B- stickers, one can include the effects of homotypic sticker attractions by setting εAA ≠ 0 and εBB ≠ 0. In this scenario, a generalization of the approach detailed above leads to the following expression for the percolation threshold:
| (14) |
In equation (14), λij is the attractive volume for the i-j interaction, i.e., , where εij ≤ 0 is the interaction energy and vij is the bond volume for the i-j interaction. Note that equation (14) reduces to the expression in equation (13) if the effects of homotypic interactions between stickers are ignored.
What if we have multiple types of stickers in our system?
The model can be further generalized to a system comprising of polymers consisting of more than two types of stickers. In this case, the percolation threshold is estimated using equation (15), written as:
| (15) |
Here, i and j are sticker type indices, λij is the attractive volume for the i-j interaction, and ni is the number of stickers of type i in each polymer. As shown in equation (15), the contribution of each sticker pair interaction is additive, weighted by the attractive volume λij. Hence, if a certain λpq term is much greater than other terms such that the percolation concentration can be approximately evaluated by only considering λpqnpnq, we can consider only stickers p and q as relevant stickers, and other stickers can be considered as spacers. This implies that there is no fixed set of stickers, and a set of stickers in one system can play the role of spacers in another system, depending on their relative contributions to the percolation threshold.
Effects of spacers
The preceding section builds on the work of Semenov and Rubinstein (141) and prescribes a quantitative, albeit mean-field model for quantifying the effects of stickers on the driving forces for phase separation and percolation. One can also use the percolation threshold as a suitable proxy for estimating the saturation concentration for phase separation providing csat ≤ cperc < cdense. Whether or not this condition is satisfied will be determined by the nature of the spacers.
The mean-field model introduced above treats spacers as phantom chains. A simple way to account for the effects of spacers is to include ad hoc spacer-specific corrections to the average volume per crosslink i.e., the value of vb. However, for realistic scenarios, one has to account explicitly for the effects of spacers. There are three possible effects of spacers that one must consider: (i) the excluded volumes (also referred to as the effective solvation volumes) of spacers (145); (ii) the contribution of auxiliary attractions between sites along spacers and specific stickers; and (iii) attractive interactions between spacers that, by definition of spacers will be weaker than sticker-sticker interactions. In the limit of strong attractions among spacers, associative polymers essentially become akin to homopolymers in poor solvents and the distinction between a stickers-and-spacers framework versus a homopolymer model becomes minimal because all the entities are equivalent to one another. Strong (vis-à-vis kBT) sticker-sticker, sticker-spacer, and spacer-spacer interactions will drive aggregation and / or precipitation into amorphous or fibrillar solids. These assemblies are distinct from the fluid-like phases that would be formed by associative polymers. Accordingly, the excluded volumes generated by spacers and their relatively weak auxiliary interactions with stickers are the main contributions that spacers make to the phase behavior of associative polymers.
The excluded volume (vex), also referred to as the effective solvation volume (ves), is the average volume per spacer site that is set aside for interactions with the surrounding volume (135) (Figure 3b). It is governed by the effective, solvent-mediated, pairwise interactions between spacer sites. If these interactions are net attractive, then vex is negative implying that the spacer sites sequester themselves from the surrounding solvent giving rise to compact spacers. Conversely, if the effective interactions are repulsive, then vex is positive implying that the spacer sites interact preferentially with the surrounding solvent, thereby giving rise to spacers that are conformationally expanded. If the spacer-solvent, solvent-solvent, and spacer-spacer interactions counterbalance one another, then vex ≈ 0 implying that spacers behave like ideal chains. In theory, this should reproduce the mean-field behavior described above since the mean-field model is based on so-called phantom spacers. However, the spacers with zero excluded volume can enhance the sticker-sticker interactions and this cooperative effect leads to phase separation being realized at concentrations that are well below the percolation threshold predicted based on Flory-Stockmayer theory, because Flory-Stockmayer theory only considers the valence of stickers and the bond formation probability, but ignores the intrinsic connectivity due to spacers (142; 143).
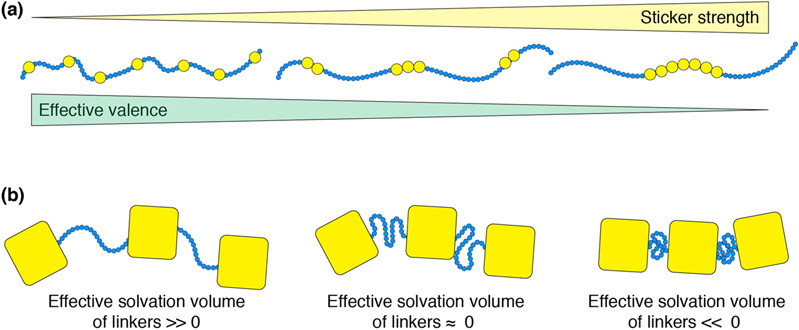
Figure 3: Schematic of sticker patterning and effective solvation volume.
(a) Three distinct sequences with identical numbers of sticker residues distributed in different arrangements. As sticker residues are clustered together, the effective sticker identity may change, such that as the number of stickers decrease the strength of each individual sticker increases. There are likely complex non-linearities in this behavior, such that the schematic here should be taken only as a qualitative description of this phenomenon. B Physical manifestation of the effective solvation volume for linkers. A positive effective solvation volume is associated with expanded and highly expanded linkers while a negative effective solvation volume leads to a collapsed and self-interacting linker. An effective solvation volume of zero implies ideal chain behavior.
Harmon et al. (145) performed lattice-based simulations intended to mimic the poly-SH3 and poly-PRM system studied by Rosen and coworkers (33; 146; 147). Simulations showed that spacers mimicking self-avoiding walks have high positive excluded volumes and the preferential interactions of these spacers with solvent will inhibit the cooperative interactions that are required to drive phase separation. Instead, the high excluded volumes lead to an upshift in the calculated percolation threshold when compared to expectations from the Flory-Stockmayer limit (142; 143). For associative polymers with high positive excluded volume spacers, percolation occurs above a percolation threshold but this is realized without phase separation implying that cperc > csat for such systems. As the spacer excluded volumes decrease, there is a stronger coupling between phase separation and percolation; this implies that the low excluded volumes of spacers enable cooperative interactions among spacers that enable concomitant density and percolation transitions.
To account theoretically for the effects of spacers that were quantified in simulations, one has to be able to calculate the signs and magnitudes of excluded volumes for each of the spacer regions. This will allow the incorporation of suitable corrections or higher-order terms due to spacer contributions into quantitative models that allow one to predict csat, cperc, and cdense directly from the sequence. Continued integration between theory and simulation should permit the development of a comprehensive model that accounts for sticker valence, sticker interaction strengths, spacer excluded volumes, and higher-order contributions to interactions among associative polymers that are due to auxiliary attractive interactions between spacer sites and specific stickers.
IDENTIFYING STICKERS VERSUS SPACERS
The identification of stickers can be performed computationally and / or experimentally. As a rule of thumb, the loss of a sticker should have a substantial impact on csat while the loss of a spacer should have minimal effects on csat. This is a simple but well-defined functional definition, but quantitatively what constitutes a substantial impact will vary across systems. A brute-force approach to identify stickers versus spacers would be full mutagenesis of every residue using alanine- or glycine-scanning and / or saturation mutagenesis that is coupled with the measurement of csat for each mutant. This would provide a systematic assessment of the contribution that each residue makes to the saturation concentration, allowing those that contribute substantially to be delineated as stickers (or falling within sticker motifs) while those that do not as spacers. On the other hand, we can perform a systematic mutation of specific residues of interest, identified a priori using bioinformatics approaches (148-151), to all possible amino acids. This directed saturation mutagenesis allows a direct comparison between two positions across equivalent sequence changes.
Rather than assessing the functional consequence of loss (or gain) of stickers, an alternative approach is the biophysical dissection of sticker-mediated intermolecular interactions. Instead of measuring csat, the early stages of assembly and / or deviations from ideal solution behavior can be measured using various methods include light scattering and fluorescence correlation spectroscopy (FCS). As an example, measuring the second virial coefficient (B2) using light scattering and / or osmotic pressure measurements as a function of sequence perturbation and changes to solution conditions provides one route to dissect how distinct residues or motifs contribute to intermolecular interaction (152).
Nuclear magnetic resonance (NMR) spectroscopy is a powerful experimental technique that provides the requisite site-specific information that can be brought to bear on identifying stickers versus spacers (16; 81; 123; 147). For IDRs, transverse relaxation rates (R2) provide information regarding local dynamics, and this may be slowed for stickers engaging in interactions with one another (153; 154). These can be detected using differences in R2 values. Chemical shift perturbations measured as a function of concentration can also provide insight into intermolecular interactions, as can cross-saturation transfer experiments. If appropriately designed experiments are performed, then paramagnetic relaxation enhancement (PRE) mediated by spin-labels provides an alternative approach to assess transient intermolecular interactions. Finally, if sufficiently strong inter-stickers interactions are present, it may be possible to detect these using intermolecular nuclear Overhauser effects (NOEs).
Many approaches used to identify stickers versus spacers will depend on the architecture of the protein / RNA molecules of interest. We define three common classes of biopolymers that comprise of stickers and spacers: folded domains, linear multivalent systems, and intrinsically disordered regions (Figure 2). Folded domains may be thought of as being analogous to rigid or deformable patchy colloids (99). Accordingly, the goal is to identify the attractive patches (stickers) on the surfaces of folded domains, the range and directionality of their attractions, any fluctuations associated with folded domains which may account for fluctuations between sticker-sticker interactions, and the nature of any spacer-mediated auxiliary interactions (151). A combination of computational, theoretical and experimental approaches that have been deployed in the context of studying the phase behavior of proteins that undergo crystallization can be adapted to identify stickers versus spacers for folded domains and quantify the interaction strengths among stickers and spacers.
Linear multivalent proteins refer to polypeptides in which multiple folded domains are connected by flexible “linker” IDRs (145). An example of such a system is the poly-SH3 + poly-PRM and the poly-SUMO + poly-SIM systems (155). For systems in which folded domains are well-defined binding modules the stickers are readily identified as binding sites on the interaction domains (SH3 / SUMO) and their cognate partners (PRM / SIM). The relative importance of specific residues in these binding sites can be assessed by mutational studies, but auxiliary interactions mediated by residues distal from these sites may also have a modulatory role (147). In these types of modular systems the flexible linkers that connect folded domains can be viewed as the spacers. The surface residues that lie outside the binding sites may also be viewed as spacers. Similar approaches can be applied to branched multivalent proteins comprising of folded oligomerization domains and IDRs, as in the case of nucleophosmin 1 (NPM1) (25) or even oligomeric proteins such as metabolic enzymes that are devoid of IDRs.
To identify and delineate stickers from spacers within IDRs we can take advantage of the maturity of methods that are now routinely brought to bear on the analysis of conformational ensembles of disordered proteins in dilute solutions. This becomes an informative exercise in the context of IDRs that lack persistent secondary or tertiary structural preferences as there is an intrinsic equivalence between intermolecular and intramolecular interactions. Degenerate interactions in the form of a network of intramolecular physical crosslinks among stickers along an IDR can lead to the partial collapse of individual molecules. Two distinct types of parameters can be determined from these single-chain simulations: parameters determined from the sequence alone, and the apparent valence of stickers and chain length. Additionally, one can obtain parameters that are governed by a combination of sequence and solution conditions and include the sticker-sticker interaction strengths and the excluded volumes of spacer regions. Both sets of parameters play a key role in determining the extent of collapse of an individual disordered protein in dilute solutions, and consequently, the extent of intermolecular interactions in sufficiently concentrated solutions. Accordingly, an assortment of methods that combine experimental and computational approaches can be used to identify and delineating stickers versus spacers in IDRs.
The combination of methods described for multivalent proteins can also be brought to bear on identifying stickers versus spacers in RNA molecules. These methods can be augmented by RNA structure prediction methods that help identify regions of complementarity that are likely to be involved in making secondary structures via base pairing and base stacking. Secondary and tertiary structure predictions are readily tested using methods such as SHAPE (156) that are sensitive to the presence of specific types of structural motifs. Recent work has shown that the ratio of purines to pyrimidines is an important determinant of the driving forces for phase separation in disordered RNA molecules (157). This observation provides a useful heuristic to quantify comparative driving forces for phase separation on the basis of the apparent valence of purine-based stickers from pyrimidine-based spacers.
Apparent valence versus effective valence of stickers:
The approaches of Semenov and Rubinstein focus primarily on the apparent valence of stickers. Recent studies have shown that the clustering / segregation of stickers along linear sequences can have a profound effect on the driving forces for phase separation. For a fixed number (apparent valence) of stickers, sequence patterning can increase or decrease the effective valence of stickers (Figure 3a). Coarse-grained simulations based on transferrable (83), phenomenological (25; 158), or learned models (159) have been developed and deployed to study the phase behavior of an assortment of protein and RNA molecules that include a combination of folded domains and disordered regions (145; 160-162). The results of simulations can be analyzed using mean-field theories and discrepancies between theoretical predictions and computational results can be used to extract the effective valence of stickers, thereby going beyond the apparent valence extracted from sequence analysis alone.
An important theoretical advance that accounts for sequence patterning effects, specifically the clustering versus segregation of charged residues, comes from Chan and coworkers (15; 163). Building on observations of the contributions of charge patterning to the dimensions of disordered proteins, Lin and Chan adapted the generalized random phase approximation originally introduced by Ermoshkin and Olvera de la Cruz for synthetic polymers (164) to incorporate sequence correlations between charged stickers and a mean-field correction to model cation-pi interactions. Their model has been used to predict differences in phase behavior for sequences that have identical numbers (apparent valence) of charged residues but are distinguished by the patterning of oppositely charged residues along the linear sequence (16). Their predictions reveal that the effective valence is lower than the apparent valence for sequences where the oppositely charged residues are segregated along the linear sequence, but the strength of sticker-sticker interactions increases substantially when compared to sequences with a more uniform distribution of charged stickers. In contrast, the effective valence is lower than the apparent valence for sequences where the oppositely charged residues are well mixed along the linear sequence. The predictions of Chan and coworkers have also been borne out in orthogonal field-theoretic simulations and in experiments based on synthetic polymers (165). These studies as well as in vitro and in cell experiments aided by sequence design approaches (34) highlight the importance of sequence patterning effects as determinants of the effective valence as opposed to the apparent valence.
The apparent and effective valence can also deviate from one another if the associative polymer of interest is characterized by conformational heterogeneity. For highly structured systems and maximally disordered systems there is likely to be a one-to-one correspondence between apparent valence and effective valence. Between these limiting scenarios, the dominant conformations in the ensembles within dilute and coexisting dense phases will govern the effective valence.
CONTRIBUTIONS OF STICKER-STICKER CROSSLINKS AND SPACER EXCLUDED VOLUMES TO STRUCTURAL AND DYNAMICAL PROPERTIES OF CONDENSATES
Condensates formed by associative polymers are not simple liquids, which are formed by spherical molecules with isotropic interactions. In contrast, liquids formed by associative polymers are characterized by physical crosslinks among highly flexible (disordered) polymers or patchy colloidal (ordered) molecules. Accordingly, these liquids are best described as network fluids (166).
The rheological properties of network fluids are governed by the extent of crosslinking, the timescales associated with making / breaking of physical crosslinks, the concentration of stickers within the dense phase, and the modulatory impact of spacers (166). Network fluids are not purely viscous liquids, but behave like elastic materials on timescales shorter than the lifetimes of crosslinks (138). On longer timescales, these fluids behave like viscous materials and therefore network fluids are in fact viscoelastic rather than purely viscous fluids. Rheological characterization of viscoelastic materials requires the measurement of dynamic moduli, which quantifies the ratio of stress to strain of the fluid under the influence of oscillatory forces (138; 162). Of direct relevance is the response of a network fluid to shear stresses and strains. If the fluid is purely viscous, then the shear strain, which refers to the deformation of the network, will lag behind the shear stress, which refers to the breaking of the network. Conversely, in a purely elastic material, the stress and strain are perfectly in phase. The phase angle between stress and strain can be quantified in terms of the dynamic modulus G*, which is a complex variable written as G* = G′ + iG″. Here, G′ and G″ are the shear storage and shear loss moduli, respectively. The storage modulus quantifies the energy stored in the network and is determined by the extent of physical crosslinking and the strengths of the crosslinks. In contrast, the loss modulus quantifies the extent of energy dissipation and is governed by the viscosity of the fluid. Values of storage and loss moduli can be measured as a function of shearing frequency and the phase angle δ is calculated as the ratio of the loss to storage modulus i.e., tan δ = (G″/ G′).
Is the distinction between viscous and viscoelastic network fluids relevant and / or of functional importance?
Nucleoli (25), nuclear speckles (167), P-granules (121; 168), and even very simple synthetic condensates (129; 157) show characteristics of multilayered, multicomponent behavior (Figure 4). Distinct layers are likely to form via different molecular networks, which may lead to different viscoelastic behaviors. Rheological measurements show that the fibrillarin-rich dense fibrillar center (DFC) of nucleoli is a bona fide viscoelastic material (25). Comparatively, the NPM1-rich granular component (GC) appears to be more of a viscous material than the DFC. Nucleoli and nuclear speckles appear to have similar organizations in that their cores are more viscoelastic than the outer layers that are more viscous. This type of architecture might have a bearing on where and when rRNA and ribosomal proteins, which experience opposing radial fluxes in nucleoli, will encounter one another and undergo ribosomal assembly (46). In contrast to nucleoli and nuclear speckles, condensates formed by essential P granule components such as MEG proteins and their cognate RNA molecules appear to have viscous cores surrounded be viscoelastic shells (121). Although rheological measurements are not available for this condensate, the inferences are drawn using data from experiments based on fluorescence recovery after photobleaching (FRAP), which might come with requisite caveats regarding their analyses (169). The coexistence with viscous protein-like liquids and apparently viscoelastic materials comprising of physically crosslinked RNA molecules is also readily observed in binary and ternary systems with simple dipeptide-rich proteins and homopolymeric RNA molecules (157).
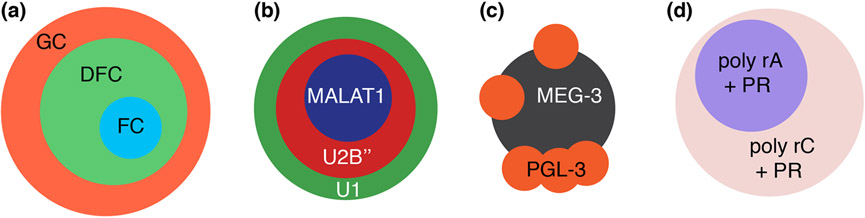
Figure 4: Four examples of multiphase assemblies formed from different components.
(a) Three-phase nucleoli assembly is readily reproduced using a simple stickers-and-spacers model, as shown by Feric et al. (b) Various distinct types of nuclear speckle architecture can also be recapitulated in a similar manner, as shown by Fei et al. (c) MEG-3 and PGL-3 form distinct phases in P-granules and in vitro, as demonstrated by Putnam et al. (d) A simple four component system (solvent, proline-arginine dipeptides, polyadenosine, polycytosine) forms two distinct dense phases due to distinct sticker-sticker strengths, as described by Boeynaems et al.
Dynamic moduli quantify the material properties of viscoelastic network fluids and these moduli are determined by the spatial organization of associative polymers with respect to one another within condensates. Spatial organization of molecules will also determine the crosslinking density within condensates. One can quantify spatial organization using distribution functions such as pair and triplet distribution functions that serve as primary descriptors of the structures of liquids. These structural descriptors revolutionized the studies of simple liquids and molecular fluids formed by small molecules. In fact, for simple liquids, Zwanzig and Mountain derived direct connections between the structures of liquids quantified in terms of pair distribution functions and dynamic moduli of these systems (170). Extensions of these approaches to connect structural descriptions of network fluids formed by associative polymers and their dynamic moduli are precisely the types of connections between structural descriptions of condensates and their material properties that are needed to understand structure-function relationships on mesoscales defined by non-stoichiometric macromolecular assemblies. Such efforts will be aided by recent advances in small angle neutron scattering (SANS) measurements that enable the direct measurement of Fourier transforms of pair distribution functions, as has been demonstrated by Mitrea et al. (45) for facsimiles of pentameric NPM1 that form condensates through heterotypic interactions with Arg-rich ligands.
THE STICKERS-AND-SPACERS MODEL IN BIOLOGICAL SYSTEMS
A variety of condensate driving systems including multivalent protein and RNA molecules appear to conform to the stickers-and-spacers architecture. The earliest demonstration of the relevance of the stickers-and-spacers model was made by Rosen and coworkers who studied the phase behavior of linear multivalent proteins in solution and anchored to membranes (33; 171). In accord with the predictions of Semenov and Rubinstein and the basic tenets of the Flory-Stockmayer theory, the valence of interaction domains was shown to contribute directly to the driving forces for phase separation and percolation / gelation. Further, in cells, multisite Tyr phosphorylation was shown to regulate the overall valence of stickers by recruiting multiple proteins with multiple SH3 and SH2 domains to the membrane anchored protein nephrin (33).
Associative polymers can form system-spanning networks, a phenomenon that is known as percolation. If phase separation and percolation are coupled (csat ≤ cperc < cdense), then the dense phase is a percolated network. In the scenario where the dense phase is a spherical droplet, it follows that the associative polymer forms a droplet-spanning network that coexists with a dilute phase of non-networked molecules. Associative polymers can also form networked solid phases, as is the case with so-called self-assembled fibrillar networks or SAFINS (172). In these cases, the associative polymers form fibrillar structures – as has been observed with several low complexity sequences – at sufficiently high concentrations. These fibrils are crosslinked to form networks that are akin to self-supporting hydrogels, and in contrast to systems that undergo LLPS through weak degenerate multivalent interactions, these systems are characterized by the acquisition of specific structural biases within the networked phase that are largely absent from polypeptides in the dilute phase.
In the context of low complexity IDPs, McKnight and coworkers identified regions (stickers) that drive the formation of cross-beta structures and enable the networking of fibrils thus giving rise to highly networked hydrogels (173-176). The Eisenberg group leveraged their ability to make microcrystals using peptide fragments to identify stickers that enable the formation of intermolecular crosslinks in the form of hydrogen bonds and zippering interactions. In their parlance, Eisenberg and coworkers refer to the stickers as LARKS (low complexity aromatic-rich kinked segments) due to their sequence composition and the fact they form cross-beta structures with a characteristic kinked topology, in contrast to standard amyloid cross-beta regions (177).
Brangwynne et al. proposed that stickers in disordered regions were likely to be SLiMs that comprise of charged, polar, and aromatic moieties (22). Drawing on the decades-old work of Burley and Petsko (178), Brangwynne et al. proposed that the three categories of residues were likely to be involved in a hierarchy of weakly polar interactions due to their intrinsic multipole moments. Charged residues such as Arg are defined by a monopole moment (charge of +1e), a finite dipole moment, and a significant quadrupole moment due to the planarity of the guanido group. In contrast, Lys has a spherically symmetric functional group and is better approximated as a point charge with a monopole moment (charge of +1e) but a negligible dipole or quadrupole moment. This would imply that Arg would be a superior sticker residue over Lys given the hierarchy of interactions it encodes. In accord with this expectation, Wang et al. (44) showed that the network of Arg-Tyr interactions derived from the high valence of Arg residues within the RNA binding domain (RBD) and equally high valence of Tyr residues within the prion-like domain (PLD) contributes directly to the driving forces for phase separation of the protein FUS. Mutations of Arg to Lys within the RBD weaken the driving forces by approximately ten-fold, as measured by the impact of these mutations on the value of csat. Tyr residues have zero net charge (zero monopole moment), but a large dipole moment due to the in-plane arrangement of the ─OH group with the planar pi system, and a significant quadrupole moment that is in accord with its aromaticity. In contrast, Phe has a near zero dipole moment and a finite quadrupole moment that is concordant with that of Tyr. Accordingly, substitution of the Tyr residues within the PLD of FUS with Phe residues causes a diminution of the driving forces for phase separation, measured again in terms of increased values for csat. The roles of hydrophobic stickers have been made clear in the work of Riback et al. (41) who quantified the effects of titrating hydrophobic residues on the driving forces for collapse and phase separation of the polyA RNA binding protein PAB1.
Recent work of Castãneda and coworkers has shown the validity of the distinctions between stickers and spacers in explaining the impact of ALS-related mutations within the protein UBQLN2 on its phase behavior (179; 180). Mutations to stickers clearly alter the driving forces for phase separation whereas mutations to spacer residues contribute to changes in material properties as measured by the recovery of fluorescence after photobleaching. Interestingly, mutations to spacer residues have minimal effects on the driving forces for phase separation, a result that is concordant with observations of Wang et al. (44) who found that changes to the PLD / RBD of FUS that lie outside the identified stickers (Tyr and Arg) impact the material properties of condensates without altering the driving forces for phase separation.
The impact of charged residues as stickers was explored in the DEAD box helicase protein DDX4 that drives the formation of nuage bodies. The work of Nott et al. (14) was the first to show the importance of clusters of positive and negatively charged residues along the linear sequence, with modest sequence changes that disrupt these charged stickers preventing phase separation in cells through a reduction in valence without changing sequence composition. They were also the first to identify the importance of complementary interactions between Lys / Arg (cationic) and Phe (aromatic) residues as drivers of phase separation. The importance of linear clustering of charged stickers was also made evident in the explorations of Pak et al. who quantified the phase behavior of a de novo designed IDR, the nephrin intracellular domain (NICD), in cells and in vitro, showing that increased linear clustering of acidic residues within NICD enhances the driving forces for phase separation and the extent of physical crosslinking aided by cationic complexing polyions (34). As for RNA molecules, mutagenesis studies, design experiments and coarse-grained simulations have demonstrated that purines are stronger stickers than pyrimidines, a result attributed to their double ring structure that gives rise to a stronger aromatic system (157).
EMERGENT VERSUS INTRINSIC STICKERS
The instantiations of the stickers-and-spacers model described thus far has focused on sequence encoded features that dictate the apparent and effective valence of stickers. These stickers are intrinsic to the molecular architecture and are directly encoded into the sequence. Hence, we refer to these as intrinsic stickers. Recently, it has become clear that hierarchical assembly processes can give rise to emergent stickers, in which oligomerization / clustering or even micro-phase separation (162) can give rise to a de novo multivalent macromolecule which itself is able to drive the formation of condensates via the type of interactions that are proposed to apply for associative polymers (Figure 5a). Clusters that serve as generators of emergent stickers can form at a relatively low molecular concentration compared with the saturation concentration associated with their constitutive monomeric components. Importantly, from a theoretical standpoint, it can be argued that the interactions that drive oligomerization / clustering are likely to be distinct from the interactions that drive coalescence of oligomers into condensates, the conversion of clusters into crystals (181), or the transformation of oligomers into fibrils (182; 183).
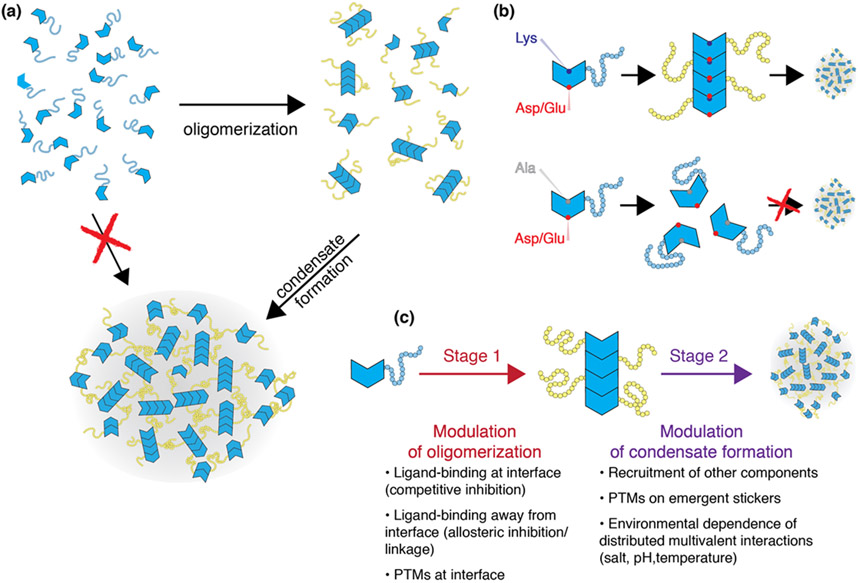
Figure 5:
(a) General model for emergent stickers formed through oligomerization. Monomeric species might lack the requisite valence to drive condensates, but oligomerization through a defined interface gives rise to multivalence of emergent stickers that drive condensate formation. (b) A schematized version of condensate regulation in the context of ARF19. The PB1 oligomerization domain drives assembly via an electrostatically mediated binding surface. Neutralization of a lysine residues abrogates oligomerization and consequently prevents condensate formation. (c) For a system that gives rise to emergent stickers, condensate formation can be regulated at two levels. An effectively binary regulation that dictates whether oligomerization occurs (modulation of valence), and a second level in which the strength of emergent stickers can be altered. Note that temporally the order in which these levels of modulation occur is irrelevant. As a tangible example, in a scenario in which IDRs phosphorylation weakens the strength of emergent stickers, the act of phosphorylation could happen before or after oligomerization. The binding of ligands, which could include other proteins, nucleic acids, small molecules, may either lead to a conformational transition that allows homotypic oligomerization or itself could drive heterotypic assembly. In principle, multiple nested layers of assembly via orthogonal interaction modes provide the foundations for arbitrarily complex regulation.
We highlight four concrete examples that anchor the idea of emergent stickers being generated from oligomers or clusters. A subset of Auxin Responsive transcription factors (ARFs) in Arabidopsis thaliana undergo oligomerization driven by a folded C-terminal PB1 (184). These linear oligomers lead to the formation of higher-order species which themselves drive the formation of condensates through an IDR-dependent assembly process. In this system, oligomerized ARFs are in effect large multivalent biopolymers that drive condensate formation through the crosslinking of stickers in IDRs whose multivalence is governed by the extent of oligomerization. If PB1 oligomerization is abrogated though a single lysine-to-alanine mutation, oligomers and consequently condensates are unable to form (Figure 5b). As a second example, the NPM1 pentamer is a prime example in which an emergent molecular species undergoes phase separation, as opposed to NPM1 monomers individually (45-47; 185). A third example is the SPOP system which has been explored in a series of elegant experiments (131; 186; 187). Finally, dimerization of HP1a/α plays a key role in forming a species with the requisite multivalence to undergo phase separation (17; 18). A synthetic system where oligomerization controls the valence of IDRs that drive condensate formation is the Corelet system designed by Bracha et al. (188). Here, the valence of IDRs appended to the oligomeric ferritin core is controlled by light and the driving forces for phase separation, as quantified by full binodals measured in living cells, is governed by the valence of the IDRs. In all of these systems the oligomerization that leads to emergent multivalence of stickers involves either evolved or designed interactions that are clearly orthogonal to the interactions that drive condensate formation through multivalence of stickers in IDRs.
A simple rationalization for the benefit of oligomerization / clustering is that helps to pre-pay some of the entropic penalty, a feature that can only be truly realized if the modes of interaction that drive oligomerization and subsequent condensate formation are distinct. This condition is well aligned with the observation that many of the proteins that undergo phase separation possess a modular architecture with distinct interaction domains (both IDRs and folded binding domains). The existence of orthogonal modes of intermolecular interaction allows the driving force for assembly to be tuned at (at least) two independent and complementary sites (Figure 5c). From a regulatory standpoint, this allows one set of regulatory systems to control on / off interactions (e.g., inhibition/promotion of oligomerization / clustering) and another to tune the molecular details of the condensate by modulating the strength of emergent stickers. While we have described oligomerization here in terms of a conventional and stoichiometric biochemical phenomenon (i.e., dimerization, polymerization, etc.) oligomerization could also itself be driven by weak multivalent interactions which would necessarily be chemically orthogonal to those that drive higher-order assembly. The multi-resolution nature of the stickers-and-spacers model is appealing as the emergence of new stickers is well described as a fractal (self-similar) phenomenon, allowing the same theoretical framework to be quantitatively applied irrespective of the molecular nature and length-scale.
CONCLUDING REMARKS
The field of intracellular phase transitions is evolving rapidly as we learn more about the molecular drivers of biomolecular condensates and the contributions of condensates to specific biological functions. Condensates appear to be ubiquitous within cells and novel cellular functions are being ascribed to newly discovered condensates. Accounts of these novel functions and details about the molecular drivers of condensates are emerging at a frenetic pace. Here, we have focused on a specific physical framework, namely, the stickers-and-spacers model, adapted from the field of associative polymers, to describe the molecular grammar that underlies the architecture, sequence-encoded driving forces, and evolution of multivalent protein and RNA molecules that drive condensate formation. Mapping of the stickers-and-spacers framework to biomacromolecules is in its infancy. However, the validity of this framework is coming into sharp focus as more accounts emerge of its utility for explaining measured phase behavior and for predicting / designing phase behavior. The availability of computational tools is further advancing our ability to dissect the interplay between spontaneous and driven processes in regulating and determining the phase behavior of mixtures of associative polymers. At this juncture, the stickers-and-spacers formalism seems like an apt modernization of classical theories developed for homopolymers that is proving to be relevant for describing phase transitions of multivalent protein and RNA molecules. The findings summarized here pave the way for understanding how the synergies of sticker and spacer interactions might be affected or modulated in multicomponent systems, which mimic naturally occurring biomolecular condensates – an important topic that merits intense study.
Predictions based on the stickers-and-spacers framework apply to systems with one or two types of multivalent protein / RNA molecules in a solvent. However, biomolecular condensates encompass hundreds of distinct types of macromolecules. A key concept that has to be generalized is that of saturation concentrations because what we have adapted thus far applies strictly to two component systems comprising of a polymer plus solvent. A recent computational study based on the LASSI simulation engine helps generalize the concept of saturation concentration by showing how obligate heterotypic interactions can give rise to apparent saturation concentrations that depend on the slopes of tie lines in multidimensional phase diagrams (158). The simulations, which are built on the stickers-and-spacers formalism, when combined with suitable experiments (189), should enable a rigorous mapping between the numbers of distinct components and the apparent saturation concentrations for each of the components. In fact, our generalizations of cperc (see equation (15)) for an arbitrary number of stickers and the findings reported by Choi et al. (158) based on the LASSI engine and by Riback et al. (189) based on experiments helps sets the stage for connecting generalized observations from simulations for multicomponent systems to theories for multicomponent systems wherein each protein / RNA component has its own set of distinct stickers that may or may not interact with stickers on other protein / RNA molecules.
ACKNOWLEDGMENTS
The US National Science Foundation (MCB-1614766), the US National Institutes of Health (5R01NS056114 and 1R01NS089932), the Human Frontier Science Program (RGP0034/2017), and the St. Jude Research Collaborative on Membraneless Organelles fund ongoing efforts in the Pappu lab. Jeong-Mo Choi is currently funded by the Basic Science Research Program (2019R1A6A1A10073887) from the Ministry of Education of the Republic of Korea through the National Research Foundation of Korea. We are grateful to Simon Alberti, Priya Banerjee, Clifford Brangwynne, Carlos Castãneda, Hue-Sun Chan, Furqan Dar, Julie Forman-Kay, Titus Franzmann, Amy Gladfelter, Tyler Harmon, Anthony Hyman, Frank Jülicher, Richard Kriwacki, Erik Martin, Tanja Mittag, Ammon Posey, Michael Rosen, Lucia Strader, Andrea Soranno, and J. Paul Taylor for numerous stimulating discussions. We dedicate this review to the memory of Suzanne Eaton, a senior group leader at the Max Planck Institute for Cell Biology (MPI-CBG) and Genetics in Dresden where the application of the stickers-and-spacers formalism matured through interactions with colleagues at the MPI. Suzanne was an inspiring scientist and human being and her legacy will continue to live on through the efforts of scientists at the MPI-CBG and the community that stretches beyond.
Footnotes
DISCLOSURSE STATEMENT
RVP is a member of the Scientific Advisory Board of Dewpoint Therapeutics Inc. This membership has not influenced the scientific content of this review. Further, the authors are not aware of any other memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Bickmore WA. 2013. Annual Reviews in Genomics Human Genetics 14:67–84 [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Tang AH, Blanpied TA. 2018. Current opinion in neurobiology 51:147–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H, DeLoache WC, Dueber JE. 2012. Metabolic engineering 14:242–51 [DOI] [PubMed] [Google Scholar]
- 4.Razin SV, Gavrilov AA, Yarovaya OV. 2010. Biochemistry. Biokhimiia 75:1307–15 [DOI] [PubMed] [Google Scholar]
- 5.Manz BN, Groves JT. 2010. Nature Reviews Molecular Cell Biology 11:342–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papusheva E, Heisenberg CP. 2010. EMBO Journal 29:2753–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenal U, Stephens C. 2002. Current opinion in microbiology 5:558–63 [DOI] [PubMed] [Google Scholar]
- 8.Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Nat Rev Mol Cell Biol 18:285–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin Y, Brangwynne CP. 2017. Science 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 10.Berry J, Brangwynne CP, Haataja M. 2018. Reports on Progress in Physics 81:046601. [DOI] [PubMed] [Google Scholar]
- 11.Walter H, Brooks DE. 1995. FEBS Letters 361:135–9 [DOI] [PubMed] [Google Scholar]
- 12.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, et al. 2009. Science 324:1729–32 [DOI] [PubMed] [Google Scholar]
- 13.Hyman AA, Weber CA, Jülicher F. 2014. Annu. Rev. Cell Dev. Biol 30:39–58 [DOI] [PubMed] [Google Scholar]
- 14.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, et al. 2015. Mol. Cell 57:936–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y-H, Forman-Kay JD, Chan HS. 2016. Physical Review Letters 117:178101. [DOI] [PubMed] [Google Scholar]
- 16.Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, et al. 2017. Proc Natl Acad Sci U S A 114:E8194–E203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Nature 547:241–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, et al. 2017. Nature 547:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Protter DSW, Rosen MK, Parker R. 2015. Molecular Cell 60:208–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brangwynne CP, Mitchison TJ, Hyman AA. 2011. Proceedings of the National Academy of Sciences USA 108:4334–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brangwynne CP. 2013. Journal of Cell Biology 203:875–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brangwynne CP, Tompa P, Pappu RV. 2015. Nature Physics 11:899–904 [Google Scholar]
- 23.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, et al. 2015. Proceedings of the National Academy of Sciences USA 112:7189–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, et al. 2015. Molecular Cell 60:220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, et al. 2016. Cell 165:1686–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei MT, Elbaum-Garfinkle S, Holehouse AS, Chen CC, Feric M, et al. 2017. Nature Chemistry 9:1118–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, et al. 2017. Molecular Cell 65:1044–55 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, et al. 2015. Cell 162:1066–77 [DOI] [PubMed] [Google Scholar]
- 29.Boke E, Ruer M, Wühr M, Coughlin M, Lemaitre R, et al. 2016. Cell 166:637–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel A, Malinovska L, Saha S, Wang J, Alberti S, et al. 2017. Science 356:753–6 [DOI] [PubMed] [Google Scholar]
- 31.Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, et al. 2018. Science 359:eaao5654. [DOI] [PubMed] [Google Scholar]
- 32.Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, et al. 2018. Cell Reports 23:3327–39 [DOI] [PubMed] [Google Scholar]
- 33.Li P, Banjade S, Cheng H-C, Kim S, Chen B, et al. 2012. Nature 483:336–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, et al. 2016. Molecular Cell 63:72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Currie SL, Rosen MK. 2017. Journal of Biological Chemistry 292:19110–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milovanovic D, Wu Y, Bian X, De Camilli P. 2018. Science 361:604–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, et al. 2015. Cell 163:123–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, et al. 2018. Molecular Cell 69:965–78. e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke KA, Janke AM, Rhine CL, Fawzi NL. 2015. Molecular Cell 60:231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, et al. 2017. EMBO Journal 36:2951–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, et al. 2017. Cell 168:1028–40 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuylen S, Blaukopf C, Politi AZ, Müller-Reichert T, Neumann B, et al. 2016. Nature 535:308–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. Cell 169:1066–77 e10 [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, et al. 2018. Cell 174:688–99. e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, et al. 2016. eLife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, et al. 2018. Nature communications 9:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitrea DM, Kriwacki RW. 2016. Cell Commun. Signal 14:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aumiller WM Jr., Keating CD. 2016. Nature Chemistry 8:129–37 [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Huang J, Lee Y, Dutta S, Yoo HY, et al. 2016. Proceedings of the National Academy of Sciences USA 113:E847–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R. 2014. eLife 3:e01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muiznieks LD, Cirulis JT, van der Horst A, Reinhardt DP, Wuite GJ, et al. 2014. Matrix Biology 36:39–50 [DOI] [PubMed] [Google Scholar]
- 52.Zwicker D, Decker M, Jaensch S, Hyman AA, Julicher F. 2014. PRoceedings of the National Academy of Sciences USA 111:E2636–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, et al. 2015. Nature Communications 6:8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. 2015. PRoceedings of the National Academy of Sciences USA 112:E5237–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, et al. 2015. eLife 4:e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguzzi A, Altmeyer M. 2016. Trends in cell biology 26:547–58 [DOI] [PubMed] [Google Scholar]
- 57.Bolognesi B, Lorenzo Gotor N, Dhar R, Cirillo D, Baldrighi M, et al. 2016. Cell Reports 16:222–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Courchaine EM, Lu A, Neugebauer KM. 2016. EMBO Journal 35:1603–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Keating CD, Rivas G. 2016. Scientific reports 6:35140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nott TJ, Craggs TD, Baldwin AJ. 2016. Nature Chemistry 8:569–75 [DOI] [PubMed] [Google Scholar]
- 61.Protter DSW, Parker R. 2016. Trends in cell biology 26:668–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su X, Ditlev JA, Hui E, Xing W, Banjade S, et al. 2016. Science 352:595–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor JP, Brown RH Jr., Cleveland DW. 2016. Nature 539:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. 2016. eLife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alberti S 2017. Current Biology 27:R1097–r102 [DOI] [PubMed] [Google Scholar]
- 66.Aumiller WM Jr., Keating CD. 2017. Advances in Colloid and Interface Science 239:75–87 [DOI] [PubMed] [Google Scholar]
- 67.Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA. 2017. Angewandte Chemie International Editon 56:11354–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falahati H, Wieschaus E. 2017. Proceedings of the National Academy of Sciences USA 114:1335–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, et al. 2017. Cell 171:148–62.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milovanovic D, De Camilli P. 2017. Neuron 93:995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, et al. 2017. Nature Communications 8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rao BS, Parker R. 2017. Proceedings of the National Academy of Sciences USA 114:E9569–e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. 2017. Cell 168:159–71.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stanek D, Fox AH. 2017. Current Opinion in Cell Biology 46:94–101 [DOI] [PubMed] [Google Scholar]
- 75.Tang SJ. 2017. Genes 8 [Google Scholar]
- 76.Weirich KL, Banerjee S, Dasbiswas K, Witten TA, Vaikuntanathan S, Gardel ML. 2017. PRoceedings of the National Academy of Sciences USA 114:2131–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Al-Husini N, Tomares DT, Bitar O, Childers WS, Schrader JM. 2018. Molecular Cell 71:1027–39.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alberti S, Saha S, Woodruff JB, Franzmann TM, Wang J, Hyman AA. 2018. Journal of Molecular Biology 430:4806–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexander EJ, Ghanbari Niaki A, Zhang T, Sarkar J, Liu Y, et al. 2018. PRoceedings of the National Academy of Sciences USA 115:E11485–e94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amaya J, Ryan VH, Fawzi NL. 2018. Journal of Biological Chemistry 293:19522–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, et al. 2018. Nature Structural and Molecular Biology 25:833–40 [DOI] [PubMed] [Google Scholar]
- 82.Cinar H, Cinar S, Chan HS, Winter R. 2018. Chemistry (Weinheim an der Bergstrasse, Germany) 24:8286–91 [DOI] [PubMed] [Google Scholar]
- 83.Dignon GL, Zheng W, Kim YC, Best RB, Mittal J. 2018. PLoS computational biology 14:e1005941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dine E, Gil AA, Uribe G, Brangwynne CP, Toettcher JE. 2018. Cell systems 6:655–63.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ditlev JA, Case LB, Rosen MK. 2018. Journal of Molecular Biology 430:4666–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Du M, Chen ZJ. 2018. Science 361:704–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dzuricky M, Roberts S, Chilkoti A. 2018. Biochemistry 57:2405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Erdel F, Rippe K. 2018. Biophysical Journal 114:2262–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, et al. 2018. Cell 173:706–19.e13 [DOI] [PubMed] [Google Scholar]
- 90.Huizar RL, Lee C, Boulgakov AA, Horani A, Tu F, et al. 2018. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kistler KE, Trcek T, Hurd TR, Chen R, Liang FX, et al. 2018. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, et al. 2018. Science 360:922–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li HR, Chiang WC, Chou PC, Wang WJ, Huang JR. 2018. Journal of Biological Chemistry 293:6090–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu H, Yu D, Hansen AS, Ganguly S, Liu R, et al. 2018. Nature 558:318–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, et al. 2018. Science 360:918–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin EW, Mittag T. 2018. Biochemistry 57:2478–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maucuer A, Desforges B, Joshi V, Boca M, Kretov DA, et al. 2018. Journal of cell science 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, et al. 2018. Molecular Cell 71:703–17.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nguemaha V, Zhou HX. 2018. Scientific reports 8:6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niewidok B, Igaev M, Pereira da Graca A, Strassner A, Lenzen C, et al. 2018. Journal of Cell Biology 217:1303–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rai AK, Chen JX, Selbach M, Pelkmans L. 2018. Nature 559:211–6 [DOI] [PubMed] [Google Scholar]
- 102.Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, et al. 2018. Science 361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sarkar J, Myong S. 2018. Methods in Molecular Biology 1814:325–38 [DOI] [PubMed] [Google Scholar]
- 104.Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, et al. 2018. Nature Communications 9:2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shan Z, Tu Y, Yang Y, Liu Z, Zeng M, et al. 2018. Nature Communications 9:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Woodruff JB. 2018. Journal of Molecular Biology 430:4762–72 [DOI] [PubMed] [Google Scholar]
- 107.Wunder T, Cheng SLH, Lai SK, Li HY, Mueller-Cajar O. 2018. Nature Communications 9:5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou HX, Nguemaha V, Mazarakos K, Qin S. 2018. Trends in Biochemical Sciences 43:499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Abbondanzieri EA, Meyer AS. 2019. Current genetics 65:691–4 [DOI] [PubMed] [Google Scholar]
- 110.Alberti S, Gladfelter A, Mittag T. 2019. Cell 176:419–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deshpande S, Brandenburg F, Lau A, Last MGF, Spoelstra WK, et al. 2019. Nature Communications 10:1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Falahati H, Haji-Akbari A. 2019. Soft matter 15:1135–54 [DOI] [PubMed] [Google Scholar]
- 113.Gasior K, Zhao J, McLaughlin G, Forest MG, Gladfelter AS, Newby J. 2019. Physical Review E 99:012411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gomes E, Shorter J. 2019. Journal of Biological Chemistry 294:7115–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hofweber M, Dormann D. 2019. Journal of Biological Chemistry 294:7137–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin Y, McCarty J, Rauch JN, Delaney KT, Kosik KS, et al. 2019. eLife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Linsenmeier M, Kopp MRG, Grigolato F, Liu D, Zurcher D, et al. 2019. Angewandte Chemie International Edition [Google Scholar]
- 118.Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, et al. 2019. Neuron 102:321–38.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Robles-Ramos MA, Lopez-Alvarez M, et al. 2019. EMBO reports 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pliss A, Levchenko SM, Liu L, Peng X, Ohulchanskyy TY, et al. 2019. Nature Communications 10:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Putnam A, Cassani M, Smith J, Seydoux G. 2019. Nature Structural and Molecular Biology 26:220–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, et al. 2019. Nature chemical biology 15:51–61 [DOI] [PubMed] [Google Scholar]
- 123.Ukmar-Godec T, Hutten S, Grieshop MP, Rezaei-Ghaleh N, Cima-Omori MS, et al. 2019. Nature Communications 10:2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, et al. 2019. Nature 566:131–5 [DOI] [PubMed] [Google Scholar]
- 125.Wang N, Liu C. 2019. Current Opinion in Genetics and Development 55:59–65 [DOI] [PubMed] [Google Scholar]
- 126.Wang Z, Zhang H. 2019. Trends in cell biology 29:417–27 [DOI] [PubMed] [Google Scholar]
- 127.Wu X, Cai Q, Shen Z, Chen X, Zeng M, et al. 2019. Molecular Cell 73:971–84.e5 [DOI] [PubMed] [Google Scholar]
- 128.Yoo H, Triandafillou C, Drummond DA. 2019. Journal of Biological Chemistry 294:7151–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, Lopez GP. 2017. Nat Chem 9:509–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, et al. 2016. EMBO Journal 35:1254–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, et al. 2018. Molecular Cell 72:19–36.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kesten H 2006. Notices of the American Mathematical Society 53:572–3 [Google Scholar]
- 133.Pappu RV, Wang X, Vitalis A, Crick SL. 2008. Arch. Biochem. Biophys 469:132–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. 2013. Proc. Natl. Acad. Sci. U. S. A 110:20075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rubinstein M, Colby RH. 2003. Polymer Physics. New York: Oxford University Press [Google Scholar]
- 136.Flory PJ. 1942. The Journal of Chemical Physics 10:51–61 [Google Scholar]
- 137.Mao S, Kuldinow D, Haataja MP, Košmrlj A. 2019. Soft matter 15:1297–311 [DOI] [PubMed] [Google Scholar]
- 138.Rubinstein M, Dobrynin AV. 1997. Trends in Polymer Science 5:181–6 [Google Scholar]
- 139.Sommer J-U. 2018. Macromolecules 51:3066–74 [Google Scholar]
- 140.Davey NE, Cyert MS, Moses AM. 2015. Cell Communication Signaling 13:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Semenov AN, Rubinstein M. 1998. Macromolecules 31:1373–85 [Google Scholar]
- 142.Stockmayer WH. 1943. The Journal of Chemical Physics 11:45–55 [DOI] [PubMed] [Google Scholar]
- 143.Flory PJ. 1941. Journal of the American Chemical Society 63:3083–90 [Google Scholar]
- 144.Prusty D, Pryamitsyn V, Olvera de la Cruz M. 2018. Macromolecules 51:5918–32 [Google Scholar]
- 145.Harmon TS, Holehouse AS, Rosen MK, Pappu RV. 2017. eLife 6:e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Banjade S, Rosen MK. 2014. eLife 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK. 2015. Proceedings of the National Academy of Sciences USA 112:E6426–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, et al. 2018. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vernon RM, Forman-Kay JD. 2019. Current Opinion in Structural Biology 58:88–96 [DOI] [PubMed] [Google Scholar]
- 150.Orlando G, Raimondi D, Tabaro F, Codice F, Moreau Y, Vranken W. 2019. Bioinformatics (Oxford, England) [DOI] [PubMed] [Google Scholar]
- 151.Wu H, Fuxreiter M. 2016. Cell 165:1055–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Petsev DN, Wu X, Galkin O, Vekilov PG. 2003. The Journal of Physical Chemistry B 107:3921–6 [Google Scholar]
- 153.Salvi N, Abyzov A, Blackledge M. 2017. Progress in nuclear magnetic resonance spectroscopy 102-103:43–60 [DOI] [PubMed] [Google Scholar]
- 154.Milles S, Salvi N, Blackledge M, Jensen MR. 2018. Progress in nuclear magnetic resonance spectroscopy 109:79–100 [DOI] [PubMed] [Google Scholar]
- 155.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, et al. 2016. Cell 166:651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Low JT, Weeks KM. 2010. Methods (San Diego, Calif.) 52:150–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boeynaems S, Holehouse AS, Weinhardt V, Kovacs D, Van Lindt J, et al. 2019. Proceedings of the National Academy of Sciences USA: 10.1073/pnas.1821038116 [DOI] [Google Scholar]
- 158.Choi J-M, Dar F, Pappu RV. 2019. PLoS computational biology 15:e1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ruff KM, Harmon TS, Pappu RV. 2015. The Journal of Chemical Physics 143:243123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Harmon TS, Holehouse AS, Pappu RV. 2018. New Journal of Physics 20:045002 [Google Scholar]
- 161.Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, et al. 2018. Journal of Biological Chemistry:jbc. RA117. 000357 [Google Scholar]
- 162.Roberts S, Harmon TS, Schaal JL, Miao V, Li K, et al. 2018. Nature materials 17:1154–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lin YH, Chan HS. 2017. Biophys J 112:2043–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ermoshkin AV, Olvera de la Cruz M. 2003. Macromolecules 36:7824–32 [Google Scholar]
- 165.McCarty J, Delaney KT, Danielsen SPO, Fredrickson GH, Shea J-E. 2019. The Journal of Physical Chemistry Letters 10:1644–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dias CS, Araújo NAM, Telo da Gama MM. 2017. Advances in Colloid and Interface Science 247:258–63 [DOI] [PubMed] [Google Scholar]
- 167.Fei J, Jadaliha M, Harmon TS, Li IT, Hua B, et al. 2017. Journal of cell science 130:4180–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jawerth LM, Ijavi M, Ruer M, Saha S, Jahnel M, et al. 2018. Physical Review Letters 121:258101. [DOI] [PubMed] [Google Scholar]
- 169.Taylor NO, Wei M-T, Stone HA, Brangwynne CP. 2019. Biophysical journal 117:1285–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zwanzig R, Mountain RD. 1965. The Journal of Chemical Physics 43:4464–71 [Google Scholar]
- 171.Banjade S, Rosen MK. 2014. eLife 3:e04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dastidar P 2019. Gels 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, et al. 2012. Cell 149:768–79 [DOI] [PubMed] [Google Scholar]
- 174.Kato M, Han TW, Xie S, Shi K, Du X, et al. 2012. Cell 149:753–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, et al. 2013. Cell 155:1049–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, et al. 2014. Science 345:1139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, et al. 2018. Science 359:698–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Burley SK, Petsko GA. 1988. Adv Protein Chem 39:125–89 [DOI] [PubMed] [Google Scholar]
- 179.Dao TP, Martyniak B, Canning AJ, Lei Y, Colicino EG, et al. 2019. Structure (London, England : 1993) 27:937–51.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang Y, Jones HB, Dao TP, Castaneda CA. 2019. Journal of Physical Chemistry B 123:3618–29 [DOI] [PubMed] [Google Scholar]
- 181.Vekilov PG. 2010. Soft matter 6:5254–72 [Google Scholar]
- 182.Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, et al. 2018. J Biol Chem 293:3734–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Šarić A, Buell AK, Meisl G, Michaels TCT, Dobson CM, et al. 2016. Nature Physics 12:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, et al. 2019. Molecular Cell 76:177–90.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mitrea DM, Grace CR, Buljan M, Yun M-K, Pytel NJ, et al. 2014. Proceedings of the National Academy of Sciences 111:4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, et al. 2016. The EMBO journal 35:1254–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pierce WK, Grace CR, Lee J, Nourse A, Marzahn MR, et al. 2016. Journal of molecular biology 428:1256–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bracha D, Walls MT, Wei M-T, Zhu L, Kurian M, et al. 2018. Cell 175:1467–80.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, et al. 2019. bioRxiv:809210 [Google Scholar]