Abstract
We tested whether post exercise ingestion of branched-chain amino acids (BCAA < 10 g) is sufficient to activate signaling associated with muscle protein synthesis and suppress exercise-induced activation of mechanisms associated with proteolysis in endurance-trained human skeletal muscle. Nine endurance-trained athletes performed a cycling bout with and without BCAA ingestion (0.1 g/kg). Post exercise ACCSer79/222 phosphorylation (endogenous marker of AMPK activity) was increased (~3-fold, P < 0.05) in both sessions. No changes were observed in IGF1 mRNA isoform expression or phosphorylation of the key anabolic markers — p70S6K1Thr389 and eEF2Thr56 — between the sessions. BCAA administration suppressed exercise-induced expression of mTORC1 inhibitor DDIT4 mRNA, eliminated activation of the ubiquitin proteasome system, detected in the control session as decreased FOXO1Ser256 phosphorylation (0.83-fold change, P < 0.05) and increased TRIM63 (MURF1) expression (2.4-fold, P < 0.05). Therefore, in endurance-trained human skeletal muscle, post exercise BCAA ingestion partially suppresses exercise-induced expression of PGC-1a mRNA, activation of ubiquitin proteasome signaling, and suppresses DDIT4 mRNA expression.
Keywords: Endurance exercise, Branched-chain amino acids, Proteolysis, Mitochondrial biogenesis
Introduction
Endurance training leads to increased skeletal muscle oxidative capacity, mitochondrial volume density, and aerobic performance. During acute aerobic exercise, various metabolites accumulate and the glycogen store is decreased in skeletal muscle. These changes activate various signaling cascades, including the AMP-activated protein kinase (AMPK)–peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α; also known as PPARGCA) pathway that plays an important role in regulating mitochondrial biogenesis. Activated PGC-1α translocates to the nucleus and regulates genes encoding mitochondrial transcription factors and other mitochondrial proteins as well as its own gene expression [1].
Aerobic training has been shown to induce muscle hypertrophy in both young and old untrained men [2]. However, a recent meta-analysis revealed that aerobic training-induced muscle hypertrophy and strength increments are significantly and negatively related to both frequency of endurance training (in the range from 1 to 5 days per week) and average duration of endurance sessions (in the range from 20 to 60 min per day) [3]. A further increase in frequency, duration and intensity of aerobic training typical for well-trained endurance athletes may not induce muscle hypertrophy or even decrease muscle mass. Indeed, high-intensity endurance training is reported to cause a decrease of the cross-sectional area of muscle fiber and muscle strength [4–6], which may have a negative effect on performance in elite endurance athletes. These changes appear to be related to a negative protein balance in endurance-trained muscle fibers [7]. The latter effect may be explained by an antagonistic relationship between the signaling pathways regulating mitochondrial biogenesis and protein synthesis [8] as well as endurance exercise-induced activation of the ubiquitin proteasome system [9–12].
Essential amino acids, in particular leucine, are broadly utilized to activate the mammalian target of rapamycin complex 1 (mTORC1), the key regulator of protein synthesis [13]. Earlier studies have demonstrated that leucine administration decreases the activities of markers of the ubiquitin proteasome system and prevents loss of soleus muscle mass during hindlimb immobilization in rats [14, 15].
Post-exercise protein-carbohydrate feeding leads to increased skeletal muscle anabolic signaling and fractional synthetic rate, reduced rate of protein breakdown after endurance exercise [16–19], and a marked increase in the insulin level in blood from ~5 to 30 mU/l. The ablation of hypoinsulinemia per se may suppress the protein breakdown rate and modulate anabolic signaling [20–22]. However, activation of the insulin signaling pathway could deactivate PGC-1α protein [23, 24] and theoretically suppress exercise-induced mitochondrial biogenesis.
Here, we tested the hypothesis that post exercise oral ingestion of branched-chain amino acids (BCAA < 10 g) that does not induce a marked insulin response is sufficient to activate signaling associated with muscle protein synthesis and suppress exercise-induced activation of signaling associated with proteolysis in endurance-trained human skeletal muscle. We administered a BCAA dose of 0.1 g/kg body mass (including 0.05 g/kg leucine) to achieve near-maximal stimulation of muscle protein synthesis [16, 25] and, on the other hand, avoid a marked rise in the insulin level in blood immediately after exercise. Feeding status substantially affects anabolic and proteolytic signaling and protein synthesis in skeletal muscle. The main objective of this investigation was to modulate post exercise anabolic and proteolytic signaling through administration of additional BCAA under normal feeding conditions typical for endurance athlethes.
Methods
Participants
Nine amateur endurance-trained athletes [18–30 years old, median (interquartile range) maximal oxygen consumption rate ( O2max) 57 (53–61) ml/kg/min, body mass 68 (66–74) kg] participated in this study. All participants were informed about the study design and provided their written consent for participation. The study was approved by the Human Ethics Committee of the Institute of Biomedical Problems (Moscow, Russia) and complied with the guidelines of the Declaration of Helsinki.
Preliminary experiments
All participants performed an incremental ramp test on an electromagnetic bicycle ergometer (Ergoselect 200; Ergoline) until exhaustion. Initial load, load increment and revolution rate were 0 W, 15 W/min, and 60–70 rpm, respectively. The pulmonary oxygen consumption rate ( O2) was measured using MetaMax 3B (Cortex). Capillary blood samples (20 µl) were obtained from a fingertip every 2 min, the blood lactate concentration evaluated using a Biosen C-line analyzer (EKF Diagnostics), and workload corresponding to the anaerobic threshold [AT] calculated at a lactate level of 4 mM. The highest O2 value for 30 s was taken as the O2max.
Main study
Each participant performed two test sessions in a random order: endurance exercise (E) and endurance exercise followed by BCAA ingestion (EA) with 1 week in between (Fig. 1). The exercise bout (70 min) consisted of a warm-up (5 min cycling at 50% AT) and intermittent exercise [(3 min cycling at 60% AT plus 2 min cycling at 95% AT) × 13]. In a previous study, muscle protein synthesis was shown to be upregulated 90–180 min after oral amino acid administration [26]. To stimulate muscle anabolic signaling over an extended period after exercise during the EA session, participants ingested 0.1 g/kg body mass of BCAA in the form of capsules (leucine, isoleucine, and valine [2:1:1], ~234 kJ; AST Sport Science) at two time-points: immediately after and 5 h after the end of the exercise session.
Fig. 1.
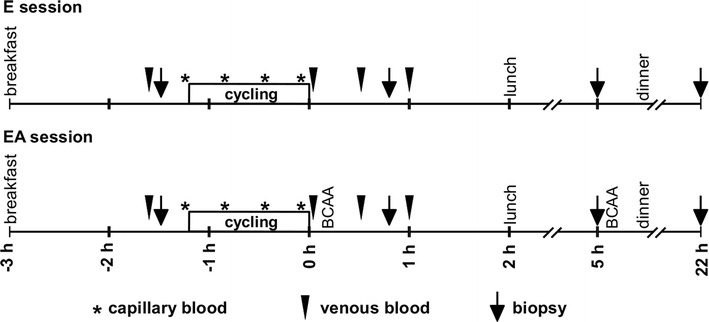
Overview of study design. Each participant performed two test sessions in a random order: endurance exercise (E) and endurance exercise followed by BCAA ingestion (EA) with 1 week in between
Participants arrived at the laboratory at 09:00 and consumed a standardized breakfast (3624 kJ; 24 g protein, 157 g carbohydrate, 15 g fat). Cycling began 1 h and 45 min after breakfast. A standardized lunch (4849 kJ; 37 g protein, 126 g carbohydrate, 67 g fat) was provided 2 h after the termination of cycling. After completing the experimental session, subjects went home and ate their usual dinner. Each subject returned to the laboratory the next morning after overnight fasting for the biopsy. Biopsy samples from m. vastus lateralis were obtained under local anesthesia (2 ml 2% lidocaine) via a microbiopsy technique [27] prior to and 40 min, 5 h, and 22 h after the termination of cycling. Muscle samples were quickly blotted with gauze to remove superficial blood, frozen in liquid nitrogen, and stored at −80 °C until further analysis. The first biopsy was performed 15 cm proximal to the condylus lateralis ossis femoris and subsequent biopsies taken 2-cm proximal to the previous one. Biopsy samples during another session were acquired from a different leg. The leg for the first biopsy was selected in a random order.
Venous blood samples were acquired from the v. intermedia cubiti via a catheter prior to, immediately after, and 30 and 60 min after the termination of cycling. Cortisol and insulin levels were evaluated via enzyme-linked immunosorbent assay (ELISA) using the ELISA-Cortisol Kit (ImmunoTek) and Insulin ELISA Kit (Mercodia). The lactate level was measured in capillary blood; samples were taken prior to, and at 20, 40 and 65 min during cycling.
RNA extraction
Frozen samples (~20 mg) were sectioned into 20-µm slices using an ultratome (Leica Microsystems) and RNA extracted using the RNeasy Mini Kit (Qiagen). RNA concentration and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific). After DNase treatment (Fermentas), cDNA was obtained from 1 μg of total RNA using the MMLV Reverse Transcriptase kit (Evrogen).
Real-time polymerase chain reaction
Real-time polymerase chain reaction (PCR) was performed using the Rotor-Gene Q cycler (Qiagen). The annealing temperature was optimized for each primer pair. The thermal profile included an initial heat denaturing step at 95 °C for 5 min, followed by 45 cycles of denaturation at 95 °C for 15 s, annealing (56–60 °C) for 30 s and extension at 72 °C for 30 s. Amplified genes were quantified via fluorescence using the EvaGreen Master Mix (Syntol). The specificity of amplification was monitored via melting curve analysis and agarose gel (1%) electrophoresis. Each sample was run in triplicate and a non-template control included in each run. Target gene mRNA expression levels were calculated using the efficiency-corrected ∆Ct method with the formula: . PCR efficiency (E) was calculated using standard curves corresponding to target and reference genes (RPLP0, GAPDH). Each standard curve included six points (diluted PCR product), with triplicate data obtained for each point. The primer sequences are shown in Table 1.
Table 1.
Primers used in this study
| Transcript | Strand | Sequence, 5′–3′ | Product size, bp |
|---|---|---|---|
| Total PGC-1α |
Forward Reverse |
CAGCCTCTTTGCCCAGATCTT TCACTGCACCACTTGAGTCCAC |
101 |
| NT-PGC-1α |
Forward Reverse |
TCACACCAAACCCACAGAGA CTGGAAGATATGGCACAT |
172 |
| TFAM |
Forward Reverse |
AGATTCCAAGAAGCTAAGGGTGATT TTTCAGAGTCAGACAGATTTTTCCA |
85 |
| TFB2 M |
Forward Reverse |
CAAGGAAGGCGTCTAAGGC AGCAGTAGGTGTGGAGGTC |
118 |
| CS |
Forward Reverse |
GAGAAGGCAGCGGTATTG AGGTAAGGGTCGGAAAGG |
196 |
| VEGFA |
Forward Reverse |
TACCTCCACCATGCCAAG GGTACTCCTGGAAGATGTC |
148 |
| TRIM63 (MURF1) |
Forward Reverse |
CTCAGTGTCCATGTCTGGAGGCCGTT GGCCGACTGGAGCACTCCTGTTTGTA |
147 |
| FBXO32 (Atrogin-1) |
Forward Reverse |
GTCCAAAGAGTCGGCAAGTC AGGCAGGTCAGTGAAGGTG |
147 |
| Myostatin |
Forward Reverse |
CATGATCTTGCTGTAACCTTCC CGATAATCCAATCCCATCC |
195 |
| CTSL |
Forward Reverse |
AAGTGGAAGGCTGCAATGGT CTCACACAGTGGGGTAGCTG |
483 |
| BNIP3 |
Forward Reverse |
TCTGGACGGAGTAGCTCCAA TTCATGACGCTCGTGTTCCT |
296 |
| IGF1-Ea |
Forward Reverse |
ATGCTCTTCAGTTCGTGTGTG GCACTCCCTCTACTTGCGTTC |
258 |
| IGF1-Ec (MGF) |
Forward Reverse |
ACCAACAAGAACACGAAGTC CAAGGTGCAAATCACTCCTA |
281 |
| DDIT4 (REDD1) |
Forward Reverse |
GGTTTGACCGCTCCACGAG ATCCAGGTAAGCCGTGTCTTC |
98 |
| RPLP0 |
Forward Reverse |
CACTGAGATCAGGGACATGTTG CTTCACATGGGGCAATGG |
77 |
| GAPDH |
Forward Reverse |
CAAGGTCATCCATGACAACTTTG GTCCACCACCCTGTTGCTGTAG |
496 |
Western blot
Frozen samples (~10 mg) were homogenized in ice-cold RIPA buffer containing protease and phosphatase inhibitors (50 mmol/l β-glycerophosphatase, 50 mmol/l NaF, 1 mmol/l Na3VO4, 20 μg/ml aprotinin, 50 μg/ml leupeptin, 20 μg/ml pepstatin and 1 mmol/l PMSF). Samples were centrifuged for 10 min at 10,000×g, at 4 °C. The protein content was analyzed with the bicinchoninic acid assay. Samples were mixed with Laemmli buffer (20 μg protein per lane) and loaded onto a 10% T polyacrylamide gel, and electrophoresis performed in the Mini-PROTEAN Tetra Cell system (Bio-Rad) at 20 mA per gel. Proteins were transferred onto nitrocellulose membranes using the Trans-Blot Turbo system (Bio-Rad) in Towbin buffer for 30 min at 25 V. Membranes were stained with Ponceau S to verify consistent loading of protein, followed by washing and incubation in 5% non-fat dry milk for 1 h. Next, membranes were incubated overnight at 4 °C with anti-p70S6K1 (sc-230), anti-phospho-FOXO1Ser256 (sc-101681), anti-p38 (sc-728), anti-phospho-AMPKα1/2Thr172 (sc-33524, all from Santa Cruz Biotechnology), anti-FOXO1 (ab52857), anti-phospho-p70S6K1Thr389 (ab2571), anti-phospho-ACCSer79/222 (ab68191), anti-phospho-eEF2Thr56 (ab115165), anti-eEF2 (ab33523), anti-phospho-p38Thr180+Tyr182 (ab4822), anti-AMPKα1/2 (ab80039) and anti-GAPDH (ab9485, all from Abcam).
The next day, membranes were incubated with HRP-linked anti-rabbit secondary antibody (Cell Signaling) for 1 h and washed with PBS–Tween 20 after each step (3 times for 5 min each). Following incubation of membranes with ECL substrate (Bio-Rad), luminescent signals were captured using the ChemiDoc Imaging System (Bio-Rad). Densitometry was performed using Image Lab 5.0 (Bio-Rad). All data are expressed as the ratio of phosphorylated protein to glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Statistical analysis
Data are expressed as median and interquartile range, since sample volumes were small (n = 9) with non-normal data distribution. To compare post exercise values with the initial levels, nonparametric ANOVA (Friedman’s test) with Dunn’s multiple comparison test was used. We investigated the differences between control (E) and experimental (EA) sessions (separately for every time-point) using the Wilcoxon matched-pairs test. The level of significance was 0.05. Statistical analysis was performed using GraphPad Prism 6 software.
Results
Physiological indices
Before cycling (immediately after baseline biopsy), the blood lactate levels were 2.6 (2.3–2.7) mmol/l in the E session and 2.4 (2.0–2.5) mmol/l in the EA session. The mean blood lactate levels during the cycling were 4.2 (3.5–5.4) and 3.5 (3.0–4.7) mmol/l in the E and EA sessions respectively, and did not differ between the sessions. The plasma insulin level was decreased immediately after cycling (0.14-fold change; P < 0.001) for all sessions, and maintained below 6 mU/l for the first hour after termination of cycling activity (Table 2). Notably, ingestion of BCAA immediately after cycling in the EA session did not lead to an increase in the insulin level (Table 2). The plasma cortisol level was not affected either by cycling or by the combination of cycling with BCAA (Table 2).
Table 2.
Blood insulin and cortisol levels before, immediately after, 30 and 60 min after the termination of intermittent cycling during the E and EA sessions
| Session | Before | After | 30 min | 60 min | |
|---|---|---|---|---|---|
| Insulin, mU/l | E | 26.7 (17.3–32.4) | 3.6* (2.0–6.5) | 5.2* (2.6–7.1) | 3.8* (2.8–6.7) |
| EA | 26.8 (15.9–43.6) | 3.80* (1.9–5.6) | 5.7 (3.9–17.8) | 5.1* (3.7–8.1) | |
| Cortisol, ng/ml | E | 155 (122–268) | 255 (201–351) | 229 (155–287) | 155 (118–245) |
| EA | 127 (97–217) | 312 (206–350) | 213 (125–240) | 134 (97–198) |
Each value represents the median and interquartile range
* Difference from the initial level (P < 0.05)
Regulation of mitochondrial biogenesis
AMP-activated protein kinase (AMPKα1/2) and p38 mitogen-activated protein kinase (p38) contents as well as p38Thr180+Tyr182 phosphorylation level were not altered during both sessions (data not shown). However, the AMPKα1/2Thr172 phosphorylation level was increased by a 1.6-fold change (P < 0.05; Fig. 2a, 5) at 40 min after termination of cycling in the EA experiment. The phosphorylation level of acetyl coenzyme A carboxylase (ACCSer79/222), an AMPK substrate used as an endogenous marker of its activity, was increased 40 min after cycling in both the E and EA sessions (by 3.5- and 3.1-fold changes, respectively, P < 0.05 for both; Fig. 2b, 5). Expression of total PGC-1α mRNA at 5 h after cycling exercise was increased to a higher extent in the E session (4.4-fold change, P < 0.05), compared to the EA session (Fig. 2c). Levels of N-truncated PGC-1α gene isoforms were increased in both the E and EA sessions (3.9- and 5.6-fold, respectively, P < 0.05 for both), with no significant differences between the sessions (Fig. 2d). In contrast, expression patterns of PGC-1α-related genes were distinct between the E and EA sessions. Expression of the mitochondrial transcription factor A (TFAM) gene was increased only after the E session by ~1.3-fold change at 22-h recovery (P < 0.05, Fig. 2e). Expression of the vascular endothelial growth factor A (VEGFA) gene increased in the E session (3.5- and 2.5-fold change at the 40-min and 5-h recovery, P < 0.05 for both, Fig. 2f), while mitochondrial transcription factor B2 (TFB2 M) and citrate synthase (CS) gene levels remained unchanged in both sessions (data not shown).
Fig. 2.
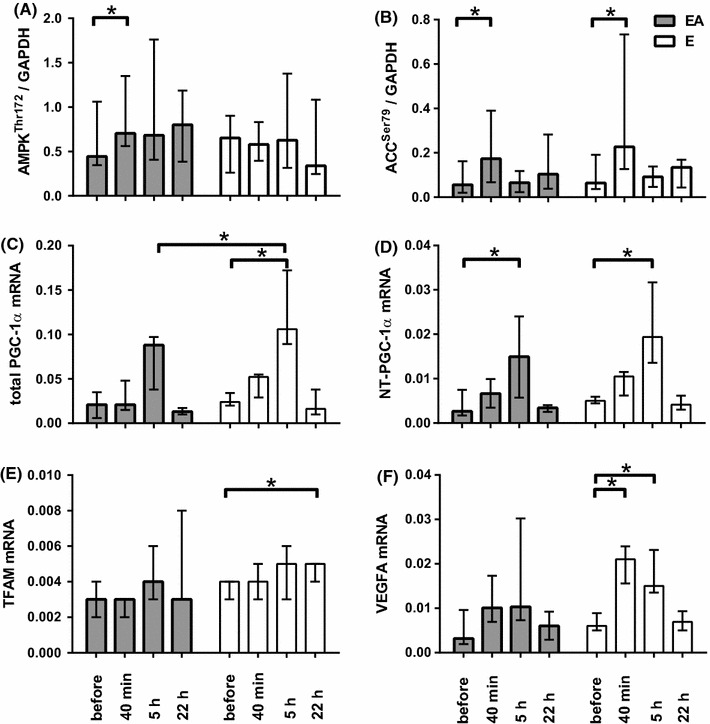
Proteins and genes related to the regulation of mitochondrial biogenesis in skeletal muscle before, 40 min, and 5 and 22 h after the termination of the endurance exercise (E) and endurance exercise followed by BCAA ingestion (EA). Phosphorylation levels of AMPKThr172 (a) and ACCSer79 (b) and mRNA abundance of total PGC-1α (c), NT-PGC-1α (d), TFAM (e), and VEGFA (f). Each target protein was normalized to GAPDH, each target mRNA was normalized to two reference genes (GAPDH and RPLP0), each value represents the median and interquartile range, *significant difference (P < 0.05)
Fig. 5.
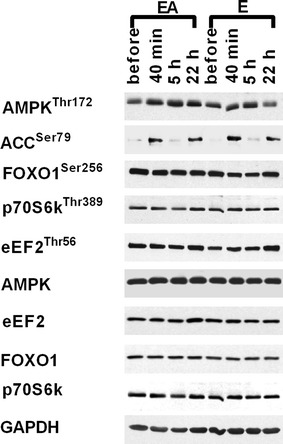
Representative immunoblots of phosphorylated proteins (AMPKThr172, ACCSer79, FOXO1Ser256, eEF2Thr56, p70S6 kThr389) and total proteins (AMPK, FOXO1, p70S6k, eEF2, GAPDH) before, 40 min, and 5 and 22 h after the termination of intermittent cycling during E and EA sessions
Regulation of proteolysis
The forkhead box O1 (FOXO1) content did not change during both sessions (data not shown). FOXO1Ser256 phosphorylation was decreased (0.83-fold change, P < 0.05, Fig. 3a, 5) up to 22 h of recovery in the E session. BCAA administration completely eliminated this cycling-induced decrease in FOXO1Ser256 phosphorylation and affected expression of the FOXO1 target genes, E3 ubiquitin ligases MURF1 and Atrogin-1. Expression of TRIM63 (tripartite motif containing 63, also known as MURF1) was increased (2.4-fold change, P < 0.05, Fig. 3b) in the E session at 40 min after termination of cycling. MURF1 mRNA levels between the E and EA sessions were significantly different (P < 0.05). Moreover, in contrast to the E session, BCAA ingestion induced a decrease (0.59-fold change, P < 0.05, Fig. 3c) in the expression of F-box protein 32 (FBXO32, also known as Atrogin-1) at 5 h after termination of cycling.
Fig. 3 .
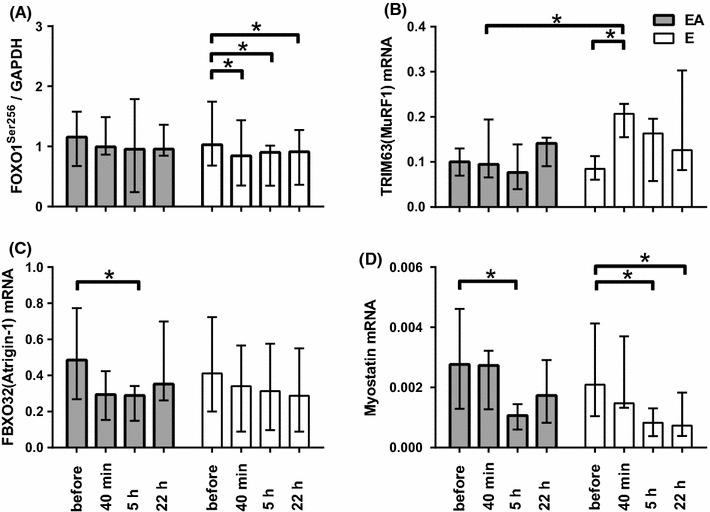
Proteins and genes related to regulation of proteolysis in skeletal muscle before, 40 min, and 5 and 22 h after the termination of endurance exercise (E) and endurance exercise followed by BCAA ingestion (EA). Phosphorylation level of FOXO1Ser256 (a) and mRNA abundance of TRIM63 [MURF1] (b), FBXO32 [Atrogin-1] (c), and Myostatin (d). Each target protein was normalized to GAPDH, each target mRNA was normalized to two reference genes (GAPDH and RPLP0), each value represents the median and interquartile range, *significant difference (P < 0.05)
Expression levels of the autophagic-lysosomal system-related genes, cathepsin L (CTSL) and BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), did not differ between the sessions (data not shown). The myostatin (MSTN) gene displayed significant decrease (more than 0.5-fold change; P < 0.05, Fig. 3d) in both exercise sessions, with no significant differences between the sessions.
Regulation of synthesis
The mRNA levels of insulin-like growth factor 1, isoform Ea (IGF1-Ea) and isoform Ec (IGF1-Ec, also known as mechano growth factor, MGF), potential regulators of myogenesis, were not different between the sessions (Fig. 4c, d). Additionally, p70 ribosomal protein S6 kinase 1 (p70S6K1) and eukaryotic elongation factor 2 (eEF2) contents remained unchanged in both sessions (data not shown). No changes were observed in the phosphorylation levels of the key anabolic markers, p70S6K1Thr389 or eEF2Thr56, between the sessions (Fig. 4a, b, 5). However, expression of DNA damage inducible transcript 4 (DDIT4, also known as REDD1), a negative regulator of mTORC1, at 40 min after termination of cycling in the EA session was lower (P < 0.05) than that in the E session (Fig. 4e).
Fig. 4.
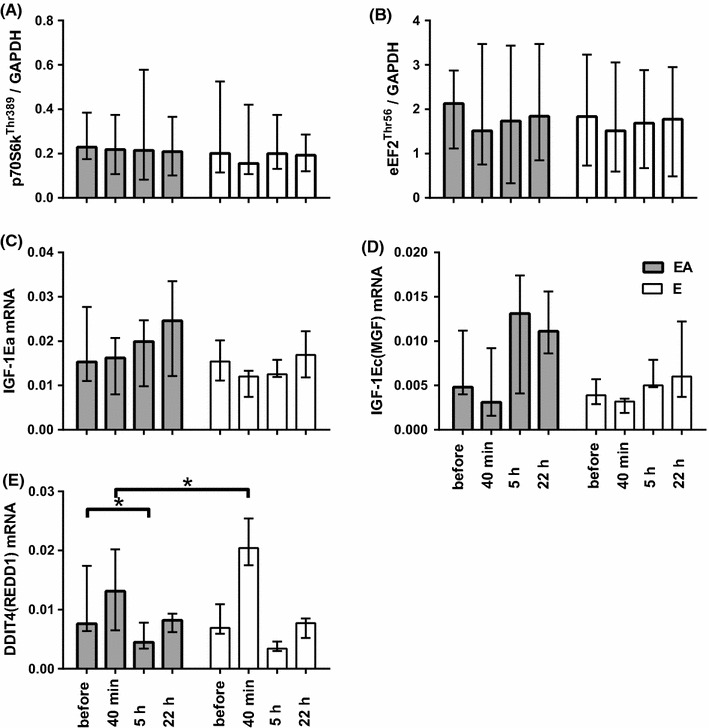
Proteins and genes related to regulation of protein synthesis in skeletal muscle before, 40 min, and 5 and 22 h after the termination of endurance exercise (E) and endurance exercise followed by BCAA ingestion (EA). Phosphorylation levels of p70S6K1Thr389 (a) and eEF2Thr56 (b), mRNA abundance of IGF1-Ea (c), IGF1-Ec [MGF] (d), and DDIT4 [REDD1] (e). Each target protein was normalized to GAPDH, each target mRNA was normalized to two reference genes (GAPDH and RPLP0), each value represents the median and interquartile range, *significant difference (P < 0.05)
Discussion
In the current study, we conducted intermittent cycling exercise tests, since this type of exercise induces a greater increase in PGC-1α and TFAM mRNA levels than continuous cycling [28]. Molecular response to acute aerobic exercise is more specific in endurance-trained skeletal muscle than untrained muscle [29–32]. Therefore, endurance-trained athletes were enrolled and only a few activated signaling proteins and genes involved in the regulation of mitochondrial biogenesis detected after the cycling bout. As expected, intermittent cycling (E session) activated AMPK (evaluated based on ACCSer79/222 phosphorylation) and increased the expression of total PGC-1α, N-truncated PGC-1α mRNA, and the PGC-1α-related genes TFAM and VEGFA. BCAA ingested after the cycling (EA session) had no effect on the exercise-induced activation of AMPK and expression of N-truncated PGC-1α mRNA, but suppressed TFAM, VEGFA, and total PGC-1α mRNA. However, the potential influence of these effects on mitochondrial biogenesis at the protein level remains to be established (Fig. 6).
Fig. 6.
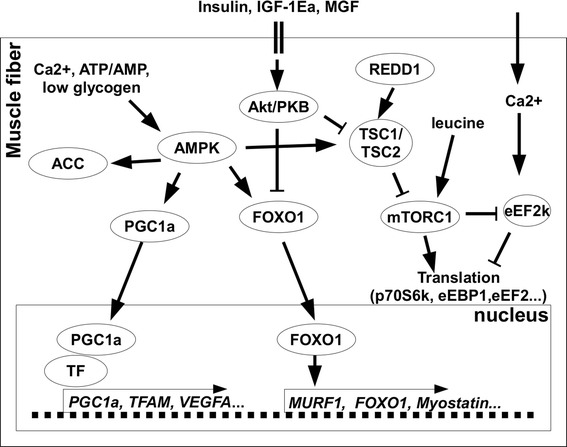
The proposed molecular mechanism for the interaction between mitochondrial biogenesis, protein synthesis and degradation in skeletal muscle of endurance trained individuals after high intensity endurance exercise combined with BCAA ingestion
We observed no changes in the levels of markers of the autophagic-lysosomal system (BNIP3 and CTSL) after exercise in both sessions. On the other hand, ubiquitin proteasome signaling was upregulated in response to E sessions, but not EA sessions. FOXO1Ser256 phosphorylation was decreased and the mRNA level of the E3 ubiquitin protein ligase, MURF1, increased after the exercise in E session. Our data are in accordance with findings from previous studies conducted on exercise-trained men [9–12]. These events may be evoked by a fall in the blood insulin level and consequent deactivation of Akt (V-Akt murine thymoma viral oncogene homolog). Indeed, suppression of insulin signaling has been shown to be associated with increased expression of E3 ubiquitin ligases in mouse skeletal muscle [33] and rat heart [34] while increase in blood insulin from ~5 to 30 mU/l induces a decrease in protein breakdown in humans [20, 22]. In our study, exercise-induced activation of ubiquitin proteasome signaling (determined based on decreased FOXO1Ser256 phosphorylation and increased MURF1 mRNA expression) was ablated following BCAA ingestion. This effect of BCAA on ubiquitin proteasome signaling is consistent with findings from studies on immobilized rat skeletal muscle [14, 15]. Expression of MURF1 is reported to be regulated via insulin [33, 34] as well as cortisol [35] and AMPK-related signaling [36, 37]. In our study, post exercise AMPK activation (evaluated based on the phosphorylation level of ACCSer79/222), as well as cortisol and insulin levels did not differ between sessions. Despite the lack of differences in the insulin levels at 30 min after exercise between sessions, several individuals in the EA group displayed insulin levels higher than 10 mU/l. We cannot exclude the possibility that this transient increase in insulin partially serves to suppress ubiquitin proteasome signaling. On the other hand, BCAA-dependent suppression of ubiquitin proteasome signaling may be related to other signaling mechanisms, for instance the activation of FOXO1-related kinase(s) and/or the inhibition of phosphatase(s). The inhibitory effect of BCAA on ubiquitin proteasome signaling supports the potential effectiveness of this approach in preventing muscle mass decrease induced by intense training in endurance athletes. Direct measurements of muscle protein breakdown are necessary to confirm this suggestion.
Amino acid administration modulates IGF1 gene expression [38, 39], anabolic signaling and the protein synthesis rate [26]. Contrary to our hypothesis, we observed no changes in IGF1-Ea and IGF1-Ec (MGF) mRNA isoform expression or phosphorylation levels of key anabolic markers (p70S6K1Thr389 and eEF2Thr56) during the EA session. Unexpectedly, BCAA administration had no effect on regulation of anabolic signaling. It might be related to metabolic perturbations in working muscle and in blood induced by intermittent endurance exercise in trained muscle. Amino acid administration under conditions of a low background blood insulin level (~5 mU/l) is reported to induce an increase in muscle protein synthesis rate without affecting the phosphorylation levels of p70S6K1Thr389 and eEF2Thr56 [21]. Therefore, we cannot exclude the possibility that BCAA administration affects muscle protein synthesis during the EA session.
DDIT4 (REDD1) may mediate attenuation of mTORC1 signaling via activation of hamartin-tuberin complex (TSC1/2), a negative regulator of mTORC1. DDIT4 mRNA and protein levels have been shown to increase in rat skeletal muscle immediately after an endurance exercise bout [40]. Murakami et al. (2011) reported that pre-exercise feeding of BCAA does not attenuate exercise-induced expression of both DDIT4 mRNA and protein in rat skeletal muscle [41]. In our experiments, DDIT4 gene expression was lower at 40 min after exercise in the EA session, compared with the E session. The discrepancies in findings between these studies may be attributable to the time of BCAA ingestion, i.e., before exercise in the study by Murakami and co-workers and just after exercise in our experiments.
Effects of BCAA in our study were observed mainly at 40 min and 5 h after the termination of exercise and first BCAA ingestion. The effect of BCAA ingestion on signaling and protein synthesis in human muscle lasts for a few hours [26, 42]. We think this finding may explain the lack of the effect of BCAA at 22 h of recovery in our work.
In conclusion, in endurance-trained human skeletal muscle, post exercise BCAA ingestion partially suppresses endurance exercise-induced expression of PGC-1a mRNA, activation of ubiquitin proteasome signaling (FOXO-1 phosphorylation, MURF1 mRNA expression), and suppresses DDIT4 mRNA expression, an inhibitor of mTORC1. Therefore, BCAA administration may present an effective tool for preventing muscle mass decrease induced by intense training in endurance athletes. Further evaluation of muscle protein synthesis and breakdown after acute exercise combined with BCAA and studies investigating the effects of long-term endurance training along with BCAA administration are necessary to assess and optimize the efficiency of this approach.
Acknowledgements
The authors are grateful to Arvo Leinsoo and Anton Bachinin for technical support and Dr. Dmitry Perfilov for tissue acquisition. This study was funded by the Russian Science Foundation (grant no. 14-15-00768, control experiment) and the Russian Foundation for Basic Research (Grant No. 14-04-01807a, BCAA experiment).
Author contributions
The study was designed by DVP, EAL and OLV. Data were collected and analyzed by EAL, TFV, EML and DVP. Data interpretation and manuscript preparation were undertaken by EAL, OLV, and DVP. All authors approved the final version of the paper.
Compliance with ethical standards
Conflict of interest
Evgeny A. Lysenko declares that he has no conflict of interest. Tatiana F. Vepkhvadze declares that she has no conflict of interest. Egor M. Lednev declares that he has no conflict of interest. Olga L. Vinogradova declares that she has no conflict of interest. Daniil V. Popov declares that he has no conflict of interest.
This study was funded by the Russian Science Foundation (Grant no. 14-15-00768, control experiment) and the Russian Foundation for Basic Research (Grant no. 14-04-01807a, BCAA experiment).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- 1.Olesen J, Kiilerich K, Pilegaard H. PGC-1alpha-mediated adaptations in skeletal muscle. Pflugers Arch. 2010;460:153–162. doi: 10.1007/s00424-010-0834-0. [DOI] [PubMed] [Google Scholar]
- 2.Konopka AR, Harber MP. Skeletal muscle hypertrophy after aerobic exercise training. Exerc Sport Sci Rev. 2014;42:53–61. doi: 10.1249/JES.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson JM, Marin PJ, Rhea MR, Wilson SM, Loenneke JP, Anderson JC. Concurrent training: a meta-analysis examining interference of aerobic and resistance exercises. J Strength Cond Res. 2012;26:2293–2307. doi: 10.1519/JSC.0b013e31823a3e2d. [DOI] [PubMed] [Google Scholar]
- 4.Kohn TA, Essen-Gustavsson B, Myburgh KH. Specific muscle adaptations in type II fibers after high-intensity interval training of well-trained runners. Scand J Med Sci Sports. 2011;21:765–772. doi: 10.1111/j.1600-0838.2010.01136.x. [DOI] [PubMed] [Google Scholar]
- 5.Kraemer WJ, Patton JF, Gordon SE, Harman EA, Deschenes MR, Reynolds K, Newton RU, Triplett NT, Dziados JE. Compatibility of high-intensity strength and endurance training on hormonal and skeletal muscle adaptations. J Appl Physiol. 1995;78:976–989. doi: 10.1152/jappl.1995.78.3.976. [DOI] [PubMed] [Google Scholar]
- 6.Terrados N, Melichna J, Sylven C, Jansson E. Decrease in skeletal muscle myoglobin with intensive training in man. Acta Physiol Scand. 1986;128:651–652. doi: 10.1111/j.1748-1716.1986.tb08026.x. [DOI] [PubMed] [Google Scholar]
- 7.Pikosky MA, Gaine PC, Martin WF, Grabarz KC, Ferrando AA, Wolfe RR, Rodriguez NR. Aerobic exercise training increases skeletal muscle protein turnover in healthy adults at rest. J Nutr. 2006;136:379–383. doi: 10.1093/jn/136.2.379. [DOI] [PubMed] [Google Scholar]
- 8.Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- 9.Stefanetti RJ, Lamon S, Wallace M, Vendelbo MH, Russell AP, Vissing K. Regulation of ubiquitin proteasome pathway molecular markers in response to endurance and resistance exercise and training. Pflugers Arch. 2015;467:1523–1537. doi: 10.1007/s00424-014-1587-y. [DOI] [PubMed] [Google Scholar]
- 10.Pagano AF, Py G, Bernardi H, Candau RB, Sanchez AM. Autophagy and protein turnover signaling in slow-twitch muscle during exercise. Med Sci Sports Exerc. 2014;46:1314–1325. doi: 10.1249/MSS.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 11.Luden N, Hayes E, Galpin A, Minchev K, Jemiolo B, Raue U, Trappe TA, Harber MP, Bowers T, Trappe S. Myocellular basis for tapering in competitive distance runners. J Appl Physiol (1985) 2010;108:1501–1509. doi: 10.1152/japplphysiol.00045.2010. [DOI] [PubMed] [Google Scholar]
- 12.Harber MP, Crane JD, Dickinson JM, Jemiolo B, Raue U, Trappe TA, Trappe SW. Protein synthesis and the expression of growth-related genes are altered by running in human vastus lateralis and soleus muscles. Am J Physiol Regul Integr Comp Physiol. 2009;296:R708–R714. doi: 10.1152/ajpregu.90906.2008. [DOI] [PubMed] [Google Scholar]
- 13.Drummond MJ, Dreyer HC, Fry CS, Glynn EL, Rasmussen BB. Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling. J Appl Physiol. 2009;106:1374–1384. doi: 10.1152/japplphysiol.91397.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baptista IL, Silva WJ, Artioli GG, Guilherme JP, Leal ML, Aoki MS, Miyabara EH, Moriscot AS. Leucine and HMB differentially modulate proteasome system in skeletal muscle under different sarcopenic conditions. PLoS ONE. 2013;8:e76752. doi: 10.1371/journal.pone.0076752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maki T, Yamamoto D, Nakanishi S, Iida K, Iguchi G, Takahashi Y, Kaji H, Chihara K, Okimura Y. Branched-chain amino acids reduce hindlimb suspension-induced muscle atrophy and protein levels of atrogin-1 and MuRF1 in rats. Nutr Res. 2012;32:676–683. doi: 10.1016/j.nutres.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Rowlands DS, Nelson AR, Phillips SM, Faulkner JA, Clarke J, Burd NA, Moore D, Stellingwerff T. Protein-leucine fed dose effects on muscle protein synthesis after endurance exercise. Med Sci Sports Exerc. 2015;47:547–555. doi: 10.1249/MSS.0000000000000447. [DOI] [PubMed] [Google Scholar]
- 17.Breen L, Philp A, Witard OC, Jackman SR, Selby A, Smith K, Baar K, Tipton KD. The influence of carbohydrate-protein co-ingestion following endurance exercise on myofibrillar and mitochondrial protein synthesis. J Physiol. 2011;589:4011–4025. doi: 10.1113/jphysiol.2011.211888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howarth KR, Moreau NA, Phillips SM, Gibala MJ. Coingestion of protein with carbohydrate during recovery from endurance exercise stimulates skeletal muscle protein synthesis in humans. J Appl Physiol. 2009;106:1394–1402. doi: 10.1152/japplphysiol.90333.2008. [DOI] [PubMed] [Google Scholar]
- 19.Ivy JL, Ding Z, Hwang H, Cialdella-Kam LC, Morrison PJ. Post exercise carbohydrate-protein supplementation: phosphorylation of muscle proteins involved in glycogen synthesis and protein translation. Amino Acids. 2008;35:89–97. doi: 10.1007/s00726-007-0620-2. [DOI] [PubMed] [Google Scholar]
- 20.Gelfand RA, Barrett EJ. Effect of physiologic hyperinsulinemia on skeletal muscle protein synthesis and breakdown in man. J. Clin. Invest. 1987;80:1–6. doi: 10.1172/JCI113033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenhaff PL, Karagounis LG, Peirce N, Simpson EJ, Hazell M, Layfield R, Wackerhage H, Smith K, Atherton P, Selby A, Rennie MJ. Disassociation between the effects of amino acids and insulin on signaling, ubiquitin ligases, and protein turnover in human muscle. Am J Physiol Endocrinol Metab. 2008;295:E595–E604. doi: 10.1152/ajpendo.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louard RJ, Fryburg DA, Gelfand RA, Barrett EJ. Insulin sensitivity of protein and glucose metabolism in human forearm skeletal muscle. J Clin Invest. 1992;90:2348–2354. doi: 10.1172/JCI116124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong T, Ning J, Yang X, Liu HY, Han J, Liu Z, Cao W. Fine-tuned regulation of the PGC-1alpha gene transcription by different intracellular signaling pathways. Am J Physiol Endocrinol Metab. 2011;300:E500–E507. doi: 10.1152/ajpendo.00225.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 25.Moore DR, Robinson MJ, Fry JL, Tang JE, Glover EI, Wilkinson SB, Prior T, Tarnopolsky MA, Phillips SM. Ingested protein dose response of muscle and albumin protein synthesis after resistance exercise in young men. Am J Clin Nutr. 2009;89:161–168. doi: 10.3945/ajcn.2008.26401. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell WK, Phillips BE, Williams JP, Rankin D, Lund JN, Smith K, Atherton PJ. A dose- rather than delivery profile-dependent mechanism regulates the “muscle-full” effect in response to oral essential amino acid intake in young men. J Nutr. 2015;145:207–214. doi: 10.3945/jn.114.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayot M, Michaud A, Koechlin C, Caron MA, LeBlanc P, Prefaut C, Maltais F. Skeletal muscle microbiopsy: a validation study of a minimally invasive technique. Eur Respir J. 2005;25:431–440. doi: 10.1183/09031936.05.00053404. [DOI] [PubMed] [Google Scholar]
- 28.Popov D, Zinovkin R, Karger E, Tarasova O, Vinogradova O. Effects of continuous and intermittent aerobic exercise upon mRNA expression of metabolic genes in human skeletal muscle. J Sports Med Phys Fitness. 2014;54:362–369. [PubMed] [Google Scholar]
- 29.Nielsen Jakob N., Mustard Kirsty J. W., Graham Drew A., Yu Haiyan, MacDonald Christopher S., Pilegaard Henriette, Goodyear Laurie J., Hardie D. Grahame, Richter Erik A., Wojtaszewski Jørgen F. P. 5′-AMP-activated protein kinase activity and subunit expression in exercise-trained human skeletal muscle. Journal of Applied Physiology. 2003;94(2):631–641. doi: 10.1152/japplphysiol.00642.2002. [DOI] [PubMed] [Google Scholar]
- 30.Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A, Spriet LL. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol. 2010;588:4795–4810. doi: 10.1113/jphysiol.2010.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu M, Stepto NK, Chibalin AV, Fryer LG, Carling D, Krook A, Hawley JA, Zierath JR. Metabolic and mitogenic signal transduction in human skeletal muscle after intense cycling exercise. J Physiol. 2003;546:327–335. doi: 10.1113/jphysiol.2002.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popov DV, Bachinin AV, Lysenko EA, Miller TF, Vinogradova OL. Exercise-induced expression of peroxisome proliferator-activated receptor gamma coactivator-1alpha isoforms in skeletal muscle of endurance-trained males. J. Physiol Sci. 2014;64:317–323. doi: 10.1007/s12576-014-0321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asp ML, Tian M, Wendel AA, Belury MA. Evidence for the contribution of insulin resistance to the development of cachexia in tumor-bearing mice. Int J Cancer. 2010;126:756–763. doi: 10.1002/ijc.24784. [DOI] [PubMed] [Google Scholar]
- 34.Paula-Gomes S, Goncalves DA, Baviera AM, Zanon NM, Navegantes LC, Kettelhut IC. Insulin suppresses atrophy- and autophagy-related genes in heart tissue and cardiomyocytes through AKT/FOXO signaling. Horm Metab Res. 2013;45:849–855. doi: 10.1055/s-0033-1347209. [DOI] [PubMed] [Google Scholar]
- 35.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. AMP-activated protein kinase agonists increase mRNA content of the muscle-specific ubiquitin ligases MAFbx and MuRF1 in C2C12 cells. Am. J. Physiol Endocrinol. Metab. 2007;292:E1555–E1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 37.Nakashima K, Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci Biotechnol Biochem. 2007;71:1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 38.Schurch MA, Rizzoli R, Slosman D, Vadas L, Vergnaud P, Bonjour JP. Protein supplements increase serum insulin-like growth factor-I levels and attenuate proximal femur bone loss in patients with recent hip fracture. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 1998;128:801–809. doi: 10.7326/0003-4819-128-10-199805150-00002. [DOI] [PubMed] [Google Scholar]
- 39.Pedrosa RG, Donato J, Jr, Pires IS, Tirapegui J. Leucine supplementation increases serum insulin-like growth factor 1 concentration and liver protein/RNA ratio in rats after a period of nutritional recovery. Appl Physiol Nutr Metab. 2013;38:694–697. doi: 10.1139/apnm-2012-0440. [DOI] [PubMed] [Google Scholar]
- 40.Hayasaka Miki, Tsunekawa Haruka, Yoshinaga Mariko, Murakami Taro. Endurance exercise induces REDD1 expression and transiently decreases mTORC1 signaling in rat skeletal muscle. Physiological Reports. 2014;2(12):e12254. doi: 10.14814/phy2.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami T, Hasegawa K, Yoshinaga M. Rapid induction of REDD1 expression by endurance exercise in rat skeletal muscle. Biochem. Biophys. Res. Commun. 2011;405:615–619. doi: 10.1016/j.bbrc.2011.01.078. [DOI] [PubMed] [Google Scholar]
- 42.Areta JL, Burke LM, Ross ML, Camera DM, West DW, Broad EM, Jeacocke NA, Moore DR, Stellingwerff T, Phillips SM, Hawley JA, Coffey VG. Timing and distribution of protein ingestion during prolonged recovery from resistance exercise alters myofibrillar protein synthesis. J Physiol. 2013;591:2319–2331. doi: 10.1113/jphysiol.2012.244897. [DOI] [PMC free article] [PubMed] [Google Scholar]


