Summary
Background
Prognosis is crucial for personalized treatment and surveillance suggestion of the resected non-small-cell lung cancer (NSCLC) patients in stage I-III. Although the tumor-node-metastasis (TNM) staging system is a powerful predictor, it is not perfect enough to accurately distinguish all the patients, especially within the same TNM stage. In this study, we developed an intelligent prognosis evaluation system (IPES) using pre-therapy CT images to assist the traditional TNM staging system for more accurate prognosis prediction of resected NSCLC patients.
Methods
20,333 CT images of 6371 patients from June 12, 2009 to March 24, 2022 in West China Hospital of Sichuan University, Mianzhu People's Hospital, Peking University People's Hospital, Chengdu Shangjin Nanfu Hospital and Guangan Peoples' Hospital were included in this retrospective study. We developed the IPES based on self-supervised pre-training and multi-task learning, which aimed to predict an overall survival (OS) risk for each patient. We further evaluated the prognostic accuracy of the IPES and its ability to stratify NSCLC patients with the same TNM stage and with the same EGFR genotype.
Findings
The IPES was able to predict OS risk for stage I-III resected NSCLC patients in the training set (C-index 0.806; 95% CI: 0.744–0.846), internal validation set (0.783; 95% CI: 0.744–0.825) and external validation set (0.817; 95% CI: 0.786–0.849). In addition, IPES performed well in early-stage (stage I) and EGFR genotype prediction. Furthermore, by adopting IPES-based survival score (IPES-score), resected NSCLC patients in the same stage or with the same EGFR genotype could be divided into low- and high-risk subgroups with good and poor prognosis, respectively (p < 0.05 for all).
Interpretation
The IPES provided a non-invasive way to obtain prognosis-related information from patients. The identification of IPES for resected NSCLC patients with low and high prognostic risk in the same TNM stage or with the same EGFR genotype suggests that IPES have potential to offer more personalized treatment and surveillance suggestion for NSCLC patients.
Funding
This study was funded by the National Natural Science Foundation of China (grant 62272055, 92259303, 92059203), New Cornerstone Science Foundation through the XPLORER PRIZE, Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), Clinical Medicine Plus X - Young Scholars Project, Peking University, the Fundamental Research Funds for the Central Universities (K.C.), Research Unit of Intelligence Diagnosis and Treatment in Early Non-small Cell Lung Cancer, Chinese Academy of Medical Sciences (2021RU002), BUPT Excellent Ph.D. Students Foundation (CX2022104).
Keywords: Self-supervised pre-training, Multi-task learning, Prognosis, CT image, Resected NSCLC
Research in context.
Evidence before this study
We searched PubMed up to July 7, 2023, for research articles containing the terms “(deep learning or convolutional neural network or artificial intelligence)” AND “(CT or MRI or PET)” AND “(prognosis or overall survival)” AND “NSCLC”, without date or language restrictions. Several studies have developed deep learning-based models to predict prognosis of NSCLC patients from CT images. However, most of these studies focused on evaluating prognosis of NSCLC patients in advanced stage. We further replaced the term “NSCLC” with “resected NSCLC in stage I-III”. We found no research been conducted to develop an AI-based model using CT images to evaluate prognosis for resected NSCLC patients in the same TNM stage, especially for the patients in stage I.
Added value of this study
To the best of our knowledge, this is the first AI-based study for predicting prognosis of stage I-III resected NSCLC patients as a complement of TNM staging system for more personalized prognosis evaluation in clinical practice. In this study, we developed an AI-based system to predict OS risk for resected NSCLC patients using self-supervised pre-training and multi-task learning, which could non-invasively obtain prognosis-related information from different patients before surgery. The IPES showed promising performance on OS risk prediction in multi-center cohorts. By adopting the IPES, the resected NSCLC patients in both stage I and stage II-III could be further stratified into low- and high-risk subgroups with significant difference. Furthermore, the IPES was able to significantly separate the resected EGFR mutant patients in stage I and II-III into low- and high-risk subgroups, respectively. Similar results were also achieved in resected EGFR wild-type patients in stage I and II-III.
Implications of all the available evidence
IPES provided a non-invasive way to obtain prognosis-related information from patients. The refined stratification of OS for resected NSCLC patients in stage I-III suggested that IPES could be helpful to identify patients who are most likely to derive benefit from corresponding treatment decision and surveillance suggestion, which showed potential to complement TNM stage in clinical practice.
Introduction
Non-small-cell lung cancer (NSCLC) is the prominent cause of cancer-related deaths worldwide. The tumor-node-metastasis (TNM) staging system for NSCLC is a vital predictor for determining treatment and prognosis.1 Pulmonary resection offers the potentially curative treatment and adjuvant therapy after complete resection is recommended in stage II-III but not stage I. However, the standard adjuvant cisplatin-based chemotherapy improves only 5.4% of 5-year overall survival (OS), according to the Lung Adjuvant Cisplatin Evaluation (LACE).2 Patients with exon 19 deletions and L858R point mutations in exon 21 are sensitive to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), but few EGFR-TKI adjuvant therapy clinical trials have shown improvement in OS.3 In addition, about 30% stage I patients experience tumor recurrence after tumor surgery while in patients with stage II-III, a portion of them free from recurrence even without adjuvant treatment.4 Therefore, it is challenging to accurately predict whether a patient will experience relapse based on clinical TNM stage alone.
Traditional statistical methods have been studied for prognosis analysis in the past, such as the Cox proportional-hazards model based on clinical and pathological characteristics.5, 6, 7, 8 However, patients with similar clinical-pathological features still exhibit varied prognosis. With the development and improvement of molecular biological techniques, many genome-based prognostic signatures including biomarkers such as favorable prognostic factors EGFR, BCL-2 and adverse prognostic factors KRAS, Ki67, HER2, and p53,9, 10, 11 have been developed for NSCLC.12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Recent studies have identified genomic factors and some other factors as potential biomarkers of OS in patients with resected NSCLC, including driver gene mutations, mRNA, miRNA, methylation in cell-free DNA, Tumor lymphocytic infiltration, PD-L1, minimal residue disease (MRD), circulating tumor cells (CTCs) and multi-omics.10,19,22, 23, 24, 25, 26, 27 However, these methods require gene sequencing of biopsied tumor tissues, leading to challenges such as a lack of overlap and repeatability, high costs, and a lack of standardized detection methods. Therefore, few of them has been widely used into real clinical utility.
Computed tomography (CT) has emerged as a non-invasive tool in cancer diagnosis and prognosis evaluation of NSCLC,28, 29, 30, 31, 32, 33, 34 due to its ability to provide scope and location information of lung lesions and morphological manifestation to guide the treatment decision.35 Related studies, including quantitative radiomic-based methods,33,34 and deep learning-based methods,28, 29, 30, 31, 32,36 have shown promising potential in extracting information from CT images. However, radiomic methods are composed of two complex steps including hand-crafted image features extraction and machine learning model training using the selected features. In contrast, deep learning-based methods can integrate feature extraction and training process into a unified step. Recently, deep learning has achieved tremendous success in CT-based medical image analysis of prognosis evaluation, patient stratification and therapeutic decision.28, 29, 30,32,37 However, previous studies exploit deep neural networks without pre-training or pre-trained on large amounts of natural images. How to make full use of the large unlabeled CT data remains challenging. In recent study, self-supervised learning-based pre-training has shown the potential to learn general representations for disease diagnosis by utilizing unlabeled data,38 which inspired us self-supervised pre-training may also have the ability to provide medical knowledge for prognosis prediction. In addition, prognosis prediction was traditionally treated as a single-task problem in medical analysis.39 However, some medical tasks, such as EGFR genotype prediction or cancer stage prediction, also have been proven to be correlated with prognosis.29 Thus, integrating prognosis-related tasks in a unified multi-task learning model may improve the prognostic prediction performance as the medical knowledge from different tasks can be shared.
In this study, we developed an intelligent prognosis evaluation system (IPES) for resected NSCLC patients in stage I-III. Our approach involved introducing a CT-based Contrastive Learning of Representations (CTCLR) framework based on self-supervised learning, which can generate a pre-trained visual encoder using large amounts of unlabeled CTs from physical examination, thereby learning rich medical knowledge from CT imaging. To the best of our knowledge, we are the first to develop a self-supervised pre-training framework for prognosis prediction. By adopting the pre-trained visual encoder from CTCLR, we then developed a multi-task learning-based network, combining prognosis prediction with other related tasks, to predict a multi-task learning-based survival risk (MTL-score) and combined it with clinical characteristics to generate a final survival risk, called IPES-score. Finally, we validated the prognosis prediction performance in both internal and external datasets and explored whether CTCLR and multi-task learning could improve the accuracy of prognosis prediction. We also validated whether patients in the same cancer stage could be further stratified into low- and high-risk subgroups with significant difference by adopting IPES-score.
Methods
Ethics statement
The study complies with the declaration of Helsinki and Chinese laws and regulations. Approval was granted by the institutional ethics committees with the approve number of 2020 (232). The study will respect the rights of participants, and written informed consent was obtained from all participants.
Study design and patients
In this study, we incorporated 20,333 CT scans of 6371 patients from five independent institutions, including West China Hospital of Sichuan University, Mianzhu People's Hospital, Peking University People's Hospital, Chengdu Shangjin Nanfu Hospital, and Guangan People's Hospital between June 12, 2009 and March 24, 2022. The detailed inclusion and exclusion criterion are presented in the appendix (see the Inclusion and exclusion criteria in the appendix and Supplementary Figure S2).
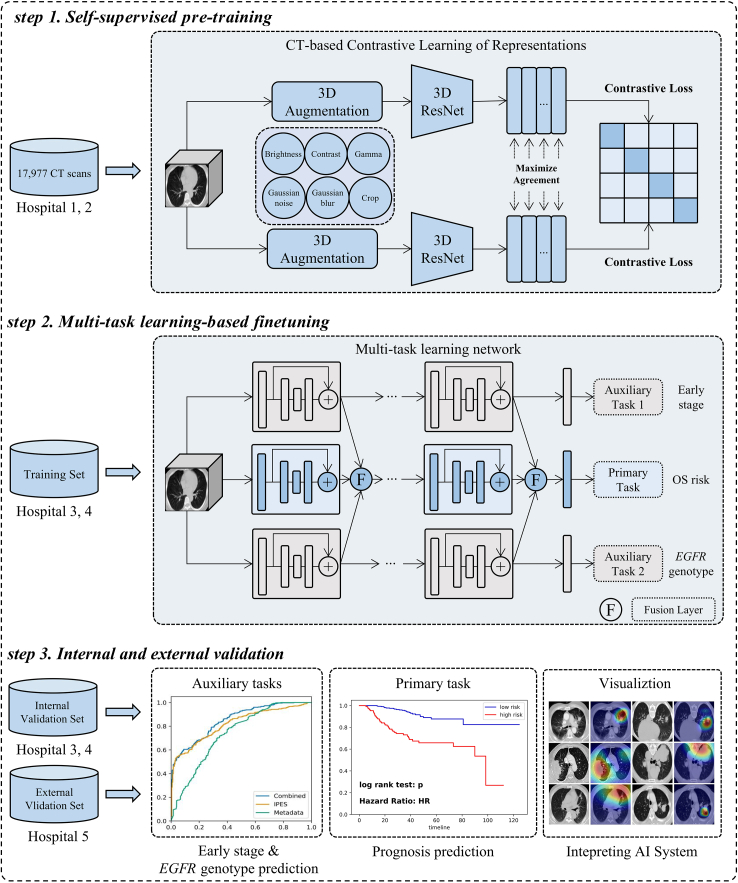
Fig. 1 displays the study design. Firstly, we incorporated patients with 17,977 CT images underwent physical examination at West China Hospital of Sichuan University and Mianzhu People's Hospital, to develop the CTCLR pre-training framework aiming at learning general visual representations from a large number of CT images without follow-up data. Then, we developed a multi-task learning network exploiting the pre-trained visual encoder from CTCLR and performed validation, using 2356 NSCLC patients with follow-up data and underwent CT examination before initial treatment. In this study, cisplatin-based chemotherapy is the main adjuvant therapy for all 2356 patients. For some patients with EGFR mutation, they also received first-generation EGFR-TKI targeted as adjuvant therapy in addition to cisplatin-based chemotherapy. The patients from Peking University People's Hospital and Chengdu Shangjin Nanfu Hospital were collected to generate the training (n = 1177) and internal validation (n = 711) sets by dividing the non-overlapping participants. Furthermore, 468 NSCLC patients from the Guangan People's Hospital were employed for the external validation set. The pre-trained multi-task learning network was fine-tuned on the training set and validated on the internal and external validation sets. To increase the model's generalizability, we incorporated all stage I-IV patients during the model development phase. For validation, we incorporated only data from stage I-III resected NSCLC patients in the internal and external sets for prognosis prediction validation. Table 1 includes detailed characteristics of the 2356 patients in the training, internal validation, and external validation sets. Of 1441 stage I-III patients underwent curative-intent surgery, with 881 at stage I and 560 at stage II-III. Of the 881 patients at stage I, 641 had EGFR mutation genotype, whereas 240 had EGFR wild-type status. Of those at stage II-III, 287 had EGFR mutation genotype and 273 had EGFR wild-type status. The detailed characteristics of the stage I-III resected NSCLC patients are further listed in the appendix (Supplementary Table S1).
Fig. 1.
Study design for the development and validation of the IPES. IPES consists of self-supervised pre-training, multi-task learning-based finetuning, and internal and external validation.
Table 1.
Patient characteristics in the training, internal validation and external validation sets.
| Training set (n = 1177) | Internal validation set (n = 711) | External validation set (n = 468) | p-value | |
|---|---|---|---|---|
| Complete resection | 0.18 | |||
| Yes | 757 (64.3%) | 474 (66.6%) | 321 (68.5%) | |
| No | 420 (35.7%) | 237 (33.4%) | 147 (31.5%) | |
| Sex | 0.26 | |||
| Male | 565 (48.0%) | 368 (51.7%) | 217 (46.3%) | |
| Female | 612 (52.0%) | 343 (48.3%) | 251 (53.7%) | |
| Age, years | 58 (48–69) | 59 (49–69) | 58 (48–67) | 0.085 |
| Smoking status | 0.96 | |||
| Former | 428 (36.3%) | 281 (39.5%) | 177 (37.8%) | |
| Never | 749 (63.7%) | 430 (60.5%) | 291 (62.2%) | |
| Cancer family history | 0.81 | |||
| Yes | 66 (5.6%) | 51 (7.1%) | 31 (6.6%) | |
| No | 1111 (94.4%) | 660 (92.9%) | 437 (93.4%) | |
| Tumor family history | 0.65 | |||
| Yes | 135 (11.4%) | 79 (11.1%) | 49 (10.4%) | |
| No | 1042 (88.6%) | 632 (88.9%) | 419 (89.6%) | |
| Histology | 0.53 | |||
| Adenocarcinoma | 1026 (87.1%) | 617 (86.7%) | 413 (88.2%) | |
| Others | 151 (12.9%) | 94 (13.3%) | 55 (11.8%) | |
| Stage | 0.15 | |||
| I | 447 (37.9%) | 270 (37.9%) | 195 (41.6%) | |
| II-III | 353 (30.0%) | 205 (28.8%) | 142 (30.3%) | |
| IV | 377 (32.0%) | 236 (33.1%) | 131 (27.9%) | |
| EGFR genotype | 0.57 | |||
| Mutant | 730 (62.0%) | 453 (63.7%) | 286 (61.1%) | |
| Wild-type | 447 (38.0%) | 258 (36.3%) | 182 (38.9%) | |
| Death status | 0.65 | |||
| Dead | 358 (30.4%) | 205 (28.8%) | 134 (28.6%) | |
| Censored | 819 (69.6%) | 506 (71.2%) | 334 (71.4%) | |
Data are n (%) or mean (SD). EGFR = epidermal growth factor receptor. Cancer family history = family history of lung cancer. Tumor family history = family history of other cancers (excluding lung cancer).
The adjusted p-value was provided in the last column, which is used to indicate the difference between the external validation set with the combined training and internal validation sets.
CT image preprocessing
In clinical practice, CT scanning protocols varies among different hospitals, thereby leading to the highly variable CT slice thickness, which has the large effect on deep learning-based methods.40 Besides, a CT image contains not only the lung parenchyma but also other distractors, which may hinder the attention of convolution neural network to the lesions. Thus, we preprocessed the CT images to reduce the search space from the whole CT scans to the region of interest (ROI) which are most likely to have lesions.
Our image pre-processing procedure is illustrated in Supplementary Figure S3, which consists of five major steps including lung mask extraction, Hounsfield Unit (HU) conversion, image resampling, intensity normalization and ROI generation. Firstly, we followed,41 to extract the left and right lung masks from the CT scan slice by slice, using ResUNet.42 Secondly, the raw CT matrix is clipped into HU with the range of [−1,000, 400]. HU is a measure of radiodensity. The same tissue of different people has the same HU range. Thirdly, all the CT images and their lung masks are resampled with the pixel spacing of [1, 1, 1], which resamples the patients’ pixels to an isomorphic resolution. Then, we linearly transformed the CT matrix from [−1,000, 400] to [0, 1] to prepare the data for our IPES. Finally, we cropped the CT along the bounding box of the lung according to the generated lung mask to obtain the ROI and resized it with the size of 64 × 256 × 256.
Development of the AI system
As Fig. 1 shown, we developed the IPES for predicting the prognosis of stage I-III resected NSCLC patients, which consisted of three steps, including self-supervised pre-training, multi-task learning-based finetuning, and internal and external validation.
In the first step, we developed CTCLR, a self-supervised pre-training framework based on contrastive learning, which learned general visual representations from large amounts of unlabeled 3D CT scans by distinguishing positive CT pairs against negative ones. Specifically, given a 3D CT image from a minibatch of N samples, the image is firstly transformed into two correlated views by using the 3D image augmentation strategies, thereby resulting in 2N augmented data points. The augmented views from the same CT image are denoted as a positive pair, whereas those from different CTs are denoted as negative pairs. Thus, we obtained one positive pair and 2(N − 1) negative examples from 2N augmented samples. Second, a state-of-the-art architecture, 3D ResNet,43 was employed as the feature extractor to extract feature representations from augmented CT images. Third, non-linear projection was conducted to project feature representations into the latent space by using the multi-layer perceptron (MLP). Finally, we exploited the normalized temperature-scaled cross entropy loss (NT-Xent),44 as the contrastive loss and applied it into 2N augmented data points to maximize the similarity of positive pairs and minimize the similarity of negative pairs. Through CTCLR, we obtained a pre-trained 3D ResNet with rich knowledge in CT imaging, which may be useful in prognosis prediction. A more detailed summary of the implementations of CTCLR is provided in the appendix (see the CT-based self-supervised pre-training framework using contrastive learning in the appendix).
In the second step, we developed a multi-task deep learning network and employed the pre-trained 3D ResNet,43 from CTCLR, as the backbone to predict patients' OS risk. The multi-task learning network consisted of three branches: two for auxiliary tasks (early-stage (stage I) and EGFR genotype prediction), and one for the primary task of predicting a survival risk score called MTL-score, where higher scores indicated increased risk of cancer progression and shorter OS. Each branch is a pre-trained 3D ResNet transferred from CTCLR. Specifically, given a 3D CT image, our multi-task learning network generated three predicted value, including MTL-score, the probability of EGFR mutation and the probability of early stage. Given the association between EGFR genotype, cancer stage, and prognosis, we hypothesized that combining the tasks and sharing feature representations could enhance the primary survival task's accuracy. Therefore, we designed a fusion layer to share features from different branches at multi-scale levels. In each fusion layer, we concatenated the feature maps from three branches as a 3D tensor and input it into two convolution layers. By adopting fusion layer at each scale, feature representations from different tasks were shared and fused, which might provide primary task with more useful prognostic knowledge. More detailed description of the proposed multi-task learning network is provided in the appendix (see the Multi-task learning network in the appendix).
To achieve a more accurate prognostic stratification and assist the traditional TNM staging system, we combined MTL-score with TNM stage using the Cox proportional hazards model to produce a final survival risk score, called IPES-score. This risk score allowed patients at the same stage to be divided into low- and high-risk subgroups, thus offering more personalized treatment and surveillance suggestion for patients.
In the third step, we evaluated the performance of our IPES on internal and external validation sets. Besides, to identify the key areas in CT images that IPES focused on for survival prediction, we employed gradient-weighted class activation mapping (Grad-CAM) algorithm,45 to visualize the model's detected characteristics.
Performance evaluation of IPES on primary task of prognosis prediction
We assessed the accuracy of IPES for predicting OS risk for stage I-III resected NSCLC patients using the Concordance index (C-index). OS was calculated from the date of intervention to the date of death. Additionally, the significance of MTL-score predicted by multi-task learning network was also validated using univariable and multivariable survival analysis with the Cox proportional hazard model to be compared with other clinical metadata. Prognostic stratification performance was assessed in the training, internal validation, and external validation sets. The cutoff IPES-score was selected in the training set by using X-tile software (Version 3.6.1), to stratify patients into low- and high-risk subgroups. The same cutoff value was used in the validation sets. Kaplan–Meier analysis and log-rank test was applied to evaluate the difference significance between the predicted low- and high-risk subgroups. We also compared the prognostic performance of TNM staging system with the IPES. Furthermore, we evaluated the accuracy improvement of IPES brought by self-supervised pre-training and multi-task learning.
Performance evaluation of IPES on auxiliary tasks
We evaluated the IPES performance on auxiliary tasks (early-stage and EGFR genotype prediction) by using the receiver operating characteristic (ROC) analysis. Evaluation metrics, including the area under the ROC curve (AUC), accuracy, sensitivity and specificity were computed. We compared IPES with clinical metadata-based model using random forest classifier. The metadata-based model used clinical factors, including age, sex, TNM stage, smoking status, histology, cancer family history and tumor family history. For early-stage prediction task, the same clinical metadata was incorporated, but TNM stage was excluded. To further validate the accuracy improvement by IPES, we combined clinical metadata with the corresponding deep learning score of IPES, using random forest classifier.
Statistical analysis
To evaluate the performance of IPES for auxiliary tasks, ROC curves were plotted. The evaluation metrics including AUC, accuracy, sensitivity, specificity with 95% CI were all computed. To evaluate the performance of IPES on primary task for prognosis prediction, C-index was computed. Kaplan–Meier analysis and log-rank test were employed to evaluate whether OS of the low- and high-risk subgroups identified by IPES had significant difference. The statistical results were considered significant when p-value was less than 0.05. Besides, the hazard ratio (HR) with the corresponding 95% confidence interval (95% CI) was computed. Univariate and multivariate survival analysis were also conducted using Cox proportional hazard model. In the multivariate survival analysis, all clinicopathological variables were used. Furthermore, the Cox proportional hazard assumptions were also evaluated. Sample size evaluation was performed using statesmodel (Version: 0.14.0) on Python (Version: 3.9.1).
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to the data in the study and had final responsibility for the decision to submit the paper for publication.
Results
A summary of the demographic variables and clinical characteristics of the training, internal validation and external validation sets is provided in Table 1. There were no statistically significant differences in these variables and characteristics between the datasets (p > 0.05).
As NSCLC tumors exhibit heterogeneous biological behavior wherein some patients experience long-term survival after surgical resection, while others exhibit early disease progression, it is crucial to design a prognostic tool for more precise, individualized survival estimates. In this study, we developed the IPES to investigate its performance in predicting OS risk of patients using self-supervised pre-training and multi-task learning.
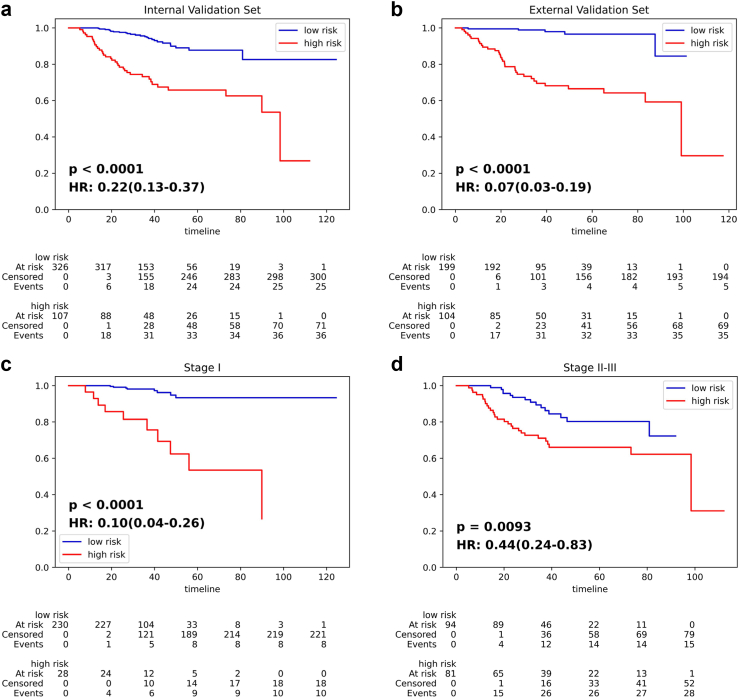
Fig. 2 displays the Kaplan–Meier survival curves that are used to stratify the NSCLC patients according to the IPES-score. Significant separation was achieved between the low- and high-risk subgroups for stage I-III resected NSCLC patients in both the internal and external validation sets (all p < 0.0001, Fig. 2(a) and (b)). The C-index of the IPES on predicting OS risk for stage I-III resected NSCLC patients was 0.806 (95% CI: 0.744–0.846) in the training set, 0.783 (95% CI: 0.744–0.825) in the internal set and 0.817 (95% CI: 0.786–0.849) in the external set (Supplementary Table S2). Furthermore, we validated the prognostic value of MTL-score predicted by IPES. Univariable and multivariable survival analysis using Cox proportional hazard model were conducted to demonstrate the significance of MTL-score compared with other clinical metadata. We found that MTL-score remained a significant prognostic predictor for OS risk prediction in patients after complete resection in stage I-III (p < 0.05; Table 2). When we combined the MTL-score with TNM stage using Cox proportional hazard model, IPES improved the C-index value from 0.733 (95% CI: 0.684–0.788) to 0.783 (95% CI: 0.744–0.825; Supplementary Table S3), which validated the prognostic value of MTL-score predicted by IPES using CT images. The Cox proportional hazard assumption evaluation indicated that IPES-score did not violate the proportional hazard assumption (p = 0.64 for training set, p = 0.40 for internal set, and p = 0.59 for external set). Furthermore, MTL-score and TNM stage demonstrated substantial statistical significance in both univariate and multivariate analyses, implying association with survival outcomes.
Fig. 2.
IPES performance on predicting OS risk of stage I-III resected NSCLC patients. (a) and (b) Kaplan–Meier curves of the low- and high-risk subgroups predicted by IPES for stage I-III resected NSCLC patients in the (a) internal validation set and (b) external validation set. (c) and (d) Kaplan–Meier curves of the low- and high-risk subgroups predicted by IPES for resected NSCLC patients in (c) stage I and (d) stage II-III in the internal validation set. In the Kaplan–Meier curves, the horizontal axis represents survival time (months) and the vertical axis represents survival probability.
Table 2.
Univariate and multivariate survival analysis for OS using Cox proportional hazards methods in the internal validation set.
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p-value | Hazard ratio (95% CI) | p-value | |
| MTL-score | 2.42 (1.78–3.29) | <0.0001 | 1.86 (1.33–2.59) | 0.00028 |
| TNM stage | 2.04 (1.53–2.72) | <0.0001 | 2.01 (1.49–2.71) | <0.0001 |
| Age | 1.06 (1.03–1.09) | <0.0001 | 1.05 (1.02–1.08) | 0.00038 |
| Sex | 1.55 (0.93–2.57) | 0.090 | 1.40 (0.68–2.90) | 0.36 |
| Smoking status | 1.12 (0.68–1.83) | 0.67 | 0.62 (0.30–1.29) | 0.20 |
| Cancer family history | 0.71 (0.26–1.95) | 0.50 | 1.17 (0.41–3.31) | 0.77 |
| Tumor family history | 1.46 (0.74–2.87) | 0.28 | 1.51 (0.75–3.05) | 0.25 |
| Histology | 1.34 (1.08–1.66) | 0.0085 | 1.26 (0.95–1.68) | 0.11 |
MTL-score = multi-task learning-based survival score. TNM = tumor-node-metastasis. CI = confidence interval.
Furthermore, we hypothesized that incorporating self-supervised pre-training and multi-task learning could improve the prognosis prediction performance. We compared the IPES with two baselines: (1) a multi-task learning network without self-supervised pre-training, (2) a single task learning network without self-supervised pre-training, both with the same backbone as IPES. Compared with the single-task learning network, multi-task learning improved the accuracy of C-index from 0.741 (95% CI: 0.695–0.798) to 0.765 (95% CI: 0.720–0.816). When we transferred the network weights from CTCLR into the down-stream multi-task learning network and fine-tuned it on the training set, the value of C-index was improved to 0.783 (95% CI: 0.744–0.825; Supplementary Table S4).
According to the guideline, postoperative treatment recommendations are different between stage I and stage II + resected patients. Extensive results showed that our IPES had good performance for the auxiliary task of predicting early-stage, which achieved AUC values of 0.804 (95% CI: 0.767–0.831) in the internal validation set and 0.837 (95% CI: 0.816–0.863) in the external validation set (Supplementary Figure S4 and Supplementary Table S5). When we combined the clinical metadata with the deep learning score predicted by IPES, the AUC values of metadata-based model could be further improved in both the internal and the external validation sets. Therefore, we further utilized IPES for prognostic prediction on stage I and II-III resected NSCLC patients respectively. For patients after complete resection in stage I, IPES was able to stratify them into two significantly different groups (p < 0.0001, HR = 0.10, 95% CI: 0.04–0.26; Fig. 2(c)), with the high-risk group showing a higher 5-year death rate of 32.1%, whereas the low-risk subgroup showed a lower 5-year death rate of 3.5% (Supplementary Table S6). The similar performance was also achieved for the patients after curative-intent surgery in stage II-III: the low- and high-risk subgroups stratified by IPES achieved significantly difference in their OS (p = 0.0093, HR = 0.44, 95% CI: 0.24–0.83; Fig. 2(d)).
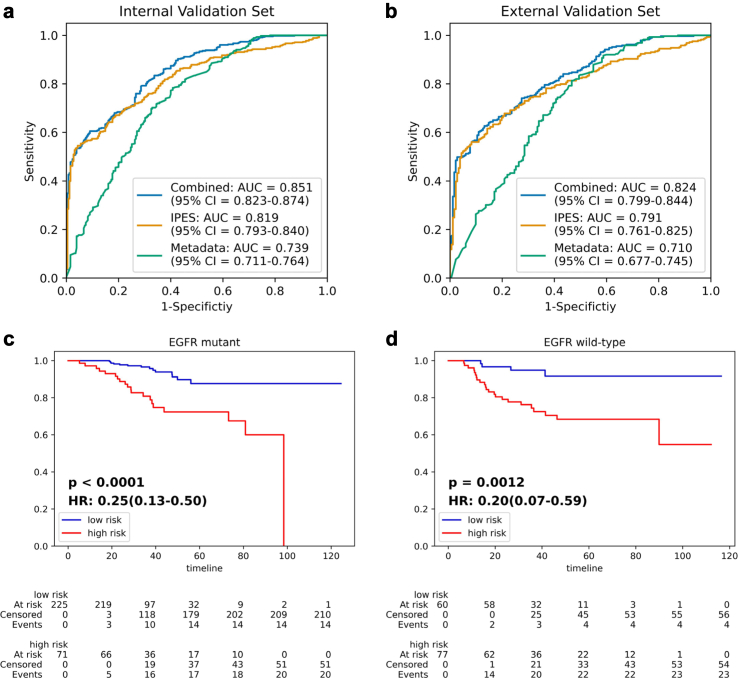
Presently, the administration of EGFR-targeted therapy is determined by EGFR genotype, which indicates the importance of EGFR genotype detection. Beyond primary task on prognosis prediction, IPES also achieved good performance on auxiliary task for EGFR genotype prediction. For predicting EGFR genotype, IPES obtained AUC values of 0.819 (95% CI: 0.793–0.840) on internal validation set and 0.791 (95% CI: 0.761–0.825) on external validation set, respectively (Fig. 3(a) and (b), Supplementary Table S7). When we combined the clinical metadata with the deep learning score predicted by IPES, the AUC values for predicting EGFR genotype could be further improved to 0.851 (95% CI: 0.823–0.874) on internal validation set and 0.824 (95% CI: 0.799–0.844) on external validation set, which proved the accuracy improvement brought by the image-based model IPES. When validated the prognosis performance of IPES in EGFR mutant and wild-type patients separately according to the IPES-score, we found that IPES achieved a significant difference in OS when comparing EGFR mutant and wild-type patients with low-versus high-risk subgroups in internal and external validation sets (Fig. 3(c) and (d) and Supplementary Figure S5). The C-index of the IPES on predicting OS risk for stage I-III resected NSCLC patients with EGFR mutation was 0.824 (95% CI: 0.777–0.868) in the training set, 0.778 (95% CI: 0.723–0.824) in the internal set and 0.803 (95% CI: 0.774–0.837) in the external set. For the stage I-III resected NSCLC patients with EGFR wild-type status, IPES achieved C-index value of 0.760 (95% CI: 0.714–0.809) in the training set, 0.762 (95% CI: 0.726–0.802) in the internal validation cohort and 0.793 (95% CI: 0.768–0.827) in the external set (Supplementary Table S8).
Fig. 3.
IPES performance on EGFR genotype prediction and prognosis analysis of stage I-III resected NSCLC patients with EGFR mutation or EGFR wild-type status. (a) and (b) ROC curves represent the EGFR genotype detection performance of IPES, metadata-based model and the combined model in the (a) internal validation set and (b) external validation set. (c) and (d) Kaplan–Meier curves of the low- and high-risk subgroups predicted by IPES for stage I-III resected NSCLC patients with (c) EGFR mutation and (d) EGFR wild-type status in the internal validation set. In the Kaplan–Meier curves, the horizontal axis represents survival time (months) and the vertical axis represents survival probability.
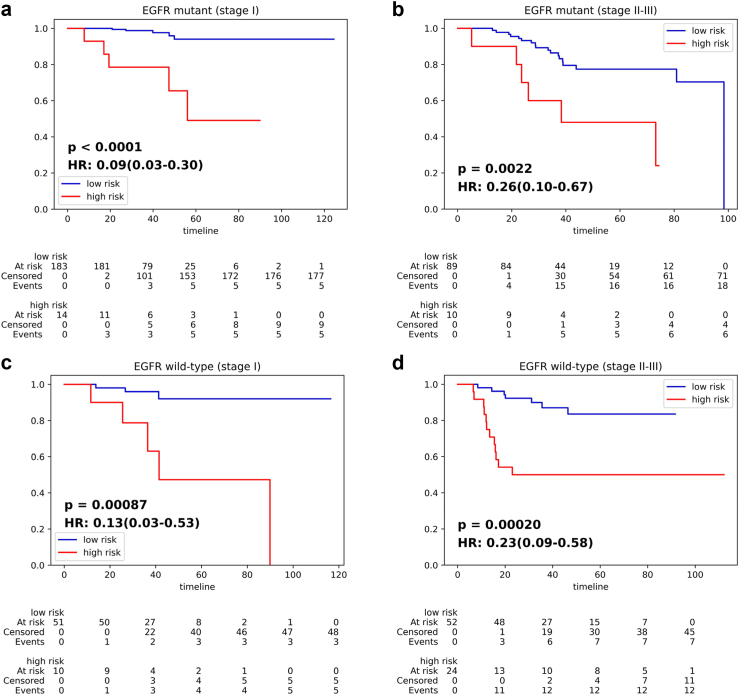
Although EGFR genotype is crucial for treatment decision of lung cancer patients, few EGFR-TKI adjuvant therapy clinical trials have shown improvement in OS. Thus, it is important to further stratify resected patients in the same stage and with the same EGFR genotype into low and high-risk subgroups to help in making accurate therapeutic decision. To further examine the prognostic stratification ability of IPES, we employed internal validation set to stratify the prognosis of the following four groups of patients according to the predicted IPES-score: (1) resected EGFR mutant patients in stage I; (2) resected EGFR mutant patients in stage II-III; (3) resected EGFR wild-type patients in stage I; (4) resected EGFR wild-type patients in stage II-III. For resected EGFR mutant patients in stage I, IPES stratified them into two significantly different groups (p < 0.0001, HR = 0.09, 95% CI: 0.03–0.30; Fig. 4(a)), with the high-risk subgroup showing a higher 5-year death rate of 35.7% and the low-risk subgroup showing a lower 5-year death rate of 2.7% (Supplementary Table S9). Similarly, the predicted low- and high-risk subgroups showed significant difference (p = 0.0022, HR = 0.26, 95% CI: 0.10–0.67; Fig. 4(b)) in resected EGFR mutant patients in stage II-III. For resected EGFR wild-type patients in stage I, all the patients were stratified into two significantly different subgroups (p = 0.00087, HR = 0.13, 95% CI: 0.03–0.53; Fig. 4(c)), with the high-risk group showing a higher 5-year death rate of 40.0% and the low-risk group showing a lower 5-year death rate of 5.9% (Supplementary Table S10). Similarly, the predicted low- and high-risk subgroups showed significant difference (p = 0.00020, HR = 0.23, 95% CI: 0.09–0.58; Fig. 4(d)) in resected EGFR wild-type patients in stage II-III. The results indicated that the IPES may be reliable for providing NSCLC patients with more individual therapeutic suggestion.
Fig. 4.
IPES performance on predicting OS risk of stage I and II-III resected NSCLC patients with EGFR mutation or EGFR wild-type status. (a) and (b) Kaplan–Meier curves of the low- and high-risk subgroups predicted by IPES for resected NSCLC patients with EGFR mutation in (a) stage I and (b) stage II-III in the internal validation set. (c) and (d) Kaplan–Meier curves of the low- and high-risk subgroups predicted by IPES for resected NSCLC patients with EGFR wild-type status in (c) stage I and (d) stage II-III in the internal validation set. In the Kaplan–Meier curves, the horizontal axis represents survival time (months) and the vertical axis represents survival probability.
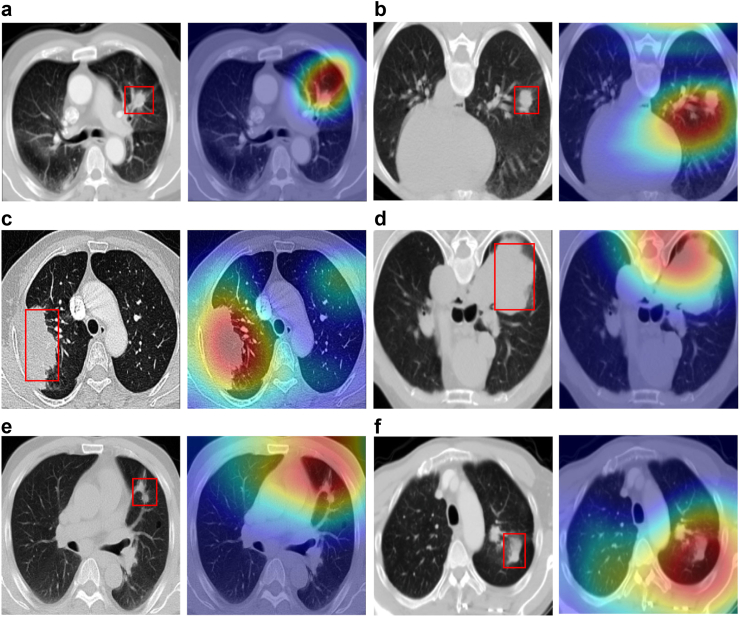
To gain further insights of which areas were mainly focused by IPES and which features contributed to the network's output, we visualized the primary branch's response in the last neural layer on the OS risk prediction task in the multi-task network. Fig. 5 shows several representative examples of original CT images and their corresponding saliency maps. We noted that features focused more on morphological features of the tumor areas, such as its boundary, shape and texture, which have been commonly used by clinicians for diagnosis and treatment decision. This finding indicated that the IPES was able to capture clinically relevant features. Moreover, in most of the cases, IPES focused more on the interaction between the tumor and its surrounding tissues, suggesting that some microenvironment changes in non-tumor tissues could also be correlated with the prognosis of patients. This attention to the non-tumor areas within the lungs could offer clinicians valuable prognostic information.
Fig. 5.
Network visualization and interpretation. Visual explanations of the areas in the CT images identified by IPES. The first column in subfigure is the original CT image, the second column in subfigure is the saliency map overlaying the original CT image. In each original CT image of (a–f), the lung tumor has been marked with a red bounding box.
Discussion
Recently, related studies on prognosis evaluation for NSCLC patients in advanced stage have been extensively investigated.29,30,34 However, the prognosis of resected NSCLC patients in stage I-III is rarely discussed, especially for the NSCLC patients in stage I. Although the TNM staging system is a powerful predictor of prognosis in NSCLC, it is not perfect enough to accurately distinguish all the patients, especially within the same TNM stage. We still need a better prognostic system to help in making therapeutic decisions. If we can divide patients of the same stage into low-risk and high-risk, we can give them a more personalized treatment and surveillance suggestion, to improve OS for patients with resected NSCLC. In this retrospective multi-institution study, we developed and validated the IPES which allows accurate prediction of OS risk for resected NSCLC patients in stage I-III based on CT images. Our findings suggested that the proposed IPES could non-invasively obtain prognosis related information from patients before surgery and identify patients into the low- and high-risk subgroups with good and poor prognosis respectively. The performance of IPES suggested that information obtained from CT images by IPES could complement TNM stage in clinical practice.
We previously built a multi-omics model for resected stage I NSCLC patients and found that increased genomic instability is significantly associated with poor prognosis.46 But such multi-omics detections are costive and need tumor samples after surgery which limited their clinical utility. In this study, we developed the IPES using CT images, which is a non-invasive and highly repeatable way to easily and economically obtain information from NSCLC patients. More importantly, IPES performed well in predicting OS risk for patients after complete resection in stage I and stage II-III, respectively. The resected NSCLC patients, whether in stage I or stage II-III, can be further stratified into low- and high-risk subgroups with significantly difference, which offers the potential to identify high-risk patients in the early-stage and low-risk patients in the locally advanced stage.
Recent study has shown that not all EGFR mutation patients can benefit from EGFR-TKI treatment after surgery.47 Patients with some molecular feature may be more sensitive to chemotherapy than EGFR-TKI even with EGFR mutations. In our study, IPES was able to significantly separate the resected EGFR mutant patients with stage I into low- and high-risk subgroups. Additionally, the similar results were also achieved in resected EGFR mutant patients with stage II-III, resected EGFR wild-type patients with stage I, and resected EGFR wild-type patients with stage II-III, which indicated that we may pick up the resected EGFR mutant patients who are not fitted for EGFR-TKI treatment by CT scan, so as to select more accurate individual treatment for such patients.
Lung cancer patients are not uniform in their biologic or genetic make-up.48 Previous research using radiomic-based methods has suggested that radiomic features of primary lung tumors can capture biological heterogeneity and exhibit variance across cancer stage.49 In this study, we developed a deep learning-based system, IPES, to extract CT features for survival risk prediction, which achieved a good performance in identifying patients with good and poor prognosis. The results demonstrated that features extrcted by IPES may also be helpful in capturing biological heterogeneity of primary lung tumors. Additionally, our visualization interpretability showed that IPES could capture morphological features of the primary tumors (Fig. 5), which suggested that our proposed system could obtain subtle features from CT images that are difficult for humans to interpret and may be relevant to prognosis.
Compared with the previous artificial intelligence-based studies, IPES predicted OS risk based on self-supervised pre-training and multi-task learning, and it achieved better performance than both the commonly used single-task model and the multi-task model trained from scratch on prognostic prediction. To the best of our knowledge, this is the first study to evaluate prognosis of NSCLC patients using self-supervised pre-training and multi-task learning. Unlike the most of studies that focused on supervised learning using large scaled labeled data, IPES learned general representations from 3D unlabeled medical images using self-supervised learning for downstream prognosis evaluation. Through contrastive learning, the proposed pre-training framework CTCLR could learn visual representations from different patients.
Furthermore, unlike current lung cancer prognosis-related tasks that rely on single-task models, we integrated prognosis-related auxiliary tasks with the primary survival prediction task in a unified multi-task learning network by sharing feature representations at multi-scale levels, which improved the generalization and accuracy performance of model. IPES demonstrated good performance for the primary task for OS risk prediction and auxiliary tasks for early-stage and EGFR genotype detection. When we combined the clinical metadata with the deep learning score predicted by IPES, the AUC values of metadata-based model could be further improved. In summary, IPES was able to predict OS risk, early-stage and EGFR genotype of NSCLC patients simultaneously, which is a great supplement to clinical practice, and the performance of IPES for survival prediction showed remarkable potential in CT-based prognostic analysis.
Several limitations exist in this study. First, although our study achieved good performance on prognosis prediction, we focused on the whole lung areas in CT images instead of the tumor regions. A simultaneous inclusion of tumor areas and whole lung regions might achieve better performance. Second, our data were collected from Asian population only. The clinicopathological characteristics might be different in different ethnicities. Third, in this study, we focused OS only, and other oncological endpoint such as disease-free survival is also critical for resected NSCLC patients in stage I-III. Fourth, other morbidities and factors, including the severity of coronary artery calcification, emphysema, muscle mass, and fat attenuation, have been established as correlated with prognosis.50, 51, 52 Integrating these factors with clinicalpathological characteristics and MTL-score may provide a more comprehensive understanding of the prognostic value of IPES. In conclusion, we developed the IPES using self-supervised pre-training and multi-task learning based on CT images, which allows refined stratification of OS for resected NSCLC patients in stage I-III. The proposed IPES could be used to help identify resected lung cancer patients in the same stage or with the same EGFR genotype who are most likely to derive benefit from corresponding treatment decision and surveillance suggestion. Compared with the traditional TNM staging system, IPES achieved better performance on distinguishing patients with good and poor prognosis, which indicates that image-based survival risk score can additionally increase the predicting accuracy of prognosis in NSCLC patients after complete surgical resection.
Contributors
GY.W., S.Z., X.L., K.W., L.Z., J.S., J.S., X.W., J.M., T.G., Z.J., C.W., and K.C. collected and analyzed the data. X.L., S.Z., K.W., L.Z., J.S., J.S., X.W., J.M., T.G., Z.J., C.W., K.C., and GY.W. conceived and supervised the project. S.Z., K.C., X.L., K.W., L.Z., J.S., J.S., X.W., J.M., T.G., Z.J., C.W., and GY.W. wrote the manuscript. All authors discussed the results, reviewed and edited the manuscript.
Data sharing statement
Data access can be requested by writing to the corresponding authors. All data and code access requests will be reviewed and (if successful) granted by the Data Access Committee.
Declaration of interests
The authors have declared no conflicts of interest.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (grant 62272055, 92259303, 92059203), New Cornerstone Science Foundation through the XPLORER PRIZE, Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), Clinical Medicine Plus X - Young Scholars Project, Peking University, the Fundamental Research Funds for the Central Universities (K.C.), Research Unit of Intelligence Diagnosis and Treatment in Early Non-small Cell Lung Cancer, Chinese Academy of Medical Sciences (2021RU002), BUPT Excellent Ph.D. Students Foundation (CX2022104). We thank members of Chen, and Wang groups for their assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102270.
Contributor Information
Kezhong Chen, Email: chenkezhong@pkuph.edu.cn.
Chengdi Wang, Email: chengdi_wang@scu.edu.cn.
Guangyu Wang, Email: guangyu.wang24@gmail.com.
Appendix A. Supplementary data
References
- 1.Goldstraw P., Chansky K., Crowley J., et al. The IASLC lung cancer staging project proposals for revision of the TNM stage grouping in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Pignon J.P., Tribodet H., Scagliotti G.V., Douillard J.Y., Chevalier T.L. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 3.Liu S.Y., Zhang J.T., Zeng K.H., Wu Y.L. Perioperative targeted therapy for oncogene-driven NSCLC. Lung Cancer. 2022;172:160–169. doi: 10.1016/j.lungcan.2022.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Frankell A.M., Dietzen M., Al Bakir M., et al. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature. 2023;616:525–533. doi: 10.1038/s41586-023-05783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis W.D., Brambilla E., Nicholson A.G., et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 6.Tsao M.S., Marguet S., Le Teuff G., et al. Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. J Clin Oncol. 2015;33:3439–3446. doi: 10.1200/JCO.2014.58.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado-Serrano J., Carnero A. Molecular radiobiology in non-small cell lung cancer: prognostic and predictive response factors. Cancers. 2022;14:2202. doi: 10.3390/cancers14092202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z.T., Wang Y.J., Zhang X.M., Zhang T.T. Pretreatment prognostic nutritional index as a prognostic factor in lung cancer: review and meta-analysis. Clin Chim Acta. 2018;486:303–310. doi: 10.1016/j.cca.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Sculier J.P., Chansky K., Crowley J.J., Van Meerbeeck J., Goldstraw P. The impact of additional prognostic factors on survival and their relationship with the anatomical extent of disease expressed by the 6th edition of the TNM Classification of Malignant Tumors and the proposals for the 7th edition. J Thorac Oncol. 2008;3:457–466. doi: 10.1097/JTO.0b013e31816de2b8. [DOI] [PubMed] [Google Scholar]
- 10.Masago K., Kuroda H., Sasaki E., et al. Association of the KRAS genotype and clinicopathologic findings of resected non-small-cell lung cancer: a pooled analysis of 179 patients. Cancer Genetics. 2022;268:1–11. doi: 10.1016/j.cancergen.2022.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 12.Hu M.M., Hu Y., He J.B., Li B.L. Prognostic value of basic fibroblast growth factor (bFGF) in lung cancer: a systematic review with meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer D.G., Kardia S.L., Huang C.C., et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 14.Tomida S., Koshikawa K., Yatabe Y., et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene. 2004;23:5360–5370. doi: 10.1038/sj.onc.1207697. [DOI] [PubMed] [Google Scholar]
- 15.Lau S.K., Boutros P.C., Pintilie M., et al. Three-gene prognostic classifier for early-stage non–small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 16.Chen H.Y., Yu S.L., Chen C.H., et al. A five-gene signature and clinical outcome in non–small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 17.Zuo S., Wei M., Zhang H., et al. A robust six-gene prognostic signature for prediction of both disease-free and overall survival in non-small cell lung cancer. J Transl Med. 2019;17:152. doi: 10.1186/s12967-019-1899-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R., Zuo S. A robust 8-gene prognostic signature for early-stage non-small cell lung cancer. Front Oncol. 2019;9:693. doi: 10.3389/fonc.2019.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu C.Q., Ding K., Strumpf D., et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D.T., Hsu Y.L., Fulp W.J., et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non–small cell lung cancer. J Natl Cancer Inst. 2011;103:1859–1870. doi: 10.1093/jnci/djr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo D.N., Wang M., Shen Z.H., Zhu J.N. A new immune signature for survival prediction and immune checkpoint molecules in lung adenocarcinoma. J Translat Med. 2020;18:118. doi: 10.1186/s12967-020-02286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner K.W., Ye Y.Q., Lin J., Vaporciyan A.A., Roth J.A., Wu X.F. Genetic variations in epigenetic genes are predictors of recurrence in stage I or II non-small cell lung cancer patients. Clin Cancer Res. 2012;18:585–592. doi: 10.1158/1078-0432.CCR-11-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K.Z., Zhao H., Shi Y.B., et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC) Clin Cancer Res. 2019;25:7058–7067. doi: 10.1158/1078-0432.CCR-19-1213. [DOI] [PubMed] [Google Scholar]
- 24.Saito M., Schetter A.J., Mollerup S., et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brambilla E., Le Teuff G., Marguet S., et al. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol. 2016;34:1223–1230. doi: 10.1200/JCO.2015.63.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin F.K., Zhu L., Shao J.B., et al. Circulating tumour cells in patients with lung cancer universally indicate poor prognosis. Eur Respir Rev. 2022;31 doi: 10.1183/16000617.0151-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi T., Zhu S., Guo H.J., et al. The impact of programmed death-ligand 1 expression on the prognosis of early stage resected non-small cell lung cancer: a meta-analysis of literatures. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.567978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu W., Jiang L., Zhang J.Y., et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. 2020;11:5228. doi: 10.1038/s41467-020-19116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Yu H., Gan Y.C., et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digital Health. 2022;4:E309–E319. doi: 10.1016/S2589-7500(22)00024-3. [DOI] [PubMed] [Google Scholar]
- 30.Deng K.X., Wang L., Liu Y.C., et al. A deep learning-based system for survival benefit prediction of tyrosine kinase inhibitors and immune checkpoint inhibitors in stage IV non-small cell lung cancer patients: a mutlicenter, prognostic study. eClinicalMedicine. 2023;51 doi: 10.1016/j.eclinm.2022.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Primakov S.P., Ibrahim A., van Timmeren J.E., et al. Automated detection and segmentation of non-small cell lung cancer computed tomography images. Nat Commun. 2022;13:3423. doi: 10.1038/s41467-022-30841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee P., Zhou M., Lee E., et al. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat Mach Intell. 2020;2:274–282. doi: 10.1038/s42256-020-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H.Y., Song Y.Q., Huang Z.K., et al. Accurate prediction of epidermal growth factor receptor mutation status in early-stage lung adenocarcinoma, using radiomics and clinical features. Asia Pac J Clin Oncol. 2022;18:586–594. doi: 10.1111/ajco.13641. [DOI] [PubMed] [Google Scholar]
- 34.Song J.D., Shi J.Y., Dong D., et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res. 2018;24:3583–3592. doi: 10.1158/1078-0432.CCR-17-2507. [DOI] [PubMed] [Google Scholar]
- 35.Henschke C.I., McCauley D.I., Yankelevitz D.F., et al. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet. 1999;354:99–105. doi: 10.1016/S0140-6736(99)06093-6. [DOI] [PubMed] [Google Scholar]
- 36.Ardila D., Kiraly A.P., Bharadwaj S., et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y.M., Zhang Z.C., Yuan Q.Y., et al. Predicting peritoneal recurrence and disease-free survival from CT images in gastic cancer with multitask deep learning: a retrospective study. Lancet Digital Health. 2022;4:E340–E350. doi: 10.1016/S2589-7500(22)00040-1. [DOI] [PubMed] [Google Scholar]
- 38.Tiu E., Talius E., Patel P., Langlotz C.P., Ng A.Y., Rajpurkar P. Expert-level detection of pathologies from unannotated chest X-ray images via self-supervised learning. Nat Biomed Eng. 2022;6:1399–1406. doi: 10.1038/s41551-022-00936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H.T., Kruger M., Wang G., et al. Vol. 11314. 2020. Multi-task learning for mortality prediction in LDCT images; pp. 541–546. (Med imaging 2020: computer-aided diagnosis). [Google Scholar]
- 40.Mayer M., Ronald J., Vernuccio F., et al. Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology. 2019;293:583–591. doi: 10.1148/radiol.2019190928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmanninger J., Prayer F., Pan J., Rohrich S., Prosch H., Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 2020;4:50. doi: 10.1186/s41747-020-00173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi Y.N., Zhang Z.J., Zhang W.C., Zhang C.R., Li W.D., Zhao T. Semantic segmentation of urban buildings from VHR remote sensing imagery using a deep convolutional neural network. Rem Sens. 2019;11:1774. [Google Scholar]
- 43.He K.M., Zhang X.Y., Ren S.Q., Sun J. 2016 IEEE conference on computer vision and pattern recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 44.Chen T., Kornblith S., Norouzi M., Hinton G. 2020 International conference on machine learning (PMLR) 2020. A simple framework for contrastive learning of visual representations; pp. 1597–1607. [Google Scholar]
- 45.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual explanations from deep network via gradient-based localization. Int J Comput Vis. 2020;128:336–359. [Google Scholar]
- 46.Chen K.Z., Yang A.R., Carbone D.P., et al. Spatiotemporal genomic analysis reveals distinct molecular features in recurrent stage I non-small cell lung cancers. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111047. [DOI] [PubMed] [Google Scholar]
- 47.Liu S.Y., Bao H., Wang Q., et al. Genomic signatures define three subtypes of EGFR-mutant stage II-III non-small-cell lung cancer with distinct adjuvant therapy outcomes. Nat Commun. 2022;12:6450. doi: 10.1038/s41467-021-26806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friberg S., Nystrom A. Cancer metastases: early dissemination and late recurrences. Cancer Growth Metastasis. 2015;8:43–49. doi: 10.4137/CGM.S31244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padole A., Singh R., Zhang E.W., et al. Radiomic features of primary tumor by lung cancer stage: analysis in BRAF mutated non-small cell lung cancer. Transl Lung Cancer Res. 2020;9:1441–1451. doi: 10.21037/tlcr-20-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Digumarthy S.R., De Man R., Canellas R., Otrakji A., Wang G., Kalra M.K. Multifactorial analysis of mortality in screening detected lung cancer. J Oncol. 2018;2018 doi: 10.1155/2018/1296246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu K.W., Khan M.S., Li T.Z., et al. AI body composition in lung cancer screening: added value beyond lung cancer detection. Radiology. 2023;308 doi: 10.1148/radiol.222937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puliti D., Mascalchi M., Carozzi F.M., et al. Decreased cardiovascular mortality in the ITALUNG lung cancer screening trial: analysis of underlying factors. Lung Cancer. 2019;138:72–78. doi: 10.1016/j.lungcan.2019.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.