Abstract
Quiescent adult stem cells are important for mammalian tissues, where they act as a reserve that supports normal tissue turnover and can mount a regenerative response following acute injuries. Quiescent stem cells are well-established in certain tissues, such as skeletal muscle, brain, and bone marrow. The quiescent state is actively maintained and is essential for long-term maintenance of stem cell pools. In this Review, we discuss the importance of actively maintaining a functional pool of quiescent adult stem cells, including hematopoietic stem cells (HSCs), skeletal- muscle stem cells (MuSCs), neural stem cells (NSCs), hair follicle stem cells (HFSCs), and mesenchymal stem cells (MSCs) such as fibro-adipogenic progenitors (FAPs) to ensure tissue maintenance and repair. We discuss the molecular mechanisms that regulate the entry into, maintenance of, and exit from the quiescent state in mice. Recent studies revealed that quiescent stem cells have a discordance between RNA and protein levels, indicating the importance of post-transcriptional mechanisms, such as alternative polyadenylation, alternative splicing, and translation repression, in the control of stem cell quiescence. Understanding how these mechanisms guide stem cell function during homeostasis and regeneration has important implications for regenerative medicine.
Introduction
Stem cells have the ability to proliferate, to self-renew (produce new stem cells), or to differentiate [G] into different cell types, thereby offering great potential for regenerative medicine1,2. First described in the 19th century, stem cells include embryonic stem cells [G], germline stem cells [G], induced pluripotent stem cells [G], and adult stem cells [G], which have key roles in the maintenance and regeneration of tissues3,4. Tissues that undergo high cell turnover, such as the blood and the epithelium of the gut, maintain populations of continuously proliferating adult stem cells and progenitor cells to generate differentiating progeny5. However, many tissues also maintain populations of stem cells in a quiescent state, to support the cycling progenitors or in reserve for tissue injuries (Figure 1a).
Figure 1: Types of quiescent stem cells and their features.
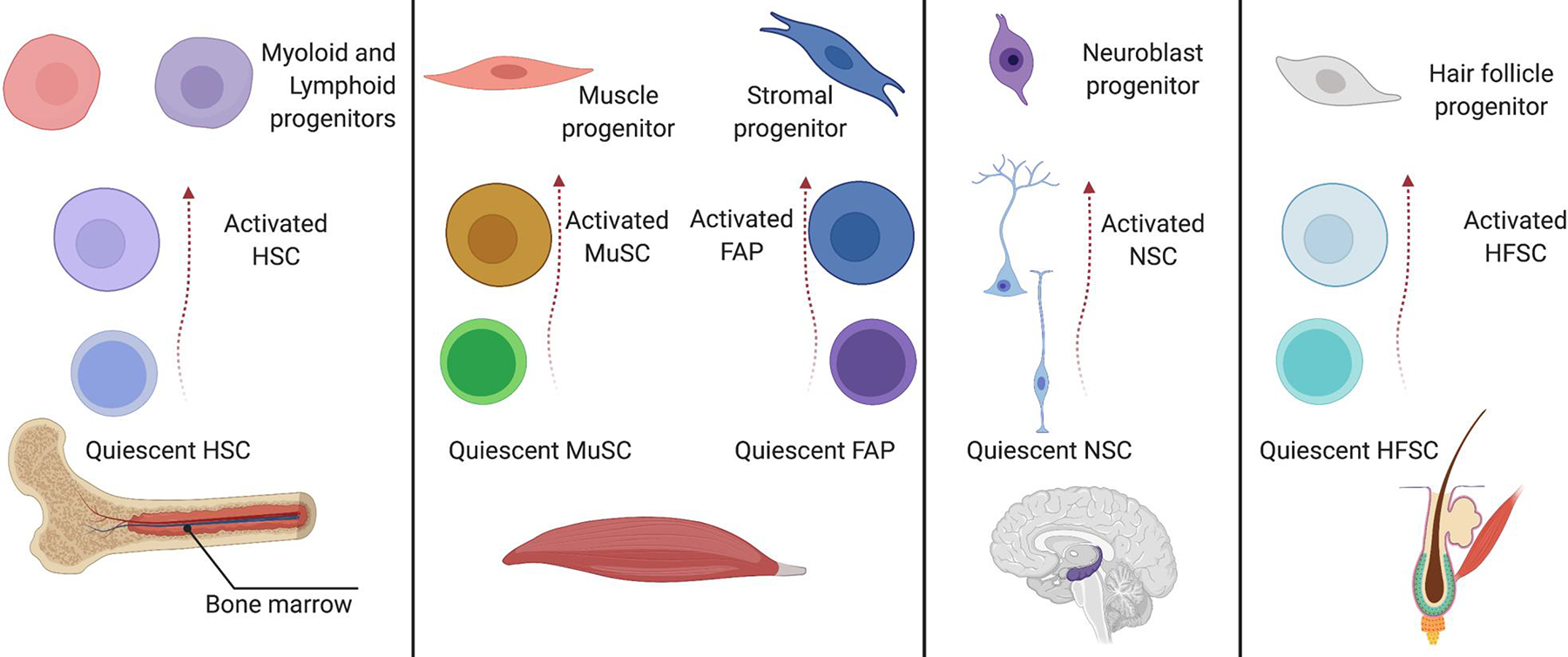
A) Populations of quiescent stem cells have been identified in several tissues. The bone marrow harbors populations of quiescent hematopoietic stem cells (HSCs) 337, which are established in mice up to six weeks postnatally60. HSCs are required to generate the progenitors that sustain the daily production of billions of blood cells and can regenerate the complete blood-cell system following transplantation or injury46. Around 6% of mouse HSCs enter the cell cycle each day, though distinct subsets display different activation parameters46,338,339. Skeletal muscle contains a population of quiescent stem cells known as skeletal-muscle stem cells (MuSCs). MuSCs are established around two months after birth in mice340. MuSCs are essential to generate the progenitors that regenerate skeletal muscles341–343. Around 0.3–1.0% of mouse MuSCs enter the cell cycle each day, depending on the muscle, and their progeny fuse with mature myofibers, though their impact on muscle homeostasis is limited195,344,345. A quiescent population of mesenchymal stem cells also called fibro-adipogenic progenitors (FAPs) supports muscle regeneration and homeostasis346,347. Around 1% of mouse FAPs enters the cell cycle daily348. The adult brain hosts populations of quiescent neural stem cells (NSCs) in the subventricular zone lining the lateral ventricles and in the subgranular zone of the dentate gyrus in the hippocampus349. NSCs are established in mice by the second week after birth161. They are required to generate the progenitors that sustain continued formation of new neurons, astrocytes and oligodendrocytes. Around 12% of mouse NSCs enter the cell cycle each day350. The hair follicles in the skin harbor populations of quiescent hair follicle stem cells (HFSCs). Hair follicles undergo cyclic rounds of growth (anagen), degeneration (catagen) and rest (telogen), termed the “hair cycle”351. Quiescent HFSCs are established by 3 weeks after birth at the end of the first hair cycle in mice36. During anagen, HFSCs are essential to generate the progenitors that sustain hair growth. They generate the progeny that will start a new hair cycle. Around 0.5% of mouse HFSCs in the bulge [G] enter the cell cycle each day during telogen352. Overview of the tissue, time of establishment and spontaneous activation rates of five common adult quiescent stem cell types: HSCs46,60,337–339, MuSCs195,340–345, FAPs346–348, NSCs161,349,350, and HFSCs36,351,352.
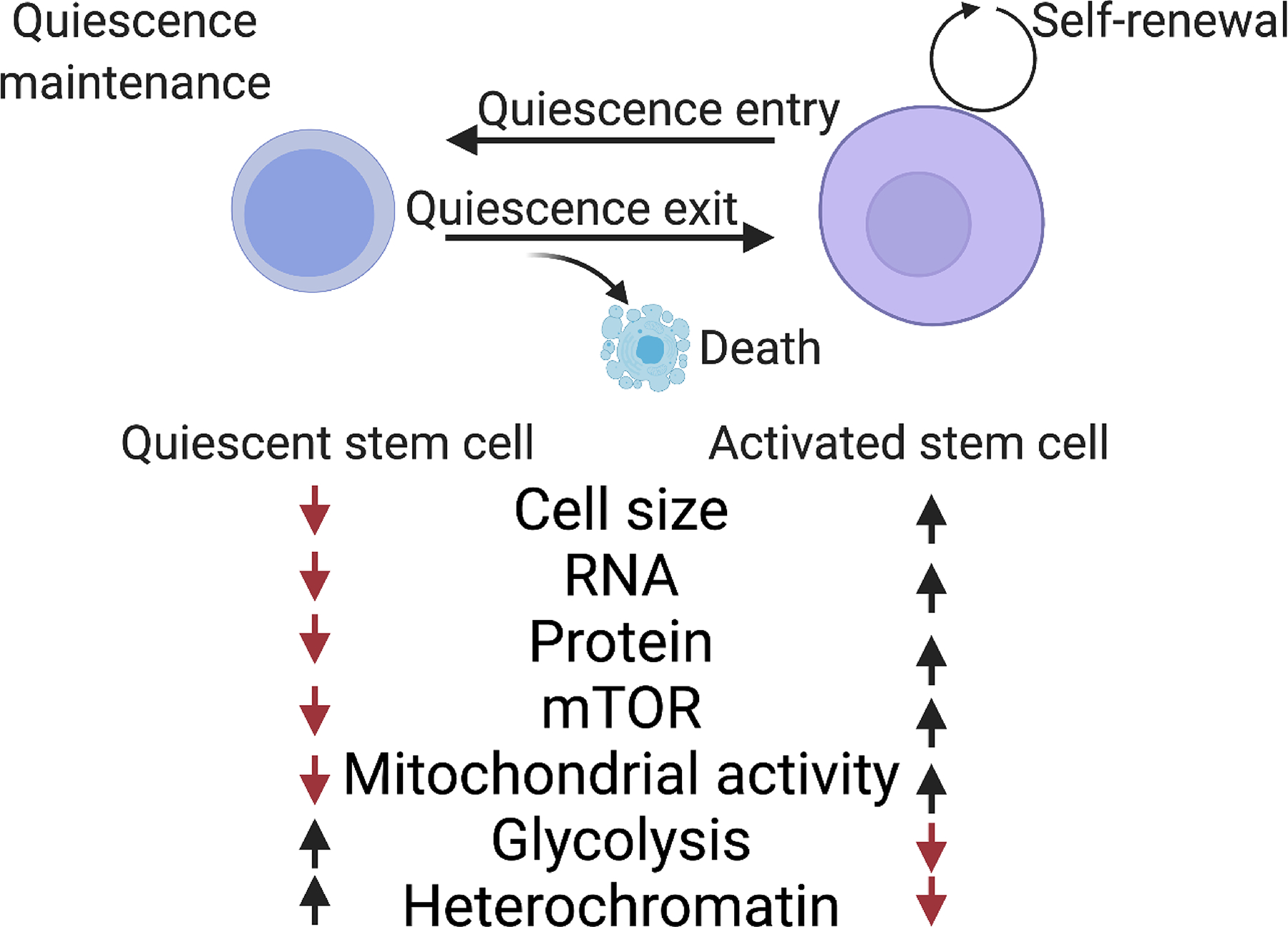
B) Quiescent stem cells display a number of distinct features compared to their activated counterparts and undergo different processes, including maintenance of the quiescent state, exit from the quiescent state (activation), and re-entry into the quiescent state (re-quiescence, or self-renewal). At any point in time, quiescent stem cells must ensure survival and may undergo apoptosis. Quiescent stem cells possess several features distinct from their activated counterparts, including small cell size, low RNA and protein content, low mTOR signaling and mitochondrial activity, high use of glycolysis and more heterochromatin, as well as stem cell type specific expression of transcription factors PAX7 (Paired Box 7, MuSCs), TCF3 and TCF4 (Transcription Factor 3 and 4, HFSCs), HOXB5 and TCF15 (Homeobox B5 and Transcription Factor 15, HSCs), and ID3 (Inhibitor of DNA Binding 3, NSCs).
Quiescence is a state of prolonged and reversible cell cycle exit, which is different from the irreversible cell-cycle exit that characterizes terminal cell differentiation or cell senescence2,6. Some differentiated cells can de-differentiate and proliferate to contribute to tissue repair (such reversible exit from the cell cycle is observed in endothelial cells and hepatocytes), but do not fit the stem cell paradigm of self-renewal7. Quiescent stem cells act as a reserve for producing progenitor cells that sustain tissue homeostasis and tissue regeneration in response to injury2,8. Among the best studied quiescent stem cell models are hematopoietic stem cells (HSCs), which are found in the bone marrow, skeletal muscle stem cells (MuSCs), neural stem cells (NSCs), hair follicle stem cells (HFSCs), and mesenchymal stem cells (MSCs), which include those in bone marrow, teeth, and the muscle-resident fibro-adipogenic progenitors (FAPs) (Figure 1a)2,9,10. Quiescent adult stem cells have recently been described in the epithelium of the gut and the mammary gland11–13, and may exist in other tissues as well. Germline stem cells in the ovary constitute a unique quiescent stem cell population that can give rise to totipotent stem cells post-fertilization.
A common feature of quiescent stem cells is their high potential for engraftment and self-renewal following transplantation compared to their cycling progeny, which is rapidly lost when these stem cells are cultured ex vivo14–18. This technical limitation poses a barrier to the expansion of quiescent stem cells for autologous stem cell transplantation therapies [G].Thus, understanding how quiescent stem cells are maintained in tissues is a topic of intense study. Quiescent stem cells have long been considered to be in an inactive state2. However, recent genetic studies have revealed many signalling pathways that must be active to maintain the quiescent state. When these pathways are disrupted, stem cells become activated prematurely and often fail to self-renew, leading to depletion of the stem cell pool and a loss of regeneration potential in vivo19–23. Moreover, many of these pathways are deregulated in ageing24 (Supplementary Table 1). Recent studies have focused on understanding how the mechanisms that control quiescence maintenance, exit, and re-entry are regulated. Much effort has gone into characterizing the transcriptional networks that regulate stem cell function and identity25–32. However, transcriptional networks are only one component of the control of stem cell function and regulation of the quiescent state33. Post-transcriptional regulatory mechanisms also play a key part. For example, quiescent MuSCs, HSCs, HFSCs, and NSCs all express genes that are crucial for stem cell activation [G] at the RNA level, but protein synthesis and accumulation are repressed to prevent activation34–37. Such mechanisms are thought to establish a poised state that enables more rapid activation when required38.
In this review, we first describe key features of quiescent stem cells and the biological importance of stem cell quiescence. We discuss the different stem cell processes that enable the function of quiescent stem cells, and many of the genes that control these processes. We then discuss recent findings of post-transcriptional regulatory mechanisms that control the quiescent state in the context of tissue homeostasis and regeneration.
Key features of quiescent stem cells
Quiescent stem cells have several features that distinguish them from their activated progeny and other proliferating stem cells (Figure 1b). This includes a lower level of activity, a reduced cell size, reduced mitochondrial content, and lower levels of biomolecules such as RNA and protein.
Gene and protein expression signatures
Typically, quiescent stem cells are smaller than non-quiescent stem cells and have low RNA and protein content39–41. This is coincides with low levels of mRNA transcription, rather than high levels of mRNA turnover, as indicated by the low levels of Ser2 phosphorylation at the C-terminal domain of RNA polymerase II — a marker of transcript elongation — in quiescent stem cells42. Nevertheless, transcription does occur in quiescent stem cells in vivo43.
Quiescent stem cells are marked by low protein and RNA levels of cyclin A2, cyclin B1, and cyclin E2, which are the key drivers of the cell cycle, and by high levels of the cyclin-dependent kinase inhibitors CDKN1A (p21 or CIP1), CDKN1B (p27 or KIP1), and CDKN1C (p57 or KIP2), which function as inhibitors of cyclins and thus block cell cycle transitions2,44,45. Cycling cells progressing through G1 normally accumulate cyclin D, which partners with cyclin-dependent kinase 4 (CDK4) or CDK6 to inhibit retinoblastoma-associated protein (Rb; also known as RB1) and enable S-phase entry46. Strikingly, cyclin D1 is expressed in quiescent HSCs and MuSCs and may function in a cell-cycle independent manner in these quiescent cells40,47.
In different stem cell types, distinct transcription factors marking the quiescent state have been identified. Quiescent MuSCs express PAX7, the level of which decreases upon MuSC activation48. Quiescent HFSCs express TCF3 (also known as TCF7L1) and TCF4 (also known as TCF7L2), which are lost upon HFSC activation49. HOXB5 and TCF15 expression mark quiescent HSCs27,50.
Epigenetic signatures
Quiescent stem cells have distinct patterns of epigenetic modifications and of heterochromatin. Quiescent MuSCs, NSCs and HFSCs can be identified in electron microscopy micrographs by their nuclei filled with dense heterochromatin51–53. Many genes in quiescent stem cells are characterized by bivalent chromatin, which includes both gene-repressive and permissive histone modifications54–57, suggesting that these genes are poised for activation38. Quiescent stem cells express regulators of histone modifications. For example, Trithorax group [G] histone methyltransferases are expressed in quiescent HSCs and MuSCs to maintain histone H3 Lys4 tri-methylation (a gene-expression permissive modification)58–60. In addition, DNA methylation patterns are dynamically regulated in quiescent HSCs and MuSCs61,62.
Low metabolic activity.
Quiescent HSCs, MuSCs, NSCs, and HFSCs all have lower energy demands than cycling cells and correspondingly low mitochondrial activity63–66 (Figure 1b). Subsets of HSCs with low mitochondrial activity are more quiescent compared to subsets of HSCs with high mitochondrial activity, and show better long term engraftment67. When HSCs and HFSCs undergo activation, they increase mitochondria biosynthesis and gradually increase oxidative phosphorylation66,67. Quiescent stem cells reside in niches with low access to oxygen68. Accordingly, they rely on glycolysis and fatty acid oxidation to satisfy their low energy demands69–72. Moreover, quiescent stem cells maintain low levels of mTOR complex 1 (mTORC1) signaling73–75, and nutrient-sensing pathways such as the AMPK and LKB1 pathways are vital for accurate balance between quiescence and activation of MuSCs and HSCs76–79.
Altered DNA damage repair.
Quiescent stem cells do not undergo DNA replication and thus have no access to high-fidelity DNA repair by homologous recombination, which generally requires a sister chtromatid as template for repair80,81. Instead, quiescent MuSCs, HSCs, HFSCs, and NSCs use the cell-cycle-independent, but error-prone process of non-homologous end-joining [G]81–85. Nevertheless, DNA damage does accumulate with age in quiescent stem cells, as shown in HSCs and MuSCs86–89. DNA damage in aged stem cells can be repaired by exposing the aged cells to a young systemic environment, due to a shared circulation after heterochronic parabiosis, or upon stem cell activation89,90. Given the importance of DNA repair, it is not surprising that deletion of the gene encoding checkpoint kinase ATR [G] results in apoptosis of activated HSCs, NSCs, and HFSCs91–93. By contrast, MuSCs lacking ATR spotaneously undergo activation in the absence of injury94. In quiescent MuSCs, ATR activity is triggered by DNA–RNA hybrids, resulting in stabilization of cyclin F and quiescence maintenance94.
Stem cell survival.
The quiescent state itself also affects the exposure to stress. For example, quiescent stem cells are protected from replication-induced DNA damage, and their low mitochondrial content limits the exposure to reactive oxygen species (ROS). Resilient subpopulations of quiescent MuSCs, HSCs, and NSCs can survive irradiation or regeneration stress95–97. Nevertheless, quiescent stem cells are still subject to a host of environmental stresses and their ability to withstand them may be limited by their low levels of gene expression and metabolism. Moreover, the complex changes that occur during stem cell activation render the cells vulnerable to damage. For example, mitochondrial biosynthesis upon activation poses additional risks for oxidative stress, and ramping up protein synthesis increases the risk of endoplasmic-reticulum stress98–103. Quiescent MuSCs and HSCs have stress response pathways, which are key for maintaining the quiescent state104,105. Whether quiescent stem cells possess unique mechanisms that actively promote survival remains largely unexplored.
Key processes related to stem cell quiescence
For quiescent stem cells to contribute to tissue homeostasis and repair, they depend on the timely execution of multiple distinct processes. During homeostasis, the stem cells maintain the quiescent state. Nevertheless, a low level of stem cell activation does occur and this contributes to normal homeostasis by supplying the progeny to sustain tissue turnover. Following tissue injury, the cells must rapidly exit the quiescent state in order to activate and enter the cell cycle to drive the repair process. When tissue repair is underway, a subset of stem cells must re-establish the quiescent stem cell pool in a process. This process of re-quiescence ensures the establishment of a stem cell reserve population for future tissue needs. During all of these processes, the cells must devote resources to survival mechanisms in the face of stress and accumulating damage.
Maintenance of stem cell quiescence
The stem cell niche triggers signaling pathways to maintain quiescence, which is instrumental to preservation of the quiescent stem cell pool. For example, a common signaling pathway for maintaining quiescence is the Delta–Notch signaling, in which Delta ligands presented by the niche trigger Notch signaling in the stem cell (Figure 2). Loss of the receptor NOTCH2 or of RBPJ, the downstream effector of Notch signaling, causes activation of MuSCs, NSCs, and HSCs19–23. Moreover, the Notch target genes HES1 and HEY1, which encode transcription factors, are key to quiescence maintenance in all three stem cell types106–108. By contrast, in HFSCs, loss of HES1 has no effect in the telogen phase of the hair growth cycle when the HFSCs are quiescent, but it severely limits the anagen phase when the HFSCs are actively cycling, suggesting that HES1 has an important role in HFSC activation109. More surprisingly, deletion of Rbpj in HFSCs does not affect quiescence or activation, but results in activation of a neighboring quiescent population of melanocyte stem cells through paracrine retinoic acid signaling110. In addition, there are signaling pathways unique to each stem cell type (Supplementary Table 2). For example, HSCs respond to thrombopoietin signals from the bone marrow niche, whereas NSCs respond to neurotransmitters.
Figure 2: Key pathways active in quiescent stem cell processes.
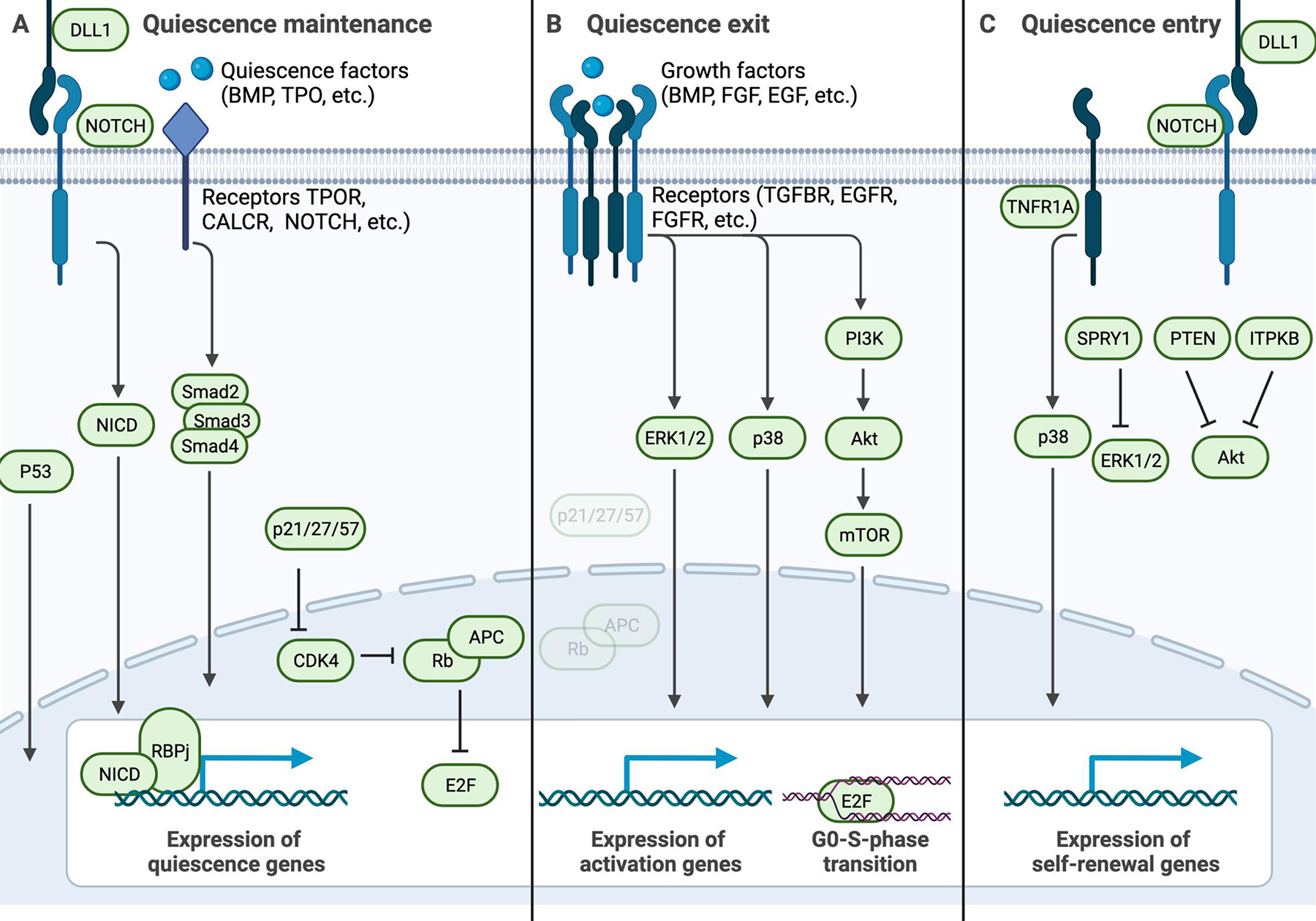
Schematic highlighting key signaling pathways that are active during quiescence maintenance, exit, or entry. A) Signals from the niche to the quiescent stem cell that maintain the quiescencent state, such as ligands for Notch receptors including DLL1 (delta-like protein 1), and quiescence factors including BMP (bone morphogenic protein), TPO (thrombopoietin), calcitonin, and COLV (collagen V) to their respective receptors NOTCH, BMPRII (bone morphogenic protein receptor 2), TPOR (thrombopoietin receptor), and CALCR (calcitonin receptor). Such receptor-mediated signaling induces gene expression patterns that maintain the quiescent state, via signalling intermediates such as NICD-RBPj, and SMAD2–4. The expression of cell-cycle inhibitors such as p21 similarly invokes quiescence maintenance via inhibition of CDK4 or CDK6, which ensures that APC/C (anaphase promoting complex/cyclosome) and Rb (retinoblastoma) block E2F and S-phase entry. B) In response to growth factors, combined with the loss of signals listed in panel (A), phosphorylation cascades such as the PI3K-AKT pathway and the p38 MAP kinase pathway induce quiescence exit and cell cycle entry. C) To exit the cell cycle and enter the quiescent state, stem cells interact with NOTCH ligands and suppress the phosphorylation cascades listed in panel (B). In activated NSCs, TNFR1 (tumor necrosis factor 1 receptor) signaling through p38 MAP kinase is key for re-entry into the quiescent state. PTEN (phosphatase and tensin homolog), MEK1 (MAPK/ERK kinase 1), and ITPKB (inositoltrisphosphate 3-kinase b) inhibit AKT, while Sprouty1 (SPRY1) suppresses ERK signaling to stop mitogenic signaling.
NICD, notch intracellular domain; RBPj, recombination signal binding protein for immunoglobulin kappa j region; CDK4/6, cyclin dependent kinase 4/6; PI3K, phosphatidylinositol-3 kinase.
In addition to signaling from the niche, quiescent stem cells use cell-intrinsic inhibitory mechanisms to prevent activation in the absence of triggering cues38 (Figure 2a). As CDK inhibitors block the G1 cyclins (cyclin D and cyclin E)111, it is not surprising that these proteins are essential to maintaining a quiescent state in stem cells (Figure 2a,3a). Deletion of the genes encoding p21, p27 or p57 result in spontaneous activation of HSCs, NSCs, MuSCs, and HFSCs12,112–119. Quiescent stem cells must avoid the restriction point, from which the cell is fully committed to complete the cell cycle. The restriction point is defined by Rb phosphorylation, which relieves the inhibition of Rb on E2F (Figure 3a)111. However, there is a short window after Rb inactivation during which stress can force the cell back into quiescence, and this window is dependent on the anaphase promoting complex/cyclosome (APC/C)120. Interestingly, the restriction point may be differentially regulated in different quiescent stem cell types. Deletion of Rb1 results in MuSC and HSC activation121–123, but has no effect in NSCs124. Deletion of APC/C results in HSC activation125, but while this lowers NSC quiescence, these cells cannot complete the cell cycle and consequently undergo apoptosis126.
Figure 3: Quiescence and the cell cycle.
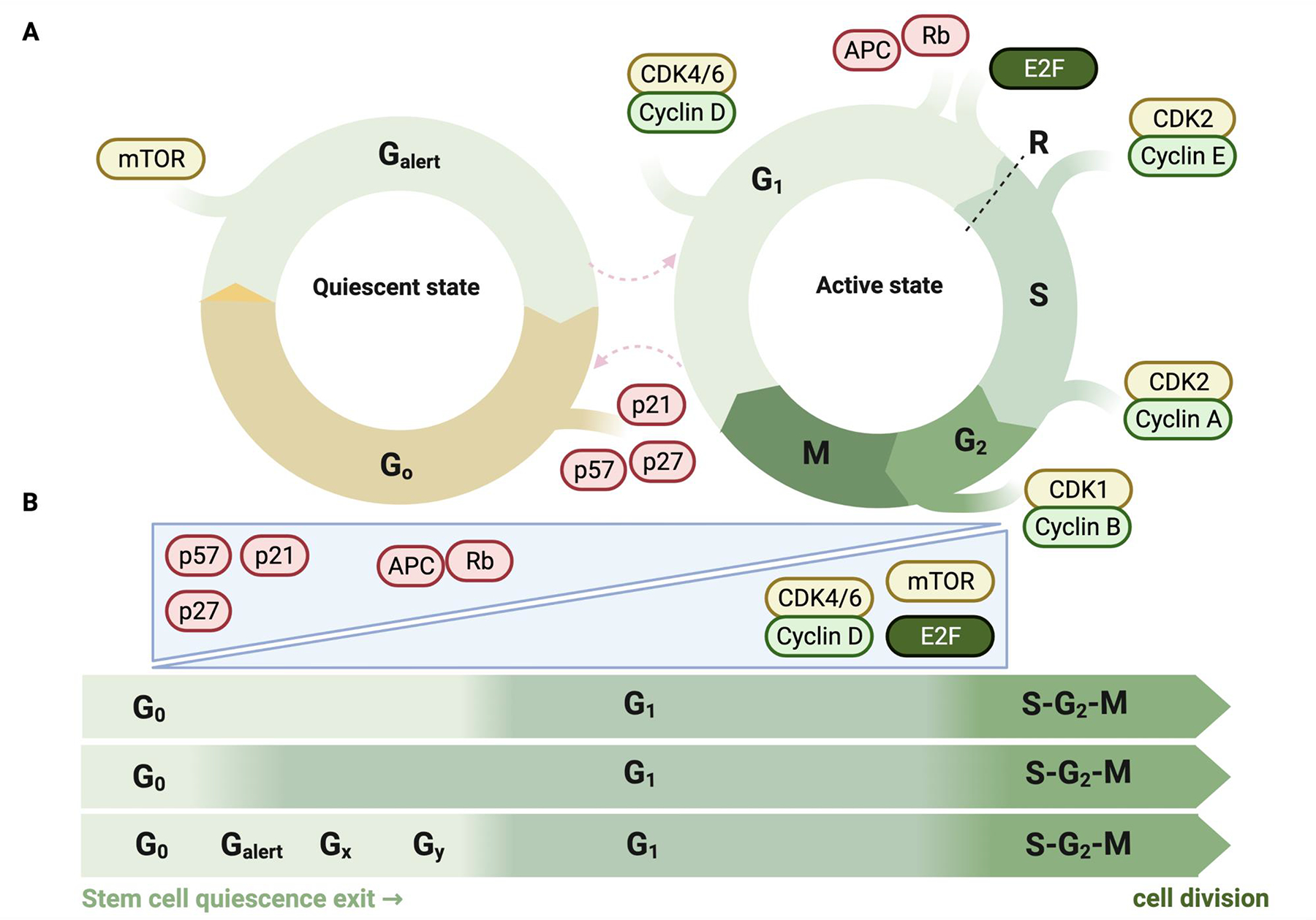
A) Progression of somatic (stem) cells through the G1, S, G2 and M phases of the cell cycle involves intricate regulation by cyclins, cyclin-dependent kinases (CDKs), and their inhibitors. Central to cell-cycle entry and exit is the phosphorylation status of the retinoblastoma (Rb) proteins. Unphosphorylated Rb is chromatin-bound and blocks E2F transcription factors from inducing key proliferation genes. In pro-growth conditions, cyclin D activates CDK4 and CDK6, which phosphorylate (P) Rb and enable transcription of E2F target genes. Somatic stem cells enter the quiescent state early in G1, as a result of the downregulation of G1 cyclins — cyclin D and cyclin E (not shown) — and the upregulation of the CDK interacting protein (CIP) and kinase inhibitory protein (KIP) family of inhibitors, including p21, p27 and p57, which inhibit CDK4 and CDK6 (not shown). Quiescent stem cells can enter an ‘Alert’ state characterized by mTOR signalling and a shorter time to cell-cycle entry. B) The first division of the activation of quiescent stem cell is considerably longer than subsequent cell divisions. It is currently unclear whether that first division consists of a long G0 phase (top), a long G1 phase (middle), or several G0 sub-phases following each other in manner similar to the conventional cell cycle phases (bottom).
Quiescent stem cells express high levels of p5388,127,128, which, in somatic cells, can trigger cell cycle exit or cell death in response to stress. Germline deletion of p53 causes aberrant activation of NSCs and HSCs127,128. In HSCs, this activation is due to the loss of the cell cycle repressors Necdin and GFI-1 (Ref.127), or owing to the indirect loss of p53 through the loss of OCIAD1, an inhibitor of p53 degradation129. In quiescent MuSCs, p53 is stabilized because Notch signaling represses MDM2 expression88. When MuSCs undergo activation, they lose Notch signaling and p53 level decreases (Figure 2). Upon activation, MuSCs that lack p53 have a higher propensity to undergo apoptosis, which can be prevented by pharmacological stabilization of p53 (Ref.88). This is in contrast to other stem cell types, where p53 stabilization induces cell death83,129–131.
Activation and exit from quiescence
Quiescent stem cells retain the capacity to undergo activation and enter the cell cycle in response to triggers such as acute injuries. Stem cell activation is disrupted during aging (BOX 1), resulting in impaired regeneration and highlighting the importance of this process for quiescent stem cell function. Activation is the result of loss of niche signals that maintain quiescence and the inhibitory signals that prevent activation, combined with activating signals and mitogenic and non-mitogenic signals that stimulate the cell cycle (Figure 2b). In preparation for cell cycle entry, the cells first complete a growth phase, which enables them to acquire sufficient biomass and volume to undergo DNA replication. Whether quiescent stem cells must overcome a size threshold to fully activate is unclear. Nevertheless, quiescent HSCs, NSCs, and MuSCs are smaller than their activated progeny and, in somatic cells there is a correlation between cell size and cell cycle speed39,41,73,132.
BOX 1: Changes or loss of stem cell quiescence during aging.
Aging disrupts many aspects of stem cell functionality and impairs tissue homeostasis and regeneration353,354. Aged quiescent stem cells (stem cells obtained from aged mice) show a functional decline, which is linked to changes in the quiescent state355–358. Strikingly, despite this cell-intrinsic dysfunction, the transcriptomes of aged neural stem cells (NSCs) and skeletal-muscle stem cells (MuSCs) are remarkably similar to the transcriptomes of cells obtained from youg mice40,54,359. This finding indicates that post-transcriptional gene regulation mechanisms have a role in age-related quiescent stem cell dysfunction.
A number of observations support this idea. Over the course of aging, there is a global switch to expression of longer mRNA isoforms in populations of quiescent HSCs360, hinting to dynamic regulation of alternative cleavage and polyadenylation of mRNA and/or of transcript stability. Aged MuSCs accumulate the alternative polyadenylation factor U1 snRNA, and as a result express longer isoforms of CD47, and show reduced proliferation361. Aged NSCs accumulate ubiquitinated proteins and protein aggregates, suggesting changes in proteostasis139,362, 343,. The E3 ubiquitin ligase FBXW7, which ensures degradation of MYC and NOTCH1 during activation out of the quiescent state, is downregulated in aged hair follicle stem cells (HFSCs)363,364. However, few studies have directly investigated the role of post-transcriptional regulation in aged quiescent stem cells or how it contributes to aging phenotypes of the quiescent stem cell. In one example, aged HFSC were shown to produce the protease ELANE, which degrades the collagen COL17A1, resulting in premature activation and depletion of the HFSC pool363. Induction of COL17A1 exprression could rescue the loss of quiescent HFSCs363. However, a better understanding of the RNA regulation in aged quiescent stem cells is needed.
It is striking that the first stem cell division upon their activation requires up to two days — considerably longer than subsequent cell divisions, which take about half a day88,133,134. It is unclear whether this extended time is due to lengthening of the G1 phase, or whether the cells transition through several uncharacterized sub-phases of quiescence, similar to the different stages of the cell cycle (Figure 3). Although the process of activation generally concludes with progression into the cell cycle, HSCs and MuSCs can progress to differentiation without first entering the cell cycle19,135.
Activation of stem cells also adjusts their metabolism to meet the energetic demands of proliferation. This adjustment includes a switch from fatty acid oxidation and glycolysis to mitochondrial respiration, which requires the biosynthesis of mitochondria69,72. In addition, MuSCs, NSCs, and HFSCs undergoing activation increase autophagy to process large biomolecules into building blocks136–140. HSCs show signs of a mitochondrial checkpoint, in which mitochondrial biosynthesis can cause stress, which in turn pushes the cell towards quiescence141–143. This process suggests the existence of quiescence checkpoints analogous to the cell cycle checkpoints, in which the cell can evaluate whether or not to progress with activation.
Stem cells are activated by growth factors (Figure 2b). MuSCs, NSCs, HSCs, and HFSCs undergoing activation trigger p38 MAP kinase signaling, and inhibition of p38 reduces activation130,144–147. Activation of HSCs, MuSCs, and HFSCs also upregulate PI3K-AKT (PKB) signaling and interfering with this pathway limits activation130,148,149. Activation of stem cells increases mTOR signaling73,150–153. In response to injuries in a different tissue, MuSCs and HSCs increase mTOR signaling to enter an “alert” state73,74 (Figure 3). MuSCs without the mTORC1 subunit regulatory-associated protein of mTOR (RAPTOR) can no longer enter the “Alert” state and show delayed activation73.
Quiescent stem cells are heterogeneous in their gene-expression levels and ability to undergo activation, which results in cells with varying degrees of “quiescence depth”154. For example, subpopulations of HSCs with low levels of CDK6 or the receptor tyrosine kinase KIT take longer to become active than subpopulations with higher levels of CDK6 or KIT134,155. MuSCs with high levels of the transcription factor PAX7 take longer to become active than MuSCs with low levels of PAX7 (Ref.156). NSCs with high levels of tyrosine-protein kinase ABL1 take longer to become active than NSCs with low levels of ABL1 (Ref.157). Quiescent stem cells can adjust quiescence depth in response to external cues. For example, distant tissue injuries can induce the more shallow Alert state in quiescent MuSCs and HSCs73,74, whereas fasting and ketosis can induce a deeper quiescent state in MuSCs, marked by delayed activation158, and norepinephrine can increase quiescence depth in HFSCs by limiting the expression of FGF18 (Ref.159). A deeper quiescent state can confer protection against certain stresses such as irradiation and chemotherapy agents2,158,160. However, it also hampers the cell’s ability to execute its function and generally correlates with impaired regeneration and impaired homeostasis during aging (BOX 1)158.
Re-entry into the quiescent state
Activated stem cells will at some point replenish the stem cell pool by undergoing re-quiescence, which involves exiting the cell cycle and reverting to the phenotype of a quiescent stem cell161–164 (Figure 1b). Generally, stem cells enter the quiescent state from the G1 phase of the cell cycle, although there is evidence of NSCs entering quiescence in G2 (Ref.165). Re-quiescence occurs in concordance with a loss of growth signals and the reconstruction of the stem cell niche163,166 (Figure 2c). Not surprisingly, deletion of the genes encoding receptors that convey niche signals for the maintenance of quiescence results in depletion of the stem cell pool19,20,167–170. Whether this is due to a defect in quiescence entry, or confounded by defects in other processes, is unclear. Conditional deletion of the gene encoding the Notch ligand Delta-like 1 (DLL-1) considerably reduces the ability of activated MuSCs to re-enter the quiescent state following muscle injury, resulting in a loss of MuSCs171.
Stem cells that enter the quiescent state must also repress the growth signaling pathways that drive activation. NSCs expressing tumor necrosis factor-α receptor 1 (TNFR1; also known as TNFRSF1A) will re-enter the quiescent state following stimulation with lipopolysaccharides, which triggers a systemic immune response172. This response depends on TNFR1-dependent phosphorylation of p38 (Figure 2b,c), and p38 inhibitors can partially abrogate re-quiescence. This function of p38 is consistent with observations in proliferating human and mouse MuSCs, in which inhibition of p38 enhances cell expansion173,174. In quiescent MuSCs, inhibition of p38 prevents cell cycle entry144, highlighting the role of cellular context on signaling pathways. Deletion of the gene encoding SPRY1, an inhibitor of ERK, abolishes MuSC re-quiescence due to increased ERK signaling175. Deletion of Mek1 or Pten, both negative regulators of PI3K, results in gradual depletion of the quiescent stem cell pool98,151,176–181. The AKT inhibitor Inositol(1,4,5)trisphosphate 3-kinase b (ITPKB) is key for HSC re-quiescence182. HSCs without ITPKB activate prematurely and fail to reestablish a population of quiescent HSCs, resulting in hematopoietic failure and anemia. This phenotype suggests that inhibition of AKT signaling is key for re-quiescence182 (Figure 2c).
Self-renewal
Self-renewal is a key process for maintaining the populations of all stem cells. Self-renewal is not equated with quiescence entry since, for example, proliferating stem cells in the epithelium of the gut undergo continuous self-renewal without passing through a quiescent state183. However, to self-renew, quiescent stem cells must go through one cell division and therefore require cell cycle entry and exit. Activated stem cells must decide whether to continue with proliferation, whether to exit the cell cycle and differentiate, or whether to re-enter the quiescent state.
Post-transcriptional quiescence control
There has been a strong interest in understanding the mechanisms that regulate stem cell quiescence. Transcriptional gene regulation has key roles in controlling the quiescent state184–188; however, differential expression of transcripts only partially determines protein abundance. In fact, in somatic cells, mRNA abundance alone often cannot explain observed protein levels189, and RNA and protein abundance often do not correlate in quiescent stem cells26,34,38,190–195. We therefore focus in this Review on regulation of stem cell quiescence by post-transcriptional mechanisms, and examine their roles in the control of quiescence maintenance, exit and entry.
RNA regulation in the nucleus
Following transcription, the pre-mRNA undergoes a series of processing steps, which can be regulated to control the amount of protein that is produced. These mechanisms include alternative polyadenylation (APA), alternative splicing, and RNA modifications such as adenosine and cytosine methylation and adenosine-to-inosine (A-I) editing. The mature mRNA can be degraded, stored, or translated. In the following sections, we discuss how these post-transcriptional mechanisms regulate processes related to stem cell quiescence (Table 1).
Table 1:
Overview of post-transcriptional mechanisms that regulate different processes in quiescent stem cells.
| Mechanism | Stem cell model | Effector | Target mRNA | Molecular effect | Effect on stem cell quiescence | Refs |
|---|---|---|---|---|---|---|
| Alternative poly-adenylation | MuSCs | U1 snRNA high | PAX3 | Shorter 3’UTRs | exit | 195 |
| MuSCs | U1 snRNA low | PAX3 | Longer 3’UTRs | maintenance | 195 | |
| FAPs | NA | PDGFRA | Intronic 3’UTRs | maintenance | 201 | |
| FAPs | NA | PDGFRA | Longer 3’UTRs | exit | 201 | |
| HSCs | NUDT21 high | Glutaminase | Shorter 3’UTRs | exit | 197 | |
| HSCs | NUDT21 low | Glutaminase | Longer 3’UTRs | maintenance | 197 | |
| HSCs | PABPN1 | NA | longer 3’UTRs | entry | 197 | |
| RNA methylation (m6A and m5C) |
MuSCs | METTL3 | NOTCH2 receptor | Stabilization | maintenance | 220 |
| HSCs | METTL3 | MYC | Stabilization | maintenance | 215,218,219 | |
| HSCs | IGF2BP2 | BMI1 | Stabilization | maintenance | 217 | |
| NSCs | METTL3 and METTL14 | NA | Global methylation | entry | 223 | |
| NSCs | FTO | SOCS5 | Destabilization | entry | 335,336 | |
| HFSCs | SUN2 | tRNA | Stabilization | exit | 224 | |
| RNA editing (A-I) |
HSCs | ADAR proteins | MDM2 | Stabilization | exit | 206 |
| HSCs | ADAR proteins | pre-miR-26 | Stabilization | exit | 206 | |
| Intron retention | MuSCs | DEK | MyoD | Intron excision | exit | 234 |
| HSCs | DEK | NA | NA | maintenance | 236 | |
| Alternative splicing | HSCs | OTT1 high | c-MPL | truncated protein | maintenance | 232 |
| HSCs | OTT1 low | c-MPL | Full-length protein | exit | 232 | |
| HSCs | PRMT5 | NA | Global isoforms | maintenance | 231 | |
| MuSCs | QKI | ITGA7 | Alternative isoforms | exit | 230 | |
| Nuclear export or import | NSCs | THOC5 | NA | NA | maintenance | 238 |
| NSCs | miR9 | NA | nuclear accumulation | maintenance | 239 | |
| RNA storage | MuSCs | DDX6 | miR31, MYF5 | Storage | maintenance | 194 |
| Oocytes | DDX6 | PI3K–AKT pathway | Storage | entry | 271 | |
| RNA degradation | MuSCs | miR489 | DEK | Degradation | maintenance | 242 |
| MuSCs | miR708 | TNS3 | Degradation | maintenance | 243 | |
| MuSCs | miR195/497 | CCND2 | Degradation | maintenance | 244 | |
| HSCs | miR21 | PDCD4 | Degradation | maintenance | 245 | |
| NSCs | miR24 | SOX11, MEIS2 | Degradation | maintenance | 249 | |
| HFSCs | miR29 | WNT, BMP pathways | Degradation | maintenance | 250 | |
| MuSCs, HSCs | DICER | NA | miRNA maturation | maintenance | 241,242 | |
| NSCs, HFSCs | DICER | NA | miRNA maturation | exit | 246–248 | |
| HSCs | miR132/212 | FOXO3 | Degradation | exit | 255 | |
| MuSCs | miR708 | NA | NA | entry | 243 | |
| MuSCs | TTP | MyoD | Degradation | maintenance | 190 | |
| MuSCs | HNRNPD1 | MMP9 | Degradation | maintenance | 261 | |
| HSCs | YTHDF2 | TAL1 | Degradation | maintenance | 216 | |
| RNA folding | MuSCs | DHX36 | GNAI2 | Translation | exit | 278 |
| HSCs | DHX36 | NA | Translation | exit | 279 | |
| Translation repression | MuSCs | STAU1 | MyoD | Repression | maintenance | 34 |
| HFSCs | MSI | SHH | Repression | maintenance | 36 | |
| MuSCs | miR206 | PAX3 | Repression | maintenance | 195 | |
| NSC | MFGE8 | mTORC1 | Global translation | maintenance | 288 | |
| MuSCs, HSCs, FAPs | RAPTOR or cMET | NA | Global translation | exit | 73,74 | |
| Translation initiation | MuSCs | eIF2a | NA | Global translation | maintenance | 273 |
| HSCs | NLE | rRNA | Ribosome biosynthesis | maintenance | 280 | |
| MuSCs | NLE | rRNA | Ribosome biosynthesis | exit | 277 | |
| HSCs | RUNX1 | ribosomal genes | Ribosome biosynthesis | maintenance | 281 | |
| Proteasomal degradation | HFSCs | SIRT7 | NFATC1 | deacetylation and degradation | exit | 300 |
| HSCs | FBW7 | MYC | Proteasomal degradation | maintenance | 294 | |
| NSCs | HUWE1 | ASCL1 | Proteasomal degradation | entry | 35 | |
| HSCs | HUWE1 | MYC | Proteasomal degradation | entry | 293 | |
| HSCs | SEL1 | RHEB | Repression of mTORC1 | maintenance | 299 | |
| NSCs | VIM | NA | localized proteasomes | exit | 304 | |
| Lysosomal degradation | NSCs | LRIG | Surface EGFR | Lysosomal degradation | maintenance, entry | 16,297 |
| HSCs | MAEA | Surface cytokine receptors | Lysosomal degradation | maintenance | 294,295 | |
| Protein phosphorylation | MuSCs | PERK | eIF2a | translation repression | maintenance, entry | 273 |
| MuSCs, HSCs, NSCs, HFSCs | PTEN | NA | NA | maintenance, entry | 98,177–179,305 | |
| MuSCs, NSCs, HSCs | PI3K–AKT | NA | NA | maintenance, entry | 130,148,149 | |
| MuSCs, NSCs, HSCs | MAPK | NA | NA | maintenance, entry | 130,144–147 | |
| Protein acetylation | HFSCs | SIRT7 | NFATC1 | deacetylation and degradation | exit | 300 |
| HFSCs | SIRT7 | NFATC1 Lys612 | degradation | exit | 300 | |
| MuSCs | MIST1 and SIRT2 | PAX7 Lys193 | DNA binding | entry | 310 | |
| Protein glycosylation | MuSCs, HSCs, NSCs | Glycosylation enzymes | NA | Global glycosylation | maintenance | 306–308 |
| MuSCs, NSCs, HSCs | NA | NOTCH receptors | Stability | maintenance | 306–308 |
FAPs, fibro-adipogenic progenitors; HFSCs, hair follicle stem cells; HSCs, hematopoietic stem cells; MuSCs, skeletal-muscle stem cells; NA, not available; NSCs, neural stem cells; UTR, untranslated region.
Alternative polyadenylation.
One of the best studied post-transcriptional regulatory mechanisms is APA, in which cells create mRNA isoforms usually with different 3’ untranslated regions (3’UTRs)196. Transcript isoforms with different 3’UTRs can be detected in multiple mouse quiescent stem cell populations, including HSCs, MuSCs, HFSCs, and NSCs195,197–200. HSCs exiting quiescence exhibit a global switch to shorter transcript isoforms197 (Figure 4). Known regulators of APA are dynamically regulated in HSCs and MuSCs195,197.
Figure 4: Post-transcriptional regulatory mechanisms in the nucleus that control stem cell quiescence.
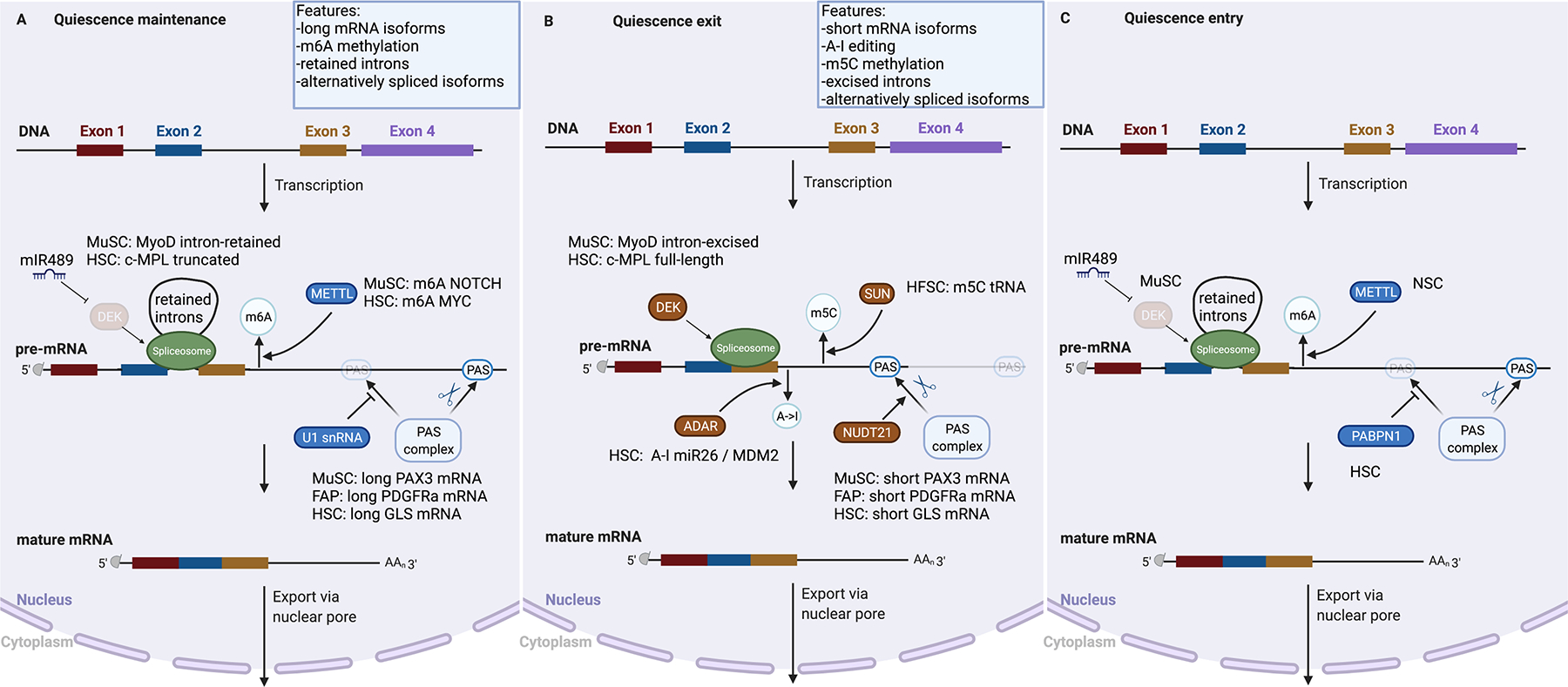
Quiescent stem cells regulate mRNA and protein levels through a variety of post-transcriptional mechanisms. Selection of a cleavage and polyadenylation site (PAS) at the pre-mRNA can be blocked by cleavage and polyadenylation (CPA) factors. For example, polyadenylate-binding nuclear protein 1 (PABPN1) and U1 small nuclear RNA (U1 snRNA) block the use of a proximal PAS and thus favour the selection of a distal PAS and production of a mRNA isoform with a longer 3’ untranslated region (3’UTR); by contrast, NUDT21 (Nudix Hydrolase 21, also known as Cleavage and Polyadenylation Specificity Factor 25 KDa Subunit) favours the selection of a proximal PAS and production of a mRNA isoform with a shorter 3’UTR. The sequence of the pre-mRNA can be modified by ADAR (adenosine deaminase RNA specific) proteins, which deaminate adenosine nucleotides into inosine (A-I editing); by METLL methyltransferases that produce N6-methyladenosine (m6A), and by the SUN2 methyltransferase, which produces 5-methylcytosine (m5C). In addition, the mRNA sequence can be modified by alternative splicing; for example, the protein DEK (DEK Proto-Oncogene) can temporarily delay intron removal. The mature capped, spliced and polyadenylated mRNA is subsequently transported to the cytoplasm.
METTL3,14, N6-adenosine-methyltransferase complex catalytic subunit 3,14; m6A, N6-Methyladenosine; m5C, 5-Methylcytosine; c-MPL, myeloproliferative leukemia protein, also known as thrombopoietin receptor; SUN2, sad1 and UNC84 domain containing 2; PAX3, paired-box 3; PDGFRa, platelet-derived growth factor receptor alpha; GLS, glutamine synthase; MDM2, mouse double minute 2 proto-oncogene; ADAR, A-I, Adenosine-to-Inosine RNA editing; NOTCH, Notch receptor; MYC, m proto-oncogene; miR, microRNA; MYOD, myogenic differentiation 1; THOC5, THO complex 5
APA of specific genes can profoundly affect quiescence. APA of PAX3 transcripts alters the propensity of MuSCs to exit quiescence195. Polyadenylation sites (PASs) in the proximal part of the PAX3 3’UTR are blocked by U1 small nuclear RNA (snRNA), leading to the production of long isoforms that are subject to translation repression by microRNAs (miRNAs) (Figure 4a). Functional knockdown of U1 snRNA results in the expression of shorter PAX3 mRNAs, increased PAX3 protein levels, and MuSC activation, whereas U1 snRNA overexpression prevents MuSC activation195. Quiescent FAPs express mRNA isoforms of platelet-derived growth factor receptor alpha (PDGFRα) that terminate at either of two PASs in the 3’UTR, or at an intronic PAS201. Transcripts generated from the intronic PAS generate a truncated protein lacking the intracellular kinase domain. Blocking the intronic PAS with antisense oligonucleotides results in increased levels of full-length mRNA and protein and in increased exit from quiescence. By contrast, blocking distal PASs leads to increased intronic-PAS choice, production of truncated mRNA and protein, blunted PDGFRα signaling, and reduced activation201. These findings show that APA affects protein levels in quiescent stem cells. This effect differs from what has been observed in somatic non-stem cells, where APA affects mRNA stability or localization202,203, and thus may be a specific feature of quiescent stem cells.
APA also regulates quiescence exit (Figure 4b). Depletion of NUDT21 (also known as cleavage and polyadenylation specificity factor subunit 5) prevents HSC activation and subsequent differentiation. During normal HSC activation, NUDT21 induces a switch towards shorter glutaminase mRNA isoforms, leading to increased glutaminase protein levels, and inhibiting glutaminase activity can similarly block HSC activation197. Diaphragm MuSCs express higher levels of PAX3 compared to MuSCs in lower hindlimb muscles, leading to increased activation rates195. Loss of PAX3 or forced lengthening of PAX3 mRNAs reduces MuSC activation195. Higher levels of PAX3 also confer stress resistance to activated MuSCs95,204. In response to stress, PAX3-expressing MuSCs have lower ROS levels, increased cell size, and higher mTORC1 signaling, and display increased activation kinetics, all suggesting these cells are in an “Alert” state95,204. FAP activation affects APA of PDGFRα201. Increasing intronic PAS choice leads to reduced proliferation and reduced fibrosis, showing that APA is key for quiescence exit. By contrast, the role of APA in quiescence entry is much less clear (Figure 4c). Depletion of the APA regulator polyadenylate-binding nuclear protein 1 (PABPN1; also known as PABP-2) in HSCs results in a global switch towards shorter mRNA isoforms and reduced serial engraftment potential, but has no effect on HSC survival197.
RNA editing.
In addition to changing mRNA 3’ ends, cells can modify individual nucleotides in the transcript. One established mechanism for nucleotide modification is RNA editing, in which adenosine is deaminated to inosine (A-I editing)205. A-I editing has been reported in quiescent HSCs and NSCs206,207. The enzymes catalysing RNA editing — ADAR (ADAR1), ADAR2 and AIMP2 — are widely expressed and their mRNAs can be found in quiescent MuSCs, NSCs and HSCs in cell atlas transcriptomics data208–210. Moreover, at least in HSCs, ADAR1 expression is dynamically regulated and increases in progenitor cells compared to HSCs211. In HSCs, overexpression of ADAR, but not of a mutant lacking enzymatic actvity, results in activation into the cell cycle (Figure 4b)206. ADAR overexpression results in editing of the primary miRNA pri-miR-26a, preventing its maturation, and of a miRNA binding sites in the 3’UTR of MDM2, leading to mRNA stabilization206. By contrast, deletion of Adar leads to quiescence maintenance and increased levels of the cell cycle inhibitor CDKN1A206. Although these experiments show that RNA editing is a key mechanism controlling HSC quiescence, it remains an open question whether ADAR also acts in a non-enzymatic manner212.
RNA methylation.
RNA can undergo extensive methylation in cells213, and this modification is often mutually exclusive with RNA editing214. In quiescent HSCs, mRNAs of an estimated 9% of the expressed genes contain N6-methyladenosine (m6A), and this fraction decreases with activation215–217. The m6A writer METTL3 is expressed in HSCs, and methylation enzymes can also be detected in other quiescent stem cell populations in cell atlas data208–210,215,216. Conditional deletion of Mettl3 from HSCs leads to quiescence exit and a gradual increase in HSC numbers215,218,219 (Figure 4a). One target of METTL3 is MYC mRNA, which has increased stability when methylated215,218. Similary, conditional loss of METTL3 in MuSCs causes an initial increase in MuSC numbers immediately after injury, suggesting enhanced activation220. Myc, well-known for its role in cell cycle progression and cell expansion, is important for quiescence maintenance in HSCs and NSCs221,222. This coincides with a loss in NOTCH mRNA methylation and decreased protein levels, while mRNA levels remain stable (suggesting decreased translation of the mRNA; see below)220. Deletion of Mettl3 or Mettl14 leads to reduced m6A-mRNA levels and delayed cell-cycle exit in embryonic NSCs, resulting in a failure to establish the quiescent NSC pool223 (Figure 4c). In HFSCs, the MYC-induced SUN2 catalyzes the formation of 5-methylcytosine (m5C) in tRNA (Figure 4b). Expression of SUN2 in HFSCs increases in anagen and its deletion prolongs quiescence maintenance, resulting in a delayed hair cycle224.
Alternative splicing.
Alternatively spliced mRNA isoforms are common in all cells210,225,226, but the observation that known splicing factors are dynamically regulated during MuSC and HSC activation indicates that alternative splicing could be an important regulatory mechanism in quiescent stem cells227–229. However, surprisingly little is known about the regulation and function of alternatively spliced isoforms in quiescent stem cells. The RNA binding protein quaking (QKI) regulates alternative splicing of cell polarity genes in MuSCs. Conditional deletion of Qki results in alternative splicing of ITGA7 and Numb, and reduced activation and assymetrical division230. Protein arginine N-methyltransferase 5 (PRMT5) controls mRNA splicing in quiescent HSCs231. Following conditional deletion of Prmt5, HSCs show a global changes in alternative splicing, increased protein synthesis and increased HSC numbers, all indicating quiescence exit231. Thrombopoietin signaling through its receptor MPL can promote either quiescence or proliferation of HSCs in a concentration-dependent manner. Sensitivity of HSCs to thrombopoietin depends on alternative splicing of the receptor MPL, which is expressed as a full-length or as a truncated, dominant-negative isoform; the RNA-binding protein OTT1 (also known as RBM15) determines the relative abundance of the two isoforms through alternative splicing232. Accordingly, by regulating the mRNA splicing of a key gene, HSCs can fine-tune their readiness to exit the quiescent state.
Splicing can also be repressed, leading to intron retention233. Transcript isoforms that retain introns accumulate in quiescent MuSCs and HSCs231,234,235. In MuSCs, overexpression of the oncogene and proliferation regulator DEK results in intron excision of intron-retaining mRNAs, which is followed by MuSC activation234 (Figure 4). Depletion of DEK blocks intron excision and prevents cell cycle entry234, indicating that intron retention is key for quiescence maintenance. Loss of DEK triggers HSC activation and diminished self-renewal236, but whether this involves altered intron retention is unknown.
Nuclear export.
Mature mRNAs are exported to the cytoplasm for translation237. Unsurprisingly, deletion of the gene encoding the mRNA export protein THO complex subunit 5 (THOC5) homolog leads to a dramatic loss of HSCs238, likely due to general toxicity of the loss of RNA export. Cells can regulate the export of specific RNAs. Quiescent NSCs in zebrafish express miR-9, which forms a complex with Argonaute (AGO) proteins, a component of the RNA-induced silencing complex, and accumulates in the nucleus239. Nuclear accumulation of miR-9 leads to quiescence maintenance, whereas cytoplasmic retention of miR-9 leads quiescence exit, although the downstream mRNA targets of miR-9, and whether any target mRNA is co-transported with it, remain unclear239.
RNA regulation in the cytoplasm
Following export into the cytoplasm, the mature mRNA can be subjected to several regulatory processes that control when and where a protein is produced. These mechanisms include RNA degradation by small RNAs, as well as RNA degradation by RNA-binding proteins. Transcripts can also be stored in mRNA granules. In the following sections, we discuss how these post-transcriptional mechanisms regulate processes related to stem cell quiescence (Figure 5).
Figure 5: Post-transcriptional regulatory mechanisms in the cytoplasm that control stem cell quiescence.
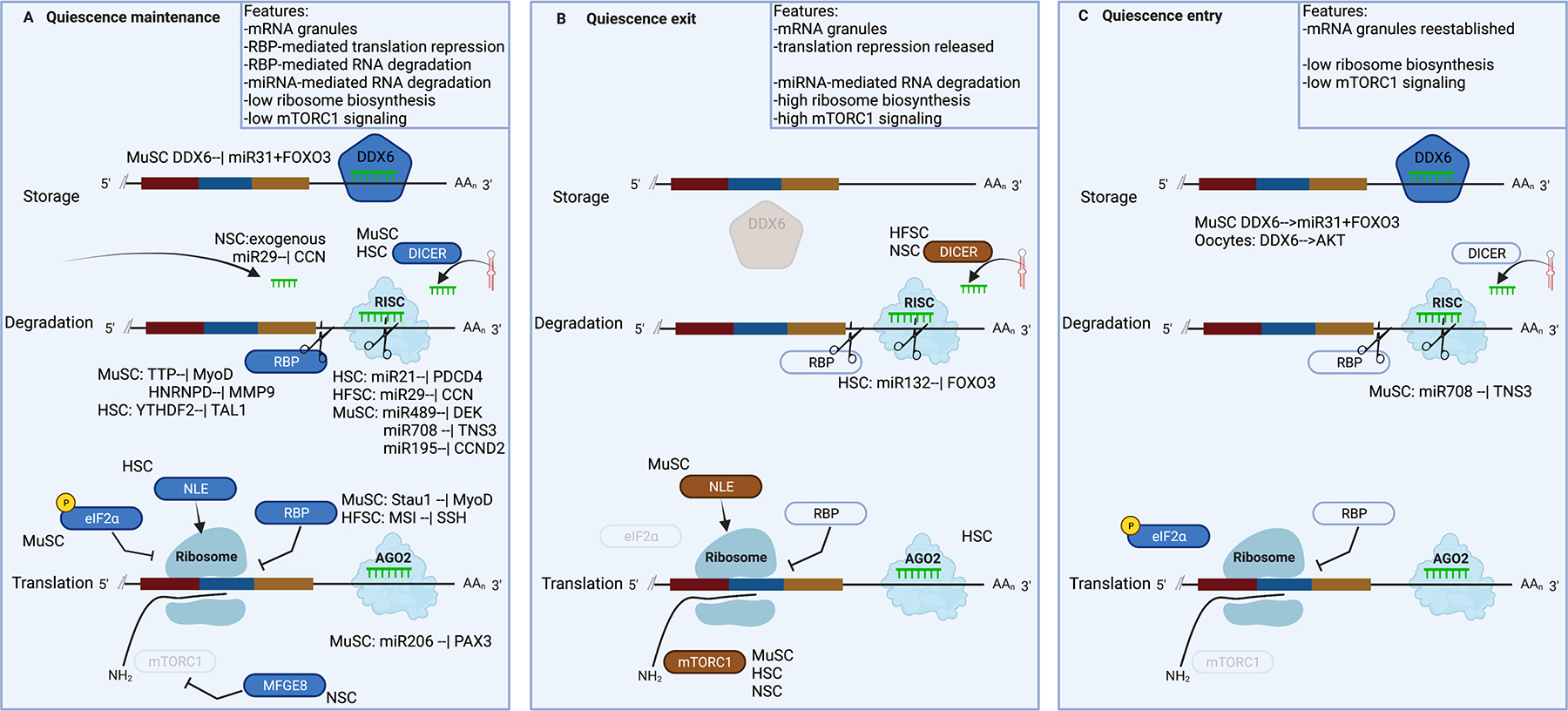
The mature mRNA can be degraded by miRNA-containing RISC complexes or RNA binding proteins (RBPs) with RNAse activity like TTP, stored in granules like DDX6-containing p-bodies, or translated by ribosomes. Translation can be regulated by RNA binding proteins like STAU1 (Staufen 1) or by mTORC signaling, and by phosphorylation of eIF2a.
DDX6, DEAD-box helicase 6; MYF5, myogenic factor 5; miR, microRNA; 3’UTR, 3’ untranslated region; DICER, double-stranded RNA-specific endoribonuclease; RISC, RNA-induced silencing complex; RBP, RNA-binding protein; MyoD, myogenic differentiation 1; HNRNPD, heterogeneous nuclear ribonucleoprotein D; MMP9, matrix metallopeptidase 9; YTHDF2, YTH N6-methyladenosine RNA binding protein 2; TAL1, T-cell acute lymphocytic leukemia protein 1; PDCD4, programmed cell death 4; DEK, DEK proto-oncogene; TNS3, tensin 3; CCND2, cyclin d2; CDC25A,B, cell division cycle 25a,b; NLE1, notchless homolog 1; eIF2a, eukaryotic translation initiation factor 2A; MSI, musashi RNA binding protein 1; SSH, slingshot protein phosphatase; PAX3, paired box 3; AGO2, argonaute 2; mTOR1, mammalian target of rapamycin 1; MFGE8, milk fat globule EGF and factor V/VIII domain containing; FOXO3, forkhead box o3; AKT, AKT serine/threonine kinase, also known as protein kinase b.
RNA degradation by microRNAs.
Mammalian cells can degrade mRNA through several classes of small RNAs, chiefly among them miRNAs. miRNAs are processed from precursors by Dicer and other ribonucleases and target mRNAs through sequence complmantarity (mostly) to 3’UTRs, in association with Argonaute proteins. miRNAs are well known to affect the quiescent state240 (Figure 5). Conditional deletion of Dicer results in activation and apoptosis in MuSCs and in apoptosis and a self-renewal defect in HSCs241,242 (Figure 5a,c). These phenotypes are at least partly due to disrupted miRNA-mediated mRNA degradation. MuSCs maintain quiescence by expressing miR-489 and miR-708, which downregulate DEK and tensin 3 (TNS3) mRNAs, respectively242,243. Overexpression of miR-489 or miR-708 prolongs quiescence, whereas their depletion triggers quiescence exit. Finally, quiescent MuSCs use miR-195 and miR-497 to destabilize mRNAs encoding the cell cycle factors CDC25A (also known as M-phase inducer phosphatase 1), CDC25B (also known as M-phase inducer phosphatase 2) and CCND2 (also known as G1/S-specific cyclin D2), thereby supporting quiescence maintenanace244. In HSCs, miR-21 decreases PDCD4 expression and function in the NFκB pathway to maintain HSC quiescence. Loss of miR-21 results in activation, whereas its overexpression prevents activation245 (Figure 5a).
In contrast to MuSCs and HSCs, deletion of Dicer has no apparent effect on quiescence maintenance in NSCs or HFSCs, but leads to a loss of activated progenitors and subsequent defects in neurogenesis or hair cycle progression, respectively246–248. Nevertheless, individual miRNAs are important for quiescence in both NSCs and HFSCs. In NSCs, miR-204 controls quiescence by enabling the degradation of the mRNAs of the pro-activation factors MEIS2 and SOX11 (Ref.249). However, quiescent NSCs acquire miR-204 from the choroid plexus and do not depend on Dicer for its synthesis249. In HFSCs, miR-29a and/or miR-29b1 destabilize cyclin mRNAs and components of the WNT signaling pathway (a potent morphogen pathway that controls cell expansion in mouse and human HFSCs, as well as in mouse MuSCs, whereas it maintains quiescence in mouse HSCs250–254), and loss of miR-29a and miR-29b1 results in activation250 (Figure 5a).
As quiescent stem cells become active, the miRNAs that prevent activation are downregulated. MuSCs undergoing activation decrease the expression of miR-195 and miR-497, resulting in increased stability of CDC25A, CDC25B and CCND2 (Ref.244). In parallel, the stem cells induce the expression of new miRNAs network that destabilizes mRNAs involved in quiescence maintenance. HSCs lacking miR-132 and miR-212 have stable FOXO3 mRNA levels and are more quiescent, show better engraftment and less apoptosis, and undergo slower activation following stimulation by lipopolysaccharides (Figure 5b)255. The transcription factor FOXO3 plays a role in maintaining and establishing the quiescent state256,257. Conversely, to establish the quiescence program, the cell needs to suppress the ongoing activation program. This may involve the degradation of specific transcripts. However, to the best of our knowledge, no studies yet have directly investigated the role of RNA degradation in the process of quiescence entry. Nevertheless, many studies of miRNAs report on decreased stem cell numbers following regeneration, hinting at a change in self-renewal and/or quiescence entry243,258,259. For example, long term depletion of miR-708 leads to apoptosis-independent reduction in MuSC numbers243 (Figure 5c).
small-RNA-independent RNA degradation.
In addition to miRNAs, quiescent stem cells use small-RNA-independent mechanisms to destabilize specific transcripts. Quiescent MuSCs express the mRNA decay activator TTP (also known as ZFP36), which degrades MyoD mRNA190 (Figure 5a). TTP loss results in MyoD mRNA accumulation and MuSC activation190. By contrast, Ttp deletion in HSCs does not affect quiescence260, possibly due to redundancy with its family members. MuSCs also express the RNA-binding protein (RBP) AUF1 (also known as HNRNPD), which targets for degradation transcripts containing AU-rich elements. MuSCs lacking AUF1 accumulate matrix metalloproteinase 9 (MMP9) mRNA, resulting in increased matrix remodeling and reduced quiescence entry261. In HSCs, deletion of the gene encoding the RBP and m6A reader YTHDF2 leads to stabilization of methylated mRNAs that encode transcription factors such as TAL1, which in turn increases the numbers of quiescent HSCs216 (Figure 5a). RBPs can also stabilize mRNAs262, for example as shown by the function of ELAV proteins on the MSI1 mRNA in NSCs263,264.
RNA storage.
Storage of mRNA is an intriguing possible mechanism of cellular memory of the activated state and priming of activation265. Processing bodies [G] (P-bodies) and RNA granules [G] store RNA and are key for stem cell differentiation and lymphocyte activation due to their regulation of differentiation and cell cycle factors266,267, yet they are poorly studied in the context of stem cell quiescence. The RNA granule protein FMRP is highly expressed in quiescent MuSCs and NSCs and its loss causes aberrant activation268–270. In quiescent MuSCs, miR-31 ensures the sequestration of its target mRNA myogenic factor 5 (MYF5; a myogenesis factor) in RNA granules containing the RNA helicase DDX6 (Figure 5a). Antagomirs against miR-31 result in loss of RNA granules and in MuSC activation194. Moreover, blocking RNA granules disassembly by inhibition of phosphatase 2A activity prevents MYF5 protein accumulation194. When stem cells undergo activation, they dissolve the RNA granules, and forced maintenance of RNA granules prolongs quiescence in MuSCs194. It is unclear whether MuSCs accumulate the MYF5 RNA in the quiescent state or during quiescence entry. In oocytes (see below), DDX6 is essential for establishing quiescence271. In the absence of DDX6, oocytes do not form RNA granules, fail to repress PI3K–AKT signaling and continuously proliferate271 (Figure 5c).
Protein synthesis and degradation
Quiescent stem cells show low levels of mRNA translation, even though the core components of the translation machinery are expressed26,75,98,272–275. Quiescent HSCs express similar levels of core rRNAs and ribosomal proteins compared to their activated counterparts98,275. Nevertheless, quiescent stem cells increase protein translation during activation26,75,98,272–274. The core translation machinery, including rRNA and ribosomal proteins, is dynamically regulated in both quiescent and activating stem cells26,276,277.
RNA folding.
To be translated, mRNAs need to be (un)folded such that the ribosome can access the start codon. 5’UTRs of mRNAs can contain regulatory elements that affect mRNA folding and accessibility, and that can affect activation of quiescent stem cells. Expression of DHX36, a helicase that unwinds RNA G-quadruplex (rG4) structures, increases with MuSC and HSC activation and its loss resulted in an activation defect278,279. In MuSCs, DHX36 specifically regulates the translation of GNAI2 mRNA by unwinding its 5′ UTR rG4 structures278.
Translation initiation.
HSCs not expressing the ribosome assembly factor Notchless protein homolog 1 (NLE1) accumulate immature ribosomal RNAs and exit quiescence280 (Figure 5a). Deletion of the transcription factor RUNX1 in HSCs similarly leads to reduced ribosome biosynthesis and protein translation and reduced HSC activation281. Among the direct targets of RUNX1 are ribosomal genes and rRNAs281. In MuSCs, eukaryotic translation initiation factor 2 alpha (eIF2α) is phosphorylated to inhibit translation273. Following the replacement of the eIF2A gene in MuSCs by a mutant gene that renders the protein resistant to phosphorylation, MuSCs undergo activation and fail to self-renew273 (Figure 5a,c).
Although phosphorylated eIF2α generally inhibits translation, certain mRNAs escape this inhibition. The 5′ UTR of the TACC3 mRNA (encoding a mitotic spindle stability factor) contains translation-inhibiting upstream open reading frames (uORFs); in stress conditions, translation of this mRNA is anabled by phospho-eIF2α-mediated readthrough of these uORFs282. Loss of TACC3 results in a MuSC activation defect, likely due to defective spindle formation282. Cytoplasmic polyadenylation element binding protein 1 (CPEB1) promotes MyoD1 protein synthesis by binding to the cytoplasmic polyadenylation elements within its 3’ UTRs to regulate MuSC activation and muscle regeneration283. In contrast to HSCs, in MuSCs, NLE1 increases early during activation, and conditional deletion of Nle1 results in accumulation of unprocessed rRNAs and in prolonged quiescence277 (Figure 5b).
The fact that manipulation of ribosome biosynthesis has immediate effects on stem-cell quiescence exit is somewhat surprising, since, at least in HSCs, there is no shortage in functional ribosomes98. A possible mechanism for these immediate effects could be the biogenesis and activity of specific, specialised ribosome subunits.
Translation repression.
Cells can repress the translation of specific mRNAs through RBPs or miRNAs284. Quiescent HFSCs express the Sonic Hedgehog (SHH) mRNA, but repress its translation through the RBP Musashi to maintain the quiescent state36. Loss of Musashi leads to SHH signaling and HFSC activation, whereas induced overexpression of Musashi prolongs telogen36 (Figure 5a). Similarly, quiescent MuSCs express the mRNA of the activation factor MyoD, but repress its translation through the RBP Staufen-134. Loss of Staufen-1 leads to MyoD translation and MuSC activation34. Staufen-1–RNA immunoprecipitation experiments revealed that its main targets are mRNA of cell cycle regulators34. Quiescent NSCs express the mRNA of the centromose protein Ninein285. The exon-junction complex [G] binds the Ninein mRNA and prevents its translation, while enabling its transport to centrosomes for localized translation285. Finally, miRNAs can repress translation by targeting the AGO2-containing RNA-induced silencing complex to mRNAs. AGO2 enables translation repression and loss of AGO2 limits HSC activation286,287. Quiescent MuSCs in hind limb muscles maintain the quiescent state through miR-206-mediated translational repression of PAX3 mRNA; loss of miR-206 results in PAX3 translation and MuSC activation195.
Translation depends on mTORC1 activity (Figure 5b). Quiescent NSCs maintain low mTOR signaling by expressing MFGE8 (also known as lactadherin)288. Loss of MFGE8 induces mTOR signaling and subsequent NSC activation, which can be rescued with the mTOR inhibitor rapamycin288. Loss of mTOR limits activation in HSCs, whereas mTOR signalling gain-of-function induces activation and limits self-renewal152. MuSCs, FAPs and HSCs can enter an alert state in response injury-induced systemic signals, in which they remain quiescent, but show active mTOR signalling and can enter the cell cycle more rapidly73,74. Deletion of Rptor (which encodes RAPTOR) or Met prevents the alert response73. Restraining the activity of the mTOR–eIF4EBP1 and mTOR–S6K1 (also known as RPS6KB1) axes is crucial for self-renewal 150,152,289. Finally, tRNA processing modulates translation; processing of tRNA-derived stress-induced small RNAs (tiRNAs) by the RNase angiogenin was shown to maintain HSC quiescence290. Transfection of one specific tRNA precursor, tiRNA-Gly-GCC, can block translation in HSCs, which leads to increased self-renewal290.
Protein degradation.
Cells can regulate protein abundance through targeted protein degradation. E3 ubiquitin ligases covalently link ubiquitin moieties to proteins to target them for proteasome-mediated degradation (see more on protein modifications below). HSCs and MuSCs undergoing activation show increased ubiquitin expression, global protein ubiquitylation and proteasome activity291,292 (Figure 6). Nevertheless, the expression of some E3 ubiquitin ligases in quiescent HSCs and NSCs declines with activation35,293–295, indicating that in addition to roles in quiescence exit, protein degradation has roles in quiescence maintenance. Deletion of the genes encoding the E3 ubiquitin ligases FBW7, HUWE1 or MAEA results in HSC activation293–295. FBW7 prevents the accumulation of MYC294, whereas MAEA targets cytokine receptors for degradation through ubiquitin-dependent internalization and autophagy294,295 (Figure 6a). In contrast to HSCs, loss of HUWE1 has no apparent effect on quiescent NSCs35.
Figure 6: Post-translational regulatory mechanisms that control stem cell quiescence.
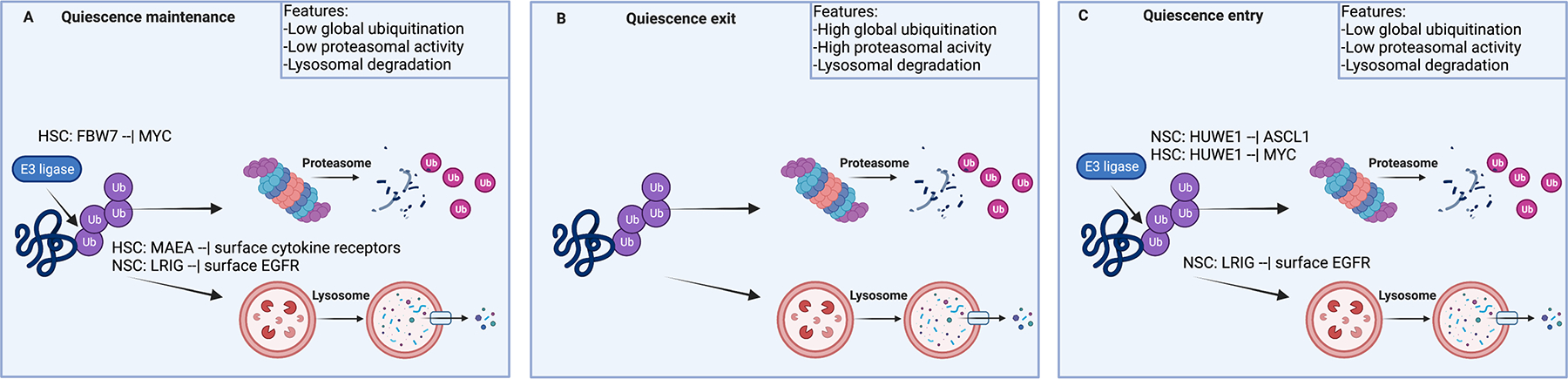
Once translated, the native protein can be modified with post-translational modifications such as phosphorylation, glycosylation, and ubiquitination, which affect protein function and turnover. Ubiquitinated proteins can be degraded by the proteasome and the lysosome.
FBW7, f-box and WD repeat domain containing 7; MYC, MYC proto-oncogene; Ub, ubiquitin; MAEA, macrophage erythroblast attacher; LRIG, leucine rich repeats and immunoglobulin like domains; EGFR, epidermal growth factor receptor; ID4, inhibitor of DNA binding 4; ASCL1, achaete-scute family BHLH transcription factor 1; SIRT7, sirtuin 7; NFATC1, nuclear factor of activated T Cells 1; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1.
Quiescent NSCs express the mRNA, but not the protein of the transcription factor ASCL1 due to immediate degradation of the newly translated protein296. The ASCL1 protein can be stabilized through binding the transcription factor E47. However, in quiescent NSCs, E47 is sequestered by ID4, rendering it unable to bind ASCL1. Loss of ID4 prevents this sequestration, causing ASCL1 accumulation and NSC activation296. Quiescent NSCs degrade epidermal growth factor receptor (EGFR) through LRIG1-dependent ubiquitylation followed by lysosomal degradation. In the absence of LRIG1, NSCs show increased EGFR signaling and activation16,297 (Figure 6a). Overexpression of constitutively active EGFR can force quiescent NSCs to undergo activatation298. Finally, deletion of the gene encoding the endoplasmic reticulum associated degradation [G] protein SEL1L results in HSC activation299. SEL1L induces degradation of the mTOR activator RHEB and loss of RHEB can rescue the SEL1L phenotype299.
In HFSCs undergoing activation, SIRT7 deacetylates the transcription factor NFATC1, which leads to its proteasomal degradation300. In the absence of SIRT7, NFATC1 is not degraded and the cells stay quiescent300 (Figure 6b). In MuSCs, deletion of the essential protoasome component RPT3 (also known as 26S proteasome regulatory subunit 6B) results in reduced proteolytic activity and proliferation and in apoptosis after injury292. Chaperone-mediated autophagy is essential for HSC and MuSC activation138,301. As discussed above, NSCs undergoing activation upregulate EGFR expression and signaling and downregulate of ID4 in order to stabilize ASCL1 (Ref.296,298). In activated MuSCs and HSCs, the cells that express high levels of ID1 show slow cycling and increased self-renewal302,303, suggesting that E47 sequestration by ID proteins could be important for quiescence exit in other stem cell populations as well.
In addition to clearing proteins involved in regulation of the quiescent state, stem cells must also clear protein aggregates. NSCs undergoing activation use the intermediate filament vimentin to localize proteasomes to protein aggregates and facilitate protein turnover, and this clearance of aggregates allows the NSCs to become active304. Loss of vimentin causes a decline in protein degradation and a delay in NSC activation304.
Stem cells entering the quiescent state must dispose of the proteins that drive activation and the cell cycle. Unsurprisingly, targeted protein degradation is therefore an important mechanism for quiescence entry. In NSCs, HUWE1 ubiquitylates the ASCL1 to induce its degradation and block transcription of cyclin D. This is key for entry into the quiescent state, as NSCs without HUWE1 fail to enter the quiescent state, resulting in NSC depletion and loss of neurogenesis35 (Figure 6c). HUWE1 enables re-quiescence also in HSCs, where it induces the degradation of N-MYC293. HSCs lacking the E3 ligase MAEA fail to reconstitute a quiescent stem cell population, suggesting a defect in quiescence entry295. NSCs without LRIG1 have more surface EGFR and are less responsive to quiescence-inducing BMPs16.
Post-translational modifications
All cells regulate protein function through post-translational modifications (PTMs), of which some of the best characterized are the histone PTMs, which have epigenetic roles. However, few studies actually examine the role of a specific modification of a specific protein in the regulation of stem cell quiescence. Loss of the phosphatase PTEN results in activation of HSCs, NSCs, HFSCs and MuSCs and gradual depletion of the stem cell pool98,177–179,305 (Figure 2c). Genetic interference with kinase signaling, such as the PI3K or MAPK pathways, affects quiescence130,144–149,176. Loss of glycosylation enzymes leads to activation of HSCs, MuSCs, and NSCs, possibly due to changes in the abundance of glycosylated Notch proteins306–308. However, none of these studies evaluate the effect of the modification on protein abundance and the quiescent state.
Histones bear many PTMs to regulate chromatin accessibility. Histone acetylation, methylation, phosphorylation and ubiquitylation have all been described in quiescent stem cells2,309. As discussed above, the gene encoding eIF2α was replaced in MuSCs with a mutant gene that encodes eIF2α that cannot be phosphorylated at Ser51, leading to MuSC activation, thereby directly demonstrating the importance of this particular PTM in stem cell quiescence273. MuSCs bearing a mutation in the gene encoding the transcription factor PAX7 rendering the protein insensitive to Lys193 acetylation show decreased quiescence entry after injury, due to changes in the protein’s DNA binding capacity310. Similarly, HFSCs bearing a mutation in the gene encoding the transcription factor NFATC1 rendering the protein insensitive to Lys612 acetylation, show decreased NFATC1 stability and quiescence exit300. Other known PTMs such as sumoylation, crotonylation, citrullination and glycosylation are yet to be studied in quiescent stem cells.
Post-transcriptional regulation of germ line quiescence
One unique stem cell model that is well-known for post-transcriptional regulation of the quiescent state is the oocyte311–315. Oocytes are adult-derived germ line stem cells that can maintain a quiescent state for decades until they are activated in response to luteinizing hormone. Upon their activation, oocytes either proceed to ovulation or die, but they never self-renew. In contrast to somatic stem cells, oocytes undergo chromatin condensation and are arrested in the prophase of the first meiosis stage. Although low levels of transcription can occur at the condensed chromatin, oocytes largely depend on post-transcriptional regulation of pre-synthesized mRNAs to regulate the quiescent state. Oocyte quiescence maintenance and exit depend on a set of key genes similar to somatic stem cell quiescence. Oocytes express high levels of CDK inhibitors and low levels of cyclins. Mice lacking p27 are infertile and have multi-oocyte follicles, consistent with a loss of quiescence316. Rb is unphosphorylated and active in oocytes and its conditional deletion results in activation and formation of teratomas317. Conditional loss of Cdk1 results in oocytes that cannot exit quiescence318.
To enter quiescence, oocytes express ZFP36L2, which degrades the mRNAs of transcription regulators to ensure transcriptional gene silencing315. Like somatic stem cells, oocytes express mRNA isoforms with different UTRs, which results in different levels of translation. However, these mRNAs are not yet polyadenylated. Instead, translation is regulated by cytoplasmic polyadenylation by CPEB1 upon activation319, which enables mRNA loading into ribosomes320. Activated oocytes undergo a shift towards isoforms with longer 3’ UTRs321. However, in contrast to somatic stem cells, this shift is caused by differences in mRNA degradation rather than by alternative cleavage and polyadenylation. Surprisingly, miRNAs, though present, are unable to repress their target genes due to lack of expression of Argonaute proteins, which explains the stability of the long 3’UTR isoforms321. Activated oocytes do degrade 20% of their transcriptome throug the activity of the m6A reader YTHDF2. Loss of YTHDF2 causes infertility due to apoptosis of activated oocytes after completion of meiosis322,323. Finally, recent work has shown that quiescent oocytes store maternal mRNAs encoding such RNA degradation factors in specialized mitochondria-associated structures marked by Zygote Arrest 1 (ZAR1)324. Although these regulatory mechanisms might be unique to the oocyte, they can serve as examples of potential new mechanisms waiting to be uncovered in the regulation of somatic stem cell quiescence.
Conclusions and future directions
Quiescent stem cells are essential for tissue homeostasis and regeneration and their dysfunction contributes to aging. Genetic loss-of-function studies revealed that stem cells actively maintain the quiescent state and recent work has begun to uncover layers of regulatory control that act post-transcriptionally to fine-tune the amount of protein that is produced. These studies reveal a dramatic and dynamic complexity beyond transcriptional profiles that underscores how stem cell quiescence and its related processes are heavily regulated, biologically complex phenomena. Moreover, it highlights the difficulties of extrapolating transcriptome data to protein levels.
Adult stem cells offer the potential to develop new cell-based therapies. The post-transcriptional regulatory mechanisms offer potential footholds to improve stem cell function in vivo. Antisense oligonucleotides can affect quiescent stem cell processes in vivo by targeting alternative polyadenylation, miRNA-dependent regulation, or translation34,195,201,239,325. This opens the door to therapeutics to enhance quiescent stem cell function, analogous to the antisense oligonucleotide technologies that are currently in clinic. Moreover, improved understanding of the post-transcriptional mechanisms that enable quiescence entry could enable interventions to improve stem cell potency following ex vivo stem cell expansion by pushing the expanded stem cells into the quiescent state for improved autologous transplantation. Allowing the cells to expand before a return to quiescence would also circumvent the barrier to gene therapies posed by the lack of homologous recombination in the quiescent state. Finally, cancer stem cells [G] can adopt a quiescent state, which renders them resistant to therapy, and these quiescent cancer stem cells are thought to contribute to post-treatment relapse326,327. Many of the signaling pathways that are important for stem cell quiescence (e.g. Notch, mTORC, p38 MAPK, and CDKN1B), also play a role in cancer stem cell quiescence326. Therefore, a better understanding of the mechanisms that regulate stem cell quiescence could help pave the way toward therapeutics that specifically target the quiescent cancer stem cell, either by activating those cells to render them sensitive to other therapies, or by prolonging quiescence to prevent activation and relapse.
While the studies discussed in this review offer a glimpse of the regulatory mechanisms of quiescence maintenance, exit, and entry, ultimately, the field needs better tools to follow cells as they undergo the different processes. The identification of specific cell surface markers for quiescent stem cells will facilitate the prospective isolation and study of these cells. The field needs better lineage tracers to track cells as they enter the quiescent state. One challenge is that under certain conditions, the first division may be asymmetric, resulting in a daughter cell that immediately enters the quiescent state. This can be followed with DNA nucleotide analogs, but the time frame may be too short to allow for genomic recombination to activate lineage tracers. One exiting area of development is DNA barcoding, which has allowed for the fate mapping and simultaneous transcriptional profiling of single HSCs after in vivo or ex vivo activation of a barcode library27,328. A further challenge in the study of stem cell quiescence is that isolation of quiescent stem cells from their native environment tends to mimic injury and leads to cell cycle entry and changes in gene expression14,43,329. Accordingly, there is a need for improved models for the study, and manipulation, of ex vivo stem cell quiescence14,18,330,331. For example, BMP4 can induce in vitro quiescence in NSCs, and the HDAC6-inhibitor Tubastatin A can induce in vitro quiescence in MuSCs21,96,186,232,239,307,332–334. Finally, the majority of studies of stem cell quiescence have been done using mice. The field needs a better understanding of stem cell quiescence in other model organisms and would benefit from a non-human primate model to enable translational research. In summary, the post-transcriptional regulation of stem cell quiescence is an exciting and open field with the potential to impact regenerative medicine and our understanding of stem cell biology.
Supplementary Material
GLOSSARY
- Differentiate:
Differentiation is the process by which a stem cell withdraws from the cell cycle and adopts specialized tissue functions, while losing stem cell functions such as the ability to self-renew
- Embryonic stem cells:
Cells derived from the blastocyst of the embryo and able to form all tissue lineages
- Germline stem cells:
Cells that can generate the haploid gametes
- Induced pluripotent stem cells:
Cells created from somatic cells through the overexpression of specific transcription factors, rendering them immortal and able to form all tissue lineages
- Adult stem cells:
Rare populations of cells that are found in the body throughout the majority of postnatal life and give rise to a limited number of mature cell types that build the tissue in which they reside
- Autologous stem cell transplantation therapies:
Procedures in which stem cells are isolated from a person and placed back following expansion and/or modification such as gene correction
- Stem cell activation:
The process by which a quiescent stem cell exits the quiescent state and enters the cell cycle
- Trithorax group:
family of proteins that modify or remodel histones to activate genes and keep them active
- Non-homologous end-joining:
A mechanism of DNA double-strand breaks repair without the need for a homologous template
- ATR:
A serine/threonine-protein kinase that senses persistent single-stranded DNA and activates a DNA damage checkpoint, leading to cell cycle arrest
- Processing bodies:
Cytoplasmic ribonucleoprotein (RNP) granules primarily composed of translationally repressed mRNAs
- RNA granules:
Non–membrane-bound organelles composed of RNA and protein
- Exon-junction complex:
Protein complex that forms on a pre-messenger RNA strand at the junction of two exons immediately after splicing
- Endoplasmic reticulum associated degradation (ERAD):
A pathway that targets misfolded proteins in the endoplasmic reticulum for ubiquitylation and proteasomal degradation
- Cancer stem cells:
Cancer cells with stem cell properties such as self-renewal; some cancer stem cells can adopt a quiescent state
- Bulge:
Morphologically distinct area in the hair follicle in between the opening of sebaceous gland and the attachment site of the arrector pili muscle that functions as the HFSC niche
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks Francois Guillemot and the other, anonymous, reviewers for their contribution to the peer review of this work.
References
- 1.Li N & Clevers H Coexistence of quiescent and active adult stem cells in mammals. Science (80-. ). 327, 542–545 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung TH & Rando TA Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orford KW & Scadden DT Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9, 115–128 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Ramalho-Santos M & Willenbring H On the Origin of the Term ‘Stem Cell’. Cell Stem Cell 1, 35–38 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Barker N, Bartfeld S & Clevers H Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell 7, 656–670 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Cho IJ et al. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Reports 12, 1190–1200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H & Watt FM Defining Adult Stem Cells by Function, not by Phenotype. Annu. Rev. Biochem. 87, 1015–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cable J et al. Adult stem cells and regenerative medicine—a symposium report. Ann. N. Y. Acad. Sci. 1462, 27–36 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An Z et al. A quiescent cell population replenishes mesenchymal stem cells to drive accelerated growth in mouse incisors. Nat. Commun. 9, 378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis EEL et al. A Quiescent, Regeneration-Responsive Tissue Engineered Mesenchymal Stem Cell Bone Marrow Niche Model via Magnetic Levitation. ACS Nano 10, 8346–8354 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Barriga FM et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell 20, 801–816.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai S et al. A Quiescent Bcl11b High Stem Cell Population Is Required for Maintenance of the Mammary Gland. Cell Stem Cell 20, 247–260.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu NY et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 19, 164–176 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Quarta M et al. An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 34, 752–759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montarras D et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science (80-. ). 309, 2064–2067 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Marqués-torrejón MÁ et al. LRIG1 is a gatekeeper to exit from quiescence in adult neural stem cells. Nat. Commun. 12, 259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacco A, Doyonnas R, Kraft P, Vitorovic S & Blau HM Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi H et al. Environmental Optimization Enables Maintenance of Quiescent Hematopoietic Stem Cells Ex Vivo. Cell Rep. 28, 145–158.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Mourikis P et al. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Bjornson CRR et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232–242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler A et al. Notch2 Signaling Maintains NSC Quiescence in the Murine Ventricular-Subventricular Zone. Cell Rep. 22, 992–1002 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Wang W et al. Notch2 blockade enhances hematopoietic stem cell mobilization and homing. Haematologica 102, 1785–1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimaki S et al. Notch1 and Notch2 Coordinately Regulate Stem Cell Function in the Quiescent and Activated States of Muscle Satellite Cells. Stem Cells 36, 278–285 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Sousa-Victor P, García-Prat L & Muñoz-Cánoves P Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 23, 204–226 (2022). [DOI] [PubMed] [Google Scholar]
- 25.De Micheli AJ et al. Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep. 30, 3583–3595.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llorens-Bobadilla E et al. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Fraticelli AE et al. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature 583, 585–589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velten L et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabezas-Wallscheid N et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169, 807–823.e19 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Machado L et al. Tissue damage induces a conserved stress response that initiates quiescent muscle stem cell activation. Cell Stem Cell 28, 1125–1135.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Joost S et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst. 3, 221–237.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zywitza V, Misios A, Bunatyan L, Willnow TE & Rajewsky N Single-Cell Transcriptomics Characterizes Cell Types in the Subventricular Zone and Uncovers Molecular Defects Impairing Adult Neurogenesis. Cell Rep. 25, 2457–2469.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Chua BA, Van Der Werf I, Jamieson C & Signer RAJ Post-Transcriptional Regulation of Homeostatic, Stressed, and Malignant Stem Cells. Cell Stem Cell 26, 138–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Morrée A et al. Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence. Proc. Natl. Acad. Sci. U. S. A. 114, E8996–E9005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urbán N et al. Return to Quiescence of mouse neural stem cells by degradation of a proactivation protein. Science (80-. ). 353, 292–295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma X et al. Msi2 Maintains Quiescent State of Hair Follicle Stem Cells by Directly Repressing the Hh Signaling Pathway. J. Invest. Dermatol. 137, 1015–1024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabezas-Wallscheid N et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 (2014). [DOI] [PubMed] [Google Scholar]
- 38.van Velthoven CTJ & Rando TA Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 24, 213–225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa MR et al. Continuous live imaging of adult neural stem cell division and lineage progression in vitro. Development 138, 1057–1068 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Brett JO et al. Exercise rejuvenates quiescent skeletal muscle stem cells in old mice through restoration of Cyclin D1. Nat. Metab. 2, 307–317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shariatmadar S et al. Electronic volume of CD34 positive cells from peripheral blood apheresis samples. Cytom. Part B - Clin. Cytom. 74, 182–188 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Freter R, Osawa M & Nishikawa SI Adult stem cells exhibit global suppression of RNA polymerase II serine-2 phosphorylation. Stem Cells 28, 1571–1580 (2010). [DOI] [PubMed] [Google Scholar]
- 43.van Velthoven CTJ, de Morree A, Egner IM, Brett JO & Rando TA Transcriptional Profiling of Quiescent Muscle Stem Cells In Vivo. Cell Rep. 21, 1994–2004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tesio M & Trumpp A Breaking the cell cycle of HSCs by p57 and friends. Cell Stem Cell 9, 187–192 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Urbach A & Witte OW Divide or Commit – Revisiting the Role of Cell Cycle Regulators in Adult Hippocampal Neurogenesis. Front. Cell Dev. Biol. 7, 55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson C, Belluschi S & Laurenti E Beyond “ to divide or not to divide ”: Kinetics matters in hematopoietic stem cells. Exp. Hematol. 92, 1–10 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Wang J et al. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood 118, 5429–5438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seale P et al. Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Lien WH et al. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat. Cell Biol. 16, 179–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauro a. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9, 493–5 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cebrián-Silla A et al. Unique Organization of the Nuclear Envelope in the Post-natal Quiescent Neural Stem Cells. Stem Cell Reports 9, 203–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuler N, Timm S & Rübe CE Hair Follicle Stem Cell Faith Is Dependent on Chromatin Remodeling Capacity Following Low-Dose Radiation. Stem Cells 36, 574–588 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Liu L et al. Chromatin Modifications as Determinants of Muscle Stem Cell Quiescence and Chronological Aging. Cell Rep. 4, 189–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lien WH et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9, 219–232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puri D, Gala H, Mishra R & Dhawan J High-wire act: The poised genome and cellular memory. FEBS J. 282, 1675–1691 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Cui K et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation Kairong. Cell Stem Cell 4, 80–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Addicks GC et al. MLL1 is required for PAX7 expression and satellite cell self-renewal in mice. Nat. Commun. 10, 4256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMahon KA et al. Mll Has a Critical Role in Fetal and Adult Hematopoietic Stem Cell Self-Renewal. Cell Stem Cell 1, 338–345 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Jones M et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J. Clin. Invest. 125, 2007–2020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkatraman A et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500, 345–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bigot A et al. Age-Associated Methylation Suppresses SPRY1, Leading to a Failure of Re-quiescence and Loss of the Reserve Stem Cell Pool in Elderly Muscle. Cell Rep. 13, 1172–1182 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Wüst S et al. Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by miR-1/133a-Mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster. Cell Metab. 27, 1026–1039.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Latil M et al. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat. Commun. 3, 903 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Wani GA et al. Metabolic control of adult neural stem cell self-renewal by the mitochondrial protease YME1L. Cell Rep. 38, 110370 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Tang Y et al. Mitochondrial aerobic respiration is activated during hair follicle stem cell differentiation, and its dysfunction retards hair regeneration. PeerJ 4, e1821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiu J et al. Using mitochondrial activity to select for potent human hematopoietic stem cells. Blood Adv. 5, 1605–1616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spencer JA et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coller HA The paradox of metabolism in quiescent stem cells. FEBS Lett. 593, 2817–2839 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flores A et al. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat. Cell Biol. 19, 1017–1026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stoll EA et al. Neural stem cells in the adult subventricular zone oxidize fatty acids to produce energy and support neurogenic activity. Stem Cells 33, 2306–2319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryall JG et al. The NAD+-dependent sirt1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodgers JT et al. MTORC1 controls the adaptive transition of quiescent stem cells from G 0 to GAlert. Nature 510, 393–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodgers JT, Schroeder MD, Ma C & Rando TA HGFA Is an Injury-Regulated Systemic Factor that Induces the Transition of Stem Cells into GAlert. Cell Rep. 19, 479–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baser A et al. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Shan T et al. Lkb1 is indispensable for skeletal muscle development, regeneration, and satellite cell homeostasis. Stem Cells 32, 2893–2907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gan B et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gurumurthy S et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659–663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakada D, Saunders TL & Morrison SJ Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitale I, Manic G, De Maria R, Kroemer G & Galluzzi L DNA Damage in Stem Cells. Mol. Cell 66, 306–319 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Mohrin M et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vahidi Ferdousi L et al. More efficient repair of DNA double-strand breaks in skeletal muscle stem cells compared to their committed progeny. Stem Cell Res. 13, 492–507 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Sotiropoulou PA et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat. Cell Biol. 12, 572–582 (2010). [DOI] [PubMed] [Google Scholar]
- 84.Barazzuol L, Ju L & Jeggo PA A coordinated DNA damage response promotes adult quiescent neural stem cell activation. PLoS Biol. 15, e2001264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cameron BD et al. Bcl2-Expressing Quiescent Type B Neural Stem Cells in the Ventricular–Subventricular Zone Are Resistant to Concurrent Temozolomide/X-Irradiation. Stem Cells 37, 1629–1639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rossi DJ et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Flach J et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu L et al. Impaired Notch Signaling Leads to a Decrease in p53 Activity and Mitotic Catastrophe in Aged Muscle Stem Cells. Cell Stem Cell 23, 544–556.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinha M et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science (80-. ). 344, 649–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beerman I, Seita J, Inlay MA, Weissman IL & Rossi DJ Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qing Y, Wang Z, Bunting KD & Gerson SL Bcl2 overexpression rescues the hematopoietic stem cell defects in Ku70-deficient mice by restoration of quiescence. Blood 123, 1002–1011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruzankina Y et al. Deletion of the Developmentally Essential Gene ATR in Adult Mice. Cell Stem Cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Onksen JL, Brown EJ & Blendy JA Selective deletion of a cell cycle checkpoint kinase (ATR) reduces neurogenesis and alters responses in rodent models of behavioral affect. Neuropsychopharmacology 36, 960–969 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salvi JS et al. ATR activity controls stem cell quiescence via the cyclin F – SCF complex. Proc. Natl. Acad. Sci. U. S. A. 119, e2115638119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scaramozza A et al. Lineage Tracing Reveals a Subset of Reserve Muscle Stem Cells Capable of Clonal Expansion under Stress. Cell Stem Cell 24, 944–957 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daynac M et al. Quiescent neural stem cells exit dormancy upon alteration of GABAAR signaling following radiation damage. Stem Cell Res. 11, 516–528 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Kaufmann K et al. A latent subset of human hematopoietic stem cells resists regenerative stress to preserve stemness. Nat. Immunol. 22, 723–734 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Signer RAJ, Magee JA, Salic A & Morrison SJ Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 508, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakada D et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miharada K, Sigurdsson V & Karlsson S Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Rep. 7, 1381–1392 (2014). [DOI] [PubMed] [Google Scholar]
- 101.Sigurdsson V & Miharada K Regulation of unfolded protein response in hematopoietic stem cells. Int. J. Hematol. 107, 627–633 (2018). [DOI] [PubMed] [Google Scholar]
- 102.Tümpel S & Rudolph KL Quiescence: Good and Bad of Stem Cell Aging. Trends Cell Biol. 29, 672–685 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Hu M et al. SRC-3 is involved in maintaining hematopoietic stem cell quiescence by regulation of mitochondrial metabolism in mice. Blood 132, 911–923 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Murakami K et al. OGT Regulates Hematopoietic Stem Cell Maintenance via PINK1-Dependent Mitophagy. Cell Rep. 34, 108579 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Gugliuzza MV & Crist C Muscle stem cell adaptations to cellular and environmental stress. Skelet. Muscle 12, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sueda R, Imayoshi I, Harima Y & Kageyama R High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33, 511–523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Z et al. Hes1 deficiency causes hematopoietic stem cell exhaustion. Stem Cells 38, 756–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Noguchi YT et al. Cell-autonomous and redundant roles of Hey1 and HeyL in muscle stem cells: HeyL requires HeS1 to bind diverse DNA sites. Development 146, dev163618 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Suen WJ, Li ST & Yang LT Hes1 regulates anagen initiation and hair follicle regeneration through modulation of hedgehog signaling. Stem Cells 38, 301–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu Z et al. Hair follicle stem cells regulate retinoid metabolism to maintain the self-renewal niche for melanocyte stem cells. Elife 9, e52712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pack LR, Daigh LH & Meyer T Putting the brakes on the cell cycle: mechanisms of cellular growth arrest. Curr. Opin. Cell Biol. 60, 106–113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matsumoto A et al. P57 Is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 9, 262–271 (2011). [DOI] [PubMed] [Google Scholar]
- 113.Zou P et al. P57 Kip2 and p27 Kip1 cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 9, 247–261 (2011). [DOI] [PubMed] [Google Scholar]
- 114.Porlan E et al. Transcriptional repression of Bmp2 by p21 Waf1/Cip1 links quiescence to neural stem cell maintenance. Nat. Neurosci. 16, 1567–1575 (2013). [DOI] [PubMed] [Google Scholar]
- 115.Andreu Z et al. The cyclin-dependent kinase inhibitor p27kip1 regulates radial stem cell quiescence and neurogenesis in the adult hippocampus. Stem Cells 33, 219–229 (2015). [DOI] [PubMed] [Google Scholar]
- 116.Furutachi S, Matsumoto A, Nakayama KI & Gotoh Y P57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 32, 970–981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee J et al. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc. Natl. Acad. Sci. U. S. A. 110, 4634–4639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chakkalakal JV et al. Early forming label-retaining muscle stem cells require p27kip1 for maintenance of the primitive state. Development 141, 1649–1659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mademtzoglou D et al. Cellular localization of the cell cycle inhibitor Cdkn1c controls growth arrest of adult skeletal muscle stem cells. Elife 7, e33337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cappell SD, Chung M, Jaimovich A, Spencer SL & Meyer T Irreversible APCCdh1 Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell 166, 167–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hosoyama T, Nishijo K, Prajapati SI, Li G & Keller C Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. J. Biol. Chem. 286, 19556–19564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Viatour P et al. Hematopoietic Stem Cell Quiescence Is Maintained by Compound Contributions of the Retinoblastoma Gene Family. Cell Stem Cell 3, 416–428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim E et al. Rb family proteins enforce the homeostasis of quiescent hematopoietic stem cells by repressing Socs3 expression. J. Exp. Med. 214, 1901–1912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Naser R et al. Role of the Retinoblastoma protein, Rb, during adult neurogenesis in the olfactory bulb. Sci. Rep. 6, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang J et al. APC/C is essential for hematopoiesis and impaired in aplastic anemia. Oncotarget 8, 63360–63369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eguren M et al. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat. Commun. 4, 2880 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Liu Y et al. p53 Regulates Hematopoietic Stem Cell Quiescence. Cell Stem Cell 4, 37–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meletis K et al. P53 Suppresses the Self-Renewal of Adult Neural Stem Cells. Development 133, 363–369 (2006). [DOI] [PubMed] [Google Scholar]
- 129.Sinha S et al. Asrij/OCIAD1 suppresses CSN5-mediated p53 degradation and maintains mouse hematopoietic stem cell quiescence. Blood 133, 2385–2400 (2019). [DOI] [PubMed] [Google Scholar]
- 130.Kim JY et al. Priming mobilization of hair follicle stem cells triggers permanent loss of regeneration after alkylating chemotherapy. Nat. Commun. 10, 3694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Farioli-Vecchioli S et al. Btg1 is required to maintain the pool of stem and progenitor cells of the dentate gyrus and subventricular zone. Front. Neurosci. 6, 124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lia Q, Rycaja K, Chena X & Tang DG Cancer stem cells and cell size: A causal link? Semin Cancer Biol 176, 139–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Benveniste P et al. Intermediate-Term Hematopoietic Stem Cells with Extended but Time-Limited Reconstitution Potential. Cell Stem Cell 6, 48–58 (2010). [DOI] [PubMed] [Google Scholar]
- 134.Laurenti E et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 16, 302–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grinenko T et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat. Commun. 9, 1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mohammad K, Dakik P, Medkour Y, Mitrofanova D & Titorenko VI Quiescence entry, maintenance, and exit in adult stem cells. Int. J. Mol. Sci. 20, 2158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.García-Prat L et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). [DOI] [PubMed] [Google Scholar]
- 138.Tang AH & Rando TA Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 33, 2782–2797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Leeman DS et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science (80-. ). 359, 1277–1283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liang R et al. Restraining Lysosomal Activity Preserves Hematopoietic Stem Cell Quiescence and Potency. Cell Stem Cell 26, 359–376.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mohrin M et al. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (80-. ). 347, 1374–1377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mohrin M, Widjaja A, Liu Y, Luo H & Chen D The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell 17, e12756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mohrin M & Chen D The mitochondrial metabolic checkpoint and aging of hematopoietic stem cells. Curr. Opin. Hematol. 23, 318–324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Troy A et al. Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38α/β MAPK. Cell Stem Cell 11, 541–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jones NC et al. The p38α/β MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 169, 105–116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frelin C et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 14, 1037–1044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kase Y, Otsu K, Shimazaki T & Okano H Involvement of p38 in Age-Related Decline in Adult Neurogenesis via Modulation of Wnt Signaling. Stem Cell Reports 12, 1313–1328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hemmati S et al. PI3K alpha and delta promote hematopoietic stem cell activation. JCI Insight 5, e125832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.García-Prat L et al. FoxO maintains a genuine muscle stem-cell quiescent state until geriatric age. Nature Cell Biology 22, 1307–1318 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Meng D, Frank AR & Jewell JL mTOR signaling in stem and progenitor cells. Development 145, dev152595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kalaitzidis D et al. MTOR complex 1 plays critical roles in hematopoiesis and pten-loss-evoked leukemogenesis. Cell Stem Cell 11, 429–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hartman NW et al. MTORC1 Targets the Translational Repressor 4E-BP2, but Not S6 Kinase 1/2, to Regulate Neural Stem Cell Self-Renewal InVivo. Cell Rep. 5, 433–444 (2013). [DOI] [PubMed] [Google Scholar]
- 153.Ren X et al. Lgr4 Deletion Delays the Hair Cycle and Inhibits the Activation of Hair Follicle Stem Cells. J. Invest. Dermatol. 140, 1706–1712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kwon JS et al. Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Rep. 20, 3223–3235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Matsuoka Y et al. Low level of C-kit expression marks deeply quiescent murine hematopoietic stem cells. Stem Cells 29, 1783–1791 (2011). [DOI] [PubMed] [Google Scholar]
- 156.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA & Tajbakhsh S A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012). [DOI] [PubMed] [Google Scholar]
- 157.Ibrayeva A et al. Early stem cell aging in the mature brain. Cell Stem Cell 28, 955–966.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Benjamin DI et al. Fasting Induces a Highly Resilient Deep Quiescent State in Muscle Stem Cells via Ketone Body Signaling. Cell Metab. 34, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shwartz Y et al. Cell Types Promoting Goosebumps Form a Niche to Regulate Hair Follicle Stem Cells. Cell 182, 578–593.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fujimaki K et al. Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Proc. Natl. Acad. Sci. U. S. A. 116, 22624–22634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Urbán N, Blomfield IM & Guillemot F Quiescence of Adult Mammalian Neural Stem Cells: A Highly Regulated Rest. Neuron 104, 834–848 (2019). [DOI] [PubMed] [Google Scholar]
- 162.Wang Z & Ema H Mechanisms of self-renewal in hematopoietic stem cells. Int. J. Hematol. 103, 498–509 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Daszczuk P et al. An Intrinsic Oscillation of Gene Networks Inside Hair Follicle Stem Cells: An Additional Layer That Can Modulate Hair Stem Cell Activities. Front. Cell Dev. Biol. 8, 595178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bottes S et al. Long-term self-renewing stem cells in the adult mouse hippocampus identified by intravital imaging. Nat. Neurosci. 24, 225–233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Otsuki L & Brand AH Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science (80-. ). 360, 99–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Giordani L, Parisi A & Le Grand F Satellite Cell Self-Renewal. Current Topics in Developmental Biology 126, 177–203 (2018). [DOI] [PubMed] [Google Scholar]
- 167.Morizur L et al. Distinct Molecular Signatures of Quiescent and Activated Adult Neural Stem Cells Reveal Specific Interactions with Their Microenvironment. Stem Cell Reports 11, 565–577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dong J et al. A neuronal molecular switch through cell-cell contact that regulates quiescent neural stem cells. Sci. Adv. 5, eaav4416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ottone C et al. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat. Cell Biol. 16, 1045–1056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morgner J et al. Integrin-linked kinase regulates the niche of quiescent epidermal stem cells. Nat. Commun. 6, 8198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang Y et al. Oscillations of Delta-like1 regulate the balance between differentiation and maintenance of muscle stem cells. Nat. Commun. 12, 1318 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Belenguer G et al. Adult Neural Stem Cells Are Alerted by Systemic Inflammation through TNF-α Receptor Signaling. Cell Stem Cell 28, 285–299.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 173.Cosgrove BD et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Charville GW et al. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports 5, 621–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shea KL et al. Sprouty1 Regulates Reversible Quiescence of a Self-Renewing Adult Muscle Stem Cell Pool during Regeneration. Cell Stem Cell 6, 117–129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Baumgartner C et al. An ERK-Dependent Feedback Mechanism Prevents Hematopoietic Stem Cell Exhaustion. Cell Stem Cell 22, 879–892.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bonaguidi MA et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Porter SN et al. Pten cell autonomously modulates the hematopoietic stem cell response to inflammatory cytokines. Stem Cell Reports 6, 806–814 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yue F et al. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat. Commun. 8, 14328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee JY et al. MTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after pten deletion. Cell Stem Cell 7, 593–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Magee JA et al. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell 11, 415–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Siegemund S et al. IP3 3-kinase B controls hematopoietic stem cell homeostasis and prevents lethal hematopoietic failure in mice. Blood 125, 2786–2797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Barker N et al. Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5+ve stem cells. Cold Spring Harb. Symp. Quant. Biol. 73, 351–356 (2008). [DOI] [PubMed] [Google Scholar]
- 184.Shokouhian M et al. Altering chromatin methylation patterns and the transcriptional network involved in regulation of hematopoietic stem cell fate. J. Cell. Physiol. 235, 6404–6423 (2020). [DOI] [PubMed] [Google Scholar]
- 185.Yi R Concise Review: Mechanisms of Quiescent Hair Follicle Stem Cell Regulation. Stem Cells 35, 2323–2330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Urbán N & Guillemot F Neurogenesis in the embryonic and adult brain: Same regulators, different roles. Front. Cell. Neurosci. 8, 396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Robinson DCL & Dilworth FJ Epigenetic Regulation of Adult Myogenesis. Current Topics in Developmental Biology 126 235–284 (2018). [DOI] [PubMed] [Google Scholar]
- 188.Kosan C & Godmann M Genetic and epigenetic mechanisms that maintain hematopoietic stem cell function. Stem Cells Int. 2016, 5178965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu Y, Beyer A & Aebersold R On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 190.Hausburg MA et al. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife 4, e03390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Grover A et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7, 11075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lechman ER et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 11, 799–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Carrelha J et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106–111 (2018). [DOI] [PubMed] [Google Scholar]
- 194.Crist CG, Montarras D & Buckingham M Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11, 118–126 (2012). [DOI] [PubMed] [Google Scholar]
- 195.de Morree A et al. Alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function. Science 366, 734–738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tian B & Manley JL Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sommerkamp P et al. Differential Alternative Polyadenylation Landscapes Mediate Hematopoietic Stem Cell Activation and Regulate Glutamine Metabolism. Cell Stem Cell 26, 722–738.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 198.Boutet SCC et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell 10, 327–336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shepard PJ et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. Rna 17, 761–772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang L, Dowell RD & Yi R Genome-wide maps of polyadenylation reveal dynamic mRNA 3’-end formation in mammalian cell lineages. Rna 19, 413–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mueller AA, Van Velthoven CT, Fukumoto KD, Cheung TH & Rando TA Intronic polyadenylation of PDGFRα in resident stem cells attenuates muscle fibrosis. Nature 540, 276–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pai AA et al. Widespread Shortening of 3’ Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS Genet. 12, e1006338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Taliaferro JM et al. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol. Cell 61, 821–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Der Vartanian A et al. PAX3 Confers Functional Heterogeneity in Skeletal Muscle Stem Cell Responses to Environmental Stress. Cell Stem Cell 24, 958–973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nishikura K A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jiang Q et al. Hyper-Editing of Cell-Cycle Regulatory and Tumor Suppressor RNA Promotes Malignant Progenitor Propagation. Cancer Cell 35, 81–94.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gal-Mark N et al. Abnormalities in A-to-I RNA editing patterns in CNS injuries correlate with dynamic changes in cell type composition. Sci. Rep. 7, 43421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tabula Muris Consortium. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.The Tabula Sapiens Consortium. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, 6594 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.XuFeng R et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc. Natl. Acad. Sci. U. S. A. 106, 17763–17768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shevchenko G & Morris KV All I’s on the RADAR: role of ADAR in gene regulation. FEBS Lett. 592, 2860–2873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zaccara S, Ries RJ & Jaffrey SR Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019). [DOI] [PubMed] [Google Scholar]
- 214.Xiang JF et al. N6-Methyladenosines Modulate A-to-I RNA Editing. Mol. Cell 69, 126–135.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 215.Lee H et al. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell Biol. 21, 700–709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Li Z et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28, 904–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yin R et al. Differential m6A RNA landscapes across hematopoiesis reveal a role for IGF2BP2 in preserving hematopoietic stem cell function. Cell Stem Cell 29, 149–159.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 218.Cheng Y et al. m6A RNA Methylation Maintains Hematopoietic Stem Cell Identity and Symmetric Commitment. Cell Rep. 28, 1703–1716.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yao QJ et al. Mettl3–Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 28, 952–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liang Y et al. METTL3-Mediated m6A Methylation Regulates Muscle Stem Cells and Muscle Regeneration by Notch Signaling Pathway. Stem Cells Int. 2021, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sheng Y et al. Role of c-Myc haploinsufficiency in the maintenance of HSCs in mice. Blood 137, 610–623 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cai C et al. c-Myc regulates neural stem cell quiescence and activation by coordinating the cell cycle and mitochondrial remodeling. Signal Transduct. Target. Ther. 6, 2020–2022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yoon KJ et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 171, 877–889.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Blanco S et al. The RNA-methyltransferase misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7, e1002403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shi Y Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670 (2017). [DOI] [PubMed] [Google Scholar]
- 226.Olivieri JE et al. RNA splicing p rograms define tissue compartments and cell types at single cell resolution. Elife 10, e70692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Farina NH et al. A role for RNA post-transcriptional regulation in satellite cell activation. Skelet. Muscle 2, 21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bowman TV et al. Differential mRNA Processing in Hematopoietic Stem Cells. Stem Cells 24, 662–670 (2006). [DOI] [PubMed] [Google Scholar]
- 229.Chen L et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science 345, 6204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Dominici C & Richard S Muscle stem cell polarity requires QKI-mediated alternative splicing of Integrin Alpha-7 (Itga7). Life Sci. Alliance 5, e202101192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tan DQ et al. PRMT5 Modulates Splicing for Genome Integrity and Preserves Proteostasis of Hematopoietic Stem Cells. Cell Rep. 26, 2316–2328.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 232.Xiao N et al. Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood 125, 941–948 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Baralle FE & Giudice J Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yue L, Wan R, Luan S, Zeng W & Cheung TH Article Dek Modulates Global Intron Retention during Muscle Stem Cells Quiescence Exit. Dev. Cell 53, 661–676 (2020). [DOI] [PubMed] [Google Scholar]
- 235.Goldstein O et al. Mapping Whole-Transcriptome Splicing in Mouse Hematopoietic Stem Cells. Stem Cell Reports 8, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chen Z et al. Nuclear DEK preserves hematopoietic stem cells potential via NCoR1/HDAC3-Akt1/2-mTOR axis. J. Exp. Med. 218, e20201974 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wickramasinghe VO & Laskey RA Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol. 16, 431–442 (2015). [DOI] [PubMed] [Google Scholar]
- 238.Mancini A et al. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol. 8, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Katz S et al. A Nuclear Role for miR-9 and Argonaute Proteins in Balancing Quiescent and Activated Neural Stem Cell States. Cell Rep. 17, 1383–1398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Treiber T, Treiber N & Meister G Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20 (2019). [DOI] [PubMed] [Google Scholar]
- 241.Guo S et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. U. S. A. 107, 14229–14234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cheung TH et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Baghdadi MB et al. Notch-Induced miR-708 Antagonizes Satellite Cell Migration and Maintains Quiescence. Cell Stem Cell 23, 859–868.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 244.Sato T, Yamamoto T & Sehara-Fujisawa A MiR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat. Commun. 5, 4597 (2014). [DOI] [PubMed] [Google Scholar]
- 245.Hu M et al. MicroRNA-21 maintains hematopoietic stem cell homeostasis through sustaining the NF-κB signaling pathway in mice. Haematologica 106, 412–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Xu Y et al. MicroRNAs are indispensable for the proliferation and differentiation of adult neural progenitor cells in mice. Biochem. Biophys. Res. Commun. 530, 209–214 (2020). [DOI] [PubMed] [Google Scholar]
- 247.Andl T et al. The miRNA-Processing Enzyme Dicer Is Essential for the Morphogenesis and Maintenance of Hair Follicles. Curr. Biol. 16, 1041–1049 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Vishlaghi N & Lisse TS Dicer- and Bulge Stem Cell-Dependent MicroRNAs During Induced Anagen Hair Follicle Development. Front. Cell Dev. Biol. 8, 338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lepko T et al. Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 38, e100481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ge M et al. miR-29a/b1 Inhibits Hair Follicle Stem Cell Lineage Progression by Spatiotemporally Suppressing WNT and BMP Signaling. Cell Rep. 29, 2489–2504.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 251.Hawkshaw NJ, Hardman JA, Alam M, Jimenez F & Paus R Deciphering the molecular morphology of the human hair cycle: Wnt signalling during the telogen–anagen transformation. Br. J. Dermatol. 182, 1184–1193 (2020). [DOI] [PubMed] [Google Scholar]
- 252.Brack AS, Conboy IM, Conboy MJ, Shen J & Rando TA A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis. Cell Stem Cell 2, 50–59 (2008). [DOI] [PubMed] [Google Scholar]
- 253.Maltzahn J, Von, Bentzinger CF & Rudnicki MA. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat. Cell. Bio. 14 186–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fleming HE et al. Wnt Signaling in the Niche Enforces Hematopoietic Stem Cell Quiescence and Is Necessary to Preserve Self-Renewal In Vivo. Cell Stem Cell 2, 274–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mehta A et al. The MicroRNA-132 and MicroRNA-212 Cluster Regulates Hematopoietic Stem Cell Maintenance and Survival with Age by Buffering FOXO3 Expression. Immunity 42, 1021–1032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gopinath SD, Webb AE, Brunet A & Rando TA FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Reports 2, 414–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Miyamoto K et al. Foxo3a Is Essential for Maintenance of the Hematopoietic Stem Cell Pool. Cell Stem Cell 1, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 258.Hu W et al. MiR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood 125, 2206–2216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Song SJ et al. The oncogenic MicroRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13, 87–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kaplan IM et al. Deletion of Tristetraprolin Caused Spontaneous Reactive Granulopoiesis by a Non–Cell-Autonomous Mechanism Without Disturbing Long-Term Hematopoietic Stem Cell Quiescence. J. Immunol. 186, 2826–2834 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chenette DM et al. Targeted mRNA Decay by RNA Binding Protein AUF1 Regulates Adult Muscle Stem Cell Fate, Promoting Skeletal Muscle Integrity. Cell Rep. 16, 1379–1390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Guallar D & Wang J RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol 9, 389–409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ratti A et al. A role for the ELAV RNA-binding proteins in neural stem cells: Stabilization of Msi1 mRNA. J. Cell Sci. 119, 1442–1452 (2006). [DOI] [PubMed] [Google Scholar]
- 264.Perrone-Bizzozero N & Bolognani F Role of HuD and other RNA-Binding proteins in neural development and plasticity. J. Neurosci. Res. 68, 121–126 (2002). [DOI] [PubMed] [Google Scholar]
- 265.Van Treeck B & Parker R Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Di Stefano B et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 25, 622–638.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Díaz-Muñoz MD & Turner M Uncovering the role of RNA-binding proteins in gene expression in the immune system. Front. Immunol. 9, 1094 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Fujita R et al. Fragile X mental retardation protein regulates skeletal muscle stem cell activity by regulating the stability of Myf5 mRNA. Skelet. Muscle 7, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Roy N et al. mRNP granule proteins Fmrp and Dcp1a differentially regulate mRNP complexes to contribute to control of muscle stem cell quiescence and activation. Skelet. Muscle 11, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Luo Y et al. Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 6, e48251(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kato Y et al. ELAVL2-directed RNA regulatory network drives the formation of quiescent primordial follicles. EMBO Rep. 20, e48251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Sharifi S, da Costa HFR & Bierhoff H The circuitry between ribosome biogenesis and translation in stem cell function and ageing. Mech. Ageing Dev. 189, 111282 (2020). [DOI] [PubMed] [Google Scholar]
- 273.Zismanov V et al. Phosphorylation of eIF2α is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell 18, 79–90 (2016). [DOI] [PubMed] [Google Scholar]
- 274.Blanco S et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Jarzebowski L et al. Mouse adult hematopoietic stem cells actively synthesize ribosomal RNA. Rna 24, 1803–1812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Gulati GS et al. Neogenin-1 distinguishes between myeloid-biased and balanced Hoxb5+ mouse long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 116, 25115–25125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Gayraud-Morel B, Le Bouteiller M, Commere PH, Cohen-Tannoudji M & Tajbakhsh S Notchless defines a stage-specific requirement for ribosome biogenesis during lineage progression in adult skeletal myogenesis. Development 145, dev162636 (2018). [DOI] [PubMed] [Google Scholar]
- 278.Chen X et al. Translational control by DHX36 binding to 5′UTR G-quadruplex is essential for muscle stem-cell regenerative functions. Nat. Commun. 12, 5043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lai JC et al. The DEAH-box helicase RHAU is an essential gene and critical for mouse hematopoiesis. Blood 119, 4291–4300 (2012). [DOI] [PubMed] [Google Scholar]
- 280.Le Bouteiller M et al. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J. Exp. Med. 210, 2351–2369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Cai X et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 17, 165–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Fujita R et al. Satellite cell expansion is mediated by P-eIF2α-dependent Tacc3 translation. Development 148, dev194480 (2021). [DOI] [PubMed] [Google Scholar]
- 283.Zeng W et al. CPEB1 directs muscle stem cell activation by reprogramming the translational landscape. Nat. Commun. 13, 947 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Das S, Vera M, Gandin V, Singer RH & Tutucci E Intracellular mRNA transport and localized translation. Nat. Rev. Mol. Cell Biol. 22, 483–504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kwon OS et al. Exon junction complex dependent mRNA localization is linked to centrosome organization during ciliogenesis. Nat. Commun. 12, 1351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Kiriakidou M et al. An mRNA m7G Cap Binding-like Motif within Human Ago2 Represses Translation. Cell 129, 1141–1151 (2007). [DOI] [PubMed] [Google Scholar]
- 287.Lu K, Nakagawa MM, Thummar K & Rathinam CV Slicer Endonuclease Argonaute 2 Is a Negative Regulator of Hematopoietic Stem Cell Quiescence. Stem Cells 34, 1343–1353 (2016). [DOI] [PubMed] [Google Scholar]
- 288.Zhou Y et al. Autocrine Mfge8 Signaling Prevents Developmental Exhaustion of the Adult Neural Stem Cell Pool. Cell Stem Cell 23, 444–452.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Signer RAJ et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 30, 1698–1703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Goncalves KA et al. Angiogenin Promotes Hematopoietic Regeneration by Dichotomously Regulating Quiescence of Stem and Progenitor Cells. Cell 166, 894–906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hidalgo San Jose L et al. Modest Declines in Proteome Quality Impair Hematopoietic Stem Cell Self-Renewal. Cell Rep. 30, 69–80.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kitajima Y et al. The Ubiquitin-Proteasome System Is Indispensable for the Maintenance of Muscle Stem Cells. Stem Cell Reports 11, 1523–1538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.King B et al. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat. Immunol. 17, 1312–1321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Thompson BJ et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205, 1395–1408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wei Q et al. MAEA is an E3 ubiquitin ligase promoting autophagy and maintenance of haematopoietic stem cells. Nat. Commun. 12, 2522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Blomfield IM et al. Id4 promotes the elimination of the pro-activation factor ascl1 to maintain quiescence of adult hippocampal stem cells. Elife 8, e48561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kobayashi T et al. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat. Commun. 10, 5446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Cochard LM et al. Manipulation of EGFR-Induced Signaling for the Recruitment of Quiescent Neural Stem Cells in the Adult Mouse Forebrain. Front. Neurosci. 15, 621076 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Liu L et al. ER associated degradation preserves hematopoietic stem cell quiescence and self- renewal by restricting mTOR activity. Blood 136, 2975–2986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Li G et al. SIRT 7 activates quiescent hair follicle stem cells to ensure hair growth in mice. EMBO J. 39, e104365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Dong S et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 591, 117–123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ono Y et al. Slow-dividing satellite cells retain long-term self-renewal ability in adult muscle. J. Cell Sci. 125, 1309–1317 (2012). [DOI] [PubMed] [Google Scholar]
- 303.Singh SK et al. Id1 Ablation Protects Hematopoietic Stem Cells from Stress-Induced Exhaustion and Aging. Cell Stem Cell 23, 252–265.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Morrow CS et al. Vimentin Coordinates Protein Turnover at the Aggresome during Neural Stem Cell Quiescence Exit. Cell Stem Cell 26, 558–568.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wang X et al. Macrophages induce AKT/β-catenin-dependent Lgr5+ stem cell activation and hair follicle regeneration through TNF. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Servián-Morilla E et al. A POGLUT 1 mutation causes a muscular dystrophy with reduced Notch signaling and satellite cell loss. EMBO Mol. Med. 8, 1289–1309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wang W et al. Notch Receptor-Ligand Engagement Maintains Hematopoietic Stem Cell Quiescence and Niche. 2280–2293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.White CW et al. Age-Related Loss of Neural Stem Cell O-GlcNAc Promotes a Glial Fate Switch through STAT3 activation. Proc. Natl. Acad. Sci. U. S. A. 117, 22214–22224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Bonitto K, Sarathy K, Atai K, Mitra M & Coller HA Is There a Histone Code for Cellular Quiescence? Front. Cell Dev. Biol. 9, 739780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Sincennes M-C et al. Acetylation of PAX7 controls muscle stem cell self-renewal and differentiation potential in mice. Nat. Commun. 12, 3253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Richter JD & Lasko P Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 3, a002758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Wang S et al. Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 180, 585–600.e19 (2020). [DOI] [PubMed] [Google Scholar]
- 313.Luong XG, Daldello EM, Rajkovic G, Yang CR & Conti M Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res. 48, 3257–3276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Clarke HJ Post-transcriptional Control of Gene Expression During Mouse Oogenesis. Results Probl Cell Differ. 55, 1–21 (2012). [DOI] [PubMed] [Google Scholar]
- 315.Dumdie JN et al. Chromatin Modification and Global Transcriptional Silencing in the Oocyte Mediated by the mRNA Decay Activator ZFP36L2. Dev. Cell 44, 392–402.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Pérez-Sanz J et al. Increased number of multi-oocyte follicles (MOFs) in juvenile p27Kip1 mutant mice: Potential role of granulosa cells. Hum. Reprod. 28, 1023–1030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Yang QE, Nagaoka SI, Gwost I, Hunt PA & Oatley JM Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice. PLoS Genet. 11, e1005355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Adhikari D et al. Cdk1, but not Cdk2, is the sole Cdk that is essential and sufficient to drive resumption of meiosis in mouse oocytes. Hum. Mol. Genet. 21, 2476–2484 (2012). [DOI] [PubMed] [Google Scholar]
- 319.Reyes JM & Ross PJ Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip. Rev. RNA 7, 71–89 (2016). [DOI] [PubMed] [Google Scholar]
- 320.Yang Y et al. Maternal mRNAs with distinct 3’ UTRs define the temporal pattern of Ccnb1 synthesis during mouse oocyte meiotic maturation. Genes Dev. 31, 1302–1307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Freimer JW, Krishnakumar R, Cook MS & Blelloch R Expression of Alternative Ago2 Isoform Associated with Loss of microRNA-Driven Translational Repression in Mouse Oocytes. Curr. Biol. 28, 296–302.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ivanova I et al. The RNA m6A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol. Cell 67, 1059–1067.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Zhao BS et al. M6 A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Cheng S et al. Mammalian oocytes store mRNAs in a mitochondria-associated membraneless compartment. Science 378, eabq4835 (2022). [DOI] [PubMed] [Google Scholar]
- 325.Than-Trong E et al. Neural stem cell quiescence and stemness are molecularly distinct outputs of the notch3 signalling cascade in the vertebrate adult brain. Development 145, dev161034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sistigu A, Musella M, Galassi C, Vitale I & De Maria R Tuning Cancer Fate: Tumor Microenvironment’s Role in Cancer Stem Cell Quiescence and Reawakening. Front. Immunol. 11, 2166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Basu S, Dong Y, Kumar R, Jeter C & Tang DG Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis. Semin. Cancer Biol. 78, 90–103 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bowling S et al. An Engineered CRISPR-Cas9 Mouse Line for Simultaneous Readout of Lineage Histories and Gene Expression Profiles in Single Cells. Cell 181, 1410–1422.e27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Jia W et al. Indispensable role of Galectin-3 in promoting quiescence of hematopoietic stem cells. Nat. Commun. 12, 2118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Martynoga B et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769–1786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Mira H et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 (2010). [DOI] [PubMed] [Google Scholar]
- 332.Genander M et al. BMP signaling and its pSMADS1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15, 619–633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Choi S et al. Corticosterone inhibits GAS6 to govern hair follicle stem-cell quiescence. Nature 592, 428–432 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Arjona M et al. Tubastatin A maintains adult skeletal muscle stem cells in a quiescent state ex vivo and improves their engraftment ability in vivo. Stem Cell Reports 17, 82–95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Cao Y et al. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet. 29, 727–735 (2020). [DOI] [PubMed] [Google Scholar]
- 336.Li L et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Renders S et al. Niche derived netrin-1 regulates hematopoietic stem cell dormancy via its receptor neogenin-1. Nat. Commun. 12, 608 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Kiel MJ et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Nakamura-Ishizu A, Takizawa H & Suda T The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141, 4656–4666 (2014). [DOI] [PubMed] [Google Scholar]
- 340.Gattazzo F, Laurent B, Relaix F, Rouard H & Didier N Distinct Phases of Postnatal Skeletal Muscle Growth Govern the Progressive Establishment of Muscle Stem Cell Quiescence. Stem Cell Reports 15, 597–611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA & Kardon G Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Lepper C, Partridge TA & Fan C-M An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Sambasivan R et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 (2011). [DOI] [PubMed] [Google Scholar]
- 344.Keefe AC et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat. Commun. 6, 7087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Pawlikowski B, Pulliam C, Betta ND, Kardon G & Olwin BB Pervasive satellite cell contribution to uninjured adult muscle fibers. Skelet. Muscle 5, 42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wosczyna MN et al. Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 27, 2029–2035.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wosczyna MN & Rando TA A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Dev. Cell 46, 135–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Joe AWB et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 12, 153–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Baser A, Skabkin M & Martin-Villalba A Neural Stem Cell Activation and the Role of Protein Synthesis. Brain Plast. 3, 27–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Lugert S et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445–456 (2010). [DOI] [PubMed] [Google Scholar]
- 351.Fuchs E & Blau HM Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 27, 532–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wang ECE, Dai Z, Ferrante AW, Drake CG & Christiano AM A Subset of TREM2 + Dermal Macrophages Secretes Oncostatin M to Maintain Hair Follicle Stem Cell Quiescence and Inhibit Hair Growth. Cell Stem Cell 24, 654–669.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 353.Goodell MA & Rando TA Stem cells and healthy aging. Science 350, 1199–1204 (2015). [DOI] [PubMed] [Google Scholar]
- 354.Brunet A et al. Ageing and rejuvenation of tissue stem cells and their niches. Nat. Rev. Mol. Cell Biol. s41580–022-00510-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Katsimpardi L & Lledo PM Regulation of neurogenesis in the adult and aging brain. Curr. Opin. Neurobiol. 53, 131–138 (2018). [DOI] [PubMed] [Google Scholar]
- 356.Ji J et al. Aging in hair follicle stem cells and niche microenvironment. J. Dermatol. 44, 1097–1104 (2017). [DOI] [PubMed] [Google Scholar]
- 357.Luinenburg DG & de Haan G MicroRNAs in hematopoietic stem cell aging. Mech. Ageing Dev. 189, 111281 (2020). [DOI] [PubMed] [Google Scholar]
- 358.Liu L & Rando TA Manifestations and mechanisms of stem cell aging. J. Cell Biol. 193, 257–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kalamakis G et al. Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell 176, 1407–1419.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 360.Wang L, Chen M, Fu H, Ni T & Wei G Tempo-spatial alternative polyadenylation analysis reveals that 3′ UTR lengthening of Mdm2 regulates p53 expression and cellular senescence in aged rat testis. Biochem. Biophys. Res. Commun. 523, 1046–1052 (2020). [DOI] [PubMed] [Google Scholar]
- 361.Porpiglia E et al. Elevated CD47 is a hallmark of dysfunctional aged muscle stem cells that can be targeted to augment regeneration. Cell Stem Cell 29 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Moore DL, Pilz GA, Araúzo-Bravo MJ, Barral Y & Jessberger S A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334–1338 (2015). [DOI] [PubMed] [Google Scholar]
- 363.Matsumura H et al. Stem cells: Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 351, aad4395 (2016). [DOI] [PubMed] [Google Scholar]
- 364.Ishikawa Y et al. Opposing functions of Fbxw7 in keratinocyte growth, differentiation and skin tumorigenesis mediated through negative regulation of c-Myc and Notch. Oncogene 32, 1921–1932 (2013). [DOI] [PubMed] [Google Scholar]
- 365.Rossi DJ et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. U. S. A. 102, 9194–9199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Yousef H et al. Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nat. Med. 25, 988–1000 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Maybury-Lewis SY et al. Changing and stable chromatin accessibility supports transcriptional overhaul during neural stem cell activation and is altered with age. Aging Cell 20, e13499 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Encinas JM et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566–579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Keyes BE et al. Nfatc1 orchestrates aging in hair follicle stem cells. Proc. Natl. Acad. Sci. U. S. A. 110, E4950–E4959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Khurana S et al. Outside-in integrin signalling regulates haematopoietic stem cell function via Periostin-Itgav axis. Nat. Commun. 7, 13500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Chen X et al. Bone Marrow Myeloid Cells Regulate Myeloid-Biased Hematopoietic Stem Cells via a Histamine-Dependent Feedback Loop. Cell Stem Cell 21, 747–760.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Nilsson SK et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 106, 1232–1239 (2005). [DOI] [PubMed] [Google Scholar]
- 373.Qian H et al. Critical Role of Thrombopoietin in Maintaining Adult Quiescent Hematopoietic Stem Cells. Cell Stem Cell 1, 671–684 (2007). [DOI] [PubMed] [Google Scholar]
- 374.Arai F et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161 (2004). [DOI] [PubMed] [Google Scholar]
- 375.Masamoto Y et al. Adiponectin Enhances Quiescence Exit of Murine Hematopoietic Stem Cells and Hematopoietic Recovery Through mTORC1 Potentiation. Stem Cells 35, 1835–1848 (2017). [DOI] [PubMed] [Google Scholar]
- 376.Frodermann V et al. Exercise reduces inflammatory cell production and cardiovascular inflammation via instruction of hematopoietic progenitor cells. Nat. Med. 25, 1761–1771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Cao Y et al. Transmembrane Protein Ttyh1 Maintains the Quiescence of Neural Stem Cells Through Ca2+/NFATc3 Signaling. Front. Cell Dev. Biol. 9, 779373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Song J et al. Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Sierra A et al. Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell 16, 488–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Tong CK et al. Axonal control of the adult neural stem cell niche. Cell Stem Cell 14, 500–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Baghdadi MB et al. Reciprocal signalling by Notch–Collagen V–CALCR retains muscle stem cells in their niche. Nature 557, 714–718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


